Abstract
Non-small cell lung cancer is recognized as the deadliest cancer across the globe. In some areas, it is more common in women than even breast and cervical cancer. Its rise, vaulted by smoking habits and increasing air pollution, has garnered much attention and resource in the medical field. The first lung cancer treatments were developed more than half a century ago. Unfortunately, many of the earlier chemotherapies often did more harm than good, especially when they were used to treat genetically unsuitable patients. With the introduction of personalized medicine, physicians are increasingly aware of when, how, and in whom, to use certain anti-cancer agents. Drugs such as tyrosine kinase inhibitors, anaplastic lymphoma kinase inhibitors, and monoclonal antibodies possess limited utility because they target specific oncogenic mutations, but other drugs that target mechanisms universal to all cancers do not. In this review, we discuss many of these non-oncogene-targeting anti-cancer agents including DNA replication inhibitors (i.e., alkylating agents and topoisomerase inhibitors) and cytoskeletal function inhibitors to highlight their application in the setting of personalized medicine as well as their limitations and resistance factors.
Key words: Non-small cell lung cancer, Personalized medicine, Pharmacogenomics, Pharmacogenetics, Drug resistance, Vinca alkaloids, Toxoids, DNA replication inhibitors
Graphical abstract
This review discusses non-oncogene-targeting anti-cancer agents including DNA replication inhibitors and cytoskeletal function inhibitors to highlight their application in personalized medicine as well as their limitations and resistance factors.
1. Introduction
Non-small cell lung cancer (NSCLC), because of its unrivaled prevalence and lethality, is by many measures the most urgent cancer-related challenge facing modern medicine. Not only is it already the most common type of cancer world-wide, increasing amounts of pollution and unhealthy life choices further drive its incidence rate upwards1. It is now estimated that NSCLC accounts for over a quarter of all cancer-related deaths in the United States with an additional 1.8 million new known cases reported worldwide on an annual basis2. Because of this, there has been a plethora of research into the deadly affliction with massive resources devoted to halt its advance. As these efforts began to mature and bear fruit, we saw the first chemotherapies developed for combating NSCLC come into use over half a century ago. Unfortunately, these early agents often elicited very low response rates, significant toxicity, and finally, great chance for relapse. Given these traditional “blind” treatments, patients generally did not survive past the first year after diagnosis3,4. Without better options and out of desperation, patients and doctors came to accept that chemotherapy was a double-edged sword in the treatment of NSCLC.
The introduction of personalized medicine was a milestone for cancer treatment as incorporating knowledge of genetic factors into determining patient-specific regimens led to astounding improvements in treatment outcome5, 6, 7, 8, 9. High failure rates of oncogene-targeting chemotherapies such as tyrosine kinase inhibitors and anaplastic lymphoma kinase inhibitors have been attributed to the treatments being mismatched to patients whose cancer lacked the drug's genetic targets10. However, the status of specific oncogenic mutations does not constrain the use of other drugs that target mechanisms common to all cancers, such as DNA replication inhibitors and cytoskeletal function inhibitors. But their lack of specialized oncogenic targets, these chemotherapies are not entirely free from the effects of genetic factors and resistance mechanisms. Sensitivity to these drugs—and thus their effectiveness—depends on individual factors, including capability to metabolize specific substances and differences in genetic physiology resulting in varying expression levels of efflux pumps and stabilizer proteins. In this review, we discuss these factors and highlight the necessity of pharmacogenomic consideration in the decision-making process for physicians when prescribing non-oncogene-targeting chemotherapies for treatment of NSCLC patients.
2. Alkylating agents
DNA replication inhibitors are a diverse category of many smaller classes of drugs aimed at preventing ravenous tumor growth by interrupting the replication of DNA. Of these, alkylating agents, topoisomerase inhibitors, antimetabolites, are among the different approaches used by various chemotherapies aimed at preventing the critical DNA replication needed for (cancerous) cell division. From as early as the nineties, gemcitabine and cisplatin were used as anti-cancer compounds for the clinical treatment of NSCLC11,12. Although gemcitabine is a topoisomerase I inhibitor instead of an alkylating agent, its discussion could arguably be made out of turn together with the alkylating agent cisplatin because the two are most often used in combination and as such, many studies apply to them both rather than individually. Because both gemcitabine and cisplatin are mainly metabolized independently of the cytochrome P450 system (cytidine deaminase for gemcitabine and cysteine S-conjugate β-lyase for cisplatin), they are excellent choices for combination therapy to avoid excessive stress on the typical hepatic enzymes that metabolize anti-cancer chemotherapies13,14. Gemcitabine works by being incorporated into the DNA in place of its normal base pairs, preventing DNA polymerase from synthesizing the entire chain while cisplatin binds to DNA to cause DNA-lesions which trigger the DNA damage response leading to apoptosis15,16. Loosely classified as a platinum-containing alkylating agent, cisplatin undergoes hydrolysis in the cytoplasm and binds to the N7 reactive center on purines in order to crosslink DNA17. Due to its toxicity, derivatives of cisplatin exist, such as carboplatin which exhibit both reduced effect and toxicity compared to cisplatin18. Gemcitabine, while also a DNA replication inhibitor, is instead an antimetabolite which disables topoisomerase I19. A prodrug dependent on activation by deoxycytidine kinase, gemcitabine allows only one additional dNTP to be added after itself before replication stalls and DNA proofreading enzymes are unable to remove it from the replication complex13.
Because DNA replication-inhibiters form DNA-level interactions as opposed to the protein-level interactions, resistance mechanisms are far different. Due to the lack of protein interaction, mutations causing conformational changes to the adenosine triphosphate (ATP) binding sites of target proteins were not identified as modifiers of drug effectiveness. With drugs that target DNA replication, the genetic components of resistance usually represent increased effectiveness or upregulation of the DNA-replication machinery, which more closely resemble intrinsic characteristics of individuals rather than definable mutations. DNA excision repair protein 1 (encoded by the ERCC1 gene) is a protein that forms a critical component of the DNA-repair/maintenance machinery whose natural activity level is implicated as an intrinsic factor affecting drug efficacy in both gemcitabine and cisplatin20,21. Ribonucleoside-diphosphate reductase large subunit 1 (encoded by the RRM1 gene), which is identified as a tumor suppressor but is also critical in deoxyribonucleotide-synthesis prior to DNA replication, is another protein whose high expression is known to blunt the treatment effects of gemcitabine and cisplatin20. Ceppi's study20 showed that in a cohort of 156 NSCLC patients, not only did RRM1 and ERCC1 expression levels exhibit a substantial degree of correlation to each other (Pearson's Rho = 0.624, P < 1 × 10−4), their combined levels of expression were inversely correlated to patient survival time when treated with gemcitabine and/or cisplatin in that treated patients with low expression survived an average of 14.9 months vs. 10.0 months (P = 0.0345) for those with high expression20. Between the two genes, ERCC1 seemed to hold more predictive power than RRM1 in that the difference in post treatment survival time between high expressers and low expressers was markedly greater when comparing ERCC1 (17.3 months in low expressers vs. 10.9 months in high expressers, P = 0.0032) than when comparing RRM1 (13.9 months vs. 10.9 months, P = 0.039)20. Furthermore, of the two drugs, cisplatin seemed to be more severely impacted by ERCC1 expression levels as the difference in post treatment survival between high expressers and low expressers was 23.0 months vs. 12.4 months (P = 1.0 × 10−4)20. In addition to these, expression of several oncogenes as well as the tumor suppressor gene P53 and topoisomerase II alpha (TOP2A) may also affect cisplatin activity16,21. These findings suggest that quantification of the mRNA of certain genes such as RRM1 and ERCC1 to determine their expression levels could yield valuable insight to developing a personalized treatment plan when considering DNA-replication targeting anti-tumor drugs such as gemcitabine and cisplatin. Other proteins that are involved with DNA replication that also play a role in treatment resistance include DNA polymerase kappa (encoded by the POLK gene) and ubiquitin-conjugating enzyme E2B (encoded by the UBE2B gene). While POLK is involved in creating a replication complex that can bypass the crosslinking disruption caused by cisplatin binding, UBE2B facilitates DNA synthesis across lesions triggered by DNA damage response22, 23, 24. Compromised mechanisms of uptake can also lead to individuals exhibiting resistance to treatment. Copper transport protein CTR1 (encoded by the SLC31A1 gene) is necessary for the uptake of cisplatin and its disruption or deficiency may lead to phenotypic resistance to cisplatin due to metabolic inability25. Furthermore, transmembrane protein 205 (encoded by the TMEM205 gene) and associated vesicular trafficking RAS-related protein 8 (encoded by the RAB8 gene) were demonstrated by Shen et al.26 to confer cisplatin resistance during overexpression26. Although the precise mechanism is unconfirmed, it is hypothesized that these proteins play a role in trafficking and localization of cisplatin. It was also found that certain cell survival/replication pathways such as nuclear factor kappa B (NF-κB) and AKT/PI3K can defend tumor cells from entering apoptosis, causing resistance in some cases to gemcitabine27,28.
Ifosfamide is another alkylating agent which was approved roughly three decades ago as a compound that is useful in treating NSCLC. As a prodrug that requires processing into its active form by cytochrome P450 enzymes, the mechanism of action is complex and not well-understood29. A study by Wang et al.30 found that in hepatoma-affected mice, ifosfamide treatment greatly caused a reduction in thioredoxin reductase, an enzyme that is often hyper-expressed in malignant tumor cells. Although the exact role(s) and mechanism(s) of the thioredoxin family are poorly-understood, the proteins are believed to be involved in defense of DNA against reactive oxygen species induced damage and are critically important in development as mouse knock-out models show embryonic lethality31. The study by Wang et al.30 found that ifosfamide reduced thioredoxin activity by 62% in solid tumors but most interestingly, those treated cells could no longer form new tumors when injected into other mice, suggesting a permanent or lasting alteration. Although the evidence shown would seem to suggest that ifosfamide achieves its anti-cancer effects as an antagonist to the critical thioredoxin pathway, the group never tested the effectiveness of ifosfamide when treating mouse models that are either externally-supplemented with thioredoxin reductase or with the TXNRD gene in hyper-expression to determine whether Ifosfamide achieved its effects dependent on antagonizing thioredoxin reductase or if the down-regulation is an unrelated effect. From the evidence present, there are at least two genetic factors to account for when considering ifosfamide as treatment for NSCLC: patient levels of P450 enzymes as well as natural expression levels of thioredoxin reductase. Judging the effectiveness and determining the dosage of ifosfamide usage in patients from a personalized medicine point of view can be significantly complicated due to the relative lack of precise pharmacogenomic information applicable to this drug.
An interesting chemotherapy that is largely physiologically regulated is cyclophosphamide. This nitrogen mustard alkylating agent is administered as a prodrug which is metabolized into its active form, phosphoramide mustard, which then crosslinks DNA strands at the guanine N7 positions in order to disrupt DNA replication32. However, this process can be disrupted by an enzyme called aldehyde dehydrogenase (encoded by the ALDH gene), by converting cyclophosphamide into carboxycyclophosphamide, which loses its ability to convert into active phosphoramide mustard33,34. Because this process only occurs in cells with low expression levels of ALDH, cyclophosphamide is both limited in use and low in overall toxicity as many of the traditional tissues that are vulnerable to the side-effects of chemotherapy exhibit high expression of ALDH. Therefore, patients presenting with naturally high expression of the ALDH gene will exhibit resistance to treatment with cyclophosphamide33,34. Similarly, it is known that due to the ability of glutathione S-transferase (GST) to facilitate the conjugation of glutathione with alkylating agents to inactivate them, it may facilitate resistance against all alkylating agents in individuals with hyperexpression of the enzyme and/or high levels of glutathione35. However, although dependent on biosynthesis pathways within the body, glutathione can also be influenced by dietary intake of its critical building blocks such as cysteine, glycine, alpha lipoic acid, vitamin D3, etc36.
3. Topoisomerase inhibitors
In the non-oncogene-targeting chemotherapy field, botanically-derived compounds have played a significant role. One of the most impactful groups is the topoisomerase I inhibitors. Camptothecin is a cytotoxic alkaloid derived from the stem of the Camptotheca acuminate tree, which gave rise to the drug irinotecan37. Taken as a prodrug, irinotecan is dependent on carboxylesterase for hydrolysis into its active metabolite form, SN-38. SN-38 attaches to the guanine base pairs surrounding the topoisomerase I cleavage complex and inactivates the topoisomerase I enzyme complex37,38. Finally, SN-38 is glucuronidated by UDP-glucuronosyltransferase 1 family, polypeptide A1 (UGT1A1) during inactivation37,38. It is commonly used in combination with cisplatin specifically for the treatment of NSCLC. Although little is known regarding the mechanisms of resistance to Irinotecan specifically and directly in regard to its treatment of NSCLC patients, several have been identified during the treatment of other types of cancer which may bear consequence to NSCLC patients as well. Metallothioneins (normally critical for metal storage and trafficking) and epidermal growth factor receptor (encoded by the EGFR gene) expression (both independently and through the Src signaling pathway) have been shown to interact with irinotecan in a self-inhibitory loop as their expression is upregulated by irinotecan and they seem to inhibit irinotecan activity in high concentrations39,40. The MAPK14/P38α pathway activates survival autophagy within the cell in response to Irinotecan to resist apoptosis41. Although the precise mechanism is unknown, it has been observed that overexpression of the heat shock protein 27 (encoded by the HSP27 gene) can cause resistance to irinotecan treatment, which can be reversed by antisense silencing of the HSP27 gene (in vitro)42. Clinically, patients with naturally higher expression of HSP27 tend to be non-responders to irinotecan treatment42. Finally, efflux pumps such as ATP-binding cassette subfamily (ABCC4 and ABCG2) have been shown to confer resistance to irinotecan in patients with high expression43,44.
An alternate semi-synthetic derivative of camptothecin is topotecan, which is commonly given with paclitaxel to treat advanced stage NSCLC. Topotecan binds the topoisomerase I enzyme complex to the lesion complex arresting it there and preventing the re-ligation of the DNA strand45, 46, 47, 48. The overabundance of arrested topoisomerase I complexes produces an apoptotic effect and eventually, their collision with the replication fork fragments the DNA45, 46, 47, 48. Known factors of genetic resistance to topotecan rely heavily on efflux pump expression. ATP-binding cassette subfamilies ABCB1, ABCC4, ABCG2 efflux pumps have all been implicated in topotecan resistance across multiple types of cancers treated but the resistance conferred by some can be countered by other drugs such as sitravatinib49, 50, 51, 52.
Etoposide is another chemotherapy approved for use in treating NSCLC but commonly presents with serious side-effects. This compound forms a bond with both the DNA and topoisomerase II enzyme complex, preventing re-ligation of the DNA and causing apoptosis-inducing breakage in a similar fashion to the mechanism of camptothecin-derived chemotherapies45,53. Resistance generally comes in two forms: either by acquired activation of cell survival factors or by efflux pumps. In patients with NSCLC, it was observed that acquired resistance to etoposide can be caused by upregulation of DNA polymerase beta (encoded by the POLB gene) which is responsible for DNA excision-based repair and neuroendocrine transcription factor NK2 homeobox 2 (encoded by the NKX2-2 gene), both of which reduce apoptotic response in cancer cells54. It was also seen in NSCLC cells that caveolae protein (encoded by the CAV1 gene) hyperexpression can promote the RAS–ERK cell cycle progression pathway, causing resistance to etoposide55. Perhaps one of the most interesting finds regarding etoposide resistance is that the R428W mutation in the P53 gene produces an aberrant protein that transcriptionally activates the tyrosyl-DNA phosphodiesterase 2 (TDP2) DNA repair gene, causing resistance to etoposide56. Efflux pump-induced resistance of etoposide is modulated by the ABCB1, major vault protein (MVP), and ABCG257, 58, 59 genes. Capicua transcriptional repressor 3 (CIC-3), a member of the CLC family, is an important component for drug sequestration as it regulates vesicular acidity; over-expression appears to correlate with etoposide resistance, possibly due to accelerated break-down of the compound through upregulating P-glycoprotein expression60. Hyperexpression of WNT5A and DKK1 in the WNT signaling pathway have also been correlated with etoposide resistance although curiously, WNT3A seems to correlate to sensitivity, which suggests that the regulation of cell cycle progression through the WNT pathway plays a role on the effects of etoposide61. The scientific picture suggested by these studies shows a rather complex and diverse web of mechanisms that are likely to produce a cumulative effect on etoposide resistance.
Another chemotherapy that targets topoisomerase II is doxorubicin, which is an anthracycline that intercalates DNA, freezing the topoisomerase II complex to prevent re-ligation and reformation of the double helix46,62,63. An additional effect that is unique to doxorubicin, however, is the occurrence of histone eviction from the duplicating chromatin, causing a myriad of issues for DNA repair genes that ultimately increase the final likelihood of apoptosis64. Common efflux pumps ABCB1, ABCB8 and ABCG2 (with a likely mutational hotspot discovered at R482 in ABCG2 that increases anthracycline efflux at the expense of Topotecan efflux), contribute to doxorubicin resistance though they may be surmountable by other chemotherapies like bafetinib65,66. Inactivating mutations in the PTEN tumor suppressor gene that drive AKT activation have been observed to cause resistance to doxorubicin although it may be reversed by treatment with berberine and/or rapamycin67,68.
4. Cytoskeleton-targeting
Due to the complexity of the mitotic process that cancerous growth depends upon, there are multiple treatment targets to exploit, and with them, possibilities for future development. In case of failure of or developed resistance against DNA-targeting therapy, cytoskeletal-targeting drugs present another opportunity. Traditionally targeting tubulin, these drugs are isolated from various plants and work by preventing the cell from completing/entering mitosis through undermining the preparations of the crucial cytoskeleton69. The pioneering drugs of the cytoskeleton-disrupting class are the vinca alkaloids derived from the Madagascan periwinkle (Catharanthus roseus). Vinblastine and vincristine are close chemical derivatives of each other differing in only one group (methyl for the former and aldehyde for the latter)69. Vinca alkaloids inhibit cellular proliferation by binding to tubulin and hindering the activities of the mitotic spindle in lower concentrations, though in higher concentrations, the entire microtubule can be destabilized70. As is typically seem in anti-cancer chemotherapies, they are metabolized by CYP3A4 and CYP3A5 with the former being generally more critical for vinblastine and the latter for vincristine71,72. The use of these vinca alkaloids for clinical treatment has been approved by the U.S. department of agriculture (USDA) as far back as 1963 due to their ability to prolong life in cancer patients however, they quickly evolved for use primarily in combination therapies (often with DNA-replication inhibitors) for NSCLC patients73, 74, 75. Of the issues that have arisen from the use of vincristine and vinblastine, among the most prominent are, unsurprisingly, cases of drug resistance and excess toxicity in patients who are genetically lacking in their the CYP3A family enzymes76, 77, 78, 79, 80. Reports of CYP3A5-deficient Japanese, Kenyan and Turkish patients suffering symptoms of excess drug toxicity from standard dosages of vincristine underline the importance of pharmacogenomic considerations even when using chemotherapies with universal targets and the rate of occurrence in various populations of patients highlights the need for population studies to improve the precision of physician guidelines77,78. Interestingly, the overexpression of HSP27 is linked to vincristine resistance79,80. HSP27 is known to inhibit caspase activation, which not only modulates apoptotic pathways but also tubulin cleavage81,82. In addition, the HSP27 protein has been shown to associate directly with tubulin and microtubules, strongly suggesting that the exact mechanism of HSP27-mediated resistance is likely to be universal to all cancer types83. In many cases where the cause of resistance is not understood, the mechanisms to circumvent resistance commonly default on the use of chemicals to aid the mechanisms of the vinca alkaloids or to increase their cytotoxicity, in essence striking a blow to the patient's resistance mechanisms rather than bypassing them84,85. However, when the gene is identified, anti-strand oligonucleotides may hold value in their ability to down-regulate or shut off the gene. Additional causes of resistance to the microtubule-destabilizing vinca alkaloids can naturally be seen in genes that modulate microtubule formation. Microtubule-associated protein 4 (encoded by the MAP4 gene) is a microtubule stabilizing protein and variation in its expression levels have been shown to correlate inversely with sensitivity to general vinca alkaloid treatment in different cell lines86. On the other hand, mutations in the β-tubulin (TUBB) gene (such as L240I) have been demonstrated specific resistance to vincristine86. Interestingly, WNT5A has shown that it can modulate vincristine sensitivity in much the same pathway as it did Etoposide although with the inclusion of glycogen synthase (encoded by the GSK3β gene)87. And finally, vincristine and vinblastine are also affected by efflux pumps ABCC1 and ABCB1, respectively but new agents like erdafitinib can ameliorate the resistance conferred88,89.
Taxoids are another family of botanical tubulin-targeting compounds which consist of paclitaxel and docetaxel. Derived from the bark of the Pacific Yew tree (Taxus brevifolia) and approved by the USDA for chemotherapy in 1993, paclitaxel targets tubulin just like the vinca alkaloids but unlike vincristine and vinblastine which destabilize and deconstruct the microtubule, the taxoids bind to microtubules to induce a hyper-stable conformation preventing them from forming the mitotic spindle needed during metaphase69. An advantage of paclitaxel is that in addition to being metabolized by the CYP3A family of enzymes like the great majority of the compounds mentioned here, it can also be metabolized by the CYP2C8 enzyme, reducing metabolic stress when used in combination therapy with other drugs, (which often rely solely on the function of CYP3A family)90. Because paclitaxel tends to result in modest improvement and low response rate, it is usually used in combination with other treatments91,92. Resistance mechanisms for paclitaxel vary. It seems that several mutations in the TUBB gene are responsible for paclitaxel irresponsiveness most likely due to alterations of the binding site/binding pocket that diminish binding affinity, however, in addition to these mutations, it has been shown that just as in DNA-replication targeted therapies, high expression of β-tubulin also leads to low response rate93, 94, 95. Specific mutations in the TUBB gene such as A185T, A248V, R306C confer resistance to paclitaxel through naturally low microtubule stability96. To add to this, mitotic centromere-associated kinesin (encoded by the MCAK gene) is a microtubule depolymerizing protein that is known to confer paclitaxel resistance when abundantly expressed97,98. Tumor suppressor Parkin confers sensitivity to the microtubule hyper-stabilizing paclitaxel via TUBB and MAP4, previously shown to increase resistance to microtubule destabilizing vinca alkaloids99. In addition to the expected interactions with efflux pumps (in this case ABCB1, ABCB4, ABCC2, and ABCC3), paclitaxel resistance was also seen in scenarios of increased expression of the Bcl-2 apoptotic regulator gene100, 101, 102. These studies suggest that sequencing for possible conformational mutations in tubulin and mRNA quantitative analysis of β-tubulin expression can be an effective way of choosing whether to use a vinca alkaloid or a taxoid like paclitaxel when treating cancer patients.
Docetaxel is a European Yew (Taxus baccata) isolated derivative of paclitaxel which enters the cell more readily and exhibits a greater retention time than Paclitaxel, allowing the dosage to be decreased103. Intracellularly, docetaxel binds reversibly to tubulin to facilitate hyper-stabilized microtubules that cannot disperse, resulting in apoptosis-inducing accumulation in the cytoplasm103,104. Because clinical use of docetaxel to treat NSCLC generally yields very low response rate, docetaxel is preferably utilized in combination therapy when used to treat NSCLC105. However, because metabolism depends on CYP3A4/5, careful consideration is required in drug pairing103,104. When combined with other chemotherapies such as cisplatin, the response rate would substantially (from ∼30% to ∼70%) improve in NSCLC patients as compared to docetaxel treatment alone106. Much of docetaxel's functions also depend on resistance mechanisms such as expression levels of drug efflux pumps (ABCB1, ABCC10 which are surmountable by ibrutinib), kinesins, and tumor suppressor gene P27107, 108, 109, 110, 111. While overexpression of the ABCB1 gene and kinesins are known to be detrimental to docetaxel efficacy, the inverse is true for PTEN (mechanism through P27)108, 109, 110. However, since P27 gene expression fluctuations in both directions have been reported to correlate with resistance to several chemotherapies without obvious connections to each other, the existence of a true causative relationship is questionable. As a direct challenge to the targeted molecular effects of taxoids, the forkhead box M1 protein (encoded by the FOXM1 gene) is capable of activating stathmin (encoded by the STMN1 gene), a protein whose role is destabilization of the microtubules, and as such, capable of conferring resistance to both docetaxel and paclitaxel112,113. As one would expect that due the universal mechanism of the taxoids, the resistance factors should be very similar as well, but surprisingly, the resistance mechanisms seen in docetaxel seem to be more widespread and scattered. Multiple outstanding factors have been identified in docetaxel resistance both in acquired and natural form. The NF-κB survival pathway as well as the interleukin 6 (encoded by the IL-6 gene) inflammatory regulators can be activated by docetaxel treatment and interfere with successful chemotherapy114. Hyper-expression of both the ribophorin II (encoded by the RPN2 gene) (role in peptide synthesis) and malate dehydrogenase 2 (encoded by the MDH2 gene) (role in citric acid cycle regulation) have shown to correlate to docetaxel resistance by unknown mechanisms115,116. Even symptoms as broad as aberrant methylation patterns which are commonly seen in cancer tissues may adversely affect docetaxel treatment success117. Extraordinarily, high levels of thioredoxins exhibited pretreatment can be correlated to docetaxel resistance, however, increases in response to chemotherapy do not yield the same118,119. This stresses the importance of establishing causation rather than accepting correlation when determining mechanisms of resistance as in this case, it is very likely that thioredoxin activity itself does not hinder the mechanism of docetaxel. Rather, it is some other factor possibly upstream that correlates to initially higher levels of thioredoxin but unlike thioredoxin, is not affected by docetaxel treatment.
5. Discussion
Although the application of personalized medicine to NSCLC has traditionally been focused on therapies designed to engage specific oncogenic mutations, pharmacogenomic considerations are not lost on other therapies that target the common themes of all cancers. However, unlike chemotherapies targeting specific driver mutations where most of the pharmacogenomic considerations go into whether a susceptible mutation is present for targeting, these non-oncogene-targeting chemotherapies are far more universal in their use. Therefore, most of the pharmacogenomic considerations that pertain to them will be avoidance of resistance factors that make them less effective or ineffective. Every patient is a unique genetic microenvironment with a multitude of (undiscovered) genetic factors which determine how quickly various drugs can be up-taken/metabolized, and how well they can carry out their intended effects once/if they reach their targets, and the rate at which they are disposed of/inactivated (Table 1). One of the most serious and lasting challenges to treatment is the complexity of natural/genetic resistance within a patient that can cause the individual to be especially resilient to the mechanisms of certain drugs. This unique personal genetic web that encompasses the tumor microenvironment is made of many components such as angiogenic factors, the various cells of the tumor stroma, immunological/inflammatory factors, etc... Forces in the tumor microenvironment that either feed tumor survival/replication or prevent the normal host tumor suppression mechanisms from their duties present quantitative challenges to all treatments. All of these factors come together to filter through the available chemotherapy options that are appropriate for treating each patient (Fig. 1). Qualitatively, however, there remains much to be explored in terms of tailoring personalized medicine regimens to unique tumor microenvironments depending on individual genetic factors120. Because these chemotherapies are not specific to oncogenic targets or their ATP-binding sites, they are naturally not susceptible to resistance from gatekeeper mutations and other changes that affect ATP binding site affinity. Therefore, the generic approach to resistance circumvention by conformationally altering the drug is usually not useful. Rather, most methods of resistance circumvention rely on employment of a second chemical, usually to block a host mechanism and/or allow the chemotherapy to increase its cytotoxicity84,85. Since efflux pumps/cassettes are often the culprit, another possibility worthy of investigation is whether changing an efflux-pump binding site (assuming it is not a critical functional group) can help them circumvent this method of resistance. Great attention and further research are needed to gain a better understanding of the pharmacogenomic factors at play to aid physicians in making more accurate predictions on the effects of certain treatments to individual patients. Further studies on additional genes and pathways that moderate the multiple components of the DNA replication machinery and cytoskeletal function/production should enhance our ability to accurately predict the treatment values of DNA replication inhibitors and cytoskeletal function inhibitors. Higher resolution understanding of proteins is also critical to decision-making. Rather than clumping proteins as those related to microtubule assembly, categorizing them by role of microtubule stabilization or destabilization can help facilitate the decision to use vinca alkyloids or taxoids for patients with different expression profiles. Microtubule destabilizers like vinca alkyloids are expected to be less effective in patients with genetic factors that support greater microtubule stability (as they must first overcome the natural hyperstability before becoming effective) and more effective in patients with lower microtubule instability (as they add to the natural instability). The opposite would be expected to be true for toxoids; however, the genes to be analyzed and genetic variations to be interpreted to determine the collective level of microtubule stability would present a far more difficult challenge to our current understanding of genetics. Furthermore, caution must be exercised when assuming causation from correlation as there is a significant amount of literature making claims that the upregulation/downregulation of a certain gene confers resistance to some chemotherapy but through an unknown mechanism. Oftentimes, certain genes can be correlated with resistance to multiple (possibly unrelated) chemotherapies, but unless the mechanism is established, we should not assume a causative relationship as changes or differences in the gene's expression can be due to upstream effects that are indirectly caused by the chemotherapy. A cumulative database of the genes known to cause resistance to chemotherapies, and their mechanisms of resistance would be a potential crown jewel to personalized treatments for NSCLC (and likely other cancers as well) (Table 2).
Table 1.
Pharmacogenomics of non-oncogene-targeting chemotherapies of non-small cell lung cancer.
| Drug name | Drug class | Metabolic pathway | Mechanism of action | Resistance |
|---|---|---|---|---|
| Cisplatin | Alkylating agent | Cysteine S-conjugate β-lyase | Binds tN7 reactive center on purine to crosslink DNA, creating lesions that trigger apoptotic pathway | ERCC1, RRM1, POLK, UBE2B, RAB8, SLC31A1, TMEM205, TOP2A, RPN2 |
| Ifosfamide | Alkylating agent | P450 family | (?) Reduction of thioredoxin reductase, likely weakening defense against ROS damage | TXNRD family |
| Cyclophosphamide | Alkylating agent | CYP2B6, CYP2C9 and CYP3A4 | Crosslinks DNA strands at the guanine N7 positions to disrupt DNA replication | ALDH, GST |
| Irinotecan | Topoisomerase I inhibitor | Carboxylesterase, UGT1A1, CYP3A4, CYP3A5 | Attaches to the guanine base pairs surrounding the topoisomerase I cleavage complex and inactivates topoisomerase I enzyme complex | Metallothioneins, EGFR, MAPK14/P38α, HSP27, ABCC4, ABCG2 |
| Topotecan | Topoisomerase I inhibitor | P450 family | Binds topoisomerase I enzyme complex to the lesion complex preventing the re-ligation of the DNA strand; overabundance of arrested topoisomerase I complexes produces apoptotic effect and collision with the replication fork fragments the DNA | ABCB1, ABCC4, ABCG2 |
| Gemcitabine | Topoisomerase I inhibitor | Cytidine deaminase | Incorporates into DNA in place of dNTP and stalls replication; DNA proofreading enzymes are unable to remove locked topoisomerase I from the replication complex | ERCC1, RRM1, NF-κB, AKT/PI3K |
| Etoposide | Topoisomerase II inhibitor | CYP3A4, CYP3A5, CYP1A2 | Bonds DNA and topoisomerase II enzyme complex, preventing re-ligation of the DNA and causing apoptosis-inducing breakage | POLB, NKX2.2, CAV1, P53:p.R428W, TDP2, ABCB1, MVP, ABCG2, CIC-3, WNT5A, DKK1 |
| Doxorubicin | Topoisomerase II inhibitor | CYP3A4, CYP2B6, CYP1B1 | Intercalates DNA, freezing topoisomerase II complex to prevent re-ligation and reformation of double helix; causes histone eviction | ABCB1, ABCB8, ABCG2, PTEN, MVP, AKT |
| Vincristine | Microtubule destabilizer | CYP3A4, CYP3A5 | Binds tubulin to prevent dimerization into microtubules; caused microtubule detachment | HSP27, MAP4, TUBB:p.L240I, WNT5A, GSK3β, ABCC1, ABCB1, MVP |
| Vinblastine | Microtubule destabilizer | CYP3A4, CYP3A5 | Binds tubulin to prevent dimerization into microtubules; caused microtubule detachment | MAP4, ABCC1, ABCB1 |
| Paclitaxel | Microtubule hyperstabilizer | CYP2C8, CYP3A4 | Binds tubulin, preventing disassembly; causes apoptosis by stopping mitosis and causing microtubule accumulation | TUBB:p.A185T, A248V, R306C, Kinesins family, ABCB1, ABCC3, ABCB4, ABCC2, BCL-2, MVP |
| Docetaxel | Microtubule hyperstabilizer | CYP3A4, CYP3A5 | Binds tubulin, preventing disassembly; causes apoptosis by stopping mitosis and causing microtubule accumulation | ABCB1, ABCC10, kinesins, P27, FOXM1, STMN1, NF-κB, IL-6, RPN2, MDH2, MVP, PTEN:p.Y68fs |
Summary of drug class, mechanism description, metabolic pathway, and genes/mutations to consider when determining usage and/or dose.
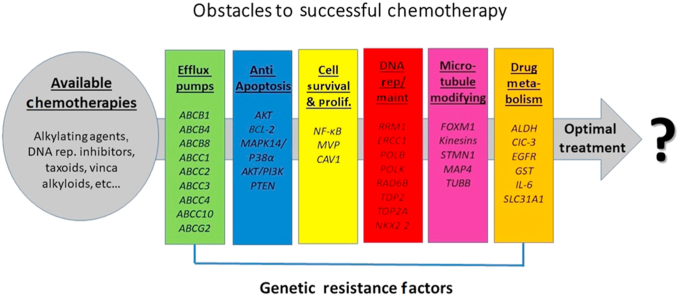
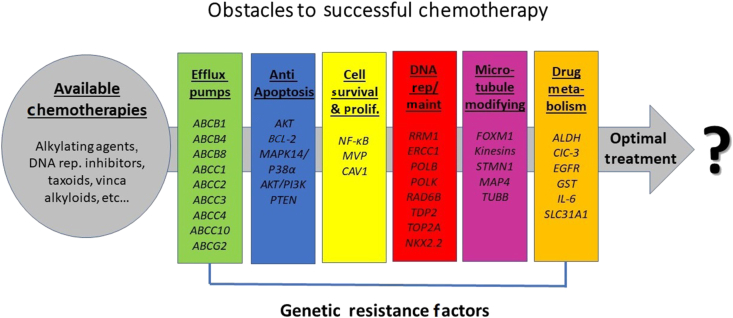
Figure 1.
The types of genetic resistance factors within each patient and the available chemotherapies choices at the physician's disposal that are appropriate for use.
Table 2.
Mechanisms of resistance against non-oncogene-targeting chemotherapies.
| Gene | Mechanism of resistance | Affected drug |
|---|---|---|
| ABCB1 | Efflux pump | Topotecan, etoposide, vincristine, vinblastine, paclitaxel, docetaxel, doxorubicin |
| ABCB4 | Efflux pump | Paclitaxel |
| ABCB8 | Efflux pump | Doxorubicin |
| ABCC1 | Efflux pump | Vincristine, vinblastine |
| ABCC2 | Efflux pump | Paclitaxel |
| ABCC3 | Efflux pump | Paclitaxel |
| ABCC4 | Efflux pump | Irinotecan, topotecan |
| ABCC10 | Efflux pump | Docetaxel |
| ABCG2 | Efflux pump | Irinotecan, topotecan, etoposide, doxorubicin |
| GSK3β | Down-regulates efflux pump ABCB1 | Vincristine |
| WNT5A | Down-regulates efflux pump ABCB1 | Etoposide, vincristine |
| AKT | Apoptosis disruption | Doxorubicin |
| BCL-2 | Apoptosis disruption | Paclitaxel |
| MAPK14/P38α | Apoptosis disruption | Irinotecan |
| AKT/PI3K | Apoptosis disruption | Gemcitibine |
| PTEN | Apoptosis disruption through AKT | Doxorubicin, docetaxel |
| NF-κB | Promotes cell survival/growth/proliferation | Gemcitibine, docetaxel |
| MVP | Indirectly promotes cell survival/growth/proliferation | Etoposide, doxorubicin, vincristine, paclitaxel, docetaxel |
| CAV1 | Indirect cell survival/growth/proliferation | Etoposide |
| RRM1 | DNA replication | Cisplatin, gemcitibine |
| ERCC1 | DNA replication/repair/maintenance | Cisplatin, gemcitibine |
| POLB | DNA replication/repair/maintenance | Etoposide |
| POLK | DNA replication/repair/maintenance | Cisplatin |
| RAD6B | DNA replication/repair/maintenance | Cisplatin |
| TDP2 | DNA replication/repair/maintenance | Etoposide |
| TOP2A | DNA replication/repair/maintenance | Cisplatin |
| NKX2.2 | DNA replication/repair/maintenance | Etoposide |
| FOXM1 | Microtubule destabilizer | Docetaxel |
| Kinesins | Microtubule destabilizer | Paclitaxel, docetaxel |
| STMN1 | Microtubule destabilizer | Docetaxel |
| MAP4 | Microtubule stabilizer | Vincristine, vinblastine |
| TUBB | Microtubule structure | Vincristine, paclitaxel |
| ALDH | Drug metabolism disruption/inactivation | Cyclophosphamide |
| CIC-3 | Drug metabolism disruption/inactivation | Etoposide |
| EGFR | Drug metabolism disruption/inactivation | Irinotecan |
| GST | Drug metabolism disruption/inactivation | Cyclophosphamide |
| IL-6 | Drug metabolism disruption/inactivation? | Docetaxel |
| SLC31A1 | Copper transport/drug uptake | Cisplatin |
The main mechanisms of drug resistance to non-oncogene-targeting chemotherapies are efflux pump removal, disruption of apoptosis/promoting cellular survival/replication, repair/maintenance of DNA, microtubule stability adjustment, and disruption or inactivation of the drug.
Furthermore, we must realize that although personalized medicine is an invaluable tool for enhancing precision health care, the ability to obtain detailed genetic information on a personal basis is a privilege that is often not available to many. In these circumstances, rather than defaulting to blind treatment, it would be greatly beneficial to have population data to guide physicians in making educated decisions on the compounds that are more likely to be effective based on the patient's ethnicity/area of ancestry11,76,77. To create such a library of common genetic profiles (for known treatment-modifying genes) based on ethnic background requires extensive data-collection and analysis on a large-scale effort that would undoubtedly be extremely costly in time and resources. Nonetheless, the benefits afforded to those around the world who don't readily have access to genetic testing would be difficult to overstate.
Despite the successes in the implementation of personalized medicine on patients afflicted with NSCLC, our knowledge on the pharmacogenomics pertaining to drugs without specific oncogenic targets still needs improvement. However, due to the prominence of NSCLC as a global health issue, there is a great deal of effort dedicated to advancing our knowledge of its treatment, which corresponds to the ever-increasing quality and precision of therapies available. Thus, although there is no end in sight to the quest to understand how unique human physiologies best interact with different chemotherapy compounds, it is a struggle than modern medicine cannot afford to shy away from.
Acknowledgments
Special thanks to the Scientific Publications Department of the Research Medical Library of The University of Texas MD Anderson Cancer Center for manuscript editing.
Author contributions
Wenxiao Jiang drafted the manuscript. Guiqing Cai was genetics-related consultant. Peter Hu was cancer-related consultant. Yue Wang was responsible for final editing of this manuscript.
Conflicts of interest
The statement of conflicts of interest is attached.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
References
- 1.Barta J.A., Powell C.A., Wisnivesky J.P. Global epidemiology of lung cancer. Ann Glob Health. 2019;85:1. doi: 10.5334/aogh.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre L.A., Siegel R.L., Jemal A. In: Ahmad A., Gadgeel S., editors. vol. 893. Springer; Cham: 2016. Lung cancer statistics; pp. 1–19. (Lung cancer and personalized medicine. Advances in experimental medicine and biology). [DOI] [PubMed] [Google Scholar]
- 3.Azzoli C.G., Baker S., Jr., Temin S., Pao W., Aliff T., Brahmer J., et al. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27:6251–6266. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiller J.H., Harrington D., Belani C.P., Langer C., Sandler A., Krook J., et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 5.Cheng L., Alexander R.E., MacLennan G.T., Cummings O.W., Montironi R., Lopez-Beltran A., et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25:347–369. doi: 10.1038/modpathol.2011.215. [DOI] [PubMed] [Google Scholar]
- 6.Chin L., Andersen J.N., Futreal P.A. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 7.Kerr K.M. Personalized medicine for lung cancer: new challenges for pathology. Histopathology. 2012;60:531–546. doi: 10.1111/j.1365-2559.2011.03854.x. [DOI] [PubMed] [Google Scholar]
- 8.Pirker R., Filipits M. Personalized treatment of advanced non-small-cell lung cancer in routine clinical practice. Cancer Metastasis Rev. 2016;35:141–150. doi: 10.1007/s10555-016-9612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W., Cai G., Hu P.C., Wang Y. Personalized medicine in non-small cell lung cancer: a review from a pharmacogenomics perspective. Acta Pharm Sin B. 2018;8:530–538. doi: 10.1016/j.apsb.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 11.Gorre M.E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P.N., et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 12.Crino L., Scagliotti G., Marangolo M., Figoli F., Clerici M., De Marinis F., et al. Cisplatin-gemcitabine combination in advanced non-small-cell lung cancer: a phase II study. J Clin Oncol. 1997;15:297–303. doi: 10.1200/JCO.1997.15.1.297. [DOI] [PubMed] [Google Scholar]
- 13.Artin E., Wang J., Lohman G.J., Yokoyama K., Yu G., Griffin R.G., et al. Insight into the mechanism of inactivation of ribonucleotide reductase by gemcitabine 5′-diphosphate in the presence or absence of reductant. Biochemistry. 2009;48:11622–11629. doi: 10.1021/bi901590q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Hanigan M.H. Role of cysteine S-conjugate β-lyase in the metabolism of cisplatin. J Pharmacol Exp Therapeut. 2003;306:988–994. doi: 10.1124/jpet.103.052225. [DOI] [PubMed] [Google Scholar]
- 15.de Sousa Cavalcante L., Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 16.Aldossary S.A. Review on pharmacology of cisplatin: clinical use, toxicity and mechanism of resistance of cisplatin. Biomed Pharmacol J. 2019;12:7–15. [Google Scholar]
- 17.Cheng L., Li C., Xi Z., Wei K., Yuan S., Arnesano F., et al. Cisplatin reacts with histone H1 and the adduct forms a ternary complex with DNA. Metallomics. 2019;11:556–564. doi: 10.1039/c8mt00358k. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira L., Caquito J.M., Jr., Rocha M.S. Carboplatin as an alternative to cisplatin in chemotherapies: new insights at single molecule level. Biophys Chem. 2018;241:8–14. doi: 10.1016/j.bpc.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Pourquier P., Gioffre C., Kohlhagen G., Urasaki Y., Goldwasser F., Hertel L.W., et al. Gemcitabine (2′,2′-difluoro-2′-deoxycytidine), an antimetabolite that poisons topoisomerase I. Clin Cancer Res. 2002;8:2499–2504. [PubMed] [Google Scholar]
- 20.Ceppi P., Volante M., Novello S., Rapa I., Danenberg K.D., Danenberg P.V., et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 21.Grenda A., Błach J., Szczyrek M., Krawczyk P., Nicoś M., Kuźnar Kamińska B., et al. Promoter polymorphisms of TOP2A and ERCC1 genes as predictive factors for chemotherapy in non-small cell lung cancer patients. Cancer Med. 2020;9:605–614. doi: 10.1002/cam4.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jha V., Ling H. Structural basis for human DNA polymerase kappa to bypass cisplatin intrastrand cross-link (Pt-GG) lesion as an efficient and accurate extender. J Mol Biol. 2018;430:1577–1589. doi: 10.1016/j.jmb.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Haynes B., Gajan A., Nangia-Makker P., Shekhar M.P. RAD6B is a major mediator of triple negative breast cancer cisplatin resistance: regulation of translesion synthesis/Fanconi anemia crosstalk and BRCA1 independence. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165561. doi: 10.1016/j.bbadis.2019.165561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders M.A., Haynes B., Nangia-Makker P., Polin L.A., Shekhar M.P. Pharmacological targeting of RAD6 enzyme-mediated translesion synthesis overcomes resistance to platinum-based drugs. J Biol Chem. 2017;292:10347–10363. doi: 10.1074/jbc.M117.792192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghaffari R., Richburg J.H. Mice with a Sertoli cell-specific knockout of the Ctr1 gene exhibit a reduced sensitivity to cisplatin-induced testicular germ cell apoptosis. Toxicol Res. 2019;8:972–978. doi: 10.1039/c9tx00142e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen D.W., Pouliot L.M., Hall M.D., Gottesman M.M. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev. 2012;64:706–721. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu C., Chen S., Guo Y., Sun C. Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-κB signaling pathway. Theranostics. 2018;8:3224. doi: 10.7150/thno.23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebrahimi S., Hosseini M., Shahidsales S., Maftouh M., Ferns G.A., Ghayour-Mobarhan M., et al. Targeting the Akt/PI3K signaling pathway as a potential therapeutic strategy for the treatment of pancreatic cancer. Curr Med Chem. 2017;24:1321–1331. doi: 10.2174/0929867324666170206142658. [DOI] [PubMed] [Google Scholar]
- 29.Torres Espindola L.M., Rojo-Serrato D., Álvaro-Heredia A., Castillejos López M.D., de Uña-Flores A., Pérez-García M., et al. Analysis of CYP450 gene allelic variants can predict ifosfamide toxicity in Mexican paediatric patients. Biomarkers. 2020;25:331–340. doi: 10.1080/1354750X.2020.1754913. [DOI] [PubMed] [Google Scholar]
- 30.Wang X., Zhang J., Xu T. Thioredoxin reductase inactivation as a pivotal mechanism of ifosfamide in cancer therapy. Eur J Pharmacol. 2008;579:66–73. doi: 10.1016/j.ejphar.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Lillig C.H., Holmgren A. Thioredoxin and related molecules—from biology to health and disease. Antioxidants Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 32.More G.S., Thomas A.B., Chitlange S.S., Nanda R.K., Gajbhiye R.L. Nitrogen mustards as alkylating agents: a review on chemistry, mechanism of action and current USFDA status of drugs. Anticancer Agents Med Chem. 2019;19:1080–1102. doi: 10.2174/1871520619666190305141458. (Formerly Curr Med Chem Anticancer Agents) [DOI] [PubMed] [Google Scholar]
- 33.Dinavahi S.S., Bazewicz C.G., Gowda R., Robertson G.P. Aldehyde dehydrogenase inhibitors for cancer therapeutics. Trends Pharmacol Sci. 2019;40:774–789. doi: 10.1016/j.tips.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Kanakry C.G., Ganguly S., Zahurak M., Bolaños-Meade J., Thoburn C., Perkins B., et al. Aldehyde dehydrogenase expression drives human regulatory T cell resistance to posttransplantation cyclophosphamide. Sci Transl Med. 2013;5:211ra157. doi: 10.1126/scitranslmed.3006960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pljesa-Ercegovac M., Savic-Radojevic A., Matic M., Coric V., Djukic T., Radic T., et al. Glutathione transferases: potential targets to overcome chemoresistance in solid tumors. Int J Mol Sci. 2018;19:3785. doi: 10.3390/ijms19123785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Zhou X., Wu W., Wang J., Xie H., Wu Z. Regeneration of glutathione by α-lipoic acid via Nrf2/ARE signaling pathway alleviates cadmium-induced HepG2 cell toxicity. Environ Toxicol Pharmacol. 2017;51:30–37. doi: 10.1016/j.etap.2017.02.022. [DOI] [PubMed] [Google Scholar]
- 37.Femke M., Goey A.K., van Schaik R.H., Mathijssen R.H., Bins S. Individualization of irinotecan treatment: a review of pharmacokinetics, pharmacodynamics, and pharmacogenetics. Clin Pharmacokinet. 2018;57:1229–1254. doi: 10.1007/s40262-018-0644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigo M.A., Jimemez A.M., Haddad Y., Bodoor K., Adam P., Krizkova S., et al. Metallothionein isoforms as double agents—their roles in carcinogenesis, cancer progression and chemoresistance. Drug Resist Updates. 2020:100691. doi: 10.1016/j.drup.2020.100691. [DOI] [PubMed] [Google Scholar]
- 40.Petitprez A., Larsen A.K. Irinotecan resistance is accompanied by upregulation of EGFR and Src signaling in human cancer models. Curr Pharmaceut Des. 2013;19:958–964. [PubMed] [Google Scholar]
- 41.Paillas S., Causse A., Marzi L., De Medina P., Poirot M., Denis V., et al. MAPK14/p38α confers irinotecan resistance to TP53-defective cells by inducing survival autophagy. Autophagy. 2012;8:1098–1112. doi: 10.4161/auto.20268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poorebrahim M., Sadeghi S., Ghanbarian M., Kalhor H., Mehrtash A., Teimoori-Toolabi L. Identification of candidate genes and miRNAs for sensitizing resistant colorectal cancer cells to oxaliplatin and irinotecan. Cancer Chemother Pharmacol. 2020;85:153–171. doi: 10.1007/s00280-019-03975-3. [DOI] [PubMed] [Google Scholar]
- 43.Tuy H.D., Shiomi H., Mukaisho K.I., Naka S., Shimizu T., Sonoda H., et al. ABCG2 expression in colorectal adenocarcinomas may predict resistance to irinotecan. Oncol Lett. 2016;12:2752–2760. doi: 10.3892/ol.2016.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berthier J., Arnion H., Saint-Marcoux F., Picard N. Multidrug resistance-associated protein 4 in pharmacology: overview of its contribution to pharmacokinetics, pharmacodynamics and pharmacogenetics. Life Sci. 2019;231:116540. doi: 10.1016/j.lfs.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Pommier Y., Leo E., Zhang H., Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosen V.M., Guerra I., McCormack M., Nogueira-Rodrigues A., Sasse A., Munk V.C., et al. Systematic review and network meta-analysis of bevacizumab plus first-line topotecan-paclitaxel or cisplatin-paclitaxel versus non-bevacizumab-containing therapies in persistent, recurrent, or metastatic cervical cancer. Int J Gynecol Cancer. 2017;27:1237–1246. doi: 10.1097/IGC.0000000000001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lv C., Liu X., Zheng Q., Chen H., Yang X., Zhong J., et al. Analysis of topoisomerase I expression and identification of predictive markers for efficacy of topotecan chemotherapy in small cell lung cancer. Thorac Cancer. 2018;9:1166–1173. doi: 10.1111/1759-7714.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bali S.K., Marion A., Ugur I., Dikmenli A.K., Catak S., Aviyente V. Activity of topotecan toward the DNA/topoisomerase I complex: a theoretical rationalization. Biochemistry. 2018;57:1542–1551. doi: 10.1021/acs.biochem.7b01297. [DOI] [PubMed] [Google Scholar]
- 49.Wu C.P., Hsiao S.H., Huang Y.H., Hung L.C., Yu Y.J., Chang Y.T., et al. Sitravatinib sensitizes ABCB1-and ABCG2-overexpressing multidrug-resistant cancer cells to chemotherapeutic drugs. Cancers. 2020;12:195. doi: 10.3390/cancers12010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaneff A., Sahores A., Gómez N., Carozzo A., Shayo C., Davio C. MRP4/ABCC4 as a new therapeutic target: meta-analysis to determine cAMP binding sites as a tool for drug design. Curr Med Chem. 2019;26:1270–1307. doi: 10.2174/0929867325666171229133259. [DOI] [PubMed] [Google Scholar]
- 51.Ricci J.W., Lovato D.M., Severns V., Sklar L.A., Larson R.S. Novel ABCG2 antagonists reverse topotecan-mediated chemotherapeutic resistance in ovarian carcinoma xenografts. Mol Cancer Therapeut. 2016;15:2853–2862. doi: 10.1158/1535-7163.MCT-15-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts J.K., Birg A.V., Lin T., Daryani V.M., Panetta J.C., Broniscer A., et al. Population pharmacokinetics of oral topotecan in infants and very young children with brain tumors demonstrates a role of ABCG2 rs4148157 on the absorption rate constant. Drug Metab Dispos. 2016;44:1116–1122. doi: 10.1124/dmd.115.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuriappan J.A., Osheroff N., de Vivo M. Smoothed potential MD simulations for dissociation kinetics of etoposide to unravel isoform specificity in targeting human topoisomerase II. J Chem Inf Model. 2019;59:4007–4017. doi: 10.1021/acs.jcim.9b00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lawson M.H., Cummings N.M., Rassl D.M., Russell R., Brenton J.D., Rintoul R.C., et al. Two novel determinants of etoposide resistance in small cell lung cancer. Cancer Res. 2011;71:4877–4887. doi: 10.1158/0008-5472.CAN-11-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui Y., Zhu T., Song X., Liu J., Liu S., Zhao R. Downregulation of caveolin-1 increased EGFR-TKIs sensitivity in lung adenocarcinoma cell line with EGFR mutation. Biochem Biophys Res Commun. 2018;495:733–739. doi: 10.1016/j.bbrc.2017.11.075. [DOI] [PubMed] [Google Scholar]
- 56.Do P.M., Varanasi L., Fan S., Li C., Kubacka I., Newman V., et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26:830–845. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipinska N., Romaniuk A., Paszel-Jaworska A., Toton E., Kopczynski P., Rubis B. Telomerase and drug resistance in cancer. Cell Mol Life Sci. 2017;74:4121–4132. doi: 10.1007/s00018-017-2573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Margiotta A.L., Bain L.J., Rice C.D. Expression of the major vault protein (MVP) and cellular vault particles in fish. Anat Rec. 2017;300:1981–1992. doi: 10.1002/ar.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zawadzka I., Jeleń A., Pietrzak J., Żebrowska-Nawrocka M., Michalska K., Szmajda-Krygier D., et al. The impact of ABCB1 gene polymorphism and its expression on non-small-cell lung cancer development, progression and therapy—preliminary report. Sci Rep. 2020;10:6188. doi: 10.1038/s41598-020-63265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Q., Liu X., Luo Z., Wang S., Lin J., Xie Z., et al. Chloride channel-3 mediates multidrug resistance of cancer by upregulating P-glycoprotein expression. J Cell Physiol. 2019;234:6611–6623. doi: 10.1002/jcp.27402. [DOI] [PubMed] [Google Scholar]
- 61.Thiago L.D., Costa E.S., Lopes D.V., Otazú I.B., Nowill A.E., Mendes F.A., et al. The Wnt signaling pathway regulates Nalm-16 B-cell precursor acute lymphoblastic leukemic cell line survival and etoposide resistance. Biomed Pharmacother. 2010;64:63–72. doi: 10.1016/j.biopha.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 62.Heart E.A., Karandrea S., Liang X., Balke M.E., Beringer P.A., Bobczynski E.M., et al. Mechanisms of doxorubicin toxicity in pancreatic β-cells. Toxicol Sci. 2016;152:395–405. doi: 10.1093/toxsci/kfw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Congras A., Caillet N., Torossian N., Quelen C., Daugrois C., Brousset P., et al. Doxorubicin-induced loss of DNA topoisomerase II and DNMT1-dependent suppression of miR-125b induces chemoresistance in ALK-positive cells. Oncotarget. 2018;9:14539. doi: 10.18632/oncotarget.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pang B., Qiao X., Janssen L., Velds A., Groothuis T., Kerkhoven R., et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y.K., Zhang G.N., Wang Y.J., Patel B.A., Talele T.T., Yang D.H., et al. Bafetinib (INNO-406) reverses multidrug resistance by inhibiting the efflux function of ABCB1 and ABCG2 transporters. Sci Rep. 2016;6:25694. doi: 10.1038/srep25694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elliott A.M., Al-Hajj M.A. ABCB8 mediates doxorubicin resistance in melanoma cells by protecting the mitochondrial genome. Mol Cancer Res. 2009;7:79–87. doi: 10.1158/1541-7786.MCR-08-0235. [DOI] [PubMed] [Google Scholar]
- 67.Liu T., Guo J., Zhang X. MiR-202-5p/PTEN mediates doxorubicin-resistance of breast cancer cells via PI3K/Akt signaling pathway. Cancer Biol Ther. 2019;20:989–998. doi: 10.1080/15384047.2019.1591674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y., Liu Y., Du X., Ma H., Yao J. Berberine reverses doxorubicin resistance by inhibiting autophagy through the PTEN/Akt/mTOR signaling pathway in breast cancer. OncoTargets Ther. 2020;13:1909. doi: 10.2147/OTT.S241632. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Zhang D., Kanakkanthara A. Beyond the paclitaxel and vinca alkaloids: next generation of plant-derived microtubule-targeting agents with potential anticancer activity. Cancers. 2020;12:1721. doi: 10.3390/cancers12071721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Snow J.W., Kao L.W., Furbee R.B. Antitubulin agents: colchicine, vinca alkaloids, and podophyllin. Crit Care Toxicol. 2016:1–23. [Google Scholar]
- 71.Zhai X., Feng Y., Liu J., Li J., Zong Y., Tuo Z., et al. Pharmacokinetic effects of capsaicin on vinblastine in rats mediated by CYP3A and Mrp2. Fundam Clin Pharmacol. 2019;33:376–384. doi: 10.1111/fcp.12448. [DOI] [PubMed] [Google Scholar]
- 72.Skiles J.L., Chiang C., Li C.H., Martin S., Smith E.L., Olbara G., et al. CYP3A5 genotype and its impact on vincristine pharmacokinetics and development of neuropathy in Kenyan children with cancer. Pediatr Blood Cancer. 2018;65 doi: 10.1002/pbc.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martino E., Casamassima G., Castiglione S., Cellupica E., Pantalone S., Papagni F., et al. Vinca alkaloids and analogues as anti-cancer agents: looking back, peering ahead. Bioorg Med Chem Lett. 2018;28:2816–2826. doi: 10.1016/j.bmcl.2018.06.044. [DOI] [PubMed] [Google Scholar]
- 74.Ferrara R., Pilotto S., Peretti U., Caccese M., Kinspergher S., Carbognin L., et al. Tubulin inhibitors in non-small cell lung cancer: looking back and forward. Expet Opin Pharmacother. 2016;17:1113–1129. doi: 10.1517/14656566.2016.1157581. [DOI] [PubMed] [Google Scholar]
- 75.Taher M.A., Nyeem M.A., Billah M.M., Ahammed M.M. Vinca alkaloid-the second most used alkaloid for cancer treatment—a review. Inter J Physiol Nutr Phys Educ. 2017;2:723–727. [Google Scholar]
- 76.Bosilkovska M., Lorenzini K.I., Uppugunduri C.R., Desmeules J., Daali Y., Escher M. Severe vincristine-induced neuropathic pain in a CYP3A5 nonexpressor with reduced CYP3A4/5 activity: case study. Clin Therapeut. 2016;38:216–220. doi: 10.1016/j.clinthera.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 77.Kayilioğlu H., Kocak U., Kan Karaer D., Percin E.F., Sal E., Tekkesin F., Isik M., et al. Association of CYP3A5 expression and vincristine neurotoxicity in pediatric malignancies in Turkish population. J Pediatr Hematol Oncol. 2017;39:458–462. doi: 10.1097/MPH.0000000000000910. [DOI] [PubMed] [Google Scholar]
- 78.Yonemori K., Ueno H., Okusaka T., Yamamoto N., Ikeda M., Saijo N., et al. Severe drug toxicity associated with a single-nucleotide polymorphism of the cytidine deaminase gene in a Japanese cancer patient treated with gemcitabine plus cisplatin. Clin Cancer Res. 2005;11:2620–2624. doi: 10.1158/1078-0432.CCR-04-1497. [DOI] [PubMed] [Google Scholar]
- 79.Soleimani A., Jalili-Nik M., Avan A., Ferns G.A., Khazaei M., Hassanian S.M. The role of HSP27 in the development of drug resistance of gastrointestinal malignancies: current status and perspectives. J Cell Physiol. 2019;234:8241–8248. doi: 10.1002/jcp.27666. [DOI] [PubMed] [Google Scholar]
- 80.Liu Z., Liu Y., Long Y., Liu B., Wang X. Role of HSP27 in the multidrug sensitivity and resistance of colon cancer cells. Oncol Lett. 2020;19:2021–2027. doi: 10.3892/ol.2020.11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sokolowski J.D., Gamage K.K., Heffron D.S., LeBlanc A.C., Deppmann C.D., Mandell J.W. Caspase-mediated cleavage of actin and tubulin is a common feature and sensitive marker of axonal degeneration in neural development and injury. Acta Neuropathol Commun. 2014;2:16. doi: 10.1186/2051-5960-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tian X., Zhao L., Song X., Yan Y., Liu N., Li T., et al. HSP27 inhibits homocysteine-induced endothelial apoptosis by modulation of ROS production and mitochondrial caspase-dependent apoptotic pathway. BioMed Res Int. 2016;2016:4847874. doi: 10.1155/2016/4847874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu X., Li J., van Marion D.M., Zhang D., Brundel B.J. Heat shock protein inducer GGA∗-59 reverses contractile and structural remodeling via restoration of the microtubule network in experimental Atrial Fibrillation. J Mol Cell Cardiol. 2019;134:86–97. doi: 10.1016/j.yjmcc.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 84.Frommann K., Appl B., Hundsdoerfer P., Reinshagen K., Eschenburg G. Vincristine resistance in relapsed neuroblastoma can be efficiently overcome by Smac mimetic LCL161 treatment. J Pediatr Surg. 2018;53:2059–2064. doi: 10.1016/j.jpedsurg.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 85.Yi Y., Gao L., Wu M., Ao J., Zhang C., Wang X., et al. Metformin sensitizes leukemia cells to vincristine via activation of AMP-activated protein kinase. J Cancer. 2017;8:2636. doi: 10.7150/jca.19873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kanakkanthara A., Teesdale-Spittle P.H., Miller J.H. Cytoskeletal alterations that confer resistance to anti-tubulin chemotherapeutics. Anticanc Agents Med Chem. 2013;13:147–158. [PubMed] [Google Scholar]
- 87.Wu F.L., Chen H.L., Hu X.W., Liang L.Y., Xu W.L. Wnt5a modulates vincristine resistance through PI3K/Akt/GSK3β signaling pathway in human ovarian carcinoma SKOV3/VCR cells. Sheng Li Xue Bao. 2019;71:415. [PubMed] [Google Scholar]
- 88.Spitzwieser M., Pirker C., Koblmüller B., Pfeiler G., Hacker S., Berger W., et al. Promoter methylation patterns of ABCB1, ABCC1 and ABCG2 in human cancer cell lines, multidrug-resistant cell models and tumor, tumor-adjacent and tumor-distant tissues from breast cancer patients. Oncotarget. 2016;7:73347. doi: 10.18632/oncotarget.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu C.P., Hung T.H., Hsiao S.H., Huang Y.H., Hung L.C., Yu Y.J., et al. Erdafitinib resensitizes ABCB1-overexpressing multidrug-resistant cancer cells to cytotoxic anticancer drugs. Cancers. 2020;12:1366. doi: 10.3390/cancers12061366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marcath L.A., Kidwell K.M., Robinson A.C., Vangipuram K., Burness M.L., Griggs J.J., et al. Patients carrying CYP2C8∗3 have shorter systemic paclitaxel exposure. Pharmacogenomics. 2019;20:95–104. doi: 10.2217/pgs-2018-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Govindan R., Szczesna A., Ahn M.J., Schneider C.P., Gonzalez Mella P.F., Barlesi F., et al. Phase III trial of ipilimumab combined with paclitaxel and carboplatin in advanced squamous non-small-cell lung cancer. J Clin Oncol. 2017;35:3449–3457. doi: 10.1200/JCO.2016.71.7629. [DOI] [PubMed] [Google Scholar]
- 92.Guo S., Zhang Y., Wu Z., Zhang L., He D., Li X., et al. Synergistic combination therapy of lung cancer: cetuximab functionalized nanostructured lipid carriers for the co-delivery of paclitaxel and 5-demethylnobiletin. Biomed Pharmacother. 2019;118:109225. doi: 10.1016/j.biopha.2019.109225. [DOI] [PubMed] [Google Scholar]
- 93.Tripathi S., Srivastava G., Sharma A. Molecular dynamics simulation and free energy landscape methods in probing L215H, L217R and L225M βI-tubulin mutations causing paclitaxel resistance in cancer cells. Biochem Biophys Res Commun. 2016;476:273–279. doi: 10.1016/j.bbrc.2016.05.112. [DOI] [PubMed] [Google Scholar]
- 94.Miyata Y., Matsuo T., Nakamura Y., Yasuda T., Ohba K., Takehara K., et al. Expression of class III beta-tubulin predicts prognosis in patients with cisplatin-resistant bladder cancer receiving paclitaxel-based second-line chemotherapy. Anticancer Res. 2018;38:1629–1635. doi: 10.21873/anticanres.12394. [DOI] [PubMed] [Google Scholar]
- 95.Kato A., Naiki-Ito A., Naitoh I., Hayashi K., Nakazawa T., Shimizu S., et al. The absence of class III β-tubulin is predictive of a favorable response to Nab-paclitaxel and gemcitabine in patients with unresectable pancreatic ductal adenocarcinoma. Hum Pathol. 2018;74:92–98. doi: 10.1016/j.humpath.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 96.Yin S., Bhattacharya R., Cabral F. Human mutations that confer paclitaxel resistance. Mol Cancer Therapeut. 2010;9:327–335. doi: 10.1158/1535-7163.MCT-09-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ganguly A., Yang H., Cabral F. Overexpression of mitotic centromere-associated kinesin stimulates microtubule detachment and confers resistance to paclitaxel. Mol Cancer Therapeut. 2011;10:929–937. doi: 10.1158/1535-7163.MCT-10-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie S., Ogden A., Aneja R., Zhou J. Microtubule-binding proteins as promising biomarkers of paclitaxel sensitivity in cancer chemotherapy. Med Res Rev. 2016;36:300–312. doi: 10.1002/med.21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang B., Ni H., Zhou Z., Li Y. Parkin enhances sensitivity of paclitaxel to NPC by arresting cell cycle. Pathol Res Pract. 2020;216:152755. doi: 10.1016/j.prp.2019.152755. [DOI] [PubMed] [Google Scholar]
- 100.Němcová-Fürstová V., Kopperová D., Balušíková K., Ehrlichová M., Brynychová V., Václavíková R., et al. Characterization of acquired paclitaxel resistance of breast cancer cells and involvement of ABC transporters. Toxicol Appl Pharmacol. 2016;310:215–228. doi: 10.1016/j.taap.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 101.Xie X., Shao X., Ma W., Zhao D., Shi S., Li Q., et al. Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures. Nanoscale. 2018;10:5457–5465. doi: 10.1039/c7nr09692e. [DOI] [PubMed] [Google Scholar]
- 102.Xu X., Jin S., Ma Y., Fan Z., Yan Z., Li W., et al. miR-30a-5p enhances paclitaxel sensitivity in non-small cell lung cancer through targeting BCL-2 expression. J Mol Med. 2017;95:861–871. doi: 10.1007/s00109-017-1539-z. [DOI] [PubMed] [Google Scholar]
- 103.Frederiks C.N., Lam S.W., Guchelaar H.J., Boven E. Genetic polymorphisms and paclitaxel-or docetaxel-induced toxicities: a systematic review. Cancer Treat Rev. 2015;41:935–950. doi: 10.1016/j.ctrv.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 104.de Weger V.A., Beijnen J.H., Schellens J.H. Cellular and clinical pharmacology of the taxanes docetaxel and paclitaxel—a review. Anti Cancer Drugs. 2014;25:488–494. doi: 10.1097/CAD.0000000000000093. [DOI] [PubMed] [Google Scholar]
- 105.Chen Y., Li J., Chen S., Zhang Y., Hu Y., Zhang G., et al. Nab-paclitaxel in combination with cisplatin versus docetaxel plus cisplatin as first-line therapy in non-small cell lung cancer. Sci Rep. 2017;7:1–7. doi: 10.1038/s41598-017-11404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li A., Wei Z.J., Ding H., Tang H.S., Zhou H.X., Yao X., et al. Docetaxel versus docetaxel plus cisplatin for non-small-cell lung cancer: a meta-analysis of randomized clinical trials. Oncotarget. 2017;8:57365. doi: 10.18632/oncotarget.17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang H., Patel A., Wang Y.J., Zhang Y.K., Kathawala R.J., Qiu L.H., et al. The BTK inhibitor ibrutinib (PCI-32765) overcomes paclitaxel resistance in ABCB1-and ABCC10-overexpressing cells and tumors. Mol Cancer Therapeut. 2017;16:1021–1030. doi: 10.1158/1535-7163.MCT-16-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang H., Wang S., Cacalano N., Zhu H., Liu Q., Xie M., et al. Oncogenic Y68 frame shift mutation of PTEN represents a mechanism of docetaxel resistance in endometrial cancer cell lines. Sci Rep. 2019;9:1. doi: 10.1038/s41598-019-38585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lucanus A.J., Yip G.W. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37:833–838. doi: 10.1038/onc.2017.406. [DOI] [PubMed] [Google Scholar]
- 110.Yin B., Lu P., Liang J., Zhang W., Xin M., Pei K., et al. The ABCB1 3435C>T polymorphism influences docetaxel transportation in ovarian cancer. J Int Med Res. 2019;47:5256–5269. doi: 10.1177/0300060519870354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sone K., Oguri T., Uemura T., Takeuchi A., Fukuda S., Takakuwa O., Maeno K., Fukumitsu K., Kanemitsu Y., Ohkubo H., Takemura M. Genetic variation in the ATP binding cassette transporter ABCC10 is associated with neutropenia for docetaxel in Japanese lung cancer patients cohort. BMC Cancer. 2019;19:1. doi: 10.1186/s12885-019-5438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin J.Z., Wang W.W., Hu T.T., Zhu G.Y., Li L.N., Zhang C.Y., et al. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett. 2020;469:481–489. doi: 10.1016/j.canlet.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 113.Khongkow P., Gomes A.R., Gong C., Man E.P., Tsang J.W., Zhao F., et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35:990–1002. doi: 10.1038/onc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deng L., Gu X., Zeng T., Xu F., Dong Z., Liu C., et al. Identification and characterization of biomarkers and their functions for docetaxel-resistant prostate cancer cells. Oncol Lett. 2019;18:3236–3248. doi: 10.3892/ol.2019.10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fujimoto D., Goi T., Koneri K., Hirono Y. RPN2 is effective biomarker to predict the outcome of combined chemotherapy docetaxel and cisplatin for advanced gastric cancer. Oncotarget. 2018;9:15208. doi: 10.18632/oncotarget.24622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ban H.S., Xu X., Jang K., Kim I., Kim B.K., Lee K., et al. A novel malate dehydrogenase 2 inhibitor suppresses hypoxia-inducible factor-1 by regulating mitochondrial respiration. PLoS One. 2016;11 doi: 10.1371/journal.pone.0162568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gómez-Miragaya J., Morán S., Calleja-Cervantes M.E., Collado-Sole A., Paré L., Gómez A., et al. The altered transcriptome and DNA methylation profiles of docetaxel resistance in breast cancer PDX models. Mol Cancer Res. 2019;17:2063–2076. doi: 10.1158/1541-7786.MCR-19-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim S.J., Miyoshi Y., Taguchi T., Tamaki Y., Nakamura H., Yodoi J., et al. High thioredoxin expression is associated with resistance to docetaxel in primary breast cancer. Clin Cancer Res. 2005;11:8425–8430. doi: 10.1158/1078-0432.CCR-05-0449. [DOI] [PubMed] [Google Scholar]
- 119.Jia J.J., Geng W.S., Wang Z.Q., Chen L., Zeng X.S. The role of thioredoxin system in cancer: strategy for cancer therapy. Cancer Chemother Pharmacol. 2019;84:453–470. doi: 10.1007/s00280-019-03869-4. [DOI] [PubMed] [Google Scholar]
- 120.Rheinbay E. The genomic landscape of advanced cancer. Nat Cancer. 2020;1:372–373. doi: 10.1038/s43018-020-0057-z. [DOI] [PubMed] [Google Scholar]