Abstract
Differentiation in the developing Drosophila eye requires synchronization of cells in the G1 phase of the cell cycle. The roughex gene product plays a key role in this synchronization by negatively regulating cyclin A protein levels in G1. We show here that coexpressed Roughex and cyclin A physically interact in vivo. Roughex is a nuclear protein, while cyclin A was previously shown to be exclusively cytoplasmic during interphase in the embryo. In contrast, we demonstrate that in interphase cells in the eye imaginal disk cyclin A is present in both the nucleus and the cytoplasm. In the presence of ectopic Roughex, cyclin A becomes strictly nuclear and is later degraded. Nuclear targeting of both Roughex and cyclin A under these conditions is dependent on a C-terminal nuclear localization signal in Roughex. Disruption of this signal results in cytoplasmic localization of both Roughex and cyclin A, confirming a physical interaction between these molecules. Cyclin A interacts with both Cdc2 and Cdc2c, the Drosophila Cdk2 homolog, and Roughex inhibits the histone H1 kinase activities of both cyclin A-Cdc2 and cyclin A-Cdc2c complexes in whole-cell extracts. Two-hybrid experiments suggested that the inhibition of kinase activity by Roughex results from competition with the cyclin-dependent kinase subunit for binding to cyclin A. These findings suggest that Roughex can influence the intracellular distribution of cyclin A and define Roughex as a distinct and specialized cell cycle inhibitor for cyclin A-dependent kinase activity.
Cell cycle progression in eukaryotes is regulated by the temporally and spatially ordered actions of a specific class of serine/threonine protein kinases (cyclin-dependent kinases [CDKs]) and their partner regulatory proteins, cyclins (for reviews, see references 26 and 32). Different cyclin-CDK complexes assemble in specific phases of the cell cycle, providing a strict and sophisticated control of cell proliferation during development. Once assembled, the activities of cyclin-CDK complexes are regulated by phosphorylation and dephosphorylation and by interaction with specific inhibitory proteins (cyclin kinase inhibitors [CKIs]; reviewed in reference 33). Both of these mechanisms contribute to the abrupt degradation of cyclins through ubiquitin-dependent proteolysis, the principal mechanism for down-regulation of cyclin protein levels (16, 19).
Recent experiments with both yeast and mammalian cells have shown that degradation of mitotic cyclins persists in G1 until the transition into S phase. In the yeast Saccharomyces cerevisiae, inactivation of the G2 cyclin Clb2 in G1 is mediated by Sic1, which binds to and inactivates Clb2-Cdc28 complexes and targets Clb2 for destruction (31). Progression from G1 into S phase occurs by phosphorylation-dependent degradation of Sic1, which is mediated by G1-specific Cln-Cdc28 kinase activity (41). Similarly, the Schizosaccharomyces pombe CKI Rum1+ binds to the mitotic Cdc2-Cdc13 complex and promotes the degradation of the Cdc13 subunit (5). Rum1 itself is targeted for degradation in G1 by Cdc2-Cig1 (2). A similar mechanism operates in metazoa (3), although the variety of cell types and the complexity of their developmental programs make elucidation of the pathways more difficult.
Cyclin A (CycA) was initially identified in marine invertebrates as a protein whose abundance correlated with mitosis (9). It was subsequently shown that injection of CycA mRNA into Xenopus oocytes could induce mitotic events such as nuclear envelope breakdown and chromatin condensation (36). CycA activity is also required for S-phase entry (13, 25, 28). In vertebrate cells, CycA associates both with Cdc2 during G2 phase and with Cdk2 during S phase (25). In contrast, immunoprecipitation experiments with stage 11 embryos indicate that Drosophila CycA is present primarily in a complex with the G2 kinase Cdc2 but that a homolog of the mammalian Cdk2 kinase, Cdc2c, associates exclusively with the G1 cyclin CycE (18). That Drosophila CycA functions in G2 is well established (17, 21, 22). However, early experiments examining ectopic CycA expression indicated that Drosophila CycA might also function during S phase (23). More recently, both genetic and cell biological data support the suggestion that CycA plays a role in S-phase progression in flies, as in other higher eukaryotes (35, 37, 38).
The Drosophila compound eye is a powerful system for the study of cell cycle control during development. The onset of patterning and differentiation in the eye is temporally and spatially coordinated with cell cycle arrest in the G1 phase of the cell cycle (27, 37). Cells arrest in G1 in the morphogenetic furrow (MF), a physical constriction in the apical surface of the eye disk epithelium; G1 cells either enter a final, synchronous S phase behind the MF or differentiate into retinal neurons. The G1 arrest is mediated in part by Roughex (Rux), a small protein of 335 amino acids with no homology to any reported protein (37). Although genetic and cell biological experiments suggest that Rux function is required to reduce CycA protein levels in G1 (35, 37, 38), there are currently no molecular or biochemical data that define a mechanism for this inhibition. In this study we present evidence for a mechanism by which Rux mediates cell cycle arrest in G1.
MATERIALS AND METHODS
Plasmid constructs.
The coding regions of the Drosophila genes rux, cycA, cycE, cdc2, and cdc2c were cloned by PCR amplification from plasmids pRX21 or pET-16b-Rux, pcDNA17, PT7T3 19U-EcoRI 2.8-typeI cDNA, pcdc2.1, and pcdc2c, respectively, with 5′ and 3′ primers designed from published sequences (21, 22, 29, 37). PCR products were subcloned into pRmHa3 (a gift from A. Orosz), pRc/CMV (Invitrogen), pM, or pVP16 (Clontech). To generate pRmHa3-Cdc2, the Cdc2 coding region was subcloned from plasmid pM-Cdc2 into pRmHa3. Rux amino (N)- and carboxy (C)-terminal deletion proteins were generated by PCR using pRX21 as a template. PCR products were subcloned into pRmHa3, pM, and pVP16 plasmids. Point mutations in Rux were generated by oligonucleotide-mediated mutagenesis using a QuickChange site-directed mutagenesis kit (Stratagene) and pRmHa3-Rux as a template. All constructs were verified by DNA sequencing. Oligonucleotide sequence information and details of plasmid construction are available upon request.
Antibodies.
Monoclonal anti-Rux antibody H6 or H9 (38) was used at a dilution of 1:50 for Western blot analysis and 1:3 or 1:7 for immunofluorescence in tissue or cells, respectively. Polyclonal anti-Rux serum was made by immunizing rabbits with a bacterially produced histidine-tagged Rux protein and was used at a dilution of 1:5,000 for Western blot analysis and at 1:300 for immunofluorescence. Monoclonal anti-CycA antibody A19 and polyclonal anti-CycA serum were generous gifts of P. O'Farrell (University of California, San Francisco). Monoclonal antibodies to CycA were used at 1:100 for Western blot analysis and at 1:25 for immunofluorescence in cells or at 1:5 in tissue. Polyclonal anti-CycA serum was used at 1:5,000 for Western blot analysis and at 1:250 for immunofluorescence. Monoclonal (Becton Dickinson) or sheep polyclonal (Research Diagnostics, Inc.) antibodies to bromodeoxyuridine (BrdU) were used at 1:100 or 1:2,000, respectively. Fluoroscein isothiocyanate- or rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used at 1:200. DNA was counterstained using propidium iodide at 2 μg/ml in RNaseA-treated samples.
Cell culture, transfections, and immunohistochemistry.
Drosophila Schneider line 2 (SL2) cells were grown, transfected, and induced as described previously (24). For immunofluorescence, SL2 cells were washed with ice-cold phosphate-buffered saline (PBS), fixed in 4% formaldehyde in PBS for 20 min at room temperature, washed with PBS, and permeabilized in 100% methanol for 10 min. Eye imaginal disks were dissected in PBS, fixed for 20 min in PLP (2% formaldehyde, 0.1 M lysine [pH 7.4], 2.5 mg of sodium metaperiodate per ml), washed in BSS (40 mM NaCl, 50 mM KCl, 12.5 mM MgSO4 · 7H2O, 5 mM CaCl2 · H2O, 1 mM Tricine, 20 mM glucose, 50 mM sucrose, 0.2% bovine serum albumin [pH 6.95]), and permeabilized in BSN (BSS plus 3% goat serum and 0.2% saponin) with 0.4% Triton X-100. Primary antibodies were incubated overnight at 4°C in BSN plus 0.4% Triton X-100. For double labeling with BrdU, disks were dissected in Drosophila Schneider's medium (Gibco) and incubated in 75 μg of BrdU per ml in Schneider's medium for 30 min. Disks were fixed in 4% formaldehyde in PBS for 15 min, further fixed for 15 min in 4% formaldehyde in PBS plus 0.6% Triton X-100, and washed twice for 15 min in PBS plus 0.6% Triton X-100. Disks were equilibrated twice for 5 min in DNase I buffer (66 mM Tris [pH 7.5], 5 mM MgCl2, 1 mM fresh 2-mercaptoethanol) and incubated for 1 h at 37°C in 150 U of DNase I (Boehringer Mannheim Biochemicals) in 0.5 ml of DNase I buffer. Samples were washed twice for 10 min in PBS plus 0.3% Triton X-100 and incubated overnight in primary antibodies. Samples were washed twice for 30 min and incubated for either 2 h at room temperature or overnight at 4°C in secondary antibody, washed, and mounted in Vectashield (Vector Laboratories) for confocal microscopy. Monoclonal anti-CycA antibodies were preabsorbed against fixed adult heads for 1 h prior to use. Images were obtained using a Bio-Rad MRC1024 confocal microscope and processed using Adobe Photoshop.
Immunoprecipitation and Western blotting.
Cells were washed in PBS, resuspended on ice in 0.2 ml of extraction buffer [10 mM HEPES (pH 7.9), 0.4 M KCl, 1 mM β-mercaptoethanol, 5% glycerol, 0.1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride], and lysed by freezing and thawing. Cellular debris was removed by centrifugation at 40,000 × g for 10 min, and lysates were precleared with 5% (vol/vol) protein A–G-Sepharose for 60 min and were then incubated with specific antibodies for 60 min. Immunoprecipitates were captured with 5% (vol/vol) protein A–G-Sepharose for 60 min, washed three times in lysis buffer, and solubilized with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Samples were resolved in an SDS–10% polyacrylamide gel, transferred to a nitrocellulose membrane (Bio-Rad) by electroblotting, and probed with appropriate antibodies. Blots were developed using enhanced chemiluminescence (Amersham) according to the manufacturer's instructions.
Mammalian transfections and luciferase assays.
CV-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) supplemented with 10% fetal calf serum and 2 mM l-glutamine. For transient transfections, cells at 70 to 80% confluence were plated in 60-mm-diameter culture dishes 48 h prior to transfection. To assess protein-protein interactions, transfection mixtures contained 1 μg of each test plasmid, 1 μg of firefly luciferase reporter plasmid (44), and 0.5 μg of the Renilla luciferase reporter plasmid pRL-TK (Promega) to normalize for transfection efficiency. For competition experiments, the total amount of plasmid DNA was kept constant (4 μg per plate) by using the appropriate empty vectors. Plasmids in 10 μl of water were mixed with 980 μl of OptiMEM (Gibco-BRL) and 20 μl of LipofectAMINE reagent (Gibco-BRL) and incubated at room temperature for 15 min. Cell culture medium was aspirated, cells were washed with DMEM, and the DNA-LipofectAMINE mixture was added to the cells. After a 6-h incubation, 1 ml of DMEM supplemented with 20% fetal calf serum was added. After a 40-h incubation, cells were lysed and luciferase activities were measured using a dual-luciferase-reporter system (Promega) and a Monolight 2001 luminometer (Analytical Luminescence Laboratory). All results are reported as firefly luciferase activity normalized to the activity of the cotransfected Renilla luciferase. Values shown are based on at least three independent transfections.
CDK assays.
Different volumes of extract prepared from SL2 cells overexpressing wild-type Rux or Rux mutant proteins were adjusted to 3 μl with extract from cells transfected with an empty vector (pRmHa3). These extracts were then mixed with 2 μl of extract prepared from cells coexpressing CycA and Cdc2 or Cdc2c. The mixtures were incubated on ice for 30 min, adjusted to 20 μl with a solution containing 50 mM HEPES (pH 7.4), 10 mM MgCl2, 5 mM MnCl2, 1 mM dithiothreitol, 10 μCi of [γ-32P]ATP, and 100 μg of histone H1 (Boehringer Mannheim) per ml, and then incubated for 30 min at 30°C. Reactions were terminated by addition of an equal volume of 2× loading buffer. Samples were fractionated by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography.
RESULTS
Rux protein contains a bipartite NLS and additional C-terminal sequences necessary for its nuclear localization.
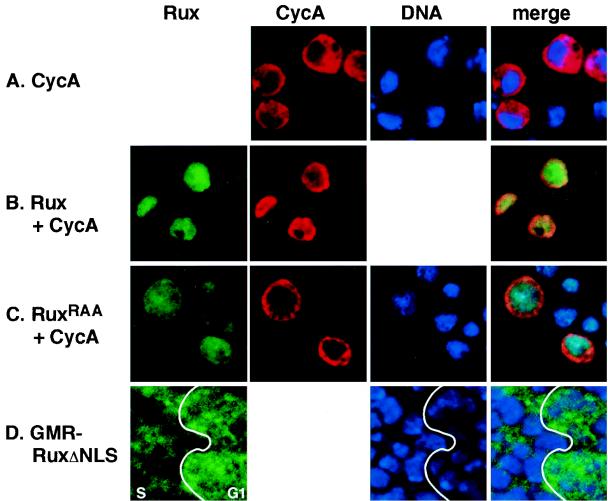
Rux is a nuclear protein in Drosophila eye disk cells (38). Similarly, transiently transfected Drosophila SL2 cells expressing Rux under the control of the metallothionein gene promoter display predominantly nuclear localization of the Rux protein, whereas CycA is predominantly cytoplasmic when it is expressed similarly (Fig. 1A and B). The Rux protein contains a sequence motif similar to the bipartite nuclear localization signals (NLS) found in many nuclear proteins (7). In Rux, this motif consists of two RKR clusters separated by 10 amino acids and is located near the C terminus of the protein (amino acids 311 to 326) (37) (Fig. 2). To show that this putative NLS is necessary for Rux nuclear localization, we separately converted each of the basic RKR clusters to RAA and tested the localization of the corresponding proteins in SL2 cells. Both mutations prevented the nuclear accumulation of the protein. Localization of one of these mutant proteins is shown in Fig. 1C. A protein in which the entire NLS was deleted, RuxΔNLS, displayed the same uniform distribution throughout the cell. These results demonstrate that the C-terminal NLS is necessary for localization of the Rux protein to the nucleus.
FIG. 1.
Nuclear localization of Rux requires the C-terminal bipartite NLS. Confocal optical sections of Drosophila SL2 cells (A to C) and eye imaginal tissue (D) ectopically expressing CycA (A to C), wild-type Rux (B), and Rux NLS mutant proteins (C and D). CycA expression is shown in red, Rux expression is shown in green, and DNA is shown in blue. (A) CycA is cytoplasmically localized in SL2 cells. (B) Wild-type Rux is localized to the nucleus in SL2 cells. Coexpression of Rux and CycA results in the translocation of CycA to the nucleus. (C) When the Rux NLS is mutated (RuxRAA), CycA remains cytoplasmic while Rux is distributed uniformly throughout the cell. In this example, the first RKR cluster in the Rux NLS has been changed to RAA. (D) Expression of the RuxΔNLS mutant protein in the eye imaginal disk. The junction between the G1 domain in the MF (G1) and the synchronous domain of S-phase cells behind the MF (S) is demarcated by a line. G1 cells show both nuclear and cytoplasmic expression of RuxΔNLS, while the mutant protein is down-regulated in the nuclei of cells reentering S phase. The anterior edge of the disk is to the right.
FIG. 2.
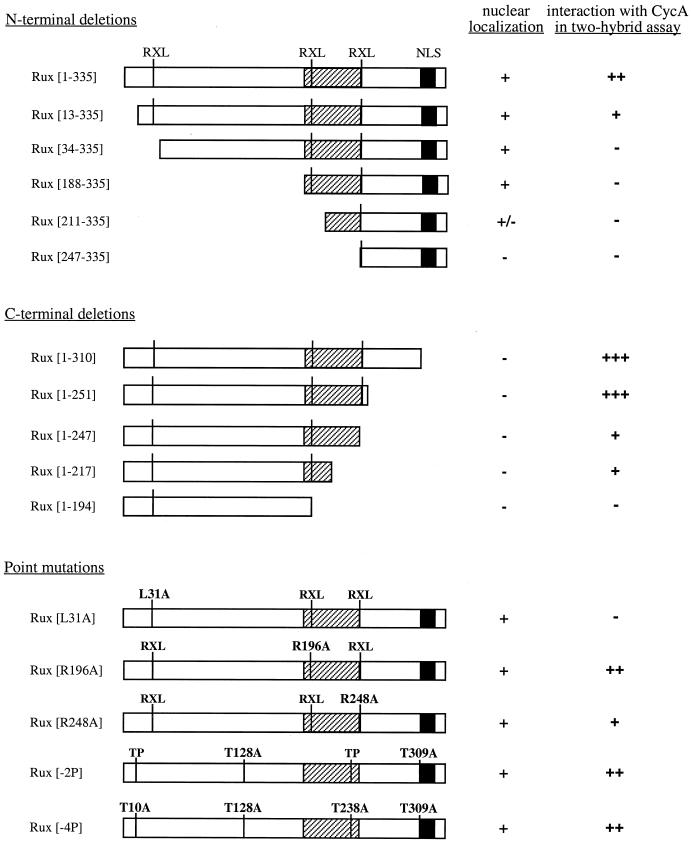
Summary of Rux mutant analysis. Rux mutants were assayed for localization in SL2 cells (nuclear localization). A + indicates that the protein is predominantly nuclear, a − indicates that the protein is uniformly distributed throughout the cell, and a +/− indicates that 80% of the cells show uniform distribution of the protein and that 20% show strictly nuclear staining. Mutants were also assessed for interaction with CycA in the mammalian two-hybrid assay. Luciferase activities relative to control values are as follows: +++, greater than 50-fold; ++, 20- to 50-fold; +, 2- to 20-fold; −, less than 2-fold. Values shown are based on at least three independent transfections. Mutant proteins had similar expression levels in SL2 cells based on Western blot and immunofluorescence analyses of SL2 and CV-1 cells (data not shown). The positions of the Rux NLS and a central region also required for nuclear localization (hatched box) are indicated, as are the positions of three RXL motifs and four consensus CDK phosphorylation sites (TP).
A similar result was obtained with transgenic flies expressing the RuxΔNLS construct under the control of the eye-specific glass multimer reporter (GMR) enhancer (GMR-RuxΔNLS). The GMR enhancer drives high levels of expression in all cells beginning in the MF and extending to the posterior edge of the disk (8, 15). At the onset of expression of GMR-RuxΔNLS in G1 cells in the MF, the mutant protein is distributed uniformly throughout the cell (Fig. 1D, “G1”), similar to the pattern of expression in SL2 cells. Behind the MF, cells that have not committed to differentiate reenter S phase synchronously (37, 43). These cells have basally localized nuclei, in contrast to the differentiating photoreceptor cells, whose nuclei are found in more apical regions of the eye disk (39). RuxΔNLS expression is lost from the nuclei of the basally localized S-phase cells, although cytoplasmic expression in these cells remains (Fig. 1D, “S”). Nuclei of differentiating cells in apical regions of the disk remain positive for RuxΔNLS expression (data not shown). This result is consistent with previous results showing a down-regulation of Rux protein levels and activity in cells ectopically expressing the G1 cyclin CycE and support the notion that Rux may be a target for CycE-mediated destruction in S-phase cells (38).
To determine whether the C-terminal bipartite NLS is sufficient for nuclear localization of the Rux protein, we created a series of N-terminally truncated constructs and determined the localizations of the corresponding proteins in transfected SL2 cells (Fig. 2). N-terminal truncations of Rux up to amino acid 187 retained nuclear localization of the protein. Deletion of sequences to amino acid 210 resulted in a loss of nuclear localization in about 80% of transformed cells, and further deletion to amino acid 246 caused a loss of nuclear localization in virtually all transformed cells. Therefore, the C-terminal bipartite NLS alone is not sufficient for nuclear localization of Rux and additional sequences located between amino acids 188 and 247 are also required.
Cytoplasmic Rux inhibits CycA-dependent S-phase and mitotic functions.
In mammalian cells, CycA is detected primarily as a nuclear protein. In contrast, immunofluorescence experiments indicate that CycA is cytoplasmically localized during interphase in both Drosophila embryos (21) and SL2 cells (Fig. 1A). Coexpression of Rux and CycA in SL2 cells resulted in translocation of CycA to the nucleus in essentially all cells (Fig. 1B), consistent with the transient nuclear accumulation of mitotic cyclins, including CycA, seen in larval and embryonic cells overexpressing Rux (14, 38). We tested the NLS-defective Rux mutants for their ability to drive CycA to the nuclei of SL2 cells. SL2 cells coexpressing any of the NLS mutants and CycA showed cytoplasmic localization of CycA (Fig. 1C and data not shown), indicating that translocation of CycA to the nucleus by Rux depends on a functional NLS.
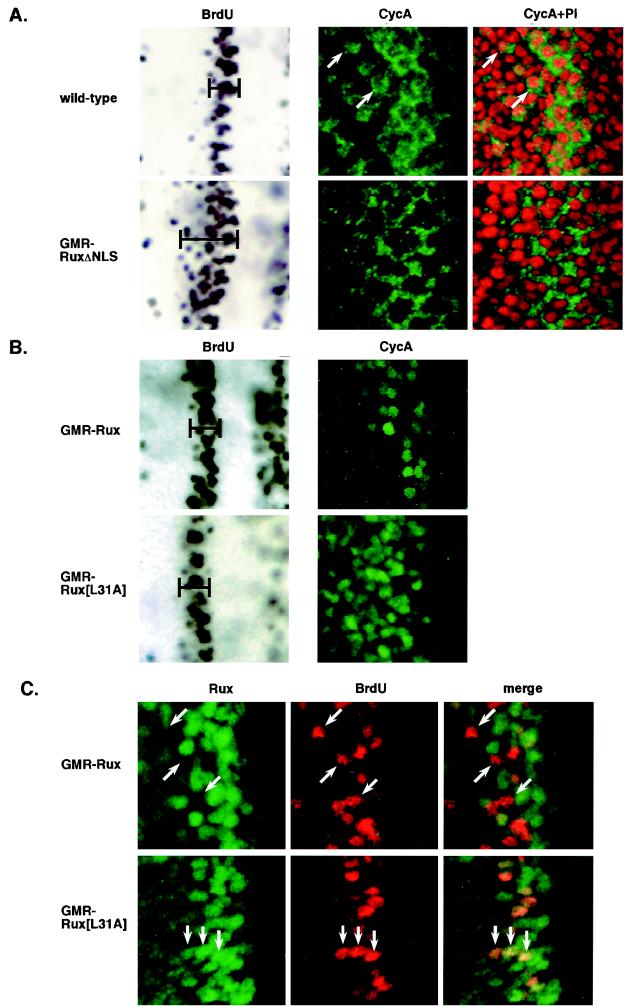
To gain further insight into the role of CycA in cell cycle progression in vivo, we examined localization of endogenous CycA protein in wild-type eye imaginal disks (Fig. 3A). CycA protein levels are low in G1 cells in the MF, while protein expression increases in cells that reenter S phase synchronously behind the MF (37). In contrast to previous results with embryos and cultured cells, optical sections through basal regions of the disk revealed that CycA is present both in the nuclei and in the cytoplasms of S-phase cells in this region, although the cytoplasmic expression is more intense. CycA protein persists in both the nuclei and the cytoplasms of small groups of cells, which are presumably in G2, to the posterior edge of the disk (Fig. 3A).
FIG. 3.
The Rux NLS and CycA-binding site are required for Rux function in vivo. (A and B) Eye imaginal disks with cells expressing the wild-type protein or GMR-RUXΔNLS, GMR-Rux, or GMR-Rux[L31A] were examined for S-phase cells by incorporation of BrdU and subsequent immunohistochemistry and for CycA (green) and DNA (propidium iodide; red) by confocal optical sectioning of fluorescently labeled samples. (C) Eye disks with cells expressing GMR-Rux or GMR-Rux[L31A] were double labeled for Rux protein (green) and S-phase cells (BrdU; red) and examined by confocal microscopy. (A) Wild-type disks display a synchronous band of S-phase cells behind the MF (bracketed region). Basal optical sections of S-phase nuclei show strong cytoplasmic expression of CycA, with lower levels of expression in the nucleus. Expression persists in small groups of cells to the posterior edge of the disk (arrows). Expression of GMR-RuxΔNLS behind the MF results in a broader band of S-phase cells in this region (compare bracketed regions) and CycA expression is exclusively cytoplasmic. (B) Expression of GMR-Rux shows a pattern of S-phase cells similar to that of the wild-type protein, and CycA is localized to the nucleus and degraded. Similarly, GMR-Rux[L31A] disks show a wild-type pattern of S-phase cells. CycA is localized to the nucleus but is stable relative to the level of expression of wild-type Rux. (C) Wild-type Rux protein is down-regulated in cells that enter S phase behind the MF. Arrows indicate examples of S-phase cells that have down-regulated Rux protein. In contrast, the Rux[L31A] mutant protein is stable in S-phase cells. Arrows show examples of double-labeled cells. The anterior edge of the disk is to the right in all panels.
We determined the physiological consequences of expressing RuxΔNLS under the control of the GMR enhancer in developing eye imaginal disks (Fig. 3A). In eye disks overexpressing the wild-type Rux protein, mitotic cyclins enter the nucleus and are rapidly degraded and no mitosis is observed (38) (Fig. 3B). In the presence of RuxΔNLS, CycA remains cytoplasmic, with no nuclear CycA detectable in basal optical sections of S-phase cells behind the MF. This result suggests that overexpression of the Rux NLS mutant protein can block translocation of endogenous CycA to the nucleus. Cells expressing RuxΔNLS reenter S phase behind the MF in an apparently normal manner. However, these S-phase nuclei are larger than wild-type nuclei and no mitotic cells are observed behind the MF (data not shown). These phenotypes are similar to those seen in cycA loss-of-function mutations (21) or in cells overexpressing wild-type Rux (38) and indicate that interaction with cytoplasmic Rux inhibits CycA mitotic functions. In addition, although cells expressing the Rux NLS mutant protein enter S phase normally, S phase persists for a longer period in these cells (Fig. 3A and B; compare bracketed regions in the BrdU-labeled panels), suggesting that S-phase progression is also disrupted when the NLS mutant protein, but not the wild-type protein, is overexpressed. This result suggests that the intracellular localization of CycA is critical for its S-phase functions.
Rux protein physically associates with CycA in Drosophila cells.
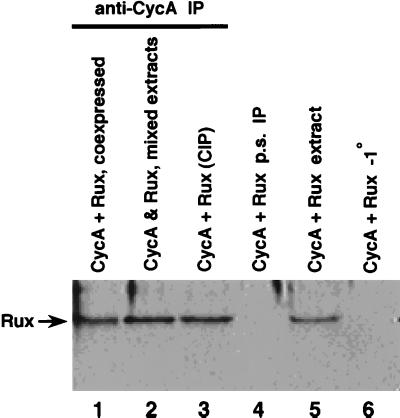
Genetic evidence (37) and the immunofluorescence experiments described above suggest a physical interaction between CycA and Rux. To address this point more directly, we performed coimmunoprecipitation experiments using extracts from SL2 cells expressing Rux and CycA proteins. Proteins were immunoprecipitated with anti-CycA polyclonal serum, size separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with anti-Rux monoclonal antibody H6 (Fig. 4). Rux protein was coimmunoprecipitated both from cells coexpressing Rux and CycA (lane 1) and from mixed extracts from cell lines individually expressing Rux or CycA (lane 2). Coimmunoprecipitation was also observed after pretreatment of the extracts with phosphatase (lane 3), suggesting that phosphorylation is not required for association between these proteins. Similar results were obtained in a reciprocal experiment, using Rux polyclonal serum for immunoprecipitation and an anti-CycA monoclonal antibody for Western blotting (data not shown). These results suggest that Rux and CycA proteins are present in a physical complex in vivo and can form a complex in cell extracts.
FIG. 4.
Rux and CycA are coimmunoprecipitated from Drosophila SL2 cells. Extracts from SL2 cells expressing Rux and/or CycA were prepared and subjected to immunoprecipitation (IP) using anti-CycA polyclonal serum. Immunoprecipitated proteins were separated by SDS-PAGE, and immunoblots were probed with anti-Rux monoclonal antibody. Lane 1, extract from cells coexpressing Rux and CycA; lane 2, extracts from cultures expressing either Rux or CycA that were mixed prior to immunoprecipitation; lane 3: extract from cells coexpressing Rux and CycA that was subjected to phosphatase treatment prior to immunoprecipitation; lane 4, preimmune serum (p.s.) control; lane 5, one-eighth of the precleared extract prior to immunoprecipitation blotted in parallel; lane 6, control immunoprecipitation without primary antibody.
Rux interacts with CycA in a two-hybrid system.
Two-hybrid analysis was used to independently confirm the Rux-CycA interaction. CycA is toxic to yeast cells (38), precluding the use of a yeast two-hybrid assay to detect interactions with CycA. We therefore used a mammalian version of a two-hybrid assay coupled with a dual-luciferase-reporter system to normalize for transfection efficiency (see Materials and Methods). Rux and CycA were fused in frame with either the yeast GAL4 DNA-binding domain (GDB; plasmid pM) or the VP16 transcriptional activation domain (plasmid pVP16). Plasmids expressing complementary pairs of proteins, GDB-Rux and VP16-CycA or GDB-CycA and VP16-Rux, were tested by cotransfection of CV-1 cells along with a pair of reporter plasmids. An interaction between the two test proteins resulted in activation of the firefly luciferase reporter via the VP16 activation domain. Results of these experiments normalized for transfection efficiency using the cotransfected Renilla luciferase reporter are shown in Table 1. These results independently confirm a physical association between Rux and CycA.
TABLE 1.
Two-hybrid interactions
| GBD fusion | VP16 fusion | Relative luciferase activitya | Fold activationb |
|---|---|---|---|
| CycA | Rux | 1,000 | 23 |
| Rux | CycA | 506 ± 42 | 23 |
| Cdc2 | CycA | 178 ± 19 | 9 |
| CycA | Cdc2c | 237 ± 33 | 9 |
| Cdc2c | CycA | 66 ± 5 | 4 |
The relative luciferase activity for each interaction was normalized to that of pM-CycA plus pVP-16-Rux, which was arbitrarily set at 1,000 U. Values are averages from at least three experiments ± standard deviations. Note that relative luciferase activity is dependent in part on the orientation of the interacting proteins in the two-hybrid vectors.
Relative to appropriate control values.
To define the region(s) of Rux required for interaction with CycA, a panel of constructs containing different regions of the Rux coding sequence was generated and tested in the mammalian two-hybrid system as described above (Fig. 2). Activation of the luciferase reporter gene was observed when Rux subclones spanning amino acids 13 to 217 were used. In contrast, no interaction was detected when a further 20 amino acids were deleted from the N terminus (Rux[33–335]). Rux mutant proteins with extended N-terminal deletions, Rux[60–335], Rux[112–335], and Rux[144–335], also showed no interaction with CycA (data not shown). Thus, the N-terminal boundary of the association domain is between amino acids 13 and 33. In addition, no detectable activation of the luciferase reporter gene was observed when plasmids with large C-terminal deletions, Rux[1–194] and Rux[1–161], were used in the assay. Constructs that failed to interact with CycA in the mammalian two-hybrid system also failed to coimmunoprecipitate with CycA from Drosophila cells (data not shown). Taken together, these results demonstrate that the interaction between Rux and CycA is dependent on both N-terminal and central regions of the Rux protein.
Rux-CycA interaction does not require CDK-directed phosphorylation of Rux.
The Rux protein contains four potential phosphorylation sites for proline-directed protein kinases (“TP” in Fig. 2), and bacterially produced Rux serves as an efficient substrate for phosphorylation by CycE-Cdc2c and CycA-Cdc2 complexes immunoprecipitated from Drosophila embryos (38). We asked whether the putative phosphorylation of Rux could directly affect its association with CycA. Coimmunoprecipitation experiments with phosphatase-pretreated extracts from SL2 cells overexpressing Rux (Fig. 4, lane 3) provided indirect evidence that CycA can bind unphosphorylated Rux. We tested two mutant constructs, Rux[−2P] and Rux[−4P], in which two (T128 and T309) and all four (T10, T128, T238, and T309) threonine residues in the CDK-directed phosphorylation sites were replaced with alanines (a gift from K. Zavitz and S. L. Zipursky). Both mutant proteins produced wild-type levels of luciferase activity when they were assayed for CycA interaction in the mammalian two-hybrid system (Fig. 2). We therefore conclude that CDK-directed phosphorylation of Rux is not required for interaction with CycA.
N-terminal conserved residue Leu-31 is critical for the association between Rux and CycA.
Genetic experiments indicate that Rux is required to inhibit CycA activity in G1 cells during development, suggesting that Rux may function as a CKI (37, 38). Recent studies show that the binding of the human CKIs p21, p27, and p57 to CycA-Cdk2 and CycE-Cdk2 is mediated by a short motif termed Cy with the minimal consensus sequence RXL (1, 4). There are three RXL sequences present within the Rux open reading frame (Fig. 2), and these motifs are completely conserved in Rux open reading frames from seven Drosophila species (S. N. Avedisov and B. J. Thomas, unpublished data). Point mutations were introduced into each of these RXL sequences, and the mutant proteins were assessed for binding to CycA in the two-hybrid system (summarized in Fig. 2). Rux[R248A] showed reduced activation of the luciferase reporter gene. Deletion of the region containing this mutation also caused a significant reduction in luciferase activity (compare Rux[1–251] and Rux[1–247]), suggesting that this RXL sequence contributes to optimal complex formation. Rux[R196A] showed wild-type levels of luciferase activity, although C-terminal deletion analysis suggested that sequences in this region also participate in CycA binding (compare Rux[1–217] to Rux[1–194]). Rux[L31A] abolished luciferase reporter activation, indicating that Leu-31 is absolutely required for association with CycA.
We examined the effect of expressing Rux[L31A] in the eye disk using the GMR enhancer (Fig. 3B and C). Entry into and progression through S phase are unaffected by expression of GMR-Rux[L31A]. Interestingly, in the presence of this mutant protein, CycA still accumulated in the nucleus but was no longer degraded. This finding suggests that the nuclear accumulation of CycA may be an indirect effect of Rux overexpression and that a direct interaction with Rux may be required for destruction of CycA protein. No mitotic cells were detected behind the MF in GMR-Rux[L31A] eye disks (data not shown). Basal optical sections from transgenic flies containing Rux[L31A] showed uniform expression of the mutant protein behind the MF. Double labeling with BrdU showed that the mutant protein was stable in cells that reenter S phase in this region (Fig. 3C). This is in contrast to the expression pattern of the wild-type protein, which is down-regulated in S-phase cells immediately posterior to the MF.
Rux inhibits CycA-dependent kinase activity in cell extracts.
In a previous study, we reported that bacterially produced glutathione S-transferase- or histidine-tagged Rux neither binds to CycA nor inhibits CycA-Cdc2 activity, although Rux itself served as an efficient substrate for phosphorylation by this complex in vitro (38). As an alternative assay for Rux inhibition of kinase activity, we examined histone H1 phosphorylation in mixed extracts from Drosophila cultured cells transiently overexpressing proteins of interest (Fig. 5). Wild-type Rux, but not the CycA-binding-deficient mutant protein Rux[L31A], was found to specifically inhibit the kinase activities of both CycA-Cdc2 and CycA-Cdc2c complexes. Furthermore, wild-type Rux itself was a good substrate for the CycA-Cdc2 kinase but not for CycA-Cdc2c. We tested the possibility that the observed decrease in kinase activity was due to the ability of Rux to compete with histone H1 as a substrate for the kinase. Rux[−4P], which lacks all four consensus CDK phosphorylation sites, was still able to inhibit CycA-Cdc2-dependent phosphorylation of histone H1, even though the mutant protein was not phosphorylated (Fig. 5, lanes 7 to 9). Rux[L31A] was also not phosphorylated in this assay, consistent with our characterization of this mutant protein as being unable to bind CycA (Fig. 2).
FIG. 5.
Rux inhibits CycA dependent kinase activity in cell extracts. The indicated volumes of extracts prepared from cells expressing wild-type (lanes 1 to 3 and 10 to 12) or mutant (lanes 4 to 9) forms of Rux were mixed with 2 μl of extract prepared from cells coexpressing CycA and Cdc2 (lanes 1 to 9) or CycA and Cdc2c (lanes 10 to 12) as described in Materials and Methods and tested for cyclin-CDK activity with histone H1 as an exogenous substrate. The reaction products were analyzed by SDS-PAGE and autoradiography. Wild-type and mutant Rux proteins were expressed at equivalent levels and migrated at the same relative size, as determined by Western blot analysis (data not shown). Increasing the volume of the Rux-containing extract from 0 to 3 μl resulted in an approximately fivefold inhibition of CycA-associated kinase activities as determined by PhosphorImager quantitation. When extract was prepared from untransfected or mocktransfected cells, a similar inhibition of endogenous CycA-CDK activity by Rux could be detected after prolonged exposure of autoradiographs (data not shown). Note that histone H1 phosphorylation products appear as a doublet in these experiments. The data are representative of at least three independent experiments.
Rux competes with CDKs for binding to CycA.
Cyclin- and CDK-binding sites have been identified or predicted in the cell cycle inhibitors p21, p27, p57, and Dacapo (4, 6, 12, 20, 30). In the case of p27, contacts with both cyclin and CDK subunits are required for optimal kinase inhibition (42). The Rux amino acid sequence lacks an apparent CDK-binding domain, and Rux immunoprecipitates from SL2 cells or embryos overexpressing Rux do not contain detectable Drosophila CDK protein (data not shown). To further examine potential interactions between CDKs, Rux, and CycA, we tested the Drosophila kinases Cdc2 and Cdc2c for interaction with cyclins and Rux in the mammalian two-hybrid assay. In control experiments, Drosophila CycE, a G1 cyclin, as expected demonstrated an interaction with Cdc2c but not with Cdc2. Surprisingly, CycA showed binding with both Drosophila kinases (Table 1), although only Cdc2 was found associated with CycA after immunoprecipitation from embryo extracts (18). In contrast, no interactions were detected between Rux and either kinase, consistent with our immunoprecipitation results.
These observations prompted us to examine the influence of Rux on CycA-CDK interaction. CycA-Cdc2 and CycA-Cdc2c interaction assays using the mammalian two-hybrid system were performed with CV-1 cells cotransfected with different amounts of pRc/CMV-Rux (see Materials and Methods). The addition of Rux had similar effects on the interaction between CycA and either of the two Drosophila CDKs (Fig. 6). Low levels of added Rux caused a small increase in CycA-CDK interaction, which was not seen when CycA was used as a competitor. A similar increase was seen in CycA-dependent histone H1 kinase activity with low levels of added Rux (11). Further increases in the level of added Rux caused binding to steadily drop to roughly one-third of the starting level, indicating competition between Rux and the kinases for binding to CycA.
FIG. 6.
Rux competes with CDKs for binding to CycA. A mammalian two-hybrid assay was performed between VP16-CycA and GDB-Cdc2 (filled squares) or GDB-CycA and VP16-Cdc2c (filled diamonds) in the presence of competitor plasmids. The averaged data ± standard deviations (error bars) from three independent experiments are shown normalized to the response without a competitor plasmid. CycA interacts with both kinases in the two-hybrid system. Rux (solid lines) competes for binding to the complex with kinetics different from those of CycA (dashed line), with approximately one-third of the complexes remaining refractory to added competitor. Control assays for binding (open symbols) contained a single fusion protein construct cotransfected with an empty vector.
DISCUSSION
Although genetic and immunohistochemical experiments indicate that Rux prevents CycA accumulation in early G1 in the developing Drosophila eye, the mechanism by which Rux functions to reduce CycA protein levels has been unclear. Using two in vivo techniques, two-hybrid analysis and coimmunoprecipitation, we have shown that Rux and CycA interact in both Drosophila and mammalian cells. Although we cannot rule out the possibility that other as yet unidentified proteins mediate the interaction between Rux and CycA, analysis of Rux point mutations as well as in vitro experiments (11) suggest that the interaction is direct. Binding of Rux to CycA both in vitro and in vivo is eliminated by a mutation in a motif, RXL, that has been shown in mammalian cells to mediate binding of a variety of proteins to CycA, including p107, p130, and the CKIs p21 and p27 (1, 4). In Rux, a single amino acid substitution in this motif is sufficient to eliminate CycA interaction in both the two-hybrid assay and Drosophila cultured cells. These data provide strong evidence that Leu-31 is part of a CycA-binding site that contains the same minimal consensus sequence seen in mammalian cell cycle inhibitors.
Although in vitro experiments indicate that Leu-31 is necessary for CycA binding, the phenotype resulting from overexpression of the Rux[L31A] mutant in the eye is unexpectedly complicated. In the presence of the mutant protein, CycA still localizes to the nucleus, both in the eye disk and in SL2 cells (Fig. 3 and data not shown). It is possible that, although Leu-31 is critical for binding to CycA in cultured cells and in vitro, residual binding occurs via one or both of the remaining two RXL sites in the protein. However, Rux mutant proteins in which all three RXL sites are eliminated still display nuclear localization of CycA in SL2 cells (data not shown). This result suggests that Rux is not directly involved in CycA nuclear import. CycA protein is stabilized in Rux[L31A] relative to expression of wild-type Rux, indicating that binding to Rux via Leu-31 may be required for degradation of CycA. Finally, mitosis does not occur in eye disks expressing Rux[L31A], a phenotype also seen in nondegradable CycA mutant proteins lacking a destruction box (34). However, in contrast to cells expressing nondegradable CycA mutant proteins, which arrest in metaphase, cells expressing Rux[L31A] arrest prior to chromosome condensation (data not shown).
The simplest explanation of these data, taken together, is that the Rux[L31A] mutant protein displays residual binding to CycA in vivo. Because the Rux[L31A] mutant protein is stable in cells that reenter the cell cycle behind the MF whereas wild-type Rux is degraded, the Rux[L31A] mutant protein is expressed to much higher levels in these S-phase cells than is the wild-type protein (Fig. 3). In addition, mutation of a second RXL motif in Rux (at position 248) showed a reduction in CycA binding in the mammalian two-hybrid system, suggesting that this second RXL site also participates in binding. It is possible that this weak residual binding coupled with the stabilization of the mutant protein in S/G2 cells leads to disruption of mitotic CycA-Cdk complexes (see below) and the observed G2 arrest. Indeed, fly transformant lines in which Rux[L31A] is expressed at lower levels than in the line analyzed here display a completely wild-type phenotype (data not shown), indicating that extremely high levels of expression of the mutant protein are required to detect these mitotic effects.
The Rux-CycA interaction occurs via a motif similar to that of characterized CKIs. However, unlike other CKIs, which typically bind both cyclin and CDK subunits, Rux does not interact with either Drosophila CDK in the two-hybrid assay. In addition, we did not observe coimmunoprecipitation of CDKs with Rux and CycA from SL2 cells expressing all three proteins (data not shown). Instead, our two-hybrid data indicate that Rux competes with CDKs for binding to CycA. Rux may do this by reducing the stability of CycA-CDK complexes or, alternatively, by preventing CDKs from binding to CycA. This conclusion is conditioned by the finding that low levels of added Rux cause a modest stimulation of CycA-CDK interaction, suggesting that the associations between these proteins may be more complex than has been suggested by a simple competition model.
In addition to the expected interaction between CycA and the G2 CDK Cdc2, we also detect an interaction between CycA and the G1 Cdk2 homolog Cdc2c. Previous experiments using stage 11 Drosophila embryos detected coimmunoprecipitation of only Cdc2 with CycA (18). Stage 11 corresponds roughly to embryonic cell cycle 16, which consists of a regulated G2 phase with no apparent G1 (10). It is possible that CycA-Cdc2c complexes are normally present in S phase at such low levels that they cannot be detected at this stage of embryonic development. Human CycA associates with Cdk1 in G2 and with Cdk2 in S phase (25). Our data suggest that the same may happen during larval cell divisions in Drosophila melanogaster. If such an interaction occurs, the activity of this complex may also be a target for regulation by Rux.
Rux is a nuclear protein both in SL2 cells and in eye imaginal disks. In Drosophila embryos, CycA is cytoplasmic during those stages of interphase when it can be detected (late S phase and G2 [21]). We found a different pattern of localization in eye disks where CycA, as in higher eukaryotes, is also present in the nuclei of S- and G2-phase cells. We have seen a similar distribution of Drosophila CDKs in S-phase cells in the developing eye using anti-PSTAIR antibodies (B. J. Thomas, unpublished observations), indicating that active CycA-Cdk complexes may be present in both cellular compartments. As a consequence, we suggest that some of the activities associated with CycA-dependent kinase complexes are likely to be regulated at the level of subcellular distribution. In support of this hypothesis, eye disks expressing the RuxΔNLS construct show an expansion in the domain of S-phase cells behind the MF compared with a similar domain in control disks, consistent with an increase in the length of S phase. This observation suggests that the subcellular localization of CycA is important for S-phase progression and is blocked by expression of the RuxΔNLS mutant protein but not by expression of wild-type Rux.
How does Rux function to reduce CycA levels in G1? We suggest that CycA normally exists in an equilibrium between nuclear and cytoplasmic fractions. In support of this notion, CycA expressed from a heat-inducible promoter in a GMR-Rux background is predominantly cytoplasmic immediately after heat shock and gradually becomes localized to the nucleus when the heat shock is removed (B. J. Thomas, unpublished observations). We suggest that in G1 cells in the MF, the level of endogenous CycA protein is very low as a consequence of the abrupt destruction of mitotic cyclins just prior to G1 arrest in the MF. In contrast, Rux is stable in these G1 cells but is absent in cells that are actively cycling (38). Thus, relatively high levels of Rux in G1 can shift the CycA subcellular distribution by binding to and effectively targeting CycA protein to the nucleus. Rux may then inhibit CycA-dependent kinase activity by preventing or disrupting the CycA-CDK interaction. Nuclear CycA is also targeted for destruction by binding with Rux, although proteolysis of CycA is apparently not required for inactivation of CycA-dependent functions (35). When cells reenter S phase behind the MF, Rux levels decline (38) and CycA reaccumulates for its S/G2 functions. This model implies that the level of Rux relative to that of CycA must be significantly higher in G1 (where inhibition of CycA occurs) than in S phase (where Rux levels are reduced).
Rux contains four consensus phosphorylation sites for CDKs, and Rux itself is a good substrate for phosphorylation by both CycE-Cdk2 and CycA-Cdc2 activities immunoprecipitated from Drosophila embryos (38) and SL2 cells (Fig. 4). We show here that phosphorylation of these sites is not required for binding to CycA. A previous study showed that the effect of ectopic Rux expression on CycA localization and stability in eye imaginal tissue could be overcome by overexpression of CycE, suggesting that Rux itself may be a target for CycE-dependent kinase activity (38). In both yeast and mammalian cells, phosphorylation of CKIs in G1 is absolutely required for their destruction by ubiquitin-mediated proteolysis (41, 42). The sequence defined in this paper as a CycA-binding site overlaps a region predicted to be important for ubiquitin-mediated degradation, suggesting that CycA may compete with the ubiquitination apparatus for binding to Rux. Indeed, the Rux[L31A] mutant protein, in which this motif is disrupted, shows increased stability in cells that reenter S phase behind the MF. It remains to be seen, however, whether Rux is phosphorylated and/or ubiquitinated in vivo. Experiments to address the role of CycE in inhibiting Rux function are in progress.
ACKNOWLEDGMENTS
We thank P. O'Farrell and A. Orosz for gifts of antibodies and reagents, K. Zavitz and S. L. Zipursky for the Rux[−2P] and Rux[−4P] constructs and the GMR-Rux[−4P] flies, and S. Panavally and A. Kuzin for help with embryo injections. We also thank M. Lichten, C. Wu, M. Lilly, and D. Wassarman for critical reading of the manuscript and M. Lichten, C. Klee, and B. Paterson for helpful discussions. We acknowledge F. Sprenger for communication of results prior to publication.
REFERENCES
- 1.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benito J, Martin-Castellanos C, Moreno S. Regulation of the G1 phase of the cell cycle by periodic stabilization and degradation of the p25rum1 CDK inhibitor. EMBO J. 1998;17:482–497. doi: 10.1093/emboj/17.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Correa-Bordes J, Gulli M-P, Nurse P. p25rum1 promotes proteolysis of the mitotic B-cyclin p56cdc13 during G1 of the fission yeast cell cycle. EMBO J. 1997;16:4657–4664. doi: 10.1093/emboj/16.15.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Nooij J C, Letendre M A, Hariharan I K. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- 7.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 8.Ellis M C, O'Neill E M, Rubin G M. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- 9.Evans T, Rosenthal E T, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 10.Foe V E. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- 11.Foley E, O'Farrell P H, Sprenger F. Rux is a cyclin-dependent kinase inhibitor (CKI) specific for mitotic cyclin-Cdk complexes. Curr Biol. 1999;9:1392–1402. doi: 10.1016/s0960-9822(00)80084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fotedar R, Fitzgerald P, Rousselle T, Cannella D, Doree M, Messier H, Fotedar A. p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity. Oncogene. 1996;12:2155–2164. [PubMed] [Google Scholar]
- 13.Girard F, Strausfeld U, Fernandez A, Lamb N J C. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 14.Gönczy P, Thomas B J, DiNardo S. roughex is a dose-dependent regulator of the second meiotic division during Drosophila spermatogenesis. Cell. 1994;77:1015–1025. doi: 10.1016/0092-8674(94)90441-3. [DOI] [PubMed] [Google Scholar]
- 15.Hay B A, Wolff T, Rubin G M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 16.King R W, Deshaies R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 17.Knoblich J A, Lehner C F. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J. 1993;12:65–74. doi: 10.1002/j.1460-2075.1993.tb05632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoblich J A, Sauer K, Jones L, Richardson H, Saint R, Lehner C F. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 19.Koepp D M, Harper J W, Elledge S J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 20.Lane M E, Sauer K, Wallace K, Jan Y N, Lehner C F, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- 21.Lehner C F, O'Farrell P H. Expression and function of Drosophila cyclin A during embryonic cell cycle progression. Cell. 1989;56:957–968. doi: 10.1016/0092-8674(89)90629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehner C F, O'Farrell P H. The roles of Drosophila cyclins A and B in mitotic control. Cell. 1990;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehner C F, Yakubovich N, O'Farrell P H. Exploring the role of Drosophila cyclin A in the regulation of S phase. Cold Spring Harbor Symp Quant Biol. 1991;56:465–475. doi: 10.1101/sqb.1991.056.01.053. [DOI] [PubMed] [Google Scholar]
- 24.Orosz A, Wisniewski J, Wu C. Regulation of Drosophila heat shock factor trimerization: global sequence requirements and independence of nuclear localization. Mol Cell Biol. 1996;16:7018–7030. doi: 10.1128/mcb.16.12.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pines J. Regulation of the G2 to M transition. In: Pagano M, editor. Cell cycle control. New York, N.Y: Springer-Verlag; 1998. pp. 57–78. [Google Scholar]
- 27.Ready D F, Hanson T E, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 28.Resnitzky D, Hengst L, Reed S I. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15:4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson H E, O'Keefe L V, Reed S I, Saint R. A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development. 1993;119:673–690. doi: 10.1242/dev.119.3.673. [DOI] [PubMed] [Google Scholar]
- 30.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 31.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 32.Sheaff R J, Roberts J M. Regulation of G1 phase. In: Pagano M, editor. Cell cycle control. New York, N.Y: Springer-Verlag; 1998. pp. 1–34. [Google Scholar]
- 33.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 34.Sigrist S J, Lehner C F. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 35.Sprenger F, Yakubovich N, O'Farrell P H. S-phase function of Drosophila cyclin A and its downregulation in G1 phase. Curr Biol. 1997;7:488–499. doi: 10.1016/s0960-9822(06)00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swenson K I, Farrell K M, Ruderman J V. The clam embryo protein cyclin A induces entry into M phase and the resumption of meiosis in Xenopus oocytes. Cell. 1986;47:861–870. doi: 10.1016/0092-8674(86)90801-9. [DOI] [PubMed] [Google Scholar]
- 37.Thomas B J, Gunning D A, Cho J, Zipursky S L. Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell. 1994;77:1003–1014. doi: 10.1016/0092-8674(94)90440-5. [DOI] [PubMed] [Google Scholar]
- 38.Thomas B J, Zavitz K H, Dong X, Lane M E, Weigmann K, Finley R L, Jr, Brent R, Lehner C F, Zipursky S L. Roughex down-regulates G2 cyclins in G1. Genes Dev. 1997;11:1289–1298. doi: 10.1101/gad.11.10.1289. [DOI] [PubMed] [Google Scholar]
- 39.Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J Embryol Exp Morph. 1985;89:313–331. [PubMed] [Google Scholar]
- 40.Truman J W, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 41.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 42.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff T, Ready D F. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J M, Wei Q, Zhao X, Paterson B M. Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4. EMBO J. 1999;18:926–933. doi: 10.1093/emboj/18.4.926. [DOI] [PMC free article] [PubMed] [Google Scholar]