ABSTRACT
The current Coronavirus Disease 2019 (COVID-19) pandemic is causing great alarm around the world. The pathogen for COVID-19 – severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – is the seventh known coronavirus to cause pneumonia in humans. While much remains unknown about SARS-CoV-2, physicians and researchers have begun to publish relevant findings, and much evidence is available on coronaviruses previously circulating in human and animal populations. In this review, we situate COVID-19 in its context as a transboundary viral disease, and provide a comprehensive discussion focused on the discovery, spread, virology, pathogenesis, and clinical features of this disease, its causative coronaviral pathogen, and approaches to combating the disease through immunotherapies and other treatments and vaccine development. An epidemiological survey revealed a potentially large number of asymptomatic SARS-CoV-2 carriers within the population, which may hamper efforts against COVID-19. Finally, we emphasize that vaccines against SARS-CoV-2, which may be developed by 2021, will be essential for prevention of COVID-19.
KEYWORDS: Human coronavirus, public health, respiratory viruses, origin, pathogenesis, sars-CoV-2
Introduction
The current global COVID-19 pandemic has caused great alarm, since its causative pathogen – severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – apparently jumped the species barrier into humans in the latter part of 20191. The causative agent of the outbreak was identified as a betacoronavirus with a genomic sequence closely related to that of the severe acute respiratory syndrome (SARS) coronavirus, and the new virus was named as SARS-CoV-2.2–4 Coronavirus disease 2019 (COVID-19; formerly known as “2019-nCoV”) has been declared a pandemic, spreading essentially to all countries and/or territories. This spread encompassed both community transmission and case clusters in countries ranging from the China to the United States, United Kingdom, Germany, France, Spain, Japan, Singapore, South Korea, Iran, and Italy.5 COVID-19-infected individuals are often asymptomatic or exhibit only mild to moderate symptoms, but the disease progresses to severe pneumonia in approximately 15% of cases, and ultimately to acute respiratory distress syndrome (ARDS) in about 5% of cases.6,7
To date, no specific treatment or vaccine for SARS-CoV-2 infection has been established, and some attention has focussed on repurposed therapeutics and/or immunotherapeutic options for supportive care.8,9
Many of the serious questions this disease poses the medical community remain unanswered. However, investigators and researchers are starting to publish data ranging from real-world clinical evidence and clinical study results to initial investigations in animal models. Furthermore, there is a wealth of evidence on other coronaviruses (CoVs) in both human and animal populations. In this review, we set out to provide a comprehensive discussion of this transboundary viral disease, covering its discovery, spread, virology, pathogenesis, and clinical features, and immunotherapies and vaccines that may be used to combat it, setting these issues within the context of evidence on previously circulating CoVs.
Historical background on CoVs
CoVs are members of the subfamily Orthocoronavirinae in the family Coronaviridae and the order Nidovirales. CoVs are classified into four genera Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV) and Deltacoronavirus (δ-CoV) according to phylogenetic relationships and genomic structures.10 The pathogen host ranges comprise only mammalian species for α-CoVs and β-CoVs, but mainly avian species as well as some mammalian species for γ-CoVs and δ-CoVs.10 CoVs are a group of enveloped, positive-strand RNA viruses. Their genomes are the largest among RNA viruses (at 26–32 kb) and their numbers of open reading frames (ORFs) vary between six and 11.11 Further novel CoVs are predicted to emerge in the future, due to their large genetic diversity and frequent genomic recombination, as well as the increasing opportunities for human-animal interface.10 Up to the present, a number of CoV infections have been seen in animal and human populations. The diseases that result from such infections range from the asymptomatic to the severe, and where symptoms are presented, these can include respiratory, enteric, hepatic, and neurological signs.12 Human coronaviruses (HCoVs) have been known pathogens since the mid-1960s. To date, there are seven known HCoVs; these comprise two α-CoVs (HCoV-229E and HCoV-NL63) and five β-CoVs (HCoV-OC43 [lineage A], HCoV-HKU1 [lineage A], severe acute respiratory syndrome CoV (SARS-CoV) [lineage B], Middle East respiratory syndrome CoV (MERS-CoV) [lineage C], and SARS-CoV-2 [lineage B])13 (Figure 1).
Figure 1.
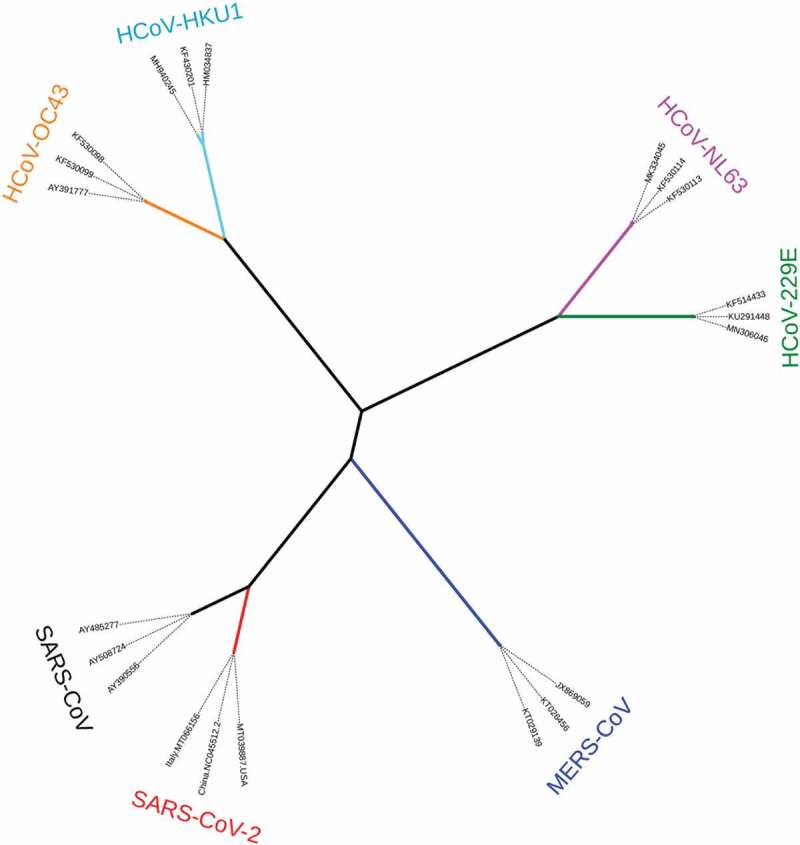
Phylogenetic relationships of human coronaviruses (HCoVs). The tree was reconstructed based on full, published genome sequences for HCoVs using the maximum-likelihood algorithm implemented in MEGA X software.14 The tree was edited using iTool version 3 software.15
HCoVs-229E and HCoVs-OC43 were first described in the 1960 s. Infection with these viruses generally causes symptoms resembling a common cold in immunocompetent individuals, but their symptoms can be more severe in infants, the elderly, and the immunocompromised.16,17 The known natural hosts are bat and palm civets for HCoVs-229E, and bats for HCoVs-OC43. HCoVs-229E and HCoVs-OC43 use human aminopeptidase N (CD13) and angiotensin-converting enzyme 2 (ACE2) as their respective receptors.18 HCoVs cause the common winter cold, and their fatality rates have been estimated at 0.5%-1.5%.19
In contrast to HCoVs-229E and HCoVs-OC43, SARS-CoV, which emerged in Guangdong Province, China, in November 2002, causes severe lung disease. It rapidly spread to 29 countries and many regions across the globe, and the total number of cases reached 8,098 – with 774 fatalities – by the end of the epidemic in July 2003. Nosocomial transmission was seen in many cases, with adverse implications for health-care workers.5,20 Infected patients exhibited atypical pneumonia characterized by diffuse alveolar damage, with the potential to progress to acute respiratory distress syndrome (ARDS). Severe ARDS-related coronaviruses (SARSr-CoVs) or SARS-like CoVs in horseshoe bats (Rhinolophus spp.) share 88–95% sequence homology with human and civet CoV isolates, which suggests that bats were probably the natural reservoir of a close ancestor of SARS-CoV and palm civets were possible intermediate hosts.21 SARS-CoV infects human alveolar type II cells and uses ACE2 as a receptor.22
HCoV-NL63 was first detected in patients with respiratory tract illness shortly after the emergence of SARS-CoV, in 2004.23 HCoV-NL63 is prevalent worldwide and appears with a seasonal distribution similar to that of other HCoVs.24 Usually, HCoV-NL63 infection causes a mild respiratory tract disease, but fatalities have been reported in infants and the elderly.25,26 In addition, HCoV-NL63 and HCoV-229E CoVs most likely emerged in the human population during two separate zoonotic transmission events.27
Both HCoV-NL63 and SARS-CoV have a similar way of binding to the cellular membrane. This is done via heparan sulfate proteoglycans, which then facilitate interaction with the ACE2.28
HKU1 (HCoV-HKU1) was first detected in 2005 and is associated with community-acquired pneumonia.29 HCoV-HKU1 induces only a mild disease in most patients; however, there is a risk of death in a minority of patients who have low hemoglobin concentrations, monocyte counts, serum albumin levels, and oxygen saturation levels.30
MERS-CoV was first reported in Jeddah, Saudi Arabia, in September 2012 and has become endemic in Middle Eastern countries and South Korea.31,32 Since its emergence, notifications for MERS-CoV to the World Health Organization (WHO) have covered 2494 laboratory-confirmed cases of infection and 858 deaths, across 27 countries.32 Population groups most at risk of a severe disease following MERS-CoV infection include elderly people, immunosuppressed individuals, and patients with cancer, chronic lung disease, or diabetes.33 MERS-CoV enters the cell through the dipeptidyl-peptidase 4 (DPP4, also known as CD26).34 The transmission routes for MERS-CoV are not fully established, but one possibility involves an initial transmission from bats to camels and other possible vector species.34
SARS-CoV-2 is the second highly pathogenic human coronavirus to emerge since the global scare caused by MERS-CoV. This novel coronavirus (initially named 2019-nCoV) emerged in Wuhan, Hubei province, China, in December 2019. A “wet” market in Wuhan – selling seafood and animals – is regarded as the likely source of the outbreak. Sequence analysis showed that the newly identified virus is related to the SARS-CoV clade1. On January 30, 2020, the WHO officially declared the 2019-nCoV (previous designation for the disease) epidemic as a Public Health Emergency of International Concern. Later, the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (ICTV) changed the name of the virus from 2019-nCoV to its current name, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), based on the phylogenetic relationship to related coronaviruses.3 The virus has rapidly been disseminated since the initial outbreak, with clear cases of human-to-human transmission enhancing its dissemination to pandemic proportions.4,5 Hospitals are confronted with a massive influx of COVID-19 patients. This creates a considerable stress for the wider health system and also quickly renders the number of ventilator-equipped beds insufficient, and the number of deaths may increase as a consequence.35,36 Globally, around five million people have been infected, of whom more than three hundred thousand have died. Infection and subsequent fatalities have occurred in almost all countries and/or territories around,5 with the latter surpassing the combined reported death toll of SARS and MERS (a current total of 1632 fatalities).
The experiences of China and South Korea may offer useful lessons. Those countries have undertaken aggressive population-based tracing, mass testing and isolation measures.37,38 This contrasts with many other countries where testing is limited only to those with more severe disease or those who are at risk of serious complications.
The three highly pathogenic HCoVs (SARS-CoV, MERS-CoV, and SARS-CoV-2) cause severe respiratory syndrome in humans. SARS-CoV and MERS-CoV have much higher fatality rates but much lower transmissibility than SARS-CoV-2. It appears that SARS-CoV-2 is less pathogenic than SARS-CoV, and much less pathogenic than MERS-CoV.39 Moreover, the majority of SARS-CoV and MERS-CoV cases were associated with nosocomial spread in hospitals, whereas COVID-19 is much more widely transmitted in the community, mostly by pre-symptomatic and asymptomatic carriers.6,40 In addition, it has been suggested that modest ACE2 expression in the upper respiratory tract might limit SARS-CoV transmissibility.41 SARS-CoV-2 has a greater kinetic binding affinity for ACE2 than SARS-CoV,39,42 and this may partially explain why SARS-CoV-2 appears to be more readily transmissible.
Virology
SARS-CoV-2 is an enveloped, non-segmented positive sense RNA virus with a genome ranging from 29,891 to 29,903 nucleotides in length. It belongs to the genus β-CoV of the subfamily Orthocoronavirinae in the Coronaviridae family. The SARS-CoV-2 genome possesses 14 ORFs encoding 27 proteins.43 The orf1ab and orf1a genes located at the 5ʹ-terminus of the genome encode the pp1ab and pp1a proteins, respectively, with 15 non-structural proteins (nsps), including nsp1 to nsp10 and nsp12 to nsp16.43 The 3ʹ-terminus of the genome contains four structural proteins (S, E, M, and N) and eight accessory proteins (3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14).43 SARS-CoV-2 has 79.6% sequence identity to SARS-CoV and 51.8% identity to MERS-CoV.1 Receptor recognition is an important step and is a key determinant of host cell and tissue tropism and cross-species infection for viruses. SARS-CoV-2 attaches to ACE2 using the spike protein (S protein) anchored in the viral envelope, similarly to SARS-CoV.1,39,42 The S protein of SARS-CoV-2 has a functional polybasic (furin) cleavage site at the boundary between the S1 (host cell receptor) and S2 (fusion of the viral and cellular membranes) subunits, which is processed during biogenesis, a novel feature which distinguishes this virus from SARS-CoV and SARSr-CoVs.44 The S1 subunit includes the receptor-binding domain (RBD), whereas the carboxyl terminal S2 portion of the molecule contains regions that drive membrane fusion. SARS-CoV-2 entry into cells depends on the ACE2 receptor and transmembrane serine protease 2 (TMPRSS2) activity, and these constitute promising candidates for anti-SARS-CoV-2 therapeutics.39 Both SARS-CoV-2 and SARS-CoV utilize ACE2 as a receptor and require the TMPRSS2 to mediate cell entry, but it is possible that differences in host-entry mechanisms account for some of the differences between the two viruses in transmission and pathogenesis. Importantly, SARS-CoV-2 is more pathogenic, at least in part because of its 10- to 20-fold greater binding affinity for ACE2 than SARS-CoV S-protein.39 Further dissemination in the kidneys, the gastrointestinal tract, or brain may be related to local ACE2 expression.45 Moreover, increased ACE2 expression and ACE2 gene polymorphisms may increase SARS-CoV-2 susceptibility and the risk of a poor COVID-19 disease outcome.19
Protease cleavage of the S protein is required for virus-cell fusion and release of genomic RNA into the cytoplasm, as well as translation, RNA replication, packaging, and ultimately the release of a virion structurally similar to that of SARS-CoV.46
Host and reservoir
Based on current knowledge of virus transmission, some speculation is possible about the reservoir and transmission route for SARS-CoV-2.
Such speculation inevitably centers on bats, as known reservoir hosts for a wide range of zoonotic viruses.47 They also host the vast majority of CoV strains known around the world today, at 91 of 100 identified phylogenetic lineages, a figure which includes β-CoVs.47
Phylogenetically proximate strains to SARS-CoV-2 have been found in two species. One species was the Malayan pangolins (Manis javanica), which hosts a strain with 90% overall nucleotide sequence identity to SARS-CoV-2.48 This strain carries a receptor-binding domain (RDB) predicted to interact with ACE2 and sharing 97.4% identity in its amino acid sequence with that of SARS-CoV-2.49 The other species was the Rhinolophus affinis. The CoV strain isolated from this bat species (BatCoV RaTG13) is 96.2% identical to SARS-CoV-2 at the whole‐genome level.1 However, the BatCoV RaTG13 spike diverges from SARS-CoV-2 in the RBD and does not have a multibasic cleavage site, which suggests that it may not bind efficiently to human ACE2.50
Assessing the evidence from these two species, the pangolin and Rhinolophus affinis-hosted CoV strains are unlikely to be the immediate ancestor of SARS-CoV-2, in view of the sequence divergences over the whole genome, although they are closely related to SARS-CoV-2.51 It thus seems likely that SARS-CoV-2 originated in bats, which acted as the reservoir before transmission to humans, either directly or via Malayan pangolins or an as-yet unknown intermediate host.1,48 Currently, no intermediate host has been identified for SARS-CoV-2.
Transmission and clinical manifestations
SARS-CoV-2 appears to be highly contagious and can spread from person to person through respiratory droplets when an infected person coughs or sneezes, and through fomites. Presymptomatic and asymptomatic carriers may also represent a significant driver of transmission.52 This was borne out in the case of the Diamond Princess, a cruise ship quarantined at the port of Yokohama, Japan, when a number of passengers were known to have developed COVID-19. Ultimately, a large number of the crew and passengers tested positive for the virus, and the proportion of asymptomatic cases was 17.9%.53 Other study has reported a high viral load shortly after symptom onset, highlighting SARS-CoV-2’s ease of transmission.54
Considering the potential for asymptomatic and pre-symptomatic transmission, many recommendations for preventing virus spread (within the community generally and to people in health care settings in particular) and “flattening the increase curve” involve testing, isolation of those who test positive, and social distancing, and many view such measures as essential.55,56 Confronted with this situation, many countries have started to develop and manufacture a wide range of vital equipment such as ventilators, medical oxygen, and personal protective equipment.
SARS-CoV-2 has an estimate basic reproduction rate (R0) of 2–6.47, higher than those of SARS-CoV (≈3) and MERS-CoV (>1), indicating a higher transmissibility than other for HCoVs; however, the exact R0 is still a subject of debate.57–59 Nevertheless, the R0 of SARS-CoV-2 is gradually decreasing with the implementation of epidemiological intervention strategies. The median incubation period for COVID-19 is estimated to be 4–5.1 days,60 and 97.5% of those who develop symptoms will do so within 11.5 days of infection.61 Accordingly, the length of the incubation period should be considered when setting the duration of quarantine or periods of active monitoring for individuals potentially exposed to COVID-19, although longer monitoring periods are regarded as the most effective measure to limit spread of SARS-CoV-2 infection.62
The majority of immunocompetent adult COVID-19 patients experience an uncomplicated condition with mild symptoms including dry cough, fever, dyspnea, and fatigue. Some patients have also reported the loss of smell and taste. The spectrum of symptoms also extends to sputum production, headache, hemoptysis, acute cardiac injury, diarrhea, dyspnea, thrombocytopenia, and lymphopenia.6,60 In more complicated cases (14–29%), developments can include acute respiratory distress syndrome (ARDS), RNAaemia, bacterial super-infections, acute kidney and cardiac injury, and death.6 Around 14% of complicated cases require hospitalization and oxygen support, and around 5% require admission to an intensive care unit.63 Older patients, men, and those with high body-mass index or comorbidities such as pulmonary, cardiac, or renal disease, diabetes, and hypertension have a greater risk of mortality, suggesting that there are specific susceptible populations for COVID-19.60,64
Pregnant women show similar clinical manifestations of COVID-19 to non-pregnant women, with COVID-19.65 Pediatric COVID-19 is mainly due to family transmission, presents with mild symptoms, and is associated with a more favorable prognosis than the adult condition.66 Interestingly, children and adolescents represent a low proportion of SARS-CoV-2 infections and do not exhibit higher viral loads than adults, and thus may not contribute significantly to virus circulation.67 The mean case-fatality rate worldwide is estimated to be about 6.7%.5 Due to the lack of a real denominator, and considering the probability of large numbers of asymptomatic carriers, mortality rates are probably overestimates. Overall, COVID-19 seems to have a much lower fatality rate than either SARS (9.6%) or MERS (37%).68,69
Pathogenesis and immune responses
The mechanisms underlying the pathogenesis of COVID-19 remain poorly understood. Common laboratory findings on admission to hospital include lymphopenia and pneumonia with characteristic and bilateral pulmonary ground-glass opacity changes on chest x-ray or computed tomography scan.4,6 Histological examination of biopsy samples from lung, liver, and heart tissue from fatal case has shown bilateral diffuse alveolar damage with cellular fibromyxoid exudates and interstitial mononuclear inflammatory infiltrates dominated by lymphocytes in the bilateral lungs.7 These pathological features greatly resemble those observed in SARS and MERS. Interestingly, clinical and histopathological findings for the upper and lower respiratory tract in animal models of SARS-CoV-2 infection resemble those in humans.70–72 Histopathologically, this model showed exudative inflammation in the initial phase progressing to diffuse alveolar damage with hemorrhage and necrosis, and a proliferative phase was reached after 1 week.70 Thus, COVID-19 onset may result in progressive respiratory failure due to alveolar damage, and death. ACE2, to which SARS-CoV-2 attaches, has a broad expression pattern with high expression in the type II alveolar cells in the lungs, as well as in the gastrointestinal system, heart, brain and kidney.73 There is already some evidence that COVID-19 can cause damage to tissues and organs other than the lung.74
Laboratory diagnosis
The genome of SARS-CoV-2 was sequenced very early during the outbreak and the full genome sequence data have been published, allowing the development of a reliable technique to diagnose COVID-19.4 WHO published the protocols for a real-time reverse transcription-polymerase chain reaction (RT-PCR) COVID-19 test targeting the SARS-CoV-2 N, E, S and RdRP viral genes. It is recommended that all patients meeting the suspected case definition be tested for COVID-19 virus regardless of whether another respiratory pathogen is suspected.1,75,76 Sample preparation and RNA extraction should be carried out at Biosafety Level 2, using a Class-2 Biosafety Cabinet and wearing PPE equipment. Collecting specimens from the upper respiratory tract (nasal, pharyngeal, and nasopharyngeal swabs or washings in ambulatory patients) and/or lower respiratory tract (sputum, endotracheal aspirate, or bronchoalveolar lavage in patients with more severe respiratory disease) is important when COVID-19 is suspected.76 Such specimens should be promptly stored and shipped at 2–8°C within 1 to 5 days for laboratory testing.76 COVID-19 virus has been detected in rectal swabs, blood, and saliva, but not in urine specimens.77 In addition to molecular testing, sequencing of strains isolated from clinical samples can be useful to monitor viral genome mutations and sequencing results can also be used to make adaptations to molecular testing.76 Additionally, several serological tests are being developed but they will require validation before they can be brought into general use to complement the RT-PCR assay.
Therapeutics and immunotherapeutics
To date, no therapeutics or vaccines against any HoCV have been approved. The current approach to COVID-19 management focuses on supportive care, bed rest, repurposing of many drugs including antiviral and antibiotic drugs, immunomodulating therapy, organ function support, respiratory support, and extracorporeal membrane oxygenation.78,79 Randomized clinical trials have been started in China, US, Japan, and Europe (such as the Solidarity and Discovery studies) with many drugs and results are expected by during 2020 (Clinicaltrials.gov).
Researchers are gaining some understanding of the adaptive immune response. Seroconversion times for total antibody (Ab), and IgM and IgG Abs have reported durations of 11, 12 and 14, days, respectively.80 Furthermore, high Ab levels have been proposed as a risk factor for critical illness, independently from older age, male gender, and relevant comorbidities.81 COVID-19 patients show decreases in helper, suppressor, and regulatory T cell counts and these decreases may be implicated in the pathological process of SARS-CoV-2 infection.7,82 Patients with severe COVID-19 show a higher neutrophil count and a lower lymphocyte count than those with a mild disease.82 Of further interest, COVID-19 patients have higher serum levels of IL-1B, IL-1RA, IL-7, IL-8, IL-9, IL-10, basic FGF, GCSF, GMCSF, IFN-γ, IP10, MCP1, MIP1A, MIP1B, PDGF, TNFα, and VEGF than healthy adults. Markers of systemic inflammation show higher levels in patients admitted to an intensive care unit (ICU) than those not admitted, which may reflect the heightened severity of patient condition.6 This evidence demonstrates that patients may experience a massive cytokine storm associated with the development of typical lung damage and the consequent ARDS when COVID-19 is at its most aggressive. Moreover, compared with SARS-CoV, which is thought to induce inadequate interferon responses, SARS-CoV-2 robustly triggers expression of numerous IFN-stimulated genes.83
Historically, convalescent plasma and immunoglobulin (IG) have been used as the fastest therapeutic option in outbreaks of emergent or reemergent infections.84
The first strategy involves an immunotherapy approach in the treatment of inflammatory disorders in which convalescent plasma (CP) is transferred to infected COVID-19 patients, and this approach may be effective in neutralizing the virus and preventing further infection.85 Preliminary data on treatment with CP show infected patients tolerate plasma from86 COVID-19-recovered patients, and suggest that CP transfer potentially improves clinical outcomes for critically ill patients.87–90 However, the optimal dose of CP, treatment time points, and the clinical benefits of this therapy require evaluation in larger, well-controlled clinical trials.
Given the variability of the virus, CP only has utility in the same geographic area and around the time it is prepared. Monoclonal antibodies (mAbs) thus represent a potential alternative for controlling the SARS-CoV-2 infection. Previous reports showed that passive transfer of mAbs protected animals challenged with various SARS-CoV isolates and SARSr-CoV.86 Another recently discovered and characterized mAb, S309, which was isolated from memory B cells of a SARS-CoV survivor and cross-neutralizes SARS-CoV and SARS-CoV-2, could potentially be used prophylactically for individuals at high risk of exposure or as a post-exposure therapy.91
Tocilizumab (Actemra), a monoclonal antibody that targets IL-6 receptors, may block detrimental effects of IL-6 on COVID-19. In a preliminary study in 21 patients with COVID-19 receiving tocilizumab on a compassionate basis, patient temperatures rapidly returned to normal in each case with marked improvement also shown for respiratory function and other symptoms. In that study, 20/21 (95%) of patients recovered and were discharged within 2 weeks from the start of tocilizumab therapy, without showing any adverse drug reactions.92
Among immunotherapy approaches to block virus attachment or entry, immunoglobulins (IVIG) are suggested to represent potential alternative effective treatments for COVID-19.93 Recent In vitro data using (IVIG) preparations (Gamunex-C and Flebogamma DIF) showed cross-reactivity against S1 protein of SARS-CoV-2.94 The utility of IVIG preparations for COVID-19 management requires further investigation. Taken together, these findings highlight the importance of treatment regimen for each phase, from measures against virus entry and replication inhibition in early infection to anti-inflammatory, immunotherapeutic, and anti-thrombotic treatments at most advanced stage of the disease.45 Thus, combining the use of immunotherapy approaches and antiviral drugs may be more effective than using either modality alone in COVID-19 management.
Advances in the development of vaccines
The early release of genome sequence of SARS-CoV-2 has triggered intense global research and development (R&D) activity to develop a vaccine against the COVID-19. Prior studies of SARS-CoV and MERS-CoV have enabled the extraordinarily rapid development of candidate vaccines that hopefully can curtail the incendiary spread of SARS-CoV-2.95 Advances in technology, vaccine platforms, clinical trial designs, and bioinformatics are assisting SARS-CoV-2 vaccine development. However, all of these platforms have advantages and disadvantages and it is not possible to predict which strategy will be faster or more successful.96 As of 8 April 2020, the global COVID-19 vaccine R&D landscape includes 115 vaccine candidates enabled by insights from the fields of SARS-CoV and MERS-CoV vaccine development and well-characterized vaccine antigen expression platforms, including those shown to be safe in humans.95,97 Multiple types of vaccine candidates are in development including inactivated whole virus, live attenuated virus, non-replicating viral vector, replicating viral vector, recombinant protein, peptide-based, virus-like particle, DNA, and RNA vaccines and have moved into phase 1 trials to evaluate their safety and immunogenicity in humans.95,97
The most advanced candidates have moved into phase 1 trials. These include a lipid nanoparticle-encapsulated modified mRNA vaccine encoding a prefusion-stabilized S protein (mRNA-1273) being developed by Moderna Therapeutics (NCT04283461), an adenovirus (Ad) type 5 vector that expresses S protein (Ad5-nCoV) being developed by CanSino Biologicals (NCT04313127), a DNA plasmid-encoding S protein delivered by electroporation (INO-4800) being developed by Inovio Pharmaceuticals (NCT04336410), and DCs modified with lentiviral vector expressing synthetic minigene based on domains of selected viral proteins, administered with antigen-specific CTLs (LV-SMENP-DC) (NCT04276896) and artificial antigen-presenting cell (aAPC) modified with lentiviral vector expressing synthetic minigene based on domains of selected viral proteins (Pathogen-specific aAPC) (NCT04299724) being developed by Shenzhen Geno-Immune Medical Institute.95,97 In addition, a simian Ad vector expressing the SARS-CoV- 2 S protein is being evaluated in a phase 1/2 clinical trial to evaluate its safety, immunogenicity, and efficacy at the University of Oxford (NCT04324606).95 A phase 3 clinical trial of an immunostimulatory vaccine approach is underway at Murdoch Childrens Research Institute University Medical Center, Netherlands (NCT04327206); in that study, Bacille Calmette-Guerin is being investigated for possible protective use against COVID-19 pathology.95 Moreover, many vaccine developers have plans to develop adjuvanted vaccines for use against COVID-19, including GlaxoSmithKine, Seqirus and Dynavax.97 A major limiting factor in the quest for identifying ideal vaccines is time.98 Given the imperative for speedy clinical trials, such vaccines could probably be available under emergency use by 2021 (Clinicaltrials.gov).
An additional mechanism facilitating viral cell entry and subsequent damage may involve antibody-dependent enhancement (ADE). Some antibodies have been shown to target one serotype of viruses but only to sub-neutralize another, leading to ADE of subsequent invading viruses.99–101 ADE has been proposed as a factor accounting for the severity of COVID-19, based on cases observed in Hubei province in China and among other places in the world, due to prior exposure to other coronaviruses.102 ADE will need to be carefully investigated for any role it plays in SARS-CoV-2, since any such role represents a potential barrier to vaccine development and antibody-based drug therapy.102
However, the goal should be to make vaccines available to the global population, which presents further challenges. In addition, there remain substantive challenges in vaccine development during a pandemic due, in part, to a need to understand the immune response and the lack of high-throughput small animal disease models.95
Conclusion
The COVID-19 pandemic is having a serious, adverse impact on public health, human society, and the economy across the globe. This review provides an overview of published information on the disease globally and our current understanding of COVID-19. SARS-CoV-2 is an emerging infection known to be a highly pathogenic human virus and probably a zoonotic agent, but otherwise, it still leaves the medical community with many unanswered questions. There are many areas where a better understanding of the disease is urgent; these cover virological, epidemiological (source of the virus and rapid global spread), and immunological (why some infected individuals showed only mild or no symptoms) issues, as well as the need to identify biomarkers to predict outcomes. Much remains unknown about SARS-CoV-2, and this creates some risk for vaccine development under the current pandemic conditions.
At the present time, the world lacks effective drugs and vaccines. There has been a significant progress in the research area from complete genome sequencing of SARS-CoV-2 to the beginning of clinical trials with COVID-19 vaccines. Until they are developed, appropriate strategies to contain the pandemic include focusing on public health measures through rapid diagnosis, contact tracing, successful containment, optimizing care for infected patients, and awareness-raising in the community to halt the spread of COVID-19.
Acknowledgments
This work was supported by grants from the Tokyo Metropolitan Government and Japan Society for the Promotion of Science. We are particularly grateful to Mr. Henry Smith for pertinent English revision of the manuscript.
Funding Statement
This work was supported by the Tokyo Metropolitan Government and the Japan Society for the Promotion of Science.
Disclosure of potential conflicts of interest
We declare no competing interests.
References
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Zhu Y, Li B, Huang CL, Chen HD, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–73. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei -Y-Y, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–69. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of V. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020; 5(4):536-544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO . Coronavirus disease (COVID-19) outbreak. Situation reports. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200518-covid-19-sitrep-119.pdf?sfvrsn=4bd9de25_4 [accessed on 19 May 2020].
- 6.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020;9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Sevestre J, et al. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med Infect Dis. 2020;34:101663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, Zhu H, Zhao W, Han Y, Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–64. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen K-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–36. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–49. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci U S A. 1967;57:933–40. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyrrell DA, Bynoe ML, Hoorn B. Cultivation of “difficult” viruses from patients with common colds. Br Med J. 1968;1:606–10. doi: 10.1136/bmj.1.5592.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lim YX, Ng YL, Tam JP, Liu DX. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4(3):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53(3):425–35. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–70. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–79. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–54. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PME, Kaandorp J, Spaargaren J, Berkhout B, et al. Identification of a new human coronavirus. Nat Med. 2004;10(4):368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milewska A, Nowak P, Owczarek K, Szczepanski A, Zarebski M, Hoang A, Berniak K, Wojarski J, Zeglen S, Baster Z, et al. Entry of Human coronavirus NL63 into the cell. J Virol. 2018;92(3):e01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oosterhof L, Christensen CB, Sengelov H. Fatal lower respiratory tract disease with human corona virus NL63 in an adult haematopoietic cell transplant recipient. Bone Marrow Transplant. 2010;45:1115–16. doi: 10.1038/bmt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konca C, Korukluoglu G, Tekin M, Almis H, Bucak İH, Uygun H, Altas AB, Bayrakdar F. The first infant death associated with human coronavirus NL63 infection. Pediatr Infect Dis J. 2017;36:231–33. doi: 10.1097/INF.0000000000001390. [DOI] [PubMed] [Google Scholar]
- 27.Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S. Surveillance of Bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol. 2017;91(5):e01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A. 2005;102:7988–93. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–95. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, Chu CM, Lee RA, Luk WK, Wong GK, Wong BH, et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 2005;192:1898–907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 32.Middle WHO . East respiratory syndrome coronavirus (MERS-CoV) factsheet. 2019. [Available from: https://www.who.int/emergencies/mers-cov/en/ accessed 26th March 2020]. 2019.
- 33.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–54. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freund Y. The challenge of emergency medicine facing the COVID-19 outbreak. Eur J Emerg Med. 2020;27(3):155. doi: 10.1097/MEJ.0000000000000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai J, Ma S, Wang Y, Cai Z, Hu J, Wei N, Wu J, Du H, Chen T, Li R, et al. Factors associated with mental health outcomes among health care workers exposed to coronavirus disease 2019. JAMA Netw Open. 2020;3:e203976. doi: 10.1001/jamanetworkopen.2020.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zastrow M. South Korea is reporting intimate details of COVID-19 cases: has it helped? Nature. 2020.doi:10.1038/d41586-020-00740-y. [DOI] [PubMed] [Google Scholar]
- 38.Pan X, Ojcius DM, Gao T, Li Z, Pan C, Pan C. Lessons learned from the 2019-nCoV epidemic on prevention of future infectious diseases. Microbes Infect. 2020;22:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. Novel Coronavirus A. Emerging in China – key questions for impact assessment. N Engl J Med. 2020;382:692–94. [DOI] [PubMed] [Google Scholar]
- 41.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, et al. Genome composition and divergence of the Novel Coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT. Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281-292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bourgonje AR, Abdulle AE, Timens W, Hillebrands JL, Navis GJ, Gordijn SJ, Bolling MC, Dijkstra G, Voors AA, Osterhaus AD, et al. Angiotensin-converting enzyme-2 (ACE2), SARS-CoV-2 and pathophysiology of coronavirus disease 2019 (COVID-19). J Pathol. 2020;10.1002/path.5471. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kupferschmidt K, Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–13. [DOI] [PubMed] [Google Scholar]
- 47.Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, et al. Global patterns in coronavirus diversity. Virus Evol. 2017;3:vex 012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020. doi: 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- 49.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, et al. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. BioRxiv. 2020. 2020. [DOI] [PubMed]
- 50.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020; 26(4):450-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuen KS, Ye ZW, Fung SY, Chan CP, Jin DY. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qian G, Yang N, Ma AHY, Wang L, Li G, Chen X, Chen X. A COVID-19 transmission within a family cluster by presymptomatic infectors in China. Clin Infect Dis. 2020;ciaa316. doi: 10.1093/cid/ciaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Day M. Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ. 2020;368:m1165. [DOI] [PubMed] [Google Scholar]
- 56.Wilder-Smith A, Freedman DO. Isolation, quarantine, social distancing and community containment: pivotal role for old-style public health measures in the novel coronavirus (2019-nCoV) outbreak. J Travel Med. 2020;27(2):taaa020. doi: 10.1093/jtm/taaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, Diao M, Yu W, Pei L, Lin Z, Chen D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: a data-driven analysis. Int J Infect Dis. 2020;93:201–04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bauch CT, Lloyd-Smith JO, Coffee MP, Galvani AP. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology. 2005;16:791–801. [DOI] [PubMed] [Google Scholar]
- 59.Choi S, Jung E, Choi BY, Hur YJ, Ki M. High reproduction number of Middle East respiratory syndrome coronavirus in nosocomial outbreaks: mathematical modelling in Saudi Arabia and South Korea. J Hosp Infect. 2018;99:162–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, et al. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. The incubation period of Coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baum SG. COVID-19 incubation period: an update. Nejm Journal Watch. 2020. https://www.jwatch.org/na51083/2020/03/13/covid-19-incubation-period-update
- 63.TeamNCPERE . Vital surveillances: the epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) – China. China CDC Weekly. 2020;2:113–22. [PMC free article] [PubMed] [Google Scholar]
- 64.Patel AB, Verma A. COVID-19 and Angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: what is the evidence? JAMA. 2020. doi: 10.1001/jama.2020.4812. [DOI] [PubMed] [Google Scholar]
- 65.Yu N, Li W, Kang Q, Xiong Z, Wang S, Lin X, Liu Y, Xiao J, Liu H, Deng D, et al. Clinical features and obstetric and neonatal outcomes of pregnant patients with COVID-19 in Wuhan, China: a retrospective, single-centre, descriptive study. Lancet Infect Dis. 2020;20(5):559-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su L, Ma X, Yu H, Zhang Z, Bian P, Han Y, Sun J, Liu Y, Yang C, Geng J, et al. The different clinical characteristics of corona virus disease cases between children and their families in China – the character of children with COVID-19. Emerg Microbes Infect. 2020;9:707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colson P, Tissot-Dupont H, Morand A, Boschi C, Ninove L, Esteves-Viera V, Gautre P, Brouqui P, Parola P, et al. Children account for a small proportion of diagnoses of SARS-CoV-2 infection and do not exhibit greater viral loads than adults. Pré-Prints IHU [COVID-IHU #3]. https://wwwmediterranee-infectioncom/pre-prints-ihu/ 2020. [DOI] [PMC free article] [PubMed]
- 68.Hui DS, Azhar EI, Kim YJ, Memish ZA, Oh MD, Zumla A. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis. 2018;18:e217–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Y, Gao GF. Towards our understanding of SARS-CoV, an emerging and devastating but quickly conquered virus. Comp Immunol Microbiol Infect Dis. 2007;30:309–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, et al. Simulation of the clinical and pathological manifestations of Coronavirus disease 2019 (COVID-19) in golden Syrian Hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020:ciaa325. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020. doi: 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Perez-Perez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020. doi: 10.1038/s41586-020-2324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santos RAS, Oudit GY, Verano-Braga T, Canta G, Steckelings UM, Bader M. The renin-angiotensin system: going beyond the classical paradigms. Am J Physiol Heart Circ Physiol. 2019;316:H958–H70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steardo L, Steardo L Jr., Zorec R, Verkhratsky A. Neuroinfection may potentially contribute to pathophysiology and clinical manifestations of COVID-19. Acta Physiol (Oxf). 2020;229(3):e13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;2525(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.WHO . Laboratory testing for coronavirus disease (COVID-19) in suspected human cases. Available online: https://www.who.int/health-topics/coronavirus/laboratory-diagnostics-for-novel-coronavirus [accessed on 1 April 2020]. 2020.
- 77.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nicola M, O’Neill N, Sohrabi C, Khan M, Agha M, Agha R. Evidence based management guideline for the COVID-19 pandemic – Review article. Int J Surg. 2020;77:206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salvi R, Patankar P. Emerging pharmacotherapies for COVID-19. Biomed Pharmacother. 2020;128:110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020.ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Li H, Li J, Yang C, Guo X, Hu Z, Chen Z, Wang S, Liu J. Elevated serum IgM levels indicate poor outcome in patients with coronavirus disease 2019 pneumonia: a retrospective case-control study. medRxiv preprint. 2020.
- 82.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020. ciaa248. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020; 27(6):883-90.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bozzo J, Jorquera JI. Use of human immunoglobulins as an anti-infective treatment: the experience so far and their possible re-emerging role. Expert Rev Anti Infect Ther. 2017;15:585–604. [DOI] [PubMed] [Google Scholar]
- 85.Casadevall A, Pirofski LA. The convalescent sera option for containing COVID-19. J Clin Invest. 2020;130:1545–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahn JY, Sohn Y, Lee SH, Cho Y, Hyun JH, Baek YJ, Jeong SJ, Kim JH, Ku NS, Yeom JS, et al. Use of convalescent plasma therapy in two COVID-19 patients with acute respiratory distress syndrome in Korea. J Korean Med Sci. 2020;35:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, Wang F, Li D, Yang M, Xing L, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020; 323(16):1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang B, Liu S, Tan T, Huang W, Dong Y, Chen L, Chen Q, Zhang L, Zhong Q, Zhang X, et al. Treatment with convalescent plasma for critically ill patients with SARS-CoV-2 infection. Chest. 2020;S0012-3692(20)30571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020; 117(17):9490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 92.Fu B, Xu X, Wei H. Why tocilizumab could be an effective treatment for severe COVID-19? J Transl Med. 2020;18:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jawhara S. Could intravenous immunoglobulin collected from recovered coronavirus patients protect against COVID-19 and strengthen the immune system of new patients? Int J Mol Sci. 2020; 21(7):2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Diez JM, Romero C, Gajardo R. Currently available intravenous immunoglobulin contains antibodies reacting against severe acute respiratory syndrome coronavirus 2 antigens. Immunotherapy. 2020;12(8):571-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diamond MS, Pierson TC. The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe. 2020;27:699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, Mayhew S. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020; 19(5):305-6. [DOI] [PubMed] [Google Scholar]
- 98.Dhama K, Sharun K, Tiwari R, Dadar M, Malik YS, Singh KP, Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020;16(6):1232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020;94(5):e02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, Harris E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358:929–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, Chen KH, Liu FT, Liu WT, Chen YM, et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451:208–14. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tetro JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


