Abstract
A pathogen inactivation step during collection or processing of clinical samples has the potential to reduce infectious risks associated with diagnostic procedures. It is essential that these inactivation methods are demonstrated to be effective, particularly for non-traditional inactivation reagents or for commercial products where the chemical composition is undisclosed. This study assessed inactivation effectiveness of twenty-four next-generation (guanidine-free) nucleic acid extraction lysis buffers and twelve rapid antigen test buffers against SARS-CoV-2, the causative agent of COVID-19. These data have significant safety implications for SARS-CoV-2 diagnostic testing and support the design and evidence-based risk assessment of these procedures.
Subject terms: Infectious diseases, Clinical microbiology, Virology
Introduction
The international response to the COVID-19 pandemic has required diagnostic testing of a vast number of patient samples worldwide for the detection of SARS-CoV-21. SARS-CoV-2 is classified as a Hazard Group 3 (HG3) pathogen in the UK by the Advisory Committee for Dangerous Pathogens due to its potential to cause severe respiratory disease; propagative work for any purpose must be carried out at containment level 3 (CL3)2, which can severely restrict testing capabilities. Due to the exceptional circumstances presented by COVID-19, non-propagative diagnostic testing may be carried out at a lower level of containment with heightened control measures and suitable, sufficient risk assessment of procedures2,3. In the UK, infectious material must be inactivated by a validated method before being handled outside a microbiological safety cabinet (MSC)2; selected inactivation methods must be both effective at reducing pathogen infectivity and compatible with downstream sample processing.
Development of rapid tests designed for near-patient or point-of-care (POC) use, including rapid RT-PCR, loop-mediated isothermal amplification (LAMP) and lateral flow immunoassay (LFIA) antigen tests, has permitted SARS-CoV-2 testing to move outside traditional laboratory settings into the community. As for all diagnostic procedures, performing these tests at POC or near-POC requires a suitable and sufficient risk assessment and implementation of appropriate control measures2–4: it is essential that the rapid testing procedure, which may involve swab stirring/mixing into an elution buffer with the potential for associated generation of aerosols, does not place individuals performing these tests at an increased risk of SARS-CoV-2 exposure. It is therefore highly desirable that rapid testing procedures include a step that reduces the titre of infectious virus present e.g. by eluting samples into an effective lysis buffer.
The chaotropic agents guanidine thiocyanate (GTC) and guanidine hydrochloride (GHCl) are commonly used to inactivate viruses prior to nucleic acid testing5 and guanidine-based lysis buffers are effective at reducing SARS-CoV-2 titre6,7. However, reagents containing GTC and GHCl are a chemical hazard in testing laboratories due to their toxicity and incompatibility with sodium hypochlorite-based disinfectants8. Their hazardous nature also limits use of GTC and GHCl buffers in home-sampling kits because of the potential for misuse by members of the public. There are now many commercial non-hazardous alternatives to guanidine-based lysis buffers; these would prove valuable additions to COVID-19 testing procedures if effective at inactivating SARS-CoV-2. Currently, there is limited information available on the efficacy of SARS-CoV-2 inactivation by either guanidine-free lysis buffers or by rapid test buffers. This study provides data on inactivation of SARS-CoV-2 by twenty-four guanidine-free molecular extraction reagents and specimen transport media, and twelve LFIA antigen test buffers.
Methods
All handling of infectious SARS-CoV-2 was performed inside Class III MSCs within a CL3 facility. SARS-CoV-2 (England/02/2020) was propagated in Vero E6 cells (Vero C1008; ATCC CRL-1586) as previously described6. Virus was used in inactivation tests at passage 2 or 3 (with virus titres of approximately 1 × 107 PFU per ml). Approximately tenfold concentrated SARS-CoV-2 virus stocks (approximately 1 × 108 PFU per ml) were produced by centrifuging SARS-CoV-2 supernatant through Amicon Ultra-15 (100 K) filters (Millipore) according to the manufacturer’s instructions.
Guanidine-free lysis buffers and LFIA tests kit buffers evaluated and the active ingredients of these products where disclosed are listed in Table 1. Inactivation testing was performed essentially as previously described6 and is depicted in Fig. 1. In brief, SARS-CoV-2 was incubated with nucleic acid extraction buffers at a volume ratio and for a contact time indicated in the manufacturer’s instructions, where these were available. LFIA buffers were tested after a 1-min treatment time, to mimic likely contact times in real-life testing scenarios. 5- and 10-min treatment times were additionally evaluated to determine the effect of longer incubation times. All tests were conducted at ambient room temperature (18–22 °C). For most products, treated samples were passed through a filtration matrix (Detergent Removal Resin [Pierce], Sephadex LH-20 [GE Healthcare], Sephacryl S-400HR [Sigma-Aldrich] or BioBeads SM2 [Biorad]) to remove cytotoxic chemicals prior to virus titration to improve the limit of detection (LOD)6. The optimum purification method for each product was assessed as previously described6, and is listed in Table 1. All experiments were carried out as three independent inactivation tests. After sample clean-up, samples were tenfold serially diluted in PBS and plated on to 96-well plates containing 2.5 × 104 Vero E6 cells. Plates were incubated for 6–7 days at 37 °C/5%CO2, then fixed and stained with crystal violet to visualise cytopathic effect (CPE). The 50% tissue culture infectious dose (TCID50) was calculated according to the Spearman-Karber method9. As a control for virus recovery, SARS-CoV-2 was mock-treated with PBS in parallel, at the same ratio as the reagent being tested. PBS-treated samples were subjected to the same filtration and titration methods as product-treated samples. Titre reduction was calculated by subtracting the mean log10 titre of tested samples from the mean log10 titre of PBS controls. A cytotoxicity control was purified (if applicable) and titrated alongside other test samples, and used to determine the LOD for each test; this control consisted of PBS in place of virus, treated with an equivalent volume of product as test samples. 95% confidence intervals for titre reductions were calculated as mean log10 titre reduction ± 1.96 standard error9. Products were tested against an unconcentrated SARS-CoV-2 stock as standard. Concentrated virus preparations were used to increase the maximum possible titre reduction for products (indicated in Table 1) that displayed high levels of cytotoxicity and/or that were previously found to reduce the titre of unconcentrated virus to below the test LOD.
Table 1.
SARS-CoV-2 inactivation by guanidine-free nucleic acid extraction reagents.
| Product name (manufacturer) | Active ingredient/s, if known* | Purification resin‡ | Reagent: virus ratio | Contact time (min) | Titre reduction, log10 TCID50/ml [± 95% CI]† | Virus detectable in TCID50 (virus detected in n/N replicates) |
|---|---|---|---|---|---|---|
| virusPHIX + (RNAssist) | 20–90% trifluoroacetamide 20–90% trimethylglycine | PDRR | 3:1 | 10 | ≥ 6.4 [6.2–6.7] | No3 |
| 30 | ≥ 7.1 [6.9–2.4] | No3 | ||||
| virusPHIX-LV (RNAssist) | 20–50% trifluoroacetamide 20–50% trimethylglycine | PDRR | 10:1# | 10 | ≥ 5.1 [4.8–5.5] | No4 |
| 30 | ≥ 6.0 [5.7–6.3] | No | ||||
| virusPHIX-P9 (RNAssist) | 2–5% trifluoroacetamide 1–3% trimethylglycine | LH20 | 3:1 | 10 | ≥ 4.4 [4.1–4.6] | Yes (1/3) |
| 30 | ≥ 4.8 [4.5–5.1] | No3 | ||||
| virusPHIX-CU (RNAssist) | Urea, choline chloride | None | 3:1 | 10 | 0.6 [0.1–1.0] | Yes (3/3) |
| 30 | 1.0 [0.6–1.4] | Yes (3/3) | ||||
| VitaPCR SARS-CoV-2 sample collection buffer (Credo Diagnostics) | 0.2% sodium hydroxide | PDRR | 10:1# | 1 | ≥ 5.0 [4.7–5.3] | Yes (3/3) |
| 5 | 6.2 [5.9–6.5] | Yes (3/3) | ||||
| MELT medium B (Mast Group) | 1–10% Triton X-100 | PDRR | 10:1# | 5 | ≥ 7.0 [6.6–7.3] | Yes (1/3) |
| MELT V1 (Mast Group) | Ethylene oxide propylene oxide copolymer mono (nonylphenyl) ether | PDRR | 10:1 | 10 | 1.4 [1.0–1.8] | Yes (3/3)1 |
| 30 | 1.9 [1.5–2.2] | Yes (3/3) | ||||
| MELT V3 (Mast Group) | Triton X-100 reduced | SM2 | 10:1 | 15 | ≥ 5.5 [5.2–5.8] | No1 |
| MELT V4 (Mast Group) | Triton CG-110 | SM2 | 10:1 | 15 | ≥ 5.5 [5.2–5.8] | No1 |
| Virus preservation solution (D-Biotech) | Citric acid < 0.1% Triton X-100 | PDRR | 10:1# | 10 | ≥ 6.0 [5.7–6.3] | Yes (2/3) |
| 30 | ≥ 5.3 [5.0–5.6] | No2 | ||||
| ID NOW COVID-19 elution buffer (Abbott) | 0.1% Triton X-100 | PDRR | 25:1# | 1 | 3.6 [3.2–4.0] | Yes (3/3) |
| 5 | 4.0 [3.6–4.4] | Yes (3/3) | ||||
| OMNIgene oral (DNA Genotek) | 1–5% SDS | S400HR | 1:1 | 10 | ≥ 4.3 [4.0–4.6] | No1 |
| COVID-19 PROmate sample preparation buffer with standard Triton X-100 (Novacyt) | 0.5% Triton X-100 | SM2 | 10:1# | 2 | 5.3 [5.0–5.7] | Yes (3/3) |
| 5 | 5.9 [5.6–6.2] | Yes (3/3) | ||||
| 10 | ≥ 6.2 [5.9–6.6] | Yes (1/3) | ||||
| COVID-19 PROmate sample preparation buffer with Triton X-100 reduced (Novacyt) | 0.5% Triton X-100 reduced | SM2 | 10:1# | 2 | ≥ 6.2 [5.9–6.6] | Yes (2/3) |
| Saliva RNA sample collector kit (Zeesan) | 5–15% sodium lauroyl sarcosinate | PDRR | 2:1 | 30 | ≥ 5.6 [5.3–5.9] | Yes (2/3) |
| Virus RNA collection Kit (Zeesan) | 5–10% sodium lauroyl sarcosinate | SM2 | 2:1 | 10 | 3.3 [2.9–3.7] | Yes (3/3) |
| 30 | 4.5 [4.3–4.8] | Yes (3/3) | ||||
| Salicovgel-1 (LGC) | Enzymatic | S400HR | 10:1 | 60 | 1.0 [0.6–1.4] | Yes (3/3) |
| Salicovgel-2 (LGC) | Enzymatic | S400 HR | 10:1 | 60 | 3.1 [2.9–3.4] | Yes (3/3) |
| 180 | ≥ 3.4 [3.2–3.7] | Yes (2/3) | ||||
| GeneFix Saliva RNA collector (IsoHelix) | Not available | S400HR | 1:1 | 10 | ≥ 5.3 [5.0–5.6] | No4 |
| BuccalFix (IsoHelix) | Not available | S400HR | 5:1# | 10 | ≥ 4.7 [4.4–5.0] | No4 |
| MicroLYSIS-RNA (Clent Life Science) | Not available | S400HR | 1:1 | 5 | 1.0 [0.6–1.4] | Yes (3/3) |
| 20 | 1.9 [1.6–2.3] | Yes (3/3) | ||||
| Coronavirus extraction reagent 2506 (Arcis Biotechnology) | Not available | None | 2:1 | 10 | ≥ 3.3 [3.0–3.6] | No5 |
| FRANKD buffer (GeneMe) | Not available | PDRR | 10:1 | 1 | 3.6 [3.3–3.9] | Yes (3/3) |
| 5 | 4.0 [3.7–4.3] | Yes (3/3) | ||||
| SAVD buffer (GeneMe) | Not available | PDRR | 4:1 | 1 | ≥ 4.5 [4.2–4.8] | No5 |
See overleaf for table footnotes.
Where identical results were obtained at multiple contact times, only the shortest contact time is shown.
*As indicated on product safety data sheets or other product literature.
†Values are given as ≥ when at least one replicate was below the limit of detection.
#Tested using concentrated virus stock.
‡PDRR: Pierce Detergent Removal Resin; SM2: Biorad SM2 BioBeads; S400HR: Sephacryl S-400HR; LH20: Sephadex LH-20.
STM specimen transport media.
Limit of detection: 10.7 log10 TCID50/mL; 20.8 log10 TCID50/mL; 31.0 log10 TCID50/mL; 42.0 log10 TCID50/mL; 51.7 log10 TCID50/mL.
Figure 1.
Inactivation testing workflow. SARS-CoV-2 suspensions are treated with guanidine-free molecular extraction reagents or LFIA buffers and mixed before incubating for the recommended contact time. If filtration is required to remove product-associated cytotoxicity, samples are added to a purification resin and centrifuged to elute and immediately titrated. If filtration steps are not required, samples are immediately titrated. Image created in Inkscape version 1.0 https://inkscape.org.
Results
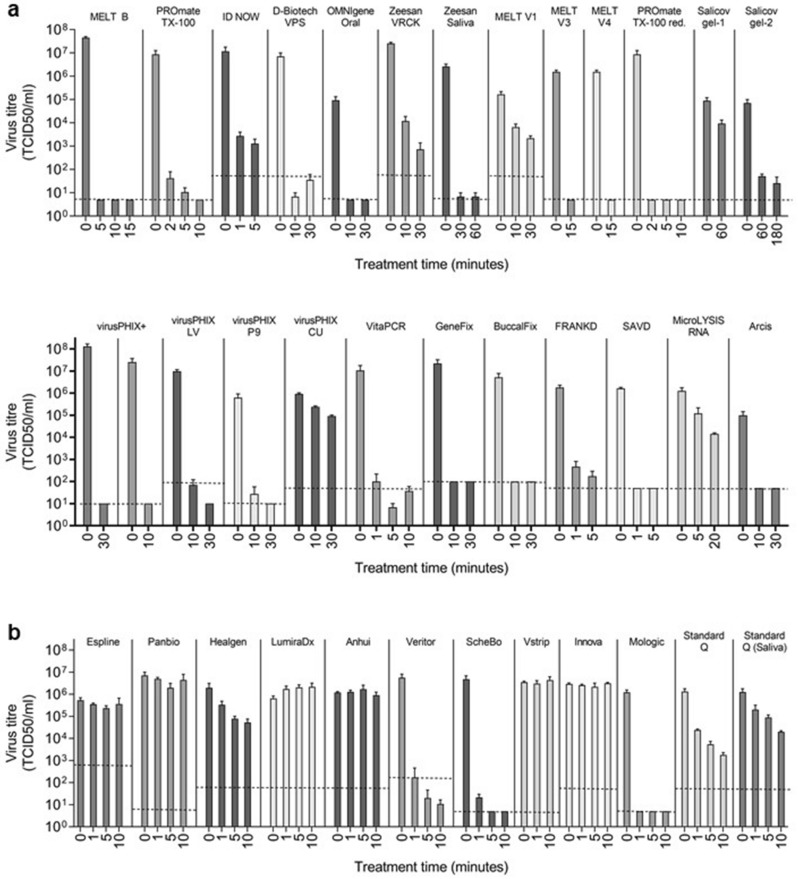
The guanidine-free lysis buffers listed in Table 1 were tested for their ability to inactivate SARS-CoV-2 at contact times determined according the manufacturer’s instructions where available. These reagents are variously marketed as appropriate for lysis of different sample types, including nasopharyngeal/oropharyngeal swabs, sputum and saliva, prior to nucleic acid extraction or as inactivating specimen transport media. A > 4.0 log10 reduction in SARS-CoV-2 titre, required by the British European standard BS EN 14476 for quantitative virucidal suspension tests9, was demonstrated for most guanidine-free nucleic acid extraction buffers tested (Table 1; Fig. 2a), including: virusPHIX+, virusPHIX-LV and virusPHIX-P9; VitaPCR Sample Collection Buffer; MELT Medium B, V3 and V4; D-Biotech Virus Preservation Solution; ID NOW COVID-19 Elution Buffer (after a 5-min treatment); OMNIgene Oral; PROmate Sample Preparation Buffer containing either Triton X-100 or Triton X-100 reduced; Zeesan Saliva RNA Sample Collector Kit buffer; Zeesan Viral RNA Collection Kit buffer (after 30 min); IsoHelix GeneFix and Buccalfix buffers; GeneMe FRANKD (after 5 min) and SAVD buffers.
Figure 2.
SARS-CoV-2 titre reductions following treatment with guanidine-free molecular extraction reagents and LFIA buffers. SARS-CoV-2 suspension was treated with guanidine-free molecular extraction reagents (a) or LFIA buffers (b), using the contact times and concentrations stated in Tables 1 and 2, respectively. SARS-CoV-2 was mock-treated with an equivalent volume of PBS in parallel. Samples were purified by methods indicated in Tables 1 and 2 to remove product-associated cytotoxicity, then titrated by TCID50 on Vero E6 cells to determine virus titre. All treatments and mock-treatments were performed in triplicate; bars show the mean of triplicate inactivation tests and error bars the standard deviation. The limit of detection for each test was determined using the cytotoxicity control for each test, and is indicated on the graph for each test by a dashed line. Where cytotoxicity of sample replicates and/or treatment times for a product differed, the highest LOD for the entire test is displayed. Variation in virus titres for PBS-treated samples between product tests was due to differences in the titre of virus stock used, the ratio of sample to product or PBS used for treatment, and differing virus recovery following sample filtration with different methods6. VRCK: Virus RNA Collection Kit; VPS: Virus Preservation System; Triton X-100 red.: Triton X-100 reduced. Graphs created in GraphPad Prism version 9 https://graphpad.com.
Of the molecular extraction reagents tested, we failed to demonstrate > 4.0 log10 reduction in SARS-CoV-2 titre for Salicovgel-1 (1.0 log10 after 1 h), Salicovgel-2 (3.1 and ≥ 3.4 log10 reduction after 1 and 3 h, respectively), MicroLYSIS-RNA (1.0 log10 reduction after the recommended 5 min contact time and 1.9 log10 reduction after 20 min), virusPHIX-CU (1.0 log10 reduction after 30 min) and Arcis Coronavirus Extraction Reagent 2506 (≥ 3.3 log10 reduction after 10 min) (Table 1; Fig. 2a).
Only three of the twelve LFIA buffers tested reduced SARS-CoV-2 titre by greater than 4 log10 (Table 2; Fig. 2b): the BD Veritor Extraction Reagent (≥ 4.5 log10 reduction after a 1-min treatment); the ScheBo SARS-CoV-2 Quick Antigen extraction buffer (5.3 log10 and ≥ 6.0 log10 reduction after 1 min and 5 min, respectively); and the Mologic Rapid Antigen Test Sample Buffer (≥ 5.4 log10 reduction after 1 min). Several LFIA buffers reduced SARS-CoV-2 titre more modestly. Standard Q Extraction Buffer from the SD Biosensor COVID-19 Ag LFIA gave a reduction of 2.9 log10 after 10 min. Maximum demonstrable titre reductions for Healgen and Standard Q Saliva LFIA buffers were 1.6 log10 and 1.8 log10, respectively, after 10 min. ESPLINE, Panbio, LumiraDx, Anhui Deepblue, Innova and Vstrip LFIA buffers had a negligible effect on infectious SARS-CoV-2 , even after a 10-min contact time (Table 2; Fig. 2b).
Table 2.
SARS-CoV-2 inactivation by rapid antigen test buffers.
| Product name (manufacturer) | Active ingredient/s, if known* | Purification resin‡ | Reagent: virus ratio | Contact time (min) | Titre reduction, log10 TCID50/ml [± 95% CI)† | Virus detectable in TCID50 (virus detected in n/N replicates) |
|---|---|---|---|---|---|---|
| ESPLINE SARS-CoV-2 extraction solution (Fujirebio) | ≥ 0.25– ≤ 0.5% cetrimonium chloride | SM2 | 2.3:1 | 1 | 0.2 [− 0.3–0.6] | Yes (3/3) |
| 5 | 0.4 [− 0.1–0.8] | Yes (3/3) | ||||
| 10 | 0.2 [− 0.2–0.6] | Yes (3/3) | ||||
| Panbio COVID-19 Ag rapid test device buffer (Abbott) | 0.49% Tween-20 | PDRR | 3:1 | 1 | 0.2 [− 0.3–0.6] | Yes (3/3) |
| 5 | 0.6 [0.1–1.0] | Yes (3/3) | ||||
| 10 | 0.2 [− 0.2–0.7] | Yes (3/3) | ||||
| Healgen coronavirus Ag rapid test cassette swab buffer (Zhejiang Orient Gene) | Not known | PDRR | 3:1 | 1 | 0.8 [0.4–1.2] | Yes (3/3) |
| 5 | 1.4 [1.0–1.8] | Yes (3/3) | ||||
| 10 | 1.6 [1.2–2.0] | Yes (3/3) | ||||
| LumiraDx SARS-CoV-2 Ag test extraction buffer (LumiraDx) | 0.5% unspecified detergent 1.00% Tween 20 | PDRR | 7.5:1 | 1 | − 0.4 [− 0.9–0.0] | Yes (3/3) |
| 5 | − 0.5 [− 0.9– − 0.1] | Yes (3/3) | ||||
| 10 | − 0.5 [− 1.0–0.0] | Yes (3/3) | ||||
| COVID-19 (SARS-CoV-2) antigen test kit extraction reagent (Anhui Deepblue Medical Technology) | Not known | None | 3:1 | 1 | 0.0 [− 0.4–0.3] | Yes (3/3) |
| 5 | 0.0 [− 0.6–03] | Yes (3/3) | ||||
| 10 | 0.1 [− 0.3–0.5] | Yes (3/3) | ||||
| Veritor extraction reagent (BD) | 0.1– < 1% Triton X-100 | PDRR | 3.25:1 | 1 | ≥ 4.5 [4.2–4.8] | No3 |
| 5 | ≥ 5.4 [5.2–5.7] | No2 | ||||
| 10 | ≥ 5.7 [5.4–6.0] | No1 | ||||
| SARS-CoV-2 quick antigen extraction buffer (ScheBo) | < 1% Triton X-100 | PDRR | 2.5:1 | 1 | 5.3 [5.1–5.6] | Yes (3/3) |
| 5 | ≥ 6.0 [5.7–6.3] | Yes (2/3) | ||||
| 10 | ≥ 6.0 [5.7–6.3] | No4 | ||||
| Vstrip COVID-19 antigen rapid test extraction buffer (Panion & BF Biotech) | < 1% Tergitol < 1.2% Methanol | PDRR | 10:1 | 1 | 0.1 [− 0.3–0.4] | Yes (3/3) |
| 10 | 0.0 [− 0.5–0.3] | Yes (3/3) | ||||
| SARS-CoV-2 antigen qualitative test extraction solution (Innova) | Not known | None | 2:1 | 1 | 0.1 [− 0.3–0.5] | Yes (3/3) |
| 5 | 0.1 [− 0.2–0.5] | Yes (3/3) | ||||
| 10 | 0.0 [− 0.5–0.4] | Yes (3/3) | ||||
| COVID-19 rapid antigen test sample buffer (Mologic) | 0.4% unspecified surfactant | PDRR | 3.5:1 | 1 | ≥ 5.4 [5.1–5.7] | No1 |
| Extraction buffer from Standard Q COVID-19 Ag Test Kit (SD Biosensor) | Not known | PDRR | 2:1 | 1 | 1.7 [1.3–2.1] | Yes (3/3) |
| 5 | 2.4 [2.0–2.8] | Yes (3/3) | ||||
| 10 | 2.9 [2.4–3.3] | Yes (3/3) | ||||
| Extraction buffer from Standard Q COVID-19 Ag Saliva test kit (SD biosensor) | Not known | PDRR | 3.5:1 | 1 | 0.8 [0.4–1.2] | Yes (3/3) |
| 5 | 1.1 [0.7–1.6] | Yes (3/3) | ||||
| 10 | 1.8 [1.4–2.2] | Yes (3/3) |
*As indicated on product safety data sheets or other product literature.
†Values are given as ≥ when at least one replicate was below the limit of detection.
‡PDRR: Pierce Detergent Removal Resin; SM2: Biorad SM2 BioBeads; Limit of detection: 11.0 log10 TCID50/mL; 21.3 log10 TCID50/mL; 32.2 log10 TCID50/mL; 40.7 log10 TCID50/mL; 51.7 log10 TCID50/mL.
Discussion
Incorporation of novel products into existing diagnostic workflows requires them to be effective at inactivating pathogens; this study has provided inactivation efficacy data for twenty-four new products against SARS-CoV-2. Many new nucleic acid extraction reagents are guanidine-free and their inclusion in diagnostic procedures in place of hazardous guanidine-based reagents has the potential to eliminate the chemical hazards associated with guanidine use. Furthermore, many new products are supplied as non-hazardous: non-hazardous inactivating specimen transport media have the potential to increase the safety of sample transport and the speed of testing processes while not posing a chemical hazard to users at the sampling stage. SARS-CoV-2 inactivation effectiveness of twelve buffers provided with LFIA tests was additionally evaluated in this study, since these are being increasingly employed as COVID-19 testing capabilities are scaled up to include home and workplace testing.
Of the products that were demonstrated to inactivate SARS-CoV-2 effectively (i.e. > 4 log10 reduction in titre following treatment), four nucleic acid extraction buffers (MELT medium B, D-Biotech Virus Preservation Solution, Abbott ID NOW Elution Buffer, PROmate Sample Preparation Buffer, and two LFIA buffers (BD Veritor and Schebo) are known to contain the non-denaturing detergent Triton X-100 as an active ingredient; we and others have previously shown that the Triton X-100 is effective at reducing SARS-CoV-2 titres by > 5–6 log106,10. Commercial products containing Triton X-100 reduced (PROmate and MELT V3), Triton CG-110 (MELT V4), SDS (OMNIgene Oral), trifluoroacetamide/trimethylglycine (virusPHIX+, -LV and -P9), sodium hydroxide (VitaPCR), sodium lauroyl sarcosinate (Zeesan Saliva RNA Collector Kit buffer) were also effective at reducing SARS-CoV-2 infectivity. Several guanidine-free products (Geneme FRANKD and SAVD buffers, GeneFix and BuccalFix) and one LFIA buffer (Mologic) were effective but information on chemical composition for these buffers has not been disclosed by the manufacturers. Enzymatic inactivation may be an alternative to chemical inactivation, and the inactivation of viruses has been previously demonstrated using proteolytic enzymes11. Virucidal activity of the enzyme-based inactivants Salicovgel-1 and Salicovgel-2 were assessed in this study; while a > 4 log10 titre reduction could not be demonstrated by either of these products, Salicovgel-2 reduced SARS-CoV-2 titre by ≥ 3 log10 after 1-h treatment, indicating potential for enzymatic products as SARS-CoV-2 inactivants. Data presented in this study will aid testing laboratories that are required to replace Triton X-100 with Triton X-100 alternatives, due to the inclusion of Triton X-100 on the European Authorisation list (Annex XIV) of the Registration, Evaluation, Authorisation and Restrictions of Chemicals (REACH) for phasing out of use12,13. In addition, several of the products shown to be effective inactivators of SARS-CoV-2 are marketed as non-hazardous specimen transport media, indicating that these may have potential for use in home-sampling kits.
LFIA tests require mixing of a test swab or specimen with the buffer followed by use of a pipette or dropper to load a test cassette; the LFIA testing procedure may therefore lead to the generation of infectious aerosols. Furthermore, the infectious titre of samples tested directly after patient sampling (as is the case for POC tests) is likely to be higher than for those that are transported from sampling sites to testing laboratories before further handling. Inactivation efficacy of POC test buffers is a key consideration for risk assessment of POC testing processes: only three of the twelve LFIA buffers tested in this study were effective at inactivating SARS-CoV-2. Several LFIA buffers tested were completely ineffective against SARS-CoV-2 and of those that did reduce virus titre, most gave modest reductions in comparison with other extraction buffers. Data presented here indicate that testing centres should not rely upon LFIA buffers to completely inactivate infectious samples and that additional control measures should be implemented to ensure the protection of test operators.
Data on the effectiveness of any inactivation step is crucial for designing and risk assessing testing procedures. Findings presented here are relevant for SARS-CoV-2 diagnostic laboratories, testing centres and mass testing programmes worldwide, providing data to support evidence-based risk assessment of testing procedures.
Acknowledgements
This work was supported by Public Health England. The views expressed are those of the authors and not necessarily those of Public Health England or the Department of Health.
Author contributions
K.D., U.A., H.B., C.B., S.R.W., N.G., R.S., T.B.K., N.H., M.J.H., M.J. and M.J.K. conducted inactivation experiments; K.D., P.S. and M.J.K. analysed data; K.D. and M.J.K. prepared the manuscript; P.A.C., C.B.B., N.W., A.D.G. and M.J.K. conceived and designed the work. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hasell J, et al. A cross-country database of COVID-19 testing. Sci. Data. 2020;7:345. doi: 10.1038/s41597-020-00688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Public Health England. COVID-19: Safe Handling and Processing for Laboratories. https://www.gov.uk/government/publications/wuhan-novel-coronavirus-guidance-for-clinical-diagnostic-laboratories/wuhan-novel-coronavirus-handling-and-processing-of-laboratory-specimens (2020).
- 3.World Health Organization. Laboratory BIOSAFETY Guidance Related to Coronavirus Disease (COVID-19)https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-(covid-19) (2020).
- 4.Centers for Disease Control and Prevention. Guidance for SARS-CoV-2 Point-of-Care Testing. https://www.cdc.gov/coronavirus/2019-ncov/lab/point-of-care-testing.html (2020). [PubMed]
- 5.Farrell, R. E. RNA Isolation Strategies. Rna Methodologies: Laboratory Guide for Isolation and Characterization, 5th Edition, 75–115. 10.1016/B978-0-12-804678-4.00003-8 (2017).
- 6.Welch SR, et al. Analysis of inactivation of SARS-CoV-2 by specimen transport media, nucleic acid extraction reagents, detergents, and fixatives. J. Clin. Microbiol. 2020;58:66. doi: 10.1128/JCM.01713-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Public Health England. COVID-19: PHE Laboratory Assessments of Inactivation Methods. https://www.gov.uk/government/publications/covid-19-phe-laboratory-assessments-of-inactivation-methods (2020).
- 8.U.S. Food and Drug Administration. Transport Media Safety Risk—Use Compatible Transport Media with SARS-CoV-2 Tests that Use Bleach—Letter to Clinical Laboratory Staff and Health Care Providers. https://www.fda.gov/medical-devices/letters-health-care-providers/transport-media-safety-risk-use-compatible-transport-media-sars-cov-2-tests-use-bleach-letter (2020).
- 9.British Standards Institution. BS EN 14476:2013+A2:2019—Chemical Disinfectants and Antiseptics. Quantitative Suspension Test for the Evaluation of Virucidal Activity in the Medical Area. Test method and requirements (Phase 2/Step 1) (2013).
- 10.Patterson EI, et al. Methods of inactivation of SARS-CoV-2 for downstream biological assays. J. Infect. Dis. 2020;222:1462–1467. doi: 10.1093/infdis/jiaa507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amtmann A, et al. Virucidal effects of various agents—including protease—against koi herpesvirus and viral haemorrhagic septicaemia virus. J. Fish. Dis. 2020;43:185–195. doi: 10.1111/jfd.13106. [DOI] [PubMed] [Google Scholar]
- 12.European Chemicals Agency. Authorisation List: List of substances included in Annex XIV of REACH. https://echa.europa.eu/authorisation-list (2021).
- 13.White R, Jobling S, Hoare SA, Sumpter JP, Parker MG. Environmentally persistent alkylphenolic compounds are estrogenic. Endocrinology. 1994;135:175–182. doi: 10.1210/en.135.1.175%JEndocrinology. [DOI] [PubMed] [Google Scholar]