Abstract
Immune system gene regulation perturbation has been found to be a major cause of the development of various types of cancer. Numbers of mechanisms contribute to gene expression regulation, thus, systematically identification of potential regulons of immune-related pathways is critical to cancer immunotherapy. Here, we comprehensively chart the landscape of transcription factors, microRNAs, RNA binding proteins and long noncoding RNAs regulation in 17 immune-related pathways across 33 cancers. The potential immunology regulons are likely to exhibit higher expressions in immune cells, show expression perturbations in cancer, and are significantly correlated with immune cell infiltrations. We also identify a panel of clinically relevant immunology regulons across cancers. Moreover, the regulon atlas of immune-related pathways helps prioritizing cancer-related genes (i.e. ETV7, miR-146a-5p, ZFP36 and HCP5). We further identified two molecular subtypes of glioma (cold and hot tumour phenotypes), which were characterized by differences in immune cell infiltrations, expression of checkpoints, and prognosis. Finally, we developed a user-friendly resource, ImmReg (http://bio-bigdata.hrbmu.edu.cn/ImmReg/), with multiple modules to visualize, browse, and download immunology regulation. Our study provides a comprehensive landscape of immunology regulons, which will shed light on future development of RNA-based cancer immunotherapies.
INTRODUCTION
Immune system dysregulation has been found to be a major cause of the development of cancer (1,2). Immunotherapy has been emerged as a promising cancer treatment strategy (3). Knowledge underlying the dysregulation of immune-related gene expression is critical for generating a robust immunity and provides novel insights into cancer immunotherapy (3,4).
Numbers of mechanisms contribute to gene expression regulation and ensure transcriptional response to external signals (5). Important new insights have been gained in identifying the regulators of gene expression through the quantitative and computational analyses (5–7). Transcription factors (TFs) are key regulators of gene expression and several TFs have been demonstrated to play key roles in immune regulation. For example, HIF and the hypoxia pathway can control innate and adaptive immunity (8). The functional roles of IRF family of TFs are becoming clearer in immune responses and oncogenesis (9). Moreover, STAT3 has dual role in tumor inflammation and immunity and is a promising target for cancer therapy (10).
In addition to TFs, gene expression is found to be regulated by other regulators, such as microRNAs (miRNAs), RNA binding proteins (RBPs) and long non-coding RNAs (lncRNAs) (11–13). MiRNAs act by negatively regulating the expression of key immune-related genes, thus contributing important logic elements to the regulatory circuitry (14,15), such as miRNA-17-92 cluster (16). Moreover, several RBPs and lncRNAs were also revealed to play fundamental roles in immune systems (17). For example, RNA binding protein ZAP functioned as both a direct antiviral restriction factor and an interferon-resolution factor (18). One recent study has uncovered the role of YTHDF3 as a negative regulator of antiviral immunity through the translational promotion of FOXO3 mRNA (19). Moreover, lncRNAs NeST, NRON and LINK-A were also found to be critical for immune response regulation (20,21). We recently proposed ImmLnc to identify lncRNA regulons and help prioritize cancer-related lncRNAs (22). However, a comprehensive repertoire of immune regulators of immune-related pathways is still lack.
To fill this gap, we systematically identified the potential regulons (including TFs, miRNAs, RBPs and lncRNAs) of immune-related pathways across 33 cancer types. We found that immune regulons are likely to be highly expressed in immune cell populations, exhibited perturbed expression in cancer and are correlated with immune cell infiltration in cancer. These immune regulons help identify cancer-related genes and further identifying cancer subtypes with distinct clinical characteristics. Comprehensive knowledge of the complete repertoire of potential regulons of immune-related pathways is an important prerequisite for understanding the architecture of the regulatory network under tumor microenvironment.
MATERIALS AND METHODS
Immune-related pathways
We first collected 17 immune-related pathways from the ImmPort project (23), which were widely used in several immune-related studies (22,24). We mapped all genes to Ensembl IDs and obtained 1811 genes in 17 pathways.
Collection of TFs, miRNAs, RBPs and lncRNAs
The TF gene list was collected from several databases and recent literature, including AnimalTFDB v3.0 (25), TRRUST v2.0 (26), Hocomoco (27), TFcheckpoint v3.0 (28), HumanTFs (29) and Ravasi et al. (30). All TF symbols were mapped to Ensembl ID. In total, we obtained 3178 TFs in our analysis (Supplementary Table S1). For miRNAs, we mapped ID to the symbol of miRBase (V22) and obtained 2449 miRNAs in total. RNA binding proteins were obtained from one of our recent study (31), which manually curated the RBPs from recently published literature. The annotations of lncRNAs were downloaded from GENCODE. Moreover, we obtained the regulator-target information from hTFtarget (32), starBase (33), miRTarBase (34) and LncRNA2Target (35).
Genome-wide mRNAs, lncRNAs and miRNAs expression across cancer types
The genome-wide mRNA, lncRNAs and miRNAs expression across cancer types were obtained from the The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/). The expression profile data were downloaded via the R package ‘TCGAbiolinks’. The fragments per kilobase of transcript per million mapped reads (FPKM) were used to measure the expression of mRNAs and lncRNAs. The annotations of lncRNAs were obtained from GENCODE and further classified into different subtypes. The mRNAs and lncRNAs with zero reads in all samples were excluded from our analyses. Based on the hg38 reference genome information from GENCODE, we obtained the expression values of 19663 protein-coding RNAs, which contain 3178 TFs and 1934 RBPs and 15 513 long non-coding RNAs.
Identification of potential immunology regulons
The regulon in our study refer to a group of regulators that can potentially regulate the immune-related pathways. To identify immune-related regulons works in varied ways, we modified the method ‘ImmLnc’ described in one of our previous studies (22). First, we calculated the partial correlation coefficient (PCC) between the expressions of candidate regulons and other genes. Second, in order to apply the method to four types of regulons with diverse functions, we changed the thresholds for screening genes as follows: (a) For TFs and RBPs which belong to protein coding genes, we excluded low expression genes with 0 in >30% samples. (b) For miRNAs, considering the lower expression level, we excluded genes with 0 in >70% samples. (c) For lncRNAs, we excluded genes with 90% quantile less than 0.1 or median = 0. Next, we computed for each regulon its activity in immune pathways based on the pre-ranked gene set enrichment analysis (GSEA). The P-value of GSEA was further converted to a RES score and regulon-pathway pairs with RES >0.995 and FDR <0.05 were selected in our analysis.
Immune cell infiltration in cancer
To explore the correlation of candidate regulons and immune regulation, we obtained the infiltration of immune cells from six data sources, including TIMER (36), EPIC (37), MCP-counter (38), xCell, QUANTISEQ and CIBERSORT (39–41). Here we mainly focused on B cell, Macrophage/Monocyte, CD4 T cell, T helper, CD8 T cell, T cell other, Endothelial, Dendritic, Treg, Granulocyte and NK cell. We first calculated the Spearman correlation coefficient (R) between the expressions of regulon and immune cell infiltrations, separately. Based on P < 0.05 & |R| > 0.3, we obtained the significantly correlated pairs of regulon-immune cell for different computational methods. When analyzing the correlations between immune cells and immune regulons, we merged the regulon-immune cell pairs (union sets) with same cell type belonging to different sources.
Prioritization of immunology regulons
Given that oncogenes induce tumor growth by influence of immune response, we considered a regulon that was enriched in more immune functions may have an extensive impact. For each regulon in one cancer, we ranked regulons based on the number of potentially regulated immune-related pathways. Furthermore, we calculated an average rank of each regulon in all cancer types and performed ‘0–1’ normalization to the average ranks for four types of regulons.
The known cancer-related TFs, RBPs and lncRNAs were collected from ‘Cancer Gene Census’ (42), ‘CancerMine’ (43) and ‘Lnc2Cancer’ (44). The known disease-related TFs and RBPs were collected from ‘DisGeNET’ (45). The known disease-related miRNAs were integrated from HMDD (46) and miR2Disease (47). The known disease-related lncRNAs were collected from ‘LncRNADisease v2.0’ (48) and ‘LncTarD’. We compared the normalized ranks of known cancer/disease-regulons with those of other candidate regulons by Wilcoxon rank-sum test.
Differential expression of regulons
To perform the differential expression analysis, cancers with more than five normal samples were analyzed. We estimated the expression pattern of all genes between normal samples and cancer samples. We used Fisher's exact test and t-test to obtain the statistical significance for the genes with 0 expressions in most samples and other genes, respectively (49). Furthermore, we calculated the ratio of 0 expression samples and fold change, which were respectively to decipher the differential direction.
Clinical relevance analysis of regulons
The clinical information including overall survival and state of 33 cancer types were collected from TCGA. We applied cox proportional hazards regression model to assess the influence power to prognosis of all genes, based on their median expression. The K–M survival curves were generated by the ‘ggsurvplot’ function with corresponding log-rank P values. For the regulons with positive beta of ‘coxph’, we defined them as prognosis-risky factors, and the negative ones were prognosis-protective factors.
Subtype classification of glioma patients
Briefly, we classified samples from LGG and GBM based on the similarity of integrated key regulons, including TFs, RBPs and lncRNAs. We firstly filtered the regulons with the following principle: (a) their immune regulations were shared between cancers; (b) they were significantly correlated with the infiltration of at least eight immune cells in cancers. We then classified samples into optimal clusters by referencing the expression of key regulons using the R package ‘ConsensusClusterPlus’. The parameters were ‘maxK = 3, reps = 50, clusterAlg = pam, distance = pearson’.
To characterize the subtypes, we compared several clinical characteristics of samples. For the immune features, we obtained immune signatures from Takahiro et al. and calculated the ssGSEA score of samples by R package ‘ssgsea’. In addition, we considered the expression levels of several immune checkpoints and their ligands from CellPhoneDB (50) and InnateDB (51). Moreover, we compared the infiltration of immune cells from TIMER and the stemness indexes from Tathiane et al. (52). For the clinical features, we used the survival information with diverse therapy.
The validation dataset of glioma patients were obtained from Chinese Glioma Genome Atlas (CGGA). We classified samples into different subtypes based on the expressions of key regulons using the same methods above. The clinical data, including mutations, recurrent and multiple treatments were annotated by CGGA.
RESULTS
The landscape of immunology regulons across cancer types
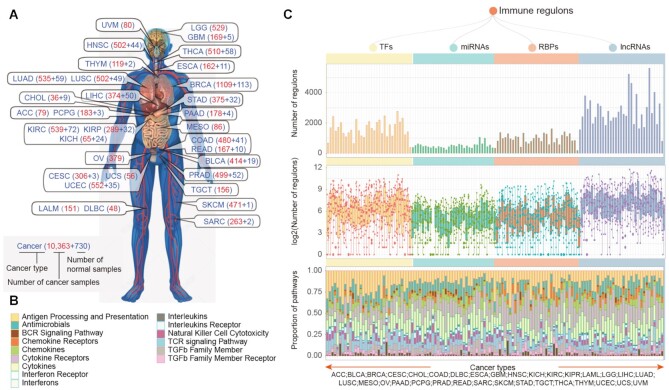
To systematically understand the regulation of immune-related pathways in cancer, we analyzed the genome-wide expression of 10,363 cancer and 730 normal samples across 33 cancer types (Figure 1A). Next, we identified the potential regulons (including TF, miRNA, RBP, and lncRNAs) for 17 immune pathways (Figure 1B) based on the ImmLnc pipeline (22). In total, there were 382–2771 TFs, 225–1061 miRNAs, 182–1620 RBPs and 797–5627 lncRNAs regulons identified across cancers (Figure 1C, top panel). We found that the proportions of TF regulons (12.02–87.20%) were higher than other regulons across cancers, suggesting that TFs might play dominant roles in regulating gene expression. In addition, we found that there were more regulons in testicular germ cell tumors (TGCT). This might be explained by the higher number of tissue-specific expressed genes or lncRNAs observed in recent studies in TGCT (53–55).
Figure 1.
The immunology regulons across cancer types. (A) Number of samples across 33 cancer types. The red and blue numbers imply the number of cancer and normal samples, separately. (B) Immune-related pathways with different colors used in this study. (C) Overview of four type regulons identified by extended ImmLnc across cancer types. The top panel showing the number of regulons enriched in immune pathways across cancers. The middle panel showing the distribution of the number of regulons enriched in each immune pathway across cancer (log2 transformed). The bottom panel showing the proportion of regulons enriched in each immune pathway across cancers.
Moreover, we calculated the numbers of regulons for immune-related pathways across cancer (Figure 1C, middle panel). We found that these pathways were potentially regulated by various numbers of regulons. Particular, antigen processing and presentation, cytokine receptors and cytokines pathways were regulated by more regulons across cancers (Figure 1C, bottom panel). On the other hand, we investigated the components of regulons and found that TFs in C2H2, bHLH, and homeobox families can potentially regulate more immune-related pathways (Supplementary Figure S1A). For miRNA regulons, there were more members from mir-154, let-7 and mir-8 families (Supplementary Figure S1B). miR-369-3p in mir-154 family has been demonstrated to play important roles in cell proliferation (56) and inflammatory response (57). miR-146a in mir-146 family plays important roles in cancer development, cell migration and invasion, as well as in innate immunity (58). We also investigated the functions of RBP regulons and found that they were involved in various types of functions, such as RNA splicing, RNA catabolic process and ribosome biogenesis (Supplementary Figure S1C and Table S2). Moreover, we found that there were more lncRNA regulons were located in intergenic regions and antisense of protein coding genes (Supplementary Figure S1D). Taken together, these results suggest that immune-related pathways were potentially regulated by various types of regulons. The comprehensive regulons landscape provided valuable resource for understanding the immune regulation in cancer.
Immunology regulons are likely to be dysregulated in cancer
Dysregulation of immune-related pathways has been linked to the development of various types of cancers. We next investigated whether the expression of potential immunology regulons were perturbed in cancer. We found that there were hundreds of regulons exhibiting differential expressions across cancers (Figure 2A, |log2FC| > 1 and FDR <0.001, Supplementary Table S3). Particularly, TFs, miRNAs and RBPs regulons were likely to exhibit higher expression in cancer. In addition, there were more miRNAs and lncRNAs specifically differentially expressed in less cancers than those of TFs and RBPs (Supplementary Figure S2A–D). These results are consistent with the observations that noncoding RNAs were with higher tissue specific expression than protein coding genes. Although some regulons might be up-regulated in some cancer types, and down-regulated in other cancer types, we found that numbers of regulons exhibited consistent up-regulation or down-regulation in more than two cancer types (Supplementary Figure S3). When compared the proportion of differentially expressed immunology regulons to all genes, we found that the immunology regulons were more likely to show expression perturbations than other genes (Figure 2A). For example, we identified 605 and 1,325 immunology lncRNAs exhibited higher or lower expressions in breast cancer; this is about 1.5-fold as high as expected proportion (P < 0.001, hypergeometric test). These results suggest that the immunology regulons exhibited widespread expression perturbations in cancer.
Figure 2.
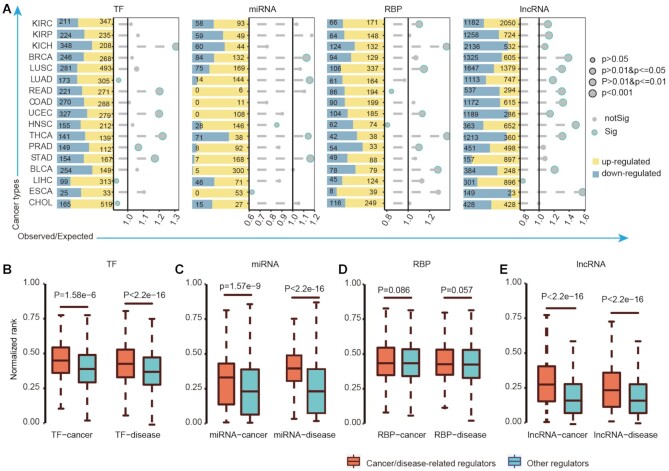
Dysregulation of immunology regulons across cancer types. (A) The histogram showing the number of significantly differentially expressed genes with FDR <0.001 and |log2FC| >1. The dot plot showing the result of hypergeometric test which evaluated the overlap of immune regulons with differentially expressed genes. The sizes of dots showing the significant levels. The x-axis represents the O/E values. The significant results were with blue frame (P-values < 0.05). (B–E) The normalized ranks of cancer/disease-related regulons (orange) and other genes (blue) which were compared by Wilcoxon's rank-sum tests. (B) for TFs; (C) for miRNAs; (D) for RBPs and (E) for lncRNAs.
We next explored to what extent the potential immunology regulons were associated with cancers. We hypothesized that if a regulon potentially regulated more pathways and observed in more cancer types, it is likely to be involved in cancer. Thus, we ranked each regulon based on the number of potentially regulated pathways in each cancer (Supplementary Table S4). The ranks across cancer types were averaged and normalized to a value between 0 and 1. We found that the cancer-/disease-related regulons (TFs, miRNAs and lncRNAs) have significantly higher ranks than others (Figure 2B–E, P < 0.001, two-sided Wilcoxon's rank-sum test). Particularly, we analyzed the top ranked 50 TFs, miRNAs, RBPs and lncRNAs in detail (Figure 3A and Supplementary Table S5) and found that they were more likely to regulate the cytokine/chemokine pathways and T cell receptor (TCR) singling pathway. To systematically investigate the correlations among regulons, we further investigated the co-expression of top-ranked regulons. We found that the regulons were positively correlated with each other in expression and the majority of correlations (96.88%) were observed in at least two cancer types (Supplementary Figure S4). In particular, there were more correlations between TFs and RBPs (Supplementary Figure S5). These results suggest that different types of regulons can potentially synergistically regulate the tumor microenvironment.
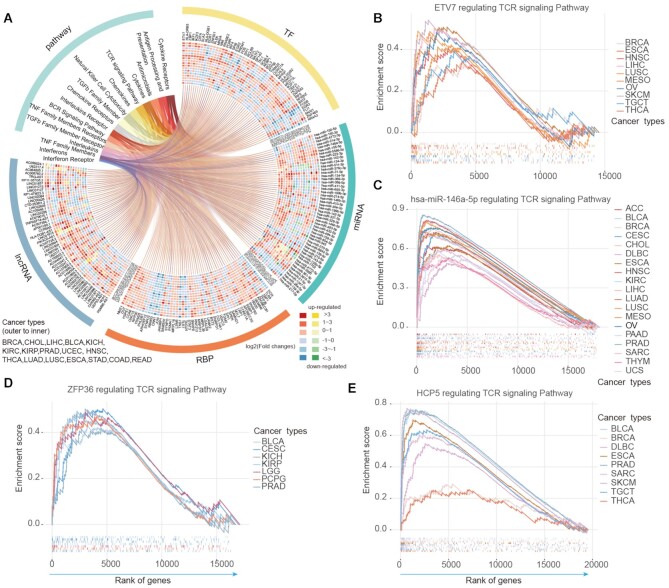
Figure 3.
Top-ranked immunology regulons across cancers. (A) The circos plot showing the top ranked 200 regulons with their enriched pathways across cancer types. For each type of regulons, we showed top 50 and the colored cells represent the corresponding differential expression level of regulons across cancers. Red and yellow representing up-regulation; blue and green representing down-regulation. (B–E) Four examples in circos plot significantly enriched in T cell receptor signaling pathway across cancer types. (B) for TF ETV7; (C) for miRNA hsa-miR-146a-5p; (D) for RBP ZFP36 and (E) for lncRNAs HCP5.
Moreover, these regulons exhibited consistent higher or lower expression across cancer types (Figure 3A). For example, we found that ETV7 exhibited higher expression in 13 cancer types (Supplementary Figure S6A) and potentially regulate TCR signaling pathway in several cancers (Figure 3B). Previous study has demonstrated that ETV7 particularly showed a strong positive correlation with CD8 + T cell infiltration in melanoma (59). Several studies have determined singling cascades associated with miR-146a-5p, such as toll-like receptor pathway and ErB pathway (60). We found that miR-146a-5p can potentially regulating TCR signaling pathways in 19 cancer types (Figure 3C) and exhibited consistent higher expression in cancer (Supplementary Figure S6B). Dynamic post-transcriptional control of RNA expression by RBPs and lncRNAs is critical during immune response. We also identified RBP ZFP36 and lncRNA HCP5 can potentially regulate TCR signaling across cancers (Figure 3D–E). While ZFP36 exhibited lower expression in 14 cancers, HCP5 exhibited higher expression in nine cancers (Supplementary Figure S6C–D). These results were consistent with previous observations that ZFP36 play important roles in regulating T-cell activation, proliferation and effector functions in mouse (61). HCP5 is also a regulatory lncRNAs involved in adaptive and innate immune response (62). Collectively, all these results suggest that the immunology regulons are more likely to be dysregulated in cancer and the regulon-pathway pairs identified in this study provide a pathway-based view to improve our understanding of their regulatory roles in cancer.
Immunology regulons exhibit high expression in immune cells
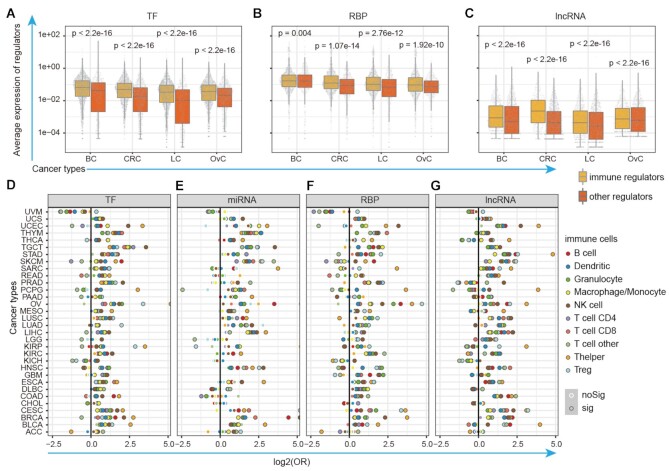
It is reasonable to assume that the immunology regulons have higher expression in immune cells as they play important roles in immune regulation. We thus investigated their expression in immune cells based on single cell sequencing transcriptome in cancer. We first calculated the averaged expression of TFs, RBPs and lncRNAs across immune cells in breast cancer (BC), colorectal cancer (CRC) lung cancer (LC) and ovarian cancer (OvC). We found that the potential immunology regulons have higher average expression levels in immune cells when compared with other genes (Figure 4A–C, P-values < 0.005, two-sided Wilcoxon's rank-sum tests). Moreover, we particularly focused on the regulons that potentially regulate T cell receptor (TCR) or B cell receptor (BCR) signaling pathways. We found that these regulons were also exhibited significantly higher expression in T or B cells (Supplementary Figure S7A–F), in particular for TFs and lncRNAs. For example, BCL11B, IKZF1, ETV7, IKZF3 and LINC00861, which potentially regulated the TCR pathways, exhibited higher expression in T cells across cancer types (Supplementary Figure S8). These results suggest that the potential immunology regulons have higher expression in immune cell populations.
Figure 4.
Immunology regulons exhibit higher expression in immune cell and correlated with immune cell infiltrations. (A–C) Distribution of average expression for immunology regulons and other genes in single immune cells of BC, CRC, LC and OvC. (A) for TFs; (B) for RBPs and (C) for lncRNAs. (D–G) The immunology regulons were likely to be enriched in gene sets that were correlated with immune cell infiltration. The x-axis represents the log-transformed odds ratio (OR) of Fisher's exact tests with diverse color representing diverse types of immune cells. The black-frame dots were with P-values <0.05 in two-sided Fisher's exact test. (D) for TFs; (E) for miRNAs; (F) for RBPs and (G) for lncRNAs.
Furthermore, it has suggested that genes whose expression is correlated with immune cell infiltration are also likely to play critical roles in immunology. We thus estimated the immune cell infiltration based on gene expression based on several computational methods, such as CIBERSORT (40), TIMER (63) and xCell (39). The associations between expression of regulons and immune cell infiltration levels were evaluated by Spearman's rank correlation coefficient (|R| > 0.3 and P < 0.05). There were more TF and RBPs correlated with immune cell infiltrations (Supplementary Figure S9). We next used the Fisher's exact test to investigate whether the immunology regulons were likely to be associated with immune cell infiltrations. We found that a significant higher proportion of immunology regulons are correlated with immune cell infiltrations in a majority of cancer types (Figure 4D–G and Table S6). In particular, the lncRNA regulons (such as HCP5 and ITGB2-AS1) are more likely to be associated with T help cells infiltration across cancer types. All these results suggest that the potential immunology regulons are likely to be associated with immune cell infiltration in cancer, further demonstrated their roles in immunology.
Association of immunology regulons with clinical outcomes
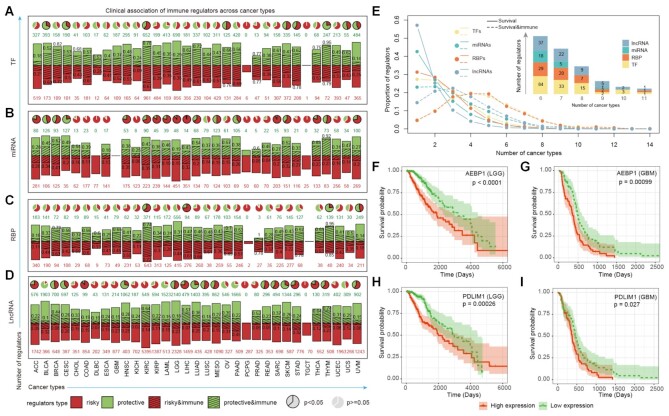
To investigate the clinical relevance of the potential immunology regulons, we first identified all TFs, miRNAs, RBPs and lncRNAs that correlated with patients’ overall survival times across cancer types with p <0.05. We found that hundreds of risky and protective regulons were identified across cancer types (Figure 5A-D, bar plots panels, Supplementary Table S7). Moreover, higher numbers of risky miRNAs than protective miRNAs were identified in cancer. For example, 451 risky and 48 protective miRNAs were identified in lower grade glioma (LGG). Next, we calculated the proportion of risky or protective immunology regulons in cancer. We found that a significant proportion of immunology regulons were likely to be correlated with patients’ survival times across cancer types (Figure 5A–D, pie plots panels).
Figure 5.
The association between immunology regulons and clinical outcome. (A–D) The association between immunology regulons and survival-related genes (log-rank test, P-value < 0.05). Bar plots representing the numbers of protective genes (green bar; green number) and risky genes (red bar; red number) across cancers, with the shadows and black numbers showing the proportions of immune-related regulons. The pie plots above showing the proportion of protective genes to risky genes. Enrichment between survival-related genes and immunology regulons was evaluated using a hypergeometric test exactly. Significant results were labeled by the black frame of pie plots (P-value < 0.05). (A) for TFs; (B) for miRNAs; (C) for RBPs and (D) for lncRNAs. (E) Distribution of regulators occur in different number of cancers. The solid and dotted lines represent survival-related genes and the survival genes which were also enriched in immune pathways. The number of four types survival-related immunology regulons shared in multiple cancers were displayed in the bar plot above. (F–I) Kaplan–Meier plots for LGG (F and H) and GBM (G and I) patients classified by the median expression levels of AEBP1 (F and G) and PDLIM1 (H and I). The survival difference between clusters was calculated by log-rank test. Red line, high-expression group; green line, low-expression group.
Next, we calculated the proportion of regulons that are correlated with survival observed in different number of cancers. We found that the majority of clinical associated regulons were observed in only one cancer type (Figure 5E, solid lines). In contrast, the majority of clinical associated immunology regulons were likely to be observed in more than one cancer (Figure 5E, dash lines). For example, several immunology TFs and RBPs (such as YBX1, TRIM38, TRIM32 and CDK2) were correlated with patients’ survival in five cancer types. Moreover, there were three immunology regulons (ZNF831, APOBEC3G and LINC00861) were correlated with survival in even 11 cancers. In particular, we identified that high expression of AEBP1 was correlated with poor survival in LGG (Figure 5F, P < 0.0001, log-rank test) and glioblastoma multiforme (GBM, Figure 5G, P = 0.00099, log-rank test). AEBP1 downregulation has been demonstrated to suppress cell proliferation and invasion by inhibiting NF-kB signaling pathway (64). Another example is PDLIM1, whose higher expression was associated with poor survival in LGG and GBM (Figure 5H–I, P = 0.00026 and 0.027, log-rank tests). PDLIM1 was a novel signaling adaptor for p75 and played important roles in glioblastoma (65). Together, these results suggest that immunology regulons were likely to be correlated with patients’ survival and provided candidate therapeutic strategies for cancer.
Cancer subtyping based on immunology regulons
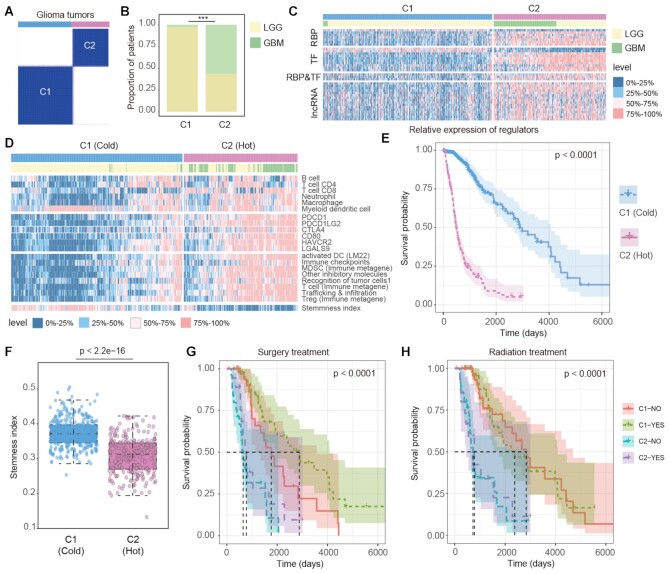
Distinct immune environment of cancer is important for immunotherapy. We next investigate to what extent the identified immunology regulons can be applied to molecular cancer subtyping. Glioma is the most prevalent type of central nervous system malignant tumor. We thus investigated the function of immunology regulons in LGG and GBM in detail. First, we identified 36 regulons that potentially regulated at least one immune-related pathway and correlated with eight immune cell infiltrations in both LGG and GBM (Supplementary Figure S10A). We found that the expressions of these regulons were significantly correlated with each other (Supplementary Figure S10A), suggesting that they might play functions in a module pattern. Next, we classified all the glioma patients (LGG and GBM) into two subtypes based on the expression of 36 regulons (Figure 6A and Supplementary Table S8). The patients in C1 subtype were mainly from LGG, but C2 subtype was formed by both LGG and GBM patients (Figure 6B). The majority of these immunology regulons were highly expressed in C2 subtype (Figure 6C).
Figure 6.
Immunology subtypes of glioma patients with distinct features. (A) The classification of glioma patients based on key regulons shared by LGG and GBM samples from TCGA. (B) The proportions of LGG and GBM samples in C1 and C2 subtypes. (C) The expression levels of key regulons in C1 and C2 subtypes. Annotation bars represent the sample subtypes and cancer types. The expression level was split into 0–25% quantile, 25–50% quantile, 50–75% quantile and 75–100% quantile. (D) The heat map showing the infiltration levels of 6 types of immune cell from TIMER, the expression of immune checkpoint genes and their ligands, the ssGSEA scores of eight immune-related signatures and the stemness indexes of samples. Annotation bars represent the sample subtypes and cancer types. (E) The Kaplan–Meier plot of glioma patients based classifications generated from consensus clustering. C2 patients showing a significantly poorer prognosis than C1 patients. (F) Distribution of stemness indexes of samples in C1 and C2 subtypes. P-value for two-sided Wilcoxon's rank-sum tests. (G–H) Kaplan–Meier plot of survival for LGG and GBM samples classified by the combinations of immunology subtypes and treatment states. The survival difference among clusters was calculated by log-rank test. (G) for surgery treatment; (H) for radiation treatment.
Next, we compared the immune microenvironments of patients in two subtypes. We found that C2 patients exhibited higher immune cell infiltrations, such as CD4 and CD8 T cells (Figure 6D). Moreover, the checkpoint genes exhibited higher expression in C2 patients (Figure 6D). We also evaluated the pathway activities in glioma patients by gene set variation analysis (GSVA). We found that patients in C2 subtype exhibited higher T cell and Treg pathway activities (Figure 6D). However, patients in C2 subtype were with relative lower stemness index (Figure 6E). These results suggest that patients in C2 were likely to be ‘hot’ tumors whereas patients in C1 tend toward a ‘cold’ tumor phenotype. Upon comparison of the survival rates among the two subtypes, we found that C2 patients had poorer prognosis than C1 patients (Figure 6F, log-rank test P < 0.0001).
To investigate whether the immunology regulons-based classification could help clinical therapy of glioma, we investigated the survival rates in the context of clinical treatment. We found that C1 patients who had received surgery treatment exhibited the best survival (Figure 6G, log-rank P < 0.0001). However, there were no differences in survival of C2 patients either received or not received surgery treatments. When considering the radiation treatment, we found that patients in C1 exhibited better survival (Figure 6H). But no differences were found between C1/C2 patients with radiation treatment or not. We next validated these results in the Chinese Glioma Genome Atlas (CGGA) dataset (Supplementary Table S9). We also revealed two subtypes with distinct immune microenvironments and survival rates (Supplementary Figures S10 and S11). Taken together, all these results suggested that the immunology regulons identified could provide useful insights for cancer classification and had independent prognostic effect in glioma.
An interactive web portal for cancer immunology regulons
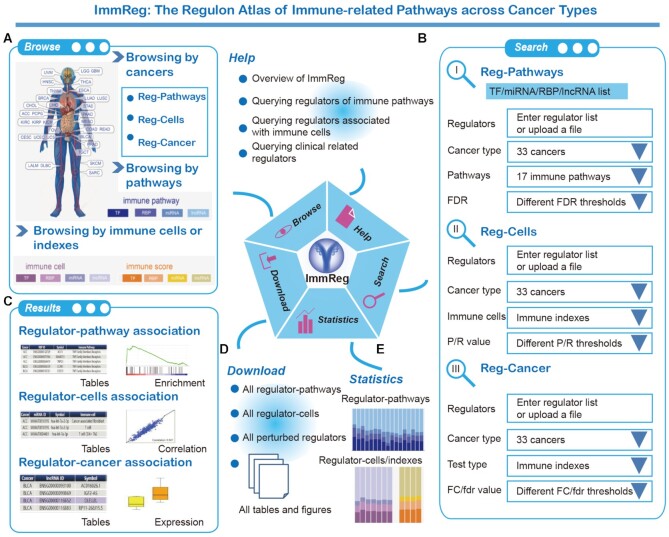
We developed a user-friendly data portal, ImmReg (http://bio-bigdata.hrbmu.edu.cn/ImmReg/), to facilitate visualizing, searching and browsing of immunology regulons data by the biomedical research community (Figure 7). Several entryways were provided for browsing and querying the regulon-pathways, regulon-cells and regulon-cancer relationships across 33 cancer types. Users can enter different browsing pages when click the human body map or bar plots in the homepage (Figure 7A). Moreover, users can enter querying pages to search regulon-pathway, regulon-cell or regulon-cancer of interest from the menus of the homepage. We provided a search section for users to query the data based on regulons, cancer types, pathways, immune cells or significant levels (Figure 7B).
Figure 7.
Overview of ImmReg web-based resource. (A) In home page, users can browse immunology regulons by cancers, pathways and immune cells or indexes. (B) The search page includes ‘Reg-Pathways’, ‘Reg-Cells’ and ‘Reg-Cancers’. Users can search TFs, miRNAs, RBPs and lncRNAs of interest by setting advanced filter sections. (C) In the result page, the regulons and related information were listed and available. By checking the detail section, users can check the enrichment plot, correlation plot and differential expression box plot. (D) Users can obtain all the immunology regulon results in the download page and there are statistics above (E).
For each browsing or querying record, we provided the tables and diagram results for users (Figure 7C). The gene set enrichment analysis (GSEA) diagram, scatter plot and box plot were all embedded in each record to display the associations between immunology regulons and immune-related pathways (Figure 7C). All the data generated in this study can be downloaded in the download page (Figure 7D–E) and figures of results can be downloaded as PNG file. The help page includes an introduction and interpretation guidance for this resource. The detail interpretation of each section was provided in Supplementary Figure S12. This web portal is a valuable resource and will be of great interest to the cancer immune community, which will provide novel insights into cancer immunotherapy.
DISCUSSION
Increasing numbers of immunology regulons are being identified in cancer. However, only a few candidates have been investigated. In the present study, we systematically identified the potential immunology regulons (including TFs, miRNAs, RBPs and lncRNAs) across cancer types based on ImmLnc method (22). Notably, there were hundreds of regulons that potentially regulate immune-related pathways in each cancer type. Moreover, we found that immune-related regulons are likely to show expression perturbations in cancer, exhibit higher expression in immune cells and their expression levels are significantly correlated with immune cell infiltrations. We believe these results and datasets allow us to comprehensive understand the potential immunology roles of regulons in cancer.
In addition to the transcriptional regulators, emerging evidence has demonstrated that cancer immunology is complicated in post-translational modifications (PTM) (66,67). We thus applied the ImmLnc pipeline to identify the protein kinase and protein phosphorylation enzymes that can potentially regulating the expression of immune-related genes. We first downloaded the protein kinase and phosphorylation enzymes from iEKPD database (68). Based on the computational pipeline, we identified numbers of protein kinase and protein phosphorylation enzymes can potentially regulate the immune-related pathways across cancer types (Supplementary Figure S13). In particular, the antigen processing and presentation, cytokine receptors and cytokines were regulated by more post-translational regulators. The identified PTM-related regulons provided ideal candidates for cancer therapy. Indeed, we found that approximate 60% of protein kinase regulons can be targeted by at least one drug in DrugBank (Supplementary Figure S13 and Table S10).
Previous studies suggest that biomarkers always exhibited tissue-specific expression patterns. Thus, we investigated the expression of immunology regulons based on public single cell sequencing datasets (69). We found that the average expressions of immunology regulons are significantly higher that other genes, suggesting that immunology regulons exhibited higher expression in immune cells. Particularly, the regulons that potentially regulated T or B cell signaling pathways also exhibited higher expression in T or B cells (Supplementary Figures S7 and S8). These finding strongly suggested their critical roles in immunology regulation. Moreover, the current pipeline is applied to bulk RNA-seq data in various cancer types. Emerging single cell sequencing data as well as computational methods for estimate the immune cell infiltrations, such as ImmCellAI (70), have increased our understanding of immune regulation in cancer (71). We posited that integration of single cell sequencing data and the ImmLnc pipeline will promote our understanding of tumoral genetic heterogeneity and immune regulation.
ImmLnc is a model-free method for identifying immune regulons in cancer. Although numbers of machine-learning methods have been proposed to identify genes in cancer (72), it is difficult to use in immune regulation. Because there were limited numbers of experimentally validated immune regulons reported in literature. To preliminarily validate the results obtained with ImmLnc, we explored whether the regulons–pathway pairs could be reproduced with different datasets of the same cancer type. We found the identified regulons–pathway pairs were significantly overlapped (Supplementary Figure S14), suggesting the pipeline is robust. We preliminarily identified the potential regulons for further experimental validation; it is a critical step to identify the targets of these regulons. Here, we used the expression correlated genes rather than direct targets because numbers of the regulons were with unknown targets. In addition, we obtained the regulator-target information from hTFtarget, starBase, miRTarBase and LncRNA2Target. We found that there were approximate 45% of predicted immunology regulon-pathway pairs with leading edge genes were enriched in predicted targets, which were significantly higher than other regulon-pathway pairs (Supplementary Figure S15). These results suggest that the correlation-based method might identify more candidate immune regulons for further functional validation. The leading edge genes identified here can also help predicting the targets of regulons. Increasing biotechnology has been proposed to identify the targets of TFs, RBPs, miRNAs or lncRNAs. For example, ChIP-Seq and CLIP-Seq were widely used for investigating the TF and RBP targets. However, the TF or RBP regulation is context specific; determining the specific targets in cancer context remains a challenge. Moreover, lncRNAs can exert their functions in many potential ways, capturing their targets is difficult. Here, we identified genes that are correlated with the expression of regulons and further investigated the enrichment in immune pathways. With the development of high throughput sequencing methods, we will be able to identify the context-specific targets of immunology regulons and further improve our understanding of their functions.
Moreover, we found the immunology regulons helps prioritizing cancer-related biomarkers and cancer subtyping. Based on the expression of immunology regulons, we identified two subtypes of glioma. Patients in C1 and C2 subtypes were with distinct clinical features (Figure 6). The composition of immune cells in tumor microenvironment is also known to affect patient prognosis (73). In this study, higher infiltration of T cells, macrophage and dendritic cells was identified in C2 subtype. It is apparent that patients in C2 subtype tend toward a ‘hot’ tumor phenotype, suggesting likely sensitive to immunotherapy. However, patients in C2 were with poor survival. These results are consistent with one of recent studies that demonstrating that glioma patients with higher immune infiltrations were with high risk scores (74). C2 patients were with higher immune cell infiltrations and were likely to be with inflammation microenvironments. The majority of C1 patients were at the primary grades (LGG) of tumors. Moreover, we further divided the patients into four groups considering the grades and immune subtypes. We found LGG-hot patients tend to have moderate CD8 T cell infiltration, EMT scores, cell cycle scores and hypoxia scores (Supplementary Figure S16). These results suggest that LGG-hot patients were likely to be the transitional subtypes from LGG to GBM. Together, these results suggested that the model based on expression of immunology regulons can be applied to predict survival outcome and immunotherapy response.
In summary, our study highlights the functional and regulatory roles of TF, miRNAs, RBPs and lncRNAs in immunology regulation. It is important to understand the function of immunology regulons in cancer. Identification of the potential regulons provided the first step for immunotherapy. Continued investigation of the immunology regulons identified here will greatly promote our understanding of the mechanism of cancer and better immunotherapies.
DATA AVAILABILITY
ImmReg is a user-friendly data portal to facilitate visualizing, searching and browsing of immunology regulons data by the biomedical research community (http://bio-bigdata.hrbmu.edu.cn/ImmReg/).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the TCGA and CGGA project to provide the omics data of multiple types of cancer.
Contributor Information
Tiantongfei Jiang, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China.
Weiwei Zhou, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China.
Zhenghong Chang, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China.
Haozhe Zou, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China.
Jing Bai, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China.
Qisen Sun, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China.
Tao Pan, Key Laboratory of Tropical Translational Medicine of Ministry of Education, College of Biomedical Information and Engineering, Hainan Women and Children's Medical Center, Hainan Medical University, Haikou 571199, China.
Juan Xu, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China.
Yongsheng Li, Key Laboratory of Tropical Translational Medicine of Ministry of Education, College of Biomedical Information and Engineering, Hainan Women and Children's Medical Center, Hainan Medical University, Haikou 571199, China.
Xia Li, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China; Key Laboratory of Tropical Translational Medicine of Ministry of Education, College of Biomedical Information and Engineering, Hainan Women and Children's Medical Center, Hainan Medical University, Haikou 571199, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31871338, 31970646, 61873075, 32060152, 32070673, 32170676]; Science and Technology special fund of Hainan Province [ZDYF2021SHFZ051]; Hainan Provincial Natural Science Foundation of China [820MS053]; Major Science and Technology Program of Hainan Province [ZDKJ202003]; HMU MarshalInitiative Funding [HMUMIF-21024]; Marshal Initiative Funding of Hainan Medical University [JBGS202103]; Hainan Province Clinical Medical Center; National Key R&D Program of China [2018YFC2000100]; Natural Science Foundation for Distinguished Young Scholars of Heilongjiang Province [JQ2019C004]; Heilongjiang Touyan Innovation Team Program; Hainan Provincial Key Laboratory of Carcinogenesis and Intervention [JCKF2021003]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of Interest Statement. None declared.
REFERENCES
- 1. Lin W.M., Fisher D.E.. Signaling and immune regulation in melanoma development and responses to therapy. Annu. Rev. Pathol. 2017; 12:75–102. [DOI] [PubMed] [Google Scholar]
- 2. Ye L., Creaney J., Redwood A., Robinson B.. The current lung cancer neoantigen landscape and implications for therapy. J. Thorac. Oncol. 2021; 16:922–932. [DOI] [PubMed] [Google Scholar]
- 3. Mellman I., Coukos G., Dranoff G.. Cancer immunotherapy comes of age. Nature. 2011; 480:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu D. CAR-T ‘the living drugs’, immune checkpoint inhibitors, and precision medicine: a new era of cancer therapy. J. Hematol. Oncol. 2019; 12:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pope S.D., Medzhitov R.. Emerging principles of gene expression programs and their regulation. Mol. Cell. 2018; 71:389–397. [DOI] [PubMed] [Google Scholar]
- 6. Kim H.D., Shay T., O'Shea E.K., Regev A.. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009; 325:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andersson R., Sandelin A., Danko C.G.. A unified architecture of transcriptional regulatory elements. Trends Genet.: TIG. 2015; 31:426–433. [DOI] [PubMed] [Google Scholar]
- 8. Palazon A., Goldrath A.W., Nizet V., Johnson R.S.. HIF transcription factors, inflammation, and immunity. Immunity. 2014; 41:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taniguchi T., Ogasawara K., Takaoka A., Tanaka N.. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001; 19:623–655. [DOI] [PubMed] [Google Scholar]
- 10. Yu H., Pardoll D., Jove R.. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 2009; 9:798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiao J., Jin X., Zhang C., Zou H., Chang Z., Han N., Li X., Zhang Y., Li Y.. Systematic analysis of enhancer regulatory circuit perturbation driven by copy number variations in malignant glioma. Theranostics. 2021; 11:3060–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Z., Yin J., Zhou W., Bai J., Xie Y., Xu K., Zheng X., Xiao J., Zhou L., Qi X.et al.. Complex impact of DNA methylation on transcriptional dysregulation across 22 human cancer types. Nucleic Acids Res. 2020; 48:2287–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., McGrail D.J., Xu J., Li J., Liu N.N., Sun M., Lin R., Pancsa R., Zhang J., Lee J.S.et al.. MERIT: systematic analysis and characterization of mutational effect on RNA interactome topology. Hepatology. 2019; 70:532–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mehta A., Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nature reviews. Immunology. 2016; 16:279–294. [DOI] [PubMed] [Google Scholar]
- 15. Xu J., Shao T., Song M., Xie Y., Zhou J., Yin J., Ding N., Zou H., Li Y., Zhang J.. MIR22HG acts as a tumor suppressor via TGFbeta/SMAD signaling and facilitates immunotherapy in colorectal cancer. Mol. Cancer. 2020; 19:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuo G., Wu C.Y., Yang H.Y.. MiR-17-92 cluster and immunity. J. Formos. Med. Assoc. 2019; 118:2–6. [DOI] [PubMed] [Google Scholar]
- 17. Zhang J., Li S., Zhang L., Xu J., Song M., Shao T., Huang Z., Li Y.. RBP EIF2S2 promotes tumorigenesis and progression by regulating MYC-mediated inhibition via FHIT-related enhancers. Mol. Ther. 2020; 28:1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwerk J., Soveg F.W., Ryan A.P., Thomas K.R., Hatfield L.D., Ozarkar S., Forero A., Kell A.M., Roby J.A., So L.et al.. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat. Immunol. 2019; 20:1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Wang X., Zhang X., Wang J., Ma Y., Zhang L., Cao X.. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. PNAS. 2019; 116:976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brahic M., Bureau J.F., Michiels T.. The genetics of the persistent infection and demyelinating disease caused by Theiler's virus. Annu. Rev. Microbiol. 2005; 59:279–298. [DOI] [PubMed] [Google Scholar]
- 21. Hu Q., Ye Y., Chan L.C., Li Y., Liang K., Lin A., Egranov S.D., Zhang Y., Xia W., Gong J.et al.. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 2019; 20:835–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y., Jiang T., Zhou W., Li J., Li X., Wang Q., Jin X., Yin J., Chen L., Zhang Y.et al.. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 2020; 11:1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhattacharya S., Andorf S., Gomes L., Dunn P., Schaefer H., Pontius J., Berger P., Desborough V., Smith T., Campbell J.et al.. ImmPort: disseminating data to the public for the future of immunology. Immunol. Res. 2014; 58:234–239. [DOI] [PubMed] [Google Scholar]
- 24. Li B., Cui Y., Diehn M., Li R.. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non-small cell lung cancer. JAMA Oncol. 2017; 3:1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu H., Miao Y.R., Jia L.H., Yu Q.Y., Zhang Q., Guo A.Y.. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019; 47:D33–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han H., Cho J.W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E.et al.. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018; 46:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kulakovskiy I.V., Vorontsov I.E., Yevshin I.S., Sharipov R.N., Fedorova A.D., Rumynskiy E.I., Medvedeva Y.A., Magana-Mora A., Bajic V.B., Papatsenko D.A.et al.. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018; 46:D252–D259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chawla K., Tripathi S., Thommesen L., Laegreid A., Kuiper M.. TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics. 2013; 29:2519–2520. [DOI] [PubMed] [Google Scholar]
- 29. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 30. Ravasi T., Suzuki H., Cannistraci C.V., Katayama S., Bajic V.B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N.et al.. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010; 140:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J., Pan T., Chen L., Wang Q., Chang Z., Zhou W., Li X., Xu G., Li X., Li Y.et al.. Alternative splicing perturbation landscape identifies RNA binding proteins as potential therapeutic targets in cancer. Mol. Ther. Nucleic Acids. 2021; 24:792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Q., Liu W., Zhang H.M., Xie G.Y., Miao Y.R., Xia M., Guo A.Y.. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genomics Proteomics Bioinformatics. 2020; 18:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H.. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014; 42:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang H.Y., Lin Y.C., Li J., Huang K.Y., Shrestha S., Hong H.C., Tang Y., Chen Y.G., Jin C.N., Yu Y.et al.. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020; 48:D148–D154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng L., Wang P., Tian R., Wang S., Guo Q., Luo M., Zhou W., Liu G., Jiang H., Jiang Q.. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019; 47:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S.. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017; 77:e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Racle J., de Jonge K., Baumgaertner P., Speiser D.E., Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017; 6:e26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautes-Fridman C., Fridman W.H.et al.. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016; 17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aran D., Hu Z., Butte A.J.. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017; 18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A.. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015; 12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H., Krogsdam A., Loncova Z., Posch W., Wilflingseder D.et al.. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome medicine. 2019; 11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A.. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018; 18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lever J., Zhao E.Y., Grewal J., Jones M.R., Jones S.J.M.. CancerMine: a literature-mined resource for drivers, oncogenes and tumor suppressors in cancer. Nat. Methods. 2019; 16:505–507. [DOI] [PubMed] [Google Scholar]
- 44. Gao Y., Shang S., Guo S., Li X., Zhou H., Liu H., Sun Y., Wang J., Wang P., Zhi H.et al.. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021; 49:D1251–D1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang Z., Shi J., Gao Y., Cui C., Zhang S., Li J., Zhou Y., Cui Q.. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019; 47:D1013–D1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y.. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009; 37:D98–D104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019; 47:D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y., Li L., Wang Z., Pan T., Sahni N., Jin X., Wang G., Li J., Zheng X., Zhang Y.et al.. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018; 46:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R.. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020; 15:1484–1506. [DOI] [PubMed] [Google Scholar]
- 51. Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., Winsor G.L., Hancock R.E., Brinkman F.S., Lynn D.J.. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013; 41:D1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Weinstein J.N., Kaminska B., Huelsken J., Omberg L., Gevaert O.et al.. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018; 173:338–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu K., Cai Y., Zhang M., Zou H., Chang Z., Li D., Bai J., Xu J., Li Y. Pan-cancer characterization of expression and clinical relevance of m(6)A-related tissue-elevated long non-coding RNAs. Mol. Cancer. 2021; 20:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lv D., Xu K., Jin X., Li J., Shi Y., Zhang M., Jin X., Li Y., Xu J., Li X.. LncSpA: LncRNA spatial Atlas of expression across normal and cancer tissues. Cancer Res. 2020; 80:2067–2071. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Q., Liu W., Liu C., Lin S.Y., Guo A.Y.. SEGtool: a specifically expressed gene detection tool and applications in human tissue and single-cell sequencing data. Brief. Bioinform. 2018; 19:1325–1336. [DOI] [PubMed] [Google Scholar]
- 56. Li P., Dong M., Wang Z.. Downregulation of TSPAN13 by miR-369-3p inhibits cell proliferation in papillary thyroid cancer (PTC). Bosn. J. Basic Med. Sci. 2019; 19:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Scalavino V., Liso M., Cavalcanti E., Gigante I., Lippolis A., Mastronardi M., Chieppa M., Serino G.. miR-369-3p modulates inducible nitric oxide synthase and is involved in regulation of chronic inflammatory response. Sci. Rep. 2020; 10:15942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pedersen I., David M.. MicroRNAs in the immune response. Cytokine. 2008; 43:391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qu H., Zhao H., Zhang X., Liu Y., Li F., Sun L., Song Z.. Integrated analysis of the ETS family in melanoma reveals a regulatory role of ETV7 in the immune microenvironment. Front. Immunol. 2020; 11:612784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aksenenko M., Palkina N., Komina A., Tashireva L., Ruksha T.. Differences in microRNA expression between melanoma and healthy adjacent skin. BMC dermatology. 2019; 19:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moore M.J., Blachere N.E., Fak J.J., Park C.Y., Sawicka K., Parveen S., Zucker-Scharff I., Moltedo B., Rudensky A.Y., Darnell R.B.. ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. eLife. 2018; 7:e33057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kulski J.K. Long noncoding RNA HCP5, a hybrid HLA class I endogenous retroviral gene: structure, expression, and disease associations. Cells. 2019; 8:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S.. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020; 48:W509–W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cheng L., Shao X., Wang Q., Jiang X., Dai Y., Chen S.. Adipocyte enhancer binding protein 1 (AEBP1) knockdown suppresses human glioma cell proliferation, invasion and induces early apoptosis. Pathol. Res. Pract. 2020; 216:152790. [DOI] [PubMed] [Google Scholar]
- 65. Ahn B.Y., Saldanha-Gama R.F., Rahn J.J., Hao X., Zhang J., Dang N.H., Alshehri M., Robbins S.M., Senger D.L.. Glioma invasion mediated by the p75 neurotrophin receptor (p75(NTR)/CD271) requires regulated interaction with PDLIM1. Oncogene. 2016; 35:1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hu X., Lin Z., Wang Z., Zhou Q.. Emerging role of PD-L1 modification in cancer immunotherapy. Am. J. Cancer Res. 2021; 11:3832–3840. [PMC free article] [PubMed] [Google Scholar]
- 67. Satpathy S., Krug K., Jean Beltran P.M., Savage S.R., Petralia F., Kumar-Sinha C., Dou Y., Reva B., Kane M.H., Avanessian S.C.et al.. A proteogenomic portrait of lung squamous cell carcinoma. Cell. 2021; 184:4348–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guo Y., Peng D., Zhou J., Lin S., Wang C., Ning W., Xu H., Deng W., Xue Y.. iEKPD 2.0: an update with rich annotations for eukaryotic protein kinases, protein phosphatases and proteins containing phosphoprotein-binding domains. Nucleic Acids Res. 2019; 47:D344–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qian J., Olbrecht S., Boeckx B., Vos H., Laoui D., Etlioglu E., Wauters E., Pomella V., Verbandt S., Busschaert P.et al.. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020; 30:745–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Miao Y.R., Zhang Q., Lei Q., Luo M., Xie G.Y., Wang H., Guo A.Y.. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv. Sci. 2020; 7:1902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Baslan T., Hicks J.. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat. Rev. Cancer. 2017; 17:557–569. [DOI] [PubMed] [Google Scholar]
- 72. Rajkomar A., Dean J., Kohane I.. Machine learning in medicine. N. Engl. J. Med. 2019; 380:1347–1358. [DOI] [PubMed] [Google Scholar]
- 73. Quail D.F., Joyce J.A.. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013; 19:1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang Z., Su G., Dai Z., Meng M., Zhang H., Fan F., Liu Z., Zhang L., Weygant N., He F.et al.. Circadian clock genes promote glioma progression by affecting tumour immune infiltration and tumour cell proliferation. Cell Prolif. 2021; 54:e12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ImmReg is a user-friendly data portal to facilitate visualizing, searching and browsing of immunology regulons data by the biomedical research community (http://bio-bigdata.hrbmu.edu.cn/ImmReg/).