ABSTRACT
Coronaviruses are single-stranded RNA viruses that cause severe respiratory, enteric, and systemic infections in a vast range of hosts, including man, fish, mammals, and avian. Scientific interest has heightened on coronaviruses after the emergence of the 2019 novel Coronavirus (SARS-CoV-2). This review provides current perspectives on morphology, genetic diversity, transmission characteristics, replication cycle, diagnostic approaches, epidemiological assessment, and prevention strategies against the SARS-CoV-2. Moreover, different potential biotherapeutics such as small drug molecules, different vaccines, and immunotherapies to control severe acute respiratory infections caused by 2019 novel coronavirus (SARS-CoV-2) are repurposed and discussed with different mechanistic approaches. The current growth trends of the SARS-CoV-2/COVID-19 outbreak globally and preventive measures are briefly discussed. Furthermore, the lessons learned from the COVID-19 outbreak, so far, concluding remarks and future directions for controlling for COVID-19, are also recommended for a safer tomorrow.
KEYWORDS: Coronavirus, COVID-19, transmission characteristics, diagnosis, epidemiology, prevention strategies, therapeutic candidates
1. Introduction
A group of people was admitted to the hospital in December 2019, having the pneumonia-like condition. All these people belonged to the seafood market of Wuhan, Hubei Province, China.1,2 The identified virus was named by World health organization (WHO) as SARS-CoV-2 (novel coronavirus 2019) on January 7, 2020, 3 and the associated disease was known as coronavirus disease (COVID-19).4,5 Until now, SARS-CoV-2 infected the millions and killed hundreds of thousands of people in almost every country in the world. No clinically approved vaccines and drugs are available for the treatment of coronavirus patients to date. Intensive research is urgently needed against coronavirus to identify potential drug targets, which will be helpful in the development of treatment/vaccine.
2. Morphology, structure and genetic diversity
Coronaviruses (CoVs) belong to a big family of viruses having single-stranded RNA. This virus can infect animals and humans, resulting in hepatic, respiratory, gastrointestinal, and neurological disorders.6 Coronaviruses are non-segmented, enveloped, RNA viruses consist of single-strand and their genome range in size from 26 to 32 kilobases, which is generally a big known Viral-RNA genome. The nucleocapsid of the virion is made up of genomic RNA and phosphorylated nucleocapsid protein (N), which is concealed inside the bilayers of phospholipid and surrounded by two kinds of spike proteins. One type of spike protein is called spike glycoprotein trimmer (S) that can be found in all coronaviruses, but the other type of hemaglutinin‐esterase (HE) exists in some coronaviruses. The membrane protein (M), which is a type III glycoprotein and the envelope protein, is found among the S proteins in the virus envelope. Corona’s name is given to viruses because of their crown-like appearances .7 The structure is presented in Figure 1.
Figure 1.

Structure presentation of Coronavirus particle.7
These viruses are further categorized into four different genera as α, β, γ, and δ coronaviruses.8 Among them, α and β coronaviruses can infect mammals, and the other two genera can infect both birds and mammals. Until now, seven different types of human coronaviruses (HCoVs) have been identified, which includes α-CoVs HCoVs-NL63, HCoVs-229E and the β-CoVs HCoVs-OC43, HCoVs-HKU1, severe acute respiratory syndrome-CoV (SARS-CoV),9 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the Middle East respiratory syndrome-CoV (MERS-CoV).10 The human-respiratory system is profoundly affected by coronaviruses. Severe acute respiratory syndrome (SARS)-CoV and the Middle East respiratory syndrome (MERS)-CoV are the previously reported outbreaks of pathogenic coronaviruses, which posed the critical problems. HCoVs-229E and HCoVs-OC43 are known since the 1960s. HCoVs-NL63 and HCoVs-HKU1 were recognized in 2004 and 2005, respectively. (MERS)-CoV was isolated in 2012. SARS-CoV-2 is responsible for the spread of COVID-19 disease. These viruses intensely affect immune-compromised individuals. Some coronaviruses can infect livestock, whales, birds, mice, bats, camels, dogs, pigs, cows, and few other wild species. Table 1 reveals the major human Coronaviruses, 11 while the animal and birds pathogenic coronaviruses are listed in Table 2.11
Table 1.
List of human coronaviruses.11
| Coronaviruses | Genus | Host | Symptoms with a mortality rate |
|---|---|---|---|
| HCoVs-229E | α | Human | Mild respiratory tract infection |
| HCoVs-NL63 | α | Human | Mild respiratory tract infection |
| HCoVs-OC43 | β | Human | Mild respiratory tract infection |
| HCoVs-HKU1 | β | Human | Pneumonia |
| SARS-CoV | β | Human | Severe acute respiratory syndrome, 11% |
| MERS-CoV | β | Human | Severe acute respiratory syndrome, 34% |
| SARS-CoV-2 | β | Human | Severe acute respiratory syndrome, 2.6% |
Table 2.
List of coronaviruses reported in animals and birds11
| Coronaviruses | Genus | Host | Symptoms with a mortality rate |
|---|---|---|---|
| MHV-A59 | β | Mouse | Acute pneumonia, lung injuries |
| TGEV/PUR46-MAD | α | Pig | Diarrhea (100% mortality rate in piglets (< 2 weeks old)) |
| SeACoV-CH/GD-01 | α | Pig | Severe and acute Diarrhea and acute vomiting |
| PRCV/ISU-1 | α | Pig | Mild respiratory tract infection |
| PEDV/ZJU-G1-2013 | α | Pig | Severe watery Diarrhea |
| Camel-α-coronavirus | α | Camel | Asymptomatic |
| Bovine CoV/ENT | β | Cow | Diarrhea |
| Canine CoV/TU336/F/2008 | α | Dog | Mild clinical signs, diarrhea |
| Feline infections peritonitis virus | α | Cat | Fever, vasculitis, and serositis |
| Equine CoV/Obihiro12-1 | β | Horse | Anorexia, leukopenia, and fever |
| Beluga whale Cov/SW1 | γ | Whale | Liver failure and pulmonary disease |
| Bulbul coronavirus HKU11 | δ | Bulbul | Respiratory disease obtained from a wild bird,s respiratory tract |
| Sparrow coronavirus HKU17 | δ | Sparrow | Respiratory disease obtained from a wild bird,s respiratory tract |
| IBV | γ | Chicken | Severe respiratory disease |
The coronaviruses of animal origin may undergo genetic-recombination and result in structurally changed coronaviruses that may prove extremely dangerous to human beings.12 The mutation rate was assessed to be 0.80–2.38 × 10−3 nucleotide substitutions per site per year in the SARS-CoV genome, and non-synonymous and synonymous substitution rates were evaluated to 1.16–3.30 × 10−3 and 1.67–4.67 × 10−3 per site per year, respectively, which are found comparable to that of other RNA viruses.13 The evolution and recombination of coronaviruses of animal origin may occur within the host or from jumping one to another species. Such evolved variants, when entered in humans, have much pathogenic capacity.14,15 According to the recent reports, two mutations of the N and S proteins, SARS-CoV-2, may explain its zoonotic transmission.16 Arrangement of the genome of 54 SARS-CoV-2 recognized two hotspots of hyper-variability at positions 8789 (synonymous variant) and 28151(Ser/Leu change) located in the poly-protein and ORF8 genes, respectively.17
3. Transmission routes and replication cycle
Various studies have been carried out to search the intermediate carrier and reservoir host, which is responsible for the spread of coronavirus to human beings. According to initial investigations, snake species have been identified as a possible reservoir of COVID-19. However, up to the present date, no proper evidence of coronavirus reservoirs has been found other than mammals and birds. COVID-19 genomic analysis exhibited 88% identity with two bat-derived severe acute respiratory syndrome-like coronaviruses.18,19 This is indicating the link of mammals between COVID-19 and humans. Person to person transmission of COVID-19 has also been reported.20 Reports of health officials demonstrated shreds of evidence of transmission within the chain of four generations as a person who was initially caught the virus from non-human source infected some-one, who transmitted it to another, who then infected to another individual, this chain is indicating human to human transmission.21 Up till now, the main transmission route has been found respiratory droplets created through coughing and sneezing; the virus can also be transmitted through contact.22 The new coronavirus has been found in feces of identified patients in the city of Wuhan and Shenzhen. In the USA, this detection indicates that the virus can replicate in the digestive tract and can exist there and can exit from the body also demonstrated the possibility of fecal and oral transmission.23 The WHO suggests that further studies required to find the chances of transmission through aerosol.24 According to one report, the mother was diagnosed with new coronavirus-pneumonia, and the newborn was found positive for viral nucleic acid in pharynx swab (after 30 h of birth), this explains that coronavirus may be responsible for neo-natal infection through mother to child transmission, but more studies needed to confirm these phenomena.25
The initial step of viral infection is the binding of the receptor of the host cell; then, it is followed by the fusion with the cell membrane. The virus primarily targets the epithelial cells of the lungs. The binding between the receptor-binding domain of spikes of virus and angiotensin-converting enzyme 2 (ACE2) receptor (cellular receptor) is responsible for the human to human transmission of SARS-CoV.19,26 Studies have demonstrated that the receptor binding site of coronavirus (COVID-19) spikes resembles SARS-CoV. Studies are suggesting that virus entry into the host cell is most likely through the ACE2 receptor.19
4. Symptoms of COVID-19 infection
The most common symptoms of COVID-19 are dry cough, tiredness, and fever. Some patients may have diarrhea, sore throat, runny nose, nasal congestion, aches, pain, and sputum (thick mucus coughed up from the respiratory tract). Some reports show that people might be got infected without showing any symptoms. Most of the people about 80% can be recovered from the COVID-19 infections without any kind of unique treatments; however, one out of six people get severely infected from this disease and feel difficulty in breathing. Aged people, medically (diabetes, heart problems, and high blood pressure) and immunology compromised people are more susceptible to developing severe illness issues such as pneumonia, severe acute respiratory syndrome, kidney failure, and death could be occurred.27 Figure 2 shows the symptoms of COVID-19 concerning the percentage.
Figure 2.
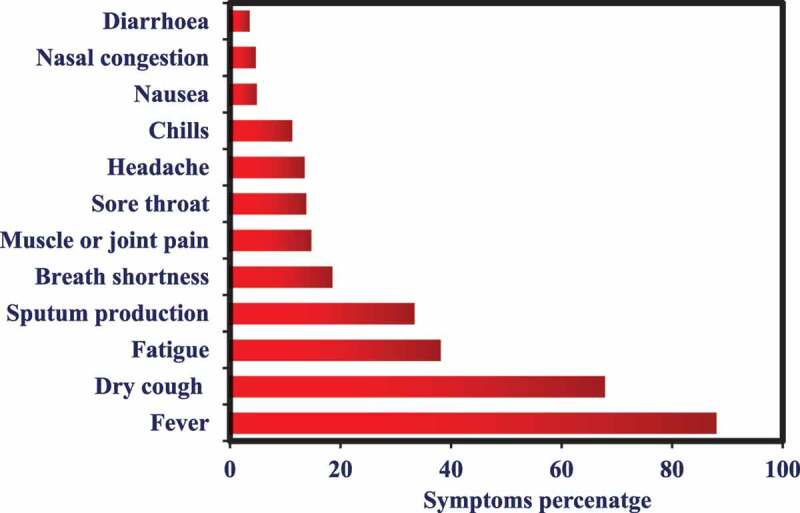
The symptoms of SARS-CoV-2 infections with respect to the rate
5. Diagnostic approaches for COVID-19 disease
The rapid and efficient diagnosis is the critical factor to control and manage the outbreak of infectious diseases. The swift and prompt investigation of virologic infections helps accurate public health surveillance, prevention, and monitoring.28 After the outbreak of COVID-19, two methods are widely employed for its diagnosis.
5.1. Genetic material detection
Genetic material detection accompanied by Cobas SARS-CoV-2 Test.29 This test is a single well dual-target protocol and is a real-time RT-PCR test. This test proposed for the qualitative investigation of nucleic-acids from SARS-CoV-2 in nasopharyngeal and oropharyngeal swab samples from the patients got infected by SARS-CoV-2. This test operates on the Cobas 6800/8800 systems and possesses full process internal, positive, and negative control. It must be noted that the negative results do not be precluded SARS-CoV-2 infection and further must not be employed for deciding patient management. Therefore, these results should be carefully evaluated by combining them with epidemiological information, patient history, and clinical observations. This test (Cobas SARS-CoV-2) must always be performed by trained and clinical laboratory officials who have sound knowledge and experience of in vitro diagnostic and real-time PCR protocols.
5.2. Serological testing
The extent of infections, estimating asymptomatic, and attack rates can be determined conclusively by serological tests such as neutralization, IIFT, and ELISA tests. The serological tests are involved in detecting antibodies and antigens in comparison to the RT-PCR assay, which identifies the viral genome. The lag period for the antibodies to appear against the virus can be generally ranged between 14 to 28 days after infection.30 Therefore, delayed detection of antibodies due to their lag period production can cause mortality due to high virus loads. Thus, serological tests can only be chosen when nucleic amplification tests are not available.31
6. Current treatment opportunities
6.1. Targeting proteins identifications
Target identification is an essential key factor in the development of drugs or the use of already existing medications for the treatment of any kind of disease. Figure 3 and Table 3 enlisted the potential target site, their role in viral infections, and drug candidates that potentially can act on the corresponding target sites in the same kind of viruses. Therefore, these drugs candidates can be evaluated for their potential efficacy against COVID-19 disease. The 3-C-like protease (3CLpro) and a papain-like protease (PLpro) play their role during the replication process and also assist in packaging inside the host-body. Therefore, the drug candidates, including ritonavir, lopinavir, HIV drugs, etc. that target 3CLpro and PLpro in other viruses can be potentially explored.32,33 RdRP is an RNA-dependent R.N.A. polymerase that catalyzes the synthesis of viral RNA, which can be blocked by employing antiviral drug candidates (Remdesivir).33 The spike protein (S protein) is a type of abundant transmembrane proteins and are responsible for COVID-19 infections in human respiratory epithelial cells by their interaction with the ACE2 (angiotensin-converting enzyme 2) receptor of human. The drug candidates that potentially inhibit the S protein fusion with the ACE2 receptor might be proved viable drugs. The antiviral drug “Arbidol” against the influenza virus has inhibitory efficacy and prevent the fusion of S protein with the ACE2 receptor of host cells and ultimately prevents the virus from penetrating the host’s body. This drug is in a clinical trial now for COVID-19.34–36 Another factor for the illness to enter into the host body is depends on TMPRSS2 protease that secreted by the host cells and plays an essential role in the priming of S protein. Thus, the camostat mesylate, a clinically approved TMPRSS2 protease inhibitor, can be an alternative to treat COVID-19.37
Figure 3.
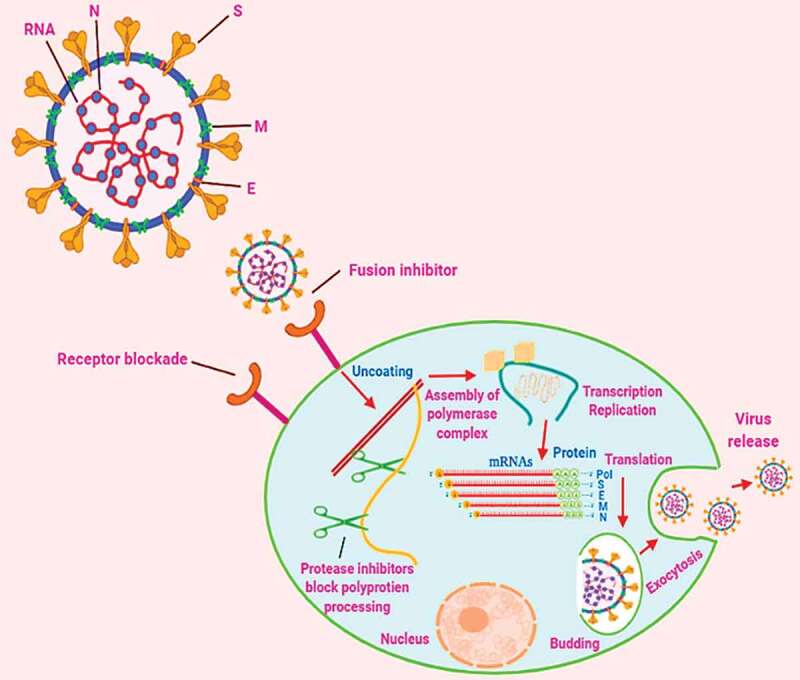
Replication process steps of coronavirus with respect to potential target-sites for repurposing therapeutic drug molecules and vaccines
Table 3.
Potential drug candidates for different targeting proteins for the treatment of SARS-CoV-2 infections
| Sr. No. | Target site | Role in viral infection | Drugs |
|---|---|---|---|
| 1 | 3CLpro | Replication process | Ritonavir, Lopinavir, HIV drugs |
| 2 | PLpro | Replication process | Ritonavir, Lopinavir, HIV drugs |
| 3 | RdRP | Catalyzes the synthesis of viral RNA | Remdesivir |
| 4 | S protein | Interaction with the ACE2 receptor |
Arbidol |
| 5 | TMPRSS2 | Priming of S protein | Camostat mesylate |
Spike (S) proteins are good contenders for vaccines. This attributes to the fact that neutralizing antibodies are oriented against S proteins. On the other hand, receptor blockade on host cells surface employing drug molecules or ligands or antibodies may prohibit the entry of viral particles. The conformational alterations induced by receptors in S proteins could be blocked employing peptides, which can prevent membrane fusion and virus entry. Further, the virus replication process can be blocked using protease inhibitors as these proteases assist in the cleaving of polyprotein of the replicase protein into functional units. Polymerase performs its role in the vicinity of cytoplasm in a very distinct membrane-bound complex systems that can be considered as potential targets for therapeutic drug molecules. As shown in the cytoplasm, viral messenger RNAs (mRNAs) constructed by discontinuous transcription with protein and each encodes designated at the right-side. Most common seventy base pair sequence is displayed in red color on the 5′ end of each mRNA. Moreover, both exocytosis, as well as the budding process, are significant and vital for the replication of the virus, which could be an operative target for the development of antiviral drugs. N is a nucleocapsid-phosphoprotein; E is membrane protein performs a part in coronavirus-assembly; S is spike-protein, and M denotes protein need for virus-budding.
6.2. Potential drug candidates’ patents for the targeting proteins of coronavirus
Figure 4 displays the potential drug candidates’ patents with respect to the targeting proteins of coronavirus as per the CAS contents collection. It can be seen that two targeting proteins PLpro and 3CLpro has attracted much attention as compared to all other targeting proteins. More number of potential therapeutic efficacy having drug molecules were widely investigated against both these proteins. This might be attributed to the extensive work done on the SARS-CoV that also has these two proteins.
Figure 4.
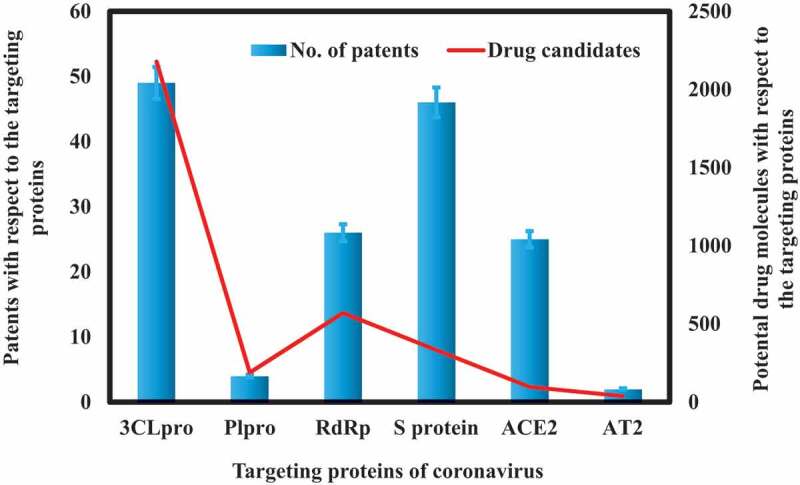
Potential drug candidates’ patents with respect to the targeting proteins of coronavirus
6.3. Potential therapeutic drug candidates for the treatment of COVID-19 disease
There is an immediate requirement for the development of potential therapeutic drug candidates for the treatment of COVID-19 disease until a vaccine can be developed. Another way is to employ existing drug candidates against SARS-CoV-2 infections based on their structure-activity relationship (SAR) and mechanism of actions. There are following drug candidates that can be potentially evaluated for their efficacy against COVID-19 (Figure 5).
Figure 5.
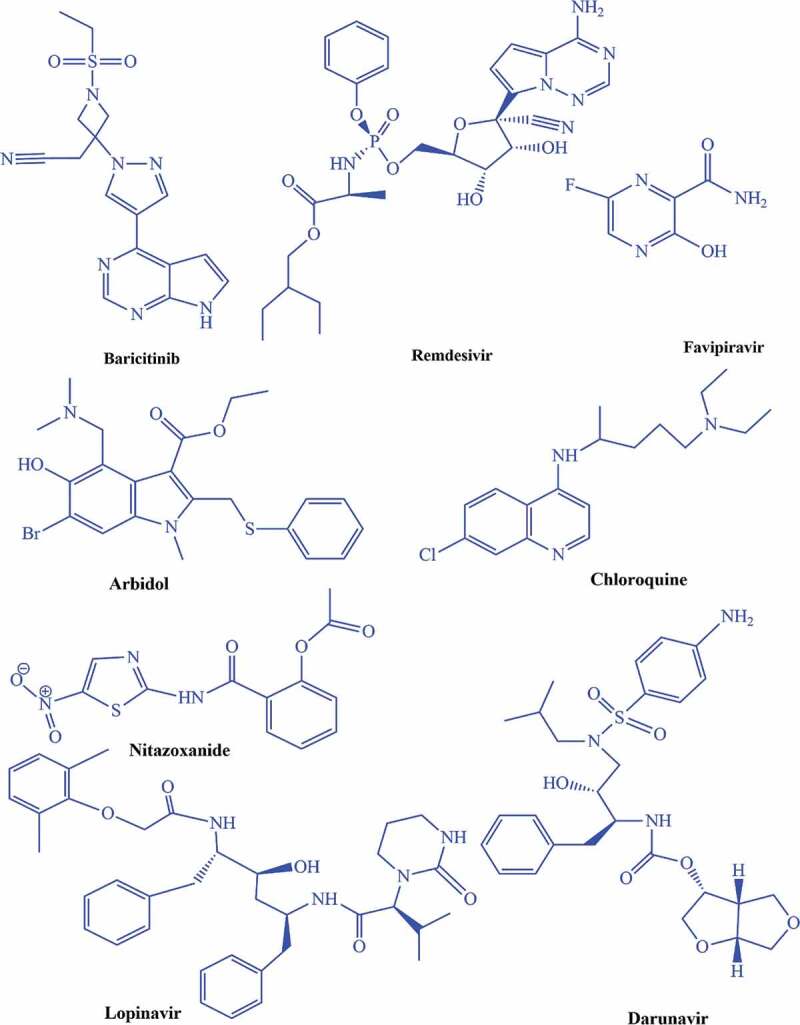
Drug candidates that can be potentially evaluated for their effectiveness against SARS-CoV-2 infections
Baricitinib drug plays its role as an inhibitor for Janus kinase (JAK), further blocking the JAK1 and JAK2,38 so it can be evaluated for SARS-CoV-2 infections to decrease the inflammation and viral entry in host body.39 Remdesivir is an antiviral drug that belongs to the nucleotides analogs and performs its antiviral action by blocking viral nucleotides synthesis, which further prohibits the viral replication process .33,40,41 The literature demonstrates that remdesivir was administered to one US patient infected by SARS-CoV-2 in late January 2020 that was improved dramatically and discharged.23 In late January 2020, Chinese medical researchers explored the efficacy of 30 drug candidates against SARS-CoV-2 and found a beneficial inhibitory action of remdesivir on SARS-CoV-2 at the cellular level. Clinically trial on remdesivir has been started in China on February 6, 2020.42 Favipiravir has potential efficacy against many RNA viruses.43,44 Favipiravir performs its role by inhibiting the viral RNA- dependent RNA polymerase (RdRp). In February 2020, this drug was employed in China for the treatment of SARS-CoV-2 infections and now has been approved for clinical trials.45 Umifenovir has been used for the treatment of influenza infections in Russia and China.46,47 On February 4, 2020, Li Lanjan, a Chinese epidemiologist, repurposed that arbidol with darunavir (employed for HIV/AIDS treatments) can be proved as a potential treatment for SARS-CoV-2 infections.48 Chloroquine is employed for the treatment of malaria.42,49 Based on its therapeutic effectiveness against viral infections, Wang et al. evaluated five FDA approved (chloroquine, nafamostat, nitazoxanide, penciclovir, ribavirin) and two antiviral drugs in vitro against the isolates of SARS-CoV-2 infections. They concluded that chloroquine demonstrated excellent efficacy for the treatment of SARS-CoV-2 infections in vitro, and it would be a promising clinical drug candidate.42,50 Lopinavir/ritonavir (LPV/r) drug combination uses for the treatment of HIV/AIDS.51 This drug combination has explored anti-coronavirus activity in January 2020 due to their proteases inhibiting efficacy that might inhibit the 3CLpro and PLpro. Lopinavir/ritonavir drug has demonstrated the inhibitory efficacy against SARS-CoV-2.52 Nitazoxanide is an antiviral and antiparasitic drug.49,53 A recent study reported by Wang et al. demonstrated that nitazoxanide drug exhibited potent inhibitory efficacy against the SARS-CoV-2 in Vero E6 cells at micromolar concentration level.42
6.4. Potential vaccine candidates for the treatment of COVID-19 disease
After the outbreak of COVID-19 illness, about 15 potential vaccine candidates are under investigation worldwide. Different technologies, such as synthetic and modified virus-like particles, nanoparticles, DNA based, and messenger RNA based, are employing for the development of these vaccines. About a one-year time frame required for the vaccines to be assessed for phase 1 clinical trials. Table 4 shows a brief outline of the development of potential vaccines against COVID-19 disease by different industrial and academic sectors.
Table 4.
A brief outline for the development of potential vaccines against SARS-CoV-2 disease
| Sr. No. | Company | Timeline | Technology | References |
|---|---|---|---|---|
| 1 | Moderna Therapeutics | 3-months to early stage | Messenger RNA | 28 |
| 2 | Inovio Pharmaceuticals | Testing on humans within the next few months | DNA based | 54,55 |
| 3 | Novavax | 3 months | Nanoparticle based | 54 |
| 4 | Queensland University | Six months | Rapid response technology | 56,57 |
| 5 | Vir Biotechnology | - | Anti-coronavirus monoclonal antibodies. | 58,59 |
| 6 | Chinese Center for Disease Control and Prevention |
One month for development and 2–3 years for availability | Inactivated virus vaccine | 60–63 |
| 7 | Tongji University |
Less than 40 days for vaccine samples | Messenger RNA technology | 64 |
| 8 | Johnson & Johnson | One year to in the market | Adenovirus-vectored technology | 65,66 |
| 9 | University of Hong Kong | One year for clinical trails | Modified nasal spray influenza vaccine | 62 |
| 10 | VIDO-InterVac | 6–8 weeks for animals and 1 year for human trails | - | 67 |
| 11 | GeoVax-BravoVax | - | Modified Vaccina Ankara-Virus Like Particles |
68 |
| 12 | Clover Biopharmaceuticals | - | Trimer-Tag© technology |
69 |
| 13 | CureVac | - | Messenger RNA technology | 70 |
| 14 | Texas Children’s Hospital | - | - | 71 |
| 15 | Codagenix | - | - | 71 |
6.5. Potential immunotherapies for the treatment of COVID-19
The SARS-CoV-2 and SARS-CoV share the same ACE2 receptor for their entry into the body of the host so that it can be a potential target site. The potential biotherapeutics and strategies employed for SARS-CoV can be used to inhibit the SARS-CoV-2. Other than therapeutics drugs and vaccines, immunotherapies are also considered a viable treatment approach. Among immunotherapies, monoclonal antibodies are very useful in blocking the entry or attachment of viral particles to the host’s cell. They are mostly recommended due to distinguishing characteristics (purity, specificity, reduced pathogenic contamination, and safer than serum therapy and intravenously immunoglobulins preparations).72,73
Other than the ACE2 receptor, spike proteins can also be potential target sites for monoclonal antibodies. Monoclonal antibodies have demonstrated impressive efficacy against the MERS-CoV and SARS-CoV by targeting their spikes proteins. Monoclonal antibodies alone or in combination with other monoclonal antibodies might be proved beneficial for neutralizing the virus by recognizing different types of epitopes on the virus surface.72,73 Furthermore, the cytokines are also a promising candidate in dealing with viruses with immunotherapies. Among cytokines, the targeting IL-6 and its receptor (IL6 R) by Siltuximab and tocilizumab monoclonal antibodies could mitigate cytokine storm-related symptoms in severe COVID-19 patients.73,74 Although a lot of research is going on in this direction, still no commercial solution is available at present. Different factors are behind that are creating hindrances in the commercialization of monoclonal antibody-based passive immunotherapy. The commercial-scale production of antibodies is expensive, time-consuming, and challenging.73,74 Therefore, designing and developing advanced expression systems as well as protein production systems are required to obtain efficient monoclonal antibodies with cost-effectiveness within a short period.73,74
7. Effect of high temperature and humidity on transmission of COVID-19
The number of factors, including temperature, humidity (climatic factors), and population density, may be responsible for the transmission of viruses.75 To estimate the intensity and end time of COVID-19, there is a need to explore the relationship between the transmission of COVID-19 and weather. According to rough observations during initial timings of COVID-19 breakout, the countries outside china with lower humidity and lower temperature, including Korea, Japan, Iran, etc. severe breakout was observed as compared to more humid and warmer countries (Thailand, Singapore, Malaysia, etc.). Natural log (ln) of an average number of cases/day from 8th-29th February is used to present the sternness of coronavirus outbreak for certain countries. Severity was found negatively related to temperature and humidity by using 14 different countries with more than 20 new cases during this period.76 Wang et al., 2020 investigated the effect of temperature and humidity on the transmission of COVID-19. After statistically analyzing data, the results showed that high humidity and temperature potentially reduce virus transmission. So by concluding this study, it is demonstrated that upon arrival of summer and rainy weather, the transmission of COVID-19 can effectively reduce in the northern hemisphere.77
8. Current growth trends of coronavirus worldwide and preventive measures
The governments of different countries havedeclared the emergency After confirmation of cases, WHO (world health organization) is giving priority to stable the specialized health care for coronavirus patients. WHO Team has supported over COVID-19 reference hospitals with various isolation centers across the world with required response and preparedness with the distribution of medical equipment, awareness-raising materials with all kinds of technical supports. The ministry of health has launched telephone helpline to respond to the queries of the public regarding the virus. People are encouraged to report to health ministry if they are having symptoms so they can be provided with proper medical care. To control the spread of coronavirus,the schools, colleges, and universities have been shut down with postponed examinations to prevent the person to person contact. All types of public gatherings, including wedding ceremonies, festivals, religious congregations, and sports festivals, have been strictly prohibited by the governments. Profiteering of hand-sanitizers and masks has also been banned. Shrines, cinemas, gyms, social clubs, and marriage halls have been shut down. Governemnets have also closed the shared borders to control the pandemic.
9. Lessons learned from the COVID-19 outbreak
As compared to the SARS outbreak, the international response to COVID-19 has been found more efficient and transparent. Various points should be learned from COVID-19 in the event of a future epidemic (Table 5). Mainly it has been recommended that viral response guidelines may have been issued 13 days earlier before the public was informed by the Chinese government.78
Table 5.
Lessons learned from the outbreak of COVID-19
| Current response issues |
Event | Consequence | Learning points |
|---|---|---|---|
| Transparency deficiency | Intimidation of doctors who initially identified COVID-19 | Information of COVID-19 cases was reported very late | To introduce fast, quick whistleblowing policies in case of global emergencies |
| Suspension of traveling | Minimum health screenings were carried out at airports and operation of flights was carried over a month after breakout | People were moving from high-risk areas to other areas without required health screening | Earlier screenings should be introduced at traveling points |
| Quarantine delay | first COVID-19 case reported on Dec 31st, 2019, but Wuhan started quarantine nearly one month later (23rd January) | Infected patients were allowed potentially to spread virus nationally and internationally | After identification of health threats, the high-risk areas should be Quarantine as soon as a possible |
| Public mislead | Rumors, speculations and wrong information spread among the public due to transparency lack | Improper precautions, racism and exceptional fear surrounding COVID-19 | Open access should be provided to all information through media and printing materials. |
| delay in announcement of Emergency | WHO declared Global health emergency on December 30th (a month later after the outbreak) | Outbreak sternness was not broadcasted and acknowledged widely which delayed the measures | Announce threat status as early as possible after the outbreak |
| Research and development | Lack of resources and funding made a delay in the development of a vaccine for the virus (COVID-19) | The death toll is rising continuously, and 430119 patients worldwide have died due to COVID-19 up till 13th June 2020, | Investments required for developing Effective required treatment. And the establishment of robust methods to beat outbreaks. Budget/funding should be allocated for research. Because only research can help to overcome such outbreaks |
10. Controlling measures for COVID-19 and future directions
The lethal effects of the current epidemic of COVID-19 can be controlled or reduced by decreasing the person-to-person contacts in society. The peoples must follow the principles of herd immunity and social distancing (Figure 6–8).
Figure 6.
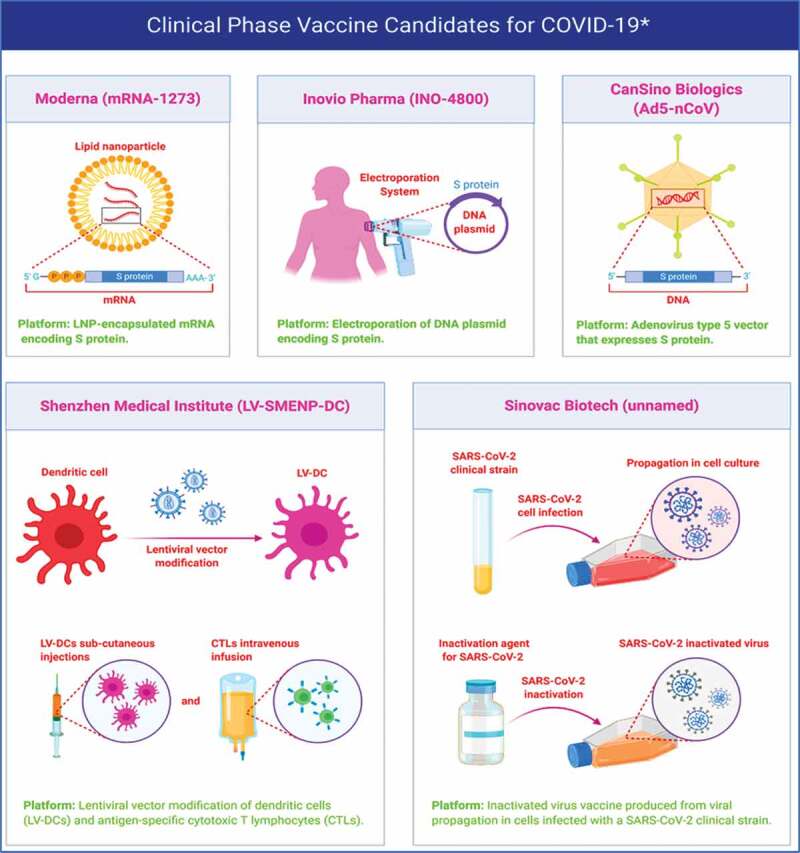
Different vaccine candidates in clinical phase for the treatment of COVID-19. The Figure was created with “BioRender.com” template and exported under the terms of premium subscription
Figure 7.

Potential repurposed monoclonal antibodies for the treatment of COVID-19. The Figure was created with “BioRender.com” template and exported under the terms of premium subscription
Figure 8.
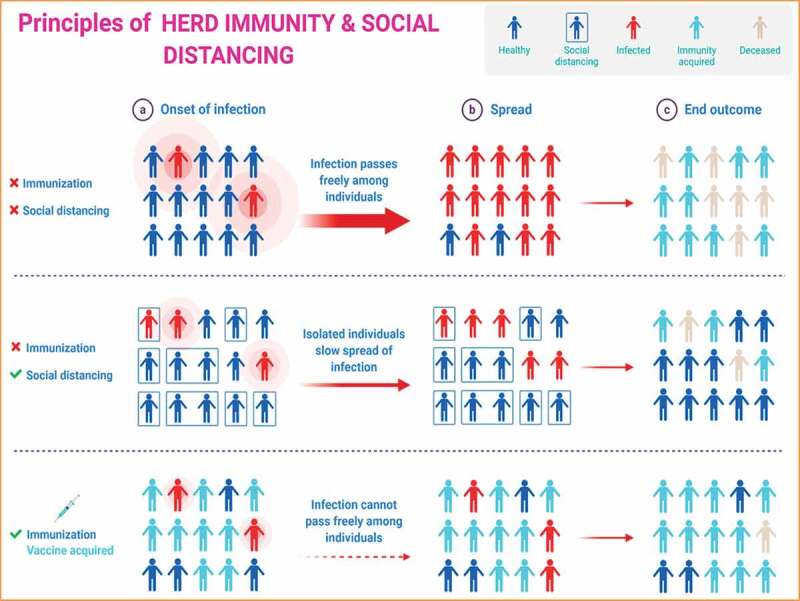
Principles of herd immunity and social distancing. The Figure was created with “BioRender.com” template and exported under the terms of premium subscription
Extraordinary efforts and responsiveness are required to reduce or protect the transmission in vulnerable residents, including children, health officials, and aged individuals. The persons such as healthcare providers, researchers, medical staff, that are dealing with public health in this pandemic must follow the strict guidelines, which had already been directed by WHO.22 Many reports suggest that the proliferation rate of SARS-CoV-2 infections are very high in individuals with weaker immunity. The elderly persons and children who suffered from COVID-19 are more susceptible to death because they have weaker immunity. Moreover, the patients already underwent from other diseases such as diabetes, blood pressure, heart disease, etc. are also more vulnerable to this pandemic, and more death rate was observed in them due to COVID-19.76 As still, no drug or vaccine is available to treat COVID-19, so only solutions possible to control and reduce this pandemic are by following the particular hygienic outlines such as washing hands regularly with some antiseptic soaps or liquid hand wash, by using sanitizers, covering the face with some surgical masks, etc. directed by WHO. Furthermore, the physical interaction with contaminated and wet objects can be considered to deal with the virus, particularly agents such as urine and fecal samples, which can function as an alternative means of transmission.79 Significant prevention measures have been implemented by China and other countries of the world to control the spread of COVID-19. These measures include the screening of travelers, limiting the traveling, and lockdown the peoples to their communities and societies. Epidemiological fluctuations in infection of COVID-19 should be examined by captivating the subclinical infections and possible means of transmission. Additionally, evolution, adaptation, and spreading of the virus among humans and likely intermediate animals and reservoirs can also be controlled by observing the above-discussed guidelines. Several questions are still to be answered, which may include but not limited to details about how many and who have been tested, what fraction of tested ones turned positive, and whether this rate remains constant or variable. Very few infant cases have been reported so far; is this because of the lack of liability/infection or lack of testing? Some basic questions would provide a framework for which more specific and detailed public health measures can be implemented.
11. Concluding remarks
This review provides current perspectives about SARS-CoV-2 on morphology, genetic diversity, transmission characteristics, replication cycle, diagnostic approaches, epidemiological assessment, and prevention strategies. It further entails global research for the development of therapeutic drug candidates vaccines and immunotherapies. The small drug molecules, vaccines, and immunotherapies repurposing efforts summarized in this work are engrossed primarily on agents presently recognized to be operative counter to other RNA viral particles such as Ebola, HCV, influenza, MERS-CoV, SARS-CoV and antimalarial as well as anti-inflammatory drugs. Moreover, the effect of elevated temperature on SARS-CoV-2 infections, causing COVID-19 summarized. Ongoing research on the development of different vaccines by different sectors described comprehensively based on its different developmental stages. Besides, possible immunotherapies based on immunoglobulins and plasma therapy were elaborated extensively, which indicated that immunotherapy could also be an efficient therapeutic option against the COVID-19 pandemic. Although the literature on this pandemic is growing rapidly, we hope this review could be proved beneficial in finding appropriate drug candidates, vaccines and immunotherapies against COVID-19 and further provide the best practice for the treatment and management of symptomatic cases.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Bogoch A, Watts A, Thomas-Bachli C, Huber MUG, Kraemer K.. Khan, Pneumonia of unknown etiology in wuhan, China: potential for international spread via commercial air travel. J. Trav. Med. 2020;27(2). doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H, Stratton CW, Tang YW. outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J. Med. Virol. 2020;92(4):401–02. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. [World Health Organization. Laboratory testing of human suspected cases of novel coronavirus (nCoV) infection: interim guidance; 2020 Jan 10. World Health Organization. https://apps.who.int/iris/handle/10665/330374. License: CC BY-NC-SA 3.0 IGO]. [Google Scholar]
- 4.Calisher C, Carroll D, Colwell R, Corley RB, Daszak RP, Drosten C, Enjuanes L, Farrar J, Field H, Golding J. Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. The Lancet. 2020;395(10226):e42–e43. doi: 10.1016/S0140-6736(20)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci. Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 6.Weiss SR, Leibowitz JL. 2011. Coronavirus pathogenesis. Advances in Virus Research. Vol. 81. 85–164. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi:10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D, Leibowitz JL. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015;206:120–33. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten C, Günther C,S, Preiser W, Van Der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Eng. J. Of Med. 2003;348(20):1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 10.Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Eng. J. Of Med. 2012;367(19):1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends in microbio. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Z, Li H, Wu X, Zhong Y, Zhang K, Zhang YP, Boerwinkle E, Fu YX. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004;4(1):21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo PC, Lau SK, Huang Y, Yuen KY. K, Coronavirus diversity, phylogeny and interspecies jumping. Exp. Bio. And Med. 2009;234(10):1117–27. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 15.Woo PC, Lau SK, Yip CC, Huang Y, Tsoi HW, Chan KH, Yuen KY. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Of Virol. 2006;80(14):7136–45. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benvenuto D, Giovannetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M. The 2019‐new coronavirus epidemic: evidence for virus evolution. J. Med. Virol. 2020;92(4):455–59. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceraolo C, Giorgi FM. Genomic variance of the 2019‐nCoV coronavirus. J. Med. Virol. 2020;92(5):522–28. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;94(7). doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, Xing F, Liu J, Yip CCY, Poon RWS. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet. 2020;395(10223):514–23. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelan AL, Katz R, Gostin LO. The novel coronavirus originating in Wuhan, China: challenges for global health governance. Jama. 2020;323(8):709–10. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- 22.Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, Fang C, Huang D, Huang LQ, Huang Q, et al. Management, f. t. Z. H. o. W. U. N. C.; Research Team, E.-B. M. C. o. C. I. E.; Promotive Association for, M.; Health, C., A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil.Medi.Res. 2020;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, et al. First Case of 2019 Novel Coronavirus in the United States. New Eng. J. Of Med. 2020;382(10):929–36. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO . How does COVID-19 Spread? Geneva: World Health Organization; 2020. [Google Scholar]
- 25.Zhu H, Wang L, Fang C, Peng S, Zhang L, Chang G, Xia S, Zhou W. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Transl.Pedi. 2020;9(1):51. doi: 10.21037/tp.2020.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaimes JA, Millet JK, Stout AE, André NM, Whittaker GR. A tale of two viruses: the distinct spike glycoproteins of feline coronaviruses. Viruses. 2020;12(1):83. doi: 10.3390/v12010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.symptoms https://www.who.int/news-room/q-a-detail/q-a-coronaviruses#:~:text=symptoms . 16 march 2020.
- 28.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurve. 2020;25(3). doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.https://www.roche.com/media/releases/med-cor-2020-03-13.htm [Accessed 2020 Mar 30].
- 30.Al Johani S, Hajeer AH. MERS-CoV diagnosis: an update. J. Of Inf.and Pub. Health. 2016;9(3):216–19. doi: 10.1016/j.jiph.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly-Cirino C, Mazzola LT, Chua A, Oxenford CJ, Van Kerkhove MD. An updated roadmap for MERS-CoV research and product development: focus on diagnostics. BMJ Global Health. 2019;4(2):e001105. doi: 10.1136/bmjgh-2018-001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen PI, Ianevski A, Lysvand H, Vitkauskiene A, Oksenych V, Bjørås M, Telling K, Lutsar I, Dampis U, Irie Y. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int. J. Of Inf. Dis. 2020;93:268–76. doi: 10.1016/j.ijid.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheahan TP, Sims AC, Leist SR, Schäfer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Comm. 2020;11(1):1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Of the Nat. Acad. Of Sci. 2017;114(2):206–14. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Therapeutic options for the 2019 novel coronavirus (2019-nCoV). [Accessed 2020 Mar 15]. https://www.nature.com/articles/d41573-020-00016-0. 2020. [DOI] [PubMed]
- 36.Linghua LI, The efficacy of lopinavir plus ritonavir and arbidol against novel coronavirus infection (ELACOI). ClinicalTrials.gov Identifier: NCT04252885, https://clinicaltrials.gov/ct2/show/NCT042528. 2020.
- 37.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lilly and incyte announce submission of NDA to FDA for oral once-daily baricitinib for treatment of moderate-to-severe rheumatoid arthritis. [Accessed 2020 Apr 1]. https://www.drugs.com/nda/baricitinib_160119.html.
- 39.Richardson P, Griffin I, Tucker C, Smith D, Oechsle O, Phelan A, Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. The Lancet. 2020;395(10223):e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–85. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo MK, Jordan R, Arvey A, Sudhamsu J, Shrivastava-Ranjan P, Hotard AL, Flint M, McMullan LK, Siegel D, Clarke MO. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7(1):43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–71. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PloS One. 2013;8(7):e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mifsud EJ, Hayden FG, Hurt AC. Antivirals targeting the polymerase complex of influenza viruses. Antivir. Res. 2019;169:104545. doi: 10.1016/j.antiviral.2019.104545. [DOI] [PubMed] [Google Scholar]
- 45.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 46.Leneva IA, Russell RJ, Boriskin YS, Hay AJ. Characteristics of arbidol-resistant mutants of influenza virus: implications for the mechanism of anti-influenza action of arbidol. Antivir. Res. 2009;81(2):132–40. doi: 10.1016/j.antiviral.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 47.The efficacy of lopinavir plus ritonavir and arbidol against novel coronavirus infection (ELACOI). [Accessed 2020 Apr 3]. https://clinicaltrials.gov/ct2/show/NCT04252885.
- 48.Ng E. Coronavirus: are cocktail therapies for flu and HIV the magic cure?. South China Morning Post. Bangkok and Hangzhou hospitals put combination remedies to the test. [Accessed 2020. Feb 4]. https://www.scmp.com/business/companies/article/3048888/could-cocktail-therapies-hiv-and-flu-be-magic-cure-new.
- 49.Guo D. Old weapon for new enemy: drug repurposing for treatment of newly emerging viral diseases. Virolo. Sini. 2020;35:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases. The Lancet Inf. Dis. 2003;3(11):722–27. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeldin RK, Petruschke RA. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J. Of Antimirob. Chem. 2004;53(1):4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- 52.Paules CI, Marston HD, Fauci AS. Coronavirus infections-more than just the common cold. Jama. 2020;323(8):707–08. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 53.White AC Jr. Nitazoxanide: a new broad spectrum antiparasitic agent. Expert rev. of anti-infec. Therapy. 2004;2:43–49. [DOI] [PubMed] [Google Scholar]
- 54.McKay BLP, Drugmakers rush to develop vaccines against china virus the wall street journal. [Accessed 2020 Mar 22]. https://www.wsj.com/articles/drugmakers-rush-to-develop-vaccines-against-china virus11579813026.
- 55.Inovio Pharmaceuticals Inc . Inovio selected by cepi to develop vaccine against new coronavirus inovio. [Accessed 2020 Mar 20]. http://ir.inovio.com/news-and-media/news/press-release-details/2020/Inovio-Selected-byCEPI-to-Develop-Vaccine-Against-New Coronavirus/default.aspx.
- 56.University of Queensland Australia . Race to develop coronavirus vaccine. [Accessed 2020 Mar 22]. https://www.uq.edu.au/news/article/2020/01/race-develop-coronavirus-vaccine.
- 57.Hennessy J Australia’s been asked to make a coronavirus vaccine at ‘unprecedented speed’ business insider. [Accessed 2020 Mar 25]. https://www.sciencealert.com/australian-scientists-asked-to-make-coronavirus-vaccine-atunprecedented-speed.
- 58.Reinicke C, An obscure biotech stock skyrockets 38% after saying it’s testing a coronavirus antibody (vir). [Accessed 2020 Mar 28]. https://markets.businessinsider.com/news/stocks/vir-biotechnology-stock-price-surgescoronavirus-antibody-tests-crispr-2020-1-1028847912.
- 59.VIR Vir biotechnology applying multiple platforms to address public health risk from wuhan coronavirus. [Accessed 2020 Mar 20]. https://investors.vir.bio/news-releases/news-release-details/vir-biotechnology-applyingmultiple-platforms-address-public.
- 60.Xinhua, China cdc developing novel coronavirus vaccine xinhua. [Accessed 2020 Mar 29]. http://www.xinhuanet.com/english/2020-01/26/c_138734908.htm.
- 61.Lee JHZW, Zhou L, Chinese scientists race to develop vaccine as coronavirus death toll jumps: South china morning post. https://www.scmp.com/news/china/society/article/3047676/numbercoronavirus-cases-china-doubles-spread-rate-accelerates.
- 62.Cheung E. China coronavirus: hong kong researchers have already developed vaccine but need time to test it, expert reveals. South china morning post. https://www.scmp.com/news/hong-kong/healthenvironment/article/3047956/china-coronavirus-hong-kong-researchers-have.
- 63.China Daily . Novel coronavirus vaccine being developed: China daily. https://www.chinadaily.com.cn/a/202001/26/WS5e2d1768a310128217273481.html.
- 64.Xinhua . China fast-tracks novel coronavirus vaccine development xinhua. [Accessed 2020 Mar 28]. http://www.xinhuanet.com/english/2020-01/28/c_138739378.htm.
- 65.Berkeley L Jr.. Us health officials fast-track coronavirus vaccine, hope to start clinical trial in three months. Cnbcmarkets. https://www.cnbc.com/2020/01/28/us-fast-tracks-coronavirus-vaccinehopes-to-start-trial-in-three-months.html.
- 66.Bursztynsky J. J&j scientific officer ‘pretty confident’ they can create coronavirus vaccine as outbreak widens. Cnbcmarkets. https://www.cnbc.com/2020/01/27/jj-pretty-confident-it-can-create-chinacoronavirus-vaccine.html.
- 67.Saskatchewan lab joins global effort to develop coronavirus vaccine. Canada: Kelly Geraldine Malone; 2020. [Accessed 2020 Mar 29]. https://www.cbc.ca/news/canada/saskatchewan/vido-intervac-working-on-coronavirus-vaccine-1.5439118 [Google Scholar]
- 68.In GeoVax . Geovax and bravovax (wuhan, china) to collaborate on development of coronavirus vaccine. [Accessed 2020 Mar 27]. https://www.geovax.com/news/geovax-and-bravovax-wuhan-china-to-collaborate-ondevelopment-of-coronavirus-vaccine.
- 69.Clover Biopharmaceuticals . Clover initiates development of recombinant subunit- trimer vaccine for wuhan coronavirus (2019-ncov). [Accessed 2020 Apr 2]. http://www.cloverbiopharma.com/index.php?m=content&c=index&a=show&catid=11&id=40.
- 70.Precision Vaccinations . Mrna vaccines can induce immune responses for 2019- ncov. [Accessed 2020 Mar 28]. https://www.precisionvaccinations.com/curevac-mrna-platform-specifically-suitable-rapidlyprovide-answer-viral-outbreak-situation-novel.
- 71.Usdin S, J&j developing coronavirus vaccine, at least nine other vaccines under development. [Accessed 2020 Mar 30]. https://www.biocentury.com/bc-extra/company-news/2020-01-27/jj-developing-coronavirus-vaccineleast-nine-other-vaccines-under-.
- 72.Shanmugaraj B, Siriwattananon K, Wangkanont K, Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac. J. Allergy Immunol. 2020;38(1):10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 73.Amin JA, Ghasemi S. The possible of immunotherapy for COVID-19: A systematic review. Int. Immunopharmacol. 2020;2:106455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu T, Zhang J, Yang Y, Zhang L, Ma H, Li Z, Zhang J, Cheng J, Zhang X, Wu G, et al. The potential role of IL-6 in monitoring coronavirus disease 2019. 2020. doi: 10.1101/2020.03.01.20029769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dalziel BD, Kissler S, Gog JR, Viboud C, Bjørnstad ON, Metcalf CJE, Grenfell BT. Urbanization and humidity shape the intensity of influenza epidemics in US cities. Sci. 2018;362(6410):75–79. doi: 10.1126/science.aat6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. New Eng. J. Of Med. 2020;382(13):1199–207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Tang K, Feng K, Lv W, High temperature and high humidity reduce the transmission of COVID-19. Available at SSRN 3551767 (2020).
- 78.Financial Times , China’s Xi Jinping knew of coronavirus earlier than first thought, 2020. [Accessed 2020. Mar 16]. https://www.ft.com/content/3da73290-5067-11ea-8841-482eed0038b1.
- 79.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeen RF, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13(9):752–61. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


