Abstract
Reducing speckle noise is an important task for improving visual and automated assessment of retinal OCT images. Traditional image/signal processing methods only offer moderate speckle reduction; deep learning methods can be more effective but require substantial training data, which may not be readily available. We present a novel self-fusion method that offers effective speckle reduction comparable to deep learning methods, but without any external training data. We present qualitative and quantitative results in a variety of datasets from fovea and optic nerve head regions, with varying SNR values for input images.
1. INTRODUCTION
Speckle noise can be a barrier to both visual assessment and automated analysis of OCT scans, which are widely used for retinal imaging. Many approaches have been proposed for reducing speckle noise. Hardware-based methods typically consist of acquiring multiple scans of the same or similar location and averaging to boost SNR; variants include multiple backscattering angles1 and joint aperture detection.2 Software-based methods include adaptive methods3-9 and variational methods.10,11 Others have proposed wavelet-based methods12-15 that seek to suppress speckle in the wavelet, curvelet,16 wave atom17 or spectral18 domains. Classic Perona-Malik gradient anisotropic diffusion filtering, which works well for additive noise, will enhance speckle rather than reducing it; however, speckle-reducing variants have been developed.19-21 Many of these traditional methods only offer moderate speckle reduction (software) or require additional acquisition time (hardware). In recent years, several learning and deep learning approaches have been proposed to better address this task. Fang et al.22 build a sparse representation dictionary from high-SNR images. Yu et al.23 proposes to use PCANet in conjunction with non-local means filtering. Ma et al.24 proposes an edge-sensitive conditional generative adversarial network (cGAN). Halupka et al.25 propose a GAN architecture with Wasserstein distance and perceptual similarity. Huang et al.26 also use a GAN structure, with the aim of simultaneous de-speckling and super-resolution. While these deep learning approaches are typically more successful than traditional methods in reducing speckle, they suffer from a need to re-train for different image appearances (such as images acquired with systems from different vendors). In this paper, we propose a non-learning method with a speckle reduction ability comparable to that of deep learning methods without requiring any external data.
Multi-atlas label fusion methods have been widely used for medical image segmentation.27 These methods rely on multiple so-called ‘atlases’ that are (often manually) labeled training samples. To segment a new test image, each atlas is deformably registered to the test image, allowing both the atlas image and the corresponding segmentation label map to be aligned to the test image. In the common space of the test image, each atlas is assigned a spatially varying weight, typically based on the residual registration error between the registered atlas and the test image. The segmentation result is then obtained through a weighted vote between the atlas labels.
In previous work, we introduced multi-atlas intensity fusion.28 This method also relies on atlases registered to a test image, but uses the weight maps to combine the atlases themselves rather than associated label maps. The outcome is a new image that represents the same anatomy as the test image using the atlases as basis functions. The weighted averaging allows noise reduction. The atlas set can be manipulated to achieve additional effects, such as the removal of lesions from the test image by using atlases that do not contain any lesions.28
In this paper, we propose a new ‘self-fusion’ technique that does not require any atlases. Instead, for each B-scan in an OCT volume, we use the neighboring B-scans as ‘atlases’. Since the entire volume is acquired through the same camera from the same eye, these B-scans offer exceptionally well-fitting atlases, in terms of both image appearance and anatomy, making registration and weight estimation very robust. The outcome is speckle reduction abilities comparable to that of hardware-based averaging without requiring multiple acquisitions. Unlike deep learning methods, no external training data is needed, as the volume serves as its own ‘atlas’.
2. METHODS
2.1. Data acquisition
One volume was acquired on a 400 kHz 1060±100 nm Axsun swept-source OCT engine (9.6 μm axial resolution in air) and sampled at 2560 × 500 × 400 pix. (spectral × lines × frames) with 4 repeated frames. OCT signal was acquired on a balanced photodiode and digitized at 2GS/s. 12 additional OCT volumes were acquired using a spectral-domain (SD-OCT) system with a 845±85 nm superluminescent diode light source (1.85 μm axial resolution in air) detected on a 4096 pix. line-scan CMOS sensor. Volumes were sampled at 4096 × 500 × 500 pix. (spectral × lines × frames) with 5 repeated frames at each position for a total of 2500 frames per volume. OCT SNR was adjusted by varying the detector exposure time from 6.7 μs, 3.35 μs, and 2 μs resulting in SNR values of 101 dB, 96 dB, and 92.5 dB, respectively. Volumes were acquired in two healthy volunteers in foveal and optic nerve head (ONH) region at each exposure setting.
On each volume, motion correction was performed by compensating for measured lateral and axial motion shifts using discrete Fourier transform registration on sequential B-scans. The repeated frames (4 for the first dataset, 5 for the second) were split into separate volumes. A single acquisition was used as the input to the denoising algorithm. The repeated acquisitions were averaged together to create the ‘ground truth’ for evaluation.
2.2. Self-fusion for OCT noise reduction
Given an input 3D OCT volume, we consider each 2D B-scan individually. For each input B-scan, we synthesize a new 2D B-scan that represents the same anatomy but with less noise. Then, we tile the synthesized B-scans together to obtain the final result, a denoised 3D OCT volume.
Let us consider an arbitrary input 2D B-scan Bi, where i indicates the slice index within the 3D OCT volume. We define a slice neighborhood by considering the set of B-scans within a radius R to Bi: NR(Bi) = {Bj∣i–R≤j≤i+R}. Then, we use the joint label fusion model of Wang et al.29 We note that in this interpretation of the multi-atlas fusion framework, we use the neighboring slices Bj ∈ NR(Bi) as the ‘atlases’. We call this method ‘self-fusion’ because, unlike traditional multi-atlas methods that rely on external atlases, our approach does not require any input other than the 3D OCT volume to be denoised. We further note that there are no ‘labels’ in our fusion approach: we rather use the intensity fusion technique, where the weight maps assigned to each atlas are used to combine the atlas intensities, rather than atlas labels.28 The result of intensity fusion is a synthesized image that represents the target image as a weighted combination of the atlas images.
Specifically, we begin the synthesis process by registering each 2D B-scan Bj ∈ NR(Bi) to the current B-scan Bi. Then, for each pixel (x, y) on each registered slice Bj, we compute a weight wj(x, y) following the joint label fusion model. The joint label fusion model takes into account both the local patch similarity between the atlas Bj and the target slice Bi, as well as the patch similarities between pairs of atlases Bj and Bk (Bj, Bk ∈ NR(Bi)). We use a 5 × 5 pixel neighborhood to compute patch similarity.
We note that we use Bi itself as an atlas to synthesize Bi. This is desirable since Bi naturally contains the most information about the anatomy represented in Bi, and it is thus highly valuable for successful fusion. However, Bi has, by definition, perfect similarity to the target image Bi, which means it would be assigned an extremely high weight that would dominate any contribution from other atlases. This would lead to a synthesis result that is practically identical to the input image. We avoid this problem by using a very high value for α, the weight of the conditioning identity matrix used to compute the atlas similarity matrix M in the joint label fusion framework.29 This makes it possible to emphasize Bi by assigning its pixels relatively high weights in the fusion, while still allowing nontrivial contribution from the other atlases in NR(Bi).
3. RESULTS
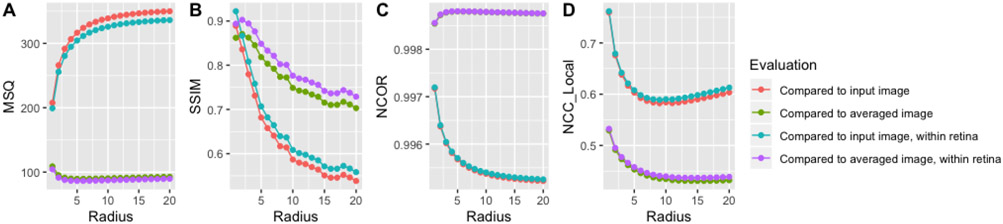
Figure 1 shows quantitative evaluation of the self-fusion results on the first dataset as a function of the neighborhood radius R. As can be expected, these metrics are similar to each other in overall trend, but the comparison to the average image performs better than the comparison to the noisy input, and the metrics improve when only considering the voxels within the retina. We note that some metrics improve with increasing radius, as the SNR becomes stronger, whereas other metrics deteriorate, likely due to the increased blurring. Based on this analysis, we choose a radius of 5 voxels for the second dataset.
Figure 1.
Quantitative evaluation for one OCT volume (first dataset) for varying radius R. A Mean square difference (MSQ), B structural similarity index (SSIM, 2-voxel radius), C image-wide normalized cross-correlation (NCC), D local (patch-based, patch radius 4 voxels) NCC. We report each metric between the self-fusion result and the input image (single acquisition) or the ground truth (4 acquisitions averaged), for the whole image or within the retina (manual mask).
Figures 2 and 3 show qualitative results for varying SNR levels, in the ONH and fovea regions respectively, in images from the second dataset. We note the good performance of our self-fusion method on even very noisy images. This is potentially useful in clinical applications for patients who may suffer from cataracts, vitreal haze, or corneal opacity. We also note the preservation of fine features, such as blood vessels and their shadows, as well as tissue layers.
Figure 2.
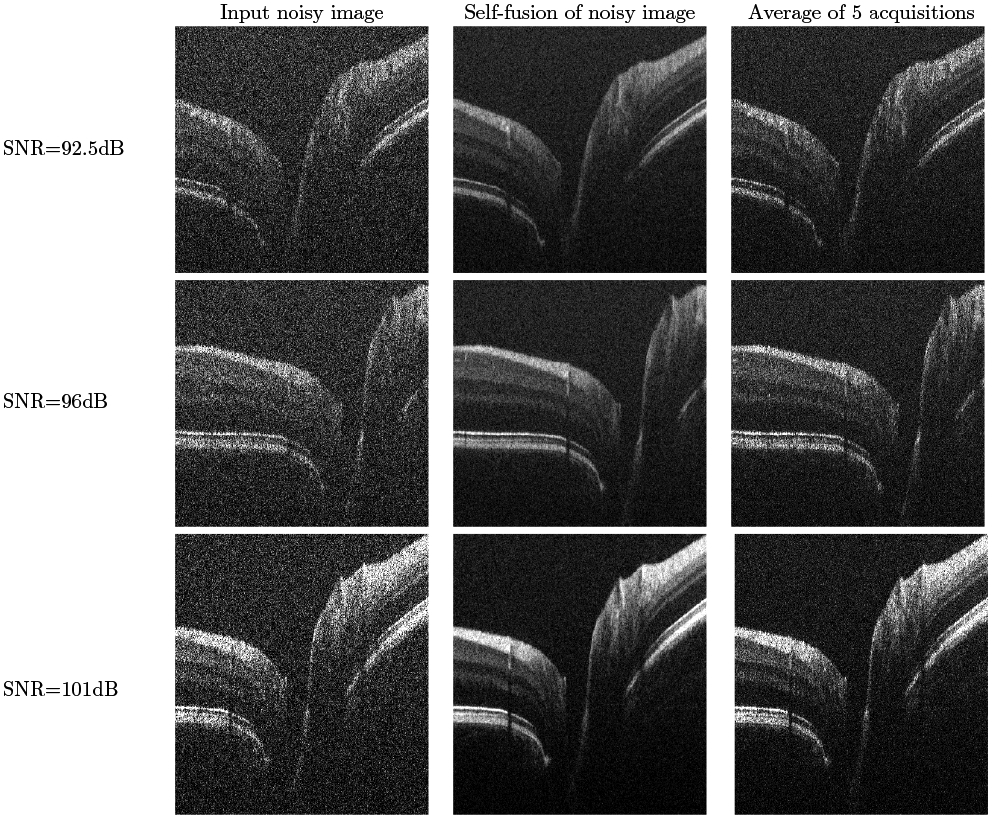
Self-fusion in the ONH. Left: input noisy image. Middle: Self-fusion of the single input noisy image. Right: average of 5 noisy images. The three rows are from SNR levels of 92.5dB, 96dB and 101dB respectively.
Figure 3.
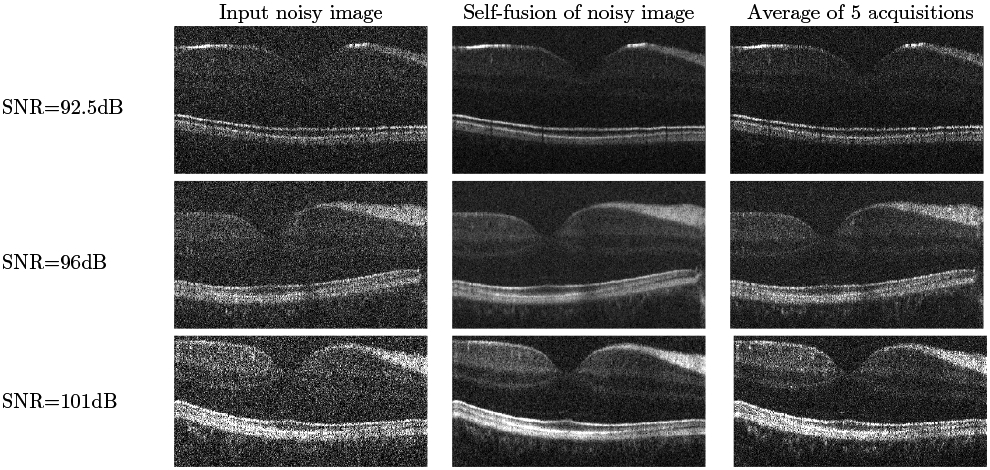
Self-fusion in the fovea. Left: input noisy image. Middle: Self-fusion of the single input noisy image. Right: average of 5 noisy images. Note the external limiting membrane is visible on the 96 dB and 101 dB self-fusion and averaged images but not on the raw single images.
Finally, we note that for many settings, the self-fusion result visually has better contrast than the average of 5 repeated frames, which is often considered the gold standard for despeckling. We hypothesize that this is likely because the self-fusion method allows us to leverage more data in the reconstruction: while the ‘ground truth’ is limited to the average of only 5 repeated frames at the same location, the self-fusion results shown in Figures 2 and 3 use a radius R of 5, which means 11 B-scans (5 on each side, and the central B-scan itself) were fused together. This is made possible because of two complementary components of self-fusion: on one hand, the deformable registration step allows data from further B-scans to be leveraged. On the other hand, the use of the joint fusion metric29 rather than simple averaging helps suppress the contribution of inappropriate data within this larger set, whether caused by registration error, image artifacts, noise or anatomical mismatch. This helps slow down the degradation caused by incorporating B-scans from increasingly distant locations. Figure 4 illustrates the filtering output for radius R in range [1..20]. We observe that the overall image quality is relatively stable even for R=20, which corresponds to 41 self-fused B-scans. While some edge blurring can be observed, especially for small features such as blood vessels, this blurring is minimal compared to simple averaging of the same number of images, even after registration. The image artifact visible on the left side of the input B-scan is noteworthy: the averaging of 4 repeated frames can only mildly softens this artifact, while self-fusion is able to alleviate it considerably.
Figure 4.
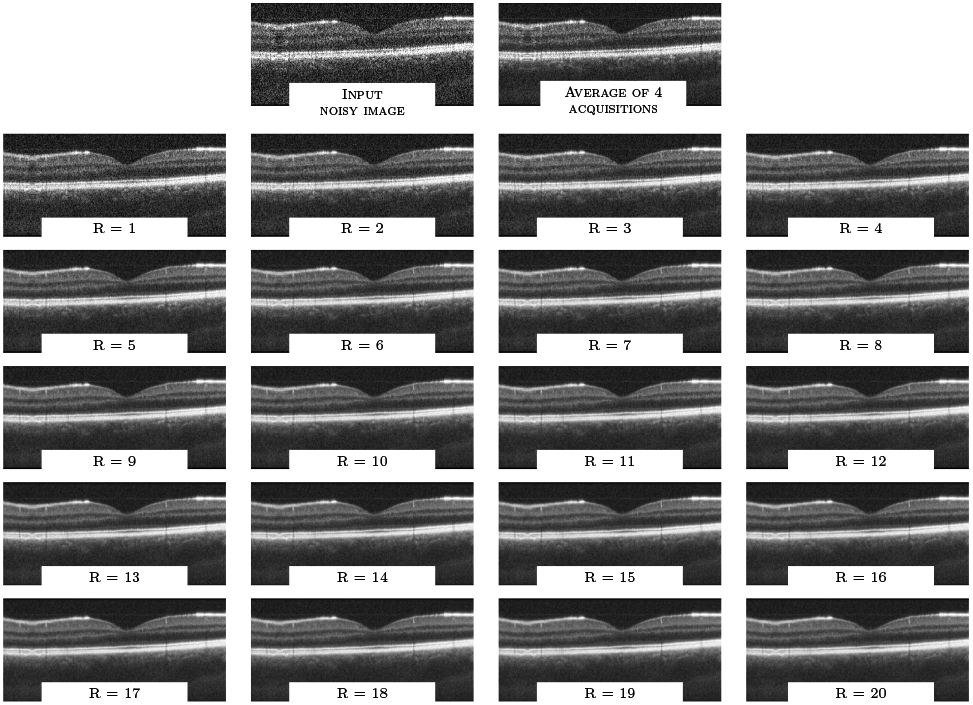
Qualitative evaluation for one OCT volume (first dataset) for varying radius R.
4. DISCUSSION AND CONCLUSION
We presented the novel self-fusion algorithm for speckle noise reduction in OCT images of the retina. The quantitative and qualitative results illustrate the performance of this method on a variety of settings. One current drawback of the method is that it is considering each B-scan independently, which may potentially cause consistency artifacts in 3D space; we plan on exploring this issue in future work.
ACKNOWLEDGMENTS
This work was supported, in part, by the NIH grant R01-NS094456 and the Vanderbilt Discovery Grant program.
Footnotes
© 2019 Vanderbilt University
REFERENCES
- [1].Desjardins AE, Vakoc BJ, Oh WY, Motaghiannezam SM, Tearney GJ, and Bouma BE, “Angle-resolved OCT with sequential angular selectivity for speckle reduction.,” Optics express (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Klein T, André R, Wieser W, Pfeiffer T, and Huber R, “Joint aperture detection for speckle reduction and increased collection efficiency in ophthalmic MHz OCT.,” Biomedical Optics Express 4, 619–634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Loupas T, McDicken WN, and Allan PL, “An adaptive weighted median filter for speckle suppression in medical ultrasonic images,” IEEE Transactions on Circuits and Systems 36(1), 129–135. [Google Scholar]
- [4].Kuan D, Sawchuk A, Strand T, and Chavel P, “Adaptive restoration of images with speckle,” IEEE Transactions on Acoustics, Speech, and Signal Processing 35(3), 373–383 (1987). [Google Scholar]
- [5].Lee J-S, “Speckle Suppression And Analysis For Synthetic Aperture Radar Images,” Optical Engineering 25(5), 255636 (1986). [Google Scholar]
- [6].Anantrasirichai N, Nicholson L, Morgan JE, Erchova I, Mortlock K, North RV, Albon J, and Achim A, “Adaptive-weighted bilateral filtering and other pre-processing techniques for optical coherence tomography.,” Computerized medical imaging and graphics : the official journal of the Computerized Medical Imaging Society 38(6), 526–539 (2014). [DOI] [PubMed] [Google Scholar]
- [7].Chen H, Fu S, Wang H, Lv H, and Zhang C, “Speckle attenuation by adaptive singular value shrinking with generalized likelihood matching in OCT.,” Journal of biomedical optics 23(3), 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- [8].Eybposh MH, Turani Z, Mehregan D, and Nasiriavanaki M, “Cluster-based filtering framework for speckle reduction in OCT images.,” Biomedical Optics Express 9(12), 6359–6373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rabbani H, Sonka M, and Abràmoff MD, “OCT Noise Reduction Using Anisotropic Local Bivariate Gaussian Mixture Prior in 3D Complex Wavelet Domain.,” Int. Journal of Biomedical Imaging 2013(5035), 417491–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yin D, Gu Y, and Xue P, “Speckle-constrained variational methods for image restoration in OCT.,” Journal of the Optical Society of America. A, Optics, image science, and vision 30(5), 878–885 (2013). [DOI] [PubMed] [Google Scholar]
- [11].Gong G, Zhang H, and Yao M, “Speckle noise reduction algorithm with total variation regularization in optical coherence tomography.,” Optics express 23(19), 24699–24712 (2015). [DOI] [PubMed] [Google Scholar]
- [12].Pizurica A, Philips W, Lemahieu I, and Acheroy M, “A versatile wavelet domain noise filtration technique for medical imaging.,” IEEE transactions on medical imaging 22(3), 323–331 (2003). [DOI] [PubMed] [Google Scholar]
- [13].Adler DC, Ko TH, and Fujimoto JG, “Speckle reduction in optical coherence tomography images by use of a spatially adaptive wavelet filter.,” Optics letters 29(24), 2878–2880 (2004). [DOI] [PubMed] [Google Scholar]
- [14].Xu J, Ou H, Sun C, Chui PC, Yang VXD, Lam EY, and Wong KKY, “Wavelet domain compounding for speckle reduction in optical coherence tomography.,” Journal of biomedical optics 18(9), 096002 (2013). [DOI] [PubMed] [Google Scholar]
- [15].Gleich D and Datcu M, “Wavelet-based SAR image despeckling and information extraction, using particle filter.,” IEEE transactions on image processing : a publication of the IEEE Signal Processing Society 18(10), 2167–2184 (2009). [DOI] [PubMed] [Google Scholar]
- [16].Jian Z, Yu L, Rao B, Tromberg BJ, and Chen Z, “Three-dimensional speckle suppression in Optical Coherence Tomography based on the curvelet transform.,” Optics express 18(2), 1024–1032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Du Y, Liu G, Feng G, and Chen Z, “Speckle reduction in optical coherence tomography images based on wave atoms.,” Journal of biomedical optics 19(5), 056009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao Y, Chu KK, Eldridge WJ, Jelly ET, Crose M, and Wax A, “Real-time speckle reduction in optical coherence tomography using the dual window method.,” Biomedical Optics Express 9(2), 616–622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu Y and Acton S, “Speckle reducing anisotropic diffusion,” IEEE Trans, on Image Processing. 1260–1270 (2002). [DOI] [PubMed] [Google Scholar]
- [20].Salinas HM and Fernández DC, “Comparison of PDE-based nonlinear diffusion approaches for image enhancement and denoising in optical coherence tomography.,” IEEE transactions on medical imaging 26(6), 761–771 (2007). [DOI] [PubMed] [Google Scholar]
- [21].Puvanathasan P and Bizheva K, “Interval type-II fuzzy anisotropic diffusion algorithm for speckle noise reduction in optical coherence tomography images.,” Optics express 17(2), 733–746 (2009). [DOI] [PubMed] [Google Scholar]
- [22].Fang L, Li S, Nie Q, Izatt JA, Toth CA, and Farsiu S, “Sparsity based denoising of spectral domain optical coherence tomography images.,” Biomedical Optics Express 3(5), 927–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yu H, Ding M, Zhang X, and Wu J, “PCANet based nonlocal means method for speckle noise removal in ultrasound images.,” PloS one 13(10), e0205390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ma Y, Chen X, Zhu W, Cheng X, Xiang D, and Shi F, “Speckle noise reduction in optical coherence tomography images based on edge-sensitive cGAN.,” Biomedical Optics Express 9(11), 5129–5146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Halupka K, Antony B, Lee M, Lucy K, Rai R, Ishikawa H, Wollstein G, Schuman J, and Garnavi R, “Retinal OCT image enhancement via deep learning,” Biomedical Optics Express , 6205–6221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang Y, Lu Z, Shao Z, Ran M, Zhou J, Fang L, and Zhang Y, “Simultaneous denoising and super-resolution of OCT images based on GAN.,” Optics express 27(9), 12289–12307 (2019). [DOI] [PubMed] [Google Scholar]
- [27].Iglesias JE and Sabuncu MR, “Multi-atlas segmentation of biomedical images: A survey.,” Medical Image Analysis 24(1), 205–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fleishman GM, Valcarcel A, Pham DL, Roy S, Calabresi PA, Yushkevich P, Shinohara RT, and Oguz I, “Joint Intensity Fusion Image Synthesis Applied to MS Lesion Segmentation.,” BrainLes, MICCAI, 43–54 (2018). [PMC free article] [PubMed] [Google Scholar]
- [29].Wang H, Suh JW, Das SR, Pluta JB, Craige C, and Yushkevich PA, “Multi-Atlas Segmentation with Joint Label Fusion,” IEEE Trans. on Pattern Analysis and Machine Intelligence 35(3), 611–623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]