Abstract
Ral GTPases have been implicated as mediators of Ras-induced signal transduction from observations that Ral-specific guanine nucleotide exchange factors associate with Ras and are activated by Ras. The cellular role of Ral family proteins is unclear, as is the contribution that Ral may make to Ras-dependent signaling. Here we show that expression of activated Ral in quiescent rodent fibroblasts is sufficient to induce activation of NF-κB-dependent gene expression and cyclin D1 transcription, two key convergence points for mitogenic and survival signaling. The regulation of cyclin D1 transcription by Ral is dependent on NF-κB activation and is mediated through an NF-κB binding site in the cyclin D1 promoter. Ral activation of these responses is likely through an as yet uncharacterized effector pathway, as we find activation of NF-κB and the cyclin D1 promoter by Ral is independent of association of Ral with active phospholipase D1 or Ral-binding protein 1, two proteins proposed to mediate Ral function in cells.
Ral proteins are small GTPases that have been implicated in the control of cell proliferation and Ras-mediated oncogenic transformation (14, 48). The two known Ral isoforms, RalA and RalB, are 85% identical and comprise a distinct family within the Ras superfamily of GTPases (10). Ral proteins are more than 50% identical to Ras, have overall structural features similar to those of Ras, but do not share any known effector or regulatory proteins with Ras (6, 14).
Like Ras GTPases, Ral proteins become biologically active upon exchange of bound GDP for GTP. This exchange is catalyzed in vivo by Ral-specific guanine nucleotide exchange factors (RalGEFs) (14). Several RalGEFs which contain carboxy-terminal Ras binding domains have been identified (14, 42). The observations that activated Ras can associate directly with RalGEFs (14) and activate the enzymatic activity of RalGEFs in vitro and in transfected cells (14, 55, 62) and that mitogen-dependent activation of Ral proteins requires Ras activation (63) have lead to the hypothesis that RalGEFs are Ras effector proteins. Consistent with this hypothesis is the observation that activation of Ral proteins appears to be required for Ras-induced oncogenic growth and morphological transformation (50, 55, 60) and induction of DNA synthesis (38). In addition, expression of RalGEFs or activated Ral proteins can cooperate with activation of other Ras effector cascades to transform cells (50, 55, 60). These observations suggest that Ral proteins may be important mediators of Ras-induced proliferative signals. However, the mechanism by which Ral may contribute to Ras signaling is unknown.
In addition to effects on proliferation, Ral has been directly implicated in receptor-mediated endocytosis (40), Src kinase activation (20), phospholipase D1 (PLD1) activation (16, 23), and regulation of the actin cytoskeleton (42). Active PLD1 (23, 36), Ral-binding protein 1 (RalBP1) (9, 25, 44), and filamin (42) have been identified as Ral-interacting proteins and may function as Ral effectors. PLD1 is constitutively associated with Ral protein in cells (23). However, activation of Ral cooperates with ADP ribosylation factor GTPases to activate PLD1, perhaps by contributing to the formation of a PLD1 activation complex (28, 35). There is some evidence that active PLD1 can contribute to proliferation (13). For example, transfected PLD1 can contribute to oncogenic transformation of fibroblasts overexpressing epidermal growth factor (EGF) receptors (34). Unlike PLD1, RalBP1 associates with Ral in a GTP-dependent manner (9, 25, 44). The functional significance of a Ral-RalBP1 interaction is unknown; however, RalBP1 contains a GTPase-activating protein (GAP) domain that has activity toward Cdc42 and Rac GTPases (9, 25, 44). This observation has led to the hypothesis that Ral may negatively regulate the activity of these GTPases. In support of this, studies using PC12 cells suggest that RalGEFs can interfere with neurite differentiation in a Rac-dependent fashion (19). However, a direct effect of Ral on Rac or Cdc42 regulation has not yet been demonstrated. Finally, the Ral-filamin interaction may influence regulation of the actin cytoskeleton. Filamin binds Ral-GTP, and Ral-GTP will induce filopodia in human melanoma cells in a filamin-dependent fashion. In contrast to a potential role of Ral upstream of Rac/Cdc42 via RalBP1, Ral-mediated generation of filopodia is likely downstream of Cdc42 activation (42).
To further elaborate the mechanism by which Ral GTPases may contribute to proliferation and transformation, we have examined the consequences of Ral activation on gene induction events that have been defined as critical convergence points for multiple mitogenic signaling cascades. We show here that activated Ral is sufficient to induce NF-κB transcription factor activity and accumulation of cyclin D1 protein. Ral activation of cyclin D1 expression is NF-κB dependent and is mediated by NF-κB binding sites in the cyclin D1 promoter. Activation of NF-κB and cyclin D1 expression by Ral is independent of Ral association with either PLD1 or RalBP1 and likely proceeds through a novel effector pathway. Ral-dependent regulation of NF-κB and cyclin D1 provides a mechanistic explanation for the positive role of Ral proteins in the regulation of proliferation and oncogenic transformation.
MATERIALS AND METHODS
Plasmids.
All Ral expression constructs encode variants of the simian RalB protein. pRK5-ralB23V, pRK5-ralB28N, pRK5-ralB23V,49N, pRK5-ralB23V(ΔCAAX), and ΔN11ralB23V were derived by site-directed mutagenesis as described elsewhere (6). All alleles were entirely sequenced after transfer to pRK5. The coding sequence for ralB23V was inserted as a BamHI fragment into pBabePuro to create pBabePuro-ralB23V. Luciferase reporter plasmids 2X NF-κB-Luc, -1745 CD1-Luc, -66 CD1-Luc, -66 CD1-κBmut-Luc, and -66 ATFmut1-Luc are described elsewhere (17, 21). To create -66 ATFmut2-Luc, the region of -66 CD1-Luc containing the activating transcription factor (ATF) binding site consensus sequence was mutated from 5′-TAACGTCACACGGACT-3′ to 5′-TcgCGTCAccCGGACT-3′ (mutated bases are in lowercase) by PCR using human cyclin D1-specific primers. To create 3X SRE-Luc, three tandem repeats of the murine c-Fos serum response element (SRE) were removed from 5X SRE-CAT (30) and inserted upstream of the firefly luciferase gene in pGL2-basic (Promega). pCMV-GFP expresses enhanced green fluorescent protein (GFP) under control of the constitutive cytomegalovirus (CMV) promoter in pCMV5 (gift from S. W. Lacey, UT Southwestern Medical Center). pCH110 constitutively expresses the lacZ gene from a simian virus 40 promoter (Pharmacia Biotech, Inc.). pDCR-ras12V, pDCR-ras12V,37G, pRSV-p65, pCMV-IKKβKM, pCEP4-IκBα SS/AA, pcDNA3-HA-NIK, pcDNA3myc-NIK (EE429/430AA), and pEGFP-p65 are described elsewhere (2, 17, 52, 54, 59).
Luciferase reporter assays.
NIH 3T3 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum. The day prior to transfection, cells were seeded at a density of 200,000 cells per 35-mm-diameter dish. Calcium phosphate precipitates were prepared by standard methods with 0.75 μg of luciferase reporter construct, 1.0 μg of pCH110, 0.2 μg of pCMV-GFP, and 6 μg of Ral expression constructs as indicated in the figures; 18 h following transfection, precipitates were replaced with DMEM–0.5% calf serum. After 24 h of incubation in low serum, lysates were prepared in luciferase lysis buffer (200 μl/plate; Promega). Firefly luciferase assays were performed as instructed by the manufacturer (Promega). β-Galactosidase activity from aliquots of the same lysates was assayed as described previously (51). Levels of reporter gene induction in transiently transfected cells were calculated by normalizing luciferase activity to β-galactosidase activity. RalB variant expression was monitored by Western analysis with ant-RalB polyclonal antibodies (Transduction Laboratories).
Immunofluorescence.
To detect effects of RalB expression on localization of p65/RelA, NIH 3T3 cells were transfected with 3 μg of pRSV-p65 together with 0.2 μg of pCMV-GFP and 3 μg of pRK5 or pRK5-ralB23V. At 24 h posttransfection, cells were fixed in 3.7% formaldehyde and then permeabilized in 0.25% Triton X-100 and 1% calf serum in phosphate-buffered saline for 1 h at room temperature. Expressed p65 was detected by incubation with anti-p65 rabbit polyclonal antibodies (Santa Cruz Biotechnology) followed by rhodamine-conjugated goat anti-rabbit immunoglobulin G (IgG). Transfected cells were detected by GFP fluorescence. Alternatively, cells were transfected with 3 μg pEGFP-p65, which can be visualized by direct fluorescence. To detect effects of RalB expression on cyclin D1 accumulation, NIH 3T3 cells were transfected with 0.2 μg of pCMV-GFP together with 6 μg of pRK5 or pRK5-ral23V; 18 h posttransfection, growth medium was replaced with serum-free DMEM, and the cells were incubated for an additional 24 h. The serum-starved cells were then fixed and permeabilized as described above. Expression of endogenous cyclin D1 was detected by incubation with anti-cyclin D1 monoclonal antibody (Upstate Biotechnology, Inc.) followed by rhodamine-conjugated goat anti-mouse IgG. To detect effects of dominant inhibitory RalB on oncogenic ras-induced cyclin D1 accumulation, NIH 3T3 cells stably expressing ras12V were transfected with 7 μg of pRK5-ralB28N. Following serum starvation as described above, cells were fixed in 3.7% formaldehyde and permeabilized in chilled acetone for 5 min at −20°C. Cyclin D1 expression was detected as described above. ralBN28 expression was detected using rabbit anti-RalB polyclonal antibodies (Transduction Laboratories) followed by fluorescein-conjugated goat anti-rabbit IgG. All images were acquired with a Zeiss fluorescence microscope at a magnification of ×40 using a chilled Argus charge-coupled device camera (Hamamatsu).
Reverse transcription (RT)-PCR analysis.
NIH 3T3 cells were transfected with 6 μg of pRK5, pRK5-ralB23V, or pDCR-ras12V; 16 h posttransfection, cells were incubated for an additional 24 h in DMEM–0.5% calf serum. Cells were then lysed, and total RNA was prepared using TRIZOL reagent (Life Technologies). Following DNase treatment, oligo(dT)-primed cDNA was prepared using a RAP-PCR kit (Stratagene). PCR amplification was carried out using primer pairs specific to mouse cyclin D1 (5′-CCATTCCCTTGACTGCCCGAG-3′ and 5′-GACCAGCCTCTTCCTCCAC-3′) and mouse β-actin (Stratagene).
EMSA.
NIH 3T3 cells were infected with replication-defective retrovirus derived from pBabePuro and pBabePuro-ralB23V packaged in Phoenix-ECO (45). Stable populations of infected cells were obtained by selection in medium-containing puromycin. Nuclear extracts for electrophoretic mobility gel shift assays (EMSA) were prepared by the method of Li et al. (32). The NF-κB site at bp −39 to −30 in the cyclin D1 promoter (CD1 NF-κB wt, 5′-TAC AGG GGA GTT TTG TTG AAG-3′) was synthesized as complementary oligodeoxyribonucleotide strands for EMSA (21, 58).
RESULTS
Ral regulation of Ras-responsive promoter elements.
Activation of Ral family GTPases has been implicated as an important step mediating oncogenic Ras-induced cellular transformation (7, 14). The mechanism by which Ral proteins may contribute to a growth-transformed phenotype has not been determined. To begin to define the role of Ral activation in growth control, we examined the consequences of Ral expression on gene regulatory responses that are downstream of Ras activation and required for oncogenic Ras-induced transformation. The activation of SRE-dependent gene expression, induction of cyclin D1 protein expression, and activation of NF-κB transcription factors have all been defined as critical events mediating Ras transformation (15, 29, 49) and are convergence points for multiple Ras-dependent signals (18, 41, 43, 64).
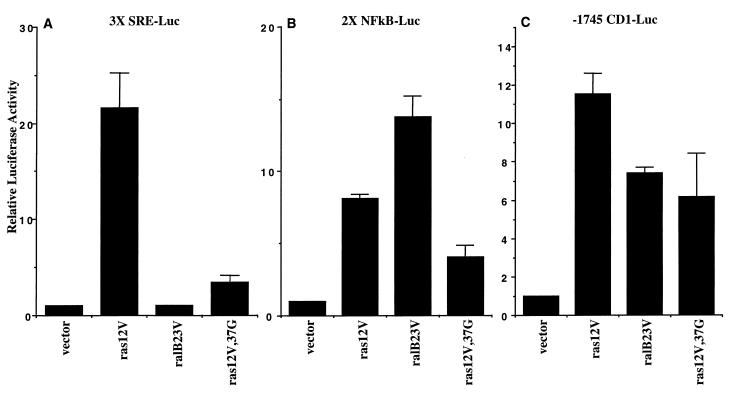
We first tested the effects of activated Ral on the regulation of luciferase reporter genes with promoter elements consisting of ternary complex factor-serum response factor binding sites, NF-κB binding sites, or the 5′ flanking sequences of the cyclin D1 gene. Consistent with previous reports (62), we found that transient expression of a GTPase-defective RalB variant (ralB23V) was not sufficient to induce expression of a luciferase gene with an upstream fusion to three tandem repeats of the c-Fos SRE (3X SRE-Luc) (Fig. 1A). In contrast, expression of ralB23V was sufficient to induce expression of luciferase reporters driven by two tandem repeats of an NF-κB binding site (2X NFκB-Luc) or the human cyclin D1 promoter (-1745 CD1-Luc). The levels of activation of these reporters by ralB23V were comparable to those observed upon expression of oncogenic Ras (ras12V) and a Ras variant (ras12V,37G) that can activate RalGEFs but not Raf1 (Fig. 1B and C). Expression of a GTPase-defective RalA variant (ralA23V) yielded results similar to those observed with ralB23V (data not shown).
FIG. 1.
Activated Ral induces expression from NF-κB and cyclin D1 promoter elements. NIH 3T3 cells were transfected with the indicated expression vectors together with luciferase reporter constructs driven by three tandem copies of the c-Fos SRE (A), two tandem copies of the NF-κB binding site from the κB promoter (B), or the 1745 5′ flanking residues of the human cyclin D1 gene (C). Relative luciferase activity is shown normalized to activities obtained with empty vector. Bars represent the standard error from the mean of average values from three independent experiments performed in duplicate.
As oncogenic Ras can lead to activation of Ral proteins through regulation of RalGEFs, it is possible that Ral contributes to oncogenic Ras regulation of 2X NFκB-Luc and -1745 CD1-Luc. To test this, we expressed ras12V,37G together with a dominant interfering variant of RalB (ralBN28). This variant inhibits activation of endogenous Ral proteins by forming unproductive complexes with RalGEFs. Inhibition of Ral activation resulted in a significant inhibition in the activation of both 2X NFκB-Luc and -1745 CD1-Luc by ras12V,37G (Fig. 2).
FIG. 2.
ralB28N inhibits ras12V,37G activation of 2X NFκB-Luc. Luciferase activity derived from the indicated reporter constructs was monitored upon expression of ras12V,37G alone or together with ralB28N. Experiments were performed as described for Fig. 1 except that relative luciferase activities were normalized to the values obtained upon expression of ras12V,37G alone.
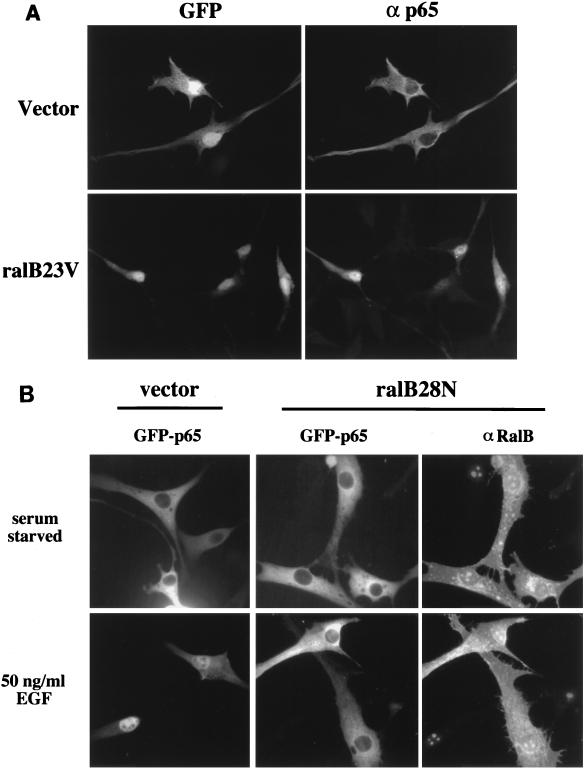
The activation of 2X NFκB-Luc by ralB23V was completely blocked upon coexpression of a dominant inhibitory IκB (IκBαSS/AA) (data not shown), suggesting that ral23V can induce nuclear accumulation of active NF-κB complexes. To test this directly, the κB transcription factor p65/RelA was expressed together with empty vector or ral23V and visualized by immunostaining with anti-p65 antibodies following serum starvation of the transfected cells. As shown in Fig. 3A, ectopically expressed p65 is predominantly cytoplasmic in serum-starved NIH 3T3 cells 24 h posttransfection, presumably due to association with endogenous IκB. In contrast, ralB23V expression is sufficient to drive accumulation of p65 in the nucleus.
FIG. 3.
Activated Ral induces nuclear accumulation of p65 RelA. (A) NIH 3T3 cells were transiently transfected with pCMV-GFP and pRSV-p65 together with pRK5 (vector) or pRK5-ralB23V (ralB23V); 24 h posttransfection, the cells were fixed and stained with anti-p65 polyclonal antibodies and rhodamine-conjugated anti-rabbit IgG. GFP fluorescence (left) is shown to indicate transfected cells; corresponding signal from ectopically expressed p65 is shown on the right. (B) NIH 3T3 cells were transiently transfected with pEGFP-p65 together with pRK5 (vector) or pRK5-ralB28N (ralB28N). Following overnight incubation in serum-free medium, the indicated cells were stimulated for 50 min with EGF (50 ng/ml). p65 was visualized by autofluorescence of fused GFP, and ralB28N was visualized with anti-RalB polyclonal antibodies and rhodamine-conjugated anti-rabbit IgG.
Cellular Ral proteins are activated by EGF in a Ras-dependent manner (63). EGF stimulation of quiescent cells can induce detectable nuclear accumulation of GFP-p65 fusion proteins (Fig. 3B). These fusion proteins have been previously documented as being responsive to signals that activate NF-κB (52). To determine if Ral activation may contribute to EGF-stimulated nuclear accumulation of p65, we transiently expressed GFP-p65 together with ralB28N. The majority of cells expressing ralB28N failed to accumulate GFP-p65 in the nucleus in response to EGF stimulation, suggesting that Ral activation is required for this response (Fig. 3B).
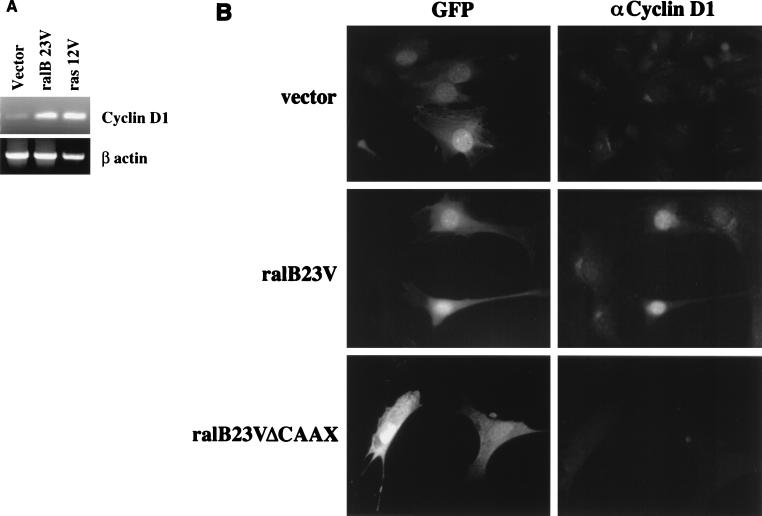
Elevation of native cyclin D1 mRNA levels and cyclin D1 protein can be detected in oncogenic ras-expressing NIH 3T3 cells (33). As ralB23V, like ras12V, was sufficient to induce -1745 CD1-Luc, we examined the consequences of ralB23V expression on induction of cellular cyclin D1. Total RNA prepared from NIH 3T3 cells transiently transfected with vector, ras12V, or ral23V was subjected to RT-PCR using primers for mouse cyclin D1 and β-actin. The levels of cyclin D1 product were significantly higher in ras12V- and ral23V-expressing cells than in the vector control in repeated experiments (Fig. 4A). In addition, expression of ral23V was sufficient to induce accumulation of native cyclin D1 protein as detected by immunofluorescence (Fig. 4B). NIH 3T3 cells stably expressing ras12V (NIH 3T3:ras12V cells) are growth transformed and exhibit constitutive serum-independent accumulation of cyclin D1 protein (33). To determine if Ral activation may contribute to this phenotype, we transiently expressed ralB28N in NIH 3T3:ras12V cells. The majority of cells expressing detectable levels of ralB28N (Fig. 5A) had reduced levels of cyclin D1 protein (Fig. 5B) compared to neighboring untransfected cells. These results suggest that Ral activation results in elevated cyclin D1 production in cells and that active Ral or RalGEF is required for chronic ras12V activation of cyclin D1 expression.
FIG. 4.
Activated Ral induces accumulation of endogenous cyclin D1. (A) NIH 3T3 cells were transiently transfected with the indicated expression vectors. After 24 h of incubation in low serum, cells were lysed and total RNA was isolated. RT-PCR was performed with primers specific to the mouse cyclin D1 and β-actin genes. Following PCR amplification, the products were separated by gel electrophoresis and visualized by ethidium bromide staining. No signal was observed in the absence of reverse transcriptase (not shown). (B) NIH 3T3 cells were transiently transfected with pCMV-GFP together with pRK5 (vector) or pRK5-ralB23V (ralB23V). Confluent, serum-starved cells were fixed and stained with anti-cyclin D1 monoclonal antibody. More than 80% of the GFP-expressing cells cotransfected with ralB23V also expressed detectable levels of cyclin D1. Less than 10% of the GFP-expressing cells cotransfected with empty vector expressed detectable levels of cyclin D1.
FIG. 5.
ralBN28 inhibits oncogenic ras-dependent cyclin D1 expression. Growth and morphologically transformed NIH 3T3 cells stably expressing H-ras12V were transiently transfected with pRK5-ralB28N. Following a 24-h incubation in the absence of serum, cells were fixed and stained with monoclonal anti-cyclin D1 and polyclonal anti-RalB antibodies. Arrowheads indicate the nuclei of ralB28N-expressing cells with reduced levels of cyclin D1 expression. Two fields of view are shown to display multiple representative cells. RalB staining is predominantly in the plasma membrane; however, the anti-RalB antibody cross-reacts with the nucleoli of untransfected cells.
Ral activation of NF-κB and cyclin D1 through a PLD1- and RLIP/RalBP1-independent effector pathway.
Ral potentially regulates Rac/Cdc42 family GTPases through a GTP-dependent association with RalBP1, a Cdc42/Rac GAP (9, 25, 44). In addition, Ral may contribute to regulation of PLD1 through direct association (23). Either of these activities conceivably couples to regulation of NF-κB and cyclin D1. For example, Rac can regulate NF-κB and cyclin D1 induction through activation of p21-activated protein kinase and other downstream effectors (17, 24, 46). In contrast to negative regulation of Rac, sequestration of RalBP1 by Ral-GTP possibly elevates basal levels of active Rac. PLD1 may couple to NF-κB and cyclin D1 through production of second messenger phosphatidic acid, lysophosphatidic acid, or diacylglycerol (13). To examine the effector dependency of Ral activation of NF-κB and cyclin D1, we tested the activity of Ral variants that uncouple association of Ral with RalBP1 versus PLD1. The Ral effector mutant ralB23V,49N has severely impaired RalBP1 binding activity but can still associate with active PLD1 (6, 23, 25, 35). Truncation of 11 amino acids from the amino terminus of Ral, a region unique to this GTPase (ΔN11ralB23V), eliminates association of Ral with PLD activity but not with RalBP1 (36). As shown in Fig. 6, neither of these mutations affects Ral-dependent induction of NF-κB-dependent gene expression. In contrast, impairing guanyl nucleotide association (ralB28N) or blocking lipid modification by a truncation of the four carboxy-terminal amino acids (ralB23VΔCAAX) blocks Ral activation of NF-κB. Similar results were observed in experiments with -1745 CD1-Luc (data not shown). Taken together, these observations suggest that Ral activation of NF-κB and cyclin D1 is independent of association with either RalBP1 or PLD1.
FIG. 6.
RalB variants uncouple Ral activation of NF-κB from association with RalBP1 or active PLD1. NIH 3T3 cells were transiently transfected with the indicated expression vectors together with 2X NF-κB-Luc. Luciferase assays were performed as described for Fig. 1. Error bars are as described for Fig. 1. A sample of each lysate was used to monitor expression of the RalB variants by Western analysis. Expression of the RalB variants from a representative experiment is shown below the graph.
Ral activation of cyclin D1 expression is NF-κB dependent.
It has recently been reported that NF-κB can directly activate the cyclin D1 promoter through NF-κB binding sites (21, 22). This introduces the possibility that Ral activation of cyclin D1 is mediated by Ral activation of NF-κB transcription factors. To test this, we examined the consequences of inhibition of NF-κB on Ral induction of cyclin D1. Expression of either a kinase-dead dominant inhibitory IκB kinase β (IKKβKM) or the NF-κB superepressor (IκBα SS/AA) (8) blocked ralB23V activation of -1745 CD1-Luc (Fig. 7A). Therefore, nuclear accumulation of NF-κB is required for activation of cyclin D1 promoter activity either downstream or in parallel to Ral activation.
FIG. 7.
NF-κB mediates a ralB23V activation of the cyclin D1 promoter. (A) NIH 3T3 cells were transfected with the -1745 CD1-Luc reporter and ralB23V alone or together with IκBα SS/AA or IKKβKM as indicated. Luciferase assays were performed as described for previous figures. (B) Cells were transfected with ralB23V together with luciferase reporter genes driven by the first 66 bp of 5′ flanking residues from the human cyclin D1 gene (-66 CD1-Luc), the same region with point mutations that disrupt the NF-κB binding site (-66 CD1-κBmut-Luc), or with point mutations that disrupt a neighboring CRE/ATF consensus binding site (-66 CD1-ATFmut1-Luc and -66 CD1-ATFmut2-Luc). (C) EMSAs using the NF-κB binding site at bp −39 to −30 of the human cyclin D1 promoter. The arrow indicates shifted complexes. Nuclear extracts were prepared from stable populations of cells expressing ralB23V or from vector control cells. NIK and NIKDN were introduced by high-efficiency transient transfection with Superfect transfection reagent (Qiagen, Valencia, Calif.) using pcDNA3-HA-NIK and pcDNA3myc-NIK (EE429/430AA).
A minimal domain of the cyclin D1 promoter that is responsive to NF-κB is contained within 66 bp of the 5′ flanking sequence of the human cyclin D1 gene. This region contains an NF-κB consensus binding site at -36 (21). As shown in Fig. 7B, ralB23V induced expression of a luciferase reporter gene coupled to this region of the cyclin D1 promoter (-66 CD1-Luc). Mutation of the NF-κB binding site, but not of an adjacent putative ATF binding site, blocked the responsiveness of this region to ral23V (Fig. 7B). In addition, nuclear extracts prepared from ral23V-expressing cells induced an electrophoretic mobility shift of the cyclin D1 promoter NF-κB binding site (Fig. 7C). Coexpression of a dominant inhibitory variant of the IKK kinase NIK (NIKDN) inhibited the mobility shift, suggesting that the observed activity is dependent on IKK activation. These observations suggest that Ral-dependent activation of cyclin D1 expression is mediated by activation of NF-κB.
Ral activation of NF-κB does not require c-Src.
Recently, it has been reported that Ral can induce the tyrosine kinase activity of c-Src through an unknown mechanism (20). Src can mediate activation of NF-κB by tumor necrosis factor in some cell types, possibly through direct phosphorylation of IκB (1). In addition, oncogenic Src leads to activation of cyclin D1 expression (31). We therefore tested the role of Src in Ral activation of NF-κB. Although weaker than activity observed in wild-type NIH 3T3 cells, expression of ralB23V in NIH 3T3 Src−/− Yes−/− cells resulted in activation of 2X NFκB-Luc to a similar extent as was observed upon expression of NIK (Fig. 8A). In addition, ral23V expression was sufficient to drive nuclear accumulation of GFP-p65 in Src−/− Yes−/− cells (Fig. 8B). Although we have not ruled out a role for Src in Ral-induced activation of NF-κB, these results suggest that Src-independent pathways are involved. In support of this, it has recently been demonstrated that Src regulates cyclin D1 expression through a cyclic AMP response element (CRE)/ATF site in the cyclin D1 promoter that is not required for Ral to regulate this promoter (reference 31 and Fig. 7B).
FIG. 8.
Ral23V can activate NF-κB in the absence of c-Src expression. (A) NIH 3T3 Src−/− Yes−/− cells were transfected with the indicated expression vectors together with 2X NFκB-Luc. Relative luciferase activity is shown normalized to activities obtained with empty vector. Error bars represent the standard error of the mean. (B) Src−/− Yes−/− cells were transfected with pEGFP-p65 together with pRK5 or pRK5-ralB23V as indicated. Ral expression and GFP-p65 were detected as described for Fig. 3.
DISCUSSION
Ral GTPases were originally identified on the basis of sequence similarity to Ras proteins (11). The discovery that RalGEF activity can be activated by oncogenic ras and that Ral activation may mediate some cellular responses to activated Ras (14, 61), has sparked considerable interest in understanding the biological role of Ral proteins. Inhibition of Ral activation, through the use of dominant inhibitory Ral variants, inhibits oncogenic ras-mediated growth transformation (38, 55, 60). Expression of active Ral and RalGEFs can contribute to the generation of a growth-transformed phenotype (55, 60). These observations suggest that at least one function of Ral is to contribute to the regulation of proliferation. Here, we have shown that expression of activated Ral (ral23V) in quiescent cells is sufficient to activate NF-κB and induce accumulation of cyclin D1 protein, two critical responses to mitogenic signals.
NF-κB and Rel family transcription factors were originally characterized as regulators of inflammatory and immune responses (3). However, it has recently become clear that NF-κB can also function to promote cell proliferation, both indirectly through induction of survival factors (4) and directly by promoting cell cycle progression through activation of cyclin D1 transcription (21, 22). NF-κB is positively regulated by a variety of mitogens in addition to stress-inducing agents and inflammatory cytokines (5). We find that expression of ral23V is sufficient to induce nuclear translocation of p65/RelA and NF-κB-dependent transcription in NIH 3T3 cells. The dominant mechanism regulating NF-κB transcriptional activity is subcellular localization. IκB association with NF-κB family members prevents nuclear accumulation of the transcription factors (56). As phosphorylation of IκB by IκB kinases causes the ubiquitination and degradation of IκB (56), the observation that Ral can induce nuclear translocation of p65/RelA suggests that an IκB kinase is activated downstream of Ral.
The mechanism by which Ral can regulate NF-κB has not been identified. However, the observation that Ral variants that uncouple association of Ral with PLD1 or RalBP1 retain full NF-κB activation activity suggests that neither PLD1 nor RalBP1 mediates this response. The cytoskeletal protein filamin has been identified as a potential Ral effector molecule that can mediate Ral-dependent reorganization of the actin cytoskeleton in human melanoma cell lines (42). We were unable to assess the role of filamin in Ral-dependent activation of NF-κB, as human melanoma cells do not respond to ras12V or ralB23V by further detectable activation of NF-κB, regardless of the presence of filamin 1 (data not shown). Ral proteins have been implicated in the regulation of clathrin-dependent endocytosis through observations that expression of either ralV23 or ralN28 inhibits endocytosis in A431 cells (40). This raises the possibility that cellular responses to Ral expression may be an indirect response to inhibition of receptor downregulation. This is particularly important to consider when examining mitogenic signal transduction cascades. Inhibition of clathrin-mediated receptor downregulation results in prolonged signaling from EGF receptors (39). In fact, EGF receptor mutants that can not be endocytosed are oncogenic (39). Although inhibition of receptor downregulation may contribute to Ral effects on transformation in some contexts, the impact of Ral on NF-κB regulation is likely to be independent of this phenomenon. First, both ral28N and ral23V inhibit endocytosis (40), while only ral23V activates NF-κB. Second, Ral regulation of endocytosis appears to occur through regulation of RalBP1 and RalBP1-associated proteins (40). Ral variants defective for RalBP1 interaction are still fully active on the NF-κB pathway.
Cyclin D1 in complex with cyclin-dependent kinases 4 and 6 (CDK4 and -6) affects cell cycle progression directly at the level of RB phosphorylation, as well as through the titration of CDK inhibitors away from cyclin E-CDK2 complexes (53). Multiple mitogenic and oncogenic signaling cascades converge to elevate cyclin D1 protein at the level of transcription and protein stabilization (12, 47). We observe that expression of activated Ral can induce transcription of the cyclin D1 promoter and accumulation of cyclin D1 protein, suggesting a mechanism by which Ral can contribute to proliferative signals. Ral activation of the cyclin D1 promoter is blocked by inhibition of NF-κB activation. In addition, the minimal NF-κB-responsive cyclin D1 promoter element is activated by Ral. Mutation of the NF-κB binding site in this element eliminates Ral responsiveness. These observations suggest that Ral regulation of cyclin D1 is at least in part a downstream consequence of Ral activation of NF-κB.
Ras regulation of both NF-κB and cyclin D1 is complex and likely occurs through multiple effector interactions. For example, expression of activated Raf kinase is sufficient to upregulate both NF-κB activity and cyclin D1 expression (27). However, Ras variants uncoupled from Raf interaction can induce both responses through Raf-independent pathways (18, 41, 64). These results are consistent with our observation that activation of Ral appears to be required for oncogenic ras-induced cyclin D1 expression. It is well established that multiple Ras effector pathways relay signals from oncogenic ras that induce growth and morphological transformation of cells (26, 37, 57). Cellular responses such as NF-κB activation and induction of cyclin D1 expression likely represent critical convergence points for growth-stimulatory signals regulated by Ras.
ACKNOWLEDGMENTS
We thank J. Frost, M. Cobb, K. Akama, J. Schmid, and A. Alberts for some of the plasmids and reagents used in this study. We thank Leslie Hasbini for excellent technical assistance.
This research was supported by NIH grant R01CA71443 and the Welch Foundation (to M.A.W.) and grant RO1CA75503 and the Pfeiffer Foundation (to R.G.P.).
D.O.H. and S.A.M. contributed equally to this work.
REFERENCES
- 1.Abu-Amer Y, Ross F P, McHugh K P, Livolsi A, Peyron J F, Teitelbaum S L. Tumor necrosis factor-alpha activation of nuclear transcription factor-kappaB in marrow macrophages is mediated by c-Src tyrosine phosphorylation of Ikappa Balpha. J Biol Chem. 1998;273:29417–29423. doi: 10.1074/jbc.273.45.29417. [DOI] [PubMed] [Google Scholar]
- 2.Akama K T, Van Eldik L J. Beta-amyloid stimulation of inducible nitric-oxide synthase in astrocytes is interleukin-1beta- and tumor necrosis factor-alpha (TNFalpha)-dependent, and involves a TNFalpha receptor-associated factor- and NFkappaB-inducing kinase-dependent signaling mechanism. J Biol Chem. 2000;275:7918–7924. doi: 10.1074/jbc.275.11.7918. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Baichwal V R, Baeuerle P A. Activate NF-kappa B or die? Curr Biol. 1997;7:R94–R96. doi: 10.1016/s0960-9822(06)00046-7. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin A S., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Bauer B, Mirey G, Vetter I R, Garcia-Ranea J A, Valencia A, Wittinghofer A, Camonis J H, Cool R H. Effector recognition by the small GTP-binding proteins Ras and Ral. J Biol Chem. 1999;274:17763–17770. doi: 10.1074/jbc.274.25.17763. [DOI] [PubMed] [Google Scholar]
- 7.Bos J L. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 1998;17:6776–6782. doi: 10.1093/emboj/17.23.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα- to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantor S B, Urano T, Feig L A. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chardin P. The ras superfamily proteins. Biochimie. 1988;70:865–868. doi: 10.1016/0300-9084(88)90226-x. [DOI] [PubMed] [Google Scholar]
- 11.Chardin P, Tavitian A. The ral gene: a new ras related gene isolated by the use of a synthetic probe. EMBO J. 1986;5:2203–2208. doi: 10.1002/j.1460-2075.1986.tb04485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehl J A, Cheng M, Roussel M F, Sherr C J. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Exton J H. New developments in phospholipase. D. J Biol Chem. 1997;272:15579–15582. doi: 10.1074/jbc.272.25.15579. [DOI] [PubMed] [Google Scholar]
- 14.Feig L A, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 15.Finco T S, Westwick J K, Norris J L, Beg A A, Der C J, Baldwin A S., Jr Oncogenic Ha-Ras-induced signaling activates NF-kappaB transcriptional activity, which is required for cellular transformation. J Biol Chem. 1997;272:24113–24116. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 16.Frankel P, Ramos M, Flom J, Bychenok S, Joseph T, Kerkhoff E, Rapp U R, Feig L A, Foster D A. Ral and Rho-dependent activation of phospholipase D in v-Raf-transformed cells. Biochem Biophys Res Commun. 1999;255:502–507. doi: 10.1006/bbrc.1999.0234. [DOI] [PubMed] [Google Scholar]
- 17.Frost J A, Swantek J L, Stippec S, Yin M J, Gaynor R, Cobb M H. Stimulation of NFκB activity by multiple signaling pathways requires PAK1. J Biol Chem. 2000;275:19693–19699. doi: 10.1074/jbc.M909860199. [DOI] [PubMed] [Google Scholar]
- 18.Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 19.Goi T, Rusanescu G, Urano T, Feig L A. Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol Cell Biol. 1999;19:1731–1741. doi: 10.1128/mcb.19.3.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goi T, Shipitsin M, Lu Z, Foster D A, Klinz S G, Feig L A. An EGF receptor/Ral-GTPase signaling cascade regulates c-Src activity and substrate specificity. EMBO J. 2000;19:623–630. doi: 10.1093/emboj/19.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttridge D C, Albanese C, Reuther J Y, Pestell R G, Baldwin A S., Jr NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinz M, Krappmann D, Eichten A, Heder A, Scheidereit C, Strauss M. NF-κB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol Cell Biol. 1999;19:2690–2698. doi: 10.1128/mcb.19.4.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Luo J Q, Urano T, Frankel P, Lu Z, Foster D A, Feig L A. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- 24.Joyce D, Bouzahzah B, Fu M, Albanese C, D'Amico M, Steer J, Klein J U, Lee R J, Segall J E, Westwick J K, Der C J, Pestell R G. Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-kappaB-dependent pathway. J Biol Chem. 1999;274:25245–25249. doi: 10.1074/jbc.274.36.25245. [DOI] [PubMed] [Google Scholar]
- 25.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis J H. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 26.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 27.Kerkhoff E, Rapp U R. Cell cycle targets of Ras/Raf signalling. Oncogene. 1998;17:1457–1462. doi: 10.1038/sj.onc.1202185. [DOI] [PubMed] [Google Scholar]
- 28.Kim J H, Lee S D, Han J M, Lee T G, Kim Y, Park J B, Lambeth J D, Suh P G, Ryu S H. Activation of phospholipase D1 by direct interaction with ADP-ribosylation factor 1 and RalA. FEBS Lett. 1998;430:231–235. doi: 10.1016/s0014-5793(98)00661-9. [DOI] [PubMed] [Google Scholar]
- 29.Ledwith B J, Manam S, Kraynak A R, Nichols W W, Bradley M O. Antisense-fos RNA causes partial reversion of the transformed phenotypes induced by the c-Ha-ras oncogene. Mol Cell Biol. 1990;10:1545–1555. doi: 10.1128/mcb.10.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee G, Gilman M. Dual modes of control of c-fos mRNA induction by intracellular calcium in T cells. Mol Cell Biol. 1994;14:4579–4587. doi: 10.1128/mcb.14.7.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee R J, Albanese C, Stenger R J, Watanabe G, Inghirami G, Haines III G K, Webster M, Muller W J, Brugge J S, Davis R J, Pestell R G. pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J Biol Chem. 1999;274:7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- 32.Li Y C, Ross J, Scheppler J A, Franza B R., Jr An in vitro transcription analysis of early responses of the human immunodeficiency virus type 1 long terminal repeat to different transcriptional activators. Mol Cell Biol. 1991;11:1883–1893. doi: 10.1128/mcb.11.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J J, Chao J R, Jiang M C, Ng S Y, Yen J J, Yang-Yen H F. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Z, Hornia A, Joseph T, Sukezane T, Frankel P, Zhong M, Bychenok S, Xu L, Feig L A, Foster D A. Phospholipase D and RalA cooperate with the epidermal growth factor receptor to transform 3Y1 rat fibroblasts. Mol Cell Biol. 2000;20:462–467. doi: 10.1128/mcb.20.2.462-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo J Q, Liu X, Frankel P, Rotunda T, Ramos M, Flom J, Jiang H, Feig L A, Morris A J, Kahn R A, Foster D A. Functional association between Arf and RalA in active phospholipase D complex. Proc Natl Acad Sci USA. 1998;95:3632–3637. doi: 10.1073/pnas.95.7.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J Q, Liu X, Hammond S M, Colley W C, Feig L A, Frohman M A, Morris A J, Foster D A. RalA interacts directly with the Arf-responsive, PIP2-dependent phospholipase D1. Biochem Biophys Res Commun. 1997;235:854–859. doi: 10.1006/bbrc.1997.6793. [DOI] [PubMed] [Google Scholar]
- 37.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 38.Miller M J, Prigent S, Kupperman E, Rioux L, Park S H, Feramisco J R, White M A, Rutkowski J L, Meinkoth J L. RalGDS functions in Ras- and cAMP-mediated growth stimulation. J Biol Chem. 1997;272:5600–5605. doi: 10.1074/jbc.272.9.5600. [DOI] [PubMed] [Google Scholar]
- 39.Mineo C, Gill G N, Anderson R G. Regulated migration of epidermal growth factor receptor from caveolae. J Biol Chem. 1999;274:30636–30643. doi: 10.1074/jbc.274.43.30636. [DOI] [PubMed] [Google Scholar]
- 40.Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, Iwamatsu A, Kishida S, Kikuchi A. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–3642. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norris J L, Baldwin A S., Jr Oncogenic Ras enhances NF-kappaB transcriptional activity through Raf- dependent and Raf-independent mitogen-activated protein kinase signaling pathways. J Biol Chem. 1999;274:13841–13846. doi: 10.1074/jbc.274.20.13841. [DOI] [PubMed] [Google Scholar]
- 42.Ohta Y, Suzuki N, Nakamura S, Hartwig J H, Stossel T P. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okazaki M, Kishida S, Hinoi T, Hasegawa T, Tamada M, Kataoka T, Kikuchi A. Synergistic activation of c-fos promoter activity by Raf and Ral GDP dissociation stimulator. Oncogene. 1997;14:515–521. doi: 10.1038/sj.onc.1200860. [DOI] [PubMed] [Google Scholar]
- 44.Park S H, Weinberg R A. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- 45.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal J C. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 47.Pestell R G, Albanese C, Reutens A T, Segall J E, Lee R J, Arnold A. The cyclins and cyclin-dependent kinase inhibitors in hormonal regulation of proliferation and differentiation. Endocrine Rev. 1999;20:501–534. doi: 10.1210/edrv.20.4.0373. [DOI] [PubMed] [Google Scholar]
- 48.Reuther G W, Der C J. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 49.Robles A I, Rodriguez-Puebla M L, Glick A B, Trempus C, Hansen L, Sicinski P, Tennant R W, Weinberg R A, Yuspa S H, Conti C J. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Viciana P, Warne P, Khwaja A, Marte B, Pappin D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Schmid J A, Birbach A, Hofer-Warbinek R, Pengg M, Burner U, Furtmuller P G, Binder B R, de Martin R. Dynamics of NF kappa B and Ikappa Balpha studied with green fluorescent protein (GFP) fusion proteins. Investigation of GFP-p65 binding to DNa by fluorescence resonance energy transfer. J Biol Chem. 2000;275:17035–17042. doi: 10.1074/jbc.M000291200. [DOI] [PubMed] [Google Scholar]
- 53.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 54.Swantek J L, Christerson L, Cobb M H. Lipopolysaccharide-induced tumor necrosis factor-alpha promoter activity is inhibitor of nuclear factor-kappaB kinase-dependent. J Biol Chem. 1999;274:11667–11671. doi: 10.1074/jbc.274.17.11667. [DOI] [PubMed] [Google Scholar]
- 55.Urano T, Emkey R, Feig L A. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 56.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 57.Vojtek A B, Der C J. Increasing complexity of the Ras signaling pathway. J Biol Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe G, Lee R J, Albanese C, Rainey W E, Batlle D, Pestell R G. Angiotensin II activation of cyclin D1-dependent kinase activity. J Biol Chem. 1996;271:22570–22577. doi: 10.1074/jbc.271.37.22570. [DOI] [PubMed] [Google Scholar]
- 59.White M A, Nicolette C, Minden A, Polverino A, van Aelst L, Karin M, Wigler M H. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 60.White M A, Vale T, Camonis J H, Schaefer E, Wigler M H. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J Biol Chem. 1996;271:16439–16442. doi: 10.1074/jbc.271.28.16439. [DOI] [PubMed] [Google Scholar]
- 61.Wolthuis R M, Bos J L. Ras caught in another affair: the exchange factors for Ral. Curr Opin Genet Dev. 1999;9:112–117. doi: 10.1016/s0959-437x(99)80016-1. [DOI] [PubMed] [Google Scholar]
- 62.Wolthuis R M, de Ruiter N D, Cool R H, Bos J L. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 1997;16:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolthuis R M, Zwartkruis F, Moen T C, Bos J L. Ras-dependent activation of the small GTPase Ral. Curr Biol. 1998;8:471–474. doi: 10.1016/s0960-9822(98)70183-6. [DOI] [PubMed] [Google Scholar]
- 64.Yang J J, Kang J S, Krauss R S. Ras signals to the cell cycle machinery via multiple pathways to induce anchorage-independent growth. Mol Cell Biol. 1998;18:2586–2595. doi: 10.1128/mcb.18.5.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]