Abstract
Quantitative traits analyzed in Genome-Wide Association Studies (GWAS) are often nonnormally distributed. For such traits, association tests based on standard linear regression are subject to reduced power and inflated type I error in finite samples. Applying the rank-based inverse normal transformation (INT) to nonnormally distributed traits has become common practice in GWAS. However, the different variations on INT-based association testing have not been formally defined, and guidance is lacking on when to use which approach. In this paper, we formally define and systematically compare the direct (D-INT) and indirect (I-INT) INT-based association tests. We discuss their assumptions, underlying generative models, and connections. We demonstrate that the relative powers of D-INT and I-INT depend on the underlying data generating process. Since neither approach is uniformly most powerful, we combine them into an adaptive omnibus test (O-INT). O-INT is robust to model misspecification, protects the type I error, and is well powered against a wide range of nonnormally distributed traits. Extensive simulations were conducted to examine the finite sample operating characteristics of these tests. Our results demonstrate that, for nonnormally distributed traits, INT-based tests outperform the standard untransformed association test, both in terms of power and type I error rate control. We apply the proposed methods to GWAS of spirometry traits in the UK Biobank. O-INT has been implemented in the R package RNOmni, which is available on CRAN.
Keywords: direct and indirect rank-based inverse normal transformation, nonnormality, omnibus test, quantitative traits, transformation, type I error rate
1 |. INTRODUCTION
In Genome-Wide Association Studies (GWAS) of continuous (quantitative) traits, the covariate-adjusted genetic effect is typically estimated by linear regression using ordinary least squares (OLS). When the residual distribution is normal, the OLS estimator is normally distributed, consistent, and efficient (Rawlings et al., 1998). However, for many complex traits, including spirometry measurements, the residual distribution is markedly nonnormal. An example is peak expiratory flow (PEF), whose residual distribution is skewed and asymmetric even when the outcome is log transformed. When the residual distribution is nonnormal, but has mean zero and finite variance, the OLS estimator remains consistent and asymptotically normal (Cameron and Trivedi, 2005). However, the discrepancy between the asymptotic and finite-sample distributions of the test statistic makes association tests based on the OLS estimator sensitive to the underlying residual distribution (Rawlings et al., 1998). Due to slower convergence of the sampling distribution in the tails, excessive sample sizes n ≫ 105 may be required to achieve nominal control of the type I error at the genome-wide significance threshold of α = 5 × 10−8. Moreover, even if the sample is sufficiently sized to protect the type I error, the OLS estimator is no longer efficient when the residual distribution is nonnormal (Serfling, 1980). Consequently, OLS-based association tests may lack power for detecting true effects. These limitations of standard association tests in finite samples (failure to control the type I error and poor power) are highlighted in our simulation studies.
The rank-based inverse normal transformation (INT) is commonly applied during GWAS of nonnormally distributed traits. INT is a nonparametric mapping that replaces sample quantiles by quantiles from the standard normal distribution. After INT, the marginal distribution of any continuous outcome is asymptotically normal. INT has the effect of symmetrizing and concentrating the residual distribution around zero. Based on the Edgeworth expansion (Lehmann, 1999), convergence of the OLS estimator’s sampling distribution is accelerated when the residual distribution is more nearly normal. Heuristically, INT improves the operating characteristics of standard association testing by increasing residual normality, which in turn allows the sampling distribution of the test statistic to converge more quickly.
We classify INT-based tests into direct and indirect methods. In the direct method (D-INT), INT is applied directly to the phenotype, and the INT-transformed phenotype is regressed on genotype and covariates. Covariates may include age, sex, and adjustments for population structure, such as genetic ancestry principal components (PCs). D-INT has been applied to GWAS of BMI (Scuteri et al., 2007), circulating lipids (Barber et al., 2010), polysomnography signals (Cade et al., 2016), and many quantitative traits in the UK Biobank (Abbott et al., 2017). In the indirect method (I-INT), the phenotype is first regressed on covariates to obtain residuals, then the INT-transformed phenotypic residuals are regressed on genotype, with or without secondary adjustment for population structure. I-INT has been applied to GWAS of gene expression (Emilsson et al., 2008; Consortium et al., 2017), serum metabolites (Kettunen et al., 2012), and spirometry measurements (Repapi et al., 2010). However, the relative performance of D-INT versus I-INT has not been studied in detail. For the nonnormal quantitative traits encountered in practice, it is unclear which of the these methods will more robustly control the type I error, and which will provide better power.
As discussed by Beasley and colleagues (Beasley et al., 2009), the question of whether INT-based methods have desirable operating characteristics in the GWAS context has not been critically evaluated. INT of the outcome in a regression model does not guarantee correct model specification. This is because standard linear regression, considered parametrically, requires normality of the residual distribution, not of the marginal distribution of the outcome (Rawlings et al., 1998). Here we systematically study the direct and indirect INT-based association tests and provide recommendations on how to apply the INT in practice. We begin by formally defining D-INT and I-INT, studying their underlying assumptions and connections. We demonstrate that if the observed trait is generated by a nonlinear, rank-preserving transformation of a latent normal trait, then INT provides an approximate inverse of the generative transformation, under the null hypothesis and supposing the covariate effects are small. Moreover, if the mean of the observed trait is linear in covariates but the residual distribution is nonnormal, then I-INT is asymptotically exact. Our derivation of I-INT agrees with recent work recommending double adjustment for covariates during INT-based association testing (Sofar et al., 2019).
Through extensive simulations covering the types of residual nonnormally often encountered in practice, we compare D-INT and I-INT with the standard untransformed association test (UAT). We find that INT-based tests robustly control the type I error and dominate the UAT in terms of power. However, neither D-INT nor I-INT was uniformly most powerful, and their relative performance depended on the underlying data generating mechanism. Since this is seldom known in practice, we next propose an adaptive omnibus test (O-INT) that synthesizes D-INT and I-INT. O-INT robustly controls the type I error, and across traits is nearly as powerful as the more effective of the component methods. We have implemented all candidate INT-based tests (D-INT, I-INT, and O-INT) in the R package RNOmni, which is available on CRAN.
We applied the UAT and the INT-based association tests to GWAS of spirometry traits in the UK Biobank (Sudlow et al., 2015). To demonstrate the power advantage provided by O-INT, we compare the results from the overall analysis (n = 292K) with those from a subgroup analysis (n = 29K) among asthmatics. All associations identified by O-INT in the asthmatic subgroup were declared significant by UAT in the overall analysis. Hence UAT and O-INT tests agree as to the importance of these loci. However, the more-powerful O-INT test was able to detect them using only a fraction (9.7%) of the sample. In both the asthmatic subgroup and the overall analysis, O-INT realized empirical efficiency and discovery gains over the UAT.
The remainder of this paper is structured as follows: In the methods section, we present all candidate association tests, theoretically study D-INT and I-INT, and propose the INT-based omnibus test (O-INT). In the simulation studies, we present evidence that INT-based association tests robustly control the type I error, whereas the UAT often does not. We demonstrate that INT-based association tests dominate the UAT in terms of power and show that O-INT is an effective compromise between D-INT and I-INT. In the data application, we compare the performance of all candidate association tests for GWAS of spirometry traits from the UK Biobank. We conclude with a discussion of the implications of our findings for quantitative trait GWAS.
2 |. STATISTICAL METHODS
2.1 |. Setting
For each of n independent subjects, the following data are observed: a continuous (quantitative) phenotype Yi, genotype gi at the locus of interest, and a p × 1 vector xi = (xi,1,…, xi,p) of covariates. In our data application, the phenotype Yi is a spirometry measurement, while the covariates include an intercept, age, sex, and genetic PCs. Let y = (Y1,…, Yn) denote the n × 1 sample phenotype vector, g the n × 1 genotype vector, and X the n × p covariate design matrix.
2.2 |. Untransformed association test
The UAT is derived from the normal linear model:
| (1) |
where ϵ ~ N(0, σ2I) is an n × 1 residual vector, βG is the genetic effect, and βX is the covariate effect. Define the error projection PX = I − X(X′X)−1X′, and the phenotypic residual eY = PXy, which is the residual after regressing y on X, or the projection of y onto the orthogonal complement of the column space of X. The efficient score for βG is , and the score statistic assessing H0 : βG = 0 is
| (2) |
which follows a distribution. Under H0, an unbiased estimate of the residual variance is given by . The Wald statistic for assessing H0 : βG = 0 takes the same form as (2), save the residual variance is estimated as , where and .
If the normal residual assumption is relaxed to allow for an arbitrary distribution with mean zero and finite variance, then TU still follows an asymptotic distribution. Although (2) is eventually valid for any continuous trait with constant residual variance, when the residual distribution exhibits excess skew or kurtosis, the sample size required for valid inference at α = 5 × 10−8 may become impractically large. Moreover, as we will show, even in samples of sufficient size for valid inference, the UAT is generally less powerful than INT-based tests.
2.3 |. Rank-based inverse normal transformation
INT is a nonparametric mapping applicable to observations from any absolutely continuous distribution. The process may be decomposed into two steps. In the first, the observations are replaced by their fractional ranks. This is equivalent to transforming the observations by their empirical cumulative distribution function (ECDF). If W is any continuous random variable with CDF FW, then the transformed random variable U = FW(W) is uniformly distributed (Casella and Berger, 2002). Since the empirical process converges uniformly to the CDF FW, in independent and identically distributed samples of sufficient size is uniformly distributed (van der Vaart, 1998). After transformation by , the observations reside on the probability scale. In the next step, these probabilities are mapped to Z-scores using the probit function Φ−1. If U is uniformly distributed, then Z = Φ−1(U) follows the standard normal distribution. Consequently, in large samples, , regardless of the initial distribution FW.
In practice, an offset is introduced to ensure that all fractional ranks are strictly between zero and one, which in turn guarantees that all Z-scores are finite. Suppose that Wi is observed for each of n independent subjects. The modified INT is:
| (3) |
Hereafter we adopt the conventional Blom offset of c = 3/8 (Beasley et al., 2009). Since other choices for c lead to Z-scores that are nearly linear transformations of one another, the choice of offset is considered immaterial.
2.4 |. Direct inverse normal transformation (D-INT)
In direct INT (D-INT), the INT-transformed phenotype is regressed on genotype and covariates according to the association model:
| (4) |
where z = INT(y) is the INT-transformed phenotype, and . Model (4) is immediately comparable with model (1), the only difference being replacement of y by z. Thus, the D-INT score statistic for assessing H0 : βG = 0 is:
| (5) |
A p-value is assigned with reference to the distribution. The score statistic estimates the residual variance as , whereas the Wald statistic estimates the residual variance as .
D-INT is adapted for data generating processes (DGPs) of the form:
| (6) |
where , and h(·) is a rank-preserving transformation. An example is the log-normal phenotype, for which h(t) = exp(t). When h(·) is non-linear, the regression function E(Yi|xi, gi) is non-linear in its parameters, and the residuals have non-additive effects. However, there exists a transformed scale on which the mean model is linear and has additive normal residuals, namely: h−1(y) = XβX + gβG + ϵD. Thus, under H0 : βG = 0, the efficient score for βG is . Since h−1(·) is seldom known, D-INT makes the approximation , where is the marginal ECDF of the phenotype Yi.
To justify this approximation, observe that under model (6), the conditional distribution of the transformed-scale residual is . The marginal and conditional CDFs of Yi are related via:
| (7) |
where F(xi, gi) is the joint CDF of xi and gi. The empirical counterpart to (7) is:
Under the complete null H0 : (βX = 0) and (βG = 0), converges to , such that the D-INT approximation is asymptotically exact. Under the standard H0 : βG = 0, the approximation is accurate when βX ≈ 0.
2.5 |. Indirect inverse normal transformation (I-INT)
In indirect INT (I-INT), the phenotype is first regressed on covariates to obtain residuals, then the INT-transformed phenotypic residuals are regressed on genotype. Specifically, I-INT is based on the association model:
| (8) |
where is the INT-transformed phenotypic residual (ie, eY = PXy), eG = PXg is the genotypic residual, which is the residual after regressing g on X, and . The I-INT score statistic for assessing H0 : βG = 0 takes the form:
| (9) |
A p-value is assigned with reference to the , distribution. The score statistic estimates the residual variance as , while the Wald statistic estimates the residual variance as , where .
Since INT is a non-linear transformation, the INT transformed phenotypic residuals are no longer orthogonal to the covariates. That is, the correlation between and the columns of X is non-zero. Consequently, secondary adjustment for covariates has been recommended Sofar et al. (2019), as in the association model:
| (10) |
We demonstrate that the score statistic from model (10), which adjusts twice for covariates, is equivalent to the score statistic from model (8), which instead adjusts for genotypic residuals.
I-INT is adapted for a DGP of the form:
| (11) |
where the residuals ε ~ fε(·) follow an arbitrary continuous distribution with mean zero and constant finite variance. Under (11), , such that under the complete null H0 : (βX = 0) and(βG = 0), the DGPs in (11) and (6) are equivalent.
To motivate I-INT, we begin by showing that the efficient score for βG from model (11) is consistently estimated by the score for βG from the model eY = eGβG + ε. Observe that, for any fε, the OLSs estimator remains consistent for βX. Thus, the profile log likelihood from (11) is consistently estimated by
| (12) |
Letting , the efficient score for βG from (11) is consistently estimated by the gradient of (12), which is .
Now consider the following model for the phenotypic residual:
| (13) |
where ε is distributed as before. The profile log likelihood for βG in (13), with αX evaluated at the consistent estimator , coincides with the profile log likelihood in (12). Moreover, the log likelihood for βG from the following model, which relates eY to the genotypic residual eG, is also identical to (12):
| (14) |
Thus, the score for βG from (14) is consistent for the efficient score for βG from (11).
Now, under H0 : βG = 0, model (14) and model (13) with αX evaluated at its least squares estimate , both reduce to eY = ε. Model (14), together with the observation that , motivates the I-INT association model in (8). Moreover, under H0 : βG = 0, the distributional assumption in (8) is asymptotically exact, with .
2.6 |. Omnibus inverse normal transformation test
As shown in the simulation studies, both D-INT and I-INT robustly controlled the type I error. However, neither D-INT nor I-INT was uniformly most powerful. We therefore propose combining the two approaches into a robust and powerful omnibus test. The omnibus statistic is constructed using the method of Cauchy aggregation, in which the p-values from dependent hypothesis tests are converted to standard Cauchy random deviates and then combined (Liu et al., 2019; Liu and Xie, 2019). Cauchy aggregation is preferred to classic approaches for combining p-values, such as Fisher’s method (Fisher, 1934), since analytical expression are available for the finite-sample distribution of a Cauchy combination of dependent p-values.
Let pD and pI denote the p-values from D-INT and I-INT. Define the O-INT statistic as:
| (15) |
where is the inverse CDF of the standard Cauchy distribution. Under H0 : βG = 0, pD and pI are each uniformly distributed, such that and are standard Cauchy. Since the Cauchy distribution is symmetric and closed with respect to convolution, the omnibus statistic is again standard Cauchy in the tail (Liu et al., 2019; Liu and Xie, 2019), even though pD and pI are in general positively correlated. The p-value of the O-INT statistic (15) with observed value tO is
3 |. SIMULATION STUDIES
3.1 |. Simulation methods
Extensive simulations were conducted to evaluate the type I error and power of the UAT and INT-based association tests (D-INT, I-INT, O-INT). Genotypes exhibiting linkage disequilibrium were randomly sampled from unrelated subjects in the UK Biobank. The genotypes were additively coded, assuming values gi ∈ {0, 1, 2}. Simulated covariates included age and sex. Age was drawn from a gamma distribution with mean 50 and variance 10, and sex was drawn independently from a Bernoulli distribution with proportion 1/2. To emulate population structure, the top three PCs of the empirical genetic relatedness matrix were included as covariates. These correspond to the leading three left singular vectors from the subject by variant genotype matrix.
For type I error simulations, a subject-specific linear predictor ηi was generated as , where xi included an intercept, age, sex, and three genetic ancestry PCs. Regression coefficients were selected such that the proportion of total phenotypic variation explained (PVE) by age and sex was 20%, and the PVE by PCs was 5%. For power simulations, the linear predictor included a contribution from genotype. The PVE by genotype or heritability, defined as h2 = Var(giβG)/Var(yi), ranged between 0.1% and 1.0%.
Phenotypes were generated either from models with additive residuals, as in yi = ηi + ϵi, or from nonlinear transformations of such models, as in yi = h(ηi + ϵi). Here, we report on four representative traits: three with additive residuals, and one with multiplicative residuals. The additive models were (1) a reference trait, with N(0, 1) residuals; (2) a skewed trait, with residuals; and (3) a kurtotic trait, with t3 residuals. In all cases, the residual distribution was centered and scaled to have mean zero and unit variance. For the multiplicative model, a log-normal phenotype was generated by exponentiating a latent normal trait: yi = exp(ηi + ϵi), where ϵi ~ N(0,1).
3.2 |. Type I error simulations
A total of R = 108 simulation replicates were performed under H0 : βG = 0 at samples size of n ∈ {103,104,105}. On each simulation replicate, the four phenotypes (normal, skewed, kurtotic, log-normal) were generated independently and tested for association with genotype by each of the four association methods (UAT, D-INT, I-INT, O-INT).
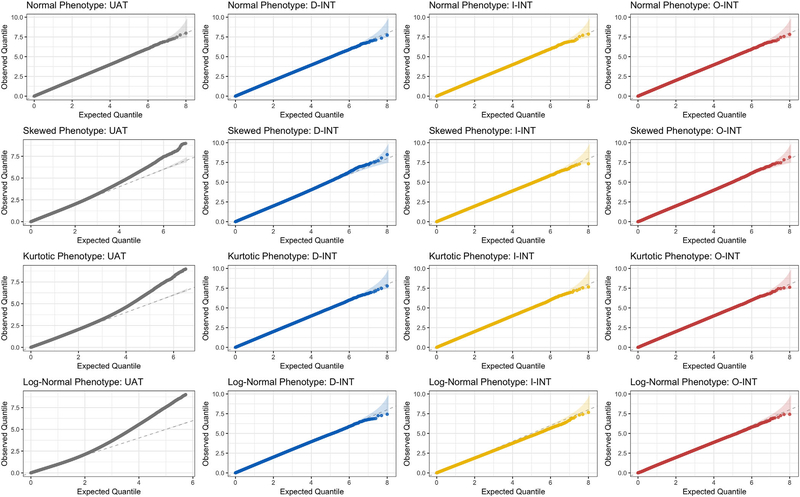
The uniform QQ plots in Figure 1 summarize the distribution of association p-values at sample size n = 103 for each combination of phenotype (row) and association test (column). All association tests performed well against the normal phenotype (row 1), providing uniformly distributed p-values. UAT (column 1) exhibited inflated type I error against all nonnormal phenotypes, although inflation attenuated with increasing sample size (Web Figures S1–2). Inflation was most severe for the log-normal phenotype (row 4), likely because the standard linear model is misspecified when the residuals have multiplicative rather than additive effects. However, inflation was still present for the skewed (row 2) and kurtotic t3 (row 3) phenotypes, for which UAT is correctly specified. In contrast, by sample size n = 103 the INT-based tests provided uniformly distributed p-values when applied to nonnormal phenotypes. Although the modeling assumptions underlying D-INT were not met for the skewed or kurtotic phenotypes, D-INT maintained the type I error across all scenarios. I-INT exhibited slight deflation against the log-normal phenotype, for which its modeling assumptions were not met. This deflation ameliorated with increasing sample size. O-INT performed well under all scenarios.
FIGURE 1.
Distribution of association p-values under the null at sample size n =103 across R =108 simulation replicates. Rows correspond to different phenotype distributions. The first phenotype has normal residuals; the second has residuals; the third phenotype has t3 residuals; and the log of the fourth phenotype has normal residuals. Columns correspond to different association tests. The first is the untransformed association test (UAT), the second is the direct INT (D-INT), the third is indirect INT (I-INT), and the fourth column is omnibus INT (O-INT). Note that this figure appears in color in the electronic version of this article, and any mention of color refers to that version
Type I error estimates at α = 10−6 and sample sizes n ∈ {103, 104, 105} are presented in Table 1. For all nonnormal phenotypes, UAT had substantially inflated type I error at sample size n = 103. This includes the skewed and kurtotic t3 phenotypes, for which UAT should provide asymptotically valid inference. Although the type I error approached its nominal level with increasing sample size, for the kurtotic and log-normal phenotypes the UAT still exhibited excess type I error at n = 105. For the nonnormal phenotypes, D-INT generally provided nearly the nominal type I error, while I-INT was slightly conservative. However, this does not imply I-INT is less powerful for these phenotypes (see power simulations). For all phenotypes and sample sizes considered, the omnibus test provided nominal control of the type I error.
TABLE 1.
Empirical type I error (×106) at α = 10−6 across R = 108 simulation replicates
| Phenotype | Test | Sample size |
||
|---|---|---|---|---|
| n = 103 | n = 104 | n = 105 | ||
| Normal | UAT | 1.04 | 0.93 | 1.03 |
| Normal | D-INT | 0.84 | 0.87 | 1.02 |
| Normal | I-INT | 0.97 | 0.93 | 1.00 |
| Normal | O-INT | 0.91 | 0.93 | 0.99 |
| Skewed | UAT | 8.03 | 1.87 | 1.43 |
| Skewed | D-INT | 1.20 | 1.10 | 1.05 |
| Skewed | I-INT | 0.67 | 0.84 | 0.89 |
| Skewed | O-INT | 1.10 | 1.01 | 0.98 |
| Kurtotic | UAT | 15.89 | 5.54 | 3.12 |
| Kurtotic | D-INT | 0.94 | 0.91 | 0.95 |
| Kurtotic | I-INT | 1.00 | 0.88 | 1.00 |
| Kurtotic | O-INT | 0.96 | 0.90 | 0.97 |
| Log-normal | UAT | 59.34 | 11.01 | 7.52 |
| Log-normal | D-INT | 0.74 | 1.02 | 1.02 |
| Log-normal | I-INT | 0.40 | 0.40 | 0.43 |
| Log-normal | O-INT | 0.55 | 0.74 | 0.76 |
Size simulations were conducted under the H0 : βG = 0 at sample sizes ranging from n = 103 to n = 105. The following association tests were evaluated: the UAT, D-INT, I-INT, and O-INT. Each test was applied to a normal phenotype, a skewed phenotype with residuals, a kurtotic phenotype with t3 residuals, and a phenotype whose log had normal residuals.
3.3 |. Power simulations
At each heritability h2 ∈ {0.1, 0.2, …, 1.0}%, a total of R = 106 power simulations were performed at sample size n = 103. On each replicate, a single randomly selected locus served as the causal locus. As before, the phenotypes were generated independently and tested for association with genotype by each of the candidate association methods. Power is considered even for the UAT, which did not consistently control the type I error, because this approach is still often applied in practice.
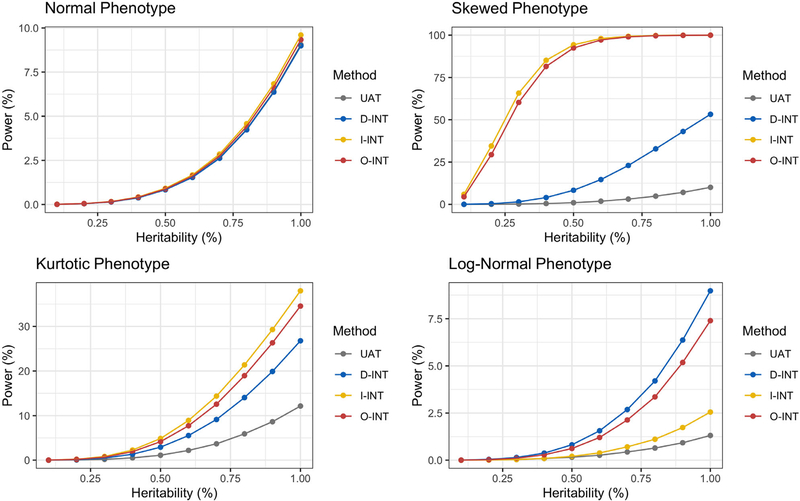
Power curves at α = 10−6 are presented in Figure 2. Relative efficiency (RE) curves, comparing the INT-based tests with UAT, are presented in Web Figure S3. RE was calculated as the ratio of the non-centrality parameters. This metric has the advantage of not depending on either α level or sample size n. For the normal phenotype, the UAT is theoretically most powerful. However, the INT-based tests were fully efficient, achieving a RE of one. Despite having inflated type I error under the null hypothesis, UAT was consistently least powerful for detecting true associations with the nonnormal phenotypes. Since the RE of the INT-based tests always exceeded one for the nonnormal phenotypes, this conclusion is expected to extend across significance levels and sample sizes. For the log normal phenotype, D-INT was most powerful, achieving twice the efficiency of the UAT. For this phenotype, the log transform is theoretically optimal. Since the log transform maps the log normal phenotype to a normal phenotype, the power of the log transform against the log normal phenotype is identical to the power of the UAT against the normal phenotype. Comparing the power of D-INT against the log normal phenotype with that of UAT against the normal phenotype, we observe that D-INT attains optimal power. For the skewed phenotype, I-INT was most powerful, achieving over five times the efficiency of the UAT, while D-INT was twice as efficient. For the kurtotic t3 phenotype, the efficiency gains provided by the INT-based tests were more modest yet still noteworthy, at around 55% for the I-INT and 35% for D-INT.
FIGURE 2.
Power curves at α =10−6 and sample size n =103 across R =106 simulation replicates. Simulations were conducted at heritabilities ranging from 0.1% and 1.0%. Gray is the UAT, blue is D-INT, I-INT, and red is O-INT. Each panel corresponds to a different phenotype. The first phenotype has normal residuals; the second has residuals; the third phenotype has t3 residuals; and the log of the fourth phenotype has normal residuals. Note that this figure appears in color in the electronic version of this article, and any mention of color refers to that version
By synthesizing D-INT and I-INT, O-INT aims to provide a test that is well powered across the residual distributions encountered in practice. As a compromise between complementary methods, the power and RE of O-INT were intermediate to those of D-INT and I-INT. However, for all phenotypes studies, O-INT performed comparably to the more efficient of D-INT and I-INT. Thus, O-INT achieves robustness to the underlying data generating mechanism with little to no loss of efficiency. In addition, we compared INT-based testing with the nonparametric Kruskal-Wallis (KW) test (Kruskal and Wallis, 1952). Unlike regression-based association tests, adjusting for covariates in the KW test is not straightforward. Yet even in the absence of covariates, INT-based testing was more powerful than the KW test.
4 |. APPLICATION TO UK BIOBANK
4.1 |. Application methods
We conducted GWAS of spirometry phenotypes within the UK Biobank. To mitigate confounding due to population structure, our study population was restricted to unrelated subjects of white, British ancestry. The phenotypes were forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), the FEV1 to FVC ratio (FEV1/FVC), and the logarithm of peak expiratory flow (lnPEF). Our analyses focused on 360 761 additively coded and directly genotyped, as opposed to imputed, autosomal SNPs, with sample minor allele frequencies (MAFs) exceeding 5%, and a per locus missingness rates of less than 10%. Covariates included an intercept, age, sex, BMI, two orthogonal polynomials in height, genotyping array, and 20 genetic PCs. Each locus was tested individually for association with the four spirometry phenotypes (FEV1, FVC, FEV1/FVC, lnPEF). The results were greedily “clumped” in PLINK (Purcell et al., 2007) using a 1000-kb radius and an r2 threshold of 0.2. The overall analysis consisted of n = 292K subjects that met all inclusion criteria. A subgroup analyses was conducted among subjects with physician diagnosed asthma (n = 29K).
4.2 |. Empirical type I error
Linkage disequilibrium score regression (LDSC) was performed to assess inflation of the association test statistics due to confounding bias (Bulik-Sullivan et al., 2015). Briefly, in LDSC the test statistic for each locus is regressed on a local measure of linkage disequilibrium. An intercept exceeding one suggests inflation, whereas an intercept falling below one suggests deflation. The results from applying LDSC in the overall sample and in the asthmatic subgroup are presented in Web Tables S4–5. Overall, there was no evidence of residual confounding due to population structure. Therefore, under the null hypothesis of no genetic effects, the marginal distribution of each spirometry trait is expected to be independent of genotype.
The empirical type I error of the association tests was assessed via a permutation analysis. Genotypes and phenotypes were first regressed on covariates to obtain residuals, then the genotypic residuals were permuted. Regressing out the effects of covariates accounts for potential confounding of the genotype-phenotype relationship. Permuting the genotypic residuals breaks the association between genotype and the spirometry traits, thereby imposing the null hypothesis of no genetic effect. Uniform QQ plots for the association p-values after permutation are presented in Web Figure S7. For all association methods (columns) and spirometry traits (rows), the p-values were uniformly distributions, suggesting nominal control of the type I error for the observed residual distributions.
4.3 |. Empirical discovery and efficiency gains
Table 2 presents the average statistics across all loci that reached genome-wide significance (α = 5 × 10−8) in the overall sample according to at least one of the association methods. Average for the asthmatic subgroup are presented in Web Table S6. The empirical efficiency gain (O-INT vs UAT) was defined as the ratio of the noncentrality parameters minus one, where the noncentrality parameters were estimated using loci that reached significance according to at least one association method:
In all cases the average statistics of the INT-based tests exceeded those of UAT, both in the overall analysis and in the asthmatic subgroup. Table 2 also presents the counts of genome-wide significant associations after LD “clumping” to reduce redundant signals. Counts for the asthmatic subgroup are presented in Web Table S6. The empirical discovery gain (O-INT vs UAT) was defined as
where nOINT is the number of associations identified by O-INT only, nUAT is the number of associations identified by UAT only (if any), and nOINT∪UAT is the number of associations identified by either O-INT or UAT. In all cases, the INT-based tests discovered more independent (at r2 = 0.2) associations with the target phenotype than UAT. All associations that reach genome-wide significance in the asthmatic subgroup, according to either UAT or the INT-based tests, reached significance according to O-INT in the overall sample.
TABLE 2.
Empirical efficiency and discovery gains for lung function GWAS in the UK Biobank (n = 292 K)
|
Average
| |||||
| Trait | UAT | D-INT | I-INT | O-INT | Efficiency Gain (%) |
| FEV1 | 56.77 | 57.55 | 64.69 | 63.59 | 12 |
| FVC | 37.14 | 46.91 | 51.47 | 50.59 | 37 |
| FEV1/FVC | 63.21 | 83.76 | 83.00 | 83.63 | 33 |
| lnPEF | 17.72 | 52.44 | 65.40 | 64.11 | 278 |
|
Significant associations
| |||||
| Trait | UAT | D-INT | I-INT | O-INT | Discovery gain (%) |
| FEV1 | 331 | 352 | 422 | 398 | 15 |
| FVC | 213 | 323 | 375 | 364 | 38 |
| FEV1/FVC | 450 | 653 | 649 | 652 | 28 |
| lnPEF | 39 | 202 | 270 | 251 | 79 |
Genome-wide significance was declared at α = 5×10−8. The average statistics are reported across all loci detected by at least one of the association tests. The empirical efficiency gain, comparing O-INT with UAT, is the ratio of the estimated noncentrality parameters minus 1. The counts of significant associations are reported after LD clumping within 1000 kb radii at r2 = 0.2 to remove redundant signals. The discovery gain, comparing O-INT with UAT, is the ratio of the number of associations uniquely identified by O-INT to the total number of associations detected.
The efficiency and discovery gains were more dramatic for those traits whose residuals were less normally distributed (Web Figure S6). However, the INT-based tests were consistently more powerful than the UAT, even when the normal residual assumption was not unreasonable. Consistent with the simulations, the power of O-INT was always intermediate between that of D-INT and I-INT. For a given trait, the number of discoveries by O-INT was generally closer to the number of discoveries by the more powerful of D-INT and I-INT.
5 |. DISCUSSION
In this paper, we have systematically investigated the utility of different INT-based association tests for GWAS of quantitative traits with nonnormally distributed residuals. We formally defined the direct (D-INT) and indirect (I-INT) INT-based tests, demonstrating that these approaches are adapted to different underlying data-generating processes. D-INT posits that the outcome could have arisen from a monotone transformation of a latent normal trait, whereas I-INT posits that the outcomes has additive but potentially nonnormal residuals. When covariate effects are small, the two approaches are approximately equivalent under the null hypothesis of no genetic effect; and in the absence of covariates, the two approaches are identical.
For nonnormally distributed quantitative traits, INT-based tests provided nominal control of the type I error by n = 103, whereas the UAT exhibited excess type I error even at n = 105. Moreover, the INT-based tests were consistently more powerful than UAT. Neither D-INT nor I-INT was uniformly more powerful. To obviate the need for choosing between them, we have proposed an adaptive omnibus test (O-INT). O-INT combines the p-values from D-INT and I-INT via Cauchy combination (Liu et al., 2019; Liu and Xie, 2019) and may easily be extended to incorporate p-values from complementary (eg, nonparametric) association tests. In simulations and data applications, O-INT provided valid and powerful inference that was robust to the underlying data generating process. As a compromise between complementary methods, O-INT cannot be expected to outperform both D-INT and I-INT. However, the performance of O-INT was similar to the more efficient of the component tests. O-INT was uniformly more powerful than UAT and is applicable whenever UAT is applicable. All INT-based tests (D-INT, I-INT, O-INT) have been implemented in the R package RNOmni, which is available on CRAN. We further demonstrated the utility of INT-based association tests through GWAS of spirometry traits from the UK Biobank.
In D-INT, the INT is applied directly to the phenotype, and the transformed phenotype is regressed on genotype and covariates. Under the complete null hypothesis of no genetic or covariate effects, D-INT is asymptotically exact, and when the covariate effects are small, D-INT holds approximately. I-INT is a two-stage procedure. Different variants of I-INT have been considered in the literature. In all approaches, the phenotype is first regressed on covariates to obtain residuals. In the second stage, the INT-transformed residuals are regressed on genotype, with or without a secondary adjustment for genetic PCs. To provide guidance on which approach to use in practice, we formally derived I-INT, starting from the assumption that the observed phenotype follows a linear regression model with a nonnormally distributed residual. Our derivations indicate that, during the second stage of I-INT, the transformed phenotypic residuals should be regressed on genotypic residuals, which are the residuals obtained by regressing genotype on covariates. This second stage is equivalent to regressing the INT-transformed phenotypic residuals on genotype while performing a secondary adjustment for covariates. Therefore, all covariates, including genetic PCs, should be included in both the first and second stage regressions. Under the standard null hypothesis of no genetic effects, I-INT is asymptotically exactand, under the complete null hypothesis of neither genetic nor covariate effects, D-INT and I-INT are asymptotically equivalent.
The use of INT does not compromise the validity of association testing, whose primary objective is to determine whether there is evidence that genotype is associated with the phenotype. Moreover, INT is useful for estimating standardized effect sizes. After INT, any absolutely continuous random variable is unitless, with mean zero, unit variance, and a common limiting distribution. Consequently, effect sizes estimated after INT are comparable across traits measured in different units and along different dimensions. Standardized effect sizes estimated via D-INT (Cade et al., 2016; Abbott et al., 2017) and via I-INT (Kettunen et al., 2012; Consortium et al., 2017) have been reported in numerous applications.
A limitation of INT-based tests is the restriction to absolutely continuous phenotypes. The INT cannot ensure asymptotic normality of a distribution with discrete probability masses. Our simulation studies and data application were restricted to common variants, those having a sample MAF exceeding 5%. For variants with lower MAF, unequal sample sizes can result in nonconstant variance across minor allele count strata. This heteroscedasticity is not remedied by INT and is likely more deleterious than residual nonnormally (Beasley et al., 2009). A future direction is to develop set-based tests that leverage the INT to improve power in rare variant association testing.
Finally, this paper has focused on GWAS of independent subjects. However, INT-based tests can be extended to the correlated data setting using linear mixed models (LMMs). We plan to develop INT-based tests for LMMs in which that correlation across related subjects is modeled via a random effect whose covariance pattern depends on the genetic relatedness matrix (Kang et al., 2010; Loh et al., 2015; Chen et al., 2016). A similar modeling strategy can accommodate longitudinal phenotypes arising from repeated measurements on the same subjects across time (Chen et al., 2019).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by F31 HL140822 (to ZM); by R35 HL135818 (to SR); and by R35 CA197449, P01-CA134294, U01-HG009088, U19-CA203654, and R01-HL113338 (to XL). The UK Biobank Resource was used in part for conducting this research (application numbers 6818 and 52008). We would like to thank the participants and researchers from the UK Biobank who contributed or collected data. We thank the referees for their insightful comments on our manuscript. We would like to thank the editor, the associate editor, and two referees for their helpful comments that have improved this paper.
Funding information
National Institutes of Health, Grant/Award Numbers: F31 HL140822, P01-CA134294, R01-HL113338, R35 CA197449, R35 HL135818, U01-HG009088, U19-CA203654
Footnotes
SUPPORTING INFORMATION
Web Appendix A, referenced in Sections 3 and 4, is available with this paper at the Biometrics website on Wiley Online Library. The association tests described in this paper (D-INT, I-INT, O-INT) are implemented in the R package RNOmni, available on CRAN at: https://cran.r-project.org/web/packages/RNOmni/index.html.
REFERENCES
- Abbott L, Bryant S, Churchhouse C, Ganna A, Howrigan D, Palmer D, Neale B, Walters R, Carey C for The Hail team. (2017) UK Biobank GWAS results, https://www.nealelab.is/uk-biobank (Accessed 2 January 2019).
- Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA et al. (2010) Genome-wide association of lipid-lowering response to statins in combined study populations. PLOS One, 5, e9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley TM, Erickson S and Allison DB (2009) Rank-based inverse normal transformations are increasingly used, but are they merited? Behavioral Genetics, 39, 580–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL and Neale BM (2015) LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics, 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade BE, Chen H, Stilp AM, Gleason KJ, Sofer T et al. (2016) Genetic associations with obstructive sleep apnea traits in Hispanic/Latino Americans. American Journal of Respiratory and Critical Care Medicine, 194, 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AC and Trivedi PK (2005) Microeconometrics, 1st edition. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Casella B and Berger R (2002) Statistical Inference, 2nd edition. Pacific Grove, CA: Duxbury/Thomson Learning. [Google Scholar]
- Chen H, Huffman J, Brody JA, Wang C, Lee S, Li Z, Gogarten SM et al. (2019) Efficient variant set mixed model association tests for continuous and binary traits in large-scale whole-genome sequencing studies. American Journal of Human Genetics, 104, 260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang C, Conomos MP, Stilp AM, Li Z, Sofer T et al. (2016) Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. American Journal of Human Genetics, 98, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J et al. (2008) Genetics of gene expression and its effect on disease. Nature, 452, 423–428. [DOI] [PubMed] [Google Scholar]
- Fisher RA (1934) Statistical Methods for Research Workers, 4th edition. Edinburgh, UK: Oliver and Boyd. [Google Scholar]
- GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)- Analysis Working Group; Statistical Methods groups-Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; et al. (2017) Genetic effects on gene expression across human tissues. Nature, 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C and Eskin E (2010) Variance component model to account for sample structure in genome-wide association studies. Nature Genetics, 42, 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Kangas AJ et al. (2012) Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nature Genetics, 44, 269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal WH and Wallis WA (1952) Use of ranks in one-criterion variance analysis. Journal of the American Statistical Association, 47, 583–621. [Google Scholar]
- Lehmann EL (1999) Elements of Large Sample Theory. Berlin, Germany: Springer. [Google Scholar]
- Liu Y, Chen S, Li Z, Morrison AC, Boerwinkle E and Lin X (2019) ACAT: a fast and powerful p value combination method for rare-variant analysis in sequencing studies. The American Journal of Human Genetics, 104, 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y and Xie J (2019) Cauchy combination test: a powerful test with analytic p-value calculation under arbitrary dependency structures. Journal of the American Statistical Association. [Preprint]. 10.1080/01621459.2018.1554485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ., Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, Patterson N and Price AL (2015) Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nature Genetics, 47, 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, De Bakker PIW, Daly MJ et al. (2007) Plink: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JO, Pantula SG and David DA (1998) Applied Regression Analysis, 2nd edition. Berlin, Germany: Springer. [Google Scholar]
- Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M et al. (2010) Genome-wide association study identifies five loci associated with lung function. Nature Genetics, 42, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J et al. (2007) Genome-wide association scan shows genetic variants in the fto gene are associated with obesity-related traits. PLOS Genetics, 3, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfling RJ (1980) Approximation Theorems of Mathematical Statistics. New York: John Wiley and Sons. [Google Scholar]
- Sofar T, Zheng X, Gogarten S, Laurie C, Grinde K, Shaffer JR et al. (2019) A fully adjusted two-stage procedure for rank-normalization in genetic association studies. Genetic Epidemiology, 43, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J et al. (2015) UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Medicine, 12, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart AW (1998) Asymptotic Statistics, 1st edition. New York: Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.