Abstract
Phosphoinositide (PI) 3-kinase is a key mediator of insulin-dependent metabolic actions, including stimulation of glucose transport and glycogen synthesis. The gene for the p85α regulatory subunit yields three splicing variants, p85α, AS53/p55α, and p50α. All three have (i) a C-terminal structure consisting of two Src homology 2 domains flanking the p110 catalytic subunit-binding domain and (ii) a unique N-terminal region of 304, 34, and 6 amino acids, respectively. To determine if these regulatory subunits differ in their effects on enzyme activity and signal transduction from insulin receptor substrate (IRS) proteins under physiological conditions, we expressed each regulatory subunit in fully differentiated L6 myotubes using adenovirus-mediated gene transfer with or without coexpression of the p110α catalytic subunit. PI 3-kinase activity associated with p50α was greater than that associated with p85α or AS53. Increasing the level of p85α or AS53, but not p50α, inhibited both phosphotyrosine-associated and p110-associated PI 3-kinase activities. Expression of a p85α mutant lacking the p110-binding site (Δp85) also inhibited phosphotyrosine-associated PI 3-kinase activity but not p110-associated activity. Insulin stimulation of two kinases downstream from PI-3 kinase, Akt and p70 S6 kinase (p70S6K), was decreased in cells expressing p85α or AS53 but not in cells expressing p50α. Similar inhibition of PI 3-kinase, Akt, and p70S6K was observed, even when p110α was coexpressed with p85α or AS53. Expression of p110α alone dramatically increased glucose transport but decreased glycogen synthase activity. This effect was reduced when p110α was coexpressed with any of the three regulatory subunits. Thus, the three different isoforms of regulatory subunit can relay the signal from IRS proteins to the p110 catalytic subunit with different efficiencies. They also negatively modulate the PI 3-kinase catalytic activity but to different extents, dependent on the unique N-terminal structure of each isoform. These data also suggest the existence of a mechanism by which regulatory subunits modulate the PI 3-kinase-mediated signals, independent of the kinase activity, possibly through subcellular localization of the catalytic subunit or interaction with additional signaling molecules.
Upon stimulation, the activated insulin receptor tyrosine kinase phosphorylates several intracellular substrates, leading to stimulation of a wide variety of metabolic and mitogenic actions (20, 37). This occurs via interaction between the phosphorylated insulin receptor substrate (IRS) proteins and a number of Src homology 2 (SH2) domain-containing proteins including Grb2, SHP2, and the class Ia phosphoinositide (PI) 3-kinase (37). A great deal of evidence has shown that PI 3-kinase plays a pivotal role in carbohydrate, lipid, and protein metabolism regulated by insulin (34). The mechanisms by which PI 3-kinase-dependent signaling mediates these metabolic effects are unclear, since these biological endpoints are quite specific for insulin, but an increase in PI 3-kinase activity associated with tyrosine-phosphorylated receptor or its substrates is a common event in hormone, growth factor, and cytokine signaling pathways (34, 37).
The class Ia PI 3-kinase consists of a regulatory subunit and a 110-kDa catalytic subunit (p110). Three isoforms of p110 (α, β, and δ) are independent gene products and have been identified as class Ia based on their ability to bind the regulatory subunits (11). Of these, both p110α and p110β have been implicated in insulin signaling, although the functional difference between them remains unclear (34). At least eight isoforms of regulatory subunit have been identified, all of which can bind to pYXXM or pYMXM motifs on IRS proteins through their SH2 domains (38). Structurally, they can be classified into two groups based on length. p85α and p85β belong to the full-length version of the regulatory subunits and consist of an SH3 domain, a Bcr homology domain flanked by two proline-rich domains, an N-terminal SH2 (nSH2) domain, an inter-SH2 (iSH2) region containing the p110-binding site, and a C-terminal SH2 (cSH2) domain (27). In addition, two truncated versions of regulatory subunits, AS53 (also known as p55α) (2, 18) and p50α (10, 19), are splicing variants derived from p85α gene. These share a common nSH2-iSH2-cSH2 structure with p85α but lack the SH3 domain, N-terminal proline-rich domain, and Bcr domain; in their place they have unique N-terminal ends consisting of 34 and 6 amino acids, respectively. Another truncated regulatory subunits is p55PIK, which is encoded by a distinct gene but has an nSH2-iSH2-cSH2 structure highly homologous to that of p85α and a 34-amino-acid N terminus similar to that of AS53 (29). In addition, it is known that p85α and AS53 (and probably p50α) have another splicing variant in which a nine-amino-acid insertion replaces aspartic acid located in the iSH2 domain close to a regulatory phosphorylation site (2). It is not clear why these multiple isoforms of regulatory subunit exist or what the physiological role of each isoform is, although differences in level of expression in different tissues suggest the existence of a specific role for each isoform. Various insulin-sensitive tissues and cells express virtually all eight regulatory subunits. p85α is usually the dominant isoform, whereas the expression levels of AS53 and p50α are variable, dependent on tissue and cell types (2, 19) and metabolic conditions (1, 22). p85β binds to IRS proteins (33) but has been reported to show little stimulation by insulin (19), and thus its effect on insulin action is controversial (34).
To explore the physiological role of the different regulatory subunits, we have expressed p85α, AS53, and p50α with or without p110α in fully differentiated L6 myotubes, using adenovirus-mediated gene transfer. We find that three different regulatory subunits mediate PI 3-kinase-dependent signals with different efficiencies and that all negatively modulate the PI 3-kinase catalytic activity to different extents, dependent on the unique N-terminal structure of each isoform. These data suggest that the balance of the regulatory subunits in cells and tissues may be necessary for appropriate physiological signaling and that changes in this balance can affect the downstream insulin actions, leading to alteration of insulin sensitivity.
MATERIALS AND METHODS
Generation of adenoviruses.
cDNAs of human p85α and AS53 were cloned as described previously (2), and the coding region of each clone was subcloned to pBluescript. An influenza virus hemagglutinin (HA) sequence tag (YPYDVPDYA) was added to each clone in place of the original stop codon to create p85α-HA and AS53-HA. A p50α-HA cDNA was created by replacing the first 34-amino-acid sequence of AS53 with the N-terminal unique sequence (MHNLQT) of p50α (10, 19). Each of these was subcloned into the pSVSPORT mammalian expression vector and digested with EcoRI and SphI. After both ends were blunted, the cDNA fragment was ligated into the SwaI site of the pAdex1CAwt cosmid cassette (25). The recombinant adenoviruses, Adex1CAp85α-HA, Adex1CAAS53-HA, and Adex1CAp50α-HA, were constructed by homologous recombination between the expression cosmid cassette and parental virus genome (25). A recombinant adenovirus encoding a mutant p85α (Adex1CAΔp85) that lacks the p110-binding site was kindly provided by Masato Kasuga (Kobe University) (31). The cDNA of mouse p110α with a c-Myc epitope tag at the N terminus was kindly provided by Lewis Cantley (Beth Israel-Deaconess Medical Center, Boston, Mass.). It was also subcloned into the SwaI site of the pAdex1CAwt cosmid cassette, followed by construction of recombinant adenovirus Adex1CAp110α. The control adenovirus, Adex1CALacZ, and the cosmid cassette were kindly provided by Izumi Saito (University of Tokyo).
Cell culture and adenovirus infection.
L6 cells were maintained in Dulbecco's modified Eagle medium (DMEM) and induced to differentiate into myotubes as previously described (36). The differentiated cells were cultured in media containing the adenoviruses for 1 h at 37°C; DMEM supplemented with fetal calf serum was added, and cells were cultured for 24 h. Cells were subjected to assays after 20 h of serum deprivation. The adenoviruses were applied at the MOI (multiplicity of infection) indicated for each experiment. Under these conditions, lacZ gene expression was observed in over 90% of L6 cells on postinfection days 1 through 4, as measured by β-galactosidase assay.
Antibodies.
Rabbit polyclonal antibodies to all isoforms of p85α (αp85pan) generated against the rat N-terminal SH2 domain of p85α were purchased from Upstate Biotechnology Inc. Rabbit polyclonal anti-p110α antibodies (αp110α) generated against a peptide corresponding to amino acids 189 to 390 of human p110α and those to p110α, -β, and -δ (αp110pan) generated against a peptide corresponding to amino acids 800 to 1039 of human p110β were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-Akt antibodies (αAkt) generated against the pleckstrin homology domain of human Akt1 were purchased from Upstate Biotechnology, while p70 S6 kinase (p70S6K)-specific antibodies (αp70S6K), generated against a peptide corresponding to positions 485 to 502 rat p70S6K, and those to GSK3α (αGSK3α), generated against a peptide corresponding to amino acids 408 to 483 of human GSK3α, were purchased from Santa Cruz Biotechnology. Rabbit polyclonal antibodies to phospho-Akt (αphospho-Akt), generated against a phosphoserine peptide corresponding to Ser473 of mouse Akt1, and those to phospho-p70S6K (αphospho-p70S6K), generated against a phosphoserine peptide corresponding to Ser411 of human p70S6K, were purchased from New England Biolabs Inc. Mouse monoclonal antiphosphotyrosine antibody 4G10 was purchased from Upstate Biotechnology. Mouse monoclonal anti-HA antibodies (αHA) generated against a peptide corresponding to the sequence YPYDVPDYA were purchased from Boehringer Mannheim Corp. Rabbit polyclonal antibodies to IRS-1 (αIRS-1) and IRS-2 (αIRS-2) were generated as previously described (17).
Immunoprecipitation and Western blotting.
After serum starvation for 20 h, cells were treated with insulin for the indicated period and then lysed with buffer A containing 25 mM Tris-HCl (pH 7.4), 2 mM Na3VO4, 10 mM NaF, 10 mM Na4P2O7, 1 mM EGTA, 1 mM EDTA, 10 nM okadaic acid, leupeptin (5 μg/ml), aprotinin (5 μg/ml), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1% Nonidet-P40. The lysates were subjected to immunoprecipitation with one of the antibodies described above and immobilized on protein A or G-Sepharose beads. After sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the lysates or immunoprecipitates were subjected to Western blotting and visualized by enhanced chemiluminescence (Boehringer Mannheim).
PI 3-kinase assay.
The immunoprecipitates with αHA, αp85α, 4G10, or αp110α were washed three times with buffer A, washed twice with PI 3-kinase reaction buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.5 mM EGTA), and suspended in 50 μl of PI 3-kinase reaction buffer containing 0.1 mg of PI (bovine liver; Avanti Polar Lipids) per ml. The reactions were initiated by adding 5 μl of MgCl2-ATP mixture (200 mM MgCl2, 200 μM ATP) containing 5 μCi of [γ-32P]ATP to the reaction mixture and incubating the mixture at 25°C for 20 min. The reactions were terminated by adding 150 μl of chloroform–methanol–11.6 N HCl (100:200:2). After addition of 120 μl of chloroform to each sample, the organic phase was separated by centrifugation and washed twice with methanol–1 N HCl (1:1). After evaporation, the pellets were resuspended in 20 μl of chloroform, spotted onto a silica gel plate, and developed in chloroform–methanol–28% ammonium hydroxide–water (43:38:5:7). The phosphorylated lipids were visualized by autoradiography.
In vitro kinase assays.
Cells were lysed with buffer A as described above, and the lysates were subjected to immunoprecipitation with αAkt and αGSK3α, followed by Akt kinase and GSK3 kinase assays (36). For the p70S6K kinase assay, cells were lysed with buffer B containing 20 mM Tris-HCl (pH 7.5), 25 mM β-glycerophosphate, 100 mM NaCl, 1 mM sodium orthovanadate, 2 mM EGTA, leupeptin (5 μg/ml), aprotinin (5 μg/ml), and 1 mM PMSF and then immunoprecipitated with αp70S6K (36). The αPKB, αGSK3α, or αp70S6K immunoprecipitates were washed and resuspended in 50 mM Tris-HCl (pH 7.5)–10 mM MgCl2–1 mM dithiothreitol, to which 50 μM ATP, 5 μCi of [γ-32P]ATP, and 1 μg of Crosstide in the Akt kinase assay, 1 μg of phospho-glycogen synthase (phospho-GS) peptide (Upstate Biotechnology) in the GSK3 kinase assay, or 1 μg of S6 peptide (32-mer peptide from the C-terminal sequence of ribosomal S6 protein; Life Technologies Inc.) in the S6 kinase assay had been added. After 20 min at 30°C, the reaction was stopped, the aliquots were spotted on squares of P-81 paper and washed with 0.5% phosphoric acid, and radioactivity was counted.
2-DG uptake assays.
2-Deoxyglucose (2-DG) uptake assays were performed as described elsewhere (36). Cells were grown in 12-well plates and infected with adenoviruses as described above. Before use in the glucose uptake assay, cells were washed three times with phosphate-buffered saline and incubated in 1 ml of serum-free DMEM for 3 h at 37°C. Cells were then washed once with Krebs-Ringer phosphate-HEPES buffer (KRHB) containing 130 mM NaCl2, 5 mM KCl2, 1.3 mM CaCl2, 1.3 mM MgSO4, 10 mM Na2HPO4, and 25 mM HEPES (pH 7.4) and incubated in 1 ml of KRHB containing 0.1% bovine serum albumin without or with insulin for 15 min at 37°C. Glucose uptake was initiated by the addition of 2-deoxy-d-[2,6-3H]glucose to a final concentration of 0.5 μCi for 5 min at 37°C and terminated by two washes with ice-cold KRHB. Cells were solubilized with 0.4 ml of 0.1% SDS and counted in a scintillation counter. Nonspecific glucose uptake was measured in the presence of 20 μM cytochalasin B and was subtracted from each assay to obtain specific uptake.
GS assays.
GS activity was measured as previously described (36). Cells were infected with adenoviruses as described above and incubated in serum-free DMEM for 20 h. They were then washed twice and incubated with KRBH without or with 100 nM insulin for 20 min. Cells were lysed with lysis buffer containing 25 mM Tris-HCl (pH 7.0), 30% glycerol, 10 mM EDTA, 100 mM KF, and 1 mM PMSF. The lysates were centrifuged, and 30 μl of the supernatant was added to 60 μl of assay mixture containing 50 mM Tris-HCl (pH 7.4), 25 mM NaF, 20 mM EDTA, glycogen (1 mg/ml), and 0.1 μCi of UDP-[14C]glucose plus 0.25 or 10 mM glucose-6-phosphate. After incubation at 30°C for 30 min, aliquots were spotted on 3MM paper (Whatman) and washed four times with ice-cold 70% ethanol, and radioactivity was counted in a scintillation counter.
RESULTS
p85α-, AS53-, and p50α-associated PI 3-kinase activity and binding to IRS proteins.
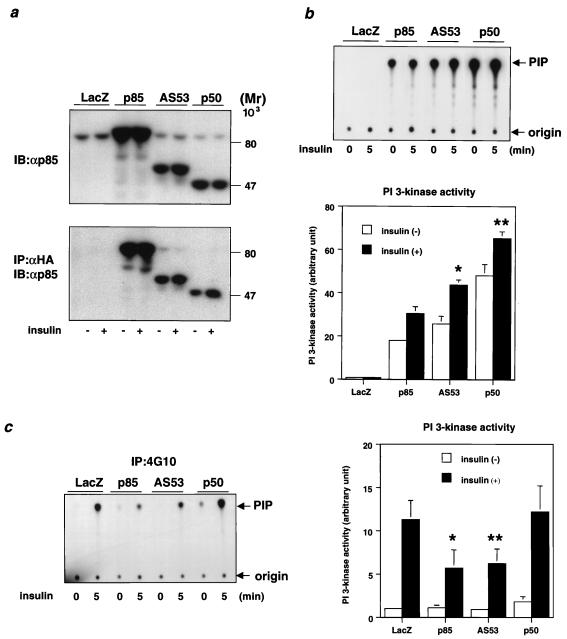
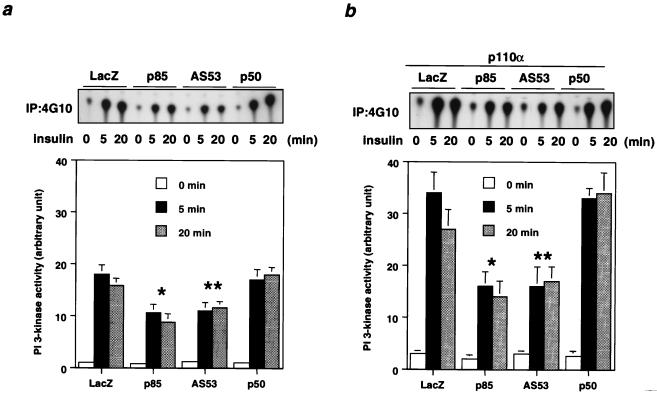
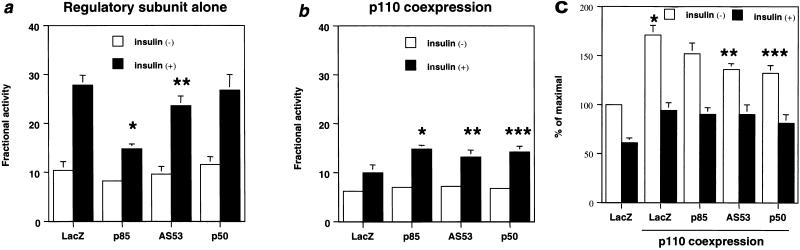
To elucidate the physiological role of each regulatory subunit of PI 3-kinase in insulin signaling, we expressed p85α, AS53, or p50α with a C-terminal HA tag in fully differentiated L6 myotubes. Using adenovirus-mediated gene transfer, we have previously shown that this results in high and efficient expression without modulating the differentiated function of cells (36). Following infection, the expression level of each introduced subunit as estimated by Western blotting following αHA immunoprecipitation was similar and at least fivefold higher than that of endogenous p85α, which is the predominant isoform in L6 myotubes, although AS53 and p50α are also detectable at a very low level (Fig. 1a). Thus, the introduced protein, rather than endogenous p85α, is the dominant regulatory form for PI 3-kinase signaling in the transfected cells. To assess PI 3-kinase activity associated with each introduced protein, cell extracts were specifically immunoprecipitated with αHA or 4G10, and the PI 3-kinase activity was estimated for each isoform. As shown in Fig. 1b, the PI 3-kinase specific activity in the αHA precipitates associated with p50α was about two or three times higher than that with p85α, and the activity associated with AS53 was intermediate between those with p85α and p50α. By contrast, PI 3-kinase activity (shown as mean ± standard deviation [SD]) in the 4G10 precipitates demonstrated that overexpression of p85α or AS53, but not p50α, significantly decreased the activity associated with tyrosine-phosphorylated proteins in response to insulin compared with the LacZ-infected control (Fig. 1c).
FIG. 1.
Transient expression of regulatory subunits and PI 3-kinase activities associated with each isoform and tyrosine-phosphorylated proteins. (a) Expression level of each regulatory subunit. Fully differentiated L6 myotubes were infected with the indicated adenoviruses at an MOI of 20 as described in Materials and Methods. After culturing in medium with 2% serum for 24 h, cells were starved for 20 h and then stimulated with 100 nM insulin for 5 min. Cell lysates were subjected to SDS-PAGE (9% gel) followed by immunoblotting (IB) with αp85pan (top) or immunoprecipitation (IP) with αHA. The immunoprecipitates were also subjected to Western blotting with αp85pan (bottom). (b) PI 3-kinase activity associated with each regulatory subunit. The αHA immunoprecipitates were subjected to PI 3-kinase assay as described in Materials and Methods. The top panel shows a representative result; each bar in the bottom panel represents the mean ± SD of the relative PI-3 kinase activity normalized for the expression level of each regulatory subunit as calculated from at least three independent experiments. (∗, P < 0.01 p85 versus AS53; ∗∗, P < 0.01 AS53 versus p50). (c) PI 3-kinase activity associated with tyrosine-phosphorylated proteins in cells expressing each regulatory subunit isoform. The immunoprecipitates were prepared using antiphosphotyrosine antibody 4G10 and subjected to a PI 3-kinase assay as described in Materials and Methods. The left panel shows a representative result; each bar in the right panel represents the mean ± SD of the relative PI-3 kinase activity calculated from at least four independent experiments (∗, P < 0.01 LacZ versus p85; ∗∗, P < 0.01 LacZ versus AS53).
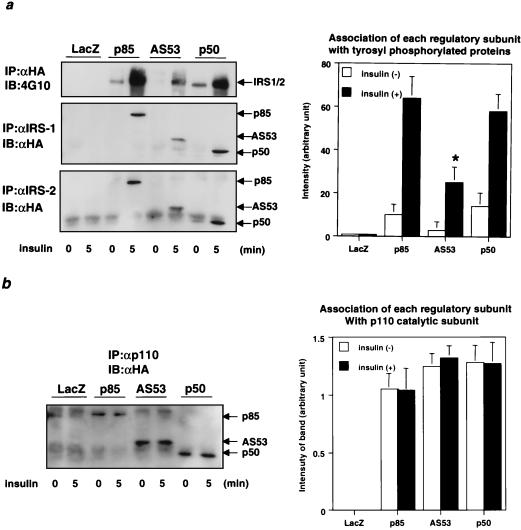
Insulin-stimulated PI 3-kinase activity depends on the critical role of the regulatory subunit to link a phosphorylated IRS protein with the p110 catalytic subunit of PI 3-kinase. To assess the interaction between each regulatory subunit and IRS proteins, we performed Western blotting with αHA after immunoprecipitation with αIRS-1 or αIRS-2. The results revealed that both p85α and p50α had a high affinity for tyrosine-phosphorylated IRS-1 and IRS-2, while AS53 had a much lower affinity for both proteins; the higher binding level of p85α and p50α than of AS53 to tyrosine-phosphorylated IRS proteins was also apparent after normalization by expression level (Fig. 2a). Similar results were observed in NIH 3T3 cells overexpressing human insulin receptor, in which regulatory subunits were overexpressed using plasmid expression vectors (data not shown). The affinity of each regulatory subunit for the p110 catalytic subunit normalized for expression level was also estimated by immunoprecipitation of cell lysates using αp110pan followed by Western blotting with αHA. p50α and AS53 had similar affinities for p110, while the binding of p85α was slightly lower, although the difference did not reach statistical significance (Fig. 2b). These data indicate that all three isoforms of regulatory subunit bind strongly to the p110 catalytic subunit and that both p85α and p50α have a higher affinity for IRS proteins than AS53. Furthermore, when overexpressed in cells, p85α and AS53 (but not p50α) inhibit phosphotyrosine-associated PI 3-kinase activity.
FIG. 2.
Affinities of each isoform of regulatory subunit for IRS proteins and p110 catalytic subunit. (a) Affinity of each isoform for tyrosine-phosphorylated proteins. Fully differentiated L6 myotubes were infected with the indicated adenoviruses at an MOI of 20. Cells were stimulated with 100 nM insulin for 5 min. Cell lysates were subjected to immunoprecipitation (IP) with αHA followed by immunoblotting (IB) with 4G10 (upper left). They were also subjected to immunoprecipitation with αIRS-1 (middle left) or αIRS-2 (lower left) followed by Western blotting with αHA. In the right panel, each bar represents the mean ± SD of the relative amount of tyrosine-phosphorylated proteins associated with each isoform normalized for its expression level calculated from the results of at least four independent experiments. (b) Affinity of each regulatory subunit isoform to p110. Cell lysates were subjected to immunoprecipitation with αp110pan followed by Western blotting with αHA. Shown are a representative result (left) and the mean ± SD of the relative amount of each isoform associated with p110 normalized for its expression level from at least four independent experiments (right).
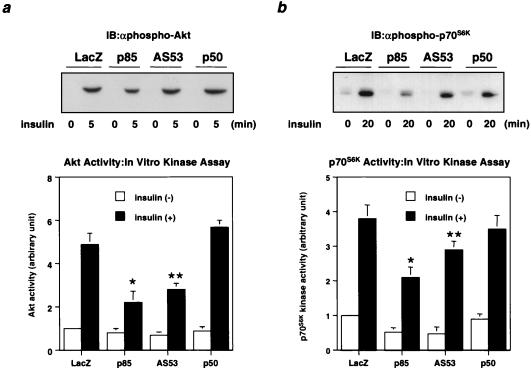
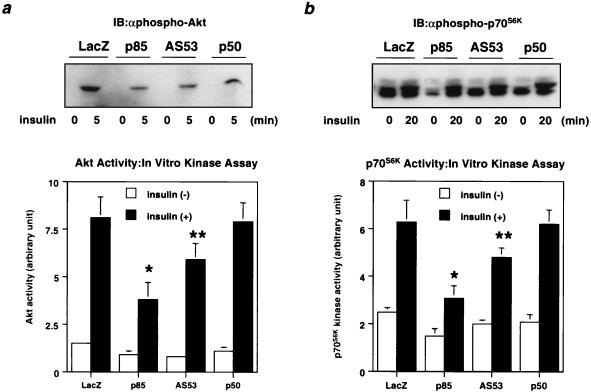
In the cascade of insulin signaling, Akt and p70S6K lie downstream of PI 3-kinase. Both Akt and p70S6K activities were decreased in cells expressing each isoform to a level comparable to the PI 3-kinase activity associated with phosphotyrosine, although the reduction of p70S6K activity by expression of each regulatory subunit was less than that of Akt (Fig. 3).
FIG. 3.
Effect of PI 3-kinase regulatory subunit isoform expression on downstream kinases. (a) Insulin-induced Akt activity in cells expressing each regulatory isoform. Fully differentiated L6 myotubes were infected with the indicated adenoviruses at an MOI of 20 and 2 days later stimulated with 100 nM insulin for 5 min as described in Materials and Methods. Cell lysates were subjected to SDS-PAGE (9% gel) followed by immunoblotting (IB) with αphospho-Akt (top) or immunoprecipitation with αAkt. The immunoprecipitates were subjected to an immune complex kinase assay. In the lower panel, each bar represents the mean ± SD of the relative Akt kinase activity calculated from at least four independent experiments (∗, P < 0.01 LacZ versus p85; ∗∗, P < 0.01 LacZ versus AS53). (b) Insulin-induced p70S6K activity in cells expressing each regulatory isoform. After a 20-min stimulation with 100 nM insulin, cell lysates were subjected to SDS-PAGE (9% gel) followed by Western blotting with αphospho-p70S6K (top) or immunoprecipitation with αp70S6K. The immunoprecipitates were subjected to an immune complex kinase assay. In the lower panel, each bar represents the mean ± SD of the relative p70S6K kinase activity calculated from at least four independent experiments (∗, P < 0.01 LacZ versus p85; ∗∗; P < 0.05 LacZ versus AS53).
Effect of each regulatory subunit on the catalytic activity of p110 PI 3-kinase.
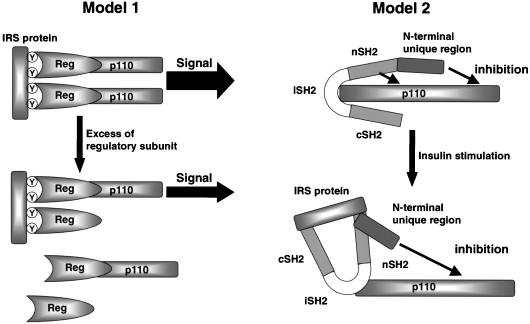
There are at least two possible mechanisms for the inhibitory effect of the regulatory subunits on the PI 3-kinase-dependent signaling. One is that overexpression of a regulatory subunit results in an increase in a monomeric form of the subunit occupying tyrosyl phosphorylation sites on IRS proteins. This would result in a secondary inhibition of insulin-stimulated PI 3-kinase activity and its downstream signaling cascade by competing for the active heterodimer (Fig. 4, left). Alternatively, it is possible that the regulatory subunit directly inhibits the catalytic activity of p110 subunit by some allosteric mechanism (Fig. 4, right).
FIG. 4.
Hypothetical models for the inhibitory mechanism of PI 3-kinase by the regulatory subunit. Model 1 shows how occupation of phosphorylation sites on IRS proteins by the monomeric form of regulatory subunits might decrease effective PI 3-kinase-mediated signals. Model 2 shows that regulatory subunits might exert a direct negative effect on p110 catalytic subunit activity.
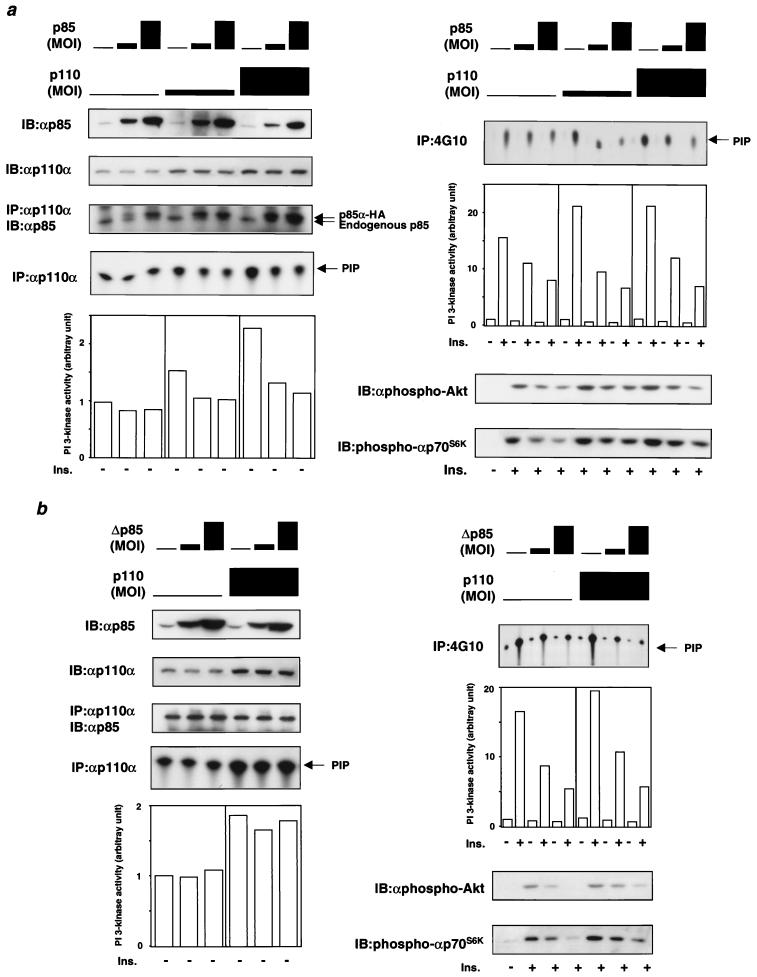
To evaluate these two alternations, we expressed p85α and p110α in various ratios and measured the PI-3 kinase activity associated with phosphotyrosine-containing proteins or the p110 catalytic subunit. We reasoned that if the inhibition was caused only by the binding of the regulatory subunits to phosphotyrosine residues on IRS proteins as a monomer, coexpression of p110 with the regulatory subunit should rescue the inhibition, and overexpression of the regulatory subunit would not affect the catalytic activity of the p110 subunit at least in the basal state. As shown in Fig. 5a, however, the PI 3-kinase activity associated with phosphotyrosine was decreased by increasing p85α expression, even when p85α was coexpressed with p110α. Coexpression of p110 and p85α produced similar effects on Akt and p70S6K activities. In the absence of exogenous p85α, both the Akt and p70S6K activities were increased by p110α expression, whereas these activities were significantly decreased by coexpression of p85α (Fig. 5a). The inhibitory effect on both kinases correlated with a decrease in phosphotyrosine protein-associated PI 3-kinase activity. On the other hand, the amount of p85 protein bound to p110α was increased by increasing p85α expression in cells expressing p110α, whereas it did not change in the absence of p110α expression (Fig. 5a, top left). These data suggest that in the absence of increased p110α expression, endogenous p110 is already saturated with the regulatory subunit, while in the presence of p110α overexpression, p110α is more abundant than the regulatory subunits and some portion of p110 exists as a monomer (at least in the absence of p85α overexpression). Under these conditions, the basal PI 3-kinase activity associated with p110 is decreased with increasing p85α expression (Fig. 5a, bottom left), supporting the hypothesis that the interaction between the regulatory subunits and the p110 subunit exerts an inhibitory effect on the catalytic activity of p110.
FIG. 5.
Effect of coexpression of p85α with p110α on PI 3-kinase activities and downstream kinases. (a) Expression of p85α decreases the PI 3-kinase activities associated with p110 and phosphotyrosine, leading to inhibition of downstream kinases from PI 3-kinase even when coexpressed p110α. Fully differentiated L6 myotubes were coinfected with the indicated adenoviruses at MOIs expressed as bars (representing, from left to right, MOIs of 0, 4, and 20, respectively) and stimulated with 100 nM insulin (Ins.) for 5 min. Cell lysates were subjected to immunoblotting (IB) with αp85α or αp110α (top two panels on left). They were also subjected to immunoprecipitation (IP) with αp110α followed by Western blotting with αp85α and an in vitro PI 3-kinase assay (middle two panels on left). The bottom left panel represents the mean of the relative PI-3 kinase activity calculated from two independent experiments. The immunoprecipitates were prepared using antiphosphotyrosine antibody 4G10 and subjected to PI 3-kinase assay (top panel on right). The middle right panel represents the mean of the relative PI-3 kinase activity calculated from two independent experiments. The bottom two panels show representative results of Western blotting with αphospho-Akt and αphospho-p70S6K. (b) Expression of a mutant p85α lacking the p110-binding site (Δp85) inhibits the PI 3-kinase activity associated with phosphotyrosine but not the activity associated with p110. Fully differentiated L6 myotubes were coinfected with the indicated adenoviruses at MOIs expressed as described above and stimulated with 100 nM insulin for 5 min. Other details are as described above.
To further confirm the existence of the direct inhibitory effect on p110 catalytic activity by the regulatory subunit, we expressed a mutant p85α lacking the p110-binding site (Δp85) in the presence of p110α expression (Fig. 5b). Overexpression of this mutant would be expected to inhibit insulin actions by occupying tyrosyl phosphorylation sites on IRS proteins but should not affect the p110 catalytic activity in the basal state by the direct inhibitory mechanism. As previously shown in other systems (14, 31), overexpression of Δp85 prominently decreased PI 3-kinase activity associated with tyrosine-phosphorylated proteins, thereby inhibiting Akt and p70S6K activities (Fig. 5b, right). However, it did not affect p110-associated PI 3-kinase activity. These data support the hypothesis that there is direct inhibition of the p110 catalytic subunit by the regulatory subunits and also suggest that an interaction between p110 and the regulatory subunit is required for this direct inhibitory mechanism.
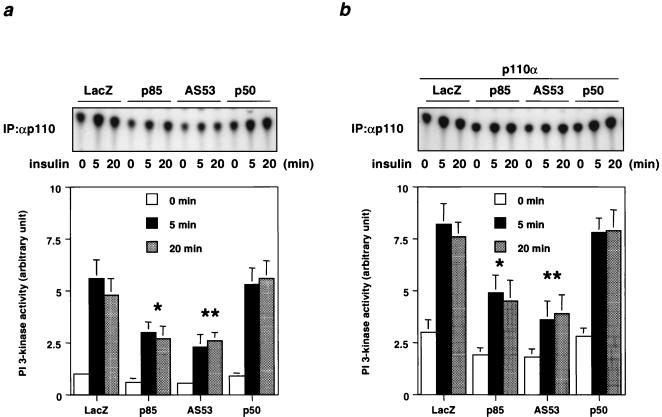
The short forms of regulatory subunit had differential effects on PI 3-kinase and downstream kinase activities. Thus, expression of AS53 reduced the PI 3-kinase activity associated with phosphotyrosine proteins with or without coexpression of p110α, whereas overexpression of p50α did not inhibit PI 3-kinase activity when expressed either alone or in the presence of overexpression of p110α (Fig. 6). Similarly, expression of AS53 or p85α decreased the PI 3-kinase activity associated with endogenous p110 (Fig. 7). As expected, overexpression of p110α increased p110-associated PI 3-kinase activity in both basal and insulin-stimulated states, and this was also inhibited by coexpression of p85α or AS53 but not p50α (Fig. 7).
FIG. 6.
Effect of coexpressing each regulatory subunit isoform with or without p110α on the PI 3-kinase activity associated with phosphotyrosine. (a) Time course of PI 3-kinase activity associated with phosphotyrosine in cells expressing each regulatory isoform. Fully differentiated L6 myotubes were infected with the indicated adenoviruses at an MOI of 20 and then stimulated with 100 nM insulin for the indicated period. Cell lysates were subjected to immunoprecipitation (IP) with 4G10 followed by a PI 3-kinase assay. Shown are a representative result (top) and the mean ± SD of the relative PI-3 kinase activity calculated from at least four independent experiments (∗, P < 0.01 LacZ versus p85; ∗∗; P < 0.01 LacZ versus AS53) (bottom). (b) Time course of PI 3-kinase activity associated with phosphotyrosine in cells coexpressing each regulatory isoform with p110α. Fully differentiated L6 myotubes were infected with the adenoviruses encoding the regulatory subunit and p110α at an MOI of 20. Following insulin stimulation, cells lysates were subjected to a PI 3-kinase assay as described above. Shown are a representative result (top) and the mean ± SD of the relative PI-3 kinase activity calculated from at least four independent experiments (∗, P < 0.01 LacZ versus p85; ∗∗, P < 0.01 LacZ versus AS53) (bottom).
FIG. 7.
Effect of coexpressing each regulatory isoform with or without p110α on the PI 3-kinase activity associated with p110. (a) Time course of PI 3-kinase activity associated with p110 in cells expressing each regulatory subunit isoform. Fully differentiated L6 myotubes were infected with the indicated adenoviruses at an MOI of 20 and then stimulated with 100 nM insulin for the indicated period. Cell lysates were subjected to immunoprecipitation (IP) with αp110α followed by a PI 3-kinase assay. Shown are a representative result (top) and the mean ± SD of the relative PI-3 kinase activity calculated from at least four independent experiments (∗, P < 0.05 LacZ versus p85; ∗∗, P < 0.05 LacZ versus AS53) (bottom). (b) Time course of PI 3-kinase activity associated with p110 in cells coexpressing each regulatory subunit isoform with p110α. Fully differentiated L6 myotubes were infected with the adenoviruses of the regulatory subunit and p110α at an MOI of 20. Lysates from insulin-treated cells were subjected to a PI 3-kinase assay. Shown are a representative result (top) and the mean ± SD of the relative PI-3 kinase activity calculated from at least four independent experiments (∗, P < 0.05 LacZ versus p85; ∗∗, P < 0.05 LacZ versus AS53) (bottom).
Again, Akt and p70S6K activities paralleled the changes in the PI 3-kinase activity associated with phosphotyrosine. Thus, Akt and p70S6K activities were increased by p110α expression compared with the LacZ control, and coexpression of p85α or AS53 reduced this enhancement by p110, although for p70S6K activity, p110α expression increased the basal activity significantly more than with Akt. Coexpression of p50α, on the other hand, had no effect on the p110 stimulation of Akt or p70S6K (Fig. 8). These results are consistent with the notion that the association of p85α and AS53 with p110 inhibits the catalytic activity of PI 3-kinase and secondarily decreases activities of downstream enzymes.
FIG. 8.
Effect of coexpressing each regulatory isoform with p110 on downstream kinases from PI 3-kinase. (a) Insulin-induced Akt activity in cells coexpressing each regulatory isoform with p110α. Fully differentiated L6 myotubes were infected with the indicated adenoviruses at an MOI of 20 and then stimulated with 100 nM insulin for 5 min. Cell lysates were subjected to SDS-PAGE (9% gel) followed by immunoblotting (IB) with αphospho-Akt (top) or immunoprecipitation with αAkt. The immunoprecipitates were subjected to an immune complex kinase assay. In the lower panel, each bar represents the mean ± SD of the relative Akt kinase activity calculated from at least four independent experiments (∗, P < 0.01 LacZ versus p85; ∗∗, P < 0.05 LacZ versus AS53). (b) Insulin-induced p70S6K activity in cells coexpressing each regulatory isoform with p110α. After a 20-min stimulation with 100 nM insulin, cell lysates were subjected to SDS-PAGE (9% gel) followed by Western blotting with αphospho-p70S6K (top) or immunoprecipitation with αp70S6K. The immunoprecipitates were subjected to an immune complex kinase assay. In the lower panel, each bar represents the mean ± SD of the relative p70S6K kinase activity calculated from at least four independent experiments (∗, P < 0.01 LacZ versus p85; ∗∗, P < 0.05 LacZ versus AS53).
The regulatory subunits modulate glucose transport and GS activity by PI 3-kinase-dependent and -independent mechanisms.
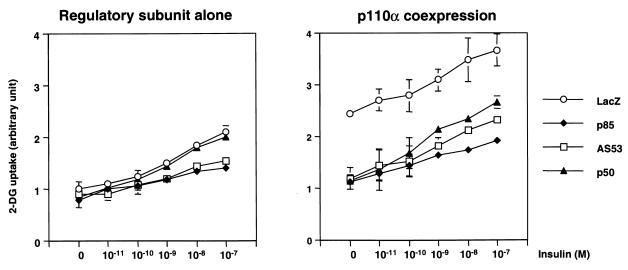
Most of insulin's major metabolic effects, including stimulation of glucose transport and glycogen synthesis in skeletal muscle, lie downstream of PI 3-kinase (3, 7, 23). When an individual regulatory subunit was expressed without the p110 catalytic subunit, both p85α and AS53 produced a significant decrease in insulin-dependent glucose transport activity (Fig. 9). By contrast, cells expressing p50α possessed glucose transport activity compared with the control level (Fig. 9). On the other hand, overexpression of p110α increased glucose transport activity, even in the basal state, to almost the same level as the maximal activity induced by insulin in the control cells (Fig. 9). As in the normal cells, coexpression of any regulatory subunit with p110α significantly decreased the enhanced glucose transport activity, and the magnitude of this decrease by p85α or AS53 coexpression was larger than that produced by p50α coexpression (Fig. 9b). It is worth noting that cells coexpressing p50α with p110α have almost the same level of PI 3-kinase, Akt, and p70S6K activities as cells expressing p110α alone but still exhibit reduced glucose transport. Thus, this inhibition of glucose transport may be due to an effect other than modulation of PI 3-kinase activity.
FIG. 9.
Effect of expressing each regulatory isoform with or without p110α on insulin-induced glucose transport activity. Cells were grown in 12-well dishes and infected with the indicated adenoviruses at an MOI of 20. One day after infection, cells were treated with the indicated concentration of insulin and subjected to 2-DG uptake assay as described in Materials and Methods. The results are expressed as the ratio to the value of untreated cells expressing LacZ. Each bar represents the mean ± SD of at least four independent experiments.
When the regulatory subunits were expressed alone, the GS activity stimulated by insulin correlated with the PI 3-kinase activity associated with phosphotyrosine proteins, i.e., was decreased in the order p85α > AS53 > p50α (Fig. 10a, left), although the difference between AS53 and p50α does not reach statistical significance. Although some studies have suggested that PI 3-kinase activity is required for GS activation (30, 39), to our surprise, expression of p110α alone dramatically decreased GS activity compared with the LacZ controls (Fig. 10a, right). This finding, however, is consistent with some recent studies indicating that expression of the wild type or an activated form of the catalytic subunit of PI 3-kinase expression can inhibit GS activity (8, 9). On the other hand, Akt activity, which has been shown to be sufficient for GS activation in L6 cells (7, 36), was increased to a level in cells expressing p110α comparable with that in cells expressing LacZ. Interestingly, in cells expressing p110α, GSK3, an enzyme immediately downstream from Akt that negatively regulates GS activity, was significantly increased in the basal state and remained at the almost same level after insulin stimulation as the basal activity in cells expressing LacZ (Fig. 10b). This increase in GSK3 activity could contribute to the poor activation of GS by insulin in cells expressing p110α.
FIG. 10.
Effect of expressing each regulatory isoform with or without p110α on GS and GSK3 activity (a and b) GS activity. One day after infection with the indicated adenoviruses at an MOI of 20, cells were starved for 20 h. They were treated with 100 nM insulin for 30 min and then subjected to GS assay as described in Materials and Methods. Each result was converted to the activity ratio determined by dividing the activity measured with 0.25 mM glucose-6-phosphate (ligand-dependent activity) by the activity measured with 10 mM glucose-6-phosphate (total activity). Each bar represents the mean ± SD of at least four independent experiments. (a) GS activity in the absence of p110α expression (∗, P < 0.01 LacZ versus p85; ∗∗, P < 0.05 LacZ versus AS53); (b) GS activity in the presence of p110α expression (∗, P < 0.01 LacZ versus p85; ∗∗, P < 0.01 LacZ versus AS53; ∗∗∗, P < 0.01 LacZ versus p50). (c) GSK3 kinase activity. One day after infection with the indicated adenoviruses at an MOI of 20, cells were starved for 20 h, then treated with 100 nM insulin for 20 min, and immunoprecipitated with αGSK3α. The immunoprecipitates were used for a kinase assay as described in Materials and Methods. The results are expressed as percentage of the maximum value for untreated cells expressing LacZ. Each bar represents the mean ± SD of at least four independent experiments (∗, P < 0.01 LacZ versus Lacz with p110; ∗∗, P < 0.01 LacZ with p110 versus AS53 with p110; ∗∗∗, P < 0.01 LacZ with p110 versus p50 with p110).
DISCUSSION
Over the past several years, a great deal of evidence indicating a central role of PI 3-kinase in the metabolic actions of insulin has accumulated (34, 37). PI 3-kinase activity has been shown to be required and, in some cases, sufficient for a variety of insulin's metabolic and mitogenic actions, including glucose transport (5, 9, 14, 21, 26), glycogen synthesis (30, 39), protein synthesis (24), and DNA synthesis (5, 9). PI 3-kinase is a heterodimer in which both regulatory and catalytic subunits occur in multiple isoforms as a result of products of different genes and alternative splicing (11, 34). It has been shown that regulatory subunits p85α, AS53/p55α, and p50α (products of the p85α gene) are involved in insulin signaling, and it has been suggested that each may have a specific physiological role (2, 19, 33). Disruption of all spliced isoforms of the p85α gene results in neonatal lethality (12), but recent studies in our lab suggest that in a heterozygous state, there is an increase in insulin sensitivity, possibly due to more efficient coupling of p85α and p110(F. Mauvais-Jarvis, K. Ueki, D. Fruman, D. Accili, L. C. Cantley, and C. R. Kahn, submitted for publication). Transgenic mice lacking only the long form of p85α exhibit hypersensitivity to insulin, suggesting compensatory and possibly even improved signaling by p50α or other short isoforms (35). Furthermore, we and others have shown that in insulin-resistant obese animals, expression of p85α in liver is decreased, while expression of AS53/p55α and p50α is greater than that in their lean littermates (1, 22). Taking into consideration the data for knockout mice, the alteration in expression of the regulatory subunits in these obese animals may be a compensatory reaction to the insulin-resistant state. These findings suggest that different splice isoforms of p85α gene may be required for normal metabolism and development and that each isoform may have certain distinct signaling characteristics. Indeed, several reports have suggested that each isoform has a distinct affinity for p110 and phosphorylated proteins (2, 19, 33), although the mechanism and the physiological implication of this are still unclear.
In this study, we have shown that in one of the tissues physiologically important for glucose metabolism, skeletal muscle, the PI 3-kinase activity associated with p50α is greater than that associated with p85α or AS53. This difference in PI 3-kinase activity associated with each regulatory subunit could be explained by the difference in affinity of each for p110 or IRS proteins. In this regard, p85α and p50α bind tyrosine-phosphorylated IRS proteins more efficiently than does AS53, while AS53 and p50α have slightly higher affinity for p110 than does p85α. Thus, it is not likely that the affinities for p110 and IRS proteins are only factors defining the PI 3-kinase activity mediated by each regulatory subunit. As noted by others (19, 21), the overall level of insulin stimulation is small in αHA or αp85pan (data not shown) precipitates, suggesting that only a small portion of the regulatory-catalytic subunit complex binds to IRS proteins and is activated by insulin.
In cells expressing p50α, the PI 3-kinase activity associated with phosphotyrosine, which tends to reflect the intensity of the signals to biological responses, is much greater than that in cells expressing p85α or AS53. Interestingly, expression of p85α or AS53, but not p50α, decreases PI 3-kinase activity and activation of the downstream kinases, Akt and p70S6K, compared to the LacZ expression control. One of the possible explanations for this finding would be that when the regulatory subunits are overexpressed without additional catalytic subunits, they occupy phosphorylation sites on IRS proteins as monomers, thereby inhibiting effective PI 3-kinase signaling by the PI 3-kinase heterodimer. Indeed, this type of competitive inhibition has been shown by overexpression of a signaling-incompetent mutant of p85, such as the p85α mutant lacking the p110-binding site (Δp85) (14, 31) or the isolated SH2 domains of p85α (32). This explains why the phosphotyrosine-associated PI 3-kinase activity in cells overexpressing p85α is less than that in control cells which express endogenous p85α. It can also explain why expression of p50α does not increase the phosphotyrosine-associated PI 3-kinase activity above the control level, despite the fact that p50α-associated PI 3-kinase activity is higher than that associated with either p85α or AS53. However, this may not be the only inhibitory mechanism of PI 3-kinase-dependent signaling by the regulatory subunits, since overexpression of p85α or AS53, but not p50α, also decreases p110-associated PI 3-kinase activity in the basal state. This effect is more pronounced in the presence of coexpression of p110, probably because endogenous p110 seems to be almost saturated with the regulatory subunits. This latter effect appears to be due to a direct inhibitory effect of the regulatory subunits on p110 catalytic activity, suggesting allosteric interactions between these subunits. Expression of Δp85 fails to inhibit basal p110 activity, even in the presence of p110α expression, supporting the existence of this allosteric inhibition.
The inhibitory effect of the regulatory subunits appears to depend on the structure of the N terminus of the molecule, since this is the only region that differs among these three isoforms. It is still unclear, however, whether the regulatory subunit inhibits the p110 catalytic activity directly or through other molecules that interact with N-terminal region of the regulatory subunit. Indeed, we have obtained several clones interacting with N-terminal unique region of AS53 using the yeast two-hybrid system (K. Ueki and C. R. Kahn, unpublished data), and there are several molecules which are known to interact with the N-terminal half of p85 (4, 13, 16, 28). Thus, it is possible that the specific protein which interacts with the N-terminal region of each regulatory subunit contributes to differential modulation of the p110 catalytic activity.
This study is the first demonstrating that the regulatory subunits can inhibit p110 activity and PI 3-kinase-dependent signaling in vivo with different efficiencies. Yu and coworkers have previously shown both inhibition and stabilization of p110 by the p85 regulatory subunit in in vitro systems (40, 41). Recently, Harpur and coworkers have shown that the p85 regulatory subunit may also exist as a homodimer through the intermolecular interaction of the N-terminal proline-rich region and the SH3 domain, or possibly intermolecular interaction of Bcr homology domains (15). This dimerization might contribute to the regulation of PI 3-kinase by p85α but cannot explain the effects of AS53 or p50α, both of which lack these regions. Regardless of mechanism, the data suggest that PI 3-kinase behaves as a classical allosteric enzyme in which the regulatory subunits negatively regulate the catalytic activity. The interaction of the regulatory subunit with tyrosine-phosphorylated proteins reduces this inhibitory effect, resulting in stimulation of PI 3-kinase activity.
With regard to the final biological effects mediated by insulin stimulation, our data are in agreement with those of Katagiri et al. (21), who found that expression of p110α increases glucose transport. This, however, has not been observed in all studies (9). The differences may be explained by the observation that p110α has different levels of stability and activity depending on whether it has an N-terminal or C-terminal tag (41). Our p110α construct has an N-terminal Myc tag, which may contribute to stability and activity of p110 observed in this study. Nonetheless, coexpression of any regulatory subunit isoform markedly reduces this p110 effect on glucose transport. This is inconsistent with the fact that coexpression of p50α with p110α has little effect on phosphotyrosine- and p110-associated PI 3-kinase activity compared to cells expressing p110α alone. These data suggest that the regulatory subunits can modulate some downstream signaling by direct effects on PI 3-kinase activity, as well as indirect mechanisms, independent of the level of PI 3-kinase activity.
One possibility for an indirect mechanism of regulation is an alteration of the subcellular distribution of the catalytic subunit and the intracellular site of PI 3-kinase activity. Indeed, recent studies have revealed that specific subcellular compartmentalization of the signaling complexes with PI 3-kinase following insulin stimulation may contribute to the unique metabolic actions by insulin (6, 17). Little is known about the mechanisms of intracellular trafficking of these molecules. However, since each IRS protein seems to have a unique trafficking characteristics upon insulin stimulation (6), the affinity of each regulatory subunit for these docking proteins may reflect the subcellular localization of PI 3-kinase, thereby regulating PI 3-kinase-dependent biological activity. Preliminary experiments in our lab using conventional cell fractionation have failed to detect significant differences in the distribution pattern of p110 or PI 3-kinase activity between cells expressing p110α alone or with coexpression of each regulatory subunit (data not shown); however, more detailed studies on this point are needed. Furthermore, it is possible that there exist several compartments with different efficiencies for different PI 3-kinase signaling events, even in the same fraction separated by the conventional fractionation method. Indeed, expression of p110 monomer results in a marked decrease GS activity, presumably through an increase in GSK3 activity. Expression of p110 or a constitutively active mutant p110 has also been shown to inhibit GS activity in 3T3-L1 adipocytes (8, 9). These results suggest that p110 monomer, usually unstable, exists in the particular compartment which is appropriate for support of glucose transport but disadvantageous for activation of GS. If this is in the case, the interaction with the various regulatory subunits might recruit p110 to compartments which can release the appropriate signals for normal insulin signaling.
In summary, our data demonstrate that the various regulatory subunits modulate PI 3-kinase-dependent signaling by at least three different mechanisms: occupation of IRS proteins by the regulatory subunit monomer, inhibition of catalytic activity, and possibly alteration of the subcellular compartment. As a result, they relay the signals from IRS proteins to PI 3-kinase with different efficiencies. These findings indicate that changes in the level of expression of each regulatory subunit can lead to major alterations in insulin signals in different insulin-responsive tissues and potentially contribute to an insulin-resistant state, such as diabetes mellitus.
ACKNOWLEDGMENTS
This work was supported by NIH grants DK 33201 and DK55545.
We thank M. Kasuga for the adenovirus encoding Δp85, L. C. Cantley for the p110 construct, I. Saito for the cosmid cassette and control adenovirus, and T. L. Bellman-Azar and J. Konigsberg for excellent secretarial assistance.
REFERENCES
- 1.Anai M, Funaki M, Ogihara T, Terasaki J, Inukai K, Katagiri H, Fukushima Y, Yazaki Y, Kikuchi M, Oka Y, Asano T. Altered expression levels and impaired steps in the pathway to phosphatidylinositol 3-kinase activation via insulin receptor substrates 1 and 2 in Zucker fatty rats. Diabetes. 1998;47:13–23. doi: 10.2337/diab.47.1.13. [DOI] [PubMed] [Google Scholar]
- 2.Antonetti D A, Algenstaedt P, Kahn C R. Insulin receptor substrate 1 binds two novel splice variants of the regulatory subunit of phosphatidylinositol 3-kinase in muscle and brain. Mol Cell Biol. 1996;16:2195–2203. doi: 10.1128/mcb.16.5.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger J, Hayes N, Szalkowski D M, Zhang B. PI 3-kinase activation is required for insulin stimulation of glucose transport into L6 myotubes. Biochem Biophys Res Commun. 1994;205:570–576. doi: 10.1006/bbrc.1994.2703. [DOI] [PubMed] [Google Scholar]
- 4.Bokoch G M, Vlahos C J, Wang Y, Knaus U G, Traynor-Kaplan A E. Rac GTPase interacts specifically with phosphatidylinositol 3-kinase. Biochem J. 1996;315:775–779. doi: 10.1042/bj3150775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheatham B, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark S F, Martin S, Carozzi A J, Hill M M, James D E. Intracellular localization of phosphatidylinositide 3-kinase and insulin receptor substrate-1 in adipocytes: potential involvement of a membrane skeleton. J Cell Biol. 1998;140:1211–1225. doi: 10.1083/jcb.140.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 8.Egawa K, Sharma P M, Nakashima N, Huang Y, Huver E, Boss G R, Olefsky J M. Membrane-targeted phosphatidylinositol 3-kinase mimics insulin actions and induces a state of cellular insulin resistance. J Biol Chem. 1999;274:14306–14314. doi: 10.1074/jbc.274.20.14306. [DOI] [PubMed] [Google Scholar]
- 9.Frevert E U, Kahn B B. Differential effects of constitutively active phosphatidylinositol 3-kinase on glucose transport, glycogen synthase activity, and DNA synthesis in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:190–198. doi: 10.1128/mcb.17.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fruman D A, Cantley L C, Carpenter C L. Structural organization and alternative splicing of the murine phosphoinositide 3-kinase p85 alpha gene. Genomics. 1996;37:113–121. doi: 10.1006/geno.1996.0527. [DOI] [PubMed] [Google Scholar]
- 11.Fruman D A, Meyers R E, Cantley L C. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 12.Fruman D A, Snapper S B, Yballe C M, Davidson L, Yu J Y, Alt F W, Cantley L C. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 13.Gout I, Dhand R, Hiles I D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 14.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson T R, Hawkins P T, Dahnd R, Clark A E, Holman G D, Waterfield M D, Kasuga M. 1-Phosphatidylinositol 3-kinase activity is required for insulin-stimulated glucose transport but not for RAS activation in CHO cells. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harpur A G, Layton M J, Das P, Bottomley M J, Panayotou G, Driscoll P C, Waterfield M D. Intermolecular interactions of the p85alpha regulatory subunit of phosphatidylinositol 3-kinase. J Biol Chem. 1999;274:12323–12332. doi: 10.1074/jbc.274.18.12323. [DOI] [PubMed] [Google Scholar]
- 16.Hunter S, Koch B L, Anderson S M. Phosphorylation of cbl after stimulation of Nb2 cells with prolactin and its association with phosphatidylinositol 3-kinase. Mol Endocrinol. 1997;11:1213–1222. doi: 10.1210/mend.11.9.9980. [DOI] [PubMed] [Google Scholar]
- 17.Inoue G, Cheatham B, Emkey R, Kahn C R. Dynamics of insulin signaling in 3T3-L1 adipocytes. Differential compartmentalization and trafficking of insulin receptor substrate (IRS)-1 and IRS-2. J Biol Chem. 1998;273:11548–11555. doi: 10.1074/jbc.273.19.11548. [DOI] [PubMed] [Google Scholar]
- 18.Inukai K, Anai M, Van Breda E, Hosaka T, Katagiri H, Funaki M, Fukushima Y, Ogihara T, Yazaki Y, Kikuchi M, Oka Y, Asano T. A novel 55-kDa regulatory subunit for phosphatidylinositol 3-kinase structurally similar to p55PIK is generated by alternative splicing of the p85alpha gene. J Biol Chem. 1996;271:5317–5320. doi: 10.1074/jbc.271.10.5317. [DOI] [PubMed] [Google Scholar]
- 19.Inukai K, Funaki M, Ogihara T, Katagiri H, Kanda A, Anai M, Fukushima Y, Hosaka T, Suzuki M, Shin B C, Takata K, Yazaki Y, Kikuchi M, Oka Y, Asano T. p85alpha gene generates three isoforms of regulatory subunit for phosphatidylinositol 3-kinase (PI 3-kinase), p50alpha, p55alpha, and p85alpha, with different PI 3-kinase activity elevating responses to insulin. J Biol Chem. 1997;272:7873–7882. doi: 10.1074/jbc.272.12.7873. [DOI] [PubMed] [Google Scholar]
- 20.Kahn C R. Banting Lecture. Insulin action, diabetogenes, and the cause of type II diabetes. Diabetes. 1994;43:1066–1084. doi: 10.2337/diab.43.8.1066. [DOI] [PubMed] [Google Scholar]
- 21.Katagiri H, Asano T, Ishihara H, Inukai K, Shibasaki Y, Kikuchi M, Yazaki Y, Oka Y. Overexpression of catalytic subunit p110alpha of phosphatidylinositol 3-kinase increases glucose transport activity with translocation of glucose transporters in 3T3-L1 adipocytes. J Biol Chem. 1996;271:16987–16990. doi: 10.1074/jbc.271.29.16987. [DOI] [PubMed] [Google Scholar]
- 22.Kerouz N J, Horsch D, Pons S, Kahn C R. Differential regulation of insulin receptor substrates-1 and -2 (IRS-1 and IRS-1) and phosphatidylinositol 3-kinase isoforms in liver and muscle of the obese diabetic (ob/ob) mouse. J Clin Investig. 1997;100:3164–3172. doi: 10.1172/JCI119872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Marchand-Brustel Y, Gautier N, Cormont M, Van Obberghen E. Wortmannin inhibits the action of insulin but not that of okadaic acid in skeletal muscle: comparison with fat cells. Endocrinology. 1995;136:3564–3570. doi: 10.1210/endo.136.8.7628394. [DOI] [PubMed] [Google Scholar]
- 24.Mendez R, Myers M G, Jr, White M F, Rhoads R E. Stimulation of protein synthesis, eukaryotic translation initiation factor 4E phosphorylation, and PHAS-I phosphorylation by insulin requires insulin receptor substrate 1 and phosphatidylinositol 3-kinase. Mol Cell Biol. 1996;16:2857–2864. doi: 10.1128/mcb.16.6.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyake S, Makimura M, Kanegae Y, Harada S, Sato Y, Takamori K, Tokuda C, Saito I. Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc Natl Acad Sci USA. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada T, Kawano Y, Sakakibara T, Hazeki O, Ui M. Essential role of phosphatidylinositol 3-kinase in insulin-induced glucose transport and antilipolysis in rat adipocytes. Studies with a selective inhibitor wortmannin. J Biol Chem. 1994;269:3568–3573. [PubMed] [Google Scholar]
- 27.Otsu M, Hiles I, Gout I, Fry M J, Ruiz-Larrea F, Panayotou G, Thompson A, Dhand R, Hsuan J, Totty N. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- 28.Pleiman C M, Hertz W M, Cambier J C. Activation of phosphatidylinositol-3′ kinase by Src-family kinase SH3 binding to the p85 subunit. Science. 1994;263:1609–1612. doi: 10.1126/science.8128248. [DOI] [PubMed] [Google Scholar]
- 29.Pons S, Asano T, Glasheen E, Miralpeix M, Zhang Y, Fisher T L, Myers M G, Jr, Sun X J, White M F. The structure and function of p55PIK reveal a new regulatory subunit for phosphatidylinositol 3-kinase. Mol Cell Biol. 1995;15:4453–4465. doi: 10.1128/mcb.15.8.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaue H, Hara K, Noguchi T, Matozaki T, Kotani K, Ogawa W, Yonezawa K, Waterfield M D, Kasuga M. Ras-independent and wortmannin-sensitive activation of glycogen synthase by insulin in Chinese hamster ovary cells. J Biol Chem. 1995;270:11304–11309. doi: 10.1074/jbc.270.19.11304. [DOI] [PubMed] [Google Scholar]
- 31.Sakaue H, Ogawa W, Takata M, Kuroda S, Kotani K, Matsumoto M, Sakaue M, Nishio S, Ueno H, Kasuga M. Phosphoinositide 3-kinase is required for insulin-induced but not for growth hormone- or hyperosmolarity-induced glucose uptake in 3T3-L1 adipocytes. Mol Endocrinol. 1997;11:1552–1562. doi: 10.1210/mend.11.10.9986. [DOI] [PubMed] [Google Scholar]
- 32.Sharma P M, Egawa K, Huang Y, Martin J L, Huvar I, Boss G R, Olefsky J M. Inhibition of phosphatidylinositol 3-kinase activity by adenovirus-mediated gene transfer and its effect on insulin action. J Biol Chem. 1998;273:18528–18537. doi: 10.1074/jbc.273.29.18528. [DOI] [PubMed] [Google Scholar]
- 33.Shepherd P R, Nave B T, Rincon J, Nolte L A, Bevan A P, Siddle K, Zierath J R, Wallberg-Henriksson H. Differential regulation of phosphoinositide 3-kinase adapter subunit variants by insulin in human skeletal muscle. J Biol Chem. 1997;272:19000–19007. doi: 10.1074/jbc.272.30.19000. [DOI] [PubMed] [Google Scholar]
- 34.Shepherd P R, Withers D J, Siddle K. Phosphoinositide 3-kinase: the key switch mechanism in insulin signalling. Biochem J. 1998;333:471–490. doi: 10.1042/bj3330471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terauchi Y, Tsuji Y, Satoh S, Minoura H, Murakami K, Okuno A, Inukai K, Asano T, Kaburagi Y, Ueki K, Nakajima H, Hanafusa T, Matsuzawa Y, Sekihara H, Yin Y, Barrett J C, Oda H, Ishikawa T, Akanuma Y, Komuro I, Suzuki M, Yamamura K, Kodama T, Suzuki H, Kadowaki T. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 36.Ueki K, Yamamoto-Honda R, Kaburagi Y, Yamauchi T, Tobe K, Burgering B M, Coffer P J, Komuro I, Akanuma Y, Yazaki Y, Kadowaki T. Potential role of protein kinase B in insulin-induced glucose transport, glycogen synthesis, and protein synthesis. J Biol Chem. 1998;273:5315–5322. doi: 10.1074/jbc.273.9.5315. [DOI] [PubMed] [Google Scholar]
- 37.Virkamaki A, Ueki K, Kahn C R. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J Clin Investig. 1999;103:931–943. doi: 10.1172/JCI6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White M F, Kahn C R. The insulin signaling system. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 39.Yamamoto-Honda R, Tobe K, Kaburagi Y, Ueki K, Asai S, Yachi M, Shirouzu M, Yodoi J, Akanuma Y, Yokoyama S, et al. Upstream mechanisms of glycogen synthase activation by insulin and insulin-like growth factor-I. Glycogen synthase activation is antagonized by wortmannin or LY294002 but not by rapamycin or by inhibiting p21ras. J Biol Chem. 1995;270:2729–29234. doi: 10.1074/jbc.270.6.2729. [DOI] [PubMed] [Google Scholar]
- 40.Yu J, Wjasow C, Backer J M. Regulation of the p85/p110alpha phosphatidylinositol 3′-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J Biol Chem. 1998;273:30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 41.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr G A, Backer J M. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]