SUMMARY
Functional tags are ubiquitous in cell biology, and for studies of one chromosomal locus, the centromere, tags have been remarkably useful. The centromere directs chromosome inheritance at cell division. The location of the centromere is defined by a histone H3 variant, CENP-A. The regulation of the chromatin assembly pathway essential for centromere inheritance and function includes posttranslational modification (PTM) of key components, including CENP-A itself. Others have recently called into question the use of functional tags, with the claim that at least two widely used tags obscured the essentiality of one particular PTM, CENP-AK124 ubiquitination (ub). Here, we employ three independent gene replacement strategies that eliminate large, lysine-containing tags to interrogate these claims. Using these approaches, we find no evidence to support an essential function of CENP-AK124ub. Our general methodology will be useful to validate discoveries permitted by powerful functional tagging schemes at the centromere and other cellular locations.
In brief
Using three gene replacement strategies, Salinas-Luypaert et al. demonstrate that CENP-AK124ub is not essential for CENP-A function at centromeres. Thus, functional tags do not mask the role of K124 when it is mutated. These strategies can be employed to interrogate posttranslational modifications at the centromere and other cellular locations.
Graphical Abstract
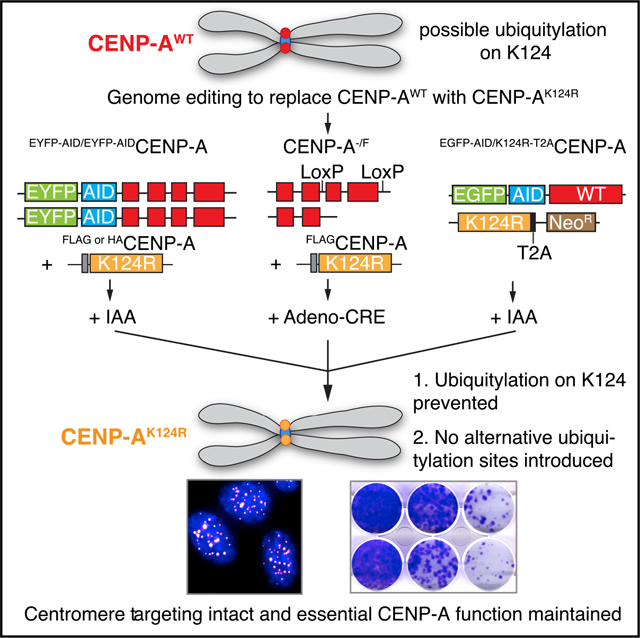
INTRODUCTION
The centromere is the chromosomal locus that recruits the kinetochore at mitosis to connect to spindle microtubules, driving accurate genome segregation (Kixmoeller et al., 2020). The histone H3 variant centromere protein A (CENP-A) (Earnshaw and Rothfield, 1985; Palmer and Margolis, 1985) assembles into nucleosomes that specify the location of the centromere epigenetically (i.e., independent of a particular DNA sequence) (Black and Cleveland, 2011), even on chromosomes that lack the typically repetitive DNA found at the centromeres of many eukaryotes, including mammals (Depinet et al., 1997; Locke et al., 2011; Logsdon et al., 2019; Schueler et al., 2001; Wade et al., 2009). One area of current major investigation is the relationship between the epigenetic (CENP-A nucleosomes) and genetic (centromere DNA repeats) components of the centromere, with molecular evidence recently emerging for centromere strengthening centromeric chromatin via direct (i.e., a special fit with CENP-A nucleosomes; Allu et al., 2019; Iwata-Otsubo et al., 2017) and indirect (i.e., through the centromere repeat-binding protein CENP-B; Dumont and Fachinetti, 2020; Fachinetti et al., 2015; Hoffmann et al., 2016, 2020; Okada et al., 2007; Otake et al., 2020) interactions. These findings are relevant to a long-standing hypothesis for centromere drive, wherein the epigenetic features rapidly evolve to suppress ever-strengthening genetic features (Henikoff et al., 2001). A second area of current major investigation is on the biochemical steps, and their regulation via posttranslational modifications (PTM), that propagate centromere chromatin (Mitra et al., 2020a), centered on deposition of CENP-A at centromeres by its specific histone chaperone, HJURP (Dunleavy et al., 2009; Foltz et al., 2009).
While not unique to the centromere field, the importance of genetically encoded functional tags to the most seminal findings over the last ~15–25 years cannot be overstated. Notably useful tags on CENP-A-related studies have included fluorescent proteins (i.e., EGFP/EYFP) (Black et al., 2004; Fachinetti et al., 2013), those appropriate for pulse-labeling approaches (i.e., the self-labeling SNAP-tag) (Bodor et al., 2013; Falk et al., 2015; Jansen et al., 2007), those that permit tandem affinity purifications out of cell lysates (Dunleavy et al., 2009; Foltz et al., 2006, Foltz et al., 2009; Nechemia-Arbely et al., 2017, 2019; Okada et al., 2006), those that confer rapid-inducible degradation (i.e., auxin-inducible degron [AID]) (Fachinetti et al., 2015; Hoffmann et al., 2016, 2020; Holland et al., 2012), and those that permit recruitment of centromeric chromatin components to an ectopic locus, such as the LacO-LacI tethering system (Barnhart et al., 2011; Bassett et al., 2012; Chen et al., 2014; Hori et al., 2013; Logsdon et al., 2015, 2019; Mendiburo et al., 2011). The use of fluorescent tags permitted the identification of the CENP-A targeting domain (CATD) within its histone fold domain, as well as the finding that the CATD confers recognition of nascent CENP-A by HJURP (Bassett et al., 2012; Black et al., 2004; Foltz et al., 2009). Also, the use of pulse labeling with the SNAP-tag defined how nascent CENP-A nucleosome assembly is restricted to mitotic exit (Bodor et al., 2013; Jansen et al., 2007). This finding has had a wide-ranging impact on the field, arguably with the most direct influence in thinking of the biochemical regulation of the process involving PTM of centromeric chromatin (Mitra et al., 2020a). Phosphorylation (French and Straight, 2019; McKinley and Cheeseman, 2014; Silva et al., 2012; Stankovic et al., 2017), ubiquitylation (Bade et al., 2014; Niikura et al., 2015), sumoylation (Liebelt et al., 2019; Mitra et al., 2020b), methylation (Hori et al., 2014), and acetylation (Shang et al., 2016) have all been proposed to occur as part of the processes that promote centromeric chromatin assembly or regulate its homeostasis in another manner (Srivastava et al., 2018).
Recent findings from Kitagawa and colleagues have called into question experiments with centromeric chromatin that utilize tags, such as EYFP, SNAP, and the Lac repressor (LacI) (Niikura et al., 2019). They initially reported that a residue outside the CATD, CENP-AK124, is ubiquitylated (K124ub) and that this is essential for CENP-A deposition at the centromere (Niikura et al., 2015). Specifically, CENP-AK124ub was claimed to be crucial for the interaction with HJURP and necessary for the epigenetic propagation mechanism that ensures the faithful inheritance of centromere location on the chromosome (Niikura et al., 2015, 2016). These findings were based on transient overexpression studies using a CENP-AK124R mutant that cannot be ubiquitylated at that particular residue and a version of CENP-AK124R with ubiquitin fused to its C terminus. Using multiple gene replacement strategies and nascent CENP-A assembly assays that included functional tags (EYFP, SNAP, and LacI-hemagglutinin [LacI-HA]), we failed to find any support for their conclusions; CENP-AK124R mutant interacts with HJURP, accumulates at centromeres, and supports kinetochore function and cell viability (Fachinetti et al., 2017). In a more recent study (Niikura et al., 2019), they counter by proposing that “EYFP tagging induces ubiquitylation at a lysine other than K124, and this ubiquitylation allows EYFP-CENP-AK124R to bind to HJURP” and that “because the SNAP tag (20 kDa) is also a larger tag than CENP-A… and has 10 lysins [sic], SNAP-CENP-A K124R, presumably, is ubiquitylated at a site different than K124” (Niikura et al., 2019). By extension, their conclusion suggests that the LacI-HA-tag may induce yet another ubiquitylation that could functionally substitute for K124ub. Nevertheless, the authors did not provide any experimental evidence or a clear explanation as to how the size or presence of lysines on the tag would cause this substitute ubiquitylation to occur. These new statements also modify their initial claims of the importance of K124 itself (Niikura et al., 2015), as they now propose that CENP-A ubiquitylation does not need to be site specific to bind to HJURP. This model is structurally inconsistent with the well-established mechanism of HJURP binding to CENP-A (Bassett et al., 2012; Hu et al., 2011), and it is unclear how a site-nonspecific ubiquitin moiety would impact CENP-A-HJURP complex formation. One possibility is that EYFP, SNAP, or LacI tagging all mask the importance of CENP-AK124.
We now report findings from rapid protein replacement strategies, including genome editing at the endogenous CENP-A locus and more traditional Cre-mediated gene excision, to test versions of CENP-AK124R that lack any additional lysine residues that could themselves be ubiquitylated, or lead to CENP-A ubiquitylation at sites other than K124.
RESULTS AND DISCUSSION
Stable HA- or FLAG-CENP-AK124R expression rescues endogenous CENP-A loss in RPE-1 cells
We first tested if stably expressed FLAG- or HA-tagged CENP-AK124R (the same short epitope tags used in Niikura et al., 2019) is able to rescue cells in which the endogenous CENP-A was rapidly degraded by an AID tag in genetically stable, diploid hTERT RPE-1 cells (Hoffmann et al., 2016). The FLAG-tag (DYKDDDDK) contains two lysine residues that, in principle, could themselves be ubiquitylated, while the HA-tag (YPYDVPDYA) does not. Importantly, FLAG-tagged CENP-AK124R was reported not to be ubiquitylated at any site, even when co-overexpressed with ubiquitin (Niikura et al., 2015, 2019). EYFP-AIDCENP-A RPE-1 cells (hereafter named EACENP-A) were transduced with retroviral vectors expressing the tagged CENP-A rescue constructs (wild type [WT] or K124R, with FLAG- or HA-tags), and single colonies surviving antibiotic selection were isolated, individually characterized, and pooled (Figure 1A). Upon addition of the auxin hormone indole-3-acetic acid (IAA) EACENP-A expressed from the endogenous CENP-A gene locus is fully and rapidly degraded (Figures 1B and 1C), as previously shown (Hoffmann et al., 2016, 2020), leaving the cells to express only the stably integrated rescue proteins (Figures 1B and S1A). We note that in our cell lines, the rescue proteins are uniformly expressed at higher levels than endogenous CENP-A (Figure S1A). HA- or FLAG-tagged CENP-A rescue proteins, both WT and K124R, target to centromeres and colocalize with CENP-C even when endogenous CENP-A is fully degraded (Figures 1B and S1B).
Figure 1. Lysine-free tagged CENP-AK124R localizes efficiently at centromeres.
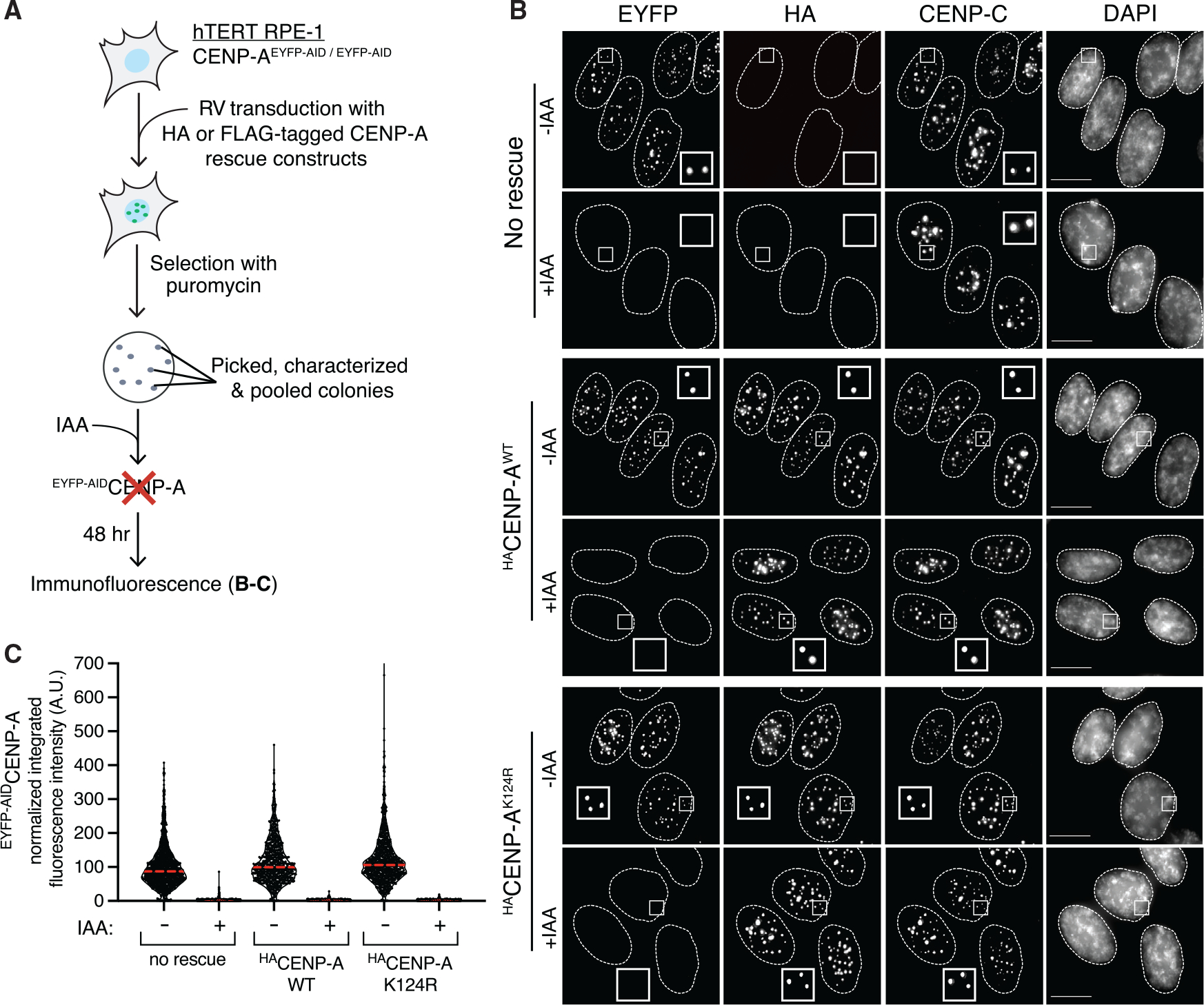
(A) Schematic of the experimental strategy for generating CENP-A rescues in hTERT RPE-1 cells with EACENP-A. EA, EYFP-AID; RV, retrovirus; IAA, indole-3-acetic acid.
(B) Representative immunofluorescence images showing the localization of EYFP-AID-tagged CENP-A and HA-tagged WT or K124R CENP-A rescues at centromeres. Cells were treated with 500 μM IAA for 48 h to degrade EACENP-A. α-CENP-C was used as a marker to determine centromere position. Nuclei are contoured with white dashed lines. Scale bar, 10 μm.
(C) Quantification of EACENP-A integrated fluorescence intensity at centromeres of n > 780 centromeres per condition in presence or absence of 500 μM IAA for 48 h. The red lines represent the median. The data are normalized to “no rescue -IAA.”
Long-term cell viability (14 days) was assessed by means of colony-formation assays (Figure 2A). EACENP-A cells with one of the four different rescue constructs (WT or K124R mutant, each tagged with either HA or FLAG), were cultured in presence or absence of IAA. After 2 weeks, the cells were fixed and stained with crystal violet (Figure 2B). The percentage of colonies formed in the presence of IAA, relative to the number of colonies formed in its absence for the same rescue, was calculated (Figure 2C). The CENP-AK124R mutant rescue constructs confer long-term cell viability after removal of EACENP-A to a similar extent as the CENP-AWT rescue constructs, regardless of the tag used (Figure 2C). Thus, in this context, the FLAG- and HA-tagged constructs provide no evidence to support the claim of impaired function of a CENP-A mutant that cannot be ubiquitylated.
Figure 2. Preventing CENP-A ubiquitylation on K124 does not interfere with long-term centromere function and viability in RPE-1 cells.
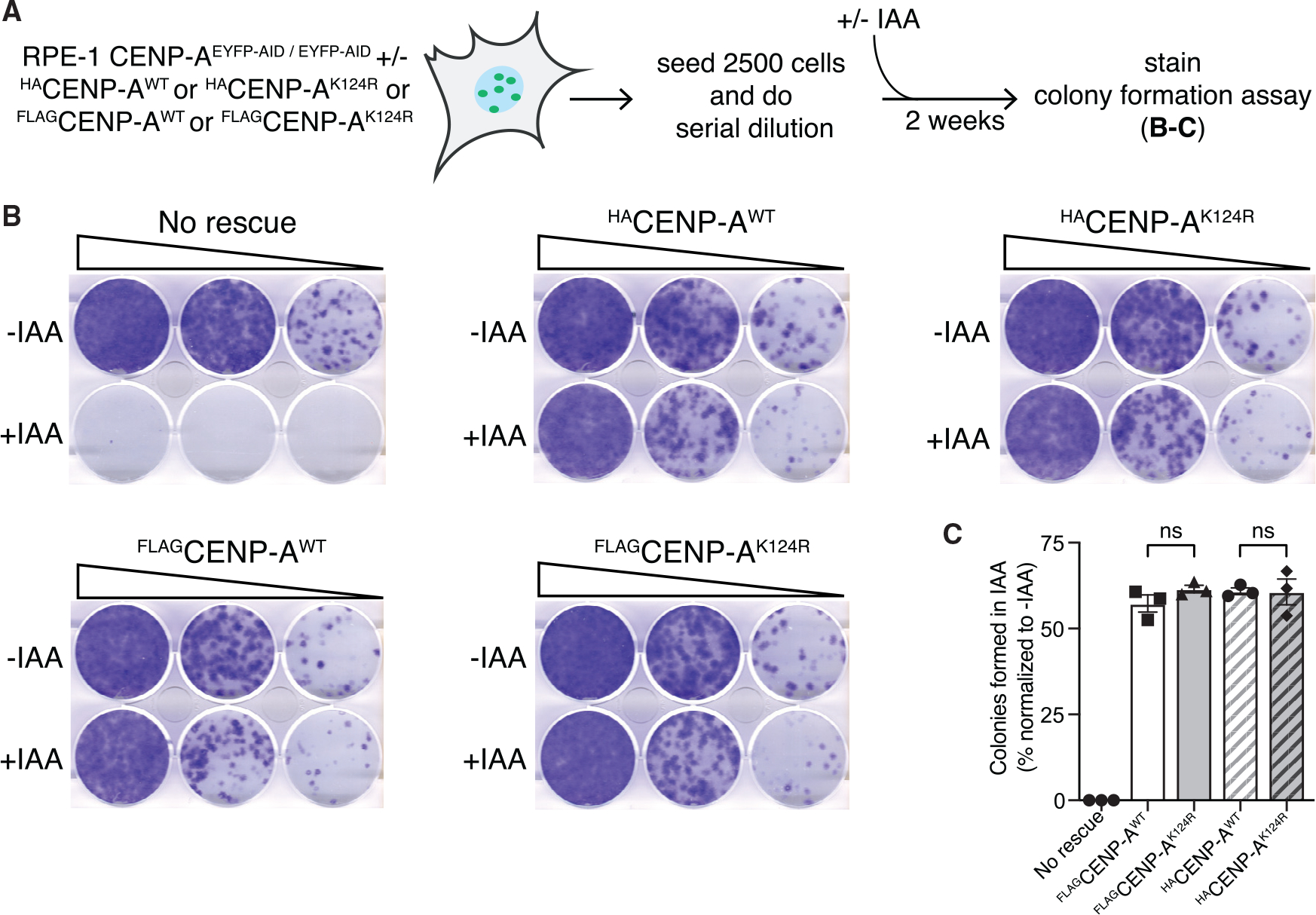
(A) Schematic of the experimental strategy for colony-formation assays.
(B) Representative images of crystal-violet-stained colonies in the indicated cell lines from the colony-formation assays described in (A). Serial dilution of cells grown for 2 weeks without (top wells) or with IAA (bottom wells).
(C) Quantification of the colony-formation assays. Bars represent the percentage of colonies formed after 2 weeks of culture with 500 μM IAA normalized to the untreated. Each dot represents one experiment, and error bars represent SEM. ns: non-significant differences, unpaired t test.
See also Figure S2.
To understand how we arrive at such different results than those reported by Kitagawa and colleagues, we designed an experiment modeled precisely after the strategy they employed (Niikura et al., 2019) using retroviral integration of blasticidin- or puromycin-resistant FLAGCENP-A WT or K124R rescues in RPE-1 cells that carry one knocked-out (−) and one floxed (F) CENP-A allele (Figure S2). The transduction of CENP-A−/F cells with an adenoviral vector expressing the Cre recombinase (Ad-Cre) leads to CENP-A gene depletion and gradual removal of CENP-A protein (Fachinetti et al., 2013). Populations of blasticidin-resistant cells (either CENP-AWT or CENP-AK124R) with no characterization, including no assessment of CENP-A expression levels (as in Niikura et al., 2019), were tested by colony formation assay after Ad-Cre treatment to score for cell viability, generating similar results as the ones reported by Niikura et al. (2019) (Figure S2B), where CENP-AWT appears to rescue much better than CENP-AK124R. However, upon characterization of single clones from these same blasticidin-resistant populations, we found that the majority do not express the rescue constructs (Figures S2C and S2D), providing a direct explanation to the increased lethality observed in the CENP-AK124R population. We note that, in contrast, most of our puromycin-resistant clones do express the rescue constructs (data not shown). Consistent with our initial conclusions in characterized clones (Figures 1 and 2), in individual blasticidin- or puromycin-resistant clones that do express the FLAGCENP-AWT or FLAGCENP-AK124R rescues, we observed that the CENP-AK124R mutant confers cell viability following Ad-Cre treatment to a similar extent as the CENP-AWT rescue construct (Figures S2E and S2F). In summary, the results obtained with the CENP-A gene depletion method match the ones generated using the CENP-A rapid protein degradation (Figure 2).
CENP-AK124R expression from the endogenous locus supports centromere function and DLD-1 cell viability
To test the CENP-AK124R mutant in a system where it is expressed from the endogenous gene locus, thus avoiding expression at levels exceeding the endogenous ones that could, in principle, somehow bypass an essential function of K124 ubiquitylation, we performed CRISPR-Cas9-based biallelic replacement of the CENP-A gene on DLD-1 cells (Figure 3A). DLD-1 cells are pseudodiploid, colorectal cancer cells with a stable karyo-type and have been previously used in studies to generate CRISPR-Cas9 tagging or replacement to define many centromere-kinetochore functions (Dumont et al., 2020; McKinley et al., 2015). The system employed is similar to the one we used previously (Fachinetti et al., 2017). One allele is EGFP-AID tagged for rapid protein degradation, while the other is mutated (K124R) or not (WT). In our current effort, we follow the coding sequence of the second allele with a T2A viral cleavage site and an antibiotic selection marker. The T2A site was chosen because it does not leave any lysine residues in the remaining C-terminal sequence that extends only 26 amino acids beyond the natural CENP-A stop site (Figure 3B). This additional sequence cannot be ubiquitylated and, due to its small size (2.65 kDa), is not expected to alter the ubiquitylation status of the endogenous CENP-A. We obtained several monoclonal lines where biallelic replacement is successful. We performed PCR genotyping and Sanger sequencing of the entire CENP-AK124R open reading frame (ORF), confirming the desired edit with no changes to the designed rescue gene product (Figures 3C, 3D, and S3A). The addition of IAA potently degrades EACENP-A but leaves the rescue proteins intact (Figures 3E and S3B).
Figure 3. Biallelic gene replacement for the expression of lysine-free tagged CENP-A K124R mutant from the endogenous CENP-A locus in DLD-1 cells.
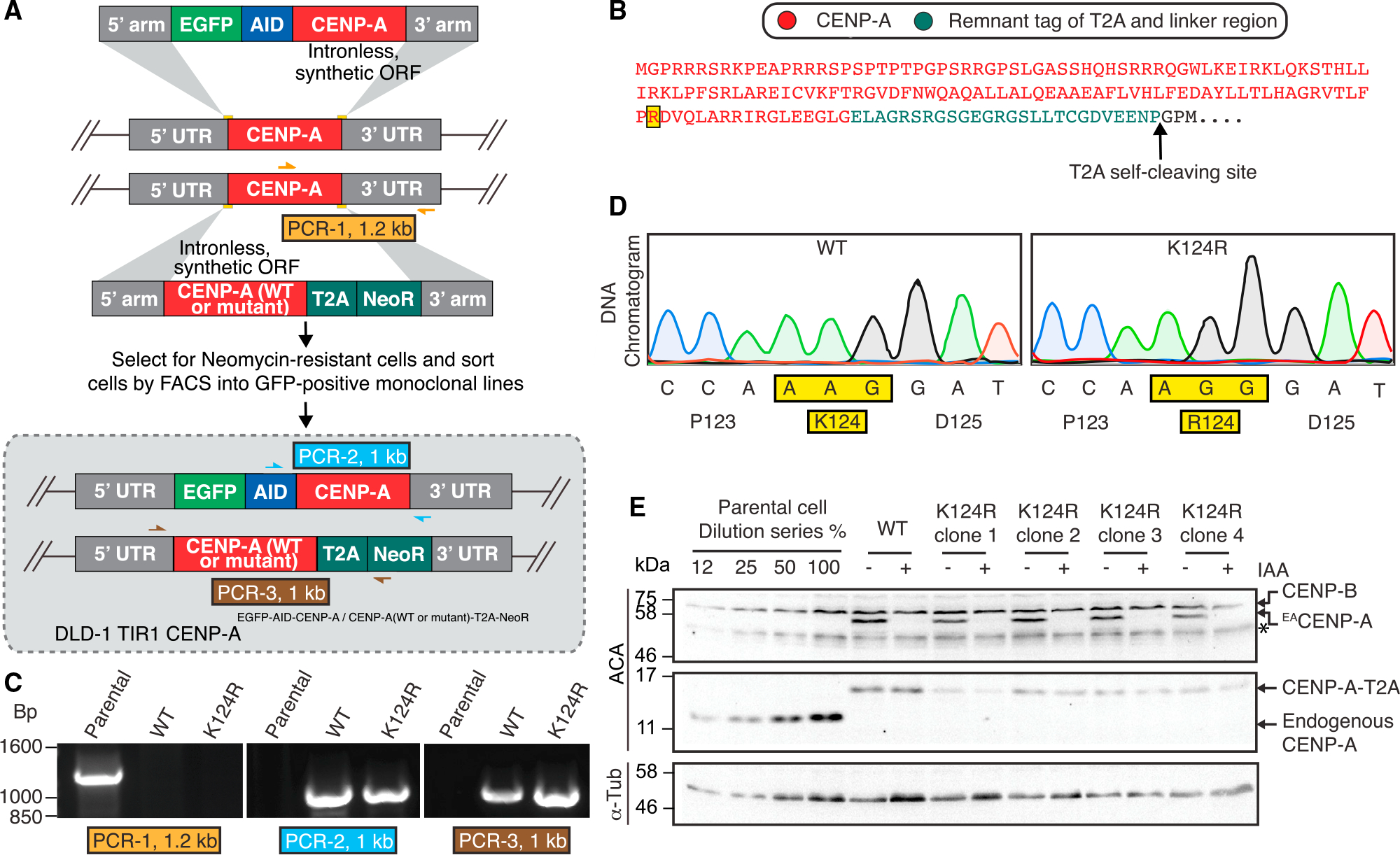
(A) Schematic of CRISPR-Cas9-mediated gene editing in DLD-1 TIR-1 cells. In this biallelic gene replacement approach, one of the endogenous alleles of the CENP-A gene is replaced with EACENP-A and the second allele with CENP-A (WT or K124R) in-frame with T2A-neomycin resistance gene (NeoR). Color-coded arrows indicate primer pairs of each PCR for genotype verification shown in (C). EA, EGFP-AID.
(B) Protein translation of the K124R-edited (highlighted in yellow) CENP-A gene at the endogenous locus with the 26-amino-acid linker and T2A remnant (2.65 kDa). The T2A self-cleavage site is indicated by the arrow. Amino acids in black are lost after cleavage.
(C) Genomic DNA was isolated from the monoclonal lines, and PCR was performed with primer pairs as shown in (A). Positive PCR-1 for parental DLD-1 TIR-1 endogenous CENP-A alleles and no PCR product for either of WT or K124R genome-edited cells. Positive PCR-2 for the endogenous CENP-A allele replaced with EACENP-A. Positive PCR-3 for the other allele of the CENP-A gene replaced with either CENP-AWT-T2A-NeoR or CENP-AK124R-T2A-NeoR.
(D) PCR-3 product from (C) was purified by agarose gel extraction and Sanger sequenced. Raw DNA sequencing data for CENP-AWT-T2A-NeoR or CENP-AK124R-T2A-NeoR (clone 4) alleles showing the mutation.
(E) Immunoblot analysis of whole-cell lysates from the indicated cells with or without 500 μM IAA treatment for 24 h to degrade EACENP-A using an anti-centromere (ACA) and anti-α-tubulin antibodies. Asterisks (*) mark unspecific bands.
See also Figure S3.
CENP-AK124R remains properly localized to the centromere after a short 24-h IAA treatment (Figures 4A, S3C, and S3D). Further, it clearly confers cell viability, similar to CENP-AWT (Figures 4B and S4A), an opposite outcome relative to the mutant α2.2 that fails to interact with HJURP (Bassett et al., 2012), does not target to centromeres, is unstable at the protein level, and thus effectively serves as a null mutation. After removal of EACENP-A with prolonged IAA treatment, CENP-AK124R localized to centromeres in similar amounts as CENP-AWT (10 days) (Figures 4C, 4D, and S4B) and confers long-term viability (14 days) (Figures 4E, 4F, and S4C), to a similar extent as the WT version of the rescue protein.
Figure 4. Lysine-free tagged CENP-AK124R, expressed from the endogenous CENP-A locus, efficiently targets centromeres and confers cell survival and centromere functions.
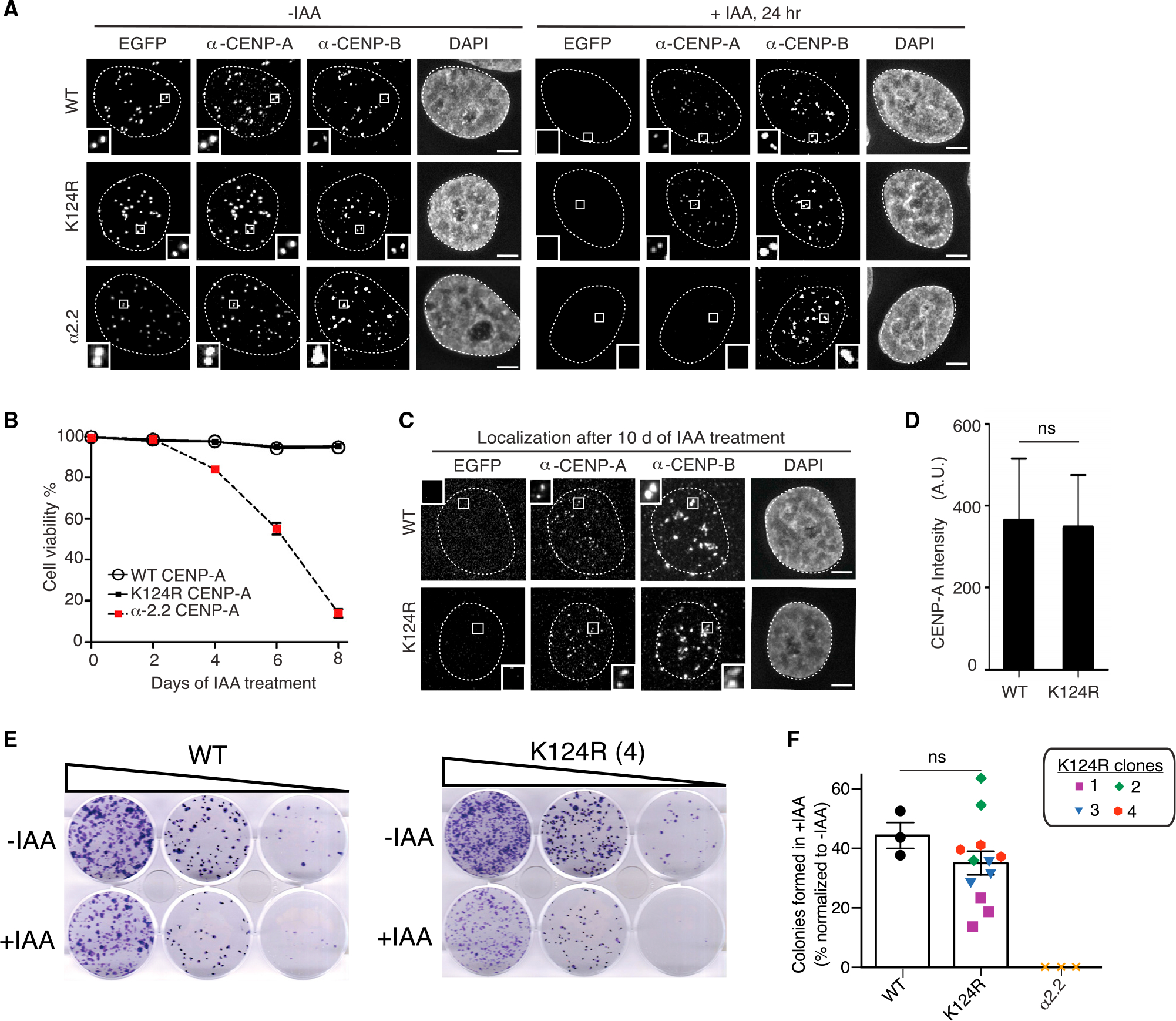
(A) Representative immunofluorescence images of CENP-AWT, CENP-AK124R (clone 4), and CENP-Aα2.2 cells showing localization of EACENP-A and WT or K124R CENP-A-T2A at centromeres. Cells were treated with 500 μM IAA for 24 h to degrade EACENP-A. α-CENP-B was used as a marker to determine centromere position. Nuclei are contoured with white dashed lines. Scale bar, 5 μm.
(B) Quantification of the percentage of viable cells expressing CENP-AWT, CENP-AK124R (clone 4), or α2.2 CENP-A upon treatment with 500 μM IAA for 8 days. Every 2 days, media with 500 μM IAA was changed. Cells were collected every other day and stained with trypan blue. Viability was calculated based on trypan blue uptake. Mean ± SEM from three independent experiments is shown for each time point.
(C) Representative images of viable cells expressing CENP-AWT or CENP-AK124R (clone 4) after 10 days of treatment with 500 μM IAA. CENP-AK124R is still present at endogenous centromeres (marked by CENP-B). Nuclei are contoured with white dashed lines. Scale bar, 5 μm.
(D) Quantitation of centromeric CENP-AWT-T2A and CENP-AK124R-T2A (clone 4) intensity after 10 days of 500 μM IAA treatment. Mean ± 95% confidence interval from three independent experiments is shown (n > 700 centromeres for each replicate). ns: non-significant differences (unpaired t test).
(E) Representative images of a colony formation assay and its quantitation in the indicated cells.
(F) Bars represent the percentage of colonies formed after 2 weeks of culture with IAA, normalized to the untreated. Each dot represents one experiment. The four CENP-AK124R clones are depicted with different symbols and colors, and error bars represent SEM. ns: non-significant differences (unpaired t test).
See also Figures S3 and S4.
CENP-AK124ub is dispensable, with or without large tags
The mechanistic basis of centromere identification and maintenance conferred by CENP-A have mostly emerged from studies using tagged proteins. Pulse labeling (SNAP), fluorescent labeling (EGFP/EYFP), LacI-LacO exogenous genomic site targeting, and AID tagging have all offered critical insights to postulate the standing model of epigenetic centromere specification. In our current effort, we found no support for the most recent explanation provided by Kitagawa and colleagues for why we have not been able to find any survival defects on cells with CENP-A that cannot be ubiquitylated at lysine 124 (K124R mutants). Their explanation is that, in the presence of commonly used tags such as the ones we used in our prior studies (Fachinetti et al., 2017), substitute ubiquitylation site(s) can be generated on CENP-A residue(s) other than K124 (Niikura et al., 2019). Our findings in two different cell backgrounds, with three independent methods that avoid any added lysines and eliminate the possibility of alternative ubiquitylation due to tag size, as demonstrated by the authors themselves (Niikura et al., 2015, 2019), refute these claims. Kitagawa and colleagues reached their conclusions with experiments that include the use of uncharacterized polyclonal cells and transient overexpression of relevant components, as in the in vivo ubiquitylation assays and mass-spectrometry-based analysis (Niikura et al., 2019), thus yielding unreliable results. Further, the report of a single gene replacement experiment lacks data to sufficiently characterize the edited locus (Niikura et al., 2019). A simple explanation for their loss of cell viability is that unintended mutations might have been introduced when the authors generated their CENP-AK124R allele and that these mutations led to a reduction/loss of expression and/or function of their rescue protein. Perhaps related to this notion, we found that CENP-AK124R clones can vary somewhat in expression, with the lowest-expressing clone (clone 1, which expresses the rescue protein at less than half of the WT clone; Figure 3E) exhibiting reduced colony formation (Figures 4F and S4C). These results demonstrate that the levels of CENP-A are critical in colony-formation assays, as markedly reduced CENP-A levels within a clone correlate with a decrease in the abundance of centromere/kinetochore components and an increase in the rate of chromosome mis-segregation (Fachinetti et al., 2013). In addition, we have shown that following retroviral integration and selection, many blasticidin-resistant clones (11 out of 16) do not express the desired constructs, and therefore, they act as null allele (Figure S2C). While the basis for the clone-to-clone variability remains unclear, this particular finding illustrates the experimental importance of careful characterization of clonal populations (crucial to determine the phenotypes from mutational studies) and the folly in drawing conclusions from uncharacterized mixtures of cells. We emphasize that we do not dispute that ubiquitylation of CENP-A is possible, but our observations are in strong opposition to the major claims from Kitagawa and colleagues (Niikura et al., 2015, 2016, 2019) that such modifications of K124 (or a substituting lysine in its absence) are essential for CENP-A nucleosome assembly at centromeres and overall centromere function, large tag or no.
Strategies for assessing the functional relevance of particular PTMs
Great interest has been laid on CENP-A PTMs since they could, in principle, impact upon CENP-A nucleosome targeting and affect centromere identity and function. CENP-A PTM sites reported to date lie outside the CATD, and while some of these residues have been shown to be important for centromere identity and function, the specific roles of some of these PTMs remain elusive and, in some cases, controversial (Bailey et al., 2013; Barra et al., 2019; Fachinetti et al., 2017; Hori et al., 2020; Niikura et al., 2015; Sathyan et al., 2017; Yu et al., 2015; Zeitlin et al., 2001). Strategies to test the importance of a specific PTM typically rely on ectopic expression or gene replacement studies that involve the usage of large tags to assess the localization, function, and dynamics of the modified protein of interest (as in the case of CENP-A). It is always possible that protein tagging may alter important characteristics or functions of the protein under investigation. The methods used here with gene replacement strategies using small tags that lack lysines and confer rapid removal of the endogenous WT protein represent a compelling alternative to this problem and can now clearly be applied at the centromere. This type of approach also supports the use of functional tags in the study of CENP-A, its partners, and centromeric chromatin. More broadly, our approach can be readily adaptable as a general methodology in biology to interrogate candidate sites of PTM in diverse cellular contexts.
Limitations of the study
The study is limited to two human cultured cell backgrounds, DLD-1 (near diploid; colon cancer) and RPE-1 (diploid; non-transformed retinal pigment epithelial). A role for CENP-AK124ub in other cell types or in the context of an animal cannot be excluded.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ben E. Black (blackbe@pennmedicine.upenn.edu).
Materials availability
Plasmids and cell lines generated in this study will be provided upon request to the Lead Contact.
Data and code availability
Data reported in this paper will be shared by the Lead Contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
Cells were cultured at 37°C in a humidified incubator at 5% CO2. The DLD-1 TIR1 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) medium containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. hTERT RPE-1 WT, CENP-AF/+ and CENP-A−/F cells were maintained in DMEM:F12 (GIBCO) medium containing 10% heat inactivated FBS (biosera), 0.123% sodium bicarbonate and 2 mM L-glutamine.
METHOD DETAILS
Constructs
For DLD-1 TIR1 stable cell lines, repair templates with WTCENP-A-T2A-NeoR and K124RCENP-A-T2A-NeoR plasmids were generated. To this end, gBlock gene fragment (IDT) T2A-NeoR was PCR amplified, restriction digested with XhoI and NheI sites and cloned into vectors with WTCENP-A-SNAP-3xHA-P2A-NeoR and K124RCENP-A-SNAP-3xHA-P2A-NeoR (Fachinetti et al., 2017) under the same sites replacing SNAP-3xHA-P2A-NeoR with T2A-NeoR. Retroviral plasmids used in the generation of RPE-1 stable cell lines were generated by cloning the full length of the human CENP-A open reading frame (WT or K124R mutant), tagged with HA or FLAG at the amino terminal, into pBabe-Puro or pBabe-Blast based vectors.
Cell line generation
DLD-1 TIR1 (Holland et al., 2012) stable cell lines were generated by replacing both CENP-A alleles with EGFP-AID-CENP-A (Fachinetti et al., 2017) and CENP-A (WT or K124R)-T2A-NeoR. To obtain biallelic replacement of CENP-A gene locus with these constructs DLD-1 TIR1 cells were cotransfected with 400 ng of each repair template and 100 ng of each sgRNA-Cas9 (Fachinetti et al., 2017) using lipofectamine 2000 (Invitrogen). After 5 days of transfection antibiotic resistance selection was performed with 750 μg/mL G418-S and after 2–3 weeks of selection GFP-positive monoclonal lines were FACS sorted into 96 well plates. EGFP-AIDCENP-A expression was assessed by immunofluorescence microscopy and immunoblot. From DLD-1 TIR1 CENP-A WT or K124R monoclonal lines, genomic DNA was isolated and verified by PCR and Sanger sequencing. hTERT RPE-1 EYFP-AIDCENP-A OsTIR1 (Hoffmann et al., 2016) and hTERT RPE-1 CENP-A−/F (Fachinetti et al., 2013) stable cell lines were generated by retroviral delivery of the HA or FLAG-tagged CENP-A (WT or K124R) transgenes. Briefly, retroviral plasmids (pBabe-Puro or pBabe-Blast based) were cotransfected with the VSV-G pseudotyping plasmid into 293-GP cells. The resulting retroviral supernatant was collected after 48 hours, mixed with 8 μg/ml polybrene and added to the RPE-1 cells in culture. Stable integration was selected with 5 μg/mL puromycin for 7 days or 10 μg/mL blasticidin for 10 days. Single clones were isolated and characterized. For the hTERT RPE-1 EYFP-AIDCENP-A OsTIR1 experiments, three clones with similar expression levels for each construct were pooled.
Immunoblot analysis
Whole cell lysates were collected from the different cell lines, treated or not with 500 μM IAA (I5148, Sigma) dissolved in ddH2O. For DLD-1 cells, proteins were separated by 15% SDS-PAGE and transferred onto PVDF membranes (GE healthcare). Immunoblot analysis was performed by blocking the membrane with 5% skim milk and probing with anti-ACA (Antibodies Incorporated, 2 μg/mL) and anti-α-tubulin (Clone DM1A, 1:5000) antibodies. Three independent gels were probed to quantify CENP-A-T2A levels using Fiji software (Schindelin et al., 2012). Proteins from RPE-1 whole cell lysates were separated by SDS-PAGE using 12% TGX gels (BioRad) or hand casted 15% gels, and transferred onto nitrocellulose membranes using the Trans-Blot Turbo transfer system (BioRad). Immunoblot analysis was performed by blocking the membrane with 5% skim milk and probing overnight at 4°C with anti-CENP-A (1:1000, Cell Signaling #2186), anti-Vinculin (1:2000, Sigma #V9264) and anti-GAPDH (1:5000, Cell Signaling #2118) antibodies.
Cell viability assay
Cell viability assay was performed in triplicate on DLD-1 TIR1 WT or K124R cell lines by seeding at 1.5 × 105 cells/well with 500 μM IAA. Cells were collected every other day at indicated time points and then Trypan Blue staining (Corning) was performed. The percentage of living cells out of total cells was calculated on a hemocytometer based on Trypan Blue uptake. After 8-day IAA treatment, surviving colonies were grown on poly-D-lysine-coated coverslips in a 6-well plate with 500 μM IAA for 2 days and subjected to immunofluorescence microscopy.
Colony formation assay
For the colony formation assays, 2500 cells were plated on 6-well plates and two 1:5 serial dilutions were performed. After 12 or 14 days, colonies were fixed for 30 minutes in methanol, washed with PBS and then stained for 15 minutes using a 1% crystal violet in 20% ethanol stain before being thoroughly washed with PBS. The plates were air-dried and scanned, and then colonies were manually counted.
Indirect immunofluorescence
For experiments involving DLD-1 TIR1 stable cell lines, cells were fixed with 4% formaldehyde in PBS for 5 minutes and quenched with 100 mM Tris pH 7.5 for 5 minutes. Cell were washed three times with 0.1% Tween in PBS each for 5 minutes and permeabilized with 0.5% Triton X-100 in PBS for 5 minutes. Permeabilized cells were washed with 0.1% Tween in PBS for 5 minutes prior to blocking in blocking buffer (2% FBS, 2% BSA, and 0.1% Tween in PBS). Primary antibody incubation with anti-CENP-A (1 μg/mL of mouse monoclonal antibody ADI-KAM-CC006-E, Enzo) and anti-CENP-B (200 μg/ml rabbit polyclonal sc-22788, Santa Cruz Biotechnology) was performed in the blocking buffer for 1 hour at room temperature. Cells were washed three times with 0.1% Tween in PBS for 5 minutes, followed by Cy3-conjugated donkey anti-mouse and Cy5-conjugated donkey anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories) incubation at 1:200 dilution. Cells were washed with 0.1% Tween in PBS for 5 minutes followed by DAPI staining and mounting with Vectashield mounting medium (Vector Laboratories). For experiments involving hTERT RPE-1 stable cell lines, cells were fixed with 4% formaldehyde in PBS for 10 minutes, then washed and permeabilized with 0.1% Triton X-100 in PBS, and blocked for 30 minutes at room temperature in blocking buffer (0.2 M glycine, 2.5% FBS and 0.1% Triton X-100). Primary antibody incubation was performed for 2 hours at room temperature in blocking buffer, with rabbit anti-GFP (PABG1, ChromoTek) 1:200, guinea pig anti-CENP-C (PD030, MBL) 1:500, and mouse anti-HA (A-M-M#07, Recombinant Antibody Platform (TAb-IP), Institut Curie) 1:200 or mouse anti-FLAG (F3165, Sigma) 1:1000 antibodies. Cells were washed three times with 0.1% Triton X-100 in PBS before incubation for 1 hour at room temperature in blocking buffer with donkey anti-mouse Alexa Fluor® 488, donkey anti-rabbit Cy3 and donkey anti-guinea pig Alexa Fluor® 647 secondary antibodies (Jackson ImmunoResearch Laboratories, 1:500). Cells were washed three times with 0.1% Triton X-100 in PBS before DAPI staining (1 μg/mL in PBS) for 10 minutes and mounting with ProLong Gold Antifade Mountant (#P36934, Invitrogen).
Image acquisition and quantification
For experiments involving DLD-1 TIR1 cells, images were captured as 0.2 μm z sections at room temperature on an inverted fluorescence microscope (Leica DMI6000B) at 100× 1.4 NA oil immersion objective lens, equipped with a charge-coupled device camera (Hamamatsu Photonics ORCA AG). Images were deconvolved using LAS-X software (Leica) and max-projected for fluorescence intensity measurements. The mean fluorescence intensity at centromeres was measured using an ImageJ macro, CRaQ v1.12, under default parameters. Plots were generated from three independent immunofluorescence experiments. For experiments involving hTERT RPE-1 cell lines imaging was performed on a DeltaVision Core system (Applied Precision) consisting of an Olympus IX71 inverted microscope equipped with a CoolSNAPHQ2 camera (Photometrics). Images were captured as 0.2 μm z sections at room temperature with a 100× 1.4 NA oil immersion objective lens (Olympus), deconvolved and 3D maximum intensity projected using DeltaVision’s softworx software. The integrated fluorescence intensity at centromeres was measured using Fiji (Giunta et al., 2021).
Ad-Cre transduction
4 ×104 hTERT RPE-1 CENP-AF/+, CENP-A−/F +/− rescue constructs cells were seeded in 12 well plates 24 hours prior of transduction. The cells were washed three times in DMEM:F12 medium containing 2% FBS and transduction with variable amount of CsCl purified Ad-Cre (1x, 2x and 4x) was carried out in 400 μL final volume of 2% FBS containing media for 4 hours. The cells were then washed extensively in medium containing 10% FBS, and left to recover for 48 hours before seeding colony formation assays.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details, which include the value of n (where n represents the number of centromeres measured) can be found in the figure legends. All statistical tests were performed using GraphPad Prism 9.0 for Mac (GraphPad Software, San Diego, California USA, https://www.graphpad.com:443/).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit polyclonal anti-GFP | ChromoTek | Cat# PABG1-20; RRID: AB_2749857 |
| Guinea pig polyclonal anti-CENP-C | MBL International | Cat# PD030; RRID: AB_10693556 |
| Mouse monoclonal anti-HA (clone 12CA5) | Recombinant Antibody Platform (TAb-IP), Institut Curie | Cat# A-M-M#07 |
| Mouse monoclonal anti-FLAG (clone M2) | Sigma-Aldrich | Cat# F3165; RRID: AB_259529 |
| Rabbit polyclonal anti-CENP-A | Cell Signaling Technology | Cat# 2186; RRID: AB_10828491 |
| Human polyclonal anti-centromere antisera (ACA) | Antibodies Incorporated | Cat# 15-235-0001; RRID: AB_2797146 |
| Mouse monoclonal anti-CENP-A (clone 3-19) | Enzo Life Sciences | ADI-KAM-CC006-E RRID: AB_2038993 |
| Rabbit polyclonal anti-CENP-B | Santa Cruz Biotechnology | Cat# sc-22788, RRID: AB_2078775 |
| Mouse monoclonal anti-αTubulin (clone DM1A) | Sigma-Aldrich | Cat# T9026; RRID: AB_477593 |
| Rabbit monoclonal anti-GAPDH (clone14C10) | Cell Signaling Technology | Cat# 2118; RRID: AB_561053 |
| Mouse monoclonal anti-Vinculin (clone hVIN-1) | Sigma-Aldrich | Cat# V9264; RRID: AB_10603627 |
| Donkey polyclonal anti-mouse - Cy3 | Jackson ImmunoResearch Laboratories | Cat# 715-165-151; RRID: AB_2315777 |
| Donkey polyclonal anti-rabbit - Cy5 | Jackson ImmunoResearch Laboratories | Cat# 711-175-152; RRID:AB_2340607 |
| Donkey polyclonal anti-mouse - Alexa Fluor® 488 | Jackson ImmunoResearch Laboratories | Cat# 715-545-150; RRID: AB_2340846 |
| Donkey polyclonal anti-rabbit - Cy3 | Jackson ImmunoResearch Laboratories | Cat# 711-165-152; RRID: AB_2307443 |
| Donkey polyclonal anti-guinea pig - Alexa Fluor® 647 | Jackson ImmunoResearch Laboratories | Cat# 706-605-148; RRID: AB_2340476 |
| Donkey polyclonal anti-human - HRP | Jackson ImmunoResearch Laboratories | Cat# 709-035-149; RRID: AB_2340495 |
| Sheep anti-mouse - HRP | GE Healthcare | Cat# NA931l; RRID: AB_772212 |
| Donkey anti-rabbit - HRP | GE Healthcare | Cat# NA934, RRID: AB_772206 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| RV-FLAG-CENP-A(WT)-Puro | This paper | N/A |
| RV-FLAG-CENP-A(K124R)-Puro | This paper | N/A |
| RV-FLAG-CENP-A(WT)-Blast | This paper | N/A |
| RV-FLAG-CENP-A(K124R)-Blast | This paper | N/A |
| RV-HA-CENP-A(WT)-Puro | This paper | N/A |
| RV-HA-CENP-A(K124R)-Puro | This paper | N/A |
| AdCre | Fachinetti et al., 2013 | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Indole-3-acetic acid (IAA) sodium salt | Sigma-Aldrich | Cat# I5148; CAS: 6505-45-9 |
| Polybrene® | Santa Cruz Biotechnology | Cat# sc-134220; CAS 28728-55-4 |
| VectaShield | Vector Laboratories | Cat# H-1000; RRID: AB_2336789 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: hTERT RPE-1 EYFP-AID/EYFP-AIDCENP-A OsTIR1 | Hoffmann et al., 2016 | N/A |
| Human: hTERT RPE-1 CENP-A−/F | Fachinetti et al., 2013 | N/A |
| Human: hTERT RPE-1 CENP-A+/F | Fachinetti et al., 2013 | N/A |
| Human: DLD-1 Flp-In T-Rex | Holland et al., 2012 | N/A |
| Human: DLD-1 TIR1 CENP-AEGFP-AID-CENP-A/CENP-A(a-2.2)-SNAP-3xHA-P2A-NeoR | Fachinetti et al., 2017 | N/A |
| Human: DLD-1 TIR1 CENP-AEGFP-AID-CENP-A/CENP-A(WT)-T2A-NeoR | This paper | N/A |
| Human: DLD-1 TIR1 CENP-AEGFP-AID-CENP-A/CENP-A(K124R)-T2A-NeoR | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pBabe-FLAG-CENP-A(WT)-PuroR | This paper | N/A |
| pBabe-FLAG-CENP-A(K124R)-PuroR | This paper | N/A |
| pBabe-FLAG-CENP-A(WT)-BlastR | This paper | N/A |
| pBabe-FLAG-CENP-A(K124R)-BlastR | This paper | N/A |
| pBabe-HA-CENP-A(WT)-PuroR | This paper | N/A |
| pBabe-HA-CENP-A(K124R)-PuroR | This paper | N/A |
| pUC19-EGFP-AID-CENP-A | Fachinetti et al., 2017 | N/A |
| pUC19-CENP-A(WT)T2A-NeoR | This Paper | N/A |
| pUC19-CENP-A(K124R)-T2A-NeoR | This Paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Fiji | Schindelin et al., 2012 | https://imagej.net/software/fiji |
| CRaQ v1.12 | Bodor et al., 2012 | http://facilities.igc.gulbenkian.pt/microscopy/macros/CRaQ_v1.12.ijm |
| Centromere intensity | Giunta et al., 2021 | Available upon request |
| GraphPad Prism | https://www.graphpad.com:443/ | RRID:SCR_002798 |
Highlights.
Preventing CENP-A ubiquitylation on K124 does not interfere with cell viability
Lysine-free tagged CENP-AK124R supports long-term centromere function
Common functional tags do not mask an essential function for K124 modification
General strategies presented for interrogating the function of specific PTM sites
ACKNOWLEDGMENTS
We thank D.W. Cleveland (UCSD) for suggestions. We thank the Recombinant Antibody Platform (TAb-IP), Institut Curie, for the HA antibody. D.F. receives salary support from the Centre National de la Recherche Scientifique (CNRS, France). This work was supported by the National Institutes of Health (NIH, USA) grant R35GM130302 (B.E.B.) and by the Agence Nationale de la Recherche (ANR, France) grants ANR-18-ERC1-0004-01 (D.F.), ANR-19-CE12-0022 (D.F.), and Labex «Cell(n)Scale » with the references ANR-10-LABX-0038 and ANR-10-IDEX-0001-02 (D.F.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109924.
REFERENCES
- Allu PK, Dawicki-McKenna JM, Van Eeuwen T, Slavin M, Braitbard M, Xu C, Kalisman N, Murakami K, and Black BE (2019). Structure of the human core centromeric nucleosome complex. Curr. Biol. 29, 2625–2639.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade D, Pauleau A-L, Wendler A, and Erhardt S (2014). The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev. Cell 28, 508–519. [DOI] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai P-J, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. (2013). Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc. Natl. Acad. Sci. USA 110, 11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, and Foltz DR (2011). HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194, 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra V, Logsdon GA, Scelfo A, Hoffmann S, Hervé S, Aslanian A, Nechemia-Arbely Y, Cleveland DW, Black BE, and Fachinetti D (2019). Phosphorylation of CENP-A on serine 7 does not control centromere function. Nat. Commun. 10, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, and Black BE (2012). HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell 22, 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, and Cleveland DW (2011). Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 144, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr., and Cleveland DW (2004). Structural determinants for generating centromeric chromatin. Nature 430, 578–582. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Rodríguez MG, Moreno N, and Jansen LET (2012). Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Curr. Protoc. Cell Biol. 55, 8.8.1–8.8.34. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Valente LP, Mata JF, Black BE, and Jansen LET (2013). Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell 24, 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, and Mellone BG (2014). CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 204, 313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, et al. (1997). Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum. Mol. Genet. 6, 1195–1204. [DOI] [PubMed] [Google Scholar]
- Dumont M, and Fachinetti D (2020). Centromere strength: just a sense of proportion. Mol. Cell. Oncol. 7, 1742063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Gamba R, Gestraud P, Klaasen S, Worrall JT, De Vries SG, Boudreau V, Salinas-Luypaert C, Maddox PS, Lens SM, et al. (2020). Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 39, e102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, and Almouzni-Pettinotti G (2009). HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, and Rothfield N (1985). Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LET, and Cleveland DW (2013). A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, and Cleveland DW (2015). DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell 33, 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Logsdon GA, Abdullah A, Selzer EB, Cleveland DW, and Black BE (2017). CENP-A Modifications on Ser68 and Lys124 are dispensable for establishment, maintenance, and long-term function of human centromeres. Dev. Cell 40, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. (2015). Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, and Cleveland DW (2009). Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137, 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR 3rd, and Cleveland DW (2006). The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469. [DOI] [PubMed] [Google Scholar]
- French BT, and Straight AF (2019). CDK phosphorylation of Xenopus laevis M18BP1 promotes its metaphase centromere localization. EMBO J. 38, e100093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S, Hervé S, White RR, Wilhelm T, Dumont M, Scelfo A, Gamba R, Wong CK, Rancati G, Smogorzewska A, et al. (2021). CENP-A chromatin prevents replication stress at centromeres to avoid structural aneuploidy. Proc. Natl. Acad. Sci. USA 118, e2015634118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, and Malik HS (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Hervé S, Cleveland DW, and Fachinetti D (2016). CENP-A is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep. 17, 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, Izquierdo HM, Gamba R, Chardon F, Dumont M, Keizer V, Hervé S, McNulty SM, Sullivan BA, Manel N, and Fachinetti D (2020). A genetic memory initiates the epigenetic loop necessary to preserve centromere position. EMBO J. 39, e105505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, and Cleveland DW (2012). Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 109, E3350–E3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang W-H, Takeuchi K, and Fukagawa T (2013). The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200, 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang W-H, Toyoda A, Misu S, Monma N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, et al. (2014). Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev. Cell 29, 740–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Cao J, Nishimura K, Ariyoshi M, Arimura Y, Kurumizaka H, and Fukagawa T (2020). Essentiality of CENP-A depends on its binding mode to HJURP. Cell Rep. 33, 108388. [DOI] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. (2011). Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, and Black BE (2017). Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 27, 2365–2373.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, and Cleveland DW (2007). Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kixmoeller K, Allu PK, and Black BE (2020). The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol. 10, 200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebelt F, Jansen NS, Kumar S, Gracheva E, Claessens LA, Verlaan-de Vries M, Willemstein E, and Vertegaal ACO (2019). The poly-SUMO2/3 protease SENP6 enables assembly of the constitutive centromere-associated network by group deSUMOylation. Nat. Commun. 10, 3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang S-P, Wang Z, Chinwalla AT, Minx P, et al. (2011). Comparative and demographic analysis of orang-utan genomes. Nature 469, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, and Black BE (2015). Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 208, 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Gambogi CW, Liskovykh MA, Barrey EJ, Larionov V, Miga KH, Heun P, and Black BE (2019). Human artificial chromosomes that bypass centromeric DNA. Cell 178, 624–639.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, and Cheeseman IM (2014). Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell 158, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, and Cheeseman IM (2015). The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol. Cell 60, 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, and Heun P (2011). Drosophila CENH3 is sufficient for centromere formation. Science 334, 686–690. [DOI] [PubMed] [Google Scholar]
- Mitra S, Srinivasan B, and Jansen LET (2020a). Stable inheritance of CENP-A chromatin: Inner strength versus dynamic control. J. Cell Biol. 219, e202005099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Bodor DL, David AF, Abdul-Zani I, Mata JF, Neumann B, Reither S, Tischer C, and Jansen LET (2020b). Genetic screening identifies a SUMO protease dynamically maintaining centromeric chromatin. Nat. Commun. 11, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechemia-Arbely Y, Fachinetti D, Miga KH, Sekulic N, Soni GV, Kim DH, Wong AK, Lee AY, Nguyen K, Dekker C, et al. (2017). Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J. Cell Biol. 216, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechemia-Arbely Y, Miga KH, Shoshani O, Aslanian A, McMahon MA, Lee AY, Fachinetti D, Yates JR 3rd, Ren B, and Cleveland DW (2019). DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nat. Cell Biol. 21, 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, and Kitagawa K (2015). CENP-A K124 ubiquitylation is required for CENP-A deposition at the centromere. Dev. Cell 32, 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, and Kitagawa K (2016). CENP-A ubiquitylation is inherited through dimerization between cell divisions. Cell Rep. 15, 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, Fang L, and Kitagawa K (2019). CENP-A ubiquitylation is indispensable to cell viability. Dev. Cell 50, 683–689.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR 3rd, Desai A, and Fukagawa T (2006). The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8, 446–457. [DOI] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, and Masumoto H (2007). CENP-B controls centromere formation depending on the chromatin context. Cell 131, 1287–1300. [DOI] [PubMed] [Google Scholar]
- Otake K, Ohzeki JI, Shono N, Kugou K, Okazaki K, Nagase T, Yamakawa H, Kouprina N, Larionov V, Kimura H, et al. (2020). CENP-B creates alternative epigenetic chromatin states permissive for CENP-A or heterochromatin assembly. J. Cell Sci. 133, jcs243303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, and Margolis RL (1985). Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol. Cell. Biol. 5, 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan KM, Fachinetti D, and Foltz DR (2017). α-amino trimethylation of CENP-A by NRMT is required for full recruitment of the centromere. Nat. Commun. 8, 14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, and Willard HF (2001). Genomic and genetic definition of a functional human centromere. Science 294, 109–115. [DOI] [PubMed] [Google Scholar]
- Shang W-H, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y, et al. (2016). Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat. Commun. 7, 13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MCC, Bodor DL, Stellfox ME, Martins NMC, Hochegger H, Foltz DR, and Jansen LET (2012). Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 22, 52–63. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Zasadzińska E, and Foltz DR (2018). Posttranslational mechanisms controlling centromere function and assembly. Curr. Opin. Cell Biol. 52, 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic A, Guo LY, Mata JF, Bodor DL, Cao X-J, Bailey AO, Shabanowitz J, Hunt DF, Garcia BA, Black BE, and Jansen LET (2017). A dual inhibitory mechanism sufficient to maintain cell cycle restricted CENP-A assembly. Mol. Cell 65, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. ; Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team (2009). Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, et al. (2015). Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev. Cell 32, 68–81. [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Barber CM, Allis CD, and Sullivan KF (2001). Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci. 114, 653–661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the Lead Contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


