Abstract
Topoisomerase IB catalyzes recombinogenic DNA strand transfer reactions in vitro and in vivo. Here we characterize a new pathway of topoisomerase-mediated DNA ligation in vitro (flap ligation) in which vaccinia virus topoisomerase bound to a blunt-end DNA joins the covalently held strand to a 5′ resected end of a duplex DNA containing a 3′ tail. The joining reaction occurs with high efficiency when the sequence of the 3′ tail is complementary to that of the scissile strand immediately 5′ of the cleavage site. A 6-nucleotide segment of complementarity suffices for efficient flap ligation. Invasion of the flap into the duplex apparently occurs while topoisomerase remains bound to DNA, thereby implying a conformational flexibility of the topoisomerase clamp around the DNA target site. The 3′ flap acceptor DNA mimics a processed end in the double-strand-break-repair recombination pathway. Our findings suggest that topoisomerase-induced breaks may be rectified by flap ligation, with ensuing genomic deletions or translocations.
The eukaryotic type IB topoisomerase family includes nuclear topoisomerase I and the topoisomerases of vaccinia and other cytoplasmic poxviruses (37). The type IB enzymes relax DNA supercoils by transiently breaking and rejoining one strand of the DNA duplex through a covalent DNA–(3′-phosphotyrosyl) enzyme intermediate. The catalytic domains of type IB topoisomerases and the λ Int family of site-specific recombinases (now called tyrosine recombinases) have a common reaction mechanism and tertiary structure, prompting the inference that they derive from a common ancestral DNA strand transferase (5, 8, 10, 14, 24, 44). Type IB topoisomerases and tyrosine recombinases consist of two domains that form a C-shaped clamp around duplex DNA. A noncatalytic N-terminal domain interacts with DNA in the major groove, whereas the catalytic C-terminal domain recognizes the minor-groove face of the cleavage site. The domains are linked by a flexible hinge that must open to allow noncovalent binding of DNA in the interdomain cleft and then close to establish the clamp.
The mechanistic and evolutionary connections between topoisomerase IB and site-specific recombinases lend currency to the long-standing hypothesis that eukaryotic topoisomerases can act as catalysts of genetic recombination. Champoux et al. envisioned a recombinogenic pathway whereby topoisomerase I would form a covalent adduct with one strand of the DNA duplex and then religate the bound DNA to a heterologous acceptor strand (1, 2, 4). Purified type IB topoisomerases catalyze such strand transfer reactions in vitro (7, 9, 34, 35). Vaccinia virus topoisomerase catalyzes excisional recombination when expressed in a heterologous system in vivo (31, 32). Similarly, overexpression of yeast Top1 in yeast elicits an increase in the frequency of illegitimate integrative recombination (53).
The cleavage-religation equilibrium of type IB topoisomerase on duplex DNA favors the noncovalently bound state. However, the equilibrium can be strongly skewed toward covalent adduct formation in vitro by modifications in the DNA target site and by drugs such as camptothecin that selectively impede religation (3, 11, 20). DNA structural alterations that induce high levels of covalent adduct formation by topoisomerase IB include naturally occurring lesions, such as nicks, gaps, base pair mismatches, abasic sites, UV-induced thymine dimers, deoxyuridine incorporation, and ribonucleotide incorporation immediately 3′ of the scissile phosphate and nicks on the complementary strand opposite to or immediately flanking the scissile phosphodiester (6, 15, 22, 23, 27). Thus, topoisomerase will catalyze “suicide” cleavage reactions in vivo in response to natural and pharmacological poisons. Covalent trapping of topoisomerase IB on DNA by camptothecin is cytotoxic. Yet treatment of human fibroblasts with subtoxic doses of camptothecin leads to a 300-fold increase in chromosomal integration of viral DNA vectors (26), suggesting that stabilization of the covalent topoisomerase-DNA intermediate is prorecombinogenic in vivo. Similar inferences are drawn from the increase in chromosomal rearrangements in yeast cells bearing a missense Top1 mutant that displays a higher level of equilibrium cleavage than wild-type Top1 (16).
In principle, there are multiple pathways to remove topoisomerase I when it binds covalently to DNA and the original 5′ OH leaving strand is no longer available for religation. These pathways can be classified as extrinsic (i.e., catalyzed by enzymes other than topoisomerase) or intrinsic (i.e., catalyzed by topoisomerase per se). An example of an extrinsic repair factor is the 3′ tyrosyl phosphodiesterase discovered by Nash and colleagues; this enzyme, the product of a single gene, can hydrolytically resolve DNA–3′-pTyr to yield DNA–3′-phosphate and Tyr (21, 51). Topoisomerase itself is capable of transferring the covalently held DNA strand to a nucleic acid acceptor other than the strand originally cleaved. The acceptor can be 5′ OH DNA, in which case the repair process is recombinogenic (7, 34, 35). The acceptor can also be 5′ OH RNA (27).
We have exploited vaccinia virus topoisomerase as a model system for the analysis of topoisomerase IB-catalyzed DNA rearrangements in vitro. The poxvirus topoisomerase is distinguished from cellular topoisomerase I by its compact size (314 amino acids), its resistance to camptothecin (39), and its site specificity in DNA transesterification. Vaccinia virus topoisomerase binds and cleaves duplex DNA at a pentapyrimidine target sequence, 5′ (T/C)CCTT↓ (38). Cleavage and religation of DNAs containing a single CCCTT target site have been examined in depth under single-turnover and equilibrium conditions (40, 41, 43, 49). Indeed, vaccinia virus topoisomerase is the only member of the type IB topoisomerase family for which a detailed kinetic scheme is available.
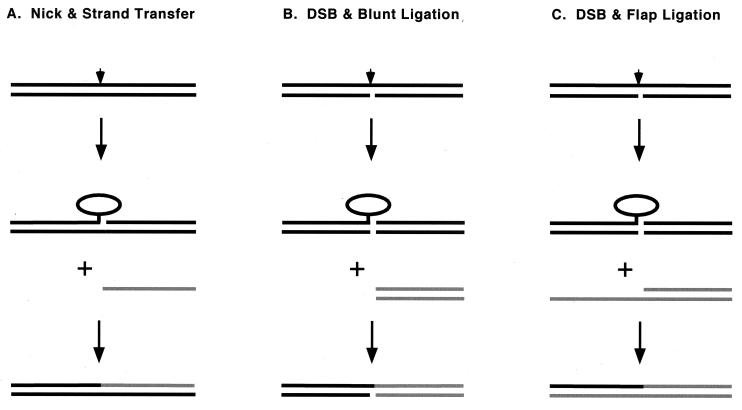
We previously delineated two pathways of intermolecular DNA joining reactions catalyzed by vaccinia virus topoisomerase: (i) sticky-end ligation after topoisomerase-induced nicking and single-strand uptake (Fig. 1A) and (ii) blunt-end ligation (Fig. 1B). These pathways differ in their requirements for DNA sequence homology. When the CCCTT-containing DNA substrate is configured such that the scissile bond is situated within 6 bp of the 3′ end of the scissile strand, cleavage is accompanied by spontaneous dissociation of the downstream portion of the incised strand (Fig. 1A). The resulting topoisomerase-DNA complex, containing a 5′ single-strand tail on the noncleaved strand, can religate to an acceptor DNA if the acceptor molecule has a 5′ OH tail complementary to that of the activated donor complex (34, 35). The topoisomerase discriminates very well between correctly and incorrectly base paired 5′ overhangs (36). Sticky-end ligation is a plausible mechanism for intrinsic repair of topoisomerase-DNA complexes covalently trapped after collision with an approaching replication fork leaves a single-strand gap 3′ of the cleavage site. A role for this homology-dependent strand transfer pathway in topoisomerase I-catalyzed recombination is suggested by the finding that a majority of in vivo recombination events catalyzed by vaccinia virus topoisomerase involve parental half-sites with several bases of sequence identity 3′ of the pentamer cleavage motif (32).
FIG. 1.
Three pathways for intermolecular DNA strand transfer by topoisomerase IB. See the text for a discussion. Topoisomerase I is depicted as an ellipse. The site of transesterification of topoisomerase to one strand of the DNA duplex is denoted by an arrowhead. The incoming DNA acceptor molecules are colored gray. The top strands of the DNAs are oriented 5′ (left) to 3′ (right).
When the CCCTT-containing substrate is configured so that the nonscissile strand has a nick opposite the scissile phosphate, topoisomerase cleavage results in dissociation of the entire downstream duplex and the formation of a covalently activated blunt-end donor complex (Fig. 1B). Topoisomerase bound covalently at a blunt CCCTT duplex end can religate to any 5′ OH blunt-end duplex acceptor regardless of its sequence (35). A DNA with a 5′ single-strand tail is not an effective acceptor for the blunt-end CCCTT donor.
Here we describe a new pathway of intermolecular strand transfer—flap ligation—that is catalyzed by vaccinia virus topoisomerase in vitro. We show that topoisomerase bound covalently to a blunt-end DNA is capable of joining the CCCTT strand to an acceptor DNA with a 3′ tail (Fig. 1C). The joining reaction occurs with high efficiency when the sequence of the 3′ tail is homologous to the topoisomerase recognition motif upstream of the scissile phosphodiester. Because the acceptor DNA mimics the 5′ resected intermediates in the double-strand-break repair (DSBR) pathway (45), we suggest that flap ligation is a plausible mechanism for rectification of topoisomerase-induced DSBs and for generating chromosomal deletions and translocations in response to topoisomerase poisons.
MATERIALS AND METHODS
Enzyme purification.
Vaccinia virus topoisomerase was expressed in Escherichia coli BL21 cells infected with bacteriophage λCE6 and then purified from a soluble bacterial lysate by phosphocellulose column chromatography (39). The protein concentration of the phosphocellulose preparation was determined by using the dye-binding method (Bio-Rad) with bovine serum albumin as the standard.
Preparation of topoisomerase cleavage substrates and strand transfer acceptors.
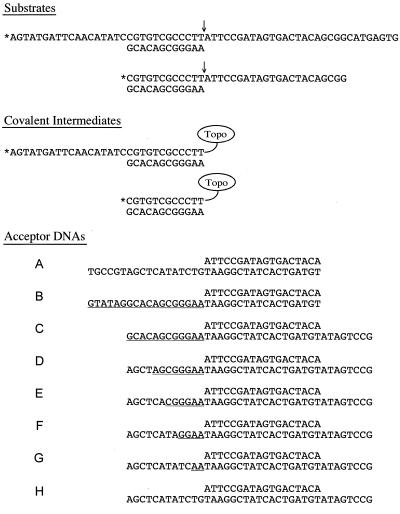
CCCTT-containing DNA oligonucleotides (60-mer and 34-mer) (Fig. 2) were 5′ end labeled by enzymatic phosphorylation in the presence of [γ-32P]ATP and T4 polynucleotide kinase and then purified by preparative electrophoresis through a 20% polyacrylamide gel containing 90 mM Tris-borate and 2.5 mM EDTA. The labeled oligonucleotides were eluted from an excised gel slice and then annealed to an unlabeled 12-mer complementary oligonucleotide(s) at a fourfold-molar excess. The unlabeled strand transfer acceptor DNAs (Fig. 2) were prepared by annealing equimolar amounts of the complementary component oligonucleotides. The annealing reaction mixtures containing 0.2 M NaCl and oligonucleotides were heated to 70°C and then slowly cooled to 22°C. The hybridized DNAs were stored at 4°C.
FIG. 2.
Substrates, covalent intermediates, and acceptor DNAs. The 60-mer–12-mer and 34-mer–12-mer substrates are shown with the cleavage site indicated by an arrow, and the 5′ 32P label is denoted by an asterisk. The covalent intermediates generated by transesterification of Tyr-274 of the topoisomerase to the scissile CCCTTpN phosphodiester are shown. The noncovalently held single strand dissociates spontaneously, thereby trapping topoisomerase at the DSB. A series of acceptor DNAs for topoisomerase-mediated strand transfer are shown. The segments of sequence homology between the 3′ tails of the acceptor DNAs and the covalent intermediates are underlined.
Topoisomerase-catalyzed strand transfer reactions.
A reaction mixture containing 50 mM Tris-HCl (pH 7.5), 25 nM 5′ labeled topoisomerase substrate (Fig. 2), and 125 nM topoisomerase was incubated at 37°C for 15 min to form the topoisomerase-activated covalent donor complex. An aliquot (20 μl) was then withdrawn and quenched by adding sodium dodecyl sulfate to 0.5% (time zero). Strand transfer was initiated by adding 1.25 μM acceptor DNA (Fig. 2), and incubation was continued at 37°C. Aliquots (20 μl) were withdrawn at the times specified and quenched immediately with sodium dodecyl sulfate. The denatured samples were digested with 10 μg of proteinase K for 60 min at 37°C. The samples were ethanol precipitated, resuspended in formamide-dye solution, and electrophoresed through a 15% polyacrylamide gel containing 7 M urea in Tris-borate-EDTA. Radiolabeled reaction products were visualized by autoradiography of the gel. The extents of formation of the recombinant strands (as percentages of the total radioactivity) were quantitated by scanning the gel with a FUJIX BAS2000 phosphorimager.
RESULTS
Covalently activated blunt donor complex.
The DNA substrate used to prepare the donor complex consisted of a 5′ 32P-labeled 60-mer strand with a centrally placed CCCTT site annealed to an unlabeled 12-mer strand to yield a 5′- and 3′-tailed duplex molecule with no base pairing 3′ of the scissile phosphodiester (Fig. 2). The transesterification reaction of topoisomerase with this substrate resulted in formation of the covalent intermediate depicted in Fig. 2. Note that the noncovalently held portion of the incised strand readily dissociated from the covalent intermediate, leaving the topoisomerase linked to a blunt duplex end. The covalent intermediate was detected (after proteinase K digestion of the reaction mixture) as a 32P-labeled DNA-peptide adduct migrating at ∼32 to 33 nucleotides during polyacrylamide gel electrophoresis (Fig. 3, lane 0). Eighty-five percent of the input 60-mer strand was converted to covalent adduct in reactions containing 125 nM topoisomerase and 25 nM substrate.
FIG. 3.
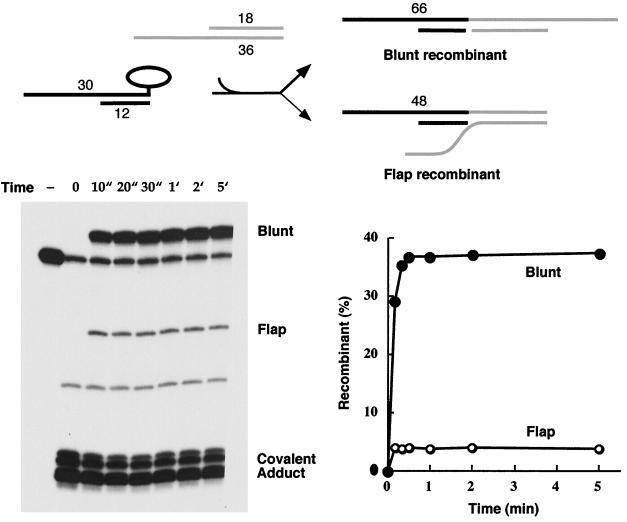
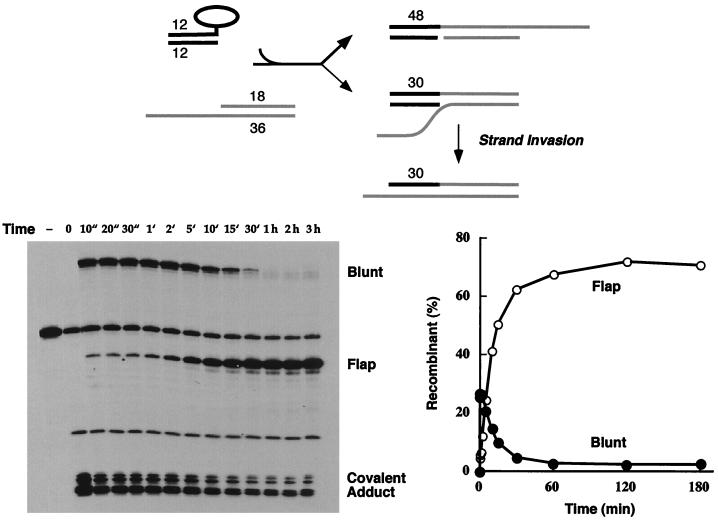
Blunt ligation is favored over heterologous flap ligation. Reaction of the 5′-tailed 3′-blunt covalent topoisomerase-DNA intermediate with acceptor A was performed as described in Materials and Methods. The reaction products were analyzed by polyacrylamide gel electrophoresis. An autoradiogram of the gel is shown in the left panel. The strand transfer reaction times are specified above the lanes. ", seconds; ′, minutes. A control reaction mixture containing the 5′ 32P-labeled 60-mer substrate and no topoisomerase was analyzed in lane −. The positions of the radiolabeled 66-mer blunt recombinant, 48-mer flap recombinant, and covalent DNA-peptide adducts are indicated. The extents of recombinant formation (as percentages of total labeled DNA) are plotted as a function of time in the right panel. The reaction pathway is illustrated at the top and discussed in the text.
Strand transfer to a blunt-end acceptor is favored over a nonhomologous 3′-tailed acceptor.
The blunt-end covalent donor complex was mixed with a 50-fold molar excess of an 18-mer–36-mer DNA molecule containing a blunt end on one side and an 18-nucleotide 3′ single-strand tail on the other side (Fig. 2, acceptor A). The sequence of the 3′ tail was designed so that there was no potential for base pairing to the scissile strand of the covalent donor complex. Note that the acceptor DNA contained two potential 5′ OH nucleophiles that could attack the covalent intermediate and displace the trapped enzyme: (i) the 5′ OH of the 36-mer strand at the blunt end and (ii) the recessed 5′ OH end of the 18-mer strand. Transfer of the 30-mer CCCTT strand to the blunt terminus should yield a recombinant molecule with a radiolabeled 66-mer CCCTT strand and a bipartite complementary strand with a nick opposite the scissile phosphodiester, whereas religation to the recessed 5′ OH should produce a “flap recombinant” with a radiolabeled 48-mer CCCTT strand (Fig. 3).
We observed that both the blunt and recessed 5′ OH ends were capable of attacking the covalent complex to generate the expected recombinant strands, but the yields of the two products differed significantly. The flap recombinant accumulated to ∼4% of the total 32P-labeled DNA, while the blunt recombinant comprised 38% of the labeled DNA (Fig. 3). Kinetic analysis showed that both strand transfer reactions were complete within 20 to 30 s and that the distribution of the products did not change with reaction times up to 3 h (Fig. 3 and data not shown). This experiment shows that vaccinia virus topoisomerase can catalyze ligation of a blunt end to a 3′ flap acceptor, but the enzyme prefers to rejoin to a blunt end if given the chance.
Homology of the 3′ acceptor flap to the donor complex enhances ligation.
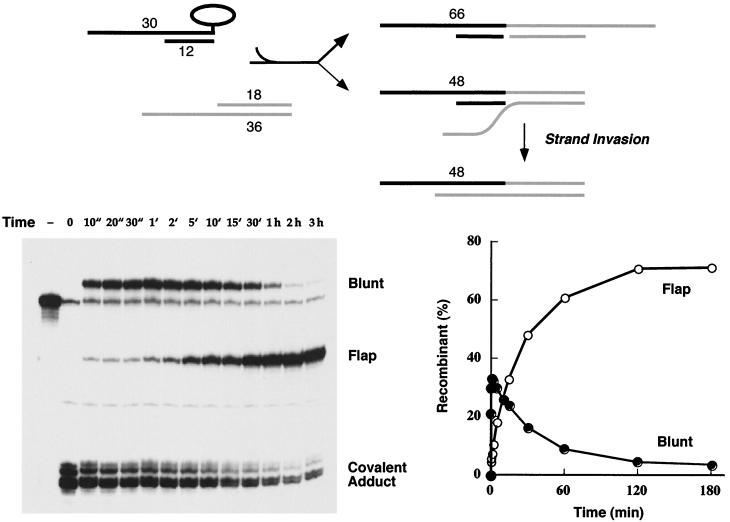
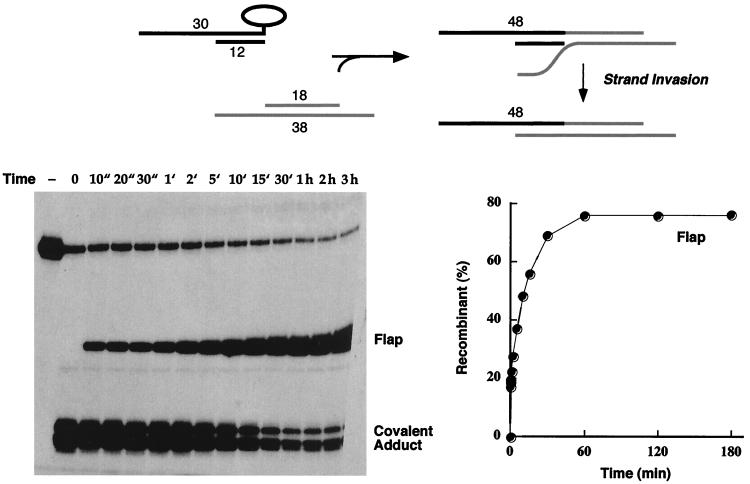
To evaluate the influence of sequence homology on the flap ligation reaction, we constructed a second 18-mer–36-mer acceptor DNA in which the sequence of the 18-nucleotide 3′ tail was complementary to that of the scissile strand of the covalent donor complex (Fig. 2, acceptor B). The blunt-end covalent intermediate reacted quickly with the homologous 3′-tailed acceptor to form a mixture of blunt and flap recombinant products (Fig. 4). As noted in the previous experiment, the blunt recombinants predominated over the flap ligation products at the earliest times (10 s to 2 min). However, strand transfer to the homologous 3′-tailed acceptor did not attain a stable equilibrium in this state but instead segued into a second reaction phase characterized by a steady accumulation of the 48-mer flap recombinant over 2 h (to an extent of 70% of the total labeled DNA) and a concomitant decay in the abundance of the blunt recombinant to less than 4% of the level of labeled DNA (Fig. 4).
FIG. 4.
Homology of the 3′ acceptor tail to the donor complex enhances flap ligation. The 5′-tailed 3′-blunt covalent topoisomerase-DNA intermediate was reacted with acceptor B, and the products were analyzed by polyacrylamide gel electrophoresis. An autoradiogram of the gel is shown in the left panel. The strand transfer reaction times are specified above the lanes. ", seconds; ′, minutes. A control reaction mixture containing the 5′ 32P-labeled 60-mer substrate and no topoisomerase was analyzed in lane −. The extents of recombinant formation (as percentages of total labeled DNA) are plotted as a function of time in the right panel. The reaction pathway is illustrated at the top and discussed in the text.
The explanation for the biphasic kinetic profile is as follows (Fig. 4). (i) The initial burst phase dominated by blunt ligation reflects a state in which topoisomerase (which is present in excess over the CCCTT target site) remains bound to the recombinant DNA products and establishes an internal cleavage-religation equilibrium that favors the covalent intermediate. (ii) The second, slow phase entails invasion of the complementary 3′ flap into the CCCTT duplex with concomitant displacement of the original nonscissile strand, thereby resulting in a new configuration of the flap recombinant in which the 30-mer CCCTT strand and the 18-mer 5′ OH nucleophile are bridged in cis by a continuous 36-mer nonscissile strand. (iii) The internal cleavage-religation equilibrium on the new flap recombinant favors the noncovalently bound state, and thus the abundance of the sealed 48-mer increases while the covalent adduct declines. (iv) The blunt recombinant is depleted by mass action as the slow strand invasion process favors the flap recombinant. The increase in the 48-mer flap recombinant strand in the interval from 2 min to 2 h was a pseudo-first order reaction. The data fit well to a single exponential with an apparent rate constant of 7 × 10−4 s−1. We surmise that the process of strand invasion was the rate-limiting step in the slow reaction phase.
3′ single-strand annealing is not necessary for efficient flap ligation.
In the experiment depicted in Fig. 4, the six nucleotides at the 3′ end of the acceptor flap were complementary to the single-stranded segment of the donor complex immediately upstream of the 12-bp duplex segment. Thus, it was conceivable that the strand invasion process was initiated by single-strand annealing 5′ of the CCCTT target site. We subsequently eliminated the potential for single-strand annealing by shortening the 5′ end of the scissile strand of the cleavage substrate to leave only a 12-bp blunt end (Fig. 2). This substrate DNA was prepared by annealing a 5′ 32P-labeled 34-mer strand to an unlabeled complementary 12-mer. Transesterification of topoisomerase to the 34-mer–12-mer substrate resulted in formation of the covalent intermediate depicted in Fig. 2. The covalent adduct was detected after proteinase K digestion as a 32P-labeled DNA-peptide adduct migrating at ∼14 to 16 nucleotides during polyacrylamide gel electrophoresis (Fig. 5, lane 0). Eighty percent of the input 34-mer strand was converted to covalent adduct.
FIG. 5.
Single-strand annealing of the acceptor 3′ tail is not necessary for efficient flap ligation. The 5′-blunt 3′-blunt covalent topoisomerase-DNA intermediate was reacted with acceptor B, and the products were analyzed by polyacrylamide gel electrophoresis. An autoradiogram of the gel is shown in the left panel. The strand transfer reaction times are specified above the lanes. ", seconds; ′, minutes. A control reaction mixture containing the 5′ 32P-labeled 34-mer substrate and no topoisomerase was analyzed in lane −. The positions of the radiolabeled 48-mer blunt recombinant, 30-mer flap recombinant, and covalent DNA-peptide adducts are indicated. The extents of recombinant formation are plotted as a function of time in the right panel. The reaction pathway is illustrated at the top and discussed in the text.
The double-blunt-end covalent intermediate reacted with acceptor B to form a mixture of 48-mer blunt and 30-mer flap recombinant products. The reaction displayed biphasic kinetics with a rapid burst of blunt ligation, followed by a slow phase of accumulation of the flap recombinant (Fig. 5). The flap recombinant comprised 70% of the labeled DNA at the reaction end point, when the blunt recombinant was reduced to less than 3%. Thus, the absence of single-strand annealing had no effect on the high efficiency of homologous flap ligation. The increase in the 30-mer flap recombinant strand in the interval from 2 min to 2 h was a pseudo-first order reaction with an apparent rate constant of 1.4 × 10−3 s−1, which is actually slightly faster than the rate of the second phase of homologous flap ligation, when single-strand annealing could occur. Base pairing of the single strands upstream of the CCCTT duplex may well impede productive strand invasion from the direction of the nick opposite the scissile phosphodiester.
Homologous flap ligation in the absence of competing reactions.
To focus exclusively on the homologous flap ligation reaction, we designed an 18-mer–38-mer acceptor DNA containing at one end a 12-nucleotide 3′ tail complementary to the scissile strand of the covalent intermediate and at the other end an 8-nucleotide 5′ single-strand tail with no potential for base pairing to the donor complex (Fig. 2, acceptor C). Because vaccinia virus topoisomerase bound covalently at a blunt end cannot transfer the covalently held CCCTT strand to a 5′ OH single-strand acceptor, the recessed 5′ OH of the 18-mer acceptor strand was the only nucleophile available for attack on the covalent intermediate. The 5′-tailed blunt-end covalent intermediate reacted with the 18-mer strand of acceptor C to form a 48-mer flap recombinant product (Fig. 6). The reaction displayed biphasic kinetics, with a rapid burst of flap recombinant formation at 10 to 30 s to the extent of 15% of the total labeled DNA, followed by a slow phase of accumulation of the flap recombinant to an endpoint of 75% after 60 min (Fig. 6). The higher amplitude of the initial burst of flap ligation reflected the absence of a competing reaction with the 5′ blunt end present in acceptors A and B used in the previous experiments. The increase in the 48-mer recombinant strand in the interval from 1 min to 1 h was a pseudo-first order reaction with an apparent rate constant of 1.7 × 10−3 s−1.
FIG. 6.
Homologous flap ligation in the absence of competing reactions. The 5′-tailed 3′-blunt covalent topoisomerase-DNA intermediate was reacted with acceptor C, and the products were analyzed by polyacrylamide gel electrophoresis. An autoradiogram of the gel is shown in the left panel. The strand transfer reaction times are specified above the lanes. ", seconds; ′, minutes. A control reaction mixture containing the 5′ 32P-labeled 60-mer substrate and no topoisomerase was analyzed in lane −. The positions of the radiolabeled 48-mer blunt flap recombinant and covalent DNA-peptide adducts are indicated. The extent of flap recombinant formation is plotted as a function of time in the right panel. The reaction pathway is illustrated at the top and discussed in the text.
Six nucleotides of homology suffice for efficient flap ligation.
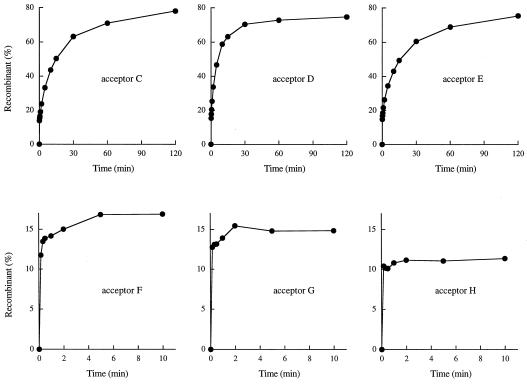
The homology length requirement for the second phase of flap ligation was investigated using a series of 18-mer–38-mer acceptors with a recessed 5′ OH nucleophile (Fig. 2, acceptors C through H). Each molecule contained a 12-nucleotide 3′ tail, the sequence of which was varied to limit the potential for base pairing with the CCCTT strand of the covalent intermediate to either 12 bp (acceptor C), 8 bp (acceptor D), 6 bp (acceptor E), 4 bp (acceptor F), or 2 bp (acceptor G) immediately 5′ of the scissile phosphodiester. A control acceptor H had no potential for base pairing with the covalent donor complex.
The rates and amplitudes of the biphasic flap recombination reaction were quite similar when the segments of homology were 12, 8, or 6 nucleotides long (Fig. 7, acceptors C to E). The yields of flap recombinant at the reaction endpoints were 75 to 80% of total labeled DNA. Thus, strand invasion through a 6-bp region including the CCCTT target site sufficed for efficient flap ligation. In contrast, the reactions of the blunt covalent intermediate with acceptors containing 12-mer tails but only 4, 2, or 0 potential bp to the scissile strand displayed a rapid initial burst of flap recombinant formation (to the extent of 11 to 17% of total DNA) but evinced no second phase of recombinant accumulation for up to 3 h (Fig. 7, acceptors F to H; data not shown). Thus, the potential for up to 4 bp of strand invasion by the 3′ tail into the CCCTT target site was not sufficient to drive the flap ligation reaction to a high yield.
FIG. 7.
Six nucleotides of homology suffice for efficient flap ligation. The 5′-tailed 3′-blunt covalent topoisomerase-DNA intermediate was reacted with acceptors C through H, and the products were analyzed by polyacrylamide gel electrophoresis. The extent of flap recombinant formation is plotted as a function of time for each acceptor as specified.
Evidence that strand invasion occurs while topoisomerase is bound to DNA.
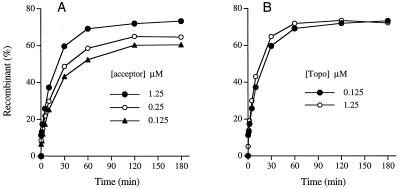
Does strand invasion occur while topoisomerase remains bound to the CCCTT target site or during a hypothetical interval that is subsequent to strand transfer and enzyme dissociation yet prior to rebinding of topoisomerase to the CCCTT motif? Given that topoisomerase is present in molar excess over the CCCTT-containing substrate, one might expect that the cleavage sites would be occupied throughout the strand transfer reaction. However, it is possible that the addition of a molar excess of acceptor DNA (to an acceptor/topoisomerase ratio of 10) might act as a nonspecific decoy for topoisomerase molecules that have catalyzed the strand transfer step. If this scenario is true, then we would expect the rate and amplitude of homology-dependent flap recombination during the second phase to be directly proportional to the amount of acceptor DNA present in the reaction. Yet we observed that reducing the concentration of acceptor C (normally 1.25 μM) by a factor of 5 or 10 had little impact on the amplitude of the second phase (Fig. 8A). The fact that the flap recombinant comprised 60% of the total DNA even at 0.125 μM acceptor C (acceptor/substrate ratio of 5 and acceptor/topoisomerase ratio of 1) highlights the inherent efficiency of flap ligation. The key point of this experiment was that the apparent rate constants (kobs) for the second phase of the reaction were hardly affected by the decrement in acceptor concentration (kobs = 1.3 × 10−3, 1.2 × 10−3, and 1 × 10−3 s−1 for reactions containing 1.25, 0.25, and 0.125 μM acceptor). Thus, we infer that a decoy function of the acceptor is not promoting the strand invasion step.
FIG. 8.
Evidence that strand invasion occurs while topoisomerase is bound to DNA. (A) Effect of acceptor concentration. Reaction mixtures containing 50 mM Tris-HCl (pH 7.5), 25 nM 5′-labeled 60-mer–12-mer substrate, and 125 nM topoisomerase were incubated at 37°C for 15 min. Strand transfer was initiated by adding 0.125, 0.25, or 1.25 μM acceptor C. (B) Effect of topoisomerase concentration. Reaction mixtures containing 50 mM Tris-HCl (pH 7.5), 25 nM 5′-labeled 60-mer–12-mer substrate, and either 0.125 or 1.25 μM topoisomerase were incubated at 37°C for 15 min. Strand transfer was initiated by adding 1.25 μM acceptor C. The extents of flap recombinant formation are plotted as a function of time.
A more critical test of the issue of topoisomerase occupancy of the target site during strand invasion was to examine the influence of topoisomerase concentration on the flap ligation reaction. The prediction was that if topoisomerase dissociated from the DNA to permit strand invasion, then an increase in the topoisomerase concentration beyond 125 nM (the standard enzyme concentration in the reactions) should inhibit the rate and extent of flap ligation to acceptor C. Yet if strand invasion occurred while topoisomerase was bound to CCCTT, we anticipated that an increase in topoisomerase concentration would have little effect. We observed that increasing topoisomerase concentration tenfold to 1.25 μM (a 50-fold excess of topoisomerase over DNA substrate) had virtually no effect on the rate and extent of flap ligation (Fig. 8B). These experiments are consistent with strand invasion with topoisomerase bound at the CCCTT target site. This implies previously unappreciated conformational flexibility of the topoisomerase-DNA clamp on the 5′ side of the scissile phosphodiester.
DISCUSSION
The present study illuminates a recombinogenic pathway of lesion repair whereby vaccinia virus topoisomerase bound covalently at a blunt DSB can ligate the covalently held strand to a recessed 5′ OH terminus of a duplex DNA acceptor with a 3′ single-strand tail. The transfer reaction occurs with exceptionally high efficiency to DNA acceptors that contain a 3′ tail complementary to the scissile strand upstream of the cleavage site. The flap ligation pathway differs from the two classes of intermolecular DNA ligation reactions described previously in the following respects. (i) Strand transfer by a covalent donor complex with a 5′ single-strand tail is entirely dependent on base pairing between the exogenous single-strand acceptor and the 5′ tail of the donor complex in the segment immediately downstream of the scissile phosphodiester. (ii) Blunt-end ligation by topoisomerase bound at a flush duplex CCCTT end is independent of the sequence of the blunt-end acceptor molecule.
The slow phase of homology-dependent flap ligation is dependent on, and likely limited by, invasion of the 3′ single-strand tail into the duplex 5′ of the covalent attachment site. The increase in flap recombinant strands reflects the establishment of a new transesterification equilibrium that favors strand closure, i.e., because the nonscissile strand of the recombinant no longer has a nick opposite the scissile phosphodiester that precipitates suicide cleavage. Thus, homology-dependent flap ligation rectifies damage on both strands of the duplex. In contrast, flap ligation without strand invasion restores only one DNA strand. The opposing nick must still be repaired to avoid reiterating the topoisomerase-induced DSB.
The rate of the second phase of the flap ligation reaction (1.7 × 10−3 s−1) is low compared to the rates of the chemical steps of transesterification (kcl, ∼0.3 s−1; krel, ∼1 s−1). Yet the reaction proceeds to a high yield over a time scale of 20 to 30 min in vitro, which is compatible with damage correction within a single round of vaccinia virus replication in vivo and, by extension to cellular topoisomerase IB, within a single cell cycle in vivo, especially when cell cycle delays or checkpoints are triggered by topoisomerase-induced DSBs (47, 52).
The homology-dependent phase of flap ligation is several orders of magnitude slower than the expected rate of spontaneous DNA branch migration under the solution conditions used in our assays, i.e., low ionic strength and no divalent cation (18). Therefore, topoisomerase bound to the CCCTT site is actually an impediment to strand invasion. The remarkable and instructive finding is that strand invasion occurs at all, given that topoisomerase IB binds circumferentially to the DNA duplex 5′ of the scissile phosphodiester (5, 24, 28, 30). For strand invasion into the CCCTT site to be feasible, the topoisomerase C-clamp around the target site must open up to allow ingress of the invading 3′ flap. Opening of the clamp would likely occur by retroflexion of the bridge segment of the polypeptide that links the N-terminal domain on the major-groove face of the DNA with the C-terminal catalytic domain on the minor-groove face (29). We infer that the opening of the clamp to allow strand invasion occurs when the topoisomerase is bound covalently to the flap recombinant because opening of the clamp by noncovalently bound topoisomerase would result in immediate dissociation of the protein from the DNA. Because the C-terminal domain is tethered covalently to the CCCTT strand, we suspect that it is the N-terminal domain that moves to open the clamp.
Our results illuminate a previously unappreciated conformational flexibility of covalently bound topoisomerase with respect to its DNA contacts on the 5′ side of the scissile phosphodiester. It had been noted before that the protein-DNA interface 3′ of the scissile phosphodiester is loose and flexible. In catalyzing the relaxation of supercoiled DNA, covalently bound topoisomerase IB releases its grip on the downstream duplex and permits rotation of the duplex around the phosphodiester opposite the scissile phosphate before resealing the backbone. About five negative supercoils are removed by vaccinia virus topoisomerase for each cleavage event (42). The DNA cleavage and religation reactions using linear substrates do not take into account any DNA rotation events, because there are no measurable changes in topology. Yet the finding that topoisomerase trapped covalently at a nick in linear duplex DNA is capable of efficient strand transfer to an exogenous nucleophile provides evidence that the noncovalently held 5′ OH DNA end is fairly mobile, i.e., that it spontaneously departs from the acceptor site while still tethered to the complex by the nonscissile strand (13).
Redinbo et al. (25) have provided important insights into the nature of the structural flexibility of DNA-bound topoisomerase IB through their analysis of multiple nonisomorphous cocrystal structures. Crystallographic flexibility of the “cap” subdomain of human topoisomerase I (25), which is analogous to the N-terminal domain of vaccinia virus topoisomerase, is consistent with our hypothesis that loosening of the N-terminal domain contacts with the CCCTT site in the covalent topoisomerase-DNA complex permits strand invasion during homology-dependent flap ligation. Our finding that 6 bp of homology allows the second phase of accumulation of flap recombinant while ≤4 bp of homology is without effect can be explained easily if the open topoisomerase clamp is unable to close when the flap branch point resides within the CCCTT target site, e.g., because of steric hindrance. It is known that modifications of the phosphate backbone of the nonscissile strand within the complementary 3′ GGGAA element adversely affect DNA binding and catalysis (6, 28).
Prior functional analyses of DNA binding and transesterification by vaccinia virus topoisomerase have revealed numerous features shared by the cellular topoisomerase IB enzymes, e.g., the circumferential protein-DNA interface (28, 30), conformational changes elicited by DNA binding (29), and the identity of the catalytic side chains at the active site (13a, 37). All of the cleavage and strand transfer reactions that we demonstrated previously for vaccinia virus topoisomerase (e.g., RNA cleavage and cyclization, hairpin formation, sticky-end DNA ligation, blunt-end DNA ligation, and strand transfer to non-nucleic acid nucleophiles) have been demonstrated for cellular topoisomerase IB. Therefore, we predict that cellular type IB topoisomerases can also catalyze flap ligation. (Testing this prediction is beyond the scope of the present study.) The flap ligation reaction has interesting implications for thinking about pathways of topoisomerase-mediated DNA rearrangements in vivo.
The proposed flap ligation pathway overlaps in part with the initial steps of the DSBR pathway of homologous recombination (45), which invokes nucleolytic processing of the DSB to generate 3′ tails that can invade a homologous duplex (48). Covalent attachment of topoisomerase on one side of the DSB protects the covalently activated terminus from nucleases (6, 33) while still allowing processing of the blunt duplex leaving group. The simple hypothesis that a 5′ to 3′ exonuclease generates the recessed 5′ terminus and the 3′ single-strand tail DSBR intermediates is complicated by the finding that Mre11, a nuclease that is clearly involved in eukaryotic DSBR, is both an endonuclease and a 3′ to 5′ exonuclease (19, 46). However, the nuclease activity of yeast Mre11 is not required for DSBR in mitotic cells (17), suggesting that mitotic cell DSBs may be processed by other nucleases that are functionally redundant with Mre11. Irrespective of the identity of the processing nucleases, it is clear that repair of topoisomerase-induced DSBs by the flap ligation pathway calls for a 5′ OH moiety on the recessed DNA strand, which could arise either via dephosphorylation of a 5′ phosphate product of endo- or exonuclease digestion or via the action of a nuclease that directly yields a 5′ OH end.
We have shown that flap ligation by vaccinia virus topoisomerase occurs with or without homology of the 3′ tail of the acceptor to the covalent donor complex. The yield of the recombinant strand is seven to eightfold higher with flap homology, but the probability of encountering an exact match at nucleotide position 6 immediately flanking the recessed 5′ OH end of the recombination partner is low (1/4,096), assuming random recession of the blunt-end leaving group. Flap sites corresponding to the topoisomerase cleavage recognition element may represent hot spots for flap ligation in vivo, but they may not comprise the majority of targets for topoisomerase-catalyzed DNA rearrangements. The hallmarks of homology-independent flap ligation (or blunt ligation) would be the presence of a topoisomerase cleavage site at only one of the two recombining half-sites and the absence of homology immediately 3′ of the scissile phosphodiester. The key features of homology-dependent flap ligation would be the occurrence of topoisomerase recognition sequences at both recombining half-sites and no homology 3′ of the scissile phosphodiester. The presence of 3′ homology implicates the sticky-end topoisomerase-catalyzed strand transfer pathway.
The chromosomal excision events directly catalyzed by vaccinia virus topoisomerase in vivo all entailed strand transfer at topoisomerase cleavage sites in parallel orientation (32). Most events (3/5) displayed 3′ homology and were therefore attributable to some manner of sticky-end ligation. One pair of partners lacked 3′ homology but displayed heterology within the 5′ pentamer recognition sites, which suggests blunt ligation as the likely mechanism. Only one event was consistent with either blunt ligation of two ends, both generated by topoisomerase-induced DSBs, or flap ligation of a topoisomerase-induced DSB to a processed end with a homologous 3′ tail. The nature of the in vivo recombination assay, which entailed robust overexpression of vaccinia topoisomerase, may have selected for events in which both recombining partners were cleaved by the topoisomerase. In contrast, a high proportion of the illegitimate integration events promoted by yeast topoisomerase I in vivo entailed activation of the chromosomal DNA donor by topoisomerase. The transfected plasmid acceptor was linearized to give a 5′ tail and converted to 5′ OH prior to transfection (53). A 5′-tailed plasmid can be filled in by DNA polymerases in the cell, which may have favored the detection of integration events with little or no 5′ resection of the plasmid.
An appealing biological scenario is that flap ligation plays a role in the recombinogenic repair of topoisomerase-induced DSBs during the interval in which checkpoint controls and apoptotic decisions are in critical balance. In such an environment, the activation of DNase II-like nucleases would directly generate the reactive 5′ OH DNA termini for flap ligation. DNase II, which is active at low pH values in vitro, plays a role in DNA degradation during apoptosis (12, 50). Apoptosis is induced in mammalian cells by the topoisomerase IB poison camptothecin and is associated with intracellular acidification. Recent studies suggest that DNase II is active in vivo even at neutral pH (12). Thus, current evidence indicates that DSBs induced by topoisomerase in response to endogenous or pharmacological poisons can induce pathways that either promote break repair or favor cell demise, presumably according to the level of induced DNA breakage. Because the damage response may generate suitable substrates for intrinsic repair of the trapped covalent adducts by intermolecular religation and flap ligation in particular, there is potential for ensuing topoisomerase-catalyzed genome rearrangements in those cells that survive.
REFERENCES
- 1.Been M D, Champoux J J. DNA breakage and closure by rat liver type I topoisomerase: separation of the half-reactions by using a single-stranded DNA substrate. Proc Natl Acad Sci USA. 1981;78:2883–2887. doi: 10.1073/pnas.78.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock P, Champoux J J, Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model or nonhomologous recombination. Science. 1985;230:954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- 3.Burgin A B, Huizenga B H, Nash H A. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 1995;15:2973–2979. doi: 10.1093/nar/23.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champoux J J, Bullock P A. Possible role for the eucaryotic type I topoisomerase in illegitimate recombination. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 655–666. [Google Scholar]
- 5.Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 6.Cheng C, Shuman S. Site-specific transesterification by vaccinia topoisomerase: role of specific phosphates and nucleosides. Biochemistry. 1999;38:16599–16612. doi: 10.1021/bi992001d. [DOI] [PubMed] [Google Scholar]
- 7.Christiansen K, Westergaard O. Characterization of intra- and intermolecular DNA ligation mediated by eukaryotic topoisomerase I: role of bipartite DNA interaction in the ligation process. J Biol Chem. 1994;269:721–729. [PubMed] [Google Scholar]
- 8.Guo F, Gopaul D N, Van Duyne G D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 9.Halligan B D, Davis J L, Edwards K A, Liu L F. Intra- and intermolecular strand transfer by HeLa DNA topoisomerase I. J Biol Chem. 1982;257:3995–4000. [PubMed] [Google Scholar]
- 10.Hickman A B, Waninger S, Scocca J J, Dyda F. Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 Å resolution. Cell. 1997;89:227–237. doi: 10.1016/s0092-8674(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 11.Kingma P S, Osheroff N. The response of eukaryotic topoisomerases to DNA damage. Biochim Biophys Acta. 1998;1400:223–232. doi: 10.1016/s0167-4781(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 12.Krieser R J, Eastman A. The cloning and expression of human deoxyribonuclease II: a possible role in apoptosis. J Biol Chem. 1998;273:30909–30914. doi: 10.1074/jbc.273.47.30909. [DOI] [PubMed] [Google Scholar]
- 13.Krogh B O, Shuman S. DNA strand transfer catalyzed by vaccinia topoisomerase: peroxidolysis and hydroxylaminolysis of the covalent protein-DNA intermediate. Biochemistry. 2000;39:6422–6432. doi: 10.1021/bi000184u. [DOI] [PubMed] [Google Scholar]
- 13a.Krogh B O, Shuman S. Catalytic mechanism of DNA topoisomerase IB. Mol Cell. 2000;5:1035–1041. doi: 10.1016/s1097-2765(00)80268-3. [DOI] [PubMed] [Google Scholar]
- 14.Kwon H J, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanza A, Tornaletti S, Rodolfo C, Scanavini M C, Pedrini A M. Human DNA topoisomerase I-mediated cleavages stimulated by ultraviolet light-induced DNA damage. J Biol Chem. 1996;271:6978–6986. doi: 10.1074/jbc.271.12.6978. [DOI] [PubMed] [Google Scholar]
- 16.Levin N A, Bjornsti M A, Fink G R. A novel mutation in DNA topoisomerase I of yeast causes DNA damage and RAD9-dependent cell cycle arrest. Genetics. 1993;133:799–814. doi: 10.1093/genetics/133.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreau S, Ferguson J R, Symington L S. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panyutin I G, Hsieh P. The kinetics of spontaneous DNA branch migration. Proc Natl Acad Sci USA. 1994;91:2021–2025. doi: 10.1073/pnas.91.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paull T T, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 20.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 21.Pouliot J J, Yao K C, Robertson C A, Nash H A. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 22.Pourquier P, Ueng L, Kohlhagen G, Mazumder A, Gupta M, Kohn K W, Pommier Y. Effects of uracil incorporation, DNA mismatches, and abasic sites on DNA cleavage and religation activities of mammalian topoisomerase I. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 23.Pourquier P, Pilon A A, Kohlhagen G, Mazumder A, Sharma A, Pommier Y. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. J Biol Chem. 1997;272:26441–26447. doi: 10.1074/jbc.272.42.26441. [DOI] [PubMed] [Google Scholar]
- 24.Redinbo M R, Stewart L, Kuhn P, Champoux J J, Hol W G J. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 25.Redinbo M R, Stewart L, Champoux J J, Hol W G J. Structural flexibility of human topoisomerase I revealed in multiple non-isomorphous crystal structures. J Mol Biol. 1999;292:685–696. doi: 10.1006/jmbi.1999.3065. [DOI] [PubMed] [Google Scholar]
- 26.Russell D W, Alexander I E, Miller A D. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiguchi J, Cheng C, Shuman S. Kinetic analysis of DNA and RNA strand transfer reactions catalyzed by vaccinia topoisomerase. J Biol Chem. 1997;272:15721–15728. doi: 10.1074/jbc.272.25.15721. [DOI] [PubMed] [Google Scholar]
- 28.Sekiguchi J, Shuman S. Vaccinia topoisomerase binds circumferentially to DNA. J Biol Chem. 1994;269:31731–31734. [PubMed] [Google Scholar]
- 29.Sekiguchi J, Shuman S. Proteolytic footprinting of vaccinia topoisomerase bound to DNA. J Biol Chem. 1995;270:11636–11645. doi: 10.1074/jbc.270.19.11636. [DOI] [PubMed] [Google Scholar]
- 30.Sekiguchi J, Shuman S. Identification of contacts between topoisomerase I and its target DNA by site-specific crosslinking. EMBO J. 1996;15:3448–3457. [PMC free article] [PubMed] [Google Scholar]
- 31.Shuman S. Vaccinia DNA topoisomerase I promotes illegitimate recombination in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:3489–3493. doi: 10.1073/pnas.86.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shuman S. Recombination mediated by vaccinia DNA topoisomerase I in Escherichia coli is sequence specific. Proc Natl Acad Sci USA. 1991;88:10104–10108. doi: 10.1073/pnas.88.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuman S. Site-specific interaction of vaccinia virus DNA topoisomerase with duplex DNA: minimal DNA substrate for strand cleavage in vitro. J Biol Chem. 1991;266:11372–11379. [PubMed] [Google Scholar]
- 34.Shuman S. DNA strand transfer reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992;267:8620–8627. [PubMed] [Google Scholar]
- 35.Shuman S. Two classes of DNA end-joining reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992;267:16755–16758. [PubMed] [Google Scholar]
- 36.Shuman S. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J Biol Chem. 1994;269:32678–32684. [PubMed] [Google Scholar]
- 37.Shuman S. Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme. Biochim Biophys Acta. 1998;1400:321–337. doi: 10.1016/s0167-4781(98)00144-4. [DOI] [PubMed] [Google Scholar]
- 38.Shuman S, Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990;265:17826–17836. [PubMed] [Google Scholar]
- 39.Shuman S, Golder M, Moss B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J Biol Chem. 1988;263:16401–16407. [PubMed] [Google Scholar]
- 40.Stivers J T, Shuman S, Mildvan A S. Vaccinia DNA topoisomerase I: single-turnover and steady-state kinetic analysis of the strand cleavage and ligation reactions. Biochemistry. 1994;33:327–339. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- 41.Stivers J T, Shuman S, Mildvan A S. Vaccinia DNA topoisomerase I: kinetic evidence for general acid-base catalysis and a conformational step. Biochemistry. 1994;33:15449–15458. doi: 10.1021/bi00255a027. [DOI] [PubMed] [Google Scholar]
- 42.Stivers J T, Harris T K, Mildvan A S. Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 43.Stivers J T, Jagadeesh G J, Nawrot B, Stec W J, Shuman S. Stereochemical outcome and kinetic effects of Rp- and Sp-phosphorothioate substitutions at the cleavage site of vaccinia type I DNA topoisomerase. Biochemistry. 2000;39:5561–5572. doi: 10.1021/bi992429c. [DOI] [PubMed] [Google Scholar]
- 44.Subramanya H S, Arciszewska L K, Baker R A, Bird L E, Sherratt D J, Wigley D B. Crystal structure of the site-specific recombinase XerD. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 46.Trujillo K M, Yuan D F, Lee E, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21446–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 47.Wan S, Capasso H, Walworth N C. The topoisomerase I poison camptothecin generates a Chk1-dependent checkpoint damage signal in fission yeast. Yeast. 1999;15:821–828. doi: 10.1002/(SICI)1097-0061(199907)15:10A<821::AID-YEA422>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 48.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wittschieben J, Shuman S. Mechanism of DNA transesterification by vaccinia topoisomerase: catalytic contributions of essential residues Arg-130, Gly-132, Tyr-136, and Lys-167. Nucleic Acids Res. 1997;25:3001–3008. doi: 10.1093/nar/25.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Y C, Stanfield G M, Horvitz H R. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- 51.Yang S W, Burgin A B, Huizenga B N, Robertson C A, Yao K C, Nash H A. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin M B, Guo B, Vanhoefer U, Azrak R G, Minderman H, Frank C, Wrzosek C, Slocum H K, Ristum Y M. Characterization of protein kinase chk1 essential for the cell cycle checkpoint after exposure of human head and neck carcinoma A253 cells to a novel topoisomerase I inhibitor BNP1350. Mol Pharmacol. 2000;57:453–459. doi: 10.1124/mol.57.3.453. [DOI] [PubMed] [Google Scholar]
- 53.Zhu J, Schiestl R H. Topoisomerase I involvement in illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]