Late in mitosis, chromatin interactions with the inner nuclear membrane (INM) are reestablished and passed to postmitotic nuclei. As shown in Ptak and Saik et al., in S. cerevisiae this process is supported by a wave of SUMOylation at the INM directed by the binding of the SUMO E3-ligase Siz2 to the VAP protein Scs2.
Abstract
In eukaryotes, chromatin binding to the inner nuclear membrane (INM) and nuclear pore complexes (NPCs) contributes to spatial organization of the genome and epigenetic programs important for gene expression. In mitosis, chromatin–nuclear envelope (NE) interactions are lost and then formed again as sister chromosomes segregate to postmitotic nuclei. Investigating these processes in S. cerevisiae, we identified temporally and spatially controlled phosphorylation-dependent SUMOylation events that positively regulate postmetaphase chromatin association with the NE. Our work establishes a phosphorylation-mediated targeting mechanism of the SUMO ligase Siz2 to the INM during mitosis, where Siz2 binds to and SUMOylates the VAP protein Scs2. The recruitment of Siz2 through Scs2 is further responsible for a wave of SUMOylation along the INM that supports the assembly and anchorage of subtelomeric chromatin at the INM and localization of an active gene (INO1) to NPCs during the later stages of mitosis and into G1-phase.
Introduction
The spatial organization of a eukaryotic genome within the nucleoplasm is influenced by multiple factors including the nuclear envelope (NE), which functions as a two-dimensional interaction surface for chromatin. The inner nuclear membrane (INM) often associates with densely packed, gene-poor, and transcriptionally inactive heterochromatin, while nuclear pore complexes (NPCs) typically engage transcriptionally active euchromatin (Ptak et al., 2014; Buchwalter et al., 2019; Misteli, 2020). However, these interactions are not static, and in multiple eukaryotes it has been shown that they are remodeled in response to gene activation, during DNA metabolism (e.g., replication or repair), and during mitosis (e.g., NE breakdown and chromosome segregation; Güttinger et al., 2009; Taddei and Gasser, 2012; Ptak et al., 2014; Ma et al., 2015; Ptak and Wozniak, 2016). In Saccharomyces cerevisiae, heterochromatin-like subtelomeric chromatin is associated with the INM, and transcriptional activation has been shown to involve localization of specific gene loci to NPCs (e.g., INO1 and GAL1-10; Egecioglu and Brickner, 2011; Taddei and Gasser, 2012). In addition, although the NE remains intact during mitosis in budding yeast, chromatin–NE interactions are also lost and reformed during mitosis (Hediger et al., 2002; Ebrahimi and Donaldson, 2008; Brickner and Brickner, 2010). This overall regulation of chromatin–NE interactions (i.e., both disruption and reformation of these contacts) is critical for proper genome organization and epigenetic inheritance during cell division (Politz et al., 2013; Champion et al., 2019; Falk et al., 2019; Poleshko et al., 2019).
As a cell progresses through mitosis, the cyclical phosphorylation/dephosphorylation of various NE proteins contributes to the disruption and later reformation of NE–chromatin interactions (Güttinger et al., 2009; Wurzenberger and Gerlich, 2011). Despite the importance of reestablishing chromatin–NE interactions in the later stages of mitosis, it is unclear whether these events are supported by specific processes, such as phosphorylation or other post-translational modifications (PTMs). A candidate for such a functional role is SUMO (small ubiquitin-like modifier) modification. SUMOylation has been linked to heterochromatin assembly and chromatin tethering in yeast and higher eukaryotes (Hari et al., 2001; Ferreira et al., 2011; Lapetina et al., 2017; Ninova et al., 2019) and the association of active genes with NPCs (Texari and Stutz, 2015; Saik et al., 2020). In addition, the SUMO E3 ligase Siz2 has been implicated in the formation of NE–chromatin interactions during interphase (Ferreira et al., 2011; Freudenreich and Su, 2016; Lapetina et al., 2017; Saik et al., 2020).
Here, we report a PTM cascade involving phosphorylation and SUMOylation that is essential for establishing NE–chromatin interactions during progression through mitosis. With anaphase onset, we find that phosphorylation induces binding of Siz2 to the VAP (Vesicle-associated membrane protein [VAMP]–Associated Protein) family member Scs2 at the INM, where it remains until cytokinesis. At the INM, Siz2 drives a wave of mitotic SUMOylation involving NE proteins, with targets that include Scs2 and subtelomeric chromatin–associated Sir4. We show that these spatially regulated SUMOylation events are required to establish the proper organization of subtelomeric chromatin at the INM and the association of the activated INO1 gene with NPCs by anaphase/telophase of mitosis. These findings establish a temporally and spatially regulated set of phosphorylation-dependent SUMOylation events that regulate the reestablishment of nuclear architecture during the late stages of mitosis.
Results
Scs2 directs mitotic SUMOylation at the NE
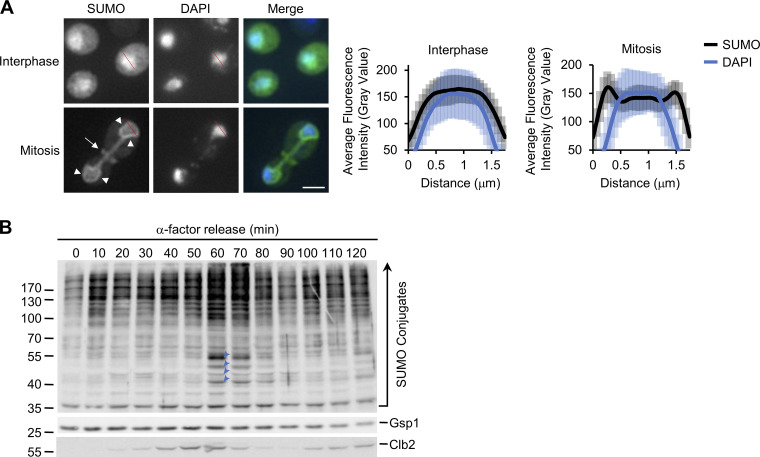
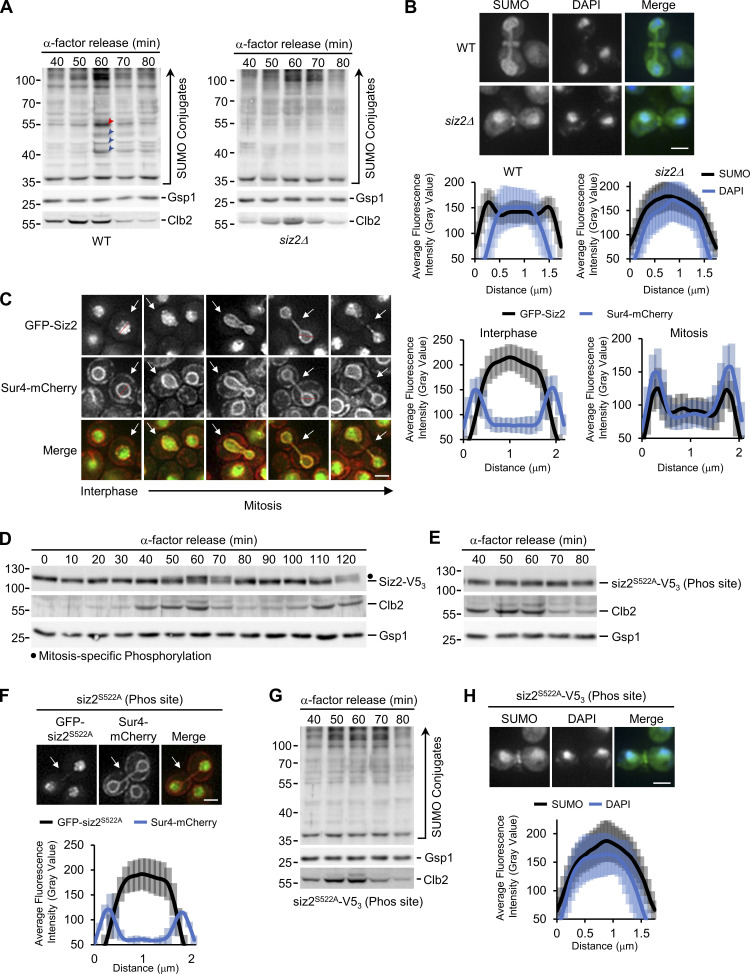
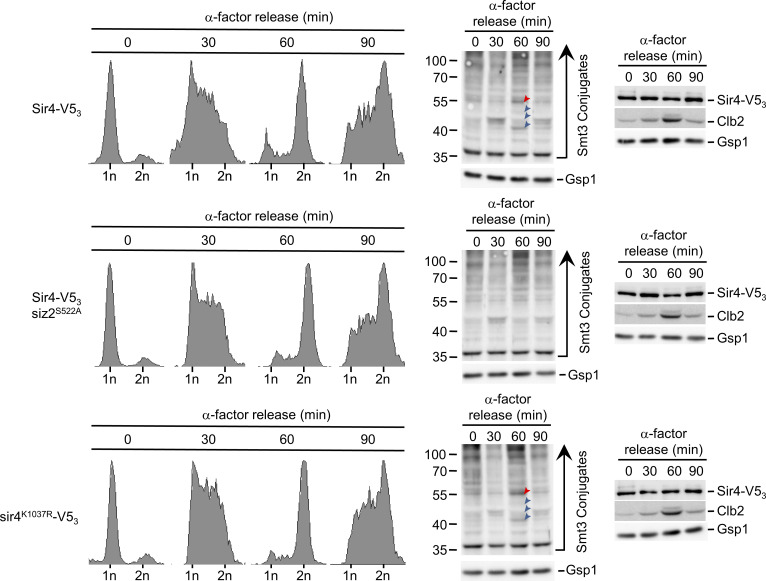
Immunofluorescence microscopy (IF) and Western blotting were performed using a SUMO-specific antibody to investigate spatial and temporal changes in cellular SUMOylation during the cell cycle in S. cerevisiae. Imaging of asynchronous IF-stained cells revealed a predominantly nuclear SUMO signal in interphase cells (unbudded and small-budded cells) and a concentration of SUMO at septin rings in mitotic cells (large budded) as previously reported. However, in mitotic cells, SUMO conjugates were also concentrated at the NE (Fig. 1 A). Coinciding with this mitotic redistribution of SUMO, Western blotting analysis in synchronized cell cultures revealed an increase in mitotic levels of various SUMOylated species that occurred just after peak levels of the cyclin Clb2 (Fig. 1 B). Noticeable among these were four clearly resolved SUMOylated species in the 40–55 kD range, which are smaller in size than the septins that are known to be SUMOylated in mitosis (Johnson and Blobel, 1999). Given that Clb2 is degraded upon anaphase onset (Irniger, 2002), the combined data are suggestive of SUMOylation events occurring in late metaphase/anaphase when SUMO is evident at the NE.
Figure 1.
Mitotic NE SUMOylation events. (A) Epifluorescence images of WT cells analyzed by anti-SUMO immunofluorescence (SUMO). Nuclear position is determined by DAPI staining. Arrowheads highlight SUMO along the NE. SUMOylated septin ring position is indicated by an arrow. Images were rendered using the unsharp mask filter in ImageJ. Nuclear levels of fluorescence were quantified using line scan intensities of equatorial optical sections through nuclei (e.g., see red lines) of interphase (unbudded or small budded) and mitotic (anaphase/telophase) cells as described in Materials and methods. Plots show average fluorescence intensity (SUMO-IF and DAPI) at multiple points along a 1.75-µm line for n = 25 nuclei per cell cycle phase. Note that the perimeter of the DAPI signal lies adjacent to the NE and peaks of SUMO-IF intensity in mitotic cells. Error bars represent SD. Bar, 2 µm. (B) Cells were arrested in G1-phase using α-factor. Following α-factor removal, cultures were sampled every 10 min and analyzed by Western blotting using antibodies directed against the proteins indicated on the right. Note, Clb2 levels peak in metaphase. Gsp1 is a loading control. Blue arrowheads highlight four prominent SUMOylated species in the 40–55-kD range that arise in mitosis and decay as cells enter G1-phase. Molecular mass markers are shown in kD.
To identify these prominent mitotic targets, we curated a list of SUMOylated proteins of similar masses (Panse et al., 2004; Wohlschlegel et al., 2004; Wykoff and O’Shea, 2005; Zhou et al., 2004; Denison et al., 2005; Hannich et al., 2005). From this list of proteins, 141 nonessential genes were selected. Western blot analysis of these null mutant strains revealed that the four mitotic SUMOylated species were absent in cells lacking the gene encoding Scs2 (Fig. 2 A). Scs2 is an ER/NE localized membrane protein of the VAP family (Loewen and Levine, 2005). Moreover, SUMOylated Scs2 was previously shown to migrate with an apparent mass of ∼55 kD (Felberbaum et al., 2012), like the most prominent of the four mitosis-specific SUMO species. Thus, SUMOylation of Scs2 would be consistent with both the size and location of the observed SUMO modifications (Fig. 1, A and B).
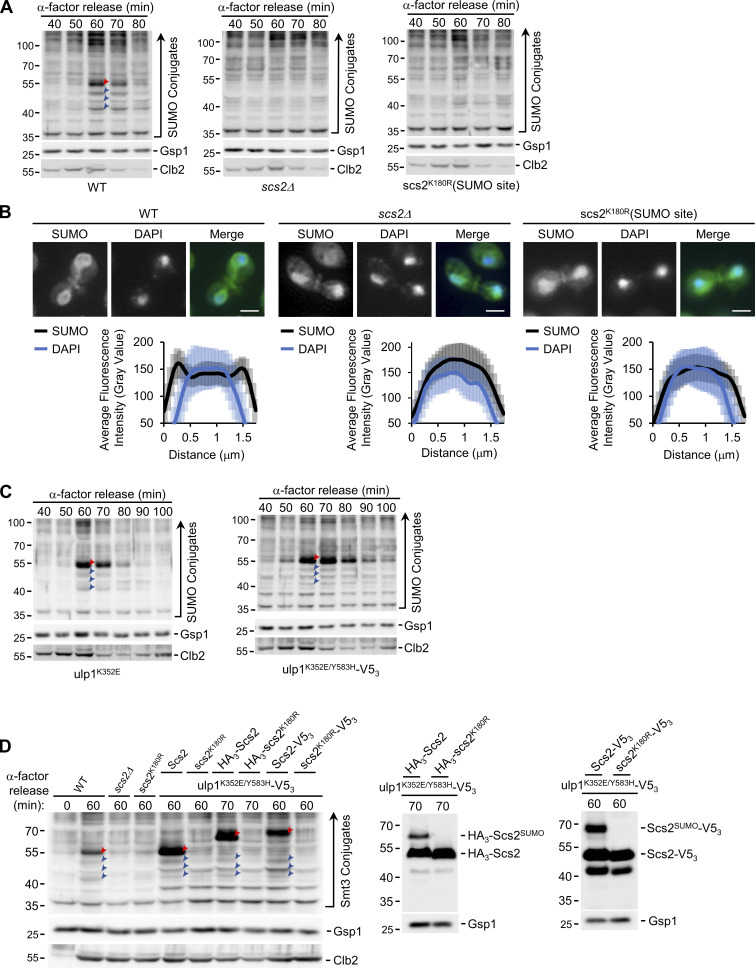
Figure 2.
Scs2 is required for, and is a target of, mitotic SUMOylation events. (A, C, and D) α-factor arrest-release assays were performed as in Fig. 1. Cell lysates from the indicated strains, at the times shown after release, were analyzed by Western blotting to detect SUMO conjugates, Clb2, and the Gsp1 load control (A, C, and D), as well as the V53 and HA3 tags (D) as specified to the right of the blot. Note in panel D, for the ulp1K352E/Y583H–V53 strains, the form of Scs2 produced in the cells examined is indicated above the lane. Red arrowheads point to SUMOylated Scs2 or SUMOylated tagged Scs2. Note that the WT SUMO conjugate profile shown in A is derived from that shown in Fig. 1 A. Blue arrowheads point to three other prominent mitotic SUMO conjugates. Molecular mass markers are shown in kD. (B) Epifluorescence images of WT, scs2Δ, and scs2K180R (SUMOylation site mutant—SUMO site) cells analyzed by anti-SUMO immunofluorescence (SUMO) and DAPI staining are shown. Imaging and quantification were performed as outlined in the Fig. 1 legend. Mitotic cells are shown (note SUMOylated septin ring). The nuclear distribution of SUMO, in relation to DAPI-stained nuclear DNA, in mitotic cells from each strain was quantified using line scans (n = 25). Note, quantification of lines scans shown in B were obtained at the same time as data shown in Fig. 1 A, and the WT data are also shown here for comparison. Error bars represent SD. Bar, 2 µm.
Other evidence establishes that the 55-kD mitotic SUMO-conjugate is Scs2-SUMO. First, this SUMO-conjugate was absent in cells lacking Scs2 (scs2Δ) or expressing a scs2 point mutation, scs2K180R (Felberbaum et al., 2012), which eliminates the known Scs2 SUMO acceptor site (Fig. 2 A; also see Fig. 2 D and Fig. S2 A). Second, in the absence of Scs2 or in strains containing the scs2K180R SUMO site mutant, SUMO accumulation at the NE during mitosis was not observed (Fig. 2 B). Third, when we examined a strain containing a mutation in the deSUMOylase Ulp1 (ulp1K352E) that was shown to increase cellular levels of SUMOylated Scs2 (Felberbaum et al., 2012), there was a marked increase in the level of the 55-kD SUMOylated protein during mitosis, consistent with it being Scs2-SUMO (Fig. 2 C). Serendipitously, we discovered a ULP1-V53 allele bearing an additional mutation (ulp1K352E/Y583H-V53) that showed further elevated levels of the 55-kD SUMO species (Fig. 2 C).
Figure S2.
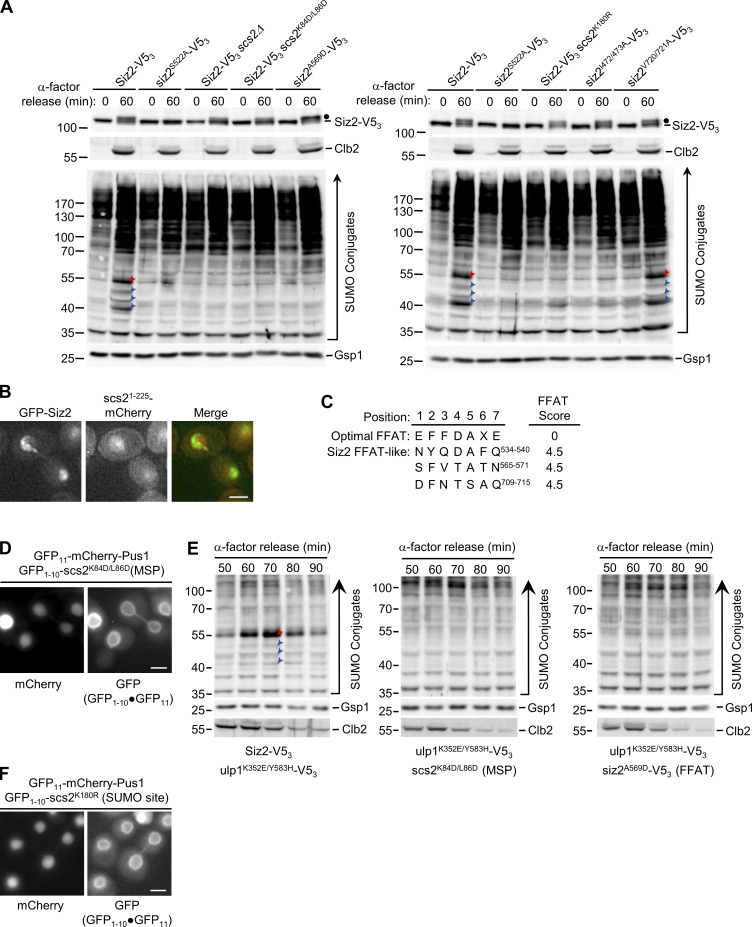
Siz2 directs mitosis-specific SUMOylation of the VAP family member Scs2. (A and E) α-factor arrest-release assays were performed as in Fig. 1 using the indicated strains. Cell lysates were analyzed by Western blotting using anti-V5 (A), SUMO, Clb2, and Gsp1 (load control; A and E) antibodies. Only the 0- and 60-min time points were analyzed in A. Red arrowheads point to SUMOylated Scs2. Blue arrowheads point to other prominent mitotic SUMO conjugates. In A, dots highlight the position of mitotically phosphorylated Siz2-V53. Molecular mass markers are shown in kD. (B) Shown are epifluorescence images of a mitotic cell producing GFP-Siz2 and scs21–225-mCherry. (C) The position, sequence, and score (Murphy and Levine, 2016) of putative Siz2 FFAT-like motifs are shown. An optimal FFAT sequence is shown for comparison. (D and F) Using the split-superfolder GFP system (see Fig. 4 A), the INM association of the scs2 MSP domain mutant (D) or the scs2 SUMO site mutant (F) were assessed in cells producing GFP1–10–scs2K84D/L86D or GFP1–10–scs2K180R and the plasmid-encoded GFP11-mCherry-Pus1 reporter. Dot represents association of the GFP fragments. Error bars represent SD. Bar, 2 µm.
Fourth, we tagged endogenous Scs2 at the N- or C-terminus with HA3 or V53 to both detect and alter the mass of Scs2 (Fig. 2 D), which would be predicted to decrease the electrophoretic mobility of the 55-kD SUMO species. Indeed, the 55-kD SUMO species was absent in mitotic WT cells producing the HA3-Scs2, and a SUMO species of ∼67 kD appeared, consistent with the predicted size of HA3-Scs2-SUMO (Fig. S1 A). However, SUMO blots also revealed that each of the four prominent mitotic SUMO species was reduced in these cells, as well as in cells producing Scs2-V53, suggesting that tagging alters Scs2 and inhibits SUMOylation. Consequently, we examined the tagged versions of Scs2 in the ulp1K352E/Y583H strain. In these cells, the four mitosis-specific SUMO species were more prominent, and as predicted, the 55-kD SUMO species was absent and was replaced with SUMO species of ∼67 and 70 kD that comigrate with SUMO-modified forms of HA3-Scs2 and Scs2-V53 (Fig. 2 D). Notably, the detected SUMOylated proteins were absent when the SUMO acceptor site of Scs2 was mutated in the fusion proteins (HA3-scs2K180R and scs2K180R-V53). Moreover, analyses of cells producing either the HA3-Scs2 or Scs2-V53 showed that the three SUMOylated species migrating faster than Scs2-SUMO were of the same size as those detected in WT or the ulp1K352E/Y583H mutant cells. Thus, they are unlikely to be proteolytic fragments of Scs2-SUMO and instead represent distinct mitotic SUMOylation targets that are dependent on Scs2 SUMOylation.
Figure S1.
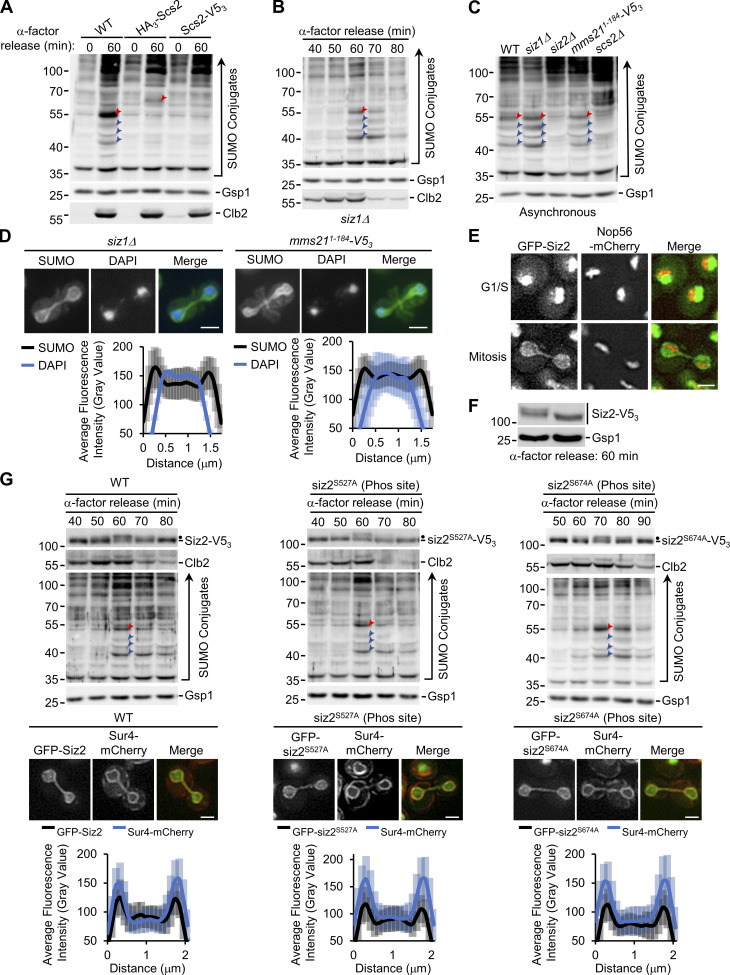
Siz2 localization and PTM. (A, B, and G) α-factor arrest-release assays were performed as in Fig. 1. Shown are WT cells and cells producing HA3-Scs2 or Scs2-V53 (A), siz1Δ cells (B), and Siz2-V53–, siz2S527A-V53–, and siz2S674A-V53–producing cells (G). Cell lysates were analyzed by Western blotting using anti-SUMO, Clb2, and Gsp1 (load control) antibodies, as well as a V5 antibody in G, as indicated. Only the 0-and 60-min time points were analyzed in A. (C) Cell lysates derived from asynchronous cultures of WT, siz1Δ, siz2Δ, and mms211–184–V53 (Mms21 derivative deficient in SUMO E3 ligase activity) cells were assessed by Western blotting using an anti-SUMO antibody to assess SUMO conjugate profiles. Gsp1 is a loading control. In A–C and G, red arrowheads point to SUMOylated Scs2 or SUMOylated tagged Scs2. Blue arrowheads point to other prominent mitotic SUMO conjugates. In G, dots highlight the position of mitotically phosphorylated Siz2-V53. Molecular mass markers are shown in kD. (D) Anti-SUMO immunofluorescence analysis of siz1Δ and mms211–184–V53 cells was performed as described in Fig. 1. DAPI staining identifies nuclear position. Imaging and quantification of the nuclear distribution of SUMO in mitotic cells (n = 25) were performed as in Fig. 1. (E) Shown are epifluorescence images of representative G1/S- and M-phase WT cells producing GFP-Siz2 and nucleolar Nop56-mCherry. Merged images show that GFP-Siz2 is largely excluded from the nucleolus. (F) WT mitotic cell lysates were isolated at 60 min after release from α-factor arrest (see Fig. 1 B). Proteins were extracted and solubilized in buffer lacking (-PPase) or containing protein phosphatase (+PPase). Samples were analyzed by Western blotting using anti-V5 and Gsp1 (loading control) antibodies. Mobility of modified Siz2-V53 is increased by phosphatase treatment. Vertical line indicates that all bands observed consist of Siz2-V53. (G) Imaging and quantification of the nuclear distribution of GFP-Siz2, GFP-siz2S527A, and GFP-siz2S674A compared with Sur4-mCherry were examined as described in Fig. 3 for cells (n = 25) in mitosis. Note, line scans show enrichment of Siz2-GFP, GFP-siz2S527A, and GFP-siz2S674A at the NE with Sur4-mCherry in these cells. Quantification of lines scans shown in G were obtained at the same time as data shown in Fig. 3 C, and the WT data are also shown here for comparison. Error bars represent SD. Bar, 2 µm. Phos, phosphorylation.
From these data, we conclude that Scs2 is an NE-associated mitotic SUMOylation target that directs mitotic SUMOylation events in a manner dependent upon its own SUMOylation.
Siz2 recruitment to the NE directs mitotic SUMOylation
In S. cerevisiae three SUMO E3 ligases, Siz1, Siz2/Nfi1, and Mms21 (Jentsch and Psakhye, 2013), guide the selection of most SUMOylation targets in actively growing cells. Anti-SUMO Western blotting of synchronized or asynchronous cell cultures showed that only cells lacking Siz2 failed to accumulate Scs2-SUMO and the smaller, prominent SUMOylated species during mitosis (Fig. 3 A; and Fig. S1, B and C). Moreover, Siz2, but not Siz1 or Mms21, was required for mitotic NE accumulation of SUMO conjugates (Fig. 3 B and Fig. S1 D). Together, these observations indicate that Siz2, as well as Scs2, is required to direct mitotic SUMOylation events at the NE.
Figure 3.
Mitotic recruitment of Siz2 to the NE directs SUMOylation events. (A, D, E, and G) α-factor arrest-release assays were performed as in Fig. 1. Cell lysates were analyzed by Western blotting to detect SUMO conjugates (A and G), Siz2-V53 (D), and siz2S522A-V53 (phosphorylation site mutant—Phos site; E), as well as Clb2 and the Gsp1 load control in the indicated strain background. In A, the position of Scs2-SUMO is indicated by a red arrowhead. Blue arrowheads point to three other prominent mitotic SUMO conjugates. In D, the dot highlights the position of mitotically phosphorylated Siz2-V53. Molecular mass markers are shown in kD. (B and H) Anti-SUMO immunofluorescence analysis of WT and siz2Δ (B) as well as siz2S522A (H) cells was performed as described in Fig. 1. DAPI staining identifies nuclear position. Imaging and quantification of the nuclear distribution of SUMO in mitotic cells (n = 25) were performed as in Fig. 1 and Fig. 2. Note, quantification of lines scans shown in panel B were obtained at the same time as data shown in Fig. 2 B, and the WT data are also shown here for comparison. (C and F) Epifluorescence images of cells producing GFP-Siz2 (C) or GFP-siz2S522A (F) along with the NE/ER marker Sur4-mCherry. Cell cycle stage of the highlighted cell (arrows) is indicated in C. Mitotic cells are shown in F. Imaging and quantification of the nuclear distribution of GFP-Siz2 (C) or GFP-siz2S522A (F) compared with Sur4-mCherry were examined using scans along a 2.1-µm line (see C, red lines), as described in the Fig. 1 legend, for cells (n = 25) in mitosis (F) or interphase and mitosis (C) as indicated. Note, line scans in C show enrichment of Siz2-GFP at the NE with Sur4-mCherry in mitotic cells. Error bars represent SD. Bar, 2 µm. Phos, phosphorylation.
Siz1 has previously been shown to target to the septin ring during M-phase, where it SUMOylates the septins (Johnson and Gupta, 2001; see Fig. S1 D). In an analogous manner, we hypothesized that the spatiotemporal recruitment of Siz2 to the NE during mitosis may direct SUMOylation of NE proteins. Upon examination of GFP-Siz2 localization in interphase cells, we observed GFP-Siz2 diffusely and in distinct puncta throughout the nucleoplasm, while partially excluded from the nucleolus (Fig. 3 C and Fig. S1 E). GFP-Siz2 puncta were also visible along the NE, including the NE adjacent to the nucleolus. Time-lapse imaging revealed that these puncta are dynamic and do not persist in mitosis (Video 1). Strikingly, as the NE elongated and cells progressed into anaphase, GFP-Siz2 accumulated at the NE, where it remained until dissolution of the NE membrane bridge that links the mother and daughter nuclei during cytokinesis (Fig. 3 C and Video 1).
Video 1.
Dynamics of GFP-Siz2 localization throughout the cell cycle. Cell cycle–dependent changes in Siz2 localization were visualized using time-lapse epifluorescence microscopy on cells producing GFP-Siz2. Images were acquired at 1.5-min intervals over a 30-min period. Highlighted are cells progressing through G1- and S-phase (yellow dots), entering mitosis (red dots), and exiting mitosis (green dots). Images were rendered using the unsharp mask filter in ImageJ and saved as a QuickTime movie.
Critical transitions throughout mitosis are driven by PTMs, including phosphorylation (Cuijpers and Vertegaal, 2018). Given the sharp transition of GFP-Siz2 from the nucleoplasm to the NE, we examined whether Siz2 was post-translationally modified during mitosis using Western blot analysis of synchronized cells (Fig. 3 D). Between 60 and 70 min after release of cells from G1-phase arrest, we observed a decrease in the electrophoretic mobility of Siz2 (tagged with V53 for detection) consistent with it being post-translationally modified. Notably, this mobility change corresponded with the peak mitotic levels and subsequent loss of Clb2, suggesting that Siz2-V53 was modified in metaphase or at the onset of anaphase (Irniger, 2002). These data indicate that a Siz2 PTM occurs within a similar time window as Siz2 NE recruitment and Scs2 SUMOylation.
Phosphoproteome analyses previously identified putative Siz2 phosphorylation sites at serine residues 522, 527, and 674 (Albuquerque et al., 2008; Holt et al., 2009). Consistent with Siz2 being phosphorylated, phosphatase treatment of mitotic cell lysates removed the slower migrating species (Fig. S1 F). By examining Siz2 electrophoretic mobility in serine to alanine point mutants, the siz2S527A-V53 and siz2S674A-V53 mutants were found to retain an electrophoretic shift during mitosis like Siz2-V53. Moreover, they showed mitotic NE association and SUMO-conjugate profiles similar to WT cells (Fig. S1 G). This was in contrast to the siz2S522A point mutation, which showed no mitotic mobility shift (Fig. 3 E; also see Fig. S2 A). The siz2S522A mutant (GFP-siz2S522A) also failed to localize to the NE during mitosis (Fig. 3 F). Furthermore, the mitotic increase in the SUMOylation of Scs2 and three other prominent SUMO species (as detected by anti-SUMO Western blotting analysis) and the NE accumulation of SUMO were not detected in the siz2S522A mutant (Fig. 3, G and H). These results suggest a phosphorylation-dependent recruitment of Siz2 to the NE that supports SUMOylation of Scs2 and several other NE proteins during mitosis.
Scs2 is required for Siz2 recruitment to the NE
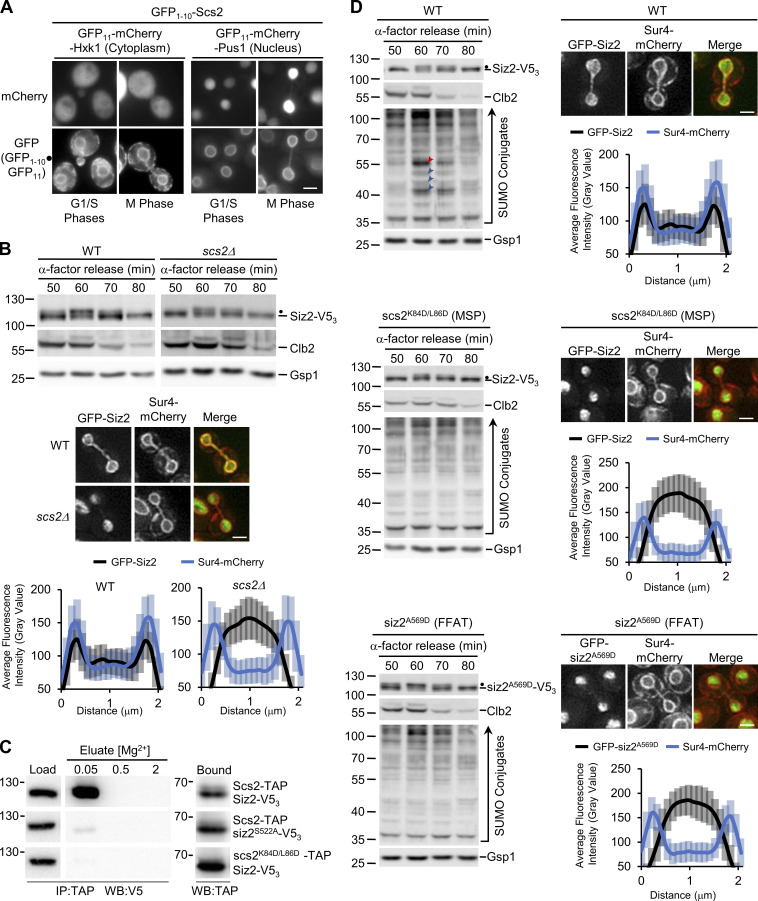
The concordant phenotypes of Scs2 and Siz2 mutants suggested to us that they may form a complex at the NE to direct mitotic SUMOylation events. Although Scs2 functions as a receptor on the ER for multiple cytoplasmic proteins (Stefan et al., 2011; Chao et al., 2014; Encinar del Dedo et al., 2017; Ng et al., 2020), our postulate would require that Scs2 is present at the INM to bind nucleoplasmic Siz2. Scs2 contains a single transmembrane region at its C-terminus that tethers it to the ER and outer nuclear membrane (Loewen and Levine, 2005), but an INM pool of Scs2 has not been described. To assess Scs2 localization, we used a split-superfolder GFP assay previously used to characterize the INM proteome (Smoyer et al., 2016). In this assay, if a GFP11-reporter resides in the same subcellular compartment as a target protein fused to GFP1–10, the two GFP fragments can assemble and fluoresce. As expected, we observed a pool of Scs2 localized at the ER/outer nuclear membrane, based on the association of GFP1–10–Scs2 with the cytoplasmic GFP11-Hxk1 reporter (Fig. 4 A). Importantly, GFP1–10–Scs2 and the nucleoplasmic GFP11-Pus1 reporter also yielded a robust NE GFP signal but no ER signal, suggesting Scs2 accesses the INM (Fig. 4 A).
Figure 4.
INM association of Scs2-Siz2 directs mitosis-specific SUMOylation events. (A) Scs2 localization was assessed using the split-superfolder GFP system. Epifluorescence images of WT cells containing GFP1–10–Scs2 and plasmid-encoded GFP11-mCherry-Hxk1(cytoplasmic) or GFP11-mCherry-Pus1 (nuclear). Localization of the reporter (mCherry) and assembled GFP1–10⋅GFP11 (GFP) in representative G1-, S-, and M-phase cells is shown. Bar, 2 µm. Dot represents association of the GFP fragments. (B and D) The indicated strains were assessed for Siz2-V53 phosphorylation and their mitotic SUMO conjugate profile by Western blot analysis as described in Fig. 1 and Fig. 2. Dots highlight the position of mitotically phosphorylated Siz2-V53. GFP-Siz2 and Sur4-mCherry imaging and quantification of the nuclear distribution were performed on mitotic cells of the specified strains as described in Fig. 3. Note, quantification of line scans shown in panels B and D was obtained at the same time as data shown in Fig. 3 C, and the WT data are also shown here for comparison. The scs2K84D/L86D mutations lie in the MSP domain (MSP) and the siz2A569 mutation within the FFAT-like motif (FFAT). Error bars represent SD. Bar, 2 µm. In D, the position of Scs2-SUMO is indicated by a red arrowhead, and blue arrowheads point to three other prominent mitotic SUMO conjugates. Molecular mass markers are shown in kD. (C) Binding of Scs2-TAP to either Siz2-V53 or siz2S522A-V53 and binding of scs2K84D/L86D-TAP to Siz2-V53 was assessed. Scs2-TAP or scs2K84D/L86D-TAP was affinity-purified from cells (IP) producing Siz2 derivatives, and bound proteins were eluted using a Mg2+ step gradient. Equivalent portions of the indicated fractions were analyzed by Western blotting (WB) to assess levels of the V5- and TAP-tagged fusions. Note, all Load, Elution, or Bound fractions shown in panel C were derived from the same Western blot.
Consistent with Scs2 acting as a receptor for Siz2 at the INM, Siz2 was phosphorylated but no longer localized to the NE during mitosis in cells lacking Scs2 (scs2Δ; Fig. 4 B) or producing a scs21–225 truncation missing the transmembrane domain (Brickner and Walter, 2004; Loewen et al., 2007; Fig. S2 B). Furthermore, immunoprecipitation (IP) from a cell lysate followed by Western blotting showed that Scs2-TAP binds Siz2-V53, but only weakly to the siz2S522A mutant that fails to associate with the NE (Fig. 4 C; siz2S522A-V53). These observations support the conclusion that Scs2 is an INM receptor for Siz2.
Scs2, like other members of the VAP family, contains an N-terminal MSP (major sperm protein) domain that binds FFAT (two phenylalanines in an acidic tract) or FFAT-like motifs within binding partners (Loewen et al., 2003; Loewen and Levine, 2005; Kaiser et al., 2005). Notably, Siz2 contains three potential FFAT-like motifs (Murphy and Levine, 2016; Fig. S2 C). Previous studies showed that K84D and L86D substitutions within the MSP motif of Scs2 specifically disrupts interactions with an FFAT-containing peptide (Kaiser et al., 2005). Since scs2K84D/L86D retains INM localization (Fig. S2 D), we tested the impact of an scs2K84D/L86D mutant on the NE localization of Siz2. To complement this approach, we also produced a mutation in Siz2 (A569D) predicted to disrupt a putative FFAT-like motif detected between residues 565–571 of Siz2 (Murphy and Levine, 2016). Both the scs2K84D/L86D and siz2A569D mutations inhibited mitosis-specific INM localization of Siz2 (Fig. 4 D). This phenotype did not appear to be due to a loss of mitotic phosphorylation of Siz2 in these strains (Fig. 4 D and Fig. S2 A).
Consistent with the defect in INM localization of Siz2, the scs2K84D/L86D and siz2A569D mutations also inhibited SUMOylation of mitotic targets, including Scs2, in both an otherwise WT background (Fig. 4 D and Fig. S2 A) and in ulp1K352E/Y583H–V53 cells (Fig. S2 E). Furthermore, we showed that the scs2K84D/L86D mutant exhibits reduced binding to Siz2 as determined by IP analysis (Fig. 4 C). On the basis of these data, we propose that an FFAT-like motif in Siz2 promotes an interaction with the MSP domain of Scs2 at the INM that is critical for Siz2-directed mitotic SUMOylation events.
A SUMO-interaction motif in Siz2 and SUMOylation of Scs2 contribute to Scs2-Siz2 association
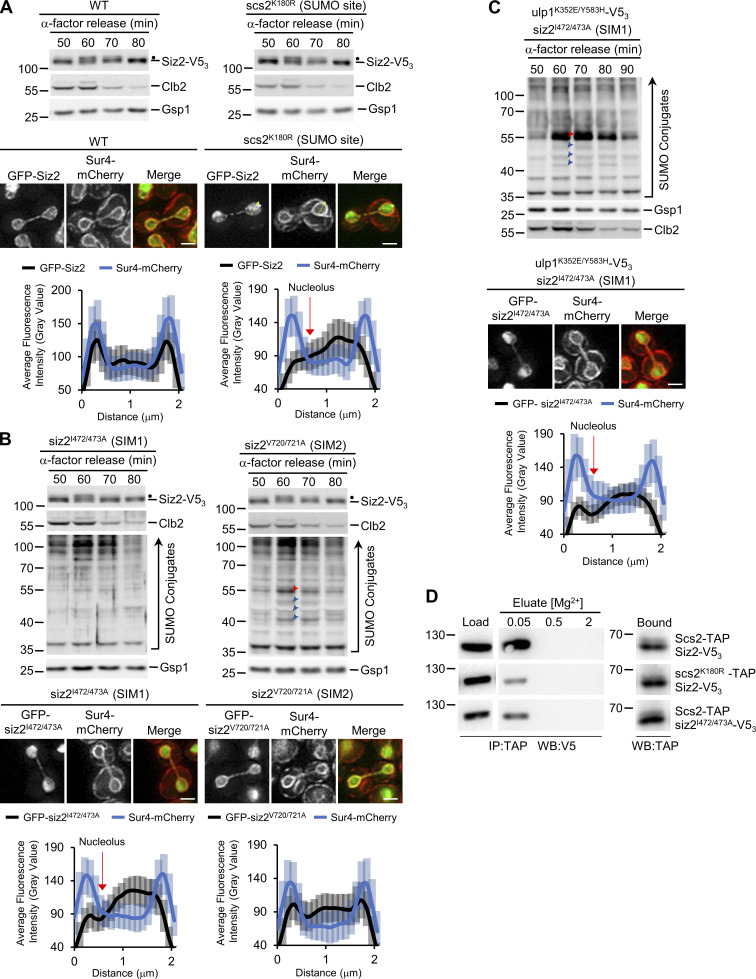
Our data show that both Siz2 phosphorylation and an FFAT–MSP domain interaction are required for Siz2 to bind and SUMOylate Scs2. This raised the possibility that Scs2 SUMOylation may stabilize its association with Siz2. To investigate this possibility, a strain producing the SUMOylation-deficient scs2K180R mutant, which retains INM localization (Fig. S2 F), was examined. In the scs2K180R mutant, Siz2 was still phosphorylated, but recruitment to the NE was reduced (Fig. 5 A and Fig. S2 A). Given that Siz2 contains two SUMO-interaction motifs (SIM domains), this result suggested that stable binding of Siz2 to Scs2 may be supported by a SUMO–SIM interaction. We tested this possibility by examining SIM1 (siz2I472/473A) or SIM2 (siz2V720/721A) mutants that would be expected to inhibit SIM function (Psakhye and Jentsch, 2012). Neither mutation changed Siz2 mitotic phosphorylation (Fig. 5 B and Fig. S2 A); however, the SIM1 mutant caused a visible reduction in mitotic NE association of Siz2 and mitotic SUMOylation of Scs2 and other Siz2 targets, while the SIM2 mutant had little effect on either (Fig. 5 B and Fig. S2 A). Of note, the mitotic SUMOylation defect of the SIM1 mutant, but not Siz2 localization, was restored in a strain also producing deSUMOylase-defective ulp1K352E/Y583H–V53, which is consistent with weak NE association and not a loss of Siz2 SUMO ligase activity (Fig. 5 C). These data demonstrate that the SIM1 motif of Siz2 contributes to Siz2 NE localization and Siz2-mediated SUMOylation during mitosis. Since SUMOylation of Scs2 is required for NE association of Siz2, a potential binding partner of the Siz2 SIM1 motif is SUMO-modified Scs2. In support of this conclusion, mutations in the Siz2 SIM1 motif (siz2I472/473A) or the Scs2 SUMO site (scs2K180R) reduced binding of Scs2 to Siz2 as determined by IP analysis (Fig. 5 D).
Figure 5.
A Siz2 SIM motif and Scs2-SUMO contribute to mitosis-specific SUMOylation. (A and B) The indicated strains, including WT and mutations in the Scs2 SUMO site (scs2K180R) or Siz2 SIMs (siz2I472/473A [SIM1] and siz2V720/721A [SIM2]) were assessed for mitotic Siz2 phosphorylation as described in Fig. 3. Dots highlight the position of mitotically phosphorylated Siz2-V53 and siz2-V53 mutants. (B and C) Mitotic SUMO conjugate profiles were assessed as described in Fig. 1 using siz2I472/473A, siz2V720/721A (B) and ulp1K352E/Y583H-V53 siz2I472/473A (C) strains. The position of Scs2-SUMO is indicated by a red arrowhead, and blue arrowheads point to three other prominent mitotic SUMO conjugates. Molecular mass markers are shown in kD. (A–C) Imaging and quantification of cells producing Sur4-mCherry along with GFP-Siz2, in a WT or scs2K180R background (A), GFP-siz2I472/473A alone (B), or in an ulp1K352E/Y583H-V53 background (C) or GFP-siz2V720/721A (B) were performed as outlined in Fig. 3. The locations of the GFP-fusions in mitotic cells were compared with Sur4-mCherry, and nuclear signal distribution was quantified using line scans. For strains exhibiting weak NE localization of the GFP-fusion, that is, in scs2K180R (A) and GFP-siz2 I472/473A (B and C) cells, each line was drawn starting at a point along the NE adjacent to the nucleolus, as exemplified by the yellow arrows in A. Nucleoli correspond to regions of reduced GFP-siz2 signal (see Fig. S1 E). Note, quantification of line scans shown in A–C was obtained at the same time as data shown in Fig. 3 C, and the WT data are also shown here for comparison. The approximate position of the nucleolus along these plots of average fluorescence intensity is indicated (red arrow, A–C). Error bars represent SD. Bar, 2 µm. (D) Binding of Scs2-TAP to Siz2-V53 or siz2I472/473A-V53 or binding of scs2K180R-TAP to Siz2-V53 was assessed. Scs2-TAP or scs2K180R-TAP was affinity-purified from cells (IP) producing Siz2 derivatives, and bound proteins were eluted using a Mg2+ step gradient. Equivalent portions of the indicated fractions were analyzed by Western blotting (WB) to assess levels of the V5- and TAP-tagged fusions. Note, all Load, Elution, or Bound fractions shown in panel D were derived from the same Western blot. These results shown here are from experiments performed in parallel with those shown in Fig. 4 C, and the same WT (Scs2-TAP/Siz2-V53) samples are shown here to allow samples shown in Fig. 4 C and Fig. 5 D to be directly compared.
The INM-localized Scs2–Siz2 complex is required for telomere tethering to the NE during mitosis
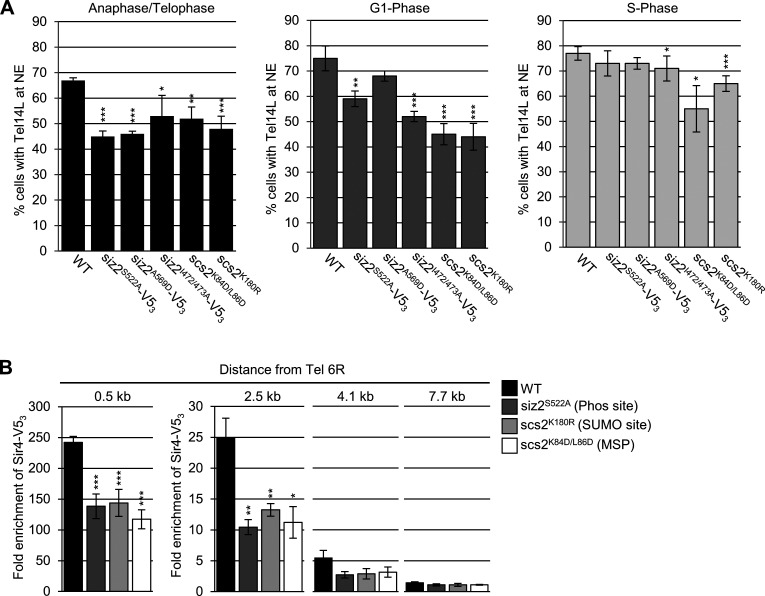
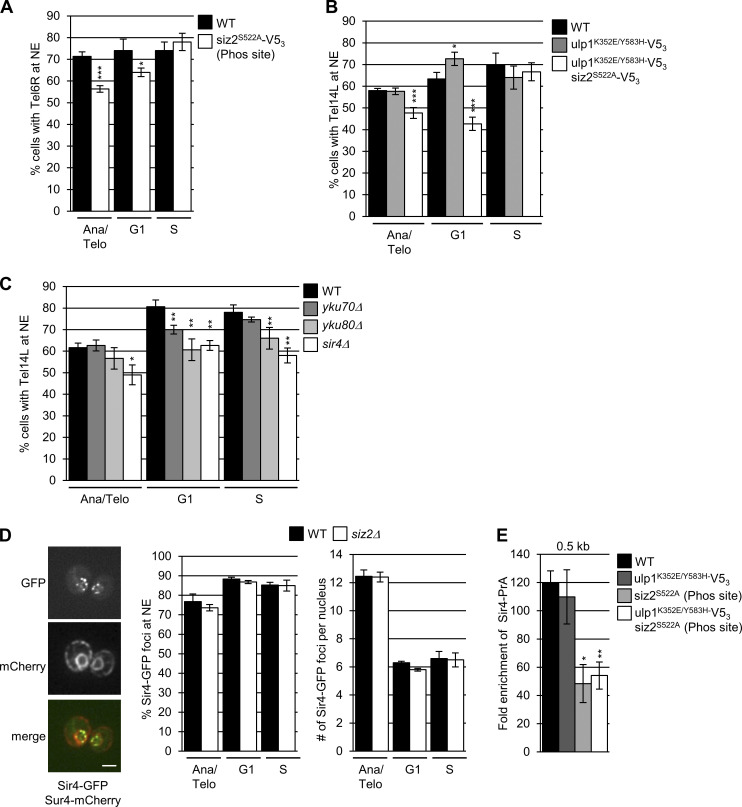
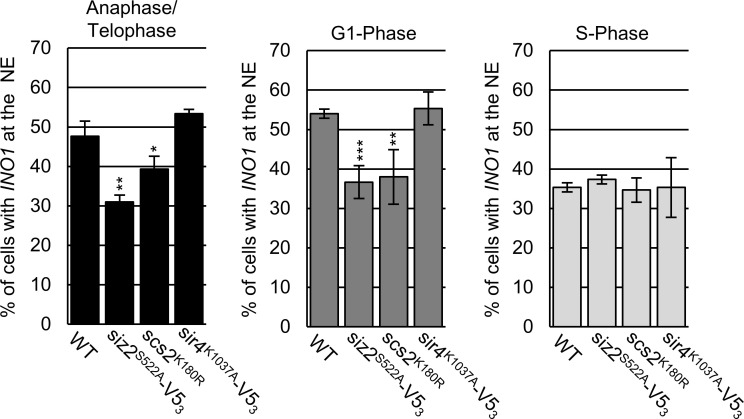
The Scs2-dependent repositioning of Siz2 from the nucleoplasm to the INM is initiated during a period of transition from metaphase to anaphase. During this time, sister chromosomes are segregated to daughter nuclei, and specific chromatin–NE interactions are reestablished as cells continue into G1-phase of the cell cycle (Hediger et al., 2002; Ebrahimi and Donaldson, 2008). To test for a role of Scs2-Siz2–mediated INM SUMOylation in establishing NE–chromatin interactions during mitosis, we examined the association of telomeres with the INM using a GFP-labeled telomere localization assay (Hediger et al., 2002). In WT cells, telomeres were detected at the NE in ∼65–75% of cells in each of the three cell cycle stages examined: anaphase/telophase, G1-phase, and S-phase. However, siz2 and scs2 point mutants that inhibit both Siz2 INM association and SUMOylation of INM targets showed decreased telomere 14L (Tel14L; Fig. 6 A) and 6R (Fig. S3 A) tethering to the NE during anaphase/telophase. Reduced Tel14L tethering persisted into G1-phase, with the exception of the siz2A569D (FFAT motif) mutation, which showed less of a defect (Fig. 6 A). By contrast, in the ulp1K352E/Y583H–V53 mutant, where Scs2-SUMO levels are elevated in mitosis and into G1-phase (Fig. 2 C), Tel14L levels at the NE are normal in M-phase and increased in G1-phase cells (Fig. S3 B). By S-phase, the NE association of Tel14L in the siz2, scs2, and ulp1 point mutants was largely normal or, in the case of scs2K180R (SUMO site mutation), their levels at the NE increased in S-phase compared with G1-phase (Fig. 6 A and Fig. S3 B). These data establish a strong correlation between Siz2 localization at the NE, Scs2-Siz2–mediated SUMOylation of INM proteins, and the tethering of Tel14L to the NE during mitosis.
Figure 6.
Telomere NE tethering during mitosis and G1-phase requires Scs2-Siz2 association and Scs2 SUMOylation. (A) Tethering of Tel14L to the NE was examined using epifluorescence imaging. The percentage of the total number of GFP-lacI/Tel14L-LacO256 foci examined that overlapped with NE-associated Sec63-mCherry signal was determined for at least three biological replicates. The graph shows the average values for various strains at the indicated points in the cell cycle. Cell cycle stage was assessed by bud size and nuclear morphology. n = 50 cells/replicate/cell cycle stage. Error bars represent SD. (B) Sir4-V53 binding to chromatin adjacent to Tel6R was assessed by ChIP and qPCR analysis using asynchronous cultures of the indicated strains. Graphs represent at least three biological replicates. Error bars represent SEM. Asterisks: significant change relative to WT using a two-tailed Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Phos, phosphorylation.
Figure S3.
INM localized Scs2-Siz2 is required for telomere tethering during mitosis and G1-phase of the cell cycle. (A–C) NE tethering of Tel6R in WT and siz2S522A-V53 cells (A), as well as NE tethering of Tel14L in WT (B and C), ulp1K352E/Y583H-V53, ulp1K352E/Y583H-V53 siz2S522A-V53 (B), yku70Δ, yku80Δ, and sir4Δ cells (C) were examined as described in Fig. 6. (D) Shown are epifluorescence images of WT cells producing Sir4-GFP and NE/ER–localized Sur4-mCherry. Merged images were used to assess the relative position of Sir4-GFP foci with respect to the NE (identified by Sur4-mCherry). Images were rendered using the unsharp mask filter in ImageJ. Bar, 2 µm. Bar graphs show the percentage of total Sir4-GFP foci at the NE (middle panel) and the average number of Sir4-GFP foci per nucleus (right panel) in WT and siz2Δ cells at the indicated cell cycle stage. Note, cells in anaphase/telophase show double the number of Sir4-GFP foci as seen in G1- and S-phase cells. Graphs in A–D represent data from at least three biological replicates. n = 50 cells/replicate/cell cycle stage. Error bars represent SD. Asterisks: significant difference relative to WT using a two-tailed Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (E) Sir4-PrA binding to chromatin adjacent to Tel6R was assessed by ChIP and qPCR analysis using asynchronous cultures of WT, ulp1K352E/Y583H-V53, siz2S522A-V53, and ulp1K352E/Y583H-V53 siz2S522A-V53 cells. Graphs represent at least three biological replicates. Error bars = SEM. Asterisks: significant change relative to WT using a two-tailed Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01. Ana/Telo, Anaphase/Telophase; Phos, phosphorylation.
Currently, the factors that contribute to telomere association with the INM during mitosis are unknown; however, various mechanisms have been described for telomere and subtelomeric chromatin tethering to the INM during other stages of the cell cycle. Key players include Sir4 and Yku70/80, which function to tether telomeres to the NE in G1- and S-phase (Taddei and Gasser, 2012; Kupiec, 2014). Sir4 and Yku80 are both SUMOylated by Siz2, with previous studies suggesting that Siz2-mediated SUMOylation directs Sir4- but not Yku80-dependent telomere tethering during G1 (Ferreira et al., 2011). Similarly, we find that Sir4, but not Yku70/80, plays a significant role in M-phase telomere tethering (Fig. S3 C). Yet, cells lacking Siz2 showed no defects in the INM association or number of Sir4-GFP foci (Lapetina et al., 2017; Fig. S3 D), suggesting that Sir4 association with the INM is not altered in the absence of Siz2. We therefore tested the alternative possibility that the Scs2–Siz2 complex supports the association of Sir4 with subtelomeric chromatin by chromatin immunoprecipitation (ChIP) analyses of Sir4 at regions adjacent to Tel6R. Similar to previous studies (Van de Vosse et al., 2013; Moradi-Fard et al., 2016), analysis of WT cells revealed Sir4-V53 bound near Tel6R, with the highest enrichment occurring near the telomere (0.5 kb), followed by a progressive decrease in association with increasing distance from the telomere (Fig. 6 B). In siz2 and scs2 mutant cells defective in mitotic SUMOylation, Sir4 enrichment was significantly reduced in all regions adjacent to Tel6R. By contrast, enhanced Scs2-Siz2–dependent mitotic SUMOylation (ulp1K352E/Y583H–V53) did not alter levels of Sir4 bound near Tel6R (Fig. S3 E). Thus, one function of the Scs2–Siz2 mitotic complex may be to mediate telomere tethering by supporting Sir4-subtelomeric chromatin association.
SUMOylation stabilizes the association of Sir4 with subtelomeric chromatin in mitosis
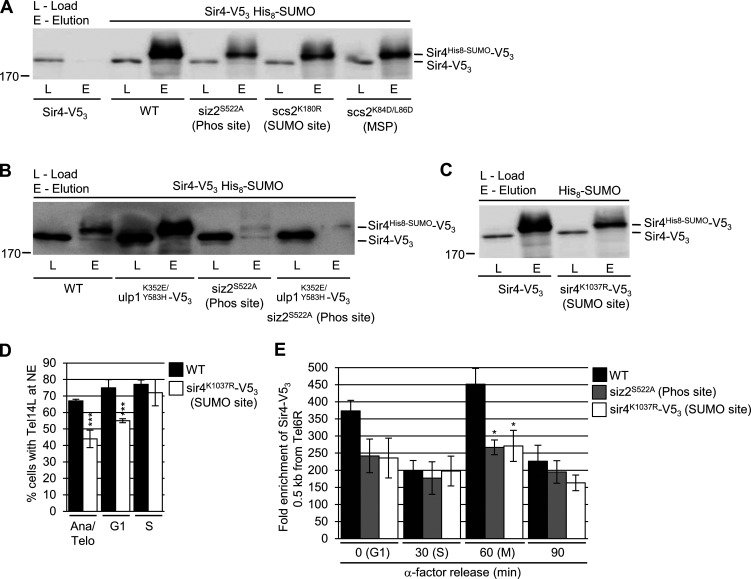
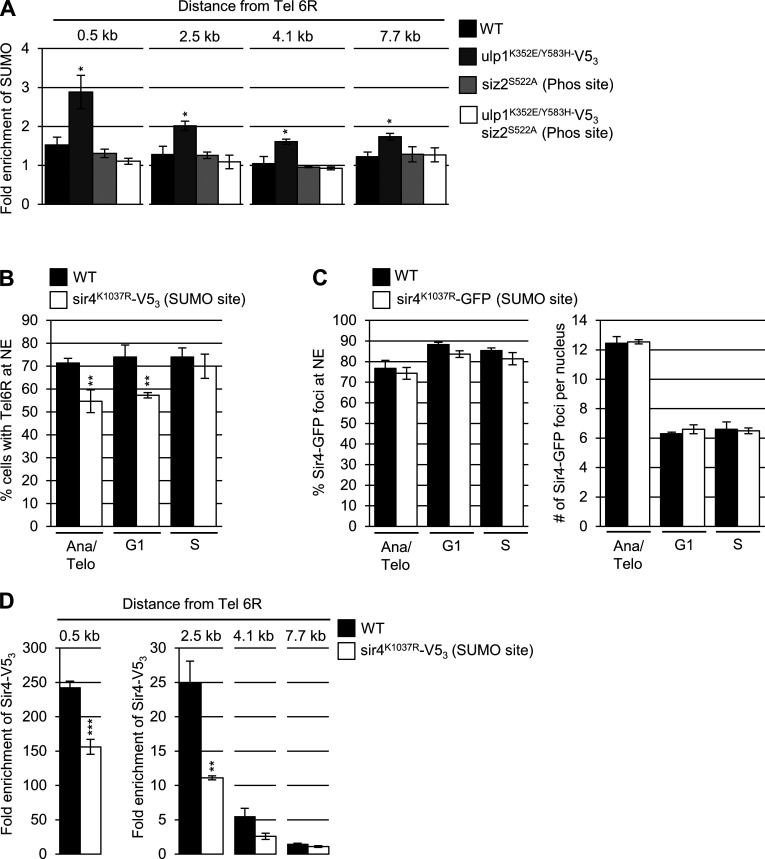
Sir4 SUMOylation is dependent on Siz2 (Ferreira et al., 2011), suggesting that formation of the mitotic Scs2–Siz2 complex may be responsible for mediating Sir4-subtelomeric association through SUMOylation. As such, we assessed Sir4-SUMO levels by examining purified His8-SUMO conjugates isolated from strains producing Sir4-V53. We found in siz2S522A, scs2K180R, or scs2K84D/L86D mutants, Sir4 SUMOylation was reduced compared with WT cells (Fig. 7 A). By contrast, enhanced Scs2-Siz2–dependent mitotic SUMOylation (ulp1K352E/Y583H–V53) showed increased SUMOylation of Sir4 (Fig. 7 B) and increased levels of SUMO at subtelomeric chromatin (Fig. S4 A), and both phenotypes were dependent on Siz2 targeting to the INM (Fig. 7 B and Fig. S4 A). These observations indicate that the targeting of Siz2 to the INM during mitosis is required for proper Sir4 SUMOylation and the association of Sir4 with subtelomeric chromatin.
Figure 7.
SUMOylation of Sir4 supports its incorporation into subtelomeric chromatin during mitosis. (A–C) His8-SUMO–conjugates were affinity purified from the indicated strains containing Sir4-V53 (A and B) or the sir4K1037R-V53 SUMO site mutant (C). Levels of Sir4 and sir4K1037R in the cell lysates (L) and Sir4-V53-SUMO and sir4K1037R-V53-SUMO in eluates (E) were examined by anti-V5 Western blotting. (D) Tethering of Tel14L to the NE in the indicated strains was assessed as described in Fig. 6. WT data are the same as that used in Fig. 6 A. Graphs represent at least three biological replicates where n = 50 cells/replicate/cell cycle stage. Error bars represent SD. (E) ChIP analysis was performed using synchronized cultures of the indicated strains at various cell cycle stages, including G1-phase (α-factor arrested cells), S-phase (30 min after α-factor release), and M-phase (60 min after α-factor release). Analysis was performed on a region 0.5 kb from Tel6R. Graphs represent at least three biological replicates. Error bars represent SEM. Asterisks: significant change relative to WT using a two-tailed Student’s t test. *, P ≤ 0.05; ***, P ≤ 0.001. Ana/Telo, Anaphase/Telophase; Phos, phosphorylation.
Figure S4.
The sir4K1037R mutation affects telomere tethering but not Sir4 NE localization. (A and D) SUMO conjugates (A), Sir4-V53, or sir4K1037R-V53 (D) bound to chromatin adjacent to Tel6R was assessed by ChIP and qPCR analysis using antibodies directed against SUMO or the V5 epitope in asynchronous cultures of the indicated strains. Graphs represent at least three biological replicates. Error bars represent SEM. Asterisks: significant change relative to WT using a two-tailed Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B) Tethering of Tel6R in WT and sir4K1037R-V53 cells was examined as described in Fig. 6 A. (C) NE localization of Sir4-GFP and sir4K1037R-GFP was assessed as described in Extended Data Fig. S3 D. Bar graphs show the percentage of total GFP foci at the NE (left panel) and the average number of foci per nucleus (right panel) at the indicated cell cycle stage. Cells in anaphase/telophase show double the number of Sir4-GFP foci as seen in G1- and S-phase cells. Note, WT data shown are the same as those in Extended Data Fig. S3 D. Graphs in B and C represent data from at least three biological replicates. n = 50 cells/replicate/cell cycle stage. Error bars represent SD. Asterisks: significant difference relative to WT using a two-tailed Student’s t test. **, P ≤ 0.01. Ana/Telo, Anaphase/Telophase; Phos, phosphorylation.
We further tested the significance of Sir4 SUMOylation by analyzing a Sir4 SUMOylation site mutant at residue K1037 within the Sir4 PAD domain (Zhao et al., 2014), a region necessary for telomere tethering activity (Andrulis et al., 2002; Taddei et al., 2004). The sir4K1037R mutant showed reduced Sir4 SUMOylation to levels similar to that detected in siz2 and scs2 point mutants (Fig. 7 C). Moreover, NE tethering of telomeres Tel14L (Fig. 7 D) and Tel6R (Fig. S4 B) were also significantly reduced in the sir4K1037R mutant cells, specifically in M- and G1-phase nuclei, but not S-phase. These tethering defects did not appear to stem from mislocalization of Sir4, as NE association of the sir4K1037R-GFP mutant protein was indistinguishable from that of Sir4-GFP (Fig. S4 C). The sir4K1037R mutant protein also showed a significantly reduced enrichment in all regions adjacent to Tel6R (Fig. S4 D). Notably, these phenotypes were similar to those observed in siz2 and scs2 point mutants exhibiting reduced Sir4 SUMOylation (Fig. 6 B and Fig. 7 A).
Finally, the defects in NE telomere tethering seen in both the siz2S522A and sir4K1037R mutant cells were restricted to M- and G1-phase (Fig. 6 A, Fig. 7 D, Fig. S3 A, and Fig. S4 B). Therefore, we examined levels of Sir4-V53 bound within a 0.5-kb region adjacent to Tel6R within synchronized cell cultures of these mutants (see Fig. S5). In WT cells, this analysis revealed an approximately twofold higher level of Sir4-V53 associated with Tel6R in M- and G1-phase cells compared with S-phase cells (Fig. 7 E). In cells where Sir4 SUMOylation is inhibited (siz2S522A or sir4K1037R mutant), S-phase levels of Sir4 bound to Tel6R subtelomeric chromatin were similar to WT; however, in M- and G1-phase cells, the association of Sir4 with Tel6R was reduced. These observations are consistent with a mitotic role for the Scs2–Siz2 complex in the SUMOylation of Sir4 and assembly of Sir4 into subtelomeric chromatin.
Figure S5.
SUMOylation contributes to Sir4 subtelomeric chromatin association during mitosis. To confirm the cell cycle stage of cultures used for cell cycle–dependent ChIP analysis (see Fig. 7 E), samples at each time point were analyzed by FACS to determine DNA content of cells in the population. The positions of 1n and 2n DNA peaks are shown. To the right, proteins derived from cell lysates harvested at the various time points were analyzed by Western blotting using anti-V5, SUMO, Clb2, and Gsp1 (load control) antibodies. Red arrowheads point to SUMOylated Scs2. Blue arrowheads point to other prominent mitotic SUMO conjugates. Molecular mass markers are shown in kD.
NPC association of active INO1 requires the Scs2–Siz2 complex
To extend our studies beyond telomeres, we examined the localization of the INO1 gene under conditions of gene activation (e.g., growth in media lacking inositol). Previous studies have shown that INO1 activation induces relocalization of the gene locus from the nucleoplasm to NPCs. This can be quantified by an increase in the frequency of cells displaying an NE-associated INO1 locus (Brickner and Walter, 2004; Light et al., 2010). Our examination of the nuclear positioning of activated INO1 during the cell cycle in WT cells revealed NE enrichment during M- and G1-phase (∼50–55% of cells) but not S-phase (Fig. 8), consistent with a previous report (Brickner and Brickner, 2010). However, in siz2S522A and scs2K180R point mutants, which inhibit SUMOylation of INM targets, the activated INO1 locus did not show increased localization at the NE during M- and G1-phase. By contrast, INO1 localization appeared normal in sir4K1037R mutant cells. These observations indicate that the temporally and spatially regulated functions of the Scs2–Siz2 complex direct the establishment of different types of NE–chromatin interactions by independent mechanisms.
Figure 8.
NPC association of activated INO1 requires the Scs2–Siz2 complex and Scs2 SUMOylation. NE localization of the activated INO1 locus was examined using epifluorescence imaging. The graphs show the percentage of total GFP-lac/INO1-LacO256 foci that colocalize with NE localized Nup49-RFP. For each indicated strain, three biological replicates were assessed. Cell cycle stage was determined using bud size and nuclear morphology. n = 50 cells/replicate/cell cycle stage. Error bars represent SD. Asterisks: significant change relative to WT using a two-tailed Student’s t test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Discussion
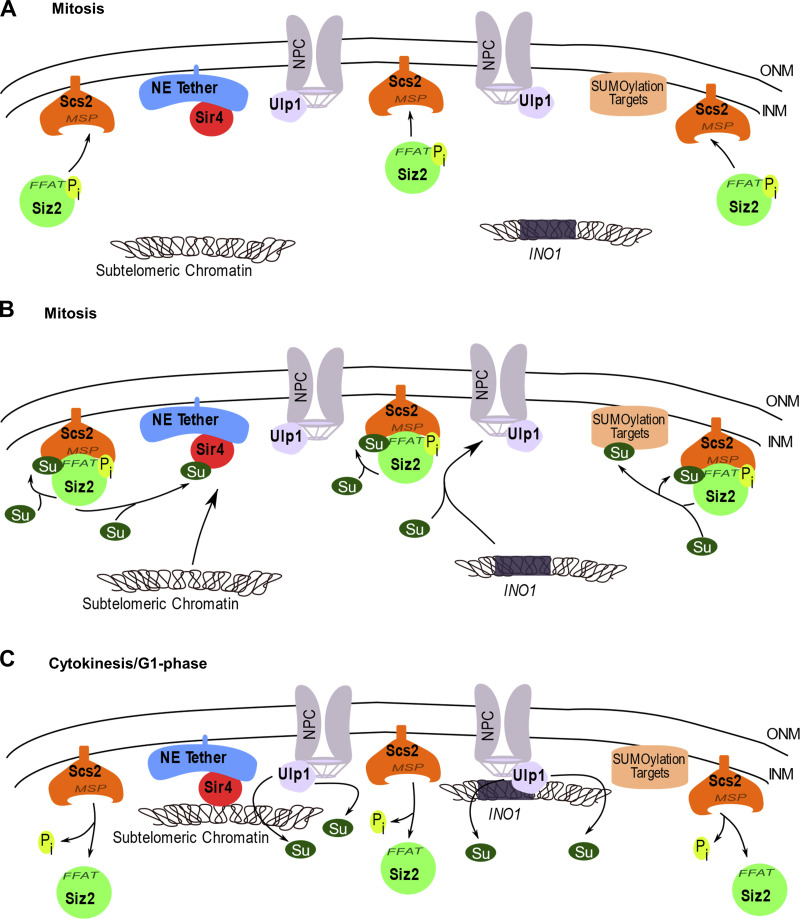
As cells move from interphase into mitosis, chromatin disassociates from the INM in preparation for distribution of the duplicated genome between two postmitotic nuclei. Beginning in anaphase and into telophase, chromatin interactions with the NE membrane are reformed, establishing interactions of the INM and NPCs with chromatin (Ebrahimi and Donaldson, 2008; Brickner and Brickner, 2010; Poleshko et al., 2013; Poleshko et al., 2019). In this work, we show that a key step in this process in S. cerevisiae is the phosphorylation-dependent binding of the SUMO E3-ligase Siz2 to the INM-localized membrane protein Scs2. The Scs2–Siz2 complex initiates multiple mitotic SUMOylation events along the nucleoplasmic face of the NE that fosters reformation of NE–chromatin interactions and the faithful inheritance of epigenetic programs by newly forming nuclei (Fig. 9).
Figure 9.
A model for Scs2-Siz2–dependent mitotic SUMOylation at the INM and its role in chromatin recruitment. (A) Early in mitosis, Siz2 as well as telomeres and activated INO1 localize within the nucleoplasm. Progression through metaphase and into anaphase is accompanied by the phosphorylation of Siz2 that directs its interaction with the integral membrane protein Scs2 at the INM. (B) Siz2 phosphorylation facilitates Scs2-Siz2 association through the Scs2 MSP domain and an FFAT-like motif in Siz2. These interactions lead to Scs2 SUMOylation and the association of Scs2-SUMO with a SIM motif in Siz2, further stabilizing the Scs2–Siz2 complex. The Scs2–Siz2 SUMO ligase complex then directs the SUMOylation of INM-associated proteins, including Sir4 and proteins associated with the activated INO1 locus, and these modifications facilitate subtelomeric chromatin association with Sir4 at the INM and activated INO1 with an NPC. (C) As cells exit mitosis and undergo cytokinesis, dephosphorylation of Siz2 and Ulp1-dependent deSUMOylation of Scs2 and other proteins along the INM occurs. These events lead to the dissolution of the Scs2–Siz2 complex, while telomeres and activated INO1 remain associated with the nuclear periphery. ONM, outer nuclear membrane; Pi, inorganic phosphate; Su, SUMO.
Spatiotemporal regulation of SUMOylation via Scs2
As cells progress through mitosis, we observed an accumulation of SUMO-conjugates along the NE that is directed by the phosphorylation of Siz2 and its subsequent NE association (Fig. 3 and Fig. 9). Our data suggest that Siz2 binding to the INM is, in part, controlled by a mechanism like that employed by Scs2 and other VAP protein family members to recruit soluble and membrane-bound cytoplasmic proteins to the ER (Brickner and Walter, 2004; Manford et al., 2012; Freyre et al., 2019). As in other cases (Goto et al., 2012; Kumagai et al., 2014; Weber-Boyvat et al., 2015; Kirmiz et al., 2018; Di Mattia et al., 2020), phosphorylation and an FFAT-like motif in Siz2 are critical for binding Scs2 (Fig. 3 and Fig. 4). However, unique to the Scs2–Siz2 complex is that its formation occurs in the nucleus and it uses a SUMO–SIM interaction to reinforce the FFAT–MSP domain interaction (Fig. 5). We envisage that a SUMOScs2–SIMSiz2 interaction functions downstream of the MSPScs2–FFAT/PhosphoSiz2 interaction to stabilize the Scs2–Siz2 complex at the INM. To our knowledge, this is the first instance where a SUMO–SIM interaction has been shown to reinforce interactions between an FFAT-like motif and an MSP-containing binding partner. Whether SUMOylation contributes to the interactions of Scs2 or other VAP family members with a larger repertoire of FFAT-containing proteins will be of future interest.
As cells exit mitosis and progress through cytokinesis, the release of Siz2 from the INM is presumed to require dephosphorylation of Siz2 and deSUMOylation of Scs2. Our data led us to conclude that Scs2 deSUMOylation is performed by Ulp1 as strains expressing the ulp1K352E point mutation show dramatically increased levels of SUMOylated Scs2 during mitosis and into interphase of the next cell cycle (Fig. 2). Since the ulp1K352E protein appears to remain competent to bind NPCs (Felberbaum et al., 2012), presumably at sites along the NPC nuclear basket (Panse et al., 2003; Zhao et al., 2004; Lewis et al., 2007; Palancade et al., 2007), we envisage that the K352E mutation may alter ulp1K352E recognition of or accessibility to INM-associated Scs2.
A mitotic role for the Scs2–Siz2 complex in NE–chromatin interactions
The abundance of SUMO-conjugates along the INM during mitosis (Fig. 1) suggests numerous proteins and a broad spectrum of NE-associated processes are temporally regulated by these SUMOylation events. This high concentration of SUMO-modified proteins at the INM would establish a two-dimensional binding surface for SIM domain–containing proteins. In this way, SUMO modifications at the INM could establish a multivalent interaction surface capable of promoting the assembly of various types of macromolecular complexes. Notably, the potential for SUMOylation to partition SIM-containing proteins near the INM parallels properties described in mammals for phase-separated PML bodies and their enhanced compartmentalization of SUMO and SIM-containing interacting partners (Van Damme et al., 2010; Banani et al., 2016; Min et al., 2019).
In this manner, as we show, the wave of mitotic NE SUMOylation supports interactions of both telomeres and the activated INO1 gene with the NE during the later stages of mitosis (Fig. 6, Fig. 7, and Fig. 8). With respect to telomeres, we further identified Sir4 as a key SUMOylation target of the Scs2–Siz2 complex (Fig. 7). Based on our analysis of the siz2S522A and sir4K1037R mutants and other mutants that inhibit Sir4 SUMOylation, we conclude that mitotic Sir4 SUMOylation supports Sir4 binding to subtelomeric chromatin and the association of this chromatin with the INM during the later stages of M- and the G1-phase of the next cell cycle (Fig. 6 and Fig. 7). Notably, reduced Sir4 SUMOylation (Fig. 7) does not alter Sir4 association with the NE (Fig. S4 C). These results, coupled with the observation that Siz2 recruitment to the INM is required for Sir4 SUMOylation (Fig. 7 A), lead us to conclude that Sir4 binding to subtelomeric chromatin during M/G1-phase occurs at the INM (Fig. 9).
Previous studies of the siz2Δ mutant have also shown that Siz2 contributes to additional interactions that support telomere association with the INM in S-phase (Ferreira et al., 2011; Lapetina et al., 2017). Among these are anchoring pathways mediated by the telomere-binding proteins Yku70 and Yku80, both of which have been shown to be SUMOylated, at least in part, by Siz2. Moreover, the siz2Δ mutation inhibited the anchoring functions of Yku80 in S-phase (Ferreira et al., 2011). When and where Siz2 SUMOylation of the Yku proteins occurs remains to be determined. Nonetheless, our data suggest that Yku70 and Yku80 SUMOylation is unlikely to contribute to telomere association with the INM during mitosis as yku70Δ and yku80Δ cells exhibit no defects in telomere association during this period of the cell cycle (Fig. S3 C).
For activated genes, we further note that the timing of Scs2-Siz2–mediated SUMOylation correlates with the observed binding of several active genes, including INO1, with NPCs in M-phase following their dissociation from the NE in the preceding S-phase (Brickner and Brickner, 2010). As such, we envisage that Scs2-Siz2–directed SUMOylation broadly functions to support binding of multiple active gene loci to NPCs, including those using transcription factors (Brickner et al., 2019) and especially those previously implicated to involve SUMOylation and Siz2 (Rosonina et al., 2010; Texari et al., 2013; Texari and Stutz, 2015; Saik et al., 2020).
Both the NE recruitment of Siz2 and SUMOylation of target proteins are transient, being largely reversed by G1-phase (Fig. 1 and Fig. 3). We interpret this to suggest that SUMO–SIM interactions formed in mitosis guide and augment additional protein–protein interactions that are maintained in interphase and support chromatin-NE associations. Moreover, the removal of these SUMO modifications may also play an important role. Interestingly, we observed that conditions that delay deSUMOylation of Scs2 and other INM proteins (ulp1K352E/Y583H mutant) led to increased INM retention of telomeres in G1-phase (Fig. S3 B). These results suggest that postmitotic deSUMOylation may relax interactions and promote the periodic switching of telomeres between NE bound and unbound states observed in interphase cells (Hediger et al., 2002).
It is also important to consider that the multiple Scs2-Siz2–dependent SUMO-conjugates detected during mitosis may reflect a broader spectrum of NE-associated processes impacted by these temporal and spatial SUMOylation events. Scs2 in particular has roles in the regulation of lipid metabolism and the expression of genes controlling inositol biosynthesis (Loewen et al., 2003; Brickner and Walter, 2004; Manford et al., 2012; Gaspar et al., 2017). Consequently, it is possible that SUMOylation of Scs2 coupled with Siz2 recruitment could further regulate Scs2 functions, including lipid metabolism, to accommodate NE expansion and the formation of two daughter nuclei. We expect that future work aimed at the identification of INM targets of Scs2-Siz2 and the consequences of blocking these specific SUMOylation events will be critical to addressing these possibilities.
Materials and methods
Yeast strains and plasmids
Yeast strains are listed in Table S1. Strains were derived from S288C (BY4741, BY4742) backgrounds except for telomere localization strains (W303) and INO1 localization strains (YEF473A). Cells were grown in 1% yeast extract, 2% bactopeptone, and 2% glucose medium or synthetic medium (per liter: 1.7 g yeast nitrogen base, 5 g ammonium acetate, 1.7 g amino acid dropout powder, and 2% glucose), as required.
Transformations were performed using a lithium acetate/polyethylene glycol method (Gietz and Woods, 2002). Gene deletions were derived from strains in the haploid Matα yeast deletion library (Invitrogen). Strains bearing gene modifications for protein fusions and amino acid substitutions were generated using a PCR-based genomic integration method (Longtine et al., 1998). To switch an MX marker gene, associated with a given gene deletion or gene encoding a tagged protein, a PCR cassette containing the appropriate MARKER (HIS3, KAN, NAT or HPH)–MX gene was used for integration. Oligonucleotides used are listed in Table S2.
PCR cassettes, used for integrating the coding sequence for the V53 tag at the 3′ end of relevant genes, were generated using the plasmid pTM1198 (Lapetina et al., 2017) as a template and oligonucleotides listed in Table S2.
A PCR cassette, used for integrating the coding sequence for the HA3 tag at the 5′ end of SCS2, was produced using the plasmid pFA6a-kanMX6-SCS2pr-3HA as a template and oligonucleotides listed in Table S2. pFA6a-kanMX6-SCS2pr-3HA was generated by replacing the GAL1 promoter, bounded by BglII/PacI restriction enzyme sites in pFA6a-kanMX6-PGAL1-3HA (Longtine et al., 1998), with a PCR cassette bounded by BglII/PacI restriction enzyme sites and containing 372 bp of the SCS2 5′ UTR (see Table S2 for oligonucleotides used).
Strains producing Sir4-eGFP, Sir4-PrA were derived from those described in Lapetina et al. (2017). Strains carrying tagged telomeres (i.e., TelXIV-L::256xlacO-TRP1 or TelXIV-L::256xlacO-TRP1) were derived from those described in Van de Vosse et al. (2013). Strains carrying a tagged INO1 locus (i.e., INO1::lacO(256)-TRP1) were derived from those described in Saik et al. (2020). Scs2-TAP was obtained from the yeast TAP tag library (Ghaemmaghami et al., 2003). PCR cassettes used to integrate the coding sequence for mCherry at the 3′ end of SUR4, NOP56, and scs21–225 were generated using the pGEM-4Z-mCherry-NAT plasmid as a template (Cairo et al., 2013) and oligonucleotides listed in Table S2.
pRS315.SMT3pr-His8-SMT3-HPH was constructed by cloning three PCR products into pRS315 (Sikorski and Hieter, 1989), including (1) 350 bp of the SMT3 5′ UTR bounded by SacI/NotI restriction enzyme sites; (2) the coding region for His8-Smt3 plus 324 bp of the SMT3 3′ UTR bounded by NotI/SalI restriction enzyme sites; and (3) the HPH-MX sequence bounded by SalI/ApaI restriction sites (New England Biolabs). This plasmid was used as a PCR template, and the resulting SMT3pr-His8-SMT3-HPH cassette was integrated at the SMT3 locus. Oligonucleotides used to produce PCR cassettes employed in plasmid construction and gene integration are listed in Table S2.
Amino acid substitutions were introduced into WT cells by site-directed mutagenesis of genomic loci using a PCR-based one-step integration method. Genomic DNA derived from SIR4-, SIZ2-, and ULP1-V53 strains, as well as a KAN-SCS2 strain, were used as the DNA templates. Mutagenic oligonucleotides employed for PCR reactions are listed in Table S2. The KAN-SCS2 strain was generated by integrating a PCR cassette, containing the KAN-MX marker gene, between nucleotides 242/241 5′ of the SCS2 start codon. Oligonucleotides employed to produce this KAN-MX cassette are listed in Table S2. All mutations were sequence verified.
All GFP-siz2 fusions were generated in two steps. A PCR cassette containing GFPS65T was integrated at the 5′ end of each siz2-V53-KAN gene to produce GFP-siz2-V53 strains (Lapetina et al., 2017). The V53-KAN region was then replaced by integration of a PCR cassette derived using genomic DNA from a SIZ2-HPH strain where the HPH-MX cassette was integrated between nucleotides 273/274 3′ of the SIZ2 stop codon as a template. The PCR cassette used consisted of 60 bp at the 3′ end of the SIZ2 ORF - 273 bp after the SIZ2 stop codon - HPH-MX - nucleotides 274–334 3′ of the SIZ2 stop codon. To generate the GFP-siz2V720/721A strain, the PCR cassette used to replace V53-KAN included nucleotides for the A720/721 codons. Regeneration of the 3′ end of the GFP-siz2 genes was confirmed by sequencing. Similarly, the V53 coding sequence was removed from siz2S522A-V53-KAN to generate siz2S522A-NAT strains. Note that genomic DNA from a SIZ2-NAT strain was used in this case. A similar strategy was used to remove the V53 tag coding sequence and the Y583H mutation from ulp1K352E/Y583H–V53 to generate ulpK352E. In this case, genomic DNA from an ULP1-KAN strain, where the KAN-MX cassette was integrated between nucleotides 359/360 3′ of the ULP1 stop codon, was used as a template for the PCR. The resulting PCR cassette used for integration consisted of 266 bp at the 3′ end of the ULP1 ORF - 359 bp after the ULP1 stop codon - HPH-MX - nucleotides 359–424 3′ of the ULP1 stop codon. Oligonucleotides employed to produce the various PCR cassettes described are listed in Table S2.
Genomic DNA from the SCS2-TAP strain was used as a template to produce a PCR cassette used to integrate the coding sequence for TAP at the 3′ end of scs2K180R and scs2K84D/L86D. Oligonucleotides used are listed in Table S2.
Strains used for split-superfolder GFP analysis were generated by first integrating a PCR cassette, consisting of NAT-CDC42pr-GFP1–10 (Smoyer et al., 2016) at the 5′ end of SCS2, scs2K180R, and scs2K84D/L86D. Oligonucleotides employed are listed in Table S2. Plasmids encoding GFP11-mCherry-Hxk1 or GFP11-mCherry-Pus1 (Smoyer et al., 2016) were then transformed into the NAT-CDC42pr-GFP1–10-SCS2, NAT-CDC42pr-GFP1–10-scs2K180R, and NAT-CDC42pr-GFP1–10-scs2K84D/L86D strains.
α-Factor arrest-release assays
All strains used in α-factor arrest-release assays were MATa bar1Δ. 25 ml YPD cultures, incubated overnight at RT, were diluted to an OD600 = 0.3 in 30 ml of YPD followed by addition of α-factor to 10 ng/ml (Sigma; T6901) and incubated at 30°C for ∼2 h 15 min to induce G1-phase arrest. Cells from an aliquot of this culture equivalent to OD600 = 1 were pelleted, resuspended in 50 µl of 2× SDS sample buffer, and incubated at ∼80°C for 15 min. This represented the 0-min time point. The remaining cells were pelleted, washed once with 1 ml of YPD, pelleted again, and resuspended in 25 ml fresh YPD to a final OD600 = 0.6, thereby removing α-factor. Cultures were grown at 30°C and sampled every 10 min as described for the 0-min time point. All lysates were subsequently sonicated (Branson Sonifier 250) and debris pelleted by centrifugation before running through an 8% SDS-PAGE gel and subsequent Western blot analysis.
For α-factor arrest-release ChIP experiments, 50-ml cultures were incubated overnight at RT, then diluted to an OD600 of 0.2 in 500 ml of YPD, and incubated for 1.5 h at 30°C followed by addition of α-factor (10 ng/ml) and incubation at 30°C for ∼2 h 15 min. An OD600 = 50 equivalent was harvested from each culture, representing the 0-h time point, and treated as described for ChIP (see below). To remove α-factor, the remaining cells were pelleted, washed twice with ddH2O, and resuspended in 300 ml fresh YPD, which was partitioned into three separate flasks. These cultures were incubated at 30°C for 30, 60, or 90 min before harvesting and subsequent treatment. Samples were then analyzed by ChIP (see below). OD600 = 1 equivalents were taken at all time points for Western blot, as described above and for FACS analysis.
Phosphatase treatment
To a 50-µl lysate sample, from the 60-min α-factor arrest-release time point using Siz2-V53 cells, 200 µl of methanol, 50 µl of chloroform, and 150 µl of ddH2O were added sequentially, followed by vortexing for 10 s after each addition. The final mixture was centrifuged for 2 min at 15,000 rpm. The resulting top layer was carefully removed, leaving the protein precipitate and the bottom layer. 300 µl of methanol was added to the remaining sample, and the mixture was vortexed for 10 s followed by centrifugation for 2 min at 15,000 rpm. Residual liquid was removed, and the resulting pellet was air-dried. The dried pellet was resuspended in 50 µl of 0.5% wt/vol SDS. To 10 µl of this sample, 10 µl of λ phosphatase buffer (New England Biolabs; B0761S), 10 µl of 10 mM MnCl2, 1 µl λ phosphatase (New England Biolabs; P0753S), and 69 µl ddH2O were added. For the -PPase sample, 1 µl λ phosphatase was replaced with 1 µl ddH2O. Reactions were incubated at 30°C for 1 h, after which 20 µl of 50% TCA was added, and the samples were incubated overnight at 4°C. Samples were then centrifuged at 15,000 rpm for 20 min at 4°C, the supernatant was removed, and the remaining pellet was air-dried. The dried pellet was resuspended in 25 µl of 2× sample buffer and heated at 80°C for ∼15 min before Western blot analysis.
Anti-SUMO immunofluorescence
5 ml YPD cultures, incubated overnight at RT, were diluted to an OD600 = 0.2 in 5 ml of fresh YPD and incubated at 30°C to an OD600 ∼0.8. To each culture, 0.6 ml of 10X phosphate buffer (1 M KH2PO4, 370 mM KOH, and 0.5 mM MgCl2) and 0.8 ml of 37% formaldehyde were added, followed by incubation at 30°C for 30 min. Cells were then pelleted and washed 2× with 1X phosphate buffer. The final pellets were resuspended in 100 µl of sorbitol-citrate buffer (100 mM K2PO4, 3.6 mM citric acid, 1.2 M sorbitol, and 0.5 mM MgCl2), and DTT was added to a final concentration of 1 mM. Cells were then pelleted and resuspended in 100 µl of sorbitol-citrate buffer supplemented with 2 mg/ml 20T zymolyase and incubated at 30°C for 20 min. Cells were then pelleted and washed 2× with 1 ml sorbitol-citrate buffer, and the final pellet was resuspended in 50 µl of sorbitol-citrate buffer. 20 µl of the cell suspension was pipetted onto a multi-well slide coated with 0.1% poly-L-lysine, and the slide was incubated at RT in a covered box lined with damp paper towels for 30 min. All subsequent incubations and washes were performed at RT in the same box. ∼20 µl of solution was used at each step, and solutions were exchanged by their aspiration off of the wells followed by the addition of fresh solution. Steps included (1) 1× PBS with Tween 20 (PBST) wash, (2) 1× addition of PBS 0.1% Triton X-100 with a 10-min incubation, (3) 2× wash with PBST, (4) 1× addition of PBST, 1% BSA with a 10-min incubation, (5) 1× addition of PBST, 1% BSA supplemented with a rabbit polyclonal anti-SUMO antibody (Wozniak Lab) at a 1:500 dilution with a 1-h incubation, (6) 10× wash with PBST, 0.1% BSA, (7) 1× addition of PBST, 1% BSA supplemented with Alexa Fluor 488 donkey anti-rabbit IgG antibody (Life Technologies; A11055) at a 1:200 dilution, and (8) 10× wash with PBST, 0.1% BSA. After the final wash, ∼3 µl of DAPI–Fluoromount-G (SouthernBiotech; 0100–20) was added to each well, and a coverslip was placed over the slide. Cells were then analyzed by epifluorescence imaging.
INO1 gene induction
Overnight synthetic media (+inositol) cultures, incubated at RT, were diluted into the fresh synthetic media (+inositol) to an OD600 = 0.2, then incubated at 30°C to an OD600 = ∼0.8. Cells were then pelleted, washed once with water, and resuspended in synthetic media lacking inositol to an OD600 = 0.5 to induce INO1 expression. Cultures were incubated at 30°C for 3 h followed by analysis by epifluorescence imaging as described below.
Epifluorescence imaging and analysis
For live cell imaging of various GFP-Siz2–producing strains, as well as strains producing split-superfolder-GFP, GFP-tagged telomeres, and Sir4-GFP, YPD or synthetic media cultures, incubated overnight at RT, were diluted to an OD600 ∼0.1, then incubated at 30°C to an OD600 ∼1, except for GFP-Siz2 strains, which were incubated at RT. GFP-tagged telomere strains were also grown in YPD supplemented with 120 µg/ml of adenine. Cells from 1 ml of culture were then pelleted and washed with 1 ml of synthetic complete medium, and the resulting pellet was suspended in ∼20 µl of synthetic complete medium. 1.5 µl of this cell suspension was then spotted onto a microscope slide coated with a 1% agarose pad consisting of 1 g of agarose per 100 ml of synthetic complete medium. All imaging was performed at RT.
Epifluorescence images of cells prepared for anti-SUMO immunofluorescence, as well as cells producing GFP–Siz2/Sur4–mCherry (Video 1), GFP–Siz2/Nop53–mCherry, GFP–Siz2/scs21–225–mCherry, or split-superfolder–GFP fusions, were acquired using an Axio Observer.Z1 microscope (Carl Zeiss Inc.) equipped with a UPlanS-Apochromat 100×/1.40 NA oil objective lens (Carl Zeiss Inc.) and an AxioCam MRm digital camera with a charge-coupled device (Carl Zeiss Inc.). Images were saved using the AxioVision software and rendered using ImageJ software (National Institutes of Health [NIH]) for display. The unsharp mask filter was used to analyze anti-SUMO immunofluorescence images (Radius [Sigma]: 1.0 pixels; Mask Weight: 0.9) as well as GFP–Siz2/Sur4–mCherry (Video 1), GFP–Siz2/Nop56–mCherry, and GFP–Siz2/scs21–225–mCherry images (Radius [Sigma]: 3.0 pixels; Mask Weight: 0.8).
Epifluorescence images of cells containing GFP–Siz2/Sur4–mCherry, GFP-tagged telomeres, Sir4-eGFP, or GFP-tagged INO1 were acquired on a DeltaVision Elite imaging system (GE Healthcare Life Sciences) with a 60×/1.42 NA oil, Plan Apo N objective (Olympus). Images were collected as 15 × 0.2-µm z-stacks using the SoftWoRx software (GE Healthcare Life Sciences; version 6.5.2), then rendered and analyzed using ImageJ (NIH). The unsharp mask filter was used to analyze GFP–Siz2/Sur4–mCherry images (Radius [Sigma]: 2.0 pixels; Mask Weight: 0.8).
Cell cycle stage was assessed based on bud size and/or nuclear morphology, specifically, G1-phase: unbudded cells, round nucleus; S phase: small-budded cells, round nucleus away from budneck; and anaphase/telophase: large-budded cells, barbell-shaped nuclei.
For line scan quantification of SUMO/DAPI and GFP–Siz2/Sur4–mCherry, images from each channel were rendered in ImageJ as described above, converted to an 8-bit image, and then combined into a single stacked image file. A line of specific length, 1.75 µm for SUMO/DAPI and 2.1 µm for GFP–Siz2/Sur4–mCherry, was drawn through individual cell nuclei, and the fluorescence intensity along the line was quantified for each channel using ImageJ. In addition, for IF quantification, each line segment was drawn to be centered upon the DAPI signal and encompass the enriched SUMO signal observed in each cell. For GFP–Siz2/Sur4–mCherry, each line was drawn to pass through two points of Sur4-mCherry signal along the NE such that the line passed through the nucleoplasmic GFP-Siz2 signal, avoiding the nucleolus where the GFP-Siz2 signal is reduced. However, as the weak NE GFP-Siz2 signal in scs2K180R and siz2I472/473A backgrounds was most apparent in the regions adjacent to the nucleolus, lines drawn through the nuclei of these cells were initiated at the NE region adjacent to the nucleolus. n = 25 for each strain and cell cycle stage analyzed. Error bars represent SD.
Localization of foci corresponding to Sir4-eGFP, GFP-tagged INO1, and GFP-tagged telomeres in anaphase/telophase cells, as well as foci corresponding to GFP-tagged telomeres in G1- and S-phase ulp1K352E/Y583H-V53 cells and corresponding WT cells, were scored positive when the focus fully or partially overlapped with the NE localized marker (i.e., Sur4-mCherry, Nup49-RFP, or Sec63-GFP). Subnuclear localization of G1- and S-phase foci of GFP-tagged telomeres was determined, as previously described (Lapetina et al., 2017), by dividing the telomere distance from the NE (TD) by the nuclear radius (r). The TD/r ratio (R) was used to group telomeres into three concentric zones of equal area. Zone 1 represents foci with ratios ≤0.184 × R (telomere at the NE); zone 2 represents foci with ratios >0.184 × R and <0.422 × R; and zone 3 represents foci with ratios ≥0.422 × R. This method was only used in cells and at cell cycle stages, where the nuclei remained spherical.
Western blot analysis
Proteins separated using SDS-PAGE were transferred to nitrocellulose membranes. Membranes were incubated in blocking buffer (Tris-buffered saline with 0.1% Tween [TBST], or PBST with 5% milk powder) for at least 1 h at RT. Fresh blocking buffer supplemented with primary antibody was then added, followed by incubation overnight at 4°C. Membranes were then washed 3× with TBST or PBST, then incubated in fresh blocking buffer supplemented with a secondary antibody-HRP conjugate and incubated for at least 1 h at RT. Membranes were then washed 3× with TBST or PBST, and proteins were visualized by chemiluminescence (Amersham; RPN2106) using an ImageQuant LAS 4000 (GE Healthcare Life Sciences) imaging system. All Western blot images were rendered using ImageJ software (NIH).
Primary antibodies used were rabbit polyclonal anti-Smt3 (Wozniak Lab), mouse monoclonal anti-V5 (AbCam; ab27671), mouse monoclonal anti-HA (Santa Cruz; sc-7392), rabbit polyclonal anti-Clb2 (Santa Cruz; sc-9071), rabbit polyclonal anti-PrA (used to probe Scs2-TAP; Sigma; P3775), and rabbit polyclonal anti-Gsp1 (Wozniak Lab). Secondary antibodies used were goat anti-rabbit IgG (H+L)-HRP conjugate (Bio-Rad; 170–6515) and goat anti-mouse IgG (H+L)-HRP conjugate (BioRad; 170–6516). Antibodies were all used at a 1:10,000 dilution.
Coaffinity purification
For coaffinity purifications (Van de Vosse et al., 2013), 50 ml YPD cultures of Scs2-TAP, scs2K84D/L86D–TAP, or scs2K180R-TAP–producing cells, grown overnight at RT, were diluted in 1 liter of fresh YPD to an OD600 ∼0.1, then incubated at 30°C to an OD600 ∼1.0. Cells were pelleted and washed once with 25 ml cold ddH2O, and the resulting pellet was extruded through a 5-ml syringe directly into a 50-ml falcon tube containing liquid nitrogen–producing cell noodles. Liquid nitrogen was removed, and the noodles were stored at −80°C. Noodles were then subjected to at least seven rounds of ball mill grinding (Reitch; PM100; 1 min 15 s, 450 rpm per round), keeping the grinding vessel cold between rounds by partial immersion in liquid nitrogen. The resulting cell powder was stored at −80°C.
To 1 g of cell powder, 2 ml of IP buffer (2 mM MgCl2, 20 mM Hepes-KOH, pH 7.4, 0.1% Tween-20, 110 mM KOAc, antifoam-B emulsion at 1:5,000 dilution, and protease inhibitors [1 complete EDTA-free pellet; Roche 05056489001]/50 ml buffer) was added. The suspension was incubated on ice for 30 min, with vortexing every 5 min. The resulting lysate was cleared by centrifugation at 1,500 g for 10 min at 4°C. 25 µl of the clarified lysate, representing the load, was added to 1 ml of ddH20, followed by TCA precipitation and resuspension of the resulting pellet in 75 µl of 2X sample buffer. 3 mg of IgG-conjugated magnetic beads (Dynabeads; Invitrogen; 143.01, Rabbit IgG; Sigma; I5006-10MG) in 100 µl of IP buffer was added to the 2 ml of clarified cell lysate, and the mixture was incubated for 1 h at 4°C with rotation. Beads were collected using a magnet and washed 10× with 1 ml of IP buffer at 4°C. Proteins bound to beads were eluted at 4°C using 0.5 ml IP buffer containing incrementally increasing concentrations of MgCl2 (0.05, 0.5, and 2 M) followed by a final elution using 0.5 ml of 0.5 M acetic acid to release the TAP fusion protein from the beads (Bound). To the 500 µl eluate fractions, 500 µl of ddH20 was added followed by TCA precipitation as described above. All samples collected were analyzed by Western blotting.
His8-SUMO pulldowns
Frozen cell powders derived from strains producing Sir4-V53 and His8-Smt3 (His8-SUMO) were produced as described for coaffinity purification. Note that all 8 M urea/phosphate buffers were made fresh just before use. To 1 g of frozen cell powder, 10 ml of resuspension buffer (8 M Urea, 50 mM NaPO4, pH 8.0, 500 mM NaCl, and 1% NP-40-Igepal) was added, and the mixture was incubated at RT with intermittent vortexing. Lysates were centrifuged at 15,000 rpm for 20 min. 25 µl of the clarified lysate was added to 1 ml ddH20, followed by TCA precipitation and resuspension in 100 µl of 2X sample buffer (Load sample). The remaining lysate was transferred to a 15-ml falcon tube, and 1 ml of a 50% slurry consisting of NiNTA agarose beads (Qiagen; 30210) in resuspension buffer was added per 10 ml of lysate. The mixture was incubated at RT with rotation for 2 h. NiNTA agarose beads were then pelleted by centrifugation at 1,000 rpm for 1 min, and the lysate was removed. The beads were then washed 3× with 6× bead volume of wash buffer (8 M Urea, 50 mM NaPO4, pH 6.3, 500 mM NaCl, and 1% NP-40-Igepal). After the last wash, 2 ml of elution buffer (8 M Urea, 50 mM NaPO4, 500 mM NaCl, and 1% NP-40-Igepal adjusted to pH 4.5) was added to the beads, and the slurry was incubated at RT with rotation for 1 h followed by centrifugation at 1,000 rpm for 1 min. The resulting eluant was collected and subjected to TCA precipitation, and the resulting pellet solubilized using 100 µl of 2X sample buffer. Load and eluate samples were then analyzed by Western blotting.
ChIP
10 ml YPD cultures, incubated overnight at RT, were diluted into 50 ml of fresh YPD to an OD600 ∼0.2 and grown at 30°C to an OD600 ∼0.8. Cells from an OD600 = 50 equivalent of each culture were pelleted, and the cells were resuspended in 50 ml of YPD 1% formaldehyde, followed by incubation at 30°C for 20 min to induce cross-linking. Glycine was added to 125 mM followed by a 5-min incubation at RT to quench cross-linking. Cells were pelleted and washed with TBS, and the resulting pellet was flash frozen using liquid nitrogen and stored at −80°C.
Cells were resuspended in 500 µl FA lysis buffer (50 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% Na deoxycholate) and disrupted by glass bead lysis at 4°C. Glass beads were removed from the lysate, and the lysates were then subjected to sonication (Branson Sonifier 250), while keeping samples on ice, to shear chromosomal DNA to an average size of ∼400 bp, as assessed by electrophoresis. Lysates were diluted by the addition of FA lysis buffer to a final volume of 1.5 ml. 500 µl, representing the Input, was then diluted 100× into a 10 mM Tris (pH 8.0), 1 mM EDTA, 1% SDS solution and subsequently reverse cross-linked as described below. The remaining lysate was incubated with 5 µl of single-stranded DNA (10 mg/ml) and 2 µl of mouse monoclonal anti-V5 antibody (Abcam; ab27671), 4 µl of rabbit polyclonal anti-PrA (Sigma; P3775) antibody, or 4 µl of rabbit polyclonal anti-Smt3 (SUMO) antibody (Wozniak Lab) for 2 h at 4°C. 30 µl of Protein G Dynabeads (Invitrogen; 10004D) in FA lysis buffer was added to each lysate followed by 1-h incubation at 4°C. Beads were collected by magnet and sequentially washed 2× with FA lysis buffer, 1× with FA lysis buffer including 500 mM NaCl, 1× with wash buffer (10 mM Tris-HCl, pH 8, 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, and 0.5% sodium deoxycholate), and 1× with TE buffer. After the final wash, chromatin was eluted from the beads using two rounds of incubation at 65°C for 10 min with 200 µl of TE 1% SDS. Input and ChIP samples were then reverse cross-linked by overnight incubation at 65°C, followed by addition of 5 µl of Proteinase K (20 mg/ml) and 1 µl of glycogen (20 mg/ml) to the samples and incubation at 37°C for 2 h. 40 µl of 5 M LiCl was then added to each sample, followed by phenol/chloroform extraction and ethanol precipitation. The resulting DNA pellets were resuspended in 50 µl TE and 5 µl of RNase A (5 mg/ml), then incubated at 37°C for 1 h followed by purification using Qiagen PCR Purification Kit (28106).
Samples were analyzed by qPCR (Makio and Wozniak, 2020). ChIP and Input DNA were used to amplify target sequences of interest using PerfeCTa SYBR green PCR mix (Quanta Biosciences; 95056–500) and an MX3000 (Agilent) instrument. The relative fold enrichment of the subtelomeric chromatin immunoprecipitated with the protein of interest was evaluated by the comparative cycle threshold (ΔΔCt) method (Livak and Schmittgen, 2001). PCR amplification of each subtelomeric region was first normalized against the amplification of the corresponding Input DNA to generate a Δcycle threshold value. Each subtelomeric region was then normalized to the amplification of a nonspecific binding control region, including either 17.1 kb from Tel6R (Sir4 ChIP) or an intergenic region in Chromosome V (SUMO ChIP), to generate ΔΔCt values. The relative fold enrichment of subtelomeric chromatin over background was given as 2-ΔΔCt, based on the assumption that the PCR reaction was 100% efficient. Oligonucleotides used for qPCR are listed in Table S2.
FACS
Cells pelleted for DNA content determination by FACS (see α-Factor arrest-release assays) were resuspended in 1 ml of 70% ethanol and incubated overnight at 4°C. Cells were then pelleted, and the pellets were washed 2× with 1 ml 50 mM Tris-HCl (pH 8.0). Final pellets were resuspended in 0.5 ml of 50 mM Tris-HCl (pH 8.0) containing RNase A (0.4 mg/ml) and incubated at 37°C for 2 h. Cells were then washed 2× with 1 ml 50 mM Tris-HCl (pH 8.0), and the final pellets were resuspended in 200 µl of a 5-mg/ml pepsin solution (5 mg pepsin, 5 µl concentrated HCl, per milliliter ddH20) and incubated at 37°C for 1 h. 1 ml of 50 mM Tris-HCl (pH 8.0) was added, and the cells were pelleted and then washed once with 1 ml of the same buffer. After wash, pellets were suspended in 250 µl propidium iodide solution (50 mM Tris-HCl, pH 8.0, and 50 µg/ml propidium iodide) and incubated overnight at 4°C. 50 µl of this cell suspension was added to 2 ml of 50 mM Tris-HCl (pH 8.0) in a round-bottom tube, and the sample was briefly sonicated at low power to resuspend cells. DNA content was then determined using a BD LSRFortessa cell analyzer (Software Version 2.0).
Statistical analysis
All graphs were generated using Excel. The average of at least three replicates is shown for all telomere tethering (n = 50 cells/replicate/cell cycle stage), INO1 localization (n = 50 cells/replicate/cell cycle stage), and ChIP data. Error bars show SD for telomere tethering and INO1 localization experiments and SEM for ChIP data. Data distribution was assumed to be normal, but this was not formally tested. Significance was assessed relative to WT using a two-tailed Student’s t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). Signal variance for GFP-Siz2, Sur4-mCherry, SUMO, and DAPI is presented as error bars showing SD (n = 25 cells/cell cycle stage).
Online supplemental material
Fig. S1 shows comparisons of global SUMOylation profiles between different tagged versions of Scs2; SUMOylation profiles and SUMO localization in different SUMO E3 ligase mutants; and SUMOylation profiles as well as siz2 mitotic localization and phosphorylation state for different Siz2 phosphomutants. Siz2 nucleolar exclusion and phosphatase treatment of Siz2 in mitotic arrest-release samples are also shown. Fig. S2 compares Siz2 phosphorylation and global SUMOylation profiles between various mutants, GFP-Siz2 localization in scs21–225 cells, as well as the INM localization of scs2 mutants. Fig. S3 shows telomere tethering, Sir4 NE localization, and Sir4 subtelomeric association in various mutants. Fig. S4 shows SUMO association with subtelomeric chromatin in various mutants, as well as telomere tethering, sir4 localization, and sir4 association with subtelomeric chromatin in the sir4K1037R mutant. Fig. S5 confirms the cell cycle stages of strains used for cell cycle–dependent ChIP shown in Fig. 7 E using FACS, SUMOylation profiles, and Clb2 levels and also confirms that Sir4-V53 levels remain stable. Video 1 shows the dynamics of GFP-Siz2 localization throughout the cell cycle. Table S1 presents the strains list. Table S2 presents the oligonucleotides list.
Supplementary Material
presents the strains list.
presents the oligonucleotides list.
Acknowledgments
We thank Dr. Michael Hendzel (University of Alberta) and members of the Aitchison, Montpetit, and Wozniak laboratories for helpful discussions.
Funding for this work is supported by the Canadian Institutes of Health Research (MOP 106502 and 36519) to R.W. Wozniak and the National Institutes of Health, USA (NCDIR: 2P41GM109824-06 and R01: 2R01GM112108-05 to J.D. Aitchison and R01GM124120 to B. Monpetit).
The authors declare no competing financial interests.
Author contributions: Conceptualization: C. Ptak, N.O. Saik, D.L. Lapetina, J.D. Aitchison, B. Monpetit, and R.W. Wozniak; Methodology: C. Ptak, N.O. Saik, and R.W. Wozniak; Investigation: C. Ptak, N.O. Saik, A. Premashankar, and D.L. Lapetina; Validation: C. Ptak and N.O. Saik; Funding acquisition: R.W. Wozniak; Project administration: R.W. Wozniak; Visualization: C. Ptak, N.O. Saik, and R.W. Wozniak; Writing—original draft: C. Ptak, N.O. Saik, and R.W. Wozniak; Writing—review and editing: C. Ptak, N.O. Saik, J.D. Aitchison, B. Monpetit, and R.W. Wozniak.
References
- Albuquerque, C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., and Zhou H.. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 7:1389–1396. 10.1074/mcp.M700468-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis, E.D., Zappulla D.C., Ansari A., Perrod S., Laiosa C.V., Gartenberg M.R., and Sternglanz R.. 2002. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 22:8292–8301. 10.1128/MCB.22.23.8292-8301.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani, S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., and Rosen M.K.. 2016. Compositional control of phase-separated cellular bodies. Cell. 166:651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, D.G., and Brickner J.H.. 2010. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol. Biol. Cell. 21:3421–3432. 10.1091/mbc.e10-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, J.H., and Walter P.. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342. 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, D.G., Randise-Hinchliff C., Lebrun Corbin M., Liang J.M., Kim S., Sump B., D’Urso A., Kim S.H., Satomura A., Schmit H., et al. 2019. The role of transcription factors and nuclear pore proteins in controlling the spatial organization of the yeast genome. Dev. Cell. 49:936–947.e4. 10.1016/j.devcel.2019.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter, A., Kaneshiro J.M., and Hetzer M.W.. 2019. Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet. 20:39–50. 10.1038/s41576-018-0063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo, L.V., Ptak C., and Wozniak R.W.. 2013. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol. Cell. 49:109–120. 10.1016/j.molcel.2012.10.017 [DOI] [PubMed] [Google Scholar]
- Champion, L., Pawar S., Luithle N., Ungricht R., and Kutay U.. 2019. Dissociation of membrane-chromatin contacts is required for proper chromosome segregation in mitosis. Mol. Biol. Cell. 30:427–440. 10.1091/mbc.E18-10-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, J.T., Wong A.K., Tavassoli S., Young B.P., Chruscicki A., Fang N.N., Howe L.J., Mayor T., Foster L.J., and Loewen C.J.. 2014. Polarization of the endoplasmic reticulum by ER-septin tethering. Cell. 158:620–632. 10.1016/j.cell.2014.06.033 [DOI] [PubMed] [Google Scholar]
- Cuijpers, S.A.G., and Vertegaal A.C.O.. 2018. Guiding mitotic progression by crosstalk between post-translational modifications. Trends Biochem. Sci. 43:251–268. 10.1016/j.tibs.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Denison, C., Rudner A.D., Gerber S.A., Bakalarski C.E., Moazed D., and Gygi S.P.. 2005. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics. 4:246–254. 10.1074/mcp.M400154-MCP200 [DOI] [PubMed] [Google Scholar]
- Di Mattia, T., Martinet A., Ikhlef S., McEwen A.G., Nominé Y., Wendling C., Poussin-Courmontagne P., Voilquin L., Eberling P., Ruffenach F., et al. 2020. FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts. EMBO J. 39:e104369. 10.15252/embj.2019104369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi, H., and Donaldson A.D.. 2008. Release of yeast telomeres from the nuclear periphery is triggered by replication and maintained by suppression of Ku-mediated anchoring. Genes Dev. 22:3363–3374. 10.1101/gad.486208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu, D., and Brickner J.H.. 2011. Gene positioning and expression. Curr. Opin. Cell Biol. 23:338–345. 10.1016/j.ceb.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar del Dedo, J., Idrissi F.Z., Fernandez-Golbano I.M., Garcia P., Rebollo E., Krzyzanowski M.K., Grötsch H., and Geli M.I.. 2017. ORP-mediated ER contact with endocytic sites facilitates actin polymerization. Dev. Cell. 43:588–602.e6. 10.1016/j.devcel.2017.10.031 [DOI] [PubMed] [Google Scholar]
- Falk, M., Feodorova Y., Naumova N., Imakaev M., Lajoie B.R., Leonhardt H., Joffe B., Dekker J., Fudenberg G., Solovei I., and Mirny L.A.. 2019. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature. 570:395–399. 10.1038/s41586-019-1275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felberbaum, R., Wilson N.R., Cheng D., Peng J., and Hochstrasser M.. 2012. Desumoylation of the endoplasmic reticulum membrane VAP family protein Scs2 by Ulp1 and SUMO regulation of the inositol synthesis pathway. Mol. Cell. Biol. 32:64–75. 10.1128/MCB.05878-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, H.C., Luke B., Schober H., Kalck V., Lingner J., and Gasser S.M.. 2011. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 13:867–874. 10.1038/ncb2263 [DOI] [PubMed] [Google Scholar]
- Freudenreich, C.H., and Su X.A.. 2016. Relocalization of DNA lesions to the nuclear pore complex. FEMS Yeast Res. 16:fow095. 10.1093/femsyr/fow095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyre, C.A.C., Rauher P.C., Ejsing C.S., and Klemm R.W.. 2019. MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol. Cell. 76:811–825.e14. 10.1016/j.molcel.2019.09.011 [DOI] [PubMed] [Google Scholar]
- Gaspar, M.L., Chang Y.F., Jesch S.A., Aregullin M., and Henry S.A.. 2017. Interaction between repressor Opi1p and ER membrane protein Scs2p facilitates transit of phosphatidic acid from the ER to mitochondria and is essential for INO1 gene expression in the presence of choline. J. Biol. Chem. 292:18713–18728. 10.1074/jbc.M117.809970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami, S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., and Weissman J.S.. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods R.A.. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87–96. 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- Goto, A., Liu X., Robinson C.A., and Ridgway N.D.. 2012. Multisite phosphorylation of oxysterol-binding protein regulates sterol binding and activation of sphingomyelin synthesis. Mol. Biol. Cell. 23:3624–3635. 10.1091/mbc.e12-04-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttinger, S., Laurell E., and Kutay U.. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10:178–191. 10.1038/nrm2641 [DOI] [PubMed] [Google Scholar]
- Hannich, J.T., Lewis A., Kroetz M.B., Li S.J., Heide H., Emili A., and Hochstrasser M.. 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280:4102–4110. 10.1074/jbc.M413209200 [DOI] [PubMed] [Google Scholar]
- Hari, K.L., Cook K.R., and Karpen G.H.. 2001. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15:1334–1348. 10.1101/gad.877901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger, F., Neumann F.R., Van Houwe G., Dubrana K., and Gasser S.M.. 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12:2076–2089. 10.1016/S0960-9822(02)01338-6 [DOI] [PubMed] [Google Scholar]
- Holt, L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., and Morgan D.O.. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 325:1682–1686. 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger, S. 2002. Cyclin destruction in mitosis: a crucial task of Cdc20. FEBS Lett. 532:7–11. 10.1016/S0014-5793(02)03657-8 [DOI] [PubMed] [Google Scholar]
- Jentsch, S., and Psakhye I.. 2013. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47:167–186. 10.1146/annurev-genet-111212-133453 [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., and Blobel G.. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147(5):981–994. . 10.1083/jcb.147.5.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S., and Gupta A.A.. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 106:735–744. 10.1016/S0092-8674(01)00491-3 [DOI] [PubMed] [Google Scholar]
- Kaiser, S.E., Brickner J.H., Reilein A.R., Fenn T.D., Walter P., and Brunger A.T.. 2005. Structural basis of FFAT motif-mediated ER targeting. Structure. 13:1035–1045. 10.1016/j.str.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Kirmiz, M., Vierra N.C., Palacio S., and Trimmer J.S.. 2018. Identification of VAPA and VAPB as Kv2 channel-interacting proteins defining endoplasmic reticulum-plasma membrane junctions in mammalian brain neurons. J. Neurosci. 38:7562–7584. 10.1523/JNEUROSCI.0893-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, K., Kawano-Kawada M., and Hanada K.. 2014. Phosphoregulation of the ceramide transport protein CERT at serine 315 in the interaction with VAMP-associated protein (VAP) for inter-organelle trafficking of ceramide in mammalian cells. J. Biol. Chem. 289:10748–10760. 10.1074/jbc.M113.528380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec, M. 2014. Biology of telomeres: lessons from budding yeast. FEMS Microbiol. Rev. 38:144–171. 10.1111/1574-6976.12054 [DOI] [PubMed] [Google Scholar]
- Lapetina, D.L., Ptak C., Roesner U.K., and Wozniak R.W.. 2017. Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes. J. Cell Biol. 216:3145–3159. 10.1083/jcb.201609049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, A., Felberbaum R., and Hochstrasser M.. 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178(5):813–827. . 10.1083/jcb.200702154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light, W.H., Brickner D.G., Brand V.R., and Brickner J.H.. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 40:112–125. 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Loewen, C.J., and Levine T.P.. 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280:14097–14104. 10.1074/jbc.M500147200 [DOI] [PubMed] [Google Scholar]
- Loewen, C.J., Roy A., and Levine T.P.. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035. 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen, C.J., Young B.P., Tavassoli S., and Levine T.P.. 2007. Inheritance of cortical ER in yeast is required for normal septin organization. J. Cell Biol. 179:467–483. 10.1083/jcb.200708205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Ma, Y., Kanakousaki K., and Buttitta L.. 2015. How the cell cycle impacts chromatin architecture and influences cell fate. Front. Genet. 6:19. 10.3389/fgene.2015.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio, T., and Wozniak R.W.. 2020. Passive diffusion through nuclear pore complexes regulates levels of the yeast SAGA and SLIK coactivator complexes. J. Cell Sci. 133:jcs237156. 10.1242/jcs.237156 [DOI] [PubMed] [Google Scholar]
- Manford, A.G., Stefan C.J., Yuan H.L., Macgurn J.A., and Emr S.D.. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23:1129–1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Min, J., Wright W.E., and Shay J.W.. 2019. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 33:814–827. 10.1101/gad.324905.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. 2020. The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell. 183:28–45. 10.1016/j.cell.2020.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi-Fard, S., Sarthi J., Tittel-Elmer M., Lalonde M., Cusanelli E., Chartrand P., and Cobb J.A.. 2016. Smc5/6 is a telomere-associated complex that regulates Sir4 binding and TPE. PLoS Genet. 12:e1006268. 10.1371/journal.pgen.1006268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S.E., and Levine T.P.. 2016. VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 1861(8 Pt B):952–961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Ng, A.Q.E., Ng A.Y.E., and Zhang D.. 2020. Plasma Membrane Furrows Control Plasticity of ER-PM Contacts. Cell Rep. 30:1434–1446.e7. 10.1016/j.celrep.2019.12.098 [DOI] [PubMed] [Google Scholar]
- Ninova, M., Fejes Tóth K., and Aravin A.A.. 2019. The control of gene expression and cell identity by H3K9 trimethylation. Development. 146:dev181180. 10.1242/dev.181180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade, B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., and Doye V.. 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 18(8):2912–2923. . 10.1091/mbc.e07-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse, V.G., Hardeland U., Werner T., Kuster B., and Hurt E.. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279:41346–41351. 10.1074/jbc.M407950200 [DOI] [PubMed] [Google Scholar]
- Panse, V.G., Küster B., Gerstberger T., and Hurt Ed. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5(1):21–27. . 10.1038/ncb893 [DOI] [PubMed] [Google Scholar]
- Poleshko, A., Mansfield K.M., Burlingame C.C., Andrake M.D., Shah N.R., and Katz R.A.. 2013. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 5:292–301. 10.1016/j.celrep.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshko, A., Smith C.L., Nguyen S.C., Sivaramakrishnan P., Wong K.G., Murray J.I., Lakadamyali M., Joyce E.F., Jain R., and Epstein J.A.. 2019. H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis. eLife. 8:e49278. 10.7554/eLife.49278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., Scalzo D., and Groudine M.. 2013. Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu. Rev. Cell Dev. Biol. 29:241–270. 10.1146/annurev-cellbio-101512-122317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye, I., and Jentsch S.. 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 151:807–820. 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- Ptak, C., and Wozniak R.W.. 2016. Nucleoporins and chromatin metabolism. Curr. Opin. Cell Biol. 40:153–160. 10.1016/j.ceb.2016.03.024 [DOI] [PubMed] [Google Scholar]
- Ptak, C., Aitchison J.D., and Wozniak R.W.. 2014. The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr. Opin. Cell Biol. 28:46–53. 10.1016/j.ceb.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina, E., Duncan S.M., and Manley J.L.. 2010. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 24:1242–1252. 10.1101/gad.1917910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saik, N.O., Park N., Ptak C., Adames N., Aitchison J.D., and Wozniak R.W.. 2020. Recruitment of an Activated Gene to the Yeast Nuclear Pore Complex Requires Sumoylation. Front. Genet. 11:174. 10.3389/fgene.2020.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. 10.1093/genetics/122.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoyer, C.J., Katta S.S., Gardner J.M., Stoltz L., McCroskey S., Bradford W.D., McClain M., Smith S.E., Slaughter B.D., Unruh J.R., and Jaspersen S.L.. 2016. Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol. 215:575–590. 10.1083/jcb.201607043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan, C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., and Emr S.D.. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144:389–401. 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Taddei, A., and Gasser S.M.. 2012. Structure and function in the budding yeast nucleus. Genetics. 192:107–129. 10.1534/genetics.112.140608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., Hediger F., Neumann F.R., Bauer C., and Gasser S.M.. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23:1301–1312. 10.1038/sj.emboj.7600144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texari, L., and Stutz F.. 2015. Sumoylation and transcription regulation at nuclear pores. Chromosoma. 124:45–56. 10.1007/s00412-014-0481-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texari, L., Dieppois G., Vinciguerra P., Contreras M.P., Groner A., Letourneau A., and Stutz F.. 2013. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol. Cell. 51:807–818. 10.1016/j.molcel.2013.08.047 [DOI] [PubMed] [Google Scholar]
- Van Damme, E., Laukens K., Dang T.H., and Van Ostade X.. 2010. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 6:51–67. 10.7150/ijbs.6.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse, D.W., Wan Y., Lapetina D.L., Chen W.M., Chiang J.H., Aitchison J.D., and Wozniak R.W.. 2013. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell. 152:969–983. 10.1016/j.cell.2013.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Boyvat, M., Kentala H., Lilja J., Vihervaara T., Hanninen R., Zhou Y., Peränen J., Nyman T.A., Ivaska J., and Olkkonen V.M.. 2015. OSBP-related protein 3 (ORP3) coupling with VAMP-associated protein A regulates R-Ras activity. Exp. Cell Res. 331:278–291. 10.1016/j.yexcr.2014.10.019 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel, J.A., Johnson E.S., Reed S.I., and Yates J.R. III. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:45662–45668. 10.1074/jbc.M409203200 [DOI] [PubMed] [Google Scholar]
- Wurzenberger, C., and Gerlich D.W.. 2011. Phosphatases: providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 12:469–482. 10.1038/nrm3149 [DOI] [PubMed] [Google Scholar]
- Wykoff, D.D., and O’Shea E.K.. 2005. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics. 4:73–83. 10.1074/mcp.M400166-MCP200 [DOI] [PubMed] [Google Scholar]
- Zhao, Q., Xie X.Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., and Ren J.. 2014. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42:W325–W330. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., Ryan J.J., and Zhou H.. 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279:32262–32268. 10.1074/jbc.M404173200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Wu C.-Y., and Blobel G.. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167(4):605–611. . 10.1083/jcb.200405168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
presents the strains list.
presents the oligonucleotides list.