Abstract
Background
Glioma is a common type of malignant brain tumor with a high mortality and relapse rate. The endosomal sorting complex required for transport (ESCRT) has been reported to be involved in tumorigenesis. However, the molecular mechanisms have not been clarified.
Methods
Bioinformatics was used to screen the ESCRT subunits highly expressed in glioma tissues from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. The function of the ESCRT subunits in glioma cells was examined in vitro. Transcriptome sequencing analyzed the target genes and signaling pathways affected by the ESCRT subunit. Finally, the relationship between m6A (N6-methyladenosine) modification and high expression of the ESCRT subunit was studied.
Results
VPS25 was upregulated in glioma tissues, which was correlated with poor prognosis in glioma patients. Furthermore, VPS25 knockdown inhibited the proliferation, blocked the cell cycle, and promoted apoptosis in glioma cells. Meanwhile, VPS25 induced a G0/G1 phase arrest of the cell cycle in glioma cells by directly mediating p21, CDK2, and cyclin E expression, and JAK-signal transducer and activator of transcription (STAT) activation. Finally, YTHDC1 inhibited glioma proliferation by reducing the expression of VPS25.
Conclusion
These results suggest that VPS25 is a promising prognostic indicator and a potential therapeutic target for glioma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-021-02304-0.
Keywords: YTHDC1, VPS25, Cell cycle, JAK–STAT, Glioma
Background
Glioma refers to a tumor originating from glial cells. It is the most common tumor in the central nervous system (CNS) and has the highest lethal rate [1]. According to the 2016 World Health Organization (WHO) classification, glioma can be classified into astrocytic tumors (astrocytoma, anaplastic astrocytoma, and glioblastoma), oligodendrogliomas, ependymomas, and mixed gliomas [2]. The 2015 annual report of the National Central Cancer Registry of China (NCCR) indicates that the morbidity and mortality of glioma patients are increasing year by year [3]. Glioma has a fast growth, easy invasion and metastasis, a high mortality, and a short average survival time. Thus, it has become a type of disease that causes great harm to human life and health [4]. At present, the traditional multi-mode therapy of surgical resection, radiotherapy, and chemotherapy has not significantly improved the prognosis of glioma patients [5]. Thus, exploring the underlying molecular mechanisms associated with glioma progression and metastasis may contribute to discovering more effective treatment methods and targets.
VPS25 is one subunit of the endosomal sorting complex required for transport (ESCRT) [6]. ESCRT is the molecular machinery in eukaryotic cells that recognizes the ubiquitinated cargo at the intraluminal vesicles (ILVs) and sorts them into late endosomes to form multivesicular bodies (MVBs). Its main function is to promote the degradation of ubiquitinated proteins, and it is also related to retrovirus budding, cell division, cell cycle, autophagy, fungi pH sensing, and transcriptional regulation [7–9]. The ESCRT system contains five complexes, termed ESCRT-0, I, II, and III, and a series of accessory proteins, such as VPS4 [10]. For example, the ESCRT-II complex comprises the VPS25, VPS22, and VPS36 subunits [11].
VPS25 regulates endosomal protein sorting by interacting with the CHMP6 subunit in the ESCRT-III complex, and the inhibition of this interaction prevents cell division and leads to cell death [12]. Studies in Drosophila have found that VPS25 is an unconventional tumor suppressor that prevents the non-autonomous hyperproliferation of cells by modulating NOTCH signaling [13, 14]. VPS25 also regulates HIV production and release through pre- and post-transcriptional mechanisms or by interacting with other ESCRT subunits [15]. Previous studies have shown that multiple ESCRT subunits are involved in tumorigenesis [16]. For example, the TSG101, which is overexpressed in most tumors, is positively associated with tumor formation [17]. VPS4A functions as a tumor suppressor in human hepatoma cells by regulating the secretion and uptake of extracellular exosomal microRNAs [18]. Current research shows that the role of ESCRT in tumorigenesis is due to the loss of transmembrane protein homeostasis and the resulting consequences on signal transduction, and it could account for a defective MVB pathway and cell cytokinesis. In addition, ESCRT is also thought to affect tumorigenesis through autophagy and exosomes [8].
In this study, we screened the high expression subunits of the ESCRT complex in glioma tissue. We found that the expression of VPS25 was upregulated in glioma and associated with poor prognosis of glioma patients. VPS25 was also necessary for glioma cells' proliferation, cell cycle, and apoptosis. Furthermore, m6A methyltransferases (METTL3 and METTL14) and m6A reader (YTHDC1) were found to regulate VPS25 expression.
Materials and methods
Patient samples
This study was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Wannan Medical College. A written informed consent was obtained from all the patients and/or their legal guardians. There were 66 glioma tissues and 14 normal brain tissues that were collected from patients diagnosed pathologically who underwent surgical resection at the Department of Neurosurgery of the First Affiliated Hospital of Wannan Medical College. Tissue samples were preserved in liquid nitrogen and stored at − 80 °C until RNA and protein extraction.
Gene expression profiling and survival analysis
The mRNA expression array data of the ESCRT complex were downloaded from the Gene Expression Omnibus (GEO) database (accession number is GSE4290), which had been published previously [19], and The Cancer Genome Atlas database (TCGA, https://portal.gdc.cancer.gov/).
We used Gene Expression Profiling Interactive Analysis 2 (GEPIA2, https://gepia2.cancer-pku.cn/) [20] to analyze the VPS25 expression data and overall survival in glioma based on the TCGA and GTEx (Genotype-Tissue Expression) projects.
Cell lines and culture
The human glioma cell lines U87MG and U251 were purchased from American Type Culture Collection (ATCC). Glioma cells were cultured in Dulbecco's modified Eagle medium–high glucose (DMEM; Hyclone, GE Healthcare Life Sciences, UT, USA) containing 10% (v/v) fetal bovine serum (FBS; Gibco, Life Technologies, NY, USA). All the cell lines were cultured in a humidified incubator with 5% CO2 at 37 °C.
Real-time qPCR analysis
TRIzol reagent (Ambion, Life Technologies, CA, USA) was used for total RNA extraction from the tissues or cells according to the manufacturer's protocol described previously [21]. The RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Lithuania, USA) was used for RNA reverse transcription. Real-time quantitative PCR was performed according to the instructions of QuantiNova™ SYBR® Green PCR Kit (Qiagen, Hilden, Germany) using the Bio-Rad CXF96 PCR system (Bio-Rad, Hercules, CA, USA). The relative gene expression of VPS25 was calculated using the 2−ΔΔCt method. GAPDH served as an internal control. The primers were synthesized by Sangon Biotech (Shanghai, China). VPS25: F-5′-TGGTGCTCGCTGGTCCTGT C-3′, R-5′-GGACTTGCTCTTATCCAACCACTCG-3′. METTL3: F-5′-TCAGCATC GGAACCAGCAAAG-3, R-5′-TCCTGACTGACCTTCTTGCTC-3′. METTL14: F-5′-GTTGGAACATGGATAGCCGC-3, R-5′-CAATGCTGTCGGCACTTTCA-3′. YTHDC1: F-5′-AGGGATCCTGAAAGGAGGGC-3, R-5′-CACTGCTGCCAGTCTC ATGG-3′.
siRNA, shRNA, and overexpression of plasmid transfection
The negative control siRNA (NC) and VPS25 siRNA (siVPS25) were synthesized by RiboBio (Guangzhou, China). The sequence of siRNA targeting the VPS25-1, VPS25-3, METTL3, METTL14 and YTHDC1 was GCACAAGGCCGAGATCATC, GTCGATCCAGATTGTATTA, CAAGTATGTTCACTATGAA, GAAGACGCCTT CATCTATT and CAAGGAGTGTTATCTTAAT. Cells (15 × 104 cells/well) were seeded in a 6-well plate and, after 24 h, were transfected with siRNA at 100 nM by riboFECT™ CP Transfection Kit (RiboBio) according to the manufacturer's instructions. For the stable silencing, shRNA lentivirus (lenti-sh-NC, lenti-sh-VPS25) was purchased from HANBIO (Shanghai, China). Lentiviruses were transfected into glioma cells for 48 h, and stable cell clones were selected for 1 week using puromycin (5 μg/ml). For overexpression, the cDNA encoding VPS25 was subcloned into the pTSB02-GFP-PURO vector and transfected into U251 cells according to the manufacturer's protocol of Lipofectamine 3000 (Invitrogen, Carlsbad, CA).
Cell proliferation and cycle assay
For the cell proliferation assay, the xCELLigence RTCA DP (ACEA Biosciences, CA, USA) was used as described previously [22]. At 24 h after transfection, cells were seeded in a 16-well E-plate (ACEA Biosciences) at a density of 8000 cells per well. The cell index was monitored by the xCELLigence system (ACEA Biosciences) in real time. In addition, the growth curve of the cell was analyzed by Excel (Microsoft, Washington, USA) or GraphPad Prism 5 (La Jolla, California, USA).
For the colony formation assay, transfected cells were seeded in a 6-well plate at 2 × 103 cells per well for 2 weeks. The cells were washed twice with PBS, fixed with 4% paraformaldehyde, and stained with 0.5% crystal violet. The colonies were photographed and analyzed statistically.
For the cell cycle assay, transfected cells were incubated for 48 h and fixed in 75% ethanol at 4 °C overnight. Then the cells were washed twice with PBS and incubated in RNase A/PI (propidium iodide) solution (100 μg/ml RNase A and 50 μg/ml PI) at 37 °C for 30 min in the dark. The DNA contents of stained cells were analyzed using a Beckman Coulter FC500MPL flow cytometer (CA, USA). The data were analyzed by FlowJo V10 software (FlowJo LLC, OR, USA).
Cell apoptosis assay
Flow cytometry with Annexin V and PI labeling was used to determine cell apoptosis. The transfected cells were incubated for 48 h and washed with PBS twice. Then cells were resuspended in 300 μl of binding buffer containing 5 μl Annexin V-FITC and 10 μl PI. After incubating for 15 min at room temperature in the dark, the cells were analyzed by a Beckman Coulter FC500MPL.
Apoptosis was detected by One Step TUNEL Apoptosis Assay Kit for TUNEL assay (Beyotime, Shanghai, China) according to the manufacturer's instructions. After TUNEL staining, the glioma cells were stained using DAPI and observed using a High-Content Imaging System (ImageXpress Micro Confocal, Molecular Devices, USA). The number of apoptotic cells is presented as a percentage of the total cells counted.
RNA sequencing analysis
U251 cells were transfected with NC (n = 2) or siVPS25 (n = 2) for 48 h. Then, total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion) following the manufacturer's protocol. The transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China). The libraries were sequenced on the Illumina sequencing platform (HiSeq™ 2500, Illumina, CA, USA). Significantly differentially expressed genes (DEGs) between NC and siVPS25 cells were identified based on fold change ≥ 1.5 and P-value ≤ 0.05 using the DEGseq functions. Functional pathway analysis of DEGs was performed using KEGG pathway enrichment analysis.
Western blots
The protein expression of p-JAK1, 2, 3 (Rabbit mAb; CST), p-STAT1, 2, 3 (Rabbit mAb; CST), CDK2 (Rabbit Polyclonal; Proteintech), cyclin E (Rabbit Polyclonal; Proteintech), p21 (Rabbit Polyclonal; Proteintech), VPS25 (Rabbit Polyclonal; Proteintech), METTL3, METTL14 and YTHDC1 (Rabbit Polyclonal; Proteintech) was detected by western blot as described previously [23, 24]. Briefly, cells were lysed by Sample Buffer, Laemmli 2 × Concentrate (Sigma, MO, USA) via the manufacturer's protocol. Next, the proteins were isolated by 10% SDS–PAGE gel and transferred to nitrocellulose membranes. After immunoblotting with primary and secondary antibodies, the protein bands were incubated with enhanced chemiluminescence (ECL) and visualized using a Tanon 5200 Chemiluminescence Imaging System (Shanghai, China).
RNA immunoprecipitation
For quantification of m6A-modified VPS25 levels, RNA immunoprecipitation (RIP) experiments were performed. First, RNA was incubated with anti-m6A (Mouse monoclonal, Abcam, MA, USA) or anti-IgG (CST) antibody for immunoprecipitation according to the standard protocol of a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA, USA). After RIP, the enrichment of m6A containing VPS25 was detected via RT-qPCR assays.
RNA m6A modification quantification
RNA m6A modification was quantified as described previously [25, 26]. Briefly, 200 ng of sample RNA was incubated with a diluted capture antibody. The m6A content of total mRNA was detected by an m6A-RNA methylation quantification kit (ab185912; Abcam, UK), following the manufacturer’s protocol. A microplate reader measured the m6A levels at a wavelength of 450 nm.
In vivo xenograft mice experiments
BALB/c nude mice (4–5 weeks old, 10 mice) were purchased from the Experimental Animal Center of Qinglongshan (Nanjing, China) and raised in pathogen-free mouse colonies. The U87MG cells (1 × 107 cells in 0.1 ml PBS) were injected subcutaneously into BALB/c nude mice. Tumor width and length were recorded every 5 days. The following formula calculated the volume of the tumors: volume = (length × width2)/2. The tumors were weighed after the mice were sacrificed. The ethics committee of the First Affiliated Hospital of Wannan Medical College approved the animal experiment.
Immunohistochemistry (IHC)
For IHC staining, the paraffin-embedded tissue sections were deparaffinized and re-hydrated. After antigen retrieval and blocking endogenous activity, the sections were incubated overnight at 4 °C with primary antibodies, followed by HRP-conjugated secondary antibody at 37 °C for 1 h. DAB chromogenic agent was used to detect the VPS25.
Statistical analyses
Data were presented as the mean ± S.D. of three independent experiments. GraphPad Prism 5 (La Jolla) was used for all the statistical analyses. Student's t-test was performed to compare the data statistically. *P < 0.05, **P < 0.01, and ***P < 0.001 were statistically significant.
Results
Screening for highly expressed ESCRT subunits in glioma
To examine the function of the ESCRT complex in glioma tumorigenesis, the expression of main ESCRT subunits was screened from two databases: TCGA and GEO (Additional file 1: Fig. S1 and Additional file 2: Fig. S2). In the TCGA database, 10 subunits were significantly increased, and 3 subunits were significantly decreased in glioma tissues compared to normal brain tissues. In the GEO database, 10 subunits were significantly increased, and 9 subunits were significantly decreased. Combining the two databases, we noticed that MVB12A, VPS25, CHMP2A, and IST1 were highly expressed in glioma (Table 1). We then analyzed the relationship between the expression of these four genes and the pathological grade of glioma. We found that a higher expression of VPS25 and CHMP2A was more frequent in tissues with advanced tumor stage from GEO and TCGA databases (Additional file 3: Fig. S3A–C).
Table 1.
Expression of ESCRT subunits in normal brain tissues and glioma tissues from TCGA and GEO database
| Complex | Proteins | TCGA | GEO | ||
|---|---|---|---|---|---|
| Expression | p-value | Expression | p-value | ||
| ESCRT-0 | STAM2 | Up | * | Up | ns |
| ESCRT-I | MVB12A | Up | *** | Up | *** |
| MVB12B | Down | ns | Down | * | |
| TSG101 | Up | ** | Down | ns | |
| VPS28 | Up | * | Up | *** | |
| VPS37B | Up | * | Up | ** | |
| VPS37C | Down | ns | Down | * | |
| VPS37D | Up | ns | Up | *** | |
| ESCRT-II | VPS25 | Up | *** | Up | *** |
| VPS36 | Down | *** | Down | *** | |
| ESCRT-III | CHMP1A | Up | ns | Up | * |
| CHMP1B | Down | *** | Down | *** | |
| CHMP2A | Up | *** | Up | *** | |
| CHMP2B | Down | ns | Down | * | |
| CHMP3 | Down | * | Down | *** | |
| CHMP4A | Up | * | Down | ns | |
| CHMP4B | Down | ns | Up | ns | |
| CHMP4C | Up | ns | Up | * | |
| CHMP5 | Up | ns | Down | *** | |
| CHMP6 | Up | ** | Up | *** | |
| CHMP7 | Up | ns | Down | ns | |
| ESCRT-others | IST1 | Up | *** | Up | *** |
| VPS4A | Down | ns | Down | * | |
| VPS4B | Down | ns | Down | *** | |
ns: no significant
*P < 0.05, **P < 0.01, ***P < 0.001
VPS25 is upregulated in glioma
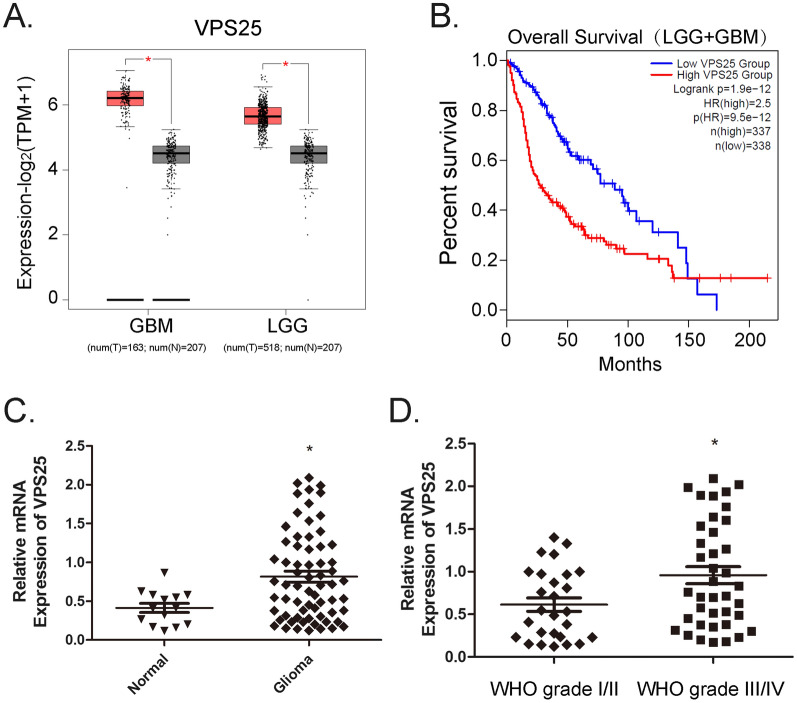
The GEPIA2 database was used to confirm the expression and clinical particularities of VPS25. The expression of VPS25 was significantly increased in glioma vs. controls (Fig. 1A). The glioma patients including both lower grade glioma (LGG) and GBM with high VPS25 expression had a shorter survival time than those with low VPS25 expression (Fig. 1B). However, high expression of VPS25 was not associated with poor outcomes in LGG or GBM. (Additional file 3: Fig. S3D). We analysis the VPS25 expression in IDH-mutated and IDH-wildtype by Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/index.jsp). In WHO II and III grade glioma, the VPS25 expression in IDH-mutated is higher than in IDH-wildtype (Additional file 3: Fig. S3E). After that, we determined the expression level of VPS25 in normal brain (n = 14) and glioma tissues (n = 66) collected from Yijishan Hospital by RT-qPCR. Compared with the normal brain tissues, the VPS25 expression was significantly increased in glioma tissues (Fig. 1C). The expression of the VPS25 protein in the GBM patient tissue is also higher than in normal tissues (Additional file 4: Figure S4). Further analysis showed that the expression of VPS25 was increased in tissues with advanced tumor stage (Fig. 1D). Taken together, these results provided evidence that VPS25 upregulation may play an important role in the development of glioma.
Fig. 1.
VPS25 is upregulated in glioma tissues. A The VPS25 expression level in GBM or LGG was higher than in normal brain tissues based on data available from the GEPIA2 database. B The overall survival of glioma patients (LGG and GBM) with high (n = 337) and low (n = 338) levels of VPS25 was plotted from GEPIA2. C VPS25 in glioma was higher than in normal brain tissues. The relative expression levels of VPS25 were determined using RT-qPCR in 66 glioma tissues and 14 normal brain tissues. D VPS25 expression levels in WHO grade III/IV were higher than in WHO grade I/II. Data of VPS25 expression in glioma tissues were from A. normal and N: non tumor samples; T: glioma; GBM: glioblastoma multiforme; LGG: lower grade glioma, * P < 0.05
Function of VPS25 in glioma cell proliferation
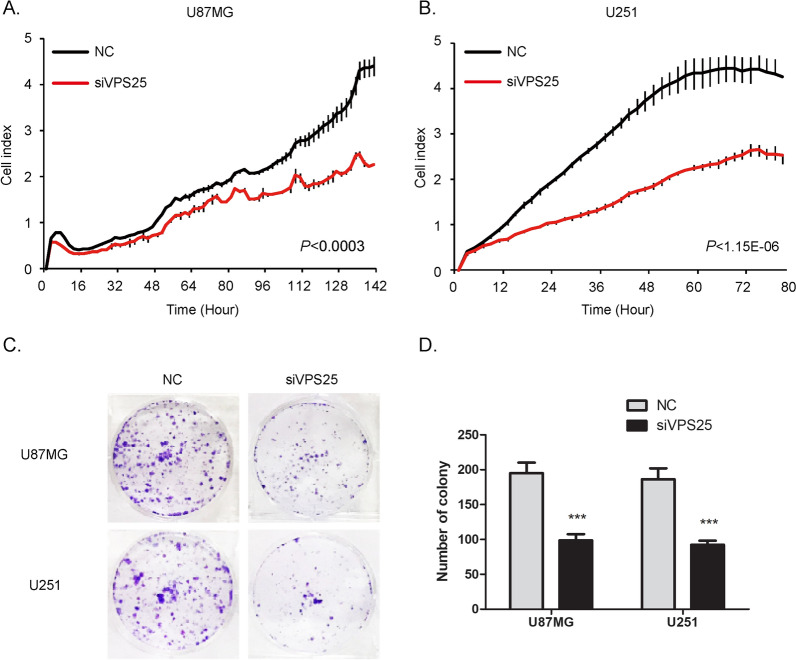
To verify the hypothesis that VPS25 functions as a candidate promoter in glioma, we examined the effect of VPS25 expression on cell proliferation and colony formation. RT-qPCR showed that VPS25 expression was significantly increased in glioma cell lines (U87MG and U251), compared with VPS25 expression measured in normal brain tissues (Additional file 5: Fig. S5A). Then, VPS25 siRNA was transfected to reduce the intracellular mRNA level of glioma. The RT-qPCR analysis revealed that transfection of glioma cells with siVPS25-3 significantly decreased the expression of VPS25 by 95% in U87MG and 83% in U251 (Additional file 5: Fig. S5B and C). Here, we employed xCELLigence RTCA DP to evaluate the influence of VPS25 on cell proliferation. Compared with the NC, glioma cells transfected with the siVPS25 showed a remarkable inhibition of cell proliferation (Fig. 2A and B). Additionally, the colony formation assay was used to assess the long-term effects of VPS25 on cell proliferation. Fewer colonies were formed in glioma cells transfected with siVPS25 than in the NC (Fig. 2C and D). These data suggest that VPS25 positively regulates glioma cell proliferation in vitro.
Fig. 2.
VPS25 regulates glioma cell proliferation. A RTCA xCELLigence assay showed that transfection of siVPS25 suppressed the proliferation of U87MG cells. B Transfection of siVPS25 suppressed proliferation of U251 cells. Experiments were performed and are presented as shown in A. C Transfection of siVPS25 suppressed the colony formation of U87MG and U251 cells. D Statistics of the colony number in C. Data are mean ± SD from three independent experiments. ***P < 0.001. NC: negative control; siVPS25: VPS25 gene silencer
Function of VPS25 in glioma cell cycle and apoptosis
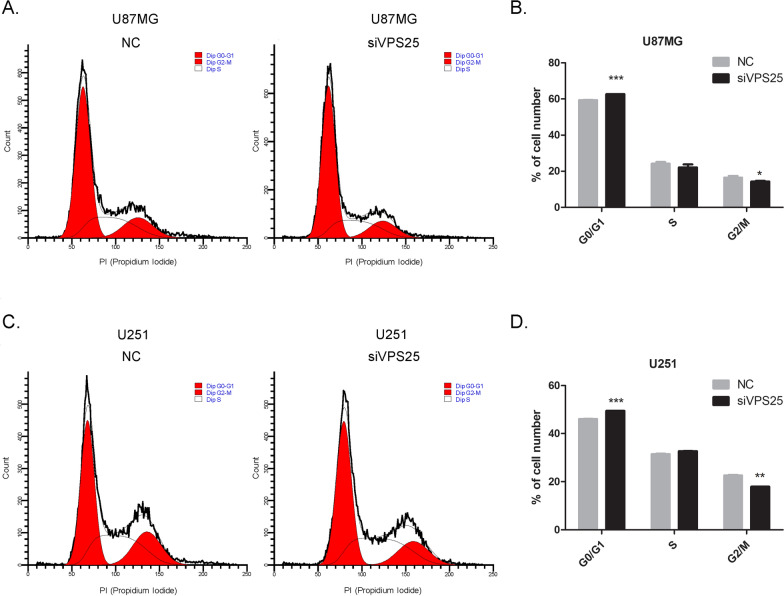
To confirm the function of VPS25 in cell proliferation, we investigated cell cycle distribution by flow cytometry. We found that the glioma cells under-expressing VPS25 were arrested at the G0/G1 phase, with a corresponding decrease in the percentage of cells in the G2/M phase (Fig. 3). This result suggested that the VPS25 knockdown (KD) inhibits glioma proliferation by blocking the cell cycle in the G0/G1 phase.
Fig. 3.
VPS25 regulates the glioma cell cycle. A The effect of VPS25 on the cell-cycle distribution of U87MG cells was monitored via flow cytometry. The VPS25-silenced U87MG cells were arrested at the G0/G1 phase of the cell cycle. B Histograms show the percentage (%) of cell populations at different stages of the cell cycle in A. C The VPS25-silenced U251 cells were arrested at the G0/G1 phase of the cell cycle. Experiments were performed and are presented as in A. D Histograms show the percentage (%) of cell populations at different stages of the cell cycle in C. Data are mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. NC: negative control; siVPS25: VPS25 gene silencer
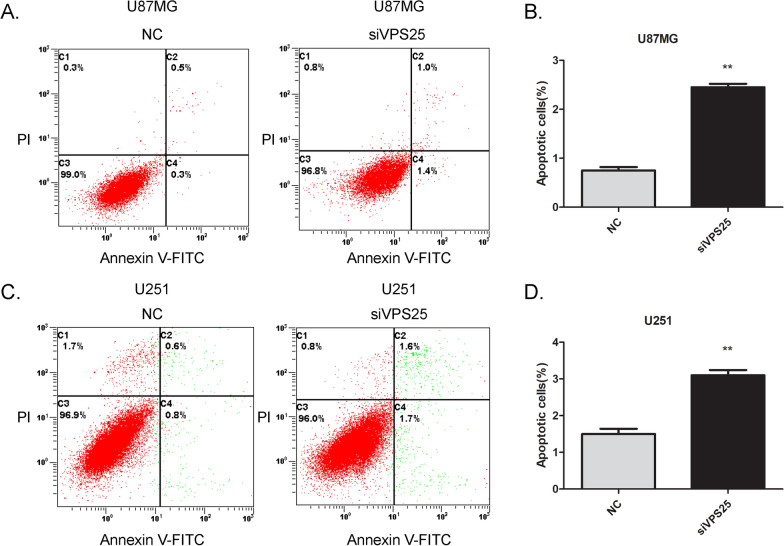
To figure out the mechanism of VPS25-regulated cell proliferation, we used flow cytometry to detect glioma cell apoptosis. By double staining Annexin V-FITC/PI in glioma cells, the proportion of apoptotic cells transfected with siVPS25 was significantly higher than in the NC group (Fig. 4). Moreover, the TUNEL assay shown glioma cells treated with siVPS25 had a higher rate of apoptosis than cells treated with NC (Additional file 6: Fig. S6). These results indicate that VPS25 has a role in glioma cell apoptosis.
Fig. 4.
VPS25 regulates glioma cell apoptosis. A The Annexin V/PI analysis was used to evaluate the effect of VPS25 expression alteration on U87MG cell apoptosis. B Percentage of apoptotic cells in A. C The Annexin V/PI analysis was used to evaluate the effect of VPS25 expression alteration on U251 cell apoptosis. Experiments were performed and are presented as in A. D Percentage of apoptotic cells in C. Data are mean ± SD from three independent experiments. **P < 0.01, NC: negative control; siVPS25: VPS25 gene silencer
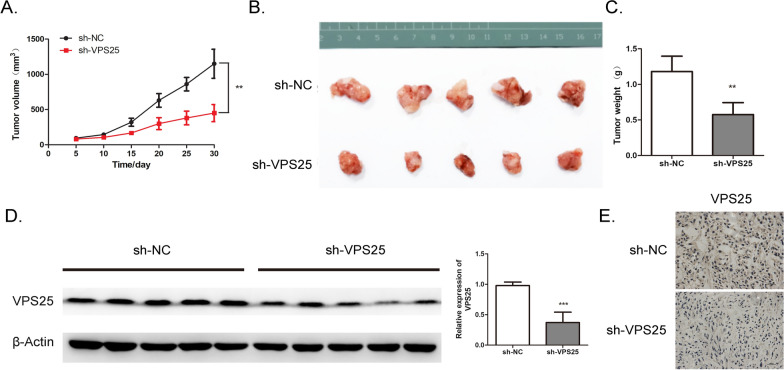
Knockdown of VPS25 represses glioma growth in vivo
To explore the role of VPS25 in glioma growth, stable VPS25-KD U251 cells were constructed using shRNA lentivirus (shVPS25). Next, a subcutaneous xenograft model was used. The growth of tumors (volume and weight) from VPS25-KD xenografts was inhibited compared with the tumors from the NC group (Fig. 5A–C). The VPS25 expression in the xenograft tumor was then examined by western blot and immunohistochemistry. As presented, VPS25 expression was downregulated in the VPS25-KD tumor group (Fig. 5D and E). In conclusion, these findings indicated that knockdown of VPS25 suppresses glioma growth in vivo.
Fig. 5.
Knockdown of VPS25 represses glioma growth in vivo. A A xenograft murine model was built using the U87MG cells transfected with VPS25 stable knockdown (sh-VPS25) or control (sh-NC). The glioma volume curves were detected in sh-VPS25 and sh-NC groups for 30 days. B Tumors were collected from nude mice (sh-VPS25 and sh-NC) at 30 days. C Statistics of the weight of the tumors in B. D The VPS25 protein expression in sh-VPS25 and sh-NC groups was detected by western blot. E The VPS25 protein expression in sh-VPS25 and sh-NC groups was detected by immunohistochemistry. The results are shown as the mean ± SD. **P < 0.01, ***P < 0.001
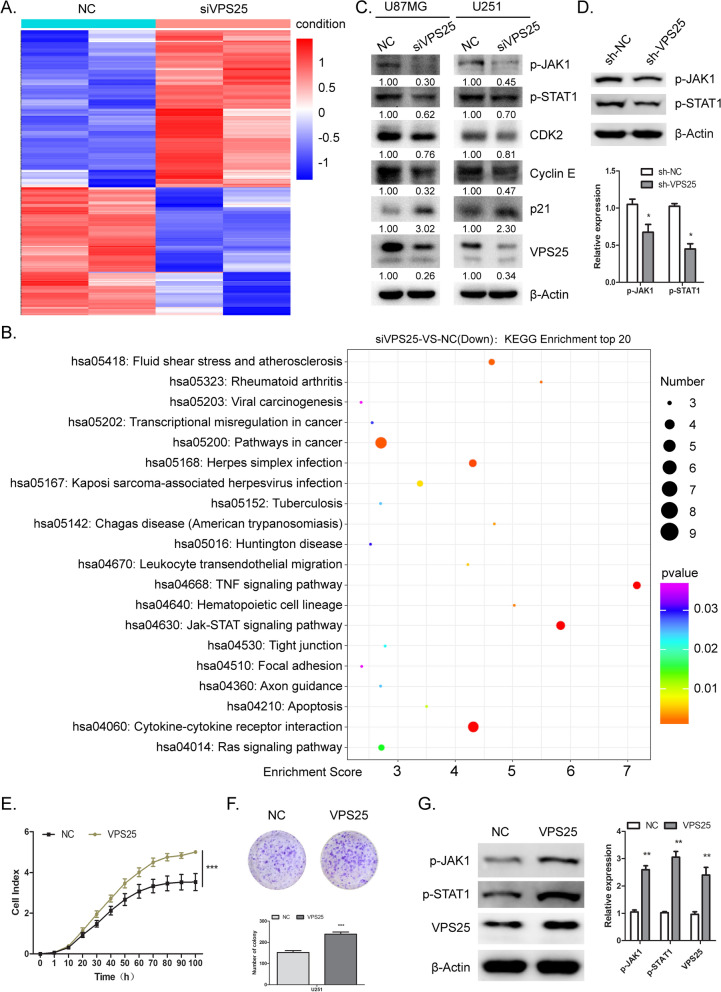
VPS25 regulates JAK-signal transducer and the activator of transcription (STAT) signaling pathway
To investigate the mechanism of VPS25 in glioma, we detected the DEG in VPS25-silenced U251 cells by RNA-seq. We identified 134 upregulated genes and 109 downregulated genes after VPS25 KD (Fig. 6A and Additional file 7: Table S1). KEGG pathway enrichment analysis showed that the TNF signaling pathway, JAK-STAT signaling pathway, cytokine-cytokine receptor interaction, and herpes simplex infection were downregulated (Fig. 6B and Additional file 8: Table S2). The JAK-STAT signaling pathway plays a significant role in the cell cycle of various cancers [27]. We then checked the activity of JAK-STAT signaling by western blot in U87MG and U251 cells, as well as the VPS25 KD tumor tissue in vivo. Phospho-JAK1 and phospho-STAT1 were decreased when VPS25 was silenced (Fig. 6C and D). However, VPS25 depletion did not affect other JAK and STAT proteins (Additional file 9: Fig. S7). After that, we detected the expression of various cell proliferation and cell cycle regulatory proteins known to be downstream targets of the JAK-STAT signaling pathway. We found that the cyclin-dependent kinase inhibitor p21 was significantly upregulated, whereas the cyclin E and cyclin-dependent kinase CDK2 were downregulated with reduced VPS25 (Fig. 6C). These results were consistent with our cell cycle data showing that VPS25 KD led to a G0/G1 phase arrest. Then we employed another independent VPS25 siRNA (siVPS25-1) to detect the function of VPS25 in glioma. VPS25 knockdown inhibited the proliferation and blocked the cell cycle. Furthermore, VPS25 knockdown reduced the expression of p-JAK1 and p-STAT1 (Additional file 10: Fig. S8).
Fig. 6.
The genes regulated by VPS25 in glioma. A Heat map representing the DEGs regulated by VPS25 KD. B KEGG pathway analysis showing the most enriched pathways of downregulated genes after VPS25 KD. C Western blotting was used to determine the levels of downstream target proteins in VPS25-silenced U87MG and U251 cells. D Western blotting was used to determine the levels of p-JAK1 and p-STAT1 in VPS25 KD tumor tissue in vivo. E RTCA xCELLigence assay showed that transfection of a VPS25-encoding plasmid promoted the proliferation of U251 cells. F Transfection of the VPS25-encoding plasmid promotes the colony formation of U251 cells. G Western blotting was used to determine the levels of p-JAK1 and p-STAT1 in VPS25 overexpressed U251 cells. β-Actin was used as a loading control. NC: negative control. siVPS25: VPS25 gene silencer; VPS25: VPS25 overexpression plasmid. *P < 0.05, **P < 0.01, ***P < 0.001
To further elucidate whether VPS25 mediates proliferation through the JAK-STAT pathway, we overexpressed VPS25 in the glioma cells. The results of the RTCA assay showed that VPS25 overexpression promoted the proliferation of U251 cells (Fig. 6E). In addition, the colony-forming ability of U251 cells was increased due to VPS25 overexpression (Fig. 6F). Furthermore, VPS25 overexpression increased the protein expression of p-JAK1 and p-STAT1, noticeably in U251 (Fig. 6G).
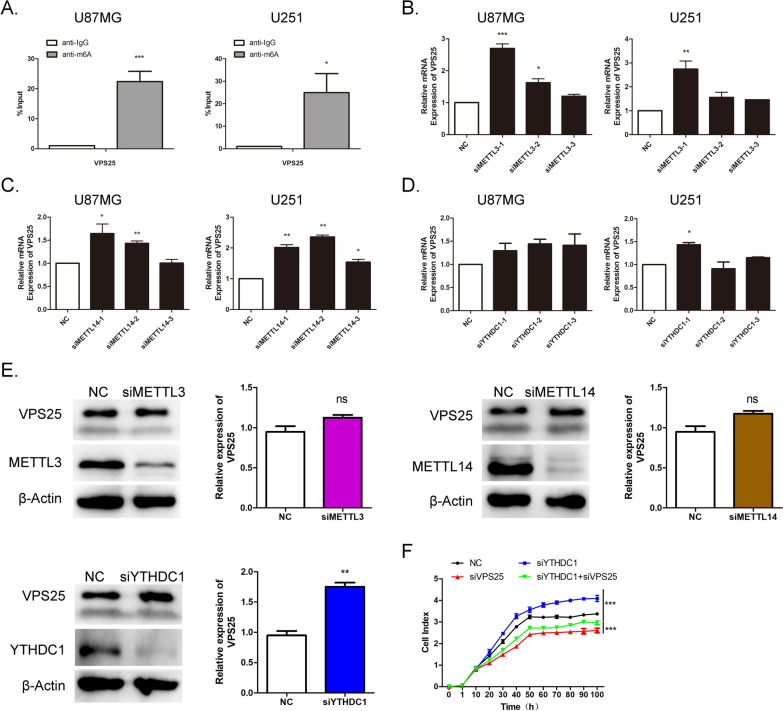
m6A modification is associated with VPS25 expression in glioma cells
Recent studies in epigenetic mechanisms have shown that m6A modification in mRNAs plays critical roles in various cancers. Therefore, we investigated whether the upregulation of VPS25 in glioma was associated with the m6A modification. According to the RMBase v2.0 database (http://rna.sysu.edu.cn/rmbase/) [28], multiple m6A modification sites were found in VPS25, and YTHDC1 is an RNA-binding protein (RBP) of VPS25. Therefore, we detected the m6A modification of VPS25 in glioma cells by m6A RIP-qPCR analysis. Compared with the IgG control, the m6A antibody levels of VPS25 in U87MG and U251 cells were increased 22- and 25-fold, respectively (Fig. 7A).
Fig. 7.
m6A modification is associated with VPS25 expression in glioma cells. A m6A RIP-qPCR analysis showed that m6A is highly enriched within the VPS25 transcript in U87MG and U251 cells. B The expression of VPS25 was detected in U87MG and U251 cells with METTL3 KD. C The expression of VPS25 was detected in U87MG, and U251 cells with METTL14 KD. D The expression of VPS25 was detected in U87MG and U251 cells with YTHDC1 KD. E Western blot was used to detect the VPS25 expression in METTL3, METTL14, or YTHDC1 KD cells. F U251 glioma cells were used to evaluate proliferation after being transfected with siVPS25 and siYTHDC1. *P < 0.05, **P < 0.01, ***P < 0.001. ns: no significant, NC: negative control. siMETTL3: METTL3 gene silencer; siMETTL14: METTL14 gene silencer; siYTHDC1: YTHDC1 gene silencer; siVPS25: VPS25 gene silencer
METTL3 and METTL14 are essential m6A methyltransferases and have been reported to be involved in tumorigenesis of glioblastoma stem cells [29]. YTHDC1 is an m6A-binding nuclear protein with a YTH domain [30]. To explore the possible mechanisms of m6A-regulated expression of VPS25, METTL3, METTL14, and YTHDC1 were knocked down in glioma cells. RT-qPCR assays in U87MG and U251 verified the KD efficiency (Additional file 11: Fig. S9A–C). The m6A quantitative analysis showed that the METTL3 and METTL14 KD decreased the total m6A modification level in glioma cells, whereas the YTHDC1 KD did not (Additional file 11: Fig. S9D). The mRNA level of VPS25 was upregulated in glioma cells with KD of METTL3 or METTL14, but it was not affected in the YTHDC1 KD cells (Fig. 7B–D). However, the protein level of VPS25 was not affected in glioma cells with KD of METTL3 or METTL14, but it was upregulated in the YTHDC1 KD cells (Fig. 7E). These data suggest that METTL3 or METTL14-mediated m6A is associated with the downregulation of VPS25 mRNA in glioma. In turn, YTHDC1 may regulate the protein expression of VPS25.
We then speculated that METTL3, METTL14, and YTHDC1 might affect the proliferation of glioma cells through VPS25. We decreased VPS25 expression in METTL3, METTL14, and YTHDC1 KD glioma cells, and a series of restoration assays were performed. KD of METTL3 and METTL14 inhibited cell proliferation, whereas KD of YTHDC1 promoted cell proliferation. Silencing VPS25 decreased the proliferation in YTHDC1-KD U251 cells, but not in METTL3 and METTL14-KD cells (Fig. 7F and Additional file 11: Fig. S9E and F). Together, these results indicate that YTHDC1 may inhibit glioma proliferation by reducing VPS25 expression.
Discussion
ESCRT acts as a complex primarily to sort ubiquitinated receptors into MVBs, which are then degraded by the lysosomes. During this process, each ESCRT subunit performs its functions concomitantly on membrane bending and scission [31]. In this study, we screened the expression of ESCRT subunits in glioma from TCGA and GEO databases. However, the expression trends of different ESCRT subunits are inconsistent. For instance, MVB12A, VPS25, CHMP2A, and IST1 were significantly upregulated in glioma, whereas VPS36 and CHMP1B were significantly downregulated (Table 1). As reported previously, ESCRT components have been implicated in both oncogenic and tumor suppressor function in various types of human cancer [17, 18]. Interestingly, VPS25 and VPS36, which belong to ESCRT-II, have opposite trends. VPS36 are under-expressed in advanced prostate cancer and have been associated with prostate cancer cell proliferation [32]. Therefore, we speculate that VPS25 may function independently of the ESCRT complex in glioma.
The ESCRT complex has been confirmed to participate in several key biological functions of cancer development, including proliferation, cell cycle, invasion, autophagy, and exosomes [8]. However, the function of VPS25 in tumors remains largely unknown. Here, we determined that the VPS25 deficiency significantly inhibited glioma cell proliferation by inducing the G0/G1 phase arrest of the cell cycle (Figs. 2 and 3). By western blot, we found that VPS25 regulates the protein expression of CDK2, cyclin E, and p21 (Fig. 6). Cyclin E binds and activates CDK2, and thus promotes transition through G1/S and G2/M phases. Meanwhile, p21 encodes a cyclin-dependent kinase inhibitor, which binds to and inhibits the activity of cyclin E-CDK2 complexes. Therefore, p21 mediates cell cycle arrest at the G1/S and G2/M phases [33]. Previous studies have shown that the RAS association domain family 1 isoform A (RASSF1A) induces cell cycle arrest in the G1 phase with observed reduced expression of cyclin E and increased levels of p21 [34]. In addition, PKD1 expression induces cell cycle arrest in G0/G1 and upregulates p21 [35]. As a component of the ESCRT-1 complex, TSG101 deficiency in cancer cells was accompanied by growth inhibition, cell cycle arrest, apoptosis, and an increased abundance of p21 mRNA and protein [36]. These data strongly suggest that VPS25 may regulate its effects on the cell cycle and proliferation of glioma cells mainly by regulating cyclin E, CDK2, and p21 expression.
We further investigated the mechanism by which VPS25 promotes glioma progression. JAK (janus kinase)-STAT (signal transducer and activator of transcription) signaling pathway is crucial in various physiological processes, including immune function, cell growth, differentiation, and hematopoiesis [37]. Studies have shown that its deregulation contributes to the emergence and progression of various cancers, including glioma [38]. Previous research showed that inhibition of the JAK-STAT signaling pathway blocked the cell cycle at the G0/G1 phase [39]. Moreover, dysregulation of the JAK-STAT signaling pathway in Drosophila VPS25 mutant tissues is associated with tumorigenesis [40]. In this study, the phosphorylation of JAK-STAT was downregulated by VPS25 KD, whereas it was upregulated by VPS25 overexpression. This finding indicates that VPS25 may regulate glioma growth through the JAK-STAT pathway.
N6-methyladenosine (m6A) is the most abundant internal modification in eukaryotic mRNAs [41]. The METTL3 and METTL14, which are the methyltransferase complex's main components, catalyzed the formation of m6A modification [42]. YTHDC1 has been identified as an m6A direct reader that recognizes m6A-modified RNAs [30]. Recent research shows that KD of METTL3 or METTL14 promotes GSC self-renewal and tumorigenesis by upregulating the expression of oncogenes, such as ADAM19 [43]. YTHDC1 also mediates nuclear export and splicing of N6-methyladenosine methylated mRNAs [44, 45]. We revealed that METTL3 or METTL14 only regulates the mRNA expression of VPS25. Furthermore, YTHDC1 only regulates VPS25 protein expression. We also demonstrated that YTHDC1 affects the proliferation of glioma cells by regulating VPS25 expression. As a nuclear m6A reader, YTHDC1 promotes PTEN mRNA degradation to increase AKT phosphorylation, thus facilitating neuronal survival after ischemia [46]. This molecular mechanism should be further investigated to elucidate whether YTHDC1 affects the JAK-STAT pathway through decreases in VPS25 mRNA stability.
Conclusions
In summary, we determined the VPS25 expression and its biological function in glioma. We found that VPS25 is overexpressed in glioma and promotes the proliferation of glioma. Additionally, VPS25 targets the p21 and JAK-STAT signaling pathways. Thus, this study not only elucidates the VPS25-dependent regulation of glioma but also provides a new putative biomarker and therapeutic target for glioma diagnosis and treatment. However, the application of VPS25 in clinical practice needs further research.
Supplementary Information
Additional file 1: Figure S1. mRNA expression of all ESCRT subunits in glioma from TCGA database. *P < 0.05, **P < 0.01, ***P < 0.001. Normal: normal brain tissue; Glioma: glioma tissue.
Additional file 2: Figure S2. mRNA expression of all ESCRT subunits in glioma from GEO database. *P < 0.05, **P < 0.01, ***P < 0.001. Normal: normal brain tissue; Glioma: glioma tissue.
Additional file 3: Figure S3. Expression of ESCRT subunits in glioma tissues based on tumor grade. (A) Expression of MVB12A, VPS25, CHMP2A, and IST1 in WHO grade II and WHO grade III/IV from GEO database. (B) Expression of VPS25 in WHO grade II and WHO grade III from TCGA database. (C) Expression of CHMP2A in WHO grade II and WHO grade III from TCGA database. (D) The overall survival of LGG or GBM patients with high and low levels of VPS25 was plotted from GEPIA2. (E) The correlation between VPS25 expression with IDH mutation status in glioma. *P < 0.05, ***P < 0.001.
Additional file 4: Figure S4. The protein expression of VPS25 in glioma. (A) The VPS25 was detected by western blot in GBM (n = 8) and normal brain tissues (n = 14). (B) Statistics of the VPS25 expression in A. N: normal brain tissues, G: GBM tissues, ***P < 0.001.
Additional file 5: Figure S5. The knockdown of VPS25 in glioma cells. (A) The relative VPS25 expression level in 2 glioma cell lines (U87MG and U251) was compared with those in 14 non tumor brain tissues. (B) The U87MG cells, which were transfected with NC and siVPS25-1, -2, -3 for 48 h, were harvested. Then RT-qPCR detected the expression of VPS25 in U87MG cells. (C) RT-qPCR detected the expression of VPS25 in U251 cells transfected with siRNA. Experiments were performed and presented as shown in B. ***P < 0.001. NC: negative control. siVPS25: VPS25 gene silencer.
Additional file 6: Figure S6. VPS25 regulates glioma cell apoptosis. (A) TUNEL assays were performed on U87MG cells, statistical analysis is shown on the right. (B) TUNEL assays were performed on U251 cells, statistical analysis is shown on the right. Scale bars, 100 μm. Data are mean ± SD from three independent experiments. * P < 0.05, NC: negative control. siVPS25: VPS25 gene silencer.
Additional file 7: Table S1. The mRNAs expression between NC and siVPS25.
Additional file 8: Table S2. Top KEGG enrichment for the down-expressed mRNAs.
Additional file 9: Figure S7. The p-JAK2,3 and p-STAT2,3 was detected by western blot in NC and siVPS25 glioma cells.
Additional file 10: Figure S8. The function of siVPS25-1 in glioma cells. (A) RTCA xCELLigence assay showed that transfection of siVPS25-1 suppressed the proliferation of U251 cells. (B) Transfection of siVPS25-1 suppressed the colony formation of U251 cells. (C) The VPS25-silenced U251 cells were arrested at the G0/G1 phase of the cell cycle. (D) The p-JAK1 and p-STAT1 was detected by western blot in NC and siVPS25-1 glioma cells. Data are mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. NC: negative control. siVPS25: VPS25 gene silencer.
Additional file 11: Figure S9. The knockdown of METTL3, METTL14, and YTHDC1 in glioma cells. (A) RT-qPCR was performed to verify the knockdown efficiency of METTL3 in U87MG and U251 cells. (B) RT-qPCR was performed to verify the knockdown efficiency of METTL14 in U87MG and U251 cells. (C) RT-qPCR was performed to verify the knockdown efficiency of YTHDC1 in U87MG and U251 cells. (D) The total m6A modification level was detected in METTL3, METTL14, and YTHDC1-knockdown cells. (E) U251 glioma cells were used to evaluate proliferation after being transfected with siVPS25 and siMETTL3. (F) U251 glioma cells were used to evaluate proliferation after being transfected with siVPS25 and siMETTL14. *P < 0.05, **P < 0.01, ***P < 0.001. ns: no significant.
Acknowledgements
We thank all the contributors for this work.
Abbreviations
- CNS
Central nervous system
- WHO
World Health Organization
- NCCR
National Central Cancer Registry of China
- ESCRT
Endosomal sorting complex required for transport
- ILVs
Intraluminal vesicles
- MVBs
Multivesicular bodies
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- NC
Negative control
- KD
Knockdown
- KEGG
Kyoto encyclopedia of genes and genomes
- m6A
N6-Methyladenosine
- RBP
RNA-binding protein
- RASSF1A
RAS association domain family 1 isoform A
- JAK
Janus kinase
- STAT
Signal transducer and activator of transcription
- GEPIA2
Gene Expression Profiling Interactive Analysis 2
- GTEx
Genotype-Tissue Expression
- ATCC
American Type Culture Collection
- DMEM
Dulbecco's modified Eagle medium
- FBS
Fetal bovine serum
- PI
Propidium iodide
- DEGs
Differentially expressed genes
- ECL
Enhanced chemiluminescence
- RIP
RNA immunoprecipitation
- GBM
Glioblastoma multiforme
- LGG
Lower grade glioma
Authors' contributions
KL and XZ designed the experiments. XZ, HY, MZ, XW, LJ and XL performed the experiments and analyzed the data. XZ wrote the manuscript. KL and XZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81802503 to H.Y., 81701557 to X.Q.L., 81870017 to Y.Y.Z., and 81772180, 82072370 to K.L.), Key Projects of Natural Science Research of Universities in Anhui Province (Grant No. KJ2020A0607 to X.C.L.), Natural Science Foundation of Anhui Province (Grant Nos. 2008085QH367 to X.L.Z. and 1908085QH380 to T.B.C.), Funding of "Peak" Training Program for Scientific Research of Yijishan Hospital, Wannan Medical College (Grant Nos. GF2019G10 to X.L.Z. and GF2019G15 to H.Y.).
Availability of data and materials
The data used to support the findings of this study are included within the article.
Declarations
Ethics approval and consent to participate
The use of patient’s tissues was reviewed and approved by the Ethics Committee of the Yijishan Hospital of Wannan Medical College. The consent was obtained from all patients before specimen collection.
Consent for publication
The publication of this manuscript has been approved by all authors.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaolong Zhu and Hui Yang contributed equally to this work
References
- 1.Grassi ES, Pantazopoulou V, Pietras A. Hypoxia-induced release, nuclear translocation, and signaling activity of a DLK1 intracellular fragment in glioma. Oncogene. 2020;39:4028–4044. doi: 10.1038/s41388-020-1273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Masui K, Kato Y, Sawada T, Mischel PS, Shibata N. Molecular and genetic determinants of glioma cell invasion. Int J Mol Sci. 2017;18:2609. doi: 10.3390/ijms18122609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park DM, Sathornsumetee S, Rich JN. Medical oncology: treatment and management of malignant gliomas. Nat Rev Clin Oncol. 2010;7(2):75–77. doi: 10.1038/nrclinonc.2009.221. [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre C, Largeau C, Michelet X, Fourrage C, Maniere X, Matic I, Legouis R, Culetto E. The ESCRT-II proteins are involved in shaping the sarcoplasmic reticulum in C. elegans. J Cell Sci. 2016;129(7):1490–1499. doi: 10.1242/jcs.178467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pieper GH, Sprenger S, Teis D, Oliferenko S. ESCRT-III/Vps4 controls heterochromatin-nuclear envelope attachments. Dev Cell. 2020;53(1):27–41. doi: 10.1016/j.devcel.2020.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilievska J, Bishop NE, Annesley SJ, Fisher PR: The Roles of ESCRT proteins in healthy cells and in disease. Current Frontiers and Perspectives in Cell Biology, Prof. Stevo Najman (Ed.), ISBN: 978-953-51-0544-2: InTech. 2012.
- 9.Buysse D, Pfitzner AK, West M, Roux A, Odorizzi G. Doa4 directly binds Snf7 to inhibit the recruitment of ESCRT-III remodeling factors. J Cell Sci. 2020;133(8):jcs241455. doi: 10.1242/jcs.241455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem Sci. 2017;42(1):42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goliand I, Nachmias D, Gershony O, Elia N. Inhibition of ESCRT-II–CHMP6 interactions impedes cytokinetic abscission and leads to cell death. Mol Biol Cell. 2014;25(23):3740–3748. doi: 10.1091/mbc.E14-08-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9(5):687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 14.Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9(5):711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Meng B, Ip NCY, Prestwood LJ, Abbink TEM, Lever AML. Evidence that the endosomal sorting complex required for transport-II (ESCRT-II) is required for efficient human immunodeficiency virus-1 (HIV-1) production. Retrovirology. 2015;12(1):72. doi: 10.1186/s12977-015-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattissek C, Teis D. The role of the endosomal sorting complexes required for transport (ESCRT) in tumorigenesis. Mol Membr Biol. 2014;31(4):111–119. doi: 10.3109/09687688.2014.894210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Ou Y, Cheng X. Role of TSG101 in cancer. Front Biosci. 2013;18:279–288. doi: 10.2741/4099. [DOI] [PubMed] [Google Scholar]
- 18.Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, Zhou R, Shang CZ, Cao J, He H. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61(4):1284–1294. doi: 10.1002/hep.27660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lixin S, Ai-Min H, Qin S, Alexander V, Yuri K, Sandra P, Antonino P, Jayant M, Jennifer W, Rolando B. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao J, Zhang M, Zhong M, Zhang Y, Lv K. MicroRNA-204, a direct negative regulator of ezrin gene expression, inhibits glioma cell migration and invasion. Mol Cell Biochem. 2014;396(1–2):117–128. doi: 10.1007/s11010-014-2148-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D, Zhou H, Liu J, Mao J. Long noncoding RNA ASB16-AS1 promotes proliferation, migration, and invasion in glioma cells. Biomed Res Int. 2019;2019:5437531. doi: 10.1155/2019/5437531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Cai J, Zhou F, Wu Z, Li D, Li Y, Xie Z, Zhou Y, Liang Y. Genome-wide screening of budding yeast with honokiol to associate mitochondrial function with lipid metabolism. Traffic. 2018;19(11):867–878. doi: 10.1111/tra.12611. [DOI] [PubMed] [Google Scholar]
- 24.Zhao W, Cui Y, Liu L, Ma X, Qi X, Wang Y, Liu Z, Ma S, Liu J, Wu J. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-Myc stability via YTHDF1-mediated m6A modification. Mol Ther Nucleic Acids. 2020;20:1–12. doi: 10.1016/j.omtn.2020.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, Pan B, He B, Pan Y, Sun H, et al. METTL14 suppresses crc progression via regulating N6- methyladenosine-dependent primary miR-375 processing. Mol Ther. 2020;28(2):599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Zhu X, Zou S, Li Y, Liang Y. Transcriptomic analysis of Saccharomyces cerevisiae upon honokiol treatment. Res Microbiol. 2017;168(7):626–635. doi: 10.1016/j.resmic.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Ashrafizadeh M, Rafiei H, Mohammadinejad R, Afshar EG, Farkhondeh T, Samarghandian S. Potential therapeutic effects of curcumin mediated by JAK/STAT signaling pathway: a review. Phytother Res. 2020;34:1745–1760. doi: 10.1002/ptr.6642. [DOI] [PubMed] [Google Scholar]
- 28.Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zheng LL, Qu LH, Yang JH. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46:D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng X, Su R, Feng X, Wei M, Chen J. Role of N6-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10(11):927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 31.Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Curr Opin Cell Biol. 2009;21(4):568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worst TS, Meyer Y, Gottschalt M, Weis CA, Von HJ, Frank C, Steidler A, Michel MS, Erben P. RAB27A, RAB27B and VPS36 are downregulated in advanced prostate cancer and show functional relevance in prostate cancer cells. Int J Oncol. 2017;50(3):920–932. doi: 10.3892/ijo.2017.3872. [DOI] [PubMed] [Google Scholar]
- 33.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 34.Thaler S, Hahnel PS, Schad A, Dammann R, Schuler M. RASSF1A mediates p21Cip1/Waf1-dependent cell cycle arrest and senescence through modulation of the Raf-MEK-ERK pathway and inhibition of Akt. Cancer Res. 2009;69(5):1748–1757. doi: 10.1158/0008-5472.CAN-08-1377. [DOI] [PubMed] [Google Scholar]
- 35.Bhunia AK, Piontek K, Boletta A, Liu L, Qian F, Xu PN, Germino FJ, Germino GG. PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell. 2002;109(2):157–168. doi: 10.1016/s0092-8674(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 36.Young TW, Rosen DG, Fang C. Mei, Nan L, Jinsong L, Xiao-Fan W, Xiaodong C: Up-regulation of tumor susceptibility gene 101 conveys poor prognosis through suppression of p21 expression in ovarian cancer. Clin Cancer Res. 2007;13(13):3848–3854. doi: 10.1158/1078-0432.CCR-07-0337. [DOI] [PubMed] [Google Scholar]
- 37.Groner B, von Manstein V. Jak Stat signaling and cancer: Opportunities, benefits and side effects of targeted inhibition. Mol Cell Endocrinol. 2017;451:1–14. doi: 10.1016/j.mce.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 38.Rani A, Murphy JJ. STAT5 in cancer and immunity. J Interferon Cytokine Res. 2016;36(4):226–237. doi: 10.1089/jir.2015.0054. [DOI] [PubMed] [Google Scholar]
- 39.Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10(3):287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodfield SE, Graves HK, Hernandez JA, Bergmann A. De-regulation of JNK and JAK/STAT signaling in ESCRT-II mutant tissues cooperatively contributes to neoplastic tumorigenesis. PLoS ONE. 2013;8(2):e56021. doi: 10.1371/journal.pone.0056021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61(4):507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Wang Q, Zhao X, Shao L, Liu G, Zheng X, Xie L, Zhang Y, Sun C, Xu R. YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 2020;11(11):977. doi: 10.1038/s41419-020-03186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. mRNA expression of all ESCRT subunits in glioma from TCGA database. *P < 0.05, **P < 0.01, ***P < 0.001. Normal: normal brain tissue; Glioma: glioma tissue.
Additional file 2: Figure S2. mRNA expression of all ESCRT subunits in glioma from GEO database. *P < 0.05, **P < 0.01, ***P < 0.001. Normal: normal brain tissue; Glioma: glioma tissue.
Additional file 3: Figure S3. Expression of ESCRT subunits in glioma tissues based on tumor grade. (A) Expression of MVB12A, VPS25, CHMP2A, and IST1 in WHO grade II and WHO grade III/IV from GEO database. (B) Expression of VPS25 in WHO grade II and WHO grade III from TCGA database. (C) Expression of CHMP2A in WHO grade II and WHO grade III from TCGA database. (D) The overall survival of LGG or GBM patients with high and low levels of VPS25 was plotted from GEPIA2. (E) The correlation between VPS25 expression with IDH mutation status in glioma. *P < 0.05, ***P < 0.001.
Additional file 4: Figure S4. The protein expression of VPS25 in glioma. (A) The VPS25 was detected by western blot in GBM (n = 8) and normal brain tissues (n = 14). (B) Statistics of the VPS25 expression in A. N: normal brain tissues, G: GBM tissues, ***P < 0.001.
Additional file 5: Figure S5. The knockdown of VPS25 in glioma cells. (A) The relative VPS25 expression level in 2 glioma cell lines (U87MG and U251) was compared with those in 14 non tumor brain tissues. (B) The U87MG cells, which were transfected with NC and siVPS25-1, -2, -3 for 48 h, were harvested. Then RT-qPCR detected the expression of VPS25 in U87MG cells. (C) RT-qPCR detected the expression of VPS25 in U251 cells transfected with siRNA. Experiments were performed and presented as shown in B. ***P < 0.001. NC: negative control. siVPS25: VPS25 gene silencer.
Additional file 6: Figure S6. VPS25 regulates glioma cell apoptosis. (A) TUNEL assays were performed on U87MG cells, statistical analysis is shown on the right. (B) TUNEL assays were performed on U251 cells, statistical analysis is shown on the right. Scale bars, 100 μm. Data are mean ± SD from three independent experiments. * P < 0.05, NC: negative control. siVPS25: VPS25 gene silencer.
Additional file 7: Table S1. The mRNAs expression between NC and siVPS25.
Additional file 8: Table S2. Top KEGG enrichment for the down-expressed mRNAs.
Additional file 9: Figure S7. The p-JAK2,3 and p-STAT2,3 was detected by western blot in NC and siVPS25 glioma cells.
Additional file 10: Figure S8. The function of siVPS25-1 in glioma cells. (A) RTCA xCELLigence assay showed that transfection of siVPS25-1 suppressed the proliferation of U251 cells. (B) Transfection of siVPS25-1 suppressed the colony formation of U251 cells. (C) The VPS25-silenced U251 cells were arrested at the G0/G1 phase of the cell cycle. (D) The p-JAK1 and p-STAT1 was detected by western blot in NC and siVPS25-1 glioma cells. Data are mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. NC: negative control. siVPS25: VPS25 gene silencer.
Additional file 11: Figure S9. The knockdown of METTL3, METTL14, and YTHDC1 in glioma cells. (A) RT-qPCR was performed to verify the knockdown efficiency of METTL3 in U87MG and U251 cells. (B) RT-qPCR was performed to verify the knockdown efficiency of METTL14 in U87MG and U251 cells. (C) RT-qPCR was performed to verify the knockdown efficiency of YTHDC1 in U87MG and U251 cells. (D) The total m6A modification level was detected in METTL3, METTL14, and YTHDC1-knockdown cells. (E) U251 glioma cells were used to evaluate proliferation after being transfected with siVPS25 and siMETTL3. (F) U251 glioma cells were used to evaluate proliferation after being transfected with siVPS25 and siMETTL14. *P < 0.05, **P < 0.01, ***P < 0.001. ns: no significant.
Data Availability Statement
The data used to support the findings of this study are included within the article.