Abstract
The Drosophila melanogaster suppressor of sable gene, su(s), encodes a novel, 150-kDa nuclear RNA binding protein, SU(S), that negatively regulates RNA accumulation from mutant alleles of other genes that have transposon insertions in the 5′ transcribed region. In this study, we delineated the RNA binding domain of SU(S) and evaluated its relevance to SU(S) function in vivo. As a result, we have defined two arginine-rich motifs (ARM1 and ARM2) that mediate the RNA binding activity of SU(S). ARM1 is required for in vitro high-affinity binding of SU(S) to small RNAs that were previously isolated by SELEX (binding site selection assay) and that contain a common consensus sequence. ARM1 is also required for the association of SU(S) with larval polytene chromosomes in vivo. ARM2 promotes binding of SU(S) to SELEX RNAs that lack the consensus sequence and apparently is neither necessary nor sufficient for the stable polytene chromosome association of SU(S). Use of the GAL4/UAS system to drive ectopic expression of su(s) cDNA transgenes revealed two previously unknown properties of SU(S). First, overexpression of SU(S) is lethal. Second, SU(S) negatively regulates expression of su(s) intronless cDNA transgenes, and the ARMs are required for this effect. Considering these and previous results, we propose that SU(S) binds to the 5′ region of nascent transcripts and inhibits RNA production in a manner that can be overcome by splicing complex assembly.
Eukaryotic protein-coding RNAs are typically transcribed as larger pre-mRNAs that are processed to a mature form. Pre-mRNA processing is coupled to transcription (7, 8, 27, 42) and involves a complex set of events including the addition of a 7-methylguanosine cap to the 5′ end, splicing to remove internal introns, and cleavage/polyadenylation of the 3′ end. Interactions between the cellular transcription and RNA processing apparatuses and between RNA processing components that assemble at various sites on the pre-mRNA are thought to facilitate the efficient production of mRNAs that are suitable substrates for translation. Incorrectly processed transcripts can be recognized as such and degraded (16, 24, 26, 40).
The Drosophila melanogaster suppressor of sable gene, su(s), encodes a protein involved in nuclear pre-mRNA metabolism. Loss-of-function su(s) mutations either suppress or enhance specific mutant alleles of a variety of unlinked genes (49). Some su(s) mutants also exhibit defects in viability and male fertility (52). Although both the su(s) gene and mutant alleles affected by su(s) have been cloned and characterized, the function of the su(s) gene product, SU(S), has been somewhat elusive. The enhanced alleles are associated with large, complex genes that cannot easily be analyzed in detail at a molecular level. More is known about the suppressed alleles, through molecular studies of vermilion (v), yellow (y), and purple (pr) (20–22, 29). The su(s)-suppressible mutations have transposon insertions near the 5′ end of the transcribed region that interrupt either the first exon or the first intron. These mutant genes produce a reduced level of RNA in su(s)+ flies, and RNA levels are elevated in su(s) mutants. The RNAs generated initiate at the normal transcription start site of the genes. Antisense transposon sequences are incorporated into the pre-mRNA and can be removed during splicing. In the case of v and y, transposon sequences are removed inefficiently by splicing at cryptic splice sites near the ends of the inserted sequences (20, 22). Previous work from this lab demonstrated that the improvement of a cryptic 5′ splice site near the beginning of a mutant v transgene to a consensus site increased RNA production from a mutant v transgene, without improving the splicing efficiency. This change also eliminated the inhibition of RNA production by SU(S) (21). While these and other studies have established a connection between SU(S)-mediated regulation of RNA levels and the efficiency of splicing complex assembly in the 5′ region of the pre-mRNA (21, 29), the mechanism by which SU(S) regulates accumulation of these RNAs has not been established. Thus, it is unclear whether SU(S) directly regulates transcription, splicing complex assembly, or pre-mRNA stability.
SU(S) is a novel, 150-kDa nuclear protein. The initial sequence analysis (51) defined two regions of SU(S) with similarity to structural motifs found in RNA processing proteins, a highly charged region in the N-terminal portion of SU(S) and an RNA recognition motif-like motif in the C-terminal region. The importance of these regions to SU(S) function has not been determined. Subsequently, two tandem copies of a CCCH zinc binding domain (57) of unknown function were identified within SU(S) (43). This motif is also found in several other eukaryotic proteins, including the Caenorhabditis elegans transcriptional repressor PIE-1 (6, 43), the mRNA destabilization protein TTP/Nup475/TIS11 (12, 32), the 35-kDa subunit of the splicing factor U2AF (43, 48), and the 30-kDa subunit of the polyadenylation factor CPSF (3–5).
Recombinant SU(S), expressed in baculovirus, binds to RNA in vitro; using a PCR-based binding site selection assay (SELEX), we isolated high-affinity RNA substrates (45). In this paper, we have delineated the RNA binding domain of SU(S) and examined its role in SU(S) function in vivo. Based on the results presented here and previous studies, we propose that SU(S) binds to the 5′ region of the nascent transcripts via arginine-rich RNA binding motifs (ARMs) and inhibits RNA production in a way that can be overcome by splicing complex assembly in the 5′ region.
MATERIALS AND METHODS
Generation and analysis of MBP-SU(S) fusion derivatives.
In mapping the RNA binding regions, a 1,084-bp ClaI-ScaI su(s) cDNA fragment that encodes the first 360 amino acids of SU(S) was used to generate smaller fragments by digestion with either appropriate restriction enzymes or exonuclease III. Subsequently, these fragments were cloned into the maltose binding protein (MBP) expression vector pPR997 (New England Biolabs). Deletions of sequences encoding the two ARMs of SU(S) were introduced using overlapping PCR mutagenesis (25). Unique XbaI and HindIII restriction sites were introduced at the site of the ARM1 and ARM2 deletions, respectively, to facilitate identification of clones containing the desired changes. Convenient restriction sites were used to introduce fragments containing either one or both deletions into pMAL- SU(S)1–434, an MBP-SU(S) fusion encoding amino acids 1 to 434. In addition, small PCR-generated fragments containing the coding region for ARM1 and ARM2 alone were cloned into pPR997. Prior to affinity purification of the fusion proteins as described previously (45), clones were sequenced to ensure that undesirable alterations were not introduced during the PCRs. RNA binding activity of the MBP-SU(S) fusions was measured by nitrocellulose filter binding and Northwestern blot assays as described previously (45). Kds were determined using SigmaPlot (Jandel Scientific). Under the conditions of these experiments, the Kd is equal to the protein concentration that results in 50% of the maximal RNA binding.
Construction of clones for germ line transformations.
An approximately 4-kb XhoI-SpeI cDNA fragment containing the su(s) coding region from either the wild type or ΔARM derivatives was cloned into XhoI-XbaI-digested plasmid pUAST (9). These clones were injected into yw embryos, and germ line transformants were isolated essentially as described by Karess (28). Standard balancer chromosomes were used to establish homozygous stocks of w+ transformant lines and to cross the transgenes into the background of su(s) mutants.
Viability studies.
GAL4-expressing stocks were obtained from the Bloomington Stock Center and crossed to stocks of transformants carrying UAS-su(s) transgenes. The expression patterns of the GAL4 drivers are described in Flybase (http://flybase.bio.indiana.edu). To examine the viability of flies ectopically expressing SU(S) under control of different GAL4 drivers, crosses were set up with a single vial for each transformant. The flies were reared at 18°C. All except one of the crosses involved balanced GAL4 driver stocks, and viability was assessed by determining the proportion of progeny that lacked the balancer chromosome. One stock, 1799, was homozygous for the heat-shock inducible GAL4 driver (hs-GAL4). Thus, all of the progeny would be expected to express the UAS-su(s) transgene. Vials yielding normal numbers of progeny were scored as 100% viable; therefore, this value in this particular cross is only an approximation.
The crosses between transformants carrying either a wild-type su(s) [su(s)wt] or su(s)ΔARM transgene and GAL4 stock 2023 were performed in the following way. Three vials, each containing five pairs of virgin females and males, were set up for each transformant line tested. Crosses were performed at 25 and at 18°C; progeny were collected and scored for 10 and 20 days, respectively.
RNA analysis.
Total RNA was isolated essentially as described previously (15), and polyadenylated RNA was purified from the total RNA preparations by using a Poly ATtract mRNA isolation kit (Promega). Northern analysis was performed as described previously (21). The probe used for Northern analysis was a ClaI/DraI su(s) cDNA fragment from plasmid p15-1, labeled by random priming (2). RNase protection experiments were performed as described in Current Protocols in Molecular Biology (2). Each reaction contained 20 μg of total RNA. The probes used were radioactively labeled, antisense RNAs of an XmaI/BamHI su(s) cDNA fragment containing portions of exons 3 and 4, and rp49 (21) was used as the internal control. The probes were labeled by in vitro transcription as described previously (21).
Protein analysis.
Whole cell extracts were prepared by grinding 25 to 30 mg of frozen adult flies in 100 μl of 1.5× sodium dodecyl sulfate (SDS) sample buffer (6× SDS sample buffer is 0.35 M Tris-Cl [pH 6.8], 10% SDS, 93 mg dithiothreitol per ml, and 30% glycerol) on ice. Samples were boiled for 10 min prior to a 10-min centrifugation at 13,000 × g. The supernatant was assayed for protein content by the Bradford method using the Bio-Rad protocol. Proteins were resolved by SDS-polyacrylamide gel electrophoresis, and Western blots were probed with a 1:1,000 dilution of an affinity-purified polyclonal antibody directed against SU(S) amino acids 42 to 146. Horseradish peroxidase-conjugated goat anti-rabbit antibody (Promega) was used as the secondary antibody, and bands were visualized by enhanced chemiluminescence detection (Amersham) as recommended by the manufacturer.
Immunocytochemistry.
Polytene chromosome squashes of third-instar larval salivary glands were prepared and immunostained as described by Ashburner (1). SU(S) was detected using a 1:400 dilution of affinity-purified polyclonal antibody, directed against SU(S) amino acids 648 to 808 (45), and a 1:50 dilution of goat anti-rabbit antibody conjugated with rhodamine as primary and secondary antibodies, respectively. HRP36 was detected using a 1:600 dilution of mouse monoclonal antibody 5cA5 (a gift from Gideon Dreyfuss) and a 1:50 dilution of goat anti-mouse secondary antibody conjugated with fluorescein isothiocyanate. Samples were examined by confocal microscopy. In double-labeling experiments, individual channel images were pseudocolored and combined using Photoshop software. For the whole mounts, salivary glands were fixed in 4.7% formaldehyde for 5 min, washed with phosphate-buffered saline, and immunostained for SU(S) in essentially the same way as the squashes except that the incubation with primary antibody was longer (overnight instead of 4 h), and a lower dilution (1:500) of the secondary antibody was used. The wild-type larvae were from strain Oregon R. The su(s) null mutant stock was su(s)R39 ras vk. Larvae expressing the su(s) transgenes were from a su(s)R39 ras vk stock with a recombinant third chromosome carrying both the hs-GAL4 driver and a su(s) transgene. The transgenic stocks also contain the third chromosome balancer TM2; thus, larvae were either homozygous or heterozygous for the GAL4 driver and the UAS (upstream activation sequence)-su(s) transgene.
RESULTS
Two ARMs mediate the specific RNA binding activity of SU(S) in vitro.
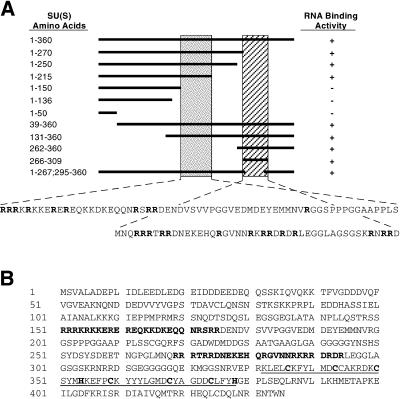
The amino acids responsible for the high-affinity RNA binding map to the N-terminal 360 amino acids of SU(S) (45). To define the RNA binding domain more precisely, we expressed cDNA fragments encoding various portions of this region in Escherichia coli as MBP fusions (see Materials and Methods). We then measured RNA binding activity of the affinity-purified fusion proteins using radioactively labeled 473-nucleotide (nt) ftz RNA as the substrate in either nitrocellulose filter binding or blot overlay binding assays as described previously (45) (Fig. 1A). These experiments defined two nonoverlapping RNA binding regions, amino acids 150 to 215 and amino acids 266 to 309. Examination of the amino acid sequence revealed that each of these regions contains a high proportion of arginine residues (Fig. 1A). This suggested that the RNA binding activity of SU(S) might involve ARMs (11, 56). To determine whether the arginine-rich regions are required for the RNA binding activity of SU(S), PCR mutagenesis was used to create precise 25-amino-acid deletions of sequences encoding residues 151 to 176 (ΔARM1) and 269 to 294 (ΔARM2) (Fig. 1B). The deletions were introduced into an MBP-su(s) cDNA clone encoding the N-terminal 434 amino acids of SU(S), which also includes the two zinc binding motifs (Fig. 1B).
FIG. 1.
(A) Delineation of the SU(S) RNA binding domain. MBP-SU(S) fusion proteins containing different portions of the first 360 amino acids of SU(S) were evaluated for binding to radioactively labeled ftz pre-mRNA by nitrocellulose filter binding or blot overlay assays. Plus symbols indicate binding; minus symbols indicate no binding. The horizontal solid line indicates the region included in each fusion protein. The shaded vertical bars indicate the two RNA binding regions defined from this analysis, and the amino acid sequence of each region is shown beneath the schematic figure. Arginine residues are indicated in bold type. (B) Amino acid sequence of the N-terminal 434 amino acids of SU(S). The amino acids deleted in the SU(S) ΔARM mutants are shown in bold type. The two zinc binding motifs are underlined, and the cysteine (C) and histidine (H) residues that are characteristic of this motif are shown in bold type.
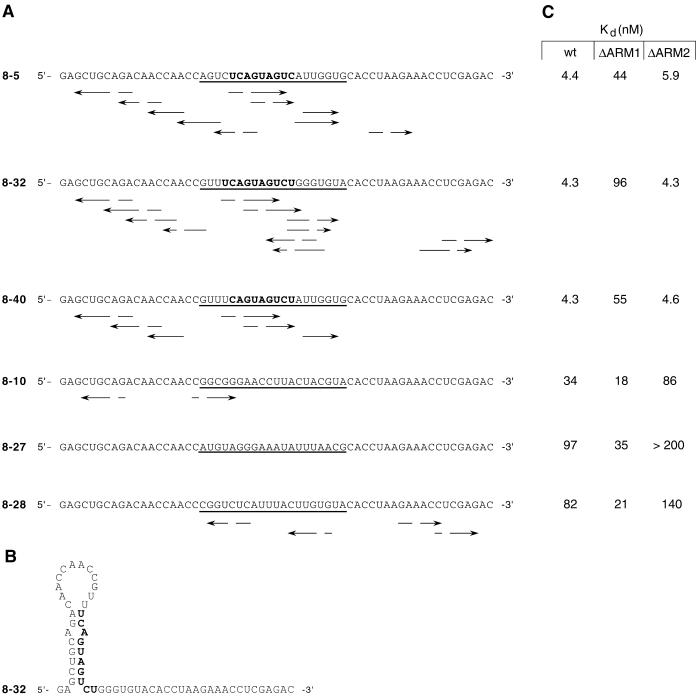
Nitrocellulose filter binding assays were used to measure the affinity of the SU(S)ΔARM fusion proteins for ftz RNA and several small RNAs that were previously isolated by SELEX (45). The small RNAs were isolated after eight rounds of SELEX as high-affinity SU(S) targets from a starting pool of 59-nt RNAs, randomized over the central 20 nucleotide positions and with invariant flanking sequences. Three of the RNAs tested were from the class of SELEX RNAs that contain a close match to the consensus sequence UCAGUAGUCU (consensus RNAs 8-5, 8-32, and 8-40 [Fig. 2A]). Each consensus RNA is capable of forming several possible stem-loops similar in structure, typically containing a mismatched base pair in the stem (Fig. 2A). The SELEX consensus sequence is predicted to base pair with sequences near the 5′ end of the RNA (Fig. 2B). The three other SELEX RNAs tested were also isolated after eight rounds of SELEX but lack the consensus sequence (nonconsensus RNAs 8-10, 8-27, and 8-28 [Fig. 2A]) and are less capable of forming stem-loops than the consensus RNAs. The nonspecific RNA binding activity of the fusion proteins was measured using a pool of 59-nt RNAs that are randomized in the central 20 positions but have the same invariant flanking sequences as the SELEX round 8 RNAs.
FIG. 2.
SELEX RNAs used in the RNA binding assays. (A) Complete nucleotide sequences of RNAs were isolated as high-affinity substrates for SU(S) from a pool of 59-nt RNAs with random sequences in the central 20 positions (round 0) after eight rounds of SELEX (45). The arrows underneath each sequence define regions of potential structures as defined by the GCG Stemloop software. The randomized region of each RNA is defined by underlining. The SELEX consensus sequence is shown in bold type. The consensus RNAs are 8-5, 8-32, and 8-40; the nonconsensus RNAs are 8-10, 8-27, and 8-28. (B) Consensus RNA 8-32 with its SELEX consensus sequence paired in stem-loop structure. The consensus sequence is shown in bold type. (C) RNA binding activities of the MBP-SU(S) fusion proteins measured in nitrocellulose filter binding experiments. Kds shown were determined from the experiments shown in Fig. 3 by using SigmaPlot and are the averages of two to four independent experiments. Under the conditions of these experiments, the Kd is equal to the protein concentration that results in 50% of the maximal RNA binding. In reactions with very low levels of binding over the range of protein concentrations tested, a maximum binding of 60% was assumed in estimating the Kd.
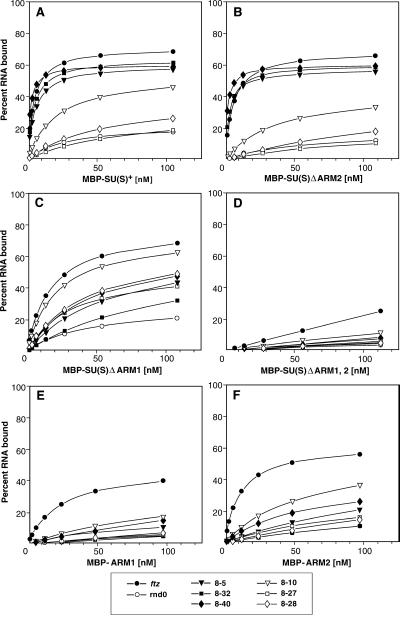
Figures 2C and 3 illustrate the results of RNA binding experiments with these MBP-SU(S) fusion proteins. Whereas full-length recombinant SU(S) purified from baculovirus bound ftz and all six SELEX round 8 RNAs with similar, high affinities (apparent Kd = 2 to 9 nM) (45), the binding properties of the MBP-SU(S) fusion proteins were more complex. The affinity of the wild-type fusion protein [MBP-SU(S)wt] for ftz and the three consensus RNAs (apparent Kd = 4 to 5 nM) was 8- to 22-fold higher than its affinity for the nonconsensus RNAs. Likewise, the fusion protein lacking ARM2 [MBP-SU(S)ΔARM2] bound consensus RNAs with apparent Kds of 4.3 to 5.9 nM and nonconsensus RNAs with 15- to 50-fold-lower affinity. On the other hand, the derivative lacking ARM1 [MBP-SU(S)ΔARM1] exhibited a 10- to 20-fold-lower affinity for consensus RNAs (apparent Kd = 44 to 96 nM) than MBP-SU(S)wt, although it bound ftz and nonconsensus RNAs with Kds ranging between 12 and 35 nM. The affinity of each of the MBP-SU(S) proteins for the randomized RNA pool was greater than 200 nM, and the derivative that lacks both ARMs [MBP-SU(S)ΔARM1,2] did not bind to any of the RNAs tested.
FIG. 3.
RNA binding activity of MBP-SU(S) ARM deletion derivatives. The nitrocellulose filter binding assay was used to analyze the binding of affinity-purified preparations of MBP-SU(S) fusion proteins to various radioactively labeled RNAs. SELEX RNAs 8-5, 8-32, and 8-40 are consensus RNAs; SELEX RNAs 8-10, 8-27, and 8-28 are nonconsensus RNAs. Sequences of the SELEX round 8 RNAs are shown in Fig. 2. Each set of binding curves shows the activity of a different MBP-SU(S) fusion protein. (A) MBP-SU(S)wt, which contains the N-terminal 434 amino acids; (B to D) fusion proteins with deletions of ARM2, ARM1, and both ARM1 and ARM2, respectively; (E and F) derivatives with the 25 amino acids of ARM1 and ARM2, respectively, fused to MBP. Under the conditions of these experiments, the Kd is equal to the protein concentration that results in 50% of maximal RNA binding.
To determine whether either of the arginine-rich regions is sufficient for RNA binding, we generated MBP fusion proteins that contained only the 25-amino-acid arginine-rich segments, ARM1 or ARM2, that were deleted in the experiments just described. MBP-ARM1 bound ftz RNA with an apparent Kd of approximately 40 nM and the SELEX RNAs with Kds of 200 nM or greater (Fig. 3E). MBP-ARM2 bound ftz with a Kd of about 10 nM and the SELEX RNAs with Kds of 60 nM or greater (Fig. 3F). Thus, it appears that amino acids outside the regions defined by the ARM deletions contribute to the high-affinity binding of SU(S) to the SELEX RNAs. Furthermore, these results provide additional confirmation that the ARM-dependent RNA binding activity is not due to electrostatic interactions.
Taken together, these experiments demonstrate that the amino acids within the regions defined by the ARM deletions mediate the in vitro RNA binding activity of SU(S). Whereas either ARM1 or ARM2 is sufficient for high-affinity binding to the larger ftz RNA, the ARM deletions differentially affect binding to the smaller SELEX RNAs. ARM1 is required for high-affinity binding to consensus RNAs, and ARM2 mediates binding to the nonconsensus RNAs. Since both the SU(S) zinc binding motifs are intact in the MBP-SU(S)ΔARM1,2 fusion protein, which does not bind RNA, these motifs, by themselves, do not mediate RNA binding in vitro.
Overexpression of SU(S) can be lethal in vivo.
Having established that the ARM deletions eliminate the in vitro RNA binding activity of SU(S), we wanted to assess the consequences of these alterations on SU(S) function in vivo. For this analysis, a wild-type full-length su(s) cDNA and derivatives with the ARM deletions described above were constructed and introduced into flies by germ line transformation. A suitable su(s) promoter fragment capable of driving expression of a su(s) cDNA had not been defined. Therefore, we ligated the su(s) coding region to a promoter fragment containing UASs that are responsive to the transcriptional activator GAL4 (9). Transformants were isolated under conditions where the UAS-su(s) transgenes were transcriptionally inactive. Subsequently, ectopic expression of the transgenes was activated by performing a genetic cross to introduce a GAL4 transgene (GAL4 driver).
Because initial experiments indicated that ubiquitous expression of a wild-type su(s) transgene was lethal (data not shown), we examined the viability of flies expressing the UAS-su(s)wt transgene under the control of six different GAL4 drivers (Table 1). This analysis was performed in the background of wild-type endogenous su(s). Analysis of the progeny recovered from these crosses demonstrated that different ectopic SU(S) expression patterns are lethal to variable degrees (Table 1). No progeny that carried both UAS-su(s)wt and the ubiquitously expressed e22c-GAL4 driver were recovered. The viability of UAS-su(s)wt transformants containing T80-GAL4, which is ubiquitously expressed in imaginal discs, varied widely, with no T80-GAL4/UAS-su(s)wt progeny being recovered in more than half of the transformant lines tested (data not shown). The variation in the severity of the viability defect most likely reflects position-dependent differences in the expression levels of the UAS-su(s)wt transgene, presumably with higher levels of expression producing more pronounced effects. In contrast to the results obtained with ubiquitously expressed GAL4 drivers, flies carrying hs-GAL4 and UAS-su(s) were recovered at the usual frequency when reared in the absence of heat shock. The other GAL4 drivers (sev-GAL4, 69B-GAL4, and 30A-GAL4), which generate restricted GAL4 expression patterns, produced relatively modest effects on viability. Thus, this analysis clearly indicates that overexpression of SU(S) is lethal. Analysis of survival at different developmental stages did not reveal a clear-cut lethal period; however, all of the developing flies died prior to pupation (data not shown).
TABLE 1.
Viability of flies ectopically expressing UAS-su(s)wt under control of different GAL4 driversa
| Line |
GAL4
|
No. of GAL4/UAS-su(s)wt lines tested | % Viability
|
||
|---|---|---|---|---|---|
| Driver | Expression pattern | Mean | Range | ||
| e22c | e22c-GAL4 | Ubiquitous | 13 | 0 | 0 |
| 2023 | sev-GAL4 | Restricted | 15 | 82 | 38–96 |
| 1799b | hs-GAL4 | Restricted | 5 | 100 | 100 |
| 1878 | T80-GAL4 | Ubiquitous | 10 | 10 | 0–90 |
| 1774 | 69B-GAL4 | Restricted | 14 | 32 | 0–100 |
| 1795 | 30A-GAL4 | Restricted | 6 | 91 | 76–104 |
Transformant lines, homozygous for a UAS-su(s)wt transgene, were crossed to GAL4 driver stocks, and the progeny were reared at 25°C. Multiple transformant lines were tested, and a single vial cross was set up for each transformant line. Viability was determined by calculating the percentage of progeny recovered that carried both transgenes relative to the expected number. Expression of the different GAL4 drivers (Bloomington Stock Center designations) has been characterized primarily in embryos and larval imaginal discs. The terms “ubiquitous” and “restricted” refer to the GAL4 expression pattern at the developmental stages or in tissues where it has been examined.
This cross was performed without heat shock induction. The 1799 stock was homozygous for the hs-GAL4 driver, whereas the other GAL4 driver stocks were heterozygous with a balancer chromosome. Thus, this cross lacks a control group of siblings, and the 100% value in this case is an estimate of viability (see Materials and Methods).
We also examined the viability of flies expressing UAS-su(s)ΔARM single- and double-deletion mutant transgenes. As was seen with the wild-type su(s) transgene, no e22c-GAL4/UAS-su(s)ΔARM progeny were recovered with any of the derivatives tested (data not shown). Thus, the lethality caused by ubiquitous, high expression depends on a region of SU(S) other than, or in addition to, the ARMs. The viability of flies carrying the wild-type and ARM deletion mutants was also examined in the background of the sev-GAL4 driver. Expression of the sev-GAL4 transgene is controlled by a hybrid promoter, consisting of the hsp70 TATA box and the sevenless enhancer. This promoter directs GAL4 expression primarily in eye discs (47). Examination of expression of a UAS-lacZ reporter gene under control of sev-GAL4 showed that this driver directs a low level of expression of a UAS promoter in other larval tissues as well (our unpublished observations). Because the viability defects produced by this driver are relatively subtle, multiple transformant lines were analyzed for each UAS-su(s) transgene. Furthermore, to distinguish defects related to UAS-su(s) expression from those caused by disruption of a gene at the site of the transgene insertion, parallel experiments were performed both at 25 and 18°C. Because GAL4 is less active at lower temperatures, viability defects related to the level of ectopically expressed SU(S) are expected to be more severe at 25 than 18°C (9), whereas defects related to gene disruption at the site of the transgene insertion are unlikely to be affected by temperature.
In these experiments, differences were observed in the viability of flies expressing the su(s)wt and various su(s)ΔARM transgenes (Table 2). Flies expressing su(s)wt were recovered at 19 to 59% of the expected frequency at 25°C and at higher frequencies at 18°C. Thus, consistent with the analysis described earlier, expression of the wild-type transgene reduces viability. On the other hand, two of the three transformant lines expressing su(s)ΔARM1 and all three lines expressing su(s)ΔARM1,2 were recovered at close to the expected frequencies at both temperatures, indicating that these transgenes do not negatively affect viability. The viability defect observed in one UAS-su(s)ΔARM1 transformant line (28A [Table 2]) appeared to be related to disruption of a gene at the site of the UAS-su(s) transgene insertion because flies expressing the transgene were recovered at similar, low frequencies at both temperatures. Two of the three lines expressing su(s)ΔARM2 were recovered at frequencies that were not significantly different from the wild-type level at 25°C. However, all three su(s)ΔARM2-expressing lines were recovered at a lower than expected frequency at 18°C. Taken together, these results indicate that the ARMs mediate one, but not the only, component of the lethal effect of overexpressing SU(S). Furthermore, the lower viability of su(s)ΔARM2-expressing flies at 18°C, the temperature at which less protein should be produced, suggests that deletion of ARM2 increases the detrimental activity of SU(S) at 18°C.
TABLE 2.
Viability of flies expressing UAS-su(s)ΔARM derivativesa
| su(s) transgene | Transformant line |
sev-GAL4/UAS-su(s) progeny
|
|||
|---|---|---|---|---|---|
| % of total
|
% Viability
|
||||
| 25°C | 18°C | 25°C | 18°C | ||
| None | NA | 55.9 (3) | 55.4 (2) | 100 | 100 |
| wt | 18A | 33.1 (12)* | 46.4 (3)*** | 59 | 84 |
| 25A | 20.9 (3)**** | 30.8 (4)*** | 37 | 56 | |
| 27A | 10.7 (8)*** | 30.5 (5)** | 19 | 55 | |
| ΔARM1 | 15B | 48.4 (5)* | 53.8 (1)* | 87 | 97 |
| 19C | 52.6 (1)* | 52.5 (4)* | 94 | 95 | |
| 28A | 35.6 (2)*** | 34.5 (4)*** | 64 | 62 | |
| ΔARM2 | 1A | 33.0 (2)*** | 27.2 (5)*** | 59 | 49 |
| 4E | 50.1 (3)* | 27.2 (7)** | 90 | 49 | |
| 40C | 39.0 (10)* | 31.2 (3)**** | 70 | 56 | |
| ΔARM1,2 | 9B | 49.4 (5)* | 49.4 (6)* | 88 | 89 |
| 10C | 54.5 (3)* | 56.7 (2)* | 97 | 102 | |
| 17A | 45.2 (2)** | 46.0 (7)* | 81 | 83 | |
Females homozygous for a UAS-su(s) transgene on the second chromosome were crossed with heterozygous males carrying a second chromosome insertion of the sev-GAL4 transgene and a second chromosome balancer marked with Cy. The vials were incubated at either 25 or 18°C. Control crosses between yw females from the injection stock, lacking a UAS-su(s) transgene, and sev-GAL4/Cy males were performed in parallel. The percentage of sev-GAL4/UAS-su(s) progeny was determined by calculating the percentage of progeny with straight wings. Viability was measured by determining the percentage of straight-winged progeny obtained from the experimental versus the control cross. Standard deviations are shown in parentheses. P values, calculated by Student's t test, indicate the probability that the number obtained from each experimental cross is the same as number from the control cross: ∗, P > 0.05 (not significantly different from the control); ∗∗, 0.05 > P < 0.01; ∗∗∗, 0.01 > P < 0.001; ∗∗∗∗, P < 0.001. NA, not applicable.
Examination of the eye color of flies carrying the hypomorphic su(s)51c15 allele, the suppressible vk allele, the UAS-su(s)wt cDNA transgene, and the hs-GAL4 driver revealed that the UAS-su(s)wt cDNA transgene did not provide su(s) function sufficient to rescue the v eye color phenotype at 25°C (data not shown). At higher temperatures, heat shock-induced expression of UAS-su(s)wt was lethal. Other GAL4-driven su(s) expression patterns either did not rescue or gave ambiguous results. Thus, this expression system was unsuitable for determining whether the ARMs are required for the function of SU(S) in regulating expression of mutant v alleles. These observations, together with the ectopic expression induced lethality, indicate that the normal regulatory activity of SU(S) is highly dependent on the level and pattern of su(s) expression.
ARM1 is required for the polytene chromosome association of SU(S).
Endogenous SU(S) is found both in the extrachromosomal compartment of the nucleus and at discrete sites on larval salivary gland polytene chromosomes (45). When polytene chromosomes from wild-type flies are stained by indirect immunofluorescence with anti-SU(S) polyclonal antibodies, a strong signal is observed at fewer than 20 chromosomal sites whereas many other sites give a weak signal. Since SU(S) is an RNA binding protein, the chromosomal localization is likely to represent SU(S) binding to nascent RNA transcripts, although it is possible that SU(S) also interacts with one or more chromatin-associated components. To test whether the ARM deletions affect the chromosomal association of SU(S), we used immunofluorescence to examine polytene chromosomes from larvae expressing wild-type and ARM deletion UAS-su(s) transgene derivatives under control of the hs-GAL4 driver, which is expressed in salivary glands even in the absence of heat shock (Fig. 4). This analysis was performed in the background of an endogenous su(s)R39 null mutant, which gives no immunofluorescence signal with the anti-SU(S) antibodies used (Fig. 4B). Polytene chromosomes were simultaneously labeled with polyclonal anti-SU(S) polyclonal antibodies and with a monoclonal antibody (5cA5) that recognizes HRP36, one of the abundant hnRNP proteins in Drosophila. In this experiment, anti-HRP36 was used solely as a positive control for the immunostaining procedure.
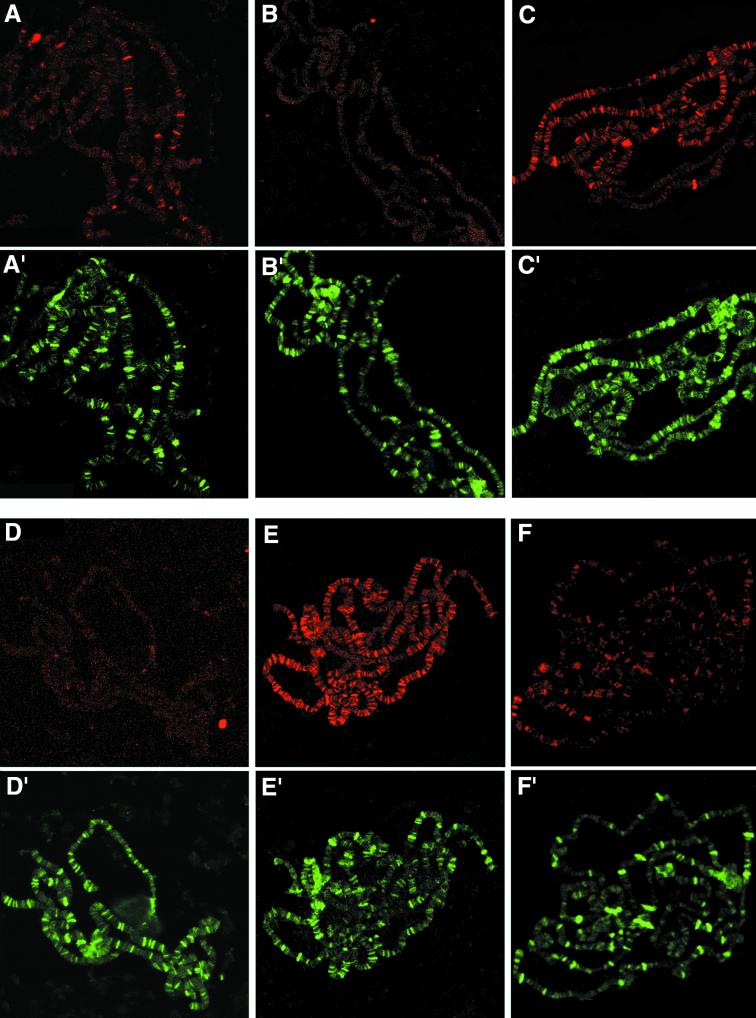
FIG. 4.
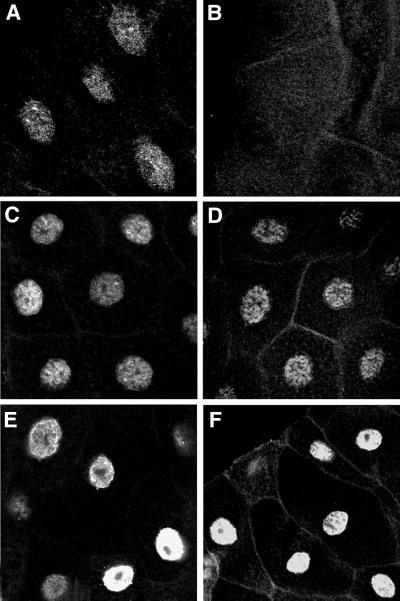
Immunolocalization of SU(S) ARM mutant derivatives on third-instar larval salivary gland polytene chromosomes. Shown are confocal images of chromosome squashes double labeled by indirect immunofluorescence with antibodies that recognize SU(S) and the positive control HRP36. The pseudocolored images indicate SU(S) in red (A to F) and HRP36 in green (A′ to F′). HRP36 is found at a much larger number of bands than endogenous SU(S), and there is little overlap between strong HRP36 sites and strong SU(S) sites. (A and A′) su(s)+; (B and B′) su(s)R39 null mutant. The remaining images were prepared from larvae grown at room temperature, expressing an SU(S) cDNA transgene under control of the hs-GAL4 driver in su(s) null mutant background: (C and C′) SU(S)wt; (D and D′) SU(S)ΔARM1; (E and E′) SU(S)ΔARM2; (F and F′) SU(S)ΔARM1,2.
A larger number of chromosomal sites stained strongly with the anti-SU(S) antibodies in chromosome squashes prepared from larvae expressing SU(S)wt (Fig. 4C) and SU(S)ΔARM2 (Fig. 4E) than in larvae lacking the transgene (Fig. 4A). The stronger signal at least in part reflects the higher level of SU(S) produced by the transgenes (see below). In contrast, no signal was observed when chromosomes expressing SU(S)ΔARM1 were stained with anti-SU(S) antibodies (Fig. 4D). Results obtained with chromosomes from transformants expressing the double-deletion derivative SU(S)ΔARM1,2 were somewhat variable. In some experiments no chromosomal association of SU(S)ΔARM1,2 was detected (data not shown), whereas in other experiments weak staining was observed. As described below, SU(S)ΔARM1,2 is expressed at very high levels in comparison to the other SU(S) derivatives. Thus, it is possible that SU(S) interacts weakly with polytene chromosomes independent of its RNA binding motifs.
One possible explanation for the failure of SU(S)ΔARM1 and SU(S)ΔARM1,2 to associate with polytene chromosomes might be that the ARM1 deletion blocks entry of these proteins into the nucleus. Immunofluorescence labeling of whole salivary gland cells demonstrated that each of the SU(S) derivatives accumulates in the nucleus as expected (Fig. 5). Another possibility is that the proteins are unstable. However, Western blot analysis (data not shown; see below) indicates that the proteins produced by the transgenes accumulate at higher levels than endogenous SU(S). Based on these results, we conclude that ARM1, which promotes binding to SELEX consensus RNAs in vitro, is required for the stable association of SU(S) with polytene chromosomes in vivo and that ARM2 alone is neither sufficient nor required for this interaction.
FIG. 5.
Nuclear localization of SU(S)ARM derivatives in salivary gland cells. Shown are confocal images of whole mounts of third-instar larval salivary glands labeled by indirect immunofluorescence with a polyclonal anti-SU(S) antibody. Like endogenous SU(S), the four SU(S) derivatives localize in the nucleus. (A) su(s)+; (B) su(s) null mutant. (C to F) Images prepared from larvae expressing various SU(S) transgenes under control of the hs-GAL4 driver in su(s) null mutant background: (C) SU(S)wt; (D) SU(S)ΔARM1; (E) SU(S)ΔARM2; (F) SU(S)ΔARM1,2.
SU(S)wt represses RNA accumulation from the su(s) cDNA transgenes.
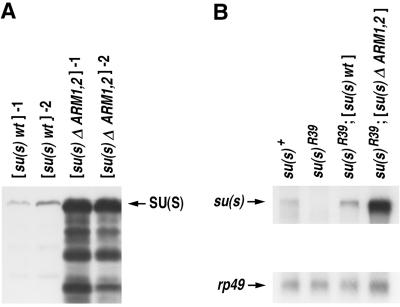
We used Western blots to compare the levels of SU(S) produced by the different transgenes. Quite unexpectedly, we found that SU(S)ΔARM1,2 accumulates at a much higher level than SU(S)wt (Fig. 6A). This higher level of mutant protein accumulation was observed with more than 10 different transformant lines (data not shown) and with three different GAL4 drivers (data not shown). Thus, the difference is not likely to be related to position-dependent variation in transcription of the transgene or tissue-specific effects. The accumulation level of SU(S) derivatives with single ARM deletions varied between transformant lines. In some transformants, SU(S)ΔARM1 or SU(S)ΔARM2 accumulated at similar levels as SU(S)wt. In other transformants, the single deletion derivatives accumulated at levels intermediate levels, i.e., higher than SU(S)wt but substantially lower than SU(S)ΔARM1,2 (data not shown).
FIG. 6.
Levels of su(s)wt and su(s)ΔARM1,2 expression. (A) Western blot of total protein (200 μg per lane) extracted from balanced stocks with a recombinant third chromosome carrying both a UAS-su(s) transgene and the hs-GAL4 driver. The blot was probed with an anti-SU(S) polyclonal antibody. (B) Northern blot of polyadenylated adult RNA (2 μg per lane) isolated from the same flies as in panel A probed with su(s) and rp49 cloned sequences.
We performed Northern blot analysis on poly(A)+ mRNA isolated from adult flies carrying the wild-type and double-mutant su(s) transgenes in the background of the su(s)R39 allele (Fig. 6B). This analysis showed that the difference also occurs at the RNA level; i.e., su(s)ΔARM1,2 transcript accumulates at a higher level than su(s)wt mRNA transcript. We reasoned that the difference in RNA levels could be due to either nucleotide sequence differences between the wild-type and mutant RNAs or a difference in the ability of the wild-type and mutant SU(S) proteins to regulate the amount of RNA generated from the transgenes. To distinguish between these possibilities, we designed a cis-trans test and performed a cross to introduce both transgenes into the same cells. If the mutant RNA accumulates at a higher level because it is inherently more stable (a cis effect), then the difference in RNA levels will also occur when both transgenes are expressed in the same cells. On the other hand, if SU(S) regulates accumulation of these RNAs (a trans effect), then the su(s)wt and su(s)ΔARM1,2 RNAs will be found at similar levels in cells that express both proteins. In the latter case, the level of the two RNAs would depend on the degree to which SU(S)ΔARM1,2 interferes with the activity of SU(S)wt.
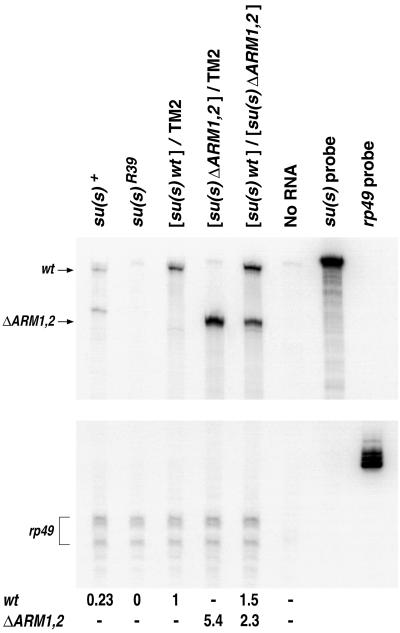
We performed RNase protection analysis to compare the levels of the su(s)wt and su(s)ΔARM1,2 RNAs in flies carrying one copy of either the wt or mutant transgene and flies carrying one copy of both transgenes (Fig. 7). This analysis was performed in the background of the su(s)R39 null mutant. To distinguish between the su(s)wt and su(s)ΔARM1,2 transcripts, we used a wild-type, antisense su(s) RNA probe that spans the ARM coding region. After hybridization and RNase treatment, su(s)wt and su(s)ΔARM1,2 transcripts generate 394- and 262-nt protected fragments, respectively. As shown in Fig. 7, the level of su(s) transcript in RNA prepared from flies carrying only the mutant transgene was fivefold higher than the level observed in flies that were carrying only the su(s)wt transgene. In contrast, in RNA prepared from flies expressing both the wild-type and ΔARM1,2 transgenes, both su(s) RNA types accumulated at a similar level. This demonstrates that SU(S) protein regulates accumulation of RNA from the UAS-su(s) cDNA transgenes and that the ARMs mediate this regulation. In flies expressing both transgenes, the level of su(s)wt RNA was 1.5-fold higher than the level of this RNA in flies carrying only the wild-type transgene. The level of su(s)ΔARM1,2 RNA was twofold lower in flies carrying both transgenes than in flies expressing only the su(s)ΔARM1,2 transgene. These intermediate levels suggest that both the wild-type and ARM deletion derivatives of SU(S) can influence RNA production from the transgenes and that there is not a clear dominance relationship between the wild-type and mutant forms of the protein.
FIG. 7.
RNase protection analysis of su(s)wt and su(s)ΔARM1,2 RNA levels. Total RNA was isolated from adult flies reared at 25°C. Flies with one copy of a transgene contained the third chromosome balancer TM2 and a recombinant third chromosome carrying a UAS-su(s) transgene and the hs-GAL4 driver. Flies with both transgenes contained a wild-type transgene on one of the third chromosomes and a mutant transgene on the other. The transgenic flies also carry the null su(s)R39 allele on the X chromosome and thus produce no endogenous su(s) protein or mRNA; 20 μg of total RNA was used in each reaction. The 447-nt hybridization probe in this experiment was prepared from a wild-type su(s) cDNA clone and spans the region that encodes the ARMs (see Materials and Methods). The su(s)wt and su(s)ΔARM1,2 protected fragments are 394 and 262 nt, respectively. Indicated beneath the lanes are the relative RNA levels. The values shown are the average of three separate experiments performed with the same RNA preparations. The standard deviations were less than 10% of the values shown.
In attempt to test whether su(s)ΔARM1,2 is capable of elevating the amount of RNA produced from endogenous su(s), we crossed the GAL4-driven UAS-su(s)ΔARM1,2 transgene into the background of a wild-type endogenous su(s) allele and performed Northern analysis with a probe that could detect only the endogenous su(s) transcript. We observed no effect of the su(s)ΔARM1,2 transgene on the level of endogenous su(s) RNA (data not shown). This could indicate that SU(S)ΔARM1,2 is not capable of affecting regulation of the endogenous su(s)+ gene. However, the absence of an effect might be because SU(S)ΔARM1,2 produced by the GAL4-driven transgene is not expressed in the appropriate tissue or at the level needed to effect expression of endogenous su(s). Thus, no definitive conclusion could be drawn from this experiment.
DISCUSSION
We have shown that two ARMs mediate the in vitro RNA binding activity and in vivo function of SU(S). ARM1 is required for high-affinity binding of SU(S) to SELEX consensus RNAs, whereas ARM2 promotes binding at roughly a 10-fold-lower affinity to SELEX nonconsensus RNAs. The CCCH zinc binding motifs, located just downstream of the ARMs, are incapable of promoting stable RNA binding in vitro, although they may influence the stability of the SU(S)-RNA interaction in vivo. The SELEX consensus RNAs contains a close match to the sequence UCAGUAGUCU, flanked by GU-rich sequences. Previous in vitro RNA footprinting experiments showed that recombinant, baculovirus-expressed SU(S) interacts with nucleotides of the consensus and GU-rich regions (45). The SELEX consensus sequence is complementary to invariant sequences near the 5′ end of the SELEX RNA. Thus, one explanation for the repeated isolation of RNAs containing this sequence is that it enables the RNA to fold into a hairpin structure recognized by SU(S), rather than representing a particular sequence that is bound by SU(S). However, the sequence composition of the stem-loop might be a factor in SU(S) binding. For example, as previously noted (45), the SELEX consensus sequence resembles the 5′ splice site consensus MAGGURAGU, where M denotes C or A, R denotes A or G, and the underlined GU is the invariant dinucleotide found at the 5′ boundary of the intron (44) and the sequence commonly found near the transcription initiation site (UCAGU) (14). Perhaps SU(S) binds to stem-loops in regions of a pre-mRNA that includes these sequences, i.e., near the cap site and/or 5′ splice site. Additional work will be required to determine the relative importance of RNA sequence versus structure in the binding activity mediated by ARM1.
The ARMs of SU(S), particularly ARM1, exhibit the usual features that are characteristic of this class of RNA binding motif. ARM-RNA binding domains are typically short, i.e., 10- to 20-amino-acid, arginine-rich regions that recognize particular structural features of RNA rather than a specific sequence (11). The most extensively studied ARM proteins, including N protein of bacteriophage lambda, Nun protein of bacteriophage HK022, and human immunodeficiency virus Tat and Rev, bind near the 5′ end of their target transcripts (34, 39, 50, 54, 55) and regulate either transcription elongation or RNA export (17–19).
Previous work has shown that several su(s)-suppressible alleles have antisense transposon insertions in the 5′ region of their genes that disrupt either the first exon or the first intron (20–22, 29). A higher level of RNA is produced by these mutant alleles in su(s) mutants than in su(s)+ flies. In our earlier studies, we proposed that the regulation of RNA accumulation by SU(S) was at the level of RNA stability, because the effect was related to splicing complex assembly, which at the time was thought to occur posttranscriptionally. However, since transcription and RNA processing are coupled, it is possible that SU(S) binding to RNA affects transcription. For example, perhaps SU(S) binding stabilizes stem-loop structure in the 5′ region of the pre-mRNA, which, in turn, pauses the elongating transcription complex. Splicing complex assembly in the 5′ region of the pre-mRNA might release the paused RNA polymerase II just as ribosome binding to a nascent transcript releases the paused prokaryotic RNA polymerase to synchronize transcription and translation (13). Suppressible mutant alleles contain transposon insertions near their 5′ regions and thus lack normal splicing signals near the transcription start sites. Expression of these insertion mutant alleles would, thus, be irreversibly repressed by SU(S) binding. This type of role for SU(S) in regulating cotranscriptional splicing complex assembly is analogous to the proposed role of SPT5 and its homologues in repressing transcription elongation to couple capping to transcription (23, 53). Studies currently under way are designed to test whether SU(S) regulates transcription or RNA stability.
In this report, we have demonstrated that SU(S) negatively regulates production of transcripts from the UAS-su(s) cDNA transgenes. Thus, we have identified the first example of regulation by SU(S) that does not involve mutant transcripts with a transposon insertion. This effect cannot be explained in terms of our previous model that SU(S) binds cryptic splice sites to promote recognition of authentic splice sites because the only intron present in these constructions is a small simian virus 40 intron, which was introduced into the 3′ untranslated region during subcloning of the cDNA into the transformation vector (9). Like the mutant v and y alleles that are negatively regulated by SU(S), the first intron of the su(s) cDNA transgenes is a long distance, 5 kb, downstream of the transcription start site. In most intron-containing pre-mRNAs, the first intron is located near the beginning of the transcribed region (41), and an intron in this position appears to be important for expression at a high level. Studies in a variety of different systems indicate that cDNA transgenes are expressed at lower levels than their genomic counterparts in vivo and that expression of a cDNA can be elevated by inclusion of an intron near the 5′ end of the gene (10, 30, 31, 37, 38, 46). The magnitude of this effect varies, depending on sequences present in the RNA and the promoter that is used to direct transgene expression (33, 35). Furthermore, in a study that examined the factors that influence intron-dependent enhancement of gene expression, the strength of the 5′ splice site was found to be important (31). Likewise, we previously showed that improvement of a cryptic 5′ splice site near the beginning of a mutant v transgene to a consensus site increased RNA production from a mutant v transgene, without improving the splicing efficiency. In addition, SU(S) does not inhibit the production of RNA from the transgene with a consensus 5′ splice site near the beginning of the transcribed region but does limit production of RNA from a similar transgene that lacks a strong 5′ splice site in this region (21). In light of these observations, it seems plausible that SU(S) is a component of the intron-dependent gene expression pathway and that studies of SU(S) function will provide insights into the general mechanism that limits expression of cDNA transgenes and antisense RNAs in vivo. The su(s) gene is nonessential, although su(s) mutations impair viability and fertility to variable degrees (36). However, our finding that overexpression of SU(S) can be lethal suggests that SU(S) functions in an essential process and that the level of SU(S) in the nucleus is an important aspect of its regulatory activity.
ACKNOWLEDGMENTS
We thank G. Dreyfuss for providing monoclonal antibody 5cA5. We thank G. Maroni and M. Peifer for critical comments on the manuscript and R. Kole for a helpful discussion. We are grateful to S. Whitfield for assistance with the figures.
This work was supported by grants MCB-950631 and MCB-9808150 from the National Science Foundation.
REFERENCES
- 1.Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Bai C, Tolias P P. Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol Cell Biol. 1996;16:6661–6667. doi: 10.1128/mcb.16.12.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai C, Tolias P P. Drosophila clipper/CPSF 30K is a post-transcriptionally regulated nuclear protein that binds RNA containing GC clusters. Nucleic Acids Res. 1998;26:1597–1604. doi: 10.1093/nar/26.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barabino S M, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 6.Batchelder C, Dunn M A, Choy B, Suh Y, Cassie C, Shim E Y, Shin T H, Mello C, Seydoux G, Blackwell T K. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes Dev. 1999;13:202–212. doi: 10.1101/gad.13.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 8.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 9.Brand A H, Manoukian A S, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 10.Buchman A R, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 12.Carballo E, Lai W S, Blackshear P J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 13.Chan C L, Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site. Mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989;264:20796–20804. [PubMed] [Google Scholar]
- 14.Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–93. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Culbertson M R. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 17.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 18.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fridell R A, Pret A M, Searles L L. A retrotransposon 412 insertion within an exon of the Drosophila melanogaster vermilion gene is spliced from the precursor RNA. Genes Dev. 1990;4:559–566. doi: 10.1101/gad.4.4.559. [DOI] [PubMed] [Google Scholar]
- 21.Fridell R A, Searles L L. Evidence for a role of the Drosophila melanogaster suppressor of sable gene in the pre-mRNA splicing pathway. Mol Cell Biol. 1994;14:859–867. doi: 10.1128/mcb.14.1.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geyer P K, Chien A J, Corces V G, Green M M. Mutations in the su(s) gene affect RNA processing in Drosophila melanogaster. Proc Natl Acad Sci USA. 1991;88:7116–7120. doi: 10.1073/pnas.88.16.7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartzog G A, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 26.Hilleren P, Parker R. mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA. 1999;5:711–719. doi: 10.1017/s1355838299990519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 28.Karess R E. P element mediated germ line transformation of Drosophila. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 2. Oxford, England: IRL Press; 1985. pp. 121–141. [Google Scholar]
- 29.Kim N, Kim J, Park D, Rosen C, Dorsett D, Yim J. Structure and expression of wild-type and suppressible alleles of the Drosophila purple gene. Genetics. 1996;142:1157–1168. doi: 10.1093/genetics/142.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohler U, Donath M, Mendel R R, Cerff R, Hehl R. Intron-specific stimulation of anaerobic gene expression and splicing efficiency in maize cells. Mol Gen Genet. 1996;251:252–258. doi: 10.1007/BF02172925. [DOI] [PubMed] [Google Scholar]
- 31.Korb M, Ke Y, Johnson L F. Stimulation of gene expression by introns: conversion of an inhibitory intron to a stimulatory intron by alteration of the splice donor sequence. Nucleic Acids Res. 1993;21:5901–5908. doi: 10.1093/nar/21.25.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai W S, Carballo E, Strum J R, Kennington E A, Phillips R S, Blackshear P J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange P, Victor M, Benecke B J. Basal level transcription of the human hsp86 gene is directed by intron-based elements. Genes Cells. 1997;2:185–194. doi: 10.1046/j.1365-2443.1997.d01-309.x. [DOI] [PubMed] [Google Scholar]
- 34.Lazinski D, Grzadzielska E, Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 35.Lee T X, Johnson L F. Pre-mRNA processing enhancer (PPE) element increases the expression of an intronless thymidylate synthase gene but does not affect intron-dependent S phase regulation. J Cell Biochem. 1998;69:104–116. [PubMed] [Google Scholar]
- 36.Lindsley D L, Zimm G G. The genome of Drosophila melanogaster. San Diego, Calif: Academic Press, Inc.; 1992. [Google Scholar]
- 37.Liu X, Mertz J E. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Mertz J E. Sequence of the polypyrimidine tract of the 3′-terminal 3′ splicing signal can affect intron-dependent pre-mRNA processing in vivo. Nucleic Acids Res. 1996;24:1765–1773. doi: 10.1093/nar/24.9.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 40.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 41.Maroni G. The organization of eukaryotic genes. Evol Biol. 1996;29:1–19. [Google Scholar]
- 42.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 43.Mello C C, Schubert C, Draper B, Zhang W, Lobel R, Priess J R. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- 44.Mount S. Messenger RNA splicing signals in Drosophila genes. In: Maroni G, editor. An atlas of Drosophila genes. New York, N.Y: Oxford University Press; 1993. pp. 333–358. [Google Scholar]
- 45.Murray M V, Turnage M A, Williamson K J, Steinhauer W R, Searles L L. The Drosophila suppressor of sable protein binds to RNA and associates with a subset of polytene chromosome bands. Mol Cell Biol. 1997;17:2291–2300. doi: 10.1128/mcb.17.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rethmeier N, Seurinck J, Van Montagu M, Cornelissen M. Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J. 1997;12:895–899. doi: 10.1046/j.1365-313x.1997.12040895.x. [DOI] [PubMed] [Google Scholar]
- 47.Ruberte E, Marty T, Nellen D, Affolter M, Basler K. An absolute requirement for both the type II and type I receptors, punt and thick veins, for dpp signaling in vivo. Cell. 1995;80:889–897. doi: 10.1016/0092-8674(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 48.Rudner D Z, Kanaar R, Breger K S, Rio D C. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci USA. 1996;93:10333–10337. doi: 10.1073/pnas.93.19.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutledge B J, Mortin M A, Schwarz E, Thierry-Mieg D, Meselson M. Genetic interactions of modifier genes and modifiable alleles in Drosophila melanogaster. Genetics. 1988;119:391–397. doi: 10.1093/genetics/119.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian T, Govindarajan R, Chinnadurai G. Heterologous basic domain substitutions in the HIV-1 Tat protein reveal an arginine-rich motif required for transactivation. EMBO J. 1991;10:2311–2318. doi: 10.1002/j.1460-2075.1991.tb07768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voelker R A, Gibson W, Graves J P, Sterling J F, Eisenberg M T. The Drosophila suppressor of sable gene encodes a polypeptide with regions similar to those of RNA-binding proteins. Mol Cell Biol. 1991;11:894–905. doi: 10.1128/mcb.11.2.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voelker R A, Huang S M, Wisely G B, Sterling J F, Bainbridge S P, Hiraizumi K. Molecular and genetic organization of the suppressor of sable and minute (1) 1B region in Drosophila melanogaster. Genetics. 1989;122:625–642. doi: 10.1093/genetics/122.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watnick R S, Gottesman M E. Binding of transcription termination protein nun to nascent RNA and template DNA. Science. 1999;286:2337–2339. doi: 10.1126/science.286.5448.2337. [DOI] [PubMed] [Google Scholar]
- 55.Watnick R S, Gottesman M E. Escherichia coli NusA is required for efficient RNA binding by phage HK022 nun protein. Proc Natl Acad Sci USA. 1998;95:1546–1551. doi: 10.1073/pnas.95.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiss M A, Narayana N. RNA recognition by arginine-rich peptide motifs. Biopolymers. 1998;48:167–180. doi: 10.1002/(SICI)1097-0282(1998)48:2<167::AID-BIP6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 57.Worthington M T, Amann B T, Nathans D, Berg J M. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc Natl Acad Sci USA. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]