Abstract
Members of the transforming growth factor β (TGF-β) family transduce signals through Smad proteins. Smad signaling can be regulated by the Ras/Erk/mitogen-activated protein pathway in response to receptor tyrosine kinase activation and the gamma interferon pathway and also by the functional interaction of Smad2 with Ca2+-calmodulin. Here we report that Smad–TGF-β-dependent transcriptional responses are prevented by expression of a constitutively activated Ca2+-calmodulin-dependent protein kinase II (Cam kinase II). Smad2 is a target substrate for Cam kinase II in vitro at serine-110, -240, and -260. Cam kinase II induces in vivo phosphorylation of Smad2 and Smad4 and, to a lesser extent, Smad3. A phosphopeptide antiserum raised against Smad2 phosphoserine-240 reacted with Smad2 in vivo when coexpressed with Cam kinase II and by activation of the platelet-derived growth factor receptor, the epidermal growth factor receptor, HER2 (c-erbB2), and the TGF-β receptor. Furthermore, Cam kinase II blocked nuclear accumulation of a Smad2 and induced Smad2-Smad4 hetero-oligomerization independently of TGF-β receptor activation, while preventing TGF-β-dependent Smad2-Smad3 interactions. These findings provide a novel cross-talk mechanism by which Ca2+-dependent kinases activated downstream of multiple growth factor receptors antagonize cell responses to TGF-β.
The transforming growth factor β (TGF-β) superfamily which includes TGF-β, activins, and bone morphogenic proteins (BMPs) regulate cell growth, motility, apoptosis, differentiation, and matrix production and can have a multifunctional role in tumorigenesis (reviewed in reference 29). Specific cell surface receptors for the TGF-β family possess intrinsic serine-threonine kinase activity, and signaling is mediated by the type I and type II receptors, which are able to form a ligand-inducible active heteromeric complex (42). Identification of the Smad family of transcription factors is beginning to reveal the mechanisms of TGF-β-mediated signaling from the cell surface to the nucleus. There are three types of Smad comprising pathway-restricted, common-mediator, and inhibitory Smads. Pathway-restricted Smad1 and Smad5 are substrates for the BMP type I receptor, whereas Smad2 and Smad3 are specific targets for TGF-β and activin receptors. Phosphorylation of Smads by the activated type I receptor kinase induces their release and subsequent association with a common mediator Smad4 (reviewed in reference 29). This oligomeric complex then translocates to the nucleus, and cooperation with DNA-binding proteins such as FAST-1 and FAST-2 directs specific transcriptional responses (12, 26, 46).
Phosphorylation of Smads is critical for regulating their activity and all pathway restricted Smads have a C-terminal SSXS motif in which the two end serines are phosphorylated by the type I receptor (1, 41). Receptor-mediated phosphorylation of the C-terminal serines in pathway-restricted Smads is required for transcriptional activity and facilitates both the association with Smad4 to form hetero-oligomers and the translocation to the nucleus (reviewed in reference 29). The MH2 domain of activated-pathway-restricted Smads comprises a transcriptional activation domain and is also responsible for interactions with Smad4 and FAST-1 (12). Smad2 is not able to bind DNA directly, and a ternary bridge complex is formed at appropriate response elements in which Smad2 interacts with Smad4 and FAST-1 that bind separately to two specific DNA sequence elements (26, 46). Other nuclear proteins that interact with Smads and influence their transcriptional activity include the fos-jun complex, Sp1, the basic helix-loop-helix leucine zipper protein TFE3, the coactivator p300/CBP, vitamin D receptor, Evi-1, the Ski oncoprotein, and the corepressor TGIF (for recent reviews, see references 30 and 36).
Recently, input from other growth factor receptor systems have also been shown to influence the Smad signaling network and to have an impact upon responsiveness to members of the TGF-β superfamily. Erk–mitogen-activated protein (MAP) kinase activated in response to epidermal growth factor (EGF) and hepatocyte growth factor can phosphorylate the linker region of BMP pathway-specific Smad1 and inhibit both nuclear translocation and transcriptional activity (24). Oncogenic ras can also repress Smad–TGF-β signaling in mammary and lung epithelial cells due to Erk-MAP kinase-mediated phosphorylation of the linker regions of Smad2 and Smad3 (25). Conversely, MEKK-1, a component of the stress-activated protein kinase pathway, selectively activates Smad2-mediated transcription in cultured endothelial cells (8). Smads and JNK signaling pathways also cooperate to generate more robust TGF-β-mediated transcriptional responses (18). Functional interaction of Smad2 with Ca2+-calmodulin can inhibit activin–TGF-β signaling, although the precise mechanisms responsible are as yet unknown (48). In the present study, we investigated whether Smad function could be controlled by the activation of cytoplasmic intracellular Ca2+-dependent kinases, more specifically the ubiquitously expressed Ca2+-calmodulin-dependent protein kinase II (Cam kinase II). We demonstrate that Cam kinase II prevents Smad2 nuclear localization and transcriptional function and concomitantly induces Smad2-Smad4 hetero-oligomerization independently of TGF-β receptor activation while completely preventing TGF-β-dependent Smad2-Smad3 hetero-oligomerization. We also present evidence that specific phosphorylation of Smad2 at novel regulatory sites occurs in response to activation of several growth factor receptor signaling pathways. Taken together, our data suggest a novel mechanism for regulating the activity of Smads through changes in cytoplasmic Ca2+ linked to the activation of intracellular Ca2+-dependent kinases that can occur through growth factors and other extracellular stimuli.
MATERIALS AND METHODS
Materials, cell lines, and transfections.
Human embryonic kidney fibroblasts (HEK-293 cells) and Cos-1 cells were obtained from the American Type Culture Collection (Rockville, Md.), and were maintained in Dulbecco modified Eagle medium containing 10% fetal calf serum. Recombinant TGF-β1 was obtained from R&D, thapsigargin was from Calbiochem, KN-93 was from Sigma, monoclonal anti-flag M2 antibody was from Sigma, rat monoclonal anti-hemagglutinin (HA) antibody was from Roche (used for Western blotting), mouse monoclonal anti-HA antibody was from Clontech (used for immunoprecipitation), polyclonal anti-green fluorescent protein (GFP) antibody was from Clontech, Smad2 antibody was from TCS Biologicals, and monoclonal anti-Cam kinase II-α antibody was from Gibco Life Technologies. Rat Cam kinase II-α and Cam kinase II1–290 cDNAs were from Tony Means and subcloned into pCMV1 as described elsewhere (19). The cDNAs for the human EGF receptor and chimeric receptors comprising the extracellular domain of the human EGF receptor connected to either the cytoplasmic domain of the mouse β-platelet-derived growth factor (PDGF) receptor (EP-R) (39), or HER2 (c-erbB2) (HER-1/2) (27) were provided by Axel Ullrich. The 3TP-lux reporter plasmids was provided by Jeffrey Wrana, the ARE-lux reporter was from Malcolm Whitman, and the cDNA for GFP with S65T mutation from Hans-Hermann Gerdes. Full-length human Smad2 (EST clone 1419-H7), human Smad3 (provided by Rik Derynck), and human Smad4 (provided by Scott Kern) were subcloned into pCMV1 incorporating C- or N-terminal HA or GFP epitopes by PCR. The type I TGF-β receptor, provided by Carl-Henrik Heldin, was subcloned into pCMV1, and the constitutively active kinase mutant was prepared by mutation of threonine 204 to aspartic acid. Site-directed mutagenesis was performed as described earlier (10), and pGEX-2T (Pharmacia) constructs were prepared by PCR (19). Cells were transfected using calcium phosphate precipitation (11) or Fugene (Roche).
RNA extraction and RT-PCR analysis.
Total RNA was extracted using the RNAzol B method (Tel-Test, Inc.). Equal amounts of total RNA (3 μg) from each sample were first reverse-transcribed into cDNAs with SuperScript II RNase H+ reverse transcriptase (RT; Gibco-BRL) and random primers. Equal amounts (1 μl) of the RT reaction (20 μl) were in turn subjected to PCR amplification for 30 cycles. To quantify PCR products comparatively and to confirm the use of equal amounts of the RNAs, we coamplified a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The amount of RT reaction used for the amplification (1 μl) was selected as being nonsaturating for the PCR product of GAPDH after 30 cycles of amplification. The sequences of primers were all designed from the published sequence of the human genes as follows: PAI-1 (sense), 5′-GTATCTCAGGAAGTCCAGCC-3′; PAI-1 (antisense), 5′-TCTAAGGTAGTTGAATCCGAGC-3′; GAPDH (sense), 5′-ACCACAGTCCATGCCATCAC-3′; and GAPDH (antisense), 5′-TCCACCACCCTGTTGCTGTA-3′.
Luciferase assays.
Cos-1 cells were transfected in six-well plates with 2 μg of 3TP-lux or ARE-lux and combinations of empty pCMV1 vector or pCMV1-Cam kinase II1–290 as indicated in the figure legends. Cells were starved in medium containing 0.5% fetal calf serum and 48 h after transfection were treated for 2 h with 1% dimethyl sulfoxide (DMSO) or 1 μM thapsigargin (100 μM stock solution in DMSO). In some instances, cells were also pretreated with the specific Cam kinase II inhibitor KN-93 at a concentration of 10 μM (10 mM aqueous stock solution). Luciferase activity was measured after 15 h of treatment with 5 ng of TGF-β using the dual-reporter system (Promega).
Phosphorylation assays.
Phosphorylation in vivo, phosphorylation in vitro of glutathione S-transferase (GST) proteins, and two-dimensional phosphoamino acid analyses were performed as described previously (19). Transfected HEK-293 cells were labeled overnight with 0.5 mCi of [32P]orthophosphate, and lysates were immunoprecipitated with 10 μg of monoclonal anti-flag M2 or anti-HA antibody, together with 20 μg of rabbit anti-mouse immunoglobulin G (IgG) to facilitate the binding of the monoclonal antibodies to protein A-Sepharose. Smad2-GST fusion proteins were prepared and phosphorylated with purified Cam kinase II as described previously (19). GST proteins included the following Smad2 sequences; Smad2-WT, residues 1 to 467; Smad2-110-WT, glycine-100 to valine-117; Smad2-227-WT, asparagine-215 to glutamine-239; Smad2-240-WT, glycine-230 to histidine-259; Smad2-260-WT, threonine-243 to tyrosine-268; and Smad2-465-WT, valine-300 to serine-467. For two-dimensional phosphoamino acid analysis, proteins were transferred to polyvinylidene difluoride (PVDF) membrane, excised, hydrolyzed in 6 M HCl (110°C, 90 min), and separated in the presence of 5 μg of unlabeled standard phosphoamino acids on a 20-by-20-cm thin-layer cellulose plate using the HTLE-7000 system (CBS Scientific). The plate was then dried, sprayed with ninhydrin to visualize phosphoamino acid standards, and exposed to X-ray film.
Phosphopeptide antisera.
Rabbits were immunized with a phosphoserine-containing peptide (E-T-S-D-Q-Q-L-N-Q-S-M-D-T-C; corresponding to sequence containing serine-240 of human Smad2) coupled to keyhole limpet hemocyanin carrier protein. Antisera were adsorbed with GST-Smad2 to remove any peptide-specific antibodies and then immunoaffinity purified on a Sepharose column containing the original phosphopeptide immunogen prepared using Sulpholink-Sepharose (Promega).
Immunofluorescence and nuclear localization assays.
Cos-1 cells were transfected on glass coverslips with 2 μg of pCMV1-Smad2-GFP together with 2 μg of empty pCMV1 vector or pCMV1-Cam kinase II1–290 as indicated in the figure legends. The cells were starved in 0.5% fetal calf serum and treated with TGF-β as indicated. Cells were then fixed in 3% paraformaldehyde, permeabilized with 0.1% Triton X-100, fluorescence visualized with mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole), and examined by confocal microscopy. Cotransfection with Cam kinase II1–290 was confirmed with anti-Cam kinase II antibody and tetramethyl rhodamine isocyanate (TRITC) secondary antibody.
Immunoprecipitation and immunoblotting.
HEK-293 cells were transiently transfected with expression constructs for Smads alone or in combination with the constitutively active type I TGF-β receptor and Cam kinase II. Cells were lysed at 4°C with 1 ml of lysis buffer (50 mM HEPES [pH 7.5] containing 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, and 50 mM sodium fluoride). Clarified lysates were then incubated with 10 μg of anti-HA or anti-FLAG M2 monoclonal antibodies, together with 20 μg of polyclonal rabbit anti-mouse IgG and 30 μl of prewashed protein A-Sepharose. After 2 h at 4°C, immunoprecipitates were washed four times with 1 ml of wash buffer (lysis buffer plus 0.1% Triton X-100). Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and exposed to film (Kodak X-Omat). For immunoblotting, proteins were transferred onto nitrocellulose and probed with anti-HA, anti-FLAG, or anti-GFP antibodies that were visualized using the ECL System (Amersham).
RESULTS
Inhibition of TGF-β-dependent transcriptional activity by Ca2+ and Cam kinase II.
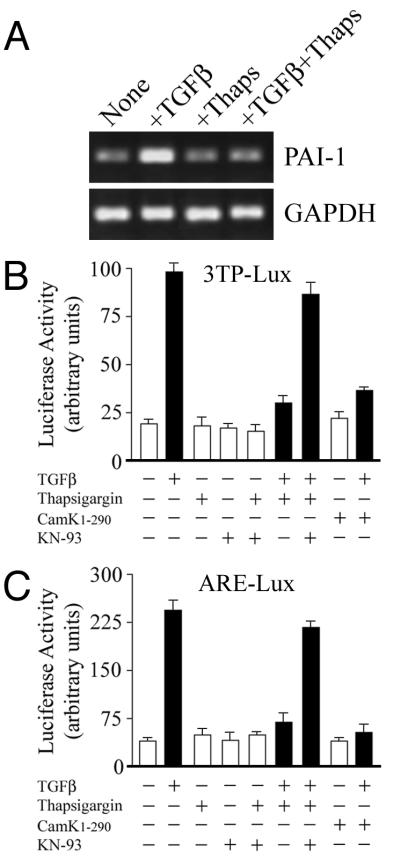
Receptor tyrosine kinases (RTKs), such as the EGF and PDGF receptor families, activate multiple signaling pathways. RTKs can also raise cytoplasmic Ca2+ levels following the recruitment of phospholipase Cγ and subsequent breakdown of phosphoinositides to inositol trisphosphate, causing release of Ca2+ from intracellular stores (reviewed in reference 38). In this study, we have investigated whether both Ca2+ signaling and Cam kinase II activated in response to RTK signaling could affect TGF-β-dependent responses. Thapsigargin, a reagent that inhibits the Ca2+-ATPase pump to raise intracellular Ca2+, completely abolished TGF-β induction of the endogenous plasminogen-activator inhibitor 1 (PAI-1) gene in Cos-1 cells (Fig. 1A). A similar response was found following transfection of Cos-1 cells with the 3TP-lux reporter, which contains TGF-β response elements for both PAI-1 and collagen type 1 linked to luciferase expression. TGF-β increased luciferase activity three- to fourfold, and this effect was completely blocked by pretreatment with thapsigargin (Fig. 1B). To confirm that this inhibition was mediated through endogenous Cam kinase II, we used a specific water-soluble inhibitor, KN-93. In the presence of KN-93, thapsigargin-mediated inhibition of transcription is almost completely restored (Fig. 1B). Further confirmation for the involvement of Cam kinase II in the thapsigargin-mediated inhibition was sought using a constitutively active form of this enzyme. The functional kinase activity of Cam kinase II is tightly regulated by a C-terminal autoinhibitory domain, and removal of this region in the Cam kinase II1–290 construct generates a constitutively activated form of enzyme that no longer requires Ca2+-calmodulin for activation (14). Cam kinase II1–290 blocked 3TP-lux reporter activation to an extent similar to that of thapsigargin (Fig. 1B). Since the 3TP-lux promoter contains three AP-1 sites, we also examined whether Cam kinase II1–290 could prevent activation of the more specific Smad–FAST-1-restricted ARE-lux reporter. ARE-lux contains DNA sequences derived from the Xenopus Mix.2 gene that convey TGF-β–activin responsiveness through complexes of Smads and FAST-1 (12). In transfected Cos-1 cells, TGF-β-induced luciferase activity through ARE-lux approximately fourfold, and this response was again completely abolished by thapsigargin and to a greater extent by coexpression with Cam kinase II1–290 (Fig. 1C). It is unlikely that Cam kinase II1–290 is mediating this effect by overall inhibition of cellular transcriptional machinery, since other promoters, such as the fos AP-1-like element can be stimulated by the same constitutively active enzyme (3). The thapsigargin-mediated inhibition could also again be reversed by the Cam kinase II inhibitor KN-93 (Fig. 1C). In control experiments, treatment of cells with KN-93 and/or thapsigargin had no effect on basal 3TP-lux and ARE-lux transcriptional activity in the absence of TGF-β (Fig. 1B and C).
FIG. 1.
Inhibition of TGF-β-dependent transcriptional responses by Cam kinase II. (A) The expression of the PAI-1 gene was monitored in Cos-1 cells by semiquantitative PCR as described in Materials and Methods. In some instances cells were pretreated with 1 μM thapsigargin in DMSO and then stimulated with 5 ng of TGF-β per ml for 15 h. Expression of GAPDH was monitored as an internal control. Cos-1 cells were transiently transfected with 2 μg of pCMV1-Cam kinase II1–290 or empty pCMV1, together with 2 μg of 3TP-lux reporter plasmid (B), or 2 μg of ARE-lux reporter plasmid (C). In some instances cells were pretreated with 1 μM thapsigargin in DMSO and then stimulated with 5 ng of TGF-β per ml for 15 h (black bars) as indicated. Some cells were also pretreated for 1 h with the specific Cam kinase II inhibitor, KN-93, at a concentration of 10 μM. Control cells, in the absence of TGF-β, were also treated with either KN-93 or KN-93 and thapsigargin.
Phosphorylation of Smad proteins by Cam kinase II.
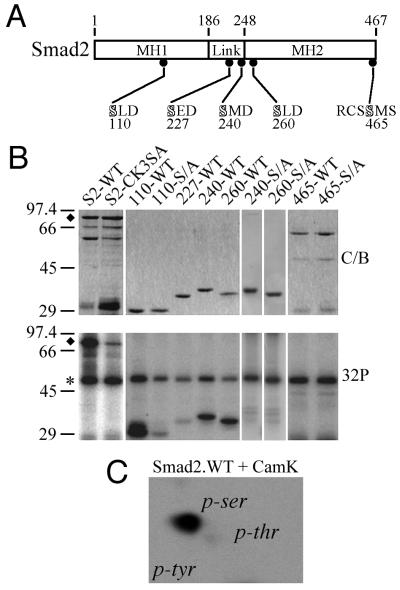
Consensus phosphorylation sites for Cam kinase II are RXXS/T, and one of these at serine-465 overlaps with a site of phosphorylation by the type I TGF-β receptor kinase (Fig. 2A). We have also recently identified an alternative Cam kinase II consensus motif, SXD, that shows a preference for a hydrophobic amino acid at the X position (20). Four SXD consensus sites are found in Smad2 at serine-110, -227, -240, and -260, and three of these fulfill the requirement for a hydrophobic residue (Fig. 2A). These sites are clustered in or around the linker region, and an additional site is found in the middle of the MH1 domain at serine-110 (Fig. 2A). Bacterially expressed GST fusion proteins spanning these consensus sites were prepared and used as in vitro substrates for purified Cam kinase II. Full-length Smad2 is efficiently phosphorylated by Cam kinase II, and this was prevented by the mutation of serine-110, -240, and -260 to alanine in the Smad2-CK3SA construct (Fig. 2B). Shorter fusion proteins representing individual sites revealed that serine-110, -240, and -260 are sites for Cam kinase II, whereas serine-465 within the RXXS consensus and serine-227 are not phosphorylated (Fig. 2B). The phosphorylation of serines-110, -240, and -260 was confirmed by individual mutation of these sites to alanine in the shorter GST fusion proteins, which were no longer targeted by Cam kinase II in vitro (Fig. 2B). Phosphorylation of Smad2 by Cam kinase II in vitro occurs only on serine residues, as determined by two-dimensional phosphoamino acid analysis (Fig. 2C).
FIG. 2.
In vitro mapping of Cam kinase II phosphorylation sites in Smad2. (A) Schematic representation of Smad2 highlighting the linker region and the highly conserved MH1 and MH2 domains. The location and sequence of putative RXXS and SXD consensus Cam kinase II sites are indicated. (B) Smad2-GST fusion proteins were prepared and phosphorylated in vitro with Cam kinase II as described in Materials and Methods. Samples were separated by SDS–12.5% PAGE, and gels were either stained with Coomassie blue (C/B) or dried and exposed to X-ray film (32P). The migration of autophosphorylated Cam kinase II (∗) and full-length Smad2-GST (⧫) are indicated. (C) Separation of phosphoamino acids in GST-S2-WT (residues 1 to 467) phosphorylated with Cam kinase II. GST-S2-WT was separated by SDS–12.5% PAGE, blotted onto PVDF membrane, and acid hydrolyzed, and phosphoamino acids were separated by thin-layer chromatography (TLC). The migration of the following phosphoamino acids is indicated: phosphoserine (p-ser), phosphothreonine (p-thr), and phosphotyrosine (p-tyr).
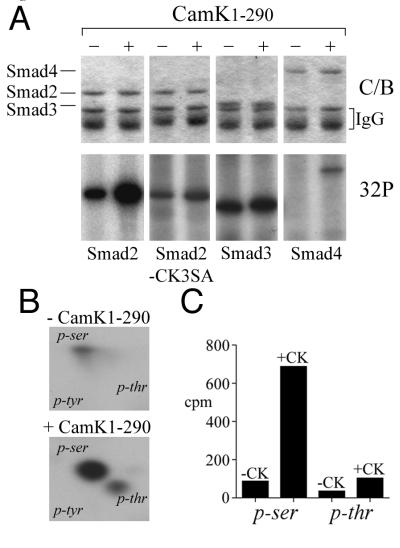
We then examined whether Cam kinase II1–290 could phosphorylate any of the pathway-restricted Smads in vivo by coexpressing epitope-tagged Smad constructs in HEK-293 cells metabolically labeled with [32P]orthophosphate. Immunoprecipitated HA-Smad2 shows a basal degree of phosphorylation that increases approximately fivefold in the presence of Cam kinase II1–290 (Fig. 3A). Phosphorylation of HA-Smad3 increases about twofold, and FLAG-Smad4, which is nonphosphorylated in the basal state, is also an in vivo substrate for Cam kinase II, although this is relatively weak compared to Smad2, and the significance of this requires further study (Fig. 3A). Mutation of serine-110, -240, and -260 to alanine in Smad2-CK3SA abolishes most of the in vivo phosphorylation by Cam kinase II1–290 (Fig. 3A). Smad2 also appears to be the preferred substrate, and two-dimensional phosphoamino acid analysis indicates that basal phosphorylation occurs predominantly on serine with some phosphorylation of threonine and that Cam kinase II1–290 significantly increases both phosphoserine and to a lesser extent phosphothreonine (Fig. 3B). Quantitation indicates that Cam kinase II1–290 increases phosphoserine by about 10-fold and phosphothreonine by approximately 3-fold and that, overall, there is 10-fold more phosphoserine (Fig. 3C). These data contrast with the presence of only phosphoserine in vitro (Fig. 2C) and suggest that Cam kinase II might also activate other kinases in vivo that can target Smad2 at specific threonines. These data might also explain why Smad2 phosphorylation is not completely prevented by the mutation of serine-110, -240, and -260 to alanine in the Smad2-CK3SA mutant and that this could be due to residual threonine phosphorylation (Fig. 3A).
FIG. 3.
Smads are substrates for Cam kinase II in vivo. HEK-293 cells were transfected with HA-Smad2, HA-Smad2-CK3SA, HA-Smad3, or FLAG-Smad4 in the presence or absence of Cam kinase II1–290 and metabolically labeled with [32P]-orthophosphate. Cell lysates were immunoprecipitated either with anti-flag M2 antibody or anti-HA antibody. (A) Immunoprecipitates were separated by SDS–10% PAGE and stained with Coomassie blue (C/B) to detect total Smad protein, or phosphoproteins were visualized by autoradiography (32P). (B) HA-Smad2 immunoprecipitates were blotted onto PVDF, HA-Smad2 proteins acid were hydrolyzed, and phosphoamino acids were separated by TLC. The migration of the following phosphoamino acids is indicated: phosphoserine (p-ser), phosphothreonine (p-thr), and phosphotyrosine (p-tyr). (C) Radioactive phosphoamino acids were scraped from the TLC plate and quantitated by Cerenkov counting.
Multiple growth factor receptors induce downstream phosphorylation of Smad2 at serine-240.
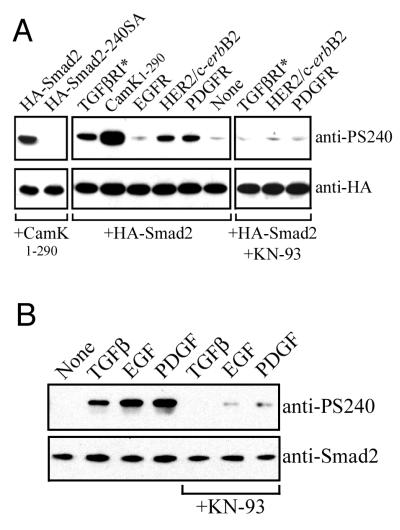
To monitor the phosphorylation status of the Cam kinase II sites in vivo, we next attempted to generate phosphopeptide-specific antisera against all three sites using Smad2 synthetic peptide immunogens. Initial screening of the resulting antisera indicated that both phosphoserine-110- and phosphoserine-260-containing peptide antisera reacted with both phosphorylated and nonphosphorylated Smad2 (data not shown). However, immunoaffinity-purified antisera raised against the phosphoserine-240-containing peptide reacted exclusively with Smad2 when coexpressed with constitutive Cam kinase II1–290 (Fig. 4A). Mutation of serine-240 to alanine in Smad2-240SA completely abolished immunoreactivity against the anti-PS240 antiserum (Fig. 4A). We then used this antiserum to examine whether other growth factor receptors could signal downstream to induce phosphorylation of the negative regulatory site at serine-240 in Smad2. In this instance, 293-HEK cells were cotransfected with HA-Smad2, together with the full-length EGF receptor, or chimeric receptors comprising the extracellular domain of the EGF receptor connected to the respective PDGF receptor (EP-R) or HER2/HER-1/2 intracellular domains. These chimeras have a functional EGF ligand binding domain and mediate cytoplasmic responses that reflect the nature of the intracellular domain (27, 39). Both PDGF receptor and HER2 coexpression induced a significant level of phosphorylation at serine-240 (Fig. 4A). The EGF receptor induced weaker phosphorylation at serine-240 and, interestingly, a significant degree of phosphorylation was seen in the presence of the constitutively active type I TGF-β receptor (Fig. 4A). Phosphorylation at serine-240 in Smad2 by the kinase domains of HER2, TGF-β receptor, and PDGF receptor was prevented by the selective Cam kinase II inhibitor KN-93, indicating that this event was mediated through endogenous Cam kinase II in HEK-293 cells (Fig. 4A). We then treated HepG2 cells with the growth factors TGF-β, EGF, and PDGF to examine whether cells expressing physiological levels of receptors for these ligands could induce phosphorylation of serine-240 on endogenously expressed Smad2. All three ligands induced a significant degree phosphorylation of serine-240 in Smad2, although in this system we found that the response with EGF was more pronounced, which may reflect differences in cell type characteristics or differential receptor expression levels (Fig. 4B). Importantly, this growth factor-mediated phosphorylation could be inhibited by KN-93, indicating that the response was likely to be mediated through downstream activation of endogenous Cam kinase II (Fig. 4B).
FIG. 4.
Phosphorylation of serine-240 occurs downstream of multiple growth factor receptors. (A) HA-tagged Smad2 was expressed in HEK-293 cells in the presence or absence of Cam kinase II1–290 and immunoprecipitated with anti-HA, and samples were probed by Western blotting against anti-HA or anti-PS240 antibodies. Equivalent expression levels were monitored in whole lysates by anti-HA Western blotting. Expression of Cam kinase II1–290 was confirmed by using specific antisera (data not shown). HA-tagged Smad2 was also expressed in HEK-293 cells in the presence or absence of the constitutively active type I TGF-β receptor (TGFβRI*), the human EGF receptor (EGFR), or chimeric receptors comprising the extracellular domain of the EGF receptor connected to the respective PDGF (EP-R) or HER2 (HER-1/2) intracellular domains. The EGF receptor and chimeric receptors were activated with 100 ng of EGF per ml for 30 min immediately prior to lysis. Samples were immunoprecipitated with anti-HA, and samples were probed by Western blotting against anti-HA or anti-PS240 antibodies. Equivalent expression levels were monitored by anti-HA blotting, and the expression of receptors was confirmed with specific antisera (not shown). In some instances, cells were pretreated for 1 h with the specific Cam kinase II inhibitor, KN-93, at a concentration of 10 μM. (B) HepG2 cells were stimulated for 30 min with the indicated growth factors. In some instances, cells were pretreated for 1 h with the specific Cam kinase II inhibitor, KN-93, at a concentration of 10 μM. Cells were lysed, and samples were probed by Western blotting against Smad2 or PS-240.
Nuclear translocation of Smad2 is prevented by Cam kinase II.
We then examined whether Cam kinase II might affect the subcellular localization of Smad2 analogous to Erk-MAP kinase preventing Smad1 and Smad2 nuclear translocation (24, 25). GFP-Smad2 was expressed in Cos-1 cells, and approximately 60% of transfected cells showed nuclear localized Smad2 following TGF-β stimulation relative to 13% in unstimulated controls (Fig. 5A). Overexpression of Cam kinase II1–290 prevented GFP-Smad2 from localizing to the nucleus (Fig. 5A). A similar, or slightly enhanced, degree of TGF-β-induced nuclear translocation was seen with Smad2-CK3SA in which Cam kinase II sites at serine-110, -240, and -260 are mutated to alanine; however, this was not prevented by overexpression of Cam kinase II1–290 (Fig. 5B).
FIG. 5.
Cam kinase II regulates the subcellular localization of Smad2-GFP. Smad2-GFP fusion proteins were expressed in Cos-1 cells, grown in 0.5% fetal calf containing medium, and stimulated 48 h posttransfection with TGF-β for 15 h. The percentage of Smad2 localized to the nucleus was determined by counting 100 immunofluorescence-positive cells and is denoted in the top left corner of each image. Cos-1 cells were also transfected with Smad2-GFP, together with Cam kinase II1–290, and the percentage of nuclear-positive cells was assessed as described above only in cells expressing both the Smad and Cam kinase. Cotransfection was confirmed with primary Cam kinase II mouse monoclonal antibody (anti-Cam kinase II) and secondary TRITC-conjugated goat anti-mouse antibody. (A) Wild-type Smad2. (B) Smad2-CK3SA.
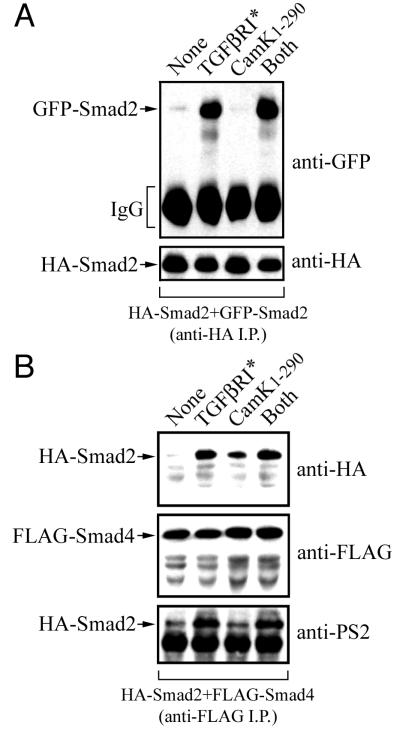
Cam kinase II induces Smad2-Smad4 interactions independently of TGF-β receptor activation while preventing TGF-β-dependent Smad2-Smad3 interactions.
Receptor-mediated phosphorylation of the C-terminal serines in pathway-restricted Smads by the TGF-β receptor kinase is required for transcriptional activity and facilitates both homo-oligomerization and the hetero-oligomerization of Smad2 with Smad3 and Smad4 (23). We therefore examined whether Cam kinase II could disrupt these interactions by coexpressing combinations of HA-, FLAG-, or GFP-tagged Smads in HEK-293 cells, together with the constitutively active type I TGF-β receptor or Cam kinase II1–290. The formation of HA-Smad2 and GFP-Smad2 homo-oligomers was identified by immunoprecipitation with anti-HA, and the induction of this interaction in the presence of the constitutively active type I TGF-β receptor was not affected by Cam kinase II1–290 (Fig. 6A). The HA-Smad2–FLAG-Smad4 hetero-oligomer was identified by anti-FLAG immunoprecipitation and is induced in the presence of the constitutively active type I TGF-β receptor (Fig. 6B). However, surprisingly, we found that Cam kinase II1–290 alone also significantly induces Smad2-Smad4 hetero-oligomerization (Fig. 6B). Similar results were obtained when Smad4 was coexpressed with Smad2-CK3SA, indicating that this effect was not due to phosphorylation at serine-110, -240, and -260 in Smad2 (data not shown). We then probed the anti-FLAG immunoprecipitates with a phosphopeptide-specific anti-PS2 antibody raised against the C-terminal TGF-β receptor phosphorylation sites in Smad2 (35). Anti-PS2 reacts only with Smad4-associated Smad2 when coexpressed with the constitutively active type I TGF-β receptor and does not react with Smad4-associated Smad2 when expressed with Cam kinase II1–290 alone (Fig. 6B). These data confirm that this induction of Smad2-Smad4 hetero-oligomerization by Cam kinase II1–290 is not due to activation of endogenous TGF-β receptors or to increased endogenous expression of TGF-β ligands.
FIG. 6.
Cam kinase II induces Smad2-Smad4 interactions independently of TGF-β receptor activation. (A) HEK-293 cells were transfected with N-terminally tagged HA-Smad2 and GFP-Smad2 in the presence or absence of the constitutively active type I TGF-β receptor (TGFβR*), constitutively active Cam kinase II (Cam kinase II1–290), or both (TGFβR* plus Cam kinase II1–290). Cell lysates were immunoprecipitated with anti-HA, and samples were probed by immunoblotting against anti-HA or anti-GFP antibodies. Expression of the TGF-β receptor and Cam kinase II was confirmed using specific antisera (data not shown). (B) HEK-293 cells were transfected with N-terminally tagged HA-Smad2 and C-terminally tagged FLAG-Smad4 in the presence or absence of the constitutively active type I TGF-β receptor (TGFβR*), constitutively active Cam kinase II (Cam kinase II1–290), or both (TGFβR* plus Cam kinase II1–290). Cell lysates were immunoprecipitated with anti-FLAG, and samples were probed by immunoblotting against anti-HA, anti-FLAG, or anti-PS2 antibodies. Expression of the TGF-β receptor and Cam kinase II was confirmed by using specific antisera (data not shown).
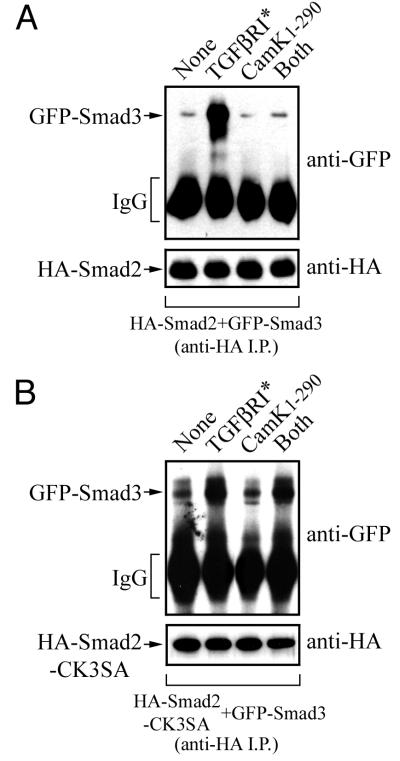
We then examined whether Smad2 and Smad3 hetero-oligomerization was affected by coexpressing HA-Smad2 and GFP-Smad3 in HEK-293 cells and monitoring interactions in anti-HA immunoprecipitates. The constitutively active type I TGF-β receptor induced the formation of Smad2-Smad3 hetero-oligomers; however, Cam kinase II1–290 completely prevented TGF-β-induced Smad2-Smad3 hetero-oligomerization (Fig. 7A). We then performed the same experiment with Smad3 and Smad2-CK3SA and found that the loss of interaction in the presence of Cam kinase II1–290 was restored (Fig. 7B). This indicates that the Smad2-Smad3 interaction can be directly regulated by Cam kinase II-dependent phosphorylation at serine-110, -240, and -260 in Smad2.
FIG. 7.
TGF-β-dependent Smad2-Smad3 interactions are prevented by Cam kinase II. HEK-293 cells were transfected with N-terminally tagged HA-Smad2 (A) or HA-Smad2-CK3SA (B), together with GFP-Smad3, in the presence or absence of the constitutively active type I TGF-β receptor (TGFβR*), constitutively active Cam kinase II (Cam kinase II1–290), or both (TGFβR* plus Cam kinase II1–290). Cell lysates were immunoprecipitated with anti-HA, and samples were probed by immunoblotting against anti-HA or anti-GFP antibodies. Expression of the TGF-β receptor and Cam kinase II was confirmed using specific antisera (data not shown).
DISCUSSION
Smads play an essential role in transmitting activin, BMP, and TGF-β signals from the cell surface to the nucleus. More recently, it is becoming apparent that Smads are also extensively regulated by direct input from a variety of receptor signaling pathways (reviewed in reference 30). In the present study, we have shown that Smad function can be directly controlled by cytoplasmic Ca2+-dependent kinases, specifically the ubiquitously expressed Ca2+-calmodulin-dependent Cam kinase II. These observations are also consistent with the recent report that Ca2+-calmodulin can regulate Smad function (48).
The exact mechanism by which phosphorylation at serine-110, -240, and -260 in Smad2 inhibits Smad–TGF-β-dependent gene transcription is likely to be complex. Important features of this process are likely to involve the location of Cam kinase II phosphorylation sites within critical Smad functional domains. The recent crystal structure of the conserved MH1 domain of Smad3 indicates that the main DNA-binding motif is an 11-residue β hairpin that is embedded within the major groove of DNA (40). Although Smad2 is not able to bind to DNA directly through this motif, its effector function in the conserved MH2 domain can be repressed by direct contact with the MH1 domain (21). The Cam kinase II site in Smad2 at serine-110 is on the edge of the β hairpin motif, and it is interesting to speculate that phosphorylation at this site could inhibit Smad effector function by stabilizing repressive interactions between the MH1 and the MH2 domains. Similarly, these interactions could also be affected by Cam kinase II phosphorylation at the serine-260 that is within the conserved MH2 domain. The third phosphorylation site at serine-240 lies within the linker region and is immediately adjacent to several Ser/Thr-Pro sites that are targets for the Ras/Erk/MAP kinase pathway (25). Phosphorylation at these sites also represses Smad2 function, and it is possible that the stoichiometry of Erk-MAP kinase modification could be potentiated by Cam kinase targeting of the adjacent serine-240 site.
The prevention of TGF-β-dependent Smad2 nuclear translocation by Cam kinase II represents an important mechanism for the control of Smad–TGF-β signaling. Recently, the presence of a distinct nuclear localization signal in the MH1 of Smad3 was shown to determine the ligand-induced nuclear translocation of Smad3 (43). Phosphorylation of Smad2 by Cam kinase II could mask this domain or, alternatively may affect interactions with proteins that regulate the export of transcription factors from the nucleus such as the nuclear export factor CRM-1 (31). In this context of the regulation of Smad2 nuclear import and export it is also important to consider the normal distribution and subcellular localization of Cam kinases. In the present study, we have used a constitutively active truncated form of the neuronal Cam kinase II-α that does not contain a nuclear localization sequence but is sufficiently small in size to be able to diffuse freely between the nucleus and cytoplasm. Multifunctional Cam kinases comprise a multigene family consisting of Cam kinase I, Cam kinase II, and Cam kinase IV, which display a common substrate specificity and are widely distributed in mammalian tissue (reviewed in reference 5). The Cam kinase II subfamily is derived from four related genes, namely, α, β, γ, and δ. The αB, δB, and γA isoforms share a common core nuclear localization sequence, and nuclear localization in vivo of αB and δB has been reported (7). Nuclear and cytoplasmic isoforms can be found in most tissues, although some regions of the brain (7) and also the heart (17) express predominantly nuclear forms of Cam kinase II. These observations raise the possibility that, under normal physiological circumstances, the degree of regulation of Smad signaling by Cam kinase and the manner in which this might be achieved is likely to be very tissue specific. In tissues that express cytoplasmic isoforms of Cam kinase II, activation of this enzyme would block TGF-β-dependent gene responses by preventing entry of the Smad transcriptional complex into the nucleus. Alternatively, in those tissues expressing nuclear forms of Cam kinase II, the TGF-β-dependent assembly and translocation from the cytoplasm would be unaffected. However, once inside the nucleus activated Cam kinase II could target the Smad complex to disrupt the Smad2-Smad3 interaction and directly affect gene responses in situ. The essential role of Smad3 in efficient Smad–TGF-β-dependent transcriptional activity has been highlighted by a number of recent gene knockout studies (4, 44, 47). Our results also suggest that inhibition of TGF-β-dependent transcriptional responses described in the present study could be explained in part by the fact that Cam kinase II prevents the TGF-β-dependent recruitment of Smad3 into the Smad transcriptional complex.
Recent accumulating evidence has also suggested a role for Ca2+ in controlling some of the actions of TGF-β (2, 22). The type I TGF-β receptor interacts with the immunophilin FKBP12 (13), a ubiquitously expressed protein that can inhibit the Ca2+-dependent protein phosphatase calcineurin (28). FKBP12 and calcineurin also control cytoplasmic calcium levels by stabilizing the inositol 1,4,5-trisphosphate and ryanodine receptors (6, 9). Interestingly, we find that activation of the TGF-β receptor can also induce the downstream phosphorylation of the regulatory Cam kinase II site at serine-240 in Smad2. This raises the possibility that this may form part of an intrinsic negative feedback inhibitory loop in which cells receive a pulsed signal input through the Smad–TGF-β system. Ca2+ signals can occur as single transients, as oscillations, or as a sustained plateau, and this triggers the differential activation and/or inhibition of downstream mediators to determine signal response, strength, and specificity (16). Cam kinase II is also thought to be a major decoder of Ca2+ oscillation since its kinase activity reflects the size and frequency of Ca2+ spikes (15). In this regard the TGF-β-dependent mobilization of intracellular Ca2+ in some cell types requires incubation times of more than 1 h (32), whereas Ca2+ mobilization can be instantaneous in response to RTK signaling. Therefore, differences in amplitude, duration, and the scheduled delivery of various growth factor stimuli would be critical in determining the final outcome of the biological response. In this regard, it will be of interest in future studies to thoroughly analyze the time course and concentration dependence of various growth factor agonists by utilizing both the anti-PS240 (inactive Smad2) and anti-PS2 (active Smad) antisera.
We have also shown that enhanced downstream Cam kinase II signaling in response to RTK-induced Ca2+ mobilization can upregulate phosphorylation at serine-240 in Smad2. These observations have important implications for normal cellular homeostasis, cellular proliferation, and also neoplastic transformation. TGF-β is a classical suppressor of cell growth, and disruption of components of the TGF-β signaling cascade, including Smads, has been demonstrated in several types of cancer. Permanent coupling of oncogenic HER2 to phospholipase Cγ activity and Ca2+ mobilization is also seen in tumor cells (34). The HER2 gene is amplified in a variety of human adenocarcinomas arising at a number of sites, including the breast, colon, and stomach (reviewed in reference 33). Therefore, the present data showing increased phosphorylation of the negative regulatory site (serine-240) in Smad2 by overexpression of the chimeric HER-1/2 protein kinases raises the intriguing possibility that an important feature of HER2-mediated oncogenic transformation could involve the shutdown of the TGF-β–Smad tumor-suppressive pathway.
In summary, we have defined a novel role for Cam kinase II activation in the regulation and differential control of Smad–TGF-β function. The molecular detail of this inhibition incorporates phosphorylation at consensus sites in Smad2, disruption of Smad protein complex formation, and effects on Smad subcellular localization. We propose that costimulatory input from multiple growth factors leading to a differential Ca2+ mobilization provides a potent cross-talk and intrinsic feedback program that will ultimately coordinate complex patterns of TGF-β-dependent gene regulation and cellular responsiveness. An important goal will be to correlate differential expression patterns and cytosolic-nuclear compartmentalization of members of the Cam kinase family with the tissue-specific integration of growth factor signals. Furthermore, it will be important to address the complex issue of spatial and temporal Ca2+ mobilization in response to TGF-β and other environmental stimuli, particularly in the context of recent reports of the direct control of specific gene transcription by nuclear Ca2+ (37). Finally, the phosphopeptide antisera generated in this study should permit a detailed analysis of the importance of Smad2 transmodulation following oncogenic RTK overexpression in human cancer.
ACKNOWLEDGMENTS
We thank Tony Means, Hans-Hermann Gerdes, Carl-Henrik Heldin, Jeffrey Wrana, Scott Kern, Rik Derynck, Malcolm Whitman, and Axel Ullrich for kindly providing cDNA constructs and Peter ten Dijke for the PS2 antiserum. We also thank Dylan Edwards and Simak Ali for helpful discussions.
This study was supported by Wellcome Trust grant 045206/Z/95/Z (A.C.), The Special Trustees of Charing Cross Hospital (A.C.), and a Livingstone Studentship from Imperial College (S.J.W.).
REFERENCES
- 1.Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana J L. TGFβR1 phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 2.Alevizopoulos A, Dusserre Y, Ruegg U, Mermod N. Regulation of the transforming growth factor β-responsive transcription factor CTF-1 by calcineurin and calcium/calmodulin-dependent protein kinase IV. J Biol Chem. 1997;272:23597–23605. doi: 10.1074/jbc.272.38.23597. [DOI] [PubMed] [Google Scholar]
- 3.Antoine M, Gaiddon C, Loeffler J P. Ca2+/calmodulin kinase types II and IV regulate c-fos transcription in the AtT20 corticotroph cell line. Mol Cell Endocrinol. 1996;120:1–8. doi: 10.1016/0303-7207(96)03806-3. [DOI] [PubMed] [Google Scholar]
- 4.Ashcroft G S, Yang X, Glick A B, Weinstein M, Letterio J J, Mizel D E, Anzano M, Greenwell-Wild T, Wahl S M, Deng C, Roberts A B. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat Cell Biol. 1999;1:260–266. doi: 10.1038/12971. [DOI] [PubMed] [Google Scholar]
- 5.Braun A P, Schulman H. The multifunctional calcium/calmodulin-dependent protein kinase: from form to function. Annu Rev Physiol. 1995;57:417–445. doi: 10.1146/annurev.ph.57.030195.002221. [DOI] [PubMed] [Google Scholar]
- 6.Brillantes A B, Ondrias K, Scott A, Kobrinsky E, Ondriasova E, Jayaraman T, Landers M, Ehrlich B E, Marks A R. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell. 1994;77:513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 7.Brocke L, Srinivasan M, Schulman H. Developmental and regional expression of multifunctional Ca2+/calmodulin-dependent protein kinase isoforms in rat brain. J Neurosci. 1995;15:6797–6808. doi: 10.1523/JNEUROSCI.15-10-06797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J D, DiChiara M R, Anderson K R, Gimbrone M A, Jr, Topper J N. MEKK-1, a component of the stress (stress-activated protein kinase/c-jun N-terminal kinase) pathway, can selectively activate Smad2-mediated transcriptional activation in endothelial cells. J Biol Chem. 1999;274:8797–8805. doi: 10.1074/jbc.274.13.8797. [DOI] [PubMed] [Google Scholar]
- 9.Cameron A M, Steiner J P, Roskams A J, Ali S M, Ronnett G V, Snyder S H. Calcineurin associated with the inositol 1,4,5-trisphosphate receptor-FKBP12 complex modulates Ca2+ flux. Cell. 1995;83:463–472. doi: 10.1016/0092-8674(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 10.Chantry A. The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J Biol Chem. 1995;270:3068–3073. [PubMed] [Google Scholar]
- 11.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Samd4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y-G, Liu F, Massagué J. Mechanism of TGFβ receptor inhibition by FKBP12. EMBO J. 1997;16:3866–3876. doi: 10.1093/emboj/16.13.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruzalegui F H, Kapiloff M S, Morfin J-P, Kemp B E, Rosenfeld M G, Means A R. Regulation of intrasteric inhibition of the multifunctional calcium/calmodulin-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:12127–12131. doi: 10.1073/pnas.89.24.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Koninck P, Schulman H. Sensitivity of Cam kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 16.Dolmetsch R E, Lewis R S, Goodnow C C, Healy J I. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 17.Edman C F, Schulman H. Identification and characterization of delta B-CaM kinase and delta C-CaM kinase from rat heart, two new multifunctional Ca2+/calmodulin-dependent protein kinase isoforms. Biochim Biophys Acta. 1994;1221:89–101. doi: 10.1016/0167-4889(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 18.Engel M E, McDonnell M A, Law B K, Moses H L. Interdependent SMAD and JNK signaling in transforming growth factor-β-mediated transcription. J Biol Chem. 1999;274:37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 19.Feinmesser R L, Gray K, Means A R, Chantry A. HER-2 is phosphorylated by calmodulin-dependent protein kinase II on a single site in the cytoplasmic tail at threonine-1172. Oncogene. 1996;12:2725–2730. [PubMed] [Google Scholar]
- 20.Feinmesser R L, Wicks S, Taverner C, Chantry A. Ca2+/calmodulin-dependent protein kinase II phosphorylates the epidermal growth factor receptor on multiple sites in the cytoplasmic tail and serine-744 within the kinase domain to regulate signal generation. J Biol Chem. 1999;274:16168–16173. doi: 10.1074/jbc.274.23.16168. [DOI] [PubMed] [Google Scholar]
- 21.Hata A, Lo R S, Wotton D, Lagna G, Massagué J. Mutations increasing autoinhibition inactivate tumour supressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 22.Ishiyama N, Shibata H, Kanzaki M, Shiozaki S, Miyazaki J, Kobayashi I, Kojima I. Calcium as a second messenger of the action of transforming growth factor-beta on insulin secretion. Mol Cell Endocrinol. 1996;117:1–6. doi: 10.1016/0303-7207(95)03726-8. [DOI] [PubMed] [Google Scholar]
- 23.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGFβ family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 25.Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFβ/Smad signaling by oncogenic ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labbé E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Dull T J, Lax J, Schlessinger J, Ullrich A. HER2 cytoplasmic domain generates normal mitogenic and transforming signals in a chimaeric receptor. EMBO J. 1989;8:167–173. doi: 10.1002/j.1460-2075.1989.tb03361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Farmer J D, Jr, Lane W S, Friedman J, Weissman I, Schreiber S L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 29.Massagué J. TGFβ signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 30.Massagué J, Chen Y-G. Controlling TGF-β signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 31.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 32.Muldoon L L, Rodland K D, Magun B E. Transforming growth factor beta and epidermal growth factor alter calcium influx and phosphatidyl turnover in rat-1 fibroblasts. J Biol Chem. 1988;263:18834–18841. [PubMed] [Google Scholar]
- 33.Peles E, Yarden Y. Neu and its ligands: from oncogene to neural factors. Bioessays. 1993;15:815–824. doi: 10.1002/bies.950151207. [DOI] [PubMed] [Google Scholar]
- 34.Peles E, Ben-Levy R, Or E, Ullrich A, Yarden Y. Oncogenic forms of the neu/HER2 tyrosine kinase are permanently coupled to phospholipase C gamma. EMBO J. 1991;10:2077–2086. doi: 10.1002/j.1460-2075.1991.tb07739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engström U, Heldin C-H, Funa K, ten Dijke P. The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
- 36.Piek E, Heldin C-H, ten Dijke P. Specificity, diversity, and regulation in TGF-β superfamily signaling. FASEB J. 1999;13:2105–2124. [PubMed] [Google Scholar]
- 37.Rogue P J, Malviya A N. EMBO workshop report: calcium signals in the cell nucleus. EMBO J. 1999;18:5147–5152. doi: 10.1093/emboj/18.19.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlessinger J, Ullrich A. Growth factor signaling by receptor tyrosine kinases. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 39.Seedorf K, Felder S, Millauer B, Schlessinger J, Ullrich A. Analysis of platelet-derived growth factor receptor domain function using a novel chimeric receptor approach. J Biol Chem. 1991;266:13828–13833. [PubMed] [Google Scholar]
- 40.Shi Y, Wang Y-F, Jayaraman L, Yang H, Massagué J, Pavletich N P. Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA binding in TGFβ signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 41.Souchelnytski S, Tamaki K, Engström U, Wernstedt C, ten Dijke P, Heldin C-H. Phosphorylation of Ser465 and Ser467 in the C-terminus of Smad2 mediates interaction with Smad4 and is required for TGFβ signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 42.Wrana J L, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGFβ receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 43.Xiao Z, Liu X, Henis Y I, Lodish H F. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc Natl Acad Sci USA. 2000;97:7853–7858. doi: 10.1073/pnas.97.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X, Letterio J J, Lechleider R J, Chen L, Hayman R, Gu H, Roberts A B, Deng C. Targeted disruption of Smad3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-β. EMBO J. 1999;18:1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 46.Zhou S, Zawel L, Lengauer C, Kinzler K W, Vogelstein B. Characterisation of human FAST-1, a TGFβ and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Y, Richardson J A, Parada L F, Graff J M. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 48.Zimmerman C M, Kariapper M S T, Mathews L S. Smad proteins physically interact with calmodulin. J Biol Chem. 1998;278:677–680. doi: 10.1074/jbc.273.2.677. [DOI] [PubMed] [Google Scholar]