Summary
αβ lineage T cells, most of which are CD4+ or CD8+ and recognize MHC I or MHC II-presented antigens, are essential for immune responses and develop from CD4+CD8+ thymocytes. The absence of in vitro models and the heterogeneity of αβ thymocytes have hampered analyses of their intrathymic differentiation. Here, combining single-cell RNA- and ATAC- (chromatin accessibility) sequencing, we identified mouse and human αβ thymocyte developmental trajectories. We demonstrated asymmetric emergence of CD4+ and CD8+ lineages, matched differentiation programs of agonist-signaled cells to their MHC specificity, and identified correspondences between mouse and human transcriptomic and epigenomic patterns. Through computational analysis of single cell data and binding sites for the CD4+lineage transcription factor Thpok, we inferred transcriptional networks associated with CD4+- or CD8+lineage differentiation, and with expression of Thpok or of the CD8+-lineage factor Runx3. Our findings provide insight into the mechanisms of CD4+ and CD8+ T cell differentiation, and a foundation for mechanistic investigations of αβ T cell development.
eToc blurb
Chopp et al use single-cell RNA- and ATAC sequencing to define developmental trajectories of mouse and human αβ thymocytes. Their findings identify differentiation programs specific of agonist-signaled cells and of CD4+ and CD8+ lineages and infer gene regulatory networks involved in cell fate decisions.
Graphical Abstract
Introduction
αβ lineage T cells, defined by the expression of an αβ T cell antigen receptor (TCR), are essential for immune responses. These cells include “conventional” T cells, recognizing peptide antigens bound to classical Major Histocompatibility Complex (MHC)-I and MHC-II molecules, and other lineages of diverse functions. αβ T cells develop in the thymus from precursors that express both CD4 and CD8 coreceptors (“double positive” [DP] thymocytes) (Carpenter and Bosselut, 2010; Rothenberg, 2019); in contrast, mature conventional T cells express only CD8 if MHC I-restricted or CD4 if MHC IIrestricted, whereas other αβ lineage cells express either coreceptor or neither.
Interactions between the αβ TCR and intrathymic MHC or MHC-like ligands determine the fate of DP thymocytes (Hogquist and Jameson, 2014; Palmer, 2003; Starr et al., 2003; Stritesky et al., 2012). Cells with moderate affinity for self MHC peptide are rescued from cell death (positive selection), and become conventional CD4+ or CD8+ single positive (SP) thymocytes and T cells (Singer et al., 2008; Taniuchi, 2018; Xiong and Bosselut, 2012). In contrast, cells with high affinity for intrathymic ligands are actively deleted (negative selection), or differentiate into diverse “agonist-selected” fates, including regulatory T (Treg) cells or precursors of CD8α+ CD8β− (CD8αα) gut intra-epithelial lymphocytes (IELp) (Li and Rudensky, 2016; McDonald et al., 2018; Ohkura et al., 2013; Ruscher and Hogquist, 2019).
The development of these thymocyte lineages and the transcriptional programs that underlie their differentiation are incompletely understood, in part because of the heterogeneity characteristic of thymocyte populations. Commitment transcription factors have been identified for specific lineages, including Thpok (encoded by Zbtb7b) for CD4+ differentiation and Runx3 for CD8+ differentiation (He et al., 2005; Sun et al., 2005; Taniuchi et al., 2002; Woolf et al., 2003). However, much remains to be learned on what controls the expression of such factors and on their target gene sets. Genetic studies are limited by the absence of suitable in vitro approaches to study αβ thymocyte differentiation, and are often confounded by pleiotropic effects or functional redundancy between paralog genes, e.g. between Runx3 and the related Runx1 molecule (Collins et al., 2009; Egawa et al., 2007; Woolf et al., 2003).
Single-cell analysis of gene expression and chromatin accessibility maps developmental trajectories with minimal bias from prior knowledge (Buenrostro et al., 2015; Papalexi and Satija, 2018), unlike population-based analyses (Mingueneau et al., 2013; Yoshida et al., 2019). However, previous scRNAseq studies of the thymus neither focused on αβ lineages nor compared the trajectories of MHC I- and MHC II-signaled thymocytes (Kernfeld et al., 2018; Lavaert et al., 2020; Le et al., 2020; Park et al., 2020), and therefore could not map the divergence of conventional CD4+ or CD8+ lineages or associate the fate of agonist-signaled thymocytes with their MHC restriction. Furthermore, while chromatin organization is a defining feature of cell identity (Sen et al., 2016), how transcriptomic changes map to the chromatin dynamics in developing αβ lineage thymocytes is unclear.
Here we leveraged single cell approaches to identify the transcriptomic and epigenomic features of αβ thymocytes. We mapped divergence branch points between conventional and agonist-selected lineages, and the asynchronous emergence of CD4+- and CD8+ lineage differentiation programs at early and late stages of thymocyte development, respectively. Integration of transcriptomic and epigenomic data enabled the inference of gene regulatory networks associated with differentiation into either lineage. Last, we documented the conservation of these transcriptomic and epigenomic patterns in human thymocytes. These developmental maps and associated regulatory networks present a resource towards deciphering the mechanisms of αβ T cell lineage differentiation.
Results
Defining signatures for the CD4+ and CD8+ transcriptomes
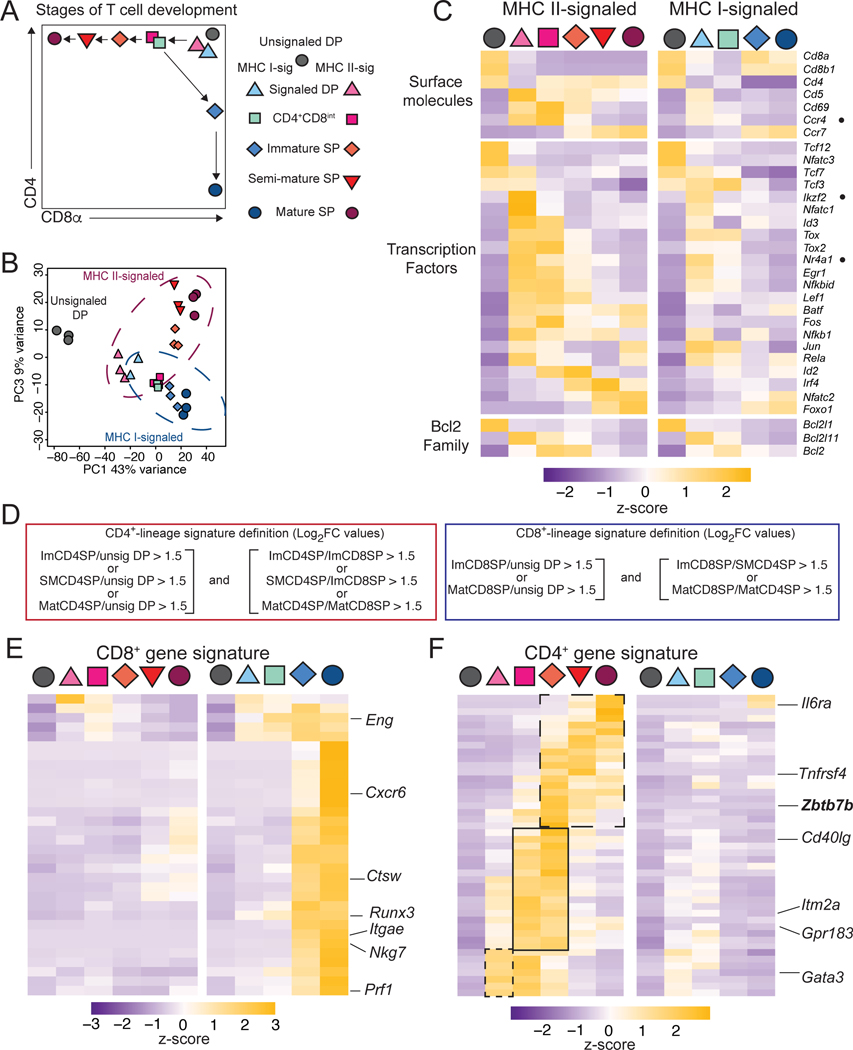
We used RNAseq to compare subsets of developing CD4+ and CD8+ T cells. We sorted cells based on expression of CD4 and CD8, CD69 (a TCR signaling marker), and MHC-I, expressed on the most mature thymocytes (Xing et al., 2016). In addition to unsignaled (CD69–) and signaled (CD69+) DP cells, we purified immature (CD69hi MHC-Ilo) CD4+CD8int and SP, semi-mature (CD69hi MHC-Ihi) SP, and mature (CD69lo MHC-Ihi) SP thymocytes (Figs. 1A and S1A–C). We used cells carrying reporter alleles for the transcription factors Thpok (encoded by Zbtb7b) and Runx3, specifically expressed in CD4+- and CD8+-lineage thymocytes, respectively (Wang et al., 2008b; Zamisch et al., 2009). These cells were obtained from chimeric mice generated by transplanting irradiated MHC II (H2Ab1)- or MHC I (B2m)-deficient hosts with bone marrow precursors carrying the reporter alleles (Fig. S1BC). Principal Component Analysis (PCA) grouped experimental replicates together (Figs. 1B and S1D). Among the first three components (PC), accounting for 79% of the total variability, PC1 was enriched in genes corresponding to lineage-independent developmental maturation (e.g. Rag1, Dntt, Il7r, S1pr1), and PC3 in genes involved in CD4+-CD8+ lineage differentiation (e.g. Zbtb7b, Cd4, Cd8a, Itgae); PC2, comprising a diverse array of genes, roughly paralleled expression of TCR-induced genes (e.g. Cd69) across thymocyte subsets (Table S1).
Figure 1: Transcriptomic analyses of thymocyte subsets.
(A) Populations sorted for RNA-seq analyses. Unsignaled (grey) MHC I- (blue) and MHC II- (red) signaled subsets are shown on a schematic CD4-CD8 expression plot.
(B) PCA displays cell subsets according to PC1 and PC3. Each symbol represents an individual biological replicate.
(C) Heatmap shows row-standardized (Z-scores of reads per million [RPM] values) mRNA expression in populations identified as defined in (A).
(D) Definition of CD4+ and CD8+ signature genes shown in (E,F).
(E, F) Heatmaps (as in [C]) of CD8+- (E) and CD4+- (F) lineage signature gene expression. CD4+ signature genes differing by their kinetics of expression are boxed (F).
See also Figure S1
Differential gene expression was observed both across developmental stages and between lineages (Fig. 1C). We defined CD4+ and CD8+-lineage signatures as sets of genes with greater expression in CD4+- or CD8+-lineage cells, respectively, relative to each other and to unsignaled DP cells (Fig. 1D and Table S1). We based signature definitions on gene expression in immature, semi-mature and mature cells of each lineage, to include genes that would be transiently expressed in lineagedifferentiating cells. Most CD8+ signature genes were expressed in both immature and mature CD8+ SP cells with little or no expression detected at previous stages of MHC I-restricted cell differentiation (Fig. 1E). In contrast, the CD4+ signature was detected earlier and included components with distinct kinetic patterns (Fig. 1F).
Unbiased transcriptomic analysis of αβ lineage thymocyte differentiation
At the signaled DP stage, there were trends towards preferential expression of CD4+-signature genes, and of Ccr4, in MHC II-signaled cells (Figs. 1C, F). However, other results suggested heterogeneity of that population. A few MHC I-signaled DP cells expressed the chemokine receptor Ccr7, which promotes cell migration from the thymic cortex to the medulla (Kwan and Killeen, 2004; Ueno et al., 2004; Yin et al., 2007), (Fig. S1E). Furthermore, expression of genes encoding transcription factors Nur77 (Nr4a1) and Helios (Ikzf2), or the proapoptotic protein Bim (Bcl2l11) suggested that the signaled DP subsets included cells targeted for negative selection (Fig. 1C) (Bouillet et al., 2002; Daley et al., 2013; Marsden and Strasser, 2003; Stritesky et al., 2013). Thus, population-based RNAseq could not unambiguously identify positive selection and lineage differentiation transcriptomic programs, prompting us to compare MHC-I and MHC II-signaled thymocytes by scRNAseq.
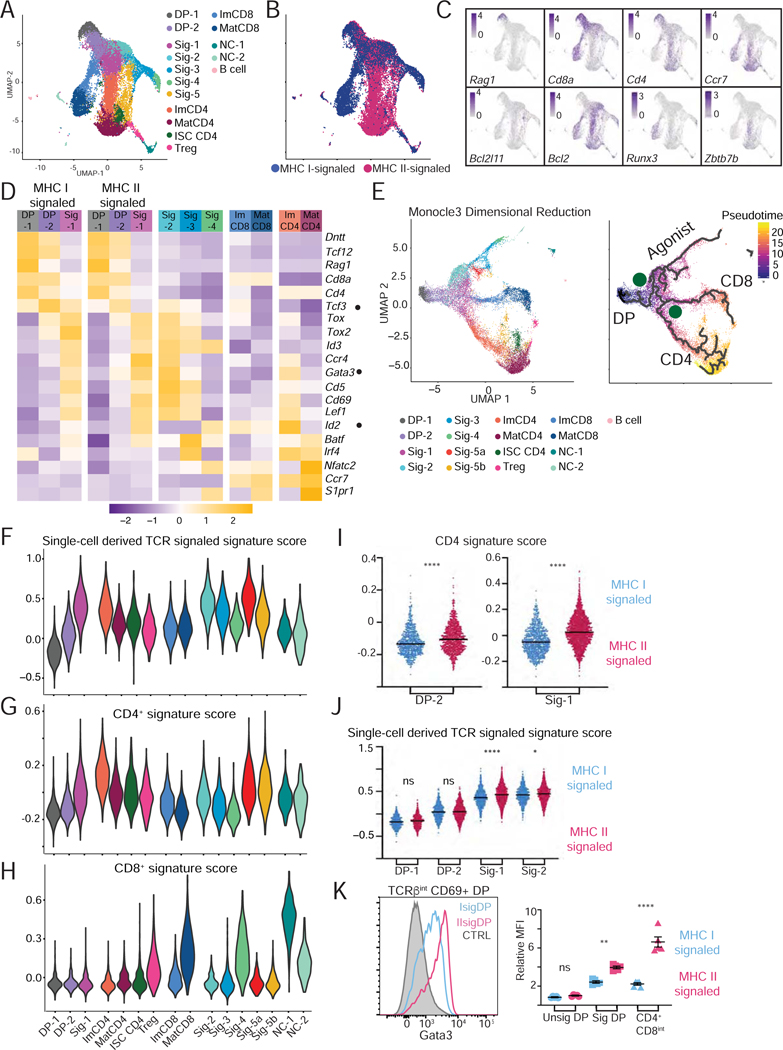
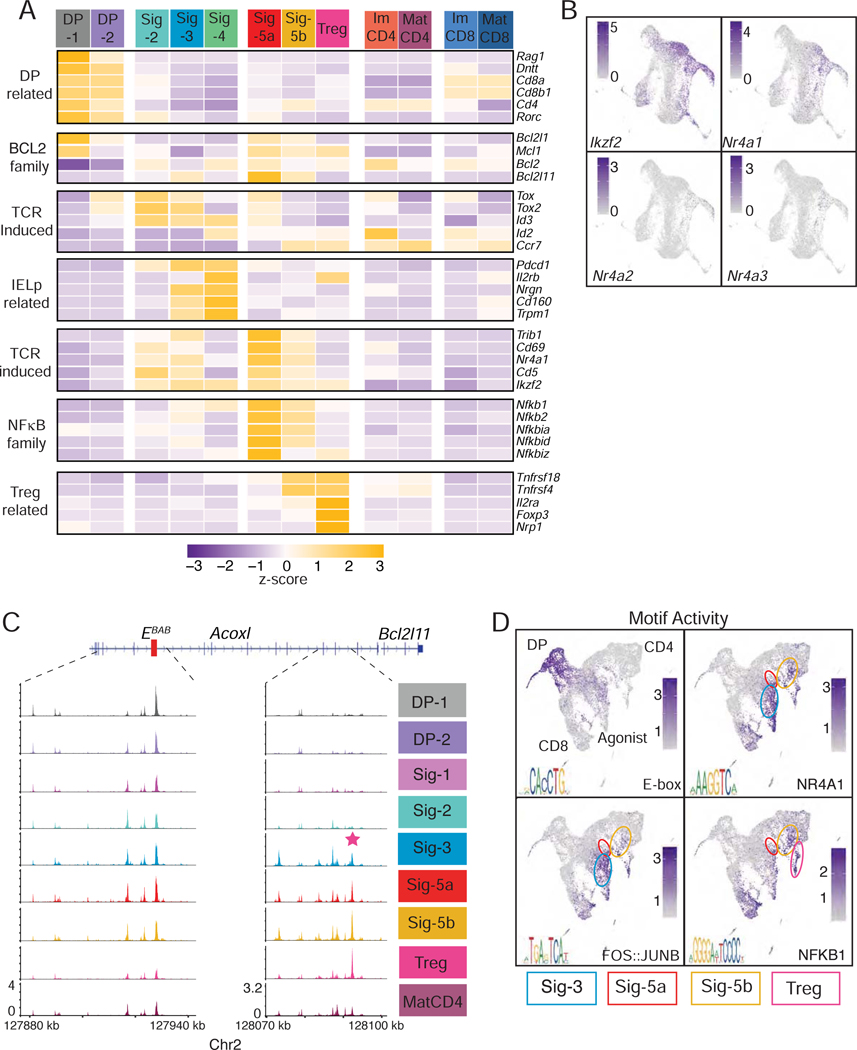
We sorted CD69lo/+ thymocytes, from either H2-Ab1−/− or B2m−/− mice, separately captured them using the 10x Genomics Chromium platform and processed them for scRNAseq (Chung et al., 2017) (Fig. S2AB, Table S2 for QC metrics). We performed two independent captures for each genotype, analyzed data with the Seurat R package (Butler et al., 2018), and integrated data from both captures to remove experimental batch effect (Stuart et al., 2019). Following PCA and dimensional reduction, unsupervised clustering distributed the 18,644 total cells into 16 clusters (Fig. 2A and S2C). We verified that clustering was not driven by batch effect between experiments (Fig. S2D); the same was true for all single cell captures reported in this study (as shown for each experiment). Clusters were largely segregated from each other when projected onto a UMAP dimensional reduction plot of the integrated data (Fig. 2A; see Table S1 for cluster-specific genes), except cluster Sig-5 which was split into two components (Sig-5a and Sig-5b, Fig. S2E). Eight clusters included both MHC I- and MHC IIsignaled cells, whereas the other eight predominantly or exclusively comprised only one (Figs. 2B and S2F). Expression of key genes was used to tentatively name MHC I- or MHC II-specific clusters along two CD4+ and CD8+ “arms” (Fig. 2A–D and Table S1, and discussed hereafter). The CD4+ arm included a cluster with features of Treg cells and another (ISC CD4) with marks of signaling by type Iinterferon, a cytokine known to contribute to gene expression in mature thymocytes (Xing et al., 2016) (Figs. 2A and S2G, and Table S1).
Figure 2: Single cell RNAseq analysis of mouse αβ lineage thymocyte selection.
(A-H) MHC I- and MHC II-signaled thymocytes (see Fig. S2AB) were analyzed by scRNAseq. Data integrates two biological replicates from each genotype.
(A) UMAP plot displays thymocytes color-coded according to their distribution into clusters (names on the right). Sig = signaled; Im = immature; Mat = mature; NC = non-conventional; ISC = interferon stimulated cluster.
(B) Same UMAP plot as in (A) with cells colored according to their MHC restriction.
(C) Scaled expression of indicated genes is shown on UMAP plots.
(D) Heatmap of average gene expression in indicated clusters (right three sets of columns) or in their MHC-I or MHC II-signaled subsets (left two sets of columns). Data is row-standardized.
(E) (Left) UMAP plot of thymocytes, generated after Monocle3-derived dimensional reduction, colorcoded by Seurat-defined clusters. (Right) developmental trajectories (thick lines) defined by pseudotime analysis is shown on cells color-coded according to their pseudotime value. DP: DP thymocytes; Agonist, CD4, and CD8 indicate the trajectories emerging at branchpoint S (selection) and L(lineage).
(F-H) Violin plots show cluster-based average expression scores of (F) TCR signal induced genes, (Table S1), and (G) CD4+- or (H) CD8+-lineage signature genes (Fig. 1D–F and Table S1).
(I) Expression score of CD4+ signature genes in DP-2 and Sig-1 cells, MHC I- (blue) vs. MHC II- (pink) signaled. **** p < 0.0001 (unpaired student’s t-test with Welch’s correction).
(J) Expression score of TCR signal induced genes in DP-1, DP-2, Sig-1, and Sig-2 cells displayed as in (I). * p < 0.01; **** p < 0.0001 (one-way ANOVA and Sidak’s Multiple Comparisons Test).
(K) Overlaid histograms (left) and mean fluorescence intensity (MFI, right) of intracellular Gata3 protein on signaled (CD69+) DP thymocytes from B2m- or H2-Ab1-deficient mice, gated as in Fig. S1E. MFI is expressed relative to CTRL (wild-type CD69– DP thymocytes). ** p < 0.005, **** p < 0.0001 (one-way ANOVA and Sidak’s Multiple Comparisons Test). Error bars show SEM.
See also Figure S2
In contrast, most shared clusters could not be unambiguously positioned along known developmental paths. Most expressed both Cd4 and Cd8a and had low expression of Ccr7, a pattern characteristic of cortical thymocytes (Fig. 2C). Cluster DP-1 showed the highest expression of Rag1, indicating that it included pre-selection cells. In contrast, others (“Sig”) exhibited marks of TCR signaling. To determine developmental relationships between these clusters, we performed pseudotime mapping using Monocle 3, which infers developmental trajectories from scRNAseq data (Trapnell et al., 2014). Verifying the consistency of Monocle and Seurat analyses, cells color-coded according to their Seurat-defined cluster were grouped together on a UMAP plot generated from the Monocle-processed data (Fig 2E, left). Pseudo-time mapping displayed the cells along an asymmetric trajectory (Fig. 2E, right) with three main endpoints corresponding to clusters Sig-4, mature CD4+ (MatCD4) and CD8+ (MatCD8) SP thymocytes, and two main branch-points, designated S (selection) and L (lineage).
Cells along the common stem sequentially mapped to clusters DP-1, DP-2, and Sig-1 (Fig. 2E). Their progression was marked by cessation of Rag1 expression, increased expression of TCR signaling targets Cd5, Cd69 and Tox (Azzam et al., 1998; Swat et al., 1993; Turka et al., 1991; Wilkinson et al., 2002) (Fig. 2C–E), and increased scores for a TCR signaling signature composed of genes induced during the DP-1 to Sig-1 transition (Fig. 2F). This signature also indicated higher TCR signaling in CD4+ than CD8+ immature clusters (consistent with previous analyses, Moran et al., 2011). Expression of pro-survival Bcl2 (Marsden and Strasser, 2003) increased along the common stem and was high in CD4+ and CD8+ arm cells, whereas pro-apoptotic Bcl2l11 (Bim) was detected only in agonist arm clusters (Fig. 2C). Thus, we reasoned that DP-2 and Sig-1 cells were signaled for positive selection, allowing us to track the emergence of lineage-specific transcriptomic patterns.
MHC-specific transcriptomic divergence in signaled double positive thymocytes
To map the divergence of lineage-specific programs, we scored each cell for CD4+-and CD8+-lineage transcriptomic signatures defined in Fig. 1D. Positive scores for the corresponding signature were found among clusters within CD4+ and CD8+ branches, with the same kinetics of appearance as in population RNAseq (Fig. 2GH). Elevated CD4+ signature scores were detected as early as the DP-2 cluster (Fig. 2G), and were higher in MHC II- than in MHC I-signaled cells (Fig. 2I); as expected, scores for that signature were low in Rag1-and Rag2-expressing (pre-selection) DP-1 thymocytes, regardless of their genotype (data not shown). The scoring asymmetry between MHC I- and MHC II-signaled cells was specific of the CD4+ signature, as it was not detected with the TCR signaling signature (Fig. 2J). It was associated with decreasing expression of Cd4, Cd8a and Cd8b1 in DP-2 and Sig-1 clusters, with similar kinetics in MHC I- and MHC II-signaled cells (Fig. 2D and S2H). The CD4+ signature included Gata3, a factor needed for CD4+-lineage differentiation (Hernandez-Hoyos et al., 2003; Pai et al., 2003) and Id2, an inhibitor of E-protein activity (Kee, 2009), but not its paralog Id3. We verified higher expression of Gata3 protein in MHC II- than MHC I-signaled DP cells (Fig. 2K).
Strong correspondence of epigenomic and transcriptomic dynamics
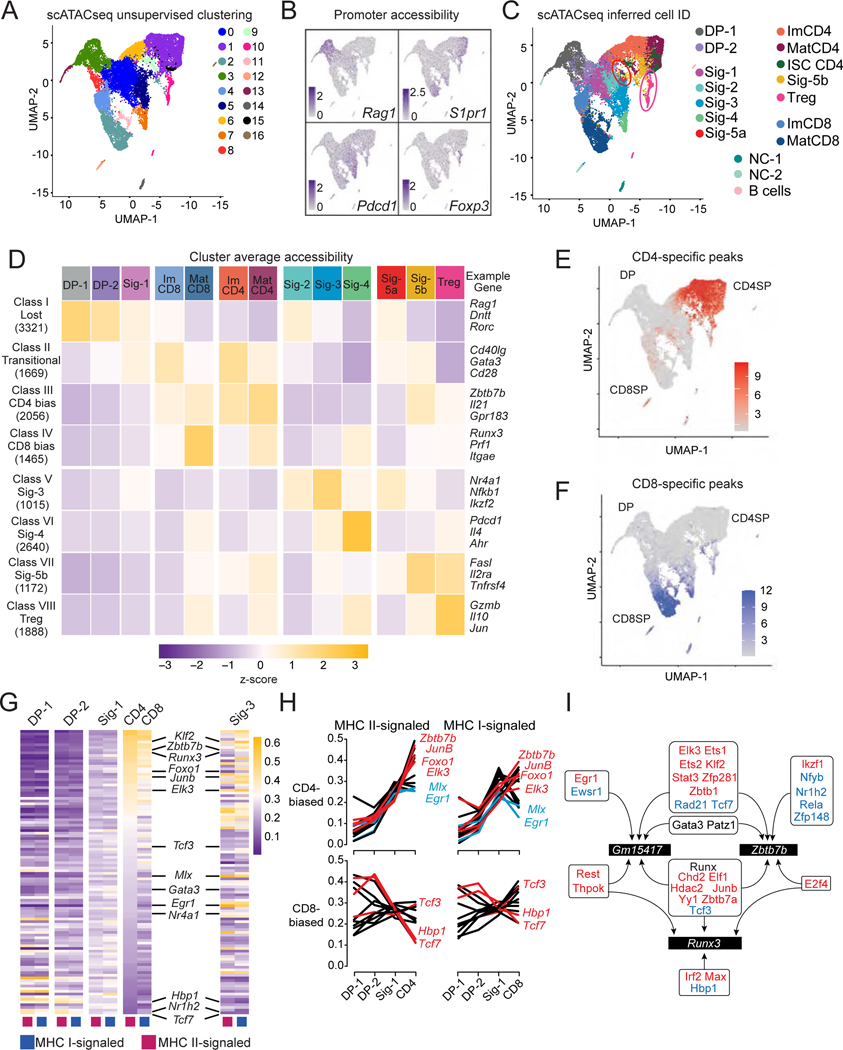
Cell differentiation is associated with changes in chromatin accessibility, which can be detected by Assay for Transposase Accessible Chromatin (ATACseq). Thus, we performed single-cell ATACseq (scATACseq) (Buenrostro et al., 2015) on MHC I- and MHC II-signaled thymocytes purified as for scRNAseq; cells were captured using 10x Genomics and data analyzed using the Signac extension of Seurat. Following normalization and dimensional reduction, we identified 17 cell clusters (Fig. 3A, Table S2), some containing both MHC I- and MHC II-signaled cells and others unique to either population (Fig. S3AB). We verified that this clustering was not simply driven by signal at and near transcription start sites by repeating the analysis after excluding sequencing reads mapping to gene promoters (Figs. S3D). To estimate gene expression in each of the scATACseq cells, we assumed, for each gene, RNA expression from high promoter accessibility (illustrated on Fig. 3B) (Stuart et al., 2019). From such an imputed transcriptome, each scATACseq cell was assigned an “inferred identity” (IID), corresponding to the scRNAseq cluster containing the best matching scRNAseq cell (Fig. 3C). Projecting IID (color-coded by scRNAseq clusters) onto the scATACseq UMAP plot grouped cells with a shared IID in a pattern overlapping that generated by scATACseq clustering (Fig. 3A, C). Such overlap between scATACseq clusters and IID groups indicated that epigenomic and transcriptomic analyses distinguished similar stages of cellular differentiation.
Figure 3: Single cell ATACseq analyses.
(A-J) MHC I- and MHC II-signaled thymocytes were prepared as in Fig. S2AB for scATACseq. Data integrates two biological replicates from each genotype.
(A) UMAP plot displays thymocytes color-coded according to their cluster distribution.
(B) UMAP plots shows scaled accessibility scores at promoter (2 kb segment upstream of transcription start site) of indicated genes.
(C) UMAP plot shows cells color-coded according to scRNAseq-based IID. NC = non-conventional. Circles denote Sig-5a (red) and Tregs (pink).
(D) Heatmap shows row-standardized accessibility of 15,226 OCR differentially accessible between any two IID-defined cell groups (columns) and classified by K-means clustering (rows). Gene assignment of OCRs performed by Homer; example genes are indicated.
(E, F) UMAP plot shows scaled accessibility scores for individual cells at CD4+- and CD8+-lineage specific peaks.
(G) Heatmap shows gene set expression scores for each of the 106 CellOracle defined transcription factor activities (TA) (rows) in MHC I- and MHC II-signaled cells from indicated clusters (columns). TA are ranked by decreasing score in MHC II-signaled ImCD4 cells.
(H) Line graphs depict the scores of the top 15 significantly CD4+-biased (top) and the 12 significantly CD8+-biased TA (bottom) across MHC II-signaled (left) and MHC I-signaled (right) DP-1, DP-2, Sig-1, ImCD4 and ImCD8 cells.
(I) Schematic showing transcription factors predicted by CellOracle and AU cell to bind the Runx3 or Zbtb7b locus (including Zbtb7b and Gm15417). Red and blue font indicate factors identified as CD4+- and CD8+-biased, respectively. Gata3, Runx factors and Patz1 (Mazr), identified as neither CD4+- nor CD8+-biased, were previously shown to bind Zbtb7b and Gm15417 (see text for references). Arrows indicate predicted binding.
See also Figures S2–3 and Tables S3–S6
To evaluate chromatin dynamics, we identified 15,226 peaks with differential accessibility across cell groups defined by a shared IID. The resulting peaks were scored across cell groups sharing the same IID; k-means clustering on those peaks generated eight clusters, called peak classes here for clarity. Accessibility at peak classes I and V-VIII was associated with DP thymocytes and agonist-signaled cells, respectively (Fig. 3D) (discussed later). In contrast, classes II-IV were preferentially accessible in ImCD4, MatCD4, ImCD8 and MatCD8 groups, although most of the variability mapped with cell maturation rather than lineage differentiation. We found similar results in population ATACseq analyses on sorted unsignaled DP, MHC I- and MHC II-signaled DP, and immature CD4+ and CD8+ SP thymocyte (data not shown).
The limited variability associated with lineage differentiation in classes II-IV (Fig. 3D) prompted us to examine if the divergence of CD4+- and CD8+-lineage transcriptomes was matched at the chromatin level. Accessibility scores for genes included in the CD8+ and CD4+ transcriptomic signatures were elevated in cells with the corresponding inferred SP thymocyte IID but remained low in cells with a signaled DP thymocyte IID (Fig. S3EF); thus, chromatin accessibility is lineage-specific in CD4+ and CD8+ SP cells. Because lineage-specific enhancers can be distant from the genes they control, we examined 1,115 chromatin regions differentially accessible in CD4+ vs. CD8+ lineage cells, regardless of gene proximity. Accessibility scores for this set were uniformly low in DP cells (Fig. 3EF). Neither the CD4+-lineage specific peaks at Cd40lg and Zbtb7b, nor the few CD8+ lineage-specific peaks upstream of Runx3 were accessible in MHC I- and MHC II-signaled DP-1 cells (Fig. S3G). Functionally identified Runx3 enhancers (Kojo et al., 2017) were similarly accessible in DP, CD4+ SP and CD8+ SP cells, whereas no lineage-specific peak was associated with Gata3 (Fig. S3G and data not shown). Thus, there was no evidence for epigenomic “poising” of pre-selection DP thymocytes towards either lineage. Last, we observed epigenomic opening at genes involved in mature T cell effector functions (Shih et al., 2014): it was largely CD4+-lineage specific at Il21 and Cd40lg (Fig. S3GH), whereas CD8+-lineage preferential peaks opened at the Ifng and Gzmb loci.
Transcriptional regulators driving CD4+ and CD8+ differentiation
We next sought insight into the transcriptional control of CD4+-CD8+ lineage differentiation, including of Zbtb7b or Runx3 expression. We used CellOracle (Kamimoto et al., 2020), which integrates transcription factor motif analysis with scRNAseq and scATACseq data to identify putative target gene sets for transcription factors. We defined target sets in DP-2 and Sig-1 clusters, that include both MHC I- and MHC II-signaled cells, for each of the 106 factors expressed in at least one of the scRNAseq-defined clusters (Table S3). Although assigned to specific factors, target gene sets are best understood as associated with transcriptional activities (TA) sharing similar DNA binding motifs, e.g. within transcription factor families. To quantify TA in differentiating cells, we scored the expression of each target set, separately in cells of each genotype, across common-stem and immature CD4+- and CD8+-lineage clusters, and in the agonist-signaled Sig-3 cluster (Table S4).
Scores for most TA were similar across genotypes in early (DP-1, DP-2 and Sig-1) clusters (Fig. 3G & Table S4), presumably because transcriptomic differences related to MHC specificity were modest at these differentiation steps compared to the broad size of the target sets inferred by CellOracle (Median gene number: 1608, Inter-Quartile Range: 799–3302). In contrast, scores for 39 TA were significantly different between ImCD4 and ImCD8 clusters (with 27 scoring higher in ImCD4 and 12 higher in ImCD8, Table S4). Some CD8+-biased TA were inhibited by TCR signaling (e.g. Tcf3 encoding E2A, inhibited by TCR-induced Id2) or involved in CD8+-lineage differentiation (Tcf7, Hbp1) (Sekkali et al., 2005; Steinke et al., 2014); all showed unchanged or decreasing activity as cells progressed from DP to CD8+ SP (Fig. 3H). In contrast, the vast majority of CD4+-biased TA increased during the DP to CD4+ SP differentiation, and comprised factors induced by TCR signaling, including Elk3, JunB (an AP-1 family member) and Egr1 (Fig. 3H). There was no correspondence between TA scores in Sig-3 and in lineage-differentiating clusters (Fig. 3G): some CD4+-biased TA scored similarly in Sig-3 and ImCD4 cells (e.g. Elk3, JunB), whereas others had lower (e.g. Zbtb7b, Foxo1) or higher (e.g. Egr1 or Mlx) scores in Sig-3 than ImCD4 cells (Fig. S3I). This indicated that thymocyte fates were associated with specific TA combinations.
CellOracle identified 58 TA putatively targeting Zbtb7b, or the non-coding RNA Gm15417 that overlaps with the Zbtb7b silencer, and 31 TA targeting Runx3 (Table S6). Zbtb7b-targeting TA included known Zbtb7b activators (Gata3) or repressors (Runx1 or Runx3); many were CD4+- or CD8+-biased, including some predicted to bind both Zbtb7b and Runx3 (Fig. 3I). Thus, computational analyses leveraging both transcriptomic and epigenomic data identify transcriptional activities putatively associated with CD4+ or CD8+-lineage differentiation, including with Zbtb7b and Runx3 expression.
Genomic and transcriptomic impacts of Thpok during CD4+ T cell differentiation
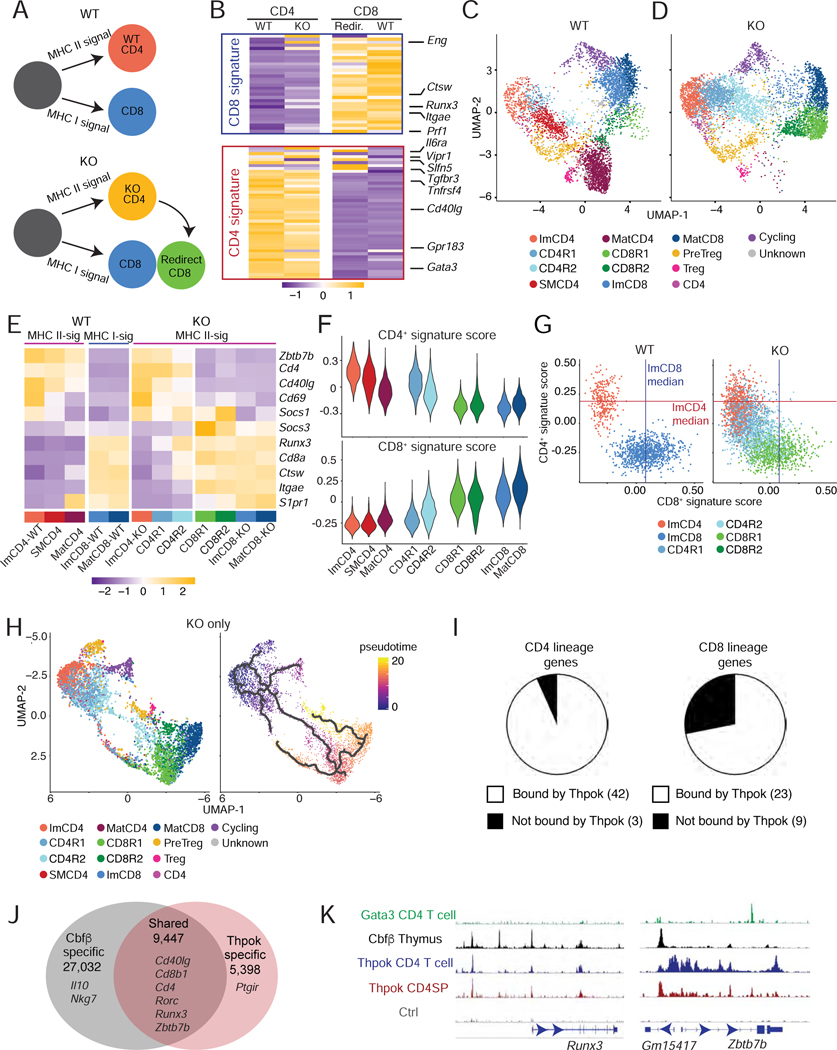
These results suggested implementation of a CD4+-specific gene regulatory network in the ImCD4 scRNAseq cluster, which scored highest for the CD4+ gene signature (Fig. 2G) and had high Zbtb7b expression (Fig. 2C, bottom right). Thus, we examined how Thpok affected gene expression in MHC II-restricted thymocytes. We first performed RNAseq on sorted populations of Thpok-sufficient (“WT”) and -deficient (“KO”) thymocytes, the latter from mice in which expression of Cd4-Cre deletes Zbtb7bfl alleles in DP cells. Both strains carried the Zbtb7bGFP bacterial artificial chromosome (BAC) reporter (Wang et al., 2008b), which results in GFP expression in MHC II-signaled cells, including in Thpok-deficient thymocytes as they become immature CD4+ SP cells and are “redirected” into CD8+ SP cells (Figs. 4A and S4A) (He et al., 2005). We compared (i) MHC II-restricted (Zbtb7bGFP+) Thpok-sufficient and -deficient immature (MHC-Ilo CD69+) CD4+ SP cells, (ii) “redirected” MHC II-restricted (Zbtb7bGFP+) CD8+ SP cells, and (iii) wild-type (MHC I-restricted) CD8+ SP cells (Figs. 4A and S4A). Zbtb7b disruption almost fully converted the transcriptome of redirected MHC II-restricted CD8+ SP cells to that of wild-type CD8+ SP cells, demonstrating that Thpok is required to maintain the CD4+ transcriptome (Fig. 4B). Immature Thpok-deficient CD4+ SP thymocytes expressed CD8+ signature genes, notably Runx3 mRNAs initiated from the CD8+-lineage-specific distal promoter (Fig. S4B) (Egawa et al., 2007).
Figure 4: Transcriptomic and genomic footprint of Thpok.
(A) Schematic development of wild-type and Thpok-deficient thymocytes. Thpok-deficient MHC-II restricted cells differentiate into immature CD4+ SP thymocytes before being re-directed into the CD8+-lineage.
(B) Heatmap shows row-standardized (Z-scored RPM values) expression of CD8+ and CD4+ signature genes (Fig. 1D–F and Table S1) in immature CD4+ SP (left) or CD8+ SP (right) Thpok-sufficient (WT) or -deficient (KO) thymocytes sorted as indicated in Fig. S4A.
(C-H) scRNAseq comparison of Thpok-sufficient (WT) and -deficient (KO) thymocytes (detailed genotype information in Fig. S4E). Data integrates two biological replicates from each genotype.
(C, D) UMAP plots displaying WT (C) and KO (D) thymocytes, color-coded by clusters.
(E) Heatmap shows row-standardized expression (Z-scores of average values) of genes in indicated clusters or genotype-defined cluster subsets (for ImCD4, ImCD8 and MatCD8 clusters).
(F) Violin plots show scores for the CD4+- and CD8+-lineage signature in cell clusters as shown in (C, D).
(G) Scatter plots show expression scores for the CD4+- and CD8+-lineage signatures in individual WT and KO cells from the indicated clusters. Lines indicate median scores for the CD8+-lineage signature in WT ImCD8 cells and for the CD4+-lineage signature in WT ImCD4 cells.
(H) Thpok KO thymocytes are shown on UMAP plot generated after Monocle3-derived dimensional reduction, and colored-coded by Seurat-defined clusters (left) or pseudotime value (right). Thick lines (right) are developmental trajectories defined by pseudotime analysis.
(I-K) ChIP-seq on sorted Zbtb7bBio/+ Rosa26BirA+ (Thpok) and Zbtb7b+/+ Rosa26BirA+ (Ctrl) CD4+ SP thymocytes.
(I) Pie charts display number of genes within the CD4+ and CD8+-lineage signatures (Fig. 1D–F and Table S1) near ChIP-seq Thpok binding sites annotated by Homer.
(J) Venn diagram depicts numbers of shared and unique binding sites (peaks) of Thpok and Cbfβ (GSE90794); relevant genes are shown.
(K) ChIP-seq traces for indicated factors on Runx3 and Zbtb7b loci. ChIP-seq data for Gata3 (CD4 T cells) from GSE20898, and for Cbfβ (thymocytes) from GSE90794.
See also Figure S4
Contrasting with these results, the expression of CD4+ signature genes in immature CD4+ SP thymocytes did not require Thpok (Fig. 4B), demonstrating that Thpok is dispensable for CD4+ lineage specification, as inferred earlier (Egawa and Littman, 2008; Wang et al., 2008b). In light of a report that Thpok acts by promoting the expression of Socs-family inhibitors of cytokine signaling (Luckey et al., 2014), we examined expression of these genes in Thpok-deficient cells. Despite de-repression of Runx3, we found that Zbtb7b disruption had no significant impact on Socs1 (Fig. S4C), whereas it increased expression of Socs3 and there was little or no expression of other Socs genes and Cish (data not shown and see below). ATACseq on CD4+ SP thymocytes indicated that Thpok is dispensable for epigenomic opening of CD4-specific peaks, including at the Il2-Il21 locus (Fig. S4D and data not shown). These experiments showed that Thpok serves to maintain transcriptomic patterns characteristic of MHC II-signaled thymocytes.
To further analyze redirection, we performed scRNAseq on (i) Thpok-sufficient and -deficient (Zbtb7bfl/fl Cd4-Cre) MHC II-signaled (i.e. expressing the Zbtb7bGFP reporter) thymocytes and (ii) wild-type CD8+ SP (MHC I-signaled) thymocytes. We captured Thpok-sufficient and -deficient cells separately, and included pre-selection CD69− DP thymocytes with each to facilitate data integration (Table S2). Low-resolution clustering and UMAP display verified dataset integration and separated preselection DP from other thymocytes (Fig. S4E–G). Higher resolution clustering on Zbtb7bGFP-expressing and wild-type CD8+ SP cells distributed WT Zbtb7bGFP+ cells into clusters similar to previously identified ImCD4 and MatCD4 clusters (Fig. 4C, Table S1), and an intermediate semi-mature (SMCD4) cluster. Thpok-deficient Zbtb7bGFP+ cells were excluded from the SMCD4 and MatCD4 clusters, but contributed to cluster ImCD4 and to clusters that did not include wild-type cells (Fig. 4DE). Among these, cluster CD4R2 showed partial loss of CD4+- and gain of CD8+-lineage features (Fig. 4F). At the single cell level, this cluster was enriched in cells expressing both Cd4 and Cd8a mRNAs, and the loss of the CD4+ signature generally preceded gain of CD8+ features (Fig. S4H and 4G). This corresponded to an abrupt transition within the CD4R2 cluster in the pseudotime analysis (Fig. 4H) and with the onset of Runx3 expression (Fig. 4E). Thus, the CD8+-lineage redirection of Thpok-deficient thymocytes follows a de-differentiation pattern, with minimal gain of CD8+-lineage features before cessation of CD4+-lineage gene expression. Similar to population RNAseq, we did not observe any reduction in Socs1 expression in Thpok-deficient clusters compared to their Thpok-sufficient counterparts, whereas expression of Socs3 was increased (Fig 4E). This challenges the idea that Thpok functions in thymocytes by promoting expression of Socs-family genes (Luckey et al., 2014).
To examine if Thpok binds genes it controls, we performed ChIPseq on CD4+ SP thymocytes from mice expressing biotinylated Thpok molecules (Ciucci et al., 2019). We identified 14,845 Thpok-binding sites (Fig. S4I), of which 8,497 (e.g. at Cd8 and Cd40lg, Fig. S4J, L) had not been found to recruit Thpok in activated CD4+ T cells (Ciucci et al., 2019). Thpok bound to most CD4+ and CD8+ signature genes (Fig. 4I). Given that Thpok binds Cd4 and Zbtb7b near Runx binding sites (Muroi et al., 2008), we compared genome-wide binding for both factors, using published thymocyte ChIPseq data for Cbfβ, a co-factor of both Runx1 and Runx3 (Fig. 4J) (Collins et al., 2009; Tenno et al., 2018). Most Thpok binding sites co-localized with Cbfβ sites, including at CD4+ and CD8+ signature genes, notably Cd40lg and, as previously noted, at Zbtb7b and Runx3 (Fig. S4K, and Kojo et al., 2017; Muroi et al., 2008). In contrast, there was no such association with Gata3 binding sites (Wei et al., 2011) (Figs. 4K and S4L).
To gain insight into Thpok function, we used CellOracle to identify transcription factors putatively recruited near Thpok binding sites in genes expressed in CD4+ lineage thymocytes. We found binding motifs for all the 106 transcription factors selected in our previous analysis (Fig. 3G and Table S5), including factors binding Zbtb7b, Gm15417 and Runx3 genes (Table S6 and Fig. 3I). These results support the conclusion that Thpok functionally cooperates with multiple transcription factors, including of the Runx family, to control gene expression in developing T cells.
Transcriptomic and epigenomic features of agonist-selected cells
We then considered the agonist arm of the scRNAseq pseudotime trajectory, diverging from lineage-differentiating cells at the S (selection) branch point and including clusters Sig-2–4, and Sig-5a. All cells along this arm were post-DP, expressing little or no Cd4, Cd8 or Rag1 (Fig. 5A). They expressed neither Ccr7 (consistent with a cortical location) nor Zbtb16 (PZLF), characteristic of iNK T and MAIT cells (Kovalovsky et al., 2008; Rahimpour et al., 2015; Savage et al., 2008) and detected in cluster NC-1 (data not shown). Agonist clusters were enriched for cells expressing pro-apoptotic Bcl2l11 (Bim), a hallmark of negative selection (Bouillet et al., 2002). Clusters Sig-2 and −5 had high scores for the TCR signaling signature (Fig. 2F) and for transcription factors Nr4a1 (Nur77) and Ikzf2 (Helios) (Fig. 5AB), which are both highly expressed in cells signaled for negative selection (Daley et al., 2013; Stritesky et al., 2013). Flow cytometric staining for Bim, Helios, Nur77, PD-1, Ccr7, CD5 and CD69 identified cell subsets corresponding to clusters DP-1, DP-2, Sig-1–3 and Sig-5a (Fig. S5A). These subsets had low levels of Ccr7, and their expression of CD4 and CD8 proteins matched the patterns from scRNAseq clusters (Fig. S5BC). In addition, we identified TCRhi CD44lo PD1+ CD122+ CD4− CD8− cells that matched cluster Sig-4 (Fig. S5A) and were similar to agonist-signaled CD8αα IELp (McDonald et al., 2018; Ruscher and Hogquist, 2019). Analysis of thymocytes from B2m- and H2-Ab1-deficient mice verified that subsets matching clusters DP-1, DP-2, Sig-1 and Sig-2 were contributed in similar proportions by both genotypes. In contrast, and similar to scRNAseq results, MHC II- and MHC I-restricted cells dominated in subsets corresponding to clusters Sig-5, and Sig-3 and −4, respectively (Fig. S5D and S2F). Expression of intra-cellular Gata3 in DP-1, DP-2 and Sig-1 subsets matched that of Gata3 mRNA in the corresponding scRNAseq clusters (Fig. S5E); consistent with Fig. 2K, it was higher in MHC II- than in MHC I-signaled cells. Bim and Nur77 expression in flow cytometric subsets also matched that of the corresponding scRNAseq clusters (Fig. S5FG). In line with its co-expression of Bim and Nur77, the Sig-5a matching subset had the highest frequency of cells staining for activated Caspase-3, an early marker of apoptosis (Fig. S5H).
Figure 5: Agonist-signaled cell developmental trajectories.
(A-B) scRNA-seq of thymocytes sorted from MHC II- and β2m-deficient mice, as in Fig. 2.
(A) Heatmap shows row-standardized gene expression (Z-scored cluster averages).
(B) Scaled expression of indicated genes is shown on UMAP plots generated as in Fig. 2A.
(C-D) scATACseq-seq of thymocytes sorted from MHC II- and β2m-deficient mice, as in Fig. 3.
(C) Genome browser tracks show average scaled scATACseq signals at Bcl2l11 for all cells sharing the indicated IID (right). Red box on DNA track indicates the EBAB enhancer, and pink star an agonist-specific OCR.
(D) Scaled “activity” for indicated transcription factor motifs shown on UMAP plots generated as in Fig. 3A. E-box identified as TCF4 motif in enrichment analyses. Circles denote Sig-4 (blue), Sig-5a (red), Sig-5b (yellow), and Treg (pink) clusters on the UMAP plot.
See also Figures S2 and S5
Expression of Bcl2l11 in clusters Sig-3 and Sig-5a was accompanied by epigenomic opening upstream of the Bcl2l11 locus in scATACseq cells with the corresponding IID (Fig. 5C); this open chromatin region (OCR) contained Nur77 binding motifs (Fig. S5I) and differed from the T cell Bcl2l11 EBAB enhancer (Hojo et al., 2019). Motif analysis in OCR found enrichment for AP1 (Fos::JunB), Nur77, NF-κB, and the E-box motif (Fig. 5D), in patterns matching the expression of the corresponding transcription factor.
Consistent with their co-clustering, clusters Sig-5a and −5b were transcriptomically similar, and both expressed NF-κB family genes (Fig. 5A). However, cells from Sig-5b had higher expression of Ccr7, suggesting a thymic medullary location; they scored lower for TCR signaling markers and Tox-family genes, and higher for Tnfrsf4 (Ox40) and Tnfrsf18 (Gitr)(Fig. 5A), two Treg cell markers (Mahmud et al., 2014). Together with Il2ra and Foxp3, these were expressed on the Treg cluster (Hemmers et al., 2019). This suggests that cluster Sig-5b includes early Treg cell precursors, despite the lack of a pseudotime connection with the Treg cluster. Of note, although flow cytometry identified a fraction of CD25+ cells (corresponding to cluster Sig-5b) expressing high levels of Bim, few cells in this population stained for activated Caspase 3 (Fig. S5H).
Conservation of transcriptomic programs between mouse and human thymus
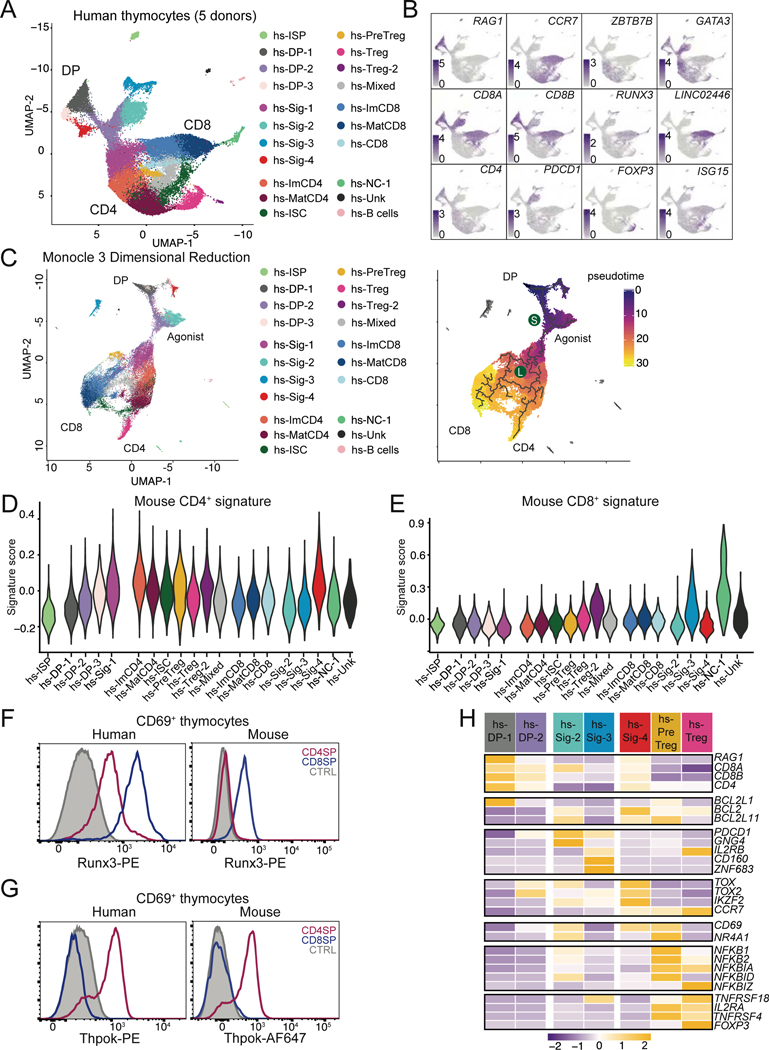
We next examined conservation of transcriptomic and epigenomic features between mouse and human thymocytes. We first performed scRNAseq, on human thymocytes obtained from infants undergoing surgery for conditions not associated with immunodeficiency. We purified CD69+ and pre-selection thymocytes from five donors, and CD4+ SP and CD8+ SP thymocytes from two of these donors (Fig. S6AB and Tables S2 and S7). Analyzing the combined 39,814 cells using the same procedures as for mouse thymocytes, we identified 21 clusters (Fig. 6A and S6CD, and Table S1). Although expression of characteristic genes helped identify human clusters, there was no strict matching between human and mouse clusters (Fig. 6AB). However, the pseudo-time analysis showed conservation of mouse developmental trajectories in human thymocytes, delineating cells with agonist-signaled properties emerging from a common stem at an early S branchpoint, and a subsequent divergence (L) of CD4+ and CD8+ lineage cells (Fig 6C). Scores for both the mouse TCR signaling and CD4+-lineage signatures followed patterns similar to those in mouse thymocytes (Fig. S6E and 6D). In contrast, scores for the mouse CD8+ signature were low in most human CD8+ SP clusters, and high only in populations of Zbtb16 expressing cells (cluster hs-NC-1), with marks of high signaling and effector differentiation (Fig. 6E). Reciprocally, a CD8+-gene signature derived from human thymocytes scored high in both mouse mature CD4+ and CD8+ clusters, suggesting it was best associated with acquisition of mature cell features (Fig. S6FG). The top differentially expressed gene in human CD8+ but not CD4+ SP cells was a long non-coding RNA located in the NK inhibitory receptor locus (LINC02446) and had no mouse ortholog (Fig. 6B). Unlike in mouse cells, expression of RUNX3 mRNA was detected in human CD4+-lineage cells, which we confirmed by intra-cellular staining and flow cytometry (Fig. 6F). In contrast, Thpok protein expression in human thymocytes was strictly specific of the CD4+-lineage, as in the mouse (Fig. 6G). Thus, these results indicated a strong human-mouse conservation of the CD4+-lineage differentiation program, unlike for the CD8+-lineage program.
Figure 6: Early divergence of agonist signaled cells and CD4 transcriptome conservation in human αβ lineage thymocytes.
(A-H) Thymocytes sorted from human donors (Table S7) were prepared for scRNAseq as in Fig. S6AB. Data integrates nine distinct captures from five individual donors.
(A) UMAP plot displaying thymocytes color-coded according to their distribution into clusters. hs denotes Homo sapiens. NC = non-conventional. Unk = unknown ISC = interferon stimulated cluster.
(B) Scaled expression of indicated genes is shown on UMAP plots generated as in (A).
(C) (Left) UMAP plot generated after Monocle3-derived dimensional reduction, colored-coded by Seurat-defined clusters as shown in (A). (Right) developmental trajectories (thick lines) defined by pseudotime analysis on cells color-coded according to their pseudotime value. DP: DP thymocytes; Agonist, CD4, and CD8 indicate the trajectories emerging at branchpoints S (selection) and L (lineage).
(D, E) Violin plots show cluster average expression scores of mouse CD4+- and CD8+-lineage signatures (Fig. 1D–F and Table S1) in cell clusters defined as in (A).
(F, G) Overlaid histograms of Runx3 (F) or Thpok (G) intra-cellular expression on CD69+ CD4+ (red) or CD8+ (blue) SP human (left) or mouse (right) thymocytes. CTRL trace is CD69– DP thymocytes. Human data representative of 3 distinct donors from 3 independent experiments. Mouse data representative of > 5 mice from >5 experiments.
(H) Row-standardized gene expression (Z-scored cluster averages) in select scRNAseq human cell clusters.
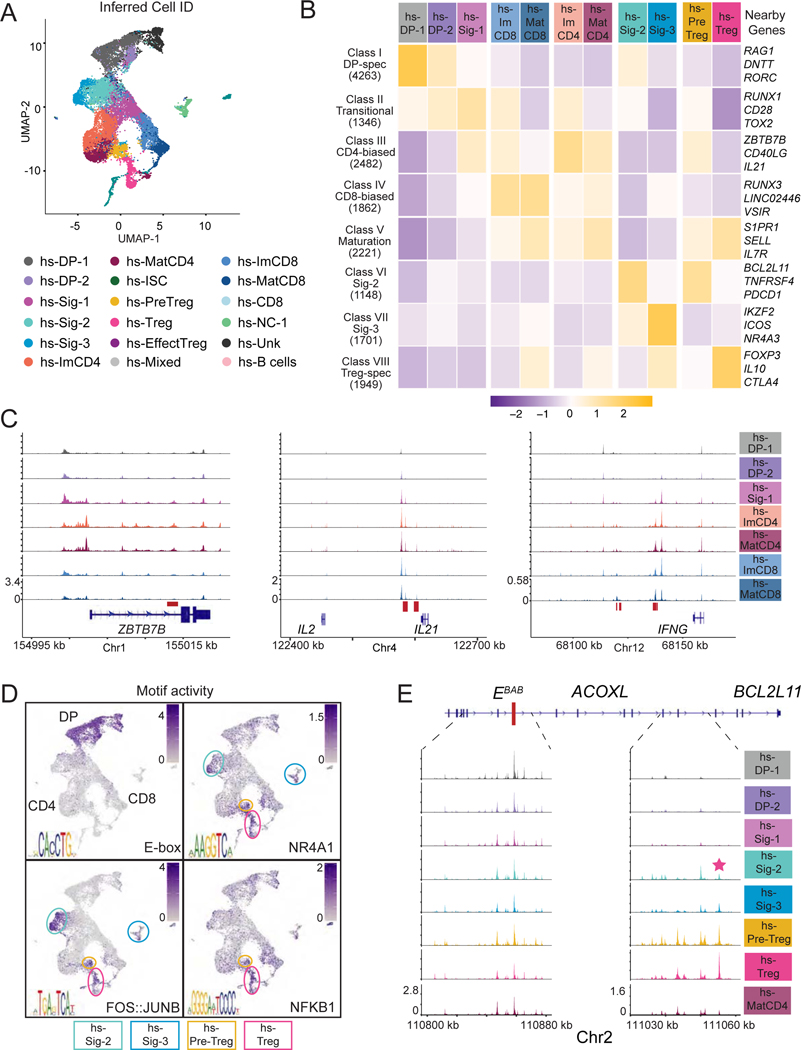
To examine chromatin dynamics, we performed scATACseq on a mix of pre-selection DP and CD69+ human thymocytes from three donor thymi (Fig. S6A and Tables S2 and S7). We identified 19 clusters, which were not driven by batch effect (Fig. S7AB). They closely overlapped with cells groups sharing a common transcriptomic IID (Fig. 7A), indicating a strong correspondence between chromatin and transcriptomic dynamics in human thymocytes, as in the mouse. We identified 16,972 peaks with differential accessibility across cell groups, distributed as in the mouse (Figs. 7B and 3D), including peaks at RAG1, ZBTB7B, IL21 and IFNG (Figs. 7C and S7E), and similar opening patterns of CD4+ and CD8+ lineage specific peaks (Fig. S7C). We identified a human “ortholog” for 11,173 of the 15,226 differentially accessible mouse OCR and found that accessibility at these human regions followed a similar pattern in human mouse cell groups (Fig. S7D). These finding suggested strong human-mouse conservation of epigenomic programs of T cell development.
Figure 7: Single cell epigenomic analysis of human thymocytes.
(A-E) Human thymocytes (Table S7) were prepared for scATACseq as in Fig. S6A. Data integrates thymocytes from three distinct donors.
(A) UMAP plot displaying thymocytes color-coded according to their inferred cell identity. hs denotes Homo sapiens. NC = non-conventional. Unk = unknown. ISC = interferon stimulated.
(B) Heatmap shows row-standardized accessibility of 16,972 OCR, selected for their differential accessibility between any two groups of cells (columns) and distributed into classes by K-means clustering (rows). Gene assignment of OCRs performed by Homer; example genes are indicated.
(C) Genome browser tracks show average scaled scATACseq signals at indicated genes (bottom) for all cells sharing the indicated IID (right). Red boxes indicate sequence-conserved OCR.
(D) Scaled “activity” for indicated transcription factor motifs shown on UMAP plots as in (A). E-box identified as TCF4 motif in enrichment analyses. Circles denote Sig-2 (cyan), Sig-3 (blue), Pre-Treg (yellow), and Treg (pink) clusters on the UMAP plot.
(E) Genome browser tracks show average scaled scATACseq signals at BCL2L11 for all cells sharing the indicated IID (right). The enhancer element EBAB is indicated by a red rectangle. The sequence conserved agonist-specific OCR is denoted by a pink star.
See also Figures S6–7 and Table S7
Cells within the agonist arm expressed TCR signaling markers, including CD69 and NR4A1, and BCL2L11 (Fig. 6H and S6H). This arm was dominated by cluster hs-Sig-2, which contained cells expressing CD8A (but not CD8B), GNG4 and PDCD1 (Fig. 6B, H); such cells were similar to human thymocytes proposed to be enriched in IEL precursors, and that also expressed TNFRSF9 and HIVEP3 (Le et al., 2020; Park et al., 2020). Combined expression of Nr4a1, Hivep3, Tnfrsf9, and Pdcd1 was most characteristic of mouse clusters Sig-3 and Sig-5a (Figs. 5B and S6I). Cluster hs-Sig-3, which was not connected to the main developmental tree, expressed ZNF683 and IL2RB (Fig. 6A, C, H), characteristic of a recently identified pattern shared between αβ and γδ lineage thymocytes (Park et al., 2020). The conservation between human and mouse agonist-signaled cells extended to chromatin accessibility and motif enrichment (Fig. 7D and 5D). This included peaks specific of agonist-selected cells near PDCD1 and IL10 in highly signaled and Treg cells, respectively (Fig. 7B and data not shown). The vicinity of BCL2L11 contained an OCR homolog to that we had identified in the mouse and previously characterized in B cells (Wood et al., 2016) (Figs. 7E and 5C).
In summary, these findings reveal transcriptomic and epigenomic programs of TCR-signaled mouse and human αβ lineage thymocytes, and map developmental trajectories of conventional (CD4+ and CD8+ lineages) and agonist-selected thymocytes, of which most, but not all, are conserved in the human thymus.
Discussion
Here we examined gene expression and chromatin accessibility at the single cell level to map the developmental trajectories of αβ thymocytes undergoing selection, including CD4+-CD8+-lineage cells and agonist-signaled thymocytes. The robustness of these developmental pathways relied on a strong correspondence between transcriptome and epigenome dynamics, and most of them were conserved between human and mouse thymocytes. Leveraging this information, we used computational approaches to build gene regulatory networks that support CD4+ and CD8+ lineage differentiation. Unlike recent thymus scRNAseq studies (Kernfeld et al., 2018; Lavaert et al., 2020; Le et al., 2020; Park et al., 2020), we separately analyzed cells based on their MHC specificity and integrated epigenomic and transcriptomic data, so as to provide insight into CD4+-CD8+ and agonist-signaled cell differentiation.
Expression of the transcription factor Thpok (Zbtb7b) seals CD4+ lineage commitment and is the pivot of CD4+-CD8+ lineage differentiation; it depends on TCR signals in immature CD4+CD8int MHC II-restricted thymocytes (Adoro et al., 2012; Liu et al., 2005). Transcription factors important for Zbtb7b expression, including Gata3, Tox, Tcf1, Bcl11b, Satb1 or E-proteins, serve many functions during both CD4+ and CD8+ T cell differentiation, confounding genetic analyses (Aliahmad and Kaye, 2008; Jones-Mason et al., 2012; Kakugawa et al., 2017; Kojo et al., 2017; Steinke et al., 2014; Wang et al., 2008b). Using computational approaches that are not hampered by such pleiotropic effects, we identified transcription factors (“CD4+-biased”) putatively associated with the emergence of the CD4+ lineage and Zbtb7b expression. The activity of most such CD4+-biased factors (evaluated by expression of their putative targets) increased as TCR signaled thymocytes underwent differentiation, consistent with the role of TCR signals in Zbtb7b expression (Adoro et al., 2012; He et al., 2008; Liu et al., 2005). Some of these factors are controlled by TCR signaling at the expression (e.g. Egr1, AP-1 [JunB]) or activity (e.g. Elk proteins) levels (Costello et al., 2004; Shao et al., 1997). Reciprocally, factors biased towards the CD8+-lineage included Tcf3-encoded E2A (Kee, 2009), which is inhibited by TCR-induced Id2, expressed in CD4+-lineage thymocytes. Among factors we identified as putatively controlling Zbtb7b and Runx3 expression, Runx molecules, Thpok, Gata3, Tcf1 (Tcf7), Mazr (Patz1), E-proteins (Tcf3) and Hbp1, actually affect Zbtb7b expression or lineage differentiation (Egawa et al., 2007; Jones-Mason et al., 2012; Muroi et al., 2008; Sakaguchi et al., 2010; Setoguchi et al., 2008; Steinke et al., 2014; Wang et al., 2008b). This supports the idea that other identified factors, whose expression or activity is controlled by TCR, contribute to trigger Zbtb7b expression upon TCR signaling.
The CD4+ or CD8+ lineage bias of transcriptional activities was detected only after lineage bifurcation, but not in MHC II- vs. MHC I-signaled thymocytes within the common stem trajectory. This suggests that the initial differentiation of TCR-signaled thymocytes is “lineage neutral”. This is in line with the “kinetic signaling” model of CD4+-CD8+ lineage commitment. This model proposes that asymmetric changes in CD4 or CD8 expression, not intrinsic differences between MHC I- and MHC II-induced TCR signals, are the primary determinant of lineage differentiation (Singer et al., 2008). The model predicts that, regardless of MHC specificity, TCR signaling in DP thymocytes terminates Cd8 expression, preventing continued TCR signaling and thereby Zbtb7b expression in MHC I-restricted thymocytes, in which TCR signaling is CD8-dependent, but not in MHC II-restricted thymocytes. Also consistent with this model is our observation that Cd8 expression decreased in TCR-signaled thymocytes, with kinetics independent of MHC specificity, although the trajectory leading from DP to CD8+ SP thymocytes did not include cell clusters with no detectable Cd8 gene expression.
However, other results from our study indicate that the transcriptomes of MHC II- and MHC I-signaled thymocytes diverged early, and independently of changes in Cd4 or Cd8 expression. Scoring for a gene signature specific of CD4+ lineage cells indicated an early divergence between MHC II- and MHC I-signaled cells. The small size of this CD4+ signature (45 genes), compared to the computationally defined gene sets defining transcriptional activities (~2,000 genes), facilitated an early detection of lineage divergence. The divergence between MHC II- and MHC I-signaled cells preceded any detectable asymmetry in Cd4 and Cd8 gene expression, as expression of Cd4 was reduced before, and at least to a similar detectable extent as that of Cd8 genes. These results support the idea of differences between MHC I- and MHC II-induced TCR signaling that do not depend on the kinetics of coreceptor gene expression. They also agree with the concept, based on multiple observations (Singer et al., 2008), that cessation of Cd8 gene expression in MHC I-signaled thymocytes contributes to prevent Zbtb7b expression.
Our study provides insight into the fate of agonist-signaled thymocytes, which include precursors of regulatory cells and cells targeted for negative selection. These cells maintained a high level of TCR signaling and expressed Ikzf2, Nr4a1 or Bcl2l11 (encoding Helios, Nur77 and Bim, respectively), unlike conventional positively selected thymocytes, which expressed Bcl2 and diverged into CD4+ and CD8+ lineages. A subset of agonist-signaled cells undergo negative selection, apoptotic cell death induced by TCR engagement (Hogquist and Jameson, 2014; Stritesky et al., 2012). Indeed, we identified an agonist-signaled subset enriched for active caspase-3, an effector of apoptosis; most of these cells are MHC II-restricted and express little Ccr7, suggesting cortical location. Although we detected little active caspase 3 among other agonist-signaled subsets, this presumably reflects the fact that apoptotic thymocytes are rapidly engulfed by thymic stromal cells and therefore escape detection. We show marked transcriptomic and epigenomic differences between mouse thymocytes signaled by MHC II- and β2-mdependent agonist ligands, and the conservation of cluster signatures across species suggests similar patterns in human thymocytes. Both pseudo-time mapping and scoring for a TCR signaling signature support the idea of an initial common stem shared by agonist-signaled and conventional thymocytes, before they diverge towards their respective fates. This shared program includes changes in expression of Rag genes, Bcl2, and multiple transcriptional regulators, including Tox and Tox2. A scenario whereby TCR engagement in DP cells initiates a differentiation program common to all signaled thymocytes fits with the differentiation of Treg cells in the medulla from immature CD4+ SP thymocytes that have been subject to positive selection (Cowan et al., 2013; Lee and Hsieh, 2009). Alternatively, other studies support the view that agonist TCR signaling, by immobilizing thymocytes and prolonging contact with ligands, triggers signaling pathways and transcription programs that rapidly diverge from those of conventional thymocytes (Au-Yeung et al., 2014; Daniels et al., 2006; McGargill et al., 2009; Melichar et al., 2013; Palmer, 2003). These observations are not inconsistent with the common stem idea, because scRNAseq-based developmental trajectories are defined independently of actual kinetics.
In summary, by integrating single cell transcriptomics and epigenomics, we defined MHC-specific developmental trajectories of conventional and agonist-signaled αβ lineage thymocytes, and documented the conservation of these features in human T cell development. Furthermore, our computational analyses assembled the elements of transcriptional circuits involved in CD4+ and CD8+ lineage differentiation, including expression of the lineage-specific factors Thpok and Runx3.
Limitations of the study
The conclusions of this study must be understood in its biological, technical, and computational context. Agonist-signaled cells targeted for negative selection are quickly engulfed by thymic macrophages and therefore under-represented in thymocyte preparations. Human data is affected by inter-individual variability and clinical history, hence the collection of samples from three to five distinct donors. Droplet-based scRNAseq and scATACseq do not detect all expressed genes or all OCR in every cell; for scRNAseq, this notably applies to low-abundancy mRNAs and small cells (including DP and most post-DP thymocytes). For computational inference of gene regulatory networks, consensus binding motifs are imperfect predictors of in vivo transcription factor DNA recognition, and can be shared by transcription factors of the same family.
Star Methods
Resource Availability
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rémy Bosselut (remy.bosselut@nih.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The common accession number for all sequence data reported in this paper is GSE148981. Specific accession numbers are listed in the key resource table. All other data and code are available upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Bim-AF647 (C34C5) | Cell Signaling Technology | Cat# 10408S, RRID:AB_2797721 |
| Anti-Bim-PE (C34C5) | Cell Signaling Technology | Cat# 12186S, RRID:AB_2797842 |
| Anti-Cleaved Caspase-3 (ASP175) (Clone D3-E9) | Cell Signaling Technology | Cat# 8788S, RRID:AB_2797665 |
| Anti-CCR7-Pe-Cy7 (Clone 4B12) | Thermofisher | Cat# 25-1971-82, RRID:AB_469652 |
| Anti-CD122-FITC (Clone TM-Beta 1) | Thermofisher | Cat# 11-1222-82, RRID:AB_465189 |
| Anti-CD25- APC-eF780 (Clone PC61.5) | Thermofisher | Cat# 47-0251-82, RRID:AB_1272179 |
| Anti-CD25-PerCP-Cy5.5 (Clone PC61.5) | Thermofisher | Cat# 45-0251-82, RRID:AB_914324 |
| Anti-CCR7-PE-Cy7 (Clone 4B12) | Thermofisher | Cat# 25-1971-82, RRID:AB_469652 |
| Anti-CD279-BV786 (Clone J43) | BD Pharmigen | Cat# 744548, RRID:AB_2742319 |
| Anti-CD4-APC (Clone GK1.5) | Thermofisher | Cat# 17-0041-82, RRID:AB_469320 |
| Anti-CD4-BV650 | BD Pharmigen | Cat# 563747, RRID:AB_2716859 |
| Anti-CD4-BV786 (Clone RM4-4) | BD Pharmigen | Cat# 563727 RRID:AB_2728707 |
| Anti-CD4-eFluor450 (Clone GK1.5) | Thermofisher | Cat# 48-0041-82, RRID:AB_10718983 |
| Anti-CD4-PerCP-Cy5.5 (Clone RM4-5) | Thermofisher | Cat# 45-0042-82, RRID:AB_1107001 |
| Anti-CD44-Alexa Fluor 700 (Clone IM7) | Thermofisher | Cat# 56-0441-82, RRID:AB_494011 |
| Anti-CD45.2-BV786 (Clone 104) | BD Pharmigen | Cat# 563686, RRID:AB_2738375 |
| Anti-CD5-BV605 (Clone 53-7.3) | BD Pharmigen | Cat# 563194, RRID:AB_2738061 |
| Anti-CD69-PE (Clone H1.253) | Thermofisher | Cat# 12-0691-82 RRID: AB_465732 |
| Anti-CD69-PerCP-Cy5.5 (Clone H1.253) | Thermofisher | Cat# 45-0691-82, RRID:AB_1210703 |
| Anti-CD8a-APC (Clone 53-6.7) | Thermofisher | Cat# 17-0081-82, RRID:AB_469335 |
| Anti-CD8a-APC-eFluor780 (Clone 53-6.7) | Thermofisher | Cat# 47-0081-82, RRID:AB_1272185 |
| Anti-CD8a-BUV395 (Clone 53-6.7) | BD Pharmigen | Cat# 563786, RRID:AB_2732919 |
| Anti-CD8a-PerCP-Cy5.5 (Clone 53-6.7) | Thermofisher | Cat# 45-0081-82, RRID:AB_1107004 |
| Anti-CD8a-PE (Clone 53-6.7) | Thermofisher | Cat# 12-0081-83, RRID:AB_465530 |
| Anti-GATA3-APC (Clone L50-823) | BD Pharmigen | Cat# 560078, RRID:AB_1645317 |
| Anti-HELIOS-Alexa Fluor 647 (Clone 22F6) | BioLegend | Cat# 137218, RRID:AB_10660750 |
| Anti-HELIOS-Pacific Blue (Clone 22F6) | BioLegend | Cat# 137220, RRID:AB_10690535 |
| Anti-MHC Class I (H-2Kb)-APC (Clone AF6-88.5.5.3) | Thermofisher | Cat# 17-5958-82, RRID:AB_1311280 |
| Anti-MHC Class I (H-2Kb)-APC (Clone AF6-88.5.5.3) | Thermofisher | Cat# 17-5958-82, RRID:AB_1311280 |
| Anti-NUR77-Alexa Fluor 647 (Clone 12.14) | Thermofisher | Cat# 51-5965-82, RRID:AB_2802306 |
| Anti-NUR77-PE (Clone 12.14) | Thermofisher | Cat# 12-5965-82, RRID:AB_1257209 |
| Anti-RUNX3-PE (Clone R3-5G4) | BD Pharmigen | Cat# 564814, RRID:AB_2738969 |
| Anti-TCRb-BUV737 (Clone H57-597) | BD Pharmigen | Cat# 612821 |
| Anti-THPOK-Alexa Fluor 647 (Clone T43-94) | BD Pharmigen | Cat# 565500, RRID:AB_2739268 |
| Anti-CD4-APC (Clone RPA-T4) | Thermofisher | Cat# 17-0049-42, RRID:AB_1272048 |
| Anti-CD4-eFluor 450 (Clone RPA-T4) | Thermofisher | Cat# 48-0049-42, RRID:AB_1272057 |
| Anti-CD69-FITC (Clone FN50) | BD Pharmigen | Cat# 560969, RRID:AB_10562195 |
| Anti-CD8-PerCp-Cy5.5 (Clone RPA-T8) | BD Pharmigen | Cat# 560662, RRID:AB_1727513 |
| Anti-RUNX3-PE (Clone R3-5G4) | BD Pharmigen | Cat# 564814, RRID:AB_2738969 |
| Anti-THPOK-PE (Clone 6/hcKrox) | BD Pharmigen | Cat# 565730 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde | Thermofisher | 28906 |
| Critical Commercial Assays | ||
| Transcription factor Staining Buffer Set | Thermofisher | 00-5523-00 |
| Cytofix/Cytoperm | BD Pharmingen | 554714 |
| Protein-A beads | Invitrogen | 10001D |
| Streptavidin beads (M280) | Invitrogen | 12205D |
| QIAshredder | QIAGEN | 79656 |
| RNeasy Plus Micro Kit | QIAGEN | 74034 |
| RNeasy Plus Mini Kit | QIAGEN | 74134 |
| 10X Chromium single cell 3’ V2 kit | 10X Genomics | 120267, 120262 |
| 10X Chromium Next GEM Single Cell 5’ Reagent Kit (v1.1) kit | 10X Genomics | 1000127, 1000165 |
| 10X Chromium Single Cell ATAC Solution (v1.0) kit | 10X Genomics | PN-1000110 |
| 10X Chromium NextGEM Single Cell ATAC Solution (V1.1) kit | 10X Genomics | 1000176, 1000162 |
| Proteinase K | Invitrogen | 25530049 |
| RNase A | Invitrogen | 1209121 |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1030 |
| Minelute PCR Purification Kit | QIAGEN | 28004 |
| QIAquick PCR Purification Kit | QIAGEN | 28104 |
| Deposited Data | ||
| Developing Mouse Thymocyte Population RNAseq | This study | GSE148973 |
| Developing Mouse Thymocyte Single-cell RNAseq | This study | GSE148977 |
| Thpok KO Thymocyte Population RNAseq | This study | GSE148974 |
| Thpok KO Thymocyte Single-cell RNAseq | This study | GSE157286 |
| Thpok KO Thymocyte Population ATACseq | This study | GSE148975 |
| Developing Mouse Thymocyte Single-cell ATACseq | This study | GSE148979 |
| Developing Human Thymocyte Single-cell RNAseq | This study | GSE148978 |
| Developing Human Thymocyte Single-cell ATACseq | This study | GSE148980 |
| CHIPseq (Thpok CD4 SP thymocytes) | This study | GSE148976 |
| CHIPseq (Thpok CD4 T Cells) | Ciucci et al. (2019) | GSE116506 |
| CHIPseq (Cbfβ) | Tenno et al. (2019) | GSE90794 |
| CHIPseq (Gata3) | Wei et al. (2011) | GSE20898 |
| Experimental Models: Organisms/Strains | ||
| Zbtb7b fl | (Wang et al., 2008a) | |
| Zbtb7b Bio | (Ciucci et al., 2019) | |
| Rosa26 BirA | (Driegen et al., 2005) | |
| NCI B6-Ly5.1/Cr (CD45.1) | Charles River | Charles River 564 |
| C57BL/6Ncr (CD45.2) | Charles River | Charles River 556 |
| B6 CD45.1 CD45.2 | Generated in house | |
| Zbtb7b GFP | (Wang et al., 2008b) | |
| Runx3 RFP | (Zamisch et al., 2009) | |
| B2m −/− | Taconic | (Zijlstra et al., 1990) |
| H2-Ab1 −/− | JAX | (Grusby et al., 1991) |
| Cd4-cre | Taconic | (Lee et al., 2001) |
| Software and Algorithms | ||
| Graphpad Prism 7.0 | graphpad.com | n/a |
| Flowjo 10.0 | flowjo.com | n/a |
| R | r-project.org | n/a |
| Seurat v 3.1 | (Butler et al., 2018) | |
| Signac v 0.2.1 and 1.0.0 | github.com/timoast/signac | |
| MACS2 (v2.2.5) | github.com/taoliu/MACS | |
| CellRanger v2.2 and 3.1 | 10X Genomics | |
| DESeq2 | (Love et al., 2014) | |
| STAR aligner (v2.4.0) | github.com/alexdobin/STAR | |
| CellOracle (v0.3.5) | github.com/morris-lab/CellOracle | |
| AUCell | (Aibar et al., 2017) | |
| Htseq (v0.11.4) | (Anders et al., 2015) | |
| Homer (v4.10) | (Heinz et al., 2010) | |
| Bowtie2 (v2.3.4) | (Langmead and Salzberg, 2012) | |
| Samtools (v1.6) | (Li et al., 2009) | |
| Picard (v2.2.8) | http://broadinstitute.github.io/picard/ | |
| Bedtools (v2.29.2) | github.com/arq5x/bedtools2 | |
| Diffbind v(3.1) | (Ross-Innes et al., 2012) | |
| Deeptools (v3.3.0) | (Ramirez et al., 2016) | |
| Monocle3 | (Cao et al., 2019) | |
| biomaRt (v2.42.0) | github.com/grimbough/biomaRt | |
| CellRanger ATAC (V1.01 and 1.1) | ||
| UCSC LiftOver | (Kent et al., 2002) | |
Experimental Model and Subject Details
Mice
Zbtb7bfl, Zbtb7bGFP, and Runx3RFPmice were described previously (Wang et al., 2008a; Wang et al., 2008b; Zamisch et al., 2009). Zbtb7bBio/+ Rosa26BirA+and Zbtb7b+/+ Rosa26BirA+ mice were described previously (Ciucci et al., 2019). Β2m−/−, Cd4-Cre (both from Taconic) and H2-Ab1−/− (Jax) mice were previously reported (Grusby et al., 1991; Lee et al., 2001; Zijlstra et al., 1990). CD45.1, CD45.2, and C57BL/6 mice from Charles River Laboratories. H2-Ab1−/− and B2m−/− mice were crossed to CD45.1 mice to obtain allelically marked strains. Mice were housed in specific pathogen-free facilities and most experiments were performed on sex-matched 6–14 week old mice. Animal procedures were approved by the NCI Animal Care and Use Committee.
Human thymic samples
Fresh human thymus samples were obtained from the pathology department of the Children’s National Medical Center in Washington, DC following cardiothoracic surgery from children with congenital heart disease not associated with immunodeficiency, as the thymic tissue is routinely removed and discarded to gain adequate exposure of the retrosternal operative field. Use of these thymus samples for this study was determined to be exempt from review by the NIH Institutional Review Board in accordance with the guidelines issued by the Office of Human Research Protections. Age and gender of donors is listed in Table S7.
Method Details
Bone marrow chimeras
Bone marrow was isolated from CD45.2 Zbtb7bGFPRunx3RFP mice, T cell-depleted with Mouse Pan T (Thy1.2) Dynabeads (ThermoFisher Scientific), and injected into lethally irradiated (900rad) wild-type, B2m−/−, or H2-Ab1−/− recipient mice, all CD45.1. Cells were sorted from thymus at least six weeks post-transplant.
Antibodies
Fluorochrome-labeled antibodies of the following mouse specificities were purchased either from Becton Dickinson PharMingen, ThermoFisher Ebiosciences, BioLegend, or Cell Signaling Technologies: CD4 (Rm4.4, Rm4.5, or GK1.5), CD8α (53–6-7), CD44 (IM7), Thpok (T43–94), Runx3 (R3–5G4 [mouse and human]), TCRβ (H57–597), CD45.1 (A20), CD45.2 (104), CD69 (H1.2F3), H2kb (MHC-I, AF6–88.5.5.3), CD103 (2E7, M290), CD25 (PCG1.5), CCR7 (4B12), CD24 (M1/69), CD5 (53–7.3), Helios (22F6), NUR77 (12.14), Bim (C34C5), CD122 (TM-Beta1), PD1 (J43), Cleaved Caspase-3 (Asp175, clone DE39), and Gata3 (L50–823). Fluorochrome-labeled antibodies of the following human specificities were purchased either from Becton Dickinson PharMingen or ThermoFisher Ebiosciences: CD4 (RPA-T4 or OKT4), CD8 (RPA-T8), CD69 (FN50), and Thpok (6/hcKrox).
Cell preparation, staining and flow cytometry
Human thymocytes were obtained by gentle teasing of thymic fragments cut from the surgery piece and filtered through a 70 μm filter. Mouse thymocytes were prepared and stained as previously described (Carpenter et al., 2012). Staining of Thpok, Gata3, and Runx3 was performed for 1 hour at room temperature on cells fixed and permeabilized with the eBioscience Transcription Staining Buffer Set (EBioscience). For Gata3 staining on signaled DP thymocytes (Fig. 2K), to ensure equal intracellular staining, we first surface stained the cells from each genotype (H2-Ab1−/− or B2m−/−) with different color fluorochromes conjugated to anti-CD45, then mixed cells prior to fixation and intra-cellular Gata3 staining. For intracellular staining of Bim, Helios, Ccr7, Gata3, and Nur77 (Fig. S5), surface stained cells were incubated for 30 minutes at 4oC in BD Cytofix/Cytoperm fixation and permeabilization solution, then incubated for 45 minutes at 4oC in 1X permeabilization buffer from the eBioscience Transcription Staining Buffer Set. Cells were then washed in 1X permeabilization buffer, then stained for 12 hours at 4oC in antibody, washed, and analyzed. For data shown in Fig. S5D, populations of cells gated as in Fig. S5A were normalized to 100,000 CD69+ thymocytes from H2-Ab1- and β2m-deficient mice. For ATACseq on wild-type and Thpok-deficient thymocytes, to ensure samples were processed equivalently and to enable simultaneous sorting of thymocytes from distinct animals, control and Thpok-deficient thymocytes were first stained with different fluorochromes conjugated to anti-CD45 for each genotype, then mixed prior to cell sorting. Purification of thymocytes was performed on a FACSAria II, a FACSViolet, or a FACSFusion (BD Biosciences). Flow cytometry data were acquired on an LSRFortessa, LSRFortessa X-20, BDFacsSymphony, or FACSCanto II, and analyzed with FlowJo (TreeStar) software. Dead cells and doublets were excluded by DAPI or Fixable Viability Dye UV staining (Invitrogen), and forward/side scatter height by width gating. For sorting, cells were not stained with viability dye, but were gated on live cells determined by size.
Population ATACseq
ATACseq and library construction were performed as previously described (Buenrostro et al., 2013). 50,000 cells were pelleted for 5 minutes at 550 x g and washed once with 1 mL of 1X pbs, then washed once with 50 μL of lysis buffer (10mM Tris-HCL [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and spun for 10 minutes at 550 x g. Nuclei pellets were resuspended in 50 μL transposition reaction buffer with 2.5 μL Tn5 transposase (FC-121–1030; Illumina). The reaction was incubated for 45 minutes at 37oC. Tagmented DNA was eluted using the MinElute Reaction Cleanup Kit (QIAGEN), and amplified with 11 PCR cycles with NEBNext High-Fidelity 2X mastermix (New England BioLabs). Libraries were purified using a QIAQuick PCR purification kit (QIAGEN) and sequenced on an Illumina HiSeq 4000 (150bp paired-end). Thymocytes were processed in three biological replicates. Raw ATACseq fastq files were trimmed with Trimmomatic (Bolger et al., 2014), and aligned to mouse genome (mm10) using Bowtie2 (v2.3.4) (Langmead and Salzberg, 2012) with X set to 2000. Low quality (MAPQ < 30) and mitochondrial reads were removed with Samtools (v1.6) (Li et al., 2009). PCR duplicates were removed with Picard (v2.2.8). Files were converted to bed with Bedtools (v2.29.2). Peaks were called using Macs2 (v2.2.4) (Zhang et al., 2008), with the parameters pvalue 1e-7 and --keep-dup all. DiffBind (v3.1, http://bioconductor.org/packages/release/bioc/html/DiffBind.html) was used for peak merging, read counting, and differential accessibility analyses (Ross-Innes et al., 2012). Homer (4.10) was used for peak annotation and motif enrichment analyses (Heinz et al., 2010). BigWig files for visualization were generated with Deeptools (3.3.0) with counts per million normalization (Ramirez et al., 2016).
Population RNAseq
RNA was extracted from thymocytes sorted according to gates shown in Fig. S1C using QIAshredder columns and RNeasy Plus Micro or Mini kit (QIAGEN). RNA samples with an RNA integrity number (RIN) > 8 as measured by bioanalyzer (Agilent) were processed for library preparation using SMARTer Ultra Low input reagent (Takara) and Nextera XT DNA (Illumina) library preparation kits. Libraries were sequenced with paired-end reads on a HiSeq 2500, HiSeq 3000, or HiSeq 4000 (Illumina). For data shown in Fig. 1, 2–3 replicates were sorted per population, all from chimeric mice as described in Fig. S1B. Data shown in Fig. 4 includes three biological replicates for each population, sorted as in Fig. S4A from control or Zbtb7bfl/fl Cd4-cre Zbtb7bGFP BAC reporter mice. In all experiments, biological replicates were processed separately from sorting to sequencing. Raw fastq files were trimmed with Trimmomatic, aligned to mouse genome (mm10) using STAR (v.2.4.0h) with mouse gencode (release 11) gtf file (Ensembl m38.86) (Dobin et al., 2013; Mudge and Harrow, 2015). Count of RNA reads and gene assignment were done with HTseq (Anders et al., 2015). DESeq2 was used for differential expression analysis on samples collected within the same experiment (Love et al., 2014). Gene expression is shown as reads per million (RPM), and heatmaps were generated with pHeatmap. Gene signatures were defined as follows. For the CD4+-signature, genes induced from DP to immature CD4SP, or semi-mature CD4SP, or mature CD4SP were combined, and only those that are also higher in immature CD4SP > immature CD8SP, or semi-mature CD4SP > immature CD8SP, or mature CD4SP > mature CD8SP were retained. Finally, only genes with > 200 RPM in at least one sequenced sample were included in the signature. For the CD8-signature, genes induced from DP to immature CD8SP or DP to mature CD8SP were combined, and only those that are also higher in semi-mature CD8SP > immature CD4SP or mature CD8SP > mature CD4SP were retained. Only genes with > 75 reads per million in at least one sequenced sample were included in the signature.
ChIP-seq
CD4 SP thymocytes (3–6 × 106) were sorted from mice carrying alleles for Thpok-biotin and Bir-A ligase (Ciucci et al., 2019) and fixed by adding 16% formaldehyde (28906, Thermo Scientific) to samples in PBS for a final concentration of 1% and incubating at 37oC for 10 min. Fixation was quenched by addition of 2M glycine (Sigma) in PBS at a final concentration of 125 mM. Cells were washed twice in cold PBS, and pellets were snap frozen in dry ice and stored at −80oC. Fixed pellets were pooled to generate samples of ~20 × 106 cells each for Thpok ChIP (three independent samples) and BirA ligase control (one sample). Fixed pellets were thawed on ice and resuspended in 2mL of cold RIPA buffer (10mM TrisHCl pH 7.6, 1mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% TritonX100, 1 Complete Mini EDTA free proteinase inhibitor (Roche)). Sonication was performed using the Covaris S220 sonicator at duty cycle 20%, peak incident power 175, cycle/burst 200 for 30 cycles of 60 seconds with 30 second pause after every cycle. Chromatin samples were clarified by centrifugation at 21,000 g at 4oC for 10 minutes, and pre-cleared with 100 μL of prewashed Dynabeads Protein-A (Invitrogen 10001D) for 1 hour at 4oC with rotation. 100 μL of prewashed Dynabeads M-280 Streptavidin (Invitrogen 11205D) were added to 1 mL of precleared chromatin followed by overnight incubation at 4oC on a rotator. Beads were washed at 4oC twice in cold RIPA buffer, twice with RIPA buffer containing 0.3M NaCl, twice with LiCl buffer (0.25 M LiCl, 0.5% Igepal-630, 0.5% sodium deoxycholate), once with TE (10 mM Tris pH 8.0, 1mM EDTA) plus 0.2% Triton X-100, and once with TE. Crosslinking was reversed by incubating the beads at 50oC for 1 hour in the presence of 0.3% SDS and 1mg/mL of Proteinase K (Invitrogen), followed by four hours at 65oC with vortexing every hour. Immunoprecipitated DNA was removed from beads and stored at −20oC until library preparation. ChIP libraries were constructed with Illumina and sequenced on an Illumina NextSeq500 (75bp single-end reads) as previously described (Canela et al., 2019).
ChIP-seq analysis
Fastq files were aligned to the mm10 genome using Bowtie2 (v2.3.4) and filtered with Samtools (v1.6), using -q 20. BigWig files for visualization were generated with Deeptools (3.3.0) with counts per million normalization. Peak calling was performed on the sorted bam files using Macs2 (v2.2.5) with default parameters and using the BirA-ligase sample, which lacks the Thpok-Biotin acceptor molecule, as the control. Consensus peakset was generated by using the Homer (V4.1.0) MergePeaks function. MergePeaks was used to identify shared and specific peak-sets between different experiments. All published ChIP-Seq datasets were re-analyzed following the criteria above.
Mouse Single cell RNAseq
For mice deficient in either B2m or H2-Ab1, 5–8 × 103 sorted CD69+ thymocytes were loaded on the 10X Chromium platform (10X genomics), and libraries were constructed using the Single Cell 3’ Reagent Kit V2 according to the manufacturer’s instructions. Two biological replicates, each with two captures (one for each genotype) were processed separately. For Thpok wild-type and deficient mice, 5–8 × 103 cells comprising a mixture of sorted Zbtb7bGFP+, CD8SP, and CD69-DP (wild-type control), and 5–8 × 103 cells comprised of a mixture of Zbtb7bGFP+ and CD69-DP (Thpok-deficient), were loaded separately on the 10X Chromium platform. Each library was constructed using the Next GEM Single Cell 5’ Reagent Kit (v1.1) according to the manufacturer’s instructions. Two biological replicates, each with two captures (one for each genotype) were processed separately. Libraries were sequenced on one NextSeq run using 26×98bp or 26×57 bp to a depth of at least 20,000 reads/cell. Sequencing files were processed, mapped to mm10, and count matrices were extracted using the Cell Ranger Single Cell Software (v 2.2.0 or 3.1.0). Further analyses were performed in R using the Seurat package (v 3.1) (Butler et al., 2018; Stuart et al., 2019), and the Monocle3 package (Cao et al., 2019; Haghverdi et al., 2018; Levine et al., 2015; Qiu et al., 2017a; Qiu et al., 2017b; Trapnell et al., 2014).
Human Single cell RNAseq
For each of the five donors, CD69+ sorted thymocytes were mixed in 75:25 ratio with unsignaled DP thymocytes prior to capture. For donors #4 and #5, CD4+ and CD8+ SP thymocytes were sorted and captured separately in addition to the CD69+:unsignaled DP mixture, so that 3 scRNAseq captures were performed from each of these two donors (Fig. S6A). For each sample, 5–12 × 103 sorted thymocytes were loaded on the 10X Chromium Platform, and libraries were constructed using the Next GEM Single Cell 5’ Reagent Kit (v1.1) according to the manufacturer’s instructions. Libraries were sequenced on one NextSeq run resulting in at least 20,000 reads/cell. Sequencing files were processed, aligned to GRCh38–3.0.0, and count matrices were extracted using the Cell Ranger Single Cell Software (v3.1.0). Further analyses were performed in R using Seurat (v3.1) and Monocle3.
Single cell RNAseq analysis
Data was pre-processed in Seurat by removing genes expressed in fewer than 2 cells and excluding cells that were outliers for number of RNA molecules, or more than 5% mitochondrial genes. The datasets were merged together and integrated following the Seurat standard integration method. Following normalization, UMAP dimensional reduction was performed using the first 30 principal components. Clustering was performed following identification of nearest neighbors, using the first 20 dimensions and a resolution of 0.85 for the B2m−/− and H2-Ab1−/− mouse data and 0.7 for the human data. Clusters predominantly comprising either cells with low RNA content, doublet cells, or human donorspecific cells, were removed and the datasets were re-clustered following dimensional reduction (resolution of 0.85 for the B2m−/− and H2-Ab1−/− mouse sample, and 0.8 for human). For analysis of Thpok-deficient and sufficient thymocytes (Figs. 4 and S4), initial clustering was performed with a resolution of 0.8. Clusters with low content RNA and non-T cell lineages were removed. Following dimensional reduction, cells were re-clustered with a resolution of 0.1 (Fig. S4F), and the cluster corresponding to DP thymocytes was removed. Following dimensional reduction, cells were re-clustered with a resolution of 1.0 (Fig. 4C).
Marker genes for each cluster were determined by the FindAllMarkers function of Seurat with a minimum Log2 fold change threshold of +0.25 (cluster of interest over all other clusters) which uses the Wilcoxon Rank Sum Test. The mouse TCR signal-induced signature included genes with a +0.5 (Log2) fold higher expression in cluster Sig-1 over cluster DP-1. Signature scores were calculated on a per cell basis by using the AddModuleScore function to score each cell for a list of genes, with the number of control features set to 10. Mouse-derived gene signatures were converted to human gene symbols using the biomaRt package (v2.42.0). For scoring of the human CD8+ gene signature on mouse cells, human gene symbols were converted to mouse gene symbols using the biomaRt package.
Pseudotime analysis
Filtered and merged datasets were imported into Monocle3 by generating a cell data set from the raw counts slot of the Seurat object. Normalization and PCA were done with the preprocess_cds command from Monocle3 using the first 100 dimensions, and batch correction was applied using the align_cds command, which utilizes the Batchelor tool (Haghverdi et al., 2018). UMAP dimensional reduction was performed using the reduce_dimension command. Cells were clustered with cluster_cells using “Louvain” with the k set to 50 for B2m−/− and H2-Ab1−/− sample (Fig. 2E), 200 for the Thpokdeficient sample (Fig. 4H), and 100 for the human sample (Fig. 6C). The trajectory graph was learned on the Monocle-derived clusters by calling learn_graph. Cells on the UMAP plot are colored by Seuratderived clusters. Pseudotime was determined using unsignaled DP cluster as the starting point for B2m−/− and H2-Ab1−/−, and human samples, and ImCD4 for Thpok-deficient sample.
Mouse Single Cell ATACseq
Nuclei were isolated, following 10X genomics instructions, from 0.5–1 × 106 CD69+ thymocytes sorted from either Β2m or H2-Ab1 deficient mice. 5,000–10,000 nuclei were loaded on the 10X chromium platform, and libraries were constructed using the 10X Chromium Single Cell ATAC Solution (v1.0) according to the manufacturer’s instructions. Libraries were sequenced on two (Replicate or one (Replicate 2) NextSeq (150 bp) runs resulting in at least 15,000 fragments per cell. CellRanger ATAC V1.01 (Replicate 1) and 1.1 (Replicate 2) was used to generate fastq files using the mkfastq command. The count command was used to filter, align to mm10, count barcodes, identify transposase cut sites, call peaks, call cells, and generate a count matrix.
Human scATACseq
CD69+ sorted thymocytes were mixed with CD69- DP sorted thymocytes from the same donor in a 75:25 ratio. All donor samples were processed and sequenced separately. Nuclei were isolated from 0.5–1 × 106 cells of the mixed sample. 5,000–10,000 nuclei were loaded on the 10X chromium platform, and libraries were constructed using the 10X Chromium NextGEM Single Cell ATAC Solution (V1.1). Libraries were sequenced on NextSeq (150bp), and CellRanger ATAC (V1.1) was used to generate fastq files using the mkfastq command. The count command was used to filter, align to hg38, count barcodes, identify transposase cut sites, call peaks, call cells, and generate a count matrix.
scATACseq filtering and peakset generation
Downstream analysis was performed using the Signac extension (v0.2.1 and 1.0.0) of Seurat (https://github.com/timoast/signac). Briefly, each of the four mouse samples was processed independently with the following criteria: Nucleosomal signal < 10, peak_region_fragments > 5,000 & < 50,000, blacklist_ratio < 0.025, pct_reads_in_peaks > 25 and TSS.enrichment > 2. Each of the three donor samples was processed as above, with the exception of peak region fragments > 5,000 & < 40,000 and blacklist_ratio < 0.001. Gene activity scores were calculated for each cell by measuring the accessibility within 2kb upstream of the transcription start site (TSS) using the FeatureMatrix command. For the mouse, the aggr command from CellRanger (V1.1) was used to call peaks on a merged file containing both the B2m or H2-Ab1-deficient samples for each independent experiment. To identify peaks common to both experiments, the Signac command GetIntersectingFeatures was run on the experiment-specific peaksets generated from the CellRanger aggr command. This resulted in 81,109 peaks common to the two experiments, which were used to generate a feature matrix of reads within these peaks for each sample. For the human, the Signac command MergeWithRegions was used to combine two samples, followed by GetIntersectingFeatures between the merged sample and the third independent sample, which generated a peakset of 62,555 common peaks.
scATACseq sample integration
The replicates for each species were merged together as in the standard Seurat integration procedure; however, term frequency-inverse document frequency (TF-IDF) normalization and singular value decomposition (SVD) linear dimensional reduction were used instead of RNA dimensional reduction procedures. To correct for experimental batch effect, we found integration anchors between the two experiments (mouse) or between donors (human) using the standard Seurat Integration procedure. We performed SVD linear dimensional reduction followed by UMAP using latent semantic indexing (LSI) reduction on the integrated data, and clustered the cells using a resolution of 0.3 (mouse) and 0.4 (human). For the human sample, low quality and doublet clusters were removed prior to re-clustering. To calculate inferred cell identify, we followed the procedure outlined in the standard integration workflow (Stuart et al., 2019). Briefly, we compared each ATACseq cell’s assumed transcriptome (gene activity) to the corresponding single-cell RNAseq dataset. For the mouse, this was the integrated RNAseq sample, and for the human, this was the integrated sample only including the mixture of CD69+ and unsignaled DP cells. The scATACseq cell was then labelled with the cluster assignment of the closest matching scRNAseq cell.
scATACseq lineage-specific chromatin accessibility
To identify lineage specific peak-sets (Figs. 3EF, S7C), we compared populations using the FindMarkers command with the likelihood ratio test, with experimental batch as a latent variable to mitigate the effect of batch on the result. For the CD4+-specific set, we took the union of peaks higher in ImCD4 than in ImCD8 or higher in MatCD4 than in MatCD8, and the opposite for CD8+-specific set. We then scored each cell for accessibility at these peaks using the AddChromatinModule command. To visualize this signature, we plotted the scores using FeaturePlot, with a minimum cutoff of “q5” and a maximum cutoff of “q95”. To identify differentially accessible chromatin regions across thymocytes (Figs. 3D, 7D), we compared peak accessibility between any two populations of cells with a shared inferred cell ID using the likelihood ratio test as before. We used Homer annotatePeaks to identify nearby transcriptional start sites for each peak. The 15,226 mouse peaks and 16,972 human peaks exhibiting differential accessibility between cell groups were classified by patterns of accessibility by k-means clustering. The number of k-means clusters to classify differentially accessible peaks into was determined by plotting the number of clusters (k) by the total within-clusters sum of squares and choosing the number for k at which the graph levels off. Pseudo-bulk genome coverage tracks were generated using the CoveragePlot command in Signac. To generate CD4+ and CD8+-transcriptome associated peak signature scores, we first found scATACseq peaks near CD4+ and CD8+ signature genes (Fig. 1D), using Homer annotatePeaks which identifies the nearest transcription start site. We then scored each cell for accessibility at these peaks using the AddChromatinModule command. To visualize this signature, we plotted the scores using FeaturePlot, with a minimum cutoff of “q5” and a maximum cutoff of “q95”. To calculate per-cell motif “activity” scores, we used the RunChromVar command within Signac (Schep et al., 2017). To find enriched motif activity within groups of cells, we used the FindAllMarkers command on the ChromVar assay using the likelihood ratio test. For comparison of human scATACseq to mouse scATACseq, the 15,226 differentially accessible peaks from the mouse scATACseq dataset were converted to human chromosomal coordinates using UCSC genome browser Lift Over tool (Kent et al., 2002), yielding 11,173 conserved regions. The Signac FeatureMatrix command was used to count reads at these coordinates, and a heatmap showing classes of these peaks across groups of cells was generated as described above.
Gene regulatory network analysis and scoring
To infer gene regulatory networks, we used the CellOracle suite (v0.3.5), which integrates scRNAseq and scATACseq to find putative target genes for a given transcription factor (Kamimoto et al., 2020). CellOracle utilizes the Cicero (Pliner et al., 2018) and Monocle3 packages to identify co-accessible regions from scATACseq. We used the SeuratWrapper command as.cell_data_set to convert the Seurat scATACseq object into a cell data set, then used the Cicero make_cicero_cds command to generate a Cicero object. We then followed the CellOracle pipeline to generate gene regulatory networks in a cluster specific manner.
CellOracle inferred target genes of 467 TFs across DP-1, DP-2, Sig-1, Sig-2, Sig-3, ImCD4 and ImCD8 clusters. We then selected 106 TFs which were expressed in at least 10% of cells in at least one of the above clusters. We pruned the target gene sets of each TF by retaining only those genes whose TF-target interaction score had a Bonferroni-adjusted p-value less than 0.1. We then used AUCell (Aibar et al., 2017) to compute the activity of each of these target gene sets across all B2m or H2-Ab1 deficient cells. We then used the AUCell Global_k1 activity threshold and declared as active cells with an AUCell gene set score above this threshold. To account for differences in gene detection between B2m or H2-Ab1 deficient cells, we computed Global_k1 activity thresholds separately for B2m and H2-Ab1 deficient cells. The output of this pipeline was a cluster-by-’TF target set’ fractional activity matrix, one for each genotype, where each row corresponded to the fraction of cells in each cluster in which the TF target set was active.
Next we determined gene sets whose fractional activities differed significantly between genotypes. For each TF and for each cluster, we assessed using Fisher test whether the fractions of active cells were different across the two genotypes. The p-values were then corrected for multiple testing using the Benjamini-Hochberg procedure in R (Benjamini and Hochberg, 1995).
Inference of Zbtb7b Gene Regulatory Networks
To identify transcription factors that cooperate with Thpok, we followed the CellOracle pipeline using Thpok ChIP-seq peaks instead of scATACseq, as outlined in the CellOracle manual, which identified gene sets for factors that bind within Thpok binding sites. For input we used the Thpok-sufficient scRNAseq sample, and focused on ImCD4, SMCD4, and MatCD4. To determine Thpok gene set activities, the Thpok gene set inferred by CellOracle was filtered to retain Thpok-target interactions with a Bonferroni-corrected p-value less than 0.1.
Quantification and Statistical Analysis
Except for deep-sequencing data, statistical significance was calculated with Prism software. Except where otherwise indicated in figure legends, error bars in graphs indicate standard error of the mean and statistical comparisons were done by a Student’s t-test or a One-Way ANOVA followed by a Sidak’s multiple comparisons test.
Supplementary Material
Mouse_population RNAseq: Top 100 genes contributing to principal components 1–3, and CD4+ and CD8+ gene signatures defined as in Fig. S1E.
Mouse_scRNAseq_B2M_MHCIIKO_1: Top twenty signature genes (rows) in mouse clusters from Fig. 2 and S2 (columns A-Q). Signature genes defined as those enriched in the indicated cluster over all other cells, including a column (R) containing the TCR signaling signature, defined as genes induced in Sig-1 from DP-1.
Mouse_scRNAseq_B2M_MHCIIKO_2: Columns A-G include all statistically enriched genes in mouse thymocyte clusters. pct.1 indicates percentage of cells within cluster expressing the gene, and pct.2 indicates percentage positive cells in all other clusters.
ThpokKO_scRNAseq_1: Top twenty signature genes (rows) in mouse clusters from Fig. 4 and S4 (columns A-N). Signature genes defined as those enriched in the indicated cluster over all other cells.
ThpokKO_scRNAseq_2: All statistically enriched genes in mouse thymocyte clusters. pct.1 indicates percentage of cells within cluster expressing the gene, and pct.2 indicates percentage positive cells in all other clusters.
Human_scRNAseq_1: Top twenty genes signature genes (rows) in human clusters (columns A-U). Signature genes defined as those enriched in the indicated cluster over all other cells.
Human_scRNAseq_2: Human orthologues of mouse gene signatures.
Human_scRNAseq_3: Human-derived CD8+ Specific genes and their mouse orthologues.
Human_scRNAseq_4: All statistically enriched genes in human thymocyte clusters. pct.1 indicates percentage of cells within cluster expressing the gene, and pct.2 indicates percentage positive cells in all other clusters.
Median and interquartile range of number of RNA molecules (UMI) and genes per cell for scRNAseq experiments, and fragments in peaks per cell for scATACseq experiments.
Gene composition of CellOracle target gene sets for each of the 106 analyzed transcription factors (tabs), as defined from scRNAseq and scATACseq analyses of B2m- and H2-Ab1-deficient thymocytes (shown in Figs. 2 and 3). The P-value computed by CellOracle represents the significance level of the gene being a target of the factor in the cluster indicated by the cell-type column. The adjusted p-values are Bonferroni-corrected, with only those targets whose adjusted p-value is less than 0.1 provided here.
AU Cell gene-set activity scores for indicated population (B-O). Delta values indicate differences between genotypes for a given cluster (B2m- and H2-Ab1-deficient). The p-values are the statistical significance of the observed Delta values, which are computed using a Fisher test for over-representation (see Methods). The q-values were computed using the Benjamini-Hochberg correction.
Gene composition of CellOracle target gene sets for each of the 106 analyzed transcription factors (tabs), as defined from scRNAseq and Thpok ChIPseq of Zbtb7b+/+ CD4+ SP thymocytes (shown in Figs. 4 and S4) The P-value computed by CellOracle represents the significance level of the gene being a target of the TF in the cluster indicated by the cell-type column. The adjusted p-values are Bonferroni-corrected, with only those targets whose adjusted p-value is less than 0.1 provided here.
Factors predicted (score 1) or not (score 0) to bind Zbtb7b, Gm15417, or Runx3 loci from CellOracle data (A) from scRNAseq and scATACseq analyses of B2m- and H2-Ab1-deficient thymocytes (Table S3) or (B) from scRNAseq and ChIPseq data from Zbtb7b+/+ CD4+ SP thymocytes (Table S5).
Donor age range and gender, and sorted population for human thymocyte analyses.
Highlights:
Distinct developmental fates induced by β2m- and MHC II-dependent agonist ligands
Asymmetric emergence of CD4+ and CD8+ differentiation programs in thymocytes
Thpok binds and controls multiple targets during CD4+-lineage commitment
High conservation of differentiation programs between mouse and human αβ thymocytes
Acknowledgements
We thank J. Paiano, A. Stanlie, and A. Nussenzweig for assistance with ChIPseq experiments; Q. Xiao, M. Balmaceno-Criss, and H. Kwak for expert animal care and genotyping; L. Notarangelo for guidance with patient samples; A. Bhandoola, B. Larsen, J. O’Shea, H-Y. Shih, T. Stuart, and M. Taves for useful discussions; Y. Zhao, the CCR Flow Cytometry Core, and the NIH high performance computing cluster staff for assistance; and J. Ashwell, M. Balmaceno-Criss, A. Bhandoola, V. Lazarevic, P. Love, L. Notarangelo, H-Y. Shih, and M. Vacchio for reading the manuscript. Support from CCR Single Cell Analysis Facility was funded by FNLCR Contract HHSN261200800001E. Sequencing was performed with the CCR Genomics Core or Frederick National Lab for Cancer Research CCR Sequencing Facility. This work used the NIH high-performance computing cluster. This work was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH, and by an NSF fellowship to L.B.C.
Footnotes
Competing Interest
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adoro S, McCaughtry T, Erman B, Alag A, Van Laethem F, Park JH, Tai X, Kimura M, Wang L, Grinberg A, et al. (2012). Coreceptor gene imprinting governs thymocyte lineage fate. EMBO J 31, 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nature methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliahmad P, and Kaye J. (2008). Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med 205, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yeung BB, Melichar HJ, Ross JO, Cheng DA, Zikherman J, Shokat KM, Robey EA, and Weiss A. (2014). Quantitative and temporal requirements revealed for Zap70 catalytic activity during T cell development. Nat Immunol 15, 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, and Love PE (1998). CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med 188, 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300. [Google Scholar]
- Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, and Strasser A. (2002). BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415, 922–926. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Huang SN, Wutz G, Tang W, Zagnoli-Vieira G, Callen E, Wong N, Day A, Peters JM, et al. (2019). Topoisomerase II-Induced Chromosome Breakage and Translocation Is Determined by Chromosome Architecture and Transcriptional Activity. Molecular cell 75, 252–266.e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AC, and Bosselut R. (2010). Decision checkpoints in the thymus. Nat Immunol 11, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AC, Grainger JR, Xiong Y, Kanno Y, Chu HH, Wang L, Naik S, dos Santos L, Wei L, Jenkins MK, et al. (2012). The transcription factors Thpok and LRF are necessary and partly redundant for T helper cell differentiation. Immunity 37, 622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, Ryu HS, Kim S, Lee JE, Park YH, et al. (2017). Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nature Communications 8, 15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, et al. (2019). The Emergence and Functional Fitness of Memory CD4(+) T Cells Require the Transcription Factor Thpok. Immunity 50, 91–105.e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Littman DR, and Taniuchi I. (2009). RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol 9, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello PS, Nicolas RH, Watanabe Y, Rosewell I, and Treisman R. (2004). Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nat Immunol 5, 289–298. [DOI] [PubMed] [Google Scholar]
- Cowan JE, Parnell SM, Nakamura K, Caamano JH, Lane PJ, Jenkinson EJ, Jenkinson WE, and Anderson G. (2013). The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+ thymocyte development. J Exp Med 210, 675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley SR, Hu DY, and Goodnow CC (2013). Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-kappaB. J Exp Med 210, 269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, and Palmer E. (2006). Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, and Littman DR (2008). ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol 9, 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Tillman RE, Naoe Y, Taniuchi I, and Littman DR (2007). The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 204, 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusby MJ, Johnson RS, Papaioannou VE, and Glimcher LH (1991). Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science 253, 1417–1420. [DOI] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nature biotechnology 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, and Kappes DJ (2005). The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833. [DOI] [PubMed] [Google Scholar]
- He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, and Kappes DJ (2008). CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmers S, Schizas M, Azizi E, Dikiy S, Zhong Y, Feng Y, Altan-Bonnet G, and Rudensky AY (2019). IL-2 production by self-reactive CD4 thymocytes scales regulatory T cell generation in the thymus. The Journal of experimental medicine 216, 2466–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, and Alberola-Ila J. (2003). GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19, 83–94. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, and Jameson SC (2014). The self-obsession of T cells: how TCR signaling thresholds affect fate ‘decisions’ and effector function. Nat Immunol 15, 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo MA, Masuda K, Hojo H, Nagahata Y, Yasuda K, Ohara D, Takeuchi Y, Hirota K, Suzuki Y, Kawamoto H, and Kawaoka S. (2019). Identification of a genomic enhancer that enforces proper apoptosis induction in thymic negative selection. Nat Commun 10, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, and Zhuang Y. (2012). E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity 36, 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakugawa K, Kojo S, Tanaka H, Seo W, Endo TA, Kitagawa Y, Muroi S, Tenno M, Yasmin N, Kohwi Y, et al. (2017). Essential Roles of SATB1 in Specifying T Lymphocyte Subsets. Cell reports 19, 1176–1188. [DOI] [PubMed] [Google Scholar]
- Kamimoto K, Hoffmann CM, and Morris SA (2020). CellOracle: Dissecting cell identity via network inference and in silico gene perturbation. bioRxiv, 2020.2002.2017.947416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL (2009). E and ID proteins branch out. Nat Rev Immunol 9, 175–184. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D. (2002). The human genome browser at UCSC. Genome research 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, and Maehr R. (2018). A Single-Cell Transcriptomic Atlas of Thymus Organogenesis Resolves Cell Types and Developmental Maturation. Immunity 48, 1258–1270 e1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojo S, Tanaka H, Endo TA, Muroi S, Liu Y, Seo W, Tenno M, Kakugawa K, Naoe Y, Nair K, et al. (2017). Priming of lineage-specifying genes by Bcl11b is required for lineage choice in post-selection thymocytes. Nat Commun 8, 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. (2008). The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol 9, 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan J, and Killeen N. (2004). CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol 172, 3999–4007. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavaert M, Liang KL, Vandamme N, Park J-E, Roels J, Kowalczyk MS, Li B, Ashenberg O, Tabaka M, Dionne D, et al. (2020). Integrated scRNA-Seq Identifies Human Postnatal Thymus Seeding Progenitors and Regulatory Dynamics of Differentiating Immature Thymocytes. Immunity 52, 1088–1104.e1086. [DOI] [PubMed] [Google Scholar]
- Le J, Park JE, Ha VL, Luong A, Branciamore S, Rodin AS, Gogoshin G, Li F, Loh Y-HE, Camacho V, et al. (2020). Single-Cell RNA-Seq Mapping of Human Thymopoiesis Reveals Lineage Specification Trajectories and a Commitment Spectrum in T Cell Development. Immunity 52, 1105–1118.e1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, and Hsieh CS (2009). Rare development of Foxp3+ thymocytes in the CD4+CD8+ subset. J Immunol 183, 2261–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. (2001). A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774. [DOI] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, and Rudensky AY (2016). T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol 16, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Taylor BJ, Sun G, and Bosselut R. (2005). Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes. J Immunol 175, 4465–4474. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey MA, Kimura MY, Waickman AT, Feigenbaum L, Singer A, and Park JH (2014). The transcription factor ThPOK suppresses Runx3 and imposes CD4(+) lineage fate by inducing the SOCS suppressors of cytokine signaling. Nat Immunol 15, 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud SA, Manlove LS, Schmitz HM, Xing Y, Wang Y, Owen DL, Schenkel JM, Boomer JS, Green JM, Yagita H, et al. (2014). Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat Immunol 15, 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden VS, and Strasser A. (2003). Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annual review of immunology 21, 71–105. [DOI] [PubMed] [Google Scholar]
- McDonald BD, Jabri B, and Bendelac A. (2018). Diverse developmental pathways of intestinal intraepithelial lymphocytes. Nat Rev Immunol 18, 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGargill MA, Ch’en IL, Katayama CD, Pages G, Pouyssegur J, and Hedrick SM (2009). Cutting edge: Extracellular signal-related kinase is not required for negative selection of developing T cells. J Immunol 183, 4838–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Ross JO, Herzmark P, Hogquist KA, and Robey EA (2013). Distinct temporal patterns of T cell receptor signaling during positive versus negative selection in situ. Science signaling 6, ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, et al. (2013). The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol 14, 619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, and Hogquist KA (2011). T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 208, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge JM, and Harrow J. (2015). Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mammalian genome : official journal of the International Mammalian Genome Society 26, 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, and Taniuchi I. (2008). Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol 9, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Ohkura N, Kitagawa Y, and Sakaguchi S. (2013). Development and maintenance of regulatory T cells. Immunity 38, 414–423. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, and Ho IC (2003). Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19, 863–875. [DOI] [PubMed] [Google Scholar]
- Palmer E. (2003). Negative selection — clearing out the bad apples from the T-cell repertoire. Nature Reviews Immunology 3, 383–391. [DOI] [PubMed] [Google Scholar]
- Papalexi E, and Satija R. (2018). Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 18, 35–45. [DOI] [PubMed] [Google Scholar]
- Park JE, Botting RA, Dominguez Conde C, Popescu DM, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, et al. (2020). A cell atlas of human thymic development defines T cell repertoire formation. Science 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. (2018). Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Molecular cell 71, 858–871.e858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C. (2017a). Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C. (2017b). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, Chen Z, Whittle B, Liu L, Fairlie DP, et al. (2015). Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med 212, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV (2019). Programming for T-lymphocyte fates: modularity and mechanisms. Genes Dev 33, 1117–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscher R, and Hogquist KA (2019). Development, ontogeny, and maintenance of TCRαβ+ CD8αα IEL. Current Opinion in Immunology 58, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Hombauer M, Bilic I, Naoe Y, Schebesta A, Taniuchi I, and Ellmeier W. (2010). The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol 11, 442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A. (2008). The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14, 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekkali B, Szabat E, Ktistaki E, Tolaini M, Roderick K, Harker N, Patel A, Williams K, Norton T, and Kioussis D. (2005). Human high mobility group box transcription factor 1 affects thymocyte development and transgene variegation. J Immunol 175, 5203–5212. [DOI] [PubMed] [Google Scholar]
- Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, et al. (2016). The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, and Taniuchi I. (2008). Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825. [DOI] [PubMed] [Google Scholar]
- Shao H, Kono DH, Chen LY, Rubin EM, and Kaye J. (1997). Induction of the early growth response (Egr) family of transcription factors during thymic selection. J Exp Med 185, 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HY, Sciume G, Poholek AC, Vahedi G, Hirahara K, Villarino AV, Bonelli M, Bosselut R, Kanno Y, Muljo SA, and O’Shea JJ (2014). Transcriptional and epigenetic networks of helper T and innate lymphoid cells. Immunol Rev 261, 23–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A, Adoro S, and Park JH (2008). Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol 8, 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, and Hogquist KA (2003). Positive and negative selection of T cells. Annual review of immunology 21, 139–176. [DOI] [PubMed] [Google Scholar]
- Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B, Kawamoto H, Zhu J, Tan K, and Xue HH (2014). TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat Immunol 15, 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky GL, Jameson SC, and Hogquist KA (2012). Selection of self-reactive T cells in the thymus. Annual review of immunology 30, 95–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky GL, Xing Y, Erickson JR, Kalekar LA, Wang X, Mueller DL, Jameson SC, and Hogquist KA (2013). Murine thymic selection quantified using a unique method to capture deleted T cells. Proc Natl Acad Sci U S A 110, 4679–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, and Bosselut R. (2005). The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol 6, 373–381. [DOI] [PubMed] [Google Scholar]
- Swat W, Dessing M, von Boehmer H, and Kisielow P. (1993). CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur J Immunol 23, 739–746. [DOI] [PubMed] [Google Scholar]
- Taniuchi I. (2018). CD4 Helper and CD8 Cytotoxic T Cell Differentiation. Annual review of immunology 36, 579–601. [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, and Littman DR (2002). Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633. [DOI] [PubMed] [Google Scholar]
- Tenno M, Kojo S, Lawir DF, Hess I, Shiroguchi K, Ebihara T, Endo TA, Muroi S, Satoh R, Kawamoto H, et al. (2018). Cbfbeta2 controls differentiation of and confers homing capacity to prethymic progenitors. J Exp Med 215, 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turka LA, Schatz DG, Oettinger MA, Chun JJ, Gorka C, Lee K, McCormack WT, and Thompson CB (1991). Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science 253, 778–781. [DOI] [PubMed] [Google Scholar]
- Ueno T, Saito F, Gray DH, Kuse S, Hieshima K, Nakano H, Kakiuchi T, Lipp M, Boyd RL, and Takahama Y. (2004). CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med 200, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, and Bosselut R. (2008a). The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29, 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, and Bosselut R. (2008b). Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol 9, 1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. (2011). Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, and Bosselut R. (2007). The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J Immunol 179, 4405–4414. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, and Kaye J. (2002). TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol 3, 272–280. [DOI] [PubMed] [Google Scholar]
- Wood CD, Veenstra H, Khasnis S, Gunnell A, Webb HM, Shannon-Lowe C, Andrews S, Osborne CS, and West MJ (2016). MYC activation and BCL2L11 silencing by a tumour virus through the large-scale reconfiguration of enhancer-promoter hubs. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, et al. (2003). Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A 100, 7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Wang X, Jameson SC, and Hogquist KA (2016). Late stages of T cell maturation in the thymus involve NF-kappaB and tonic type I interferon signaling. Nat Immunol 17, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, and Bosselut R. (2012). CD4-CD8 differentiation in the thymus: connecting circuits and building memories. Curr Opin Immunol 24, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Ladi E, Chan SW, Li O, Killeen N, Kappes DJ, and Robey EA (2007). CCR7 expression in developing thymocytes is linked to the CD4 versus CD8 lineage decision. J Immunol 179, 7358–7364. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, Desland F, Chudnovskiy A, Mortha A, Dominguez C, et al. (2019). The cis-Regulatory Atlas of the Mouse Immune System. Cell 176, 897–912.e820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamisch M, Tian L, Grenningloh R, Xiong Y, Wildt KF, Ehlers M, Ho IC, and Bosselut R. (2009). The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med 206, 2685–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, and Jaenisch R. (1990). Beta 2-microglobulin deficient mice lack CD4–8+ cytolytic T cells. Nature 344, 742–746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mouse_population RNAseq: Top 100 genes contributing to principal components 1–3, and CD4+ and CD8+ gene signatures defined as in Fig. S1E.
Mouse_scRNAseq_B2M_MHCIIKO_1: Top twenty signature genes (rows) in mouse clusters from Fig. 2 and S2 (columns A-Q). Signature genes defined as those enriched in the indicated cluster over all other cells, including a column (R) containing the TCR signaling signature, defined as genes induced in Sig-1 from DP-1.
Mouse_scRNAseq_B2M_MHCIIKO_2: Columns A-G include all statistically enriched genes in mouse thymocyte clusters. pct.1 indicates percentage of cells within cluster expressing the gene, and pct.2 indicates percentage positive cells in all other clusters.
ThpokKO_scRNAseq_1: Top twenty signature genes (rows) in mouse clusters from Fig. 4 and S4 (columns A-N). Signature genes defined as those enriched in the indicated cluster over all other cells.
ThpokKO_scRNAseq_2: All statistically enriched genes in mouse thymocyte clusters. pct.1 indicates percentage of cells within cluster expressing the gene, and pct.2 indicates percentage positive cells in all other clusters.
Human_scRNAseq_1: Top twenty genes signature genes (rows) in human clusters (columns A-U). Signature genes defined as those enriched in the indicated cluster over all other cells.
Human_scRNAseq_2: Human orthologues of mouse gene signatures.
Human_scRNAseq_3: Human-derived CD8+ Specific genes and their mouse orthologues.
Human_scRNAseq_4: All statistically enriched genes in human thymocyte clusters. pct.1 indicates percentage of cells within cluster expressing the gene, and pct.2 indicates percentage positive cells in all other clusters.
Median and interquartile range of number of RNA molecules (UMI) and genes per cell for scRNAseq experiments, and fragments in peaks per cell for scATACseq experiments.
Gene composition of CellOracle target gene sets for each of the 106 analyzed transcription factors (tabs), as defined from scRNAseq and scATACseq analyses of B2m- and H2-Ab1-deficient thymocytes (shown in Figs. 2 and 3). The P-value computed by CellOracle represents the significance level of the gene being a target of the factor in the cluster indicated by the cell-type column. The adjusted p-values are Bonferroni-corrected, with only those targets whose adjusted p-value is less than 0.1 provided here.
AU Cell gene-set activity scores for indicated population (B-O). Delta values indicate differences between genotypes for a given cluster (B2m- and H2-Ab1-deficient). The p-values are the statistical significance of the observed Delta values, which are computed using a Fisher test for over-representation (see Methods). The q-values were computed using the Benjamini-Hochberg correction.
Gene composition of CellOracle target gene sets for each of the 106 analyzed transcription factors (tabs), as defined from scRNAseq and Thpok ChIPseq of Zbtb7b+/+ CD4+ SP thymocytes (shown in Figs. 4 and S4) The P-value computed by CellOracle represents the significance level of the gene being a target of the TF in the cluster indicated by the cell-type column. The adjusted p-values are Bonferroni-corrected, with only those targets whose adjusted p-value is less than 0.1 provided here.
Factors predicted (score 1) or not (score 0) to bind Zbtb7b, Gm15417, or Runx3 loci from CellOracle data (A) from scRNAseq and scATACseq analyses of B2m- and H2-Ab1-deficient thymocytes (Table S3) or (B) from scRNAseq and ChIPseq data from Zbtb7b+/+ CD4+ SP thymocytes (Table S5).
Donor age range and gender, and sorted population for human thymocyte analyses.
Data Availability Statement
The common accession number for all sequence data reported in this paper is GSE148981. Specific accession numbers are listed in the key resource table. All other data and code are available upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Bim-AF647 (C34C5) | Cell Signaling Technology | Cat# 10408S, RRID:AB_2797721 |
| Anti-Bim-PE (C34C5) | Cell Signaling Technology | Cat# 12186S, RRID:AB_2797842 |
| Anti-Cleaved Caspase-3 (ASP175) (Clone D3-E9) | Cell Signaling Technology | Cat# 8788S, RRID:AB_2797665 |
| Anti-CCR7-Pe-Cy7 (Clone 4B12) | Thermofisher | Cat# 25-1971-82, RRID:AB_469652 |
| Anti-CD122-FITC (Clone TM-Beta 1) | Thermofisher | Cat# 11-1222-82, RRID:AB_465189 |
| Anti-CD25- APC-eF780 (Clone PC61.5) | Thermofisher | Cat# 47-0251-82, RRID:AB_1272179 |
| Anti-CD25-PerCP-Cy5.5 (Clone PC61.5) | Thermofisher | Cat# 45-0251-82, RRID:AB_914324 |
| Anti-CCR7-PE-Cy7 (Clone 4B12) | Thermofisher | Cat# 25-1971-82, RRID:AB_469652 |
| Anti-CD279-BV786 (Clone J43) | BD Pharmigen | Cat# 744548, RRID:AB_2742319 |
| Anti-CD4-APC (Clone GK1.5) | Thermofisher | Cat# 17-0041-82, RRID:AB_469320 |
| Anti-CD4-BV650 | BD Pharmigen | Cat# 563747, RRID:AB_2716859 |
| Anti-CD4-BV786 (Clone RM4-4) | BD Pharmigen | Cat# 563727 RRID:AB_2728707 |
| Anti-CD4-eFluor450 (Clone GK1.5) | Thermofisher | Cat# 48-0041-82, RRID:AB_10718983 |
| Anti-CD4-PerCP-Cy5.5 (Clone RM4-5) | Thermofisher | Cat# 45-0042-82, RRID:AB_1107001 |
| Anti-CD44-Alexa Fluor 700 (Clone IM7) | Thermofisher | Cat# 56-0441-82, RRID:AB_494011 |
| Anti-CD45.2-BV786 (Clone 104) | BD Pharmigen | Cat# 563686, RRID:AB_2738375 |
| Anti-CD5-BV605 (Clone 53-7.3) | BD Pharmigen | Cat# 563194, RRID:AB_2738061 |
| Anti-CD69-PE (Clone H1.253) | Thermofisher | Cat# 12-0691-82 RRID: AB_465732 |
| Anti-CD69-PerCP-Cy5.5 (Clone H1.253) | Thermofisher | Cat# 45-0691-82, RRID:AB_1210703 |
| Anti-CD8a-APC (Clone 53-6.7) | Thermofisher | Cat# 17-0081-82, RRID:AB_469335 |
| Anti-CD8a-APC-eFluor780 (Clone 53-6.7) | Thermofisher | Cat# 47-0081-82, RRID:AB_1272185 |
| Anti-CD8a-BUV395 (Clone 53-6.7) | BD Pharmigen | Cat# 563786, RRID:AB_2732919 |
| Anti-CD8a-PerCP-Cy5.5 (Clone 53-6.7) | Thermofisher | Cat# 45-0081-82, RRID:AB_1107004 |
| Anti-CD8a-PE (Clone 53-6.7) | Thermofisher | Cat# 12-0081-83, RRID:AB_465530 |
| Anti-GATA3-APC (Clone L50-823) | BD Pharmigen | Cat# 560078, RRID:AB_1645317 |
| Anti-HELIOS-Alexa Fluor 647 (Clone 22F6) | BioLegend | Cat# 137218, RRID:AB_10660750 |
| Anti-HELIOS-Pacific Blue (Clone 22F6) | BioLegend | Cat# 137220, RRID:AB_10690535 |
| Anti-MHC Class I (H-2Kb)-APC (Clone AF6-88.5.5.3) | Thermofisher | Cat# 17-5958-82, RRID:AB_1311280 |
| Anti-MHC Class I (H-2Kb)-APC (Clone AF6-88.5.5.3) | Thermofisher | Cat# 17-5958-82, RRID:AB_1311280 |
| Anti-NUR77-Alexa Fluor 647 (Clone 12.14) | Thermofisher | Cat# 51-5965-82, RRID:AB_2802306 |
| Anti-NUR77-PE (Clone 12.14) | Thermofisher | Cat# 12-5965-82, RRID:AB_1257209 |
| Anti-RUNX3-PE (Clone R3-5G4) | BD Pharmigen | Cat# 564814, RRID:AB_2738969 |
| Anti-TCRb-BUV737 (Clone H57-597) | BD Pharmigen | Cat# 612821 |
| Anti-THPOK-Alexa Fluor 647 (Clone T43-94) | BD Pharmigen | Cat# 565500, RRID:AB_2739268 |
| Anti-CD4-APC (Clone RPA-T4) | Thermofisher | Cat# 17-0049-42, RRID:AB_1272048 |
| Anti-CD4-eFluor 450 (Clone RPA-T4) | Thermofisher | Cat# 48-0049-42, RRID:AB_1272057 |
| Anti-CD69-FITC (Clone FN50) | BD Pharmigen | Cat# 560969, RRID:AB_10562195 |
| Anti-CD8-PerCp-Cy5.5 (Clone RPA-T8) | BD Pharmigen | Cat# 560662, RRID:AB_1727513 |
| Anti-RUNX3-PE (Clone R3-5G4) | BD Pharmigen | Cat# 564814, RRID:AB_2738969 |
| Anti-THPOK-PE (Clone 6/hcKrox) | BD Pharmigen | Cat# 565730 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde | Thermofisher | 28906 |
| Critical Commercial Assays | ||
| Transcription factor Staining Buffer Set | Thermofisher | 00-5523-00 |
| Cytofix/Cytoperm | BD Pharmingen | 554714 |
| Protein-A beads | Invitrogen | 10001D |
| Streptavidin beads (M280) | Invitrogen | 12205D |
| QIAshredder | QIAGEN | 79656 |
| RNeasy Plus Micro Kit | QIAGEN | 74034 |
| RNeasy Plus Mini Kit | QIAGEN | 74134 |
| 10X Chromium single cell 3’ V2 kit | 10X Genomics | 120267, 120262 |
| 10X Chromium Next GEM Single Cell 5’ Reagent Kit (v1.1) kit | 10X Genomics | 1000127, 1000165 |
| 10X Chromium Single Cell ATAC Solution (v1.0) kit | 10X Genomics | PN-1000110 |
| 10X Chromium NextGEM Single Cell ATAC Solution (V1.1) kit | 10X Genomics | 1000176, 1000162 |
| Proteinase K | Invitrogen | 25530049 |
| RNase A | Invitrogen | 1209121 |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1030 |
| Minelute PCR Purification Kit | QIAGEN | 28004 |
| QIAquick PCR Purification Kit | QIAGEN | 28104 |
| Deposited Data | ||
| Developing Mouse Thymocyte Population RNAseq | This study | GSE148973 |
| Developing Mouse Thymocyte Single-cell RNAseq | This study | GSE148977 |
| Thpok KO Thymocyte Population RNAseq | This study | GSE148974 |
| Thpok KO Thymocyte Single-cell RNAseq | This study | GSE157286 |
| Thpok KO Thymocyte Population ATACseq | This study | GSE148975 |
| Developing Mouse Thymocyte Single-cell ATACseq | This study | GSE148979 |
| Developing Human Thymocyte Single-cell RNAseq | This study | GSE148978 |
| Developing Human Thymocyte Single-cell ATACseq | This study | GSE148980 |
| CHIPseq (Thpok CD4 SP thymocytes) | This study | GSE148976 |
| CHIPseq (Thpok CD4 T Cells) | Ciucci et al. (2019) | GSE116506 |
| CHIPseq (Cbfβ) | Tenno et al. (2019) | GSE90794 |
| CHIPseq (Gata3) | Wei et al. (2011) | GSE20898 |
| Experimental Models: Organisms/Strains | ||
| Zbtb7b fl | (Wang et al., 2008a) | |
| Zbtb7b Bio | (Ciucci et al., 2019) | |
| Rosa26 BirA | (Driegen et al., 2005) | |
| NCI B6-Ly5.1/Cr (CD45.1) | Charles River | Charles River 564 |
| C57BL/6Ncr (CD45.2) | Charles River | Charles River 556 |
| B6 CD45.1 CD45.2 | Generated in house | |
| Zbtb7b GFP | (Wang et al., 2008b) | |
| Runx3 RFP | (Zamisch et al., 2009) | |
| B2m −/− | Taconic | (Zijlstra et al., 1990) |
| H2-Ab1 −/− | JAX | (Grusby et al., 1991) |
| Cd4-cre | Taconic | (Lee et al., 2001) |
| Software and Algorithms | ||
| Graphpad Prism 7.0 | graphpad.com | n/a |
| Flowjo 10.0 | flowjo.com | n/a |
| R | r-project.org | n/a |
| Seurat v 3.1 | (Butler et al., 2018) | |
| Signac v 0.2.1 and 1.0.0 | github.com/timoast/signac | |
| MACS2 (v2.2.5) | github.com/taoliu/MACS | |
| CellRanger v2.2 and 3.1 | 10X Genomics | |
| DESeq2 | (Love et al., 2014) | |
| STAR aligner (v2.4.0) | github.com/alexdobin/STAR | |
| CellOracle (v0.3.5) | github.com/morris-lab/CellOracle | |
| AUCell | (Aibar et al., 2017) | |
| Htseq (v0.11.4) | (Anders et al., 2015) | |
| Homer (v4.10) | (Heinz et al., 2010) | |
| Bowtie2 (v2.3.4) | (Langmead and Salzberg, 2012) | |
| Samtools (v1.6) | (Li et al., 2009) | |
| Picard (v2.2.8) | http://broadinstitute.github.io/picard/ | |
| Bedtools (v2.29.2) | github.com/arq5x/bedtools2 | |
| Diffbind v(3.1) | (Ross-Innes et al., 2012) | |
| Deeptools (v3.3.0) | (Ramirez et al., 2016) | |
| Monocle3 | (Cao et al., 2019) | |
| biomaRt (v2.42.0) | github.com/grimbough/biomaRt | |
| CellRanger ATAC (V1.01 and 1.1) | ||
| UCSC LiftOver | (Kent et al., 2002) | |