Abstract
We deduced the structure of the mouse profilin II gene. It contains five exons that can generate four different transcripts by alternative splicing. Two transcripts encode different profilin II isoforms (designated IIa and IIb) that have similar affinities for actin but different affinities for polyphosphoinositides and proline-rich sequences. Profilins IIa and IIb are also present in humans, suggesting that all mammals have three profilin isoforms. Profilin I is the major form in all tissues, except in the brain, where profilin IIa is most abundant. Profilin IIb appears to be a minor form, and its expression is restricted to a limited number of tissues, indicating that the alternative splicing is tightly regulated. Western blotting and whole-mount in situ hybridization show that, in contrast to the expression of profilin I, the expression level of profilin IIa is developmentally regulated. In situ hybridization of adult brain sections reveals overlapping expression patterns of profilins I and IIa.
Changes in both cell shape and motility in response to extracellular signals require mechanical forces from the actin cytoskeleton. The formations of lamellipodia, focal contacts, the contractile ring during cytokinesis, and neuronal growth cone motility depend on the controlled polymerization-depolymerization of actin filaments (7, 40). The regulation of these processes is accomplished by many actin binding proteins that act at different points in time and space, depending on the extracellular signals (44).
Profilin is a small, ubiquitous actin binding protein, thought to be a key regulatory of actin dynamics in living cells (8, 41). In vitro experiments have shown that profilin acts as an actin monomer-sequestering protein when barbed ends of filaments are capped (e.g., by gelsolin). When the capping protein is removed, polymerization of actin can occur, and actin-profilin complexes can add to the fast-growing ends, thereby enhancing actin polymerization in the presence of thymosin β4 (22, 34).
Profilin can bind other molecules including phosphatidylinositol 4,5-bisphosphate (PIP2) (28) and proline-rich domains of several proteins like the vasodilator-stimulated phosphoprotein (VASP), Enabled (Ena), mammalian Ena (Mena), aczonin, the Wiskott-Aldrich syndrome protein (WASP), its neural homologue N-WASP, and mammalian Diaphanous (p140mDia) (1, 2, 13, 38, 45, 46, 48). In lower eukaryotes, many other proline-rich proteins have been identified as profilin ligands, such as formin-homology proteins (47). The actin-related protein-2/3 complex (Arp2/3 complex), composed of seven different polypeptides, was originally identified as a profilin binding complex (29).
Until 1993, only one profilin isoform was known to be present in mammalian cells. Honoré and coworkers (20) discovered a second profilin gene in a random cDNA cloning project. The open reading frame predicted a protein with a high similarity to profilin I (62.1% identity), and therefore the protein was designated profilin II. Northern blot analysis showed that the expression levels of the two profilins were complementary in many tissues. Profilin I expression was highest in placenta, lung, liver, and kidney, while profilin II transcripts were most abundant in brain, skeletal muscle, and kidney. However, three transcripts of different lengths that hybridized with the coding as well as noncoding regions of the profilin II cDNA were observed for profilin II.
In 1995, two papers reported conflicting data on the biochemical characterization of profilin II. We purified and characterized profilin II from bovine brain, taking advantage of the low expression of profilin I and high expression of profilin II in this organ (25). In an independent study, Gieselmann and coworkers analyzed the human recombinant profilin II form, produced in Escherichia coli (14). We found that bovine profilins I and II have similar affinities for actin and that profilin I has a higher affinity for PIP2 while profilin II binds more strongly to poly-l-proline. We subsequently showed that profilin II recruits VASP from bovine brain extracts, while profilin I does not. In contrast, profilin I prefers binding to PIP2 over poly-l-proline in a competition assay (26). The results of Gieselmann and coworkers (14) were strikingly different and showed that human profilins I and II have similar affinities for PIP2 and poly-l-proline, though profilin I has a five-times-higher affinity for actin.
The data that we present in this paper show that the two groups worked with different profilin II isoforms (designated profilin IIa and profilin IIb) that are generated by alternative splicing. The different carboxy-terminal parts confer different biochemical properties on the proteins. The genomic sequence of profilin II reveals a complex gene structure. In addition to the four exons necessary for the formation of the two profilin isoforms, a fifth (noncoding) exon is present, potentially giving rise to truncated profilin forms. The profilin IIb message is detected in only a few tissues and is present at much lower levels than is profilin IIa. Furthermore, we provide evidence that the expression patterns of profilin I and of profilin IIa are different during development and that profilin IIa is the major form in neural tissues. In the adult brain, expression of profilin I and that of profilin II are overlapping.
MATERIALS AND METHODS
Recombinant DNA methods.
Total RNAs from various adult mouse and rat tissues and from mouse developmental stages were prepared by the guanidinium isothiocyanate method (11). Oligo(dT)-primed reverse transcriptions (RT) were carried out using the Superscript Preamplification system (mouse) or Thermoscript R (rat) (both from GIBCO BRL). PCRs were performed with Taq DNA polymerase (GIBCO BRL) using the primers listed in Table 1. The amplified products were separated on 2% agarose gels and stained with ethidium bromide. The intensities of the bands were quantified with the Gel Pro Analyzer (Techtum) package. The same amount of cDNA and the same PCR protocol were used to amplify β-actin as a control. 5′ rapid amplification of cDNA ends (RACE) of the profilin II cDNA was done using mouse brain Marathon RACE-ready cDNA (Clontech) and the Advantage Klen Taq Polymerase mix (Clontech) according to the manufacturer's instructions. The first amplification was carried out with the adapter primer AP1 (Clontech) and the gene-specific primer exon 3 reverse (Table 1). The second, nested amplification was performed using primer AP2 (Clontech) and the gene-specific primer exon 2 reverse.
TABLE 1.
Sequences of oligonucleotide primers used in the various experiments
| Procedure and primer name | Sequence |
|---|---|
| PCR (expt in Fig. 2) | |
| Human and rat profilin II forward | GGGCTCGACCATGGCCGGTTGGCAGAGCTACG |
| Human and rat profilin IIa reverse | CCTAGCAGCTAGAACCCAGAGTCTCTC |
| Human and rat profilin IIb reverse | GGGAGAGGCTGCTTATACATCAGACCTC |
| Mouse exon 1A forward (E1Af) | CCGCCATTGTCGGCTACTGC |
| Mouse exon 1B forward (E1Bf) | CTGGATCTGGTTCGGTTGTG |
| Mouse exon 3 reverse (E3r) | GAGCAATTTTCCCCTAATAC |
| Mouse exon 4 reverse (E4r) | CGTACAGAGGCATGCACT |
| Mouse β-actin forward | GTGGGCCGCTCTAGGCACCAA |
| Mouse β-actin reverse | CTCTTTGATGTCACGCACGATTTC |
| 5′ RACE | |
| Exon 3 reverse | GAGCAATTTTCCCCTAATAC |
| Exon 2 reverse | GTCAAACCGTTGGTAAAGAA |
Nucleotide sequencing of genomic DNA and mouse PCR products was performed with the Thermo Sequenase Dye Terminator Cycle Sequencing premix kit (Amersham). Sequence reaction products were resolved on an ABI Prism 377 Automated Sequencer (Applied Biosystems). Nucleotide sequence analysis was performed using the GCG software package (Genetics Computer Group). Dideoxy DNA sequencing on human and rat PCR products was done using the T7 sequencing kit from Pharmacia on Wizard (Promega)-purified plasmid DNA with M13 forward and reverse primers.
To clone the profilin II gene, we screened a P1-derived artificial chromosome library containing mouse genomic DNA (United Kingdom HGMP Resource Centre, Cambridge, United Kingdom) using a 32P-labeled NcoI-PstI fragment of a profilin II mouse expressed sequence tag (EST) clone (AA032658, obtained from the IMAGE consortium). One positive clone was identified and digested with EcoRI and BamHI. Fragments hybridizing with the profilin II cDNA were subcloned into pBluescript KS(+).
Biochemical methods.
We cloned the profilin cDNAs in the NcoI/BamHI sites of the pEt11d vector. This does not result in additional amino acids in the expressed proteins. The proteins were expressed in E. coli strain MC1061 harboring pT7POL26 (32). When the cells reached an optical density at 600 nm of 1, we added IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM, and the cells were induced for 3 to 4 h. Cell pellets were resuspended in 20 mM Tris-HCl (pH 8.1)–1 mM EDTA–5 mM dithiothreitol (buffer A) supplemented with protease inhibitors. Subsequently, we lysed the cells with a French press and cleared the lysate by ultracentrifugation for 1 h at 100,000 × g. We loaded the cleared lysate on a poly(l-proline)–Sepharose column, and after washing the column with buffer A, we eluted the proteins with increasing concentrations of urea (3, 5, and 8 M in buffer A). Bovine profilin IIa was purified from bovine brain (25). α-Actin was purified from rabbit skeletal muscle as described previously (35, 43). Cys-375–pyrene-labeled actin was prepared according to the protocol of Brenner and Korn (6).
We determined the dissociation constant for the α-actin–profilin interaction in the presence of gelsolin-capped filaments as described by Pantaloni and Carlier (34). Time course experiments were carried out with 10 μM α-actin (10% pyrene labeled) and 5 μM profilin using a Hitachi F-4500 spectrophotometer. The excitation and emission wavelengths were 365 and 388 nm, respectively. Polymerization was started by adding MgCl2 and KCl (to final concentrations of 2 and 100 mM, respectively) to the preincubated α-actin–profilin sample in G buffer (5 mM Tris-HCl [pH 7.7], 0.2 mM ATP, 0.2 mM dithiothreitol, 0.1 mM CaCl2).
The affinity of the profilins for polyproline was assayed with surface plasmon resonance (BIACORE X). A VASP-derived peptide [biotin-CGPPPPPGPPPPPGPPPPPGL-OH, (GP5)3] was coupled to the streptavidin-coated sensor chip, and different concentrations of profilin were passed over the chip. Response units were measured for each concentration. The concentration at which half of the maximal response (Rmax/2) is obtained is a measure of affinity (21).
We carried out PIP2 binding as described earlier (18, 26). Briefly, 5 μM profilin was incubated for 30 min on ice with different concentrations of PIP2 micelles as indicated in the legend to Fig. 6. Subsequently, the sample was applied to a Millipore filter with a molecular weight cutoff of 30,000 and centrifuged for 1 min at 4°C. The flowthrough was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
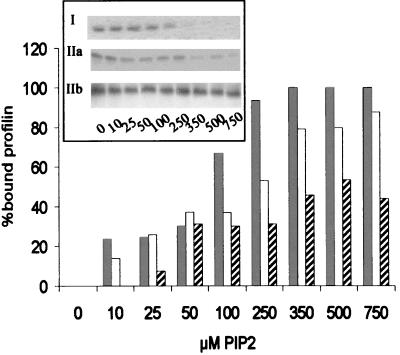
FIG. 6.
Profilin isoforms have different affinities for PIP2. The bars represent the percentages of bound profilin at the PIP2 concentrations indicated. These values were calculated from the amounts of nonbound profilin found in the flowthrough after microfiltration. Values for human profilin I (grey bars), rat profilin IIa (white bars), and human profilin IIb (hatched bars) are shown. The average of two experiments is shown. The inset shows SDS-polyacrylamide gels of the flowthrough fractions.
Western blotting and in situ hybridization on tissue sections.
Dissected organs were homogenized in radioimmunoprecipitation assay buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 1% (vol/vol) Triton X-100, 0.5% (wt/vol) deoxycholate, 0.1% (wt/vol) SDS, 1 μM pepstatin, 1 mM phenylmethylsulfonyl fluoride, 0.3 μM aprotinin, 1 μM leupeptin, 5 μM E-64, 1 mM EDTA, 1 mM sodium vanadate, 50 mM sodium fluoride, 2 mM levamisole, and 30 mM sodium pyrophosphate, and homogenates were centrifuged for 30 min at 100,000 × g. Protein concentrations were determined using the bicinchoninic acid assay (Pierce). Sixty-five micrograms of total protein of each lysate was loaded on SDS–15% polyacrylamide gels. SDS-PAGE and Western blotting were done by standard techniques. The polyclonal antibodies used are G117 for profilin I and G124 for profilin II and a goat anti-rabbit horseradish peroxidase conjugate as secondary antibody. The signal was developed with the Renaissance chemiluminescence reagent (Dupont-NEN). Concentration series of pure profilin I and profilin IIa (indicated in Fig. 7) were used to determine the relative amount of each profilin in the different lysates, by comparing the signal intensities of the Western blots.
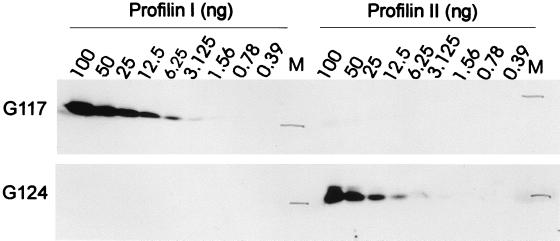
FIG. 7.
Antibodies G117 and G124 distinguish between profilin I and profilin II on Western blots. Decreasing amounts (as indicated) of human profilin I and bovine profilin IIa were used to test the specificity of the antibodies. Based on the intensities on these Western blots, we determined relative amounts of profilin in the lysates (Fig. 8A, B, and D) by performing the Western blottings simultaneously.
In situ hybridization of sections (3) was performed with digoxigenin-dUTP (Boehringer Mannheim)-labeled antisense or sense riboprobes specific for the 3′ untranslated regions of mouse profilin I (49) and profilin II. For whole-mount in situ hybridization, probes were prepared by in vitro transcription of linearized template DNA using digoxigenin-labeled nucleotides. We used the mouse EST sequences coding for either profilin I or profilin II (AA871094 or AA032658, respectively) as DNA templates. In situ hybridization was done as described previously (19).
RESULTS
Comparing EST sequences leads to the identification of two profilin II isoforms.
The conflicting data reported for bovine brain and recombinant human profilin II were attributed to the use of profilin II proteins from different species. In a search for the mouse and rat profilin II cDNAs, we screened mouse and rat EST databases (National Center for Biotechnology Information) using amino acid sequences from both human and bovine profilin II. We found several EST sequences that are very similar to the bovine profilin II isoform. Sequencing mouse EST AA032658 identified a 1,762-bp cDNA sequence containing a 25-bp-long 5′ untranslated region, an open reading frame of 423 bp ending with a TAG stop codon, and a 1,314-bp-long 3′ untranslated region. The open reading frame encodes a putative mouse profilin II of 140 amino acids, which is identical to bovine profilin II (Fig. 1). Interestingly, embedded in the 3′ region was an additional short open reading frame coding for a polypeptide which showed 81% identity to the corresponding COOH-terminal part of human profilin II (pfn2, GenBank accession no. NM_002628) (20) and only 59% to that of bovine profilin II (25). The human profilin EST HS101130 (Fig. 1) is similar to the mouse EST identified above containing potential coding information for two different COOH termini.
FIG. 1.
Alignment of the sequences of the various profilin isoforms and translated cDNAs and ESTs from humans, rats, and mice. The sequences are human (Hum) profilin I (pfn1; GenBank accession no. NM_005022) (24), human profilin IIa (this work; accession no. AF228738), human profilin IIb (pfn2; accession no. NM_002628) (this work and reference 20), the translated Rat IIa+b long transcript (RatIIa, accession no. AF228736, and Rat IIa+b, accession no. AF228737), and MouseIIa+b (this work) (also derived from EST AA032658). HS101130 is from human cells, and AA942886 is a combination of two overlapping rat ESTs (AA942886 and H32106). Profilins IIa and IIb differ only in the underlined region; residues different in the IIb sequences are shaded. An asterisk denotes a stop codon, X denotes an unidentified residue, and a dash denotes a gap; the arrow indicates where the sequences diverge as a result of alternative splicing (also Fig. 3).
These results suggest the possible generation of two profilin II isoforms by alternative splicing. We will refer to these forms as profilin IIa, which is the homologue of the protein purified from bovine brain (25), and profilin IIb, which is similar to the human protein described by Honoré et al. (20).
To verify the presence of alternatively spliced transcripts within the same organism, we performed a series of PCRs on cDNA libraries from various human, rat, and mouse tissues using specific primers for the respective isoforms (for primers, see Table 1). We cloned and sequenced several of these transcripts, including human and rat profilin IIa and human and mouse profilin IIb (Fig. 1). The human profilin IIb transcript corresponds exactly to the human clone described earlier by Honoré et al. (20) and shows 95% identity with the mouse homologue that is six amino acids longer (146 amino acids versus 140 amino acids).
In addition, using the profilin IIb primers we consistently amplified from all human, rat (data not shown), and mouse (Fig. 2A) tissues examined a longer transcript. These longer forms have sequences very similar to those of the mouse EST (AA032658) described above and contain profilin IIa coding information (sequences indicated as IIa+b in Fig. 1). Translation of the mRNA from which this PCR product is derived will result in the profilin IIa isoform. Interestingly, the PCR product for profilin IIb is present in only some mouse tissues, i.e., liver, kidney, and skin (additionally, it was present in human placenta [data not shown]; this tissue was not probed in the mouse). This indicates that formation of the profilin IIb transcript is regulated in a tissue-dependent manner. Note that the profilin IIb message is absent from the brain (see also below). Based on these quantitative RT-PCR experiments, within a given tissue, we estimate that the mouse profilin IIa message in liver and skin is 7- to 8-fold, and in kidney 20-fold, more abundant than the profilin IIb message.
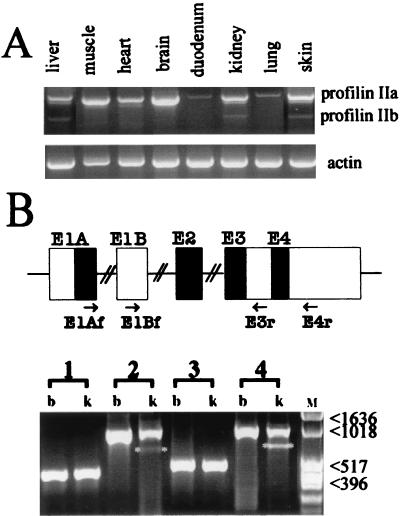
FIG. 2.
Profilin IIa and IIb transcripts in mouse tissues. (A) Quantitative RT-PCR on mRNA isolated from the indicated mouse tissues. The forward and reverse primers used are E1Af and E4r, respectively (see Table 1 for primer sequences and panel B for location). The short PCR product (approximately 690 bp) is an amplification of the profilin IIb mRNA. The longer transcript of approximately 960 bp contains coding information for profilin IIa, a noncoding region, and information for the C-terminal region of profilin IIb (also Fig. 1). β-Actin amplification was used as the control. (B) RT-PCR analysis of alternative spliced transcripts of the mouse profilin II gene (lower panel). First-strand cDNAs prepared from brain (b) and kidney (k) were subjected to PCR using primer combinations E1Af and E3r (lanes 1), E1Af and E4r (lanes 2), E1Bf and E3r (lanes 3), and E1Bf and E4r (lanes 4). Primer sequences are listed in Table 1, and their locations are indicated in the drawing. Profilin II transcripts spliced to exon 3 (corresponding to mRNA-a or -c [Fig. 4]) were detectable as strong bands in both brain and kidney. Transcripts spliced to exon 4 (corresponding to mRNA-b or -d and marked with asterisks) were found weakly expressed in kidney. M, molecular size marker (numbers are in base pairs).
Additionally, we characterized the 5′ end of mouse profilin II transcripts, using 5′ RACE with adult mouse brain cDNA as template. We found two amplification products of different lengths (data not shown). The shorter product (216 bp) was identical to the 5′ end of the sequenced EST clone (AA032658) and did not further extend the 5′ untranslated region. Surprisingly, sequence analysis of the longer RACE clone (278 bp) revealed the existence of a novel transcript in which the 5′ untranslated region and the nucleotides encoding the first 44 amino acids of profilin II were replaced by a new sequence of 214 bp (see below).
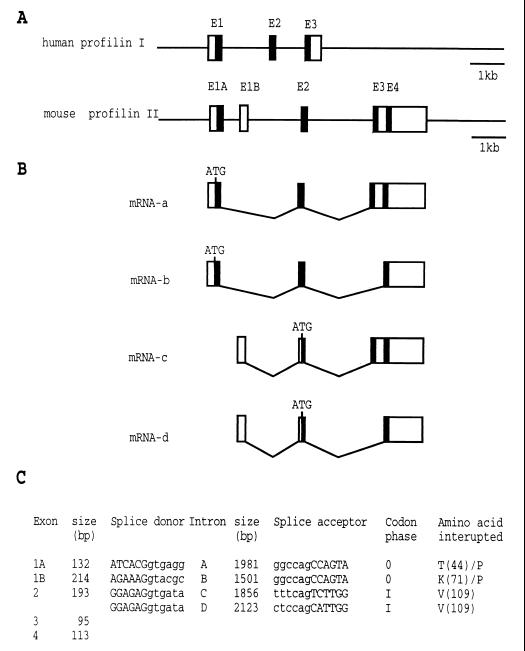
The exon-intron organization of the mouse profilin II gene gives rise to at least four alternatively spliced messages.
To obtain the genomic sequence of the mouse profilin II gene, we screened a mouse P1-derived artificial chromosome library with an 833-bp-long NcoI-PstI fragment derived from the EST clone AA032658. We isolated one clone, digested it with BamHI and EcoRI, and subcloned fragments hybridizing with the cDNA probe into pBSII KS(+). Sequence analysis of the plasmid inserts showed that the mouse profilin II gene spans over 7.3 kb including 1.6 kb of 5′- and 1 kb of 3′-flanking regions (Fig. 3 and 4). Taking into account the mouse ESTs and RACE sequences described above, we predict five exons, flanked by consensus splice signals (Fig. 4C). This could theoretically result in four types of mRNA (Fig. 4B). The first type (represented by mRNA-a), spliced from exons 1A, 2, and 3, encodes profilin IIa. mRNA-b contains the sequence of exons 1A, 2, and 4 and codes for profilin IIb. mRNA-c and -d begin with exon 1B followed by exons 2 and 3 and exons 2 and 4, respectively. The nucleotide sequence of exon 1B is identical to the one of the longer RACE clone described above, strongly suggesting that the splice products mRNA-c and -d do exist.
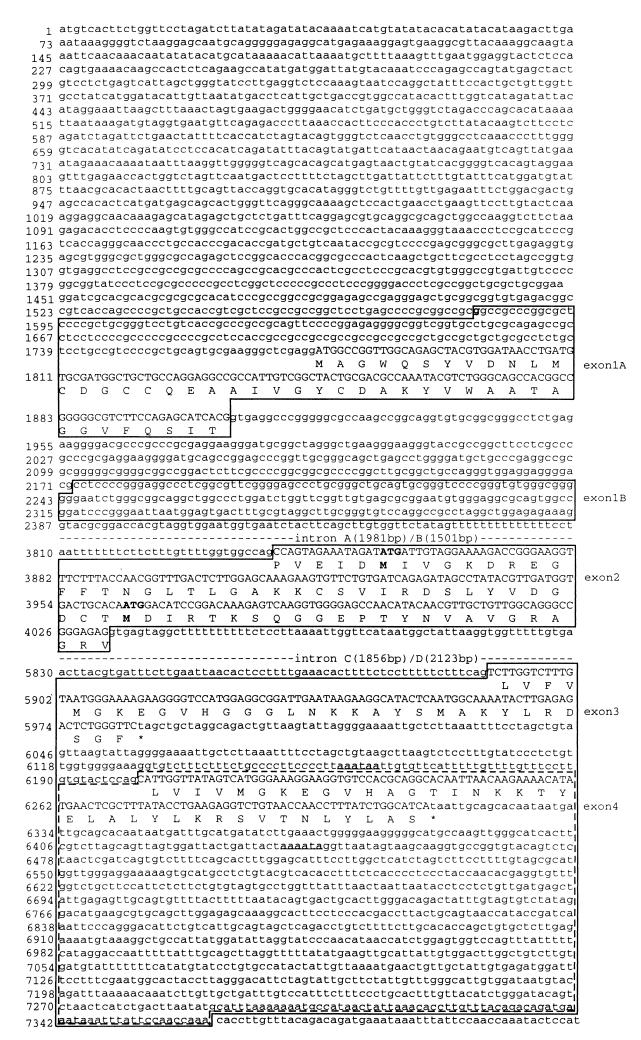
FIG. 3.
Nucleotide sequence of the mouse profilin II gene (GenBank accession no. AF237680). Exons 1A, 1B, 2, and 3 are boxed. Exon 4 is located inside exon 3 and is bordered by dashed lines. Coding sequences of exons and corresponding amino acid residues are in capital letters. Noncoding sequences of exons, intron sequences, and the 5′- and 3′-flanking sequences are in lowercase. Numbering (left) of the nucleotides includes intron sequences. Intron A is between exon 1A and exon 2, intron B is between exon 1b and exon 2, intron C is between exon 2 and exon 3, and intron D is between exon 2 and exon 4. The potential transcription initiation nucleotide for exon 1A is in boldface. The putative translation initiation codons and the corresponding methionines in exon 2 are in boldface. The two consensus polyadenylation signals are in boldface and underlined.
FIG. 4.
Exon-intron organization of the mouse profilin II gene and alternatively spliced transcripts. (A) The mouse profilin II 10,000-bp genomic sequence is shown as a thin horizontal line. Exons (E) are indicated by boxes. Filled boxes represent coding sequences, and open boxes represent noncoding sequences. Note that exon 4 (E4) is located in the 3′ untranslated region of exon 3 (E3). For comparison, the human profilin I gene structure (sequence in AC004771.1) is also given. (B) A diagram showing the four alternatively spliced profilin II transcripts (mRNA-a to -d). mRNA-a and -b code for profilin IIa and profilin IIb, respectively. The initiation codon ATG in exon 1A is indicated. mRNA-c and -d start with exon 1B, are spliced to exon 2, and then are spliced to exon 3 or exon 4, respectively. A putative translation initiation codon (ATG) in exon 2 is shown. (C) Information on sequences of splice donor and acceptor sites and intron and exon length; for the location of the introns, see Fig. 3.
To confirm the existence of mRNA-c and -d, we performed RT-PCR with forward primers from exon 1B and reverse primers from exons 3 and 4 (Table 1). The results were unambiguous (lanes 3 and 4 in Fig. 2B). In brain and kidney tissues, a cDNA corresponding to mRNA-c is present, whereas one corresponding to mRNA-d (lacking exon 3) is present in kidney tissue. Comparing the ethidium bromide staining intensities of the amplified fragments revealed that transcripts without sequences of exon 3 are present in much lower amounts than those having exon 3 sequences (Fig. 2B). Nevertheless, our data indicate that four different mRNAs can be produced from the profilin II gene.
mRNA-c and -d both contain an open reading frame starting in exon 2. At present, we have no evidence that these transcripts result in functional proteins, but we note that the flanking region of the second in-frame ATG (nucleotide positions 3963 to 3965 [Fig. 3]) matches the consensus sequence for translation initiation sites (23). The proteins predicted from mRNA-c and -d are 55 and 61 amino acids long and have calculated molecular weights of 5,905 and 6,611, respectively.
Profilins IIa and IIb have different biochemical properties.
To compare the profilin II isoforms biochemically, we expressed the recombinant rat IIa and human IIb proteins in E. coli and purified them by poly-l-proline affinity chromatography (see Materials and Methods). Bovine profilin IIa and recombinant human profilin I were used as a reference in all experiments.
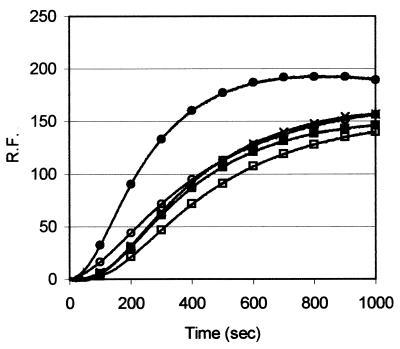
We determined the dissociation constant for the α-actin–profilin interaction for the different profilin isoforms using a classical sequestration assay with capped filaments (34). All recombinant forms have similar Kd values for α-actin: human profilin I, 0.4 μM; human profilin IIb, 0.6 μM; rat profilin IIa, 1 μM (Table 2). These are consistent with values reported previously for the nonrecombinant forms of profilins I and IIa (25, 34). In addition, in a time course polymerization experiment we observed no major differences between the different profilins (Fig. 5), i.e., recombinant human profilins I and IIb and rat profilin IIa decreased the rate of α-actin polymerization and reduced the total amount of F-actin to a similar extent as did profilin IIa purified from bovine brain.
TABLE 2.
Binding characteristics of profilin IIa and profilin IIb for actin (GP5)3 and PIP2
| Profilin | Kd for actin (μM) | (GP5)3Rmax/2a at μM | PIP2 bindingc |
|---|---|---|---|
| Human profilin I | 0.4 | NVb | ++++ |
| Human profilin IIb | 0.6 | NV | + |
| Rat profilin IIa | 1.0 | 0.3 | ++ |
| Bovine profilin IIa | 0.7 | 0.5 | ++ |
The interaction with peptide (GP5)3 was measured assuming a stoichiometry of two profilin molecules for one peptide (21).
NV, no value; Rmax/2 was not reached at the highest concentration used, 200 and 127 μM for profilin I and profilin IIb, respectively.
+ to ++++, lowest to highest level of binding, respectively.
FIG. 5.
The profilin isoforms have similar effects on actin polymerization. We monitored the change in pyrene fluorescence (relative fluorescence [R.F.]) after induction of polymerization with 2 mM MgCl2 and 0.1 M KCl of 10 μM actin alone (●) or in the presence of 5 μM human profilin I (○), human profilin IIb (□), rat profilin IIa (■), or bovine profilin IIa (×).
We used surface plasmon resonance technology to compare the affinities of the profilin isoforms for peptide (GP5)3, corresponding to the proline-rich sequence in VASP. As a measure for affinity, we determined the molar concentration of profilin needed to reach half of the maximal response (for details of the procedure, see reference 21). This concentration for rat profilin IIa is 0.3 to 0.5 μM (Table 2), in agreement with previous data obtained for bovine profilin IIa, where we found a concentration of 0.5 μM with a stoichiometry of two profilin molecules for one peptide (21). The high-affinity interaction is due to cooperative binding of the two profilin molecules to one peptide. For recombinant human profilin IIb and profilin I, we consistently found a low response (data not shown), in agreement with previous observations for bovine profilin I and different types of poly-l-proline binding studies (26, 37). The fact that the response unit values were low, even at the highest concentrations tested, indicates that the affinity of isoforms I and IIb for proline-rich sequences is much lower than the affinity of profilin IIa.
We assayed PIP2 binding in a microfiltration experiment. The flowthrough, representing the nonbound part of profilin, was analyzed by SDS-PAGE, and the amount of protein was quantified (Fig. 6, inset). From these data, the percentage of bound profilin can be calculated (Fig. 6). Less profilin IIa than profilin I was associated with the PIP2 micelles, and binding of profilin IIb to PIP2 was even more reduced. The results obtained for recombinant profilin IIa are similar to those described previously for bovine profilin IIa (26). Thus, both profilin II forms have a reduced affinity for PIP2 compared to that of profilin I (Table 2).
Profilin IIa is mainly expressed in neural tissues.
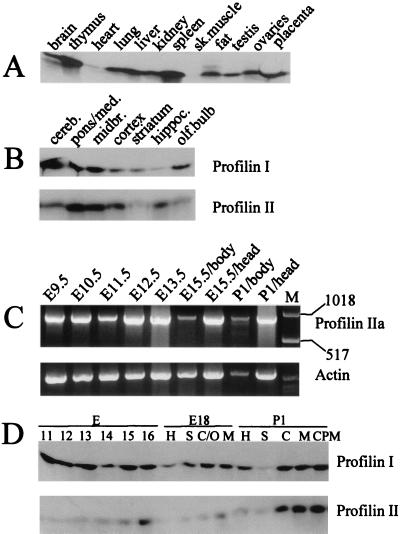
Western blotting experiments were performed with polyclonal rabbit antibodies G117 and G124, raised against bovine profilin I and bovine profilin IIa, respectively. G124 does not discriminate between profilins IIa and IIb, but given our RT-PCR results (see above and Fig. 8C), we probed profilin IIa expression in brain tissue. G117 and G124 displayed no or very weak cross-reactivity with the other isoform (Fig. 7). Based on this concentration series, we determined the amounts of profilin I or IIa in the different tissues examined.
FIG. 8.
Western blot analysis and RT-PCR of mouse tissues and brain regions. (A) Adult mouse organs probed with polyclonal anti-profilin I antibody G117. sk., skeletal. (B) Adult mouse brain regions: cerebellum (cereb.), pons/medulla (pons/meds), midbrain (midbr.), cortex, striatum, hippocampus (hippoc.), and olfactory bulb (olf.bulb) probed with polyclonal antibodies G117 and G124 against profilin I and profilin IIa, respectively. (C) Quantitative RT-PCR, using primers E1Af and E4r, of profilin messages at the indicated stages of mouse development. Only the long transcript encoding profilin IIa is observed. Lane M contains size markers (1,018-bp upper marker and 517-bp lower marker). β-Actin amplification was used as the control. (D) Developmental stages of mouse brain prepared from total brains of E11 to E16 embryos and different regions of E18 and P1 brains. H, hippocampus; S, striatum; C/O, cortex and olfactory bulb; M, midbrain; C, cortex; CPM, cerebellum, pons, and medulla.
Profilin I was found in relatively high concentrations in all tissues tested except for skeletal muscle and heart (Fig. 8A). In contrast, using this approach, profilin IIa was detectable only in brain (reference 50 and data not shown). Since both profilin I and profilin IIa are expressed in the brain, we probed different adult brain regions and found that both isoforms are present in all regions of the brain tested. Profilin IIa is expressed weakly in the striatum and the olfactory bulb and more strongly in the cerebellum, pons and medulla, midbrain, cortex, and hippocampus (Fig. 8B).
We also investigated profilin isoform expression during mouse development. RT-PCR, with profilin IIb-specific primers (amplifying the profilin IIb message and/or the long transcript coding for profilin IIa [see above]), of RNA isolated from various mouse embryonic stages revealed the presence of solely the long transcript, indicating that only profilin IIa is significantly expressed (Fig. 8C). We next addressed differences in profilin I and profilin IIa expression during brain development by Western blotting. Embryonic heads at stages E11 to E16 and E18 and P1 brain regions were dissected (Fig. 8D). The expression level for profilin I is constant during embryogenesis (0.004 to 0.008% of total protein) and does not increase after birth, while profilin IIa expression is clearly upregulated from stage E14 on and increases until after birth (0.004 to 0.075% of total protein). The antibodies hardly detected profilin IIa in earlier stages of development.
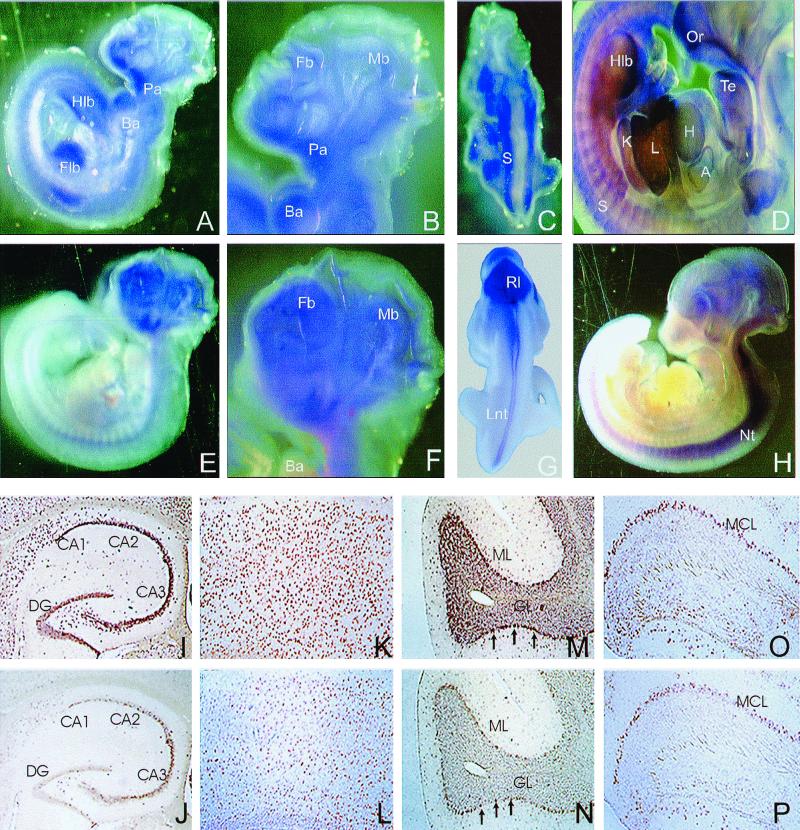
To analyze profilin IIa expression at these earlier stages, we performed in situ hybridization on mouse E8.5 to E11.5 whole-mount embryos (Fig. 9A to H). Whereas profilin I mRNA was found in most parts of the embryo, even at the early stage E8.5, no profilin IIa mRNA could be detected at stages E8.5 and E9.5 (data not shown), although the message could be amplified by RT-PCR (Fig. 8C). For E10.5 embryos, profilin I was seen in the forebrain and midbrain, the pharyngeal and branchial arches, and the four limb buds (Fig. 9A and B). AT E11.5, profilin I expression was prominent in most fetal organs, including heart, liver, and kidney (Fig. 9D). In contrast, profilin IIa expression was limited to the developing brain and the neural tube (Fig. 9E to H). Profilins I and IIa stained different structures on the dorsal sides of the embryos. The somites expressed profilin I (Fig. 9C), whereas profilin IIa was expressed in the rhombic lip and the lateral region of the neural tube (Fig. 9G).
FIG. 9.
In situ hybridization of whole-mount mouse embryos and adult mouse brain sections. (A to H) Expression patterns of profilin I (A to D) and profilin II (E to H) during early embryonic development. (A, B, E, and F) Lateral view of E10.5 embryos. Profilin I (A and B) is found in the midbrain (Mb), forebrain (Fb), branchial arches (Ba), pharyngeal arches (Pa), forelimb bud (Flb), and hind limb bud (Hlb). Profilin II expression (E and F) was detected in the entire brain region and the neural tube. (C and G) Dorsal view of E10.5 embryos shows profilin I expression in the somites (S) and profilin II in the rhombic lip (Rl) and the lateral neural tube (Lnt). (D and H) Lateral views of E11.5 embryos showing profilin I expression (D) in the olfactory region (Or), tongue epithelium (Te), heart (H), atrium (A), liver (L), kidney (K), somites, and hind limb bud and profilin II expression (H) in the developing brain and the neural tube (Nt). (I to P) Overlapping expression of profilin I and profilin II transcripts in adult mouse brain. Coronal (I to L) or sagittal (M to P) paraffin sections were hybridized with digoxigenin-labeled antisense riboprobes specific for profilin I (I, K, M, and O) or profilin II (J, L, N, and P). (I and J) Profilins I and II are expressed in the dentate gyrus (DG) and the CA regions of the hippocampus. (K and L) Strong expression of profilins was detected in all layers of the cortex. (M and N) Intensive signals were observed in the Purkinje cells (arrows) of cerebellum. Cells in both the molecular (ML) and granular (GL) layers were positive for profilins I and II. (O and P) Main olfactory bulb; strong staining was seen in the mitral cell layer (MCL).
We also employed in situ hybridization to compare the expression patterns of profilins I and IIa on tissue sections from 2-month-old mouse brains. In agreement with the Western blotting data, both profilin I and profilin IIa were detected in various regions of the central nervous system including the cerebrum, cerebellum, and olfactory bulb (Fig. 9I to P). Identical expression patterns of profilins were observed in the dentate gyrus and the cornus ammonis (CA) regions, in the hippocampus (Fig. 9I and J), and in the cortical layers of the forebrain (Fig. 9K and L). In the cerebellum, Purkinje cells were highly positive for both profilin I and profilin IIa (Fig. 9M and N, arrows). Coexpression of profilin I and profilin IIa was also detected in the cells of the molecular and the granular layers (Fig. 9M and N). In the olfactory bulb, the strongest signals were observed in the granule cells of the mitral cell layer (Fig. 9O and P).
DISCUSSION
The five exons in the profilin II gene are used to form four different messages.
In this paper, we report the profilin II gene sequence and deduce the exon-intron structure. At least four different mRNAs are generated from this gene by alternative splicing. Two of these are translated to different profilin II isoforms with distinct biochemical properties. Thus, our results clarify conflicting observations previously reported for bovine (25, 26) and human (14) profilin II which were due to the use of two different isoforms, designated here profilins IIa and IIb, respectively. The two other messages potentially code for shorter proteins lacking the NH2-terminal parts of profilins IIa and IIb. These fragments await further characterization, but we note that these polypeptides, if they adopt a three-dimensional structure similar to that in profilin, would possess a large hydrophobic surface, suggesting that they are associated with another protein or are membrane bound. In addition, these truncated forms lack some of the key residues involved in poly-l-proline binding (e.g., W3, Y6, and W31; numbering is without the initiator methionine that is posttranslationally removed) (4, 5, 30, 31) and actin binding (e.g., F59, V60, S71, R74, E82, and T84) (39; also discussed in reference 33), suggesting that these truncated forms do not bind these two ligands.
Profilins IIa and IIb are differentially expressed.
We observed ubiquitous expression of profilin I using Western blotting and RT-PCR, suggesting that it is the major form in almost all tissues. The profilin IIa message can also be detected in most tissues using a sensitive RT-PCR. The protein is, however, only abundantly expressed in the brain, where it is the major isoform in some regions, in which it is present at two- to fivefold-higher levels than those of profilin I. In the adult brain, we did not observe regions expressing either profilin IIa or profilin I separately. The two profilins are relatively highly expressed in the cortical layers of the forebrain, in the CA regions, and in the dentate gyrus, where Mena is also strongly expressed (32). The occurrence of profilin IIb is rare. It is not expressed during embryogenesis, but the mRNA is present in mouse kidney, skin, and liver. This restricted expression pattern indicates that generation of this splice variant is tightly regulated.
The expression pattern of profilin IIa suggests a neuronal function.
During all stages of brain development, profilin I expression is constant. Profilin IIa mRNA can first be detected at E9.5 to E10. The protein level steadily increases until after birth. Strong profilin IIa expression is restricted to brain structures, which suggests a specific function for this isoform in central nervous system and neural tube development. The neural tube closure defects of Mena−/− profilin I−/+ mice indicate an important role of profilin in the migration of neuroepithelial cells (27). Since profilin IIa expression starts after neural tube closure, it cannot compensate for profilin I during this process.
The higher in vitro affinity of profilin IIa for proline-rich peptides derived from VASP (21, 26), EVL (Ena/VASP-like protein [26a]), and Mena [and probably Mena(+)] (A. Lambrechts, V. Jonckheere, and C. Ampe, unpublished data) suggests that profilin IIA is a preferred partner of Ena/VASP proteins. Interestingly, expression of EVL in brain starts around E15, and the neuron-specific splice variant of Mena, Mena(+), shows a maximum expression level between E15 and P1 (27), which correlates with profilin IIa expression. These data suggest a function for profilin IIa in modulating actin reorganization at specific points during brain development.
Additionally, profilin II may be involved in regulation of synaptic vesicle trafficking, based on the profilin immunostaining at presynaptic sites and binding of profilin to several synaptic proteins (12, 50). Aczonin, a protein containing a proline-rich region and concentrated at active zones of presynaptic sides, binds more strongly to profilin IIa than to profilin I (46). Other partners of profilin II in neuronal tissues are protein isoforms encoded by the SMN gene and responsible for spinal muscular atrophy. These candidate proteins for determining loss of motoneurons were reported to associate preferentially with profilin II (15). Currently, it is not known whether both isoforms of profilin II or only profilin IIa can associate with SMN protein.
The three profilin isoforms have different biochemical properties.
Profilins are small proteins, which bind multiple ligands. Therefore, they are thought to act at the crossroads of different signaling pathways toward the actin cytoskeleton (41). The presence of two or three different isoforms with different biochemical properties in a cell allows better fine-tuning of signaling to the actin system. For each of the three profilin isoforms, we observed similar Kds for α-actin, but obviously the natural partners of profilin in nonmuscle cells are β- and γ-actin. The previously reported lower affinity of profilin IIb for α-actin (14) may arise from averaging Kd values derived from different assays using capped and noncapped filaments. The similar affinities correlate well with conservation of profilin residues in the interface with actin (33, 39).
Although it is generally accepted that basic residues in proteins mediate the interaction with PIP2, their identity remains elusive, as conflicting data were reported previously for mutated profilins from invertebrates and mammals (18, 42). Recent studies have proposed that residues from the NH2- and COOH-terminal α helices are implicated in PIP2 binding (5, 9, 10). The latter result is consistent with our recent data that suggest a role for amino acids Arg135 and Arg136 of human profilin I in binding to PIP2. Mutating Arg136 to Asp results in reduced PIP2 affinity (A. Lambrechts, V. Jonckheere, D. Dewitte, J. Vandekerckhove, and C. Ampe, unpublished results). Interestingly, profilin IIa has this amino acid change, which may explain why this form has a reduced affinity for PIP2 compared to that of profilin I. Profilin IIb has, however, an arginine at position 136 but also an aspartic acid residue at position 138, which may cause the lower binding affinity of this isoform.
The elucidation of two crystal structures with different modes of binding of profilin I to proline-rich peptides (30, 31) complicates interpretation of the data on (GP5)3 binding of the various profilin isoforms. It is clear that the high-affinity binding that we observe for a dimer of profilin IIa molecules is cooperative (21). The reduced affinity of profilins I and IIb may then be due to inefficient dimer formation, i.e., the profilins bind independently from each other, consistent with the lack of contact between the profilin molecules in the profilin I dimer l-Pro10 crystal structure (30). Alternatively, data from the profilin IIb crystal structure (33) suggest an aromatic extension of the binding pocket by an additional stacking interaction and a hydrogen bond with poly-l-proline by tyrosine 29 (serine in profilin I) which is found in both profilin II isoforms. The lower affinity of profilin IIb for proline-rich sequences (this study and reference 14) may then be due to negative effects exerted by its C-terminal part. We note that the aromatic character of the C-terminal residue, known to be important in contacting prolines (4, 5, 33), is conserved in profilin IIa but not in profilin IIb.
Our data indicate that each of the three isoforms has a different combination of binding affinities to the ligands PIP2 and (GP5)3, which serve as models for polyphosphoinositol lipids and proteins from the Ena/VASP family, respectively. Profilins may use these different ligand binding properties for modulating actin polymerization in a different manner. Indeed, the effect on actin interaction with these ligands is very different. PIP2 inhibits the interaction with actin (28), whereas a ternary complex can be formed among poly-l-proline, profilin (I or II), and actin (21, 25, 36). In the case of profilin IIa, dimerization on (GP5)3 results in an enhanced desequestration of actin from the pool bound to thymosin β4 and in increased actin polymerization (21). In addition to a modulatory role, these ligands may recruit either isoform to different subcellular locations depending on the cellular context, the available ligands, and the concentration of the profilin isoforms.
ACKNOWLEDGMENTS
Anja Lambrechts and Attila Braun contributed equally to this paper.
We thank F. Van Roy for use of the DNA sequencing facility for some of the profilin clones. We acknowledge Leen Van Troys for critically reading the manuscript and Griet Vandeweghe and Daniel Broekaert for assistance with additional experiments. Part of this work was carried out in the laboratory of Frank Gertler (Massachusetts Institute of Technology, Cambridge), and we acknowledge his contribution.
A.L. was the recipient of a travel grant from the Fund for Scientific Research-Flanders (FWO-Vlaanderen, Belgium) to work at MIT. C.A. is a research associate of the Fund for Scientific Research-Flanders. This work was supported by grants G004497, G006096, and G022598 of the Fund for Scientific Research-Flanders to C.A. and J.V.; a grant from the “Geneeskundige Stichting Koningin Elisabeth” to C.A.; GOA grants 12051296 and 120C1797 to J.V.; and a grant of the Swedish Cancer Foundation to R.F.
ADDENDUM IN PROOF
A paper describing the profilin II gene structure and the alternative splice variants profilins IIa and IIb by Di Nardo et al. is in press in J. Cell. Sci. The nomenclature used for the two profilin isoforms is the same as in this paper.
REFERENCES
- 1.Ahern-Djamali S M, Bachmann C, Hua P, Reddy S K, Kastenmeier A S, Walter U, Hoffmann F M. Identification of profilin and src homology 3 domains as binding partners for Drosophila enabled. Proc Natl Acad Sci USA. 1999;96:4977–4982. doi: 10.1073/pnas.96.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anton I M, Lu W, Mayer B J, Ramesh N, Geha R S. The Wiskott-Aldrich syndrome protein-interacting protein (WIP) binds to the adaptor protein Nck. J Biol Chem. 1998;273:20992–20995. doi: 10.1074/jbc.273.33.20992. [DOI] [PubMed] [Google Scholar]
- 3.Aszodi A, Chan D, Hunziker E, Bateman J F, Fassler R. Collagen II is essential for the removal of the notochord and the formation of intervertebral discs. J Cell Biol. 1998;143:1399–1412. doi: 10.1083/jcb.143.5.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorkegren C, Rozycki M, Schutt C E, Lindberg U, Karlsson R. Mutagenesis of human profilin locates its poly(L-proline)-binding site to a hydrophobic patch of aromatic amino acids. FEBS Lett. 1993;333:123–126. doi: 10.1016/0014-5793(93)80388-b. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkegren-Sjogren C, Korenbaum E, Nordber P, Lindberg U, Karlsson R. Isolation and characterization of two mutants of human profilin I that do not bind poly(L-proline) FEBS Lett. 1997;418:258–264. doi: 10.1016/s0014-5793(97)01376-8. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S, Korn E. On the mechanism of actin monomer-polymer subunit exchange at steady state. J Biol Chem. 1983;258:5013–5020. [PubMed] [Google Scholar]
- 7.Carlier M-F. Control of actin dynamics. Curr Opin Cell Biol. 1998;10:45–51. doi: 10.1016/s0955-0674(98)80085-9. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson L, Nystrom L E, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 9.Cedergren-Zeppezauer E S, Goonesekere N C, Rozycki M D, Myslik J C, Dauter Z, Lindberg U, Schutt C E. Crystallization and structure determination of bovine profilin at 2.0 A resolution. J Mol Biol. 1994;240:459–475. doi: 10.1006/jmbi.1994.1461. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhary A, Chen J, Gu Q M, Witke W, Kwiatkowski D J, Prestwich G D. Probing the phosphoinositide 4,5-bisphosphate binding site of human profilin I. Chem Biol. 1998;5:273–281. doi: 10.1016/s1074-5521(98)90620-2. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Faivre-Sarrailh C, Lena J Y, Had L, Vignes M, Lindberg U. Location of profilin at presynaptic sites in the cerebellar cortex; implication for the regulation of the actin polymerization state during axonal elongation and synaptogenesis. J Neurocytol. 1993;22:1060–1072. doi: 10.1007/BF01235749. [DOI] [PubMed] [Google Scholar]
- 13.Gertler F, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- 14.Gieselmann R, Kwiatkowski D J, Janmey P A, Witke W. Distinct biochemical characteristics of the two human profilin isoforms. Eur J Biochem. 1995;229:621–628. doi: 10.1111/j.1432-1033.1995.tb20506.x. [DOI] [PubMed] [Google Scholar]
- 15.Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch J W, Buchmeier S, Jockusch B M, Jockusch H. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with SMN in nuclear gems. J Biol Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 16.Goldschmidt-Clermont P J, Machesky L M, Baldassare J J, Pollard T D. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science. 1990;247:1575–1578. doi: 10.1126/science.2157283. [DOI] [PubMed] [Google Scholar]
- 17.Goldschmidt-Clermont P J, Kim J W, Machesky L M, Rhee S G, Pollard T D. Regulation of phospholipase C-gamma 1 by profilin and tyrosine phosphorylation. Science. 1991;251:1231–1233. doi: 10.1126/science.1848725. [DOI] [PubMed] [Google Scholar]
- 18.Haarer B K, Petzold A S, Brown S S. Mutational analysis of yeast profilin. Mol Cell Biol. 1993;13:7864–7873. doi: 10.1128/mcb.13.12.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogan B, Beddington R, Constantini F, Lacy E. Immunocytochemistry of whole mount embryos. In: Hogan B, Beddington R, Constantini F, Lacy E, editors. Manipulation of the mouse embryo. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 340–367. [Google Scholar]
- 20.Honoré B, Madsen P, Andersen A H, Leffers H. Cloning and expression of a novel human profilin variant, profilin II. FEBS Lett. 1993;330:151–155. doi: 10.1016/0014-5793(93)80262-s. [DOI] [PubMed] [Google Scholar]
- 21.Jonckheere V, Lambrechts A, Vandekerckhove J, Ampe C. Dimerization of profilin II upon binding the (GP5)3 peptide from VASP overcomes the inhibition of actin nucleation by profilin II and thymosin β4. FEBS Lett. 1999;447:257–263. doi: 10.1016/s0014-5793(99)00293-8. [DOI] [PubMed] [Google Scholar]
- 22.Kang F, Purich D, Southwick F. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J Biol Chem. 1999;274:36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwiatkowski D J, Bruns G A. Human profilin. Molecular cloning, sequence comparison, and chromosomal analysis. J Biol Chem. 1988;263:5910–5915. [PubMed] [Google Scholar]
- 25.Lambrechts A, Van Damme J, Goethals M, Vandekerckhove J, Ampe C. Purification and characterization of bovine profilin II; actin, poly(L-proline) and inositolphospholipid binding. Eur J Biochem. 1995;230:281–286. [PubMed] [Google Scholar]
- 26.Lambrechts A, Verschelde J-L, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Lambrechts, A. I., A. Kwiatkowski, L. M. Lanier, J. E. Bear, J. Vandeckerckhove, C. Ampe, and F. B. Gertler. PKA phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3-domains. J. Biol. Chem., in press. [DOI] [PubMed]
- 27.Lanier L, Gates M, Witke W, Menzies S, Wehman A, Macklis J, Kwiatkowski D, Soriano P, Gertler F. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 28.Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- 29.Machesky L M, Atkinson S J, Ampe C, Vandekerckhove J, Pollard T D. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahoney N M, Janmey P A, Almo S C. Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- 31.Mahoney N M, Rozwarski D A, Fedorov E, Fedorov A A, Almo S C. Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat Struct Biol. 1999;6:666–671. doi: 10.1038/10722. [DOI] [PubMed] [Google Scholar]
- 32.Mertens N, Remaut E, Fiers W. Tight transcriptional control mechanism ensures stable high-level expression from T7 promoter-based expression plasmids. Bio/Technology. 1995;13:175–179. doi: 10.1038/nbt0295-175. [DOI] [PubMed] [Google Scholar]
- 33.Nodelman I M, Bowman G D, Lindberg U, Schutt C E. X-ray structure determination of human profilin II: comparative structural analysis of human profilins. J Mol Biol. 1999;294:1271–1285. doi: 10.1006/jmbi.1999.3318. [DOI] [PubMed] [Google Scholar]
- 34.Pantaloni D, Carlier M-F. How profilin promotes actin filament assembly in the presence of thymosineβ4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 35.Pardee J, Spudich J. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 36.Perelroizen E C, Marchand J-B, Blanchoin L, Didry D, Carlier M-F. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- 37.Petrella E C, Machesky L M, Kaiser D A, Pollard T D. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 1996;35:16535–16543. doi: 10.1021/bi961498d. [DOI] [PubMed] [Google Scholar]
- 38.Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockush B, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schutt C, Myslik J, Rozycki M, Goonesekere N, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 40.Small J V, Rottner K, Kaverina I, Anderson K I. Assembling an actin cytoskeleton for cell attachment and movement. Biochim Biophys Acta. 1998;1404:271–281. doi: 10.1016/s0167-4889(98)00080-9. [DOI] [PubMed] [Google Scholar]
- 41.Sohn R H, Goldschmidt-Clermont P J. Profilin: at the crossroads of signal transduction and the actin cytoskeleton. Bioessays. 1994;16:465–472. doi: 10.1002/bies.950160705. [DOI] [PubMed] [Google Scholar]
- 42.Sohn R H, Chen J, Koblan K S, Bray P F, Goldschmidt-Clermont P J. Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J Biol Chem. 1995;270:21114–21120. doi: 10.1074/jbc.270.36.21114. [DOI] [PubMed] [Google Scholar]
- 43.Spudich J, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 44.Stossel T P, Hartwig J H, Janmey P A, Kwiatkowski D J. Cell crawling two decades after Abercrombie. Biochem Soc Symp. 1999;65:267–280. [PubMed] [Google Scholar]
- 45.Suetsugu S, Miki H, Takenawa T. Distinct roles of profilin in cell morphological changes: microspikes, membrane ruffles, stress fibers, and cytokinesis. EMBO J. 1998;17:6516–6526. doi: 10.1016/s0014-5793(99)01086-8. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Kibschull M, Laue M M, Lichte B, Petrasch-Parwez E, Kilimann M W. Aczonin, a 550-kD putative scaffolding protein of presynaptic active zones, shares homology regions with Rim and Bassoon and binds profilin. J Cell Biol. 1999;147:151–162. doi: 10.1083/jcb.147.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wasserman S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B, Narumiya S. P140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widada J S, Ferraz C, Liautard J P. Total coding sequence of profilin cDNA from Mus musculus macrophage. Nucleic Acids Res. 1989;17:2855. doi: 10.1093/nar/17.7.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witke W, Podtelejnikov A V, Di Nardo A, Sutherland J D, Gurniak C B, Dotti C, Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]