Abstract
The bacteriophages have been explored at a huge scale as a model system for their applications in many biological-related fields. Jumbo phages with a large genome size from 200 to 500 kbp were not previously assigned a great value, and characterized by complex structures coupled with large virions with a wide variety of hosts. The origin of most of the jumbo phages was not well understood; however, many other prominent features have been discovered recently. In the current review, we strive to unearth the most advanced characteristics of jumbo phages, particularly their significance and structural organization that holds immense value to the viral life cycle. The unique characteristics of jumbo phages are the basis of variations in different types of phages concerning their organization at the genomic level, virion structure, evolution, and progeny propagation. The presence of tRNA and additional translation-related genes along with chaperonin genes mark the ability of these phages for being independent of host molecular machinery enabling them to have wide host options. A large number of jumbo phages have been isolated from various sources through advanced standard screening methods. The current review has summarized the available data on jumbo phages and discussed the genome orientation of jumbo phages, translational machinery, diversity and evolution of jumbo phages. In the studies conducted, jumbo phages possessed special additional genes that helps to reduce the dependence of jumbo phages on their hosts. Furthermore, their genomes might have evolved from smaller genome phages.
Keywords: bacteriophages, tRNA, large genome, wide host range, chaperonin
Introduction
Bacteriophages bearing tails with genome size beyond 200 kbp are termed jumbo bacteriophages.1 Many studies have tried to explore the new jumbo phages that infect bacteria of different species.2–7 They are also different from other bacteriophages not only because of size but also due to evolutionary path, genetic orientation, structure, and progeny transmission.8 Thus, they attained scientist’s attention to isolate and characterize the new species of jumbo phages, as well as detailed elucidation about their biological mechanisms.9 Specific factors in the evolution of these phages are needed to explore the tendency toward gigantism. There are only a few numbers of tailed bacteriophages which have such large genome size.10 Mostly, predicted proteins of jumbo bacteriophages do not show similarity to protein sequences available in databases. The genome size of jumbo phages is too large to compare traditionally with smaller phages, but functions can be assigned to genes based on evaluation from smaller phages through comparative analysis.8 Comparisons have been done to check the genome similarity as it might be suggested that these have been evolved from small genome phages possibly due to limitations posed by the capsid size of the genome.11 N1M2 was selected as an example to further investigate by phylogenetic and genomic analyses and it was found to be similar to jumbo phage-related detached to Pseudomonas OBP. It was studied previously and exhibited to be possessing likeness with phiKZ-like jumbo phages.2
The ICTV’s Bacterial and Archaeal Viruses Subcommittee has now classified tailed phages into the following structural groups according to genomic data. Three new families of myoviruses have been officially approved Ackermannviridae, Chaseviridae, Herelleviridae; two for the siphoviruses, Demerecviridae and Drexlerviridae, and one of podoviruses, Autographiviridae.12 This is a latest taxonomy of tailed bacteriophages. According to this classification cultured jumbo phages that are reported in the literature until now are Myo and Sipho viruses (Table 1), suggesting that jumbo phages spread independently in different phage groups.13 Jumbo phages showed many variations in their head and tail morphologies and hair-like projections and long whiskers from both the head and tail sheath present in some cases.14
Table 1.
General Characteristics of Jumbo Bacteriophages
| Phage | Host | Family | Viron Size (nm) | Genome Size (nt) | No. of ORFs | No. of tRNA | Sample Source | Accession No. | References | |
|---|---|---|---|---|---|---|---|---|---|---|
| Head | Tail | |||||||||
| G | Bacillus megaterium | Myoviridae | 160 | 453 | 497,513 | 675 | 20 | NA | NC_023719 | Donelli et al 1975 |
| vB_CsaM_GAP32 | Cronobacter sakazakii | Myoviridae | 115 | 118 | 358,663 | 545 | 26 | Wastewater | NC_019401 | Abbasifar et al 2014 |
| 121Q | Escherichia coli | Myoviridae | 116 | 115 | 348,532 | 611 | 7 | Sewage | NC_025447 | Ackermann and Nguyen 1983 |
| PBECO 4 | Escherichia coli | Myoviridae | 132 | 125 | 348,113 | 551 | 6 | River water | NC_027364 | Kim et al 2013 |
| K64-1 | Klebsiella pneumoniae | Myoviridae | NA | NA | 346,602 | 64 | NA | NA | NC_027399 | Pan et al 2015 |
| vB_KleM-RaK2 | Klebsiella sp. | Myoviridae | 123 | 128 | 345,809 | 534 | 5 | NA | NC_019526 | Šimoliūnas et al 2013 |
| 201phi2-1 | Pseudomonas chlororaphis | Myoviridae | 129 | 200 | 316,674 | 461 | 1 | Soil | NC_010821 | Thomas et al 2008 |
| PhiPA3 | Pseudomonas aeruginosa | Myoviridae | 100 | 185 | 309,208 | 375 | 5 | Sewage | NC_028999 | Monson et al 2011 |
| Phabio | Pseudomonas fluorescens | Myoviridae | 134 | 174 | 309,157 | 469 | 3 | Soil | MF042360 | Joanna et al 2017 |
| Xoo-sp13 | Xanthomonas oryzae pv. oryzae | Myoviridae | 60 | 121 | 309,023 | 401 | 28 | Soil | MN047793 | Nazir et al 2020 |
| OBP | Pseudomonas fluorescens | Myoviridae | 119 | 191 | 284,757 | 309 | 4 | Compost | NC_016571 | Cornelissen et al 2012b |
| Lu11 | Pseudomonas putida | Myoviridae | 124 | 200 | 280,538 | 391 | 0 | Soil | NC_017972 | Adriaenssens et al 2012 |
| phiKZ | Pseudomonas aeruginosa | Myoviridae | 145 | 160 | 280,334 | 306 | 6 | Sewage | NC_004629 | Mesyanzhinov et al 2002 |
| Noxifer | Pseudomonas fluorescens | Myoviridae | 130 | 191 | 278,136 | 334 | 4 | Soil | MF063068 | Joanna et al 2017 |
| CcrColossus | Caulobacter crescentus | Myoviridae | 292x95 | 65 | 279,967 | 448 | 28 | Surface water | NC_019406 | Meczker et al 2014 |
| KTN4 | Pseudomonas aeruginosa | Myoviridae | 130 | 168 | 279,593 | 368 | 6 | Irrigated fields | KU521356.1 | Danis-Wlodarczyk et al 2016 |
| vB_EamM_Deimos-Minion | Erwinia amylovora | Myoviridae | NA | NA | 273,501 | 326 | NA | Tree | KU886225.1 | Esplin et al 2017 |
| vB_EamM_Special G | Erwinia amylovora | Myoviridae | NA | NA | 273,224 | 324 | 0 | Branches and blossums | KU886222.1 | Esplin et al 2017 |
| vB_EamM_RAY | Erwinia amylovora | Myoviridae | NA | NA | 271,182 | 319 | 1 | Tree | KU886224.1 | Esplin et al 2017 |
| vB_EamM_Simmy50 | Erwinia amylovora | Myoviridae | NA | NA | 271,088 | 321 | 1 | Bark | KU886223.1 | Esplin et al 2017 |
| Ea35-70 | Erwinia amylovora | Myoviridae | NA | NA | 271,084 | 318 | 1 | Soil | NC_023557 | Yagubi et al 2014 |
| PA7 | Pseudomonas aeruginosa | Myoviridae | NA | NA | 266,743 | 341 | NA | Mudflat | JX233784.1 | Kwan et al 2006 |
| phiR1-37 | Yersinia enterocolitica | Myoviridae | 138 | 383 | 262,391 | 367 | 5 | Sewage | NC_016163 | Kiljunen et al 2005 |
| vB_EamM_Yoloswag | Erwinia amylovora | Myoviridae | NA | NA | 259,700 | 334 | NA | Tree | KY448244.1 | Esplin et al 2017 |
| PaBG | Pseudomonas aeruginosa | Myoviridae | 136 | 220 | 258,139 | 308 | NA | Lake water | NC_022096 | Sykilinda et al 2014 |
| vB_BpuM_BpSp | Bacillus pumilus | Myoviridae | 137 | 192 | 255,569 | 318 | 0 | Soil | KT895374 | Yuan et al 2016a, b |
| N1M2 | Klebsiella aerogenes | Myoviridae | 113 | 158 | 253,367 | 257 | 24 | Fecal | MN642089 | Rhea Lewis et al 2020 |
| P-SSM2 | Prochlorococcus | Myoviridae | 115 | 123 | 252,401 | 334 | 1 | Seawater | NC_006883 | Sullivan et al 2005 |
| AR9 | Bacillus subtilis | Myoviridae | NA | NA | 251,042 | 291 | 1 | NA | NC_031039 | Lavysh et al 2016 |
| ValKK3 | Vibrio alginolyticus | Myoviridae | NA | NA | 248,088 | 390 | NA | Marine sediment | NC_028829 | Lal et al 2016 |
| nt-1 | Vibrio natriegens | Myoviridae | NA | NA | 247,511 | 405 | 28 | Marine sediment | NC_021529 | Comeau et al 2014 |
| VH7D | Vibrio harveyi | Myoviridae | NA | NA | 246,964 | 327 | NA | Seawater | NC_023568 | Luo et al 2015 |
| phi-pp2 | Vibrio parahaemolyticus | Myoviridae | 90x50 | 110 | 246,421 | 383 | 30 | Aquaculture waterway | JN849462.1 | Lin and Lin 2012 |
| vB_EamM_Kwan | Erwinia amylovora | Myoviridae | NA | NA | 246,390 | 285 | 8 | NA | NC_031010 | Esplin et al 2017 |
| vB_EamM_Asesino | Erwinia amylovora | Myoviridae | NA | NA | 246,291 | 277 | NA | NA | NC_031107 | NA |
| vB_EamM_ChrisDB | Erwinia amylovora | Myoviridae | NA | NA | 244,840 | 277 | 11 | Tree | NC_031126 | Esplin et al 2017 |
| KVP40 | Vibrio parahaemolyticus | Myoviridae | 140x70 | NA | 244,834 | 381 | 30 | Marine sediment | NC_005083 | Miller et al 2003 |
| phiEaH2 | Erwinia amylovora | Siphoviridae | NA | NA | 243,050 | 262 | NA | Soil | NC_019929 | Dömötör et al 2012 |
| vB_EamM_Stratton | Erwinia amylovora | Siphoviridae | NA | NA | 243,953 | 276 | 12 | Tree | KX397373.1 | Esplin et al 2017 |
| vB_EamM_Machina | Erwinia amylovora | Myoviridae | NA | NA | 241,654 | 272 | 9 | Tree | KX397370.1 | Esplin et al 2017 |
| vB_EamM_Caitlin | Erwinia amylovora | NA | NA | NA | 241,147 | 271 | 7 | Tree | NC_031120 | Esplin et al 2017 |
| vB_EamM_Parshik | Erwinia amylovora | Myoviridae | NA | NA | 241,050 | 271 | 10 | Tree | KX397371.1 | Esplin et al 2017 |
| vB_EamM_Huxley | Erwinia amylovora | Myoviridae | NA | NA | 240,761 | 271 | 9 | Tree | NC_031127 | Esplin et al 2017 |
| SPN3US | Salmonella enterica | Myoviridae | NA | NA | 240,413 | 264 | 2 | Chicken feces | NC_027402 | Lee et al 2011 |
| VP4B | Vibrio harveyi | Myoviridae | NA | NA | 236,053 | 212 | NA | Ocean | KC131130.1 | NA |
| vB_EamM_Joad | Erwinia amylovora | Myoviridae | NA | NA | 235,374 | 245 | NA | Tree | MF459647.1 | Esplin et al 2017 |
| 65 | Aeromonas salmonicida | Myoviridae | NA | NA | 235,229 | 437 | 16 | NA | NC_015251 | Petrov et al 2010 |
| vB_EamM_RisingSun | Erwinia amylovora | Myoviridae | NA | NA | 235,108 | 243 | NA | Tree | MF459646.1 | Esplin et al 2017 |
| vb_AbaM_ME3 | Acinetobacter baumanni | Myoviridae | NA | NA | 234,900 | 326 | 4 | Wastewater | KU935715.1 | Buttimer et al 2016 |
| Aeh1 | Aeromonas hydrophila | Myoviridae | NA | NA | 233,234 | 352 | 27 | NA | NC_005260 | Chow and Rouf 1983 |
| S-SSM7 | Synechococcus | Myoviridae | NA | NA | 232,878 | 319 | 5 | Seawater | NC_015287 | Sullivan et al 2010 |
| Xoo-sp14 | Xanthomonas oryzae pv. oryzae | Myoviridae | NA | NA | 232,104 | 251 | 0 | Soil | MT939492 | Nazir et al 2020 |
| CC2 | Aeromonas hydrophila | Myoviridae | NA | NA | 231,743 | 427 | 9 | Sewage | NC_019538 | Shen et al 2012 |
| phiRSL1 | Ralstonia solanacearum | Myoviridae | 150 | 138 | 231,255 | 343 | 3 | Soil | NC_010811 | Yamada et al 2010 |
| vB_EamM_Phobos | Erwinia amylovora | Myoviridae | NA | NA | 229,501 | 247 | NA | Tree | NC_031043 | Esplin et al 2017 |
| ACG-2014f isolate Syn7803C90 | Synechococcus | Myoviridae | NA | NA | 228,143 | 292 | NA | Ocean | NC_026927 | Gregory et al 2016 |
| phiAS5 | Aeromonas salmonicida | Myoviridae | 121 x 71 | 98 | 225,268 | 343 | 24 | River water | NC_014636 | Kim et al 2012 |
| CR5 | Cronobacter sakazakii | Myoviridae | NA | NA | 223,989 | 231 | NA | Soil | NC_021531 | Lee et al 2016 |
| vB_EamM_EarlPhillipIV | Erwinia amylovora | Myoviridae | NA | NA | 223,935 | 241 | NA | Tree | NC_031007 | Esplin et al 2017 |
| RSL2 | Ralstonia solanacearum | Myoviridae | NA | NA | 223,932 | 224 | NA | Soil | NC_028950 | Bhunchoth et al 2016 |
| Rogue | Caulobacter crescentus | Siphoviridae | 205x60 | 319 | 223,720 | 350 | 23 | Surface water | NC_019408 | Meczker et al 2014 |
| RSF1 | Ralstonia solanacearum | Myoviridae | NA | NA | 222,888 | 230 | NA | Soil | NC_028899 | Bhunchoth et al 2016 |
| PX29 | Aeromonas salmonicida | Myoviridae | NA | NA | 222,006 | 330 | 25 | NA | NC_023688 | Petrov et al 2010 |
| SP-15 | Bacillus subtilis | Myoviridae | NA | NA | 221,908 | 317 | NA | Soil | NC_031245 | Taylor and Thorne 1963 |
| Karma | Caulobacter crescentus | Siphoviridae | 205x61 | 314 | 221,828 | 353 | 26 | Surface water | NC_019410 | Meczker et al 2014 |
| PAU | Sphingomonas paucimobilis | Myoviridae | NA | NA | 219,372 | 295 | 7 | Silkworms | NC_019521 | White and Suttle 2013 |
| Swift | Caulobacter crescentus | Siphoviridae | 219x63 | 295 | 219,216 | 343 | 27 | Surface water | NC_019411 | Meczker et al 2014 |
| 0305phi8-36 | Bacillus thuringiensis | Myoviridae | 95 | 486 | 218,948 | 246 | 0 | NA | NC_009760 | Serwer et al 2007 |
| Magneto | Caulobacter crescentus | Siphoviridae | 211x58 | 293 | 218,929 | 347 | 27 | Surface water | NC_019407 | Meczker et al 2014 |
| PhiEaH1 | Erwinia amylovora | Siphoviridae | NA | NA | 218,339 | 241 | NA | Aerial tissue | NC_023610 | Meczker et al 2014 |
| S-CAM7 | Synechococcus | Siphoviridae | NA | NA | 216,121 | 266 | NA | NA | NC_031927 | NA |
| phiCbK | Caulobacter crescentus | Siphoviridae | 205x56 | 300 | 215,710 | 338 | 26 | Surface water | NC_019405 | Meczker et al 2014 |
| EL | Pseudomonas aeruginosa | Myoviridae | 140 | 200 | 211,215 | 201 | NA | NA | NC_007623 | Hertveldt et al 2005 |
| S-SKS1 | Synechococcus | Siphoviridae | NA | NA | 208,007 | 281 | 11 | Seawater | NC_020851 | NA |
| phiN3 | Sinorhizobium | Myoviridae | NA | NA | 206,710 | 402 | 6 | NA | NC_028945.1 | NA |
| vB_EamM_Y3 | Erwinia amylovora | Myoviridae | NA | NA | 261,365 | 338 | 0 | Soil | KY984068.1 | Buttimer et al 2018 |
| XacN1 | Xanthomonas citri | Myoviridae | NA | NA | 384,670 | 592 | 56 | Soil | AP_018399.1 | Yoshikawa et al 2018 |
| Joad | Erwinia | Myoviridae | 143 | 206 | 235,374 | 246 | 0 | Apple blossom | MF459647.1 | Arens et al 2018 |
| RisingSun | Erwinia | Myoviridae | 143 | 206 | 235,108 | 244 | 0 | Apple blossom | MF459646.1 | Arens et al 2018 |
| Atu_ph07 | Agrobacterium tumefaciens | Myoviridae | 146x 152 | 136 | 490,380 | 714 | 33 | Soil | MF403008.1 | Attai et al 2018 |
| RP12 | Ralstonia solanacearum | Myoviridae | 120 | 180 | 279,845 | 289 | 1 | Soil | AP017924.1 | Matsui et al 2017 |
| RP31 | Ralstonia solanacearum | Myoviridae | 120 | 180 | 276,958 | 287 | 1 | Soil | AP017925.1 | Matsui et al 2017 |
| JA11 | Dickeya solani | Myoviridae | NA | NA | 255,356 | 325 | 0 | Sewage | MH389777.1 | Day et al 2018 |
| JA13 | Dickeya solani | Myoviridae | NA | NA | 254,061 | 326 | 0 | Sewage | MH460460.1 | Day et al 2018 |
| JA29 | Dickeya solani | Myoviridae | NA | NA | 253,323 | 324 | 0 | Sewage | MH460461.1 | Day et al 2018 |
| JA33 | Dickeya solani | Myoviridae | NA | NA | 255,356 | 325 | 0 | Sewage | MH460462.1 | Day et al 2018 |
| AD1 | Dickeya solani | Myoviridae | NA | NA | 261,658 | 332 | 0 | Sewage | MH460463.1 | Day et al 2018 |
| Bonaishi | Vibrio coralliilyticus | Myoviridae | 120 | 190 | 303,340 | 301 | 0 | Seawater | MH595538.1 | Jacquemot et al 2018 |
| MIJ3 | Pseudomonas aeruginosa | Myoviridae | 130 | 140 | 288,170 | 417 | 12 | Equine livery yard | LR588166.1 | Imam et al 2019 |
| Psa21 | Pseudomonas syringae | Myoviridae | NA | NA | 305260 | 420 | 8 | Kiwi orchard | MK552327.1 | Frampton et al 2019 |
| SPFM | Salmonella spp. | Myoviridae | Multiple | Multiple | 233 to 242 kb | Multiple | Multiple | Multiple | Multiple | Thanki et al 2019 |
| DM | Erwinia amylovora | Myoviridae | Multiple | Multiple | 271 to 275kb | Multiple | Multiple | Multiple | Multiple | Sharma et al 2019 |
| PTm1 | Tenacibaculum maritimum | Myoviridae | 120 | 150 | 224,680 | 308 | NA | Seawater | AP019524.1 | Kawato et al 2020 |
| PTm5 | Tenacibaculum maritimum | Myoviridae | NA | NA | 226,876 | 306 | NA | Seawater | AP019525.1 | Kawato et al 2020 |
| PA5oct | Pseudomonas aeruginosa | Myoviridae | 131 | 136 | 286,783 | 465 | 12 | Sewage | MK797984.1 | Cédric Lood et al 2020 |
Until now, over 150 jumbo phages have been isolated since the discovery of phages (GenBank database, last accessed 1 June 2021). More than 85% isolated in just past 4 to 5 years, as research on bacteriophages revitalized and advanced by high throughput sequencing techniques. Their genomes were completely sequenced, exhibiting various remarkable aspects of their biology.15,16 These could be isolated through already in-use methods like agar diffusion and filtration. Due to the large genome size and capsid, jumbo phages might not be passed from the membrane and removed with bacteria.17,18 This can be the possible reason for the isolation of only a few jumbo phages until now. Additionally, we discussed only about cultured jumbo phages reported in the literature and have genome size larger than 200 kbp, phages approaching genome size 200 kbp will not be discussed in this review. We are trying to discuss the latest aspects related to jumbo bacteriophages including their general characteristics, diversity, and evolution in this review.
Hosts and Distribution
Jumbo phages were extracted from a variety of atmospheres ranging from marine sediments, water to soil constituted habitats like composts, animal feces, silkworms, and plant tissues (Table 1). But most recurrently, these phages were isolated from aquatic environments which provides ease in some of their functions like diffusion and infection to host bacteria which explains their frequency in such environments.8,19
Jumbo phages have been identified from gammaproteo bacteria, betaproteo bacteria, alphaproteo bacteria, zetaproteo bacteria, bacteroidetes, cyanobacteria, sporulating firmicutes, and actinobacteria. Currently, mostly jumbo phages present in databases are infecting gammaproteo bacteria.8 Most of the time these phages were isolated from a variety of gram-negative bacteria, while only eleven jumbo phages were extracted from gram-positive, the host is a common one from Bacillus spp.20 It is still needed to investigate whether the affinity of these eleven phages to Bacillus is because of their structural compatibility or just because of the discovery of a small number of jumbo phages studied currently.8,21 Further studies shed light on a phenomenon that why these four phages have an attraction toward gram-positive strain.
Genome Structure and Orientation
The most remarkable property that distinguishes the jumbo phages from smaller phages is their large genome size. The first ever jumbo phage with the biggest known size so far is phage G with capsid size 160 nm, tail 453 nm in length, and genome size 497 kbp.22 PhiKZ phage particles possess large sized hexagonal heads of about 120 nm and contractile tail of about 180×20 nm in size.23 Due to their large capsids, jumbo phages can regulate larger genomes compared to the phages having smaller capsids.16 It has been observed that the genome of phage G is only smaller just by 87 kbp than Mycoplasma genitalium, the genome of the smallest known bacterium.24 Big size enable the jumbo genomes to carry a lot of genes that normally are not possible to hold by smaller genome size phages. For instance, all reported jumbo phages possess more genome replication and nucleotide metabolism facilitating genes while some jumbo phages possessed more than one paralogous gene for DNA polymerase and RNA polymerase.25 The RNA Polymerase (RNAP) enzymes mostly are multi-subunit RNAP which are encoded by the phage genomes, while some of those have been observed in phage virions.8 The structural fragments of RNAP constitute multiple subunits and early gene transcription could be mediated by injecting subunits to host bacteria even before the expression of phage and host RNAP.26 Detailed transcriptomic-based analysis of this phage’s injection to the host highlights the fact that the expression of phage genes is only dependent upon the phage’s RNAP and cellular machinery rather than the host’s RNAP.27 These phages also have more enzymes and proteins to take on the defensive mechanism of a host such as glycoside hydrolase, chitinase, and many others.28
Majority of the phage genes are not well studied so far to comprehend their true and complete function. But some advanced research conducted in this field has unlocked unique and specific biological functions of cellular machinery including a conserved protein tubulin homolog which supplements a proteinaceous nucleus-like compartment that provides space and separates phage DNA.29,30 This arrangement facilitates proper phage and bacterial host infection and an optimal rate of reproduction. This spindle-like structure presents physical protection against the bacterial immune system and offers phage with a broad level of resistance.31
A few available reports showed that jumbo phages can synthesize their own NAD+ essential for the enzymes involved in phage DNA replication and regulation.32,33 Jumbo phages specify different diverse mechanisms like methylotransferases and incorporation of uracil instead of thymine in the genome that helps them to evade restriction attacks as a result of DNA modification. Furthermore, tRNAs help to get over the host defense mechanisms such as those by utilizing the endoRNases that create hindrance in translation by cleaving tRNAs.13
Translational Machinery
Growing shreds of evidence indicate that jumbo phages possess a large number of tRNA and the enzymes involved in the translation process.34 tRNAs are stable molecules, and many of these phages retain more than one tRNA gene and can have up to 28 tRNAs (Table 1). The sequences of these tRNAs are different from host tRNAs. For instance, PhiAS5 holds 56 tRNA genes that could give rise to anticodon sequences of 16 variable amino acids.35 During phage infection, host tRNA molecules are compromised, at this stage, translation is mediated by phage-encoded tRNAs. It is also known that some phages belong to the Myoviridae family encoding tRNA genes have broad host range.36 Therefore, it is suggested that phages with a large number of tRNAs have a wide host range.37 Phage XacN1 showed a wider host range by infecting nine Xanthomonas citri strains than the other phages that did not encode tRNAs like Siphovirus phage Cp1 and podovirus phage Cp2.34
Another important enzyme tRNA synthetase was observed to be present in more than one jumbo phage like Yersinia phage ΦR1-37 and many others.38 Different tRNAs in the phage genome are present to facilitate the translational process. The tRNAs are especially abundant to regulate the translation of proteins that comprise structure and smooth the translational performance of phage-specific genes.39 Mostly, the number of tRNAs increases as genome length becomes large.40 Jumbo phages have an average of 15 tRNAs per genome which is distinct but related to their hosts. Generally, Jumbo phages encode a larger number of translation-related genes than those encoded by smaller phages.41 Therefore, numerous genes encoded in viral genomes appear to signify an increased level of independence on host translational machinery. These genes rectify and provide an alternative for host genes that are necessary for the life cycle, which display jumbo phages are usually not dependent upon host proteins which are usually required for smaller phage genomes.31 This kind of less dependence of jumbo phages on host bacteria proves their importance as a complete functional unit and opens up new avenues to gather more genetic information from bacteria through horizontal gene transfer.42,43
Genetic Characterization
Giant phiKZ-like bacteriophages belong to myoviruses, which include Pseudomonas aeruginosa phiKZ, EL, OBP, and Pseudomonas chlororaphis 201phi2-1 that are found to have circular genomes.44,45 These genomes are packaged inside the capsid through a head-full packaging mechanism ensuring that the entire interior space of the viral protein head is filled with DNA in the course of the DNA packaging. After genome sequencing of the giant phages, it is concluded that their genomes encode structural proteins of capsid and tail, RNA polymerases,26,46 chaperonins,45,47 and inner body proteins.48 Virus inner body possessed an internal proteinaceous structure, specific for jumbo phages.49 Its function is to protect the DNA inside the capsid, helping in phage infection and form the depot of phage proteins. During infection to bacterial cell, some proteins are co-injected required to build the imperative machinery for the transcription of early genes.48
Some members of this class of viruses which are diverse as far as their phylogenetic organization is concerned, usually can give rise to two enzymes. One is non-virion DNA-dependent RNA polymerase (RNAPs) and virion RNAP.9,48 Contrary to many known virus-encoded enzymes, the enzymes from the virus of this category consist of enzymes of multi-subunit nature and are related to cellular organisms. The Jumbo RNAPs usually do not have conserved subunits needed for enzyme-oriented assembly as previously studied enzymes.50 Their working mechanism is also different in various aspects. For instance, the promoter recognition steps are different from mechanisms usually adapted by various cellular enzymes. This could be explained by an example of a non-virion RNAP from phage AR9 which needs uracil on the promoter region to start promoter-oriented transcription originating from ssDNA.9 A common ancestor has been found in the case of multisubunit RNAP from jumbo phage which makes them a different subgroup among the much-stretched group of jumbo phages.51
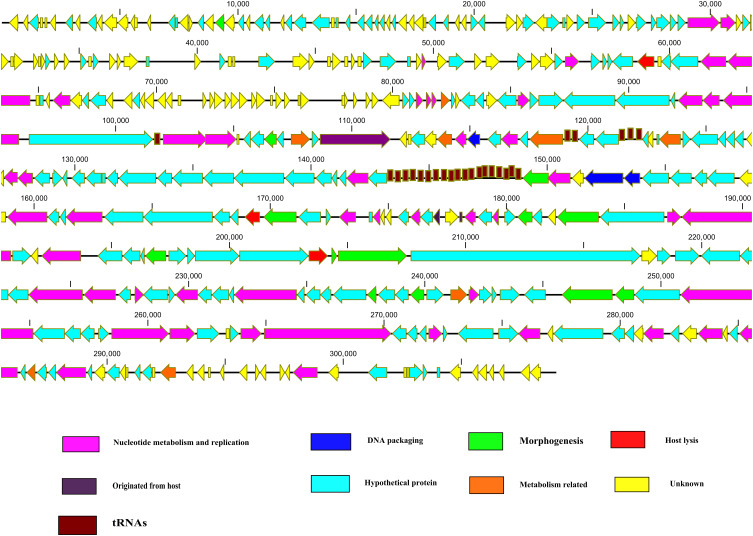
Another jumbo phage Atu-ph07 has 714 ORFs in its genome. Among 714 ORFs 214 were hypothetical proteins and 390 ORFs found no homologs. Proteins with assigned functions based on similarity with conserved regions were found to be 110. The predicted proteins with assigned functional annotations share similarities with different proteins from bacteria and bacteriophage at the sequence level.52 These genes with annotated functionality are found to give rise to various kinds of proteins. Some of these proteins are involved in nucleotide metabolism, while some of these are lysis proteins, structural and in the metabolism of tRNA, while some genes are found to encode proteins regulating the DNA replication (Figure 1). Further characterization and analysis of proteins discover the relation of proteins with those of 16T4 core proteins, which are homologous and similar to RAK2-like phages. Applying the tandem mass spectrometry, most structural proteins were confirmed experimentally and 112 more proteins were predicted of virion-associated type. The study concludes that phage is very lytic and the mode of its interaction with the host reflects application as a biocontrol agent.52
Figure 1.
Schematic representation of the dsDNA genome of jumbo phages. Jumbo phage genes related to specific function scattered throughout the genome. Jumbo phages also contained a large number of tRNAs. Putative ORFs are presented as arrows, with predicted functions where available. Proposed modules are based on predicted functions. Turquoise, hypothetical protein; yellow, unknown; pink, nucleotide metabolism and replication; green, morphogenesis; red, lysis; blue, DNA packaging; brown, tRNA. The map was drawn with CLC Genomics main Workbench version 3.6.1 (CLC bio, Aarhus, Denmark).
Another study was conducted to characterize the novel jumbo phage which is lytic along with head fiber-like appendages. The full genome characterization of two phages (PTm5 and PTm1) of this type were found to be 226,876 and 224,680 bp in size respectively, with a genome compactness of about 29.7% and encoding 306 and 308 ORFs respectively. The sequences were found to be 99.5% identical between PTm5 and PTm1, highlighting the striking similarity between both of these phages.53
The genetic similarity was observed in predicted ORFs between PTm1 and PTm5 belonging to Tenacibaculum maritimum, PT24 from tenacibaculum, and phage PAU from Paucimobilis till 15.0 to 16.6%. The close relation between these phages (PT24, PTm1, PAU, PTm5) was determined based on sequence similarity at the terminase large subunit genes through phylogenetic analysis. This phylogenetic analysis-based similarity was found to be more than those jumbo myoviruses which have equal genome sizes. Such genomes of equal size but differing at the phylogenetic level are ten in number.53
A most recent study analyzed the genetic analysis of PALS2 and found its genome of 268,746 bp enabling it to be classified as a jumbo phage and its genome contained about 279 ORFs and 1 tRNA encoding asparagine while many of its predicted genes whose proteins cannot be assigned a function.54
Gene Expression System
The smaller phages normally possess a modular genome structure, and genes having associated functions form clusters. However, the genes having alike functions in jumbo phage genomes are scattered in all over the genome (Figure 1).38,41,55 Efficient production of phage progeny required the timely expression of phage genes. Different phages adopt different strategies for the timely expression of genes. Similar to the small-genome phage, ΦKZ, a jumbo phage, genes are transcribed in a typical pattern, promptly by the phage-encoded RNAP.26 Contrarily, the transcriptions of phage ΦR1-37 genes do not follow the typical pattern and expressed throughout the infection process by the phage-encoded RNAPs.27 Notably, in both strategies, phage genes regulate under phage-encoded RNAPs instead of host RNAPs.
A research was conducted to find out some of the neglected, unexplored features of DNA transcription, recombination, and replication of jumbo phages.13 It also sheds light on the virion maturation system of jumbo viruses that infect bacteria. A new enzyme was found to have an important role in controlling host cellular machinery and its orientation is protein modifying as far as its nature of action is concerned. Mechanisms were found to deal with the bacterial immune system by keeping the focus on host–virus interactions (R-M modification system, CRISPR–Cas systems).56,57 For example, effector activation is based on NAD and cyclic nucleotide mechanism of protection from superinfection during the process of pseudolysogeny. Overall, this research concludes that many proteins in jumbo viruses reflect the living systems observed in prehistoric replicons.13
According to Gaun (2020), most genes found in jumbo phages have not been studied well so far as their proper function is concerned. But now, some of the current research has highlighted the rare biological features which include conserved protein homologs which integrate a proteinaceous nucleus-like section that resides and separates phage DNA.13 Phages encoded a tubulin like protein PhuZ that possessed dynamic instability. The spindle of tubulin protein displays the instability at the dynamic level and positions the nucleus of phage within the bacterial host during infection of phage for efficient reproduction.31 The shell extends protection at the physical level for safety of phage genome from potential assault DNA targeting the bacterial immune system in this providing it huge resistance.31 CRISPR is an emerging tool for gene editing applied in broad fields and applications from biotechnology to medical-related fields. The CRISPR–Cas systems of jumbo phages can turn off genes encoding host transcription factors and translational genes. This mechanism of tuning the genes off offers a part of a larger interaction network that intercepts translation to redirect biosynthesis to phage-encoded functions.32 The CRISPR system enables bacteria to defend themselves from bacteriophage by presenting its adaptive immunity.13 But DNA modification and synthesis of anti CRISPR proteins coupled with some other mechanisms protect jumbo phages against the CRISPR–Cas system. A jumbo phage was found to be able to evade the CRISPR–Cas system.58
Genetic analysis and characterization of PALS2 indicate the involvement of discovered genes in DNA repair, replication, metabolism of nucleotides, and multi-subunit RNA polymerase encoding genes, which is a genetic trait and a common characteristic of a jumbo virus. PALS2 was found to be like PhiKZ–like a virus in the result of comparative genomic analysis. The comparative genomic analysis also showed similarity to typical jumbo phages rather than staphylococcus phages.54 The Cas protein from bacteria is usually unable to approach virus nucleic acid during infection. But changing the location of an important restriction enzyme EcoR1 via engineering inside the proteinaceous compartment paves the way of targeting the virus and ensures the safety of the host cell. ΦKZ is also vulnerable to cas13a-CRISPR Cas enzyme that targets the virus nucleic acid. This study concludes that pseudomonas jumbo viruses defend themselves from quite a range of DNA targeting nucleases via making a protein barrier encapsulating their genomes.29,30,59,60
Moreover, as being virulent, jumbo phages possessed some extra host lysis genes. They also possessed DNA replication, transcription, and nucleotide metabolism genes which help them to wider host range and reduce their dependence on the host.8,61–64 Jumbo phages should be comprehensively characterized first. Furthermore, a large number of proteins of jumbo phages are hypothetical with unclear functions. These proteins could have any side effects is not predicted yet.9,56,65 Therefore, the application of jumbo phages needs to reach a compromise between host specificity and optimal treatment.31 In summarizing the whole story, further research of jumbo phages will unveil more attractive characteristics that useful for phage biology. They are also enriched in evolutionary perspectives and intensify the phage usage in diversified applications.
Evolutionary Aspects
The evolutionary status of jumbo phages has not been understood in well manner because of non-availability of sufficient number of jumbo phages and their high level of diversity at genomic level. Till now, some phages like ΦKZ-like phages66 and T4-like phages67 are classified on the basis of morphological similarity and range of host. Moreover, no clear evidence has been observed for the classification of jumbo phages. Because of less similarity with already discovered phages, jumbo phages have been described as new genetic lineage. Yuan and Gao had classified 93 jumbo phages using amino acid sequences of terminase large subunits in five singletons and 11 clusters.8
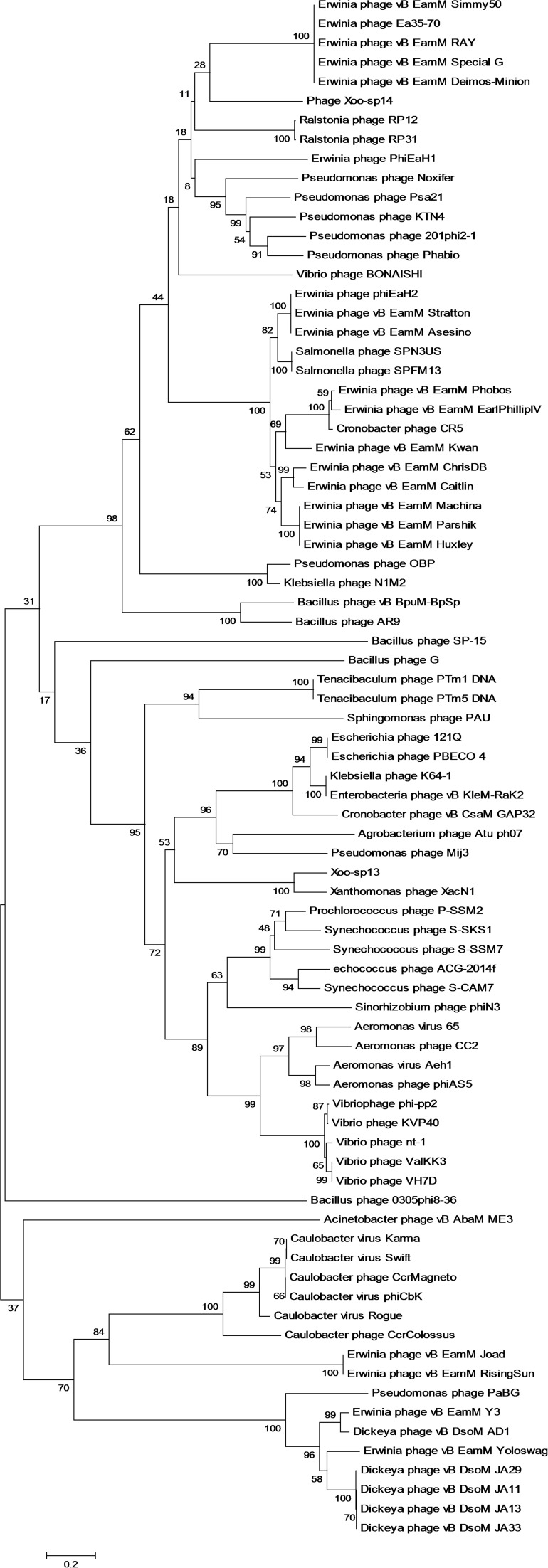
This study performed a phylogenetic analysis of terminase large subunits of 152 jumbo phages and showed that these jumbo phages could be classified into 17 clusters and 8 singletons (Figure 2). As per phylogenetic analysis some phages that classified as ΦKZ including phage OBP and phage Lu11, now classified differently in this study. According to core gene analysis of the jumbo phages, some phages should be classify differently as they previously classified as T4-like phages, eg ΦPAS5 which shares only 26% genes with T4-like phages and classified as T4-like phage. Contrarily, phage ΦPAS5 and Aeh1, which share 98% of their genes, have been classified in the same cluster in this study. The jumbo phages related to the same cluster tend to infect the host strains from similar genus and isolated from the same ecological environment. This study has assigned some phages to different clusters contrary to previous classification, on the basis of phylogenetic analysis.
Figure 2.
Phylogenetic analysis of jumbo phages. The evolutionary history was inferred using the neighbor-joining method. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved amino acid sequences of terminase large subunits from 152 jumbo phages. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA5.
The diverse foundation of jumbo phages is marked by very small level of similarity or no match at all among phages of different clusters. And phages from similar clusters are highly alike to each other exhibiting great degree or relatedness. As suggested in previous studies, evolution of jumbo phages might be a result of extracting novel genetic segments by smaller genome phages and increasing genome sizes over a certain period of time to give them their present shape.16 Essential genes imperative for phage life cycle are found in both small phages as well as jumbo phages as was concluded in the result of analysis of core genes.35,68 Genetic analysis of phage 0305Φ8-36 shed light on the origin that it has come into existence as the result of fusion of two viral genomes of ancestral nature by dint of the horizontal exchange of a genome module (block of genes) throughout the evolutionary course.69 Whereas the main stream number of the jumbo phages are thought to to get their genes from their respective hosts via horizontal gene transfer to formulate larger genomes.65
Keeping the jumbo phages with unclear method of propagation on one side, many of the large dsDNA viruses comprising phycodnaviruses, asfarviruses, poxviruses, iridoviruses, and ascoviruses are characterized as nucleocytoplasmic large dsDNA viruses,70 and gigantic sized viruses which normally infect amoeba, comprising Mollivirussibericum, faustoviruses, mimiviruses, marseilleviruses, pandoraviruses, and pithoviruses.71 The replication cycles of aforementioned giant and large dsDNA viruses consist of the occurrence inside the host cytoplasm of viral factories which give rise to the progeny viruses.72 These viral factories were imagined to be the origin and starting point of the contemporary eukaryotic nucleus.71 Jumbo phages tend to display parallel replication physiognomies to the eukaryotic NCLDVs. The protein of tubulin-like nature PhuZ of phage 20182-1 can arrange a spindle apparatus and spot the phage genomic DNA to the mid-cell area of the bacterial host. Consequently, the encapsidated DNA arranges a rosette-like assembly encircled via a huge DNA-based mass, that looks alike to the viral factory of NCLDVs till some extent.73 PhuZ related proteins have also been observed in the genomic structure of many jumbo phages coupled with phage genomes possessing near about 200 kbp. Evolutionary analysis of homologous proteins of PhuZ via genetic means marks similarity of jumbo phages with genome near 200 kbp, but is found to be different from the phages of small genome and the cellular microorganisms.8 Although phages having smaller genomes do not give rise to tubulin-like protein inside their own genomes, but they also facilitate engagement of the tubulin-like protein from the host bacteria to enable replication of the phage genome.74 Development of viral factory-related assemblies via large viruses and jumbo phages paves the way for the creation of a podium to quintessence virus genomes, virus replication, host proteins required for replication, and associated proteins which assist in protecting viruses from host defenses,72 which in turn can smooth the process of virus propagation. Setting the features of forming viral factories aside, NCLDVs coupled with giant viruses from amoeba possess more genes affiliated with nucleotide metabolism, genome replication, and some other biochemical processes.75 Even though NCLDVs, jumbo phages, and giant viruses of amoeba inclined to reflect many similar features, they are spaced far apart as far as evolutionary similarities are concerned.8 The jumbo phages have been observed more closely related to the archaea and bacteria, while the NCLDVs close evolutionary similarity is associated with the eukaryotes.76–80
Conclusion
Isolation and discovery of jumbo viruses has greatly enriched our understanding of biological entity diversity and evolution. They have been isolated from diverse environments and showed high genetic diversity. However, genome sizes and their non-modular structures, scattered genes throughout the genome related to specific function, presence of RNAPs in phage virion that control the gene expression, and distance between jumbo phages and smaller phages are the prominent features that differentiate jumbo phages from the smaller-genome phages. Furthermore, genome similarity, infection and propagation mechanisms are more divergent characteristics that jumbo phages showed among each other. Several areas need to be investigated deeply for a greater understanding of the jumbo phages. Although research showed that jumbo phages evolve from small phages but show bundle of differences. Mostly, functions of jumbo phages genes were predicted by using bioinformatics analysis. So, functional analysis of these genes will provide great understanding about phage interactions and evolution. Origin and evolution of jumbo phages need to be investigated more so that we can understand the origin and evolution of these cellular biological entities.
Funding Statement
This research was supported by a grant from The National Key Research and Development Program of China (2018YFA0903000), the National Natural Science Foundation of China (81672001), and China MoST Emergency Project on COVID-19 (2020YFC0840800).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work and declare that this research has been conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Nazir A, Dong Z, Liu J, et al. Genomic analysis of bacteriophage Xoo-sp13 infecting Xanthomonas oryzae pv. oryzae. Arch Virol. 2021;166(4):1263–1265. doi: 10.1007/s00705-021-04985-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelissen A, Hardies SC, Shaburova OV, et al. Complete genome sequence of the giant virus OBP and comparative genome analysis of the diverse ϕKZ-related phages. J Virol. 2012;86(3):1844–1852. doi: 10.1128/JVI.06330-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JM, LaBarre BA, Hewson I. Characterization of Trichodesmium-associated viral communities in the eastern Gulf of Mexico. FEMS Microbiol Ecol. 2013;84(3):603–613. doi: 10.1111/1574-6941.12088 [DOI] [PubMed] [Google Scholar]
- 4.Luo Z-H, Yu Y-P, Jost G, Xu W, Huang X-L. Complete genome sequence of a giant Vibrio bacteriophage VH7D. Mar Genomics. 2015;24:293–295. doi: 10.1016/j.margen.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 5.Kim SG, Jun JW, Giri SS, et al. Isolation and characterisation of pVa-21, a giant bacteriophage with anti-biofilm potential against Vibrio alginolyticus. Sci Rep. 2019;9(1):1–10. doi: 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SG, Giri SS, Yun S, et al. Genomic characterization of bacteriophage pEt-SU, a novel phiKZ-related virus infecting Edwardsiella tarda. Arch Virol. 2020;165(1):219–222. doi: 10.1007/s00705-019-04432-5 [DOI] [PubMed] [Google Scholar]
- 7.Weintraub ST, Mohd Redzuan NH, Barton MK, et al. Global proteomic profiling of Salmonella infection by a giant phage. J Virol. 2019;93(5):e01833–01818. doi: 10.1128/JVI.01833-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan Y, Gao M. Jumbo bacteriophages: an overview. Front Microbiol. 2017;8:403. doi: 10.3389/fmicb.2017.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavysh D, Sokolova M, Minakhin L, et al. The genome of AR9, a giant transducing Bacillus phage encoding two multisubunit RNA polymerases. Virology. 2016;495:185–196. doi: 10.1016/j.virol.2016.04.030 [DOI] [PubMed] [Google Scholar]
- 10.Koonin EV, Yutin N. Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv Virus Res. 2019;103:167–202. [DOI] [PubMed] [Google Scholar]
- 11.Van Etten JL. Lesser Known Large dsDNA Viruses. Vol. 328. Springer Science & Business Media; 2008. [Google Scholar]
- 12.Turner D, Kropinski AM, Adriaenssens EM. A roadmap for genome-based phage taxonomy. Viruses. 2021;13(3):506. doi: 10.3390/v13030506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer M, Anantharaman V, Krishnan A, Burroughs AM, Aravind L. Jumbo phages: a comparative genomic overview of core functions and adaptions for biological conflicts. Viruses. 2021;13(1):63. doi: 10.3390/v13010063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buttimer C, Born Y, Lucid A, Loessner MJ, Fieseler L, Coffey A. Erwinia amylovora phage vB_EamM_Y3 represents another lineage of hairy Myoviridae. Res Microbiol. 2018;169(9):505–514. doi: 10.1016/j.resmic.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 15.Brandes N, Linial M. Giant viruses—big surprises. Viruses. 2019;11(5):404. doi: 10.3390/v11050404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrix R. Jumbo bacteriophages. In: Lesser Known Large dsDNA Viruses. Springer; 2009:229–240. [Google Scholar]
- 17.Serwer P, Hayes SJ, Thomas JA, Hardies SC. Propagating the missing bacteriophages: a large bacteriophage in a new class. Virol J. 2007;4(1):1–5. doi: 10.1186/1743-422X-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Etten JL, Lane LC, Dunigan DD. DNA viruses: the really big ones (giruses). Annu Rev Microbiol. 2010;64:83–99. doi: 10.1146/annurev.micro.112408.134338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day A, Ahn J, Salmond GP. Jumbo bacteriophages are represented within an increasing diversity of environmental viruses infecting the emerging phytopathogen, Dickeya solani. Front Microbiol. 2018;9:2169. doi: 10.3389/fmicb.2018.02169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lood C, Danis‐Wlodarczyk K, Blasdel BG, et al. Integrative omics analysis of Pseudomonas aeruginosa virus PA5oct highlights the molecular complexity of jumbo phages. Environ Microbiol. 2020;22(6):2165–2181. doi: 10.1111/1462-2920.14979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devoto A, Santini J, Olm M, et al. Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat Microbiol. 2019;4:693–700. doi: 10.1038/s41564-018-0338-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drulis-Kawa Z, Olszak T, Danis K, Majkowska-Skrobek G, Ackermann H-W. A giant Pseudomonas phage from Poland. Arch Virol. 2014;159(3):567–572. doi: 10.1007/s00705-013-1844-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krylov V, Bourkaltseva M, Pleteneva E, et al. Phage phiKZ—the first of giants. Viruses. 2021;13(2):149. doi: 10.3390/v13020149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser CM, Gocayne JD, White O, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270(5235):397–404. doi: 10.1126/science.270.5235.397 [DOI] [PubMed] [Google Scholar]
- 25.Thomas JA, Hardies SC, Rolando M, et al. Complete genomic sequence and mass spectrometric analysis of highly diverse, atypical Bacillus thuringiensis phage 0305ϕ8–36. Virology. 2007;368(2):405–421. doi: 10.1016/j.virol.2007.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceyssens P-J, Minakhin L, Van den Bossche A, et al. Development of giant bacteriophage ϕKZ is independent of the host transcription apparatus. J Virol. 2014;88(18):10501–10510. doi: 10.1128/JVI.01347-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leskinen K, Blasdel BG, Lavigne R, Skurnik M. RNA-sequencing reveals the progression of phage-host interactions between φR1-37 and Yersinia enterocolitica. Viruses. 2016;8(4):111. doi: 10.3390/v8040111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill JJ, Berry JD, Russell WK, et al. The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genom. 2012;13(1):1–21. doi: 10.1186/1471-2164-13-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendoza SD, Nieweglowska ES, Govindarajan S, et al. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature. 2020;577(7789):244–248. doi: 10.1038/s41586-019-1786-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornuault JK, Moineau S. A jumbo formation in the viral game plan. CRISPR J. 2020;3(1):14–17. doi: 10.1089/crispr.2020.29082.jco [DOI] [PubMed] [Google Scholar]
- 31.Guan J, Bondy-Denomy J, Margolin W. Intracellular organization by jumbo bacteriophages. J Bacteriol. 2020;203(2):e00362–e00320. doi: 10.1128/JB.00362-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JY, Li Z, Miller ES, Henkin TM. Vibrio phage KVP40 encodes a functional NAD + salvage pathway. J Bacteriol. 2017;199(9):e00855–e00816. doi: 10.1128/JB.00855-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertani B, Ruiz N, Slauch JM. Function and biogenesis of lipopolysaccharides. EcoSal Plus. 2018;8(1). doi: 10.1128/ecosalplus.ESP-0001-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa G, Askora A, Blanc-Mathieu R, et al. Xanthomonas citri jumbo phage XacN1 exhibits a wide host range and high complement of tRNA genes. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-22239-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JH, Son JS, Choi YJ, et al. Complete genome sequence and characterization of a broad-host range T4-like bacteriophage phiAS5 infecting Aeromonas salmonicida subsp. salmonicida. Vet Microbiol. 2012;157(1–2):164–171. doi: 10.1016/j.vetmic.2011.12.016 [DOI] [PubMed] [Google Scholar]
- 36.Enav H, Béja O, Mandel-Gutfreund Y. Cyanophage tRNAs may have a role in cross-infectivity of oceanic Prochlorococcus and Synechococcus hosts. ISME J. 2012;6(3):619–628. doi: 10.1038/ismej.2011.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad AA, Ogawa M, Kawasaki T, Fujie M, Yamada T. Characterization of bacteriophages Cp1 and Cp2, the strain-typing agents for Xanthomonas axonopodis pv. citri. Appl Environ Microbiol. 2014;80(1):77–85. doi: 10.1128/AEM.02310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skurnik M, Hyytiäinen HJ, Happonen LJ, et al. Characterization of the genome, proteome, and structure of yersiniophage ϕR1-37. J Virol. 2012;86(23):12625–12642. doi: 10.1128/JVI.01783-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiljunen S, Hakala K, Pinta E, et al. Yersiniophage ϕR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology. 2005;151(12):4093–4102. doi: 10.1099/mic.0.28265-0 [DOI] [PubMed] [Google Scholar]
- 40.Al-Shayeb B, Sachdeva R, Chen L-X, et al. Clades of huge phages from across Earth’s ecosystems. Nature. 2020;578(7795):425–431. doi: 10.1038/s41586-020-2007-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mesyanzhinov VV, Robben J, Grymonprez B, et al. The genome of bacteriophage φKZ of Pseudomonas aeruginosa. J Mol Biol. 2002;317(1):1–19. doi: 10.1006/jmbi.2001.5396 [DOI] [PubMed] [Google Scholar]
- 42.O’Donnell M, Langston B, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizuno CM, Guyomar C, Roux S, et al. Numerous cultivated and uncultivated viruses encode ribosomal proteins. Nat Commun. 2019;10(1):1–11. doi: 10.1038/s41467-019-08672-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hertveldt K, Lavigne R, Pleteneva E, et al. Genome comparison of Pseudomonas aeruginosa large phages. J Mol Biol. 2005;354(3):536–545. doi: 10.1016/j.jmb.2005.08.075 [DOI] [PubMed] [Google Scholar]
- 45.Danilova YA, Belousova VV, Moiseenko AV, Vishnyakov IE, Yakunina MV, Sokolova OS. Maturation of pseudo-nucleus compartment in P. aeruginosa, infected with giant phiKZ phage. Viruses. 2020;12(10):1197. doi: 10.3390/v12101197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yakunina M, Artamonova T, Borukhov S, Makarova KS, Severinov K, Minakhin L. A non-canonical multisubunit RNA polymerase encoded by a giant bacteriophage. Nucleic Acids Res. 2015;43(21):10411–10420. doi: 10.1093/nar/gkv1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lecoutere E, Ceyssens PJ, Miroshnikov KA, et al. Identification and comparative analysis of the structural proteomes of ϕKZ and EL, two giant Pseudomonas aeruginosa bacteriophages. Proteomics. 2009;9(11):3215–3219. doi: 10.1002/pmic.200800727 [DOI] [PubMed] [Google Scholar]
- 48.Thomas JA, Rolando MR, Carroll CA, et al. Characterization of Pseudomonas chlororaphis myovirus 201ϕ2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology. 2008;376(2):330–338. doi: 10.1016/j.virol.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokolova O, Shaburova O, Pechnikova E, et al. Genome packaging in EL and Lin68, two giant phiKZ-like bacteriophages of P. aeruginosa. Virology. 2014;468:472–478. doi: 10.1016/j.virol.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 50.Orekhova M, Koreshova A, Artamonova T, Khodorkovskii M, Yakunina M. The study of the phiKZ phage non-canonical non-virion RNA polymerase. Biochem Biophys Res Commun. 2019;511(4):759–764. doi: 10.1016/j.bbrc.2019.02.132 [DOI] [PubMed] [Google Scholar]
- 51.Sokolova ML, Misovetc I, Severinov V. Multisubunit RNA polymerases of jumbo bacteriophages. Viruses. 2020;12(10):1064. doi: 10.3390/v12101064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Attai H, Boon M, Phillips K, Noben J-P, Lavigne R, Brown PJ. Larger than life: isolation and genomic characterization of a jumbo phage that infects the bacterial plant pathogen, Agrobacterium tumefaciens. Front Microbiol. 2018;9:1861. doi: 10.3389/fmicb.2018.01861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawato Y, Istiqomah I, Gaafar AY, et al. A novel jumbo Tenacibaculum maritimum lytic phage with head-fiber-like appendages. Arch Virol. 2020;165(2):303–311. doi: 10.1007/s00705-019-04485-6 [DOI] [PubMed] [Google Scholar]
- 54.Lee Y, Son B, Cha Y, Ryu S. Characterization and genomic analysis of PALS2, a novel Staphylococcus jumbo bacteriophage. Front Microbiol. 2021;12:395. doi: 10.3389/fmicb.2021.622755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Šimoliūnas E, Kaliniene L, Truncaitė L, et al. Klebsiella phage vB_KleM-RaK2—A giant singleton virus of the family Myoviridae. PLoS One. 2013;8(4):e60717. doi: 10.1371/journal.pone.0060717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi: 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 57.Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;11(10):675–687. doi: 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- 58.Malone LM, Warring SL, Jackson SA, et al. A jumbo phage that forms a nucleus-like structure evades CRISPR–Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol. 2020;5(1):48–55. doi: 10.1038/s41564-019-0612-5 [DOI] [PubMed] [Google Scholar]
- 59.Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. Phage ϕC2 mediates transduction of Tn 6215, encoding erythromycin resistance, between Clostridium difficile strains. MBio. 2013;4(6):e00840–00813. doi: 10.1128/mBio.00840-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kortright KE, Chan BK, Koff JL, Turner PE. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 2019;25(2):219–232. doi: 10.1016/j.chom.2019.01.014 [DOI] [PubMed] [Google Scholar]
- 61.Imam M, Alrashid B, Patel F, et al. vB_PaeM_MIJ3, a novel jumbo phage infecting Pseudomonas aeruginosa, possesses unusual genomic features. Front Microbiol. 2019;10:2772. doi: 10.3389/fmicb.2019.02772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wojtus JK, Frampton RA, Warring S, Hendrickson H, Fineran PC, Putonti C. Genome sequence of a jumbo bacteriophage that infects the kiwifruit phytopathogen Pseudomonas syringae pv. actinidiae. Microbiol Resource Announce. 2019;8(22):e00224–00219. doi: 10.1128/MRA.00224-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nazir A, Dong Z, Liu J, et al. Sequence analysis of a jumbo bacteriophage, Xoo-sp14, that infects Xanthomonas oryzae pv. oryzae. Microbiol Resource Announce. 2020;9(48):e01072–01020. doi: 10.1128/MRA.01072-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nazir A, Dong Z, Liu J, et al. Isolation, characterization, and genome sequence analysis of a novel lytic phage, Xoo-sp15 infecting Xanthomonas oryzae pv. oryzae. Curr Microbiol. 2021;78:1–9. doi: 10.1007/s00284-020-02263-1 [DOI] [PubMed] [Google Scholar]
- 65.Burkal’tseva M, Krylov V, Pleteneva E, et al. Phenogenetic characterization of a group of giant φKZ-like bacteriophages of Pseudomonas aeruginosa. Russ J Genet. 2002;38(11):1242–1250. doi: 10.1023/A:1021190826111 [DOI] [PubMed] [Google Scholar]
- 66.Krylov V, Cruz DD, Hertveldt K, Ackermann H-W. “φKZ-like viruses”, a proposed new genus of myovirus bacteriophages. Arch Virol. 2007;152(10):1955–1959. doi: 10.1007/s00705-007-1037-7 [DOI] [PubMed] [Google Scholar]
- 67.Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J. 2010;7(1):1–19. doi: 10.1186/1743-422X-7-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller ES, Heidelberg JF, Eisen JA, et al. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol. 2003;185(17):5220–5233. doi: 10.1128/JB.185.17.5220-5233.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hardies SC, Thomas JA, Serwer P. Comparative genomics of Bacillus thuringiensis phage 0305φ8-36: defining patterns of descent in a novel ancient phage lineage. Virol J. 2007;4(1):1–17. doi: 10.1186/1743-422X-4-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117(1):156–184. doi: 10.1016/j.virusres.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 71.Forterre P, Gaïa M. Giant viruses and the origin of modern eukaryotes. Curr Opin Microbiol. 2016;31:44–49. doi: 10.1016/j.mib.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 72.Netherton CL, Wileman T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr Opin Virol. 2011;1(5):381–387. doi: 10.1016/j.coviro.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kraemer JA, Erb ML, Waddling CA, et al. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell. 2012;149(7):1488–1499. doi: 10.1016/j.cell.2012.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muñoz-Espín D, Daniel R, Kawai Y, et al. The actin-like MreB cytoskeleton organizes viral DNA replication in bacteria. Proc Natl Acad Sci. 2009;106(32):13347–13352. doi: 10.1073/pnas.0906465106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Legendre M, Bartoli J, Shmakova L, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci. 2014;111(11):4274–4279. doi: 10.1073/pnas.1320670111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selvarajan Sigamani S, Zhao H, Kamau YN, Baines JD, Tang L. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J Virol. 2013;87(12):7140–7148. doi: 10.1128/JVI.00311-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rixon FJ, Schmid MF. Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr Opin Virol. 2014;5:105–110. doi: 10.1016/j.coviro.2014.02.003 [DOI] [PubMed] [Google Scholar]
- 78.Koonin EV, Krupovic M, Yutin N. Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses. Ann N Y Acad Sci. 2015;1341(1):10. doi: 10.1111/nyas.12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mutsafi Y, Fridmann-Sirkis Y, Milrot E, Hevroni L, Minsky A. Infection cycles of large DNA viruses: emerging themes and underlying questions. Virology. 2014;466:3–14. doi: 10.1016/j.virol.2014.05.037 [DOI] [PubMed] [Google Scholar]
- 80.Colson P, De Lamballerie X, Yutin N, et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol. 2013;158(12):2517–2521. doi: 10.1007/s00705-013-1768-6 [DOI] [PMC free article] [PubMed] [Google Scholar]