Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) plays a major role in adipogenesis. PPARγ binds to DNA as a heterodimer with retinoid X receptor (RXR), and PPARγ-RXR can be activated by ligands specific for either receptor; the presence of both ligands can result in a cooperative effect on the transactivation of target genes. How these ligands mediate transactivation, however, remains unclear. PPARγ is known to interact with both the p160/SRC-1 family of coactivators and the distinct, multisubunit coactivator complex called DRIP. A single DRIP subunit, DRIP205 (TRAP220, PBP), binds directly to PPARγ. Here we report that PPARγ and RXR selectively interacted with DRIP205 and p160 proteins in a ligand-dependent manner. At physiological concentrations, RXR-specific ligands only induced p160 binding to RXR, and PPARγ-specific ligands exclusively recruited DRIP205 but not p160 coactivators to PPARγ. This selectivity was not observed in interaction assays off DNA, implying that the specificity of coactivator binding in response to ligand is strongly influenced by the allosteric effects of DNA-bound heterodimers. These coactivator-selective effects were also observed in transient-transfection assays in the presence of overexpressed p160 or DRIP coactivators. The results suggest that the cooperative effects of PPARγ- and RXR-specific ligands may occur at the level of selective coactivator recruitment.
Peroxisome proliferator-activated receptor γ (PPARγ) plays pivotal roles in mediating adipocyte differentiation and in modulating insulin sensitivity. Overexpression of PPARγ in fibroblasts and myoblasts drive these cells to differentiate into adipocytes (34, 45). In addition, PPARγ inhibits the growth of several human tumor cell lines in culture in response to various synthetic PPARγ ligands (2, 14, 35). For example, PPARγ is able to induce the growth arrest and differentiation of human liposarcoma cells (10, 46).
PPARγ is a member of the nuclear hormone receptor superfamily. Like other nuclear receptors, PPARγ is comprised of an amino-terminal ligand-independent transactivation region (AF-1) (50), a central DNA-binding domain (DBD), and a carboxy-terminal ligand-binding domain (LBD) that contains a second, ligand-dependent transactivation surface (AF-2). Typical of many nuclear receptors, PPARγ associates with DNA targets as a heterodimer with the 9-cis-retinoic acid receptor, RXR; transactivation requires the high-affinity binding of PPARγ- or RXR-specific ligands to their respective receptors in the context of the heterodimer. The first natural PPAR-responsive element (PPRE) was identified in the promoter of the acyl coenzyme A (acyl-CoA) oxidase gene (13, 48), and the analyses of all identified PPREs have revealed a consensus sequence, 5′-AACTAGGNCA A AGGTCA-3′ (39). The properties of the PPRE, including an extented 5′ half-site and an adenine as the spacing nucleotide between two half-sites, contribute to the discrimination between PPRE and other direct repeat (DR-1)-type nuclear receptor response elements (19, 26, 29, 49). Further characterization of PPAR-RXR binding to the consensus PPRE has revealed a polarity in binding, such that PPAR and RXR occupy the 5′ and 3′ half-sites, respectively (12); this polarity is the opposite of that observed for other nuclear receptor-RXR heterodimers.
PPARγ, and its related subtypes, PPARα and PPARδ, were originally discovered as orphan receptors, but this was followed by the relatively rapid identification of both natural and synthetic ligands (for a review, see reference 51). The natural ligands for PPARγ include a number of fatty acid and eicosanoid derivatives. In addition, the J-series of prostagladins derived from PGD2 have also been established as potent PPARγ ligands (53). The terminal metabolite 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) binds and activates PPARγ at micromolar concentrations and is currently the best-characterized, naturally occurring PPARγ ligand (16, 28). The components of oxidized low-density lipoprotein, such as 13-hydroxyoctadecadienoic acid and 15-hydroxyeicosatetraenoic acid, are also considered as naturally occurring PPARγ ligands (23). A class of antidiabetic agents known as thiazolidinediones (TZDs) were the first identified synthetic compounds that bound with high affinity to PPARγ (31). TZDs are able to induce PPRE-directed transactivation in adipocytes and can promote adipocyte differentiation (24, 27). More importantly, the potency of TZDs to bind PPARγ is closely related to their glucose-lowering activity in rodents, suggesting that the antidiabetic effects of TZDs occur primarily through PPARγ (1, 31, 52). Recently, a series of tyrosine-based PPARγ agonists, exemplified by GW1929, were reported (8, 9, 22). These compounds are non-TZD antidiabetic drugs and are among the most potent PPARγ agonists. For example, GW1929 exhibited equal antihyperglycemic activity at a concentration 1,000-fold lower than troglitazone in ZDF rats (3, 22), and this activity closely matches their differences in PPARγ affinity and transactivation. Other structurally diverse PPARγ agonists have also been described. Hypolipidemic agents, such as the fibrate analogue GW2331, have been shown to have activity on PPARγ.
Besides PPARγ-specific ligands, PPARγ-mediated responses can also be elicited through RXR-specific ligands. Unlike many other nuclear receptors, RXR is viewed as a permissive partner for PPARγ, whereby the former can activate transcription in response to RXR-specific ligands in the context of the PPARγ-RXR heterodimer (for a review, see reference 30). Overexpression of PPARγ and RXR leads to transcriptional activation from a PPRE in response to 9-cis-retinoic acid, and simultaneous administration of both PPAR- and RXR-specific ligands to transfected cells results in an additive induction of reporter gene expression (19, 26, 29). Moreover, RXR agonists can enhance the sensitivity of diabetic and obese mice to insulin, demonstrating the ability of RXR to activate PPAR-RXR signaling pathway in vivo (36).
The mechanisms by which nuclear receptors regulate transcription from target genes are not yet fully elucidated. However, compelling evidence indicates that ligand binding results in a conformational change within the receptor that permits the dissociation of corepressors, such as NCoR and SMRT, that bridge the binding of histone deacetylases, and the concomitant association of coactivators, such as the p160 class that link histone acetyltransferases (HAT) such as CBP/p300 and PCAF to the receptor (reviewed in references 17 and 20). Using an immobilized vitamin D3 receptor (VDR) LBD affinity column, we previously isolated a complex of at least 15 VDR interacting proteins (DRIPs), ranging in size from 30 to 250 kDa, from Namalwa B-cell nuclear extracts (41, 42). These proteins selectively bind as a complex to VDR in a 1,25(OH)2D3-dependent manner. A single subunit, DRIP205 (also known as TRAP220), anchors the other 14 proteins comprising the DRIP complex to nuclear receptors. DRIP is essentially identical to the thyroid receptor-interacting TRAP-SMCC complex (15, 21) and the ARC coactivator complex (37). Importantly, the mouse homologue of DRIP205 was also identified as a PPARγ-binding protein (PBP-PPARBP) in a yeast two-hybrid screen using the PPARγ LBD as a bait (55). Overexpression of PBP moderately enhanced PPARγ-mediated transcriptional induction in a ligand-dependent manner, suggesting a regulatory role for the DRIP complex in PPARγ-mediated transactivation.
The DRIP complex contains several subunits found within the RNA polymerase II (Pol II)-interacting mediator-SRB complexes. We have recently demonstrated that DRIP is physically distinct from the p160-CBP complex (40). This suggests that p160-CBP coactivators might act initially to remodel nucleosomes through histone modifying activities and that DRIP, perhaps through mediator-SRB subunits, might function by directly targeting RNA Pol II holoenzyme to promoters. Moreover, the fact that both the p160 and DRIP coactivators utilize the same surface within the receptor LBD (i.e., the AF-2) suggests that they do not interact simultaneously with nuclear receptors, and a key question is what factors are influencing the recruitment by the receptor of one complex over the other.
In the work presented here, we report that PPARγ and RXR in the context of a DNA-bound heterodimer play distinct roles in recruiting DRIP versus p160 coactivators. We find that PPARγ and RXR selectively interacted with DRIP205 and p160 proteins in a ligand-dependent manner, where RXR-specific ligands only induced p160 binding to RXR, and PPARγ-specific ligands exclusively recruited DRIP205 but not p160 coactivators to PPARγ. This selectivity was not observed in interaction assays off DNA, implying that the specificity of the coactivator binding in response to ligand is strongly influenced by allosteric effects of DNA-bound heterodimers. These results also suggest that the cooperativity of PPARγ- and RXR-specific ligands on transactivation may occur at the level of coactivator recruitment and provides an explanation for how one complex is recruited over the other despite the similarities they appear to share in the resident determinants for nuclear receptor interaction.
MATERIALS AND METHODS
Expression and purification of GST fusion proteins.
Glutathione S-transferase (GST) fusion proteins were expressed in BL21 cells by induction with 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 22°C. Bacterial pellets were resuspended in phosphate-buffered saline containing 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 0.5 mM leupeptin. Cell suspensions were sonicated and centrifuged at 20,000 × g for 20 min. Supernatants were incubated with glutathione-Sepharose beads for 1 h at 4°C. Proteins bound to the beads were eluted with elution buffer (3 mg of reduced glutathione per ml, 0.1 M KCl, 20 mM Tris, 0.2 mM EDTA, 20% glycerol, 1 mM DTT, 0.5 mM PMSF, 0.05% NP-40, and 0.5 mM leupeptin). In order to ensure equal amount of proteins used for electrophoretic mobility shift assay (EMSA) eluted proteins were quantified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
EMSAs.
For all EMSAs, the DNA probe used was derived from the acyl-CoA oxidase PPRE. Complementary oligonucleotides (top strand, 5′-AGCTGGACCAGGACAA-3′) were annealed, followed by 32P-end labeling with T4 polynucleotide kinase. In vitro-translated PPARγ and RXR or baculovirus-expressed RXR (32) was mixed and incubated in the presence of various ligands on ice for 10 min in 18 μl of 1× binding buffer (20 mM Tris, pH 8.0; 1 mM EDTA; 50 mM KCl; 0.05% NP-40, 10% glycerol), 2 mM DTT, and 50 μg of poly(dI-dC) per ml. Labeled probe (typically 20,000 cpm per reaction) and purified coactivators as GST fusion proteins were added sequentially. After incubation for 30 min at room temperature, reaction mixtures were loaded on an 8% polyacrylamide nondenaturing gel and separated in 0.5× Tris-borate-EDTA at 4°C. Gels were dried prior to autoradiography.
Cell culture and nuclear extract preparation.
Namalwa cells (American Type Culture Collection) were cultured in 4-liter spinner flasks and maintained in RPMI medium supplemented with 5% fetal bovine serum, 5% calf serum and 300 μg of glutamine per ml. Nuclear extracts were prepared by the method described by Dignam et al. (11). NIH 3T3 cells were maintained in Eagle's minimal essential medium medium supplemented with 10% fetal bovine serum.
Transient-transfection assays.
NIH 3T3 cells were plated at a density of 5 × 105 cells/60-mm plate 24 h prior to transfection. One plate of cells was cotransfected with 5 μg of a PPRE-luc reporter (44), 1 μg each of cytomegalovirus (CMV)-RXR and CMV-PPAR expression plasmids, 2 μg of β-galactosidase expression vector, and the indicated amounts of coactivator expression vector. Carrier DNA was used to ensure that equal amounts of DNA were transfected in each plate. After 16 h, transfected cells were treated with ligands as described in the figure legends or with dimethyl sulfoxide (DMSO) for 24 h. Treated cells were harvested and lysed, and extracts were assayed for luciferase activity by dilution in cell culture lysis reagent (Promega) and measurement in 100 μl of luciferase assay reagent (Promega) in a luminometer. The luciferase activity of each sample was normalized by the level of β-galactosidase activity. Each transfection was carried out in duplicate and repeated at least three times.
GST pulldown assay.
For GST-PPARγ-LBD and DRIP complex interactions, 40 μg of GST-PPARγ-LBD fusion proteins immobilized on beads were incubated with 10−5 M GW1929 or DMSO in GST-binding buffer (20 mM Tris-HCl [pH 7.9], 180 mM KCl, 0.2 mM EDTA, 0.05% NP-40, 0.5 mM PMSF, 1 mM DTT) containing 1 mg of bovine serum albumin (BSA) for 2 h at 4°C. Immobilized PPARγ-LBD was incubated with approximately 2 mg of Namalwa nuclear extracts containing 200 mM KCl in the presence of 10−5 M GW1929 or DMSO for 16 h. Bound proteins were washed six times with 1 ml of washing buffer (GST-binding buffer containing 0.1% NP-40) and eluted by incubation with GST-binding buffer containing 0.1% NP-40 and 0.15% of Sarkosyl (Sigma) at 4°C for 20 min. Eluted proteins were separated by SDS-PAGE and visualized by silver nitrate staining. For GST-PPARγ and GRIP-1, SRC-1, ACTR, or DRIP205 interactions and GST-RXRα and GRIP-1, SRC-1, ACTR, or DRIP205 interactions, 15 μg of GST fusion protein immobilized on beads was incubated with GST-binding buffer containing 3.5 mg of BSA per ml at 4°C for 2 h. GST fusion proteins were then incubated with 0.5 μl of [35S]methionine-labeled in vitro-translated DRIP205 or GRIP-1 in the presence of the ligands, as described in the legend to each figure, at 4°C for 2 h. After three washes, the samples were resolved by SDS-PAGE, and the gels were dried and exposed to X-ray film.
RESULTS
Coactivators bind PPARγ in response to a range of PPARγ-specific compounds.
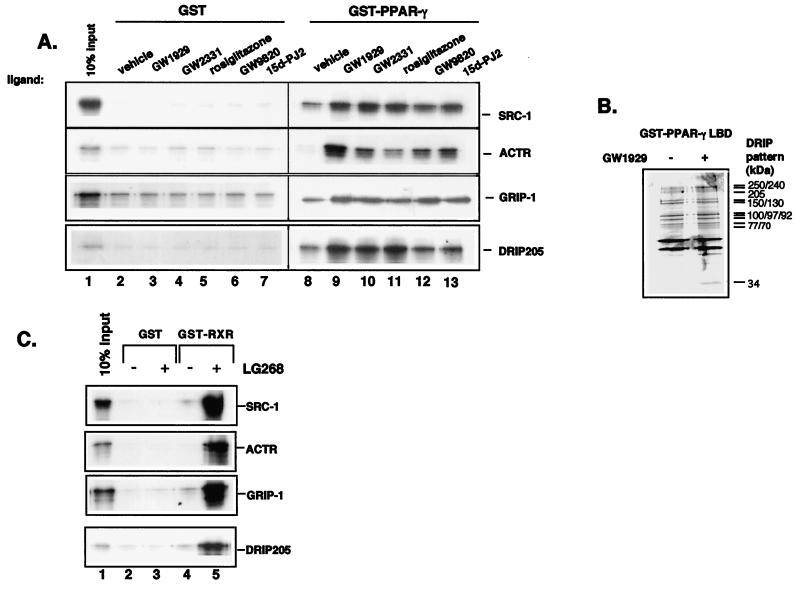
We were initially interested in ascertaining the efficacies of various PPARγ ligands on coactivator binding. To do so, we examined the abilities of DRIP205 or three different p160 coactivators, SRC-1, ACTR, and GRIP-1, to interact with full-length PPARγ in response to a natural ligand, 15d-PGJ2, two antidiabetic agents (rosiglitazone and GW1929), and two hypolipidemic agents (GW9820 and GW2331). Using GST–full-length PPARγ as the bait, all of the tested compounds enhanced the in vitro binding of SRC-1, ACTR, and GRIP-1 to PPARγ to a similar extent (Fig. 1A, rows 1 to 3, lanes 9 to 13). In contrast, only the two antidiabetic agents and one hypolipidemic compound (GW2331) exhibited potent effects in inducing PPARγ-DRIP205 interaction (Fig. 1A, row 4, lanes 9 to 11). The DRIP205 interaction is likely to represent the binding of the entire multisubunit DRIP complex, since the PPARγ-LBD pulled down the entire complex from nuclear extracts, albeit in a ligand-independent manner (Fig. 1B). We also did not observe strong ligand stimulation of DRIP205 binding using GST-PPARγ-LBD as a bait (data not shown), suggesting that some regions beyond the LBD also play critical roles in mediating ligand effects in recruiting DRIP205 and other coactivators.
FIG. 1.
(A) Inducible interactions between PPARγ and coactivators by various PPAR ligands. GST (lanes 2 to 7) or GST-PPARγ (lanes 8 to 13) was incubated with in vitro-translated 35S-labeled SRC-1 (row 1), ACTR (row 2), GRIP-1 (row 3), and DRIP205 (row 4) in the presence or absence (lanes 2 and 8) of 10−5 M concentrations of the indicated ligands. The input (10%) of each in vitro-translated protein is indicated in lane 1. (B) Interaction of PPARγ-LBD and the multisubunit DRIP coactivator complex from Namalwa B-cell nuclear extracts. Immobilized GST-PPARγ LBD was incubated with a Namalwa B-cell nuclear extract in the presence of vehicle or 10−5 M GW1929. Bound proteins were eluted with N-lauroyl Sarkosine. Eluted proteins were separated by SDS–7.5% PAGE and visualized by silver nitrate staining. (C) Ligand-inducible interactions between RXR and coactivators. GST (lanes 2 and 3) or GST-RXR (lanes 4 and 5) was incubated with in vitro-translated 35S-labeled SRC-1 (row 1), ACTR (row 2), GRIP-1 (row 3), or DRIP205 (row 4) in the absence (lanes 2 and 4) or presence (lanes 3 and 5) of 10−6 M LG268. Lane 1 in each row represents 10% of the input in vitro-translated proteins.
Since PPARγ binds to DNA as a heterodimer with RXR, we also examined the effect of LG268, an RXR-specific ligand, on DRIP205 and p160 coactivator binding to GST-RXR. LG268 was able to induce the binding of all the tested coactivators to RXR (Fig. 1C). The induction of p160 protein binding to RXR appeared more pronounced relative to DRIP205, suggesting that RXR might have an intrinsic preference for p160 coactivators over the functionally and structurally distinct DRIP205.
DRIP and p160 coactivators exhibit highly selective binding to PPARγ and RXR in the context of a DNA-bound heterodimer.
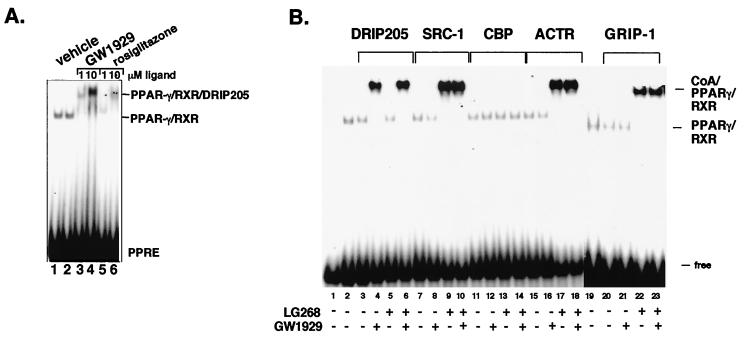
Gel EMSAs were carried out to determine whether the ability of the various PPARγ-specific ligands to induce DRIP205 binding observed in the GST pulldown assays could be recapitulated in the form of an PPARγ-RXR heterodimer bound to the acyl-CoA oxidase PPRE (13). Initially, we compared two doses of two antidiabetic agents, rosiglitazone and GW1929 (Fig. 2A). The results indicate that the minimum concentration of rosiglitazone required for inducing DRIP205 binding, manifested in the form of a supershift of the bound PPARγ-RXR heterodimer, was 10−5 M, whereas GW1929 was at least 10-fold more potent than rosiglitazone (lane 3 versus lane 6). Since the antidiabetic effect of GW1929 is known to be 2 orders of magnitude more potent than rosiglitazone (3), it is tempting to speculate that these compounds' biological activities are directly related to their abilities to recruit the DRIP complex.
FIG. 2.
Ligand-selective binding of coactivators to DNA-bound PPARγ-RXR heterodimers. (A) Baculovirus-expressed RXR and in vitro-translated PPARγ were combined and incubated without ligand (lanes l and 2) or in the presence of indicated concentrations of GW1929 (lane 3 and 4) or rosiglitazone (lanes 5 and 6). Receptors were combined with a radiolabeled PPRE probe and GST-DRIP205 (lanes 2 to 6) and DNA-bound complexes separated by EMSA. The PPARγ-RXR heterodimer and the coactivator-heterodimer complexes are indicated. (B) Baculovirus-expressed RXR and in vitro-translated PPARγ were incubated without ligand (lanes 3, 7, 11, 15, and 20) or in the presence of 10−5 M GW1929 (lanes 4, 8, 12, 16, and 21 ), 10−8 M LG268 (lanes 5, 9, 13, 17, and 22), or both ligands (lanes 6, 10, 14, 18, and 23). These samples were then combined with the radiolabeled PPRE probe and either GST-DRIP205 (lanes 3 to 6), GST-SRC-1 (lanes 7 to 10), GST-CBP (lanes 11 to 14), GST-ACTR (lanes 15 to 18), or GST-GRIP-1 (lanes 20 to 23) and subjected to EMSA. The PPARγ-RXR heterodimer and the coactivator-heterodimer complexes are indicated.
EMSA was then used to examine the effects of one PPARγ-specific and one RXR-specific ligand on their respective abilities to induce coactivator binding in the context of the PPARγ-RXR heterodimer bound to DNA. As shown in Fig. 2B, no coactivator associated with PPARγ-RXR in the absence of ligands (lanes 3, 7, 11, and 15). We chose to use the PPARγ-specific compound GW1929, since it induced strong binding of DRIP205 to the heterodimer in Fig. 2A. Remarkably, 10−5 M GW1929 selectively induced DRIP205 binding to the PPARγ-RXR heterodimer (lane 4), whereas it failed to induce binding to three p160 coactivators, SRC-1, ACTR, and GRIP-1 (lanes 8, 16, and 21). Conversely, the addition of 10−8 M LG268 had no effect on DRIP205 binding (lanes 5 and 6) but induced strong binding of SRC-1 (lanes 9 and 10), ACTR (lanes 17 and 18), and GRIP-1 (lanes 22 and 23) to the heterodimer. No CBP binding was observed in the presence of either ligand alone or both ligands added simutaneously (lanes 11 to 14). Therefore, in the context of DNA binding, RXR and PPARγ ligands exhibited strong selectivity in inducing two distinct classes coactivators to associate with the heterodimer.
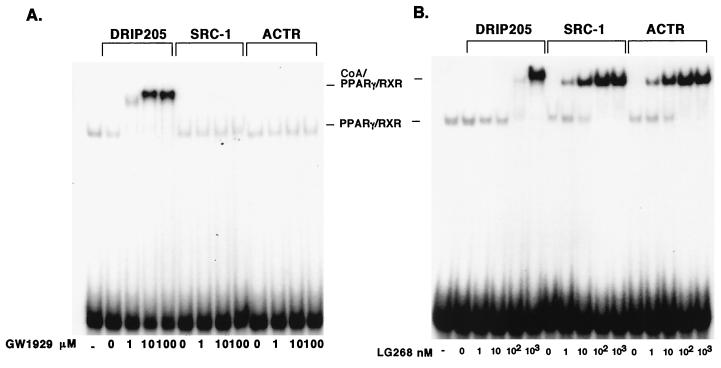
We also compared dose titrations of GW1929 and LG268 on DRIP205, SRC-1, and ACTR binding to the DNA-bound PPARγ-RXR heterodimer. GW1929 induced DRIP205 binding in a dose-dependent manner, starting at 10−6 M (Fig. 3A). No SRC-1 or ACTR binding was observed with GW1929, even at the highest concentration used (10−4 M). In contrast, LG268 was able induce SRC-1 or ACTR binding at concentrations as low as 10−9 M (Fig. 3B). The induction appeared to reach a plateau at concentrations higher than 10−7 M. It is noteworthy that high doses of LG268 also resulted in the binding of DRIP205 to the heterodimer (Fig. 3B).
FIG. 3.
Ligand titrations of PPARγ-RXR coactivator interactions. (A) GW1929 titration of DRIP205 binding to PPARγ-RXR. Purified RXR was mixed with in vitro-translated PPARγ and incubated on ice in the presence of the indicated concentrations of GW1929; the DNA probe and GST-DRIP205, GST-SRC-1, or GST-ACTR were added to samples and subjected to EMSA. (B) LG268 titration. Binding and EMSA conditions were exactly as described for panel A.
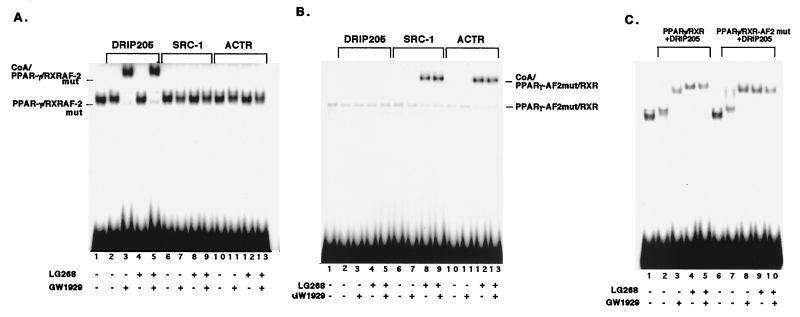
In order to ascertain the relative and specific contributions RXR and PPARγ play in recruiting coactivators, we performed an EMSA with receptors carrying point mutations in helix 12 of their AF-2's that have been previously shown to eliminate their ability to bind coactivators and transactivate PPARγ AF-2 (L466A and L467A) and RXR AF-2 (M454A and L455A) (43). The RXR AF-2 mutant completely abolished the binding of SRC-1 and ACTR to the PPARγ-RXR heterodimer but had no effect on DRIP205 binding (Fig. 4A). Conversely, when the PPARγ AF-2 mutant was used, no DRIP205 binding was detected, while SRC-1 and ACTR could bind to the heterodimer (Fig. 4B). These results indicate that the AF-2 of RXR and PPAR are directly responsible for recruiting p160 coactivators and DRIP205, respectively, in response to their cognate ligands.
FIG. 4.
AF2 mutations abolish coactivator binding in a receptor-restrictive manner. (A) RXR AF-2 mutation abolishes SRC-1 and ACTR, but not DRIP205, binding to PPARγ-RXR. In vitro-translated wild-type PPARγ and AF-2 mutated RXR (M454A and L455A) were combined and incubated on ice for 10 min in the absence of ligands (lanes 2, 6, and 10) or in the presence of 10−5 M GW1929 (lanes 3, 7, and 11), 10−8 M LG268 (lanes 4, 8, and 12), or both ligands (lanes 5, 9, and 13). DNA probe and GST-DRIP205 (lanes 2 to 5), GST–SRC-1 (lanes 3 to 9), or GST-ACTR (lanes 10 to 13) were added to the samples, and the indicated complexes were resolved by EMSA. (B) The PPARγ AF-2 mutation abolishes DRIP205, but not SRC-1 or ACTR binding to PPARγ-RXR. Purified RXR and in vitro-translated AF-2 mutated PPARγ (L466A and L467A) were combined, and assays were carried out as described for panel A. (C) Comparison of DRIP205 binding to PPARγ-RXR and PPARγ-RXR AF-2 mutant at high LG268 concentrations. PPARγ was incubated with either wild-type RXR (lanes 2 to 5) or AF-2 mutated RXR (lanes 7 to 10), and assays were carried out as described above, except that 10−6 M LG268 was used.
We also used the AF2 mutants to test whether high concentrations of LG268 that induced DRIP205 binding to the PPARγ-RXR heteodimer observed in Fig. 3B was due to the so-called phantom ligand effect (43). Here, we examined the effects of nanomolar concentrations of LG268 on the binding of DRIP205 to the PPARγ-RXR AF-2 mutant (Fig. 4C). When the RXR AF-2 mutant was used, the mutated heterodimer could bind DRIP205 to an extent similar to that of the wild type (Fig. 4C; lanes 4, 5, 9, and 10), indicating that high levels of LG268 do not induce the direct binding of DRIP205 to RXR but rather that the binding of LG268 may lead to a conformational change in PPARγ, consequently inducing the binding of DRIP205 to PPARγ.
Binding of DRIP205 to PPARγ-RXR occurs through NR box 1.
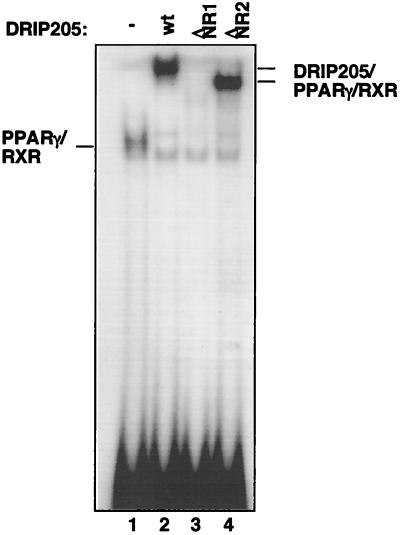
Two closely spaced LXXLL signature motifs were previously identified within the DRIP205 sequence (40, 54). These motifs, referred to as NR 1 and NR 2, are located at residues 589 to 593 and 630 to 634, respectively. We recently showed that NR box 2 is necessary for directing DRIP205 binding to VDR (40). Similar results have been reported for interactions with TR (54). The opposite appears to be the case for estrogen receptor (ER) interactions with DRIP205 (4). Elimination of NR1 by either point mutation or deletion completely abolished DRIP205 binding to ER, whereas NR2 had little effect on the DRIP205-ER interaction. Here, we asked which NR box is required for DRIP205 binding to PPARγ-RXR. Two DRIP205 deletion fragments containing either NR1 (residues 527 to 604) or NR2 (residues 604 to 774) were expressed as GST proteins and were incubated with PPARγ-RXR in the presence of GW1929 and resolved by EMSA. As shown in Fig. 5, the loss of NR box 1 completely eliminated the binding of DRIP205 to the DNA-bound heterodimer, while deletion of NR2 retained wild-type affinity for PPARγ-RXR (lanes 2 to 4). This indicates that, in contrast to VDR and TR, NR box 1 plays a central role in directing DRIP205 binding to PPARγ-RXR. Considering that RXR-VDR (and RXR-TR) and PPARγ-RXR have opposite polarities in the context of how they bind DNA, we speculate that the NR box requirements for DRIP205 binding may be determined by the polarity of the heterodimer.
FIG. 5.
NR box 1 of DRIP205 directs the binding of DRIP205 to PPARγ-RXR. Baculovirus-expressed RXR and in vitro-translated PPARγ were combined in the presence of 10−5 M GW1929 and 10−8 M LG268. PPRE probe and GST-DRIP205 (residues 527 to 774; lane 2), GST-DRIP205ΔNR1 (residues 604 to 774; lane 3), or GST-DRIP205 ΔNR2 (residues 527 to 604; lane 4) were then added, and the indicated complexes were resolved by EMSA.
p160 and DRIP205 coactivators cannot co-occupy PPARγ-RXR.
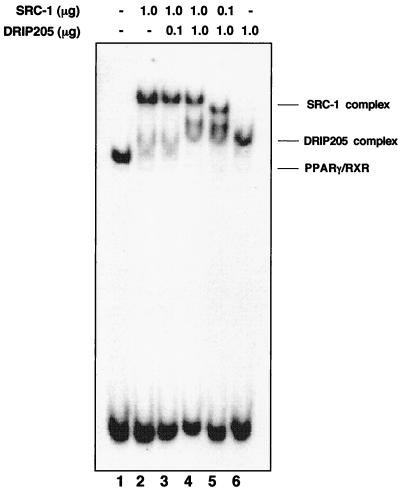
The finding that DRIP205 and p160 coactivators are able to selectively bind to PPARγ and RXR, respectively, raises the question of whether these two coactivators can interact simultaneously with the PPARγ-RXR heterodimer on DNA or whether they compete for binding. To address this, we coincubated various amounts of DRIP205 and SRC-1 with the PPARγ-RXR heterodimer in the presence of both ligands. When equal amounts of both coactivators were used, SRC-1 and DRIP205 (a fragment smaller than the one used in previous experiments to resolve differences in mobilities between the two coactivators) bound to the heterodimer in the presence of both LG268 and GW1929 as distinct complexes, represented by the formation of two distinguishable supershifts, albeit with stronger binding from SRC-1 (Fig. 6, lane 4). Consistent with the ability of SRC-1 to outcompete DRIP205 for binding to the heterodimer, a 1:10 ratio of SRC-1 and DRIP205 bound the PPARγ-RXR heterodimer to similar extents (lane 5), and a 10:1 ratio of SRC-1 to DRIP205 did not permit an association of DRIP with the PPARγ-RXR heterodimer at all (lane 3). Thus, SRC-1 appears to have a higher affinity for the PPARγ-RXR heterodimer than does DRIP205, so that at equivalent levels, SRC-1 would preferentially bind, at least in the presence of ligands for both receptors. Moreover, at all ratios of both coactivators used here, separate supershifted species were always observed (i.e., lanes 4 and 5), rather than the appearance of a slower, unique supershift that would presumably represent the co-occupancy of both coactivators. Therefore, the binding of SRC-1 and DRIP205 do not appear to occur simultaneously on each resident receptor subunit of the heterodimer.
FIG. 6.
SRC-1 excludes DRIP205 binding to PPARγ-RXR. Baculovirus-expressed RXR and in vitro-translated PPARγ were combined in the absence of ligands (lane 1) or in the presence of both 10−5 M GW1929 and 10−8 M LG268 (lanes 2 to 5), together with the indicated amounts of both SRC-1 and DRIP205. Complexes were then resolved by EMSA. Note that a truncated DRIP205 construct was used in this experiment to resolve the two coactivator complexes.
Selectivity of coactivator utilization in vivo.
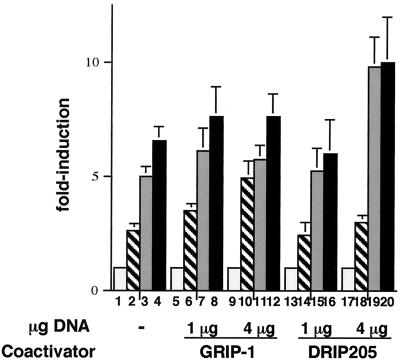
To determine whether the selective ligand effects on coactivator recruitment seen in the in vitro EMSA assays could be reflected in cells at the level of transactivation in response to either PPARγ- or RXR-specific ligands, we carried out cotransfection experiments with either DRIP205 or GRIP-1 overexpressing plasmids. As shown in Fig. 7, when 4 μg of either coactivator DNA and (PPRE)3-tk-Luc reporter plasmids were used to transfect NIH 3T3 cells in the presence of the PPARγ-specific GW1929 ligand, luciferase expression was further potentiated almost twofold by DRIP205 (lane 3 versus lane 19); this potentiation was not observed with the p160 coactivator GRIP-1 (lane 3 versus lane 11). When the same experiment was carried out with RXR-specific ligand LG101305 (a compound that is structurally related to LG268 and is able to induce p160 binding to PPARγ-RXR in the same concentration range as LG268, [W.Y. and L.P.F., data not shown]), GRIP-1, but not DRIP205, was able to potentiate transactivation by PPARγ-RXR from the PPRE-directed reporter (lane 2 versus lane 10 and lane 2 versus lane 18, respectively). These effects on transactivation essentially recapitulate the results of the EMSA assays and support the notion that the two distinct ligands induce the recruitment of distinct coactivators to each partner in the PPARγ-RXR heterodimer.
FIG. 7.
DRIP205 but not GRIP-1 enhances transactivation in response to a PPARγ-specific ligand, and GRIP-1 but not DRIP205 enhances the response to an RXR-specific ligand. NIH 3T3 cells were transfected with a reporter containing three copies of the acyl-CoA oxidase PPRE cloned upstream of thymidine kinase promoter-luciferase and expression vectors for PPARγ, RXR-α, DRIP205, or GRIP-1. The amounts of coactivators are indicated. Transfected cells were treated with 10−5 M GW1929, 10−6 M LG101305, or both ligands for 24 h. Luciferase activity was normalized based on β-galactosidase activity. The results are expressed as the fold induction over the control. Each value represents the mean of three independent experiments. Bars: □, Vehicle; ▧, LG101305; ░⃞, GW1929; ▪, GW1929 plus LG101305.
DISCUSSION
PPARγ- and RXR-specific ligands lead to a cooperative effect on PPAR-mediated transactivation in vivo and in vitro (29, 36). In mouse models of non-insulin-dependent diabetes mellitus and obesity, either RXR- or PPARγ-specific ligands alone functioned as insulin sensitizers and significantly reduced fasting glucose levels after 14 days of treatment. Combination treatment with both LG268 and rosiglitazone, albeit at submaximal doses, exhibited an even more potent effect. A molecular mechanism for this cooperativity, however, has not to this point been elucidated. Since facilitated recruitment of coactivators has been considered a general model for transcriptional synergy (see review in reference 5), we wished to examine this in the context of RXR- and PPARγ-specific ligands by dissecting their effects on coactivator recruitment. Utilization of the EMSA allowed us to assess the effects of these ligands on the recruitment of nuclear receptor coactivators on PPARγ-RXR heterodimers bound to a specific response element sequence. Our results clearly show that liganded PPARγ and RXR each exhibited a preference for two distinct classes of coactivators. The binding of PPARγ-specific ligands resulted in exclusive association of DRIP205; conversely, LG268, an RXR-selective ligand, led to only p160 (SRC-1, ACTR, and GRIP-1) binding. Furthermore, inactivation of PPARγ and RXR AF-2 domains eliminated the binding of DRIP205 and p160, respectively. Taken together, our data support the hypothesis that the observed synergy manifested by PPARγ- and RXR-specific ligands is a result of the recruitment of two distinct coactivator complexes that might be working in concert during transcriptional initiation (see below).
In contrast to the DNA binding results with PPARγ-RXR heterodimers, our initial GST pulldown assays in solution demonstrated no such selectivity of coactivator recruitment, such that either liganded RXR or liganded PPARγ was able to interact with p160 and DRIP205. These results argue for decisive conformational differences induced by heterodimerization and/or DNA binding. The crystal structure of rosiglitazone-bound PPARγ-RXR is significantly different from that of rosiglitazone-bound PPARγ homodimer (18, 38). A hydrogen bond between rosiglitazone and Q286 of PPARγ was observed in the homodimer but not in the PPARγ-RXR heterodimer. The rosiglitazone side chain showed a different gauche conformation in the heterodimer than in the homodimer. Moreover, the pyridyl nitrogen of rosiglitazone formed a hydrogen bond with a water molecule within the ligand pocket in the heterodimer that is not seen in the homodimer. Clearly, such conformational changes resulting from heterodimerization could conceivably alter the ability of the receptors in binding coactivators.
The crystal structures have also revealed the conformational difference between rosiglitazone-bound and a tyrosine-based molecule (GI262570)-bound PPARγ-RXR heterodimers. The binding of GI262570 to PPARγ-RXR resulted in additional hydrophobic interactions from a benzophenone group that were not available in the presence of rosiglitazone. As a result, the ligand pocket is 40% occupied by GI262570 but only 25% occupied by rosiglitazone. Accordingly, we found that GW1929, a tyrosine-based PPARγ ligand that is closely related to GI262570 (T. Willson, personal communication), was at least 10 times more potent in inducing DRIP205 binding to PPARγ-RXR heterodimer than rosiglitazone (Fig. 2A). Thus, the ability of PPARγ ligands to induce coactivator binding correlates with their interaction with PPARγ.
Interestingly, although 10−8 M LG268 exclusively induced the binding of p160 proteins to PPARγ-RXR, higher concentrations of the ligand (10−6 or 10−5 M) resulted in the binding of DRIP205 and p160 as well. A mutation of RXR AF-2 had little effect on LG268-induced binding of DRIP205 to heterodimer, while the analogous mutation in PPARγ completely abolished DRIP205 binding. These results suggest that the induction of DRIP205 binding to PPARγ-RXR by the RXR ligand is not directly through the AF-2 of RXR but rather may lead to a favorable conformational change in PPARγ that allows the binding of DRIP205 to PPARγ AF-2. Schulman et al. reported that the ability of RXR ligands to act synergistically with PPARγ ligands did not require the former's hormone-dependent activation function (43). Furthermore, they found that the binding of LG268 to RXR altered the protease sensitivity of PPARγ in vitro, indicating a phantom effect of liganded RXR on PPARγ conformation. It is therefore possible that at these pharmacological doses, both receptors can recruit DRIP, perhaps contributing to the cooperative effects of both ligands observed in reporter assays.
It is noteworthy that micromolar concentrations of PPARγ ligand are required for inducing DRIP205 binding, while RXR ligand induces p160 protein binding in the nanomolar range. The crystal structure of the PPARγ-RXR heterodimer has revealed that the ligand-binding pocket of PPARγ (1,440 Å3) is much larger than that of RXR (470 Å3) (18). As a result, 9-cis-retinoic acid and rosiglitazone occupy 75 and 25%, respectively, of the available pockets in their LBDs. The larger ligand-binding pocket of PPARγ may also allow the binding of structurally diverse compounds with rather low affinity. For example, many naturally occurring fatty acids, such as linoleic acid, linolenic acid, arachidonic acid, eicosapentaenoic acid, and 15d-PGJ2, have been shown to bind PPARγ at micromolar concentrations (reviewed in reference 51). The concentrations of free fatty acids in normal human serum are in the range required to activate PPAR, although the effective concentrations of fatty acids within cells are difficult to ascertain (25).
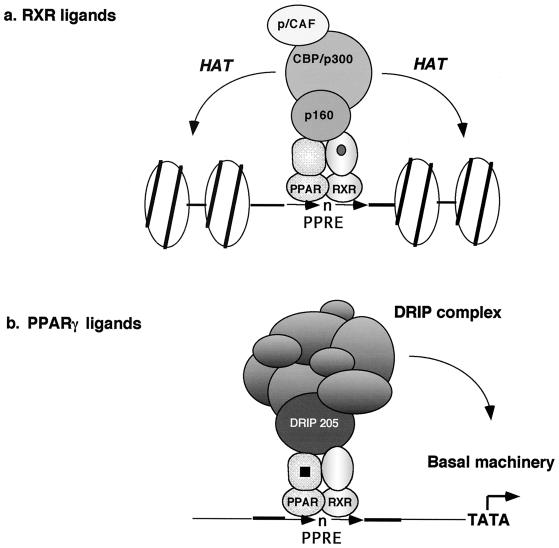
The differential selectivity of coactivator binding by PPARγ-RXR suggests that two coactivator complexes could be simultaneously bound by each subunit of the heterodimer in the presence of both ligands. If true, this would provide an attractive mechanism for cooperative effects observed with both ligands in mouse models of non-insulin-dependent diabetes (36). However, we observed no co-occupancy of p160 and DRIP205 to PPARγ-RXR under a variety of gel shift conditions. Instead, two distinct coactivator complexes were found in the presence of SRC-1 and DRIP205, albeit with different relative affinities for their specific receptor (Fig. 6). Similar results were reported when both thyroid hormone and 9-cis-retinoic acid stimulated the binding of DRIP205 (TRAP220) or p160 proteins to a DNA-bound TR-RXR heterodimer (47). While we cannot rule out technical limitations of the EMSA that restrict our ability to detect both coactivators simultaneously bound to the heterodimer, it is possible that steric hindrance impedes co-occupancy and that each coactivator acts independently to stimulate the transcription (Fig. 8). Another possibility is that the p160 and DRIP complexes mediate transcription in a sequential fashion (17). p160 members are structurally related to each other and bridge HAT-containing CPB/p300 and PCAF complexes to the receptor. They appear to mediate gene transcription by acetylating histone tails (and other factors), resulting in a remodeling of chromatin (reviewed in reference 33). On the other hand, the DRIP complex is structurally and functionally distinct from the p160 coactivators, and no HAT activity has been detected within DRIP complex (41). More importantly, the DRIP complex shares subunits with RNA Pol II holoenzyme and is able to associate with Pol II in the presence of receptor and ligand, suggesting that the DRIP complex may act to facilitate the recruitment of Pol II holoenzyme by creating a binding surface (7). Thus, the p160 and DRIP complexes appear to be functionally complementary and may work sequentially to stimulate gene transcription. Indeed, our results indicate that SRC-1 has a higher affinity for DNA-bound PPARγ-RXR heterodimer than DRIP205 (Fig. 6), suggesting that at steady state in the presence of both RXR- and PPARγ-specific ligands, p160 binding will outcompete the DRIP complex. In addition, the binding of DRIP205 to PPARγ requires higher concentrations of ligand than comparable binding of p160 coactivators to RXR. Thus, at steady state, initial coactivator binding should be the p160 complex to RXR, which could then be followed by the subsequent dissociation of this coactivator (6) and recruitment of the DRIP complex to PPARγ.
FIG. 8.
Model for the selective recruitment of coactivator complexes by PPARγ- and RXR-specific ligands. See text for details.
A question that arises from such a model is why a single ligand is sufficient to induce transactivation mediated by PPARγ-RXR. Endogenous PPARγ ligands such as fatty acids or their metabolites, and RXR ligands such as 9-cis-retinoic acid may reside in cells. Thus, these naturally occurring ligands would lead to a basal activity often observed for PPARγ without the addition of exogenous ligand. Conceivably, the exogenous addition of a synthetic PPARγ ligand would potentiate the transactivation of a responsive reporter gene by enodgenous RXR ligands through an enhancement of DRIP complex recruitment. Conversely, the addition of a synthetic RXR ligand would result in an increased recruitment of p160 coactivators and consequently potentiate the effect of endogenous PPAR ligands. Clearly, a role for the selective recruitment of coactivator complexes by specific ligands must now be considered in light of the results presented here.
ACKNOWLEDGMENTS
We thank B. Forman, M. Lazar, and I. Schulman for plasmids; T. Willson and R. Heyman for PPARγ and RXR ligands, respectively; and B. Forman, T. Willson, and R. Heyman for valuable suggestions and discussions.
This work was supported by grants from the NIH (DK45460 and DK 52621) to L.P.F. W.Y. was supported by the Endocrine Research Training Program (DK07313).
REFERENCES
- 1.Berger J, Bailey P, Biswas C, Cullinan C A, Doebber T W, Hayes N S, Saperstein R, Smith R G, Leibowitz M D. Thiazolidinediones produce a conformational change in peroxisomal proliferator-activated receptor-gamma: binding and activation correlate with antidiabetic actions in db/dbmice. Endocrinology. 1996;137:4189–4195. doi: 10.1210/endo.137.10.8828476. [DOI] [PubMed] [Google Scholar]
- 2.Brockman J A, Gupta R A, Dubois R N. Activation of PPARγ leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology. 1998;115:1049–1055. doi: 10.1016/s0016-5085(98)70072-1. [DOI] [PubMed] [Google Scholar]
- 3.Brown K K, Henke B R, Blanchard S G, Cobb J E, Mook R, Kaldor I, Kliewer S A, Lehmann J M, Lenhard J M, Harrington W W, Novak P J, Faison W, Binz J G, Hashim M A, Oliver W O, Brown H R, Parks D J, Plunket K D, Tong W Q, Menius J A, Adkison K, Noble S A, Willson T M. A novel N-aryl tyrosine activator of peroxisome proliferator-activated receptor-gamma reverses the diabetic phenotype of the Zucker diabetic fatty rat. Diabetes. 1999;48:1415–1424. doi: 10.2337/diabetes.48.7.1415. [DOI] [PubMed] [Google Scholar]
- 4.Burakov D, Wong C W, Rachez C, Cheskis B J, Freedman L P. Functional interactions between the estrogen receptor and DRIP205, a subunit of the heteromeric DRIP coactivator complex. J Biol Chem. 2000;275:20928–20934. doi: 10.1074/jbc.M002013200. [DOI] [PubMed] [Google Scholar]
- 5.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 7.Chiba N, Suldan Z, Freedman L P, Parvin J D. Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J Biol Chem. 2000;275:10719–10722. doi: 10.1074/jbc.275.15.10719. [DOI] [PubMed] [Google Scholar]
- 8.Cobb J E, Blanchard S G, Boswell E G, Brown K K, Charifson P S, Cooper J P, Collins J L, Dezube M, Henke B R, Hull-Ryde E A, Lake D H, Lenhard J M, Oliver W, Jr, Oplinger J, Pentti M, Parks D J, Plunket K D, Tong W Q. N-(2-Benzoylphenyl)-l-tyrosine PPARγ agonists. 3. Structure-activity relationship and optimization of the N-aryl substituent. J Med Chem. 1998;41:5055–5069. doi: 10.1021/jm980414r. [DOI] [PubMed] [Google Scholar]
- 9.Collins J L, Blanchard S G, Boswell G E, Charifson P S, Cobb J E, Henke B R, Hull-Ryde E A, Kazmierski W M, Lake D H, Leesnitzer L M, Lehmann J, Lenhard J M, Orband-Miller L A, Gray-Nunez Y, Parks D J, Plunkett K D, Tong W Q. N-(2-Benzoylphenyl)-l-tyrosine PPARγ agonists. 2. Structure-activity relationship and optimization of the phenyl alkyl ether moiety. J Med Chem. 1998;41:5037–54. doi: 10.1021/jm980413z. [DOI] [PubMed] [Google Scholar]
- 10.Demetri G D, Fletcher C D, Mueller E, Sarraf P, Naujoks R, Campbell N, Spiegelman B M, Singer S. Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-gamma ligand troglitazone in patients with liposarcoma. Proc Natl Acad Sci USA. 1999;96:3951–3956. doi: 10.1073/pnas.96.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiRenzo J, Soderstrom M, Kurokawa R, Ogliastro M H, Ricote M, Ingrey S, Horlein A, Rosenfeld M G, Glass C K. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. Mol Cell Biol. 1997;17:2166–2176. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 14.Elstner E, Muller C, Koshizuka K, Williamson E A, Park D, Asou H, Shintaku P, Said J W, Heber D, Koeffler H P. Ligands for peroxisome proliferator-activated receptor gamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci USA. 1998;95:8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. 15-Deoxy-delta12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 17.Freedman L P. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 18.Gampe R T J, Montana V G, Lambert M H, Miller A B, Bledsoe R K, Milburn M V, Kliewer S A, Willson T A, Xu E H. Asymmetry in the PPAR-gamma/RXR-alpha structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 19.Gearing K L, Gottlicher M, Teboul M, Widmark E, Gustafsson J A. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci USA. 1993;90:1440–1444. doi: 10.1073/pnas.90.4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- 21.Gu W, Malik S, Ito M, Yuan C X, Fondell J D, Zhang X, Martinez E, Qin J, Roeder R G. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 22.Henke B R, Blanchard S G, Brackeen M F, Brown K K, Cobb J E, Collins J L, Harrington W W, Jr, Hashim M A, Hull-Ryde E A, Kaldor I, Kliewer S A, Lake D H, Leesnitzer L M, Lehmann J M, Lenhard J M, Orband-Miller L A, Miller J F, Mook R A, Jr, Noble S A, Oliver W, Jr, Parks D J, Plunket K D, Szewczyk J R, Willson T M. N-(2-Benzoylphenyl)-l-tyrosine PPARγagonists. 1. Discovery of a novel series of potent antihyperglycemic and antihyperlipidemic agents. J Med Chem. 1998;41:5020–5036. doi: 10.1021/jm9804127. [DOI] [PubMed] [Google Scholar]
- 23.Huang J T, Welch J S, Ricote M, Binder C J, Willson T M, Kelly C, Witztum J L, Funk C D, Conrad D, Glass C K. Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 24.Ibrahimi A, Teboul L, Gaillard D, Amri E Z, Ailhaud G, Young P, Cawthorne M A, Grimaldi P A. Evidence for a common mechanism of action for fatty acids and thiazolidinedione antidiabetic agents on gene expression in preadipose cells. Mol Pharmacol. 1994;46:1070–1076. [PubMed] [Google Scholar]
- 25.Jungling E, Kammermeier H. A one-vial method for routine extraction and quantification of free fatty acids in blood and tissue by HPLC. Anal Biochem. 1988;171:150–157. doi: 10.1016/0003-2697(88)90136-4. [DOI] [PubMed] [Google Scholar]
- 26.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc Natl Acad Sci USA. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kletzien R F, Clarke S D, Ulrich R G. Enhancement of adipocyte differentiation by an insulin-sensitizing agent. Mol Pharmacol. 1992;41:393–398. [PubMed] [Google Scholar]
- 28.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 29.Kliewer S A, Umesono K, Noonan D J, Heyman R A, Evans R M. Convergence of 9-cis-retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leblanc B P, Stunnenberg H G. 9-cis-retinoic acid signaling: changing partners causes some excitement. Genes Dev. 1995;9:1811–1816. doi: 10.1101/gad.9.15.1811. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 32.Lemon B D, Fondell J D, Freedman L P. Retinoid X receptor-vitamin D3 receptor heterodimers promote stable preinitiation complex formation and direct 1,25-dihydroxyvitamin D3-dependent cell-free transcription. Mol Cell Biol. 1997;17:1923–1937. doi: 10.1128/mcb.17.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemon B D, Freedman L P. Nuclear receptor cofactors as chromatin remodelers. Curr Opin Genet Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 34.Mandrup S, Lane M D. Regulating adipogenesis. J Biol Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- 35.Mueller E, Sarraf P, Tontonoz P, Evans R M, Martin K J, Zhang M, Fletcher C, Singer S, Spiegelman B M. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 36.Mukherjee R, Davies P J, Crombie D L, Bischoff E D, Cesario R M, Jow L, Hamann L G, Boehm M F, Mondon C E, Nadzan A M, Paterniti J R, Jr, Heyman R A. Sensitization of diabetic and obese mice to insulin by retinoid X receptor agonists. Nature. 1997;386:407–410. doi: 10.1038/386407a0. [DOI] [PubMed] [Google Scholar]
- 37.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 38.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 39.Palmer C N, Hsu M H, Griffin H J, Johnson E F. Novel sequence determinants in peroxisome proliferator signaling. J Biol Chem. 1995;270:16114–16121. doi: 10.1074/jbc.270.27.16114. [DOI] [PubMed] [Google Scholar]
- 40.Rachez C, Gamble M, Chang C P, Atkins G B, Lazar M A, Freedman L P. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol Cell Biol. 2000;20:2718–2726. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 42.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulman I G, Shao G, Heyman R A. Transactivation by retinoid X receptor-peroxisome proliferator-activated receptor gamma (PPARγ) heterodimers: intermolecular synergy requires only the PPARγ hormone-dependent activation function. Mol Cell Biol. 1998;18:3483–3494. doi: 10.1128/mcb.18.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao D, Rangwala S M, Bailey S T, Krakow S L, Reginato M J, Lazar M A. Interdomain communication regulating ligand binding by PPAR-gamma. Nature. 1998;396:377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 45.Tontonoz P, Hu E, Spiegelman B M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 46.Tontonoz P, Singer S, Forman B M, Sarraf P, Fletcher J A, Fletcher C D, Brun R P, Mueller E, Altiok S, Oppenheim H, Evans R M, Spiegelman B M. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid X receptor. Proc Natl Acad Sci USA. 1997;94:237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Treuter E, Johansson L, Thomsen J S, Warnmark A, Leers J, Pelto-Huikko M, Sjoberg M, Wright A P, Spyrou G, Gustafsson J A. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- 48.Tugwood J D, Issemann I, Anderson R G, Bundell K R, McPheat W L, Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 1992;11:433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varanasi U, Chu R, Huang Q, Castellon R, Yeldandi A V, Reddy J K. Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J Biol Chem. 1996;271:2147–2155. doi: 10.1074/jbc.271.4.2147. [DOI] [PubMed] [Google Scholar]
- 50.Werman A, Hollenberg A, Solanes G, Bjorbaek C, Vidal-Puig A J, Flier J S. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor gamma (PPARγ). Differential activity of PPARγ1 and −2 isoforms and influence of insulin. J Biol Chem. 1997;272:20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- 51.Willson T M, Brown P J, Sternbach D D, Henke B R. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 52.Willson T M, Cobb J E, Cowan D J, Wiethe R W, Correa I D, Prakash S R, Beck K D, Moore L B, Kliewer S A, Lehmann J M. The structure-activity relationship between peroxisome proliferator-activated receptor gamma agonism and the antihyperglycemic activity of thiazolidinediones. J Med Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 53.Yu K, Bayona W, Kallen C B, Harding H P, Ravera C P, McMahon G, Brown M, Lazar M A. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 54.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Y, Qi C, Jain S, Rao M S, Reddy J K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]