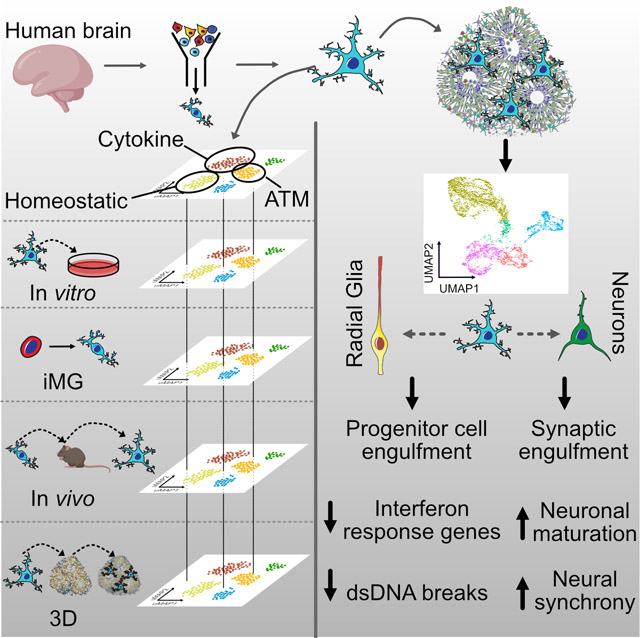
SUMMARY
Microglia are resident macrophages in the brain that emerge in early development and respond to local environment by altering their molecular and phenotypic states. Fundamental questions about microglia diversity and function during development remain unanswered as we lack experimental strategies to interrogate their interactions with other cell types and responses to perturbations ex vivo. We compared human microglia states across culture models, including cultured primary and pluripotent stem cell-derived microglia. We developed a ‘report card’ of gene expression signatures across these distinct models to facilitate characterization of their responses across experimental models, perturbations, and disease conditions. Xenotransplantation of human microglia into cerebral organoids allowed us to characterize key transcriptional programs of developing microglia in vitro and reveal that microglia induce transcriptional changes in neural stem cells and decreases interferon signaling response genes. Microglia additionally accelerate the emergence of synchronized oscillatory network activity in brain organoids by modulating synaptic density.
Keywords: Microglia, microglia culture, iPSC, iMG, organoids, human microglia, organoid activity, neuro-immune, induced microglia, interferon response, double-stranded DNA breaks
Graphical Abstract
eTOC
Our “microglia report card” reveals that human microglia selectively regulate different biological gene cluster modules in an environment-dependent fashion. The presence of microglia in the brain organoids protects against double-stranded DNA breaks and attenuates gene response on interferon signaling pathway. Additionally, microglia accelerate neural network synchronization in organoids.
INTRODUCTION
Microglia are the resident macrophages of the brain and have been shown to respond to many viral pathogens to protect neurons and glia from pathogen-induced damage (Garden 2002, Retallack, Di Lullo et al. 2016, Bortolotti, Gentili et al. 2019). Mutations in microglia-related genes have been implicated in a wide range of neurological (Rademakers, Baker et al. 2011, Oosterhof, Chang et al. 2019), neuropsychiatric (Needleman and McAllister 2012, Sekar, Bialas et al. 2016), and neurodegenerative diseases (Krasemann, Madore et al. 2017). Moreover, microglia have been shown to adopt disease-associated transcriptional states in a wide variety of disease environments (Keren-Shaul, Spinrad et al. 2017, Li, Cheng et al. 2019, Masuda, Sankowski et al. 2019). Finally, microglia transcriptomes have been shown to vary substantially across species (Geirsdottir, David et al. 2019), suggesting that in order to dissect the contributions of microglia to neurodevelopmental and disease processes in the human brain may require human cell-based experimental models. However, it is unclear which models can best recapitulate the molecular architecture of human microglia subpopulations, as microglia are known to undergo substantial changes in gene expression upon isolation from the brain parenchyma, as well as across culture models (Gosselin, Skola et al. 2017). A comprehensive molecular map of microglia diversity across experimental models would enable data-driven decisions about the most relevant experimental systems.
By harnessing single cell transcriptomics, we sought to create a comprehensive ‘report card’ for human microglia during development. We found that three-dimensional neural tissue environment presents the strongest transcriptomic correlations of microglia to their in vivo counterparts. In particular, xenotransplantation of microglia into brain organoids that are largely absent of myeloid lineage cells allows a systematic comparison of neurodevelopmental processes in the presence or absence of microglia. Interestingly, we found widespread expression of genes involved in type I interferon response in organoid radial glia, which becomes attenuated in the presence of microglia. In contrast, neurons appear transcriptionally invariant to microglia transplantation, but instead undergo synaptic remodeling and an accelerated maturation of network properties. Together, our work highlights molecular plasticity of human microglia across culture models, underscores the potential shortcomings of microglia derived from pluripotent stem cells in recapitulating in vivo signatures, and reveals transcriptional responses of the developing human neural tissue to the presence of microglia.
RESULTS
Microglia “Report Card” Across Culture Models
Efficient culture models of human microglia are required to advance our understanding of normal brain development and neuroimmune interactions in disease states. Unfortunately, transcriptomic and morphological changes in cultured microglia (Gosselin, Skola et al. 2017) represents a significant challenge for human microglia-related studies. To characterize what microglia subtypes are preserved and how they change across different models, we generated new single cell RNA sequencing data from two in vitro models and combined it with a previously published humanized mouse model engrafted with human iPSC-derived microglia (iMG) (Svoboda, Barrasa et al. 2019). To avoid microglia exposure to peripheral blood-derived secreted factors that were shown to induce quick changes in gene expression in cultured microglia (Gosselin, Skola et al. 2017), we used a serum-free culture media that has been shown to promote microglia survival in vitro (Bohlen, Bennett et al. 2017, Montilla, Zabala et al. 2020). We purified primary microglia using magnetic beads binding CD11b and cultured the cells as a monolayer culture in serum-free conditions supplemented with IL34, TGFβ2 and cholesterol to provide necessary trophic conditions for microglia survival (Bohlen, Bennett et al. 2017) (Figure 1A, D; Materials and Methods). Throughout the culture duration, microglia remained ramified and motile (Video S1). After eight days in culture, microglia expressed Iba1, a broad macrophage marker, and P2RY12, a microglia-specific marker that has been previously reported to be downregulated in ex vivo conditions (Gosselin, Skola et al. 2017) (Figure S1A). Next, we explored the conservation of microglia signatures across common experimental models, including microglia that were induced from pluripotent stem cells using a recently established protocol (Abud, Ramirez et al. 2017) (Figure 1B, E; Figure S1, Materials and Methods). We also analyzed recently published data from induced microglia that had been transplanted into in vivo mouse brain (Svoboda, Barrasa et al. 2019) (Figure 1C, F, Figure S1). To perform an unbiased comparison of microglia states in the native brain environment and in culture, and to determine whether microglia heterogeneity is preserved in vitro, we compared these combined scRNAseq datasets to primary human microglia from the BRAIN Initiative Cell Consensus Network (BICCN) that was combined with scRNAseq data from the developing human hippocampus (Zhong, Ding et al. 2020). For the primary clustered microglia cells that were in silico purified from samples ranging from gestational week (GW) 16 to GW27, we excluded clusters that were enriched for gene markers of B cells (CD79B, IGHM, IGKC), natural killer cells (NKG7, PRF1, KLRB1) and dividing cells (Figure S2A). After iteratively clustering the resulting microglia, we identified five molecularly unique clusters of microglia, all of which expressed high levels of pan-macrophage marker AIF1 (Figure S2B–C). Cluster 1 was characterized by high expression of LGALS1, LGALS3, APOC1, FTL and SPP1, recapitulating a microglia subtype that was described previously (Hammond, Dufort et al. 2019, Li, Cheng et al. 2019). In accordance with these reports, we call this microglia cluster Axon Tract-Associated microglia (ATM). Cluster 2 expressed high levels of chemokines and cytokines (CCL3, CCL4, CXCL8, IL1B), and we call it “cytokine-associated microglia”. Cluster 3 had a gene signature consisting of heat shock proteins and immediate early genes (JUN, RHOB, HSPA1A, DNAJB1) that has been previously shown to be induced by enzymatic digestion of primary brain tissue (Marsh, Kamath et al. 2020), therefore, we call it “Ex vivo activated” microglia. Cluster 4 had a higher expression of genes associated with mature homeostatic microglia (CX3CR1, P2RY12, P2RY13, VSIR), therefore we termed it “Homeostatic” microglia. Finally, a small cluster 5 was defined by gene signature (F13A1, MRC1, LYVE1) consistent with perivascular macrophages (PVM) (Zeisel, Munoz-Manchado et al. 2015). We additionally performed sex-related differentially expressed gene analysis among microglia from the “Homeostatic” and “ATM” clusters (Table S1, Figure S2J).
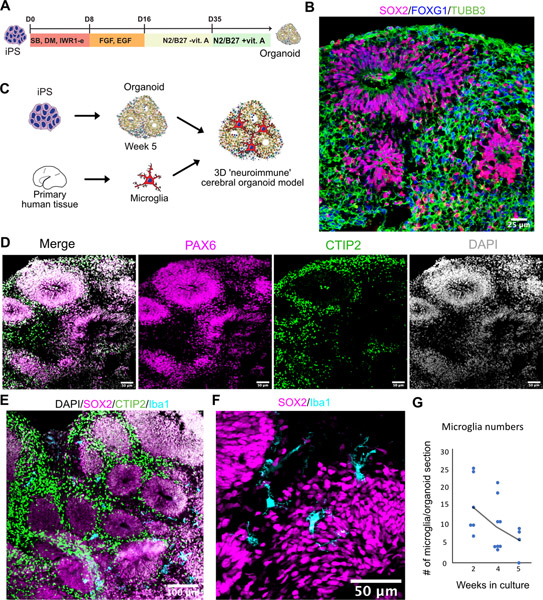
Figure 1. Developing Brain Microglia “Report Card” Across Culture Models.
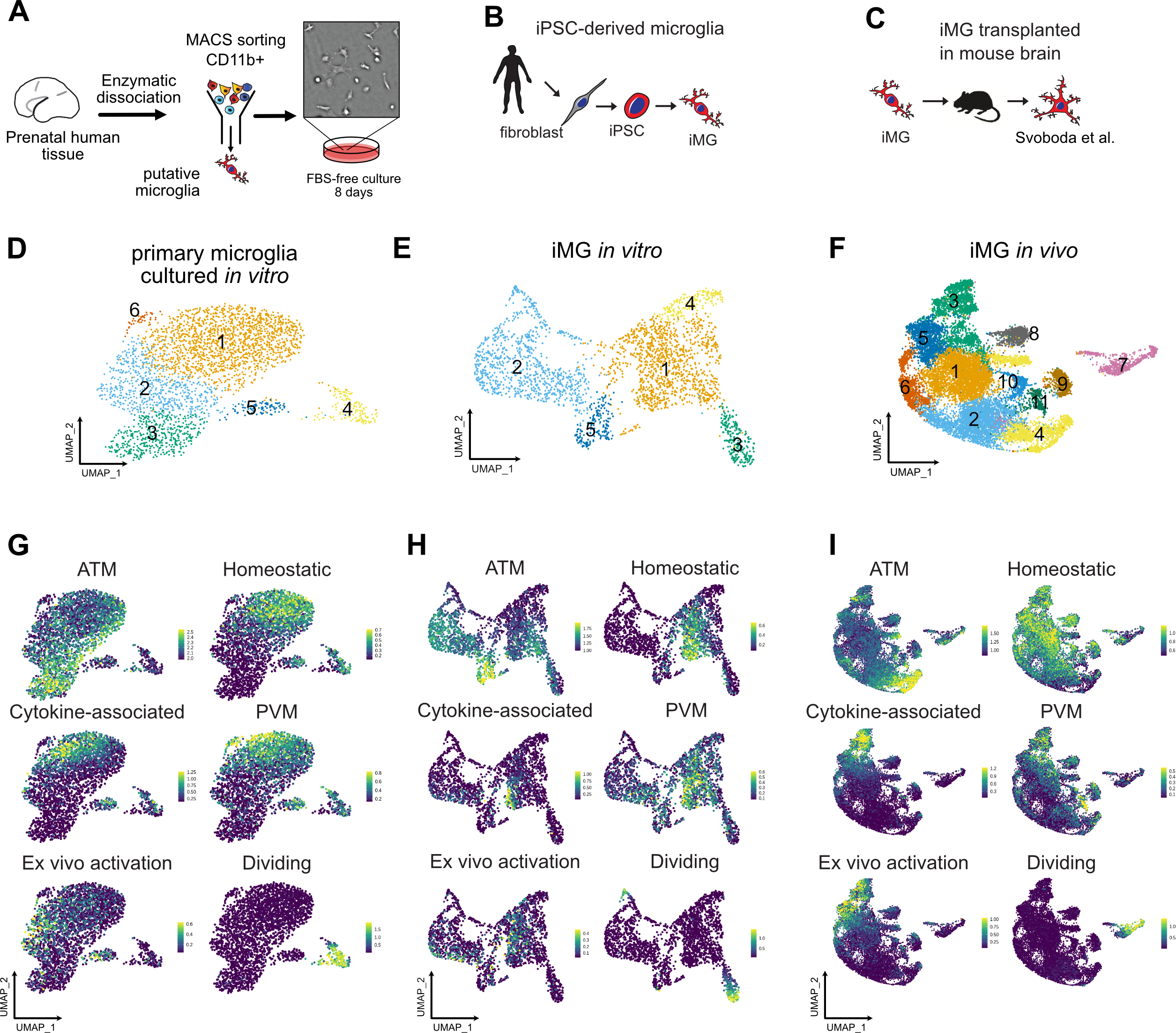
A. Primary human microglia were extracted from the mid-gestation human brain. The tissue was enzymatically dissociated with papain, MACS-sorted with Cd11b magnetic beads-conjugated antibody and cultured in defined media for 8 days. B. Microglia from induced pluripotent stem cells as a reliable microglia source. N=1 (iCell Microglia, cat#01279). C. A schematic for induced microglia transplanted into the mouse brain (from (Svoboda, Barrasa et al. 2019). D. UMAP plot of 2,970 microglia after 8 days of in vitro culture reveals five microglia clusters. N=1 brain sample (GW23 primary cortex). See also Figure S1. E. scRNAseq identifies six molecular clusters among induced microglia cells. 2,659 induced microglia (iMG) cells from N=1 experiment were used. F. scRNAseq identifies eight molecular clusters among induced microglia cells “cultured” in the mouse brain. 15,971cells from N=4 mouse brain samples 60 days post-injection were used, as previously described (Svoboda, Barrasa et al. 2019). G, H, I. Scores for individual microglia clusters for primary human microglia from Figures S1–2 were calculated and projected onto gene expression space for each microglia model. Individual genes used to calculate model scores are in Figure S2. ATM – axon tract associated microglia (with high expression of LGALS/LGALS3, APOC1, SPP1); homeostatic microglia – cluster with high expression of CX3CR1, P2RY12/P2RY13, VSIR, IFNGR1; cytokine-associated microglia – cluster with high expression of CCL3/CCL4, CXCL8, IL1B, TNF; PVM – perivascular macrophages (characterized by high expression of F13A1, MRC1, LYVE1, LYZ); Ex vivo activation – microglia cluster with high expression of genes associated with cell stress during enzymatic cell dissociation (JUN, HSPA6/HSPA1A, DUSP1, FOS); dividing cells – cells characterized by cell cycle genes (TOP2A, MKI67, CENPF).
To facilitate the comparison of different microglia subtypes across the models, we created a score for each transcriptionally-defined microglia subtype based on an eigengene for the top 15 differentially expressed cluster markers (Figure S3), in addition to a dividing cell signature, and projected these scores on the individual scRNAseq clusters for each condition (Figure 1G–I). In accordance with previous reports (Gosselin, Skola et al. 2017), primary microglia cultured in vitro had lower expression levels for the homeostatic signature (Figure 1G). Importantly, certain microglia subtypes clustered separately from each other on the UMAP even after eight days in culture, including the two distinct clusters of homeostatic and ATM microglia. However, while cluster 1 of in vitro cultured microglia became simultaneously associated with the homeostatic and PVM microglia signature, suggesting that in vitro cultured microglia may attenuate some of the brain-associated specification and become more peripheral macrophage-like, some degree of separation between these two populations remained. This is consistent with previous findings that both the tissue environment and cell ontology drive microglia specification and potentially explains what microglia signatures can revert to their brain identity after being maintained in ex vivo conditions (Bennett, Bennett et al. 2018). In contrast, microglia derived from iPSC cells had a fully overlapping signature for both homeostatic microglia and peripheral macrophages. These two populations became well separated again upon transplantation into the mouse brain, suggesting that the initial minimal patterning cocktail to derive iMG may still lack some of the signaling molecules that are unique to the brain parenchyma environment.
Consistent with previous reports, we found that human iMG transplanted into mouse brain recapitulated many of the primary cell signatures and develop transcriptomic similarities with microglia in the human brain (Figure 1I, Figure S1C), suggesting that the neural tissue environment likely provides cues that support normal microglia identity (Bohlen, Bennett et al. 2017). Individual module scores projected onto the UMAP revealed high preservation of microglia from the ATM cluster (Figure 1I). In contrast to the mouse brain transcriptomic data where this cluster is transiently present in early postnatal period of mouse development and then disappears in the later time points (Hammond, Dufort et al. 2019), our analysis suggests that human microglia retain this signature even in the adult mouse brain. Interesting, this cluster is characterized by high expression of APOE and TREM2, among others – two genes that are implicated in Alzheimer’s disease. Another cluster that was strongly preserved between primary and in vivo transplanted microglia was cytokine-associated module that was previously proposed to be specifically enriched in the human brain (Geirsdottir, David et al. 2019). Together, this analysis revealed that while in vitro culturing decreases some of the key homeostatic microglia genes, it retains gene module specificity between different cell clusters. This specification is maintained and further improved by xenotransplantation into the mouse brain. Two clusters (ATM and cytokine-associated) from the in vivo transplanted microglia potentially uncover gene module signatures that in the human microglia are maintained based on cell-intrinsic programs instead of relying on brain tissue environment.
‘Neuroimmune’ Organoid Model of Human Brain Development
The role of microglia in the early stages of human neurogenesis and neuronal maturation remains a long-standing question due to the lack of experimental models. Given the extreme transcriptional plasticity of microglia and their sensitivity to the cell environment, we wondered whether seeding them into brain organoids could rescue the transcriptomic signatures of cultured microglia. In vitro differentiation protocols of human pluripotent stem cells are becoming widely adopted to derive cultured models of developing brain (Figure 2A–B,D, Figure S4A) (Pasca 2018). Unlike neurons, astrocytes, and oligodendrocytes, mature microglia do not emerge robustly in organoids derived by standard protocols but can be reconstituted by transplantation (Ormel, Vieira de Sa et al. 2018). Combining 3D cortical organoids with exogenously derived microglia offers an opportunity to dissect the roles of this cell type in brain development. To this end, we differentiated human cortical brain organoids using standard protocols (Pasca, Sloan et al. 2015), and at 5 weeks of differentiation, we introduced human microglia purified from mid-gestation cortical tissue specimens (Figure 2C). We selected this time period for transplantation because this is when the organoids are transferred to the final maintenance media formulation that remains unchanged for the rest of the culture. Moreover, microglia do not actively proliferate in the organoids, while non-microglia progenitor cells do, therefore, we did not want to dilute the total proportion of microglia to non-microglia cells in the organoids. The resulting neuro-immune organoids were maintained in the organoid maintenance media without any addition of trophic factors for microglia survival, as our data suggest that different cell types present in the organoid express factors necessary for microglia survival, including IL34 in excitatory neurons, CSF1 in radial glia and TGFβ in dividing progenitor cells (https://mg-models.cells.ucsc.edu). Over the course of several days, microglia invaded the organoid (Videos S2 and S3) and transformed their morphology from ameboid to ramified state that could be detected even 5 weeks after engraftment (Figure 2E–G, Figure S4B–D, Movie S2). Similar results were obtained with microglia derived from iPSC (Figure S4E–G). Microglia were predominantly located around rosettes with Sox2-positive neural stem cells, consistent with their role in the neurogenic niche of the developing mammalian brain (Figure S4H) (Cunningham, Martinez-Cerdeno et al. 2013). The presence of microglia did not induce any macroscopic structural changes to the organoids (Figure S4J). We did not detect any Iba1-positive cells in any of the organoids that were not transplanted with microglia. Thus, the ‘neuroimmune’ organoid offers an opportunity to explore the long-term impact of microglia on human neurogenesis and neuronal maturation.
Figure 2. ‘Neuroimmune’ Organoid Model of Human Brain Development.
A. Cortical organoids were generated and pattern using dual SMAD and Wnt inhibition. After day 35 in culture, cortical organoids were moved into a maintenance media containing vitamin A and were maintained in the same media for the duration of the experiment. B. Cortical organoids express forebrain marker FOXG1, radial glia marker Sox2 and pan-neuronal marker TUBB3. C. Organoids were generated from iPSC cells and after switching to maintenance media at five weeks were combined with primary human microglia to generate neuro-immune organoids. D. Cortical organoids express markers for cortical radial glial PAX6 and cortical layer 5 neuronal marker CTIP2. E. Microglia distribute within the organoid. F. Iba1-positive microglia cells are ramified and distributed between and around rosettes. G. Microglia numbers monotonically decrease over the five-week co-culture time.
See also Figure S4.
A time-lapse imaging assay demonstrated that microglia were ramified with highly active motile processes, consistent with their surveillance function under homeostatic conditions (Figure 3A) (Nimmerjahn, Kirchhoff et al. 2005). We also detected a subset of phagocytic microglia identified by the presence of phagocytic cups – protrusions of plasma membrane engulfing nuclei of neighboring cells (Figure 3B). 3D reconstruction revealed that Sox2-positive progenitor cells can be found encapsulated by these phagocytic cups. Microglia additionally made contacts with Tuj1-positive neuronal fibers (Figure 3C). To examine whether microglia were actively phagocytosing synaptic material, consistent with their role in vivo, we performed additional staining for the microglial lysosome marker, CD68. We detected synaptic material that was co-localized with microglial lysosomes (Figure 3D), and the overall volume of the engulfed synapse material was comparable to what is seen in the mouse brain (Figure 3E) (Li, Chiou et al. 2020).
Figure 3. Microglia engrafted in organoids are motile and phagocyte progenitor cells and synaptic material.
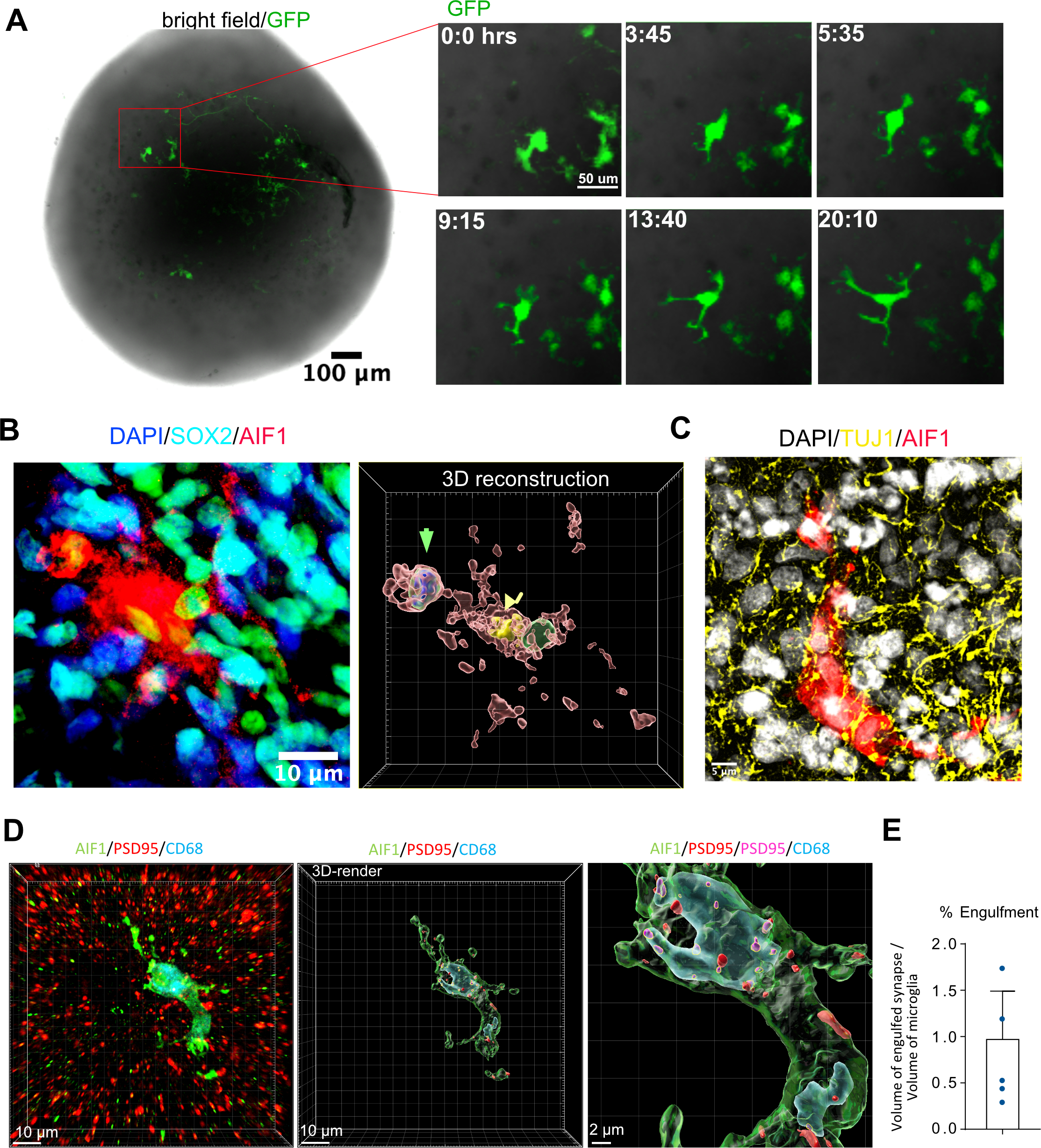
A. Microglia are ramified and motile in the organoid. For time-lapse imaging, acutely purified microglia were labeled with adenovirus CMV-GFP and then engrafted into organoids. 7 days after integration, the organoid was imaged for 24 hours to demonstrate microglia’s motile surveilling behavior. Virally labeled microglia were used only for the imaging experiments. B. Microglia in organoids form phagocytic cups containing Sox2-positive nuclei. Left – immunofluorescence labeling of a microglia cell (AIF1, red) and Sox2-positive organoid-resident radial glial cells (cyan). Right – 3D reconstruction of Sox2-positive nucleus (green arrowhead) and microglia nucleus (yellow nucleus). C. Engrafted microglia (AIF1, red) surrounded by organoid-resident neuronal fibers (labeled with tuj1, yellow). D. Microglia (labeled with AIF1, in red) in an organoid stained with lysosomal marker CD68 (in blue) demonstrates postsynaptic material (psd95; red) located inside of the lysosomes (psd95 overlapped with CD68; magenta). E. Volume of engulfed synapses normalized to the microglia volume.
Microglia in the 3D organoid environment
To investigate whether organoids can be used to preserve the homeostatic signatures of primary microglia or mature iMG, we cultured primary human microglia in the organoids for 2 weeks before performing scRNAseq analysis (Figure 4A–B, Figure S5). After filtering non-microglia cells and iterative re-clustering, we identified 13 molecularly unique clusters. Our data suggest that while the organoid environment increases the “homeostatic” score compared to the 2D in vitro cultured microglia, it also leads to an increase of the cytokine-associated signature to levels observed in primary uncultured microglia (Figure 4C). This cytokine-associated signature has been previously reported to be human-specific (Geirsdottir, David et al. 2019), and a recent transcriptomic analysis of primary human microglia (Kracht, Borggrewe et al. 2020), together with our own data, confirms the existence of this transcriptomic signature in normal microglia without any prior immune stimulation. In contrast to the human brain, mouse microglia expression signature of chemokines and IL1B, CCL3 and CCL4 (also known as macrophage inflammatory protein-1b, or Mip-1b) emerge in the aged brain (Hammond, Dufort et al. 2019). Our comparative analysis reveals that the mouse brain environment induces only moderate expression of the cytokine gene module, while human brain organoids, representing a spectrum of cell types of the developing human brain, are sufficient to restore the chemokine signature in microglia to the levels seen in the developing human brain. Together, these data suggest that the chemokine signature may be specifically associated with the human brain environment and is significantly downregulated in the mouse brain, reflecting the dynamic inducible nature of this microglia subtype/state (Figure 4C, Figure S5C).
Figure 4. Microglia transplanted into organoids recapitulate primary human microglia.
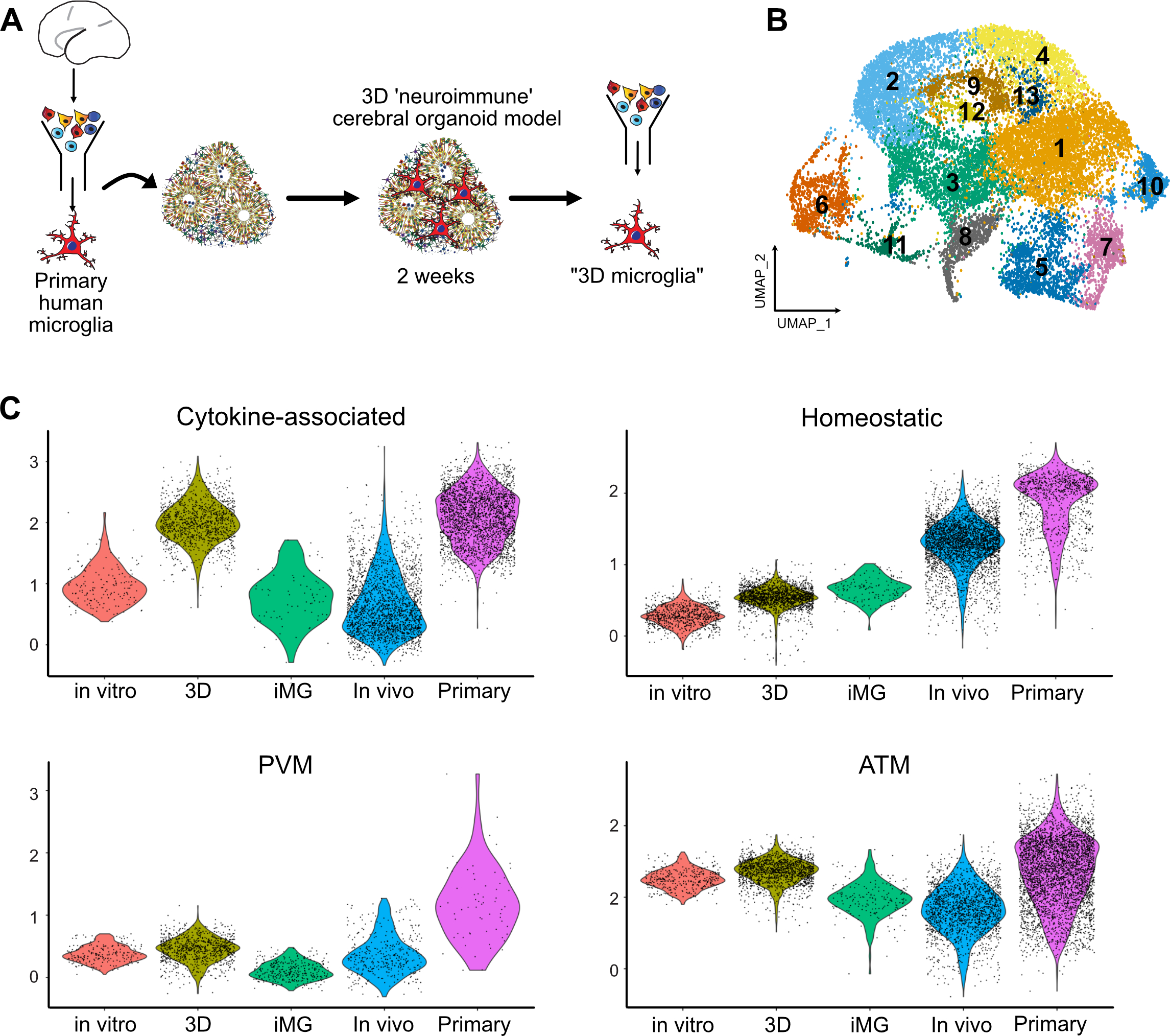
A. Primary human microglia from primary human GW23 brain were purified using MACS Cd11b beads and added to cortical organoids. Neuro-immune organoids were allowed to develop for two weeks, and then microglia cells were re-purified using CD11b magnetic beads for downstream scRNAseq processing. B. 17,705 microglia cells purified from organoids formed 13 clusters. C. Cross-model comparison of primary human microglia cultured in vitro (“in vitro”), primary human microglia engrafted in cortical organoids (“3D”), induced microglia (“iMG”), induced microglia transplanted into mouse brain (“In vivo”), and primary human microglia from mid-gestation from the BICCN dataset.
See also Figure S5.
In addition to the main signatures of the primary microglia (Figure 4C), we detected several populations of microglia that were only present in the culture environment. Similar to the 2D cultured cells, we identified a small population of microglia associated with the type I interferon response signaling gene signature (cluster 12, Figure S5B). We also identified a cluster with high expression of metallothionein gene family (MT1G, MT1H, MT2A) that was absent in any other model conditions. Metallothioneins are small proteins that are rich in cysteine residues, can bind multiple metals under physiological conditions and are involved in heavy metal detoxification and redox regulation in the cells. In the brain, metallothionein expression is protective in response to injury and counteracts the effects of pro-inflammatory cytokines (Penkowa, Carrasco et al. 1999, Chung, Leung et al. 2009). High expression of these genes in organoid-cultured microglia may reflect their response to the cell stress of the organoid brain environment by upregulating the oxidate cell stress response (Bhaduri, Andrews et al. 2020).
Human Microglia Reduce Cell Stress and Attenuate Interferon Response Genes
Given the strong transcriptomic signature that would be predicted to be protective, we wondered how the presence of microglia influence different cell types in the organoids. To systematically identify which transcriptomic signatures are differentially regulated among neural cells in the presence of microglia, neuroimmune organoids generated from three different iPS lines were dissociated at 5 weeks after transplantation and profiled using scRNAseq. We identified the major cell types, including neurons, radial glia, intermediate progenitors and dividing cells (Figure S6A–D). After selecting for cells of forebrain identity (FOXG1-positive), iterative clustering revealed the major cell types of the neuronal lineage, radial glia, intermediate progenitors, and dividing cells (Figure 5A, Figure S6E). Cells derived from control and neuroimmune organoids, as well as cells from all three organoid lines were found evenly distributed across all clusters (Figure 5B).
Figure 5. Microglia differentially regulate gene expression profile in radial glia and neurons from cortical organoids.
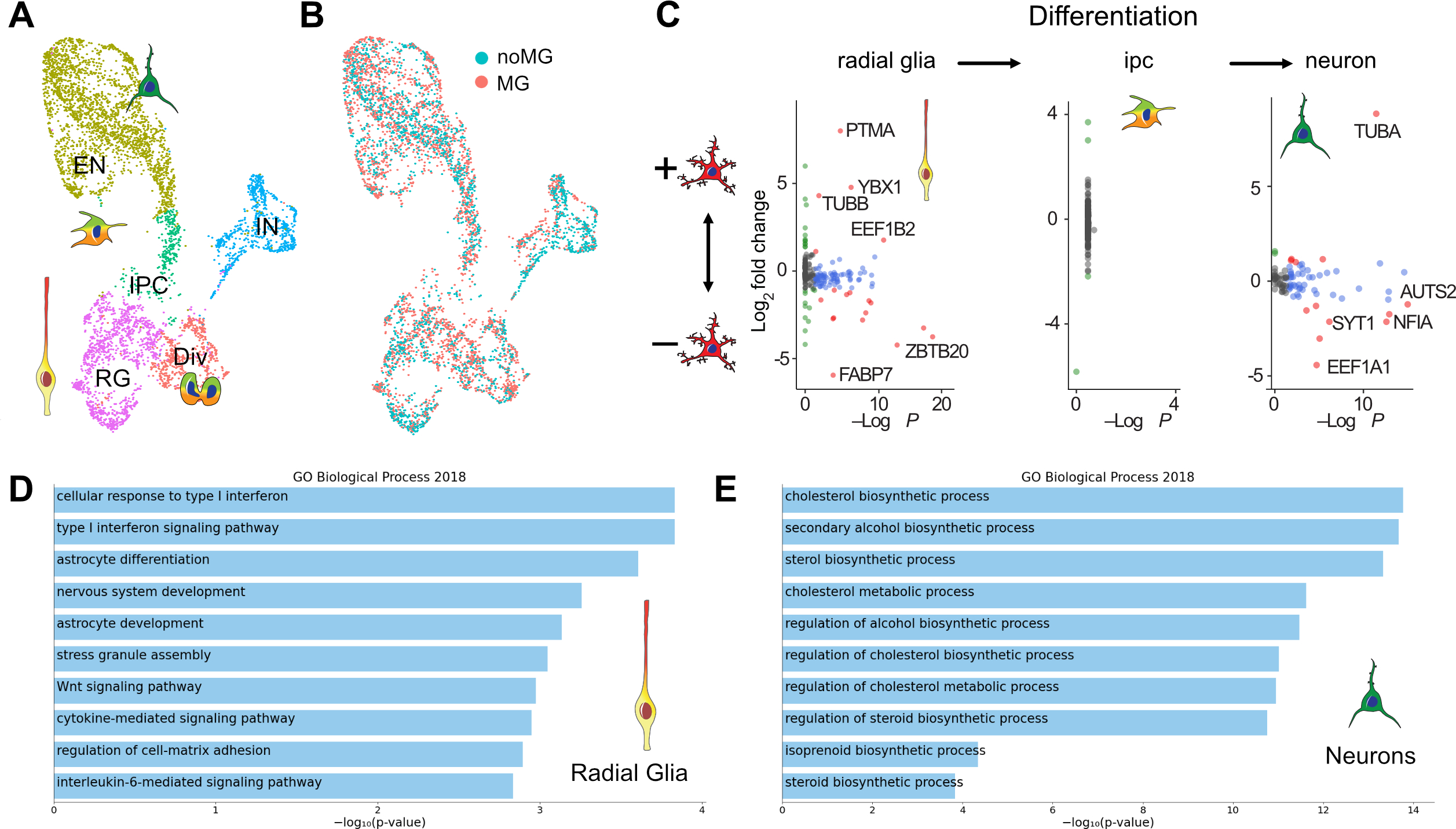
A. scRNAseq identifies main cell types in week 10 organoids. UMAP plot colored by clusters representing cell types. EN – excitatory neurons; IN – inhibitory neurons; IPC – intermediate progenitor cells; RG – radial glia cells; Div – dividing cells. B. UMAP of 5,507 cells colored by microglia condition. C. Differentially expressed genes between control organoids and organoids transplanted with microglia. D. GO term analysis of the most affected genes in neurons with an adjusted p-value cut off of 0.05. Neurons upregulated genes in a lipid biosynthesis pathway. E. Radial glia from organoids transplanted with microglia downregulate genes in the interferon response pathway.
To uncover microglia-induced transcriptomic changes in each major cell type, we first calculated the amount of variance in expression level attributable to individual variables, and selected genes with expression variation significantly contributed to by experimental condition (microglia-transplanted vs control organoids). Differential gene expression analysis revealed that radial glia and dividing cells are two top responding cell types to microglia (Figure 5C, Table S2). Among radial glial cells, Gene Set Enrichment Analysis Enrichr (Chen, Tan et al. 2013) identified top biological processes associated with the presence of microglia, including significant reduction of genes involved in metabolism as well as cellular response to interferon (Figure 5D). In contrast, fewer changes were identified in neurons. Among them, we observed genes involved in a cholesterol biosynthesis and acetyl-CoA pathway that were uniformly upregulated in response to the presence of microglia (Figure 5E, Figure S7D). Although we did not detect significant differences in individual gene expression in neurons with and without microglia, we noted some trends in the expression of two genes associated with neuronal differentiation/maturation. Specifically, genes enriched in immature neurons, such as NEUROD2, were detected at a higher average abundance in neurons derived from non-transplanted organoids. In contrast, genes typically expressed in more mature neurons, such as NRXN1, were modestly enriched in neurons derived from microglia-transplanted organoids (Figure S7E).
By focusing on the top differentially expressed genes from the interferon signaling pathway in radial glia in the neuroimmune organoids, we identified several genes that were markedly decreased in the presence of microglia (Figure 6A, Figure S7A). Interferon Regulatory Factor 1 (IRF1), Interferon Alpha-Inducible Protein 6 (IFI6), Interferon Induced Transmembrane Protein 3 (IFITM3) and Human Leukocyte Antigen E (HLA-E) were among the top genes driving the GO Biological Process analysis, and in the profiled organoids, this difference was specific to radial glial cells. We validated this difference by performing immunostaining in rosettes from neuro-immune organoids (Figure 6B). Co-staining with IFITM3 and Sox2 revealed a small, but statistically significant decrease at the level of IFITM3 levels in the presence of microglia, consistent with the transcriptomic data (Figure 6B–C, Figure S7F–H). While the role of these changes at the level of radial glia cells in the human brain remains to be determined, it is interesting to note that IFITM3, one of the top interferon response genes identified in our analysis, is robustly expressed in the human, but not mouse, radial glia cells (Figure S7B,C).
Figure 6. Human microglia attenuate type I Interferon response and decrease double-stranded DNA breaks in radial glia.
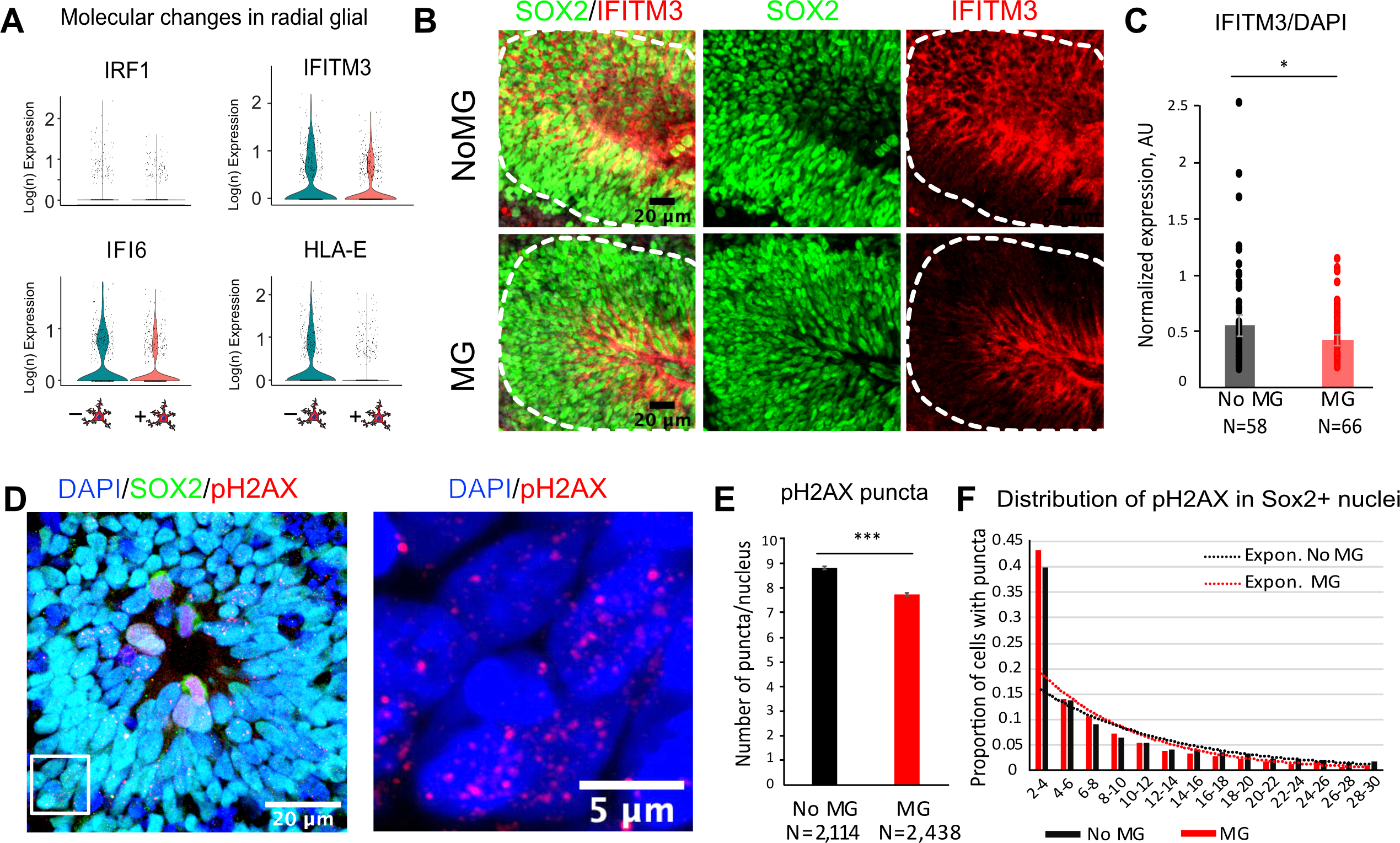
A. Individual differentially expressed genes in organoids with and without microglia driving pathway analysis from Fig. 5D. B. Immunofluorescence validation of IFITM3, Interferon Induced Transmembrane Protein 3, in rosettes from organoids with and without microglia. IFITM3 fluorescence signal was averaged across the rosette area (outlined in white). C. IFITM3 has a small but statistically significant decrease in rosettes from organoids with microglia. p-value <0.05. N are individual rosettes for each condition, quantified from sections from three independent organoids per condition. D. An example of a rosette labeled with Sox2 for radial glial cells and a marker for double-stranded DNA breaks, pH2AX. A zoomed in window is an example of cell nuclei with pH2AX puncta. E. Averaged number of pH2AX puncta per cell nucleus. Cells nuclei with 2–30 pH2AX-positive puncta were used for analysis. p-value < 0.001. F. Distribution of pH2AX number of puncta between conditions. Sox2-positive nuclei from microglia-transplanted organoids had a larger proportion of cells with fewer number of puncta. Organoids without microglia had consistently higher proportion of cells with the number of puncta over 10.
See also Figure S7.
Although predominantly associated with immune defenses against various pathogenic stimuli, including viral infections, interferon response is implicated in a general response to cellular stress, including DNA damage (Brzostek-Racine, Gordon et al. 2011). Since our culture condition did not involve any viral challenge, we hypothesized that the interferon response pathway may be activated in response to cell stress induced by the organoids culture paradigm (Bhaduri, Andrews et al. 2020). To test this hypothesis, we performed additional staining with pH2AX, a marker for double stranded DNA breaks, and quantified resulting puncta using CellProflier (Jones, Kang et al. 2008). Within the cell nuclei stained with Sox2 and pH2AX, we excluded the ones with high level of pH2AX, suggestive of dividing stem cells in the process of chromosome segregation (Turinetto and Giachino 2015), and only quantified cells with 2–30 pH2AX-positive puncta (Figure 6D–F). On average, the number of puncta per cell was greater in the absence of microglia. Distribution of nuclei with pH2AX-positive puncta revealed a trend with higher proportion of cells skewed towards larger number of puncta per nucleus in the control organoids, suggesting that a small, but significant and consistent increase in double-stranded DNA breaks in control organoids.
Microglia Facilitate the Maturation of Neural Networks During Brain Development
It has been previously shown that microglia influence early network activity through physical contacts and secretion of secreted factors. In the scRNAseq data, we detected a small, but statistically significant change in the expression of genes implicated in synaptic transmission. We sought to explore whether microglia might influence functional network maturation (Trujillo, Gao et al. 2019). To determine the effect of microglia on the maturation of neural networks, we transplanted microglia into 15 weeks old organoids, when the majority of neuronal types are expected to be present. We recorded spontaneous neural activity from the surface of the organoids using multi-electrode arrays in neuroimmune and control organoids 5 weeks later (Figure 7A). In both conditions, we detected spike activity with characteristic waveforms for single-unit activity (Figure 7B). Excitingly, we discovered increased synchronization and frequency of oscillatory bursts in neuroimmune organoids compared to control (Figure 7C–E), consistent with accelerated network maturation in organoids (Trujillo, Gao et al. 2019) (Rigato, Buckinx et al. 2011, Parkhurst, Yang et al. 2013). To test the hypothesis that microglia involved in synaptic refinement, we performed double immunostaining for vGlut1 and PSD95 and revealed a significant reduction in synaptic puncta, consistent with the role of microglia in synaptic remodeling, and we were able to detect synaptic material within microglia (Schafer, Lehrman et al. 2012, Weinhard, di Bartolomei et al. 2018) (Figure 7F–G, Video S4). These findings suggest that microglia integrate into cerebral organoids, can be maintained within organoids for long-term culture, and contribute to development of functional neural networks.
Figure 7. Microglia Facilitate the Maturation of Neural Networks During Brain Development.
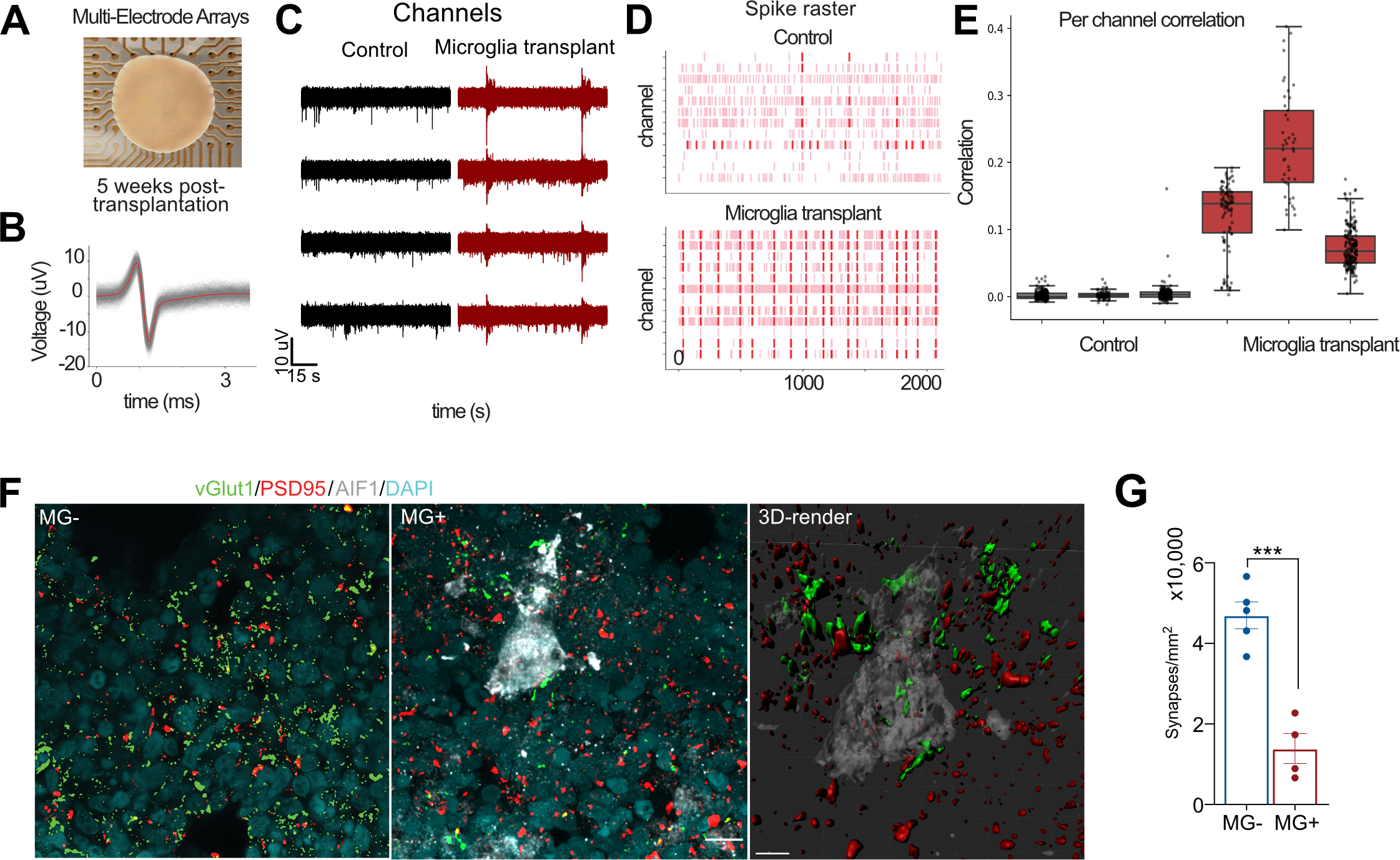
A. Organoids transplanted with microglia were used for MEA recordings on a 64-electrode plate (Axion Biosystems). B. An example of a single unit event. Individual events or “spikes” were overlaid on top of each other (gray), and the mean of these events is shown in red. C. Microglia transplantation (in red) increases pairwise correlation between MEA electrodes compared with control (in black) conditions. Each point represents one channel pair. Correlations are determined by binarizing activity in 40 ms bins. D. Microglia transplantation promotes synchronized burst activity in cerebral organoids. Each pink line represents a spike that crossed the noise threshold (+/− 6x standard deviation), with red lines representing 10 or more spikes per second. E. Correlation in activity between channels is increased in response to microglia transplantation. Each dot represents a single channel. F. Microglia-transplanted organoids show reduced density of synapses. G. Synapses were quantified based on co-localization of presynaptic marker vGlut1 and postsynaptic marker psd95. The recordings were performed across three organoid lines with at least three fields of view per section. Data points represent average of each organoid, and the bar chart represents average of all fields of view for each condition. Error bar represents standard error mean. P-value < 0.001.
DISCUSSION
We created a comprehensive comparison atlas among primary and iPSC-induced human microglia under different culture conditions including 2D culture and iMG transplanted into mouse brains. We compared strategies for microglia culture that could serve as experimental systems for functional characterization of human microglia, including two-dimensional culture and transplantation into cerebral organoids. To make this resource useful for the scientific community, we make it available via an interactive single cell browser (https://cells-test.gi.ucsc.edu/?ds=mg-models). We show that while the in vivo mouse brain environment significantly enhances fidelity of native microglia states, several important differences between iMG and primary microglia still remain. First, we show that a number of cytokine and chemokine genes is better preserved in human-specific brain environment. It has been recently reported that the chemokine and cytokine signatures distinguish between mouse and human microglia (Geirsdottir, David et al. 2019), and we now show that the same gene modules remain downregulated in the microglia transplanted in the mouse brain. This finding suggests that the native human brain environment may provide instructive cues for microglia development, perhaps reflecting deeper differences between the two species and further highlighting the effect of cell-cell interactions in shaping transcriptional and functional difference. In contrast, ATM microglia cluster is maintained in different culture conditions, including mouse brain, suggesting that this microglia type may represent a stable microglia subtype.
Our research was motivated by two major questions: first, can brain organoids serve as a better culture environment for microglia, and second, can neuro-immune organoids become a more accurate model of the developing brain? To address these questions, we performed a primary microglia transplantation into the organoids followed by a scRNAseq analysis of both microglia and neural cells. Furthermore, we systematically characterized a neuro-immune model of 3D brain organoids transplanted with primary human microglia, and we show that microglia integrate into the organoids and affect brain development. By performing scRNAseq analysis, we demonstrated that microglia transplanted in cerebral organoids had the closest resemblance to their in vivo counterparts. Furthermore, by comparing gene expression profiles between transplanted and control organoids, we detected subtle but statistically significant transcriptional differences in a cell type-dependent manner. In radial glia, the presence of microglia reduces genes associated with interferon response and their downstream effectors. Interestingly, expression of interferon genes is specific to radial glia in the human, but not mouse brain, and activation of interferon pathway regulates proliferation of human neural progenitors (Pereira, Medina et al. 2015). We attribute these changes to dsDNA damage that is enriched in long neural genes in neural stems and progenitor cells (Wei, Chang et al. 2016). Interestingly, increased DNA damage associated with increased neural progenitor proliferation has been associated with stress-susceptible genes, including the ones implicated in autism spectrum disorder pathogenesis (Wang, Wei et al. 2020). While downstream effects of these gene expression differences and their connection to the pathogenesis of neurodevelopmental disorders are outside of the scope of this study, our work provides a roadmap for future hypothesis-based investigations into microglia-brain interactions.
In addition to identifying broad gene expression changes in microglia-containing organoids, our study reveals a role of microglia in neural circuit development. We detect an increase in network-level synchronized activity in microglia-containing organoids. This increase has been reported in maturing cortical organoids and was characteristic of developing cortical networks. While several studies on microglia depletion in adult mouse brain have revealed a role of microglia in suppressing spontaneous activity and reducing seizures (Badimon, Strasburger et al. 2020, Merlini, Rafalski et al. 2020, Wu, Li et al. 2020), the role of microglia in early circuit formation may differ from adult circuits. In the postnatal mice, depletion of microglia leads to deficient synaptic pruning, accumulation of immature synapses and weakening of functional connectivity (Zhan, Paolicelli et al. 2014). Consistent with the role of microglia in phagocytic engulfment, we detected synaptic material inside of the microglia cells and overall reduction of synaptic puncta in microglia-transplanted organoids. However, we cannot exclude the role of microglia that is not associated with direct phagocytosis, such as release of soluble factors (Cheadle, Rivera et al. 2020). Together, we report a new neuro-immune model that can be used to investigate cell interactions between microglia and different cell types in the developing brain and serve for disease modeling and amenable to therapeutic screenings.
Limitations of the Study
Microglia number and cellular properties are tightly controlled in various brain regions. It has been previously shown that microglia from different brain regions differ in density, distribution, metabolism and phagocytic/pruning behavior (Lawson, Perry et al. 1990, Tan, Yuan et al. 2020). In our neuro-immune model, we noticed varying transplantation efficiency, with different organoids having different number of transplanted microglia. However, we did not notice any difference between different organoids at the level of single-cell transcriptome or protein expression levels. Our study is limited to only cortical microglia transplanted into cortical organoids, therefore performing transplantations using microglia isolated from different brain regions as well as employ organoids representing regional brain niches would be of great interest in the future. Additionally, while human organoids provide a good model for the early brain development, they have not yet reach neural circuitry resembling a mature brain. In this study, we focused a relatively short time period when neural circuits are just being developed. It would be important to see how the absence of microglia influence at later time points. It has been previously reported that the spontaneous network formation displayed regular oscillatory events over their maturation trajectory (Trujillo, Gao et al. 2019). More work is needed to determine long-lasting microglia effects on different cell types in the organoids.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tomasz J. Nowakowski, tomasz.nowakowski@ucsf.edu.
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Single-cell RNA-seq data for non-human data have been deposited at GEO (GEO: GSE180945) and are publicly available as of the date of publication. Newly generated single-cell RNA-seq data from de-identified human subjects has been deposited at Synapse, (https://www.synapse.org/#!Synapse:syn26009957) and require an authorized user login. Any questions should be referred to the Lead Contact. Re-analyzed data from other available sources is listed in the Deposited Data section of Reagents and Resource. Accession numbers for all data are listed in the key resources table. Microscopy data reported in this paper will be shared by the Lead Contact upon request.
All data analysis has been done by using previously published pipelines with DOIs listed in the key resource table. A detailed description of the computational processing and parameters is provided in Methods Details.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD68 | Abcam | Abcam Cat# ab955, RRID:AB_307338 |
| CTIP2 | Abcam | Cat# ab18465, RRID:AB_2064130 |
| EOMES | R&D Systems | Cat# AF6166, RRID:AB_10569705 |
| FOXG1 | Abcam | Cat# ab196868, RRID:AB_2892604 |
| Iba1 | Synaptic Systems | Cat# 234 004, RRID:AB_2493179 |
| Iba1 | Wako | Cat# 019-19741, RRID:AB_839504 |
| IFITM3 | Proteintech | Cat# 11714-1-AP, RRID:AB_2295684 |
| Ki67 | Agilent | Cat# M724001-2, RRID:AB_2631211 |
| P2RY12 | Sigma Aldrich | Cat# HPA014518, RRID:AB_2669027 |
| PAX6 | BioLegend | Cat# 901301, RRID:AB_2565003 |
| pH2AX | Cell Signaling Technologies | Cat# 9718, RRID:AB_2118009 |
| psd95 | Thermo Fisher | Cat# 51-6900, RRID:AB_2533914 |
| SATB2 | Santa Cruz Biotechnology | Cat# sc-81376, RRID:AB_1129287 |
| SOX2 | Santa Cruz Biotechnology | Cat# sc-17320, RRID:AB_2286684 |
| Tuj1 | Biolegend | Cat# 80120, RRID:AB_2313773 |
| VGLUT1 | Millipore Sigma | Cat# MAB5502, RRID:AB_262185 |
| Bacterial and Viral Strains | ||
| AV-CMV-GFP | Vector Biolabs | 1060 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Antibiotic-Antimycotic | Gibco | 15240-062 |
| B27 supplement w/o Vitamin A | Gibco | 12587-001 |
| B27 w/ Vitamin A | Gibco | 17504-001 |
| BDNF | Alomone | b250 |
| CD lipid concentrate | Thermo Fisher | 11905031 |
| CellTracker CM-DiI | Thermo Fisher | C7000 |
| DAPI Fluoromount | Southern Biotech | 0100-20 |
| DMEM/F12 with Glutamax | Gibco | 10565042 |
| Dnase | Sigma Aldrich | DN25 |
| Donkey Serum | Jackson Immuno | 017-000-121 |
| Dorsomorphin | Sigma-Aldrich | P5499 |
| DPBS | Gibco | 14190250 |
| Eagle’s basal medium | UCSF Media Production Core | N/A |
| EGF | Peprotech | 100-47 |
| FBS | HyClone | SH30071.03 |
| Fibronectin | Corning | 354008 |
| FGFb | Peprotech | 100-18B |
| GlutaMax Supplement | Thermo Fisher Scientific | 35050061 |
| HBSS | UCSF Media Production Core | N/A |
| Human Insulin | Sigma | I2643-50MG |
| IL34 | Peprotech | 200-34 |
| Insulin-Transferrin-Selenium (ITS-G) (100×) | Thermo Fisher Scientific | 41400045 |
| IWR1 -ε | Cayman Chemical | 13659 |
| Laminin | Thermo Fisher | 23017015 |
| Matrigel | Fisher Scientific | 354234 |
| N2 supplement | Gibco | 17502-048 |
| Neurobasal | Gibco | 21103049 |
| Non-essential Amino Acids (NEAA) | Thermo Fisher Scientific | 11140050 |
| PBS-EDTA, pH 7.5 | Lonza | BE02-017F |
| Trypsin, 2.5% | Gibco | 15090046 |
| Trypsin, 0.25% | Thermo Fisher | 25200056 |
| Critical Commercial Assays | ||
| 10X Chromium V2 | 10X Genomics | PN-120237 |
| 10X Chromium V3 | 10X Genomics | PN-1000092 |
| Cd11b magnetic beads | Miltenyi Biotec | 130-049-601 |
| Experimental Models: Cell Lines | ||
| Human iPS cell line 28126 (male) | Gilad Lab (Gallego Romero et al., 2015) | |
| Human iPS cell line “WTC-11” (male) | Conklin Lab (Bershteyn et al., 2017; Kreitzer et al., 2013) | CVCL_Y803 |
| Human iPS cell line “1323-4” (female) | Conklin Lab (Matsumoto et al., 2013) | CVCL_0G84 |
| Human iPS cell line “WTC-10” (male) | Conklin Lab (Bershteyn et al., 2017; Kreitzer et al., 2013) | |
| Human iMG | Fujifilm Cellular Dynamics | 01279 |
| Software and Algorithms | ||
| ImageJ (Fiji) | Schindelin et al., 2012 | https://imagej.net/Fiji |
| CellRanger v3.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Seurat v3.0 | Satija Lab (Butler et al., 2018a) | https://satijalab.org/seurat/ |
| deMULTIplex | Gartner Lab (McGinnis et al., 2019) | https://github.com/chris-mcginnis-ucsf/MULTI-seq |
| CellProfiler 3.0 | McQuin, Goodman et al. 2018 | https://cellprofiler.org/ |
| CellBender | (Fleming, Marioni et al. 2019) | https://github.com/broadinstitute/CellBender |
| UCell | (Andreatta and Carmona 2021) | https://github.com/carmonalab/UCell |
| Deposited data | ||
| Primary microglia BICCN scRNAseq | Brain Initiative Cell Census Network | NeMO archive: RRID:SCR_015820 |
| Primary microglia in vitro culture scRNAseq | This study | Synapse: syn26009957 |
| Primary microglia in organoids scRNAseq | This study | Synapse: syn26009957 |
| Induced microglia scRNAseq | This study | GEO: GSE180945 |
| Organoids scRNAseq | This study | GEO: GSE180945 |
| Microglia in mouse brain scRNAseq | (Svoboda, Barrasa et al. 2019) | GEO: GSE139194 |
| Other | ||
| 6-well Ultralow Attachment Plates | Corning | 3471 |
| 40 μm cell strainer | Corning | 352340 |
| LS columns | Miltenyi Biotec | 130-042-401 |
| glass-bottom 24 well plates | Cellvis | P24-1.5H-N |
| glass-bottom six-well plate | Mattek | P06G-1.5-10-F |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary human samples
Deidentified primary tissue samples were collected with previous patient consent in strict observance of the legal and institutional ethical regulations.
Protocols were approved by the Human Gamete, Embryo, and Stem Cell Research Committee (institutional review board) at the University of California, San Francisco.
Sample size estimation
Single cell RNA sequencing experiments on organoids with and without microglia were performed on three organoid cell lines, two individual organoids per condition to account for variability between different lines and organoids within the same batch. Cell counts for each individual experiment are identified in the text and figure legends.
Immunofluorescence-based imaging quantifications were performed on a minimum of three individual organoids per condition. Sample size and error bar designation are displayed in figure legends for appropriate experiments.
How subjects/samples were allocated to experimental groups
For the organoids in the presence and absence of microglia, all organoids from the experimental cohort were grown on the same plate and were randomly assigned to each experimental group.
Health/immune status
All human-derived samples came from apparently normal specimens without any known abnormalities.
Culture conditions for in vitro systems
Primary human microglia
Prenatal human microglia were purified from primary human cortical brain tissue from mid-gestation (gestational week 18–23) samples using magnetic-activated cell sorting kit with CD11b magnetic beads (Miltenyi Biotec, 130–049-601) following manufacturer’s instructions. Briefly, primary brain tissue was minced to 1mm2 pieces and enzymatically digested in 10 ml of 0.25% trypsin reconstituted from 2.5% trypsin (Gibco, 15090046) in DPBS (Gibco, 14190250) for 30 mins at 37 °C. 0.5 ml of 10 mg/ml of Dnase (Sigma Aldrich, DN25) was added in the last 5 minutes of dissociation. After the enzymatic digestion, tissue was mechanically triturated using a 10 ml pipette, filtered through a 40 μm cell strainer (Corning 352340), pelleted at 300×g for 5 minutes and washed twice with DBPS. Dissociated cells were resuspended in MACS buffer (DPBS with 1 mM EGTA and 0.5% BSA) with addition of DNAse and incubated with CD11b antibody for 15 minutes on ice. After the incubation, cells were washed in a 10 ml of MACS buffer and loaded on LS columns (Miltenyi Biotec, 130–042-401) on the magnetic stand. Cells were washed 3 times with 3 ml of MACS buffer, then the column was removed from the magnetic field and microglia cells were eluted using 5 ml of MACS buffer. Cells were pelleted, re-suspended in 1 ml of culture media and counted. We routinely obtained 1×10^6 of microglia cells per MACS purification.
Primary human microglia culture
Microglia were cultured on glass-bottom 24 well plates (Cellvis, P24–1.5H-N) pre-coated with 0.1 mg/ml of poly-d-lysine (Sigma Aldrich, P7280) for 1 hr and 1:200 laminin (Thermo Fisher, 23017015) and 1:1,000 fibronectin (Corning, 354008) for 2 hrs. Microglia were plated at 1,5×10^5 cells/well and maintained in culture media containing 66% (vol/vol) Eagle’s basal medium, 25% (vol/vol) HBSS, 2% (vol/vol) B27 (Thermo Fisher, 17504001), 1% N2 supplement (Thermo Fisher, 17502001), 1% penicillin/streptomycin, and GlutaMax (Thermo Fisher, 35050061) additionally supplemented with 100 ng/ml IL34 (Peprotech, 200–34), 2 ng/ml TGFβ2 (Peprotech,100–35B), and 1x CD lipid concentrate (Thermo Fisher, 11905031) for 5–8 days. For single cell RNA sequencing capture, cultured cells were washed with DPBS, incubated with 0.25% trypsin (Thermo Fisher, 25200056) for 10 mins at 37°C. Trypsin was inactivated by addition of DPBS with 10% FBS, the cells were centrifuged at 300x at 4°C for 5 mins and washed with DPBS twice for a total of 3 washes.
Induced microglia
Induced microglia cells were generated from human iPSC cells under defined conditions as previously described (Abud, Ramirez et al. 2017) and were a kind gift from Fujifilm Cellular Dynamics (iCell Microglia, 01279, catalog # RC1110).
Organoid generation
Cerebral organoids were generated based on a previously published method (Pasca, Sloan et al. 2015) with several modifications. Briefly, hiPSCs cultured on Matrigel were dissociated into clumps using 0.5 mM EDTA in calcium/magnesium-free PBS and transferred into ultra-low attachment 6-well plates in neural induction media (GMEM containing 20% (v/v) KSR, 1% (v/v) penicillin-streptomycin, 1% (v/v) non-essential amino acids, 1% (v/v) sodium pyruvate, and 0.1mM 2-mercaptoethanol). For the first nine days, neural induction media was supplemented with the SMAD inhibitors SB431542 (5 μM) and dorsomorphin (2 μM), and the Wnt inhibitor IWR1-endo (3 μM), with a media exchange performed every three days. Additionally, the Rho Kinase Inhibitor Y-27632 (20 μM) was added during the first six days of neural induction to promote survival. Between days 9–25, organoids were transferred to a neural differentiation media (1:1 mixture of Neurobasal and DMEM/F12 containing 2% (v/v) B27 without vitamin A, 1% N2, 1% (v/v) non-essential amino acids, 1% (v/v) Glutamax, 1% (v/v) antibiotic/antimycotic, 0.1mM 2-mercaptoethanol) supplemented with FGFb (10 ng/mL) and EGF (10 ng/mL). Between days 25–35, organoids were maintained in neural differentiation media without FGF or EGF. From Day 35 onward, organoids were maintained in neural differentiation media containing B27 with vitamin A with media exchanges every 2–3 days.
Microglia-organoid engraftment and co-culture
Microglia from mid-gestation cortical tissue were MACS-purified and immediately added to week 5 or week 15 organoids in 6-well plates at 1×10^5 microglia cells/organoid and kept off the shaker overnight. The next day, the plates were returned to the shaker and maintained following the usual organoid maintenance protocol.
Authentication of cell lines used
All iPS cell lines used in this work has been karyotyped and regularly tested for mycoplasma. Before experiments, cortical organoids were validated for the expression of relevant brain cortex markers (Foxg1, Pax6) and presence of rosettes reminiscent of ventricular zone to confirm their cortical identity.
METHOD DETAILS
Time-lapse imaging of microglia in 2D culture
Time lapse imaging experiment for primary cultured microglia was started on day 6 of culture and continued for 24 hrs in environmental control chamber (5%CO2, 37°C and relative humidity of 70%). Imaging data were acquired using a Leica DMI-8 inverted widefield microscope, with 20x objective magnification at 0.55NA (air), and 0.4NA condensor on a Hamamatsu Flash-4 LT camera (6.5 μm pixels). The cells were held at a constant 37°C, 5% CO2 using the Okolab stage-top incubator (H101-K-Frame). Each frame was acquired with 50 ms camera exposure. Imaging was done for a period of 24 hours at 27-minute time intervals.
Time-lapse imaging of microglia in organoids
For long-term tracking of microglia engrafted into the organoids, we achieved GFP expression with adenoviral infection. We incubated MACS-purified microglia with adenovirus CMV-GFP (E1/E5, Vector Biolabs, Cat. 1060). Briefly, freshly isolated cells were incubated with the virus for 1 hour, then washed with PBS three times to remove the viral particles, and then engrafted into organoids as previously described. Sparse GFP expression of GFP was observed after 24 hours in culture. Microglia were allowed to engraft into the organoid for seven days and then were imaged using confocal Leica SP8, 10x lens every 20 minutes for 20 hours. Image Z-stacks were projected as averages. GFP-labeled microglia were used only for the imaging experiments for Figure 3A. Unlabeled microglia were used for the rest of the functional and transcriptomic evaluations.
Labeling of microglia for short-term tracking (related to Video S2) was done using CellTracker CM-DiI (Thermo Fisher, C7000). Acutely purified microglia were labeled with the cell tracker for 30 mins and then added to an organoid embedded in matrigel in a glass-bottom six-well plate (Mattek, P06G-1.5–10-F). The imaging was performed using Leica SP8 for the next 24 hours to demonstrate microglia moving towards the organoid surface and engrafting into the organoid.
Organoid single-cell capture for single-cell RNA sequencing
Two organoids per experimental condition were cut into 1 mm2 pieces and enzymatically digested with papain digestion kit (Worthington, LK003163) with the addition of DNase for 1 hr at 37°C. Following enzymatic digestion, organoids were mechanically triturated using a P1000 pipette, filtered through a 40 um cell strainer test tube (Corning 352235), pelleted at 300xg for 5 minutes and washed twice with DBPS. Cells were counted and barcoded with MULTI-seq indices (McGinnis, Patterson et al. 2019) for multiplexing. Three organoid lines with and without microglia were combined and captured on two lanes of 10x Genomics using Chromium single cell 3’ reagent kit (v3 Chemistry) following the manufacturer’s protocol.
Single cell RNA sequencing library preparation
Single cell RNA-seq libraries were generated using the 10x Genomics Chromium 3’ Gene Expression Kit. Briefly, single cells were loaded onto chromium chips with a capture target of 10,000 cells per sample. Libraries were prepared following the provided protocol and sequenced on an Illumina NovaSeq with a targeted sequencing depth of 50,000 reads per cell. BCL files from sequencing were then used as inputs to the 10X Genomics Cell Ranger pipeline.
Immunofluorescence
Cells cultured on glass-bottom well plates were fixed in 4% PFA for 10 minutes. Blocking and permeabilization were performed in a blocking solution consisting of 10% normal donkey serum, 1% Triton X-100, and 0.2% gelatin for 1 hour. Primary and secondary antibodies were diluted and incubated in the blocking solution. Cell cultures were incubated with primary antibodies at the room temperature for 1 hour, washed 3x with washing buffer (0.1% Triton X-100 in PBS), and incubated with secondary antibodies for 1 hour at the room temperature. Images were collected using Leica SP8 confocal system with 20x air objective and 63x oil objective and processed using ImageJ/Fiji and Illustrator.
Organoid samples were fixed in 4% PFA for 1 hour. Tissue sections were cryopreserved in OCT/30% sucrose (1:1) and cryosectioned at 20 μm or 40 μm (for synaptic density staining) thickness. Heat-induced antigen retrieval was performed in 10mM sodium citrate (pH=6.0) for 10 min in boiling-hot solution. Blocking and permeabilization were performed in a blocking solution consisting of 10% normal donkey serum, 1% Triton X-100, and 0.2% gelatin for 1 hour. Primary and secondary antibodies were diluted and incubated in the blocking solution. Primary antibodies used in this study included: rabbit Iba1 (1:500, Wako, 019–19741) guinea pig Iba1 (1:500, Synaptic Systems, 234 004), mouse VGLUT1 (1:200, Millipore Sigma MAB5502), rabbit psd95 (1:350, Thermo Fisher, 51–6900), sheep EOMES (1:200, R&D Biosystems, AF6166), rabbit FOXG1(1:1000, Abcam, ab18259), mouse Tuj1 (1:300, Biolegend, MMS-435P), goat SOX2 (1:200, Santa Cruz Biotechnology, SC-17320), rabbit Pax6 (1:200, Biolegend, 901301), mouse Ki67 (1:200, Dako, MIB-1), rat CTIP2 (1:500, Abcam, ab18465), mouse SATB2 (1:250, Santa Cruz Biotechnology, SC-81376), mouse CD68 (1:100, Abcam, ab955), rabbit pH2AX (1:50, Cell Signalling Technologies, S139), rabbit IFITM3 (1:50, Proteintech, 11714–1-AP), rabbit P2RY12 (1:500, Sigma, HPA014518). Cryosections were incubated with primary antibodies at 4°C overnight, washed 3x with washing buffer (0.1% Triton X-100 in PBS). Secondary antibodies were species-specific AlexaFluor secondary antibodies (1:500). Images were collected using Leica SP8 confocal system with 20x air lens (0.75 NA) and 63x oil lens (1.40 NA) and processed using ImageJ/Fiji and Affinity Designer software.
Axion recordings
For MEA recordings, MACS-purified primary cortical microglia were transplanted into week 15 organoids, and the neuro-immune organoid cultures were allowed to mature for additional 5 weeks. Multielectrode array recordings were conducted using the Axion Maestro Pro system 64 low-impedance PEDOT electrodes with 300 μm electrode spacing. Briefly, organoids were transferred to a 64-channel multielectrode plate and equilibrated at 37 °C, 5% CO2 for at least 15 minutes prior to recording. Spontaneous neural activity was recorded for 40 mins. Firing events were defined as events with threshold amplitude of more than six standard deviations of the background noise. The event amplitude ranged between 8 μV to over 60 μV for some events. The majority of events consisted of multi-unit events and demonstrated waveforms characteristic for a firing neuron. The recordings at 12,500 samples/second were performed in the same media used for organoid growth. After recording, organoids were detached from the MEA plate and kept on the shaker.
QUANTIFICATION AND STATISTICAL ANALYSIS
Single cell RNA sequencing Analysis
For preprocessing of scRNA-seq data, CellRanger was used to create a cell by gene matrix which was then processed using Cellbender (Bernstein, Fong et al. 2019) to remove ambient RNA from every run and Solo (Fleming, Marioni et al. 2019) for doublet detection and removal with default parameters. A minimum of 500 unique genes, and 20% mitochondrial cutoff were used to remove low quality cells from all datasets. The data analyzed in this study were produced through the Brain Initiative Cell Census Network (BICCN: RRID:SCR_015820). To in silico sort microglia from all brain cells of the BICCN datasets, we selected cells expressing 10 or more cumulative UMI counts of the following genes: CCL3L3, CCL4, C3, CCL3, PLEK, FOLR2, ITGAX, SPP1, CSF3R, BIN2, DHRS9, CD74, CD69, IL1B, CX3CR1, OLR1, CH25H, FCGR2A, ADORA3, LAPTM5, P2RY12, IRF8, AIF1. scRNAseq data from microglia from the developing human hippocampus (Zhong, Ding et al. 2020) was in silico filtered by selecting a cluster representing immune cells. After the two data sources were combined, the resulting dataset was further filtered for non-microglial cells after dimensional reduction based on preliminary clustering and marker genes that were not exhibiting canonical signatures, or other cell types. The SCTransform (Hafemeister and Satija 2019) workflow was used and then PCA was computed on the residuals for input into Harmony (Korsunsky, Millard et al. 2019) for batch correction. The parameters of Harmony were set to use the top 20 principal components with theta set to 10. Uniform manifold approximation and projection (UMAP) (Becht, McInnes et al. 2018) embeddings and neighbors for Leiden clustering (Traag, Waltman et al. 2019) used the 10 components outputted from Harmony. Organoid demultiplexing and doublet filtering was done through deMULTIplex (McGinnis, Patterson et al. 2019). Pearson correlation was calculated on the intersection of the shared genes between datasets which averaged Pearson residuals for each cluster. Organoid cells were batch corrected using default parameters of the SCTransform integration workflow. UCell (Andreatta and Carmona 2021) was used to calculate gene signature scores using the top 15 genes ordered by logFC from each primary cluster calculated with a Wilcoxon rank sum test. The clusters from Leiden clustering with the highest signature score per each condition were used for joint comparison across conditions. For joint scoring of multiple conditions, all datasets were normalized using SCTransform together and the corrected normalized expression values were used for input into UCell.
Differential Expression
MAST (Finak, McDavid et al. 2015) was used on log normalized raw counts was used for all differential expression tests. Wilcoxon ranked sum test was used on Pearson residuals from SCTransform for increased sensitivity on microglia treated organoids. Volcano plots were set to have a threshold of Bonferroni corrected p-value of 0.05. Differentially expressed sex genes were calculated per specific cluster using Wilcoxon rank sum test on the log normalized counts. MALAT1 and mitochondrial genes were removed from visualization but remain in the Table S2.
Synapse quantification
Organoid confocal images were acquired with a Leica SP8 system. For synapse quantification, three different optical fields per organoid were imaged. For each optical field, 15 μm Z-stack (0.5μm Z-step) were collected using a 63X/1.40 oil objective. Synapse determination was based on the colocalization between Vglut1 (1:200; Millipore Sigma MAB5502) and PSD95 (1:350; Invitrogen 51–6900) as previously described (Lehrman, Wilton et al. 2018) using ImageJ software. Briefly, the background was subtracted with a rolling bar radius of 10 pixels. Then, threshold was applied to every channel in order to distinguish synaptic puncta from background and generate two new binary images with the synaptic markers. Colocalization was determined overlaying both binary images with the synaptic markers. Finally, synaptic puncta were determined using the function “analyze particles”.
For volume quantification, all the volumes of synapse, lysosome and microglia are retrieved from Imaris based on the 3D-rendered structures.
IFITM3 quantification
Microglia were engrafted into week-5 organoids and were cultured as described above for seven days before being fixed. IFITM3 values were measured using ImageJ. Freehand selection tool was used to outline Sox2-positive rosettes, and then mean value intensities were measured for the three channels (DAPI, Sox2, IFITM3). IFITM3 fluorescence values were normalized to the DAPI fluorescence values to account for different number of cells in each region of interest. A two-tailed Student’s t-test was used to determine significance.
pH2AX quantification
Microglia were engrafted into week-5 organoids and were cultured as described above for seven days before being fixed. pH2AX puncta were quantified using the CellProfiler 4.1.3 software (McQuin, Goodman et al. 2018). The “Speckle Counting” example pipeline was modified to produce robust identification of puncta. First, Sox2+ nuclei were identified using a diameter range of 20–100 pixels, threshold strategy “Global”, and threshold method “Otsu.” The pH2AX channel was first enhanced and then masked, and puncta were subsequently counted using a diameter range of 1–10 pixels, threshold strategy “Adaptive”, and threshold method “Otsu.” A parent-child relationship was then assigned to group pH2AX puncta by Sox2+ nucleus, and the CellProfiler output was exported to Microsoft Excel. Nuclei with 0–1 or >30 puncta were discarded, to avoid counting cells with high pH2AX signal that were actively dividing. The average number of pH2AX puncta per Sox2+ nucleus was calculated in organoid conditions with and without microglia. Then, nuclei were binned to generate a histogram of total pH2AX puncta within each bin in each condition. A two-tailed T-test was used to determine significance.
Axion data analysis
Raw data from the Axion recordings were read into Matlab then converted to numpy arrays for further processing in Python. Spikes were counted using a standard deviation threshold of 6 and converted to an array of spike times for each individual channel. Bursting activity was determined by the presence of 10 or more spikes on a given channel within one second. Next, recording time was divided into 40 ms bins and spike events were binarized based on whether or not there was at least one spike within a given time bin. Pairwise correlation between channels was determined by calculating the Pearson correlation between these binarized spike times.
Supplementary Material
Video S3. 3D-rendered microglia engraftment into organoids. Related to Figure 2E.
Video S4. Microglia with internalized synaptic material. Related to Figure 7.
Video S1. MACS-purified cortical microglia cultured in 2D. Time-lapse microscopy of cultured microglia after 8 days in culture. Images were taken every 20 mins for 24 hours. Related to Figure 1A.
Video S2. Cell-tracker-labeled microglia engraft into organoids within 24 hours in culture. Primary human microglia were purified and labeled with Cell Tracker. Microglia then were added to a dish with an organoid and were recorder for 24 hours. 3D rendering is presented in Video S3. Related to Figure 2E.
Table S2: Differentially expressed genes in organoids with and without microglia. Related to Figure 5.
Table S1: Differentially expressed genes between male and female microglia. Related to Figure 1.
HIGHLIGHTS.
Microglia culture models differentially attenuate and preserve gene signatures
Brain organoid microenvironment preserves microglial homeostatic state
Genes in the interferon response pathway are attenuated in the presence of microglia
Microglia decrease double stranded DNA breaks and cell stress in radial glia
Acknowledgments
We thank Drs. Aparna Bhaduri and Arnold Kriegstein for generously facilitating the access to BICCN datasets. We thank all members of the Nowakowski, Mehta and Piao laboratories for helpful discussions and comments throughout this project. This study was supported by gifts from Schmidt Futures and the William K. Bowes Jr. Foundation, Simons Foundation grant (SFARI 491371 to T.J.N.), Chan Zuckerberg Biohub Intercampus Investigator Award (to T.J.N), and NRSA F32 1F32MH118785 (to GP).
Footnotes
Declaration of Interests
The authors declare no competing interests.
ADDITIONAL RESOURCES
Figures were created using Affinity Designer software and BioRender (BioRender.com).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, Caraway CA, Fote GM, Madany AM, Agrawal A, Kayed R, Gylys KH, Cahalan MD, Cummings BJ, Antel JP, Mortazavi A, Carson MJ, Poon WW and Blurton-Jones M (2017). “iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases.” Neuron 94(2): 278–293 e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M and Carmona SJ (2021). “UCell: robust and scalable single-cell gene signature scoring.” bioRxiv: 2021.2004.2013.439670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badimon A, Strasburger HJ, Ayata P, Chen X, Nair A, Ikegami A, Hwang P, Chan AT, Graves SM, Uweru JO, Ledderose C, Kutlu MG, Wheeler MA, Kahan A, Ishikawa M, Wang YC, Loh YE, Jiang JX, Surmeier DJ, Robson SC, Junger WG, Sebra R, Calipari ES, Kenny PJ, Eyo UB, Colonna M, Quintana FJ, Wake H, Gradinaru V and Schaefer A (2020). “Negative feedback control of neuronal activity by microglia.” Nature 586(7829): 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F and Newell EW (2018). “Dimensionality reduction for visualizing single-cell data using UMAP.” Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Bennett ML, Yaqoob F, Mulinyawe SB, Grant GA, Hayden Gephart M, Plowey ED and Barres BA (2018). “A Combination of Ontogeny and CNS Environment Establishes Microglial Identity.” Neuron 98(6): 1170–1183 e1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein N, Fong N, Lam I, Roy M, Hendrickson DG and Kelley DR (2019). “Solo: doublet identification via semi-supervised deep learning.” bioRxiv: 841981. [DOI] [PubMed] [Google Scholar]
- Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, Jung D, Schmunk G, Haeussler M, Salma J, Pollen AA, Nowakowski TJ and Kriegstein AR (2020). “Cell stress in cortical organoids impairs molecular subtype specification.” Nature 578(7793): 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB and Barres BA (2017). “Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures.” Neuron 94(4): 759–773 e758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotti D, Gentili V, Rotola A, Caselli E and Rizzo R (2019). “HHV-6A infection induces amyloid-beta expression and activation of microglial cells.” Alzheimers Res Ther 11(1): 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostek-Racine S, Gordon C, Van Scoy S and Reich NC (2011). “The DNA damage response induces IFN.” J Immunol 187(10): 5336–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle L, Rivera SA, Phelps JS, Ennis KA, Stevens B, Burkly LC, Lee WA and Greenberg ME (2020). “Sensory Experience Engages Microglia to Shape Neural Connectivity through a Non-Phagocytic Mechanism.” Neuron 108(3): 451–468 e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR and Ma’ayan A (2013). “Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool.” BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RS, Leung YK, Butler CW, Chen Y, Eaton ED, Pankhurst MW, West AK and Guillemin GJ (2009). “Metallothionein treatment attenuates microglial activation and expression of neurotoxic quinolinic acid following traumatic brain injury.” Neurotox Res 15(4): 381–389. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V and Noctor SC (2013). “Microglia regulate the number of neural precursor cells in the developing cerebral cortex.” J Neurosci 33(10): 4216–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS and Gottardo R (2015). “MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data.” Genome Biol 16: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming SJ, Marioni JC and Babadi M (2019). “CellBender remove-background: a deep generative model for unsupervised removal of background noise from scRNA-seq datasets.” bioRxiv: 791699. [Google Scholar]
- Garden GA (2002). “Microglia in human immunodeficiency virus-associated neurodegeneration.” Glia 40(2): 240–251. [DOI] [PubMed] [Google Scholar]
- Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, Balic A, Giladi A, Sheban F, Dutertre CA, Pfeifle C, Peri F, Raffo-Romero A, Vizioli J, Matiasek K, Scheiwe C, Meckel S, Matz-Rensing K, van der Meer F, Thormodsson FR, Stadelmann C, Zilkha N, Kimchi T, Ginhoux F, Ulitsky I, Erny D, Amit I and Prinz M (2019). “Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program.” Cell 179(7): 1609–1622 e1616. [DOI] [PubMed] [Google Scholar]
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH and Glass CK (2017). “An environment-dependent transcriptional network specifies human microglia identity.” Science 356(6344). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C and Satija R (2019). “Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression.” Genome Biol 20(1): 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, Marsh SE, Saunders A, Macosko E, Ginhoux F, Chen J, Franklin RJM, Piao X, McCarroll SA and Stevens B (2019). “Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes.” Immunity 50(1): 253–271 e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P and Carpenter AE (2008). “CellProfiler Analyst: data exploration and analysis software for complex image-based screens.” BMC Bioinformatics 9: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M and Amit I (2017). “A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease.” Cell 169(7): 1276–1290 e1217. [DOI] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR and Raychaudhuri S (2019). “Fast, sensitive and accurate integration of single-cell data with Harmony.” Nat Methods 16(12): 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracht L, Borggrewe M, Eskandar S, Brouwer N, Chuva de Sousa Lopes SM, Laman JD, Scherjon SA, Prins JR, Kooistra SM and Eggen BJL (2020). “Human fetal microglia acquire homeostatic immune-sensing properties early in development.” Science 369(6503): 530–537. [DOI] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, Greco DJ, Smith ST, Tweet G, Humulock Z, Zrzavy T, Conde-Sanroman P, Gacias M, Weng Z, Chen H, Tjon E, Mazaheri F, Hartmann K, Madi A, Ulrich JD, Glatzel M, Worthmann A, Heeren J, Budnik B, Lemere C, Ikezu T, Heppner FL, Litvak V, Holtzman DM, Lassmann H, Weiner HL, Ochando J, Haass C and Butovsky O (2017). “The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases.” Immunity 47(3): 566–581 e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P and Gordon S (1990). “Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain.” Neuroscience 39(1): 151–170. [DOI] [PubMed] [Google Scholar]
- Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, Walker AJ, Heller MD, Umemori H, Chen C and Stevens B (2018). “CD47 Protects Synapses from Excess Microglia-Mediated Pruning during Development.” Neuron 100(1): 120–134 e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, Marschallinger J, Yu G, Quake SR, Wyss-Coray T and Barres BA (2019). “Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing.” Neuron 101(2): 207–223 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chiou B, Gilman CK, Luo R, Koshi T, Yu D, Oak HC, Giera S, Johnson-Venkatesh E, Muthukumar AK, Stevens B, Umemori H and Piao X (2020). “A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding.” EMBO J 39(16): e104136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SE, Kamath T, Walker AJ, Dissing-Olesen L, Hammond TR, Young AMH, Abdulraouf A, Nadaf N, Dufort C, Murphy S, Kozareva V, Vanderburg C, Hong S, Bulstrode H, Hutchinson PJ, Gaffney DJ, Franklin RJM, Macosko EZ and Stevens B (2020). “Single Cell Sequencing Reveals Glial Specific Responses to Tissue Processing & Enzymatic Dissociation in Mice and Humans.” bioRxiv: 2020.2012.2003.408542. [Google Scholar]
- Masuda, Sankowski TR, Staszewski O, Bottcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grun D, Priller J, Stadelmann C and Prinz M (2019). “Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution.” Nature 566(7744): 388–392. [DOI] [PubMed] [Google Scholar]
- McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS, Werb Z, Chow ED and Gartner ZJ (2019). “MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices.” Nat Methods 16(7): 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, Wiegraebe W, Singh S, Becker T, Caicedo JC and Carpenter AE (2018). “CellProfiler 3.0: Next-generation image processing for biology.” PLoS Biol 16(7): e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini M, Rafalski VA, Ma K, Kim KY, Bushong EA, Rios Coronado PE, Yan Z, Mendiola AS, Sozmen EG, Ryu JK, Haberl MG, Madany M, Sampson DN, Petersen MA, Bardehle S, Tognatta R, Dean T Jr., Acevedo RM, Cabriga B, Thomas R, Coughlin SR, Ellisman MH, Palop JJ and Akassoglou K (2020). “Microglial Gi-dependent dynamics regulate brain network hyperexcitability.” Nat Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montilla A, Zabala A, Matute C and Domercq M (2020). “Functional and Metabolic Characterization of Microglia Culture in a Defined Medium.” Front Cell Neurosci 14: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman LA and McAllister AK (2012). “The major histocompatibility complex and autism spectrum disorder.” Dev Neurobiol 72(10): 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F and Helmchen F (2005). “Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo.” Science 308(5726): 1314–1318. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, Ounadjela JR, Shuga J, Wang X, Lim DA, West JA, Leyrat AA, Kent WJ and Kriegstein AR (2017). “Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex.” Science 358(6368): 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhof N, Chang IJ, Karimiani EG, Kuil LE, Jensen DM, Daza R, Young E, Astle L, van der Linde HC, Shivaram GM, Demmers J, Latimer CS, Keene CD, Loter E, Maroofian R, van Ham TJ, Hevner RF and Bennett JT (2019). “Homozygous Mutations in CSF1R Cause a Pediatric-Onset Leukoencephalopathy and Can Result in Congenital Absence of Microglia.” Am J Hum Genet 104(5): 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel PR, Vieira de Sa R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, Johansen LE, van Dijk RE, Scheefhals N, Berdenis van Berlekom A, Ribes Martinez E, Kling S, MacGillavry HD, van den Berg LH, Kahn RS, Hol EM, de Witte LD and Pasterkamp RJ (2018). “Microglia innately develop within cerebral organoids.” Nat Commun 9(1): 4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, Hempstead BL, Littman DR and Gan WB (2013). “Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor.” Cell 155(7): 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA and Pasca SP (2015). “Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture.” Nat Methods 12(7): 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca SP (2018). “The rise of three-dimensional human brain cultures.” Nature 553(7689): 437–445. [DOI] [PubMed] [Google Scholar]
- Penkowa M, Carrasco J, Giralt M, Moos T and Hidalgo J (1999). “CNS wound healing is severely depressed in metallothionein I- and II-deficient mice.” J Neurosci 19(7): 2535–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Medina R, Baena M, Planas AM and Pozas E (2015). “IFN gamma regulates proliferation and neuronal differentiation by STAT1 in adult SVZ niche.” Front Cell Neurosci 9: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, Lash J, Wider C, Wojtas A, DeJesus-Hernandez M, Adamson J, Kouri N, Sundal C, Shuster EA, Aasly J, MacKenzie J, Roeber S, Kretzschmar HA, Boeve BF, Knopman DS, Petersen RC, Cairns NJ, Ghetti B, Spina S, Garbern J, Tselis AC, Uitti R, Das P, Van Gerpen JA, Meschia JF, Levy S, Broderick DF, Graff-Radford N, Ross OA, Miller BB, Swerdlow RH, Dickson DW and Wszolek ZK (2011). “Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids.” Nat Genet 44(2): 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J, Pollen AA, Mandel-Brehm C, Nowakowski TJ, Kriegstein AR and DeRisi JL (2016). “Zika virus cell tropism in the developing human brain and inhibition by azithromycin.” Proc Natl Acad Sci U S A 113(50): 14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigato C, Buckinx R, Le-Corronc H, Rigo JM and Legendre P (2011). “Pattern of invasion of the embryonic mouse spinal cord by microglial cells at the time of the onset of functional neuronal networks.” Glia 59(4): 675–695. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA and Stevens B (2012). “Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner.” Neuron 74(4): 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Schizophrenia C Working Group of the Psychiatric Genomics, Daly MJ, Carroll MC, Stevens B and McCarroll SA (2016). “Schizophrenia risk from complex variation of complement component 4.” Nature 530(7589): 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda DS, Barrasa MI, Shu J, Rietjens R, Zhang S, Mitalipova M, Berube P, Fu D, Shultz LD, Bell GW and Jaenisch R (2019). “Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain.” Proc Natl Acad Sci U S A 116(50): 25293–25303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YL, Yuan Y and Tian L (2020). “Microglial regional heterogeneity and its role in the brain.” Mol Psychiatry 25(2): 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag VA, Waltman L and van Eck NJ (2019). “From Louvain to Leiden: guaranteeing well-connected communities.” Sci Rep 9(1): 5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, Domissy A, Vandenberghe M, Devor A, Yeo GW, Voytek B and Muotri AR (2019). “Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development.” Cell Stem Cell 25(4): 558–569 e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V and Giachino C (2015). “Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions.” Nucleic Acids Res 43(5): 2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wei PC, Lim CK, Gallina IS, Marshall S, Marchetto MC, Alt FW and Gage FH (2020). “Increased Neural Progenitor Proliferation in a hiPSC Model of Autism Induces Replication Stress-Associated Genome Instability.” Cell Stem Cell 26(2): 221–233 e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW and Schwer B (2016). “Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells.” Cell 164(4): 644–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhard L, di Bartolomei G, Bolasco G, Machado P, Schieber NL, Neniskyte U, Exiga M, Vadisiute A, Raggioli A, Schertel A, Schwab Y and Gross CT (2018). “Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction.” Nat Commun 9(1): 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Li Y, Wei Y, Bosco DB, Xie M, Zhao MG, Richardson JR and Wu LJ (2020). “Microglial depletion aggravates the severity of acute and chronic seizures in mice.” Brain Behav Immun 89: 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J and Linnarsson S (2015). “Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq.” Science 347(6226): 1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, Vyssotski AL, Bifone A, Gozzi A, Ragozzino D and Gross CT (2014). “Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior.” Nat Neurosci 17(3): 400–406. [DOI] [PubMed] [Google Scholar]
- Zhong S, Ding W, Sun L, Lu Y, Dong H, Fan X, Liu Z, Chen R, Zhang S, Ma Q, Tang F, Wu Q and Wang X (2020). “Decoding the development of the human hippocampus.” Nature 577(7791): 531–536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S3. 3D-rendered microglia engraftment into organoids. Related to Figure 2E.
Video S4. Microglia with internalized synaptic material. Related to Figure 7.
Video S1. MACS-purified cortical microglia cultured in 2D. Time-lapse microscopy of cultured microglia after 8 days in culture. Images were taken every 20 mins for 24 hours. Related to Figure 1A.
Video S2. Cell-tracker-labeled microglia engraft into organoids within 24 hours in culture. Primary human microglia were purified and labeled with Cell Tracker. Microglia then were added to a dish with an organoid and were recorder for 24 hours. 3D rendering is presented in Video S3. Related to Figure 2E.
Table S2: Differentially expressed genes in organoids with and without microglia. Related to Figure 5.
Table S1: Differentially expressed genes between male and female microglia. Related to Figure 1.
Data Availability Statement
Single-cell RNA-seq data for non-human data have been deposited at GEO (GEO: GSE180945) and are publicly available as of the date of publication. Newly generated single-cell RNA-seq data from de-identified human subjects has been deposited at Synapse, (https://www.synapse.org/#!Synapse:syn26009957) and require an authorized user login. Any questions should be referred to the Lead Contact. Re-analyzed data from other available sources is listed in the Deposited Data section of Reagents and Resource. Accession numbers for all data are listed in the key resources table. Microscopy data reported in this paper will be shared by the Lead Contact upon request.
All data analysis has been done by using previously published pipelines with DOIs listed in the key resource table. A detailed description of the computational processing and parameters is provided in Methods Details.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD68 | Abcam | Abcam Cat# ab955, RRID:AB_307338 |
| CTIP2 | Abcam | Cat# ab18465, RRID:AB_2064130 |
| EOMES | R&D Systems | Cat# AF6166, RRID:AB_10569705 |
| FOXG1 | Abcam | Cat# ab196868, RRID:AB_2892604 |
| Iba1 | Synaptic Systems | Cat# 234 004, RRID:AB_2493179 |
| Iba1 | Wako | Cat# 019-19741, RRID:AB_839504 |
| IFITM3 | Proteintech | Cat# 11714-1-AP, RRID:AB_2295684 |
| Ki67 | Agilent | Cat# M724001-2, RRID:AB_2631211 |
| P2RY12 | Sigma Aldrich | Cat# HPA014518, RRID:AB_2669027 |
| PAX6 | BioLegend | Cat# 901301, RRID:AB_2565003 |
| pH2AX | Cell Signaling Technologies | Cat# 9718, RRID:AB_2118009 |
| psd95 | Thermo Fisher | Cat# 51-6900, RRID:AB_2533914 |
| SATB2 | Santa Cruz Biotechnology | Cat# sc-81376, RRID:AB_1129287 |
| SOX2 | Santa Cruz Biotechnology | Cat# sc-17320, RRID:AB_2286684 |
| Tuj1 | Biolegend | Cat# 80120, RRID:AB_2313773 |
| VGLUT1 | Millipore Sigma | Cat# MAB5502, RRID:AB_262185 |
| Bacterial and Viral Strains | ||
| AV-CMV-GFP | Vector Biolabs | 1060 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Antibiotic-Antimycotic | Gibco | 15240-062 |
| B27 supplement w/o Vitamin A | Gibco | 12587-001 |
| B27 w/ Vitamin A | Gibco | 17504-001 |
| BDNF | Alomone | b250 |
| CD lipid concentrate | Thermo Fisher | 11905031 |
| CellTracker CM-DiI | Thermo Fisher | C7000 |
| DAPI Fluoromount | Southern Biotech | 0100-20 |
| DMEM/F12 with Glutamax | Gibco | 10565042 |
| Dnase | Sigma Aldrich | DN25 |
| Donkey Serum | Jackson Immuno | 017-000-121 |
| Dorsomorphin | Sigma-Aldrich | P5499 |
| DPBS | Gibco | 14190250 |
| Eagle’s basal medium | UCSF Media Production Core | N/A |
| EGF | Peprotech | 100-47 |
| FBS | HyClone | SH30071.03 |
| Fibronectin | Corning | 354008 |
| FGFb | Peprotech | 100-18B |
| GlutaMax Supplement | Thermo Fisher Scientific | 35050061 |
| HBSS | UCSF Media Production Core | N/A |
| Human Insulin | Sigma | I2643-50MG |
| IL34 | Peprotech | 200-34 |
| Insulin-Transferrin-Selenium (ITS-G) (100×) | Thermo Fisher Scientific | 41400045 |
| IWR1 -ε | Cayman Chemical | 13659 |
| Laminin | Thermo Fisher | 23017015 |
| Matrigel | Fisher Scientific | 354234 |
| N2 supplement | Gibco | 17502-048 |
| Neurobasal | Gibco | 21103049 |
| Non-essential Amino Acids (NEAA) | Thermo Fisher Scientific | 11140050 |
| PBS-EDTA, pH 7.5 | Lonza | BE02-017F |
| Trypsin, 2.5% | Gibco | 15090046 |
| Trypsin, 0.25% | Thermo Fisher | 25200056 |
| Critical Commercial Assays | ||
| 10X Chromium V2 | 10X Genomics | PN-120237 |
| 10X Chromium V3 | 10X Genomics | PN-1000092 |
| Cd11b magnetic beads | Miltenyi Biotec | 130-049-601 |
| Experimental Models: Cell Lines | ||
| Human iPS cell line 28126 (male) | Gilad Lab (Gallego Romero et al., 2015) | |
| Human iPS cell line “WTC-11” (male) | Conklin Lab (Bershteyn et al., 2017; Kreitzer et al., 2013) | CVCL_Y803 |
| Human iPS cell line “1323-4” (female) | Conklin Lab (Matsumoto et al., 2013) | CVCL_0G84 |
| Human iPS cell line “WTC-10” (male) | Conklin Lab (Bershteyn et al., 2017; Kreitzer et al., 2013) | |
| Human iMG | Fujifilm Cellular Dynamics | 01279 |
| Software and Algorithms | ||
| ImageJ (Fiji) | Schindelin et al., 2012 | https://imagej.net/Fiji |
| CellRanger v3.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Seurat v3.0 | Satija Lab (Butler et al., 2018a) | https://satijalab.org/seurat/ |
| deMULTIplex | Gartner Lab (McGinnis et al., 2019) | https://github.com/chris-mcginnis-ucsf/MULTI-seq |
| CellProfiler 3.0 | McQuin, Goodman et al. 2018 | https://cellprofiler.org/ |
| CellBender | (Fleming, Marioni et al. 2019) | https://github.com/broadinstitute/CellBender |
| UCell | (Andreatta and Carmona 2021) | https://github.com/carmonalab/UCell |
| Deposited data | ||
| Primary microglia BICCN scRNAseq | Brain Initiative Cell Census Network | NeMO archive: RRID:SCR_015820 |
| Primary microglia in vitro culture scRNAseq | This study | Synapse: syn26009957 |
| Primary microglia in organoids scRNAseq | This study | Synapse: syn26009957 |
| Induced microglia scRNAseq | This study | GEO: GSE180945 |
| Organoids scRNAseq | This study | GEO: GSE180945 |
| Microglia in mouse brain scRNAseq | (Svoboda, Barrasa et al. 2019) | GEO: GSE139194 |
| Other | ||
| 6-well Ultralow Attachment Plates | Corning | 3471 |
| 40 μm cell strainer | Corning | 352340 |
| LS columns | Miltenyi Biotec | 130-042-401 |
| glass-bottom 24 well plates | Cellvis | P24-1.5H-N |
| glass-bottom six-well plate | Mattek | P06G-1.5-10-F |