Shinnakasu et al. show that the glycan engineering immunogens of SARS-CoV-2 spike receptor-binding domain (RBD) elicited higher proportions of the core-RBD-specific germinal center (GC) B cells and antibodies, thereby manifesting significant neutralizing activity not only for SARS-CoV-2 but also for more broad SARS-related viruses.
Abstract
Broadly protective vaccines against SARS-related coronaviruses that may cause future outbreaks are urgently needed. The SARS-CoV-2 spike receptor-binding domain (RBD) comprises two regions, the core-RBD and the receptor-binding motif (RBM); the former is structurally conserved between SARS-CoV-2 and SARS-CoV. Here, in order to elicit humoral responses to the more conserved core-RBD, we introduced N-linked glycans onto RBM surfaces of the SARS-CoV-2 RBD and used them as immunogens in a mouse model. We found that glycan addition elicited higher proportions of the core-RBD–specific germinal center (GC) B cells and antibody responses, thereby manifesting significant neutralizing activity for SARS-CoV, SARS-CoV-2, and the bat WIV1-CoV. These results have implications for the design of SARS-like virus vaccines.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by the β-coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; herein called CoV-2), is a global health crisis. Coronavirus entry into host cells is mediated by the virus S protein, which forms trimeric spikes on the viral surface. The entry receptor for CoV-2 and SARS-CoV (herein called CoV-1) is the human cell-surface angiotensin converting enzyme 2 (ACE2), and the receptor-binding domain (RBD) of the spike from both of these viruses binds ACE2 with high affinity. Hence, the RBD is the primary target for neutralizing antibodies and has become a promising vaccine candidate (Dai and Gao, 2021; Walls et al., 2020).
Although the mutation rate of coronaviruses is low when compared with other viruses such as influenza or HIV, certain mutations in the S protein of CoV-2 have emerged in the setting of the rapidly spreading pandemic. One such mutation, D614G, which has now spread and become a dominant strain worldwide, turned out not to affect the overall neutralizing ability of patient sera (Korber et al., 2020), therefore, at least to some extent, reducing concerns about reinfection with CoV-2 variants.
However, given that prior coronavirus epidemics (e.g., CoV-1 and Middle East respiratory syndrome–CoV) have occurred due to zoonotic coronaviruses crossing the species barrier (Wacharapluesadee et al., 2021), the potential for the emergence of similar viruses in the future poses a significant threat to global public health, even in the face of effective vaccines for current viruses. For instance, bat SARS-like coronavirus, WIV1-CoV (WIV1; Wec et al., 2020), sharing 77% amino acid identity in S proteins to CoV-2, has been shown to be able to infect human ACE2-expressing cells (Ge et al., 2013; Menachery et al., 2016). However, sera from CoV-2–infected individuals exhibited very limited cross-neutralization of WIV1, except for rare individuals with low levels of neutralizing antibodies (Garcia-Beltran et al., 2021). This observation suggests that, although generation of broadly neutralizing antibodies is possible, current infection and vaccines are unlikely to provide protection against the emergence of novel SARS-related viruses.
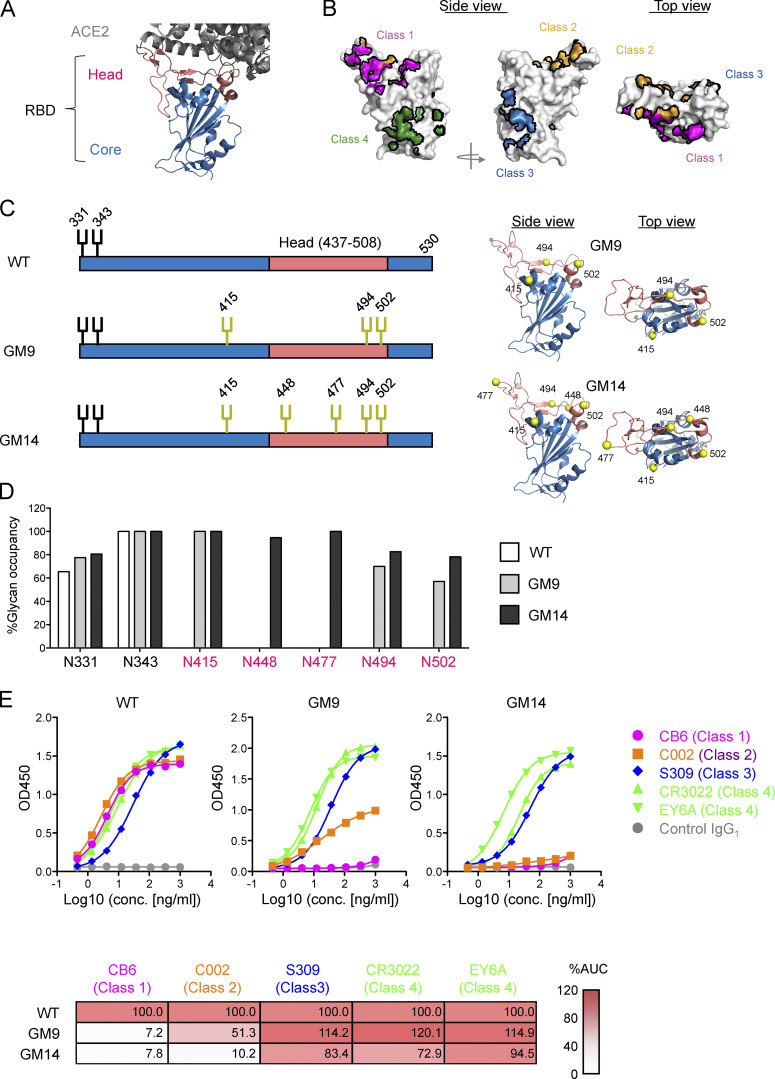
The CoV-2 RBD is composed of two regions (Fig. 1 A). The core-RBD consists of a central β sheet flanked by α-helices (indicated in blue in Fig. 1 A) and presents a stable folded scaffold for the receptor-binding motif (RBM; residues 437–508; indicated in red in Fig. 1 A; Shang et al., 2020), which encodes ACE2 binding and receptor specificity (herein, the core-RBD and RBM-encoded regions are called the core- and head-RBD subdomains, respectively; Shang et al., 2020). Initial analysis of convalescent sera from CoV-2–infected individuals demonstrated that many neutralizing antibodies (class 1 and 2) recognize the head-RBD subdomain (Fig. 1 B; Barnes et al., 2020a). However, in terms of broadly protective responses, the following three lines of evidence suggest that the core-RBD subdomain could be a more promising target. First, sequences for this subdomain are well conserved between CoV-1 and CoV-2 (Fig. S1 A). Second, nonbiased deep mutational scanning studies showed that the core-RBD subdomain has more mutational constraint for protein expression than the head-RBD subdomain (Starr et al., 2020). Finally, a recent study showed that, among mAbs recognizing the core-RBD from CoV-1–infected individuals in the 2003 SARS outbreak, there exist potent cross-neutralizing antibodies for CoV-2 and the bat SARS-like virus WIV1. Moreover, several CoV-2–infected individual–derived mAbs targeting the core-RBD of CoV-2 exhibited cross-neutralizing activity against CoV-1 and other sarbecoviruses (Brouwer et al., 2020; Jette et al., 2021 Preprint; Liu et al., 2020a).
Figure 1.
Design of CoV-2 RBD glycan engineering mutants. (A) Structure of RBD of CoV-2 S (Protein Data Bank accession no. 6YZ5) that engages the ACE2 ectodomain (Protein Data Bank accession no. 6M0J). Head and core subdomains are colored in red and blue, respectively. ACE2 is colored in gray. (B) Epitopes of representative anti–CoV-2 mAbs. (C) Schematic illustration of glycosylation sites in CoV-2 RBD WT, GM9, and GM14. The native and additional glycosylation sites are shown in black and yellow, respectively. Ribbon models of GM9 and GM14 are shown on the right. The yellow spheres are the introduced N-glycan sites. (D) Glycan occupancy at N-linked glycosylation sites of CoV-2 RBD WT, GM9, and GM14 determined by LC/MS. The bars indicate the percentage of glycan occupancy for each site. (E) ELISA binding of previously reported mAbs against CoV-2 RBD (CB6, C002, S309, CR3022, and EY6A) to CoV-2 RBD WT, GM9, and GM14. Anti–Candida albicans human IgG1 mAb 23B12 was used for a negative control. Heatmap shows the percentage of binding (the value of the area under the curve [AUC] in ELISA for CoV-2 RBD WT is set at 100% for each mAb). Representative results from three independent experiments are shown. conc., concentration.
Figure S1.
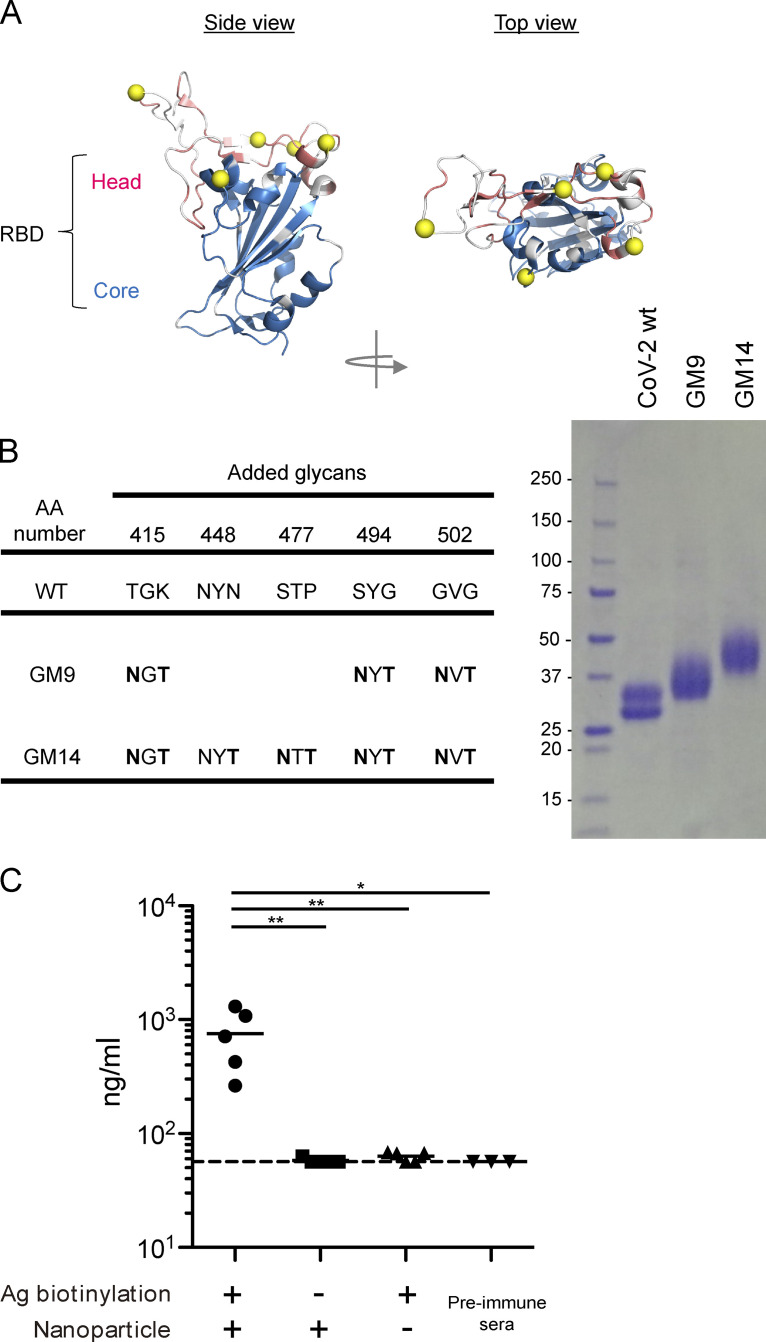
Design and expression of antigens, and effective induction of anti–CoV-2 RBD antibodies after immunization with nanoparticle antigen. (A) RBD-ACE2 complex. Non-conserved residues between CoV-1 and CoV-2 are colored in white. (B) The parental amino acid sequences and introduced NXT sequons (left). SDS-PAGE of RBD WT, GM9, and GM14 (right). (C) ELISA plots for CoV-2 RBD WT probe recognition of sera from respective biotin (+) RBD/Streptavidin nanoparticles, biotin (−) RBD/Streptavidin nanoparticles, or only biotin (+) RBD-immunized mice 3 wk after primary immunization or preimmune mice. The CR3022 mouse IgG1 mAb (Invivogen) was used as a standard. Representative of two independent experiments. Horizontal lines indicate mean values. Biotin (+) RBD/Streptavidin nanoparticles (n = 5); biotin (−) RBD/Streptavidin nanoparticles (n = 5); only biotin (+) RBD (n = 5); preimmune sera (n = 3). Dotted lines indicate detection limit. Horizontal lines indicate mean values; each symbol indicates one mouse. *, P < 0.05; **, P < 0.01; unpaired Student’s test. Ag, antigen.
One of the potential obstacles to targeting the core-RBD subdomain for vaccine development is that the epitopes in this subdomain seem to be immuno-subdominant (Dai and Gao, 2021; Liu et al., 2020b). To circumvent this problem, possibly directing immune responses to ordinarily immuno-subdominant epitopes, and providing protection against SARS-related CoV, at least two approaches can be considered: (1) altering the immunogen surface through targeted point mutations and deletions (Impagliazzo et al., 2015; Jardine et al., 2013; Valkenburg et al., 2016); and (2) introducing N-linked glycans believed to shield the neighboring epitopes by means of the NxS/T sequons (Duan et al., 2018; Eggink et al., 2014). To facilitate the immune responses to the core-RBD subdomain, we introduced glycans into the CoV-2 head-RBD subdomain. Here, we demonstrated that the glycan engineering facilitated the elicitation of potent cross-neutralizing antibodies toward CoV-1, WIV1, SARS related SHC014-CoV (SHC014), and Pangolin CoV GX-P2V (PaGX; clade 1 sarbecoviruses; Ge et al., 2013; Lam et al., 2020; Menachery et al., 2016). Thus, glycan engineering onto the RBD could provide one of the promising designs for SARS-related virus vaccines.
Results
Design of CoV-2 RBD glycan mutants
To engineer the CoV-2 head-RBD subdomain (Fig. 1 A), we introduced N-linked glycosylation sites (NxT motif) in this subdomain (Barnes et al., 2020b), which contains essential residues for ACE2 engagement (Fig. 1, A and C). The already established mAbs recognizing CoV-2 RBD are classified into four types; as shown in Fig. 1 B, class 1/2 and 3/4 recognize epitopes in the head-RBD and core-RBD subdomains, respectively. Five potential sites for the introduction of NxT sequons (GM14) were identified based on the following criteria: (1) surface residues on the class 1 or 2 epitope; (2) residues outside the class 3 and 4 epitopes in the core-RBD subdomain (Fig. 1 C; Barnes et al., 2020b); (3) residues within, or conformationally close to, the nonconserved patches of the head-RBD subdomain (Fig. S1 A); (4) NxT mutations not expected to disrupt the overall structure of RBD; and (5) not expected to reduce RBD protein expression or stability (Starr et al., 2020). Although the N415 site is somewhat distal from the head-RBD subdomain (Fig. 1 C), this site was already reported to be a part of the epitope recognized by the class 1 mAb CB6 (Shi et al., 2020). We designed two glycan mutants, GM9 and GM14, that have three and five additional glycosylation sites, respectively. Structural modeling predicted that these additional glycans would prevent antigen recognition by class 1 and 2 antibodies (Barnes et al., 2020b; Yuan et al., 2020).
The glycan mutants were coupled with 6xHis-Avi-tag at the C terminus and expressed in mammalian Expi293 cells. They exhibited a higher molecular weight than WT RBD in SDS-PAGE (Fig. S1 B). The glycan occupancy of each introduced glycosylation sites was determined by liquid chromatography/mass spectrometry (LC/MS). All added glycosylation sites had occupancies >75% except the added N494 and N502 glycans from GM9, which showed 70% and 57%, respectively (Fig. 1 D). The process of N-glycosylation in the ER is affected by conformational constraints (Breitling and Aebi, 2013). The somewhat lower glycan occupancy at N494 and N502 could be due to limited accessibility of oligosaccharyl transferases to these asparagine residues, which are positioned in close proximity to each other.
We then investigated whether the glycan modifications prevent binding of class 1/2 antibodies but maintain antigenic epitopes for class 3/4 antibodies. In ELISA, all tested antibodies were strong binders to WT RBD (Fig. 1 E). mAb CB6, a stereotypic class 1 neutralizing antibody, completely failed to bind to either of the glycan mutants (GM9 and GM14). Although the binding of mAb C002, a class 2 antibody (Robbiani et al., 2020), to RBD was abolished by GM14, this mAb still bound weakly to GM9, indicating that the five mutations are required for complete inhibition of class 2 mAb C002. In contrast, core-RBD subdomain targeting mAb S309 (class 3; Pinto et al., 2020), mAb CR3022 (class 4; Yuan et al., 2020), and mAb EY6A (class 4; Zhou et al., 2020) bound to WT RBD, GM9, or GM14 at a similar level, indicating that the introduced mutations for additional glycosylation do not affect overall structure of the core-RBD subdomain (Fig. 1 E).
Glycan engineering of the CoV-2 head-RBD generated more antibodies toward the core-RBD with higher affinity
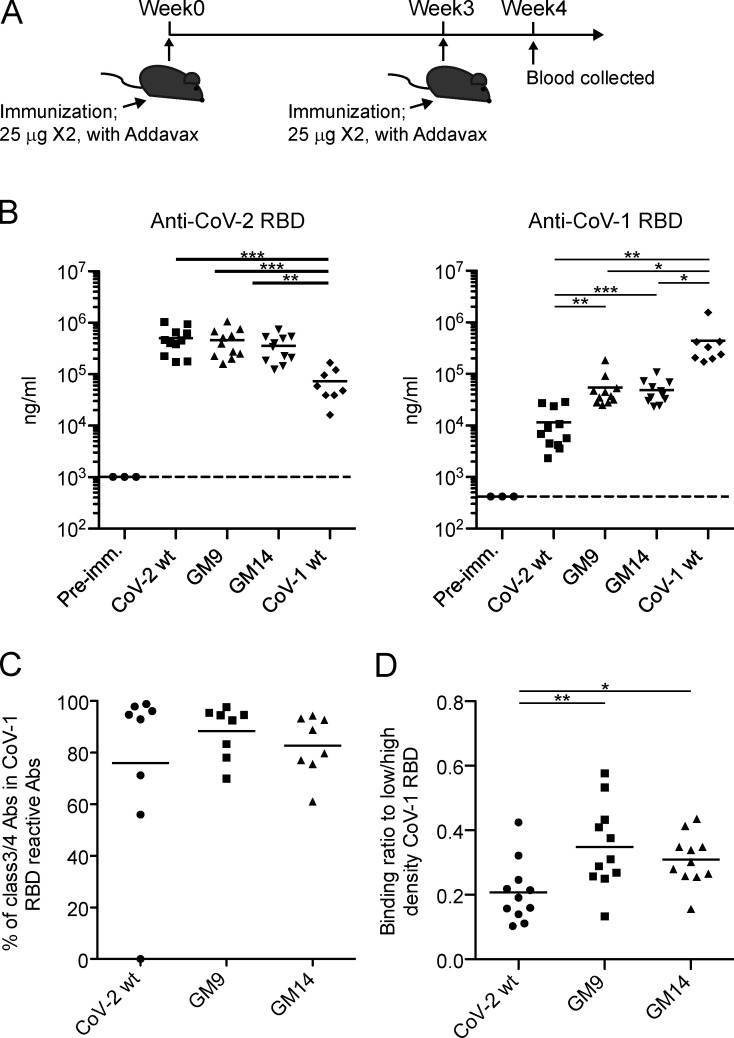
To overcome the limited immunogenicity of the relatively small CoV-2 RBD, we multivalently displayed it on streptavidin polystyrene nanoparticles. Consistent with a previous report (Walls et al., 2020), in contrast to soluble monomeric RBD, the particulate RBD gave rise to more robust primary antibody responses (Fig. S1 C). These particulate antigens (WT RBD, GM9, or GM14) were injected intramuscularly into BALB/c mice with AddaVax adjuvant. The mice were boosted with respective nonparticulate antigens 3 wk after priming (Fig. 2 A). We tested sera taken at 7 d after boosting for reactivity to CoV-2 RBD. Since, like alum, AddaVax dominantly induces IgG1-type antibody in the mouse (Sato et al., 2020), we measured the IgG1 response. To first address the possibility that the mice may have mounted an immune response toward neo-epitopes generated by glycan engineering that the parental CoV-2 RBD probe would not detect, we examined serum and germinal center (GC) B cell responses from the respective immunization protocols using immunogen-matched glycosylation mutant and parental probes. ELISA analysis indicated similar RBD-specific serum titers between the probe sets; by flow cytometry analysis, ∼10 or 15% of GC B cells bound to only immunogen-matched glycosylation mutant probes (Fig. S2, A and B). Hence, to some extent, B cells recognizing neo-epitopes appear to emerge upon GM9 or GM14 immunization. Nevertheless, 80–90% of the responding cells were reactive to the parental CoV-2 RBD probe; thus, we usually used the parental CoV-2 RBD probe in our assays.
Figure 2.
Glycan mutants of CoV-2 RBD immunogen effectively elicited CoV-1 RBD–specific antibodies. (A) Schematic overview of the experimental design. (B) CoV-2 RBD (left) or CoV-1 RBD (right) –specific IgG1 antibody levels in sera from preimmune (Pre-imm.) mice or CoV-2 RBD WT, GM9, GM14, or CoV-1 RBD WT–immunized mice were measured by ELISA. Samples were pooled from three independent experiments. Preimmune sera (n = 3); CoV-2 RBD WT (n = 11); GM9 (n = 11); GM14 (n = 11); CoV-1 RBD WT (n = 8). (C) Class 3/4 type serum antibodies from CoV-2 RBD WT, GM9, or GM14–immunized mice were measured by epitope-blocking ELISA with a mixture of CR3022/EY6A/S309 human IgG1 mAbs. CoV-2 RBD WT (n = 8); GM9 (n = 8); GM14 (n = 8). Samples were pooled from two independent experiments. (D) Affinity measurements were determined by ELISA, expressed as the binding ratio to low density/high density of plate-bound CoV-1 RBD protein. Samples were pooled from three independent experiments. CoV-2 RBD WT (n = 11); GM9 (n = 11); GM14 (n = 11). The CR3022 mouse IgG1 mAb (Invivogen) was used as a standard. Dotted lines indicate detection limit. Horizontal lines indicate mean values; each symbol indicates one mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test (A, B, and D). Abs, antibodies.
Figure S2.
Antibody response of mice immunized with CoV-2 RBD WT, GM9, GM14, and CoV-1 RBD WT. (A) ELISA plots for CoV-2 RBD WT and GM9-GM14 probes of sera from GM9 or GM14-immunized mice. Sera were collected at 3 wk after primary immunization. Representative results from three independent experiments are shown. (B) The representative FACS plots of GC B cells from dLNs after immunization are shown. Cells were gated for antigen-binding IgG+ GC B cells (CD138−B220+IgG+GL7+Fas+). The graph shows the percentage of positive cells for RBD WT binding among GM9- or GM14-binding cells. GM9 (n = 4); GM14 (n = 4). (C) CoV-1 head-RBD subdomain specific serum antibodies from CoV-2 RBD WT–, GM9-, or GM14-immunized mice were measured by epitope-blocking ELISA with S230 human IgG1 mAb as shown in Fig. 2 C. ELISA binding of S230 human mAb against CoV-1 head-RBD to plate-coated CoV-1 RBD (left). Percentages of CoV-1 head-RBD subdomain specific serum antibodies in whole CoV-1 RBD reactive antibodies (right). Samples were pooled from two independent experiments. The CR3022 mouse IgG1 mAb (Invivogen) was used as a standard. CoV-2 RBD WT (n = 8); GM9 (n = 8); GM14 (n = 8). (D) Serum neutralization against authentic CoV-2. A mixture of 100 TCID50 virus and serially diluted, heat-inactivated plasma samples (twofold serial dilutions starting from 1:40 dilution) were incubated at 37°C for 1 h before being placed on VeroE6-TMPRSS2 cells seeded in 96-well plates. After culturing for 4 d, cells were fixed with formalin and stained with crystal violet solution. Cutoff dilution index with >50% cytopathic effect was presented as microneutralization titer. Microneutralization titer of the sample below the detection limit (1:40 dilution) was set at 20. (E) Pseudovirus assay using VSV-ΔGluc carrying CoV-2 S K417N/E484K/N501Y. Horizontal lines indicate mean values; each symbol indicates one mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s test (D and E). Ab, antibody.
The IgG1 from mice immunized with glycan mutants displayed similar reactivity to CoV-2, assessed by ELISA, to that by WT RBD immunization (Fig. 2 B, left). In regard to the reactivity to CoV-1, GM9-, or GM14-immunized mice exhibited about eightfold higher than WT RBD immunization (Fig. 2 B, right). Given the sequence conservation of the RBD-core subdomain between CoV-2 and CoV-1 ( Fig. S1 A), it is most likely that the enhanced reactivity to CoV-1 by GM9 or GM14 immunization is due to generation of more IgG1 toward the conserved RBD-core subdomain of CoV-2. Supporting this possibility, when we used CoV-1 RBD-coated ELISA, CoV-1–reactive IgG1 elicited by GM9 or GM14 immunization was ∼90 or 85% inhibited by pretreatment of verified class 3/4 anti-CoV-2 core-RBD antibodies (mixtures of mAb S309, mAb CR3022, and mAb EY6A), respectively, like WT RBD immunization (Fig. 2 C). Instead, when we used class 1 mAb (mAb S230) toward the CoV-1 head-RBD subdomain (Rockx et al., 2008), we couldn’t detect inhibition (Fig. S2 C). Thus, it is reasonable to conclude that cross-reactive antibodies to CoV-1, mainly targeted to the conserved RBD-core subdomain, are generated with low levels even though by WT CoV-2 RBD immunization, and their generation is facilitated by GM9 or GM14 immunization.
Next, we characterized the affinity of cross-reactive IgG1 elicited by GM9 or GM14 immunization. The affinity of IgG1 for CoV-1 RBD was measured using an affinity ELISA that was modified from the nitrophenyl system (Wang et al., 2015); this assay measures the ratio of high-affinity to all-affinity binding IgG1. As shown in Fig. 2 D, in contrast to WT RBD immunization, cross-reactive IgG1 elicited by GM9 or GM14 immunization had significantly higher affinity. Together, glycan engineering of the CoV-2 head-RBD subdomain facilitates the generation of core-RBD subdomain specific antibodies with higher affinity.
Glycan engineering of the CoV-2 head-RBD elicited cross-neutralizing antibodies against CoV-1 and other related coronaviruses
To determine neutralizing activity, we majorly employed a vesicular stomatitis virus (VSV)–based pseudovirus method (Nie et al., 2020) and calculated half-maximal neutralization titers (NT50s). CoV-2 WT RBD, GM9, or GM14 immunization gave rise to similar neutralizing activity to CoV-2. GM9 or GM14 immunization manifested somewhat increased or decreased activity, respectively, compared with CoV-2 WT RBD immunization (Fig. 3 A). By using virus neutralizing assay, this tendency by GM9 or GM14 immunization was also observed (Fig. S2 D).
Figure 3.
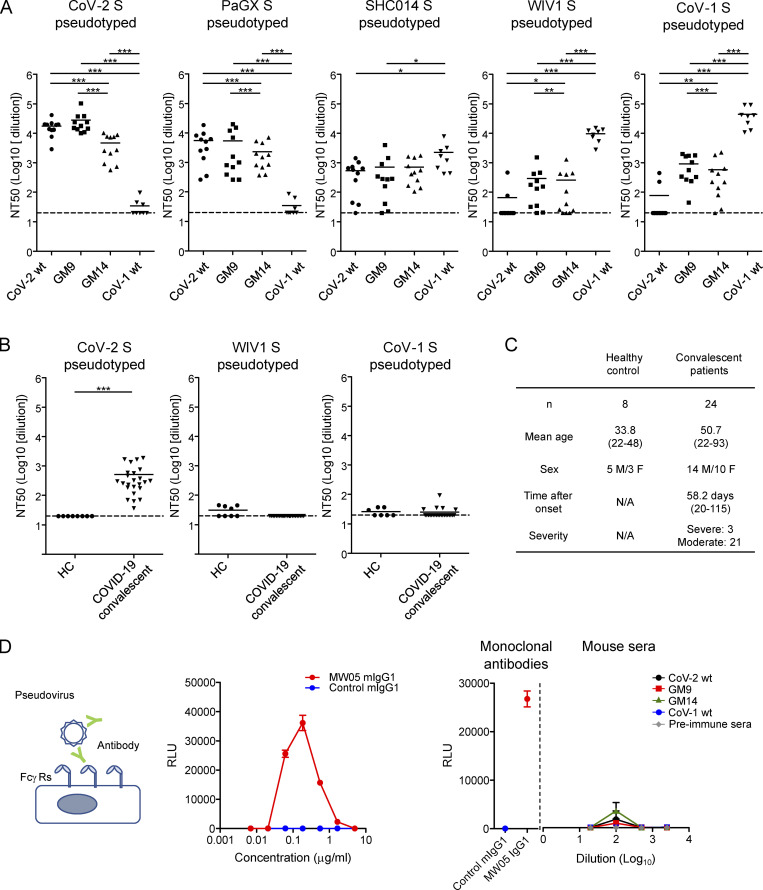
Glycan engineering of the CoV-2 head-RBD elicited cross-neutralizing activities against CoV-1 and WIV1. (A) BALB/c mice were prime-boost-immunized with CoV-2 RBD WT, GM9, GM14, or CoV-1 RBD WT as shown in Fig. 2. Sera were collected 7 d after boost and preincubated with CoV-2 S, PaGX S, SHC014 S, WIV1 S, or CoV-1 S–pseudotyped VSVΔG-luc for 1 h. The mixture was incubated with VeroE6 TMPRESS2 cells overnight. CoV-2 RBD WT (n = 11); GM9 (n = 11); GM14 (n = 11); CoV-1 RBD WT (n = 8). (B) Neutralization activities of plasma samples from prepandemic healthy donors (HC, n = 8) or convalescent COVID-19 patients (n = 24) against CoV-2, CoV-1, and WIV1 S–pseudotyped VSVΔG-luc. (C) Donor information. (D) ADE assay. Schematic representation of ADE assay (left). ADE of infection of Raji cells by MW05 mouse IgG1 mAb (middle). CoV-2 S–pseudotyped VSVΔG-luc was preincubated with different concentrations of MW05 mouse IgG1 mAb, or control mouse IgG1, and then added onto Raji cells. The luciferase activity was measured at 16 h after infection. Serially diluted sera from immunized mice showed no significant ADE activity (right). MW05 mouse IgG1 and irrelevant mouse IgG1 (0.1 µg/ml) were used for positive and negative control, respectively. CoV-2 WT (n = 4); GM9 (n = 5); GM14 (n = 5); CoV-1 WT (n = 4); preimmune sera (n = 3). Representative results from two or three independent experiments are shown (A–D). Data are mean ± SEM (D). Dotted lines in the graphs (NT50 = 20) represent the lower limit of detection (A and B). *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test (A and B).
For neutralization activity toward CoV-2 variants, we used a VSV-based pseudovirus carrying K417N/E484K/N501Y mutation derived from B.1.351 variant (Hoffmann et al., 2021), demonstrating that even CoV-2 WT RBD immunization evoked somewhat higher neutralization activity, compared with using WT CoV-2 (Fig. S2 E). Given that human sera upon mRNA vaccination showed reduced neutralization activity against viruses containing an E484K mutation (Chen et al., 2021), it is possible that our immunization regimen may generate more resistant sera. Alternatively, this might be attributable to differences between mouse and human immune systems.
Then we measured cross-neutralization activity against SARS-related viruses. Toward CoV-1, antisera elicited by GM9 or GM14 immunization possessed neutralizing activity with ∼15- or 10-fold higher potency, respectively, than CoV-2 WT RBD immunization, assessed by the pseudovirus method (Fig. 3 A). Similarly, these sera exhibited substantial neutralizing activity against WIV1, SHC014, and PaGX (Figs. 3 A and S3; Menachery et al., 2015; Lam et al., 2020; Menachery et al., 2016). Importantly, the levels of cross-neutralization we observed after GM9 or GM14 immunization were similar to convalescent plasma samples from CoV-2–infected individuals for their neutralizing activity of CoV-2, analyzed by the same laboratory in the same assay format (Fig. 3, B and C). Given that the major epitopes of the CoV-1–reactive antibodies elicited by GM9 or GM14 immunization are located in the core-RBD subdomain (Fig. 2 C), it is most likely that these anti–core-RBD antibodies contribute to cross-neutralization of CoV-1, WIV1, SHC014, and PaGX.
Figure S3.
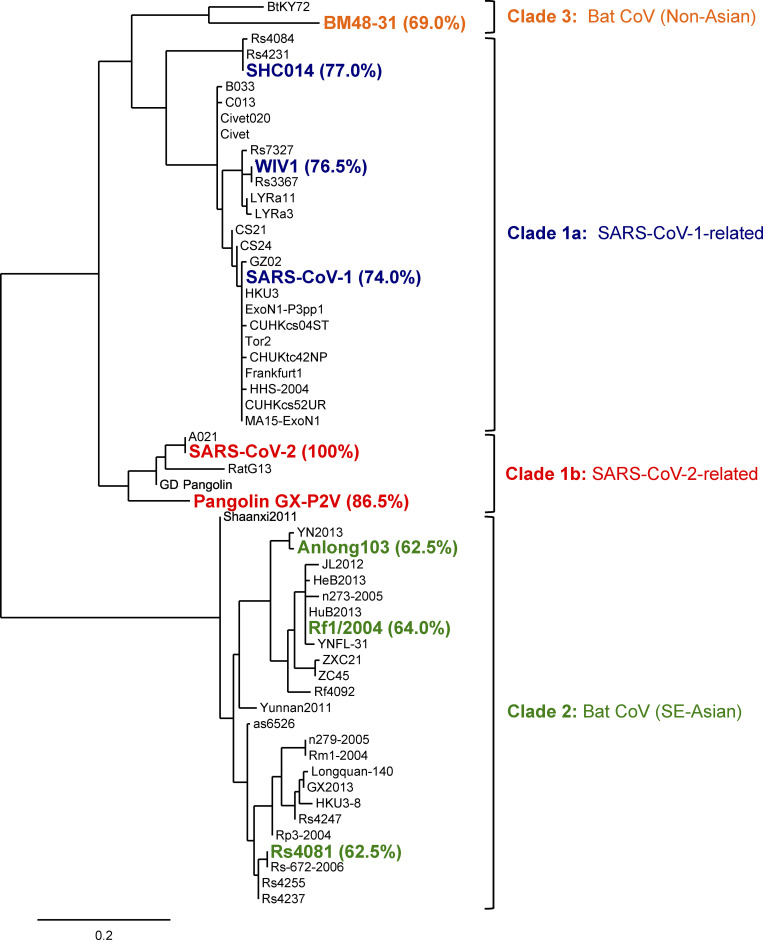
Phylogenetic tree of RBD sequences from representative sarbecoviruses. The tree was constructed by Phylogeny.fr with Jones–Taylor–Thornton substitution model. The scale bar represents phylogenetic distance of 0.2 amino acid substitutions per site. Sarbecovirus RBDs used in bead-based flow-cytometric assays and biolayer interferometry (Fig. 5) are shown in bold. The percentages of sequence identity of RBDs from these viruses compared with CoV-2 RBD (aa 331–529) are denoted in parentheses. SE-Asian, Southeast Asian.
Previous studies of CoV-1 and other viruses have suggested that antibody-dependent enhancement (ADE) of infection might take place after vaccination (Smatti et al., 2018). In the case of dengue virus, in vitro modeling of ADE has attributed enhanced pathogenesis to FcγR-mediated viral entry, rather than canonical viral receptor–mediated entry (Bournazos et al., 2020). To examine this possibility, VSV-based pseudovirus was incubated with serially diluted sera from immunized mice and then inoculated on Raji, a human B lymphoma cell line that is often used to assess ADE activity of SARS-CoVs (Wang et al., 2020). As a positive control for this assay, we showed that infection of the CoV-2 S pseudovirus was robustly enhanced by addition of anti–CoV-2 RBD MW05 chimeric human/mouse IgG1 antibody (Fig. 3 D, middle; Wang et al., 2020). In this experimental setting, like sera from mice immunized with CoV-2 WT RBD, sera from GM9- or GM14-immunized mice did not show significant ADE activity (Fig. 3 D, right).
Reactivity profiles of cross-reactive GC B cells
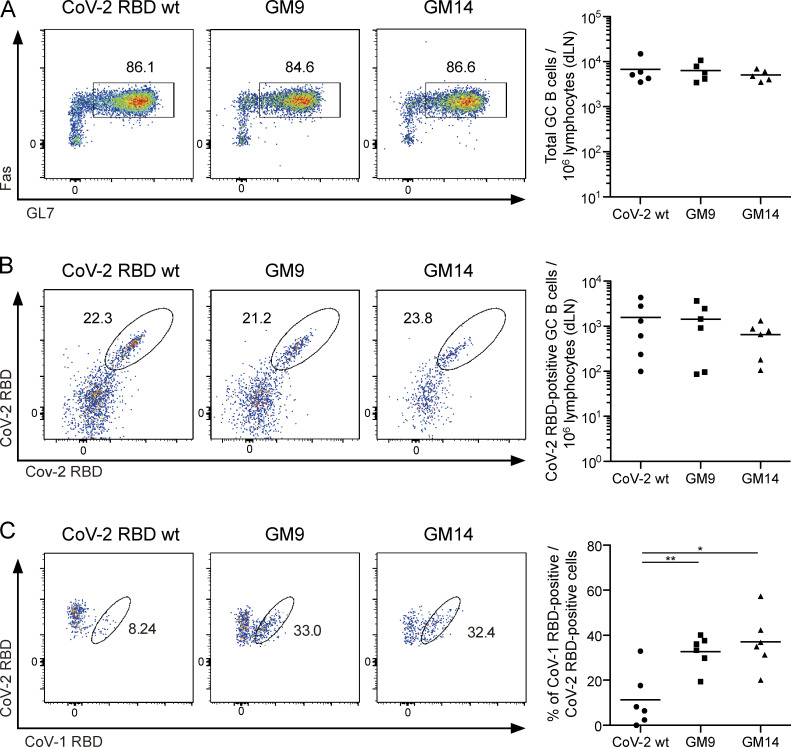
To complement the above serological characterization, we assessed the reactivity of GC B cells isolated after priming. As demonstrated by FACS analysis, WT RBD, GM9, or GM14 immunization gave rise to similar levels of overall GC B cells (Fig. 4 A). Among the CoV-2+ cells, the frequency of GC B cells positive for both CoV-2 and CoV-1 probes was increased by immunization of GM9 or GM14, compared with WT RBD immunization (Fig. 4, B and C).
Figure 4.
Immunization with CoV-2 GM antigens leads to the activation of CoV-1 and CoV-2 cross-reactive GC B cells. (A) Representative flow cytometry plots analyzing GC B cells (CD138−B220+IgD−IgM−GL7+Fas+) from draining lymph nodes (dLNs) of mice 3 wk after primary immunization with each antigen. The plots are representative of findings from two independent experiments (left). Quantification of absolute numbers of GC B cells (right) from multiple mice in one experiment. Results are representative of two independent experiments. CoV-2 RBD WT (n = 5); GM9 (n = 5); GM14 (n = 5). (B) Representative flow cytometry plots analyzing CoV-2 RBD–binding dLN GC B cells (CD138−B220+IgD−IgM−GL7+Fas+CoV-2 RBD+). Plots are representative of findings from two independent experiments (left). Quantification of absolute numbers of GC B cells (right) from multiple mice in one experiment. Results are representative of two independent experiments. CoV-2 RBD WT (n = 6); GM9 (n = 6); GM14 (n = 6). (C) Cross-reactivity of CoV-2 RBD-binding dLN GC B cells (CD138−B220+IgD−IgM−GL7+Fas+CoV-2 RBD [APC]+ CoV-2 RBD [PE-Cy7]+) with CoV-1 RBD assessed by flow cytometry. Plots are representative of findings from two independent experiments (left). The frequency of CoV-1 and CoV-2 RBD cross-reactive GC B cells (right) from multiple mice in one experiment. Results are representative of two independent experiments. CoV-2 RBD WT (n = 6); GM9 (n = 6); GM14 (n = 6). Horizontal lines indicate mean values; each symbol indicates one mouse. *, P < 0.05; **, P < 0.01; unpaired Student’s t test.
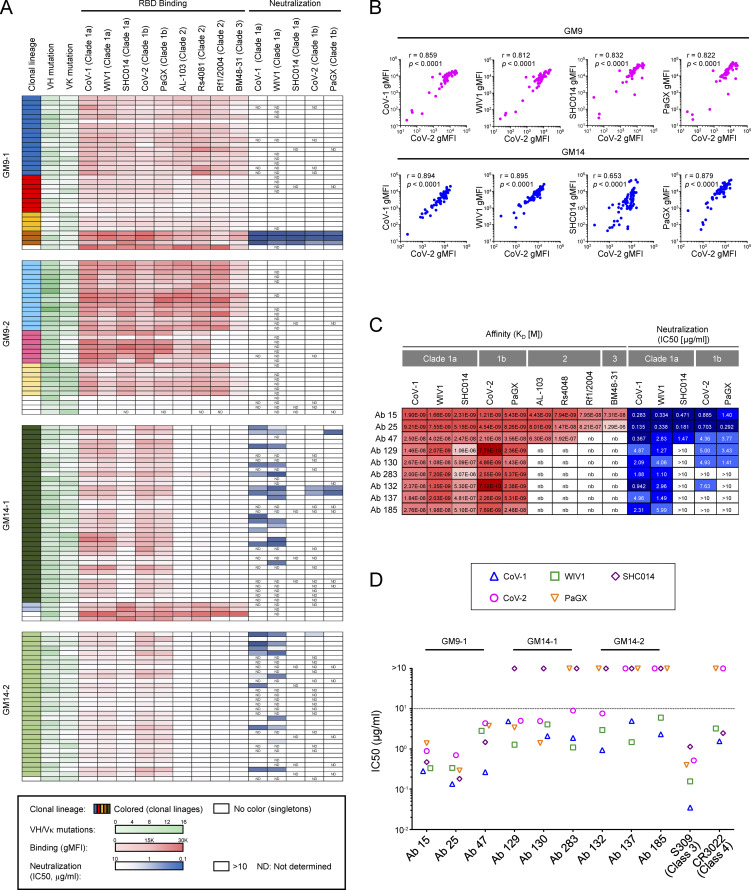
To examine cross-reactivity profiles for each CoV-2+/CoV-1+ GC B cell, we characterized mAbs from single cell sorted GC B cells from four individual mice (GM9-1, GM9-2, GM14-1, or GM14-2). Sequence analysis revealed that, in each mouse, >85% of the isolated mAbs were members of expanded clonal lineages. Particularly in GM14-1 or GM14-2 mice, almost all the mAbs were encoded by the same combination of VH/VK with different DH and JK segments: GM14-1 (VH14-3-DH3-2-JH4 /VΚ4-111-JΚ2), and GM14-2 (VH14-3-DH2-3-JH4/ VV4-111-JK1; Table S1).
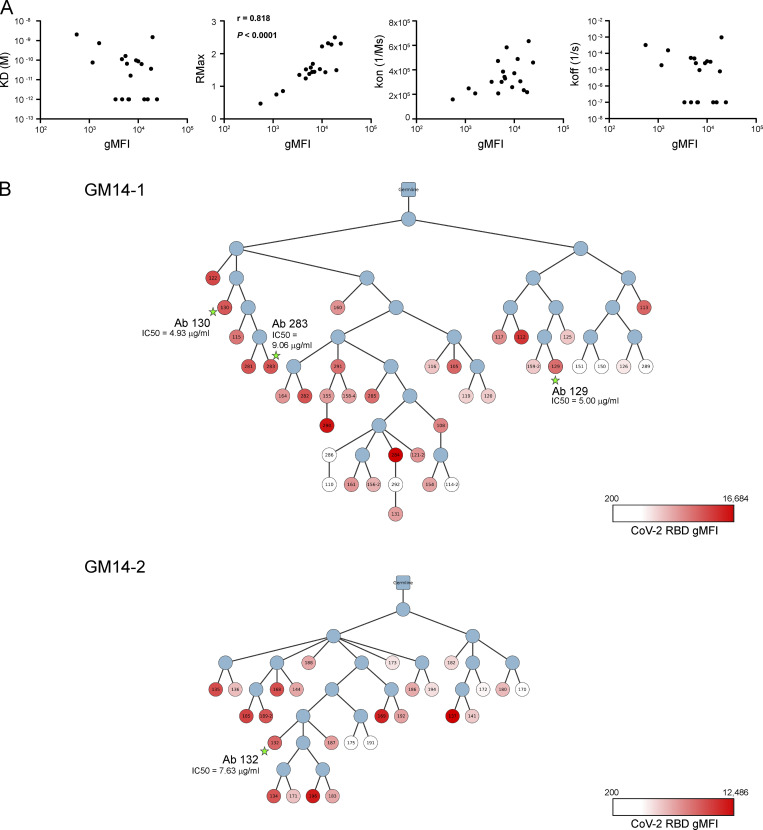
Among all the analyzed mAbs, many of them showed blockade of their binding to CoV-2 RBD by authentic class 4 mAbs, except that mAbs derived from VH14-1/VK8-27 gene pairing were blocked by authentic class 3 mAb (Table S1). For assessment of the breath of recognition to sarbecovirus RBDs by the above mAbs, we assigned a relative binding index to each antibody by using flow cytometry–based measurement (Fig. 5 A; Bajic et al., 2019; Leach et al., 2019); the relative binding indexes represented well RMax of bivalent IgG antibodies measured by Octet kinetics assay (Fig. S4 A). We selected RBDs from distantly related clade 2 (bat CoV Anlong-103 [AL-103], bat CoV Rs4081 [Rs4081], and bat CoV Rf1/2004 [Rf1/2004]) and clade 3 (bat CoV BM48-31 [BM48-31]) (Lau et al., 2020; Letko et al., 2020), in addition to clade 1 (CoV-1, WIV1, SHC014 as clade 1a, and CoV-2, PaGX as clade 1b) (Fig. S3). As expected, almost all the isolated mAbs from GM9-1, GM9-2, GM14-1, or GM14-2 mice displayed binding reactivity to clade 1a and 1b RBDs. The relative binding indexes were well correlated between CoV-1/WIV1/SHC014/PaGX and CoV-2 in GM9- or GM14-immunized mice (Fig. 5 B). In regard to clade 2 and 3 RBDs, some of the isolated mAbs from GM9-1 or GM9-2, and only a few from the GM14-1 mouse, exhibited their significant binding reactivity (Fig. 5 A).
Figure 5.
Binding properties of GC-derived mAbs. (A) Clonality, VH and VK mutations, binding to S RBDs of CoV-1, WIV1, SHC014, CoV-2, PaGX, AL-103, Rs4081, Rf1/2004, and BM48-31, neutralizing activity against CoV-1, WIV1, SHC014, CoV-2, and PaGX pseudoviruses of mAbs derived from CoV-1 and CoV-2 RBD cross-reactive GC B cells of mice immunized with GM9 (GM9-1 and GM9-2) or GM14 (GM14-1 and GM14-2). (B) Correlation between mAb binding to CoV-2 RBD and other sarbecovirus RBDs. Binding of mAbs with individual RBDs was measured by bead-based flow-cytometric assays. The binding signals to respective RBDs (gMFI) are plotted in the graph. Spearman’s rank correlation coefficients and P values are shown. (C) Binding affinity of human Fab fragments with RBD proteins determined by biolayer interferometry. The equilibrium dissociation constants (KD[M]) and IC50s (μg/ml) of individual clones are shown. (D) Neutralizing activity (IC50 [μg/ml]) of representative cross-reactive mAbs and mouse IgG2c recombinant antibodies carrying the variable regions of previously isolated S304 (class 3) and CR3022 (class 4) human anti-RBD antibodies. Results are representative of at least two independent experiments (A–D). Ab, antibody.
Figure S4.
Reactivity and phylogenetic analysis of mAbs. (A) Correlation between gMFI from bead-based flow-cytometric assay and Rmax from biolayer interferometry (Octet) assay. gMFI and dissociation constant [KD(M)], maximal R (RMax), association rate (kon; 1/ms), and dissociation rate (koff) of bivalent mAb-CoV-2 RBD interactions. Spearman coefficient and P value were calculated by Prism software. (B) The phylogenetic tree of clonal lineages of GC-derived clones from GM14-1 and GM14-2 mice. The root is unmutated germline ancestor. The blue circles are inferred sequences. The white and red colors of the circles indicate the intensity of CoV-2 RBD binding in bead-based flow-cytometric assay (Table S1). The circles with stars are clones with high neutralization activity against CoV-2 (Fig. 5 C).
Then we evaluated the neutralization activities of each mAb toward CoV-1, WIV1, SHC014, CoV-2, or PaGX by the aforementioned VSV-based pseudotype assays (Fig. 5 A and Table S1). Because of the large number of antibodies, we screened and focused on nine mAbs using the following two criteria (Fig. 5 C). First, the neutralization activities are <10 µg/ml half-maximal inhibitory concentration (IC50) toward at least two types of viruses among five. Second, the highest potency toward these viruses is <3 µg/ml IC50. As shown in Fig. 5 D, like authentic mAb S309 or CR3022, these mAbs possessed higher IC50s for CoV-2 than those observed for CoV-1 or WIV1. In VH14-3-DH3-2-JH4/VK4-111-JK2 (GM14-1) or VH14-3-DH2-3-JH4/VK4-111-JΚ1 (GM14-2) lineage, mAbs with similar or higher binding indexes to that of the mAb (antibodies 129, 130, 283, or 132) for CoV-2 (Fig. 5 C) do not possess high potency of neutralization activity toward CoV-2 (Fig. S4 B and Table S1). Thus, antibody neutralization potency is likely governed at least in part by factors beyond binding affinity.
Elicitation of cross-reactive long-lived plasma cells and memory B cells
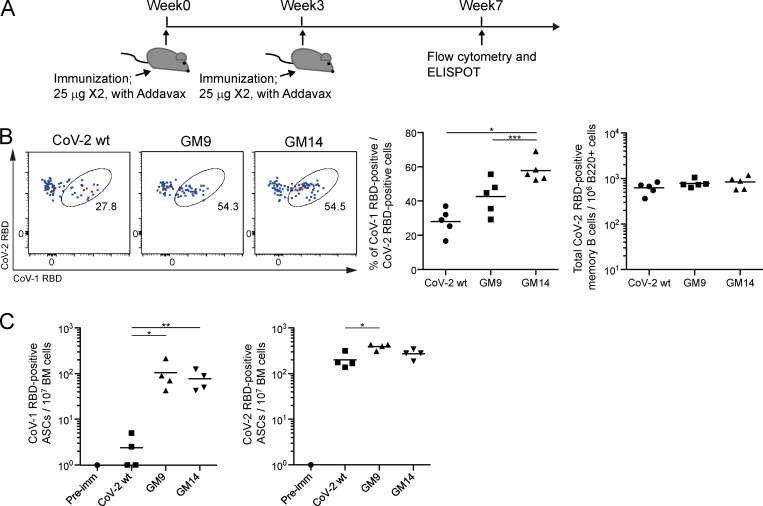
Most successful vaccine approaches rely on the generation of memory B cells and long-lived plasma cells. As shown in Fig. 6 A, analysis was performed at 7 wk. At this time, CoV-2+ class-switched IgG type memory B cells were similarly generated by WT RBD, GM9, or GM14 immunization, although GM9 and GM14 gave rise to more CoV-1+CoV-2+ cells (Fig. 6 B). Furthermore, as assessed by CoV-1–specific antibody-secreting cells (ASCs) in the bone marrow, GM9 or GM14 immunization induced more CoV-1+ ASC (Fig. 6 C), together indicating that both cross-reactive memory compartments are more efficiently generated by our immunization protocol using GM9 or GM14, compared with WT RBD immunization.
Figure 6.
Generation of cross-reactive memory B cells and long-lived plasma cells in mice immunized with GM9 or GM14. (A) Schematic overview of the experimental design. (B) The cross-reactivity of CoV-2 RBD-specific memory B cells (CD138−B220+IgG+CD38+GL7− CoV-2 RBD[APC]+ CoV-2 RBD[PE-Cy7]+) with CoV-1 RBD from spleen of mice 1 mo after boost immunization with each antigen was assessed by flow cytometry. The plots are representative of finding from two independent experiments (left). Frequency of CoV-1 and CoV-2 RBD cross-reactive memory B cells (middle) and quantification of absolute numbers of total CoV-2 RBD-specific memory B cells (right) from multiple mice in one experiment. Results are representative of two independent experiments. CoV-2 RBD WT (n = 5); GM9 (n = 5); GM14 (n = 5). (C) ELISPOT analysis of CoV-1 RBD– (left) or CoV-2 RBD– (right) specific IgG1 ASC responses of bone marrow (BM) cells from preimmune (Pre-imm) mice, or mice 1 mo after boost immunizations with each antigen. Results are representative of two independent experiments. Preimmune (n = 1); CoV-2 RBD WT (n = 4); GM9 (n = 4); GM14 (n = 4). Horizontal lines indicate mean values; each symbol indicates one mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
Discussion
Despite its potency in eliciting neutralizing antibodies for CoV-2, the native CoV-2 RBD vaccine was incapable of eliciting significant neutralizing activity for CoV-1 or WIV1 in our mouse model. These mouse data, together with the recent human evidence that convalescent sera from CoV-2–infected individuals lacked cross-neutralization activity for WIV1, highlight the need to develop broadly protective interventions (next-generation vaccines) to prevent future coronavirus pandemics (Garcia-Beltran et al., 2021). In this context, we hypothesized that the well conserved core-RBD subdomain could be a promising target for inducing broadly neutralizing antibodies. However, this subdomain seems to be immuno-subdominant in its natural configuration. Hence, we introduced glycans into the normally immuno-dominant head-RBD subdomain to focus the immune response more to the core-RBD. Here, we showed that glycan engineering was capable of eliciting cross-neutralizing responses toward PaGX, SHC014, WIV1, and CoV-1, due to increased generation of anti-core RBD-specific antibodies, therefore providing one of the frameworks for immunogen design for SARS-related virus vaccines. It should be mentioned that the S protein has several conserved epitopes outside of RBD, which can be recognized by cross-neutralizing antibodies (Song et al., 2021; Sauer et al., 2021; Wang et al., 2021). Therefore, in order to develop more broadly protective interventions, designs might be necessary to induce cross-reactive antibodies against both the core-RBD and conserved regions outside RBD.
One of the potential weak points of glycan engineering is that this method seems to rely on the expense of decreased neutralization of authentic CoV-2, potentially causing suboptimal protection against the original virus. Glycan addition has been currently thought to redirect and/or select the B cell receptors (BCRs) with the most fitness for the modified epitopes rather than simply masking or obscuring epitopes (Duan et al., 2018). For instance, added glycans nearest to the class 1 or 2 site in the head-RBD subdomain may have restricted the approach angles from which epitope 1/2–specific BCR could access these sites. Hence, it might be possible to find out the appropriate glycan combination on the head-RBD subdomain that selects the class 1/2 BCR with highest neutralizing potency while suppressing overall immuno-dominance of the head-RBD subdomain. Indeed, in the case of HIV vaccination, glycan fine-tuning on vaccines is successfully developed to select rare broadly neutralizing antibody VRC01 (Duan et al., 2018).
As discussed above, glycan addition was initially intended as a method to mitigate off-target immune responses. Nevertheless, the risk for creating undesired neo-epitopes seems to coexist in this method, too. In fact, in contrast to GM9 (three additional mutants), GM14 (five additional mutants) generated more GC B cells recognizing such neo-epitopes (Fig. S2 B), which might account for causing the reduced neutralizing activity against native CoV-2 (Fig. 3 A).
As an initial step for glycan engineering on CoV-2 RBD, here we used simple criteria for its design. Despite enhancement of the immune responses to the conserved core-RBD subdomain by current glycan engineering, the CoV-2+Cov-1− (likely recognizing the head-RBD subdomain) populations were still largely remaining, indicating that the head-RBD subdomain still acts as antigenic immuno-dominant. Taking account of the above several aspects, more extended glycan combination studies are required to suppress immuno-dominance of the head-RBD subdomain, to select the class 1/2 BCR with high neutralizing potency, and to minimize generation of the undesired neo-epitopes.
How might the data from this study inform SARS-related vaccine strategy? CoV-1–infected individuals, albeit rare ones, possess conserved RBD-directed cross-neutralizing antibodies, some of which recognize the overlapping site with the authentic CR3022 epitope (class 4) in the core-RBD subdomain (Wec et al., 2020). Similarly, mice can produce core-RBD–directed antibodies predominantly recognizing overlapping site on the authentic class 4 mAb epitope, and some of them manifest cross-neutralization. Both human and mouse antibodies are likely to require GC processes to acquire their cross-neutralizing activities. Such similar properties of the antibodies highly suggest that the core-RBD could be one of the useful components for SARS-related virus vaccine and that the mouse data from this study are applicable to human vaccine development.
Materials and methods
Mice
BALB/c mice were purchased from SLC Japan and maintained under specific pathogen–free conditions. Sex-matched 7–8-wk-old mice were used for all experiments. All animal experiments were conducted in accordance with the animal experiment guidelines of Osaka University.
Human subjects
Human blood samples were collected at Tokyo Shinagawa Hospital, and plasma was isolated using Vacutainer CPT Tubes (BD Bioscience). The study protocol was approved by the National Institute of Infectious Diseases Ethic Review Board for Human Subjects, the ethics committees of Tokyo Shinagawa Hospital, and Osaka University. All participants provided written informed consent in accordance with the Declaration of Helsinki.
Vaccine design
The mammalian expression constructs for parental and glycan mutant rRBD (pCAGGS-CoV-2 RBDwt-His-Avi, pCAGGS-GM9-His-Avi, and pCAGGS-GM14-His-Avi) encode aa 331–529 of SARSCoV-2 S, Middle East respiratory syndrome S protein–derived signal peptide (MIHSVFLLMFLLTPTESYVD) at the N terminus, and a 6XHis-Avi-tag (HHHHHHGLNDIFEAQKIEWHE) at the C terminus. For glycan masking, we introduced NxT sequons in the RBD surface residues shown in Fig. S1. Mutations expected to disrupt the structure of RBD or with the low glycosylation score (<0.5) by NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/) were avoided. The candidate constructs were transfected into Expi293F cells to examine whether added glycosylation prevented RBD expression. Antigenicity of the GM mutants was evaluated by ELISA using previously published anti-RBD antibodies.
Recombinant protein expression and purification
The plasmids for the recombinant CoV-2 RBD (RefSeq accession no. NC_045512.2) and GM mutants, CoV-1 RBD (GenBank accession no. AY278741.1), and SARS-related CoV RBDs (PaGX, GenBank accession no. MT072864.1; WIV1, GenBank accession no. KF367457.1; SHC014, GenBank accession no. AGZ48806.1; bat CoV AL103, GenBank accession no. ARI44799.1; Rs4081, GenBank accession no. KY417143.1; Rf1/2004, GenBank accession no. DQ412042.1; and BM48-31, RefSeq accession no. YP_003858584.1) were transiently transfected into Expi293F cells (Thermo Fisher Scientific) with the ExpiFectamine 293 Transfection Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. After 3–4 d, supernatants were collected and passed through a 0.45-µm filter. The recombinant proteins were purified from supernatants by Talon metal affinity resin (Takara) according to the manufacturer’s protocol. The elute from the resin was concentrated using an Amicon Ultra 4 10,000 NWML, in which the elution buffer was exchanged with PBS. For nanoparticle antigens and probes, post-translational biotinylation of rRBD and GM mutants was performed in culture via coexpression of the BirA enzyme by cotransfection of a BirA-Flag expression vector (Addgene; Leach et al., 2019). Cells were maintained in culture supplemented with 100 µM biotin (Sigma-Aldrich) after transfection as described above.
For preparation of ELISA or ELISPOT antigens, mammalian expression constructs of CoV-2 RBD WT, GM9, GM14, and CoV-1 RBD containing a thrombin cleavage site (LVPRGS) in front of the C-terminal 6XHis-Avi-tag were transiently transfected into Expi293F cells. Purified proteins were treated with thrombin using a Thrombin kit (Merck) according to the manufacturer’s protocol. The cleaved 6XHis-AviTag and thrombin were removed by Talon metal affinity resin (Takara) and Benzamidine Sepharose (GE), respectively. Purity or biotinylation efficiency of recombinant proteins was confirmed by SDS-PAGE.
Anti–CoV-2 S RBD mAbs
DNA fragments encoding heavy and κ or λ chain variable regions from previously published anti–CoV-2 mAbs (Pinto et al., 2020; Shi et al., 2020; Yuan et al., 2020; Zhou et al., 2020) were synthesized (Eurofins) and cloned into human IgG1, Igκ, and Igλ expression vectors, respectively. The heavy and light chain expression vectors were cotransfected into Expi293F. Respective mAbs were purified with Protein G Sepharose (GE) according to the manufacturer’s protocol.
N-linked glycan occupancy analysis by LC/MS
Sample preparation was performed by referring to a previous report (Tajiri-Tsukada et al., 2020). Briefly, the sample protein (10 µg) was treated using MPEX PTS Reagents (GL Sciences). The reduced and carboxymethylated protein was digested with 2 µg of chymotrypsin (1 µg/ml; Promega) at 37°C for 3 d. The resulting peptides were desalted using an Oasis HLB μElution plate (Waters), and dried and dissolved in 50 µl of 0.1% formic acid solution. The sample solution was analyzed by LC/MS using the parallel acquisition mode using higher-energy collisional dissociation (HCD) and electron-transfer/HCD. The LC/MS system and HCD-MS/MS conditions were in accordance with our previous report (Tajiri-Tsukada et al., 2020). The electron-transfer/HCD-MS/MS condition was as follows: 23% of supplemental activation collision energy, m/z 2 of isolation window, and 100 ms of maximum injection time. The peptide identifications were performed by database searching using BioPharma Finder 3.1 (Thermo Fisher Scientific). The search parameters were as follows: a mass tolerance of ±5 ppm, confidence score of >80, and identification type of MS2. Carboxymethylation (+58.005 dalton) was set as a static modification of Cys residues. A human glycan database stored in the software was used for glycopeptide identifications. The integrated peak area of the multiple precursor ions from each glycopeptide was calculated, and the glycan occupancy (%) was calculated by the following formula:
Nanoparticle coating
Streptavidin-coated 0.11-µm nanoparticles (Bangs Laboratories) were washed twice with PBS, and 150 µg nanoparticles were incubated with 50 µg biotinylated proteins in 80 µl PBS per one mouse for 5 h at 4°C. Coating efficiency was measured by flow cytometry.
Immunization
At 7–8 wk of age, each antigen group was vaccinated with a prime immunization, and 3 wk later, mice were boosted with a second vaccination. Prior to inoculation, immunogen suspensions were gently mixed 2:1 vol/vol with AddaVax adjuvant (Invivogen) to reach a final concentration of 0.4 mg/ml antigen. Mice were injected intramuscularly using a 29 G × 1/2 needle syringe (Terumo) with 60 µl per injection site (120 µl total) of immunogen under isoflurane anesthesia. For prime immunization, antigens were used as nanoparticles, and for boost immunization, antigens were used as monomeric proteins.
Flow cytometry
Single-cell suspensions were prepared from inguinal and iliac lymph nodes or spleen. Inguinal and iliac lymph nodes were collected and pooled for individual mice. Detection of CoV-1 RBD, CoV-2 RBD WT, GM9, and GM14-specific B cells was performed using biotinylated rRBD prelabeled with fluorophore-conjugated streptavidin. To exclude induced 6XHis-, AviTag-, biotin-, and streptavidin-specific B cells, samples were prestained with decoy probe. Cell samples were analyzed using an Attune NxT flow cytometer (Thermo Fisher Scientific) and sorted using a FACS Aria II cell sorter (BD Bioscience). APC-eFluor780-B220, eFluor450-GL7, and FITC-IgM antibodies and APC-streptavidin and PerCP-Cy5.5-streptavidin were purchased from Thermo Fisher Scientific. BV510-IgD, FITC-IgG1 (for IgG mix), FITC-IgG2a (for IgG mix), FITC-IgG3 (for IgG mix), FITC-CD95, BV786-CD38, BV711-CD38, BV786-CD138, and CD16/32 (as Fc blocker) antibodies and PE-streptavidin and PE-Cy7-streptavidin were purchased from BD Bioscience. Alexa Fluor 700-IgD and FITC-IgG2b (for IgG mix) antibodies and BV510-streptavidin and 7-AAD (as viability dye) were purchased from BioLegend. Data were analyzed using FlowJo software V10 (Tree Star).
BCR cloning and antibody expression
CoV-1 and CoV-2 RBD cross-reactive GC B cells were single-cell sorted from a lymph node of mice 3 wk after primary immunization. Cloning and expression of antibodies were performed as described previously (Inoue et al., 2021) with the following modifications. PCR-amplified Igγs and Igκ V(D)J transcripts were cloned into the mouse Igγ2c/Igκ-expression vector (this vector replaces the human IgG1 and Igκ constant regions of pVITRO1-dV-IgG1/κ [Addgene #52213; Dodev et al., 2014] with mouse IgG2c and Igκ constant regions, respectively), or human Fab-expression vector (this vector replaces the human IgG1 constant regions of pVITRO1-dV-IgG1/κ with His-tag) using the seamless ligation cloning extract method. mAbs were expressed using the Expi293 Expression System (Thermo Fisher Scientific) and purified from the culture supernatants of Expi293F cells by Protein G Sepharose (GE) for full antibodies or by Talon metal affinity resin (Takara) for His-tagged Fab antibodies.
Flow cytometry analysis of mAb binding to the antigen
Anti-mouse Igκ microparticles and negative control particles (BD CompBeads) were mixed at a 1:1 ratio. The beads were then mixed with 250 ng of purified mAbs (mIgG2c/mIgκ) for 20 min, washed with PBS containing 2% FBS, and labeled with PE-conjugated antigen for 20 min. Binding capacity of mAbs to antigen was assessed by flow cytometry (BD FACSCanto II and FACS Aria II).
ELISA
Nunc Maxisorp Immuno plates (Thermo Fisher Scientific) were coated with each rRBD (100 ng/50 µl). Blocking was performed using BlockingOne solution (Nacalai Tesque). Plates were sequentially incubated with serially diluted serum samples or mAb for standard and IgG1-HRP detection antibody (Southern Biotech). Detection was performed using KPL SureBlue TMB Microwell Peroxidase Substrate (SeraCare). The CR3022 mouse IgG1e3 mAb (Invivogen) was used as a standard.
For mAb-based epitope-blocking ELISA, CoV-1 RBD– or CoV-2 RBD–coated plates for serum samples or cloned mAbs, respectively, were prepared. After blocking step to block sites in the well not occupied by rRBDs, the following were added alone or in a mixture: CR3022 (class 4), EY6A (class 4), and/or S309 (class 3) human IgG1 mAbs (150 ng/50 µl each) or S230 (anti-CoV-1 head-RBD [class 1 type]) human IgG1 mAb (150 ng/50 µl; Absolute Antibody Ltd.). After incubation at room temperature for 3 h, the plate was washed with PBS with 0.05% Tween20, and then serially diluted serum samples or cloned mAbs were added and incubated at 37°C for 1 h. After three washes, HRP-conjugated goat anti-mouse IgG1 (Southern Biotech) for serum samples or IgG2c (Southern Biotech) for cloned mAb samples were added and incubated for 1 h at room temperature. Occupancy of class 3/4 or class 1 type antibodies in whole anti CoV-1 RBD antibodies was calculated by subtraction of titers measured by epitope-blocking ELISA and conventional serum titer ELISA and then shown as a percentage. The CR3022 mouse IgG1e3 mAb (Invivogen) was used as a standard. For detection of human mAbs used as blocking antibody, HRP-conjugated goat anti-human IgG (Southern Biotech) was used.
For affinity ELISAs, serially diluted sera were incubated on plates coated with 1 μg/ml CoV-1 RBD protein (low density) or 8 μg/ml CoV-1 RBD (high density). The affinity of CoV-1 RBD–specific IgG1 was expressed as a ratio of binding to low-density:high-density CoV-1 RBD–coated plates as in a previous report (Wang et al., 2015). The CR3022 mouse IgG1e3 mAb (Invivogen) was used as a standard.
ELISPOT assay
Plates with a cellulose membrane bottom were coated with CoV-1 RBD or CoV-2 RBD (100 ng/50 µl). 2 × 107 cells/ml and 1:3 serial dilutions of bone marrow cells were added to the wells and incubated in 200 µl RPMI-1640 medium supplemented with 10% FBS, 50 µM 2-ME, and 2 mM sodium pyruvate for 5 h at 37°C under 5% CO2. After washing with PBS with 0.05% Tween20, goat anti-mouse IgG1 Ab was added to the wells followed by addition of alkaline phosphatase-labeled anti-goat IgG Ab. Spots were visualized by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium substrate (Promega) and counted.
Pseudovirus assay
Preparation of SARS CoV S protein-pseudotyped VSVΔG-luc has been described elsewhere (Tani et al., 2010; Yoshida et al., 2021). In brief, HEK293T cells were transfected with expression plasmids for respective CoV S proteins (CoV-2 Wuhan strain S, CoV-2 Wuhan S K417N/E484K/N501Y mutant, PaGX S, SHC014 S, CoV-1 S, and WIV1 S) by usingTransIT-LT1 (Mirus) according to the manufacturer’s instructions. At 24 h after transfection, VSVΔG-luc virus (multiplicity of infection = 0.1) was inoculated onto the transfectants. After 2 h incubation, cells were washed with DMEM and were further cultivated for an additional 24–48 h. Cell-free supernatant was harvested and used for the neutralizing assay as described previously (Nie et al., 2020). Mouse sera and human plasma samples were incubated at 56°C for 30 min and then serially diluted from 1/20 in culture medium. CoV-1 S-pseudotyped VSVΔG-luc was incubated with different dilutions of mouse sera, human plasma, or recombinant antibodies for 1 h at 4°C, and then inoculated onto a monolayer culture of VeroE6-TMRPSS2 (JCRB1819; NIBION) in a 96-well plate. At 16 h after inoculation, cells were washed with PBS and then lysed with luciferase cell culture lysis reagent (Promega). After centrifugation, cleared cell lysates were incubated with firefly luciferase assay substrate (Promega) in 96-well white polystyrene plates (Corning). Luciferase activity was measured by GloMax Discover luminometer (Promega). NT50 or IC50 was calculated by Prism software (GraphPad).
ADE assay
Raji cells (American Type Culture Collection) were maintained in RPMI-1640 (Fujifilm) containing 10% heat-inactivated FBS. For the expression vector of human/mouse chimera MW05, the variable regions from heavy and light chains of human antibody clone MW05 (Wang et al., 2020) were synthesized to be fused with mouse IgG1 and mIgκ constant regions, respectively. MW05 mouse IgG1 was prepared as described above. ADE of infection was assayed as described elsewhere (Wang et al., 2020). Briefly, CoV-2 S pseudotyped VSVΔG was preincubated with different concentration of MW05 mouse IgG1 or control mouse IgG1 (Southern Biotech), or mouse sera in culture medium. After 1 h incubation, the mixture was added onto Raji cells (0.1 million/well) in a 96-well plate. Then cells were cultivated for 16 h. Luciferase activity of infected cells was measured as above.
Virus neutralization assay
A mixture of 100 TCID50 of CoV-2 Wuhan strain WK-521 (2019-nCoV/Japan/WK-521/TY/2020 [National Institute of Infectious Diseases, Pathogen Genomics Center, Japan]) and serially diluted, heat-inactivated plasma samples (twofold serial dilutions starting from 1:40 dilution) were incubated at 37°C for 1 h before being placed on VeroE6-TMPRSS2 cells seeded in 96-well flat-bottom plates (TPP). VeroE6-TMPRSS2 cells were maintained in low glucose DMEM (Fujifilm) containing 10% heat-inactivated FBS, 1 mg/ml geneticin (Thermo Fisher Scientific), and 100 U/ml penicillin/streptomycin (Thermo Fisher Scientific) at 37°C supplied with 5% CO2. After culturing for 4 d, cells were fixed with 20% formalin (Fujifilm) and stained with crystal violet solution (Sigma-Aldrich). Cutoff dilution index with >50% cytopathic effect was presented as microneutralization titer. Microneutralization titer of the sample below the detection limit (1:40 dilution) was set as 20.
Biolayer interferometry assay
The kinetics of mAb binding to antigen was determined with the OctetRED96e system (ForteBio) at 30°C with shaking at 1,000 rpm. Biotinylated-RBD proteins from CoV-1, WIV1, SHC014, CoV-2, PaGX, AL-103, Rs4081, Rf1/2004, and BM48-31 were loaded at 6 µg/ml in 1× kinetics buffer (0.1% BSA and 0.02% Tween-20 in PBS) for 900 s onto streptavidin biosensors (ForteBio) and incubated with serially diluted Fab antibodies (100, 33.3, 11.1, and 0 nM for CoV-1, WIV1, CoV-2, PaGX, AL-103, Rf4081, Rf1/2004, and BM48-31 and 900, 300, 100, and 0 nM for SHC014) for 120 s, followed by immersion in 1× kinetics buffer for 300 s of dissociation time. The binding curves were fit in a 1:1 (CoV-1, WIV1, CoV-2, PaGX, AL-103, Rf4081, Rf1/2004, and BM48-31) or 2:1 (SHC014) binding model, and the dissociation constant values were calculated by Octet Data Analysis software (ForteBio).
Phylogenetic analysis of antibody clones
For each cell, V, D, J genes and CDR3 assignment were conducted on the full-length BCR sequences using Igblast and the ImMunoGeneTics mouse references. Clones were defined separately for each experiment, on the basis of their heavy chain only, using the DefineClones function of the Change-O package, and 38/38 cells of GM14#1 and 26/31 cells of GM14#2 were considered the same clone. Clonal tree reconstruction was thus performed for these two major clones using the Alakazam (both Change-O and Alakasam are part of the Immcantation analysis framework; Gupta et al., 2015). Finally, reconstructed clonal trees were plotted using Cytoscape.
Phylogenetic analysis of RBD of sarbecoviruses
RBD regions of selected sarbecovirus Spike proteins were extracted based on multiple sequence alignment of the entire Spike proteins calculated by Multiple Alignment using Fast Fourier Transform (Katoh et al., 2019) with the L-INS-i algorithm. A phylogenetic tree was constructed following a workflow offered by Phylogeny.fr (Dereeper et al., 2008) by using the Jones–Taylor–Thornton substitution model with 100 times bootstrapping.
Statistics
Statistical analyses were performed by a two-tailed unpaired and paired Student’s t test using GraphPad Prism software. P values <0.05 were considered significant. Error bars denote mean ± SEM.
Online supplemental material
Fig. S1 shows the conserved residues between CoV-1 and CoV-2, design and characterization of GM9 and GM14, and the effective induction of anti-RBD IgG1 by multivalent nanoparticles of CoV-2 RBD WT (related to Fig. 1). Fig. S2 shows that immunization of GM9 and GM14 induces antibodies targeting the conserved regions of RBD through GC reaction (related to Fig. 2) and serum neutralizing activity against authentic CoV-2 (Wuhan) and CoV-2 S K417N/E484K/N501Y pseudovirus. Fig. S3 shows phylogeny of RBD sequences from representative sarbecoviruses. Fig. S4 shows correlation between geometric mean fluorescence intensity (gMFI) from bead-based flow-cytometric assay and Rmax from biolayer interferometry (Octet) assay (related to Fig. 5 B), and phylogeny of representative clonal lineages of GC-derived clones from GM14-1 and GM14-2 mice (related to Fig. 5 D). Table S1 lists information on mAbs generated from GC B cells of mice immunized with GM9 or GM14 (related to Fig. 5 A).
Supplementary Material
lists information on mAbs generated from GC B cells of mice immunized with GM9 or GM14 (related to Fig. 5 A).
Acknowledgments
We thank Chie Kawai (Immunology Frontier Research Center, Osaka University) for her technical assistance; Nana Iwami, Soichiro Haruna, and Marwa Ali (Immunology Frontier Research Center, Osaka University) for their contributions to plasmid construction and protein expression; and Junya Suzuki and Yoko Hiruta (National Institute of Health Sciences) for their technical assistance for LC/MS analysis. We also thank Peter Burrows for reading the manuscript and critical comments.
This work was supported by Japan Society for the Promotion of Science KAKENHI (JP19H01028 to T. Kurosaki) and by Japan Agency for Medical Research and Development (JP20fk0108104 to Y. Takahashi and T. Kurosaki; JP21fk0108534 to Y. Takahashi and T. Kurosaki).
Author contributions: R. Shinnakasu, S. Sakakibara, H. Yamamoto, S. Moriyama, N. Hashii, and T. Inoue performed the experiments and analyzed the data; R. Shinnakasu, S. Sakakibara, K. Yamashita, and T. Kurosaki designed the experiments; C. Ono, A. Yamanaka, Y. Adachi, T. Onodera, T. Sato, M. Shinkai, R. Suzuki, Y. Matsuura, and Y. Takahashi provided essential reagents; P. Wang and K. Yamashita performed the structural studies; N. Sax and K. Yamashita performed bioinformatics analyses; and R. Shinnakasu, S. Sakakibara, and T. Kurosaki wrote the manuscript.
References
- Bajic, G., Maron M.J., Adachi Y., Onodera T., McCarthy K.R., McGee C.E., Sempowski G.D., Takahashi Y., Kelsoe G., Kuraoka M., and Schmidt A.G.. 2019. Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe. 25:827–835.e6. 10.1016/j.chom.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. 2020a. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 588:682–687. 10.1038/s41586-020-2852-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, C.O., West A.P. Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. 2020b. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 182:828–842.e16. 10.1016/j.cell.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos, S., Gupta A., and Ravetch J.V.. 2020. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 20:633–643. 10.1038/s41577-020-00410-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling, J., and Aebi M.. 2013. N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5:a013359. 10.1101/cshperspect.a013359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. 2020. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 369:643–650. 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. 2021. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 27:717–726. 10.1038/s41591-021-01294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, L., and Gao G.F.. 2021. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 21:73–82. 10.1038/s41577-020-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper, A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue, Web Server):W465-9. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodev, T.S., Karagiannis P., Gilbert A.E., Josephs D.H., Bowen H., James L.K., Bax H.J., Beavil R., Pang M.O., Gould H.J., et al. 2014. A tool kit for rapid cloning and expression of recombinant antibodies. Sci. Rep. 4:5885. 10.1038/srep05885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, H., Chen X., Boyington J.C., Cheng C., Zhang Y., Jafari A.J., Stephens T., Tsybovsky Y., Kalyuzhniy O., Zhao P., et al. 2018. Glycan Masking Focuses Immune Responses to the HIV-1 CD4-Binding Site and Enhances Elicitation of VRC01-Class Precursor Antibodies. Immunity. 49:301–311.e5. 10.1016/j.immuni.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink, D., Goff P.H., and Palese P.. 2014. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 88:699–704. 10.1128/JVI.02608-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran, W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., et al. 2021. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 184:476–488.e11. 10.1016/j.cell.2020.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 503:535–538. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., and Kleinstein S.H.. 2015. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 31:3356–3358. 10.1093/bioinformatics/btv359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. 2021. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 184:2384–2393.e12. 10.1016/j.cell.2021.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo, A., Milder F., Kuipers H., Wagner M.V., Zhu X., Hoffman R.M., van Meersbergen R., Huizingh J., Wanningen P., Verspuij J., et al. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 349:1301–1306. 10.1126/science.aac7263 [DOI] [PubMed] [Google Scholar]
- Inoue, T., Shinnakasu R., Kawai C., Ise W., Kawakami E., Sax N., Oki T., Kitamura T., Yamashita K., Fukuyama H., and Kurosaki T.. 2021. Exit from germinal center to become quiescent memory B cells depends on metabolic reprograming and provision of a survival signal. J. Exp. Med. 218:e20200866. 10.1084/jem.20200866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine, J., Julien J.P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.S., MacPherson S., Jones M., et al. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science. 340:711–716. 10.1126/science.1234150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette, C.A., Cohen A.A., Gnanapragasam P.N.P., Muecksch F., Lee Y.E., Huey-Tubman K.E., Schmidt F., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C., et al. 2021. Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. bioRxiv. (Preprint posted April 26, 2021) 10.1101/2021.04.23.441195 [DOI]
- Katoh, K., Rozewicki J., and Yamada K.D.. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20:1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group . 2020. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 182:812–827.e19. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 583:282–285. 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- Lau, S.K.P., Luk H.K.H., Wong A.C.P., Li K.S.M., Zhu L., He Z., Fung J., Chan T.T.Y., Fung K.S.C., and Woo P.C.Y.. 2020. Possible Bat Origin of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 26:1542–1547. 10.3201/eid2607.200092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, S., Shinnakasu R., Adachi Y., Momota M., Makino-Okamura C., Yamamoto T., Ishii K.J., Fukuyama H., Takahashi Y., and Kurosaki T.. 2019. Requirement for memory B-cell activation in protection from heterologous influenza virus reinfection. Int. Immunol. 31:771–779. 10.1093/intimm/dxz049 [DOI] [PubMed] [Google Scholar]
- Letko, M., Marzi A., and Munster V.. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5:562–569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., et al. 2020a. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 53:1272–1280.e5. 10.1016/j.immuni.2020.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Xu W., Xia S., Gu C., Wang X., Wang Q., Zhou J., Wu Y., Cai X., Qu D., et al. 2020b. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct. Target. Ther. 5:282. 10.1038/s41392-020-00402-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery, V.D., Yount B.L. Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., et al. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21:1508–1513. 10.1038/nm.3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery, V.D., Yount B.L. Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., et al. 2016. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 113:3048–3053. 10.1073/pnas.1517719113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. 2020. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 15:3699–3715. 10.1038/s41596-020-0394-5 [DOI] [PubMed] [Google Scholar]
- Pinto, D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 583:290–295. 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- Robbiani, D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. 2020. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 584:437–442. 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx, B., Corti D., Donaldson E., Sheahan T., Stadler K., Lanzavecchia A., and Baric R.. 2008. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J. Virol. 82:3220–3235. 10.1128/JVI.02377-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, R., Makino-Okamura C., Lin Q., Wang M., Shoemaker J.E., Kurosaki T., and Fukuyama H.. 2020. Repurposing the psoriasis drug Oxarol to an ointment adjuvant for the influenza vaccine. Int. Immunol. 32:499–507. 10.1093/intimm/dxaa012 [DOI] [PubMed] [Google Scholar]
- Sauer, M.M., Tortorici M.A., Park Y.J., Walls A.C., Homad L., Acton O.J., Bowen J.E., Wang C., Xiong X., de van der Schueren W., et al. 2021. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol. 28:478–486. 10.1038/s41594-021-00596-4 [DOI] [PubMed] [Google Scholar]
- Shang, J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., and Li F.. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature. 581:221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. 2020. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 584:120–124. 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- Smatti, M.K., Al Thani A.A., and Yassine H.M.. 2018. Viral-Induced Enhanced Disease Illness. Front. Microbiol. 9:2991. 10.3389/fmicb.2018.02991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. 2021. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 12:2938. 10.1038/s41467-021-23074-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. 2020. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 182:1295–1310.e20. 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajiri-Tsukada, M., Hashii N., and Ishii-Watabe A.. 2020. Establishment of a highly precise multi-attribute method for the characterization and quality control of therapeutic monoclonal antibodies. Bioengineered. 11:984–1000. 10.1080/21655979.2020.1814683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, H., Shiokawa M., Kaname Y., Kambara H., Mori Y., Abe T., Moriishi K., and Matsuura Y.. 2010. Involvement of Ceramide in the Propagation of Japanese Encephalitis Virus. J. Virol. 84:2798–2807. 10.1128/JVI.02499-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkenburg, S.A., Mallajosyula V.V., Li O.T., Chin A.W., Carnell G., Temperton N., Varadarajan R., and Poon L.L.. 2016. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci. Rep. 6:22666. 10.1038/srep22666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee, S., Tan C.W., Maneeorn P., Duengkae P., Zhu F., Joyjinda Y., Kaewpom T., Chia W.N., Ampoot W., Lim B.L., et al. 2021. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 12:972. 10.1038/s41467-021-21240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. 2020. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell. 183:1367–1382.e17. 10.1016/j.cell.2020.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.T., Maamary J., Tan G.S., Bournazos S., Davis C.W., Krammer F., Schlesinger S.J., Palese P., Ahmed R., and Ravetch J.V.. 2015. Anti-HA Glycoforms Drive B Cell Affinity Selection and Determine Influenza Vaccine Efficacy. Cell. 162:160–169. 10.1016/j.cell.2015.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Peng Y., Wang R., Jiao S., Wang M., Huang W., Shan C., Jiang W., Li Z., Gu C., et al. 2020. Characterization of neutralizing antibody with prophylactic and therapeutic efficacy against SARS-CoV-2 in rhesus monkeys. Nat. Commun. 11:5752. 10.1038/s41467-020-19568-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., van Haperen R., Gutiérrez-Álvarez J., Li W., Okba N.M.A., Albulescu I., Widjaja I., van Dieren B., Fernandez-Delgado R., Sola I., et al. 2021. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat. Commun. 12:1715. 10.1038/s41467-021-21968-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec, A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. 2020. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 369:731–736. 10.1126/science.abc7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Ono C., Hayashi H., Fukumoto S., Shiraishi S., Tomono K., Arase H., Matsuura Y., and Nakagami H.. 2021. SARS-CoV-2-induced humoral immunity through B cell epitope analysis in COVID-19 infected individuals. Sci. Rep. 11(1). 10.1038/s41598-021-85202-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. 2020. Structural basis of a shared antibody response to SARS-CoV-2. Science. 369:1119–1123. 10.1126/science.abd2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., Lin Y.C., Cheng C.Y., Cheng S.H., Huang Y.C., et al. 2020. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 27:950–958. 10.1038/s41594-020-0480-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists information on mAbs generated from GC B cells of mice immunized with GM9 or GM14 (related to Fig. 5 A).