Abstract
A 1969 report that described biochemical and activity properties of the three eukaryotic RNA polymerases revealed Pol III as highly distinguishable, even before its transcripts were identified. Now known to be the most complex, Pol III contains several stably-associated subunits referred to as built-in transcription factors (BITFs) that enable highly efficient RNA synthesis by a unique termination-associated recycling process. In vertebrates, subunit RPC7(α/β) can be of two forms, encoded by POLR3G or POLR3GL, with differential activity. Here we review promoter-dependent transcription by Pol III as an evolutionary perspective of eukaryotic tRNA expression. Pol III also provides nonconventional functions reportedly by promoter-independent transcription, one of which is RNA synthesis from DNA 3′-ends during repair. Another is synthesis of 5′ppp-RNA signaling molecules from cytoplasmic viral DNA in a pathway of interferon activation that is dysfunctional in immunocompromised patients with mutations in Pol III subunits. These unconventional functions are also reviewed, including evidence that link them to the BITF subunits. We also review data on a fraction of the human Pol III transcriptome that evolved to include vault RNAs and snaRs with activities related to differentiation, and in innate immune and tumor surveillance. The Pol III of higher eukaryotes does considerably more than housekeeping.
INTRODUCTION
The idea that eukaryotes might have ‘evolved’ two distinct RNA polymerase (Pol) types, one to synthesize the G + C-rich ribosomal-RNA, genes known to be repetitive and localized in nucleoli, and another Pol to synthesize the rest of the RNA, was met by identification and characterization of Pols I, II and III more than fifty years ago (1). The cellular Pols in bacteria, archaea and eukarya derive from a common lineage whose multiple subunits have conserved sequence, structure and mechanisms of action (2). Although archaea share many features with bacteria including polycistronic genes and a single Pol to transcribe all RNA types, the polymerase composition and associated general transcription factors (GTFs) are more similar to eukaryotes (3) (Figure 1). Beyond this, the combinatorial nature of promoters, enhancers, and the GTFs and activators that interact with them are also more expansive in eukaryotes than archaea (3). However, a more profound reorganization accompanied evolution of the eukaryl system, the division of transcription to three Pols. Most important to this review, this included not only a dedicated Pol that was specialized for tRNA synthesis but also the reorganization of genomic tRNA sequences as individual genes.
Figure 1.
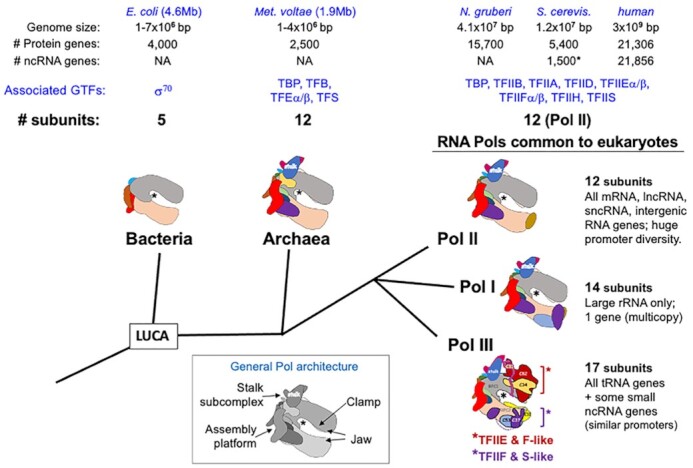
Evolutionary distinction of the RNA Polymerases. Phylogenetic tree of the multisubunit Pols. The DNA-binding clefts of the cartoon depictions of the Pols are indicated by an * in the active center. Columns above the cartoon representations of the Bacteria, Archaea and Eukarya Pols are headed by species names (Escherichia coli, Methanococcus voltae, Naegleria gruberi, Sachharomyces cerevisiae), followed by genome characteristics, associated GTFs (general transcription factors) and the number of Pol subunits. NA stands for not available. LUCA refers to last universal common ancestor. The right side illustrates the three Pols common to eukaryotes and their features as discussed in the text. Note that two subcomplexes referred to in the text as built-in transcription factors (BIFTs) with homology to GTFs TFIIE and TFIIF, are organized on Pol III as two subcomplexes on either side of the cleft, indicated by brackets. Inset: general architecture of Pol is depicted; see (2).
In this Survey, we review distinguishing features of the Pol III system including evolution of its transcriptome in animal cells. We also attempt to pull together information relevant to understanding novel Pol III functions in innate immunity and DNA repair, including properties that support these activities.
Early and new discoveries reveal distinguishing features and unique activities of Pol III
The initial characterization of catalytic properties demonstrated that Pol III differed from Pols II and I in that it did not require denaturation (strand melting) of the calf thymus DNA substrate for optimal template activity (1). Differential sensitivity of Pol II and Pol III to α-amanitin allowed characterization of cellular tRNA and 5S rRNA synthesis as well as adenovirus associated small RNAs (4,5, and references therein). Two other properties now appear relevant to its unconventional function in immunity. First, it was documented that the substantial majority, ∼65%, of Pol III activity was found to reside in the cytoplasmic fraction of cellular extract compared to 12–16% of Pols I and II in the same fractions (of multiple cell types) (6). Second, Pol III was 11- to 16-fold more active with poly(dA:dT) relative to calf thymus DNA as template (7).
These properties appear to be relevant because Pol III was discovered as a cellular sensor of A + T-rich DNA in the cytoplasm (8,9) that protects against heritable forms of childhood and adult immunodeficiency (10). In this function, Pol III produces transcripts that act more like signaling molecules than typical ncRNAs, in which the nascent 5′-triphosphate end (5′pppN) is the most important feature rather than the sequence per se. Moreover, the limited data available suggest that this Pol III activity utilizes a novel transcription initiation mechanism not yet understood.
A second recently discovered unconventional activity for Pol III is the programmed RNA synthesis that occurs during the process of DNA double strand break repair (11). This serves a critical function of protecting the 3′-overhang DNA information during resection and replacement of the 5′ DNA strand (11).
We go on to detail small ncRNAs transcribed by Pol III and their connections to the immune system or other activities unique to higher eukaryotes. For example, both Pol III-transcribed Vault and snaR ncRNAs are processed to miRNAs that exert downstream effects on mRNA profiles with function in cellular differentiation-cancer axis (12,13). Throughout, we try to link unique features of Pol III with its newly discovered RNA synthesis activities and how they contribute to these functions. Prior to this however, we consider Pol evolution and properties of Pol III that make it unique for a process referred to as termination-associated reinitiation-recycling that reflects its specialized activities.
Evolution of the eukaryotic Pols
The Pols from bacteria to human are comprised of subunits that can be classified into three major functional groups: assembly platform subunits, catalytic subunits that make up the core, and auxiliary subunits (reviewed in 2). Archaeal Pol and eukarya Pol II share a highly homologous 12 subunit architecture (2) ‘evolved’ from an ancestral five subunit bacterial structure (Figure 1). Some subunits were added to the assembly platform and others to the core. Notable are subunits of the stalk sub-complex, as these supported integration of GTFs used by archaea and eukaryotes (2) (Figure 1 and inset).
Eukaryl promoters are more complex in combinatorial architecture as compared to archaeal promoters. This may reflect that eukarya contain more TBP-associated factors (TAFs), some of which contact promoter elements, as compared to archaea. Archaea also lack some GTFs that appear in eukaryotes such as TFIIA, TFIIF, TFIIH (Figure 1), as well as an expansively large number, ∼1600, of transcriptional activators (14) that work with Pol II to control a wide range of gene-specific transcription. Thus, archaeal Pol and eukaryl Pol II are similar while the control elements of genes they transcribe and associated factors differ in complexity.
In all eukaryotes, Pols I, II and III transcribe different gene classes (15) (Figure 1); some plants also contain specialized homologs of Pol II known as Pols IV and V that serve gene silencing functions (16,17). Pol I is limited to the synthesis of rRNA from a single medium size gene (10–15 kb) that is usually present in many tandem copies at one to a few chromosomal loci, totaling up to a few hundred gene copies (e.g. yeast and human). The rRNAs accumulate to high levels in part due to much longer half-life relative to mRNA. Pol III is responsible for synthesis of the tRNAs, which in yeasts, human and many other model eukaryotes occurs from 200–500 genes. The Pol III transcriptome from yeast to human includes additional essential small noncoding (nc) RNAs, usually ∼30–50 copies of 5S rRNA plus a few other ‘single copy’ ncRNA genes (Figure 2A, B). As discussed later, the Pol III transcriptome expanded in higher eukaryotes.
Figure 2.
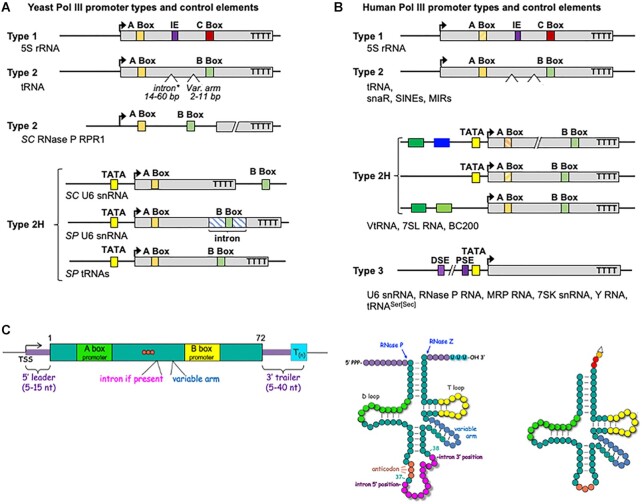
The different promoter types used to direct transcription of Pol III-dependent genes in yeast and human. (A) The two promoter types of yeast are depicted along with the hybrid subtype 2H, as suggested by (41). Type 1 is conserved from yeast to human and limited to 5S rRNA genes. Type 2 promoters contain an A Box and B Box, the distance between which varies as tRNA genes contain extra arms of variable length, and a subset contain introns of variable length. Type 2H (hybrid) promoters are similar to type 2 with an A-Box followed by a B-Box and also contain an upstream TATA element at −30. Bent arrows indicate the transcription start sites (TSS). In yeast (SC, Saccharomyces cerevisiae, SP Schizosaccharomyces pombe) type 2H have unique variations of the A- and B- Box seen in type 2. For SC RPR1 (RNase P RNA) the promoter elements are found upstream of what becomes the mature RNA species. For U6 the B Box is outside the mature RNA in SC and within an intron in SP. Not shown is the SC scR1 gene schematic encoding SRP RNA which has a TATA beginning at −31 relative to +1 of scR1. (B) The three promoter types in human are depicted along with the 2H subtypes which vary in upstream control elements (41), notable for 7SL, BC200 and VtRNAs (colored boxes). The type 3 promoter is present in higher eukaryotes (not found in yeast) consisting of a proximal sequence element (PSE), a TATA box and a distal sequence element (DSE). A type 3 promoter type, comprised of elements that direct assembly of the transcription complex exclusively from upstream of the TSS, is not found in yeast. Regardless of the variable types of promoters that direct initiation, the control element that terminates transcription by the Pols III from yeast to human, is a short oligo(T) tract on the non-template strand. (C) Left: A schematized tRNA gene is illustrated with its variable hallmark features, color-coded according to the tRNA cloverleaf models to the right. These include the Pol III terminator, the 5′pppN TSS = transcription start site, 5′ leader, 3′ trailer, variable arm and intron between the A and B Boxes. Note that three orange circles represent the anticodon. Middle: General cloverleaf model of a nascent precursor-tRNA with an intron and variable arm and other segments color-coded with the gene diagram. The RNase P and RNase Z cleavage sites are indicated, as are the 5′ppp and 3′OH ends. A tract of Us at the 3′ end that results from termination is the sequence-specific binding site for the La protein, pre-tRNA chaperone/maturation factor. Right: Representation of a corresponding mature tRNA. Following splicing and processing, CCA is added to the matured 3′ end, after which the amino acid is attached (open triangle).
Pol II transcribes the vast domain of the remainder of the DNA to produce all coding mRNAs and a number of long and short ncRNAs, which in human may be much more numerous than the protein genes (18). Thus, by gene count, Pol II transcribes the large majority and much greater diversity of promoter types. This is likely true in the representative of the primordial eukaryote ancestor, Naegleria gruberi, supported by analysis of its 41 Mb genome (Figure 1) (19,20). Although details of its Pol system have not been noted previously, the predicted 16 000 genes of N. gruberi include 12 subunits for Pol II, multiple subunits for TFIIA, E, F and H as well as 11 TAFs (Figure 1). Thus, as represented by N. gruberi, the ancient Pol II system was associated with more GTFs than Archaeal Pol, and accordingly therefore, the potential for greater combinatorial regulatory gene control (Figure 1). The N. gruberi gene repertoire also includes homologs of the Pol I- and Pol III-specific subunits (not shown).
Although the primordial eukaryote also contained the Pol I and Pol III systems (Figure 1), the particular benefit(s) of this arrangement over a single Pol with expanded diversity of GTFs and promoter elements is not readily apparent and intriguing to consider. Evidence supports the idea that Pol I and Pol III have each been specialized with enzymatic and other characteristics for efficient differential transcription of their specific gene classes (21,22). Analysis of 25 yeast species genomes supports the idea that rDNA promoter sequences have been coevolving with the Pol I system at higher mutation rates relative to the Pol III and Pol II system machineries (23, 21 and references therein). Other analyses led to surprising findings for Pol III (23,24). The relatively high evolutionary rate of Pol III could not be explained by any adaptive theory (24). This suggested that a theory to account for some aspects of Pol III evolution would remain to be proposed.
What may have led to a larger, more complex and highly specialized 17-subunit Pol III in the Eukarya lineage? In retrospect, there would appear no reason a priori that the ancestral Pol II system alone, represented by N. gruberi if it existed in the absence of Pols III and would not have been able to handle an additional few hundred tRNA and rRNA genes as part of its transcriptome. These additional genes may be likened to certain Pol II-highly transcribed gene classes, such as the multicopy U1-U5 snRNAs and similar others that use specialized promoters and transcription termination mechanisms, for example, the high output histone gene repeats, as well as numerous other multicopy gene families. Thus, a perspective to consider is a potential advantage to a new lineage, of a dedicated specialized Pol (III) with control of a few hundred tRNA genes which encode multiple copies of individual anticodon decoders.
Individualization of tRNA genes and a dedicated Pol III are co-equal attributes of the Eukarya
The genomic organization of tRNA genes sets the eukarya apart from bacteria and archaea. tRNA synthesis directed by internal promoters of dispersed independent transcription units is not found in the absence of Pol III. At the time described, the adaptors that were proposed to transfer amino acids dictated by the sequence of nucleic acid template had not yet been discovered as tRNAs (25). To deal with degeneracy in a triplet genetic code, the Wobble hypothesis emerged (26). Synonymous codons are now more appropriately considered redundant, known for extra information capacity separate from dictating the amino acid sequence of a polypeptide (reviewed in 27). Ample evidence indicate that biased use of synonymous codons in mRNAs is coordinated by tRNA wobble modifications as part of ‘tunable’ genetic responses to various stresses (28), and comprise up to 10% of a transcriptome (reviewed in 29). The redundancy component of the code is relatively large with much capacity for secondary information of a variety of types (28–30).
A fully internal promoter can support fluidity of tRNA gene placement within the genome and also contribute to copy-number (gene dosage) adjustable tRNAomes not afforded to genes with conventional upstream promoters. One can envision that the Pol III system could empower its collective tRNA genes as a dynamic force of genetic modifiers of mRNA-encoded genes (30) that could help emerging species exploit the redundancy component of the genetic code. The extraordinary diversity of eukaryotic tRNAomes, which range from ∼100 individual tRNA genes in simple eukaryotes, to >104 in some complex animals (31), reflects the potential for and evidence of their expansion.
A monocistronic tRNA gene structure could also provide more uniform tRNA processing. The A-Box and B-Box promoter elements overlap with conserved motifs in tRNA (Figure 2C). This gene structure provides each emergent transcript with key functional hallmarks: 5′ppp followed by self-folding D- and T-stem loop structures (32), and a 3′-oligo(U) motif that serves as a termination signal for Pol III (Figure 2C) and high-affinity binding site for the eukaryote-ubiquitous La protein pre-tRNA maturation factor (33).
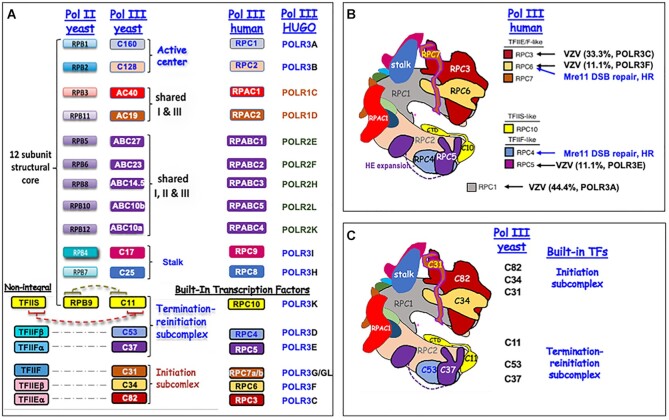
Pol III was specialized with properties suited to the short gene transcripts it synthesizes in promoter-dependent manner (22). It was noted in 2008 that Pols I and III contain stable integral subunits that are homologs of key transcription factors in the Pol II system (34), and these were appropriately referred to as ‘built-in’ transcription factors (BITFs) (35,36) (Figure 1). For Pol III the BITFs participate in initiation, elongation and termination (37,38, and refs therein). We note that it is generally accepted that the BITFs do not function precisely as, nor would fit a strict definition of, a TF. The benefits of BITF subunits that enable Pol III termination-associated reinitiation-recycling for abundant RNA synthesis from tRNA genes can account for the relative complexity of Pol III (Figure 1). The six Pol III BITFs are indicated in Figure 3A, C.
Figure 3.
Schematic structural representation of Pol III. (A) Comparison of subunit compositions of Pol II and Pol III (yeast and human) with emphasis on the built-in transcription factors (BITFs) of Pol III. The human gene nomenclature is provided for the homologs under the column headed by HUGO. Note that humans have two alternate isoforms of RPC7 encoded by POLR3G and POLR3GL. (B) A structure model of human Pol III based on cryo-EM, modeled after Girbig et al. (51). HE expansion refers to the extra purple globular domain of RPC5 attached by a linker (dashed line) which is specific to higher eukaryotes. The subunits targeted by mutations found in VZV-susceptible patients (10) or in yeast-two hybrid studies for DSB repair (11) are indicated as discussed in the text indicated by arrows. Numbers in parentheses indicate the percentage of POLR3-unique VZV-susceptible alleles identified have been reported to date in the designated gene. (C) As in (B). the structure of Pol III is shown but with yeast nomenclature. The BITFs are indicated as such.
GENERAL AND SPECIFIC FEATURES OF THE POL III-DEPENDENT GENE TRANSCRIPTION SYSTEM
Even with its BITFs, Pol III cannot efficiently target its substrate genes nor initiate transcription with the start site precision required of short ncRNAs. Similar to other Pols, gene-specific initiation by Pol III occurs by a promoter-dependent, and TF-mediated mechanism, the components of the latter of which vary with the gene type (below) (39–41). Type 1 and 2 promoters are referred to as internal because they reside between the transcription start site (TSS) and termination site, and for 5S and tRNAs, classic type 1 and 2, are represented in the functional RNA, while the type 3 promoter (not found in yeast) resides entirely upstream of the TSS (Figure 2A, B). Because type 1 and 2 control elements overlap with highly conserved motifs at the RNA level (see below), assessing their conservation for transcription function required careful comparative analyses (42) (for early reviews see 43,44).
The three subunit initiation factor, TFIIIB (TBP, Brf1/2, Bdp1) serves to bring Pol III to its target genes and direct the TSS. TFIIIB is recruited by type 1 and 2 promoters by the multisubunit TFIIIC, and to type 3 promoters by the multisubunit TF, SNAPc (below) (for arrangement of these TFs on their target gene promoters see figure two in 45). TFIIIB serves additional critical function after recruiting Pol III to its target genes (46). It bends DNA at −30 upstream of the TSS and works with Pol III in isolating a single strand of the DNA duplex for loading into the active site, in proximity to the initiating nucleoside-triphosphate in preparation for RNA synthesis (46,47). It should be noted that the transcription initiation factor activity of TFIIIB requires all of its subunits; neither alone is considered a TF in the Pol III system. Thus, for type 2, initiation of RNA synthesis requires preplacement of TFIIIB on DNA upstream of TSS guided by a TFIIIC subunit bound to the A-Box, followed by IIIB recruitment of Pol III and positioning at/near the TSS (48).
The type 1 promoter is exclusively used by 5S rRNA genes, consisting of A-Box, intermediate and C-Box elements (Figure 2A, B). Type 2 promoters control the numerous tRNA and other ncRNA genes, consisting of A-Box, B-Box and terminator elements, the former two of which may vary in sequence among genes along with the A-B Box and B-terminator distances, exhibiting plasticity (Figure 2A) attributable to properties of TFIIIC (see 49 and refs therein). In addition, posttranscriptional processing can contribute to the plasticity of function as in the case of the gene for the RNA subunit of RNase P in S. cerevisiae (Figure 2A). A promoter type termed 2-hybrid (2H) exist in yeast and higher eukaryotes with A- and B-box elements but with additional upstream elements (Figure 2A, B). The few TATA elements upstream of S. cerevisiae tRNA genes apparently compensate for unusually long A-to-B Box distance (due to long intron length) that otherwise weaken promoter strength, which may also apply to the yeast U6 gene (50) (Figure 2A). The A and B Box sequences overlap with the D and T stem-loops of tRNAs, respectively, whereas 5′ leader and 3′ trailer sequences are removed during tRNA maturation (Figure 2C).
Evolutionary appearance of a type 3 promoter, absent in yeast but found in higher eukaryotes, accompanied and may have contributed to expansion of the Pol III transcriptome. A type 3 promoter assembles the transcription complex via control elements located exclusively upstream of the TSS. The proximal sequence element (PSE) of type 3 promoter appears to have co-evolved with subunits of the SNAPc TF that recognizes it (51). In this case, the PSE and TATA upstream elements exclusively direct Pol III to initiate synthesis of type 3 ncRNAs (Figure 2B). Interestingly, tRNASer(Sec) genes contain D and T loop sequences that appear as A and B boxes, but utilize a type 3 promoter that directs initiation, and has been conserved through evolution (52). This type 3 promoter produces the only eukaryotic pre-tRNA that lacks a 5′ leader; rather, the 5′pppG at the TSS is the +1 nucleotide of the mature tRNA (53,54).
Higher eukaryotic Pol III transcriptomes contain additional ncRNA genes not found in yeast, such as 7SK, Y RNAs, VtRNAs, snaRs and high copies of SINEs most of the latter of which are usually repressed but can be conditionally activated (below). The type 2H promoters (40,41), use a TATA-rich element similar to type 3; however these rely on A- and B-boxes as principal determinants of TFIIIB placement but can also use additional upstream elements for tissue-specific or conditional activation (Figure 2B).
A distinguishable feature of Pol III is that unlike at classic Pols II promoters and bacterial σ54 type promoters, Pol III initiation does not require ATP hydrolysis (e.g. of a TFIIH/helicase-like factor) to ‘melt’ the duplex DNA, i.e., open a transcription bubble to isolate and secure the template strand in the catalytic site. Before detailing this, we should note that this may be an important feature of cytoplasmic-Pol III activity in its innate immunity function to be reviewed in a later section.
TFIIIB-Pol III when positioned on A + T-rich DNA, undergoes an ordered DNA strand melting process, utilizing Pol III subunits C34 and C37 and TFIIIB subunits (37,38). Next is selection of a template-base-pairing NTP and the de novo phosphodiester bond that initiates 5′ppp-RNA synthesis (55) (see 47,48 and references therein). Pol III most often selects purine-NTP for initiation and if similar to other pols, this would likely be directed by stacking influences of purine at the minus-1 position in the template DNA (56,57). Thus, after TFIIIB and Pol III are in place, the TSS will most often be a purine preceded by a pyrimidine at minus-1.
Pol III has a relatively simple and efficient mode of termination compared to Pols I and II. In addition to autonomous termination of RNA synthesis and dissociation upon encountering a tract of 5Ts in the non-template strand (5As in the template), TFIIIB can readily support reinitiation by Pol III for repeated cycles of RNA synthesis (58, and refs therein). Such analyses use templates, or Pol III, immobilized on beads or other support, from which the terminated/released RNA to be analyzed can be readily separated from the bound material, although gel filtration or filter-binding has also been used (59–62).
The termination mechanism unique to Pol III generates a short run of 3′ terminal uridines in the transcripts, to which the La protein readily associates because U(n)U-3′OH is recognized by its high affinity binding site (63). Thus nascent Pol III transcripts are effectively ‘handed off’ to La protein, which serves as a molecular chaperone (64). For vertebrate Pol III, a minimal of 4Ts is sufficient as a terminator which represents an intrinsic species-specific difference in the termination mechanism and which appears to correlate with the oligo(U) length requirement for high affinity binding to La protein (65 and refs therein). Precursor-tRNAs are the most numerous, abundant and diverse Pol III transcripts. They also undergo the most complex maturation pathway consisting of sequential RNA cleavage events and several modifications before carrier-mediated export to the cytoplasm (66,67). La uses nuclear retention activity as part of its molecular chaperone function. Pre-tRNAs and other Pol III transcripts eventually lose La protein by dissociation or processing regardless of whether they ultimately reside in the cytoplasm or nuclei (64).
Termination-associated reinitiation-recycling by Pol III requires a subset of its BITFs
To sustain high molar excess of tRNAs relative to ribosomes for efficient translation during cellular growth and proliferation, Pol III would have to initiate transcription at rates many fold relative to Pol I (68). Important distinguishing features of Pol III transcription were determined early. (i) after assembly on the DNA, the preinitiation complex (PIC) is highly stable to dissociation, (ii) the PIC can direct multiple rounds of transcription (recycling) by Pol III and (iii) TFIIIB is a key PIC component, common to all promoters (59,69–71). Alluded to earlier, this is a major difference from the Pol II system in which the GTFs were shown to disassemble the PIC-promoter complex upon productive elongation (72). The contribution of Pol III to recycling (62) began to be articulated as it was appreciated to contain stably-integral subunits that are homologs of transcription initiation factors in the Pol II system (34), referred to as built-in transcription factors (35,see 36) (Figure 3A, B). The BITF subunits indeed actively participate in initiation and elongation (37,38, and references therein,73), as well as termination (62,65,74–81).
It should be noted that much focus on the termination-associated reinitiation-recycling process has been on the BITFs that were principally linked to elongation and termination (62,65,74,75,77–81). These are the C37/53 heterodimer (human RPC4/5), and the C11 subunit (RPC10). As indicated in Figure 3A, C37 and C53 are related to TFIIF subunits and C11 homologous to TFIIS in their C-terminal RNA 3′ cleavage domains and to Pol II subunit RPB9, and RPA12.2 in their N-terminal domains. For a more comprehensive description of C11 and its role in Pol III recycling see (81). In contrast to the C37/53 and C11 BITFs, the roles of the heterotrimeric initiation complex BITFs in Pol III recycling per se have have been much less studied.
In summary, the basis of high efficiency Pol III recycling activity as currently understood is comprised of three components, (i) a highly stable PIC, (ii) up to six stably-associated BITFs organized in two subcomplexes and (iii) an enzymatic-allosteric mechanism contributed in part by the highly mobile C11 subunit (yeast nomenclature, human RPC10, Figure 3) in addition to core components. This is coordinated in part by movements of the initiation subcomplex BITFs and associated opening of the RPC1 clamp (Figure 1, inset), an allosteric element of the dsDNA binding site. In this model, the clamp is key to ready transition between termination and reinitiation (51,81).
Structural and assembly related aspects of Pol III provide new insights including into POLR3G/GL RPC7α/β and tumor suppressor, Maf1
The first cryo-EM structures of the human Pol III complex were recently solved in apo-enzyme form and as elongation complexes with DNA and RNA in the active site (36,51,82,83). The collective work of four research groups advanced multiple aspects of Pol III. These include a potential basis of tumor suppression by Maf1 related to activity of POLR3G/RPC7α-dominant cancer and a mode of auto-inhibition by POLR3G. Others include dynamics of RPC10 whose transitions could contribute to coordination of termination and reinitiation. In one state, the CTD RNA cleavage hairpin of RPC10 was observed inserted into the pore and funnel of Pol III reaching toward the RNA 3′ end of the elongation complex, in position to monitor termination or other events (51,83). An intricacy of RPC7 was revealed, threading through the three-subunit initiation subcomplex bridging the stalk and the Pol III core module, and also connecting to the clamp (51,82,83). An iron−sulfur (Fe−S) cluster in the RPC6 subunit of the heterotrimer, absent from the yeast subunit, appears to guide the N-terminal loop of RPC7 toward the clamp and the DNA exit tunnel (51,82,83). Evidence suggests that the Fe-S cluster not only stabilizes the heterotrimer but can allow Pol III to sense oxidative stress and respond via regulated transcription output (see 83). Human RPC5 includes a large expansion relative to yeast that includes four winged-helix (WH) domains specific to higher eukaryotes, that appears to have co-evolved with SNAPc factors required for type 3 promoter function (51). One domain of the expansion was found in two conformations, swung out so that the WHs were in position to interact with the DNA just upstream of TFIIIB in the modeled PIC (51). In other work on RPC5 it was found to be important for assembly and stability of human Pol III in living cells (36).
Also striking is a regulatory link uncovered between Rbs1, a Pol III assembly factor, and Rpb10, the small subunit common to all three Pols (84). Control of Rbs1 levels and activity is intricate with potential to coordinate Pol III assembly and nuclear import with other events (85), suggesting possible links to the regulation of cytoplasmic-Pol III in human cells.
Insights into RPC7α and its paralog RPC7β are notable. Remarkably, vertebrate Pol III exists in two distinct isoforms, Pol IIIα and Pol IIIβ because the ∼32 kDa subunit is derived from paralogous genes, POLR3G and POLR3GL which produce RPC7α/RPC32α and RPC7β/RPC32β respectively (86). Affinity-purified Pol IIIα and Pol IIIβ containing RPC7α or RPC7β were comparably competent for in vitro transcription of all three promoter type genes (86). The study documented expression of RPC7α and RPC7β in embryonic stem (ES) cells whereas RPC7α stoichiometry increased with cellular transformation (86). Importantly, RPC7α was associated with characteristics of tumorigenesis, not proliferation per se but Pol II transcriptome shifts, mRNAs known to promote cell survival, tumor growth, metastasis, and decreased tumor suppressors (86).
Landmark studies made early connections, and others continue to expand links between Pol III, associated factors and the biology of cellular differentiation and cancer (87–91). Deregulation of Pol III in cancer was comprehensively reviewed as part of a broader perspective (92). Deborah Johnson's group found that inhibition of Pol III activity by its general repressor Maf1 promotes differentiation of mouse embryonic stem cells reflected by induction of mRNA differentiation-specific genes (91) and also showed that repression either by knock-down of Brf1 or treatment with the small molecule inhibitor of Pol III, ML-60218 (ML6 hereafter) also promoted differentiation, consistent with tumor suppressor activity of MAF1 (90). The three methods led to overlapping changes in RNA-seq profiles, although each with a specific signature (91). White et al. examined the POLR3G and GL subunits in a prostate cancer cell model. POLR3G depletion by ML6 promoted differentiation along a pathway of what we will refer to as cellular identity, whereas its over-expression opposed this pathway toward proliferative growth. However beyond this conclusion, White et al. discovered that ML6 depleted POLR3G with the same differentiation and cancer cell inhibition effects as POLR3G depletion by siRNA (93).
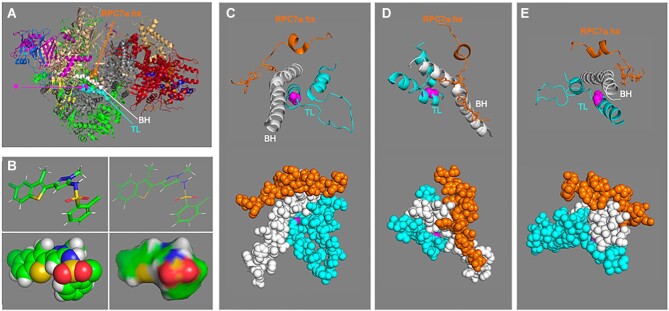
A highly conserved region of the C-terminal tail of RPC7α was found inserted into the empty DNA binding cleft and RNA exit channel of the hPol III apoenzyme, that is expected to be ejected by Brf1/2 components of TFIIIBα/β during formation of a PIC as an autoinhibitory mechanism (82). This novel mode of autoinhibition is unique among those for Pols I and II (94,95 and refs). Examination of the hPol III apo-enzyme structure reveals the conserved short α-helix of RPC7α autoinhibitory motif over the kinked region of the bridge helix (BH) of RPC1 which itself rides directly over the trigger loop (TL)-helix residue that comprises the highly conserved presumed target site of ML6, Gly-1045 of human Pol III (96) (Figure 4).
Figure 4.
Structure model of human apo-Pol III presumed amino acid target of inhibitor ML-60218 (ML6). Based on cryo-EM structure PDB 7D59 reported by Wang et al. (82), all viewed and produced with PyMOL Molecular Graphics system. (A) The features of interest are as follows: bridge helix (BH), and trigger loop (TL) regions of RPC1 are shown as spheres in white and cyan respectively while the RPC7α helix (hx) is in orange. This color coding is carried through in panels C-E. In this model the target glycine, G1045 of RPC1 is shown in magenta in sphere representation (indicated by arrow asterisk) and the rest of the 17-subunit model is in cartoon. RPC1 is colored grey; the color code follows that in figure 3B for the Pol III-specific subunits with the shared subunits in green. (B) Four representations of the inhibitor ML-60218 (C19H15Cl2N3O2S2, molecular mass: 452.4 Da (https://pubchem.ncbi.nlm.nih.gov/compound/RNA-Polymerase-III-Inhibitor). The top two are stick and line representations, and the bottom are sphere and surface, all in the same rotational view. (C−E) Zoomed in views of different rotations of the human Pol III structure in panel A after removal of other residues to better reveal features of the close-ups. The top parts show cartoon views as in A and the bottoms show the same view in sphere representation.
Although the region that lies over the BH is conserved in RPC7α and RPC7β these paralogs differ elsewhere. First, the region in RPC7β preceding the short helix has a 10-amino acid deletion (not shown). Second, the N-terminal region of RPC7α binds the coiled coils of the DNA binding clamp of RPC1 using Tyr-12 and Phe-21, blocking the Maf1 binding site, which are not conserved in RPC7β; this suggests that Pol IIIβ would be more sensitive to repression by the MAF1 tumor suppressor (51). This led to a hypothesis that RPC7α can protect against Maf1-mediated inhibition of Pol III, -a link connecting selection of cancer-associated RPC7α and its inability to interact with the MAF1 tumor suppressor (51).
The cumulative data suggest that connections among ML6, POLR3G, POLR3GL and MAF1 are intricate. The presumed ML6 binding site is near the α-amanitin binding site between the BH and TL of Pol II (97 and references therein). The BH and TL are key elements central to the transcription mechanism and adjacent to numerous point mutations in Pol III (RPC1) termination mutants (98). ML6 inhibits transcription from a nonspecific template that does not require Pol III TFs, and cytoplasmic Pol III (8,9). Although this would be completely consistent with autoinhibition mediated by RPC7α, cumulative observations are also consistent with a small molecule such as ML6 that binds the TL-BH region, to affect post-initiation steps in the transcription mechanism.
CYTOPLASMIC POL III TRANSCRIPTION OF A+T-RICH DNA FUNCTIONS IN INNATE IMMUNITY
A large number of gene products comprise a cellular innate immune system that uses a variety of molecular pattern recognition receptors (PRR) that evolved to detect signs of infectious agents referred to as pathogen associated molecular patterns (PAMPs) a subset of which are on nucleic acids (99). The RIG-I like receptors (RLRs), comprised of RIG-I, MDA5 and related LGP2 are cytoplasmic and detect PAMPs on RNA. Once activated, RLRs initiate a signaling cascade that leads to a type 1 interferon (IFN) response activating > 100 IFN-stimulated genes (ISGs) whose products offset early and late viral functions and promote cellular homeostasis (100). RIG-I and MDA5 exhibit distinct specificity characteristics. RIG-I is preferentially activated by 5′ppp-containing RNAs, most robustly a 5′pppN that is blunt-end base-paired, whereas MDA5 is preferentially activated by long double strand RNA (reviewed in 101,102). Prior to 2009, it was known that a PRR specific for A + T-rich DNA existed but was unidentified. This led two groups to purify the activity required as the sensor of cytoplasmic high A + T-rich DNA, and characterize the pathway, which unexpectedly revealed that it was dependent on the RNA sensor RIG-I (8,9). The groundbreaking work identified A + T-rich DNA sensing was principally by cytoplasmic Pol III (cy-Pol III, Figure 5). Both groups demonstrated that Pol III activity from cytosolic extract produces the immunostimulatory RNA, with the 5′pppN hallmark of a nascent Pol III transcript as the essential activating component of RIG-I (Figure 5) (8,9).
Figure 5.
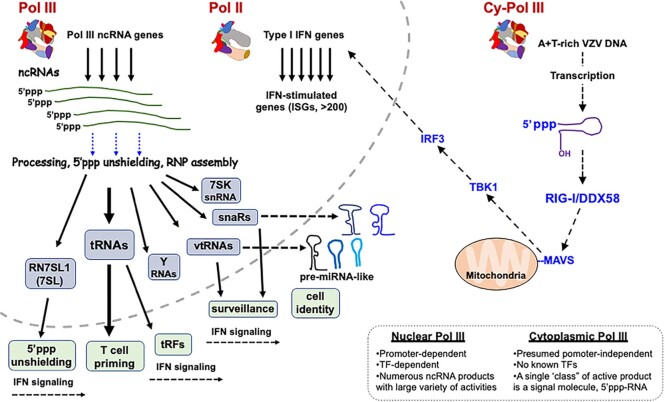
Nuclear Pol III and cy-Pol III have distinct activities in immune and other functions. The nuclear and cytoplasmic compartments are illustrated as separated by the curved dashed line. In the nucleus, Pol III transcribes ncRNA genes which go through gene-specific maturation events, including association with general and RNA-specific proteins. Removal of the 5′ppp occurs by the triphosphatase DUSP11, although other mechanisms are involved in shielding the 5′ppp end of nascent Pol III transcripts including binding by La and other proteins (see text). Some of the ncRNAs Pol III transcriptome activities in immune function are shown enclosed in green, including T cell priming by tRNAs, tRF generation from tRNA genes, RIG-I activation by 7SL/SRP RNA unshielding. Activities referred to as surveillance and cell identity involving the snaR and VtRNAs discussed in the text are also indicated. The dashed black arrows in the nuclear compartment indicate crosstalk with Pol II gene regulation such as snaR association with NF90/interleukin-3 factor and 7SK snRNA involvement in regulation of immune response genes. Furthermore, crosstalk to the Pol II transcriptome can occur via micro-RNAs generated after nuclear export of the parent vtRNAs and snaRs which can target mRNAs. In the cytoplasm, cy-Pol III transcribes A + T-rich DNA into a direct-activating RIG-I ligand, which signals through the mitochondrial-tethered MAVS, leading to Pol II-mediated transcriptional induction of IFN and ∼100 IFN-stimulated genes (ISGs). The insert lists general features of the contrasting Pol III transcription systems.
Characterization of this cy-Pol III activity revealed that short DNA of 50–100 base pairs was sufficient if it was of high A + T content, and experimental observations suggested that its synthesis was not attributed to a known promoter-dependent initiation mechanism (8,9). This specificity fit with a promoter-independent mechanism rather than isolated regions of A + T-richness, e.g. found upstream of a number of Pol III-transcribed genes. This was examined by an approach in which 72 bp DNAs of 82% and 22% A + T were compared; despite a stretch of 80% A + T including AAAATAA in the 22% A + T DNA, it was inactive in the cy-Pol III pathway (103). In addition, TBP, an integral subunit of TFIIIB and therefore required by all Pol III promoters, was not an active component of cy-Pol III (8). It has long been known that purified Pol III utilizes (dA:dT)(n) and is a preferred template relative to more complex DNA as substrate to monitor its activity independent of TFIIIB (7). Thus, cy-Pol III is certainly known to transcribe (dA:dT)(n) without promotors or TFIIIB. In any case, the mechanism remains unresolved and further studies are needed.
Consistent with any model of initiation, siRNA-mediated knockdown of the Pol III catalytic subunits, RPC1 and RPC2 decreased the IFN response to poly(dA-dT) (9). However, siRNAs against RPC3 and RPC7 mRNAs that encode initiation BITFs (Figure 3) did not impair the IFN response (9). While this was consistent with promoter-independent initiation, and the target mRNAs were shown to be decreased, the extent to which RPC3 and RPC7 proteins were depleted if any after siRNA treatment was not reported (9).
Children and adult patients with severe immunodeficiency against the A + T-rich varicella zoster virus (VZV) is attributed to debilitating pathogenic mutations in POLR3C and POLR3F that encode RPC3 and RPC6 respectively. In each case, the patients’ immune cells could not mount proper response to VZV nor A + T-rich DNA specifically (10). A mechanistic link between these point mutations in the alleles and an effect on the molecular biology of cy-Pol III is unknown. Of the nine Pol III alleles attributed with causing debilitating immunodeficiency to VZV, four are in POLR3A (none in the active center) and five in the BITFs, three in POLR3C and one each in 3F and 3E (Figure 3C) all as heterozygous mutations.
Mapping of VZV-associated mutations onto hPol III structures revealed the three resolved for RPC3 involved intra- and inter-subunit contacts with RPC6, consistent with a view that these may disrupt integrity of the heterotrimer initiation complex (36,51,83). Three of the mutations in POLR3A/RPC1 were surface exposed and the fourth was predicted to disrupt assembly of the heterotrimer complex (36,51).
The cumulative data appear most consistent with a promoter-independent mechanism of cy-Pol III transcription that nonetheless require the BITF heterotrimer and heterodimer subunits for RNA synthesis in the A + T-rich anti-VZV pathway (Figure 3). Transcription initiation by Pol III is preceded by concerted actions of specific BITF initiation subunits (55) (also see 47,48 and refs therein). The yeast homologs of RPC3 and RPC6 BITFs function in dsDNA loading into the active center followed by strand-melting as a prerequisite to de novo transcription initiation (37,38), although RPC3, aka hRPC62, has also been characterized (104,105) (below). Nonetheless, it remains possible that an unknown promoter or factor may direct cy-Pol III to A + T DNA or otherwise facilitate initiation (10).
The VZV-related heterozygous (dominant) mutations in Pol III lead to a conditional phenotype
The history is that several otherwise healthy children were reported to suffer severe complications of VZV infection which is normally a self-limited disease (chicken pox) (103). These are heterozygous single nucleotide mutations in either POLR3A/RPC1 or POLR3C/RPC3) (103). Thorough investigations including cDNA rescue experiments in patient-derived cells documented that the mutations were pathogenic for the innate immune function of cy-Pol III (103). Additional patients with severe disease response to VZV were later found to harbor other heterozygous mutations in POLR3A, 3E and 3F encoding RPC1, RPC5 and RPC6 (106,107). Their immune cells were also impaired for response to poly(dA-dT) and to VZV (reviewed in 10).
Pol III-related VZV susceptibility is a conditional phenotype, manifested only after infection. This is in contrast to Pol III-related disorders, hypomyelinating leukodystrophy (HLD), Wiedemann-Rautenstrauch syndrome (WDRTS) and others (108). Such a difference is consistent with the apparent activities involved. The overt phenotypes of HLD and WDRTS are likely evidence of disrupted tRNA and other ncRNA gene expression during development (109–111). Yet, despite dominance of the VZV-associated heterozygous alleles in immune function, they do not otherwise confer overt phenotype (10).
Dominant inheritance of heterozygous mutations is also seen for TBK1 (TANK-binding kinase-1) as noted in childhood Herpes simplex virus-1 (HSV-1) encephalitis (112). Multiple IFN-inducing signaling pathways converge on Tbk1, including cy-Pol III (Figure 5). Heterozygous dominant inheritance is also observed for numerous other immunity genes, in some cases with disease manifested conditionally (113).
After reports of cy-Pol III as immune sensor of A + T-rich DNA, it was also implicated in other viral and bacterial infections (reviewed in 114). There may be multiple sensing pathways in which cy-Pol III participates and has been overlooked. Thus far however, VZV represents the clearest case demonstrating cy-Pol III’s specific and unconventional function as a direct cytoplasmic sensor of A + T-rich DNA.
The VZV-related mutations raise interesting questions for molecular, cell and structural biologists, biochemists, geneticists, and public health scientists. In some affected adults VZV was reactivated as herpes zoster, the incidence of which steadily increases with age (115). The mechanisms that maintain high cytoplasmic levels of cy-Pol III, and how the pathogenic dominant mutations may affect the transcription initiation process are unknown. The prevalence of potential pathogenic mutations should be considered given the global immunization state and activation rate of herpes zoster (115). These are outstanding issues.
NUCLEAR POL III AND IMMUNITY; CANONICAL AND NONCANONICAL ROLES OF tRNA GENES
tRNA genes dominate the Pol III transcriptome. The tRNA gene transcripts follow a complex intranuclear maturation pathway consisting of multiple RNA processing steps and modifications prior to export to the cytoplasm (66,67,116). Mature tRNAs carry various modifications that affect their canonical function in translation (117).
Rapidly dividing T cells require high rates of translation and upregulation of tRNA gene transcription, referred to as T cell priming (Figure 5) (118). tRNA synthesis is under control of the general inhibitor MAF1, which interferes with the TFIIIB-binding site on Pol III (92). The p65 subunit of NF-kB TF associates with TFIIIB on tRNA genes accompanied by increased transcription in activated macrophages (119). Downregulation of select tRNAs, increased MAF1 promoter binding, and altered chromatin topology accompanied macrophage differentiation (120).
tRNA genes are the source of tRNA-derived fragments (tRFs) which are generated by endonucleolytic cleavage of mature or precursor-tRNAs by a number of ribonucleases. As such tRFs have adopted a range of noncanonical activities (reviewed in 121) including mRNA stabilization, modulation of translation by base-pairing to select target mRNAs, and other mechanisms (121).
Work in B, T and monocyte cells documented tRF activities in multiple immune-related pathways (Figure 5) (122 and refs therein). Viruses can hijack tRFs. Infection by Respiratory syncytial virus (RSV) enhances tRF formation, including tRF5-GluCTC which silences antiviral APOER2 (123). Another tRF signals the host immune system during Mycobacterium bovis infection. Macrophages infected with M. bovis produced and exported tRF5-HisGUG in vesicles which could then stimulate IFN in recipient cells (124).
Recent analyses suggest that some tRNA genes can modulate tRFs as their product (125). This is important on its own and also because it suggests regulatable function separate from mRNA decoding (125).
RIG-I/DDX58 on guard for 5′ppp-RNA detection, -nascent Pol III transcripts can tip the balance
Based on the number of immune pathway genes involved, the most critical RNA modification in self versus non-self-discrimination is of the 5′-terminal nucleotide of mRNA (126). It is important to note that self vs. non-self-discrimination underlies pervasive health issues that extend beyond infectious diseases, involving inflammatory cytokines to autoimmunity and cancer (127,128). A broad surveillance system targets defective and/or non-self RNAs that serves to prevent disease (129). As noted, RIG-I is the primary cytosolic sensor of 5′ppp-RNAs which include cy-Pol III transcripts that mimic ‘uncapped’ viral RNA (Figure 5) as well as authentic uncapped short viral 5′-leader (vle)RNAs and some mRNAs made by viral Pols. In some cases, Pol III 5′ppp-RNAs such as 7SL/RN7SL1/2 activate RIG-I (below).
Transcripts synthesized by RNA Pols initiate with nucleoside-triphosphate that remains as 5′pppN until action by another enzyme(s). Some viral Pols, e.g., Vaccinia, a poxvirus, SARS-CoV-2, have an associated m7Gppp capping enzyme that modifies 5′pppN as the elongating transcript emerges from the polymerase, similar to Pol II, whereas other viral Pols nor Pols I and III do not have such activity. Pol III synthesizes about 15-molar excess of pre-tRNAs relative to the pre-rRNA molecules synthesized by Pol I (68), a huge amount of 5′pppN- of potential immunoreactivity. These 5′ppp-RNAs are dealt with in a variety of ways. Dual-specificity phosphatase-11 (DUSP11) is an RNA 5′-triphosphatase (130). DUSP11 is concentrated in nuclei and dephosphorylates the large amounts of 5′ppp-RNA produced by Pol III (131).
Host cell mRNAs undergo additional m7GpppN cap modifications by 2′-O-ribose methylation of the first and second transcribed nucleotides, whereas some viral RNAs lack this modification or bear naked 5′pppN. Prominent among ISGs are the abundant IFIT1 and IFIT5 proteins that exhibit 5′-termini-specific discriminatory activity and function as anti-viral factors (100). These IFITs sequester viral RNAs as non-self, preventing their engagement with host translation initiation factors. In this way IFIT1 and IFIT5 promote the expression and translation of ISG self mRNAs (100). However, they work after IFN induction.
By contrast, RIG-I is a primary sentry specialized to recognize non-self RNAs with 5′pppN and robustly activate the IFN response. As a constitutive 5′ppp-RNA sensor, the chronic challenge to RIG-I is that it must distinguish against a multitude of self-RNA molecules very well most of the time, when there is no infection, to not activate the system. When it fails, IFN responses occur unchecked and health declines. Therefore apparently, this is why RIG-I exists in an autoinhibited state (132,133). As part of the means to manage this balance, high affinity RNA binding onto RIG-I is not sufficient for its activation, a subsequent allosteric event must occur for signaling activity (134,135). Relevant is that gene mutations to MDA5 and RIG-I cause Singleton-Merten syndrome (SMS), characterized as interferonopathies (136,137) and as monogenic links to classic autoimmune disorders (138). Biochemical investigation of the mutations in RIG-I indicate that they impair ability to discriminate against self-RNA, specifically to dissociate from self-RNA, thus to retain propensity for the signaling-active state (134,135).
La protein can shield 5′pppN-RNAs from innate immune sensors
The nuclear envelope separates 5′pppN-RNAs produced by Pol III from their otherwise potential immunoreactivity with the cytoplasmic sensor RIG-I (Figure 5). The tRNAs are the most numerous Pol III transcripts and would thus have the greatest potential to activate RIG-I. DUSP11-mediated dephosphorylation of pre-tRNA leaders after release from tRNA would be a likely way of deactivating these potential immunostimulatory RNAs (139). A conserved short basic motif in nonphosphorylated La protein is known to recognize and bind the 5′pppN of nascent Pol III transcripts (reviewed in 64). The specificity and affinity of this interaction is decreased after gamma phosphate methylation as occurs for U6 and 7SK snRNAs (140,141). Deletion of La from mouse brain three weeks after birth was soon followed by disrupted pre-tRNA processing, activation of multiple immune genes, inflammation and neurodegeneration (142,143).
Nonphosphorylated La accumulates in the cytoplasm (144). Several studies have found La bound to short 5′ppp-viral leader vleRNAs including of RSV and others (detailed in 64). Shielding of the 5′ppp-vleRNA by La appears to allow RSV to circumvent RIG-I detection, delaying an early IFN response (145).
Conversely, La was shown to serve as a positive factor in antiviral defense (146). La supported the IFN response against Sendai virus whereas its depletion led to a marked decrease in IFN and increased infection (147). Co-immunoprecipitations that suggested La can bind RIG-I together with its ligand, led to a model in which La can enhance RIG-I interactions with target RNA to support antiviral immunity (147).
AN EVOLVING INTERFACE OF A POL III ncRNA SURVEILLANCE TRANSCRIPTOME
Multiple novel and alternative activities for some Pol III ncRNAs have been noted as evidence of an evolving complexity (148). In this section we review data on Pol III transcriptome activity in innate immune pathways that constitute a type of surveillance.
Short interspersed elements (SINEs) are high copy number retroposon families (149,150). These can produce Pol III ncRNAs, including in response to stress and viral infection and may be involved in the responses (151–154). The human Alu and the murine B1 SINEs were derived from an ancestral 7SL (SRP) RNA (encoded by human RN7SL1/2 genes) whereas the B2 SINEs were derived from tRNAs. Based on Pol III and TF occupancies, several hundred of the one-million Alu SINEs appear ready for transcription as ncRNA genes (155,156) although at much lower levels relative to tRNA genes (157). However, Pol III Alu transcription increases massively after Adenovirus infection, and in response to environmental stressors (151–153). Infection by MHV68 (murine gammaherpesvirus) induces SINE transcription and stimulation of the NF-κB pathway although in this case, coopted by the virus (158).
The lengthy oligo(A) tracts at the 3′ ends of Alu and other SINE DNA suggest that their Pol III transcripts were ‘modified’ by the nonconventional poly-A polymerase of the nuclear surveillance machinery (159) prior to retroposition. As such this modification is likely molecular fossil evidence of productive interaction of these highly successful retroposons with innate RNA surveillance systems.
The tracing of Alu SINE amplifications through ancestral lineages indicates that a relative small number of active ‘source’ genes spawn new retropositions (149,150). The success of a SINE source gene requires activities beyond transcription, which likely involves competing pathways of RNA 3′ end formation/metabolism and competence for specific protein binding (160–162 and references therein) (see 163). New SINE offspring often have sequence-specific and copy-number characteristics (150). These and the genomic loci into which a SINE retroposon inserts may influence future potential. A case in point are the Alu-derived snaR genes, derived from one or more Alu SINE (164 and refs therein).
The snaRs (small NF90/ILF3-associated RNAs) are synthesized from a family of sequence-related short ncRNA genes. SnaRs are bound to nuclear factor-90 (NF90)/ILF3 (interleukin enhancer-binding factor-3). SnaR genes contain 20–30 members (155). SnaR association with NF90, also referred to as NF110 had suggested a link to innate immunity. NF110 refers to a C-terminal extended isoform (aka ILF3) which was characterized as an enhancer-binding factor involved in transcription of interleukin-2 (IL2) and IL13 (165) and coactivator of c-fos which regulates cytokines and growth factors (166).
A genomics approach that integrated Pol III occupancy and the paralogous subunits POLR3G and POLR3GL, uncovered snaR genes as highly sensitive to POLR3G in proliferating cells. Concomitant decrease in POLR3G and snaR-A gene activity, accompanied by increase in POLR3GL was demonstrated for differentiated immune cell types. Reciprocally, coordinate expression of POLR3G and snaR-A gene activity was found in human cancers (Van Bortle et al., 2021 bioRxiv preprint server). Association of snaR ncRNA with cancer had been noted (167,168, and references therein). A snaR-A RNA was recently discovered to produce noncanonical miRNA (miR-snaR) by a Drosha-independent processing pathway (13) consistent with the reported short half-life (15 min) of snaR-A (169). The miR-snaR pathway requires nuclear export by XPO5 (13). The miR-snaR was increased in cancer cells, was shown associated with Ago proteins and to target NME1 mRNA, a known cancer metastasis inhibitor (13).
Production of miR-snaR by processing of the primary snaR transcript is reminiscent of other Pol III transcripts. The microRNA-like RNAs, svRNA4 and miR-886 are derived from VtRNA1-1 and VtRNA2-1 respectively (12). miR-886 was also uncovered in the Drosha-independent screen (13). The intricacy of involvement of these Pol III transcripts in their specific pathways is notable (12). As larger transcripts snaRs minimally interact with NF90, and some enter a processing pathway and become effectors of mRNA expression. The vtRNA1-1 was identified is a prime RNA that interacts with and regulates the autophagy receptor p62/SQSTM1 (170).
Vault-associated RNAs (vtRNAs) were discovered as subunits of vault RNPs (171,172). VtRNA 1–1, 1–2, 1–3 genes contain type-2 promoters and conserved upstream elements (172). VtRNA2-1/nc886 was identified as cord blood lymphocyte-derived ncRNA-3 (173) and later as a potential pre-miRNA (157,174). Multiple lines of evidence indicate that nc886 is distinguished by several criteria including lack of vault RNP association (175–182). Remarkably for a Pol III gene, its locus undergoes allele-specific epigenetic maternal imprinting with DNA methylation transcriptional control (177,178) (179,181 and refs therein).
Data suggests that the vtRNAs independently interface with the innate immune system (Figure 5). PKR (protein kinase activated by RNA), is a cytoplasmic dsRNA PRR important for balancing self and non-self RNA detection (126), and can be negatively regulated by vtRNA2-1/nc886 (175,183–186). This suggested that nc886 ncRNA contributes to balancing immune and inflammatory responses, i.e., immunosurveillance (reviewed in 179,187). Some data suggest that nc886 can suppress the IFN response initiated by the cy-Pol III pathway (188). Other nc886 activities are consistent with anti-viral functions (182,186). For example, vtRNAs 1–1, 1–2 and 1–3 were bound to RIG-I after Kaposi's sarcoma-associated herpesvirus infection (189). In uninfected cells, the 5′pppN of the vtRNAs were dephosphorylated by the 5′ triphosphatase, DUSP11. However, the infected cells expressed decreased levels of DUSP11, retention of 5′pppN by vtRNAs and their RIG-I mediated activation of immune response (189). This is evidence that the vtRNAs together with DUSP11 are used in defense against viral infection.
As noted, vtRNAs are endonucleolytically processed into smaller RNAs that go on to regulate genetic programs by targeting mRNAs, similar to miRNAs (Figure 5) (182,190). An activity derived from vtRNA1.1 is regulated processing into svRNA4, key in the differentiation of keratinocytes into epidermal cells. The posttranscriptional cytosine methylation of vtRNA1.1 by NSUN2 determines differential binding by the SRSF2 protein, which controls the processing to svRNA4 (12). Thus cytosine methylation determines whether the cells will be maintained in an undifferentiated state or go on to differentiate (12). This intricate pathway of vtRNA1.1 metabolism is involved in determination of cell identity, i.e. a differentiated cell type that carries distinct identity markers. As noted, vtRNA2-1/nc886 produces small microRNA-like RNAs with tumor-suppressor and cancer-promoting activity (13,181). Enriched association with Ago protein and identification of mRNA targets with complementarity to the short svRNA, snc-886-3p establishes nc886 as a targeting molecule with relevance in prostate cancer outcome (191).
The snaRs and vtRNAs resemble the secondary structures of the conserved Y RNA family of Pol III transcripts (192,193). Y RNAs are stably bound by Ro-60 as Ro-Y RNPs which are involved in multiple aspects of RNA surveillance and immunosurveillance (193). Ro-60 is a conserved protein and autoantigen in patients including with neonatal lupus in which the RNP may be pathogenic (193).
Although some tumors appear to express ISGs at high levels and this is linked to poor patient outcome (194), researchers can attempt to utilize the innate immune response for good. The 7SL RNA (RN7SL1) of the signal recognition particle, SRP, was packaged and secreted in exosomes which upon uptake by breast cancer cells, stimulated ISGs (195). Upregulation of Pol III activity by c-Myc (196) increased 7SL RNA levels above SRP protein subunit binding capacity and the excess was packaged into exosomes (197). This ‘unshielded’ 5′pppG of the naked 7SL RNA activated RIG-I in the recipient cells (195). These observations were recently utilized to enhance the efficacy of chimeric antigen receptor (CAR) T cells for treatment of solid tumors (198). These CAR-T cells engineered for ectopic expression of 7SL RNA in vesicles, activated the PRR-mediated innate immune response in the microenvironment of the solid tumor they were programmed to target (198). It would appear that the ‘unshielded’ 7SL RNA served as an adjuvant.
Transcripts whose sequences map to 5S rRNA pseudogenes RNA5SP141, RNA5SP243 and RNA5SP99 were bound to RIG-I after HSV-1 infection. In uninfected cells, the RNA5SP141 transcripts were nuclear but redistributed to the cytoplasm after HSV-1 infection, where they activated RIG-I (199). However, although the pseudogene RNAs were presumed to be Pol III transcripts, evidence to address this was not presented, notable because Pol II can have pervasive effects on Pol III genes (200). tRFs, derived from the Pol III transcriptome, can also effect mRNA-mediated posttranscriptional control in higher eukaryotes (121).
In summary, the vtRNA and snaR gene families produce ncRNAs that are limited to vertebrate and primate Pol III transcriptomes, respectively. Evidence reviewed above indicate that these ncRNAs are enriched for interactions with factors involved in innate immunity and cancer. Notably, Pol III transcripts from these gene families undergo processing into mi-RNAs that are loaded into argonaut (Ago) that target mRNAs by which downstream genetic programs flow. The vtRNAs have involvement in anti-viral activity, regulation of the PKR self-RNA surveillance factor, and in controlling cell fate decision toward identity as a differentiated cell or to remain as undifferentiated. Emerging data indicate that snaR expression is strongly linked to the Pol III content and/or expression of the paralogous POLR3G/RPC7α and POLR3GL/RPC7β subunits which themselves have been independently linked to cellular differentiation versus maintenance of the undifferentiated state (see above). Recognition of self vs non-self is an identity activity. Determination of a differentiated cell type is an identity activity, as much as it is typified by distinct identity markers.
Pol III ACTIVITY AT DNA 3′-ENDS FOR REPAIR DURING HOMOLOGOUS RECOMBINATION
A template is used for homology-directed repair of damaged DNA at a double strand break (DSB) as a source of genetic information. In a simplistic overview, repair begins with binding of the MRN complex (MRE11, NBS1, and RAD50) to the DSB. CtIP then binds and activates the MRN complex to nucleolytically resect the DNA followed by additional activities that produce a long 3′-end ssDNA that ‘invades’ the homologous template to guide repair. Although formation of RNA-DNA hybrids at DSBs had implicated Pol III during homologous recombination (HR), multiple outstanding issues were recently resolved (11). The 3′ overhang DNA directs Pol III synthesis of nascent RNA to form the RNA−DNA hybrid, protecting the DNA and promoting repair. Inhibition of Pol III, knockdown of RPC1 or RPC7 prevented RNA-DNA hybrid formation and led to genetic loss at the DSB (11). The authors demonstrated that recruitment of Pol III to the DSB is facilitated by the MRN complex. Specifically, in addition to reciprocal immunoprecipitation, they used yeast two hybrid screening to demonstrate that MRE11 independently binds to RPC4 and RPC6, two of the BITFs but not other Pol III subunits (Figure 3C) nor Pol II or Pol I subunits, whereas no other MRN subunit was found capable of interacting with the Pol III subunits (11).
Several Pol III-specific subunits were examined by additional methods and support a functional Pol III holoenzyme at DSB sites. Also, Pol III was observed with the MRN complex at DSBs in euchromatin and heterochromatin, and not only in vicinity of Pol III-transcribed genes (11). Thus, all together the data indicate that Pol III activity synthesizes the RNA of the RNA−DNA hybrid that protects the DNA at DSBs.
Initiation on 3′ overhang DNA is a form of promoter-independent transcription. This and the presumed promoter-independent transcription initiation by cy-Pol III may reflect properties of the BITF subunits. It may therefore be worth further considering Pol III involvement in these processes as they may share other features. For example, it had been known that Pol III can initiate transcription from dsDNA with a 3′ overhang, from which it synthesizes RNA of RNA−DNA hybrids longer than expected (201). Specifically, by this mode of initiation, RNA-DNA hybrid formation was favored relative to TFIIIB-dependent initiation and was associated with RNA synthesis that extended beyond the Pol III termination signal, oligo(T) (201). Although Pol III paused at its terminator, the extensively base-paired hybrid apparently prevented release of the RNA and Pol III therefore resumed elongation (201). This fits with the long RNA-DNA hybrids found at DSB sites.
CONCLUDING REMARKS
The Pol III transcription system that emerged in eukaryotes has characteristics that sets it apart from Pol I and Pol II, notably its BITF subunits which enable efficient reinitiation onto stable TFIIIB initiation complexes. These likely also enable Pol III for its newly discovered functions that appear to use promoter-independent initiation, innate immune signaling in the cytoplasm and RNA−DNA formation during DNA repair.
Another unique characteristic is the evolution in vertebrates of cell type preferential use of one or another of two paralogous subunits, RPC7α or RPC7β encoded by divergent (human) genes POLR3G and POLR3GL. These subunits occupy Pol III to different extents in different cells and although attributed with activities that remain to be precisely defined, they appear to support the undifferentiated state accompanied by cell proliferation (and cancer), versus a cellular pathway toward differentiation, identity respectively.
The minimal Pol III transcriptome is dominated by tRNAs and includes a few other single copy ncRNAs. This was expanded in higher eukaryotes, evolved from retroposons as ∼25 snaR genes, and a family of 4 vtRNA genes collectively involved in modes of self vs. non-self-discrimination activities that contribute to immune and possibly tumor surveillance.
Most notably, both snaRs and vtRNAs give rise to Ago-associated miRNAs that can alter the mRNA transcriptome with profound outcome. In this way a vtRNA has been demonstrated active in a undifferentiated state versus cellular differentiation pathway. This is somewhat reminiscent of RPC7α and RPC7β subunits. Emerging data on snaRs suggest that this gene family may reveal similar activities.
Pol III of higher eukaryotes is clearly more than a housekeeping enzyme. Future experiments related to data from recent structures of human Pol III can examine the new functions (see pp 6–7). We expect that such research will advance insights into many aspects of a new era of Pol III that is still in its infancy.
ACKNOWLEDGEMENTS
We thank Patricia Chan for help with N. Gruberi.
Contributor Information
Alan C Kessler, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, 20892 USA.
Richard J Maraia, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, 20892 USA.
FUNDING
Division of Intramural Research of the National Institute of Child Health and Human Development [ZIA HD000412-34 to R.M.]. Funding for open access charge: U.S. Department of Health and Human Services, National Institutes of Health; Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Conflict of interest statement. None declared.
REFERENCES
- 1. Roeder R.G., Rutter W.J.. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969; 224:234–237. [DOI] [PubMed] [Google Scholar]
- 2. Werner F., Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011; 9:85–98. [DOI] [PubMed] [Google Scholar]
- 3. Blombach F., Matelska D., Fouqueau T., Cackett G., Werner F.. Key concepts and challenges in archaeal transcription. J. Mol. Biol. 2019; 431:4184–4201. [DOI] [PubMed] [Google Scholar]
- 4. Gurdon J., Brown D.D.. The transcription of 5S DNA injected into xenopus oocytes. Dev. Biol. 1978; 67:346–356. [DOI] [PubMed] [Google Scholar]
- 5. Sklar V.E., Roeder R.G.. Transcription of specific genes in isolated nuclei by exogenous RNA polymerases. Cell. 1977; 10:405–414. [DOI] [PubMed] [Google Scholar]
- 6. Jaehning J.A., Roeder R.G.. Transcription of specific adenovirus genes in isolated nuclei by exogenous RNA polymerases. J. Biol. Chem. 1977; 252:8753–8761. [PubMed] [Google Scholar]
- 7. Sklar V.E.F., Roeder R.G.. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase III from the mouse plasmacytoma, MOPC 315. J. Biol. Chem. 1976; 251:1064–1073. [PubMed] [Google Scholar]
- 8. Chiu Y.H., Macmillan J.B., Chen Z.J.. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009; 138:576–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K.A., Hornung V.. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009; 10:1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carter-Timofte M.E., Paludan S.R., Mogensen T.H.. RNA polymerase III as a gatekeeper to prevent severe VZV infections. Trends Mol. Med. 2018; 24:904–915. [DOI] [PubMed] [Google Scholar]
- 11. Liu S., Hua Y., Wang J., Li L., Yuan J., Zhang B., Wang Z., Ji J., Kong D. RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell. 2021; 184:1314–1329. [DOI] [PubMed] [Google Scholar]
- 12. Sajini A.A., Choudhury N.R., Wagner R.E., Bornelöv S., Selmi T., Spanos C., Dietmann S., Rappsilber J., Michlewski G., Frye M.. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat. Commun. 2019; 10:2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stribling D., Lei Y., Guardia C.M., Li L., Fields C.J., Nowialis P., Opavsky R., Renne R., Xie M.. A noncanonical microRNA derived from the snaR-A noncoding RNA targets a metastasis inhibitor. RNA. 2021; 27:694–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 15. Vannini A., Cramer P.. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell. 2012; 45:439–446. [DOI] [PubMed] [Google Scholar]
- 16. Ream T.S., Haag J.R., Wierzbicki A.T., Nicora C.D., Norbeck A.D., Zhu J.K., Hagen G., Guilfoyle T.J., Pasa-Tolić L., Pikaard C.S.. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell. 2009; 33:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haag J.R., Pikaard C.S.. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011; 12:483–492. [DOI] [PubMed] [Google Scholar]
- 18. Kopp F., Mendell J.T.. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018; 172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koonin E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010; 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fritz-Laylin L.K., Prochnik S.E., Ginger M.L., Dacks J.B., Carpenter M.L., Field M.C., Kuo A., Paredez A., Chapman J., Pham J.. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010; 140:631–642. [DOI] [PubMed] [Google Scholar]
- 21. Viktorovskaya O.V., Schneider D.A.. Functional divergence of eukaryotic RNA polymerases: unique properties of RNA polymerase I suit its cellular role. Gene. 2015; 556:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maraia R.J., Rijal K.. Structural biology: a transcriptional specialist resolved. Nature. 2015; 528:204–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carter R., Drouin G.. The evolutionary rates of eukaryotic RNA polymerases and of their transcription factors are affected by the level of concerted evolution of the genes they transcribe. Mol. Biol. Evol. 2009; 26:2515–2520. [DOI] [PubMed] [Google Scholar]
- 24. Drouin G., Carter R.. Evolution of eukaryotic RNA polymerases. eLS. 2010; 10.1002/9780470015902.a0022872. [DOI] [Google Scholar]
- 25. Crick F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958; 12:138–163. [PubMed] [Google Scholar]
- 26. Crick F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966; 19:548–555. [DOI] [PubMed] [Google Scholar]
- 27. Liu Y., Yang Q., Zhao F.. Synonymous but not silent: the codon usage code for gene expression and protein folding. Annu. Rev. Biochem. 2021; 90:375–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Begley U., Dyavaiah M., Patil A., Rooney J.P., Direnzo D., Young C.M., Conklin D.S., Zitomer R.S., Begley T.J.. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007; 28:860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chan C., Pham P., Dedon P.C., Begley T.J.. Lifestyle modifications: coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 2018; 19:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maraia R.J., Iben J.R.. Different types of secondary information in the genetic code. RNA. 2014; 20:977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2015; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leamy K.A., Yamagami R.A.-O.H.O.O., Yennawar N.H.A.-O.h.o.o.X., Bevilacqua P.C.A.-O.h.o.o.. Single-nucleotide control of tRNA folding cooperativity under near-cellular conditions. Proc. Natl Acad. Sci. U.S.A. 2019; 116:23075–23082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maraia R.J., Arimbasseri A.G.. Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon-Codon Use. Biomolecules. 2017; 7:27. [Google Scholar]
- 34. Cramer P., Armache K.J., Baumli S., Benkert S., Brueckner F., Buchen C., Damsma G.E., Dengl S., Geiger S.R., Jasiak A.J.et al.. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 2008; 37:337–352. [DOI] [PubMed] [Google Scholar]
- 35. Kuhn C.D., Geiger S.R., Baumli S., Gartmann M., Gerber J., Jennebach S., Mielke T., Tschochner H., Beckmann R., Cramer P.. Functional architecture of RNA polymerase I. Cell. 2007; 131:1260–1272. [DOI] [PubMed] [Google Scholar]
- 36. Ramsay E.P., Abascal-Palacios G., Daiss J.L., King H., Gouge J., Pilsl M., Beuron F., Morris E., Gunkel P., Engel C.et al.. Structure of human RNA polymerase III. Nat. Commun. 2020; 11:6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abascal-Palacios G., Ramsay E.P., Beuron F., Morris E., Vannini A.. Structural basis of RNA polymerase III transcription initiation. Nature. 2018; 553:301–306. [DOI] [PubMed] [Google Scholar]
- 38. Vorlander M.K., Khatter H., Wetzel R., Hagen W.J.H., Muller C.W.. Molecular mechanism of promoter opening by RNA polymerase III. Nature. 2018; 553:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schramm L., Hernandez N.. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002; 16:2593–2620. [DOI] [PubMed] [Google Scholar]
- 40. Lata E., Choquet K., Sagliocco F., Brais B., Bernard G., Teichmann M.. RNA polymerase III subunit mutations in genetic diseases. Front Mol Biosci. 2021; 8:696438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dieci G., Conti A., Pagano A., Carnevali D. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta. 2013; 1829:296–305. [DOI] [PubMed] [Google Scholar]
- 42. Lee Y., Erkine A.M., Van Ryk D.I., Nazar R.N.. In vivo analyses of the internal control region in the 5S rRNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1995; 23:634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geiduschek E.P., Tocchini-Valentini G.P.. Transcription by RNA polymerase III. Ann. Rev. Biochem. 1988; 57:873–914. [DOI] [PubMed] [Google Scholar]
- 44. Geiduschek E.P., Kassavetis G.A.. McKnight S., Yamamoto K.. Transcriptional Regulation. 1992; NY: Cold Spring Harbor Laboratory Press; 247–280. [Google Scholar]
- 45. Yeganeh M., Hernandez N.. RNA polymerase III transcription as a disease factor. Genes Dev. 2020; 34:865–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kassavetis G.A., Kumar A., Letts G.A., Geiduschek E.P.. A post-recruitment function for the RNA polymerase III transcription- initiation factor IIIB. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Han Y., Yan C., Fishbain S., Ivanov I., He Y.. Structural visualization of RNA polymerase III transcription machineries. Cell Discov. 2018; 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vorländer M.K., Jungblut A., Karius K., Baudin F., Grötsch H., Kosinski J., Müller C.W.. Structure of the TFIIIC subcomplex τA provides insights into RNA polymerase III pre-initiation complex formation. Nat. Commun. 2020; 11:4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nagarajavel V., Iben J.R., Howard B.H., Maraia R.J., Clark D.J.. Global ‘bootprinting’ reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res. 2013; 41:8135–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dieci G., Yukawa Y., Alzapiedi M., Guffanti E., Ferrari R., Sugiura M., Ottonello S.. Distinct modes of TATA box utilization by the RNA polymerase III transcription machineries from budding yeast and higher plants. Gene. 2006; 379:12–25. [DOI] [PubMed] [Google Scholar]
- 51. Girbig M., Misiaszek A.D., Vorländer M.K., Lafita A., Grötsch H., Baudin F., Bateman A., Müller C.W.. Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states. Nat. Struct. Mol. Biol. 2021; 28:210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu X.M., Zhou X., Carlson B.A., Kim L.K., Huh T.L., Lee B.J., Hatfield D.L.. The zebrafish genome contains two distinct selenocysteine tRNA[Ser]sec genes. FEBS Lett. 1999; 454:16–20. [DOI] [PubMed] [Google Scholar]
- 53. Lee B.J., Kang S.G., Hatfield D. Transcription of Xenopus selenocysteine tRNA Ser (formerly designated opal suppressor phosphoserine tRNA) gene is directed by multiple 5′- extragenic regulatory elements. J. Biol. Chem. 1989; 264:9696–9702. [PubMed] [Google Scholar]
- 54. Lee B.J., de la Pena P., Tobian J.A., Zasloff M., Hatfield D. Unique pathway of expression of an opal suppressor phosphoserine tRNA. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kassavetis G.A., Letts G.A., Geiduschek E.P.. A minimal RNA polymerase III transcription system. EMBO J. 1999; 18:5042–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zecherle G.N., Whelen S., Hall B.D.. Purines are required at the 5′ ends of newly initiated RNAs for optimal RNA polymerase III gene expression. Mol. Cell. Biol. 1996; 16:5801–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Basu R.S., Warner B.A., Molodtsov V., Pupov D., Esyunina D., Fernández-Tornero C., Kulbachinskiy A., Murakami K.S.. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 2014; 289:24549–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arimbasseri A.G., Rijal K., Maraia R.J.. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2014; 5:e27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Setzer D.R., Brown D.D.. Formation and stability of the 5 S RNA transcription complex. J. Biol. Chem. 1985; 260:2483–2492. [PubMed] [Google Scholar]
- 60. Maraia R.J., Kenan D.J., Keene J.D.. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol. 1994; 14:2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cabart P., Lee J., Willis I.M.. Facilitated recycling protects human RNA polymerase III from repression by Maf1 in vitro. J. Biol. Chem. 2008; 283:36108–36117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Landrieux E., Alic N., Ducrot C., Acker J., Riva M., Carles C.. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006; 25:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Teplova M., Yuan Y.-R., Ilin S., Malinina L., Phan A.T., Teplov A., Patel D.J.. Structural basis for recognition and sequestration of UUU-OH 3′-termini of nascent RNA pol III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006; 21:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Maraia R.J., Mattijssen S., Cruz-Gallardo I., Conte M.R.. The LARPs, La and related RNA-binding proteins: structures, functions and evolving perspectives. Wiley Interdiscip Rev. RNA. 2017; 8:e1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang Y., Intine R.V., Mozlin A., Hasson S., Maraia R.J.. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ cleavage activity increase 3′terminal oligo(U) length and La-dependent tRNA processing. Mol. Cell. Biol. 2005; 25:621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kessler A.C., Almeida G.S., Alfonzo J.D.. The role of intracellular compartmentalization on tRNA processing and modification. RNA Biol. 2017; 15:554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hopper A.K., Nostramo R.T.. tRNA processing and subcellular trafficking proteins multitask in pathways for other RNAs. Front. Genet. 2019; 10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Moir R.D., Willis I.M.. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim. Biophys. Acta. 2013; 1829:361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bogenhagen D.F., Wormington W.M., Brown D.D.. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982; 28:413–421. [DOI] [PubMed] [Google Scholar]
- 70. Lassar A.B., Martin P.L., Roeder R.G.. Transcription of class III genes: formation of preinitiation complexes. Science. 1983; 222:740–748. [DOI] [PubMed] [Google Scholar]
- 71. Kassavetis G.A., Braun B.R., Nguyen L.H., Geiduschek E.P.. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990; 60:235–245. [DOI] [PubMed] [Google Scholar]
- 72. Zawel L., Kumar K.P., Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995; 9:1479–1490. [DOI] [PubMed] [Google Scholar]
- 73. Kassavetis G.A., Prakash P., Shim E.. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J. Biol. Chem. 2010; 285:2695–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chedin S., Riva M., Schultz P., Sentenac A., Carles C.. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998; 12:3857–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iben J.R., Mazeika J.K., Hasson S., Rijal K., Arimbasseri A.G., Russo A.N., Maraia R.J.. Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III. Nucleic Acids Res. 2011; 39:6100–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu C.C., Lin Y.C., Chen H.T.. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol. Cell. Biol. 2011; 31:2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Arimbasseri A.G., Maraia R.J.. Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III. Mol. Cell. Biol. 2013; 33:1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rijal K., Maraia R.J.. RNA polymerase III mutants in TFIIFα-like C37 cause terminator readthrough with no decrease in transcription output. Nucleic Acids Res. 2013; 41:139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Arimbasseri A.G., Maraia R.J.. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol. Cell. 2015; 58:1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mishra S., Maraia R.J.. RNA polymerase III subunits C37/53 modulate rU:dA hybrid 3′ end dynamics during transcription termination. Nucleic Acids Res. 2019; 47:310–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mishra S., Hasan S.H., Sakhawala R.M., Chaudhry S., Maraia R.J.. Mechanism of RNA Polymerase III termination-associated reinitiation-recycling conferred by the essential function of the N terminal-and-Linker domain of the C11 subunit. Nat. Commun. 2021; 12:5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang Q., Li S., Wan F., Xu Y., Wu Z., Cao M., Lan P., Lei M., Wu J.. Structural insights into transcriptional regulation of human RNA polymerase III. Nat. Struct. Mol. Biol. 2021; 28:220–227. [DOI] [PubMed] [Google Scholar]
- 83. Li L., Yu Z., Zhao D., Ren Y., Hou H., Xu Y.. Structure of human RNA polymerase III elongation complex. Cell Res. 2021; 31:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cieśla M., Turowski T.W., Nowotny M., Tollervey D., Boguta M.. The expression of Rpb10, a small subunit common to RNA polymerases, is modulated by the R3H domain-containing Rbs1 protein and the Upf1 helicase. Nucleic Acids Res. 2020; 48:12252–12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Turowski T.W., Boguta M.. Specific Features of RNA Polymerases I and III: Structure and Assembly. Front. Mol. Biosci. 2021; 8:680090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Haurie V., Durrieu-Gaillard S., Dumay-Odelot H., Da Silva D., Rey C., Prochazkova M., Roeder R.G., Besser D., Teichmann M.. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4176–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. White R.J., Stott D., Rigby P.W.J.. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989; 59:1081–1092. [DOI] [PubMed] [Google Scholar]
- 88. White R.J., Trouche D., Martin K., Jackson S.P., Kouzarides T.. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996; 382:88–90. [DOI] [PubMed] [Google Scholar]
- 89. Johnson S.A., Dubeau L., Johnson D.L.. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J. Biol. Chem. 2008; 283:19184–19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Palian B.M., Rohira A.D., Johnson S.A., He L., Zheng N., Dubeau L., Stiles B.L., Johnson D.L.. Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLos Genet. 2014; 10:e1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen C.Y., Lanz R.B., Walkey C.J., Chang W.H., Lu W., Johnson D.L.. Maf1 and repression of RNA polymerase III-mediated transcription drive adipocyte differentiation. Cell Rep. 2018; 24:1852–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Willis I.M., Moir R.D.. Signaling to and from the RNA polymerase III transcription and processing machinery. Annu. Rev. Biochem. 2018; 87:75–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Petrie J.L., Swan C., Ingram R.M., Frame F.M., Collins A.T., Dumay-Odelot H., Teichmann M., Maitland N.J., White R.J.. Effects on prostate cancer cells of targeting RNA polymerase III. Nucleic Acids Res. 2019; 47:3937–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Heiss F.B., Daiß J.L., Becker P., Engel C.. Conserved strategies of RNA polymerase I hibernation and activation. Nat. Commun. 2021; 12:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Aibara S., Dienemann C., Cramer P.. Structure of an inactive RNA polymerase II dimer. Nucleic Acids Res. 2021; 49:10747–10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wu L., Pan J., Thoroddsen V., Wysong D.R., Blackman R.K., Bulawa C.E., Gould A.E., Ocain T.D., Dick L.R., Errada P.et al.. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell. 2003; 2:256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu X., Farnung L., Wigge C., Cramer P.. Cryo-EM structure of a mammalian RNA polymerase II elongation complex inhibited by α-amanitin. J. Biol. Chem. 2018; 293:7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rijal K., Maraia R.J.. Active center control of termination by RNA polymerase III and tRNA gene transcription levels in yivo. PLos Genet. 2016; 12:e1006253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tan X., Sun L., Chen J., Chen Z.J.. Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 2018; 72:447–478. [DOI] [PubMed] [Google Scholar]
- 100. Schneider W.M., Chevillotte M.D., Rice C.M.. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014; 32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hur S. Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol. 2019; 37:349–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yao H., Dittmann M., Peisley A., Hoffmann H.H., Gilmore R.H., Schmidt T., Schmidt-Burgk J., Hornung V., Rice C.M., Hur S.. ATP-dependent effector-like functions of RIG-I-like receptors. Mol. Cell. 2015; 58:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ogunjimi B., Zhang S.Y., Sorensen K.B., Skipper K.A., Carter-Timofte M., Kerner G., Luecke S., Prabakaran T., Cai Y., Meester J.et al.. Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. J. Clin. Invest. 2017; 127:3543–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ayoubi L.E., Dumay-Odelot H., Chernev A., Boissier F., Minvielle-Sébastia L., Urlaub H., Fribourg S., Teichmann M.. The hRPC62 subunit of human RNA polymerase III displays helicase activity. Nucleic Acids Res. 2019; 47:10313–10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang Z., Roeder R.G.. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997; 11:1315–1326. [DOI] [PubMed] [Google Scholar]
- 106. Carter-Timofte M.E., Hansen A.F., Mardahl M., Fribourg S., Rapaport F., Zhang S.Y., Casanova J.L., Paludan S.R., Christiansen M., Larsen C.S.et al.. Varicella-zoster virus CNS vasculitis and RNA polymerase III gene mutation in identical twins. Neurol. Neuroimmunol. Neuroinflamm. 2018; 5:e500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Carter-Timofte M.E., Hansen A.F., Christiansen M., Paludan S.R., Mogensen T.H.. Mutations in RNA Polymerase III genes and defective DNA sensing in adults with varicella-zoster virus CNS infection. Genes Immun. 2019; 214-223:214–223. [DOI] [PubMed] [Google Scholar]
- 108. Coulombe B., Derksen A., La Piana R., Brais B., Gauthier M.S., Bernard G.. POLR3-related leukodystrophy: how do mutations affecting RNA polymerase III subunits cause hypomyelination. Fac Rev. 2021; 10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yee N.S., Gong W., Huang Y., Lorent K., Dolan A.C., Maraia R.J., Pack M.. Mutation of RNA polymerase III subunit rpc2/polr3b leads to deficiency of the RNA cleavage subunit, Rpc11/Polr3k, and disrupts zebrafish digestive system development. PLoS Biol. 2007; 5:2484–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang X., Gerber A., Chen W.Y., Roeder R.G.. Functions of paralogous RNA polymerase III subunits POLR3G and POLR3GL in mouse development. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:15702–15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Merheb E., Cui M.H., DuBois J.C., Branch C.A., Gulinello M., Shafit-Zagardo B., Moir R.D., Willis I.M.. Defective myelination in an RNA polymerase III mutant leukodystrophic mouse. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2024378118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Herman M., Ciancanelli M., Ou Y.H., Lorenzo L., Klaudel-Dreszler M., Pauwels E., Sancho-Shimizu V., Pérez de Diego R., Abhyankar A., Israelsson E.et al.. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 2012; 209:1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Casanova J.L., Abel L.. The human genetic determinism of life-threatening infectious diseases: genetic heterogeneity and physiological homogeneity. Hum. Genet. 2020; 139:681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ramanathan A., Weintraub M., Orlovetskie N., Serruya R., Mani D., Marcu O., Stepensky P., Weisblum Y., Djian E., Shaag A.et al.. A mutation in POLR3E impairs antiviral immune response and RNA polymerase III. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:22113–22121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Dooling K.L., Guo A., Patel M., Lee G.M., Moore K., Belongia E.A., Harpaz R.. Recommendations of the advisory committee on immunization practices for use of herpes zoster vaccines. MMWR Morb. Mortal. Wkly. Rep. 2018; 67:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Blewett N.H., Maraia R.J.. La involvement in tRNA and other RNA processing events including differences among yeast and other eukaryotes. Biochim. Biophys. Acta. 2018; 1861:361–372. [DOI] [PubMed] [Google Scholar]
- 117. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021; 22:375–392. [DOI] [PubMed] [Google Scholar]
- 118. Reverendo M., Arguello R.J., Polte C., Valecka J., Camosseto V., Auphan-Anezin N., Ignatova Z., Gatti E., Pierre P.. Polymerase III transcription is necessary for T cell priming by dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:22721–22729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Graczyk D., White R.J., Ryan K.M.. Involvement of RNA polymerase III in immune responses. Mol. Cell. Biol. 2015; 35:1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Van Bortle K., Phanstiel D.H., Snyder M.P.. Topological organization and dynamic regulation of human tRNA genes during macrophage differentiation. Genome Biol. 2017; 18:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Su Z., Wilson B., Kumar P., Dutta A.. Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annu. Rev. Genet. 2020; 54:47–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yu X., Xie Y., Zhang S., Song X., Xiao B., Yan Z.. tRNA-derived fragments: mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics. 2021; 11:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Deng J., Ptashkin R.N., Chen Y., Cheng Z., Liu G., Phan T., Deng X., Zhou J., Lee I., Lee Y.S.et al.. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 2015; 23:1622–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Pawar K., Shigematsu M., Sharbati S., Kirino Y.. Infection-induced 5′-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol. 2020; 18:e3000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Torres A.G., Reina O., Stephan-Otto Attolini C., Ribas de Pouplana L.. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:8451–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Schlee M., Hartmann G.. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016; 16:566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Dunker W., Ye X., Zhao Y., Liu L., Richardson A., Karijolich J.. TDP-43 prevents endogenous RNAs from triggering a lethal RIG-I-dependent interferon response. Cell Rep. 2021; 35:108976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Willbanks A., Wood S., Cheng J.X.. RNA epigenetics: fine-tuning chromatin plasticity and transcriptional regulation, and the implications in human diseases. Genes (Basel). 2021; 12:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wolin S.L., Maquat L.E.. Cellular RNA surveillance in health and disease. Science. 2019; 366:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Burke J.M., Sullivan C.S.. DUSP11 - an RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells. RNA Biol. 2017; 14:1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Choi J.H., Sullivan C.S.. DUSP11 and triphosphate RNA balance during virus infection. PLoS Pathog. 2021; 17:e1009145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S.. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011; 147:423–435. [DOI] [PubMed] [Google Scholar]
- 133. Rawling D.C., Fitzgerald M.E., Pyle A.M.. Establishing the role of ATP for the function of the RIG-I innate immune sensor. Elife. 2015; 4:e09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Dickey T.H., Song B., Pyle A.M.. RNA binding activates RIG-I by releasing an autorepressed signaling domain. Sci. Adv. 2019; 5:eaax3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Devarkar S.C., Schweibenz B., Wang C., Marcotrigiano J., Patel S.S.. RIG-I uses an ATPase-powered translocation-throttling mechanism for kinetic proofreading of RNAs and oligomerization. Mol. Cell. 2018; 72:355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kato H., Oh S.W., Fujita T.. RIG-I-like receptors and type I interferonopathies. J. Interferon Cytokine Res. 2017; 37:207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ferreira C.R., Crow Y.J., Gahl W.A., Gardner P.J., Goldbach-Mansky R., Hur S., de Jesús A.A., Nehrebecky M., Park J.W., Briggs T.A.. DDX58 and classic singleton-merten syndrome. J. Clin. Immunol. 2019; 39:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Demirkaya E., Sahin S., Romano M., Zhou Q., Aksentijevich I.. New horizons in the genetic etiology of systemic lupus erythematosus and lupus-like disease: monogenic lupus and beyond. J Clin Med. 2020; 9:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Burke J.M., Bass C.R., Kincaid R.P., Sullivan C.S.. Identification of tri-phosphatase activity in the biogenesis of retroviral microRNAs and RNAP III-generated shRNAs. Nucleic Acids Res. 2014; 42:13949–13962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Fan H., Goodier J.L., Chamberlain J., Engelke D.R., Maraia R.J.. 5′ Processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell Biol. 1998; 18:3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bhattacharya R., Perumal K., Sinha K., Maraia R., Reddy R.. Methylphosphate cap structure in small RNAs reduces the affinity of RNAs to La protein. Gene Expr. 2002; 10:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gaidamakov S., Maximova O.A., Chon H., Blewett N.H., Wang H., Crawford A.K., Day A., Tulchin N., Crouch R.J., Morse H.C. 3rdet al.. Targeted deletion of the gene encoding the La protein SSB autoantigen in B cells or cerebral cortex causes extensive tissue loss. Mol. Cell. Biol. 2014; 34:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Blewett N.H., Iben J.R., Gaidamakov S., Maraia R.J.. La deletion from mouse brain alters pre-tRNA metabolism and accumulation of pre-5.8S rRNA, with neuron death and reactive astrocytosis. Mol. Cell. Biol. 2017; 37:e00588-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Intine R.V., Tenenbaum S.A., Sakulich A.S., Keene J.D., Maraia R.J.. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol. Cell. 2003; 12:1301–1307. [DOI] [PubMed] [Google Scholar]
- 145. Bitko V., Musiyenko A., Bayfield M.A., Maraia R. J., Barik S.. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J. Virol. 2008; 82:7977–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Liu Y., Tan H., Tian H., Liang C., Chen S., Liu Q.. Autoantigen La promotes efficient RNAi, antiviral response, and transposon silencing by facilitating multiple-turnover RISC catalysis. Mol. Cell. 2011; 44:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Mahony R., Broadbent L., Maier-Moore J.S., Power U.F., Jefferies C.A.. The RNA binding protein La/SS-B promotes RIG-I-mediated type I and type III IFN responses following Sendai viral infection. Sci. Rep. 2017; 7:14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Park J.L., Lee Y.S., Kunkeaw N., Kim S.Y., Kim I.H., Lee Y.S.. Epigenetic regulation of noncoding RNA transcription by mammalian RNA polymerase III. Epigenomics. 2017; 9:171–187. [DOI] [PubMed] [Google Scholar]
- 149. Schmid C., Maraia R.. Transcriptional regulation and transpositional selection of active SINE sequences. Curr. Opin. Genet. Dev. 1992; 2:874–882. [DOI] [PubMed] [Google Scholar]
- 150. Konkel M.K., Walker J.A., Hotard A.B., Ranck M.C., Fontenot C.C., Storer J., Stewart C., Marth G.T., Batzer M.A.. Sequence analysis and characterization of active human Alu subfamilies based on the 1000 Genomes Pilot Project. Genome Biol Evol. 2015; 7:2608–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Panning B., Smiley J.R.. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol. Cell. Biol. 1993; 13:3231–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Liu W.-M., Chu W.-M., Choudary P.V., Schmid C.W.. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995; 23:1758–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Allen T.A., Von Kaenel S., Goodrich J.A., Kugel J.F.. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004; 11:816–821. [DOI] [PubMed] [Google Scholar]
- 154. Tucker J.M., Glaunsinger B.A.. Host noncoding retrotransposons induced by DNA viruses: a SINE of infection. J. Virol. 2017; 91:e00982-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Oler A.J., Alla R.K., Roberts D.N., Wong A., Hollenhorst P.C., Chandler K.J., Cassiday P.A., Nelson C.A., Hagedorn C.H., Graves B.J.et al.. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010; 17:620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Conti A., Carnevali D., Bollati V., Fustinoni S., Pellegrini M., Dieci G.. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res. 2015; 43:817–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Canella D., Praz V., Reina J.H., Cousin P., Hernandez N.. Defining the RNA polymerase III transcriptome: genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010; 20:710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Karijolich J., Abernathy E., Glaunsinger B.A.. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-kappaB pathway. PLoS Pathog. 2015; 11:e1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Roy-Engel A.M. A tale of an A-tail: the lifeline of a SINE. Mob Genet Elements. 2012; 2:282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Maraia R. The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucl. Acids Res. 1991; 19:5695–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Maraia R.J., Chang D.Y., Wolffe A.P., Vorce R.L., Hsu K.. The RNA polymerase III terminator used by a B1-Alu element can modulate 3′ processing of the intermediate RNA product. Mol. Cell. Biol. 1992; 12:1500–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sarrowa J., Chang D.-Y., Maraia R.J.. The decline in human Alu transposition was accompanied by an asymmetric decrease in SRP9/14 binding to dimeric Alu RNA and increased expression of small cytoplasmic Alu RNA. Mol. Cell. Biol. 1997; 17:1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Roy-Engel A.M., Salem A.H., Oyeniran O.O., Deininger L., Hedges D.J., Kilroy G.E., Batzer M.A., Deininger P.L.. Active Alu element “A-tails”: size does matter. Genome Res. 2002; 12:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Parrott A.M., Tsai M., Batchu P., Ryan K., Ozer H.L., Tian B., Mathews M.B.. The evolution and expression of the snaR family of small non-coding RNAs. Nucleic Acids Res. 2011; 39:1485–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Kiesler P., Haynes P.A., Shi L., Kao P.N., Wysocki V.H., Vercelli D. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J. Biol. Chem. 2010; 285:8256–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995; 14:4905–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Lee J., Jung J.H., Chae Y.S., Park H.Y., Kim W.W., Lee S.J., Jeong J.H., Kang S.H.. Long noncoding RNA snaR regulates proliferation, migration and invasion of triple-negative breast cancer cells. Anticancer Res. 2016; 36:6289–6295. [DOI] [PubMed] [Google Scholar]
- 168. Lee J., Park H.Y., Kim W.W., Lee S.J., Jeong J.H., Kang S.H., Jung J.H., Chae Y.S.. Biological function of long noncoding RNA snaR in HER2-positive breast cancer cells. Tumour Biol. 2017; 39:1010428317707374. [DOI] [PubMed] [Google Scholar]
- 169. Parrott A.M., Mathews M.B.. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007; 35:6249–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Horos R., Büscher M., Kleinendorst R., Alleaume A.M., Tarafder A.K., Schwarzl T., Dziuba D., Tischer C., Zielonka E.M., Adak A.et al.. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019; 176:1054–1067. [DOI] [PubMed] [Google Scholar]
- 171. Kickhoefer V.A., Searles R.P., Kedersha N.L., Garber M.E., Johnson D.L., Rome L.H.. Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J. Biol. Chem. 1993; 268:7868–7873. [PubMed] [Google Scholar]
- 172. Stadler P.F., Chen J.J., Hackermüller J., Hoffmann S., Horn F., Khaitovich P., Kretzschmar A.K., Mosig A., Prohaska S.J., Qi X.et al.. Evolution of vault RNAs. Mol. Biol. Evol. 2009; 26:1975–1991. [DOI] [PubMed] [Google Scholar]
- 173. Mrázek J., Kreutmayer S.B., Grässer F.A., Polacek N., Hüttenhofer A.. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007; 35:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Seal R.L., Chen L.L., Griffiths-Jones S., Lowe T.M., Mathews M.B., O’Reilly D., Pierce A.J., Stadler P.F., Ulitsky I., Wolin S.L.et al.. A guide to naming human non-coding RNA genes. EMBO J. 2020; 39:e103777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Nandy C., Mrázek J., Stoiber H., Grässer F.A., Hüttenhofer A., Polacek N.. Epstein-barr virus-induced expression of a novel human vault RNA. J. Mol. Biol. 2009; 388:776–784. [DOI] [PubMed] [Google Scholar]
- 176. Lee K., Kunkeaw N., Jeon S.H., Lee I., Johnson B.H., Kang G.Y., Bang J.Y., Park H.S., Leelayuwat C., Lee Y.S.. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. 2011; 17:1076–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Treppendahl M.B., Qiu X., Søgaard A., Yang X., Nandrup-Bus C., Hother C., Andersen M.K., Kjeldsen L., Möllgård L., Hellström-Lindberg E.et al.. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood. 2012; 119:206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Romanelli V., Nakabayashi K., Vizoso M., Moran S., Iglesias-Platas I., Sugahara N., Simon C., Hata K., Esteller M., Court F.et al.. Variable maternal methylation overlapping the nc886/vtRNA2-1 locus is locked between hypermethylated repeats and is frequently altered in cancer. Epigenetics. 2014; 9:783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Lee Y.S. A novel type of non-coding RNA, nc886, implicated in tumor sensing and suppression. Genomics Inform. 2015; 13:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kunkeaw N., Lee Y.S., Im W.R., Jang J.J., Song M.J., Yang B., Park J.L., Kim S.Y., Ku Y., Kim Y.et al.. Mechanism mediated by a noncoding RNA, nc886, in the cytotoxicity of a DNA-reactive compound. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:8289–8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Fort R.S., Garat B., Sotelo-Silveira J.R., Duhagon M.A.. vtRNA2-1/nc886 produces a small RNA that contributes to its tumor suppression action through the microRNA pathway in prostate cancer. Noncoding RNA. 2020; 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Hahne J.C., Lampis A., Valeri N.. Vault RNAs: hidden gems in RNA and protein regulation. Cell. Mol. Life Sci. 2021; 78:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Lee K.S., Shin S., Cho E., Im W.K., Jeon S.H., Kim Y., Park D., Frechet M., Chajra H., Jung E.. nc886, a non-coding RNA, inhibits UVB-induced MMP-9 and COX-2 expression via the PKR pathway in human keratinocytes. Biochem. Biophys. Res. Commun. 2019; 512:647–652. [DOI] [PubMed] [Google Scholar]
- 184. Golec E., Lind L., Qayyum M., Blom A.M., King B.C.. The noncoding RNA nc886 regulates PKR signaling and cytokine production in human cells. J. Immunol. 2019; 202:131–141. [DOI] [PubMed] [Google Scholar]
- 185. Ma H., Wang M., Zhou Y., Yang J.J., Wang L.Y., Yang R.H., Wen M.J., Kong L.. Noncoding RNA 886 alleviates tumor cellular immunological rejection in host C57BL/C mice. Cancer Med. 2020; 9:5258–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Calderon B.M., Conn G.L.. A human cellular noncoding RNA activates the antiviral protein 2′-5′-oligoadenylate synthetase 1. J. Biol. Chem. 2018; 293:16115–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Lee Y.S., Kunkeaw N., Lee Y.S.. Protein kinase R and its cellular regulators in cancer: An active player or a surveillant. Wiley Interdiscip. Rev. RNA. 2020; 11:e1558. [DOI] [PubMed] [Google Scholar]
- 188. Lee Y.S., Bao X., Lee H.H., Jang J.J., Saruuldalai E., Park G., Im W.R., Park J.L., Kim S.Y., Shin S.et al.. Nc886, a novel suppressor of the type I interferon response upon pathogen intrusion. Int. J. Mol. Sci. 2021; 22:2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Zhao Y., Ye X., Dunker W., Song Y., Karijolich J.. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat. Commun. 2018; 9:4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Persson H., Kvist A., Vallon-Christersson J., Medstrand P., Borg A., Rovira C.. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009; 11:1268–1271. [DOI] [PubMed] [Google Scholar]
- 191. Fort R.S., Matho C., Geraldo M.V., Ottati M.C., Yamashita A.S., Saito K.C., Leite K.R.M., Mendez M., Maedo N., Mendez L.et al.. Nc886 is epigenetically repressed in prostate cancer and acts as a tumor suppressor through the inhibition of cell growth. BMC Cancer. 2018; 18:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Hendrick J.P., Wolin S.L., Rinke J., Lerner M.R., Steitz J.A.. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol. Cell. Biol. 1981; 1:1138–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Boccitto M., Wolin S.L.. Ro60 and Y RNAs: structure, functions, and roles in autoimmunity. Crit. Rev. Biochem. Mol. Biol. 2019; 54:133–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Weichselbaum R.R., Ishwaran H., Yoon T., Nuyten D.S., Baker S.W., Khodarev N., Su A.W., Shaikh A.Y., Roach P., Kreike B.et al.. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18490–18495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Nabet B.Y., Qiu Y., Shabason J.E., Wu T.J., Yoon T., Kim B.C., Benci J.L., DeMichele A.M., Tchou J., Marcotrigiano J.et al.. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017; 170:352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Campbell K.J., White R.J.. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb. Perspect. Med. 2014; 4:a018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Gomez-Roman N., Grandori C., Eisenman R.N., White R.J.. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003; 421:290–294. [DOI] [PubMed] [Google Scholar]
- 198. Johnson L.R., Lee D.Y., Eacret J.S., Ye D., June C.H., Minn A.J.. The immunostimulatory RNA RN7SL1 enables CAR-T cells to enhance autonomous and endogenous immune function. Cell. 2021; 184:4981–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Chiang J.J., Sparrer K.M.J., van Gent M., Lassig C., Huang T., Osterrieder N., Hopfner K.P., Gack M.U.. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat. Immunol. 2018; 19:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Gerber A., Ito K., Chu C.S., Roeder R.G.. Gene-specific control of tRNA expression by RNA polymerase II. Mol. Cell. 2020; 78:765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Campbell F.E. Jr, Setzer D.R. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 1992; 12:2260–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]