Abstract
Telomere shortening can cause detrimental diseases and contribute to aging. It occurs due to the end replication problem in cells lacking telomerase. Furthermore, recent studies revealed that telomere shortening can be attributed to difficulties of the semi-conservative DNA replication machinery to replicate the bulk of telomeric DNA repeats. To investigate telomere replication in a comprehensive manner, we develop QTIP-iPOND - Quantitative Telomeric chromatin Isolation Protocol followed by isolation of Proteins On Nascent DNA - which enables purification of proteins that associate with telomeres specifically during replication. In addition to the core replisome, we identify a large number of proteins that specifically associate with telomere replication forks. Depletion of several of these proteins induces telomere fragility validating their importance for telomere replication. We also find that at telomere replication forks the single strand telomere binding protein POT1 is depleted, whereas histone H1 is enriched. Our work reveals the dynamic changes of the telomeric proteome during replication, providing a valuable resource of telomere replication proteins. To our knowledge, this is the first study that examines the replisome at a specific region of the genome.
INTRODUCTION
Telomeres are crucial for maintaining chromosome structure and genome stability. In humans, telomeric DNA consists of 5′-TTAGGG-3′/5′-CCCTAA-3′ repeats with an overall length of 5000–15 000 nucleotides. Telomeric DNA is bound by a large number of proteins functioning in different processes occurring at the chromosome ends. Most prominent is the six subunits containing shelterin protein complex consisting of TRF1 (telomeric repeat binding factor 1), TRF2 (telomeric repeat binding factor 2), POT1 (protection of telomeres 1), TPP1 (also known as ACD, adrenocortical dysplasia protein homolog), TIN2 (TRF1-interacting nuclear factor 2) and RAP1 (repressor-activator protein 1) (1). In addition to the shelterin components, several hundred proteins associate with telomeres (2–4). Experimental manipulation and genetic data linking mutant gene products to telomere syndromes have revealed important functions for several of these proteins.
Telomere shortening in telomerase-negative somatic cells triggers a DNA damage response (5). This leads to cellular senescence providing a powerful tumor-suppressive mechanism as it limits the proliferation of cells in precancerous lesions (6). On the other hand, premature telomere shortening can lead to a number of degenerative disorders giving rise to bone marrow failure, pulmonary fibrosis and cancer (7–11). In these telomere syndromes, telomere maintenance by either semi-conservative DNA replication or telomerase is defective.
Telomeres pose the following challenges to the DNA replication machinery: First, unwinding of the parental DNA during telomere replication exposes the G-rich single stranded DNA which can form highly stable G-quadruplex (G4) structures and other stable secondary structures involving guanine bases. G4 structures can cause stalling or collapse of replication forks, requiring specialized helicases including WRN, RTEL1 and BLM for unwinding (12–17). Second, the t-loop in which the telomeric 3′ overhang is tucked into the double stranded part of the telomeric DNA (18) needs to be unwound during telomere replication by the RTEL1 helicase (19,20). Third, the long noncoding RNA TERRA is transcribed from a large number of chromosome ends (21–24). TERRA can form RNA/DNA hybrid structures post transcription (25) which interfere with DNA replication if not removed by RNase H, the THO complex, RNA surveillance factors or the splicing factors SFPQ and NONO (26–31). Finally, replication origins are present in the subtelomeric regions but replication initiates only rarely within the telomeric repeats (14,15,32). Therefore, replication at telomeres is mostly unidirectional and stalled forks may not be rescued from converging forks that come from the end of chromosomes.
In order to specifically characterize the replication machinery that overcomes the many hurdles posed at telomeres, we combined QTIP (Quantitative Telomeric chromatin Isolation Protocol) (4,33,34) to purify telomeric chromatin with iPOND (isolation of Proteins On Nascent DNA) (35,36), which enriches proteins associated with replicating DNA. We identified a large set of proteins that are specifically enriched at telomeres during their replication and demonstrate crucial functions for several of these proteins. Moreover, we found that the single strand telomeric DNA binding protein POT1 is depleted from replication forks consistent with a POT1-to-RPA switch at telomere replication forks (37). On the other hand, histone H1 variants are present during telomere replication and are depleted from telomeres only later when telomeric chromatin matures. Our study provides comprehensive insights into the dynamic changes of telomeric proteins during telomere replication. The resource provided by our work will broaden our understanding of genome instability caused by telomere replication defects in cancer and telomere diseases.
MATERIALS AND METHODS
Cell lines
HEK293E cells: A subclone of the Human Embryonic Kidney (HEK) epithelium suspension cell line (female origin) expressing the EBNA-1 protein was cultured in EX-CELL® 293 Serum-Free Medium (Merck, 14571C) containing 4 mM GlutaMAX supplement (ThermoFisher, 35050061) at 37°C and 5% CO2. Stable cell lines expressing endogenous FLAG-TRF1 and FLAG-TRF2 (heterozygous) were generated by retroviral infection and selection in puromycin-containing medium.
HeLa-Long cells: A super-telomerase population of HeLa cells (with ∼33 kb long telomeres) expressing hTERT from the LTR (long terminal repeat) promoter and human telomerase RNA from the U1 promoter generated by retroviral infection (38). The parental HeLa cell line (female origin) was obtained from ATCC. Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Tet System Approved FBS (Clontech), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) at 37°C and 5% CO2.
CRISPR/Cas9 gene editing and transfections
HEK 293E cl75 was generated by transfecting pSpCas9(BB)-2A-Puro (PX459) plasmid expressing gRNAs targeting the genomic region proximal to the start codon of TRF1 (TRF1-gRNA: 5′-GCGAGCCATTTAACATGGCGG-3′) or TRF2 (TRF2-gRNA: 5′-TCTATCATGGCCGCGGGAGC-3′) and non-linearized repair template plasmid. The repair template was amplified from genomic DNA of HEK293E cells. It contained 600 bp homology regions flanking both sites of the start codons of TRF1 or TRF2. Three tandemly repeated FLAG tags, an EcoRI restriction site, the Kozak sequence and linker base pairs were added by overlapping PCR. The PCR fragments were cloned into TOPO Zero Blunt for Sequencing (Invitrogen). Cells were transfected in a six-well plate containing 2 ml DMEM/FBS, 500 μl Opti-MEM (Gibco) and 10 μl Lipofectamine 2000 (Invitrogen) with 3 μg DNA of gRNA plasmid and 3 μg repair template. Cells were split the next day and selected with 1 μg/mL puromycin (Invitrogen) for 4 days. Single-cell clones were obtained by limited dilution and genomic DNA was extracted with the Wizard genomic DNA Purification System (Promega) for screening. PCR with primers flanking the region of insertion was performed and screened for the presence of the FLAG-tags. PCR fragments were digested with EcoRI to distinguish homo- from heterozygosity of the edited locus. PCR products were subcloned into the TOPO Zero Blunt plasmid and sequenced. Positive cell clones were expanded.
Cell cycle analysis
HEK293E cells were harvested, washed with PBS and centrifuged for 3 min at 300 g at room temperature (RT). Pellets were fixed by adding dropwise cold 70% EtOH under vortexing to a final concentration of 1 million cells per ml and stored at 4°C overnight. Cells were washed with PBS once and permeabilized in 0.5% Triton X-100 at a final concentration of 10 million cells per ml for 20 min. Cells were washed with PBS twice and were stained with PBS containing DAPI (2 μg/ml) incubating for 10 min at 37°C. Cells were transferred to FACS tubes and cell cycle distribution was determined by flow cytometry (LSRII (SORP)).
QTIP-iPOND
The procedures that were employed to analyze the telomeric replisome by QTIP-iPOND are schematically depicted in Figure 1A, B and Supplementary Figure S1A. Cells were pulsed with 10 μM EdU for 10 min and split. EdU pulse samples were directly crosslinked by adding formaldehyde (FA) (Applichem, A0877) to 1% FA/ml and fixed for 15 min at RT. Cells of thymidine (Thy) chase samples were washed with Thy-containing medium (10 μM Thy) and centrifuged for 5 min at 300 g, and resuspended in Thy-containing medium. Cells were incubated for 4 h and crosslinked as above. After crosslinking, glycine (pH 2.5) was added to a final concentration of 125 mM for 5 min to quench the crosslinking reaction. Cells were washed with 1× PBS three times and centrifuged for 5 min at 900 g in between. Cells (10 million cells/ml) were permeabilized with permeabilization buffer (1× PBS containing 0.5% Triton-X-100) for 30 min at RT and cell debris washed with cold 1× PBS containing 0.5% BSA and centrifuged for 5 min at 900 g, and then with cold 1× PBS and centrifuged at the same speed. Pellets were resuspended in Click reaction cocktail (1x PBS pH 7.4, 10 μM Biotin azide [TRCB391600, Toronto Research Chemicals], 10 mM sodium ascorbate [A7631, Sigma-Aldrich], 2 mM CuSO4 [C1297, Sigma]) and incubated for 2 h at RT in the dark and kept in the dark for subsequent steps. Pellets were washed once with cold 1× PBS containing 0.5% BSA and twice with cold 1× PBS. Pellets were then resuspended in LB3 buffer (10 mM Tris–HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.25% Sodium lauroyl sarkosinate, cOmplete-EDTA-free protease inhibitor cocktail [Roche]) at a final concentration of 10 million cells/ml and sonicated for 7 min on ice using a Branson Tip sonicator (30% power, 20 s constant pulse, and 20 s pause). Cell lysates were centrifuged for 10 min at 16 800 g at 4°C to remove insoluble materials. Next, extracts were dialyzed in a dialysis tube (Thermo Scientific, SnakeSkin Dialysis Tubing, 30K MWCO) against IP buffer (50 mM Tris–HCl pH 8.0, 600 mM NaCl, 10 mM EDTA pH 8.0, 0.75% Triton X-100) with 5× of the sample volume twice (5 h and overnight).
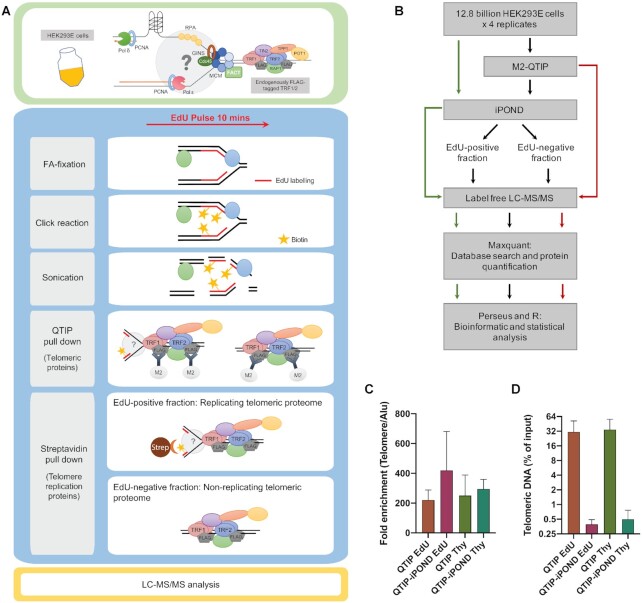
Figure 1.
Experimental scheme of QTIP-iPOND. (A) Upper panel: In the HEK293E cells, TRF1 and TRF2 were endogenously tagged with FLAG-tags. Lower panel: To identify proteins near telomere replication forks, we combined QTIP with iPOND. Upon purification of telomeric chromatin with QTIP, the replicating telomeric proteome was separated from the non-replicating telomeric proteome using streptavidin beads. FA: formaldehyde; Strep: streptavidin. (B) Flow chart of the QTIP-iPOND experiment. 12.8 billion cells were used for each replicate, which was analyzed by QTIP-iPOND (9.6 billion cells for EdU pulse and Thy chase), iPOND (1.6 billion cells for EdU pulse and Thy chase) and QTIP (1.6 billion cells for FLAG-tagged TRF1/2 HEK293E and non-tagged WT HEK293E). (C) Telomeric DNA recovery. The data was obtained by dot blot of three biological replicates. (D) Fold enrichment of telomeric DNA over Alu element. The data was obtained by dot blot of three biological replicates. Data are represented as mean + standard deviation.
ANTI-FLAG M2 Affinity Agarose Gel (Sigma) (1.6 ml slurry beads/billion cells) was added to the extract for TRF1/TRF2 immunoprecipitation (IP) overnight at 4°C. The next day, beads were washed 5 times with IP buffer. Elution was performed 5 times with IP buffer containing 100 μg/ml of FLAG peptide (Sigma) incubating 30 min on a wheel for each elution. For the QTIP-only experiments without subsequent iPOND purification step, the elution was performed with IP buffer containing FLAG peptide without adding Triton X-100 (Figure 1B). The eluted samples were further concentrated using Centrifugal Filters (Amicon® Ultra-15 10K). For the second IP (iPOND purification step), Dynabeads MyOne Streptavidin C1 beads (Invitrogen) (1 ml beads/ 5 billion cells) were added and incubated for 20 h at 4°C. The next day, beads were washed twice with IP buffer (QTIP-iPOND) or LB3 buffer (iPOND only), once with 500 mM NaCl and twice with LB3 buffer at RT. Samples were reverse crosslinked using 2x Laemmli buffer (150 mM Tris–HCl pH 6.8, 4% SDS, 100 mM DTT, 20% glycerol, bromophenol blue) for 30 min at 95°C and were loaded on 10% Mini-PROTEAN TGX Precast Protein Gels (BioRad) for MS analysis.
MS analysis
In-gel digestion and LC–MS/MS analyses were performed by the proteomics core facility at EPFL which followed a previously published protocol with minor modifications (4). In brief, SDS-PAGE gel lanes were sliced into 8 fractions. Samples were washed twice in 50% ethanol and 50 mM ammonium bicarbonate (AB, Sigma-Aldrich) for 20 min, and dried by vacuum centrifugation. Reduction was then performed with 10 mM dithioerythritol (Merck-Millipore) for 1 h at 56°C. After washing-drying the samples as above described, an alkylation step was performed with 55 mM Iodoacetamide (Sigma-Aldrich) for 45 min at 37°C in the dark. Samples were washed-dried again and digested overnight at 37°C using modified mass spectrometry grade trypsin (Trypsin Gold, Promega) at a concentration of 12.5 ng/μl in 50 mM AB and 10 mM CaCl2. Resulting peptides were extracted in 70% ethanol, 5% formic acid (Merck-Millipore) twice for 20 min with permanent shaking. Samples were further dried by vacuum centrifugation and stored at –20°C.
Peptides were desalted on C18 StageTips (39) and dried by vacuum centrifugation prior to LC–MS/MS injection. Samples were resuspended in 2% acetonitrile (Biosolve), 0.1% formic acid and nano-flow separations were performed on a Dinoex Ultimate 3000 RSLC nano UPLC system (Thermo Fischer Scientific) on-line connected with a Lumos Orbitrap Mass Spectrometer (Thermo Fischer Scientific). A capillary precolumn (Acclaim Pepmap C18, 3 μm-100Å, 2 cm × 75μm ID) was used for sample trapping and cleaning. Analytical separations were then performed on a 50 cm long capillary column (75 μm ID; in-house packed using ReproSil-Pur C18-AQ 1.9 μm silica beads; Dr Maisch) at 250 nl/min over a 90-min biphasic gradient. Data Dependent mode was used for MS acquisitions (m/z window: 375–1500) where parent ions were detected in the Orbitrap at a resolution of 240 000 (at m/z 200) while daughter ions were detected on the Linear Ion Trap at low resolution (Rapid mode and Max. injection time of 20 ms). Only charge states from 2 to 6 were selected for fragmentation using HCD mode with (collision energy value of 30).
Identifications were performed using MaxQuant (version 1.6.2.10) (40,41) incorporating the Andromeda search engine (42) against the UniProt human database containing 73 920 sequences (release 2019_01; www.uniprot.org). Streptavidin sequence was added manually. A concatenated decoy database of common contaminant sequences generated by MaxQuant was used to determine the false discovery rate and exclude false positive hits. Both peptide and protein identifications were filtered at 1% FDR relative to hits against the decoy database built by reversing protein sequences. The minimal peptide length was seven amino acids, at least two peptides were required for protein identification and a minimum of two ratio counts was required to quantify proteins. Carbamidomethylation (C) was set as a fixed modification, whereas oxidation (M), acetylation (protein N-term) and phosphorylation (STY) were considered as variable modifications. LFQ intensities generated by MaxQuant were used for further analysis.
Dot blot
Reverse crosslinked DNA was purified by NucleoSpin Gel and PCR Clean-up with buffer NTB (Macherey-Nagel) and eluted in 100 μl pre-warmed H2O (65°C). Next, DNA was denatured for 10 min at 95°C and chilled on ice for 10 min. DNA was blotted onto a Hybond N+ nylon membrane (GE Healthcare) using a BioRad dot blot apparatus. The membranes was UV-crosslinked. For telomere and Alu signal detection, the Hybond N+ nylon membrane was denatured with 0.5 M NaOH, 1.5 M NaCl for 15 min and neutralized with 0.5 M Tris–HCl pH 7.0, 1.5 M NaCl for 10 min. The membrane was first blocked in Church buffer (1% BSA, 1 mM EDTA pH 8.0, 0.5 M phosphate buffer pH 7.2 [0.5 M Na2HPO4, 0.17% H3PO4], 7% SDS) for 1 h at 65°C and incubated with a 32P-labeled telomeric probe overnight at 65°C. The telomeric probe was prepared by random labeling with α-32P-dCTP and cold dTTP, dATP and dGTP using the RadPrime DNA Labeling System (Invitrogen). The template mixture of 300 bp to 1 kb long telomeric DNA fragments had been synthesized by PCR using telomeric DNA oligonucleotides ((TTAGGG)5, (CCCTAA)5). After hybridization, the membrane was washed with 1× SSC containing 0.5% SDS 3 times for 30 min and exposed to a phosphorimager screen overnight. Radioactive signal was detected with Amersham typhoon (serial number 76150124, firmware version 208, FPGA version 2, software version 1.1.0.7, Cytiva) and the intensity was quantified with AIDA Image Analyzer software version 4.06.034 (Elysia-Raytest). Next, the telomeric probe was removed by washing the membrane 3 times for 20 min with boiling 0.1× SSC containing 1% SDS. Then the membrane was prehybridized with Church buffer at 55°C and hybridized with a 32P-labeled Alu probe (5′-TGATCCGCCCGCCTCGGCCTCCCAAAGTG-3′) overnight at 55°C. The Alu probe had been 5′-end-labeled with γ-32P-ATP using T4 Polynucleotide Kinase (NEB). The next day, the membrane was washed 3 times for 30 min with 1× SSC containing 0.5% SDS and exposed to a phosphorimager screen for 2 days. Radioactive signal was again detected with Amersham typhoon (Cytiva) and the intensity was quantified with AIDA Image Analyzer software version 4.06.034 (Elysia-Raytest).
Immunoblots
For protein detection, samples were resuspended in 2× Laemmli buffer and denatured for 5 min at 95°C. Samples were fractionated on 7.5% Mini-PROTEAN TGX Precast Protein Gels (BioRad) and wet-transferred onto Amersham Protran 0.2 NC nitrocellulose Western blotting membranes (GE Healthcare). Membranes were blocked with 3% BSA in 1× PBS containing 0.1% Tween 20 (1× PBST) for 1 hour at RT and incubated with primary antibodies overnight at 4°C. The next day, the membrane was washed three times for 15 min with 1× PBS containing 0.1% Tween 20. HRP-conjugated secondary antibodies: anti-mouse IgG HRP-conjugated (W4021, Promega) or anti-rabbit IgG HRP-conjugated (W4011, Promega) in combination with ECL spray (Advansta) were used to reveal the signal on a Fusion FX (Vilber) detector. The following primary antibodies were used: anti-Vinculin (ab129002, Abcam, diluted 1:10,000 in 1x PBST containing 5% milk); anti-TRF1 (sc-6165-R, Santa Cruz, diluted 1:2000 in 1× PBST containing 3% BSA).
Calcium phosphate transfection of siRNA
siGENOME SMARTpools from Horizon Discovery at a final concentration of 5.45 nM were used. HeLa-Long cells were transfected in 10 cm dishes at 30–40% confluency using a calcium phosphate precipitation protocol. 0.5 ml transfection mix was prepared containing 125 mM CaCl2, 60 nM siRNA and 1× HBSS pH 7.4 (50 mM HEPES, 280 mM NaCl, 1.19 mM Na2HPO4•2H2O, 10 mM KCl), incubated 10 min at RT and added to the cells covered with 5 ml antibiotic-free DMEM/10% FBS. The cells were harvested 72 h post transfection.
Telomere fluorescence in situ hybridization (FISH) on metaphase spreads
HeLa-Long cells were treated with 50 ng/ml demecolcine for 2 h. Supernatants were collected and the remaining cells were trypsinized. Trypsinized cells were merged with supernatants and spun down for 5 min at 300 g. Pellets were resuspended in 0.056 M KCl and swollen for 7 min at 37°C. Swollen cells were spun down for 3 min at 200 g and the supernatant was decanted. The cells were resuspended in fixative (75% methanol, 25% acetic acid) and stored at 4°C overnight. To obtain metaphase spreads, the cell suspensions were dropped on glass slides, incubated 1 min at 70°C in a wet chamber and air-dried overnight. For FISH staining, slides were rehydrated in 1× PBS for 5 min, treated with 4% formaldehyde in 1× PBS for 5 min, washed 3 times with 1× PBS for 5 min, and dehydrated with increasing amounts of ethanol (70%, 95%, 100%) for 5 min at each concentration. Air-dried slides were placed on coverslips with 70 μl hybridization mix (10 mM Tris–HCl pH 7.4, 70% formamide, 0.5% blocking reagent [11096176001, Roche], 89 μM Cy3-OO-(CCCTAA)3 PNA probe [F1002, PNA Bio]) and denatured at 80°C for 3 min. Hybridization took place for 3 h in a light-protected humified chamber at RT. The slides were washed twice for 15 min in 70% formamide/10 mM Tris–HCl pH 7.4 and three times for 5 min in 0.1 M Tris–HCl pH 7.4/0.15 M NaCl/0.08% Tween-20 with 0.1 μg/ml DAPI in the second wash. The slides were dehydrated with increasing amounts of ethanol as described above, mounted in Vectashield embedding medium and stored at -20°C. Images were taken with a Zeiss-Axioplan microscope using a 100x objective.
Quantitative RT-PCR
Cells were directly lysed in 6-well plates and total RNA was isolated using the NucleoSpin RNA isolation kit (Macherey-Nagel) according to the manufacturer's protocol with three DNase treatments. RT-qPCR was performed on a 7900HT Fast Real-Time System (Applied Biosystems) using the Luna Universal OneStep RT-qPCR kit (NEB) according to the manufacturers protocol in a 384-well reaction plate. No-reverse transcription controls were included for each RNA sample and primer mix and no-template controls for each primer mix. All experiments were performed in two technical replicates. Relative expression levels were calculated using the 2–ΔΔCt method. GAPDH RNA was used as a reference for normalization.
Quantification and statistical analysis
For MS data analysis, the MaxQuant output table ‘proteinGroups.txt’ was processed to assess the significance of outlier ratios using Perseus (43) and home-made programs written in Perl (v5.18.2) and RStudio (Version 1.2.1335). 1732 nuclear proteins were first extracted according to annotations from Gene Ontology (http://geneontology.org) (44) and The Human Protein Atlas (https://www.proteinatlas.org) (45), which contained 22 481 reference proteins that have been shown to localize to the nucleus and/or nucleolus. The ratio between two datasets was normalized to the LFQ intensity value of TRF1 and was then calculated by dividing the number of LFQ intensity values. Only proteins identified in at least 3 of the 4 datasets were included in the statistical analysis in Perseus using a two-tailed t-test with Benjamini-Hochberg FDR correction (46) with thresholds of 0.05, 0.01, 0.005 and 0.001 on the adjusted P-value and the background variance parameter s0 as 3. Graphical display was made by RStudio and Prism 8. Other statistical analyses in this study were done by unpaired t-test with Welch's correction using Prism 8 as specified in figure legends and the results section.
RESULTS
QTIP-iPOND purification of telomeric chromatin during replication
To examine the dynamic changes of the telomeric proteome that occur during replication, we developed QTIP-iPOND, which combines two previously established techniques in a consecutive manner: In the first step, crosslinked telomeric chromatin is purified by QTIP using antibodies against TRF1 and TRF2 (4,34). In the second step, the purified telomeric chromatin is further fractionated by iPOND (35,36) which separates the telomeric chromatin present at DNA replication forks from non-replicating telomeric chromatin (Figure 1A). We took advantage of HEK293E suspension cells to expand large scale cultures. Moreover, we endogenously FLAG-tagged TRF1 and TRF2 in HEK293E cells. This allowed immunoprecipitation of telomeric chromatin during QTIP purification with anti-FLAG antibodies and elution of purified telomeric chromatin with excess FLAG peptide without denaturing telomeric chromatin. To label nascent DNA at replication forks, we incubated cells with EdU (5-ethynyl-2′-deoxyuridine) - a thymidine (Thy) analog - for 10 min. In the EdU pulse experiment, cells were fixed directly after EdU incubation. EdU-labeled nascent DNA was further covalently linked with biotin-azide using a click reaction. After cell lysis, we first purified telomeric chromatin using anti-FLAG beads targeting the FLAG-tagged TRF1 and TRF2 and telomeric chromatin was eluted with excess FLAG peptide. In the second step, the telomeric chromatin associated with nascent replicating DNA was separated from non-replicating telomeric chromatin using streptavidin beads targeting the biotinylated EdU-labeled DNA. Moreover, we carried out a Thy chase experiment in which the EdU-containing medium was replaced by Thy-containing medium after EdU incorporation. For the chase, cells were harvested 4 h after Thy addition (Supplementary Figure S1A; see Materials and Methods for details).
Four biological replicates, in which 12.8 billion cells were used each time, were performed. Each replicate was analyzed by QTIP-iPOND, iPOND and QTIP. For the QTIP-iPOND analyses, the EdU-positive fraction (EdU(+)), which contained telomeric chromatin during replication, and the EdU-negative fraction (EdU(−)), which contained non-replicating telomeric chromatin, were analyzed using LC–MS/MS (Figure 1B). In order to obtain similar amounts of peptides in the EdU(+) and EdU(−) fractions, we estimated protein quantities on Coomassie-stained protein gels and used only 4% of the EdU(−) sample for LC–MS/MS analysis. For iPOND, the EdU pulse and Thy chase fractions were analyzed. For QTIP, fractions pulled down by FLAG-tagged TRF1 and TRF2 were analyzed and compared to pull-downs from cells having untagged TRF1 and TRF2. To evaluate the enrichment and recovery of telomeric chromatin in the QTIP and QTIP-iPOND samples, we performed dot blot hybridization with a telomeric probe and a probe for Alu elements, which are dispersed with more than one million copies throughout the human genome, thus representing non-telomeric regions. Upon QTIP, telomeric DNA was enriched approximately 250-fold over Alu element-containing DNA (Figure 1C). The recovery of telomeric DNA was around 30% (Figure 1D). In the QTIP-iPOND samples, approximately 0.45% of telomeric DNA was recovered. This low yield was expected as upon harvest, only 18% of the 293E cells were in S phase (Supplementary Figure S1B). Furthermore, only a subset of S phase telomeres was expected to be labeled during the 10 min EdU pulse. Alu repeat elements were very low in the QTIP samples (0.14% average) and barely detectable after QTIP-iPOND (0.001%) (Figure 1D, Supplementary Figure S1C and S1D). These results indicated a strong enrichment of telomeric over contaminating chromatin. The large-scale experiments gave sufficient material to allow analysis by mass spectrometry.
Proteomic analysis reveals proteins that are enriched at telomeres during DNA replication
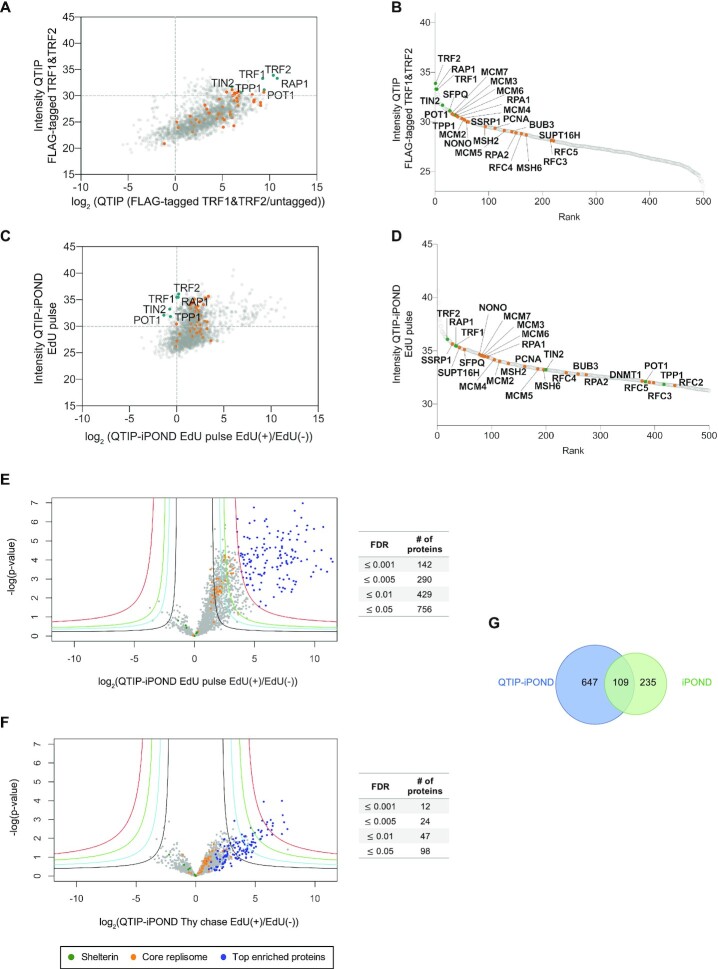
We first analyzed the intensity of telomeric proteins in telomeric QTIP fractions obtained upon purification via the FLAG epitope present on endogenously FLAG-tagged TRF1 and TRF2 cells. When compared to chromatin obtained from negative control cells with untagged TRF1 and TRF2, the shelterin components were the most abundant proteins as expected (Figure 2A and B). In addition, several DNA replication proteins were enriched in the telomeric chromatin (orange dots in Figure 2A and B). Next, we attempted to identify the proteins enriched near telomeric replication forks. Therefore, we compared by label-free quantification (LFQ) the intensity of proteins present in EdU-positive versus EdU-negative fractions (EdU(+)/EdU(−)) obtained upon QTIP-iPOND (Figure 2C and D) from the EdU pulse labeled cells. Since telomeric proteins were purified by pulling down TRF1 and TRF2, and TRF1 is present at non-replicating as well as replicating telomeres (15), we normalized protein amounts to TRF1. 1732 nuclear proteins were identified. We first examined 40 core replication proteins which have known functions for genome-wide DNA replication and were identified here as well as in several replisome purification approaches before (47) (see orange dots in Figure 2C and D). 35 of the core replication proteins were enriched more than 2.8-fold (log2(1.5)) in the EdU(+) over the EdU(−) telomeric chromatin fractions (Figure 2C and E). These enrichment factors are comparable to the ones reported for iPOND samples (36,48,49), indicating the successful enrichment of replication proteins in the EdU(+) fractions upon QTIP-iPOND purification.
Figure 2.
Protein distribution upon QTIP-iPOND. (A) Comparison of enrichment of core replication proteins to shelterin components by QTIP. (B) Rank of 500 top enriched proteins in QTIP EdU pulse samples. (C) Enrichment of core replication proteins with respect to shelterin components upon QTIP-iPOND in EdU pulse EdU(+) samples. (D) Rank of 500 top enriched proteins in QTIP-iPOND EdU pulse EdU(+) samples. Intensity: label-free quantification (LFQ) intensity. (E) Comparison of QTIP-iPOND enriched proteins in EdU(+) versus EdU(−) samples in the EdU pulse experiment. (F) Comparison of QTIP-iPOND enriched proteins in EdU(+) versus EdU(−) samples in the Thy chase experiment. Red curves: FDR = 0.001; green curves: FDR = 0.005; cyan curves: FDR = 0.01; black curves: FDR = 0.05; s0 = 3. (A–F) Orange dots represent core replication proteins; dark green dots represent shelterin components; blue dots represent the 142 top enriched proteins with FDR ≤ 0.001 in QTIP-iPOND EdU pulse samples. (G) Comparison of QTIP-iPOND (756 proteins) and iPOND (344 proteins) EdU pulse enriched proteins.
Next, we determined which proteins were >2.8-fold enriched in EdU(+)/EdU(−) and had an FDR (false discovery rate) ≤0.05 in the QTIP-iPOND EdU pulse samples (Figure 2E and Supplementary Table S1). 756 proteins fulfilled these criteria of which 142 had an FDR ≤0.001. Importantly, the enrichment of these proteins was also strongly reduced in the Thy chase EdU(+) samples (Figure 2F and Supplementary Table S2), confirming that these proteins were specifically enriched at telomeres during their replication. We also searched the Contaminant Repository for Affinity Purification (CRAPome) database with the here identified proteins (50). Out of the 142 top enriched proteins with FDR ≤0.001, only nine proteins were present in the CRAPome list. Out of the 756 enriched proteins with FDR ≤0.05, an additional 72 proteins were present in the contamination list, including MCM3, PCNA, NONO, SFPQ and several others, which we expected to be present at telomere replication forks. Thus, we did not exclude factors from the CRAPome list. Overall, this analysis supported the specific enrichment of replicating telomeric chromatin by QTIP-iPOND.
We also compared the telomeric to the general replisome. Therefore, we performed iPOND analyses upon EdU pulse and Thy chase (Figure 1B). 344 proteins were enriched with an FDR ≤0.05 in iPOND during the EdU pulse. Of these, roughly one third (109 proteins) were also enriched in QTIP-iPOND EdU pulse (Figure 2G) indicating different compositions of chromatin during replication at telomeres versus the bulk of the DNA. There was no overlap between the proteins present in the iPOND EdU pulse and QTIP-iPOND Thy chase fractions. This result confirms that replication proteins were removed from EdU upon Thy chase.
Of note, we observed upon replacement of EdU by thymidine in the Thy chase experiments a slower recovery of DNA replication in QTIP-iPOND than in iPOND only samples. We suspect that the repeated pelleting and resuspension of cells in order to wash out the EdU before addition of thymidine may have induced cellular stress, which affected replication forks at telomeres for longer periods of time than at non-telomeric sites. We observed a 3.2-fold enrichment of PCNA in EdU pulse EdU(+)/Thy chase EdU(+) in iPOND samples against only a 1.9-fold enrichment in the QTIP-iPOND chromatin (Supplementary Tables S1-S3). Therefore, the majority of replication proteins and the 142 top enriched proteins were still enriched at telomeres when we compared the EdU(+) fractions of EdU pulse versus Thy chase in QTIP-iPOND samples, though to a much lesser extent (compare Supplementary Figure S2A and Figure 2E). As expected, the EdU(−) fractions obtained in QTIP-iPOND, which mainly contained the non-replicating telomeric proteome, showed no striking differences between the EdU pulse and the Thy chase (Supplementary Figure S2B).
QTIP-iPOND detects core replisome components at telomeres
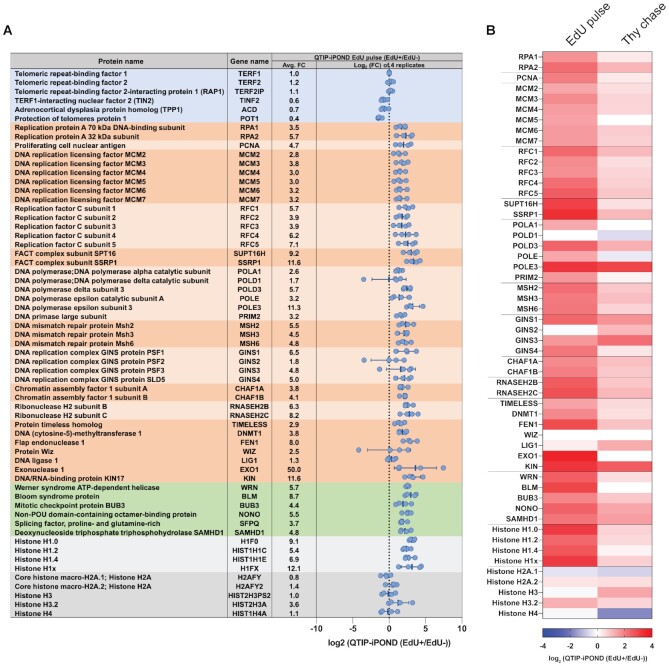
40 proteins that represent core replication proteins are listed in Figure 3, including RPA subunits, PCNA, MCM proteins, RFC proteins, FACT complex, MSH proteins, DNA polymerases and others. The enrichment of these proteins was similar in all four biological replicates of QTIP-iPOND (Figure 3A) and was largely decreased in the Thy chase sample (Figure 3B), demonstrating that the enrichment of these proteins at telomere replication forks was reproducible and significant.
Figure 3.
List of selected proteins identified in the QTIP-iPOND experiments. (A) Proteins in the blue background are shelterin components; proteins in the dark/light orange background are core replication proteins; proteins in the green background are telomere replication proteins; proteins in the dark/light grey background are histones. Each blue dot indicates the log2(EdU(+)/EdU(−)) in one replicate. Avg. FC: the average fold change of EdU(+)/EdU(−). Data are represented as mean ± standard deviation. (B) Heatmap of replication proteins and histones shown in (A) indicating the log2(EdU(+)/EdU(−)) in the QTIP-iPOND EdU pulse and Thy chase.
Moreover, we identified proteins that had been shown to function specifically in telomere replication (Figure 3). This includes WRN (12), BLM (17), NONO and SFPQ (29), and BUB3 (51). SAMHD1 was also enriched at telomere replication forks, which has been recently identified by QTIP in oncogene-transformed human lung fibroblasts (HLFs) and is important for telomere integrity upon TRF1 depletion (52). RTEL1 was not detected in both QTIP-iPOND and QTIP (Supplementary Tables S1 and S4), possibly due to its low abundance at telomeres. Overall, these results indicated that telomeric replication proteins were specifically purified by QTIP-iPOND.
POT1 is partially depleted from replication forks at telomeres
When we compared the abundance of proteins of EdU-positive and EdU-negative QTIP-iPOND fractions the values had been normalized to TRF1. Thus, it was not unexpected that the shelterin components TRF2 and Rap1 did not change in abundance relative to TRF1 (Figure 3A). Interestingly, however, TIN2, TPP1 and most noticeably POT1 were less abundant in the replicating versus non-replicating telomeric chromatin fractions, with POT1 being reduced by 60% (Figure 3A). Thus, our data provide direct experimental evidence that POT1 is removed from telomere replication forks. Previous findings already suggested that POT1 is not required for semiconservative replication of telomeres. Deletion of POT1 causes rapid telomere elongation that is led by the homologous recombination machinery (33). Depletion of recombination proteins suppressed the rapid telomere elongation phenotype of POT1 deficient cells while not causing immediate telomere maintenance defects. Thus, it seemed that replication of telomeres is occurring normally in the absence of POT1 and RAD51 (33). On the other hand, we found that not only RPA but also the TERRA- and single stranded telomeric DNA-binding protein hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1) are enriched in the QTIP-iPOND EdU(+) fraction (Supplementary Table S1). Both of these proteins have been implicated to regulate POT1 binding during S phase (37). According to the model of Flynn et al., hnRNPA1 displaces RPA from telomeric ssDNA in late S phase during which TERRA levels are low. Upon TERRA increase after replication, hnRNPA1 is released from telomeric DNA associating with TERRA and liberating the telomeric DNA for POT1 (37). Our results support the model that POT1 is replaced at telomeres during DNA replication by RPA and hnRNPA1.
Histone H1 variants are present during telomere replication
In our iPOND samples, the levels of core histone proteins remained unchanged during and after DNA replication (Supplementary Figure S3), which is consistent with previous reports (35,53,54). Our QTIP-iPOND analyses indicated that also at telomeres, histones H2A, H3 and H4 are similarly abundant during and after replication though slight variations were observed in individual experiments (Figure 3). Very strikingly, however, we identified that four histone H1 variants were strongly enriched in the EdU(+) over EdU(−) QTIP-iPOND fractions (Figure 3A and B). Their abundance also decreased in the Thy chase samples (Figure 3B). In contrast, histone H1 variants were depleted in the iPOND EdU pulse samples (Supplementary Figure S3), which is consistent with previous nascent chromatin purification experiments (49,53,54).
Previous studies have shown that at telomeres, nucleosomes are closely spaced (55–57). Furthermore, histone H1 was not detected in telomeric chromatin suggesting that it may be depleted from telomeres (3). On the other hand, it was observed that the depletion of histone H1 increases the amount of telomere sister chromatid exchanges (T-SCEs) and telomere length in murine embryonic stem cells (58). Our results appear to be consistent with both of these observations, indicating that histone H1 variants are enriched at telomeres only during their replication. Thus, histone H1 may play critical roles for maintaining telomere stability upon synthesis.
Telomerase subunits and nucleolar proteins are detected near telomere replication forks
From the 142 top enriched proteins, 139 were specifically enriched in QTIP-iPOND but not in iPOND (Figure 4 (highlighted with red background) and Supplementary Figure S4). Interestingly, three of the four H/ACA ribonucleoprotein complex subunits, DKC1, NHP2 and GAR1, which associate with the telomerase RNA moiety hTR, were identified. Although we did not detect hTERT in our MS samples, which may be due to technical limitations, the identification of three telomerase subunits suggests that the telomerase complex or subcomplexes may travel with telomere replication forks as suggested previously but never directly demonstrated (59,60). Of note, EdU is not a substrate for telomerase (61), indicating that in QTIP-iPOND we did not purify telomeric chromatin during elongation by telomerase. However, we cannot exclude the possibility that telomerase subunits were recruited to the replicating chromatin only once the semi-conservative replication machinery reached the end of telomeric repeats.
Figure 4.
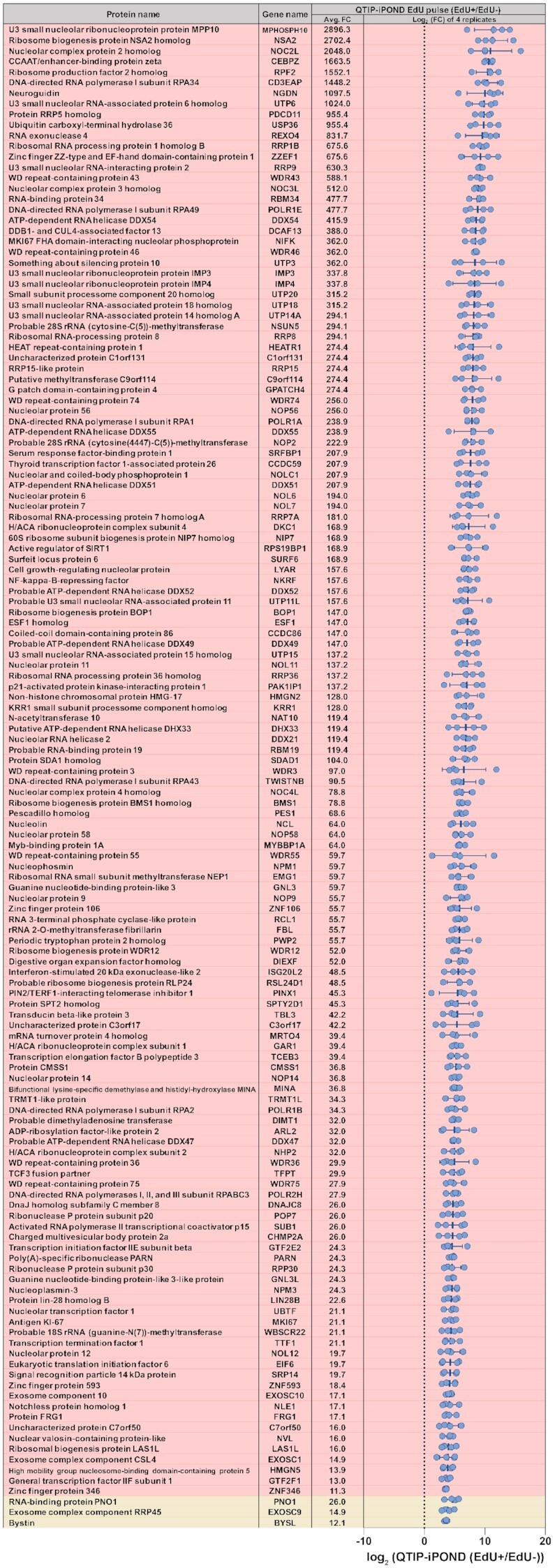
List of the 142 proteins that were top-enriched by QTIP-iPOND (EdU pulse EdU(+) samples). The list indicates the 142 proteins for which the FDR is below 0.001. Each blue dot indicates the log2(EdU(+)/EdU(−)) ratio of one replicate. Proteins in the red background are specifically enriched in telomere replication; proteins in the light-yellow background were enriched by both, QTIP-iPOND and iPOND (EdU pulse). Avg. FC: the average fold change of EdU(+)/EdU(−). Data are represented as mean ± standard deviation.
We also identified nucleolar proteins with known functions related to telomere biology. This includes the Guanine nucleotide-binding proteins GNL3L and GNL3 (also known as nucleostemin) which stabilize and destabilize TRF1, respectively (62–64). The enrichment of both proteins suggests a tight regulation of TRF1 at telomeres during replication. Another identified protein is NOLC1 (nucleolar and coiled-body phosphoprotein 1) which has been reported to interact with TRF2 and regulate its nuclear and nucleolar localization (65). DDX21 has been shown to interact with WRN and bind TERRA, probably by interacting with G-quadruplex structures in TERRA (66–68). Besides, PINX1, a Pin2/TRF1-interacting protein which mediated the accumulation of TRF1 in nucleoli and also interacts with TRF1 in the nucleoplasm enhancing TRF1 binding to telomeres (69), was identified, as well as PARN, a poly(A)-specific ribonuclease which is involved in hTR maturation, the regulation of shelterin and RTEL1 transcript expression. PARN deficiency is associated with the Høyeraal–Hreidarsson (HH) syndrome (70–72).
Telomere fragility assay reveals the requirement of proteins identified by QTIP-iPOND for efficient telomere replication
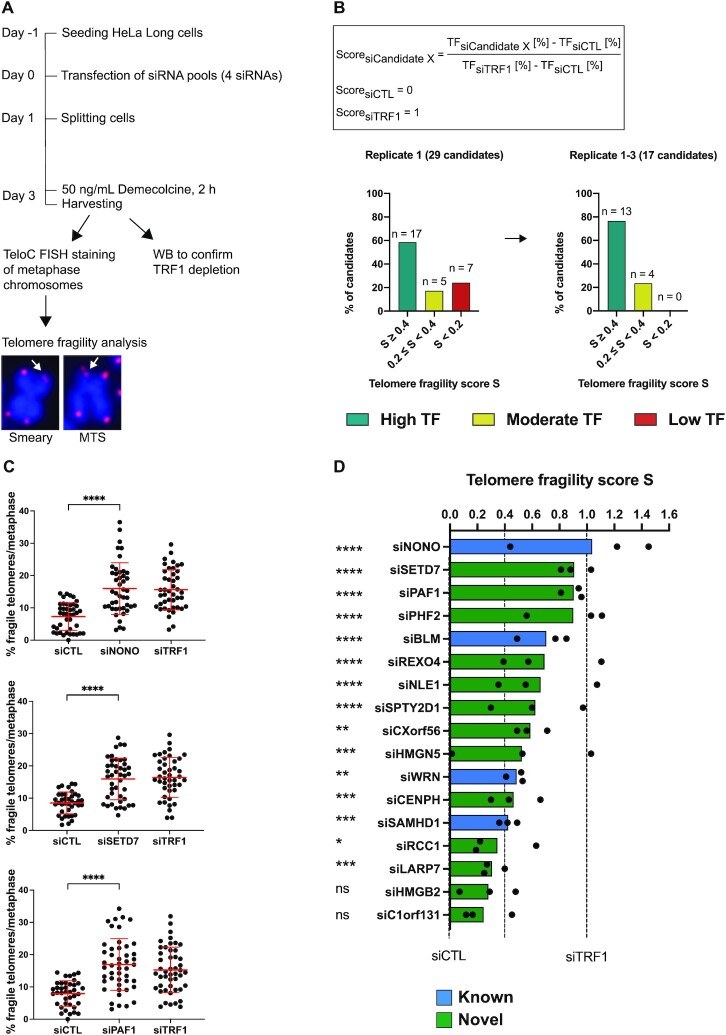
To assess the functional relevance of identified proteins for telomere replication, we chose 24 proteins enriched in QTIP-iPOND that were not present in iPOND-only fractions and which had not been previously linked to telomere biology. We depleted these proteins from HeLa-Long cells (which have an average telomere length of 33 kb) using siRNA pools, with each pool consisting of 4 different siRNAs targeting the same candidate (Figure 5 and Supplementary Figure S5A). After siRNA transfection, telomere morphology of metaphase chromosomes was analyzed by fluorescence in situ hybridization with a telomere specific probe (Figure 5A). Telomeres that showed smeary or multiple telomeric signals upon gene depletion were recognized as fragile telomeres and inferred to have telomere replication defects (15,73). Apart from the positive control siTRF1, we depleted four additional known telomere replication proteins (BLM, WRN, NONO and SAMHD1) that were present in our QTIP-iPOND samples (12,14,29,52,74). Of note, SFPQ, the functional partner of NONO, was depleted in our siRNA screen as well (Supplementary Figure S5A). However, its depletion did not substantially increase telomere fragility which is consistent with previous literature (29). Based on the levels of telomere fragility of the non-targeting control and TRF1 depletion that were assessed in each individual experiment, a score was assigned to each candidate (Figure 5B). The non-targeting control was given a score of 0 and TRF1 depletion a score of 1 (Figure 5B). Out of the 29 depleted proteins, 17 (59%) gave a telomere fragility score that was higher than 0.4 (high telomere fragility), 5 (17%) of them scored between 0.2 and 0.4 (moderate telomere fragility), and 7 (24%) of them were scored below 0.2 (low telomere fragility) in the first experiment (Figure 5B and Supplementary Figure S5). We repeated the fragility analysis for the 17 proteins that had scores higher than 0.4 (Figure 5C, Supplementary Figure S6 and S7A). Thirteen (76%) of them (including the four positive controls BLM, WRN, NONO and SAMHD1) remained in the same high fragility category (score ≥ 0.4) in experiments carried out in triplicate (Figure 5B–D). The depletion level of the mRNAs for all 17 proteins was determined by RT-qPCR, showing a reduction of mRNA levels by 54–95%. In addition, TRF1 depletion was confirmed by western blot analysis for each experiment (Supplementary Figure S7B and C). Altogether, we could confirm a significant effect of 11 novel telomere replication proteins in protecting telomeres from fragility.
Figure 5.
Candidate validation through telomere fragility analysis. (A) Experimental outline of siRNA screen for telomere fragility using HeLa-Long cells (with an average telomere length of 33 kb). MTS: multiple telomeric signals. Pixel size of FISH images was reduced 4-fold for illustration. (B) Telomere fragility score calculation and score for tested candidates. The percentage of fragile telomeres per metaphase was determined for each depleted protein candidate and compared to TRF1-depleted cells (siTRF1) and control transfected cells that had been transfected with a non-targeting siRNA (siCTL). A score of 1 corresponds to the amount of telomere fragility detected in TRF1-depleted cells and a score of 0 to the amount of telomere fragility detected in the negative control (siCTL). TF: telomere fragility. (C) Representative data of 3 biological replicates of siRNA screen for the top three hits. Percentage of fragile telomeres per metaphase is shown. Red bars represent the mean and standard deviation. (D) Telomere fragility score for three biological replicates with 39–51 metaphases analyzed per condition in total. Blue and green indicate factors that have known or novel functions in counteracting telomere fragility, respectively. (C, D) Statistical analysis was done by unpaired t-test with Welch's correction to compare the percentage of fragile telomeres per metaphase of siCandidate X to siCTL. * <0.05, ** <0.01, *** <0.001, **** <0.0001.
Next, we analyzed the 11 validated candidates by gene ontology (44,75). The biological processes to which the 11 novel telomere replication proteins have previously been classified include nucleosome organization, cell cycle regulation, histone modification, and RNA processing (Table 1). The validated candidates include histone modifiers and chaperones (SETD7, PAF1, PHF2 and SPTY2D1) which might have roles in modifying the chromatin environment at replicated telomeres, an RNA helicase (REXO4) that might play roles in regulating TERRA while telomere replication takes place, and LARP7 whose fission yeast analog Lar7 has important functions in telomerase biogenesis and in repressing transcription of telomeric non-coding RNAs (76,77). In addition, telomere shortening was observed in LARP7-deficient Alazami syndrome patients and knockdown of LARP7 in human cells decreased telomerase enzymatic activity (78). Altogether, our results support the notion that a large fraction of proteins identified by QTIP-iPOND protects telomeres from fragility, indicating that they sustain telomere replication, further validating QTIP-iPOND as a method. The small amounts of TRF1 and TRF2 which may be present at non-telomeric regions did not prevent the identification of crucial components of the telomeric replisome.
Table 1.
GO of the 11 newly identified telomere replication proteins
| Biological process (Gene ontology) | Gene name | |
|---|---|---|
| Nucleosome organization | GO:0034728 | PAF1, CENPH, SPTY2D1 |
| Regulation of cell cycle | GO:0051726 | PAF1, HMGN5, RCC1, NLE1 |
| Histone modification | GO:0016570 | PAF1, SETD7, PHF2 |
| RNA processing | GO:0006396 | PAF1, REXO4, LARP7 |
| Unclassified | CXorf56 | |
Network of telomere replication proteins
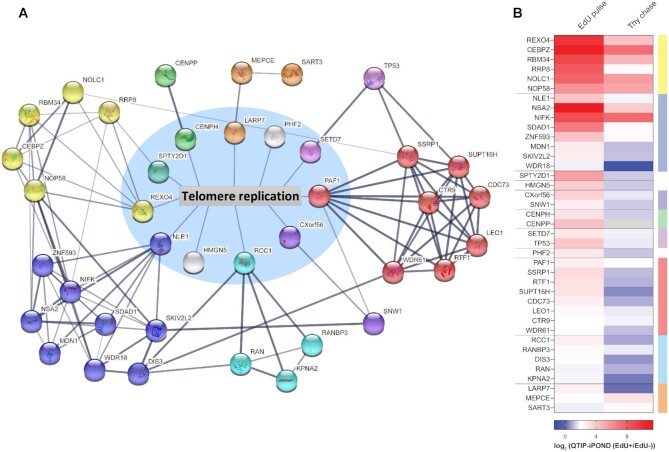
To further investigate if known interactors of the newly identified telomere replication proteins might also participate in telomere replication, we took advantage of the STRING database (https://string-db.org, v11.0b) (79). We included in the analysis the 10 most confident interactors of each of the 11 novel telomere replication proteins that protected telomeres from fragility. Only proteins directly linked to the target proteins by physical interaction were included in the analysis. Among these proteins, 28 proteins were enriched in our QTIP-iPOND EdU(+) samples (Figure 6 and Supplementary Figure S8) and only two of these were also enriched in the iPOND EdU pulse fractions (TP53 and KPNA2). We represented each factor as a node. Proteins showing extensive interactions were defined as groups and color-coded (Figure 6A). While some groups formed highly interactive networks, suggesting that they work in a complex (or in the same pathway), others showed only one or two interactions. Strikingly, the groups further connected with each other forming an extensive network (Figure 6). A total of 94 links were detected between the 39 proteins, which are significantly more interactions than expected from random events (expected link number: 10, PPI enrichment P-value: < 10–16).
Figure 6.
Interactome of telomere replication proteins. (A) Network of proteins that is composed of groups of validated candidates. Proteins inside the circle with a blue background are here validated telomere replication proteins. Proteins outside the blue circle are the most confident hits provided by STRING enriched in QTIP-iPOND EdU pulse EdU(+). Nodes with the same color indicate proteins in the same group. (B) Heatmap of proteins in the network upon QTIP-iPOND EdU pulse and Thy chase. Color code on the right corresponds to the color of nodes in (A). Note that the white baseline is at 2.
Interestingly, factors participating in nucleosome organization and RNA surveillance were particularly enriched. For instance, all six components of the PAF1 complex (PAF1, CTR9, CDC73, LEO1, RTF1 and WDR61) which has diverse roles in transcription regulation, RNA processing and histone modification (the red group in Figure 6) (80) were enriched in the QTIP-iPOND EdU(+) sample. Among them, PAF1, CTR9, RTF1 and WDR61 are functionally related to histone methylation (80). WDR61 (also named SKI8) also belongs to the SKI complex which is involved in RNA decay (81). The other SKI complex component, SKI2, was also significantly enriched in the QTIP-iPOND EdU(+) samples. WDR61 is also linked to the exoribonuclease DIS3 (blue node in Figure 6) which participates in RNA decay (82). Moreover, PAF1 connects with the transcription regulator SNW1 in the CXorf56 group (purple), which in turn is linked to the ATP-dependent RNA helicase SKIV2L2 (blue). The levels of TERRA are regulated during the cell cycle, with a prominent decrease in S phase (83). The removal of TERRA from telomere replication forks is important for preventing collisions with TERRA RNA-DNA hybrids (21,25–27,31,84,85). However, the mechanisms behind the regulation of TERRA levels during S phase remain ill-defined. The network of proteins discovered in this analysis reveals possible regulators of TERRA during telomere replication.
In our analysis, SSRP1 (red), a member of the FACT complex which participates in DNA replication, is linked to NOLC1 (yellow), a nucleolar protein that is known to interact with TRF2 (65). Both NOLC1 and SSRP1 also physically interact with histone H1 in human CEM cells (65,86). Since histone H1 was highly enriched in our QTIP-iPOND EdU(+) samples, it is possible that NOLC1 and SSRP1 associate with telomere replication forks via H1.
Histone modifiers have been shown to associate with nucleosomes during DNA replication and regulate the reestablishment of epigenetic marks on newly assembled histones (87). Additional interactors that are involved in the regulation of nucleosome organization identified in our analysis include the two histone methyltransferases SETD7 (purple) and RRP8 (yellow) and the two histone chaperones SPTY2D1 (cyan) and SART3 (orange) (88–91). While the two novel telomere replication proteins PHF2 (a lysine-specific demethylase) and HMGN5 (a nucleosome-binding protein) did not have any interactor identified in this analysis, they are both functionally related to nucleosome organization (92,93). With 5 of the 11 novel telomere replication proteins (PAF1, PHF2, SPTY2D1, SETD7 and HMGN5) and their interactors functioning in nucleosome organization (80,88,91–93), our data support the notion that nucleosome organization and histone methylation in particular, play important roles in telomere replication.
Although there is no known interaction between REXO4 and NLE1, the two groups (the yellow and blue nodes, respectively) are extensively connected, suggesting that they may cooperate. The existence of these extensive interactions within the network supports the notion that the list of telomere replication proteins that we identified in QTIP-iPOND is highly specific. The list should serve as a useful resource to investigate the roles and cooperation of these proteins in telomere replication.
DISCUSSION
The successful replication of telomeres is crucial for maintaining genome stability and preventing cancer and telomere syndromes, yet the molecular mechanisms are not fully understood. Here, we developed QTIP-iPOND to investigate telomere replication in a comprehensive manner. The method allowed the comparison of replicating and non-replicating telomeric proteomes as the telomeric chromatin obtained by the QTIP step was further fractionated into EdU(+) and EdU(−) telomeric fractions using the iPOND protocol (Figure 1). The pulse-chase experiment that was classically employed in iPOND experiments causes pronounced replication stalling at telomeres. Therefore, our design circumnavigated this problem and was able to reveal the dynamic changes of proteins that occur at telomeres during replication.
In our quadruplicate experiment, we identified the majority of core replication proteins and known telomere replication factors using a stringent selection threshold (2.8-fold change and FDR ≤ 0.05) (Figures 2 and 3). We identified 756 proteins near telomere replication forks of which 647 were specifically present at telomeres but not at non-telomeric replication forks. Of the 647 proteins, we validated 29 proteins using telomere fragility analysis (Figure 5). 76% of them showed moderate to high telomere fragility when depleted. This result provides functional evidence that QTIP-iPOND allows the specific identification of telomere replication proteins. Eleven proteins that showed strong telomere fragility upon depletion had not been linked to replication before. In addition to general replication proteins, we identified 142 proteins which were very highly and significantly enriched near telomere replication forks (FDR < 0.001), of which only three were detected by iPOND (Figure 4). This supports the notion that the telomeric replisome requires a large set of specialized factors. We also identified telomerase subunits enriched in the replicating telomeric proteome, suggesting that the telomerase complex is present. Live cell imaging has demonstrated that telomerase makes during S phase thousands of transient telomere interactions that last less than one second (94,95). Longer lasting interactions occur only when telomerase engages with the telomeric 3′ overhang during telomere elongation. However, we expect that active telomerase-mediated telomere elongation is not captured by QTIP-iPOND as EdU inhibits telomerase in vitro (61). Thus, it seems that the detection of telomerase subunits in replicating chromatin is due to transient short-lived interactions with telomeres.
Interestingly, among the top enriched proteins were several that preferentially localize to nucleoli. hTERT has been found to associate with nucleoli during S phase (96,97). Many nucleolar proteins such as PINX1, PARN, nucleostemin and GNL3L have also been reported to interact with shelterin components and participate in the regulation of telomere length (64,69–71,98). Furthermore, six of the nine novel telomere replication proteins that we verified in our study (SETD7, PHF2, REXO4, NLE1, SPTY2D1 and CENPH) show nucleolar localization according to the gene ontology resource (44,75). Since highly repetitive sequences are an important feature of both telomeric DNA and ribosomal DNA (rDNA), it is possible that they share a group of proteins for DNA replication. For instance, BLM has been shown to bind to both telomere replication forks and to the non-transcribed region of rDNA which is the initiation location of replication (99). The detailed mechanisms by which these novel telomere replication factors contribute to telomere maintenance need to be unraveled in the future. Overall, our results suggest crosstalk between telomeres and nucleoli. It is tempting to speculate that these structures contribute to telomere replication by their propensity to undergo phase separation.
Linker histone H1 is the least well understood histone protein (100). Several studies in different species have suggested that histone H1 participates in regulating the timing of replication, especially at late replicating regions (101,102). Surprisingly, we observed that histone H1 is enriched at telomeres during replication, whereas it is depleted from mature telomeric chromatin (3) (Figure 3). Since previous studies showed an increase of T-SCE and an elongation of telomere length in H1-depleted murine embryonic stem cells (58), our results suggest that histone H1 specifically contributes to telomere stability during replication. H1 removal in replicated telomeric chromatin should subsequently enable the formation of compacted telomeric chromatin displaying a short nucleosomal periodicity (55–57) for which a columnar packing has been proposed (103). We do not know the identity of the factors that remove H1 from replicated telomeric chromatin. Intriguingly, we identified HMGN5 enriched in replicating telomeric chromatin and its depletion increased telomere fragility. HMGN5 binds nucleosomes counteracting linker histone-mediated chromatin compaction (93,104). It will be interesting to test how HMGN5 and H1 cooperate at replicating telomeres. We also identified the chromatin assembly factor 1 (CAF-1) subunits CHAF1A and CHAF1B enriched at replicating telomeres. CAF-1 acts as H3/H4 histone chaperone and is responsible for replication-dependent nucleosome assembly. Its importance at telomeres has been demonstrated in Arabidopsis in which loss of CAF-1 function caused dramatic telomere shortening (105).
Our data show that POT1 is depleted from replication forks at telomeres (Figure 3). Previous studies suggested roles of POT1 in semi-conservative DNA replication. Overexpression of POT1 mutants which are prevalent in T cell lymphoma in human fibrosarcoma HT1080 cells increased telomere fragility indicative of DNA replication defects (106). On the other hand, it has been proposed that POT1 is replaced by RPA at telomeres in S phase (37). Our data support the notion that POT1 is replaced by RPA for telomere replication. Therefore, we suspect that the telomere fragility elicited upon overexpression of POT1 mutants was due to indirect roles of POT1 for telomere maintenance, which may relate to its function in suppressing homology directed repair at telomeres, which in turn promotes telomere fragility (33,73).
We built a network according to our validated new telomere replication proteins and their interactors at telomere replication forks (Figure 6). The network reveals that proteins that are involved in nucleosome organization and RNA surveillance are enriched in the QTIP-iPOND EdU(+) samples. Histone disassembly and reassembly are important processes during DNA replication. It has been shown that post-translational modifications are inherited, while histone modifiers and histone chaperons are needed to maintain the modification marks (87,107). Our result discovers several potential candidates that may be involved in histone modifications and nucleosome regulation at telomeres. Besides, TERRA plays an important role in telomere maintenance (21). However, the level of TERRA remains low in the S phase, implying that it is regulated to avoid the possible hazards caused by collisions with DNA-RNA hybrids (21,25–27,83–85). In our analysis, we found several proteins or protein complexes that have previously been associated with RNA decay or transcription regulation.
To conclude, QTIP-iPOND enabled the discovery of the dynamic changes that occur in the telomeric proteome to mediate telomere maintenance. QTIP-iPOND not only identified the core components of the replicative machinery, but also the large set of protein components which ensures that telomere replication occurs smoothly at all chromosome ends. Our data provides a comprehensive resource and describes a sophisticated machinery that orchestrates hundreds of proteins to accomplish telomere replication, which will be useful to determine how defects in telomere replication cause disease.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (108) partner repository with the dataset identifier PXD018712.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Lingner lab for discussions and sharing reagents; Galina Glousker, Suna In, Eftychia Kyriacou and Rita Valador Fernandes for comments on the manuscript; Romain Hamelin, Florence Armand and Diego Chiappe at the Proteomics Core Facility at EPFL for mass spectrometry analysis; the Proteomic Expression Core Facility at EPFL for HEK293E cells; the Flow Cytometry Core Facility at EPFL for technical advice; the Gene Expression Core Facility at EPFL for technical advice and maintenance of the qPCR instrument; and the Bioimaging and Optics Platform at EPFL for training and maintenance of the Zeiss-Axioplan microscope.
Author contributions: G.L. and J.L. conceived the study initially. C.-Y.G.L., A.C.N., T.L. and A.V. planned and carried out the experiments that are shown in the paper, G.L. constructed the FLAG-tagged cell line and carried out pilot experiments, and C.-Y.G.L. and J.L. wrote the manuscript with contributions from A.C.N.
Notes
Present address: Aleksandra Vančevska, The Francis Crick Institute, London, UK.
Present address: Gérald Lossaint, Institut du Cancer de Montpellier, Montpellier, France.
Contributor Information
Chih-Yi Gabriela Lin, Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland.
Anna Christina Näger, Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland.
Thomas Lunardi, Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland.
Aleksandra Vančevska, Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland.
Gérald Lossaint, Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland.
Joachim Lingner, Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation (SNSF) [310030_184718]; SNSF-funded National Centre of Competence in Research RNA and Disease Network [182880]; Innovative Training Network (ITN) (aDDRess) from the European Commission's Seventh Framework Programme [812829]; EMBO long-term postdoctoral fellowship [ALTF 106-2018 to C.-Y.G.L.]; PhD fellowship from the Boehringer-Ingelheim Fonds [to A.C.N.]; EPFL. The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. de Lange T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018; 52:223–247. [DOI] [PubMed] [Google Scholar]
- 2. Bartocci C., Diedrich J.K., Ouzounov I., Li J., Piunti A., Pasini D., Yates J.R., Lazzerini Denchi E.. Isolation of chromatin from dysfunctional telomeres reveals an important role for ring1b in NHEJ-mediated chromosome fusions. Cell Rep. 2014; 7:1320–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Déjardin J., Kingston R.E.. Purification of proteins associated with specific genomic loci. Cell. 2009; 136:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grolimund L., Aeby E., Hamelin R., Armand F., Chiappe D., Moniatte M., Lingner J.. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat. Commun. 2013; 4:2848. [DOI] [PubMed] [Google Scholar]
- 5. Fagagna F. d’Adda di, Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P.. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003; 426:194–198. [DOI] [PubMed] [Google Scholar]
- 6. Maciejowski J., de Lange T.. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017; 18:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Armanios M. Syndromes of telomere shortening. Annu. Rev. Genomics Hum. Genet. 2009; 10:45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glousker G., Lingner J.. When telomerase causes telomere loss. Dev. Cell. 2018; 44:281–283. [DOI] [PubMed] [Google Scholar]
- 9. Glousker G., Touzot F., Revy P., Tzfati Y., Savage S.A.. Unraveling the pathogenesis of Hoyeraal-Hreidarsson syndrome, a complex telomere biology disorder. Br. J. Haematol. 2015; 170:457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kong C.M., Lee X.W., Wang X.. Telomere shortening in human diseases. FEBS J. 2013; 280:3180–3193. [DOI] [PubMed] [Google Scholar]
- 11. Sarek G., Marzec P., Margalef P., Boulton S.J.. Molecular basis of telomere dysfunction in human genetic diseases. Nat. Struct. Mol. Biol. 2015; 22:867–874. [DOI] [PubMed] [Google Scholar]
- 12. Crabbe L. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004; 306:1951–1953. [DOI] [PubMed] [Google Scholar]
- 13. Deng Z., Glousker G., Molczan A., Fox A.J., Lamm N., Dheekollu J., Weizman O.-E., Schertzer M., Wang Z., Vladimirova O.et al.. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal–Hreidarsson syndrome. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3408–E3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drosopoulos W.C., Kosiyatrakul S.T., Schildkraut C.L.. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J. Cell Biol. 2015; 210:191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vannier J.-B., Pavicic-Kaltenbrunner V., Petalcorin M.I.R., Ding H., Boulton S.J.. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012; 149:795–806. [DOI] [PubMed] [Google Scholar]
- 17. Yang Z., Takai K.K., Lovejoy C.A., de Lange T.. Break-induced replication promotes fragile telomere formation. Genes Dev. 2020; 34:1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 19. Sarek G., Kotsantis P., Ruis P., Van Ly D., Margalef P., Borel V., Zheng X.-F., Flynn H.R., Snijders A.P., Chowdhury D.et al.. CDK phosphorylation of TRF2 controls t-loop dynamics during the cell cycle. Nature. 2019; 575:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vannier J.-B., Pavicic-Kaltenbrunner V., Petalcorin M.I.R., Ding H., Boulton S.J.. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012; 149:795–806. [DOI] [PubMed] [Google Scholar]
- 21. Azzalin C.M., Lingner J.. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015; 25:29–36. [DOI] [PubMed] [Google Scholar]
- 22. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 23. Feretzaki M., Renck Nunes P., Lingner J.. Expression and differential regulation of human TERRA at several chromosome ends. RNA. 2019; 25:1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Porro A., Feuerhahn S., Delafontaine J., Riethman H., Rougemont J., Lingner J.. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014; 5:5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feretzaki M., Pospisilova M., Valador Fernandes R., Lunardi T., Krejci L., Lingner J.. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature. 2020; 587:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M.. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014; 5:5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S., Bender K., Luke B.. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013; 20:1199–1205. [DOI] [PubMed] [Google Scholar]
- 28. Luke B., Panza A., Redon S., Iglesias N., Li Z., Lingner J.. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008; 32:465–477. [DOI] [PubMed] [Google Scholar]
- 29. Petti E., Buemi V., Zappone A., Schillaci O., Broccia P.V., Dinami R., Matteoni S., Benetti R., Schoeftner S.. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat. Commun. 2019; 10:1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfeiffer V., Crittin J., Grolimund L., Lingner J.. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013; 32:2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernandes R.V., Feretzaki M., Lingner J.. The makings of TERRA R-loops at chromosome ends. Cell Cycle. 2021; 20:1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drosopoulos W.C., Deng Z., Twayana S., Kosiyatrakul S.T., Vladimirova O., Lieberman P.M., Schildkraut C.L.. TRF2 mediates replication initiation within human telomeres to prevent telomere dysfunction. Cell Rep. 2020; 33:108379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glousker G., Briod A., Quadroni M., Lingner J.. Human shelterin protein POT1 prevents severe telomere instability induced by homology-directed DNA repair. EMBO J. 2020; 39:e104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Majerská J., Redon S., Lingner J.. Quantitative telomeric chromatin isolation protocol for human cells. Methods. 2017; 114:28–38. [DOI] [PubMed] [Google Scholar]
- 35. Sirbu B.M., Couch F.B., Cortez D.. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 2012; 7:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sirbu B.M., McDonald W.H., Dungrawala H., Badu-Nkansah A., Kavanaugh G.M., Chen Y., Tabb D.L., Cortez D.. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 2013; 288:31458–31467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flynn R.L., Centore R.C., O’Sullivan R.J., Rai R., Tse A., Songyang Z., Chang S., Karlseder J., Zou L.. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011; 471:532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cristofari G., Lingner J.. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006; 25:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rappsilber J., Mann M., Ishihama Y.. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007; 2:1896–1906. [DOI] [PubMed] [Google Scholar]
- 40. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 41. Cox J., Michalski A., Mann M.. Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom. 2011; 22:1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011; 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- 43. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 44. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A.et al.. Tissue-based map of the human proteome. Science. 2015; 347:1260419–1260419. [DOI] [PubMed] [Google Scholar]
- 46. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995; 57:289–300. [Google Scholar]
- 47. Cortez D. Proteomic analyses of the eukaryotic replication machinery. Methods in Enzymology. 2017; 591:Elsevier; 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Somyajit K., Gupta R., Sedlackova H., Neelsen K.J., Ochs F., Rask M.-B., Choudhary C., Lukas J.. Redox-sensitive alteration of replisome architecture safeguards genome integrity. Science. 2017; 358:797–802. [DOI] [PubMed] [Google Scholar]
- 49. Wessel S.R., Mohni K.N., Luzwick J.W., Dungrawala H., Cortez D.. Functional analysis of the replication fork proteome identifies BET proteins as PCNA regulators. Cell Rep. 2019; 28:3497–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.-P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y.et al.. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods. 2013; 10:730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li F., Kim H., Ji Z., Zhang T., Chen B., Ge Y., Hu Y., Feng X., Han X., Xu H.et al.. The BUB3-BUB1 complex promotes telomere DNA replication. Mol. Cell. 2018; 70:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Majerska J., Feretzaki M., Glousker G., Lingner J.. Transformation-induced stress at telomeres is counteracted through changes in the telomeric proteome including SAMHD1. Life Sci. Alliance. 2018; 1:e201800121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alabert C., Bukowski-Wills J.-C., Lee S.-B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A.. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014; 16:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lopez-Contreras A.J., Ruppen I., Nieto-Soler M., Murga M., Rodriguez-Acebes S., Remeseiro S., Rodrigo-Perez S., Rojas A.M., Mendez J., Muñoz J.et al.. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013; 3:1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Makarov V.L., Lejnine S., Bedoyan J., Langmore J.P.. Nucleosomal organization of telomere-specific chromatin in rat. Cell. 1993; 73:775–787. [DOI] [PubMed] [Google Scholar]
- 56. Tommerup H., Dousmanis A., de Lange T.. Unusual chromatin in human telomeres. Mol. Cell. Biol. 1994; 14:5777–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Galati A., Magdinier F., Colasanti V., Bauwens S., Pinte S., Ricordy R., Giraud-Panis M.-J., Pusch M.C., Savino M., Cacchione S.et al.. TRF2 controls telomeric nucleosome organization in a cell cycle phase-dependent manner. PLoS One. 2012; 7:e34386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Murga M., Jaco I., Fan Y., Soria R., Martinez-Pastor B., Cuadrado M., Yang S.-M., Blasco M.A., Skoultchi A.I., Fernandez-Capetillo O.. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 2007; 178:1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Greider C.W. Regulating telomere length from the inside out: the replication fork model. Genes Dev. 2016; 30:1483–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miller K.M., Rog O., Cooper J.P.. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006; 440:824–828. [DOI] [PubMed] [Google Scholar]
- 61. Zeng X., Hernandez-Sanchez W., Xu M., Whited T.L., Baus D., Zhang J., Berdis A.J., Taylor D.J.. Administration of a nucleoside analog promotes cancer cell death in a telomerase-dependent manner. Cell Rep. 2018; 23:3031–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meng L., Hsu J.K., Zhu Q., Lin T., Tsai R.Y.L.. Nucleostemin inhibits TRF1 dimerization and shortens its dynamic association with the telomere. J. Cell Sci. 2011; 124:3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhu Q., Yasumoto H., Tsai R.Y.L.. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol. Cell. Biol. 2006; 26:9279–9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu Q., Meng L., Hsu J.K., Lin T., Teishima J., Tsai R.Y.L.. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J. Cell Biol. 2009; 185:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yuan F., Li G., Tong T.. Nucleolar and coiled-body phosphoprotein 1 (NOLC1) regulates the nucleolar retention of TRF2. Cell Death Discov. 2017; 3:17043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lachapelle S., Gagné J.-P., Garand C., Desbiens M., Coulombe Y., Bohr V.A., Hendzel M.J., Masson J.-Y., Poirier G.G., Lebel M.. Proteome-wide identification of WRN-interacting proteins in untreated and nuclease-treated samples. J. Proteome Res. 2011; 10:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McRae E.K.S., Davidson D.E., Dupas S.J., McKenna S.A.. Insights into the RNA quadruplex binding specificity of DDX21. Biochim. Biophys. Acta BBA - Gen. Subj. 2018; 1862:1973–1979. [DOI] [PubMed] [Google Scholar]
- 68. de Silanes I.L., Alcontres M.S., Blasco M.A.. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 2010; 1:33. [DOI] [PubMed] [Google Scholar]
- 69. Yoo J.E., Oh B.-K., Park Y.N.. Human PinX1 mediates TRF1 accumulation in nucleolus and enhances TRF1 binding to telomeres. J. Mol. Biol. 2009; 388:928–940. [DOI] [PubMed] [Google Scholar]
- 70. Benyelles M., Episkopou H., O’Donohue M., Kermasson L., Frange P., Poulain F., Burcu Belen F., Polat M., Bole-Feysot C., Langa-Vives F.et al.. Impaired telomere integrity and rRNA biogenesis in PARN-deficient patients and knock-out models. EMBO Mol. Med. 2019; 11:e10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moon D.H., Segal M., Boyraz B., Guinan E., Hofmann I., Cahan P., Tai A.K., Agarwal S.. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat. Genet. 2015; 47:1482–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tummala H., Walne A., Collopy L., Cardoso S., de la Fuente J., Lawson S., Powell J., Cooper N., Foster A., Mohammed S.et al.. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 2015; 125:2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Glousker G., Lingner J.. Challenging endings: how telomeres prevent fragility. Bioessays. 2021; 43:e2100157. [DOI] [PubMed] [Google Scholar]
- 74. Zimmermann M., Kibe T., Kabir S., de Lange T.. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014; 28:2477–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019; 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mennie A.K., Moser B.A., Nakamura T.M.. LARP7-like protein Pof8 regulates telomerase assembly and poly(A)+TERRA expression in fission yeast. Nat. Commun. 2018; 9:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Collopy L.C., Ware T.L., Goncalves T., Kongsstovu S., Yang Q., Amelina H., Pinder C., Alenazi A., Moiseeva V., Pearson S.R.et al.. LARP7 family proteins have conserved function in telomerase assembly. Nat. Commun. 2018; 9:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Holohan B., Kim W., Lai T.-P., Hoshiyama H., Zhang N., Alazami A.M., Wright W.E., Meyn M.S., Alkuraya F.S., Shay J.W.. Impaired telomere maintenance in Alazami syndrome patients with LARP7 deficiency. BMC Genomics. 2016; 17:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P.et al.. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Van Oss S.B., Cucinotta C.E., Arndt K.M.. Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem. Sci. 2017; 42:788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhu B. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005; 19:1668–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Robinson S., Oliver A., Chevassut T., Newbury S.. The 3′ to 5′ exoribonuclease DIS3: from structure and mechanisms to biological functions and role in human disease. Biomolecules. 2015; 5:1515–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Porro A., Feuerhahn S., Reichenbach P., Lingner J.. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 2010; 30:4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pan X., Chen Y., Biju B., Ahmed N., Kong J., Goldenberg M., Huang J., Mohan N., Klosek S., Parsa K.et al.. FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci. Rep. 2019; 9:19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Silva B., Pentz R., Figueira A.M., Arora R., Lee Y.W., Hodson C., Wischnewski H., Deans A.J., Azzalin C.M.. FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 2019; 10:2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kalashnikova A.A., Winkler D.D., McBryant S.J., Henderson R.K., Herman J.A., DeLuca J.G., Luger K., Prenni J.E., Hansen J.C.. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 2013; 41:4026–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Petruk S., Sedkov Y., Johnston D.M., Hodgson J.W., Black K.L., Kovermann S.K., Beck S., Canaani E., Brock H.W., Mazo A.. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012; 150:922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J.et al.. Regulation of p53 activity through lysine methylation. Nature. 2004; 432:353–360. [DOI] [PubMed] [Google Scholar]
- 89. Long L., Thelen J.P., Furgason M., Haj-Yahya M., Brik A., Cheng D., Peng J., Yao T.. The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J. Biol. Chem. 2014; 289:8916–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K.et al.. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008; 133:627–639. [DOI] [PubMed] [Google Scholar]
- 91. Osakabe A., Tachiwana H., Takaku M., Hori T., Obuse C., Kimura H., Fukagawa T., Kurumizaka H.. Vertebrate Spt2 is a novel nucleolar histone chaperone that assists in ribosomal DNA transcription. J. Cell Sci. 2013; 126:3123–1332. [DOI] [PubMed] [Google Scholar]
- 92. Baba A., Ohtake F., Okuno Y., Yokota K., Okada M., Imai Y., Ni M., Meyer C.A., Igarashi K., Kanno J.et al.. PKA-dependent regulation of the histone lysine demethylase complex PHF2–ARID5B. Nat. Cell Biol. 2011; 13:668–675. [DOI] [PubMed] [Google Scholar]
- 93. Furusawa T., Rochman M., Taher L., Dimitriadis E.K., Nagashima K., Anderson S., Bustin M.. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 2015; 6:6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Laprade H., Querido E., Smith M.J., Guérit D., Crimmins H., Conomos D., Pourret E., Chartrand P., Sfeir A.. Single-molecule imaging of telomerase RNA reveals a recruitment-retention model for telomere elongation. Mol. Cell. 2020; 79:115–126. [DOI] [PubMed] [Google Scholar]
- 95. Schmidt J.C., Zaug A.J., Cech T.R.. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell. 2016; 166:1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jády B.E., Richard P., Bertrand E., Kiss T.. Cell cycle-dependent recruitment of telomerase RNA and cajal bodies to human telomeres. Mol. Biol. Cell. 2006; 17:944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tomlinson R.L., Ziegler T.D., Supakorndej T., Terns R.M., Terns M.P.. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006; 17:955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhou X.Z., Lu K.P.. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell. 2001; 107:347–359. [DOI] [PubMed] [Google Scholar]
- 99. Schawalder J., Paric E., Neff N.F.. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol. 2003; 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Brockers K., Schneider R.. Histone H1, the forgotten histone. Epigenomics. 2019; 11:363–366. [DOI] [PubMed] [Google Scholar]
- 101. Andreyeva E.N., Bernardo T.J., Kolesnikova T.D., Lu X., Yarinich L.A., Bartholdy B.A., Guo X., Posukh O.V., Healton S., Willcockson M.A.et al.. Regulatory functions and chromatin loading dynamics of linker histone H1 during endoreplication in Drosophila. Genes Dev. 2017; 31:603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Thiriet C., Hayes J.J.. Linker histone phosphorylation regulates global timing of replication origin firing. J. Biol. Chem. 2009; 284:2823–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Fajkus J., Trifonov E.N.. Columnar packing of telomeric nucleosomes. Biochem. Biophys. Res. Commun. 2001; 280:961–963. [DOI] [PubMed] [Google Scholar]
- 104. Rochman M., Postnikov Y., Correll S., Malicet C., Wincovitch S., Karpova T.S., McNally J.G., Wu X., Bubunenko N.A., Grigoryev S.et al.. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates yranscription. Mol. Cell. 2009; 35:642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mozgová I., Mokroš P., Fajkus J.. Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell. 2010; 22:2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pinzaru A.M., Hom R.A., Beal A., Phillips A.F., Ni E., Cardozo T., Nair N., Choi J., Wuttke D.S., Sfeir A.et al.. Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. 2016; 15:2170–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Reverón-Gómez N., González-Aguilera C., Stewart-Morgan K.R., Petryk N., Flury V., Graziano S., Johansen J.V., Jakobsen J.S., Alabert C., Groth A.. Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Mol. Cell. 2018; 72:239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (108) partner repository with the dataset identifier PXD018712.