Abstract
The protein K-Ras functions as a molecular switch in signaling pathways regulating cell growth. In the human mitogen-activated protein kinase (MAPK) pathway, which is implicated in many cancers, multiple K-Ras proteins are thought to assemble at the cell membrane with Ras effector proteins from the Raf family. Here we propose an atomistic structural model for such an assembly. Our starting point was an asymmetric guanosine triphosphate-mediated K-Ras dimer model, which we generated using unbiased molecular dynamics simulations and verified with mutagenesis experiments. Adding further K-Ras monomers in a head-to-tail fashion led to a compact helical assembly, a model we validated using electron microscopy and cell-based experiments. This assembly stabilizes K-Ras in its active state and presents composite interfaces to facilitate Raf binding. Guided by existing experimental data, we then positioned C-Raf, the downstream kinase MEK1 and accessory proteins (Galectin-3 and 14-3-3σ) on and around the helical assembly. The resulting Ras–Raf signalosome model offers an explanation for a large body of data on MAPK signaling.
The protein K-Ras is a crucial molecular switch in signaling pathways that regulate cell proliferation, differentiation and survival1. Gain-of-function mutations in three closely related members of the Ras family, K-Ras, H-Ras and N-Ras, are found in approximately 25% of all human cancers, with mutations in K-Ras being particularly common2. In many cases, oncogenic mutations in Ras are associated with hyperactivation of the MAPK signaling pathway3. Dysregulation of the MAPK pathway has also been implicated in a host of hyperproliferative developmental disorders termed RASopathies4. Inhibiting oncogenic or aberrant Ras is thus of great clinical interest5,6. Our structural understanding of how K-Ras activates downstream MAPK signaling, however, has been incomplete.
Ras is active when guanosine triphosphate (GTP) bound and inactive when guanosine diphosphate (GDP) bound; conversions between these states are catalyzed by guanine nucleotide-exchange factors (GEFs) and GTPase-activating proteins (GAPs), respectively7. Active and inactive Ras conformations differ primarily at the ‘switch’ regions involved in effector binding8,9. Despite some early evidence that Ras can form oligomers10, activated Ras has long been thought to act in MAPK signaling as a monomeric membrane anchor for C-Raf or other effectors from the Raf kinase family (with Ras itself being anchored to the plasma membrane by its lipidated C-terminal tail). A growing body of data now indicate that Ras proteins dimerize11–15, that their dimerization may depend on GTP binding16 and that Ras dimerization may be critical for Raf activation17–20.
A notable fraction of membrane-bound, GTP-bound Ras proteins have been found to organize into relatively immobile ‘nanoclusters’, each with about eight members and a radius of roughly 100 Å (refs. 21,22). Ras dimerization appears to be crucial to nanocluster formation19, and it is possible that nanoclusters comprise networks of loosely interacting, lower-order Ras structures (such as monomers and dimers); structurally well-defined, higher-order oligomers; or a mixture of both. It is known that Galectin-3 (Gal-3) is crucial for K-Ras nanoclustering and signaling23, as is Galectin-1 (Gal-1) for H-Ras24. Raf kinases colocalize with nanoclustered Ras proteins; it has been shown that such colocalized effectors generate most of the downstream MAPK signal22,25, and that Ras activity can be suppressed by proteins that disrupt this clustering19,26. Structural information about nanoclusters is sparse, however, because they are difficult to reconstitute in vitro and the resolution of cellular imaging is limited.
In the work reported here, we used unbiased molecular dynamics simulations to inform the construction of an atomistic structural model of a K-Ras nanocluster. We term our proposed structure a Ras–Raf signalosome because it is a higher-order hetero-oligomer (containing multiple monomers of K-Ras, C-Raf, Gal-3 and other proteins) with well-defined structural features and is reminiscent of ‘signalosomes’ that underlie the activities of other signaling systems27. We first generated a GTP-mediated asymmetric (GMA) dimer model by directly simulating the association of two K-Ras monomers; subsequent mutagenesis experiments strongly supported this model. The asymmetric dimer model was then extended to a higher-order oligomer model by adding monomers in a head-to-tail fashion, resulting in a compact helical K-Ras assembly. This assembly promotes the stability and accessibility of active K-Ras and creates composite interfaces that facilitate Raf binding. Guided by existing experimental data and further simulations, we then positioned C-Raf dimers, the downstream kinase MEK1, and the accessory proteins Gal-3 and 14–3-3σ around the K-Ras core. Our Ras–Raf signalosome model, which synthesizes a large body of data on the MAPK pathway, may extend to other Ras and Raf isoforms and provides a structural framework to inform further investigations of MAPK signaling.
Results
We first summarize key structural features of our Ras–Raf signalosome model for convenience before detailing its construction and resulting findings.
The signalosome model centers on a K-Ras helical assembly.
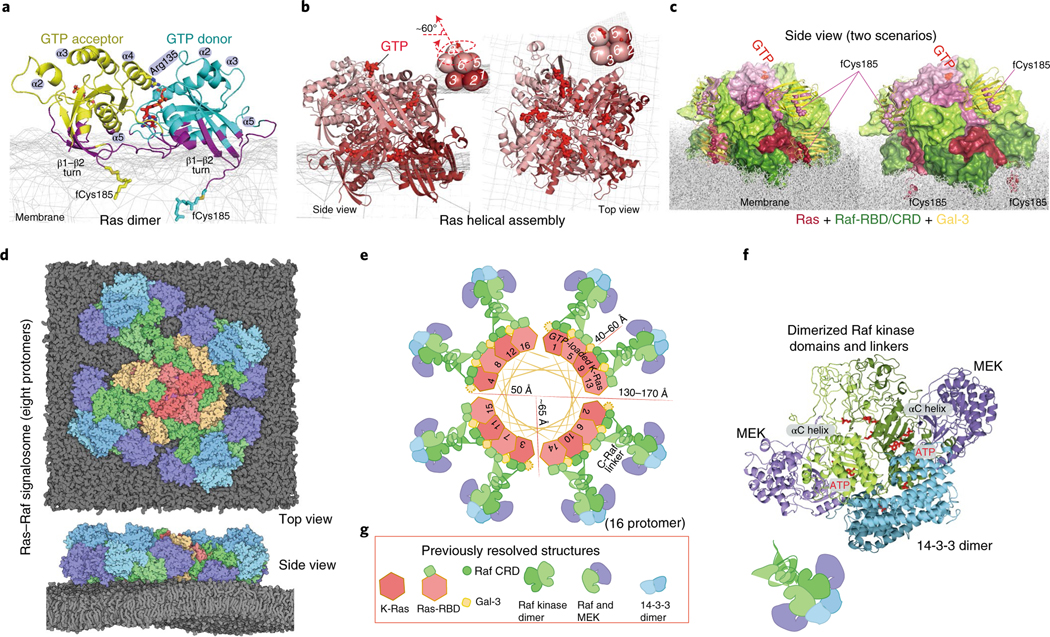
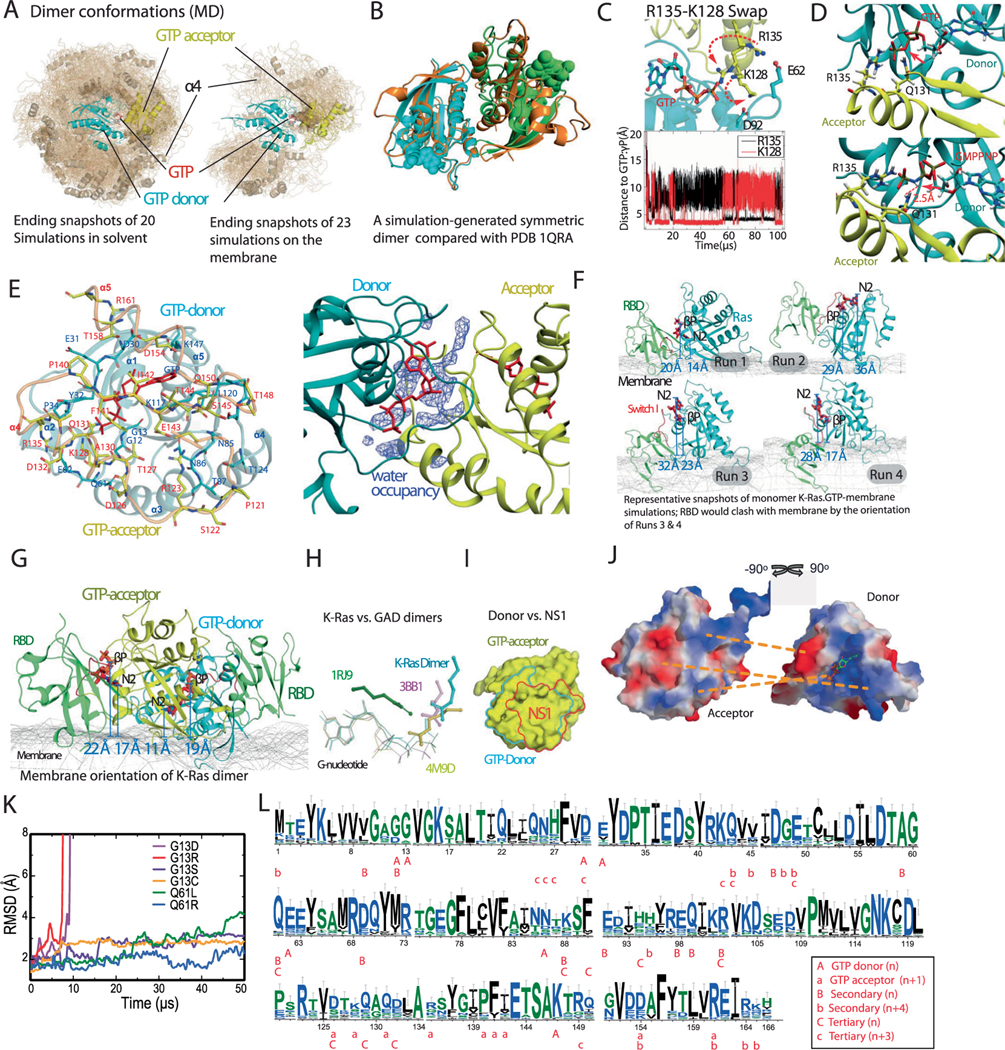
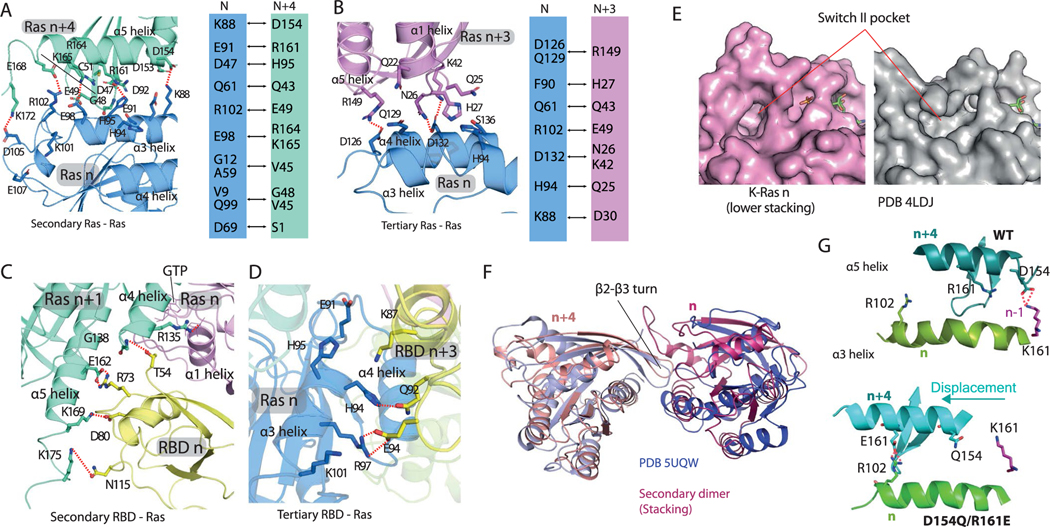
In brief, we obtained our Ras–Raf signalosome model by proposing a GMA K-Ras dimer model (Fig. 1a), extending it to a K-Ras helical assembly (Fig. 1b) and adding further components. The final model is anchored to the plasma membrane, and the Ras-binding domains (RBDs) and cysteine-rich domains (CRDs) of C-Raf interact directly with the K-Ras helical assembly (Fig. 1c), essentially covering the outside of the assembly. The four K-Ras molecules at the base of the helical assembly contact the membrane. We considered two possible scenarios with respect to the farnesylated Cys185 (fCys185) residues28 of the K-Ras proteins at the base tier. In one, the farnesyl groups at the base tier are capped by membrane-bound Gal-3 molecules (we will discuss in detail below the motivation for this aspect of the model) and in another, which is more consistent with the current understanding of the role fCys185 in membrane-anchoring K-Ras, these fCys185 residues are inserted into the membrane. Our model was stable in both scenarios in our simulations. In both scenarios, Gal-3 molecules cap the farnesyl groups of all the K-Ras molecules beyond the base tier, in line with experimental data23.
Fig. 1 |. Outline of the structural model of a Ras–Raf signalosome.
a, The GMA K-Ras dimer model. Residues contacting the membrane (mesh) are shown in purple; the GTP at the interface is in red. b, The (eight-protomer) K-Ras helical assembly with illustrative cartoons. c, The K-Ras helical assembly (pink), decorated with C-Raf RBD and CRD domains (green) and Gal-3 proteins (yellow). Two scenarios—one with Gal-3 at the base tier (left) and one without (right)—are shown, with the same orientation. d, The signalosome model on the membrane. The eight-protomer model contains four catalytic units at the periphery. e, Diagram of the signalosome model with a helical wheel representing the Ras assembly at the center. The Gal-3 molecules associated with the base-tier K-Ras (1–4) are outlined by dashed lines to indicate that Gal-3 is not structurally required at the base tier. To illustrate how the signalosome could grow beyond eight protomers, 16 protomers are shown. f, One catalytic unit centered on a Raf kinase dimer. ATP and phosphorylated Raf residues (pSer338, pTyr341 and pSer621) are in red. g, Previously resolved structures used as building blocks in the modeling: the K-Ras monomer85 (PDB 4DSN), H-Ras–Raf RBD complex47 (PDB 4G0N), C-Raf CRD86 (PDB 1FAR), Gal-3 (ref. 87) (PDB 3ZSM), C-Raf kinase domain dimer64 (PDB 3OMV), B-Raf–MEK1 complex65 (PDB 4MNE) and 14–3-3σ dimer bound to C-Raf phosphopeptide63 (PDB 4IEA).
Unstructured linkers connect the CRDs of C-Raf proteins to their kinase domains, which dimerize at the periphery of the structure (Fig. 1d,e), in complex with MEK1 kinases and 14–3-3 dimers (Fig. 1f). The signalosome model, which is structurally open-ended, can accommodate a variable number of K-Ras and Raf molecules. We anticipate that the signalosome is regulated by GAP and GEF at the cell membrane and thus cannot grow beyond the reach of membrane-anchored GEF.
In this report, we mostly focus on an eight-protomer signalosome as an example system. The signalosome (Supplementary Video 1, Fig. 1d and Extended Data Fig. 1a; atomic coordinates in Supplementary Dataset 1) produces a millimolar local concentration of C-Raf (Supplementary Note and Analyses), which would be sufficient to ensure the dimerization-dependent activation of C-Raf29. The model incorporates seven previously resolved single- or double-domain structures of the constituent proteins (Fig. 1g). The following sections describe in greater detail our construction of this signalosome model, which was informed by existing experimental data, new experiments conducted as part of the current study, and previous experience with molecular dynamics simulations of protein–protein and protein–small molecule association30–32.
A GMA dimer of active K-Ras.
We first performed 20 simulations (680 μs total simulation time) of two GTP-bound K-Ras proteins (Protein Data Bank (PDB) ID 4DSN) in aqueous solvent (Extended Data Fig. 2a, left). In one of the simulations, two K-Ras proteins formed a symmetric dimer that is almost identical (Extended Data Fig. 2b) to the crystal structure of a widely discussed symmetric Ras dimer that uses the α4–α5 helices as the dimer interface13,20, illustrating the ability of the simulations to recapitulate experimentally observed interactions for this system. (We will discuss this and other existing Ras dimer models in relation to our signalosome model later in this article.) In another simulation, two K-Ras proteins formed stable interactions mediated in part by a bound GTP (Supplementary Video 2). This model is new and compelling because it provides a direct explanation for the GTP dependence of K-Ras dimerization16. Hereafter we will refer to this model as the GMA dimer model.
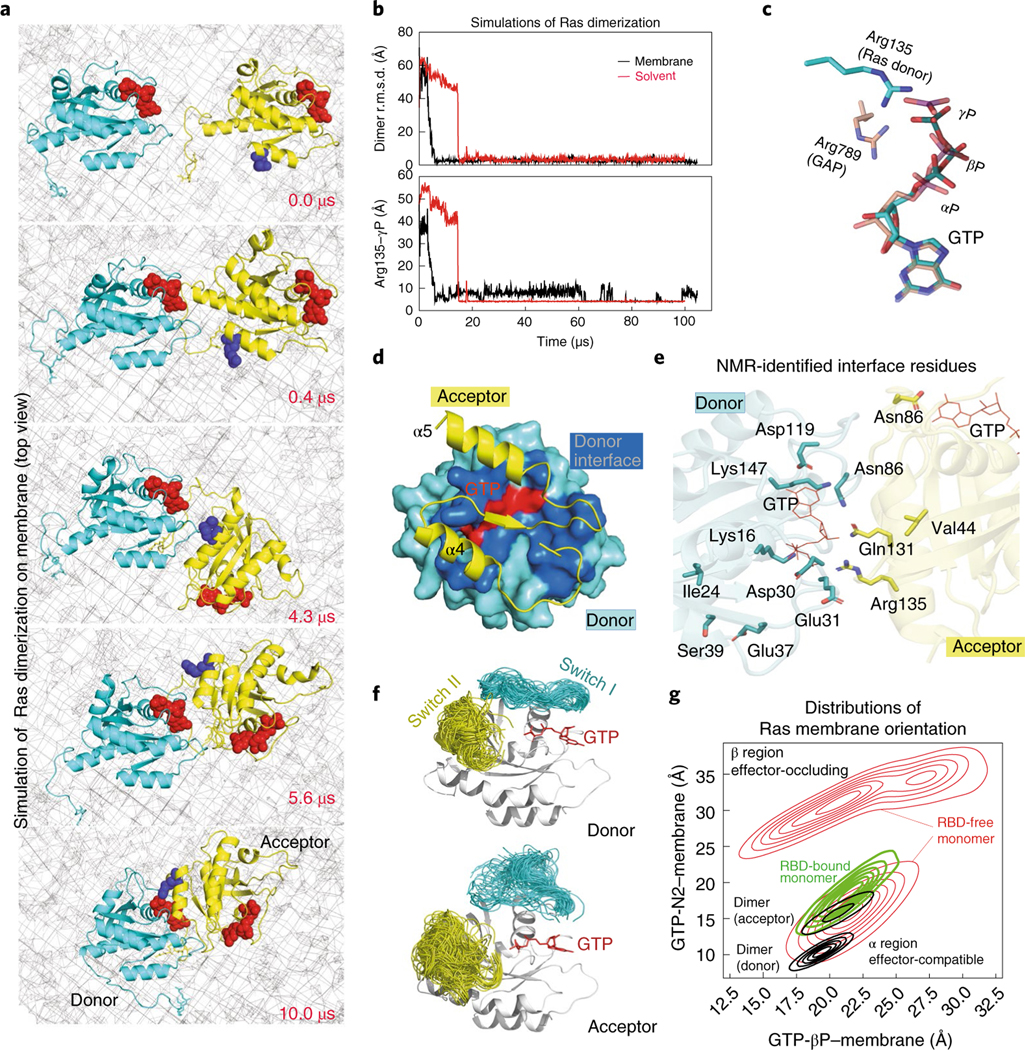
Because K-Ras dimerization occurs at the membrane, we then performed 23 simulations (363 μs total simulation time) of two GTP-bound K-Ras proteins anchored to the membrane by their fCys185 residues (Extended Data Fig. 2a, right). In one of these membrane simulations (Fig. 2a and Supplementary Video 3), the K-Ras proteins also formed the GMA dimer; the structure is virtually identical to that obtained from the solvent simulations (Fig. 2b, upper panel). The GMA dimer (atomic coordinates in Supplementary Dataset 2) remained stable (Fig. 2b).
Fig. 2 |. The GMA dimer model of K-Ras.
a, Snapshots, taken at the simulation times shown, of the GTP-mediated dimerization of membrane-anchored K-Ras proteins (top view, membrane shown as a mesh). b, Upper panel, the root mean squared deviation (r.m.s.d.) of the two K-Ras proteins in the solvent and in the membranous simulation of K-Ras dimerization with respect to the final GMA K-Ras dimer model (the snapshot at 100 μs from the simulation shown in a). Lower panel, the Arg135 distance to the GTP γ-phosphate (γP) at the dimer interface in the simulations. c, Comparison of the GTP–Arg135 (or Lys128) interaction with the GAP ‘arginine finger’. d, The dimer interface; only part of the acceptor is shown. e, All 12 NMR-identified residues16 involved in K-Ras dimerization, shown with respect to the GMA dimer interface. f, The conformations of the switch I and II regions in simulations of the GMA dimer. g, Distributions of K-Ras membrane orientations in simulations of a K-Ras monomer, an RBD-bound K-Ras monomer and the GMA dimer in the membrane environment; the orientations are mapped to a 2D space defined by the distances of the GTP β-phosphate (βP) and the GTP amine nitrogen of the guanine ring (N2) to the membrane plane (Extended Data Fig. 2d,e). The α and β regions correspond to the α and β orientations40, respectively; in the former, the α4 and α5 helices contact the membrane and, in the latter, the β strands do.
At the GMA dimer interface, a key interaction occurs between the GTP γ-phosphate of one K-Ras protein and Arg135 or Lys128 of the other (Fig. 2b,c and Extended Data Fig. 2c). Hereafter, we will refer to the former K-Ras protein as the GTP donor (or, more briefly, the donor) and the latter as the GTP acceptor (or simply the acceptor). Unlike the ‘arginine finger’ interaction between Ras and GAP, whereby an arginine mediates electron transfer from the γ-phosphate to the β-phosphate33 and catalyzes GTP hydrolysis34, the GMA dimer model has Arg135 or Lys128 interacting only with the γ-phosphate (Fig. 2c), hindering electron transfer and thus GTP hydrolysis.
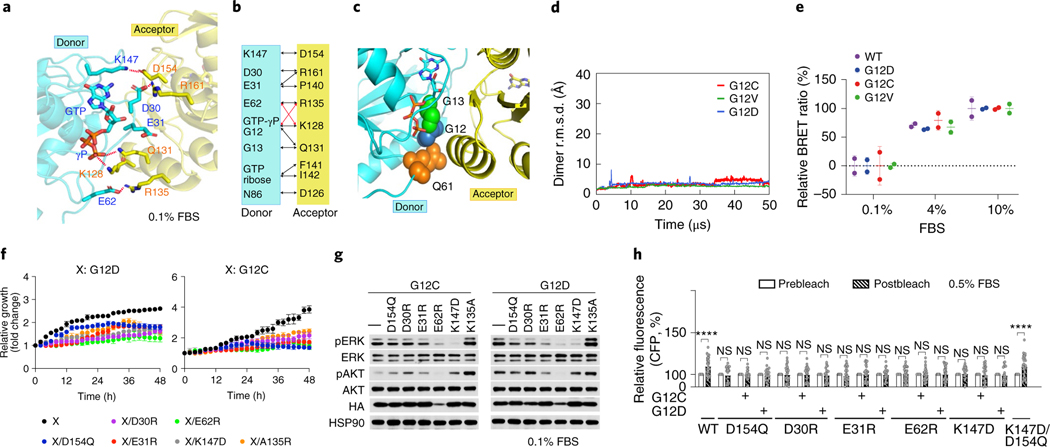
The GMA dimer interface has a buried surface area of roughly 1,750 Å2 and pronounced electrostatic complementarity (Extended Data Fig. 2j). The donor interface involves parts of the switch I region and the β4–α3, β5–α4 and β6–α5 loops. The acceptor interface primarily involves the α4 and α5 helices and the β6 strand (Fig. 2d) and largely overlaps with the interface for NS1 (Extended Data Fig. 2i), a synthetic protein that disrupts Ras dimerization19. The GMA dimer is consistent with existing nuclear magnetic resonance (NMR) spectroscopy data: residues that exhibited broadening of 15N-HSQC spectra on sample dilution16 are predominantly located at the dimer interface (Fig. 2e and Supplementary Analyses). The interface is mostly polar (Extended Data Fig. 2e), suggesting modest dimerization affinity. Several salt bridges (for example, Asp154–Lys147, Arg161–Asp30/Glu31 and Glu62–Lys128/Arg135) are present at the interface (Fig. 3a,b).
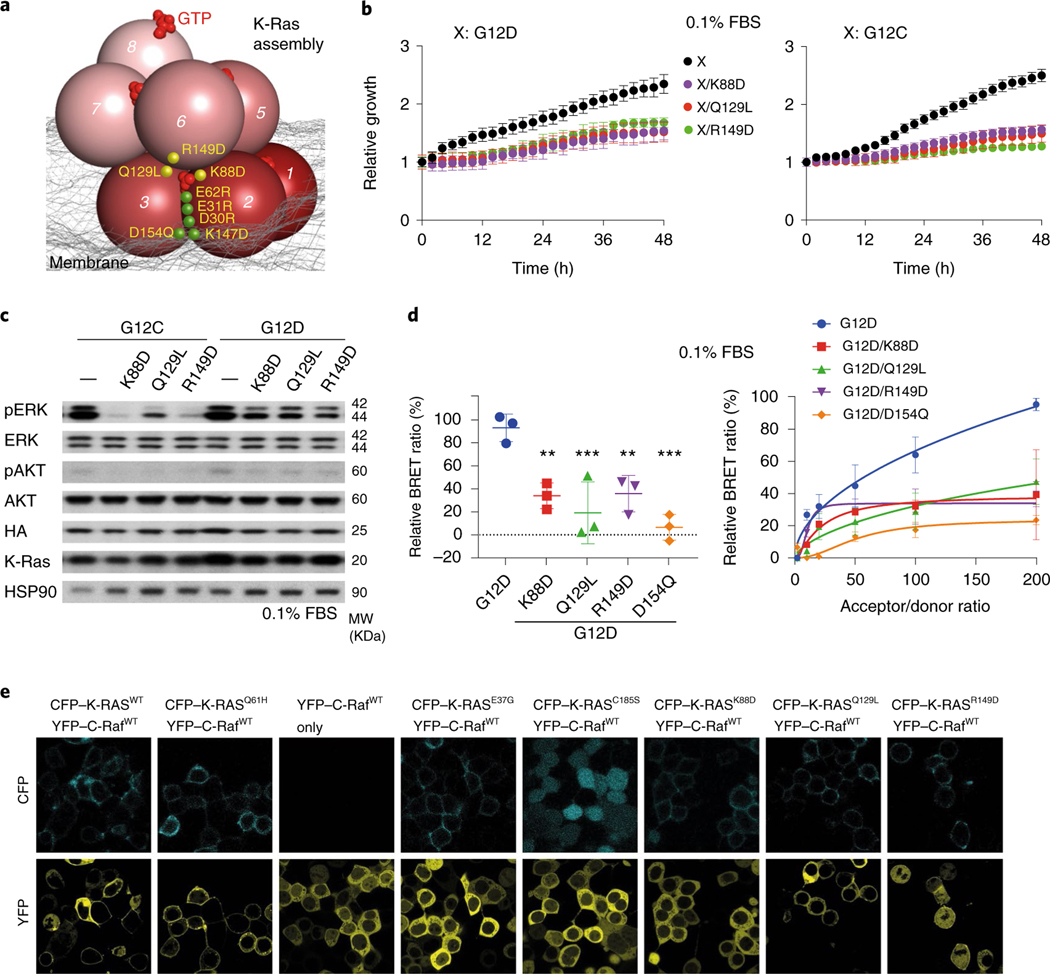
Fig. 3 |. Mutagenesis validation of the GMA dimer interface.
a, The residues involved in the salt bridges at the dimer interface. b, Residue contacts at the dimer interface (black and red lines). Red lines highlight E62–K128 and GTP-γP–R135 contacts, which alternate with E62–R135 and γP–K128 contacts. c, G12, G13 and Q61 oncogenic mutation sites at the GMA dimer interface. d, The stability of the G12C, G12D and G12V GMA dimers in simulations in terms of Cα r.m.s.d. e, BRET signals, which provide an indication of K-Ras assembly, for wild-type (WT) K-Ras and three oncogenic mutants at three different FBS concentrations. f, Growth rates of K-Raslox/K-RASMUT cells shown as confluence values measured by IncuCyte. G12C or G12D was in the background of the tested mutations. Representative images at the endpoint are shown in Extended Data Fig. 4a. Cells were kept in 0.1% FBS (see Extended Data Fig. 4b for curves obtained in 0.5%, 1% and 10% FBS). Data are shown as mean ± s.d. (n = 3 biologically independent experiments). g, Phosphorylation of ERK and AKT in K-Raslox/K-RASMUT cells. Cells were lysed after incubation for 48 h. See Extended Data Fig. 4c for the phosphorylation at other FBS levels. These results are representative of three independent experiments with similar results. h, CFP emission in HEK293T cells cotransfected with CFP- and YFP-fused K-Ras (wild type and various mutants), where binding between the CFP- and YFP-fused proteins would bring the fluorescent tags into close proximity, leading to an increased CFP emission after YFP bleaching. Assays were repeated three times. Data are shown as mean ± s.e.m. WT, n = 35; D154Q, n = 22; G12C/D154Q, n = 18; G12D/D154Q, n = 20; D30R, n = 30; G12C/D30R, n = 26; G12D/D30R, n = 23; E31R, n = 32; G12C/E31R, n = 27; G12D/E31R, n = 23; E62R, n = 34; G12C/E62R, n = 33; G12D/E62R, n = 36; K147D, n = 25; G12C/K147D, n = 26; G12D/K147D, n = 22 and K147D/D154Q, n = 31 (****P < 0.0001 by two-way ANOVA; NS, not significant).
The GMA dimer stabilizes the active state of K-Ras.
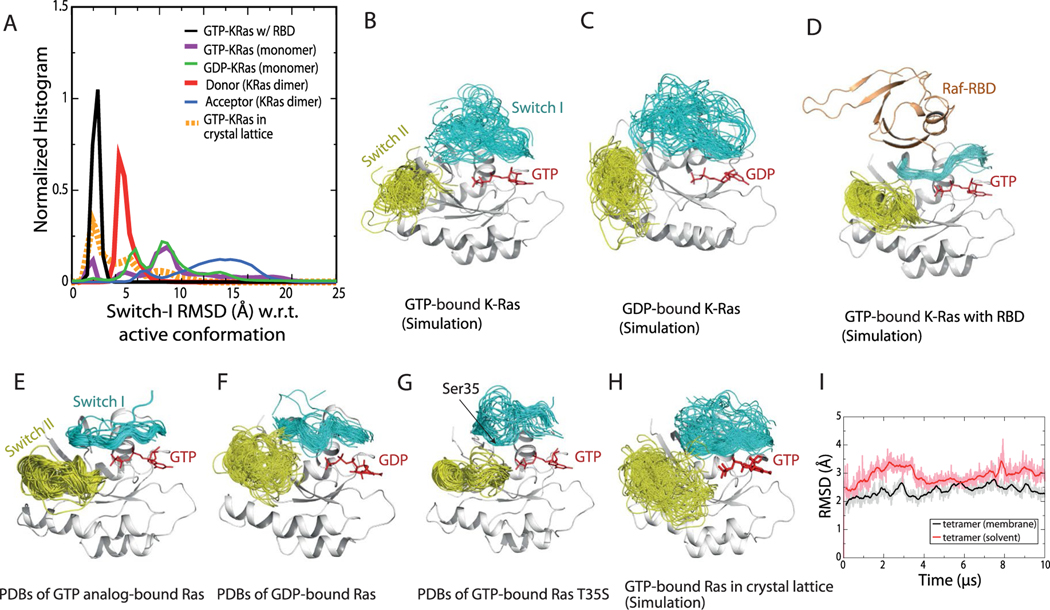
The GMA dimer may promote RBD binding by stabilizing the active Ras conformation and favoring membrane orientations that accommodate the RBD. It has been shown that GTP binding favors the active conformation35 but does not fully stabilize it9,36–39. In our simulations, although the switch I region visited the active conformation with greater frequency in a GTP-bound K-Ras monomer than in a GDP-bound one (Extended Data Fig. 3a), the inactive conformation was prevalent in both (Extended Data Fig. 3b,c). In contrast, in simulations of the GMA dimer, the switch I region of the donor (but not that of the acceptor) was largely stabilized in active-like conformations (Fig. 2f and Extended Data Fig. 3a).
Our GMA dimer model also constrains the membrane orientation of K-Ras in a way that is consistent with previous NMR and simulation data40–42. In the GMA dimer formed on the membrane (Fig. 1a), the donor is in the α orientation40 and the acceptor is in a similar orientation (Fig. 2g). Both the donor and acceptor orientations are compatible with RBD binding (Extended Data Fig. 2g). This contrasts with monomeric K-Ras, which mostly adopted the β membrane orientation that occludes RBD binding (Fig. 2g and Extended Data Fig. 2f).
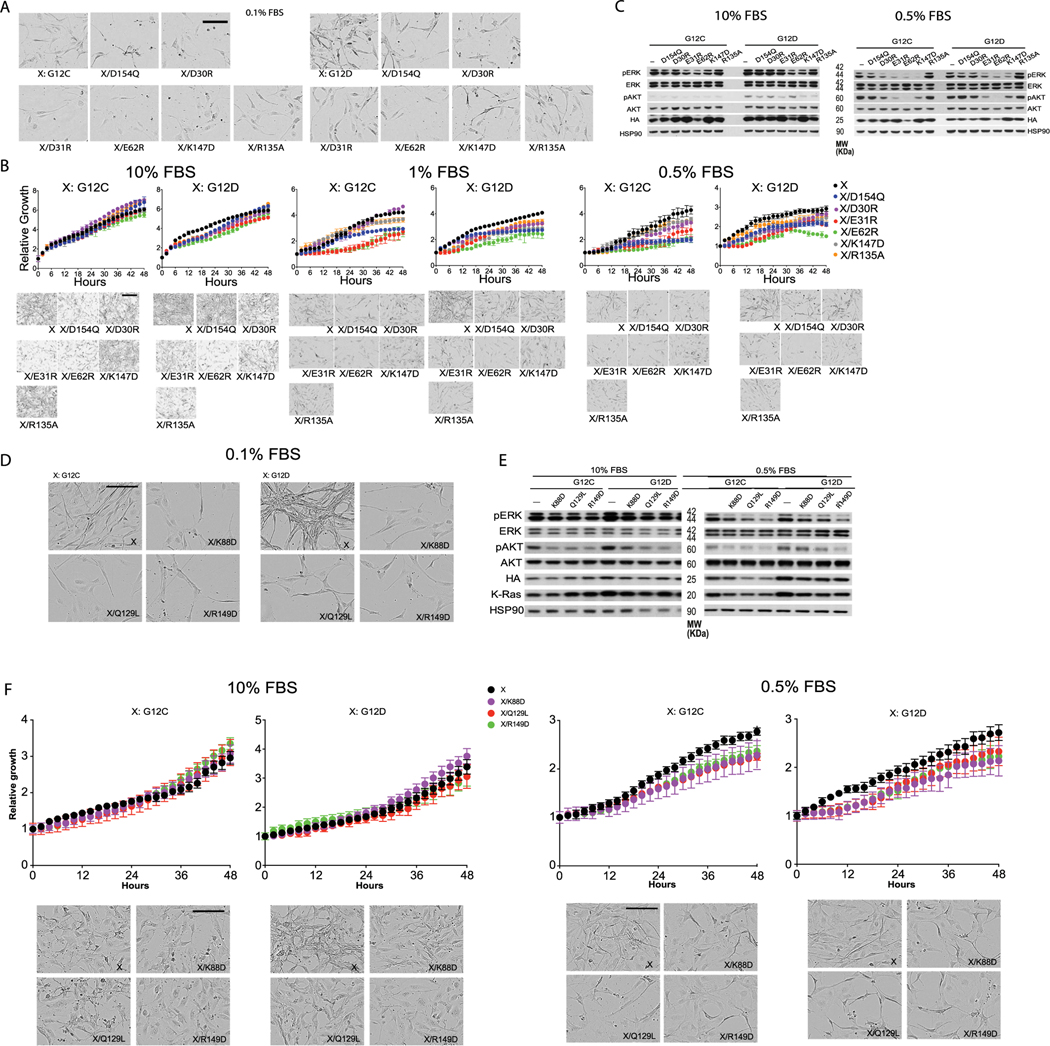
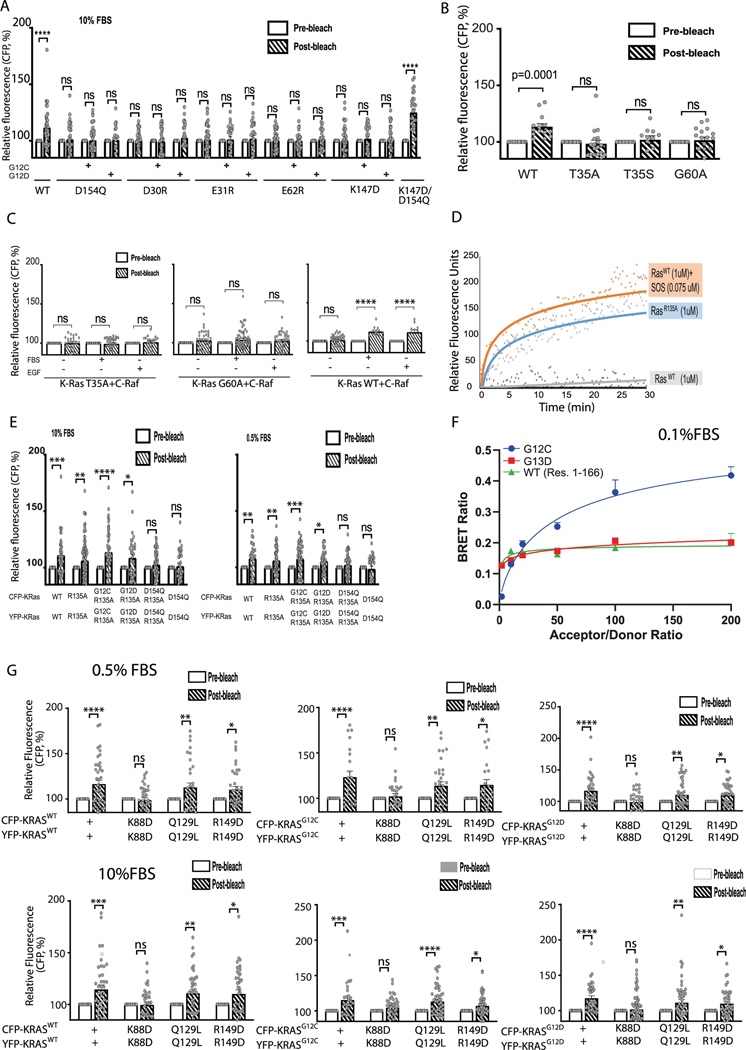
The common oncogenic K-Ras mutations are at or adjacent to the GMA dimer interface (Fig. 3c). We simulated GMA dimers with G12V, G12D, G12C, G13C, G13S, Q61L and Q61R and found that the dimer was not disrupted by these mutations (Fig. 3d and Extended Data Fig. 2j), likely because the GMA dimer interface can sterically accommodate the mutations. Although not quantified by the simulations, these mutations likely exert subtle differential effects on GMA dimerization. By contrast, the G13R and G13D mutations to bulkier residues did disrupt the GMA dimer in simulations (Extended Data Fig. 2k); this suggests that these two mutants may differ from the G12 and Q61 mutants in their signaling mechanism. Our bioluminescence resonance energy transfer (BRET) experiment showed that indeed the wild type and the G12C, G12V and G12D mutants form assemblies in cells (Fig. 3e), but the G13D mutant does not (Extended Data Fig. 5f).
Experimental validation of the GMA dimer model.
A K-Raslox/K-RASMUT inducible system—wherein cell lines generated from Ras-less mouse embryonic fibroblasts (MEFs)43 are dependent on exogenous K-Ras expression for proliferation—has been reported in a previous study20, along with a cell-based fluorescence resonance energy transfer (FRET) platform, to probe K-Ras–K-Ras interactions. In both experiments, K-Ras signaling is constitutive with introduction of oncogenic K-Ras mutants such as G12D or G12C. Because the G12D and G12C mutations are compatible with the dimer (Fig. 3d,e), we used these two systems to test the GMA dimer. The MEF and FRET experiments showed that D154Q and R161E mutations in the background of either G12C or G12D impair cell fitness by disrupting K-Ras–K-Ras interactions. The GMA dimer model is also consistent with those findings, because Asp154 and Arg161 are involved in key salt bridges at the GMA dimer interface (Fig. 3a).
We used the MEF and FRET systems to test residues (Asp30, Glu31, Glu62 and Lys147) involved in the GMA dimer interface (Fig. 3a,b). We found that D30R, E31R, E62R and K147D mutations each impaired cell growth (Fig. 3f and Extended Data Fig. 4b) and reduced ERK phosphorylation (Fig. 3g and Extended Data Fig. 4c). The mutations also disrupted the FRET signal of K-Ras–K-Ras interactions (Fig. 3h and Extended Data Fig. 5a). We further found that a D154Q/K147D double mutation, which was predicted to restore the interaction, indeed recovered the FRET signal (Fig. 3h and Extended Data Fig. 5a). FRET analysis also showed that mutations disrupting the active conformation of Ras, such as T35A, T35S38 and G60A44, also disrupted K-Ras–K-Ras interactions (Extended Data Fig. 5b,c), supporting the connection between GMA dimerization and the active conformation. The substantial set of FRET and MEF results collectively are highly consistent with and provide strong support for the GMA dimer model. Of these results, we find the observed compensatory effect of D154Q and K147D mutations especially compelling.
Extrapolation of the GMA dimer into a K-Ras helical assembly.
Adding K-Ras proteins to the GMA dimer in a head-to-tail fashion extends it into a left-handed helical assembly, which is highly compact and yet does not exhibit any steric clashes (Fig. 1b). In this assembly, each K-Ras protein serves as both a donor and an acceptor (with the exception of the ‘head’ serving only as a donor and the ‘tail’ serving only as an acceptor). For convenience, we number the K-Ras proteins in the assembly from 1 to n starting from the head (and incrementing by one from a donor to its acceptor). The helical assembly consists of four K-Ras proteins per turn and has a radius of roughly 50 Å and a pitch of roughly 40 Å, with virtually no unoccupied space between turns or in the center. We placed the K-Ras assembly on the membrane, with the axis of the helical assembly at an approximately 60° angle to the membrane surface (Fig. 1b, left), which maximizes membrane contact with the base tier. We refer to the tier comprising K-Ras 1–4 as the base tier and the four highest-numbered K-Ras proteins as the top tier. The helical assembly can grow in a straight tower shape, but the base tier alone, without further assembly, is stable on the membrane in our simulations (Extended Data Fig. 3i). In the helical assembly, the RBD interface of each K-Ras protein is exposed and available for Raf recruitment. Further, the C and N termini of the K-Ras proteins are positioned outward at the surface of the assembly, allowing the C-terminal tail to interact with other proteins and for fluorescent proteins to fuse with the N termini45 without disrupting the assembly.
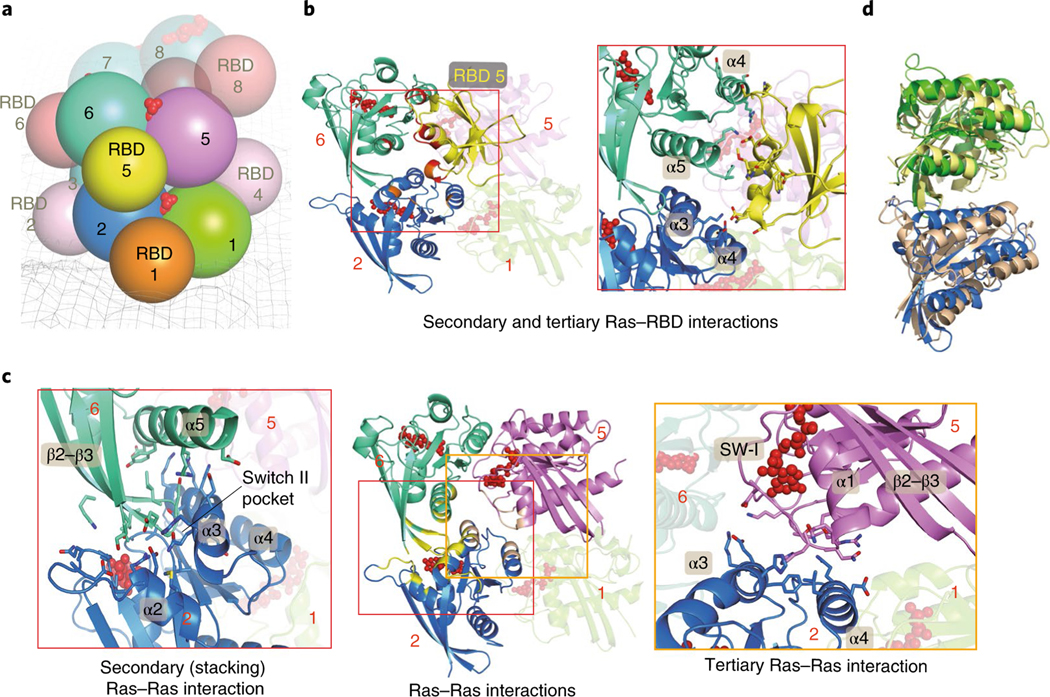
A K-Ras protein at position n in our helical assembly model engages in multivalent interactions (Fig. 4a), making contact with up to six other K-Ras proteins: GTP-mediated primary interactions with K-Ras n − 1 and n + 1, secondary (stacking) interactions with K-Ras n − 4 and n + 4 along the axis of the helical assembly, and tertiary interactions with K-Ras n − 3 and n + 3 (Fig. 4c and Extended Data Fig. 6a,b). The secondary and tertiary interactions are predominantly polar, with a buried surface area of roughly 1,450 Å2 and 660 Å2, respectively (Extended Data Fig. 6a,b). Crystallographic and simulation data provide independent support for the secondary interaction. In one of the 20 simulations of the association of two K-Ras molecules (Extended Data Fig. 2a), the secondary (stacking) dimer formed and remained stable (Fig. 4d). Further, the secondary dimer structure closely resembles a K-Ras dimer seen in crystal packing (for example, in PDB 5UQW; Extended Data Fig. 6f). The secondary interaction places the β2–β3 hairpin of K-Ras n + 4 into what is known as the ‘switch II pocket’ of K-Ras n, which is exploited by covalent inhibitors of K-Ras G12C46.
Fig. 4 |. K-Ras–K-Ras and K-Ras–RBD interactions in the signalosome model.
a, Cartoon representation of the relative positions of K-Ras proteins and RBD domains in the model; the membrane is shown as a mesh and GTP is shown in red. b, Left, the primary (with K-Ras 5), secondary (with K-Ras 6) and tertiary (with K-Ras 2) RBD–Ras interactions (of RBD 5); a fourth K-Ras (K-Ras 1) is shown for geometric context. The secondary and tertiary interface residues (and GTP) are colored in red and orange. Right, close-up of the Ras–RBD interactions. c, Center, three K-Ras proteins engaged in primary, secondary and tertiary interactions with one another; the interface residues are shown in yellow and GTP is shown in red. Left and right, close-ups of the secondary (stacking) and tertiary Ras–Ras interactions, respectively. d, Superposition of a K-Ras dimer generated by simulation of K-Ras dimerization in aqueous solution and the secondary K-Ras dimer interactions in the helical assembly model.
Experimental validation of the helical assembly.
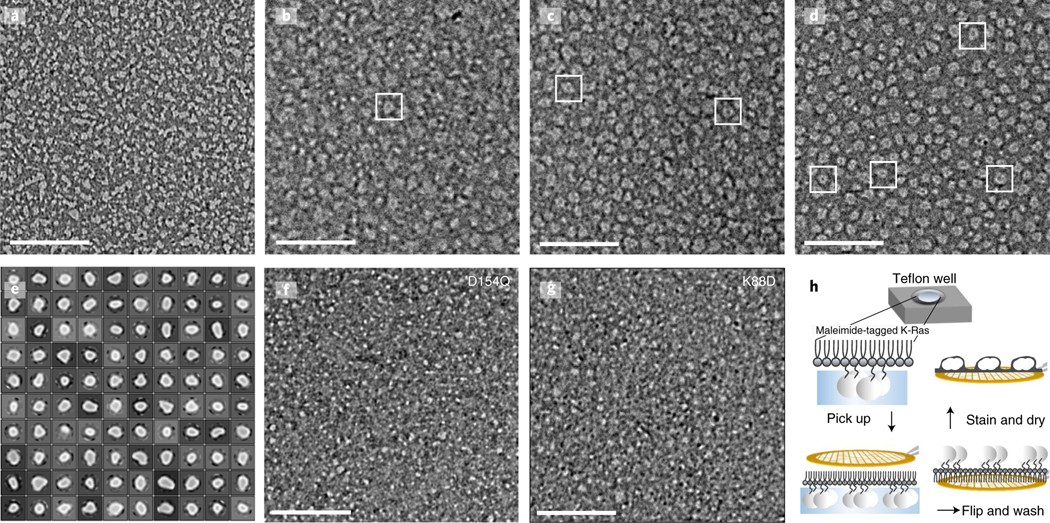
To experimentally validate the K-Ras helical assembly, we first used negative-stain electron microscopy to image K-Ras particles in a reconstituted system. In these experiments, full-length K-Ras proteins were tethered to a lipid monolayer by Cys185, forming chemical bonds with maleimide lipids. The K-Ras proteins were then applied to a grid, stained and imaged (Fig. 5h). The K-Ras particles form in a maleimide lipid-dependent manner (Fig. 5a–d), consistent with the notion that K-Ras assembly requires K-Ras membrane localization. Classification of 73,282 visualized particles into 100 classes revealed a range of sizes (Fig. 5e). The variability of particle size is consistent with the notion that the helical assembly can accommodate different numbers of K-Ras molecules, but particle heterogeneity prevented reconstruction of the 3D structures. Consistent with the MEF FRET (Fig. 3f–h) and BRET (Fig. 6d) analyses, the D154Q mutation disrupted particle formation (Fig. 5f). Particle formation was also disrupted by K88D (Fig. 5g), a mutation that disrupted the FRET and BRET signals of K-Ras assembly in cells (Fig. 6d and Extended Data Fig. 5g). These mutagenesis data indicate that, at least partially, the helical assemblies underlie the K-Ras particles we visualized. Based on our model, a subtle yet important difference between the effects of D154Q and K88D is expected: D154Q is located at the primary interface, and it should thus disrupt GMA dimers, whereas K88D is at the secondary interface, and it should thus not interfere with GMA dimerization. This prediction is consistent with the negative-stain images, in which the D154Q particles generally appear to be smaller than the K88D ones (Fig. 5f,g) under identical conditions.
Fig. 5 |. Electron microscopy images with negative stain of K-Ras assembly.
a–d, K-Ras wild type at a 10 μM concentration; maleimide lipids were used at the following percentages: 0% (a), 5% (b), 10% (c) and 20% (d). e, 2D class average of the K-Ras wild-type particles. f, D154Q K-Ras at a 10 μM concentration with 20% maleimide lipids. g, K88D K-Ras at a 10 μM concentration with 20% maleimide lipids. h, Workflow of the experiment. After incubation with the monolayer lipid overnight, the K-Ras particles were moved to a carbon-coated copper grid, washed intensively, negatively stained and examined by electron microscopy. Scale bars, 200 nm.
Fig. 6 |. Mutagenesis validation of the secondary and tertiary K-Ras–K-Ras interactions.
a, Illustration of the locations of the tested mutations, including those at the primary interface shown in Fig. 3f–h (green) and those at the secondary and tertiary interfaces (yellow). b, Growth rates of K-Raslox/K-RASMUT cells with K88D, Q129L and R149D mutations in cis with either G12C or G12D. Representative images at the endpoint are shown in Extended Data Fig. 4d. See Extended Data Fig. 4f for results obtained in 0.5% and 10% FBS. Data are shown as mean ± s.d. (n = 3 biologically independent experiments). c, Phosphorylation of ERK and AKT in K-Raslox/K-RASMUT cells. See Extended Data Fig. 4e for the phosphorylation in 0.5% and 10% FBS. These results are representative of three independent experiments with similar results. d, Left, endpoint BRET measurements for cells with K88D, Q129L and R149D in the background of G12D, compared with the G12D mutation only; D154Q is included as a positive control. K-Ras construct (residues 1–166) lacking the membrane-anchoring hypervariable region was also used as a control (Extended Data Fig. 5f). The BRET signals were normalized to the maximum signal of cells with only G12D. An acceptor/donor expression plasmid ratio of 200:1 was chosen for comparing the K-Ras mutants. The average of three technical replicates was calculated for each of the biological triplicates and subjected to one-way ANOVA followed by Dunnett’s post tests (**P < 0.01, ***P < 0.001). Right, cotransfection of increasing ratios of donor and acceptor plasmids, which allows discrimination between specific and nonspecific (random collision) protein–protein interactions. e, Fluorescence imaging of Raf localization of K-Ras mutants K88D, Q129L and R149D; K-Ras wild type and Q61H are used as positive controls, and E37G, C185S and K-Ras-free cells are used as negative controls. Assays were repeated three times and 8–10 regions of interest were imaged each time.
We also evaluated mutations predicted to disrupt the K-Ras helical assembly using the MEF BRET and FRET systems. K-Ras-dependent MEFs with K88D, which was predicted to disrupt the secondary interface, and Q129L and R149D, which were predicted to disrupt the tertiary K-Ras–K-Ras interface (Fig. 6a), slowed proliferation (Fig. 6b and Extended Data Fig. 4f) and impaired ERK phosphorylation (Fig. 6c and Extended Data Fig. 4e). To supplement the FRET experiment, we confirmed using BRET, which allows better quantification, that K88D, Q129L and R149D mutations in the background of G12D disrupt K-Ras assembly (Fig. 6d). Although they are distal to the Raf interface of K-Ras, K88D, Q129L and R149D mutations also hindered K-Ras recruitment of C-Raf to the cell membrane (Fig. 6e), suggesting that the K-Ras assemblies play an important role in K-Ras–Raf interaction.
Multivalent interactions between C-Raf and the K-Ras assembly.
Using a cocrystal structure of the C-Raf RBD and H-Ras (PDB 4G0N)47 as a template, we docked a C-Raf RBD to each K-Ras of the helical assembly. The docked RBDs did not clash with any K-Ras proteins, nor with one another. The RBD that primarily interacts with K-Ras n, which we refer to as RBD n, forms a secondary contact with K-Ras n + 1 and a tertiary contact with K-Ras n − 3 (Fig. 4b and Extended Data Fig. 6c,d). These secondary and tertiary Ras–RBD interactions may confer an advantage on the helical assembly over a K-Ras monomer in terms of RBD binding, and, conversely, RBD binding may help stabilize the K-Ras assembly.
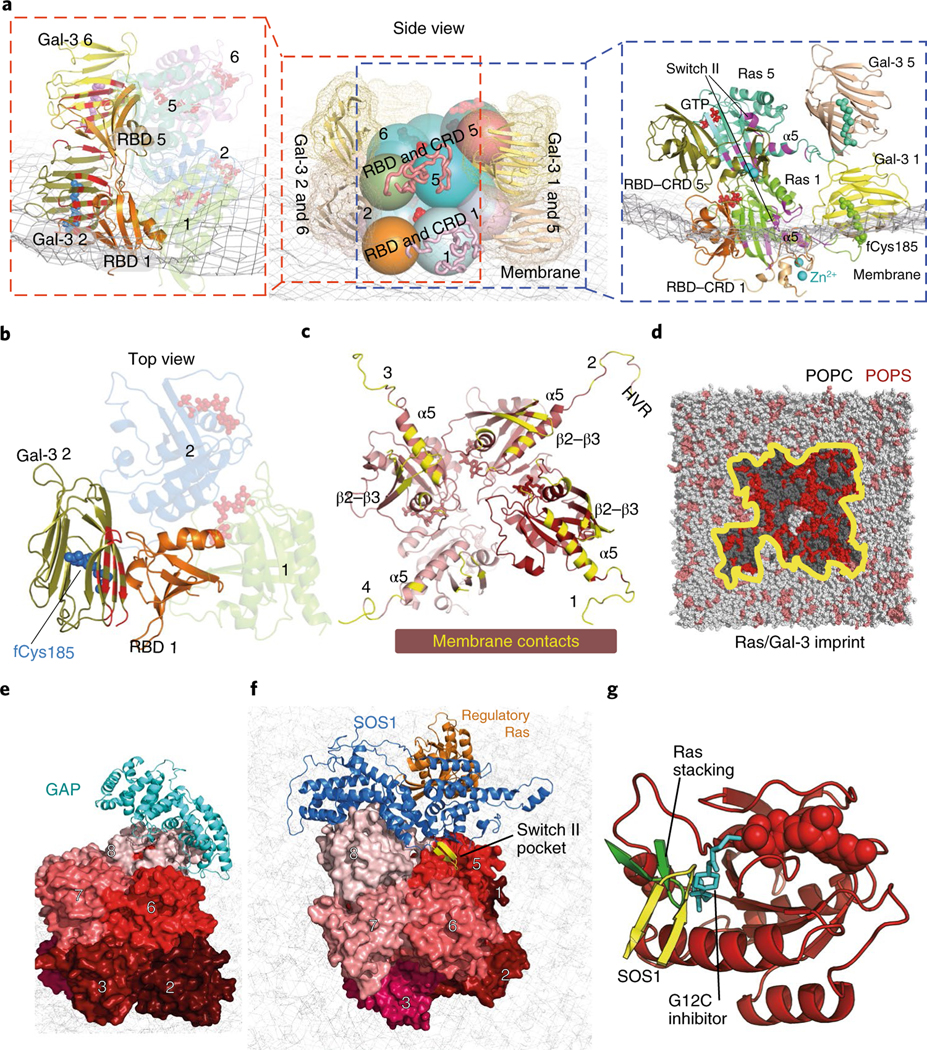
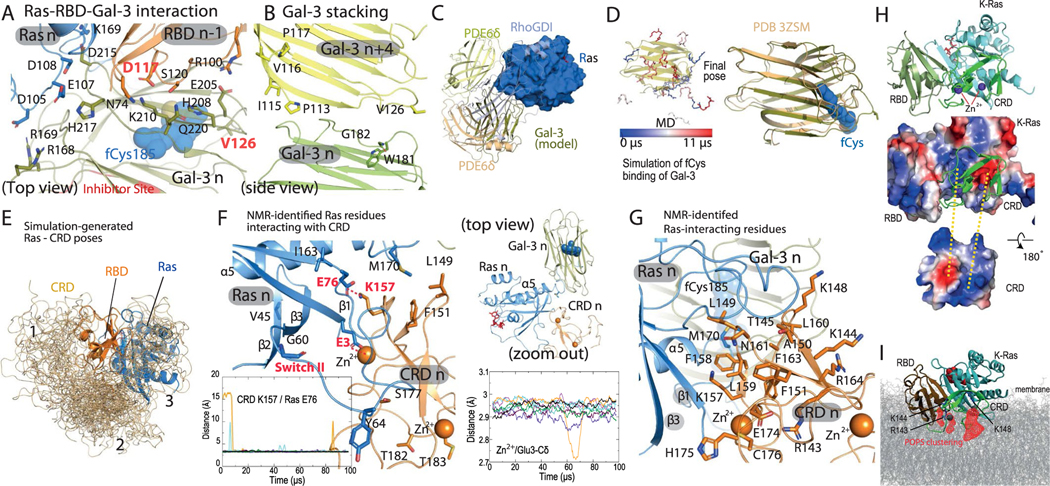
The 52-residue zinc-coordinated Raf CRD also interacts with Ras48 and imparts specificity to Ras–Raf binding49. Informed by experimental data and extensive simulations of a CRD tethered to a structure of a K-Ras–RBD complex (Supplementary Analyses), we positioned the CRD at the C terminus of the α5 helix and β strands of K-Ras (Fig. 7a), with an interface of roughly 1,400 Å2. The CRD also contacts the switch II region of K-Ras (Extended Data Fig. 7f); this is consistent with switch II mutations (G60A and Y64W) hindering the CRD binding of H-Ras50. NMR and other analyses have identified a set of CRD and Ras residues involved in the Ras–CRD interaction48,51–53; these residues are either part of the modeled Ras–CRD interface or near it (Extended Data Fig. 7f,g). By our model, a CRD interacts with a K-Ras and an adjacent RBD with extensive electrostatic complementarity (Extended Data Fig. 7h). We added a CRD to each K-Ras protein in the helical assembly. In the base tier, both the RBDs and CRDs interact extensively with the membrane (Fig. 7a).
Fig. 7 |. Ras interactions with Gal-3 and Raf RBD and CRD domains.
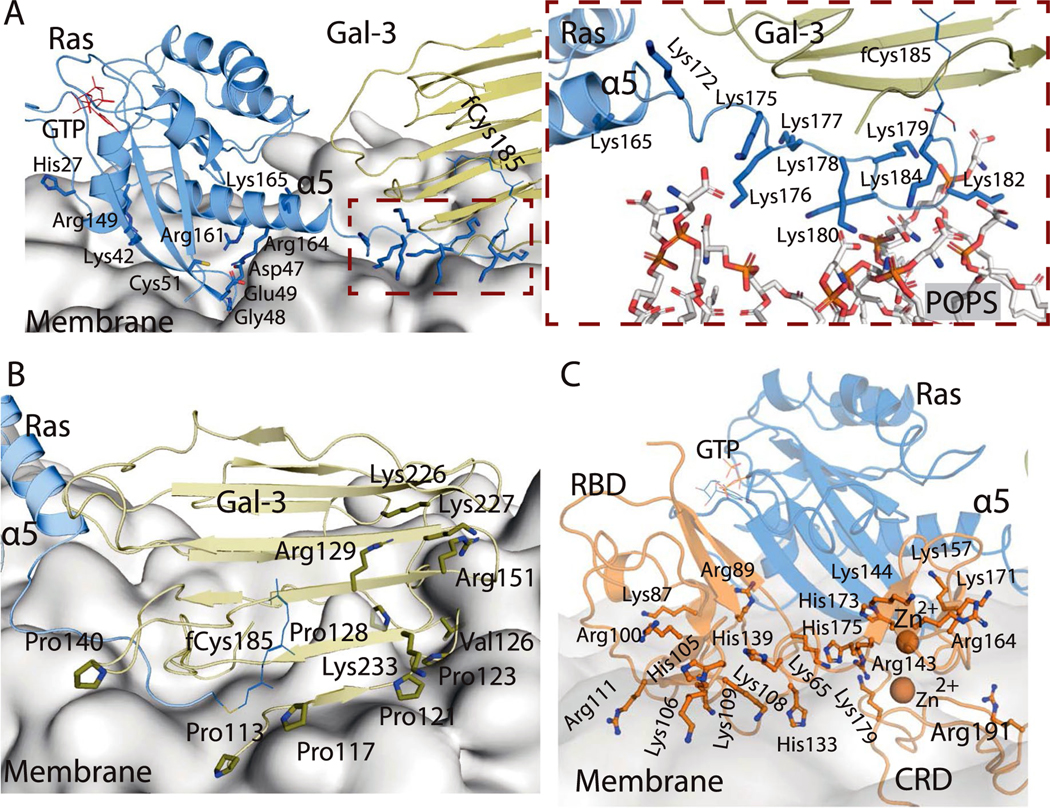
a, Side views of the interactions of two stacking K-Ras proteins with Gal-3 and C-Raf RBD and CRD domains. Center, relative positions of K-Ras, Gal-3, and Raf RBD and CRD domains. K-Ras and RBDs are shown as spheres, CRDs are shown as ribbons and Gal-3 proteins are shown as ribbons surrounded by mesh. Left, the K-Ras proteins with RBDs and Gal-3 proteins in cartoon representation; the galectin-bound fCys185 residues of K-Ras are shown in blue and purple, and Gal-3–RBD interfaces are marked in red. Right, positions of CRD domains and Gal-3 proteins with respect to RBD-bound K-Ras. Ras–CRD interfaces are marked in purple. Also labeled is the α2 helix in the switch II region of K-Ras involved in the interaction with the CRD. b, Top view of the RBD interaction with Gal-3. Two K-Ras proteins, which form a GTP-mediated dimer, are shown as faded images. The fCys185 of K-Ras 2 is shown. c, K-Ras membrane-contacting residues (yellow) in the base tier. d, Membrane imprint of the K-Ras and Gal-3 proteins of the base tier, which is enriched in POPS lipids. e, GAP interacting with the tail K-Ras (K-Ras 8) of the helical assembly. f, SOS1 interacting with the tail K-Ras. In the secondary SOS1–K-Ras interaction, the SOS1 helical hairpin (yellow) is inserted into the switch II pocket of K-Ras 5. g, Three interactions using the switch II pocket: the secondary SOS1–K-Ras interaction, the K-Ras–K-Ras stacking involving the β2–β3 turn (green) and covalent G12C inhibitor binding (cyan).
Gal-3 plays important roles in the signalosome.
The mechanism of K-Ras membrane localization is commonly thought to be insertion of fCys185. Consistent with this notion, the C-terminal tails of K-Ras molecules at the base tier of the helical assembly contact the membrane, and thus their fCys185 residues can be readily buried in the membrane. For the higher tiers, the C-terminal tails are lifted from the membrane, and the fCys185 residues in our model are capped by Gal-3 molecules. This is motivated by the notion that Gal-3, which can cap the farnesyl group of fCys185, mediates membrane localization of K-Ras54 and is essential for K-Ras nanoclustering23,55,56, as is Gal-1 for H-Ras nanoclustering24,57,58.
Since galectins associate with the lipid bilayer59 and have been shown to anchor Ras to the membrane57, we also considered another scenario in which all fCys185 residues, including those in the base tier, interact with Gal-3 proteins and Gal-3 proteins interact with and localize the K-Ras assembly to the membrane. Although this scenario departs from the current understanding of how fCys185 interacts with the membrane, we find it compelling in that it provides an explanation for the importance of Gal-3 dimerization in facilitating K-Ras nanoclustering60. The Ras–Raf signalosome model was stable in simulations with or without base-tier Gal-3 molecules (Extended Data Fig. 1c). Further experimental elucidation will be needed to fully establish the role of Gal-3 in the signalosome.
In the presence of C-Raf RBDs and CRDs surrounding the K-Ras helical assembly, the unoccupied space is limited but can accommodate the Gal-3 molecules. Existing mutagenesis and dimerization data on Gal-3 and Gal-1 with respect to Ras nanoclustering, and extensive simulations (Supplementary Note and Analyses), led us to a model in which Gal-3 n (the Gal-3 protein bound to the fCys185 residue of K-Ras n) interacts with RBD n − 1 (Fig. 7a,b). Any pair of Gal-3 n and n + 4 proteins in the model are stacked parallel to the K-Ras stacking, with the C-terminal tail of K-Ras n + 4 positioned near the interface, contacting both Gal-3 proteins (Fig. 7a,d and Extended Data Fig. 7b). As in the GMA dimer, K-Ras proteins in the base tier of the helical assembly adopt the α-like orientation, contacting the membrane with their α5 helices and β2–β3 hairpins (Fig. 7c), and Gal-3 proteins interact with the membrane extensively (Fig. 7a and Extended Data Fig. 8a,b). This model of K-Ras–Gal-3 interaction resembles resolved K-Ras–PDEδ structures61 (see the Supplementary Analyses for a functional comparison of Gal-3 and PDEδ). From this model, by simply removing the four Gal-3 molecules at the base tier and embedding the fCys185 residues, we obtained a model that is consistent with the generally accepted view that fCys185 embedding in membrane is crucial to the membrane anchoring of K-Ras. With or without Gal-3 at the base tier, the model was stable overall in our simulation (Extended Data Fig. 1b).
The C-Raf linker and a fuzzy signalosome structure.
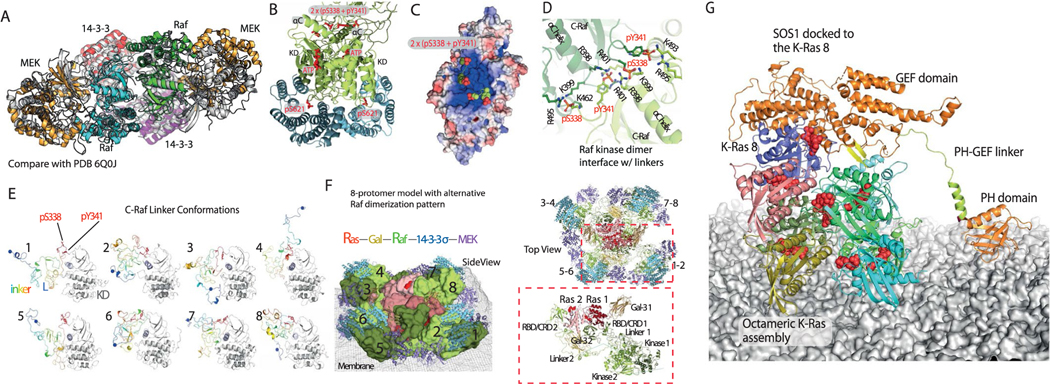
In C-Raf activation in the MAPK pathway, a 14–3-3 dimer acts as a scaffold that binds to and stabilizes a C-Raf kinase domain dimer62. C-Raf contains a loop C-terminal to the kinase domain that is phosphorylated at Ser621 and binds to 14–3-3 in C-Raf activation. There are multiple proteins in the 14–3-3 family; in the modeling, we incorporated 14–3-3σ into the Ras–Raf signalosome. Each C-Raf kinase domain also binds to and phosphorylates MEK1 kinase. We separately modeled the C-Raf kinase domain dimer complexed with a 14–3-3σ dimer and two MEK1 kinase domains (Fig. 1f) using crystal structures63–65 that were available at the time we were constructing the model (Fig. 1g) (Supplementary Note and Analyses). The resulting model is overall highly consistent with subsequently reported cryo-EM structures of the Raf–14–3-3 heterotetramer (Extended Data Fig. 9a)66,67. C-Raf protein contains a roughly 200-residue-long, unstructured linker between its CRD and kinase domains. We modeled such linkers to connect the RBD and CRD domains to the kinase domains (Supplementary Note and Analyses). We obtained a diverse set of conformations with transient secondary structures (Extended Data Fig. 9e). We then grafted a linker in a simulation-generated conformation onto each CRD in the model and then grafted onto each linker pair a C-Raf kinase domain dimer bound to 14–3-3σ and MEK1 (Fig. 1f). Incorporating this model into the larger model then produced our full signalosome model. The unstructured linkers introduced ‘fuzziness’ to the signalosome, which is often seen in signalosome complexes68.
GEF and GAP regulate the size of the Ras–Raf signalosome.
GAPs, such as RasGAP, catalyze the hydrolysis of GTP in Ras to revert it to its GDP-bound state7. Ras nanoclustering has been suggested to attenuate RasGAP-catalyzed GTP hydrolysis55,56 and to sequester Ras from GAP binding23,57. Consistent with these results, RasGAP is sterically prevented from interacting with all but the tail K-Ras protein in the helical assembly. Based on an H-Ras–p120GAP structure33, GAP can dock to a tail K-Ras protein without steric clashes, provided that the K-Ras protein is not bound to Raf (Fig. 7e). GAP can then convert the tail K-Ras protein to the GDP-bound state, preventing any additional K-Raf protein from joining the helical assembly and promoting dissolution of the signalosome. Compared to the other K-Ras molecules in the signalosome, the tail K-Ras is most exposed and likely the least stable. We thus anticipate that, under GAP regulation, a signalosome will eventually disassemble by one tail-position K-Ras falling off at a time.
Based on a structure of H-Ras in complex with the GEF protein SOS1 (ref. 69), an SOS1 protein in its active conformation can also dock to the tail K-Ras protein (Fig. 7f). By ensuring that the tail K-Ras protein is in the GTP-bound state, ready to receive another K-Ras protein into the signalosome, SOS1 may help maintain and grow the signalosome. SOS1 is activated in part by an allosteric GTP-bound Ras protein70, which our model accommodates. The model predicts, moreover, that if an active SOS1 protein docks to the tail K-Ras protein n, it additionally engages K-Ras n − 3 in a secondary interaction in which a helical hairpin of SOS1 is inserted into the switch II pocket of K-Ras n − 3 (Fig. 7f,g). This secondary Ras–SOS1 interaction may contribute to the specificity of SOS1 in regulating MAPK signaling. One consequence of this SOS1 interaction is that it may place an upper limit on the height (and thus overall size) of the signalosome, since SOS1 is membrane-anchored by its pleckstrin homology domain and thus has only limited reach from the membrane.
Discussion
The Ras–Raf signalosome model we propose here entails a host of multivalent interactions. Although each interaction is weak on its own, together they give rise to a stable signalosome that provides a favorable composite interface for Raf recruitment. Signalosome formation is likely a nucleation-like process, which should yield a switch-like signal. The low affinity of the GMA dimer means that the signalosome cannot form without the membrane, which promotes GMA dimerization by raising the local concentration of K-Ras and restricting its orientation in favor of dimerization and signalosome formation (Fig. 2g).
Two (the α and β) membrane orientations of monomeric K-Ras have been observed (Fig. 2g)40, and they appear to be associated with different K-Ras assemblies. In addition to the widely discussed symmetric dimer13,20 that has been observed using crystallography and was also generated by our simulation (Extended Data Fig. 2b), a number of Ras dimer models71–73 and a pentamer model have been previously proposed15. Several crystal and NMR structures of a K-Ras–CRD complex, which differ from our model, have also recently been reported74–76. Notably, whereas the GMA dimer model imposes the α orientation of K-Ras on the membrane, these alternative dimer and pentamer models all impose the β orientation. We speculate that these differences may result from differences in the local environment—such as membrane composition and Gal-3 concentration—and may lead to distinct downstream signals.
Proteins downstream of Ras may also affect signalosome formation. Certain small-molecule inhibitors of B-Raf give rise to ‘paradoxical activation’ of Raf by inducing C-Raf–B-Raf heterodimerization and even C-Raf homodimerization64 and also promote Ras nanoclustering by inducing more, but not larger, Ras nanoclusters in cells77. Consistent with this surprising B-Raf inhibitor effect, our model suggests a scenario in which enhanced Raf dimerization promotes nucleus formation and, in turn, signalosome formation. Similarly, 14–3-3 proteins may promote Raf dimerization and in turn Ras assembly, enabling a feedback loop in MAPK signaling.
The K-Ras helical assembly provides a possible explanation for why wild-type K-Ras acts as a tumor suppressor in cells driven by oncogenic K-Ras mutants78,79. Wild-type K-Ras, which is predominantly GDP bound and cannot serve as a GTP donor, may inhibit the growth of a helical assembly by capping it at the tail position. This scenario is consistent with the D154Q K-Ras mutant being inactive as a tumor suppressor20, since the D154Q mutation at the acceptor interface may prevent the mutant from assuming the tail position. This rationale leads us to predict that mutations at the donor interface (for example, K147D and R161E) should not disrupt the tumor suppressor activity, as these mutants should still be able to assume the tail position and cap helical assemblies of oncogenic K-Ras mutants.
It is worth noting that the K-Ras helical assembly is almost certainly not applicable to PI3K activation (Supplementary Analyses) or to Raf signaling involving KSR pseudokinase as a scaffold, as in this case K-Ras dimerization may not be required80. Moreover, many proteins not incorporated in our signalosome model (for example, SHOC2, ref. 81; SUR-8, ref. 82; Aurora kinase A83; and SHP2 phosphatase84) are involved in Ras signaling and may, for example, prime K-Ras for the signalosome without being part of it. In many respects, however, findings on H-Ras nanoclustering echo those on K-Ras45, and our signalosome model is consistent with the structures of other Ras and Raf proteins (such as N-Ras, H-Ras, A-Raf and B-Raf) involved in MAPK signaling. With local alterations, our model may be extendable to those Ras and Raf proteins and provide a framework for understanding their overlapping, yet distinct roles in MAPK signaling.
Methods
Molecular dynamics simulations.
Simulation protocol and force field parameters.
Molecular dynamics simulations were performed on the special-purpose supercomputer Anton 2 (ref. 88). The simulated systems ranged in size from around 37,000 to 1,740,000 atoms, and were parameterized using the TIP3P model89 for water molecules, the Amber99SB*-ILDN force field90–95 for proteins and the CHARMM36 force field96 for lipids. Specialized force field parameters were used for farnesylated cysteine97, phosphorylated serine and tyrosine98, and GDP and GTP99. Simulations of the disordered C-Raf linker by itself were exceptional in that the simulation systems were parameterized using a99SB-disp100 and the TIP4P water model101. a99SB-disp is a variant of the Amber force field developed to work together with TIP4P for accurate simulations of disordered (as well as ordered) protein states. Due to concerns about force field inaccuracy for Zn2+ ions, distance restraints were applied between CRD zinc finger residues and Zn2+ ions in simulations that included the zinc-coordinated Raf CRD domain. Molecular dynamics simulations were performed in the NPT ensemble with constant pressure (1 bar) and constant temperature (310 K) imposed by a Martyna–Tuckerman–Klein Nosé–Hoover chain coupling scheme102, which was implemented using a multigrator scheme103 with a relaxation time of 10 ps. Initial velocities were sampled from the Boltzmann distribution. Water molecules and all bond lengths to hydrogen atoms were constrained using an in-house implementation104 of M-SHAKE105. Production simulations (for data collection) were launched after energy minimization and nanosecond-timescale molecular dynamics simulation with harmonic position restraints on backbone atoms.
The protein backbone atoms were restrained to their initial positions using an initial harmonic potential with a force constant of 10 kcal mol−1 Å−2 for 10–500 ns as an equilibration step; the force constant was linearly reduced to zero over the course of the equilibration step. The van der Waals and short-range electrostatic interactions were cut off at 10 Å for simulations without membrane and 12 Å for simulations with membrane. Long-range electrostatic forces were calculated in k-space using a grid-based method with Gaussian spreading106 to the grid every 7.5 fs. The simulation time step was 1 fs for the equilibration stage and 2.5 fs for production simulations; the r-RESPA integration method107 was used, with long-range electrostatics evaluated every 7.5 fs.
System preparation.
Structures derived from the PDB were back-mutated into the wild type unless stated otherwise, and missing atoms and residues were built in. The system of interest was placed at the center of a simulation box that was cubic for solvent simulations and orthorhombic for membranous simulations, with a separation greater than 20 Å from any periodic image. Explicitly represented water molecules were added to fill the system, and Na+ and Cl− ions were included to maintain physiological salinity (150 mM) and to obtain a neutral total charge for the system. Residue protonation states corresponded to pH 7.
In membranous simulations, a phosphotidylcholine (POPC) lipid bilayer with roughly 30% phosphotidylserine (POPS) in the inner layer proximal to the proteins was used. The model membrane was built from a neutral POPC lipid membrane by replacing 15% (molar) POPC with negatively charged POPS lipids. (The POPC lipids to replace were all taken from the inner bilayer but otherwise chosen at random.) This POPS fraction was chosen to mimic the abundance of anionic lipids in the mammalian plasma membrane20,108,109, in which approximately 10% of all lipids are POPS species, with other anionic species, such as phosphoinositides, bringing the total content of anionic lipids up to 15%. The POPS lipids were introduced only in the intracellular leaflet, which is where anionic lipids are almost exclusively found in cell membranes. The lipid content was thus around 30% POPS and 70% POPC in the intracellular leaflet and 100% POPC in the extracellular leaflet.
Simulations of K-Ras dimerization.
We performed 20 simulations, each 10 μs long, of two GTP-bound K-Ras molecules (PDB 4DSN) in aqueous solvent. In addition, we performed 23 unbiased simulations, each at least 2 μs long, of two GTP-bound K-Ras proteins anchored to the membrane by their fCys185 residues.
FRET assays.
Full details of our FRET assay are available elsewhere20. Briefly, human embryonic kidney 293T (HEK293T) cells were cotransfected with paired cyan fluorescent protein (CFP)- and yellow fluorescent protein (YFP)-fused KRAS constructs under the conditions of 0.5 and 10% FBS in four-well chambered cover glass (Lab-Tek). After 36–48 h, live cell imaging was performed using a Confocal/Multiphoton Zeiss LSM880 microscope. To examine the GTP-dependent Ras–Raf interaction, cotransfection of CFP-K-RasWT and YFP-C-Raf, CFP-K-RasT35A and YFP-C-Raf, CFP-K-RasG60A and YFP-C-Raf, or CFP-K-RasG13D and YFP-C-Raf plasmids was conducted. After 24 h of transfection, cells underwent 22 h of serum starvation, followed by epidermal growth factor (EGF) (10 ng ml−1) exposure for 30 min, and then were subjected to microscopy. Data were collected from three biological repeats, and 10–12 different cells in different fields from the same coverslip were selected for microscopy. Quantitation was done using ZEN software (Zeiss).
Preparation of GMP-PNP-loaded K-Ras protein for electron microscopy studies.
K-Ras (1–188) wild-type protein was expressed and purified as described previously110. Point mutations were generated using the GeneArt Site-Directed Mutagenesis System (Life Technologies). K-Ras proteins (100 μM) in buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 15 mM EDTA and 200 mM (NH4)2SO4) were loaded with 200 μM GMP-PNP at 4 °C overnight. The reaction was terminated by buffer exchange to 20 mM HEPES pH 8.0, 150 mM NaCl, 200 μM GMP-PNP, 10 mM MgCl2 with Zeba Spin Desalting Columns (Thermo Scientific). GMP-PNP loading was verified by back extraction of nucleotide using 6 M urea and evaluation of nucleotide peaks by high-performance liquid chromatography using an ion-exchange column as described previously110.
Preparation of lipid monolayers.
Phospholipids obtained from Avanti Polar Lipids were mixed according to the following proportions in chloroform:methanol (3:1, vol/vol): 10% DOPS, 10% DOPE, 60% Egg-PE and 20% PE-MCC111. Assembly of protein on lipid monolayers was patterned following previous methods112. Briefly, droplets (15 μl) of K-Ras (10 μM) in buffer were added to Teflon depression wells (3 mm in diameter and 0.5 mm in depth). 0.5 μl of 1 mM lipid solution was then delivered to the surface of the droplet. The wells were incubated in an airtight humidified chamber at 4 °C overnight. The lipid monolayers at the air–liquid interface were collected by hydrophobic carbon-coated grids (catalog no. CF400-Cu, Electron Microscopy Sciences). After washing with buffer three times, the grids were blotted and stained with uranyl formate solution (1% wt/vol) three times.
Transmission electron microscopy data collection and processing.
Stained grids were examined in an FEI Tecnai G2 Spirit Biotwin Transmission Electron Microscope operated at 120 kV with a magnification of 13,000 times. Images were collected with a pixel size of 0.78 nm. RELION 3.0 was used for all image processing113. For wild-type K-Ras reconstructions, 73,282 particles were computationally selected from 185 micrographs and subjected to 2D classification into 100 classes.
Raf localization imaging.
Raf localization was evaluated by cotransfection of plasmids expressing CFP–K-RasK88D and YFP–C-Raf, CFP–K-RasQ129L and YFP–C-Raf, and CFP–K-RasR149D and YFP–C-Raf. Constructs were designed as previously reported23. Cells were cultured in 10% FBS. Then, 36 h after transfection, cells were subjected to microscopy. Images were selected from three biological replicates, and 10–20 different cells in different fields from the same coverslip were used for analysis. Imaging was done using ZEN software (Zeiss).
BRET assays.
Construct design.
N-terminal fusions of KRAS to either mNeonGreen or NanoLuc were constructed in a pcDNA3.1(+) vector (Invitrogen) as reporters for BRET assays. KRAS point mutations were introduced either by site-directed mutagenesis (QuikChange, Agilent) or by replacing the KRAS gene with a synthetic DNA string (Geneart) carrying the desired mutation. All sequences were confirmed by sequencing.
Transfection.
HEK293T cells were cultured in DMEM complete medium (10% FBS) and seeded in white 96-well clear-bottom plates (Corning) 24 h before transfection. Cotransfections of the reporter plasmids were carried out with TransIT-293 (Mirus) using the manufacturer’s suggested protocol, using ratios as indicated in the figures. Then, 24 h after transfection, medium was exchanged to DMEM with no phenol red (Life Technologies), containing varying amounts of FCS (0.1%, 4.0% and 10.0%).
BRET measurements and data analysis.
Here, 48 h after transfection, BRET measurements were taken on a Victor 3 Multilabel Plate Reader after the addition of 10 μl of 32 μM coelenterazine 400a (Cayman Chemical) resulting in a 2.9 μM final concentration. Emission of mNeonGreen and NanoLuc was observed for 2 s at 535 ± 25 and 460 ± 25 nm, respectively. Expression of both reporters was monitored by measuring mNeonGreen (excitation 485 nm, emission 535 nm) before the addition of luciferase substrate and measuring total luminescence directly after the BRET measurements for 0.3 s.
BRET ratios were calculated based on the following equation for each transfected well:
The average of three technical replicates was calculated for each of the biological duplicates or triplicates after removal of obvious outliers and subjected to statistical analysis. Statistical analyses were performed using one-way ANOVA followed by Dunnett’s post tests (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Plasmids used in the FRET experiments.
pcDNA3-CFP (catalog no. 13030), pcDNA3-YFP (catalog no. 13033) and pBABEpuro-CRAF (catalog no. 51124) plasmids were purchased from Addgene. Full-length fragments of KRASWT and CRAF were inserted into vectors containing CFP or YFP to obtain CFP-KRASWT, YFP-KRASWT and YFP-CRAF constructs. KRASG12C, KRASG12D, KRASG13D, KRASD30R, KRASE31R, KRAST35A, KRAST35S, KRASG60A, KRASE62R, KRASR135A, KRASK147D, KRASD154Q, KRASG12C/D30R, KRASG12D/D30R, KRASG12C/E31R, KRASG12D/E31R, KRASG12C/E62R, KRASG12D/E62R, KRASG12C/R135A, KRASG12D/R135A, KRASR135A/D154Q, KRASG12C/K147D, KRASG12D/K147D and KRASK147D/D154Q mutants were generated by site-directed mutagenesis using PfuUItra II Hotstart PCR Master Mix (catalog no. 600850–51). The sequences were confirmed by sequencing.
Generation of K-Raslox/K-RASMUT cells.
Full details of our generation of K-Raslox/K-RASMUT cells are available elsewhere20. Briefly, K-RASD154Q, K-RASD30R, K-RASE31R, K-RASE62R, K-RASK147D and K-RASA135R mutations, in cis with either a G12C or G12D mutation, were created by point mutagenesis from pBABE HA-tagged K-RASWT retroviral plasmid (provided by C. Der, Addgene plasmid no. 75282). Retroviruses were generated by cotransfection of pBABE plasmids together with pAmpho plasmid into HEK293T cells using FuGENE HD Transfection Reagent (Promega). The retroviruses were transduced into H-Ras−/−; N-Ras−/−; K-Raslox/lox MEFs43 followed by 2 weeks of puromycin selection (1 μg ml−1) in DMEM supplemented with 10% FBS, 100 μg ml−1 penicillin and 100 units ml−1 streptomycin. To obtain K-Raslox/K-RASMUT clones, we then cultured cells in the presence of 4-hydroxytamoxifen (4OHT) (Sigma, 600 nM) for another 2 weeks to achieve complete deletion of endogenous K-Ras alleles.
Growth assessment by IncuCyte.
Cells (1 × 103) were seeded in 96-well plates in 150 μl of DMEM complete medium. The following day, 10% FBS medium was replaced by cell starvation medium (1%, 0.5% or 0.1% FBS) after two washes with PBS. Plates were incubated in the IncuCyte Zoom for real-time imaging, with three fields imaged per well under ×10 magnification every 2 h. Data were analyzed using the IncuCyte Confluence v.1.5 software, which quantified cell surface area coverage as confluence values. IncuCyte experiments were performed in triplicate. A single representative growth curve is shown for each condition.
Western blot analysis.
Cells were lysed in RIPA lysis buffer (catalog no. 89900, Thermo Fisher) supplemented with protease and phosphatase inhibitor cocktail tablets (Roche). The antibodies used for western blotting included those against HA-Tag (6E2) (Cell Signaling, catalog no. 2367, 1:1,000), HSP90 (H114) (Santa Cruz Biotech, catalog no. sc-7947, 1:1,000), phosphorylated Akt (Ser473) (Cell Signaling, catalog no. 4060, 1:1,000), Akt (Cell Signaling, catalog no. 9272, 1:1,000), phosphorylated ERK1/2 (Cell Signaling, catalog no. 4370, 1:1,000), ERK1/2 (Cell Signaling, catalog no. 4695, 1:1,000), phosphorylated S6 (Ser235/236) (Cell Signaling, catalog no. 4858, 1:1,000), S6 ribosomal protein (Cell Signaling, catalog no. 2217, 1:1,000), antirabbit IgG, HRP-linked secondary antibody (Cell Signaling, catalog no. 7074P2, 1:2,000), ECL Sheep anti-Mouse IgG, HRP-linked secondary antibody (GE Healthcare, catalog no. NA931V, 1:4,000) and ECL donkey antirabbit IgG and HRP-linked secondary antibody (GE Healthcare, catalog no. NA934V, 1:2,000).
Nucleotide-exchange assay.
Our SOS1-mediated Ras nucleotide-exchange assay was performed at Icagen. The purified K-Ras mutant R135A was diluted in assay buffer (40 mM HEPES pH 7.5, 1.5 μM Mant-GDP, 10 mM MgCl2, 0.05% CHAPS and 0.01% NP-40) to a final concentration of 1 μM. The exchange reaction was triggered by adding purified human SOS1 (residues 564–1049, 0.05 μM) to the reaction mix. Kinetic readings were taken to measure fluorescence (excitation at 360 nm, emission at 450 nm) for 30 min at 30-s intervals using a 384-well plate reader.
Bioinformatics analysis.
The K-Ras sequences used for evolution analysis were compiled using protein–protein BLAST searches in the NCBI nonredundant protein database, with the human K-Ras sequence (residues 1–166) as reference. The initial search generated 20,000 sequences with a minimal sequence identity of 29.84%. K-Ras sequences of mammals, birds and fish were then extracted from the initial search results based on taxonomy IDs. The sequence pool was then filtered based on the following rules: (1) duplicate entries or near-identical (sequence identity greater than 95%) entries were removed; (2) sequences with large (greater than 25 amino acids) insertions or deletions compared to human K-Ras were removed; and (3) sequences labeled as ‘partial’, ‘synthetic’ or ‘predicted’ were removed. After the filtering, 334 sequences remained and were used for multiple-sequence alignment. Sequence alignment of these sequences was obtained from the NCBI server using default settings, and sequence logos were generated using the online tool WebLogo3. The final figures were manually adjusted from the WebLogo3 result to show only the residues corresponding to human K-Ras (residues 1–166).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
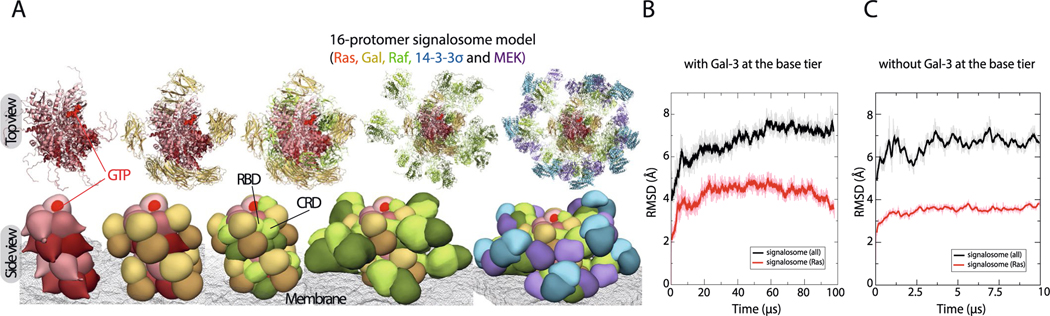
Extended Data Fig. 1 |. Construction of the Ras-Raf signalosome model in 16-protomer form.
a. Top and side views of the stepwise addition (left to right) of Gal-3 (yellow), C-Raf (green), 14–3-3σ (blue), and MEK1 (violet) to the K-Ras helical assembly (red), producing the 16-protomer signalosome model. The membrane is shown as a mesh. Each C-Raf KD dimer binds to a 14–3-3σ dimer and two MEK1 kinases. b. The Cα atom RMSD (w.r.t. the starting structure) of a membrane-anchored 8-protomer Ras-Raf signalosome and its K-Ras octamer core in a 100-μs simulation. As shown, the signalosome— especially the K-Ras core—was stable in the course of the simulation. c. Similar to B, the Cα atom RMSDs of a membrane-anchored 8-protomer Ras-Raf signalosome without Gal-3 at the base tier.
Extended Data Fig. 2 |. The GTP-mediated K-Ras dimer model and its orientation on the membrane.
a. Left panel: A collection of snapshots of the 20 simulations starting from two separated K-Ras proteins in solvent. The snapshots are aligned to one K-Ras protein therein (cyan). The GMA dimer model is highlighted. The α4 helix is shown in cartoon representation. Snapshots 10 μs apart were taken from the 20 simulations (which have an aggregate simulation time of 680 μs). Right panel: A collection of snapshots of the 23 simulations of two GTP-bound K-Ras proteins on the membrane; snapshots 10 μs apart were taken from the 23 simulations (which have an aggregate simulation time of 210 μs). Only in one solvent and one membrane simulation was the GMA dimer reached, and in both cases the dimer remained stable. b. A stable dimer (orange) generated from one of the 20 simulations of K-Ras/K-Ras association in solvent, which is consistent with the crystal structures of Ras symmetric dimer using the α4-α5 interface (for example PDB 1QRA, cyan and green). c. Upper panel: Acceptor (yellow) interaction with the γ-phosphate of the GTP donor (cyan) in a simulation of a GMA K-Ras dimer. The Arg135 of the acceptor interacts with the γ-phosphate, while the Lys128 interacts with the Glu62 of the donor, and vice versa. Snapshots at 40 μs and 60 μs from the simulation of K-Ras dimerization on the membrane are shown; the motions of Arg135 and Lys128 are shown by dashed red arrows. Lower panel: Distances from Arg135 (black) and Lys128 (red) to the γ-phosphate. d. Comparison of GTP interactions and GMPPNP interactions with the acceptor residues at the GMA dimer interface. e. Interface residues of the GMA dimer (left) and water occupancy map (right, blue) at the interface. f. Snapshots at 10 μs from four independent runs of free GTP-bound K-Ras monomer (cyan) on the membrane. To illustrate the relationship between the membrane orientation and RBD binding of K-Ras, the C-Raf RBD (green) is positioned on each snapshot based on the Ras-RBD structure (PDB 4G0N). Shown are the distance of the β phosphorus (βP) and the amine nitrogen of the guanine ring (N2) from the plane of the membrane surface; we use these distances to describe the membrane orientation of a Ras protein (Fig. 2g). The Run 1 snapshot is compatible with RBD binding, which corresponds to the Ras membrane orientation of the lower red contours in Fig. 2g in the main text. The snapshot from Run 2 corresponds to the right center of the top red contours; this orientation of K-Ras on the membrane positions the RBD away from the membrane. In the Run 3 and Run 4 snapshots, RBD binding leads to a severe steric clash with the membrane; this corresponds to the left center of the top red contour. g. Snapshot at 100 μs from the simulation of the GMA dimer formation on the membrane, with two C-Raf RBDs (green) added based on the Ras-RBD pose; the RBDs do not clash with the membrane. The membrane orientation of the donor and acceptor Ras proteins correspond to the lower and upper black contours, respectively, in Fig. 2g in the main text. h. Examples of G nucleotide–mediated dimerization, in which an arginine interacts with a nucleotide phosphate at the dimerization interface. The Toc34 homodimer114 (PDB 3BB1), the Ffh-FtsY heterodimer115 (PDB 1RJ9), and the adenylosuccinate synthetase homodimer (PDB 4M9D) are compared to the GMA K-Ras dimer model (cyan); the host G proteins (not shown) were aligned in this comparison. i. The binding site of the synthetic monobody NS119 (red) and the donor site (cyan) of an acceptor in the GMA dimer are shown on a Ras protein (yellow). The two binding sites largely overlap. j. The electrostatic complementarity at the GMA dimer interface, with contacting areas at the interface connected by dashed lines. k. Cα RMSD of the GMA dimer of the wild type and various oncogenic mutants in simulations. l. Sequence conservation of K-Ras proteins in various species (see Materials and Methods). A graphical representation of sequence alignment of representative K-Ras proteins in evolution. The residues involved in the K-Ras/K-Ras interactions in the K-Ras helical assembly are labeled. The 32 K-Ras sequences by GenBank IDs are: OLS17184.1, OLS30914.1, OLS23071.1, XP_020603162.1, XP_023347765.1, XP_023347766.1, XP_003378992.1, XP_003377451.1, GBC14959.1, XP_027484897.1, QBM87817.1, XP_013758736.1, EHB13737.1, OQV11758.1, XP_027289840.1, NP_001356715.1, XP_027290137.1, XP_027290057.1, XP_027289839.1, XP_027289566.1, XP_029436565.1, ELK24704.1, XP_027290375.1, XP_023664743.1, XP_021777811.1, XP_021247616.1, XP_025064324.1, XP_006127724.1, NP_001243091.1, RDD38924.1, KPM02442.1, OTF69949.1.
Extended Data Fig. 3 |. Stability of the active switch i and switch ii conformations.
a. Distributions of the switch I conformation of K-Ras in various simulations. Normalized histograms of the switch I backbone root-mean-square deviation (RMSD) with respect to the active switch I conformation in PDB 4DSN are shown. b. Conformations of the switch I (residues 30–38, cyan) and switch II (residue 60–76, yellow) regions in simulations of the GTP-bound K-Ras monomer. c. Conformations of the switch I and II regions in simulations of GDP-bound K-Ras monomer. d. Conformations of the switch I and II regions in simulations of GTP-bound K-Ras bound with C-Raf RBD. e. Crystal structures of Ras bound with GTP or GTP analogs, with the switch I and switch II regions highlighted. The PDB entries included are 1AGP, 1CLU, 1CTQ, 1GNP, 1GNR, 1HE8, 1JAH, 1JAI, 1K8R, 1LF0, 1LFD, 1NVU, 1NVV, 1NVW, 1NVX, 1P2S, 1P2T, 1P2U, 1P2V, 1PLJ, 1PLK, 1QRA, 1RVD, 1ZW6, 2C5L, 2RGA, 2RGB, 2RGC, 2RGD, 2RGE, 2RGG, 2UZI, 2VH5, 3DDC, 3GFT, 3I3S, 3K8Y, 3L8Y, 3L8Z, 3LBH, 3LBI, 3LBN, 3OIU, 3OIV, 3OIW, 3RRY, 3RRZ, 3RS0, 3RS2, 3RS3, 3RS4, 3RS5, 3RS7, 3RSO, 3TGP, 3V4F, 4DLR, 4DLS, 4DLT, 4DLU, 4DLV, 4DLW, 4DLX, 4DLY, 4DLZ, 4DSN, 4DSO, 4DST, 4EFL, 4EFM, 4EFN, 4G0N, 4G3X, 4K81, 4L9W, 4NMM, 4NYI, 4NYJ, 4NYM, 4RSG, 4XVQ, 4XVR, 5B2Z, 5B30, 5P21, 6Q21, 121 P, 421 P, 521 P, 621 P, 721 P, 821 P, and 221 P. f. Crystal structures of Ras loaded with GDP. The PDB entries included are 1AA9, 1CRP, 1CRQ, 1CRR, 3LO5, 1IOZ, 1LF5, 1PLL, 1Q21, 1WQ1, 1XD2, 1XJ0, 1ZVQ, 2CE2, 2CLD, 2Q21, 2QUZ, 2X1V, 3CON, 3KUD, 4DSU, 4EPR, 4EPT, 4EPV, 4EPW, 4EPX, 3EPY, 4L8G, 4L9S, 4LDJ, 4LPK, 4LRW, 4LUC, 4LV6, 4LYF, 4LYH, 4LYJ, 4M1O, 4M1S, 4M1T, 4M1W, 4M1Y, 4M21, 4M22, 4OBE, 4PZY, 4PZZ, 4Q01, 4Q02, 4Q03, 4Q21, 4QL3, 4TQ9, 4TQA, 4WA7, and 5F2E. g. Crystal structures of the T35S Ras mutant loaded with GTP or GTP analog, with the switch I and switch II regions highlighted. The PDB entries included are 1IAQ, 2LCF, 2LWI, 3KKM, and 3KKN. The hydroxyl of Thr35 in the switch I region in wild-type Ras coordinates the Mg2+ ion bound to the GTP, and the T35S mutation is known to disrupt the switch I active conformation. The switch I inactive conformations sampled by the simulations (Supplementary Fig. 3B and C) are broadly consistent with the inactive conformations in T35S structures116. h. Conformations of the switch I and II regions in simulations of 24 copies of GTP-bound K-Ras in a crystal lattice (of PDB 3GFT). i. The Cα atom RMSDs of a GMA K-Ras tetramer (w.r.t. the starting structure) in a 10-μs simulation with membrane and in a 10-μs simulation in water solvent. As shown, the membrane helps stabilize the tetramer structure.
Extended Data Fig. 4 |. Cellular validation of the K-Ras signalosome interfaces.
Panels A–C here refer to the experiments testing mutations at the GMA dimer interface, and panels D–F refer to the experiments testing secondary/tertiary K-Ras interfaces. a. Representative cell images at the endpoint of the experiment testing mutations at the GMA dimer interface (Fig. 3f); scale bar: 50 μm. b. Growth rates of K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations in 10%, 1%, and 0.5% FBS medium, represented by the confluence value assessed by IncuCyte. Representative pictures at the endpoint are shown in the bottom panels (scale bar: 50 μm). Data are shown as mean +/− standard deviation (n = 3 biologically independent experiments). c. Phosphorylation of ERK and AKT in K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations. Cells were lysed after 48 hours incubation in 0.5% or 10% FBS, as indicated, and analyzed by Western blot. These results are representative of three independent experiments with similar results. d. Representative cell images at the endpoint of the experiment testing mutations at the secondary and tertiary K-Ras/K-Ras interfaces (Fig. 6b); scale bar: 50 μm. e. Phosphorylation of ERK and AKT in K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations. Cells were lysed after 48 hours incubation in 0.5% or 10% FBS as indicated and analyzed by Western blot. These results are representative of three independent experiments with similar results. f. Growth rates of K-Raslox/K-RASMUT cells expressing the indicated mutations in cis with either G12C or G12D mutations to test the secondary and tertiary K-Ras/K-Ras interfaces in the background of either G12C or G12D, shown as confluence values measured by IncuCyte. Representative images at the end point are also shown. Cells were kept in 0.5% or 10% FBS. Data are shown as mean +/− standard deviation (n = 3 biologically independent experiments).
Extended Data Fig. 5 |. FRET and BRET validation of K-Ras signalosome interfaces.
a. CFP emission of wild-type K-Ras and various K-Ras mutants to test the K-Ras/K-Ras GMA dimer. HEK293T cells were co-transfected with CFP-K-Ras and YFP-K-Ras, serum starved, and stimulated with 10% FBS. Each K-Ras construct is labeled under its respective plot. Assays were repeated three times. Error bars represent mean ± S.E.M. WT: n = 42; D154Q: n = 33; G12C/D154Q: n = 35; G12D/D154Q: n = 20; D30R: n = 32; G12C/D30R: n = 30; G12D/D30R: n = 28; E31R: n = 42; G12C/E31R: n = 26; G12D/E31R: n = 28; E62R: n = 33; G12C/E62R: n = 32; G12D/E62R: n = 22; K147D: n = 36; G12C/K147D: n = 34; G12D/K147D: n = 38; K147D/D154Q: n = 29. (**** denotes P < 0.0001 by two-way ANOVA; ns stands for not significant). b. CFP emission of wild-type K-Ras and T35 and G60 mutants to test K-Ras association. Assays were repeated three times. Error bars represent mean ± S.E.M. WT: n = 14; T35A: n = 18; T35S: n = 11; G60A: n = 17. (P value was calculated by two-way ANOVA; ns stands for not significant). c. CFP emission of wild-type K-Ras and T35 and G60 mutants with C-Raf to monitor Ras-Raf binding at 10% FBS or 10 ng mL−1 EGF. Assays were repeated three times. Error bars represent mean ± S.E.M. T35A (FBS): n = 36; T35A (No FBS): n = 27; T35A (No FBS + EGF): n = 21; G60A (FBS): n = 55; G60A (No FBS): n = 38; G60A (No FBS + EGF): n = 37; WT (FBS): n = 12; WT (No FBS): n = 37; WT (No FBS + EGF): n = 11. (**** denotes P < 0.0001 by two-way ANOVA; ns stands for not significant). d. Nucleotide exchange assay of wild-type K-Ras with (orange) and without (gray) SOS1, and R135A without SOS1 (blue). e. CFP emission of wild-type K-Ras and R135A mutants. Assays were repeated three times. Error bars represent mean ± S.E.M. In 10% FBS condition, WT: n = 44; R135A: n = 82; G12C/R135A: n = 50; G12D/R135A: n = 34; D154Q/R135A: n = 51; D154Q: n = 33. In 0.5% FBS condition, WT: n = 25; R135A: n = 47; G12C/R135A: n = 44; G12D/R135A: n = 34; D154Q/R135A: n = 42; D154Q: n = 26. (* denotes P < 0.05, ** denotes P < 0.01, *** denotes P < 0.001, **** denotes P < 0.0001 by two-way ANOVA; ns stands for not significant). f. BRET signal as an indicator of K-Ras assembly for G12C and G13D mutants, and for the K-Ras construct lacking the membrane-anchoring HVR tail (residues 1–166). Co-transfection of increasing ratios of donor and acceptor plasmids enables discrimination between specific and non-specific (random collision) protein-protein interactions. G. CFP emission of wild-type K-Ras and K88D, Q129L, and R149D mutants. Assays were repeated three times. Error bars represent mean ± S.E.M. In 0.5% FBS condition, WT: n = 43; K88D: n = 38; Q129L: n = 34; R149D: n = 37; G12C: n = 22; G12C/K88D: n = 29; G12C/Q129L: n = 34; G12C/R149D: n = 23; G12D: n = 37; G12D/K88D: n = 34; G12D/Q129L: n = 44; G12D/R149D: n = 38. In 10% FBS condition, WT: n = 40; K88D: n = 35; Q129L: n = 40; R149D: n = 42; G12C: n = 27; G12C/K88D: n = 41; G12C/Q129L: n = 46; G12C/R149D: n = 57; G12D: n = 39; G12D/K88D: n = 67; G12D/Q129L: n = 49; G12D/R149D: n = 42. (* denotes P < 0.05, ** denotes P < 0.01, *** denotes P < 0.001, **** denotes P < 0.0001 by two-way ANOVA; ns stands for not significant).
Extended Data Fig. 6 |. Structural details of secondary and tertiary Ras-Ras and Ras-RBD interactions.
a. Key contacts of the secondary (stacking) Ras-Ras interaction of K-Ras n (blue) and K-Ras n + 4 (green). A series of salt bridges form intermittently in MD simulations between the Lys88, Glu91, Glu98, Arg102, and Asp105 residues on the α3 helix of K-Ras n and the Asp154, Arg161, Lys165, Glu168, and Lys172 residues, respectively, on the α5 helix of K-Ras n + 4. b. Key contacts at the interface of K-Ras n + 3 (purple) and K-Ras n (blue). c. Key contacts of the secondary Ras-RBD interaction of C-Raf RBD n (yellow) with K-Ras n + 1 (green), which is the GTP acceptor of K-Ras n (purple), the primary binding partner of the RBD. The RBD also interacts with the HVR of K-Ras n + 1. Residues 1–53 of C-Raf are structurally unresolved but are likely to make additional interactions at this secondary Ras-RBD interface. d. Key contacts of the tertiary Ras-RBD interaction of Raf RBD n + 3 (yellow) and K-Ras n (blue). e. Unoccupied Switch-II pocket in a crystal structure of K-Ras (PDB 4LDJ) compared with the Switch-II pocket of K-Ras n in the stacking interaction. f. Superposition of stacking (n and n + 4) K-Ras proteins from the signalosome model with a crystal dimer of K-Ras (PDB 5UQW); in both cases the dimer interaction is mediated by a β2- β3 turn inserting into a Switch-II pocket. G. Comparison of D154Q/R161E with the WT in K-Ras oligomer simulations. As shown, the double mutation disrupts the D154-K147 salt bridge at the GMA dimer (n/n-1), and as compensation, allows an E161-R102 salt bridge in stacking (n/n + 4) by shifting the relative position of the α3 and α5 helices.
Extended Data Fig. 7 |. Structural details of interactions of K-Ras, Gal-3 and the C-Raf CRD.
a. Gal-3 n (olive) binds to the HVR fCys185 of Ras n (blue) and to C-Raf RBD n − 1 (orange). The thiodigalactoside inhibitor binding site of Gal-3117 (PDB 4JC1) is shown in red. b. Gal-3 stacking; Gal-3 n (green) and Gal-3 n + 4 (yellow) are stacked in parallel with K-Ras stacking. c. Position of Gal-3 (olive) with respect to K-Ras (blue), in comparison with that of PDE6δ (orange) with respect to farnesylated K-Ras61 (PDB 5TAR), PDE6δ (yellow) with respect to farnesylated Rheb118 (PDB 3T5G), and RhoGDI (cyan) with respect to geranylgeranylated CDC42119 (PDB 1DOA). The Gal-3 position somewhat resembles the PDE6δ complex in the overall pose. The insert shows the simulation-generated Gal-3 binding pose of fCys with the Gal-3 crystal structure (PDB 3ZSM) overlaid for comparison. d. Left: snapshots of the simulation in which fCys enters the Gal-3 (yellow) pocket, with color coding on fCys indicating simulation time; right: the final Gal-3/fCys model superimposed on the Gal-3 structure used to initiate the simulation (PDB 3ZSM). e. Final snapshots from 55 simulations of the three-domain system of the CRD (light orange) tethered to a K-Ras-bound RBD (orange). The snapshots were aligned with respect to the K-Ras protein. Of the 55 simulations, 24 were in solvent (80 μs in aggregate), and the other 31 were with the RBD and the membrane (117 μs in aggregate). The simulation-generated CRD poses are grouped into three clusters: 1) clashing with the GMA K-Ras dimer; 2) clashing with the membrane if applied to the GMA K-Ras dimer on the membrane (Fig. 1a); and 3) near the C-terminal of the α5 helix and the β1, β2, and β3 strands of K-Ras. Only cluster 3 is compatible with the helical assembly of K-Ras; cluster 2 is similar to recently reported structures74–76. f. Details of the CRD-Ras interface of a CRD pose selected from the third cluster. NMR-identified Ras residues involved in Ras-CRD interaction are shown. The Glu3 residue on the β1 strand of the K-Ras protein interacts stably with a CRD-bound Zn2+ ion (lower right panel). The Lys157(CRD)/Glu76(K-Ras) distance in simulations is shown to indicate the stability of the Ras-CRD pose (lower left panel). The upper right panel shows the relative positions of K-Ras (blue), CRD (orange), Gal-3 (olive), and GTP (red) in a top view. g. CRD residues identified by an NMR study51 as part of the K-Ras interface. K-Ras M170, fCys185, and CRD Zn2+ ions are also shown. h. The surfaces of RBD-bound K-Ras and CRD colored by their electrostatic potential (calculated using PyMOL). A cartoon image is used to show the orientation of the Ras-CRD complex. The CRD-bound Zn2+ ions are shown in purple. I. A C-Raf CRD at the base tier with RBD and K-Ras. Arg143, Lys144, Lys148, and the two coordinating Zn2+ ions of the CRD are shown, together with the occupancy map of POPS lipids in contact with the three amino acids shown in red mesh.
Extended Data Fig. 8 |. Membrane interface of K-Ras, Gal-3, and C-Raf.
a. Positively charged residues of a base-tier K-Ras protein proximal to the membrane in the signalosome model. The inset shows the direct interaction between the HVR lysines of a K-Ras protein at the base tier and the phosphotidylserine lipids. b. Positively charged residues and prolines of Gal-3 proximal to the membrane at the base tier. A Val126 residue that has been suggested to be involved in Gal-3 membrane interaction23 is also shown. c. Positively charged residues of the C-Raf RBD and CRD and the two CRD-bound Zn2+ ions proximal to the membrane.
Extended Data Fig. 9 |. Additional interactions between Raf, 14-3-3σ, and MEK1.
a. Superposition of the structural model (gray) of the Raf/MEK/14-3-3 complex with the separately resolved cryo-EM structure (PDB 6Q0J). b. Interaction between the Raf kinase domain dimer (green) and 14–3-3σ dimer (cyan). The linker residues pSer338 and pTyr341 are located between the two αC helices of the kinase dimer, and pSer621 is bound to a 14–3-3σ protein. c. Electrostatic potential surface of the C-Raf kinase domain dimer; the region of the two αC helices, which is where pSer339 and pTyr341 (shown) are located, is highly positively charged. d. Interaction of phosphorylated residues pSer338 and pTyr341 with the positively charged residues of the two αC helices of the C-Raf KD dimer. e. Samples of conformations of the C-Raf linker (rainbow colors) in the Ras-Raf signalosome model. The C-Raf KD (gray) is attached to the linker for reference. f. Top and side views of an alternative eight-protomer signalosome model, wherein C-Raf n and C-Raf n + 1 form a dimer, as opposed to the model shown in Fig. 1d and e, wherein C-Raf n and C-Raf n + 4 form a dimer. The color scheme and protein representation are similar to those in Fig. 1d. The inset shows that in this alternative model a C-Raf linker (linker 2) has to circumvent a Gal-3 protein (Gal-3 2) to engage the other C-Raf linker in a C-Raf dimer. G. An illustration of how the GEF domain of SOS1 can reach the K-Ras 8. The modeling was based on PDB entry 1NVU for the K-Ras/SOS1 pose, and PDB entry 1XD4 for the structures of the PH domain and the PH-GEF linker; the linker orientation with respect to the PH and the GEF domains was manually adjusted and energetically minimized in the modeling. The helical hairpin of SOS1 is shown in yellow.
Supplementary Material
Acknowledgements
We thank M. Eastwood for helpful discussions, P. Ayaz for a critical reading of the manuscript, K. Yuh for assistance with videos, J. McGillen and B. Frank for editorial assistance, and C. Scholl, J.V. Schaefer and J. Binz for their support with the BRET experiments. This work was supported in part by a Stand Up To Cancer (SU2C)-American Cancer Society Lung Cancer Dream Team Translational Research Grant (no. SU2C-AACR-DT17-15 to P.A.J.), the Gadzooks Fund (to P.A.J.), the Cammarata Family Foundation Research Fund (to P.A.J.), the Giovanni Armenise-Harvard Foundation, the Lung Cancer Research Foundation, the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 101001288) (to C.A.), the US Department of Defense (W81XWH-16-1-0106), Hale Center and Pinard Family (DFCI), the National Institutes of Health (NIH 5R01CA244341), the Cancer Prevention and Research Institute of Texas (RP170373) (to K.D.W.) and Krebsliga Schweiz grants (nos. KFS 4147-02-2017 and KFS-529002-2021-R) (to A.P.). SU2C is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. P.A.J. has served as a consultant for and has received sponsored research funding from AstraZeneca, Mirati Therapeutics and Araxes Pharmaceuticals.
Footnotes
Competing interests
P.A.J. reports grants from AstraZeneca, Boehringer Ingelheim, Daiichi Sankyo, Eli-Lilly, Takeda Oncology, Astellas, PUMA and Revolution Medicines and personal fees from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Eli-Lilly, Chugai, Ignyta, Loxo Oncology, SFJ Pharmaceuticals, Voronoi, Daiichi Sankyo, Biocartis, Novartis, Sanofi Oncology, Takeda Oncology, Mirati Therapeutics, Trasncenta, Silicon Therapeutics, Syndax, Bayer, Esai, Allorion Therapeutics, Accutar Biotech and AbbVie outside the submitted work; P.A.J. also receives postmarketing royalties from a DFCI-owned patent on EGFR mutations issued and licensed to Lab Corp. K.D.W. receives or has received grant funding from Revolution Medicines and Astellas outside the submitted work. He also is on the scientific advisory board for Vibliome Therapeutics. The remaining authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41594-021-00667-6.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41594-021-00667-6.
Peer review information Nature Structural & Molecular Biology thanks John Hancock, Francesco Gervasio and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Anke Sparmann and Florian Ullrich were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41594-021-00667-6.
Data availability
Atomic coordinates of the structural model of the eight-protomer Ras–Raf signalosome are available as Supplementary Dataset 1, and atomic coordinates of the GMA K-Ras dimer on a membrane are available as Supplementary Dataset 2. Due to the large size of the molecular dynamics trajectories (listed in Supplementary Table 1), they are available upon request (for noncommercial use) by contacting trajectories@deshawresearch.com. Source data are provided with this paper.
Code availability
The molecular dynamics simulations were performed using the Anton 2 supercomputer (the simulation code we used is specialized to Anton 2, but codes for performing molecular dynamics simulations are widely available).
References
- 1.Wennerberg K, Rossman KL & Der CJ The Ras superfamily at a glance. J. Cell Sci 118, 843–846 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Hobbs GA, Der CJ & Rossman KL RAS isoforms and mutations in cancer at a glance. J. Cell Sci 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan TL et al. Differential effector engagement by oncogenic KRAS. Cell Rep. 22, 1889–1902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tidyman WE & Rauen KA The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev 19, 230–236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prior IA, Lewis PD & Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simanshu DK, Nissley DV & McCormick F. RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherfils J. & Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev 93, 269–309 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Wittinghofer A. & Pal EF The structure of Ras protein: a model for a universal molecular switch. Trends Biochem. Sci 16, 382–387 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Wittinghofer A. & Vetter IR Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem 80, 943–971 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Santos E, Nebreda AR, Bryan T. & Kempner ES Oligomeric structure of p21 ras proteins as determined by radiation inactivation. J. Biol. Chem 263, 9853–9858 (1988). [PubMed] [Google Scholar]
- 11.Dementiev A. K-Ras4B lipoprotein synthesis: biochemical characterization, functional properties, and dimer formation. Protein Expr. Purif 84, 86–93 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Chen M, Peters A, Huang T. & Nan X. Ras dimer formation as a new signaling mechanism and potential cancer therapeutic target. Mini-Rev. Med. Chem 16, 391–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Güldenhaupt J. et al. N-Ras forms dimers at POPC membranes. Biophys. J 103, 1585–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werkmüller A, Triola G, Waldmann H. & Winter R. Rotational and translational dynamics of Ras proteins upon binding to model membrane systems. Chem. Phys. Chem 14, 3698–3705 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Sarkar-Banerjee S. et al. Spatiotemporal analysis of K-Ras plasma membrane interactions reveals multiple high order homo-oligomeric complexes. J. Am. Chem. Soc 139, 13466–13475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muratcioglu S. et al. GTP-dependent K-Ras dimerization. Structure 23, 1325–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inouye K, Mizutani S, Koide H. & Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem 275, 3737–3740 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Nan X. et al. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl Acad. Sci. USA 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer-Smith R. et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol 13, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrogio C. et al. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Plowman SJ, Muncke C, Parton RG & Hancock JF H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl Acad. Sci. USA 102, 15500–15505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian T. et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol 9, 905–914 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Shalom-Feuerstein R. et al. K-Ras nanoclustering is subverted by overexpression of the scaffold protein Galectin-3. Cancer Res. 68, 6608–6616 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belanis L, Plowman SJ, Rotblat B, Hancock JF & Kloog Y. Galectin-1 is a novel structural component and a major regulator of H-Ras nanoclusters. Mol. Biol. Cell 19, 1404–1414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plowman SJ, Ariotti N, Goodall A, Parton RG & Hancock JF Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Mol. Cell. Biol 28, 4377–4385 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutton MN et al. DIRAS3 (ARHI) blocks RAS/MAPK signaling by binding directly to RAS and disrupting RAS clusters. Cell Rep. 29, 3448–3459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell 153, 287–292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright LP & Philips MR Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res 47, 883–891 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F. & Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Shan Y. et al. How does a drug molecule find its target binding site? J. Am. Chem. Soc 133, 9181–9183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan Y. et al. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat. Struct. Mol. Biol 21, 579–584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plattner N, Doerr S, De Fabritiis G. & Noé F. Complete protein-protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat. Chem 9, 1005–1011 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Scheffzek K. et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Ahmadian MR, Stege P, Scheffzek K. & Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol 4, 686–689 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Vetter IR & Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Geyer M. et al. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry 35, 10308–10320 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Ito Y. et al. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry 36, 9109–9119 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR & Wittinghofer A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc. Natl Acad. Sci. USA 98, 4944–4949 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Araki M. et al. Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers. J. Biol. Chem 286, 39644–39653 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazhab-Jafari MT et al. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl Acad. Sci. USA 112, 6625–6630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF & McCammon JA Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem 50, 674–684 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Prakash P, Zhou Y, Liang H, Hancock JF & Gorfe AA Oncogenic K-Ras binds to an anionic membrane in two distinct orientations: a molecular dynamics analysis. Biophys. J 110, 1125–1138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drosten M. et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung YJ, Carter M, Zhong JM & Hwang YW Mutagenesis of the H-ras p21 at glycine-60 residue disrupts GTP-induced conformational change. Biochemistry 34, 3470–3477 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y. & Hancock JF Ras nanoclusters: versatile lipid-based signaling platforms. Biochim. Biophys. Acta 1853, 841–849 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Ostrem JM, Peters U, Sos ML, Wells JA & Shokat KM K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fetics SK et al. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu C-D et al. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J. Biol. Chem 270, 30274–30277 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Okada T. et al. The strength of interaction at the Raf cysteine-rich domain is a critical determinant of response of Raf to Ras family small GTPases. Mol. Cell. Biol 19, 6057–6064 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drugan JK et al. Ras interaction with two distinct binding domains in Raf-1 may be required for Ras transformation. J. Biol. Chem 271, 233–237 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Williams JG et al. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem 275, 22172–22179 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Winkler DG et al. Identification of residues in the cysteine-rich domain of Raf-1 that control Ras binding and Raf-1 activity. J. Biol. Chem 273, 21578–21584 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Thapar R, Williams JG & Campbell SL NMR characterization of full-length farnesylated and non-farnesylated H-Ras and its implications for Raf activation. J. Mol. Biol 343, 1391–1408 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Levy R, Biran A, Poirier F, Raz A. & Kloog Y. Galectin-3 mediates cross-talk between K-Ras and Let-7c tumor suppressor microRNA. PLoS ONE 6, e27490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elad-Sfadia G, Haklai R, Balan E. & Kloog Y. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J. Biol. Chem 279, 34922–34930 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Ashery U. et al. Spatiotemporal organization of Ras signaling: rasosomes and the galectin switch. Cell. Mol. Neurobiol 26, 469–493 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paz A, Haklai R, Elad-Sfadia G, Ballan E. & Kloog Y. Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 20, 7486–7493 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Rotblat B. et al. Galectin-1 (L11A) predicted from a computed galectin-1 farnesyl-binding pocket selectively inhibits Ras-GTP. Cancer Res. 64, 3112–3118 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Lukyanov P, Furtak V. & Ochieng J. Galectin-3 interacts with membrane lipids and penetrates the lipid bilayer. Biochem. Biophys. Res. Commun 338, 1031–1036 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Yang R-Y, Hill PN, Hsu DK & Liu F-T Role of the carboxyl-terminal lectin domain in self-association of galectin-3. Biochemistry 37, 4086–4092 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Dharmaiah S. et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl Acad. Sci. USA 113, E6766–E6775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tzivion G, Luo Z. & Avruch J. A dimeric 14–3-3 protein is an essential cofactor for Raf kinase activity. Nature 394, 88–92 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Molzan M. et al. Stabilization of physical RAF/14–3-3 interaction by cotylenin A as treatment strategy for RAS mutant cancers. ACS Chem. Biol 8, 1869–1875 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Hatzivassiliou G. et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Haling JR et al. Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling. Cancer Cell 26, 402–413 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Park E. et al. Architecture of autoinhibited and active BRAF-MEK1–14-3–3 complexes. Nature 575, 545–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondo Y. et al. Cryo-EM structure of a dimeric B-Raf:14–3-3 complex reveals asymmetry in the active sites of B-Raf kinases. Science 366, 109–115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu H. & Fuxreiter M. The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell 165, 1055–1066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D. & Kuriyan J. The structural basis of the activation of Ras by SOS. Nature 394, 337–343 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Margarit SM et al. Structural evidence for feedback activation by Ras·GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Prakash P. et al. Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers. Sci. Rep 7, 40109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jang H, Muratcioglu S, Gursoy A, Keskin O. & Nussinov R. Membrane-associated Ras dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem. J 473, 1719–1732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee KY et al. Two distinct structures of membrane-associated homodimers of GTP- and GDP-bound KRAS4B revealed by paramagnetic relaxation enhancement. Angew. Chem. Int. Ed. Engl 59, 11037–11045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cookis T. & Mattos C. Crystal structure reveals the full Ras:Raf interface and advances mechanistic understanding of Raf activation. Preprint at bioRxiv 10.1101/2020.07.28.225938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran TH et al. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun 12, 1176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fang Z. et al. Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane. Proc. Natl Acad. Sci. USA 117, 12101–12108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cho K-J et al. Raf inhibitors target Ras spatiotemporal dynamics. Curr. Biol 22, 945–955 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Zhang Z. et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet 29, 25–33 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Singh A, Sowjanya AP & Ramakrishna G. The wild-type Ras: road ahead. FASEB J. 19, 161–169 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Lavoie H. et al. MEK drives BRAF activation through allosteric control of KSR proteins. Nature 554, 549–553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M. & McCormick F. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol. Cell 22, 217–230 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Li W, Han M. & Guan KL The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev. 14, 895–900 (2000). [PMC free article] [PubMed] [Google Scholar]
- 83.Umstead M, Xiong J, Qi Q, Du Y. & Fu H. Aurora kinase A interacts with H-Ras and potentiates Ras-MAPK signaling. Oncotarget 8, 28359–28372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dance M, Montagner A, Salles JP, Yart A. & Raynal P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell. Signal 20, 453–459 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Maurer T. et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl Acad. Sci. USA 109, 5299–5304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mott HR et al. The solution structure of the Raf-1 cysteine-rich domain: a novel Ras and phospholipid binding site. Proc. Natl Acad. Sci. USA 93, 8312–8317 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saraboji K. et al. The carbohydrate-binding site in galectin-3 is preorganized to recognize a sugarlike framework of oxygens: ultra-high-resolution structures and water dynamics. Biochemistry 51, 296–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shaw DE et al. Anton 2: raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer. In Proc. International Conference for High Performance Computing, Networking, Storage and Analysis (SC14) 41–53 (IEEE, 2014). [Google Scholar]
- 89.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926–935 (1983). [Google Scholar]
- 90.Joung IS & Cheatham TE III Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 112, 9020–9041 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, Cieplak P. & Kollman PA How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem 21, 1049–1074 (2000). [Google Scholar]
- 92.Duan Y. et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem 24, 1999–2012 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Hornak V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Best RB & Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Phys. Chem. B 113, 9004–9015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindorff-Larsen K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klauda JB et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khoury GA, Thompson JP, Smadbeck J, Kieslich CA & Floudas CA Forcefield_PTM: ab-initio charge and AMBER force field parameters for frequently occurring post-translational modifications. J. Chem. Theory Comput 9, 5653–5674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Steinbrecher T, Latzer J. & Case DA Revised AMBER parameters for bioorganic phosphates. J. Chem. Theory Comput 8, 4405–4412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meagher KL, Redman LT & Carlson HA Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem 24, 1016–1025 (2003). [DOI] [PubMed] [Google Scholar]
- 100.Robustelli P, Piana S. & Shaw DE Developing a molecular dynamics force field for both folded and disordered protein states. Proc. Natl Acad. Sci. USA 115, E4578–E4766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abascal JLF & Vega C. A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys 123, 234505 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Martyna GJ, Tobias DJ & Klein ML Constant pressure molecular dynamics algorithms. J. Chem. Phys 101, 4177–4189 (1994). [Google Scholar]
- 103.Lippert RA et al. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J. Chem. Phys 139, 164106 (2013). [DOI] [PubMed] [Google Scholar]
- 104.Lippert RA et al. A common, avoidable source of error in molecular dynamics integrators. J. Chem. Phys 126, 046101 (2007). [DOI] [PubMed] [Google Scholar]
- 105.Kräutler V, Van Gunsteren WF & Hünenberger PH A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem 22, 501–508 (2001). [Google Scholar]
- 106.Shan Y, Klepeis JL, Eastwood MP, Dror RO & Shaw DE Gaussian split Ewald: a fast Ewald mesh method for molecular simulation. J. Chem. Phys 122, 054101 (2005). [DOI] [PubMed] [Google Scholar]
- 107.Tuckerman M, Berne BJ & Martyna GJ Reversible multiple time scale molecular dynamics. J. Chem. Phys 97, 1990–2001 (1992). [Google Scholar]
- 108.Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J 294, 1–14 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Meer G, Voelker DR & Feigenson GW Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu J. et al. KRAS G12C drug development: discrimination between switch II pocket configurations using hydrogen/deuterium-exchange mass spectrometry. Structure 25, 1442–1448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gureasko J. et al. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol 15, 452–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shen QT et al. Bowl-shaped oligomeric structures on membranes as DegP’s new functional forms in protein quality control. Proc. Natl Acad. Sci. USA 106, 4858–4863 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gasper R, Meyer S, Gotthardt K, Sirajuddin M. & Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell Biol 10, 423–429 (2009). [DOI] [PubMed] [Google Scholar]
- 115.Egea PF et al. Substrate twinning activates the signal recognition particle and its receptor. Nature 427, 215–221 (2004). [DOI] [PubMed] [Google Scholar]
- 116.Shima F. et al. Structural basis for conformational dynamics of GTP-bound Ras protein. J. Biol. Chem 285, 22696–22705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bum‐Erdene K. et al. Investigation into the feasibility of thioditaloside as a novel scaffold for Galectin‐3‐specific inhibitors. ChemBioChem 14, 1331–1342 (2013). [DOI] [PubMed] [Google Scholar]
- 118.Ismail SA et al. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol 7, 942–949 (2011). [DOI] [PubMed] [Google Scholar]
- 119.Hoffman GR, Nassar N. & Cerione RA Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100, 345–356 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.