Abstract
Background
Brain tumors, whether primary or secondary, have limited therapeutic options despite advances in understanding driver gene mutations and heterogeneity within tumor cells. The cellular and molecular composition of brain tumor stroma, an important modifier of tumor growth, has been less investigated to date. Only few studies have focused on the vasculature of human brain tumors despite the fact that the blood-brain barrier (BBB) represents the major obstacle for efficient drug delivery.
Methods
In this study, we employed RNA sequencing to characterize transcriptional alterations of endothelial cells (EC) isolated from primary and secondary human brain tumors. We used an immunoprecipitation approach to enrich for EC from normal brain, glioblastoma (GBM), and lung cancer brain metastasis (BM).
Results
Analysis of the endothelial transcriptome showed deregulation of genes implicated in cell proliferation, angiogenesis, and deposition of extracellular matrix (ECM) in the vasculature of GBM and BM. Deregulation of genes defining the BBB dysfunction module was found in both tumor types. We identified deregulated expression of genes in vessel-associated fibroblasts in GBM.
Conclusion
We characterize alterations in BBB genes in GBM and BM vasculature and identify proteins that might be exploited for developing drug delivery platforms. In addition, our analysis on vessel-associated fibroblasts in GBM shows that the cellular composition of brain tumor stroma merits further investigation.
Keywords: blood-brain barrier, brain metastasis, glioblastoma, insulin receptor, vessel-associated fibroblasts
Key Points.
Brain tumor vasculature shows deregulated expression of transporters, which could be exploited to deliver drugs across the BBB into tumor tissue.
Brain tumor stroma contains fibroblast-like cells.
Importance of the Study.
Despite therapeutic advances, the prognosis for both primary and secondary brain tumors is poor. The role of tumor stromal cells in tumor growth and response to therapy is increasingly recognized; yet, the underlying composition and accompanying molecular alterations are largely unknown. Furthermore, although the blood-brain barrier (BBB) is compromised in brain malignancies, tumor vasculature clearly hinders the penetration of drugs into brain tumor tissue. Herein we characterize changes in the BBB of glioblastoma and lung cancer brain metastases. We identify proteins expressed by the tumor vasculature that could be leveraged for drug delivery into the tumor. In addition, we have identified the presence of previously unrecognized fibroblast-like cells in tumor stroma.
Therapeutic options are limited for primary and secondary brain malignancies. Glioblastoma (GBM) is the most common primary brain tumor in adults. Secondary tumors of extracranial origins (metastasis) often arise from tumors of breast, lung, and skin origin. While GBM is the deadliest primary brain tumor, life-threatening brain metastases (BM) develop in cancer patients despite significant advances in managing extracranial primary tumors.1 Many studies have contributed to understanding the inter- and intra-tumor heterogeneity of GBM cells and have identified drivers for BM. The important role of tumor stromal cells in tumor growth and therapy is increasingly recognized2; however, stromal composition and molecular alterations have been less explored. Several studies have focused on immune cells in brain malignancies, which exhibit disease-specific phenotypes and activation profiles.3 Another stromal component, the tumor vasculature, supports the viability and metabolism of malignant cells. It has been hypothesized that by blocking tumor angiogenesis, new blood vessel growth into tumor tissue, would quickly inhibit tumor progression.4 Anti-angiogenic therapies inhibiting the master regulatory vascular endothelial growth factor (VEGF) pathway combined with standard therapy, concurrent temozolomide, and radiotherapy followed by maintenance temozolomide, have yielded disappointing results without improvement in overall survival for GBM patients.5 It is increasingly recognized that tumor cells from many tumor types, including brain malignancies, hijack the existing vasculature of nonmalignant tissue by vascular co-option, and thus are not entirely dependent on angiogenesis for their growth.6
The blood-brain barrier (BBB) of normal brain vasculature acts as a protective barrier that controls brain nutrition and homeostasis. Endothelial cells (EC) possess distinct characteristics that restrict the passage of blood-borne molecules into the brain (closed endothelial cell-cell junctions), while ensuring delivery of nutrients (expression of various solute carrier [SLC] transporters) and protecting the brain from xenobiotics (expression of ATP-binding cassette [ABC] transporters).7 In addition, normal brain endothelium restricts leukocyte passage into brain parenchyma. Although the BBB is compromised in brain malignancies, tumor vasculature clearly hinders the penetration of drugs into GBM and BM tissue.8–10 In clinical practice, the compromised integrity of the BBB, which is permeable to small gadolinium-based contrast agents, is visualized with contrast-enhanced MRI. As the BBB is a multicomponent structure, vascular permeability to contrast-enhancing agents or other blood-borne molecules is not necessarily paralleled by improved delivery of small lipid-soluble drugs, which are substrates for ABC transporters into the brain parenchyma.11 Furthermore, GBM patients have a significant tumor burden that is not visualized by contrast-enhanced MRI, suggesting that portions of the tumor possess a relatively intact BBB.10 Interestingly, the prognosis of medulloblastoma has been linked to the BBB function. Specifically, vasculature of Wnt-pathway medulloblastoma (favorable prognosis) lacks a BBB, whereas sonic-hedgehog-driven medulloblastoma (poor prognosis) possesses an intact BBB.12
Genome-wide molecular characterization of GBM and BM has changed the diagnostic approach for brain malignancies. Therapeutic options, however, remain slim, mostly due to poor penetration of generic and targeted therapeutics into the brain.1,9 In addition, it is difficult to predict the success rate of a compound based on preclinical pharmacokinetic studies performed in rodent models due to qualitative and quantitative differences in the expression of ABC transporters between rodent and human BBB.13,14 Thus, a better understanding of vascular changes in human brain malignancies could lead to improved therapeutic targeting of tumors.
Numerous studies have investigated the expression of individual genes and proteins within brain tumor vasculature, but only few have used a systems biology approach to investigate transcriptional alterations in GBM vasculature.5,15 In BM, such studies are absent. In this study, we aimed to characterize the transcriptome of EC isolated with an immunoprecipitation approach (CD31+ cells) from primary GBM and BM of non–small-cell lung cancer, the most frequent secondary brain tumor.16
Materials and Methods
Human Tissue Samples
Autopsy control material was isolated from human cortex. Specimens were taken from subjects without any evidence of cerebral neoplasia after a maximum postmortem interval of 24 h. Sampling complied with guidelines of the Kantonale Ethikkommission (KEK) Zürich (BASEC-Nr. Req-2018-0073). Samples of GBM and lung adenocarcinoma metastases as well as tissue from epilepsy resection surgery material were collected with the permission of the KEK (BASEC-Nr. 2019-02027) and the patient’s approval. Surgical biopsies of GBM were diagnosed at the Institute of Neuropathology and Molecular Pathology, University Hospital Zürich.
Isolation of EC From Brain Tissue
Tissue was mechanically dissociated using scissors, followed by enzymatic dissociation. To enrich the samples for EC, we performed a negative selection by adding anti-CD15 antibody (Ab), and anti-CD45 Ab conjugated Dynabeads. CD45 and CD15-positive cells were removed using a magnetic particle concentrator rack followed by incubation with anti-CD31 Ab Dynabeads. CD31-positive EC were lysed and RNA was isolated using the Qiagen AllPrep DNA/RNA/Protein Mini Kit (Qiagen, 80004) according to the manufacturer’s instructions. RNA samples were stored at −80°C until subjected to RNA sequencing (RNA-seq).
RNA-Seq
Libraries were prepared using the SMARTer Stranded Total RNA-seq Pico Input Mammalian Kit (Clontech Laboratories). Samples were sequenced with an Illumina HiSeq4000, single-end, with a read length of 125 bp. Sequencing depth was 30-50 Mio reads. RNA-seq of samples was carried out at the Functional Genomics Center Zurich (FGCZ) at the University of Zürich and ETH.
Immunohistochemistry
Immunohistochemistry was performed on cryosections. Primary antibodies used in the study are listed in Supplementary Table 1. All secondary antibodies made in donkey conjugated to fluorophores suitable for multiple labeling were purchased from Jackson Immunoresearch. Imaging was performed using a SP5 laser scanning confocal microscope (Leica). For image processing, Imaris (Bitplane) software was used. Quantification of fluorescence images was performed using FIJI (ImageJ) software.
Bioinformatics Analysis of RNA-Seq Data
Raw reads in fastq sequencing files were trimmed and reads were then aligned against the human reference genome (GRCh38.p10, annotation release 89, 2017-05-31). Number of reads and gene biotypes of each sample are listed in Supplementary Table 2. Normalization and differential gene expression analysis were performed on the counts data using R (v.3.5.3) and the DESeq function of the DESeq2 package for R, with default parameters. Differential expression of genes was considered significant at adjusted P value <.05. Gene set overrepresentation analysis of gene ontology (GO) biological process gene sets was performed using the enrichGO implemented in the clusterProfiler package.
To determine cell-type-specific enrichment in either GBM or BM, we used smaller cell-type-specific gene sets, applying gene set enrichment analysis (GSEA). To determine whether samples were successfully enriched with CD31-positive EC (list of EC marker genes was extracted from Figure 2A of Butler et al.17), we compared our samples to bulk brain samples available in the GTEx project (https://gtexportal.org/home/). Differential gene expression analysis compared each GTEx brain tissue type with our control and tumor RNA-seq tissues by fitting a linear model using limma. GSEA was used to calculate an enrichment score for the EC marker gene list among or between each GTEx brain tissue type and our control and tumor RNA-seq tissues.
Experimental details on EC isolation, immunohistochemistry, and bioinformatics analysis are given in the Supplementary Material.
Data Availability
RNA-seq data, both raw data and gene-by-sample matrix of raw counts, were deposited in Gene Expression Omnibus (GEO) under accession number GSE159851.
All data used to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Material.
Results
Isolation and RNA-seq of Brain EC
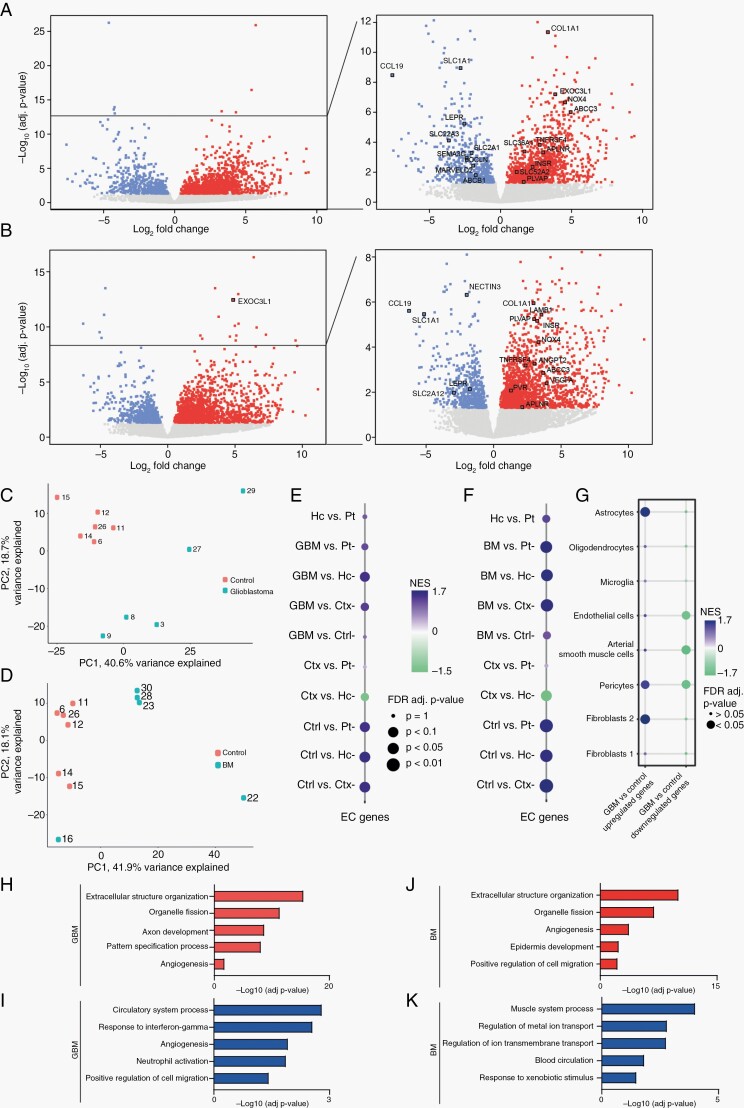
In order to investigate changes in the endothelium of brain tumors, we performed RNA-seq of CD31+ EC isolated from GBM and adenocarcinoma BM as well as non-tumorous brain tissue from epilepsy surgery or non-neoplastic autopsy cases (Supplementary Table 3, Supplementary Figures 1 and 2A). RNA-seq reads were aligned to the Homo sapiens genome, assembly GRCh38. Unsupervised clustering of the dataset showed samples grouping according to disease (GBM vs control and BM vs control) (Supplementary Figure 2B, C). When comparing GBM and control samples, we detected 1773 differentially expressed genes (DEG), and when comparing adenocarcinoma BM and control samples, we detected 2074 DEG (false discovery rate [FDR] <0.05) (Figure 1A, B, Supplementary Tables 4 and 5). Principal component analysis (PCA) using the 1000 most variable genes showed that the first principal component (PC) accounts for variability within tumor samples, whereas the second PC accounts for differences between tumor and control samples (Figure 1C, D).
Fig. 1.
Differently expressed genes (DEG) in CD31+ EC fraction isolated from GBM, BM, and normal brain tissue derived by RNA-seq. Volcano plot showing DEG in GBM (A) and BM (B) samples compared to controls. Upregulated genes—in red, downregulated genes—in blue, deregulated genes—in gray. PCA of GBM and control samples (C) and BM vs control samples (D). Dot plot showing a significant enrichment score of 25 core EC genes17 in control (Ctrl) (E, F), GBM (E), and BM (F) samples compared to healthy brain GTEx expression data (Ctx: cortex, Hc: hippocampus; Pt: putamen). (G) Dot plot showing the enrichment score of cell-type-specific genes18 in DEG in CD31+ fraction isolated from GBM tissue. GO overrepresentation analysis of the biological process of DEG in GBM (H, I) and in BM (J, K) vessels. Red and blue color bars show significantly regulated biological processes among upregulated and downregulated genes, respectively. Abbreviations: BM, brain metastasis; EC, endothelial cells; GBM, glioblastoma; GO, gene ontology; PCA, principal component analysis.
We next used the GSEA to determine whether our dataset was enriched for core EC genes. We compared the expression of 25 core EC genes17 in our dataset with the expression in total tissue isolated from healthy hippocampus (Hc), cortex (Ctx), and putamen (Pt).19 A significant positive enrichment score showed that we had successfully enriched our tumor and control samples for ECs compared to total brain tissue (Figure 1E, F, Supplementary Figures 3 and 4).
EC are difficult to isolate from other cells of the neurovascular unit (NVU), including mural cells, perivascular fibroblasts, and astrocytes. The presence of these cell types could alter the transcriptomic profile of samples. GSEA revealed that upregulated DEG in GBM samples compared to controls were enriched for marker genes of astrocytes and a subtype of fibroblasts (fibroblast type 2). Downregulated DEG in GBM samples were enriched for marker genes of mural cells and EC (Figure 1G). This finding suggests the altered cellular composition of samples enriched for EC isolated from normal and GBM blood vessels. Cellular composition of BM samples was not altered significantly compared to control samples (Supplementary Figure 2D).
In GBM samples, GO enrichment analysis revealed extracellular matrix (ECM), cell cycle-associated gene sets within the significantly upregulated gene sets (Figure 1H, Supplementary Table 6). Significantly downregulated gene sets in GBM samples contained angiogenesis and vascular development associated gene sets as well as response to interferon gamma and neutrophil activation (Figure 1I, Supplementary Table 6). Analysis of DEG in lung adenocarcinoma BM samples revealed upregulated gene sets similar to GBM, ie, related to the organization of ECM, cell division, and angiogenesis (Figure 1J, Supplementary Table 7). Downregulated gene sets were associated with muscle cell contraction and ion transport (Figure 1K, Supplementary Table 7). Interestingly, in GBM samples, a gene set related to angiogenesis was significantly enriched within both up- and downregulated genes (Figure 1H, I).
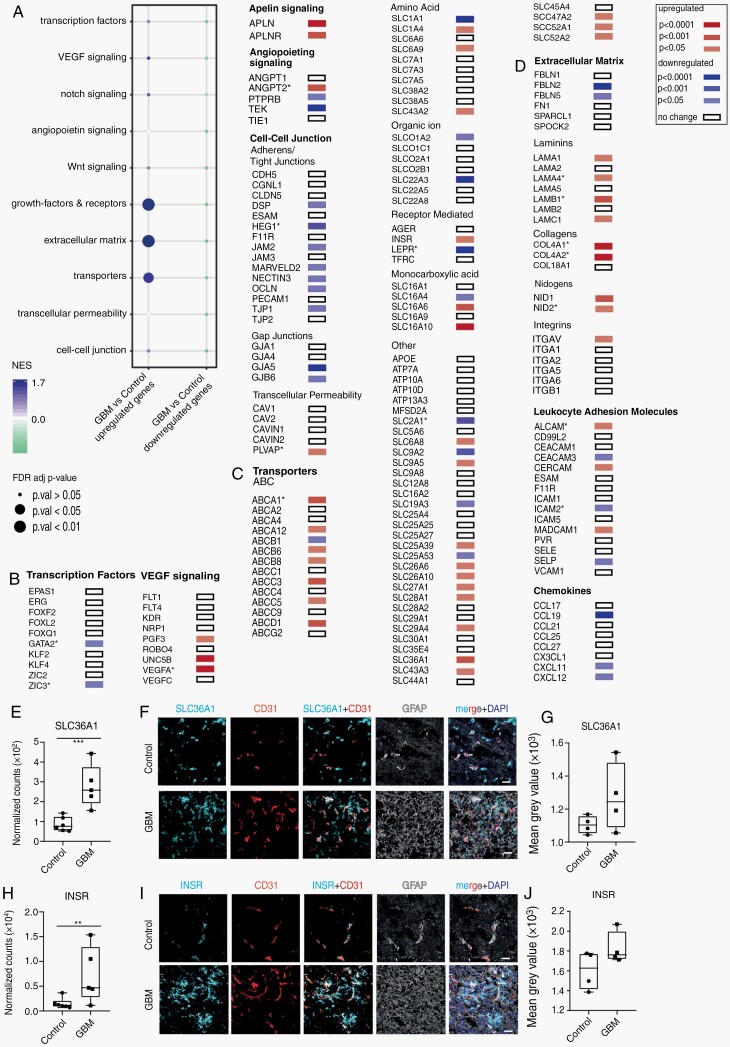
Deregulated Expression of BBB Genes of GBM Vasculature
Using the Ivy Glioblastoma Atlas Project (IVY GAP) RNA-seq data obtained from anatomically different GBM regions,20 we found that expression of DEG in GBM blood vessels varies according to anatomical region (Supplementary Figures 5–7, Supplementary Table 8, for a detailed description, see Supplementary Material). We next investigated which BBB-related genes and signaling pathways are deregulated in GBM EC compared to control EC. As previously published, GBM vasculature shows deregulation of signaling pathways implicated in angiogenesis (VEGF-, angiopoietin-, and apelin) (Figure 2A, B). In addition, several angiogenic EC marker genes are upregulated in GBM vasculature—COL4A1, COL4A2, LAMB1 (Figure 2). Expression of PLVAP (plasmalemma vesicle-associated protein) is upregulated in GBM vasculature (Figure 2B), which is associated with loss of the BBB phenotype and increased vessel permeability.21 Consistent with reported increased vascular permeability, GBM EC showed reduced mRNA expression of endothelial junctional proteins occludin (OCLN), ZO-1 (TJP1), JAM 2 (JAM2), tricellulin (MARVELD2), Heart of Glass (HEG1) (Figure 2B). In our dataset, we detected reduced expression of several arterial EC marker genes (eg, GJA5, SEMA3G, BMX)18,22 in GBM vasculature (Figure 1A, Supplementary Table 4). This could reflect changes in the organization of the vascular network in GBM and/or loss of endothelial zonation in the arterial-capillary-vein axis. Our analysis showed profound changes in the expression of various transporters by GBM vasculature (Figure 2A, C). GBM EC express higher levels of lipid transporters ABCD1 and ABCA1. In addition, several other ABC transporters from the “C” family, previously implicated in multidrug resistance, ABCC3 (MRP3) and ABCC5 are upregulated in GBM vasculature (Figures 1A and 2C), whereas ABCB1 expression (P-glycoprotein) is downregulated. Interestingly, GBM EC show upregulation of mitochondrial transporters ABCB6, ABCB8, SLC25A39, and SLC25A53 encoded by the nucleus (Figure 2C). Several SLC transporters involved in pH and cell volume regulation (SLC4A3, SLC4A8, SLC9A5) are upregulated in GBM vasculature. Expression of several amino acid transporters (SLC16A10, SLC36A1, SLC43A2) is higher in GBM vasculature (Figure 2C). Neurotransmitter transporters expressed by EC, SLC1A1, and SLC22A3 (encoding for OCT3), have lower expression in GBM vasculature (Figure 2C). SLC1A1 expression is higher in leading edge and infiltrating tumor regions (IVY GAP data, Supplementary Figure 2A, C), indicating possible loss of SLC1A1 expression by EC in the microvascular proliferation (MVP) region. Surprisingly, we found reduced expression of several glucose transporters, including SLC2A1 (encoding for GLUT1) (Figures 1A and 2C). The reduced expression of the SLC2A1 mRNA in GBM EC is puzzling given that GLUT1 is the main glucose transporter at the BBB and angiogenic EC are glycolytic.23,24 In addition, GBM growth is dependent on glycolysis, which is elevated 3-fold compared to normal brain.25 Consistent with our data on the reduced mRNA expression of SLC2A1, a recent proteome study of GBM vessels reported a 50% reduction of GLUT1 protein levels in GBM vasculature.14 In addition, GBM vasculature shows reduced expression of several transporters that deliver substrates important for energy metabolism (SLCA6A8, SLC19A3, transporting creatine and thiamine, respectively) (Figure 2C). The expression of nucleoside transporters SLC28A1 and SLC29A4, and nucleobase transporter SLC43A3 is higher in GBM vasculature (Figure 2C). Out of three known riboflavin transporters, two, SLC52A1 and SLC52A2, are upregulated in GBM vasculature. In addition, GBM vasculature shows deregulated expression of transporters that are mainly expressed by other vascular cells (eg, fibroblasts and mural cells) and vessel-associated astrocytes (Supplementary Figure 8A). These transporters are implicated in various cellular processes such as acid-base homeostasis (SLC4A3, SLC4A8) and glucose transport (SLC2A4, SLC2A12). Another striking feature of GBM vasculature is deregulated expression of genes encoding for ECM (Figure 2A, D, Supplementary Figure 8B), possibly reflecting an altered cellular phenotype of EC (eg, angiogenic, COL4A1, COL4A2, LAMB1) and/or an altered cellular composition of vasculature (eg, the presence of fibroblast-like perivascular cells, discussed above).
Fig. 2.
Changes in the expression of genes implicated in BBB function in GBM vasculature. (A) Dot plot of GSEA analysis of key pathways regulating BBB development and function (defined in Corada et al.)26 in GBM vasculature. (B-D) Differential expression of selected genes linked to the development of brain vasculature and BBB function in GBM vasculature. The gene list was generated using BBB marker genes published by Corada et al.,26 brain vascular cell-enriched genes identified by Vanlandewijck et al.,18 and the BBB dysfunction module identified by Munji et al.27 Upregulated genes are in red, downregulated genes are in blue, White box—no change in gene expression between the GBM and normal brain vessels. An asterisk marks genes previously reported deregulated in GBM vasculature. mRNA expression levels of SLC36A1 (E) and INSR (H) in GBM and control samples. Immunohistochemical detection of SLC36A1 (F) and INSR (I), all in cyan, in GBM, and control tissue. All samples were co-stained for CD31 (endothelium, in red) and GFAP (in gray). Cell nuclei are visualized with DAPI (in dark blue, F and I). Quantification of SLC36A1 (G) and INSR (J) immunohistochemistry staining. Statistical test: E, H—FDR corrected t-test. **P < .01, ***P < .001; Scale bar—30 µm. Abbreviations: BBB, blood-brain barrier; ECM, extracellular matrix; FDR, false discovery rate; GBM, glioblastoma; GSEA, gene set enrichment analysis; LAM, leukocyte adhesion molecules.
We did not detect alterations in the expression of ICAM-1 and VCAM-1, two pro-inflammatory molecules mediating leukocyte transmigration via endothelium (Figure 2D). Expression of MADCAM-1 and ALCAM was found to be higher (Figure 2D), the latter reported previously in GBM vasculature.28,29
We investigated protein expression of two upregulated genes, SLC36A1 (encoding for a proton-dependent glycine/GABA exchanger) and INSR (encoding for an insulin receptor [IR]) (Figures 1A, 2B, C and 3E, H), which have been exploited as a drug delivery platform for chemo- and biotherapeutics.30,31 Both genes are preferentially overexpressed in the MVP region in GBM (Supplementary Figure 5F, J). SLC36A1 protein expression showed high variability between individual GBM samples (Figure 3F, G), whereas INSR expression showed a higher variability in control samples (Figure 3I, J).
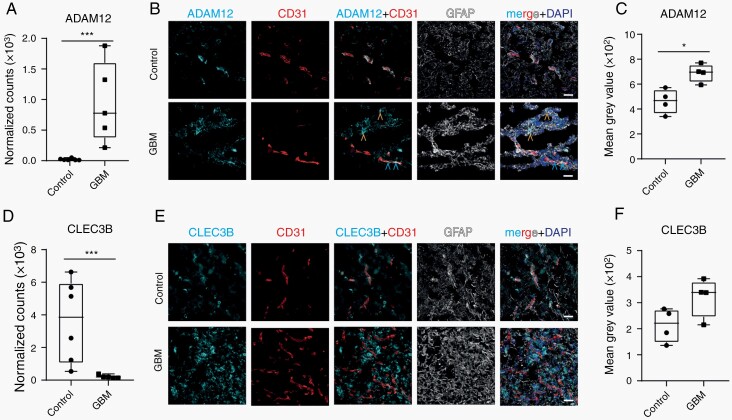
Fig. 3.
Expression of perivascular fibroblast marker genes in normal brain and GBM vasculature. (A) mRNA expression level of ADAM12 in GBM and control samples. (B) Representative image of ADAM12 (in cyan) expression, co-stained with vessels (CD31, in red) and GFAP (in gray) in GBM and control brain tissue. Blue arrowheads point to ADAM12-positive perivascular cells and yellow arrowheads point to ADAM12-positive tumor cells. (C) Quantification of ADAM12 staining around CD31-positive vessels. (D) RNA expression level of CLEC3B in GBM and control samples. (E) Representative image of CLEC3B (in cyan), co-stained with vessels (CD31, in red) and GFAP (in gray) in GBM and control tissue. (F) Quantification of CLEC3B staining around CD31-positive vessels. Cell nuclei are visualized with DAPI (in dark blue, B and E). Statistical tests: A, D—FDR corrected t-test and C, F—two-tailed unpaired t-test. Data distribution was analyzed using the Q-Q plot. *P < .05, ***P < .001. Scale bar—30 µm. Abbreviations: FDR, false discovery rate; GBM, glioblastoma.
We next compared DEG in GBM vasculature from our dataset with published data where gene expression alterations were identified in GBM vasculature using different approaches (Supplementary Figure 9, Supplementary Table 9, for a detailed description, see Supplementary Material).5,15 Dusart et al. identified human malignancy-associated EC changes in silico5 and out of these 42 malignancy-associated EC genes, 41 genes were expressed in our dataset, and 9 genes were significantly upregulated in GBM samples compared to controls (Supplementary Figure 9A, Supplementary Table 9). Dieterich et al. analyzed the transcriptome of vessels isolated by laser microdissection from nonmalignant brain, low-grade glioma, and GBM using Affymetrix microarrays.15 Twenty-four genes out of 78 identified by Dieterich et al. upregulated in GBM vessels compared to low-grade glioma and control vessels, were significantly upregulated also in our dataset (Supplementary Figure 9E, F).
Vessel-Associated Fibroblasts Surround GBM Blood Vessels
Cell-type enrichment analysis revealed other vessel-associated cell types (astrocytes, fibroblasts) than EC in the CD31+-enriched fraction of GBM samples (Figure 1G). In addition, a GO-gene set analysis of DEG in GBM EC preferentially expressed in MVP region showed profound changes in the ECM composition (Supplementary Figure 5I). Many of these ECM proteins are expressed in mouse brain by perivascular fibroblasts (COL1A1, COL3A1, COL6A3).18 In our dataset, perivascular fibroblast marker genes (ADAM12, DPEP1, FBLN2, CLEC3B) were deregulated (Figures 1A and 3A, D, Supplementary Figure 10, Supplementary Table 4). In mouse brain, upregulated genes Adam12 and Dpep1, as well as downregulated genes Fbln2 and Clec3b are markers for perivascular fibroblast subpopulation 2 (FB2) and 1 (FB1), respectively18 (Supplementary Figure 10A–D). Next, we used immunohistochemical stainings to investigate perivascular fibroblast markers in normal brain and GBM vasculature. In GBM samples, ADAM12 immunolabeling was detected around vessels and in tumor tissue, but in control tissue only around vessels (Figure 3B). Quantification of intensity of ADAM12 staining in vessel proximity showed a significant upregulation of ADAM12 expression in GBM samples compared to controls (Figure 3C). CLEC3B Immunohistochemistry showed diffuse positivity throughout tumor tissue in contrast to localized staining along CD31+ vessels in control samples (Figure 3E). However, quantification of CLEC3B expression at the protein level in the vicinity of blood vessels did not reflect results seen at the RNA level (Figure 3F). We next explored the IVY GAP in situ hybridization (ISH) data for expression data of brain perivascular fibroblast markers. Expression of several fibroblast markers could be detected adjacent to blood vessels (Supplementary Figure 10G–L). Thus, we detect cells expressing perivascular fibroblast marker genes in GBM vasculature.
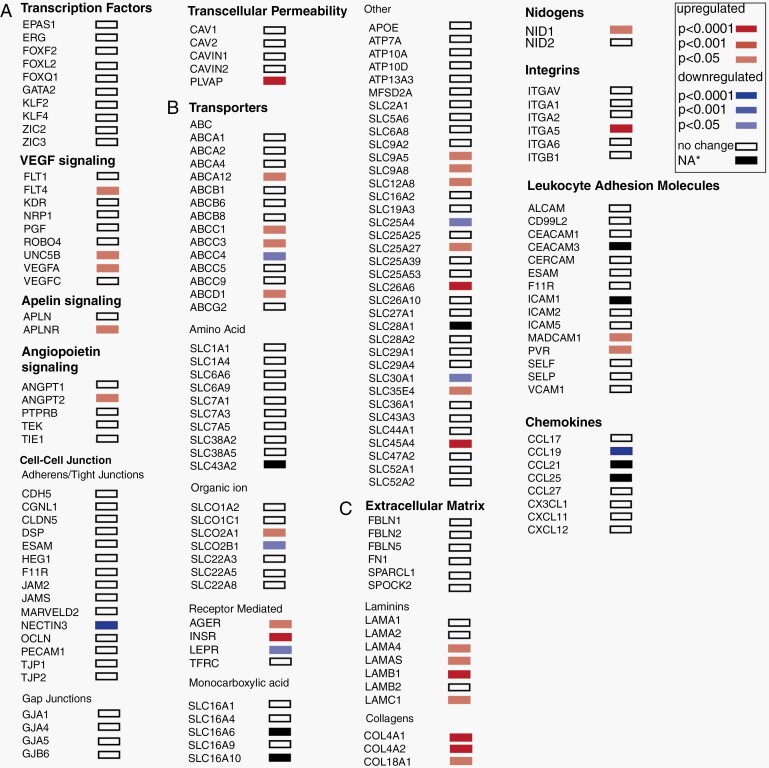
Deregulated Expression of BBB Genes of BM Vasculature
We investigated the expression of genes regulating BBB development and function in BM EC similar to that for the GBM EC (Figure 4). Expression of angiogenesis genes (VEGFA, ANGPT2) (Figure 4A, C) and genes encoding other ECM proteins (laminins, nidogens, collagens) is higher (Figure 4C, Supplementary Figure 8D) in BM vessels than in control vessels. There were fewer changes in the expression level of genes encoding cell-cell junction proteins, compared to GBM endothelium. We only detected decreased expression of NECTIN-3, a gene encoding a junctional protein supporting endothelial integrity in vitro (Figure 4A). The expression of influx- and efflux transporters is altered in BM vessels compared to normal vessels (Figure 4B). Similar to GBM vessels, expression of the gene encoding for the multidrug resistance protein ABCC3 is increased. Expression of SLC transporters such as SLCO2A1 (organic anion transporter), SLC26A6 (anion exchanger), SLC35E4 (nucleotide sugars, for glycosylation) is higher in BM than in control EC (Figure 4B). BM EC show upregulated genes encoding for Na/H+ exchangers involved in intracellular pH homeostasis (SLC9A5 and SLC9A8) and choline transport SLC44A4. Expression of the gene SLCO2B1, encoding a transporter (OATP2B1) for bile salts, steroid conjugates, thyroid hormone, and many drugs (eg, statins) is downregulated by BM EC.
Fig. 4.
Changes in the expression of genes implicated in blood-brain barrier (BBB) function in adenocarcinoma brain metastasis (BM) vasculature. (A-C) Differential expression of selected genes linked to brain vasculature development and BBB function in BM vasculature. The selected gene list was generated as described in Figure 2. Upregulated genes are in red, downregulated genes are in blue. White box—no change in gene expression between the BM and normal brain vessels. *Black box—the P value was not calculated by DESeq2. The log2 fold changes for these genes are: SLC43A2—2.14, SLC16A6—3.26, SLC16A10—4.52, SLC28A1—4.27, CEACAM3—2.13, ICAM1—0.93, TICAM1—1.03, CCL25—not applicable.
Interestingly, mRNA levels of several SLC transporters expressed by vessel-associated astrocytes are deregulated (Supplementary Figure 8C). We detected significant downregulation of SLC1A2 (EAAT2/GLT-1), an astrocyte-specific principal glutamate transporter. NMDA (N-methyl-d-aspartate) receptor-mediated signaling was shown to promote colonization and growth of brain cancer cells in the brain.32 Thus, downregulation of SLC1A2 can contribute to cancer growth.
BM EC, similar to GBM EC show upregulation of INSR (Figure 4A) and no alterations in ICAM-1 and VCAM-1 expression. Expression of mRNA encoding for MADCAM-1, a leukocyte cell adhesion receptor involved in regulating passage and retention of leukocytes was upregulated in BM EC (Figure 4C). In addition to MADCAM-1, PVR, encoding for an adhesion receptor for natural killer cells, showed upregulation in BM vasculature (Figure 4C). Another similarity between GBM and BM vasculature is the reduced expression of CCL19, a chemokine regulating T-cell trafficking and activation (Figure 4C).
Common Deregulated Pathways in GBM and BM Vasculature
We next compared DEG in GBM and BM vasculature identified in our study (Supplementary Figure 11). GO overrepresentation analysis using genes upregulated in BM and GBM EC showed deregulation of genes implicated in angiogenesis and ECM deposition Supplementary Figure 11A–D, Supplementary Table 9). We next investigated whether GBM and adenocarcinoma metastasis EC showed deregulation of a generic “BBB dysfunction module” 27 (Supplementary Figure 11H–K). The BBB dysfunction module genes were significantly enriched in both datasets (Supplementary Figure 11J, K). Thus, both GBM and BM blood vessels show alterations in genes defining the “BBB dysfunction module” indicating changes in cellular and ECM composition and cell quiescence.
Discussion
We performed genome-wide transcriptomics using RNA-seq in EC isolated from normal human brain, primary GBM, and brain lung cancer metastases in order to explore differences in tumor vasculature. An additional aim was to compare vascular changes specific for tumors of CNS origin (gliomas) to tumors of CNS-extrinsic origin (metastases).
Our data point to profound differences in ECM composition in brain tumor vessels compared to normal blood vessels. Brain tumor vessels upregulated the expression of genes encoding for laminins, collagens, nidogens, and integrins. EC and mural cells secrete vascular basement membrane. In addition, we detected upregulation of genes encoding for collagens and matrix modifying enzymes (eg, LOXL2) not expressed by these cells in normal brain. In peripheral organs, fibroblasts deposit ECM rich in collagen in an organ- and region-specific manner.33 Brain parenchyma is devoid of fibroblasts; however, brain vasculature harbors a specific type of fibroblast—“vessel-associated fibroblast.” 18 It is therefore plausible that these cells become activated in malignant conditions with deposition of ECM in brain tumor vasculature similar to that described in other tumors.34 In the mouse, two populations of vessel-associated fibroblasts (FB1 and FB2) have been defined based on their gene expression profile.18 We found evidence that the GBM EC transcriptome was enriched for vessel-associated FB type 2 genes. Immunohistochemical analysis also showed an increased expression of ADAM12 (a marker for FB2) along GBM vessels. Cancer-associated fibroblasts (CAF) are the most prevalent cell type in many cancers. In the GBM microenvironment, these cells have not been reported and thus their function is not discussed.2 However, Clavreul et al. have reported the presence of GBM-associated stromal cells with phenotypic and functional properties similar to those of CAF.35 The cellular origin of these diploid cells is not known but it is plausible that these cells originate from vessel-associated fibroblasts. Interestingly, comparison of gene expression profiles from low- and high-ADC GBMs led to the identification of these 13 genes higher expressed in the high-ADC (poor survival) group.36 Eight out of these 13 genes (COL1A1, COL3A1, COL6A3, COL11A1, CYP1B1, DCN, LUM, MYLK) are enriched in or are marker genes for vessel-associated fibroblasts in the mouse brain.18 Single-cell RNA-seq studies performed on GBM have detected tumor cells as well as other cell types including myeloid cells and EC. However, fibroblasts have not been previously reported. CAF not only regulate cancer cell proliferation and migration but also responsiveness to therapy.34 In addition to limiting drug access by an excessive ECM deposition, CAF mediate resistance to various anti-cancer therapies, including immunotherapy.34 Understanding the origin, heterogeneity, and role of fibroblasts in GBM tissue might lead to additional therapeutic strategies to target brain tumors.
The BBB hinders the transport of most drugs into the brain parenchyma, posing an obstacle to therapeutic intervention. Although tumor vessels are thought to have compromised BBB, therapeutic targeting of primary and secondary brain tumors remains challenging. Upregulated ABCC3 (MRP3) is found in both GBM and BM vasculature, which is associated with resistance to cisplatin, methotrexate, vincristine, and epipodophyllotoxin chemotherapy.37 Expression of MRP3 in GBM tissue and GBM EC has been reported previously.38 Surprisingly, we found the downregulated expression of ABCB1 (P-glycoprotein) in GBM vasculature, also reported by Dusart et al.5 Interestingly, in a recent proteomic study of GBM vasculature, Bao et al. reported reduced protein level of ABCB1.14 GBM vasculature upregulates expression of genes encoding amino acid (SLC16A10, SLC36A1, SLC43A2 [LAT 4]), nucleobase and nucleoside transporters (SLC28A1, SLC29A4, SLC43A3), which could indicate altered metabolic requirements of GBM EC or metabolic cooperation between EC and tumor cells. Regardless, GBM endothelium may possess an increased capacity to transport nucleobase and nucleoside antimetabolites, commonly used anti-cancer drugs or amino acid-based prodrugs. Of note, SLC43A3 was defined as a BBB dysfunction module gene in mice.27
A subset of SLC transporters is important for cellular uptake of cancer therapeutics.39 We have found deregulation of the expression in brain tumor vasculature of several SLC transporters implicated in drug transport (eg, SLC22A3, SLC36A1, SLC28A1, SLC29A4, SLC43A3). Interestingly, we have detected upregulated expression of SLC36A1 mRNA in GBM EC. SLC36A1 is a low-affinity, high-capacity amino acid transporter that transports several CNS therapeutics (eg, vigabatrin) and diagnostic substrates (eg, 5-ALA).31 Immunohistochemical analysis showed that SLC36A1 was expressed by GBM tumor cells as well as endothelium. Protein expression of SLC36A1 by GBM EC showed high variability. Accordingly, further studies should be performed to characterize the variable SLC36A1 expression by GBM EC. Since GBM tissue also showed high expression of SLC36A1, it could potentially be used as a drug shuttle platform into tumor cells.
Receptor-mediated transcytosis by brain EC has been used to deliver biopharmaceutical agents into the brain.40 INSR is expressed at high levels by brain EC.18 The humanized anti-IR antibody HIRMAb shows a good brain penetration41 and HIRMAb-protein fusions have been tested to deliver several large therapeutic proteins into the brain (eg, glial cell derived neurotrophic factor, erythropoietin). Our analysis identified upregulation of INSR mRNA in GBM and BM EC, suggesting that INSR-mediated transcytosis might be exploited to deliver biopharmaceuticals to brain tumors.
Immunotherapy of brain tumors holds therapeutic promise.42 Clinical effects of T-cell-based checkpoint inhibition therapy have been observed in patients with BM from melanoma and non–small-cell lung cancer.43 However, a clinical benefit has not yet been observed in patients with primary brain tumors.44 Tumor cell heterogeneity may partially account for the distinct clinical response between GBM and BM. However, the distinct immune microenvironment3 and vascular properties might contribute as well. We could not detect any increase in transcript levels of various leukocyte adhesion molecules implicated in leukocyte transmigration via brain endothelium (ICAM1, VCAM1). However, MADCAM-1 expression was upregulated in GBM and BM EC. GBM vasculature also upregulated ALCAM, as previously reported.28,29 CCL19, a chemoattractant to CCR7-expressing T cells was downregulated by GBM and BM vasculature. Decreased expression of chemotactic factors that promote immune cell infiltration is reported in IDH-1-mutated tumors.42 Thus, understanding alterations in brain tumor vasculature could facilitate the development of successful immunotherapeutic strategies.
In summary, we have identified common and distinct changes in the BBB of GBM and adenocarcinoma BM (Supplementary Figure 12A–C). We identified SLC transporters and receptors more abundantly expressed by tumor vasculature, which could be exploited for efficient drug delivery across the BBB into the tissue (Supplementary Figure 12D). In addition, we have detected fibroblast-like cells in GBM tissue (Supplementary Figure 12D). Future studies should be directed toward understanding the role of these cells in GBM growth and treatment.
Funding
This work was supported by the Swiss Cancer League to A.K. and M.C.N. (grant KLS-3848-02-2016), Jubiläumsstiftung von Swiss Life, the Leducq Foundation (grant 14CVD02), the Swiss National Science Foundation (grants 31003A_159514, 310030_188952), the Synapsis Foundation and the Choupette Foundation (grant 2019-PI02), and the Swiss Heart Foundation to A.K.
Supplementary Material
Acknowledgments
Imaging was performed with equipment at the Center for Microscopy and Image Analysis, University of Zurich. Library preparation and RNA sequencing were performed by the Functional Genomics Center Zürich, University of Zurich and ETH. We thank S. Hornemann for discussions.
Conflict of interest statement. The authors declare that they have no competing interests.
Author contribution. A.K. and M.C.N. conceived the study. J.S. performed the experiments. J.S. and A.K. analyzed and interpreted the data. T.W. and L.H analyzed the bioinformatics data. M.D. supervised bioinformatics analysis. E.J.R., F.V., M.C.N., and L.R. analyzed the clinical data. J.S., T.W., and A.K. wrote the manuscript, all authors commented on the manuscript. A.K. supervised the study.
References
- 1. Suh JH, Kotecha R, Chao ST, Ahluwalia MS, Sahgal A, Chang EL. Current approaches to the management of brain metastases. Nat Rev Clin Oncol. 2020;17(5):279–299. [DOI] [PubMed] [Google Scholar]
- 2. Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO. Multidimensional communication in the microenvirons of glioblastoma. Nat Rev Neurol. 2018;14(8):482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Friebel E, Kapolou K, Unger S, et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell. 2020;181(7):1626–1642.e20. [DOI] [PubMed] [Google Scholar]
- 4. Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182–1186. [DOI] [PubMed] [Google Scholar]
- 5. Dusart P, Hallstrom BM, Renne T, Odeberg J, Uhlen M, Butler LM. A systems-based map of human brain cell-type enriched genes and malignancy-associated endothelial changes. Cell Rep. 2019;29(6):1690–1706.e4. [DOI] [PubMed] [Google Scholar]
- 6. Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR. Vessel co-option in cancer. Nat Rev Clin Oncol. 2019;16(8):469–493. [DOI] [PubMed] [Google Scholar]
- 7. Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. Establishment and dysfunction of the blood-brain barrier. Cell. 2015;163(5):1064–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ostermann S, Csajka C, Buclin T, et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin Cancer Res. 2004;10(11):3728–3736. [DOI] [PubMed] [Google Scholar]
- 9. Vivanco I, Robins HI, Rohle D, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2(5):458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarkaria JN, Hu LS, Parney IF, et al. Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol. 2018;20(2):184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mihajlica N, Betsholtz C, Hammarlund-Udenaes M. Pharmacokinetics of pericyte involvement in small-molecular drug transport across the blood-brain barrier. Eur J Pharm Sci. 2018;122:77–84. [DOI] [PubMed] [Google Scholar]
- 12. Phoenix TN, Patmore DM, Boop S, et al. Medulloblastoma genotype dictates blood brain barrier phenotype. Cancer Cell. 2016;29(4):508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uchida Y, Ohtsuki S, Katsukura Y, et al. Quantitative targeted absolute proteomics of human blood-brain barrier transporters and receptors. J Neurochem. 2011;117(2):333–345. [DOI] [PubMed] [Google Scholar]
- 14. Bao X, Wu J, Xie Y, et al. Protein expression and functional relevance of efflux and uptake drug transporters at the blood-brain barrier of human brain and glioblastoma. Clin Pharmacol Ther. 2020;107(5):1116–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dieterich LC, Mellberg S, Langenkamp E, et al. Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFβ2 in vascular abnormalization. J Pathol. 2012;228(3):378–390. [DOI] [PubMed] [Google Scholar]
- 16. Davis FG, Dolecek TA, McCarthy BJ, Villano JL. Toward determining the lifetime occurrence of metastatic brain tumors estimated from 2007 United States cancer incidence data. Neuro Oncol. 2012;14(9):1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Butler LM, Hallstrom BM, Fagerberg L, et al. Analysis of body-wide unfractionated tissue data to identify a core human endothelial transcriptome. Cell Syst. 2016;3(3):287–301.e3. [DOI] [PubMed] [Google Scholar]
- 18. Vanlandewijck M, He L, Mäe MA, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554(7693):475–480. [DOI] [PubMed] [Google Scholar]
- 19. GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puchalski RB, Shah N, Miller J, et al. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360(6389):660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carson-Walter EB, Hampton J, Shue E, et al. Plasmalemmal vesicle associated protein-1 is a novel marker implicated in brain tumor angiogenesis. Clin Cancer Res. 2005;11(21):7643–7650. [DOI] [PubMed] [Google Scholar]
- 22. Kalucka J, de Rooij L, Goveia J, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180(4):764–779.e20. [DOI] [PubMed] [Google Scholar]
- 23. De Bock K, Georgiadou M, Schoors S, et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154(3):651–663. [DOI] [PubMed] [Google Scholar]
- 24. Pardridge WM, Boado RJ, Farrell CR. Brain-type glucose transporter (GLUT-1) is selectively localized to the blood-brain barrier. Studies with quantitative western blotting and in situ hybridization. J Biol Chem. 1990;265(29):18035–18040. [PubMed] [Google Scholar]
- 25. Oudard S, Arvelo F, Miccoli L, et al. High glycolysis in gliomas despite low hexokinase transcription and activity correlated to chromosome 10 loss. Br J Cancer. 1996;74(6):839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corada M, Orsenigo F, Bhat GP, et al. Fine-tuning of sox17 and canonical Wnt coordinates the permeability properties of the blood-brain barrier. Circ Res. 2019;124(4):511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Munji RN, Soung AL, Weiner GA, et al. Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood-brain barrier dysfunction module. Nat Neurosci. 2019;22(11):1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Samaha H, Pignata A, Fousek K, et al. A homing system targets therapeutic T cells to brain cancer. Nature. 2018;561(7723):331–337. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Samaha H, Pignata A, Fousek K, et al. Retraction note: a homing system targets therapeutic T cells to brain cancer. Nature. 2019;567(7746):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coloma MJ, Lee HJ, Kurihara A, et al. Transport across the primate blood-brain barrier of a genetically engineered chimeric monoclonal antibody to the human insulin receptor. Pharm Res. 2000;17(3):266–274. [DOI] [PubMed] [Google Scholar]
- 31. Thwaites DT, Anderson CM. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br J Pharmacol. 2011;164(7):1802–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeng Q, Michael IP, Zhang P, et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature. 2019;573(7775):526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muhl L, Genové G, Leptidis S, et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun. 2020;11(1):3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valkenburg KC, de Groot AE, Pienta KJ. Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin Oncol. 2018;15(6):366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clavreul A, Etcheverry A, Tétaud C, et al. Identification of two glioblastoma-associated stromal cell subtypes with different carcinogenic properties in histologically normal surgical margins. J Neurooncol. 2015;122(1):1–10. [DOI] [PubMed] [Google Scholar]
- 36. Pope WB, Mirsadraei L, Lai A, et al. Differential gene expression in glioblastoma defined by ADC histogram analysis: relationship to extracellular matrix molecules and survival. AJNR Am J Neuroradiol. 2012;33(6):1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kool M, van der Linden M, de Haas M, et al. MRP3, an organic anion transporter able to transport anti-cancer drugs. Proc Natl Acad Sci U S A. 1999;96(12):6914–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Calatozzolo C, Gelati M, Ciusani E, et al. Expression of drug resistance proteins Pgp, MRP1, MRP3, MRP5 and GST-pi in human glioma. J Neurooncol. 2005;74(2):113–121. [DOI] [PubMed] [Google Scholar]
- 39. Hu C, Tao L, Cao X, Chen L. The solute carrier transporters and the brain: physiological and pharmacological implications. Asian J Pharm Sci. 2020;15(2):131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freskgård PO, Urich E. Antibody therapies in CNS diseases. Neuropharmacology. 2017;120:38–55. [DOI] [PubMed] [Google Scholar]
- 41. Boado RJ, Zhang Y, Zhang Y, Wang Y, Pardridge WM. GDNF fusion protein for targeted-drug delivery across the human blood-brain barrier. Biotechnol Bioeng. 2008;100(2):387–396. [DOI] [PubMed] [Google Scholar]
- 42. Ratnam NM, Gilbert MR, Giles AJ. Immunotherapy in CNS cancers: the role of immune cell trafficking. Neuro Oncol. 2019;21(1):37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goldberg SB, Gettinger SN, Mahajan A, et al. Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol. 2016;17(7):976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reardon DA, Brandes AA, Omuro A, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the checkmate 143 phase 3 randomized clinical trial. JAMA Oncol. 2020;6(7):1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data, both raw data and gene-by-sample matrix of raw counts, were deposited in Gene Expression Omnibus (GEO) under accession number GSE159851.
All data used to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Material.