Abstract
Many biological systems are composed of diverse single cells. This diversity necessitates functional and molecular single-cell analysis. Single-cell protein analysis has long relied on affinity reagents, but emerging mass-spectrometry methods (either label-free or multiplexed) have enabled quantifying >1,000 proteins per cell while simultaneously increasing the specificity of protein quantification. Here we describe the Single Cell ProtEomics (SCoPE2) protocol, which uses an isobaric carrier to enhance peptide sequence identification. Single cells are isolated by FACS or CellenONE into multiwell plates and lysed by Minimal ProteOmic sample Preparation (mPOP), and their peptides labeled by isobaric mass tags (TMT or TMTpro) for multiplexed analysis. SCoPE2 affords a cost-effective single-cell protein quantification that can be fully automated using widely available equipment and scaled to thousands of single cells. SCoPE2 uses inexpensive reagents and is applicable to any sample that can be processed to a single-cell suspension. The SCoPE2 workflow allows analyzing ~200 single cells per 24 h using only standard commercial equipment. We emphasize experimental steps and benchmarks required for achieving quantitative protein analysis.
Introduction
Biological systems, such as the tissues of multicellular organisms, are composed of diverse cell types and states1-4. This cellular diversity is well appreciated and has motivated the fruitful development of numerous analytical approaches for analyzing individual cells2-5. Indeed over the last decade, multiplexed approaches for single-cell transcriptomics have scaled to detecting the transcripts from thousands of genes across many thousands of single cells5. Single-cell transcriptomics methods are proving useful in understanding fundamental and clinical problems, such as the interaction of cancer and immune cells6 and cancer drug resistance7. Despite this progress, the pervasive posttranscriptional regulation across human tissues8 cannot be characterized based on nucleic acids analysis alone and has motivated methods for analyzing proteins in single cells9.
Traditionally, single-cell protein analysis has relied primarily on affinity reagents, such as antibodies and aptamers3, while the powerful mass-spectrometry (MS) methods that afford comprehensive quantification of cellular proteomes have been limited to the analysis of bulk samples composed of many cells10-14. Recently, new MS methods have been developed for quantifying thousands of proteins in individual human cells as reviewed in refs. 15,16. These single-cell MS methods hold much potential to facilitate the characterization of molecular mechanisms of health and disease9.
Development of SCoPE2
Single Cell ProtEomics (SCoPE2) is a second-generation method enabled by concepts and approaches introduced by its first-generation method (Single Cell ProtEomics by Mass Spectrometry; SCoPE-MS17) and new ones introduced with SCoPE218.
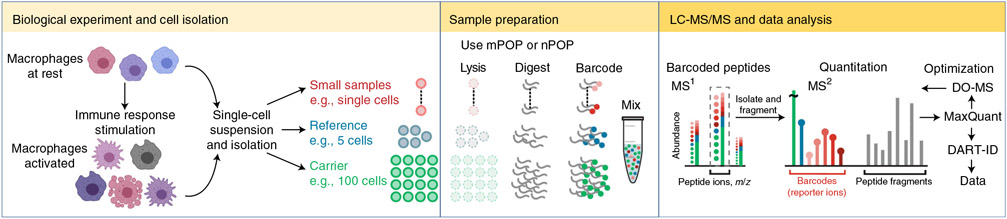
The key feature of SCoPE2 is the isobaric carrier concept19 (Fig. 1). Tandem mass tags (TMTs) are used to label proteins from a carrier sample and a reference sample as well as the single cells. The ratio of cells for each of these labeled samples is, for example, 100:5:1. The 16-plex TMTpro allows analysis of 12 single cell samples at a time, as two TMT labels ideally should be skipped due to isotopic impurities of the tag used for labeling the carrier. The proteins are identified using peptides generated from all the cells that are lysed in this mix, and the amount of each protein labeled with each TMT label is estimated from the amount of isobaric tag present in the spectrum.
Fig. 1 ∣. Single-cell proteomics with SCoPE2.
Single cells differ in biologiucal functions and in molecular composition, as illustrated here with macrophages. The cells from such heterogeneous populations can be converted into single-cell suspensions, individual cells isolated by FACS sorting, CellenONE or another method. The isolated single cells are lysed, the proteins digested and the resulting peptides labeled with TMTs. All of these stages can be automated and performed in parallel by either mPOP and nPOP. The labeled peptides are mixed and analyzed by LC-MS/MS with parameters optimized by DO-MS. The confidence in correct pepide identification can be further increased by tools, such as DART-ID.
Labeling peptides from individual cells with isobaric mass tags and combining them with ‘isobaric carrier’ peptides derived from a larger number of cells results in less surface adsorption loss of single-cell peptides and enhanced peptide sequence identification due to the presence of a higher concentration of peptide fragments19,20. Importantly, the isobaric carrier approach allows experiments to be designed to maximize either depth of proteome coverage or to maximize copies of ions sampled per peptide19. The isobaric carrier approach has been adopted by multiple laboratories for ultrasensitive MS analysis of single cells and other small samples (reviewed in ref. 15).
To enable inexpensive and robust single-cell proteomics, SCoPE2 built upon key ideas of SCoPE-MS and introduced many technological and analytical improvements, including the data analytics for optimizing experimental designs and enhancing data interpretation18.
One of the requirements of this system is that cell lysis is done by approaches that obviate clean-up and associated sample losses. SCoPE-MS achieved this using adaptive focused acoustics17, while SCoPE2 uses a freeze–heat lysis method called Minimal ProteOmic sample Preparation (mPOP)21. Because mPOP requires smaller volumes, it is possible to perform the freeze–heat cycle that enables lysis of cells in multiwell plate formats; this allows high-throughput, automation of the sample preparation steps18,21,22.
Recently, we introduced a droplet method, nano-ProteOmic sample Preparation (nPOP), that enabled an automated and simultaneous preparation of hundreds of single cells in 20 nl droplets23. As a whole, the second-generation method, SCoPE2, has increased throughput and quantitative accuracy while lowering the cost and the barriers to adoption15,18.
We also enhanced the means of normalizing single-cell quantification data by the inclusion of a five-cell reference channel, prepared in bulk and common across all SCoPE2 sets from an experiment18. The reference is usually a diluted sample from the same cell population that is used for the carrier, and is used for the purpose of normalizing for MS run-to-run variation. Such variation is induced by difference in the position of apex sampling, ionization and/or fragmentation efficiencies.
Improved data analytics in SCoPE2
Some SCoPE2 spectra allow quantification of the corresponding peptides but do not contain enough peptide fragments to support confident sequence identification. To help recover these additional peptides, we introduced the DART-ID software available from dart-id.slavovlab.net. It implements a false discovery rate (FDR) controlled Bayesian update of peptide confidence of identification by employing informative features of peptides, for example, its aligned retention time across many liquid chromatography tandem mass spectrometry (LC-MS/MS) runs24.
To optimize instrumentation settings for SCoPE2 sample analysis, we developed an approach to data-driven optimization of MS (DO-MS)25. This approach is implemented by extendable Shiny interface freely available from do-ms.slavovlab.net. DO-MS allows the analysis and optimization of LC-MS/MS experiments, with particular utility for ultrasensitive proteomics as employed in SCoPE218,25.
In this protocol, we provide a detailed description of the principles and experimental steps that allow adoption and application of SCoPE2 to different systems. We have already published software tools24,25 and experimental guidelines19 to aid in the adoption of SCoPE2. However, fulfilling its potential for making quantitative measurements requires a comprehensive guide to designing, implementing and benchmarking single-cell MS analysis. Here, we aim to provide such a guide, emphasizing the principles that enable quantitative SCoPE2 analysis and tailoring it to different experimental constraints and scientific aims.
Applications of SCoPE2
Any samples of primary tissues or cell cultures that can be prepared as a suspension of single cells can be analyzed by the SCoPE2 protocol described here. The single-cell suspensions can be prepared by methods used for single-cell RNA-seq. While most methods should be applicable to preparing single cells for SCoPE2 analysis, some methods, such as protease treatments, might affect cell surface proteins. Furthermore, the SCoPE2 protocol described here and its principles for ultrasensitive analysis may be applied successfully to other small samples, such as biopsies.
Comparison with other methods for single-cell protein analysis
Single-cell methods for investigating protein levels have existed for decades in the form of technologies employing affinity reagents (such as antibodies) and fluorescent proteins3. These classical approaches either employ antibodies against epitopes of interest, or use modified cells expressing fluorescent fusion proteins or reporters for a protein of interest. These methods therefore require antibodies that vary in their specificity, or engineering fluorescent fusion proteins or reporters that may influence the activity of a protein of interest or its modified host cell3. Both approaches are limited in the number of proteins that can be analyzed to about one to ten target proteins. This limit has been relaxed to ~50–100 target proteins by advanced methods barcoding the affinity reagents. An example of such barcoding includes cytometry by time of flight (CyTOF), which uses antibodies conjugated to rare-earth metals26. Other examples include approaches such as the RNA expression and protein sequencing assay (REAP-seq) and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)that use DNA oligo-linked antibodies, and permit higher multiplexing, as well as the possibility of obtaining both single-cell level RNA and protein data3. The limited specificity of affinity reagents can be mitigated by targeting multiple epitopes per proteins, as implemented by proximity extension and ligation assays27,28, or by using additional features of the protein, as implemented by single-cell western blots29. These advances have enabled approaches based on affinity reagents to quantify ~100 proteins per single cell. However highly specific antibodies and antibody validation remain required and sometimes challenging steps for these approaches.
MS-based approaches offer an alternative, bringing the promise of quantifying orders of magnitude more proteins, without the time and expense of obtaining and qualifying specific antibodies, or the potential to inadvertently disrupt normal protein function through generation of fusion fluorescent proteins9. Two main types of MS methods have been introduced, with some examples listed in Table 1: (i) multiplexed approaches employing isobaric carriers, including SCoPE-MS17 and SCoPE218,30 discussed here, and label-free approaches31-35. The label-free approaches often seek to miniaturize traditional proteomic sample preparation to enable processing of individual single cells with minimal sample losses due to the sub-microliter sample preparation volumes used by approaches such as nanodroplet processing in one pot for trace samples (nanoPOTS)36,37 and oil air droplet (OAD)38. Recently, some of these approaches have been automated39; for comprehensive reviews, see refs. 15,16. Each single-cell lysate is then digested and analyzed individually by LC-MS/MS, usually using MS1-based quantification. In contrast, the SCoPE2 protocol presented here sought to minimize losses during chromatography by using an isobaric carrier and to avoid the losses inherent in sample cleanup procedures through the use of clean lysis. Importantly, the SCoPE2 approach uses sample multiplexing, so that protein identification can be performed on material pooled from multiple cells, rather than a single cell, with TMTpro 16-plex reagents permitting multiplexed analysis of 12–14 single cells per experiment. Detailed reviews of current MS methods to conduct single-cell proteomics can be found in refs. 15,16.
Table 1 ∣.
Examples of MS methods used for analyzing proteins in single cells
| Sample preparation |
Cell types | No. of single cells |
Total number of proteins |
MS instruments | Ref. |
|---|---|---|---|---|---|
| Isobaric carrier | |||||
| mPOP | U937 monocytes and macrophages | 1,490 | 3,042 | Q Exactive | 18 |
| nanoPOTS | C10, RAW and SVEC cells | 61 | 1,225 | Q Exactive plus hybrid quadrupole-Orbitrap | 37 |
| nPOP | HeLa and U937 cells | 938 | 2,756 | Q Exactive | 23 |
| Label-free | |||||
| nanoPOTS | HeLa, human motor neurons (MN) and interneurons (IN) | 6 | 1,012 (MN) 1,085 (IN) |
Orbitrap Tribrid Eclipse with FAIMS | 34 |
| T-SCP | HeLa cells | 430 | 1,536 | timsTOF SCP | 35 |
| OAD | HeLa, mouse oocytes | 6 | 51 from HeLa 355 from oocyte | Orbitrap Elite | 38 |
The table summarizes examples of label-free and multiplexed (isobaric carrier) methods, highlighting the sample preparation method, total number of successfully analyzed cells, and total number of proteins quantified across many single cells; the number of proteins quantified per cell is smaller. Some studies were not included because of space limitations.
Experimental design
Design considerations familiar to scRNAseq practitioners are also applicable to SCoPE2 experiments40. Those parameters in common include the scale of the experiment and distributing populations of interest across batches to minimize the impact of batch effects. Similarly, many of the downstream types of data analysis steps, such as batch correction, imputation and dimensionality reduction, are similar to those used for processing scRNAseq data. Indeed, packages developed for performing procedures such as single-cell multi-omics dataset alignment and pseudotime or trajectory inference are compatible with single-cell proteomics. For example, we used Conos41 for joint analysis of RNA and proteins18.
We needed to implement slight adaptations to Conos to import SCoPE2 data. Similarly, other methods might need minor adjustments. Furthermore, methods developed for scRNA-seq do not take advantage of specific features to single-cell proteomics data15,42. These design considerations are common to many different forms of single-cell proteomics data analysis and are discussed extensively elsewhere40,43. There are, however, some parameters that are specific to, or should be considered differently for, SCoPE2-based single-cell proteomics analysis (Table 1).
Single-cell isolation: similar to scRNAseq, it is necessary to disaggregate complex samples or tissues prior to SCoPE2 analysis in order to obtain a single-cell suspension that can then be further processed, for example, by FACS or CellenONE. While enzymatic treatment such as the use of trypsin or accutase is common for cell disaggregation methods generally, this can influence the surface proteome, so the method of choice may differ between scRNAseq and SCoPE2 sample preparation depending on the sample and experimental question.
Single-cell population selection: enrichment of rare, or specific, subpopulations of interest is often performed for scRNAseq, but it is especially important for SCoPE2 experiments. It is challenging for SCoPE2 experiments to reach the very high numbers of cells that current droplet-based scRNAseq methods can yield. Enriching particular subpopulations of interest is a useful strategy to help mitigate this issue, but will influence the selection of carrier and reference proteome composition.
Carrier and reference composition: single-cell sampling for SCoPE2 experiments can allow for enrichment of specific subpopulations of interest. However, when performing SCoPE2 experiments in a data-dependent manner, the carrier and reference channels should be an even mix of the populations of interest or as close to it as is achievable. Uneven representation in the carrier or reference could result in failure to detect proteins only represented in particular subpopulations. In the SCoPE2 manuscript, we studied proteomic heterogeneity in macrophages and monocytes. Therefore, both the carrier and reference were an even mixture of the selected cell types. If we had wanted to study a third cell type, we would have again evenly mixed the cell types, with 33% of each cell type contributing to the carrier and the reference.
One potential mechanism to alleviate this issue is to spike specific peptides of interest into both carrier and reference channels; synthetic peptides can be obtained from many vendors, including Thermo Fisher. Ideally, the spiked-in peptides should be added to the cells during the isolation step prior to mPOP or right before digestion. They should be added at the level of tens of thousands of molecules per single cell, to accurately reflect the endogeneous peptide levels in a single cell. Such spike-ins will ensure that those peptides are sent for MS2 analysis. For example, using carrier peptides enriched for posttranslational modifications (e.g., phosphorylation) or adding synthetic peptides may enable single-cell posttranslational modification analysis as previously suggested9. However, if the abundance of these peptides in the single cells is very low, they may not be quantified in the single cells.
This challenge may be partially mitigated by increasing the MS2 accumulation time in order to increase the chances of quantifying those peptides in the single cells. While spiking in synthetic peptides also adds cost to performing SCoPE2 experiments, it may be well justified by the increased probability of analyzing proteins and modifications of special biological interest.
Synthetic peptides can also be used to perform quality control of digestion and labeling. To this end, the peptides should be added directly to the wells containing isolated single cells. Because these peptides can be designed to contain cleavage sites, they can be used to diagnose the efficiency of digestion for each single cell.
Carrier abundance: while a 100-cell carrier has proven suitable for the majority of our experiments, this is a parameter that will likely require adjusting depending on the cells of interest and their proteome abundance. Optimization of carrier abundance is discussed in the protocol. Higher carrier abundance can allow identifying peptides with shorter ion accumulation times and, thus, analyzing more peptides per unit time. However, the shorter accumulation times will reduce the ion copies sampled from single-cell proteins and thus will reduce quantitative accuracy and increase missing data in single cells. An extensive discussion of considerations for carrier abundance and optimization for different experimental designs can be found in Specht and Slavov (2020) (ref. 19).
Single or dual carrier selection: the use of a single carrier channel maximizes the number of single cells that can be analyzed per SCoPE2 run. Depending on the experimental design, however, the use of a second carrier channel can provide an internal control. In this way, each SCoPE2 set would contain two carrier channels labeled with two different TMT labels along with labeled single cells. We describe such an example here with the 100xMaster samples that are used for LC-MS/MS optimization prior to running SCoPE2 sets. The 100xMaster samples contain bulk cellular material that is prepared in the same manner as the carrier and referenc; it is a bulk standard diluted so that each injection contains cell concentrations representing a SCoPE2 set, i.e., 100–200 carrier cells and ‘single cell’ channels, upon injection into the mass spectrometer.
Channel selection for carrier: the isobaric tag used to label the carrier peptides should be chosen to minimize its influence on samples labeled with other tags. This includes two considerations. First, the isotopic contaminations of the tag should affect as few samples as possible, which is the case for the lightest and the heaviest tags since their isotopic contaminants affect only one sample. Second, ideally the tag should not be mDa away from another tag (e.g., a C-N pair tag) since these tags become challenging to resolve when used for samples of very different abundance. These considerations favor the use of 126 with TMT 11-plex (since 131 is a C-N pair) and 126 or 134N with TMTpro 16-plex since 134C is not used for the 16-plex.
LC-MS/MS system suitability: before conducting a SCoPE2 experiment, an LC-MS/MS system can require substantial optimization to allow it to successfully obtain meaningful SCoPE2 data. We recommend the use of dilute 100xMaster standards, diluted to single-cell levels to optimize instrument performance without the biological variation inherent to true single-cell samples. Details on 100xMaster generation and guidelines for instrument optimization are provided in the SCoPE2 protocol below.
Positive and negative controls within single-cell sets: we recommend adding controls within each SCoPE2 experiment. In a typical experiment, we omit adding a single cell to negative control wells and add diluted bulk cell lysate to positive control wells. Bulk cells that are diluted down to single-cell levels and then undergo digestion and/or labeling with the single cells within the 384-well plates are useful positive controls in diagnosing both digestion and labeling efficiencies. Positive controls allow evaluation of sample preparation independent of cell isolation and are particularly useful when the quality of the single cells or their isolation is uncertain. Negative controls receive all reagents in the same manner as the single-cell wells and thus provide a benchmark for the background noise. They are particularly useful for evaluating potential problems with sample preparation, such as cross-labeling.
Level of expertise needed to implement the protocol
The sample handling portions of this protocol should be approachable for a biochemist or cell biologist with cell culture and molecular biology experience. It is recommended that users not experienced in sample preparation for proteomics consult with an individual with experience in this area to avoid common mistakes such as polymer contamination deriving from plasticware or improperly handled buffers.
The LC-MS/MS system requires the engagement of an experienced mass spectrometrist early in the project to ensure that the instrumentation is performing at a level suitable for single-cell analysis. Many core facilities will not be able to perform this analysis without extensive planning and engagement. Due to the range of samples processed, facility workflows are typically optimized for robustness as opposed to sensitivity. Due to the extremely low abundance of single-cell samples and carryover from more abundant samples run previously on the LC-MS/MS system, significant optimization may be required to adapt workflows, and guidelines for this process are described in the protocol.
Data analysis steps specific to SCoPE2 single-cell proteomics experiments are presented, for example, quality control metrics and identification of failed wells. Downstream analysis including batch correction, dimensionality reduction and differential expression falls outside the scope of this protocol, but will be familiar to users with experience in scRNAseq data processing.
Limitations
The current sample preparation method described here is robust and uses equipment readily available to many laboratories. However, while the volumes used for sample preparation are an order of magnitude lower than those used in the original SCoPE-MS protocol17, they are still orders of magnitude larger than those used for droplet-based scRNAseq sample processing. Reduced sample volumes will significantly reduce sample losses to surfaces during processing. Alternative approaches for preparing single cells for MS analysis, such as OAD or nanoPOTS, allow for reducing sample processing volumes to 200 nL36, a five- to tenfold reduction on the 1–2 μL used here. The droplet sample preparation (nPOP) allows further reduction (100-fold) of the sample preparation volumes to 20 nL23. If desired, these sample preparation methods can be easily incorporated within the SCoPE2 framework. mPOP has been scaled to simultaneously process hundreds of single cells23.
Currently a major bottleneck for all single-cell MS methods is the large number of LC-MS/MS runs required to analyze thousands of single cells. The multiplexing afforded by SCoPE2 reduced the number of runs needed by over tenfold, but the number remains large and likely represents the single largest expense for the analysis, particularly for investigators working through a core facility18. Improved barcoding strategies, for example, the newly announced TMTpro reagents from Thermo Scientific, allows at least 50% increase in throughput by reducing the number of required LC-MS/MS runs by approximately one-third for analysis of the same number of single cells. A strategy that can further increase multiplexing is the use of both TMT 11plex and TMTpro 16plex reagents. This strategy offers tradeoffs discussed below. The possibilities and limitations of increasing multiplexing further are discussed in ref. 15.
Co-isolation and co-fragmentation of ions limits the quantitative accuracy of isobaric labeling methods, including SCoPE2. This limitation can be mitigated by performing quantification at the MS3 level at the cost of sensitivity loss. Another solution would be to use complement ions, which are peptide fragments with the balancer portion of the TMT still attached. In our laboratory, we aim to mitigate co-isolation by sampling peptides at the apex of the elution peaks to reduce co-isolation, along with using narrow isolation windows. The optimization of the apex targeting is facilitated by DO-MS and has the additional benefit of increasing the number of ion copy samples per unit time and, thus, sensitivity and quantification accuracy25.
Another limitation originates from the tendency of current workflows to sample only a small fraction of ions available for analysis. Such sampling has been highly successful in bulk proteomics where only short accumulation times are typically required to sample individual peptides. The ability to sample ions more completely, e.g., by accumulating ions in parallel and thus for longer times (as exemplified by the PASEF mode of the timsTOF instruments44) can substantially enhance the sensitivity and the accuracy of quantification. A tradeoff of the timsTOF instruments at present is that their resolution does not permit the level of sample multiplexing possible with Orbitrap instruments and TMT-based isobaric labeling reagents, thus limiting the number of cells that can be analyzed per LC/MS-MS run.
These limitations and how they may be mitigated by future developments are discussed in more details in refs. 45,46. These developments will further improve the sensitivity, throughput and robustness of single-cell protein analysis.
Materials
Biological materials
Isolated primary cells, or cell lines grown in culture. Multiple cell lines can be used for preparation of initial SCoPE2 100xMaster samples. In our laboratory, 100xMaster samples typically contain U937 (ATCC CRL-1593.2, RRID: CVCL_0007) and Jurkat (ATCC, RRID: CVCL_0367) cells, grown in RPMI-1640 medium, supplemented with 10% bovine serum and penicillin/streptomycin. To generate a small-scale SCoPE2 experiment that is featured within this manuscript, we used U937 and HeLa cells (RRID: CVCL_0030) ! CAUTION Cell lines should be checked to ensure they are authentic, and free of mycoplasma contamination.
Reagents
▲ CRITICAL All solutions should be prepared with LC-MS/MS-grade water and reagents. Use of lower-quality reagents can result in contamination of solutions with polymers and compromise LC-MS/MS detection of peptides of interest. Plasticware used for storing solvents should be rinsed with ethanol or isopropanol prior to use, and solutions should be used within 1 week.
Water, Optima LC-MS/MS grade (Fisher Scientific, cat. no. W6-1)
Acetonitrile (for buffer preparation), Optima LC-MS/MS grade (Fisher Scientific, cat. no. A955-1) ! CAUTION Acetonitrile is a flammable liquid that can irritate the eyes, skin, respiratory tract and central nervous system, and can cause liver or kidney injuries. Wear personal protective equipment when using acetonitrile. This chemical should be handled under a chemical fume hood.
Acetonitrile (for TMT preparation), anhydrous, 99.8% (Sigma Aldrich, cat. no. 271004-100ML)
Triethylammonium bicarbonate (TEAB), 1 M pH 8.5 (Sigma Alrich, cat. no. T7408100ML)
Formic acid, Pierce, LC-MS/MS grade (Thermo Fisher Scientific, cat. no. 85178) ! CAUTION Formic acid is a flammable liquid that can cause serious eye damage or skin burns. Wear personal protective equipment, keep away from any heat, and use in a well-ventilated area.
TMTs, TMTpro 16plex Label Reagent Set, 1 × 5 mg (Thermo Fisher Scientific, cat. no. A44520). We recommend using the 16plex reagents owing to their higher throughput; however, if you wish to use 11plex reagents, they are: TMTs, TMT10plex Isobaric label reagent set plus TMT11-131C (Thermo Fisher Scientific, cat. no. A34808)
Hydroxylamine (HA), 50% wt/vol (Sigma, cat. no. 467804-50ML) ! CAUTION HA can cause skin irritation; use appropriate protective equipment.
Trypsin, Trypsin Gold Mass Spectrometry Grade (Promega, cat. no. V5280) ! CAUTION Different sources of trypsin can vary in their purity. Less pure trypsin will negatively impact SCoPE2 results. ! CAUTION Trypsin is chemical that can cause skin, respiratory and eye irritation. Use under a chemical fume hood with personal protective equipment.
Benzonase nuclease (Sigma Aldrich, cat. no. E1014-25KU)
MassPREP peptide mixture (Waters, cat. no. 186002337) ▲ CRITICAL The MassPREP peptide mixture represents a simple mixture of nine nontryptic peptides used for passivation of plasticware. It could be substituted with a range of retention time standards or similar mixtures.
Phosphate-buffered saline (PBS), 10×, pH 7.4, RNase-free (Thermo Fisher Scientific, cat. no. AM9625)
Equipment
PCR plate, 384-well, standard (Thermo Fisher Scientific, cat. no. AB1384) ▲ CRITICAL Different sources of PCR plate can have high levels of polymer contamination rendering them unsuitable for SCoPE2 sample preparation. If using plates from another source, the levels of polymer contamination should be assessed prior to beginning the experiment.
Adhesive PCR plate foils (Thermo Fisher Scientific, cat. no: AB0626)
PCR tubes: TempAssure 0.2 mL PCR 8-tube strips (USA Scientific, cat. no. 1402-3900) ▲ CRITICAL As described for 384-well plates, different plasticware sources can have high levels of polymer contamination, and this should be assessed before using different PCR strips.
Glass autosampler inserts, 9 mm (Thermo Fisher Scientific, cat. no. C4010-630) ▲ CRITICAL The use of glass rather than plastic for sample storage greatly reduces sample losses. The use of autosampler vial inserts permits the SCoPE2 samples to be resuspended and injected into the instrument in smaller (1 μL) volumes, again minimizing losses.
9 mm clear glass screw thread vials (Thermo Fisher Scientific, cat. no. 60180-509)
9 mm autosampler vial screw thread caps (Thermo Fisher Scientific, cat. no. C5000-51B)
384-well PCR machine with heated lid, e.g., C1000 Touch with 384-well module (Bio-rad, cat. no. 1851138)
96-well PCR machine with heated lid, e.g., T100 Thermal Cycler (Bio-rad, cat. no. 1861096); if using this model, the use of the tube support ring (Bio-rad, cat. no. 1862000) is recommended; if only a 384-well PCR machine is available, all steps requiring PCR tubes and a 96-well PCR machine can be accomplished using the 384-well plates in the 384-well PCR machine
Water bath sonicator, e.g., 2.8 L ultrasonic cleaner with digital timer (VWR, cat. no. 97043964)
Plate spinner, e.g., PlateFuge microcentrifuge (Benchmark Scientific, Model C2000). This plate spinner does not offer speed control as it is used to collect liquid at the bottom of a well, rather than for pelleting material
PCR tube spinner, e.g., 16-place microcentrifuge for 0.2 mL tubes (USA Scientific, cat. no. 2621-0016)
Autosampler vial spinner, e.g., myFuge 5 (MTC Bio, cat. no. C2595). This centrifuge does not offer speed control as it is used to collect liquid at the bottom of the autosampler vial, rather than for pelleting material
Vortex, e.g., Analog vortex mixer (VWR, Model 58816-121)
Mantis microfluidic liquid handler (Formulatrix) ! CAUTION If using a different liquid dispensing robot/handler, it is important to check if it is compatible with the 100% acetonitrile that the TMT reagents are in, and also that the plasticware used does not introduce polymer contamination into the samples.
Mantis microfluidic chips, low-volume silicone chips (Formulatrix, cat. no. MCLVS12) ▲ CRITICAL These chips are suitable for use with aqueous solutions, and are used for all dispensing steps aside from TMT reagent dispensing as they are not recommended for high solvent concentrations.
Mantis microfluidic chips, low-volume 3PFE chips (Formulatrix, cat. no. MCLVPR2) ▲ CRITICAL These chips are suitable for high solvent concentrations and are used solely for dispensing TMT reagents during SCoPE2 sample preparation.
Mantis PCR plate adapter with wide conical pins for automated plate handling (Formulatrix, cat. no. 232400)
LC-MS/MS system (e.g., Q-Exactive with Nanospray Flex Ion Source, Thermo Scientific)
nanoLC System (e.g., Dionex UltiMate 3000 UHPLC, Thermo Scientific)
25 cm × 75 μm IonOpticks Aurora Series UHPLC column (IonOpticks, cat. no. AUR225075C18A) ▲ CRITICAL Good chromatography is crucial for obtaining high-quality SCoPE2 data. Columns need to have sharp chromatographic resolution and, additionally, need to be able to tolerate the neutralized TMT in genuine SCoPE2 samples. We have had success with these columns. Columns from other suppliers may also work well, but will require testing.
In-source blower elbow (Idex Health & Science, part no. P-432)
Active background ion reduction device (ABIRD; ESI Source Solutions, cat. no. ABFLEXTM) ▲ CRITICAL We recommend using this device if you are using an ion source that is open to the room, for example, the Nanospray Flex Ion Source.
nanoLC column heater and controller, e.g., Bufferfly heater/controller PST-BPH-20, PSTCHC (Phoenix S&T)
Software
MaxQuant software (v1.6.7 or newer), available at https://www.maxquant.org with free registration47,48. Other software (e.g., Proteome Discoverer, Comet, and FragPipe) can be used with minor adjustments of DART-ID and DO-MS software to the output of these different search engines. Note that TMTpro 16plex modifications are not included with this release, but an updated modifications . xml file including these forms part of Supplementary Data 1
DART-ID software, freely available from https://dart-id.slavovlab.net/24. The DART-ID software allows FDR-controlled and improved peptide identification based on a Bayesian approach that updates peptide PEPs using informative peptide features, e.g., retention times. Other software packages can also enhance peptide identification using informative peptide features49,50
DO-MS software, freely available from https://do-ms.slavovlab.net/25. The DO-MS software al- lows visualization of MS run features, which is the basis for specifically diagnosing problems and optimizing data acquisition ▲ CRITICAL For optimal use, the ‘Peak features’ option must be enabled in the MaxQuant analysis options.
Reagent setup
Tandem mass tags
Following Thermo Fisher recommendations, 5 mg of TMT powder should be resuspended in 200 μL of anhydrous acetonitrile, which produces a stock concentration of 85 mM. Thermo Fisher recommends storing unused TMT reagents at −20 °C with desiccant for a period of 1 week. We have found that storing aliquotted TMT labels at −80 °C works as well. ▲ CRITICAL Use of alternative (non-anhydrous) acetonitrile can result in loss of TMT reactivity and poor downstream labeling.
Equipment setup
Instruments
As no cleanup is performed on SCoPE2 samples, material can accumulate rapidly on the heated capillary during SCoPE2 experiments, requiring cleaning and recalibration on a more regular basis than desirable. Minor modifications to the exterior of the mass spectrometer and run method avoid this by ensuring that material eluting from the LC during sample loading does not enter the instrument.
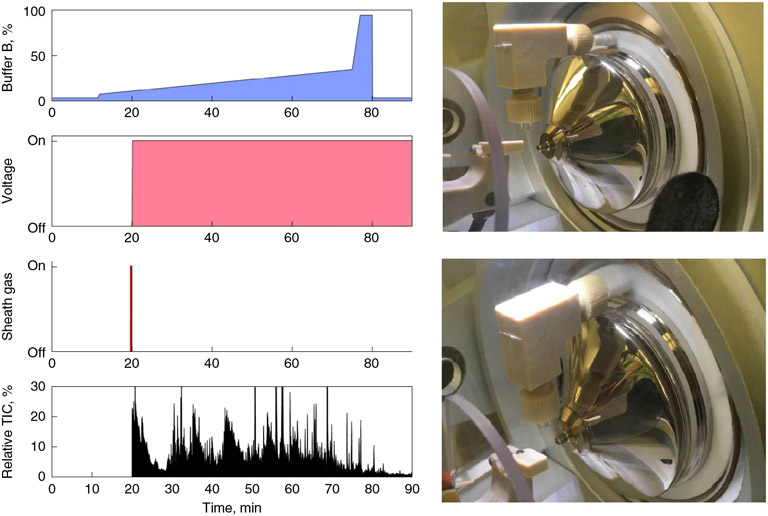
On Orbitrap instruments, the ion source is used with the ion sweep cone removed. An inexpensive blower elbow is attached to the sheath gas outlet above the heated capillary, and aimed at the capillary outlet. The attachment of this blower elbow to a Q-Exactive-type instrument is shown in Fig. 2. This is used in combination with a modified run method in Xcalibur. This approach switches through several tune files that regulate the spray voltage and sheath gas flow rate. We highlight our LC-MS/MS run method for SCoPE2 samples below illustrating these switches. During the 20 min loading period, a first tune file supplies no voltage to the source, causing material eluting from the column to collect on the emitter. For the final 20 s of loading, a second tune file keeps the voltage off, while applying sheath gas. The sheath gas is directed at the emitter tip through the blower elbow attached above, removing the accumulated material. Finally, the method switches to a normal tune method, representing the latest instrument calibration where the voltage is applied and sheath gas turned off. Spectra are only recorded when this final tune file is applied. As no spectra are recorded for the first two tune files, these do not require updating during regular instrument calibration and maintenance.
Fig. 2 ∣. LC-MS/MS setup for SCoPE2 experiments.
The left panels show the typical gradient parameters used for SCoPE2 runs. Nonstandard portions of the gradient include turning the voltage off during the initial phase to the gradient to reduce the contamination of the heated capillary. Material collected on the emitter tip is then removed with sheath gas briefly directed at the tip through a blower elbow. After this point, the voltage is applied and scan data collected. The right panels show the attachment of the blower elbow to a Q-Exactive classic instrument.
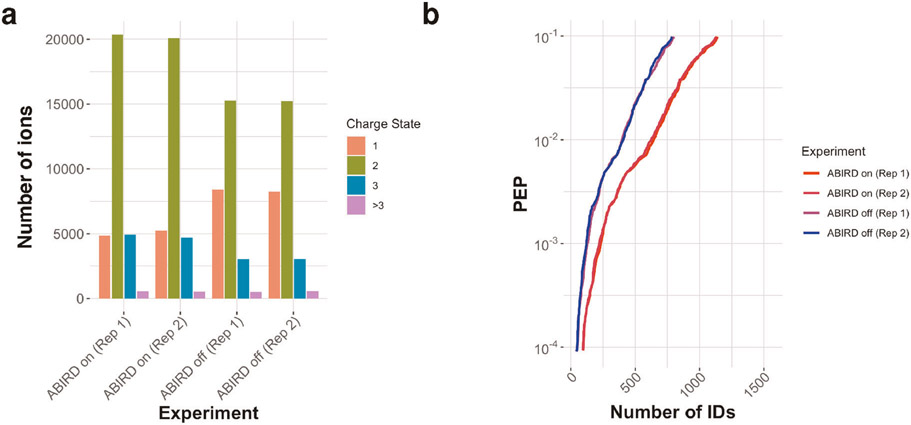
If the laboratory atmosphere has a high background ion levels, these may pose difficulties for low-abundance sample analysis, especially with a source open to the room atmosphere such as the flex ion source. In such case, the level of contaminant ions can be reduced significantly by using the ABIRD (Extended Data Fig. 1). The ABIRD directs HEPA-filtered air towards the source, thus reducing background ions considerably. The benefits of using an ABIRD are pronounced only when the atmosphere is contaminated.
Liquid chromatography
Performance of the nanoLC is crucial for obtaining high-quality quantification data from SCoPE2 samples. The key criterium for column selection is short full-width half-maximum peak widths. However, some commercial columns appear to poorly tolerate the high levels of neutralized TMT and HA in the SCoPE2 samples, and this should also be assessed. The 25 cm × 75 μm IonOpticks Aurora series column meets both these criteria, and these columns typically offer a lifetime of several months for SCoPE2 sample analysis. The column is enclosed in a nanoLC column heater, with the temperature set to 60 °C. In our setup, the nanoLC system is operated without a trapping column since trapping columns can contribute to losses of limited samples such as SCoPE2 samples. The nanoLC system should be plumbed to minimize dead volumes and transfer lengths since those contribute to peak broadening.
Autosampler parameters
The general settings for the Dionex Ultimate WPS-3000 autosampler are shown below:
| Parameter | Setting |
|---|---|
| Draw speed | 0.050 |
| Draw delay | 5.000 |
| Dispense speed | 2.000 |
| Dispense delay | 2.000 |
| Dispense to waste speed | 4.000 |
| Sample height | 0.000 |
| Puncture depth | 8.000 |
| Wash volume | 50.000 |
| Wash speed | 4.000 |
User-defined pickup method
The settings for running a user-defined pickup method on the Dionex UltiMate WPS-3000 autosampler are shown below. This pickup method optimizes sample loading time and, thus, overall sample throughput. However, it is not essential, and the predefined uLPickup method can be used. Inject mode is set to UserProg. Reagent A is set to autosampler location G1. This location in the autosampler must contain buffer A or water.
| Command | Parameters |
|---|---|
| UdpDraw | ReagentAVial, 5 [μL], 0.2 [μL/s], 5 [mm] |
| UdpMixWait | 5 [s] |
| UdpDispense | Drain, 0.000, 2 [μL/s], 5 [mm] |
| UdpInjectValve | Load |
| UdpDraw | SampleVial, Sampler.Volume, 0.2 [μL/s], 0 [mm] |
| UdpMixWait | 5 [s] |
| UdpDispense | Drain, 0.000, 2 [μL/s], 5 [mm] |
| UdpDraw | ReagentAVial, 2.4 [μL], 0.2 [μL/s], 5 [mm] |
| UdpMixWait | 5 [s] |
| UdpDispense | Drain, 0.000, 2 [μL/s], 5 [mm] |
| UdpInjectValve | Inject |
| UdpInjectMarker | |
| UdpMixWait | 5 [s] |
| UdpMixNeedleWash | 200 [μL] |
LC-MS/MS run parameters
The run method parameters shown below are what is used on the author’s Thermo Q-Exactive (classic) Instrument. These settings will vary for individual instruments (even from the same model), and for different experimental designs. However, these may serve as a convenient starting point for optimization. Details on how to optimize the run method for the users system are given in the protocol.
| Parameter | Setting |
|---|---|
| Spray voltage | 1,800-2,500 V |
| Capillary temperature | 250 °C |
| Full-scan MS range | 450-1,600 m/z |
| MSI resolution (m/z 200) | 70,000 |
| Maximum injection time | 100 ms |
| AGC target | 1 × 106 |
| TopN | 7 |
| Precursor charge state | 2-5+ |
| (N)CE | 33 |
| MS2 resolution (m/z 200) | 70,000 |
| MS2 accumulation time | 300 ms |
| MS2 AGC target | 5 × 104 |
| MS2 AGC minimum | 2 × 104 |
| Isolation width | 0.7 m/z |
| Isolation offset | 0.3 m/z |
| Charge exclusion | Unassigned, 1, ≥4 |
| Dynamic exclusion | 30 s |
These parameters may need a slight adjustment for the new line of Orbitrap instruments, such as Exploris and Eclipse. A primary advantage of the newer Orbitraps is that they need less transient time to offer the high resolution needed by SCoPE2 (60,000–70,000). This advantage does not appreciably change the data acquisition parameters, because the speed of SCoPE2 analysis is usually limited by the time needed for ion accumulation rather than the transient time needed to achieve high resolution18,19. Thus, we expect that most parameters outlined in the table above remain a good starting point for the newer Orbitrap instruments.
With Exploris and Eclipse, we recommend doubling the resolution for MS2 scans because such an increase of the resolution increases the signal-to-noise ratio at no cost18, that is, it does not slow down the rate of acquiring MS2 scans. These theoretical expectations have been consistent with results from our collaborations with colleagues using the latest Exploris and Eclipse Orbitrap instruments.
We have recently begun performing SCoPE2 analysis with a timsTOF system, and our preliminary data show much promise of the system. The higher sensitivity of the TOF detector reduces the size of the isobaric carrier needed to achieve the designed tradeoff of throughput and copy number sampling from the single cells19. We hope that our parameters and guidelines are a useful starting point, and we recommend that each laboratory optimize their MS systems for single-cell analysis, as each instrument might need slightly different parameters.
Liquid chromatography
Gradient details are shown below, for an example LC gradient for performing SCoPE2 on a Thermo Q-Exactive (classic) Instrument with a Dionex 3000 nanoLC. Further gradient optimization may be required depending on the use case. Buffer A is 0.1% formic acid in HPLC-grade water; Buffer B is 0.1% formic acid in 80% acetonitrile/20% HPLC-grade water. The gradient is run without a trap column at a constant 200 nL/min flow rate. Typical gradients and the voltage and sheath-gas switching can also be found in Fig. 2.
| Time (min) | % B |
|---|---|
| 0.0 (Start) | 4.0 |
| 11.5 | 4.0 |
| 12.0 | 8.0 |
| 75.0 | 35.0 |
| 77.0 | 95.0 |
| 80.0 | 95.0 |
| 80.1 | 4.0 |
| 95.0 (End) | 4.0 |
Mantis liquid dispenser
The liquid handler will require calibrating for the 384-well plates used for preparing SCoPE2 samples, as per the instruments normal method. 384-well and 96-well plates benefit significantly from use of the PCR plate adaptor (see ‘Equipment’), and we strongly recommend its use. A practice dispense of 1 μl of water in a spare plate prior to using the system for SCoPE2 processing is recommended to visually confirm plate alignment and liquid handler calibration prior to use.
Software setup
MaxQuant
MaxQuant can be downloaded freely from maxquant.org, following free registration. For users unfamiliar with standard MaxQuant usage, an annual summer school is offered. Videos from past years can be found on YouTube, and documentation is linked to from maxquant.org.
Where search parameters used for SCoPE2 analysis deviate from the MaxQuant defaults, these are given in Table 2. There are two profiles of settings used for analysis of SCoPE2 data with MaxQuant. The first (listed under ‘Variable search parameters’) is used to determine labeling efficiency with TMT or TMTpro reagents. The second is for searching SCoPE2 data or 100xMaster data. Custom modifications are required for the variable search and are included with the modifications.xml file in Supplementary Data 1. This should be used to replace the modifications.xml file that comes with MaxQuant and can be found in the MaxQuant directory, in /bin/conf/. If you wish to generate these custom search modifications yourself, simply duplicate the TMT or TMTpro 126 N-terminal and lysine modifications and change the label type from isobaric label to standard. If using TMTpro reagents, MaxQuant 1.6.7 does not come with these in its library, and they have been included in the modifications.xml file. A mqpar.xml file is also included that can be loaded into MaxQuant, which preloads TMTpro isobaric labels.
Table 2 ∣.
Maxquant settings
| Parameter name | Setting |
|---|---|
| Variable search parameters | |
| Group-specific paramters | |
| → Type | Standard |
| → Variable modifications (All) | Acetyl (K) Acetyl (N-term) |
| → Variable modifications: 11plex ONLY | Variable TMT10plex N-terma Variable TMT10plex Lysa |
| → Variable modifications: 16plex ONLYb | Variable TMTpro16plex N-terma,b Variable TMTpro16plex Lysa,b |
| All other parameters default | |
| SCoPE2 search parameters | |
| Group-specific paramters | |
| → Type | Reporter ion MS2: 11plex TMT or 16plex TMTprob |
| → Variable modifications | Oxidation (M) Acetyl (Protein N-term) |
| → Fixed modifications | None |
| Global parameters | |
| → Identification | PSM FDR: 1.00 (if using DART-ID) Protein FDR: 1.00 (if using DART-ID) |
| → Advanced identification | Uncheck Second peptides |
| → Advanced | Check Calculate peak properties |
| All other parameters default |
MaxQuant adjustment from default settings for using MaxQuant to assess TMT labeling efficiency (Variable search) or for using MaxQuant to analyze SCoPE2 or 100xMaster data.
Custom modifications named per the included modifications.xml file.
TMTpro modifications are not included in the 1.6.7 release of MaxQuant. If you wish to search TMTpro 16plex data, a modifications .xml file and mqpar file to load the TMTpro reagents into Maxquant 1.6.7 are included in Supplementary Data 1.
The isobaric tags of TMT contain isotopic impurities, which affect SCoPE2 quantification as they do all TMT workflows. Because of the impurities, a portion of each sample is labeled with a tag whose mass is different (mostly by ±1 Da) from the intended mass of the tag. The influence of such isotopic impurities can be modeled with a linear superposition model and computationally mitigated by performing a deconvolution. This is a standard part of many workflows, including the MaxQuant one, and we recommend inputting the TMT batch-specific impurities supplied by the manufacturer for achieving optimal correction. With MaxQuant, this option can be accessed via the ‘Group-specific parameters’, and then the ‘Type’ tab, Reporter ion MS2 will first need to be selected. Then, the isotope correction factors can be imported directly into the software, using the ‘Import’ button. In the context of isobaric carriers, the impurities have an additional impact: the impurities of the tag used for the abundant carrier peptides may have a disproportionate impact on a couple of neighboring channels, and thus we recommend not using these channels for single-cell samples.
DART-ID
The DART-ID24 project website, which contains detailed installation and usage instructions, is at https://dart-id.slavovlab.net/.
DO-MS
The DO-MS25 project website, which contains detailed installation and usage instructions, is at https://do-ms.slavovlab.net/.
Procedure
▲ CRITICAL We recommend the use of the newer TMTpro 16plex or 18plex reagents for their higher throughput and reduced cost per single cell. If you wish to use 11plex reagents, sections where the protocol deviates will be indicated with ‘11plex’.
Controlled experiments have demonstrated that the freeze–heat lysis method, named mPOP, is as efficient as classical urea lysis21. mPOP is used in this procedure.
Generation of 100xMaster and 100xCarrier Samples ● Timing 1-2 d
▲ CRITICAL Refer to Table 3 for recommended labeling strategy for 100xMaster and 100xCarrier samples.
Table 3 ∣.
Recommended 100xMaster and 100xCarrier-only sample design
| 100xMaster | 100xCarrier | TMT label |
|---|---|---|
| 5,000 cells, cell type A | 5,000 cells, cell type A | 126 |
| 5,000 cells, cell type B | 5,000 cells, cell type B | 127N |
| Unused | Unused | 127C |
| Unused | Unused | 128N |
| 100 cells, cell type A | Unused | 128C |
| 100 cells, cell type B | Unused | 129N |
| 100 cells, cell type A | Unused | 129C |
| 100 cells, cell type B | Unused | 130N |
| 100 cells, cell type A | Unused | 130C |
| 100 cells, cell type B | Unused | 131N |
| Unused | Unused | 131C |
Master standards diluted to 1× allow convenient testing and optimization of the LC-MS/MS system without the variability seen in true single-cell samples. The use of a carrier-only sample allows testing for sample carryover in the single-cell channels, and detection of low-level contaminants that could not be identified by using ‘blank’ runs. The table lists 11plex TMT reagents, and the section from 128C to 131C could be extended if using 16plex TMTpro reagents, keeping the layout for 126-131C and adding additional 100 cells, cell type A or B in 132-134N.
-
Obtain single-cell suspensions of mammalian cell lines of interest in ice-cold PBS. Two cell types from the same species will be used for the samples consistuting a 100xMaster standard. Hereafter, the cell types are referred to as samples A and B.
▲ CRITICAL STEP The exact means by which these cell lines are prepared as single-cell suspensions will vary depending on the cell lines of interest. The cell lines chosen for single-cell analysis should ideally comprise the carrier and reference material as well. If obtaining large amounts of the single cells of interest for the carrier or reference is a problem, then a cell line from the same species that is as similar as possible to the single cells of interest should be chosen instead. For the 100xMasters, two cell types from the same species (e.g., for human cells, U-937 and Jurkat) should be chosen in order to better diagnose instrument suitability, which will be described later in the text.
-
Count the cells (a hemocytometer is sufficient), and resuspend 40,000 of each cell type in separate PCR tubes in a volume of 20 μL of HPLC-grade water, resulting in 2,000 cells/μL.
▲ CRITICAL STEP There is a question here about whether it is worth doing experiments to optimize the number of cells used. We have performed bicinchoninic acid protein content assays on different cell types lysed via mPOP. The bicinchoninic acid data confirmed the estimated protein levels per single cell between two technical replicates per cell type. HeLa cells are among the largest cell types: we extracted 0.5 ng of protein per cell, which agrees with the expected total protein per HeLa cell51. Additionally, Jurkats were among the smallest cells we tested, which was confirmed by the scaled protein content, of ~0.1 ng of protein per cell. Therefore, if we extract protein from 40,000 HeLa cells, we would expect a protein yield of ~20,000 ng; likewise, from 40,000 Jurkats, we would expect 4,000 ng. If the cell types of interest are of dissimilar size, such as Jurkat and HeLa cells, we recommend mixing cell types of dissimilar size based on expected protein amount per cell, rather than pure cell number. This allows for an even number of peptides generated from each cell type in the carrier and reference material.
-
Freeze PCR tubes containing cells at −80 °C for a minimum of 30 min.
■ PAUSE POINT The tubes can be frozen for up several months without any change to the result.
Transfer PCR tubes to a thermocycler with a heated lid, and heat to 90 °C for 10 min (set lid to 105 °C), holding the sample at 12 °C when the heating cycle has completed.
Once the samples have cooled to 12 °C, add 1 μL of benzonase nuclease diluted to ≥5 units/μL.
Briefly vortex the samples for 3 s, followed by centrifugation in a bench-top PCR tube spinner for 3 s.
Sonicate the samples for 10 min in a water bath sonicator, and place the tubes on ice.
- Once on ice, supplement both samples with 4 μL of a master mix containing:
Component Amount Final concentration per tube Trypsin Gold (200 ng/μL) 1.3 μL 10 ng/μL TEAB pH 8.5 2.5 μL 100 mM HPLC-grade water 0.2 μL Not applicable Briefly vortex the tubes to mix, followed by brief centrifugation to collect liquid.
Transfer the samples to a thermocycler, and digest for 3 h at 37 °C (heated lid set to 52 °C).
-
For 100xMasters: add 3.2 μL of sample A to each of four PCR tubes (one for the carrier channel, three for the ‘single-cell’ channels). Do the same with four new PCR tubes for sample B.
For 100xCarrier: add 3.2 μL of sample A to one PCR tube. Do the same with a new PCR tube for sample B.
▲ CRITICAL STEP Larger volumes of cells can be used. Adjust the volume of subsequent reagent additions proportionally to the volume of the input digested sample.
Following the layout detailed in Table 3, add 1.6 μL of 85 mM TMT label to each of the tubes. For example, for a 100xMaster, the four PCR tubes containing sample A should be labeled with 126, 128C, 129C and 130C.
Briefly vortex the tubes; spin down to collect liquid.
Incubate tubes at room temperature (18–22 °C) for 1 h.
Add 0.7 μL of 1% HA (vol/vol from 50% stock HA indicated in ‘Materials’) to each tube.
Briefly vortex to mix, followed by centrifugation to collect liquid.
Incubate tubes at room temperature for 30 min.
-
For 100xMasters: combine both carrier sample tubes (126, 127N) in a single glass insert. Dilute the remaining samples comprising the ‘single-cell’ samples to 25 μL, and add 0.5 of each to the glass insert. This produces dilutions for the carrier and ‘single-cell’ samples as described in Table 3. Mix well by pipetting.
For 100xControls: combine both tubes into a glass insert, mixing well by pipetting.
-
Divide the 100xMaster or 100xControl samples into five aliquots of equal volume. These each contain 20 injections of material.
▲ CRITICAL STEP If different aliquot sizes are required, adjust the subsequent resuspension volumes proportionally, keeping the volume at 1 μl per injection.
Use a speedvac to reduce the samples to dryness. These can be stored dry at −80 °C until required.
-
To use an aliquot, resuspend in 20 μL of 0.1% formic acid. See the following section for guidance on LC-MS/MS optimization.
? TROUBLESHOOTING
LC-MS/MS optimization using 100xMaster Samples ● Timing 1+ d
▲ CRITICAL While it is possible that optimization could be accomplished in a couple of days with a well-running LC-MS/MS system set up as described in instrument setup, deviation from this setup may result in significant increases in the amount of time required for instrument optimization.
The key elements to optimize are efficient sample delivery to the mass spectrometer and sampling peptide-like ions as close to their elution peak apex as possible. The latter both maximizes signal intensity and minimizes co-isolation with other ions that can compromise MS2-based quantitation using TMT.
While several software packages can be employed to help optimize these parameters, we recommend using MaxQuant and DO-MS as described previously25. To generate all plots within the DO-MS dashboard, users must set up MaxQuant using the parameters found in Table 2.
Optimizing data acquisition
▲ CRITICAL Given that input amounts in SCoPE2 samples are lower than for typical bulk proteomics preparations, optimizing chromatography to maximize the number of ions of each species delivered per unit time is of primary importance.
-
22Optimize the chromatography. To do this, plot the peak widths, both at base and at full-width half max, for different elution conditions. Parameters that can be changed include:
- Gradient length
- Gradient steepness
- Column chemistry
- LC plumbing
- The mobile phases
-
23Optimize sampling so that you are preferentially looking at ions collected at the elution peak apex. There are several interrelated instrument parameters that directly impact on ion sampling: MS2 fill times, top N, and AGCmin. By adjusting the length of time that the instrument collects ions at the MS2 level and changing the number of ion species that are selected for MS2 analysis, it is possible to alter the point in the elution profile where the ion is sampled.
- Establish an Apex Offset performance baseline by running two replicate 1× injections of the master sample.
- Search these 1× injections using MaxQuant (sample MaxQuant parameters provided in Supplementary Data 1).
- Load the MaxQuant output into DO-MS, and navigate to the ‘Ion Sampling’ dashboard tab.
- The Apex Offset plot window presents the temporal distance between when each ion was selected for MS2 analysis and that ion’s elution apex, i.e., when the collected ions were most abundant. In general, it is preferable to bias MS2 sampling of an ion as close to its elution apex as possible.
- If the Apex Offset distribution is biased toward early sampling, increase the length of the duty cycle by increasing MS2 fill times or increasing the number of ions selected for MS2 analysis (top N).
- If the Apex Offset distribution is biased toward sampling ions after their elution apices, decrease the MS2 fill time or top N. In practice, it will not be possible to target all peptide-like ions in a complex mixture at their elution apices.
-
24
Look for coisolation of precursors. Co-eluting precursors remains a problem for all data-dependent acqusition method of isobarically labeled peptides. Peptides should be sampled as purely as possible for MS2 fragmentation. If two or more peptides are sampled and sent for MS2 analysis, then those peptides will contribute to reporter ion intensity (RII), which reduces accuracy of single-cell quantitation. A straightforward metric for assessing MS2 spectral purity, or the degree to which coisolation of multiple precursors prior to precursor fragmentation is occurring, is generated by MaxQuant as precursor ion fraction (PIF). A distribution of PIF values on a per-experiment basis can be found in the Peptide Identification tab of DO-MS.
Carrier channel assessment
▲ CRITICAL It is important to determine the degree of labeling of the carrier channel peptides, as any unlabeled peptides in this channel could be labeled by unquenched single-cell sample barcodes upon pooling of the carrier channel, reference channel, control well samples and single-cell samples.
-
25Before combining the bulk prepared carrier channel with your TMT-labeled single-cell samples, a 1× injection of the carrier material should be run separately to assess labeling efficiency as well as missed cleavage rate.
- If only 1% of the peptides in the carrier channel are subject to this cross-labeling, then the unlabeled material is equivalent to the peptide input of a single cell.
- Labeling efficiency of the carrier channel can be assessed using a custom-defined modification within MaxQuant, shown in Table 2 under ‘Variable search parameters’. This modification accounts for the mass of a TMT label on either the N-terminus or any lysine residues present in a given sequence, and will allow the fraction of labeling sites that were successfully labeled to be determined. The results are quantified and visualized by a dedicated tab of DO-MS.
- A high number of missed cleavage sites in the carrier channel input indicates suboptimal digestion conditions and can impair peptide identification.
- If either the labeling efficiency is low (samples should be at least 99% labeled) or the missed cleavage rate is high (the amount of miscleaved peptides should be <15%), a new carrier channel should be prepared if possible.
- Relabeling the carrier channel is often not a successful course of action owing to the presence of HA from the previous quenching reaction.
Metrics for evaluating single-cell quantification
▲ CRITICAL Many factors may undermine data quality, including the possibility of inefficient cell isolation (sorting), poor protein digestion, incomplete TMT label quenching, low TMT reactivity due to partial hydrolysis, and background contamination. Thus, we strongly recommend using built-in controls to benchmark SCoPE2 data quality.
-
26Determine the distributions of reporter ion intensities (RIIs) for each label, and normalize by those of the most abundant sample’s label. From these data, estimate the relative peptide input in each channel.
- The median intensity value in the distributions of relative reporter ion intensities (RRIIs) in the single-cell channels should be 100-fold less than the normalized median RII in the carrier channel, if the carrier channel contains digested peptides from 100 cells.
- Channels that contain median RRII equivalent to those found in the control wells may be failed wells.
-
Channels that contain median RRII equivalent to the median RRII in the reference channel may not have been properly quenched, allowing for cross-labeling of abundant material from the carrier channel during sample combination.A plot of RRII distributions can be found in DO-MS’ Single-Cell Diagnostics tab.
-
27
A correlation matrix of RIIs is another useful metric that often can detect major problems in sample preparation. To prepare a correlation matrix for assessing whether cross-labeling has occurred, remove the following items from the experimental dataset: peptide spectral matches below a chosen confidence cut off, reverse matches, and contaminants. Then, plot the pairwise correlations between vectors of RIIs for each channel, and display the information as a correlation matrix.
In general, the RIIs for cells of one type should correlate more highly with one another than with cells of another type or control wells. Such a correlation matrix can be found in DO-MS’ Single-Cell Diagnostic tab.
SCoPE2 sample preparation ● Timing 1–2+ d
Preparation of SCoPE2 sets can be divided into three key stages: (i) initial preparation of 384-well plates containing sorted single cells and control wells for the system under investigation; (ii) preparation of the carrier and reference channel material in bulk; and (iii) preparation of the 384-well plates containing single-cell material, and its combination with the carrier and reference material to generate completed SCoPE2 sets (Table 4). A single 384-well plate, along with its carrier and reference material, can be sorted and processed in 1–2 d. Additional 384-well plates can be processed faster with ~2 h of hands-on time per plate.
Table 4 ∣.
Automated and manual options for performing the SCoPE2 protocol
| Protocol step | Automated options | Manual options |
|---|---|---|
| Cell isolation (22-25) | FACS or CellenONE | Manual cell pickinga |
| Cell lysis (40-42) | Liquid handler (e.g., Mantis) | Multi- or single-channel pipette |
| Protein digestion (43-46) | Liquid handler (e.g., Mantis) | Multi- or single-channel pipette |
| TMT labeling (48-54) | Liquid handler (e.g., Mantis) | Multi- or single-channel pipette |
| Pooling samples (55-57) | Not used | Single-channel pipette |
| Loading samples (65) | Autosampler | Not used |
| LC-MS/MS (65) | Xcalibur or timsControl | Not used |
Placing the 384-well plates with sorted cells into the liquid handlers and the pooled SCoPE2 samples into the autosamplers is performed manually.
Live single cells can be picked under a microscope with a pipette as previously described17.
SCoPE2 sample preparation part I: isolating single cells ● Timing 1 h 20 min +
▲ CRITICAL Isolating single cells can be accomplished by FACS or by CellenONE as demonstrated in this protocol. When using CellenONE, samples can be prepared by nPOP as described in ref. 23. We have successfully used Aria II, Aria III and Sony MA900 sorters. Other single-cell dispensers, such as Namocell, might also be usable, but we do not have direct experience with them.
Ideally, single-cell isolation/sorting should be evaluated by simple and direct means, e.g., by using colorimetric assays. Its success should be maximized by ensuring stable spray and good alignment for FACS sorters and low static electricity for CellenONE. If the isolation process is not well validated, the use of positive control well (containing cell lysate diluted to single-cell level) is strongly encouraged.
-
28
Add 1 μL of HPLC-grade water, supplemented with 25 fmol per peptide per μL of Waters MassPrep to each well of a 384-well PCR plate. Seal the plates, and then centrifuge for 20 s (subsequent use of a plate spinner in this manner will be marked ‘spin down’). These plates can either be used immediately or stored at −80 °C until required.
▲ CRITICAL STEP While the use of a liquid dispensing robot is strongly recommended, it is not essential for this step.
■ PAUSE POINT The plates can be stored at −80 °C for at least 1 week.
-
29
Prepare a disaggregated suspension of unfixed cells, washed twice with 1× ice-cold PBS, and resuspended in ice-cold PBS. Exactly how this is achieved will vary depending on the cell type of interest. Some examples are given below:
Suspension cells: centrifuge to remove cell culture medium, and wash with, and then resuspend in, ice-cold 1× PBS.
Adherent cells: trypsinize or remove cells from a dish surface by scraping. Centrifuge to remove cell culture medium. Then wash with, and resuspend in, ice-cold 1× PBS.
-
30
Distribute single cells into the 384-well plates prepared earlier. A suitable method is the use of a cell sorter, with care to ensure that the sorted population represents single live cells of interest.
▲ CRITICAL STEP As described in the section on experimental design, the distribution of particular cells on and between different 384-well plates should include no-cell controls (negative control samples). If multiple cell treatments are analyzed, these should be distributed equally across the plates to avoid pairing cell types and batch effects.
The wells on the outer edge of the 384-well plate may experience edge effects during heating and incubation steps, leading to more rapid evaporation. You may wish to omit these wells from use.
-
31
After isolating single cells into the plates, seal and spin down the plates, and then transfer to a −80 °C freezer as rapidly as possible.
■ PAUSE POINT The plates can be stored at −80 °C for at least 1 week until you have processed all the cells that you are going to take forward. Ideally all the cells of interest should be prepared in a single batch.
SCoPE2 sample preparation part II: carrier and reference channel processing ● Timing 1 d
▲ CRITICAL The number of cells used for isobaric carrier can be determined based on the principles and tradeoffs established from controlled experiments19. For the default size of 100–200-cell carrier, you need ~11,275 cells to prepare the carrier and the reference per 384-well plate and fewer cells can be used if needed. If cells are not limited, we recommend retaining ~22,000 cells per 384-well plate being prepared. This allows for a 200-cell carrier and five-cell reference channel per SCoPE2 set, and also minimizes dilution or small-volume pipetting steps. Cells should be combined according to the carrier/reference design, washed in PBS, then resuspended in 11 μL of HPLC-grade water in a PCR tube and stored at −80 °C until the carrier and reference channel processing stage of the protocol.
We strongly recommend that the carrier and reference material is prepared as a single batch, sufficient for the number of 384-well plates being prepared for the SCoPE2 experiment. This is especially important (i.e., essential) for the reference material. The numbers given in the steps below will result in sufficient carrier/reference material to prepare a single 384-well plate of SCoPE2 samples and should be scaled up accordingly for increased numbers of plates.
-
32
Remove the PCR tube containing the sample from the −80 °C freezer, and transfer to a PCR machine as rapidly as possible.
-
33
Heat the sample to 90 °C for 10 min (heated lid at 105 °C), then allow it to cool to 12 °C, and spin down using a PCR tube spinner.
-
34
Transfer the sample to a water bath sonicator, and sonicate for 5 min at room temperature, and spin down.
-
35To the sample, add 2.2 μL of HPLC-grade water containing the following:
Component Amount Final concentration per tube Trypsin Gold 50 ng/μL 8.33 ng/μL TEAB pH 8.5 500 mM 83.33 mM Benzonase nuclease 1.2 units 0.2 units -
36
Vortex and spin down the sample PCR tube.
-
37
Incubate the sample in a 96-well PCR machine at 37 °C (heated lid 52 °C) for 3 h.
-
38
After digestion, spin down the sample, and split into two equal 6.6 μL volumes, each containing 11,000 cells in separate PCR tubes.
-
39
To one sample, add 3.3 μL of 126 TMT label (85 mM). This will become the carrier material. To the other sample, add 3.3μL of 127N TMT label (85 mM). This will become the reference material. This step is identical whether 11plex TMT or TMTpro 16plex reagents are used. In both cases, the 126 and 127N labels are used.
-
40
Spin down the two samples, and incubate at room temperature for 1 h.
-
41
Add 1.65 μL of 0.5% HA diluted in HPLC-grade water to each of the two samples. Then vortex the tubes and spin down.
-
42
Incubate the tubes at room temperature for 30 min.
-
43
Dilute the 127N-labeled tube containing the reference material by taking 2 μL of the reference and mixing it with 78 μL of HPLC-grade water. Then, the carrier and diluted reference material can be mixed 1:1 and subsequently further diluted so that carrier and reference concentrations are 100–200 cells/μL and 5 cells/μL, respectively. (For example: combine 8 μL of carrier with 8 μL of diluted reference, then add 22 μL of HPLC-grade water to obtain 200 cells/μL carrier and 5 cells/μL reference.) ▲ CRITICAL STEP If preparing more than one 384-well plate as part of a SCoPE2 experiment, it is recommended to prepare carrier and reference material in bulk, and prepare the carrier and reference channel material in SCoPE2 set into aliquots, so they can be used per 384-well plate. The volumes described in this section can be scaled up relative to cell numbers.
-
44
Vortex and spin down the combined carrier/reference material. Store at −80 °C until required in the following section. Consider not diluting the mixed carrier and reference material if not using right away.
■ PAUSE POINT The carrier/reference material can be stored at −80 °C for at least 2 weeks.
-
45
Assess the quality of the carrier material prior to combining it with single-cell samples in the following section III. Such quality checks include miscleavage rate and labeling efficiency, which should be <15% and >99%, respectively. Additionally, overlabeling should be assessed. This can be done in MaxQuant, by specifying TMT Pro labels as a variable modification on histidine, serine, threonine and tyrosine, and as a fixed modifications on primary amines (N-terminus and lysine).
In our experience, overlabeling has not been a major problem with the SCoPE2 protocol. Nonetheless, we evaluated the degree of overlabeling for the data generated for this protocol, both the single-cell and bulk samples. We performed a MaxQuant search in which TMT Pro labels were specified as a variable modification on histidine, serine, threonine and tyrosine, and as a fixed modifications on primary amines (N-terminus and lysine); data are available at doi: 10.6084/m9.figshare.15145092. We found very few peptides with TMT labeling on histidine, serine, threonine or tyrosine. After applying standard filters used by the SCoPE2 data pipeline (keeping peptides that have posterior error probability (PEP) ≤ 0.02 and PIF ≥ 0.9 and eliminating reverse hits and potential contaminants), we find a total of 225 unique overlabeled sequences, amounting to ~3% of all peptide sequences. When evaluating each SCoPE2 run individually, the amount of overlabeled and confidently identified peptides ranges from 0.1% to 2.92% of the total run.
? TROUBLESHOOTING
SCoPE2 sample preparation part III: single-cell processing SCoPE2 set generation ● Timing 8 h (2 h hands-on) per 384-well plate
▲ CRITICAL For the following steps, a single 384-well plate should be prepared at a time.
-
46
Remove the 384-well plate from the −80 °C freezer, and transfer to a 384-well PCR machine as rapidly as possible.
-
47
Set the PCR machine to heat the 384-well plate to 90 °C for 10 min with the heated lid set to 105 °C, followed by cooling the plate to 12 °C. Spin down the plate.
-
48
Transfer the 384-well plate to a water bath sonicater. Sonicate for 5 min at room temperature, and spin down.
-
49While sonication is being performed, prepare 100 μL of a master mix per 384-well plate, which contains the following:
Component Amount Final concentration per tube Trypsin Gold 50 ng/μL 8.33 ng/μL TEAB pH 8.5 500 mM 83.33 mM Benzonase nuclease 1.2 units 0.2 units ▲ CRITICAL STEP The use of alternative sources of trypsin requires optimization, as some suppliers have higher levels of contamination and perform poorly for SCoPE2.
-
50
Using a liquid handler (e.g., Mantis), dispense 0.2 μL of master mix into each well of the 384-well plate.
▲ CRITICAL STEP If using manual liquid dispensing, use of larger volumes is recommended to reduce handling errors.
-
51
After adding the master mix to all wells of the 384-well plate, seal the plate, vortex for 5 s and spin down.
-
52
Incubate the 384-well plate in a 384-well PCR machine at 37 °C, with the heated lid set to 52 °C, for 3 h.
-
53
After digestion, spin down the plate.
-
54
As illustrated in Table 5, a typical layout for a SCoPE2 experiment has a 126-labeled carrier channel, 127N-labeled reference and 127C left blank. This leaves eight channels available for labeling single cells or control wells from 128N-131C. If using TMTpro 16plex reagents, the SCoPE2 layout remains similar, and is shown in Table 6. Remove the 85 mM stocks of TMT labels from 128N-131C from the −80 °C freezer, warm to room temperature and dilute in anhydrous acetonitrile to 22 mM.
▲ CRITICAL STEP The placement of carrier, reference and empty channels in these designs helps prevent contamination of single-cell data due to isotopic contamination of these TMT labels from the much more abundant carrier and reference channels. The extra empty channel included with the 16plex TMTpro is due to the current higher level of isotopic contamination with these reagents compared with 11plex TMT.
-
55
Using a liquid dispensing robot (e.g., Mantis), add 0.5 μL of the diluted TMT reagents to the single cells using the 3PFE chips suitable for high solvent concentrations.
▲ CRITICAL STEP Ensure equal distribution of the different TMT tags so that sets contain one each of samples labeled from 128N to 131C. These can, for example, be prepared in columns on the 384-well plate.
-
56
After adding the TMT reagents to all wells of the 384-well plate, seal the plate, vortex for 5 s and spin down.
-
57
Incubate the plate at room temperature for 1 h.
-
58
Add 0.2 μl of 0.5% HA diluted in HPLC-grade water to each well of the 384-well plate using the liquid dispensing robot.
-
59
After adding the 0.5% HA to all wells of the 384-well plate, seal the plate, vortex for 5 s and spin down.
-
60
Incubate the plate at room temperature for 30 min.
-
61
Remove the combined carrier and reference material prepared in the previous carrier and reference channel processing step from the −80 °C freezer.
-
62
For each SCoPE2 set, take 1 μL of the carrier/reference material and add it to the first single-cell well of the 384-well plate (128N).
-
63
Pass the combined carrier/reference material through each of the remaining single-cell wells (128C–131C) to generate a complete SCoPE2 set containing carrier, reference and eight single-cell or control samples.
▲ CRITICAL STEP Combining the samples in this way helps to minimize losses when handling the single-cell material, as the much more abundant carrier material will disproportionately experience the losses.
-
64
Take 5 μL of 50% acetonitrile prepared in HPLC-grade water, and pass it through the same single-cell wells, combining with the carrier/reference/single-cell material in the final well. This step can help recover any remaining material from the single-cell wells.
-
65
Transfer SCoPE2 sets to individual glass autosampler inserts.
-
66
Place the autosampler inserts into a speedvac, and dry down completely.
-
67
Once dried, if not running immediately, store the dried SCoPE2 sets at −80 °C until required.
■ PAUSE POINT The dried sets can be stored for at least 1 week.
-
68
When ready for LC-MS/MS analysis, resuspend SCoPE2 sets in 1.2 μL of 0.1% formic acid in HPLC-grade water.
-
69
Place the glass autosampler inserts in glass autosampler vials, capped using screw-thread caps.
-
70
Vortex the capped autosampler vials for 5 s, then centrifuge briefly in a vial spinner to collect the sample at the bottom of the glass autosampler insert. Visually inspect to ensure the sample is at the bottom of the insert, and then place into the autosampler.
-
71
Inject 1 μL of each SCoPE2 set for LC-MS/MS analysis.
▲ CRITICAL STEP Instrument and LC method parameters for analysis of individual SCoPE2 were optimized in previous section on LC-MS/MS optimization using 100xMaster Standards. Use these optimized parameters.
▲ CRITICAL STEP Only 1 μL of the 1.2 μL sample is injected to allow for potential evaporation, or inefficient sample pickup by the autosampler.
? TROUBLESHOOTING
Table 5 ∣.
SCoPE2 sample labeling strategy
| 126 | 127N | 127C | 128N | 128C | 129N | 129C | 130N | 130C | 131N | 131C |
| Carrier | Reference | Empty | SC | SC | SC | SC | SC | SC | SC | SC |
126 and 127N are used for the carrier and reference, which are prepared in bulk as described in the ‘Carrier and reference channel processing’ section of the protocol and are subsequently diluted. 128N-131C are used for labeling SCoPE2 single-cell and control samples. 127C is unused due to isotopic contamination from 126.
Table 6 ∣.
SCoPE2 sample labeling strategy using 16plex TMTpro reagents
| 126 | 127N | 127C | 128N | 128C | 129N | 129C | 130N | 130C | … | 134N |
| Carrier | Reference | Empty | Empty | SC | SC | SC | SC | SC | … | SC |
Sample layout and isobaric reagent usage is essentially the same as when using 11plex TMT reagents. 131N to 133C are not shown, but can also be used for single-cell or control samples. The higher level of isotopic carryover from 127N with some TMT reagent lots means that 127C and 128N are both unused due to isotopic contamination from 126 and 127N. However, the total number of single-cell channels that can be used per SCoPE2 set increases from 8 to 12 in comparison with 11plex reagents.
Raw data processing ● Timing 1+ h
-
72
Search MS spectra for quantified peptides. Raw files corresponding to the SCoPE2 sets constituting a SCoPE2 experiment can be analyzed using a variety of proteomics search engines. For the purpose of accessibility and simplicity, we will focus on settings appropriate to searching the data with the freely available software MaxQuant. All parameters are specified in the search parameters table.
-
73Optional: use additional features to enhance MS data interpretation. The SCoPE2 pipeline uses the DART-ID software to update the confidence of peptide identification, e.g., update the PEPs in the evidence.txt file generated by MaxQuant. The DART-ID software uses a rigorous Bayesian model to incorporate retention time information in evaluating confidence of peptide identification24. For a more complete explanation of the DART-ID software and its usage, please see https://dart-id.slavovlab.net. Once installed, DART-ID can be run by editing the default configuration file provided to include the path to your evidence file and the path to your desired output location, for example:
input: a. /path/to/your_search_results/evidence.txt output: b. /path/to/output_folder/ Then, run DART-ID in the python programming environment using the following command, specifying the path to the configuration file, for example: dart_id -c path/to/config/example_config_file.yaml
Evaluating quality of single-cell preparation ● Timing 1+ d
● CRITICAL The quality of sample preparation should be evaluated for every single cell. The inclusion of negative control wells allows background signal to be characterized. Below, we describe data analysis that can be implemented using the SCoPE2 GitHub52 or Zendo53 repositories. This SCoPE2 code was used to develop the scp Bioconductor package54,55, which can also be used to analyze the data.
-
74Filter the peptide data to improve the quality of peptide and protein identifications.
- Remove peptides with FDR >1%. Sometimes, it may be desirable to filter out peptides with PEP >0.02
- Remove peptides belonging to proteins with FDR >1%
- Remove reverse and contaminant peptides (as annotated by MaxQuant)
- Remove peptides with precursor ion fraction (PIF) <0.8
-
Remove peptides with >10% RII of the carrier channel RII? TROUBLESHOOTING
-
75Perform the necessary data transformations.
- To control for differences in the sampling of elution profiles in different LC-MS/MS runs, a common reference was used as a denominator for all single-cell quantification. This means that the RII for every peptide in every single cell or control was divided by the RII from the reference channel from the same peptide.
- Then, to control for differences in sample loading, the relative peptide RIIs (relative to reference) for each single cell or control were divided by their median RII.
- Then, to put all peptides on the same scale, the relative peptide reporter intensities for each peptide across all single cells and controls were divided by their median value.
- Protein quantification was determined by taking the median peptide value for peptides belonging to said protein (denoted by MaxQuant as Leading Razor Protein).
-
76
Determine whether the expected signal level was obtained. The preparation of single cells can be evaluated by looking for the expected amount of ions observed. The expectation is that well-prepared cells yield RIIs roughly proportional with the intensity from the carrier or reference channels (so ratios of 200:1 or 5:1, respectively) and that control wells and poorly prepared single cells yield values less than that ratio (200:0.1 or 5:0.1, for example).
-
77
Determine whether the quantification was consistent. Additionally, the preparation of single cells can be evaluated by looking for the consistency of peptide quantification within those peptides coming from the same proteins. This is captured by the coefficient of variation (CV) statistic, the expectation being that well-prepared cells yield peptides with consistent quantification if they come from the same proteins (typical interquartile range of protein CVs for a successful single cell would be 0.2–0.4) and, thus, lower CV than the control wells or poorly prepared single cells.
-
78
Identify failed wells. For every single cell and control, plot the median RRII versus the median CV. Failed single cells and negative controls should cluster apart from successfully prepared single cells.
-
79Identify batch effects. As in any experiment, multiple parts of SCoPE2 are subject to potential batch effects. Experiments should be designed to make sure experimental conditions do not correlate with potential batch effects. For SCoPE2, potential batch effects include:
- 384-well plate (i.e., do not process different conditions on separate 384-well plates)
- Plate location (i.e., edge effects)
- Chromatography (i.e., LC-MS/MS column, or run-to-run variability)
- TMT label (i.e., differences in labeling efficiency due to contaminated reagent)
Data availability recommendations
▲ CRITICAL In addition to following conventional practices for deposition of raw MS data and search results, we make the following recommendations for data availability to facilitate data reuse.
-
80Prepare data files from intermediate steps of data processing. We usually provide at least three files in comma separated values (csv) format as follows:
- Peptides-raw.csv: peptides × single cells at 1% FDR and including peptides identified by DART-ID. The first two columns list the corresponding protein identifiers and peptide sequences, and each subsequent column corresponds to a single cell
- Proteins-processed.csv: proteins × single cells at 1% FDR, imputed (k-nearest neighbors, k = 3) and batch corrected (ComBat package in R)
- Cells.csv: annotation × single cells. Each column corresponds to a single cell, and the rows include relevant metadata, such as cell type if known, measurements from the isolation of the cell (e.g., from index sorting by FACS), and derivative quantities, i.e., RRI, CVs, reliability
These files can be included with the raw file deposition, and as supplementary data provided with a manuscript.
Troubleshooting
Troubleshooting advice can be found in Table 7.
Table 7 ∣.
Troubleshooting table
| Step | Problem | Cause | Solution |
|---|---|---|---|
| LC-MS system | |||
| 21 | Apex offset distribution not centered around 0 | Consistently sampling peptides before or after their elution apices | Adjust the length of your duty cycle by changing the number of peptides submitted for MS2 (top N setting) and/or the MS2 fill times. See ref. 25 for examples and guidelines |
| Increased pressure trace | Undigested protein or nucleic acids obstructing column | If using the recommended ionOpticks column, remove column, examine the end for obstructions under a light microscope, and trim end just beyond obstructions using a diamond cutter | |
| Multiple MS2 scans/peptide | Broad elution peak | Aging LC column or free space in the LC plumbing | |
| 65 | No signal from a SCoPE sample injection | Autosampler needle may not have aspirated fully the expected 1 μL injection, resulting in an injection of air | Spin down sample and ensure that liquid is settled evenly at bottom of tube and/or service the autosampler |
| 68 | Low number of identified peptides | Poor sample delivery, lower-than-expected carrier, contamination, suboptimal parameters or anothe problem | This can be caused by many factors. Look at the DO-MS report (do-ms.slavovlab.net) to specifically diagnose the problem |
| Sample | |||
| 39 | Poor labeling efficiency | TMT levels may be hydrolyzed or conditions may not be optimal for TMT labeling | Check your TMT stock and ensure the postdigest pH is ~8 |
| 68 | Relative reporter ion ratios do not match expected values | Sample input may not match expected input amount, or TMT labels may not have been properly quenched | Use the positive controls to diagnose the problem |
| Low correlations between cell-type-based RII ratios for single-cell channels and carrier channels (if using an experimental design that features two carrier channels) | Possible cross-labeling of samples | Ensure that labeling and quenching conditions match those described here and that HA is still active. Additionally, ensure that the isobaric carrier is well labeled. Unlabeled carrier peptides may be labeled by residual labels used for the single-cell channels if the labels have not been fully quenched | |
| Analysis | |||
| 39 | Poor labeling efficiency | Suboptimal labeling conditions or hydrolyzed TMT | Check your TMT stock39 and ensure the postdigest pH is ~8 |
The authors delivered a workshop covering SCoPE2 experimental design, sample preparation and data analysis at the SCP2019 conference on single-cell proteomics, held in Boston in June 2019. Videos from this workshop are available on YouTube and may be of use to those seeking further clarification or troubleshooting56.
Timings
Preparatory work
Steps 1–21, production of 100xMaster and 100xCarrier samples: 1–2 d
Steps 22–27, system suitability and optimization: 1+ d
Single-cell work
Step 28, plate preparation: 20 min
Steps 29–31, cell isolation: 1+ h
Steps 32–67, sample preparation: 8 h (2 h hands-on) per 384-well plate
Steps 68–71, LC/MS analysis: 95 min per TMT set
Steps 72–80, initial data analysis: 1+ d
Sample preparation and LC/MS analysis will be repeated multiple times as required to achieve the desired experiment depth. Up to 48 SCoPE sets can be prepared per 384-well plate using 11plex TMT reagents, or 32 SCoPE2sets with 16plex TMTpro depending on the exact experimental design
Anticipated results
The SCoPE2 method was initially applied to the study of U937 monocyte cells differentiating to macrophage-like cells, which provided an example of how the quantified proteins can be used for biological analysis18. Here, we exemplify the described SCoPE2 protocol (using cellenONE for cell isolation) with a smaller and simpler experiment (analyzing six SCoPE2 sets consisting of single HeLa and U937 cells) so that we can focus on key technical benchmarks that will be helpful for new groups adopting the SCoPE2 workflow.
The success and the problems of data acquisition can be analyzed by the corresponding DO-MS report. The full report consisting of many dozens of plots can be found as supplemental material, and Fig. 3 shows a few plots demonstrating expected results. The distribution of precursor intensities in Fig. 3a is a useful benchmark to evaluate problems with sample pickup (which manifests as sporadic runs with very low precursor intensities) or poor sample delivery to the instrument (which manifests with decline in the precursor abundances relative to a reference standard). To make this comparison meaningful, DO-MS compares distributions composed of the same peptides across all runs25.
Fig. 3 ∣. Evaluating data acquisition and interpretation using diagnostic plot generated by DO-MS.
a, Distributions of intensities for precursors quantified in all displayed experiments. The similar medians and shapes indicate successful and uniform delivery of the SCoPE2 samples to the MS instrument. This distribution and many additional distributions are generated by DO-MS, for example, the distribution of all detected precursors, which contains many less abundant precursor ions. The corresponding dataset can be found at https://doi.org/10.6084/m9.figshare.15060774.v1. b, The distributions of times for accumulating ions for MS2 scans indicate that most ions were accumulated for maximum time allowed (300 ms) and thus the size of the isobaric carrier and the choice of AGC max did not limit the sampling of peptides from the single cells19. c, Distributions of times between the apexes of elution peaks and the time when they were sampled for MS2 scans. The small deviations from the apex indicate good apex sampling, and larger devisions would require further optimization as described previously25. d, Number of MS2 scans and PSMs at different levels of confidence. The PSM confidence is quantified by the PEP computed by MaxQuant as well as by the PEP updated by DART-ID. These and many other plots are automatically generated by DO-MS25.
Suboptimal choice of instrument parameters might result in short MS2 accumulation times and, thus, in sampling only a few copies from the single-cell peptides19,20,25. To evaluate and control this possibility, DO-MS displays the distributions of MS2 accumulation times as shown in Fig. 3b. As expected for our six runs, the isobaric carrier did not limit MS2 accumulation times, and peptides selected for MS2 scans were accumulated for the maximum time specified in our instrument methods, 300 ms.
To maximize the copy number of ions sampled for quantification and the purity of MS2 spectra, SCoPE2 aims to sample elution peaks close to the apex18. To help achieve this goal, DO-MS displays the distributions of apex offsets as shown in Fig. 3c. In the six example runs included here, most ions were sampled within a few seconds from the apex without systematic bias of early or late sampling. If such biases are present, follow the recommendations outlined in ref. 25 to mitigate them.
DO-MS evaluates the number of identified peptides in the context of total number of MS2 scans taken, the total number of peptide-spectrum-matched (PSMs), in addition to the number of confident PSMs based on spectra alone and based on both spectra and DART-ID. The example plot in Fig. 3 shows that most runs acquired ~9,000 MS2 scans (which is close to the number expected for full duty cycles), and a large fraction of these MS2 spectra were assigned sequences with high confidence, ~20% based on spectra alone and ~40% based on spectra and retention time.
Before evaluating the accuracy of protein quantification by SCoPE2, we evaluate whether the RRIIs follow the expected trends. In particular, negative control wells shown have low RIIs (low background) and the single-cell RIIs should be about 1/x the RIIs of the isobaric carrier, where x is the number of cells in the isobaric carrier. These expected results are illustrated in Fig. 4a with data from five single HeLa cells and five single U937 cells. The RIIs for the HeLa cells are larger because of their larger cell sizes. Most reporter ions are not detected in the negative control, indicating that background noise is below the limit of detection of our system.
Fig. 4 ∣. Evaluating protein quantification results from SCoPE2 analysis.
a, Distributions of relative reporter ion intensities for all samples from a single SCoPE2 set. The samples include an isobaric carrier, a reference 5 single HeLa cells, five single U937 cells, a negative control well (Ctr−) and a positive control well (Ctr+) from diluted U937 cell lysate. Two of the TMT labels (between the reference and the single cells) are not used because they are affected by isotopic impurities of the TMT labels used for the carrier and the reference. The bin marked by >−3.5 corresponds mostly to RI intensities below the detection limit (missing values). The spread of the single-cell RI distributions corresponds to differences in the relative protein levels among the single cells and the isobaric carrier/reference. b, Distributions of CVs quantify the consistency of protein quantification based on different peptides originating from the same protein. Lower CVs for the single cells correspond to higher consistency of quantification compared with the CVs of the negative control wells, which correspond to noise. c, PCA of 126 single cells and bulk samples. The PCA is based on 1,756 proteins with ~1,000 proteins quantified per single cell. b and c are generated by the SCoPE2 pipeline53.
In a large-scale experiment, some single-cell samples may not provide useful data because of problems during cell isolation or sample preparation. Such samples can be identified based on the low consistency of quantification derived from different peptides as shown in Fig. 4b. This diagnostic plot is generated by the SCoPE2 analysis pipeline18,53.
Low-dimensional projections, such as the principal component analysis (PCA) shown in Fig. 4b, are frequently used to summarize single-cell data, and the degree to which similar samples cluster together may reflect the discriminatory power of the data. However, samples may be separated by PCA because of technical signals, such as batch effects or incomplete normalization of the size difference between HeLa and U937 cells, Fig. 4a. To evaluate whether the separation along the first principal component (PC1) is indeed capturing cell-type-specific differences in protein abundance, we strongly recommend including bulk samples, as in the PCA displayed in Fig. 4c. We expect single cells and bulk samples from the same cell type to cluster close to each other and far away from clusters corresponding to different cell types, as observed in Fig. 4c.
Extended Data
Extended Data Fig. 1 ∣. The ABIRD may suppress contaminant ions and enhance peptide identification.
ABIRD may suppress contaminant ions and enhance peptide identification. Replicate injections of a 1× standard were analyzed with the ABIRD on or off. a, The replicates with ABIRD on had a reduced number of +1 ions (likely corresponding to contaminants) and an increased number of higher-charge-state ions, which are likely to correspond to peptides. b, With the ABIRD on, the number of identified peptides is increased across all confidence levels (PEP).
Supplementary Material
Acknowledgements
We thank A. Chen, and J. Neveu for assistance, discussions and constructive comments. This work was funded by a New Innovator Award from the NIGMS from the National Institutes of Health to N.S. under Award Number DP2GM123497, an Allen Distinguished Investigator award through The Paul G. Allen Frontiers Group to N.S., a Seed Networks Award from CZI CZF2019-002424 to N.S., through a Merck Exploratory Science Center Fellowship, Merck Sharpe & Dohme Corp. to N.S., and a Thermo Scientific Tandem Mass Tag Research Award to E.E. Funding bodies had no role in data collection, analysis and interpretation.
Footnotes
Competing interests
The authors declare no competing interests.
Code availability
The SCoPE2 pipeline used here is available at https://github.com/SlavovLab/SCoPE2 and via the SCoPE2 website (https://scope2.slavovlab.net/mass-spec/protocol).
Extended data is available for this paper at https://doi.org/10.1038/s41596-021-00616-z.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596-021-00616-z.
Data availability
All data are available at MassIVE MSV000087041 and via the SCoPE2 website: https://scope2.slavovlab.net/mass-spec/protocol. Datasets associated with certain figures have been uploaded to figshare: Fig. 3: Slavov, Nikolai (2021): Evaluating data acquisition and interpretation using diagnostic plots generated by DO-MS (https://doi.org/10.6084/m9.figshare.15060774.v1); Fig. 4: Slavov, Nikolai (2021): Principal Component Analysis (PCA) of quantification results from SCoPE2 analysis (https://doi.org/10.6084/m9.figshare.15060789.v1); Extended Data Fig. 1: Slavov, Nikolai (2021): The ABIRD device may suppress contaminant ions and enhance peptide identification (https://doi.org/10.6084/m9.figshare.15060846.v1) and Slavov, Nikolai (2021): Variable TMT search indicates very low level of over labeled peptides during SCoPE2 analysis (https://doi.org/10.6084/m9.figshare.15145092.v1). Source data are provided with this paper.
References
- 1.Symmons O & Raj A What’s luck got to do with it: single cells, multiple fates, and biological non-determinism. Mol. Cell 62, 788–802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul I, White C, Turcinovic I & Emili A Imaging the future: the emerging era of single-cell spatial proteomics. FEBS J. 2020). [DOI] [PubMed] [Google Scholar]
- 3.Levy E & Slavov N Single cell protein analysis for systems biology. Essays Biochem. 10.1042/EBC20180014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regev A et al. Science forum: the human cell atlas. eLife 6, e27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziegenhain C et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Engblom C et al. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science 358 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaffer SM et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franks A, Airoldi E & Slavov N Post-transcriptional regulation across human tissues. PLoS Comput. Biol 13, e1005535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slavov N Unpicking the proteome in single cells. Science 367, 512–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Washburn MP, Wolters D & Yates JR Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol 19, 242–247 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Altelaar AFM, Munoz J & Heck AJR Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet 14, 35–48 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Cravatt BF, Simon GM & Yates Iii JR The biological impact of mass-spectrometry-based proteomics. Nature 450, 991 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S & Heck AJ Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc 4, 484–494 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Muntel J et al. Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time. J. Proteome Res 18, 1340–1351 (2019). PMID: 30726097. [DOI] [PubMed] [Google Scholar]
- 15.Slavov N Single-cell protein analysis by mass spectrometry. Curr. Opin. Chem. Biol 60, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly RT Single-cell proteomics: progress and prospects. Mol. Cell. Proteomics 19, 1739–1748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budnik B, Levy E, Harmange G & Slavov N SCoPE-MS: mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biology 19, 161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Specht H et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 10.1186/s13059-021-02267-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Specht H & Slavov N Optimizing accuracy and depth of protein quantification in experiments using isobaric carriers. J. Proteome Res 20, a8802021–887 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung TK et al. Defining the carrier proteome limit for single-cell proteomics. Nat. Methods 18, 76–83 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Specht H et al. Automated sample preparation for high-throughput single-cell proteomics. Preprint at bioRxiv 10.1101/399774 (2018). [DOI] [Google Scholar]
- 22.Marx V A dream of single-cell proteomics. Nat. Methods 16, 809–812 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Leduc A, Huffman RG & Slavov N Droplet sample preparation for single-cell proteomics applied to the cell cycle. Preprint at bioRxiv 10.1101/2021.04.24.441211 (2021). [DOI] [Google Scholar]
- 24.Chen AT, Franks A & Slavov N DART-ID increases single-cell proteome coverage. PLOS Comput. Biol 15, 1–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huffman G, Chen AT, Specht H & Slavov N DO-MS: data-driven optimization of mass spectrometry methods. J. Proteome Res 10.1021/acs.jproteome.9b00039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albayrak C et al. Digital quantification of proteins and mRNA in single mammalian cells. Mol. Cell 61, 914–924 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Darmanis S et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 14, 380–389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes AJ et al. Single-cell western blotting. Nat. Methods 11, 749–755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Specht H, Emmott E, Perlman DH, Koller A & Slavov N High-throughput single-cell proteomics quantifies the emergence of macrophage heterogeneity. Preprint at bioRxiv. 10.1101/665307 (2019). [DOI] [Google Scholar]
- 31.Virant-Klun I, Leicht S, Hughes C & Krijgsveld J Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol. Cell. Proteomics 15, 2616–2627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lombard-Banek C, Moody SA & Nemes P Single-cell mass spectrometry for discovery proteomics: quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angew. Chem. Int. Ed 55, 2454–2458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cong Y et al. Improved single-cell proteome coverage using narrow-bore packed nanoLC columns and ultrasensitive mass spectrometry. Anal. Che 92, 2665–2671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong Y et al. Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell. Chem. Sci 12, 1001–1006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunner A-D et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Preprint at bioRxiv 10.1101/2020.12.22.423933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun 9, 882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou M et al. High-throughput single cell proteomics enabled by multiplex isobaric labeling in a nanodroplet sample preparation platform. Anal. Chem 91, 13119–13127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z-Y et al. Nanoliter-scale oil-air-droplet chip-based single cell proteomic analysis. Anal. Chem 90, 5430–5438 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Liang Y et al. Fully automated sample processing and analysis workflow for low-input proteome profiling. Anal. Chem 93, 1658–1666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafzi A, Moutinho C, Picelli S & Heyn H Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc 13, 2742–2757 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Barkas N et al. Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat. Methods 16, 695–698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marx V A dream of single-cell proteomics. Nat. Methods 10.1038/s41592-019-0540-6 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Grün D & van Oudenaarden A Design and analysis of single-cell sequencing experi- ments. Cell 163, 799–810 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Meier F et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Specht H & Slavov N Transformative opportunities for single-cell proteomics. J. Proteome Res 17, 2563–2916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slavov N Driving single cell proteomics forward with innovation. J. Proteome Res 10.1021/acs.jproteome.1c00639 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized ppb- range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Tyanova S, Temu T & Cox J The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc 11, 2301 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Yu S-H, Kyriakidou P & Cox J Isobaric matching between runs and novel PSM-level normalization in MaxQuant strongly improve reporter ion-based quantification. J. Proteome Res 19, 3945–3954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fondrie WE & Noble WS mokapot: fast and flexible semisupervised learning for peptide detection. J. Proteome Res 20, 1966–1971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milo R, Jorgensen P, Moran U, Weber G & Springer M BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 38, D750–D753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Specht H et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity. GitHub github.com/SlavovLab/SCoPE2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Specht H et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Zenodo 10.5281/zenodo.4339954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanderaa C & Gatto L Replication of single-cell proteomics data reveals important computational challenges. Preprint at bioRxiv. 10.1101/2021.04.12.439408 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Vanderaa C & Gatto L Mass spectrometry-based single-cell proteomics data analysis. Bioconductor http://www.bioconductor.org/packages/release/bioc/html/scp.html (2020). [Google Scholar]
- 56.SCP2019 Workshop on single-cell proteomics August. 2019. http://workshop2019.single-cell.net [Google Scholar]
Related links
Key references using this protocol
- Specht H et al. Genome Biol. 22, 50 (2021): 10.1186/s13059-021-02267-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik B et al. Genome Biol. 19, 161 (2018): https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1547-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht H & Slavov NJ Proteome Res. 20, 880887 (2021): https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
▲ CRITICAL In addition to following conventional practices for deposition of raw MS data and search results, we make the following recommendations for data availability to facilitate data reuse.
-
80Prepare data files from intermediate steps of data processing. We usually provide at least three files in comma separated values (csv) format as follows:
- Peptides-raw.csv: peptides × single cells at 1% FDR and including peptides identified by DART-ID. The first two columns list the corresponding protein identifiers and peptide sequences, and each subsequent column corresponds to a single cell
- Proteins-processed.csv: proteins × single cells at 1% FDR, imputed (k-nearest neighbors, k = 3) and batch corrected (ComBat package in R)
- Cells.csv: annotation × single cells. Each column corresponds to a single cell, and the rows include relevant metadata, such as cell type if known, measurements from the isolation of the cell (e.g., from index sorting by FACS), and derivative quantities, i.e., RRI, CVs, reliability
These files can be included with the raw file deposition, and as supplementary data provided with a manuscript.
All data are available at MassIVE MSV000087041 and via the SCoPE2 website: https://scope2.slavovlab.net/mass-spec/protocol. Datasets associated with certain figures have been uploaded to figshare: Fig. 3: Slavov, Nikolai (2021): Evaluating data acquisition and interpretation using diagnostic plots generated by DO-MS (https://doi.org/10.6084/m9.figshare.15060774.v1); Fig. 4: Slavov, Nikolai (2021): Principal Component Analysis (PCA) of quantification results from SCoPE2 analysis (https://doi.org/10.6084/m9.figshare.15060789.v1); Extended Data Fig. 1: Slavov, Nikolai (2021): The ABIRD device may suppress contaminant ions and enhance peptide identification (https://doi.org/10.6084/m9.figshare.15060846.v1) and Slavov, Nikolai (2021): Variable TMT search indicates very low level of over labeled peptides during SCoPE2 analysis (https://doi.org/10.6084/m9.figshare.15145092.v1). Source data are provided with this paper.