Abstract
One of two general pathways of mRNA decay in the yeast Saccharomyces cerevisiae occurs by deadenylation followed by 3′-to-5′ degradation of the mRNA body. Previous results have shown that this degradation requires components of the exosome and the Ski2p, Ski3p, and Ski8p proteins, which were originally identified due to their superkiller phenotype. In this work, we demonstrate that deletion of the SKI7 gene, which encodes a putative GTPase, also causes a defect in 3′-to-5′ degradation of mRNA. Deletion of SKI7, like deletion of SKI2, SKI3, or SKI8, does not affect various RNA-processing reactions of the exosome. In addition, we show that a mutation in the SKI4 gene also causes a defect in 3′-to-5′ mRNA degradation. We show that the SKI4 gene is identical to the CSL4 gene, which encodes a core component of the exosome. Interestingly, the ski4-1 allele contains a point mutation resulting in a mutation in the putative RNA binding domain of the Csl4p protein. This point mutation strongly affects mRNA degradation without affecting exosome function in rRNA or snRNA processing, 5′ externally transcribed spacer (ETS) degradation, or viability. In contrast, the csl4-1 allele of the same gene affects rRNA processing but not 3′-to-5′ mRNA degradation. We identify csl4-1 as resulting from a partial-loss-of-function mutation in the promoter of the CSL4 gene. These data indicate that the distinct functions of the exosome can be separated genetically and suggest that the RNA binding domain of Csl4p may have a specific function in mRNA degradation.
Gene expression is a process that can be regulated at multiple steps. One important control point is the regulation of the decay rate of mRNA. In recent years two general pathways of mRNA decay in the yeast Saccharomyces cerevisiae have been characterized and some of the proteins required for these pathways have been identified (reviewed in reference 32). Both general pathways of mRNA degradation in yeast involve the shortening of the poly(A) tail as the initial step. Subsequently, the mRNA can be degraded in either the 5′-to-3′ direction or the 3′-to-5′ direction. Degradation in the 5′-to-3′ direction involves removal of 7mGDP from the 7mGpppN cap structure by the Dcp1p decapping enzyme. Following decapping the remaining mRNA is rapidly digested by the 5′-to-3′ exonuclease Xrn1p. In the 3′-to-5′ pathway the mRNA is degraded by a complex of 3′-to-5′ exonucleases named the exosome (17).
Although the pathways of mRNA decay in other eukaryotes have not been worked out in detail, it appears likely that both 5′-to-3′ and 3′-to-5′ degradation of mRNA operates in other eukaryotes (reviewed in reference 32). Although both general pathways of mRNA decay are probably conserved in other eukaryotes, it appears likely that some organisms, cell types, or mRNAs may preferentially use one or the other pathway. Indeed, yeast cells appear to degrade the majority of their mRNA through the 5′-to-3′ pathway. It has been estimated that in wild-type cells the 5′-to-3′ degradation of the PGK1 mRNA is approximately twofold faster than 3′-to-5′ degradation (25).
The 3′-to-5′ mRNA degradation pathway requires the exosome (17). The exosome is a protein complex that contains multiple 3′-to-5′ exoribonucleases and RNA binding proteins (1, 23). The exosome is also required for a number of nuclear RNA 3′ processing reactions (2, 7, 22, 34). The exosome is located both in the nucleus and in the cytoplasm (1, 19, 23, 37). Therefore, it appears likely that the exosome contains the nuclease(s) that carries out both the mRNA degradation and the RNA processing reactions for which it is required. This raises several questions such as how the exosome recognizes the wide variety of its substrates and why it processes some RNAs to shorter forms while it completely degrades others. One possible explanation is that the exosome requires specific additional proteins to act on the various substrates. Candidates for these proteins include the products of the MTR4 and SKI2 genes. The MTR4 gene encodes a nuclear RNA helicase that is required for the processing of a variety of RNA species by the exosome (2, 13, 34), and SKI2 encodes a homologous cytoplasmic RNA helicase that is required for 3′-to-5′ mRNA degradation (10, 17).
The SKI genes were first identified because mutations in them cause a “superkiller” phenotype (31). Ski2p was later found to be in a cytoplasmic complex that also contains Ski3p and Ski8p (10), and all three proteins are required for 3′-to-5′ mRNA decay by the exosome (17). Similarly, Ski6p was found to be one of the 3′-to-5′ exonucleases in the exosome (23), and mutations in SKI6 inhibit 3′-to-5′ degradation of mRNA (17). Therefore mutations in at least four SKI genes affect 3′-to-5′ degradation of mRNA. In contrast, SKI1 encodes the Xrn1p enzyme involved in 5′-to-3′ mRNA decay (18).
Superkiller strains are yeast strains that are especially effective in killing other yeast strains. Many yeast strains contain killer viruses that encode a secreted toxin and kill strains not infected by the virus. The killer virus replicates as an RNA molecule that lacks both a cap and a 3′ poly(A) tail (reviewed in references 35 and 36). In these respects this RNA is similar to cellular mRNA that has been deadenylated and decapped and is about to be completely degraded. The superkiller phenotype is obtained when either pathway of mRNA degradation is blocked. One simple explanation of these observations is that in the absence of either degradation pathway the uncapped unadenylated killer toxin mRNA is more stable and therefore more killer toxin protein is produced per killer toxin mRNA. Mutations in the SKI2, SKI3, SKI6, SKI7, and SKI8 genes also result in increased production of luciferase activity from unadenylated luciferase RNA introduced into yeast by electroporation (7, 8, 21). It was proposed that this increased luciferase production is caused by a reduced ability of the translation machinery of ski mutant cells to distinguish between polyadenylated and unadenylated mRNA. In this case, the defect in 3′-to-5′ mRNA degradation might be a secondary effect of the defect in translation. Alternatively, the difference in luciferase production in the electroporation experiments might be a result of the decreased degradation of mRNA in ski mutants. The latter possibility is supported by the observation that the increased luciferase production is at least partially due to an increased functional half-life of the luciferase mRNA (7, 8, 21).
To further characterize the role of the SKI genes, we have analyzed the effects of the ski4 and ski7 mutations on 3′-to-5′ mRNA degradation. The possible roles of Ski4p and Ski7p in mRNA decay are largely unexplored. The SKI4 gene remains unidentified, and beyond the superkiller phenotypes they produce little is known about the effect of ski4 mutations. SKI7 has recently been cloned and was found to encode a putative GTPase (8). Mutations in the SKI7 gene resemble ski2, ski3 and ski8 mutations in two aspects. First, deletion of any of these four genes does not have a detectable effect on growth (except at very low temperatures, e.g., 8°C). Second, all four deletions affect the yield of luciferase activity when capped but unadenylated luciferase mRNA is introduced into yeast cells by electroporation (8, 21). In contrast to the common phenotypes of ski2, ski3, ski7, and ski8 mutants is the observation that the Ski2p, Ski3p, and Ski8p proteins form a complex. This complex does not appear to include Ski7p, nor is Ski7p required for complex formation (10). The latter observation may suggest that the roles of Ski7p and the Ski2p-Ski3p-Ski8p complex might be different.
In the experiments described here we address the roles of both Ski4p and Ski7p in cellular mRNA degradation. Both proteins are required for 3′-to-5′ degradation of yeast mRNA. We have also identified the SKI4 gene and found that it is allelic to CSL4, which encodes one of the subunits of the exosome. The ski4-1 mutation appears to block the degradation of mRNA by the exosome but has little or no effect on other exosome reactions, suggesting possible roles for Csl4p/Ski4p in 3′-to-5′ degradation of mRNA.
MATERIALS AND METHODS
Yeast strains.
The genotypes of the strains used are in Table 1. The ski4-1 strain 2373 was generously supplied by R. Wickner. The CSL4 gene from this strain was amplified by PCR using primers oRP943 (ATGAGCTTATGGTACGGCATG) and oRP944 (GTTTAATCACGTTCCCGCTTC). The products from two independent PCRs were gel purified and sequenced directly using the same oligonucleotides. Sequence analysis from −80 of the start codon to +100 of the stop codon revealed eight nucleotide differences compared to the sequence deposited in the yeast data base (SGD; http://genome-www.stanford.edu /Saccharomyces/; G-3-5 to C, T-5 to C, G194 to A, T199 to C, G258 to A, C363 to T, T540 to C, and G758 to A, numbered from the AUG start codon). These changes result in two changes in the predicted amino acid sequence as detailed in Results. The mutation that alters glycine 253 also results in the destruction of a recognition site for restriction endonuclease StyI.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| 2373 | MATa ura3 ski4-1 | |
| BM3-40A | MATa leu2 lys2 ura3 his4-580 ade3 cep1Δ cyh2 cry1 ade2-1 tyr1 SUP4-3 csl4-1 [CEP1/LEU2/ADE3] | 5 |
| R95-1-1 | MATa leu2 lys2 ura3 his4-580 ade3 cep1Δ cyh2 cry1 ade2-1 tyr1 SUP4-3 [CEP1/LEU2/ADE3] | 5 |
| p170 | MATa trp1 leu2Δ1 ura3 his3Δ200 gal2 gal108 csl4::HIS3::GAL::csl4 | 1 |
| 6000 | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 hbs1Δ::NEO | Research Genetics |
| BY4742 | MATα leu2Δ0 ura3Δ his3Δ1 lys2Δ0 | Research Genetics |
| V15E4 | MATa/MATa leu2-98/leu2-98 cry1R/CRY1 ade2-101/ade2-101 HIS3/his3-200 ura3-52/ura3-52 lys2-801/lys2-801 can1R/CAN1 trp1-1/TRP1 CYH2/cyh2RCSL4/CSL4::mTn-3xHA-LacZ [Cir0] | 27 |
| yRP840 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 16 |
| yRP841 | MATa leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 16 |
| yRP1195 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski2Δ::LEU2 | 17 |
| yRP1070 | MATa leu2-3,112 his4-539 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG dcp1Δ::URA3 | 6 |
| yRP1204 | MATa leu2-3,112 his4-539 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski6Δ::URA3 [ski6-100/LYS2] | 17 |
| yRP1340 | MATa leu2-3,112 his4-539 lys2-201 trp1 ura3 dcp1-2::TRP1 | 14 |
| yRP1346 | MATa leu2-3,112 his4-539 lys2-201 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG dcp2Δ::TRP1 | 14 |
| yRP1501 | MATα leu2-3,112 lys2-201 trp1 ura3-52 dcp2-7::URA3 | 14a |
| yRP1515 | MATα leu2-3,112 his4-539 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1-2::TRP1 | Dunkley and Parker |
| yRP1516 | MATa leu2-3,112 his4-539 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp2-7::URA3 | Dunkley and Parker |
| yRP1533 | MATα trp1 leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski7Δ::NEO | This study |
| yRP1534 | MATα leu2-3,112 his4-539 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski2Δ::LEU2 ski3Δ::TRP1 ski7Δ::NEO ski8Δ::URA3 | This study |
| yRP1535 | MATa leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1-2::TRP1 ski4-1 | This study |
| yRP1536 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1-2::TRP1 ski7Δ::NEO | This study |
| yRP1537 | MATa leu2-3,112 his4-539 lys2-201 trp1 ura3-52 dcp2-7::URA3 ski4-1 | This study |
| yRP1538 | MATα leu2-3,112 his4-539 lys-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp2-7::URA3 ski7Δ::NEO | This study |
| yRP1539 | MATa leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski4-1 | This study |
| yRP1540 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski4-1 | This study |
| yRP1541 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski4-1 [URA3] | This study |
| yRP1542 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski4-1 [CSL4/URA3] | This study |
| yRP1543 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG [URA3] | This study |
| yRP1544 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG [CSL4/URA3] | This study |
| yRP1545 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski4-1 [csl4-G253E/URA3] | This study |
| yRP1570 | MATα leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski4-1::GFP | This study |
| yRP1571 | MATα leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG SKI4::GFP | This study |
| R314-3B | MATa ade2-1 his4-3 lys20trp1aSUP4-3tsura3 CSL4::GFP | This study |
| R318 | MATa/MATα leu2-98/leu2-98 cry1R/CRY1 ade2-101/ade2-101 HIS3/his3-200 ura3-52/ura3-52 lys2-801/lys2-801 can1R/CAN1 trp1-1/TRP1 CYH2/cyh2RCSL4/csl4-1::mTn-3xHA-LacZ cep1::URA3/CEP1 [Cir0] [URA3/CSL4] | This study |
The ski4-1 mutation was introduced into the yRP840 genetic background by repeated backcrossing. In these backcrosses the segregation of the ski4-1 allele was determined either by Northern blotting or by PCR amplification of the CSL4 gene followed by either sequencing or StyI digestion.
The ski7Δ mutation was introduced into the yRP840 genetic background by PCR amplification and homologous recombination. The ski7Δ::NEO cassette from a ski7Δ strain obtained from Research Genetics (record no. 1852) was amplified using oligonucleotides oRP922 (TAGCGTCCTCAGCTGTCAC) and oRP923 (GTGTCACAATCTGCTCCCG). The PCR product was gel purified and used to transform (R. Agatep, R. D. Kirkpatrick, D. L. Parchaliuk, R. A. Woods, and R. D. Gietz, Technical tips online [http://tto.trends.com], 1998) a yRP840/yRP841 diploid. Transformants were selected on yeast extract-peptone-dextrose plates containing 150 mg of Geneticin/liter. The resulting diploid transformants were sporulated, and ski7Δ::NEO haploids were selected. Successful gene disruption was confirmed by PCR analyses of the resulting haploids.
The mutation in csl4-1 was identified by PCR amplification of csl4-1 DNA using oligonucleotides 40-519 (CGACACTTATGGAGAATTCG) and 40-1053b (TTCCGTACCGTACTGTGGGC) followed by direct DNA sequencing of the purified PCR product. To confirm that this substitution caused the csl4-1 phenotype, the C-172T mutation was introduced into a wild-type CSL4/SKI4 strain by “pop-in/pop-out” gene replacement (28). A CSL4/SKI4 integration plasmid analogous to pRB289 except containing the C-172T substitution was constructed as described previously (5). The plasmid was then cleaved with NruI and introduced into a wild-type CSL4/SKI4 strain (pop-in); URA3+ transformants were selected, and correct integration at the CSL4/SKI4 locus was verified by PCR of genomic DNA. The C-172T mutation was detected by the presence of a linked BglII restriction fragment length polymorphism (RFLP) located at position −115. One of the C-172T integrants was grown nonselectively and plated on 5-fluoroorotic acid to select plasmid pop-outs, which were then screened for retention of the mutation by BglII analysis of PCR-amplified genomic DNA. (The BglII RFLP by itself has no detectable phenotype.)
The CSL4::lacZ reporter strain V15E4 was obtained from M. Snyder (27), and the URA3 marker was changed to LEU2 (5). The C-172T mutation (csl4-1) was introduced by pop-in–pop-out recombination as described above. A cep1::URA3 gene disruption in the V15E4 genetic background was obtained by one-step gene replacement as described previously (4). Haploid segregants of csl4-1::LacZ and cep1::URA3 were obtained by sporulating the parental diploids. A CSL4/SKI4 plasmid (pRB306) (5) was introduced into the diploids before sporulation to provide CSL4/SKI4 function to cls4-1::LacZ segregants.
Double-mutant strains were made by crossing the respective single-mutant strains, sporulating the resulting diploids, and dissecting tetrads.
Plasmids.
pRP1000 (i.e., pCSL4) was constructed by digestion of pRB289 with XbaI and XhoI (5) and isolation of the 1.5-kb fragment. This fragment was ligated into CEN URA3 plasmid pRS416 (29), also digested with XbaI and XhoI. The resulting plasmid was sequenced to confirm its identity. The sequence proved identical to the one deposited in SGD. Two independent constructions of this plasmid were tested and gave identical results.
pRP1001 (i.e., pcsl4-G253E) was constructed from pRP1000 by site-directed mutagenesis as described by Kunkel et al. (20) using the antisense oligonucleotide oRP977 (GCTCTGGCGAACACGACCTCAAGGTCATTCC [mutation in boldface]). This mutation is identical to the one in ski4-1 that changes G253 to E and destroys a StyI site. Resulting plasmids were screened using digestion with StyI. Sequencing was used to confirm that the plasmid contained the desired mutation and no other mutations. Two independent constructions of this plasmid were tested and gave identical results.
The plasmids encoding MFA2pG and PGK1pG have been described previously (13, 24). They were introduced for the experiment shown in Fig. 1 into strain 2373 using standard transformation procedures (Agatep et al., http://tto.trends.com). For the experiment shown in Fig. 9 the MFA2pG plasmid was introduced into csl4-1 strain R95-1-1, which already contained plasmid pAP2, and into isogenic CSL4 strain BM3-40a (5).
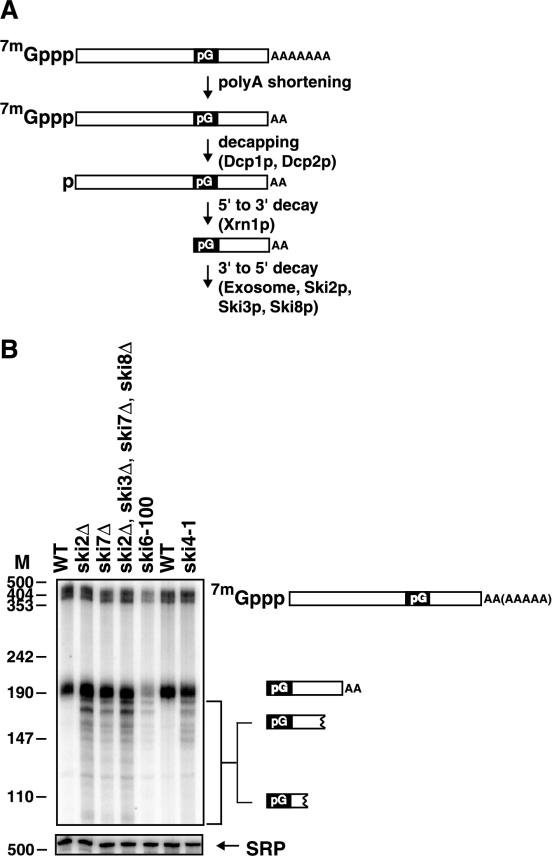
FIG. 1.
The ski4-1 and ski7Δ mutations affect the metabolism of a degradation intermediate of MFA2pG mRNA. (A) The degradation of MFA2pG mRNA through the 5′-to-3′ pathway is initiated by decapping, which requires Dcp1p and Dcp2p. Decapping is followed by digestion by the 5′-to-3′ exonuclease Xrn1p. This 5′-to-3′ exonuclease cannot proceed through a stable secondary structure formed by the poly(G) insert. The resulting poly(G)-to-3′-end fragment is therefore degraded by the exosome in a process that requires Ski2p, Ski3p, and Ski8p. (B) The metabolism of the poly(G)-to-3′-end fragment of MFA2pG is altered in ski4-1 and ski7Δ strains. Shown is a polyacrylamide Northern blot of the indicated strains probed with an oligonucleotide that hybridizes just 3′ of the poly(G) insert of MFA2pG mRNA. The cartoons to the right of the Northern blot indicate the identities of the various species detected. The first five lanes (from the left) contain RNA from strains containing the MFA2pG gene integrated into the genome grown in YEP containing 2% Gal. The MFA2pG reporter was introduced on a plasmid into a ski4-1 strain. To maintain this plasmid, the ski4-1 strain used for the last lane was grown in medium lacking uracil but containing 2% Gal. The wild-type (WT) strain in the sixth lane was grown in the same medium supplemented with uracil. All strains were grown at 30°C with the exception of the ski6-100 strain, which was grown at 24°C and shifted to 37°C for 2 h. The length of HpaII fragments of pUC18 is given in nucleotides. M, nucleotides.
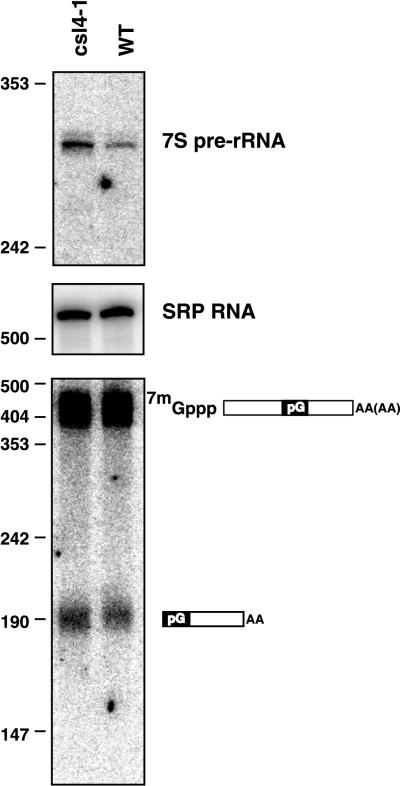
FIG. 9.
The csl4-1 mutation affects rRNA processing but not mRNA degradation. Shown are polyacrylamide Northern blots probed for 3′ extended precursors of 5.8S rRNA (top), the RNA subunit of the SRP (middle), or MFA2pG mRNA (bottom). The indicated strains were grown at 30°C in medium lacking leucine and uracil and containing 2% sucrose and 2% Gal. The length of HpaII fragments of pUC18 is given in nucleotides. Numbers on the left indicate nucleotides. WT, wild type.
RNA isolation and analyses.
mRNA and fragment half-lives were determined in duplicate as described by Jacobs Anderson and Parker. (17). The half-lives given in Fig. 2 are for the experiment shown. A duplicate experiment gave identical half-lives. Similarly, the half-lives given in Fig. 4 are for the experiment shown, and a duplicate experiment yielded essentially the same results (wild type, 5 min; ski7Δ strain, 4 min; ski4-1 strain, 3 min; dcp1-2 strain, 7 min; dcp1-2 ski7Δ strain, 38 min; dcp1-2 ski4-1 strain, >90 min). RNA isolation and Northern blotting were done as described previously (34). The oligonucleotides used as probes were as follows: MFA2pG, oRP140 (ATATTGATTAGATCAGGAATTCC); PGK1pG, oRP141 (AATTGATCTATCGAGGAATTCC); signal recognition particle (SRP), oRP100 (GTCTAGCCGCGAGGAAGG); 7S pre-rRNA, oJA003 (TGAGAAGGAAATGACGCT); 5.8S rRNA, oRP924 (TTTCGCTGCGTTCTTCATC); pre-U4 snRNA, oRP768 (CAGTCCCTTTGAAAGAATGAAT); U4 snRNA, oRP756 (CGGACGAATCCTCACTGATA); 5′ externally transcribed spacer (ETS), oRP993 (CGAACGACAAGCCTACTCG).
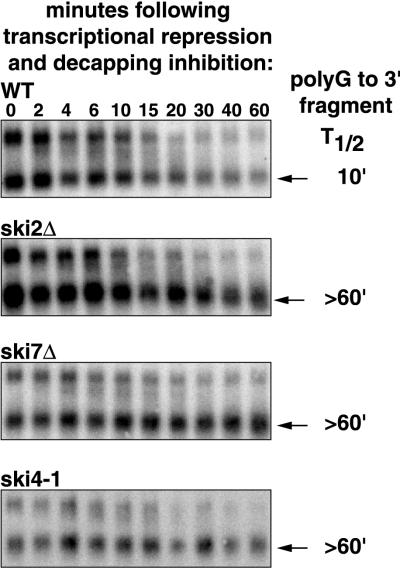
FIG. 2.
The ski4-1 and ski7Δ mutations stabilize a degradation intermediate of MFA2pG mRNA. Shown are agarose Northern blots of the indicated strains with the poly(G)-to-3′-end degradation intermediate of the MFA2pG mRNA (arrows). Each strain was grown in YEP containing 2% Gal. At the beginning of the experiment glucose and cycloheximide were added to inhibit transcription from the Gal promoter and the decapping enzyme, respectively. Samples were harvested at the indicated times after the beginning of this treatment and analyzed. The signals were quantitated using a phosphorimager and corrected for loading using the SRP RNA. Half-lives were calculated and are indicated on the right.
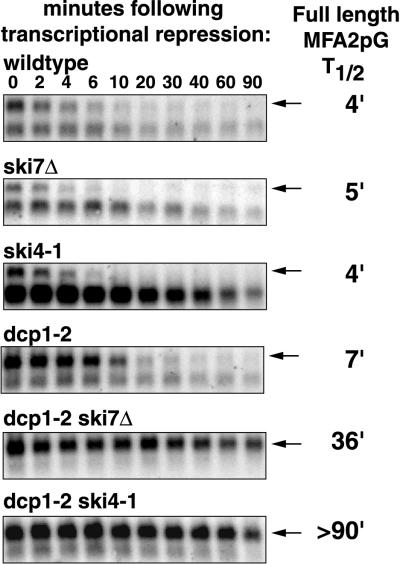
FIG. 4.
The ski4-1 and ski7Δ mutations inhibit 3′-to-5′ degradation of the MFA2pG mRNA. In a dcp1-2 strain at the restrictive temperature mRNA degradation occurs (almost) exclusively 3′ to 5′. Shown are agarose Northern blots of the indicated strains with the MFA2pG mRNA (arrows). Each strain was grown in YEP containing 2% Gal at 24°C and incubated for 1 h at 37°C. After this incubation, glucose was added to inhibit transcription from the Gal promoter. Samples were harvested at the indicated times after the addition of glucose and analyzed. The signals were quantitated using a phosphorimager and corrected for loading using the SRP RNA. Half-lives were calculated and are indicated on the right.
Localization of Csl4p/Ski4p.
The GFP-CSL4 allele was constructed by inserting DNA encoding green fluorescent protein (GFP)-Bex1 (3) into the naturally occurring SphI site at codon 2 of CSL4/SKI4. The GFP-Bex1 insert was generated by PCR using primers containing in-frame SphI sites. The GFP-CSL4 allele was introduced into yeast by pop-in/pop-out recombination as described above, replacing the endogenous gene. Localization of GFP fused to Ski4-1p was done using a ski4-1 strain as the starting strain for the pop-in/pop-out recombination. The pop-out recombination yielded isogenic SKI4::GFP and ski4-1::GFP, depending on the site of resolution of the Holliday junction. SKI4::GFP and ski4-1::GFP isolates were identified by PCR and StyI digestion as described above. The steady-state MFA2pG mRNA phenotypes of SKI4::GFP and ski4-1::GFP were indistinguishable from those of SKI4 and ski4-1 strains, respectively.
For microscopy, GFP-CSL4 cells were grown in yeast extract-peptone (YEP) medium containing 2% glucose to a cell density of 0.5 × 107 to 1 × 107 cells/ml and fixed by the addition of 1/10 volume of 37% formaldehyde. After continued incubation for 45 min under growth conditions, the fixed cells were washed once in standard phosphate-buffered saline (PBS), incubated on ice for 15 min in ethanol containing 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml, washed twice with PBS, resuspended in PBS, and kept on ice. Microscopy was performed either with a Nikon microscope equipped with epifluorescence optics and charge-coupled device (CCD) camera (Santa Barbara Instrument Group) or with a Leica microscope and Hamamatsu CCD camera. Collected images were adjusted for contrast and brightness and colorized using Adobe Photoshop.
RESULTS
The ski4-1 and ski7Δ lesions affect the metabolism of an mRNA fragment.
In order to determine whether the ski4 and ski7 mutants affected 3′-to-5′ mRNA degradation, we first examined the effect of the ski4-1 allele and a ski7Δ mutant on the degradation of an mRNA fragment. In these experiments we used two reporter mRNAs, PGK1pG and MFA2pG. Each of these mRNAs contains a poly(G) tract inserted into its 3′ untranslated region. This poly(G) tract forms a stable secondary structure that effectively blocks 5′-to-3′ degradation by Xrn1p (24, 26). Thus, 5′-to-3′ degradation of these mRNAs produces an intermediate that stretches from the 5′ end of the poly(G) tract to the 3′ end of the mRNA (Fig. 1A). This fragment is normally degraded by the 3′-to-5′ mRNA degradation pathway (17). Therefore, mutants that are defective in 3′-to-5′ decay accumulate higher levels of this fragment and show a slow digestion from the 3′ end, leading to the appearance of a ladder phenotype (17). As shown in Fig. 1B, both the ski4-1 allele and the ski7Δ mutant cause alterations in the amount and structure of the MFA2pG fragment similar to those seen in the ski2Δ mutant known to affect 3′-to-5′ mRNA turnover (similar data for PGK1pG not shown). This observation strongly suggests that Ski4p and Ski7p are involved in 3′-to-5′ degradation of mRNA.
If Ski7p acts in the same pathway as Ski2p, Ski3p, and Ski8p, then the deletion of the SKI7 gene and deletion of the SKI2, SKI3, or SKI8 gene should not have additive effects. As shown in Fig. 1B the phenotype of the ski2Δ ski3Δ ski7Δ ski8Δ quadruple mutant is no more severe than the phenotype of the ski2Δ single-mutant strain (additional data not shown). This observation suggests that the Ski7p protein acts in the same pathway of 3′-to-5′ degradation of mRNA as Ski2p, Ski3p, and Ski8p.
In order to confirm that the ski4 and ski7 lesions affected 3′-to-5′ degradation of the poly(G)-to-3′-end mRNA fragment, we directly measured its decay rate. In this experiment we monitored the loss of the decay fragment over time after blocking the production of new fragment. We utilized a combination of cycloheximide and glucose to inhibit production of fragment. Cycloheximide is known to inhibit decapping rapidly (6, 17), while glucose represses transcription from the Gal promoter. We used a ski2Δ strain as a control in this experiment, because SKI2 was previously shown to be required for the normal 3′-to-5′ degradation of the poly(G)-to-3′-end fragment (17). As shown in Fig. 2, like the ski2Δ lesion, both the ski4-1 and ski7Δ lesions caused a decrease in the rate at which the MFA2 fragment is degraded, leading to an increase in its half-life from about 10 min to more than 60 min. Similar results were obtained for the PGK1pG mRNA (data not shown), indicating that this effect is not specific for the MFA2 mRNA. Therefore, like Ski2p, Ski3p, and Ski8p, Ski4p and Ski7p are likely to affect the 3′-to-5′ degradation of a wide variety of mRNAs.
The ski4-1 and ski7Δ lesions are synthetically lethal with mutations affecting decapping.
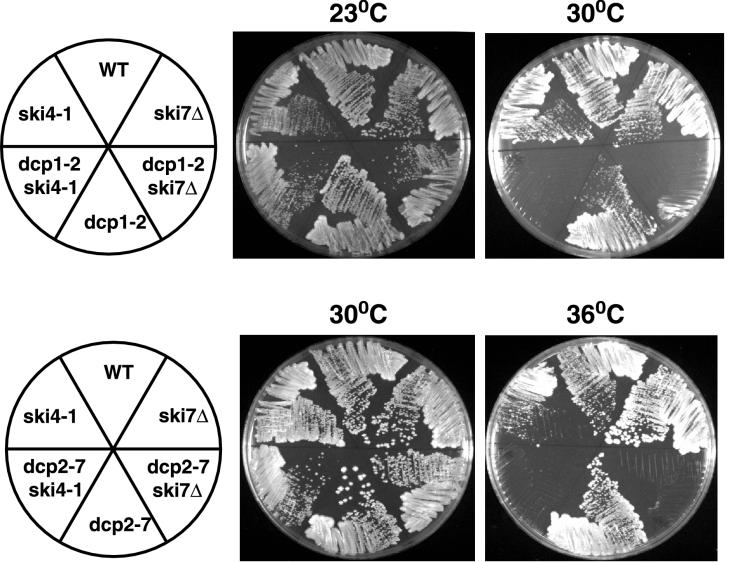
The above results suggested that the ski4-1 and ski7Δ lesions affected 3′-to-5′ mRNA degradation. One property of mutations affecting this process is that they are synthetically lethal with lesions that block the other major decay pathway of decapping and 5′-to-3′ degradation (17; T. Dunckley and R. Parker, personal communication). In order to test if ski4-1 and ski7Δ mutations are synthetically lethal with mutations blocking 5′-to-3′ decay, we crossed the ski4-1 and ski7Δ strains to dcp1Δ and dcp2Δ strains, which are defective in decapping. In all four crosses we observed a reduced viability of spores and failed to recover any double mutants. This suggests that the ski4-1 and ski7Δ mutations are synthetically lethal with the dcp1Δ and dcp2Δ alleles. We confirmed this conclusion by crossing both the ski4-1 and ski7Δ strains to strains containing temperature-sensitive alleles of either DCP1 or DCP2 (i.e., dcp1-2 and dcp2-7 [14a, 30]). Because the 5′-to-3′ decay pathway is not essential in our strain background, these temperature-sensitive alleles of DCP1 and DCP2 do not by themselves affect viability but are synthetically lethal at the restrictive temperature with mutations affecting 3′-to-5′ decay of mRNA (14a, 17). In crosses of dcp1-2 or dcp2-7 strains with ski4-1 or ski7Δ strains we were able to recover double mutants by germinating the spores at low temperature (i.e., 23°C). In each case these double mutants were unable to grow at the temperature restrictive for the relevant dcp mutation (Fig. 3). This conditional lethality supports a role of the Ski4p and Ski7p proteins in 3′-to-5′ degradation of mRNA.
FIG. 3.
Strains containing either ski4-1 or ski7Δ in combination with dcp1-2 or dcp2-7 are not able to grow under conditions restrictive for the decapping defect. Shown are plates of YEP–2% glucose containing the indicated strains grown at the indicated temperatures.
The ski4-1 and ski7Δ lesions affect the rate of 3′-to-5′ mRNA degradation.
The dcp1-2 ski4-1 and dcp1-2 ski7Δ double-mutant strains allowed us to examine if the ski4-1 and ski7Δ alleles affect the 3′-to-5′ decay of a full-length mRNA by comparing them to a dcp1-2 strain. In a dcp1-2 strain mRNA degradation at the restrictive temperature occurs (almost) exclusively through the 3′-to-5′ pathway (17). To measure mRNA decay rates in the double mutants, these strains were grown at the permissive temperature and shifted to a restrictive temperature for 1 h. Subsequently, transcription of the reporter MFA2pG mRNA was inhibited by addition of glucose, allowing monitoring of the loss of mRNA over time. Both the ski4-1 and ski7Δ mutations cause a severe decrease in the rate of degradation of the MFA2pG mRNA under these conditions (Fig. 4). The half-life of the MFA2pG mRNA increased from 7 min in the dcp1-2 strain to 36 and more than 90 min in the dcp1-2 ski7Δ and dcp1-2 ski4-1 strains, respectively. This implies that the SKI4 and SKI7 gene products are required for the 3′-to-5′ degradation of mRNA.
Based on all of these results we conclude that the Ski4p and Ski7p proteins are required for 3′-to-5′ mRNA degradation as are the components of the exosome and the Ski2p, Ski3p, and Ski8p proteins.
The SKI4 gene is identical to the CSl4 gene.
In order to understand the function of the Ski4p protein in 3′-to-5′ degradation, it was important to examine the properties of the encoded gene product. The SKI4 gene has not been cloned. However, we hypothesized that the SKI4 gene might be identical to the CSL4 gene for two reasons. First, the mapping of the ski4-1 allele placed it within a few map units of the KEX2 gene (31), which is about 10 kb from the CSL4 gene. Second, the Csl4p protein is a component of the exosome (1), and therefore defects in this protein might be expected to affect 3′-to-5′ mRNA degradation.
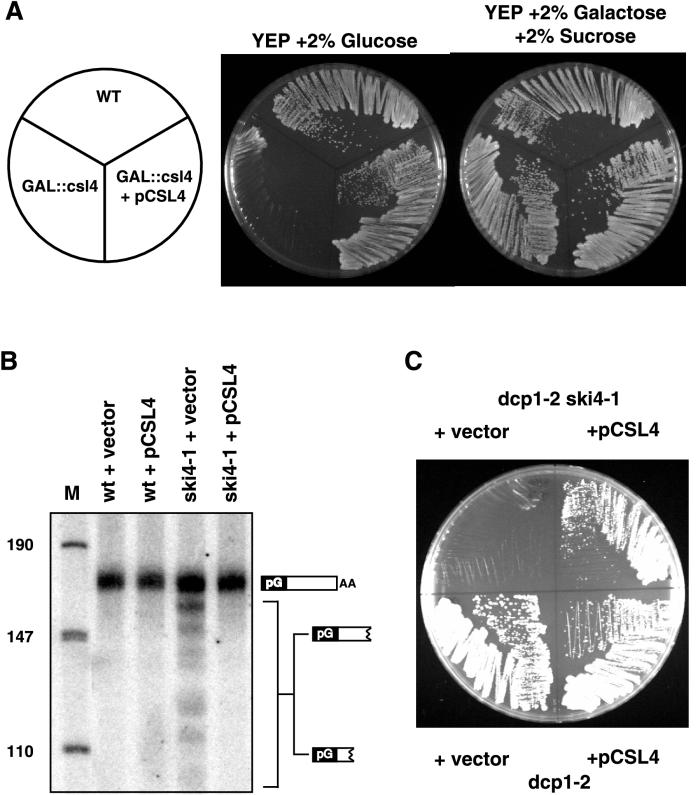
In order to test if the CSL4 gene was identical to the SKI4 gene, we first determined if a plasmid carrying the wild-type CSL4 gene would complement the defect in mRNA decay in a ski4-1 strain. We constructed a low-copy-number plasmid containing CSL4 and introduced it into a GAL::csl4 strain. The GAL::csl4 gene is not expressed in the presence of glucose. Since the CSL4 gene is an essential gene, the GAL::csl4 strain cannot grow on plates containing glucose (1). The low-copy-number CSL4 plasmid complemented this conditional growth defect (Fig. 5A), indicating that the plasmid carried a functional CSL4 gene. As shown in Fig. 5B, this plasmid also complements the mRNA decay defect of a ski4-1 strain, indicating that SKI4 and CSL4 are the same gene. In addition the same plasmid complements the temperature-sensitive growth defect of a dcp1-2 ski4-1 strain (Fig. 5C). These results, in combination with the prior linkage data strongly argue that the ski4-1 allele represents a mutation in the CSL4 gene.
FIG. 5.
The ski4-1 mutation is complemented by a wild-type (WT) CSL4 gene. (A) Plates of YEP containing either 2% glucose or 2% Gal and 2% sucrose. The GAL::csl4 strain fails to grow on plates containing glucose but grows on plates containing Gal and sucrose, because the only copy of the essential CSL4 gene has been placed under the control of the Gal promoter. This conditional growth defect is corrected by pCSL4. (B) Polyacrylamide Northern blot of RNA from wild-type and ski4-1 strains containing either pCSL4 or the empty vector. The blot was probed with an oligonucleotide that detects the poly(G)-to-3′-end fragment of PGK1pG mRNA. The detected species are indicated with cartoons on the right. The length of HpaII fragments of pUC18 is givne in nucleotides. M, nucleotide marker lane. (C) Plates containing YEP–2% glucose. The indicated strains were grown at 33°C.
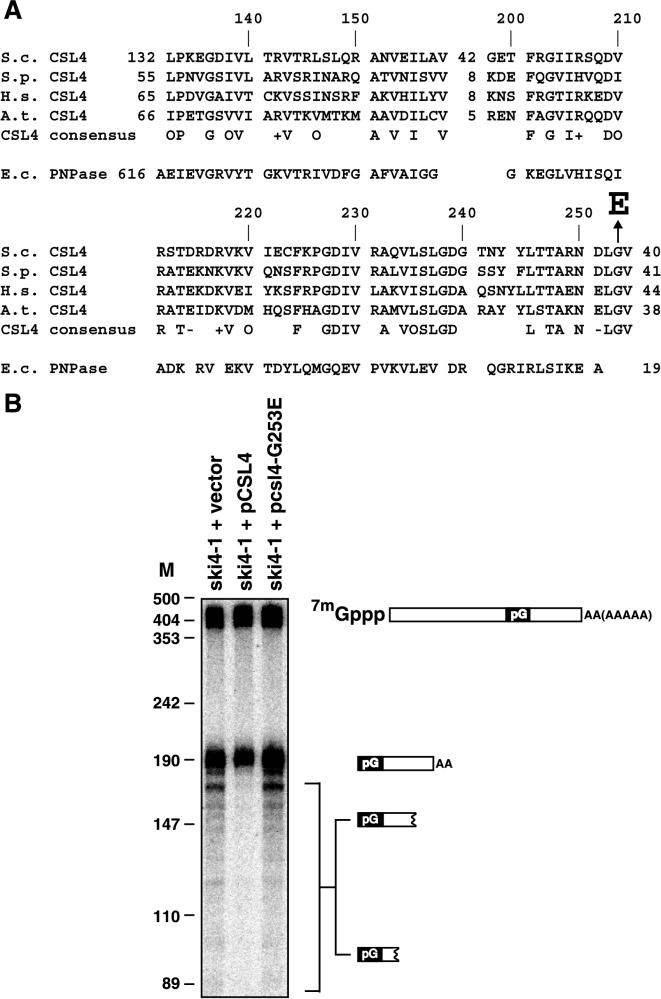
To further test the hypothesis that ski4-1 and csl4 are allelic and to identify the specific sequence change in ski4-1, we amplified the CSL4 gene from the ski4-1 strain by PCR. Sequence analysis of two independent PCRs each revealed that there were two amino acid residues changed in Csl4p. One of these (R65K) is a conservative substitution in the N-terminal part of the Csl4p. This N-terminal domain is apparently not conserved in any of the Csl4p homologs. The other amino acid change (G253E) affects a glycine that is conserved in Csl4p homologs from mammals, Schizosaccharomyces pombe, and plants and is part of the S1 RNA binding domain (Fig. 6A). We suspected that the latter change is responsible for the phenotypes of ski4-1. We tested this hypothesis by introducing a coding sequence change producing the G253E change into a plasmid containing a wild-type CSL4 gene. This pcsl4-G253E plasmid no longer was able to complement the RNA phenotype of a ski4-1 strain (Fig. 6B). Therefore, we conclude that the G253E change is responsible for the mRNA degradation defect of ski4-1. Based on the mapping, complementation, and sequencing data we conclude that the ski4-1 allele represents a mutation in the CSL4 gene.
FIG. 6.
A mutation in the conserved S1 RNA binding domain of Csl4p is responsible for the ski4-1 phenotypes. (A) Alignment of the S1 RNA binding domain of Csl4p (S.c.) with putative homologs from S. pombe (S.p.) (accession no. T41654), Homo sapiens (H.s.) (5), and Arabidopsis thaliana (A.t.) (deduced from genomic sequence under accession no. AB009048). Included is a consensus of residues conserved in all four CSL4 sequences (O, hydrophobic residue, I, L, V, or M; +, positively charged residue R, K, or H; −, negatively charged residue D or E). Also included is the alignment of the S1 RNA binding domain from Escherichia coli (E.c.) PNPase, which is the only S1 RNA binding domain for which the structure has been determined (11). The alignment was generated using a hidden Markov model for the S1 RNA binding domain (I. S. Mian, personal communication). The Arabidopsis sequence was added manually based on BLAST results after removal of putative introns. Arrow, G-to-E change in the product of ski4-1 and csl4-G253E (B) Polyacrylamide Northern blot probed with an oligonucleotide that detects the MFA2pG species indicated to the right. A ski4-1 strain was transformed with the indicated plasmids. The resulting strains were grown at 30°C in medium lacking uracil and containing 2% Gal. The length of HpaII fragments of pUC18 is given in nucleotides. M, nucleotides.
The ski4-1 allele genetically separates the functions of the exosome in rRNA processing and mRNA decay.
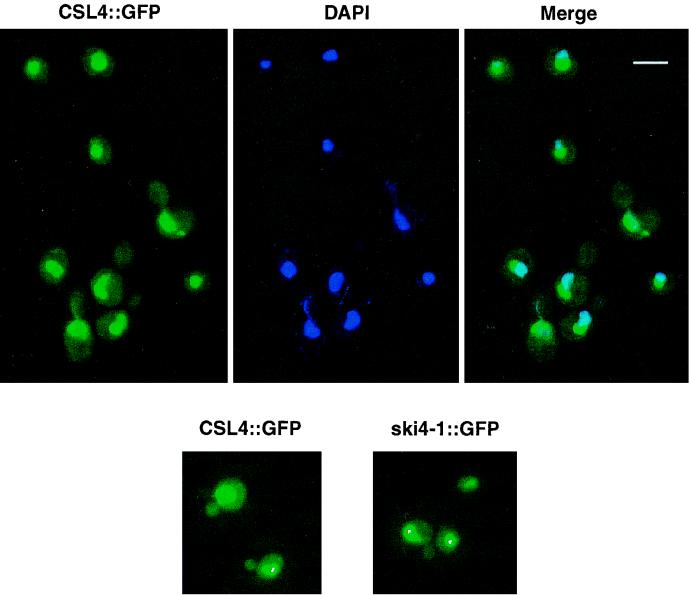
The Csl4p/Ski4p protein is a component of the exosome and is essential for viability (1, 5). Depletion of this protein from cells leads to a variety of defects including defects in 5.8S rRNA and U4 snRNA processing (1) (data not shown). Moreover, Csl4p/Ski4p copurifies with two different complexes that presumably correspond to the nuclear and cytoplasmic exosomes (1). That Csl4p/Ski4p is required for 5.8S rRNA processing reactions explains why CSL4/SKI4 is an essential gene. All these data suggest that Csl4p/Ski4p functions in both the nucleus and cytoplasm; however, we sought to confirm this hypothesis by localizing the Csl4p/Ski4p protein. We generated an N-terminal fusion of GFP to Csl4p/Ski4p. The GFP::CSL4 gene was integrated into the genome at the CSL4 locus under the control of the CSL4 promoter and 3′ untranslated region (see Materials and Methods). The resulting strains containing GFP-CSL4 as the sole source of Csl4p were viable and showed no detectable mRNA phenotype, indicating that the fusion protein was functional. The nucleus was highly fluorescent, and some fluorescence was also observed in the cytoplasm (Fig. 7). Within the nucleus, most of the fluorescence was in a region adjacent to the region stained with DAPI. This region of high fluorescence most likely corresponds to the nucleolus. We conclude that Csl4p/Ski4p is localized to the nucleus and to the cytoplasm, which is consistent with previous data on the function of the CSL4 gene and purification of the exosome (1). Therefore the observation that the ski4-1 mutation affects mRNA degradation by the exosome without affecting other reactions carried out by the exosome (see below) cannot be explained by Csl4p/Ski4p being present only in the cytoplasmic exosome. In addition, we localized GFP fused to the Ski4-1p. The localization of this mutant protein was indistinguishable from that of the wild-type GFP-Ski4p (Fig. 7). We conclude that the G253E mutation does not grossly alter the subcellular distribution of Csl4p/Ski4p.
FIG. 7.
Csl4p/Ski4p is localized in the nucleus and the cytoplasm. A gene encoding a GFP-CSL4 fusion protein was integrated into the genome at the CSL4 locus. This yielded GFP fused to wild-type Csl4p/Ski4p or to the mutant Ski4-1p as indicated. Cells were fixed and stained for DNA using DAPI. Bar, 5 μm.
The ski4-1 allele of CSL4/SKI4 is not lethal, and the cells grow normally under a variety of conditions, as do other mutants defective solely in 3′-to-5′ degradation of mRNA (e.g., the ski2Δ mutant). However, temperature-sensitive mutations in core exosome components, such as the ski6-100 mutation, and depletion of core exosome components, such as Csl4p, are known to inhibit several essential RNA-processing events. These events include 3′ trimming of 5.8S rRNA and U4 snRNA precursors (1, 2, 22, 34) (data not shown). In addition, depletion of Csl4p and other exosome components leads to accumulation of the 5′ ETS of the rRNA primary transcript (2). Given this, we determined if the ski4-1 allele of CSL4 causes a defect in either 5.8S rRNA or U4 snRNA processing or in the degradation of the 5′ ETS. As shown in Fig. 8, we observe no accumulation of either the 5′ ETS or 3′-extended forms of 5.8S rRNA or U4 snRNA in the ski4-1 strain. These data suggest that the ski4-1 lesion might disrupt the function of the Csl4p/Ski4p protein in mRNA decay but not its function in RNA processing.
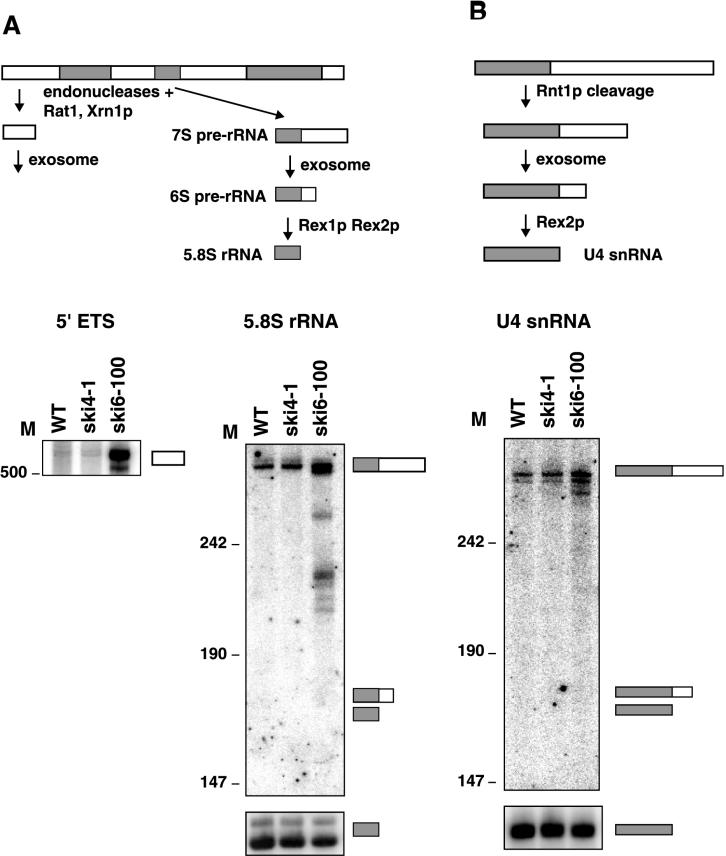
FIG. 8.
The ski4-1 mutation does not affect all Csl4p functions. (A) 5.8S rRNA processing and 5′ ETS degradation are not affected by ski4-1. Shown is a diagram of part of the processing pathway of the 35S pre-rRNA. This processing pathway yields the 5′ ETS (white box) and 5.8S rRNA (gray box), which are substrates for the exosome. The 35S pre-rRNA also yields 18S and 25S rRNA and several other spacer fragments, which are not shown. Also shown is a polyacrylamide Northern blot probed for 5′ ETS (left) and 3′ extended precursors of 5.8S rRNA (top right) and reprobed for the mature 5.8S rRNA (bottom right). The wild-type (WT) and ski4-1 strains were grown at 30°C. The ski6-100 strain was grown at 24°C and incubated for 1 h at 37°C. (B) U4 snRNA processing is not affected by ski4-1. Same as panel A except that U4 snRNA probes were used. The length of HpaII fragments of pUC18 is given in nucleotides. M, nucleotides.
The defect of ski4-1 in mRNA degradation but not in RNA processing can be explained in two ways. First, it is possible that the ski4-1 mutation effectively lowers the exosome activity and that mRNA degradation requires more activity than RNA processing. This hypothesis seems unlikely, because partial-loss-of-function mutations in other exosome components such as those encoded by mtr3-1, rrp4-1, ski6-2, and ski6-100 disrupt both mRNA degradation and RNA processing (1, 2, 17, 22, 34). Second, it is possible that the G253E substitution in the product of ski4-1 specifically affects mRNA degradation. To try to distinguish between these two possibilities, we characterized the defect in a second allele of CSL4/SKI4, csl4-1. The CSL4 gene was initially identified because the csl4-1 mutation is synthetically lethal with a deletion of the CEP1 gene, which encodes a transcription factor. The csl4-1 locus was amplified by PCR and sequenced. The only nucleotide change observed was a C-to-T substitution at position −172 of the CSL4 5′-flanking DNA. We verified that this mutation gives rise to the cep1Δ synthetic lethality phenotype of csl4-1 by introducing the same mutation into a wild-type CSL4 strain and testing synthetic lethality with cep1Δ by tetrad analysis. No viable double mutants were recovered in this cross. The csl4-1 mutation is a substitution in a putative Reb1p binding site in the promoter region of CSL4/SKI4. This substitution lowered the expression of a CSL4-LacZ fusion protein fivefold (data not shown; see Discussion). The identity of the specific sequence change, combined with the reduction in LacZ reporter activity, indicates that csl4-1 introduces a partial loss of function mutation in the promoter that reduces the expression of Csl4p and presumably also reduces the number of functional exosome complexes.
The csl4-1 mutation leads to a reproducible increase in the level of the 7S precursor to the 5.8S rRNA, indicating that the processing of 5.8S rRNA is affected by the csl4-1 mutation (Fig. 9). We did not observe any intermediates between 7S pre-rRNA and 5.8S rRNA in the csl4-1 strain like those seen in the ski6-100 strain (Fig. 8) and cells depleted of Csl4p (1). The signals for the 7S pre-rRNA were quantitated and normalized for the amount of RNA loaded using the RNA subunit of the SRP. The SRP RNA is not known to be processed by the exosome, and previous analysis has shown that its levels are not affected by various exosome mutations (34). This experiment revealed that 7S pre-rRNA was about 2.5-fold more abundant in the csl4-1 strain than in an isogenic wild-type strain. This result was confirmed by repeating the experiment with either the U6 snRNA or the RNA subunit of RNase MRP as the loading control. Like the SRP RNA subunit, these two RNAs have previously been shown not to be affected by exosome mutations (34). These results indicate that processing of 5.8S rRNA is affected by the csl4-1 mutation but that this processing is still relatively efficient. In contrast to the defect seen for 5.8S rRNA processing, we did not observe a defect in the degradation of the MFA2pG fragment under the same conditions (Fig. 9). However, we cannot exclude the possibility that csl4-1 has a small effect on 3′-to-5′ mRNA decay. In either case, these data show that a simple partial-loss-of-function mutation in CSL4/SKI4 affects rRNA processing more severely than mRNA degradation. The combination of the effects of csl4-1 and ski4-1 suggests that the ski4-1 mutation separates the function of Csl4p/Ski4p in mRNA degradation from its other functions.
The SKI7 gene is specifically required for mRNA degradation by the exosome.
The exosome is required both for 3′-to-5′ degradation of mRNA and for various RNA-processing reactions. In contrast, the Ski2p, Ski3p, and Ski8p proteins are specifically required for 3′-to-5′ degradation of mRNA. Given this we tested whether a ski7Δ strain showed defects in 5′-ETS degradation or processing of 5.8S rRNA or U4 snRNA. As shown in Fig. 10, these reactions are not affected by the ski7Δ mutation. We therefore conclude that Ski7p is specifically required for mRNA degradation by the exosome.
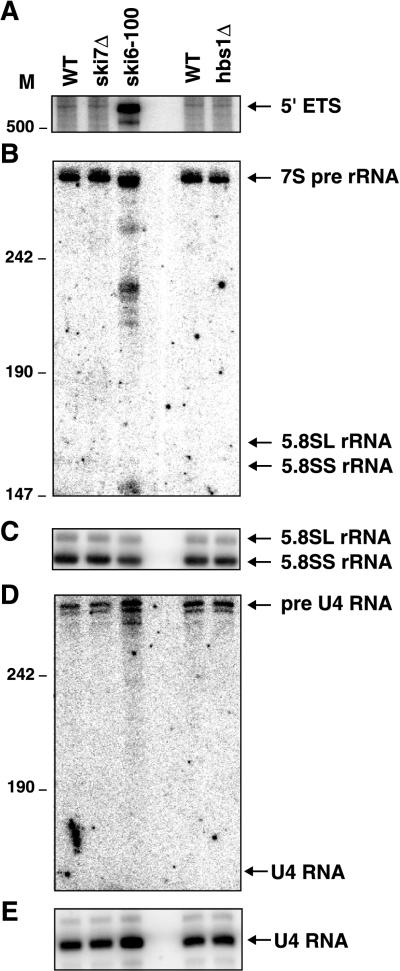
FIG. 10.
The ski7Δ and hbs1Δ mutations do not affect 5.8S rRNA or U4 snRNA processing or 5′ ETS degradation. Shown are polyacrylamide Northern blots. The wild type (WT), ski7Δ, and hbs1Δ strains were grown at 30°C in YEP medium containing 2% Gal. The ski6-100 strain was grown in the same medium at 24°C and incubated at 37°C for 1 h. (A) A Northern blot was probed for the 5′ ETS. (B) The Northern blot in panel A was stripped and reprobed for 3′ extended precursors of 5.8S rRNA. (C) The Northern blot in panel B was stripped and reprobed for 5.8S rRNA. (D) Northern blot probed for 3′ extended precursors of U4 snRNA. (E) The Northern blot in panel D was stripped and reprobed for U4 snRNA. M, nucleotides.
One possible reason why Ski7p is not required for RNA-processing reactions by the exosome is that another protein may replace Ski7p in this reaction. This would be similar to what has been found for Ski2p. Ski2p is only required for mRNA degradation, and Mtr4p, a close homolog of Ski2p, is required for all other exosome-mediated reactions. Interestingly, the HBS1 gene codes for a protein that is homologous to Ski7p. We therefore tested whether an hbs1Δ mutation affected 5′-ETS degradation or processing of 5.8S rRNA or U4 snRNA. As shown in Fig. 10, none of these reactions are significantly affected by a deletion of the HBS1 gene, suggesting that Hbs1p does not function in exosome-mediated RNA-processing reactions.
DISCUSSION
The Ski4p and Ski7p proteins are required for mRNA degradation.
Multiple observations indicate that both Ski4p and Ski7p are required for the degradation of mRNA through the 3′-to-5′ pathway. First, both ski4-1 and ski7Δ mutations result in a decrease in the 3′-to-5′ decay rate of mRNA. This is evident for full-length mRNA under conditions where the 5′-to-3′ decay pathway is blocked in trans, as well as for the poly(G)-to-3′-end fragment whose 5′-to-3′ degradation is blocked in cis. Second, both ski4-1 and ski7Δ mutations are synthetically lethal with mutations that block decapping. Third, the ski4-1 mutation is a mutation in the CSL4/SKI4 gene and the CSL4/SKI4 gene product is a component of the exosome (1) which is known to be required for 3′-to-5′ degradation of mRNA (17). Fourth, the ski7Δ mutation does not act synergistically with ski2Δ, ski3Δ, and ski8Δ mutations, indicating that all four mutations likely affect the same pathway.
Several functions for the Ski proteins have been proposed. We have previously suggested that the Ski proteins, with the exception of Ski1p, function directly in 3′-to-5′ mRNA degradation (17, 33, 34). An alternative explanation that has been proposed is that the Ski proteins function in ribosome biogenesis in the nucleolus, such that ski mutants contain altered ribosomes that have a reduced ability to discriminate between polyadenylated and nonadenylated mRNA molecules (7, 8, 21). According to this intriguing hypothesis, the altered translation of mRNAs would affect mRNA degradation as a secondary effect.
Several observations now combine to argue that the Ski2p, Ski3p, Ski7p, and Ski8p proteins and the exosome function directly in cytoplasmic mRNA degradation. First, the exosome has the required 3′-to-5′ exonucleolytic activity (22, 23). Second, the exosome and the Ski2p and Ski3p proteins are known to be localized to the cytoplasm (1, 10, 19, 23, 37). Third, the ski2Δ, ski3Δ, ski7Δ, ski8Δ, and exosome mutations all affect the degradation of a poly(G)-to-3′-end mRNA fragment, which is no longer translated at the time it is degraded (17). Fourth, the ski2Δ, ski3Δ, ski7Δ, ski8Δ, and ski4-1 mutations affect mRNA degradation without affecting any other known function of the exosome (17, 36). The simplest explanation of all of these observations is that the exosome is the catalytic complex that degrades mRNA 3′ to 5′ in the cytoplasm and that the Ski2p, Ski3p, Ski7p and Ski8p proteins function to allow or promote this exosome function. In this view the increased luciferase expression from electroporated unadenylated mRNA in ski mutants may reflect a competition between exosome-mediated degradation and entry into the translatable pool.
The role of Ski7p in mRNA degradation.
Ski7p has significant sequence similarity to translation elongation factor EF1A (8). However, it is not clear whether Ski7p has a role in translation. Interestingly, the sequence similarity of Ski7p and EF1A appears to be limited to their GTPase domains. The similarity between Ski7p and EF1A is reminiscent of what has been observed for Snu114p and EF2. EF2 is another GTPase involved in translation elongation, and Snu114p is very similar to EF2. In this case the homology is not limited to the GTPase domain but covers all of EF2 (15). Despite this extensive homology, Snu114p is part of the U5 snRNP particle and functions in pre-mRNA splicing, with no apparent role in translation (15). This argues that despite similarity to EF1A, Ski7p may not function in translation.
Ski7p belongs to the family of GTPases whose members can bind GTP and hydrolyze it to GDP. This transition from the GTP-bound state to the GDP-bound state is often associated with large conformational changes in the protein, resulting in alterations of protein-interacting and/or RNA-interacting surfaces (11). Therefore, one hypothesis is that Ski7p functions to bring two or more macromolecules together and uses GTPase activity as a source of energy for a conformational change. Under this hypothesis possible interacting molecules would be the substrate mRNA, Ski2p, Ski3p, Ski8p, and the exosome. Previous studies have failed to detect an interaction between Ski7p and the Ski2p-Ski3p-Ski8p complex (10). However, it appears likely that the interactions of Ski7p are dependent on whether it is in a GTP-bound state or a GDP-bound state, and therefore an interaction may only be detectable in vitro in the presence of GDP, GTP, or a GTP analog.
Why is Ski7p required for mRNA degradation by the exosome but not for 5.8S rRNA processing by the exosome? One possibility is that a different protein substitutes for Ski7p in 5.8S rRNA processing. This would be similar to what has been found for Ski2p and Mtr4p (see reference 33 for a review on the possible roles of Ski2p and Mtr4p). The most likely candidate for a second GTPase required for exosome function appeared to be Hbs1p. Hbs1p and Ski7p are more similar to each other over a longer stretch of both proteins than either protein is to other GTPases (8; our unpublished observations). However, an hbs1Δ strain does not have an obvious growth defect, as might be expected for a strain defective in rRNA processing, nor does it have an obvious defect in the processing of 5.8S rRNA or U4 snRNA or the degradation of the 5′ ETS of the 35S pre-rRNA. Therefore, it appears unlikely that Hbs1p plays a role in RNA processing by the exosome.
Implications for the function of Csl4p/Ski4p in mRNA degradation.
The observation that the ski4-1 mutation affects mRNA degradation by the exosome without affecting other functions of the exosome is strong support for a direct role of the exosome in mRNA degradation. There are several explanations why the one amino acid change in Csl4p could affect mRNA degradation specifically. One appealing model is that Csl4p/Ski4p is an RNA binding protein that specifically binds mRNA molecules that are substrates for the exosome. The mutation in the RNA binding domain of CSL4/SKI4 in the ski4-1 allele would alter the specificity or affinity of Csl4p/Ski4p for mRNA under this hypothesis. Alternatively the ski4-1 mutation might disrupt a protein interaction between Csl4p/Ski4p and some other protein. This protein-protein interaction would be required for mRNA degradation by the exosome. Interestingly, the ski4-1 mutation does not affect the degradation of the 5′ ETS of the pre-rRNA. Therefore the mutation disrupts an mRNA-specific interaction and not an interaction involved in distinguishing degradation substrates of the exosome from processing substrates.
Synthetic lethality of csl4-1 and cep1Δ mutations.
The csl4-1 mutation was originally identified in a screen for mutations that were synthetically lethal with the cep1Δ mutation. The CEP1 gene encodes the kinetochore protein/transcription factor CP1/Cbf1p (5). The identification of the csl4-1 mutation as a mutation in the CSL4/SKI4 promoter sheds light on the synthetic lethality of this mutation with a cep1Δ mutation. The csl4-1 mutation likely disrupts the binding of transcription factor Reb1p to the CSL4 promoter. Disruption of this binding is insufficient to affect the viability of the yeast strains (5). The CSL4/SKI4 promoter also contains a binding site for the general regulatory factor Cep1p. Disruption of this binding by deletion of the CEP1 gene or the Cep1p binding site also is insufficient to affect viability (5). However, we hypothesize that disruption of both Reb1p binding and Cep1p binding reduces expression of the essential CSL4/SKI4 gene so much that it no longer is sufficient to support viability. The importance of Reb1p and Cep1p binding was tested by analyzing the expression of a CSL4-LacZ fusion protein. The CSL4-LacZ reporter gene in this case contained either a wild-type CSL4 promoter or a promoter containing the csl4-1 mutation. This analysis showed that the csl4-1 mutation lowers the expression of the CSL4-LacZ fusion by about fivefold. Moreover, the cep1Δ mutation further lowers the expression of the reporter gene driven by the csl4-1 promoter to 29-fold below wild-type levels (R. E. Baker, unpublished results). Thus, the synthetic lethality of csl4-1 and cep1Δ mutations is likely the result of a strongly reduced expression of the essential CSL4/SKI4 gene.
ACKNOWLEDGMENTS
We thank Melanie Oakes and Masayasu Nomura for advice on nucleolar localization. We are grateful to I. Saira Mian for communicating the results of the S1 RNA binding domain hidden Markov modeling prior to publication, Reed Wickner for the ski4-1 strain, Mike Snyder for the CSL4-lacZ strain, and Phil Mitchell and David Tollervey for the gal::csl4 strain. We are grateful to members of the Parker laboratory for helpful comments on this work.
This work was supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ to 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson M T, Tjioe I M, Lorincz M C, Parks D R, Herzenberg L A, Nolan G P. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell using engineered green fluorescent proteins. Proc Natl Acad Sci USA. 1996;93:8508–8511. doi: 10.1073/pnas.93.16.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker R E, Masison D C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker R E, Harris K, Zhang K. Mutations synthetically lethal with cep1 target S. cerevisiae kinetochore components. Genetics. 1998;149:73–85. doi: 10.1093/genetics/149.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beelman C A, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 7.Benard L, Carroll K, Valle R C, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benard L, Carroll K, Valle R C P, Masison D C, Wickner R B. The Ski7 antiviral protein is an EF1-α homolog that blocks expression of non-poly(A) mRNA in Saccharomyces cerevisiae. J Virol. 1999;73:2893–2900. doi: 10.1128/jvi.73.4.2893-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourne H R. GTPases everywhere! In: Dickey B F, Birnbaumer L, editors. Handbook of experimental pharmacology. Berlin, Germany: Springer-Verlag; 1993. pp. 3–15. [Google Scholar]
- 10.Brown J T, Bai X, Johnson A W. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bycroft M, Hubbard T J, Proctor M, Freund S M, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 12.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Dunckley, T., M. Tucker, and R. Parker. Two related proteins, Edc1p and Edc2p, stimulate decapping in Saccharomyces cerevisiae. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 15.Fabrizio P, Laggerbauer B, Lauber J, Lane W S, Luhrmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs Anderson J S, Parker R P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson A W, Kolodner R D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita N, Goebl M, Yanagida M. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol Cell Biol. 1991;11:5839–5847. doi: 10.1128/mcb.11.12.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 21.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Decoying the cap− mRNA degradation system by a double-stranded RNA virus and poly(A)− mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 24.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 25.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poole T L, Stevens A. Structural modifications of RNA influence the 5′ exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1997;235:799–805. doi: 10.1006/bbrc.1997.6877. [DOI] [PubMed] [Google Scholar]
- 27.Ross-Macdonald P, Coelho P S, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung K H, Sheehan A, Symoniatis D, Umansky L, Heidtman M, Nelson F K, Iwasaki H, Hager K, Gerstein M, Miller P, Roeder G S, Snyder M. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 28.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 29.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toh-E A, Wickner R B. “Superkiller” mutations suppress chromosomal mutations affecting double-stranded RNA killer plasmid replication in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1980;77:527–530. doi: 10.1073/pnas.77.1.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tucker M, Parker R. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu Rev Biochem. 2000;69:571–595. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 33.van Hoof A, Parker R. The exosome: a proteasome for RNA? Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- 34.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickner R B. Prions and RNA viruses of Saccharomyces cerevisiae. Annu Rev Genet. 1996;30:109–139. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 36.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanchin N I, Goldfarb D S. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol Cell Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]