Abstract
Higher-order chromatin packing serves as a structural barrier to the recognition and repair of genomic lesions. The initiation and outcome of the repair response is dictated by a highly coordinated yet complex interplay between chromatin modifying enzymes and their cognate readers, damage induced chemical modifications, nucleosome density, transcriptional state, and cell cycle dependent availability of DNA repair machinery. The physical and chemical properties of the DNA lesions themselves further regulate the nature of ensuing chromatin responses. Here we review recent discoveries across these various contexts, where chromatin regulates the homology-guided double-strand break repair mechanism, homologous recombination, and also highlight the key knowledge gaps vital to generate a holistic understanding of this process and its contributions to genome integrity.
Keywords: Homologous recombination, Chromatin, BRCA, R-loops, telomeres
INTRODUCTION
Central to the maintenance of genomic stability is the ability of cells to repair lethal DNA double-strand breaks (DSBs) or damaged replication forks in an error-free manner by homologous recombination (HR). HR is a multi-step process that entails DSB resection to generate 3’single stranded (ss)-DNA, followed by a homology search, templated DNA synthesis and finally resolution of the repair intermediate. Emerging studies discussed here illustrate how the initiation and nature of homology repair are dictated by the chromatin environment. We also highlight examples where BRCA proteins, key mediators of HR and tumor suppression, contribute to maintaining chromatin integrity. Understanding the molecular intricacies of this multifaceted chromatin-HR interaction brings forth possibilities of decoding disease etiologies and developing targeted therapies.
γH2AX-MDC1: an apical notch of chromatin response to DSBs
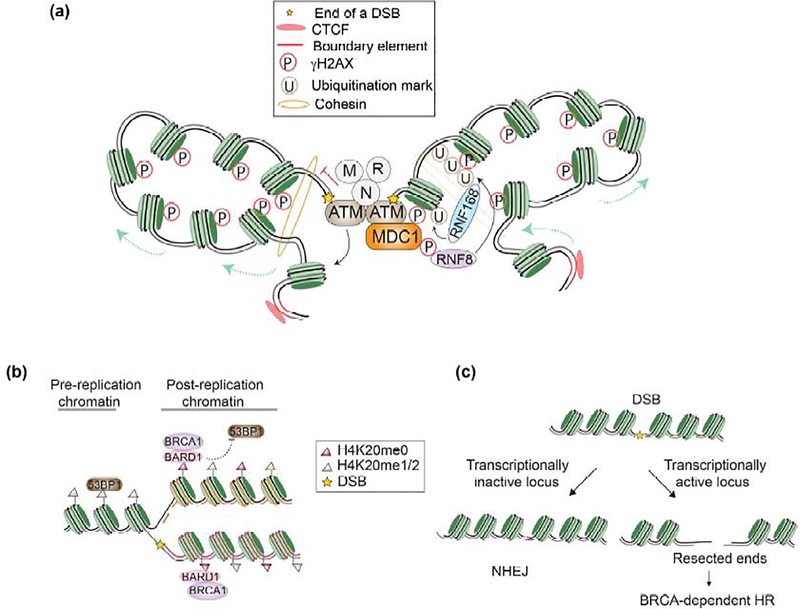
Irrespective of the damaging source, one of the earliest chromatin responses to a DSB is the phosphorylation of serine139 in the histone variant H2AX, referred to as γH2AX[1], by one of three phosphatidylinositol 3-kinase-like kinases (PI3KK): ATM, DNA-PK and ATR. Although dispensable for the initial recognition of DSB by repair proteins, γH2AX is critical for the conglomeration of repair complexes in the vicinity of DSBs[2]. This is achieved via rapid spreading of the γH2AX mark over megabase pairs (Mbp) of the chromatin by a positive feedback loop mechanism. The initial γH2AX mark, which likely arises by the binding of ATM to break sites, is recognized by MDC1 which then recruits the end-resection machinery, MRN complex (MRN11-Rad50-NBS1)[3]. NBS1 in turn recruits and activates ATM, which then generates the γH2AX mark across several Mbp of chromatin. Analogous to Nbs1, Ku80 and ATRIP mediate damage recruitment of DNA-PK and ATR, respectively,[4]. How γH2AX spreading events on chromatin are exclusive to ATM and cannot be established by DNA-PK and ATR kinases remains unclear[5].
A recent study demonstrated the contribution of chromatin loop extrusion to rapid establishment of γH2AX territories across millions of base pairs in the genome. ATM phosphorylates H2AX on both sides of the break as the chromatin is threaded unidirectionally through the cohesin ring. Looping stalls at the topologically associated domains (TADs), highlighting how the predefined chromatin structure contributes to DNA repair signalling[6••]. In the future, it would be informative to examine if DSB repair is regulated differently at regions closer to the TAD boundaries, the identities of chromatin motors mediating this response, and if and how ATM activity contributes to this extrusion differentially from DNA-PK or ATR kinase.
Chromatin modifications that regulate DSB repair pathway choices
Once associated with chromatin, MDC1 serves as a lynchpin for several downstream repair signalling events. ATM-mediated phosphorylation of MDC1 serves to recruit the E3 ubiquitin (Ub) ligase, RNF8, to the DNA lesion[7,8]. RNF8 in conjunction with K63-ubiquitin specific E2 ligase, Ubc13, was reported to catalyze polyubiquitination of H1 linker histone and L3MBTL2, which serve to recruit the E3 Ub ligase RNF168[9,10]. RNF168 in concert with E2 ligase, UbcH5c, catalyzes the mono ubiquitination (mUb) of H2A/H2AX at K13/K15[11]. The studies identifying a role for RNF168 in DSB repair response also proposed that this E3 ligase can catalyze K63 linked PolyUb[12,13]. However, the substrates RNF168 polyubiquitinates need further examination in the light of recent findings[9,11] (Fig.1a).
Figure 1.
(a) Initial events of DSB response signaling on chromatin. Loop extrusion promotes bidirectional spreading of ATM-catalyzed γH2AI marks within the boundary elements. (b) Recognition of H4K20me0 by BARD1-BRCA 1 complex on the nascent nucleosome precludes 53BP1 binding at DSBs. (c) Transcriptionally active chromatin preferentially gets repaired by BRCA-dependent HR.
An intricate network of chromatin modifications and cell cycle states modulate the fate of downstream events emanating from ubiquitinated chromatin. A critical event in dictating the fate of DSBs is the process of resection which involves nuclease mediated generation of 3’ ss-DNA for homology search. Resection is contingent on the eviction of nucleosomes at the break site by RSC and SWI/SNF remodelers[14]. In the absence of resection, the two broken DNA ends are ligated via Ku-DNAPK-XRCC4-ligase IV through a process called non-homologous end joining (NHEJ). In contrast, resection of DSBs tips the balance towards homology-directed synthesis and repair via BRCA-mediated HR. The master chromatin regulator of this resection is the scaffolding protein p53-binding protein 1 (53BP1). RNF168-catalyzed H2AK15Ub serves as a binding platform for the ubiquitination-dependent recruitment (UDR) domain of 53BP1[15]. Damage induced assembly of 53BP1 with Rif1SHLD1–3-Rev7 complex on chromatin serves as a regulatory node to prevent accumulation of BRCA1 and the downstream resecting nucleases[16].
Effective 53BP1 binding to chromatin is established only on nucleosomes that carry both H2AK15Ub and H4K20me2 marks[15,17]. H4K20me½ marks are abundant on old histones and these are enriched on the chromatin during the G1 and M phase of the cell cycle. In contrast, the obligate BRCA1 binding partner BARD1 binds via its ankyrin repeat to mononucleosomes with H4K20me0, a signature of histones that are newly incorporated during replication[18,19••]. These observations highlight the ingenuity with which the chromatin biases the repair pathway choice towards HR in S/G2 and NHEJ in G1 (Fig.1b).
Different BRCA1 complex may have distinct chromatin recruitment signatures
The BRCA1 protein exists in multiple distinct complexes[20] and it is unclear if 53BP1 equally regulates their recruitment. Based on the available evidence, 53BP1 complex serves as a steric block to MRN-CtIP activity. By the virtue of preventing 53BP1 occupancy on chromatin, BRCA1 promotes MRN-CtIP mediated resection[21]. Resection can be further facilitated by the ubiquitination of H2AK125/127/129 by the BRCA1-BARD1 complex. These Ub marks serve to recruit the Swr1-like remodeler, SMARCAD1, which is proposed to displace the 53BP1 bound nucleosomes [22,23]. Deubiquitinase, USP48 was recently shown to antagonize this BRCA1-BARD1 E3-ligase activity [24]. How selectivity of this ubiquitin mediated chromatin reorganization is restricted to 53BP1 bound nucleosomes remains to be determined. Recruitment of BRCA1 complexes can be regulated by multiple other factors and this indeed seems to be the case. The BRCA1-A is a six member-complex consisting of BRCA1 and its associated proteins, RAP80, BRCC36, Abraxas, MERIT40 and BRCC45. The Ub-interacting motif (UIM) domain of RAP80 mediates localization of this complex to damaged chromatin via binding to K63-linked PolyUb chains, albeit the identity of chromatin associated protein carrying these K63 chains is still obscure[8,25,26]. In contrast to the canonical function of BRCA1 in promoting HR, the BRCA1-A complex was shown to negatively regulate resection[27,28]. This intriguing observation was supported by subsequent studies showing that loss of BRCA1-A complex and not 53BP1 imparted resistance to the topoisomerase I poison, topotecan or Poly (ADP-ribose) polymerase inhibitors (PARPi) in ATM-deficient cells[29]. More recent proteome analysis of camptothecin-damaged fork revealed enrichment of the BRCA1-A complex upon ATM inhibition[30•]. Together these studies suggest that analogous to 53BP1 at DSBs, BRCA1-A may compete with HR-mediated repair at broken replication forks. Enrichment of H4K20me0 on the replication fork may mediate preferential involvement of the BRCA1-A complex over 53BP1 in this anti-resection response. How chromatin marks at replication forks change upon ATM deficiency, what mediates RNF168 recruitment in the absence of ATM and how the BRCA1-A complex counteracts resection remain open questions. Additionally, chromatin modifications that regulate recruitment of the BRCA1-B (BRCA-FANCJ-TopBP1) and BRCA1-P (BRCA1-PALB2-BRCA2-RAD51) complexes remain enigmatic. Recent studies suggest a role for RNF168 in bypassing BRCA1 function in Rad51 loading, albeit how the ubiquitinated chromatin mediates recruitment of PALB2-BRCA2-RAD51 is unclear[31,32•].
Transcriptionally active chromatin preferentially recruits HR proteins
Cells have evolved multiple levels of regulation to promote HR during the S/G2 phase of the cell cycle given that S-phase nucleosomes still carry equal proportions of new and old histones with the H4K20me0 and H4K20me2 signatures, respectively. The lysine acetyltransferase 60kDa Tat-Interactive Protein (TIP60/KAT5) represents an important chromatin directed determinant of repair mechanism. TIP60 dependent histone H4 and H2A acetylation has been shown to reduce 53BP1 damage site localization, promoting BRCA-mediated HR[33,34]. Consistent with an open chromatin state dictating this competition, inducible transcription at a defined locus increased BRCA1, resection and Rad51 delivery while reducing 53BP1, establishing direct evidence that active transcription is a key chromatin determinant that preferentially deploys HR at DSBs[33]. Subsequent genome wide studies lend further support to this notion by revealing that genomic loci prone to homology-mediated repair are enriched for SETD2-dependent H3K36me3, a chromatin mark for transcriptional elongation (Fig.1c)[35,36]. While a HR-permissive environment at loci with active transcription may ensure the integrity of gene bodies, it would be informative to track the fate of such sites if they are damaged in the G1 phase of the cell cycle.
Unique contributions of BRCA proteins in resolving R-loop chromatin
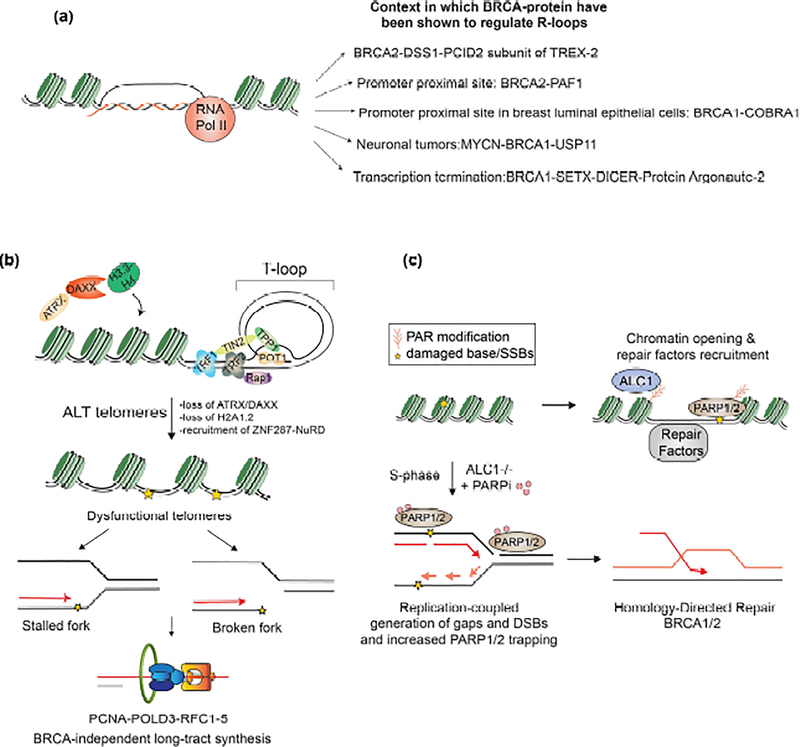
Beyond post-translational modifications, the presence of nascent RNA at transcriptionally active regions can also present physical challenges that necessitate resolution by the BRCA1 and 2 proteins. Notable amongst those is emerging evidence that highlights the roles of BRCA proteins in resolving R-loop, a three-stranded nucleic acid structure, wherein a RNA hybridizes with a template DNA displacing the non-templated strand. One of the initial indications came from studies showing that BRCA2 loss in cells depleted of messenger ribonucleoprotein biogenesis and export proteins resulted in an increased accumulation of R-loops. Intriguingly, no increase in R-loops was observed upon depletion of Rad51 suggesting a HR-independent function of BRCA2 in mediating this response[37]. BRCA1 and BRCA2 have also been implicated in releasing aberrantly stalled RNA PolII and hence preventing intergenic R-loop accumulation[38–41]. Based on available evidence, the functions of BRCA1 and BRCA2 in resolving R-loops appear to be mostly non-epistatic. The function of BRCA1 in limiting these aberrant R-loops, however, seems to be subject to the genetic background[39,40]. R-loops have also been attributed physiological functions, particularly in terminating transcription on a subset of highly transcribed genes[42]. BRCA1 is reported to protect ss-DNA at R-loops that accumulate on transcriptional termination sites[43]. More recently it was demonstrated that BRCA1 forms a complex with helicase SETX, RNAse type III enzyme DICER and Protein argonaute-2 in a R-loop dependent manner and promotes the generation of single-stranded, DNA-damage-associated small RNA (sdRNA). sdRNA, via the PALB2-Rad52 complex, in turn protects ss-DNA breaks generated by R-loops at transcriptional pause sites[44••]. This study also helped unravel the mystery of how non-coding RNAs could contribute to DNA repair signalling[45]. A recent study also uncovered a role for RNA-DNA hybrids in protecting resection generated 3’ss-DNA, albeit the molecular details of how these structures are resolved for subsequent steps of RAD51 loading remains to be explored[46]. Together, these findings have set the stage for appreciating the roles of BRCA proteins in tumor suppression beyond DSB repair and replication fork protection (Fig.2a). Future investigations are essential to clarify if common themes exist across various genetic settings for recognition and resolution of R-loop by the BRCA1 and BRCA2 proteins.
Figure 2.
(a) Different contexts in which BRCA proteins resolve R-loops. (b) Altered chromatin environment at ALT telomeres instigate homology-directed telomeric DNA synthesis. RF’Cl-5 loads the PCNA clamp onto damaged telomeres. DNA polymerase δ relies on PoID3 interaction with PCNA to execute kilobases of DNA synthesis at damaged ALT telomeres in a homology directed repair mechanism that is BRCA-independent. (c) Model showing the proposed basis for PARPi hypersensitivity in ALC 1-deficient BRCA mutant cells. Nucleosome sliding by ALC1 cooperates with PARP activity to generate chromatin accessibility for lesions generated by base damage. DNA lesions in ALC1 deficient cells result in replication-coupled gaps and DSBs, that accentuate PARPi trapping and increase reliance on BRCA-dependent HR.
Chromatin architecture dictates telomeric integrity and lengthening mechanisms
A special chromatin context where regulation of DSB repair pathways becomes essential is telomere maintenance. Inactivation of end-joining responses at telomeres is achieved by a six-member protective complex called shelterin[47]. A key component of this complex necessary to preclude DNA repair responses is TRF2, which not only re-models telomeric ends into t-loop to sequester the reactive DNA ends from recombining, but also suppresses ATM and RNF168 signalling (Fig. 2b)[48–50]. Strikingly, recent studies in pluripotent stem cells revealed that the functions of TRF2 in averting toxic end-joining are dispensable in this cell type[51•,52•]. Although the molecular details for this unique rewiring in stem cells needs to be delineated, it was shown that TRF2 loss upregulates the ZSCAN4 cluster which possibly along with other genes ensures continued telomere lengthening that subverts telomeric fusions[52].
BRCA2 along with Rad51 also contribute to maintaining telomeric chromatin architecture by mediating the recruitment of Telomeric-repeat-containing RNA, a central component in protecting the chromosomal ends[53••]. Alterations in the chromatin landscape at telomeres can attenuate shelterin binding. This is exemplified in the settings of cancers that rely on telomerase independent-telomere length maintenance mechanism called Alternative lengthening of telomeres (ALT). ALT telomeres are characterized by interspersed repeats that vary from the canonical 5′-TTAGGG-3′ tandem repeats. These sequences serve as binding sites for NR2C/F class orphan nuclear receptor, which in turn can recruit zinc finger protein ZNF287 and the nucleosome remodelling histone deacetylation (NuRD) complex, displacing shelterin and accentuating replication stress[54,55]. Further alterations in the chromatin environment at ALT telomeres arise due to the frequent loss of function mutations in histone 3.3 chaperone, Alpha thalassemia/mental retardation syndrome X-linked chromatin remodeler (ATRX) or its interacting protein, Death domain-associated protein (DAXX). Irrespective of telomere length, loss of ATRX/DAXX was shown to result in telomeric chromatin de-condensation resulting in replication dysfunction and necessitating homology-directed repair mechanisms as the only viable means of survival[56,57•]. ATRX deficiency also correlates with the transient loss of H2A1.2, resulting in frequent fork stalling and replication stress at ALT telomeres[58]. Chromatin assembly factor HIRA was proposed to compensate for ATRX function in promoting homology-synthesis at ALT telomeres[59]. Induction of damage in this altered chromatin environment induces ALT-like features, including homology-directed telomere clustering, synthesis of telomeric DNA and frequent association of telomeres with promyelocytic leukemia bodies (APBs)[60]. APBs display liquid-liquid phase separation properties which promote telomere clustering and hence can facilitate homology repair and recombination[61,62]. Intriguingly, homology-directed synthesis at ALT telomere can happen in a BRCA½-independent manner and can extend along the entire telomere length of >50 kbp[63] (Fig.2b). This non-canonical synthesis resembles the long-tract break-induced repair pathway in yeast and employs a unique replisome that does not involve many canonical S-phase replication proteins[63–65]. R-loops were also shown to be pivotal in triggering recombinogenic activity and hence telomere maintenance specifically in ALT cells[66]. Together, these studies highlight how chromatin alterations can necessitate non-canonical homology repair mechanisms.
Communication between PAR-dependent chromatin remodelling and HR
Chromatin accessibility represents another scenario that necessitates coordination with HR to maintain genome stability. Recent findings demonstrate that PARP dependent chromatin remodellfing is essential for the proliferation of BRCA-deficient cells. PARPi are synthetically lethal with HR-deficiency and to date four different PARPi are FDA-approved for the regimen of HR-deficient breast, ovarian, pancreatic and prostate cancers[67–69]. PARP enzymes are one of the earliest sensors of different genomic lesions and DNA binding allosterically activates their catalytic ability to PARylate histones[70]. Chromatin PARylation serves as a platform to recruit downstream repair proteins involved in a myriad of repair pathways. Additionally, PAR modification results in chromatin de-condensation, a discovery made nearly 40 years ago[71] yet its implications in the viability and PARPi response of HR-deficient cells remained unclear. Recent studies identified the PAR-binding nucleosome sliding enzyme, Amplified in Liver Cancer 1 (ALC1) as a key determinant of viability and PARPi responses in BRCA-mutant cells[72–74]. The enhanced PARPi therapeutic window upon ALC1 loss helped mitigate several clinical known resistance mechanisms. ALC1 is the only SNF2 ATPase with a PAR-binding macrodomain, which recognizes tri-ADP ribose with a 10nM Kd[75–77]. This high binding affinity could permit ALC1 to alter chromatin at a low damage level when PARylation is limiting. Perhaps this explains why unlike PARP1 which is essential for resolving damage to a myriad of damaging agents, ALC1 deficiency imparts sensitivity to only base and nucleotide damaging agents. ALC1 cooperates with PARP activity in creating chromatin accessibility to endogenous base lesions. Persistent accumulation of damaged bases could instigate the increased genesis of replication-coupled single-strand gaps and DSBs, lesions that trap PARP1 and PARP2 enzymes, creating a hyper-reliance on HR[73••,74••] (Fig.2c). These findings uncover chromatin accessibility to base damage as a mechanistically distinct aspect of PARPi response. Examining the nature of endogenous base lesions in BRCA-mutant cells may help inform on intrinsic events that precipitate genetic instability in HR-deficient cancers.
The studies discussed here highlight how communication between chromatin and HR underline several fundamental processes including DNA repair, replication, transcription, chromosome structure maintenance, as well as responses to targeted therapy. These findings also illustrate that chemical and physical alterations to chromatin modulate HR not only at local chromosomal loci but also can make the repair choice cell-cycle and cell-type specific. It is noteworthy that much of our recent knowledge has emerged from the implementation of advanced methodologies such as CRISPR editing technology, quantitative chromatin proteomics, genome-wide deep sequencing and chromosome conformation capture techniques to address several long-standing questions in the DSB repair biology. Collectively, these findings and emerging methodologies will be pivotal to develop a comprehensive understanding of how chromatin alterations are temporally regulated to coordinate homology directed repair and how disease specific alterations in chromatin directed factors impact genome integrity.
ACKNOWLEDGEMENTS
RAG is supported by NIH grants GM101149, CA138835 and CA17494, the Penn Center for Genome Integrity, the Basser Center for BRCA, a V Foundation Team Convergence Award, and a Gray Foundation Team Science Award. PV was supported by the Ann and Sol Schreiber Mentored Investigator Award (Ovarian Cancer Research Fund Alliance) and Pilot funds from the Ovarian Cancer Translational Center for Excellence (UPenn).
Footnotes
DISCLOSURE
R.A.G is a co-Founder and Scientific Advisory Board member for RADD Pharmaceuticals and JAMM Therapeutics. P.V. declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM: DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998, 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 2.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A: Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 2003, 5:675–679. [DOI] [PubMed] [Google Scholar]
- 3.Stewart GS, Wang B, Bignell CR, Taylor AMR, Elledge SJ: MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003, 421:961–966. [DOI] [PubMed] [Google Scholar]
- 4.Falck J, Coates J, Jackson SP: Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434:605–611. [DOI] [PubMed] [Google Scholar]
- 5.Caron P, Choudjaye J, Clouaire T, Bugler B, Daburon V, Aguirrebengoa M, Mangeat T, Iacovoni JS, Álvarez-Quilón A, Cortés-Ledesma F, et al. : Non-redundant Functions of ATM and DNA-PKcs in Response to DNA Double-Strand Breaks. Cell Reports 2015, 13:1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arnould C, Rocher V, Finoux A-L, Clouaire T, Li K, Zhou F, Caron P, Mangeot PE, Ricci EP, Mourad R, et al. : Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 2021, 590:660–665. •• Using high-throughput chromosome conformation capture and ChIP-seq methods this work establishes that rapid spreading of the γH2AX mark across millions of base pairs is achieved by unidirectional loop extrusion on either side of DNA DSBs.
- 7.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J: RNF8 Ubiquitylates Histones at DNA Double-Strand Breaks and Promotes Assembly of Repair Proteins. Cell 2007, 131:887–900. [DOI] [PubMed] [Google Scholar]
- 8.Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J: RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007, 131:901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, et al. : Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 2015, 527:389–393. [DOI] [PubMed] [Google Scholar]
- 10.Nowsheen S, Aziz K, Aziz A, Deng M, Qin B, Luo K, Jeganathan KB, Zhang H, Liu T, Yu J, et al. : L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nat Cell Biol 2018, 20:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattiroli F, Vissers JHA, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK: RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150:1182–1195. [DOI] [PubMed] [Google Scholar]
- 12.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. : The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 2009, 136:420–434. [DOI] [PubMed] [Google Scholar]
- 13.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. : RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell 2009, 136:435–446. [DOI] [PubMed] [Google Scholar]
- 14.Peritore M, Reusswig K-U, Bantele SCS, Straub T, Pfander B: Strand-specific ChIP-seq at DNA breaks distinguishes ssDNA versus dsDNA binding and refutes single-stranded nucleosomes. Mol Cell 2021, doi: 10.1016/j.molcel.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Fradet-Turcotte A, Canny MD, Escribano-Díaz C, Orthwein A, Leung CCY, Huang H, Landry M-C, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. : 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 2013, 499:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirman Z, de Lange T: 53BP1: a DSB escort. Genes Dev 2020, 34:7–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botuyan MV, Lee J, Ward IM, Kim J-E, Thompson JR, Chen J, Mer G: Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 2006, 127:1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saredi G, Huang H, Hammond CM, Alabert C, Bekker-Jensen S, Forne I, Reverón-Gómez N, Foster BM, Mlejnkova L, Bartke T, et al. : H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. Nature 2016, 534:714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakamura K, Saredi G, Becker JR, Foster BM, Nguyen NV, Beyer TE, Cesa LC, Faull PA, Lukauskas S, Frimurer T, et al. : H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat Cell Biol 2019, 21:311–318. •• Ankyrin repeats of BARD1 bind the H4K20me0 marks on nascent chromatin. This promotes HR over NHEJ during the S/G2 phase of cell cycle.
- 20.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM: Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev 2006, 20:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunting SF, Callén E, Wong N, Chen H-T, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. : 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010, 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Densham RM, Garvin AJ, Stone HR, Strachan J, Baldock RA, Daza-Martin M, Fletcher A, Blair-Reid S, Beesley J, Johal B, et al. : Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat Struct Mol Biol 2016, 23:647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo CSY, van Toorn M, Gaggioli V, Dias MP, Zhu Y, Manolika EM, Zhao W, van der Does M, Mukherjee C, Gonçalves JGS, et al. : SMARCAD1-mediated active replication fork stability maintains genome integrity. Science Advances 2021, 7:eabe7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uckelmann M, Densham RM, Baas R, Winterwerp HHK, Fish A, Sixma TK, Morris JR: USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nat Commun 2018, 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA: RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007, 316:1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ: Abraxas and RAP80 Form a BRCA1 Protein Complex Required for the DNA Damage Response. Science 2007, 316:1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman KA, Greenberg RA: The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 2011, 286:13669–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM: RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes & Development 2011, 25:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balmus G, Pilger D, Coates J, Demir M, Sczaniecka-Clift M, Barros AC, Woods M, Fu B, Yang F, Chen E, et al. : ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat Commun 2019, 10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura K, Kustatscher G, Alabert C, Hödl M, Forne I, Völker-Albert M, Satpathy S, Beyer TE, Mailand N, Choudhary C, et al. : Proteome dynamics at broken replication forks reveal a distinct ATM-directed repair response suppressing DNA double-strand break ubiquitination. Mol Cell 2021, 81:1084–1099.e6. • Proteome profiling highlighted key differences in broken and stalled replication forks. Additionally, this study suggested a unique role for ATM in mediating HR-dependent fork restart by suppressing the recruitment of anti-resection BRCA1-A complex and DSBs associated ubiquitin signaling at broken replication fork.
- 31.Luijsterburg MS, Typas D, Caron M-C, Wiegant WW, van den Heuvel D, Boonen RA, Couturier AM, Mullenders LH, Masson J-Y, van Attikum H: A PALB2-interacting domain in RNF168 couples homologous recombination to DNA break-induced chromatin ubiquitylation. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zong D, Adam S, Wang Y, Sasanuma H, Callén E, Murga M, Day A, Kruhlak MJ, Wong N, Munro M, et al. : BRCA1 Haploinsufficiency Is Masked by RNF168-Mediated Chromatin Ubiquitylation. Mol Cell 2019, 73:1267–1281.e7. • The study shows that unlike 53BP1, RNF168 loss did not rescue embryonic lethality of BRCA1Δ11/Δ11. Furthermore, RNF168 loss abrogated PALB2 damage localization and conferred tumorigenesis in BRCA+/Δ11 settings, supporting a role for chromatin ubiquitin pathway in bypassing Rad51 loading function of BRCA1.
- 33.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA: Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 2013, 20:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert J-P, Roques C, Pandita RK, Paquet E, Herst P, Gingras A-C, Pandita TK, et al. : The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol Cell 2016, 62:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sørensen CS, Petersen NHT, Sorensen PHB, Lukas C, Bartek J, et al. : LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat Struct Mol Biol 2012, 19:803–810. [DOI] [PubMed] [Google Scholar]
- 36.Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, et al. : Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 2014, 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A: BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014, 511:362–365. [DOI] [PubMed] [Google Scholar]
- 38.Shivji MKK, Renaudin X, Williams ÇH, Venkitaraman AR: BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Rep 2018, 22:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herold S, Kalb J, Büchel G, Ade CP, Baluapuri A, Xu J, Koster J, Solvie D, Carstensen A, Klotz C, et al. : Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature 2019, 567:545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Chiang H-C, Wang Y, Zhang C, Smith S, Zhao X, Nair SJ, Michalek J, Jatoi I, Lautner M, et al. : Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat Commun 2017, 8:15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, Ahmed K, Constantinou S, Renaudin X, Lee M, Aebersold R, et al. : A Class of Environmental and Endogenous Toxins Induces BRCA2 Haploinsufficiency and Genome Instability. Cell 2017, 169:1105–1118.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ: R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 2014, 516:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. : BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 2015, 57:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hatchi E, Goehring L, Landini S, Skourti-Stathaki K, DeConti DK, Abderazzaq FO, Banerjee P, Demers TM, Wang YE, Quackenbush J, et al. : BRCA1 and RNAi factors promote repair mediated by small RNAs and PALB2–RAD52. Nature 2021, 591: 665–670. •• Identified a new small RNA species which in complex with BRCA1 and RNAi proteins mediates repair of endogenous DNA break that emerge owing to R-loops at transcription pause sites.
- 45.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F: Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S, Hua Y, Wang J, Li L, Yuan J, Zhang B, Wang Z, Ji J, Kong D: RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell 2021, 184:1314–1329.e10. [DOI] [PubMed] [Google Scholar]
- 47.de Lange T: Shelterin-Mediated Telomere Protection. Annu Rev Genet 2018, 52:223–247. [DOI] [PubMed] [Google Scholar]
- 48.Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR 3rd, Denchi EL: A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 2013, 494:502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denchi EL, de Lange T: Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448:1068–1071. [DOI] [PubMed] [Google Scholar]
- 50.Doksani Y, Wu JY, de Lange T, Zhuang X: Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 2013, 155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ruis P, Van Ly D, Borel V, Kafer GR, McCarthy A, Howell S, Blassberg R, Snijders AP, Briscoe J, Niakan KK, et al. : TRF2-independent chromosome end protection during pluripotency. Nature 2021, 589:103–109. • See annotation to ref [52]
- 52. Markiewicz-Potoczny M, Lobanova A, Loeb AM, Kirak O, Olbrich T, Ruiz S, Lazzerini Denchi E: TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature 2021, 589:110–115. • [51–52] reveal that unlike most other somatic cell types studied to-date, telomeres in pluripotent stem cells do not undergo end-joining upon loss of TRF2.
- 53. Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L, Lingner J: RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 2020, 587:303–308. •• RAD51 was shown to catalyze RNA-DNA hybrid formation and mediate localization of TERRA RNA to telomeres.
- 54.Conomos D, Stutz MD, Hills M, Neumann AA, Bryan TM, Reddel RR, Pickett HA: Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. Journal of Cell Biology 2012, 199:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marzec P, Armenise C, Pérot G, Roumelioti F-M, Basyuk E, Gagos S, Chibon F, Déjardin J: Nuclear-receptor-mediated telomere insertion leads to genome instability in ALT cancers. Cell 2015, 160:913–927. [DOI] [PubMed] [Google Scholar]
- 56.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. : Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011, 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li F, Deng Z, Zhang L, Wu C, Jin Y, Hwang I, Vladimirova O, Xu L, Yang L, Lu B, et al. : ATRX loss induces telomere dysfunction and necessitates induction of alternative lengthening of telomeres during human cell immortalization. EMBO J 2019, 38:e96659. • Revealed that ATRX loss leads to telomeric chromatin decompaction and hence telomere dysfunction irrespective of cell type, leaving homology-directly synthesis as the only viable means of telomere length maintenance.
- 58.Kim J, Sun C, Tran AD, Chin P-J, Ruiz PD, Wang K, Gibbons RJ, Gamble MJ, Liu Y, Oberdoerffer P: The macroH2A1.2 histone variant links ATRX loss to alternative telomere lengthening. Nature Structural & Molecular Biology 2019, 26:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoang SM, Kaminski N, Bhargava R, Barroso-González J, Lynskey ML, García-Expósito L, Roncaioli JL, Wondisford AR, Wallace CT, Watkins SC, et al. : Regulation of ALT-associated homology-directed repair by polyADP-ribosylation. Nat Struct Mol Biol 2020, 27:1152–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho NW, Dilley RL, Lampson MA, Greenberg RA: Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 2014, 159:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Zhao R, Tones J, Liu M, Dilley RL, Chenoweth DM, Greenberg RA, Lampson MA: Nuclear body phase separation drives telomere clustering in ALT cancer cells. Molecular Biology of the Cell 2020, 31:2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min J, Wright WE, Shay JW: Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev 2019, 33:814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA: Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roumelioti F-M, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD, Gagos S: Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep 2016, 17:1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anand RP, Lovett ST, Haber JE: Break-induced DNA replication. Cold Spring Harb Perspect Biol 2013, 5:a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM: RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 2014, 5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. : Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434:917–921. [DOI] [PubMed] [Google Scholar]
- 68.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T: Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature 2005, 434:913–917. [DOI] [PubMed] [Google Scholar]
- 69.Rose M, Burgess JT, O’Byrne K, Richard DJ, Bolderson E: PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Frontiers in Cell and Developmental Biology 2020, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pascal JM: The comings and goings of PARP-1 in response to DNA damage. DNA Repair 2018, 71:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P: Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A 1982, 79:3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blessing C, Mandemaker IK, Gonzalez-Leal C, Preisser J, Schomburg A, Ladurner AG: The Oncogenic Helicase ALC1 Regulates PARP Inhibitor Potency by Trapping PARP2 at DNA Breaks. Mol Cell 2020, 80:862–875.e6. [DOI] [PubMed] [Google Scholar]
- 73. Hewitt G, Borel V, Segura-Bayona S, Takaki T, Ruis P, Bellelli R, Lehmann LC, Sommerova L, Vancevska A, Tomas-Loba A, et al. : Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol Cell 2021, 81:767–783.e11. •• See annotation to ref [74]
- 74. Verma P, Zhou Y, Cao Z, Deraska PV, Deb M, Arai E, Li W, Shao Y, Puentes L, Li Y, et al. : ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells. Nature Cell Biology 2021, 23:160–171. •• Ref [70–71] discovered that loss of ALC1 mediating nucleosome sliding renders PARP inhibitor hypersensitivity in BRCA-mutant cells. The inability of ALC1 deficient cells to process damaged base lesions was shown to be the mechanistic basis for the altered drug efficacy.
- 75.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. : Poly(ADP-ribose)-Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science 2009, 325:1240–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. : Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A 2009, 106:13770–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh HR, Nardozza AP, Möller IR, Knobloch G, Kistemaker HAV, Hassler M, Harrer N, Blessing C, Eustermann S, Kotthoff C, et al. : A Poly-ADP-Ribose Trigger Releases the Auto-Inhibition of a Chromatin Remodeling Oncogene. Mol Cell 2017, 68:860–871.e7. [DOI] [PubMed] [Google Scholar]