C1qbp promotes epigenetic and transcriptional regulation of effector CD8+ T cell differentiation through up-regulation of OXPHOS.
Abstract
Early-activated CD8+ T cells increase both aerobic glycolysis and mitochondrial oxidative phosphorylation (OXPHOS). However, whether and how the augmentation of OXPHOS regulates differentiation of effector CD8+ T cell remains unclear. Here, we found that C1qbp was intrinsically required for such differentiation in antiviral and antitumor immune responses. Activated C1qbp-deficient CD8+ T cells failed to increase mitochondrial respiratory capacities, resulting in diminished acetyl–coenzyme A as well as elevated fumarate and 2-hydroxyglutarate. Consequently, hypoacetylation of H3K27 and hypermethylation of H3K27 and CpG sites were associated with transcriptional down-regulation of effector signature genes. The effector differentiation of C1qbp-sufficient or C1qbp-deficient CD8+ T cells was reversed by fumarate or a combination of histone deacetylase inhibitor and acetate. Therefore, these findings identify C1qbp as a pivotal positive regulator in the differentiation of effector CD8+ T cells and highlight a metabolic-epigenetic axis in this process.
INTRODUCTION
CD8+ cytotoxic T lymphocytes (CTLs) are crucial players that eliminate intracellular pathogens and cancer cells (1, 2). Early after infection, CD8+ T cells bifurcately differentiate into either terminal/short-lived effector cells (TE/SLECs; KLRG1+CD127−) or memory precursor effector cells (MPECs; KLRG1−CD127+) characterized by different expression levels of killer cell lectin-like receptor G1 (KLRG1) and interleukin-7R𝛂 (IL-7R𝛂) (CD127) (3, 4). Following the elimination of invading pathogens, the major population of CD8+ T cells, SLECs, mostly die by apoptosis. In contrast, the minor population, MPECs, survive to further differentiate into a heterogenous pool of memory CD8+ T cells. SLECs and MPECs have distinct transcriptional profiling (5). Their differentiations are initiated by the pioneer transcription factors including Batf/Irf4 and Runx3 and are coordinately instructed by a series of transcription factors including the following pairs: T-bet (encoded by Tbx21)/Eomes, Blimp-1 (encoded by Prdm1)/Bcl-6, Id2/Id3, and Zeb2/Zeb1 (1, 2, 6). Not all these transcription factor–encoding genes are preferentially expressed in either SLECs or MPECs, which suggests that additional mechanism may contribute to their activities to control cell fates (7).
Within naïve CD8+ T cells, bivalent modifications (H3K4me3+ H3K27me3+) are deposited at the promoters of genes encoding transcription factors that are crucial for initiating an effector program, such as T-bet and Irf4 (2, 7–9). Upon T cell activation, these bivalent loci quickly lose the occupancy by H3K27me3 and acquire a permissive histone methylation signature (H3K4me3+ H3K27me3−) to allow rapid transcription of proeffector genes. During the differentiation of naïve CD8+ T cells into SLECs and MPECs, deposition of H3K27me3 by Ezh2/PRC2, H3K9me3 by Suv39h1, or DNA methylation by Dnmt3a contributes to the silencing of memory- and stemness-associated gene expression and thus maintains effector T cell identity (10–12). However, how the epigenetic remodeling is triggered remains largely unknown.
Quiescent naïve CD8+ T cells primarily rely on mitochondrial oxidative phosphorylation (OXPHOS) to maintain their survival. Upon T cell activation, CD8+ T cells increase both OXPHOS and aerobic glycolysis to fulfill the bioenergetic and biosynthetic demands (13–15). Effector CD8+ T cells reduce OXPHOS but maintain higher glycolysis to support proliferation and effector function, in part via histone acetylation of Ifng locus (16). In contrast, memory CD8+ T cells switch back to OXPHOS fueled by fatty acid oxidation, although different subsets may use different sources of fatty acid (17–20). However, whether OXPHOS, especially its transient increase, contributes to epigenetic regulation of differentiation and function of effector CD8+ T cells has not been uncovered.
C1qbp, also known as p32, gC1qR, or HABP1, has been originally identified as a protein associated with pre-mRNA splicing factor ASF/SF2 (21). The C1qbp protein, primary localized in the mitochondrial matrix but also present at the cell surface, cytosol, and nucleus, exhibits a doughnut-shaped trimer (22, 23). C1qbp is required for mitochondrial OXPHOS because it functions as an RNA binding protein to facilitate mitochondrial translation (24, 25). In this study, we present evidence of a critical role for C1qbp in the differentiation of effector CD8+ T cells. Mice with C1qbp deficiency in T cells exhibited substantial cell-intrinsic defect in differentiation of effector CD8+ T cells after acute lymphocytic choriomeningitis virus (LCMV) infection. Activated C1qbp-deficient CD8+ T cells were not able to increase OXPHOS resulting in diminished acetyl–coenzyme A (CoA) as well as elevated 2-hydroxyglutarate (2-HG) and fumarate. Consequently, hypoacetylation of H3K27 and hypermethylation of H3K27 and CpG sites were associated with transcriptional down-regulation of effector signature genes in C1qbp-deficient CD8+ T cells. Furthermore, pretreatment of wild-type (WT) or C1qbp-deficient CD8+ T cells with fumarate or combination of histone deacetylase inhibitor tubastatin A and acetate (TA/Ac) respectively reversed their capacities to differentiate into effector CD8+ T cells. Our data suggest that C1qbp is essential for the differentiation of effector CD8+ T cells through a metabolic-epigenetic axis.
RESULTS
Activated C1qbp-deficient CD8+ T cells fail to increase mitochondrial OXPHOS
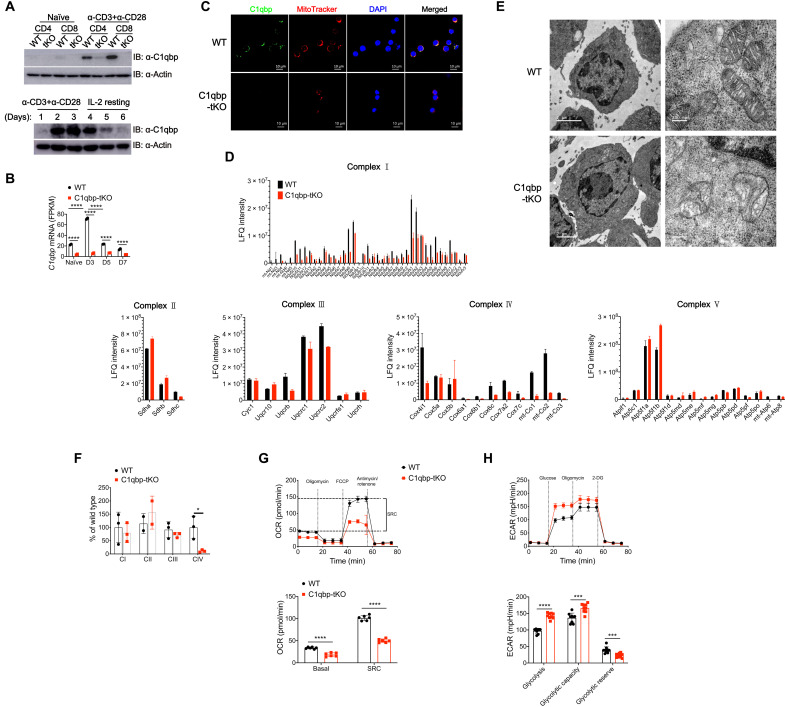
Previous reports have showed that C1qbp binds to mitochondrial mRNAs and facilitates the translations and therefore promotes mitochondrial OXPHOS (24, 25). To investigate the role of C1qbp in coupling metabolism to T cell differentiation, we crossed mice with loxP-flanked C1qbp alleles with transgenic mice containing a Cre-recombinase gene driven by the distal promoter of the lymphocyte protein tyrosine kinase (Lck) gene (dLck-Cre) to generate mice with T cell–specific C1qbp deficiency (C1qbpfl/fl dLck-Cre, called C1qbp-tKO hereinafter) (fig. S1A). Unlike other cell types that constitutively express C1qbp (25, 26), WT but not C1qbp-deficient T cells transiently induced the expression of C1qbp protein upon stimulation with anti-CD3 and anti-CD28 antibodies and lost the expression when the stimulation was removed (Fig. 1A). Similarly, C1qbp mRNA expression in antigen-specific CD8+ T cells was transiently induced with the expression peak at day 3 after infection with LCMV Armstrong strain (Fig. 1B). By using immunofluorescence microscopy, we observed that almost all C1qbp proteins localized to mitochondria in activated T cells, which is consistent with previously published studies (Fig. 1C) (24–26). To assess the consequences of C1qbp deficiency, we next isolated mitochondria from in vitro–activated CD8+ T cells and measured protein levels of mitochondrial proteins by liquid chromatography–tandem mass spectrometry (LC-MS/MS). As expected, the protein levels of all mitochondrial DNA (mtDNA)–encoded proteins except ATP8 (encoded by mt-Atp8) were much lower in activated C1qbp-deficient CD8+ T cells than in WT counterparts (Fig. 1D and table S1). These data suggested that C1qbp deficiency resulted in impaired new synthesis of mitochondrial electron transport chain proteins in activated T cells, consistent with previous studies.
Fig. 1. C1qbp deficiency results in failure to increase mitochondrial OXPHOS in CD8+ T cells upon activation.
Naïve CD4+ or CD8+ T cells purified from WT and C1qbp-tKO mice were stimulated in vitro with plate-bound anti-CD3ε (3 μg/ml) and anti-CD28 (3 μg/ml) for 2 days (A and C to F) or 1 day (G and H). (A) Immunoblot (IB) analysis of C1qbp protein in naïve and activated WT or C1qbp-tKO T cells (top row) and C1qbp protein in WT CD8+ T cells activated by anti-CD3ε and anti-CD28 followed by IL-2 resting for indicated days (bottom row). (B) Normalized expression level of C1qbp mRNA in naïve WT and C1qbp-deficient P14 CD8+ T cells and those from recipient mice at days 3 (D3), 5, and 7 after infection with the LCMV Armstrong strain (n = 2 per group). FPKM, expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced. (C) Confocal micrographs of activated WT or C1qbp-tKO CD8+ T cells stained with anti-C1qbp (green), MitoTracker (red), and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bars, 10 μm. (D) Quantification of protein levels of mitochondrial respiratory chain complexes in activated WT or C1qbp-tKO CD8+ T cells by mass spectrum (n = 3 per group). SRC, spare respiratory capacity; LFQ, label-free quantification. (E) EM images of mitochondria in activated WT or C1qbp-tKO CD8+ T cells [scale bars, 2 μm (left) and 200 nm (right)]. (F) The activity of each complex was measured using isolated mitochondria as described in Materials and Methods (n = 2 to 3 per group). (G and H) Seahorse extracellular flux analysis of OCR (G) and ECAR (H) of WT or C1qbp-tKO CD8+ T cells 1 day after stimulation [n = 6 (G) or 9 (H) wells per group]. Each symbol represents an individual mouse or individual well of cells; small horizontal lines indicate the means (±SD). *P < 0.05, ***P < 0.001, and ****P < 0.0001 (two-tailed unpaired Student’s t test). Data are representative of two (D and E) or three (A, C, and F to H) independent experiments (error bars, SD). FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; 2-DG, 2-deoxyglucose.
During T cell activation, WT CD8+ T cells displayed increased mitochondrial mass and membrane potential. Activated C1qbp-deficient CD8+ T cells showed comparable increase in mitochondrial mass but raised mitochondrial membrane potential much less than WT counterparts did (fig. S1B). We next sought to examine the mitochondrial ultrastructure by electron microscopy (EM). We found that activated WT and C1qbp-deficient CD8+ T cells have comparable numbers of mitochondria (fig. S1C). The mitochondria in activated WT CD8+ T cells displayed filamentous or tubular shape and had obvious and tight cristae structures, whereas those in activated C1qbp-deficient CD8+ T cells exhibited fragmented shape but lacked cristae structure (Fig. 1E). To investigate the role of C1qbp in OXPHOS of CD8+ T cells, we next measured the activities of the electron transport chain complexes in activated CD8+ T cells by spectrophotometric assays. Although some subunits in complex I were also declined, only the activity of complex IV was markedly reduced in activated C1qbp-deficient CD8+ T cells (Fig. 1F). Mitochondria are essential hubs of metabolic activity that couple the tricarboxylic acid (TCA) cycle to OXPHOS to generate maximum adenosine triphosphate (ATP) (27). Upon T cell activation, CD8+ T cells increase both OXPHOS and glycolysis (14, 15). Thus, we analyzed the metabolic consequence of C1qbp ablation in naïve or in vitro–activated CD8+ T cells by using Seahorse Extracellular Flux Analyzer. Given that naïve CD8+ T cells did not express C1qbp protein, the capacities of glycolysis and OXPHOS in naïve CD8+ T cells were not affected by C1qbp deficiency, indicated by the extracellular acidification rate (ECAR) and the oxygen consumption rate (OCR) (fig. S1, D and E). As expected, both glycolysis (ECAR) and OXPHOS (OCR) were enhanced in WT CD8+ T cells after T cell activation (Fig. 1, G and H). In contrast, C1qbp-deficient CD8+ T cells were not able to increase the OXPHOS rates at both basal and spare respiratory capacities (SRCs) after T cell activation (Fig. 1G). ECAR rates at glycolysis and glycolytic capacity were increased more in activated C1qbp-deficient CD8+ T cells than in WT counterparts (Fig. 1H), consistent with previous reports that loss of C1qbp shifted energy metabolism from OXPHOS toward glycolysis (24, 26). Collectively, these results demonstrated that C1qbp was required for increasing mitochondrial OXPHOS in CD8+ T cells upon T cell activation through assisting in synthesis of mtDNA-encoded electron transport chain proteins.
Antiviral effector CD8+ T cell differentiation intrinsically requires C1qbp
We next sought to explore the role of C1qbp in T cell development and homeostasis. The percentages and numbers of thymocyte populations and peripheral T cells were comparable between C1qbp-tKO mice and their WT littermates (fig. S2, A to D). Furthermore, the C1qbp-tKO mice and WT mice had similar compartments of CD44hiCD62Lhi central memory and CD44hiCD62Llo effector memory T cells (fig. S2, C and D). Last, the percentages and numbers of regulatory T cells in spleen and lymph nodes were also comparable between C1qbp-tKO mice and their WT littermates (fig. S2, E and F). Together, these data suggested that C1qbp was dispensable for thymocyte development and peripheral T cell homeostasis.
As C1qbp expression was induced upon T cell activation, we next examined whether it affected T cell activation and proliferation in turn. To address this issue, we isolated naïve CD8+ T cells from WT and C1qbp-tKO mice and stimulated them with anti-CD3 and anti-CD28 antibodies. The up-regulation of activation surface markers, including CD25, CD44, CD69, and CD71, was comparable between WT and C1qbp-deficient cells until 48 hours after stimulation (fig. S3A). However, we observed one division later in proliferation of C1qbp-deficient T cells than in WT cells at 72 hours after in vitro activation (fig. S3A). It has been reported that persistent in vitro IL-2 stimulation promoted effector CTL differentiation (28); thus, we next investigated whether C1qbp regulated in vitro effector CTL differentiation. We purified naïve CD8+ T cells from WT and C1qbp-tKO mice, cocultured them in the presence of anti-CD3 and anti-CD28 antibodies, and then removed them from these stimuli and recultured them in different concentrations of IL-2. The expansion of C1qbp-deficient T cells was indeed less than that of WT counterparts, and the production of effector cytokine interferon-γ (IFN-γ) was also lower in C1qbp-deficient T cells than in WT counterparts (fig. S3, B and C). Together, these data suggested that C1qbp was critical for proliferation and effector function of CD8+ T cells upon in vitro activation.
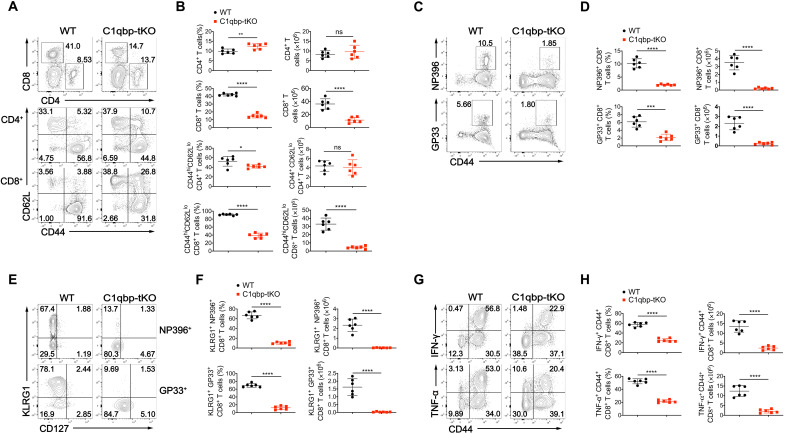
To investigate the physiology role of C1qbp in antiviral T cell response, we infected C1qbp-tKO mice and WT littermates with LCMV Armstrong strain as an acute infection model. At day 8 after infection, we analyzed T cell and B cell responses by flow cytometry. WT mice mounted robust frequency and number of CD8+ T cells, most of which were CD44hiCD62Llo effector T cells, but the abundance of both total and effector CD8+ T cells were much lower in their C1qbp-tKO counterparts after such infection (Fig. 2, A and B). In contrast, the abundance of effector CD4+ T cells, germinal center (GC) B cells, and plasma cells were comparable between C1qbp-tKO mice and WT littermates (Fig. 2, A and B, and fig. S3D). Moreover, C1qbp-tKO mice generated much fewer LCMV-specific CD8+ T cells than WT counterparts did after acute LCMV infection, as shown by the major histocompatibility complex class I (MHC-I) tetramer-positive cells (Fig. 2, C and D). Most of the LCMV-specific CD8+ T cells in WT mice presented as KLRG1+CD127− SLECs, but only a very small proportion of the LCMV-specific CD8+ T cells did so in C1qbp-tKO mice (Fig. 2, E and F). We then assessed the production of antiviral IFN-γ and tumor necrosis factor–𝛂 (TNF-𝛂) as an important function of effector CD8+ T cells. The production of these effector cytokines was greatly impaired in C1qbp-tKO mice at day 8 after infection (Fig. 2, G and H). To further assess the consequences of the defective effector CD8+ T cell differentiation in C1qbp-tKO mice, we measured viral serum load at day 5 after infection with the LCMV Armstrong strain. The viral titers in serum of C1qbp-tKO mice were slightly higher than those in WT litter mates (fig. S3E). We also used the chronic LCMV clone 13 (C13) model as an additional investigation of viral clearance in C1qbp-tKO mice. WT but not C1qbp-tKO mice experienced substantial weight loss from day 7 after LCMV C13 infection (fig. S3F). In addition, the viral control of C13 was compromised in C1qbp-tKO mice at least at day 20 after infection (fig. S3G). Together, these data demonstrated that C1qbp was required for the generation of robust antiviral effector CD8+ T cell response.
Fig. 2. C1qbp ablation in T cells leads to defective antiviral effector CD8+ T cell response.
(A) Flow cytometry of total splenocytes (top row) and splenic 2CD4+ (middle row) and splenic CD8+ (bottom row) T cells from WT and C1qbp-tKO mice at day 8 after infection with the LCMV Armstrong strain. (B) Frequency and total number of splenic CD4+ or CD8+ (among splenocytes), CD44hiCD62Llo effector CD4+ or CD8+ T cells (among splenic CD4+ or CD8+ T cells) from mice as in (A) (n = 6 per group). (C) Flow cytometry of splenic CD8+ T cells from mice as in (A). (D) Frequency (among splenic CD8+ T cells) and total number of NP396–404 (NP396)–specific (top row) and GP33–41 (GP33)–specific (bottom row) CD44+ CD8+ cells from mice as in (A) (n = 6 per group). (E) Flow cytometry of splenic NP396-specific (top row) and GP33-specific (bottom row) CD44+ CD8+ T cells from mice as in (A). (F) Frequency [among NP396-specific (top row) or GP33-specific (bottom row) CD44+ CD8+ T cells] and total number of NP396- or GP33-specific KLRG1+ effector CD8+ T cells in spleens of mice as in (A) (n = 6 per group). (G) Flow cytometry of splenic CD8+ T cells from mice as in (A). (H) Frequency (among splenic CD8+ T cells) and total number of IFN-γ+ (top row) or TNF-𝛂+ (bottom row) CD44+ CD8+ T cells in spleens of mice as in (A) (n = 6 per group). Each symbol represents an individual mouse; small horizontal lines indicate the means (±SD). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant (two-tailed unpaired Student’s t test). Data are representative of three independent experiments (error bars, SD).
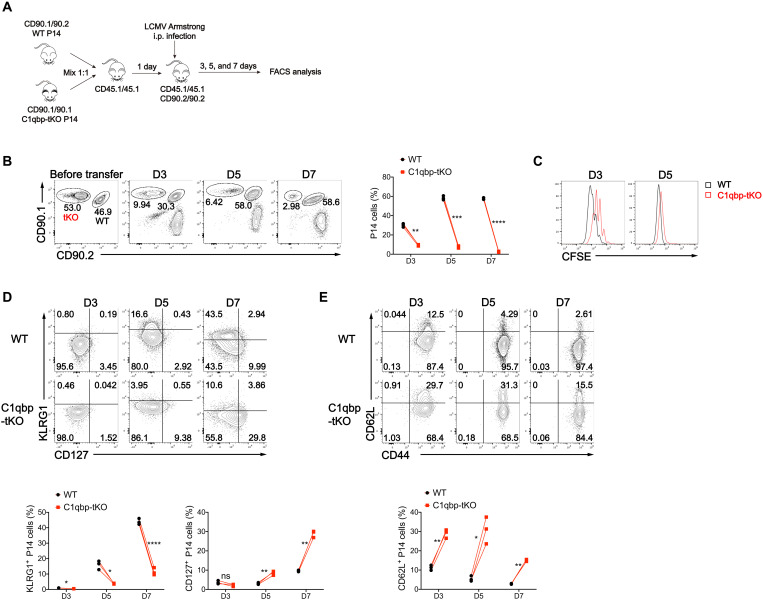
We next investigated whether C1qbp regulated effector CD8+ T cell differentiation in a CD8+ T cell–intrinsic manner. To address this issue, we crossed C1qbpfl/fl dLck-Cre mice with P14 mice (which have transgenic expression of a T cell antigen receptor specific for the epitope of LCMV glycoprotein amino acids 33 to 41, presented by the MHC-I molecule H2-Db) to generate C1qbp-tKO P14 mice. We isolated naïve CD8+ T cells from WT and C1qbp-tKO P14 mice, mixed them equally, and adoptively transferred them into B6.SJL-recipient mice, followed by subsequent infection of the host mice with the LCMV Armstrong strain. The donor cells were distinguished from the host cells by congenic CD90 expression and analyzed at different time points as indicated (Fig. 3A). We observed a significantly impaired accumulation of C1qbp-deficient antigen-specific cells compared with WT counterparts as early as day 3 after infection, accompanied by notably delayed cell division (Fig. 3, B and C). The difference between C1qbp-sufficient and C1qbp-deficient cells increased from 3-fold at day 3 to 20-fold at day 7 after infection (Fig. 3B). Among WT P14 CD8+ T cells, KLRG1+CD127− SLECs appeared around day 5 after infection and progressively increased in frequency and number until the peak of expansion (7 to 8 days after infection). In contrast, the formation of SLECs was markedly impaired within C1qbp-deficient P14 CD8+ T cells at these time points (Fig. 3D). However, the frequency of KLRG1−CD127+ MPECs was much higher in C1qbp-deficient P14 CD8+ T cells than in WT counterparts (Fig. 3D). In addition, the down-regulation of naïve or memory marker CD62L was also impaired in in C1qbp-deficient P14 CD8+ T cells compared with WT counterparts (Fig. 3E). Collectively, these results revealed that C1qbp was required for antiviral effector CD8+ T cell responses in a cell-intrinsic manner.
Fig. 3. C1qbp is intrinsically required for proliferation and differentiation of effector CD8+ T cells.
(A) Schematic of P14 cotransfer experiments. i.p., intraperitoneal; FACS, fluorescence-activated cell sorting. (B) Flow cytometry of splenic CD8+ T cells (left) and frequency of donor P14 CD8+ T cells (among splenic CD8+ T cells) (right) from B6.SJL recipient mice given transfer of naïve WT (CD90.1/CD90.2) and C1qbp-deficient (CD90.1/CD90.1) P14 CD8+ T cells, followed by infection with the LCMV Armstrong strain and analysis at indicated days after infection as in (A) (n = 3 per group). (C) Proliferation of donor WT and C1qbp-deficient P14 CD8+ T cells from recipient mice as in (A), indicated by flow cytometry of carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution. (D and E) Flow cytometry of donor WT and C1qbp-deficient P14 CD8+ T cells and frequency of KLRG1+ (D, bottom left) or CD127+ (D, bottom right) or CD62L+ (E) P14 CD8+ T cells (among donor P14 CD8+ T cells) from recipient mice as in (A) (n = 3 per group). Each symbol represents an individual mouse; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (two-tailed paired Student’s t test). Data are representative of three independent experiments (error bars, SD).
C1qbp deficiency results in distinct alteration of metabolites in activated CD8+ T cells
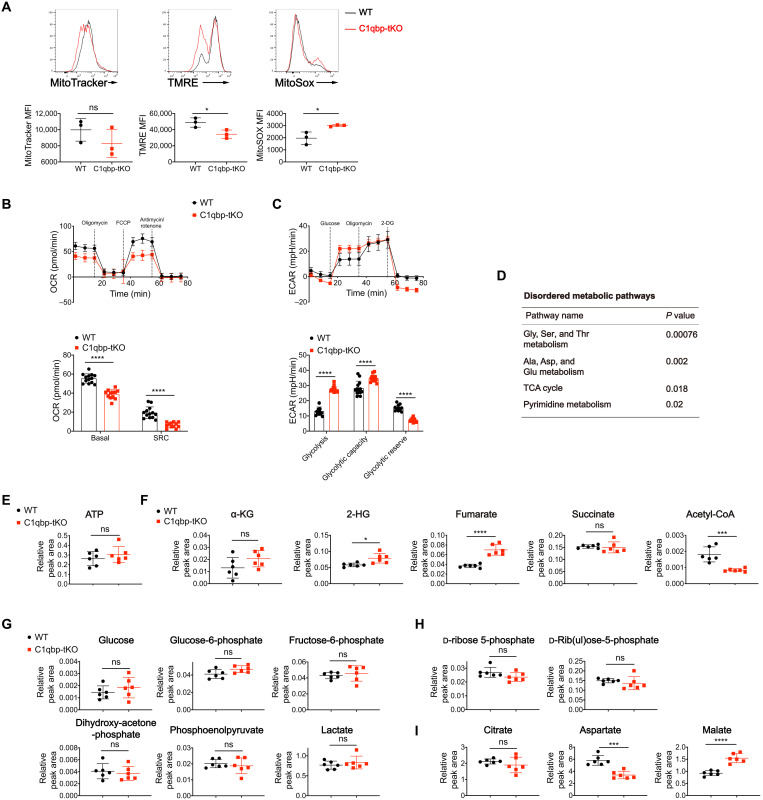
As we observed disrupted mitochondrial electron transport chain and impaired OXPHOS in C1qbp-deficient CD8+ T cells upon in vitro activation, we then analyze the in vivo metabolic consequence of C1qbp ablation in activated CD8+ T cells after acute LCMV infection. Similar to in vitro observation, lower mitochondrial membrane potential was observed in activated CD8+ T cells from C1qbp-tKO mice at day 4.5 after infection, whereas mitochondrial mass was unaltered (Fig. 4A), which is consistent with a previous report that CD8+ T cells with higher mitochondrial membrane potential have more capacity to differentiate into SLECs (29). The production of mitochondrial reactive oxygen species (ROS) was higher in activated CD8+ T cells from C1qbp-tKO mice than in counterparts from WT mice, although mitochondrial ROS was shown to be required for T cell activation through activation of nuclear factor of activated T cells (NFAT) (Fig. 4A) (30). Furthermore, in extracellular flux assays, activated CD8+ T cells from C1qbp-tKO mice at day 8 after acute LCMV infection exhibited lower OCR at both basal respiratory capacity and SRC than WT counterparts did, consistent with the observation from in vitro activated CD8+ T cells (Fig. 4B). Similar to the in vitro observation, the ECAR at glycolysis and glycolytic capacity but not at glycolytic reserve was increased in activated CD8+ T cells from C1qbp-tKO mice compared to counterparts from WT littermates (Fig. 4C). Together, these data verified that C1qbp was essential for mitochondrial OXPHOS in activated CD8+ T cells under physiological conditions of viral infection.
Fig. 4. C1qbp deficiency decreases acetyl-CoA and elevates fumarate and 2-HG in activated CD8+ T cells.
(A) Mitochondrial mass, mitochondrial membrane potential, and mitochondrial superoxide production were measured by MitoTracker Deep Red staining, tetramethylrhodamine ethyl ester perchlorate (TMRE) staining, and MitoSOX Red staining, respectively, in activated CD8+ T cells from WT or C1qbp-tKO mice at day 4.5 after infection with the LCMV Armstrong strain (n = 3 per group) . MFI, mean fluorescence intensity. (B and C) Seahorse extracellular flux analysis of OCR (B) and ECAR (C) of activated (CD44+) CD8+ T cells sorted from WT and C1qbp-tKO mice at day 8 after infection with the LCMV Armstrong strain (n = 12 to 14 wells per group). (D to I) Metabolomic analysis of WT and C1qbp-deficient P14 CD8+ T cells sorted from B6.SJL recipient mice respectively, at day 5 after infection with the LCMV Armstrong strain, was performed by mass spectrum. (D) Enrichment of the dysregulated metabolic pathways. (E) Relative abundance of ATP. (F) Relative abundance of α-KG, 2-HG, fumarate, succinate, and acetyl-CoA. (G to I) Relative abundance of metabolites in glycolytic (G), pentose phosphate pathway (H), and TCA cycle (I) (n = 6 per group). Each symbol represents an individual mouse; small horizontal lines indicate the means (±SD). *P < 0.05, ***P < 0.001, and ****P < 0.0001 (two-tailed unpaired Student’s t test). Data are representative of two independent experiments (error bars, SD).
To gain insight into the potential metabolic changes caused by defective OXPHOS in C1qbp-deficient CD8+ T cells during early effector T cell differentiation after acute LCMV infection, we isolated adoptively transferred P14 CD8+ T cells from recipient mice at day 5 after infection and performed metabolomic analysis by using high-performance LC-MS/MS (HPLC-MS/MS) (table S2). The metabolomic analysis revealed that there were substantial changes in metabolites in TCA cycle, nucleotide, and amino acid metabolisms in C1qbp-deficient P14 CD8+ T cells compared to their WT counterparts (Fig. 4D). Although C1qbp-deficient P14 CD8+ T cells displayed diminished OXPHOS, their production of ATP was similar to that of WT counterparts, probably compensated by the increased glycolysis (Fig. 4E). Upon T cell activation, metabolic pathways are not only harnessed to accommodate increased bioenergetic and biosynthetic demands but also obligated to provide metabolites as donors, substrates, cofactors, or antagonists for DNA- and chromatin-modifying enzymes (31). For instance, acetyl-CoA and S-adenosylmethionine (SAM) are donors for acetyl group of histone acetylation and methyl group of DNA/histone methylation, respectively. A TCA cycle intermediate, 𝛂-ketoglutarate (𝛂-KG), functions as a critical cofactor for both histone and DNA dioxygenases, including JmjC domain–containing histone lysine demethylases (KDMs) and the ten-eleven translocation (TET) family of 5-methylcytosine (5mC) hydroxylases (31). In contrast, the TCA cycle intermediates that are structurally similar to 𝛂-KG, including succinate and fumarate, as well as 2-HG, can competitively inhibit 𝛂-KG–dependent dioxygenases including KDMs and TETs (32, 33). These metabolites were shown to be accumulated in cells deficient in Uqcrfs1, which encodes for the Rieske iron-sulfur protein, an essential subunit of the mitochondrial complex III (34). Similarly, the concentrations of 2-HG and fumarate were notably increased in C1qbp-deficient P14 CD8+ T cells, whereas the concentrations of 𝛂-KG and succinate were comparable between C1qbp-deficient and C1qbp-sufficient P14 CD8+ T cells (Fig. 4F). Nicotinamide adenine dinucleotide (NAD+) is needed for the activity of the Sirtuin family of histone deacetylases (31). Although NAD+/NADH (reduced form of NAD+) ratio in C1qbp-deficient P14 CD8+ T cells was similar to that in WT counterparts, the concentration of acetyl-CoA in C1qbp-deficient P14 CD8+ T cells was much lower than in WT counterparts (Fig. 4F and fig. S4A) probably because of lower activity of pyruvate dehydrogenase (24, 26). The intermediates in glycolysis and pentose phosphate pathway were intact in C1qbp-deficient P14 CD8+ T cells (Fig. 4, G and H). Aspartate, whose biosynthesis is supported by mitochondrial respiration, is necessary for de novo nucleotide synthesis and cellular proliferation (35, 36). As expected, both aspartate and nucleotides were substantially diminished in C1qbp-deficient P14 CD8+ T cells, in agreement with the impaired proliferation (Fig. 4I and fig. S4B). Together, these data demonstrated that impaired mitochondrial OXPHOS caused by C1qbp ablation in CD8+ T cells resulted in a great deal of dysregulated metabolites including those play key roles in deposition of epigenetic marks.
C1qbp orchestrates the epigenetic program of effector CD8+ T cells
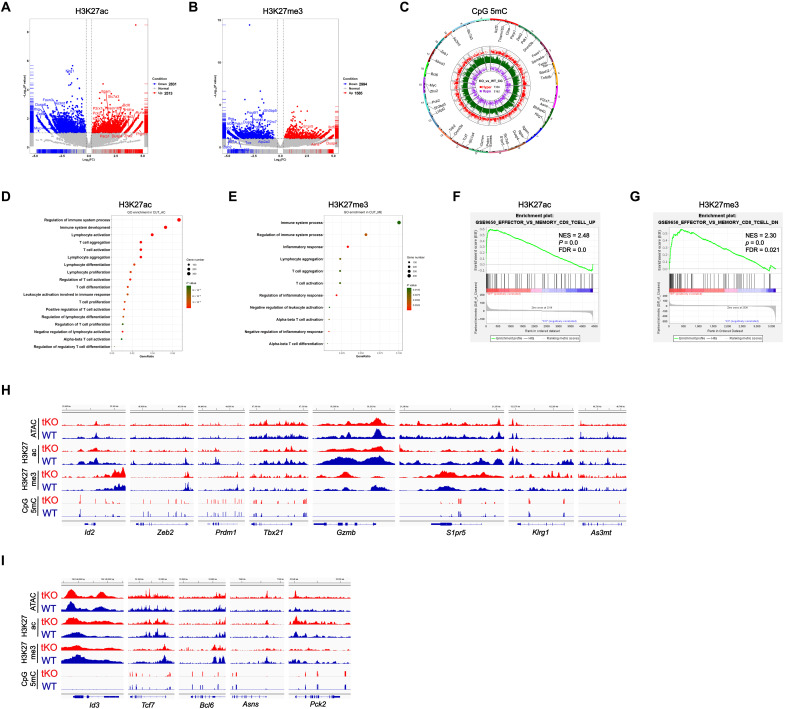
Some metabolites play critical roles in epigenetic regulation by serving as substrates, donors, cofactors, and antagonists for chromatin- and DNA-modifying enzymes, which provides potential cross-talk between metabolism and differentiation program (31). Such metabolites as acetyl-CoA, 2-HG, and fumarate were either decreased or increased in C1qbp-deficient CD8+ T cells, so we next sought to determine whether the epigenetic profiling was altered by C1qbp deficiency in CD8+ T cells after acute LCMV infection. To this end, at day 5 after LCMV Armstrong infection, we sorted C1qbp-sufficient or C1qbp-deficient P14 CD8+ T cells from recipient mice that received separate transfer before infection. Then, we performed Cleavage Under Targets and Tagmentation (CUT&Tag), a chromatin profiling strategy (37), to characterize genome-wide profiling of histone modifications including H3K27ac and H3K27me3. We also performed multiplexed reduced representation bisulfite sequencing (mRRBS) to dissect genome-wide profiling of DNA methylation at CpG sites. We then combined analysis of histone modifications and CpG methylation with accessible chromatins obtained by assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) to infer regions with active or repressed transcription. We were able to map a total of 32,373 and 51,266 H3K27ac peaks, 9536 and 19,128 H3K27me3 peaks, and 17,219 and 21,812 ATAC-seq peaks in C1qbp-sufficient and C1qbp-deficient P14 CD8+ T cells, respectively (fig. S5A). More than one-quarter of H3K27ac and H3K27me3 modifications and more than half of the ATAC-seq peaks were localized to promoter regions (fig. S5B). The proportionalities of the peak distributions were comparable between C1qbp-sufficient and C1qbp-deficient P14 CD8+ T cells (fig. S5B). Furthermore, there was also no global change in H3K27ac and H3K27me3 modifications between the two genotypes (fig. S5C). In contrast, there were remarkably higher levels of 5mC in genome-wide CpG sites (CpG 5mC) in C1qbp-deficient P14 CD8+ T cells than in WT counterparts (fig. S5, D to F).
We next determined consensus peaks that contained differentially modified regions (DMRs; differences in H3K27ac, or H3K27me3, or CpG 5mC depositions) and differentially accessible regions (DARs) from ATAC-seq between WT and C1qbp-deficient P14 CD8+ T cells. 5344 H3K27ac DMRs, 4579 H3K27me3 DMRs, 10,498 CpG 5mC DMRs, and 2063 DARs were identified between the two genotypes (Fig. 5, A to C, and fig. S5G). These histone DMRs and DARs were annotated to the nearest gene transcriptional start site (TSS) (fig. S5, H and I). Gene Ontology (GO) analysis revealed that genes containing DMRs of H3K27ac or H3K27me3 were associated with T cell activation and differentiation (Fig. 5, D and E). Gene set enrichment analysis (GSEA) further uncovered that gene loci with DMRs of H3K27ac, H3K27me3, or CpG 5mC were significantly enriched in effector and memory CD8+ T cell pathway. Moreover, effector signature genes in C1qbp-deficient P14 CD8+ T cells showed less H3K27ac occupancy and more H3K27me3 occupancy and CpG methylation, relative to C1qbp-sufficient P14 CD8+ T cells (Fig. 5, F and G, and fig. S5J).
Fig. 5. C1qbp regulates the epigenetic program of effector CD8+ T cells.
(A and B) Volcano plots illustrating DMRs of H3K27ac (A) and H3K27me3 (B) between WT and C1qbp-deficient P14 CD8+ T cells from recipient mice at day 5 after infection with the LCMV Armstrong strain (n = 2 per group). FC, fold change; FDR, false discovery rate; NES, normalized enrichment score. (C) CpG 5mC DMRs circos of CD8+ T cells as in (A) (n = 2 per group). (D and E) GO analysis of the DMRs of H3K27ac (D) and H3K27me3 (E) in the CD8+ T cells as in (A) (n = 2 per group). (F and G) GSEA of indicated signature genes from the ranked list of DMRs of H3K27ac (F) and H3K27me3 (G) in WT versus C1qbp-deficient P14 CD8+ T cells as in (A) (n = 2 per group). (H and I) Integrative Genomics Viewer (IGV) showing DARs from ATAC-seq, DMRs of H3K27ac, H3K27me3, and CpG 5mC depositions at the effector signature (H) and memory signature (I) gene loci with the indicated scale between WT (blue) and C1qbp-deficient (red) P14 CD8+ T cells as in (A) (n = 2 per group). Data shown contain the union of significant consensus peaks identified across two independent biological replicates.
To gain deep insight into the differential epigenetic profiles between C1qbp-sufficient and C1qbp-deficient P14 CD8+ T cells, we examined individual patterns of H3K27ac, H3K27me3, CpG 5mC, and ATAC-seq peaks in several crucial and well-studied effector and memory signature genes. At effector signature genes including Zeb2, Id2, Prdm1, Tbx21, Gzmb, S1pr5, and Klrg1, C1qbp-deficient P14 CD8+ T cells contained much less H3K27ac depositions than WT counterparts (Fig. 5H). In contrast, there were substantial more H3K27ac depositions at a few memory signature genes including Id3, Asns, and Pck2 in C1qbp-deficient P14 CD8+ T cells than in WT cells (Fig. 5I). On the other hand, H3K27me3 depositions were increased at a few effector signature genes including Id2, Gzmb, S1pr5, and Klrg1 but were decreased at some memory signature genes including Id3, Tcf7, Bcl6, Asns, and Pck2 within C1qbp-deficient P14 CD8+ T cells (Fig. 5, H and I). Notably, DNA methylation at CpG sites within genes (Id2, Prdm1, and Tbx21) encoding several key transcription factors that foster effector CD8+ T cell differentiation was strongly enhanced in C1qbp-deficient P14 CD8+ T cells (Fig. 5H). Although there were notable changes in histone modifications especially H3K27ac between C1qbp-sufficient and C1qbp-deficient P14 CD8+ T cells, chromatin accessibility was barely changed and augmented only at very few loci including Tcf7, Asns, and Pck2 (Fig. 5, H and I), consistent with a previous study that chromatin accessibility profiles were very similar between SLECs and MPECs (38). Collectively, these data suggested that most of notable and substantial changes in H3K27ac and H3K27me3 depositions as well as CpG methylation in C1qbp-deficient P14 CD8+ T cells preferentially occurred at genes that play critical roles in effector and memory CD8+ T cell differentiation.
C1qbp promotes the dynamic transcriptional program of effector CD8+ T cells
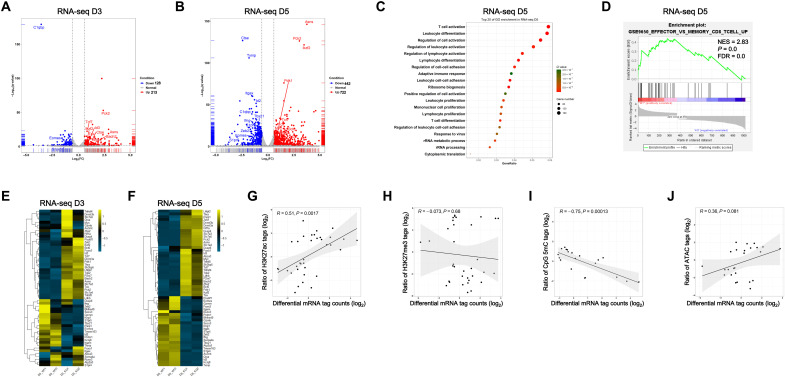
To explore the mechanism by which C1qbp regulates effector CD8+ T cell differentiation, we sorted WT or C1qbp-deficient P14 CD8+ T cells at days 3, 5, and 7 after LCMV Armstrong infection from recipient mice that received transfer before infection. Then, we conducted RNA sequencing (RNA-seq) analysis of these cells together with the naïve counterparts to characterize the dynamic transcriptional changes in CD8+ T cells in response to acute LCMV infection. To minimize skewing in gene expression due to defective effector T cell differentiation in C1qbp-deficient CD8+ T cells, at day 7 after LCMV Armstrong infection, we sorted KLRG1− P14 CD8+ T cells for RNA-seq analysis. Like its protein level, C1qbp mRNA expression in P14 CD8+ T cells was transiently induced with the expression peak at day 3 after infection (Fig. 1B). In line with this, there were very few differences in transcriptomes of naïve C1qbp-sufficient and C1qbp-deficient P14 CD8+ T cells (fig. S6A). The numbers of differentially expressed genes (DEGs) were increased over time until day 5 after infection (Fig. 6, A and B, and fig. S6, A and B). At day 7 after infection, some key DEGs remained even after subset normalization (fig. S6B). GO analysis revealed that the pathways of T cell activation and differentiation were gradually enriched until day 5 after infection (Fig. 6C and fig. S6C). GSEA further demonstrated that C1qbp-sufficient but not C1qbp-deficient CD8+ T cells were enriched with genes up-regulated in effector CD8+ T cell differentiation (Fig. 6D).
Fig. 6. C1qbp is required for the dynamic transcriptional program of effector CD8+ T cells.
(A and B) Volcano plots illustrating DEGs in the WT and C1qbp-deficient P14 CD8+ T cells from recipient mice at day 3 (A) and day 5 (B) after infection with the LCMV Armstrong strain (n = 2 per group). (C) GO analysis of DEGs in WT and C1qbp-deficient P14 CD8+ T cells as in (B). rRNA, ribosomal RNA. (D) GSEA of indicated signature genes from the ranked list of DEGs in WT versus C1qbp-deficient P14 CD8+ T cells as in (B). (E and F) Heatmaps of differentially expressed effector signature and memory signature genes in the WT and C1qbp-deficient CD8+ T cells from recipient mice at days 3 (E) and 5 (F) after infection with the LCMV Armstrong strain. (G to J) The ratio of sequence tags for H3K27Ac (G), H3K27me3 (H), CpG 5mC (I), and ATAC (J) correlated with the log2 fold change in mRNA expression between WT and C1qbp-deficient CD8+ T cells as in (B).
Although virus-specific CD8+ T cells did not phenotypically differentiate into effector or memory precursor at day 3 after infection, slightly but substantially differential expression in several key effector/memory signature genes (including Id2, Id3, and Tcf7) was already evident between C1qbp-sufficient and C1qbp-deficient P14 CD8+ T cells (Fig. 6E). More differentially expressed effector/memory signature genes emerged at day 5 after infection. The genes (including Zeb2, Id2, Tbx21, and Prdm1) encoding crucial transcription factors that promote terminal differentiation of effector CD8+ T cells were down-regulated in C1qbp-deficient CD8+ T cells compared to WT counterparts (Fig. 6F). Conversely, the genes (including Id3, Tcf7, Bcl6, Bach2, and Zeb1) encoding transcription factors that foster the formation and maintenance of memory CD8+ T cells were all up-regulated in C1qbp-deficient CD8+ T cells (Fig. 6F). Furthermore, expression of functional effector genes including Ifng, Gzmb, Klrg1, and S1pr5 was also reduced, while other memory marker genes including Asns and P2rx7 were increased in C1qbp-deficient CD8+ T cells (Fig. 6F). Although C1qbp expression was already declined at day 5 after infection, some of these effector/memory signature genes including Zeb2, Id2, Tbx21, Prdm1, Id3, and Tcf7 maintained such differential expression at day 7 after infection, even after subset normalization (fig. S6D). Collectively, these results demonstrated that C1qbp was required for inducing the transcriptional program of effector CD8+ T cells and for repressing that of memory CD8+ T cells.
We next sought to determine the correlation between differential gene expression and differential histone modifications of effector/memory signature genes (8). We correlated log2 fold change in mRNA expression at day 5 after infection between C1qbp-sufficient and C1qbp-deficient P14 CD8+ T cells with log2 ratios of the normalized H3K27ac, H3K27me3, CpG 5mC, or ATAC-seq tag counts. We observed that the differential expression of effector/memory signature genes significantly positively correlated with differential H3K27ac modification, slightly positively correlated with differential accessibility, markedly negatively correlated with differential CpG methylation, and poorly negatively correlated with differential H3K27me3 modification (Fig. 6, G to J). Together, our data suggested that C1qbp might promote transcriptional program of effector CD8+ T cells by establishing the corresponding epigenetic program.
Metabolite supplementations alter effector CD8+ T cell differentiation
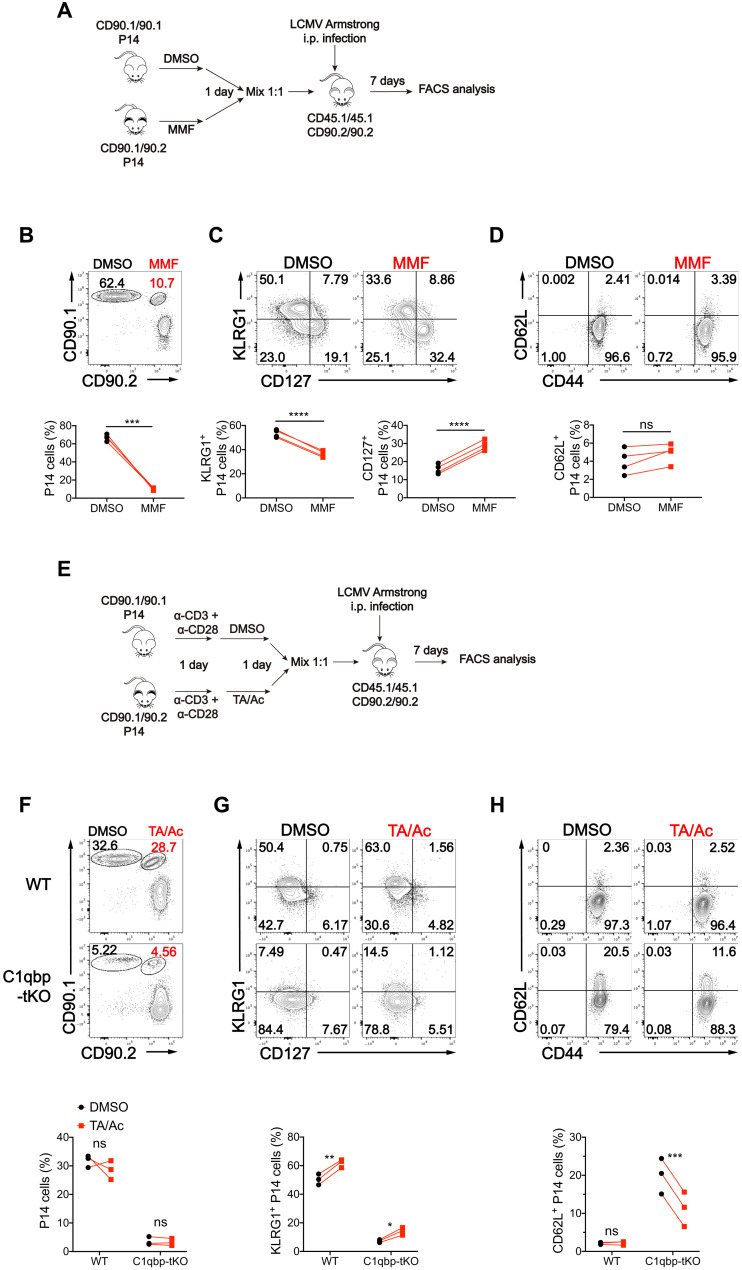
As we observed aberrant epigenetic and transcriptional programs associated with dysregulation of metabolites in C1qbp-deficient CD8+ T cells after acute LCMV infection, we next sought to investigate whether the changed metabolites were the causal factors for the defective effector CD8+ T cell differentiation. Like 2-HG, both fumarate and succinate are also competitive inhibitors of 𝛂-KG–dependent dioxygenases including KDMs and TETs (32). It has been reported that pretreatment of S-2-HG enantiomer suppresses effector CD8+ T cell differentiation but enhances memory CD8+ T cell differentiation (39). Given that the increase of fumarate was stronger than that of 2-HG in C1qbp-deficient CD8+ T cells, we next determined whether fumarate had similar effect to S-2-HG on CD8+ T cell differentiation. To this end, we first treated naïve WT CD8+ T cells with or without membrane-permeable form of fumarate [monomethyl fumarate (MMF)] before in vitro activation. Similar to S-2-HG treatment, MMF treatment remarkably suppressed the production of effector cytokines IFN-γ and TNF-𝛂 in activated CD8+ T cells (fig. S7, A and B). We then pretreated WT P14 CD8+ T cells with or without MMF and cotransferred treated and untreated cells into B6.SJL recipient mice, followed by subsequent infection of the host mice with the LCMV Armstrong strain (Fig. 7A). Fumarate pretreatment markedly restrained expansion of P14 CD8+ T cells in response to acute LCMV infection (Fig. 7B). Notably, fumarate pretreatment noticeably inhibited the commitment of P14 CD8+ T cells to KLRG1+CD127− SLEC differentiation but strongly enhanced the commitment of P14 CD8+ T cells to KLRG1−CD127+ MPEC differentiation (Fig. 7C). However, unlike S-2-HG treatment, fumarate pretreatment did not increase expression of CD62L (Fig. 7D). Together, these data suggested that pretreatment of WT CD8+ T cells with fumarate mimicked to some extent the defect of effector T cell proliferation and differentiation in C1qbp-deficient CD8+ T cells after acute LCMV infection, which was not completely similar to S-2-HG treatment.
Fig. 7. MMF inhibits effector CD8+ T cell differentiation and TA/Ac partially rectifies the defective effector differentiation of C1qbp-deficient CD8+ T cells.
(A) Schematic of MMF-treated P14 experiments. CD90.1/90.1 or CD90.1/90.2 P14 cells were cultured in vitro with dimethyl sulfoxide (DMSO) or MMF respectively for 1 day, and then 1:1 mixed and transferred into CD45.1/45.1 recipient mice, followed by infection with the LCMV Armstrong strain. Flow cytometry analysis was done at day 7 after infection. (B to D) Flow cytometry of splenic CD8+ T cells (B) and the donor P14 CD8+ T cells (C and D) as in (A). Frequency of the donor P14 cells (among splenic CD8+ T cells) (B), KLRG1+ P14 or CD127+ P14 subsets (among donor P14 cells) (C), and CD62L+ P14 subsets (among donor P14 cells) (D) as in (A) (n = 4 per group). (E) Schematic of TA/Ac-treated P14 experiments. CD90.1/90.1 or CD90.1/90.2 WT or C1qbp-deficient P14 cells were activated in vitro with plate-bound anti-CD3ε (1 μg/ml) and anti-CD28 (1 μg/ml) for 1 day and cultured with DMSO or TA/AC in the presence of anti-CD3ε and anti-CD28 for another day, and then were 1:1 mixed and transferred into CD45.1/45.1 recipient mice, followed by infection with LCMV Armstrong strain. Flow cytometry analysis was done at day 7 after infection. (F to H) Flow cytometry of splenic CD8+ T cells (F) and the donor P14 CD8+ T cells (G and H) as in (E). Frequency of the donor P14 cells (among splenic CD8+ T cells) (F), KLRG1+ P14 subsets (among donor P14 cells) (G), and CD62L+ P14 subsets (among donor P14 cells) (H) as in (E) (n = 3 per group). Each symbol represents an individual mouse; small horizontal lines indicate the means (±SD). *P < 0.05, **P < 0.01, and ***P < 0.001, ****P < 0.0001 (two-tailed paired Student’s t test). Data are representative of three independent experiments (error bars, SD).
There was remarkable decrease in acetyl H3K27 in the proeffector gene loci associated with decreased cellular acetyl-CoA concentration in C1qbp-deficient CD8+ T cells. It has been reported that combination of histone deacetylase inhibitor (TA) and Ac increased histone acetylation as well as expression of effector molecules in CD8+ T cells (40, 41). The previous studies prompted us to test whether TA plus Ac (TA/Ac) treatment could rectify the defective effector CD8+ T cell differentiation in C1qbp-deficient CD8+ T cells. To address this, we first determined the effect of TA/Ac treatment during in vitro effector CTL differentiation. TA/Ac treatment only slightly potentiated TNF-𝛂 production in WT CD8+ T cells but fully restored the production of both IFN-γ and TNF-𝛂 in C1qbp-deficient CD8+ T cells (fig. S7, C to F). We also pretreated WT P14 CD8+ T cells with or without TA/Ac and then cotransferred treated and untreated WT or C1qbp-deficient P14 CD8+ T cells into B6.SJL recipient mice, followed by subsequent infection of the host mice with the LCMV Armstrong strain (Fig. 7E). At day 7 after infection, in agreement with the previous study (40), the proliferation of neither WT nor C1qbp-deficient P14 CD8+ T cells was affected, but the differentiation of KLRG1+CD127− SLECs in both WT and C1qbp-deficient P14 CD8+ T cells was slightly but significantly enhanced by TA/Ac pretreatment (Fig. 7, F and G). Furthermore, TA/Ac pretreatment also promoted the down-regulation of CD62L in C1qbp-deficient P14 CD8+ T cells (Fig. 7H). These data suggested that decreased acetyl-CoA was one of the causal factors for the defect of effector T cell differentiation rather than for the defect of proliferation in C1qbp-deficient CD8+ T cells.
T cell receptor (TCR) triggers CD8+ T cells to produce up to millimolar of S-2-HG under physiological oxygen conditions through a hypoxia-inducible factor 1𝛂 (Hif-1𝛂)–dependent mechanism (39). As both 2-HG production and Hif1a mRNA expression were increased in C1qbp-deficient CD8+ T cells (Figs. 4F and 6F), we next investigated whether Hif-1𝛂 was involved in the defective effector CD8+ T cell differentiation. To address this, we crossed C1qbpfl/fl Hif1afl/fl mice with mice carrying a transgene encoding Cre recombinase under control of the Cd4 enhancer-promoter (CD4-Cre) to generate C1qbpfl/fl Hif1afl/fl CD4-Cre mice (C1qbp-Hif1a-DtKO, called DtKO hereinafter). Following infection with LCMV Armstrong strain, the total and CD44hiCD62Llo effector CD8+ T cells in DtKO mice were slightly but significantly increased compared with those in C1qbp-tKO mice, although the frequencies and numbers of total and KLRG1+CD127− SLECs in DtKO mice were similar to those in C1qbp-tKO mice (fig. S8, A to D). Moreover, the expression of effector cytokines including IFN-γ and TNF-𝛂 was partially restored in DtKO mice (fig. S8, E and F). These data indicated that Hif-1𝛂–induced S-2-HG accumulation might also account for the impaired effector differentiation in C1qbp-deficient CD8+ T cells. Together, these results suggested that dysregulated metabolites including 2-HG, fumarate, and acetyl-CoA all contributed to the defective effector T cell differentiation in C1qbp-deficient CD8+ T cells.
C1qbp ablation in T cells dampens antitumor immunity
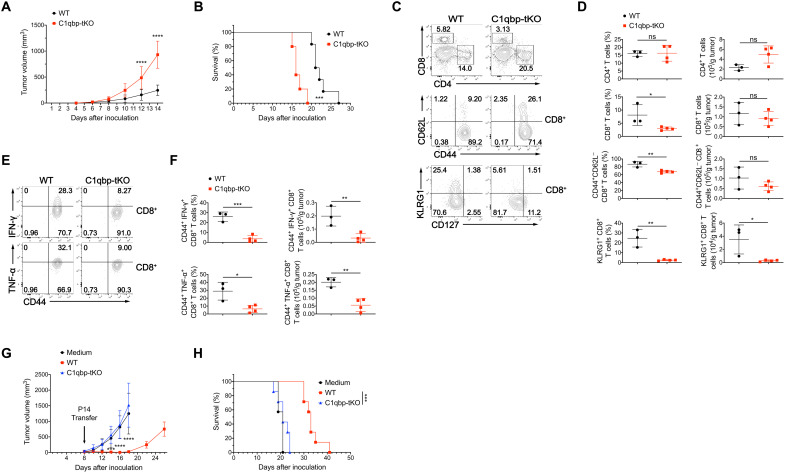
In addition to antiviral immunity, CD8+ cytotoxic T cells also play a central role in antitumor immunity through the capacity to kill malignant cells. To explore the effect of C1qbp in antitumor T cell response, we challenged WT and C1qbp-tKO mice with B16F10 melanoma cells. Compared to WT mice, C1qbp-tKO mice displayed a drastically increased tumor burden with shorten survival (Fig. 8, A and B). B16F10-challenged C1qbp-tKO mice had a moderately decreased frequency of tumor-infiltrating CD44+CD62L− effector CD8+ T cells and a nearly complete loss of KLRG1+ effector CD8+ T cells (Fig. 8, C and D). In contrast, the percentages and numbers of tumor-infiltrating CD4+ T cells in WT and C1qbp-tKO mice were comparable (Fig. 8, C and D). Moreover, the frequencies and numbers of tumor-infiltrating IFN-γ– or TNF-𝛂–producing effector CD8+ T cells were markedly reduced in C1qbp-tKO mice (Fig. 8, E and F). Thus, these results suggested that C1qbp facilitated CD8+ T cell–mediated antitumor immunity.
Fig. 8. T cell–specific deletion of C1qbp impairs antitumor immunity.
(A and B) Tumor growth (A) and survival curves (B) of WT and C1qbp-tKO mice injected subcutaneously with 2 × 105 B16F10 melanoma cells (WT, n = 6; C1qbp-tKO, n = 5). (C) Flow cytometry of tumor-infiltrating lymphocytes (TILs) (top row), TIL CD8+ T cells (middle and bottom rows) from WT and C1qbp-tKO mice at day 12 after subcutaneous injection with 2 × 105 B16F10 melanoma cells. (D) Frequency and total number of TIL CD4+ or CD8+ T cells (among TILs), CD44+CD62L− or KLRG1+ effector CD8+ T cells (among TIL CD8+ T cells) from mice as in (C) (WT, n = 3; C1qbp-tKO, n = 4). (E) Flow cytometry of TIL CD8+ T cells from mice as in (C). (F) Frequency (among TIL CD8+ T cells) and total number of IFN-γ+ (top row) or TNF-𝛂+ (bottom row) TIL effector CD8+ T cells from mice as in (C) (WT, n = 3; C1qbp-tKO, n = 4). (G and H) Tumor growth (G) and survival curves (H) of B16F10-GP33–bearing B6 mice received adoptive transfer of WT P14 cells, C1qbp-deficient P14 cells, or medium as control at day 8 after inoculation (n = 7 per group). Each symbol represents an individual mouse; small horizontal lines indicate the means (±SD). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 [two-way analysis of variance (ANOVA) (A and G), two-tailed unpaired Student’s t test (D and F), or log-rank (Mantel-Cox) test of survival curve (B and H)]. Data are representative of two (G and H) or three (A to F) independent experiments.
To explore the cell-intrinsic role of C1qbp on antigen-specific CD8+ T cells in antitumor immunity, we adoptively transferred WT or C1qbp-deficient P14 CD8+ T cells into B6 mice bearing LCMV GP33-expressing B16F10 melanoma cells. Mice transferred with WT P14 CD8+ T cells gained a strong ability to suppress tumor growth and to prolong survival, whereas the adoptive transfer of C1qbp-deficient P14 CD8+ T cells had nearly no effect on tumor control and mouse survival (Fig. 8, G and H). Together, our data demonstrated that T cell–specific C1qbp deficiency attenuated CD8+ T cell–mediated antitumor immunity.
DISCUSSION
Early-activated CD8+ T cells simultaneously increase both aerobic glycolysis and mitochondrial OXPHOS (14, 15). The roles of glycolysis on effector CD8+ T cells and OXPHOS on memory CD8+ T cells have been well established (13). However, whether and how mitochondrial OXPHOS regulates the differentiation of effector CD8+ T cells is still not fully understood. In this study, through the use of a combination of genetic, metabolomic, epigenetic, and transcriptomic approaches, we have revealed a critical role of C1qbp in the differentiation of effector CD8+ T cells in antiviral and antitumor immune responses. We have shown that C1qbp deficiency in T cells intrinsically impaired the differentiation of effector CD8+ T cells and was associated with a failure to increase mitochondrial respiratory capacities. Last, we have also shown that C1qbp promoted the epigenetic and transcriptional programs of effector CD8+ T cells by controlling production of metabolites including acetyl-CoA, fumarate, and 2-HG (fig. S9).
C1qbp is an evolutionally conserved protein involved in many biological processes because it is able to interact with a diverse array of cellular, plasma, and microbial proteins (23). C1qbp, also known as gC1qR, has been identified as a receptor for globular heads of C1q, the initiator of the classical complement pathway (42). MPECs rather than SLECs from mice lacking C1q (C1qa−/−) had impaired mitochondrial SRC, but C1qa−/− mice exhibited aberrantly stronger LCMV-specific effector CD8+ T cell response at day 8 after LCMV Armstrong infection (43). This phenotype is opposite to that of C1qbp-tKO mice, suggesting that C1q restrains the differentiation of effector CD8+ T cells through a C1qbp-independent pathway.
Early-activated CD8+ T cells increase both aerobic glycolysis and mitochondrial OXPHOS but then reduce OXPHOS and maintain higher glycolysis after differentiation into effector CD8+ T cells (14, 15). Glycolysis promotes differentiation of effector CD8+ T cells through a phosphoinositide 3-kinase–lactate dehydrogenase A positive-feedback circuit (44) and enhances Ifng expression via histone acetylation of Ifng locus (16). Although it has been reported that mitochondrial OXPHOS is required for antigen-specific T cell expansion and is also essential for memory CD8+ T cell formation (17, 30, 45, 46), the role of the transient increase of OXPHOS is still elusive. Here, we revealed that C1qbp-mediated augmentation of OXPHOS was essential for epigenetic regulation of the differentiation of effector CD8+ T cells. Activated CD8+ T cells have greater glycolytic and OXPHOS requirements than activated CD4+ T cells and CD8+ T cells are more affected by impaired OXPHOS (45), which is consistent with our observation that C1qbp deletion resulted in selective defect in differentiation of effector CD8+ T cells but not of effector CD4+ T cells.
Metabolomic analysis demonstrated that acetyl-CoA was reduced whereas fumarate and 2-HG were elevated in C1qbp-deficient P14 CD8+ T cells compared with WT counterparts after acute LCMV infection. In accord with the reduction of acetyl-CoA in C1qbp-deficient CD8+ T cells, significant decrease of H3K27Ac in effector signature genes was observed. Furthermore, TA/Ac pretreatment fully rectified in vitro effector CD8+ T cell differentiation and partially restored in vivo differentiation but not proliferation of C1qbp-deficient effector CD8+ T cells. Thus, our results indicated that differentiation of effector CD8+ T cells uncoupled from proliferation was promoted by histone acetylation. Although fumarate and 2-HG can both act as antagonists to 𝛂-KG–dependent dioxygenase including KDMs and TETs (32, 33), we observed more H3K27me3 depositions at only a few effector signature genes in C1qbp-deficient CD8+ T cells. Nevertheless, we detected a significant global increase of CpG methylation in C1qbp-deficient CD8+ T cells. CpG hypermethylation within genes (Id2, Prdm1, and Tbx21) encoding several master transcription factors that drive effector CD8+ T cell differentiation was observed in C1qbp-deficient P14 CD8+ T cells. These results suggested that transcriptional down-regulation of effector signature genes could be caused by combined effect of reduced H3K27Ac and increased H3K27me3 and CpG methylation, which might be due to decrease of acetyl-CoA and increase of fumarate and 2-HG, respectively. S-2-HG alters CD8+ T cell differentiation through modulation of histone and DNA demethylation (39). Whether fumarate suppressed the differentiation of effector CD8+ T cells mainly through inhibition of TET-mediated DNA demethylation or KDM5-mediated histone demethylation (47, 48), or any other mechanism (49), needs to be further investigated. Oral dimethyl fumarate (DMF) has been approved for the treatment of relapsing-remitting multiple sclerosis, which is characterized by preferential reduction of CD8+ T cells (50). Thus, our data provided a complementary mechanism for the therapeutic effects of DMF treatment.
In summary, we have found a critical function for C1qbp as a positive regulator in effector CD8+ T cell differentiation. We have also shown that C1qbp facilitated establishing epigenetic and transcriptional programs of effector CD8+ T cells by controlling metabolites coupled to OXPHOS. Therefore, we provide a mechanistic link between mitochondrial metabolism and effector CD8+ T cell differentiation. In addition, our findings suggest that these metabolite supplementations or targeting OXPHOS may have important application in T cell therapies of cancer, infectious diseases, and autoimmune diseases.
MATERIALS AND METHODS
Mice
C57BL/6 J (#000664), B6.SJL (#002014), Hif1afl/fl (#007561), dLck-Cre (#012837), CD4-Cre (#022071), and P14 TCR transgenic (#004694) mice were from the Jackson Laboratory. C1qbpfl/fl (B6;129- C1qbptm1/Nju) mice were purchased from the Model Animal Research Center of Nanjing University and were backcrossed to C57BL/6 J for six generations. All mice were maintained in specific pathogen–free facilities of Xiamen University Laboratory Animal Center. All animal protocols were approved by members of the Institutional Animal Care and Use Committee of Xiamen University.
Antibodies
For T cell activation, anti-mouse CD3ε (clone 145-2C11; catalog number 100359) and anti-mouse CD28 (clone 37.51; catalog number 102121) were purchased from BioLegend. For T cell purification, biotin-conjugated antibodies were purchased from BioLegend and listed as follows: anti-mouse CD4 (clone GK1.5; catalog number 100404), anti-mouse CD11b (clone M1/70; catalog number 101204), anti-mouse CD11c (clone N418; catalog number 117304), anti-mouse CD44 (clone IM7; catalog number 103004), anti-mouse CD45R/B220 (clone RA3-6B2; catalog number 103204), anti-mouse Ly-6G/Ly-6C(Gr-1) (clone RB6-8C5; catalog number 108404), anti-mouse TCRγ/δ (clone GL3; catalog number 118103), anti-mouse NK-1.1 (clone PK136; catalog number 108704), and anti-mouse TER-119/erythroid cells (clone TER-119; catalog number 116204). For flow cytometry and cell sorting, anti-mouse CD3e (clone 145-2C11; catalog numbers 11-0031-82 and 25-0031-82), anti-mouse CD4 (clone RM4-5; catalog number 48-0042-82 and clone GK1.5; catalog number 47–0041-82), anti-mouse CD8a (clone 53-6.7; catalog number 47-0081-82, 48-0081-82 and 45-0081-82), anti-mouse CD44 (clone IM7; catalog number 48-0441-82), anti-mouse CD45.1 (clone A20; catalog number 48-0453-82), anti-mouse CD45.2 (clone 104; catalog number 47-0454-80), anti-mouse CD69 (clone H1.2F3; catalog number 17-0691-82 and 11-0691-82), anti-mouse CD71 [clone R17217 (RI7 217.1.4); catalog number 12-0711-81], anti-mouse CD90.2 (clone 53-2.1; catalog number 17-0902-82), anti-mouse CD127 (clone A7R34; catalog number 48-1271-82), anti-mouse CD154(CD40L) (clone MR1; catalog number 17-1541-82), anti-mouse CD278(ICOS) (clone 7E.17G9; catalog number 25-9942-82), anti-mouse Foxp3 (clone FJK-16 s; catalog number 17-5773-82), anti-mouse Granzyme B (clone NGZB; catalog number 25-8898-82), anti-mouse immunoglobulin D (IgD) [clone 11-26c (11-26); catalog number 48-5993-82], streptavidin (catalog number 25-4317-82), and anti-mouse TNF-𝛂 (clone MP6-XT22; catalog number 17-7321-82 and 12-7423-41) were purchased from eBioscience. Anti-mouse CD25 (clone PC61; catalog numbers 102008 and 102016), anti-mouse CD45R/B220 (clone RA3-6B2; catalog numbers 103236 and 103212), anti-mouse CD90.1 (clone OX-7; catalog number 202524), anti-mouse CD127 (clone A7R34; catalog number 135007), anti-mouse KLRG1 (clone 14C2A07; catalog number 368614), and anti-mouse PD1 (clone 29F.1A12; catalog number 135214) were purchased from BioLegend. Anti-mouse CD44 (clone IM7; catalog number 553133 and 559250), anti-mouse CD62L (clone MEL-14; catalog number 560516), anti-mouse CD95 (clone Jo2; catalog number 554258), anti-mouse CD138 (clone 281-2; catalog number 553713), anti-mouse T and B cell activation antigen (clone GL7; catalog number 561529), and anti-mouse IFN-γ (clone XMG1.2; catalog numbers 554412 and 554413) were purchased from BD Biosciences. For Western blots, anti–β-actin (8H10D1; 3700), anti-C1QBP (D7H12; 6502), and anti-histone H3 (D2B12; 4620) were from Cell Signaling Technology. Anti-H3K27ac (ab4729) were from Abcam. Anti-H3K27me3 (07-449) was from Millipore Sigma. HRP Goat Anti-Mouse IgG (H+L) (AS003) and HRP Goat Anti-Rabbit IgG (H+L) (AS014) were from ABclonal.
Flow cytometry and cell sorting
All samples were collected from adult (8- to 12-week-old) mice. Single-cell suspension of lymphoid organs was prepared by mashing through a cell strainer. After treatment with red blood lysis buffer, cell suspensions were surface stained with fluorochrome-conjugated antibodies in flow cytometry buffer [phosphate-buffered saline (PBS) + 0.5% bovine serum albumin (BSA) + 0.05% NaN3]. Intracellular cytokine staining was performed after stimulation with phorbol 12-myristate 13-acetate (50 ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich) in the presence of GolgiStop for 4 hours. Cells were incubated with antibodies against cell surface markers and then fixed and permeabilized with BD Cytofix/Cytoperm buffer. Cells were then stained with antibodies against indicated cytokines. Intracellular Foxp3 was stained after cell surface staining. Samples were fixed and permeabilized with Foxp3 staining buffer according to the manufacturer’s manual (00-5523; eBioscience). Samples were incubated for 45 to 60 min at 4°C with fixation/permeabilization buffer and washed with 1× permeabilization buffer. Samples were incubated for another 45 to 60 min with antibody to Foxp3 (FJK-16 s; eBioscience) in permeabilization buffer. All samples were analyzed on BD LSRFortessa or ACEA NovoCyte flow cytometers, and data analysis was performed using FlowJo software. For cell sorting, cell suspensions were stained with fluorochrome-conjugated antibodies in sorting buffer [PBS supplemented with 1 mM EDTA, 25 mM Hepes, and 1% fetal bovine serum (FBS)] and then sorted with a BD Aria sorter.
For carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution assay, cells were labeled with 1 or 10 μM CFSE in PBS at 37°C for 10 min. Then, the cells were washed with PBS before culture or transfer.
To measure mitochondrial ROS, mass, and potential, CD8+ T cells were incubated with 5 μM MitoSOX Red (Life Technologies), 200 nM MitoTracker Deep Red (Life Technologies), or 100 nM TMRE (tetramethylrhodamine, ethyl ester, perchlorate) (Molecular Probes) for 30 min at 37°C, followed by surface staining.
Cell culture
Naïve CD8+ T cells were purified (>95% purity) by negative selection using BeaverBeads Streptavidin from red blood cell–lysed single-cell suspensions of pooled spleen and lymph nodes. For in vitro activation, naïve CD8+ T cells were stimulated with plate-bound anti-CD3ε (3 μg/ml) and anti-CD28 (3 μg/ml) in the presence of recombinant murine IL-2 (20 ng/ml) in RPMI 1640 medium with 10% FBS, 1 mM glutamine, 1 mM sodium pyruvate, 10 mM Hepes, 50 μM β-mercaptoethanol, penicillin (50 U/ml), and streptomycin (50 μg/ml).
For in vitro effector CTL differentiation, naïve CD8+ T cells were activated with plate-bound anti-CD3ε (1 μg/ml) and anti-CD28 (1 μg/ml) for 2 days and were subsequently removed from TCR signal and recultured in the presence of IL-2 for 4 days to generate effector cells.
For in vitro fumarate treatment, naïve CD8+ T cells were culture in medium with vehicle [dimethyl sulfoxide (DMSO)] or 100 μM MMF for 24 hours. Then, cells were washed and stimulated with plate-bound anti-CD3ε (1 μg/ml) and anti-CD28 (1 μg/ml) in the presence of recombinant murine IL-2 (20 ng/ml) in culture medium for 3 days.
For in vitro TA and sodium Ac treatment, naïve CD8+ T cells were stimulated with plate-bound anti-CD3ε (1 μg/ml) and anti-CD28 (1 μg/ml) in the presence of recombinant murine IL-2 (20 ng/ml) in culture medium for 2 days. Then, cells were resuspended and cultured in medium with recombinant murine IL-2 (20 ng/ml) and vehicle (DMSO) or TA (50 μM) (Selleckchem)/Ac (10 mM) (Sigma-Aldrich) for 24 hours. After treatment, cells were resuspended and cultured in medium with recombinant murine IL-2 (20 ng/ml) for another 2 days.
Virus preparation and titration
Both Armstrong and C13 stains of LCMV were propagated in BHK-21 cells. The titration of LCMV in stocks and serum samples were quantified by plaque assay or VL4–green fluorescent protein (GFP) staining assay. Briefly, Vero cells were seeded in a flat 96-well plate. The stock of LCMV or indicated serum samples was diluted and applied to infect Vero cells for about 20 hours. Then, the cells were fixed with 4% paraformaldehyde, followed by VL4-GFP staining for 2 hours at room temperature. The stained cells were then counted under a fluorescent microscope.
T cell isolation, adoptive transfer, and LCMV infection
For P14 T cell transfer, CD90.1+ (Thy1.1+) WT P14 CD8+ T cells were purified by negative selection using BeaverBeads Streptavidin. Then, 5 × 105 cells (for day 3 after infection), 5 × 104 cells (for day 5), and 3 × 104 cells (for day 7) were transferred into recipient mice by intravenous injection into tails 1 day before infection. Each mouse was infected with 2 × 105 or 5 × 105 plaque-forming units of the LCMV Armstrong strain by bilateral intraperitoneal injection for experiments at day 8 or 3, respectively.
For fumarate treatment, naïve WT P14 CD8+ T cells were culture in the medium with vehicle (DMSO) or 100 μM MMF for 24 hours. Vehicle and MMF-treated cells were then mixed 1:1 and transferred intravenously into recipient mice followed by infection with the LCMV Armstrong strain.
For TA and sodium Ac treatment, WT and C1qbp-deficient P14 CD8+ T cells were stimulated with plate-bound anti-CD3ε (1 μg/ml) and anti-CD28 (1 μg/ml) in the presence of recombinant murine IL-2 (20 ng/ml) in culture medium. Culture medium was then replaced with fresh medium containing vehicle (DMSO) or TA (10 μM) (Selleckchem)/Ac (5 mM) (Sigma-Aldrich) at day 1 after activation. At day 2, vehicle or TA/Ac-treated WT or C1qbp-deficient cells were mixed 1:1 and transferred intravenously into recipient mice followed by infection with the LCMV Armstrong strain.
Immunoblot analysis
CD8+ T cells were collected and lysed using NP-40 lysis buffer containing 20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 5 mM EDTA (pH 8.0), 5 mM Na4P2O7, 1 mM Na3VO4, 5 mM NaF, and protease inhibitor cocktail (Thermo Fisher Scientific). The cell lysates were diluted with SDS loading buffer and were denatured by being boiled for 10 min. Samples were then separated by SDS–polyacrylamide gel electrophoresis, followed by electrotransfer to polyvinylidene difluoride membranes (Merck Millipore). Then, the membranes were blocked with 5% nonfat milk in PBS with 0.1% Tween 20 (PBST) and incubated at 4°C overnight with primary antibodies. After washing with PBST, membranes were incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour at room temperature and were then developed with an enhanced chemiluminescence detection system (RPN 2235; GE Healthcare).
Confocal microscopy assay
Naïve CD8+ T cells were purified as described above and then stimulated with plate-bound anti-CD3ε (1 μg/ml) and anti-CD28 (1 μg/ml) for 48 hours, followed by staining with MitoTracker Deep Red (Life Technologies). Then, the cells were transferred to a 24-well plate with poly-l-lysine slides and fixed with ice-cold methanol, permeabilized with PBST, and then blocked with PBST supplemented with 1% BSA, 10% normal goat serum, and 0.3 M glycine. The cells were then incubated with anti-mouse C1qbp/gC1qR antibody (5 μg/ml; Abcam, ab24733) overnight at 4°C, followed by staining with goat anti-rabbit IgG-AF488 (1:200; Cell Signaling Technology) and 4′,6-diamidino-2-phenylindole. Confocal fluorescence images were acquired with a Zeiss LSM 780 confocal microscope, and images were processed and analyzed using ZEN 2 software.
Transmission EM
WT and C1qbp-deficient CD8+ T cells were stimulated with plate-bound anti-CD3ε (3 μg/ml) and anti-CD28 (3 μg/ml) in the presence of recombinant murine IL-2 (20 ng/ml) for 72 hours. Then, the cells were fixed with 2.5% glutaraldehyde overnight at 4°C, rinsed with phosphate buffer, and postfixed with 1% osmium tetroxide. Next, the samples were rinsed and dehydrated in ethanol and acetone. Then, samples were embedded in spurr resin. Last, 70-nm ultrathin sections were stained with 2% uranyl Ac and lead citrate and were visualized using a Hitachi HT-7800 transmission electron microscope.
Mitochondria mass spectrometry
CD8+ T cells were stimulated with plate-bound anti-CD3ε (3 μg/ml) and anti-CD28 (3 μg/ml) in the presence of recombinant murine IL-2 (20 ng/ml) for 48 hours. Then, cells were collected and washed with PBS. Mitochondria were isolated using the Cell Mitochondria Isolation Kit (Beyotime Biotechnology) following the manufacturer’s instructions. Briefly, cells were lysed on ice for 10 min followed by homogenate using the glass homogenizer. Then, samples were centrifuged at 600g for 10 min at 4°C. The supernatants were transferred to new tubes and were centrifuged at 11,000g for 10 min at 4°C. The sediments, the isolated mitochondria of the cells, were resuspended with 8 M urea and sonicated for lysis. Samples were analyzed by LC-MS/MS using Bruker timsTOF Pro (Bruker).
Activity of mitochondrial respiratory complex
Respiratory complex activity was measured using the Mitochondrial Respiratory Complex Activity Assay Kit (SolarBio) following the manufacturer’s instructions. Briefly, WT and C1qbp-deficient P14 CD8+ T cells were stimulated with plate-bound anti-CD3ε (1 μg/ml) and anti-CD28 (1 μg/ml) in the presence of recombinant murine IL-2 (20 ng/ml) for 48 hours. Cells were then lysed and mitochondria were collected. The absorbance was measured using the Cary 60 UV-Vis Spectrophotometer (Agilent).
Extracellular flux analysis
OCRs and ECARs were measured using a Seahorse XFe96 extracellular flux analyzer (Agilent Technologies). Briefly, OCR of CD8+ T cells were analyzed under basal conditions and after sequential injections of 1 μM oligomycin A (APExBIO Technology), 2 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (Selleckchem), and 0.1 μM rotenone (Selleckchem) with 1 μM antimycin A (Abcam). ECAR was analyzed under basal conditions and after sequential injections of 10 mM glucose (Sigma-Aldrich), 1 μM oligomycin A, and 50 mM 2-deoxyglucose (Sangon Biotech).
Metabolite measurements and analysis
WT and C1qbp-deficient P14 CD8+ T cells were sorted from recipient mice at day 5 after LCMV Armstrong infection. Then, cells were washed once with 0.9% NaCl. Dry cell pellets were frozen in liquid nitrogen. Following collection of all samples over multiple days, samples were thawed on ice and suspended in 1 ml of ice-cold 80% methanol with vortex for 2 min. Then, samples were sonicated for 5 min at 4°C. Following this, cells were centrifuged at 18,000g for 10 min at 4°C. The supernatant was transferred to fresh tubes and dried. Samples were analyzed by HPLC-MS/MS using AB SCIEX QTRAP 5500. The liquid chromatography with SCIEX ExionLC AD was prepared, and all chromatographic separations were performed with a Millipore ZIC-pHILIC column (5 μm, 2.1 mm by 100 mm internal dimensions, PN: 1.50462.0001). The column was maintained at 40°C, and the injection volume of all samples was 2 μl. The mobile phase consisted of 15 mM ammonium Ac and ammonium hydroxide (3 ml/liter; > 28%) in LC-MS grade water (mobile phase A) and LC-MS grade 90% (v/v) acetonitrile–HPLC water (mobile phase B) run at a flow rate of 0.2 ml/min. The analysts were separated with the following gradient program: 95% B held for 2 min, increased to 45% B in 13 min, held for 3 min, and the post time was set 4 min. The gradient was as follows: 95% B for 2 min, then to 45% B within 13 min (linear gradient), then maintained for 3 min, then to 95% B directly, and maintained for 4 min. The flow rate was 0.2 ml/min. The QTRAP mass spectrometer used a Turbo V ion source. The ion source was run in negative mode with a spray voltage of −4500 V, Gas1 40 psi (~275.7 kPa), Gas2 50 psi (~344.7 kPa), and Curtain gas 35 psi (~241.3 kPa). Metabolites were measured using the multiple reactions monitoring mode. The relative amounts of metabolites were analyzed by MultiQuant Software Software (AB SCIEX).
Melanoma mouse model
WT or C1qbp-tKO mice (8 to 10 weeks old) were inoculated subcutaneously with B16F10 cells (2 × 105) in the dorsal area. Tumor size was measured over time and calculated as length × (width2)/2. Survival was recorded each day. Mice with tumor size reaching 2000 mm3 were euthanized and counted as mortality.
Cellular therapy
B16F10-GP33 cells (2 × 105) were implanted subcutaneously into C57BL/6 mice. At day 8 after implantation, tumor-bearing mice with similar tumor size were divided into three groups randomly and received with plain RPMI 1640, in vitro–activated WT or C1qbp-deficient P14 CD8+ T cells (3 days post in-vitro activation 1 million cells per mouse). Cells were washed with plain RPMI 1640 before they were transferred into B16-GP33–bearing mice at day 8 post tumor implantation After T cell transfer, tumor size was measured every 2 days.
Cytosine methylation analysis
DNA methylation was measured using the MethylFlash Global DNA Methylation (5-mC) ELISA Easy (Colorimetric) Kit (Epigentek) following the manufacturer’s instructions. Briefly, DNA was isolated from sorted P14 cells using a QIAamp DNA mini kit (Qiagen). DNA concentrations were measured using the NanoDrop 2000 (Thermo Fisher Scientific). One hundred nanograms of DNA was loaded per well of a MethylFlash kit. The absolute quantity of methylcytosine in samples was obtained through the generation of a standard curve of known concentrations of methylcytosine. The absorbance was read on a VersaMax ELISA microplate reader (Molecular Devices).
RNA-seq analysis
Cells were lysed with RLT Buffer from a Qiagen RNeasy Mini kit (Qiagen). Cell lysates were stored at −80°C until RNA was extracted. RNA isolations were performed using the Qiagen RNeasy Mini kit (Qiagen) following the manufacturer’s protocol. RNA quality and quantity were measured using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies). mRNA libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. To select complementary DNA fragments of preferentially 250 to 300 base pairs (bp) in length, the library fragments were purified with an AMPure XP system (Beckman Coulter, Beverly). Then, USER Enzyme (NEB) was used with size-selected fragments and polymerase chain reaction (PCR) products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on the cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina NovaSeq platform, and 150-bp paired-end reads were generated. Raw data (raw reads) of Fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy N, and low-quality reads from raw data. At the same time, Q20, Q30, and GC content of the clean data was calculated. All the downstream analyses were based on the clean data with high quality. Reads were then aligned to the mm10 genome. Index of the reference genome was built using Hisat2(version 2.0.5) and paired-end clean reads were aligned to the reference genome using Hisat2. We selected Hisat2 as the mapping tool because Hisat2 can generate a database of splice junctions based on the gene model annotation file and thus a better mapping result than other nonsplice mapping tools. FeatureCounts (version 1.5.0) was used to count the read numbers mapped to each gene. Then, FPKM of each gene was calculated on the basis of the length of the gene and reads count mapped to this gene. FPKM, expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced, considers the effect of sequencing depth and gene length for the read count at the same time and is currently the most commonly used method for estimating gene expression levels. Differential expression analysis of two groups (two biological replicates per condition) was performed using DESeq2 (version 1.16.1). DESeq2 provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Genes with an adjusted P < 0.05 found by DESeq2 were assigned as differentially expressed. GO enrichment analysis of DEGs was implemented by the clusterProfiler (version 3.16.1), in which gene length bias was corrected. GO terms with corrected P < 0.05 were considered significantly enriched by DEGs. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (www.genome.jp/kegg/). We used clusterProfiler to test the statistical enrichment of differential expression genes in KEGG pathways. GSEA was done using the Broad Institute GSEA-P (version 4.1.0). Briefly, the gene list output from edgeR was ranked by calculating a rank score of each gene as −log10(P value) × sign(FC), in which FC is fold change {expressed as log2[expression in knockout (KO)/expression in WT} and the sign depends on whether the gene is up-regulated or down-regulated. A preranked GSEA was done using 1000 permutations and the immunologic pathway database.
Assay for transposase-accessible chromatin with high-throughput sequencing
ATAC-seq was performed as previously described (51). A total of 5 × 104 P14 CD8+ T cells were sorted from the spleens of mice at day 5 after infection with the LCMV Armstrong strain. The unfixed nuclei of these cells were tagged using tn5 transposase (TruePrep DNA Library Prep Kit V2 for Illumina) for 30 min at 37°C, and the resulting library fragments were generated by 10 to 12 PCR cycles, and lastly sequenced on Illumina Nova-PE150. Two biological replicates were processed per biological sample. The pair-end reads were mapped to the mm10 mouse genome using Bowtie2 (version 2.3.4.3). All unmapped reads, nonuniquely mapped reads, and PCR duplicates were removed by SAMTools. Resulting SAM files were converted to BAM files by SAM tools. For peak calling, parameters used were MACS2 (version 2.2.7.1). Visualization of peak distribution along genomic regions of interested genes was performed with the Integrative Genomics Viewer (IGV). Genes with a peak within 3 kb of the TSS were considered target genes. An area of 3 kb surrounding each TSS was selected to get ATAC-seq profiles using deepTools (version 3.4.3). The peaks were annotated to the nearest gene TSS using ChIPseeker (version 1.22.1)—for regions with one-to-many mappings for a given transcript, the most highly expressed transcript across KO and WT CD8+ T cells was used to reduce annotation to be one to one. The consensus peakset was defined first for each condition based on the replicates; we used Irreproducible Discovery Rate (IDR) (version 2.0.3) to capture consensus regions. The DMR was performed with DESeq2 (version 1.22.2) with the read counts normalized by fitting the data to a local smoothed dispersion fit to better capture the observed dispersion-mean relationship. Differential analysis was then performed to determine statistically significant DMRs, defined as those with P value less than 0.1 and fold change greater than 1.2. For downstream analysis, we normalized the read counts by computing the numbers of reads per kilobase of bin per million of reads sequenced. The analysis and visualization of data were performed using custom R scripts.
CUT&Tag
CUT&Tag assay was performed using the Hyperactive In-Situ ChIP Library Prep Kit for Illumina (Vazyme) following the manufacturer’s instructions. Briefly, P14 CD8+ T cells were sorted from the spleens of mice at day 5 after infection with the LCMV Armstrong strain. A total of 5 × 104 to 10 × 104 cells were tagged with concanavalin A–coated magnetic beads and then incubated with 1 μg of primary antibody [rabbit polyclonal anti-Histone H3 (acetyl K27) antibody (ab4729, Abcam), rabbit polyclonal anti-Histone H3 (trimethyl K27) antibody (07-449, Millipore), rabbit polyclonal anti-Histone H3 (trimethyl K9) antibody (ab8898, Abcam), and Rabbit IgG Isotype Control (3900, CST)] overnight at 4°C with slow rotation. After incubated with secondary antibody (1 μg) [goat anti-rabbit IgG H&L (ab6702, Abcam)] at room temperature for 1 hour, cells were incubated with 0.04 μM Hyperactive pG-Tn5 Transposon and then resuspended in tagmentation buffer for tagmentation. DNA was purified and amplified by 15 to 17 PCR cycles to generate the libraries. Libraries were sequenced on Illumina Nova-PE150, and 150-bp paired-end reads were generated. All raw data were quality-trimmed to a minimum phred score of 20 using trimmomatic (version 0.39). Apparent PCR duplicates were removed using Picard MarkDuplicates (version 2.23.3). All reads produced by CUT&Tag of acetyl-H3K27 and trimethyl-H3K27 were aligned to the mm10 mouse genome using Bowtie2 (version 2.3.4.3) with options: –local–very-sensitive-local–no-unal–nomixed–no-discordant–phred33 -I 10 -X 700. Sequence tags were aligned to the genome and then subsequently analyzed by MACS2 (version 2.2.7.1) to detect genomic regions enriched for multiple overlapping DNA fragments (peaks) that we considered to be putative binding sites. For peak calling, parameters used were macs2 callpeak -t input_file –q 0.05 –f BAMPE –keep-dup all –n out_name. Peaks with a FDR lower than 5% were saved to detect chromosomal regions for further analyses. Visualization of peak distribution along genomic regions of interested genes was performed with IGV. Genes with a peak within 3 kb of the TSS were considered target genes. The analysis and software for peak annotation, DMR selection, and the other analysis were same as the ATAC-seq analysis.
DNA methylation analysis
To evaluate genome-wide DNA methylation status, mRRBS was performed. Briefly, P14 cells were sorted from the spleens of mice infected with LCMV A4 at day 5. Then, genomic DNA was isolated using the QIAamp DNA mini kit (Qiagen). DNA concentration was measured using the Qubit DNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, CA, USA). A total amount of 1.5 μg of genomic DNA spiked with moderate lambda DNA was handled by Msp I, followed by end repair and adenylation. Cytosine-methylated barcodes were ligated to DNA as per manufacturer’s instructions. Then, these DNA fragments were treated with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research). The library was constructed by Novogene Corporation (Beijing, China). Subsequently, paired-end sequencing of sample was performed on an Illumina platform (Illumina, CA, USA). Library quality was assessed on the Agilent Bioanalyzer 2100 system.
The library was sequenced on the Illumina NovaSeq platform. Image analysis and base calling were performed with an Illumina CASAVA pipeline and lastly generated 150-bp paired-end reads. First of all, we used FastQC (fastqc_v0.11.5) to perform basic statistics on the quality of the raw reads. Then, those reads sequences produced by the Illumina pipleline in FASTQ format were preprocessed through fastp (fastp 0.20.0). The remaining reads that passed all the filtering steps was counted as clean reads, and all subsequent analyses were based on this. Last, we used FastQC to perform basic statistics on the quality of the clean data reads. Bismark software (version 0.16.3) was used to perform alignments of bisulfite-treated reads to a reference genome (-X 700 --dovetail). The reference genome was firstly transformed into bisulfite-converted version (C-to-T and G-to-A converted) and then indexed using bowtie2. Clean reads were also transformed into fully bisulfite-converted versions (C-to-T and G-to-A converted) before being aligned to the similarly converted versions of the genome in a directional manner. Sequence reads that produce a unique best alignment from the two alignment processes (original top and bottom strands) were then compared to the normal genomic sequence, and the methylation state of all cytosine positions was inferred. The same reads that aligned to the same regions of genome were regarded as duplicated ones. The sequencing depth and coverage were summarized using deduplicated reads. The results of methylation extractor (bismark_methylation_extractor, -- no_overlap) were transformed into bigWig format for visualization using IGV browser. The sodium bisulfite nonconversion rate was calculated as the percentage of cytosine sequenced at cytosine reference positions in the lambda genome. Methylated sites were identified with a binomial test using the methylated counts (mC), total counts (mC + umC), and the nonconversion rate (r). Sites with FDR-corrected P < 0.05 were considered as a methylated site. To calculate the methylation level (ML) of the sequence, we divided the sequence into multiple bins, with a bin size of 10 kb. The sum of methylated and unmethylated read counts in each window was calculated. ML for each window or C site shows the fraction of methylated Cs. Differentially methylated regions were identified using the DSS software. The core of DSS is a new dispersion shrinkage method for estimating the dispersion parameter from Gamma-Poisson or beta-binomial distributions. According to the distribution of differentially methylated regions through the genome, we defined the genes related to differentially methylated regions as genes whose gene body region [from TSS to transcription end site (TES)] or promoter region (2 kb upstream from the TSS) has an overlap with the differentially methylated regions.
Statistical analysis
All data were analyzed by a paired or unpaired t test or two-way analysis of variance (ANOVA) with GraphPad Prism 8.0. A log-rank (Mantel-Cox) test was used for comparison of survival curves.
Acknowledgments
We thank S. Lin, C. Zhang, and all members of the Xiao and Liu laboratories for discussion and technical assistance. We thank L. Hong, X. Sun, X. Ma, C. Zhang, M. Zhu, C. Xie, L. Yao, C. Wu at the Core Facility of Biomedical Sciences, Xiamen University and S. Wu at the Xiamen University Laboratory Animal Center for technical assistance.
Funding: This study was supported by the National Natural Science Foundation of China (31770955 and 31570883 to N.X.), 1000 Young Talents Program of China (to N.X.), and the Fundamental Research Funds for the Central Universities of China-Xiamen University (20720150065 to N.X. and 20720190101 to Q.L.).
Author contributions: X.Z. and K.L. designed and executed the experiments, analyzed the data, and prepared the manuscript. H.F. and Q.Z. analyzed high-throughput sequencing data under the supervision of Q.L. X.G., F.L., S.Z., X.W., and Y.N. performed the experiments. Y.H., S.-H.L., W.-H.L., and C.X. provided materials and study advice. N.X. conceived the study, designed the experiments, analyzed the data, and wrote the manuscript with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: ATAC-seq, CUT&tag, RNA-seq, and mRRBS datasets have been deposited in the Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo/query/acc.cgi) with accession numbers GSE176369, GSE176368, GSE176310, GSE182519, respectively. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Kaech S. M., Cui W., Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Zander R., Khatun A., Schauder D. M., Cui W., Transcriptional and epigenetic regulation of effector and memory CD8 T cell differentiation. Front. Immunol. 9, 2826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaech S. M., Tan J. T., Wherry E. J., Konieczny B. T., Surh C. D., Ahmed R., Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Joshi N. S., Cui W., Chandele A., Lee H. K., Urso D. R., Hagman J., Gapin L., Kaech S. M., Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best J. A., Blair D. A., Knell J., Yang E., Mayya V., Doedens A., Dustin M. L., Goldrath A. W.; The Immunological Genome Project Consortium , Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat. Immunol. 14, 404–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang J. T., Wherry E. J., Goldrath A. W., Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 15, 1104–1115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu B., Zhang K., Milner J. J., Toma C., Chen R., Scott-Browne J. P., Pereira R. M., Crotty S., Chang J. T., Pipkin M. E., Wang W., Goldrath A. W., Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russ B. E., Olshanksy M., Smallwood H. S., Li J., Denton A. E., Prier J. E., Stock A. T., Croom H. A., Cullen J. G., Nguyen M. L. T., Rowe S., Olson M. R., Finkelstein D. B., Kelso A., Thomas P. G., Speed T. P., Rao S., Turner S. J., Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8+ T cell differentiation. Immunity 41, 853–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henning A. N., Roychoudhuri R., Restifo N. P., Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 18, 340–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray S. M., Amezquita R. A., Guan T., Kleinstein S. H., Kaech S. M., Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity 46, 596–608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pace L., Goudot C., Zueva E., Gueguen P., Burgdorf N., Waterfall J. J., Quivy J. P., Almouzni G., Amigorena S., The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359, 177–186 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Ladle B. H., Li K. P., Phillips M. J., Pucsek A. B., Haile A., Powell J. D., Jaffee E. M., Hildeman D. A., Gamper C. J., De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc. Natl. Acad. Sci. U.S.A. 113, 10631–10636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L., Romero P., Metabolic control of CD8+ T cell fate decisions and antitumor immunity. Trends Mol. Med. 24, 30–48 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Ma E. H., Verway M. J., Johnson R. M., Roy D. G., Steadman M., Hayes S., Williams K. S., Sheldon R. D., Samborska B., Kosinski P. A., Kim H., Griss T., Faubert B., Condotta S. A., Krawczyk C. M., DeBerardinis R. J., Stewart K. M., Richer M. J., Chubukov V., Roddy T. P., Jones R. G., Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity 51, 856–870.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Levine L. S., Hiam-Galvez K. J., Marquez D. M., Tenvooren I., Madden M. Z., Contreras D. C., Dahunsi D. O., Irish J. M., Oluwole O. O., Rathmell J. C., Spitzer M. H., Single-cell analysis by mass cytometry reveals metabolic states of early-activated CD8+ T cells during the primary immune response. Immunity 54, 829–844.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng M., Yin N., Chhangawala S., Xu K., Leslie C. S., Li M. O., Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Windt G. J. W., Everts B., Chang C.-H., Curtis J. D., Freitas T. C., Amiel E., Pearce E. J., Pearce E. L., Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Sullivan D., van der Windt G. J. W., Huang S. C.-C., Curtis J. D., Chang C.-H., Buck M. D., Qiu J., Smith A. M., Lam W. Y., DiPlato L. M., Hsu F.-F., Birnbaum M. J., Pearce E. J., Pearce E. L., Memory CD8+ T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui G., Staron M. M., Gray S. M., Ho P. C., Amezquita R. A., Wu J., Kaech S. M., IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell 161, 750–761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan Y., Tian T., Park C. O., Lofftus S. Y., Mei S., Liu X., Luo C., O’Malley J. T., Gehad A., Teague J. E., Divito S. J., Fuhlbrigge R., Puigserver P., Krueger J. G., Hotamisligil G. S., Clark R. A., Kupper T. S., Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krainer A. R., Mayeda A., Kozak D., Binns G., Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66, 383–394 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Jiang J., Zhang Y., Krainer A. R., Xu R. M., Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl. Acad. Sci. U.S.A. 96, 3572–3577 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghebrehiwet B., Geisbrecht B. V., Xu X., Savitt A. G., Peerschke E. I. B., The C1q receptors: Focus on gC1qR/p33 (C1qBP, p32, HABP-1)1. Semin. Immunol. 45, 101338 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Fogal V., Richardson A. D., Karmali P. P., Scheffler I. E., Smith J. W., Ruoslahti E., Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol. Cell. Biol. 30, 1303–1318 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yagi M., Uchiumi T., Takazaki S., Okuno B., Nomura M., Yoshida S., Kanki T., Kang D., p32/gC1qR is indispensable for fetal development and mitochondrial translation: Importance of its RNA-binding ability. Nucleic Acids Res. 40, 9717–9737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotoh K., Morisaki T., Setoyama D., Sasaki K., Yagi M., Igami K., Mizuguchi S., Uchiumi T., Fukui Y., Kang D., Mitochondrial p32/C1qbp is a critical regulator of dendritic cell metabolism and maturation. Cell Rep. 25, 1800–1815.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Geltink R. I. K., Kyle R. L., Pearce E. L., Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pipkin M. E., Sacks J. A., Cruz-Guilloty F., Lichtenheld M. G., Bevan M. J., Rao A., Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32, 79–90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sukumar M., Liu J., Mehta G. U., Patel S. J., Roychoudhuri R., Crompton J. G., Klebanoff C. A., Ji Y., Li P., Yu Z., Whitehill G. D., Clever D., Eil R. L., Palmer D. C., Mitra S., Rao M., Keyvanfar K., Schrump D. S., Wang E., Marincola F. M., Gattinoni L., Leonard W. J., Muranski P., Finkel T., Restifo N. P., Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 23, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sena L. A., Li S., Jairaman A., Prakriya M., Ezponda T., Hildeman D. A., Wang C. R., Schumacker P. T., Licht J. D., Perlman H., Bryce P. J., Chandel N. S., Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chisolm D. A., Weinmann A. S., Connections between metabolism and epigenetics in programming cellular differentiation. Annu. Rev. Immunol. 36, 221–246 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Xiao M., Yang H., Xu W., Ma S., Lin H., Zhu H., Liu L., Liu Y., Yang C., Xu Y., Zhao S., Ye D., Xiong Y., Guan K. L., Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S. H., Ito S., Yang C., Wang P., Xiao M. T., Liu L. X., Jiang W. Q., Liu J., Zhang J. Y., Wang B., Frye S., Zhang Y., Xu Y. H., Lei Q. Y., Guan K. L., Zhao S. M., Xiong Y., Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anso E., Weinberg S. E., Diebold L. P., Thompson B. J., Malinge S., Schumacker P. T., Liu X., Zhang Y., Shao Z., Steadman M., Marsh K. M., Xu J., Crispino J. D., Chandel N. S., The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 19, 614–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan L. B., Gui D. Y., Hosios A. M., Bush L. N., Freinkman E., Vander Heiden M. G., Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birsoy K., Wang T., Chen W. W., Freinkman E., Abu-Remaileh M., Sabatini D. M., An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaya-Okur H. S., Wu S. J., Codomo C. A., Pledger E. S., Bryson T. D., Henikoff J. G., Ahmad K., Henikoff S., CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott-Browne J. P., Lopez-Moyado I. F., Trifari S., Wong V., Chavez L., Rao A., Pereira R. M., Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45, 1327–1340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tyrakis P. A., Palazon A., Macias D., Lee K. L., Phan A. T., Velica P., You J., Chia G. S., Sim J., Doedens A., Abelanet A., Evans C. E., Griffiths J. R., Poellinger L., Goldrath A. W., Johnson R. S., S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu J., Villa M., Sanin D. E., Buck M. D., O’Sullivan D., Ching R., Matsushita M., Grzes K. M., Winkler F., Chang C.-H., Curtis J. D., Kyle R. L., Van Teijlingen Bakker N., Corrado M., Haessler F., Alfei F., Edwards-Hicks J., Maggi L. B. Jr., Zehn D., Egawa T., Bengsch B., Klein Geltink R. I., Jenuwein T., Pearce E. J., Pearce E. L., Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C., Zhu B., Son Y. M., Wang Z., Jiang L., Xiang M., Ye Z., Beckermann K. E., Wu Y., Jenkins J. W., Siska P. J., Vincent B. G., Prakash Y. S., Peikert T., Edelson B. T., Taneja R., Kaplan M. H., Rathmell J. C., Dong H., Hitosugi T., Sun J., The transcription factor Bhlhe40 programs mitochondrial regulation of resident CD8+ T cell fitness and functionality. Immunity 51, 491–507.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghebrehiwet B., Lim B. L., Peerschke E. I., Willis A. C., Reid K. B., Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular "heads" of C1q. J. Exp. Med. 179, 1809–1821 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling G. S., Crawford G., Buang N., Bartok I., Tian K., Thielens N. M., Bally I., Harker J. A., Ashton-Rickardt P. G., Rutschmann S., Strid J., Botto M., C1q restrains autoimmunity and viral infection by regulating CD8+ T cell metabolism. Science 360, 558–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu K., Yin N., Peng M., Stamatiades E. G., Shyu A., Li P., Zhang X., Do M. H., Wang Z., Capistrano K. J., Chou C., Levine A. G., Rudensky A. Y., Li M. O., Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371, 405–410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarasenko T. N., Pacheco S. E., Koenig M. K., Gomez-Rodriguez J., Kapnick S. M., Diaz F., Zerfas P. M., Barca E., Sudderth J., DeBerardinis R. J., Covian R., Balaban R. S., DiMauro S., McGuire P. J., Cytochrome c oxidase activity is a metabolic checkpoint that regulates cell fate decisions during T cell activation and differentiation. Cell Metab. 25, 1254–1268.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan H., Yang K., Li Y., Shaw T. I., Wang Y., Blanco D. B., Wang X., Cho J. H., Wang H., Rankin S., Guy C., Peng J., Chi H., Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sciacovelli M., Goncalves E., Johnson T. I., Zecchini V. R., da Costa A. S., Gaude E., Drubbel A. V., Theobald S. J., Abbo S. R., Tran M. G., Rajeeve V., Cardaci S., Foster S., Yun H., Cutillas P., Warren A., Gnanapragasam V., Gottlieb E., Franze K., Huntly B., Maher E. R., Maxwell P. H., Saez-Rodriguez J., Frezza C., Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 537, 544–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arts R. J. W., Novakovic B., Ter Horst R., Carvalho A., Bekkering S., Lachmandas E., Rodrigues F., Silvestre R., Cheng S.-C., Wang S.-Y., Habibi E., Goncalves L. G., Mesquita I., Cunha C., van Laarhoven A., van de Veerdonk F. L., Williams D. L., van der Meer J. W. M., Logie C., O’Neill L. A., Dinarello C. A., Riksen N. P., van Crevel R., Clish C., Notebaart R. A., Joosten L. A. B., Stunnenberg H. G., Xavier R. J., Netea M. G., Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 24, 807–819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kornberg M. D., Bhargava P., Kim P. M., Putluri V., Snowman A. M., Putluri N., Calabresi P. A., Snyder S. H., Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer C. M., Crabtree-Hartman E. C., Lehmann-Horn K., Cree B. A. C., Zamvil S. S., Reduction of CD8+ T lymphocytes in multiple sclerosis patients treated with dimethyl fumarate. Neurol. Neuroimmunol. Neuroinflamm. 2, e76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 and S2