Graphical abstract
Keywords: RBD mutations, ACE2 binding, Antibody binding, Binding free energy, π-π and π-cation interactions, Hydrogen bonds
Abstract
SARS-CoV2 mutants B.1.1.7, B.1.351, and P.1 contain a key mutation N501Y. B.1.135 and P.1 lineages have another mutation, E484K. Here, we decode the effect of these two mutations on the host receptor, ACE2, and neutralizing antibody (B38) recognition. The N501Y RBD mutant binds to ACE2 with higher affinity due to improved π-π stacking and π-cation interactions. The higher binding affinity of the E484K mutant is caused due to the formation of additional hydrogen bond and salt-bridge interactions with ACE2. Both the mutants bind to the B38 antibody with reduced affinity due to the loss of several hydrogen-bonding interactions. The insights obtained from the study are crucial to interpret the increased transmissibility and reduced neutralization efficacy of rapidly emerging SARS-CoV2 VOCs.
1. Introduction
SARS-CoV2 emerged initially from a local seafood market in Wuhan, China, is now a pandemic that causes severe outbreaks in more than 216 countries. During the outbreak, genetic diversification of the virus under different selection biases leads to several SARS-CoV2 genomic variants [1]. SARS-CoV2 is a positive-stranded RNA virus enclosed within a viral envelope [2] where three structural proteins, E, M, and spike glycoprotein, are embedded [2]. Spike protein forms a homo-trimeric large clover-shaped protrusion that mediates viral entry to the host cell through the human ACE2 receptor [3]. Each spike monomer consists of the S1 and S2 domains. Three S1 domains associate to form the ectodomain and the S2 domains entangle to create the stalk, transmembrane, and small intracellular domains [2], [4]. The receptor-binding domain (RBD) of the S1 binds to the peptidase domain (PD) of the ACE2 receptor to open up the S1/S2 and S2′ cleavage sites. Cleavage by the host proteases mediates the fusion of the viral membrane to the host membrane [4], [5]. Due to its role in host receptor recognition, spike protein is under positive selection pressure to produce SARS-CoV2 variants with increased transmissibility and infection rate [6]. Spike protein is also the target for vaccine and immunogenic therapy development as there exist many immunodominant solvent-exposed epitopes that are readily accessible by antibody pool [7]. Thus, tracking the SARS-CoV2 spike variants, particularly the RBD variants, is very important to identify mutant viral strains with higher transmissibility and the ability to cause immune invasion.
Data indicate that the viral genome acquires ∼ 2–3 mutations per month [8]. Although most of the mutations purge out from the population, few of them are fitness-enhancing mutations that alter the antigenic potential of SARS-CoV2, which require focused attention. Using a large-scale genomic screening pipeline, my group previously identified two spike mutants, V367F and S494P, with enhanced human ACE2 binding ability [9]. Later, it was demonstrated that the S494P caused a 3–5 fold decrease in neutralization titer [10] and was reported in many cases in UK, USA, and Mexico [11]. Gan et al. extended the screening on 16,083 sequences and identified several spike mutants with enhanced receptor recognition abilities. These include N440K, S443A, G476S, E484R, and G502P, which cluster near known human ACE2 recognition sites, Lys31 and Lys353 [12]. Mutations that occur within the RBM (Receptor Binding Motif; residues 438‐506) of the RBD deserve special attention since they can alter human ACE2 binding affinity and decrease neutralization by several mAbs. To date, many significant spike RBD mutants were identified with altered ACE2 recognition and antigenic properties. N439K, L452R, and Y453F showed an increase in ACE2 receptor binding ability. Among these mutants, the N439K exhibits resistance to several mAbs and even escapes some polyclonal responses [13]. While, in vitro study demonstrates reduced sensitivity of the L452R RBD mutant to BNT162b2 mRNA vaccine-elicited antibodies [14]. On the other hand, an ELISA-based ACE2/RBD inhibition assay reports Y453F RBD mutant does not decrease established humoral immunity from previously infected individuals or affect the neutralizing antibody response [15]. On the otherhand, G446V, S477N, G485R, and F490S RBD mutants demonstrated ∼ 3–5 fold decrease in neutralization titer for few sera [16]. G446V RBD mutation was reported to reduce ACE2 binding affinities [17], but S477N mutation strengthen the binding between SARS-CoV2 spike RBD and hACE2 [18]. Analysis of the crystal structure of G485R RBD mutant-ACE2 complex (PDB ID: 7LO4) reveals that the residue 485 is not directly interacts with hACE2. G485R mutation leads to a rotation in the loop, affecting some interacting residues without significantly reducing the affinity. The F490S mutation on the other hand showed limited effects on ACE2 binding affinity. Table 1 summarizes a list of RBD mutants and escape variants along with their effects on ACE2 binding and antibody recognition.
Table 1.
List of RBD mutants and escape variants along with their effects on ACE2 binding and antibody recognition.
| RBD mutants | Effect on ACE2 binding with respect to wild-type | Effect on neutralizing antibody and immunity |
|---|---|---|
| E340K | Decrease [19] | Resistance to a broadly reactive NAb, S309 [19] |
| N343A | Loss of ACE2 binding [19] | Impaired neutralizing antibody generation [20] |
| T345I | Limited effect [17] | Efficient binding of all NAbs [17] |
| R346S | Limited effect [19] | Resistant to C135, but retained full sensitivity to both C121 and C144 [21] |
| V367F | Increase [22] | Low binding affinity in HLA-A01:01, HLA-B07:02, and HLA-B35:01 compared to the wild type [23] |
| R408I | Limited effect [24] | decrease the binding affinity of S protein to the CR3022 antibody [24] |
| K417N | Moderately decrease [25] | Resistance to plasma samples, as the NAb titers were approximately 2–4 times lower than those for the wildtype [19] |
| K417T | Moderately decrease [25] | Capable of antibody escape [17] |
| N439K | Increase [13] | Confers resistance against several neutralizing monoclonal antibodies, escapes some polyclonal responses [13] |
| N440Y | Limited effect [17] | Disrupted fewer NAb interactions [17] |
| G446V | Decrease [17] | Demonstrated escape to COVA2-15 and C135 mAbs [26] |
| N450K | Limited effect [19] | Immune escape the neutralization by monoclonal antibodies and human convalescent sera [27] |
| L452R | Increase [28] | confer escape from HLA-A24-restricted cellular immunity [28] |
| Y453F | Increase [15] | Confer escape from HLA-A24-restricted cellular immunity [28] |
| A475V | Decrease [17] | Less binding to multiple class I antibodies [17] |
| G476S | Decrease [29] | Shortened a linear B cell epitope length and even abolished the discontinuous B cell epitope [23] |
| S477N | Increase [25] | resistant to neutralization by multiple monoclonal antibodies [27] |
| S477G | Moderate increase [30] | Conferred resistance to sera [27] |
| T478I | Unaltered [31] | Reduced neutralization by monoclonal antibodies and human convalescent sera [27] |
| E484K | Increase [25] | Less sensitive to neutralization by convalescent human sera, evades antibody neutralization elicited by infection or vaccination [32] |
| E484Q | Limited effect [33] | Limited effect on immune invasion [14] |
| F486L | Decrease [34] | Resistance to plasma samples, as the NAb titers were approximately 2–4 times lower [19] |
| F490S | Limited effect [19] | Predicted to be markedly resistant to neutralization by LY-CoV555 [35] |
| Q493L | Beneficial [36] | Possible vaccine escape [11] |
| S494P | Increase [9] | Reduces antibody neutralization of convalescent and post-immunization sera [37] |
| N501Y | Increase [25] | 272 convalescent sera showed reduced binding of anti-RBD IgG to N501Y [38] |
| Y505W | Increase [17] | Efficiently binds to different Nabs [17] |
WHO declared several SARS-CoV2 mutants as variants of concern (VOC) as they cause sustained disease outbreak across the globe. Currently, there are four variants of concerns: Alpha variant (B.1.1.7; RBD mutations: N501Y, A570D), Beta (B.1.351; RBD mutations: K417N, E484K, and N501Y), Gamma (P.1, B.1.1.28.1; RBD mutations: K417N/T, E484K, and N501Y) and Delta (B.1.617.2; RBD mutations: L452R, T478K) [39]. (https://viralzone.expasy.org/9556) Among them, lineages B.1.1.7, B.1.351, and P.1 contain a key common RBM mutation, N501Y, which was experimentally shown to increase the ACE2 binding affinity [40]. In pseudoviruses carrying the N501Y mutation, a 10-fold decrease in efficacy was reported during the neutralization of mRNA vaccine‐elicited mAbs [41]. However, mouse‐adapted SARS‐CoV‐2 N501Y strain can be effectively neutralized by vaccine‐elicited sera [42].
Recently, computational and experimental approaches have been adopted to understand the mechanism of increased transmissibility and the immune invasion ability of the N501Y spike mutant. Ali et al. reported minor enhancement in the RBD-ACE2 interaction energies upon N501Y mutations using the molecular dynamics simulations. However, the length scale of the simulation is very short [43]. Khan et al. did not observe any significant enhancement in binding affinity when Y501 binds with ACE2 compared to N501 using Protein-Protein docking [44]. Verma et al., on the other hand, performed structural modeling using computation mutagenesis followed by energy minimization and binding free energy calculations using MM/GBSA (Molecular Mechanics/Generalized Born Surface Area) method to probe the effect of N501Y RBD mutation on the ACE2 binding affinity. They observed an enhancement of 7 kcal/mol in ACE2 binding energy upon N501Y mutations [45]. Notably, these modeling approaches qualitatively evaluate the binding affinity as they inherently ignore the effect of mutations on non-local interactions. Recently, Luan et al. performed 185 ns equilibrium simulation and free energy calculation using the FEP (Free energy perturbation) method to evaluate the effect of N501Y mutation on ACE2 binding [46]. An enhancement of −0.81 kcal/mol in the ACE2 binding free energy was reported for the N501Y mutant, which was attributed to the formation of favorable interactions with Tyr41 and Lys353 of ACE2. However, surface plasmon resonance (SPR) binding assays revealed over ten times increment in binding affinity for Y501 RBD with ACE2, in comparison to the wild-type N501Y due to the formation of two new hydrogen bonds with the side chains of Asp38 and Lys353 of ACE2, in addition to the formation of a π stacking interaction between Tyr501 of RBD and Tyr41 of ACE2. Notably, the large change in binding affinity is atypical to a single amino acid mutation, indicating mutation-induced remodeling of the RBD-ACE2 interface, which is challenging to probe accurately using computational approaches.
Two other rapidly emerging SARS-CoV2 variants (B.1.135 and P.1) contain another crucial RBM mutation, E484K. The variant of interest P.2, first reported in Rio de Janeiro and then rapidly widespread in the northeast region of Brazil, contains only the E484K spike mutation [16]. This mutation itself and in combination with the N501Y mutation significantly enhance the ACE2 binding affinity, evident from SPR data [40]. Initial modeling studies suggest enhancement of ACE2 binding affinity for E484K mutant due to the formation of additional hydrogen bond involving Lys484 of mutant RBD with ACE2 and gain in average solvation energy [12], [44].
E484K mutation may be accountable for evasion from neutralizing antibodies [47], [48]. Recently, in-vitro micro-neutralization assays revealed a significant reduction in neutralization efficiency for the recombinant (r)SARS-CoV-2 virus with E484K mutation compared to the control USA-WA1/2020 strain on 34 sera collected from different study participants [48]. Also, the E484K variant caused a 3·4-fold decrease in the neutralization titer in five individuals who received two doses of the Pfizer–BioNTech vaccine [48]. Recently, native Spike-targeted monoclonal antibodies (mAbs) were developed by Regeneron and Eli Lilly which was given emergency approval by the FDA [49], [50], [51]. Recent data suggest that N501Y mutation does not significantly alter the binding affinity with one of the mAb, Bamlanivimab [51]. However, the E484K RBD mutation diminishes its interaction with Bamlanivimab in vitro [52].
A molecular-level insight is urgently required to quantitate the effect of both the N501Y and E484K RBD mutations on receptor recognition and therapeutic monoclonal antibody recognition. This information is highly essential to interpret the higher infectivity rates of SARS-CoV2 VOCs and efficacy of mutant strains on therapeutics mAbs, neutralization and vaccines elicited sera. The effect of a single mutation on the protein–protein interface is very difficult to probe computationally due to high conformational complexity. Large-scale conformational changes are generally associated with the long-timescale phenomenon. Understanding the effect of mutation on the entire RBD-ACE2 interface requires extensive computation that accounts for mutation-induced large-scale conformational dynamics as well as long-distant allosteric effects. Here, we critically decode the role of both the N501Y and E484K spike mutations on ACE2 and neutralization antibody recognition using extensive all-atom molecular dynamics simulation complimented with binding free energy simulations.
2. Materials and Method
2.1. Preparation of mutant spike-ACE2, and spike-antibody complexes
Recently, my group refined the crystal structure of the SARS-CoV2 RBD-ACE2 complex (PDB ID: 6M0J [53]) using extensive molecular dynamics simulation, which was used as a starting structure in the present study [9]. The E484K and N501Y mutant complexes were built from the wild-type SARS-CoV2 RBD-ACE2 complex by mutating the glutamate to lysine at 484th residue and asparagine to tyrosine at the 501st residue, respectively, in the spike RBD region.
The recently resolved crystal structure of the SARS-CoV2 spike RBD complexed with a neutralizing antibody (B38) was considered for the study (PDB ID:7BZ5) [54]. The wild-type SARS-CoV2 RBD from the RBD-ACE2 complex was aligned on the RBD-B38 crystal structure, and then the aligned RBD complexed with the B38 antibody was considered as the wild-type RBD-B38 docked complex. The E484K-B38 and N501Y-B38 complexes were prepared by carrying out the specific mutations on the wild-type SARS-CoV2 RBD-B38 complex using the mutagenesis toolkit of Visual Molecular Dynamics (VMD) [55].
2.2. Equilibrium simulations of wild-type and mutant SARS-CoV2 spike RBD-ACE2 and RBD-antibody complexes
All the simulations were performed using GROMACS 2018.1 [56], [57] packages using the AMBER99SB-ILDN force field [58]. All the complexes were first energy minimized in vacuo to remove any bad contacts. Then each complex was immersed in a triclinic box so that the minimum distance between any protein atom and box walls was > 10 Å. The box dimensions for wild-type and mutant SARS-CoV2 RBD-ACE2 complexes were 100 × 100 × 180 Å3, and for wild-type and mutant SARS-CoV2 RBD-B38 complexes, the box dimensions were 100 × 100 × 190 Å3. Each box was solvated with TIP3P (Transferable intermolecular potential 3 point) water model, and an appropriate number of counter ions were added to neutralize the charge of each system. Then, 500 steps of energy minimization using the steepest descent algorithm were carried out for each system, followed by 10 ns of position-restrained dynamics where the protein backbone dynamics were restrained. At the same time, water molecules were allowed to move freely. After that, a 2 ns simulation in NVT (canonical) ensemble was carried out for each complex at 298 K, followed by another 2 ns simulation in NPT (Isothermal–isobaric) ensemble where both the proteins and solvent molecules were allowed to move freely. Finally, 500 ns of production simulations were performed in the NPT ensemble. All the simulations were carried out under periodic boundary conditions. The temperature was kept constant by coupling to a Nosé–Hoover thermostat with a coupling time constant of 0.1 ps. The pressure was maintained at 1 bar through coupling to the isotropic Parrinello–Rahman barostat with the time constant for coupling set to 2 ps. Electrostatic interactions were calculated using the PME method with default values for grid spacing.
2.3. Calculation of the potential of mean force (PMF) for wild-type and mutant spike RBD to ACE2 and B38 antibody
The optimized wild-type and mutant (E484K and N501Y) SARS-CoV2 RBDs complexed with ACE2 and B38 were immersed in a triclinic box filled with TIP3P water such that the minimum distance between any protein atom and box edges was > 10 Å. The box dimensions for wild-type and mutant SARS-CoV2 RBD-ACE2 complexes were 100 × 100 × 200 Å3, and for wild-type and mutant SARS-CoV2 RBD-B38 complexes, the box dimensions were 100 × 100 × 190 Å3. In Z-direction, the box length was chosen in a way that it was greater than the double of final pull distance. Charges of each system were neutralized by adding an appropriate number of counterions. Each system was then minimized with 500 steps using the steepest descent algorithm. Then, 10 ns position restrained dynamics were performed where the proteins were restrained while water molecules were allowed to move freely. This was followed by 2 ns equilibration at 298 K in the NVT ensemble, and 2 ns NPT simulation, performed using the same simulation protocol mentioned in the equilibrium simulation section.
The potential of mean force (PMF) for pulling the spike RBD (Wild-type and mutants) from the ACE2 or B38 was computed using the umbrella sampling techniques. RBD was pulled from the ACE2/B38 protein binding interface along the Z-direction in an interval of 1 Å with an umbrella force constant of 500 kJ.mol−1.nm2. In each umbrella window, 2 ns equilibration was performed, followed by 3 ns production run in NPT ensemble using the same thermostat, barostat, and associated coupling parameters, mentioned above. Twenty four windows were considered for each case to sample the entire reaction coordinate. A weighted histogram analysis method (WHAM) [59] was used to construct the PMF profile. Sufficient overlap among all the windows was confirmed by histogram analysis. For each system, at least three independent umbrella simulations were performed.
3. Results
3.1. Interactions of wild-type, N501Y and E484K mutant RBDs with ACE2: Insights from equilibrium simulations
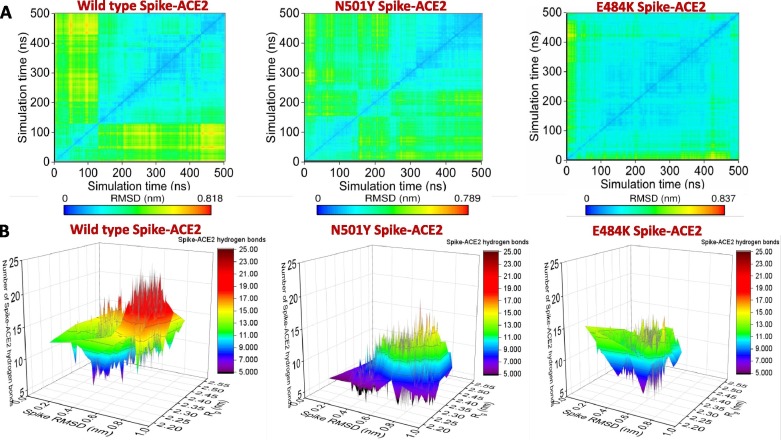
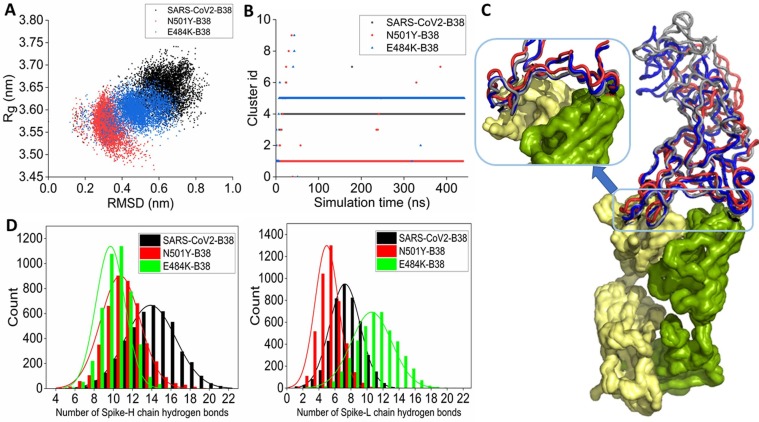
The RBD structure was built using the sequence of SARS-CoV2 samples collected from the Wuhan seafood market (isolate = Wuhan-Hu-1; Accession NC_045512, Version NC_045512.2), referred to as wild-type in the text. Equilibrium molecular dynamics simulations have been used to underscore the effect of the E484K and N501Y RBD mutations on ACE2 recognition. A root mean square deviation (RMSD) based clustering of the simulation trajectories for the three systems (Wild-type RBD-ACE2, N501Y RBD-ACE2, and E484K-ACE2) is shown in Fig. 1 A. Upon complexed with ACE2, the wild-type RBD is comparatively stable with two evident conformational clusters. The larger cluster contains RBD conformations observed during the 120–500 ns simulation timescale.
Fig. 1.
(A) Root mean square deviation (RMSD) based clustering of the SARS-CoV2 spike RBD-ACE2 complexes obtained from the simulation trajectories for the three systems (Wild-type RBD-ACE2, N501Y RBD-ACE2, and E484K-ACE2) is shown. (B) 3-D representation of the conformational dynamics obtained from the simulations of all the three complexes in terms of the root mean square deviation (RMSD), the radius of gyration (Rg) space of RBD, and RBD-ACE2 hydrogen bonds.
The N501Y mutation in spike RBM induces dynamics in the complex. Three different conformational clusters are evident. Conformations from the first 120 ns of the simulation are clubbed together in a cluster. The second cluster contains conformations observed during the 120–200 ns timescale. All the conformations from the last 300 ns are clubbed into the third cluster. On the other hand, the E484K RBD mutation significantly stabilizes the complex. All the conformations are clubbed into a single cluster.
3-D representations of the conformational dynamics obtained from the simulations for all the three complexes are shown in Fig. 1B. In the root mean square deviation (RMSD) and radius of gyration (Rg) space, wild-type RBD dynamics are confined rather narrowly compared to the N501Y mutant when complexed with the ACE2. In this conformational ensemble, the RBD-ACE2 hydrogen bonding varies greatly. On average, there are 11–13 RBD-ACE2 hydrogen bonds. Two populations are evident, one with 7–9 RBD-ACE2 hydrogen bonds and the other one with 17–19 RBD-ACE2 hydrogen bonds. In contrast, N501Y mutant RBD forms less number of interfacial hydrogen bonds. A small proportion of the complex conformations are observed with 7–9 hydrogen bonds, while the rest of the complex conformations are characterized by ∼ 11–13 RBD-ACE2 hydrogen bonds. The conformational space sampled during the molecular dynamics simulations by the E484K spike mutant is more confined in RMSD and Rg space, reconfirming high stabilization of the complex. The average number of interfacial hydrogen bonds ranges between 13 and 15 for most of the sampled conformations. A small conformational state with reduced number of RBD-ACE2 hydrogen bonds (7–9 hydrogen bonds) is also noticed.
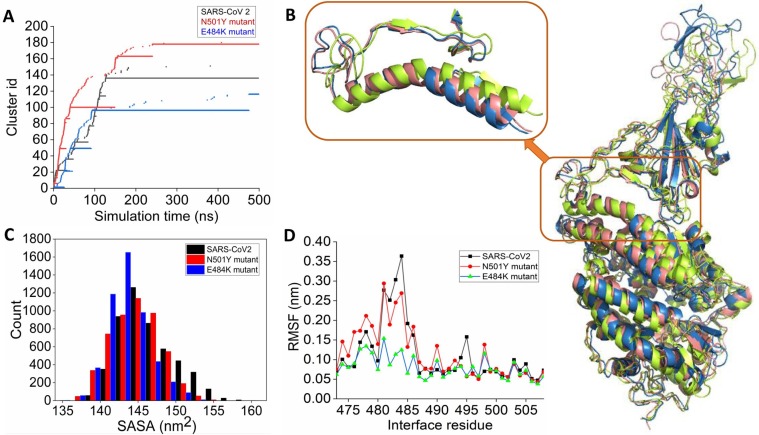
Further RMSD based clustering is used to identify the most populated solution structure of wild-type, N501Y, and E484K RBD complexed with ACE2. The wild-type complex visited 151 conformational clusters during the simulation using a 1.2 Å RMSD cut-off. The 136th cluster is the most populated, span over 120–500 ns during the simulation (Fig. 2 A). N501Y mutation increases the number of clusters visited during the simulation. A total of 180 clusters are observed (Fig. 2A). Many small conformational clusters are evident from the simulation trajectory. Among them, the 178th cluster is the most populated one. E484K RBD-ACE2 complex visited the least number of clusters during the simulation, further indicating high stabilization of the complex. The 96th cluster is the most populated one. The alignment of the average complex structure from the most populated clusters for the three complexes is shown in Fig. 2B. Overall the RBD-ACE2 complex structures remain very similar, apart from the fluctuations of several loop regions. The binding interface has been zoomed in the inset. The wild-type and E484K RBD-ACE2 complexes show similar interfacial packing. However, the N501Y RBD mutation alters interfacial packing. Notably, the N-terminal loopy overhang region that packs the interfacial helical region around the peptidase domain of the ACE2 changes its conformation such that it loses contacts with the ACE2. Analysis of the solvent-accessible surface area (SASA) of RBD reveals that the interfacial areas packed between the mutants (N501Y and E484K) spike RBD and ACE2 are higher than the wild-type RBD. The E484K RBD is packed tightly to the ACE2 PD surface during the simulation (Fig. 2C). A notable observation is that there is a significant reduction of the complex population with higher SASA (i.e., low RBD-ACE2 interface area) in the case of both the mutants (Fig. 2C). This signifies tight interfacial packing between RBD and ACE2 in the case of both N501Y and E484K mutants. Root mean square fluctuations also reveal that E484K mutation highly stabilized the RBM upon complexation with the ACE2. The region from 480 to 486 is highly stabilized in E484K mutated RBD compared to the wild-type. N501Y mutation in the RBD also mildly stabilizes this region (Fig. 2D).
Fig. 2.
(A) Time-evolution of conformational clusters evident from the RMSD based clustering of simulation trajectory of wild-type (black), N501Y (red), and E484K (blue) RBD complexed with ACE2. (B) The alignment of the average complex structure from the most populated clusters for the three systems is shown. Wild-type, N501Y, and E484K RBD complexed with ACE2 are colored as deep salmon, greenish-yellow, and blue, respectively. (C) Distribution of the solvent-accessible surface area (SASA) of wild-type and mutant RBDs obtained from the simulations of three RBD-ACE2 complexes. (D) Root mean square fluctuations (RMSF) of the receptor binding motif obtained from the simulations of RBD-ACE2 complexes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. E484K and N501Y mutants show higher binding affinity to hACE2 compared to wild-type RBD: Evidences from free-energy calculations
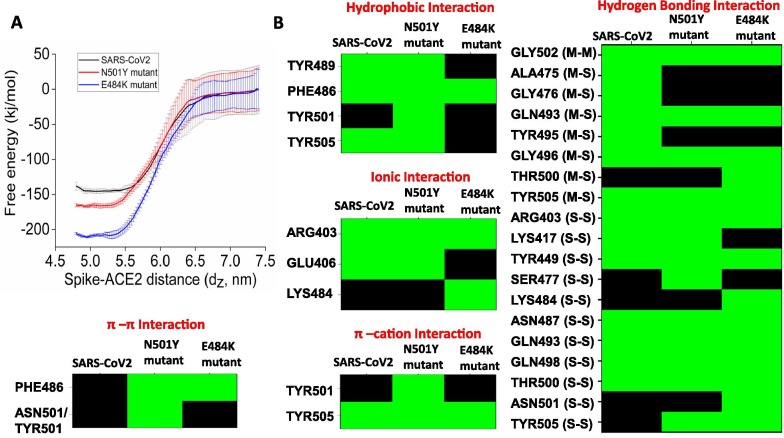
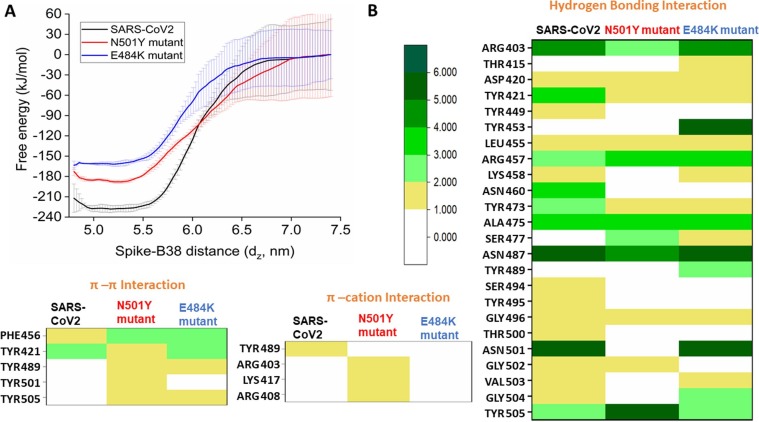
Equilibrium simulations indicate that both N501Y and E484K mutations stabilize the RBM upon complexation with ACE2. The potential of mean force (PMF) has been computed to evaluate the effect of stabilization of mutant RBDs on ACE2 binding free-energy. The free energy for complex formation (ΔGbind) is obtained by calculating the unbinding energy of wild-type and mutant RBDs from the RBD-ACE2 complexes using the umbrella sampling method. Wild-type RBD binds to the ACE2 with an affinity of −144 kJ/mol. The N501Y mutant RBD interacts more strongly with the ACE2. The calculated binding free energy is −165 kJ/mol. The E484K mutant RBD showed a remarkably higher affinity towards ACE2 with the calculated binding free energy of −210 kJ/mol (Fig. 3 A).
Fig. 3.
(A) Potential of mean force for the binding of wild-type (black), N501Y (red), and E484K (blue) RBD to ACE2. (B) Color-coded representation of interfacial interactions involved in ACE2 recognition by the wild-type and mutant spike RBDs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. E484K and N501Y mutations alter the interaction profiles of RBD-ACE2 interface
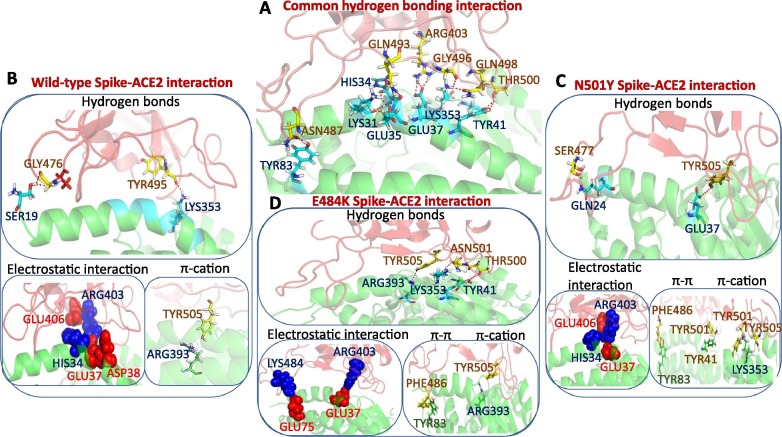
We then decode the gain of the ACE2 binding energy for mutant RBDs in terms of interactive features. Fig. 3B represents the colored coded representation of interfacial interactions involved in ACE2 recognition by spike RBDs for all three cases, and Fig. 4 A, B, C, and D show the mapping of those interactions on the RBD-ACE2 interface for wild-type and mutant spike RBDs. Recently, Chakraborty et al. showed that the Phe486 is the highest energetic contributor for forming the spike-ACE2 complex using the MM/GBSA method [9]. This interaction is also preserved for mutants too. The residue interacts with the Met82 of ACE2 using van der Waals interactions. TYR489 is another significant energetic contributor that interacts with Phe486. This interaction is evident in the wild-type and N501Y mutant but lost upon E484K mutation. The E484K mutation disrupts most of the hydrophobic interactions involved in ACE2 recognition. Upon N501Y mutation, the Tyr501 gains van der Waals contacts with the ACE2 interface. Fig. 4A represents the common interfacial hydrogen bonds are evident in all the three RBD-ACE2 complexes. Hydrogen bonding interactions involving the Gln493, Arg403, Gly496, Gln498, and Thr500 present at the middle and C-terminal loop of the RBM remain preserved in all the three RBD-ACE2 complexes.
Fig. 4.
(A) Display of common hydrogen-bonding interactions present in all the three RBD-ACE2 complexes. Protein is rendered in cartoon mode and residues forming hydrogen bonding interactions are shown in stick mode. Unique interactions present in the wild-type RBD-ACE2 complex (B), N501Y RBD-ACE2 complex (C), and E484K RBD-ACE2 complex (D) are shown. RBD and ACE2 are colored in red and green, respectively. Residues forming hydrogen bonds, π-π, and π-cation interactions are shown in stick mode. Residues involved in electrostatic interactions are shown in the sphere representation. Positive and negatively charged residues are colored in blue and red, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In addition, wild-type RBD forms three unique hydrogen bonds between the Ala475, Gly476, and Tyr495 of the RBM and Ser19, Lys353 of ACE2 (Fig. 4B), which are absent in both the mutants. N501Y mutant RBD forms a specific hydrogen bond involving Ser477 of the RBD. The gain in binding affinity for the N501Y mutant to the ACE2 is due to the improved π-π and π-cation interactions. Mutation of asparagine to tyrosine at the 501st position allows formation for a π-cation interaction with the Lys353 and a π-π stacking interaction with Tyr41 of ACE2 (Fig. 4C).
The E484K mutation allows Lys484 of the RBM to form specific hydrogen bonds with ACE2. In addition, the particular residue is involved in salt-bridge interaction with Glu75 of ACE2. These high-affinity interactions allow E484K mutant RBD to be firmly bound to the ACE2 interface. This intense encounter of E484K RBD to ACE2 allows the remodeling of few interfacial residues. The Tyr505 forms hydrogen bonding interactions with Arg393, Asn501 forms interactions with Lys353, and the Thr500 also forms hydrogen-bonding interactions with Tyr41 of ACE2 (Fig. 4D). The Phe486 of RBD creates additional π-π stacking interaction with Tyr83 of ACE2, which is also present in the N501Y RBD-ACE2 complex.
The work highlights that the E484K and N501Y are gain-of-function mutants. By specific modulation of intermolecular hydrogen bonds, π-π stacking, and π-cation interactions, both the mutants bind to the host receptor with increased affinity.
3.4. Interactions of wild-type, N501Y and E484K mutant RBDs with a neutralizing antibody, B38: Insights from the equilibrium simulations
Recently, four human-origin monoclonal neutralizing antibodies (mAB) were identified from a convalescent patient [54]. Among them, the B38 antibody binds to the spike RBD and competes with the ACE2. The crystal structure of the B38-RBD complex was also resolved at high resolution [54]. The complex structure is used to explore the effect of both the E484K and N501Y RBM mutations on the B38 monoclonal antibody recognition.
In the 2-D space defined by RMSD and Rg, the wild-type RBD-B38 complex occupies a distinct space compared to the two mutant-B38 complexes. As a result, the conformational ensemble sampled during the simulation timescale for the wild-type SARS-CoV2 RBD-B38, E484K RBD-B38, and N501Y RBD-B38 is different with almost no overlap (Fig. 5 A). This altered conformational dynamics can result from differential interactions of mutant spikes with the heavy (H) and light (L) chains of the antibody.
Fig. 5.
(A) 2-D scatter representation of the conformational ensemble of wild-type RBD-B38 (black), N501Y RBD-B38 (red), and E484K RBD-B38 (blue) complexes obtained from the equilibrium simulation in RMSD and Rg space. (B) Time-evolution of conformational clusters of wild-type (black), N501Y (red), and E484K (blue) RBD complexed with B38. (C) The alignment of the average complex structure from the most populated clusters for the three systems is shown. Wild-type, N501Y, and E484K RBD are colored gray, red, and blue. B38 is represented in surface mode. (D) The distribution of the number of hydrogen bonds between the wild-type and mutant RBDs and the H-chain (left) and L-chain (right) of the antibody. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Further, cluster analysis has been performed to identify the most populated solution structure of all the three RBD-antibody complexes. The 4th, 5th, and 1st cluster conformations are the most stable during the simulation timescale for wild-type RBD-B38, E484K RBD-B38, and N501Y RBD-B38, respectively (Fig. 5B). Therefore, the representative conformation from each cluster has been identified for each complex, aligned, and shown in Fig. 5C. The loopy overhang of the RBM, which is in contact with the H and L chain of the B38 antibody, is highly stable without any noticeable deformation. However, the distal loop regions of the RBD show high flexibility. A closer look at the contact regions reveals the difference in the recognition mechanism (Fig. 5C, inset). The RBM forms many contacts with both the H and L chains, and E484K mutant RBM shows very similar protein–protein contacts with the binding interface of the B38 antibody. However, the RBM of the N501Y mutant spike loses critical contact with the H-chain of the antibody due to the upward curvature of a loop region. (Inset of the Fig. 5C). Wild-type SARS-CoV2 RBD forms a higher number of hydrogen bonds with the H-chain. Both the mutants show a significant reduction in the number of hydrogen bonds involving the H-chain of the antibody (Fig. 5D). Particularly, the E484K RBD loses a higher number of hydrogen bonding interactions during the simulation. The wild-type RBD forms a lower number of hydrogen bonding interactions with the L-chain than the H-chain. N501Y mutation further decreases the hydrogen bonding interaction with the L-chain. But the E484K mutant RBD shows an increase in hydrogen bonding interactions with the L-chain during the simulation. However, the distribution is broad with a reduction in the occurrence frequency, indicating the transient nature of these hydrogen-bonding interactions.
3.5. Binding affinity predictions of wild-type, E484K and N501Y RBD to B38 antibody using potential of mean force calculations
Furthermore, the binding affinities of the wild-type and mutant RBDs with the antibody have been calculated using umbrella sampling method. Astonishingly, the potential of mean force (PMF) profiles reveals that the binding affinity is maximum with the B38 mAB for wild-type RBD, and mutations reduce the binding affinity. Notably, the E484k mutant showed the least binding affinity to the B38 mAb (Fig. 6 A). Then, the pharmacophoric feature for antibody recognition has been decoded for both the wild-type and mutant RBDs, and the results are shown in Fig. 6B. The N501Y mutant RDB gains few π-π stacking and π-cation interactions during antibody recognition. However, the loss of binding affinity is primarily due to the loss of crucial hydrogen bonding interactions. Asn501 in wild-type RBD forms six hydrogen bonds with the B38 antibody involving both the sidechain and mainchain donors and acceptors. Mutation of the residue with tyrosine leads to the complete loss of the entire hydrogen bonding network. Apart from this specific loss, Tyr449, Lys458, Asn460, Ser494, Tyr495, Thr500, Val503, and Gly504 also lose their hydrogen-bonding interactions with the B38 mAb. Thus, the loss of many hydrogen-bonding interactions with the antibody accounts for the reduced binding affinity of the N501Y mutant. The E484K mutant RBD loses all the π-cation interactions in addition to the loss of several hydrogen-bonding interactions with the B38 antibody. Tyr449, Asn460, Ser494, Tyr495, Thr500, and Gly502 lose hydrogen bonding interactions with the antibody for the E484K mutant.
Fig 6.
(A) Potential of mean force for the binding of wild-type (black), N501Y (red), and E484K (blue) RBD to B38 antibody. (B) Color-coded representation of interfacial interactions involved in the B38 antibody recognition by the wild-type and mutant spike RBDs. The frequency of interactions is scaled according to the color bar. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussions
The present study explores the effect of two critical mutations, E484K and N501Y, observed in few recently emerging variants of concern of SARS-CoV2 on host receptor recognition and immune invasion. These two mutations occur at the receptor-binding motif (RBM), which is involved in host receptor recognition as well as binds to the class I antibody. Previous modeling and simulation studies showed contradicting results for N501Y mutant on ACE2 recognition [43], [44], [45]. Notably, these modeling approaches are based on protein–protein docking and short simulations, which inherently ignore the mutation-induced allosteric changes in the RBM interface and the RBM dynamics, which accounts for the observed disparity. The extensive free energy simulation performed here complements the experimental data and suggests 21 kJ/mol enhancement in ACE2 binding free energy over the wild-type RBD for N501Y mutant. The extensive simulations also reveal that the N501Y mutations change the local interactions involving π stacking interaction between Tyr501 of RBD and Tyr41 of ACE2 [20]. The study suggests mutation induced remodelling of the entire RBD-ACE2 interface. Three hydrogen bonds observed between the Ala475, Gly476, and Tyr495 of the RBM and Ser19, Lys353 of ACE2, observed in the wild-type RBD, are absent for Y501 RBD mutant. Also, asparagine mutation to tyrosine at the 501st position allows formation for a π-cation interaction with the Lys353. Not only the interaction pattern, but also the N501Y mutation allows stabilization and close packing of the RBD with ACE2 interface.
On the other hand, how the E484K mutation alters the ACE2 recognition is yet to be understood at the structural level. Equilibrium simulation and free energy calculations suggest that mutation of glutamic acid with lysine at the 484th position of the RBM allows the formation of specific hydrogen bonds with ACE2 and a salt-bridge interaction with Glu75 of ACE2. Apart from this local alteration, long-distance allosteric changes allows formation of additional hydrogen bonding interactions with ACE2. The Phe486 of RBD creates a new π-π stacking interaction with Tyr83 of ACE2, which is also present in the N501Y RBD-ACE2 complex.
Three therapeutic monoclonal antibodies, Bamlanivimab (LY-CoV555) from Eli Lilly, Casirivimab (REGN10933), and Imdevimab (REGN10987) from Regeneron, were given emergency approval by US FDA for COVID19 [49], [50], [51]. Recent data suggest that N501Y mutation does not significantly alter the binding affinity with Bamlanivimab [51]. However, the E484K mutation diminishes its interaction with Bamlanivimab in vitro [52]. Wang et al. recently evaluated the susceptibility of the 28 pseudoviruses expressing many spike single and multiple variations to neutralization by 12 mAbs (11 of them are anti-RBD) and observed that the E484K mutation is resistant to class II mAbs, while in combination with K417N and N501Y it showed resistance to class I and II mAbs [60]. Thus it is important to evaluate the effect of both E484K and N501Y mutations on mAbs recognition, particularly against anti-RBD mAbs. The B38 antibody binds to the spike RBM and outcompetes ACE2. Therefore, it was chosen to decode the effect of both the mutations on mAbs recognition. The extensive simulation and free energy data suggest that the E484K mutation showed the highest reduction in binding affinity, while N501Y showed a moderate reduction in binding affinity. Further analysis reveals the structural rationale behind the observed reduction in binding affinity. Asn501 in wild-type RBD participates in a hydrogen-bonding network involving six hydrogen bonds with the B38 antibody. Mutation of the residue leads to the complete destruction of the hydrogen bonding network. In addition, N501Y mutant lose several additional hydrogen bonding interactions with the B38 mAB. The E484K mutant RBD loses all the π-cation interactions in addition to the loss of several hydrogen-bonding interactions with the B38 mAb. Data suggest that N501Y mutant RDB gains few π-π stacking and π-cation interactions during antibody recognition which accounts for less reduction in binding affinity for this mutant with B38 mAB.
Results obtained from this study provide crucial insight into the efficacy of therapeutic mAbs on SARS-CoV2 variants harboring the E484K and N501Y mutations. Recent contact analysis of 57 PDB structures of RBD-antibodies reveals E484 is one of the highly frequent contacts with mAbs [37]. These indicate that the observed loss of binding affinity to the B38 mAb is possibly applicable to other anti-RBD mAbs. The structural insights into the loss of epitopic potential of spike RBM upon E484k and N501Y mutation provide a crucial guideline for future therapeutic antibody designing against SARS-CoV2 variants of concerns.
CRediT authorship contribution statement
Sandipan Chakraborty: Conceptualization, Methodology, Software, Writing – original draft, Writing – review & editing, Investigation.
Declaration of Competing Interest
The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The author gratefully acknowledges Microsoft AI for Health program for providing Azure cloud capabilities to carry out the research (Microsoft AI for Health COVID-19 grant ID: 00011000243).
References
- 1.Ghosh S., Chakraborty S., Yang H. Phylogenomics Analysis of SARS-CoV2 Genomes Reveals Distinct Selection Pressure on Different Viral Strains. Biomed Res. Int. 2020;2020:1–8. doi: 10.1155/2020/5746461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020;7:344. doi: 10.3389/fmolb.2020.605236. https://www.frontiersin.org/article/10.3389/fmolb.2020.605236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y.u., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. C.-19 G.U.K. (COG-U. Consortium, SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21(2):73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava S., Banu S., Singh P., Sowpati D.T., Mishra R.K. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. J. Biosci. 2021;46:22. doi: 10.1007/s12038-021-00145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem. Biophys. Res. Commun. 2021;534:374–380. doi: 10.1016/j.bbrc.2020.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R., Chen J., Gao K., Wei G.-W. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics. 2021;113:2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan H.H., Twaddle A., Marchand B., Gunsalus K.C. Structural Modeling of the SARS-CoV-2 Spike/Human ACE2 Complex Interface can Identify High-Affinity Variants Associated with Increased Transmissibility. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N., De Marco A., Li K., Bassi J., O’Toole A., Pinto D., Colquhoun R.M., Culap K., Jackson B., Zatta F., Rambaut A., Jaconi S., Sreenu V.B., Nix J., Zhang I., Jarrett R.F., Glass W.G., Beltramello M., Nomikou K., Pizzuto M., Tong L., Cameroni E., Croll T.I., Johnson N., Di Iulio J., Wickenhagen A., Ceschi A., Harbison A.M., Mair D., Ferrari P., Smollett K., Sallusto F., Carmichael S., Garzoni C., Nichols J., Galli M., Hughes J., Riva A., Ho A., Schiuma M., Semple M.G., Openshaw P.J.M., Fadda E., Baillie J.K., Chodera J.D., Rihn S.J., Lycett S.J., Virgin H.W., Telenti A., Corti D., Robertson D.L., Snell G. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira I.A.T.M., Kemp S.A., Datir R., Saito A., Meng B., Rakshit P., Takaori-Kondo A., Kosugi Y., Uriu K., Kimura I., Shirakawa K., Abdullahi A., Agarwal A., Ozono S., Tokunaga K., Sato K., Gupta R.K. I.S.-C.-2 G.C. CITIID-NIHR BioResource COVID-19 Collaboration, T.G. to P.J. (G2P-J. Consortium, SARS-CoV-2 B.1.617 Mutations L452R and E484Q Are Not Synergistic for Antibody Evasion. J. Infect. Dis. 2021;224:989–994. doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayarri-Olmos R., Rosbjerg A., Johnsen L.B., Helgstrand C., Bak-Thomsen T., Garred P., Skjoedt M.-O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Focosi D., Maggi F. Neutralising antibody escape of SARS-CoV-2 spike protein: Risk assessment for antibody-based Covid-19 therapeutics and vaccines. Rev. Med. Virol. 2021;31(6) doi: 10.1002/rmv.v31.610.1002/rmv.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshpande A., Harris B.D., Martinez-Sobrido L., Kobie J.J., Walter M.R. Epitope Classification and RBD Binding Properties of Neutralizing Antibodies Against SARS-CoV-2 Variants of Concern. Front. Immunol. 2021;12:2185. doi: 10.3389/fimmu.2021.691715. https://www.frontiersin.org/article/10.3389/fimmu.2021.691715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mejdani M., Haddadi K., Pham C., Mahadevan R. SARS-CoV-2 receptor-binding mutations and antibody contact sites. Antib. Ther. 2021;4:149–158. doi: 10.1093/abt/tbab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi C., Sun X., Lin Y., Gu C., Ding L., Lu X., Yang Z., Zhang Y., Ma L., Gu W., Qu A., Zhou X., Li X., Xu J., Ling Z., Xie Y., Lu H., Sun B. Comprehensive mapping of binding hot spots of SARS-CoV-2 RBD-specific neutralizing antibodies for tracking immune escape variants. Genome Med. 2021;13:164. doi: 10.1186/s13073-021-00985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azad T., Singaravelu R., Taha Z., Jamieson T.R., Boulton S., Crupi M.J.F., Martin N.T., Brown E.E.F., Poutou J., Ghahremani M., Pelin A., Nouri K., Rezaei R., Marshall C.B., Enomoto M., Arulanandam R., Alluqmani N., Samson R., Gingras A.-C., Cameron D.W., Greer P.A., Ilkow C.S., Diallo J.-S., Bell J.C. Nanoluciferase complementation-based bioreporter reveals the importance of N-linked glycosylation of SARS-CoV-2 S for viral entry. Mol. Ther. 2021;29(6):1984–2000. doi: 10.1016/j.ymthe.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M., Robbiani D.F., Rice C.M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junxian O., Zhonghua Z., Ruixue D., Jing Z., Shan Z., Xiaowei W., Wendong L., Yi R., Lilian C., Qiaoshuai L., Lu L., Donald S., James C., Jianguo W., Gong Z., Qiwei Z., Tom G. V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity. J. Virol. 2021;95:e00617–e621. doi: 10.1128/JVI.00617-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu W., Wang M., Yu D., Zhang X. Variations in SARS-CoV-2 Spike Protein Cell Epitopes and Glycosylation Profiles During Global Transmission Course of COVID-19. Front. Immunol. 2020;11:2222. doi: 10.3389/fimmu.2020.565278. https://www.frontiersin.org/article/10.3389/fimmu.2020.565278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C., Jin X., Chen X., Qiu L., Leng Q., Qiu T. Antigenic Evolution on a Global Scale Reveals the Potential Natural Selection of Severe Acute Respiratory Syndrome-Coronavirus 2 by Pre-existing Cross-Reactive T-Cell Immunity. Front. Microbiol. 2021;12:931. doi: 10.3389/fmicb.2021.599562. https://www.frontiersin.org/article/10.3389/fmicb.2021.599562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton M.I., MacGowan S.A., Kutuzov M.A., Dushek O., Barton G.J., van der Merwe P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10 doi: 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez E., Haycroft E.R., Adair A., Mordant F.L., O’Neill M.T., Pymm P., Redmond S.J., Lee W.S., Gherardin N.A., Wheatley A.K., Juno J.A., Selva K.J., Davis S.K., Grimley S.L., Harty L., Purcell D.F.J., Subbarao K., Godfrey D.I., Kent S.J., Tham W.-H., Chung A.W. Simultaneous evaluation of antibodies that inhibit SARS-CoV-2 variants via multiplex assay. JCI Insight. 2021;6 doi: 10.1172/jci.insight.150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S., Whelan S.P.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3):477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., Yue Y., Shimizu R., Ito J., Torii S., Yonekawa A., Shimono N., Nagasaki Y., Minami R., Toya T., Sekiya N., Fukuhara T., Matsuura Y., Schreiber G., Ikeda T., Nakagawa S., Ueno T., Sato K. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng S., Sobitan A., Rhoades R., Liu D., Tang Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 2021;22:1239–1253. doi: 10.1093/bib/bbaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh A., Steinkellner G., Köchl K., Gruber K., Gruber C.C. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci. Rep. 2021;11:4320. doi: 10.1038/s41598-021-83761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upadhyay V., Lucas A., Panja S., Miyauchi R., Mallela K.M.G. Receptor binding, immune escape, and protein stability direct the natural selection of SARS-CoV-2 variants. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D., Boon A.C.M., Presti R.M., O’Halloran J.A., Kim A.H.J., Deepak P., Pinto D., Fremont D.H., Crowe J.E., Corti D., Virgin H.W., Ellebedy A.H., Shi P.-Y., Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S. N.I.C. Team SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorg. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han P., Su C., Zhang Y., Bai C., Zheng A., Qiao C., Wang Q., Niu S., Chen Q., Zhang Y., Li W., Liao H., Li J., Zhang Z., Cho H., Yang M., Rong X., Hu Y., Huang N., Yan J., Wang Q., Zhao X., Gao G.F., Qi J. Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants. Nat. Commun. 2021;12:6103. doi: 10.1038/s41467-021-26401-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurini E., Marson D., Aulic S., Fermeglia A., Pricl S. Molecular rationale for SARS-CoV-2 spike circulating mutations able to escape bamlanivimab and etesevimab monoclonal antibodies. Sci. Rep. 2021;11:20274. doi: 10.1038/s41598-021-99827-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delgado Blanco J., Hernandez-Alias X., Cianferoni D., Serrano L., Kolodny R. In silico mutagenesis of human ACE2 with S protein and translational efficiency explain SARS-CoV-2 infectivity in different species. PLOS Comput. Biol. 2020;16(12):e1008450. doi: 10.1371/journal.pcbi.1008450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alenquer M., Ferreira F., Lousa D., Valério M., Medina-Lopes M., Bergman M.-L., Gonçalves J., Demengeot J., Leite R.B., Lilue J., Ning Z., Penha-Gonçalves C., Soares H., Soares C.M., Amorim M.J., Diamond M.S. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLOS Pathog. 2021;17(8):e1009772. doi: 10.1371/journal.ppat.1009772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu L.u., Chu A.-H., Zhang R.R., Chan W.-M., Ip J.D., Tsoi H.-W., Chen L.-L., Cai J.-P., Lung D.C., Tam A.R., Yau Y.-S., Kwan M.-W., To W.-K., Tsang O.-Y., Lee L.-Y., Yi H., Ip T.-C., Poon R.-S., Siu G.-H., Mok B.-Y., Cheng V.-C., Chan K.H., Yuen K.-Y., Hung I.-N., To K.-W. The impact of spike N501Y mutation on neutralizing activity and RBD binding of SARS-CoV-2 convalescent serum. EBioMedicine. 2021;71:103544. doi: 10.1016/j.ebiom.2021.103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S., Sharma V., Sarkar S., Ludhiadch A. Battle against the pandemic: Emergence of SARS-CoV2 variants and global challenge. Travel Med. Infect. Dis. 2021;44 doi: 10.1016/j.tmaid.2021.102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G., Ramos V., Da Silva J., Xu J., Colbert R.A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P.J., Casellas R., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C. Nussenzweig, mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.-F., Li J., Zhang N.-N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D.-Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.-F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science (80-.) 2020:369. doi: 10.1126/science.abc4730. 1603 LP – 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali F., Kasry A., Amin M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med. Drug Discov. 2021;10 doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cell. Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma J., Subbarao N. Insilico study on the effect of SARS-CoV-2 RBD hotspot mutants’ interaction with ACE2 to understand the binding affinity and stability. Virology. 2021;561:107–116. doi: 10.1016/j.virol.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luan B., Wang H., Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595:1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Paesen G.C., Lopez-Camacho C., Slon-Campos J., Hallis B., Coombes N., Bewley K., Charlton S., Walter T.S., Skelly D., Lumley S.F., Dold C., Levin R., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S., James W., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., García-Sastre A., Schotsaert M., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Chernet R.L., Eaker L.Q., Ferreri E.D., Floda D.L., Gleason C.R., Kleiner G., Jurczyszak D., Matthews J.C., Mendez W.A., Mulder L.C.F., Russo K.T., Salimbangon A.-B., Saksena M., Shin A.S., Sominsky L.A., Srivastava K. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2(7):e283–e284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuccori M., Ferraro S., Convertino I., Cappello E., Valdiserra G., Blandizzi C., Maggi F., Focosi D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline. MAbs. 2020;12(1):1854149. doi: 10.1080/19420862.2020.1854149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurt A.C., Wheatley A.K. Neutralizing Antibody Therapeutics for COVID-19. Viruses. 2021;13(4):628. doi: 10.3390/v13040628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H., Zhang Q., Wei P., Chen Z., Aviszus K., Yang J., Downing W., Jiang C., Liang B.o., Reynoso L., Downey G.P., Frankel S.K., Kappler J., Marrack P., Zhang G. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res. 2021;31(6):720–722. doi: 10.1038/s41422-021-00496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widera M., Wilhelm A., Hoehl S., Pallas C., Kohmer N., Wolf T., Rabenau H.F., Corman V.M., Drosten C., Vehreschild M.J.G.T., Goetsch U., Gottschalk R., Ciesek S. Limited Neutralization of Authentic Severe Acute Respiratory Syndrome Coronavirus 2 Variants Carrying E484K In Vitro. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q.i., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 54.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science (80-.) 2020:368. doi: 10.1126/science.abc2241. 1274 LP – 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 56.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.v26:1610.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 57.Kutzner C., Páll S., Fechner M., Esztermann A., de Groot B.L., Grubmüller H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019;40:2418–2431. doi: 10.1002/jcc.26011. [DOI] [PubMed] [Google Scholar]
- 58.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinforma. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hub J.S., de Groot B.L., van der Spoel D. g_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J. Chem. Theory Comput. 2010;6(12):3713–3720. doi: 10.1021/ct100494z. [DOI] [Google Scholar]
- 60.Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., Liao H., Feng Y., Li X., Shi X., Zhang Z., Zhang F., Ding Q., Zhang T., Wang X., Zhang L. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]