Dear Editor,
Our auditory organ, the cochlea, resides in the ventral portion of the inner ear, and its sensory epithelium, the organ of Corti, contains hair cells (HCs) and supporting cells (SCs), which are both descendants of the same progenitors. HCs are prone to damage by multiple ototoxic factors, and how to regenerate damaged HCs using key genes involving cochlear development is of importance [1, 2]. Cochlear progenitor cells express Sox2 and proliferate before embryonic day 12.5 (E12.5) in the mouse but become quiescent in an apical-to-basal gradient between E12.5 and E14.5 [3]. The development of cochlear HCs and SCs is regulated by many key genes including Atoh1 (atonal bHLH transcription factor 1) and other signaling pathways [4–6].
Prox1 is a homeobox transcriptional factor, homologous to the Drosophila Prospero, and is first detectable at approximately E14.5 but is restricted to two SC subtypes, pillar cells (PCs) and Deiters’ cells (DCs), at the perinatal stage, and Prox1 expression gradually declines postnatally, becoming undetectable at around postnatal day 14 (P14) [7]. Conditional deletion of Prox1 has been found to cause a reduction in the size of cristae and disorganized neuronal fibers in the cochlea [8]. However, the molecular mechanisms underlying Prox1 functions and its genome-wide DNA-binding sites in the cochlea remain unknown; this is partly because traditional chromatin immunoprecipitation-sequencing (ChIP-Seq) is not suitable for cochlear tissues, which harbor only a limited number of Prox1+ cells. An alternative to ChIP, CUT&RUN (Cleavage Under Targets and Release Using Nuclease) maps chromatin features via binding of a specific antibody to the target protein and cutting out target protein-bound DNA fragments, which are released into the supernatant [9, 10]. Without cross-linking, CUT&RUN minimizes problems with epitope masking and other artefacts. The improved signal-to-noise ratio and high efficiency allow it to generate high-quality data from low starting numbers of cells.
Here, we successfully mapped 1638 binding sites of Prox1 using the CUT&RUN technique and fresh cochlear epithelium collected from P1 wild-type mice. The results of functional enrichment analysis suggested several Prox1 functions, one being cell-cycle regulation, and a role of Prox1 in regulating the pool size of cochlear progenitors was supported by in vivo genetic evidence: ectopic Prox1 overexpression in the otocyst caused cochlear shortening. We have elucidated, for the first time, the in vivo DNA-binding sites of Prox1 in the mouse cochlea and have provided a key finding of a role of Prox1 in cell-cycle regulation by using genetic gain-of-function analysis. Our work facilitates future mechanistic studies of other transcription factors expressed in rare tissues such as the cochlea.
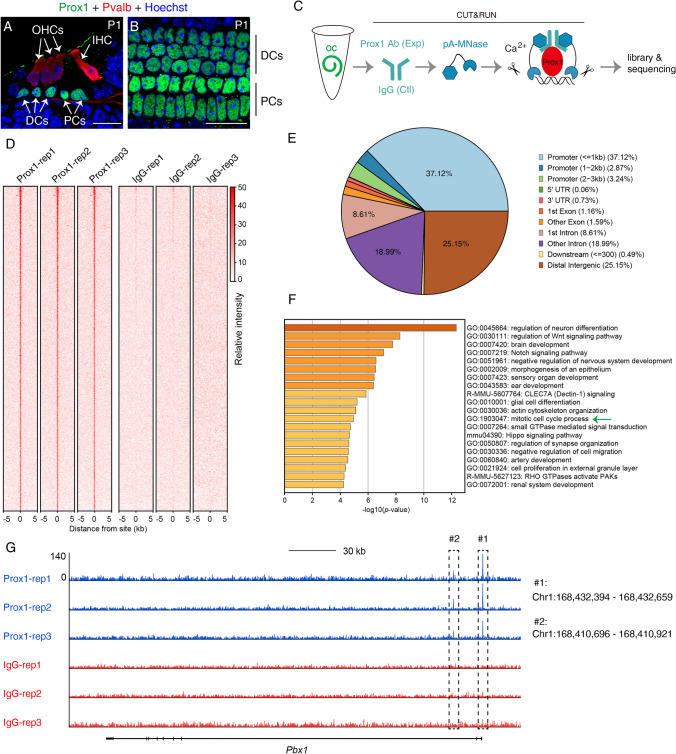
Prox1 is abundantly expressed in two SC subtypes, PCs and DCs, in the organ of Corti at P1 (Fig. 1A, B). Considering the extremely limited numbers of Prox1+ cells present in the cochlea, we applied CUT&RUN [9, 10], a highly sensitive tool for analyzing protein-DNA binding, to map the genome-wide binding sites of Prox1 in PCs and DCs. We dissected out fresh tissues from the organ of Corti region and immediately used the samples in CUT&RUN assays in which Prox1 antibody (experimental group) and mouse IgG (control group) were applied in parallel (Fig. 1C); each group included three replicates, with four cochlear samples (from 2 mice) included per replicate. Here, spiral ganglion neurons were also noted to express Prox1 at P1, but they were excluded from the study because our aim was to selectively identify genuine Prox1-binding sites in PCs and DCs. In the three replicates in the experimental group, 1638 binding sites were captured, whereas minimal signal was detected in the three replicates in the control group (Fig. 1D). Supplemental Table 1 lists all identified Prox1-binding sites. The binding sites were highly enriched in promoters, with approximately 43% of the peaks located at promoter regions (Fig. 1E). Intriguingly, Prox1 itself was also included among the 1638 binding sites, which suggests a potential autoregulation mechanism. Gene-function enrichment analysis revealed multiple functions of Prox1 targets, including sensory organ development, ear development, Notch signaling, and mitotic cell-cycle progression (green arrow in Fig. 1F). Collectively, our results showed that we had successfully captured the genome-wide binding sites of endogenous Prox1 in a limited sample of cochlear cells by using CUT&RUN.
Fig. 1.
Genome-wide search for Prox1-binding sites in neonatal PCs and DCs at P1. A, B Co-staining of Prox1 and parvalbumin (Pvalb) in a cryosection (A) and a whole-mount (B) cochlear sample. Pvalb, a pan-HC marker, is expressed in both IHCs and OHCs. Prox1 is exclusively expressed in PCs and DCs. IHC, inner hair cell; OHCs, outer hair cells; DCs, Deiters’ cells; PCs, pillar cells.; scale bars, 20 μm. C Key CUT&RUN experimental procedures. D Peak distribution of Prox1-binding sites (all 1638 binding sites detected) in the mouse genome (mm10) in three control (mouse IgG) and three experimental (Prox1 antibody) groups. E Summary of distribution of Prox1-binding sites among distinct genomic regions. F GO analysis of genes associated with Prox1-bound peaks. G The gene Pbx1 was selected for visualizing peaks in three control (IgG, red) and experimental (Prox1 antibody, blue) replicates.
To further validate the identified binding sites of Prox1, we generated a new conditional Prox1-overexpressing knock-in mouse model: Rosa26-CAG-Loxp-stop-Loxp-Prox1.3*V5/+ (Rosa26-LSL-Prox1/+ for short; Figs 2A and S1). Pax2Cre is active in the otocyst and serves as the driver for turning on the expression of ectopic Prox1 harboring the V5-tag at its C-terminus [11]. Pax2Cre+; Rosa26-LSL-Prox1/+ mice constituted the experimental group, and Rosa26-LSL-Prox1/+ mice were the control group. At E9.5, Sox2+ otocyst cells in the control group did not express ectopic Prox1 and were V5– (Figs 2B and S2A), whereas numerous Sox2+ otocyst cells in the experimental group were V5+ (arrows in Figs 2C and S2B). Together, these results confirmed that ectopic Prox1 was successfully and permanently induced in otocyst cells.
Fig. 2.
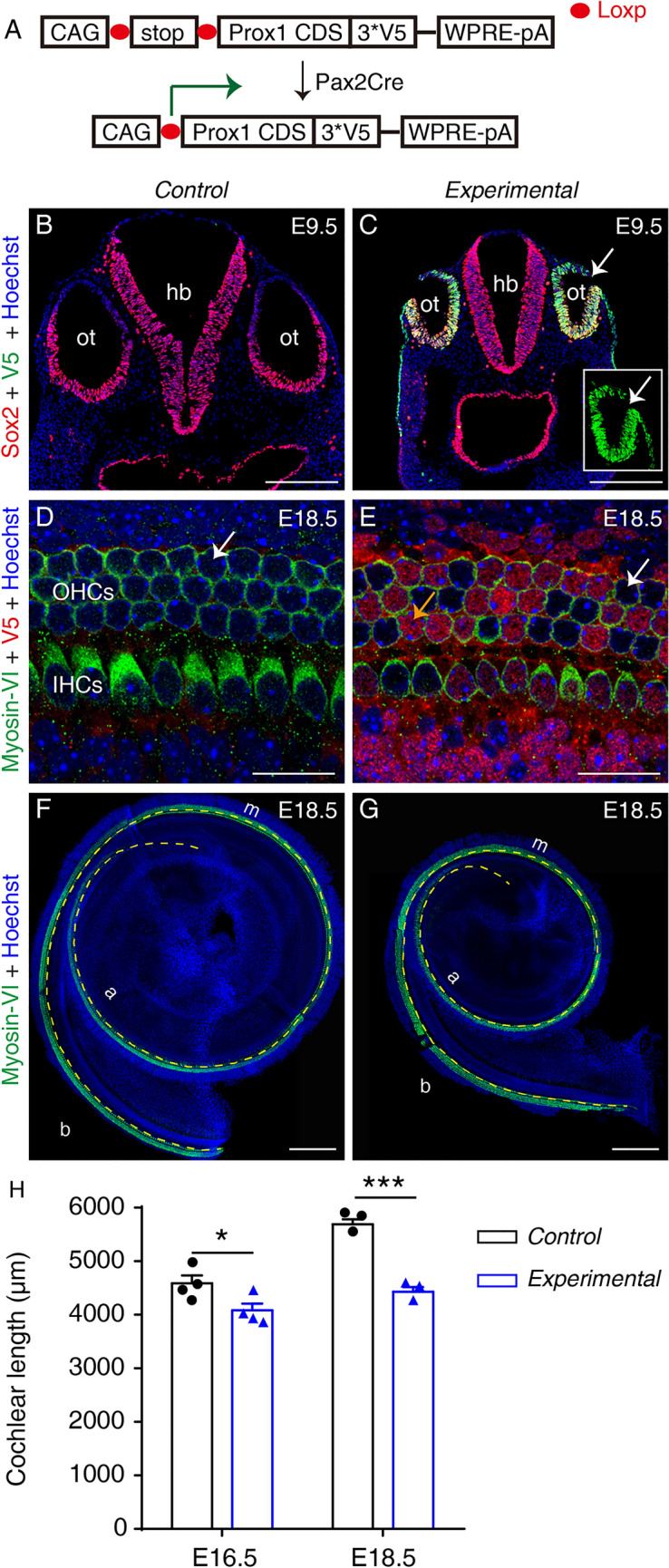
Ectopic Prox1 expression causes cochlear shortening without disrupting HC differentiation by E18.5. A Design of conditional Prox1-overexpression mouse model. Pax2Cre turns on ectopic Prox1 in mouse otocyst cells. B, C Double staining of Sox2 and V5 (Prox1) in the otocyst (ot) in a control Rosa26-LSL-Prox1/+ mouse (B) and an experimental Pax2Cre+; Rosa26-LSL-Prox1/+ mouse (C). Ectopic V5-tagged Prox1 is detectable in experimental but not control otocyst cells (Sox2+). Inset in (C) is the same otocyst (arrow in C) but visualized in V5 panel only. D, E Double staining of Myosin-VI and V5 (Prox1) in whole-mount cochlear samples of a control (D) and an experimental (E) mouse. At E18.5, no HCs in cochlear samples from control mice express V5-tagged Prox1 (arrow in D), whereas in the cochlear sample from an experimental mouse, numerous HCs and other cells (descendants of Pax2+ progenitors) are V5+ (orange and white arrows in E indicate V5+ and V5– HCs). The weak red signal in D is background and the control sample is negative for V5. F, G Cochlear samples from a control (F) and an experimental (G) mouse stained for Myosin-VI, a pan-HC marker (yellow dashed line between the inner and outer hair cells is used to measure cochlear length). H Comparison of cochlear length between control and experimental mice at E16.5 and E18.5. At both ages, the cochlear duct is significantly shorter in experimental than in control mice. Data are presented as means ± SEM. *P <0.05, ***P <0.001. IHCs, inner hair cells; OHCs, outer hair cells; b, basal; m, middle; a, apical; ot, otocyst; hb, hindbrain; scale bars, 200 μm (B, C, F, G), and 20 μm (D, E).
Prox1 was previously shown to antagonize HC formation partially by blocking Atoh1 activity [12, 13]; thus, we initially expected HCs to be absent from the experimental group or to be present at a lower density than in the control group. Unexpectedly, HC differentiation was normal in both groups (Fig. 2D, E). At E18.5, all Myosin-VI+ HCs in the control group were V5– (white arrow in Fig. 2D), while both Myosin-VI+/V5+ HCs (orange arrow in Fig. 2E) and Myosin-VI+/V5– HCs (white arrow in Fig. 2E) were present in the experimental group. We did not analyze samples at older ages because the experimental mice did not survive after birth. Nevertheless, our results supported the conclusion that HC fate specification and early differentiation were normal at birth, but additional genetic models will be required to elucidate the long-term effects of permanent Prox1 overexpression in HCs.
Although general HC fate specification was normal following ectopic Prox1 expression, the cochlea was significantly shorter in the experimental group than in the control group: at E18.5, cochlear length was 5691.7 ± 93.5 μm in the control group, but only 4432.0 ± 85.5 μm in the experimental group (n = 3 mice/group; P <0.001) (Fig. 2F–H). To further confirm this phenotype, we measured cochlear length in both groups at E16.5, and again found that the cochlea was longer (P <0.05) in the control group (4589.3 ± 146.7 μm; n = 4 mice) than in the experimental group (4083.5 ± 125.3 μm; n = 4 mice) (Fig. 2H). The statistical difference was greater at E18.5 likely because the cochlear duct undergoes extension at late embryonic ages [14].
How can we interpret the shorter cochlear duct and fewer HCs when Prox1 is overexpressed in otocysts? Otic epithelial cells, which are progenitors of the cochlear and vestibular organs, are highly proliferative and express no or very low Prox1 (below the detection threshold). Therefore, our first speculation is that otic epithelial cells are very sensitive to ectopic Prox1 and become less proliferative, causing decreased total cell numbers in the otocyst. This speculation was supported by the smaller size of the otocyst in the experimental than in the control group, as visualized by the analysis of both transverse (Fig. 2B, C) and sagittal (Figs. S2A and S2B) sections. The smaller otocyst would subsequently contain a decreased pool of progenitors that would develop into the cochlear duct. Thus, the cochlear duct would become shorter.
Our second speculation was that ectopic Prox1 also leads to a precocious cell-cycle exit of cochlear progenitor cells. To test this hypothesis, we gave EdU to mice at E12.5, and analyzed cochlear samples at E18.5 (Fig. S2C–D’’). In contrast to our prediction, there was no statistical difference in the percentage of EdU+ HCs between the control and experimental groups (Fig. S2E). This suggests that the expression of p27, a gene critical for the cell-cycle exit of cochlear progenitors [3], is not significantly affected, which agrees with the finding that no Prox1-binding sites are located near the p27 locus. Thus, our data suggest that the first speculation might account for the shortened cochlea. In other words, ectopic Prox1 caused smaller otocysts and subsequently the initial cochlear progenitor pool was decreased but the proliferation capacity of cochlear progenitors at E12.5 did not significantly change.
In one study, Prox1 protein was reported to be undetectable in the cochlea until E14.5 when it is expressed in a basal-to-apical gradient [7]. However, the opposite pattern was described in another report [8]. This difference may be due to the sensitivities of different methods of detection. We speculate that Prox1 might be expressed at a low level (below the detection threshold in immunostaining) in cochlear progenitors before E12.5 or between E12.5 and E14.5. Prox1 expression at such a low level might not be present in the nucleus and could be diffusely distributed in the cytoplasm, as in the case of its homolog Prospero in Drosophila neural stem cells [15]. Lastly, among the Prox1-targeted genes relevant to cell-cycle regulation (Supplemental Table 2), which gene might be the key target of Prox1? Pbx1 is expressed in the cochlea and is regulated by Six1 [16]. The Pbx1 sequences bound by Prox1 included the transcription start site (#1 in Fig. 1G) and an element downstream of the promoter (#2 in Fig. 1G) that was shown to be also bound by Six1 (Fig. 1G). Therefore, Pbx1 is highly likely to be directly regulated by Prox1. Pbx1 appears to play a general role in promoting cell proliferation, because Pbx1–/– mice display hypoplasia in multiple organs, including the pancreas [17]. The detailed genetic interactions among Prox1 and Pbx1 warrant further investigation.
In summary, we have demonstrated that CUT&RUN can be successfully used to search for the genome-wide binding sites of transcription factors in limited numbers of cells, such as cochlear cells. Furthermore, the Prox1-binding sites that we identified were supported by the results of our in vivo Prox1 genetic gain-of-function analysis. We believe that these methods will serve as powerful tools for deciphering the gene-regulatory networks that control the development of small organs or tissues such as the cochlea.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Qian Hu and the Optical Imaging Facility at the Institute of Neuroscience (ION) for support with image analysis, and the Embryology Department of the Animal Center at ION, Chinese Academy of Sciences, for in vitro fertilization. The Rosa26-LSL-Prox1/+ knock-in mouse was generated by Beijing Biocytogen Co., Ltd. We thank Dr. Steve Henikoff from the Fred Hutchinson Cancer Research Center for kindly providing pA-MNase. This research was supported by the State Key Laboratory of Molecular Developmental Biology (2019-MDB-KF-12), the National Key R&D Program of China (2017YFA0103901), the Strategic Priority Research Program of the Chinese Academy of Science (XDB32060100), the National Natural Science Foundation of China (81771012), a Shanghai Municipal Science and Technology Major Project (2018SHZDZX05), and the Innovative Research Team of High-Level Local Universities in Shanghai (SSMU-ZLCX20180601).
Footnotes
Zhengnan Luo and Jixiang Zhang have contributed equally to the work.
Contributor Information
Falong Lu, Email: fllu@genetics.ac.cn.
Zhiyong Liu, Email: Zhiyongliu@ion.ac.cn.
References
- 1.Xiong H, Lai L, Ye YY, Zheng YQ. Glucose protects cochlear hair cells against oxidative stress and attenuates noise-induced hearing loss in mice. Neurosci Bull. 2021;37:657–668. doi: 10.1007/s12264-020-00624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson PJ, Huarcaya Najarro E, Sayyid ZN, Cheng A. Sensory hair cell development and regeneration: Similarities and differences. Development. 2015;142:1561–1571. doi: 10.1242/dev.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YS, Liu F, Segil N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development. 2006;133:2817–2826. doi: 10.1242/dev.02453. [DOI] [PubMed] [Google Scholar]
- 4.Tateya T, Sakamoto S, Ishidate F, Hirashima T, Imayoshi I, Kageyama R. Three-dimensional live imaging of Atoh1 reveals the dynamics of hair cell induction and organization in the developing cochlea. Development 2019, 146: dev177881. [DOI] [PubMed]
- 5.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 6.Jovanovic S, Milenkovic I. Purinergic modulation of activity in the developing auditory pathway. Neurosci Bull. 2020;36:1285–1298. doi: 10.1007/s12264-020-00586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, Hayashi T. Expression of Prox1 during mouse cochlear development. J Comp Neurol. 2006;496:172–186. doi: 10.1002/cne.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritzsch B, Dillard M, Lavado A, Harvey NL, Jahan I. Canal cristae growth and fiber extension to the outer hair cells of the mouse ear require Prox1 activity. PLoS One. 2010;5:e9377. doi: 10.1371/journal.pone.0009377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 2017, 6: e21856. [DOI] [PMC free article] [PubMed]
- 10.Meers MP, Bryson TD, Henikoff JG, Henikoff S. Improved CUT&RUN chromatin profiling tools. Elife 2019, 8: e46314. [DOI] [PMC free article] [PubMed]
- 11.Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- 12.Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirjavainen A, Sulg M, Heyd F, Alitalo K, Ylä-Herttuala S, Möröy T, et al. Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Dev Biol. 2008;322:33–45. doi: 10.1016/j.ydbio.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Montcouquiol M, Kelley MW. Development and patterning of the cochlea: From convergent extension to planar polarity. Cold Spring Harb Perspect Med 2020, 10: a033266. [DOI] [PMC free article] [PubMed]
- 15.Lai SL, Doe CQ. Transient nuclear Prospero induces neural progenitor quiescence. Elife 2014, 3: e03363. [DOI] [PMC free article] [PubMed]
- 16.Li J, Zhang T, Ramakrishnan A, Fritzsch B, Xu J, Wong EYM, et al. Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium. Nucleic Acids Res. 2020;48:2880–2896. doi: 10.1093/nar/gkaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SK, Selleri L, Lee JS, Zhang AY, Gu XY, Jacobs Y, et al. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat Genet. 2002;30:430–435. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.