Abstract
p53 is an important mediator of the cellular stress response with roles in cell cycle control, DNA repair, and apoptosis. 53BP2, a p53-interacting protein, enhances p53 transactivation, impedes cell cycle progression, and promotes apoptosis through unknown mechanisms. We now demonstrate that endogenous 53BP2 levels increase following UV irradiation induced DNA damage in a p53-independent manner. In contrast, we found that the presence of a wild-type (but not mutant) p53 gene suppressed 53BP2 steady-state levels in cell lines with defined p53 genotypes. Likewise, expression of a tetracycline-regulated wild-type p53 cDNA in p53-null fibroblasts caused a reduction in 53BP2 protein levels. However, 53BP2 levels were not reduced if the tetracycline-regulated p53 cDNA was expressed after UV damage in these cells. This suggests that UV damage activates cellular factors that can relieve the p53-mediated suppression of 53BP2 protein. To address the physiologic significance of 53BP2 induction, we utilized stable cell lines with a ponasterone A-regulated 53BP2 cDNA. Conditional expression of 53BP2 cDNA lowered the apoptotic threshold and decreased clonogenic survival following UV irradiation. Conversely, attenuation of endogenous 53BP2 induction with an antisense oligonucleotide resulted in enhanced clonogenic survival following UV irradiation. These results demonstrate that 53BP2 is a DNA damage-inducible protein that promotes DNA damage-induced apoptosis. Furthermore, 53BP2 expression is highly regulated and involves both p53-dependent and p53-independent mechanisms. Our data provide new insight into 53BP2 function and open new avenues for investigation into the cellular response to genotoxic stress.
p53 is mutated in more than 50% of human cancers and plays a pivotal role in mediating cellular responses to stress signals such as DNA damage and hypoxia (24, 25). Loss of p53 function leads to defects in apoptosis, cell cycle arrest, and DNA repair, all of which lead to an increase in genomic instability (2, 8, 9, 20, 25). p53 protein can transcriptionally activate, as well as repress, a number of important target genes (24, 25, 28). Furthermore, p53 also appears to have important transcription-independent functions (2, 3, 6, 17, 20, 32). Precisely how, and in what context, p53 uses this diverse array of mechanisms to regulate these critical processes is not clear and is currently under intensive investigation.
In addition to covalent modifications of p53, protein-protein interactions play a major role in modulating p53 function (21, 24, 25). An example is the p53-MDM2 autoregulatory feedback loop that returns p53 to its low basal levels via proteasomal degradation after DNA damage (10, 12, 13, 17, 22, 37, 40). Many different p53-interacting proteins have been identified, though the functional significance of some of these interactions remains to be determined (24, 25). Understanding the complex pathways defined by p53-interacting proteins is therefore important for understanding the cellular response to stress signals.
This report examines the function of the p53-interacting protein, 53BP2, following DNA damage (18, 30). 53BP2 was originally identified by its ability to interact with wild-type (but not mutant) p53 in a yeast two-hybrid assay (18, 38). The full-length cDNA was subsequently isolated from a human cDNA library and contains an open reading frame encoding a protein migrating at approximately 165 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE (30). The crystal structure solution (2.2Å) of the p53-53BP2 complex has revealed that the SH3 domain and fourth ankyrin repeat of 53BP2's carboxy terminus binds two adjacent but discontinuous portions of an evolutionarily conserved surface on the p53 core domain to which most tumorigenic mutations map (5, 11). Indeed, two frequent p53 mutations in human cancer (R248 and R273) involve surface residues that are important p53-53BP2 intermolecular contact sites (11). However, the functional significance of this interaction remains to be determined.
The precise cellular function(s) of 53BP2 are not well characterized. 53BP2 can functionally interact with the p53 pathway by enhancing transcriptional activation of p53-reporter constructs and the endogenous p53-target gene p21 (19). Furthermore, 53BP2 can inhibit cell growth, impede cell cycle progression at G2/M, and induce apoptosis (30, 41). It has also been suggested that 53BP2 can partially suppress E1A and ras-mediated transformation of rat embryo fibroblasts (19). Together, these observations suggest that 53BP2 may be a potent modulator of cell growth and survival. As such, it would be predicted that 53BP2 protein levels would change in response to cellular stress. However, to date no information is available regarding the physiologic role of 53BP2 in response to cellular damage.
In this report, we describe several observations that suggest 53BP2 functions as part of the cellular response to genotoxic damage. First, 53BP2 protein levels increase following UV irradiation-induced DNA damage by a p53-independent mechanism. Second, 53BP2 expression is suppressed in unstressed cells by a p53-dependent mechanism. Third, 53BP2 expression sensitizes cells to apoptosis and decreases clonogenic survival following UV irradiation. Fourth, attenuation of 53BP2 induction enhances clonogenic survival following UV irradiation.
MATERIALS AND METHODS
Cell culture and cell lines.
Cells were grown in Dulbecco modified Eagle medium (DMEM; CellGro) with 10% (vol/vol) heat-treated fetal calf serum and 290 μg of l-glutamine, 100 U of penicillin, and 100 μg of streptomycin per ml and maintained in logarithmic growth at 37°C in 5% CO2. 041TR cells (8) were additionally maintained in 2 μg of tetracycline, 600 μg of G418, and 50 μg of hygromycin B per ml. 293/53BP2, 293/LZ, HT/53BP2, and HT/LZ cells were additionally maintained in 600 μg of G418 and 500 μg of Zeocin per ml. The pIND-53BP2 expression vector was constructed using standard techniques (34) by cloning the 53BP2 cDNA (30) into the BamHI-XbaI cloning site of the ecdysone-inducible expression vector pIND (Invitrogen). The HT/53BP2 cell line was constructed by cotransfecting the pIND-53BP2 expression vector and the pVgRXR vector (Invitrogen) into HT1080 cells and selecting cotransfectants in G418-Zeocin-containing media. Cells were subcloned and screened for 53BP2 protein expression by Western blot after ponasterone A induction. HT/LZ cells were similarly constructed using the pINDLacZ vector (Invitrogen). The 293/53BP2 cell line was constructed by stably transfecting pIND-53BP2 into EcR293 cells (Invitrogen). Stable transfectants were selected in G418-Zeocin-containing media, subcloned, and screened for 53BP2 protein expression by Western blot after ponasterone A induction. 293/LZ cells were similarly constructed using a pINDLacZ vector. Li-Fraumeni syndrome (LFS) fibroblasts were obtained from Michael Tainsky, M. D. Anderson Cancer Center, Houston, Tex. p21−/− and p53−/− mouse embryo fibroblasts (MEFs) were obtained from Al Fornace, National Cancer Institute, Bethesda, Md. XPA+/− MEFs were obtained from Kiyoji Tanaka, Osaka University, Osaka, Japan. All other cell lines were obtained from the American Type Culture Collection.
Antibodies and reagents.
Rabbit polyclonal anti-53BP2 (Rab-1) was raised against a GST-53BP2 fusion protein (18). A study on production and characterization of the anti-53BP2 mouse monoclonal antibody DX547 will be published elsewhere (D. J. O'Connor and X. Lu, manuscript in preparation). Anti-p53 mouse monoclonal antibody 1801 was from Santa Cruz Biotech, anti-tubulin mouse monoclonal antibody was from Sigma, anti-hsc70 mouse monoclonal antibody was from Alan Krensky, Stanford University, Calif., and goat anti-mouse and goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies were from Accurate.
Western blotting.
Total cellular lysates were prepared from subconfluent cultures and quantitated as previously described (8). Using standard techniques (34), lysates were resolved by SDS–6% PAGE, transferred to polyvinylidene difluoride membrane (Boehringer-Mannheim), blocked with 5% nonfat milk, probed with the specified primary antibody, and visualized with the appropriate HRP-conjugated secondary antibody and enhanced chemiluminescence (Pierce SuperSignal). Bands were quantitated in the linear range of X-ray film on a Bio-Rad scanning densitometer (GS-710) using Molecular Analysis software and normalized to a tubulin or hsc70 signal detected after reprobing the membrane.
Northern blotting.
Total RNA was single-step extracted from cells utilizing TRIzol reagent (Gibco-BRL)-chloroform, precipitated with isopropanol, dissolved in diethyl pyrocarbonate-treated H2O, and quantitated. Using standard techniques (34), total RNA was resolved on a 1.5% formaldehyde-agarose gel, transferred to nylon membrane, and hybridized with a 32P-labeled 53BP2 cDNA probe. The probe was prepared by isolating and purifying the BamHI-XbaI fragment (30) and labeled with random primers extended by Klenow fragment in the presence of deoxynucleoside triphosphates (dNTPs) and [32P]dCTP utilizing the RadPrime DNA Labeling System (Gibco-BRL). Images were captured, quantitated on a Bio-Rad phosphorimager, and normalized to a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or actin signal detected after stripping and reprobing the membrane.
Apoptosis and survival assays.
UV irradiation was delivered using a 15-W germicidal UV lamp (254 nm). Flow cytometry was performed on a Becton-Dickinson FACScan with 10,000 events counted per sample, as previously described (8). Nuclear morphology was determined by Hoechst staining and fluorescence microscopy as previously described (8). For clonogenic survival assays, exponentially growing cells were plated and exposed to UV irradiation from 0 to 25 J/m2. After culture for 12 days, cells were rinsed, fixed with methanol, and stained with methylene blue. Colonies with >50 cells were scored, and the plating efficiency and surviving fractions for each dose were calculated.
53BP2 antisense oligonucleotides.
Morpholino-oligonucleotides were introduced into cells by an osmotic delivery system. First, 20 μM 53BP2 antisense or control morpholino-oligonucleotide solutions were prepared in Osmotic Delivery Solution (GENE TOOLS) at a 680 μl final volume per 9.6 cm2. Then, immediately prior to delivery, the culture medium was completely aspirated from confluent HT1080 cells. Morpholino-oligonucleotides prepared in Osmotic Delivery Solution were then added, and the cells were incubated at 37°C with frequent gentle mixing for a maximum of 15 min. Oligonucleotide solutions were completely aspirated, and the cells were rinsed twice with 10 volumes of warmed DMEM and then incubated in complete medium at 37°C for at least 5 h prior to assaying them for phenotype changes. FITC-Dextran (GENE TOOLS) was used as a positive delivery control and revealed approximately 90% efficiency as determined by fluorescence microscopy. Morpholino-oligonucleotides were synthesized, purified, and analyzed by high-pressure liquid chromatography (HPLC) per GENE TOOLS criteria.
The 53BP2 antisense morpholino-oligonucleotide sequence used was 5′-ACATCAAACATTCGCTCATTATCCG-3′. The control morpholino-oligonucleotide sequence used was 5′-CCTCTTACCTCAGTTACAATTTATA-3′.
RESULTS
53BP2 levels increase following UV irradiation-induced DNA damage.
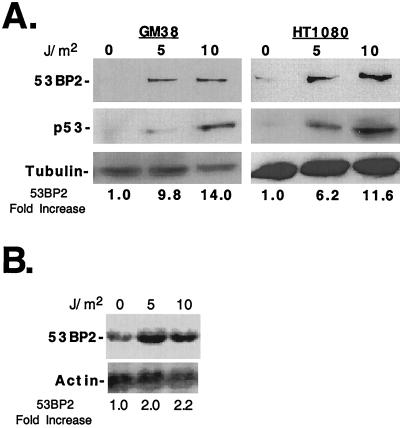
Overexpression of a 53BP2 transgene can arrest cell growth (30), induce apoptosis (41), and enhance p53 transactivation of the cell cycle inhibitor p21 (19), all of which are potential cellular responses to stress. However, using antibodies that can readily detect ectopically expressed 53BP2 protein in cells, we found that endogenous 53BP2 protein levels were very low in a variety of normal human tissues (Genotope, Inc.) (data not shown), a finding consistent with prior reports (30, 41). Therefore, we wondered if cellular stress, such as DNA damage, could signal a physiologic increase in endogenous 53BP2 levels. After UV irradiation of GM38 early-passage primary human skin fibroblasts, 53BP2 protein increased 9.8-fold after 5 J/m2 and 14-fold after 10 J/m2, as determined by quantitative Western blotting (Fig. 1A, left panels). We also UV irradiated HT1080 human fibrosarcoma cells and found that 53BP2 protein increased 6.2-fold after 5 J/m2 and 11.6-fold after 10 J/m2 (Fig. 1A, right panels). As expected, endogenous wild-type p53 was induced after UV irradiation in GM38 and HT1080 cells since both cell types contain wild-type p53 (Fig. 1A, middle panels). By Northern blotting, there was at least a 2.2-fold increase in 53BP2 mRNA levels normalized to actin (Fig. 1B). These results demonstrate that 53BP2 levels increase in response to UV-induced DNA damage in GM38 and HT1080 cells.
FIG. 1.
53BP2 levels increase following UV irradiation-induced DNA damage. (A) Western blots of lysates (30 μg of total protein per lane) prepared from GM38 cells (left panels) and HT1080 cells (right panels) 24 h after UV irradiation with 0, 5, or 10 J/m2. The fold increase of 53BP2 protein levels compared to no-UV-irradiation levels are indicated below, normalized for tubulin. 53BP2 was detected with monoclonal antibody DX547; p53 was detected with monoclonal antibody 1801. (B) Northern blot of total RNA (15 μg/lane) prepared from HT1080 cells 24 h after UV irradiation at the indicated doses, probed for 53BP2, stripped, and reprobed for actin. The fold increase of 53BP2 mRNA is indicated below, normalized for actin.
53BP2 expression is suppressed by a wild-type p53 gene in unstressed cells.
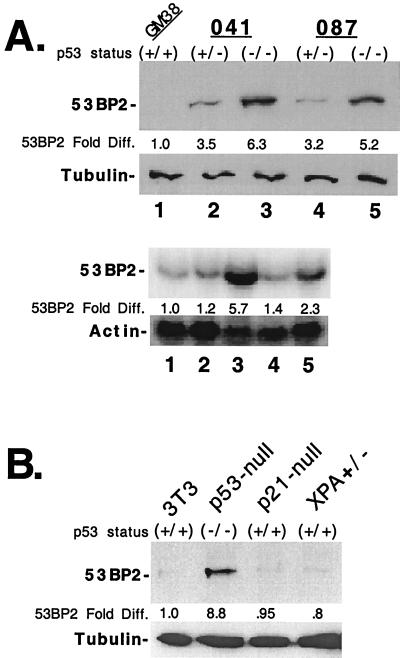
Although 53BP2 levels increased after UV-induced DNA damage, it remained unclear what regulated 53BP2 steady-state levels. Therefore, we sought to further characterize what factors influenced endogenous 53BP2 levels in unstressed cells. Several observations suggested that the p53 pathway could be involved. First, 53BP2 interacts with wild-type (but not mutant) p53 protein (11, 18, 38). Second, 53BP2 can functionally interact with the p53 pathway to enhance p53 transactivation (19). Thus, we wondered if endogenous 53BP2 levels were influenced by the mutational status of p53. We therefore determined 53BP2 levels in human skin fibroblasts derived from individuals with LFS with different p53-inactivating mutations (4, 26, 36, 42). LFS 041+/− cells are heterozygous for a frameshift mutation at one allele that results in a truncated and unstable p53 protein (26). LFS 087+/− cells are heterozygous for a missense mutation at codon 248 (Arg to Trp) (4). That mutation disrupts sequence-specific DNA binding, as well as 53BP2 protein binding in vitro, and is frequently found in tumors (4, 11). Homozygous p53-inactivated derivatives from both of these cell lines were obtained after spontaneous immortalization and loss of p53 heterozygosity in tissue culture (26, 36, 42). By quantitative Western blot, we found that LFS cells, both heterozygous and homozygous for p53 mutations, expressed higher levels of 53BP2 protein than wild-type p53 containing normal primary human skin fibroblasts (GM38) (Fig. 2A, upper panels). In fact, LFS cells mutated at both p53 alleles (lanes 3 and 5) expressed more 53BP2 protein than cells mutated at one allele (lanes 2 and 4) when normalized for tubulin (Fig. 2A, upper panels). By quantitative Northern blot, 53BP2 mRNA levels were also increased in cells mutated at both p53 alleles when normalized for actin (Fig. 2A, lower panels). Compared to GM38 cells, 041−/− cells had 5.7-fold more mRNA and 087−/− cells had 2.3-fold more mRNA (lanes 3 and 5, respectively). 53BP2 mRNA levels were modestly increased (up to 1.4-fold) in cells heterozygous for p53 mutations (lanes 2 and 4). These results demonstrate that endogenous levels of 53BP2 are suppressed by the presence of a wild-type p53 gene in unstressed fibroblasts. This is in contrast to the elevated levels of 53BP2 seen after UV irradiation induced DNA damage.
FIG. 2.
53BP2 expression in cell lines containing wild-type or mutant p53. (A) The top panels show Western blots of lysates (30 μg of total protein per lane) prepared from human 041 and 087 LFS skin fibroblasts heterozygous for wild-type p53 (+/−) (lanes 2 and 4, respectively) or homozygous for p53 mutations (−/−) (lanes 3 and 5, respectively) and from normal primary human skin fibroblasts (GM38) homozygous for wild-type p53 (+/+) (lane 1). The fold differences, compared to GM38 cells, of 53BP2 protein normalized for tubulin are indicated below. 53BP2 protein was detected with monoclonal antibody DX547. The bottom panels show Northern blots of total RNA (15 μg/lane) prepared from cells as indicated above, probed for 53BP2, stripped, and reprobed for actin. The fold differences, compared to GM38 cells, of 53BP2 mRNA normalized for actin are indicated below. (B) Western blots of lysates (30 μg of total protein per lane) prepared from mouse fibroblasts homozygous for wild-type p53 (3T3, p21-null, and XPA+/−) or from p53 knockout mouse fibroblasts (p53-null). The fold differences, compared to 3T3 cells, of 53BP2 protein normalized for tubulin are indicated below. 53BP2 detected with rabbit antisera Rab-1.
To demonstrate that suppression of endogenous 53BP2 protein was not unique to LFS fibroblasts, we determined 53BP2 protein levels in MEFs derived from wild-type p53 or p53-null mice using a rabbit anti-53BP2 antibody (Rab-1). By quantitative Western blot, MEFs from p53-null mice had >8.5-fold-higher levels of 53BP2 protein, normalized for tubulin, compared to MEFs with wild-type p53 from p21-null or XPA+/− mice or from NIH 3T3 fibroblasts containing wild-type p53 (Fig. 2B). These results suggest that in mouse fibroblasts, 53BP2 protein levels are also suppressed in a p53-dependent manner.
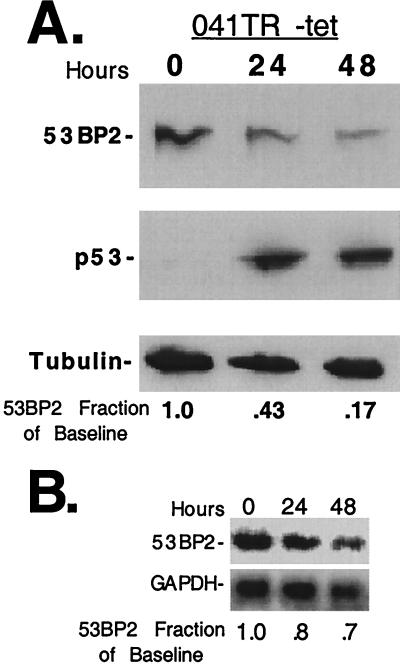
To further confirm the role of p53 in suppressing 53BP2 protein, we examined 53BP2 levels after induction of a tetracycline-regulated wild-type p53 gene in a p53-null human fibroblast cell line (041TR) (8). By utilizing a p53-inducible cell line, we could determine how p53 affects 53BP2 levels in the absence of cellular damage. p53-null LFS 041−/− fibroblasts express high levels of 53BP2 protein (Fig. 2A) and are the parent cell line of 041TR cells (1, 8). As expected in the absence of p53 expression, 041TR cells express high levels of 53BP2 protein as seen by Western blot (Fig. 3A, time zero). However, after p53 expression was induced by withdrawing tetracycline, 53BP2 protein decreased to 9 to 17% of baseline levels by 48 h (Fig. 3A and data not shown). 53BP2 transcript decreased to 48 to 70% of baseline levels by 48 h, as determined by Northern blotting after tetracycline withdrawal (Fig. 3B and data not shown). Reduction of 53BP2 was not simply due to induction of apoptosis as determined by nuclear morphology at 48 h (<15% apoptotic nuclei). Lack of significant apoptosis after p53 expression in the absence of cellular damage has been previously described in 041TR cells (8). These results demonstrate that wild-type p53 can suppress 53BP2 expression in the absence of cellular damage in 041TR cells.
FIG. 3.
Expression of wild-type p53 in the absence of cellular damage decreases 53BP2 levels. (A) Western blot of lysates (30 μg of total protein per lane) prepared from 041TR cells at the indicated times after tetracycline withdrawal. The decreases in 53BP2 protein, as a fraction of baseline levels determined at time zero, are indicated below, normalized for tubulin. 53BP2 was detected with monoclonal antibody DX547; p53 was detected with monoclonal antibody 1801. (B) Northern blot of total RNA (15 μg/lane) prepared from the 041TR cells above at the indicated time points, probed for 53BP2, stripped, and reprobed for GAPDH. The decreases in 53BP2 mRNA, as a fraction of baseline levels determined at time zero, are indicated below, normalized for GAPDH.
Increased 53BP2 protein levels after UV irradiation involves a p53-independent component.
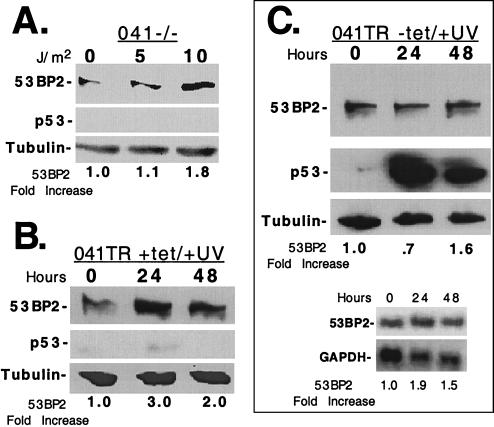
Although maintenance of low steady-state levels of 53BP2 required an intact p53 pathway (Fig. 2 and 3), UV irradiation resulted in increased 53BP2 levels despite the presence of DNA damage-induced p53 (Fig. 1). This suggested that p53-independent pathways participated in the damage-induced increase of 53BP2 protein. To examine this, we UV irradiated p53-null 041−/− LFS cells. In the absence of cellular damage, endogenous 53BP2 protein is overexpressed at baseline in these fibroblasts compared to normal primary GM38 fibroblasts and p53-heterozygous LFS 041+/− fibroblasts (Fig. 2A). After at least 10 J/m2, 53BP2 protein further increased 1.8-fold when assayed at 24 h by Western blotting (Fig. 4A). As expected, no p53 protein was detected after UV irradiation (Fig. 4A, middle panel). We also UV irradiated 041TR cells without p53 induction (plus tetracycline) (Fig. 4B). In noninduced 041TR cells, 20 J/m2 of UV irradiation further increased endogenous 53BP2 protein at least 3.0-fold at 24 h, as determined by Western blotting (Fig. 4B). 53BP2 transcript levels increased at least twofold at 24 h, as determined by Northern blotting (data not shown). These results demonstrate that at least a component of 53BP2 induction after UV irradiation is mediated by a p53-independent mechanism in 041−/− and 041TR fibroblasts.
FIG. 4.
p53-independent mechanisms participate in the UV irradiation induction of 53BP2. (A) Western blot of lysates (15 μg of total protein per lane) prepared from 041−/− cells 24 h after UV irradiation with 0, 5, or 10 J/m2. The fold increase of 53BP2 protein compared to no-UV-irradiation lysates are indicated below, normalized for tubulin. (B) Western blot of equivalent amounts of lysates prepared from 041TR cells maintained in 2 μg of tetracycline per ml at the indicated times after UV irradiation at 20 J/m2. The fold increases of 53BP2 protein compared to the levels at time zero are indicated below, normalized for tubulin. (C) The upper panels show Western blots of equivalent amounts of lysates prepared from 041TR cells at the indicated times after tetracycline withdrawal. At time zero, cells were UV irradiated with 20 J/m2. The fold increases of 53BP2 protein compared to levels at time zero are indicated below, normalized for tubulin. 53BP2 protein was detected with monoclonal antibody DX547; p53 was detected with monoclonal antibody 1801. The lower panels show Northern blots of total RNA (15 μg/lane) prepared from the 041TR cells above at the indicated time points, probed for 53BP2, stripped, and reprobed for GAPDH. The fold increases of 53BP2 mRNA compared to the levels at time zero are indicated below, normalized for GAPDH.
UV irradiation-induced DNA damage inhibits p53-mediated suppression of 53BP2 protein.
To further characterize the finding that 53BP2 levels are suppressed in undamaged cells but increased in damaged cells, we used the 041TR cell line with regulated p53 expression. Since 53BP2 levels increased after UV irradiation by a p53-independent mechanism, we reasoned that expression of p53 after UV damage would not result in a decrease of endogenous 53BP2 protein (as shown in Fig. 3). At time zero, tetracycline was withdrawn to express p53 and the cells were UV irradiated with 20 J/m2. 53BP2 protein levels decreased only slightly at 24 h (70% of baseline) but increased 1.6-fold by 48 h despite the presence of p53 protein (Fig. 4C, upper panels). 53BP2 transcript levels modestly increased, as demonstrated by Northern blotting (Fig. 4C, lower panels). These results are in contrast to p53 expression in the absence of cellular damage; 53BP2 protein was reduced to 17% of the baseline after p53 expression without UV irradiation but increased 1.6-fold when p53 was expressed after UV irradiation (Fig. 3 versus Fig. 4C). These results suggest that UV irradiation activates cellular factors that can relieve p53-mediated suppression of 53BP2 protein in 041TR fibroblasts.
Expression of 53BP2 sensitizes cells to UV irradiation-induced apoptosis.
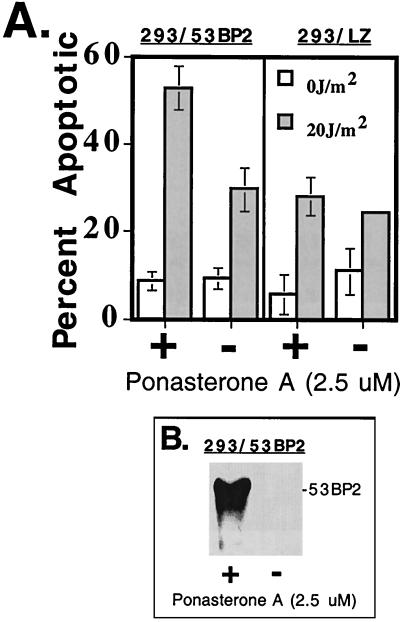
To examine how the cellular response to UV irradiation is modulated by 53BP2 expression, we utilized a human 293 cell line (293/53BP2) stably transfected with a 53BP2 cDNA under the control of a ponasterone A-inducible promoter (Fig. 5A). 53BP2 was first expressed for 24 h with ponasterone A in the absence of cellular damage, as shown by Western blotting (Fig. 5B). Cells were then UV irradiated (20 J/m2), and the percentage of apoptotic cells was determined by examining nuclear morphology 48 h later. Conditional expression of 53BP2 protein resulted in a 1.8-fold increase in apoptotic cells after UV irradiation at 48 h compared to 293/53BP2 cells that had been mock induced prior to UV irradiation (Fig. 5A, left panel). This was specific for 53BP2 expression and not ponasterone A exposure since a similar increase in apoptosis was not seen in control 293 cells (293/LZ) that had been constructed with a ponasterone A-inducible LacZ cDNA (Fig. 5A, right panel). These results were consistent with flow cytometry on propidium iodide-stained cells. Conditional expression of 53BP2 protein prior to UV irradiation resulted in a 1.6-fold increase in sub-G0 cells compared to 293/53BP2 cells that had been mock induced prior to UV irradiation. A similar increase in sub-G0 cells was not seen in control 293/LZ cells. These results demonstrate that, in 293/53BP2 cells, 53BP2 expression can lower the apoptotic threshold to UV irradiation-induced DNA damage.
FIG. 5.
Expression of 53BP2 sensitizes cells to UV irradiation-induced apoptosis. (A) Percentage of 293/53BP2 and 293/LZ cells with apoptotic nuclear morphology 48 h after UV irradiation (shaded bars) or no UV irradiation (open bars). Cells induced to express 53BP2 or β-galactosidase (+) were treated with 2.5 μM ponasterone A for 24 h prior to UV irradiation. Mock-induced cells (−) were treated with an equivalent volume of control carrier (ethanol). Shown are the means and standard deviations from triplicate plates. (B) Western blot using anti-53BP2 monoclonal antibody DX547 of equivalent amounts of lysates prepared from 293/53BP2 cells induced for 24 h with 2.5 μM ponasterone A (+) or ethanol (−).
Expression of 53BP2 decreases clonogenic survival after UV irradiation-induced DNA damage.
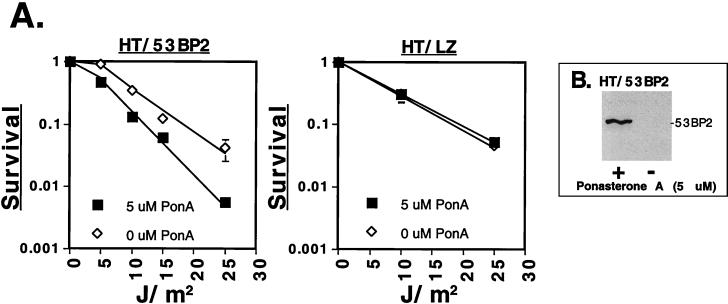
To demonstrate that the functional consequences of 53BP2 expression were not specific to 293 cells, we constructed a human HT1080 cell line (HT/53BP2) stably transfected with a 53BP2 cDNA under the control of a ponasterone A-inducible promoter. Furthermore, to demonstrate that 53BP2 expression could affect cellular survival, we determined the long-term clonogenic potential after UV irradiation (Fig. 6A, left graph). 53BP2 was first expressed for 48 h with ponasterone A in the absence of cellular damage, as shown by Western blotting (Fig. 6B). Cells were then replated, allowed to reattach for 8 h, and then UV irradiated with 0 to 25 J/m2. The resultant survival curve demonstrated decreased clonogenic potential compared to HT/53BP2 cells mock induced prior to UV irradiation (Fig. 6A, left graph). This was specific for 53BP2 expression and not ponasterone A exposure since a similar reduction in clonogenic potential was not seen in control HT1080 cells (HT/LZ) which had been constructed with a ponasterone A-inducible LacZ cDNA (Fig. 6A, right graph). These results demonstrate that expression of 53BP2 can decrease clonogenic survival after UV irradiation.
FIG. 6.
Expression of 53BP2 decreases clonogenic survival in response to UV irradiation. (A) The left graph shows the surviving fraction of HT/53BP2 cells after induction of 53BP2 expression with 5 μM ponasterone A (■) or ethanol control carrier (⋄) for 48 h prior to UV irradiation at the indicated doses. Shown are the means and standard deviations from triplicate plates. The right graph shows the surviving fraction of HT/LZ cells after the induction of LacZ expression with 5 μM ponasterone A (■) or ethanol control carrier (⋄) for 48 h prior to UV irradiation at the indicated doses. Colonies were fixed, stained, and counted after 12 days. (B) Western blot using anti-53BP2 monoclonal antibody DX547 of equivalent amounts of lysates prepared from HT/53BP2 cells induced for 48 h with 5 μM ponasterone A (+) or ethanol (−).
Attenuation of 53BP2 induction enhances clonogenic survival following UV irradiation induced DNA damage.
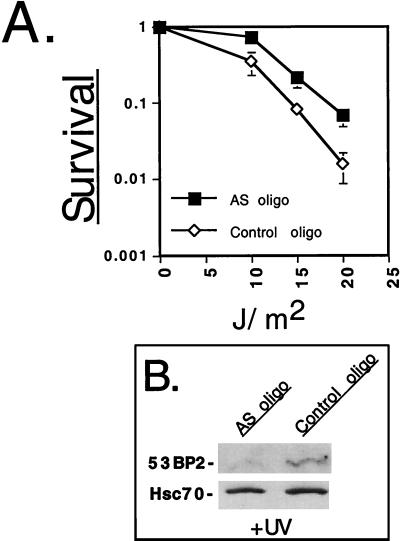
Since endogenous 53BP2 was induced by UV irradiation (Fig. 1) and enforced 53BP2 expression promoted UV damage-induced cell death (Fig. 5 and 6), we wondered if the abrogation of 53BP2 induction would result in enhanced resistance to UV-induced cell death. To examine this, we utilized a 53BP2 antisense oligonucleotide (AS-oligo) to inhibit endogenous 53BP2 expression and then determined the clonogenic potential following UV irradiation (Fig. 7A). AS-oligo or control-oligonucleotide (control-oligo) was introduced into HT1080 cells by osmotic delivery, and the cells were incubated in complete medium for 5 h prior to beginning subsequent experiments. To confirm attenuation of 53BP2 UV induction, AS-oligo- and control-oligo-treated cells were UV irradiated, and 53BP2 protein levels were determined at 24 h by quantitative Western blotting (Fig. 7B). After treatment with 10 J/m2, AS-oligo-treated cells had a 2.7-fold reduction in 53BP2 protein levels compared to control-oligo-treated cells normalized to hsc70. To determine the clonogenic potential, AS-oligo- and control-oligo-treated cells were first counted, replated, and allowed to reattach and then UV irradiated with 0 to 20 J/m2. The resultant survival curves demonstrated that AS-oligo-treated cells had enhanced resistance to UV irradiation compared to control-oligo-treated cells (Fig. 7A). These results demonstrate that inhibiting the UV-induced increase of endogenous 53BP2 levels with an antisense oligonucleotide can promote survival in HT1080 cells. Together with the observation that ectopically expressed 53BP2 decreases survival and promotes apoptosis following UV irradiation (Fig. 5 and 6), these results suggest that 53BP2 plays a physiologic role in mediating the apoptotic response to UV irradiation-induced DNA damage.
FIG. 7.
Attenuation of 53BP2 induction enhances clonogenic survival following UV irradiation. (A) Surviving fraction of HT1080 cells after introduction of AS-oligo (■) or control-oligo (⋄). Oligonucleotide-treated cells were incubated for 5 h, followed by counting, replating, and UV irradiation at the indicated doses. Shown is a representative experiment with the means and standard deviations from triplicate plates. (B) Western blot using anti-53BP2 monoclonal antibody DX547 of equivalent amounts of lysates (50 μg of total protein per lane) prepared from HT1080 cells after the introduction of AS-oligo or control-oligo. Oligonucleotide treated cells were incubated and UV irradiated in parallel with the clonogenic assay. Lysates were prepared 24 h after UV irradiation. Anti-hsc70 monoclonal antibody was used to confirm equal loading.
DISCUSSION
We have demonstrated that 53BP2 is a DNA damage-inducible protein that functions to promote DNA damage-induced apoptosis. Additionally, the regulation of 53BP2 expression is complex and involves both p53-dependent and p53-independent mechanisms. Our findings define a physiologic role for 53BP2 not previously described.
Our data argue strongly that the proapoptotic and growth-inhibitory effects of 53BP2 are part of the normal cellular stress response to genotoxic insults. First, although it may not happen in all cell types, we found that in normal diploid primary human skin fibroblasts (GM38), as well as in HT1080 cells, endogenous 53BP2 levels increased even after exposure to low levels (5 J/m2) of UV irradiation (Fig. 1). Similarly, DNA damage caused by the chemotherapy agent doxorubicin also increased 53BP2 protein levels in these cells (unpublished observations). Second, conditional expression of a 53BP2 cDNA enhanced apoptosis, as well as attenuated clonogenic survival after UV irradiation (Fig. 5 and 6). Third, attenuation of endogenous 53BP2 induction using a 53BP2 antisense oligonucleotide enhanced cellular survival following UV irradiation-induced DNA damage (Fig. 7). It remains to be determined if 53BP2's growth-inhibitory effects and enhancement of damage-induced apoptosis requires an intact p53 pathway. Several lines of evidence suggest that p53 might participate. First, 53BP2 expression can enhance p53 transactivation of the endogenous cell cycle inhibitor p21 (19), as well as MDM-2 and BAX-promoter-luciferase reporter constructs (L. H. Rohde, Y. Ao, and L. Naumovski, manuscript in preparation). Second, 53BP2's proapoptotic and growth-inhibitory functions appear to be disabled in a p53 mutant background, as suggested by the elevated endogenous 53BP2 levels found in the p53 mutant cells we examined (Fig. 2). An example of p53 inactivation disabling proapoptotic function is seen with Myc-induced apoptosis (7, 15, 39). However, apoptosis can also be mediated by p53-independent mechanisms (2), and 53BP2's precise role remains to be further clarified.
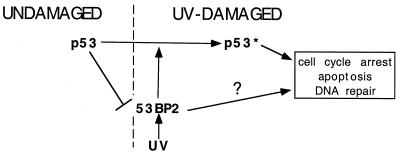
53BP2 levels increased after UV irradiation-induced DNA damage in cells with wild-type or mutant p53 (Fig. 1 and 4). This suggests, that at least in the cell lines we examined, a component of 53BP2 induction after UV irradiation is mediated by p53-independent mechanisms. In contrast, low steady-state levels of endogenous 53BP2 are maintained by p53-dependent mechanisms. This is suggested by the following observations. First, at least in the human and mouse cell types we examined, elevated 53BP2 levels were found in cells with p53-inactivating mutations (Fig. 2). Second, 53BP2 levels decreased in p53-null 041TR cells after expression of a tetracycline-regulated p53 gene (Fig. 3). Whether this reduced 53BP2 expression is mediated directly by p53 or indirectly via p53-mediated cell cycle arrest remains to be determined. However, 53BP2 levels did not decrease in 041TR cells if p53 was expressed after UV irradiation (Fig. 4C). Thus, it appears that, at least in 041TR cells, UV irradiation-induced DNA damage activates cellular factors that can relieve p53-mediated suppression of 53BP2 levels (Fig. 8).
FIG. 8.
Model of 53BP2 pathways. In undamaged cells, p53 mediates the suppression of 53BP2 levels. UV damage can relieve this suppression and increase 53BP2 levels. Through unknown mechanisms, 53BP2 can inhibit cell growth and promote damage-induced apoptosis after UV irradiation, as well as enhance p53 transactivation and cell cycle arrest. Lines with arrowheads denote positive regulation; the slashed line denotes negative regulation. “p53*” denotes the DNA damage-activated state.
Our observations suggest that the regulation of 53BP2 is potentially complex. A wide range in 53BP2 mRNA levels has been reported in different tumor cell lines, but the uncharacterized genetic differences between those different tumor cells makes interpretation of those findings problematic (27). Transcriptional or posttranscriptional mechanisms could mediate the changes in 53BP2 mRNA levels we observed by Northern blotting. Additionally, we cannot exclude p53-dependent transcriptional activation of other genes that could modulate 53BP2 levels because the p53 mutants examined in this report are also defective for sequence-specific DNA binding (5). Posttranslational mechanisms may also be important determinants of 53BP2 protein levels. At least in normal human tissues, 53BP2 protein is only detectable in low amounts, yet 53BP2 mRNA is readily detectable (data not shown) (18, 30, 41). Furthermore, changes in the magnitude of 53BP2 protein levels were not always reflected by similar changes in magnitude of 53BP2 mRNA levels (Fig. 1 to 3). Consistent with these observations is the fact that 53BP2 contains at least three distinct regions with high PEST scores which would be predicted to target 53BP2 for rapid intracellular proteolysis (16, 31, 41). Since 53BP2 appears to be a potent inhibitor of cellular survival and promotes damage-induced apoptosis, it is conceivable that multiple regulatory mechanisms may exist to control its expression, and we are currently investigating these possibilities.
The mechanism of 53BP2 function remains unclear. Although 53BP2's ability to bind the p53 sequence-specific DNA-binding domain would be predicted to inhibit transactivation (11), the opposite appears to be true (19). Indeed, 53BP2 appears to be a predominantly cytosolic protein and thus unable to interact with damage-activated p53 in the nucleus (19, 30, 41). If 53BP2 interacts with p53 in the cytoplasm, then perhaps it serves as a p53 chaperone. As such, it could affect p53 modifications, conformation, or intracellular localization. Others have reported that 53BP2 expression does not affect p53 levels (19). Likewise, we have been unable to demonstrate increased p53 levels in our 53BP2 regulatable cell lines under the conditions we have examined thus far (data not shown). Interestingly, 53BP2 contains a novel nuclear import sequence within the first three ankyrin repeats (33). Furthermore, a green fluorescent protein–carboxy-terminal-53BP2 fusion protein localizes to the nucleus (41). The significance of these findings remains unknown but it leaves open the possibility that 53BP2, or fragments of the protein, could have a nuclear function.
Although it remains to be determined why 53BP2 steady-state levels are negatively affected by an intact p53 pathway, several possibilities can be envisioned. Since 53BP2 can enhance p53 function (19), perhaps one possibility is that p53-mediated suppression of 53BP2 steady-state levels affords a level of feedback control. An example of p53 negatively affecting the levels of a protein that can enhance p53 function is seen with the p53-ARF feedback loop (23, 35). p53-null cells have increased steady-state levels of ARF; however, ectopic expression of p53 in such cells decreases ARF levels (35). Another possible reason for p53 to suppress 53BP2 steady-state levels is that p53 could act upstream of 53BP2 to modulate its functions, namely, growth inhibition and damage-induced apoptosis. In this regard, the potential interactions between 53BP2 and other cell death-regulating proteins such as Bcl-2 (30) and the p65/RelA subunit of NF-κB (41) are most intriguing. In undamaged cells, p53 could suppress 53BP2 expression to maintain it at low levels and thus attenuate 53BP2 function. Cell damage signals would then relieve this suppression, allowing 53BP2 to interact with Bcl-2, p65/RelA, or other proteins (14, 29, 30, 41) to modulate their functions.
Our data that 53BP2 is a DNA damage-inducible protein provides direct evidence that 53BP2 is part of the normal cellular response to genotoxic stress. Furthermore, our findings that 53BP2 lowers the apoptotic threshold and inhibits survival after UV irradiation suggests it is involved in damage-induced apoptosis. Importantly, our demonstration that attenuation of endogenous 53BP2 induction enhances clonogenic survival following UV irradiation argues that 53BP2 plays a physiologic role in the cellular damage response; this underscores the importance that genetic models will play in further elucidating 53BP2 function. Finally, our observations that 53BP2 levels are modulated by both p53-dependent and -independent mechanisms support the notion that the regulation of 53BP2 is complex. These results open new avenues for investigation into the cellular response to stress and add another gene product to the complex homeostatic mechanisms that govern cellular survival.
ACKNOWLEDGMENTS
This work was supported by a Walter V. and Idun Y. Berry Foundation Fellowship to C.D.L., by NRSA grant HL09552 to L.H.R., and by Public Health Service Grant CA76316 from the National Cancer Institute to L.N.
We thank Nataya Boonmark and Mint Sirisawad for excellent technical support and Jingshu Guo for help with the oligonucleotide design. We thank Philip Hanawalt and Gilbert Chu for critical reading of the manuscript.
REFERENCES
- 1.Agarwal M L, Agarwal A, Taylor W R, Stark G R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates S, Vousden K H. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett M, MacDonald K, Chan S, Luzio J, Simari R, Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. doi: 10.1126/science.282.5387.290. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff F Z, Yim O S, Pathak S, Grant G, Siciliano M J, Giovanella B C, Strong L C, Tainsky M A. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: aneuploidy and immortalization. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 5.Cho Y, Gorina S, Jeffrey P, Pavletich N. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 6.Del Sal G, Ruaro E, Utrera R, Cole C, Levine A, Schneider C. Gas1-induced growth suppression requires a transactivation-independent p53 function. Mol Cell Biol. 1995;15:7152–7160. doi: 10.1128/mcb.15.12.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evan G, Littlewood T. A matter of life and death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 8.Ford J M, Hanawalt P C. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 9.Ford J M, Hanawalt P C. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman D A, Wu L, Levine A J. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorina S, Pavletich N. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 12.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z, Kumar S, Howley P, Livingston D. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 13.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387. [DOI] [PubMed]
- 14.Helps N, Barker H, Elledge S J, Cohen P. Protein phosphatase 1 interacts with p53BP2, a protein which binds to the tumour suppressor p53. FEBS Lett. 1995;377:295–300. doi: 10.1016/0014-5793(95)01347-4. [DOI] [PubMed] [Google Scholar]
- 15.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh J-K, Chan F, O'Connor D, Mittnacht S, Zhong S, Lu X. Rb regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 18.Iwabuchi K, Bartel P L, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwabuchi K, Li B, Massa H F, Trask B J, Takayasu D, Fields S. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem. 1998;273:26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 20.Janus F, Albrechtsen N, Dornreiter I, Wiesmuller L, Grosse F, Deppert W. The dual role model for p53 in maintaining genomic integrity. Cell Mol Life Sci. 1999;55:12–27. doi: 10.1007/s000180050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayaraman L, Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by MDM2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 23.Kamijo T, Weber J, Zambetti G, Zindy F, Roussel M, Sherr C. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko L, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 25.Levine A. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 26.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 27.Mori T, Okamoto H, Takahashi N, Ueda R, Okamoto T. Aberrant overexpression of 53BP2 mRNA in lung cancer cell lines. FEBS Lett. 2000;465:124–128. doi: 10.1016/s0014-5793(99)01726-3. [DOI] [PubMed] [Google Scholar]
- 28.Murphy M, Ahn J, Walker K, Hoffman W, Evans R, Levine A, George D. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y. APCL, a central nervous system-specific homologue of adenomatous polyposis coli tumor suppressor, binds to p53-binding protein 2 and translocates it to the perinucleus. Cancer Res. 2000;60:101–105. [PubMed] [Google Scholar]
- 30.Naumovski L, Cleary M L. The p53-binding protein 53BP2 also interacts with Bcl2 and impedes cell cycle progression at G2/M. Mol Cell Biol. 1996;16:3884–3892. doi: 10.1128/mcb.16.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 32.Ruaro E, Collavin L, Del Sal G, Haffner R, Oren M, Levine A, Schneider C. A proline-rich motif in p53 is required for transactivation-independent growth arrest as induced by Gas1. Proc Natl Acad Sci USA. 1997;94:4675–4680. doi: 10.1073/pnas.94.9.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachdev S, Hoffman A, Hannink M. Nuclear localization of IkBα is mediated by the second ankyrin repeat: the IkBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol Cell Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 35.Stott F, Bates S, James M, McConnell B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tainsky M, Bischoff F, Strong L. Genomic instability due to germline p53 mutations drives preneoplastic progression toward cancer in human cells. Cancer Metastasis Rev. 1995;14:43–48. doi: 10.1007/BF00690210. [DOI] [PubMed] [Google Scholar]
- 37.Tao W, Levine A. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thukral S, Blain G, Chang K, Fields S. Distinct residues of human p53 implicated in binding to DNA, simian virus 40 large T antigen, 53BP1 and 53BP2. Mol Cell Biol. 1994;14:8315–8321. doi: 10.1128/mcb.14.12.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner A, Kokontis J, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 40.Weber J, Taylor L, Roussel M, Sherr C, Bar-Sagi D. Nucleolar Arf sequesters MDM2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Hori M, Takahashi N, Kawabe T, Kato H, Okamoto T. NF-kB subunit p65 binds to 53BP2 and inhibits cell death induced by 53BP2. Oncogene. 1999;18:5177–5186. doi: 10.1038/sj.onc.1202904. [DOI] [PubMed] [Google Scholar]
- 42.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]