Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Predicting signature, IgG, Non-structural/accessory protein, Outcome
Highlights
-
•
By taking advantage of a newly developed SARS-CoV-2 proteome microarray, IgG responses of 1,034 patients upon admission against 20 SARS-CoV-2 proteins were analyzed.
-
•
The magnitude of IgG antibodies against 8 non-structural proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, and NSP14) and 2 accessory proteins (ORF3b and ORF9b) possessed significant predictive power for patient death, even after further adjustments for potential confounding factors.
-
•
IgG responses to all of these 10 non-structural/accessory proteins were also associated with the severity of disease, and differential kinetics and serum positive rate of these IgG responses were confirmed in COVID-19 patients of varying severities.
-
•
The AUCs for these IgG responses, determined by computational cross-validations, were between 0.62 and 0.71.
Abstract
Introduction
The COVID-19 global pandemic is far from ending. There is an urgent need to identify applicable biomarkers for early predicting the outcome of COVID-19. Growing evidences have revealed that SARS-CoV-2 specific antibodies evolved with disease progression and severity in COIVD-19 patients.
Objectives
We assumed that antibodies may serve as biomarkers for predicting the clinical outcome of hospitalized COVID-19 patients on admission.
Methods
By taking advantage of a newly developed SARS-CoV-2 proteome microarray, we surveyed IgG responses against 20 proteins of SARS-CoV-2 in 1034 hospitalized COVID-19 patients on admission and followed till 66 days. The microarray results were further correlated with clinical information, laboratory test results and patient outcomes. Cox proportional hazards model was used to explore the association between SARS-CoV-2 specific antibodies and COVID-19 mortality.
Results
Nonsurvivors (n = 955) induced higher levels of IgG responses against most of non-structural proteins than survivors (n = 79) on admission. In particular, the magnitude of IgG antibodies against 8 non-structural proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, and NSP14) and 2 accessory proteins (ORF3b and ORF9b) possessed significant predictive power for patient death, even after further adjustments for demographics, comorbidities, and common laboratory biomarkers for disease severity (all with p trend < 0.05). Additionally, IgG responses to all of these 10 non-structural/accessory proteins were also associated with the severity of disease, and differential kinetics and serum positive rate of these IgG responses were confirmed in COVID-19 patients of varying severities within 20 days after symptoms onset. The area under curves (AUCs) for these IgG responses, determined by computational cross-validations, were between 0.62 and 0.71.
Conclusions
Our findings might have important implications for improving clinical management of COVID-19 patients.
Introduction
The coronavirus disease 2019 (COVID-19), the emerging infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and first identified in December 2019, has quickly become the greatest crisis of global public health and social development in our times [1]. As of August 22, 2021, there has been 211.28 million confirmed cases and 4.42 million patients death from SARS-CoV-2 infection worldwide [2]. Similar to SARS-CoV and MERS-CoV, SARS-CoV-2 belongs to the beta-coronavirus genus and its genome encodes 4 major structural proteins (S, spike; E, envelope; M, membrane; N, nucleocapsid), 15 non-structural proteins (Nsp1-10 and Nsp12-16), and 8 accessory proteins [3]. Among these, the S protein, consisting of a N-terminal S1 peptide with receptor binding domain (RBD) and a C-terminal S2 subunit, plays an essential role in viral attachment, fusion, and entry into the target cells which express the viral-binding receptor angiotensin-converting enzyme 2 (ACE2) [4]. There has been rapidly growing serological evidence that IgM, IgG, and IgA antibodies against S or N proteins of SARS-CoV-2 evolve rapidly in the serum of both asympomatic and symptomatic COVID-19 infections within one week after infection or onset of symptoms [5], [6], [7], [8]. Moreover, these antibodies elevated with disease progression and severity in symptomatic COIVD-19 patients [9]. Therefore, anti-SARS-CoV-2 specific antibodies may involve in the pathogenesis and affect the disease progression. However, the immunogenicity of most of the non-structural/accessory proteins has not been elucidated, and the clinical relevance, jointly with dynamics of nonstructural/accessory proteins in COVID-19 patients are still poorly understood.
In this study, we assumed that levels of anti-SARS-CoV-2 IgG antibodies may help predict the prognosis and outcome of patients with COVID-19. Proteome microarray technology has been confirmed as a mature and repeatable assay, which has been widely used in serological analysis of various diseases [10], [11], [12]. To enable the global understanding of SARS-CoV-2 specific IgG responses and their application, we constructed a proteome microarray with 20 out of the 28 predicted proteins of SARS-CoV-2 [6], [13]. Clinical serum specimens were analyzed on the SARS-CoV-2 proteome microarray, which can provide a high-throughput assay for 12 samples on each microarray and a rapid turnaround time of assay results (within 5 h after sample collection).
1034 patients hospitalized for confirmed COVID-19 disease at Tongji hospital from the day of hospitalization to the day of discharge or death were enrolled in this study. Serum IgG profiles for 1034 patients with COVID-19 on admission were probed using the SARS-CoV-2 proteome microarray. The microarray results were further correlated with laboratory biomarkers of disease severity and comorbidities, and with death of each patient, whose known clinical outcomes collected from electronic medical records. We found that the magnitude IgG responses to most of non-structural/accessory proteins are powerful predicting signatures for the COVID-19 death, independent of other biomarkers of laboratory and clinical severity factors, which might provide potential biomarkers for accurately monitoring disease progression and predicting clinical outcome.
Materials and methods
Patient information and data source
1056 confirmed COVID-19 patients were recruited from Tongji Hospital, Wuhan, China, between 17 February 2020 and 28 April 2020. COVID-19 was diagnosed based on positive SARS-CoV-2 nucleic acid test from respiratory tract specimens or based on clinical diagnosis with clinical symptoms and imaging features of pneumonia on chest computed tomographic (CT) according to the fifth version of COVID-19 diagnostic and treatment guideline, published by the National Health Commission of China (NHCC) [14]. Demographic information, medical history, comorbidities, signs and symptoms, chest CT, laboratory findings during hospitalization, and clinical outcomes were collected from electronic medical records. Among these, laboratory biomarkers related with disease severity factors such as the blood routine (leucocytes, lymphocytes, platelets, and neutrophils), liver and kidney functions (aspartate aminotransferase, alanine aminotransferase, lactate dehydrogenase, and creatinine), coagulation function (D-dimer) and inflammatory biomarkers (C-reactive protein, procalcitonin) were performed by automated analyzers according to the manufacturers’ instructions. The level of IL-2R in serum was measured by an automatic solid-phase two-site chemiluminescent immunometric assay via IMMULITE 1000 Analyzer (Siemens, Germany). Serum IL-6 was measured by an electro-chemiluminescence method (Roche Diagnostics, Switzerland).
Serum specimens were collected from each patient on admission and were stored at −80 °C until use. Serum detection based on proteome microarray and data analysis were performed during April 2020 to March 2021. After excluding 22 individuals with more than three missing anti-SARS-CoV-2 antibody indicators, a total of 1034 eligible participants (524 females and 510 males) with available data from serum proteome microarray and their clinical outcomes were used for the final analysis. Among 1034 eligible participants, some of whom had serial serum samples and were collected for a total of 2977 samples.
Ethics statement
The study was approved by the Ethical Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China (IRB ID:TJ-C20200128).
Protein microarray fabrication
The microarray used for serum IgG profiling was prepared as described previously [6], [13]. ACE2-Fc is ACE2 with a human Fc tag, which can be combined with anti-human secondary antibodies and used as a positive control in the microarray. 20 proteins of SARS-CoV-2 with indicated concentrations, along with the negative (GST: Glutathione S-transferase, Biotin-control, and eGFP: enhanced green fluorescent protein) and positive controls (Human IgG and ACE2-Fc), were printed in quadruplicate on PATH substrate slide (Grace Bio-Labs, USA) to generate identical arrays in a 2 × 7 subarray format using Super Marathon printer (Arrayjet, UK). The prepared protein microarrays were incubated in blocking buffer (3% BSA in 1 × PBS buffer with 0.1% Tween 20) for 3 h, and then stored at −80 °C until use.
Microarray-based serum analysis
The protein microarrays stored at −80 °C were warmed to room temperature before detection and were performed to probe all available seral samples. A 14-chamber rubber gasket was mounted onto each slide to create individual chambers for the 14 identical subarrays. Serum samples were diluted 1:200 in PBS containing 0.1% Tween 20 and a total of 200 μL of diluted serum or buffer only (negative controls) was incubated with each subarray for 2 h at 4 °C. The arrays were washed with 1 × PBST and bound antibodies were detected by incubating with Cy3-conjugated goat anti-human IgG (Jackson ImmunoResearch, USA), which were diluted 1: 1000 in 1 × PBST, and incubated at room temperature for 1 h. The microarrays were then washed with 1 × PBST and dried by centrifugation at room temperature and scanned by LuxScan 10 K-A (CapitalBio, China) with the parameters set as 95% laser power/PMT 480 for IgG. Data of fluorescent intensity (FI) from each microarray was extracted by GenePix Pro 6.0 software (Molecular Devices, USA). The result of FI for each serum response to each protein was defined as the median of the foreground subtracted by the median of background for each spot and then averaged the triplicate spots for each protein. The result of the protein-specific antibody in the serum was expressed as log2(FI).
Statistical analysis
Shapiro-Wilk test was used to test data normality. Two-tailed t-test was conducted to test difference in means between survivor and nonsurvivor groups, Mann-Whitney U test was performed to test difference in skewed parameters. Chi-square tests or Fisher's exact test, when appropriate, was used for categorical variables. IgG responses against each protein were categorized into 3 groups (T1: first tertile, T2: second tertile, T3: third tertile) according to tertiles distribution (Supplementary Table 1). Cox proportional-hazards model was performed to estimate the hazard ratios (HRs) and 95% confidence intervals (CIs) of COVID-19 mortality for individual levels of protein-specific IgG responses categorized into tertiles according to distributions. The lowest tertile (T1) was considered as the reference group. Both age and sex were included in Model 1. In Model 2, we further adjusted hypertension (yes/no), diabetes (yes/no), lymphopenia (<1.1, ≥1.1, ×10^9/L), increased alanine aminotransferase (<40, ≥41, U/L), and increased lactate dehydrogenase (<214, ≥214, U/L). Linear trend p-values were calculated by modeling the median value of each antibody tertiles as a continuous variable in the adjusted models. Spearman's rank correlation analysis was performed to explore the correlations between virus-specific IgG responses and laboratory results in the study population. The principal component analysis (PCA) based on the 20 proteins of SARS-CoV-2 specific IgG responses was used to optimize the type of data and extract principal components (PCs). SARS-CoV-2 protein-specific IgG responses with factor loadings over 0.7 on a particular PC were regarded as main contributors of it. PCs were categorized into 3 groups (T1, T2, and T3) according to tertiles distribution: <-1.60, −1.60–1.08, and ≥ 1.08 for PC1; <-0.10, −0.10–0.94, and ≥ 0.94 for PC2; <-0.49, −0.49–0.66, and ≥ 0.66 for PC3; <-0.43, −0.43–0.50, and ≥ 0.50 for PC4, respectively. Each PC was modeled into the Cox proportional-hazards models as tertiles to evaluate the association with anti-SARS-CoV-2 specific IgG responses and the COVID-19 mortality.
In addition, the results of antibodies were classified as two groups of the high levels (≥median) and low levels (<median) based on the medians of IgG responses to each protein and further correlated these results with on day 66 mortality of all involved COVID-19 patients by Kaplan-Meier survival curve and log-rank test. Loess regression was used to establish the kinetics of SARS-CoV-2 specific antibodies. Cluster analysis was performed with pheatmap package of R. SAS (version 9.4), R (version 4.0.0), and SPSS (version 23.0) were used to conduct statistical analysis, when applicable. Two-sided statistical tests were considered to be significant at a p value below 0.05.
Computational cross-validations of the prediction efficacy for clinical outcome
The receiver operating characteristic curve was conducted for the prediction of COVID-19 survival and death. The IgG response to 10 proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, ORF3b and ORF9b) was explored as 10 potential biomarkers for predicting clinical outcome during the discovery phase. Further, 1000 times of computational cross-validations were conducted in the validation. For each cross-validation procedure, 477 survivors and 39 nonsurvivors were randomly selected as the training set, and the rest of the samples were treated as the testing set (478 survivors and 40 nonsurvivors). The area under curve (AUC) of these IgG antibodies for predicting COVID-19 death was calculated by R (version 4.0.0).
Results
Characteristics of the study population
1034 participants, having available serum microarray results and consisting of 955 survivors and 79 nonsurvivors, were enrolled in this study. Baseline characteristics of participated patients based on electronic medical records were analyzed as Table 1. The median age of all enrolled patients was 63 years old (IQR, 51–71). The median intervals from onset of symptoms to hospital admission, from onset of symptoms to recovery, and from onset of symptoms to death were 13 days (IQR, 8–21), 41 days (IQR, 33–52), and 32 days (IQR, 25–39), respectively. The median length of all COVID-19 patients’ hospital stay was 24 days (IQR, 15–35). 37% patients with COVID-19 had hypertension and 18.5% with diabetes. 30.7% patients had lymphopenia, while increased levels of lactate dehydrogenase and alanine aminotransferase were detected in 43% and 25.4% patients, respectively. Consistent with previous reports [15], [16], nonsurvivors were more likely to be male, and older than survivors (p < 0.001). Higher proportion of abnormal laboratory results and shorter hospitalization time were observed in nonsurvivors than those of survivors (p < 0.001).
Table 1.
Baseline characteristics of participated COVID-19 patients.
| All patients | Survivors | Nonsurvivors | p value | |
|---|---|---|---|---|
| N | 1034 | 955 | 79 | |
| Age, median (IQR), years | 63(51–71) | 62(51–70) | 68(59–78) | <0.001 |
| Female, n (%) | 524(50.7) | 491(51.4) | 33(41.8) | 0.10 |
| Time from onset to admission, Median (IQR), days | 13(8–21) | 13(8–22) | 11(5–19) | 0.03 |
| Length of hospital stay, Median (IQR), days | 24(15–35) | 25(16–35) | 18(9–26) | <0.001 |
| Time from onset to outcome, Median (IQR), days | 40(33–52) | 41(33–52) | 32(25–39) | <0.001 |
| Comorbidity, n (%) | ||||
| Hypertension | 383(37.0) | 355(37.2) | 28(35.4) | 0.76 |
| Diabetes | 191(18.5) | 173(18.1) | 18(22.8) | 0.30 |
| Coronary heart disease | 68(6.6) | 57(6.0) | 11(13.9) | 0.006 |
| Chronic obstructive pulmonary disease | 6(0.6) | 3(0.3) | 3(3.8) | 0.007 |
| Cerebrovascular disease | 44(4.3) | 37(3.9) | 7(8.9) | 0.07 |
| Chronic liver disease | 21(2.0) | 19(2.0) | 2(2.5) | 0.67 |
| Chronic renal disease | 23(2.2) | 20(2.1) | 3(3.8) | 0.41 |
| Cancer | 45(4.4) | 35(3.7) | 10(12.7) | 0.001 |
| Laboratory results, n (%) | ||||
| Lymphopenia, <1.1 × 10^9/L | 294(30.7) | 234(26.4) | 60(83.3) | <0.001 |
| Neutrophilia, ≥6.3 × 10^9/L | 181(18.9) | 125(14.1) | 56(77.8) | <0.001 |
| Thrombocytopenia, ≥350 × 10^9/L | 64(6.7) | 62(7.0) | 2(2.7) | 0.16 |
| Leukocytosis, ≥9.5 × 10^9/L | 146(15.2) | 98(11.1) | 48(65.8) | <0.001 |
| Increased lactate dehydrogenase, ≥214 U/L | 405(43.0) | 342(39.3) | 63(88.7) | <0.001 |
| Increased alanine aminotransferase, ≥41 U/L | 239(25.4) | 217(24.9) | 22(31.0) | 0.26 |
| Increased aspartate aminotransferase, ≥40 U/L | 129(13.7) | 101(11.6) | 28(40.0) | <0.001 |
| Increased creatinine, ≥104 μmol/L | 57(6.3) | 39(4.7) | 18(26.1) | <0.001 |
| Increased C-reactive protein, ≥3mg/L | 330(45.9) | 289(42.7) | 41(97.6) | <0.001 |
| Increased procalcitonin, ≥0.05 ng/ml | 159(29.3) | 122(24.3) | 37(92.5) | <0.001 |
| Increased D-dimer, ≥0.5 mg/L | 361(59.4) | 302(55.1) | 59(98.3) | <0.001 |
| Increased IL2R, >710 U/mL | 67(16.2) | 57(14.4) | 10(55.6) | <0.001 |
| Increased IL6, >7 ng/L | 98(23.5) | 82(20.6) | 16(88.9) | <0.001 |
Data were shown as medians (IQR) or number (%), respectively. IQR: inter-quartile ranges. Two-tailed t-test was conducted to test difference in means between survivor and nonsurvivor groups, Mann-Whitney U test was performed to test difference in skewed parameters. Chi-square tests or Fisher's exact test, when appropriate, was used for categorical variables.
Nonsurvivors produce higher levels of IgG responses against most of non-structural proteins than survivors
To establish the association of anti-SARS-CoV-2 IgG antibodies with COVID-19 survival and death, serum collected from each involved patients on admission was used for microarray-based serum analysis. Based on the FI value extracted from the proteome microarray for each serum sample of 1034 patients, we first compared IgG profiles against 20 proteins of SARS-CoV-2 (Table 2). There was no statistical difference of the levels of either anti-S or N IgG antibodies between nonsurvivors and survivors. However, higher levels of IgG responses against 15 proteins, namely, E, NSP1, NSP2, NSP4, NSP5, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, NSP15, NSP16, ORF3b and ORF9b, were induced in nonsurvivors than those of survivors. Our results indicate that the magnitude of IgG responses against most of non-structural proteins of SARS-CoV-2 might predict the prognosis and outcome of COVID-19.
Table 2.
Comparison of SARS-CoV-2 specific IgG responses ([log2(FI)]) between survivors and nonsurivivors.
| Proteins | All | Survivors | Nonsurvivors | p |
|---|---|---|---|---|
| S1 | 13.9(13.0–14.4) | 13.9(13.0–14.4) | 13.6(12.0–14.6) | 0.3 |
| S2 | 9.1(8.4–9.6) | 9.1(8.4–9.6) | 9.0(8.1–9.7) | 0.34 |
| N | 10.3(9.2–11.1) | 10.3(9.2–11.2) | 9.8(7.9–10.6) | <0.001 |
| N-Nter | 13.2(12.3–13.8) | 13.2(12.3–13.8) | 13.1(11.6–13.7) | 0.1 |
| N-Cter | 13.4(12.5–14.0) | 13.4(12.6–14.0) | 13.3(11.8–14.2) | 0.68 |
| E | 5.5(4.6–6.8) | 5.5(4.6–6.8) | 5.8(4.9–7.6) | 0.04 |
| NSP1 | 8.2(7.5–9.1) | 8.1(7.4–9.0) | 9.0(8.4–9.6) | <0.001 |
| NSP2 | 6.6(5.6–7.8) | 6.5(5.6–7.7) | 7.1(6.0–8.2) | 0.01 |
| NSP4 | 7.9(7.4–8.7) | 7.9(7.3–8.7) | 8.2(7.8–9.4) | <0.001 |
| NSP5 | 5.5(4.9–6.2) | 5.5(4.9–6.2) | 5.8(5.1–6.7) | 0.01 |
| NSP7 | 9.4(8.8–10.0) | 9.4(8.8–10.0) | 9.9(9.6–10.4) | <0.001 |
| NSP8 | 7.8(6.8–9.0) | 7.6(6.7–8.9) | 8.8(8.0–9.2) | <0.001 |
| NSP9 | 8.7(8.0–9.5) | 8.7(8.0–9.5) | 9.4(8.6–9.8) | <0.001 |
| NSP10 | 6.3(5.3–7.6) | 6.2(5.3–7.4) | 7.2(6.5–8.0) | <0.001 |
| RdRp | 8.1(7.4–9.3) | 8.0(7.4–9.2) | 9.2(8.5–9.6) | <0.001 |
| NSP14 | 7.4(6.7–8.4) | 7.3(6.6–8.3) | 8.3(7.7–9.1) | <0.001 |
| NSP15 | 7.1(6.2–8.4) | 7.1(6.1–8.3) | 7.7(6.6–9.1) | 0.02 |
| NSP16 | 7.1(6.3–8.2) | 7.0(6.3–8.2) | 7.7(6.6–8.9) | 0.004 |
| ORF3a | 5.2(4.0–6.6) | 5.3(4.0–6.6) | 4.6(3.4–5.7) | 0.001 |
| ORF3b | 8.7(8.0–9.6) | 8.6(8.0–9.6) | 9.6(9.1–9.9) | <0.001 |
| ORF6 | 3.7(0.0–4.9) | 3.7(0.0–4.9) | 3.4(0.0–4.7) | 0.3 |
| ORF7b | 6.4(5.4–7.2) | 6.4(5.5–7.2) | 5.6(4.8–6.8) | <0.001 |
| ORF9b | 8.0(7.5–8.8) | 8.0(7.4–8.7) | 8.4(7.9–9.5) | <0.001 |
FI: Fluorescence Intensity. Mann-Whitney U test was conducted to test difference between survivor and nonsurvivor groups.
IgG responses against 10 non-structural/accessory proteins positively correlate with COVID-19 mortality risk
To assess the relationship of the magnitude of IgG antibodies with the mortality risk of COVID-19 patients, the HRs (95% CIs) for the mortality risk associated with the levels of IgG responses against different proteins of SARS-CoV-2 were categorized into tertiles (Table 3). We first analyzed the effects of age and gender on the disease death as model 1. After adjusting for age and gender, we found that IgG responses to 10 proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, ORF3b and ORF9b) were significantly positively associated with the COVID-19 mortality, whereas negative significant association was observed between N, ORF3a, and ORF7b- specific IgG responses and the death. Previous studies reported that comorbidities and laboratory biomarkers related with the function of important organs also might be the risk factors of the COVID-19 death [16], [17]. Therefore, we further adjusted the association for hypertension, diabetes, lymphopenia, increased alanine aminotransferase and lactate dehydrogenase as shown in model 2. Interestingly, IgG responses to 10 proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, ORF3b and ORF9b) were also significantly positively associated with the mortality risk of COVID-19 (Table 3).
Table 3.
Hazard ratio (95 %CI) for COVID-19 mortality according to tertiles of anti-SARS-CoV-2 specific IgG responses.
| Proteins | Model |
Tertile of proteins [log2(FI)] |
p trend | ||
|---|---|---|---|---|---|
| T1 | T2 | T3 | |||
| N | Model 1 | 1 | 0.63(0.38–1.05) | 0.40(0.22–0.73) | 0.002 |
| Model 2 | 1 | 0.79(0.46–1.34) | 0.73(0.39–1.37) | 0.52 | |
| E | Model 1 | 1 | 1.07(0.59–1.92) | 1.25(0.72–2.20) | 0.41 |
| Model 2 | 1 | 1.11(0.59–2.09) | 1.25(0.68–2.29) | 0.56 | |
| NSP1 | Model 1 | 1 | 3.05(1.38–6.71) | 3.76(1.77–8.03) | 0.0006 |
| Model 2 | 1 | 2.84(1.21–6.63) | 3.10(1.38–6.99) | 0.02 | |
| NSP2 | Model 1 | 1 | 0.96(0.53–1.75) | 1.30(0.75–2.26) | 0.3 |
| Model 2 | 1 | 0.76(0.39–1.45) | 1.18(0.66–2.11) | 0.64 | |
| NSP4 | Model 1 | 1 | 1.90(0.98–3.66) | 2.06(1.09–3.90) | 0.03 |
| Model 2 | 1 | 2.74(1.29–5.85) | 2.60(1.24–5.46) | 0.03 | |
| NSP5 | Model 1 | 1 | 1.08(0.60–1.95) | 1.48(0.85–2.57) | 0.15 |
| Model 2 | 1 | 1.15(0.60–2.22) | 1.79(0.98–3.27) | 0.07 | |
| NSP7 | Model 1 | 1 | 4.43(1.85–10.62) | 4.94(2.10–11.64) | 0.0003 |
| Model 2 | 1 | 4.01(1.52–10.53) | 4.28(1.67–10.98) | 0.008 | |
| NSP8 | Model 1 | 1 | 2.71(1.23–5.98) | 3.91(1.84–8.32) | 0.0002 |
| Model 2 | 1 | 2.34(0.99–5.52) | 3.20(1.42–7.21) | 0.009 | |
| NSP9 | Model 1 | 1 | 1.92(0.92–4.01) | 3.28(1.65–6.54) | 0.0003 |
| Model 2 | 1 | 1.40(0.64–3.07) | 2.69(1.29–5.61) | 0.005 | |
| NSP10 | Model 1 | 1 | 3.55(1.46–8.59) | 5.36(2.28–12.60) | <0.0001 |
| Model 2 | 1 | 3.19(1.22–8.38) | 4.89(1.92–12.46) | 0.0005 | |
| RdRp | Model 1 | 1 | 2.17(1.00–4.69) | 3.57(1.74–7.32) | 0.0002 |
| Model 2 | 1 | 2.31(1.02–5.20) | 2.80(1.30–6.02) | 0.02 | |
| NSP14 | Model 1 | 1 | 1.75(0.79–3.85) | 3.49(1.70–7.14) | 0.0001 |
| Model 2 | 1 | 1.37(0.59–3.19) | 2.65(1.23–5.71) | 0.007 | |
| NSP15 | Model 1 | 1 | 1.03(0.56–1.90) | 1.40(0.80–2.45) | 0.2 |
| Model 2 | 1 | 0.85(0.44–1.65) | 1.23(0.68–2.22) | 0.49 | |
| NSP16 | Model 1 | 1 | 0.91(0.49–1.70) | 1.52(0.87–2.64) | 0.09 |
| Model 2 | 1 | 0.71(0.36–1.39) | 1.40(0.78–2.50) | 0.28 | |
| ORF3a | Model 1 | 1 | 1.03(0.63–1.68) | 0.50(0.27–0.92) | 0.04 |
| Model 2 | 1 | 1.35(0.79–2.29) | 0.69(0.35–1.33) | 0.53 | |
| ORF3b | Model 1 | 1 | 1.63(0.77–3.43) | 3.20(1.66–6.17) | 0.0001 |
| Model 2 | 1 | 1.68(0.76–3.70) | 2.69(1.34–5.38) | 0.02 | |
| ORF7b | Model 1 | 1 | 0.60(0.35–1.03) | 0.45(0.26–0.81) | 0.005 |
| Model 2 | 1 | 0.79(0.45–1.39) | 0.71(0.39–1.30) | 0.2 | |
| ORF9b | Model 1 | 1 | 1.66(0.87–3.15) | 2.02(1.11–3.68) | 0.02 |
| Model 2 | 1 | 1.72(0.86–3.43) | 2.11(1.11–4.04) | 0.03 | |
FI: Fluorescence Intensity, CI: confidence interval, T1: first tertile, T2: second tertile, T3: third tertile. The tertiles cutoffs of IgG responses ([log2(FI)]) against each protein were shown in Supplementary Tables 1. Cox proportional-hazards model was performed to estimate the hazard ratios (HRs) and 95% CIs, and linear trend p-values were calculated by modeling the median value of each antibody tertiles as a continuous variable.
Model 1: Adjusted for age and sex.
Model 2: Additional adjustment for hypertension, diabetes, lymphopenia, increased alanine aminotransferase, and increased lactate dehydrogenase.
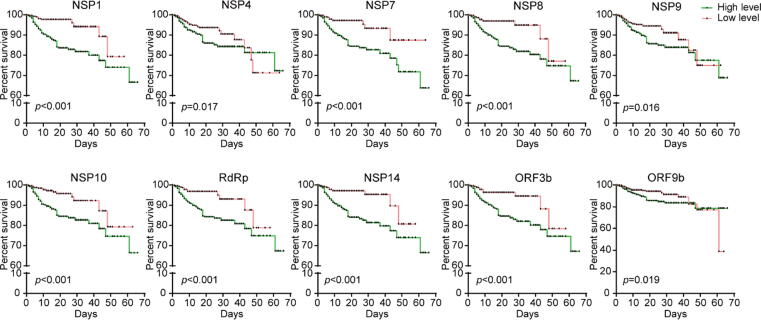
The Kaplan-Meier survival curve also supported that COVID-19 patients with higher levels of specific IgG responses against NSP1 (log2FI ≥ 8.2), NSP4 (log2FI ≥ 7.9), NSP7 (log2FI ≥ 9.4), NSP8 (log2FI ≥ 7.8), NSP9 (log2FI ≥ 8.7), NSP10 (log2FI ≥ 6.3), RdRp (log2FI ≥ 8.1), NSP14 (log2FI ≥ 7.4), ORF3b (log2FI ≥ 8.7), and ORF9b (log2FI ≥ 8.0) had higher morality risk after admission, respectively (Fig. 1).
Fig. 1.
Kaplan-Meier survival curves of patients with high and low levels of IgG to 10 non-structural/accessory proteins. 1034 hospitalized COVID-19 patients were detected for IgG responses against 20 proteins of SARS-CoV-2 on admission and followed till 66 days. Based on the median level of IgG responses to each protein, patients were classified as both high and low level groups after admission. Kaplan-Meier survival curves of patients with high (green) and low (red) levels of IgG antibodies to each protein, and Log-rank test was used to analyze the difference between two groups.
To further establish the association among IgG responses to different proteins with the outcome of COVID-19, we further conducted principal component analyses (PCs) and screened hypothetical new variables that account for the variance as much as possible, in order to reduce the dimension of data and the complexity of data with the least loss of original information. The HRs (95 %CIs) for the COVID-19 mortality according to PCs tertiles were presented in Table 4. Four PCs with eigenvalues > 1 were extracted, accounting for 71.95% of the total variance. Of four PCs, we found that only PC1 had the statistical association with the COVID-19 mortality (p trend = 0.004, Table 4), whatever adjusting age and sex, or further for hypertension, diabetes, lymphopenia, increased alanine aminotransferase and lactate dehydrogenase. In addition, the total variance of PC1 was 43.26%, which was the most important influencing factor among the four principal components (Table 5). Interestingly, the IgG responses to 10 proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, ORF3b and ORF9b) were still the main contributor to PC1 (Table 5), in line with our above mentioned findings. Our findings indicated that the IgG responses to 10 proteins were the most important for predicting disease outcome.
Table 4.
Hazard ratio (95 %CI) for COVID-19 mortality according to tertiles of principal components of anti-SARS-CoV-2 specific IgG responses.
| Proteins |
Tertile of principal components |
p trend | ||
|---|---|---|---|---|
| T1 | T2 | T3 | ||
| PC1 | ||||
| Model 1 | 1.00 | 2.17(1.05–4.51) | 2.79(1.40–5.59) | 0.004 |
| Model 2 | 1.00 | 1.66(0.76–3.65) | 2.24(1.07–4.68) | 0.03 |
| PC2 | ||||
| Model 1 | 1.00 | 0.70(0.43–1.13) | 0.31(0.16–0.61) | <0.001 |
| Model 2 | 1.00 | 0.89(0.53–1.51) | 0.62(0.31–1.25) | 0.20 |
| PC3 | ||||
| Model 1 | 1.00 | 0.69(0.42–1.14) | 0.48(0.26–0.88) | 0.01 |
| Model 2 | 1.00 | 0.82(0.47–1.41) | 0.72(0.38–1.39) | 0.30 |
| PC4 | ||||
| Model 1 | 1.00 | 0.70(0.40–1.21) | 0.98(0.59–1.65) | 0.91 |
| Model 2 | 1.00 | 0.94(0.52–1.72) | 1.24(0.71–2.16) | 0.47 |
PC: principal component, FI: Fluorescence Intensity, CI: confidence interval, T1: first tertile, T2: second tertile, T3: third tertile. The tertiles cutoffs of PCs were < -1.60, −1.60–1.08, and ≥ 1.08 for PC1; <-0.10, −0.10–0.94, and ≥ 0.94 for PC2; <-0.49, −0.49–0.66, and ≥ 0.66 for PC3; <-0.43, −0.43–0.50, and ≥ 0.50 for PC4. Cox proportional-hazards model was performed to estimate the hazard ratios (HRs) and 95% CIs, and linear trend p-values were calculated by modeling the median value of each antibody tertiles as a continuous variable.
The main contributors are NSP1, NSP2, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, NSP15, NSP16, ORF3b, and ORF9b for PC1; S1, N, N-Nter, and N-Cter for PC2; ORF7b for PC3.
Model 1: Adjusted for age and sex.
Model 2: Additional adjustment for hypertension, diabetes, lymphopenia, increased alanine aminotransferase, and increased lactate dehydrogenase.
Table 5.
Factor loadings of 20 proteins of anti-SARS-CoV-2 specific IgG responses among the study participants.
| Variables | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| S1 | 0.26 | 0.87 | −0.15 | −0.06 |
| S2 | 0.36 | 0.59 | −0.05 | 0.27 |
| N | 0.15 | 0.87 | 0.07 | −0.07 |
| N-Nter | 0.26 | 0.90 | −0.08 | −0.07 |
| N-Cter | 0.39 | 0.83 | −0.12 | −0.08 |
| E | 0.67 | −0.07 | 0.45 | −0.29 |
| NSP1 | 0.87 | −0.13 | −0.13 | 0.09 |
| NSP2 | 0.78 | −0.04 | 0.27 | −0.17 |
| NSP4 | 0.87 | −0.10 | −0.06 | 0.11 |
| NSP5 | 0.65 | 0.01 | 0.49 | −0.12 |
| NSP7 | 0.78 | −0.05 | −0.18 | 0.14 |
| NSP8 | 0.79 | −0.16 | −0.21 | 0.13 |
| NSP9 | 0.72 | −0.13 | −0.22 | 0.25 |
| NSP10 | 0.77 | −0.19 | −0.28 | 0.19 |
| RdRp | 0.81 | −0.12 | −0.21 | 0.00 |
| NSP14 | 0.89 | −0.05 | 0.07 | −0.20 |
| NSP15 | 0.78 | −0.09 | 0.15 | −0.17 |
| NSP16 | 0.81 | −0.04 | 0.29 | −0.21 |
| ORF3a | −0.20 | 0.29 | 0.50 | 0.47 |
| ORF3b | 0.85 | −0.13 | −0.13 | 0.09 |
| ORF6 | 0.17 | 0.04 | 0.23 | 0.67 |
| ORF7b | 0.18 | −0.02 | 0.72 | 0.19 |
| ORF9b | 0.78 | −0.05 | −0.10 | 0.03 |
| Eigen values | 9.95 | 3.609 | 1.79 | 1.198 |
| Total variance (%) | 43.263 | 15.691 | 7.784 | 5.207 |
| Cumulative variance (%) | 43.263 | 58.954 | 66.737 | 71.945 |
PC: principal component. The principal component analysis was used to optimize the type of data and extract PCs. Bold values denote factor loading > 0.7 are deemed to be statistically significant.
In addition, previous studies have established the associations between COVID-19 death with several laboratory measurements, such as lymphocyte count, procalcitonin, C-reactive protein, lactate dehydrogenase, D-dimer, IL-2R, IL-6, and ferritin [15], [16], [17]. Linear correlation between SARS-CoV-2 specific IgG responses with these biomarkers was further analyzed (Table 6). Interestingly, the IgG responses to 10 proteins were positively correlated with most of these biomarkers, but negatively associated with the lymphocyte count. Taken together, our results confirmed that the IgG responses to 10 non-structural/accessory proteins were positively correlated with the mortality risk of COVID-19.
Table 6.
Correlations between the levels of anti-SARS-CoV-2 specific IgG responses and other laboratory biomarkers related with severity factors.
| PCT | CRP | LYMPH | LDH | DD | lL-2R | IL-6 | Ferritin | |
|---|---|---|---|---|---|---|---|---|
| NSP1_IgG | ||||||||
| rs | 0.19** | 0.21** | −0.16** | 0.17** | 0.31** | 0.18** | 0.09 | 0.26** |
| NSP4_IgG | ||||||||
| rs | 0.09* | 0.14** | −0.09** | 0.10** | 0.21** | 0.10* | 0.02 | 0.19** |
| NSP7_IgG | ||||||||
| rs | 0.19** | 0.22** | −0.17** | 0.19** | 0.31** | 0.14** | 0.08 | 0.26** |
| NSP8_IgG | ||||||||
| rs | 0.12** | 0.19** | −0.15** | 0.16** | 0.31** | 0.11* | 0.12* | 0.20** |
| NSP9_IgG | ||||||||
| rs | 0.12** | 0.17** | −0.09** | 0.12** | 0.17** | 0.07 | 0.07 | 0.19** |
| NSP10_IgG | ||||||||
| rs | 0.12** | 0.21** | −0.15** | 0.15** | 0.31** | 0.16** | 0.13** | 0.28** |
| RdRp_IgG | ||||||||
| rs | 0.17** | 0.19** | −0.15** | 0.14** | 0.31** | 0.13** | 0.11* | 0.24** |
| NSP14_IgG | ||||||||
| rs | 0.15** | 0.17** | −0.15** | 0.16** | 0.27** | 0.17** | 0.11* | 0.24** |
| ORF3b_IgG | ||||||||
| rs | 0.15** | 0.18** | −0.14** | 0.16** | 0.29** | 0.12* | 0.06 | 0.23** |
| ORF9b_IgG | ||||||||
| rs | 0.12** | 0.12** | −0.07* | 0.11** | 0.19** | 0.04 | 0.01 | 0.20** |
Spearman's rank correlation analysis was performed to explore the correlations. *p < 0.05, **p < 0.01. PCT: procalcitonin; CRP: C-reactive protein; LYMPH: lymphocyte count; LDH: lactate dehydrogenase; DD: D-dimer; IL-2R: interleukin-2 receptor; IL-6: interleukin-6.
IgG responses against 10 non-structural/accessory proteins are associated with the severity of COVID-19 disease
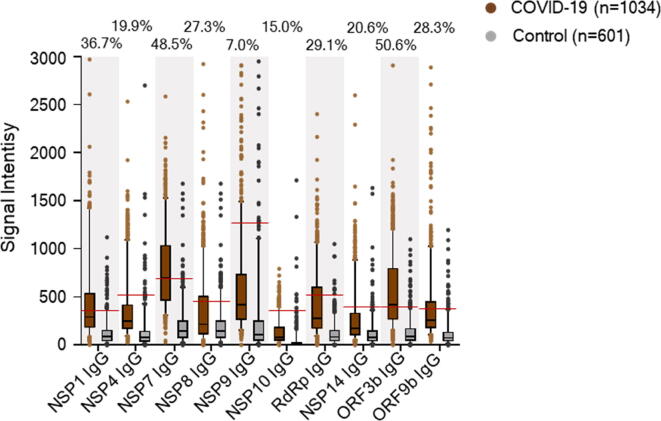
To assess the role of IgG to 10 non-structural/accessory proteins for the prediction of the clinical outcome, signal intensities and serum positive rates of IgG antibodies against 10 non-structural/accessory proteins in 1034 COVID-19 patients were compared with those of 601 healthy human serum controls. The cut-off value was set as mean + 2SD of the control group, and positive rates was calculated for each protein. Interestingly, COVID-19 patients had stronger signal intensities of serum IgG responses to all of these 10 proteins than healthy controls (Fig. 2). In addition, the serum positive rates of IgG antibodies in COVID-19 patients ranged from 7.0% to 50.6%, varying with different proteins. ORF3b, NSP7, and NSP1 specific IgG antibodies listed the top three of the serum positive rates in COVID-19 patients (Fig. 2).
Fig. 2.
Comparison of signal intensities and positive rates of IgG antibodies between COVID-19 patients and healthy controls. We surveyed IgG responses against 20 proteins of SARS-CoV-2 in 1034 hospitalized COVID-19 patients on admission. IgG responses to 10 non-structural/accessory proteins were compared between 1034 COVID-19 patients and 601 healthy serum controls. IgG responses were depicted as the boxplot according to the signal intensity of each serum sample on the proteome microarray. Data were represented by the median and 5th-95th percentile. The cut-off values of IgG antibody to each protein were set as mean + 2SD of the control group (n = 601) and shown as the red line. The positive rates of IgG antibodies to each protein in the patient groups were labeled on the figure.
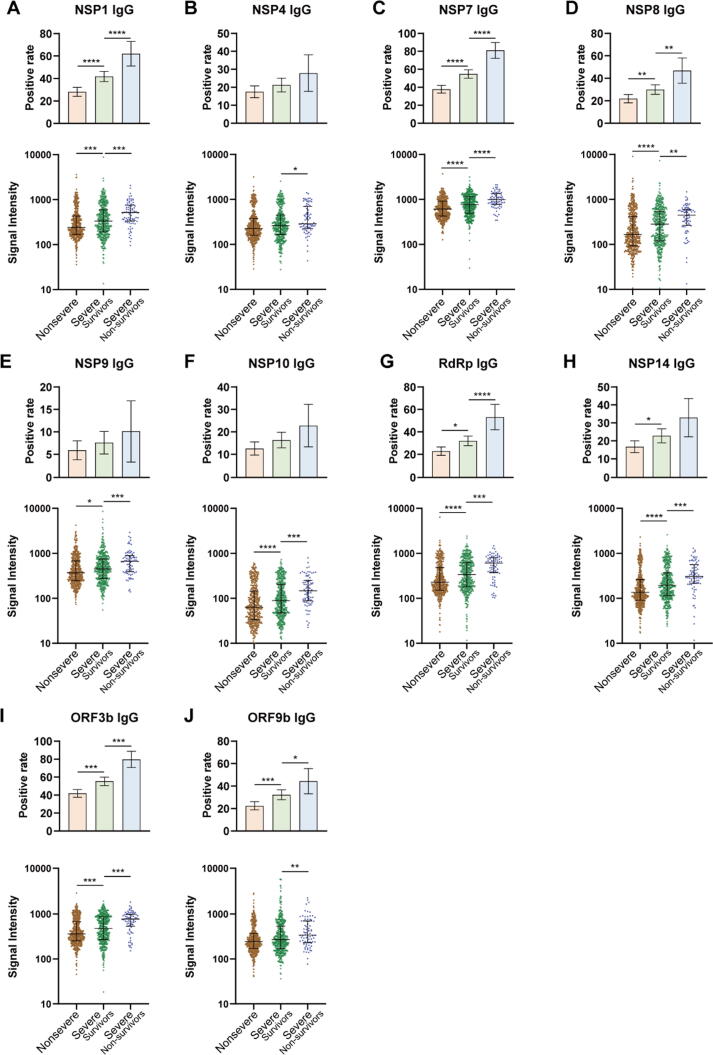
To further explore the association of IgG antibodies with the severity of illness, 1034 COVID-19 patients included in this study were divided into three groups: non-severe (n = 508), severe-survivors (n = 447), and severe-nonsurvivors (n = 79). Both the serum positive rate and the signal intensity of IgG responses were compared among these groups (Fig. 3). Interestingly, severe-nonsurvivors had higher serum positive rates of NSP1, NSP7, NSP8, RdRp, ORF3b and ORF9b specific IgG antibodies than severe-survivors and the non-severe group. In addition, the overall signal intensities for the 10 protein-specific IgG antibodies were higher in severe-nonsurvivors than those of severe-survivors (Fig. 3). These results suggested that the IgG responses of 10 non-structural/accessory proteins were also associated with the disease severity and might be effective predictors of disease prognosis.
Fig. 3.
Comparison of IgG responses of 10 non-structural/accessory proteins among different severities of patients. 1034 hospitalized COVID-19 patients were detected for IgG responses against 20 proteins of SARS-CoV-2 on admission. 1034 COVID-19 patients included in this study were divided into three groups: non-severe (n = 508), severe-survivors (n = 447), and severe-nonsurvivors (n = 79). Serum positive rate and signal intensity of IgG responses to NSP1 (A), NSP4 (B), NSP7 (C), NSP8 (D), NSP9 (E), NSP10 (F), RdRp (G), NSP14 (H), ORF3b (I), and ORF9b (J) were compared among different groups. For the positive rate analysis, error bar was given as the 95% confidential interval, and χ2 test was used to calculate p values. For the signal intensity analysis, the middle line was set as the median value; the upper and lower hinges were the values of 75% and 25% percentile, and Kruskale Wallis test and post-hoc test (Dunn-Bonferroni) were conducted to calculate p values. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
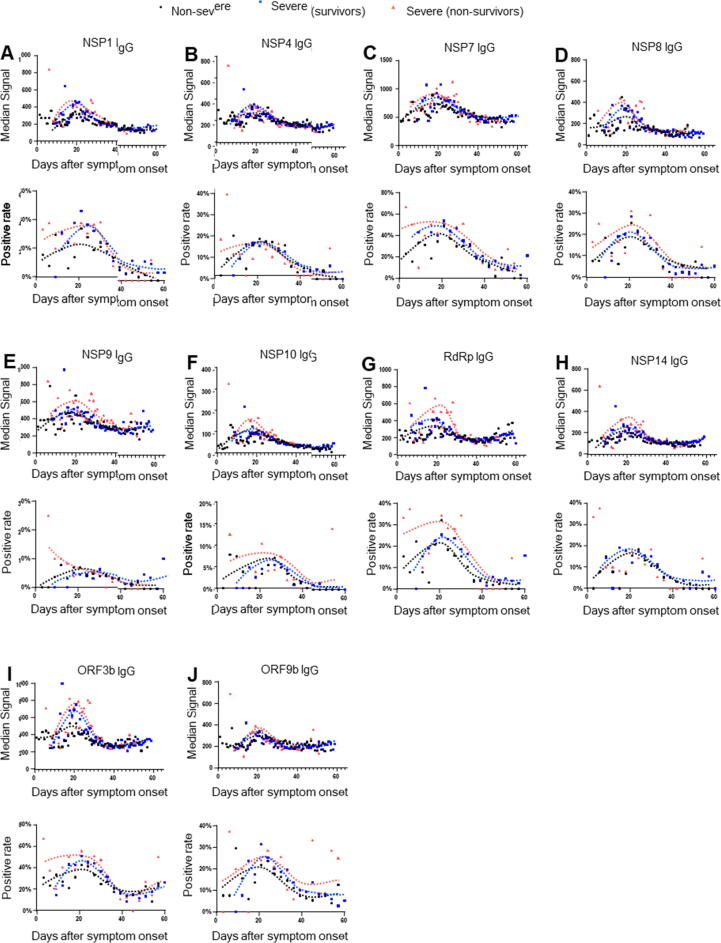
IgG responses to 10 non-structural/accessory proteins peak within 20 days after onset
To explore the detection time of IgG responses for the prediction, we further established the dynamic of IgG responses to 10 non-structural/accessory proteins from 0 to 60 days after onset, using 2977 seral samples from 1034 COVID-19 patients. Overall, the signal intensity and serum positive rate of the 10 protein-specific IgG antibodies increased persistently with the time after the symptom onset, peaked about 20 days later, and then declined gradually (Fig. 4). Interestingly, severe-nonsurvivors had a stronger signal intensity and higher serum positive rate than non-severe and severe-survivors. Our results indicated that detection of these antibodies within 20 days after the symptom onset might be used to predict the prognosis of disease.
Fig. 4.
The dynamics of IgG responses to 10 non-structural/accessory proteins between different groups. 2977 seral samples from 1034 COVID-19 patients were used. All seral samples were collected when the patients were on admission and during the hospital stay. The patients were divided into three groups: non-severe (n = 508), severe-survivors (n = 447), and severe-nonsurvivors (n = 79). The black, blue and red line showed the trends of signal intensities and positive rate at different time points for 10 specific IgG antibodies in non-severe, severe-survivors and severe-nonsurvivors, respectively. Signal intensity and serum positive rate of IgG responses to NSP1 (A), NSP4 (B), NSP7 (C), NSP8 (D), NSP9 (E), NSP10 (F), RdRp (G), NSP14 (H), ORF3b (I), and ORF9b (J) were compared among different groups. For signal intensity analysis, samples were grouped per day and the points with sample number<4 were excluded. For positive rate analysis, samples were grouped per three days.
Validation models confirm high prediction efficacy of IgG antibodies for clinical outcome
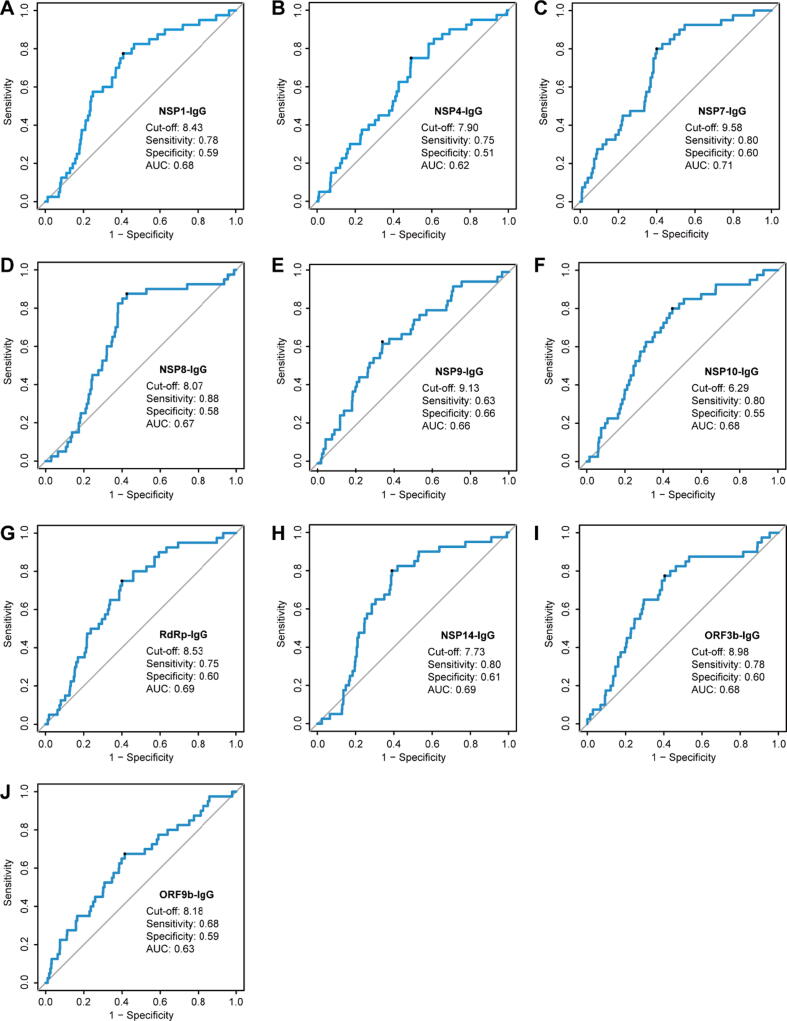
It is a common practice to validate “potential biomarker” by independent sample cohort. However, it is very difficult to collect new COVID-19 serum samples in China. To assure the reliability of our finding, we performed computational cross-validation based on the large sample cohort, by following protocols as established previously [11]. Interestingly, the AUCs of the IgG responses to 10 proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, ORF3b and ORF9b) for predicting COVID-19 death ranged from 0.62 to 0.71 (Fig. 5). Among these, NSP7, RdRp, and NSP14 specific IgG responses listed the top three with high AUC values.
Fig. 5.
Computational cross-validations of IgG responses to 10 non-structural/accessory proteins for the prediction efficacy. AUC: area under curve. 1034 hospitalized COVID-19 patients were detected for IgG responses against 20 proteins of SARS-CoV-2 on admission. The prediction efficacy was determined by a computational cross-validation. The receiver operating characteristic curve was conducted for the prediction of COVID-19 survival and death, and 1000 times of computational cross-validations were conducted. For each cross-validation procedure, 477 survivors and 39 non-survivors were randomly selected as the training set. The rest of the samples were treated as the testing set (478 survivors and 40 non-survivors). The average cutoff values were shown.
Discussion
In this study, we demonstrated that early IgG responses to 10 non-structural/accessory of SARS-CoV-2, namely, NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, NSP14, ORF3b, and ORF9b were significantly positively correlated with the mortality risk and the severity of COVID-19 patients. Especially, we also confirmed that all of these SARS-CoV-2 specific IgG responses are powerful predicting signatures for early predicting clinical outcome. Our findings have important indications for medical intervention and better control of the COVID-19 pandemic.
Firstly, we established a rapid and high-throughput assay platform based on proteome microarrays to measure IgG responses against 20 SARS-CoV-2 proteins in the serum of COVID-19 patients. After analyzing 1034 hospitalized patients, we found that the clinical outcome of COVID-19 patients is associated with high levels of IgG responses to 10 non-structural/accessory proteins of SARS-CoV-2 at presentation. Importantly, our observations indicated that SARS-CoV-2 specific IgG responses are predictive of COVID-19 mortality, independently of demographics and comorbidities, as well as routine clinical biomarkers of disease severity. In particular, we found that IgG antibodies against 8 non-structural proteins (NSP1, NSP4, NSP7, NSP8, NSP9, NSP10, RdRp, and NSP14) and 2 accessory proteins (ORF3b and ORF9b) were predictors of death after adjusting for the demographic features and comorbidities. The AUCs for the 10 non-structural/accessory protein-specific IgG responses ranged from 0.62 and 0.71, which were slightly lower than that of several severity indicators, such as PCT, CRP, and LDH [18]. However, these IgG responses against 10 non-structural/accessory proteins as potential biomarkers for predicting clinical outcome have not been reported before. In addition, only 1 μL of serum can be used in each round and 12 serum samples can be tested at one time. Early IgG antibody measurements based on our established serum proteome microarray analysis as predictors of mortality, therefore, raise the importance of using antibody levels for rapidly improving clinical management, treatment decisions and rational allocation of medical resources in short supply during the process of dealing with the COVID-19 pandemic.
Although the function of each non-structural/accessory proteins of the SARS-CoV-2 is not yet fully understood, their protein sequences are highly similar to those of SARS-CoV. Most non-structural proteins always locate in the core of virion and play important roles in the pathogenesis. For example, RdRp, also called NSP12 of SARS-CoV, can catalyze the synthesis of viral RNA and plays an important role in the replication and transcription cycle of the virus [19], [20]. RdRp itself performs the polymerase reaction with limited efficiency, whereas NSP7 and NSP8 as co-factors can significantly stimulate its polymerase activity [19]. Previous studies based on cryogenic electron microscopy (cryo-EM) indicated that the viral polymerase (RdRp-NSP7-NSP8 complex) might be an excellent target for developing new therapeutics of SARS and COVID-19 [20], [21]. NSP1 of the SARS-CoV may promote viral gene expression and immune escape by affecting interferon-mediated signal transduction [22]. NSP4 is a multichannel membrane protein, which is an essential protein for viral replication [23]. NSP9 plays a role of dimeric ssRNA binding protein during viral replication [24], [25]. NSP10 interacts with NSP14 and regulates ribose-2′-O-MTase activities involved in mRNA capping [25], [26], [27]. In this study, the levels of non-structural/accessory protein-specific IgG antibodies were positively correlated with routine clinical biomarkers of the disease severity (procalcitonin, C-reactive protein, lactate dehydrogenase, D-dimer, IL-2R, and IL-6), but inversely related with the lymphocyte count. Previous studies also confirmed that the massive release of inflammatory mediators such as C-reactive protein, procalcitonin, and D-dimer [17], [28], [29], as well as inflammatory cytokines such as IL-6 and IL-2R in severe/critical COVID-19 patients [30] might result in the excessive inflammatory response and acute lung injury, further exacerbating disease progression. In addition, low cycle threshold (Ct) values mean high viral load [31]. Most of the IgG response to 10 non-structural/accessory proteins were negatively with Ct values (Supplementary Table 2). Therefore, nonsurvivors might result in more deaths of virus-infected cells and larger release of viral components from the dying cells than survivors, especially within 20 days after the symptom onset. Consequently, more comprehensive interaction between viral non-structural/accessory proteins and the immune system of nonsurvivors might result in stronger IgG responses to these proteins as evidenced in this study, which underlines the scientific background of these IgG responses as predicting signatures for the clinical outcome.
Moreover, some studies reported that treatment of COVID-19 patients with convalescent plasma was effective [32], [33], whereas others did not observe the positive results [34], [35]. Several patients developed chills, rashes, shortness of breath, cyanosis, and severe dyspnea after treatment with convalescent plasma [36], which might be related to the antibody-dependent enhancement (ADE). In our study, we observed high levels of IgG antibodies against these non-structural proteins and accessory proteins that increased the risk of death and severity of COVID-19 patients. These IgG responses might play a detrimental role during SAS-CoV-2 infections, which might raise concerns about the ADE for these proteins. To mitigate the potential risks of ADE with convalescent plasma therapy, plasma should be encouraged to purify from donated convalescent plasma to enrich for neutralizing antibodies. It is also very important for monitoring the levels of these IgG antibodies in convalescent plasma before being used for treatment, in order to avoid the risks of ADE caused by non-neutralizing antibodies against these non-structural/accessory proteins. Both S1 and N proteins are highly immunogenic, which elicit strong IgG, IgM and IgA responses and S1 specific antibodies mainly play a protective role. Most of the COVID-19 subunit vaccines, such as mRNA-1273 [37] and BNT162b2 [38], are designed based on the S protein. Although survivors tended to induce a higher level of S1 and N IgG antibodies than that of nonsurvivors, no significant association of both S and N IgG responses with the mortality risk was observed. The safety of these vaccines in phase III clinical trials [37], [38] also corroborates our findings. Therefore, S1 and N specific IgG response are not suitable predictors of the risk of COVID-19 mortality. In addition, only the weak association between anti-SARS-CoV-2 IgM responses and the risk of COVID-19 death was observed in our study (data not shown). The relationship of IgA responses with clinical outcome remains to be investigated.
In conclusion, we provided a novel application of SARS-CoV-2 proteome microarray to detect serum IgG responses for early predicting COVID-19 death. Our results demonstrate that high level of IgG responses against 8 non-structural proteins and 2 accessory proteins on admission increased the COVID-19 mortality risk. Our research might improve clinical management and guide the development of effective medical interventions and vaccines by deeply understanding of the pathogenesis of COVID-19.
Ethics statement
The study was approved by the Ethical Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology , Wuhan, China ( IRB ID : TJ-C20200128 ).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Prof. H. Eric Xu (Shanghai Institute of Materia Medica) for providing RdRp protein. We also thank Healthcode Co., Ltd., Hangzhou Bioeast biotech Co., Ltd. and Vacure Biotechnology Co.,Ltd. for providing the proteins.
Funding/Support
This work was supported by grants from Wuhan Bureau of Science and Technology (No. 2020020601012218) and the Fundamental Research Funds for the Central Universities (HUST COVID-19 Rapid Response Call No. 2020kfyXGYJ040).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.11.014.
Contributor Information
Feng Wang, Email: wangfeng@tjh.tjmu.edu.cn.
Sheng-ce Tao, Email: taosc@sjtu.edu.cn.
Xiong-lin Fan, Email: xlfan@hust.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhu N.a., Zhang D., Wang W., Li X., Yang B.o., Song J., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Coronavirus disease (COVID-2019) situation reports. 2021; Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed 29 August, 2021.
- 3.Wu Aiping, Peng Yousong, Huang Baoying, Ding Xiao, Wang Xianyue, Niu Peihua, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls Alexandra C., Park Young-Jun, Tortorici M. Alejandra, Wall Abigail, McGuire Andrew T., Veesler David. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Jun, Liang Boyun, Chen Cunrong, Wang Hua, Fang Yaohui, Shen Shu, et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei Qing, Li Yang, Hou Hong‐yan, Wang Feng, Ouyang Zhu‐qing, Zhang Yandi, et al. Antibody dynamics to SARS-CoV-2 in asymptomatic COVID-19 infections. Allergy. 2021;76(2):551–561. doi: 10.1111/all.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelzang EH, Loeff FC, Derksen NIL: Development of a SARS-CoV-2 Total Antibody Assay and the Dynamics of Antibody Response over Time in Hospitalized and Nonhospitalized Patients with COVID-19. J Immunol 2020; 205:3491–3499. [DOI] [PubMed]
- 8.Li Kening, Huang Bin, Wu Min, Zhong Aifang, Li Lu, Cai Yun, et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-19943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch K.L., Whitman J.D., Lacanienta N.P., Beckerdite E.W., Kastner S.A., Shy B.R., et al. Magnitude and kinetics of anti-SARS-CoV-2 antibody responses and their relationship to disease severity. Clin Infect Dis. 2021;72:301–308. doi: 10.1093/cid/ciaa979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Shu, Liu Yuming, Chen Jing, Shu Hong, Shen Siyun, Li Yin, et al. Autoantibody signature in hepatocellular carcinoma using seromics. J Hematol Oncol. 2020;13(1) doi: 10.1186/s13045-020-00918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Lina, Wang Jingfang, Li Jianfang, Zhang Hainan, Guo Shujuan, Yan Min, et al. Identification of Serum Biomarkers for Gastric Cancer Diagnosis Using a Human Proteome Microarray. Mol Cell Proteomics. 2016;15(2):614–623. doi: 10.1074/mcp.M115.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Jiaoyu, Bi Lijun, Zhou Lin, Guo Shu-juan, Fleming Joy, Jiang He-wei, et al. Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep. 2014;9(6):2317–2329. doi: 10.1016/j.celrep.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Jiang He-wei, Li Yang, Zhang Hai-nan, Wang Wei, Yang Xiao, Qi Huan, et al. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Government of the People's Republic of China. New coronavirus pneumonia diagnosis and treatment plan (Fifth Edition). 2020; Available at: http://www.nhc.gov.cn/yzygj/s7653p/202002/d4b895337e19445f8d728fcaf1e3e13a.shtml. Accessed May 18, 2020.
- 15.Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al: Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 2020; 368:m1091. [DOI] [PMC free article] [PubMed]
- 16.Zhou Fei, Yu Ting, Du Ronghui, Fan Guohui, Liu Ying, Liu Zhibo, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Caizheng, Lei Qing, Li Wenkai, Wang Xiong, Liu Wei, Fan Xionglin, et al. Clinical Characteristics, Associated Factors, and Predicting COVID-19 Mortality Risk: A Retrospective Study in Wuhan. China Am J Prev Med. 2020;59(2):168–175. doi: 10.1016/j.amepre.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Feng, Hou Hongyan, Wang Ting, Luo Ying, Tang Guoxing, Wu Shiji, et al. Establishing a model for predicting the outcome of COVID-19 based on combination of laboratory tests. Travel Med Infect Dis. 2020;36:101782. doi: 10.1016/j.tmaid.2020.101782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subissi Lorenzo, Posthuma Clara C., Collet Axelle, Zevenhoven-Dobbe Jessika C., Gorbalenya Alexander E., Decroly Etienne, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan G., Liming Y., Yucen H., Fengjiang L., Zihe R. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(eabb7498) doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jauregui Andrew R., Savalia Dhruti, Lowry Virginia K., Farrell Cara M., Wathelet Marc G., Li Kui. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS ONE. 2013;8(4):e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelini Megan M., Akhlaghpour Marzieh, Neuman Benjamin W., Buchmeier Michael J., Moscona Anne. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4(4) doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miknis Zachary J., Donaldson Eric F., Umland Timothy C., Rimmer Ryan A., Baric Ralph S., Schultz L. Wayne. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol. 2009;83(7):3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snijder E.J., Decroly E., Ziebuhr J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouvet Mickaël, Debarnot Claire, Imbert Isabelle, Selisko Barbara, Snijder Eric J., Canard Bruno, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A. 2012;109(24):9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo X, Zhou W, Yan X, Guo T, Wang B, Xia H, et al: Prognostic Value of C-Reactive Protein in Patients With Coronavirus 2019. Clin Infect Dis 2020; 71:2174–2179. [DOI] [PMC free article] [PubMed]
- 29.Liappis Angelike P., Gibbs Kevin W., Nylen Eric S., Yoon Bona, Snider Richard H., Gao Baochong, et al. Exogenous procalcitonin evokes a pro-inflammatory cytokine response. Inflamm Res. 2011;60(2):203–207. doi: 10.1007/s00011-010-0255-8. [DOI] [PubMed] [Google Scholar]
- 30.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G: Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 2020; 58:1021–1028. [DOI] [PubMed]
- 31.Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, et al: Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill 2020; 25. [DOI] [PMC free article] [PubMed]
- 32.Liu STH, Lin HM, Baine I, Wajnberg A, Gumprecht JP, Rahman F, et al: Convalescent plasma treatment of severe COVID-19: a propensity score-matched control study. Nat Med 2020; 26:1708–1713. [DOI] [PubMed]
- 33.Shen Chenguang, Wang Zhaoqin, Zhao Fang, Yang Yang, Li Jinxiu, Yuan Jing, et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020;323(16):1582. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pathak E.B. Convalescent plasma is ineffective for covid-19. BMJ. 2020;371:m4072. doi: 10.1136/bmj.m4072. [DOI] [PubMed] [Google Scholar]
- 35.Simonovich Ventura A., Burgos Pratx Leandro D., Scibona Paula, Beruto María V., Vallone Marcelo G., Vázquez Carolina, et al. A Randomized Trial of Convalescent Plasma in Covid-19 Severe Pneumonia. N Engl J Med. 2021;384(7):619–629. doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., et al. Effect of Convalescent Plasma Therapy on Time to Clinical Improvement in Patients With Severe and Life-threatening COVID-19: A Randomized Clinical Trial. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Widge Alicia T., Rouphael Nadine G., Jackson Lisa A., Anderson Evan J., Roberts Paul C., Makhene Mamodikoe, et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N Engl J Med. 2021;384(1):80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polack Fernando P., Thomas Stephen J., Kitchin Nicholas, Absalon Judith, Gurtman Alejandra, Lockhart Stephen, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.