Abstract
Persistent antigen induces a dysfunctional CD8 T cell state known as “exhaustion” characterized by PD-1 expression. Nevertheless, exhausted CD8 T cells retain functionality through continued differentiation of progenitor into effector cells. However, it remains ill-defined how CD8 T cell effector responses are sustained in situ. Here we show using the mouse chronic LCMV infection model that CX3CR1+ CD8 T cells contain a T-bet-dependent TIM3− PD-1lo subpopulation that is distinct from the TIM3+ CX3CR1+ PD-1+ proliferative effector subset. The TIM3− CX3CR1+ cells are quiescent and express a low but significant level of the transcription factor TCF-1, demonstrating similarity to TCF-1hi progenitor CD8 T cells. Furthermore, following the resolution of LCMV viremia, a substantial proportion of TCF-1+ memory-like CD8 T cells show evidence of CX3CR1 expression during the chronic phase of the infection. Our results suggest a subset of the CX3CR1+ exhausted population demonstrates progenitor-like features that support the generation of the CX3CR1+ effector pool from the TCF-1hi progenitors and contribute to the memory-like pool following resolution of viremia.
Keywords: CD8 T cells, exhaustion, progenitor, T-bet, TOX
Introduction
Upon encountering antigen, naive CD8 T cells undergo robust clonal expansion and acquire effector functions for host protection against pathogens and tumors. However, in the context of persistent antigen exposure during chronic viral infections, they are subject to alternative differentiation known as T cell exhaustion, which is characterized by continued expression of inhibitory receptors, decreased cytokine secretion, and blunted proliferation (1, 2). Nevertheless, exhausted CD8 T cells are not inert and can actively control antigen burden (3). This function is in part facilitated by a progenitor-progeny relationship within exhausted CD8 T cells that facilitates sustainable generation of functional effector cells. However, our understanding of the progenitor cell dynamics that supports this continuous differentiation process is limited.
The TCF-1hi (CXCR5+ Ly-108/SLAMF6+) progenitor exhausted CD8 T cells (TPEX) give rise to TCF1− effector-like cells, of which a substantial fraction expresses the inhibitory receptor/exhaustion marker TIM3 (4–8). CX3CR1 is also expressed in a subset of TCF-1− effector-like cells, which exhibit enhanced proliferative activity and cytolytic function compared to CX3CR1− cells (9, 10). Since TPEX cells are quiescent in situ and mainly present in the white pulp of the spleen and are thus physically separated from red-pulp predominant TCF-1lo/– cells (4), it is possible that proliferative CX3CR1+ CD8 T cells are supported by a non-TPEX intermediate progenitor population. Prior work identified T-bet expressing exhausted CD8 T cells as having progenitor-like features with low intrinsic turnover (11), however their relationship to TCF-1hi TPEX cells remains unclear as a vast majority of TPEX express T-bet at a low level. The CX3CR1+ population is enriched for T-bet expressing cells compared to TCF-1hi TPEX cells (4, 5), suggesting that they could be composed of a progenitor-like subpopulation even though they lack cells with high TCF-1 expression.
In this work, we found using single-cell RNA-seq (scRNA-seq) that CX3CR1+ cells are composed of two clusters. Cells present in one of the CX3CR1+ cell clusters are quiescent, highly enriched for expression of T-bet, and distinguished from the proliferating cluster of CX3CR1+ cells by TIM3 expression. The quiescent cluster also resembles TPEX cells by basal expression of Tcf7 and Bach2, the transcription factor genes highly expressed in TPEX. Slow replenishment of the total CX3CR1+ cells by CX3CR1− cells, dependency on the transcription factor T-bet as well as trajectory based on scRNA-seq diffusion pseudotime analysis suggest that TIM3− CX3CR1+ cells serve as upstream precursors for the TIM3+ CX3CR1+ effector-like cells. By contrast, the resistance of a vast majority of TIM3+ CX3CR1− cells to T-bet deletion and CD4 T cell-depletion suggests that they are independent of CX3CR1+ cells or develop through an alternative pathway. Furthermore, a substantial fraction of TCF-1hi memory-like cells that persist after the resolution of viremia are marked by a history of Cx3cr1 expression during high viral burden and display continued expression of CX3CR1 and the exhaustion-associated gene TOX. These results collectively suggest multiple trajectories of CD8 T cell differentiation dynamics under exhausting conditions and the development of memory-like cells that persist after the resolution of viremia (12–14).
Materials and Methods
Mice and Infection
Male C57BL/6N and B6-CD45.1 mice were purchased from Charles River Laboratory. Prdm1-EYFP, CD8 (E8I)-Cre, Tbx21F, EomesF, Cx3cr1CreER, Rosa26LSL-tdT mice were originally obtained from The Jackson Laboratory. Bach2F mice are described previously (15). All mice were housed in a specific pathogen-free facility at Washington University in St. Louis, and were used for infection at 8 to 12 weeks of age unless stated otherwise. All experiments were performed according to a protocol approved by Washington University’s Institutional Animal Care and Use Committee (IACUC). Stocks of LCMV were made by propagating virus by infection of BHK21 cells, followed by determination of titer in culture supernatants by plaque assay on Vero cells. For LCMV infection, mice were infected with 2 × 105 plaque-forming units (PFU) of LCMV-Armstrong via the intraperitoneal route or 2 × 106 PFU of LCMV-clone 13 by intravenous injection. For the quantification of plasma viral load, RNA was extracted from 10 μl of plasma using Trizol (Thermo Fisher). Before RNA extraction a spike-in of exogenous control ERCC RNA (Thermo Fisher) was carried out and used to normalize viral loads following qPCR as described previously (16, 17).
Bulk RNA-Seq
Polyclonal PD-1+ CD8 T cells, which are enriched for LCMV-antigen-specific CD8 T cells, were purified and sorted based on indicated cell surface markers on 16 dpi of LCMV-c13 from Prdm1-YFP mice as described previously (6, 18). Briefly, CD8+ T cells were first enriched from splenocytes using the Dynabeads FlowComp Mouse CD8 T cell kit (Thermo Fisher) followed by surface staining. We then sorted Prdm1-EYFP− TIM3− CX3CR1− cells, which corresponded to TCF-1hi CD8 TPEX cells, and divided Prdm1-EYFP+ cells into TIM3− CX3CR1+, TIM3+ CX3CR1+, and TIM3+ CX3CR1− populations. Total RNA was extracted from 20,000–50,000 sorted cells using Direct-Zol Micro Kit (Zymo) according to the manufacturer’s instructions. cDNA synthesis and amplification were performed with Next Ultra RNA Library Preparation Kit (NEB). Libraries were sequenced on a HiSeq3000 (Illumina) in single-read mode, with a read length of 50 nucleotides producing ~25 million reads per sample. Sequencing reads were mapped to the NCBI mm9 mouse genome assembly using STAR with default parameters, and mapping rates were higher than 90%. Transcripts with 4 TPM at least in one sample were initially filtered, followed by principal component analysis, unsupervised clustering, and profiling of subpopulation-specific gene expression using Phantasus and limma packages (V.15.1, https://artyomovlab.wustl.edu/phantasus/). The raw data has been deposited to NCBI GEO.
scRNA-seq
scRNA-seq libraries were prepared using the 10X Single Cell Immune Profiling Solution Kit (v1 Chemistry), according to the manufacturer’s instructions. Briefly, FACS sorted cells were washed once with PBS + 0.04% BSA. Following reverse transcription and cell barcoding in droplets, emulsions were broken and cDNA purified using Dynabeads MyOne SILANE followed by PCR amplification (98 ºC for 45 sec; 14 cycles of 98 ºC for 20 sec, 67 ºC for 30 sec, 72 ºC for 1 min; 72 ºC for 1 min). For gene expression library construction, 50 ng of amplified cDNA was fragmented and end-repaired, double-sided size selected with SPRIselect beads, PCR amplified with sample indexing primers (98 ºC for 45 sec; 14 cycles of 98 ºC for 20 sec, 54 ºC for 30 sec, 72 ºC for 20 sec; 72 ºC for 1 min), and double-sided size selected with SPRIselect beads. The prepared scRNA libraries were sequenced on an Illumina HiSeq 4000 to a minimum sequencing depth of 25,000 reads/cell using the read lengths 28bp Read1, 8bp i7 Index, 91bp Read2. Reads were aligned to the mm10 reference genome and quantified using cellranger count (10X Genomics, version 3.1.0). Filtered gene-barcodes matrices containing only barcodes with UMI counts passing threshold for cell detection were used for further analysis.
Additional analysis was performed using Seurat (version 3.1.2) (19). Cells with less than 200 genes detected or greater than 5% mitochondrial RNA content were excluded from analysis. For clustering, raw UMI counts were log normalized and variable genes identified based on a variance stabilizing transformation. We assigned scores for S and G2/M cell cycle phase based on previously defined gene sets (20) using the CellCycleScoring function. Scaled z-scores for each gene were calculated using the ScaleData function and regressed against the S phase score and G2/M phase score to reduce clustering based on cell cycle. Scaled z-scores for variable genes were used as input into PCA. Clusters were identified using shared nearest neighbor (SNN) based clustering based on the first 10 PCs with k = 15 and resolution = 0.23. The same principal components were used to generate the UMAP projections (21, 22), which were generated with a minimum distance of 0.1 and 30 neighbors. Expression of selected genes was plotted using log normalized gene expression values.
Pseudotime and diffusion map analysis of scRNA-seq data was performed using destiny (version 3.2.0) (23). The normalized, log-transformed transcript count matrix was used to create a diffusion map using the DiffusionMap function with default parameters. Cells were plotted using the first 3 components of the diffusion map. A randomly selected cell from the TCF1+ cluster was used as the root cell for diffusion psuedotime computation using the DPT function with default parameters.
Treatments
For depletion of CD4 T cells, 200 micrograms of anti-CD4 (GK1.5, Leinco) was injected on −1 and +1 dpi. For administration of Tamoxifen (TAM, Sigma), 10 mg/mL solutions were prepared in Corn Oil (Sigma) and 1 mg gavaged orally.
Cell Preparation, Cell Staining, and Flow Cytometry
Single-cell suspensions of splenocytes were prepared by manual disruption with frosted glass slides. Lungs were minced with scissors and digested with Collagenase D (Sigma) and DNase I (Sigma) with agitation for 1 hour at 37C followed by enrichment of lymphocytes by a 40/70 Percoll gradient. Absolute live cell counts were determined by Trypan-blue exclusion using Vi-Cell (Beckman Coulter). Tetramer staining was performed using iTag-PE and APC LCMV gp33–41 and gp276–286 (MBL international). The following monoclonal antibodies were purchased from Biolegend unless otherwise indicated: FITC-conjugated anti-CD45.2 (104); PerCP-Cy5.5 conjugated anti-CD8β (YTS156.7), anti-CD4 (GK1.5), anti-Ki-67 (16A8), PerCP-eFlour710-conjugate anti-Eomes (eBioscience, Dan11mag), APC-conjugated CD366 (TIM-3, RMT3–23), anti-TNF-α (MP6-XT22), anti-granzyme B (QA16A02), anti-T-bet (4B10), anti-CX3CR1 (SA011F11), Alexa 700-conjugated anti-CD44 (IM7), anti-CD45.2 (104), BV421-conjugated anti-TIM3 (RMT3–23), BV605-conjugated CD4 (GK1.5), anti-CD45.1 (A20), anti-CX3CR1(SA011F11), BV711-conjugated anti-CD4 (RM4–5), PE-conjugated anti-IFN-γ (XMG1.2), anti-PE-Cy7-conjugated anti-PD-1 (29F.1A12) PE-Dazzle594-conjugated anti-B220 (RA3–6B2) and BUV395-conjugated anti-CD8 (53–6.7, BD) Anti-TCF-1 antibody (Cell Signaling, C63D9) and detected by Alexa 488-conjugated Donkey anti-rabbit polyclonal IgG (Thermo Fisher, Cat. #R37118). Staining for transcription factors was performed using the Foxp3 staining kit (eBioscience) according to the manufacturer’s instructions.
For intracellular cytokine staining, splenocytes were cultured in RPMI-1640 supplemented with 10% fetal bovine serum in the presence of 1 ug/ml of LCMV-gp33–41 peptide (Genscript) and 5 ug/ml of Brefeldin A (Biolegend) for 4 hours. Cells were stained for surface makers and then subject to LIVE/DEAD Aqua staining (Thermo Fisher) for 30 minutes at 4 ºC before being fixed with 4% PFA for 10 minutes at room temperature. Cells were then washed twice with 0.03% saponin in 2% FBS/PBS before being stained with the indicated antibodies in 0.3% Saponin in 2% FBS/PBS for 20 min at 4 ºC.
BrdU incorporation assay was performed using the APC BrdU Flow Kit (BD Biosciences) according to the manufacturer’s instruction with one-time intraperitoneal injection of 1 mg of BrdU followed by tissue collection after 12 hours.
Stained samples were analyzed with BD FACS LSR Fortessa, X20, or Symphony A3 or sorted on Aria II or III. Data was processed with FlowJo Software (FlowJo. LLC).
Infection-matched Adoptive Transfer
SLAMF6+, SLAMF6− TIM3− CX3CR1+ or SLAMF6− TIM3+ CX3CR1+ PD-1+ CD8 T cells were sorted from C57BL/6 mice on 21 dpi with LCMV-c13 and 3 × 105 CFSE-labeled cells were intravenously transferred into infection-matched CD45.1 congenic recipients. Splenocytes from the recipient mice were harvested 14 days after transfer and donor-derived CD8 T cells were analyzed by flow cytometry following magnetic bead-enrichment with a MojoSort™ Mouse CD8 T Cell Isolation Kit (Biolegend).
For transfer of memory CD8 T cells, CD44+ CX3CR1+ or CD44+ CX3CR1− CD8 T cells were sorted from the spleen of mice that had been infected with LCMV-c13 >120 days prior. The sort-purified memory CD8 T cells that contained 30,000 gp33-tetramer+ CD8 T cells were transferred to uninfected CD45.1 congenic recipients and infected on the next day. We quantitated donor-derived CD8 T cells in the recipients without LCMV-Arm infection 24 hours after transfer and donor cell expansion was calculated by dividing donor cell number in infected recipients on 6 dpi by that in uninfected control gp33-tetramer+ CD8 T cells.
Statistical analysis
P-values were calculated with an unpaired two-tailed Student’s t-test or Mann-Whitney U-test for two-group comparisons and by one-way ANOVA for multi-group comparisons with the Tukey post hoc test using Prism 8 software. *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001.
Results
CX3CR1 defines two populations of exhausted CD8 T cells with distinct properties
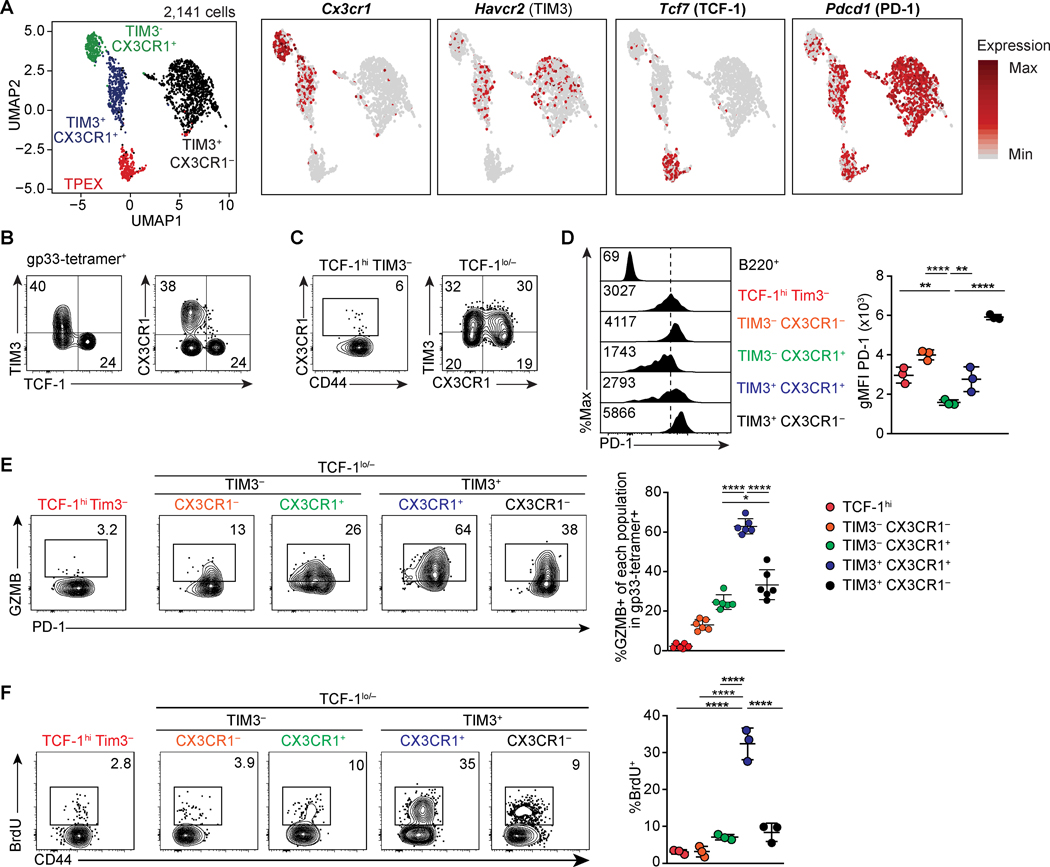
We first sought to identify additional heterogeneity in the CX3CR1+ subset, given that they are enriched for T-bet expressing cells (11). To this end, we performed scRNA-seq analysis of LCMV gp33-specific CD8 T cells during the chronic phase of LCMV-c13 infection in C57BL/6 mice. Uniform Manifold Approximation and Projection (UMAP) of the data demonstrated 4 major clusters of exhausted CD8 T cells, including the previously defined TCF-1hi (encoded by Tcf7) and TIM3 (Havcr2)+ subsets and also two distinct clusters of Cx3cr1+ cells segregated by expression of Havcr2 (Fig. 1A, Fig. S1A). Consistent with the scRNA-seq data and prior reports (10, 24), approximately 40% of LCMV-gp33-specific CD8 T cells in mice infected with LCMV-clone 13 (LCMV-c13) expressed CX3CR1 in the chronic phase, and its expression was seen exclusively in TCF-1lo/– cells (Fig. 1B, Fig. S1B). The frequency of CX3CR1+ cells was notable in both TIM3− and TIM3+ cells confirming the presence of two unique CX3CR1+ populations (Fig. 1C) We also observed a TIM3− CX3CR1− TCF1lo/– population, however, this subset was not a focus for the rest of this study (Fig. 1C).
FIGURE 1. CX3CR1 marks two distinct subsets of exhausted T cells.
(A) UMAP analysis of scRNA-seq data of 2,141 gp-33 specific CD8 T cells on 21 dpi with LCMV-c13. Right panels show the expression of indicated genes. (B-C) Flow cytometry showing expression of TIM3, TCF-1, CX3CR1, and CD44 in LCMV gp33-specific CD8 T cells 4 weeks after infection of C57BL/6 mice with LCMV-c13. Numbers indicate percentages of gated cells in each parental population shown in the Figure. Data are representative of > 5 independent experiments with >20 mice in total. (D) Expression of PD-1 measured by flow cytometry in the indicated population of LCMV-gp33-specific CD8 T cells on 22 dpi, quantified in the right panel. Data are representative of 3 independent experiments with n=3 mice each. (E) GZMB expression ex vivo as measured by flow cytometry of LCMV gp33-specific CD8 T cells in the indicated subpopulations on 22 dpi with LCMV-c13. Data are representative of two independent experiments with n=4–6 mice each. (F) BrdU incorporation as measured by flow cytometry of LCMV gp33-specific CD8 T cells in the indicated subpopulations on 22 dpi with LCMV-c13 12 hours after intraperitoneal administration of 1 mg BrdU. Data are representative of three independent experiments with n=3 mice each.
The amount of surface PD-1 expression is known to correlate with the extent of exhaustion, with higher levels corresponding to more severe exhaustion and unresponsiveness to the checkpoint blockade (11, 25, 26). Notably, the surface PD-1 level in TIM3− CX3CR1+ cells was even lower than that in TCF-1hi TPEX cells (Fig. 1D). PD-1 was elevated in the TIM3+ fraction of CX3CR1+ cells, suggesting that TIM3+ CX3CR1+ cells are more differentiated or in a more activated state than TCF-1hi and TIM3− CX3CR1+ cells. Accordingly, expression of Granzyme B (GZMB) was significantly higher in TIM3+ CX3CR1+ cells compared to TIM3− CX3CR1+ cells while GZMB expression was barely detectable in TCF-1hi cells (Fig. 1E).
We next examined cell cycle states between the two CX3CR1+ populations by measuring BrdU incorporation, which directly reflects DNA synthesis during the S-phase of the cell cycle, by pulse labeling in the chronic phase (22 dpi) of LCMV-c13 infection (Fig. 1F). We confirmed the quiescence of TCF-1hi cells as <5% of cells incorporated BrdU over 12-hours following one injection of BrdU. TIM3+ CX3CR1+ cells exhibited the highest incorporation of BrdU (~30%) while BrdU incorporation by the TIM3− CX3CR1+ cells was significantly lower than the TIM3+ counterpart, which was also reflected in Ki-67 expression (Fig. S1C). However, at an earlier time point on 8 dpi, cells in all subpopulations, including TPEX and TIM3− CX3CR1+ cells, were active in cell cycle and upregulated Nur77 (Fig. S1C, D). Notably, expression of Nur77 in TIM3− CX3CR1+ cells was slightly lower (Fig S1D), suggesting that they equally or slightly weakly received a TCR stimulus following infection. These results highlight distinct characteristics between the two subpopulations of CX3CR1+ PD-1+ CD8 T cells. While TIM3+ CX3CR1+ cells are actively proliferating effector cells similar to the cells described in previous studies (9, 10), TIM3− CX3CR1+ cells appear progenitor-like with phenotypic similarities to TPEX with regards to low to moderate cell cycle activity and low expression of PD-1 and GZMB and are thus distinct from TIM3+ CX3CR1+ cells.
Effector cells are typically found in both blood and tissue at higher frequencies compared to memory-like cells (27). To examine the migratory behavior of CX3CR1+ cells, we performed in vivo labelling of T cells by an intravenous injection of anti-CD45.2 which allowed us to discriminate between tissue resident (CD45.2−) and circulating (CD45.2+) antigen-specific CD8 T cells present in the lung. In contrast to TIM3+ CX3CR1− cells that were predominantly localized in the lung parenchyma, distribution of both CX3CR1+ subpopulations was significantly enriched in the circulating compartment (Fig. S1E). Within the CX3CR1+ populations, more TIM3+ cells were found in the tissues compared to TIM3− cells, suggesting that TIM3+ CX3CR1+ cells preferentially migrate or are retained in tissue parenchyma, and are likely key mediators of effector function.
TIM3− CX3CR1+ cells can give rise to TIM3+ CX3CR1+ effectors
To examine the development and dynamics of each subset, we next performed a time-course analysis. In the early phase of the acute phase response on 5 dpi, the TIM3+ CX3CR1− subset was dominant, however, CX3CR1+ cells were present with uniformly intermediate TIM3 expression without a clear segregation between TIM3− and TIM3+ populations (Fig. S2A, B). TIM3− CX3CR1+ cells emerged around 8 dpi and they continued to increase in numbers until 15 dpi possibly as a subset of antigen-specific CD8 T cells receive weaker TCR stimulus and slow down their proliferation during this period (Figs. S1C, D and S2A, B).
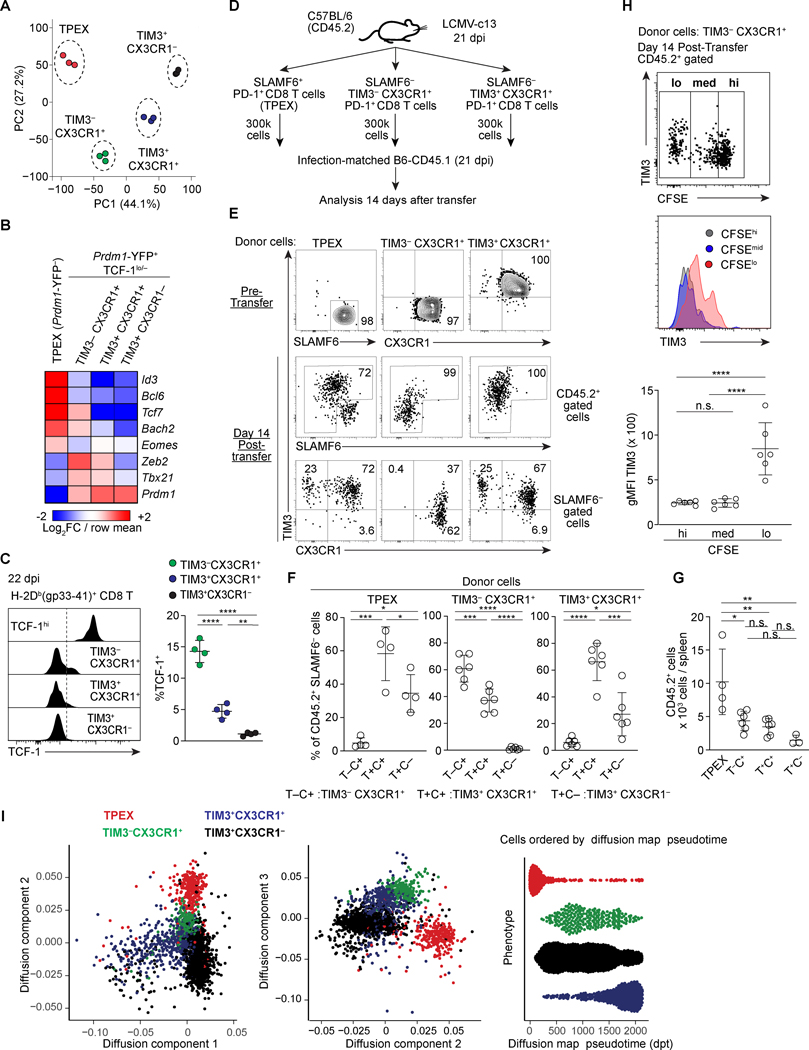
To gain additional insight into the two CX3CR1+ populations of CD8 T cells, we purified the two subsets of CX3CR1+ PD-1+ CD8 T cells based on TIM3 expression as well as TPEX and terminally exhausted (TIM3+ CX3CR1−) CD8 T cells for population-based RNA-seq on 16 dpi. Confirming our scRNA-seq data, principal component analysis and K-means clustering showed that the two CX3CR1+ populations have distinct gene expression patterns (Fig. 2A and Fig. S3A). A gene set enrichment analysis (GSEA) for differentially expressed genes between the two CX3CR1+ populations showed significant representation of pathways associated with cell division, MYC target genes, IL2-STAT5 signaling and GTPase effectors, consistent with effector functions, high proliferative activity and tissue migration (Fig. S3B). This result was supported by a gene-level analysis showing Gzmb, Prdm1, Ifng, Prf1, Lag3, Entpd1 (CD39) and Tigit being higher in TIM3+ CX3CR1+ compared to the TIM3− CX3CR1+ populations (Fig. S3C). In addition, K-means clustering showed that many genes that are differentially expressed between the CX3CR1+ populations were similarly expressed between TPEX and TIM3− CX3CR1+ cells (Fig. S3A, Clusters 2 and 5), suggesting that there is transcriptomic similarity between TPEX and TIM3− CX3CR1+ cells. Among genes encoding transcription factors, the two CX3CR1+ populations expressed high Tbx21 compared to the TPEX subset that is marked by high expression of Tcf7, Id3, Bcl6 and Bach2. It is notable that these TCF-1hi population-specific factors were also expressed at lower but significant levels in the TIM3− CX3CR1+ population, while the TIM3− CX3CR1+ population expressed higher amounts of Zeb2, which cooperates with T-bet (28, 29), than TIM3+ CX3CR1+ cells and other PD-1+ CD8 T cells (Fig. 2B). Consistently, TCF-1 protein was also expressed in a fraction of gp33-specific TIM3− CX3CR1+ CD8 T cells, and the frequency of TCF-1+ cells was substantially reduced in TIM3− CX3CR1+ cells (Figs. 1B, 2C).
FIGURE 2. TIM3− CX3CR1+ cells are progenitor-like cells upstream of TIM3+ CX3CR1+ proliferating effector CD8 T cells.
(A) Principal Component Analysis (PCA) plot derived from population-level RNA-seq analysis of the indicated subpopulations of PD-1+ CD8 T cells on 16 dpi. The populations were defined using Prdm1-YFP mice as Prdm1-YFP− (TCF-1hi) and subpopulations of Prdm1-YFP+ (TCF-1lo/–) with a combination of CX3CR1 and TIM3. (B) Heat map showing expression of indicated genes by indicated subsets of PD-1+ CD8 T cells. Expression values were averaged from three independent samples for each population and color-coded based on row mean and fold-changes. (C) Flow cytometry plots showing expression of TCF-1 in the indicated subsets of H-2Db (gp33–41)+ CD8 T cells 22dpi LCMV-c13 infection. Data in the right panel are representative of 2 independent experiments with n=3–4 mice each. (D) Experimental schematic of infection-matched adoptive transfer. (E-G) Flow cytometry showing expression of SLAMF6, CX3CR1 and TIM3 in sorted donor CD45.2+ PD-1+ CD8 T cells prior to transfer (top), and of the donor-derived (CD45.2) PD-1+ CD8 T cells 14 days after transfer. Frequencies of cells in each population and total cell yield from each donor population were quantified in (E-F). (H) CFSE dilution and expression of TIM3 of donor cells derived from TIM3− CX3CR1+ cells. Expression of TIM3 in CFSElo, CFSEmed and CFSEhi subpopulations is shown by a histogram overlay and geographical mean fluorescence intensity (gMFI). (I) Pseudotime and diffusion map analysis of scRNA-seq data from Fig. 1A performed using destiny package. Data shown in (D-G) are pooled from two independent experiments with n=4–6 mice recipient mice analyzed for each donor transfer and shown as shown as mean ± SD. Statistical analysis by one way ANOVA was performed on data in (G).
To assess the differentiation potentials of CX3CR1+ CD8 T cells, we transferred CFSE-labeled TIM3− and TIM3+ CX3CR1+ and SLAMF6+ TIM3− (TPEX) CD8 T cells isolated from LCMV-c13-infected C57BL/6 mice (CD45.2) into infection-matched congenic CD45.1 mice and analyzed donor-derived cells 14 days after transfer (Fig. 2D). Approximately 40% of donor-derived TIM3− CX3CR1+ cells upregulated TIM3 (Fig. 2E, F), and TIM3 upregulation preferentially occurred in CFSElo cells (Fig. 2H). In addition, the majority of CFSEmid and a fraction of CFSElo cells remained as TIM3− (Fig. 2E, H). TIM3+ CX3CR1+ cells barely generated TIM3− cells (Fig. 2E-G). However, expansion of both TIM3− and TIM3+ CX3CR1+ cells was 2–3-fold lower compared to TPEX cells, which gave rise to more differentiated cell types. Inefficient differentiation to CX3CR1− cells from the transferred TPEX could be affected by timing or a complication in trafficking or cell death in a setting of infection-matched adoptive transfer. It is also notable that expansion of TIM3+ CX3CR1+ cells is substantially compromised upon adaptive transfer although they were highly proliferating in situ. To gain additional insights, we treated LCMV-infected mice with anti-PD-L1 antibodies, which releases TPEX from PD-1-mediated suppression and specifically drives their expansion (Fig. S2D, E) (4). After three doses of anti-PD-L1, we detected proportional ratios between TIM3− and TIM3+ CX3CR1+ cells, suggesting that proliferating TPEX cells give rise to both populations in situ (Fig S2D, E). These results collectively indicate that TIM3− CX3CR1+ cells can be derived from TPEX and differentiate into TIM3+ CX3CR1+ cells. This result was also supported by diffusion pseudotime analysis of scRNA-seq data showing a trajectory of TPEX differentiation towards TIM3+ CX3CR1+ cells via a TIM3− CX3CR1+ intermediate (Fig. 2H). These results suggest that TIM3− cells are positioned at an intermediate between TPEX and upstream of TIM3+ cells in terms of developmental hierarchy within the CX3CR1+ PD-1+ CD8 T cells.
The loss of CD4 help via IL-21 reduces the absolute numbers of CX3CR1+ CD8 T cells during the chronic phase of the response to LCMV-c13 infection (10). In CD4-depleted LCMV-c13 infected mice, we did not observe a reduction in the number of CX3CR1+ CD8 T cells on 8 dpi at the peak of the antigen-specific CD8 T cell response (Fig. S2A, C). However, in the chronic phase on 22 dpi, the number of CX3CR1+ CD8 T cells in CD4-depleted mice was substantially reduced in both TIM3− and TIM3+ subpopulations, which is consistent with prior results (10) (Fig. S2A, C). Thus, CD4 T cell help appears to be required for maintenance of the CX3CR1+ cells rather than their generation. Interestingly, the loss of TIM3− CX3CR1+ CD8 T cells preceded that of TIM3+ cells, with a ~15-fold reduction of the former population compared to a ~4-fold reduction of the latter on 15 dpi. The severe loss of TIM3+ CX3CR1− cells followed by TIM3+ CX3CR1+ cells could reflect their developmental relationship, or highlight differential sensitivity to IL-21 for their maintenance.
T-bet is critical for maintenance of CX3CR1+ exhausted CD8 T cell populations
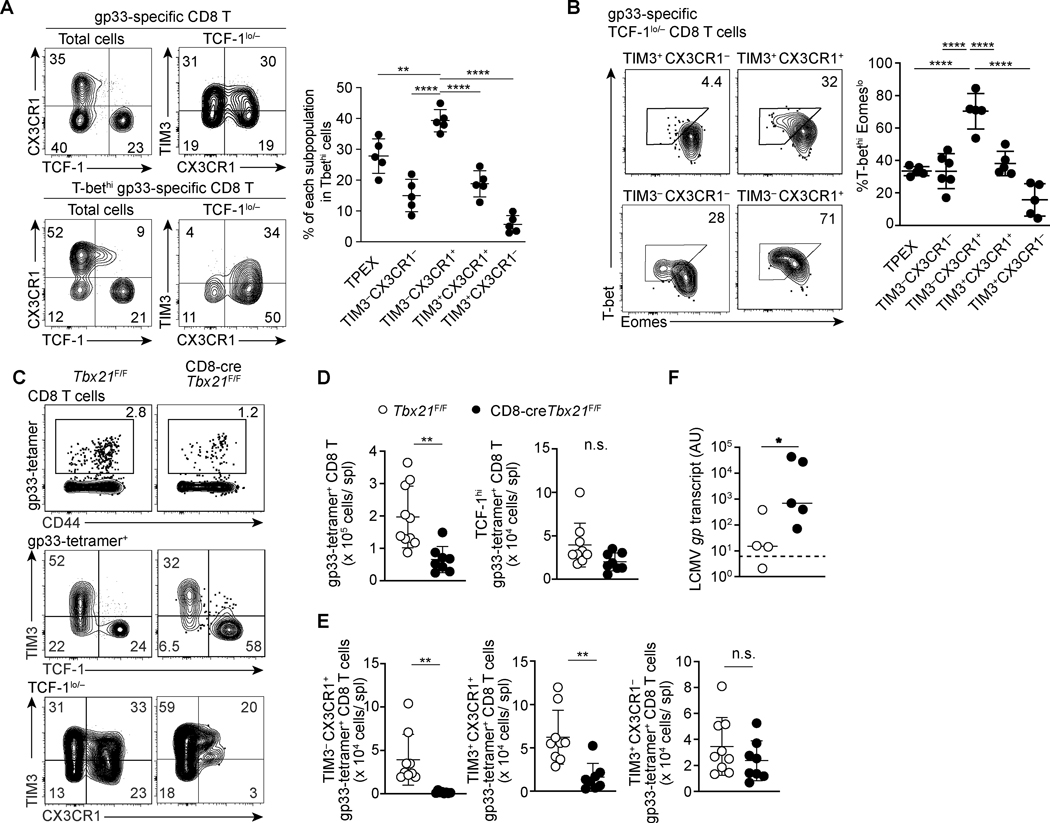
To more rigorously test this possibility that the TIM3− CX3CR1+ population overlaps the previously identified T-bethi progenitor population (11), we first examined the proportion of each subset with T-bethi cells by flow cytometry. Indeed, TIM3− CX3CR1+ cells comprised approximately 40% of total T-bethi cells followed by the TCF-1hi population (30%) (Fig. 3A). Co-staining for T-bet and EOMES showed a marked difference in T-bet and EOMES expression between the two CX3CR1+ populations (Fig. 3B). Notably, the TIM3− CX3CR1+ subset was predominantly composed of T-bethi cells (~70%) compared to the rest of exhausted CD8 T cell subsets, in which a majority of cells were EOMEShi but T-betlo (Fig. 3B). Together with low PD-1 expression, basal expression of TPEX-associated transcription factors, low proliferative activity, and capacity for differentiation to TIM3+ effector/exhausted cells, our data suggest that TIM3− CX3CR1+ CD8 T cells overlap with the previously reported T-bethi progenitor cells (11).
FIGURE 3. T-bet is critical for maintenance of CX3CR1+ exhausted CD8 T cell populations.
(A) Flow cytometry plots showing expression of indicated molecules within total and T-bethi gp33-specific CD8 T cells 22dpi LCMV-c13 infection. Data are representative of 3 independent experiments with n=4–6 mice each. (B) Flow cytometry plots showing expression of T-bet and Eomes in the indicated subsets of gp33-specific CD8 T cells in the spleen on 22 dpi with LCMV-c13 infection. Data are representative of 3 independent experiments with n=5 mice each. (C-E) Flow Cytometry showing expression of TCF-1, CX3CR1, and TIM3 by gp33-specific CD8 T cells in control Tbx21F/F and CD8-cre Tbx21 F/F mice on 29 dpi with LCMV-c13. Representative flow cytometry plots are shown in (C) with frequencies of gated cells in each parental population, and statistical analyses are shown in (D) and (D). Data are shown as mean ±SD and are pooled from 3 independent experiments with n>8 mice per genotype. Dots in the graphs in (A), (C) and (D) indicate individual mice. Data in (A), (C) and (D) are shown as mean ± SD. Statistical differences in (A) were tested using one-way ANOVA with a Tukey post-hoc test. For (C) student’s t-test was used. And for (D) Mann-Whitney test was used. (F) LCMV-gp mRNA abundance in plasma as assessed by qRT-PCR in Tbx21F/F and CD8-cre Tbx21F/F mice on 100 dpi with LCMV-c13. Data are pooled from two independent experiments with n=4 (Tbx21F/F) and n=5 (CD8-cre+ Tbx21F/F) mice per genotype in total. Horizontal bars indicate median for samples in each genotype and the statistical differences were assessed by Mann-Whitney U-test. The dotted line indicates the limit of detection.
The loss of T-bet in T cells during chronic infection has been associated with more severe exhaustion (11, 30). To determine whether T-bet is required for TPEX, T-bethi TIM3− CX3CR1+ cells or both, we analyzed CD8 T cell responses to LCMV-c13 infection in mice with CD8 T cell-specific Tbx21 deletion. During the chronic phase of LCMV infection (29 dpi), frequencies of total LCMV gp33-specific CD8 T cells in the spleen were reduced by 2.5-fold in CD8-cre Tbx21F/F mice compared to control cre(–) Tbx21F/F mice (Fig. 3C, D). The absolute number of TPEX cells did not significantly change by CD8 T cell-specific deletion of Tbx21. In contrast, the reduction of total antigen-specific CD8 T cells was attributed to marked reduction of CX3CR1+ CD8 T cells, most notably to that of TIM3− CX3CR1+ cells that expressed the highest amounts of T-bet among all fractions (Fig. 3B, D). We also observed a decreased frequency and absolute number of TIM-3+ CX3CR1+ effector-like cells, which could reflect loss of input from TIM3− CX3CR1+ cells, or defective differentiation of TPEX cells. Consistent with previous studies (11, 31, 32), ex vivo stimulation of splenocytes from Tbx21 deficient mice showed preserved granzyme B expression, with decreased IFN-γ expression in CD8 T cells, which likely contributes to defective viral control (Fig. S4A-D). To determine the impact of the loss of CX3CR1+ CD8 T cells on viral control, we tracked LCMV-c13 infected CD8-cre+ Tbx21F/F and control Tbx21F/F mice for 100 days, by which time the majority of immune-competent mice resolve viremia (33). While control mice were able to resolve viremia, Tbx21-deficient mice still demonstrated significant viremia (Fig. 3E). These results indicate that T-bet is required for the maintenance of CX3CR1+ cells despite a minimal impact on TCF-1hi cells.
Eomes and Bach2 antagonistically control differentiation of TPEX into effector-like exhausted CD8 T cells
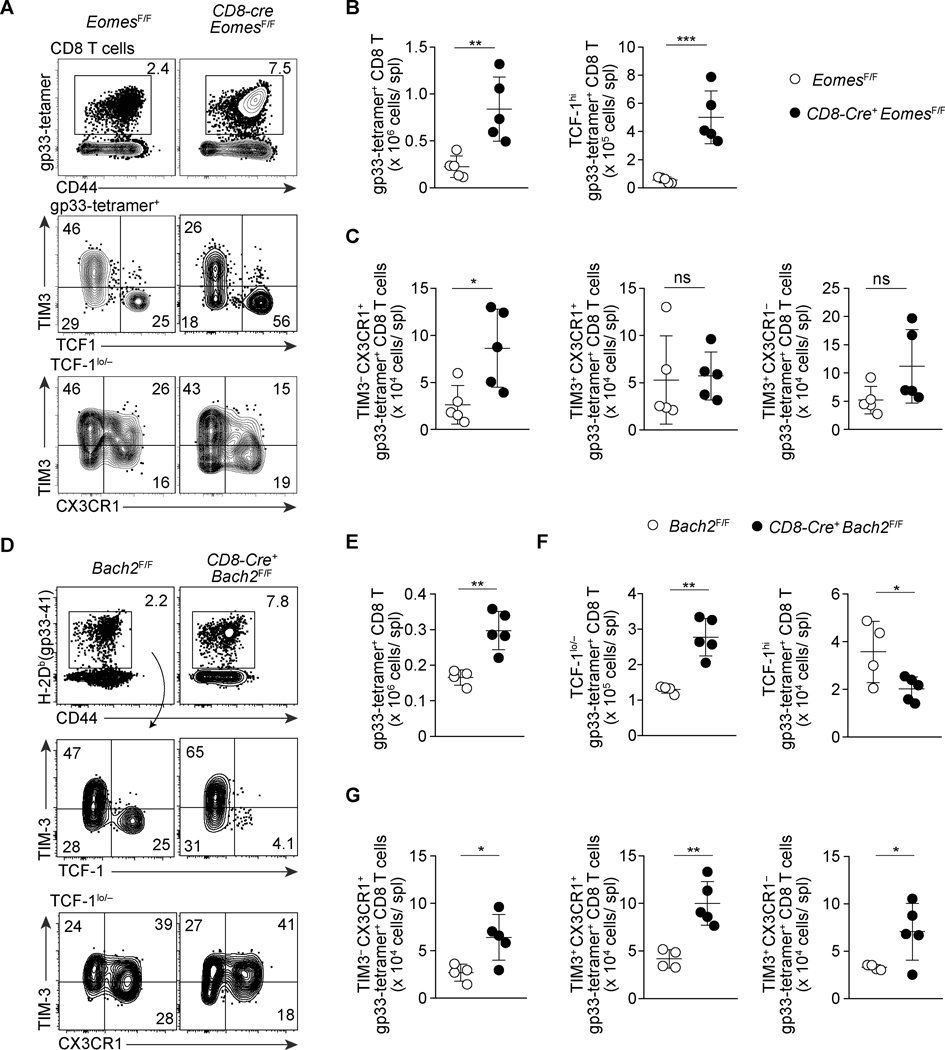
We next defined the relationship between the TCF-1hi and CX3CR1+ populations by examining the function of transcription factors that are highly expressed in and potentially important for differentiation of the TPEX population. Based on our gene expression data (Fig. 2B), we first examined the role of Eomes in the differentiation of CD8 T cells during the chronic phase of LCMV infection. Eomes expression has been associated with severe exhaustion and its loss results in reduced antigen-specific CD8 T cells numbers (11, 34). We thus anticipated that Eomes deletion would result in a reduction in TCF-1hi cells. Contrary to our prediction, in CD8-cre+EomesF/F mice, there was a 3-fold increase in both frequency and absolute numbers of total LCMV-gp33-specific CD8 T cells (Fig. 4A, B). We suspect our result is discrepant with prior results of Eomes deficiency possibly due additional deletion of Eomes in CD4 T cells using CD4-cre in prior studies, whereas the increased proportion of TPEX is consistent with the reported reduced Prdm1 expression in antigen-specific CD8 T cells (11, 35). This increase was most attributable to a >7-fold increase in TPEX cells (Fig. 4B). In addition, TIM3− CX3CR1+ CD8 T cells were increased by 3-fold (Fig. 4A, C). However, we did not observe a change in the total TCF-1lo/– PD-1+ CD8 T cells or TIM3+ CX3CR1+ CD8 T cells, and within TCF-1lo/– cells TIM3 cells were increased in number (Fig. 4A, C). Increased numbers of the two quiescent subpopulations of exhausted CD8 T cells in Eomes deficiency suggest that EOMES promotes the development of TPEX cells after T cell activation and/or represses the differentiation of progenitor-like CD8 T cells, including TPEX and TIM3− CX3CR1+ cells, to downstream effector cells.
FIGURE 4. Opposing roles of EOMES and BACH2 in the differentiation of progenitor-like subsets of exhausted CD8 T cells.
(A-C) Flow Cytometry showing expression of CD44, TCF-1, CX3CR1, and TIM3 of gp33-tetramer+ CD8 T cells in the spleen of control EomesF/F and CD8-cre EomesF/F mice on 28 dpi with LCMV-c13. Representative flow cytometry plots are shown in (A) with frequencies of gated cells in each parental population, and statistical analyses are shown in (B) and (C). Data are representative of 3 independent experiments with n=4–6 mice per genotype in each experiment. (D-G) Flow Cytometry showing expression of TCF-1, CX3CR1, and TIM3 of gp33-tetramer+ CD8 T cells in Bach2F/F and CD8-Cre+Bach2F/F mice 30 days after LCMV-c13 infection. Representative flow cytometry plots are shown in (D) with frequencies of gated cells in each parental population, and statistical analyses are shown in (E)-(G). Data are pooled from two independent experiments with n=4 (Bach2F/F) and n=5 (CD8-Cre+Bach2F/F) in total. All statistical analysis plots are shown as mean ± SD with statistics performed using an unpaired t-test.
As a consequence of altered differentiation, turnover of downstream effector cells derived from TPEX cells or TIM3− CX3CR1+ cells may be impaired and fitness of these effector cells could be compromised in Eomes-deficient mice. To test this possibility, we first examined IFN-γ and TNF-α production by CD8 T cells following stimulation of splenocytes with LCMV-specific peptides ex vivo, which showed no difference between Eomes-deficient and control CD8 T cells (Fig. S4E, F). However, we observed significant reduction of GZMB expression in Eomes-deficient antigen-specific CD8 T cells (Fig. S4G, H). Consistent with the defects in differentiation of PD-1+ CD8 T cells, the majority of CD8-cre EomesF/F mice failed to resolve viremia on ≥100 dpi (Fig. S4I). These results together indicate unique roles of the two T-box proteins, T-bet and EOMES, in CD8 T cell responses to persisting antigen, and a specific role for T-bet in the maintenance of CX3CR1 expressing cells.
Bach2 is another transcription factor highly expressed specifically in TPEX cells. CD8 T cell-specific deletion of Bach2 resulted in a similar increase in the number of gp33-specific CD8 T cells to Eomes deletion (Fig. 4D-G). However, the overall increase was attributable to TCF-1lo/– cells with a marked reduction in the frequency of TPEX as well as absolute numbers. In TCF-1lo/– cells, contrary to Eomes deficient mice, the most notable increase was seen in the TIM3+ CX3CR1+ proliferating population. These results indicate that, although both EOMES and BACH2 are highly expressed in the TPEX population, their roles may be antagonistic with BACH2 protecting TPEX cells from differentiation into the downstream effector cells.
CX3CR1+ CD8 T cells are largely self-sustainable with limited input from TPEX cells
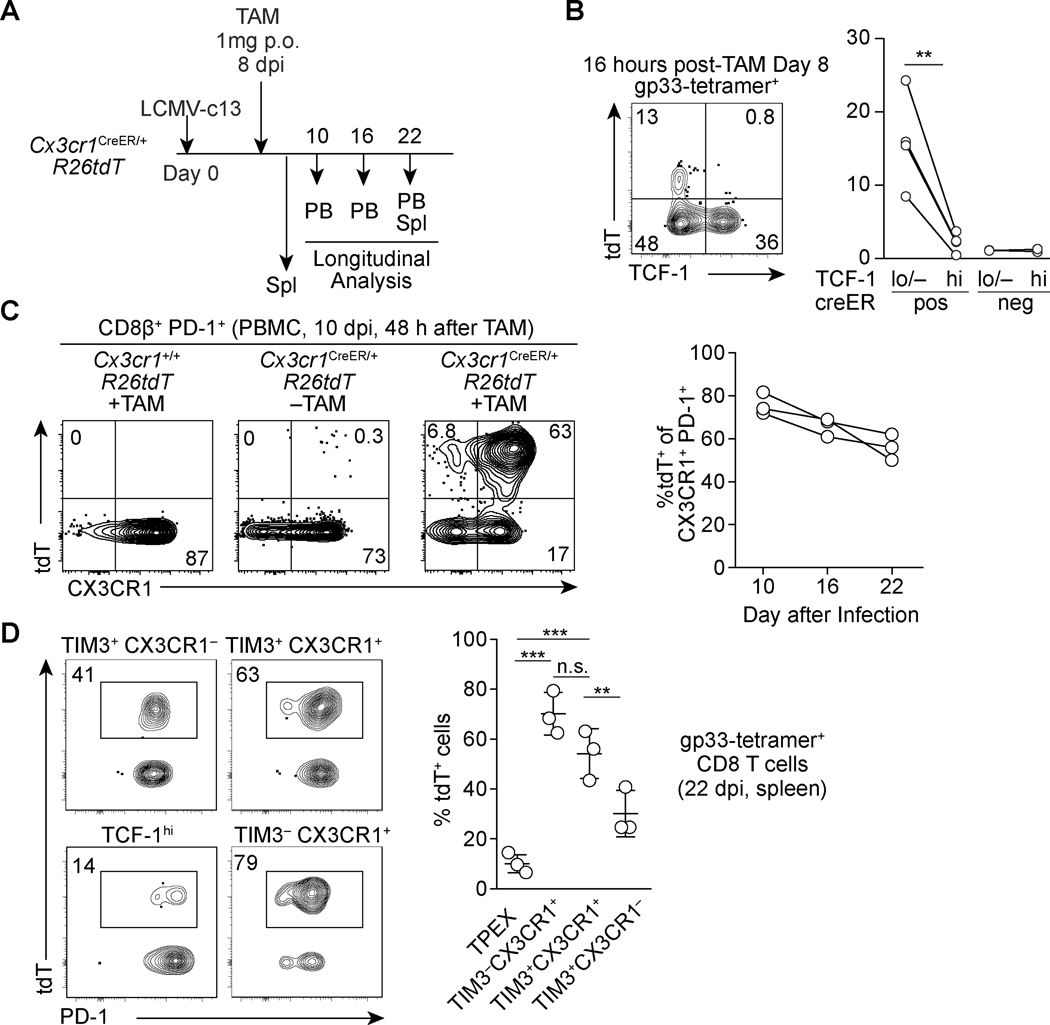
We next sought to measure the turnover of the CX3CR1+ cells by TCF-1hi TPEX-derived cells by performing genetic-labeling of CX3CR1+ cells by utilizing the Cx3cr1-creERT2 allele crossed to the ROSA26-loxP-stop-loxP-tdTomato (R26tdT) reporter (Fig. 5A). 16 hours after one dose of tamoxifen (TAM) on 8 dpi showed labeling of 10~25% of TCF-1lo/– CD8 T cells in contrast to little labeling of TPEX cells in the spleen, demonstrating specific, low background labeling of TPEX cells by the TAM-inducible cre activity (Fig. 5B). 48 hours after TAM administration, approximately 80% of CX3CR1+ CD8 T cells in the peripheral blood became tdT-positive (Fig. 5C). Given that TPEX cells can differentiate into downstream effector and exhausted CD8 T cells, we expected to observe robust displacement of tdT+ cells by TPEX-derived tdT− cells within the CX3CR1+ populations over the course of the antiviral response. However, over a two-week chase period, when TAM is no longer bioavailable in vivo (36, 37), the frequencies of tdT+ cells in the CX3CR1+ CD8 T cells were preserved with only a modest decay to 60% on 22 dpi (Fig. 5C). In the spleen, both TIM3− and TIM3+ CX3CR1+ cells retained high percentages of tdT+ cells, while ~30% of TIM3+ CX3CR1− cells were labeled with tdT (Fig. 5D), suggesting some transition from CX3CR1+ into CX3CR1− cells. These results highlight a slow turnover of CX3CR1+ cells, which could reflect a long-population half-life, or be consistent with a relatively minor contribution of TPEX cells to the pool of TIM3− and TIM3+ CX3CR1+ exhausted CD8 T cell pool.
FIGURE 5. Lineage tracing reveals stability and persistence of CX3CR1+ PD-1+ CD8 T cells.
(A) Experimental schematic of tamoxifen administration to Cx3cr1creERT2/+ R26tdT mice infected with LCMV-c13 and analysis. (B) Flow cytometry plots showing expression of tdT and TCF-1 in gp33-specific CD8 T cells 16 hours following tamoxifen administration on 8 dpi. Data from replicates are shown in the right panel. Data are representative of 2 independent experiments n=3–4 mice each. A paired t-test for the statistical analysis. (C) A representative flow cytometry plots showing expression of tdT and CX3CR1 in PD-1+ CD8 T cells in peripheral blood mononuclear cells, and frequencies of tdT+ cells within CX3CR1+ PD-1+ CD8 T cells in the PBMC in Cx3cr1CreER/+ R26tdT mice at indicated time points after infection and tamoxifen administration. Data are representative of 2 independent experiments with n=3 mice each. (D) Flow cytometry plots showing expression of the tdT reporter in each of gp33-specific CD8 T cell subpopulations in the spleen of Cx3cr1creERT2/+ R26tdT mice on 22 dpi. Data are representative of 2 independent experiments with n=3 mice each.
CX3CR1+ cells exhibit longevity following viral clearance with capability to respond to antigen rechallenge
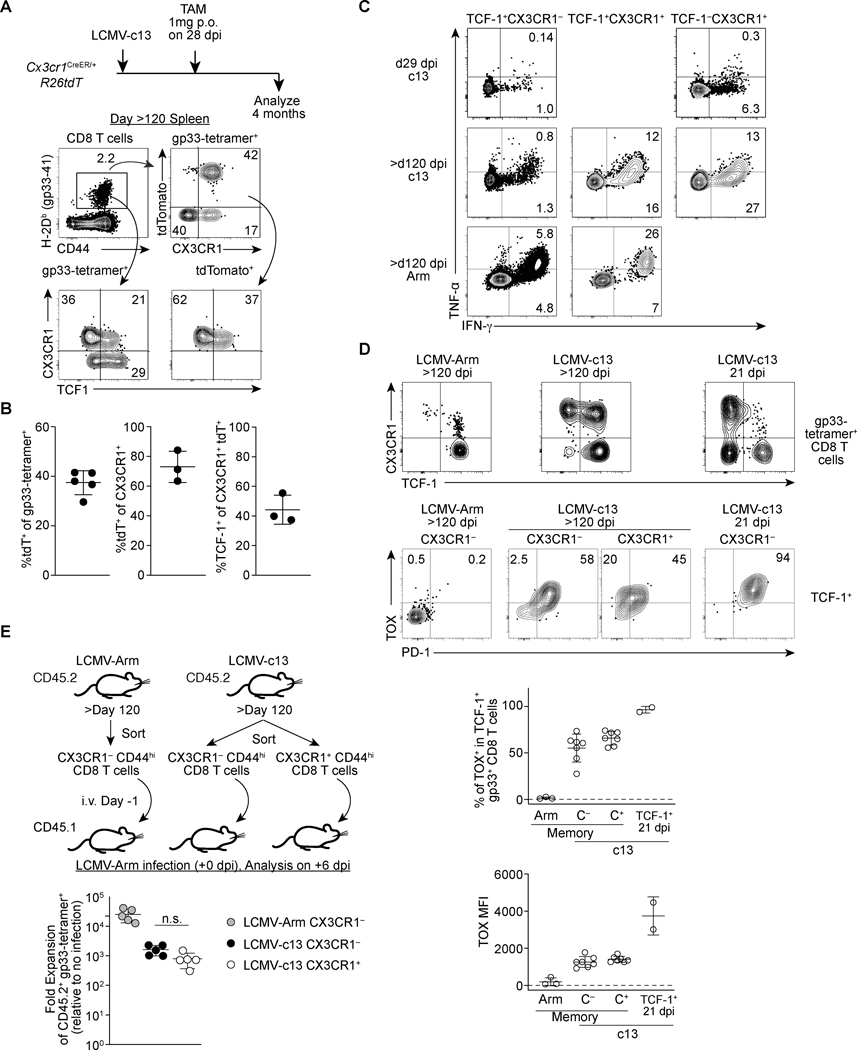
In immunocompetent mice, LCMV-c13 viremia is eventually resolved by both T and B cell responses around 3 months post-infection (16, 33, 38). We next sought to determine the long-term fate of CX3CR1+ cells as the TIM3− CX3CR1+ subset expressed low but detectable levels of the memory-related factors, Tcf7. To this end, we administered TAM to Cx3cr1-creERT2 R26tdT mice during the chronic phase of infection on 28 dpi and chased the tdT-labeled cells for 120 days. Similar to acute LCMV infection, we were able to detect the persistence of LCMV-gp33-specific CD8 T cells expressing intermediate levels of PD-1 in the peripheral blood mononuclear cells and in the spleen of LCMV-c13-immune mice (Fig. 6A). These cells likely correspond to “chronic memory” cells or “memory-like” cells that were previously reported (14, 39). In contrast to LCMV-Arm-immune mice, in which TCF-1hi memory CD8 T cells were mostly CX3CR1−, a substantial fraction of gp33-specific chronic memory CD8 T cells in LCMV-c13-infected mice showed an effector-memory phenotype with expression of CX3CR1 (Fig. 6A). TAM administration to LCMV-c13-infected Cx3cr1-creERT2 fate-mapping mice on 28 dpi resulted in marking of ~40% of LCMV-gp33-specific CD8 chronic memory T cells on 120 dpi (Fig. 6A, B). Almost all tdT+ cells continued to express surface CX3CR1 and ~40% of the tdT+ cells expressed the memory transcription factor TCF-1 at the levels slightly lower than the CX3CR1− chronic memory cells (Fig. 6A, B). The majority (~80%) of CX3CR1+ chronic memory CD8 T cells were also tdT+ and thus were derived from CX3CR1+ cells during the labeling period (Fig. 6B). In contrast to PD-1+ CD8 T cells during high antigen burden, the CX3CR1+ and CX3CR1− chronic memory CD8 T cells were able to produce considerable IFN-γ following stimulation with LCMV-gp33 ex vivo (Fig. 6C). Notably, approximately 50% of IFN-γ+ cells also expressed TNF-α, indicating partial restoration of poly-functionality in these chronic memory CD8 T cell populations (Fig. 6C). The transcription factor TOX has been closely linked to CD8 T cell exhaustion in the context of antiviral and anti-tumor immunity and is implicated in programming the unique genetic and epigenetic signatures associated with chronic antigen stimulation (40–44). Interestingly, the vast majority of chronic memory CD8 T cells continued to express TOX (Fig. 6D). Their TOX expression was lower than that of TCF-1+ progenitors during active infection, which mirrors continued expression of low PD-1 by chronic memory CD8 T cells (39). Furthermore, both CX3CR1+ and CX3CR1− chronic memory CD8 T cells from LCMV-c13-infected mice were capable of secondary expansion following adoptive transfer to congenic WT mice followed by LCMV-Arm infection (Fig. 6E). Since CX3CR1+ TCF-1+ cells barely expressed TIM3 after the resolution of viremia (data not shown), it is likely that a substantial fraction of CX3CR1+ chronic memory CD8 T cells is derived from the TCF-1lo/– TIM3− CX3CR1+ cells or a minor population of TPEX, which had turned on expression of Cx3cr1 as they initiated differentiation towards TCF-1lo/– cells. These results collectively indicate a substantial fraction of antigen-experienced TCF-1+ cells are derived from exhausted T cells that initiated differentiation away from TPEX, suggesting the plasticity for long-term persistence, polyfunctionality and secondary expansion after chronic exposure to antigen and Tcf7 downregulation.
FIGURE 6. CX3CR1+ CD8 T cells during the chronic phase give rise to TCF-1+ chronic memory cells following the resolution of viremia.
(A-B) Flow cytometry plots showing expression of tdTomato (tdT), TCF-1, CX3CR1 and TIM3 in gp33-specific CD8 T cells in the splenocytes of LCMV-c13-infected Cx3cr1creERT2/+ R26tdT mice treated with Tamoxifen on 28 dpi and analyzed on >120 dpi. Representative data from two independent experiments with n=3 mice is shown. Flow plots are shown in (A) and replicates from one experiment are shown as mean ± SD. (B). (C) Expression of IFN-γ and TNF-α in indicated subsets of CD8 T cells in the spleen of 29 dpi LCMV-c13-infected mice, >120 dpi LCMV-Armstrong-infected mice, or >120 dpi LCMV-c13-infected mice following stimulation with LCMV-gp33 peptide ex vivo for four hours. Data are representative of two independent experiments with n=XX mice each (D) Expression of TCF-1 and CX3CR1 in LCMV-gp33-specific splenic CD8 T cells (top) and expression of PD-1 and TOX in TCF-1+ LCMV-gp33-specific CD8 T cells at indicated time point after LCMV-Arm or LCMV-c13 infection. Frequencies of TOX+ cells and levels of TOX expression are shown with mean ± SD. (E) Schematic of adoptive transfer experiment to assess the capability of secondary expansion of CX3CR1+ and CX3CR1− CD8 T cells that were sorted from LCMV Arm and LCMV-c13 infected mice. The right panel shows data from replicates. Data are representative from 2 independent experiments with n=2 donor mice and n=5 recipient mice per experiment. Fold expansion was calculated by fold change of gp33-specific CD8 T cells followed by normalization to no-infection control.
Discussion
The mechanisms underlying the maintenance of functional CD8 T cell responses in the context of persistent antigen exposure remains unresolved. A substantial body of work has identified various subsets of exhausted CD8 T cells with distinct phenotypic and gene expression profiles. However, how these populations interact and ultimately promote control of viral or tumor burden remains an active area of investigation. Here, we identify a TIM-3 negative subset of PD-1lo CD8 T cells within the CX3CR1+ population which is highly enriched for T-bethi cells. This population is quiescent, exhibits low TCF-1 expression, but also demonstrates features of effector-like cells. These results highlight underappreciated heterogeneity within exhausted CD8 T cells and provide a more complete framework for understanding the dynamics of T cell responses to chronic antigen.
Prior work has established that TCF-1hi TPEX cells act as progenitors for BLIMP-1+ effector-like and terminally exhausted CD8 T cells. In comparison, an analogous relationship was described between T-bethi cells (progenitor) and Eomeshi cells (progeny) which become fully exhausted as Eomes expression elevates (11, 30). However, the nature of the T-bethi progenitor cells has remained unclear since TCF-1hi TPEX cells are predominantly T-betlo EOMEShi (4). Two recent papers described a subset of PD-1+ CD8 T cells enriched for CX3CR1 expression and T-bet (9, 10). These cells are highly proliferative and express effector molecules, which contributes to control of chronic viral infection and tumors. They also overlap with another subpopulation of exhausted CD8 T cells, Ly108 (SLAMF6)− CD69− TEX-int cells, which are proposed to be a transitory population between TPEX and terminally exhausted cells (45). Our work has identified a TIM3-negative subpopulation within the CX3CR1+ population, which is almost exclusively T-bethi cells and distinct from the transitory populations in their cell cycle status and gene expression. The TIM3− cells are phenotypically similar to TPEX with their quiescence, basal expression of TCF-1 and low levels of effector gene expression. Since each study used different markers to define populations and phenotypic changes, further work will be necessary to reconcile the discrepancies between the studies. In addition, while not a focus of this study, we identified a TIM3− CX3CR1− TCF1lo/– population whose function and development remain yet to be determined. This population was resistant to loss of CD4 T cell help and exhibited low proliferation. We speculate this could be an intermediate to TIM3+ CX3CR1− cells, which may develop from TPEX or naive CD8 T cells independent of CX3CR1+ cells.
A key question that remains unsettled is the developmental trajectory and nature of the progenitor-progeny relationships between the exhausted CD8 T cell subsets. The external cues that drive this process remain incompletely defined, however, a role for type I interferons and IL-12 in promoting differentiation has been proposed (6, 46, 47).This issue has been largely studied in the setting of adoptive transfer of various subsets to congenic recipients. Indeed, adoptively transferred TPEX cells exhibit higher proliferative potential than TCF-1− or CXCR5− PD-1+ CD8 T cells and can give rise to TIM3+ differentiated progeny (4–6). However, it should be noted that the behavior of transferred cells may not faithfully reflect the kinetics of this transition in situ due to inefficient engraftment of transferred cells that are removed from their native microenvironment. This issue is also seen in long-term hematopoietic stem cells, which show limited contribution of homeostatic hematopoiesis although they are capable of robust expansion and reconstitution of cells in multiple lineages (48–50). This may also be the case in exhausted CD8 T cells as exemplified by loss of quiescence of TPEX cells upon adoptive transfer and limited proliferative activity of TIM3+ CX3CR1+ cells in our study and Tex-int cells in another study (45). Here, we used an in vivo lineage-tracing approach to overcome the limitation, and found that displacement of pulse-labeled CX3CR1+ PD-1+ CD8 T cells by newly generated cells derived from TPEX or naive CD8 T cells occurs slowly with a half-life longer than a few weeks. This result suggests that, although transferred TPEX cells robustly give rise to CX3CR1+ and CX3CR1− effector/exhausted CD8 T cells, they are mostly quiescent in situ and do not rapidly replace downstream differentiated cells. This is consistent with low BrdU incorporation and KI67 expression as well as a pseudotime modeling of our scRNA-sequence data. We recognize two caveats which need to be considered. First, we cannot rule out the possibility that a small fraction of TCF-1hi cells that were labeled with tdT, instead of TCF-1lo/– CX3CR1+ cells, replenish the CX3CR1+ CD8 T cell pool. However, this result also suggests that the vast majority of TPEX except for those with poised Cx3cr1 expression are dormant with limited contribution to TCF-1lo/– cells in situ, which is largely consistent with our interpretation. Secondly, future work is necessary to evaluate TIM3− CX3CR1+ cells recruitment to TIM3+ CX3CR1+ cells which is technically limited due to the lack of a specific cre driver. Nevertheless, given the similarity to TPEX, TIM3− CX3CR1+ cells may function as intermediate progenitors for TIM3+ CD8 T cells with limited self-renewing capabilities, like short-term HSPCs, which are still dependent on largely dormant LT-HSCs but mainly support homeostasis of hematopoiesis (48, 49).
The antigen-specific cells present in the host after resolution of viremia no longer demonstrate high expression of PD-1 and have been termed as “chronic memory” (14). Using our CX3CR1 lineage-tracing approach we were able to track cells labelled during the chronic phase of the infection, and found that a substantial portion of TCF-1+ chronic memory cells are labelled by previous expression of CX3CR1 cells. The labelled cells are exclusively found in CX3CR1+ chronic memory cells, suggesting earlier activation of Cx3cr1 imprints its continued expression, which is consistent with trajectories of CX3CR1+ cells and TIM3− CX3CR1− TCF-1lo/– cells are mostly segregated during chronic phase. While we cannot formally rule out that these labelled cells were derived from a small proportion of labelled TPEX cells, it is unlikely this would be the case based on a much larger fraction of TCF-1+ chronic memory cells being labeled by tdT. A previous study demonstrated that memory cells that have experienced chronic antigen stimulation retain persistent PD-1 expression (39). Likewise, we found that TCF-1+ cells generated through LCMV-c13 infection retain persistent TOX expression, similar to effector memory cells or anti-CMV “memory” CD8 T cells in humans (12, 13, 51, 52). These findings suggest that cells other than TPEX or central memory-like cells possess the capacity to retain or re-express TCF-1 under certain conditions and highlighted additional heterogeneity of TCF-1+ CD8 memory T cells that may be imprinted by the kinetics of antigen stimulation or inflammatory milieu. An equivalent type of memory T cell following cancer elimination is likely (53), which may be important to prevent relapse. It remains to be determined how this memory cell type differs from CX3CR1− memory cells through the acquisition of imprinted identity during the effector phase. This knowledge will potentially transform into methods that enhance long-term immunity against chronic viral infection or cancers.
Supplementary Material
Fig. S1. Characterization of CX3CR1 expressing CD8 T cells in infection
Fig. S2. Development and dynamics of CX3CR1 positive subsets of exhausted CD8 T cells
Fig. S3. Profiling of differentially expressed genes between distinct subpopulations of PD-1+ CD8 T cells following LCMV-c13 infection by bulk RNA-seq
Fig. S4. Characterization of CD8 T cell-specific Tbx21- and Eomes-deficient mice during chronic LCMV infection
Key Points.
CX3CR1+ PD-1+ CD8 T cells contain a quiescent subset with progenitor-like features.
TIM3−CX3CR1+ cells are intermediates during a transition of TPEX to CX3CR1+ cells.
TCF-1+ TOXlo memory-like cells are derived from CX3CR1+ exhausted CD8 T cells.
Acknowledgements
We thank M. Colonna and M. Cella for LCMV stocks, K. Kim and G. Randolph for Cx3cr1-creERT2 mice, C. Fuji and T. Yangdon for the maintenance of the mouse colony, and C-S. Hsieh for discussion and critical reading of the manuscript.
Funding sources: This study was supported by NIH grants R01AI130152-01A1 and R03AI139875-01 (to T.E.), T32HL007317 (to S.R.), T32GM007200 (to S.R. and D.J.V.), and K08CA230188 (A.T.S). T.E. is a Scholar of the Leukemia and Lymphoma Society (www.lls.org). A.T.S. was supported by a Bridge Scholar Award from the Parker Institute for Cancer Immunotherapy, a Career Award for Medical Scientists from the Burroughs Wellcome Fund, and the Cancer Research Institute Technology Impact Award.
Declaration of Interests
A.T.S. is a scientific founder of Immunai. A.T.S. received funding support from 10x Genomics and Arsenal Biosciences.
Abbreviation used:
- dpi
days post infection
- HSC
hematopoietic stem cells
- LCMV
Lymphocytic choriomeningitis virus
- LCMV-Arm
LCMV-Armstrong
- LCMV-c13
LCMV clone 13
- LT-HSC
Long-term HSC
- PB
peripheral blood
- PCA
Principal Component Analysis
- R26tdT
ROSA26-loxP-stop-loxP tdT reporter
- RNA-seq
RNA sequencing
- scRNA-seq
single cell RNA-seq
- TAM
tamoxifen
- tdT
tandem dimer Tomato
- TPEX
progenitor exhausted T cells
- UMAP
Uniform Manifold Approximation and Projection
Footnotes
Data Availability: RNA-seq data are deposited to NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE145910 and GSE148497. Data will be made publicly accessible upon acceptance of the manuscript.
References:
- 1.Gallimore A, Hombach J, Dumrese T, Rammensee HG, Zinkernagel RM, and Hengartner H. 1998. A protective cytotoxic T cell response to a subdominant epitope is influenced by the stability of the MHC class I/peptide complex and the overall spectrum of viral peptides generated within infected cells. Eur. J. Immunol 28: 3301–3311. [DOI] [PubMed] [Google Scholar]
- 2.Zajac AJ, Murali-Krishna K, Blattman JN, and Ahmed R. 1998. Therapeutic vaccination against chronic viral infection: the importance of cooperation between CD4+ and CD8+ T cells. Curr. Opin. Immunol 10: 444–449. [DOI] [PubMed] [Google Scholar]
- 3.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, Bosinger SE, Paiardini M, Chahroudi A, Vanderford TH, Estes JD, Lifson JD, Derdeyn CA, and Silvestri G. 2016. CD8(+) Lymphocytes Are Required for Maintaining Viral Suppression in SIV-Infected Macaques Treated with Short-Term Antiretroviral Therapy. Immunity 45: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, Sharpe AH, Freeman GJ, Germain RN, Nakaya HI, Xue H-H, and Ahmed R. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, Pradervand S, Thimme R, Zehn D, and Held W. 2016. T Cell Factor 1-Expressing Memory-like CD8+ T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45: 415–427. [DOI] [PubMed] [Google Scholar]
- 6.Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, Wu W, Lang PA, Gattinoni L, McGavern DB, and Schwartzberg PL. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Science immunology 1: eaai8593–eaai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, Zotos D, Fenix KA, Atnerkar A, Preston S, Chipman JG, Beilman GJ, Allison CC, Sun L, Wang P, Xu J, Toe JG, Lu HK, Tao Y, Palendira U, Dent AL, Landay AL, Pellegrini M, Comerford I, McColl SR, Schacker TW, Long HM, Estes JD, Busslinger M, Belz GT, Lewin SR, Kallies A, and Yu D. 2016. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nature Publishing Group 17: 1187–1196. [DOI] [PubMed] [Google Scholar]
- 8.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, and Ye L. 2016. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature 537: 412–428. [DOI] [PubMed] [Google Scholar]
- 9.Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin J-X, Konieczny BT, Im SJ, Freeman GJ, Leonard WJ, Kissick HT, and Ahmed R. 2019. Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity 51: 1043–1058.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, and Cui W. 2019. CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51: 1028–1042.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, and Wherry EJ. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338: 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galletti G, De Simone G, Mazza EMC, Puccio S, Mezzanotte C, Bi TM, Davydov AN, Metsger M, Scamardella E, Alvisi G, De Paoli F, Zanon V, Scarpa A, Camisa B, Colombo FS, Anselmo A, Peano C, Polletti S, Mavilio D, Gattinoni L, Boi SK, Youngblood BA, Jones RE, Baird DM, Gostick E, Llewellyn-Lacey S, Ladell K, Price DA, Chudakov DM, Newell EW, Casucci M, and Lugli E. 2020. Two subsets of stem-like CD8+ memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol 21: 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine T, Perez-Potti A, Nguyen S, Gorin J-B, Wu VH, Gostick E, Llewellyn-Lacey S, Hammer Q, Falck-Jones S, Vangeti S, Yu M, Smed-Sörensen A, Gaballa A, Uhlin M, Sandberg JK, Brander C, Nowak P, Goepfert PA, Price DA, Betts MR, and Buggert M. 2020. TOX is expressed by exhausted and polyfunctional human effector memory CD8+ T cells. Sci Immunol 5. [DOI] [PubMed] [Google Scholar]
- 14.Wherry EJ, Barber DL, Kaech SM, Blattman JN, and Ahmed R. 2004. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc. Natl. Acad. Sci. U. S. A 101: 16004–16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kometani K, Nakagawa R, Shinnakasu R, Kaji T, Rybouchkin A, Moriyama S, Furukawa K, Koseki H, Takemori T, and Kurosaki T. 2013. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity 39: 136–147. [DOI] [PubMed] [Google Scholar]
- 16.Chou C, Verbaro DJ, Tonc E, Holmgren M, Cella M, Colonna M, Bhattacharya D, and Egawa T. 2016. The Transcription Factor AP4 Mediates Resolution of Chronic Viral Infection through Amplification of Germinal Center B Cell Responses. Immunity 45: 570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCausland MM, Yusuf I, Tran H, Ono N, Yanagi Y, and Crotty S. 2007. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. The Journal of Immunology 178: 817–828. [DOI] [PubMed] [Google Scholar]
- 18.Fooksman DR, Nussenzweig MC, and Dustin ML. 2014. Myeloid cells limit production of antibody-secreting cells after immunization in the lymph node. J. Immunol 192: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. 2019. Comprehensive Integration of Single-Cell Data. Cell 177: 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin J-R, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CGK, Kazer SW, Gaillard A, Kolb KE, Villani A-C, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jané-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, and Garraway LA. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW. 2018. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. [DOI] [PubMed] [Google Scholar]
- 22.McInnes L, Healy J, and Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [stat.ML]. [Google Scholar]
- 23.Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C, and Buettner F. 2016. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32: 1241–1243. [DOI] [PubMed] [Google Scholar]
- 24.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, Manos M, Gjini E, Kuchroo JR, Ishizuka JJ, Collier JL, Griffin GK, Maleri S, Comstock DE, Weiss SA, Brown FD, Panda A, Zimmer MD, Manguso RT, Hodi FS, Rodig SJ, Sharpe AH, and Haining WN. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol 20: 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, and Rosenberg SA. 2009. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114: 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blackburn SD, Crawford A, Shin H, Polley A, Freeman GJ, and Wherry EJ. 2010. Tissue-specific differences in PD-1 and PD-L1 expression during chronic viral infection: implications for CD8 T-cell exhaustion. J. Virol 84: 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung YW, Rutishauser RL, Joshi NS, Haberman AM, and Kaech SM. 2010. Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J. Immunol 185: 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan T, Dominguez CX, Amezquita RA, Laidlaw BJ, Cheng J, Henao-Mejia J, Williams A, Flavell RA, Lu J, and Kaech SM. 2018. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+T cell fates. J. Exp. Med 215: 1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, and Others. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med 212: 2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali M-AA, Intlekofer AM, Boss JM, Reiner SL, Weinmann AS, and Wherry EJ. 2011. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat. Immunol 12: 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, and Reiner SL. 2008. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science 321: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, and Rao A. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med 206: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, and Ahmed R. 2003. Viral Persistence Alters CD8 T-Cell Immunodominance and Tissue Distribution and Results in Distinct Stages of Functional Impairment. J. Virol 77: 4911–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, He Y, Hao J, Ni L, and Dong C. 2018. High Levels of Eomes Promote Exhaustion of Anti-tumor CD8+ T Cells. Front. Immunol 9: 2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, Yu S, Kurachi M, Herati RS, Vella LA, Baxter AE, Wu JE, Khan O, Beltra J-C, Giles JR, Stelekati E, McLane LM, Lau CW, Yang X, Berger SL, Vahedi G, Ji H, and Wherry EJ. 2019. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 51: 840–855.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson CH, Gamper I, Perfetto A, Auw J, Littlewood TD, and Evan GI. 2014. The kinetics of ER fusion protein activation in vivo. Oncogene 33: 4877–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson SP, Langan-Fahey SM, Johnson DA, and Jordan VC. 1991. Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab. Dispos 19: 36–43. [PubMed] [Google Scholar]
- 38.Harker JA, Lewis GM, Mack L, and Zuniga EI. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 334: 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Utzschneider DT, Legat A, Fuertes Marraco SA, Carrié L, Luescher I, Speiser DE, and Zehn D. 2013. T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat. Immunol 14: 603–610. [DOI] [PubMed] [Google Scholar]
- 40.Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, Eisenberg V, Wohlleber D, Steiger K, Merkler D, Delorenzi M, Knolle PA, Cohen CJ, Thimme R, Youngblood B, and Zehn D. 2019. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571: 265–269. [DOI] [PubMed] [Google Scholar]
- 41.Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, Attanasio J, Yan P, George SM, Bengsch B, Staupe RP, Donahue G, Xu W, Amaravadi RK, Xu X, Karakousis GC, Mitchell TC, Schuchter LM, Kaye J, Berger SL, and Wherry EJ. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, Zamarin D, Walther T, Snyder A, Femia MR, Comen EA, Wen HY, Hellmann MD, Anandasabapathy N, Liu Y, Altorki NK, Lauer P, Levy O, Glickman MS, Kaye J, Betel D, Philip M, and Schietinger A. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571: 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo H, Chen J, González-Avalos E, Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO, Zhang W, Onodera A, Wu C-J, Lu L-F, Hogan PG, Bhandoola A, and Rao A. 2019. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. U. S. A 116: 12410–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao C, Sun H-W, Lacey NE, Ji Y, Moseman EA, Shih H-Y, Heuston EF, Kirby M, Anderson S, Cheng J, Khan O, Handon R, Reilley J, Fioravanti J, Hu J, Gossa S, Wherry EJ, Gattinoni L, McGavern DB, O’Shea JJ, Schwartzberg PL, and Wu T. 2019. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol 20: 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltra J-C, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, Casella V, Ngiow SF, Khan O, Huang YJ, Yan P, Nzingha K, Xu W, Amaravadi RK, Xu X, Karakousis GC, Mitchell TC, Schuchter LM, Huang AC, and Wherry EJ. 2020. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52: 825–841.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danilo M, Chennupati V, Silva JG, Siegert S, and Held W. 2018. Suppression of Tcf1 by inflammatory cytokines facilitates effector CD8 T cell differentiation. Cell Rep. 22: 2107–2117. [DOI] [PubMed] [Google Scholar]
- 47.Snell LM, MacLeod BL, Law JC, Osokine I, Elsaesser HJ, Hezaveh K, Dickson RJ, Gavin MA, Guidos CJ, McGaha TL, and Brooks DG. 2018. CD8+ T Cell Priming in Established Chronic Viral Infection Preferentially Directs Differentiation of Memory-like Cells for Sustained Immunity. Immunity 49: 678–694.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho Y-J, Klein A, Hofmann O, and Camargo FD. 2014. Clonal dynamics of native haematopoiesis. Nature 514: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, and Rodewald H-R. 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518: 542–546. [DOI] [PubMed] [Google Scholar]
- 50.Busch K, and Rodewald H-R. 2016. Unperturbed vs. post-transplantation hematopoiesis: both in vivo but different. Curr. Opin. Hematol 23: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hensel N, Gu Z, Sagar D. Wieland, Jechow K, Kemming J, Llewellyn-Lacey S, Gostick E, Sogukpinar O, Emmerich F, Price DA, Bengsch B, Boettler T, Neumann-Haefelin C, Eils R, Conrad C, Bartenschlager R, Grün D, Ishaque N, Thimme R, and Hofmann M. 2021. Memory-like HCV-specific CD8+ T cells retain a molecular scar after cure of chronic HCV infection. Nat. Immunol 22: 229–239. [DOI] [PubMed] [Google Scholar]
- 52.Heim K, Binder B, Sagar D.Wieland , Hensel N, Llewellyn-Lacey S, Gostick E, Price DA, Emmerich F, Vingerhoet H, Kraft ARM, Cornberg M, Boettler T, Neumann-Haefelin C, Zehn D, Bengsch B, Hofmann M, and Thimme R. 2020. TOX defines the degree of CD8+ T cell dysfunction in distinct phases of chronic HBV infection. Gut gutjnl-2020–322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan Y, Cao S, Liu X, Harrington SM, Bindeman WE, Adjei AA, Jang JS, Jen J, Li Y, Chanana P, Mansfield AS, Park SS, Markovic SN, Dronca RS, and Dong H. 2018. CX3CR1 identifies PD-1 therapy-responsive CD8+ T cells that withstand chemotherapy during cancer chemoimmunotherapy. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Characterization of CX3CR1 expressing CD8 T cells in infection
Fig. S2. Development and dynamics of CX3CR1 positive subsets of exhausted CD8 T cells
Fig. S3. Profiling of differentially expressed genes between distinct subpopulations of PD-1+ CD8 T cells following LCMV-c13 infection by bulk RNA-seq
Fig. S4. Characterization of CD8 T cell-specific Tbx21- and Eomes-deficient mice during chronic LCMV infection