Abstract
N6-methyladenosine (m6A) modification is the most extensively studied epigenetic modification due to its crucial role in regulating an array of biological processes. Herein, Bsu06560, formerly annotated as an adenine deaminase derived from Bacillus subtilis 168, was recognized as the first enzyme capable of metabolizing the epigenetic nucleoside N6-methyladenosine. A model of Bsu06560 was constructed, and several critical residues were putatively identified via mutational screening. Two mutants, F91L and Q150W, provided a superiorly enhanced conversion ratio of adenosine and N6-methyladenosine. The CRISPR-Cas9 system generated Bsu06560-knockout, F91L, and Q150W mutations from the B. subtilis 168 genome. Transcriptional profiling revealed a higher global gene expression level in BS-F91L and BS-Q150W strains with enhanced N6-methyladenosine deaminase activity. The differentially expressed genes were categorized using GO, COG, KEGG and verified through RT-qPCR. This study assessed the crucial roles of Bsu06560 in regulating adenosine and N6-methyladenosine metabolism, which influence a myriad of biological processes. This is the first systematic research to identify and functionally annotate an enzyme capable of metabolizing N6-methyladenosine and highlight its significant roles in regulation of bacterial metabolism. Besides, this study provides a novel method for controlling gene expression through the mutations of critical residues.
INTRODUCTION
Modifications of cellular nucleic acids are the predominant mechanisms of epigenetic gene regulation and play a crucial role in regulating various vital biological processes. Among myriads of nucleic acid modifications, methylation at the sixth position of the purine ring in adenine, designated as N6-methyladenosine (m6A) in RNA and N6-methyl-2′-deoxyadenosine (6mA) in DNA, has been identified as the most prominent modification in recent years with the development and utilization of high-sensitivity detection technologies. The m6A is the most abundant and prevalent RNA modification in multiple RNA types, including mRNAs, rRNAs and long non-coding RNAs in eukaryotes, prokaryotes, and certain viruses (1–3). In mammalian cells, m6A is dynamically mediated by ‘Writers’ (methyltransferase, such as METTL3-METTL14 complexes), removed by ‘Erasers’ (demethylases, including FTO and ALKBH5), and recognized by ‘Readers’ (m6A binding proteins, such as YTHDC1/2) (4–8). Although the abundance of m6A mRNA modification is lower in bacteria than in mammals (m6A/A ratio: 0.02–0.28% versus 0.1–1.79%), the m6A modification on mRNA has a critical role in the regulation of bacterial respiration, amino acid metabolism, and stress response (3). Nevertheless, the mechanism of how m6A regulates cellular functions in bacteria is unclear (3,9–11). The characterization of physiological functions of m6A and 6mA has gained traction in recent years since m6A regulates RNA maturity and is strongly associated with embryonic stem cell differentiation and cancer initiation and progression. Besides, m6A regulators may represent potential targets in cancer therapy (1,2,4,6,8,12–17). However, the lack of methods for specific single-site detection or monitoring of m6A and 6mA modification in vivo has limited analysis of the biological functions and origin of m6A and 6mA methylation in bacteria (18–20).
Isotopic labeling coupled with ultrasensitive mass spectrometry has recently indicated that in multiple mammalian cells, the bulk of genomic 6mA predominantly originates from free N6-methyladenosine through the purine salvage pathway. In this pathway, N6-methyladenosine is mis-incorporated by DNA polymerases, indicating that purine metabolism can modulate the levels of genomic 6mA (21). The salvage pathways of purine nucleotides are essential and highly conserved in animals and microorganisms, and are significantly more energy-efficient than the de novo synthetic pathways (22,23). As a result, purine metabolism has attracted much interest due to recent advances in understanding tumorigenesis and targeted molecular therapies of cancer (23–25). The adenyl deaminase family proteins play a central role in the deamination of adenosine monophosphate (AMP), adenosine, and adenine in the salvage of purine bases to inosine monophosphate (IMP), inosine, and hypoxanthine, respectively (26–28). The adenosine deaminases (ADA) are essential in eukaryotes and catalyze the conversion of adenosine and 2′-deoxyadenosine to inosine and 2′-deocyinosine, respectively (29). Moreover, the adenine deaminase (ADE) and AMP deaminase (AMPD) catalyze the conversion of adenine and AMP to hypoxanthine and IMP, respectively (28,30). The IMP bridges the interconversion of purine nucleotides and maintains the cellular pools of adenylate and guanylate precursors for RNA and DNA synthesis (22,26,27). Adenosine deaminase-like (ADAL) proteins are novel paralogues of the adenyl deaminase family previously isolated and purified from rat liver in 2006 (28,31). Notably, the ADALs exhibit deamination activity towards N6-substituted purine nucleotide analogs, especially N6-methyladenosine monophosphate (N6-mAMP) and N6-methyl-2′-deoxyadenosine monophosphate (N6-mdAMP) (28,31,32). This protein has also been purified and characterized in humans and identified as ADAL isoform 1 (ADAL1, or human N6m-AMP deaminase or hsMAPDA). Moreover, ADAL1 can specifically catalyze the removal of alkyl groups from various N6- and O6-substituted purine/2-aminopurine nucleotide monophosphates. However, its activity is sensitive to modifications in the phosphate moiety (33). The N6-mAMP deaminase (MAPDA), a new ADAL from Arabidopsis thaliana, has been recently identified to function as a specific deaminase on N6-mAMP for non-canonical nucleotides metabolism (34). Moreover, the deamination of N6-mAMP via MAPDA catalysis can protect RNA from m6A mis-incorporation (34). Enzymes Bh0637 and Bsu06560 facilitate the recycling of N6-methyladenine to form hypoxanthine, which is used in the purine salvage pathway and is currently annotated as N6-methyladenine deaminases (6-MAD) (35). Besides N6-methyladenine, 6-MAD also accepts 6-chloropurine, 6-methoxypurine, and 6-methylmercaptopurine as substrates (35).
Adenosine is a key metabolic and immune checkpoint regulator in the purine metabolism, both in extracellular and intracellular systems (36). The extracellular nucleosides and adenosine have roles in several systems, including neurotransmission in the central nervous system, blood flow regulation, platelet activation, immunomodulation, vascular remodeling, and cell growth and proliferation (36–38). Moreover, extracellular adenosine mediates its physiologic and pharmacologic effects via interaction with a family of four transmembrane G-protein-coupled cell surface adenosine receptors. However, adenosine can either be directly inactivated on the cell surface via inosine to hypoxanthine through sequential activities of adenosine deaminase (ADA) and purine nucleoside phosphorylase (PNP) or via cellular uptake through specific equilibrative and concentrative nucleoside transporters (36,39–41). The increased adenosine accumulation due to insufficient enzymatic activity of adenosine deaminase is strongly immunosuppressive. Moreover, extracellular adenosine can interfere with the T cell receptor (TCR) signaling pathway when bonded to the adenosine receptor 2a (A2aR) expressed by effector T cell by elevating intracellular cyclic adenosine monophosphate (cAMP) and activating protein kinase A (PKA), leading to the activation of C-terminal SRC (CSK), thus reducing the levels of phosphorylated ZAP-70 and the signaling downstream of TCR activation (42–45). The ADA1, an isoform of human adenosine deaminase, is an essential regulator that maintains the concentration of adenosine and 2′-deoxyadenosine both in intracellular and extracellular processes. The congenital defect of ADA1 leads to severe combined immunodeficiency (SCID) (28,45,46). The ADA1 exerts its non-enzymatic functions mainly by binding to membrane-anchoring proteins, such as A2aR, thus modulating the immune system, including mediating co-stimulatory signals and promoting T-cell proliferation and differentiation (45–48). The extracellular adenosine metabolism has been widely explored and established recently as a tractable therapeutic target for conceptual and clinical cancer treatment (36). However, consideration few studies have assessed the new pathways linked to intracellular adenosine metabolism and compartmentalization of the adenosine system, thus limiting the development of more effective adenosine-based therapeutics (36). Besides, the role of epigenetic nucleosides, N6-methyladenosine, in purine metabolism is often overlooked. Therefore, future studies should elucidate the role of purinergic enzymes, especially epigenetic nucleosides-related enzymes, in the coordinated adenosine metabolism and signaling control.
There is insufficient evidence about the biological association between the N6-methyladenosine metabolic pathway and m6A modification. Although certain enzymes are involved in the deamination of N6−mAMP or N6-methyladenine, to the best of our knowledge, no specific N6-methyladenosine deaminase involved in the deamination of epigenetic nucleoside N6-methyladenosine or N6-methyl-2′-deoxyadenosine has been characterized (Supplementary Figure S1). Although few adenosine deaminase (ADAs) are involved in the removal of various substituents at the sixth position of a purine nucleoside, the underlying mechanism and biological effects of these reactions remain elusive (49,50). This study revealed that Bsu06560 can metabolize the epigenetic nucleoside N6-methyladenosine. To date, there have been no reports of Bsu06560 crystal structure. Herein, a structural model of Bsu06560 was developed to investigate the underlying regulatory mechanism of N6-methyladenosine via Bsu06560. This study revealed that Phe91 and Gln150 play a crucial role in N6-methyl-purine nucleosides targeted via Bsu06560. Compared with the wild-type Bsu06560, the F91L mutant facilitated ∼8-fold enhancement in the conversion ratio of N6-methyladenosine over N6-methyladenine while Q150W mutant enabled ∼7-fold increase in the conversion ratio of N6-methyladenine over adenine. The CRISPR/Cas9 mediated gene editing was used to introduce F91L and Q150W mutation and knock out Bsu06560 in B. subtilis 168 chromosome. Furthermore, comparing the transcriptome profiles between the variants and wild-type strains revealed that the mutant strains had a higher gene expression level with enhanced N6-methyladenosine deaminase activity. Moreover, functional and pathway enrichment analyses indicated that N6-methyladenosine metabolism is associated with numerous biological processes, including transcription, translation, metabolites regulations, and cellular signal transduction. Collectively, for the first time, this study highlighted the crucial roles of N6-methyladenosine nucleoside metabolism in the regulation of gene expression induced by editing only a few key residues of nucleoside deaminase. Therefore, these results suggest that nature may adopt multifunctional enzymes from the general purine metabolic pathway for epigenetic N6-methyl purine metabolism by evolving and modulating only a few key residues directly involved in recognition instead of using specific enzymes. This new strategy of controlling adenosine and N6-methyladenosine metabolism and intracellular gene expression regulation may provide insights into the system biology to address the challenging global gene expression regulation. Therefore, this study can provide a prudent driver of the evolution of multifunctional enzymes and assist in the rational drug design targeting the epigenetic N6-methyladenosine metabolism.
MATERIALS AND METHODS
Materials and general methods
N6-methyladenine, N6-methyladenosine, and N6-methyl-2′-deoxyadenosine were obtained from Ark Pharm (USA). Hypoxanthine, inosine, and other organic compounds were acquired from Alfa-Aesar (China). Heavy-oxygen water (H2O18) was purchased from Aladdin (China). All inorganic salts were obtained from Sinopharm Chemical Reagent (China). All antibiotics were sourced from Sangon Biotech (China). The DNA primers were synthesized from Gene Create in ultra-page grade (Wuhan, China). Ni-NTA resin was purchased from GenScript (China). Sorbitol and mannitol were obtained from VWR International, LLC. (USA). DNA and protein concentrations were quantified by NanoDrop 2000 ONEc (Thermofisher Scientific, USA). The ICP-AES analysis was performed by IRIS Intrepid II XSP (ThermoFisher Scientific, USA) through standard procedure.
Cloning, iron-free protein overexpression and purification of Bsu06560
The plasmid containing the DNA fragments encoding the full-length Bsu06560 (NCBI Reference Sequence: NP_388538.1) was codon-optimized and ordered from GENEWIZ (China). The target gene was cloned into a modified pMCSG19 vector, encoding a maltose-binding protein (MBP) tag at the N-terminus with a 6 × His tag and a TVMV protease site (51). The recombinant plasmid with the gene for Bsu06560 was transformed into Escherichia coli BL21 (DE3)-pRK1037 competent cells (52). A single colony was grown in LB medium supplemented with 50 μg/ml kanamycin and 50 μg/ml ampicillin at 37°C and induced with N-terminal His tag at 25°C in the presence of 1 mM isopropyl-β-thiogalactoside (IPTG) when the OD600 reached ∼0.6. The iron-specific chelator 2,2′-dipyridyl was added (200 μM) to prevent ferrous oxidative damage of the active site by sequestering iron before induction (53,54). The MnCl2 with the final concentration of 1 mM was added before induction. The overnight cell cultures were obtained via centrifugation at 3500 × g and stored at −80°C to obtain the cell extract. Purification was performed at 4ºC. Briefly, cells were dissolved in the lysis buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl, then lysed via sonication. The supernatant was collected after centrifugation at 12 000 × g, 4°C for 40 min, then applied to Ni-NTA resin. The loaded resin was washed with 15 volumes of wash buffer containing 25 mM imidazole, then eluted by a buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl and 250 mM imidazole. The recombinant protein was dialyzed with lysis buffer at 4°C overnight. The buffer was changed thrice. The purified protein was concentrated to 10 mg/ml and flash-frozen using liquid nitrogen, then stored at −80°C. Protein expression was confirmed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (over 95%). The protein concentration was optimized by measuring the A280 using an extinction coefficient of 90 105 M−1 cm−1 obtained from the ProtParam tool (55).
Activity assays and time-course analyses
Activity assays for Bsu06560 were conducted in a 0.25 ml reaction mixture containing 20 mM HEPES buffer (pH 7.5), 25 μM substrates and 20 μM purified bsu06560 at 30°C. The reaction was stopped by heating at 95°C for 10 min after 120 min of incubation. The time course analyses were conducted in a 0.2 ml reaction mixture containing 20 mM HEPES buffer (pH 7.5), 10 μM substrates and 20 μM purified bsu06560 at 30°C. The reaction was stopped by adding 50 μl methyl alcohol and heating at 95°C at various incubation time-points (0.5, 5, 15, 30, 60, 150, 300 and 450 min). The reaction mixture above was then centrifuged at 30 000g for 20 min. The supernatant (50 μl) was subjected to LC-HRMS analysis. For the initial activity screening of Bsu06560 mutants, the assays were similar to activity assays described above except that 25 μM substrates were used and incubation took 120 min. Similarly, for competition experiments of different substrates, assays were the same as activity assays above except that 25 μM of each substrate was used and incubation took 120 min. LC-HRMS was used to profile the substrates and confirming the products. The High-performance liquid chromatography (HPLC) with detection by High-Resolution mass spectrometry (HRMS) was conducted using a Thermo Fisher Scientific UltiMate-3000 chromatography system coupled with the Orbitrap Elite Hybrid Ion Trap-Orbitrap Mass Spectrometer. The associated Thermo Fisher Xcalibur software package was used for data collection and analysis. Assay mixtures were quenched and flowed through an Agilent ZORBAX Extended-C18 column (4.6 × 50 mm, 1.8 μm particle size) at 0.2 ml/min. The binary phases had solvent A (0.1% methane acid solution) and solvent B (methanol). The chromatographic gradient separation phase had 2–30% of solvent B (0–8 min), 100% solvent B (10–12 min), and 2% solvent B (12–15 min). An electrospray ionization (ESI-MS) (in the positive mode) was used to detect the substrates. Substrate concentrations were determined by comparing the integral areas of specific compounds with an external standard curve.
Modeling and molecular docking
The Iterative Threading ASSEmbly Refinement (I-TASSER) server with default settings was used for Bsu06560 modeling (56–59). As in I-TASSER result, the structures of atuADE (adenine deaminase from Agrobacterium tumefaciens, PDB ID 3NQB, resolution, 2.21 Å) from the RCSB protein data bank (www.rcsb.org) were used as the main template for the modelling. The QMEANDisCo (QMEAN Version: 4.2.0) (60) and ProQ3 servers (61,62) were used to predict the quality of the model. The QMEANDisCo Global score was 0.54 ± 0.05. The ProQ3 values were 0.616, 0.634, 0.531 and 0.560 for ProQ2D, ProQRosCenD, ProQRosFAD and ProQ3D, respectively. The protein preparation wizard of the Maestro suite (of Schrodinger Inc.) was used to prepare the protein structures for molecular docking analysis. All the water molecules were removed, while hydrogen atoms were added, then the enzyme structures were subjected to restrained minimization using OPLS_2005 force field (63). The above-prepared structures of Bsu06560 receptor grids were generated in the proposed active sites using the Receptor Grid Generation module (Glide) at default settings. An inner-box (10 × 10 × 10 Å3) and an outer-box (20 × 20 × 20 Å3) were used to define the receptor grids, taking the centroid of the metals as the reference point. The OPLS_2005 force field was used to assess the forces and energies. All substrates were constructed in Chem3D (CambridgeSoft) and prepared with LigPrep in Schrodinger suite using OPLS_2005 force field at default settings. The initial docking analysis was performed via standard precision mode of Ligand Docking (Glide) at default settings. The docked poses of each substrates were visually inspected. The docked poses of best Dock Score with proper orientation to catalytic metals were used. The hydroxide tetrahedral intermediate of adenosine, adenine, N6-methyladenosine and N6-methyladenine were used to obtain a more precise docking (extra precision mode) as previously reported (35). The best-docked pose of N6-methyladenosine intermediate was similar to that of adenosine. However, the Dock Score of N6-methyladenosine was significantly higher than that of N6-methyladenine (DS = –10.655 versus –6.529). The N6-methyladenine had a similar docked model as previously reported for bh0637. Hydrogen bonds form between protonated N-1 and N-6 with the side chain of Glu232 and Asp283 (Supplementary Supplementary Figure S7A). The docked model of adenosine had an additional cation-π interaction between pentacircle of purine and the side chain of Arg252 (Supplementary Figure S7C) (64).
Isotope doping assay
Isotope doping assays of Bsu06560 were performed in a 0.1 ml reaction mixture containing 20 mM HEPES buffer (pH 7.5), 25 μM N6-methyladenosine and 20 μM purified bsu06560 with various ratios of heavy oxygen water (0–80%) at 30°C. The reaction was stopped by heating at 95°C for 10 min after 2 h of incubation. The reaction mixture above was then centrifuged at 30 000 × g for 20 min at 4°C. The supernatant (50 μl) was subjected to LC-HRMS analysis as described above.
Bsu06560 mutation
All single-site mutations were constructed using the standard PCR protocol via the Phusion polymerase (New England Biolabs). Primer pairs containing codons for target residues were designed for the given site of mutagenesis. The linear plasmid was digested with DpnI, purified using DNA clean-up Kit (Axygen, USA), then transformed into Escherichia coli DH5α competent cells. Every single colony of mutants was confirmed via sequencing, then expressed and purified using the iron-free expression protocol described above.
QM/MM calculations
The QM/MM calculations were performed using default parameters using the QSite program in the Schrodinger suite (65–68). The calculations used a QM region with N6-methyladenosine. The QM region is neutral and contains 38 lacvp* atoms. Density functional theory (DFT) calculations were performed at the B3LYP/lacvp* level in the QM region. The OPLS-2005 protein molecular mechanics force field was used to represent the MM region. Relative energies were obtained by performing single-point calculations of each mutant.
CRISPR/Cas9-mediated gene editing
CRISPR/Cas9-mediated mutagenesis of B. subtilis 168 was conducted using a dual plasmid system as previously described with some modifications (69,70). For gene-specific Cas9-mediated cleavage, two sgRNAs were designed to target the mutation site of Bsu06560 (yerA) gene in B. subtilis 168 genome. For Cas9-mediated gene knockout, two sgRNAs were designed to target either end of the first 285 bp of Bsu06560 encoding gene. The CRISPOR server was used to generate sgRNA sequences and predict guide activity and potential off-targets (71). Twenty bp synthetic complementary sgRNA primers (sgRNA84RV-S and sgRNA84RV-AS, sgRNA155RV-S and sgRNA155RV-AS) were annealed, and the double-stranded sgRNA module was cloned into BsmBI-digested pHYT-BsmBI-sgRNA vector to assemble the sgRNA plasmids (pHYT-sgRNA84 and pHYT-sgRNA155, Supplementary Table S2.). A ∼2 kb donor DNA template containing the 1 kb N-terminus and 1 kb C-terminus was developed from the mutation or knockout site for proper homology-directed repair (HDR). For the construction of pHYT-sgRNA-HRA for mutation, the donor DNA template was PCR-amplified using the B. subtilis 168 genome as a template (primers: pHYT-sgRNA-HRA-FWD and pHYT-sgRNA-HRA-REV, Supplementary Table S2), and cloned into pHYT-sgRNA vector after the sgRNA module sequencing via Gibson assembly (vector was linearized by pHYT-sgRNA-HRA2-REV and pHYT-sgRNA-HRA2-FWD, Supplementary Table S2). The site mutation of donor DNA templates was then introduced as described above, obtaining plasmids pHYT-sgRNA84-HRA91 and pHYT-sgRNA-HRA150. For the construction of pHYT-sgRNA-HRAko for knockout, pHYT-sgRNA84-HRA91 plasmid was linearized with 5-yerA-ko-I and 3-yerA-ko-I, then cyclized via Gibson assembly.
The electroporation method was used for B. subtilis 168 transformations as previously described by Xue et al. with minor modifications (72). A total of 200 ng plasmid was transformed per 100 μl competent cells, followed by recovery in RM broth (LB containing 0.5 M sorbitol and 0.38 M mannitol) at 37°C overnight. The competent cell of B. subtilis 168 prepared via Xue's method was transformed with SpCas9-carrying plasmid pHT01-Cas9 to construct BS168-Cas9. A single clonal BS168-Cas9 was then transformed with pHYT-sgRNA-HRA containing donor DNA template and sgRNA transcription module. The transformation mixtures were spread on an LB agar plate containing chloramphenicol (10 μg/ml) and tetracycline (15 μg/ml). The recombinant transformants were confirmed via colony PCR. The single clone was grown in LB medium supplemented with chloramphenicol (10 μg/ml) and tetracycline (15 μg/ml) at 37°C. For expression of Cas9 and sgRNA, 0.5 mM isopropyl-β-thiogalactoside (IPTG) was added into the culture when the OD600 reached ∼0.6. The cultured cells were serially diluted and spread on an LB agar plate containing chloramphenicol (10 μg/ml) and tetracycline (15 μg/ml) after 12 h of induction. The successful mutations were confirmed using PCR and Sanger DNA sequencing (GenScript, China). For plasmid curing, the strains with successful gene editing containing pHT01-Cas9 and pHYT-sgRNA-HRA vector were grown in LB medium under antibiotic-free condition. Complete plasmid curing was confirmed when the mutant strain grew only in antibiotic-free LB agar plate (no growth in chloramphenicol or tetracycline LB agar plate), and verified by PCR.
RNA library preparation and Illumina sequencing
Triplicate samples of a single clone of modified B. subtilis 168 and unmodified B. subtilis 168 were grown in antibiotic-free LB medium at 37°C. The cell cultures were then diluted with fresh LB medium to OD600 ∼0.02 to a total volume of 15 ml, then centrifuged until the OD600 reached ∼0.8 (early exponential stage, EES). The cell pellets were resuspended using 1 ml RNAlater (ThermoFisher Scientific, USA, LOT. AM7020) and flash-frozen with liquid nitrogen. The samples were stored in dry ice and shipped to GENEWIZ (Suzhou, China) for further analyses, including RNA extraction, rRNA depletion, library preparation, Illumina sequencing and data analysis. The total RNA extracted from each sample was quantified and qualified using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and NanoDrop 2000 (ThermoFisher Scientific Inc., USA). Gel electrophoresis (1% agarose formaldehyde) was used as the integrity test. The total RNA (1 μg) with a RIN value above 9.0 was used for ribosomal RNA (rRNA) depletion and library preparation. Transcriptome sequencing (RNA-seq) was performed on the Illumina HiSeq 4000 platform. Raw data were processed using Cutadapt (version 1.9.1) (73) to ensure data quality for further analyses. Clean data were aligned to reference genome (NCBI, NC_000964.3) using software Bowtie2 (v2.2.6) (74). For expression analysis, the transcripts in Fasta format were converted from known gff annotation file and properly indexed. The HTSeq (v0.6.1p1) was used to estimate gene expression levels from the pair-end clean data with the file as a reference gene file. The DESeq2 Bioconductor package (75) was used for differential gene transcription analysis between two samples (three biological replicates). The Padj of genes was set at <0.05 to detect differentially expressed genes. The GOSeq (v1.34.1) was used to identify Gene Ontology (GO) terms that annotate a list of enriched genes with a significant P-value <0.05. The top GO was used to plot DAG. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to enrich significantly differentially expressed genes, and the 30 pathways with the most significant enrichment (lowest Q-value) were identified. Each bacterial strain had three biological repeats.
Quantitative analysis of the m6A/A ratio and m6dA/dA ratio via LC-HRMS
For RNA extraction, four samples of a single clone of modified B. subtilis 168 cells were grown and collected as described above. The cell pellets were resuspended with 1 ml RNAlater (ThermoFisher Scientific, USA, LOT. AM7020) and flash-frozen using liquid nitrogen, then stored at −80°C for further use. The RNAlater was removed via centrifugation after RNA extraction. The cell pellets were dissolved in 0.2 ml buffer lysis (30 mM Tris pH 6.2, 10 mM EDTA, 0.1 mg/ml lysozyme, DEPC-treated water), then incubated at 37°C for 30 min. Subsequently, 1 ml of TRIzol reagent (ThermoFisher Scientific Inc., USA) was added and mixed well before adding 0.2 ml chloroform. The mixture was centrifuged at 15 000 × g, 4°C for 15 min, then the aqueous phase was transferred to a different RNase-free tube. About 0.3 ml of saline solution (0.6 M NaCl, 0.4 M sodium citrate) and 1 ml of isopropyl ethanol were then added, and the mixture was stored at −20°C for 2 h. The mixture was then centrifuged at 15 000 × g, 4°C for 20 min. The pellet containing the total RNA was washed twice using 1 ml of 70% (v/v) ice-cold ethanol. Finally, the RNA pellet was air-dried and dissolved in 200 μl of RNase-free water. The concentration and quality of RNA were analyzed using NanoDrop 2000 ONEc (Thermofisher Scientific, USA). For quantitative analysis of N6-methyladenosine/adenosine ratio, total-RNA were digested into single nucleosides as previously described with minor modifications (3). Total-RNA (5 μg) was digested using nuclease P1 (10 U) in 200 μl of nuclease buffer (25 mM of NaCl and 2.5 mM of ZnCl2) at 37°C for 2 h, then NH4HCO3 (1 M, 10 μl) and alkaline phosphatase (2.5 U, NEB) added. The reaction mixture was then centrifuged at 30000 × g for 20 min at 4°C. The supernatant (50 μl) was then subjected to LC-HRMS analysis as previously described. For genome extraction, three samples of single clone modified B. subtilis 168 cells were grown and collected as described above. EasyPure Genomic DNA Kit (Transgen, China) was used for the extraction of genomic DNA. The enzymatic digestion of DNA was performed as previously described with some modifications (76). Briefly, 1 μg of genomic DNA was dissolved in 17 μl nuclease-free H2O and denatured at 95°C for 5 min, put on ice for 2 min, then incubated with S1 nuclease (10 U, TaKaRa) at 37°C for 16 h. About 0.1× volume of 1 M NH4HCO3 and alkaline phosphatase (2.5 U, NEB) were added, then the samples were incubated at 37°C for 6 h. The reaction mixture was then centrifuged at 30 000 × g for 20 min at 4°C. The supernatant was subjected to LC-HRMS analysis as described earlier.
Quantitative analysis of cellular purine triphosphates
The cellular purine triphosphates of B. subtilis 168 were extracted and quantitated as previously described with some modifications (77–79). For metabolites extraction, three samples of a single clone of modified B. subtilis 168 cells were grown and collected as described above, and 200 ml of cells were washed three times with 5 ml extraction buffer containing 0.04 M potassium fluoride. The cell pellets were resuspended with 1 ml extraction buffer with addition of 200 μg lysozyme. Then the suspensions were incubated at 37°C for 30 min. To complete cell lysis, 2 ml methanol/acetonitrile (1:1, v/v) was added to the suspension, and the resulting suspensions were sonicated in an ice bath for 15 min. Then the samples were spun in a micro-centrifuge at 30 000 × g for 30 min at 4°C and the supernatants were collected. The soluble extracts were incubated with 100 μg protease K for 120 min at 30°C, and subsequently filtered via ultra-filtration tubes (3 kDa molecular weight cutoff, Millipore, USA). The flow through was concentrated in a freeze dryer (Alpha 1–2 L D-plus, Christ, Germany) and resuspended in 0.1 ml 5% methanol, 95% water. Then the above samples were centrifuged at 30 000 × g for 20 min at 4°C, and supernatant was subjected to LC–MS/MS analysis. Purine metabolites were quantified using the Shimadzu LC-MS/MS system (LC30A HPLC coupled to 8050 triple-quadrupole mass spectrometer, SHIMADZU) in negative mode. The samples were quenched and flowed through an Agilent ZORBAX Extended-C18 column (4.6 × 50 mm, 1.8 μm particle size) at a flow rate of 0.2 ml/min. The binary phases were solvent A (10 mM ammonium acetate, pH 9.5) and solvent B (methanol). The chromatographic gradient separation phase had 3–10% of solvent B (0–10 min), 100% solvent B (11–15 min), and 3% solvent B (15–20 min). The purine nucleoside triphosphates were analysed by using retention time and multiple reaction monitoring (MRM) of the transition m/z 506 > 426 for ATP, 520 > 440 for N6-methyladenosine triphosphate (N6-mATP), and 507 > 427 for ITP. The cellular deoxycytidine triphosphate (dCTP, MRM m/z 466 > 386) was utilized as an internal reference.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
The total RNA of triplicate samples from RNA extraction described above were reverse-transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser (TAKARA Bio Inc., Cat. #RR047A) following the manufacturer's protocol. The cDNA was then diluted to 200 ng/μl for qPCR assay with each gene-specific primer (see Supplementary Table S4) using Hieff qPCR SYBR Green Master Mix (Yeasen, China) on the Bio-Rad CFX96 real-time system. Reactions were performed as follows: at 95°C for 1 min, 35 cycles at 95°C for 10 s, 55°C for 20 s and 72°C for 30 s. The rpoB gene was used as the internal control. The relative expression level of mRNA was calculated via the relative quantification (Fold Change = 2−ΔΔCt) method.
Quantification and statistical analysis
All experiments were performed in triplicates or more. GraphPad Prism 7.0 was used to calculate statistical significances via one-Way ANOVA (Dunnett's multiple comparisons test at 95% confidence level, two-sided) or unpaired t-test with triplicate values and P values <0.05 indicating statistical significance. The number of replicates and P values are shown in the respective figure legends.
RESULTS
Identification of Bsu06560 as an enzyme capable of metabolizing the N6-methyladenosine
Compared to the N6-methyladenine and N6-mAMP deaminase, the key difference in N6-methyladenosine deaminase might be attributed to the binding preference for the ribose ring of N6-methyladenosine. To identify the potential N6-methyladenosine deaminases, we searched for the m6A-adenyl deaminases with binding preference for adenosine-like structure. The sequence of known m6A-adenyl-deaminases (human ADAL, MAPDA, 6-MADs) were aligned with bacterial, human, and murine adenine deaminase (ADE)/adenosine deaminase (ADA)/AMP deaminase (AMPD) using ClustalW,(80) to capture the crucial factors affecting N6-methyladenosine deamination and facilitate enzyme. Since adenyl-deaminases evolve independently from two different ancestral enzymes with very different metal-binding sites, these two families of adenyl-deaminases were independently analyzed (28,30). The sequence alignment analysis of ADALs and ADAs revealed that two families shared highly conserved residues crucial for ADA activity since both of their substrates contained the ribose ring structure (Supplementary Figure S2 and Supplementary Table S1). However, some hydrophilic residues were highly conserved in ADALs but not in ADAs, which were responsible for the phosphate portion of the ligand (N6-mAMP) (81). The hydrophilic residues might be caused by the natural evolution of enzymes for the specific deamination of N6-mAMP. These findings explain why ADALs exhibit no activity on any N6- or O6-substituted purine nucleoside (33).
The 6-MADs, another family of proteins, have a binuclear metal center, which is distinct from the single metal center deaminases mentioned above and is annotated as N6-methyladenine deaminases (53). Herein, the sequences of 6-MADs (Bsu06560 and Bh0637) were aligned with those of ADAs and ADEs to assess the factors that discriminate adenine from adenosine (Supplementary Figure S3 and Supplementary Table S1). The Bsu06560 exhibited a homology of 20%, 22%, and 19% with human ADA (hsADA), mouse ADA (mmADA), and E. coli ADA (ecADA), respectively. Moreover, Bh0637 had 18%, 17%, and 16% of homology with hsADA, mmADA and ecADA, respectively. Further sequence alignment analysis revealed that the residues Glu67, Val159, Val333, Glu364, Leu382, and Glu498 of Bsu06560 were identical to conserved sites of adenosine deaminases (Supplementary Figure S3) but did not exist in adenine deaminases (30). These evidences indicate that Bsu06560 might prefer adenosine-like substrates instead of adenine-like substrates.
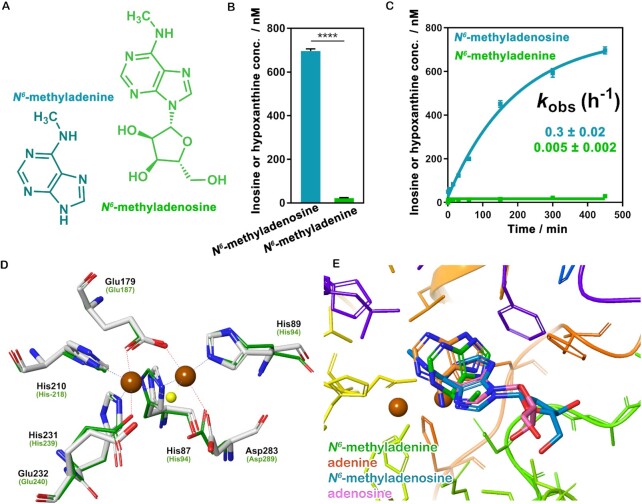
To test our hypothesis, Bsu06560 was expressed in E. coli BL21(prk1037), then the protein was purified with over 95% purity (Supplementary Figure S4). The inductively coupled plasma atomic emission spectrometry (ICP-AES) revealed that manganese was the predominant metal and contained approximately two equivalents per protein, consistent with the previous report (35). The Liquid Chromatograph-High Resolution Mass Spectrometry (LC-HRMS) assays were then used to investigate the substrate preference of the Bsu06560 enzyme. Intriguingly, Bsu06560 displayed remarkably increased deamination activity for both N6-methyladenosine and adenosine, as predicted by the sequence analyses above (Figure 1A–C). Notably, the yield of N6-methyladenosine deamination products was ∼28 folds higher than that of N6-methyladenine for the same reaction time (450 min, Figure 1B). The time-course analysis also demonstrated that Bsu06560 provided 60-fold higher kobs against the N6-methyladenosine than the N6-methyladenine (Figure 1C). Taken together, these in vitro studies suggest that Bsu06560 has a remarkably higher substrate preference for the N6-methyladenosine over the N6-methyladenine.
Figure 1.
Bsu06560 capable of metabolizing the N6-methyladenosine and the N6-methyladenine. (A) Chemical formula of N6-methyladenosine and the N6-methyladenine. (B) Deamination activities of Bsu06560 with different substrates (10 μM). The integral areas of different chromatographic peaks in LC-HRMS were used to determine the concentration of the product. Bsu06560 exhibited a remarkable deamination activity for N6-methyladenosine than N6-methyladenine. Mean values were plotted (n = 3). The error bars represent the ±SEM. Unpaired t-test (two-tailed): ****P < 0.0001. (C) Time courses for different substrates (10 μM) deaminated by Bsu06560. The data points were best fitted using a single exponential equation to follow the formation of inosine or hypoxanthine (GraphPad Prism software). The kobs value for inosine and hypoxanthine formation was estimated using the one-phase association model. Mean values were plotted (n = 3). The error bars represent the ±SEM. (D) Superimposition of the bimetal site of the Bsu06560 homologous model (green line) and the template atuADE (PDB code: 3NQB, white stick). The dashed lines indicate the polar interactions between residues and bimetal core, and μ-1,3-carboxylate glutamic (Glu179) and hydroxide (water, shown in yellow ball) bridges. The residue labels for Bsu06560 are shown using the label for corresponding residues in atuADE given in parentheses. The brown dots represent metals. (E) The binding site of N6-methyladenine, adenine, N6-methyladenosine, adenosine (lowest energy for each). The brown dots indicate metals.
Modeling and molecular docking analyses
The unique biochemical properties of Bsu06560 with the substrate-specificity suggested different targeting and recognition mechanisms for m6A-adenyl substrates compared to those of its m6A-adenyl deaminase orthologues. To further elucidate the molecular recognition mechanism of Bsu06560, a model of Bsu06560 was constructed using Iterative Threading ASSEmbly Refinement (I-TASSER) server (56–59) owing to low similarity (<30%) to the known crystal structure of proteins. For I-TASSER analysis, adenine deaminase atuADE from Agrobacterium tumefaciens was used as the main template for modeling (23% identity and E-value = 1 × 10−23, Atu4426 PDB code: 3NQB, Supplementary Figure S5). The overall alignment of the Bsu06560 model with atuADE is shown in Supplementary Figure S6. A conserved binuclear metal center was apparent in the chasm of the Bsu06560 model at a distance of 4 Å, well overlapped with the aligned residues from atuADE (Figure 1D and Supplementary Figure S5). Similar to those in typical binuclear non-haem iron-dependent frog M ferritin, a μ-1,3-carboxylate Glutamic (Glu-179) and a hydroxide (water) bridges were formed in the binuclear metal core of Bsu06560 (Figure 1D) (82–84). Superposition of Bsu06560 and atuADE indicated that the residues surrounding twin metals were highly conserved, and each metal was coordinated by two Histidine and one Glutamic/Aspartic residues (His-210/His-231/Glu-232, His-87/His-89/Asp-283) (Figure 1D and Supplementary Figure S6) (53). The previous studies revealed binuclear metal core is essential for deamination activity and is involved in the catalytic mechanism of atuADE (53), indicating that Bsu06560 has an analogical catalytic pathway with atuADE.
Molecular docking studies of Bsu06560 with N6-methyladenosine or N6-methyladenine revealed that the flexible binding pocket of Bsu06560 might attributed to the substrate-specificity. Furthermore, substrate docking studies were conducted with N6-methyladenine, adenine, N6-methyladenosine, adenosine, N6-methyl-2′-deoxyadenosine, deoxyadenosine, N6-mAMP, AMP, N6-mdAMP and dAMP using the Schrodinger suite (Schrödinger, LLC). Notably, the Dock Score (DS) of adenosine structure-like substrates, such as adenosine (DS = –7.43), N6-methyladenosine (DS = –7.12), N6-methyl-2′-deoxyadenosine (DS = –6.06) and 2′-deoxyadenosine (DS = –4.51), were relatively higher than those of N6-methyladenine (DS = –4.26) and adenine (DS = -3.11). These substantial increases in Dock Score of adenosine structure-like substrates might be due to hydrogen bonds formed between the residues (Asp283, Gly284 and Ser255) of su06560 and the ribose ring of adenosine structure-like substrates (Figure 1E and Supplementary Figure S7). However, N6-mAMP, AMP, N6-mdAMP and dAMP could not be docked in the catalytic site with the appropriate pose, consistent with the interpretation that Bsu06560 belongs to the enzyme family evolved from different ancestral enzymes compared to the ADAL family.
Encouraged by the results of molecular docking studies, we further examined the substrate scope of wild-type Bsu06560. Notably, besides N6-methyladenosine, adenosine, N6-methyladenine and adenine, Bsu06560 also accepted N6-methyl-2′-deoxyadenosine and 2′-deoxyadenosine as its substrates to provide the deamination products (Supplementary Figures S8 and S11–S17). However, Bsu06560 induced no catalysis in N6m-AMP and AMP (Supplementary Figures S8 and S18-S20). Moreover, Bsu06560 afforded higher conversion for the adenosine, which yielded ∼6.5-fold higher inosine for the same reaction time (120 min) than that of N6-methyladenosine (Supplementary Figure S8). For adenine analogues, Bsu06560 also preferred to deaminate non-methylated adenine to N6-methyladenine with ∼35 folds higher yield of hypoxanthine (Supplementary Figure S8). The products accumulated linearly in a short time (15 min) (Supplementary Figure S9A). As a result, the catalytic rates of Bsu06560 were estimated via linear fitting the time-course analysis for substrates. The catalytic rates of WT Bsu06560 with N6-methyladenosine, adenosine, N6-methyladenine and adenine were 4.22, 44.49, 0.48 and 4.73 nM/min, respectively. The conversion ratios of different substrates showed a similar result. Bsu06560 exhibited a remarkable deamination activity for adenosine and N6-methyladenosine than adenine and N6-methyladenine within 120 min (Supplementary Figure S9B). The competitive experiments of Bsu06560 showed that N6-methyladenosine had a much higher conversion ratio than N6-methyladenine (Supplementary Figures S10A and S21). Similarly, adenosine had a significantly higher conversion ratio than adenine in the adenosine and adenine mixture (Supplementary Figures S10B and S22). Taken together, these results indicate that Bsu06560 is a multifunctional deaminase that prefers nucleosides, rather than a N6-methyladenine deaminase annotated before.
Identifying the origin of oxygen atom at C-6 position of inosine from the Bsu06560 deamination
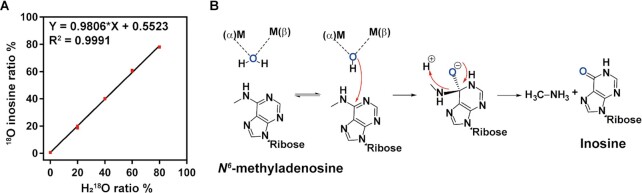
Considering the crucial biological function of m6A demethylation (7,85,86), the m6A recognition mechanism of Bsu06560 with the substituted aminopurines could be essential in maintaining the level of m6A-substituted purines in vivo. For Bsu06560-catalyzed deamination processes, the key chemical conversions occur by eliminating the N-6 amino group from the purine portion and subsequently incorporation of the oxygen atom at the C-6 position in the inosine products. This study investigated the origin of this oxygen atom. To mechanistically understand the m6A deamination mechanism, an isotope doping assay was performed to examine the oxygen atom source of 6′-hydroxyl of inosine. The isotope doping experiment was conducted by mixing heavy oxygen water (H218O) into the Bsu06560 reaction mixture at various proportions (0–80%) of total volume. The heavy oxygen-labeled inosine with two more atomic mass was detected and quantitated using LC-HRMS (Supplementary Figure S23). With the increasing proportion of heavy oxygen water in the reaction mixture, the ratio of heavy oxygen labeled inosine was also produced linearly with total inosines (Figure 2A). This result implies that water molecules are the source of the oxygen atom at the C-6 position of inosine from the Bsu06560 deamination. Moreover, the production of methylamine from N6-methyladenine deamination was also demonstrated via gas chromatograph-mass spectrometry (GC–MS; Supplementary Figure S24).
Figure 2.
Catalytic mechanism of the Bsu06560 deamination. (A) Isotope doping characterization of products during Bsu06560 deamination. The incorporation of 18O into inosine from Bsu06560 deamination had a significant linear relation (R2 = 0.9991) as the ratio of heavy oxygen water (H218O) in the reaction system increased. Quantification of 18O-inosine is shown as a ratio of total inosine product. Mean values were plotted (n = 3). The error bars represent the ±SEM. (B) Proposed catalytic mechanism of Bsu06560 deamination. Re-face of N6-methyladenosine had a nucleophilic attack by hydroxide from water. The reaction was conducted in water phase with Bsu06560 at 30°C, pH 7.5. Red arrows represent the electron transfer process.
Based on the model and biochemical experiments, the deamination mechanism of Bsu06560 was proposed and described in Figure 2B, which presents a similar catalytic mechanism as E. coli ADE (53). As Bsu06560 exhibited a conserved binuclear metal center coordinated by four histidines, a water molecule formed a bridge with two metal ions and transferred a proton to the neighboring coordinating residues. The N-1 of purine was polarized, making the C-6 more electrophilic. The bridging hydroxide was poised for nucleophilic attack at the re-face of the C-6. The nucleophilic attack of hydroxide occurred with the proton transfer from the adjacent residue to N-1 of purine, forming a tetrahedral intermediate. Subsequently, the tetrahedral intermediate collapsed with the leaving of methylamino. In this proposed mechanism, the methylation on the N-6 position of aminopurines exhibited minimal electronic effect and a slight steric hindrance to the nucleophilic attack of C-6 in the catalytic deamination process (87). Thus, we speculated that the recognition of N6-methyl-purine substrates from non-substituted aminopurines might mainly depend on the preferential interactions with various substrates, instead of a catalytic kinetics-prioritized process. However, further investigations are warranted to elucidate a more comprehensive mechanism.
Insight into m6A recognition mechanism and enhancement of N6-methyladenosine recognition via structure-guided engineering
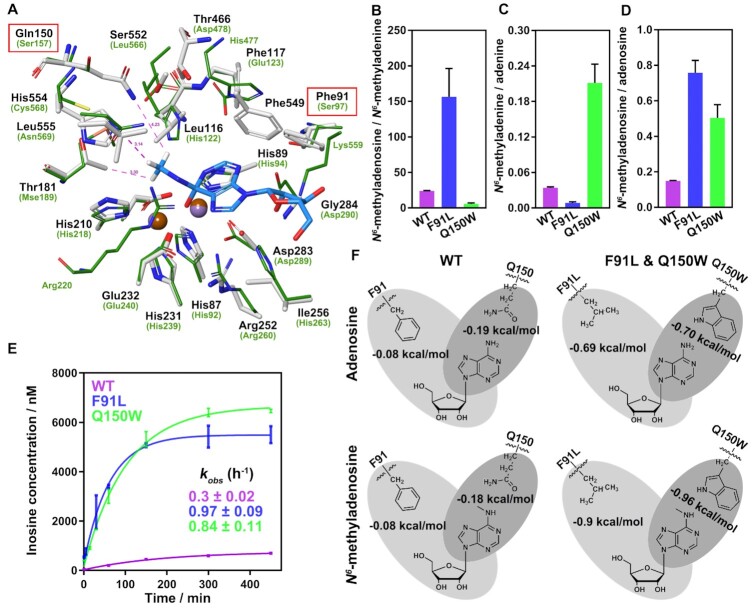
To date, only a few studies have reported on the comprehensive mechanism of the recognition of N6-methyl-purine substrates by Bsu06560 (35). Herein, the hydroxide tetrahedral intermediate of N6-methyladenosine and N6-methyladenine were used for more precise docking to examine the critical factors affecting the recognition of N6-methyl-purine substrates (Figure 3A and Supplementary Figure S25). Compared to the known adenine deaminase atuADE, the major differences in the substrate-binding pockets were found in the residues around the ribose ring and N6-methyl group of N6-methyladenosine (Figure 3A, Supplementary Figures S6 and S25). Residues Phe91 and Phe549 of Bsu06560 may form noncovalent interactions with the ribose ring of N6-methyladenosine based on the docking results, which did not exist in atuADE. The superimposition of the Bsu06560 model and the adenine deaminase atuADE also indicated that Ile256 and Gly284 of Bsu06560, aligned with His263 and Asp290 of atuADE, and elimination of Lys559 from Bsu06560, resulted in fewer clashes with ribose ring and created space for larger molecules (Figure 3A and Supplementary Figure S25). Moreover, the residues close to the N6-methyl group of N6-methyladenosine, Leu116, Phe117, Gln150, Thr181, Thr466, Ser552, His554 and Leu555 in Bsu06560 ensued larger volume and higher hydrophobicity for N6-methyl group than the correspondingly aligned residues in atuADE, thus enhancing m6A recognition by Bsu06560 (Figure 3A and Supplementary Figure S25).
Figure 3.
Biochemical and computational characterization of purified Bsu06560 mutants. (A) Superimposition of the N6-methyl surrounding residues for the high energy intermediate of N6-methyladenosine in Bsu06560 homologous model (white stick) and the adenine deaminase (atuADE, renamed Atu4426, PDB code: 3NQB, green line). Brown and violet balls represent metals in Bsu06560 and atuADE, respectively. Another metal ion in atuADE was hidden due to its unnecessary activity. Pink dotted lines indicate the distance between atoms (angstrom). (B–D) Product ratios of WT, Q150W and F91L mutant in deamination reaction at the 120 min. The substrates were saturated with enzymes. The F91L and Q150W mutants had a higher ratio than wild-type protein in product amount of N6-methyladenosine/N6-methyladenine and N6-methyladenine/adenine. Low concentrations of substrates (10 μM) were used where the enzyme was excess (20 μM). The integral areas of chromatographic peaks of LC-HRMS were used to determine the concentration of products. Mean values were plotted (n = 3). The error bars represent the ± SEM. (E) Deamination time courses for the Bsu06560 mutants for N6-methyladenosine. Mean values were plotted (n = 3). The error bars represent the ± SEM. The mutants Q150W and F91L of Bsu06560 had a substantially increased deamination activity for N6-methyladenosine. The data points were best fitted using a single exponential equation to follow the formation of inosine or hypoxanthine (GraphPad Prism software). The kobs value for inosine formation was estimated using the one-phase association model. (F) Simplified scheme of interactions between key residues of Bsu06560 and its mutants with N6-methyladenosine and adenosine. Non-bonded interactions are indicated in gray, and relative energies of interaction are provided in kcal/mol.
To further elucidate the role of these interactions in regulating the N6-methyladenosine recognition by Bsu06560, the Schrodinger suite was used to scan and predict the affinity of the residues mentioned above to obtain the interaction parameters for each Bsu06560 mutation (Dataset S1). The results of residue screening indicated that Phe91, Gln150, Gly284, Phe549, and His554 in Bsu06560 were essential for the binding affinity of N6-methyladenosine (Figure 3A, Supplementary Figure S25 and Dataset S1). To obtain the possible mutation patterns valuable for site-mutation verification, we evaluated each interaction screening result and selected the mutation patterns with the highest ‘Δ Affinity’ and ‘Δ Stability’. Several mutation patterns with the highest ‘Δ Affinity van der Waals forces’ were also selected through experience. Next, the His554, Gln150, Phe91 and Phe549 were discretely mutated based on the screening results, then the capacities of the resulting mutants were tested in N6-methyl-purine deamination (Supplementary Figures S26A and S26B). The Phe549 mutants (F549G and F549M) had similar deamination activity to that of wild-type Bsu06560, whereas the His554 mutants (H554F, H554M and H554W) had a reduced deamination activity than the original Bsu06560 (Supplementary Figures S26A and S26B). This study then tested the conversion ratio of different substrates for Bsu06560 in substrate-saturated and enzyme-saturated conditions. Notably, the single amino-acid mutation of Phe91 to Leu91 in Bsu06560 saturated conditions (10 μM of substrates) drastically enhanced the N6-methyl-purine deamination activity of the wild-type Bsu06560 with ∼8-fold higher conversion ratio of N6-methyladenosine over N6-methyladenine (Figure 3B, C and Supplementary Figure S26C). Moreover, the Q150W mutant protein remarkably increased the conversion ratio of N6-methyladenine over its corresponding non-substituted purine (adenine), with ∼7 fold higher than that of wild-type Bsu06560 (Figure 3C and Supplementary Figure S26C). These results reveal that the Phe91 residue of Bsu06560 are essential for ribose ring recognition, whereas the Gln150 residue of Bsu06560 was essential for N6-methyl recognition. These two mutations both significantly increased the conversion ratio of N6-methyladenosine than adenosine (Figure 3D and Supplementary Figure S26C). However, the conversion ratio of N6-methyladenosine and adenosine in substrate-saturated conditions (250 μM of substrates) was not significantly changed as in low concentration (Supplementary Figures S26D and S26E). This could be because the catalytic speed of each enzymatic cycle for a substrate is similar when substrates are adequate. This result is consistent with our isotope doping experiment that substrate recognition of Bsu06560 mainly depends on affinity rather than catalytic kinetics. The time-course analysis further demonstrated that overall deamination reactivity for N6-methyladenosine significantly increased in both F91L and Q150W mutants than in wild-type Bsu06560 (Figure 3E). Thus, Phe91 and Gln150 may play a crucial role in targeting adenosine and N6-methyl-purine via Bsu06560.
Next, we investigated the crucial role of Phe91 and Gln150 residues of Bsu06560 in N6-methyl-purine recognition using Quantum mechanics/molecular mechanics (QM/MM) calculations. Briefly, the QSite (65,66) module of Schrodinger suite was employed through a tight coupling of Jaguar (67) suite for QM region simulation and Impact (68) was used for MM simulations. To begin the QM/MM analysis, the preliminary structures were obtained from results of docking studies with N6-methyladenosine and adenosine, where the single-mutant of Bsu06560 underwent structural optimization. The final QM/MM energies were obtained using the single-point energy calculation based on the optimized structures. As anticipated, the F91L and Q150W mutants achieved lower energies than the wildtype protein with both N6-methyladenosine and adenosine (Figure 3F). Remarkably, the effect of substitution of Gln150 with Trp150 yielded a stabilization increase of 0.78 kcal/mol and 0.51 kcal/mol for N6-methyladenosine and adenosine, respectively. F91L also resulted in a stabilization increase of 0.82 kcal/mol and 0.61 kcal/mol for N6-methyladenosine and adenosine, respectively (Figure 3F). Therefore, the substitution of Gln150 with Trp might produce a more non-polar hydrophobic region suitable for methyl group and F91L mutant cause less steric hindrance to the ribose portion of adenosine and N6-methyladenosine (Figure 3A, F and Supplementary Figure S25). Thus, our biochemical characterization of Bsu06560 variants and QM/MM results supported that Phe91 and Gln150 play important roles in adenosine and N6-methyladenosine recognition and deamination via Bsu06560.
In vivo mutations highlight the association between m6A metabolism and bacterial gene regulations
To further determine the effects of Bsu06560, a dual-plasmids Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) system was used to knockout the Bsu06560 encoding gene (gene yerA) and edit B. subtilis 168 genome in vivo (70). In this system, an RNA-guided Streptococcus pyogenes-originated Cas9 (SpCas9) was encoded by one of the plasmids (pHT01-Cas9). Meanwhile, the other plasmid (pHYT-sgRNA-HRA) provided a sgRNA containing 20 bp complementary sequence and a donor DNA template containing a region homologous to the chromosome along with the desired mutation site, which was complementary to the target region where the double-strand break (DSB) should occur and subsequently repaired via homology-directed repair (HDR). Notably, the off-target editing in B. subtilis was relatively sparse owing to impaired non-homologous end joining (NHEJ) machinery during DNA replication and a much smaller genome of B. subtilis (69,88–90). After gene editing, Sanger sequencing analyses confirmed that the first 284 bp of Bsu06560 encoding gene were knocked out, resulting in frame shift of yerA gene and inactivation of Bsu06560 (Supplementary Figure S27). The other two mutant strains of Bsu06560 on B. subtilis 168 chromosome, Phe91 and Gln150, were successfully substituted with Leu and Trp, respectively (Supplementary Figure S28). Plasmid curing was confirmed via colony PCR. The B. subtilis 168 Bsu06560-knockout strain and mutant strains, Phe91Leu and Gln150Trp, were designated as BS-KO, BS-F91L and BS-Q150W, respectively.
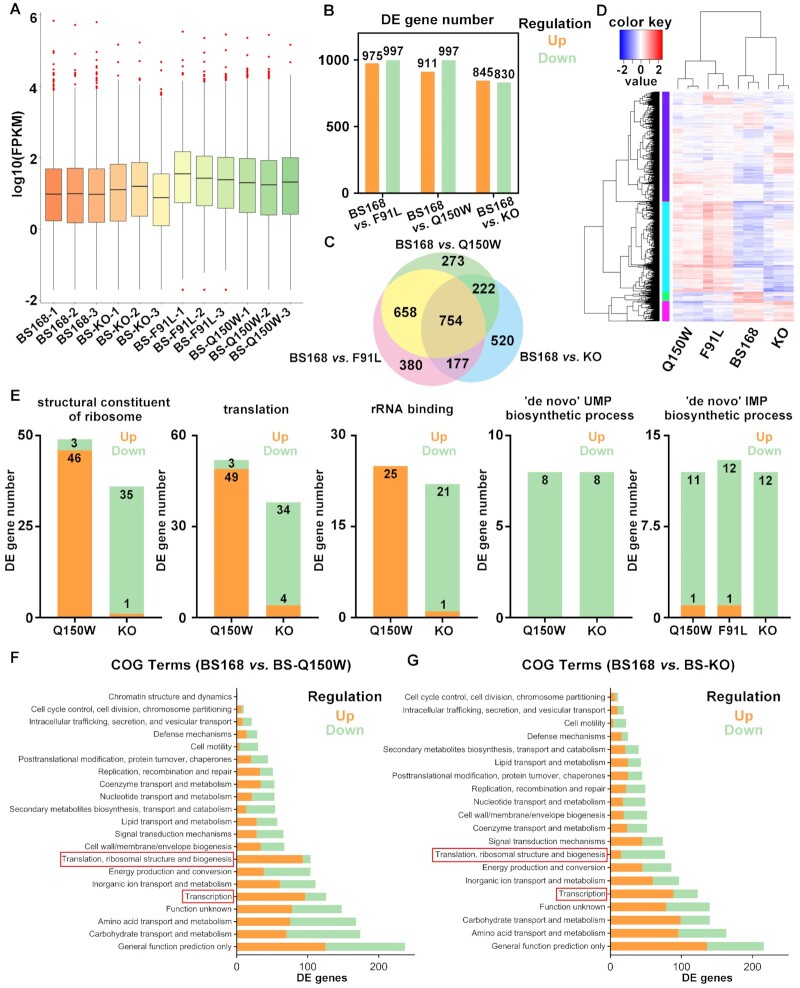
This study used Illumina high-throughput RNA sequencing for transcriptome analysis to investigate the global impact of N6-methyladenosine nucleoside deamination in B. subtilis metabolism at the molecular level. Single colony (in triplicate) of wild-type B. subtilis 168 (BS168), BS-KO, BS-F91L and BS-Q150W were cultured in antibiotic-free LB medium until the OD600 reached 0.8 (early exponential stage, EES). The cells were collected for RNA sequencing. Notably, over 98.9% of clean reads were aligned to B. subtilis 168 reference genome (Supplementary Table S3 and Dataset S2). Pearson correlation and Principal Component Analysis (PCA) indicated a high degree of reproducibility and consistency among the three biological replicates (Supplementary Figure S29).
To analyze global gene expression levels, fragments per kilobases per million reads (FPKM) were used. Over 99% of genes were positively expressed in all tested samples (FPKM > 0.1). The BS-F91L and BS-Q150W mutants had a higher gene expression level than the wild-type and Bsu06560-knockout strains (Figure 4A). Analysis of differential gene expression profiles with wild-type BS168 revealed that 975, 911 and 845 genes were significantly upregulated in BS-F91L, BS-Q150W and BS-KO, respectively. Moreover, 830 genes in BS-KO, 997 genes in BS-F91L and BS-Q150W mutants were downregulated (significant condition: FC ≤ 0.5; P < 0.05; false discovery rate (Padj) ≤ 0.05) (Figure 4B, C and Supplementary Figure S30). A total of 754 genes were present in all three groups, accounting for 16.6% of all B. subtilis 168 genes (Figure 4C). Over 70% of differentially expressed (DE) genes were overlapped between BS168 vs. BS-F91L and BS168 vs. BS-Q150W groups (Figure 4C). The complete list of differential expressed transcripts is summarized in Datasets S3 to S5. The DE transcripts were clustered in both horizontal and vertical directions using hierarchical clustering and rough set methods. All DE genes were grouped into six clusters (Figure 4D and Dataset S6). The expression levels of the metabolic pathway-related genes cluster were significantly higher in BS-F91L and BS-Q150W mutants than the wild-type BS168 and Bsu06560-knockout strains (Figure 4D and Dataset S6). Combined with the previous finding, these results implying that BS-F91L and BS-Q150W mutants might significantly reduce the m6A modification in RNA of B. subtilis 168 since they can substantially convert N6-methyladenosine to inosine, thus possibly leading to the activation of numerous genes. This assumption is consistent with the fundamental role of m6A decoration in the regulation of gene expression in mammals (1,9,12,91–93). From the above results and previous researches about the ADAL deaminases, we also assume that Bsu06560 could also act as a regulator in purine metabolism that maintain the concentration of N6-methyladenosine.
Figure 4.
Transcriptomic profiling of BS168 (WT), BS-KO, BS-F91L and BS-Q150W strains. (A) Global gene expression analysis of all RNA sequencing samples. The BS-F91L and BS-Q150W strains had higher gene expression levels than wild-type. (B) Number of DE genes in BS168 versus BS-KO, BS168 versus BS-F91L and BS168 vs. BS-Q150W groups identified via RNA sequencing. (C) Overlap of the DE genes in BS168 versus BS-KO, BS168 versus BS-F91L and BS168 versus BS-Q150W groups. (D) Hierarchical clustering of DE genes in wild-type and variant strains (red and blue represent up-regulated and down-regulated genes, respectively). (E) Gene expression patterns in several GO enrichment items. The BS-Q150W and BS-KO strains showed contrasting expression patterns in ‘structural constituent of ribosome’, ‘translation’ and ‘rRNA binding’ items, and similar expression patterns in ‘‘de novo’ UMP biosynthetic process’ and ‘‘de novo’ IMP biosynthetic process’. (F) The COG classification of the DE genes in BS168 vs. BS-Q150W group. (G) The COG classification of the DE genes in BS168 versus BS-KO group. (F, G) The BS-Q150W and BS-KO strains with contrasting expression patterns in ‘Translation, ribosomal structure and biogenesis’ term and similar expression patterns in ‘Transcription’ term.
This study further analyzed N6-methyladenosine and adenosine concentration in the total RNA of BS168 and its mutants. The results showed that the N6-methyladenosine (m6A)/adenosine (A) ratio of total RNA was significantly decreased in BS-F91L and BS-Q150W (0.026% in WT, 0.019% in BS-F91L and 0.016% in BS-Q150W, Supplementary Figure S31A), while it was significantly increased in the BS-KO (0.03%). The changes in m6A/A ratio in vivo were consistent with the enhanced activity of Bsu06560 mutants for N6-methyladenosine (Supplementary Figure S31). Moreover, the BS-Q150W strain showed a slightly lower m6A/A ratio in total RNA than Bsu06560 F91L when Q150W mutant had a higher recognition ability than the F91L mutant in vitro. Furthermore, this study increased the N6-methyladenosine or adenosine concentration in the growth medium of BS168 to test the m6A/A ratio of RNA of BS168 in different growth conditions. The m6A/A ratio of total RNA slightly increased compared with that after addition of N6-methyladenosine into the growth medium (Supplementary Figure S31C). However, the increased m6A/A ratio was not significantly different in the two growth conditions with highly varied N6-methyladenosine concentrations (10 μM versus 1 mM). An indication that some other N6-methyladenosine-related regulators or alternating nucleosides metabolic pathways can maintain the concentration of N6-methyladenosine within the cells. Besides, this study further quantified the concentration of adenosine triphosphate (ATP) and N6-methyladenosine triphosphate (N6-mATP) within B. subtilis 168 and its mutants (Supplementary Figure S32). The results showed that the ATP levels were not significantly changed among these samples (Supplementary Figure S32A). However, N6-mATP was only detected in BS-KO strains, whereas had almost no signal in wild-type, BS-F91L and BS-Q150W strains (Supplementary Figure S32B). This could be the relative low concentration of N6-mATP in these strains than BS-KO, thus showed no reproducibility on our LC–MS/MS setup. These results can also indirectly explain the important roles of N6-methyladenosine-related regulators, especially Bsu06560, that can regulate the concentration of purine metabolites in cellular nucleotides pool. Previous research showed that N6-mAMP deaminase (MAPDA or ADAL) can prevent other cellular processes from excess N6-mATP (Supplementary Figure S33) (34), consistent with our assumption that Bsu06560 acts as an N6-methyladenosine regulator in purine metabolism.
Taken together, these results indicate that Bsu06560 can regulate N6-methyladenosine incorporation into RNA from the cellular RNA salvage pathway, which could be crucial for B. subtilis 168 and might be pertinent to the control of nearly half of gene expressions. This study also determined the N6-methyl-2′-deoxyadenosine (m6dA)/deoxyadenosine (dA) ratio in genome of BS168 and its variants. The ratio of m6dA/dA was considerably reduced in BS-Q150W strain than in the BS168. In contrast, the m6dA/dA ratio did not significantly change in the BS-F91L strain (Supplementary Figure S31B). Previous isotope doping research showed that several genomic m6dA originates from N6-methyladenosine, which is processed via the nucleoside-salvage pathway and mis-incorporated by DNA polymerases (21). Thus, this is a reasonable result that Bsu06560 Q150W mutant performed better in N6-methyladenosine deamination than F91L mutant in vitro, rather better than wild-type BS168. However, the decreased m6dA/dA ratio in BS-KO strain could be due to the coordination of expression level of some specific genes changing after Bsu06560 knockout (Supplementary Figure S31B).
Furthermore, gene ontology (GO), Clusters of Orthologous Groups (COG) analysis, and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analyses were performed to analyze the biological processes and pathways that were differentially expressed in BS168 and its mutants. First, GO analysis was independently performed on BS168 versus BS-KO, BS168 versus BS-F91L and BS168 versus BS-Q150W groups (Supplementary Figure S34–S36 and Dataset S7–S9). Three domains were enriched: biological process (BP), molecular function (MF), and cellular component (CC). The most significantly enriched BP terms in BS168 vs. BS-KO group (Supplementary Figure S36) were ‘translation’, ‘phosphoenolpyruvate-dependent sugar phosphotransferase system’, ‘negative regulation of transcription, DNA-templated’, and ‘response to toxic substance’. The most enriched GO terms were ‘structure constituent of ribosome’, ‘kinase activity’, ‘rRNA binding’ in MF domain, and ‘cytosol’, ‘protein-DNA complex’, ‘outer membrane-bounded periplasmic space’ in CC domain. Moreover, the absence of Bsu06560 caused significant differences in the expression of various genes distributed across the whole genome. Most enriched GO terms were associated with central-dogma-related processes and ribosomal-related factors, such as ‘structural constituent of ribosome’, ‘translation’, ‘negative regulation of transcription, DNA-templated’ and ‘‘de novo’ IMP biosynthetic process’. These results demonstrate that the Bsu06560 protein is an important regulator in B. subtilis.
The other two groups shared many enriched GO terms but differed from the BS168 vs. BS-KO group. The most significantly enriched BP terms in BS168 vs. BS-F91L group (Supplementary Figure S34) were cellular processes and metabolic processes, including ‘protein transport’, ‘polysaccharide biosynthetic process’, ‘‘de novo’ IMP biosynthetic process’, ‘tricarboxylic acid cycle’ and ‘histidine biosynthetic process’. The most significantly enriched GO terms were ‘heme binding’, ‘ATPase activity, coupled to transmembrane movement of substances’, ‘phosphopantetheine binding’ in MF domain, and ‘extracellular region’, ‘cell wall’ in CC domain. Several DE genes in the BS168 versus BS-Q150W group were enriched in the same categories as BS168 versus BS-F91L group ("ribosome binding’, ‘extracellular region’, ‘protein transport’, ‘de novo’ IMP biosynthetic process’, ‘tricarboxylic acid cycle’, ‘histidine biosynthetic process’ and so on), except for ‘translation’, ‘phosphoenolpyruvate-dependent sugar phosphotransferase system’, ‘cell redox homeostasis’ in BP domain, ‘integral component of plasma membrane’, ‘cytosolic large ribosomal subunit’, ‘ribosome’ in CC domain, and ‘structural constituent of ribosome’, ‘rRNA binding’, ‘transferase activity’ in MF domain (Supplementary Figure S35). Similar to the knockout set, many enriched GO terms in these two mutant sets were also associated with central-dogma-related processes and ribosomal-related factors (Supplementary Figure S34 to S36). Considering the enhancement of Bsu06560 deamination activity for N6-methyladenosine, it's a reasonable speculation that the reduction of N6-methyladenosine concentration can influence numerous biological processes, especially central-dogma related processes.
Therefore, this study further compared the expression features of the GO terms enriched in both BS-KO and BS-Q150W groups. Most DE genes in the structural constituent of ribosome (GO:0003735), translation (GO:0006412) and rRNA binding (GO:0019843) terms were up-regulated in the BS-Q150W strain and down-regulated in BS-KO strain (Figure 4E). These three GO terms were closely related to the translation process in central-dogma. In contrast, the transcript levels of DE genes showed opposite regulatory results in strain with the enhancement of N6-methyladenosine deamination activity compared with the Bsu06560-deficient strain, indicating that N6-methyladenosine molecular is essential in the regulation of translation-related processes within B. subtilis. Furthermore, nearly all transcript levels of DE genes were down-regulated both in mutant strain and deficient strain in ‘de novo’ UMP and ‘de novo’ IMP biosynthetic processes. Given our assumption that Bsu06560 serves as a deaminase within the pathway of purine metabolism, it's quite reasonable that changes or defects in Bsu06560 can influence the gene expression patterns in its metabolic pathways. These results also prove that Bsu06560 is an important regulator of purine metabolism.
Subsequently, the top five enriched categories in BP were used as central components of the directed acyclic graph (DAG) (Supplementary Figure S37–S39). The DAG results revealed that DE genes were mainly enriched in cellular metabolic process and nucleoside metabolism, further supporting the hypothesis on the fundamental function of Bsu06560. Then, the DE genes were aligned against the COG database (94) and classified into 21 categories based on COG annotation (Figure 4F, G and Supplementary Figure S40). In the Bsu06560-knockout group, the ‘Amino acid transport and metabolism (E)’ had 96 up-regulated DE genes and 67 down-regulated DE genes (Figure 4G). Moreover, the ‘Transcription (K)’ group had significantly more up-regulated DE genes (89 genes) than down-regulated DE genes (34 genes, Figure 4G). However, the ‘Translation, ribosomal structure, and biogenesis (J)’ category had significantly more down-regulated (62 genes) than up-regulated genes (15 genes, Figure 4G). These results were consistent with the above GO analysis results, implied that Bsu06560 played an important role in the transcription and translation processes. Especially, the defects of Bsu06560 can impede the expression of related genes in translation and ribosomal biogenesis processes (Figure 4E). Furthermore, the ‘General functional prediction only (R)’ was the largest group in both BS168 versus BS-F91L and BS168 versus BS-Q150W groups, followed by ‘Carbohydrate transport and metabolism (G)’, ‘Amino acid transport and metabolism (E)’ and ‘Function unknown (S)’. The ‘Transcription (K)’ and ‘Translation, ribosomal structure, and biogenesis (J)’ categories had more up-regulated DE genes than down-regulated genes in both BS168 vs. BS-F91L and BS168 versus BS-Q150W groups (Figure 4F and Supplementary Figure S40). In the BS168 versus BS-F91L group, 106 of 131 and 56 of 73 DE genes were up-regulated in the ‘Transcription (K)’ and ‘Translation, ribosomal structure, and biogenesis (J)’ categories, respectively (Supplementary Figure S40). Moreover, 96 of 126 and 93 of 104 DE genes in the BS168 versus BS-Q150W group were up-regulated in the ‘Transcription (K)’ and ‘Translation, ribosomal structure, and biogenesis (J)’ categories, respectively (Figure 4F). Notably, these two mutants were more active in translation and ribosomal biogenesis processes, contrary to the Bsu06560-knockout group. Transcription and translation are closely coupled processes in the main flow of gene expression. Therefore, these results indicate that the enhancement of Bsu06560 activity promotes gene expression in B. subtilis, consistent with our global gene expression findings.
Besides, the KEGG analysis can identify the most essential biochemical metabolic pathways and signal transduction pathways associated with DE genes. Herein, KEGG analysis enriched 619, 681 and 690 DE genes in BS168 versus BS-KO, BS168 versus BS-F91L and BS168 versus BS-Q150W groups, respectively, and were classified into five parts of pathways (Supplementary Figure S41–S43 and Dataset S10–S12). The most abundant KEGG pathways in the BS168 versus BS-KO group were: biosynthesis of secondary metabolites, followed by biosynthesis of amino acids, ribosome, quorum sensing, starch and sucrose metabolism and cysteine and methionine metabolism (Supplementary Figure S43). The most enriched KEGG pathways in the BS168 versus BS-F91L group were biosynthesis of secondary metabolites, followed by ABC transporters, quorum sensing, pyruvate metabolism, citrate cycle (TCA cycle), histidine metabolism, and bacterial chemotaxis (Supplementary Figure SSupplementary Figure S41). The most abundant KEGG pathways in the BS168 versus BS-Q150W group were metabolic pathways, followed by microbial metabolism in diverse environments, carbon metabolism, ribosome, quorum sensing, starch, and sucrose metabolism and phosphotransferase system (PTS) (Supplementary Figure S42). Other interesting pathways enriched in BS168 versus BS-F91L and BS168 versus BS-Q150W groups included glyoxylate and dicarboxylate metabolism, inositol phosphate metabolism, non-ribosomal peptide structures, terpenoid backbone biosynthesis, and biosynthesis of siderophore group non-ribosomal peptides (Supplementary Figures S41 and S42). Overall, most DE genes in Bsu06560-defected and Bsu06560-mutation groups were enriched in the metabolic pathways and biosynthesis of metabolites, and others in metabolism categories related to the metabolism of basic components. Particularly, all the three comparative groups had DE genes enriched in the quorum-sensing category. This is the first report of such an association.
Quantitative real-time PCR analysis reveals the important roles of Bsu06560 in the regulation of central-dogma related genes
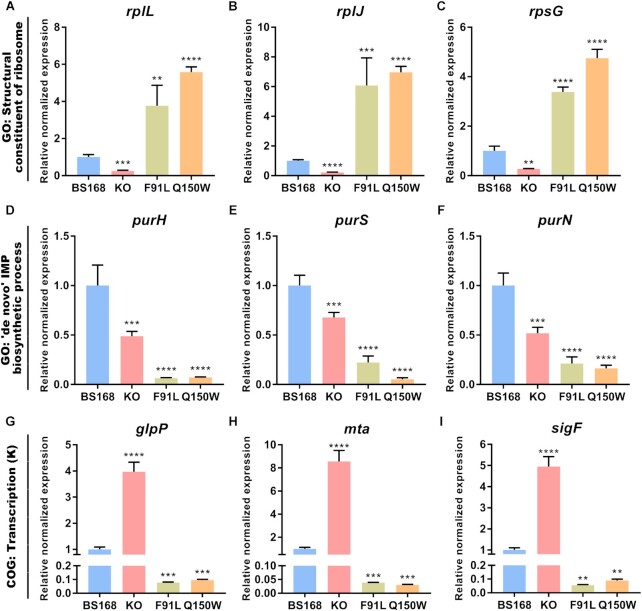
To validate the reproducibility and repeatability of DE genes identified from transcriptome sequencing, this study selected 50 genes from previous enrichment analysis, most of which were central-dogma related terms. The correlation was measured via scatter plots of log2 fold changes between RNA-Seq and RT-qPCR results. A high correlation was observed between the RNA-Seq and RT-qPCR results (Supplementary Figure S44). The RNA-Seq results showed that most of these genes were significantly differentially expressed and were consistently up-regulated or down-regulated with the gene expression profiling based on RNA-Seq, suggesting that the DE genes obtained from transcriptome sequencing were reliable. Furthermore, several attractive genes showed consistent expression trends (Figure 5). In structural constituent of ribosome term (Figure 5A to C), the expression of rplL, rplJ and rpsG genes were significantly higher in the BS-F91L and BS-Q150W strains than in the wild-type strain. In contrast, these genes were substantially down-regulated in the Bsu06560-knockout strain. In ‘de novo’ IMP biosynthetic process term (Figure 5D–F and Supplementary Figures S45A and S45B), purH, purS, purN, purD and purM genes were significantly down-regulated in all the three variants, especially in the BS-F91L and BS-Q150W strains. Moreover, in the COG ‘Transcription (K)’ term (Figure 5G to I and Supplementary Figures S45C and S45D), the expression of glpP, mta, yvdE, sigF and melR genes were significantly higher in the Bsu06560-knocked strain than in the wild-type strain. However, these genes were significantly down-regulated in N6-methyladenosine deamination-enhanced strains (BS-F91L, BS-Q150W). These results were consistent with the RNA-Seq data, verifying that the RNA-Seq was reliable. The significantly different expression pattern of B. subtilis 168 variants also highlighted the special roles of Bsu06560 protein, indicating that its mutations or defects can influence several genes, especially those related to central-dogma processes.
Figure 5.
Validation of transcriptome sequencing results via RT-qPCR. The DE genes of variants showed different expression patterns in different GO and COG categories. (A–C) Expression levels of rplL, rplJ, and rpsG genes involved in the ‘structural constituent of ribosome’ GO term. The BS-F91L and BS-Q150W strains had higher gene expression levels than BS168. However, BS-KO strain was down-regulated among these genes. (D–F) Expression levels of purH, purS and purN genes involved in the ‘‘de novo’ IMP biosynthetic process’ GO term. The purH, purS and purN genes were down-regulated in the BS-KO, BS-F91L and BS-Q150W strains. (G–I) Expression levels of glpP, mta and sigF genes involved in the ‘Transcription (K)’ COG term. The BS-KO strain had significantly higher expression levels than BS168. In contrast, these genes were significantly downregulated in BS-F91L and BS-Q150W strains. The rpoB gene was used as an internal control. The relative quantity of gene expression (fold change, Y-axis) of each gene was calculated via the comparative 2−ΔΔCt method. All gene expression levels were normalized to that of the BS168. The average of three biological replicates was used (n = 3). The error bars represent ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus BS168.
DISCUSSION
This study identified a multifunctional adenosine deaminase Bsu06560 capable of deaminating N6-methyladenosine using bioinformatics approaches and biochemical characterizations. Modeling, molecular docking, and mutational screening of Bsu06560 revealed that Phe91 and Gln150 play a critical role in N6-methyl-purine targeting via Bsu06560. The protein variant F91L increased the conversion ratio of N6-methyladenosine by ∼8-fold than N6-methyladenine, while Q150W enhanced the conversion ratio of N6-methyladenine by ∼7-fold than adenine, compared to the wild-type Bsu06560. These results further confirmed the imperative role of Phe91 and Gln150 in regulating the deamination activity of Bsu06560 for N6-methyl-purines, which was also supported by the QM/MM calculations. Furthermore, to explore the function of Bsu06560 in vivo, the dual-plasmids CRISPR-Cas9 system was successfully applied for genome editing of gram-positive B. subtilis 168 and created BS-KO, BS-F91L and BS-Q150W mutant strains. Differential gene expression analyses revealed that BS-F91L and BS-Q150W strains had a considerably higher expression level than the wild-type and Bsu06560-knockout B. subtilis 168, possibly due to the decreased m6A mis-incorporation rate in RNA. The GO, COG, KEGG and RT-qPCR analyses summarized the relevant gene functions and biological pathways, indicating that Bsu06560 is closely associated with nucleoside metabolism, influencing transcription, translation, metabolites regulations, and cellular signal transduction (Supplementary Figure S46). To the best of our knowledge, this is the first study to unveil the association between N6-methyladenosine metabolism and bacterial processes at the transcriptomic level.
These results also suggested that wild-type Bsu06560 accepts both N6-methyl purines and non-substituted aminopurines as alternative substrates to yield the deamination products, whereas providing higher activities for non-substituted aminopurines substrates than their corresponding N6-methyl purines (Supplementary Figure S8). However, these results differed from the previous work, which annotated Bsu06560 as an N6-methyladenine deaminase (35). These differences may be due to the discrimination in the UV spectrum used for hypoxanthine detection in the previous study. This study used LC-HRMS to precisely monitor the formation of products. Additionally, previous work did not test the Bsu06560 deamination activities for adenosine and N6-methyladenosine. Herein, Bsu06560 provided higher activities for ribose-containing adenosine and N6-methyladenosine than their corresponding adenine bases. Although the turnover of non-substituted aminopurines was much higher than that of N6-methyl purines in the biological system, it could be reasonable because such difference in activity of Bsu06560 may reflect its biological function to maintain the homeostasis between N6-methyl purines and non-substituted aminopurines via an activity-differentiation based strategy. Moreover, the comparison of the adenosine and the adenine deaminases in previous work illustrated the evolution of the adenine deaminase family in which the approximate partition of specificity and catalysis between structurally distinct regions was a convenient device for their evolution and thus might generate diversity in a combinatorial manner (30). As a result, Bsu06560 is highly similar to the Agrobacterium tumefaciens adenine deaminase (atuADE, Supplementary Figure S6), but it functions better performed as an adenosine deaminase rather than an adenine deaminase.
The analysis of the m6A/A ratio of BS168 and its variants showed that Bau06560 could serve as a multifunctional enzyme to maintain the concentration balance of adenosine and N6-methyladenosine. Besides, its deaminase activity for N6-methyladenosine can protect cellular processes from excess N6-methyl purines (Supplementary Figure S33). Similarly, several previous studies on ADAL (34) sharing the same family of Bsu06560, and some enzymes related to m6A mis-incorporation pathways showed similar results. For instance, previous studies showed that the N6-methyladenosine nucleotide in nucleotides pool can increase the error rate of RNA synthesis via RNA polymerase, which affects the transcription process (95). As a result, nature may only need to adopt a multifunctional enzyme, such as Bsu06560, from the general purine metabolic pathway for epigenetic N6-methyl purine metabolism by evolving and modulating only specific critical residues, which is highly efficient and avoiding the need for the specific type of enzyme evolution. Notably, the function of Bsu06560 to deaminate adenosine is conserved in many eukaryotes and prokaryotes, suggesting that Bsu06560 could be essential for normal physiological processes of B. subtilis. Therefore, numerous biological pathways may respond to genome editing of these key residues, including F91L and Q150W mutations in Bsu06560 (Figure 4B and C). However, it is often difficult to ascertain the critical residues of proteins for protein engineering. Moreover, the method that regulates intracellular adenosine-related enzyme activities through key residues mutation could help in evaluating adenosine metabolism due to its receptor-independent manner. Therefore, these mechanistic insights may enhance rational drug design targeting both adenosine and epigenetic N6-methyladenosine metabolism.
Moreover, the global gene expression analysis of B. subtilis 168 and its variants showed that the F91L and Q150W mutants exhibited significantly higher gene expression levels than the wide-type and Bsu06560-knockout strain (Figure 4A). Meanwhile, the F91L and Q150W mutants had a significantly lower m6A/A ratio than the wild-type strain. In contrast, the m6A/A and N6-mATP/ATP ratio were significantly elevated in Bsu06560-knockout strain (Supplementary Figures S31A and S32B). These results highlighted the notion that the RNA metabolism processes might be crucial for the regulation of gene expression pattern, although could not be directly verified due to experimental constraints. However, previous studies on the similar N6-methly deaminase (ADALs) have shown that the MAPDA, an N6-AMP deaminase from Arabidopsis thaliana, can act as a protection factor by preventing the random re-entry of non-canonical nucleotides stemming from degraded RNA into newly synthesized RNA (34). In another research, the results of isotope doping experiments also indicated that N6-mATP, processed via the nucleotide salvage pathway from the free N6-methyladenosine, can be mis-incorporated into genomic DNA by DNA polymerases through ADAL knockdown (21). These conclusions agree with the model that Bsu06560 act as a regulator in cellular purine metabolism (Supplementary Figure S33).
Furthermore, enhancement of N6-methyladenosine deamination activity of Bsu06560 by mutating only one codon influenced gene expression at multiple layers, including promotion of transcription and translation of genes, regulation of small molecular metabolites, cellular signal transduction, and substance transportation. The GO analysis showed that the BS-Q150W mutant exhibited a significant association with transcription and translation processes compared with BS-F91L (Supplementary Figures S34 and S35). To a certain extent, these results were consistent with the findings of activity studies of Bsu06560 mutants, indicating that Q150W has a higher N6-methyladenosine deamination activity than F91L. Remarkably, a batch of DE genes was annotated to the quorum-sensing pathway, which could be used to orchestrate bacterial gene expression machinery that underlies collective behaviors (Supplementary Figure S41 to S43) (96). A recent study revealed that microbes utilize a process of cell-cell communication, quorum sensing to initiate and perpetuate infectious diseases and escape defense systems in eukaryotes, increasing bacterial resistance to antibiotics (97). Therefore, these results may provide insights into the regulation of silencing genes, providing a new strategy for discovering and developing anti-bacterial drugs. Moreover, in the environment information processing category, 76 DE genes of the BS-F91L group were enriched in the ABC transporters pathway, and 20 DE genes of the BS-Q150W group were enriched in the phosphotransferase system (PTS) pathway (Supplementary Figures S41 and S42). ATP-binding cassette (ABC) transporters in membranes of prokaryotes and eukaryotes play crucial roles in biological processes, including nutrient uptake, pathogenicity, and innate immunes escape (98). Recent studies have revealed that the engineering of ABC transporters may control the plasma membrane permeability of microorganisms and regulate the energy system within cells, providing strategies for optimizing industrial strains growth, substrate utilization, and strain productivity (99,100).
The RT-qPCR analysis revealed that several fascinating DE genes had highly consistency in expression levels. The genes rplL and rplJ (Figure 5A and B), constituting rplJL operon, are essential in Bacillus subtilis and encode the L10(L12)4 complex, which can bind to a kink-turn structure motif in 23S rRNA during ribosome biogenesis or target mRNA for regulation (101,102). When L10(L12)4 bind to rplJL leader transcript, the transcription is auto-regulated via a transcription attenuation mechanism, where rplJL mRNA binds to its product, L10(L12)4 complex, and stabilizes an anti-anti-terminator structure of the transcription, thereby preventing the formation of the anti-terminator structure, and promoting transcription termination (102,103). When rplJL operon does not bind to L10(L12)4 complex, the formation of the anti-terminator structure of the mRNA leader allows transcription readthrough (102). Therefore, the specific conversion of different secondary structures within the leader transcript is critical in L10(L12)4 recognition. The m6A modification can regulate the reconfiguration of RNA structures, thus affecting RNA–protein interactions via ‘m6A-switch’ mechanisms (6,104). Besides, m6A is an abundant RNA modification in B. subtilis (3). Therefore, m6A modification could be the reason that initiate the structure conversion of rplJL operon. Interestingly, the gene expression patterns of the variants were consistent with our assumption. The F91L and Q150W mutants had much lower m6A modification ratio, promoting the formation of anti-terminator structure and increasing the expression of rplL and rplJ (Figure 5A and B). In contrast, Bsu06560-defected strain had a higher m6A modification ratio and showed the opposite effect for rplJL operon, thus repressing its transcription. However, more critical and direct proofs are needed to ascertain the assumption. Moreover, the gene rpsG (Figure 5C), encoding the ribosomal protein S7 in B. subtilis and playing an essential role in translation (101,105), showed similar expression patterns as rplL and rplJ. The genes purH, purS, purN, purD and purM (Figure 5D to F and Supplementary Figures S45A and S45B) were involved in regulating purR regulon and encoding purine biosynthetic enzymes within the de novo purine biosynthesis pathway (106,107). All variants exhibited significantly lower gene expression levels, especially in the N6-methyladenosine deamination enhanced strains, than the wild-type strain. These results indicate the important roles of Bsu06560 in purine biosynthesis processes and purine metabolism. Furthermore, the genes glpP, mta, yvdE and melR encoded transcriptional regulators that could regulate a batch of genes (108–111). These genes were all enriched in the COG ‘Transcription (K)’ term and were highly expressed in the Bsu06560-disrupted strains (Figure 5G to I and Supplementary Figures S45C and S45D). In contrast, these genes had a significantly low expression in Bsu06560 mutant strains. Similarly, the gene sigF (sigma factor σF), the first forespore-specific transcription factor in B. subtilis controlling genes required for the early stages of prespore development (112), showed opposite expression patterns in Bsu06560-disrupted and mutated strains. The sigF is active only in the prespore, and its activation in the prespore is crucial for successful sporulation because it regulates other sigma factors of sporulation (113–115). These results reveal that Bsu06560 can play an important role in transcription and cell development. However, more evidence is needed to explain the results.
Taken together, the transcriptional profiling and RT-qPCR results indicate that N6-methyladenosine metabolism by Bsu06560 deaminase plays a significant role in translation, transcription, metabolite regulations and biosynthesis, transportation and cellular signal transduction. Moreover, N6-methyladenosine metabolism can elicit a significant influence on bacterial biosynthesis of several bioactive molecules, which were completely novel and promising in synthetic biology. Notably, only one codon altered in Bsu06560 gene of B. subtilis 168 genome can regulate numerous genes. An illustration that the molecular docking results and discovered key residues are crucial for the deamination activity of Bsu06560. Adenosine metabolism has been explored and translated as a tractable therapeutic target for cancer treatment. However, the lack of the distinction and association between adenosine receptor-dependent and -independent pathways of adenosines limits the development of more effective adenosine-based therapeutics (36). The method described in this study (fine-tuning the catalytic performance of enzymes through key mutations) can provide adenosine-based therapeutics independent of the adenosine receptors.
Bacillus subtilis is a well-studied model organism for molecular research and an important host for industrial production. Therefore, understanding the N6-methyladenosine molecular metabolism involved in bacterial posttranscriptional modifications may facilitate the development of novel engineering approaches for the global optimization of metabolism, which may result in more extensive regulation compared with those achieved by merely varying few enzymes abundance via controlling enzymes expression pathways (116). Indeed, effective gene expression and regulation are essential for industrial applications (117). The previously employed expressional regulatory systems were mainly based on variable regulatory elements, including gene copy number and promoters (118–120), or synthetic RNA devices, including siRNAs and the CRISPR-Cas9/CRISPRi system (70,121). However, most of these systems were established for single or very few gene expressions (117). Fine-modulation of epigenetic nucleotides-related enzymes can result in a batch variation of gene expression, globally regulating metabolic pathways and numerous cellular processes. Therefore, this study can provide insights for optimizing metabolic engineering strategies, improving the performance of industrial strains, and the bacterial signal transduction process, which have not yet been associated with the metabolism of N6-methyladenosine nucleoside.
DATA AVAILABILITY
Raw and processed RNA-seq data can be obtained in the GEO database under accession code GSE179533. Model of Bsu06560 can be obtained in ModelArchive database after publication.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to Guimin Zhang's group (College of Life Science, Hubei University, Wuhan, Hubei, China) for kindly providing the plasmids of pHT01-Cas9 and pHYT-BsmBI-sgRNA and Xiangdong Chen's group (College of Life Science, Wuhan University, Wuhan, Hubei, China) for kindly providing the Bacillus subtilis (strain 168) strain and beneficial help.
Contributor Information
Zhuoran Jiang, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
Chao Wang, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
Zixin Wu, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
Kun Chen, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
Wei Yang, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
Hexiang Deng, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
Heng Song, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
Xiang Zhou, The Institute of Advanced Studies, and Key Laboratory of Biomedical Polymers-Ministry of Education, College of Chemistry and Molecular Sciences, Wuhan University, 40072 Wuhan, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFA0903200 to H.S.]; National Natural Science Foundation of China [91753201 and 21721005 to X.Z. and 31900033 to H.S]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M.et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 2. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deng X., Chen K., Luo G.Z., Weng X., Ji Q., Zhou T., He C.. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015; 43:6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frye M., Harada B.T., Behm M., He C.. RNA modifications modulate gene expression during development. Science. 2018; 361:1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang H., Weng H., Chen J.. 2020) m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 37:270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L.. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2047–E2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 9. Hoernes T.P., Clementi N., Faserl K., Glasner H., Breuker K., Lindner H., Huttenhofer A., Erlacher M.D.. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016; 44:852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trinquier A., Durand S., Braun F., Condon C.. Regulation of RNA processing and degradation in bacteria. Biochim. Biophys. Acta Gene Regul. Mech. 2020; 1863:194505. [DOI] [PubMed] [Google Scholar]
- 11. Vargas-Blanco D.A., Shell S.S.. Regulation of mRNA stability during bacterial stress responses. Front. Microbiol. 2020; 11:2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen K., Zhao B.S., He C.. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol. 2016; 23:74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K.et al.. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Konno M., Taniguchi M., Ishii H.. Significant epitranscriptomes in heterogeneous cancer. Cancer Sci. 2019; 110:2318–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Y., Hsu P.J., Chen Y.S., Yang Y.G.. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018; 28:616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiao C.L., Zhu S., He M., Chen D., Zhang Q., Chen Y., Yu G., Liu J., Xie S.Q., Luo F.et al.. N(6)-methyladenine DNA modification in the human genome. Mol. Cell. 2018; 71:306–318. [DOI] [PubMed] [Google Scholar]
- 18. Hao Z., Wu T., Cui X., Zhu P., Tan C., Dou X., Hsu K.W., Lin Y.T., Peng P.H., Zhang L.S.et al.. N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell. 2020; 78:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z., Zhao P., Xia Q.. Epigenetic methylations on N6-adenine and N6-adenosine with the same Input but different output. Int. J. Mol. Sci. 2019; 20:2931–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson C., Chen P.J., Miao Z., Liu D.R.. Programmable m(6)A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat. Biotechnol. 2020; 38:1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Musheev M.U., Baumgartner A., Krebs L., Niehrs C.. The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 2020; 16:630–634. [DOI] [PubMed] [Google Scholar]
- 22. Nyhan W.L. Nucleotide Synthesis via Salvage Pathway. eLS. 2014; 10.1002/9780470015902.a0001399.pub3. [DOI] [Google Scholar]
- 23. Pedley A.M., Benkovic S.J.. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 2017; 42:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robak P., Robak T.. Older and new purine nucleoside analogs for patients with acute leukemias. Cancer Treat. Rev. 2013; 39:851–861. [DOI] [PubMed] [Google Scholar]
- 25. Yoshida G.J. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015; 34:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murray A.W. The biological significance of purine salvage. Annu. Rev. Biochem. 1971; 40:811–826. [DOI] [PubMed] [Google Scholar]
- 27. Berg M., Van der Veken P., Goeminne A., Haemers A., Augustyns K.. Inhibitors of the purine salvage pathway: a valuable approach for antiprotozoal chemotherapy. Curr. Med. Chem. 2010; 17:2456–2481. [DOI] [PubMed] [Google Scholar]
- 28. Maier S.A., Galellis J.R., McDermid H.E.. Phylogenetic analysis reveals a novel protein family closely related to adenosine deaminase. J. Mol. Evol. 2005; 61:776–794. [DOI] [PubMed] [Google Scholar]
- 29. Maguire M.H., Lukas M.C., Rettie J.F.. Adenine nucleotide salvage synthesis in the rat heart; pathways of adnosine salvage. Biochim. Biophys. Acta. 1972; 262:108–115. [DOI] [PubMed] [Google Scholar]
- 30. Ribard C., Rochet M., Labedan B., Daignan-Fornier B., Alzari P., Scazzocchio C., Oestreicher N.. Sub-families of alpha/beta barrel enzymes: a new adenine deaminase family. J. Mol. Biol. 2003; 334:1117–1131. [DOI] [PubMed] [Google Scholar]
- 31. Schinkmanova M., Votruba I., Holy A.. N6-methyl-AMP aminohydrolase activates N6-substituted purine acyclic nucleoside phosphonates. Biochem. Pharmacol. 2006; 71:1370–1376. [DOI] [PubMed] [Google Scholar]
- 32. Schinkmanová M., Votruba I., Shibata R., Han B., Liu X., Cihlar T., Holý A.. Human N6-methyl-AMP/DAMP aminohydrolase (abacavir 5′-monophosphate deaminase) is capable of metabolizing N6-substituted purine acyclic nucleoside phosphonates. Collect. Czech. Chem. Commun. 2008; 73:275–291. [Google Scholar]
- 33. Murakami E., Bao H., Mosley R.T., Du J., Sofia M.J., Furman P.A.. Adenosine deaminase-like protein 1 (ADAL1): characterization and substrate specificity in the hydrolysis of N(6)- or O(6)-substituted purine or 2-aminopurine nucleoside monophosphates. J. Med. Chem. 2011; 54:5902–5914. [DOI] [PubMed] [Google Scholar]
- 34. Chen M., Urs M.J., Sanchez-Gonzalez I., Olayioye M.A., Herde M., Witte C.P.. m(6)A RNA degradation products are catabolized by an evolutionarily conserved N(6)-methyl-AMP deaminase in plant and mammalian cells. Plant Cell. 2018; 30:1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kamat S.S., Fan H., Sauder J.M., Burley S.K., Shoichet B.K., Sali A., Raushel F.M.. Enzymatic deamination of the epigenetic base N-6-methyladenine. J. Am. Chem. Soc. 2011; 133:2080–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boison D., Yegutkin G.G.. Adenosine metabolism: emerging concepts for cancer therapy. Cancer Cell. 2019; 36:582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Virgilio F., Sarti A.C., Falzoni S., De Marchi E., Adinolfi E.. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat. Rev. Cancer. 2018; 18:601–618. [DOI] [PubMed] [Google Scholar]
- 38. Ralevic V., Burnstock G.. Receptors for purines and pyrimidines. Pharmacol. Rev. 1998; 50:413–492. [PubMed] [Google Scholar]
- 39. Yegutkin G.G. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014; 49:473–497. [DOI] [PubMed] [Google Scholar]
- 40. Hasko G., Antonioli L., Cronstein B.N.. Adenosine metabolism, immunity and joint health. Biochem. Pharmacol. 2018; 151:307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boswell-Casteel R.C., Hays F.A.. Equilibrative nucleoside transporters-A review. Nucleosides. Nucleotides. Nucleic. Acids. 2017; 36:7–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Apasov S.G., Blackburn M.R., Kellems R.E., Smith P.T., Sitkovsky M.V.. Adenosine deaminase deficiency increases thymic apoptosis and causes defective T cell receptor signaling. J. Clin. Invest. 2001; 108:131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cassani B., Mirolo M., Cattaneo F., Benninghoff U., Hershfield M., Carlucci F., Tabucchi A., Bordignon C., Roncarolo M.G., Aiuti A.. Altered intracellular and extracellular signaling leads to impaired T-cell functions in ADA-SCID patients. Blood. 2008; 111:4209–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cekic C., Linden J.. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016; 16:177–192. [DOI] [PubMed] [Google Scholar]
- 45. Tardif V., Muir R., Cubas R., Chakhtoura M., Wilkinson P., Metcalf T., Herro R., Haddad E.K.. Adenosine deaminase-1 delineates human follicular helper T cell function and is altered with HIV. Nat. Commun. 2019; 10:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Franco R., Pacheco R., Gatell J.M., Gallart T., Lluis C.. Enzymatic and extraenzymatic role of adenosine deaminase 1 in T-cell-dendritic cell contacts and in alterations of the immune function. Crit. Rev. Immunol. 2007; 27:495–509. [DOI] [PubMed] [Google Scholar]
- 47. Pacheco R., Martinez-Navio J.M., Lejeune M., Climent N., Oliva H., Gatell J.M., Gallart T., Mallol J., Lluis C., Franco R.. CD26, adenosine deaminase, and adenosine receptors mediate costimulatory signals in the immunological synapse. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:9583–9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martinez-Navio J.M., Climent N., Pacheco R., Garcia F., Plana M., Nomdedeu M., Oliva H., Rovira C., Miralles L., Gatell J.M.et al.. Immunological dysfunction in HIV-1-infected individuals caused by impairment of adenosine deaminase-induced costimulation of T-cell activation. Immunology. 2009; 128:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Porter D.J., Spector T.. Alternative substrates for calf intestinal adenosine deaminase. A pre-steady-state kinetic analysis. J. Biol. Chem. 1993; 268:2480–2485. [PubMed] [Google Scholar]
- 50. Sideraki V., Wilson D.K., Kurz L.C., Quiocho F.A., Rudolph F.B.. Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation. Biochemistry. 1996; 35:15019–15028. [DOI] [PubMed] [Google Scholar]
- 51. Nallamsetty S., Kapust R.B., Tozser J., Cherry S., Tropea J.E., Copeland T.D., Waugh D.S.. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr. Purif. 2004; 38:108–115. [DOI] [PubMed] [Google Scholar]
- 52. Lebendiker M., Danieli T.. Walls D., Loughran S.T.. Protein Chromatography: Methods and Protocols. 2011; Totowa, NJ: Humana Press; 281–293. [Google Scholar]
- 53. Kamat S.S., Bagaria A., Kumaran D., Holmes-Hampton G.P., Fan H., Sali A., Sauder J.M., Burley S.K., Lindahl P.A., Swaminathan S.et al.. Catalytic mechanism and three-dimensional structure of adenine deaminase. Biochemistry. 2011; 50:1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kamat S.S., Holmes-Hampton G.P., Bagaria A., Kumaran D., Tichy S.E., Gheyi T., Zheng X., Bain K., Groshong C., Emtage S.et al.. The catalase activity of diiron adenine deaminase. Protein Sci. 2011; 20:2080–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Elisabeth Gasteiger C.H., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A.. The Proteomics Protocols Handbook. 2005; Totowa, NJ: Humana Press Inc. [Google Scholar]
- 56. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008; 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roy A., Kucukural A., Zhang Y.. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010; 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y.. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015; 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang J., Zhang Y.. 2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43:W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Studer G., Rempfer C., Waterhouse A.M., Gumienny R., Haas J., Schwede T.. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics. 2020; 36:2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Uziela K., Shu N., Wallner B., Elofsson A.. ProQ3: improved model quality assessments using Rosetta energy terms. Sci. Rep. 2016; 6:33509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Uziela K., Menendez Hurtado D., Shu N., Wallner B., Elofsson A.. ProQ3D: improved model quality assessments using deep learning. Bioinformatics. 2017; 33:1578–1580. [DOI] [PubMed] [Google Scholar]
- 63. Shivakumar D., Williams J., Wu Y., Damm W., Shelley J., Sherman W.. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010; 6:1509–1519. [DOI] [PubMed] [Google Scholar]
- 64. Ma J.C., Dougherty D.A.. The cationminus signpi interaction. Chem. Rev. 1997; 97:1303–1324. [DOI] [PubMed] [Google Scholar]
- 65. Murphy R.B., Philipp D.M., Friesner R.A.. A mixed quantum mechanics/molecular mechanics (QM/MM) method for large-scale modeling of chemistry in protein environments. J. Comput. Chem. 2000; 21:1442–1457. [Google Scholar]
- 66. Murphy R.B., Philipp D.M., Friesner R.A.. Frozen orbital QM/MM methods for density functional theory. Chem. Phys. Lett. 2000; 321:113–120. [Google Scholar]
- 67. Bochevarov A.D., Harder E., Hughes T.F., Greenwood J.R., Braden D.A., Philipp D.M., Rinaldo D., Halls M.D., Zhang J., Friesner R.A.. Jaguar: a high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013; 113:2110–2142. [Google Scholar]
- 68. Banks J.L., Beard H.S., Cao Y., Cho A.E., Damm W., Farid R., Felts A.K., Halgren T.A., Mainz D.T., Maple J.R.et al.. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 2005; 26:1752–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. So Y., Park S.Y., Park E.H., Park S.H., Kim E.J., Pan J.G., Choi S.K.. A highly efficient CRISPR-Cas9-mediated large genomic deletion in Bacillus subtilis. Front Microbiol. 2017; 8:1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lu Z., Yang S., Yuan X., Shi Y., Ouyang L., Jiang S., Yi L., Zhang G.. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis. Nucleic Acids Res. 2019; 47:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Concordet J.P., Haeussler M.. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018; 46:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xue G.-P., Johnson J.S., Dalrymple B.P.. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J. Microbiol. Methods. 1999; 34:183–191. [Google Scholar]
- 73. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011; 2011; 17:3. [Google Scholar]
- 74. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U.et al.. N(6)-methyladenine DNA Modification in Glioblastoma. Cell. 2018; 175:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bajad S.U., Lu W., Kimball E.H., Yuan J., Peterson C., Rabinowitz J.D.. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A. 2006; 1125:76–88. [DOI] [PubMed] [Google Scholar]
- 78. Jayaraj R.L., Narchi H., Subramanian R., Yuvaraju P.. Development and validation of LC-MS/MS method for quantification of ATP, ADP and AMP in dried blood spot, liver and brain of neonate mice pups. Results Chem. 2021; 3:100172–100179. [Google Scholar]
- 79. Wilson P.M., Labonte M.J., Russell J., Louie S., Ghobrial A.A., Ladner R.D.. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res. 2011; 39:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thompson J.D., Higgins D.G., Gibson T.J.. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wu B., Zhang D., Nie H., Shen S., Li Y., Li S.. Structure of Arabidopsis thaliana N(6)-methyl-AMP deaminase ADAL with bound GMP and IMP and implications for N(6)-methyl-AMP recognition and processing. RNA Biol. 2019; 16:1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hwang J., Krebs C., Huynh B.H., Edmondson D.E., Theil E.C., Penner-Hahn J.E.. A short Fe-Fe distance in peroxodiferric ferritin: control of Fe substrate versus cofactor decay?. Science. 2000; 287:122–125. [DOI] [PubMed] [Google Scholar]
- 83. Schwartz J.K., Liu X.S., Tosha T., Theil E.C., Solomon E.I.. Spectroscopic definition of the ferroxidase site in M ferritin: comparison of binuclear substrate vs cofactor active sites. J. Am. Chem. Soc. 2008; 130:9441–9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jasniewski A.J., Que L. Jr. Dioxygen activation by nonheme diiron enzymes: diverse dioxygen adducts, high-valent intermediates, and related model complexes. Chem. Rev. 2018; 118:2554–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G.et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bartosovic M., Molares H.C., Gregorova P., Hrossova D., Kudla G., Vanacova S.. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017; 45:11356–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Smith M.B. Organic Chemistry: An Acid-Base Approach. 2010; CRC Press. [Google Scholar]
- 88. Ayora S., Carrasco B., Cardenas P.P., Cesar C.E., Canas C., Yadav T., Marchisone C., Alonso J.C.. Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol. Rev. 2011; 35:1055–1081. [DOI] [PubMed] [Google Scholar]
- 89. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang K., Duan X., Wu J.. Multigene disruption in undomesticated Bacillus subtilis ATCC 6051a using the CRISPR/Cas9 system. Sci. Rep. 2016; 6:27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Visvanathan A., Somasundaram K.. mRNA traffic control reviewed: N6-methyladenosine (m(6) A) takes the driver's seat. Bioessays. 2018; 40:1700093. [DOI] [PubMed] [Google Scholar]
- 92. Nye T.M., van Gijtenbeek L.A., Stevens A.G., Schroeder J.W., Randall J.R., Matthews L.A., Simmons L.A.. Methyltransferase DnmA is responsible for genome-wide N6-methyladenosine modifications at non-palindromic recognition sites in Bacillus subtilis. Nucleic Acids Res. 2020; 48:5332–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tian S., Lai J., Yu T., Li Q., Chen Q.. Regulation of gene expression associated with the N6-methyladenosine (m6A) enzyme system and its significance in cancer. Front. Oncol. 2020; 10:623634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Galperin M.Y., Wolf Y.I., Makarova K.S., Vera Alvarez R., Landsman D., Koonin E.V.. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021; 49:D274–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Potapov V., Fu X., Dai N., Correa I.R. Jr, Tanner N.A., Ong J.L.. Base modifications affecting RNA polymerase and reverse transcriptase fidelity. Nucleic Acids Res. 2018; 46:5753–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Papenfort K., Bassler B.L.. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016; 14:576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kalia V.C., Patel S.K.S., Kang Y.C., Lee J.K.. Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol. Adv. 2019; 37:68–90. [DOI] [PubMed] [Google Scholar]
- 98. Rice A.J., Park A., Pinkett H.W.. Diversity in ABC transporters: type I, II and III importers. Crit. Rev. Biochem. Mol. Biol. 2014; 49:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Khakhina S., Johnson S.S., Manoharlal R., Russo S.B., Blugeon C., Lemoine S., Sunshine A.B., Dunham M.J., Cowart L.A., Devaux F.et al.. Control of Plasma Membrane Permeability by ABC Transporters. Eukaryot Cell. 2015; 14:442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Qi Y., Liu H., Chen X., Liu L.. Engineering microbial membranes to increase stress tolerance of industrial strains. Metab. Eng. 2019; 53:24–34. [DOI] [PubMed] [Google Scholar]
- 101. Kobayashi K., Ehrlich S.D., Albertini A., Amati G., Andersen K.K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P.et al.. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yakhnin H., Yakhnin A.V., Babitzke P.. Ribosomal protein L10(L12)4 autoregulates expression of the Bacillus subtilis rplJL operon by a transcription attenuation mechanism. Nucleic Acids Res. 2015; 43:7032–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Iben J.R., Draper D.E.. Specific interactions of the L10(L12)4 ribosomal protein complex with mRNA, rRNA, and L11. Biochemistry. 2008; 47:2721–2731. [DOI] [PubMed] [Google Scholar]
- 104. Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T.. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Akanuma G., Nanamiya H., Natori Y., Yano K., Suzuki S., Omata S., Ishizuka M., Sekine Y., Kawamura F.. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J. Bacteriol. 2012; 194:6282–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Weng M., Nagy P.L., Zalkin H.. Identification of the Bacillus subtilis pur operon repressor. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:7455–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bera A.K., Zhu J., Zalkin H., Smith J.L.. Functional dissection of the Bacillus subtilis pur operator site. J. Bacteriol. 2003; 185:4099–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Darbon E., Servant P., Poncet S., Deutscher J.. Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpFK expression. Mol. Microbiol. 2002; 43:1039–1052. [DOI] [PubMed] [Google Scholar]
- 109. Schonert S., Seitz S., Krafft H., Feuerbaum E.A., Andernach I., Witz G., Dahl M.K.. Maltose and maltodextrin utilization by Bacillus subtilis. J. Bacteriol. 2006; 188:3911–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Miethke M., Schmidt S., Marahiel M.A.. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J. Bacteriol. 2008; 190:5143–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Morabbi Heravi K., Watzlawick H., Altenbuchner J.. The melREDCA operon encodes a utilization system for the raffinose family of oligosaccharides in Bacillus subtilis. J. Bacteriol. 2019; 201:e00109-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Overkamp W., Kuipers O.P.. Transcriptional profile of Bacillus subtilis sigF-mutant during vegetative growth. PLoS One. 2015; 10:e0141553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Phillips Z.E., Strauch M.A.. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci.: CMLS. 2002; 59:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Do J.H., Nagasaki M., Miyano S.. The systems approach to the prespore-specific activation of sigma factor SigF in Bacillus subtilis. Biosystems. 2010; 100:178–184. [DOI] [PubMed] [Google Scholar]
- 115. Fengos G., Iber D.. Prediction stability in a data-based, mechanistic model of sigmaF regulation during sporulation in Bacillus subtilis. Sci. Rep. 2013; 3:2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Liu Y., Li J., Du G., Chen J., Liu L.. Metabolic engineering of Bacillus subtilis fueled by systems biology: Recent advances and future directions. Biotechnol. Adv. 2017; 35:20–30. [DOI] [PubMed] [Google Scholar]
- 117. Gu Y., Xu X., Wu Y., Niu T., Liu Y., Li J., Du G., Liu L.. Advances and prospects of Bacillus subtilis cellular factories: from rational design to industrial applications. Metab. Eng. 2018; 50:109–121. [DOI] [PubMed] [Google Scholar]
- 118. Cress B.F., Trantas E.A., Ververidis F., Linhardt R.J., Koffas M.A.. Sensitive cells: enabling tools for static and dynamic control of microbial metabolic pathways. Curr. Opin. Biotechnol. 2015; 36:205–214. [DOI] [PubMed] [Google Scholar]
- 119. McNerney M.P., Watstein D.M., Styczynski M.P.. Precision metabolic engineering: The design of responsive, selective, and controllable metabolic systems. Metab. Eng. 2015; 31:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Radeck J., Kraft K., Bartels J., Cikovic T., Dürr F., Emenegger J., Kelterborn S., Sauer C., Fritz G., Gebhard S.et al.. The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. Journal of Biological Engineering. 2013; 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A.. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013; 41:7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed RNA-seq data can be obtained in the GEO database under accession code GSE179533. Model of Bsu06560 can be obtained in ModelArchive database after publication.