Abstract
We describe experiments to compare the activities of two Drosophila homeodomain proteins, Bicoid (Bcd) and an altered-specificity mutant of Fushi tarazu, Ftz(Q50K). Although the homeodomains of these proteins share a virtually indistinguishable ability to recognize a consensus Bcd site, only Bcd can activate transcription from natural enhancer elements when assayed in both yeast and Drosophila Schneider S2 cells. Our analysis of chimeric proteins suggests that both the homeodomain of Bcd and sequences outside the homeodomain contribute to its ability to recognize natural enhancer elements. We further show that, unlike the Bcd homeodomain, the Ftz(Q50K) homeodomain fails to recognize nonconsensus sites found in natural enhancer elements. The defect of a chimeric protein containing the homeodomain of Ftz(Q50K) in place of that of Bcd can be preferentially restored by converting the nonconsensus sites in natural enhancer elements to consensus sites. Our experiments suggest that the biological specificity of Bcd is determined by combinatorial contributions of two important mechanisms: the nonconsensus site recognition function conferred by the homeodomain and the cooperativity function conferred primarily by sequences outside the homeodomain. A systematic comparison of different assay methods and enhancer elements further suggests a fluid nature of the requirements for these two Bcd functions in target selection.
An important question in molecular biology concerns the specificity of the actions of regulatory proteins such as transcription factors. This question is particularly important for homeodomain-containing proteins not only because of the vital biological roles they play but also because of their special properties in DNA recognition. A homeodomain is an evolutionarily conserved 60-amino-acid domain found in many proteins that control a wide spectrum of essential biological processes, ranging from mating type specification in Saccharomyces cerevisiae to embryonic pattern formation in animals (21, 48). The diverse and specific biological functions conferred by homeodomain proteins seemingly contrast with their DNA binding properties. Most homeodomain proteins bind to short DNA sequences of only 6 bp, often with a common TAAT core followed immediately by two bases that confer specificity (28, 51, 52). In addition, a number of homeodomain proteins can bind to similar DNA sequences in vitro but exhibit different biological functions in vivo (6, 14, 22, 24, 35).
Previous studies have suggested that the biological specificity of homeodomain proteins may come from at least two distinct sources: DNA binding and transcription control (6). Cooperative binding to multiple sites within an enhancer element can increase the DNA binding specificities of homeodomain proteins, thus increasing their target selectivity (4, 37). DNA binding cofactors can also increase the DNA binding activities of homeodomain proteins, further increasing their DNA binding specificities and selectivity (10, 11, 54, 55, 61, 62). More recent studies suggest that regulatory cofactors can further increase the biological specificity of homeodomain proteins by modulating their ability to activate or repress transcription (31, 32). However, individual homeodomain proteins are likely to utilize their own unique combinations of strategies governing their biological specificities, each requiring systematic experimental analyses.
Bicoid (Bcd), a Drosophila homeodomain protein, controls the development of the anterior structures in early embryos by activating target genes required for embryonic pattern formation (5, 15, 17, 20, 42). Several target genes that directly respond to Bcd function in the embryo have been identified (17, 43), including hunchback (hb) and knirps (kni), which are the earliest zygotic genes activated by maternally derived Bcd (1, 29, 39). Bcd represents an important family of related proteins that contain a signature lysine residue at the 50th position of their homeodomains (referred to as K50 homeodomains). Previous studies suggest the 50th position of a homeodomain plays a critical role in determining DNA binding specificity (26, 51); this residue is located within the homeodomain's third helix, which makes most of the specific contacts with DNA (23). Like Bcd, members of the K50 homeodomain family also play an important role in development in various organisms. For example, the mammalian pituitary homeobox protein (Pitx2) is involved in determining left-right asymmetry during embryonic development, and mutations in Pitx2 cause human Reiger syndrome (34, 41, 44, 49, 64). However, relatively little is known about the molecular mechanisms governing proper target selection by members of this important family of homeodomain proteins. The study described in this report was designed to help understand how Bcd selects its natural targets for transcription control.
In this study, we take advantage of a derivative of another Drosophila homeodomain protein, Ftz(Q50K), which has a glutamine-to-lysine change at the 50th position of the homeodomain of Ftz (Fushi tarazu protein) (40). Previous studies have shown that, despite its Bcd-like DNA binding specificity in vitro (40), Ftz(Q50K) fails to activate natural Bcd targets in Drosophila (47). We hypothesized that Ftz(Q50K) lacks important function(s) that are conferred by Bcd and required for proper target selection. By analyzing chimeric proteins generated from Bcd and Ftz(Q50K) in both yeast and Drosophila Schneider S2 cells, we demonstrate that both the homeodomain of Bcd and sequences outside the homeodomain contribute to its ability to recognize natural targets. We further show that, unlike the Bcd homeodomain, the Ftz(Q50K) homeodomain fails to recognize nonconsensus DNA sites found in the natural enhancer elements. In addition, the defect of a chimeric protein containing the homeodomain of Ftz(Q50K) in place of that of Bcd is preferentially restored when all the nonconsensus sites in natural enhancer elements are converted to consensus sites. Our results suggest that proper target selection by Bcd is facilitated combinatorially by two important functions of Bcd: recognition of nonconsensus sites by the homeodomain and a cooperative DNA binding function conferred primarily by the sequences outside the homeodomain. We also describe a systematic comparison of different assay methods, which reveals differential requirements for Bcd sequences and their conferred functions in target selection.
MATERIALS AND METHODS
Plasmid construction.
The plasmids used are listed in Tables 1 and 2.
TABLE 1.
Plasmids used in transcriptional-activation studies
| Gene | Plasmid
|
|||
|---|---|---|---|---|
| Insect vector | Yeast vector | Notes | Source | |
| Reporter genes | ||||
| hb-CAT | pCZ3005 | This study | ||
| kni-CAT | pCZ3006 | This study | ||
| hb(6A)-CAT | pCZ3007 | This study | ||
| kni(6A)-CAT | pCZ3008 | This study | ||
| hb-lacZ | pMA630R | 16 | ||
| kni-lacZ | pTA123 | This study | ||
| hb(6A)-lacZ | pCZ3004 | This study | ||
| kni(6A)-lacZ | pTA170 | This study | ||
| lexAOP-lacZ | pJP167 | J. Pearlberg | ||
| hb-lacZ(2μm) | pCZ3015 | This study | ||
| kni-lacZ(2μm) | pCZ3017 | This study | ||
| hb(6A)-lacZ(2μm) | pCZ3016 | This study | ||
| kni(6A)-lacZ(2μm) | pCZ3018 | This study | ||
| Activator genes | ||||
| HA-Bcd | pFY403 | This study | ||
| HA-Ftz(Q50K) | pCZ2079 | This study | ||
| HA-Ftz(Q50K)-VP16 | pCZ2087 | This study | ||
| HA-Bcd-Ftz(Q50K)HD-VP16 | pCZ2088 | This study | ||
| HA-Ftz-BcdHD-VP16 | pCZ2089 | This study | ||
| HA-Bcd-VP16 | pCZ2090 | This study | ||
| LexA-Bcd | pCZ99 | LEU2 marker | This study | |
| LexA-Ftz(Q50K) | pCZ97 | LEU2 marker | This study | |
| LexA-Bcd-Ftz(Q50K)HD | pCZ100 | LEU2 marker | This study | |
| LexA-Ftz-BcdHD | pCZ2038 | LEU2 marker | This study | |
| Bcd-VP16 | pMA1226 | LEU2 marker | 38 | |
| Ftz(Q50K)-VP16 | pCZ2035 | LEU2 marker | This study | |
| Bcd-Ftz(Q50K)HD-VP16 | pCZ2036 | LEU2 marker | This study | |
| Ftz-BcdHD-VP16 | pCZ2037 | LEU2 marker | This study | |
TABLE 2.
Plasmids used in biochemical studies
| Gene | Plasmid | Notes | Source |
|---|---|---|---|
| Bcd site | |||
| kni enhancer | pCZ72 | This study | |
| hb enhancer | pMAX1 | 37 | |
| hb(6A) enhancer | pCZ3003 | This study | |
| For homeodomain expression | |||
| GST-BcdHD | pCZ10 | 13 | |
| GST-Ftz(Q50K)HD | pCZ57 | 13 | |
| For in vitro translation | |||
| LexA-Bcd | pCZ95 | SP6 promoter | This study |
| LexA-Ftz(Q50K) | pCZ90 | SP6 promoter | This study |
| LexA-Bcd-Ftz(Q50K)HD | pCZ96 | SP6 promoter | This study |
| LexA-Ftz-BcdHD | pCZ2029 | SP6 promoter | This study |
| Bcd | BcdTN3 | SP6 promoter | 16 |
| Ftz(Q50K) | pFY1002 | SP6 promoter | This study |
| Bcd-Ftz(Q50K)HD | pFY1005 | SP6 promoter | This study |
| Ftz-BcdHD | pCZ2034 | SP6 promoter | This study |
(i) Constructs for in vitro translation. The DNA fragment encoding the Ftz(Q50K) sequence was generated by a PCR-mediated mutagenesis procedure using pActftz (25) as the template. The resulting PCR product was cloned into the EcoRI site of pMA1222 (37) to generate pCZ90. The replacement of the Bcd homeodomain by the Ftz(Q50K) homeodomain involved a multistep process. Briefly, StuI and BclI sites were first introduced into the ends of the Bcd homeodomain in pCZ95 by PCR-mediated mutagenesis. The Ftz(Q50K) homeodomain sequence was amplified by PCR from pCZ90 and inserted into the StuI-BclI site of pCZ95 to create pCZ96. PCZ2029 was constructed from pCZ95 by replacing the sequences flanking the Bcd homeodomain progressively with the corresponding Ftz sequences. BcdTN3 was described previously (16). pFY1002, pFY1005, and pCZ2029 were derived from pHB6 (18) with the frog globin mRNA leader upstream of each coding sequence.
(ii) Activator gene constructs for expression in yeast and S2 cells. The LexA fusion expression vectors used in yeast study were based on a Leu2 2μm plasmid, AAH5 (2). The DNA fragments containing LexA-Bcd, LexA-Ftz(Q50K), LexA-Bcd-Ftz(Q50K)HD, and LexA-Ftz-BcdHD were taken as HindIII-HindIII fragments from pCZ95, pCZ90, pCZ96, and pCZ2029, respectively, and cloned into AAH5 to generate pCZ99, pCZ97, pCZ100, and pCZ2038. The VP16 fusion expression plasmids were also based on AAH5, and the VP16 acidic activation domain was attached to the carboxyl terminus of Bcd or Ftz(Q50K) sequence. pFY403 bears the gene that encodes a modified Bcd protein with a hemagglutinin (HA) tag (MAYPYDVPDYAH) fused to its fourth codon. pFY403 was generated from pAc5.1/V5-HisC (Invitrogen), and the expression of the Bcd protein was controlled by the constitutive Drosophila actin 5c promoter, pCZ2079, which expresses an HA-tagged Ftz(Q50K) protein, was constructed with the Ftz(Q50K) coding sequence PCR amplified from pCZ97. pCZ2087, pCZ2088, pCZ2089, and pCZ2090 contain HA-tagged coding sequences for Ftz(Q50K)-VP16, Bcd-Ftz(Q50K)HD-VP16, Ftz-BcdHD-VP16, and Bcd-VP16, respectively.
(iii) Reporter genes. pTA123 was constructed by two steps. First, a 64-bp kni enhancer element was isolated as a KpnI-XhoI fragment from pKni-128 (kindly provided by H. Jackle) and cloned into pBluescript KS(−) (Stratagene) to generate pTA115. pTA123 was then produced by inserting a 300-bp PvuII-XhoI fragment from pTA115 into pLR1Δ1Δ2μ (59). pMA630R was described previously (38). The conversion from hb to hb(6A) was carried out in a stepwise manner by PCR-mediated mutagenesis using pMAX1 as the original template (37): X3s was initially mutated to a consensus site in pTA119; subsequently, X1 and X2 were converted to consensus sites in pCZ3003. Both pTA119 and pCZ3003 are bacterium vectors derived from pBluescript KS(−). To construct pCZ3004, an XhoI-XbaI (Klenow filled-in) fragment from pCZ3003 was inserted into the XhoI-SmaI fragment of pLR1Δ1Δ2μ. The kni(6A) sequence was generated by annealing two complementary 72-bp oligonucleotides (Operon). The resulting duplex was cloned into the EcoRI site of pGEM-7Zf(−) (Promega) to create pTA156. To construct pTA170, the kni(6A) element was excised from pTA156 as an XhoI-SmaI fragment and inserted into the XhoI-SmaI fragment of pLR1Δ1Δ2μ. pJP167, provided by J. Pearlberg, contains two LexA sites upstream of GAL1-lacZ reporter genes and was described previously (38). All the reporter genes carried on the replicating plasmids pCZ3015, pCZ3016, pCZ3017, and pCZ3017 are based on pLR1Δ1 (59). The chloramphenicol acetyltransferase (CAT) reporter gene constructs were generated from pG1-TATA- CAT, which contains the adenovirus E1b TATA box upstream of the CAT gene (33). To construct pCZ3005, the hb enhancer element was isolated from pMAX-1 as an XhoI-XbaI (Klenow filled-in) fragment and cloned into the XhoI-XbaI (Klenow filled-in) fragment of pG1-TATA-CAT. pCZ3006 was constructed by inserting a 300-bp PvuII-XhoI fragment from pTA115 into the XhoI-XbaI (Klenow filled-in) fragment of pG1-TATA-CAT. pCZ3007 and pCZ3008 contain modified hb(6A) and kni(6A) enhancer elements, respectively, upstream of the CAT reporter gene.
(iv) Other constructs. pCZ10 and pCZ57, which express Bcd and Ftz(Q50K) homeodomains in bacteria, are described elsewhere (13). pCZ72 contains the kni enhancer element placed between the KpnI site and the BamHI site of PGEM-7Zf(−). pMAX1 contains the hb enhancer element located between the HindIII and BamHI sites (37).
Immunoprecipitation.
All the proteins were expressed from the Sp6 promoter and generated in a TNT coupled reticulocyte system according to the instructions of the manufacturer (Promega). The relative amount of each protein generated was estimated by evaluating the incorporation of [35S]methionine. Immunoprecipitation was performed essentially as described previously (37). A modification was made in which protein A-Sepharose beads (Amersham Pharmacia Biotech) replaced Staphylococcus aureus cells for the precipitation step in order to lower background levels. Briefly, the proteins were incubated, precipitated with antibodies against LexA (a gift kindly provided by M. Ptashne's laboratory) and protein A-Sepharose beads, washed four times with 10×-volume wash buffer, and separated in sodium dodecyl sulfate–10% polyacrylamide gels. The gels were dried and visualized with a Molecular Dynamics PhosphorImager system. The incubation and washing was done in buffer B (10 mM Tris · Cl [pH 7.5], 1 mM EDTA, 150 mM NaCl, 0.1% NP-40) with 0.2% milk. The expression plasmids were purified using a Qiagen miniprep kit and dissolved in diethyl pyrocarbonate-treated water.
Transient-transfection experiments.
Drosophila S2 cells (Invitrogen) were grown at 25°C in DES expression medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco). The cells were seeded in 60-mm-diameter tissue culture plates at 4 × 106/plate 24 h before transfection. Transfection was performed by the calcium phosphate coprecipitation method by following a protocol from Gibco. Each plate was transfected with 1 μg of expression vector, 1 μg of reporter vector, and 1 μg of copia-lacZ internal control plasmid (12). Salmon sperm DNA (Invitrogen) was included as carrier DNA to bring the total amount of DNA to 10 μg. The cells were harvested 48 h after transfection, and cell lysates were prepared by a freeze-thaw method (3). The transfection efficiency was determined by monitoring the β-galactosidase activity, and the amounts of lysates used in the CAT assay and Western blotting were normalized accordingly. The CAT assay was performed according to the method of Ausubel et al. (3). For Western blotting, cell lysates were separated on sodium dodecyl sulfate–10% polyacrylamide gels and transferred to cellulose membranes. The membranes were blotted with an anti-HA monoclonal antibody (HA.11; Babco; 1:600 final dilution) and imaged by enhanced chemiluminescence (Amersham Pharmacia Biotech).
Homeodomain expression.
The Bcd homeodomain and the Ftz(Q50K) homeodomain were expressed in bacteria and purified as described previously (13). Aliquots of dialyzed proteins were stored at −80°C. The concentration of active homeodomain was measured by gel shift assay using 5 × 10−6 M A1 site and further confirmed by Scatchard plot analysis. The stock concentrations for the Bcd homeodomain and the Ftz(Q50K) homeodomain were 7 × 10−8 and 5 × 10−8 M, respectively.
Gel shift assays.
The radioactively labeled 122-bp kni probe and 300-bp hb probe for the gel shift assay were isolated as an XbaI-SacI fragment from pCZ72 and a HindIII-XbaI fragment from pMAX1, respectively. In our experiments, the probes were diluted to a final concentration of 6 × 10−11 M. The oligonucleotides used for making A1, X1, and X3s have been described elsewhere (13). For gel shift analysis of either purified homeodomains or in vitro translation proteins (see Fig. 3E for exceptions), the concentration of each probe was 5 × 10−9 M. Binding reactions were performed in 30 μl of BB buffer (15 mM HEPES [pH 7.5], 1 mM EDTA, 0.5 mM dithiothreitol, 40 mM KCl) containing 0.1 μg of poly(dI-dC)/μl on ice for 20 min. After the addition of 4 μl of 30% Ficoll, the samples were loaded onto 6% (for in vitro translation protein) or 8% (for purified homeodomains) native polyacrylamide gels with 0.5× Tris-borate-EDTA. The dried gels were analyzed with the PhophorImager system. For Kd measurements, poly(dI-dC) was not included in the reaction mixtures. To generate the binding curves of the Bcd homeodomain on hb and kni, site occupancy of the Bcd homeodomain at different protein concentrations was calculated as described previously (8).
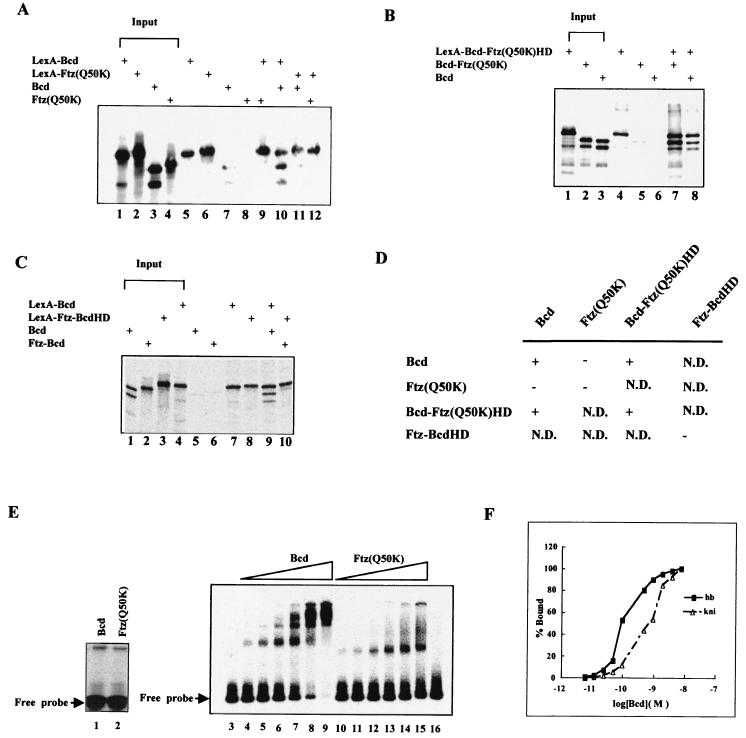
FIG. 3.
Bcd protein sequences outside the homeodomain participate in protein-protein interaction. (A to C) Shown are coimmunoprecipitation assay results. In vitro-generated and radioactively labeled proteins were incubated and then precipitated with antibodies against LexA (see Materials and Methods for details). The input represents one-fifth of the amount of proteins used in the coimmunoprecipitation assays. +, present. (D) Summary of coimmunoprecipitation assay results. +, interaction; −, no interaction; N.D., not determined. (E) Gel shift assays using in vitro-translated full-length Bcd and Ftz(Q50K). The experiments shown in lanes 3 to 6 used a modified hb enhancer element (∼1.5 × 10−11 M) with six consensus sites (Fig. 6A). Lanes 3 and 16 represent experiments with no translation lysate and 10 μl of luciferase translation lysate added, respectively. The experiments in lanes 1 and 2 were performed on a single-Bcd-site probe (∼10−8 M), permitting an estimate of the amounts of the active proteins (1 μl of translation lysates used). According to this estimate, similar amounts of active proteins were used in the experiments shown in lanes 4 to 15: 0.25, 0.5, 1, 2, 4, and 8 μl of Bcd translation lysates for lanes 4 to 9, respectively; 0.3, 0.6, 1.25, 2.5, 5, and 10 μl of Ftz(Q50K) translation lysates for lanes 10 to 15, respectively. (F) Residual cooperativity of Bcd homeodomain on hb and kni enhancer elements. Shown are binding curves of the recombinant Bcd homeodomain on the hb and kni enhancer elements measured in a gel shift assay. The occupancy of the Bcd homeodomain on DNA sites was calculated as follows: the sum of the amount of shifted complex × number of protein molecules in the complex/total number of Bcd sites on the probe (8). % Bound, fraction of maximal binding. The results in this figure are consistent with those published previously (8). The cooperativity provided by the homeodomain is very modest compared to that of full-length Bcd: it takes about a 70-fold increase in Bcd homeodomain concentration to achieve from 5 to 95% binding to the hb enhancer element, in contrast to less than a 4-fold increase for full-length Bcd to achieve similar binding (37, 63) (see panel E).
Yeast strains, β-galactosidase liquid assays, and Western blotting.
Yeast strains were generated by integrating the reporter plasmids into the URA3 locus of CY26 (matα his3Δ200 lys2-801 ura3-52 ade2-101 trp1Δ1 leu2-Δ1), a strain kindly provided by J. Peterson. The determination of copy number was performed as described previously (38). Only strains with single-copy integration were used for further experiments. The effector plasmids were introduced into the resulting yeast strains by the lithium acetate method, and at least three independent transformants were assayed for β-galactosidase activities (59). Normally, less than 20% variation between transformants was observed. For Western blotting, transformed yeast cells were grown in 50 ml of synthetic medium lacking leucine and with 2% glucose. The cells were harvested at an optical density at 600 μm of ∼0.6, washed once, and resuspended in 0.1 ml of extraction buffer (200 mM Tris-Cl [pH 8.0], 400 mM (NH4)2SO4, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride). The cells were disrupted with an equal volume of 0.45-mm-diameter glass beads, and the supernatants were collected after centrifugation. For Western blotting, the primary antibody was a rabbit anti-LexA antibody (1:600 final concentration) from M. Ptashne's laboratory, and the secondary antibody was a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad; 1:2,000 final concentration).
RESULTS
Ftz(Q50K) fails to activate transcription from natural enhancer elements.
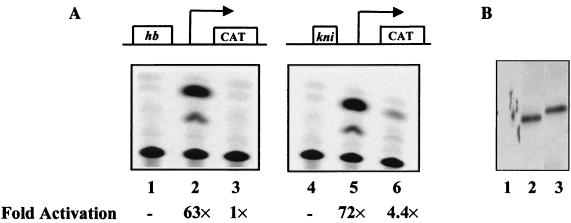
Figure 1 shows the results of our transient- transfection assays of Drosophila Schneider S2 cells analyzing the ability of Bcd and Ftz(Q50K) to activate transcription from natural enhancer elements. Two reporter genes were used in our assays, hb-CAT and kni-CAT, which contain a 250-bp enhancer element from hb and a 60-bp enhancer element from kni, respectively. Our experiments demonstrate that, unlike Bcd (Fig. 1A, lanes 2 and 5), Ftz(Q50K) fails to activate transcription efficiently from either natural enhancer element (Fig. 1A, lanes 3 and 6). This reflects a functional difference between these two proteins, because they both accumulated to similar levels in Drosophila S2 cells (Fig. 1B). These results further confirm the results of a previous study demonstrating that Ftz(Q50K), despite its ability to recognize a consensus Bcd site efficiently (40), fails to activate transcription from natural enhancer elements in Drosophila embryos (47).
FIG. 1.
Ftz(Q50K) fails to activate transcription from natural enhancer elements in Drosophila S2 cells. (A) Transient-transfection assays. Two reporter genes, hb-CAT and kni-CAT, were used in transient-transfection assays to determine the activity of HA-tagged Bcd and Ftz(Q50K). Shown are CAT assay results from cells cotransfected with effector plasmids bearing the gene encoding either no protein (lanes 1 and 4), Bcd (lanes 2 and 5), or Ftz(Q50K) (lanes 3 and 6). (B) Western blot analysis. Antibodies against HA were used in the Western blot assay. Lanes 1 to 3 represent results using Drosophila cell lysates containing no activator, Bcd, and Ftz(Q50K), respectively.
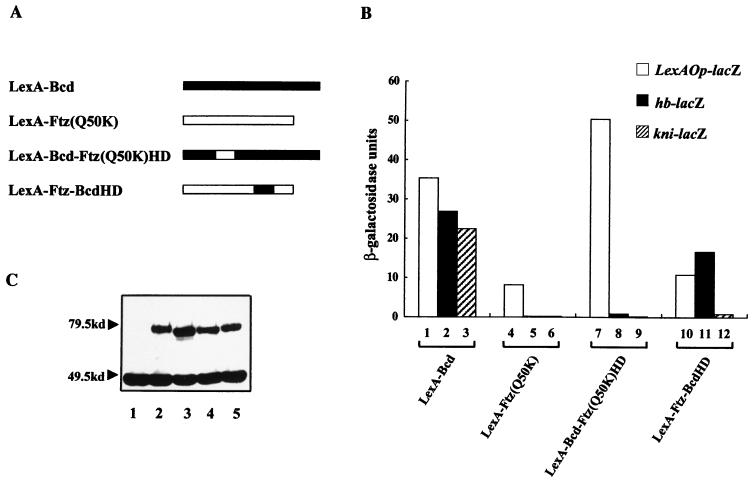
To further analyze the intrinsic properties of Bcd and Ftz(Q50K), as well as those of their chimeric derivatives (see below), we conducted transcriptional-activation assays in yeast cells (see the description of experiments conducted in Schneider cells below). For these experiments, we used three integrated single-copy lacZ reporter genes containing upstream either the hb enhancer element (hb-lacZ), the kni enhancer element (kni-lacZ), or two LexA binding sites (lexAOp-lacZ). The lexAOp-lacZ reporter gene was used because it permitted an independent analysis of the activities of our proteins, which were fused to the DNA binding domain of LexA (residues 1 to 87), to activate transcription through another DNA binding specificity. Our experiments, shown in Fig. 2B, demonstrate that, unlike LexA-Bcd (lanes 2 and 3), LexA-Ftz(Q50K) fails to activate transcription from both hb and kni enhancer elements (lanes 5 and 6). The inability of LexA-Ftz(Q50K) to activate transcription from the natural enhancer elements reflects a functional defect in recognizing these enhancer elements because the protein accumulated in yeast cells (Fig. 2C, lane 2) and, more importantly, can activate transcription from LexA sites (Fig. 2B, lane 4).
FIG. 2.
Chimeric proteins assayed in yeast cells reveal important functions of Bcd. (A) Schematic diagram of the activators used in our study. The exchanged homeodomains in the two hybrid molecules are shown as open and solid boxes. The DNA binding domain of LexA is not shown in this diagram because it is present in all of the proteins. (B) Activation assay results. Different activator proteins were assayed for their abilities to activate transcription from the integrated reporter genes hb-lacZ, kni-lacZ, and LexAOp-lacZ. Shown are β-galactosidase activities obtained from these assays. (C) Western blot assay using antibodies against LexA. Lanes 1 to 5 represent results using yeast cell lysates containing no activator, LexA-Ftz(Q50K), LexA-Bcd, LexA-Bcd-Ftz(Q50K)HD, and LexA-Ftz-BcdHD, respectively. A nonspecific band at ∼46 kDa, which has been reported previously (46), can be used as an internal control.
Multiple regions of Bcd are required for efficient natural target gene selection.
To identify defects associated with Ftz(Q50K) and thus important functions conferred by Bcd for natural-target selection, we generated and tested the following two chimeric proteins: LexA-Bcd-Ftz(Q50K)HD, containing the homeodomain of Ftz(Q50K) with the rest of the protein sequences from Bcd, and LexA-Ftz-BcdHD, containing the homeodomain of Bcd within the Ftz framework (Fig. 2A). Our transcriptional-activation experiments (Fig. 2B) demonstrate that multiple regions of Bcd, including both the homeodomain and sequences outside the homeodomain, contribute to efficient activation from the natural enhancer elements. First, unlike LexA-Bcd (lane 3), both chimeric proteins fail to support efficient activation from the kni enhancer element (lanes 9 and 12). In addition, LexA-Bcd-Ftz(Q50K)HD fails to activate transcription from the hb enhancer element (lane 8). Interestingly, transcriptional activation by both chimeric proteins from the kni enhancer element is affected more severely than that from the hb enhancer element (Fig. 2B, lanes 8 and 9 and lanes 11 and 12; also see below). The decreased activity observed with these proteins reflects their inability to efficiently recognize the natural enhancer elements, because all our proteins accumulated to comparable levels in cells (Fig. 2C, lanes 4 and 5) and can activate transcription from LexA sites (Fig. 2B, lanes 7 and 10).
Protein sequences of Bcd, but not Ftz(Q50K), outside their homeodomains confer protein-protein interaction function.
Our previous studies suggested that protein sequences of Bcd flanking its homeodomain facilitate efficient cooperative DNA recognition through a direct interaction between Bcd molecules (37, 63). To determine whether sequences of Ftz outside its homeodomain can confer a similar protein-protein interaction function, we conducted a coimmunoprecipitation assay (Fig. 3A). We also analyzed the two chimeric proteins described above in the coimmunoprecipitation assay (Fig. 3B and C). All proteins were generated and radioactively labeled in an in vitro translation system. For each experiment, two proteins, one of which was fused to LexA, were incubated and precipitated using antibodies against LexA (see Materials and Methods for details).
Figure 3D summarizes our coimmunoprecipitation results. As reported previously (63), Bcd molecules can interact with each other in the coimmunoprecipitation assay (Fig. 3A, lane 10, and C, lane 9). However, Ftz(Q50K) molecules fail to interact with each other in the same assay (Fig. 3A, lane 12). Ftz(Q50K) also fails to interact with a Bcd molecule (Fig. 3A, lane 9). In addition, the analysis of various combinations of derivatives further demonstrates that the protein interaction function of Bcd is associated with the sequences outside its homeodomain (summarized in Fig. 3D). Most notably, Bcd-Ftz(Q50K)HD, which contains the homeodomain of Ftz(Q50K) in place of that of Bcd, can self-associate (Fig. 3B, lane 7) and can interact with Bcd (Fig. 3B, lane 8). Taken together, our experiments identify a self-association defect of Ftz(Q50K) and Ftz-BcdHD, suggesting that their inability to activate efficiently from natural enhancer elements is caused, at least in part, by such a defect (see below).
To further determine the importance of the protein-protein interaction function in cooperative DNA recognition, we carried out a DNA binding assay using in vitro-translated full-length Bcd and Ftz(Q50K) proteins on an enhancer element with consensus Bcd sites. Our results (Fig. 3E) show that, at low concentrations, both proteins bind to the enhancer element similarly (lanes 3 and 4 and 10 to 12). In contrast, the binding profiles for these proteins are dramatically different at higher concentrations. While Bcd quickly forms complexes with multiple protein molecules and depletes the free probe (Fig. 3E, lanes 7 to 9), Ftz(Q50K) forms only smeary complexes in an incremental manner and fails to deplete the free probe (lanes 13 to 15). These results demonstrate that, unlike Bcd, which shows a highly cooperative binding profile (37), Ftz(Q50K) fails to bind to the enhancer element cooperatively. Together, our experiments suggest that cooperativity facilitated by the self-association function of Bcd, but not Ftz(Q50K), contributes to proper target selection.
Differential requirements for Bcd sequences in recognizing different enhancers.
Our experiments (Fig. 2B) reveal an interesting difference between the requirements of the hb and kni enhancer elements for Bcd sequences. Most strikingly, LexA-Ftz-BcdHD, which contains the homeodomain of Bcd with the rest of the sequences from Ftz, can activate transcription from the hb enhancer element efficiently (Fig. 2B, lane 11). In contrast, this protein is virtually nonfunctional from the kni enhancer element (lane 12). Although the primary protein-protein interaction and cooperative DNA binding functions of Bcd are conferred by sequences outside its homeodomain (37, 63) (Fig. 3E), the homeodomain can provide a residual cooperative DNA binding function (Fig. 3F) (8). Such residual cooperativity is more evident on the hb enhancer element than on the kni enhancer element (Fig. 3F). We propose that this difference is responsible for the reduced dependence on Bcd sequences outside its homeodomain in recognizing the hb enhancer element.
The homeodomain of Bcd, but not Ftz(Q50K), can recognize both consensus and nonconsensus sites.
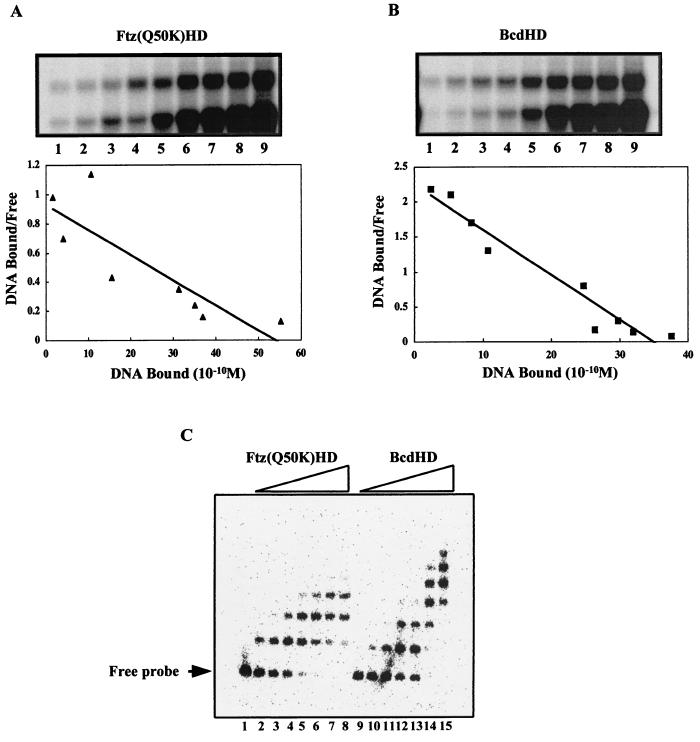
To further understand the contributions of the homeodomain to the ability of Bcd to activate transcription from natural enhancer elements, we analyzed in more detail the DNA binding properties of the homeodomains of Bcd and Ftz(Q50K). It has been shown that the Ftz(Q50K) homeodomain can recognize a consensus Bcd site, TAATCC, with high affinity (40). Our measurements suggest that both the Bcd and Ftz(Q50K) homeodomains bind to this DNA sequence with comparable affinities (Fig. 4A and B). The estimated Kd values for the interactions of the Bcd and Ftz(Q50K) homeodomains with a consensus Bcd site were 2.3 ± 0.6 × 10−10 and 5.5 ± 1.1 × 10−10 M, respectively. However, when these homeodomains were analyzed for their ability to recognize the natural kni enhancer element in a gel shift assay, fewer Bcd sites appeared to be occupied by the Ftz(Q50K) homeodomain than by the Bcd homeodomain (Fig. 4C).
FIG. 4.
Bcd and Ftz(Q50K) homeodomains have comparable affinities to a consensus Bcd site but bind to the kni enhancer element differently. (A and B) Gel shift assays for a Scatchard analysis to determine the affinities of recombinant Bcd and Ftz(Q50K) homeodomains for the consensus site A1. The DNA concentrations used in these analyses were 6 × 10−11, 1.2 × 10−10, 1.8 × 10−10, 2.4 × 10−10, 6.0 × 10−10, 1.2 × 10−9, 1.8 × 10−9, 2.4 × 10−9, and 4.8 × 10−9 M for lanes 1 to 9, respectively. The Kd values given in the text represent averages of results of three independent assays. (C) Results of gel shift assays of recombinant Bcd and Ftz(Q50K) homeodomains on the natural kni enhancer element. At low concentrations, the bindings of these proteins appear similar (lanes 2 to 3 and 10 to 11). However, at higher concentrations of the proteins, complexes containing more protein molecules were obtained for the Bcd homeodomain than for the Ftz(Q50K) homeodomain (lanes 7 to 8 and 14 to 15). See the text for further details. Since the kni enhancer element contains all nonconsensus sites, we do not know exactly which of them is recognized by the Ftz(Q50K) homeodomain. Two potential candidate sites in the kni enhancer element deviate from the consensus site only modestly because they still contain a TAAT core: TAATCG and TAATCT. The Bcd homeodomain exhibits little cooperativity on the kni enhancer element (Fig. 3F); therefore, the protein-DNA complexes observed here represent primarily progressive independent recognition of different Bcd sites.
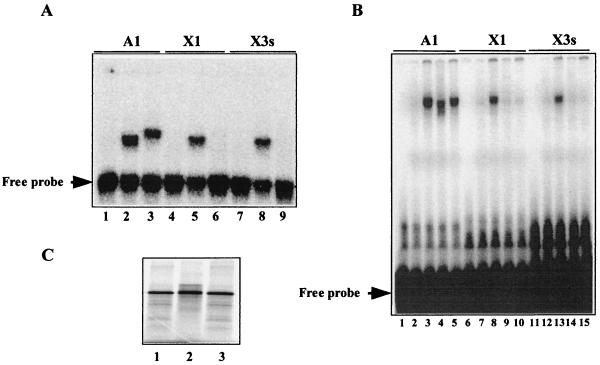
Both kni and hb enhancer elements contain Bcd sites that deviate from the TAATCC consensus, including sequences that do not have a TAAT core. Our experiments (Fig. 4C) suggest that the Ftz(Q50K) homeodomain may be defective in recognizing nonconsensus Bcd sites. To test this idea directly, we conducted gel shift experiments investigating the abilities of the homeodomains of Bcd and Ftz(Q50K) to bind to different individual sites. In our assay, we chose three different types of naturally occurring Bcd sites: TAATCC (A1), TAAGCT (X1), and TGATCC (X3s). While A1 represents a consensus site, X1 and X3s represent nonconsensus sites, each lacking a TAAT core. Our gel shift experiments (Fig. 5A) show that the Bcd homeodomain can bind to all three sites efficiently (lanes 2, 5, and 8). In contrast, the Ftz(Q50K) homeodomain can bind efficiently only to A1 (lane 3) but undetectably to X1 and X3s (lanes 6 and 9) in the same assay. Interestingly, the protein-DNA complex containing the Ftz(Q50K) homeodomain has a mobility different from that of the complex containing the Bcd homeodomain; similar mobility differences between different homeodomains have been reported previously (60).
FIG. 5.
Nonconsensus sites are recognized by the Bcd homeodomain but not the Ftz(Q50K) homeodomain. Shown are gel shift assay results of either recombinant homeodomains (A) or in vitro-translated full-length proteins (B) on three different types of Bcd sites. A1 contains a consensus sequence, TAATCC; X1 and X3s contain nonconsensus sequences, TAAGCT and TGATCC, respectively. For the experiments shown in panel A, the proteins used were none (lanes 1, 4, and 7), the Bcd homeodomain (lanes 2, 5, and 8), and the Ftz(Q50K) homeodomain (lanes 3, 6, and 9). For the experiments shown in panel B, the proteins used were none (lanes 1, 6, and 11), control lysate with luciferase translated (lanes 2, 7, and 12), lysate containing LexA-Bcd (lanes 3, 8, and 13), lysate containing LexA-Ftz(Q50K) (lanes 4, 9, and 14), and lysate containing LexA-Bcd-Ftz(Q50K)HD (lanes 5, 10, and 15). (C) Full-length proteins generated in an in vitro translation system. Lanes 1 to 3, LexA-Bcd, LexA-Ftz(Q50K), and LexA-Bcd-Ftz(Q50K)HD, respectively. See Materials and Methods for further details.
To confirm that the Bcd and Ftz(Q50K) homeodomains confer their respective individual site specificities to full-length proteins that had been used in our activation assays (Fig. 2), we conducted gel shift experiments using the following proteins: LexA-Bcd, LexA-Ftz(Q50K), and LexA-Bcd-Ftz(Q50K)HD. Our experiments (Fig. 5B) demonstrate that LexA-Ftz(Q50K) and LexA-Bcd-Ftz(Q50K)HD, both of which contain the homeodomain of Ftz(Q50K), fail to bind to X1 and X3s (lanes 9, 10, 14, and 15) despite their normal ability to bind to the consensus site A1 (lanes 4 and 5). Together, our results show that, unlike the Bcd homeodomain, the Ftz(Q50K) homeodomain is unable to efficiently recognize nonconsensus sites found in natural enhancer elements.
Modified enhancer elements preferentially restore activity to a chimeric protein containing the Ftz(Q50K) homeodomain.
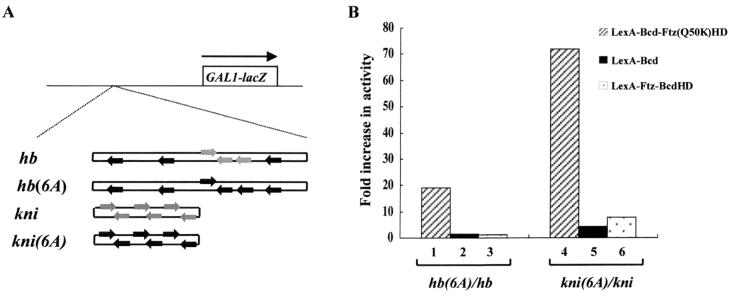
We hypothesized that the failure of LexA-Ftz(Q50K) and LexA-Bcd-Ftz(Q50K)HD to activate transcription from natural enhancer elements is caused, at least in part, by their inability to recognize nonconsensus sites in these enhancer elements. To test this hypothesis, we generated two modified enhancer elements by converting all the nonconsensus sites to consensus sites [hb(6A)-lacZ and kni(6A)-lacZ (Fig. 6A)]. Our transcriptional-activation experiments demonstrate that the activity of LexA-Bcd-Ftz(Q50K)HD, which contains the homeodomain of Ftz(Q50K) and is unable to recognize nonconsensus sites, is dramatically and preferentially increased on the modified enhancer elements (Fig. 6B, lanes 1 and 4). In contrast, the activity of LexA-Bcd (Fig. 6B, lanes 2 and 5) and LexA-Ftz-BcdHD (lanes 3 and 6), both containing the homeodomain of Bcd, is only modestly increased on the modified enhancer elements. These results demonstrate that the inability of the Ftz(Q50K) homeodomain to recognize nonconsensus sites is one underlying defect of LexA-Bcd-Ftz(Q50K)HD in activating transcription from natural enhancer elements. Our results also show that LexA-Ftz(Q50K) remains inactive from these modified enhancer elements (Fig. 6), suggesting that, in addition to its inability to recognize nonconsensus sites, this protein has other functional defects, e.g., inability to self-associate and to bind DNA cooperatively (Fig. 3A and E).
FIG. 6.
Modified enhancer elements preferentially restore the activity of a defective protein. (A) Reporter genes hb and kni represent natural enhancer elements, whereas hb(6A) and kni(6A) are modified enhancer elements with all nonconsensus sites (shaded arrows) converted to consensus sites (solid arrows). (B) Ratios of the β-galactosidase activities obtained from the hb(6A)-lacZ and kni(6A)-lacZ reporters to those from the hb-lacZ and kni-lacZ reporters. The experiments show that, relative to LexA-Bcd (lanes 2 and 5) and LexA-Ftz-BcdHD (lanes 3 and 6), the activity of LexA-Bcd-Ftz(Q50K)HD, which cannot recognize nonconsensus sites (Fig. 5), is preferentially restored by the modified enhancer elements (lanes 1 and 4). β-Galactosidase units from the hb-lacZ reporter were 27, <0.2, 1, and 17 for LexA-Bcd, LexA-Ftz(Q50K), LexA-Bcd-Ftz(Q50K)HD, and LexA-Ftz-BcdHD, respectively; from the kni-lacZ reporter, they were 23, <0.2, <0.2, and 0.8; from the hb(6A)-lacZ reporter, they were 42, 0.3, 19, and 19; and from the kni(6A)-lacZ reporter, they were 94, 0.3, 14, and 6.3.
A strong activation domain fails to restore activity to nonfunctional proteins.
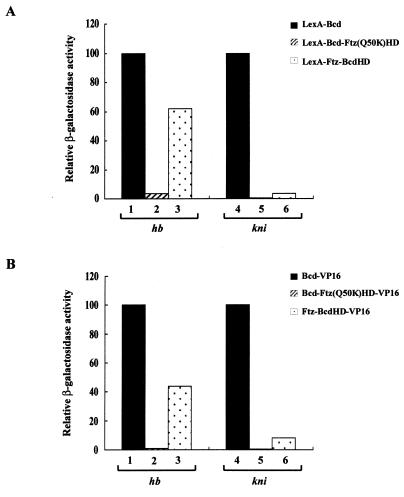
Recent studies have suggested that the activation potentials of homeodomain proteins, at least some Q50 homeodomain proteins, can help define their functional specificities. For example, it was shown that the biological function of Ultrabithorax (Ubx) was altered to mimic that of Antennapedia (Antp) by attaching to Ubx the strong activation domain VP16 (31). In addition, it has been suggested that different activation domains of a transcription factor may be utilized in different DNA binding contexts (56). To further determine whether our inactive proteins fail to activate transcription due to a lack of efficient activation functions, we fused the strong activation domain VP16 (45, 53) to them and tested their activities on different reporters. Our results show that, as expected (38), VP16 can increase the activity of Bcd on all the enhancer elements analyzed (e.g., Bcd-VP16 is 10 and 2.8 times more active than LexA-Bcd on hb-lacZ and kni-lacZ reporters, respectively). However, VP16 fails to change the relative activities of all our proteins from the natural enhancer elements (Fig. 7; compare the similar profiles of the two graphs). These experiments further support the idea that the defects associated with the inactive proteins reflect primarily their inability to recognize specific enhancer elements rather than their activation potentials (38a).
FIG. 7.
The strong activation domain VP16 fails to restore activity to inactive proteins. Shown are activities of LexA (A) and VP16 (B) fusion proteins on hb-lacZ and kni-lacZ reporters in yeast cells. The activity of LexA-Bcd and Bcd-VP16 was assigned a value of 100 for each reporter (see the legends to Fig. 6 and 8 for β-galactosidase units). A comparison of panels A and B reveals that VP16 fails to change the relative activities of other proteins, indicating that VP16 cannot restore activity to these proteins. The results further suggest that our proteins are defective in recognizing the natural enhancer elements, as opposed to lacking a functional activation domain.
Transcriptional-activation assays in Schneider cells further illustrate the importance of Bcd functions in natural-target recognition.
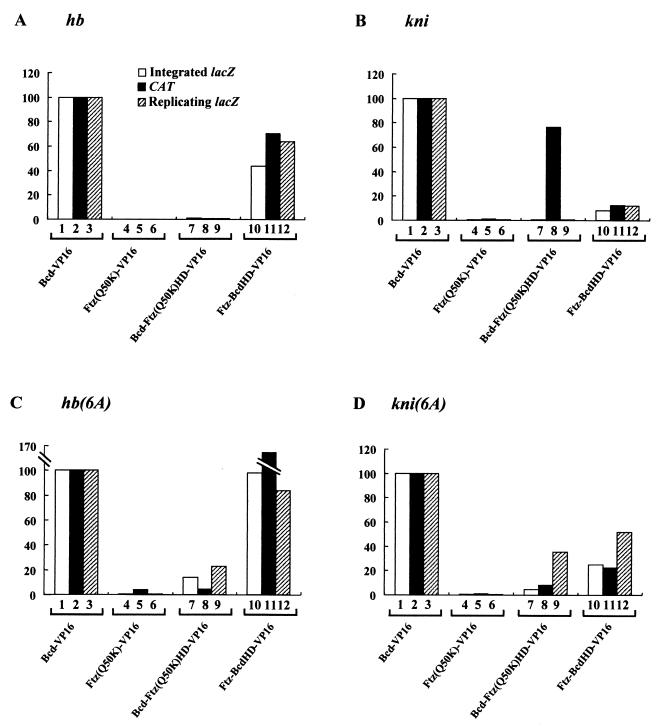
To further corroborate our findings with yeast cells, we analyzed four different proteins [Bcd-VP16, Ftz(Q50K)-VP16, Bcd-Ftz(Q50K)HD-VP16, and Ftz-BcdHD-VP16] for their abilities to activate CAT reporter genes containing either natural or modified enhancer elements in Schneider cells. All our proteins were fused to the strong activation domain VP16 in order to specifically measure their target recognition functions (see above). Our transcriptional-activation assays conducted with Schneider cells provide a strong general agreement with our findings with yeast cells (Fig. 8; compare open and solid bars) (see below for one major exception). These results further demonstrate the importance of Bcd sequences and their conferred functions in natural-target selection.
FIG. 8.
Comparison of four VP16 fusion activators in three different assay systems. Shown are activities of various VP16 fusion activators on three different reporters: integrated lacZ reporters in yeast cells, CAT reporters in Schneider cells, and lacZ reporters carried on replicating 2μm plasmids in yeast cells. For each class of reporters, four different enhancer elements were tested: the natural hb and kni enhancer elements and their modified derivatives. The comparison shows a general good agreement of the activator behaviors in different assays; see the text for discussions of major exceptions. The activity of Bcd-VP16 for each reporter was assigned a value of 100. β-Galactosidase units for this activator in yeast assays were 278, 65, 380, and 243 on integrated hb-lacZ, kni-lacZ, hb(6A)-lacZ, and kni(6A)-lacZ reporters, respectively; on the replicating reporters, they were 361, 127, 578, and 370.
Interestingly, our comparative analysis reveals one major difference between results with yeast and Schneider cells. In particular, Bcd-Ftz(Q50K)HD-VP16 activates transcription efficiently from the kni enhancer element in Schneider cells (Fig. 8B, lane 8) while it is virtually inactive in yeast cells (Fig. 8B, lane 7). We noted that our assays conducted with yeast and Schneider cells were different in terms of the reporter gene status: integrated single-copy reporter genes in yeast versus reporter genes carried on plasmids in Schneider cells. To determine whether such a difference in reporter gene status dictates the activity profiles in these two assay systems, we analyzed the four VP16 fusion activators on reporter genes that were carried on replicating plasmids in yeast. Although higher absolute activity was obtained in our plasmid reporter assays (see the legend to Fig. 8), as expected, because of the multiple copies of the reporter genes, the activity profiles are in general agreement with those obtained in single-copy integrated reporter assays (Fig. 8; compare open and shaded bars). In particular, Bcd-Ftz(Q50K)HD-VP16 remained inactive on the kni enhancer element in yeast (Fig. 8B, lane 9), suggesting that host differences can influence the requirements for Bcd sequences and their conferred functions in target selection (see below for further discussions).
DISCUSSION
Our experiments show that two K50 homeodomain proteins, Bcd and Ftz(Q50K), which have similar affinities to a consensus TAATCC site (Fig. 4), exhibit distinct abilities in mediating transcriptional activation from natural enhancer elements (Fig. 1 and 2). This observation exemplifies a puzzle underlying target selection by homeodomain proteins: why do homeodomain proteins behave differently in vivo while sharing similar or identical DNA binding specificities? We suggest that the recognition of nonconsensus sites represents an essential biochemical function that helps define biological specificity. This idea is supported by our experiments demonstrating that the activity of LexA-Bcd-Ftz(Q50K)HD, which contains the Ftz(Q50K) homeodomain and fails to bind to nonconsensus sites (Fig. 5), can be preferentially restored by converting the natural nonconsensus sites to consensus sites (Fig. 6b). Nonconsensus sites are also found in the hb enhancer elements from other fly species (7, 36). Previous studies have shown that efficient activation by homeodomain proteins requires a minimal number of recognition sites (19, 30, 38, 43), reflecting their intrinsically weak properties. Thus, nonconsensus sites found in natural enhancers, depending on their architectures (e.g., number and type of sites), are expected to either merely modulate transcription levels or act as specificity-defining elements.
Because of their critical role in mediating Bcd function, it is important to understand how nonconsensus sites are recognized by the Bcd homeodomain. Our chemical-footprint experiments with the consensus site A1 and the nonconsensus site X1 suggest that the Bcd homeodomain can establish different sets of contacts with different recognition sequences (13). Our experiments further suggest that Arg 54 of the Bcd homeodomain makes a base-specific contact with the fourth-position guanine (underlined) unique to X1 (TAAGCT). In the Ftz(Q50K) homeodomain, the 54th position contains methionine. However, an arginine residue artificially introduced in the 54th position of Ftz(Q50K) fails to confer an X1 recognition ability on the protein (13). We suggest that both the homeodomain framework and specific residues play important roles in nonconsensus-site recognition. In this context, it is interesting to note that complexes containing Ftz(Q50K) and Bcd homeodomains exhibit slightly different mobilities in electrophoresis (Fig. 5A). The analysis of several other natural K50 homeodomains further reveals that the ability to recognize all tested nonconsensus sites is unique to the Bcd homeodomain (13). We propose that the nonconsensus site recognition function of the Bcd homeodomain is a noncoincidental property that defines a unique biological specificity for Bcd.
Our present study also further underscores the importance of protein-protein interaction between Bcd molecules in natural-target selection. Such a protein interaction function, which is conferred by Bcd sequences outside its homeodomain (Fig. 3A to D), is responsible primarily for its cooperative DNA binding activity (63) (Fig. 3E). Interestingly, the hb and kni enhancer elements exhibit different requirements for the protein interaction function. In particular, Ftz-BcdHD, which contains the Bcd homeodomain in the framework of Ftz, can efficiently activate transcription from the hb enhancer element (Fig. 2B, lane 11) while it is virtually inactive on the kni enhancer element (lane 12). We propose that a residual cooperativity function conferred by the Bcd homeodomain (Fig. 3F) (8), while insufficient on the kni enhancer element, contributes to the chimeric protein's ability to recognize the hb enhancer element. We note that the hb and kni enhancer elements have architectural differences in both Bcd site composition and alignments. The hb enhancer element contains three dispersed perfect TAATCC consensus sites, in addition to at least three centrally located, tightly linked nonconsensus sites (Fig. 6A). In contrast, the kni enhancer element contains symmetrically arranged and tightly linked sites that do not match the TAATCC consensus (Fig. 6A). Exactly how these architectural features determine the different requirements for Bcd functions remains to be determined.
Our results suggest that both the cooperativity and nonconsensus site recognition functions of Bcd contribute combinatorially to target selection. Interestingly, the degree of reliance on these two functions can be influenced not only by enhancer architecture (see above) but also by the host factor(s). In particular, Bcd-Ftz(Q50K)HD-VP16 can activate transcription from the kni enhancer elements in Schneider cells but not in yeast (Fig. 8B, lanes 7 to 9). This difference is unlikely to be due to the reporter gene status, because this protein fails to activate the kni-lacZ reporter gene in yeast regardless of whether it is integrated or carried on a replicating plasmid (Fig. 8B, lanes 7 and 9). It is possible that a factor(s) present in Schneider cells but absent from yeast can influence the activity of this derivative on the kni enhancer element (but not on the hb enhancer element). Although a cofactor for Bcd has also been proposed previously (27), its identity remains elusive; interestingly, a recent study suggests that Bcd activity can be potentiated modestly by a Drosophila protein called Chip (50). Our systematic comparison of different assay systems also reveals that, in many instances, dependence on Bcd functions is reduced on reporter genes carried on plasmids, presumably because they are more accessible to activators than are integrated reporters. For example, Ftz-BcdHD-VP16 shows a higher relative activity on plasmid reporters containing the hb, kni, and kni(6A) enhancer elements than on integrated reporters (Fig. 8A, B, and D; compare lanes 10 and 12). Similarly, Bcd-Ftz(Q50K)HD-VP16 has a higher relative activity on hb(6A)-lacZ and kni(6A)-lacZ plasmid reporters than on the integrated reporters (Fig. 8C and D; compare lanes 7 and 9). Together, these results illustrate a fluid nature of the requirements for Bcd functions in target selection, a process reflective of an efficient interaction between the activator and specific enhancers in physiological environments.
Extensive studies of Q50 homeodomain proteins have produced two contrasting models to explain how their biological specificities are achieved (6). Both models center on the existence of cofactors, but the roles of these cofactors differ. The first model, referred to as the coselector model, suggests that cofactors selectively interact with different homeodomain proteins to enhance their DNA binding specificities. The second model, referred to as the widespread-binding model, proposes that, although most Q50 homeodomain proteins recognize similar or identical targets in vivo, cofactors can modulate the regulatory activities of these DNA-bound proteins. The latter model is supported by in vivo cross-linking experiments (9, 57, 58) and a recent finding that a Ubx derivative with a strong activation function gains a novel biological specificity (31). Although our present studies focus on the K50 homeodomain protein Bcd, nonconsensus site recognition most likely also plays an important role, to various extents, in target selection by all homeodomain proteins.
ACKNOWLEDGMENTS
We thank J. Manley, M. Levine, C. Peterson, M. Ptashne, S. Triezenberg, H. Jackle, J. Pearlberg, and K. Han for providing various materials used in this study and B. Aronow and members of this laboratory for discussion and/or comments on the manuscript.
This work has been supported in part by NIH grants (R01 GM52467 and P30 ES06096).
Chen Zhao and Vrushank Dave contributed equally to this work.
REFERENCES
- 1.Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- 2.Ammerer G. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Beachy P A, Varkey J, Young K E, von Kessler D P, Sun B I, Ekker S C. Cooperative binding of an Ultrabithorax homeodomain protein to nearby and distant DNA sites. Mol Cell Biol. 1993;13:6941–6956. doi: 10.1128/mcb.13.11.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggin M D, McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 7.Bonneton F, Shaw P J, Fazakerley C, Shi M, Dover G A. Comparison of bicoid-dependent regulation of hunchback between Musca domestica and Drosophila melanogaster. Mech Dev. 1997;66:143–156. doi: 10.1016/s0925-4773(97)00100-7. [DOI] [PubMed] [Google Scholar]
- 8.Burz D S, Rivera-Pomar R, Jackle H, Hanes S D. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr A, Biggin M D. A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. EMBO J. 1999;18:1598–1608. doi: 10.1093/emboj/18.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan S-K, Jaffe J, Capovilla M, Botas J, Mann R S. The DNA binding specificity of ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 11.Chang C-P, Shen W-F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 12.Colgan J, Wampler S, Manley J L. Interaction between a transcriptional activator and transcription factor IIB in vivo. Nature. 1993;362:549–553. doi: 10.1038/362549a0. [DOI] [PubMed] [Google Scholar]
- 13.Dave V, Zhao C, Yang F, Tung C-S, Ma J. Reprogrammable recognition codes in Bicoid homeodomain-DNA interaction. Mol Cell Biol. 2000;20:7673–7684. doi: 10.1128/mcb.20.20.7673-7684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desplan C, Theis J, O'Farrell P H. The Drosophila developmental gene engrailed encodes a sequence specific DNA binding activity. Nature. 1985;318:630–635. doi: 10.1038/318630a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driever W. The Bicoid morphogen: concentration dependent transcriptional activation of zygotic target genes during early Drosophila development. In: McKnight S L, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1221–1250. [Google Scholar]
- 16.Driever W, Ma J, Nusslein-Volhard C, Ptashne M. Rescue of bicoid mutant Drosophila embryos by Bicoid fusion proteins containing heterologous activating sequences. Nature. 1989;342:149–154. doi: 10.1038/342149a0. [DOI] [PubMed] [Google Scholar]
- 17.Driever W, Nüsslein-Volhard C. Bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 18.Driever W, Siegel V, Nüsslein-Volhard C. Autonomous determination of anterior structures in the early Drosophila embryo by the bicoid morphogen. Development. 1990;109:811–820. doi: 10.1242/dev.109.4.811. [DOI] [PubMed] [Google Scholar]
- 19.Ekker S C, von Kessler D P, Beachy P A. Differential DNA sequence recognition is a determinant of specificity in homeotic gene action. EMBO J. 1992;11:4059–4072. doi: 10.1002/j.1460-2075.1992.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frohnhöfer H G, Nüsslein-Volhard C. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature. 1986;324:120–125. [Google Scholar]
- 21.Gehring W J, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 22.Gehring W J, Muller M, Affolter M, Percival-Smith A, Billeter M, Qiang Y-Q, Otting G, Wuthrich K. The structure of the homeodomain and its functional implications. Trends Genet. 1990;6:323–329. doi: 10.1016/0168-9525(90)90253-3. [DOI] [PubMed] [Google Scholar]
- 23.Gehring W J, Qian Y Q, Billeter M, Furukubo-Tokunaga K, Schier A F, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 24.Gibson G, Schier A, LeMotte P, Gehring W. The specificities of sex combs reduced and Antennapedia are determined by a distinct portion of each protein that includes the homeodomain. Cell. 1990;62:1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- 25.Han K, Levine M, Manley J. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- 26.Hanes S, Brent R. A genetic model for interaction of the homeodomain recognition helix with DNA. Science. 1991;251:426–430. doi: 10.1126/science.1671176. [DOI] [PubMed] [Google Scholar]
- 27.Hanes S, Riddihough G, Ish-Horowicz D, Brent R. Specific DNA recognition and intersite spacing are critical for action of the Bicoid morphogen. Mol Cell Biol. 1994;14:3364–3375. doi: 10.1128/mcb.14.5.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 29.Ingham P W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- 30.Jaynes J B, O'Farrell P H. Activation and repression of transcription by homeodomain-containing proteins that bind a common site. Nature. 1988;336:744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, McGinnis W. Activity regulation of Hox proteins, a mechanism for altering functional specificity in development and evolution. Proc Natl Acad Sci USA. 1999;96:6802–6807. doi: 10.1073/pnas.96.12.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Veraksa A, McGinnis W. A sequence motif distinct from Hox binding sites controls the specificity of a Hox response element. Development. 1999;126:5581–5589. doi: 10.1242/dev.126.24.5581. [DOI] [PubMed] [Google Scholar]
- 33.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 34.Lin C R, Kioussi C, O'Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte J C, Rosenfeld M G. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 35.Lin L, McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- 36.Lukowitz W, Schroder C, Glaser G, Hulskamp M, Tautz D. Regulatory and coding regions of the segmentation gene hunchback are functionally conserved between Drosophila virilis and Drosophila melanogaster. Mech Dev. 1994;45:105–115. doi: 10.1016/0925-4773(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 37.Ma X, Yuan D, Diepold K, Scarborough T, Ma J. The Drosophila morphogenetic protein Bicoid binds DNA cooperatively. Development. 1996;122:1195–1206. doi: 10.1242/dev.122.4.1195. [DOI] [PubMed] [Google Scholar]
- 38.Ma X, Yuan D, Scarborough T, Ma J. Contributions to gene activation by multiple functions of Bicoid. Biochem J. 1999;338:447–455. [PMC free article] [PubMed] [Google Scholar]
- 38a.Nasiadka A, Grill A, Krause H M. Mechanisms of regulating target gene selection by the homeodomain-containing protein Fushi tarazu. Development. 2000;127:2965–2976. doi: 10.1242/dev.127.13.2965. [DOI] [PubMed] [Google Scholar]
- 39.Nusslein-Volhard C. Determination of the embryonic axis of Drosophila. Dev Suppl. 1991;1:1–10. [PubMed] [Google Scholar]
- 40.Percival-Smith A, Muller M, Affolter M, Gehring W J. The interaction with DNA of wild type and mutant fushi tarazu homeodomains. EMBO J. 1990;9:3967–3974. doi: 10.1002/j.1460-2075.1990.tb07617.x. . (Corrigendum, 11:382, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piedra M E, Icardo J M, Albajar M, Rodriguez-Rey J C, Ros M A. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- 42.Rivera-Pomar R, Jackle H. Trends Genet. 12:478–483. 1996. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. [DOI] [PubMed] [Google Scholar]
- 43.Rivera-Pomar R, Lu X, Taubert H, Perrimon N, Jackle H. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature. 1995;376:253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- 44.Ryan A K, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Pena J, Sabbagh W, Greenwald J, Choe S, Norris D P, Robertson E J, Evans R M, Rosenfeld M G, Izpisua Belmonte J C. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 45.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 46.Samson M L, Jackson-Grusby L, Brent R. Gene activation and DNA binding by Drosophila Ubx and abd-A proteins. Cell. 1989;57:1045–1052. doi: 10.1016/0092-8674(89)90342-5. [DOI] [PubMed] [Google Scholar]
- 47.Schier A F, Gehring W J. Functional specificity of the homeodomain protein fushi tarasu: the role of DNA-binding specificity in vivo. Proc Natl Acad Sci USA. 1993;90:1450–1454. doi: 10.1073/pnas.90.4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott M P, Tamkan J W, Hartzell G W. The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 49.Semina E V, Reiter R, Leysens N J, Alward W L M, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, Carey J C, Murray J C. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 50.Torigoi E, Bennani-Baiti I M, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc Natl Acad Sci USA. 2000;97:2686–2691. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Treisman J, Gönczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 52.Treisman J, Harris E, Wilson D, Desplan C. The homeodomain: a new face for the helix-turn-helix? Bioessays. 1992;14:145–150. doi: 10.1002/bies.950140302. [DOI] [PubMed] [Google Scholar]
- 53.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 54.van Dijk M A, Murre C. Extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 55.Vershon A K, Johnson A D. A short, disordered protein region mediates interactions between the homeodomain of the yeast α2 protein and the MCM1 protein. Cell. 1993;72:105–112. doi: 10.1016/0092-8674(93)90054-t. [DOI] [PubMed] [Google Scholar]
- 56.Vigano M A, Di Rocco G, Zappavigna V, Mavilio F. Definition of the transcriptional activation domains of three human HOX proteins depends on the DNA-binding context. Mol Cell Biol. 1998;18:6201–6212. doi: 10.1128/mcb.18.11.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walter J, Biggin M D. DNA binding specificity of two homeodomain proteins in vitro and in Drosophila embryos. Proc Natl Acad Sci USA. 1996;93:2680–2685. doi: 10.1073/pnas.93.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walter J, Dever C, Biggin M D. Two homeo domain proteins bind with similar specificity to a wide range of DNA sites in Drosophila embryos. Genes Dev. 1994;8:1678–1692. doi: 10.1101/gad.8.14.1678. [DOI] [PubMed] [Google Scholar]
- 59.West R W, Yocum R R, Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984;4:2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson D S, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a Paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 61.Xue D, Tu Y, Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993;261:1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]
- 62.Yu Y, Li W, Su K, Yussa M, Han W, Perrimon N, Pick L. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 63.Yuan D, Ma X, Ma J. Sequences outside the homeodomain of Bicoid are required for protein-protein interaction. J Biol Chem. 1996;271:21660–21665. [PubMed] [Google Scholar]
- 64.Zhu L, Marvin M J, Gardiner A, Lassar A B, Mercola M, Stern C D, Levin M. Curr. Biol. 9:931–938. 1999. Cerberus regulates left-right asymmetry of the embryonic head and heart. [DOI] [PubMed] [Google Scholar]