Summary
The acoel worm Hofstenia miamia, which can replace tissue lost to injury via differentiation of a population of stem cells, has emerged as a new research organism for studying regeneration. To enhance the depth of mechanistic studies in this system, we devised a protocol for microinjection into embryonic cells that resulted in stable transgene integration into the genome and generated animals with tissue-specific fluorescent transgene expression in epidermis, gut, and muscle. We demonstrate that transgenic Hofstenia are amenable to the isolation of specific cell types, investigations of regeneration, tracking of photoconverted molecules, and live imaging. Further, our stable transgenic lines revealed insights into the biology of Hofstenia, including a high-resolution three-dimensional view of cell morphology and the organization of muscle as a cellular scaffold for other tissues. Our work positions Hofstenia as a powerful system with multiple toolkits for mechanistic investigations of development, whole-body regeneration, and stem cell biology.
Graphical Abstract
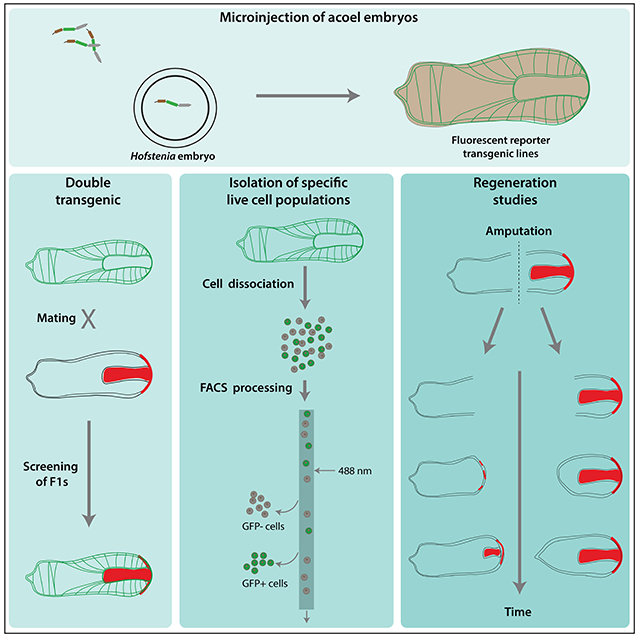
eTOC
Ricci et al. achieve stable transgenesis in the xenacoelomorph Hofstenia miamia, a model for the study of regeneration. Fluorescently-labeled tissues reveal morpho-anatomy at high resolution, including close associations of muscle fibers with epidermis and gut. Transgenic H. miamia worms are amenable to many downstream analyses including live imaging.
Introduction
New research organisms hold the potential to reveal novel biological insights into phenomena that cannot be studied via established model systems. Numerous animal species have been recently utilized in laboratory research, e.g., short-lived killifish for studying aging, hardy water bears for studying tolerance to extreme environments, or the anemone Aiptasia for studying coral bleaching (Lehnert, Burriesci and Pringle, 2012; Goldstein, 2018; Hu and Brunet, 2018). The lack of genetic tools in emerging systems can limit the depth of molecular and cellular insight, however, advances in genome-scale sequencing technology and tools for genetic manipulations (e.g., via CRISPR/CAS9) are releasing this constraint (Ikmi et al., 2014; Wudarski et al., 2017; Minor et al., 2019). Here, we developed a method for transgenesis in the acoel worm, Hofstenia miamia (Corrêa, 1960), which has emerged as a new research organism for studying regeneration and stem cell biology.
In contrast to the limited regeneration capacities of vertebrates, many invertebrate species regenerate robustly, reconfiguring entire body axes and replacing virtually any missing cell type. Notably, some of these highly regenerative invertebrates also harbor large populations of multipotent or pluripotent stem cells as adult animals (Hemmrich et al., 2012; Gahan et al., 2016; Wang et al., 2018; Kassmer et al., 2020). Hofstenia is capable of this “whole-body” regeneration and harbors adult stem cells referred to as “neoblasts”(Srivastava et al., 2014). A sequenced genome, transcriptomic resources, and robust and systemic RNA interference (RNAi) have enabled functional studies of regeneration and stem cells in Hofstenia (Srivastava et al., 2014; Raz et al., 2017; Gehrke et al., 2019; Tewari et al., 2019; Ramirez et al., 2020), and we sought to leverage the biology of this system further by developing tools for transgenesis.
Hofstenia miamia, a.k.a. the three-banded panther worm (Achatz et al., 2013; Jondelius et al., 2019), belongs to an enigmatic phyletic lineage, the Xenacoelomorpha (comprising acoels, xenoturbellids and nemertodermatids), which is a deep-diverging bilaterian lineage, thought to be either the sister lineage to all other bilaterians, or the sister group to ambulacrarians (echinoderms + hemichordates) (Figure 1A) (Ruiz-Trillo et al., 1999, 2002, 2004; Philippe et al., 2007; Sempere et al., 2007; Cannon et al., 2016; Ruiz-Trillo and Paps, 2016; Kapli and Telford, 2020). Thus, acoels are important for understanding the evolution of major features of bilaterians and have been the focus of many recent studies (Raz et al., 2017; Gavilán et al., 2019; Duruz et al., 2020; Hayes et al., 2020; Hulett et al., 2020; Sakagami et al., 2021). Moreover, despite the debate surrounding their phylogenetic position, acoels are evolutionarily distantly related to other regenerative model systems such as cnidarians, planarians, and sea squirts. Therefore, development of more genetic tools in acoels has the potential to advance the study of an important yet understudied animal group as well as to inform the evolution of pathways for development, regeneration, and stem cell biology. Here, we demonstrate that random transgenesis is efficient in the acoel Hofstenia miamia, powers biological insight, and allows multiple downstream applications.
Figure 1: Hofstenia miamia is a highly regenerative species with accessible and manipulable embryos.
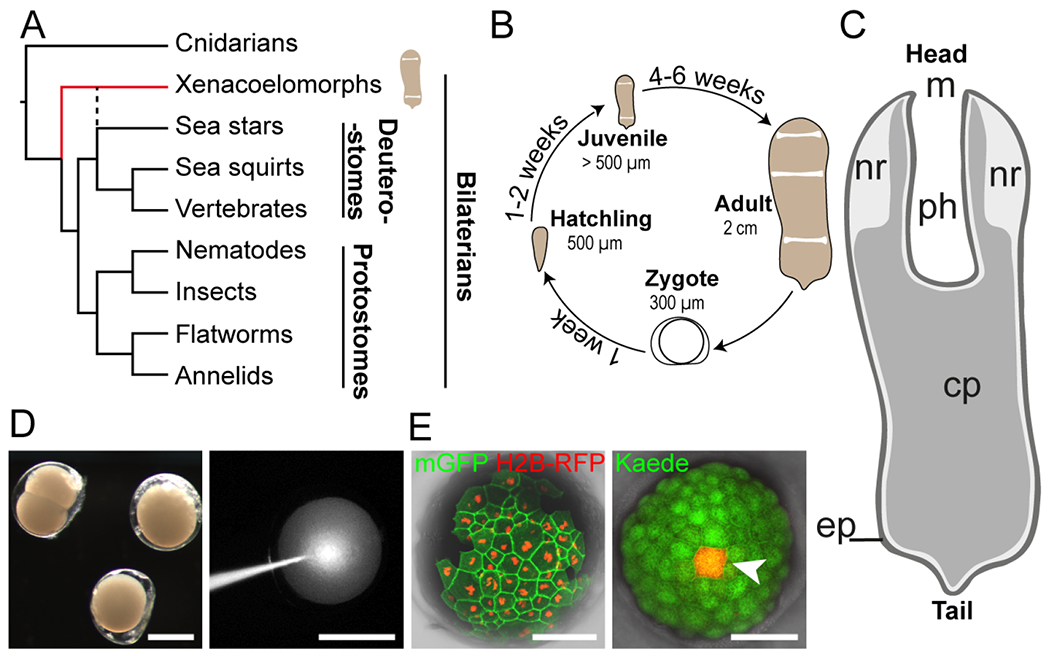
(A) Simplified phylogenetic tree of metazoans showing placement of Hofstenia within xenacoelomorphs (red line). Dashed black line indicates the alternative position of xenacoelomorphs, as sister group to ambulacrarians (e.g., sea stars); (Philippe et al., 2019; Kapli and Telford, 2020; Kapli et al., 2021). (B) Life cycle of Hofstenia miamia in lab conditions. (C) An adult worm schematic drawing showing major structures: m, mouth; nr, neural condensation (Hulett, Potter and Srivastava, 2020); ph, pharynx; cp, central parenchyma; ep, epidermis. (D) Hofstenia embryos are amenable to microinjections. Left, live embryos at 2-cell and zygote stages; right, injection of fluorescent dextran into a zygote. Scale bars, 200 μm. (E) Translation of foreign mRNAs. Left, detection of nuclear-localized H2B-RFP and membrane-bound GFP (mGFP), 1.5 days after single blastomere injection of a 2-cell stage embryo; right, single cell photoconversion of Kaede (white arrowhead), 2 days after injection of Kaede mRNA into a zygote. Scale bars, 100 μm.
Design
With this study, we aimed to generate a toolkit that would enable in depth study of cell/molecular mechanisms in a bilaterian species capable of extensive regeneration. Furthermore, because Hofstenia is now a transgenically-enabled xenacoelomorph species, these tools would facilitate access to the biology of a major yet understudied animal lineage. To achieve these objectives, we first confirmed that Hofstenia embryonic cells were able to translate exogenous mRNAs coding for fluorescent proteins. Then, taking advantage of the recently sequenced genome of Hofstenia, we cloned the promoter regions of various genes, including markers of muscle (troponin), digestive cells (prosaposin), and anterior epidermis (a Hofstenia-specific gene we refer to as epiA), in a backbone designed for meganuclease-assisted transgenesis (Renfer et al., 2010). We generated multiple, stable transgenic lines, with strong, tissue-specific fluorescence. The resulting strains revealed, at high resolution, the morpho-anatomy of putatively ectodermal, mesodermal, and endodermal cell types in Hofstenia. Additionally, these animals were amenable to studies of regeneration, to efficient fluorescence activated cell sorting (FACS), and to photoconversion-based assays. Overall, our work positions Hofstenia as a powerful system for in vivo studies, studies of organogenesis, regeneration, and embryogenesis.
Results
Embryonic microinjection and transgenesis in Hofstenia
We first sought to deliver nucleic acids into embryonic cells in Hofstenia. The life cycle of Hofstenia is completed within two months, under ideal laboratory conditions (Figure 1B, see animal culture in methods), and adults produce an average of four embryos a day that hatch within 8-9 days as juvenile worms, resembling the overall morphology of the adults (Figure 1C) (Kimura et al., 2021). Embryos are deposited on the substrate as zygotes, with each embryo enveloped by a transparent egg “shell”. We found that this tough outer covering of the embryo can be penetrated by a sharp quartz microinjection needle (Figure 1D, Table S1). As a first step toward transgenesis, we assessed the ability of Hofstenia cells to receive and effectively translate foreign mRNA molecules. We injected single blastomeres at early embryonic stages (2- and 4-cell embryos) with in vitro synthesized mRNAs encoding fluorescent proteins (FPs) with a range of green and red emission spectra. Whereas mCherry and mRuby3 mRNA injections resulted in faint and rapidly extinguished fluorescence, eGFP, Kaede, mRFP, and TagRFP-T offered optimal brightness and stability. We found that subcellular localization signals added to the injected mRNA (nuclear and plasma membrane) targeted the FPs to the appropriate cellular compartments. Green fluorescent protein Kaede could be efficiently converted, at single cell resolution, to emanate red fluorescence in Hofstenia embryos injected with Kaede-encoding mRNA (Figure 1E). Furthermore, we found that the Hofstenia genome could be edited specifically via CRISPR-Cas9 upon injection of Cas9 enzyme and guide RNAs (Figure S1). Given the low efficiency of CRISPR-based knock-ins reported in other systems, we next focused on meganuclease-based approaches to generate transgenic fluorescent reporter lines in order to label specific cell types in Hofstenia.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-TagRFP | Invitrogen | Cat#R10367; RRID, AB_10563941 |
| Hofstenia specific Ab anti-Tropomyosin | GenScript | N/A (Available from the Lead Contact upon request) |
| Bacterial and virus strains | ||
| Biological samples | N/A | |
| Chemicals, peptides, and recombinant proteins | ||
| Hoescht 33342 | Invitrogen | Cat#H3570 |
| Instant Ocean sea salt | Instant Ocean | SS15-10 |
| Dextran, Fluorescein, 10,000 MW | Invitrogen | Cat#D1820 |
| Agarose SeaKem® GTG™ | Lonza | Cat#50070 |
| 1X SceI Buffer | Benchmade | Cat#15140122 |
| Tricaine (MS222) | Sigma-Aaldrich | Cat#E10521 |
| DAPI | EMD Millipore | Cat#268298 |
| I-SceI enzyme | NEB | Cat#R0694S |
| Critical commercial assays | ||
| NucleoSpin® Gel and PCR Clean-Up kit | Mascherey-Nagel | Cat#740609 |
| Deposited data | ||
| Gene Sequences | NCBI GenBank |
OK087533 (epiA) OK087534 (psap) OK087535 (tnn) |
| Experimental models: Cell lines | N/A | |
| Experimental models: Organisms/strains | ||
| Hofstenia miamia | The animals used in this study are derived from random matings of the progeny of the original 120 worms collected in Bermuda in 2010. | N/A (Available from the Lead Contact upon request) |
| Oligonucleotides | N/A | |
| Recombinant DNA | ||
| Plasmid tnn::TagRFP-T | Generated for this paper; deposited to Addgene | Addgene ID: 176425 |
| Plasmid tnn::Kaede | Generated for this paper; deposited to Addgene | Addgene ID: 176423 |
| Plasmid psap::Kaede | Generated for this paper; deposited to Addgene | Addgene ID: 176427 |
| Plasmid psap::TagRFP-T | Generated for this paper; deposited to Addgene | Addgene ID: 176428 |
| Plasmid epiATagRFP-T | Generated for this paper; deposited to Addgene | Addgene ID: 176430 |
| Software and algorithms | N/A | |
| Other | ||
| quartz needles | Sutter Instrument | Cat#QF100-70-10 |
| Nunc™ bottom glass dishes | Thermo Scientific™ | Cat#150682 |
Establishment of stable transgenic lines
We identified genes encoded in the Hofstenia genome that, 1) either based on homology or on known fluorescent in situ hybridization (FISH) data were likely to show cell type specific expression, and 2) were expressed at high levels in transcriptome sequencing data and in FISH experiments. This yielded three genes as candidates for generating transgenic Hofstenia: a Hofstenia-specific gene expressed in epidermal cells in the anterior (epidermis anterior, epiA), a homolog of prosaposin expressed in the gut (prosaposin, psap), and a homolog of troponin expressed in muscle (troponin, tnn) (Figure S2). Given that epidermis, gut, and muscle are usually derived from ectodermal, endodermal, and mesodermal tissues, respectively, in bilaterians, these genes are putative markers of these three tissue types in Hofstenia. We cloned regulatory regions of the candidate genes into a backbone plasmid with restriction sites for the I-SceI endonuclease flanking the transgenic cassette (Figure 2A), a system that has been used successfully in multiple systems including cnidarians, protostomes, and vertebrates (Thermes et al., 2002; Deschet et al., 2003; Ogino et al., 2006; Renfer et al., 2010; Minor et al., 2019).
Figure 2: Transgene expression reveals structure and cellular anatomy of epidermal, digestive, and muscle tissues.
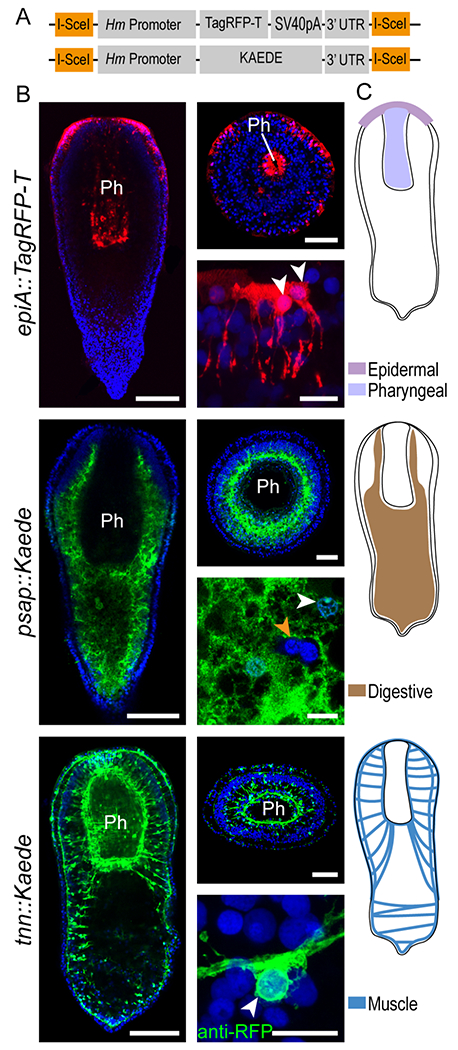
(A) Schematic of the two transgenic cassettes used in this study. Orange boxes indicate a recognition site for the I-SceI Meganuclease enzyme. (B) Expression of fluorescent reporter genes under control of tissue-specific Hofstenia promoters (name of the transgene indicated on the left) in fixed, juvenile worms. Left column: whole worm, mid-body confocal section, as observed in G1 and G2 animals. Right column, top: cross section, at the pharynx level; bottom: high magnification images of transgene-expressing cells. DAPI (nuclei, blue); Kaede (green); TagRFP-T (red); except for tnn::Kaede right bottom picture, anti-RFP : immunodetection of TagRFP-T. White arrowheads point to nuclei of transgene-expressing cells; orange arrowhead indicate nuclei of cells embedded in digestive tissue and not expressing the transgene. Scale bars: whole animal, 100 μm; cross section, 50 μm; close-up, 10 μm. (C) Schematics depicting the transgene expression as observed in G1 and G2 generations, ph, Pharynx. Images are representative of at least ten observed samples. Images and sketches of whole worms are all oriented anterior side up. See also figures S2, S4, S5 and video S1.
After determining the range of DNA concentrations offering high transgene expression and low lethality (Table 1), we co-injected large batches of Hofstenia embryos at the zygote and 2-cell stages with the I-SceI enzyme and plasmid DNA at the optimal concentration (10-25 ng/μl) (for a troubleshooting guide for microinjections in Hofstenia, refer to Table S1). Next, we screened embryos regularly to detect fluorescence. We found that around 40% of the embryos exhibited fluorescence in at least one cell during development. Upon hatching, G0 worms injected with and expressing the tnn or the epiA constructs showed mosaic expression of the transgene. Notably, embryos injected with the psap construct hatched into worms with their entire digestive tissue labeled (Figure S2), consistent with the presence of a syncytial digestive system, which has been reported for some acoels (Smith, 1981; Gavilán et al., 2019). Shortly after hatching, we sorted G0 animals based on the presence of transgene expression and the intensity of fluorescence: only those exhibiting strong fluorescence and the largest area of expression were raised to adulthood and screened regularly for continued transgene expression. Putative adult transgenic worms were then mated to wild type animals or to each other, when possible. Screening of fluorescence patterns in G1 animals suggests that at least 1% (and up to 3%) of the injected G0 animals experienced stable integration of the transgene into the genome with transmission through the germline (Table 1). Additionally, we observed obvious, discrete levels of fluorescence intensity within the G1 progeny of G0 crossed to wild type animals, suggesting multiple insertion events. G1 transgenic worms were crossed together to obtain G2 animals, which tended to show higher levels of fluorescence, likely due to a higher number of transgene copies, and/or due to homozygous transgenic loci. For this study, we focused on three transgenic lines: epiA::TagRFP-T, tnn::Kaede and psap::Kaede. Additionally, we generated tnn::TagRFP-T and psap::TagRFP-T lines to facilitate imaging in fixed specimens using an anti-TagRFP antibody.
Table 1: Efficiency of transgenesis.
The survival rate for DNA concentration ranges superior to 25 % was calculated from various batches of at least 50 injected embryos each.
| Injected DNA concentration (ng/μl) | Survival Rate (%) | GO expression (%) | Germline transmission (%) |
|---|---|---|---|
| 10-25 | 90* | 36-57 | 1-3* |
| 40-50 | 50-60 | ND | ND |
| 75-100 | 10-20 | ND | ND |
| >100 | 5-10 | ND | ND |
Calculated from 2 different batches, including zygotes and 2-cell stage injected embryos. (n = 3/97 and n = 2/182).
Correspondence of transgene fluorescence and mRNA expression patterns
Overall, transgene expression driven by the promoters of the marker genes recapitulated the corresponding mRNA expression detected via FISH. In the case of tnn and psap lines, the transgene expression patterns in G1 animals (and subsequent generations) matched FISH results perfectly (Figure 2B, 2C, Figure S2), with the entire muscle and digestive compartments being labelled, respectively, at all life stages. In contrast, the epiA expression pattern varied across individuals, both in transgenic worms and in FISH experiments. Although an overall anterior-posterior gradient was observed in juvenile worms in the epidermis in addition to expression in the pharynx, the number and the size of labelled patches of cells were highly variable between animals in both FISH and transgenic animals (Figure 2B, 2C, Figure S2). As animals grew and matured, epiA mRNA continued to be expressed in the epidermis, but became increasingly restricted in the anterior and was detectable, by FISH, in adult worms in a ring of cells located peripherally to the mouth and in two ventrolateral lines (Figure S2). Additionally, during growth, pharyngeal expression of epiA was progressively lost, as a new region of expression appeared ventrally, in cells forming the gonopore, in both transgenic animals and FISH experiments (Figure S2, Figure S3B). In adult transgenic worms, the epiA expression pattern was similar to that detected via FISH, except for the absence of the two ventrolateral lines (Figure S3B).
Organ structure and cellular anatomy revealed by transgenic Hofstenia
We used high resolution imaging to understand the structure of epidermal, gut, and muscle tissues. In addition to studying fixed transgenic worms of G1 and G2 generations, we also utilized G0 animals, where mosaic labeling enabled us to visualize single cells.
Classically, epidermis in invertebrates forms an epithelial tissue, with a flat basal surface. Hofstenia EpiA+ cells instead exhibited a large number of thin processes (approximately 10 μm in length and 0.5 μm in thickness), protruding at the basal end of the tissue and projecting internally (Figure 2B, Figure S2B, Supp. Video 1). These observations are consistent with electron microscopy studies of epithelial cell morphology in another acoel species, Convoluta pulchra (Tyler and Rieger, 1999). The apical surface of Hofstenia EpiA+ cells showed abundant ciliature and appeared polygonal, consistent with prior reports (Todt and Tyler, 2007) (Figure S2B). In addition, epiA expression was also detected in subsets of cells forming the inner wall of the pharynx (Figure 2B, Figure S2B, S5A). These pharyngeal epiA+ cells appeared to be contiguous with the mouth epidermis and resembled the morphology of the epidermal epiA+ cells in the body wall (Figure 5B, 5E, Figure S5A).
Figure 5: Muscle fibers closely contact epidermal, pharyngeal, and digestive cells.
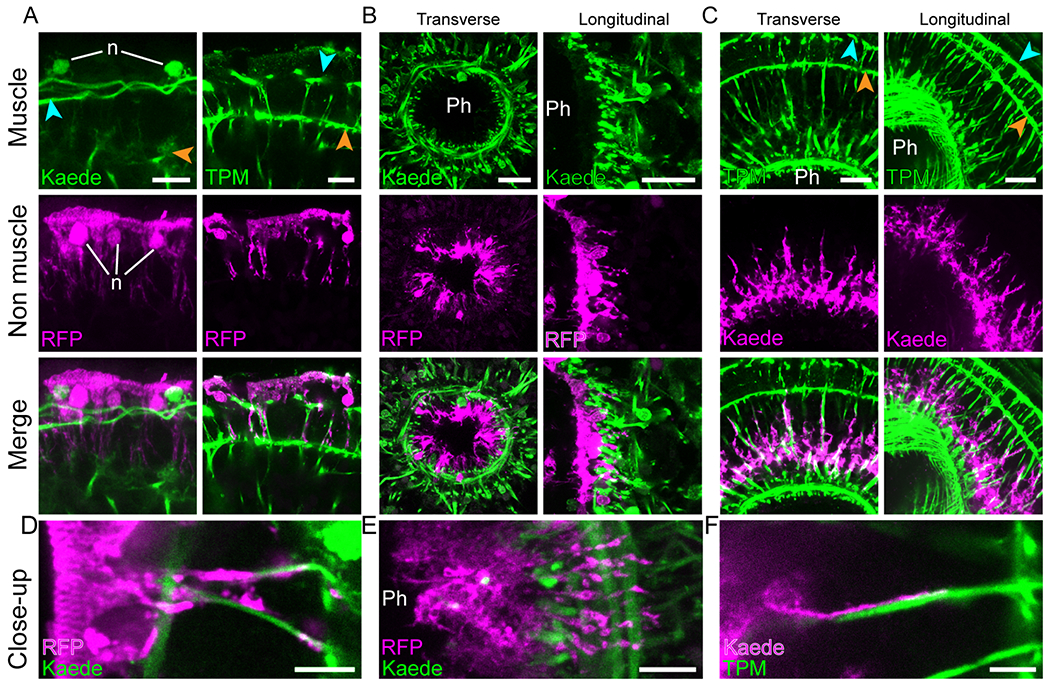
Dual labeling of muscles with epidermal (A, D), pharyngeal (B, E) and digestive (C, F) tissues. (A) Interaction of epidermal cells with peripheral muscle fibers (left) and ramifications of parenchymal muscle fibers (right). (B) Pharyngeal cells projecting ramifications through pharyngeal muscle net. Cross (left) and longitudinal (right) sections of the pharynx. (C) Digestive tissue projecting ramifications along parenchymal muscle fibers as seen in cross (left) and longitudinal (right) sections. (D, E and F) High magnification images showing the close interaction between muscle fibers and epidermal, pharyngeal and digestive cell ramifications, respectively. Blue and orange arrowheads point to peripheral and body wall muscles, respectively. Images are in pseudo-colors with green indicating muscle detected either as Kaede fluorescence or anti-Tropomyosin immunolabeling (TPM) and magenta indicating epiA+ or psap+ cells detected either as Kaede fluorescence or TagRFP-T immunolabeling (RFP); detection method indicated at bottom left corner. Except for TPM, fluorescent labeling originates from transgene expression. Images are representative of at least ten samples. Ph, pharynx lumen; n, nuclei. Scale bars, A, 10 μm; B and C, 20 μm; D, E, F, 5 μm. See also figure S5 and video S7.
Psap::Kaede animals displayed an extensive area of transgene expression along the anterior-posterior axis, from the head (surrounding the statocyst) down to the tip of the tail, in the interior of the worms (Figure 2B, Figure S2C). We also observed that psap− cells were present within the mass of psap+ tissue (Figure 2B, Figure S2C), and, additionally, that large portions of psap+ tissue were going through the pharyngeal sphincter to the pharynx lumen in fixed specimens, suggesting that Hofstenia is able to partially evaginate its digestive tissue (Figure S2C).
Tnn transgenic lines revealed both global muscular architecture and the anatomy of individual muscle fibers at single cell resolution in Hofstenia. Individual muscle cells, regardless of their directionality, displayed overall similar morphology: an elongated main fiber displaying terminal ramifications, with a small cell body located in a slightly shifted position relatively to the axis of the fiber, consistent with observations based on FISH (Raz et al., 2017) (Figure 2B, Figure S4A to S4E).
We elaborate further on the properties of epidermal, digestive, and muscle tissue, their interactions, and the utility of the corresponding transgenic lines in the following sections.
A toolkit enabled by transgenic animals
We evaluated the amenability of our various transgenic lines to new approaches for future research. First, we aimed to sort and isolate live cells from G2 tnn transgenic animals, via fluorescence activated cell sorting (FACS). We developed a protocol to identify and isolate Kaede+ cells from a single adult tnn::Kaede worm (Figure 3A). We found that the green fluorescence observed was specific to transgenic animals as there was very low background fluorescence in wild type worms. We tested the specificity of the cell isolation procedure by applying a custom anti-Tropomyosin (TPM) antibody (Hulett et al., 2020) to our cell suspension. We observed that the Kaede-gated cells exhibited both green fluorescence and reactivity to anti-TPM, indicating successful and specific isolation of muscle cells (Figure 3B, Figure S3A). Satisfyingly, only a small fraction of the control Kaede-gated cells showed reactivity to the antibody, and none of them exhibited green fluorescence.
Figure 3: Transgenesis in Hofstenia enables a vast array of experimental tools for studying regeneration and stem cell biology.
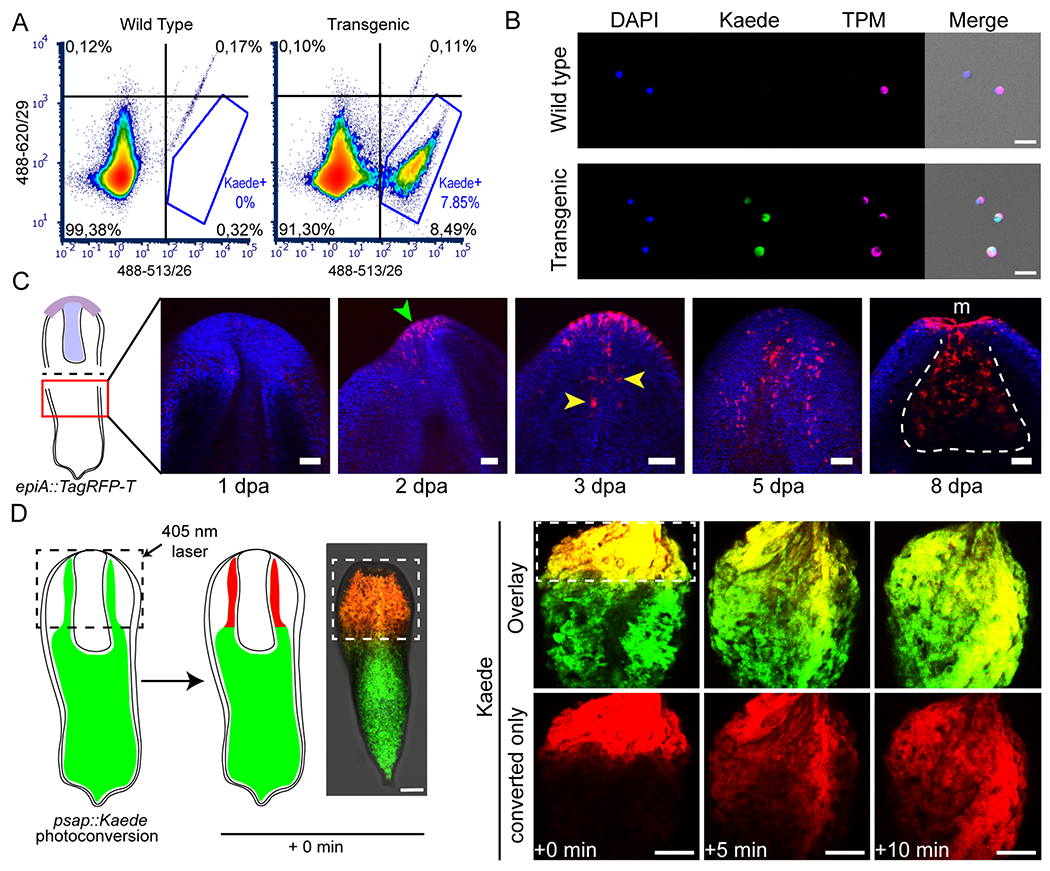
(A, B) Isolation of specific live cell populations using FACS on tnn::Kaede transgenic animals; (A) Fluorescence plots showing cell density for Kaede fluorescence (quadrant and Kaede gate percentages are calculated over a total of 76880 and 87157 singlets for wild type and transgenic samples, respectively); (B) anti-TPM immunocytochemistry on FACS-isolated cells originating from the whole cell (wild type), and the Kaede+ (transgenic) populations. Scale bars, 20 μm. FACS plot is representative of 3 biological replicates; immunocytochemistry was performed on two technical replicates of tnn::Kaede dissociated cells. (C) Regeneration of anterior structures following worm bisection (see sketch on the left) in epiA::TagRFP-T transgenics. Regeneration time points are indicated under each image, as days post amputation (dpa). Green and yellow arrowheads point at de novo expression of TagRFP-T in epidermal and pharyngeal cells, respectively. White dotted line delineates the regenerated pharynx, m, mouth. Scale bars, 50 μm. Six biological replicates were used for each time point (see figure S3). (D) Photoconversion of Kaede-expressing cells in live psap::Kaede animals (dotted squares outline the photoconverted area); left, schematic of the experiment; right, quick expansion of the photoconverted Kaede area (red) over time in a live worm (as observed in 3 biological replicates). Time elapsed since photoconversion is indicated at the bottom. Scale bars, 100 μm. See also figures S1 and S3.
Next, to assess the feasibility of studying regeneration using transgenic Hofstenia, we asked if transgenes would be re-expressed in new cells that form during regeneration. We took advantage of the anteriorly-restricted expression pattern of the epiA transgene and performed mid-body bisection of G1 animals to remove all anterior tissues including brain and pharynx. During regeneration, the anterior fragments, which lacked posterior/tail tissues, retained the epiA expression pattern as prior to the bisection (Figure S3B). Posterior fragments, which lacked anterior/head tissues regained transgene expression over time, with expression first observable in the epidermis two days post amputation (Figure 3C, Figure S3B). Although we failed to detect epiA+ pharyngeal cells immediately upon amputation in these fragments, expression of the transgene became visible in the regenerating pharynx three days post amputation.
Finally, we sought to determine if Kaede-expressing animals were amenable to photoconversion. Following embedding of psap::Kaede juveniles in agarose gel, we exposed them to 405 nm wavelength light, in order to regionally photoconvert the digestive tissue. Photoconverted Kaede initially localized in a small, bright-red area, which, immediately after the photoconversion, started to expand while decreasing in brightness (Figure 3D). This result, i.e. the spread of converted red fluorescent protein outside of the area that was targeted for conversion, provides evidence that the Hofstenia gut actually functions as a syncytium. Furthermore, together with our ability to photoconvert Kaede in single cells (Figure 1E), this ability to study live adult worms suggests that photoconversion approaches can be utilized for cell tracking in Hofstenia.
The structure and formation of musculature
The robust fluorescent labeling of muscle in the tnn::Kaede line provides an opportunity to study the formation of muscle cells during development and to determine the anatomy of musculature in detail. First, we fixed tnn::Kaede hatchlings in order to acquire fine details of muscle morphology in Hofstenia. Overall, we found that musculature is organized in four major interconnected compartments (Figure 4A), consistent with previous observations in acoels (Tyler and Rieger, 1999; Todt, 2009). Starting from the outside of the body, we observed : i) an external layer of strictly longitudinal fibers with ramifications at their tips (the peripheral muscles) (Figure 4A, Figure S4A), running from the mouth towards the posterior, but not reaching the tip of the tail (Figure 4A, Figure S4B); ii) a tight network comprising longitudinal, circular and oblique fibers (the body wall muscles), also with ramifications, located beneath the epidermis, all along the A/P axis; iii) a set of highly ramified fibers projecting through the parenchyma (the parenchymal muscles) and connecting body wall muscles to each other or connecting body wall muscle to pharynx muscle (Figure 4A, Figure S4C) and iv) a dense, three-layered, basket-shaped network (Figure 4A, Figure S4D, S4E) of longitudinal and circular fibers delimiting the pharynx (the pharyngeal muscles).
Figure 4: A muscle-specific reporter line reveals a robust network of interconnected muscle systems.
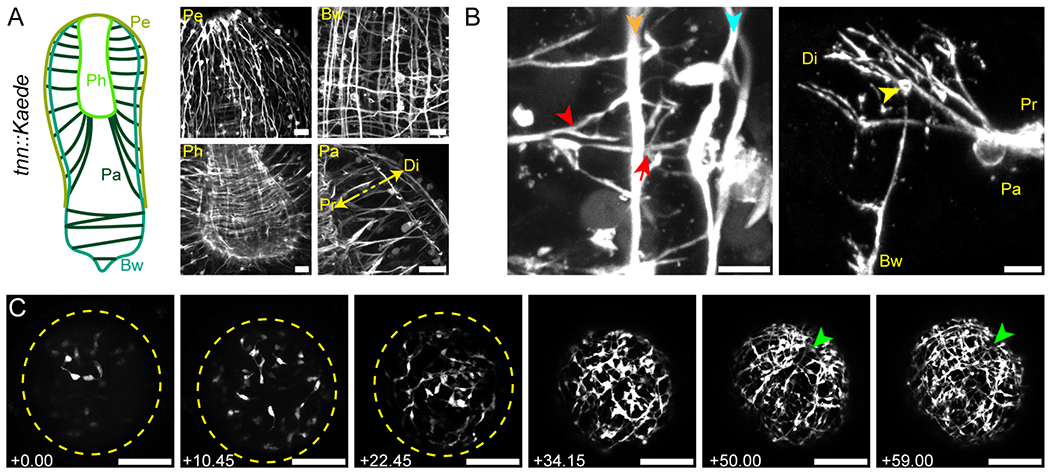
(A) Major muscle structures of Hostenia. Left, schematic showing the overall organization of Hofstenia muscles. Pe, peripheral; Bw, body wall; Pa, parenchymal; and Ph, pharyngeal muscles. Right, close-up images of the different muscle categories. The double, yellow arrow indicates the proximal (Pr) and distal (Di) sides of the parenchymal muscles. Scale bars, 20 μm. (B) Muscle-muscle interactions. Left, proximal (red arrowhead) and distal (red arrow) ramifications of a parenchymal muscle fiber, connecting it to body wall (orange arrowhead) and peripheral (blue arrowhead) muscle fibers, respectively; right, overview of two orthogonal fibers connecting through ramified projections as observed in a mosaic, G0 animal. The yellow arrowhead indicates the thickened extremity of the body wall muscle fiber. Pa, bw, single parenchymal and body wall muscle fibers, respectively, Pr, Di, proximal and distal sides of the parenchymal fiber, respectively. Images are representative of at least ten observed samples. Scale bars, left, 5 μm; right, 10 μm. (C) Birth and organization of the muscular network during a time-lapse movie of embryonic development in a tnn::Kaede G2 embryo. Left to right, Z-stack projections a single embryo, between 4 to 6 days after first cleavage. Time points (left bottom corner, hours and minutes indicated as hh.mm) indicate the time elapsed since the first z-stack. Yellow dashed lines outline the embryo at early stages of tnn expression; green arrowheads indicate the position of the future mouth. Full myogenesis time-lapse imaging was conducted on one embryo as a representative of at least ten embryos observed over time. Scale bars, 100 μm. See also figure S4, and videos S2 to S6.
Closer examination of the parenchymal muscle highlighted the interlocking of muscle in Hofstenia, providing a three-dimensional view in the whole animal of connections that had been detected via electron microscopy in another acoel (Tyler and Rieger, 1999). In the anterior region of the animal, where the pharynx is located, parenchymal muscles contacted pharyngeal muscle proximally (towards the interior of the animal), and body wall and peripheral muscle distally (towards the periphery of the animal) (Figure 4B). The contact between parenchymal muscle and body wall/peripheral muscle was mediated by multiple ramifications of the muscle fiber. As parenchymal muscle fibers approached body wall muscle, they formed ramifications, connecting with the longitudinal body wall muscle. These ramifications became further branched as they approached and made contact with the peripheral muscle. In the posterior, parenchymal muscles connected body wall muscle on opposite sides of the worm via ramifications similar to those observed in the anterior (Figure S4C). Using 3D reconstruction tools, we were able to visualize more clearly how the two kind of fibers, parenchymal, and body wall (longitudinal) interact at their extremities through ramified interdigitations. In particular, 3D rendering of G1/G2 tnn::Kaede animals allowed us to clearly determine the spatial arrangement of the parenchymal, body wall and peripheral muscles (Supp. Videos 2 and 3). Additionally, G0 animals showed that the ramifications of a single parenchymal fiber interdigitate with those of a body wall fiber (Supp. Videos 4 and 5), suggesting that parenchymal fibers do not randomly establish contact, but rather preferentially connect to ramifications at the tips of body wall muscle fibers.
Next, we developed approaches to live-image transgenic embryos (Figure 4A, Figure S4F). We focused on a subset of G2 transgenic worms that showed enhanced fluorescence relative to G0 and G1 animals. Tnn::Kaede G1 and G2 embryos started expressing Kaede around 3.5 days post laying, consistent with the timeline of muscle development (Kimura et al., 2021) and allowing in vivo time lapse imaging of the birth of muscle cells and the development of the musculature (Figure 4C, Supp. Video 6). Early Kaede expression was first seen in a small number of fast-moving cells with round cell bodies, from which emanated short and labile plasma membrane projections. These cells increased in number due to rapid cell division (Supp. Video 6). At around 4 days post laying, these cells became more stationary and their projections increased in length, establishing contact with projections from other muscle cells and beginning to resemble contractile muscle fibers. Between 4.5 to 6 days post laying, muscle fibers progressively organized into a regular grid that formed the body wall muscle. Eventually, at around 6 days post laying, with only the body wall muscle visible, embryos started rotating within their capsules, thus, making time-lapse impossible. The structures formed by the peripheral, parenchymal and pharyngeal muscles could only be observed with certainty in later stages of development, suggesting that the body wall muscle network develops first during embryogenesis.
A muscle scaffold for epidermal and digestive cells
The unique morphology of Hofstenia epidermis, with cells that project cytoplasmic processes toward the interior of the worm, the presence of a syncytial gut, and the interlocking organization of its muscular system, raised the question of how these tissues are organized spatially relative to each other. To address this, we took advantage of our transgenic lines and of our custom made anti-TPM antibody and performed dual labeling of muscle with epidermal, pharyngeal, and digestive cells, either by crossing G1 transgenics together, or by immunolabeling of muscles in epiA::TagRFP-T and psap::TagRFP-T transgenic lines.
EpiA::TagRFP-T X tnn::Kaede double transgenic animals (Figure S5A) provided a high resolution view of the interaction of muscle and epidermis, expanding on previous observations of contact between these tissues in another acoel (Tyler and Rieger, 1999). First, we saw that both epidermal cells and peripheral muscles had their cell bodies located in the same plane, slightly distal relative to the plane defined by the peripheral muscle fibers. Second, we found that the epidermal processes emerged starting at the plane containing peripheral muscle fibers and extended proximally till they reached the body wall muscle grid (Figure 5A, D). In the anterior two thirds of the animal, where parenchymal muscles connect to both pharyngeal and body wall muscles, epidermal processes were always associated with the distal-most ramifications of parenchymal muscle fibers, in the narrow space located between body wall and peripheral muscle fibers (Figure 5D). In the posterior region of the worms, which lacks peripheral muscles, epidermal cell processes were not in contact with parenchymal fiber ramifications, but rather contacted the body wall grid alone (Supp. Video 7).
Given their shared morphological features with epidermal cells, we also looked at the interaction of pharyngeal epiA+ cells with pharynx muscles (Figure 5B, E). Although the high density of the pharyngeal muscle network did not allow clear observation of muscle fiber ramifications, we found that pharyngeal epiA+ cells exhibited narrow processes, similar to the epidermal cell processes, projecting through the net of pharyngeal muscles, towards the parenchymal space.
Because Hofstenia parenchymal muscles traverse a space that is occupied with the digestive syncytium, we interrogated the spatial arrangement between these two tissues. TPM immunolabeling in psap::Kaede animals revealed a close association of parenchymal muscle fibers with digestive tissue in both anterior and posterior regions of the animal (Figure 5C, F; Figure S5B). The syncytial gut exhibited on its distal edges thin, radial processes that aligned closely with parenchymal muscle fibers, reaching out to the body wall muscle grid. Together, these observations suggest that musculature, and parenchymal muscle in particular, has a prominent role in providing mechanical support, and potentially information to other tissues.
Discussion
Transgenesis in Hofstenia via random insertion
In this manuscript, we present the establishment of transgenesis in a xenacoelomorph species, the acoel Hofstenia miamia. Injection of a linearized transgenic cassette, comprising a fluorescent reporter gene flanked by promoter and 3’UTR regions of Hofstenia genes (Figure 2A), resulted in the transient expression of the reporter gene in a large proportion of the injected embryos, and, subsequently in the stable integration of the transgene into the genome. With a rate of germline transmission of the transgene ranging between 1-3% of the injected animals (Table 1), we report that non-targeted transgenesis in Hofstenia has similar success rate to other invertebrate research organisms (Renfer et al., 2010; Backfisch et al., 2013; Wudarski et al., 2017). In particular, given the length of Hofstenia life cycle (Figure 1B) we demonstrate the ability to generate stable G2 transgenic lines within 6 months.
The activity of the transgene promoters recapitulated expression driven by their wild type counterparts, showing a high level of specificity of transgenesis in Hofstenia. The absence of epiA::TagRFP-T expression in the ventrolateral lines of adult worms (Figure S2D,S3B) represents the sole discrepancy detected between the transgenic and the wild type promoters. It is possible that the promoter we cloned did not include the regulatory elements needed for expression in this domain. We also demonstrate that Hofstenia transgenics are suitable for FACS-mediated isolation of specific live cell populations (Figure 3A), prolonged live imaging of embryonic development (Figure 4C, Figure S4F) and monitoring of the regeneration process (Figure 3B). Furthermore, the use of Kaede as a reporter gene allowed us to differentially label a small region of tissue and trace the photoconverted protein (Figure 3C). Together, our results show the significant potential of transgenic Hofstenia for the study of acoel biology, and particularly for in vivo and live imaging applications.
Cell biology in muscle and epidermis
The function of muscle cells in regeneration has recently been emphasized in planarians (Witchley et al., 2013; Scimone et al., 2017; Scimone et al., 2020), where they are hypothesized to guide progenitor cells to their correct location, likely by establishing a chemical coordinate system, through release of signaling molecules (Position Control Genes, PCGs; e.g., Wnt and TGF-beta ligands). Quite remarkably, despite the ancient divergence of acoels and planarians (550 mya), Hofstenia body wall muscles seem to share, at least partially, this instructive role as they express homologs of PCGs that are required for patterning of tissue during regeneration, and, as in planarians, PCG expression is triggered in muscle by regeneration (Raz et al., 2017; Ramirez et al., 2020). Our results show a high degree of interconnection between the different muscle compartments (Figure 4). In particular, parenchymal muscle appears central to global muscle organization, as it connects, pharyngeal muscle to body wall and peripheral muscle, making Hofstenia muscle an undisrupted network. Therefore, we suggest that parenchymal muscle could participate in tissue renewal and/or regeneration 1) by facilitating the role of body wall muscles in those processes, e.g., by delivering positional and differentiation signals, or, 2) by providing physical support for stem cell homing and progenitor cell migration along the parenchymal fiber axis.
Transgenically-labeled epidermal cells revealed a unique morphology for this tissue in Hofstenia, with cytoplasmic processes projecting toward the interior of the animal. Parenchymal muscle fibers made close contacts with these epidermal processes, as well as with pharyngeal and digestive processes, suggesting that the muscular system provides mechanical support for these tissues (Figure 5–6). In this regard, it would be interesting to investigate further the unique anatomy of Hofstenia and the role of its muscles, in particular by determining if and how muscle is associated with other tissues or cell types (e.g., stem cells, neurons, gland cells), and if, like planarians, acoel muscles are a source of extracellular matrix components (Cote et al., 2019).
Figure 6: Anatomical model for Hofstenia miamia.
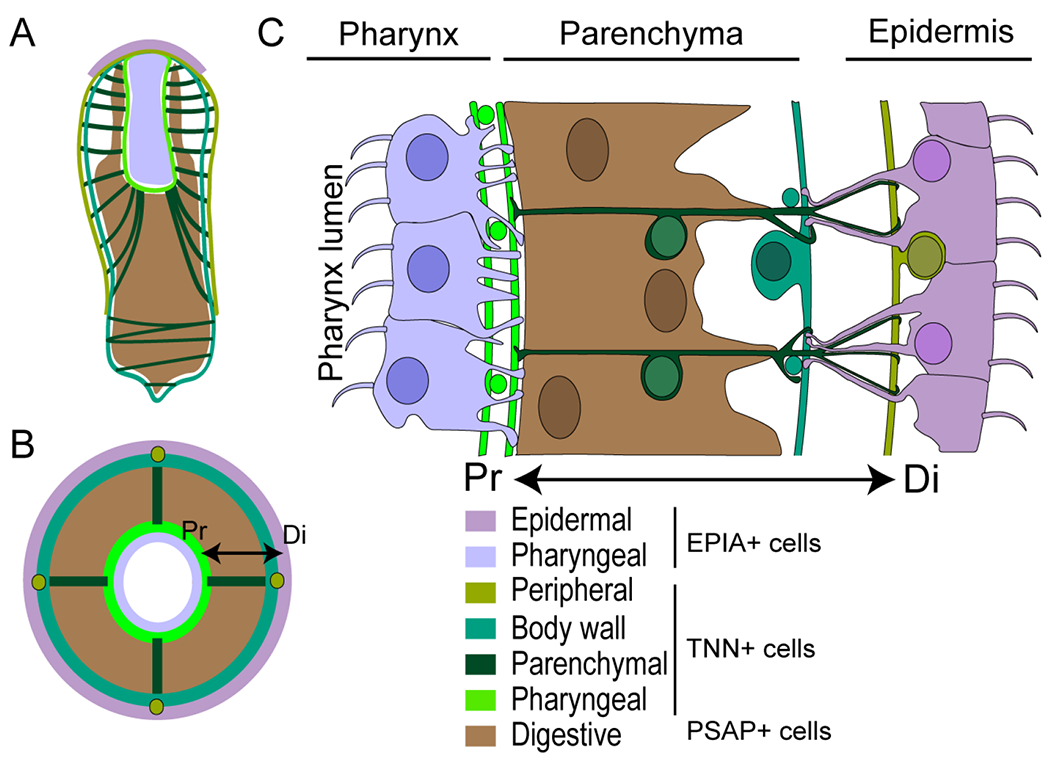
Sketches of Hofstenia juvenile along a coronal (A) and a cross (B) section depicting the arrangement of epidermis, gut and muscles. (C) Drawing showing the detail of the cell type distribution as observed in a longitudinal section at the pharynx level. The double arrow indicates the proximal (Pr) and distal (Di) ends of the parenchymal muscle.
Prospects
For future mechanistic studies of regeneration and for understanding the diversity of regenerative and developmental mechanisms that emerged across evolution, it will be important to develop additional transgenic tools in Hofstenia. Our finding that CRISPR/Cas9 mediated genome editing is feasible in Hofstenia (Figure S1) will enable targeted and conditional transgenesis. Such tools, in combination with FACS, transcriptome- and genome- wide sequencing, and other techniques will decipher how injury differentially affects the biology and behavior of certain cell populations in time and space. For instance, the mediation of the immediate local wound response could be studying by testing regeneration specific elements, such as putative enhancers identified by ATAC-seq, using simple fluorescent reporter lines. In parallel, the production of stem cell (neoblast) specific reporter lines should reveal the downstream effects of injury, ranging from cell differentiation, cell proliferation, to organogenesis. Altogether, our results show that Hofstenia will be a great system for insights into animal evolution and regeneration.
Limitations of the study
In the course of this work, we identified two main challenges for future use of our methods to make transgenic Hofstenia. First, we noted that some genomic regions were resistant to PCR amplification, likely due to some regions of the genome having low GC content (around 30%) and/or numerous “AT” repeats. This can be addressed by paying particular attention to the DNA sequence being targeted for use in a transgenesis construct. Second, egg-release cannot be artificially induced in Hofstenia. Instead, adult Hofstenia lay fertilized embryos randomly, throughout the day. Therefore, before each session of injection, embryos have to be collected manually from multiple culture tanks, then sorted, in order to identify the earliest stages.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mansi Srivastava (mansi@oeb.harvard.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement. Plasmids with transgene constructs generated in this study are available from Addgene.
Data and code availability
Gene sequences in this study are deposited to GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The laboratory population of H. miamia adults represent many generations derived from 120 sexually reproducing worms collected from Bermuda in 2010 (Srivastava et al., 2014). The embryos used in this study were the progeny of random matings of these worms, Adult Hofstenia were cultured at 21°C in plastic boxes containing 1 Liter of filtered artificial sea water with 37 parts per thousand Instant Ocean sea salt (AFSW). Twice a week, boxes were cleaned, AFSW renewed, and animals were fed freshly-hatched artemia. Embryos were laid spontaneously, in clutches, on the plastic substrate.
METHOD DETAILS
Identification and cloning of regulatory elements of Hofstenia genes
Full length transcript sequences of troponin, psap and epiA genes were identified in the Hofstenia transcriptome and the corresponding genomic loci were determined via BLAST (Gehrke et al., 2019). Primers were designed to amplify regions located 5’ to the ATG and 3’ to the STOP codon from genomic DNA (Genbank Genome accession SCFE00000000) (Supp. Table S2). The plasmid from Renfer et al. (Renfer et al., 2010) was used as the backbone for transgenesis. After digestion, Hofstenia promoters were directly cloned into the backbone. In our Kaede lines, 3’UTR regions were directly cloned into vectors already containing Hofstenia promoters and Kaede CDS. In our TagRFP-T lines, 3’UTRs were combined to the TagRFP-T CDS using the Gibson assembly Master Mix (Cat#E2611, New England BioLabs Inc). The resulting fragment was then amplified via PCR and digested prior ligation into a vector containing only Hofstenia promoter region. Following cloning, plasmid isolation and sequencing were performed to assess proper fragment insertion.
mRNA injection solution
The coding sequences for various FPs, including a T7 or SP6 promoter sequence in 5’ where obtained from various plasmids (including commercial plasmids, in Lab plasmids, and gifts from other labs). mRNA synthesis and capping was performed with 1 μg of purified, linearized plasmid, using the mMessage mMachine® kit (Cat#AM1344 and AM1340 Thermofisher Scientific), and followed by the addition of a poly(A) tail, with the Poly(A) Tailing Kit (Cat# AM1350, Invitrogen™). Next, capped and tailed mRNA where purified following the ammonium acetate/phenol-chloform method provided by the mMessage mMachine® kit. Following purification, RNA were resuspended in 4 to 5 μl of nuclease free water, quantified with a Nanodrop ND-1000 spectrophotometer, and stored in 1 μl aliquots, at −80 °C. Prior to injection, RNA tubes were thawed on ice, then diluted with nuclease free water and Fluorescein Dextran (Cat#D1820, Invitrogen, final concentration : 1.25 μg/μl) at a final concentration ranging from 500 ng/μl to 3 μg/ μl. A quartz needle (Cat#QF100-70-10, Sutter Instrument) was back-loaded with 0.75 μl of the injection solution and manipulated with Narishige MMN1 and MO202U micro-manipulators. Injections were performed under a Leica MZ10F Stereomicroscope, using a Pico-liter Microinjector (Cat# PLI-90A, Warner Instruments).
DNA injection solution
Prior to injection, 4 μg of the desired plasmid was fully digested with 4 μl of I-SceI enzyme (Cat#R0694S), in a 50 μl reaction with 1X Cutsmart buffer, left 1 to 2 hrs at 37 °C, followed by 20 minutes of heat inactivation at 65 °C. The digested plasmid was loaded in a 0.8% agarose SeaKem® GTG™ (Cat#50070, Lonza) gel. The fragment corresponding to the transgenic cassette was cut from the gel, purified with the NucleoSpin® Gel and PCR Clean-Up kit (Cat#740609, Mascherey-Nagel), eluted in 15-30 μl of elution buffer and quantified with a NanodropND-1000 spectrophotometer. The injection solution was prepared in a volume of 10 μl, by assembling the following reagents in nuclease-free water, in order to obtain the given final concentration: digested DNA (10-25 ng/μl); Fluorescein Dextran (D1820, Invitrogen; 1.25 μg/μl); 1X I-SceI Buffer (Renfer et al., 2010); I-SceI enzyme (Cat#R0694S, NEB; 0.375 U/μl). Injections were performed with the same method and same apparatus as for mRNA.
Embryo collection and microinjection dish
Embryos were collected with a glass pipette from culture boxes, then sorted under a stereomicroscope, in order to keep only the desired developmental stages (mainly zygotes and 2-cell stage embryos). Prior to each injection session, an injection dish was prepared using a plastic mold covered with square-base pins (side and length around 300 μm). The mold was deposited into a plastic culture dish (60 X 15 mm) half filled with melted agarose (1.2 % in MilliQ H2O) and left until full solidification, then gently removed, thus generating individual wells for the embryos. The injection dish was filled with AFSW and sorted embryos were transferred to the injection dish, then gently disposed into the wells for injection (one embryo per well). Following injection, embryos were collected from the wells with a glass pipette, transferred into a clean plastic dish, containing AFSW with Pennicillin and Streptavidin antibiotics (Cat#15140122, Thermofisher Scientific) at 5U/ml and placed in a 23 °C incubator. AFSW was only replaced 5 days after the injection.
Transgenic embryo and animal care
Shortly prior hatching, embryos were screened for transgene expression by observing fluorescence with a Leica MZ10F stereomicroscope. Embryos exhibiting transgene expression were placed into individual wells within 24-well plates. After hatching, juveniles were transferred into new plates, with freshly made AFSW. From that point on until they reached adulthood, transgenic worms were transferred to clean plates and AFSW (without antibiotics), and fed (L-type Brachionus rotifers) twice a week. Over the course of their growth, animals were screened regularly to monitor transgene expression. Eventually, worms that were more likely to have integrated the transgene (based on brightness and area of expression) were placed into 6-well plates (each filled with 8 ml of AFSW), also with bi-weekly water renewals and feedings. For G0 (as well as for the following generations) animals, sexually mature worms were crossed with wild type animals or with other transgenic worms. Their progeny was screened and taken care of as described above.
Immunohistochemistry (IHC), FISH and imaging
Animals were relaxed for five minutes in AFSW containing 0.5 mg/ml of Tricaine and with adjusted pH (8-8.5). This solution was then replaced by the fixation solution: 4% Paraformaldehyde (PFA) in AFSW, with 0.05% Triton X-100. Animals were fixed either 3hrs at room temperature (RT) or overnight (O/N) at 4°C, with constant agitation. Following fixation, FISH procedure was performed as described previously (Srivastava et al., 2014). For IHC, fixed worms were briefly washed three times in 1X Phosphate Buffer Saline with 0.1% Triton X-100 (PBSTx), then blocked for 2 hrs at RT in PBSTx with 1% Bovine Serum Albumin (BSA). Primary antibody incubation was performed in renewed blocking solution, at 4°C, during 48-72 hrs, with constant agitation. Long washes in PBSTx were repeated 5-6 times along the day, before incubation (48-72 hrs) with secondary antibodies, in PBSTx. Samples were then washed extensively in PBSTx, and counterstained with 4’,6-diamidino-2-phenylindole (DAPI), (268298, EMD Millipore), at 2 μg/ml, for 2 hrs, at RT. Primary antibodies used: anti-TagRFP (Cat#R10367, Invitrogen) at the dilution of 1:1000, custom made anti-HmTropomyosin at the concentration of 1 μg/ml. Secondary antibodies used : Goat anti-rabbit, Alexa Fluor® 568 (Cat#ab175471, Abcam); Goat anti-rabbit, Alexa Fluor® 488 (Cat#ab150077, Abcam). Imaging was performed with a Leica SP8 confocal microscope. Post-acquisition treatment of confocal images was performed with Fiji; in particular, 3D rendering images were made with the 3D Viewer plugin. For primer information, refer to the supplementary Table S2.
Regeneration assays
One month old G2 epiA::TagRFP-T transgenic worms were relaxed in AFSW with 0.5 mg/ml Tricaine, then bisected with a razor blade at the level of the middle stripe. Following amputation, fragments were rinsed 3 times in AFSW without Tricaine, then placed into AFSW within 12-well plates, and left to regenerate at 23°C in an incubator. AFSW was changed daily until fixation in 4% PFA for imaging.
FACS assay and Immunocytochemistry (ICC)
Cell dissociation of transgenic and wild type worms was done by placing one adult worm in a 2ml tube, replacing AFSW with calcium- and magnesium-free AFSW (CMFSW) with 1% horse serum, and pipetting until full dissociation. Dissociated cells were filtered using a 40 μm cell strainer into a clean 2ml tube, then cleaned from debris by centrifugation (5 min., 500 g, at 4 °C) over a 4% BSA cushion in CMFSW. The resulting pellet was resuspended into CMFSW with 1% horse serum and placed on ice before labeling with Hoescht 33342 (Cat#H3570, Invitrogen; 10 mg/ml final) and propidium iodide (Cat#P3566, Invitrogen; 1 mg/ml final) for 30 min. Sorting was done using a Beckman Coulter MoFlo Astrios EQ Cell Sorter. For ICC, sorted cells were collected into 2ml tubes with 1 ml of CMFSW and centrifuged (5 min., 500 g, at 4 °C). The pellets were resuspended in 150 μl of CMFSW, then deposited onto lysine-coated coverslips, at the bottom of a 12-well plate (from that point on, all incubations were performed in the dark), and allowed to adhere for 1 hour. Fixation was performed for 20 min. with 4% PFA in CMFSW and followed by 3 quick washes with PBSTx. Samples were blocked with PBSTx + 10% horse serum, then incubated with primary antibody (2 μg/ml) in block, 2 hours at RT. After 3 PBSTx washes, samples were incubated with the secondary antibody for 2 hours, at RT. Finally, samples were washed 3 times with PBSTx, incubated in Hoescht 33342 (1 mg/ml) and mounted on microscopy slides for imaging.
Live imaging of embryos
Embryos were placed into Nunc™ bottom glass dishes (Cat#150682, Thermo Scientific™) containing 4 ml of AFSW and imaged with a Leica SP8 confocal. For time-lapse imaging, Pennicillin and Streptavidin antibiotics (1 Cat#5140122, Thermofisher Scientific) at 5U/ml were added to the water.
CRISPR/CAS9 genome editing
A target site for CAS9 enzyme was identified with Geneious (https://www.geneious.com) in the Hofstenia GRP78 locus. Primers were designed in order to synthesize a single guide RNA (sgRNA) corresponding to this target site (Supp. Table S2), using the MEGAscript™ T7 Transcription Kit (Cat#AM1334, Invitrogen™) (Zhang and Reed, 2016). sgRNA was then purified with phenol-chloroform followed by isopropanol / ammonium acetate precipitation, and resuspended in nuclease free water. Embryos were injected with a mixture of sgRNA at 600 ng/μl, CAS9 enzyme at 1μg/μl (Cat#1074181, Integrated DNA Technologies) and fluorescein dextran, then left to develop until pre-hatchling stage. Genomic DNA from individual embryos was isolated with the NucleoSpin® Tissue XS kit (Cat#740901, Macherey-Nagel). A PCR reaction was performed using the isolated genomic DNA with primers flanking the CAS9 target site (Supp. Table S2). The T7E1 assay (Cat#M0302, NEB) was then applied as in (Sato et al., 2018) to the PCR product (half of the product being treated with T7 endonuclease, in the other half, T7E1 was replaced by water) which was run on an agarose gel to assess for multiple band detection. Control bands were then extracted from the gel and sequenced in order to assess for the presence of indels at the target site.
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical analyses were utilized in this study. Sample sizes reflect the number of animals observed in each experiment.
Supplementary Material
Supp. Video 1: Morphology of epidermal cells, related to Figure 2. 360° rotation of a 3D image, reconstructed from confocal z-stack, showing a patch of epiA::TagRFP-T expressing cells in a G0 fixed juvenile. White arrowhead points to cilia, on the apical side; white arrow points to the epidermal processes.
Supp. Video 2: Interlocking of parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the X axis, of a 3D image, reconstructed from confocal z-stack of a G2 tnn::Kaede juvenile worm. Colored arrows point to the different muscle categories. PA, PE, BW, parenchymal, peripheral and body wall muscles, respectively.
Supp. Video 3: Interlocking of parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the Y axis, of a 3D image, reconstructed from confocal z-stack of a G2 tnn::Kaede juvenile worm. Colored arrows point to the different muscle categories. PA, PE, BW, parenchymal, peripheral and body wall muscles, respectively.
Supp. Video 4: Interlocking of single parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the X axis, of a 3D image, reconstructed from confocal z-stack of a G0 tnn::TagRFP-T juvenile worm. PA, BW, parenchymal, and body wall muscles, respectively.
Supp. Video 5: Interlocking of single parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the Y axis, of a 3D image, reconstructed from confocal z-stack of a G0 tnn::TagRFP-T juvenile worm. PA, BW, parenchymal, and body wall muscles, respectively.
Supp. Video 6: Emergence of the musculature during embryogenesis, related to Figure 4. Time-lapse imaging of a G2 tnn::Kaede embryo over 63hrs of development, from 3.5 to 6 days post laying. Time stamp, hh::mm::ss.
Supp. Video 7: Interaction of posterior epidermis with body wall muscle, related to Figure 5. 360° rotation of a 3D image, reconstructed from confocal z-stack, showing a patch of epiA::TagRFP-T (red) expressing cells over the body wall muscles, revealed by anti-TPM IHC (green) in a G1 epiA::TagRFP-T juvenile worm. Nuclei are counterstained with DAPI (blue).
Highlights.
Stable transgenesis was achieved in the xenacoelomorph Hofstenia miamia
Transgenic worms enable many tools for studying regeneration and development
Live imaging revealed formation of muscle during embryogenesis
Muscle serves as a scaffold for epidermal and gut tissues
Acknowledgements
We thank Dr. Mark Martindale for guidance on embryonic microinjections and Dr. Seth Donoughe for help with making injection molds. We also thank Vanessa Poirier and Hafsa Sadiq for substantial help with plasmids and transgenic animal care, Dr. Andrew Gehrke for help with CRISPR experiments, and Kate Sheridan and other members of the Srivastava Lab for help with embryo collection. L.R. and M.S. were supported by grants awarded to M.S. by the Searle Scholars Program (SSP-2016-1494), the Richard and Susan Smith Family Foundation, and the National Institutes of Health (1R35GM128817).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare that they do not have any competing interests.
References
- Achatz JG, Chiodin M, Salvenmoser W, Tyler S and Martinez P (2013) “The Acoela: on their kind and kinships, especially with nemertodermatids and xenoturbellids (Bilateria incertae sedis),” Organisms, diversity & evolution, 13(2), pp. 267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backfisch B, Veedin Rajan VB, Fischer RM, Lohs C, Arboleda E, Tessmar-Raible K and Raible F (2013) “Stable transgenesis in the marine annelid Platynereis dumerilii sheds new light on photoreceptor evolution,” Proceedings of the National Academy of Sciences of the United States of America, 110(1), pp. 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JT, Vellutini BC, Smith J 3rd, Ronquist F, Jondelius U and Hejnol A (2016) “Xenacoelomorpha is the sister group to Nephrozoa,” Nature, 530(7588), pp. 89–93. [DOI] [PubMed] [Google Scholar]
- Corrêa DD (1960) “Two New Marine Turbellaria from Florida,” Bulletin of marine science, 10(2), pp. 208–216. [Google Scholar]
- Cote LE, Simental E and Reddien PW (2019) “Muscle functions as a connective tissue and source of extracellular matrix in planarians,” Nature communications, 10(1), p. 1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschet K, Nakatani Y and Smith WC (2003) “Generation of Ci-Brachyury-GFP stable transgenic lines in the ascidian Ciona savignyi,” Genesis. Wiley Online Library, 35(4), pp. 248–259. [DOI] [PubMed] [Google Scholar]
- Duruz J, Kaltenrieder C, Ladurner P, Bruggmann R, Martìnez P and Sprecher SG (2020) “Acoel single-cell transcriptomics: cell type analysis of a deep branching bilaterian,” Molecular biology and evolution, doi: 10.1093/molbev/msaa333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan JM, Bradshaw B, Flici H and Frank U (2016) “The interstitial stem cells in Hydractinia and their role in regeneration,” Current opinion in genetics & development, 40, pp. 65–73. [DOI] [PubMed] [Google Scholar]
- Gavilán B, Sprecher SG, Hartenstein V and Martinez P (2019) “The digestive system of xenacoelomorphs,” Cell and tissue research, 377(3), pp. 369–382. [DOI] [PubMed] [Google Scholar]
- Gehrke AR, Neverett E, Luo Y-J, Brandt A, Ricci L, Hulett RE, Gompers A, Ruby JG, Rokhsar DS, Reddien PW, et al. , (2019) “Acoel genome reveals the regulatory landscape of whole-body regeneration,” Science, 363(6432). doi: 10.1126/science.aau6173. [DOI] [PubMed] [Google Scholar]
- Goldstein B (2018) “The Emergence of the Tardigrade Hypsibius exemplaris as a Model System,” Cold Spring Harbor protocols, 2018(11). doi: 10.1101/pdb.emo102301. [DOI] [PubMed] [Google Scholar]
- Hayes MJ, Zakrzewski A-C, Levine TP and Telford MJ (2020) “Nucleus–Plasma Membrane Contact Sites Are Formed During Spermiogenesis in the Acoel Symsagittifera roscoffensis,” Contact. SAGE Publications Inc, 3, p. 2515256420926354. [Google Scholar]
- Hemmrich G, Khalturin K, Boehm A-M, Puchert M, Anton-Erxleben F, Wittlieb J, Klostermeier UC, Rosenstiel P, Oberg H-H, Domazet-Loso T, et al. , (2012) “Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity,” Molecular biology and evolution, 29(11), pp. 3267–3280. [DOI] [PubMed] [Google Scholar]
- Hu C-K and Brunet A (2018) “The African turquoise killifish: A research organism to study vertebrate aging and diapause,” Aging cell, 17(3), p. e12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett RE, Potter D and Srivastava M (2020) “Neural architecture and regeneration in the acoel Hofstenia miamia,” Proceedings of the Royal Society B: Biological Sciences, p. 20201198. doi: 10.1098/rspb.2020.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikmi A, McKinney SA, Delventhal KM and Gibson MC (2014) “TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis,” Nature communications, 5(1), p. 5486. [DOI] [PubMed] [Google Scholar]
- Jondelius U, Raikova OI and Martinez P (2019) “Xenacoelomorpha, a Key Group to Understand Bilaterian Evolution: Morphological and Molecular Perspectives,” in Pontarotti P (ed.) Evolution, Origin of Life, Concepts and Methods. Cham: Springer International Publishing, pp. 287–315. [Google Scholar]
- Kapli P, Natsidis P, Leite DJ, Fursman M, Jeffrie N, Rahman IA, Philippe H, Copley RR and Telford MJ (2021) “Lack of support for Deuterostomia prompts reinterpretation of the first Bilateria,” Science advances, 7(12). doi: 10.1126/sciadv.abe2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapli P and Telford MJ (2020) “Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha,” Science advances, 6(50). doi: 10.1126/sciadv.abc5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassmer SH, Langenbacher AD and De Tomaso AW (2020) “Integrin-alpha-6+ Candidate stem cells are responsible for whole body regeneration in the invertebrate chordate Botrylloides diegensis,” Nature communications, 11(1), p. 4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura JO, Ricci L and Srivastava M (2021) “Embryonic development in the acoel Hofstenia miamia,” Development, 148(13). doi: 10.1242/dev.188656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert EM, Burriesci MS and Pringle JR (2012) “Developing the anemone Aiptasia as a tractable model for cnidarian-dinoflagellate symbiosis: the transcriptome of aposymbiotic A. pallida,” BMC genomics, 13, p. 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor PJ, Clarke DN, Andrade López JM, Fritzenwanker JH, Gray J and Lowe CJ (2019) “I-SceI Meganuclease-mediated transgenesis in the acorn worm, Saccoglossus kowalevskii,” Developmental biology, 445(1), pp. 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, McConnell WB and Grainger RM (2006) “Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease,” Mechanisms of development, 123(2), pp. 103–113. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Martinez P, Riutort M and Baguñà J (2007) “Acoel flatworms are not platyhelminthes: evidence from phylogenomics,” PloS one, 2(8), p. e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Poustka AJ, Chiodin M, Hoff KJ, Dessimoz C, Tomiczek B, Schiffer PH, Müller S, Domman D, Horn M, et al. , (2019) “Mitigating Anticipated Effects of Systematic Errors Supports Sister-Group Relationship between Xenacoelomorpha and Ambulacraria,” Current biology: CB, 29(11), pp. 1818–1826.e6. [DOI] [PubMed] [Google Scholar]
- Ramirez AN, Loubet-Senear K and Srivastava M (2020) “A regulatory program for initiation of Wnt signaling during posterior regeneration,” Cell Reports, 32(9), 108098. [DOI] [PubMed] [Google Scholar]
- Raz AA, Srivastava M, Salvamoser R and Reddien PW (2017) “Acoel regeneration mechanisms indicate an ancient role for muscle in regenerative patterning,” Nature communications, 8(1), p. 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfer E, Amon-Hassenzahl A, Steinmetz PRH and Technau U (2010) “A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis,” Proceedings of the National Academy of Sciences of the United States of America, 107(1), pp. 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I and Paps J (2016) “Acoelomorpha: earliest branching bilaterians or deuterostomes?,” Organisms, diversity & evolution, 16(2), pp. 391–399. [Google Scholar]
- Ruiz-Trillo I, Paps J, Loukota M, Ribera C, Jondelius U, Baguna J and Riutort M (2002) “A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians,” Proceedings of the National Academy of Sciences, pp. 11246–11251. doi: 10.1073/pnas.172390199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Riutort M, Fourcade HM, Baguñà J and Boore JL (2004) “Mitochondrial genome data support the basal position of Acoelomorpha and the polyphyly of the Platyhelminthes,” Molecular phylogenetics and evolution, 33(2), pp. 321–332. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Riutort M, Littlewood DT, Herniou EA and Baguña J (1999) “Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes,” Science, 283(5409), pp. 1919–1923. [DOI] [PubMed] [Google Scholar]
- Sakagami T, Watanabe K, Ikeda R and Ando M (2021) “Structural analysis of the statocyst and nervous system of Praesagittifera naikaiensis, an acoel flatworm, during development after hatching,” Zoomorphology, doi: 10.1007/s00435-021-00521-9. [DOI] [Google Scholar]
- Sato M, Kosuke M, Koriyama M, Inada E, Saitoh I, Ohtsuka M, Nakamura S, Sakurai T, Watanabe S and Miyoshi K (2018) “Timing of CRISPR/Cas9-related mRNA microinjection after activation as an important factor affecting genome editing efficiency in porcine oocytes,” Theriogenology, 108, pp. 29–38. [DOI] [PubMed] [Google Scholar]
- Scimone ML, Atabay KD, Fincher CT, Bonneau AR, Li DJ and Reddien PW (2020) “Muscle and neuronal guidepost-like cells facilitate planarian visual system regeneration,” Science, 368(6498). doi: 10.1126/science.aba3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Cote LE and Reddien PW (2017) “Orthogonal muscle fibres have different instructive roles in planarian regeneration,” Nature, 551(7682), pp. 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Martinez P, Cole C, Baguñà J and Peterson KJ (2007) “Phylogenetic distribution of microRNAs supports the basal position of acoel flatworms and the polyphyly of Platyhelminthes,” Evolution & development, 9(5), pp. 409–415. [DOI] [PubMed] [Google Scholar]
- Smith JPS (1981) “Fine-structural observations on the central parenchyma in Convoluta sp,” Hydrobiologia, 84(1), pp. 259–265. [Google Scholar]
- Srivastava M, Mazza-Curll KL, van Wolfswinkel JC and Reddien PW (2014) “Whole-body acoel regeneration is controlled by Wnt and Bmp-Admp signaling,” Current biology: CB, 24(10), pp. 1107–1113. [DOI] [PubMed] [Google Scholar]
- Tewari AG, Owen JH, Petersen CP, Wagner DE and Reddien PW (2019) “A small set of conserved genes, including sp5 and Hox, are activated by Wnt signaling in the posterior of planarians and acoels,” PLoS genetics, 15(10), p. e1008401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J and Joly J-S (2002) “I-SceI meganuclease mediates highly efficient transgenesis in fish,” Mechanisms of development, 118(1–2), pp. 91–98. [DOI] [PubMed] [Google Scholar]
- Todt C (2009) “Structure and evolution of the pharynx simplex in acoel flatworms (Acoela),” Journal of morphology, 270(3), pp. 271–290. [DOI] [PubMed] [Google Scholar]
- Todt C and Tyler S (2007) “Ciliary receptors associated with the mouth and pharynx of Acoela (Acoelomorpha): a comparative ultrastructural study,” Acta zoologica . Wiley Online Library, 88(1), pp. 41–58. [Google Scholar]
- Tyler S and Rieger RM (1999) “Functional morphology of musculature in the acoelomate worm, Convoluta pulchra (Plathelminthes),” Zoomorphology. Springer, 119(3), pp. 127–142. [Google Scholar]
- Wang IE, Wagner DE and Reddien PW (2018) “Clonal Analysis of Planarian Stem Cells by Subtotal Irradiation and Single-Cell Transplantation,” Methods in molecular biology , 1774, pp. 479–495. [DOI] [PubMed] [Google Scholar]
- Witchley JN, Mayer M, Wagner DE, Owen JH and Reddien PW (2013) “Muscle cells provide instructions for planarian regeneration,” Cell reports, 4(4), pp. 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudarski J, Simanov D, Ustyantsev K, de Mulder K, Grelling M, Grudniewska M, Beltman F, Glazenburg L, Demircan T, Wunderer J, et al. , (2017) “Efficient transgenesis and annotated genome sequence of the regenerative flatworm model Macrostomum lignano,” Nature communications, 8(1), p. 2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L and Reed RD (2016) “Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns,” Nature communications, 7, p. 11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Video 1: Morphology of epidermal cells, related to Figure 2. 360° rotation of a 3D image, reconstructed from confocal z-stack, showing a patch of epiA::TagRFP-T expressing cells in a G0 fixed juvenile. White arrowhead points to cilia, on the apical side; white arrow points to the epidermal processes.
Supp. Video 2: Interlocking of parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the X axis, of a 3D image, reconstructed from confocal z-stack of a G2 tnn::Kaede juvenile worm. Colored arrows point to the different muscle categories. PA, PE, BW, parenchymal, peripheral and body wall muscles, respectively.
Supp. Video 3: Interlocking of parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the Y axis, of a 3D image, reconstructed from confocal z-stack of a G2 tnn::Kaede juvenile worm. Colored arrows point to the different muscle categories. PA, PE, BW, parenchymal, peripheral and body wall muscles, respectively.
Supp. Video 4: Interlocking of single parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the X axis, of a 3D image, reconstructed from confocal z-stack of a G0 tnn::TagRFP-T juvenile worm. PA, BW, parenchymal, and body wall muscles, respectively.
Supp. Video 5: Interlocking of single parenchymal and body wall muscle fibers, related to Figure 4. 360° rotation, along the Y axis, of a 3D image, reconstructed from confocal z-stack of a G0 tnn::TagRFP-T juvenile worm. PA, BW, parenchymal, and body wall muscles, respectively.
Supp. Video 6: Emergence of the musculature during embryogenesis, related to Figure 4. Time-lapse imaging of a G2 tnn::Kaede embryo over 63hrs of development, from 3.5 to 6 days post laying. Time stamp, hh::mm::ss.
Supp. Video 7: Interaction of posterior epidermis with body wall muscle, related to Figure 5. 360° rotation of a 3D image, reconstructed from confocal z-stack, showing a patch of epiA::TagRFP-T (red) expressing cells over the body wall muscles, revealed by anti-TPM IHC (green) in a G1 epiA::TagRFP-T juvenile worm. Nuclei are counterstained with DAPI (blue).
Data Availability Statement
Gene sequences in this study are deposited to GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


