SUMMARY
AMPA receptors (AMPARs) mediate the majority of excitatory neurotransmission. Their surface expression, trafficking, gating, and pharmacology are regulated by auxiliary subunits. Of the two types of TARP auxiliary subunits, type I TARPs assume activating roles, while type II TARPs serve suppressive functions. We present cryo-EM structures of GluA2 AMPAR in complex with type II TARP γ5, which reduces steady-state currents, increases single-channel conductance, and slows recovery from desensitization. Regulation of AMPAR function depends on its ligand-binding domain (LBD) interaction with the γ5 head domain. GluA2-γ5 complex shows maximum stoichiometry of two TARPs per AMPAR tetramer, being different from type I TARPs but reminiscent of the auxiliary subunit GSG1L. Desensitization of both GluA2-GSG1L and GluA2-γ5 complexes is accompanied by rupture of LBD dimer interface, while GluA2-γ5 but not GluA2-GSG1L LBD dimers remain 2-fold symmetric. Different structural architectures and desensitization mechanisms of complexes with auxiliary subunits endow AMPARs with broad functional capabilities.
Keywords: cryo-EM, iGluR, AMPA receptor, TARP, γ5, GSG1L
Graphical Abstract
eTOC Blurb
Klykov et al. solve structures of AMPA receptor in complex with auxiliary subunits type II TARP γ5 and GSG1L in closed and desensitized states, with stoichiometry of two auxiliary subunits per receptor tetramer. GluA2-γ5 and GluA2-GSG1L desensitization is accompanied by rupture of LBD dimer interface but follows different structural mechanisms.
INTRODUCTION
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) represent the fastest types of ionotropic glutamate receptors (iGluRs) that mediate the majority of excitatory neurotransmission in the central nervous system (Traynelis et al., 2010). AMPARs are tetrameric ion channels that have a three-layer Y-shaped architecture with an overall two-fold rotational symmetry (Sobolevsky et al., 2009). The layer of four amino-terminal domains (ATDs) at the top of the Y is connected by four linkers to the layer of four ligand-binding domains (LBDs) in the middle of the receptor. Each LBD is comprised of two polypeptide stretches (S1 and S2) and responsible for agonist, competitive antagonist, and positive allosteric modulator binding. Each of the four transmembrane domains (TMDs) at the bottom of the Y is linked to the corresponding LBD by three flexible linkers and includes three transmembrane helices (M1, M3, and M4) and a re-entrant pore loop (M2) between M1 and M3. Four TMDs assemble a cation-selective ion channel with its pore lined by M2 and M3 and the gate formed by the M3 helices bundle crossing.
The majority of neuronal AMPARs function as central elements of synaptic complexes being surrounded by auxiliary subunits, which regulate receptor trafficking, scaffolding, stability, turnover, and physiological responses (Jackson and Nicoll, 2011; Kamalova and Nakagawa, 2021; Twomey et al., 2019). Transmembrane AMPA Receptor Regulatory Proteins (TARPs) are the most abundant AMPAR-bound auxiliary subunits (Chen et al., 2000; Kato et al., 2008; Kato et al., 2007; Tomita et al., 2005; Tomita et al., 2003), originally named “γ-subunits” based on their sequence homology to γ1 calcium channel subunit (Jay et al., 1990). Of eight identified γ-subunits, γ1 and γ6 occur largely in skeletal and cardiac muscles and do not regulate AMPARs, while the other six represent TARPs that belong to type I (γ2, γ3, γ4, and γ8) or type II (γ5 and γ7) based on their functional properties and amino acid sequence similarity (Jackson and Nicoll, 2011; Kato et al., 2008; Tomita et al., 2003). Typically type I TARPs increase AMPAR surface expression, slow down desensitization and deactivation kinetics, increase channel open probability and steady-state current amplitude (Ben-Yaacov et al., 2017; Carrillo et al., 2020; Chen et al., 2000; Kato et al., 2008; Kato et al., 2007; Priel et al., 2005; Soto et al., 2007; Soto et al., 2009; Tomita et al., 2005; Tomita et al., 2003). Structures of two type I TARP representatives, γ2 (or stargazin) and γ8, have been determined, with the maximum observed stoichiometry of four TARPs γ2 per one AMPAR tetramer (4:1) (Chen et al., 2017; Herguedas et al., 2019; Twomey et al., 2016, 2017a; Twomey et al., 2018; Zhao et al., 2016).
Similar to type I TARPs, type II TARPs increase the ion channel conductance and attenuate polyamine block of calcium-permeable AMPARs (Shelley et al., 2012; Soto et al., 2009). However, other functional properties of AMPARs are regulated differently by type II TARPs compared to type I TARPs. While some of γ7 functional effects are reminiscent of type I TARPs, γ5 displays entirely dissimilar properties: it reduces or does not alter AMPAR surface expression, does not slow the rates of desensitization and deactivation, and decreases the affinity to agonist glutamate (Glu), channel open probability and steady-state current amplitude (Kato et al., 2008; Kato et al., 2007; Soto et al., 2009). Both type II TARPs are spread across the brain; the highest levels of γ5 are detected in Bergmann glia, while γ7 is enriched in Purkinje cells and cerebellum (Kato et al., 2008; Kato et al., 2007). Distinct regulation of AMPARs by type II TARPs defines their unique role in synaptic physiology and disease (Drummond et al., 2013; Jackson and Nicoll, 2011; Kamalova and Nakagawa, 2021; Twomey et al., 2019) but the structural basis of type II TARP function has remained unknown.
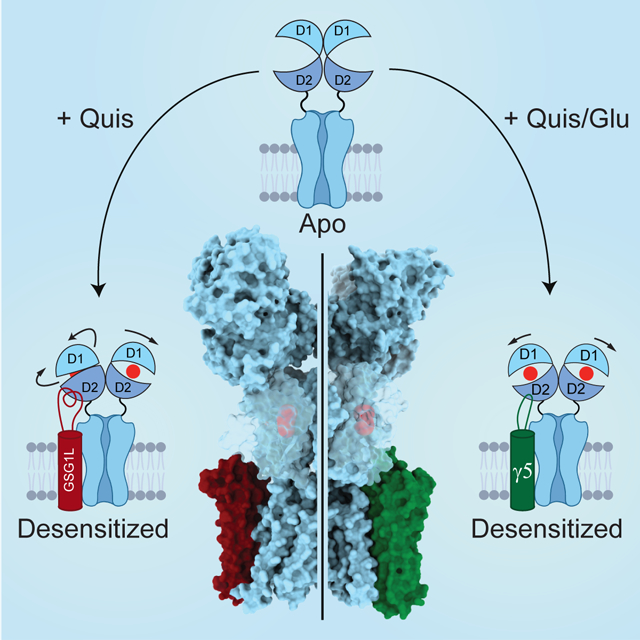
Here we employ cryo-EM to solve structures of GluA2 AMPAR in complex with type II TARP γ5. The structures display a stoichiometry of only two TARPs per AMPAR tetramer (2:1), which is different from the 4:1 maximum stoichiometry for type I TARPs but reminiscent of another auxiliary subunit GSG1L (germ cell-specific gene 1-like). GSG1L, which is enriched in anterior thalamic nuclei, involved in long-term potentiation, synapse formation, and plasticity, determines seizure susceptibility, and serves a generally suppressive function alike type II TARPs (Gu et al., 2016; Kamalova et al., 2021; Mao et al., 2017; McGee et al., 2015; Schwenk et al., 2012; Shanks et al., 2012; Twomey et al., 2017a, b). While the closed-state structures of GluA2-γ5 and GluA2-GSG1L complexes appear similar, their desensitized-state structures are different and determine distinct desensitization mechanisms. During desensitization, LBD dimers of the AMPAR-TARP complex retain their 2-fold rotational symmetry and the ones of the AMPAR-GSG1L complex become asymmetric.
RESULTS AND DISCUSSION
Functional characterization and cryo-EM
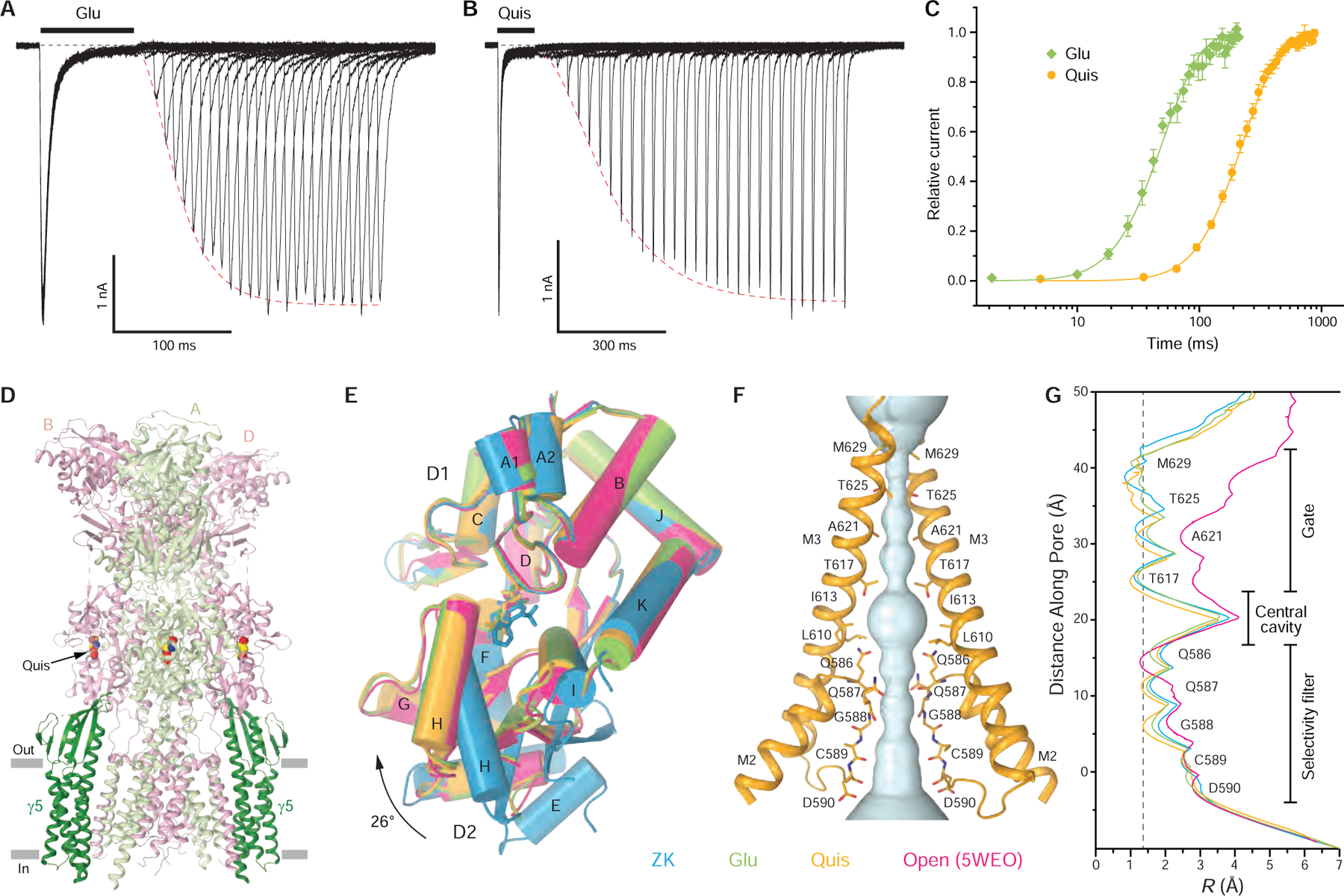
To readily assemble GluA2-γ5 complex in HEK 293 cells, we utilized a covalent fusion construct design, where the N-terminus of γ5 was covalently linked to the C-terminus of GluA2 (modified calcium-permeable rat GluA2flip subunit with Q586 at the Q/R-site, see Methods), and tested its function using patch-clamp recordings (Figure 1A). At −60 mV membrane potential, 2-ms application of 3 mM Glu elicited a rapidly activating inward current that quickly (τDeact = 2.13 ± 0.15 ms, n = 8) decayed to zero, mainly as a result of receptor deactivation (Figure 1A, black trace). Prolonged, 1-s Glu application elicited an inward current that decayed in the continuous presence of Glu more slowly (τDes = 9.25 ± 0.81 ms, n = 7), apparently due to receptor desensitization (Figure 1A, blue trace). Desensitization was nearly eliminated when Glu was applied in the presence of the positive allosteric modulator cyclothiazide (CTZ) (Figure 1A, green trace). As an estimate of the fraction of non-desensitized channels, we calculated the ratio of the steady-state current in the continuous presence of Glu (ISS) and the maximal current amplitudes in the presence of CTZ (IMax), ISS / IMax = 0.0091 ± 0.0029 (n = 6). The much smaller value of ISS / IMax for GluA2-γ5 compared to GluA2 alone (ISS / IMax = 0.042) (Yelshanskaya et al., 2017) is consistent with the reduction of steady-state currents reported for AMPARs co-expressed with γ5 (Kato et al., 2008) and suggests that γ5 favors non-conducting states of the receptor.
Figure 1. Functional and structural characterization of GluA2-γ5.
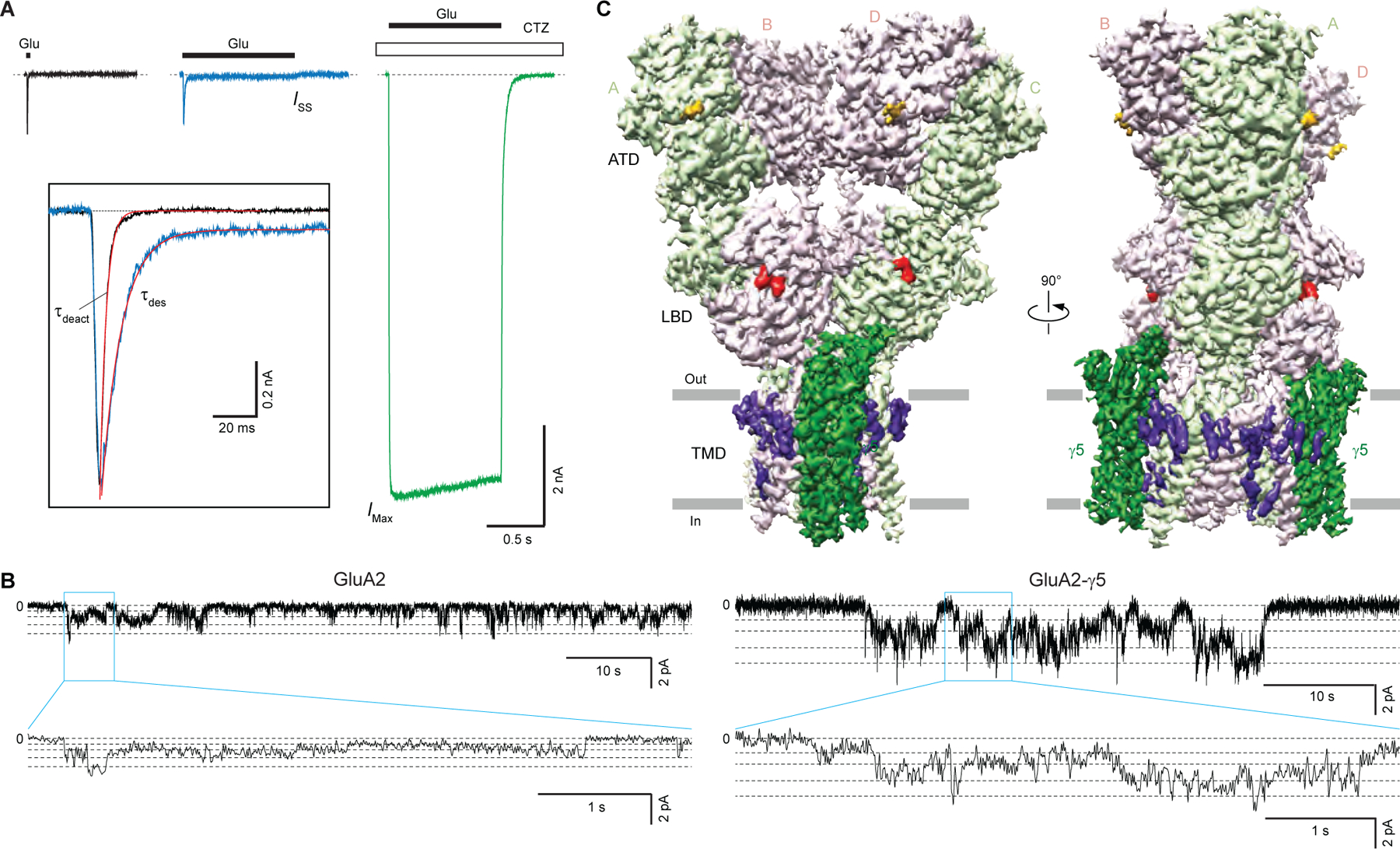
(A) Representative whole-cell currents recorded at −60 mV membrane potential from HEK 293 cell expressing GluA2-γ5 in response to 2-ms (black) or 1-s (blue) applications of 3 mM Glu alone or application of Glu in the continuous presence of 30 μM CTZ (green). The inset shows normalized currents in response to 2-ms and 1-s applications of Glu alone fitted using single exponentials (red curves).
(B) Representative single-channel currents recorded at −100 mV potential in the continuous presence of 10 mM Glu and 100 μM CTZ from GluA2 (left) and GluA2-γ5 (right) reconstituted into lipid bilayers (upper traces), with expanded views of the regions indicated by the blue boxes (lower trace). Horizontal dashed lines indicate different conductance levels.
(C) 3D cryo-EM reconstruction of GluA2-γ5, with density for GluA2 subunits colored in light green and pink, γ5 in dark green, lipids in purple, antagonist ZK200775 in red, and carbohydrates in orange.
See also Figures S1 and S3, Table S1.
To verify the functionality of purified GluA2-γ5, we reconstituted it into planar lipid bilayers and recorded single-channel currents in the presence of 10 mM Glu and 100 μM CTZ. Consistent with previous observations for AMPA receptors expressed alone or in the presence of TARPs (Carrillo et al., 2020; Prieto and Wollmuth, 2010; Rosenmund et al., 1998; Shelley et al., 2012; Smith and Howe, 2000; Swanson et al., 1997; Tomita et al., 2005), GluA2-γ5 exhibited multiple conductance levels (Figure 1B). Fitting of the GluA2-γ5 current amplitude distribution with Gaussians identified four conductance levels: 9.74 ± 0.89 pS (level 1), 20.18 ± 1.63 pS (level 2), 34.16 ± 2.58 pS (level 3) and 58.16 ± 2.58 pS (level 4) (n = 5). In line with previous observations for AMPARs co-expressed with γ5 (Shelley et al., 2012; Soto et al., 2009), the conductance values for GluA2-γ5 were higher than the corresponding values measured for GluA2 alone, in the absence of auxiliary subunits: 6.68 ± 0.74 pS (level 1), 14.50 ± 0.78 pS (level 2), 23.83 ± 1.77 pS (level 3) and 33.07 ± 1.34 pS (level 4) (n = 4). Single-channel activity of GluA2-γ5 in our experiments was higher than the activity of GluA1-γ5 in experiments of Shelley et al. (Shelley et al., 2012), marked by the presence of the fourth conductance level in our recordings that was not detected in the experiments of Shelley et al. The increased activity in our experiments could be due to differences in the probability of maximal pore opening between GluA1 and GluA2 as well as the presence of CTZ in our experiments and its absence in experiments of Shelley et al. However, the similarity between the first three conductance levels for GluA2-γ5 in our experiments and those for GluA1-γ5 reported by Shelley et al. (9.4, 21.7 and 38.1 pS) points to the high consistency of our recordings with the recordings made from native cell membranes. Our functional data, therefore, confirm that the GluA2-γ5 fusion construct is a faithful representation of the corresponding synaptic complex, where GluA2 and γ5 are not covalently linked.
We purified GluA2-γ5, added the competitive antagonist ZK200775 (ZK), and subjected this sample to single-particle cryo-EM. As a result, we reconstructed a 3D map of the entire complex to an overall 3.6 Å resolution (Figure 1C), with the core of the molecule resolved at ~3 Å resolution (Figure S1, Table S1). ATD and micelle signal subtraction improved density for the LBD-TMD region to the overall resolution of 3.3 Å (Figure S1F, Table S1). The maps were of sufficient quality to build the model of the entire γ5 and most of the GluA2 subunits, excluding the S2-M4 linkers and loops connecting M1 to M2 and M4 to γ5, which were not represented clearly in cryo-EM density. The map also showed densities for ZK molecules bound to LBDs, annular lipids preferentially located on the extracellular side of the TMD, and carbohydrates at the predicted glycosylation site N355 (Figure 1C).
Competitive antagonist ZK-bound structure of GluA2-γ5
The ZK-bound GluA2-γ5 complex (GluA2-γ5ZK) has a typical Y-shaped GluA2 tetramer in the center with two γ5 molecules assembled between protomers A/B and C/D on the periphery of the GluA2 TMD (Figure 2, A-C). Each γ5 subunit comprises a bundle of four α-helices (TM1-TM4), with an extracellular head domain, formed by polypeptides connecting TM1 to TM2 and TM3 to TM4, which includes a five-stranded β-sheet positioned atop the bundle (Figure 2D). Despite each one of four GluA2 subunits was covalently linked to γ5, no density for bound γ5 was observed between GluA2 protomers A/D and B/C, indicating that the two corresponding auxiliary subunits are likely disordered. Since these disordered subunits did not form an additional layer of density, which was previously observed below the TMD for GluA2-γ2 (Twomey et al., 2016) and GluA2-GSG1L (Twomey et al., 2017b) fusion constructs solubilized in DDM (n-dodecyl-β-d-maltopyranoside) detergent, they likely reside in the membrane but adapt random orientations and therefore do not become visible during 3D reconstruction. Compared to DDM, digitonin, which was used in the present study, either stabilizes the assembly of AMPA receptor with γ2 and CNIH3 subunits to reveal their maximal 4:1 stoichiometry (Nakagawa, 2019; Twomey et al., 2017a; Zhao et al., 2016) or keeps the unfolded auxiliary subunits as parts of the TMD density.
Figure 2. Architecture of GluA2-γ5 in complex with competitive antagonist ZK200775.
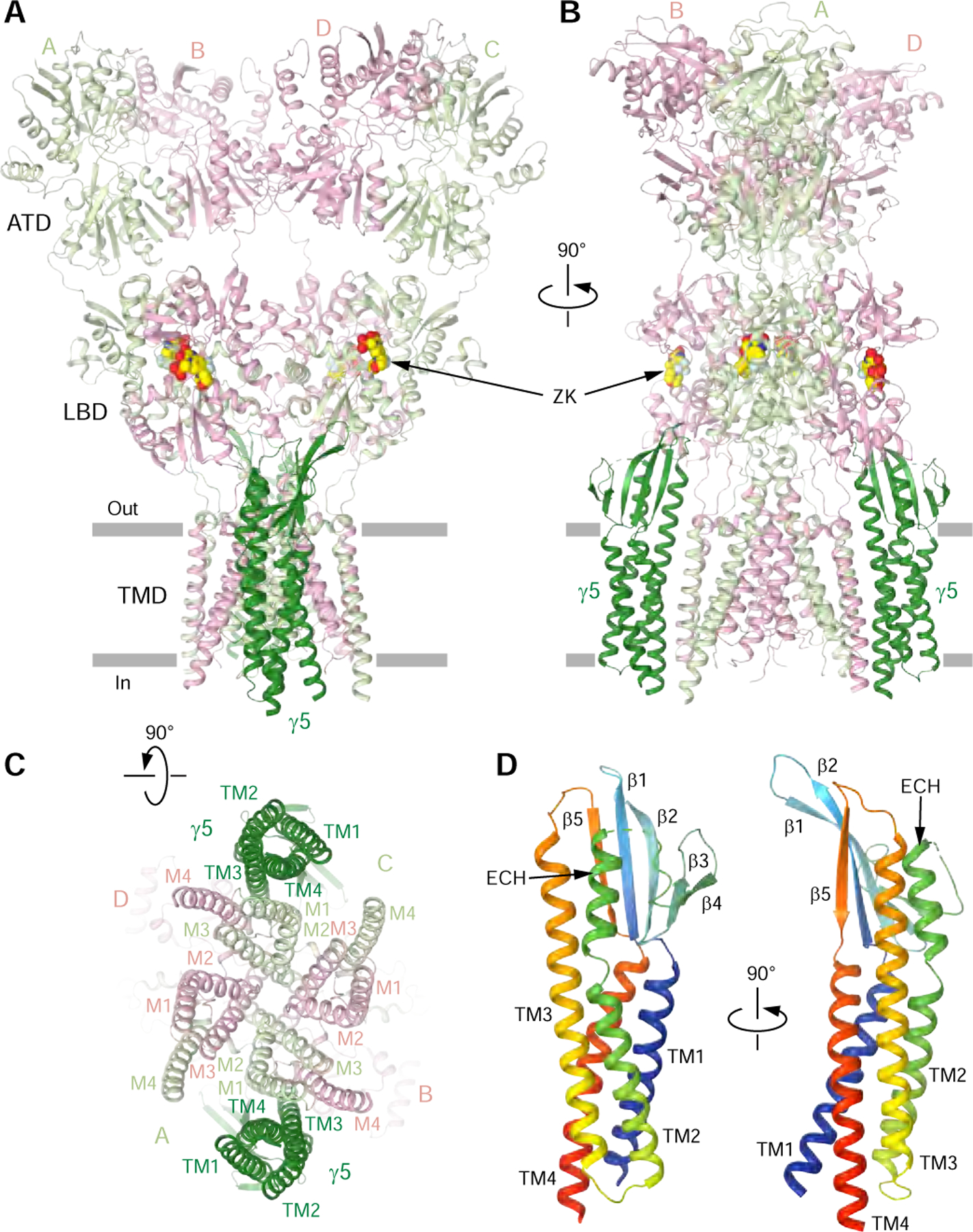
(A-C) GluA2-γ5 structure viewed parallel to the membrane (A and B) or intracellularly (C) with GluA2 subunits colored in light green and pink and γ5 in dark green. Molecules of ZK are shown as space-filling models.
(D) Rainbow-colored (from blue N-terminus to red C-terminus) structure of γ5 with the secondary structure elements labeled.
See also Figure S2.
Why γ5 and GSG1L, even in the stabilizing detergent digitonin, do not show the maximal 4:1 stoichiometry and show the 2:1 stoichiometry instead? To answer this question, we used a 90° rotation to copy two additional auxiliary subunits that show 2:1 stoichiometry to the two vacant positions to mimic the 4:1 stoichiometry of GluA2-γ2 complex (Figure S2). Compared to GluA2-γ2 complex, in which the experimental subunits show no clashes at these two positions (Figure S2A), the β1-β2 and TM3-β5 loops of the hypothetical γ5 subunits create severe clashes with the lower lobes of the LBDs (Figure S2B). Similarly, the TM3-β5 loops of the hypothetical GSG1L subunits clash with the C-terminal portions of K helices in GluA2 subunits A and C (Figure S2D). In addition, the β1-β2 loops of the hypothetical GSG1L subunits, which miss 45 residues in close proximity to E helices in GluA2 subunits B and D, would unlikely avoid clashes if the missing 45 residues are present in the model. Therefore, the unavoidable clashes of the hypothetical subunits with the LBDs strongly suggest that assembly of GluA2-γ5 and GluA2-GSG1L complexes can only occur with the 2:1 maximal stoichiometry (Twomey et al., 2017a, b) and is different from the assembly of AMPA receptor complexes with γ2 and CNIH3, which show 4:1 maximal stoichiometry (Chen et al., 2017; Nakagawa, 2019; Twomey et al., 2016, 2017a; Twomey et al., 2018; Zhao et al., 2019; Zhao et al., 2016). In this regard, γ8, which was structurally captured only at two positions equivalent to γ5 and GSG1L, shows no significant clashes at two hypothetical positions (Figure S2C). However, 23 and 9 residues missing in the β1-β2 and TM3-β5 loops, respectively, are likely to create clashes with the LBD and thus preclude the 4:1 stoichiometry. We therefore hypothesize that γ8 has the 2:1 maximal stoichiometry, similar to γ5 and GSG1L.
Similar to other TARPs, the primary contacts between GluA2 and γ5 in GluA2-γ5ZK are mediated by the transmembrane helices M1 of GluA2 subunits A/C, M4 of GluA2 subunits B/D, and TM3 and TM4 of γ5 (Figure 2C). The extracellular head domain of γ5 projects its β1-β2 loop towards the lower lobes of GluA2 subunits A/C LBDs. The proximity of the residues at the tip of this loop (Q872/43, Q874/45; numbering in the fusion construct/numbering in γ5; Figure 3) to the H helix residues of the LBD (K695, S696, K697) suggests their interactions, although the exact nature of these interactions remains uncertain due to poorly resolved side chains. A similar situation was observed for GSG1L, where the β1-β2 loop is longer and part of it has not been resolved in the cryo-EM density (Twomey et al., 2017b). GSG1L also stabilizes non-conducting states of AMPA receptor and similarly to γ5 lacks electrostatic interaction between the β4-TM2 loop and LBD, which is characteristic for the interaction of γ2 with AMPA receptor subunits B/D (Twomey et al., 2016; Zhao et al., 2016). The β4-TM2 loop in type I TARPs γ2 and γ8 includes the extracellular helix (ECH), which runs nearly parallel to the membrane (Figure 3). GSG1L completely lacks the ECH and has a longer TM2 instead, while ECH in γ5 runs perpendicular to the membrane and parallel to TM3. Correspondingly, TM3 in γ5 and GSG1L is longer than in γ2 and γ8 (Figure 3).
Figure 3. Structural and sequence comparison of AMPA receptor auxiliary subunits.
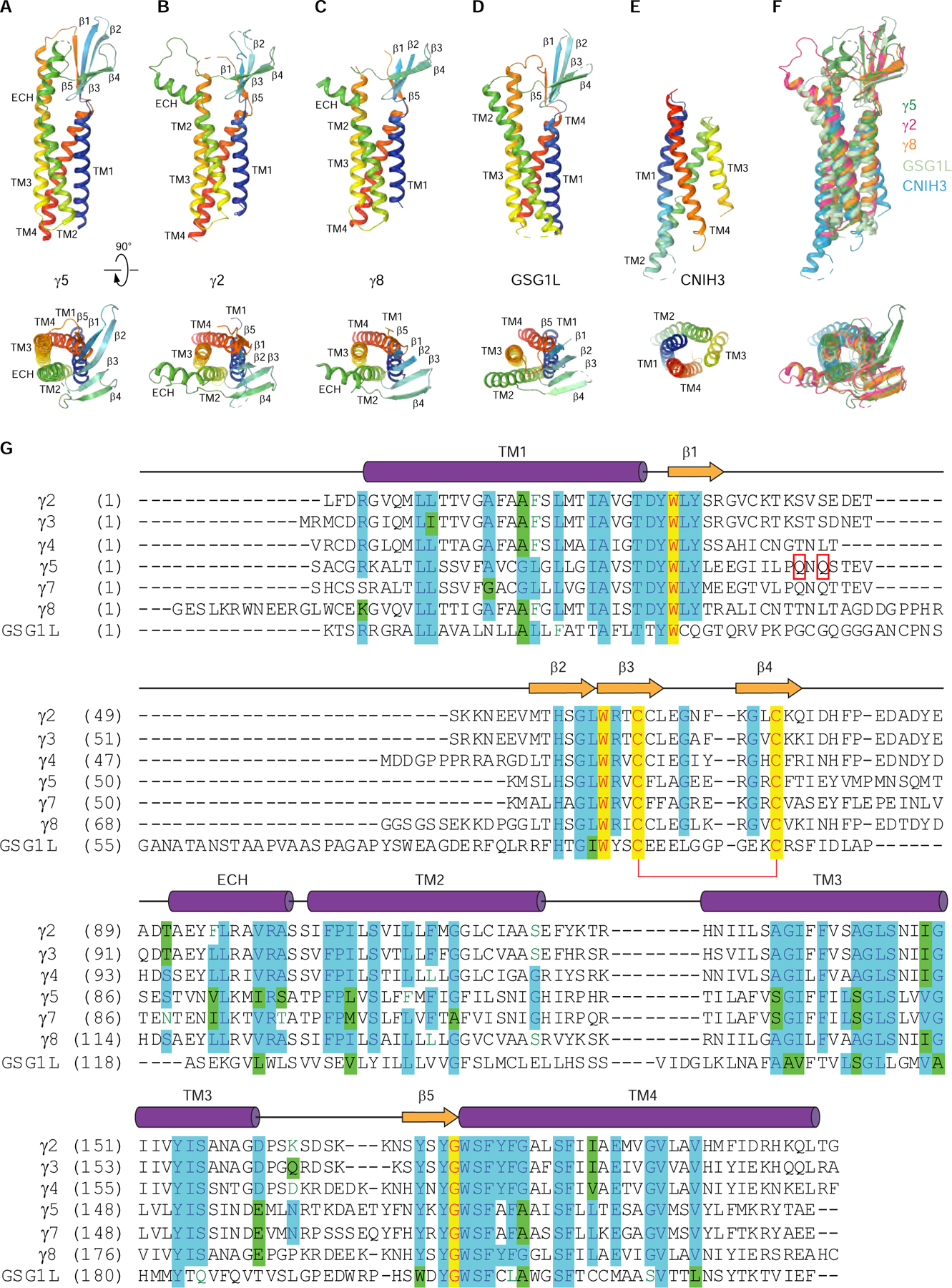
(A-E) Rainbow-colored (from blue N-terminus to red C-terminus) structures of auxiliary subunits γ5 (A, this study), γ2 (B, PDB ID: 5WEO), γ8 (C, PDB ID: 6QKC), GSG1L (D, PDB ID: 5WEL) and CNIH3 (E, PDB ID: 6PEQ) viewed parallel to the membrane (top row) or extracellularly (bottom row).
(F) Superposition of γ5 (blue), γ2 (red), γ8 (orange), GSG1L (cyan) and CNIH3 (green).
(G) Amino acid sequence alignment of AMPA receptor auxiliary subunits γ2 (Uniprot ID Q88602), γ3 (Q9JJV5), γ4 (Q9JJV4), γ5 (Q8VHW4), γ7 (P62956), γ8 (Q8VHW5) and GSG1L (D3Z7H4). The secondary structure is shown for γ2 above the sequence alignment as cylinders (α-helices), arrows (β-strands) or lines (loops). Completely conserved residues are highlighted in yellow. Mostly conserved residues are highlighted in blue (or green for homologous residues). Conserved cysteines forming a disulfide bridge between β3 and β4 are connected by a red bracket. Residues in the β1-β2 loop that are in close proximity to residues K695 and K697 in GluA2 LBD are indicated by red rectangles. The C-terminal residues are excluded.
Desensitized-state structures of GluA2-γ5
To gain insight into desensitization of GluA2-γ5, we determined structures of this complex in the presence of full agonists Glu and quisqualate (Quis). In patch-clamp experiments, GluA2-γ5 deactivation after 2-ms application of Quis was about two times slower (τDeact = 4.15 ± 0.25 ms, n = 6) in comparison to Glu, while the extent of desensitization as a result of the prolonged 1-s application of Quis was nearly the same (ISS / IMax = 0.0078 ± 0.0030, n = 6) and its rate was almost two times faster (τDes = 5.09 ± 0.33 ms, n = 11) (Figure S3A). When GluA2-γ5 was reconstituted into planar lipid bilayers, Quis evoked single-channel currents that similar to Glu (Figure 1B) showed four conductance levels, with higher conductance values than GluA2 alone (Figure S3B): 9.89 ± 1.65 pS (level 1), 23.29 ± 3.27 pS (level 2), 35.92 ± 1.76 pS (level 3) and 49.07 ± 2.12 pS (level 4) (n = 5). Recovery from desensitization was monitored using a two-pulse protocol (Figure 4A–B) and proceeded more than four-times slower after desensitization in the presence of Quis (τRecDes = 130 ± 4 ms, n = 10) compared to Glu (τRecDes = 29.8 ± 1.9 ms, n = 6) (Figure 4C). The corresponding time constants were larger than the ones for the receptor alone (τRecDes = 121 ± 7 ms, n = 5 for Quis and τRecDes = 15.3 ± 1.1 ms, n = 14 for Glu) (Figure S3F), indicating that γ5 slows the receptor’s recovery from desensitization, especially when activated by Glu. We determined cryo-EM structures of GluA2-γ5 both in the presence of Quis (GluA2-γ5Quis, Figure S4) and Glu (GluA2-γ5Glu, Figure S5). Since the two structures are nearly identical (Figure S6), we will mainly focus on GluA2-γ5Quis as it has a slightly higher resolution.
Figure 4. Desensitized-state structures of GluA2-γ5 in complex with glutamate and quisqualate.
(A-B) Two-pulse protocols monitoring recovery from desensitization for GluA2-γ5 activated by Glu (A) and Quis (B). Red dashed lines are fits of current recovery with the Hodgkin-Huxley equation.
(C) Mean recovery from desensitization measured using the protocols illustrated in (A-B) for GluA2-γ5 activated by Glu (green) and Quis (orange). The curves through the points are fits with the Hodgkin-Huxley equation. Error bars represent SEMs.
(D) Structure of GluA2-γ5Quis viewed parallel to the membrane with GluA2 subunits A and C colored in light green, B and D in pink, and γ5 in dark green. Molecules of Quis are shown as space-filling models.
(E) Superposition of LBD monomers from GluA2-γ5ZK (blue), GluA2-γ5Glu (green), GluA2-γ5Quis (orange), and GluA2-γ2Glu+CTZ (pink; PDB ID: 5WEO) based on the upper lobe D1. Relative movement of the lower lobe D2 is illustrated with black arrow.
(F) Pore-forming domains M2 and M3 in GluA2-γ5Quis with the residues lining pore shown as sticks. Only two (A and C) of four subunits are shown, with the front and back subunits (B and D) omitted for clarity. The pore profile is shown as a space-filling model (light blue).
(G) Pore radius for GluA2-γ5ZK (blue), GluA2-γ5Glu (green), GluA2-γ5Quis (orange), and GluA2-γ2Glu+CTZ (pink) calculated using HOLE. The vertical dashed line denotes the radius of a water molecule, 1.4 Å.
See also Figures S4–S6, Table S1.
GluA2-γ5Quis has the same overall architecture as GluA2-γ5ZK (Figure 4D). However, each LBD clamshell adapts a strikingly different conformation, being 26° more closed in GluA2-γ5Quis compared to GluA2-γ5ZK (Figure 4E). Similarly, closed LBD clamshells were observed in GluA2-γ5Glu as well as in previously reported desensitized- and open-state structures (Chen et al., 2017; Twomey et al., 2017a, b) (Figure 4E). Such closure of the LBD clamshell is considered to be the principal conformational change that drives iGluR gating (Armstrong and Gouaux, 2000; Twomey and Sobolevsky, 2018). At the same time, the extracellular part of the ion channel pore in GluA2-γ5Quis is hydrophobically sealed by the gate residues T617, A621, T625, and M629 (Figure 4F). The pore radius in the gate region of GluA2-γ5Quis is similar to GluA2-γ5ZK and GluA2-γ5Glu as well as in previously published closed- and desensitized-state structures (Chen et al., 2017; Herguedas et al., 2019; Nakagawa, 2019; Sobolevsky et al., 2009; Twomey et al., 2017a, b; Yelshanskaya et al., 2016; Zhao et al., 2016) but significantly smaller than the pore radius in the gate region of open-state structures (Chen et al., 2017; Twomey et al., 2017a) (Figure 4G). Taken together, the fully-activated closed LBD clamshells and non-conducting ion channel pore suggest that GluA2-γ5Quis and GluA2-γ5Glu represent the desensitized state of GluA2-γ5 complex.
Structures of GluA2-GSG1L in the closed and desensitized states
In contrast to GluA2 complexes with type I TARP γ2, which show 4:1 maximal stoichiometry and favor conducting states, GluA2-γ5 shows 2:1 maximal stoichiometry and favors non-conducting states. We, therefore, assessed whether the type II TARP γ5 is structurally more similar to another AMPA receptor auxiliary subunit GSG1L, which favors non-conducting states (Schwenk et al., 2012; Shanks et al., 2012; Twomey et al., 2017a, b). To be able to compare structural data in more detail, we solved GluA2-GSG1L complexes in the closed and desensitized states to a higher resolution than reported before (Twomey et al., 2017a, b) (Figure S7A–I). Comparison of the new and previously published structures illustrates their similar conformations (Figure S7J–K) but the higher resolution of new structures (4.2 Å versus 6.1 Å for GluA2-GSG1Lapo and 4.3 Å versus 8.4 Å for GluA2-GSG1LQuis) makes them more reliable for structural analysis. The new GluA2-GSG1Lapo and GluA2-GSG1LQuis structures have non-conducting ion channel pores, while the LBD clamshells in GluA2-GSG1Lapo are 27° more open than the ones in GluA2-GSG1LQuis, confirming that the structures represent the closed and desensitized states, respectively.
Different desensitization mechanisms of GluA2-γ5 and GluA2-GSG1L
Based on crystal structures of isolated LBD, desensitization was proposed to occur through rupture of the upper D1-D1 interface (Armstrong et al., 2006; Sun et al., 2002). Thus, in the open state, the D1-D1 interface remains intact as in the closed state, while the agonist-induced closure of individual LBD clamshells results in separation of the D2 lobes and opening of the ion channel pore (Twomey and Sobolevsky, 2018). The upper D1-D1 interface rupture in the desensitized state allows for individual LBD clamshells to remain in the closed, active conformation but brings the bottom, channel-facing portions of the dimer close together, releasing the strain on the linkers connecting LBD to TMD and allowing the channel to adapt the closed non-conducting state (Twomey and Sobolevsky, 2018). This view was supported by low-resolution full-length structures of GluA2-GSG1LQuis and GluA2-γ2Quis (Chen et al., 2017; Twomey et al., 2017b). Comparing structures of LBD dimers in different gating conformations (Figure 5A–F), the rupture of the D1-D1 interface is also obvious in the higher-resolution structures of GluA2-γ5Quis (Figure 5C), GluA2-GSG1LQuis (Figure 5D), and GluA2-γ5Glu (Figure 5E). Strikingly, the desensitized-state LBD dimers of GluA2-γ5 maintain their two-fold rotational symmetry, which is typical for the closed- and open-state dimers (Figure 5A,B,F). In contrast, the desensitized-state LBD dimers of GluA2-GSG1L lose their two-fold symmetry, signified by the appearance of a side cleft between the dimer protomers (Figure 5D).
Figure 5. State-dependent architectures of LBD dimers.
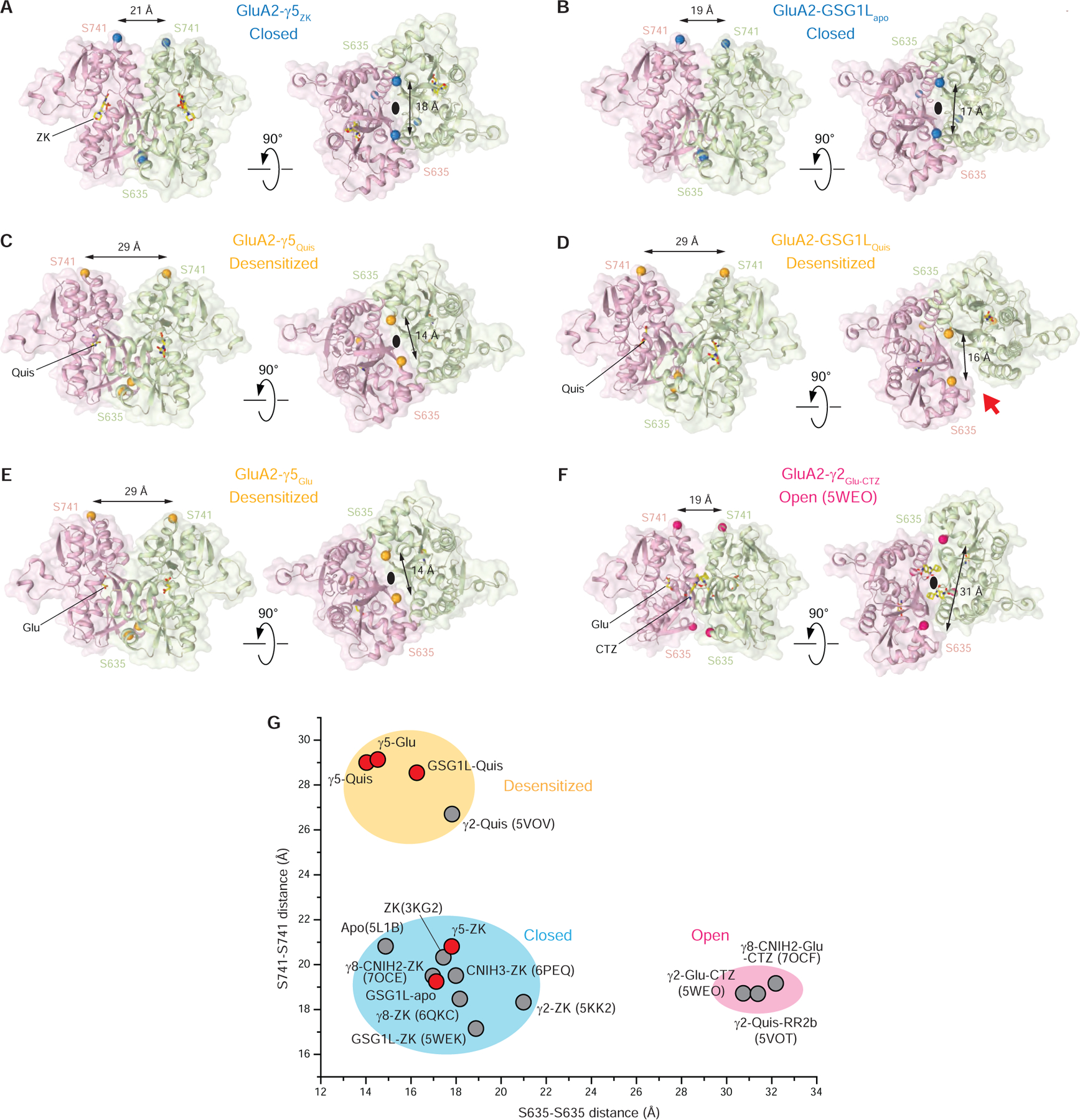
(A-F) LBD dimers from GluA2-γ5ZK (A), GluA2-GSG1Lapo (B), GluA2-γ5Quis (C), GluA2-GSG1LQuis (D), GluA2-γ5Glu (E) and GluA2-γ2Glu-CTZ (F) viewed from the side (left) and from the bottom (right) with GluA2 subunits B (D) colored pink, C (A) colored light green, and ligand molecules shown in stick representation. Cα atoms of S635 and S741 are shown as blue (closed state), orange (desensitized state) and hot pink (open state) spheres with cross-dimer distances between them indicated. The black ovals indicate the 2-fold rotational symmetry of the dimers. The red arrow points to the cleft between the GluA2-GSG1LQuis protomers signifying the loss of the LBD-dimer 2-fold rotational symmetry.
(G) Cross-dimer distances between S635 and S741 for structures reported in this study (red circles) and published previously (grey circles; PDB accession codes are given in parentheses). Clusters of the cross-dimer distances for the closed, desensitized and open states are highlighted with blue, orange and pink ovals.
Such an apparent difference in the structural mechanisms of GluA2-γ5 and GluA2-GSG1L desensitization does not alter the pattern of LBD dimer dimensions that characterize AMPAR conformational states. Indeed, separation of the upper (D1-D1) and lower (D2-D2) lobes of LBD dimers measured as distances between Cα atoms of S741 and S635, respectively, characterize closed states by short S741-S741 and S635-S635 distances, desensitized states by long S741-S741 and short S635-S635 distances, and open states by short S741-S741 and long S635-S635 distances (Figure 5C). Despite the drastic structural difference in the symmetrical organization of the desensitized-state dimers (Figure 5A–B), the desensitized-state cluster displays smaller variation in the D1-D1 and D2-D2 separations than the closed-state cluster (Figure 5C). The values of the S741-S741 and S635-S635 distances for non-symmetric LBD dimers of GluA2-GSG1LQuis are in between the values for two-fold symmetrical LBD dimers of the desensitized-state structures of GluA2-γ5 and GluA2-γ2 (Chen et al., 2017) (Figure 5C). It appears that both type I and type II TARPs promote desensitization with 2-fold symmetrical changes in the LBD, while in the presence of GSG1L, the LBD-dimers lose their 2-fold symmetry.
The difference in the structural organization of the closed- and desensitized-state LBD dimers (Figure 5) triggers different overall transformations of the full-length GluA2-γ5 and GluA2-GSG1L structures during closed-to-desensitized state transition (Figure 6, Supplementary Movie 1). Commonly, during desensitization GluA2-γ5 and GluA2-GSG1L structures become shorter and their ATD layers rotate clockwise as rigid bodies when viewed extracellularly. In detail, however, the shortening of the receptor is much stronger for GluA2-γ5 (9 Å versus 4 Å), while the ATD layer rotation is stronger for GluA2-GSG1L (17° versus 10°). Since the ATD layer moves as a rigid body, the changes in the receptor height are reflected in changes in the vertical distance (H) between the LBD layer center of mass (COM) and the center of the pore gate (Figure 6C). Similar to desensitization of GluA2-γ5 and GluA2-GSG1L, the opening of the AMPAR complex with type I TARP γ2 is accompanied by a reduction in H (Figure 6D). In contrast to AMPAR complexes with type II TARP γ5 and GSG1L, desensitization of the complex with type I TARP γ2 leads to increased H (Figure 6D). Based on measurements of H, the auxiliary subunits appear to have different effects on the height of the receptor in the closed state, with TARPs γ2 and γ5 making the receptor substantially taller, GSG1L slightly taller, and TARPs γ8 much shorter. The predicted change in receptor height may depend on its subunit composition and the type of bound ligand but more likely is dependent on the type of receptor interaction with the head domain of its auxiliary subunit. Indeed, the headless CNIH3 (Figure 3) does not alter the closed-state H (Figure 6D).
Figure 6. Different closed to desensitized state transformations of GluA2-γ5 and GluA2-GSG1L complexes and elevation of LBD over TMD in the presence of different auxiliary subunits.
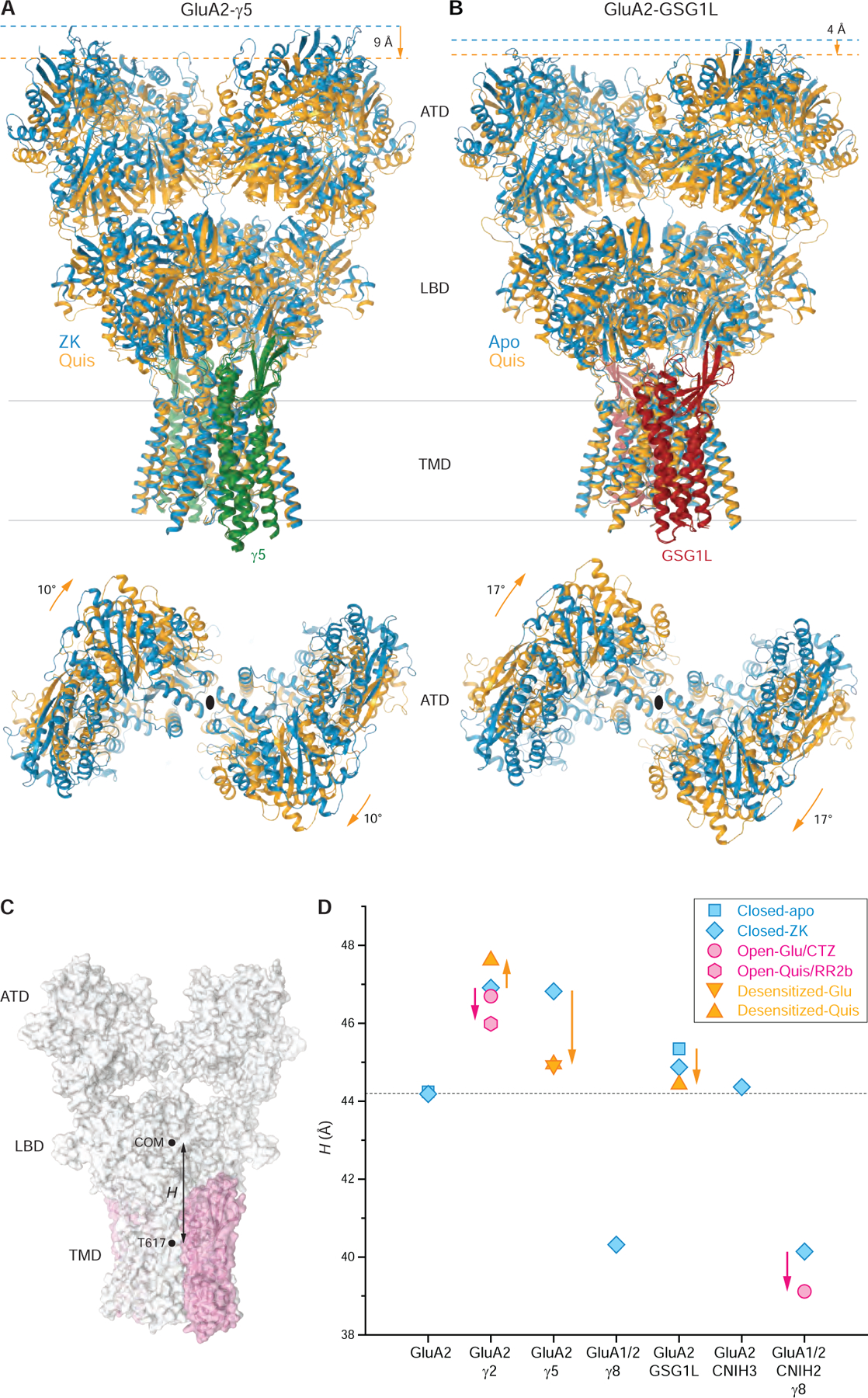
(A-B) TMD-based superposition of GluA2-γ5 (A) and GluA2-GSG1L (B) in the closed (blue) and desensitized (orange) states viewed parallel to the membrane (top) or extracellularly (bottom), with γ5 shown in green and GSG1L in red. Shortening of the receptor and ATD rotation that signify the desensitized to closed state conversion are indicated by orange arrows. Note, in the closed state, GluA2-γ5 is ~4 Å taller than GluA2-GSG1L.
(C) Schematic illustrating the measurement of LBD over TMD elevation (H) as a distance between the center of mass (COM) for the LBD layer and the T617 Cα coordinates averaged over four AMPA receptor subunits.
(D) LBD over TMD elevation for GluA2 alone and AMPA receptor complexes with different auxiliary subunits in the closed (blue), desensitized (orange) and open (pink) states. Measurements are for the same structures as in Figure 5G, including the structures reported in this study, GluA2-γ5ZK, GluA2-GSG1Lapo, GluA2-γ5Glu, GluA2-γ5Quis, and GluA2-GSG1LQuis, as well as the previously published structures GluA2apo (PDB ID: 5L1B), GluA2ZK (PDB ID: 3KG2), GluA2-γ2ZK (PDB ID: 5KK2), GluA2-γ8ZK (PDB ID: 6QKC), GluA2-CNIH3ZK (PDB ID: 6PEQ), GluA2-GSG1LZK (PDB ID: 5WEK), GluA2-γ2Quis (PDB ID: 5VOV), GluA2-γ2Glu+CTZ (PDB ID: 5WEO), GluA1-GluA2-γ8-CNIH2ZK (PDB ID: 7OCE), GluA1-GluA2-γ8-CNIH2Glu+CTZ (PDB ID: 7OCF) and GluA2-γ2Quis-RR2b (PDB ID: 5VOT). Changes in H accompanying transitions from the closed to desensitized and open states are indicated by orange and pink arrows, respectively.
See also Figure S7.
Interaction of the γ5 head domain with GluA2 LBD alters AMPAR function
To study the role of the γ5 head domain in regulation of AMPAR function, we inspected the closed- and desensitized-state GluA2-γ5 structures and confirmed that the β1-β2 loop of the head domain is in contact with the C-terminal end of H-helix in the LBD of subunits A and C (Figure 7A). On the receptor side, this contact involves lysines K695 and K697, which we mutated to alanines and tested function of the resulting receptor alone (GluA2Mut) and its fusion with γ5 (GluA2Mut-γ5) using whole-cell patch-clamp recordings (Figure 7B). The mutations did not change ISS / IMax or recovery from desensitization for the receptor alone (ISS / IMax = 0.042 ± 0.011, n = 7 and τRecDes = 15.9 ± 0.7 ms, n = 5 for GluA2Mut versus ISS / IMax = 0.042 ± 0.009, n = 22 and τRecDes = 15.3 ± 1.1 ms, n = 14 for GluA2; Figure S3F). However, the mutations reversed the reduction of steady-state currents caused by γ5 (ISS / IMax = 0.029 ± 0.009, n = 8 for GluA2Mut-γ5 versus ISS / IMax = 0.0091 ± 0.0029, n = 6 for GluA2-γ5; compare blue traces in Figure 1A and Figure 7B), without changing the time constants of desensitization and deactivation (Figure S3F). On the other hand, the recovery of GluA2Mut-γ5 from desensitization measured using the two-pulse protocol (τRecDes = 49.7 ± 3.0 ms, n = 6; Figure 7C) became ~1.7 times slower than that for GluA2-γ5 (τRecDes = 29.8 ± 1.9 ms, n = 6) and ~3.2 times slower than that for the receptor alone (Figure 7D, Figure S3F). Interestingly, one of the mutated lysines, K697, is part of the KGK motif, which when substituted with aspartate almost completely abolished the effects of γ2 on GluA2 receptor function (Dawe et al., 2016), apparently due to altered interaction of subunits B and D LBDs with the electronegative motif in the γ2 β4-TM2 loop (Twomey et al., 2016; Zhao et al., 2016). This electronegative motif in the γ2 β4-TM2 loop is highly conserved across type I TARPs, while type II TARPs γ5 and γ7 are missing this motif (Figure 3G) and instead interact with subunits A and C using their β1-β2 loop (Figure 7A).
Figure 7. Interaction of the γ5 head domain with GluA2 LBD and structural mechanisms of AMPAR regulation by auxiliary subunits.
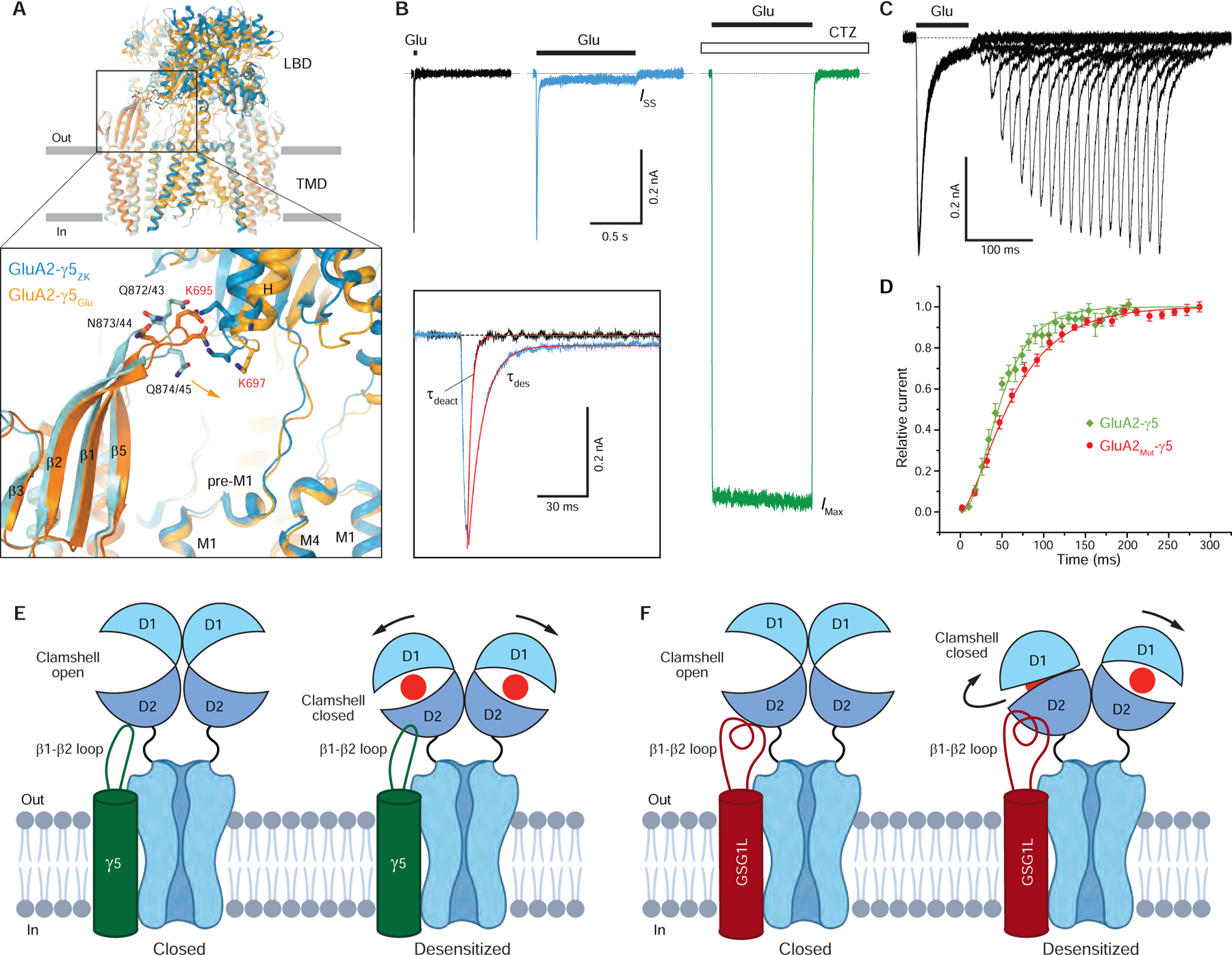
(A) TMD-based superposition of GluA2-γ5 in the closed and desensitized states. Only LBD and TMD layers are shown, with ATD layer omitted. GluA2 and γ5 are in blue and cyan for GluA2-γ5ZK and light and dark orange for GluA2-γ5Glu, respectively. The inset shows a close up view of the γ5 head domain-LBD interaction, with the residues at the interface shown as sticks. Lysines K695 and K697 mutated to alanines in GluA2Mut-γ5 are labeled red. Residues in γ5 are numbered according to the fusion construct/auxiliary subunit alone (see Figure 3G).
(B) Representative whole-cell currents recorded at −60 mV membrane potential from HEK 293 cell expressing GluA2Mut-γ5 in response to 2-ms (black) or 1-s (blue) applications of 3 mM Glu alone or application of Glu in the continuous presence of 30 μM CTZ (green). The inset shows normalized currents in response to 2-ms and 1-s applications of Glu alone fitted using single exponentials (red curves).
(C) Two-pulse protocol monitoring recovery from desensitization for GluA2Mut-γ5 activated by Glu.
(D) Mean recovery from desensitization for GluA2Mut-γ5 and GluA2-γ5 activated by Glu measured using the two-pulse protocol. The curves through the points are fits with the Hodgkin-Huxley equation. Error bars represent SEMs.
(E-F) Mechanisms of AMPAR receptor modulation by type II TARP γ5 (E) and GSG1L (F). Compared to GluA2-γ5, the β1-β2 loop in GluA2-GSG1L is much longer and interacts not only with the C-terminal part of LBD helix H but also with its central and N-terminal regions, which are more distal from the TMD. As a result of these interactions, in the continuous presence of agonist Glu (red circles), GSG1L stabilizes the desensitized state stronger than γ5 and GluA2-GSG1L LBD dimers lose their 2-fold symmetry upon desensitization. LBD dimers in GluA2-γ5 remain 2-fold symmetrical. Following these changes in LBD during desensitization, GluA2-γ5 undergoes stronger shortening, while ATD layer of GluA2-GSG1L undergoes stronger rotation.
Conclusion
Compared to type I TARP γ2 and CNIH3, which interact with AMPARs at the maximal stoichiometry of four TARPs per one AMPAR tetramer, only two type II TARP γ5 or GSG1L subunits can interact with one AMPAR (Figures 2A–C and S2). This interaction does not involve the β4-TM2 loop, which is used by type I TARPs to interact with LBDs of AMPAR subunits B and D. Instead, γ5 and GSG1L employ the β1-β2 loop to interact with LBDs of AMPAR subunits A and C. In GluA2-γ5 complex, the β1-β2 loop appears to act as a desensitized state-stabilizing latch (Figure 7E), which interacts with the C-terminal part of LBD helix H, reduces steady-state currents (Figure 1A) and slows recovery from desensitization (Figure 4A–C, Figure S3F). Similarly, the β1-β2 loop appears to play an important role in stabilizing the desensitized state in GluA2-GSG1L complex. In GluA2-GSG1L, however, the β1-β2 loop is much longer and interacts not only with the C-terminal part of LBD helix H (including K695) but also with its central and N-terminal regions (including R684, E688 and R692), which are more distal from the TMD (Figure 7F) (Twomey et al., 2017b). As a result of these interactions, which are a likely cause of stronger stabilization of the desensitized state by GSG1L compared to γ5, the lower lobes of LBDs in subunits A and C in the desensitized state of GluA2-GSG1L are pushed sideways and LBD dimers lose their 2-fold rotational symmetry. This change in the LBD dimer architecture defines the different character of conformational changes in GluA2-GSG1L complex compared to GluA2-γ5 complex where LBD dimers remain 2-fold symmetrical. Thus, in contrast to GluA2-GSG1L complex, which during desensitization becomes 4 Å shorter and its ATD layer rotates by 17°, GluA2-γ5 complex undergoes 9 Å shortening and its ATD layer rotates by 10° (Figure 6A, Supplementary Movie 1). Therefore, the unique structural features of type II TARP γ5 and GSG1L auxiliary subunits define their distinct regulation of AMPAR function.
Limitations of the study
One of the main effects of γ5 on GluA2 function appears to be the increase in single-channel conductance (Figures 1B and S3C–D). This effect implies changes in the GluA2 channel pore that are caused by the presence of γ5 and are likely induced by interactions between GluA2 and γ5 TMDs that are not present in other AMPA receptor-auxiliary subunit complexes. In the present study, we have only solved structures of GluA2-γ5 complexes in the closed and desensitized states, in which the GluA2 channel pore adapts non-conducting conformation. To better understand the mechanism of γ5 action and the molecular bases of the increase in single-channel conductance, one would need to solve GluA2-γ5 structures in the open conducting state.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for the resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alexander Sobolevsky (as4005@cumc.columbia.edu).
Materials Availability
Materials listed in key resources under “Recombinant DNA” and “Oligonucleotides” should be requested from the Lead Contact, Alexander Sobolevsky (as4005@cumc.columbia.edu).
Data and Code Availability
Cryo-EM maps have been deposited to the EMDB with the accession codes EMD-24750 (GluA2-γ5ZK, full length), EMD-24751 (GluA2-γ5ZK, LBD-TMD), EMD-24752 (GluA2-γ5Glu, full length), EMD-24748 (GluA2-γ5Glu, LBD-TMD), EMD-24753 (GluA2-γ5Quis, full length), EMD-24754 (GluA2-γ5Quis, LBD-TMD), EMD-24755 (GluA2-GSG1Lapo, full length), EMD-24756 (GluA2-GSG1LQuis, full length) and EMD-24749 (GluA2-GSG1LQuis, LBD-TMD). Coordinates have been deposited to the PDB with the accession codes 7RZ4 (GluA2-γ5ZK, full length), 7RZ5 (GluA2-γ5ZK, LBD-TMD), 7RZ6 (GluA2-γ5Glu, full length), 7RYY (GluA2-γ5Glu, LBD-TMD), 7RZ7 (GluA2-γ5Quis, full length), 7RZ8 (GluA2-γ5Quis, LBD-TMD), 7RZ9 (GluA2-GSG1Lapo, full length), 7RZA (GluA2-GSG1LQuis, full length) and 7RYZ (GluA2-GSG1LQuis, LBD-TMD).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Expression of the GluA2-γ5 and GluA2-GSG1L protein was performed in HEK 293S GnTI− cells (ATCC) that were cultured in the Freestyle 293 expression medium (GIBCO) at 37°C and 5% CO2. Baculoviruses for infecting HEK 293S GnTI− cells were produced in Sf9 cells (GIBCO) that were cultured in the Sf-900 III SFM media (GIBCO) at 27°C. HEK 293 cells for patch-clamp experiments were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium, supplemented with 10% fetal bovine serum.
METHOD DETAILS
Constructs for large-scale protein expression
The fusion construct GluA2-γ5 was prepared by introducing a GTG linker between GluA2 and γ5, where GluA2 represents a modified rat GluA2flip subunit (Uniprot ID: P19491) with Q at the Q/R-site (Q586), which was previously called GluA2* (Yelshanskaya et al., 2014) and used for making GluA2-γ2 and GluA2-GSG1L fusion constructs (Twomey et al., 2017a, b). γ5 is a C-terminally truncated mouse subunit (Cacng5, Uniport Q8VHW4, residues Ser2-Glu207), for which DNA was provided by Dr. Berndt Fakler. The GluA2-GSG1L fusion construct was made similarly to GluA2-γ5 (Twomey et al., 2017a, b). Both GluA2-γ5 and GluA2-GSG1L were introduced into BacMam vector for baculovirus-based protein expression in mammalian cells (Goehring et al., 2014), with the C-terminal thrombin cleavage site (LVPRG), followed by eGFP and streptavidin affinity tag (WSHPQFEK).
Protein expression and purification
GluA2-γ5 bacmid and baculovirus were made using standard methods (Goehring et al., 2014). The P1 and P2 viruses were produced in Sf9 cells (GIBCO, 12659017) and added to HEK 293S GnTI− cells (ATCC, CRL-3022) incubated in Free style media at 37°C and 5% CO2. At the time of transduction, the cells were supplemented with 2 nM ZK200775 (ZK, Tocris, 2345) and 0.1 mM kynurenic acid (Sigma, K3374). 12 hours post-transduction, the cells were supplemented with 10 mM sodium butyrate and the temperature was changed to 30°C. 72 hours post-transduction, the cells were harvested by low-speed centrifugation (5,500 g, 10 min), washed with 1X phosphate buffer saline (PBS, pH 8.0), and pelleted again (5,500 g, 15 min). The cell pellet was resuspended in ice-cold lysis buffer, containing 150 mM NaCl, 20 mM Tris pH 8.0, 1 mM β-mercaptoethanol (βME), 0.8 μM aprotinin, 2 μg/ml leupeptin, 2 μM pepstatin A, and 1 mM phenylmethylsulfonyl fluoride (PMSF). For GluA2-γ5ZK structure, the lysis buffer and all the following buffers were supplemented with 2 μM ZK. Cells were subsequently lysed using a Misonix Sonicator with a preset program (6 cycles of 15 s “on” at the amplitude of 8 followed by 15 s “off”; this program was repeated three times for optimal cell lysis) under constant stirring on ice. The lysate was centrifuged (9,900 g, 15 min) to remove unbroken cells and cell debris, and the supernatant was subjected to ultracentrifugation (186,000 g, 40 min) to pellet the cell membranes. The membrane pellet was mechanically homogenized and solubilized for 3 hours at 4°C in the buffer containing 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM βME, 0.1% cholesteryl hemisuccinate Tris salt (CHS, Anatrace, 4193482) and 1% DDM (Anatrace, 4211929). Insoluble material was removed by ultracentrifugation (186,000 g, 40 min), the supernatant was added to streptavidin-linked resin (2 ml resin per 1L of the initial cell culture) and the mixture was rotated for 10–14 hours at 4°C. The protein-bound resin was washed with 25 ml of the buffer containing 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 0.005% CHS and 0.05% digitonin (Cayman Chemical Company, 14952), and the protein was eluted using the same buffer supplemented with 2.5 mM D-desthiobiotin. To remove eGFP and streptavidin affinity tag, the eluted protein was subjected to thrombin digestion (1:100 w/w) for 1.5 hours at 22°C. The digest reaction was injected into Superose 6 10/30 GL size-exclusion chromatography column (GE Healthcare) equilibrated with the buffer containing 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 0.005% CHS and 0.05% digitonin. The tetrameric GluA2-γ5 peak fractions were pooled, concentrated to ~4 mg/ml, and used for cryo-EM sample preparation. All the steps, unless otherwise noted, were performed at 4°C.
The GluA2-GSG1L expression and purification was carried out as described earlier (Twomey et al., 2017a, b). Briefly, the protein was expressed in GnTI− cells and extracted by solubilizing membranes in the buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, and 1% Digitonin. Then the protein was purified by affinity chromatography using streptavidin-linked resin and digested with thrombin (1:200 w/w) to remove eGFP and streptavidin affinity tag. Finally, the protein was purified by size exclusion chromatography using Superose 6 10/300 GL column pre-equilibrated with the buffer containing 150 mM NaCl, 20 mM Tris-HCl pH 8.0, and 0.05% Digitonin. The tetrameric peak fractions were combined, concentrated, and used for cryo-EM sample preparation. All the steps, unless otherwise noted, were performed at 4°C.
Cryo-EM sample preparation and data collection
For preparing GluA2-γ5 samples, we used 300-mesh R1.2/1.3 commercial Ultra Au Foil Au/Au grids (EMS, Morrisville, NC, Q350AR1.3A) or in-house Au/Au grids prepared as described in the literature (Russo and Passmore, 2014). Briefly, the in-house grids were made by first coating C-flat (EMS, Morrisville, NC) CF-1.2/1.3–2Au holey carbon grids with ~50 nm of gold using an Edwards Auto 306 evaporator. Subsequently, an Ar/O2 plasma treatment (6 minutes, 50 watts, 35.0 sccm Ar, 11.5 sccm O2) was used to remove the carbon with a Gatan Solarus (model 950) Advanced Plasma Cleaning System. The grids were again plasma treated with Gatan Solarus system (H2/O2, 20 s, 10 watts, 6.4 sccm H2, 27.5 sccm O2) or glow discharged with PELCO easyGlow cleaning system (Ted Pella, 30 s, 15 mA) immediately before sample application to make their surfaces hydrophilic. Purified protein was supplemented with 0.1 mM ZK for the closed state, 1 mM Quis (Tocris) or 25 mM Glu for the desensitized state and incubated for 30 min on ice. An FEI Vitrobot Mark IV (Thermo Fischer Scientific) was used to plunge-freeze the grids after application of 3 μl protein solution at 4°C, 100% humidity, with the blot time of 5 sec, wait time of 15 sec, and the blot force of 5. The grids were imaged using 300-kV Titan Krios microscope equipped with a Gatan K3 direct electron detection camera and post-column GIF Quantum energy filter with the slit set to 20 eV.
For GluA2-γ5ZK, three datasets (9,875, 5,887, and 11,234 micrographs, with the total of 26,996 micrographs) were collected in the counting mode, with the pixel size of 1.1 Å (81,000× nominal magnification) across the defocus range of −1.0 to −2.0 μm. The total dose of 62.5 e−Å−2 was attained by using the dose rate of ~16.0 e−pixel−1s−1 across 50 frames of the 2.5-sec total exposure time. For GluA2-γ5Quis, two datasets (11,544 and 7,530 micrographs, with the total of 19,074 micrographs) were collected in the counting mode, with the pixel size of 1.069 Å (81,000× nominal magnification) across the defocus range of −0.8 to −2.0 μm. The total dose of 50 e−Å−2 was attained for the first dataset by using the dose rate of ~16.0 e−pixel−1s−1 across 50 frames of the 2.6-sec total exposure time. The total dose of 51.1 e−Å−2 was attained for the second dataset by using the dose rate of ~16.0 e−pixel−1s−1 across 40 frames of the 2.0-sec total exposure time. For GluA2-γ5Glu, two datasets (10,573 and 3,053 micrographs, with the total of 13,626 micrographs) were collected in the counting mode across the defocus range of −0.8 to −2.0 μm, with the pixel size of 1.083 Å for the first dataset and 1.076 Å for the second dataset. The total doses of 51.18 e−Å−2 for the first dataset and of 50.6 e−Å−2 for the second dataset were attained by using the dose rate of ~16.0 e−pixel−1s−1 across 40 frames of the 2-sec total exposure time.
The grids for GluA2-GSG1L were prepared similarly to grids for GluA2-γ5. For the desensitized-state structure, the protein concentrated to ~ 3 mg/ml was supplemented with 1 mM Quis and incubated for 30 min on ice. An FEI Vitrobot Mark IV (Thermo Fischer Scientific) was used to plunge-freeze the grids after application of 3 μl protein solution to gold-coated C-flat CF-1.2/1.3–2Au 200 mesh holey carbon grids (Protochips, Morrisville, NC) at 4°C, 100% humidity, with the blot time of 3 sec, wait time of 20 sec, and the blot force of 3. For GluA2-GSG1Lapo and GluA2-GSG1LQuis, 10,824 and 10,207 micrographs were collected, respectively, in the counting mode across the defocus range of −1.0 to −2.0 μm, with the pixel size of 0.83 Å. The total dose of 58.0 e−Å−2 was attained by using the dose rate of ~16.0 e−pixel−1s−1 across 50 frames of the 2.5-sec total exposure time.
Image processing
For GluA2-γ5ZK, the initial processing was carried out using Relion 3.1 (Zivanov et al., 2018) (Figure S1). Frame alignment was done using MotionCor2 (Zheng et al., 2017). CTF estimation was performed using Gctf (Zhang, 2016) on non-dose-weighted micrographs, while subsequent data processing was done on dose-weighted micrographs. For each data set, ~3,000 particles were picked manually (Figure S1A) to generate 2D classes (Figure S1B) that were used as templates to autopick 2,923,261 particles from the first dataset, 2,361,920 particles from the second dataset, and 1,788,042 particles from the third dataset. Each dataset was processed separately until the particle cleanup and polishing as described below. For each dataset, the extracted particles were 4×4 binned and subjected to 3D classification into 10 classes. Cryo-EM map of GluA2-γ2 (EMDB: EMD-7959) low-pass filtered to 40 Å was used as an initial reference. Particles representing the best 3D classes (188,091 particles for the first dataset, 165,861 particles for the second dataset, and 358,619 particles for the third dataset) were re-extracted without binning, cleaned up by 3D classification, CTF refined and polished separately for each dataset. Cleaned and polished particles were combined to create a pool of 512,286 particles (131,817 particles from the first dataset, 165,861 particles from the second dataset, and 214,608 particles from the third dataset), which was refined, cleaned up by multiple rounds of 2D and 3D classification without sampling, micelle subtracted and refined using both regular and CTF refinements. The final 3D reconstruction of GluA2-γ5ZK included 162,741 particles and had the resolution of 3.6 Å (Figure S1D–E), estimated using the Gold standard Fourier shell correlation (GSFSC=0.143) criterion on masking-effect-corrected FSC curves calculated between two independent half-maps (Chen et al., 2013; Scheres, 2012). The local resolutions were estimated with unfiltered half-maps using ResMap (Kucukelbir et al., 2014) in Relion 3.1 and EM density maps were visualized using UCSF Chimera (Pettersen et al., 2004). The final refinement was run with C2 symmetry, while 3D classifications were always run with the C1 symmetry. For LBD-TMD-focused classification and refinement, 512,286 of the joined unbinned particles were signal-subtracted in Relion using a soft mask for the LBD-TMD region, which removed the signal for the ATD and the detergent micelle. Refinement and post-processing of the subtracted particles yielded a map with 3.6-Å resolution. Further 3D classification into 3 classes with T = 50 and a soft mask for the LBD-TMD region identified the best class of 111,527 particles, which yielded a map at 3.3 Å resolution (Figure S1F–G).
For GluA2-γ5Quis, the processing pipeline is illustrated in the Figure S4. Each dataset was processed separately until the particle cleanup and polishing. For the first dataset, the initial processing in Relion 3.1 (see above) resulted in 3,079,354 autopicked particles that were 6×6 binned and transferred to cryoSPARC (Punjani et al., 2017), where they were cleaned by several rounds of heterogeneous classification using GluA2-γ5ZK map as a reference. Further cleanup by 2D classification yielded 498,353 particles, which were exported back to Relion. These particles were unbinned and subjected to several rounds of regular and CTF refinement. The second dataset was processed similarly, yielding 1,931,696 autopicked particles and 278,551 particles exported from cryoSPARC back to Relion. Next, the particles from two datasets were joined (776,904 particles in total) and subjected to regular and CTF refinement and several rounds of 3D classification without sampling, yielding the total of 430,295 particles in the best 3D classes. These particles were used as an initial pool for reconstructing maps of the full-length and LBD-TMD part of the protein (see below). For the full-length protein, several additional rounds of 3D classification without sampling and regular refinements resulted in selection of 127,928 particles that yielded the final map at 4.2 Å resolution (Figure S4B–C). For LBD-TMD, the particles were signal subtracted as described above for GluA2-γ5ZK and the final map at 4.1 Å resolution was obtained using 96,823 particles (Figure S4D–E).
For GluA2-γ5Glu, data processing was similar to GluA2-γ5Quis. Each dataset was processed separately until the particle cleanup and polishing. For the first dataset, the initial processing in Relion 3.1 resulted in 2,648,017 autopicked particles that were 4×4 binned and transferred to cryoSPARC, where they were cleaned by several rounds of heterogeneous classification using GluA2-γ5Quis map as a reference. Further cleanup by 2D classification yielded 290,149 particles that were exported back to Relion. These particles were unbinned and subjected to several rounds of regular and CTF refinement followed by 3D classification without sampling and polishing, resulting in a final pool of 112,593 particles. The second dataset was processed similarly, yielding 1,064,539 of autopicked particles and 119,725 particles exported from cryoSPARC back to Relion. These particles were unbinned and subjected to 3D classification without sampling, which identified the best 79,946 particles of the first dataset. Next, the particles from two datasets were joined (the total of 192,539 particles) and subjected to CTF and regular refinement. These particles were used as an initial pool for reconstructing maps of the full-length and LBD-TMD portion of the protein (see below). For the full-length protein, several additional rounds of 3D classification without sampling and regular refinements resulted in selection of 130,204 particles that yielded the final map at 4.4 Å resolution (Figure S5A–C). For LBD-TMD, the particles were signal subtracted as described above for GluA2-γ5ZK and the final map at 4.4 Å resolution was obtained using the final set of 99,056 particles (Figure S5D–F).
For GluA2-GSG1Lapo, data processing was carried out in Relion 3.1 similarly to GluA2-γ5ZK. Frame alignment was done using MotionCor2. CTF estimation was performed using Gctf on non-dose-weighted micrographs, while subsequent data processing was done on dose-weighted micrographs. Initially, ~2,500 particles were picked manually to generate 2D classes that were used as templates to autopick 1,515,005 particles, which were 4×4 binned and subjected to 3D classification with GluA2-GSG1L map (EMDB: EMD-8688) as a reference. Particles representing the best classes were unbinned, refined and polished. Polished particles were further cleaned by multiple rounds of 3D classification without sampling. Refinement, CTF refinement and post-processing for the final set of 124,571 particles yielded a map of GluA2-GSG1Lapo at the 4.15 Å resolution (Figure S7A–C).
For GluA2-GSG1LQuis, the data were processed in Relion 3.1 similarly to GluA2-GSG1Lapo. The template-based autopicking yielded 1,627,160 particles that were 4×4 binned and subjected to 3D classification with GluA2-GSG1L map (EMDB: EMD-8688) as a reference. Particles representing the best classes were unbinned, refined and polished. Polished particles were further cleaned by multiple rounds of 3D classification without sampling. Refinement, CTF refinement and post-processing for the final set of 118,416 particles yielded a map of GluA2-GSG1LQuis at the 4.26 Å resolution (Figure S7D–F). For LBD-TMD-focused classification and refinement, the total of 292,845 unbinned, re-extracted and cleaned particles were signal-subtracted using a soft mask for the LBD-TMD region. The signal-subtracted particles were refined with the soft mask and further 3D classified. The final 3D reconstruction included 113,568 particles and yielded a 4.15-Å resolution map after refinement and post-processing (Figure S7G–I).
Model building and refinement
For GluA2-γ5ZK, the open-state structure of GluA2-γ2 (PDB ID: 5WEO) was used as a guide to build GluA2 into the cryo-EM density map. To guide model building of mouse γ5, its homology model was created in Swiss-Model (Waterhouse et al., 2018) using GSG1L structure (PDB ID: 5VHZ) as a template. The resulting initial model of GluA2-γ5ZK was adjusted and finalized in Coot (Emsley and Cowtan, 2004) using cryo-EM density as a guide. Structural models of GluA2-γ5Glu and GluA2-γ5Quis were built in Coot using cryo-EM density and the model of GluA2-γ5ZK as guides. Structural models of GluA2-GSG1Lapo and GluA2-GSG1LQuis were built in Coot using cryo-EM density and the models of GluA2-GSG1L (PBD IDs: 5VHZ and 5WEL) as guides. The resulting models were real space refined in Phenix (Afonine et al., 2012) and visualized in Chimera (Pettersen et al., 2004) or Pymol (DeLano, 2002).
Patch-clamp recordings
DNA encoding GluA2-γ5 fusion (described in the Construct section), its K695A/K697A mutant (GluA2Mut-γ5) and the K695A/K697A mutant of rat GluA2flip with Q at the Q/R-site (GluA2Mut) were introduced into a pIRES plasmid for expression in eukaryotic cells that were engineered to produce green fluorescent protein via a downstream internal ribosome entry site (Yelshanskaya et al., 2014). HEK 293 cells grown on glass coverslips in 35 mm dishes were transiently transfected with 1–5 μg of plasmid DNA using Lipofectamine 2000 Reagent (Invitrogen). Recordings were made 24 to 96 hours after transfection at room temperature. Currents from whole cells, typically held at a −60 mV potential, were recorded using Axopatch 200B amplifier (Molecular Devices, LLC), filtered at 5 kHz, and digitized at 10 kHz using low-noise data acquisition system Digidata 1440A and pCLAMP software (Molecular Devices, LLC). The external solution contained (in mM): 140 NaCl, 2.4 KCl, 4 CaCl2, 4 MgCl2, 10 HEPES pH 7.3 and 10 glucose; 7 mM NaCl was added to the extracellular activating solution containing 3 mM Glu to increase solution exchange speed rate. The internal solution contained (in mM): 150 CsF, 10 NaCl, 10 EGTA, 20 HEPES pH 7.3. Rapid solution exchange was achieved with a two-barrel theta glass pipette controlled by a piezoelectric translator. Typical 10–90% rise times were 200–300 μs, as measured from junction potentials at the open tip of the patch pipette after recordings. Data analysis was performed using Origin 9.1.0 software (OriginLab Corp.). Recovery from desensitization recorded using the two-pulse protocol was fitted with the Hodgkin-Huxley equation(Robert and Howe, 2003): I = (Imax1/m − (Imax1/m − 1)×exp(−t/τRecDes))m, where I is the peak current at a given interpulse interval, t, Imax is the peak current at long interpulse intervals, τRecDes is the recovery time constant and m is an index that corresponds to the number of kinetically equivalent rate-determining transitions that contribute to the recovery time course.
Planar lipid bilayer recordings
Planar lipid bilayers measurements were performed as described previously (Cao et al., 2013). Briefly, planar lipid bilayers were formed from a 30 mM solution of synthetic lipid mix 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-glycero-3-phosphoglycine (POPG) at a 3:1:1 ratio (Anatrace) in n-decane (Sigma-Aldrich). The solution was used to paint a bilayer in an aperture of ~250 μm diameter in a Meca chip (Nanion). Each cavity in the chip contains an individual integrated Ag/AgCl-microelectrode. Bathing solutions of 150 mM KCl, 0.02 mM MgCl2, and 20 mM HEPES (pH 7.2). All reagents (Sigma-Aldrich) were ultrapure (>99%). Bilayer capacitances were in the range of 7–15 pF.
The purified protein (10 ng/ml) was added to the bilayer-forming lipid mix (1 volume of protein to 1 volume of the lipid mix) and incubated for 30 minutes at 37°C. After the bilayers had been formed by painting on a Meca chip (Nanion), they did not show any single-channel activity. Only after the incubated protein-lipid mix was added by painting, the unitary currents were recorded using an Orbit mini device (Nanion). Data were low-pass filtered at 20 kHz and digitized at 1.22 kHz controlled by pClamp10.3 software (Molecular Devices). Single-channel conductance events, all-points histograms, and other parameters were identified and analyzed with Clampfit10.3 software (Molecular Devices). Independent of the presence of the auxiliary subunit and not affecting the conductance values, the channel open probability was changing significantly during and between different experiments, reflecting the commonly observed “modal” behavior of AMPA receptors (Figure S3G) (Prieto and Wollmuth, 2010; Shelley et al., 2012; Zhang et al., 2014). For analysis of single-channel currents, we used the high open probability mode. In the high open probability mode, it was much easier to spot the recordings where more than one channel was incorporated into the lipid bilayer, especially since all our recordings were made in the presence of 100 μM CTZ. Only recordings with no more than four conductance levels and no more than four peaks in the amplitude histograms respectively (Figure S3C–E) as well as the conductance values that resembled conductance levels reported previously (Carrillo et al., 2020; Prieto and Wollmuth, 2010; Rosenmund et al., 1998; Shelley et al., 2012; Smith and Howe, 2000; Soto et al., 2009; Swanson et al., 1997; Tomita et al., 2005) were subjected to single-channel analysis. All recordings with more than one channel incorporated into lipid bilayer were discarded from the analysis. All experiments were performed at room temperature.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis (Figures 4A–C, 7D and S3) was performed using Origin 9.1.0 (OriginLab). Statistical significance was calculated using One-Way ANOVA followed by Fisher’s least significant difference test. In all figure legends, n represents the number of independent biological replicates. All quantitative data were presented as mean ± SEM.
Supplementary Material
Supplementary Movie 1. Closed-to-desensitized state structural transitions in GluA2-γ5 and GluA2-GSG1L, Related to Figures 6 and 7E–F. This movie shows cryo-EM densities and the corresponding molecular models for full-length and ATD- and micelle-subtracted GluA2-γ5 and GluA2-GSG1L complexes as well as morphs between their closed (GluA2-γ5ZK and GluA2-GSG1Lapo) and desensitized (GluA2-γ5Quis and GluA2-GSG1LQuis) states, respectively. To illustrate changes in the contact between the β1-β2 loop of γ5 and LBD during closed to desensitized state transition, the surface of the tip of the β1-β2 loop (residues K695, S696, and K697) are colored red. Lipids are shown as stick models and molecules of Quis as space-filling models. Movie is made using ChimeraX (Pettersen et al., 2021).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tris | Fisher scientific | Cat# BP152–1 |
| NaCl | Fisher scientific | Cat# BP358–212 |
| KCl | Sigma | Cat# 7300 |
| CaCl2 | Sigma | Cat# 499609 |
| MgCl2 | Fisher scientific | Cat# BP214500 |
| HEPES | Sigma | Cat# H3375 |
| Glucose | Sigma | Cat# G8769 |
| CsF | Sigma | Cat# 198323 |
| EGTA | Sigma | Cat# 324626 |
| POPC | Avanti | Cat# 850457 |
| POPG (PG) | Avanti | Cat# 840457 |
| POPE (PE) | Avanti | Cat# 850757 |
| L-Glutamate | Sigma | Cat# 49621 |
| ZK200775 (ZK) | Tocris | Cat# 2345 |
| Kynurenic acid | Sigma | Cat# K3374 |
| Phosphate buffer saline | Sigma | Cat# 17202 |
| Quisqualate | Tocris | Cat# 0188 |
| Cyclothiazide (CTZ) | Tocris | Cat# 0713 |
| Cholesteryl hemisuccinate | Anatrace | Cat# 4193482 |
| Digitonin | Cayman Chemical Company | Cat#14952 |
| Desthiobiotin | Sigma | Cat #D1411 |
| PMSF | Acros Organics | Cat# 215740500 |
| 2-Mercaptoethanol (BME) | Acros Organics | Cat# 125470100 |
| Thrombin | Haematologic Technologies | Cat# HCT-0020 |
| Fetal bovine serum | Gibco | Cat# 16140071 |
| Sf-900 III SFM | Gibco | Cat# 12658027 |
| Freestyle 293 expression medium | Gibco | Cat# 12338018 |
| Sodium butyrate | ACROS Organics | Cat# 263191000 |
| Deposited Data | ||
| Coordinates of GluA2-γ5ZK FL | This paper | PDB ID: 7RZ4 |
| Cryo-EM map of GluA2-γ5ZK FL | This paper | EMDB: EMD-24750 |
| Coordinates of GluA2-γ5ZK LBD-TMD | This paper | PDB ID: 7RZ5 |
| Cryo-EM map of GluA2-γ5ZK LBD-TMD | This paper | EMDB: EMD-24751 |
| Coordinates of GluA2-γ5Glu FL | This paper | PDB ID: 7RZ6 |
| Cryo-EM map of GluA2-γ5Glu FL | This paper | EMDB: EMD-24752 |
| Coordinates of GluA2-γ5Glu LBD-TMD | This paper | PDB ID: 7RYY |
| Cryo-EM map of GluA2-γ5Glu LBD-TMD | This paper | EMDB: EMD-24748 |
| Coordinates of GluA2-γ5Quis FL | This paper | PDB ID: 7RZ7 |
| Cryo-EM map of GluA2-γ5Quis FL | This paper | EMDB: EMD-24753 |
| Coordinates of GluA2-γ5Quis LBD-TMD | This paper | PDB ID: 7RZ8 |
| Cryo-EM map of GluA2-γ5Quis LBD-TMD | This paper | EMDB: EMD-24754 |
| Coordinates of GluA2-GSG1Lapo FL | This paper | PDB ID: 7RZ9 |
| Cryo-EM map of GluA2-GSG1Lapo FL | This paper | EMDB: EMD-24755 |
| Coordinates of GluA2-GSG1LQuis FL | This paper | PDB ID: 7RZA |
| Cryo-EM map of GluA2-GSG1LQuis FL | This paper | EMDB: EMD-24756 |
| Coordinates of GluA2-GSG1LQuis LBD-TMD | This paper | PDB ID: 7RYZ |
| Cryo-EM map of GluA2-GSG1LQuis LBD-TMD | This paper | EMDB: EMD-24749 |
| Cryo-EM map of GluA2-γ2 | Twomey et al., 2018 | EMDB: EMD-7959 |
| Cryo-EM map of GluA2-GSG1L | Twomey et al., 2017b | EMDB: EMD-8688 |
| Coordinates of GluA2-γ2 | Twomey et al., 2017a | PDB ID: 5WEO |
| Coordinates of GluA2-SG1L | Twomey et al., 2017b | PDB ID: 5VHZ |
| Coordinates of GluA2-GSG1L | Twomey et al., 2017a | PDB ID: 5WEL |
| GluA2 bound to GSG1L | Twomey et al., 2017a | PDB ID: 5WEM |
| Glutamate receptor GluA2 in Apo state | Yelshanskaya et al., 2016 | PDB ID: 5L1B |
| Glutamate receptor in complex with competitive antagonist ZK | Sobolevsky et al., 2009 | PDB ID: 3KG2 |
| Fully occupied GluA2 AMPA receptor - TARP complex | Zhao et al., 2016 | PDB ID: 5KK2 |
| GluA2 bound to antagonist ZK and GSG1L | Twomey et al., 2017a | PDB ID: 5WEK |
| Structure of AMPA receptor-TARP complex | Chen et al., 2017 | PDB ID: 5VOV |
| Structure of AMPA receptor-TARP complex | Chen et al., 2017 | PDB ID: 5VOT |
| Coordinates GluA1/2 in complex with auxiliary subunit gamma-8 | Herguedas et al., 2019 | PDB ID: 6QKC |
| Coordinates of GluA2 in complex with its auxiliary subunit CNIH3 | Nakagawa, T., 2019 | PDB ID: 6PEQ |
| Experimental Models: Cell Lines | ||
| HEK293S GnTI− | ATCC | Cat#CRL-3022 |
| Sf9 | Gibco | Cat#12659017 |
| Recombinant DNA | ||
| GluA2-GSG1L | Twomey et al., 2017a | N/A |
| Oligonucleotides | ||
| GluA2-γ5 Amplification primer, 5’ AAACGAATGAAGGGAACCGGTAGCGCCTGTGGGAGGAAGGCCCTG 3’ | This paper | N/A |
| GluA2-γ5 Amplification primer, 5’ CGCGCGGCACCAGTCCACCGGTGCAGGGAGATGATGACATCTGGTC 3’ | This paper | N/A |
| Software and Algorithms | ||
| Origin 9.1.0 software | OriginLab | https://www.originlab.com/demodownload.aspx |
| pCLAMP software | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite#gref |
| gCTF | Zhang, 2016 | http://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| Motioncor2 | Zheng et al., 2017 | http://msg.ucsf.edu/em/software/motioncor2.html |
| RELION 3.1 | Zivanov et al., 2018 | http://www2.mrc-lmb.cam.ac.uk/relion/ |
| cryoSPARC | Punjani et al., 2017 | http://cryosparc.com |
| UCSF Chimera | Pettersen et al., 2021 | https://www.cgl.ucsf.edu/chimera/ |
| Pymol (Schrödinger) | DeLano, 2002 | http://www.pymol.org |
| PHENIX | Afonine et al., 2012 | https://www.phenix-online.org/ |
| COOT | Emsley et al., 2004 | http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| Swiss-Model | Waterhouse et al., 2018 | https://swissmodel.expasy.org/ |
| Other | ||
| Ultra Au Foil Au/Au grids | EMS | Cat # Q350AR1.3A |
| CF-1.2/1.3–2Au 200 mesh holey carbon grids | Protochips | Cat#CF-1.2/1.3–2Au |
| Gold wire | Ted Pella, Inc. | Cat#21–10 |
| Size Exclusion Superose 10/300 column | GE Healthcare | Cat# 17-5172-01 |
Highlights.
Desensitized and closed-state structures of GluA2 with type II TARP γ5 and GSG1L
γ5 and GSG1L head domains define 2:1 stoichiometry of their complexes with GluA2
LBD dimer interface ruptures during GluA2-γ5 and GluA2-GSG1L desensitization
Desensitized LBD dimers remain 2-fold symmetric in GluA2-γ5 but not in GluA2-GSG1L
ACKNOWLEDGMENTS
We thank R. Grassucci and Y.-C. Chi (Columbia University Cryo-Electron Microscopy Center), H. Kuang and E. Chua (New York Structural Biology Center/ National Center for CryoEM Access and Training), C. W. Hecksel (Stanford Linear Accelerator Center/National Accelerator Laboratory) for help with microscope operation and data collection and Kirill D. Nadezhdin for help with data processing. Some of this work was performed at the Columbia University Cryo-Electron Microscopy Center. Some of this work was performed at the National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247) and NY State Assembly Majority. Some of this work was performed at the Stanford-SLAC Cryo-EM Center (S2C2), which is supported by the National Institutes of Health Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129541). A.I.S. was supported by the NIH (R01 CA206573, R01 NS083660, R01 NS107253) and NSF (1818086).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, and Gouaux E (2000). Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, and Gouaux E (2006). Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127, 85–97. [DOI] [PubMed] [Google Scholar]
- Ben-Yaacov A, Gillor M, Haham T, Parsai A, Qneibi M, and Stern-Bach Y (2017). Molecular Mechanism of AMPA Receptor Modulation by TARP/Stargazin. Neuron 93, 1126–1137. [DOI] [PubMed] [Google Scholar]
- Cao C, Zakharian E, Borbiro I, and Rohacs T (2013). Interplay between calmodulin and phosphatidylinositol 4,5-bisphosphate in Ca2+-induced inactivation of transient receptor potential vanilloid 6 channels. J Biol Chem 288, 5278–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo E, Shaikh SA, Berka V, Durham RJ, Litwin DB, Lee G, MacLean DM, Nowak LM, and Jayaraman V (2020). Mechanism of modulation of AMPA receptors by TARP-gamma8. J Gen Physiol 152, e201912451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, and Nicoll RA (2000). Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408, 936–943. [DOI] [PubMed] [Google Scholar]
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, and Henderson R (2013). High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhao Y, Wang Y, Shekhar M, Tajkhorshid E, and Gouaux E (2017). Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell 170, 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe GB, Musgaard M, Aurousseau MR, Nayeem N, Green T, Biggin PC, and Bowie D (2016). Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Neuron 89, 1264–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002). The PyMOL Molecular Graphics System (San Carlos, CA, USA: DeLano Scientific; ). [Google Scholar]
- Drummond JB, Tucholski J, Haroutunian V, and Meador-Woodruff JH (2013). Transmembrane AMPA receptor regulatory protein (TARP) dysregulation in anterior cingulate cortex in schizophrenia. Schizophr Res 147, 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Goehring A, Lee CH, Wang KH, Michel JC, Claxton DP, Baconguis I, Althoff T, Fischer S, Garcia KC, and Gouaux E (2014). Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc 9, 2574–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Mao X, Lussier MP, Hutchison MA, Zhou L, Hamra FK, Roche KW, and Lu W (2016). GSG1L suppresses AMPA receptor-mediated synaptic transmission and uniquely modulates AMPA receptor kinetics in hippocampal neurons. Nat Commun 7, 10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herguedas B, Watson JF, Ho H, Cais O, Garcia-Nafria J, and Greger IH (2019). Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma8. Science 364, eaav9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, and Nicoll RA (2011). The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay SD, Ellis SB, McCue AF, Williams ME, Vedvick TS, Harpold MM, and Campbell KP (1990). Primary structure of the gamma subunit of the DHP-sensitive calcium channel from skeletal muscle. Science 248, 490–492. [DOI] [PubMed] [Google Scholar]
- Kamalova A, Futai K, Delpire E, and Nakagawa T (2021). AMPA receptor auxiliary subunit GSG1L suppresses short-term facilitation in corticothalamic synapses and determines seizure susceptibility. Cell Rep 34, 108732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalova A, and Nakagawa T (2021). AMPA receptor structure and auxiliary subunits. J Physiol 599, 453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Siuda ER, Nisenbaum ES, and Bredt DS (2008). AMPA receptor subunit-specific regulation by a distinct family of type II TARPs. Neuron 59, 986–996. [DOI] [PubMed] [Google Scholar]
- Kato AS, Zhou W, Milstein AD, Knierman MD, Siuda ER, Dotzlaf JE, Yu H, Hale JE, Nisenbaum ES, Nicoll RA, et al. (2007). New transmembrane AMPA receptor regulatory protein isoform, gamma-7, differentially regulates AMPA receptors. J Neurosci 27, 4969–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Gu X, and Lu W (2017). GSG1L regulates the strength of AMPA receptor-mediated synaptic transmission but not AMPA receptor kinetics in hippocampal dentate granule neurons. J Neurophysiol 117, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee TP, Bats C, Farrant M, and Cull-Candy SG (2015). Auxiliary Subunit GSG1L Acts to Suppress Calcium-Permeable AMPA Receptor Function. J Neurosci 35, 16171–16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T (2019). Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science 366, 1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A, Kolleker A, Ayalon G, Gillor M, Osten P, and Stern-Bach Y (2005). Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 2682–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto ML, and Wollmuth LP (2010). Gating modes in AMPA receptors. J Neurosci 30, 4449–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Robert A, and Howe JR (2003). How AMPA receptor desensitization depends on receptor occupancy. J Neurosci 23, 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, and Stevens CF (1998). The tetrameric structure of a glutamate receptor channel. Science 280, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Russo CJ, and Passmore LA (2014). Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, et al. (2012). High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74, 621–633. [DOI] [PubMed] [Google Scholar]
- Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR 3rd, et al. (2012). Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep 1, 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley C, Farrant M, and Cull-Candy SG (2012). TARP-associated AMPA receptors display an increased maximum channel conductance and multiple kinetically distinct open states. J Physiol 590, 5723–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, and Howe JR (2000). Concentration-dependent substate behavior of native AMPA receptors. Nat Neurosci 3, 992–997. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, and Gouaux E (2009). X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto D, Coombs ID, Kelly L, Farrant M, and Cull-Candy SG (2007). Stargazin attenuates intracellular polyamine block of calcium-permeable AMPA receptors. Nat Neurosci 10, 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto D, Coombs ID, Renzi M, Zonouzi M, Farrant M, and Cull-Candy SG (2009). Selective regulation of long-form calcium-permeable AMPA receptors by an atypical TARP, gamma-5. Nat Neurosci 12, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, and Gouaux E (2002). Mechanism of glutamate receptor desensitization. Nature 417, 245–253. [DOI] [PubMed] [Google Scholar]
- Swanson GT, Kamboj SK, and Cull-Candy SG (1997). Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci 17, 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, and Bredt DS (2005). Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature 435, 1052–1058. [DOI] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, and Bredt DS (2003). Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol 161, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R, et al. (2010). Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62, 405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, and Sobolevsky AI (2018). Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry 57, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, and Sobolevsky AI (2016). Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science 353, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, and Sobolevsky AI (2017a). Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature 549, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, and Sobolevsky AI (2017b). Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron 94, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, and Sobolevsky AI (2019). Structural and functional insights into transmembrane AMPA receptor regulatory protein complexes. J Gen Physiol 151, 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Vassilevski AA, and Sobolevsky AI (2018). Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors. Neuron 99, 956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Li M, and Sobolevsky AI (2014). Structure of an agonist-bound ionotropic glutamate receptor. Science 345, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Mesbahi-Vasey S, Kurnikova MG, and Sobolevsky AI (2017). Role of the Ion Channel Extracellular Collar in AMPA Receptor Gating. Sci Rep 7, 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Singh AK, Sampson JM, Narangoda C, Kurnikova M, and Sobolevsky AI (2016). Structural bases of noncompetitive inhibition of AMPA subtype ionotropic glutamate receptors by antiepileptic drugs. Neuron 91, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Devi SP, Tomita S, and Howe JR (2014). Auxiliary proteins promote modal gating of AMPA- and kainate-type glutamate receptors. Eur J Neurosci 39, 1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Swensen AC, Qian WJ, and Gouaux E (2019). Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 364, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Yoshioka C, Baconguis I, and Gouaux E (2016). Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature 536, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1. Closed-to-desensitized state structural transitions in GluA2-γ5 and GluA2-GSG1L, Related to Figures 6 and 7E–F. This movie shows cryo-EM densities and the corresponding molecular models for full-length and ATD- and micelle-subtracted GluA2-γ5 and GluA2-GSG1L complexes as well as morphs between their closed (GluA2-γ5ZK and GluA2-GSG1Lapo) and desensitized (GluA2-γ5Quis and GluA2-GSG1LQuis) states, respectively. To illustrate changes in the contact between the β1-β2 loop of γ5 and LBD during closed to desensitized state transition, the surface of the tip of the β1-β2 loop (residues K695, S696, and K697) are colored red. Lipids are shown as stick models and molecules of Quis as space-filling models. Movie is made using ChimeraX (Pettersen et al., 2021).
Data Availability Statement
Cryo-EM maps have been deposited to the EMDB with the accession codes EMD-24750 (GluA2-γ5ZK, full length), EMD-24751 (GluA2-γ5ZK, LBD-TMD), EMD-24752 (GluA2-γ5Glu, full length), EMD-24748 (GluA2-γ5Glu, LBD-TMD), EMD-24753 (GluA2-γ5Quis, full length), EMD-24754 (GluA2-γ5Quis, LBD-TMD), EMD-24755 (GluA2-GSG1Lapo, full length), EMD-24756 (GluA2-GSG1LQuis, full length) and EMD-24749 (GluA2-GSG1LQuis, LBD-TMD). Coordinates have been deposited to the PDB with the accession codes 7RZ4 (GluA2-γ5ZK, full length), 7RZ5 (GluA2-γ5ZK, LBD-TMD), 7RZ6 (GluA2-γ5Glu, full length), 7RYY (GluA2-γ5Glu, LBD-TMD), 7RZ7 (GluA2-γ5Quis, full length), 7RZ8 (GluA2-γ5Quis, LBD-TMD), 7RZ9 (GluA2-GSG1Lapo, full length), 7RZA (GluA2-GSG1LQuis, full length) and 7RYZ (GluA2-GSG1LQuis, LBD-TMD).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


