Abstract
Background
As vaccines against SARS-CoV-2 are now being rolled out, a better understanding of immunity to the virus, whether from infection, or passive or active immunisation, and the durability of this protection is required. This will benefit from the ability to measure antibody-based protection to SARS-CoV-2, ideally with rapid turnaround and without the need for laboratory-based testing.
Methods
We have developed a lateral flow POC test that can measure levels of RBD-ACE2 neutralising antibody (NAb) from whole blood, with a result that can be determined by eye or quantitatively on a small instrument. We compared our lateral flow test with the gold-standard microneutralisation assay, using samples from convalescent and vaccinated donors, as well as immunised macaques.
Findings
We show a high correlation between our lateral flow test with conventional neutralisation and that this test is applicable with animal samples. We also show that this assay is readily adaptable to test for protection to newly emerging SARS-CoV-2 variants, including the beta variant which revealed a marked reduction in NAb activity. Lastly, using a cohort of vaccinated humans, we demonstrate that our whole-blood test correlates closely with microneutralisation assay data (specificity 100% and sensitivity 96% at a microneutralisation cutoff of 1:40) and that fingerprick whole blood samples are sufficient for this test.
Interpretation
Taken together, the COVID-19 NAb-testTM device described here provides a rapid readout of NAb based protection to SARS-CoV-2 at the point of care.
Funding
Support was received from the Victorian Operational Infrastructure Support Program and the Australian Government Department of Health. This work was supported by grants from the Department of Health and Human Services of the Victorian State Government; the ARC (CE140100011, CE140100036), the NHMRC (1113293, 2002317 and 1116530), and Medical Research Future Fund Awards (2005544, 2002073, 2002132). Individual researchers were supported by an NHMRC Emerging Leadership Level 1 Investigator Grants (1194036), NHMRC APPRISE Research Fellowship (1116530), NHMRC Leadership Investigator Grant (1173871), NHMRC Principal Research Fellowship (1137285), NHMRC Investigator Grants (1177174 and 1174555) and NHMRC Senior Principal Research Fellowships (1117766 and 1136322). Grateful support was also received from the A2 Milk Company and the Jack Ma Foundation.
Keywords: SARS-CoV-2, Neturalising antibodies, Lateral flow assay, Point of care test
Research in context.
Evidence before this study
Widespread deployment of efficacious COVID-19 vaccines has contributed to much lower disease rates in many wealthy countries, but most countries and especially low and middle-income countries (LMICs) are facing new waves of SARS-CoV-2 infection driven by factors including limited access to vaccines, variable efficacy for some of the available vaccines, and the emergence of viral variants of concern (VOCs). There are a number of laboratory-based methods for measuring SARS-CoV-2 immunity in individuals and populations, but there is an unmet need for methods that can be used at the point of care to support improved control efforts including quarantine, vaccination, and boosting as required. We searched PubMed up to 11 October 2021 using the search terms (“SARS-CoV-2” OR “COVID-19”) AND ("neutralizing antibody") AND (“lateral flow” OR “point of care”), and also manually searched references in the relevant papers to identify preprints. Twenty-nine published studies and four preprints were identified in this search, but none of the studies report a deployable, visual point of care test for measuring protective levels of SARS-CoV-2 protection.
Added value of this study
We developed a lateral flow, point of care device to measure SARS-CoV-2 neutralising antibodies, in contrast to other point of care tests that measure SARS-CoV-2 binding antibodies. The COVID-19 NAb-testTM uses the principle of inhibition of virus-receptor binding by neutralising antibodies in serum (similar to the C-PassTM laboratory-based test for neutralising antibodies). A visual semi-quantitative readout of neutralising antibody level is assessed against a pre-determined reference level (similar to interpretation for the VisitectTM CD4 T-cell tests). The device is suitable for use with whole blood including fingerprick samples and can be readily modified to incorporate VOCs as required. It is also capable of detecting neutralising antibodies in non-human samples, may prove useful in the context of assessing zoonotic SARS-CoV-2 infections.
Implications of all the available evidence
COVID-19 vaccination programs are currently focused on expanding coverage in eligible populations as quickly as possible within the constraint of vaccine availability, without adjusting for prior immunity or exposure, and largely without validation of individual or population-based immune responses to the different vaccines. With SARS-CoV-2 becoming endemic, long-term control programs including protection of high-risk individuals and health care workers, and freedom of movement within and between countries, will benefit from availability of standardised methods for validating immunity versus susceptibility, including that to new and/or prevalent VOCs. Our test provides the first point-of-care tool for measuring COVID-19 surrogate neutralising antibodies and provides a new option for immunity screening that can support enhanced COVID-19 vaccination and control programs, particularly in time critical settings, while improving resource utilisation, especially in LMICs where laboratory-based testing is difficult or impossible for many to access.
Alt-text: Unlabelled box
Introduction
Despite rapid progress, it is unlikely that COVID-19 will be eradicated in the near future and at best, the hope is that COVID-19 infections can be brought to a level of control where life can return to a “post-COVID” normality. A key part of this solution will be widespread vaccination, together in some circumstances, with testing to determine who is protected, whether through prior infection or vaccination, and how durable that protection is, both on a population level and on an individual basis. Developing timely and cost-effective tests to measure this protection will become a vital tool in the fight against COVID-19.
Antibody responses to viral infections and/or vaccines are typically determined using serological tests such as ELISAs or point-of-care (POC) assays, including lateral flow tests [1,2] and interferometry [3]. These assays measure virus-specific antibody responses, that include immunoglobulin subclasses such as IgG, IgM or IgA. Such assays are widely available for the detection of anti-SARS-CoV-2 antibodies, with variable levels of specificity and sensitivity [4]. For some viruses such as hepatitis A and hepatitis B, where the protective epitopes are highly immunodominant and demonstrate minimal variation, total antibody tests provide a satisfactory correlate of immunity. However, for SARS-CoV-2 and many other viruses, the combination of adaptive mutations to enhance receptor binding affinity, immune selection and at least moderate error rates during replication can lead to the rapid selection of variants that may escape the antibody response (akin to “antigenic drift” in influenza viruses). This is exemplified by the recent emergence of viral variants of concern (VOC) such as the B.1.351 lineage (beta variant) first detected in South Africa. Some of these variants are less efficiently neutralised by antibodies induced by prior infections and by vaccines [5]. This may require not only the development of modified vaccines, but also the development of serological tests that can detect and quantify the dominant neutralising antibodies (NAb) responses to newly emerging strains.
Neutralising antibodies are emerging as a strong correlate of protection [6,7] but are only a subset of the total polyclonal response. The dominant target of NAb in SARS-CoV-2 is the Receptor Binding Domain (RBD), located at the tip of the S1 domain of the spike (S) protein, which is responsible for the high affinity binding of the virus to its receptor Angiotensin-Converting Enzyme-2 (ACE2) present on human epithelial and endothelial cells [8,9]. For this reason, antibodies against the RBD that block its interaction with ACE2 are the most direct mechanism for neutralising the virus and indeed, one study reported that at least 90% of all SARS-CoV-2 NAb in convalescent plasma were RBD reactive [8]. However, not all RBD-specific antibodies are capable of blocking the RBD-ACE2 interaction and neutralising virus infectivity. It is therefore important to determine not only whether an individual has antibodies against SARS-CoV-2 RBD, demonstrating a prior encounter with infectious virus or vaccination, but also whether they have NAb that are likely to protect from subsequent infection.
Plaque-reduction neutralisation and SARS-CoV-2 microneutralisation assays are the gold standards for measuring virus NAb [7]. These assays involve incubating SARS-CoV-2 with serial dilutions of serum and assessing residual infectivity in the sample. A major difficulty is that these assays require appropriate biocontainment laboratories, highly trained staff, and typically take several days for results [7]. Pseudovirus neutralisation assays are widely used alternatives that incorporate SARS-CoV-2 spike as the surface entry protein for a reporter virus that can be used in a BSL2/PC2 culture laboratory [10]. Another assay that has been developed for use in a PC2 laboratory is the plate-based ‘surrogate Virus Neutralisation Test’ (sVNT or C-PASS) that requires no cell culture [11]. This FDA-approved test relies on the ability of NAb to bind to RBD and prevent its interaction with ACE2, thereby, inhibiting the signal which is measured by a colorimetric change in a cell-free ELISA-based assay. As the ability to block RBD-ACE2 binding is the major mechanism for neutralisation, the competitive ELISA is a surrogate for determining NAb activity [12]. This assay has been rapidly adopted around the world as a more reliable test for neutralisation than mere detection of anti-RBD antibodies, and several versions of this type of assay have now been published [13], [14], [15]. While the ability to run the assay in a PC2 laboratory in just a few hours is a major advantage compared to microneutralisation assays, nonetheless, it still has a number of drawbacks. In particular, it requires a venous blood sample and preparation of plasma, and transport to a dedicated laboratory with trained laboratory staff, which means long delays between testing and results. This delay may be particularly problematic in isolated areas or low-middle income countries where access to laboratories is often limited or impossible and reproducibility between labs may be an added challenge. Early prototype point-of-care tests that use the same principle of RBD-ACE2 inhibition to measure NAb have recently been published [16,17] or shared on a non-peer reviewed pre-print server [18] but those tests appear to be reliable only for relatively high levels of antibodies, and/or monoclonal antibodies, and have not demonstrated the equivalence of plasma and whole blood that are essential for use at POC and are therefore not deployable at this stage. Thus, we sought to develop a whole-blood POC test that provides comparable results to sVNT or microneutralisation assays, in a rapid, easy to use format.
Our study describes the development of a prototype lateral flow POC assay, called the COVID-19 NAb-testTM, that uses a similar principle to the sVNT/C-PASS assay, except signal is detected on a nitrocellulose test strip. The assay is completed within 20-30 minutes and is compatible with the commercially available Axxin AX-2XS reader, providing the potential for fully quantitative results and linkage to laboratory information systems. We test the ability of NAb from convalescent or vaccinee samples, versus seasonal hCoV samples, to neutralise the RBD-ACE2 interaction in our assay and compared this to the neutralisation observed in the gold-standard microneutralisation assay. We also examine the utility of our test with samples from non-human primates, as well as vaccinee whole blood samples, and finally, whether alternative RBDs (such as RBDs from VOCs) may be used as required. The COVID-19 NAb-testTM provides a potential solution for POC testing of COVID-19 antibody neutralisation that can be applied in many settings in both high- and low-middle income countries.
Methods
Study Design
The prototype COVID-19 NAb-test™ went through iterative development at a time when there were very low rates of community transmission and COVID-19 vaccination in Australia. We therefore tested the ability of successive versions of the assay to detect SARS-CoV-2 NAb using COVID+ convalescent patient or vaccinee samples and COVID- controls, as well as inoculated macaque samples, drawn from historical studies where there was ethical approval for this secondary use. Prototype test results were compared to results from gold-standard viral neutralisation assays to assess the correlation between the COVID-19 NAb-test™ and microneutralisation assay (Fig. 1).
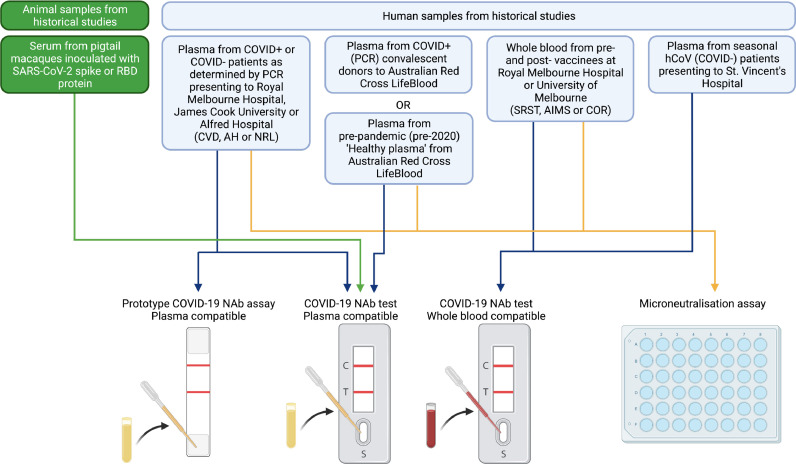
Fig. 1.
Study flow diagram. Historical samples likely to contain SARS-CoV-2 neutralising antibodies, and negative controls, from human participants or experimental macaques, were used to evaluate the various prototype stages during development of a novel lateral flow based neutralising anti-SARS-CoV-2 antibody assay. CVD, AH, NRL, SRST, AIMS and COR are historical sample sources (described in Table S1). Created with BioRender.com.
Ethics
Human experimental work was conducted according to the Declaration of Helsinki Principles and the Australian National Health and Medical Research Council Code of Practice. All participants provided written informed consent prior to the study. The study was approved by the Alfred Hospital (#280/14), The University of Melbourne (#2056761, #2056689, #1442952, #1955465, #202121198153983 and #2020-20782-12450-1), Melbourne Health (HREC/68355/MH-2020, HREC/66341/MH-2020) and James Cook University (H7886) Human Research Ethics Committees. The vaccinee samples are collected under ethics number 2021-21198-15398-3 through the University of Melbourne. Seasonal coronavirus samples collected under approval of the St Vincent's Hospital Research Ethics Committee (2020/ETH01429).
Animal studies and related experimental procedures were approved by the University of Melbourne Animal Ethics Committee (no. 1714193, no. 1914874). Macaque studies and related experimental procedures were approved by the Monash University Animal Ethics Committee (no. 23997).
Participants
All human specimen materials were considered infectious and hazardous and handled using standard biosafety procedures. Plasma: Blood specimen was collected in a lavender, blue or green top Vacutainer® collection tube containing EDTA, citrate or sodium heparin (respectively), using venepuncture. Plasma was separated by centrifugation. Plasma was then carefully withdrawn and decanted into a new pre-labelled tube. Serum: Blood specimen was collected in a red top Vacutainer® collection tube without the presence of coagulants. Blood was allowed to clot, and the serum separated using centrifugation. The serum was then carefully withdrawn and decanted into a new pre-labelled tube. Specimens were frozen at -20°C for longer term storage and tested as soon as possible after thawing (up to 3 days). For frozen samples, more than 4 freeze-thaw cycles were avoided. Prior to testing, frozen specimens were brought to room temperature slowly and gently mixed. Samples containing visible particulate matter was clarified by centrifugation before testing. Samples demonstrating gross lipaemia, gross haemolysis or turbidity were discarded to avoid interference on result interpretation. Whole blood: Samples of blood were obtained by venepuncture (stored for up to 24 hours at 2-8°C as required), or fingerprick (used immediately).
Plasma samples including COVID-19 acute/convalescent and vaccinated samples, were obtained from: The Alfred Hospital, University of Melbourne, James Cook University, Australian Red Cross Lifeblood, The Royal Melbourne Hospital. Patient samples who were PCR positive for human common cold coronaviruses (hCoV-NL63, O229E, OC43 or HKU1) and PCR negative for SARS-CoV-2 were obtained from St. Vincent's Hospital and have been published previously on a pre-print server [19]. Some samples obtained from The Royal Melbourne Hospital Department of Microbiology were part of a previously published patient cohort [4], patients C1, C2 from the Burnet Institute and COVID-negative blood and plasma samples were provided by the Australian Red Cross Lifeblood from “healthy” pre-pandemic (pre-2020) donors. Samples were collected by venepuncture into heparin tubes. Whole blood was provided from those tubes, and plasma was collected after centrifugation.
Due to low rates of both community transmission of SARS-CoV-2 and COVID-19 vaccination in Australia during the period of this work, historical patient samples were included on an ad hoc basis, based on availability, with no exclusion criteria. Convalescent patients were included in the positive cohort following a positive COVID PCR test and healthy controls were included on the basis of a negative COVID PCR test, or obtained from samples collected before the onset of the COVID-19 pandemic (pre-COVID controls). Vaccinee samples were stratified on the basis of 0, 1 or 2 doses. All un-vaccinated controls reported no prior positive tests for SARS-CoV-2 infection. Patient and control samples were not matched. Participant data is summarised in Table S1.
Macaque samples
Macaque serum samples were derived from stored samples from macaques that had been immunised with SARS-CoV-2 spike or RBD proteins, as described in detail in a recent study [20]. Macaque data is summarised in Table S2.
Reagents
Double stranded DNA encoding the truncated SARS-CoV-2 RBD (N334-P527) of the Hu-1 reference strain (NC_045512.2) with C-terminal AVI-tag was synthesised (IDT) and cloned into the pHLSec expression vector. The protein was expression by transient transfection of Expi293F cells (RRID:CVCL_D615; ThermoFisher Scientific) using Expifectamine transfection kits (Cat. No. A14525; ThermoFisher Scientific) [21]. Expressions were harvested on day 6, and the protein purified by Ni-NTA (Pierce). SARS-CoV-2 RBD-avi protein was then enzymatically biotinylated using recombinant BirA protein. Biotinylated SARS-CoV-2 RBD-biotin was further purified by size-exclusion chromatography using a HiLoad Superdex 75 16/600 (Cytiva). Biotinylation was found to be at least 95% efficient by streptavidin gel-shift.
Variants (N439K, S477N/I, and N501Y, E484K, K417N triple (beta) mutant) of SARS-CoV-2 RBD were expressed in Expi293F cells stably transfected with BirA ligase at 34°C and supplemented with L-Biotin. Four days after transfection tissue culture supernatants were clarified and target proteins were purified by IMAC using Talon metal affinity resin (Clontech Laboratories) following the manufacturer's recommendations. The eluted proteins were subject to gel filtration using a Superdex 200 16/600 column (GE Healthcare) with PBS as the liquid phase. Fractions corresponding to monomeric RBD were pooled and concentrated in Amicon Ultra 30kDa devices (Merck) prior to use.
Double stranded DNA encoding the truncated extracellular domain of human ACE2 (aa Ser19-Ser740, accession BAB40370) was fused by a Gly-Gly-Gly-Gly-Ser linking sequence to human IgG1-Fc region (Thr223-Lys447; Eu numbering, Accession no. AXN93652.1), was expressed in Expi293F cells as above, and purified using Protein A sepharose (Cytiva) followed by size-exclusion chromatography using a HiLoad Superdex 200 16/600 (Cytiva).
Anti-RBD mAb [22,23] were kindly provided by Dr Adam Wheatley; these antibodies have previously been validated by binding to anti-SARS-CoV-2 RBD and anti-SARS-CoV-2 SPIKE protein by ELISA; ELISA to other human corona viruses (no cross-reactivity detected); as well as live virus neutralisation assays.
Anti-biotin-gold (40 nm, Cat. No. MAB40) was purchased from BBISolutions, UK; anti-glycophorin A (Epiclone; Cat. No. 00992311) from Seqiris, Australia.
Equipment
The Axxin AX-2XS instrument reader was obtained from Axxin, Melbourne, Australia.
Production of Lateral flow device
The device cassette consists of a plastic housing (Nanjing BioPoint Diagnostics, PR China) with loading wells and a window to read results. Within the cassette is a nitrocellulose membrane strip (Sartorius, Germany), with, at one end, a sample loading pad, a mixing pad containing: 1) biotinylated RBD protein complexed with colloidal gold-conjugated anti-biotin antibody (BBI Solutions, UK); 2) colloidal gold-conjugated chicken IgY as a control and reference standard. The nitrocellulose membrane strip has three stripes, the first of which is a recombinant chimeric protein consisting of the extracellular domain of human ACE2 fused to human-IgG1-Fc domain line (test line 1), a control/reference line (Ref Line) coated with anti-chicken IgY antibody (BBI Solutions, UK) and a total anti-RBD antibody line (test Line 2) coated with recombinant RBD protein.
Test methods
NAb Lateral Flow assay
The test specimen (15 µl plasma) is dispensed into the sample well of the test cassette, and three drops of running buffer (phosphate buffered saline pH 7.4, 0.5% Tween20, 0.05% Sodium azide) are added to well B of the test cassette. For the test modified for whole blood, the test specimen (30 µl) and running buffer are added to the same well. In both cases, the specimen mixes with the colloidal gold RBD conjugates upon sample addition and during migration by capillary action in the mixing pad, and then migrate along the nitrocellulose membrane. Anti-SARS-CoV-2 RBD antibodies, if present in the specimen, will bind to the RBD-biotin-Au conjugates. If no antibody is present, or if the antibodies bind to RBD protein but without capacity to interfere with ACE2 binding (i.e., lack of neutralisation), then the maximum amount of RBD-gold will bind to the ACE2 line (test line 1). If NAb are present in the specimen, less of the immunocomplex will bind to the ACE2 protein. In either case, as the sample continues to flow, the chicken IgY-gold will bind to the anti-chicken IgY line (control/reference line), and next, additional SARS-CoV-2 conjugate will flow to the RBD line (test line 2), where binding of antibody-antigen complexes to the same or related antigen on the test strip will indicate the presence of antibodies to SARS-CoV-2 RBD that may or may not be neutralising because ACE2 is not involved in this interaction. This indicates a SARS-CoV-2 RBD antibody positive test result.
The intensity of the control/reference line can be adjusted by varying the amount of anti-chicken IgY on the nitrocellulose and/or the amount of chicken IgY-gold at the time of manufacture, in order to reflect the best estimates for protective levels of antibody against SARS-CoV-2 infection. The RBD-biotin can be expressed using any variant sequence of RBD and is then titrated to determine an equivalent level of binding for the inhibition assay.
Interpretation of test result
-
a)
Valid assay In addition to the presence of the reference line, if only test line 1 is developed and is more intense than the reference line, which is calibrated to reflect a defined level of intensity in test line 1, this result indicates levels of neutralising anti-RBD antibody in the sample are below the amount deemed to be neutralising based on the reference line. If test line 1 is less intense than the reference line, or very faint or invisible, this suggests that there is an adequate level of neutralising antibody present in the sample. Whilst the reference line can be adjusted based on best clinical data, 50% of the maximum test line 1 represents a reasonable change in intensity to be determined with confidence by eye. Test line 2 may develop indicating the presence of anti-RBD antibody, indicating prior exposure to SARS-CoV-2 infection or RBD-containing vaccine, but this is not necessarily neutralising. These readings can be performed at a semi-quantitative level by eye as greater than or less than the reference line, as used in the visual interpretation of the Visitect® CD4 T-cell tests, or quantitatively with a wide dynamic range using an instrument such as the Axxin AX-2XS instrument.
-
b)
Invalid assay If the C Line does not develop, the assay is invalid regardless of color development of the test line 1 and test line 2 as indicated below. In such cases, the assay should be repeated with a new device.
Microneutralisation assay
Microneutralisation assays represent the gold-standard assessment of SARS-CoV-2 antibody neutralisation, and were performed as previously described [24,25]. SARS-CoV-2 isolate CoV/Australia/VIC01/2020 [26] was passaged in Vero cells and stored at −80°C. Plasma was heat inactivated at 56°C for 30 min, then serially diluted 1:20 to 1:10,240 before the addition of 100 TCID50 of SARS-CoV-2 in MEM/0.5% BSA and incubation at room temperature for 1 h. Residual virus infectivity in the plasma/virus mixtures was assessed in quadruplicate wells of Vero cells incubated in serum-free media containing 1 μg/ml of Tosyl phenylalanyl chloromethyl ketone (TPCK) trypsin at 37°C and 5% CO2; viral cytopathic effect was read on day 5. The NAb titre was calculated using the Reed–Muench method, as previously described [27]. A microneutralisation titre of 40 has been considered a reasonable cut-off for protection in previous studies [6].
Analysis and Statistics
To compare the accuracy of the index test (50% inhibition on the COVID-19 NAb-test™) to the reference standard (microneutralisation titre of > 40), sensitivity and specificity were calculated from data derived from the final, whole blood compatible iteration of the test. Samples missing microneutralisation data or anti-RBD antibody titre were excluded from regression analyses. Regression lines were plotted using a semi-log non-linear fit, or a saturation curve with Hill-slope, as appropriate and plotted with 95%CI bands. R2 goodness-of-fit are displayed on graphs. Data was found to be neither normal nor log-normally distributed by Anderson-Darling, D'Agostino & Pearson, Shapiro-Wilk, Kolmogorov-Smirnov tests, and therefore non-parametric significance tests were used. For comparison between multiple groups, Kruskal-Wallis one-way ANOVA with Dunn's multiple comparison test was employed. Wilcoxon signed-rank test was used to compare paired data, and Mann-Whitney test to compare unpaired data. Infection and/or vaccination status of donors was available to assessors of lateral flow and microneutralisation tests, however, where experiments were designed to compare results between these tests, results from lateral flow test were not known to assessors of microneutralisation assays, and vice-versa. Sample size for testing the plasma compatable prototype was determined from our previous experience developing the Visitect® CD4 T-cell tests [28]. Sample size for proof-of-principle testing the whole blood compatible prototype was determined with regard to limited access to vaccinee samples during the study period. All statistical analyses were performed with GraphPad Prism 9.0.
Data handling
The data generated on the Axxin AX-2XS reader were exported (to a USB drive) and viewed in Microsoft Excel 365. Where necessary, raw band intensity data from the samples were converted to % inhibition in Excel [using the formula % inhibition = (1 - sample intensity/average intensity of COVID-19 negative samples) x100]. Data were then plotted in GraphPad Prism 9.0. Microneutralisation or anti-RBD antibody titres provided by collaborators in Excel spreadsheets were plotted against COVID-19 NAb-test™ data in Prism.
Role of Funders
Funders did not play any role in study design, data collection, data analyses, interpretation, or writing of this report.
Results
Development of a lateral flow assay to measure RBD-ACE2 inhibition
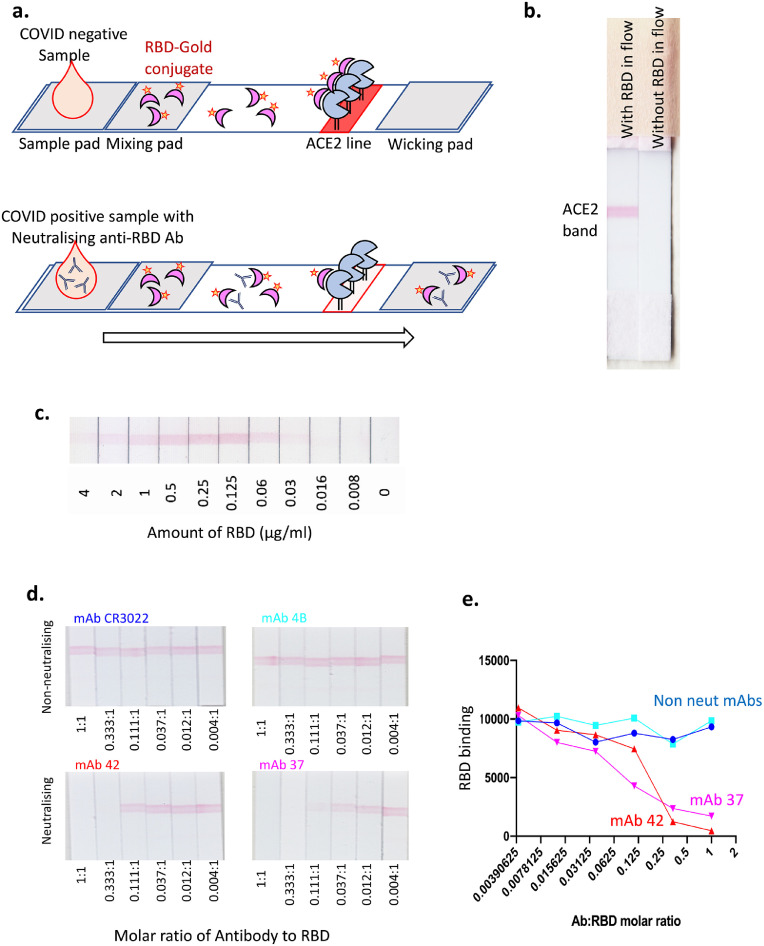
The sVNT assay is a competition ELISA assay to measure the presence of NAb in serum or plasma based on their ability to interfere with SARS-CoV-2 RBD binding to ACE2 [11]. Using the same principle of ACE2-RBD binding interference, we developed a rapid, lateral flow sVNT assay to measure NAb in biological samples at the point of care. We tested several different forms of RBD and ACE2 protein, with one or the other bound to the nitrocellulose strip that forms the stationary phase of the lateral flow assay. The approach ultimately adopted, because it provided the clearest signal and widest dynamic range, was to use a fusion protein of human ACE2 and human IgG1-Fc immobilised on the strip as the solid phase, and a complex of RBD-biotin bound to anti-biotin-colloidal gold (RBD-Au) as the mobile phase, passing over the immobilised ACE2-Fc through capillary action in the device flow (Fig. 2a). After initial exploratory experiments (Figure S1), we settled on ACE2-Fc at 2 mg/ml (0.5 µg of ACE2-Fc per 5 mm wide test strip) and detected a clear visible signal from binding of RBD-Au at 1 µg/ml RBD (Fig. 2b). Titration of the RBD-biotin before mixing with this fixed amount of anti-biotin gold, yielded a clear visual signal over a range of concentrations from 0.06-2 µg/ml (Fig. 2c), with evidence of a “prozone” effect at the highest RBD-biotin concentrations and a linear titration at lower concentrations, allowing us to identify the appropriate level of RBD-biotin that would be readily titratable upon inhibition by NAb. Importantly, the appropriate concentration of RBD-biotin must be determined for each batch of RBD-biotin to ensure assay performance. Using this format, we established an optimal concentration of 2 mg/ml ACE2-Fc to coat the assay strip and a maximum of 1 μg/ml RBD-biotin mixed with the anti-biotin gold. As a proof of principle, we tested the assay using four different anti-RBD mAb: two that were known to inhibit RBD-ACE2 binding [neutralising; clones 42 and 37 [22]] and two that were non-neutralising [clones CR3022 [23] and 4B]. These results provided a clear dose response curve showing near complete inhibition of the visual ACE2-RBD-Au signal with the neutralising mAb while the non-neutralising mAb had no clear impact on the strength of the signal (Fig. 2d). The intensities of the test lines were also quantitatively assessed on an Axxin AX-2X strip reader and a clear dose-dependent decline in signal strength was observed with increasing amounts of the neutralising mAb (Fig. 2e). Thus, we demonstrate a lateral flow test that is able to detect neutralising anti-RBD Ab levels by measuring the disruption of the ACE2-RBD interaction.
Fig. 2.
Development of a lateral flow assay to detect NAb. a. A schematic depicting our lateral flow COVID-19 NAb assay design. b. A photograph of two lateral flow strips side by side, with the left one showing a red band when RBD-Au has been included in the flow. c. Titration of RBD-Au run at a range of concentrations across the strips. d. and e. Pre-incubation of non-neutralising (CR3022, 4B) and neutralising (42, [37]) anti-RBD-specific monoclonal antibodies with RBD-Au at molar ratios as depicted. e. Graph represents intensity of RBD-Au interaction with immobilised Fc-ACE2 (red test bands) measured by Axxin strip reader.
Testing COVID-19 patient samples
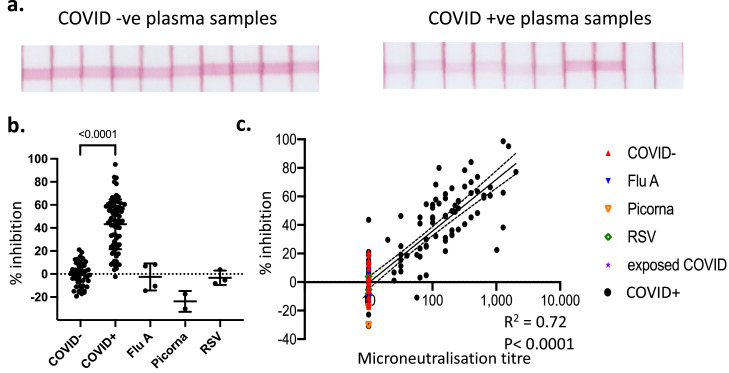
Having established that the assay can detect the presence of NAb, we tested 79 COVID-19 patient plasma samples and 47 COVID-19 negative control samples, as well as samples from patients with other viral infections (Fig. 3) [25,29]. As expected, the pre-COVID-19 healthy control samples showed minimal fluctuation in the intensity of the RBD-ACE2 signal, within a range of ±20% around the mean (Fig 3a). In contrast, COVID-19 patient samples showed a broad range of inhibition in the intensity of the RBD-ACE2 test line, with some samples almost completely inhibiting the signal (Fig. 3a). These samples were measured using the Axxin strip reader and the % inhibition within the COVID-19+ samples relative to the COVID-19- samples was calculated as: % inhibition = (1 - sample intensity/average intensity of COVID-19-ve samples) x100. Samples from patients with a confirmed COVID-19 diagnosis (COVID-19+) had a median inhibition of 47% (95%CI of 41 to 54%) compared to 0% (95%CI of -3.9 to 2.8%) for samples from COVID-19- patients (Fig. 3b). Samples taken from patients with other virus infections including RSV, influenza and picornaviruses all showed no inhibition greater than 20% compared to control COVID-19 negative samples (Fig. 3b). We plotted these inhibition values against the neutralising titres from the same samples determined using a SARS-CoV-2 microneutralisation assay, revealing a strong correlation with neutralisation (R2=0.72, p<0.0001; semi-log regression) (Fig. 3c). Collectively, our assay has the sensitivity and dynamic range to distinguish between strong, weak and absent NAb levels with good correlation to more labour intensive laboratory tests.
Fig. 3.
Testing the lateral flow assay on COVID- and COVID+ plasma samples. a. Ten representative COVID-19 -ve and COVID-19 +ve plasma samples. b. Tests of 47 COVID-19 -ve, 79 COVID-19 +ve, and 9 non-COVID virally infected individuals were converted from stripe intensity from each sample to a % inhibition based on the formula % inhibition = (1 – (sample intensity/average intensity of COVID-19-ve samples)) x100. Plotted as % inhibition on y axis. Mean±SD. P-value <0.0001, Kruskal-Wallis with Dunn's multiple comparison test. n = 79 (COVID+) / 47 (COVID-) / 9 (non-COVID virus infection) biological replicates. c. Readings from lateral flow strips were plotted against readings for same samples derived from virus microneutralisation assay. Semi-log regression with 95%CI (dashed lines).
Development and testing of a prototype assay for POC use
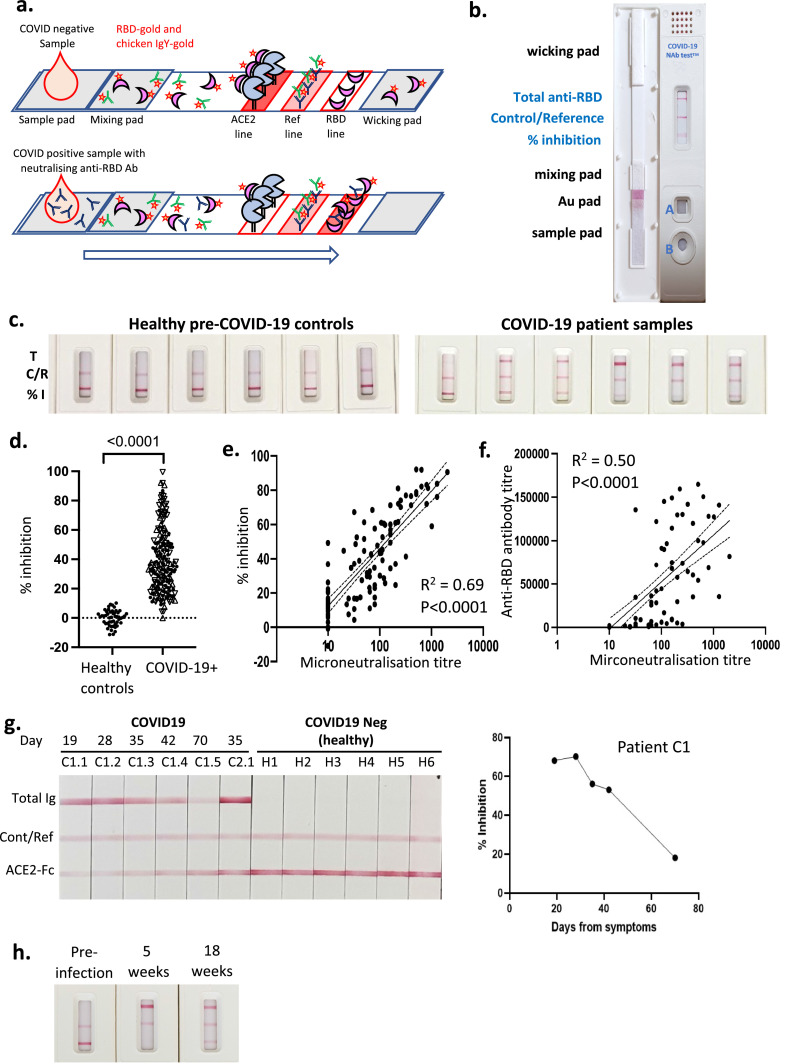
To facilitate intended use of the test in a POC setting, the assay strip configuration was modified to allow the use of a self-contained cartridge (Fig. 4). In this configuration, a small volume of plasma is added to well A, where the sample rehydrates the RBD-Au complex and begins to migrate into the mixing pad. Three drops of running buffer (buffered saline) are then added to well B, and the buffer chases the plasma and RBD-Au complex mixture along the length of the test strip within the closed cassette (Fig. 4a and b). The sample pad also contains a defined amount of gold-conjugated chicken IgY, which serves both as an assay running control and internal reference material. The sample then reaches the immobilised ACE2-Fc line, where the amount of RBD-gold complex binding will be inversely related to the amount of NAb in the sample. Two additional lines have been incorporated on these strips. The reference line is a stripe of anti-chicken IgY that binds the chicken IgY-gold, serving as a control to show that the test has run successfully and also providing a reference line for comparison to the ACE2-RBD-Fc test line for both semi-quantitative and quantitative measurement of NAb. For visual (semi-quantitative) estimation, the reference line represents the intensity associated with a predetermined amount of inhibition (eg. 50% as used in these examples). The intensity of the line can be adjusted at manufacture by varying the amount of anti-chicken IgY and/or chicken IgY-gold that are added, allowing a direct visual comparison between the test and reference lines to assess whether a sample has sufficient levels of NAb. This approach is similar to the visual reference lines incorporated in the POC Visitect® CD4 T-cell tests that have cutoffs of either 350 or 200 CD4 T-cells / µl (Omega Diagnostics, UK) [28]. For quantitative estimation (using the Axxin strip reader), the % inhibition for samples is calculated as: % inhibition = (1 – (test line intensity/[2x reference line intensity])) x100. The third line is striped with RBD protein, which forms a double-antigen sandwich with RBD-gold and anti-RBD antibody derived from the sample being tested. This line therefore indicates the presence of total anti-RBD antibody, regardless of whether or not it is neutralising and capable of blocking RBD-ACE2 interaction, and is analogous to many total antibody tests currently available for SARS-CoV-2. Our prototype assay was used to examine 60 samples from healthy pre-COVID controls and 200 samples from COVID-19 patients, examples of which are shown in (Fig. 4c). The data showed a high degree of consistency across the control samples and a wide range of inhibition from 0-95% in COVID-19 patient samples (Fig. 4d) calculated using the formula above. Samples from patients with a confirmed COVID-19 diagnosis (COVID-19+) had a median inhibtion of 35% (95%CI of 32 to 38%) compared to 1% (95%CI of -0.25 to 3.3%) for samples from healthy controls. When the data were plotted against NAb titre data from the same samples determined by microneutralisation assay, a clear correlation was observed (R2 = 0.69, P<0.0001; semi-log regression) (Fig. 4e). In contrast, when microneutralisation titres were plotted against total anti-RBD titres (on a subset of samples from Fig. 3e where this data was available) the correlation was much less clear (R2 = 0.5, P<0.0001; semi-log regression), consistent with the concept that total anti-RBD antibody titres are not necessarily reflective of NAb titres (Fig. 4f). Hence, these results provide a clear demonstration for the need for a rapid POC test that correlates closely with gold-standard lab-based neutralisation assays.
Fig. 4.
Development of a prototype test cassette for measuring Nab against SARS-CoV-2. a. and b. A schematic and a photograph depicting our prototype lateral flow assay cartridge design. c. 6 representative healthy pre-COVID-19 samples and 6 representative COVID-19 patient samples as run on the prototype lateral flow cartridge. d. Conversion of stripe intensity from each sample to a % inhibition based on the formula % inhibition = (1 - sample intensity/ average intensity of COVID-19-ve samples) x100. n = 200 (COVID-19+) / 60 (Healthy controls) biological replicates. P-value <0.0001 from Mann-Whitney test. e. Readings from lateral flow strips were plotted against readings for same samples derived from virus microneutralisation assay. Semi-log regression with 95%CI (dashed lines), n = 126 biological replicates. f. 93 of the samples from e. where anti-RBD antibody levels were available were plotted against microneutralisation titres for the same samples. Semi-log regression with 95%CI (dashed lines). g. Samples from an individual that were collected from 3 to 10 weeks. Six COVID-19 -ve samples were also run for comparison. Results from patient C1 are plotted as % inhibition vs days from symptom onset. h. Pre- and post-infection samples from a symptomatic patient at indicated timepoints.
Previous reports have suggested that NAb levels may decrease quite rapidly, within weeks to months of infection [30], [31], [32]. Accordingly, tests that can measure NAb may be useful to confirm adequate levels of NAb at regular intervals after infection or vaccination. We examined a series of longitudinal samples from a single subject (C1) beginning 3 weeks after PCR-confirmed diagnosis of infection with SARS-CoV-2 (Fig. 4g). Over the period of 3-10 weeks, the degree of ACE2-RBD inhibition shown in the test line can be visually observed to decline in the images of the strips, and using the Axxin reader it is shown to decline from ∼70% to 18%, in parallel with declining levels of total anti-RBD Ig. In contrast, a single sample (C2.1) from subject C2 shows high levels of total anti-RBD Ig but negligible (3%) ACE2-RBD inhibition (Fig. 4g). This suggests that this subject had limited NAb titres despite the presence of measurable total anti-RBD antibodies, although it is possible that this subject had other NAb specificities that do not target the RBD-ACE-2 interaction. The six healthy controls used in this experiment (H1-H6) showed no anti-RBD antibody and no detectable inhibition. Next, we examined serum samples from an individual with symptomatic, PCR-confirmed SARS-CoV-2 infection where a pre-infection sample was also available, which formally shows that a response was not detectable pre-infection (Fig. 4h). The sample taken 5 weeks after symptom onset showed seroconversion for anti-RBD and very strong RBD-ACE2 inhibition, with some decline in anti-RBD and RBD-ACE2 inhibition observed at the 18-week timepoint. Thus, the results from our lateral flow assay are consistent with the accepted behaviour of NAb levels over time.
Serum samples from SARS-CoV-2 immunised macaques
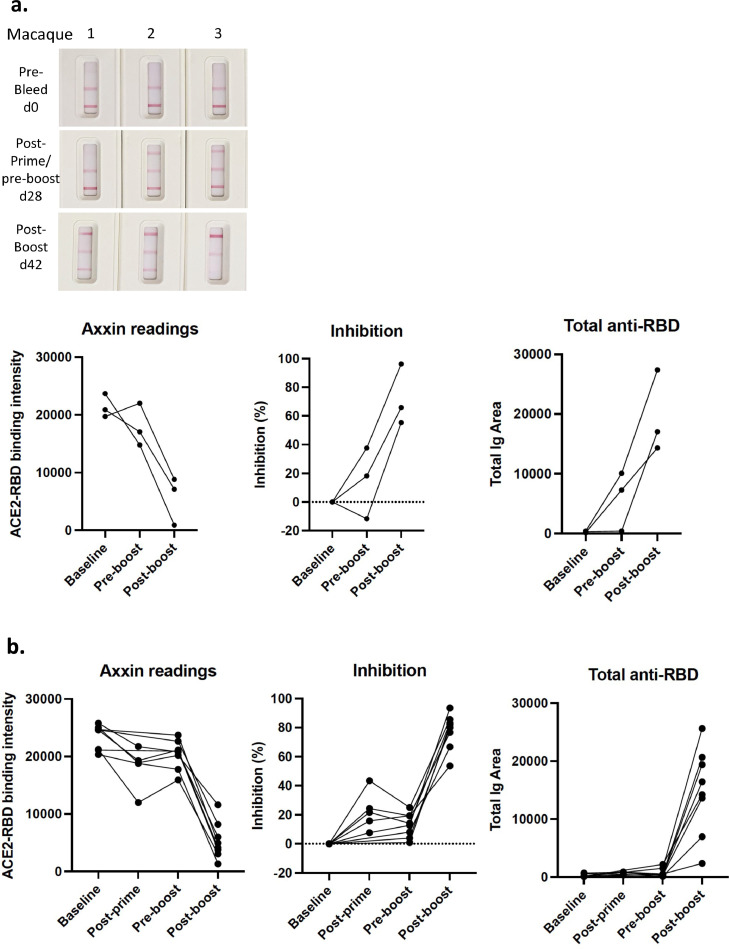
Next we examined longitudinal tests samples from macaques that had been immunised with experimental SARS-CoV-2 spike vaccines that induced high titre NAb responses [20]. Results in Fig. 5a are shown from 3 macaques primed with spike protein vaccine with Addavax adjuvant, and boosted with soluble RBD protein on day 21, and from 8 additional macaques (Fig. 5b) primed with whole spike protein (day 0) and boosted with spike protein (day 21), as described [20]. Blood samples were taken prior to the prime (day 0), post-prime (day 14; five animals only), prior to the boost (day 21), and post-boost (day 42). Graphs depict from left to right: raw Axxin readings; % inhibition (relative to pre-bleed baseline for each animal); and total anti-RBD results (Fig. 5a and b). While some evidence of RBD-ACE2 inhibition was observed with the post-prime/pre-boost samples (Fig. 5a and b), the post-boost (day 42) samples all showed clear RBD-ACE2 inhibition ranging from 50% to 95%. The total anti-RBD line also became more prominent in all animals following the boost (Fig. 5a and b). Given that NAb responses are induced by most current SARS-CoV-2 vaccines, we anticipate that similar results will be derived from humans that are immunised with SARS-CoV-2 vaccines, and these studies demonstrate that this assay can provide clear evidence of total anti-RBD antibodies and NAb from vaccine recipients. It is also noteworthy that these data demonstrate that our assay works with non-human samples and may also be valuable for assessing SARS-CoV-2 protection in animal models and SARS-CoV-2 exposure in other species.
Fig. 5.
Analysis of plasma samples from immunised macaques. a. Representative images of test results from longitudinal samples from three macaques immunised with SARS-CoV-2 spike protein (prime), RBD protein (boost) (left) and data plotted to compare % inhibition or anti-RBD antibody shown at baseline (day 0), pre-boost (day 28) and post-boost (day 42). Graphs depict raw Axxin data readings linked for each animal (left); % inhibition relative to baseline reading linked for each animal (middle); and total anti-RBD reading linked for each animal (right). n = 3 biological replicates. b. Samples from a different cohort of eight macaques immunised with SARS-CoV-2 spike protein (prime), and spike protein (boost), shown at baseline (day 0), post-prime (day 14), pre-boost (day 28) and post-boost (day 42). Graphs depict data as described in a. n = 8 biological replicates. Samples from macaques were derived as previously described [20].
Neutralisation testing for viral variants
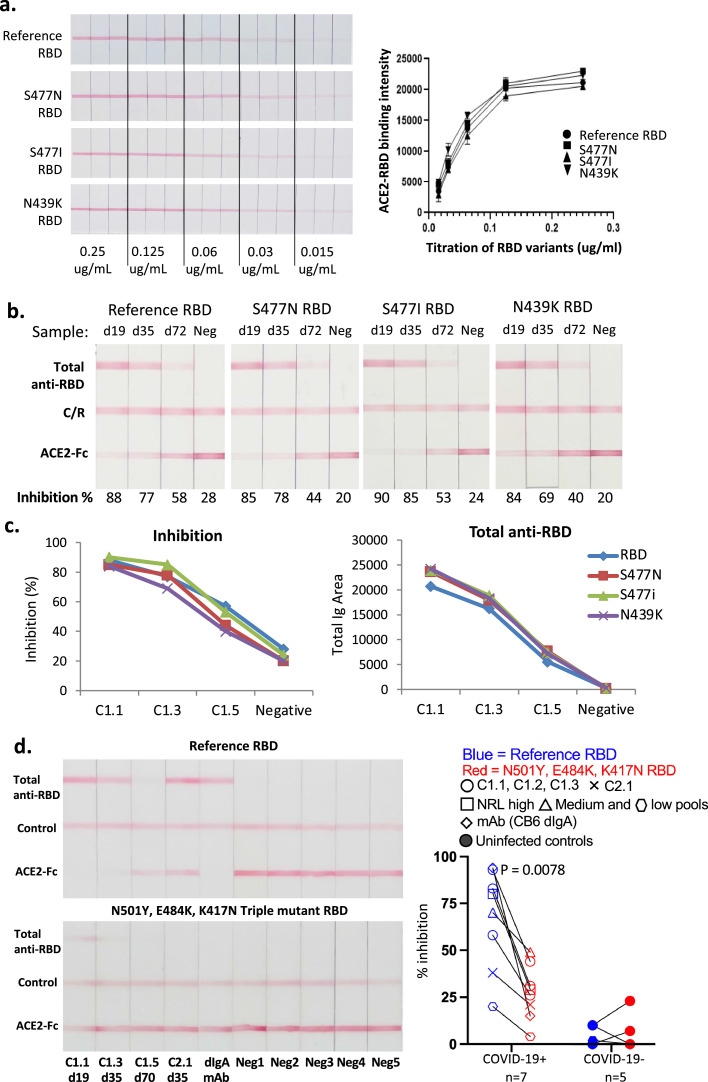
In addition to waning levels of antibody over time, the long-term efficacy of antibody-based protection to COVID-19 from vaccination or prior infection is being challenged by the emergence of variants of concern (VOC), with single and multiple mutations in the spike protein and especially in RBD that may affect binding affinity to ACE2, transmissibility (R0) and/or susceptibility to NAb. Testing of protection may therefore require consideration of circulating VOCs. Our test was designed to facilitate the substitution of variant RBD proteins by the use of RBD-biotin and anti-biotin gold nanoparticles, and we evaluated four different versions of RBD-biotin in our assay (Fig. 5), choosing variants observed during second wave infections in June 2020 that have been reported to have varying sensitivity to some mAbs [33,34]. The original (reference) RBD sequence and variants S477N, S477I and N439K were expressed with biotin tags, purified and titrated before mixing with a fixed amount of anti-biotin gold (Fig. 6). Each of the four RBDs showed essentially identical titration curves for binding of RBD-biotin-Au complexes to ACE2 (Fig. 6a). We next assessed the susceptibility of each variant to inhibition by patient antibodies, again using longitudinal plasma samples from patient C1 (infected in February 2020). As shown in Fig. 6b and c, the level of inhibition for all variants was around 90% at week 3 after symptom onset, reducing to around 80% at 5 and 50% at 10 weeks for each variant, with only some subtle differences. The level of total antibody measured with each variant RBD-biotin-Au was also similar at each time (Fig. 6c). We also tested some samples from individuals infected with the original strain of SARS-CoV-2 against NAb-testTM devices prepared with either original, or a N501Y, E484K, K417N triple-mutant RBD derived from the B.1.351 South African or ‘beta’ variant of concern (Fig. 6d). Our COVID-19 NAb-testTM results utilising the triple-mutant RBD are consistent with previous reports using gold-standard neutralising assays, revealing a sharp decline in neutralising activity against the beta variant RBD compared to the reference RBD. These experiments demonstrate the utility of our assay for the detection and assessment of NAb responses to RBDs from variants of SARS-CoV-2 through substitutions of the corresponding recombinant RBD-biotin forms.
Fig. 6.
The test cassette can be adapted to work with variant RBDs. a. Original (reference) RBD and 3 variant RBD (S477N; S477I; N439K) gold conjugates were titrated on ACE2 strips, in triplicate. Red line depicts direct binding between RBD and ACE2. Each variant showed a similar titration curve. b. Longitudinal samples from patient C1 were titrated against strips in the presence of reference RBD or the 3 variants. c. Data from B plotted to compare % inhibition, relative to the reference line, against each different RBD or anti-RBD antibody. d. Serum samples from COVID-19 convalescent and uninfected controls were run in parallel in the presence of reference RBD or an RBD equivalent to that of the beta variant (N501Y, E484K, K417N triple mutant RBD). These include longitudinal samples from donor C1, as well as NRL samples which are donor plasma samples obtained from Biomex, Germany, consisting of pools of 4 individual samples with high (593-790), medium (145-222) and low (<10) NAb titres derived from the National Serology Reference Laboratory, Australia. CB6 dIgA is a neutralising monoclonal antibody raised against the reference RBD. n = 7 (convalescent) / 5 (unifected) biological replicates. P value = 0.0078 determined by Wilcoxon signed-rank test.
Compatibility with whole blood for use at point of care
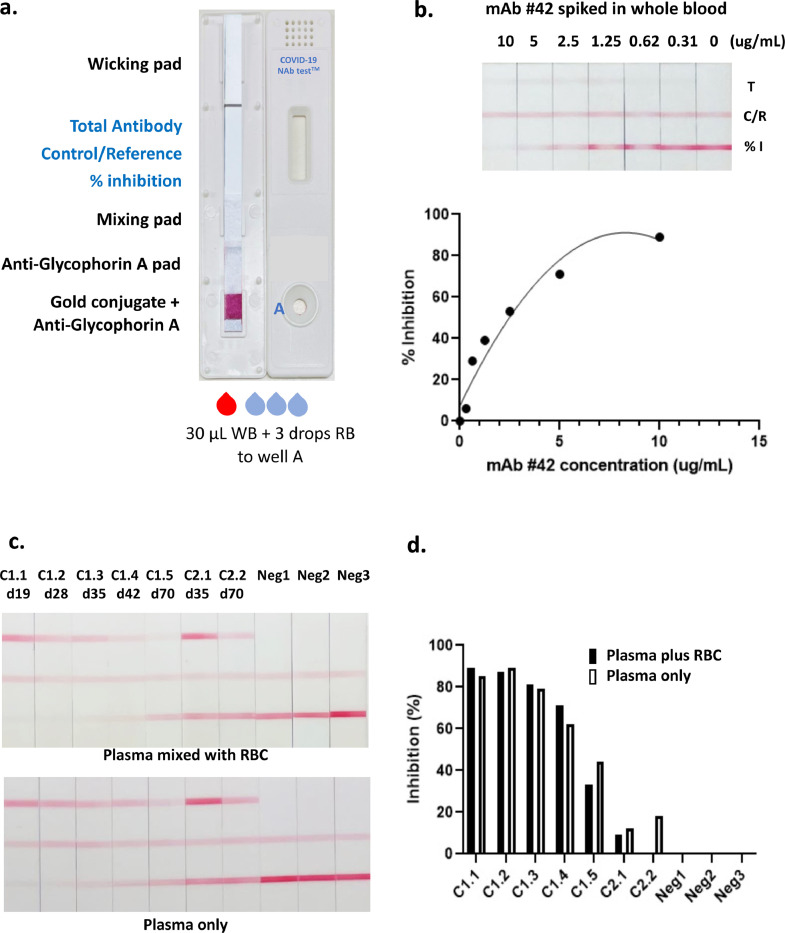
Lateral flow tests are suitable for use at POC, but for blood-based tests they must function with the use of whole blood rather than plasma or serum. To achieve this, we further modified the test by addition of anti-glycophorin A antibodies to the sample pad along with the RBD-Au complexes. Anti-glycophorin A agglutinates red blood cells, while white blood cells, plasma and the RBD-Au complexes are able to flow as normal; the white blood cells are subsequently retarded at the interface of the nitrocellulose membrane [35]. We also used a single well cartridge as we found this gave better separation from the trapped red blood cells to the test area of the strip (Fig. 7a). Fig. 7b shows the results of test strips using whole blood from a COVID-negative donor, spiked with varying levels of neutralising mAb 42. This showed good correlation of ACE2-RBD inhibition versus mAb concentration, providing proof of concept for the compatibility of the test format with whole blood. We noted that the total anti-RBD antibody line was fainter than expected with these samples, probably a consequence of a single specificity mAb compared to polyclonal antibodies, including various isotypes, present in plasma samples. To provide further proof of concept for using whole blood versus plasma, longitudinal samples from subject C1 were tested on the device strips shown in Fig. 6A using either 15 µl of plasma, or 15 µl of plasma mixed with 15 µl of packed red blood cells from a COVID-19 negative donor, emulating 30 µl of a whole blood sample with 50% hematocrit (Fig. 7c and d). The visual appearance and quantitative readout of the plasma and reconstituted whole blood samples were very similar for both the level of inhibition and the level of total anti-RBD (Fig. 7c and d), which suggests that the COVID-19 NAb-testTM should have utility for true POC use.
Fig. 7.
Development of a test cassette that measures NAb from whole blood. a. An image of the cartridge used for testing whole blood, incorporating anti-glycophorin-A pad, and a single well for sample loading. WB; whole blood, RB; running buffer b. Whole blood was spiked with varying concentrations of neutralising mAb 42 and tested on strips incorporating anti-glycophorin A which captures red blood cells. Graph depicting % inhibition (relative to the reference line, from Axxin strip reader) versus amount of mAb 42 spiked into sample. c. Longitudinal plasma samples from donor C1 were either reconstituted with an equal volume of packed red blood cells and run as 30 µl whole blood, or run as 15 µl plasma only. Two longitudinal samples from donor C2 and three COVID-19 negative samples also shown. d. Graph depicting % inhibition (relative to the reference line, from Axxin strip reader) from data shown in c., comparing plasma added to red blood cells, or plasma only.
Confirmation of neutralising Abs in immunised humans
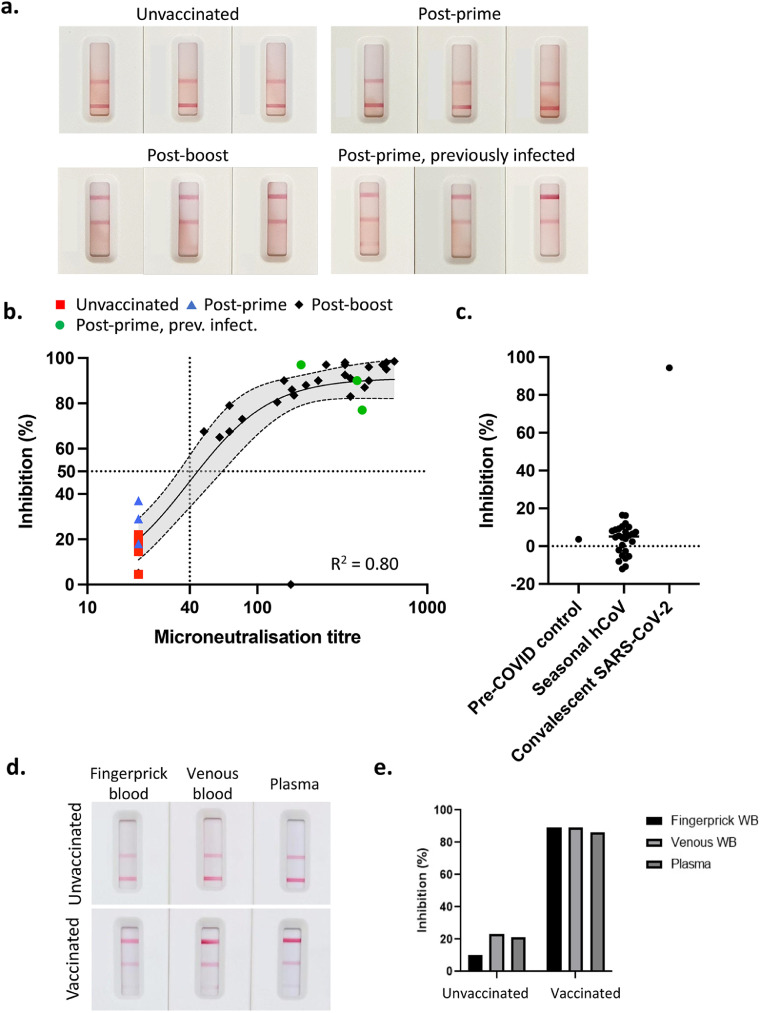
To validate our test with a sample of post-(Pfizer) vaccination humans, we tested a cohort of vaccinees at least 2 weeks following the second (boost) dose, compared to a group of unvaccinated individuals (red), using the whole blood test (Fig. 8a and b). There was a clear correlation (R2 = 0.80, p<0.0001; saturation curve with Hill-slope) between the results from our assay and microneutralisation, where >50% inhibition at the RBD-ACE2 test line was exhibited by 26 out of 27 samples with a microneutralisation titre of greater than 40 (95%CI of sensitivity of 79.1% to 99.8%) which we consider represents a reasonable lower limit for protection against SARS-CoV-2 [6]. Using these cut-offs for inhibition, specificity was considered to be 100% (38/38; 95%CI of 88.6% to 100.0%). There was one outlier in the cohort of post-boost individuals with microneutralisation titre of 157 that failed to show inhibition in our RBD-ACE2 test line, despite showing a clear reading for total anti-RBD antibody (Fig. 8b). This sample may reflect an individual who generated NAb that target parts of the spike other than the RBD.
Fig. 8.
Assessment of NAb in post-vaccinee samples using whole blood. a. Representative images of test results from human venous blood samples from (top left) unvaccinated, (top right) post-prime vaccinated (Pfizer, mRNA spike vaccine), (lower left) post-boost vaccinated and (lower right) post-prime vaccinated following previous infection with SARS-CoV-2. b. Inhibition readings from the Axxin strip reader from samples from groups described in a. were plotted against readings for the same samples derived from virus microneutralisation assays. n = 40 biological replicates; saturation curve with Hill-slope and 95%CI (dashed lines). c. 26 serum samples from patients with seasonal hCoV (HCoV-NL63, O229E, OC43 or HKU1; PCR negative for SARS-CoV-2) were compared to a pre-COVID healthy control and convalescent SARS-CoV-2 patient sample. d. Samples from fingerprick whole blood were compared to venous whole blood and plasma from two individuals, one unimmunised and uninfected and one previously primed and boosted with the Pfizer vaccine (22 days post-boost). e. Inhibition readings from the Axxin strip reader are depicted in the graph. WB; whole blood.
Six samples from vaccinees after a single dose were also included in this study (Fig. 8a and b). Three of these samples (blue) showed little or no inhibition of RBD-ACE2 binding in our test, and no microneutralisation activity above the background titre of 20 and were thus very similar to unvaccinated controls. This reflects the fact that naïve individuals require a prime and boost to develop a strong NAb response. Interestingly, however, the other three post-prime samples (green) showed strong inhibition of RBD-ACE2 binding in our test, which correlated with strong microneutralisation titres. This group were from a study of people who had previously been infected with SARS-CoV-2 and have subsequently been immunised. Additionally, we also tested serum from patients with seasonal hCoV (HCoV-NL63, O229E, OC43 or HKU1, and PCR negative for SARS-CoV-2). These samples showed little to no inhibition when compared to pre-COVID or SARS-CoV-2 convalescent controls (Fig. 8c). This highlights the ability of our test to detect NAb regardless of whether they are induced by infection, vaccination, or a combination thereof; but not seasonal hCoV infection.
Lastly, to ensure that fingerprick-derived whole blood works as effectively as venous whole blood, we compared fingerprick-derived whole blood versus venous derived whole blood versus plasma from samples derived from an unvaccinated individual and a vaccinated individual (Fig. 8d and e). These data demonstrate that samples run on our assay, whether it be from finger prick, venous whole blood or plasma, provides similar results to conventional neutralisation tests, confirming that this test can be easily used at point of care with a simple finger prick blood sample.
Discussion
As an increasing number of approved COVID-19 vaccines are now being rolled out around the globe, the hope is that individuals who have developed immunity will contribute to population-level herd immunity, and facilitate a return to a pre-COVID-19-like existence. However, we currently lack widely-deployable tools to answer the critical questions of who is actually immune, and for how long immunity lasts, and whether their immunity will protect them from new variants of SARS-CoV-2. Tests that detect viral RNA do not provide this information, nor do tests that measure total anti-virus antibody, whether they are ELISA or lateral flow-based. As we and others have shown, the existence of antibodies that can bind to viral proteins, including RBD-specific antibodies, does not necessarily demonstrate the existence of NAb. It is generally accepted that to be protective, NAb need to target the spike protein, and in SARS-CoV-1, antibodies against other structural proteins (E, M and N) were unable to mediate protective immunity [36]. Furthermore, at least 90% of neutralising activity is focussed on the RBD of the spike protein, and functions predominantly by preventing interaction with cellular receptor ACE2 [8].
It is not yet clear what titre of NAb is adequate for protection against SARS-CoV-2. Studies in macaques involving transfer of SARS-CoV-2 NAb from immune to naïve animals suggest that NAb titres of approximately 50 (based on pseudovirus assay) or total anti-RBD ELISA titres of approximately 100 are protective against infection, although the assessment was based on a limited number of animals [37]. As such, we anticipate updating our assay as further data comes to light. Furthermore, several studies have shown that NAb titres following infection wane with time, which means regular testing for immunity will likely be required. Published estimates vary, but most studies agree that neutralising antibody levels begin to decline within a few weeks of infection. One study of 149 individuals following COVID-19 disease showed that one third of these individuals had neutralising titres below 1:50 by roughly 39 days after symptom onset [38]. Depending on the maximum antibody titre, which is typically linked to the severity of infection, it may take between 4 months and 1 year for the level of NAb to drop below detection limits [39].
Scenarios such as national or international travel and border control, verification of vaccination status, risk stratification of quarantine requirements, screening of donors for convalescent plasma or antibody therapy, or returning to work or school, may require regular assessment of immune status. A rapid (20 minute) test using fingerprick whole blood overcomes the limitations associated with current tests, including requiring a venous blood draw, laboratory equipment and slow turnaround with results taking a minimum of three hours. Portable strip-reading instruments such as the Axxin AX-2XS used here are commercially available and can be deployed in non-laboratory locations such as airports and aged care or educational facilities, and the use of dedicated readers (and potentially even smartphone Apps) can provide a quantitative assessment of NAb levels. Even in the absence of a dedicated instrument, a semi-quantitative assessment by eye, visually comparing the intensity of the test line to the reference line, should provide sufficient information on whether an individual has an acceptable level of immunity. This approach has been successfully deployed for determining CD4 T-cell levels in the semi-quantitative point of care Visitect® CD4 tests [28].
An additional risk that threatens to reduce the durability of immune protection is posed by the emergence of mutant strains of SARS-CoV-2, including the B.1.1.7 lineage (also known as the alpha variant), the B.1.351 lineage (beta variant), B.1.1.28 lineage (gamma variant) and B.1.617.2 (delta variant) that are considered to be variants of concern (VOCs). Each of these VOCs are more infectious, and potentially induce more severe disease. While they carry mutations throughout various viral proteins, the key mutations appear to be those in the RBD region (N501Y in B.1.1.7; N501Y, E484K and K417N in B.1.351; N501Y, E484K and K417T in B.1.1.28, and L452R and T478K in B.1.617.2) that markedly increase ACE2 binding affinity of the virus (N501Y) or impact recognition by antibody (E484K; K417N/T; L452R). Several studies have shown that these variants, particularly B.1.351 and B1.1.28, are more difficult to neutralise, effectively lowering NAb titres by up to 33-fold in convalescent plasma and 8.3 fold in individuals vaccinated with the Pfizer and Moderna vaccines [5]. Neutralising antibody assays will need to incorporate these variants to accurately determine antibody-based protection based on the currently and locally circulating strains, helping to guide strategies for boosters to address waning immunity and/or VOCs. The ability to interchange RBDs carrying relevant mutations as required (Fig. 5), further highlights an important advantage of our assay, especially compared to the measurement of total S- or RBD-specific antibody or IgG which is unlikely to reflect neutralisation escape. Moreover, the technology could theoretically be adapted in future iterations to simultaneously measure against multiple mutants by using multiple parallel strips. Our longitudinal analysis of serum samples from macaques that had been immunised with experimental spike protein vaccines, as well as whole blood samples from humans immunised with spike mRNA vaccines, demonstrated that our assay provides a clear readout of anti-RBD antibody as well as NAb following immunisation (Figs. 4 and 7).
Limitations of this study include the underlying premise that we will need to test people from time to time to assess their antibody status and risk of infection. While this is likely to be valuable, it should be acknowledged that even if/when antibody titres fall, it is possible that memory B and T cells will provide rapid and effective protection upon re-exposure. Therefore, while antibody titres may decline, that does not necessarily mean that the individual is fully susceptible to disease. Indeed, repeat testing with an easy and rapid POC test has the potential to exaggerate fear of re-infection risk. Furthermore, it remains to be determined what the policy response will be for individuals showing low to undetectable Ab, although in the case of hepatitis B virus immunisation for high-risk workers, repeated booster immunisations are sometimes recommended for those who do not show clear seroconversion. While 90% of NAb are directed toward the RBD, inhibiting its binding to ACE2, it is possible that NAb against other parts of the S protein may be prominent in some individuals, yielding a false negative or under-estimate of immunity using our assay, and indeed we observed one such sample in our study of vaccinee samples. Conversely, it may be reasonable to assume that RBD-specific antibody neutralisation measured with the COVID-19 NAb-testTM will represent a minimum estimate of immunity to the variant included in a particular version of the test. Our samples did not evenly cover all demographics, however this is unlikely to impact on interpretation in our study which is primarily focussed on whether the COVID-19 NAb-testTM can detect NAb in serum or whole blood samples with correlation against gold standard microneutralization, regardless of their origin. Definitive validation and verification of the COVID-19 NAb-testTM will require further studies in various populations, including individuals infected with different variants of SARS-CoV-2, but such studies should be performed with tests manufactured under ISO13485 quality standards rather than the test development prototypes used here.
SARS-CoV-2 continues to spread and threatens to disrupt human activity for years to come. Fortunately, we already have vaccines that are proving to be effective at reducing disease that are now being rolled out in many countries, but countering that, many countries have low vaccine coverage and ongoing high rates of infection, raising the potential for immune-mediated selection of escape variants. It is possible that immunity tests may become a normal part of life, at least in some circumstances such as international travel, as we adjust to life with SARS-CoV-2 as an endemic virus. Our test should contribute to more widespread access to immunity testing for many populations, including those in resource-poor areas with limited access to laboratories and equipment for more conventional virus neutralisation tests or ELISA-based inhibition assays.
Contributors
Investigation; TSF, HV, NAG, SZ, MC, FLM, SLG, LK, PE
Resources; SR, RC, IB, HED, THON, DLD, ACC, PMH, LCR, ZM, KS, KK, BDW, DAW, HXT, AKW, JAJ, SJK, GM
Conceptualisation and Supervision; DIG, DFJP, DAA
Writing - original draft; DIG and DAA
TSF, HV, DFJP, DIG and DAA have verified the raw data.
All authors have read and approved the final version of the manuscript.
Data Sharing
Data reported in this article, after de-identification, is available upon request for three years after publication.
Declaration of Competing Interest
A provisional patent covering the COVID-19 NAb-testTM test and underlying technology has been submitted through The University of Melbourne.
Dr. Fulford reports a patent Australian Provisional Patent Application 2021901011 (filed 7 April 2021) “Point of care lateral flow test for COVID19 detection” pending. Mr. Van reports grants from Government of the State of Victoria, during the conduct of the study; In addition, Mr. Van has a patent Australian Provisional Patent Application 2021901011 (filed 7 April 2021) “Point of care lateral flow test for COVID19 detection” pending. Dr. Gherardin reports a patent Australian Provisional Patent Application 2021901011 (filed 7 April 2021) “Point of care lateral flow test for COVID19 detection” pending. Ms. Zheng reports grants from Government of the State of Victoria, during the conduct of the study; In addition, Ms. Zheng has a patent Australian Provisional Patent Application 2021901011 (filed 7 April 2021) “Point of care lateral flow test for COVID19 detection” pending. Mr. Ciula has nothing to disclose. Prof. Drummer has nothing to disclose. Mr. Redmond has nothing to disclose. Dr. Tan has nothing to disclose. Ms. Boo has nothing to disclose. Dr. Center has nothing to disclose. Dr. Li has nothing to disclose. Dr. Grimley has nothing to disclose. Dr. Wines reports grants from Australian Govt, Medical Research Future Fund, grants from NHMRC, National Health and Medical Research Council GNT1145303, Australia, during the conduct of the study; In addition, Dr. Wines has a patent drafted for use of ACE2-Fc pending. Dr. Nguyen has nothing to disclose. Ms. Mordant has nothing to disclose. Dr. Ellenberg has nothing to disclose. Dr. Rowntree has nothing to disclose. Lukasz Kedzierski has nothing to disclose. Prof. Cheng reports that he is a member of government advisory committees advising on COVID policy. Prof. Doolan has nothing to disclose. Prof. Matthews reports grants from Curran Foundation, during the conduct of the study; grants from Gilead sciences, grants from Abbvie Inc outside the submitted work. Dr. Bond has nothing to disclose. Prof. Hogarth reports grants from NHMRC, grants from MRFF, during the conduct of the study; In addition, Prof. Hogarth has a patent Antiviral pending. A/Prof. McQuilten reports grants from Medical Research Future Fund, during the conduct of the study. Prof. Subbarao has nothing to disclose. Prof. Kedzierska has nothing to disclose. Dr. Juno has nothing to disclose. Dr. Wheatley has nothing to disclose. Prof. Kent has nothing to disclose. Prof. Williamson has nothing to disclose. Prof. Purcell reports grants from Victorian Government DHHS, grants from Medical Research Future Fund, other from Jack Ma Foundation, during the conduct of the study; In addition, Prof. Purcell has a patent PCT/AU2021/050839 pending. Prof. Anderson reports grants from Government of the State of Victoria, during the conduct of the study; other from Nanjing BioPoint Diagnostic Technology, outside the submitted work; In addition, Prof. Anderson has a patent Australian Provisional Patent Application 2021901011 (filed 7 April 2021) “Point of care lateral flow test for COVID19 detection” pending. Prof. Godfrey reports grants from Victorian Government DHHS, and from Australian Research Council, during the conduct of the study; grants from Medical Research Future Fund, other from Jack Ma Foundation, grants from National Health and Medical Research Council, outside the submitted work; In addition, Prof. Godfrey has a patent Australian Provisional Patent Application 2021901011 (filed 7 April 2021) “Point of care lateral flow test for COVID19 detection” pending.
Funding and Acknowledgements
We are grateful to the volunteers who provided samples for study. The authors would like to thank the Australian Red Cross Lifeblood for the supply of specimens from COVID-19 convalescent plasma donors and ‘pre-COVID’ controls. Australian Governments fund the Australian Red Cross Lifeblood to provide blood, blood products and services to the Australian community. The authors gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program received by the Burnet Institute. This work was supported by grants from the Department of Health and Human Services (DHHS) of the Victorian State Government; the Australian Research Council (ARC; CE140100011, CE140100036) the National Health and Medical Research Council of Australia (NHMRC; 1113293, 2002317 and 1116530), and Medical Research Future Fund (MRFF) Awards (2005544, 2002073, 2002132). THON and AKW were supported by an NHMRC Emerging Leadership Level 1 Investigator Grants (1194036), HV was supported by an NHMRC APPRISE Research Fellowship (1116530), KK was supported by the NHMRC Leadership Investigator Grant (1173871), DLL was supported by a NHMRC Principal Research Fellowship (1137285), KS and DAW are supported by NHMRC Investigator Grants (1177174 and 1174555). KS also received support from the A2 Milk Company and KS, DP and DG received support from the Jack Ma Foundation. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. DIG and SJK were supported by NHMRC Senior Principal Research Fellowships (1117766 and 1136322).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103729.
Contributor Information
David A. Anderson, Email: david.anderson@burnet.edu.au.
Dale I. Godfrey, Email: godfrey@unimelb.edu.au.
Appendix. Supplementary materials
References
- 1.Guo J., Chen S., Tian S., Liu K., Ni J., Zhao M., et al. 5G-enabled ultra-sensitive fluorescence sensor for proactive prognosis of COVID-19. Biosens Bioelectron. 2021;181 doi: 10.1016/j.bios.2021.113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yun S., Ryu J.H., Jang J.H., Bae H., Yoo S.H., Choi A.R., et al. Comparison of SARS-CoV-2 antibody responses and seroconversion in COVID-19 patients using twelve commercial immunoassays. Ann Lab Med. 2021;41(6):577–587. doi: 10.3343/alm.2021.41.6.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murillo A.M.M., Tomé-Amat J., Ramírez Y., Garrido-Arandia M., Valle L.G., Hernández-Ramírez G., et al. Developing an Optical Interferometric Detection Method based biosensor for detecting specific SARS-CoV-2 immunoglobulins in Serum and Saliva, and their corresponding ELISA correlation. Sens Actuators B. 2021;345 doi: 10.1016/j.snb.2021.130394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond K., Nicholson S., Lim S.M., Karapanagiotidis T., Williams E., Johnson D., et al. Evaluation of serological tests for SARS-CoV-2: Implications for serology testing in a low-prevalence setting. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 6.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 7.Khoury D.S., Wheatley A.K., Ramuta M.D., Reynaldi A., Cromer D., Subbarao K., et al. Measuring immunity to SARS-CoV-2 infection: comparing assays and animal models. Nat Rev Immunol. 2020;20(12):727–738. doi: 10.1038/s41577-020-00471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020 doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4) doi: 10.1016/j.cell.2020.03.045. 894-904 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217(11) doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I., Tiu C., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 12.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 13.Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5(19) doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrnes J.R., Zhou X.X., Lui I., Elledge S.K., Glasgow J.E., Lim S.A., et al. Competitive SARS-CoV-2 serology reveals most antibodies targeting the spike receptor-binding domain compete for ACE2 binding. mSphere. 2020;5(5) doi: 10.1128/mSphere.00802-20. e00802-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker S.N., Chokkalingam N., Reuschel E.L., Purwar M., Xu Z., Gary E.N., et al. SARS-CoV-2 assays to detect functional antibody responses that block ACE2 recognition in vaccinated animals and infected patients. J Clin Microbiol. 2020;58(11) doi: 10.1128/JCM.01533-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan E., Frew E., Cooper J., Humphrey J., Holden M., Mand A.R., et al. Use of lateral flow immunoassay to characterize SARS-CoV-2 RBD-specific antibodies and their ability to react with the UK, SA and BR P.1 variant RBDs. Diagnostics. 2021;11(7):1190. doi: 10.3390/diagnostics11071190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J.J., Zhang N., Richardson S.A., Wu J.V. Rapid lateral flow tests for the detection of SARS-CoV-2 neutralizing antibodies. Expert Rev Mol Diagn. 2021;21(4):363–370. doi: 10.1080/14737159.2021.1913123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake D.F., Roeder A.J., Kaleta E., Jasbi P., Periasamy S., Kuzmina N., et al. Development of a rapid point-of-care test that measures neutralizing antibodies to SARS-CoV-2. medRxiv. 2021 doi: 10.1016/j.jcv.2021.105024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phetsouphanh C., Darley D., Wilson D.B., Howe A., Ling Munier C.M., Patel S.K., et al. Immunological dysfunction persists for 8 months following initial mild-moderate SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1038/s41590-021-01113-x. 2021.06.01.21257759. [DOI] [PubMed] [Google Scholar]
- 20.Tan H.X., Juno J.A., Lee W.S., Barber-Axthelm I., Kelly H.G., Wragg K.M., et al. Immunogenicity of prime-boost protein subunit vaccine strategies against SARS-CoV-2 in mice and macaques. Nat Commun. 2021;12(1):1403. doi: 10.1038/s41467-021-21665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez-Lombardi R., Nevoltris D., Luthra A., Schofield P., Zimmermann C., Christ D. Transient expression of human antibodies in mammalian cells. Nat Protoc. 2018;13(1):99–117. doi: 10.1038/nprot.2017.126. [DOI] [PubMed] [Google Scholar]
- 22.Wheatley A.K., Pymm P., Esterbauer R., Dietrich M.H., Lee W.S., Drew D., et al. Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain. Cell Rep. 2021:37. doi: 10.1016/j.celrep.2021.109822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 25.Koutsakos M., Rowntree L.C., Hensen L., Chua B.Y., van de Sandt C.E., Habel J.R., et al. Integrated immune dynamics define correlates of COVID-19 severity and antibody responses. 2021;2(3):100208. [DOI] [PMC free article] [PubMed]
- 26.Caly L., Druce J., Roberts J., Bond K., Tran T., Kostecki R., et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med J Aust. 2020;212(10):459–462. doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luchters S., Technau K., Mohamed Y., Chersich M.F., Agius P.A., Pham M.D., et al. Field performance and diagnostic accuracy of a low-cost instrument-free point-of-care CD4 test (Visitect CD4) performed by different health worker cadres among pregnant women. J Clin Microbiol. 2019;57(2) doi: 10.1128/JCM.01277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhi S., Tayler N., Hoang T., Ballard S.A., Graham M., Rojek A., et al. Multi-site assessment of rapid, point-of-care antigen testing for the diagnosis of SARS-CoV-2 infection in a low-prevalence setting: a validation and implementation study. Lancet Reg Health. 2021;9 doi: 10.1016/j.lanwpc.2021.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5(12):1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 32.Marot S., Malet I., Leducq V., Zafilaza K., Sterlin D., Planas D., et al. Rapid decline of neutralizing antibodies against SARS-CoV-2 among infected healthcare workers. Nat Commun. 2021;12(1):844. doi: 10.1038/s41467-021-21111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3) doi: 10.1016/j.chom.2021.02.003. 463-76.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184(5) doi: 10.1016/j.cell.2021.01.037. 1171-87 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham M.D., Haile B.A., Azwa I., Kamarulzaman A., Raman N., Saeidi A., et al. Performance of a novel low-cost, instrument-free plasma separation device for HIV viral load quantification and determination of treatment failure in people living with HIV in Malaysia: a diagnostic accuracy study. J Clin Microbiol. 2019;57(4) doi: 10.1128/JCM.01683-18. e01683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., et al. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590(7847):630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau E.H.Y., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.W., Chan W.H., Chiu S.S., et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat Commun. 2021;12(1):63. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.