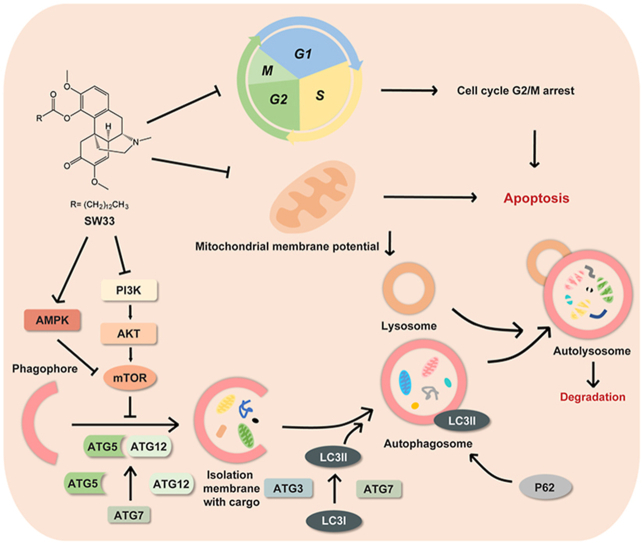
Abstract
Glioblastoma multiforme (GBM) in the central nervous system is the most lethal advanced glioma and currently there is no effective treatment for it. Studies of sinomenine, an alkaloid from the Chinese medicinal plant, Sinomenium acutum, showed that it had inhibitory effects on several kinds of cancer. Here, we synthesized a sinomenine derivative, sino-wcj-33 (SW33), tested it for antitumor activity on GBM and explored the underlying mechanism. SW33 significantly inhibited proliferation and colony formation of GBM and reduced migration and invasion of U87 and U251 cells. It also arrested the cell cycle at G2/M phase and induced mitochondria-dependent apoptosis. Differential gene enrichment analysis and pathway validation showed that SW33 exerted anti-GBM effects by regulating PI3K/AKT and AMPK signaling pathways and significantly suppressed tumorigenicity with no obvious adverse effects on the body. SW33 also induced autophagy through the PI3K/AKT/mTOR and AMPK/mTOR pathways. Thus, SW33 appears to be a promising drug for treating GBM effectively and safely.
Key words: SW33, GBM, G2/M phase, Apoptosis, Autophagy, mTOR, Anti-inflammation, Safety
Graphical abstract
Sinomenine ester derivative SW33 significantly inhibited glioblastoma multiforme (GBM) by arresting cell cycle at G2/M phase, promoting apoptosis in mitochondria-dependent manner and inducing autophagy via PI3K/AKT/mTOR and AMPK/mTOR pathways with a good safety profile in vivo and in vitro.
1. Introduction
Glioma is the most common intracranial cancer. It is very difficult to treat and has high mortality1. According to the 2007 World Health Organization (WHO), based on histopathology, gliomas can be classified into grades Ⅰ, II, III, and IV. Grades I and II are low grade gliomas (LGGs), such as astrocytomas or oligodendroglioblastomas, while grade III and IV refers to high grade gliomas (HGGs), like anaplastic astrocytomas or glioblastoma multiforme (GBM)2. The main therapies in clinic for GBM are surgery, chemotherapy and radiation treatments3. However, the prognosis of glioma is poor, especially for GBM which accounts for about 50% of all gliomas. The five-year survival rate among patients with GBM is only 5%4,5. Temozolomide (TMZ) is the major chemotherapeutic drug for GBM treatment6, but its antitumor activity can be counteracted by MGMT, a DNA repair enzyme, leading to drug resistance in patients with a high level of MGMT. TMZ is a cytotoxic monofunctional alkylating agent with serious side effects on the body7,8; therefore, more safer and effective drugs that are need to be discovered.
Nature products extracted from plants have potential for treating cancers with low toxicity. The alkaloid, sinomenine (SIN), 7,8-didehydro-4-hydroxy-3,7-dimethoxy-17-methylmorphinan-6-one (C19H23NO4), was extracted from the rhizome of the Chinese medicinal plant, Sinomenium acutum (Thunb.) Rehd. et Wils. and S. acutum (Thunb.) Rehd. et Wils. Var. cinereum Rehd. et Wils. SIN possesses anti-inflammatory activity and has been used to treat rheumatoid diseases in humans9, 10, 11. It was also reported that SIN has antitumor effects12, such as lung cancer13, gastric cancer14, prostate cancer15, and breast cancer16. However, SIN has some disadvantages, such as adverse gastrointestinal reactions, short biological half-life, and unstable physicochemical properties17. Molecular modifications have been considered as an option to solve these problems. Acylation at the position-4 hydroxyl group was considered as an effective way to reduce adverse effects and improve stability while preserving its therapeutic benefits17. By screening a series of SIN derivatives, we found sino-wcj-33 (SW33, C33H51NO5), which had been given by acylating the 4-hydroxyl with a 14-carbon carboxylic acid, had higher anti-GBM activity than the parent compound with greater safety.
In this study, we investigated the anti-GBM effects of SW33 both in vivo and in vitro, and evaluated its safety in a mouse model of GBM. It was found that SW33 significantly inhibited proliferation, migration, invasion, and colony formation of the GBM cell lines U87 and U251. It also induced mitochondrial-dependent apoptosis and arrested the cell cycle at the G2/M phase. In addition, our results showed that SW33 might induce autophagy of GBM cells by regulating the PI3K/AKT/mTOR and AMPK/mTOR pathways. SW33 also exhibited a good safety profile. All these results suggested that SW33 might be a more effective and less cytotoxic chemotherapeutic agent for the treatment of GBM.
2. Materials and methods
2.1. Cell culture
The human GBM cell lines U87 and U251 (American Type Culture Collection, ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, CA, USA) with 10% fetal bovine serum (FBS, Gibco, New Zealand). U87 and U251 cells were cultured in a humidified 5% CO2 incubator at 37 °C.
3D cell cultures were performed in culture medium containing different concentration of SW33 using Matrigel matrix (BD, San Jose, CA, USA) in a 96-well microplate according to the manufacturers’ instructions.
2.2. Source of SW33 and antitumor activity
SW33 was kindly provided by Prof. Tengfei Ji (Institute of Materia Medica, CAMS & PUMC, Beijing, China). SW33 was obtained as a yellow transparent liquid with 98% purity that was dissolved in dimethyl sulfoxide (DMSO, Merck KGaA, Darmstadt, Germany) to give a 50 mmol/L stock solution. SIN was purchased from MedChemExpress (Monmouth Junction, NJ, USA). The IC50 of SW33 and inhibition of SIN were measured with a viable cell counting kit 8 (CCK-8, Beyotime, China) assay. U87 and U251 cells were seeded into 96 well plates at 3 × 103 per well and cultured for 24 h at 37 °C. The medium was removed and replaced with medium containing 0–25 μmol/L SW33 and 0.01–1 mmol/L SIN incubation at 37 °C was continued for 24, 48, and 72 h. Viable counts were determined using the CCK-8 protocol with absorbance readings at 450 nm measured with a SpectraMax M5 plate-reader (Molecular Devices, Sunnyvale, CA, USA) and IC50 values were calculated on GraphPad Prism 7.
2.3. DNA synthesis assay
The Cell-Light Apollo 567 in vitro imaging kit (RiboBio, Guangzhou, China) was used in accordance with the manufacture's protocols. U87 and U251 cells were incubated with 0, 4, 8 and 12 μmol/L SW33 for 24 and 48 h, and ethynyl deoxyuridine (EdU) was then added to each well and incubation continued for an additional 2 h. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature and permeablized with 0.5% Trixon-X 100 for 10 min at room temperature. Afterwards, the cells were stained with the fluor Apollo 567 and Hoechst 33342 for 30 min individually18. Stained cells were scanned by high content imaging system (Cellomics ArrayScan VTI, Thermo Fisher Scientific, Carlsbad, CA, USA).
2.4. Lactate dehydrogenase assay (LDH)
Lactate dehydrogenase released from cells was measured using the LDH cytotoxicity assay kit (Dojindo, Japan). U87 and U251 cells were seeded into 96 well plates at 1 × 105/well and incubated with SW33 at 0, 4, 8 and 12 μmol/L for 24 and 48 h. After treatment, the medium was removed from each well, replaced with 100 μL of detection solution containing a tetrazolium salt and cells were incubated at room temperature for 30 min. The reaction was stopped by adding 50 μL stop solution to each well and absorbance was measured at 490 nm19.
2.5. Migration and invasion assays
The effect of SW33 on migration and invasion of U87 and U251 cells was determined by using a Transwell insert (Corning Costar, USA; #3422). Cells were detached, centrifuged, and resuspended in DMEM at 5 × 104 cells/mL, then 200 μL of cell suspension was used for the migration and invasion assays. For the invasion assay, before seeding cells into the insert, the bottom was coated with a 1:7 dilution of Matrigel (Corning Biocoat, USA) and subsequent steps were the same as the migration assay. After incubation for 19 h under migration conditions and 24 h for invasion, the cells passing through the insert membrane were fixed with 4% paraformaldehyde for 15 min, and then stained with 1% crystal violet for 20 min20. The inserts were removed, stained and photographed under a microscope (Nikon, Japan).
2.6. Colony formation assay
Soft agar was dissolved in sterile water to make a 3.5% stock solution and autoclaved. 2 mL of 0.7% agar diluted with DMEM containing 10% FBS was added to the wells of a 6-well plate as the bottom layer. After solidification, 1 mL of 0.35% agar containing 3000 cells (U251 or U87) diluted with SW33-containing medium at a final concentration of 0, 4, 8 and 12 μmol/L was added as the top layer. After cultured at 37 °C for 4 weeks, the colonies were stained with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) and counted21.
2.7. Mitochondrial membrane potential assay
U251 and U87 cells cultured in 60 mm dishes were treated with 0, 4, 8 and 12 μmol/L SW33 for 24 and 48 h. Cells were harvested, washed, and stained with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide (JC-1) at 37 °C for 30 min. Mitochondrial membrane potential of the cells was detected by BD Accuri C6 Plus Flow Cytometer (BD, San Diego, CA, USA)22.
2.8. Apoptosis assay
Apoptosis rate of U87 and U251 cells was detected using a PI/Annexin V-FITC kit (TransGen Biotech, Beijing, China). Cells cultured in 60 mm dishes were treated with 0, 4, 8 and 12 μmol/L SW33 for 24 and 48 h. Then, cells were harvested, washed with phosphate-buffered saline (PBS) and stained with propidium iodide (PI) and Annexin V-FITC for 15 min23. Flow cytometry was used to detect and quantify early and late apoptotic cells and necrotic cells. Apoptosis rate was analyzed by Flow Jo_V10 Software (Tristar, CA, USA).
2.9. Cell cycle assay
U251 and U87 cells cultured in 60 mm dishes were treated with 0, 4, 8 and 12 μmol/L SW33 for 24 and 48 h. Cells were harvested, washed with PBS and fixed in 70% ethanol overnight at 4 °C. Then, cells were washed with PBS for 3 times. After centrifugation at 200×g, 4 °C for 10 min, the supernatant was discarded and cells were resuspended and incubated with PI solution at 37 °C for 30 min24. Cell cycles were detected by BD Accuri C6 Plus Flow Cytometer (BD, San Diego, CA, USA) and data were analyzed by Flow Jo_V10 Software (Tristar, CA, USA).
2.10. RNA sequencing and determination of differential gene enrichment
U251 cells were incubated with 10 μmol/L SW33 for 24 and 48 h, then lysed in TRIzol reagent (Life Technologies, USA) and prepared for next generation sequencing by Novogene Corporation (Beijing, China). Total RNA was isolated and qualified by NanoPhotometer and Agilent 2100 bioanalyzer to ensure purity and integrity. NEB libraries were built and qualified, then, pooled and sequenced by Illumina. GO and KEGG pathway analysis was performed by Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/). Node genes were identified by MCC using cytoHubba and visualized by Cytoscape 3.6.1.
2.11. Autophagy assay by infection with adenovirus HBAD mRFP-GFP-LC3
U251 cells were cultured in 60 mm dishes to 50% confluence for 24 h before infection. Viruses used in infection were calculated according to Eq. (1):
| (1) |
U251 cells were infected with HBAD-mRFP-GFP-LC3 (HANBIO, Shanghai, China) at 37 °C for 8 h, the virus-containing medium was discarded and replaced with fresh DMEM containing 10% FBS for 48 h. Infected cells were seeded in 96-well plates at 3103 per well and incubated with 0, 4, 8, and 12 μmol/L SW33 for 24 h25. And then, cells were photographed by fluorescence microscope (Nikon, Japan).
2.12. Protein detection by Western blotting
Cells were lysed in RIPA buffer (Beyotime, Shanghai, China) containing protease inhibitor cocktail and phosphatase inhibitor cocktail (CWBIO, Beijing, China) on ice for 30 min. Cell lysates were centrifuged at 12,000×g for 15 min at 4 °C and supernatant was collected. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride (PVDF) membranes, and blocked in 5% skim milk for 2 h. The PVDF membranes were incubated overnight with primary antibodies at 4 °C. After washing 5 times with TBST, PVDF membranes were incubated with HRP-conjugated secondary antibodies (7074, anti-rabbit IgG; 7076, anti-mouse IgG, Cell Signaling Technology, Danvers, USA). The list of antibodies is given in Table 1. Chemical signals were visualized using super ECL (Applygen, Beijing, China) with the Tanon 4600 Imaging System (Tanon, Beijing, China).
Table 1.
Information of antibodies.
| Antibody | Catalogue | Dilution | Company |
|---|---|---|---|
| GAPDH | 10494-1-AP | 1:5000 | Proteintech, Europe |
| P53 | 2524 | 1:1000 | Cell Signaling Technology, USA |
| P21 | 2947 | 1:1000 | Cell Signaling Technology, USA |
| P-CDC2 | 9114 | 1:1000 | Cell Signaling Technology, USA |
| CDC2 | 9116 | 1:1000 | Cell Signaling Technology, USA |
| Aurora A | 10297-1-AP | 1:1000 | Proteintech, Europe |
| P-CDC25C | 9529 | 1:1000 | Cell Signaling Technology, USA |
| CDC25C | 4688 | 1:1000 | Cell Signaling Technology, USA |
| P-PLK1 | 9062 | 1:1000 | Cell Signaling Technology, USA |
| PLK1 | 4513 | 1:1000 | Cell Signaling Technology, USA |
| CCNB1 | 55004-1-AP | 1:1000 | Proteintech, Europe |
| BCL-2 | ab194583 | 1:1000 | Abcam, Cambridge, UK |
| BAX | 5023 | 1:1000 | Cell Signaling Technology, USA |
| PARP | 9542 | 1:1000 | Cell Signaling Technology, USA |
| Cleaved-PARP | 5625 | 1:1000 | Cell Signaling Technology, USA |
| Caspase9 | 9508 | 1:1000 | Cell Signaling Technology, USA |
| Cleaved-caspase9 | 7237 | 1:1000 | Cell Signaling Technology, USA |
| Caspase3 | 9662 | 1:1000 | Cell Signaling Technology, USA |
| Cleaved-caspase3 | 9664 | 1:500 | Cell Signaling Technology, USA |
| P-mTOR | 5536 | 1:1000 | Cell Signaling Technology, USA |
| mTOR | 2983 | 1:1000 | Cell Signaling Technology, USA |
| Beclin1 | 3495 | 1:1000 | Cell Signaling Technology, USA |
| P62 | 8025 | 1:1000 | Cell Signaling Technology, USA |
| LC3B | 3868 | 1:1000 | Cell Signaling Technology, USA |
| ATG3 | 3415 | 1:1000 | Cell Signaling Technology, USA |
| ATG5 | 12994 | 1:1000 | Cell Signaling Technology, USA |
| ATG7 | 8558 | 1:1000 | Cell Signaling Technology, USA |
| ATG12 | 4180 | 1:1000 | Cell Signaling Technology, USA |
| P-PI3K | 4228 | 1:1000 | Cell Signaling Technology, USA |
| PI3K | 4257 | 1:1000 | Cell Signaling Technology, USA |
| P-AKT | 4060 | 1:1000 | Cell Signaling Technology, USA |
| AKT | 4691 | 1:1000 | Cell Signaling Technology, USA |
| P-AMPK | 2535 | 1:1000 | Cell Signaling Technology, USA |
| AMPK | 5831 | 1:1000 | Cell Signaling Technology, USA |
2.13. Nude mouse GBM xenograft model
All animal experiments were conducted in accordance with the principles of the NIH Guide for the Care and Use of Laboratory Animals and approved protocols of the Institute of Materia Medica, CAMS & PUMC, Beijing, China. Female BALB/c-nu nude mice (16–18 g) were bought from Vital River Laboratory Animal Technology (Beijing, China) and housed at the Institute of Materia Medica animal barrier facility. Xenografting was done by subcutaneous implantation of U87 cells (1 × 107/0.1 mL) into the right flanks of the mice. After 12 days, when the tumor volume had reached about 100 mm3, the mice were randomly divided into three groups (n = 6) and treated with SW33 once a day for three weeks (30 and 60 mg/kg, i.g.). Body weight and tumor volume were recorded every two days. The tumor volume (V, mm3) was calculated by Eq. (2):
| (2) |
where a represents length and b represents width, in mm. At the end of the experiment, the mice were euthanized and blood was collected by capillary from the medial canthus of the eye. Complete blood count (CBC) was measured using a hematology analyzer (MEK-7222K, Nihon Kohden). The weight of the tumor tissues and the heart, liver, spleen, kidney, lung and brain were measured.
2.14. Cytokine analysis by ELISA
Blood was collected and coagulated at room temperature for 30 min. After centrifuged at 1000×g for 10 min, the supernatant was carefully collected and level of IL-2, IL-6 and TNFα was checked by ELISA according to the manufacturer's instructions26. Levels of IL-2, IL-6 and TNFα were determined with absorbance readings at 450 nm measured with a SpectraMax M5 plate-reader (Molecular Devices, Sunnyvale, CA, USA).
2.15. Statistical analysis
The results are presented as mean ± standard deviation (SD). Statistical significance between different groups was determined by one-way-ANOVA using GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA, USA), and P < 0.05 was considered statistically significant.
3. Results
3.1. SW33 inhibited proliferation of human GBM cell lines U87 and U251
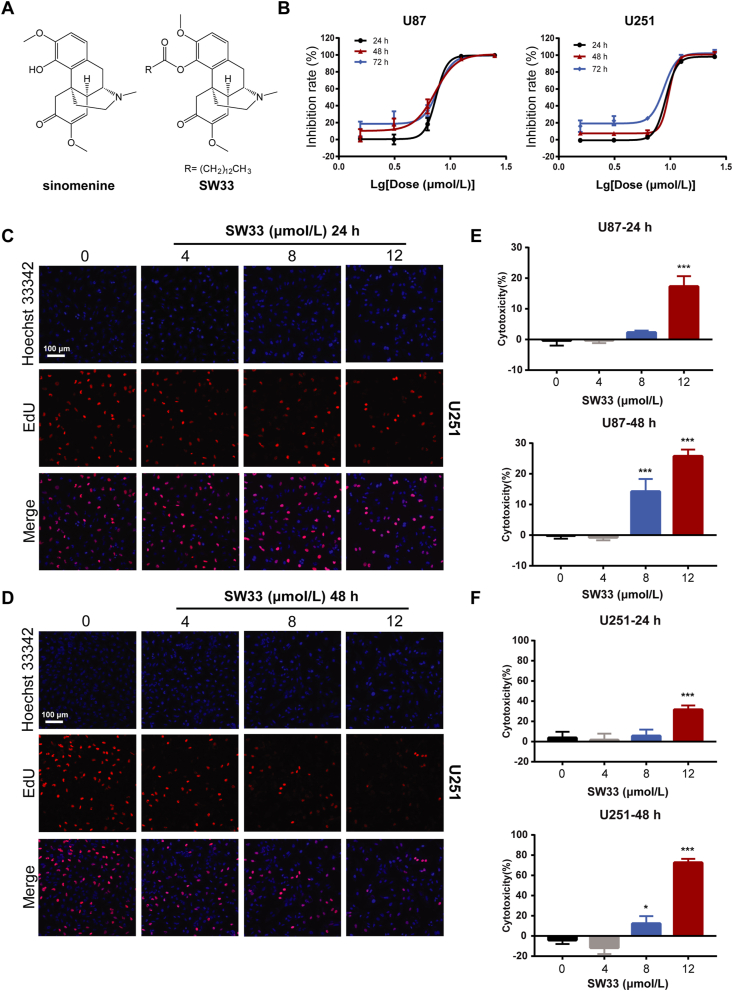
SW33 is a derivative of the natural plant compound, SIN, synthesized by acylation of the 4-hydroxyl with a long-chain carboxylic acid (Fig. 1A), which significantly improved its lipid solubility. To determine if SW33 can inhibit the growth of GBM, U87 and U251 cells were incubated with increasing concentrations of SW33 and checked by different assays. The CCK-8 assay showed that the IC50 of SW33 on U87 and U251 at 48 h was 7.43 and 8.46 μmol/L, respectively (Fig. 1B), and that SW33 inhibited the growth of GBM cells in a dose- and time-dependent manner (Supporting Information Fig. S1A and S1B). We also tested inhibitory effect of SIN on U87 and U251 cells, and the results showed that the antitumor effect of SW33 was much better than SIN (Fig. S1C). The inhibitory effect of SW33 on proliferation of U87 and U251 cells was also measured by DNA synthesis assay using ethynyl deoxyuridine, EdU. Results of the assay showed that SW33 significantly inhibited DNA synthesis of U87 (Fig. S1D and S1E) and U251 (Fig. 1C and D) in a time- and dose-dependent manner. When cells are injured, excess lactate dehydrogenase (LDH) is released, which can be quantified and used as an indicator of the toxicity of external substances on cells. To determine if SW33 was toxic to U87 and U251 cells, the LDH assay was performed and showed that SW33 had a toxic effect on U87 and U251 cells, especially at 12 μmol/L for 48 h (Fig. 1E and F). Taken together, these data prove that SW33 can inhibit the growth of U87 and U251 cells.
Figure 1.
SW33 inhibited viability and proliferation of U87 and U251 cells. (A) Structures of SIN and SW33. (B) IC50 of SW33 on U87 and U251 cells at 24, 48, and 72 h. SW33 inhibited DNA synthesis in U251 cells at 24 (C) and 48 h (D) as measured by ethynyl deoxyuridine (EdU) incorporation. Scale bar = 100 μm. (E) Toxicity of SW33 on U87 cells at 24 and 48 h by LDH assay. (F) Toxicity of SW33 on U251 cells at 24 and 48 h by LDH assay. Data are expressed as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. Control.
3.2. SW33 inhibits migration, invasion and colony formation of GBM cells
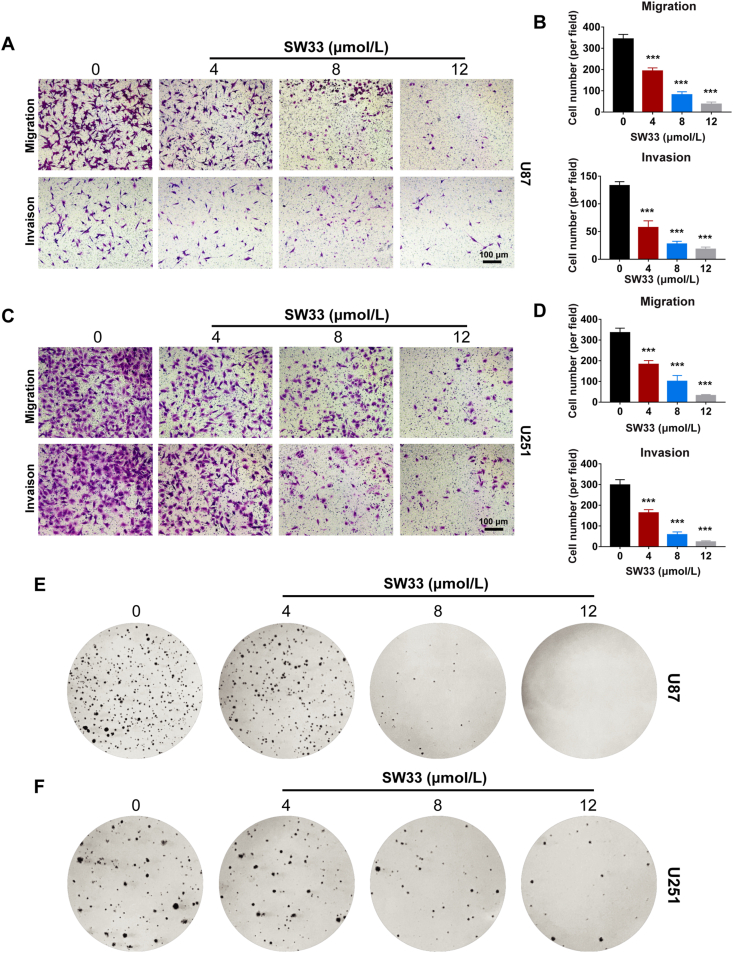
Migration and invasion ability of tumor cells are important indicators to evaluate the malignancy of cancers in vitro. SIN was reported to inhibit migration and invasion of prostate and breast cancer cells27,28. To assess the ability of SW33 to prevent GBM migration and invasion in vitro, Transwell assays were carried out. SW33 significantly reduced migration and invasion of U87 and U251 cells in a dose-dependent manner (Fig. 2A, B and 2C, D). A soft agar assay was used to evaluate the effect of SW33 on colony formation of GBM cells. As the concentration of SW33 was increased, the colony numbers of U87 and U251 cells in soft agar were decreased (Fig. 2E and F), demonstrating that SW33 could inhibit colony formation ability of GBM cells. Taken together, these results are consistent with the hypothesis that SW33 blocks tumorigenesis of GBM cells by inhibiting their migration, invasion and colony-forming abilities.
Figure 2.
SW33 inhibited migration, invasion and colony formation of U87 and U251 cells. (A) and (B) Transwell assays showed SW33 inhibited migration and invasion ability of U87 cells. Scale bar = 100 μm. (C) and (D) Transwell assays showed SW33 inhibited migration and invasion ability of U251 cells. Scale bar = 100 μm. (E) SW33 inhibited colony formation of U87 cells by soft agar assay. (F) SW33 inhibited colony formation of U251 cells by soft agar assay. Data are expressed as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. Control.
3.3. SW33 may arrest cell cycle at G2/M phase
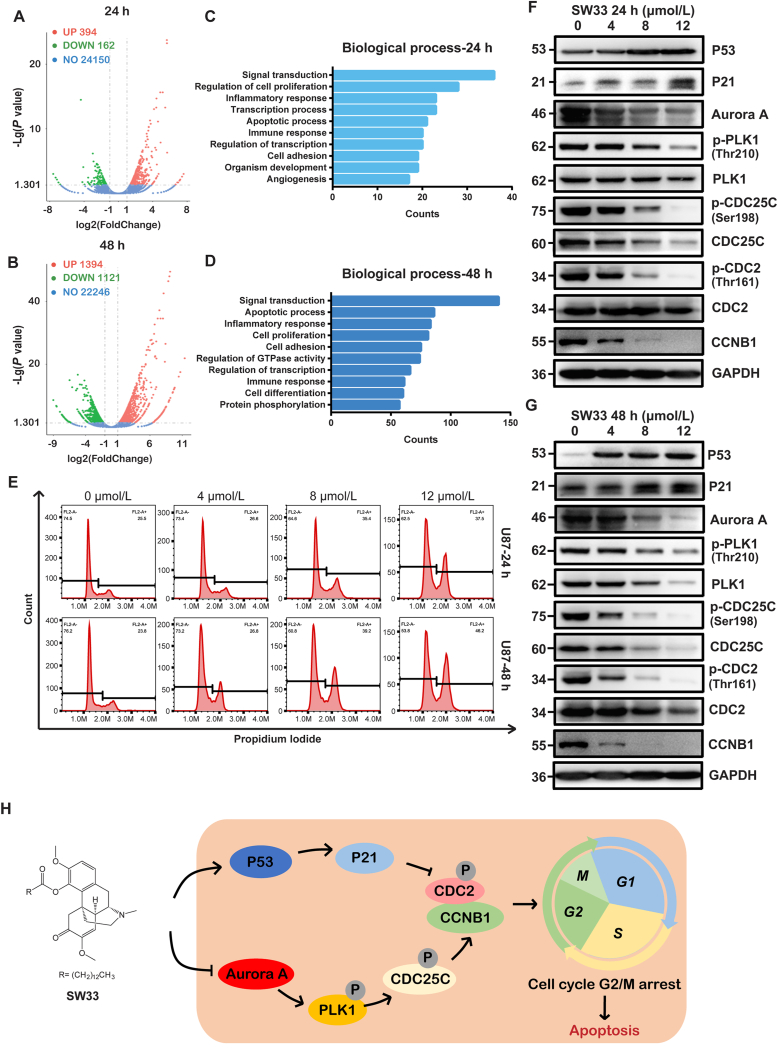
To further explore the mechanism of SW33's effects against GBM, total RNA was extracted from U251 cells treated with 10 μmol/L SW33 for 24 and 48 h and used for RNA-Seq assay. The results revealed 394 up-regulated and 162 down-regulated genes in the 24-h treatment group compared to control cells (differential regulation was defined as fold change>2 and P < 0.05). The 48-h treatment group had 1394 differentially up-regulated genes and 1121 down-regulated genes (Fig. 3A and B and Supporting Information Fig. S2B). The Venn diagram intersection of the two groups revealed 452 differentially expressed genes in common (Fig. S2A). Differential genes enriched according to gene ontology (GO) included those involved in signal transduction, cell proliferation, inflammatory responses and apoptosis as the main ones differentially regulated at both 24 and 48 h in the biology progress (BP) analysis (Fig. 3C and D). By subjecting the 452 differentially expressed genes in common after 24 and 48 h of SW33 treatment to analysis with the MCC algorithm of cytoHubba, the top 50 node genes were identified for further analysis. As shown in Supporting Information Fig. S3C, SW33 may affect the progress of mitosis by acting at the G2/M phase, which suggested an effect of SW33 on cell cycle at all time. BP analysis showed that SW33 significantly influenced cell proliferation and apoptosis (Fig. 3C and D). SW33 inhibited cell proliferation at all time of the cell cycle in 48-h-treated cells, while the apoptotic process was dominant in cells exposed to SW33 for 48 h, which may be a result of continuous cell cycle arrest.
Figure 3.
SW33 induced cell cycle arrest at G2/M phase in U87 and U251 cells. Visualization of differential genes by volcano plot of U251 cells treated with SW33 for 24 (A) and 48 h (B). Red points for up-regulated genes, green for down-regulated genes, and blue for unchanged genes (|log2FoldChange|>1, P < 0.05). Top ten enriched biological processes at 24 (C) and 48 h (D). (E) Flow cytometry analysis showed that SW33 arrested cell cycle at G2/M phase in U87 cells. Protein level of G2/M checkpoint by Western blotting at 24 (F) and 48 h (G). Expression of P53 and P21 was up-regulated while expression of Aurora A, p-PLK1, PLK1, p-CDC25C, CDC25C, p-CDC2, CDC2 and CCNB1 was down-regulated. (H) Schematic model for mechanism of cell cycle arrest at G2/M by SW33. Experiments were performed in triplicate.
To show the effect of SW33 on the cell cycle in G2/M phase, U87 and U251 cells treated with SW33 for 24 or 48 h were labeled with PI and analyzed by flow cytometry. Results show that SW33 arrested the cell cycle at the G2/M phase (Fig. 3E and Fig. S3A). In addition, the expression of proteins involved in the G2/M checkpoint was detected by Western blotting, and it was found that P53 and P21 were up-regulated while expression of CCNB1, p-CDC2 and CDC2 was reduced (Fig. 3F, G and Fig. S3B), which suggested that SW33 may utilize a P53/P21 pathway to arrest cell cycle at G2/M. The expression of Aurora A, p-PLK1, PLK1, p-CDC25C and CDC25C was also decreased, which inhibits the activity of the CCNB1/CDC2 complex and results in G2/M arrest (Fig. 3F, G and Fig. S3B). Overall, these results suggest that SW33 arrested the cell cycle at the G2/M phase by regulating the P53/P21/CCNB1/CDC2 pathway and the Aurora A/PLK1/CDC25C/CCNB1/CDC2 pathway (Fig. 3H).
3.4. SW33 induced apoptosis of GBM cells by reducing mitochondrial membrane potential
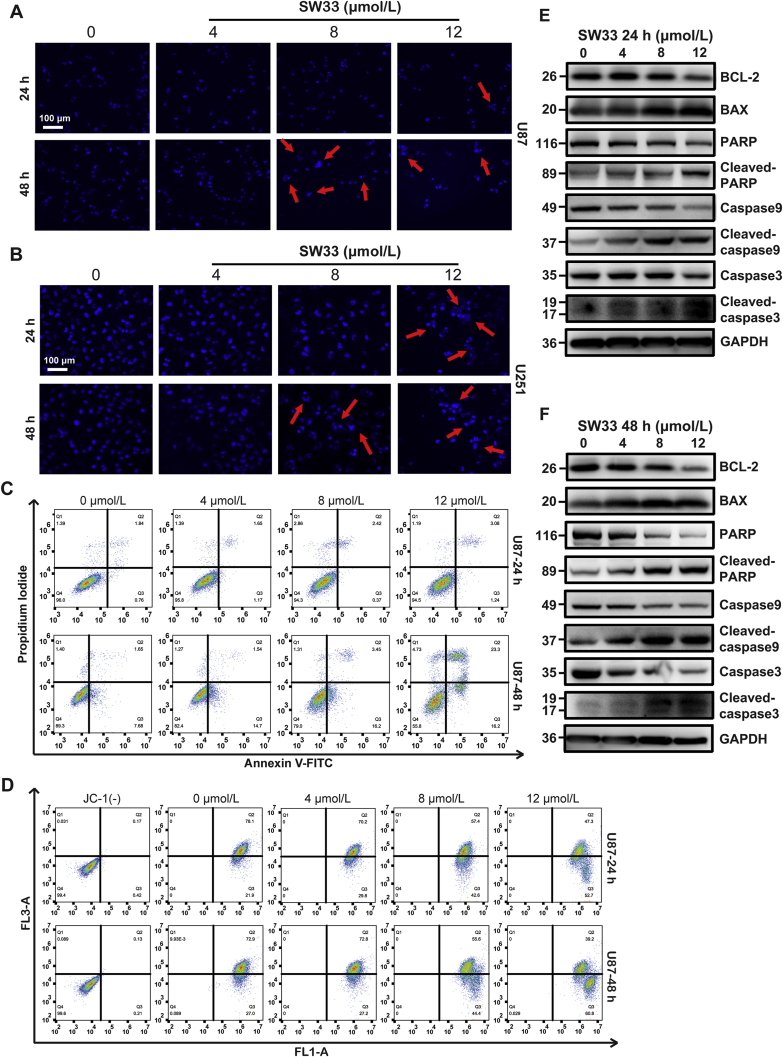
DNA staining with Hoechst 33342 showed that the nuclear morphology of the U87 and U251 cells was significantly changed by 48 h incubation with 8 and 12 μmol/L SW33. The chromatin was gathered to one side of the nuclear membrane, which means these cells were at an early stage of apoptosis (Fig. 4A and B). In other cells, the DNA was fragmented with different sized pieces of irregular shape, indicative of a later stage of apoptosis. After incubation with SW33, U87 and U251 cells were stained with PI and Annexin-V-FITC and apoptosis was analyzed by flow cytometry. SW33 increased the percentage of both early and late apoptotic cells (Fig. 4C and Supporting Information Fig. S4A).
Figure 4.
SW33-induced apoptosis of U87 and U251 cells via mitochondrial disruption. Morphological changes in nuclei of U87 (A) and U251 (B) treated with SW33. Scale bar = 100 μm. (C) Flow cytometry using PI/Annexin V-FITC double stain showed SW33 induced apoptosis of U87 at 24 and 48 h. (D) Flow cytometry analysis using 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide (JC-1) stain showed that SW33 reduced mitochondrial membrane potential of U87 at 24 and 48 h. Western blotting detection of proteins associated with apoptosis in U87 cells treated with 0, 4, 8, 12 μmol/L SW33 for 24 (E) and 48 h (F). Expression of BAX was up-regulated while ratios of cleaved PARP/PARP, cleaved caspase 9/caspase 9, cleaved caspase 3/caspase 3 and expression of BCL-2 were reduced. Experiments were performed in triplicate.
Mitochondrial membrane potential (MMP, Δψm) can reflect the status of mitochondrial function. In apoptotic cells, MMP is reduced. To further explore whether SW33-induced apoptosis was associated with change of MMP in U87 and U251 cells, these cells were stained with the membrane permeant fluorescent dye, 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide (JC-1), that shows a wavelength shift depending on MMP. Results showed that the MMP of U87 and U251 cells was decreased in a time- and dose-dependent manner after incubation with 4, 8, and 12 μmol/L SW33 (Fig. 4D and Fig. S4B). We also evaluated the expression of proteins associated with apoptosis by Western blotting. In SW33-treated U87 and U251 cells, the ratios of cleaved-PARP/PARP, cleaved-caspase 9/caspase 9, cleaved-caspase 3/caspase 3 and the protein level of BCL-2 were decreased while expression of BAX was increased, which suggested that the SW33-induced apoptosis was linked to the decrease of MMP (Fig. 4E, F and Fig. S4C). Together, these results support the hypothesis that SW33 induced apoptosis of U87 and U251 cells by disrupting mitochondrial function.
3.5. SW33 induced autophagy through regulation of the mTOR-mediated pathway
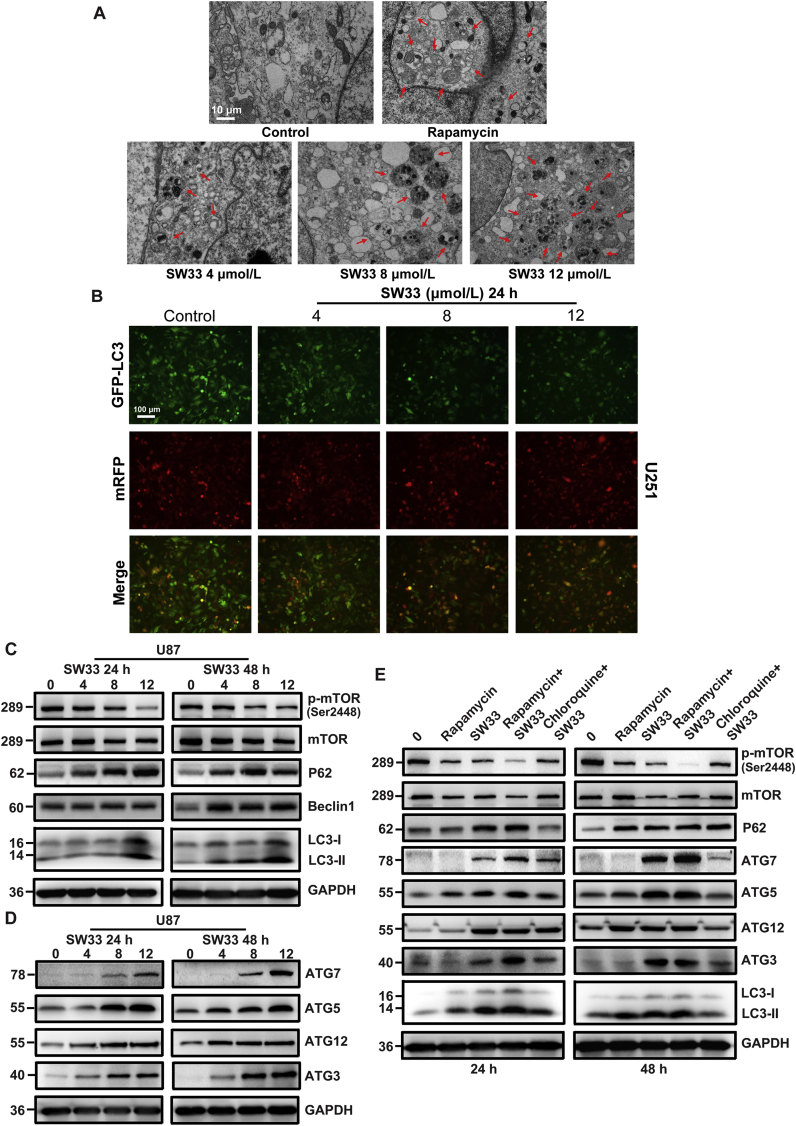
To determine if SW33 can induce autophagy in GBM, U251 cells were treated with SW33 for 24 h and morphology was examined by transmission electron microscopy. As a positive control, the cells were also incubated with rapamycin. After treatment with SW33, U251 showed significant autophagy similar to rapamycin. Compared to untreated controls, the formation of autophagosomes and autolysosomes was observed in U251 cells treated with either rapamycin or SW33, and many more autolysosomes were seen after 24 h incubation with 8 and 12 μmol/L SW33 (Fig. 5A). In addition, to assess the effect of SW33 on the dynamic process of autophagosome and lysosome fusion, the mRFP-GFP-LC3 dual fluorescent autophagy indicator system was used to track changes in LC3 and autophagy flux. Results showed that the ratio of GFP to mRFP was reduced in cells treated with 4, 8, and 12 μmol/L SW33, indicating that the drug promoted fusion of lysosomes and autophagosomes resulting in formation of autolysosomes (Fig. 5B). These results above suggested that SW33 promoted the process of phagosomes and lysosomes fusion resulting in cell autophagy. To check proteins associated with autophagy induced by SW33, Western blotting was carried out. These proteins included p-mTOR, mTOR, Beclin1, P62, LC3-I to LC3-II and a series of ATG proteins. As shown in Fig. 5C and Supporting Information Fig. S5A, the ratio of p-mTOR to mTOR was reduced whereas the protein level of LC3-II (marker of autophagy) was increased in a dose-dependent manner after treatment with increasing concentrations of SW33, indicating that the drug induced an autophagic response. Expression of Beclin1 and P62 was increased, which promoted the formation of phagophores and autophagosomes. The levels of ATG3 and ATG7 protein were also increased, which accelerated the conversion of LC3-I to LC3-II. ATG5, ATG7 and ATG12 were increased, further enhancing formation of autophagosomes (Fig. 5D and Fig. S5B).
Figure 5.
SW33 induced autophagy by regulating mTOR signaling pathway. (A) Transmission electron microscopy showing increase in phagosomes and phagolysosomes in U251 cells with increasing SW33 concentration. Scale bar = 10 μm. (B) mRFP-GFP-LC3 dual fluorescent autophagy indicators showed the ratio of GFP to mRFP was reduced in U87 cells as concentration of SW33 was increased. Scale bar = 100 μm. (C) and (D) Proteins associated with autophagy in SW33-treated U87 cells quantitated by Western blotting. Ratio of p-mTOR/mTOR was decreased and the levels of P62, Beclin1, LC3-II, ATG7, ATG5, ATG12 and ATG3 increased. (E) Autophagy protein levels in U87 cells treated with SW33, rapamycin and chloroquine for 24 and 48 h. p-mTOR/mTOR was consistently reduced after treatment with SW33 and rapamycin. A greater decrease of p-mTOR/mTOR was seen with combined SW33 and rapamycin treatment. Chloroquine reversed the SW33-mediated reduction in p-mTOR/mTOR. The level of P62 and LC3-II, as well as ATG7, ATG5, ATG12 and ATG3, increased after treatment with SW33 and rapamycin, and this was also reversed by chloroquine. Experiments were performed in triplicate.
To further explore whether SW33 induced mTOR-mediated autophagy, U87 and U251 cells were pretreated with the mTOR inhibitor rapamycin, and the autophagy inhibitor chloroquine, and the relevant proteins were checked by Western blotting. Results showed that the ratio of p-mTOR/mTOR was consistently reduced after treatment with either SW33 or rapamycin and this reduction was increased by combined treatment with SW33 plus rapamycin. Chloroquine reversed the reduction in p-mTOR/mTOR ratio by SW33. The levels of P62 and LC3-II were increased after treatment with SW33 and rapamycin, which again was reversed by chloroquine as well as ATG3, ATG5, ATG7 and ATG12 (Fig. 5E and Fig. S5C). Together, these results suggest that SW33 induces autophagy by regulating the mTOR-mediated pathway in GBM cells.
3.6. SW33 induced autophagy of GBM cells by regulating the PI3K/AKT/mTOR and AMPK/mTOR signaling pathways
Our results confirmed that SW33 induced autophagy by regulating mTOR-mediated pathway in GBM cells. To further explore how SW33 induced mTOR-mediated autophagy, RNA-Seq analysis was carried out in the cells treated with SW33. The KEGG enrichment analysis using the RNA-Seq results showed that PI3K/AKT pathway might be the most important signaling pathway involving in autophagy induced by SW33 (Fig. 6A and B). To confirm results from RNA-Seq, the protein levels of p-PI3K, PI3K, p-AKT and AKT were checked by Western blotting. Results showed that the expression of p-PI3K/PI3K, p-AKT/AKT was reduced in U87 and U251 cells in a dose dependent manner (Fig. 6C and D), which suggested that PI3K/AKT pathway was involved in autophagy of cells induced by SW33. It is well known that AMP-activated protein kinase (AMPK) is a key energy sensor and regulates cellular metabolism to maintain energy homeostasis which is important in promoting autophagy29. Furthermore, AMPK expression was checked by Western blotting and the results showed that expression of p-AMPK was increased in the cells treated by SW33 in dose-dependent manner. Taken together, we can conclude that SW33 may induce autophagy of GBM cells by regulating PI3K/AKT/mTOR and AMPK/mTOR signaling pathways (Fig. 6E).
Figure 6.
SW33 induced autophagy of GBM cells by regulating PI3K/AKT and AMPK signaling pathways. KEGG pathway analysis of differentially expressed genes at 24 (A) and 48 h (B). (C) and (D) Protein levels of p-PI3K, PI3K, p-AKT, AKT, p-AMPK, AMPK in SW33-treated U87 and U251 cells detected by Western blotting. The ratio of p-PI3K/PI3K and p-AKT/AKT decreased while p-AMPK/AMPK increased. (E) Diagram of autophagy induced by SW33 via PI3K/AKT/mTOR and AMPK/mTOR signaling pathways. Experiments were performed in triplicate.
3.7. SW33 inhibited growth of glioblastoma in vivo with good safety profile
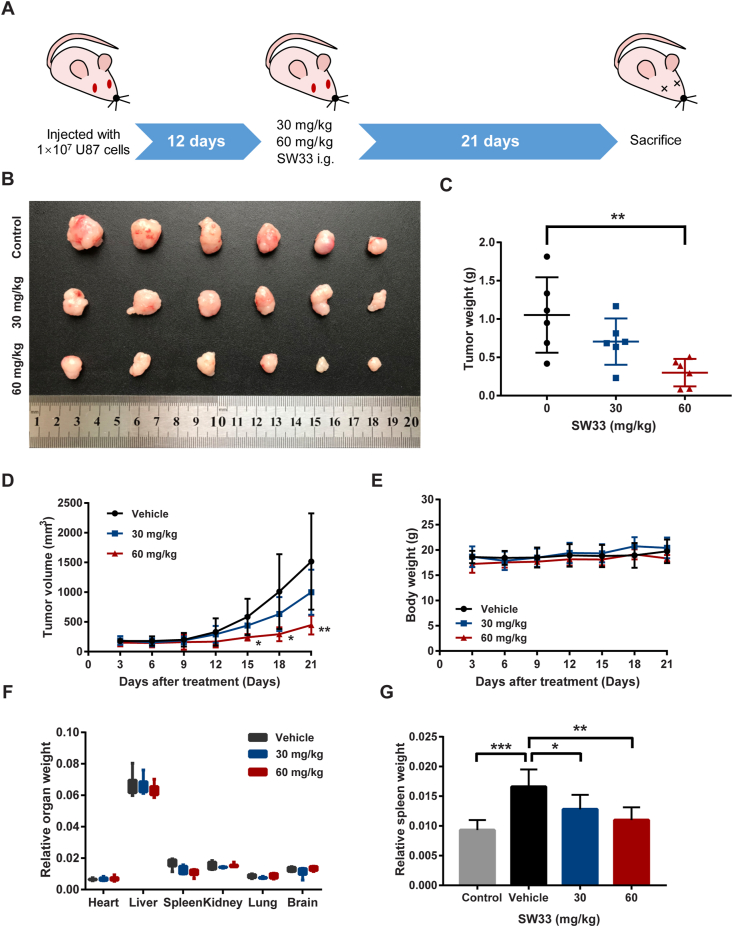
To validate the anti-GBM effect of SW33 in vivo, cells were injected s.c. into the left flank of each nude mouse. When the size of the tumor was about 100 mm3, mice were treated with 30 and 60 mg/kg SW33 (0.1 mL/mouse, i.g.) for 21 days (Fig. 7A). Compared to vehicle, tumor growth was inhibited by SW33 in a dose-dependent manner (Fig. 7B). The mass and volume of tumors was significantly decreased after administration of 60 mg/kg SW33 for 21 days (Fig. 7C and D). There was no significant change in body weight (Fig. 7E) or in relative weight of organs including heart, liver, kidney, lung and brain (Fig. 7F) during 21 days of administration. There was also no obvious change in the appearance and morphology of vital organs compared to treatment with vehicle. The spleen is the primary immune organ of the body and compared to healthy mice, the weight of spleens in tumor-bearing mice was much greater (Fig. 7G). SW33 treatment restored the spleens to the normal weight, which suggested SW33 may reduce the inflammatory reaction of the body and alleviated the splenomegaly. The change in number of white blood cells (WBCs) also further proved this (Supporting Information Fig. S6). No obvious systemic toxicity was observed in physiological blood tests (Table 2 and Fig. S6). To further verify the anti-inflammation effect of SW33, inflammation factors IL-2, IL-6 and TNFα were tested by ELISA. In tumor-bearing mice, the level of serum IL-2, IL-6 and TNFα was significantly increased, which can be reversed by SW33. In addition, to further validate the inhibitory effect of SW33 on GBM, growth of GBM cells in a 3D matrix tumor like microenvironment was assessed. Results showed that SW33 could inhibit the growth of cells in a 3D matrix tumor like microenvironment (Supporting Information Fig. S7). Taken together, these results indicated that SW33 inhibited tumor growth and reduced inflammation in vivo with a good safety profile.
Figure 7.
SW33 suppressed glioma growth in vivo. (A) Schematic diagram of GBM xenograft mouse model treated with SW33 in vivo. (B) Image of tumors in different groups treated with SW33 for three weeks. (C) Tumor weight after three weeks of treatment with SW33. (D) Tumor volume during administration of SW33. (E) Body weight during administration of SW33. (F) Relative organ weights after three weeks of treatment with SW33. (G) Changes in spleen weight of different groups after SW33 treatment for three weeks. Data are expressed as mean ± SD (n = 6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (D: vs. Vehicle).
Table 2.
Blood biochemical testing.
| Indicator | Control | Vehicle | 30 mg/kg | 60 mg/kg |
|---|---|---|---|---|
| WBC | 6.7 ± 1.10 | 25.07 ± 9.23∗∗∗ | 8.97 ± 3.28∗∗∗ | 7.45 ± 2.54∗∗∗ |
| RBC | 11.27 ± 1.20 | 9.71 ± 1.92 | 11.85 ± 0.83 | 12.5 ± 0.92∗ |
| HGB | 160.4 ± 19.20 | 140.5 ± 29.45 | 171.3 ± 14.29 | 179.4 ± 16.17∗ |
| HCT | 46.41 ± 5.50 | 41.13 ± 8.95 | 49.83 ± 3.71 | 51.5 ± 4.43 |
| MCV | 41.18 ± 1.04 | 42.2 ± 1.47 | 42.08 ± 0.98 | 41.22 ± 1.41 |
| MCH | 14.23 ± 0.44 | 14.45 ± 0.35 | 14.48 ± 0.35 | 14.36 ± 0.69 |
| MCHC | 345.7 ± 8.96 | 342.7 ± 14.50 | 343.7 ± 6.13 | 348.4 ± 4.22 |
| RDW | 16.77 ± 1.20 | 16.48 ± 1.93 | 15.53 ± 0.72 | 15.94 ± 0.87 |
| PLT | 850.1 ± 83.8 | 559.8 ± 159.4∗∗ | 576.3 ± 143.4 | 600.8 ± 23.82 |
| PCT | 0.51 ± 0.06 | 0.36 ± 0.10∗ | 0.36 ± 0.09 | 0.36 ± 0.03 |
| MPV | 5.96 ± 0.23 | 6.38 ± 0.23 | 6.32 ± 0.27 | 6.13 ± 0.34 |
| PDW | 11.63 ± 0.67 | 13.27 ± 0.82∗ | 12.97 ± 0.82 | 12.58 ± 0.93 |
| LYM | 1.67 ± 0.75 | 3.75 ± 2.37 | 1.9 ± 0.80 | 1.86 ± 0.85 |
| MON | 0.77 ± 0.49 | 6.25 ± 4.32∗∗ | 2.72 ± 1.27 | 1.46 ± 1.14∗ |
| NEUT | 4.09 ± 1.24 | 6.27 ± 4.47 | 4.07 ± 1.61 | 4.32 ± 1.18 |
| EOS | 0.17 ± 0.20 | 0.1 ± 0.15 | 0.25 ± 0.22 | 0.22 ± 0.18 |
| BAS | 0 ± 0 | 0.03 ± 0.05 | 0.03 ± 0.05 | 0.02 ± 0.04 |
| LYM% | 20.3 ± 6.49 | 24.43 ± 6.62 | 21.17 ± 5.68 | 22.15 ± 6.62 |
| MON% | 4.0 ± 3.27 | 37.4 ± 11.86∗∗ | 30.17 ± 10.42 | 21.53 ± 14.35 |
| NEUT% | 59.9 ± 13.65 | 37.13 ± 10.58∗ | 44.75 ± 8.89 | 53.65 ± 9.78 |
| EOS% | 1.28 ± 1.34 | 0.92 ± 1.75 | 1.88 ± 1.51 | 2.73 ± 2.40 |
| BAS% | 0.04 ± 0.07 | 0.12 ± 0.13 | 0.13 ± 0.13 | 0.03 ± 0.07 |
WBC, white blood cell count, 103/mm3; RBC, white blood cell count, 106/mm3; HGB, hemoglobin, g/L; HCT, hematocrit, %; MCV, mean corpuscular volume, fL; MCH, mean corpuscular hemoglobin, pg; MCHC, mean corpuscular hemoglobin concentration, g/L; RDW, red blood cell volume distribution width, %CV; PLT, platelet, 103/mm3; PCT, plateletocrit, %; MPV, mean platelet volume, fL; PDW, platelet distribution width, %; LYM, lymphocyte count; MON, monocyte count, 103/mm3; NEUT, neutrophil count, 103/mm3; EOS, eosinophil count, 103/mm3; BAS, basophil count, 103/mm3; LYM%, lymphocyte ratio, %; MON%, monocyte ratio, %; NEUT%, neutrophil ratio, %; EOS%, eosinophil ratio, %; BAS%, basophil ratio, %.
Data are mean ± SD, n = 6; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
GBM is the most malignant (WHO grade IV), aggressive and frequent subtype of human brain cancer occurring in the central nervous system. Up to now, TMZ has been the only chemotherapy drug approved for treatment of GBM. Drug resistance to TMZ can be observed in most patients with GBM. In this study, a novel compound, SW33, was obtained by modifying the plant alkaloid, SIN, through acylating the 4-hydroxyl with a 14-carbon carboxylic acid. We found that SW33 significantly inhibited proliferation, migration, invasion and colony formation of GBM cells. The drug also arrested the GBM cell cycle in G2/M phase, caused mitochondrial-dependent apoptosis and induced cell autophagy through the PI3K/AKT/mTOR and AMPK/mTOR pathways. Thus, SW33 can inhibit GBM by inducing both apoptosis and autophagy.
Natural products usually have low antitumor activity, but good safety characteristics. SIN is a natural product extracted from S. acutum which has been used as an anti-rheumatic drug. In recent years, SIN and its derivatives were reported to have potent antitumor activity, but the effective concentration was in the millimolar range15,28,30. Here, we modified the structure of SIN by acylating the 4-hydroxyl to produce a new anticancer compound, SW33. In this study, we reported that SW33 inhibited GBM both in vivo and in vitro, and revealed the underlying mechanisms. We calculated that the IC50 of SW33 was 7.43 μmol/L for U87 cells and 8.76 μmol/L for U251 cells, which was much lower than that of SIN. Therefore, SW33 has better anti-GBM activity than SIN.
SIN was reported to inhibit cell proliferation and induce apoptosis in many kinds of cancers31,32, and arrest the cell cycle at the G0/G1 phase33,34. Here, we found that, similar to SIN, SW33 significantly induced apoptosis in human GBM cell lines. However, our data showed that SW33 arrested the cell cycle at the G2/M phase. The CCNB1/CDC2 complex is the key mediator of the G2/M checkpoint, and binding of CDC2 to CCNB1 is required for the cell cycle to move into the next step of mitosis35. After treatment of SW33, the expression of p-CDC2, CDC2 and CCNB1 was reduced in GBM cells. PLK1 (polo-like kinase) is considered to be a key regulator for entry into mitosis and PLK1-dependent CDC25C phosphorylation is required for normal cell cycle progression from G2/M phase36. SW33 reduced protein level of p-PLK1 and p-CDC25C. Aurora A, the upstream regulator of PLK1 was also found to be reduced by SW33 in GBM cells. The expression of P53 and its transcriptional target, P21, was increased after the treatment with SW33, which indicated that G2/M arrest can also be regulated through the P53/P21 pathway. Collectively, these results suggest that SW33 may arrest the GBM cell cycle at the G2/M checkpoint by regulating Aurora A/PLK1/CDC25C and P53/P21 signaling pathways and reducing the activity of p-CDC2 and CCNB1, leading to apoptosis of GBM cells.
Our data showed that SW33 reduced MMP in a time- and dose-dependent manner and induced apoptosis by disrupting mitochondrial function. The dysfunction of mitochondria leads to release of cytochrome c, activation of caspase 9/3, and subsequent cleavage of PARP37, which were also verified by our results. The ratios of cleaved-PARP/PARP, cleaved-caspase 9/caspase 9, cleaved-caspase 3/caspase 3 were all decreased. Expression of BCL-2 was also reduced, while expression of BAX was increased in SW33-treated GBM cells. These outcomes are consistent with the idea that SW33 induces apoptosis via mitochondrial inhibition.
Autophagy is a normal cellular process for digesting and recycling unwanted cell components that occurs in many biological activities in response to stress and to preserve viability. Autophagy is closely associated with the cell cycle and there is a mutual interaction and regulation between them38,39. Given the connection between autophagy and cell cycle, we examined whether SW33 affected autophagy and observed alteration in the regulatory pathways in cells treated with SW33. PI3K/AKT/mTOR40, MAPK/mTOR41 and AMPK/mTOR29 have been widely reported to activate autophagy, and PI3K/AKT was found to be involved in the stimulation of autophagy by SIN in melanoma and renal carcinoma cells31,42. Our results demonstrated that SW33 may induce autophagy through PI3K/AKT/mTOR in GBM. Our data also showed that SW33 induced autophagy via the AMPK/mTOR pathway, which suggested that SW33 regulated both the PI3K/AKT/mTOR and AMPK/mTOR pathways.
SW33 has potent anti-GBM activity with a high degree of safety in vivo. Administration of 60 mg/kg SW33 significantly inhibited glioblastoma growth in a nude mouse xenograft model. There was no obvious rash or gastrointestinal reactions in the SW33-treated mice. Blood testing showed slight improvement in red blood cells (RBC) with no side effects on hemoglobin (HGB) and hematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), RDW (red blood cell volume distribution width), mean platelet volume (MPV), or platelet distribution width (PDW). Compared to controls injected with vehicle only, the platelet (PLT), plateletocrit (PCT) and platelet distribution width (PDW) changed significantly, which might be caused by tumor infiltration into bone marrow, immune reactions, hypersplenism, or platelet dysfunction. Although SW33 did not eliminate thrombocytopenia in tumor-bearing mice, it had no noticeable side effects on platelets. In the immune system, SW33 has no effect on numbers of lymphocytes, neutrophils, eosinophils or basophils while inhibits the level of monocytes.
The results from RNA-Seq analysis showed SW33 played an important role in regulating inflammation and immune response (Fig. S2C). Inflammation is important in tumorigenesis43. It has been demonstrated that cancers can be promoted by continuous stimulation of inflammation, and many cancer patients have a history of chronic inflammatory disease44,45. While the normal function of inflammation is to eliminate aberrant cells such as tumors, but the inflammatory conditions are typically seized upon and exploited by the tumor to promote its own growth and progression towards metastasis46. The spleen is the central organ for regulating the inflammatory immune response47. Enlargement of the spleen with increases in leukocyte numbers and monocyte ratio can be observed in tumor-bearing mice in the late stages of tumor growth which might be caused by inflammatory reaction and hyperimmune responses. SW33 inhibited growth of GBM in vivo and restored the spleen to its normal volume and weight (Fig. 7G). Monocytes are circulating leukocytes which play an important role in both innate and adaptive immunity, primarily through the inflammatory immune defense mechanism48. Monocyte were found to be recruited to the tumor during GBM initiation49. These peripheral monocytes may be attracted to the tumor microenvironment and act as promoters for tumor evolution through secretion of cytokines, such as TNFα, IL-1, and IL-650,51. Blood biochemical tests showed a significant increase in white blood cell (WBC) and percent monocytes (MON%) in the tumor group compared to control group and this phenomenon can be reversed by SW33, which may exert inhibitory effects on GBM by reducing inflammatory reactions and splenomegaly. Inflammation factors IL-2, IL-6 and TNFα were also decreased by SW33 (Supporting Information Fig. S8). IL-2 is a type of signal molecule in the immune system that regulates the activity of lymph cells which have immune functions. In the immune response, IL-2 acts as a growth factor to stimulate T cell expansion. IL-2 can stimulate both effector T cells and regulatory T cells, which play important roles in the immune system52. IL-6 has pleiotropic effects, but its continuous disorder will lead to the occurrence and development of various autoimmune and chronic inflammatory diseases. TNFα has cytotoxic effects on a variety of tumor cells, and is also closely related to diseases, such as inflammation and fever, arthritis, sepsis and multiple sclerosis53. Though SW33 might inhibit inflammation and immune reaction to some extent, the detailed interactions in the mechanism of SW33's anti-inflammatory effects and inflammation-related immune reactions are still unclear and need further study.
5. Conclusions
The anticancer compound, SW33, was obtained through acylation at the 4-position of the hydroxyl group of SIN. It was shown to inhibit proliferation, migration, invasion and colony formation of human glioblastoma cell lines. SW33 arrested the cell cycle at the G2/M phase, induced apoptosis via mitochondrial dysfunction and induced autophagy through regulation of the PI3K/AKT/mTOR and AMPK/mTOR pathways. SW33 reduced inflammation and restored the spleen in mice to normal size with an excellent safety profile. In conclusion, all these results suggest that SW33 should be considered as a promising drug for treating GBM.
Acknowledgments
This research was funded by Beijing Natural Science Foundation (7212157, China), CAMS Innovation Fund for Medical Sciences (2016-I2M-3-007, China), National Natural Science Foundation of China (81703536, 81803584, 81703565, China), and Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025, 2018ZX09711001-012, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.05.027.
Contributor Information
Jinhua Wang, Email: wjh@imm.ac.cn.
Tengfei Ji, Email: jitf@imm.ac.cn.
Guanhua Du, Email: dugh@imm.ac.cn.
Author contributions
Xiangjin Zheng: investigation, data curation, writing original draft; Wan Li: validation, formal analysis; Huanli Xu: methodology; Jinyi Liu: project administration; Liwen Ren: software; Yihui Yang: data curation; Sha Li: data curation; Jinhua Wang: conceptualization, funding acquisition; Guanhua Du: Supervision, funding acquisition; Tengfei Ji: resources.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ostrom Q.T., Gittleman H., Stetson L., Virk S.M., Barnholtz-Sloan J.S. Epidemiology of gliomas. Cancer Treat Res. 2015;163:1–14. doi: 10.1007/978-3-319-12048-5_1. [DOI] [PubMed] [Google Scholar]
- 2.Chen R., Smith-Cohn M., Cohen A.L., Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush N.A., Chang S.M., Berger M.S. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40:1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 4.Alexander B.M., Cloughesy T.F. Adult glioblastoma. J Clin Oncol. 2017;35:2402–2409. doi: 10.1200/JCO.2017.73.0119. [DOI] [PubMed] [Google Scholar]
- 5.Ostrom Q.T., Bauchet L., Davis F.G., Deltour I., Fisher J.L., Langer C.E., et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanan M.I., Underhill D.A., Eisenstat D.D. Targeting epigenetic pathways in the treatment of pediatric diffuse (high grade) gliomas. Neurotherapeutics. 2017;14:274–283. doi: 10.1007/s13311-017-0514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parney I.F., Chang S.M. Current chemotherapy for glioblastoma. Cancer J. 2003;9:149–156. doi: 10.1097/00130404-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Whitelaw B.C. How and when to use temozolomide to treat aggressive pituitary tumours. Endocr Relat Cancer. 2019;26:545–552. doi: 10.1530/ERC-19-0083. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Zhao C., Ma Q., Li Y. Sinomenine retards LPS-elicited inflammation via down-regulating CCAT1 in HaCaT cells. Life Sci. 2019;233:116703. doi: 10.1016/j.lfs.2019.116703. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X.X., Peng C., Zhang H., Qin L.P. Sinomenium acutum: a review of chemistry, pharmacology, pharmacokinetics, and clinical use. Pharm Biol. 2012;50:1053–1061. doi: 10.3109/13880209.2012.656847. [DOI] [PubMed] [Google Scholar]
- 11.Zhu R.L., Zhi Y.K., Yi L., Luo J.F., Li J., Bai S.S., et al. Sinomenine regulates CD14/TLR4, JAK2/STAT3 pathway and calcium signal via α7nAChR to inhibit inflammation in LPS-stimulated macrophages. Immunopharmacol Immunotoxicol. 2019;41:172–177. doi: 10.1080/08923973.2019.1568451. [DOI] [PubMed] [Google Scholar]
- 12.Gao L.N., Zhong B., Wang Y. Mechanism underlying antitumor effects of sinomenine. Chin J Integr Med. 2019;25:873–878. doi: 10.1007/s11655-019-3151-2. [DOI] [PubMed] [Google Scholar]
- 13.Jiang S., Gao Y., Hou W., Liu R., Qi X., Xu X., et al. Sinomenine inhibits A549 human lung cancer cell invasion by mediating the STAT3 signaling pathway. Oncol Lett. 2016;12:1380–1386. doi: 10.3892/ol.2016.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan H., Zhang J., Li F., Li W., Wang H. Sinomenine exerts antitumour effect in gastric cancer cells via enhancement of miR-204 expression. Basic Clin Pharmacol Toxicol. 2019;125:450–459. doi: 10.1111/bcpt.13285. [DOI] [PubMed] [Google Scholar]
- 15.Xu F., Li Q., Wang Z., Cao X. Sinomenine inhibits proliferation, migration, invasion and promotes apoptosis of prostate cancer cells by regulation of miR-23a. Biomed Pharmacother. 2019;112:108592. doi: 10.1016/j.biopha.2019.01.053. [DOI] [PubMed] [Google Scholar]
- 16.Song L., Liu D., Zhao Y., He J., Kang H., Dai Z., et al. Sinomenine inhibits breast cancer cell invasion and migration by suppressing NF-κB activation mediated by IL-4/miR-324-5p/CUEDC2 axis. Biochem Biophys Res Commun. 2015;464:705–710. doi: 10.1016/j.bbrc.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y.S., Han J.Y., Iqbal O., Liang A.H. Research advances and prospects on mechanism of sinomenin on histamine release and the binding to histamine receptors. Int J Mol Sci. 2018;20:70. doi: 10.3390/ijms20010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., Liu J., Fu W., Zheng X., Ren L., Liu S., et al. 3-O-Acetyl-11-keto-β-boswellic acid exerts anti-tumor effects in glioblastoma by arresting cell cycle at G2/M phase. J Exp Clin Cancer Res. 2018;37:132. doi: 10.1186/s13046-018-0805-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F., Liu Z., Sun H., Li C., Wang W., Ye L., et al. PCC0208017, a novel small-molecule inhibitor of MARK3/MARK4, suppresses glioma progression in vitro and in vivo. Acta Pharm Sin B. 2020;10:289–300. doi: 10.1016/j.apsb.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou K., Zhang C., Yao H., Zhang X., Zhou Y., Che Y., et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17:105. doi: 10.1186/s12943-018-0849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Li L., Liu S., Zhao Y., Wang L., Du G., et al. FOXC1 promotes melanoma by activating MST1R/PI3K/AKT. Oncotarget. 2016;7:84375–84387. doi: 10.18632/oncotarget.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sung D.K., Chang Y.S., Kang S., Song H.Y., Park W.S., Lee B.H. Comparative evaluation of hypoxic-ischemic brain injury by flow cytometric analysis of mitochondrial membrane potential with JC-1 in neonatal rats. J Neurosci Methods. 2010;193:232–238. doi: 10.1016/j.jneumeth.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Li J., He D., Wang B., Zhang L., Li K., Xie Q., et al. Synthesis of hydroxycinnamic acid derivatives as mitochondria-targeted antioxidants and cytotoxic agents. Acta Pharm Sin B. 2017;7:106–115. doi: 10.1016/j.apsb.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J., Li Q., Cai L., Zhu Z., Guan J., Wang C., et al. RBM17 controls apoptosis and proliferation to promote glioma progression. Biochem Biophys Res Commun. 2018;505:20–28. doi: 10.1016/j.bbrc.2018.09.056. [DOI] [PubMed] [Google Scholar]
- 25.Lu Z.M., Ren Y.D., Yang L., Jia A., Hu Y., Zhao Y., et al. Inhibiting autophagy enhances sulforaphane-induced apoptosis via targeting NRF2 in esophageal squamous cell carcinoma. Acta Pharm Sin B. 2021;11:1246–1260. doi: 10.1016/j.apsb.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X., Wu Y., Ye L., Wang Y., Zhang K., Wang L., et al. Aspirin alleviates endothelial gap junction dysfunction through inhibition of NLRP3 inflammasome activation in LPS-induced vascular injury. Acta Pharm Sin B. 2019;9:711–723. doi: 10.1016/j.apsb.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu F., Li Q., Wang Z., Cao X. Sinomenine inhibits proliferation, migration, invasion and promotes apoptosis of prostate cancer cells by regulation of miR-23a. Biomed Pharmacother. 2019;112:108592. doi: 10.1016/j.biopha.2019.01.053. [DOI] [PubMed] [Google Scholar]
- 28.Song L., Liu D., Zhao Y., He J., Kang H., Dai Z., et al. Sinomenine reduces growth and metastasis of breast cancer cells and improves the survival of tumor-bearing mice through suppressing the SHh pathway. Biomed Pharmacother. 2018;98:687–693. doi: 10.1016/j.biopha.2017.12.065. [DOI] [PubMed] [Google Scholar]
- 29.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Jiao Y., Liu Y., Zhang M., Wang Z., Li Y., et al. Sinomenine hydrochloride inhibits the metastasis of human glioblastoma cells by suppressing the expression of matrix metalloproteinase-2/-9 and reversing the endogenous and exogenous epithelial–mesenchymal transition. Int J Mol Sci. 2018;19:844. doi: 10.3390/ijms19030844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Z., Zheng L., Liu X., Xing W., Liu X. Sinomenine inhibits the growth of melanoma by enhancement of autophagy via PI3K/AKT/mTOR inhibition. Drug Des Devel Ther. 2018;12:2413–2421. doi: 10.2147/DDDT.S155798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Liu C., Tan T., Li S., Tang S., Chen X. Sinomenine sensitizes human gastric cancer cells to cisplatin through negative regulation of PI3K/AKT/Wnt signaling pathway. Anticancer Drugs. 2019;30:983–990. doi: 10.1097/CAD.0000000000000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H., Yin P., Shi Z., Ma Y., Zhao C., Zheng J., et al. Sinomenine, a COX-2 inhibitor, induces cell cycle arrest and inhibits growth of human colon carcinoma cells in vitro and in vivo. Oncol Lett. 2016;11:411–418. doi: 10.3892/ol.2015.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X., Maimaiti M., Jiao Y., Meng X., Li H. Sinomenine induces G1-phase cell cycle arrest and apoptosis in malignant glioma cells via downregulation of sirtuin 1 and induction of p53 acetylation. Technol Cancer Res Treat. 2018;17 doi: 10.1177/1533034618770305. 1533034618770305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor W.R., Stark G.R. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 36.Gheghiani L., Loew D., Lombard B., Mansfeld J., Gavet O. PLK1 activation in late G2 sets up commitment to mitosis. Cell Rep. 2017;19:2060–2073. doi: 10.1016/j.celrep.2017.05.031. [DOI] [PubMed] [Google Scholar]
- 37.Cui L., Bu W., Song J., Feng L., Xu T., Liu D., et al. Apoptosis induction by alantolactone in breast cancer MDA-MB-231 cells through reactive oxygen species-mediated mitochondrion-dependent pathway. Arch Pharm Res. 2018;41:299–313. doi: 10.1007/s12272-017-0990-2. [DOI] [PubMed] [Google Scholar]
- 38.Li H., Peng X., Wang Y., Cao S., Xiong L., Fan J., et al. Atg5-mediated autophagy deficiency in proximal tubules promotes cell cycle G2/M arrest and renal fibrosis. Autophagy. 2016;12:1472–1486. doi: 10.1080/15548627.2016.1190071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azzopardi M., Farrugia G., Balzan R. Cell-cycle involvement in autophagy and apoptosis in yeast. Mech Ageing Dev. 2017;161:211–224. doi: 10.1016/j.mad.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Heras-Sandoval D., Pérez-Rojas J.M., Hernández-Damián J., Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014;26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q., Wang X., Cao S., Sun Y., He X., Jiang B., et al. Berberine represses human gastric cancer cell growth in vitro and in vivo by inducing cytostatic autophagy via inhibition of MAPK/mTOR/p70S6K and Akt signaling pathways. Biomed Pharmacother. 2020;128:110245. doi: 10.1016/j.biopha.2020.110245. [DOI] [PubMed] [Google Scholar]
- 42.Deng F., Ma Y.X., Liang L., Zhang P., Feng J. The pro-apoptosis effect of sinomenine in renal carcinoma via inducing autophagy through inactivating PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2018;97:1269–1274. doi: 10.1016/j.biopha.2017.11.064. [DOI] [PubMed] [Google Scholar]
- 43.Neagu M., Constantin C., Caruntu C., Dumitru C., Surcel M., Zurac S. Inflammation: a key process in skin tumorigenesis. Oncol Lett. 2019;17:4068–4084. doi: 10.3892/ol.2018.9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao C.P., Booker R.C., Brosseau J.P., Chen Z., Mo J., Tchegnon E., et al. Contributions of inflammation and tumor microenvironment to neurofibroma tumorigenesis. J Clin Invest. 2018;128:2848–2861. doi: 10.1172/JCI99424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nadeem M.S., Kumar V., Al-Abbasi F.A., Kamal M.A., Anwar F. Risk of colorectal cancer in inflammatory bowel diseases. Semin Cancer Biol. 2020;64:51–60. doi: 10.1016/j.semcancer.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Dominguez C., David J.M., Palena C. Epithelial–mesenchymal transition and inflammation at the site of the primary tumor. Semin Cancer Biol. 2017;47:177–184. doi: 10.1016/j.semcancer.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrea L., Di Somma C., Muscogiuri G., Tarantino G., Tenore G.C., Orio F., et al. Nutrition, inflammation and liver–spleen axis. Crit Rev Food Sci Nutr. 2018;58:3141–3158. doi: 10.1080/10408398.2017.1353479. [DOI] [PubMed] [Google Scholar]
- 48.Kratofil R.M., Kubes P., Deniset J.F. Monocyte conversion during inflammation and injury. Arterioscler Thromb Vasc Biol. 2017;37:35–42. doi: 10.1161/ATVBAHA.116.308198. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z., Feng X., Herting C.J., Garcia V.A., Nie K., Pong W.W., et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 2017;77:2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu S., Rodabaugh K., Martinez-Maza O., Watson J.M., Silberstein D.S., Boyer C.M., et al. Stimulation of ovarian tumor cell proliferation with monocyte products including interleukin-1, interleukin-6, and tumor necrosis factor-alpha. Am J Obstet Gynecol. 1992;166:997–1007. doi: 10.1016/0002-9378(92)91379-o. [DOI] [PubMed] [Google Scholar]
- 51.Brailo V., Vucicevic-Boras V., Lukac J., Biocina-Lukenda D., Zilic-Alajbeg I., Milenovic A., et al. Salivary and serum interleukin 1 beta, interleukin 6 and tumor necrosis factor alpha in patients with leukoplakia and oral cancer. Med Oral Patol Oral Cir Bucal. 2012;17:e10–e15. doi: 10.4317/medoral.17323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pipkin M.E., Sacks J.A., Cruz-Guilloty F., Lichtenheld M.G., Bevan M.J., Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.