Abstract
Coronavirus (which is also known as COVID-19) is severely impacting the wellness and lives of many across the globe. There are several methods currently to detect and monitor the progress of the disease such as radiological image from patients’ chests, measuring the symptoms and applying polymerase chain reaction (RT-PCR) test. X-ray imaging is one of the popular techniques used to visualise the impact of the virus on the lungs. Although manual detection of this disease using radiology images is more popular, it can be time-consuming, and is prone to human errors. Hence, automated detection of lung pathologies due to COVID-19 utilising deep learning (Bowles et al.) techniques can assist with yielding accurate results for huge databases. Large volumes of data are needed to achieve generalizable DL models; however, there are very few public databases available for detecting COVID-19 disease pathologies automatically. Standard data augmentation method can be used to enhance the models’ generalizability. In this research, the Extensive COVID-19 X-ray and CT Chest Images Dataset has been used and generative adversarial network (GAN) coupled with trained, semi-supervised CycleGAN (SSA- CycleGAN) has been applied to augment the training dataset. Then a newly designed and finetuned Inception V3 transfer learning model has been developed to train the algorithm for detecting COVID-19 pandemic. The obtained results from the proposed Inception-CycleGAN model indicated Accuracy = 94.2%, Area under Curve = 92.2%, Mean Squared Error = 0.27, Mean Absolute Error = 0.16. The developed Inception-CycleGAN framework is ready to be tested with further COVID-19 X-Ray images of the chest.
Keywords: COVID19, Deep Learning, Transfer Learning, CycleGAN, Radiological image processing
1. Introduction
Coronavirus-2 (SARS-CoV-2) causes lung disease and by fast spreading affects the well-being of the global population. Identifying on-time the COVID-19 cases and monitoring the disease progress could prevent spreading it in the society and speed up treatment process [1]. Recently, polymerase chain reaction (RT-PCR) testing method employed to detect COVID-19 and identify SARS-CoV-2 ribonucleic acid (RNA) from respiratory specimens. The RT-PCR testing currently is the one of the top standards in detecting COVID-19, but it is very time-consuming, painful, and bit complicated manual process. Healthcare staff are required to engage in close contact with individuals to perform RT-PCR tests which may increase the risk of COVID-19 virus infection [1]. There are some other disadvantages in applying RT-PCR as well including, insufficient numbers of available test kits, costs, threats to the healthcare staff safety, and waiting time of test results [2].
An alternative method in identifying COVID-19 are chest imaging investigations, such as X-ray or computed tomography (CT) imaging which are analyzed by specialist radiologists to identify visual indicators associated with COVID-19 biological infection [2]. Early studies showed patients showed abnormalities in patients’ chest radiology images in COVID-19 infection such as ground-glass opacity, bilateral or interstitial abnormalities in chest X-Ray and CT scans. The radiology scans required to analysis by radiologist to identify COVID-19 from the images. Artificial intelligence (AI) based tools could improve the performance of the radiologist analysis. The AI and machine learning algorithms recently demonstrate remarkable progress in image-recognition tasks [2].
Wang et al. [2] have developed the COVID-Net, which is a deep convolutional neural network (DCNN) method to identify patients with COVID-19 from X-ray imaging. The applied chest radiography images dataset contains COVID-19, viral and bacterial pneumonia, and other infection classes. The overall accuracy obtained by their method is 83.5% in four classes and 92.4% overall accuracy for binary classes (i.e., COVID-19, and non-COVID groups). Hemdan et al. [3] have developed the COVIDX-Net AI algorithm to assist radiologists in identifying COVID-19 through chest X-rays. They introduced seven deep learning models involving VGG19, MobileNetV2, DenseNet201, ResNetV2, InceptionResNetV2, Xception and InceptionV3 applied these models in the COVIDX-Net algorithm. The results indicated that the effectiveness of the VGG19 and DenseNet was similar in terms of measuring F1-scores (0.91 and 0.89 respectively). On the other hand, Sethy and Behera [4] proposed a deep learning technique in X-ray scans database for the same issue by achieving an accuracy of 95.38%. Ozturk et al. [5] presented DarkNet or DarkCOVIDNet model and the proposed model has provided diagnoses for 3 classes, including positive COVID-19 cases, all types of pneumonia, and healthy classes. The model has been trained for binary classification, including COVID-19 and normal cases as well. The accuracy of 3-class classification is 87.02% and 98.08% for two classes.
CoroNet consists of a convolution neural network (CNN) has been introduced by Khan, Shah, et al. [6] for detecting COVID-19. The experimental results have an overall accuracy of 89.6% from X-ray and 95% from CT radiology images for four classes including bacterial pneumonia, COVID-19, other pneumonias, and normal. It also trained and evaluated three classes including COVID-19, pneumonia, and normal (or benign) cases. There is some other research in the same area that based on CT scans image analysis. For example, COVNet introduced by Li et al. [7] uses ResNet50 to classify COVID-19 from 4356 chest CT images and DeepPneumonia introduced by Song et al. [8] classifies patients into three groups: COVID-19, bacterial pneumonia, and healthy.
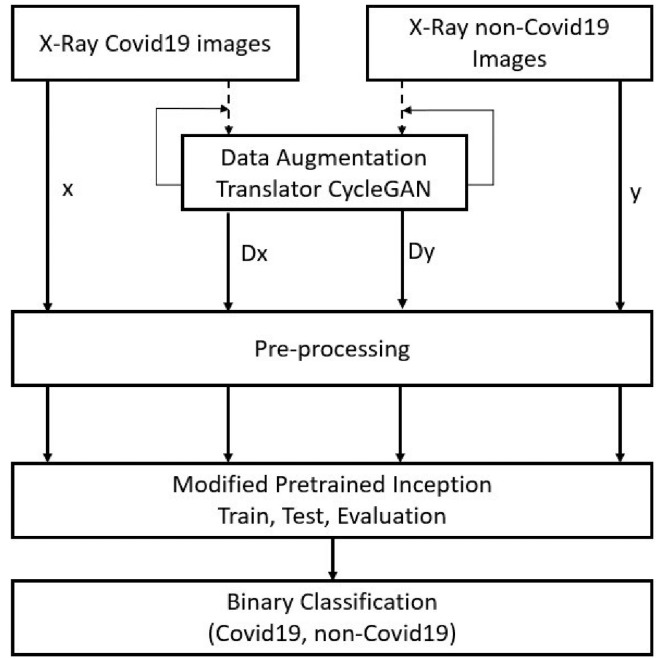
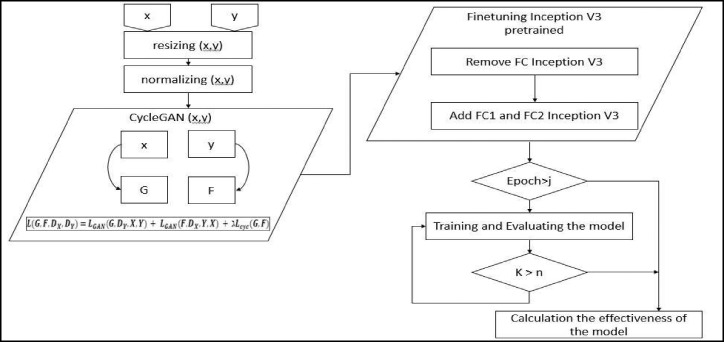
Although many deep learning techniques have been proposed to detect COVID-19 accurately [9,10,11,12,13,14], it is still challenging to detect it accurately using x-ray images alone. The current main issue in applying deep learning methods is that there are limited number of publicly available COVID-19 chest image databases available. The big data is needed to train the deep convolutional networks and generalize the developed models. To solve the limited availability of COVID-19 chest X-ray databases and for considering use of deep learning models to detect COVID-19, a new CycleGAN-Inception approach is proposed in this paper. Fig. 1 shows the proposed model's framework in COVID-19 automated detection. The Extensive COVID-19 X-ray and CT Chest Images Dataset [15] has been used for validation of the proposed model however for the training set we used the dataset in addition augmented data by CycleGAN has added to training dataset to train the proposed model. The database has collected from several COVID-19 radiological databases including the “Extensive-COVID-19-X-ray-and-CT-Chest-Images Dataset” [16] and the “Chest-Xray-Dataset” [17].
Fig. 1.
Proposed CycleGAN-Inception model. All COVID-19 (stream X) and non-COVID-19 (stream Y) X-ray images are from the selected databases. Dx and Dy data flow are the translated data by CycleGAN to deal with the data limitation problem.
For better performance of the proposed model, CycleGAN [18] has been used to generate and augment data by translating COVID-19 images to normal images and normal to COVID-19 images. The GAN methods have been previously used to produce new training images [19], refine synthetic image [20] and improve brain segmentation [21].
As shown in Fig. 1, data flow has four steps. Stream (x) presents all COVID-19 original X-ray image data from collected databases and stream (y) provides all non-COVID-19 original X-ray image data from the selected databases. The Dx and Dy are the generated translated data by CycleGAN. All original and generated X-ray image data are fed to the pre-processing section of the proposed model to undergo normalization, resizing, and centralization. The pre-processing section processes data to make it more suitable to feed the data to the proposed pre-trained Inception V3. The proposed Inception V3 pre-trained model is modified and fine-tuned by freezing fully connected layers and adding two fine-tuned fully contacted layers. The primary utility and significance of this work are listed as follows:
-
•
A novel data augmentation approach is developed to generate images with COVID-19 and normal characteristic using CycleGAN.
-
•
The developed novel model is trained and evaluated using original and translated image data using CycleGAN.
-
•
The newly designed model can identify positive COVID-19 cases from chest X-ray radiological images effectively and with high accuracy.
The rest of the paper is organized as the following: In Section 2 the structure and concepts of the proposed CycleGAN-Inception model is presented. Section 3 provides the experimental configuration, evaluation metrics, and collected data. Section 4 describes discussion and results of the trained and evaluated the CGAN-Inception proposed model and demonstrates its effectiveness. Finally, Section 5 presents the summary of the paper.
2. The proposed CycleGAN-Inception model
The CycleGAN-Inception model is developed to categorize chest X-ray images with or without COVID-19 characteristics (Fig. 1). The proposed approach has two steps: (i) data augmentation technique, the model generates extra X-ray images by applying CycleGAN introduced by Zhu et al. [18]. Then all data transfer to image pre-processing section for normalization and resizing input data. In the next step, all data transfer to the proposed and modified Inception V3 pre-trained model. More details of the components of the proposed model were described in the following subsections.
2.1. Data augmentation non COVID-19 to COVID-19 CycleGAN
A CycleGAN was introduced by Zhu et al. [18] as a model of training the deep CNNs in an image-to-image translation format. It also identifies dissimilar other generative adversarial networks developed by Goodfellow et al. [22] for image translation, allowing the CycleGAN to learn mapping between one image domain and another through an unsupervised approach [18]. The CycleGANs facilitates learning mapping from one area (X) to another area (Y), without identifying the matched training pairs. It is also often used in data augmentation. Sandfort et al. [23] evaluated the CycleGAN for data augmentation purpose by training CycleGAN in converting contrast CT scan images into non-contrast scans. GANs have been used in the past for data extension to produce unique training images set for classification purpose [19], refine synthetic images [20] and improve brain segmentation [21].
Since labelled medical imaging data can be both difficult and expensive to obtain, access to big medical data is essential in achieving an accurate and robust deep learning algorithm. Standard data augmentation is a routinely performed process which is used to increase generalizability. However, the GANs could offer a novel approach of data extension. Therefore, the newly trained CycleGAN with COVID-19 X-ray radiological images is applied in this work to augment existing models using non-contrast images.
Zhu et al. [18] and Godard et al. [24] used cycle consistency loss to supervise CNN training. Zhu et al. [18] have introduced a parallel loss to drive G and F with same coherence. Their model consists of mapping G: X→Y and F: Y→X by introducing adversarial discriminators of Dx and Dy. To distinguish between images {x} and translated images {F(y)}, Dx and to discriminate between {y} and {G(x)} Dy are applied. For more information about the principles and concepts of it the reader could refer to the Zhu et al. [18]. As a summary the definitions of components are describing mathematically as follows.
The adversarial losses are definition in Eq. (1):
| (1) |
In This formula, G generates images G(x) that appears like images from field Y and Dy observes between translated samples G(x) and original samples y.
The cycle consistency loss is defined as following in Eq. (2) as per an image y from domain Y, G and F are satisfying backward cycle consistency:
| (2) |
The cycle consistency loss is formulated (see Eq. (3)) as follow:
| (3) |
Therefore,
| (4) |
here conducts the relation of the two objectives. CycleGAN has been applied in the proposed model to translation of non-COVID-19 to COVID-19 images and vice versa as following:
The discriminators Dx and Dy are CNNs that read an image and classify it as true or false (real or fake). “True” is indicated by an output close to 1 and “false” as close to 0. The proposed architecture consists of five convolutional layers, which generate a single logit to detect whether the image is real or not. The architecture does not include a fully connected layer. The convolutional layers are then supported by batch normalization, and the rectified linear activation function (ReLU) for hidden units.
2.2. Proposed finetuned pretrained Inception V3
In reality, the datasets are often insufficient to train the deep learning algorithms and obtaining the labelled data can be costly. Transfer learning techniques are used to achieve higher performance of the machine learning algorithms by using labelled data or getting knowledge from similar domains [25]. It can implement the knowledge that has been realized in earlier settings. There are also two popular transfer learning strategies: deep feature extraction and fine-tuning [26,27].
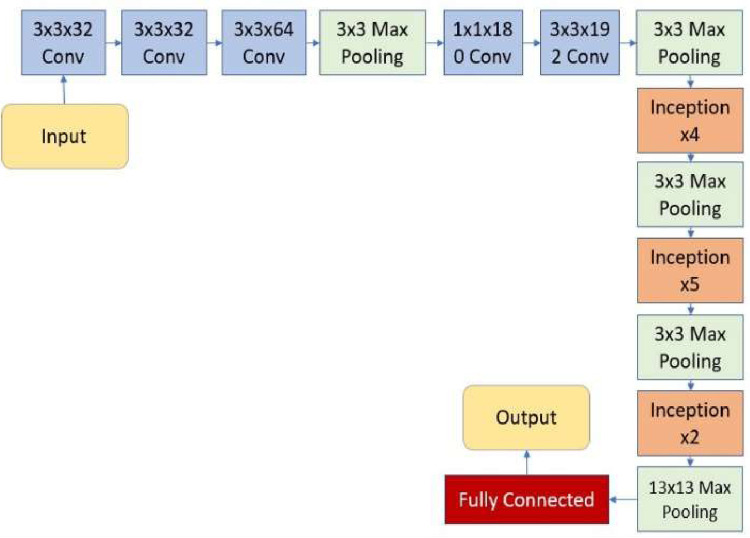
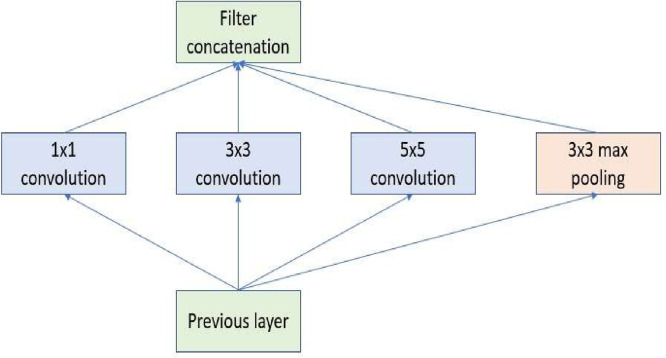
Transfer learning trains a task similar to the deep neural network. The initial layers of the pre-training network are modified if needed. The fine-tuning parameters can be applied to the model's final layers for learning the features of the new dataset. The pre-trained model retrains with a new, smaller, dataset, and the weights of the model are refined according to the new tasks. Fine-tuning takes place by backpropagation with labels. The learning to transmit approach is more efficient than the effort required to train an original, new neural network. The parameters in the newly developed neural network are not established from scratch. DL algorithms can achieve a higher function or performance for many issues -however, they rely upon large amounts of data and a longer training timeframe [28,29]. Therefore, reusing pre-trained models for similar tasks can be very beneficial. Accordingly, the Inception V3 [30,31] has been used as a pre-trained model and was finetuned with the proposed dataset in this work. A finetuned Inception V3 is modified and applied as a pre-trained Inception to classify COVID-19 images from collected and generated data by CycleGAN. Inception V3 assembles the sparse convolution kernels into fewer dense sub-convolution kernel groups. The structure of the Inception V3 and core unit of Inception is shown in Figs. 2 and 3 .
Fig. 2.
Architecture of Inception V3 [31].
Fig. 3.
Core unit of the Inception module [30].
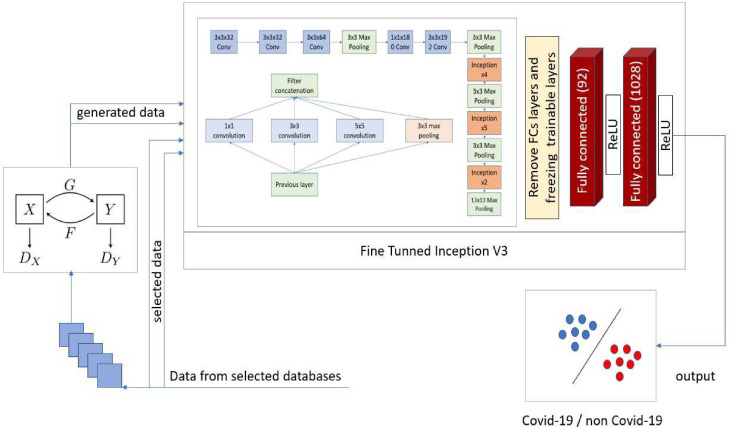
The proposed model architecture is shown in Fig. 4 . We removed the Softmax layers and fully connected layers in Inception V3 and froze trainable layers. Then two fully connected layers of size 92 and 1028, and activation ReLU are included to train the model.
Fig. 4.
The proposed CycleGAN-Inception model architecture to detect COVID-19.
2.3. The proposed algorithm
The algorithm used in our proposed model is displayed in Fig. 5 and described below:
Algorithm 1.
Proposed CycleGAN-Inception algorithm.
| 1: | x ← input COVID-19 images |
| 2: | y ← input non COVID-19 images |
| 3: | procedure pre-processing (x,y) |
| 4: | x,y ← resize (x,y) |
| 5: | x,y ← normalize (x,y) |
| 6: | end procedure pre-processing |
| 7: | procedure CycleGAN (x,y) |
| 8: | x,y ← G, F |
| 9: | L(G,F,DX,DY)=LGAN(G,DY,X,Y)+LGAN(F,DXY,X)+λLcyc(G,F) |
| 10: | end procedure CycleGAN |
| 11: | procedure Finetune_Inception (n=10, j=50, FC1 = 92, FC2 = 1028, activation = ReLU) |
| 12: | remove FC layers Inception V3 |
| 13: | add_layer (FC1, activation) |
| 14: | add_layer (FC2, activation) |
| 15: | build model CycleGAN_Inception |
| 16: | for k ← 0, n do |
| 17: | for epoch ← 0, j do |
| 18: | train CycleGAN_Inception (x, y, Dx, Dy) |
| 19: | evaluate CycleGAN_Inception (x, y, Dx, Dy) |
| 20: | end for |
| 21: | end for |
| 22: | end procedure Finetune_Inception |
Fig. 5.
Flowchart of the proposed algorithm.
3. Experimental configuration and databases
We have performed the experiments using an Intel Core i7 computer with 3.3 GHz and 16 GB memory. The suggested algorithm was implemented by Python software v3.6 [32] with Keras [33] and TensorFlow [34] libraries. Using Keras library allows us to develop the prototypes quickly and the library supports both the CNN and the recurrent neural networks.
3.1. Databases
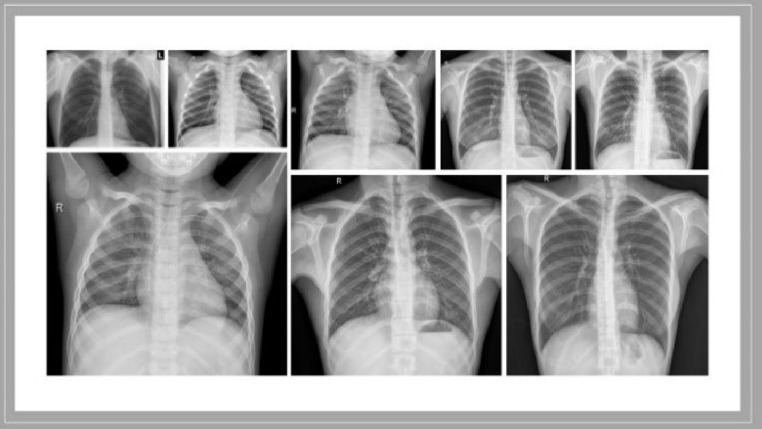
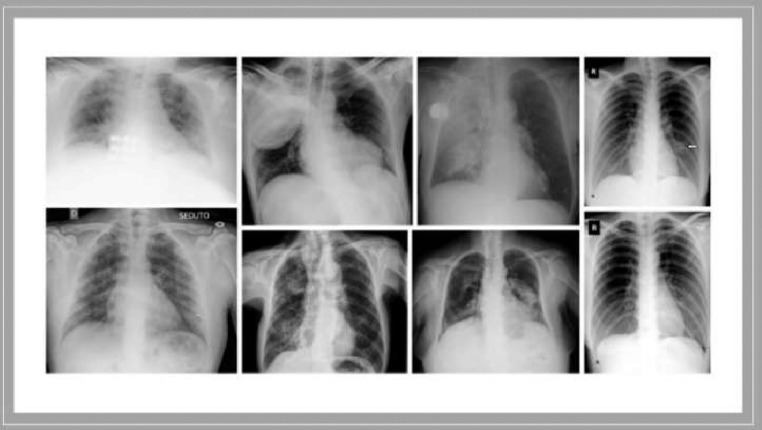
The “Extensive COVID-19 X-ray and CT Chest Images” Database were used in this work to train and develop our model [15]. The database consists of two folders: (1) 5500 normal and 4044 COVID-19 X-ray chest scans. (2) 2628 normal and 5427 COVID-19 CT scans. Only the X-ray images from the database were used in our study. Samples of COVID-19 and non-COVID-19 X-rays images are presented in Figs. 6 and 7 . The augmented data have been used to train the model in this work.
Fig. 6.
Samples of non-COVID-19 X-ray images [15].
Fig. 7.
Samples of COVID-19 X-ray images [15].
3.2. Evaluation metrics
Evaluation of obtained results is an important step in machine learning development. Estimation of the effectiveness and efficiency of the proposed model is done by training and testing datasets which are divided into 10 folds according to k-fold cross validation [35]. Cross validation is computationally intensive. It is used to develop the automated model using the training, testing and validation data. The dataset D is equally divided into k disjoint subsets. It uses (k-1) dataset for training purposes and a dataset for the purpose of testing by repeatedly training the algorithm k times.
The performances of the new model were evaluated using accuracy, area under curve, mean squared error, mean absolute error based on the confusion Matrix (Ouchicha et al.; Bradley, 1997).
The confusion matrix includes four parts, true positive (TP), false positive (FP), false negative (FN), and true negative (TN).
The accuracy (ACC) is defined as correct predictions split by the total number of input data in testing and training sets.
| (5) |
Mean absolute error (Ismael & Şengür) represents the disparity between original scores and predicted scores. It judges how removed the predictions are from the true output. Mean squared error (MSE) is the average of the square disparity between the original scores and predicted scores.
The area under curve (AUC) refers to the area under the receiver operating characteristic curve (ROC) of FP rate versus TP rate at various points from 0 to 1 [36].
4. Results and discussion
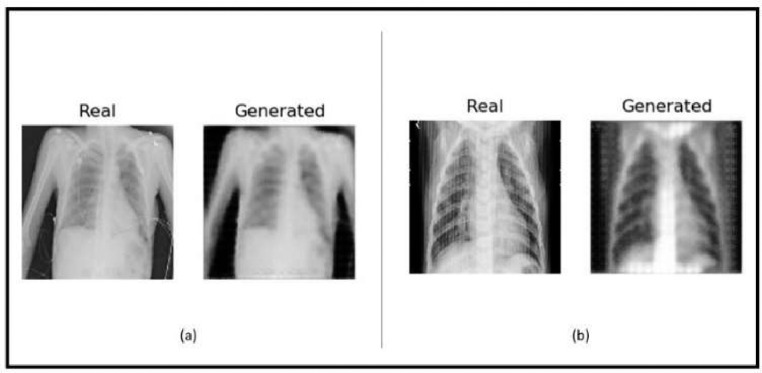
The obtained results indicate that our proposed CycleGAN-Inception can accurately detect positive COVID-19 cases and differentiate between COVID-19 andnon-COVID-19 cases using radiographic images. The sample X-ray images generated by CycleGAN: (a) a sample of real and generated images from class non-covid19 to covid19. (b) a sample of real and generated images from class covid19 to non-covid19 is shown in Fig. 8 .
Fig. 8.
Sample chest X-ray images generated by CycleGAN: (a) a sample of real and generated images from class non-covid19 to covid19. (b) a sample of real and generated images from class covid19 to non-covid19.
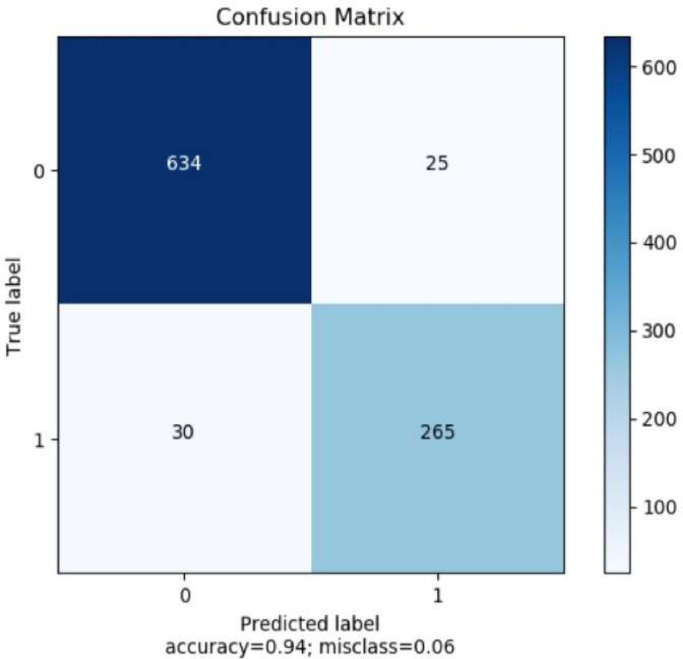
The ACC, AUC, MSE and MAE obtained by the proposed CycleGAN-Inception V3 model with k-fold cross-validation (k = 10) method is displayed in Table 1 . The proposed model is trained and tested with 50 epochs. We have obtained an average accuracy of 94.2% and AUC of 92.2%. In Fig. 9, the TP, TN, FP, and FN of the test set have been calculated and the average number of TP, FN, TN, FP images for ten runs are displayed in the figure. From the total number of 9544 images of the selected original dataset 954 images have been selected as test dataset based on the K-fold cross (K = 10) validation.
Table 1.
Average performance results from CycleGAN-Inception model.
| ACC (%) | AUC (%) | MSE | MAE |
|---|---|---|---|
| 94.2 | 92.2 | 0.27 | 0.16 |
Fig. 9.
Confusion matrix of trained and evaluated proposed model for two classes.
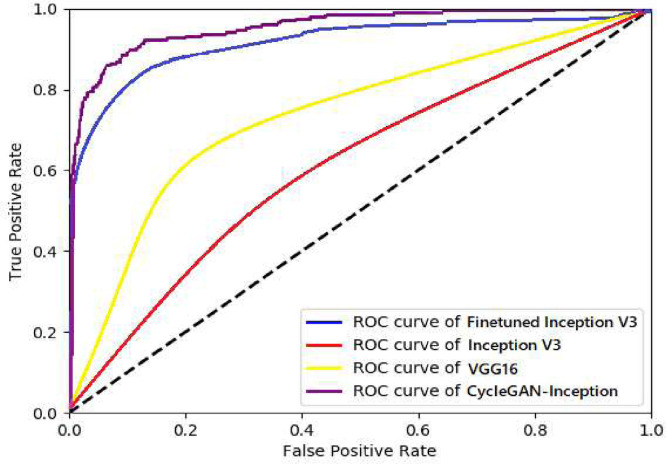
The developed model has compared with other transfer learning models namely Inception V3, VGG16 and finetuned Inception V3 using the same database. The results displayed in Table 2 demonstrate that our model outperformed the previous baseline models. The ROC curves obtained for four models including the proposed CycleGAN-Inception, Inception V3, Finetuned Inception V3, and VGG16 are shown in Fig. 10 .
Table 2.
Summary of comparison of proposed model with other transfer learning models.
| No | Model | ACC (%) | AUC (%) |
|---|---|---|---|
| 1 | Inception V3 | 68 | 62 |
| 2 | VGG16 | 82 | 75 |
| 3 | Fine tuned Inception V3 | 91 | 88 |
| 4 | CycleGAN-Inception | 94.2 | 92.2 |
Fig. 10.
ROC curves obtained for four models including the proposed CycleGAN-Inception, Inception V3, Finetuned Inception V3, and VGG16.
The comparison of our technique with the gold-standard approach for automated detection of COVID-19 from chest X-ray radiological scans is shown in Table 3 . It illustrates that we have accomplished the highest accuracy and AUCs. The advantages of our method are as follows:
-
1.
Proposed novel augmentation method. Hence, can be employed for smaller database also.
-
2.
Obtained highest classification performance (Accuracy = 94.2% and AUC = 92.2%).
-
3.
Developed model is accurate and robust as we have employed ten-fold cross-validation strategy.
-
4.
Generated system is simple and faster as we have used transfer learning method.
Table 3.
Comparing of the proposed model's results with the state-of-the-art models’ results.
| Authors | Approaches | Results |
|---|---|---|
| Wang et al. [2] | COVID-Net |
ACC = 83.5% for 4-classes, ACC = 92.4% for 3-classes. |
| Hemdan et al. [3] | COVIDX-Net | F1-scores = 0.91 and 0.89 of 2-classed (i.e., COVID-19 and normal cases) |
| Sethy and Behera[4] | ResNet50+SVM | ACC = 95.33% |
| Ozturk et al. [5] | DarkNet | ACC = 87.02% of multi-class ACC = 98.08% of 2-classes. |
| Khan et al. [6] | ]oroNet | ACC = 89.6% and 95% for 4 classes. |
| The proposed model | CycleGAN-Inception | ACC = 94.2% and AUC = 92.2% |
The limitation of our method is that we have used only one public database. Our model needs to be tested with more diverse database.
5. Conclusions
The aim of this paper was to develop a novel robust AI algorithm to detect COVID-19 using x-ray images automatically. Our proposed approach is simple, robust and more accurate. For doing this matter, we divided the model in two sections. In the first phase we applied CycleGAN technology for COVID-19 images which was the first time applied for COVID-19 X-ray image to translate COVID-19 X-ray image to normal once and vice versa. In the second phase we developed new deep learning model to classify the images in two normal and problematic classes. Therefore, we identified a new COVID-19 diagnosis system using deep learning technique and new framework was developed by integrating CycleGAN and finetuned Inception V3 X-ray transfer learning model. The proposed algorithm has trained, tested, and evaluated using the “Extensive COVID-19 X-ray Chest Images” Dataset. A CycleGAN is applied to the proposed model as an unsupervised technique for data augmentation. The pre-trained Inception V3 deep convolutional network is modified by removing fully connected layer and adding two new fully connecting layers. The whole process is trained and evaluated. The obtained results demonstrate the effectiveness of the algorithm with MSE = 0.27, MAE = 0.16, AUC = 92.2% and accuracy = 94.2%. Future work could validate the proposed model with more diverse big datasets and evaluate the performance in COVID-19 diagnosis. In future we may plan to explore the possibility of using our developed model to diagnose pulmonary edema, asthma, pericarditis, heart failure and pneumonia in addition to COVID-19 using chest x-ray images. Our new model can be used to diagnose other diseases using other imaging modalities as well.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Edited by Maria De Marsico
References
- 1.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemdan, E.E.D., Shouman, M.A., & Karar, M.E. (2020). Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv:2003.11055.
- 4.Sethy P.K., Behera S.K. Detection of coronavirus disease (covid-19) based on deep features. Preprints. 2020;2020030300:2020. https://pdfs.semanticscholar.org/9da0/35f1d7372cfe52167ff301bc12d5f415caf1.pdf [Google Scholar]
- 5.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Zhao H., Jie Y., Wang R. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. MedRxiv. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2020 doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. Covid-caps: a capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recognit. Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barua P.D., Muhammad Gowdh N.F., Rahmat K., Ramli N., Ng W.L., Chan W.Y., Kuluozturk M., Dogan S., Baygin M., Yaman O., Tuncer T., Wen T., Cheong K.H., Acharya U.R. Automatic COVID-19 detection using exemplar hybrid deep features with X-ray images. Int. J. Environ. Res. Public Health. 2021;18(15):8052. doi: 10.3390/ijerph18158052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharifrazi D., Alizadehsani R., Roshanzamir M., Joloudari J.H., Shoeibi A., Jafari M., Acharya U.R. Fusion of convolution neural network, support vector machine and Sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomed. Signal Process. Control. 2021;68 doi: 10.1016/j.bspc.2021.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, J., Xie, Y., Li, Y., Shen, C., & Xia, Y. (2020). Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv:arXiv:2003.12338, 27.
- 14.Zhang Y.D., Zhang Z., Zhang X., Wang S.H. MIDCAN: a multiple input deep convolutional attention network for Covid-19 diagnosis based on chest CT and chest X-ray. Pattern Recognit. Lett. 2021;150:8–16. doi: 10.1016/j.patrec.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walid, E.S., & Fathi, A.E.S. (2020). Extensive COVID-19 X-ray and CT chest images dataset 10.17632/8h65ywd2jr.3
- 16.Khan, S.H., Sohail, A., & Khan, A. (2020). COVID-19 Detection in chest X-ray images using a new channel boosted CNN. arXiv:2012.05073. [DOI] [PMC free article] [PubMed]
- 17.Cohen, J.P., Morrison, P., Dao, L., Roth, K., Duong, T.Q., & Ghassemi, M. (2020). Covid-19 image data collection: prospective predictions are the future. arXiv:2006.11988.
- 18.Zhu J.Y., Park T., Isola P., Efros A.A. Proceedings of the IEEE International Conference on Computer Vision. 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks. [Google Scholar]
- 19.Antoniou, A., Storkey, A., & Edwards, H. (2017). Data augmentation generative adversarial networks. arXiv preprint arXiv:1711.04340.
- 20.Shrivastava A., Pfister T., Tuzel O., Susskind J., Wang W., Webb R. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Learning from simulated and unsupervised images through adversarial training. [Google Scholar]
- 21.Bowles, C., Chen, L., Guerrero, R., Bentley, P., Gunn, R., Hammers, A., Dickie, D.A., Hernández, M.V., Wardlaw, J., & Rueckert, D. (2018). Gan augmentation: augmenting training data using generative adversarial networks. arXiv:1810.10863.
- 22.Goodfellow Ian, Pouget-Abadie Jean, Mehdi Mirza, Xu Bing, Warde-Farley David, Ozair Sherjil, Courville Aaron, Bengio Yoshua. Generative adversarial nets. Advances in neural information processing systems. 2014;27 [Google Scholar]
- 23.Sandfort V., Yan K., Pickhardt P.J., Summers R.M. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-52737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godard C., Mac Aodha O., Brostow G.J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Unsupervised monocular depth estimation with left-right consistency. [Google Scholar]
- 25.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009;22(10):1345–1359. [Google Scholar]
- 26.Bargshady G., Zhou X., Deo R.C., Soar J., Whittaker F., Wang H. The modeling of human facial pain intensity based on temporal convolutional networks trained with video frames in HSV color space. Appl. Soft Comput. 2020;97 [Google Scholar]
- 27.Kaya A., Keceli A.S., Catal C., Yalic H.Y., Temucin H., Tekinerdogan B. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron. Agric. 2019;158:20–29. doi: 10.1016/j.compag.2019.01.041. [DOI] [Google Scholar]
- 28.Bargshady G., Zhou X., Deo R.C., Soar J., Whittaker F., Wang H. Enhanced deep learning algorithm development to detect pain intensity from facial expression images. Expert Syst. Appl. 2020;149 [Google Scholar]
- 29.Soar J., Bargshady G., Zhou X., Whittaker F. Proceedings of the International Conference on Smart Homes and Health Telematics. 2018. Deep learning model for detection of pain intensity from facial expression. [Google Scholar]
- 30.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Going deeper with convolutions. [Google Scholar]
- 31.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision. [Google Scholar]
- 32.Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17(1):57–61. [PubMed] [Google Scholar]
- 33.Ketkar N. Deep Learning with Python. Springer; 2017. Introduction to keras; pp. 97–111. [DOI] [Google Scholar]
- 34.Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M. Tensorflow: a system for large-scale machine learning. Proceedings of the 12th Symposium on Operating Systems Design and Implementation; Savannah, GA; 2016. [Google Scholar]
- 35.Bengio Y., Grandvalet Y. No unbiased estimator of the variance of k-fold cross-validation. J. Mach. Learn. Res. 2004;5(Sep):1089–1105. [Google Scholar]
- 36.Powers D.M. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. J. Mach. Learn. Technol. 2011;2(1):37–63. [Google Scholar]