Abstract
Infectious diseases are a leading cause of global morbidity and mortality, and the threat of infectious diseases to human health is steadily increasing as new diseases emerge, existing diseases reemerge, and antimicrobial resistance expands. The application of imaging technology to the study of infection biology has the potential to uncover new factors that are critical to the outcome of host-pathogen interactions and to lead to innovations in diagnosis and treatment of infectious diseases. This article reviews current and future opportunities for the application of imaging to the study of infectious diseases, with a particular focus on the power of imaging objects across a broad range of sizes to expand the utility of these approaches.
Keywords: imaging, infection, microbiology, biofilms, microscopy
1. INTRODUCTION
Many scientists and physicians became excited about the natural world when they saw something that fascinated them. Images tell a story and make one wonder; they lead to the generation of hypotheses and experimentation that reveal the workings of natural life. As new imaging modalities are created, we can see with new eyes, which expands our wonder and in turn increases our knowledge.
The application of imaging to biomedicine has led to pioneering advances in science and clinical care. However, even though they are the second-leading cause of death in the world, infectious diseases have historically lagged other fields in imaging sciences. This is despite the potential for imaging to allow rapid diagnosis of infections and exciting breakthroughs in the biology of both hosts and pathogens. Moreover, imaging of infections provides the opportunity to investigate lesions that are due to catastrophic levels of inflammation and that can be easily distinguished from healthy tissue.
In this review, I address the current state and potential of imaging infectious processes with a focus on imaging across a broad range of sizes, from the scale of molecules to cells to tissues to whole animals. I discuss strengths and limitations of existing techniques. I highlight future opportunities in imaging infectious diseases that are expanding the field and promise to provide key insights into infectious processes that would be impossible to achieve through existing strategies.
2. WHOLE-ANIMAL IMAGING
Multiple imaging modalities enable investigators to image an entire animal. These technologies have provided information on anatomic changes that occur during infection, revealing the distribution of infection throughout the body and tracing the trafficking of molecules from one point within the host to another. Magnetic resonance imaging (MRI) is one such modality. MRI measures the response of atomic nuclei within the body to high-frequency radio waves upon exposure to a strong magnetic field, resulting in images of internal organs. Computed tomography (CT) is a form of cross-sectional imaging in which the motion of an X-ray source and detectors is controlled by a computer that processes the data and generates an image. In infection biology, MRI and CT have been most powerful when combined with nuclear medicine–based technologies such as single-photon emission CT (SPECT) and positron emission tomography (PET). SPECT and PET use tracers or probes labeled with high-energy-emission radionuclides. In SPECT imaging, a gamma camera rotates around the subject while taking pictures so as to create a cross-sectional image. With PET imaging, compounds labeled with positron-emitting isotopes are introduced to the body and gamma rays produced by the isotopes are detected and converted into images, allowing for visualization of a precise location within the body. These technologies can be applied to three-dimensional and relatively unaltered samples. Thus, they can be used for both preclinical and clinical imaging and enable longitudinal experiments so that the progression of an infection can be monitored.
A powerful application of SPECT involves targeting the site of bacterial localization within the body using probes that either are metabolized by bacterial pathogens or target the pathogens directly. Imaging bacteria directly allows investigators to detect and study infection prior to tissue damage. For example, 125I-FAU is a SPECT probe that can be phosphorylated by bacterial thymidine kinases and accumulates at sites of bacterial colonization. This provides positional information regarding the site of infection. This strategy has revealed bacterial burdens in visible tissue lesions in a Mycobacterium tuberculosis infection model and highlighted the heterogeneity of microbial colonization in these lesions (20). The 125I-FAU probe is generalizable to numerous infections owing to the presence of thymidine kinase in most bacteria (74). Bacterium-specific molecules can also be directly labeled as a strategy to image bacterial localization within whole animals. For instance, DOTA-Bodipy-NCS-labeled lipopolysaccharide (LPS) has been radiolabeled with 111In and followed with SPECT-CT and fluorescence microscopy. This exciting demonstration of the combined application of fluorescence imaging and SPECT-CT revealed that LPS accumulates early in the liver in mice treated with intravenous LPS (23). In a final example, the metal-binding properties of microbial siderophores have been exploited for PET imaging through the use of 68Ga to label siderophores acquired by Aspergillus fumigatus in a rat model of infection (73).
PET is a powerful strategy to image sites of infection, and the strength of this technology is most evident in diagnostic applications. PET can detect hypoxic lung lesions in a mouse model of M. tuberculosis infection (36). PET combined with 18F-labeled maltohexaose (MH18F) has been used to image bacteria in vivo with sensitivity and specificity that are orders of magnitude higher than those of other PET probes such as fluorodeoxyglucose (18FDG). Moreover, unlike 18F-FDG, 6″-18F-fluoromaltotriose appears to be specific to bacteria because it targets the maltodextrin transporter that is expressed in these organisms. This unique combination of high specificity and high sensitivity for bacteria makes MH18F a promising compound to improve the diagnosis of bacterial infections (60), as has been exemplified by the diagnosis of Pseudomonas aeruginosa in a clinically relevant mouse model of wound infection (33). Using a creative strategy, Zhang et al. (102) applied a radiofluorinated analog of para-aminobenzoic acid (2-[18F]F-PABA) as an efficient alternative substrate for Staphylococcus aureus dihydropteroate synthase. 2-[18F]F-PABA rapidly accumulates in S. aureus but not in host cells, distinguishing between infected and sterile sites. Incorporation of d-amino acids into peptidoglycan is unique to bacteria, and this fact has been exploited for PET imaging. d-[3-11C]Alanine and the dipeptide d-[3-11C]alanyl-d-alanine accumulate in a variety of both gram-positive and gram-negative pathogens, including S. aureus and P. aeruginosa, and specifically label bacteria during acute bacterial myositis, osteomyelitis, and pneumonia (66). In addition to these directed approaches, more-unbiased approaches have succeeded in uncovering new molecules that have value as imaging probes. Screening an in silico library for substrates of essential metabolic pathways in bacteria revealed fluorine-labeled analogs that were developed as PET-based imaging tracers. Using this approach, Ordonez et al. (63) found that PABA, mannitol, and sorbitol are selectively incorporated into bacterial cells. These fluorine-labeled analogs enable the rapid detection and differentiation of infection sites from sterile inflammation in mice.
In addition to exploiting bacterium-specific processes to label invading microbes, one can label antibacterial molecules that target the bacterial cells to localize infection. Bacteria have been labeled through the application of cationic peptides that are conjugated to 68Ga-NOTA-UBI-29-41. These molecules accumulate at sites of S. aureus infection, and the amount of signal of the probe positively correlates with the magnitude of infection (96). In an antibody-based method, immuno-PET (antibody-targeted PET) with [64Cu]NODAGA-labeled Yersinia-specific polyclonal antibodies targeting the outer membrane protein YadA have been used to rapidly detect systemic Yersinia enterocolitica infections (100). In addition, antibiotics have evolved to be extraordinarily specific to microbes, and this specificity can be exploited to image bacterial infection, as has been shown with trimethoprim, which is taken up by live bacteria. In a rodent myositis model, [18F]fluoropropyl-trimethoprim tracks to live bacterial infection, and this molecule also demonstrates impressive biodistribution in a primate model (84). [11C]Rifampin is also an outstanding tool for PET- and CT-based targeting of M. tuberculosis lesions in humans (Figure 1a). Collectively, these strategies underscore the value of using the specificity of antibacterial molecules to target bacteria for imaging.
Figure 1.
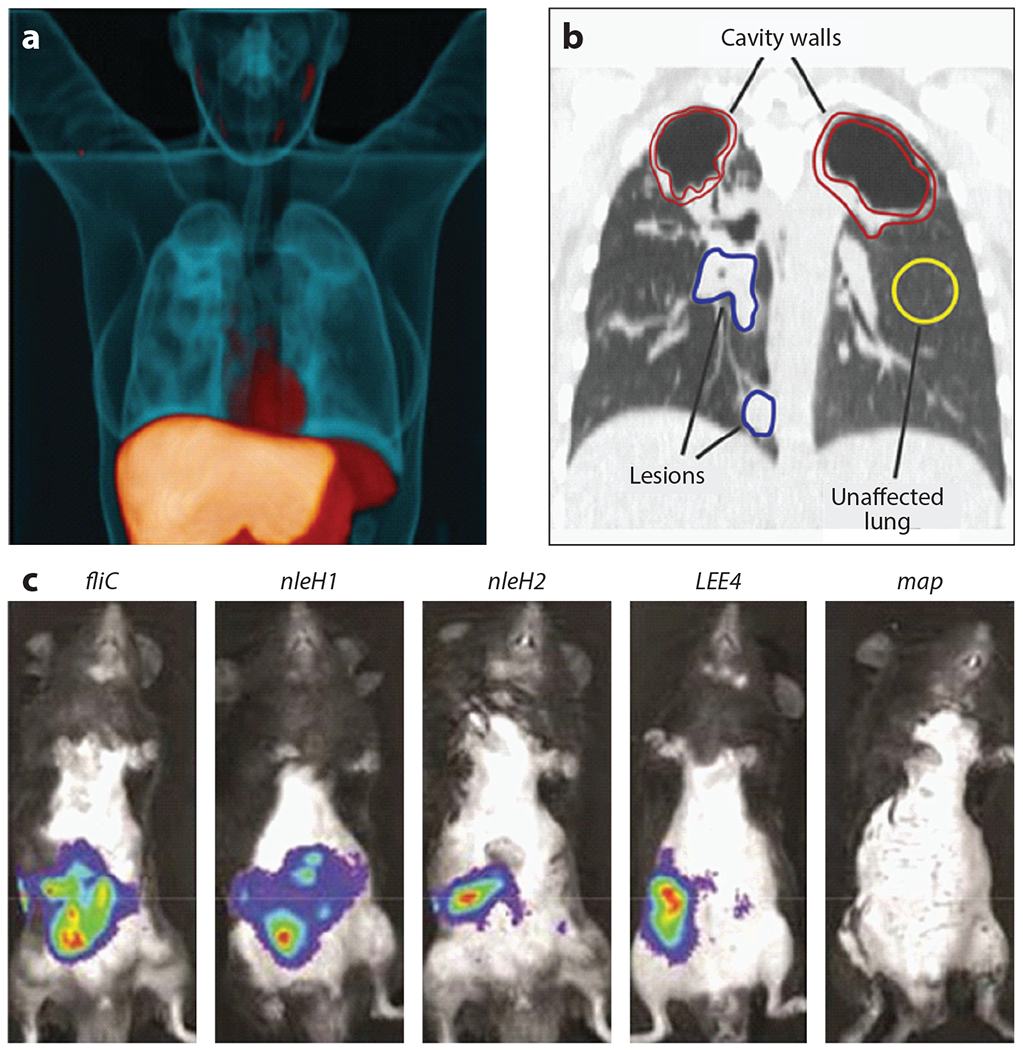
Whole-animal imaging in the study of infectious diseases. (a) Three-dimensional [11C]rifampin PET-CT image of a human patient infected with Mycobacterium tuberculosis. The 11C-labeled rifampicin (orange) is imaged using PET. The CT image is blue. Adapted with permission from Reference 62. (b) The [11C]rifampicin-associated PET signal can be quantified in a coronal CT section from the same patient. Volumes of interest are indicated. Adapted with permission from Reference 63. (c) Bacterial gene expression in a murine model of EPEC. These IVIS images show differences in expression of genes encoding EPEC effector proteins during infection. Adapted with permission from Reference 59. Abbreviations: CT, computed tomography; EPEC, enteropathogenic Escherichia coli; IVIS, in vivo imaging system; PET, positron emission tomography.
Infection often leads to considerable inflammation. Therefore, an alternative strategy to image infection in whole animals is to use probes that target host inflammatory processes. CT alone is a useful tool to study bone destruction during the pathogenesis of osteomyelitis (12), and combined PET and CT has also been very powerful in this regard. Vascular adhesion protein 1 (VAP-1) is an endothelial protein that is presented on the cell surface during inflammation. 68Ga-labeled 1,4,7,10-tetraazacyclododecane-N′,N″,N″′,N″″-tetraacetic acid-peptide (68Ga-DOTAVAP-P1) targets to VAP-1 and detects inflammation in healing bones and during S. aureus osteomyelitis (46). 68Ga-citrate and 68Ga-Cl are also used to image S. aureus bone infections (47). The sensitivity of FDG PET/CT is low. Therefore, FDG-labeled white blood cells can increase specificity for periprosthetic infection imaging (2, 24). Formylpeptide receptors (FPRs) are expressed on neutrophils and are critical for the migration of these neutrophils to the site of infection. The FPR-specific peptide N-cinnamoyl-F-(D)L-F-(D)L-F (cFLFLF) conjugated with a polyethylene glycol (PEG) moiety and labeled with [64Cu]CuCl2 enables imaging of infiltrating neutrophils during Klebsiella pneumonia lung infection (52). Additional tools can be borrowed from the field of cancer biology, where PET imaging is used to visualize IFN-γ, which is a potent cytokine produced in response to a variety of infectious agents (30). Reagents for the visualization of TNF-α have also been developed (28).
In addition to MRI, CT, and PET/SPECT, two-photon microscopy has enhanced intravital imaging studies, enabling high-resolution imaging of physiologically relevant tissues. These intravital approaches have been used to visualize infection events ranging from HIV infection of T cells within the lymph nodes (85) to phagocyte migration to sites of S. aureus infection (1, 92) to Borrelia burgdorferi infection of the skin and joints (5) to infiltration of alveolar macrophages and CD11b-positive dendritic cells to the sites of Bacillus anthracis lung infection. One of the limitations of multiphoton microscopy is motion during image acquisition. To counteract movement of the chest, a method of employing parenchymal tissue motion correction through computational analysis has been utilized (25).
Luminescence and fluorescence can be applied to image host and bacterial gene expression in whole animals. This approach is often referred to by the name of the instrument most commonly used for these studies, in vivo imaging system (IVIS). IVIS is routinely applied to infectious disease imaging. In one demonstration of how this approach is applied to infection biology, enteropathogenic Escherichia coli (EPEC)-infected whole intestines were assessed. This revealed that different bacterial effector proteins are expressed at distinct times during infection, providing information regarding which of these factors are important during colonization versus infection and highlighting the value of imaging bacterial gene expression within infected mice (59) (Figure 1c). Chemical probes that produce either luminescence or fluorescence upon binding to specific factors can also be powerful tools for the study of host-pathogen interactions. Examples include molecules that detect labile iron accumulation during Acinetobacter baumannii pneumonia (3) or hydrogen peroxide accumulation at the host-microbe interface (42).
A relatively new approach to enhance the value of imaging whole animals and complex tissues is termed CLARITY (clear lipid-exchanged, acrylamide-hybridized rigid imaging/immunostaining/in situ-hybridization-compatible tissue-hydrogel). The method was first described by Chung et al. (17), and numerous adaptations have since been developed to increase its utility. Briefly, CLARITY replaces tissue lipids with a stabilizing hydrogel (e.g., an acrylamide-based gel), giving the tissue a transparent appearance, while maintaining structural integrity and localization of proteins and nucleic acids. Removal of lipids results in increased penetration of macromolecules such as antibodies and decreases light scattering, improving the application for studies of deep tissue. CLARITY has been applied to adult zebrafish infected with Mycobacterium marinum, enabling observations of fluorescent signals from host and bacterial reporters to increase insight into the host-pathogen interaction (18). CLARITY and its adaptations [i.e., passive clarity technique (PACT) and microbial identification after PACT (MiPACT)] are also used for imaging of intact organs and other biological samples, as shown for murine lungs infected with M. marinum (18) and sputum isolated from cystic fibrosis patients (21). The use of optically transparent animal models, such as zebrafish and the nematode Caenorhabditis elegans, affords unique insights useful for monitoring host-pathogen interactions with high-resolution microscopic techniques. The use of transgenic zebrafish that express stable, lineage-specific fluorescent markers in immune cell populations, combined with the genetic tractability of zebrafish, has afforded critical insights into the recruitment of immune cells to sites of infection and cellular processes including phagocytosis and efferocytosis (37, 94).
3. TISSUE IMAGING
Perhaps one of the oldest methods of imaging infection is tissue staining followed by microscopic analysis, or traditional tissue pathology. Staining with relatively nonspecific stains, such as hematoxylin-eosin (H&E), continues to be a powerful approach to assess tissue damage and inflammation and identify pathogens. Recently, tissue imaging has benefitted from innovations that provide molecular information and dramatically expand the information obtained from a single tissue section.
Immunohistochemistry involves the use of antibodies to image targets of interest. Multiple techniques can be applied to formalin-fixed, paraffin-embedded tissue sections, and almost all of these require antibodies for antigen detection and mapping. Traditional immunohistochemistry involves the use of a single antibody against one target. However, recent advancements now enable multiple-marker imaging. Multiplexed immunohistochemistry (mIHC) combines fluorescent and chromogenic staining with automated whole-slide imaging and integrated whole-slide image analysis. mIHC therefore enables investigators to detect numerous proteins and characterize thousands of cells directly in the tissue (7). Tissue-based cyclic immunofluorescence (t-CyCIF) involves assembly of a 16-antibody panel and has been used to quantify and localize immune cells expressing immune checkpoint regulators (22). Codetection by indexing (CODEX) is a multiplexed imaging approach whereby DNA-bar-coded antibodies allow iterative visualization of each label and the subsequent imaging of dozens of markers. CODEX has tremendous potential for infection biology owing to its ability to provide quantitative characterization of tissue architecture in normal and infected tissue samples (32). Imaging mass cytometry (IMC) is an analytical technique that does not involve traditional cytometry and is therefore perhaps misnamed. Instead, IMC takes advantage of the mass-resolving power of inductively coupled plasma mass spectrometers to detect and image heavy-metal-conjugated antibodies in tissue sections. IMC allows the detection of numerous targets in a single tissue. This is perhaps best demonstrated by immunodetection of a 34-antibody panel in human snap-frozen tissue sections, which reveals the tissue architecture and spatial distribution of various cell types in the human fetal intestine (34). IMC revealed information regarding the fetal immune response, providing evidence for the generation of memory-like CD4+ T cells in the fetal intestine that is indicative of foreign antigen exposure (51). Many of these multiplexed approaches have not yet been applied to infection biology, but given the considerable power of these approaches, it is only a matter of time before this gap is traversed.
Imaging mass spectrometry (IMS) uses mass spectrometry to obtain molecular data from tissue sections that can be converted to images. IMS can be applied to a variety of analytes, from individual elements to intact proteins. Laser ablation inductively coupled plasma IMS (LA-ICP-IMS) is used to image elemental distributions within biological samples, whereas metabolites, lipids, peptides, and proteins can be interrogated using matrix-assisted laser desorption/ionization (MALDI) IMS (90) (Figure 2). Existing MALDI-IMS approaches were recently complemented by the introduction of MALDI-2. In MALDI-2, MALDI is combined with laser-induced postionization of analytes to enable improved coverage of molecular features (6, 8). For deeper insight into biological systems, IMS approaches are commonly employed in tandem with additional imaging modalities, thereby creating a need for accurate registration of images generated from separate tissue sections. Such registration has been achieved through nondestructive autofluorescence microscopy prior to IMS, and images obtained from H&E staining of adjacent tissue sections are registered subsequently (68). One exciting, recent advancement in the field of IMS is the ability to combine imaging modalities in a process termed data-driven image fusion. Through this process, effective spatial resolution of the lower-resolution modality (e.g., IMS when compared to H&E microscopy) can be increased several fold (72). A further in-depth discussion of integrated imaging can be found in Section 6 of this review.
Figure 2.
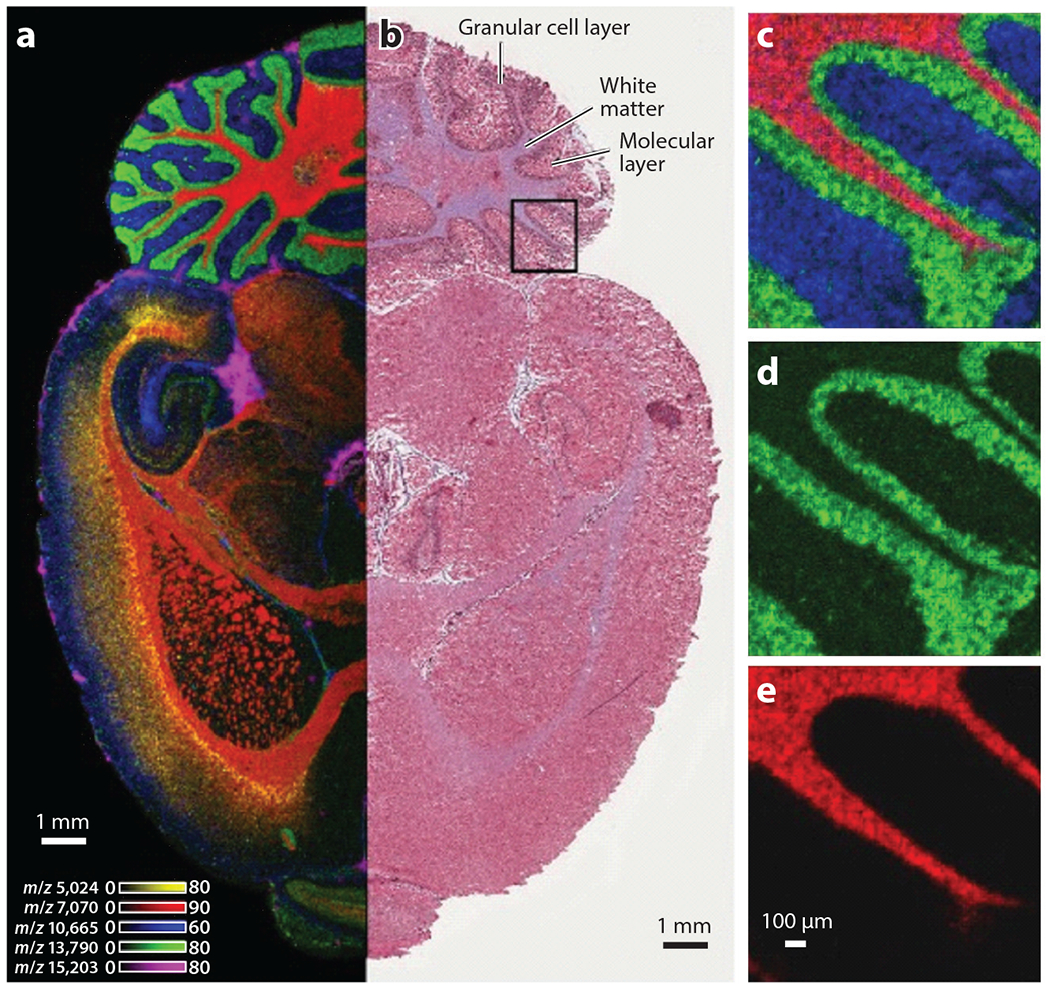
IMS provides molecular information at microscopic resolution. (a) A transverse section of a rat brain imaged using ultrahigh-throughput MALDI-IMS. Each color represents a different analyte, as indicated by the listed mass/charge. (b) Corresponding H&E-stained region of the same brain. Boxed region was imaged at 10-μM resolution, and individual ion images are shown in panels c–e. Abbreviations: H&E, hematoxylineosin; IMS, imaging mass spectrometry; MALDI, matrix-assisted laser desorption/ionization. Adapted with permission from Reference 90.
Because of the ability of IMS to assess the spatial distribution of analytes of interest in tissue, this approach has been applied to various aspects of infection. In one study, LA-ICP-IMS revealed distribution patterns of various metals in proximity to S. aureus renal abscesses and revealed heterogeneity of metal distribution across infected organs (13). Of note, in the same study a staphylococcal protein (δ-hemolysin) in infected tissue was imaged in an unbiased approach via MALDI-IMS, a pioneering example of using IMS to image bacterial protein production in host tissue. A subsequent study confirmed the nonhomogeneous nature of the host-pathogen interaction by highlighting heterogeneous abundance of staphylococcal siderophores in relation to organ-wide iron distributions using a combination of MALDI-IMS and LA-ICP-IMS (71). MALDI-IMS is not restricted to bacterial factors but has also been used to monitor the host response to systemic S. aureus infections (4). Other pathogens for which MALDI-IMS has been effective for interrogating the host-pathogen interaction include Francisella novicida (83), Salmonella enterica Typhimurium (40), Burkholderia mallei (31), and hepatitis B virus (65), highlighting the utility and power of this approach.
4. CELL IMAGING
The power of imaging the interaction between microbial pathogens and eukaryotic cells is perhaps most obvious when considering microscopic images of microbe-host encounters at the cellular level. This is particularly so when considering the application of fluorescence microscopy and electron microscopy (EM) to the biology of intracellular pathogens. The capabilities afforded by these methods, many of which are directly relevant to observation of the host-pathogen interface, have expanded massively.
There are countless examples of the value of fluorescence microscopy and EM to the study of infection biology. For instance, these technologies have revealed that Bacillus cereus nonhemolytic enterotoxin (NHE) and hemolysin BL (HBL) operate synergistically to induce inflammation and are both sensed by the inflammasome (27). Confocal microscopy has demonstrated that the positioning of splenic marginal zone CD169+ macrophages affects the rapidity of antibacterial responses (70). In another example, the increased magnification afforded by high-resolution fluorescence imaging of the autophagy response against M. marinum infection has been exploited in combination with the optical transparency of zebrafish (39). Structured-illumination microscopy (SIM) has been used to study synergy between influenza and bacterial infections caused by Streptococcus pneumoniae or S. aureus at resolutions previously not possible using standard confocal technologies. Rowe et al. (76) visualized influenza virus directly interacting with the bacterial surface, supporting the model that bacterium-influenza synergy occurs at the initiation of disease causation. These findings are examples of the many cellular interactions that would not have been revealed without the power of visual observation.
Innovations in sample preparation for fluorescence imaging have opened the door for further investigation beyond classical methods. For instance, a physical mechanism termed one-dimensional membrane wetting revealed that Neisseria meningitidis causes plasma membrane remodeling along meningococcal type IV pili fibers. This established one-dimensional membrane wetting as a new process involved in the interaction of cells with their environment (15). Fluorescence lifetime imaging microscopy (FLIM) revealed that the lifetimes of bacteria depend on their location within host cells, leading researchers to propose that this technology has the potential to replace z-stack imaging for identifying intracellular bacteria. Cell DIVE is an imaging modality that takes advantage of high-resolution, multiplexed tissue immunofluorescence (MxIF) methods and provides single-cell-level quantitation of fluorescent signals. It has been applied to fixed archived monkeypox virus–induced inflammatory skin lesions, where it conserves tissue and provides biomarker identification and colocalization data (80, 89).
EM has revealed fundamental information about cell structure and organization. Through advances in sample preparation such as vitrification, researchers can preserve the structure of cells. Moreover, the advent of three-dimensional imaging by electron tomography (ET) and enhanced methods to process data have revolutionized the application of EM to the study of microbial life. EM has been used to study the efficacy of antimicrobial peptides such as human β-defensins 2 (BD-2) and 3 (BD-3). Rat ischemic skin flaps were infected with P. aeruginosa and exposed to BD-2 and BD-3, and catheter segments were analyzed by scanning EM (SEM). This work is a strong demonstration of the application of SEM to in vivo infection models (11). EM is also a valuable tool for studying intracellular bacterial pathogens. For example, quantitative three-dimensional EM has revealed that Chlamydia trachomatis divides by binary fission and undergoes a sixfold reduction during population expansion. This seminal discovery demonstrated that the reticulate body of C. trachomatis can control the timing of the conversion of the reticulate body to the elementary body without an external signal (48) (Figure 3). In an outstanding application of both fluorescence and EM to infection biology, ubiquitin was found to recognize M. tuberculosis surface protein Rv1468c, a previously unidentified ubiquitin-binding protein that also encodes a eukaryotic-like ubiquitin-associated (UBA) domain. This study described host xenophagy triggered by binding of ubiquitin to a microbial protein as a strategy employed by M. tuberculosis to restrict microbial load within cells (14).
Figure 3.
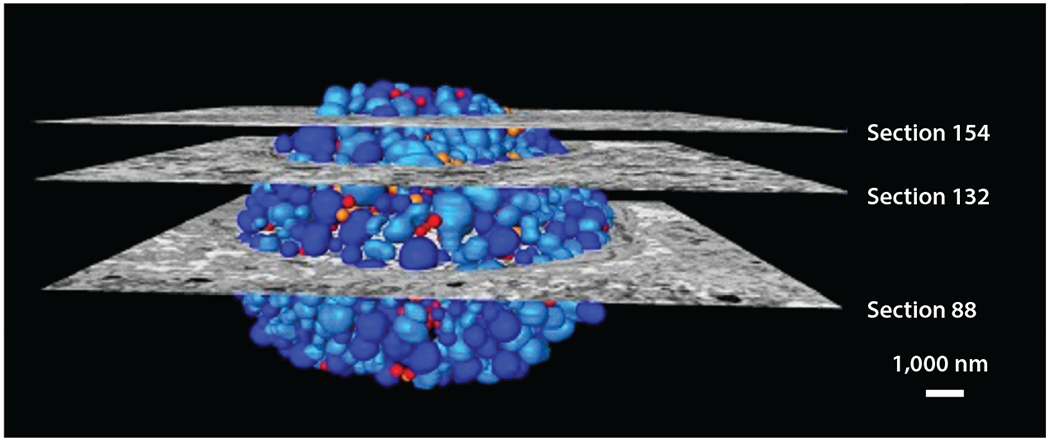
Three-dimensional scanning electron microscopy reconstruction of a chlamydial inclusion in a Chlamydia trachomatis–infected HeLa cell. Sections indicate micrographs, and colors indicate reticulate body (dark blue), dividing reticulate body (light blue), inclusion body (orange), and elementary body (red). Adapted with permission from Reference 48.
5. MOLECULAR IMAGING
EM-based techniques are beginning to reveal the structure of intact molecular machines within cells in their native context. These techniques have enabled the collection of structural information, are revealing the macromolecular organization of unmodified cellular environments (54), and are increasingly applied to host-pathogen interactions (16). Beautiful images of bacterial molecular machines have been generated, including but not limited to the S. aureus 50S-RsfS complex together with the crystal structure of the uL14-RsfS complex, which has provided information regarding the mechanism of staphylococcal ribosome shutdown (43). Cryo-EM reconstructions of the Helicobacter pylori Cag type IV secretion system core complex revealed that assembly of this molecular machine is dependent on incorporation of interwoven species-specific components (86) (Figure 4). Atomic structures of P. aeruginosa Pf4, a symbiotic filamentous viral prophage, have revealed that liquid crystalline droplets form phase-separated occlusive compartments around the bacteria, leading to increased bacterial survival. Tarafder et al. (93) suggest that this may be a conserved strategy by which secreted filamentous molecules improve bacterial survival in stressful environments. Pathogen–host cell interactions can also be imaged. Images of vaccinia virus on mammalian cells reveal cell surface–induced changes in the virus (19).
Figure 4.
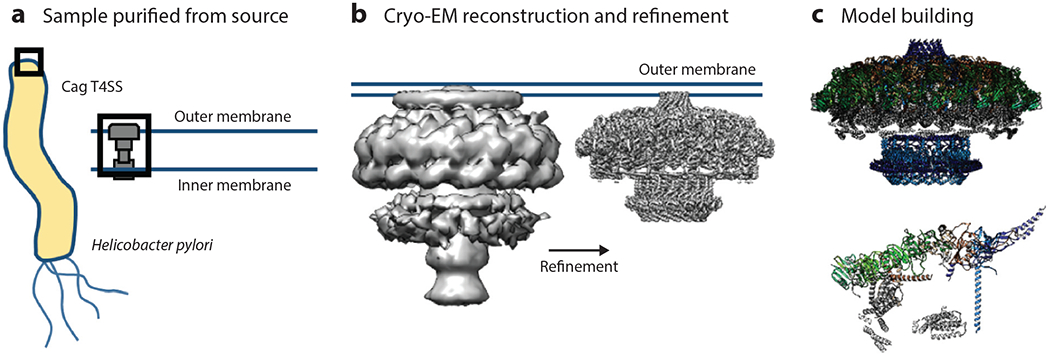
(a) The Helicobacter pylori Cag T4SS was purified directly from the bacterium for use in structural analysis. (b) A three-dimensional volume was reconstructed using the T4SS particles and was subsequently used to refine subcomplexes of the T4SS to near-atomic resolution. (c) These high-resolution reconstructions were then used to model several of the individual proteins that contribute to the structure of the T4SS (top), leading to the identification of the asymmetric unit (bottom). Abbreviations; EM, electron microscopy; T4SS, type IV secretion system. Figure adapted from image created by Drs.Michael Sheedlo and Borden Lacy.
Cryo-EM can inform drug targeting against bacterial pathogens. For example, single-particle cryo-EM elucidated five structural states of the A. baumannii ribosome, including the 70S, 50S, and 30S forms, informing future drug design (57). A technique known as plunge-freezing cryo-EM has shown that pathogenic mycobacteria produce a thick capsule under unperturbed conditions. This capsule layer comprises arabinomannan, α-glucan, and oligomannosyl-capped glycolipids. This information can be used to target this shield around the organism (79). Single-particle cryo-EM has elucidated the structure of the A. baumannii AdeB resistance–nodulation–cell division multidrug efflux pumps. These proteins were embedded in lipidic nanodiscs, enabling a resolution of 2.98 Å and providing information regarding the mechanism by which these pumps export antibiotics (91). Cryo-EM and electron cryotomography (cryo-ET) are also powerful techniques for the study of the structural biology of viruses. In a single study, cryo-EM and cryo-ET were used to determine the structure of the Ebola virus nucleocapsid within intact viruses and recombinant nucleocapsid-like assemblies (98). Such studies have not only provided information regarding the biology of viruses but also promoted the development of virus-based resources for biotechnology (55).
6. INTEGRATION STRATEGIES FOR IMAGING ACROSS A RANGE OF SIZES
The imaging methods discussed thus far, though common in their purpose of visualization, have each led to unique results. However, scientific investigation is strengthened through the combined use of distinct approaches aimed at a single goal. In this regard, integrated imaging approaches have expanded the information collected from a single experiment into a full representation of all relevant scales. This widens the lens through which hypotheses can be developed and strengthens the conclusions drawn from imaging studies.
Cryo-EM and crystallography have been combined to enable researchers to benefit from the near-atomic structures provided by crystallography with the ability to study large biological complexes at lower resolution. Combining these modalities enables investigators to image macromolecular structures that cannot be crystallized and also to study dynamic cellular processes (75). Various computational and image analysis methods have been developed that allow investigators to study the conformational flexibility of biological machines. Examples include the combination of cryo-EM and X-ray crystallography (95) along with algorithms to place three-dimensional atomic structures into scaled cryo-EM microscopy maps (78). Wang et al. (99) prepared a complex of cowpea mosaic virus saturated with a Fab fragment of a monoclonal antibody against the virus. They combined cryo-EM and X-ray crystallography to define a physical footprint of the Fab on the capsid surface and the orientation and position of the Fab.
Transmission EM (TEM) and SEM are often used in combination with light microscopy. P. aeruginosa biofilms in a mouse model of corneal infection were imaged using slit lamp microscopy in combination with light microscopy, confocal microscopy, and EM to interrogate the host-microbe interactions (81). Correlative light and electron microscopy (CLEM) combines the advantages of three-dimensional live cell imaging with ultrastructural analysis. CLEM allows investigators to use fluorescence microscopy to identify and follow cell processes in a monolayer and serial SEM to study cellular structure. CLEM has been applied to study M. tuberculosis in primary human lymphatic endothelial cells as well as HIV-1 in human monocyte-derived macrophages (77). Correlative electron and fluorescence microscopy has also been applied to individual HIV particles bound to mammalian cell surfaces and has been used to image microtubule end structures bound to mal3p in fission yeast (45). One challenge with these approaches is the fact that radiation can damage tissue. To circumvent this, a correlative fluorescence light microscopy–EM approach was developed that uses light microscopy to search for the structures of interest and allows EM to zoom in on these features. With this approach, the total dose of radiation spent on locating regions of interest is minimized, reducing damage to radiation-sensitive samples. This technique has potential for infection biology (82). CLEM has been modified to facilitate correlative light and scanning electron microscopy (CLSEM), use of gold mesh grids for epithelial cell monolayers, and imaging of infection. CLSEM has been applied to trigger invasion by S. enterica and Listeria monocytogenes, as well as the enterocyte attachment and effacement phenotype of EPEC. This highlights the ability of this technique to facilitate research into understanding how bacterial pathogens interact with the apical side of polarized epithelial cells (44). miniSOG (miniature singlet oxygen generator) is a protein fusion strategy that involves correlations of fluorescent data with EM data after fixation without the need for exogenous ligands, probes, or permeabilizing detergents. miniSOG employs a 106–amino acid fluorescent flavoprotein engineered from Arabidopsis phototropin 2. Illumination of miniSOG generates singlet oxygen that catalyzes the polymerization of diaminobenzidine into a reaction product resolvable by EM. miniSOG preserves ultrastructural information and allows three-dimensional protein localization via ET or serial section block-face SEM (87).
Cryo-EM can also be merged with fluorescence light microscopy to provide movies of dynamic cellular processes as well as still pictures at molecular resolution. Integration of cryo-EM and fluorescence imaging modalities is often referred to as correlative microscopy, and this strategy benefits from both the high resolution provided by cryo-EM and the dynamic nature of optical microscopy (101). Protocols for correlated cryogenic fluorescent light microscopy (cryo-fLM), cryo-EM, and cryo-ET (i.e., cryo-CLEM) of virus-infected or -transfected mammalian cells have been established (35) (Figure 5). In a remarkable integration of these approaches, a platform has been developed for three-dimensional cryogenic superresolution and focused ion beam–milled block-face EM across entire frozen cells to facilitate superresolution fluorescence and EM. These and other emerging technologies have the potential to revolutionize infection biology (38) (Figure 5).
Figure 5.
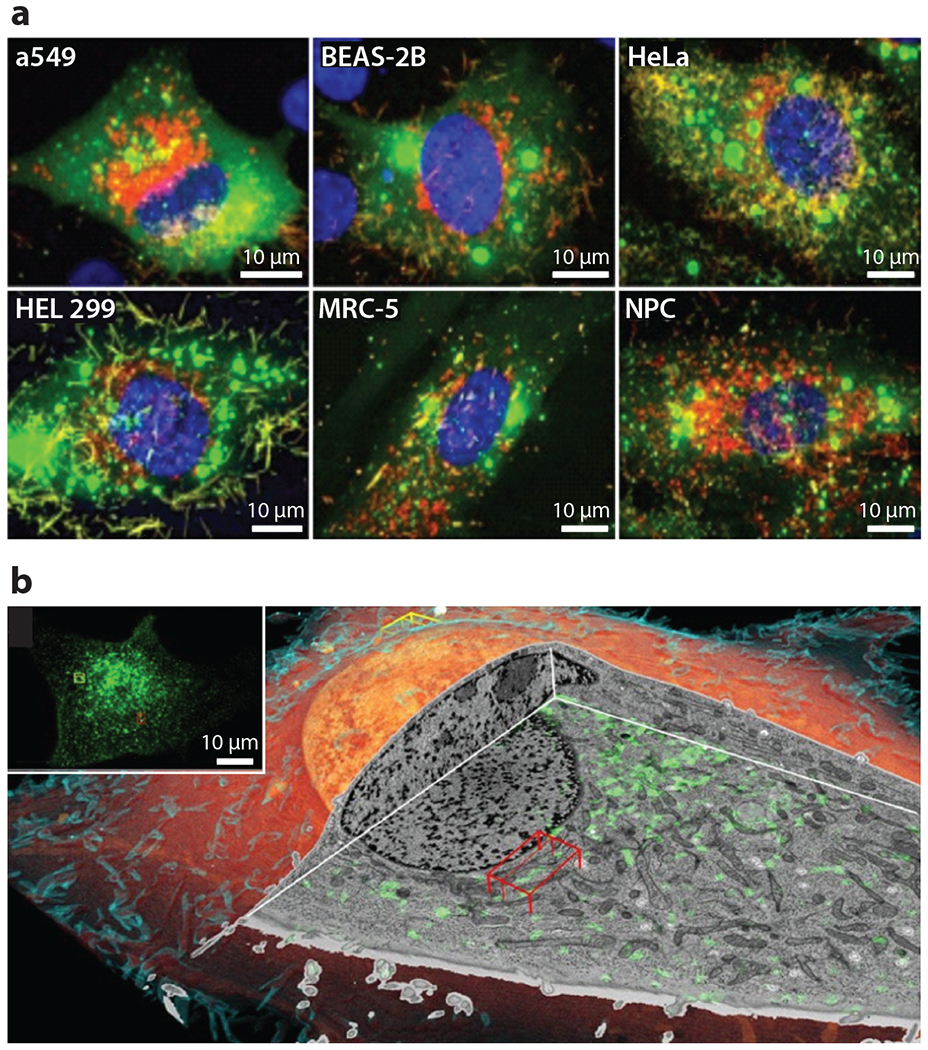
Fluorescence and correlative microscopy. (a) Fluorescence light microscopy using spinning-disk confocal and laser scanning confocal to determine the permissivity of cell lines to RSV. The images are stained for RSV fusion protein (red), nucleocapsid (green), and nucleus (blue). Adapted with permission from Reference 35. (b) Volume rendering of cryo-FIB milling coupled to SEM of a SUM159 cell. The inset is a correlative cryo–three-dimensional SIM image where transferrin is colored green to show the endolysosomal compartments. Abbreviations: FIB, focused ion beam; RSV, respiratory syncytial virus; SEM, scanning electron microscopy; SIM, structured illumination microscopy. Adapted with permission from Reference 38.
Correlative X-ray CT-steered serial block-face SEM (SBF-SEM) and TEM have been combined to provide ultrastructural images of the most used model of trichuriasis, Trichuris muris (61) (Figure 6a). In a remarkable example of imaging integration, four modalities were merged—three molecular imaging methods [PET, SPECT, and fluorescence molecular imaging (FMI)] and one anatomic imaging modality (CT)—all in the same animal and with multiple tracers. Such image integration has significant potential for the study of infectious diseases (53).
Figure 6.
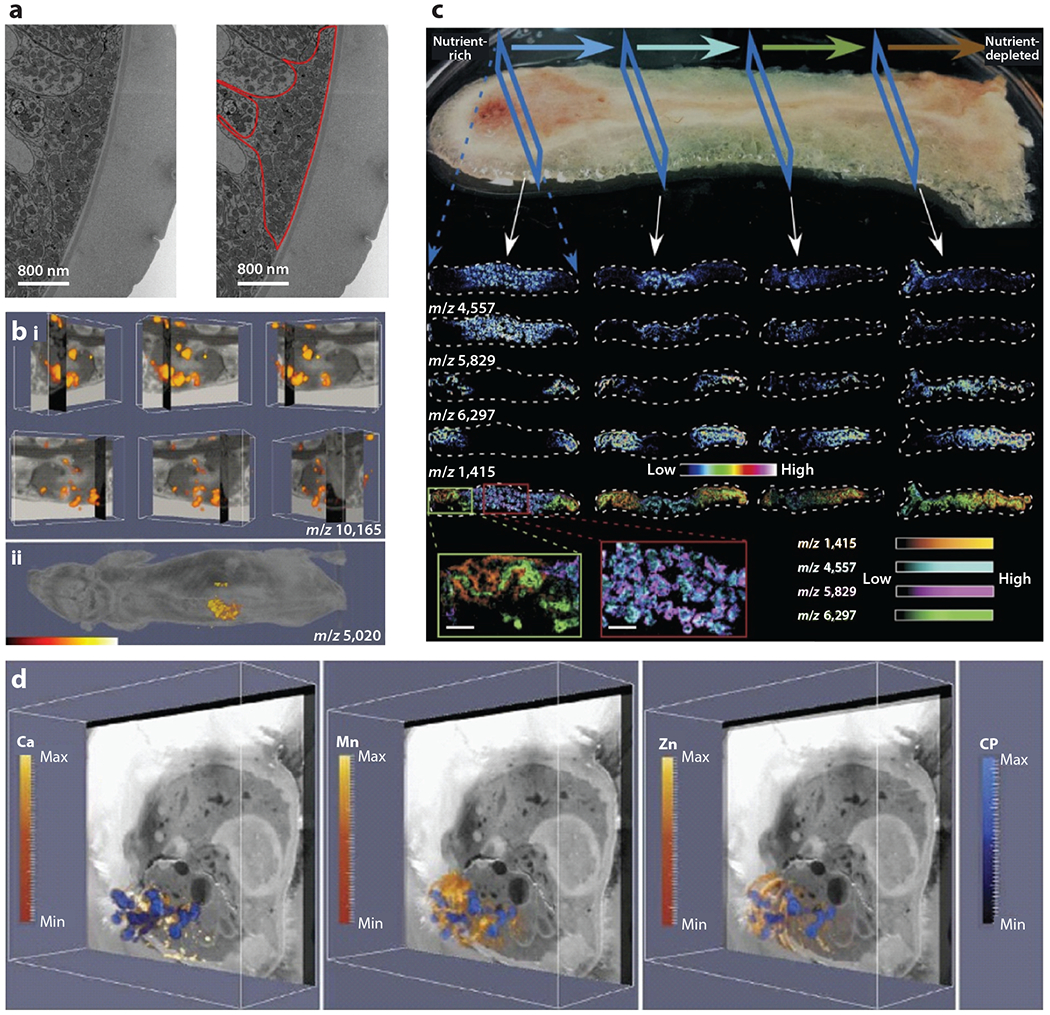
Integrating imaging modalities for infection biology. (a) Electron micrograph of Trichuris muris using a correlative workflow that employs CT-guided SBF-SEM. This image highlights cuticular inflations. A large accumulation of mitochondria is outlined in red. Adapted with permission from Reference 61. (b) IMS integrated with MRI of a Staphylococcus aureus–infected mouse shows masses in the kidney (i) or whole animal (ii) that correlate with a subunit of the innate immune protein calprotectin. Adapted with permission from Reference 4. (c) MALDI-IMS images of a Pseudomonas aeruginosa biofilm. The horizontal arrows at the top depict the flow of nutrients, and the individual mass images represent analytes that are affected by the nutrient gradient in the community. Blue dashed arrows orient the reader to the orientation of the image, and white arrows show where within the biofilm the individual column of images came from. Adapted with permission from Reference 97. (d) Integration of MALDI-IMS, LA-ICP-IMS, and block-face images of an S. aureus–infected mouse showing the localization of calcium, manganese, zinc, and calprotectin. Adapted with permission from Reference 13. Abbreviations: CP, calprotectin; CT, computed tomography; ICP, inductively coupled plasma; IMS, imaging mass spectrometry; LA, laser ablation; MALDI, matrix-assisted laser desorption/ionization; MRI, magnetic resonance imaging; SBF, serial block-face; SEM, scanning electron microscopy.
IMS has also been integrated with other imaging modalities. Pioneering work integrated spatially resolved MALDI-IMS ion images of whole mouse heads with high-resolution MRI (88). MALDI-IMS and MRI were also merged to interrogate the innate immune response to systemic S. aureus infections (4) (Figure 6b). This work was expanded to combine MALDI-IMS, LA-ICP-IMS, IVIS, and MRI in a single experiment to study the impact of staphylococcal infection on metalloprotein distribution, elemental reorganization, and bacterial gene expression during systemic infection (Figure 6d). These experiments once again revealed the significant levels of heterogeneity in analyte distribution that exist during infection, at both the host and bacterial level (13).
7. IMAGING MODALITIES APPLIED TO MICROBIAL COMMUNITIES
Microbes often live in diverse, dynamic, heterogeneous, three-dimensional communities. Imaging these communities is challenging, particularly without disrupting the three-dimensional information contained within these biofilms (58). A variety of strategies have been used to image microbial biofilms, including many of those covered above.
Numerous investigators have used SEM to study microbial communities. Campos-Silva et al. (10) imaged S. aureus biofilms in vivo in Galleria mellonella larvae, employing toothbrush bristles as an abiotic surface to mimic a medical device. Confocal laser scanning microscopy has become a standard technique to study microbial communities. Imaging biofilm formation of the model organism Bacillus subtilis has once again highlighted phenotypic heterogeneity within microbial communities (29). Multiphoton and spinning-disk microscopes provide new options for in situ imaging of microbial communities. Light sheet fluorescence microscopy has strong optical resolution, good sectioning capabilities, and high speed and therefore has considerable potential for infection biology imaging and microbiology. Light sheet fluorescence microscopy enables rapid acquisition of three-dimensional images over large fields of view and over long periods of time and can be applied to bacterial biofilms (67). Biofilm formation can also be studied using a microfluidic system in combination with epifluorescence microscopy, thus allowing for live imaging of biofilm formation and maintenance over time in a dynamic and complex environment. This system has perhaps been best used to study biofilm formation by S. aureus (56).
Other, less conventional, imaging modalities have also been applied to studies of intact microbial communities. Imaging can provide powerful information about elemental distribution and heterogeneity in microbial communities. Scanning transmission X-ray microscopy mapped the spatial distribution of iron species throughout P. aeruginosa biofilms and revealed a complex distribution of this element (41). Nanoscale multiparametric imaging of living bacteria revealed that zinc strongly increases cell wall rigidity and activates the adhesive function of S. aureus SasG (26). Thermal atomic force microscopy revealed nanomechanical properties of internal structures within bacteria, as has been applied to B. anthracis spores. This work employed a nanosurgical sectioning method to cut an individual spore, exposing its internal structure (49). Raman microscopy can also facilitate determination of the distribution of microorganisms and their metabolites inside biofilms (69).
The inherent strengths of IMS make the technique valuable for studying microbial systems. IMS can reveal molecular patterns in homogeneous or mixed microbial communities, or during interactions with the host (9). Workflows have been created that enable characterization of microbial ecosystems including Pseudomonas putida F1 and 5hewanella oneidensis MR-1. MALDI-IMS, secondary ion mass spectrometry (SIMS), and SEM of both single-species and multispecies drip flow biofilms provide incredible insight into biofilm formation, microbe-microbe interactions, and microbial processes (50). SIMS, borrowed from the field of geobiology, is another powerful tool for defining isotope and elemental abundance (64). Finally, as described above for tissues, LA-ICP-IMS can be combined with MALDI-IMS to study elemental distributions in microbial communities and to image the associated bacterial factors that respond to these changes (97) (Figure 6c).
8. CHALLENGES AND FUTURE PERSPECTIVES
The relative paucity of imaging studies in infection biology, as compared to fields like cancer biology and neuroscience, has slowed the pace of discovery in infectious disease research. The lag in development is primarily due to a few aspects of infectious disease research. First, it requires higher biosafety levels. Many institutions are not willing or able to allow the use of certain instruments to image biohazardous samples if those same instruments will subsequently be used to image noninfected or immunocompromised samples. Second, greater sensitivity is required to image smaller objects such as microbes. Finally, because of the diversity of microbial species, it is difficult to develop tools such as probes and dyes that are broadly useful across the entire field. Despite these challenges, technological and computational innovations are leading to impressive improvements in sensitivity and specificity and the ability to integrate imaging across a range of sizes. Moreover, global events are increasing the focus on infectious diseases and the microbiome so that greater resources are being directed to this area. As more researchers enter the field, infection biology should become a driving influence behind the advancement of imaging sciences, and in turn imaging sciences will provide unprecedented insights into host-microbe interactions.
ACKNOWLEDGMENTS
I would like to thank Caitlin Murdoch, Clare Laut, Valeria Reyes-Ruiz, Andy Weiss, Nichole Maloney, Sydney Drury, and Anderson Miller for critical review of this document. Work in our laboratory has been supported by NIH R01 grants (AI145992, AI069233, AI073843, AI101171, AI138581, and AI150701).
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abtin A, Jain R, Mitchell AJ, Roediger B, Brzoska AJ, et al. 2014. Perivascular macrophages mediate neutrophil recruitment during bacterial skin infection. Nat. Immunol 15:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aksoy SY, Asa S, Ozhan M, Ocak M, Sager MS, et al. 2014. FDG and FDG-labelled leucocyte PET/CT in the imaging of prosthetic joint infection. Eur. J. Nucl. Med. Mol. Imaging 41:556–64 [DOI] [PubMed] [Google Scholar]
- 3.Aron AT, Heffern MC, Lonergan ZR, Vander Wal MN, Blank BR, et al. 2017. In vivo bioluminescence imaging of labile iron accumulation in a murine model of Acinetobacter baumannii infection. PNAS 114(48):12669–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attia AS, Schroeder KA, Seeley EH, Wilson KJ, Hammer ND, et al. 2012. Monitoring the inflammatory response to infection through the integration of MALDI IMS and MRI. Cell Host. Microbe 11:664–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belperron AA, Mao J, Bockenstedt LK. 2018. Two photon intravital microscopy of Lyme Borrelia in mice. Methods Mol. Biol 1690:279–90 [DOI] [PubMed] [Google Scholar]
- 6.Bien T, Perl M, Machmuller AC, Nitsche U, Conrad A, et al. 2020. MALDI-2 mass spectrometry and immunohistochemistry imaging of Gb3Cer, Gb4Cer, and further glycosphingolipids in human colorectal cancer tissue. Anal. Chem 92:7096–105 [DOI] [PubMed] [Google Scholar]
- 7.Blom S, Paavolainen L, Bychkov D, Turkki R, Maki-Teeri P, et al. 2017. Systems pathology by multiplexed immunohistochemistry and whole-slide digital image analysis. Sci. Rep 7:15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt SL, Klopfenstein N, Wang S, Winfree S, McCarthy BP, et al. 2018. Macrophage-derived LTB4 promotes abscess formation and clearance of Staphylococcus aureus skin infection in mice. PLOS Pathog. 14:e1007244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron SJS, Takats Z. 2018. Mass spectrometry approaches to metabolic profiling of microbial communities within the human gastrointestinal tract. Methods 149:13–24 [DOI] [PubMed] [Google Scholar]
- 10.Campos-Silva R, Brust FR, Trentin DS, Macedo AJ. 2019.Alternative method in Galleria mellonellalarvae to study biofilm infection and treatment. Microb. Pathog 137:103756. [DOI] [PubMed] [Google Scholar]
- 11.Casal D, Iria I, Ramalho JS, Alves S, Mota-Silva E, et al. 2019. BD-2 and BD-3 increase skin flap survival in a model of ischemia and Pseudomonas aeruginosa infection. Sci. Rep 9:7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassat JE, Hammer ND, Campbell JP, Benson MA, Perrien DS, et al. 2013.Asecreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13:759–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassat JE, Moore JL, Wilson KJ, Stark Z, Prentice BM, et al. 2018. Integrated molecular imaging reveals tissue heterogeneity driving host-pathogen interactions. Sci. Transl. Med 10:eaan6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chai Q, Wang X, Qiang L, Zhang Y, Ge P, et al. 2019. A Mycobacterium tuberculosis surface protein recruits ubiquitin to trigger host xenophagy. Nat. Commun 10:1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charles-Orszag A, Tsai FC, Bonazzi D, Manriquez V, Sachse M, et al. 2018. Adhesion to nanofibers drives cell membrane remodeling through one-dimensional wetting. Nat. Commun 9:4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury S, Happonen L, Khakzad H, Malmstrom L, Malmstrom J. 2020. Structural proteomics, electron cryo-microscopy and structural modeling approaches in bacteria-human protein interactions. Med. Microbiol. Immunol 209:265–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, et al. 2013. Structural and molecular interrogation of intact biological systems. Nature 497:332–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cronan MR, Rosenberg AF, Oehlers SH, Saelens JW, Sisk DM, et al. 2015. CLARITY and PACT-based imaging of adult zebrafish and mouse for whole-animal analysis of infections. Dis. Model. Mech 8:1643–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cyrklaff M, Linaroudis A, Boicu M, Chlanda P, Baumeister W, et al. 2007. Whole cell cryo-electron tomography reveals distinct disassembly intermediates of vaccinia virus. PLOS ONE 2:e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis SL, Be NA, Lamichhane G, Nimmagadda S, Pomper MG, et al. 2009. Bacterial thymidine kinase as a non-invasive imaging reporter for Mycobacterium tuberculosis in live animals. PLOS ONE 4:e6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePas WH, Starwalt-Lee R, Van Sambeek L, Ravindra Kumar S, Gradinaru V, Newman DK. 2016. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7:e00796–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du Z, Lin JR, Rashid R, Maliga Z, Wang S, et al. 2019. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat. Protoc 14:2900–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duheron V, Moreau M, Collin B, Sali W, Bernhard C, et al. 2014. Dual labeling of lipopolysaccharides for SPECT-CT imaging and fluorescence microscopy. ACS Chem. Biol 9:656–62 [DOI] [PubMed] [Google Scholar]
- 24.Dumarey N, Egrise D, Blocklet D, Stallenberg B, Remmelink M, et al. 2006. Imaging infection with 18F-FDG–labeled leukocyte PET/CT: initial experience in 21 patients. J. Nucl. Med 47:625–32 [PubMed] [Google Scholar]
- 25.Fiole D, Deman P, Trescos Y, Mayol JF, Mathieu J, et al. 2014. Two-photon intravital imaging of lungs during anthrax infection reveals long-lasting macrophage-dendritic cell contacts. Infect. Immun 82:864–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Formosa-Dague C, Speziale P, Foster TJ, Geoghegan JA, Dufrene YF. 2016. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. PNAS 113:410–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox D, Mathur A, Xue Y, Liu Y, Tan WH, et al. 2020. Bacillus cereus non-haemolytic enterotoxin activates the NLRP3 inflammasome. Nat. Commun 11:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M, Lola CM, Wang M, Miller KD, Sledge GW, et al. 2010. Synthesis of carbon-11-labeled tricyclic necroptosis inhibitors as new potential PET agents for imaging of tumor necrosis factor alpha (TNF-α). Appl. Radiat. Isot 68:1950–58 [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Betancur JC, Yepes A, Schneider J, Lopez D. 2012. Single-cell analysis of Bacillus subtilis biofilms using fluorescence microscopy and flow cytometry. J. Vis. Exp 2012(6):e3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson HM, McKnight BN, Malysa A, Dyson G, Wiesend WN, et al. 2018. IFNγ PET imaging as a predictive tool for monitoring response to tumor immunotherapy. Cancer Res. 78:5706–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaros TG, Blancett CD, Bell TM, Natesan M, Ulrich RG. 2015. Serum biomarkers of Burkholderia mallei infection elucidated by proteomic imaging of skin and lung abscesses. Clin. Proteom 12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, et al. 2018. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174:968–81.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gowrishankar G, Hardy J, Wardak M, Namavari M, Reeves RE, et al. 2017. Specific imaging of bacterial infection using 6″-18F-fluoromaltotriose: a second-generation PET tracer targeting the maltodextrin transporter in bacteria. J. Nucl. Med 58:1679–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo N, van Unen V, Ijsselsteijn ME, Ouboter LF, van der Meulen AE, et al. 2020. A 34-marker panel for imaging mass cytometric analysis of human snap-frozen tissue. Front. Immunol 11:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hampton CM, Strauss JD, Ke Z, Dillard RS, Hammonds JE, et al. 2017. Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat. Protoc 12:150–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harper J, Skerry C, Davis SL, Tasneen R, Weir M, et al. 2012. Mouse model of necrotic tuberculosis granulomas develops hypoxic lesions. J. Infect. Dis 205:595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harvie EA, Huttenlocher A. 2015. Neutrophils in host defense: new insights from zebrafish. J. Leukoc. Biol 98:523–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, et al. 2020. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367:eaaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosseini R, Lamers GE, Hodzic Z, Meijer AH, Schaaf MJ, Spaink HP. 2014. Correlative light and electron microscopy imaging of autophagy in a zebrafish infection model. Autophagy 10:1844–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hulme HE, Meikle LM, Wessel H, Strittmatter N, Swales J, et al. 2017. Mass spectrometry imaging identifies palmitoylcarnitine as an immunological mediator during Salmonella Typhimurium infection. Sci. Rep 7:2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter RC, Hitchcock AP, Dynes JJ, Obst M, Beveridge TJ. 2008. Mapping the speciation of iron in Pseudomonas aeruginosa biofilms using scanning transmission X-ray microscopy. Environ. Sci. Technol 42:8766–72 [DOI] [PubMed] [Google Scholar]
- 42.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar EP. 2018. Acinetobacter baumannii OxyR regulates the transcriptional response to hydrogen peroxide. Infect. Immun 87(1):e00413–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khusainov I, Fatkhullin B, Pellegrino S, Bikmullin A, Liu WT, et al. 2020.Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach. Nat. Commun 11:1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kommnick C, Lepper A, Hensel M 2019. Correlative light and scanning electron microscopy(CLSEM) for analysis of bacterial infection of polarized epithelial cells. Sci. Rep 9:17079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. 2011. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol 192:111–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lankinen P, Makinen TJ, Poyhonen TA, Virsu P, Salomaki S, et al. 2008 . 68Ga-DOTAVAP-P1 PET imaging capable of demonstrating the phase of inflammation in healing bones and the progress of infection in osteomyelitic bones. Eur. J. Nucl. Med. Mol. Imaging 35:352–64 [DOI] [PubMed] [Google Scholar]
- 47.Lankinen P, Noponen T, Autio A, Luoto P, Frantzen J, et al. 2018. A comparative 68Ga-citrate and 68Ga-chloride PET/CT imaging of Staphylococcus aureus osteomyelitis in the rat tibia. Contrast Media Mol. Imaging 2018:9892604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JK, Enciso GA, Boassa D, Chander CN, Lou TH, et al. 2018. Replication-dependent size reduction precedes differentiation in Chlamydia trachomatis. Nat. Commun 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li AG, Burggraf LW, Xing Y. 2016. Nanomechanical characterization of Bacillus anthracis spores by atomic force microscopy. Appl. Environ. Microbiol 82:2988–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, Dunham SJB, Ellis JF, Lange JD, Smith JR, et al. 2018. A versatile strategy for characterization and imaging of drip flow microbial biofilms. Anal. Chem 90:6725–34 [DOI] [PubMed] [Google Scholar]
- 51.Li N, van Unen V, Abdelaal T, Guo N, Kasatskaya SA, et al. 2019.Memory CD4+ T cells are generated in the human fetal intestine. Nat. Immunol 20:301–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Locke LW, Chordia MD, Zhang Y, Kundu B, Kennedy D, et al. 2009.A novel neutrophil-specific PET imaging agent: cFLFLFK-PEG-64Cu. J. Nucl. Med 50:790–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y, Yang K, Zhou K, Pang B, Wang G, et al. 2014. An integrated quad-modality molecular imaging system for small animals. J. Nucl. Med 55:1375–79 [DOI] [PubMed] [Google Scholar]
- 54.Lucic V, Rigort A, Baumeister W. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol 202:407–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luque D, Caston JR. 2020. Cryo-electron microscopy for the study of virus assembly. Nat. Chem. Biol 16:231–39 [DOI] [PubMed] [Google Scholar]
- 56.Moormeier DE, Bose JL, Horswill AR, Bayles KW. 2014. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 5:e01341–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morgan CE, Huang W, Rudin SD, Taylor DJ, Kirby JE, et al. 2020. Cryo-electron microscopy structure of the Acinetobacter baumannii 70S ribosome and implications for new antibiotic development. mBio 11:e03117–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neu TR, Manz B, Volke F, Dynes JJ, Hitchcock AP, Lawrence JR. 2010. Advanced imaging techniques for assessment of structure, composition and function in biofilm systems. FEMS Microbiol. Ecol 72:1–21 [DOI] [PubMed] [Google Scholar]
- 59.Nguyen M, Rizvi J, Hecht G. 2015. Expression of enteropathogenic Escherichia coli map is significantly different than that of other type III secreted effectors in vivo. Infect. Immun 83:130–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ning X, Seo W, Lee S, Takemiya K, Rafi M, et al. 2014. PET imaging of bacterial infections with fluorine-18-labeled maltohexaose. Angew. Chem. Int. Ed. Engl 53:14096–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Sullivan JDB, Cruickshank SM, Starborg T, Withers PJ, Else KJ. 2020. Characterisation of cuticular inflation development and ultrastructure in Trichuris muris using correlative X-ray computed tomography and electron microscopy. Sci. Rep 10:5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ordonez AA, Wang H, Magombedze G, Ruiz-Bedoya CA, Srivastava S, et al. 2020. Dynamic imaging in patients with tuberculosis reveals heterogeneous drug exposures in pulmonary lesions. Nat. Med 26:529–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ordonez AA, Weinstein EA, Bambarger LE, Saini V, Chang YS, et al. 2017. A systematic approach for developing bacteria-specific imaging tracers. J. Nucl. Med 58:144–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orphan VJ, House CH. 2009. Geobiological investigations using secondary ion mass spectrometry: microanalysis of extant and paleo-microbial processes. Geobiology 7:360–72 [DOI] [PubMed] [Google Scholar]
- 65.Park ES, Lee JH, Hong JH, Park YK, Lee JW, et al. 2014. Phosphatidylcholine alteration identified using MALDI imaging MS in HBV-infected mouse livers and virus-mediated regeneration defects. PLOS ONE 9:e103955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parker MFL, Luu JM, Schulte B, Huynh TL, Stewart MN, et al. 2020. Sensing living bacteria in vivo using d-alanine-derived 11C radiotracers. ACS Cent. Sci 6:155–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parthasarathy R 2018. Monitoring microbial communities using light sheet fluorescence microscopy. Curr. Opin. Microbiol 43:31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patterson NH, Tuck M, Lewis A, Kaushansky A, Norris JL, et al. 2018. Next generation histology-directed imaging mass spectrometry driven by autofluorescence microscopy. Anal. Chem 90:12404–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patzold R, Keuntje M, Anders-von Ahlften A. 2006. A new approach to non-destructive analysis of biofilms by confocal Raman microscopy. Anal. Bioanal. Chem 386:286–92 [DOI] [PubMed] [Google Scholar]
- 70.Perez OA, Yeung ST, Vera-Licona P, Romagnoli PA, Samji T, et al. 2017. CD169+ macrophages orchestrate innate immune responses by regulating bacterial localization in the spleen. Sci. Immunol 2:eaah5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perry WJ, Spraggins JM, Sheldon JR, Grunenwald CM, Heinrichs DE, et al. 2019. Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. PNAS 116:21980–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perry WJ, Weiss A, Van de Plas R, Spraggins JM, Caprioli RM, Skaar EP 2020. Integrated molecular imaging technologies for investigation of metals in biological systems: a brief review. Curr. Opin. Chem. Biol 55:127–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrik M, Franssen GM, Haas H, Laverman P, Hortnagl C, et al. 2012. Preclinical evaluation of two 68Ga-siderophores as potential radiopharmaceuticals for Aspergillus fumigatus infection imaging. Eur. J. Nucl. Med. Mol. Imaging 39:1175–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pullambhatla M, Tessier J, Beck G, Jedynak B, Wurthner JU, Pomper MG. 2012. [125I]FIAU imaging in a preclinical model of lung infection: quantification of bacterial load. Am. J. Nucl. Med. Mol. Imaging 2:260–70 [PMC free article] [PubMed] [Google Scholar]
- 75.Rossmann MG, Morais MC, Leiman PG, Zhang W. 2005. Combining X-ray crystallography and electron microscopy. Structure 13:355–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW. 2019. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat. Microbiol 4:1328–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russell MR, Lerner TR, Burden JJ, Nkwe DO, Pelchen-Matthews A, et al. 2017. 3D correlative light and electron microscopy of cultured cells using serial blockface scanning electron microscopy. J. Cell Sci 130:278–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryzhkova MN, Tarasova LA. 1975. [Pathogenesis and treatment of manganese induced Parkinsonism]. Gig. Tr. Prof. Zabol 1975:31–34 (In Russian) [PubMed] [Google Scholar]
- 79.Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, et al. 2010. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLOS Pathog 6:e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sapermsap N, Li DD, Al-Hemedawi R, Li Y, Yu J, et al. 2020. A rapid analysis platform for investigating the cellular locations of bacteria using two-photon fluorescence lifetime imaging microscopy. Methods Appl. Fluoresc 8:034001. [DOI] [PubMed] [Google Scholar]
- 81.Saraswathi P, Beuerman RW. 2015. Corneal biofilms: from planktonic to microcolony formation in an experimental keratitis infection with Pseudomonas aeruginosa. Ocul. Surf 13:331–45 [DOI] [PubMed] [Google Scholar]
- 82.Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. 2007. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol 160:135–45 [DOI] [PubMed] [Google Scholar]
- 83.Scott AJ, Post JM, Lerner R, Ellis SR, Lieberman J, et al. 2017. Host-based lipid inflammation drives pathogenesis in Francisella infection. PNAS 114:12596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sellmyer MA, Lee I, Hou C, Weng CC, Li S, et al. 2017. Bacterial infection imaging with [18F]fluoropropyl-trimethoprim. PNAS 114:8372–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sharaf R, Mempel TR, Murooka TT. 2016. Visualizing the behavior of HIV-infected T cells in vivo using multiphoton intravital microscopy. Methods Mol. Biol 1354:189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sheedlo MJ, Chung JM, Sawhney N, Durie CL, Cover TL, et al. 2020. Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex. eLife 9:e59495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, et al.2011.A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLOS Biol 9:e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, et al. 2008. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat. Methods 5:57–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sood A, Sui Y, McDonough E, Santamaria-Pang A, Al-Kofahi Y, et al. 2020. Comparison of multiplexed immunofluorescence imaging to chromogenic immunohistochemistry of skin biomarkers in response to monkeypox virus infection. Viruses 12:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spraggins JM, Rizzo DG, Moore JL, Noto MJ, Skaar EP, Caprioli RM. 2016. Next-generation technologies for spatial proteomics: integrating ultra-high speed MALDI-TOF and high mass resolution MALDIFTICR imaging mass spectrometry for protein analysis. Proteomics 16:1678–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Su CC, Morgan CE, Kambakam S, Rajavel M, Scott H, et al. 2019. Cryo-electron microscopy structure of an Acinetobacter baumannii multidrug efflux pump. mBio 10:e01295–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Surewaard BGJ, Thanabalasuriar A, Zeng Z, Tkaczyk C, Cohen TS, et al. 2018. α-Toxin induces platelet aggregation and liver injury during Staphylococcus aureus sepsis. Cell Host Microbe 24:271–84.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tarafder AK, von Kugelgen A, Mellul AJ, Schulze U, Aarts D, Bharat TAM. 2020. Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. PNAS 117:4724–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Torraca V, Masud S, Spaink HP, Meijer AH. 2014. Macrophage-pathogen interactions in infectious diseases: new therapeutic insights from the zebrafish host model. Dis. Model. Mech 7:785–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venien-Bryan C, Li Z, Vuillard L, Boutin JA. 2017. Cryo-electron microscopy and X-ray crystallography: complementary approaches to structural biology and drug discovery. Acta Crystallogr. F 73:174–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vilche M, Reyes AL, Vasilskis E, Oliver P, Balter H, Engler H. 2016.68Ga-NOTA-UBI-29-41 as a PET tracer for detection of bacterial infection. J. Nucl. Med 57:622–27 [DOI] [PubMed] [Google Scholar]
- 97.Wakeman CA, Moore JL, Noto MJ, Zhang Y, Singleton MD, et al. 2016. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Commun 7:11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wan W, Kolesnikova L, Clarke M, Koehler A, Noda T, et al. 2017. Structure and assembly of the Ebola virus nucleocapsid. Nature 551:394–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang GJ, Porta C, Chen ZG, Baker TS, Johnson JE. 1992. Identification of a Fab interaction footprint site on an icosahedral virus by cryoelectron microscopy and X-ray crystallography. Nature 355:275–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wiehr S, Warnke P, Rolle AM, Schutz M, Oberhettinger P, et al. 2016. New pathogen-specific immunoPET/MR tracer for molecular imaging of a systemic bacterial infection. Oncotarget 7:10990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang P 2013. Correlative cryo-electron tomography and optical microscopy of cells. Curr. Opin. Struct. Biol 23:763–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Z, Ordonez AA, Wang H, Li Y, Gogarty KR, et al. 2018. Positron emission tomography imaging with 2-[18F]F-p-aminobenzoic acid detects Staphylococcus aureus infections and monitors drug response. ACS Infect. Dis 4:1635–44 [DOI] [PMC free article] [PubMed] [Google Scholar]


