Abstract
The p89/xeroderma pigmentosum complementation group B (XPB) ATPase-helicase of transcription factor IIH (TFIIH) is essential for promoter melting prior to transcription initiation by RNA polymerase II (RNAPII). By studying the topological organization of the initiation complex using site-specific protein-DNA photo-cross-linking, we have shown that p89/XPB makes promoter contacts both upstream and downstream of the initiation site. The upstream contact, which is in the region where promoter melting occurs (positions −9 to +2), requires tight DNA wrapping around RNAPII. The addition of hydrolyzable ATP tethers the template strand at positions −5 and +1 to RNAPII subunits. A mutation in p89/XPB found in a xeroderma pigmentosum patient impairs the ability of TFIIH to associate correctly with the complex and thereby melt promoter DNA. A model for open complex formation is proposed.
RNA polymerase II (RNAPII) is the multisubunit enzyme that synthesizes eukaryotic mRNA. The two largest RNAPII subunits, Rpb1 and Rpb2, which are homologous to the β′ and β subunits of prokaryotic RNA polymerases, possess the catalytic activity of the enzyme (5, 71). Rpb1 contains a repeated heptapeptide in its carboxyl-terminal domain (CTD) that becomes highly phosphorylated during the passage from initiation to elongation of transcription (8). Initiation of transcription by RNAPII in vitro requires a set of general transcription initiation factors including TATA-binding protein (TBP), transcription factor IIB (TFIIB), TFIIE, TFIIF, and TFIIH (21, 44). TBP binds to the TATA element of promoters and induces an ∼90° bend in the DNA helix (27, 30). TFIIB associates with the TBP-promoter complex (1, 37) and makes promoter contacts on each side of the DNA bend centered on the TATA box (6, 31). TFIIF, which is composed of two subunits called RNAPII-associated proteins 74 and 30 (RAP74 and RAP30), tightly binds to RNAPII and allows the binding of the enzyme to a TBP-TFIIB-promoter complex (4, 16). TFIIE is also composed of two subunits, TFIIE56 and TFIIE34 (25, 41), that stabilize the association of RNAPII with the preinitiation complex (50). A complex containing TBP, TFIIB, TFIIE, TFIIF, and RNAPII is capable of initiating transcription on a premelted linear template (23, 45, 61) and on a negatively supercoiled template (46, 62, 64). Both TFIIE and TFIIF have been shown to play a role in the TFIIH-independent melting of the promoter DNA around the transcription initiation site (TIS) (23, 45). TFIIE is also required for the association of TFIIH with the preinitiation complex and the regulation of TFIIH activities (35, 39, 40). A complex containing TBP, TFIIB, TFIIE, TFIIF, TFIIH, and RNAPII is capable of melting promoter DNA in a region between nucleotides −9 and +2 of a linear template (22, 24, 26, 69). Open complex formation requires the hydrolysis of the β-γ bond of ATP by the helicase-ATPase activity of TFIIH (22, 24).
Mammalian TFIIH is a nine-subunit complex responsible both for the melting of the template DNA prior to initiation (54, 57) and during promoter escape (19, 36) and for the phosphorylation of the RNAPII CTD (13, 35, 56). The two largest subunits of TFIIH, p89 and p80, are ATP-dependent, single-stranded DNA (ssDNA) helicases encoded by the xeroderma pigmentosum (XP) complementation group B (XPB) and D (XPD) genes (11, 52–54, 67). The CTD kinase is composed of three subunits, called the cyclin-dependent kinase (CDK)-activating kinase (CAK), that contain the kinase-cyclin pair cdk7-cyclin H and the RING-H2 finger protein MAT1 (14, 58, 59). The development by Tirode et al. (63) of a system allowing the reconstitution of TFIIH from cloned subunits, producing either the full complex (rIIH9) or subcomplexes lacking CAK (rIIH6) with wild-type or mutated forms of the helicases, has been invaluable in analyzing the function of this important factor. The rIIH6 complex lacking CAK is active in ATP-dependent formation of the open complex and supports a reduced level of transcription in vitro. By comparing rIIH6 carrying wild-type helicases and rIIH6 containing mutations in either the p89/XPB or p80/XPD helicase, Tirode et al. (63) obtained evidence supporting the notion that the p89/XPB helicase is essential for open complex formation and transcription in vitro, whereas the p80 helicase was found to be not essential but rather to stimulate these reactions. Notably, p89/XPB and p80/XPD have been implicated in nucleotide excision repair in addition to RNAPII transcription (60). Mutations in these polypeptides are associated with rare genetic disorders such as xeroderma pigmentosum, Cockayne's syndrome (CS), and trichothiodystrophy (TTD).
Characterizing the molecular organization of the preinitiation complex and its isomerization before initiation is essential for understanding transcriptional mechanisms and regulation. Evidence obtained using both prokaryotic and eukaryotic systems indicates that the promoter DNA is wrapped around RNA polymerase en route to initiation (5). Robert et al. (51) analyzed the topological organization of preinitiation complexes assembled using RNAPII and various combinations of the general initiation factors including TBP, TFIIB, TFIIE, and TFIIF (wild type and mutated) in the absence of TFIIH to determine the structure of putative intermediates in the formation of the preinitiation complex. The results support a model in which the promoter DNA is progressively wrapped around the polymerase prior to initiation. The binding of TBP and TFIIB initiates DNA wrapping by inducing a bend, or kink, into the DNA helix. TFIIE and TFIIF, which both exist as α2β2 heterotetramers in solution (3, 15, 41) and in the preinitiation complex (51), tighten the DNA wrap around RNAPII, probably by inducing a second important bend in the promoter around the TIS (17, 28). Tight DNA wrapping around RNAPII was shown to minimally require a fragment of RAP74 that contains both the RAP30-binding domain (66) and a region necessary for the formation of homomeric interactions by RAP74, that is, one capable of maintaining TFIIF as a heterotetramer (51). This RAP74 homomeric interaction region (HIR1), which is composed of amino acids 172 to 205, is also important for transcription initiation in vitro (32, 33, 67).
Using a highly sensitive technique for photo-cross-linking proteins to specific sites along promoter DNA, we have analyzed the molecular organization of preinitiation complexes assembled with various combinations of the general initiation factors and RNAPII either in the absence or in the presence of ATP. Our results provide important information on the structure of the preinitiation complex, the mechanism of open complex formation, and the roles of the various general initiation factors and RNAPII in transcription initiation.
MATERIALS AND METHODS
Protein factors.
Recombinant TBP, TFIIB, RAP30, RAP74 (full length and deletion mutants), TFIIE56 (α), TFIIE34 (β) (full length and deletion mutants), and RNAPII were prepared as described previously (42, 51). TFIIH isolated from HeLa cells was purified as previously described (18), except for modifications in the last two purification steps. The heparin 5PW 0.4 M KCl-eluted fraction was dialyzed against 50 mM Tris-HCl (pH 7.9)–50 mM KCl–0.5 mM dithiothreitol, 0.1 mM EDTA–8.7% glycerol and then loaded on a phenyl 5PW column (0.75 by 7.5 cm; flow rate, 0.6 ml/min). After a 0.9 M KCl wash, TFIIH was eluted with a 15-ml linear gradient from 0.9 to 0 M ammonium sulfate, and the peak fractions containing the TFIIH activity were loaded on a hydroxylapatite column (0.75 by 7.5 cm; flow rate, 0.4 ml/min) equilibrated with a solution containing 10 mM potassium phosphate (pH 6.0), 0.01 mM CaCl2, 0.5 mM dithiothreitol, and 8.7% glycerol. TFIIH was then eluted using a linear 0.2 to 0.6 M PO4 buffer gradient, and fractions were stored at −90°C in aliquots. Under these conditions, we obtained highly purified TFIIH at a concentration of ∼5 mg/ml from 50 × 109 cells.
Mutated TFIIH was immunopurified from patient cell extracts as previously described (2). Briefly, human lymphoblastoid cell lines GM2252 [derived from patient XP11BE; IIH-XPB(C-A)] and GM1855 (derived from patient XP11BE's mother; IIH-XPBwt) were grown in suspension in RPMI 1650 medium (Life Technologies, Inc.) supplemented with 10% fetal calf serum. Whole cell extract was fractionated on a heparin-Ultrogel column (Sepracor), and the active fractions were then immunopurified using protein A-agarose beads (Pharmacia, Uppsala, Sweden) cross-linked to p44-specific antibody 1H5 (2). After extensive washing, the resin was eluted using p44 peptide in the presence of insulin. After dialysis, 10 mg of highly purified TFIIH was stored in aliquots at a concentration of 25 mg/ml.
Recombinant TFIIH (rIIH9) and recombinant XPB [rXPBwt, rXPB(GKT), and rXPB(C-A)] were purified as previously described from Sf9 cells infected with baculoviruses expressing His-p89/XPB, p89/XPB, p80/XPD, p62, p52, p44, p34, cdk7, cyclin H, and/or MAT1 (63).
N3R photo-cross-linking and immunoprecipitation.
Synthesis of the photoreactive nucleotide N3R-dUMP, preparation of the photoprobes, and conditions for binding reactions were as described elsewhere (49) except that 25 U of DNase I and 800 U of nuclease S1 were used for the nuclease treatments. For each photoprobe, the concentration of poly(dI-dC) in the binding reactions was optimized so as to favor specific over nonspecific binding. A typical reaction with all factors contained 200 ng each of TBP, TFIIB, RAP30, RAP74, TFIIE34, TFIIE56, and purified RNAPII and 50 ng of TFIIH as specified in the figure legends. UV irradiation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of radiolabeled photo-cross-linking products were as described elsewhere (49). To identify the cross-linked polypeptides, the products of some cross-linking reactions were immunoprecipitated using specific antibodies directed against the various subunits of the factors as previously described (49). Some cross-linking reactions contained 1 mM ATP, ATPγS, or GTP (see figure legends).
Characterization of purified TFIIH.
TFIIH, either purified from HeLa cells or immunopurified from patient cells using the p44 antibody 1H5, was analyzed on SDS-gels stained with Coomassie blue or revealed by Western blotting using antibodies directed against the recombinant subunits. In vitro transcription reactions using a template containing the adenovirus major late promoter were performed in the presence of purified general transcription factors TBP, TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH, as well as RNAPII, as previously described (2). Helicase activities were measured using a standard assay (2). The substrate was obtained by annealing an oligonucleotide corresponding to fragment 6219–6255 of single-stranded M13mp19(−) DNA to single-stranded M13mp19(+) DNA. The resulting heteroduplex was digested with EcoRI and then extended to 21 and 20 bp, respectively, with Klenow polymerase in the presence of dTTP and [α-32P]ATP.
RESULTS
Photo-cross-linking of a TBP-TFIIB-TFIIF-RNAPII-TFIIE-TFIIH complex along promoter DNA.
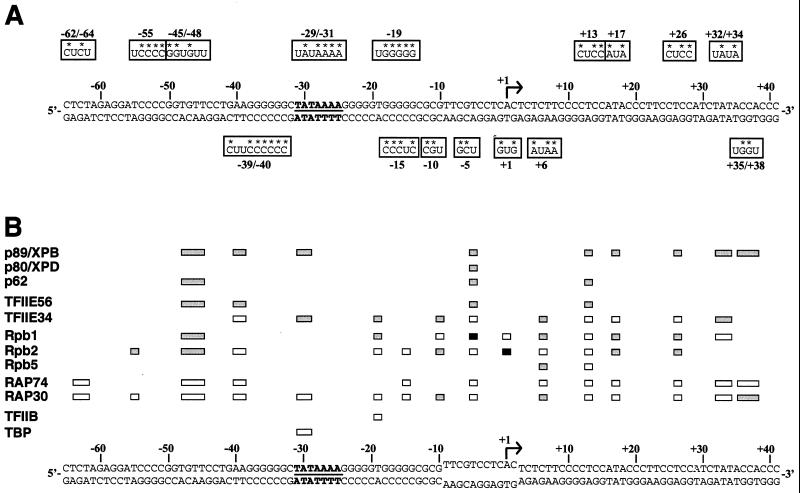
We have previously used site-specific protein-DNA photo-cross-linking to determine the molecular organization of a preinitiation complex assembled on promoter DNA in the presence of TBP, TFIIB, TFIIE, TFIIF, and RNAPII (6, 10, 17, 50, 51). We have now extended our topological analysis to a preinitiation complex containing TFIIH in addition to the other general initiation factors and RNAPII. As we have previously described, the photo-cross-linking reactions were performed in both the presence and absence of TBP in order to be able to assess the specificity of the cross-linking signals. Photo-cross-linking signals that are significantly weaker in the absence of TBP are considered as specific because the omission of TBP always had the same effect as using a photoprobe with a mutated TATA element (49). Sixteen photoprobes in which one (10 of 16), two (5 of 16), or three (1 of 16) photoreactive nucleotides are placed at specific locations along the promoter DNA were used in our analysis (Fig. 1A). Probes with more than three photonucleotides (e.g., photoprobes −25/−31, −56/−61, and −14/−2 [51]) are not included here because they did not provide sufficient resolution. As summarized in Fig. 1B, the association of TFIIH with the complex induces a number of additional cross-links of the transcription machinery along the promoter DNA, including that of RAP30 to positions −10, +6, and +35/+38, Rpb2 to positions −55, −45/−48, −10, +17, and +26, Rpb1 to positions −45/−48, −19, +6, +17, and +26, Rpb5 to position +6, and TFIIE34 to positions −29/−31, −19, −10, +6, and −32/+34. TFIIE56, a factor that was not cross-linked along promoter DNA in the absence of TFIIH, probably because it is positioned at a distance from the promoter DNA, is now cross-linked to several positions (positions −45/−48, −39/−40, −5, and +13) in the presence of TFIIH. Together, these results indicate that the association of TFIIH with the complex tethers some components, mainly RNAPII and TFIIE subunits, to the promoter DNA, supporting the idea that TFIIH tightens the DNA wrap around the enzyme.
FIG. 1.
Cross-linking of TBP, TFIIB, TFIIF (RAP74 and RAP30), TFIIE (TFIIE56 and TFIIE34), RNAPII (Rpb1, Rpb2, and Rpb5) and TFIIH (p89/XPB, p80/XPD, and p62) along promoter DNA. (A) Photoprobes derived from the adenovirus major late promoter. For each photoprobe, positions of the photoreactive (U) and radiolabeled (*) nucleotides are indicated. (B) Summary of cross-linking data. Specific cross-links that do (gray boxes) and do not (open boxes) require the presence of TFIIH are indicated. Black boxes indicate the new cross-links induced in the presence of ATP. A cross-linking signal was considered specific when its intensity was significantly higher in reactions containing TBP than in those lacking TBP (see text). A number of photoprobes that place the photonucleotide either upstream of position −62/−64 or downstream of position +35/+38 have also been used, but no significant cross-linking signals were obtained.
We have also obtained the cross-linking of the three largest subunits of TFIIH to a number of positions from −45 to +35. Our results are summarized in Fig. 1B, and representative examples are shown in Fig. 2A. p89/XPB cross-links downstream of the TIS (positions +13, +17, +26, +32/+34, and +35/+38), between the TATA box and the TIS (position −5), and to the TATA box and upstream of it (positions −29/−31, −39/−40, and −45/−48). p80/XPD cross-links weakly to position −5. The p62 subunit of TFIIH cross-links weakly upstream of the TATA element (position −45/−48), to position −5, and downstream of the TIS (position +13). Significantly, in the absence of ATP the two DNA helicases of TFIIH approach the promoter DNA in the region where the ATP-dependent melting of the promoter is to occur (e.g., between positions −9 and +2). Because p89/XPB also contacts the promoter DNA outside the −9/+2 region, our results suggest that this helicase of TFIIH functions by holding the DNA in at least two distinct regions simultaneously, with one being the section of DNA to be unwound. Because the cross-linking of p89/XPB is both stronger and more extensive than that of p80/XPD, our results are consistent with the notion that p89/XPB is the TFIIH helicase involved in promoter melting before transcription initiation.
FIG. 2.
Cross-linking of TFIIH subunits along promoter DNA. (A) Cross-linking of TFIIH subunits upstream of the TATA element, downstream of the TIS, and to position −5. Photo-cross-linking experiments using photoprobes −39/−40, −5, and +13 were performed with TFIIB, TFIIE, TFIIF, TFIIH, and RNAPII in either the presence (+) or absence (−) of TBP. In these experiments, a truncated form of RAP74 [RAP74(1-217)], which migrates at ∼35 kDa, was used to facilitate visualization of the cross-linking signals in the 50- to 100-kDa region of the gels. (B) Identification of affinity-labeled TFIIH subunits. Cross-linked polypeptides were immunoprecipitated (IP) with specific antibodies directed against recombinant p89/XPB and p80/XPD and analyzed by SDS-PAGE. The immunoprecipitated polypeptides comigrated with p89/XPB and p80/XPD controls that were electrophoresed on the gel and revealed by silver staining. p62 was identified according to its mobility in the cross-linking gels, which is indistinguishable from that of the silver-stained subunit but slightly different from that of TFIIE56. (C) Cross-linking of recombinant TFIIH. TFIIH reconstituted from its cloned subunits (rIIH9) was included in cross-linking experiments using photoprobe +13. p89/XPB carries a His tag (His-p89), and its mobility is slightly slower than that of the natural subunit.
The identification of the three largest subunits of TFIIH in our cross-linking gels relies on a number of criteria. Affinity-labeled p89/XPB and p80/XPD were immunoprecipitated with specific antibodies directed against recombinant subunits (Fig. 2B). Highly purified TFIIH was also loaded on several of our cross-linking gels. The gel was cut in parts; the part containing purified TFIIH was silver stained, while that containing the cross-linked products was autoradiographed. Under our gel conditions, silver-stained and radiolabeled bands corresponding to p89/XPB, p80/XPD, and p62 comigrated (Fig. 2B). In addition, we performed a cross-linking experiment using photoprobe +13 in the presence of rIIH9, in which p89/XPB is tagged with six histidine residues. Compared to natural TFIIH, the mobility of affinity-labeled p89/XPB was slightly lower when we used rIIH9 with His-tagged p89/XPB in our experiments (Fig. 2C).
The HIR1 domain of RAP74 is necessary for promoter contacts by p89/XPB both upstream of the TATA box and in the −9/+2 region.
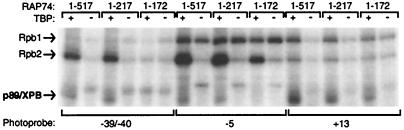
Previous results have indicated that the HIR1 region of RAP74 (amino acids 172 to 205) is important for both tight wrapping of the promoter DNA in the preinitiation complex (51) and efficient transcription in vitro (32, 33, 66). We decided to analyze the role of RAP74 in the establishment of the promoter contacts by TFIIH subunits. Cross-linking experiments were performed with all of the general initiation factors and RNAPII in the presence of various RAP74 deletion mutants [RAP74(1-517), RAP74(1-217), and RAP74(1-172)]. In these experiments we used RAP74(1-217) instead of RAP74(1-205) as the minimal fragment containing HIR1 for convenience, but both deletion mutants provided basically indistinguishable results in our cross-linking experiments (data not shown). As shown in Fig. 3 and summarized in Table 1, in the presence of RAP74(1-172) p89/XPB cross-linked to the region downstream of the TIS between nucleotides +13 and +35. However, in the presence of either RAP74(1-517) (full length) or RAP74(1-217), which contain HIR1 in addition to the RAP30-binding domain, the additional promoter contacts by p89/XPB in the −30/−45 region are obtained (the result using photoprobe −39/−40 is shown as an example in Fig. 3). This result suggests that DNA wrapping around RNAPII causes the juxtaposition of the DNA helices upstream of the TATA box and downstream of the TIS, thereby allowing p89/XPB to cross-link to both regions simultaneously. Strikingly, the contact by p89/XPB in the −9/+2 region (e.g., position −5) also requires the presence of HIR1 in the RAP74 fragments [RAP74(1-217) and RAP74(1-517)] (Fig. 3). This result suggests that DNA wrapping induced by TFIIF is required to position the p89/XPB helicase of TFIIH in contact with the DNA region from −9 to +2 that will be its substrate during open complex formation.
FIG. 3.
Cross-linking of p89/XPB using various RAP74 deletion mutants. Cross-linking experiments using photoprobes −39/−40, −5, and +13 were performed with TFIIB, TFIIE, TFIIH, RNAPII, RAP30, and RAP74(1-517), RAP74(1-217), and RAP74(1-172)] in the presence (+) or the absence (−) of TBP. RAP74(1-172) does not contain the homomeric interaction region 1 (HIR1), while RAP74(1-217) and RAP74(1-517) do. A summary of p89/XPB cross-linking using the RAP74 deletion mutants is shown in Table 1.
TABLE 1.
Summary of p89/XPB cross-linking in the presence of RAP74 deletion mutantsa
| Photoprobe | Specific cross-linking of p89/XPB in the presence of:
|
||
|---|---|---|---|
| RAP74(1-517) | RAP74(1-217) | RAP74(1-172) | |
| −62/−64 | − | − | − |
| −55 | − | − | − |
| −45/−48 | + | + | − |
| −39/−40 | + | + | − |
| −29/−31 | + | + | − |
| −19 | − | − | − |
| −15 | − | − | − |
| −10 | − | − | − |
| −5 | + | + | − |
| +1 | − | − | − |
| +6 | − | − | − |
| +13 | + | + | + |
| +17 | + | + | + |
| +26 | + | + | + |
| +32/+34 | + | + | + |
| +35/+38 | + | + | + |
Examples are shown in Fig. 3.
Two distinct domains of TFIIE34 are involved in the positioning of TFIIH in the preinitiation complex.
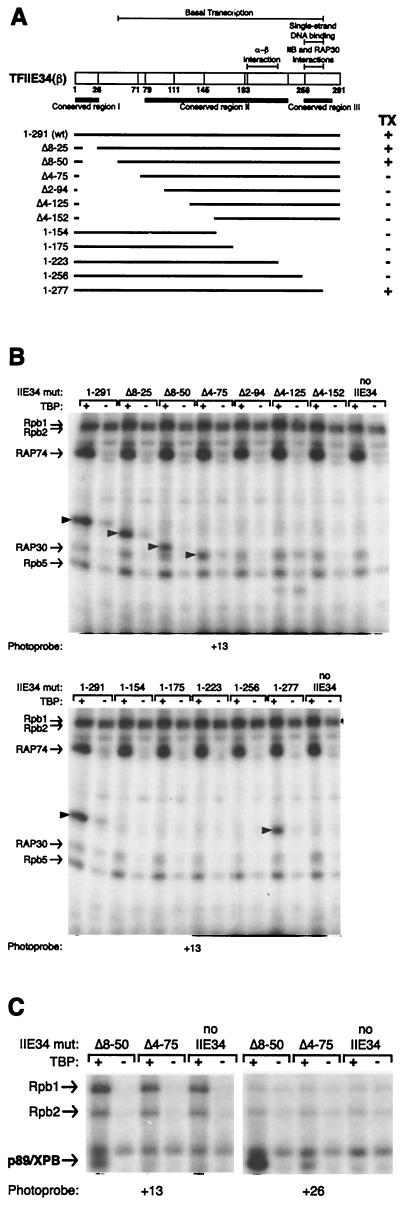
We have previously localized the two TFIIE34 molecules of the TFIIE heterotetramer along promoter DNA in the preinitiation complex (50, 51). Here we have determined the promoter contacts by TFIIE in the presence of TFIIH (Fig. 1). We next used a series of TFIIE34 deletion mutants to define the minimal domain capable of cross-linking along promoter DNA. The various mutants used in our analysis are represented in Fig. 4A, and the TFIIE34 cross-linking data are summarized in Table 2. The central region of TFIIE34 spanning amino acids 76 to 277 constitutes the minimal fragment essential for the cross-linking of the small TFIIE subunit along the promoter DNA (Fig. 4B shows two representative cross-linking gels). Although TFIIE34(Δ4-75) can associate with the preinitiation complex, this mutant does not support basal transcription activity in vitro (42).
FIG. 4.
Cross-linking of p89/XPB using various TFIIE34 deletion mutants. (A) Linear representation of TFIIE34 wild type (wt) and deletion mutants. The ability to support basal transcription (TX) in vitro is indicated by a plus sign. (B) Cross-linking experiments using photoprobe +13 were performed with TBP, TFIIB, TFIIE, TFIIF, and RNAPII in the presence of the various TFIIE34 deletion mutants (mut). Positions of the cross-linked TFIIE34 fragments are indicated by arrows on the gel. A summary of TFIIE34 deletion mutant cross-linking is provided in Table 2. TFIIE34(Δ4-75) cross-links have been confirmed by immunoprecipitation with an antibody raised against recombinant TFIIE34. (C) Cross-linking reactions using photoprobes +13 and +26 were performed as for panel B except that TFIIH was included in the reactions. A summary of p89/XPB cross-linking using the TFIIE34 deletion mutants is provided in Table 3.
TABLE 2.
Summary of TFIIE34 deletion mutant cross-linkinga
| Photoprobe | Specific cross-linking
|
||||||
|---|---|---|---|---|---|---|---|
| TFIIE34(1-291) | TFIIE34(1-277) | TFIIE34(1-256) | TFIIE34(Δ8-25) | TFIIE34(Δ8-50) | TFIIE34(Δ4-75) | TFIIE34(Δ2-94) | |
| −39/−40 | + | + | − | + | + | + | − |
| −5 | + | + | − | + | + | + | − |
| +13 | + | + | − | + | + | + | − |
| +26 | + | + | − | + | + | + | − |
Examples are shown in Fig. 4B.
We next analyzed the cross-linking of TFIIH subunits in reactions assembled with the various TFIIE34 deletion mutants. The crude data are summarized in Table 3, and some representative gels are shown in Fig. 4C. Deletion mutant TFIIE34 (Δ4-75) supported only the cross-linking of p89/XPB to the TATA box and immediately upstream of it (positions −29/−31 and −39/−40) and downstream of the TIS (positions +26, +32/+34, and +35/+38), whereas a fragment with a shorter deletion (e.g., amino acids 8 to 50) supported the cross-linking of p89/XPB to all specific positions (from −45/−48 to −29/−31, from +13 to +35/+38, and at −5). Interestingly, TFIIE34(Δ4-75) is inactive in basal transcription reactions, whereas TFIIE34(Δ8-50) is fully active (42). These results indicate that TFIIE34(Δ4-75) allows the association of TFIIH with the preinitiation complex, but that the contacts are limited to the regions where the upstream and downstream helices cross in the wrapped DNA structure. A fragment carrying additional amino acids [e.g., TFIIE34(Δ8-50)] fully supports all promoter contacts by TFIIH, including that at nucleotide −5, indicating that these promoter contacts are essential for transcription initiation.
TABLE 3.
Summary of p89/XPB cross-linking in the presence of TFIIE34 deletion mutants
| Photoprobe | Specific cross-linking of p89/XPB in the presence of:
|
|||||
|---|---|---|---|---|---|---|
| TFIIE34(1-291) | TFIIE34(1-277) | TFIIE34(Δ8-25) | TFIIE34(Δ8-50) | TFIIE34(Δ4-75) | TFIIE34(Δ2-94) | |
| −62/−64 | − | − | − | − | − | − |
| −55 | − | − | − | − | − | − |
| −45/−48 | + | − | + | + | − | − |
| −39/−40 | + | − | + | + | + | − |
| −29/−31 | + | − | + | + | + | − |
| −19 | − | − | − | − | − | − |
| −15 | − | − | − | − | − | − |
| −10 | − | − | − | − | − | − |
| −5 | + | − | + | + | − | − |
| +1 | − | − | − | − | − | − |
| +6 | − | − | − | − | − | − |
| +13 | + | − | + | + | − | − |
| +26 | + | − | + | + | + | − |
| +32/+34 | + | − | + | + | + | − |
| +35/+38 | + | − | + | + | + | − |
Examples are shown in Fig. 4C.
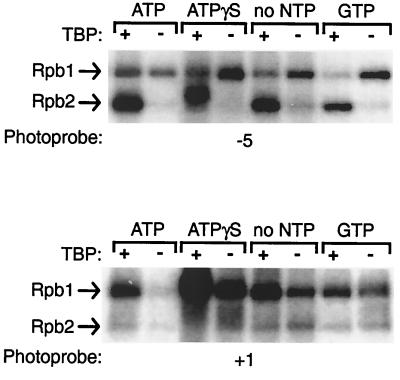
New promoter contacts by RNAPII subunits at positions −5 and +1 are induced by ATP.
TFIIH, mainly through its p89/XPB DNA helicase, is essential for the melting of promoter DNA between −9 and +2 prior to transcription initiation and during promoter clearance (22, 24, 63). Our cross-linking data indicate that both helicases make promoter contacts in the −9/+2 region in addition to other regions (Fig. 1). To analyze open complex formation, we have compared the cross-linking of preinitiation complexes assembled with all of the general initiation factors and RNAPII in the presence or the absence of ATP. Surprisingly, the addition of ATP resulted in only two modifications in the cross-linking of components of the transcription machinery along promoter DNA. In the presence of ATP, Rpb1 cross-links specifically to position −5 and Rpb2 cross-links to positions +1 (Fig. 1). The addition of GTP or ATPγS instead of ATP does not support the cross-linking of Rpb1 to position −5 and Rpb2 to position +1 (Fig. 5). In addition, and significantly, the two new promoter contacts induced by the presence of ATP are to the template strand of the promoter DNA. These results indicate that promoter melting between nucleotides −9 and +2, which is catalyzed by TFIIH in an ATP-dependent manner, tethers the template strand of the DNA to the surface of the catalytic center of the polymerase.
FIG. 5.
Nucleotide requirement for the new promoter contacts by RNAPII in the −9/+2 region. The cross-linking experiments using photoprobes −5 and +1 were performed in the presence of ATP, ATPγS, or GTP or in the absence of ribonucleoside triphosphate (NTP).
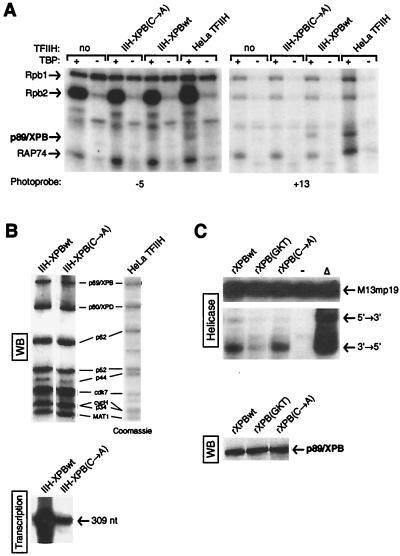
TFIIH with a mutation in p89/XPB associated with XP does not associate properly with the preinitiation complex.
Mutations in p89/XPB and p80/XPD found in patients with XP and/or CS have been shown to impair the transcription functions of TFIIH. One of these patients (XP11BE) carries a C-A transversion in the last intron of the XPB gene which generates a splice mutation at the RNA level (68). TFIIH was previously purified from lymphoblastoid cells of patient XP11BE. Cells from this XP/CS patient are heterozygous for the XPB mutation and express only the mutated (paternal) allele (2, 68). This mutated TFIIH is severely impaired in both promoter melting and in vitro transcription, whereas it is fully capable to function in a DNA helicase assay and to phosphorylate the RNAPII CTD (2). Equivalent amounts of TFIIH isolated from HeLa cells, cells from patient XP11BE [IIH-XPB(C-A)], and cells from patient XP11BE's mother (IIH-XPBwt) were included in our cross-linking experiments (Fig. 6). Although p89/XPB cross-linked to positions +13 and −5 when we used HeLa TFIIH and IIH-XPBwt, we did not obtain the cross-linking of TFIIH subunits to promoter DNA when we used IIH-XPB(C-A) (Fig. 6A). Longer exposures of the cross-linking gels failed to reveal promoter contacts by IIH-XPB(C-A). As shown in Fig. 6B and C, the C-A mutation does not affect the DNA helicase activity of XPB [compare rXPBwt to rXPB(C-A) and rXPB(GKT), which is mutated in the ATP-binding site] but impairs the transcription activity of the TFIIH complex containing the mutated XPB [compare IIH-XPBwt to IIH-XPB(C-A)] as observed previously (2). This result indicates that the TFIIH from patient XP11BE does not associate properly with the preinitiation complex.
FIG. 6.
Cross-linking of TFIIH with a mutation in p89/XPB associated with XP. (A) Cross-linking experiments using photoprobes −5 and +13 were performed in the absence or presence of TFIIH isolated either from HeLa cells or from patient cells with either a wild-type (IIH-XPBwt) or mutated [IIH-XPB(C-A)] TFIIH. (B) Runoff transcription activity of the various TFIIHs. Immunopurified wild-type (IIH-XPBwt) and mutated [IIH-XPB(C-A)] TFIIH were analyzed using Western blotting (WB) and in vitro transcription assays (Transcription). An SDS-polyacrylamide gel containing highly purified HeLa TFIIH is included for comparison. Positions of the TFIIH subunits and runoff transcript (309 nucleotides [nt]) are indicated. (C) DNA helicase activity of the various XPBs. rXPB, either wild type (rXPBwt), with a mutated ATP-binding site [rXPB(GKT)], or carrying the C-A mutation [rXPB(C-A)], was analyzed in a standard helicase assay. The positions of the heteroduplex substrate and the products of both 5′-3′ and 3′-5′ unwinding reactions are shown. −, reaction performed in the absence of XPB; Δ, reaction heated to serve as a positive control. rXPBs were analyzed by Western blotting (WB).
DISCUSSION
Topological organization of a TBP-TFIIB-TFIIF-RNAPII-TFIIE-TFIIH complex on promoter DNA.
A model for the molecular organization of the preinitiation complex is shown in Fig. 7A. This model includes a number of features that account for both our cross-linking data and additional data obtained in different laboratories (see below). The main feature of the model is that promoter DNA is tightly wrapped around RNAPII. DNA bending and wrapping during transcription initiation is supported by a large body of evidence (5). First, four important general transcription initiation factors, TBP, TFIIB, RAP30, and TFIIE34, and RNAPII have been implicated in DNA bending. The binding of TBP to the TATA element of promoters induces DNA bending by ∼90° (27, 30). The DNA bend in the TBP-promoter complex is further stabilized by the binding of TFIIB (1, 37). The domain of RAP30 required to support transcription initiation in vitro contains a cryptic DNA-binding domain which is structurally similar to the winged helix-turn-helix DNA-binding domains of linker histone H5, suggesting a role in DNA wrapping (20). The central core domain of TFIIE34 was recently shown to also contain a winged helix motif (43). The structure of the RNAPII elongation complex has revealed an important DNA bend in the region of the TIS (47). Second, RNAPII was found to cross-link to the promoter DNA from nucleotides −39/−40 to +13 in the absence of TFIIH (51) and from nucleotides −55 to +32/+34 in the presence of TFIIH (this report). These promoter regions represent about 18 and 30 nm, respectively, of B-form DNA and are longer than the longest dimension of RNAPII (14 nm) (9). This observation alone argues against promoter DNA being in a linear conformation in the preinitiation complex. Third, electron micrographs of preinitiation complexes lacking TFIIH allow the direct visualization of DNA wrapping in the complex and were used to estimate that 50 ± 20 bp of DNA is consumed within the structure, fully supporting the notion of tight DNA bending and wrapping (17, 28). Fourth, if promoter DNA is tightly wrapped around RNAPII, upstream and downstream promoter DNA segments will be found to closely approach one another near the cross-point of the loop. Using RAP74 deletion mutants in our cross-linking experiments, we have previously obtained evidence that one molecule each of RAP74 and RAP30 simultaneously contact promoter DNA in the −40/−60 and +10/+30 regions, indicating that the two helices are juxtaposed (51). Here, we provide evidence that p89/XPB also makes simultaneous contacts with upstream and downstream promoter regions. Promoter contacts by p89/XPB in both promoter regions require the presence of both TFIIE and TFIIF, two factors that are necessary for tight DNA wrapping in the preinitiation complex. Fifth, TFIIA, a factor that tightly binds to TBP and is located in a region centered immediately upstream of the TATA element from positions −42 to −30 in the context of a TBP-TFIIA-promoter complex (6, 31), also cross-links to nucleotide +26 in the context of a TBP-TFIIA-TFIIB-TFIIF-RNAPII-TFIIE promoter complex. The cross-linking of TFIIA to position +26 requires the presence of both TFIIE and TFIIF, and direct protein-protein interactions of TFIIA with RAP74, TFIIE56, and TFIIE34 have been reported (70; M. F. Langelier, D. Forget, A. Rojas, Z. F. Burton, and B. Coulombe, unpublished data). Sixth, tight DNA bending and wrapping around TFIID (38) and the prokaryotic RNA polymerase open complex (48), not to mention nucleosomes, have been reported, indicating that DNA bending and wrapping may constitute a fundamental transcription mechanism.
FIG. 7.
(A) Proposed structure for the preinitiation complex containing TBP, TFIIB, TFIIF (F74 and F30), TFIIE (E56 and E34), TFIIH, and RNAPII. The relative positions of the various factors and RNAPII are as predicted from our cross-linking data and published observations from a number of laboratories (see Discussion). In the front view, the complex is shown with the promoter DNA between the TATA element and the TIS being placed in the direction of the view. (B) A model for the mechanism of promoter melting by TFIIH. TFIIH is in yellow, and its p89/XPB subunit is in orange. For a detailed description, see Discussion.
In the model (Fig. 7A), TFIIE and TFIIF are present in the complex as α2β2 heterotetramers. The heterotetrameric structure is maintained by homomeric interactions of TFIIE34 and RAP74 (e.g., RAP30-RAP74-RAP74-RAP30 and TFIIE56-TFIIE34-TFIIE34-TFIIE56), consistent with the results of in vitro binding assays (51; Y. Ohkuma, unpublished data). The TFIIE and TFIIF subunits were positioned so that they can account for both our cross-linking data and the protein-protein interactions determined in various laboratories (5, 21, 44). TBP binds to TFIIB and TFIIE; TFIIB binds to TFIIE and TFIIF; RNAPII binds to TBP, TFIIE, TFIIF, and TFIIH; TFIIF binds to TFIIE; and TFIIE binds to TFIIH. The structure of RNAPII has been modeled to account for both the dimension of the yeast enzyme and the presence of a 2.5-nm channel that can accommodate the promoter DNA and can exist in either an open or closed conformation (7, 9, 47, 72). Finally, the structure of TFIIH is according to electron microscopy determination (55). TFIIH has been positioned in such a way that cdk7 is in the vicinity of the CTD of the largest RNAPII subunit (10), while other parts can make promoter contacts upstream of the TATA element, downstream of the TIS, and between the TATA element and the TIS at position −5, as determined by our photo-cross-linking experiments.
Proposed mechanism of promoter melting by the p89/XPB ATPase/helicase of TFIIH.
In Fig. 7B, we propose a model for promoter melting by p89/XPB that accounts for our cross-linking results. In the presence of RAP74(1-172), a mutant that supports neither tight DNA wrapping nor efficient basal transcription in vitro, p89/XPB makes promoter contacts only downstream of the TIS between positions +13 and +35/+38 (Fig. 7B, top panel). Under these conditions, the addition of ATP does not induce new promoter contacts by RNAPII in the promoter region where DNA melting is to occur (e.g., from −9 to +2). When RAP74(1-217), a mutant that contains the HIR1 domain necessary for tight DNA wrapping and efficient transcription initiation in vitro, is used, p89/XPB makes additional promoter contacts upstream of the TATA box between positions −29/−31 and −45/−48, presumably because tight DNA wrapping brings this promoter region closer to that between +13 and +35/+38 (Fig. 7B, middle panel). The additional promoter contact by p89/XPB at position −5, where the promoter DNA is to be melted in the presence of ATP, also requires the presence of RAP74(1-217) and larger mutants. The entry of TFIIH into the complex, which requires the presence of TFIIE, further tightens the DNA wrap around RNAPII. The fragment of TFIIE34 that is minimally required for both the accurate positioning of TFIIH in the complex and the tight wrapping of promoter DNA is also minimally required for transcription initiation in vitro. These findings support the notion that tight DNA wrapping destabilizes the DNA helix immediately upstream of the TIS, thereby inducing sufficient unwinding of the DNA strands to allow for the binding of p89/XPB to its single-stranded DNA substrate (34). At this stage, p89/XPB makes promoter contacts on both sides of a DNA bend centered on the TIS. ATP hydrolysis by the ATPase activity of p89/XPB is predicted to induce a conformational change in the helicase that pulls the template strand of the DNA further away from its partner strand in order to catalyze open complex formation, thereby causing additional contacts by the template strand of the DNA to RNAPII (Fig. 7B, bottom panel). Whether promoter melting by p89/XPB from −9 to +2 is achieved in a single step that requires the hydrolysis of one molecule of ATP or proceeds progressively in steps of one or a few (two to three) base pairs at a time in a process that requires the hydrolysis of several molecules of ATP is not known.
A model for DNA unwinding by the prokaryotic 3′-5′ DNA helicase PcrA has been deduced from the structure of the helicase-DNA complex (65). In this model, called the inchworm model, the helicase works as a monomer that can simultaneously bind ssDNA and double-stranded DNA (dsDNA). In the absence of ATP, the helicase is bound to ssDNA but not to dsDNA. Upon binding ATP, the protein undergoes a conformational change and the duplex region is bound by the helicase, with a concomitant unwinding of several base pairs at the junction. Finally, following ATP hydrolysis, the protein returns to its initial conformation as the protein translocates along the ssDNA by one base and releases the duplex DNA. Although PcrA is involved in DNA repair and rolling circle replication, our proposed mechanism for promoter melting has a number of similarities with the inchworm model. In both models, the helicase is monomeric (as opposed to multihomomeric DNA helicases such as T7 DNA helicase), possesses distinct binding domains for ssDNA and dsDNA, and requires a conformational change induced by ATP to exert its function. Although we do not have experimental evidence to support the occurrence of a conformational change in p89/XPB upon ATP binding or hydrolysis, the mechanism of action of several DNA helicases necessitates a conformational change in the protein (34).
Recently, Kim et al. published a detailed analysis of protein-DNA interactions in the RNAPII preinitiation complex containing TFIIH (29). In sharp contrast to the results presented here, these authors have shown that (i) the entry of TFIIH into the preinitiation complex does not substantially alter protein-DNA interactions by RNAPII and the general initiation factors, (ii) the promoter contacts by TFIIH are made by only one of its nine subunits (ERCC3/p89/XPB) and are located exclusively downstream of the transcription bubble, and (iii) the addition of ATP induces changes in TFIIH-DNA interactions downstream of the transcription bubble region. On the basis of these results, Kim et al. (29) have proposed a model for the mechanism of promoter melting by TFIIH in which, in the presence of ATP, the IIH ERCC3 (XPB) helicase rotates the DNA segment downstream of the transcription bubble relative to the rotationally fixed upstream interactions, thereby inducing melting of the transcription bubble region. In their study, however, Kim et al. (29) analyzed transcription complexes that were washed with the detergent Sarkosyl prior to UV cross-linking. The treatment of transcription complexes with the same concentration of Sarkosyl has previously been shown to alleviate the requirement for TFIIH, ATP, and downstream promoter sequences in transcription assays (A. Dvir, R. Conaway, and J. Conaway, personal communication), indicating that Sarkosyl disrupts a number of protein-protein and/or protein-DNA interactions that are normally important for transcription initiation. The disruption of regulatory interactions may also be responsible for the hyperactivity of Sarkosyl-washed complexes in phosphorylating the CTD of RNAPII before transcription initiation observed by Kim et al. (29). Most likely, the promoter contacts by p89/XPB in the region of the transcription bubble are among these interactions that are lost following Sarkosyl treatment. Similarly, the changes in promoter contacts by TFIIH downstream of the initiation site in the presence of ATP may reflect the faulty association of TFIIH with the initiation complex after Sarkosyl treatment.
An XPB mutation associated to a severe form of XP affects the positioning of TFIIH in the preinitiation complex.
In addition to playing a role in RNAPII transcription, the DNA helicases of TFIIH participate in nucleotide excision repair. Mutations in p89/XPB and p80/XPD are associated with rare genetic disorders including XP, CS, and TTD. However, the strong heterogeneous clinical features observed in these patients cannot be explained solely by defects in nucleotide excision repair. A form of TFIIH isolated from a XP/CS patient with a C-A mutation in the XPB gene was found to be significantly impaired in both transcription initiation and promoter melting, whereas its 3′-5′ helicase, ATPase, and CTD kinase activities are not affected (2, 68). Notably, the same C-A mutation does not impair DNA melting in nucleotide excision repair but rather affects 5′ incision formation (12). Our results suggest that the mutated TFIIH [IIH-XPB(C-A)] is deficient in promoter melting because the positioning of its p89/XPB helicase onto promoter DNA is impaired (Fig. 6). One interpretation of our results is that the C-A mutation in p89/XPB affects the accuracy of positioning and/or the stability of the association of TFIIH with the promoter DNA in such a way that some of its activities are preserved. Promoter melting (and possibly 5′ incision formation in nucleotide excision repair), however, which requires the accurate and stable association of TFIIH in order to make the promoter contacts necessary to completely unwind the DNA in the region of the TIS (and possibly to promote 5′ incision during nucleotide excision repair), is specifically affected in this mutated TFIIH. This conclusion implies that promoter melting and CTD phosphorylation have different topological requirements, suggesting the possibility that TFIIH is repositioned following transcription initiation in order to phosphorylate the CTD and support promoter clearance. A detailed analysis of the molecular organization of early elongation complexes is now required to test this intriguing possibility and to advance our understanding of transcriptional mechanisms.
ACKNOWLEDGMENTS
We thank Diane Bourque and Vincent Trinh for the computer-generated models, and we thank Will Home and Diane Forget for critical reading of the manuscript. We are also grateful to our colleague Zachary Burton for helpful suggestions and for providing the TFIIF deletion mutants.
This work was supported by grants from the Medical Research Council of Canada and the Cancer Research Society, Inc. (to B.C.); the Human Frontier Science Program (grant RG0193/97M), INSERM, CNRS, and Association pour la Recherche sur le Cancer (to J.M.E.); and the Ministry of Education, Science and Culture of Japan and the Core Research for Evolutional Science and Technology (to Y.O.). B.C. is a junior research scholar of the Fonds de la Recherche en Santé du Québec. M.D. and F.C. hold studentships from the NSERC and the Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Bagby S, Kim S J, Maldonado E, Tong K I, Reinberg D, Ikura M. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 2.Coin F, Bergmann E, Tremeau-Bravard A, Egly J M. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 1999;18:1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conaway J W, Conaway R C. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J Biol Chem. 1989;264:2357–2362. [PubMed] [Google Scholar]
- 4.Conaway R C, Garrett K P, Hanley J P, Conaway J W. Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors alpha and beta gamma promote entry of polymerase into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulombe B, Burton Z F. DNA bending and wrapping around RNA polymerase: a revolutionary model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulombe B, Li J, Greenblatt J. Topological localization of the human transcription factors IIA, IIB, TATA box-binding protein, and RNA polymerase II-associated protein 30 on a class II promoter. J Biol Chem. 1994;269:19962–19967. [PubMed] [Google Scholar]
- 7.Cramer P, Bushnell D A, Fu J, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 8.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 9.Darst S A, Edwards A M, Kubalek E W, Kornberg R D. Three-dimensional structure of yeast RNA polymerase II at 16 A resolution. Cell. 1991;66:121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- 10.Douziech M, Forget D, Greenblatt J, Coulombe B. Topological localization of the carboxyl-terminal domain of RNA polymerase II in the initiation complex. J Biol Chem. 1999;274:19868–19873. doi: 10.1074/jbc.274.28.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drapkin R, Reardon J T, Ansari A, Huang J C, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 12.Evans E, Moggs J G, Hwang J R, Egly J M, Wood R D. Mechanism of open complex and dual incision formation by human excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feaver W J, Gileadi O, Li Y, Kornberg R D. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 14.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 15.Flores O, Ha I, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990;265:5629–5634. [PubMed] [Google Scholar]
- 16.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z F, Reinberg D. The small subunit of transcription factor IIF recruits RNA polymerase II into the pre-initiation complex. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forget D, Robert F, Grondin G, Burton Z F, Greenblatt J, Coulombe B. RAP74 induces promoter contact by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerard M, Fischer L, Moncollin V, Chipoulet J M, Chambon P, Egly J M. Purification and interaction properties of the human RNA polymerase B (II) general transcription factor BTF2. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 19.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 20.Groft C M, Uljon S N, Wang R, Werner M H. Structural homology between the RAP30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly. Proc Natl Acad Sci USA. 1998;95:9117–9122. doi: 10.1073/pnas.95.16.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holstege F C, Fiedler U, Timmers H T. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holstege F C, Tantin D, Carey M, van der Vliet P C, Timmers H T. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holstege F C, van der Vliet P C, Timmers H T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 25.Inostroza J, Flores O, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of general transcription factor IIE. J Biol Chem. 1991;266:9304–9308. [PubMed] [Google Scholar]
- 26.Jiang Y, Triezenberg S J, Gralla J D. Defective transcriptional activation by diverse VP16 mutants associated with a common inability to form open promoter complexes. J Biol Chem. 1994;269:5505–5508. [PubMed] [Google Scholar]
- 27.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 28.Kim T K, Lagrange T, Wang Y H, Griffith J D, Reinberg D, Ebright R H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T K, Ebright R H, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor TFIIH. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 31.Lagrange T, Kim T K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei L, Ren D, Burton Z F. The RAP74 subunit of transcription factor IIF has a similar role in isomerization of the initiation and the elongation complexes. Mol Cell Biol. 1999;19:8372–8382. doi: 10.1128/mcb.19.12.8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei L, Ren D, Finkelstein A, Burton Z F. Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol Cell Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 35.Lu H, Zawel L, Fisher L, Egly J M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 36.Moreland R J, Tirode F, Yan Q, Conaway J W, Egly J M, Conaway R C. A role for the TFIIH XPB DNA helicase in promoter escape by RNA polymerase II. J Biol Chem. 1999;274:22127–22130. doi: 10.1074/jbc.274.32.22127. [DOI] [PubMed] [Google Scholar]
- 37.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 38.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 39.Ohkuma Y, Hashimoto S, Wang C K, Horikoshi M, Roeder R G. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIEα. Mol Cell Biol. 1995;15:4856–4866. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohkuma Y, Roeder R G. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 41.Ohkuma Y, Sumimoto H, Horikoshi M, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II: purification and characterization of general transcription factor TFIIE. Proc Natl Acad Sci USA. 1990;87:9163–9167. doi: 10.1073/pnas.87.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto T, Yamamoto S, Watanabe Y, Ohta T, Hanaoka F, Roeder R G, Ohkuma Y. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEβ subunit. J Biol Chem. 1998;273:19866–19876. doi: 10.1074/jbc.273.31.19866. [DOI] [PubMed] [Google Scholar]
- 43.Okuda M, Watanabe Y, Okamura H, Hanaoka F, Ohkuma Y, Nishimura Y. Structure of the central core domain of TFIIEβ with a novel double-stranded DNA-binding surface. EMBO J. 2000;19:1346–1356. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orphanides G, Lagrange T, Reinberg D. The general initiation factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 45.Pan G H, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 46.Parvin J D, Sharp P A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 47.Poglitsch C L, Meredith G D, Gnatt A L, Jensen G J, Chang W-H, Fu J, Kornberg R D. Electron crystal structure of an RNA polymerase II transcription elongation complex. Cell. 1999;98:791–798. doi: 10.1016/s0092-8674(00)81513-5. [DOI] [PubMed] [Google Scholar]
- 48.Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert, F., and B. Coulombe. The use of site-specific protein-DNA photo-crosslinking to analyze the molecular organization of the RNA polymerase II initiation complex. Methods Mol. Biol., in press. [DOI] [PMC free article] [PubMed]
- 50.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 51.Robert F, Douziech M, Forget D, Egly J M, Greenblatt J, Burton Z F, Coulombe B. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol Cell. 1998;2:341–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H J, Egly J M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 53.Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers J H J, Egly J M. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription repair factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers J H J, Chambon P, Egly J M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 55.Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly J M. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 56.Serizawa H, Conaway R C, Conaway J W. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor delta from rat liver. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serizawa H, Conaway R C, Conaway J W. Multifunctional RNA polymerase II initiation factor delta from rat liver. Relationship between carboxyl-terminal domain kinase, ATPase, and helicase activities. J Biol Chem. 1993;268:17300–17308. [PubMed] [Google Scholar]
- 58.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 59.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 60.Svejstrup J Q, Vichi P, Egly J M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 61.Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 62.Timmers H T M. Transcription initiation by RNA polymerase II does not require hydrolysis of the β-γ phosphoanhydride bond of ATP. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tirode F, Busso D, Coin F, Egly J M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 64.Tyree C M, George C P, Lira-Devito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 65.Velankar S S, Soultanas P, Dillingham M S, Subramanya H S, Wigley D B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 66.Wang B Q, Burton Z F. Functional domains of human RAP74 including a masked polymerase binding domain. J Biol Chem. 1995;270:27035–27044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z G, Buratowski S, Svejstrup J Q, Feaver W J, Wu X H, Kornberg R D, Donahue T F, Friedberg E C. The yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription. Mol Cell Biol. 1995;15:2288–2293. doi: 10.1128/mcb.15.4.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weeda G, van Ham R C A, Vermeulen W, Bootsma D, van der Eb A J, Hoeijmakers J H J. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990;62:777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- 69.Yan M, Gralla J D. Multiple ATP-dependent steps in RNA polymerase II promoter melting and initiation. EMBO J. 1997;16:7457–7467. doi: 10.1093/emboj/16.24.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yokomori K, Verrijzer C P, Tijan R. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc Natl Acad Sci USA. 1998;95:6722–6727. doi: 10.1073/pnas.95.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 72.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Crystal structure of Thermus aquaticus core RNA polymerase at a 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]