Macrophages are highly versatile and plastic immune cells that are localized in nearly all organs of the body and contribute to a plethora of physiological and pathological processes in situ. Beside their roles as major players in the “first line of defense” under inflammatory conditions, macrophages are known to participate in tissue homeostasis maintenance. Therefore, these cells are capable of removing cell debris and secreting cytokines and growth factors influencing cells in their local microenvironment and vice versa. The eye, which represents one of the most sophisticated organs in the body of vertebrates, harbors multiple macrophage populations that are localized in and adapted to different compartments. Microglia, the resident immune cells of the central nervous system (CNS) including the retina, are mainly restricted to the retina itself and the optic nerve, whereas the cornea, ciliary body and choroid contain different myeloid cell types with distinct tasks to fulfill. In comparison to brain microglia (bMG) or other CNS-associated macrophages (CAMs), that were extensively studied in mice and humans (Goldmann et al., 2016; Masuda et al., 2019), ocular macrophages (oMacs) are far less understood in points of their exact origin, fate and heterogeneity. To address this issue, recent studies applied state-of-the-art fate mapping approaches to identify the exact embryonic origin of the oMacs and single-cell transcriptomics to dissect the myeloid landscape in multiple eye compartments under homeostatic and pathological conditions (O’Koren et al., 2019; Wieghofer et al., 2021). Here, we would like to recapitulate the most important developments in fate mapping and single-cell analysis leading to these findings and delineate emerging technologies that may further fuel the research in myeloid cell biology in the brain and eye.
Over the recent years, the concept of the origin of tissue-resident macrophages was confronted with new findings challenging the previously established dogmata. It was in the 1970’s when van Furth and colleagues described the “mononuclear phagocyte system” as a model for the hierarchy of myeloid cells. Here, tissue-resident macrophages were considered to emerge from monocytes and promonocytes circulating in the blood stream and originating from precursors in the bone marrow. The origin of macrophages seemed to be established as in vivo studies using bone marrow chimeras showed an engraftment of donor cells into various tissues. In the 2000’s, studies aiming at determining the progenitor cells of microglia in the brain provided evidence that the engraftment of green fluorescent protein-labeled Ly6Chigh monocytes only occurred in mice undergoing preconditioning by whole-body irradiation that is accompanied by disturbances in the blood-brain-barrier function (Mildner et al., 2007). Therefore, irradiation leads to an influx of monocytes which does not necessarily occur in vivo. But where do macrophages come from? In the case of microglia, this question was elegantly answered using the Runx1MER-Cre-MER:Rosa26-YFP fate mapping system (Ginhoux et al., 2010). Here, the authors confirmed the previous hypothesis that microglia are derived from the primitive hematopoiesis in the yolk sac and seed the CNS in an early stage of prenatal development. Subsequently, the development of microglial cells was shown to be dependent on erythromyeloid precursors (EMP) giving rise to intermediate developmental stages, termed A1 and A2 progenitors, exhibiting differences in their CX3CR1 expression (Kierdorf et al., 2013). Taken together, these studies broke the paradigm of a supposed continuous influx of peripheral monocytes into the CNS and established a prenatal origin of microglia in the brain. Today, due to advancements in fate mapping techniques, it is commonly accepted that the majority of organs harbors macrophage cell populations predominantly originating either from early EMPs in the extra-embryonic yolk sac (e.g., microglia) or late EMPs that subsequently colonize the fetal liver (e.g., alveolar macrophages or Kupffer cells) to give rise to the second wave of fetal hematopoiesis.
The eye consists of different compartments fulfilling specific functions in the process of visual perception. In addition, these compartments display multiple microenvironments and therefore harbor different subsets of myeloid cells. Previous studies using bone marrow chimeras proposed that macrophages in the eye originate from the adult hematopoiesis in the bone marrow and are constantly replenished by circulating monocytes. However, considering the developments in the field of myeloid cell research and fate mapping techniques, recent studies reinvestigated this important question (O’Koren et al., 2019; Wieghofer et al., 2021).
In two separate studies, our group and others used multiple fate mapping systems to provide evidence for a prenatal origin of myeloid cells in the eye. By applying the Runx1MER-Cre-MER:Rosa26-YFP model, O’Koren et al. (2019) demonstrated that the administration of 4-hydroxytamoxifen at E7.5 leads to a similar amount of YFP+ cells in the retina and the brain at 8 weeks postpartum, indicating a yolk sac-derived origin as in the brain. These findings were supported by us using the Cx3cr1CreER:Rosa26-YFP mouse model, which was shown to consistently label bMG in an embryonic fate mapping approach (Goldmann et al., 2016). Taking advantage of this model in the context of the eye, our results clearly point towards a prenatal origin of retinal microglia (rMG) (Wieghofer et al., 2021). In addition, we extended our analysis towards myeloid cell populations in the ciliary body and the cornea. Here, we were able to show for the first time that cells targeted by embryonic pulse labeling were present in the ciliary body and cornea as well (Wieghofer et al., 2021). To address the question whether circulating monocytes from the definitive hematopoiesis contribute to the myeloid cell pool in different eye compartments, we used a fate mapping approach in adult mice by injecting tamoxifen into Cx3cr1CreER:Rosa26-YFP mice at the age of 6–8 weeks. Retinal microglia showed a very low turnover that is reminiscent of the longevity of bMG or CAMs (Goldmann et al., 2016, Wieghofer et al., 2021). Interestingly, we found that ciliary body macrophages also showed relatively stable YFP-labeling indicating the longevity of these cells whereas the percentage of YFP+ cells in the cornea significantly dropped over the course of time (Wieghofer et al., 2021), which is comparable to the findings in choroidal macrophages (O’Koren et al., 2019). These results were further supported by the use of Ccr2-RFP mice, where short-lived monocytes are specifically labeled, and Flt3Cre:Rosa26-YFP to target multipotent hematopoietic progenitor cells of the fetal and adult hematopoiesis. In both models, the highest recombination rates were present in the cornea and to a smaller extent in the ciliary body, whereas the recombination in the retina was negligible (Wieghofer et al., 2021). In summary, these studies redefined our understanding of the ontogeny of oMacs in different compartments of the eye and highlighted the different kinetics by which these macrophages are replenished by circulating monocytes in vivo (Figure 1A).
Figure 1.
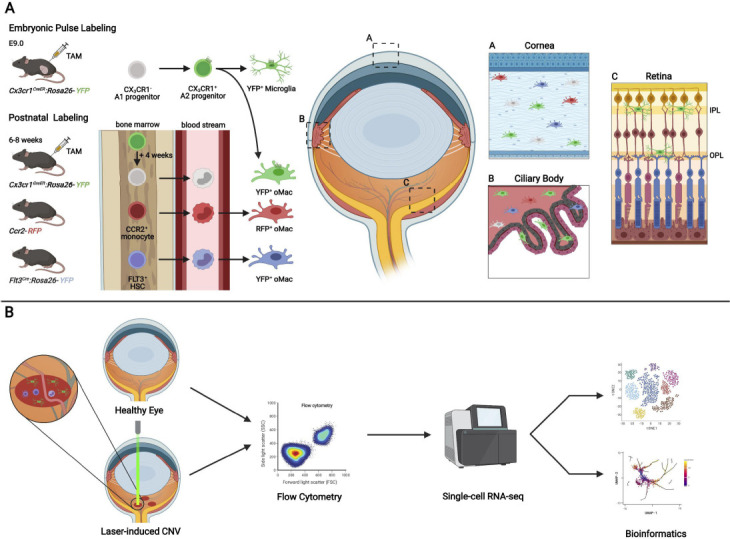
Heterogeneity of ocular macrophages in health and disease.
(A) Embryonic pulse labeling of CX3CR1+A2 progenitors in the Cx3cr1CreER:Rosa26-YFP line led to the identification of YFP+(green) retinal microglia and ocular macrophages (oMac) in the cornea and ciliary body. Postnatal labeling of CX3CR1+ cells in adult mice to conditionally target long-lived resident tissue macrophages (green) allows to investigate the turnover with peripheral blood monocytes (grey), which lose their YFP-signal in the course of 4 weeks after tamoxifen administration. Under homeostatic conditions a steady replenishment was present in the cornea, negligible in the ciliary body and absent in the retina, confirmed by other constitutive active reporter mouse models, including Ccr2-RFP(red) mice labeling monocytes and Flt3Cre:Rosa26-YFP(blue) mice labeling multipotent FLT3+ hematopoietic stem cells (HSC). (B) Workflow of purifying myeloid cells by flow cytometry in the laser-induced model of choroidal neovascularization compared to controls for the subsequent analysis by single-cell RNA-seq and bioinformatics. The figure was created with BioRender.com.
Another driving force behind the recent discoveries in myeloid cell biology is the advent of single-cell technologies like single-cell RNA sequencing (scRNA-seq) or cytometry by time-of-flight. These technologies provide the unique opportunity to study heterogenous cell populations in an unbiased and multiplexed way. By using these technologies, recent studies provided new insights into the innate immune landscape in the CNS under homeostatic and pathological conditions. In the brain, for the first time, microglial subpopulations were defined across different anatomical locations and time points during development and disease. In addition, distinct microglia subsets in humans that partially overlap with microglia clusters in mice were identified in this study (Masuda et al., 2019). In disease context, scRNA-seq was used to create an atlas of the innate immune system in experimental autoimmune encephalomyelitis, a popular model for multiple sclerosis (Jordão et al., 2019). Here, the authors delineated a core signature of genes expressed by CAMs including Mrc1, Pf4, Ms4a7, Cbr2 and Stab1 and the presence of multiple disease-associated cell clusters (Jordão et al., 2019). In another study, myeloid cell immune states were characterized in several mouse models of neuroinflammation and neurodegeneration on a protein level using cytometry by time-of-flight (Ajami et al., 2018). Noteworthily, single-cell analysis revealed the presence of similar resident myeloid cell clusters under neuroinflammatory and neurodegenerative conditions. However, different disease conditions were distinguished by unique cytokine profiles and signaling pathways (Ajami et al., 2018). Hence, although different types of perturbations may trigger different molecular pathways, these processes seem to converge on a subset of conserved myeloid cell phenotypes with specialized, either beneficial or detrimental, functions in CNS pathology.
To characterize the myeloid cell heterogeneity in multiple eye compartments including the retina, ciliary body and cornea, we used scRNA-seq to decipher the myeloid cell landscape at single-cell resolution (Figure 1B) (Wieghofer et al., 2021). We identified several cell clusters corresponding to different myeloid cell types like microglia or other macrophages and found a significant enrichment of these clusters in different compartments of the eye (Wieghofer et al., 2021). Microglial cell clusters exhibited the homeostatic gene expression signature including Tmem119 and P2ry12 (Wieghofer et al., 2021), thereby confirming the previous results from O’Koren and colleagues (O’Koren et al., 2019). Furthermore, we were able to show that bMG and rMG exhibit strong transcriptional similarities (Wieghofer et al., 2021), which may be explained by their shared ontogeny and environment as these factors are decisive for macrophage identity. Contrarily, cornea macrophages clearly separated from microglia clusters with strong similarities to bone marrow-derived monocytes reflecting the aforementioned differences in postnatal turnover (Wieghofer et al., 2021). To investigate the role of distinct myeloid subsets in retinal pathology, we used scRNA-seq in a laser-induced model of choroidal neovascularization (CNV), a commonly used mouse model for neovascular age-related macular degeneration (Wieghofer et al., 2021). Here, we identified multiple disease-associated cell clusters being enriched at different time points after laser treatment and therefore reflecting successive changes in transcriptional profiles at different disease stages. Disease-associated microglia (DAM) were the dominant myeloid cell type in CNV lesions, as shown in CNV-induced Cx3cr1CreERT2:Rosa26-tdTomato mice, and characterized by the lower abundance of homeostatic genes and the concomitant upregulation of glycolytic enzymes like Gapdh which may indicate an altered microglial metabolism (Wieghofer et al., 2021). To get deeper insights into the spatiotemporal kinetics underlying these transcriptional changes, a trajectory analysis was performed. Interestingly, cells at day 7 after CNV-induction were more similar to the homeostatic signature than cells at day 3 which may be explained by the fact that day 3 may represent the more acute phase of CNV. In another study, single-cell profiling of microglia in a model of photoreceptor degeneration revealed the presence of DAM with the downregulation of homeostatic genes and the upregulation of Lgals3, Spp1 and Lpl (O’Koren et al., 2019). Interestingly, immunofluorescence revealed a significant enrichment of these cell clusters in the subretinal space, which is devoid of any immune cells under physiological conditions (O’Koren et al., 2019; Wieghofer et al., 2021). Of note, previous studies in the brain highlighted the importance of DAM clusters as potential targets for myeloid cell-based interventions in neurodegenerative and neuroinflammatory diseases (Ajami et al., 2018; Jordão et al., 2019; Masuda et al., 2019). As bulk and scRNA-seq showed that rMG and bMG are transcriptionally very similar, it is tempting to hypothesize that the DAM clusters identified in ocular diseases may overlap with DAMs in the brain (O’Koren et al., 2019; Wieghofer et al., 2021). Thus, this approach may help us to identify molecular signatures, that are conserved across diseases in the brain and the eye and may therefore facilitate the development of drugs intended to reverse the pathological alterations in myeloid cells.
In conclusion, recent advances in fate mapping and single-cell analysis ultimately led to a completely new understanding of the ontogeny and heterogeneity of myeloid cells in the CNS including oMacs. Concerning fate mapping techniques, we need to keep in mind that the current models are limited by their cell type specificity. Upcoming studies applying binary transgenesis as recently shown for CAMs (Kim et al., 2021) could represent an interesting opportunity to study particular myeloid subsets with the possibility of more cell type-specific manipulations.
Single-cell transcriptomics identified a remarkable heterogeneity of oMacs, which is even increased under pathological conditions (O’Koren et al., 2019; Wieghofer et al., 2021). As a next step, it is necessary to link different cell clusters to their exact localization in the intact tissue. Therefore, the rapidly developing field of spatial transcriptomics including in situ sequencing methods like spatially-resolved transcript amplicon readout mapping (STARmap) may harbor promising tools (Wang et al., 2018). In addition, mapping transcriptional signatures to proteomic, epigenetic and metabolomic profiles may provide unknown insights into the molecular processes in cells during health and disease. Finally, the translation of findings in mice into the human setting will be decisive for the validation of transcriptionally distinct subpopulations (Masuda et al., 2019). As a potential target, SPP1 was not only found to be upregulated in microglia in the CNV-model of neovascular age-related macular degeneration and in macrophages in human CNV membranes but also its inhibition lead to an increased CNV size in mice (Schlecht et al., 2021). As SPP1 is known to excert proangiogenic effects and acts as a chemoattractant on macrophages, factors promoting both angiogenesis and macrophage recruitment such as SPP1 could become the center of attraction in the future. In summary, the aforementioned novel technological advancements will fuel the ongoing research in the field of oMacs and illuminate the path for the development of more targeted therapeutics from which patients with vision-threatening retinal diseases will benefit.
The authors would like to thank Bahareh Ajami (Oregon Health & Science University (OHSU), Department of Molecular Microbiology & Immunology (MMI), Portland, USA) and Clemens Lange (Eye Center, Medical Center, University of Freiburg, Germany) for the critical discussion of the manuscript. We apologize to all those authors whose works were not properly cited due to space constraints. The figure was created using BioRender.com.
Additional file: Open peer review report 1 (82.5KB, pdf) .
Footnotes
Copyright license agreement: The Copyright License Agreement has been signed by both authors before publication.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Joao N. Duarte, University of Copenhagen, Denmark.
References
- 1.Ajami B, Samusik N, Wieghofer P, Ho PP, Crotti A, Bjornson Z, Prinz M, Fantl WJ, Nolan GP, Steinman L. Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat Neurosci. 2018;21:541–551. doi: 10.1038/s41593-018-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldmann T, Wieghofer P, Jordão MJC, Prutek F, Hagemeyer N, Frenzel K, Amann L, Staszewski O, Kierdorf K, Krueger M, Locatelli G, Hochgerner H, Zeiser R, Epelman S, Geissmann F, Priller J, Rossi FMV, Bechmann I, Kerschensteiner M, Linnarsson S, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. 2016;17:797–805. doi: 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jordão MJC, Sankowski R, Brendecke SM, Sagar null, Locatelli G, Tai YH, Tay TL, Schramm E, Armbruster S, Hagemeyer N, Groß O, Mai D, Çiçek Ö, Falk T, Kerschensteiner M, Grün D, Prinz M. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363:eaat7554. doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 5.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Hölscher C, Müller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F, et al. Microglia emerge from erythromyeloid progenitors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 6.Kim JS, Kolesnikov M, Peled-Hajaj S, Scheyltjens I, Xia Y, Trzebanski S, Haimon Z, Shemer A, Lubart A, Van Hove H, Chappell-Maor L, Boura-Halfon S, Movahedi K, Blinder P, Jung S. A binary Cre transgenic approach dissects microglia and CNS border-associated macrophages. Immunity. 2021;54:176–190. doi: 10.1016/j.immuni.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grün D, Priller J, Stadelmann C, Prinz M. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–392. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 8.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Brück W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 9.O’Koren EG, Yu C, Klingeborn M, Wong AYW, Prigge CL, Mathew R, Kalnitsky J, Msallam RA, Silvin A, Kay JN, Bowes Rickman C, Arshavsky VY, Ginhoux F, Merad M, Saban DR. Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity. 2019;50:723–737. doi: 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlecht A, Zhang P, Wolf J, Thien A, Rosmus DD, Boneva S, Schlunck G, Lange C, Wieghofer P. Secreted phosphoprotein 1 expression in retinal mononuclear phagocytes links murine to human choroidal neovascularization. Front Cell Dev Biol. 2021;8:618598. doi: 10.3389/fcell.2020.618598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava FA, Deisseroth K. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361:eaat5691. doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieghofer P, Hagemeyer N, Sankowski R, Schlecht A, Staszewski O, Amann L, Gruber M, Koch J, Hausmann A, Zhang P, Boneva S, Masuda T, Hilgendorf I, Goldmann T, Böttcher C, Priller J, Rossi FM, Lange C, Prinz M. Mapping the origin and fate of myeloid cells in distinct compartments of the eye by single-cell profiling. EMBO J. 2021;40:e105123. doi: 10.15252/embj.2020105123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


