Abstract
Members of the cullin and RING finger ROC protein families form heterodimeric complexes to constitute a potentially large number of distinct E3 ubiquitin ligases. We report here that the highly conserved C-terminal sequence in CUL1 is dually required, both for nuclear localization and for modification by NEDD8. Disruption of ROC1 binding impaired nuclear accumulation of CUL1 and decreased NEDD8 modification in vivo but had no effect on NEDD8 modification of CUL1 in vitro, suggesting that ROC1 promotes CUL1 nuclear accumulation to facilitate its NEDD8 modification. Disruption of NEDD8 binding had no effect on ROC1 binding, nor did it affect nuclear localization of CUL1, suggesting that nuclear localization and NEDD8 modification of CUL1 are two separable steps, with nuclear import preceding and required for NEDD8 modification. Disrupting NEDD8 modification diminishes the IκBα ubiquitin ligase activity of CUL1. These results identify a pathway for regulation of CUL1 activity—ROC1 and the CUL1 C-terminal sequence collaboratively mediate nuclear accumulation and NEDD8 modification, facilitating assembly of active CUL1 ubiquitin ligase. This pathway may be commonly utilized for the assembly of other cullin ligases.
Ubiquitin-mediated proteolysis plays a key role in regulating the levels of a large number of proteins involved in diverse cellular processes. Through a cascade of enzymes involving ubiquitin activation (E1), conjugation (E2), and ligation (E3), the ubiquitin-dependent protein degradation pathways catalyze the formation of polyubiquitin chains onto substrate proteins via isopeptide bonds. Polyubiquitinated substrates are then rapidly delivered to and degraded by the 26S proteasome (6, 7, 12). While E1 and E2 both represent structurally related proteins that are relatively well characterized biochemically, the E3 ubiquitin ligases, generally defined as having both a ubiquitin ligase activity and a substrate targeting function, comprise a potentially large number of diverged multisubunit protein complexes. Elucidating the molecular nature and regulation of E3s has become critically important to our understanding of regulated proteolysis and is currently the focus of intensive investigation.
The three best-characterized E3 activities involve HECT (homologous to E6AP carboxy terminus) domain proteins represented by E6-AP (10), the anaphase-promoting complex (APC or cyclosome) that consists of at least 12 subunits and is required for both entry into anaphase and exit from mitosis (18, 51), and the SCF complexes, which consist of SKP1, CDC53 or its homologue, Cullin-1, and a distinct F box protein (1, 4, 42). A core subunit of SCF, cullin or CDC53, was first identified, through genetic analysis, in Caenorhabditis elegans and yeast as being encoded by a gene whose functional loss caused defects in G1 regulation (19, 29, 47). Several CDC53- or CUL1-dependent SCF complexes have been identified in both yeast and mammalian cells that function to ubiquitinate many phosphorylated substrate proteins, indicating that a similar SCF mechanism is evolutionarily conserved across species (recently reviewed in references 2, 20, and 36). A subunit of the mitotic APC E3 complex, APC2, was found to contain a limited sequence similarity to CDC53 and cullins (50, 52), further underscoring an important and conserved role for cullin proteins in ubiquitin-mediated proteolysis.
Recently a family of closely related RING finger proteins, represented by ROC1 (regulator of cullins, also known as Rbx1 for RING-box protein, or Hrt1) was identified (reviewed in references 2 and 51). ROC1 shares a high degree of sequence similarity to another APC subunit, APC11, and functions as an essential subunit of CUL1 and CDC53 ubiquitin ligases to catalyze ubiquitination of the phosphorylated inhibitor of the NF-κB transcription factor, IκBα, G1 cyclin Cln2, and the CDK inhibitor Sic1 (17, 33, 41, 43, 45). Deficiency of yeast ROC1 can be functionally rescued by mammalian ROC1 and ROC2 but not by yeast APC11 (17, 33, 41), demonstrating an evolutionary conservation and functional specificity for the ROC gene family. ROC1 and ROC2 commonly interact with all cullins to constitute multiple active ubiquitin ligases, while APC11 specifically interacts and constitutes active ligase with cullin-related APC2 (33; Ohta et al., unpublished data), suggesting the in vivo existence of a potentially large number of heterodimeric cullin-ROC ubiquitin ligases. More recently, several large RING finger proteins with otherwise diverse structures and functions were linked to ubiquitination (14, 26), suggesting a broad and general function of RING fingers in inducing E3 ligase activity.
Genetic and biochemical studies with yeast cells have identified a covalent modification of the CDC53 protein by a ubiquitin-like protein, RUB1 (related to ubiquitin), via an E1-like bipartite RUB1-activating enzyme (22, 24). In mammalian cells, catalyzed via a similar bipartite E1 activity and E2-Ubc12 (34), the RUB1 homologue, NEDD8 (neural precursor cell-expressed and developmentally down-regulated), forms a covalent conjugate with most, if not all, cullins (9, 23, 34). In fact, cullins appear to be the major cellular substrates of NEDD8 modification (15, 22, 24, 34). NEDD8 is a member of a group of ubiquitin-like proteins that are covalently linked to their substrates mostly, if not only, monomerically by E1 activating and E2 conjugating activities distinct from that utilized in ubiquitination (8, 13). Conjugation of the mammalian ubiquitin-related UCRP (ubiquitin cross-reactive protein) appears to target conjugates to the cytoskeleton (25), and conjugation of another mammalian ubiquitin-like protein, SUMO-1 (small ubiquitin-related modifier, also known as sentrin, PIC1, UBL1, and GMP1), was found to target cytosolic RanGAP1 to the nuclear pore complex (27, 30), to antagonize ubiquitin-mediated degradation of IκBα (3), and to activate transcriptional activity of p53 (5, 40). These observations indicate that in contrast to polyubiquitination, covalent modification by these ubiquitin-like proteins plays regulatory, rather than proteolytic, roles.
The regulation and biochemical significance of cullin-RUB1 or -NEDD8 conjugation are not clear at present. The RUB1 gene and its E1 (ENR2 and UBA3) activating and E2 (UBC12) conjugating genes are not essential for budding yeast growth. The defect in CDC53-RUB1 conjugation, however, is synthetically lethal, with a temperature-sensitive mutation of cdc34 that enhances the phenotype of the cdc4, cdc53, and skp1 mutations (22, 24), leading to the suggestion that RUB1 may play a regulatory role, albeit nonessential, in regulating ROC-cullin ubiquitin ligase activity in budding yeast. Disruption of NEDD8 modification in both mammalian and fission yeast cells, however, significantly reduced the level of in vitro SCF ubiquitin ligase activity of CUL1 and resulted in a cellular lethality (32, 35, 37, 39), indicating an essential function of NEDD8 modification. NEDD8 modification of both yeast CDC53 and human CUL2 is dependent on the presence of ROC1 (16). The mechanism by which ROC1 activates RUB1 and NEDD8 modification of cullins is unclear. In this paper, we report that the highly conserved cullin C-terminal sequence and binding with ROC1 are both required for efficient nuclear accumulation of cullin. Blocking of CUL1 nuclear localization reduced its NEDD8 modification, resulting in a significant loss of IκBα ubiquitin ligase activity of CUL1 both in vitro and in vivo. These results identify a pathway for regulation of CUL1 activity—ROC1 binds to CUL1 and probably collaborates with its C-terminal sequence to promote nuclear localization of CUL1, thereby facilitating NEDD8 modification and assembly of active CUL1 ubiquitin ligase in the nucleus. Three elements involved in the activation of CUL1 ligase activity—ROC1 binding, NEDD8 modification, and the C-terminal sequence—are shared by or conserved in most, if not all, cullins, suggesting that this pathway may be commonly utilized in the assembly and activation of other cullin ligases.
MATERIALS AND METHODS
Plasmids.
Full-length mammalian cullin cDNAs were described previously (31). Human ROC1 cDNA was described previously (33). Cullin mutations were introduced by site-directed mutagenesis using the Quick-Change kit (Stratagene) and verified by DNA sequencing. Human NEDD8 cDNA (21) was amplified from a HeLa cell cDNA library by PCR and subcloned into the pcDNA3 mammalian expression vector with a Myc epitope tag.
Cell lines, culture conditions, and cell transfection.
Human U2OS and 293T cells used in this study were cultured in Dulbecco modified Eagle medium, supplemented with 10% fetal bovine serum in a 37°C incubator with 5% CO2. Cell transfections were carried out using the Lipofectamine (Gibco-BRL) or Effectene (QIAGEN) transfection reagents according to the manufacturer's instructions (Gibco-BRL) (for U2OS cells) or calcium-phosphate buffer (for 293T cells). For each transfection, 2.5, 5, or 15 μg of total plasmid DNA was used for each six-well, 60- or 100-mm dish, respectively.
Antibodies, immunochemistry, and indirect immunofluorescence.
Procedures for [35S]methionine metabolic labeling, immunoprecipitation, and immunoblotting have been described previously (11). Indirect immunofluorescence was previously described in detail (54). Antibodies to human CUL1 and SKP1 (31) and to human ROC1 (33) were previously described. Monoclonal antihemagglutinin (α-HA) (12CA5; Boehringer-Mannheim), affinity-purified polyclonal α-HA (sc-805; Santa Cruz), α-Myc (9E10; NeoMarker), α-FLAG (clone M2, F3165; Sigma), α-T7 (no. 69522; Novagen), rhodamine red- and fluorescein isothiocyanate-conjugated donkey secondary antibodies (Jackson ImmunoResearch Laboratories), and a rabbit polyclonal antibody to NEDD8 (Catalog no. 210-194; Alexis Biochemicals, San Diego, Calif.) were purchased commercially.
In vitro NEDD8 assays.
[35S]methionine-labeled proteins were prepared in vitro with T7 RNA polymerase using a TNT-coupled reticulocyte lysate system according to the manufacturer's recommendation (Promega). Equal amounts of ROC1 and CUL1 plasmid DNA (0.3 μg) were mixed and were transcribed and translated together. After incubation for 90 min at 30°C, the reaction mixture was aliquoted, diluted with the NP-40 buffer, and immunoprecipitated with either HA or Myc antibody (11). Immunoprecipitates were washed three times with the same NP-40 buffer, boiled in sodium dodecyl sulfate (SDS) sample buffer containing 0.1 M dithiothreitol (DTT), and electrophoresed on 7.5 to 15% gradient denaturing polyacrylamide gels followed by autoradiography.
In vitro and in vivo ubiquitin ligase activity assays.
The procedure for an in vitro assay of the ROC- and cullin-associated ubiquitin ligase activity was essentially the same as has been previously described (33, 45). Purified rabbit E1 ubiquitin-activating enzyme was purchased from Affinity Research Products (Exeter, United Kingdom). His-tagged E2 human UbcH5C was purified using nickel beads (QIAGEN). Ub was prepared by subcloning full-length Ub as a fusion protein with a six-His tag and protein kinase C recognition site (LRRASV) and was purified with nickel beads (45). For ubiquitination assays, transfected HA-tagged wild-type or mutant CUL1 immunocomplexes were immunoprecipitated with α-HA antibody. Immunocomplexes immobilized on protein A agarose beads were washed and added to a Ub ligation reaction mixture (30 μl) that contained 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 0.75 μg of Ub, 60 ng of E1, 300 ng of E2, and 1.2 μg of phosphorylated glutathione S-transferase (GST)–IκBα1–54. Reaction mixtures were incubated at 37°C for 60 min, terminated by adding 30 μl of 2× Laemmli loading buffer and boiling for 4 min, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) followed by autoradiography to visualize the ubiquitinated IκBα ladders. For substrate preparation, 36 μg of purified glutathione S-transferase-IκBα (residues 1 to 54) was phosphorylated with an HA immunocomplex derived from 293T cells transfected with HA-tagged, constitutively active IKKβS177E/S181E kinase. The reaction was carried out in the presence of 20 μCi of [γ-32P]ATP at 37°C for 20 min in a total volume of 60 μl of kinase buffer (50 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 5 μM ATP, 2 mM NaF, 10 nM okadaic acid, and 0.6 mM DTT).
For the in vivo IκBα ubiquitination assay, 293T cells on a 100-mm dish were transfected with plasmids expressing HA-CUL1 (5 μg), Myc-β-TrCP (3 μg), Myc-ROC1 (3 μg), SKP1 (5 μg), HA-IKKβS177E/S181E (3 μg), FLAG-IκBα (1 μg), and HA-Ub (5 μg). The total amount of plasmid DNA in each transfection was adjusted to total 25 μg with a pcDNA3 empty vector when needed. Twenty hours after transfection, cells were treated with the proteasome inhibitor MG132 (25 μM) for 4 h. Cells were then collected, pelleted by centrifugation, lysed in 200 μl of preboiled SDS-lysis buffer (50 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 1% SDS, 1 mM DTT), and further boiled for an additional 10 min. Lysates were clarified by centrifugation at 14,000 rpm on a microcentrifuge (5415C; Eppendorf) for 10 min. The supernatant was diluted 10-fold with 0.5% NP-40 buffer and immunoprecipitated with anti-FLAG antibody (3.6 μg). Immunoprecipitates were washed three times and resolved on a 7.5 to 15% gradient SDS-PAGE, followed by immunoblotting with anti-HA antibody (1 μg/ml) or anti-FLAG antibody (1 μg/ml).
RESULTS
The C-terminal sequence is required for nuclear localization of CUL1.
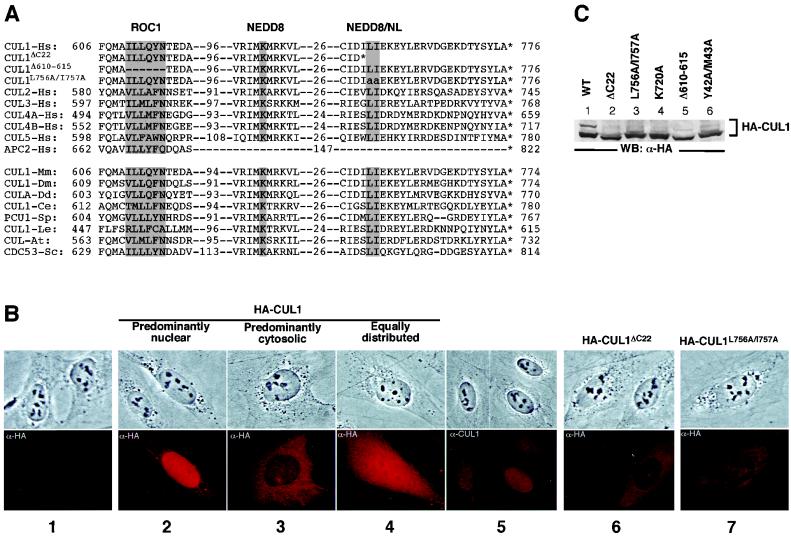
The carboxyl terminus is the most highly conserved region in cullin family members from different organisms (19) (Fig. 1A). In experiments investigating the functional significance of this highly conserved domain, we noticed that deletion of the CUL1 C terminus resulted in an altered subcellular localization. Subcellular localization of wild-type human CUL1 in transfected U2OS cells exhibited heterogeneous patterns (Fig. 1B), localized predominantly to the nucleus in most cells (81.9%) (Table 1) and predominantly to the cytoplasm in a small percentage of cells (6.5%), and equally distributed in both the nucleus and the cytoplasm in the remaining cells (11.6%). Similar heterogeneous distribution with predominant nuclear localization was also seen for endogenous CUL1 (Fig. 1B, column 5). Whether this heterogeneity reflects a dynamic trafficking of CUL1 protein or a cell cycle-dependent regulation has not been investigated. Deletion of 22 C-terminal amino acid residues (CUL1ΔC22) significantly impaired the nuclear accumulation of CUL1, resulting in a predominantly cytoplasmic accumulation of CUL1 in most cells (63.2%) and a reduction of predominantly nuclear localization from 81.9 to 14.5% of the cell population (Table 1 and Fig. 1B, column 6). To identify the residues required for CUL1 nuclear accumulation, we mutated several highly conserved residues and examined their effect on subcellular localization by indirect immunofluorescence. Mutation of two adjacent hydrophobic residues, Leu756 and Ile757 (CUL1L756A/I757A), reduced the nuclear accumulation of CUL1 from 81.9% of cells to 23.6% and increased the population of cells with predominantly cytoplasmic CUL1 staining from 6.5 to 41.2% (Table 1 and Fig. 1B, column 7). These results indicate that mutations at Leu756 and Ile757 significantly impaired nuclear accumulation of CUL1 but less severely than the C-terminal 22-residue deletion, suggesting that additional residue(s) in this region may also contribute to nuclear accumulation of CUL1.
FIG. 1.
The C-terminal sequence is required for nuclear localization of CUL1. (A) Amino acid sequence comparison of three functional domains, the ROC1 binding domain, the NEDD8 conjugation site, and the C-terminal sequence involved in both NEDD8 modification and nuclear localization (NEDD8/NL), of six human cullins, human APC2, and CUL1 from eight different organisms. Hs, Homo sapiens (human); Mm, Mus musculus (house mouse); Dm, Drosophila melanogaster (fruit fly); Dd, Dictyostelium discoideum (slime mold); Ce, C. elegans (worm); Sp, Schizosaccharomyces pombe (fission yeast); Le, Lycopersicon esculentum (tomato); At, Arabidopsis thaliana (mouse-ear cress); Sc, Saccharomyces cerevisiae (budding yeast). Shadowed residues denote mutations that were characterized in this study. Asterisks indicate the stop codon. (B) Subcellular localization of the CUL1 protein was examined by indirect immunofluorescence in untransfected U2OS cells or U2OS cells transiently transfected with plasmid DNA expressing HA-tagged wild-type and mutant CUL1. (C) Levels of CUL1 expression for wild-type and mutant proteins were examined by Western blot analysis of transfected U2OS cells.
TABLE 1.
Subcellular localization of CUL1 proteinsa
| Mutation | Results (no. of cells/%)
|
Total no. of cells counted | ||
|---|---|---|---|---|
| Predominantly nuclear | Predominantly cytosolic | Equally distributed | ||
| None | 176/81.9 | 14/6.5 | 25/11.6 | 215 |
| ΔC22 | 28/14.5 | 122/63.2 | 43/22.3 | 193 |
| L756A/I757A | 43/23.6 | 75/41.2 | 64/35.2 | 182 |
| K720A | 186/85.3 | 4/1.8 | 28/12.8 | 218 |
| Δ610–615 | 18/7.9 | 163/71.5 | 47/20.6 | 228 |
| Y42A/M43A | 182/73.7 | 10/4.0 | 55/22.3 | 247 |
U2OS cells were transfected with plasmids expressing HA-tagged wild-type or mutant CUL1. Thirty-six hours after transfection, cells were fixed and stained with a monoclonal antibody recognizing HA (12CA5). At least 180 positive cells were counted for each transfection, and subcellular distribution of HA-tagged CUL1 was scored into three categories: predominantly nuclear, predominantly cytosolic, and equally distributed.
To determine whether the conserved C termini of other cullins are also required for nuclear localization, we introduced similar double point mutations at the conserved Leu and Ile residues into human CUL2 (CUL2L725A/I726A), CUL5 (CUL5L760A/I761A), and mouse CUL4A (mCUL4AL739A/I740A) and examined their effect on subcellular localization. Like CUL1, the wild-type CUL2, mCUL4A, and CUL5 are localized predominantly to the nucleus in most cells and to the cytoplasm in a smaller population (data not shown). Mutations at the adjacent Leu and Ile residues reduced nuclear accumulation of all three cullins (data not shown). These results demonstrate that the C-terminal sequence contains an evolutionarily conserved element required for efficient nuclear localization of most, if not all, cullin proteins. Notably, the APC2 protein lacks this sequence, suggesting a distinct mechanism for regulation of APC ligase activity.
The transfected cullins described in this study—wild-type CUL1, the CUL1ΔC22 and CUL1L756A/I757A mutants, a NEDD8 modification mutant (CUL1K720A), a ROC1 binding mutant (CUL1Δ610–615), and a SKP1 binding mutant (CUL1Y42A/M43A)—were expressed at similar levels as determined by direct immunoblotting of the total cell lysate derived from each transfected cell population with anti-HA antibody (Fig. 1C, lanes 1 to 6). These results exclude the possibility that these point mutations or small internal deletions changed CUL1 protein stability or expression levels, resulting in an altered subcellular localization. From these experiments, we conclude that the C-terminal sequence of human CUL1 contains a signal critically important for CUL1's nuclear accumulation. Notably, a slow-migrating and less intense band was detected in cells expressing wild-type CUL1 (Fig. 1C, lane 1) but was not seen or was substantially reduced in all five CUL1 mutants (lanes 2 to 6). This band was later confirmed as NEDD8-modified CUL1 (see below), suggesting a potential link between CUL1 modification by NEDD8 and its nuclear import and binding with ROC1 and SKP1. Experiments to investigate these possible links are described below.
Blocking nuclear accumulation of CUL1 reduces its modification by NEDD8.
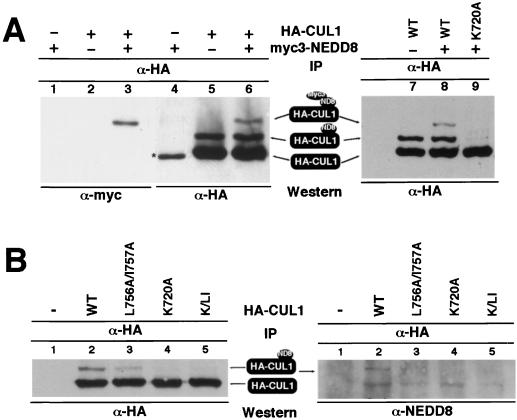
The carboxyl terminus of CDC53 was previously shown to be required for RUB1 modification of yeast CDC53 (22). The NEDD8 conjugation site in human CUL2 has been mapped to Lys residue 689 (46), corresponding to Lys720 in human CUL1 (Fig. 1A). The function of the C-terminal sequence in promoting nuclear accumulation of cullin, while distinct from the NEDD8 conjugation site, suggests that nuclear localization and RUB1 or NEDD8 modification may be two related events. To test this possibility, we first sought to confirm the NEDD8 modification site in human CUL1. Transfected wild-type, HA-tagged CUL1 was resolved into two forms as determined by coupled immunoprecipitation and immunoblotting (IP-Western) (Fig. 2A, lane 5), likely corresponding to unmodified and NEDD8-conjugated CUL1, respectively. Coexpression with Myc3-NEDD8 resulted in the appearance of a third, even slower-migrating band (lane 6). When the same α-HA precipitate was immunoblotted with α-Myc antibody, the slowest-migrating form, but not the other two bands, was detected (lane 3), confirming that it corresponds to Myc3-NEDD8-conjugated CUL1. Mutation of Lys residue 720 (HA-CUL1K720A) abolished both slow-migrating forms without affecting the fast-migrating band (lane 9), confirming that the two slow-migrating forms correspond to NEDD8- and Myc3-NEDD8-conjugated CUL1 and that Lys720 is the major, if not the only, site for modification by NEDD8 in CUL1.
FIG. 2.
The CUL1 C-terminal sequence is required for NEDD8 modification. (A) 293T cells were transiently transfected with plasmid vectors expressing Myc3-NEDD8 and wild-type or mutant CUL1 proteins as indicated. Lysates were prepared 24 h after transfection from each transfected cell population and immunoprecipitated with α-HA antibody. The precipitates were separated by SDS-PAGE and immunoblotted with α-HA or α-Myc antibodies. The asterisk in lane 4 indicates a nonspecific band cross-reacting with the monoclonal HA antibody. A polyclonal α-HA antibody that avoids this cross-reacting polypeptide was used in subsequent experiments. (B) 293T cells were transiently transfected with plasmid vectors expressing wild-type or mutant CUL1 proteins as indicated. Lysates were prepared 24 h after transfection from each transfected cell population and immunoprecipitated with α-HA antibody. The precipitates were separated by SDS-PAGE and immunoblotted with α-NEDD8 antibody (right panel), and then the same filter was stripped and reblotted with α-HA antibody. K/LI refers to a triple mutation in CUL1 (K720A/L756A/I757A). ND8, NEDD8.
To determine whether nuclear accumulation of CUL1 and its modification by NEDD8 may regulate each other, we examined NEDD8 modification of mutant CUL1 defective in nuclear localization and the subcellular localization of mutant CUL1 lacking the NEDD8 modification site. HA-tagged wild-type or mutant CUL1 that was defective in nuclear localization (L756A/I757A) or at the NEDD8 modification site (K720A), or their combination (referred to as K/LI), was transiently transfected into cultured 293T cells. NEDD8 modification of transfected CUL1 was determined by direct immunoblotting of HA immunoprecipitates with either α-HA antibody or α-NEDD8 antibody (Fig. 2B). Mutations in the CUL1 C-terminal sequence that impaired CUL1 nuclear localization (HA-CUL1L756A/I757A) consistently caused a considerable decrease, but not disruption, of NEDD8-conjugated CUL1 (Fig. 2B, lanes 2 and 3; Fig. 6A, lanes 2 and 6, Fig. 7C, lanes 5 and 6; and Fig. 7D, lanes 3 and 4).
FIG. 6.
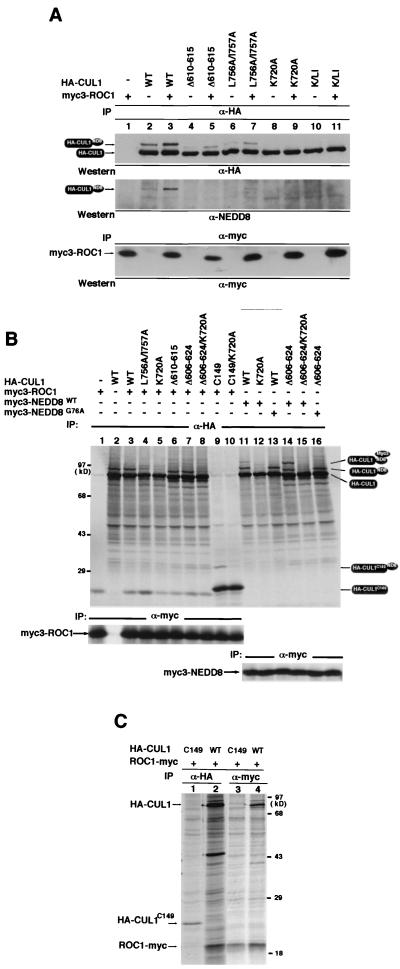
ROC1 promotes NEDD8 modification of CUL1 in vivo but not in vitro. (A) 293T cells were transiently cotransfected with plasmids expressing Myc-tagged ROC1 and HA-tagged wild-type or mutant CUL1 proteins as indicated. Twenty-four hours after transfection, lysates were prepared from each transfected cell population and immunoprecipitated with α-HA or α-myc antibodies. The α-HA precipitates were separated by SDS-PAGE and immunoblotted with α-HA antibody, and then the same filter was stripped and reblotted with α-NEDD8 antibody. The α-Myc precipitates after SDS-PAGE were blotted with α-Myc to verify the expression of Myc-ROC1. (B) In vitro assay for modification of CUL1 by NEDD8. Wild-type or mutant CUL1 proteins were synthesized together in vitro with either Myc3-ROC1 or Myc3-NEDD8 through a coupled in vitro transcription and translation system (TNT; Promega) in rabbit reticulocyte lysate in the presence of [35S]methionine. HA-CUL1, Myc3-ROC1, and Myc3-NEDD8 proteins were immunoprecipitated using HA and Myc antibodies, respectively, and resolved by SDS-PAGE. Note that the addition of ROC1 (lane 3) or mutation in the ROC1 binding region of CUL1 (Δ610–615 and Δ606–624, lanes 6 and 7) had no obvious effect on NEDD8 modification of CUL1 in vitro. (C) CUL1-ROC1 binding assay. Wild-type CUL1 and a CUL1 deletion mutant containing the C-terminal 149 residues were cotransfected with ROC1-Myc into 293T cells. CUL1-ROC1 binding was determined reciprocally by immunoprecipitation with either HA or Myc antibodies.
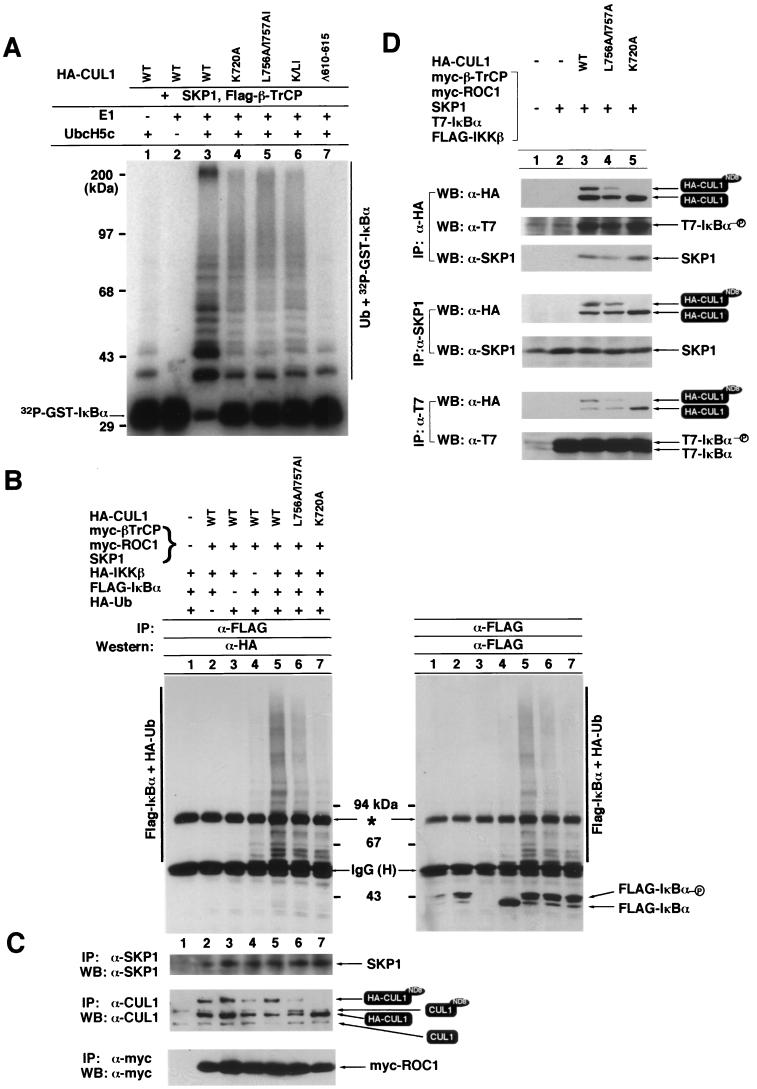
FIG. 7.
NEDD8 modification is required for IκBα ubiquitin ligase activity of CUL1. (A) In vitro IκBα ubiquitination. Purified IκBα was phosphorylated with IKKβ and incubated with α-HA immunocomplexes derived from 293T cells cotransfected with plasmids expressing SKP1, FLAG-tagged β-TrCP, and HA-tagged wild-type or various mutant CUL1 proteins as indicated. Reactions were incubated at 37°C for 60 min, terminated by adding 30 μl of 2× Laemmli loading buffer, boiled for 4 min, and resolved by SDS-PAGE followed by autoradiography to visualize the ubiquitinated IκBα ladders. (B) In vivo IκBα ubiquitination. 293T cells were transfected with the indicated plasmids, including constitutively active IKKβS177E/S181E. Twenty hours after transfection, cells were treated with the proteasome inhibitor MG132 for 4 h prior to cell lysis. Cells were lysed into a 1% SDS-containing buffer and boiled for 10 min. Lysates were then diluted to 0.1% SDS and immunoprecipitated with anti-FLAG antibody, and washed immunoprecipitates were resolved by SDS-PAGE, followed by immunoblotting with anti-HA or anti-FLAG antibodies. Exposure time was 1 min. The asterisk indicates a nonspecific protein precipitated by the anti-FLAG antibody. (C) The levels of ectopically expressed SKP1, Myc-ROC1, HA-CUL1, and NEDD8 modification were examined by IP-Western blotting. Because the HA-Ub plasmid was included in the transfection, the level of ectopically expressed HA-CUL1 was determined by immunoprecipitation with an α-CUL1 antibody that detected both endogenous CUL1 and CUL1-NEDD8 conjugates, as well as transfected HA-CUL1 and HA-CUL1-NEDD8 conjugates. (D) Interaction of SKP1 and IκBα with wild-type and mutant CUL1s. 293T cells were transfected with the indicated plasmids. Whole cell lysates were prepared using NP-40 lysis buffer and immunoprecipitated with the indicated antibodies, followed by SDS-PAGE and immunoblotting.
NEDD8 modification is not required for CUL1 nuclear localization.
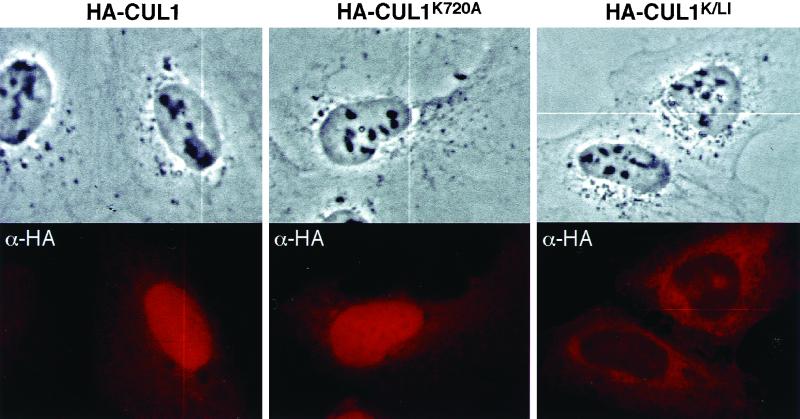
We next examined subcellular localization of a mutant CUL1K720A protein, harboring a mutation at its NEDD8 modification site. Compared with wild-type HA-CUL1 transfected in parallel, mutation at the NEDD8 modification site (K720A), in contrast to the mutations in the C-terminal sequence, had no significant effect on CUL1's nuclear localization (Fig. 3, middle column, and Table 1). CUL1 mutated in both the C-terminal and NEDD8 conjugation sites (CUL1K/LI) was still blocked from accumulating in the nucleus (Fig. 3, right column). Taken together, these observations indicate that NEDD8 modification is not required for nuclear accumulation of CUL1.
FIG. 3.
NEDD8 modification is not required for nuclear localization of CUL1. Subcellular localization of proteins was examined in transiently transfected U2OS cells for wild-type CUL1, mutant CUL1 lacking the NEDD8 modification site (K720A), and CUL1 with a mutated NEDD8 modification site and C-terminal residues (K720A/L756A/I757A).
Mapping the ROC1 binding sequence in CUL1.
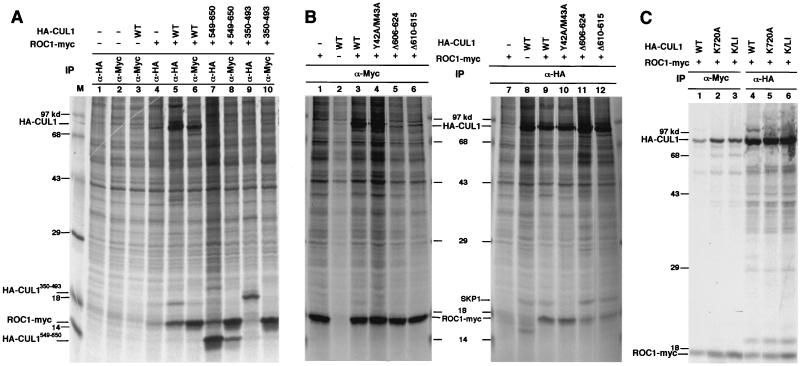
Both endogenously and ectopically expressed ROC1 protein localized mainly to the nucleus, with lower levels present in the cytoplasm (data not shown). We noticed an increase of nuclear accumulation of cullin proteins when they were cotransfected with ROC1 but not with APC11 (data not shown), raising the possibility that ROC1 may facilitate or be required for nuclear accumulation of cullins. To test this possibility directly, we first attempted to map the ROC1 binding sequence in order to identify a ROC1-binding-deficient mutant CUL1. Through a series of deletion analyses, we mapped a sequence in human CUL1, between residues 549 and 650, that is sufficient for binding with ROC1 as determined by reciprocal α-HA-CUL1 and α-ROC1-Myc immunoprecipitation (Fig. 4A, lanes 5 to 8). A comparison of the intensity of coprecipitated ROC1-Myc shows that the ROC1 binding affinity of CUL549–650 is very similar to that of wild-type CUL1 (Fig. 4A, compare lanes 5 and 7), indicating that CUL549–650 contains most, if not all, ROC1 binding activity. As a control, another CUL1 fragment containing residues 350 to 493 exhibited no detectable binding with cotransfected ROC1-Myc (lanes 9 and 10).
FIG. 4.
Mapping the ROC1 binding region in CUL1. 293T cells transiently transfected with the indicated plasmids expressing Myc-tagged ROC1 and two HA-tagged CUL1 fragments or several mutant CUL1s were pulse-labeled with [35S]methionine. Lysates prepared from transfected cells were immunoprecipitated with either α-Myc or α-HA antibody and resolved by SDS-PAGE, followed by autoradiography. (A) Mapping the ROC1 binding region in CUL1. (B) Characterization of ROC1 binding mutant CUL1s. (C) NEDD8 modification and nuclear localization of CUL1 are not required for CUL1-ROC1 binding.
Further deletion analyses within this region identified two internal deletion mutants, CUL1Δ606–624 (Fig. 4B, lanes 5 and 11) and CUL1Δ610–615 (lanes 6 and 12), that significantly reduced but did not completely disrupt the binding with ROC1 as determined by reciprocal immunoprecipitations. As a control, disruption of SKP1 association by the mutations at Tyr42 and Met43 (HA-CUL1Y42A/M43A) did not affect the binding of CUL1 with ROC1, as determined reciprocally by either anti-HA (Fig. 4B, lane 10) or anti-SKP1 (data not shown) immunoprecipitation, consistent with the previous finding that ROC1 and SKP1 bind independently to two separate regions in CUL1 (33). Six amino acid residues deleted in CUL1 (CUL1Δ610–615), which resulted in a disruption of ROC1 binding, are highly conserved among members of cullins from different organisms (Fig. 1A), suggesting that this sequence may be required for binding of ROC1 to other cullin proteins as well.
We also determined the effect of disruption of NEDD8 modification and mutation in the C-terminal sequence of CUL1 on CUL1's ability to bind ROC1 (Fig. 4C). Neither the mutation in the site for modification by NEDD8 (CUL1K720A; Fig. 4C, lanes 2 and 5), the C-terminal mutation (CUL1L756A/I757A; data not shown), nor their combination (CUL1K/LI; Fig. 4C, lanes 3 and 6) in CUL1 affects the binding of CUL1 with ROC1, indicating that binding of ROC1 to CUL1 is not dependent on CUL1's nuclear localization or NEDD8 modification.
ROC1 is required for efficient CUL1 nuclear localization.
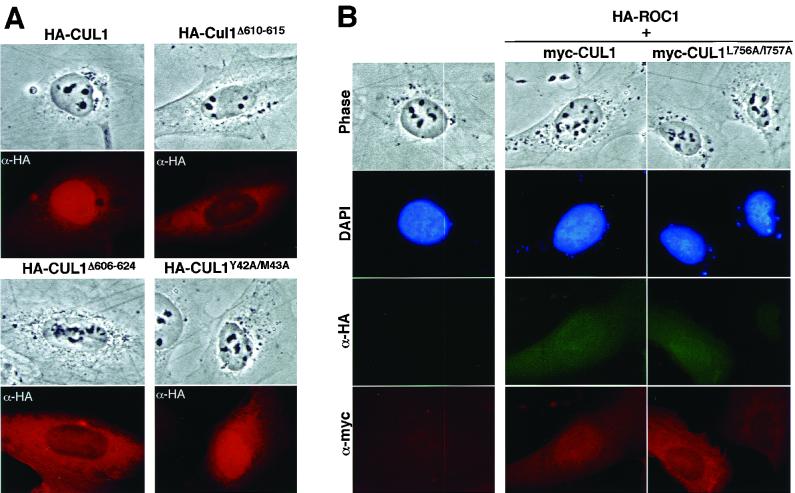
Mapping of ROC1 binding sequences and identification of ROC1 binding mutants of CUL1 allowed us to determine whether ROC1 is involved in nuclear accumulation of CUL1. Both CUL1Δ610–615 and CUL1Δ606–624 are severely impaired from entering the nucleus (Fig. 5A). The percentage of cells with predominantly nuclear staining was reduced from 81.9% for wild-type CUL1 to 7.9% for CUL1Δ610–615, and predominantly cytoplasmic staining increased from 6.5 to 71.5% (Table 1), an even more pronounced disruptive effect than that caused by deletion of the C-terminal 22 residues. Since both CUL1Δ610–615 and CUL1Δ606–624 mutants retain a residual low level of ROC1 binding activity (Fig. 4), such a prominent disruptive effect on nuclear accumulation of CUL1 associated with decreased ROC1 binding suggests the possibility that ROC1 might not act alone to promote CUL1 nuclear accumulation and that additional unidentified factor(s) may collaborate with ROC1 in this process in a way that may or may not act in a stoichiometric mode in promoting nuclear accumulation of CUL1. Disruption of the binding of CUL1 with SKP1 (Y42A/M43A), on the other hand, had only a slight effect on CUL1's nuclear accumulation (Fig. 5A). The percentage of cells with predominantly nuclear CUL1 staining was reduced from 81.9% for the wild type to 73.7% for CUL1Y42A/M43A (Table 1). Whether this result reflects a minor contribution of SKP1 to the nuclear accumulation of CUL1 has not been determined. Both CUL1Δ606–624 and CUL1Δ610–615 still bind with SKP1 as well as wild-type CUL1 does (Fig. 4B, lanes 11 and 12), and both can be modified efficiently by NEDD8 in vitro (Fig. 6B), arguing against the possibility that impairment of nuclear accumulation of CUL1 by mutations at the ROC1 binding site is caused by an overt protein conformational change or aggregation, as opposed to a loss of ROC1 binding. These observations suggest that ROC1 is required for efficient nuclear localization of CUL1.
FIG. 5.
ROC1 is required for nuclear accumulation of CUL1. (A) Subcellular localization of wild-type and mutant CUL1 proteins with disrupted ROC1 binding (Δ610–615 and Δ606–624) or SKP1 binding (Y42A/M43A) was examined in transiently transfected U2OS cells. (B) The CUL1 C-terminal sequence and ROC1 are both required to mediate CUL1 nuclear accumulation. U2OS cells were transiently cotransfected with plasmids expressing HA-ROC1 with either wild-type or C-terminal mutant CUL1. Subcellular localization of both transfected ROC1 and CUL1 protein was examined by indirect immunofluorescence with either α-HA or α-Myc antibody. Cell nuclei were stained with 4′,6′-diamidino-2-phenylindole. Note that overexpression of ROC1 is unable to promote nuclear accumulation of C-terminal mutant CUL1.
Requirement of both ROC1 and C-terminal sequence for nuclear accumulation of CUL1.
The finding that mutation of CUL1 either in the ROC1 binding region or in the C-terminal sequence impaired CUL1 nuclear accumulation led us to determine whether these two signals are functionally redundant or collaborative. In a small fraction of transfected cells expressing very high levels of cullins, we noticed a cytoplasmic staining that was more intense than nuclear staining (data not shown). In cells cotransfected with ROC1, however, CUL1 was mainly localized to the nucleus even when they were expressed at high levels (Fig. 5B). These observations are consistent with the finding that ROC1 promotes, and may even act as a rate-limiting factor for, nuclear accumulation of cullins. When the C-terminal mutant CUL1 was coexpressed with ROC1, however, it remained localized predominantly in the cytoplasm despite high levels of ROC1 expression (Fig. 5B). In most cells examined, we noticed that nuclear staining of HA-ROC1 appeared to be lower in cells expressing C-terminally mutated cullins than in cells expressing wild-type cullins, suggesting that mutations in the C-terminal sequence of cullins not only impaired their nuclear accumulation but also resulted in a cytoplasmic retention of their associated ROC1. These results, together with the observation that the mutant CUL1Δ610–615 that is deficient in ROC1 binding is impaired from entering the nucleus despite containing an intact C-terminal sequence (Fig. 5A), suggest that the cis-acting C-terminal sequence and trans-acting ROC1 binding are both needed for nuclear accumulation of cullin.
ROC1 promotes NEDD8 modification of CUL1 in vivo.
The finding that ROC1 promotes cullin nuclear accumulation led us to determine the role of ROC1 in regulating NEDD8 modification of cullins. HA-tagged wild-type or mutant CUL1 was either singly transfected or cotransfected with Myc-ROC1 into 293T cells. Similar levels of Myc-ROC1 protein were expressed in each transfection, as verified by α-Myc IP-Western blotting (Fig. 6A, bottom panel). Modification of transfected HA-CUL1 by NEDD8 was determined by immunoprecipitation with α-HA antibody followed by immunoblotting with either α-HA or α-NEDD8 antibody. Cotransfection with ROC1 increased the levels of NEDD8-conjugated CUL1 (Fig. 6A, compare lanes 2 and 3), indicating that ROC1 can promote modification of CUL1 by NEDD8 in vivo. More intense NEDD8-conjugated CUL1 was seen with the α-HA antibody than with the α-NEDD8 antibody due to the higher affinity of the HA antibody. Mutation in the ROC1 binding region (HA- CUL1Δ610–615) substantially reduced the levels of NEDD8-conjugated CUL1 (Fig. 6A, compare lanes 2 and 4). Consistent with residual ROC1 binding activity in the CUL1Δ610–615 mutant (Fig. 4B), overexpression of ROC1 also exhibited a promoting effect on NEDD8 modification of CUL1Δ610–615, but the level of NEDD8-conjugated CUL1Δ610–615 is substantially lower than that of NEDD8-conjugated wild-type CUL1 (Fig. 6A, compare lanes 3 and 5). Similarly, overexpression of ROC1 also increased NEDD8 modification of the CUL1 C-terminal mutant (CUL1L756A/I757A) (Fig. 6A, compare lanes 6 and 7), but the level of NEDD8-conjugated CUL1L756A/I757A is greatly reduced from that of wild-type CUL1, both with or without coexpression of ROC1 (Fig. 6A, compare lanes 6 and 7 with lanes 2 and 3). NEDD8 modification was completely abolished by the mutation at Lys720 (CUL1K720A) and was not detectably restored by the overexpression of ROC1 (lanes 8 and 9). These results demonstrate that ROC1 promotes NEDD8 modification in vivo and are consistent with a recent report that RUB1 or NEDD8 modification of both yeast CDC53 and human CUL2 in yeast or insect cells is dependent on the presence of ROC1 (16).
In vitro NEDD8 modification requires the CUL1 C-terminal sequence but not ROC1.
Mutations both in the CUL1 C-terminal sequence and in the ROC1 binding region impaired nuclear accumulation of CUL1 and decreased modification of CUL1 by NEDD8, suggesting a link between CUL1 nuclear accumulation of NEDD8 modification. These findings also raised the question of whether the C-terminal sequence and ROC1 are involved in two apparently separate events: promoting CUL1 nuclear accumulation and promoting NEDD8 modification. To address this question, we examined in vitro NEDD8 modification of wild-type and mutant CUL1 in rabbit reticulocyte lysate. Consistent with previous reports (23, 34), in vitro synthesized wild-type HA-CUL1 migrates as two forms: a fast-migrating major band corresponding to unmodified CUL1 and a slow-migrating minor form corresponding to NEDD8-conjugated CUL1 (Fig. 6B, lane 2). Mutation at Lys720 abolished the slow-migrating form without affecting the fast-migrating band (lane 5), confirming the NEDD8 modification in this assay. Mutation in the C-terminal sequence of CUL1 (L756A/I757A) greatly reduced levels of NEDD8-conjugated CUL1 (lane 4), indicating that the C-terminal sequence is required for both CUL1 nuclear accumulation and NEDD8 modification.
In contrast, mutations in the ROC1 binding region, both CUL1Δ610–615 (Fig. 6B, lane 6) and CUL1Δ606–624 (lane 7), had no detectable effect on NEDD8 modification, nor did addition of ROC1 have any visible effect on the levels of CUL1-NEDD8 conjugates (lane 3). Introduction of a mutation at the NEDD8 conjugation site, Lys720, into the ROC1-binding mutant CUL1Δ606–624 (CUL1Δ606–624/K720A) completely abolished modification by NEDD8 (lane 8), confirming NEDD8 modification on the CUL1Δ606–624 mutant. To further confirm ROC1-independent NEDD8 modification of CUL1 in vitro, we examined in vitro NEDD8 modification of a deletion mutant CUL1 containing only the C-terminal 149 residues (CUL1C149), which exhibits undetectable ROC1 binding activity (Fig. 6C). In vitro-translated HA-CUL1C149 migrates as two bands, a major 22-kDa form and a less intense 30-kDa form (Fig. 6B, lane 9). Introduction of the K720A (HA-CUL1C149/K720A) mutation abolished the 30-kDa form without affecting the 22-kDa band, indicating that the 30-kDa form corresponded to NEDD8-modified HA-CUL1C149 (Fig. 6B, lane 10).
In vitro-translated CUL1 was also modified by similarly in vitro-translated NEDD8, as seen by the appearance of a third slow-migrating HA-CUL1 band after mixing of in vitro-translated Myc3-NEDD8 with wild-type HA-CUL1 (Fig. 6B, lane 11) but not with the HA-CUL1K720A mutant (lane 12). In vitro-translated Myc3-NEDD8 was efficiently conjugated with the ROC1-binding-deficient HA-CUL1Δ606–624 mutant (Fig. 6B, lane 14) but not with a double HA-CUL1Δ606–624/K720A mutant (lane 15), further confirming ROC1-independent NEDD8 modification of CUL1 in vitro. As in the maturation and conjugation of ubiquitin, the C terminus of newly synthesized human NEDD8 contains five additional residues, Gly-Gly-Leu-Arg-Glu, following the conserved Gly76, that are cleaved during maturation in order to expose the Gly76 for conjugation. It was shown previously that Gly76 of NEDD8 is necessary for conjugation to other proteins (15). Mutating NEDD8 Gly76 to alanine (Myc3-NEDD8G76A) completely abolished the conjugation of NEDD8 with both wild-type CUL1 (lane 13) and the ROC1-binding-deficient CUL1Δ606–624 mutant (lane 16) without any detectable effect on the conjugation of both CUL1 proteins with the wild-type NEDD8 present in the reticulocyte lysate. From these results, we conclude that ROC1 is not required for NEDD8 modification in vitro.
NEDD8 modification is required for efficient CUL1 ubiquitin ligase activity.
To determine the functional consequence of CUL1 nuclear accumulation and NEDD8 modification, we examined in vitro ubiquitin ligase activity of mutant CUL1 mutated in the C-terminal sequence, in the NEDD8 modification site, and in ROC1 binding using IκBα as a substrate. HA-tagged wild-type or mutant CUL1 was cotransfected with SKP1 and the IκBα-targeting F box protein β-TrCP. The ubiquitin ligase activity of CUL1 was precipitated with an α-HA antibody and assayed using phosphorylated IκBα as a substrate. As previously reported (33, 45), the wild-type CUL1 immunocomplex exhibited high levels of IκBα ubiquitination activity that is dependent on both E1 and E2-Ubc5 (Fig. 7A, lanes 1 to 3). Mutations in the C-terminal sequence (L756A/I757A) (lane 5), at the NEDD8 modification site of CUL1 (K720A) (lane 4), or their combination (K/LI) (lane 6) substantially reduced the ubiquitin ligase activity of CUL1. A substantial decrease, but not complete abolition, of ligase activity by the disruption of NEDD8 modification indicates that NEDD8 modification is important, but not absolutely required, for CUL1 ubiquitin ligase activity. A mutation in the ROC1 binding region (Δ610–615) almost completely abrogated the ligase activity of CUL1 (Fig. 7A, lane 7). The more disruptive effect on the ligase activity by the mutation in the ROC1 binding sequence than by the mutation at the NEDD8 modification site indicates that in addition to promoting nuclear accumulation and NEDD8 modification, ROC1 also has an additional function in activating CUL1 ligase activity.
To further confirm the reduction of ubiquitin ligase activity by the mutations affecting CUL1's nuclear accumulation and modification by NEDD8, we established a condition to assay in vivo IκBα ubiquitination by CUL1 and determined the in vivo IκBα ubiquitin ligase activity of wild-type and mutant CUL1. FLAG-IκBα was cotransfected with a constitutively active mutant IκBα kinase (IKKβS177E/S181E), β-TrCP, ROC1 and SKP1, CUL1, and HA-tagged ubiquitin. Twenty hours after transfection, cells were treated with the proteasome inhibitor MG132, lysed in a preboiled SDS (1%) lysis buffer, and boiled for another 10 min. Whole cell lysate was then diluted with NP-40 buffer to reach a final concentration of 0.1% SDS, immunoprecipitated with α-FLAG antibody, resolved by SDS-PAGE, and immunoblotted with α-HA or with α-FLAG antibodies. A high-molecular-weight smear characteristic of ubiquitination was detected on both immunoblots (Fig. 7B, lane 5) but was not detected when HA-Ub (lane 2) or FLAG-IκBα (lane 3) was omitted from transfection, indicating that the high-molecular-weight smear corresponds to in vivo ubiquitinated IκBα. The appearance of ubiquitinated IκBα was not seen when CUL1, β-TrCP, ROC1, and SKP1 were omitted (lane 1), confirming an SCFβ-TrCP-dependent ubiquitination (44, 48, 49). Dropout of IKKβ from transfection substantially reduced the level of phosphorylated IκBα (Fig. 7B, right panel, compare lane 4 with lanes 2 and 5 to 7) and the level of ubiquitination of IκBα (lane 4), confirming the dependency of IκBα ubiquitination on its phosphorylation by IKKβ (28, 53). A low level of IκBα ubiquitination was still visible when IKKβ was omitted from transfection, which is probably attributable to a residual level of endogenous IKKβ. The levels of ectopically expressed SKP1, ROC1, and CUL1, the reduction of NEDD8 modification by the L756A/I757A mutation, and the disruption of NEDD8 modification by the K720A mutation were confirmed by sequential immunoprecipitation and immunoblotting (Fig. 7C).
Consistent with the in vitro assay, the in vivo IκBα ubiquitin ligase activity of CUL1 was reduced by both the L756A/I757A (Fig. 7B, lane 6) and the K720A (lane 7) mutations. In contrast to the in vitro assay in which the CUL1L756A/I757A and the CUL1K720A mutants exhibited similarly reduced levels of ligase activity, we have consistently observed a more pronounced effect of the K720A mutation than of the L756A/I757A mutation on the in vivo IκBα ubiquitin ligase activity of CUL1. In fact, under this in vivo assay condition, the level of ubiquitinated IκBα with NEDD8-deficient CUL1K720A is close to that seen in transfection without the IKKβ kinase (comparing lanes 4 and 7). The basis for this difference between in vivo and in vitro assays has not been determined, but it could be caused by the higher levels of the substrate (phosphorylated IκBα) in vivo than in vitro. This observation also suggests that the in vivo assay may be more sensitive than the in vitro assay in detecting the change of CUL1 ubiquitin ligase activity.
Modification by NEDD8 is not required for the assembly of the SCF complex.
Diminishing of CUL1 ubiquitin ligase activity by mutations disrupting NEDD8 modification led us to ask whether a deficiency in NEDD8 modification impairs the assembly of the SCF complex or impedes interaction of ligase with the substrate. HA-tagged wild-type or mutant CUL1 was cotransfected with SKP1, ROC1, β-TrCP, and IκBα. Interaction of CUL1 with SKP1 and IκBα was examined by IP-Western blotting (Fig. 7D). The wild type and both the L756A/I757A and K720A mutants of CUL1 coimmunoprecipitated similar amounts of SKP1. SKP1 bound to both NEDD8-modified and unmodified forms of CUL1. The ratio of these two forms of CUL1 present in SKP1 immunocomplexes was similar to that seen in CUL1 immunocomplexes, suggesting that SKP1 does not preferentially associate with either form of CUL1. Consistently, SKP1 immunocomplexes derived from cells transfected with wild-type and mutant CUL1 contained similar amounts of CUL1. These results indicate that disruption of NEDD8 modification did not affect CUL1-SKP1 interaction. Likewise, the wild type and both the L756A/I757A and K720A mutants of CUL1 coimmunoprecipitated similar amounts of IκBα, and reciprocally, IκBα immunocomplexes contained perhaps even higher levels of mutant CUL1K720A than wild-type CUL1. These results indicate that a deficiency of NEDD8 modification did not affect the assembly of SCF complexes, nor did it impede the interaction of SCF complexes with the substrate.
DISCUSSION
In this report we present evidence suggesting a novel pathway for regulation of CUL1 ubiquitin ligase—ROC1 and the CUL1 C-terminal sequence mediate CUL1 nuclear accumulation, promoting CUL1 modification by NEDD8 and ligase activation (Fig. 8). This pathway contains four previously unrecognized features. First, the CUL1 C-terminal sequence is required for both nuclear accumulation and NEDD8 modification. Second, ROC1 binding per se is not required for CUL1 modification by NEDD8, but it indirectly promotes NEDD8 modification by promoting CUL1 nuclear accumulation. Third, CUL1 nuclear accumulation facilitates its modification by NEDD8. Last, NEDD8 modification is important for efficient CUL1 ubiquitin ligase activity in vivo.
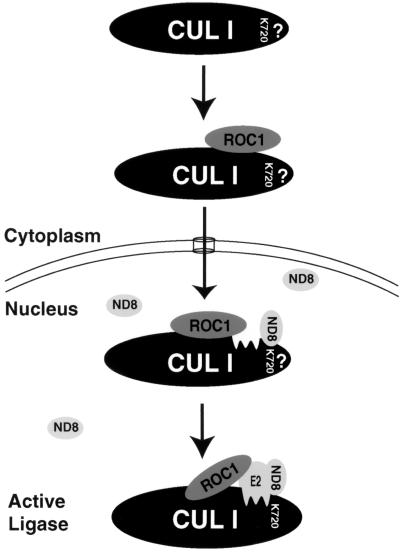
FIG. 8.
A model for cullin ubiquitin ligase activation. Cytoplasmically located CUL1 associates with ROC1 and is promoted to enter into and accumulate in the nucleus. CUL1 nuclear accumulation also requires a C-terminal sequence in CUL1 through an unknown mechanism (represented by a question mark). Once inside the nucleus, CUL1 becomes covalently modified with NEDD8 (ND8), resulting in efficient CUL1 ubiquitin ligase activity. NEDD8 modification is presumed to induce a conformational change in CUL1 to facilitate or stabilize CUL1-ROC1-E2 binding.
Dual function of the CUL1 C-terminal sequence in nuclear accumulation and NEDD8 modification.
The site for NEDD8 modification has been mapped to Lys689 in human CUL2 (46), corresponding to Lys720 in human CUL1 or Lys760 in yeast CDC53. Deletion of the C-terminal 21 residues (794 to 814) from yeast CDC53, downstream of Lys760, abrogates RUB1 modification (22). In agreement with these reports, we have confirmed that a mutation at Lys720 of human CUL1 abolished its NEDD8 modification (Fig. 2A) and found that deletion of or mutation in the C-terminal 22 residues (Fig. 1, 2, and 6) of human CUL1 severely reduced NEDD8 modification. Hence, the very C-terminal sequence does not contain the site of, but may play a regulatory role for, NEDD8 modification of CUL1 or CDC53.
Unexpectedly, we found that deletion of the C-terminal 22 residues or mutations at two conserved amino acids in this 22-residue region impaired nuclear accumulation of CUL1. The mechanism underlying the function of the C-terminal sequence in mediating CUL1 nuclear accumulation is unknown at present. This sequence has not been determined to be involved in the interaction with any known cullin-interacting proteins, such as SKP1, ROC1, or CDC34. The C-terminal sequences of different cullins are highly conserved and do not resemble the classical nuclear localization signal (NLS), which is characterized by a cluster of basic residues, as exemplified by the NLS of the SV40 large T antigen (PKKKRKV). The C-terminal 22-amino-acid sequence of CUL1 has an acidic pI of 4.2 and contains only three scattered basic residues, K759, R764, and a nonconserved K769. On the other hand, there are eight hydrophobic residues that are highly conserved in other cullins (Fig. 1A), and mutation of two of these conserved hydrophobic residues blocks nuclear accumulation of CUL1 and three other cullins examined (data not shown). Although it is formally possible that the cullin C-terminal sequence contains a nonclassical autonomous NLS to mediate import of cullin into the nucleus, this seems to be unlikely, given that mutant CUL1s defective in ROC1 binding, while retaining the intact C-terminal sequence, are impeded from nuclear accumulation.
Blocking nuclear accumulation of CUL1, by mutations either in the C-terminal region of CUL1 or in the ROC1 binding region, is associated with defective NEDD8 modification of CUL1. Although blocking of CUL1 nuclear accumulation could sufficiently explain the severe reduction of NEDD8 modification by the deletion of or mutation in the C-terminal sequence, our in vitro assay suggests that the CUL1 C-terminal sequence additionally contains another yet-to-be-identified activity required for NEDD8 modification. Mutations in the C-terminal sequence of CUL1 substantially reduced its conjugation by NEDD8 in vitro in a rabbit reticulocyte lysate (Fig. 6B). That the mutation at the NEDD8 modification site (CUL1K720A) has no detectable effect on CUL1 nuclear accumulation argues against the possibility that NEDD8 modification plays any significant role in CUL1 nuclear accumulation. We speculate that a NEDD8 conjugation activity binds to the C-terminal sequence of CUL1 and may act to retain CUL1 in the nucleus. Mutation in the CUL1 C-terminal sequence, but not the mutation at the NEDD8 modification site, disrupts this binding, resulting in a defect in both NEDD8 modification and nuclear retention. Clearly, the function of the cullin C-terminal sequence, including the binding with additional yet-to-be-identified cullin-interacting protein(s), needs to be further investigated.
ROC1 promotes NEDD8 modification by facilitating nuclear accumulation of CUL1.
Overexpression of ROC1 increased the levels of NEDD8-conjugated CUL1, and conversely, mutations in the ROC1 binding region substantially reduced the levels of NEDD8-CUL1 conjugates (Fig. 6A). These results indicate a function of ROC1 in promoting NEDD8 modification of CUL1 and are consistent with and provide additional support for a recent report that reached a similar conclusion (16). Kamura et al. demonstrated that in vitro conjugation of yeast CDC53 with yeast glutathione S-transferase-RUB1 depends on the coexpression of Rbx1 (ROC1) with CDC53. The mechanism by which ROC1 promotes RUB1 or NEDD8 modification of cullins was unclear, but it was suggested that ROC1 may recruit the RUB1-, NEDD8-conjugating enzyme E2-Ubc12 to cullins or induce conformational changes in cullins to allow the access of Ubc12 to the RUB1 or NEDD8 modification site (16). Our results suggest that ROC1 promotes NEDD8 modification of CUL1 by promoting CUL1 nuclear accumulation, but ROC1 per se is not required for NEDD8 modification. Four lines of evidence collaboratively support this conclusion. First, mutation in the CUL1 C-terminal sequence that impaired its nuclear accumulation had no detectable effect on CUL1-ROC1 association (Fig. 4C), suggesting that ROC1 can form a complex with CUL1 in the cytoplasm. Second, overexpression of ROC1 increased CUL1 nuclear accumulation and NEDD8 modification (Fig. 6A). Third, mutations in CUL1 disrupting ROC1 binding resulted in a decreased nuclear accumulation and NEDD8 modification of CUL1. Last, in vitro modification of CUL1 by NEDD8 was not affected by the addition of ROC1 or by mutations in CUL1 disrupting ROC1 binding (Fig. 6B) and can occur efficiently using a 149-residue C-terminal CUL1 fragment devoid of ROC1 binding capability (Fig. 6B and C). From these results, we conclude that ROC1 is not required for NEDD8 modification in vitro.
How ROC1 promotes CUL1 nuclear accumulation is not clear. Nuclear accumulation of a protein is the result of several regulated processes, including nuclear import, retention, and export. There is the possibility that ROC1 could bind to CUL1 in the cytoplasm and escort CUL1 into the nucleus. Alternatively, ROC1 could act as a nuclear retention factor for nuclear localization of CUL1. ROC1 alone, however, is not sufficient to localize CUL1 in the nucleus. Nuclear accumulation of mutant CUL1 that is defective in the C-terminal sequence, while retaining the intact ROC1 binding activity, is impaired (Fig. 3) and cannot be restored by the overexpression of ROC1 (Fig. 5B). These observations suggest that nuclear accumulation of cullin involves at least two separate signals: a cis-acting C-terminal sequence and a trans-acting ROC1 factor. Whether these two signals function collaboratively (e.g., one promotes nuclear import and one functions in nuclear retention) or in a regulatory manner (e.g., ROC1 may facilitate the binding of an importin receptor to the C-terminal sequence of CUL1) remains to be determined. It also should be pointed out that our results do not suggest that promoting nuclear accumulation and NEDD8 modification of cullin are the main functions of ROC1 as an essential subunit of cullin ligases. This is made evident by the observation that mutant CUL1 that is defective in ROC1 binding (CUL1Δ610–615) loses virtually all its IκBα ubiquitin ligase activity, whereas mutant CUL1 that is defective in nuclear accumulation and/or NEDD8 modification retains a considerably higher amount of ligase activity (Fig. 7).
Regulation of cullin ligase activity by nuclear localization and NEDD8 modification.
Deficiency in NEDD8 modification results in a significant reduction of CUL1 ubiquitin ligase activity (32, 35, 37, 39) (Fig. 7). It is not entirely clear how nuclear accumulation of CUL1 facilitates its modification by NEDD8 and how NEDD8 modification results in activation of CUL1 ubiquitin ligase. Both the NEDD8-activating E1 protein AXR1 (38) and NEDD8 itself (15) are localized primarily in the nucleus, suggesting the possibility that nuclear accumulation may provide CUL1 with access to a NEDD8 conjugating activity. ROC1 promotes association of CDC34 with CDC53/CUL1 when coexpressed in insect cells (43), and CDC34 is localized predominantly in the nucleus (Y.Z. and Y.X, unpublished observation), suggesting the possibility that ROC1-promoted nuclear accumulation and NEDD8 modification may facilitate or stabilize the binding of E2 with CUL1 in the nucleus, leading to the assembly of active CUL1 ubiquitin ligase. The requirement of nuclear accumulation and NEDD8 modification for optimal CUL1 ligase activity toward IκBα, a cytoplasmic protein, argues against the idea that nuclear accumulation and NEDD8 modification play a role in substrate targeting. We suggest that the assembly of active CUL1 ligase is not coupled with and is likely separated from the access to its substrate. Our results demonstrate that activation of CUL1 ligase requires a nuclear event and implies a requirement for, and potentially a regulatory step associated with, nuclear export of activated CUL1 ubiquitin ligase.
Notably, three elements involved in the regulation of CUL1 nuclear localization and ligase activation—ROC1 binding, NEDD8 modification, and the requirement of the C-terminal sequence—are shared by or highly conserved in most, if not all, cullins. We suggest that this pathway may be commonly utilized in the assembly and activation of other cullin ligases.
ACKNOWLEDGMENTS
We thank Al Baldwin for providing the GST-IκBα expression vector, Zhen-Qiang Pan for providing the HA-IKKβ kinase expression vector, and Jen Michel for helpful discussion during the course of this work and critical reading of the manuscript.
T.O. is supported in part by the First Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan. Y.X. is a recipient of the American Cancer Society Junior Faculty Award and is a Pew Scholar in Biomedical Science. This study was supported by Public Health Service grants CA65572 and CA68377 to Y.X.
REFERENCES
- 1.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Deshaies R J. SCF and cullin/RING H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 3.Desterro J M, Rodriguez M S, Hay R T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 4.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 5.Gostissa M, Hengstermann A, Fogal V, Snady P, Schwarz S E, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6467. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershko A. Role of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 7.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 8.Hochstrasser M. There's the Rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 9.Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 10.Huibregtse J M, Schneffner M, Beaudenon S, Howley P M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins C W, Xiong Y. Immunoprecipitation and immunoblotting in cell cycle studies. In: Pagano M, editor. Cell cycle: material and methods. New York, N.Y: Springer-Verlag; 1995. pp. 250–263. [Google Scholar]
- 12.Jentsch S. The ubiquitin-conjugating system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 13.Jentsch S, Ulrich H D. Ubiquitous deja vu. Nature. 1998;395:321–323. doi: 10.1038/26335. [DOI] [PubMed] [Google Scholar]
- 14.Joazeiro C A P, Wing S S, Huang H-K, Leverson J D, Hunter T, Liu Y-C. The tyrosine kinase negative regulator c-Cbl as a RING-type E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 15.Kamitani T, Kito K, Nguyen H P, Yeh E T H. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 16.Kamura T, Conrad M N, Yan Q, Conaway R C, Conaway J W. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, Harper J W, Conaway J W. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 18.King R W, Deshaies R J, Peters J-M, Kirscher M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 19.Kipreos E T, Lander L E, Wing J P, He W-W, Hedgecock E M. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 20.Koepp D M, Harper J W, Elledge S J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Yoshida Y, Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- 22.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to van Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci USA. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeb K R, Hass A L. Conjugates of ubiquitin cross-reactive protein distribute in a cytoskeletal pattern. Mol Cell Biol. 1994;14:8408–8419. doi: 10.1128/mcb.14.12.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to the nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis T. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 29.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel J, Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:439–445. [PubMed] [Google Scholar]
- 32.Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCFSKP2 toward p27Kip1. Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- 33.Ohta T, Michel J J, Schottelius A J, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 34.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD-ligating system for cullin 4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osaka F, Saeki M, Katayama S, Aida N, Toh-E A, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, Kato S. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't skp the F-box hypothesis. Trends Genet. 1998;768:236. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 37.Podust V N, Bronell J E, Glasheva T B, Luo R S, Wang C, Coggins M B, Pierce J W, Lightcap E S, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pozo J C, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- 39.Read M A, Brownell J E, Gladysheva T B, Hottelet M, Parent L A, Coggins M B, Pierce J W, Podust V N, Luo R S, Chau V, Palombella V J. Nedd8 modification of cul-1 activates SCFβ-TrCP-dependent ubiquitination of IκBα. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seol J H, Feldman R M R, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skowyra D, Craig K, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 43.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 44.Spencer E, Jiang J, Chen Z J. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan P, Fuches S Y, Angus A, Wu K, Gomez C, Ronai Z, Pan Z-Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 46.Wada H, Yeh E T, Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- 47.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 48.Winston J T, Strack P, Beer-Romero P, Chu C Y, Elledge S J, Harper J W. The SCFβ-TrCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning A M, Andersen J S, Mann M, Mercurio F, Bem-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 50.Yu H, Peters J-M, King R W, Page A M, Hieter P, Kirschner M W. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 51.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 52.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J R, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 53.Zandi E, Karin M. Bridging the gap: composition, regulation, and physiological function of the IκBα kinase complex. Mol Cell Biol. 1999;19:4547–4551. doi: 10.1128/mcb.19.7.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]