Abstract
Verticillium wilt is a severe plant disease that causes massive losses in multiple crops. Increasing the plant resistance to Verticillium wilt is a critical challenge worldwide. Here, we report that the hemibiotrophic Verticillium dahliae-secreted Asp f2-like protein VDAL causes leaf wilting when applied to cotton leaves in vitro but enhances the resistance to V. dahliae when overexpressed in Arabidopsis or cotton without affecting the plant growth and development. VDAL protein interacts with Arabidopsis E3 ligases plant U-box 25 (PUB25) and PUB26 and is ubiquitinated by PUBs in vitro. However, VDAL is not degraded by PUB25 or PUB26 in planta. Besides, the pub25 pub26 double mutant shows higher resistance to V. dahliae than the wild-type. PUBs interact with the transcription factor MYB6 in a yeast two-hybrid screen. MYB6 promotes plant resistance to Verticillium wilt while PUBs ubiquitinate MYB6 and mediate its degradation. VDAL competes with MYB6 for binding to PUBs, and the role of VDAL in increasing Verticillium wilt resistance depends on MYB6. Taken together, these results suggest that plants evolute a strategy to utilize the invaded effector protein VDAL to resist the V. dahliae infection without causing a hypersensitive response (HR); alternatively, hemibiotrophic pathogens may use some effectors to keep plant cells alive during its infection in order to take nutrients from host cells. This study provides the molecular mechanism for plants increasing disease resistance when overexpressing some effector proteins without inducing HR, and may promote searching for more genes from pathogenic fungi or bacteria to engineer plant disease resistance.
Ectopically expressed VDAL in Arabidopsis and Gossypium hirsutum alleviates the degradation of a positive disease response factor MYB6 through its interaction with PUB25 and PUB26 E3 ligases.
Introduction
The plant immune system confers effective protection against diverse pathogens. The activation of this system requires pathogen-derived molecules, which are classified into two categories (Jones and Dangl, 2006, 2017; Zhou and Zhang, 2020; Wang et al., 2020b). The conserved pathogen-associated molecular patterns (PAMPs) are molecules belong to the first category, which are perceived by specific plant pattern-recognition receptors (PRRs) at the cell surface (Kunze et al., 2004; Zipfel et al., 2006; Sun et al., 2013; Xue et al., 2020; Zhou and Zhang, 2020; Wang et al., 2020b; Manhaes et al., 2021). This PRR-initiated immunity, also known as PAMP-triggered immunity (PTI), generally occurs upon the initial contact of the pathogen with the plant cell, thus constituting the first layer of the immune response (Afzal et al., 2011; Singh et al., 2014; Bigeard et al., 2015; Zhou and Zhang, 2020; Wang et al., 2020b; Manhaes et al., 2021). PTI occurs quickly upon the perception of PAMPs when pathogens infect plants, and causes a series of rapid immune responses, such as a reactive oxygen species (ROS) burst, the activation of mitogen-activated protein kinase (MAPK) activity, callose accumulation, the induced expression of disease resistance genes, and changes in hormone levels (Boller and Felix, 2009; Boller and He, 2009; Qi et al., 2018; Yang et al., 2020; Wang et al., 2020b). The second category is named for the pathogen secreted effectors that are delivered into plant cells by various mechanisms (Liu et al., 2013; Rajamuthiah and Mylonakis, 2014; Stotz et al., 2014; Cui et al., 2015; Betsuyaku et al., 2018). Although some effectors reprogram the transcriptional profile in the host cells to create the niche permissive for infection (Boch et al., 2009; Boch and Bonas, 2010; Christian et al., 2010), most effectors suppress PTI by directly engaging specific host proteins via physical interactions or biochemical modifications with unique enzymatic activity (Block et al., 2014; Lin et al., 2016). To resist invasion of pathogen effectors, plants employ a defense response known as effector-triggered immunity (ETI), often accompanied by a hypersensitive response (HR), at the infection site, to limit pathogen infection (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Zhou and Zhang, 2020; Wang et al., 2020b). ETI and PTI can reinforce each other to increase the plant resistance to pathogens (Ngou et al., 2021; Yuan et al., 2021a, 2021b).
Verticillium dahliae is a soil-borne hemibiotrophic fungal pathogen that causes Verticillium wilt in more than 200 host species, including important economic crops (Klosterman et al., 2009; Njoroge et al., 2009; Wheeler and Johnson, 2016). Verticillium wilt causes significant economic losses to agriculture worldwide. For instance, cotton (Gossypium hirsutum) production in many areas of the world is severely threatened by this disease (Zhao et al., 2014; Han et al., 2019). As Arabidopsis thaliana can also be infected by this fungal pathogen (Veronese et al., 2003; Qin et al., 2018), it is often used as a model plant to study the molecular mechanism underlying Verticillium wilt resistance.
Verticillium dahliae infection is characterized by the association of fungal hyphae with the plant’s woody vascular tissues, where fungicides cannot readily reach, which may account for the difficulty in controlling Verticillium wilt (Vallad and Subbarao, 2008; Zhao et al., 2014; Deng et al., 2015). As one of the fungal pathogen weapons, secreted proteins are thought to play critical roles in the pathogenicity of V. dahliae (Klosterman et al., 2011; Lo Presti et al., 2015). These secreted proteins usually act as effectors that are employed by pathogens to overcome plant defense. Exposure to these proteins often leads to PTI or ETI in the host cells (Dodds et al., 2004; Doehlemann et al., 2009; Lo Presti et al., 2015). Verticillium dahliae secretes more than 700 putative effectors, including more than 100 small cysteine-rich potential effectors (Klosterman et al., 2009, 2011), but only a few have been functionally studied. For example, the tomato cell-surface-localized immune receptor Ve1 is activated by the V. dahliae-secreted protein Avirulence on Ve1 (AVe1) to trigger plant immune responses (Fradin et al., 2009; Deng et al., 2015). Effector isochorismatase (VdIsc1) hydrolyses isochorismate [the direct precursor of salicylate (SA)] to suppress SA-mediated defense (Liu et al., 2014), while the effector V. dahliae secretory protein 41 (VdSCP41) interacting with plant-specific transcription factors (TFs) CALMODULIN BINDING PROTEIN 60 (CBP60g) and SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 further inhibits transcriptional activity of CBP60g (Qin et al., 2018). Verticillium dahliae-secreted protein polysaccharide deacetylase (PDA1) promotes virulence through deacetylating chitin oligomers (Gao et al., 2019). An ethylene-inducing xylanase (EIX)-like protein, VdEIX3, from V. dahliae, can be recognized by a leucine-rich repeat receptor-like protein, NbEIX2, from Nicotiana benthamiana (Yin et al., 2021). The cotton ghBAK1, the ortholog of the Arabidopsis BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), is required for cotton resistance to Verticillium wilt (Gao et al., 2013). Another study found that infiltration of V. dahliae secreted protein PevD1 to cotyledons enhances cotton resistance and the defense response to the V. dahliae infection (Bu et al., 2014).
Protein ubiquitination, a posttranslational modification, is required for many signaling pathways involved in numerous essential cellular processes including plant growth, development, and stress response in eukaryotic cells (Kong et al., 2015; You et al., 2016; Yang et al., 2017; Ye et al., 2018; Zhou et al., 2018; Xiao et al., 2020). Protein ubiquitination reaction is catalyzed by three enzymes, the ubiquitin (Ub)-activating E1, the Ub-conjugating E2, and the Ub-ligase E3 (Yu et al., 2016), and this modification is also critical for defense responses in plants (Dombrecht et al., 2007; Tong et al., 2017; Furniss et al., 2018; Gao et al., 2021). E3 ligases that dictate the specificity of the substrate for ubiquitination are highly diverse (Kosarev et al., 2002), and can be classified into four groups, including Homologous to E6-associated protein C-Terminus, Really Interesting New Gene (RING), U-box, and Cullin-RING ligase (Stone, 2014; Liao et al., 2017). Plant U-box (PUB) E3 ligases in Arabidopsis have 64 members (Mudgil et al., 2004). All PUBs contain a high conserved U-box domain, which is required for interaction with E2 protein (Pringa et al., 2001) and essential for PUB ligase activity (Ohi et al., 2003; Zeng et al., 2004). Only a limited number of PUBs have been functionally studied in plants. For example, PUB12 and PUB13 participate in ABA signal pathway by degrading the key negative ABA coreceptor ABA INSESSITIVE 1 (Kong et al., 2015). PUB10 and PUB2 as well as PUB4 play roles in jasmonic acid (JA) and cytokinin responses, respectively (Jung et al., 2015; Wang et al., 2017b). Since PUB12 and PUB13 were also found to target FLAGELLIN-SENSITIVE 2 (FLS2) for its degradation (Lu et al., 2011), more and more lines of evidence indicate that PUBs play important roles in plant disease resistance. PUB22, PUB23, and PUB24 appear to negatively regulate plant PTI, and the mutants defective in the three corresponding genes exhibit higher PAMP-induced oxidative burst and inhibition of bacterial growth (Stegmann et al., 2012). A recent study indicates that PUB25 and PUB26 target and degrade nonactivated BOTRYTIS-INDUCED KINASE 1 (BIK1) to negatively regulate immunity in Arabidopsis (Wang et al., 2018a). These two E3 ligases also promote Arabidopsis freezing tolerance by degrading the cold signaling negative regulator MYB15 (Wang et al., 2019). PUB25 and PUB26 negatively regulate petal growth in a spatial- and temporal-specific manner (Li et al., 2021a). GhPUB17 from cotton plays a negative role in plant resistance to V. dahliae, and its E3 ligase activity can be inhibited by a cyclophilin homolog GhCyP3 (Qin et al., 2019). In other plant species, PUB ligases were found also to be linked to biotic stress (Ishikawa et al., 2014; Jiao et al., 2017).
TFs are important for gene expression. According to different DNA binding domains (BDs), they are divided into different families, such as MYC, MYB, Bzip, and bHLH family (Pabo and Sauer, 1992). The MYB TFs constitute the largest TF family among all eukaryotic organisms (Riechmann et al., 2000). The N-terminus of MYB TF contains a repetitive sequence consisting of an MYB domain of about 52 amino acids that can form three α helix structures. The second and third α helix of each MYB domain can form helix-turn-helix, which can bind to the DNA trench (Ogata et al., 1994). According to the number of MYB domains, MYB TFs can be divided into four categories, called 1R (R1/2, R3-MYB), 2R (R2R3-MYB), 3R (R1R2R3-MYB), and 4R (Dubos et al., 2010). Most of MYBs in plants belong to 2R. There are 138 R2R3-MYBs identified in Arabidopsis (Katiyar et al., 2012). Acting as transcription activators or inhibitors, MYB TFs influence plant primary and secondary metabolism, cell fate and properties, plant growth and development, and plant adaptation to different stresses (Preston et al., 2004; Zhong et al., 2007; McCarthy et al., 2009; Dubos et al., 2010; Liu et al., 2015; Deng et al., 2020). In flg22-treated Arabidopsis plants, the expression of MYB12, a positive regulator in flavanone synthesis, was upregulated, while the expression of MYB4 was downregulated, suggesting an antagonistic effect between two MYBs to coordinate plant immune response (Schenke et al., 2011; Jin et al., 2014). MYB96 modulates plant immune responses by regulating salicylic acid (SA) biosynthesis (Seo and Park, 2010). MYB30 is involved in regulating plant HR (Marino et al., 2013). In an early study, the DNA binding motif of MYB6 or MYB7 in Arabidopsis was determined (Roger and Li, 1995). The expression of MYB6 was induced by JA or SA treatment in Arabidopsis (Yanhui et al., 2006). However, whether MYB6 is involved in regulating the immunity of Arabidopsis to V. dahliae or plays other biological roles has not been reported yet.
To test the function of V. dahliae secreted proteins, we generated transgenic Arabidopsis and cotton plants ectopically expressing the V. dahliae-secreted Asp f2-like protein (VDAL). These transgenic plants exhibited significant resistance to this fungal pathogen infection, without any growth or developmental defects. Previously, several effectors such as PsCRN161 from Phytophthora sojae, SSB (Xoc), Harpin (xoo) from Xanthomonas oryzae pv. oryzicola and the class II hydrophobin ThHyd1 from Trichoderma harzianum were reported to increase plant resistance to pathogens without retarding plant growth when overexpressed in plants; however, the molecular mechanisms are largely unknown (Peng et al., 2004; Rajput et al., 2015; Cao et al., 2018; Yu et al., 2020). In this study, we show that VDAL interacts with the E3 ligases PUB25 and PUB26, but was not degraded in Arabidopsis, while PUB25 and PUB26 target a positive regulator of plant disease resistance MYB6 for its degradation. Our results suggest that VDAL competes with MYB6 to bind PUB25 and PUB26, and increases the accumulation of MYB6, thus improving disease resistance in Arabidopsis. These results suggest that plants not only use ETI and PTI to produce ROS (Zhou and Zhang, 2020; Wang et al., 2020b), but also have evolved a strategy to utilize effectors such as VDAL to resist V. dahliae infection without the sacrifice of cell death, or that pathogens take advantage of the effectors not killing host cells immediately in order to fully exploit the nutrients of host cells. VDAL may represent an ideal candidate gene for improving Verticillium wilt resistance in crops without affecting plant growth or yields.
Results
Identification of VDAL, a protein secreted by V. dahliae
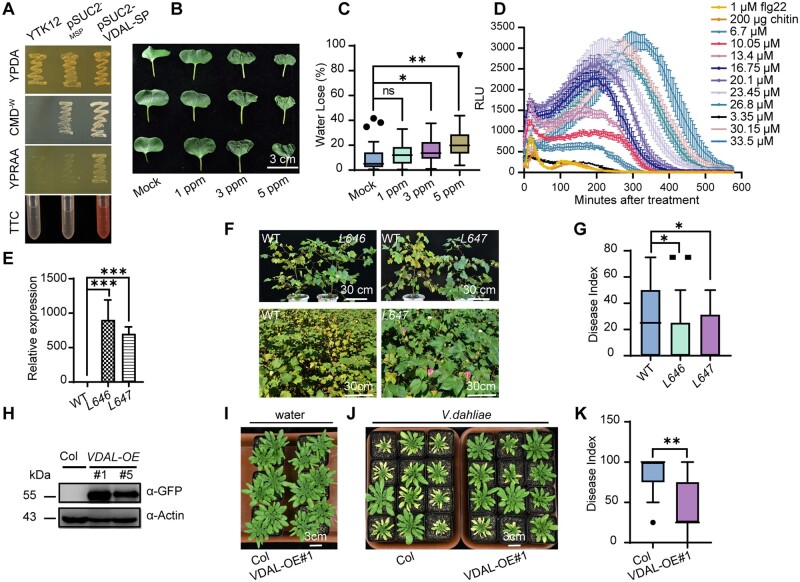
In an effort to identify proteins secreted by V. dahliae that might be responsible for Verticillium wilt symptoms in plants, we identified a protein named VDAL from the V. dahliae strain Vd991, which is homologous to human Pra1 and Asp f2 (Supplemental Figure S1A). Asp f2 is present in many fungi, including animal fungal pathogens such as Aspergillus fumigatus (Supplemental Figure S1A). Sequence analysis revealed that VDAL contains the putative Zn2+-binding motif HARxH, which is present in many metalloproteases, and a secretion signal peptide (SP) in its first 23 residues (Supplemental Figure S1B). VDAL is very conserved in some fungi such as Fusarium, Colletotrichum, Metarhizium, and Purpureocillium (Supplemental Figure S2; Supplemental File S1), which may imply a general role as an effector. We performed an invertase secretion assay to investigate whether this secretion SP is functional. Specifically, we fused the sequence encoding the putative VDAL SP with the sucrose invertase sequence in pSUC2-MSP and introduced the resulting construct into yeast strain YTK12, which is defective in invertase activity (SUC2), to generate the pSUC2-VDAL-SP fusion protein (Supplemental Figure S1C). As expected, this strain and a control strain harboring the empty vector (pSUC2−MSP) both grew robustly on CMD-W Trp dropout medium containing 0.1% glucose and 2% sucrose (Figure 1A). However, only strain YTK12-VDAL-SP gained the ability to grow on YPRAA medium with raffinose as the sole carbon source, indicating that invertase was secreted by this strain (Figure 1A). In agreement with this result, strain YTK12-VDAL-SP reduced 2, 3, 5-triphenyltetrazolium chloride (TTC) in the medium into red formazan (Figure 1A), further confirming that invertase is secreted by VDAL SP. To validate the secreting activity of VDAL SP, we deleted and mutated the critical amino acids of VDAL SP, in proteins named SP-mut-1 to SP-mut-5, respectively (Supplemental Figure S1D), and performed the invertase secretion assay to investigate whether these mutated SPs are functional. The N-terminus of either P. sojae Avr1b or Magnaporthe oryzae Mg87 was used as a positive or negative control, respectively (Fang et al., 2016). Except for the SP-mut-2, which was only a little weaker than the wild-type, the secreting activity of the mutated SPs was greatly compromised, suggesting that SP has secreting ability (Supplemental Figure S1E).
Figure 1.
Plants overexpressing VDAL are resistant to Verticillium wilt caused by V. dahliae. A, Test for functionality of the SP of VDAL. CMD−W medium for positive clone screening; YPRAA medium for verification of secretion activity on invertase; TTC assay for the test of secreted invertase activity. Note that only the strain expressing the VDAL SP fusion gained the ability to catabolize raffinose, and reduce TTC into red formazan. Both YPRAA and TTC indicate the successful secretion of the invertase. B, VDAL protein effect on leaf wilt. Petioles were soaked in different concentration of VDAL protein solution for 24 h, and then photos were taken. C, Water loss during leaf wilting. Leaf weight was measured at the beginning and at 12 h after posttreatment. D, Luminol-based assay of ROS burst with increasing concentrations of VDAL protein. Flg22 and chitin were used as positive controls. E, The expression level of VDAL in two corresponding transgenic cotton lines. F and G, Assessment of VDAL transgenic cotton line tolerance to Verticillium wilt caused by V. dahliae in the field. F, Upper: two transgenic lines (L646 and L647) and the wild-type plants were removed from the same field infected with V. dahliae, and replanted in the pots for taking photos; Lower: photos for the wild-type cotton plants and transgenic L647 plants growing in the same field infected with V. dahliae. G, Statistical analysis of the disease index in the above field condition, count with at least 25 plants. H, Immunoblot analysis of VDAL in two transgenic A. thaliana lines overexpressing VDAL. Total proteins of 10-day-old seedlings were extracted and detected with anti-GFP antibodies. Actin was an equal loading control. I–K, Assay of VDAL transgenic Arabidopsis plant resistance to Verticillium wilt caused by V. dahliae. The plants grown in green house for 2 weeks were dipped into the V. dahliae spore suspension or water for 5 min. Photos were taken at 20 dpi. K, Statistical analysis of the disease index of VDAL-oe #1 and Col, count with at least 15 plants. *, **, and *** in (C), (E), (G), and (K) represent significant difference (P < 0.05), highly significant difference (P < 0.01), and extremely significant difference (P < 0.001), respectively, Student’s t- test. The experiments were repeated independently three times with similar results.
Since VDAL is an M35-like metalloprotease that might have zinc-binding activity, we tested its zinc-binding activity. Like Pra1, VDAL could bind zinc in vitro (Supplemental Figure S1, F and G). To determine whether VDAL causes leaf wilting, we dipped cotton leaf petioles into a solution of VDAL-HIS recombinant protein that was expressed and purified from Escherichia coli. Because of the failure of purifying the full-length VDAL protein in E. coli, the SP was removed from VDAL protein in this study. Incubation with 3 or 5 ppm of the VDAL-HIS solution for 12 h led to obvious leaf wilting, while the same treatment of mock treatment (just the buffer that was used to dissolve VDAL-HIS protein) did not cause any visible wilting. We recorded water loss after 12 h of incubation and photographed the leaves after 24 h of incubation; the quantification indicated that the water loss in VDAL-HIS treatment was significantly higher than in the mock treatment (Figure 1, B and C). The wilted leaves failed to recover after being transferred to water, suggesting that cell death occurred in these leaves due to the VDAL treatment.
We reasoned that the VDAL-incubated leaves might have exhibited an HR to VDAL and overproduced ROS. We, therefore, measured ROS production in VDAL-treated Arabidopsis leaves. Bacterial flagellin and fungal chitin are well-known PAMPs that induce extracellular ROS bursts (Wang et al., 2020b). We detected ROS production in Arabidopsis leaves treated with VDAL using a luminol-based assay, with the bacterial flagellin flg22 and chitin used as positive controls. As shown in Figure 1D and Supplemental Figure S1H, similar to flg22 and chitin, VDAL elicited a ROS burst, with higher levels of ROS produced as its concentration increased. Together, these results indicate VDAL is a pathogenic elicitor protein that can cause cotton leaf wilting and ROS burst.
Plants expressing VDAL are resistant to infection by V. dahliae
The presence of a functionally secreted SP in VDAL suggests that this protein is secreted into the extracellular space or even into host cells by V. dahliae. Some secreted proteins such as the bacterial harpins (Peng et al., 2004), Verticillium Ave1 (Castroverde et al., 2016, 2017), SSB protein from Xanthomonas oryzae pv. oryzicola (Xoc, SSBXoc) (Cao et al., 2018) and the Crinkler (CRN) effector PsCRN161 from P. sojae (Rajput et al., 2015) have been used to activate plant immunity via stable expression in transgenic plants without inducing an HR. Except for PsCRN161, which can suppress cell death induced by other elicitors (Rajput et al., 2015), all other abovementioned effectors can induce HR like VDAL when applied on plants. We generated transgenic cotton (G. hirsutum) lines L646 and L647 stably expressing VDAL (Figure 1E) and examined their tolerance to wilt disease in the field. The most transgenic plants did not show any growth or developmental phenotypes compared to nontransgenic wild-type plants in the normal field. However, the transgenic plants exhibited increased tolerance to Verticillium wilt, with lower disease index than the wild-type under field conditions (Figure 1, F and G). We also tested different VDAL transgenic cotton lines and obtained similar results (Supplemental Figure S1I).
We then generated transgenic Arabidopsis plants to examine the potential role of VDAL in triggering plant immune responses. Of the various transgenic Arabidopsis lines generated, we selected two lines (#1 and #5) for further study. VDAL protein was readily detected in both transgenic lines by immunoblotting (Figure 1H). Like in cotton, the expression of VDAL in Arabidopsis did not alter plant morphology or other traits such as growth, leaf color or the timing and rate of reproduction under normal growth conditions (Supplemental Figure S1J). We then examined the susceptibility of these plants to V. dahliae infection. We carefully removed 2–3-week-old seedlings from the soil, washed the roots to remove any attached soil, and incubated them in a 1 × 106 conidia mL−1 spore suspension of V. dahliae strain Vd991 for 5 min. We transferred the inoculated plants to new pots filled with soil and observed the development of disease symptoms. During the first 7 days postinoculation (dpi), all plants resumed growth and no disease symptoms were observed. At 15 dpi, the leaves of inoculated wild-type plants began to become discolored, and no plants survived for >30 dpi (Figure 1J). In contrast, plants expressing VDAL did not show serve discernable symptoms at 15 dpi, and leaf discoloration did not appear until 20 dpi. Quantification of disease symptoms by determining the disease index (Figure 1K) of the plants at 20 dpi indicated that the VDAL transgenic lines exhibited significantly weaker disease symptoms than wild-type plants. There was no difference between Col and VDAL transgenic lines at 20 dpi after water treatment (Figure 1I). Thus, both cotton and Arabidopsis plants expressing VDAL enhanced resistance to Verticillium wilt.
VDAL interacts with the U-box Ub E3 ligases PUB25 and PUB26
To determine the mechanism of VDAL-induced Verticillium wilt resistance, we examined the subcellular localization of VDAL by expressing a VDAL-GFP fusion protein in Arabidopsis protoplasts. Robust expression of the fusion protein was detected at 16 h after transfection. The distribution pattern of VDAL-GFP indicated that VDAL localized to the cytosol, with some dots of signal observed (Supplemental Figure S1K). A similar localization pattern was observed in VDAL-GFP overexpression Arabidopsis plants (Supplemental Figure S1L).
To identify the host proteins targeted by VDAL, we transiently expressed a VDAL-MYC fusion protein in Arabidopsis protoplasts for 16 h, and incubated total proteins extracted from transfected cells with agarose beads coated with MYC-specific antibodies for 2.5 h. After extensive washing, the eluted bound proteins were concentrated and analyzed by SDS-PAGE, followed by silver staining to visualize proteins coimmunoprecipitated by VDAL-MYC (Supplemental Figure S3A). Mass spectrometry analysis identified several coimmunoprecipitated proteins, including PUB26 (Supplemental Data Set 1). PUB25 and PUB26 are two closely homologous U-box proteins. Previous studies found that PUB25 and PUB26 target nonactivated BIK1 during plant immunity and MYB15 during the freezing response (Wang et al., 2018a, 2019), thus we focused on the relationship between VDAL and these two E3 ligases.
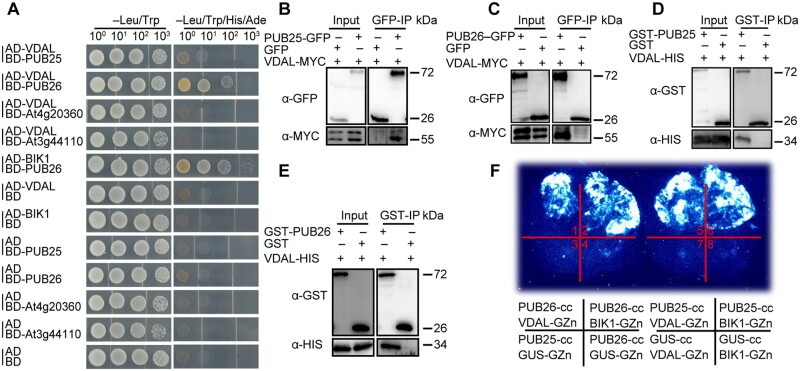
To determine whether VDAL directly interacts with these two U-box proteins, we performed a yeast two-hybrid (Y2H) assay using VDAL fused to the activation domain (AD) of the yeast transcriptional activator Gal4, and either PUB25 or PUB26 fused to its DNA-BD (Li et al., 2020). We constructed a series of yeast strains by transforming yeast Gold cells with various combinations of plasmids, including a pair that expressed fusions of BIK1 and PUB26, two established interacting proteins (Wang et al., 2018a). As expected, cells expressing the BD-PUB26 and AD-BIK1 fusions were able to grow on the reporter medium used to measure interactions (Figure 2A). Interactions were detected between VDAL and PUB25 or PUB26 (Figure 2A). In contrast, no interaction was detected in yeast strains coexpressing the fusion proteins with two Arabidopsis proteins (AT4g20360, a plastid localized EF-Tu translation elongation factor, and AT3g44110, a cochaperon DNAJ protein), and only the DNA-binding motif or the activation motif (Figure 2A), indicating that VDAL specifically interacts with PUB25 and PUB26.
Figure 2.
VDAL interacts with PUB25 and PUB26, two predicted plant U-box Ub E3 ligases. A, VDAL interaction with PUB25 and PUB26 in Y2H assay. The interaction of BD-PUB26 and AD-BIK1 is used as a positive control. BD-At4g20360 and BD-At3g44110 are used as negative controls. Yeast cells were grown on different media for 4 days. B and C, VDAL interaction with PUB25 (B) and PUB26 (C) in CoIP assay. VDAL-MYC and PUB25-GFP or PUB26-GFP plasmids were cotransfected into Arabidopsis protoplasts and incubated for 16 h. Total proteins were extracted and immunoprecipitated with anti-GFP beads, then immunoblotting analysis was carried out with anti-MYC and anti-GFP antibodies. D and E, VDAL interaction with PUB25 (D) and PUB26 (E) in the pull-down assay. Proteins purified from E. coli were immunoprecipitated with anti-GST beads and detected with anti-GST and anti-HIS antibodies. F, VDAL interaction with PUB25 and PUB26 in the split firefly LCI assay. Different plasmids combinations were transiently expressed in tobacco leaves for 72 h, then images were collected by a CCD camera.
We further verified the interactions between VDAL and the two Ub ligases by coimmunoprecipitation (CoIP), pull-down, and firefly luciferase complementation imaging (LCI) assays. We subjected total proteins from protoplasts transiently expressing VDAL-MYC and PUB25-GFP, or PUB26-GFP to CoIP assays using beads coated with anti-GFP antibodies. The protein corresponding to VDAL-MYC was readily detected in lysates coexpressing these two proteins. In contrast, these proteins were not detected in lysates from cells coexpressing GFP and VDAL-MYC (Figure 2, B and C). These results confirm the specific interactions between VDAL and the PUBs. Similar results were obtained in a pull-down assay. We purified GST-PUB25, GST-PUB26, GST, and VDAL-HIS from E. coli. Pull-down assays using GST beads indicated that GST-PUB25 or GST-PUB26, but not GST, was able to pull down VDAL-HIS (Figure 2, D and E). Robust interactions were also observed in the LCI assays. Strong luminescence was detected only in leaf sections expressing both VDAL and the PUB25 and PUB26 fusion proteins (Figure 2F;Supplemental Figure S3, B and C). Here, BIK1 and PUB26 were used as the positive control, and PUB25 or PUB26 and GUS were used as the negative controls (Figure 2F).
To identify which fragment of VDAL interacts with full-length PUB25 and PUB26 and their ARM domains, we designed two types of deletion mutants of VDAL (Supplemental Figure S3D). VDAL (1-59) interacted with full-length PUB26 but not PUB25, whereas VDAL (60-297) interacted with both full-length PUB proteins (Supplemental Figure S3E). The other deletion mutations did not interact with the full-length PUBs or their ARM domains (data not shown). Taken together, these results indicate that VDAL directly interacts with the Ub ligases PUB25 and PUB26 both in plant cells and in vitro.
VDAL is a substrate of PUB25 and PUB26 but is not degraded in planta
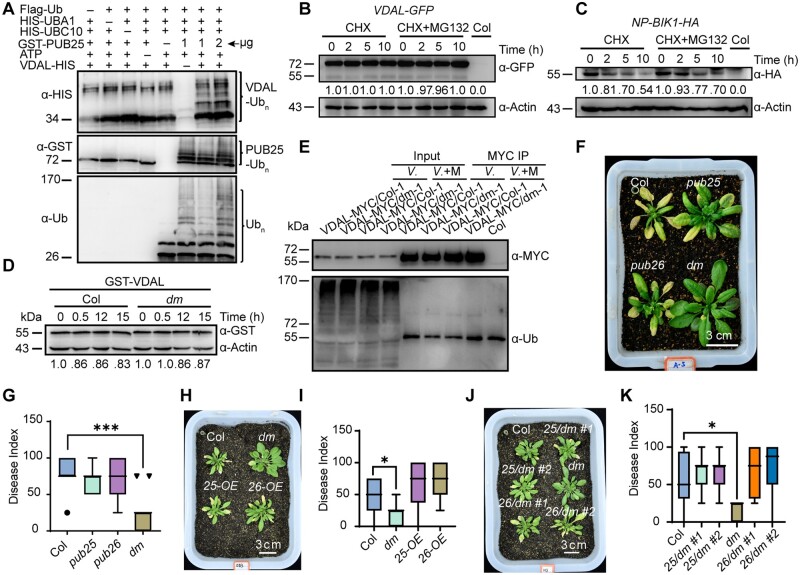
Given that VDAL directly interacts with PUB25 and PUB26, we investigated whether VDAL is a substrate of these Ub ligases. We first confirmed that the recombinant PUB25 and PUB26 proteins had Ub ligase activity. Such activity was entirely dependent on the presence of E1, E2, and one of the two E3 ligases (Supplemental Figure S3, F and G), thus confirming that the recombinant PUB25 and PUB26 were enzymatically active (Wang et al., 2018a, 2019). The addition of VDAL-HIS or GST-VDAL into the reaction led to the production of VDAL-HIS or GST-VDAL ladders, a key characteristic of ubiquitination reaction (Figures 3A;Supplemental Figure S3H). Again, the generation of ubiquitinated VDAL in this reaction required E1, E2 and PUB25, or PUB26, and reactions missing any of these components failed to ubiquitinate VDAL.
Figure 3.
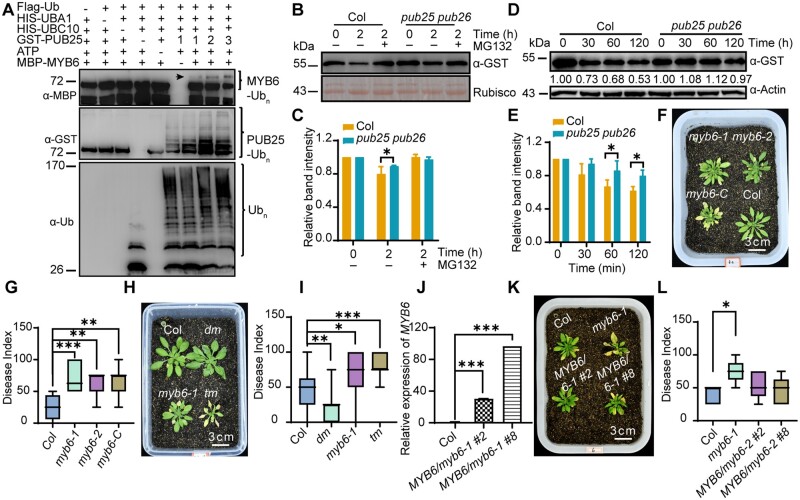
VDAL is a putative substrate of PUB25 in vitro, and PUB25 and PUB26 negatively regulate plant resistance to Verticillium wilt. A, PUB25 ubiquitination of VDAL in vitro. Different ubiquitination reaction systems with 40 mM ATP from different proteins purified from E. coli were incubated at 30°C for 3 h. The VDAL ubiquitination and PUB25 ubiquitination were detected by anti-HIS and anti-GST antibodies, respectively. The total ubiquitination signal was detected with anti-Ub antibodies. The numbers indicate the amount of GST-PUB25 added in the reaction. B, Degradation pattern of VDAL in the presence of CHX with or without MG132. The 12-day-old VDAL-GFP seedlings were treated with CHX or CHX with MG132 for different times. Total proteins were extracted. Immunoblotting analysis was carried out with anti-GFP antibodies. Actin was an equal loading control. The relative protein level was quantified with ImageJ. C, Degradation pattern of BIK1 in the presence of CHX with or without MG132. The 12-day-old BIK1-HA seedlings were treated with CHX or CHX with MG132 for different times. Total proteins were extracted. Immunoblotting analysis was carried out with anti-HA antibodies. Actin was an equal loading control. The relative protein level was quantified with ImageJ. D, VDAL degradation in Col and pub25 pub26 dm. Total proteins extracted from 12-day-old pub25 pub26 and Col seedlings were incubated with equal GST-VDAL at 25°C for different time. Immunoblotting analysis was carried out with anti-GST antibodies. Actin was an equal loading control. The relative protein level was quantified with ImageJ. E, Assessment of VDAL ubiquitination in planta. The 12-day-old VDAL-MYC seedlings were treated with V. dahliae or V. dahliae combined MG132 for 12 h. Total extracted proteins were immunoprecipitated with beads coated with anti-MYC antibodies. Immunoblotting analysis was carried out with anti-MYC and anti-Ub antibodies. F, pub25 pub26 dm resistance to Verticillium wilt compared with the wild-type or pub25 and pub26 single mutant. The plants grown in the greenhouse for 2 weeks were dipped into the V. dahliae spore suspension for 5 min. Photos were taken at 20 dpi. G, Statistical analysis of the disease index of Col, pub25, pub26 and pub25 pub26 dm in F, count with at least 15 plants. H, Comparison of PUB25 and PUB26 overexpression transgenic line susceptibility to Verticillium wilt caused by V. dahliae with that of the wild-type. The plants grown in the greenhouse for 2 weeks were dipped into the V. dahliae spore suspension for 5 min. Photos were taken at 15 dpi. I, Statistical analysis of the disease index of PUB25-OE, PUB26-OE, dm, and Col in (H), count with at least 15 plants. J, Comparison of PUB25 and PUB26 transgenic complementation lines with Col during Verticillium wilt caused by V. dahliae. The plants grown in the greenhouse for 2 weeks were dipped into the V. dahliae spore suspension for 5 min. Photos were taken at 20 dpi. K, Statistical analysis of the disease index of plants indicated in (J), count with at least 15 plants. *, **, and *** in (G), (I), and (K) represent significant difference (P < 0.05), and extremely significant difference (P < 0.001), respectively, one-way ANOVA. The experiments were repeated independently three times with similar results.
Next, we determined whether VDAL is degraded in planta. First, we treated transgenic Arabidopsis plants stably expressing VDAL-GFP with the protein synthesis inhibitor CHX or CHX combined with the 26S proteasome inhibitor MG132. Immunoblot analysis with anti-GFP antibodies indicated that VDAL-GFP accumulation did not change after treatment with CHX or CHX+MG132 for various periods of time (Figure 3B), but the positive control BIK1-HA was degraded (Figure 3C). We then investigated whether PUB25 and PUB26 would modulate VDAL degradation in a cell-free assay. We purified GST-VDAL from E. coli and added it to total proteins extracted from wild-type or pub25 pub26 double mutant (dm) plants in the presence of 10 mM ATP. We did not detect apparent differences in GST-VDAL protein levels in protein extracts from wild-type versus dm plants (Figure 3D). These results suggest that VDAL is a substrate of PUB25 and PUB26 but is highly stable in planta. We hypothesized that VDAL may not be ubiquitinated in planta. We overexpressed VDAL-MYC in Col and pub25 pub26 dm and detected the ubiquitination level of VDAL in Col versus pub25 pub26 dm after infection with V. dahliae or V. dahliae+MG132 for 12 h. The total ubiquitination was easily detected in input (Figure 3E), but we did not detect any ubiquitination of VDAL in both Col and pub25 pub26 dm in output (Figure 3E). To further confirm that VDAL cannot be ubiquitinated in plant cells, we used P62-agarose matrix to enrich total ubiquitinated proteins from VDAL-MYC, or from Col plants as a negative control, after treatment with V. dahliae and MG132. We could easily detect the total ubiquitination in both input and output, but not detect any ubiquitination of VDAL through MYC antibodies (Supplemental Figure S3I). These results suggest that VDAL is not ubiquitinated and degraded by PUB25 or PUB26 in planta.
PUB25 and PUB26 negatively regulate plant resistance to Verticillium wilt
Given that overexpressing VDAL increases plant resistance to Verticillium wilt and that PUB25 and PUB26 interact with VDAL, we wanted to know whether PUB25 and PUB26 are involved in plant resistance to V. dahliae infection. Analysis of GUS expression driven by the PUB25/PUB26 promoters indicated that they are expressed throughout the plant (Supplemental Figure S4, A and B). We also noticed that the subcellular localization of PUB25/PUB26-GFP fusion protein in Arabidopsis protoplasts was similar to that of VDAL-GFP (Supplemental Figures S1K and S4, C–E). We crossed the pub25 and pub26 single mutants obtained a pub25 pub26 dm, and then confirmed that the expression of both genes was disrupted in the dm (Supplemental Figure S4, F–H). Inoculation experiments showed that the pub25 pub26 dm was significantly more resistant to V. dahliae than pub25 or pub26 (Figure 3, F and G), and that wild-type plants were more susceptible to V. dahliae than the pub25 pub26 dms and the single mutants. Under infection conditions, the disease index of pub25 pub26 plants was significantly lower than that of both wild-type plants and the pub25 or pub26 single mutants (Figure 3, F and G). Notably, the V. dahliae resistance of pub25 pub26 was comparable to that of transgenic plants expressing VDAL (Supplemental Figure S4, I and J).
To further explore the roles of PUB25 and PUB26 in the plant response to V. dahliae, we obtained transgenic plants overexpressing PUB25 or PUB26 fused with Flag tag (PUB25-OE and PUB26-OE plants; Supplemental Figure S4K). After inoculation, the disease index analysis showed that the PUB25-OE and PUB26-OE plants were more susceptible than the wild-type to infection by V. dahliae (Figure 3, H and I). We also obtained transgenic complemented lines in which the PUB25 and PUB26 genomic sequence was fused with FLAG and MYC tag driven by the PUB25 or PUB26 native promoter, respectively, in the pub25 pub26 background (Supplemental Figure S4, L and M). Under inoculation conditions, the complemented lines largely lost the V. dahliae resistance of the dm, as indicated by the Verticillium-sensitive phenotype and disease index (Figure 3, J and K), confirming that the V. dahliae resistance of the dm was caused by pub25 pub26 mutation. Taken together, these results indicate that PUB25 and PUB26 are negative regulators of plant immunity against infection caused by V. dahliae.
PUB25 and PUB26 target MYB6 for ubiquitination and degradation
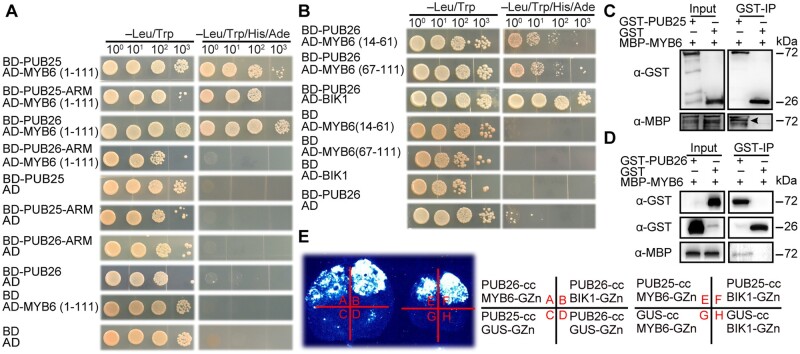
To explore the mechanism underlying the improved V. dahliae resistance of the pub25 pub26 mutant, we screened an Arabidopsis Y2H library using the ARM domain of PUB26 as bait to identify its target proteins in Arabidopsis. We identified one candidate interacting protein: MYB6 (Supplemental Data Set 2). The biological function of MYB6 in Arabidopsis has not been explored in detail. We verified the interaction of these proteins in a Y2H assay. As the full length and C-terminus (112-237) of MYB6 had autoactivation activity in yeast harboring the AD or BD vector, we did not test it further. The full-length PUB25 and its ARM domain interacted with the N-terminus of MYB6, which harbors two MYB domains (1-111). Full-length PUB26, but not its ARM domain, interacted with the N-terminus (1-111) of MYB6 (Figure 4A). Further deletion analyses indicated that both MYB repeat domains of MYB6, including MYB6 (14-61) and MYB6 (67-111), interacted with PUB26 (Figure 4B). In contrast, no interaction was detected in yeast strains coexpressing fusion proteins with only the DNA-binding motif or the activation motif, indicating that these interactions are specific.
Figure 4.
PUB25/26 interact with Arabidopsis TF MYB6. A, MYB6 MYB domain (1-111) interaction with full-length PUB25 and PUB26, and the ARM domain of PUB25 in Y2H assay. The interaction between BD-PUB26 and AD-BIK1 is used as a positive control. Yeast cells were grown on different medium for 4 days. B, Shorter MYB domain (14-61 and 67-111) of MYB6 interaction with full-length PUB26 in Y2H assay. The interaction of BD-PUB26 and AD-BIK1 is a positive control. C and D, MYB6 interaction with PUB25 and PUB26 in pull-down assay. Proteins were immunoprecipitated with anti-GST beads, and detected with anti-GST and anti-MBP antibodies. E, MYB6 interaction with PUB25 and PUB26 in the split firefly LCI assay. Different plasmid combinations were expressed in tobacco leaves for 72 h, then images were determined by a CCD camera.
We further examined the interactions between MYB6 and the two Ub ligases by performing CoIP, pull-down and LCI assays. However, we did not detect the expression of MYB6 in Arabidopsis protoplasts. We then purified GST-PUB25, GST-PUB26, GST, and MBP-MYB6 from E. coli. Pull-down assays using GST beads indicated that GST-PUB25 or GST-PUB26, but not GST, was able to pull down MBP-MYB6 (Figure 4, C and D). Robust interactions were also observed in the LCI assay (Figure 4E), as indicated by strong luminescence only in leaf sections expressing both MYB6 and PUB25 or PUB26 (Supplemental Figure S5, N and O). These results indicate that MYB6 interacts with both Ub ligases in planta and in vitro.
To determine whether MYB6 is a target of PUB25 and PUB26, we performed ubiquitination assays in vitro using MYB6 and both PUB25 and PUB26 isolated from E. coli and found that MYB6 was ubiquitinated by both PUB25 and PUB26 (Figure 5A;Supplemental Figure S5A). Next, we investigated whether PUB25 and PUB26 modulate the degradation of MYB6 in planta. Analysis of transgenic Arabidopsis plants harboring the MYB6 promoter fused to GUS indicated that MYB6 is widely expressed in plants (Supplemental Figure S5B). We generated transgenic Arabidopsis plants that stably expressed MYB6 fused with MYC, GFP, or Flag tag. MYB6 gene expression was higher in transgenic lines than in Col (Supplemental Figure S5C), but we did not detect any accumulation of MYB6 using anti-MYC, anti-GFP, or anti-FLAG antibodies whether the plants were treated with V. dahliae, treated with V. dahliae+MG132 or not treated (Supplemental Figure S5, D and E, only MYB-Flag is shown). However, when MYB6-Flag was expressed in the pub25 pub26 dm, MYB6 was easily detected using anti-Flag antibodies (Supplemental Figure S5, D and E). These results suggest that MYB6 is not stable in the wild-type but is stable in pub25 pub26.
Figure 5.
MYB6 is a target of PUB25/PUB26 and positively regulates plant resistance to Verticillium wilt. A, PUB25 ubiquitination of MYB6 in vitro. Different ubiquitination reaction systems with 40 mM ATP composed of different proteins purified from E. coli were incubated at 30°C for 3 h. The ubiquitination of MYB6 and PUB25 were detected by anti-MBP and anti-GST antibodies, respectively. The total ubiquitination signal was detected with anti-Ub antibodies. The numbers indicated 1/2/3 μg GST-PUB25. B, 26S proteasome inhibitor MG132 treatment effect on MYB6 protein stability. Total proteins extracted from 12-day-old seedlings of Col and pub25 pub26 were incubated with equal GST-MYB6 with or without 50 μM MG132 at 25°C for 2 h. Immunoblotting analysis was carried out with anti-GST antibodies. Actin was an equal loading control. C, Relative protein level in (B). The abundance of GST-MYB6 at 0 h was set as a reference to calculating relative MYB6 intensity of various treatment. D, PUB25 and PUB26 influence on MYB6 degradation. Total proteins extracted from 12-day-old Col and pub25 pub26 seedlings were incubated with equal GST-MYB6 at 25°C for different times. Immunoblotting analysis was carried out with anti-GST antibodies. Actin was an equal loading control. E, Relative protein level in (D). The abundance of GST-MYB6 at 0 h was set as a reference to calculating relative MYB6 intensity of different time. F, myb6 mutant susceptibility to Verticillium wilt compared to the wild-type. The plants grown in the greenhouse for 2 weeks were dipped into the V. dahliae spore suspension for 5 min. Photos were taken at 15 dpi. G, Statistical analysis of the disease index of Col, myb6-1, myb6-2, and myb6-C in (F), count with at least 15 plants. H, myb6 mutant and myb6 pub25 pub26 tm susceptibility to V. dahliae. The plants grown in the greenhouse for 2 weeks were dipped into the V. dahliae spore suspension for 5 min. Photos were taken at 15 dpi. I, Statistical analysis of the disease index of plants indicated in (H), count with at least 15 plants. J, Expression level of MYB6 in their corresponding transgenic complement lines. K, MYB6 transgenic complementation lines disease index to Verticillium wilt. The plants grown in the greenhouse for 2 weeks were dipped into the V. dahliae spore suspension for 5 min. Photos were taken at 15 dpi. L, Statistical analysis of the disease index of plants indicated in (K), count with at least 15 plants. *, **, and *** in (G), (I), (J), and (L) represent significant difference (P < 0.05), highly significant difference (P < 0.01), and extremely significant difference (P < 0.001), respectively, one-way ANOVA. The experiments were repeated independently three times with similar results.
We then performed a cell-free assay to determine whether the stability of MYB6 is regulated by PUB25 and PUB26. First, we added GST-MYB6 into cell extracts from wild-type and pub25 pub26 plants with or without the 26S proteasome inhibitor MG132 and observed that MG132 inhibited the degradation of GST-MYB6 in the presence of ATP (Figure 5, B and C). We then incubated purified GST-MYB6 with total proteins extracted from wild-type or pub25 pub26 plants in the presence of 10 mM ATP for different periods of time and detected changes in protein levels. GST-MYB6 degraded much more slowly in pub25 pub26 versus the wild-type (Figure 5, D and E). These results suggest that PUB25 and PUB26 modulate the stability of MYB6 via the 26S proteasome.
myb6 is susceptible to and suppresses the resistance of pub25 pub26 to V. dahliae
The finding that MYB6 was ubiquitinated by PUB25 and PUB26 suggests that it is involved in plant resistance to V. dahliae. We examined this hypothesis using an inoculation experiment. We obtained three MYB6 mutants (myb6-1, myb6-2, and myb6-Cas9) (Supplemental Figure S5, F–I) and the pub25 pub26 myb6-1 triple mutant (tm; Supplemental Figure S5J). Under normal growth conditions, the mutants did not exhibit any discernible phenotypes in terms of growth, development, or reproduction compared to the wild-type. However, under V. dahliae infection, the three myb6 mutants were more susceptible to V. dahliae infection than the wild-type (Figure 5, F and G) and the pub25 pub26 dm. The V. dahliae susceptibility of the myb6 pub25 pub26 tm was similar to that of myb6 (Figure 5, H and I). Under our experimental conditions, the disease index of the myb6 pub25 pub26 tm was approximately 75, which is similar to the disease index (65–75) of the myb6 mutants, whereas the disease indices of the wild-type and pub25 pub26 were approximately 25 and 50, respectively (Figure 5, H and I). pub25 pub26 overexpressing MYB6-Flag showed similar or slightly higher levels of disease resistance than pub25 pub26, which was more resistant than the wild-type (Supplemental Figure S5, K and L). These results suggest that MYB6 acts at downstream of PUB25 and PUB26 in response to V. dahliae infection.
To confirm that the enhanced susceptibility of the myb6 mutants to V. dahliae infection was caused by MYB6 mutation, we introduced MYB6 genomic DNA driven by the MYB6 promoter into the myb6-1 mutant background (Figure 5J). Inoculation experiments showed that two transgenic myb6 lines had comparable or even a little higher level of disease resistance than the wild-type (Figure 5, K and L). These results suggest that MYB6 positively regulates plant resistance to wilt caused by V. dahliae and acts at downstream of PUB25 and PUB26.
VDAL competes with MYB6 for interactions with PUB25 and PUB26
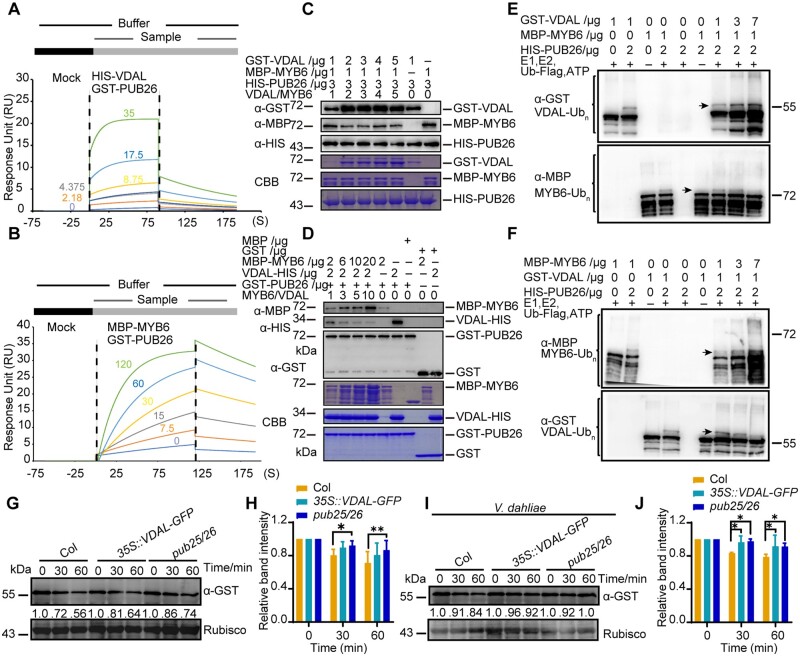
Given that both VDAL and MYB6 interacted with PUB25 and PUB26, we explored why overexpressing VDAL increased plant resistance to V. dahliae infection. We hypothesized that VDAL might compete with MYB6 for interacting with PUB25 and PUB26, thus hijacking these proteins to reduce the ubiquitination and degradation of MYB6. We performed the Biacore assay (Yan et al., 2018) to measure the affinity of HIS-VDAL and MBP-MYB6 for GST-PUB26 using proteins purified from E. coli. The anti-GST antibodies were immobilized onto a CM5 chip. After the baseline was established, the first sample (GST-PUB26 in Figure 6, A and B and Supplemental Figure S6B, GST in Supplemental Figure S6A) was overlaid on the chip, followed by the second sample (HIS-VDAL in Figure 6A and Supplemental Figure S6A, MBP-MYB6 in Figure 6B and MBP in Supplemental Figure S6B), and incubated for various periods of time. Strong interaction signals were detected between HIS-VDAL and GST-PUB26, between MBP-MYB6 and GST-PUB26. The interaction signal Response Unit values between HIS-VDAL and GST-PUB26 and between MBP-MYB6 and GST-PUB26 were comparable (Figure 6, A and B). However, the association rate (ka in Supplemental Table S1), disassociation rate (kd in Supplemental Table S1) and affinity (KD in Supplemental Table S1) of HIS-VDAL and GST-PUB26 were higher, suggesting that HIS-VDAL and GST-PUB26 associate more rapidly and have a stronger affinity than MBP-MYB6 and GST-PUB26. As expected, no interaction signals were detected between HIS-VDAL and GST or between GST-PUB26 and MBP (Supplemental Figure S6, A and B; Supplemental Table S1). These results suggest that the association between VDAL and PUB25 or PUB26 may lead to the occupation of the PUBs, thereby providing favorable conditions for the accumulation of MYB6 to help the plant resist infection by V. dahliae. Similar results were obtained in a subsequent competitive pull-down assay. The interaction between MBP-MYB6 and HIS-PUB26 decreased with increasing amounts of GST-VDAL (Figure 6C) and the interaction between VDAL-HIS and GST-PUB26 decreased with increasing amounts of MBP-MYB6 (Figure 6D).
Figure 6.
VDAL and MYB6 are competitive relationships in interacting with PUB26. A and B, Affinity characterization of HIS-VDAL (A) or MBP-MYB6 (B) to GST-PUB26. The anti-GST antibodies were immobilized onto CM5 chip. After baseline was established, the first sample GST-PUB26 injected over the chip was followed by the second sample HIS-VDAL (A) or MBP-MYB6 (B) for certain time with different concentration. The signals generated were subtracted from reference channel and then analyzed with kinetics fitting. The numbers represent the different concentrations (μg mL−1) of the second sample. C, Influence of VDAL on the interaction between PUB26 and MYB6 in the competitive pull-down assay. First, HIS-PUB26 was immunoprecipitated with anti-HIS beads, then GST-VDAL and MBP-MYB6 were added into reaction together with different concentration for 2 h. After fully washing for nonspecific proteins, proteins from beads were detected with anti-GST, anti-HIS, and anti-MBP antibodies. D, MYB6 inhibition of the interaction between PUB26 and VDAL in a competitive pull-down assay. First, GST-PUB26 was immunoprecipitated with anti-GST beads, then VDAL-HIS and MBP-MYB6 were added into reaction with different concentration. Pull-down proteins were detected with anti-GST, anti-HIS, and anti-MBP antibodies. E and F, Competition of VDAL and MYB6 for ubiquitination by PUB26. Different ubiquitination reaction systems with 40 mM ATP composed of different proteins purified from E. coli were incubated at 30°C for 3 h. The ubiquitinated VDAL and MYB6 were detected by anti-GST or anti-MBP antibodies. The total ubiquitination signal was detected with anti-Ub antibodies. G–J, Comparison of MYB6 degradation rate among Col, VDAL-OE, and pub25 pub26 dm. Total proteins extracted from 12-day-old Col, VDAL-OE, and pub25 pub26 seedlings treated with or without V. dahliae for 12 h were incubated with equal GST-MYB6 at 25°C for different times. Immunoblotting analysis was carried out with anti-GST antibodies. Actin was an equal loading control. The relative protein level was quantified with ImageJ. H, It is the relative protein level in (G), (J) is the relative protein level in (I).
Given that VDAL competes with MYB6 for binding to PUBs, we performed a competitive ubiquitination in vitro assay using purified VDAL-HIS, GST-PUB25, HIS-PUB26, and MBP-MYB6 proteins. The ubiquitination level of MBP-MYB6 decreased with increasing amounts of VDAL (Figures 6E;Supplemental Figure S6C) and vice versa (Figure 6F;Supplemental Figure S6D). Next, we performed a cell-free assay using Arabidopsis seedlings with or without V. dahliae infection to measure the degradation status of MYB6. We added GST-MYB6 protein into cell extracts from wild-type, VDAL-OE and pub25 pub26 seedlings with or without V. dahliae infection in the presence of ATP for different periods of time. GST-MYB6 was degraded at a similar rate or slightly more rapidly in the VDAL-OE line compared to pub25 pub26, but at the fastest speed in the wild-type in both the presence and absence of V. dahliae infection (Figure 6, G–J). These results suggest that VDAL competes with MYB6 for interactions with PUB25 and PUB26. Therefore, MYB6 might escape from PUB25- and PUB26-mediated degradation to enhance plant resistance to V. dahliae infection in planta.
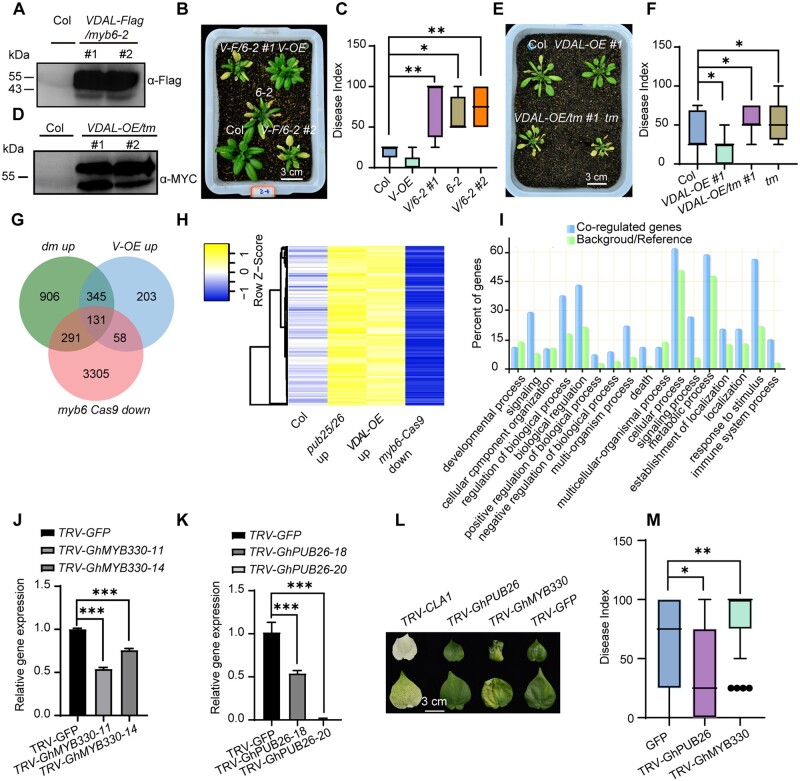
To further confirm that resistance gained by overexpression of VDAL is lost by introducing the myb6 mutation, we overexpressed VDAL fused with a Flag tag in myb6-2 (Figure 7A) and compared the disease resistance of these plants versus myb6-2. Overexpressing VDAL did not increase the resistance of myb6-2 to V. dahliae infection (Figure 7, B and C). To further verify this notion in planta, we generated transgenic lines stably expressing VDAL-MYC in the myb6 pub25 pub26 tm background (VDAL-OE/tm) to examine its potential role in triggering plant immune responses (Figure 7D). After inoculation, VDAL-OE/tm exhibited similar levels of susceptibility to V. dahliae to the pub25 pub26 myb6-1 tm, as indicated by disease index (Figure 7, E and F). These results suggest that VDAL itself does not have a direct effect on increasing resistance to V. dahliae infection; this effect likely depends on MYB6.
Figure 7.
VDAL competes with MYB6 for PUB25/26-mediated degradation in plants. A, Accumulation of VDAL protein in two myb6-2 transgenic lines. Total proteins extracted from 12-day-old Col and VDAL-Flag/myb6-2 seedlings were detected with anti-Flag antibodies. B, Phenotype comparison of disease symptoms in the wild-type (Col), VDAL overexpression line (Col, V-OE), myb6-2 (6-2), myb6-2 overexpressing VDAL lines 1 and 2 (V/6-2 #1, V/6-2 #2). Photos were taken at 15 dpi. C, Statistical analysis of the disease index of plants indicated in (B), count with at least 15 plants. D, VDAL-MYC overexpression in pub25 pub26 myb6-1 tms. Total proteins extracted from 12-day-old Col and VDAL-MYC/pub25 pub26 myb6-1 seedlings were detected with anti-MYC antibodies. E, Susceptibility of VDAL-OE/tm to V. dahliae compared to that of myb6 and Col. The plants grown in the greenhouse for 2 weeks were dipped into the V. dahliae spore suspension for 5 min. Photos were taken at 20 dpi. F, Statistical analysis of the disease index of plants indicated in (E), count with at least 15 plants. G and H, Transcriptome analysis of the myb6-Cas9, VDAL-OE, and pub25/26 dm by RNA-Seq after treatment with V. dahliae spores (1 × 106 conidia mL−1) for 12 h. (G) Venn diagrams showing the overlapped DEGs (downregulated in myb6 Cas9 and upregulated in pub25 pub26 and VDAL-OE). H, The heat map was drawn according to the expression levels in the myb6-Cas9, VDAL-OE, and pub25/26 in (G). I, Enrichment analysis of overlapping genes among myb6 downregulated and pub25 pub26 and VDAL-OE upregulated genes. J, Relative expression level of GhMYB330 in the VIGS cotton. K, Relative expression level of GhPUB26 in the VIGS cotton. L and M, Influence of knock down of GhMYB330 on cotton susceptibility to V. dahliae and knock down of GhPUB26 on cotton resistance to V. dahliae. The plants grown in the greenhouse for 3 weeks were injected 2 mL V. dahliae spore suspension (1 × 106 conidia mL−1) through stem near cotyledon. Photos were taken at 20 dpi, count with at least 25 plants. * and ** in (C), (F), (J), (K), and (M) represent significant difference (P < 0.05) and highly significant difference (P < 0.01), One-way ANOVA. The experiments were repeated independently three times with similar results.
VDAL and PUB25/PUB26 regulate plant resistance to wilt caused by V. dahliae
As both pub25 pub26 plants and plants stably expressing VDAL exhibited similar levels of resistance to V. dahliae infection, we reasoned that VDAL and PUB25/PUB26 might function via a similar molecular mechanism. More importantly, we demonstrated that MYB6 can escape from PUB25- and PUB26-mediated degradation to enhance plant resistance to V. dahliae infection in VDAL overexpression plants. These findings prompted us to explore the transcriptome profiles of plants with various genotypes. We treated 10-day-old seedlings with 106V. dahliae spores for 12 h, isolated total RNAs from two biological replicates, and subjected them to RNA-deep sequencing on the Illumina NovaSeq 6000.
We generated 150-bp high-quality trimmed paired-end reads and mapped them to the Arabidopsis genome (TAIR10) using HISAT2 (Kim et al., 2019) with default settings. We normalized raw counts and performed differential gene expression analyses in Col, myb6-Cas9, pub25 pub26, or VDAL-OE lines using DESeq2 (Love et al., 2014), respectively. Expression levels were compared for each gene in pub25 pub26 dm, VDAL-OE, and myb6-Cas9 with Col, respectively (P-value < 0.05). A total of 4,146, 1,673, and 737 genes were upregulated in myb6-Cas9, pub25 pub26, or VDAL-OE, respectively, while 3,785, 994, and 402 genes were downregulated in myb6-Cas9, pub25 pub26, or VDAL-OE, respectively (Figure 7G;Supplemental Figure S7A), compared with the wild-type. Note that mutation in the MYB6 resulted in dramatic change of the transcriptome profile (Supplemental Data Set 3).
The genes significantly induced by V. dahliae in myb6-Cas9, pub25 pub26, or VDAL-OE defined as differentially expressed genes (DEGs) were chosen for further analysis. Of the overlapping DEGs, 476 genes were higher in both VDAL-OE and pub25 pub26; 189 genes were upregulated in VDAL-OE but downregulated in myb6-Cas9; 422 genes were upregulated in pub25 pub26 but downregulated in myb6-Cas9; 131 genes were downregulated in myb6-Cas9 and upregulated in both VDAL-OE and pub25 pub26 (Figure 7, G and H for heatmap; Supplemental Data Set 4). Similarly, 197 genes were downregulated in both VDAL-OE and pub25 pub26, and 123 DEGs were upregulated in myb6-Cas9 and downregulated in pub25 pub26; 34 genes were upregulated in myb6-Cas9 and downregulated in VDAL-OE; and only 18 genes were upregulated in myb6-Cas9 and downregulated in VDAL-OE and pub25 pub26 (Supplemental Figure S7, A and B for heatmap). The high number of overlapping DEGs between pub25 pub26 and VDAL-OE suggests that they function in the same pathway.
To illustrate the biological function of these overlapping DEGs, we performed a GO (Gene Ontology) analysis using the online tool (Tian et al., 2017). These DEGs belong to different categories. The 131 overlapping genes among myb6 downregulated and pub25 pub26 and VDAL-OE upregulated genes were highly enriched in biological processes such as in response to stimulus, signaling, and immune process (Figure 7I). We constructed Venn diagrams and heatmap diagrams showing the expression patterns of common DEGs in different samples using the online tool jvenn and heatmapper, respectively.
However, we did not find any difference in chitin-activated MPK3, MPK4, and MPK6 (Wan et al., 2004; Supplemental Figure S7, C–E), and the expression of the PAMP-responsive gene ARABIDOPSIS NONRACE SPECIFIC DISEASE RESISTANCE GENE/HAIRPIN-INDUCED GENE-LIKE 10 (NHL10; a marker gene in the MPK-mediated defense pathway) (Sheikh et al., 2016; Supplemental Figure S7F), the PTI marker gene FLG22-INDUCED RECEPTOR-LIKE KINASE 1 (FRK1; At2g19190) (Asai et al., 2002; Supplemental Figure S7G), the SA marker gene PATHOGENESIS RELATED 1 (PR1; Uknes et al., 1992) did not show apparent difference under V. dahliae treatment among different plants (Supplemental Figure S7H). These results indicate that VDAL, PUB25, and PUB26 are involved in plant resistance to V. dahliae by functioning in similar immunity pathways that are different from chitin-, Flg22-, or SA-mediated pathways (Wang et al., 2020b), but they also individually regulate some other genes. Our results also suggest that the enhanced resistance of pub25 pub26 to V. dahliae is likely due to the accumulation of MYB6.
The fact that both the transgenic cotton and Arabidopsis had enhanced resistance to V. dahliae infection prompted us to explore whether this mechanism was similar between these two plants. We silenced the homologous genes (GhPUB26 and GhMYB330) of Arabidopsis PUB26 and MYB6 in island cotton by Virus-Induced Gene Silencing (VIGS; Sarde et al., 2019; Figure 7, K and L). Here chloroplastos alterados 1 (CLA1) was used as a positive reporter control for silencing efficiency. The inoculation experiment showed that the knock down of GhPUB26 conferred cotton more resistance to V. dahliae, while the knock down of GhMYB330 caused cotton to be more susceptible to V. dahliae (Figure 7, M and N) compared with the GFP control. These results suggest that the transgenic cotton plants overexpressing VDAL have a similar mechanism for disease resistance to V. dahliae as the VDAL transgenic Arabidopsis plants.
Discussion
The soil-borne hemibiotrophic fungal pathogen V. dahliae secretes more than 700 proteins (Klosterman et al., 2011), but only a few have been identified as effectors. In this study, we demonstrated that VDAL is a secretory elicitor protein that can cause leaf wilting and ROS burst when the protein was inoculated in leaves, which is very similar to bacterial Harpins (Choi et al., 2013). Some effectors produced by hemibiotrophic fungi such as V. dahliae (such as VdSCP41 and VdIsc1) inhibit plant immunity, whereas others (such as AVe1 and VDAL) increase plant immunity when overexpressed in plant cells (Castroverde et al., 2016). It was found that overexpressing Ave1 in Ve1(−) tomato induced the expression of defense genes, suggesting that Ave1 in the cytosol triggers plant immunity independently of the Ve1 receptor (Castroverde et al., 2016). The current findings suggest that VDAL can increase the Verticillium wilt resistance when overexpressed in both cotton and Arabidopsis.
Previous studies have found that several effectors from different pathogens can directly hijack host 26S proteasomes, thus inhibiting the degradation of immune-related proteins for their successful infection. For example, HopM1 from Pseudomonas syringae destabilizes A. thaliana HopM interactors, belonging to the ARF GEF proteins (AtMINs) in a proteasome-dependent manner (Nomura et al., 2006, 2011). However, HopM1, HopAO1, HopA1, and HopG1 were found to be the putative proteasome inhibitors, which makes it a little more complex to explain the function of HopM1 for promoting degradation of AtMINs (Ustun et al., 2016). HopM1 can directly interact with some E3 Ub ligases and proteasome subunits, suggesting its roles as a potential proteasome inhibitor (Ustun et al., 2016). Phytophthora infestans effector AVR3a stabilizes potato U-box E3 ligase CMPG1, and suppresses the degradation of CMPG1, thus preventing infestin 1-mediated host cell death during the biotrophic phase (Ustun et al., 2016). The rice blast fungus M. oryzae effector AvrPiz-t inhibits the rice RING E3 Ub ligase APIP6 (AvrPiz-t Interacting Protein 6) for enhancing the susceptibility of rice to M. oryzae (Park et al., 2012). Interestingly, APIP6 ubiquitinates AvrPiz-t in vitro, and both APIP6 and AvrPiz-t are degraded when coexpressed in the transient assays (Park et al., 2012). Furthermore, the effector AvrPtoB is an E3 ligase that can use the host 26S proteasome to directly target host immunity-related proteins such as the SA receptor NPR1 (Janjusevic et al., 2006; Chen et al., 2017), tomato protein kinase Fen (Rosebrock et al., 2007; Ntoukakis et al., 2009), FLS2 (Goehre et al., 2008), and CERK1 (Gimenez-Ibanez et al., 2009) for their degradation to subvert plant immunity.
Different from the aforementioned effectors that mainly target the host E3 ligases or are themselves E3 ligases targeting host proteins to disturb the plant immunity, our results indicate that VDAL targets and interferes with two negative E3 ligases PUB25 and PUB26 in plant immunity, but is not degraded in vivo, and also very stable in vitro. Cell free assays indicated that there is no significant difference in the degradation rate of VDAL protein in Col and pub25 pub26 dm. We found that the localization patterns of VDAL-GFP in protoplasts and transgenic plants were similar to that of PUB25/26-GFP in protoplasts, which provides a basis for them to work together. Plants overexpressing VDAL and the pub25 pub26 dm showed similar resistance to V. dahliae infection, that is, more resistant than the wild-type, suggesting that they genetically function in the same pathway. Transgenic plants overexpressing either PUB25 or PUB26 were more susceptible to V. dahliae infection than wild-type plants. These findings indicate that PUB25 and PUB26 play negative roles in plant resistance to V. dahliae, which are similar to their roles in response to the fungal pathogen Botrytis cinerea and the nonvirulent bacterial strain P. syringae pv. tomato (Pto) DC3000 hrcC− (Wang et al., 2018a).
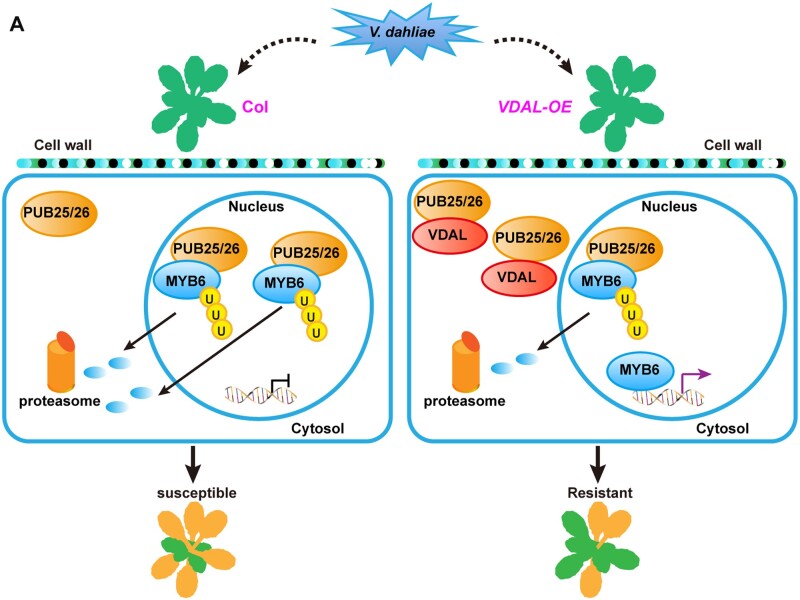
Furthermore, we found that VDAL is not ubiquitinated in plants, but could be ubiquitinated in vitro, suggesting that VDAL mainly hijacks PUBs to prevent their roles in degradation of MYB6, which leads to MYB6 protein accumulation and disease resistance (Figure 8A). One possibility is that VDAL is localized in some unknown big granule structures that may protect VDAL from degradation by PUB25 and PUB26. This phenomenon needs further exploration in the future. As PUB25 and PUB26 also target the positive regulator of BIK1 in plant immune response for its degradation (Wang et al., 2018a), we speculate that the BIK1 would be less affected by PUB25 and PUB26 in VDAL transgenic plants, thus increasing disease resistance, which needs further study. Genetics analysis indicated that myb6 mutants were more susceptible to V. dahliae than the wild-type, while the pub25 pub26 myb6 tm showed a similar susceptibility to V. dahliae as the myb6 single mutant. The pub25 pub26 dm overexpressing MYB6 was more resistant to V. dahliae than the wild-type. MYB6 protein is hard to be detected in the wild-type overexpressing MYB6, but accumulated to a high level in pub25 pub26 mutant. However, the disease resistance is comparable between the pub25 pub26 and wild-type overexpressing MYB6, implying that the presence of MYB6 is necessary, but not sufficient for increasing disease resistance. These findings indicate that PUB25 and PUB26 directly target MYB6 and negatively regulate MYB6 stability during the pathogen response and that MYB6 is a positive regulator in resistance for V. dahliae disease. Notably, plants overexpressing VDAL in the myb6 mutant background showed similar susceptibility to V. dahliae as the myb6 single mutant, suggesting that increasing resistance to V. dahliae infection by overexpressing VDAL depends on MYB6. Our Biacore and competitive pull-down assays suggested that VDAL interacts with PUB25 and PUB26, thus interfering with their interactions with MYB6 and reducing MYB6 degradation, thereby leading to increased resistance to V. dahliae infection.
Figure 8.
A proposed model for VDAL in increasing resistance to V. dahliae via interacting with E3 ligases PUB25 and PUB26. A, In wild-type plants, PUB25 and PUB26 ubiquitinate the TF MYB6, and mediate its degradation through the 26S proteasome. In VDAL-overexpressing plants, VDAL competitively binds PUB25 and PUB26, which results in reducing the degradation of MYB6. As a positive regulator, the accumulated MYB6 increases disease resistance.
In this study, our results showed that VDAL possess a SP that has secreting activity. This is consistent with the notion that effectors are characterized by N-terminal SPs that can lead them to the secretory pathway (Sperschneider et al., 2017; Wang et al., 2021). The VDAL homologs widely exist in other fungi. We speculate that VDAL is secreted into the extracellular space as an elicitor to attack plants, or even into host cells. There are two possible explanations for VDAL roles inside host cells: as a hemibiotrophic fungal pathogen, V. dahliae might use VDAL to target PUB25 PUB26 in order to accumulate more MYB6, thus does not kill the plant cells immediately during its infection; alternatively, plants might have evolved a new mechanism to exploit VDAL to fight against V. dahliae.
We determined the kinase activity of MAPK3, MAPK4, and MAPK6 in the VDAL transgenic plants and myb6 mutant after treatment with chitin or V. dahliae and did not find any obvious difference. The transcript accumulation measured by RT-qPCR analyses indicates that the expression of some marker genes such as the MPK-mediated marker gene NHL10 (Sheikh et al., 2016), the PTI marker gene FRK1 (Asai et al., 2002), and the SA marker gene PR1 (Uknes et al., 1992) were not changed in the VDAL transgenic plants and myb6 mutants. However, we found that the expression of some disease-related genes such as those encoding MAPKKK protein, NAC, and WRKY TFs (see Supplemental Data Sets 3 and 4) was changed in VDAL plants and myb6 mutants. These results suggest that the MYB6-mediated plant immune response does not function through the well-studied traditional pathways such as systemic acquired resistance and HR during bacterial infection (Zhou and Zhang, 2020; Wang et al., 2020b). However, we cannot exclude a possible role for BIK1 in plant resistance to Verticillium wilt as BIK1 is targeted by PUB25 and PUB26 (Wang et al., 2018a), and BIK1 plays critical roles in both ETI and PTI for plant resistance to P. syringae pv. tomato DC3000 strain (Ngou et al., 2021; Yuan et al., 2021a, 2021b).
When plants are infected by pathogens, plant growth is usually retarded due to the growth-defense trade-off (Zhou and Zhang, 2020; Zhou et al., 2020b). In this study, Arabidopsis and cotton plants overexpressing VDAL did not show any growth or developmental defects, but they showed increased resistance to V. dahliae. We found that GhMYB330 and GhPUB26 in cotton have the similar physiological functions during V. dahliae infection as MYB6 and PUB26 in Arabidopsis, which suggests that the mechanism of plant disease resistance caused by overexpression of VDAL is conserved. A recent study involving the successful cloning of the glutathione S-transferase gene Fhb7 from Triticum aestivum indicated that the horizontal transfer of this gene from a fungus underlies Fusarium head blight resistance in wheat (Wang et al., 2020a). Therefore, VDAL represents a good candidate gene from V. dahliae for improving plant resistance to V. dahliae infection without yield penalty when overexpressed in plants. Although many effector proteins such as Harpins (Peng et al., 2004), SSBXoc (Cao et al., 2018), and PsCRN161 (Rajput et al., 2015) are found to increase the disease resistance when expressed in plants without affecting plant development and growth, their molecular mechanisms are not well understood. Our results provide a molecular basis for these outcomes and highlight the potential for using genes from pathogens to improve crop pathogen resistance (Miao et al., 2020; Li et al., 2021b).
Materials and methods
Plant materials and growth conditions
All transgenic A. thaliana lines and T-DNA insertion mutants used in this study are in the Columbia-0 (Col-0) ecotype background. The T-DNA insertion mutants pub25 (AT3G19380 and SALK-147032C), pub26 (AT1G49780, CS351943, and GK308D07), myb6-1 (AT4G09460, SALK_074789C) and myb6-2 (AT4G09460, CS403018) were obtained from the Arabidopsis Biological Resource Center (ABRC). Homozygous lines were obtained by PCR using T-DNA specific and the gene-specific primers. Pro35S:VDAL-GFP, Pro35S:VDAL-MYC, Pro35S:MYB6-Flag, Pro35S:PUB25-GFP, and Pro35S:PUB26-GFP were introduced into Col by the floral-dip method (Clough and Bent, 1998) to generate overexpression transgenic lines. For complementation lines of myb6-1, MYB6 genomic DNA including the native promoter and coding region were amplified and cloned into PCAMBIA1301 vector (ProMYB6:MYB6), then transferred into myb6-1 mutant. Pro35S:PUB25-Flag, Pro35S:PUB26-Flag, ProPUB25:PUB25-Flag (Wang et al., 2018a) and ProPUB26:PUB26-MYC were described previously (Wang et al., 2019)
Arabidopsis seeds were sown on Murashige and Skoog (MS) medium containing 2% sucrose and 0.8% agar. Then 7- or 10-day seedlings were transferred to soil and grown under short-day (12-h light/12-h dark) or long-day (16-h light/8-h dark) conditions in a growth room at 20–22°C.
Vector construction
For constructing Pro35S:VDAL-GFP/Flag/MYC, the cDNA of V. dahliae VDAL was amplified and cloned into pCAMBIA vector driven by 35S promoter. For Pro35S:MYB6-MYC/Flag/GFP, Pro35S:PUB25-GFP and Pro35S:PUB26-GFP, the cDNA of MYB6, PUB25, or PUB26 from Col was cloned into pCAMBIA vector driven by 35S promoter and fused to MYC, Flag, or GFP tag, respectively. For ProMYB6:MYB6, the promoter and genomic DNA of MYB6 from Col genomic DNA were cloned into pCAMBIA1301 vector. For Pro35S:PUB25-ccLUC, Pro35S:PUB26-ccLUC, Pro35S:MYB6-GZnLUC, Pro35S:VDAL-GZnLUC, Pro35S:BIK1-GZnLUC, Pro35S:GUS-GZnLUC, Pro35S: GUS-ccLUC, the CDS of PUB25, PUB26, MYB6, VDAL, BIK1, or GUS was cloned into pCAMBIA-ccLUC and pCAMBIA-GZnLUC, respectively, and fused to ccLUC and GZnLUC, respectively. For BD-PUB25, BD-PUB26, AD-MYB6s, AD-VDAL, or AD-BIK1, the CDS of PUB25, PUB26, MYB6, VDAL, or BIK1 was cloned into pGBKT7 or pGADT7, respectively. For GST-PUB25, GST-PUB26, HIS-PUB26, GST-MYB6, MBP-MYB6, GST-VDAL, HIS-UBA1 (E1), and HIS-UBC10 (E2), the CDS of PUB25, PUB26, MYB6, VDAL, E1, or E2 was cloned into pGEX-4T-1, pET30a, or pMAL-c5X, respectively, and fused to GST, HIS, or MBP tag, respectively.
Phylogenetic analysis
Protein sequences were obtained from National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) through protein BLAST. Full-length protein sequences were aligned, and the phylogenetic tree was produced by the neighbor-joining method in MEGA version 7.0 through default settings.
Pathogen culture and infection assay
Verticillium dahliae strain Vd991 was grown on potato dextrose agar medium at room temperature (25°C) in the dark for 1–2 weeks. The spore suspension solution was prepared at 1 × 106 conidia mL−1. Two-week-old Arabidopsis seedlings were removed carefully from pots and washed with water. The roots were dipped into the spore suspension solution for 5 min, and then replanted in new prepared soil, and cultured at 23°C in a 12-h-light/12-h-dark cycle for phenotyping. Disease index and pictures were recorded or taken on 10–20 dpi (Fradin et al., 2011).
Disease index of verticillium wilt caused by V. dahliae
The disease index of Arabidopsis caused by V. dahliae was recorded on 15–25 dpi. Since one of the distinguishing phenotypes, after infected by V. dahliae, was the leaves turned to yellow abnormally and rapidly. According to the severity of the condition (proportion of leaves turned to yellow), disease was divided into five levels. No disease after incubation was defined as level 0; less than one-fourth of leaves infected was defined as level 1; less than half of leaves infected was defined as level 2; less than three-fourth of leaves infected was defined as level 3; and more than three-fourth of leaves was defined as level 4. , where k is the disease level of Arabidopsis; n is the number of Arabidopsis. Data are presented as means ± sd, and analyzed by one-way ANOVA performed with GraphPad Prism version 8 software. At least three biological replicates were used to perform each of the experiments, each replicate with at least 15 plants, and all the experiments were repeated for three times, with consistent results.
Confirmation of the secreted SP of VDAL in yeast
Functional validation of the VDAL SP was performed using the yeast invertase secretion assay as described previously with slight modification (Oh et al., 2009). Briefly, the different forms of VDAL SP were introduced into pSUC2T7M13ORI (pSUC2-MSP) vector using EcoR1 and Xho1 restriction sites (pSUC2-VDAL). Yeast YTK12 strains were transformed with 500 ng pSUC2-VDAL-SP or pSUC2-MSP using the lithium acetate method (Modrof et al., 2003). Transformed yeast cells were plated on selection CMD-W medium (0.67% yeast N base without amino acids, Trp dropout supplement, 2% sucrose, 0.1% glucose, and 2% agar). Positive clones were transferred to new CMD-W medium and YPRAA medium (1% yeast extract, 2% peptone, 2% raffinose, and 2% agar, pH 5.8) for testing invertase secretion. Invertase enzymatic activity was assayed by TTC (Schenke et al., 2011) reduction assay. Untransformed YTK12 strain, YTK12 carrying the pSUC2-MSP, and pSUC2-VDAL-SP vector were cultured in 5 mL YPDA liquid medium (2% peptone, 1% yeast extract, 2% glucose, pH 5.8) over night at 30°C. Total yeast cells were collected and washed by ddH2O. Then the pellet was resuspended with 0.1% colorless dye TTC at 30°C for 1–2 h and color change was obtained in room temperature.
Identification of VDAL-interacting proteins
To identify VDAL-targeted proteins in Arabidopsis, VDAL protein complex was purified as described previously with slight modification (Li et al., 2014). VDAL-MYC was expressed in Arabidopsis protoplasts. Total proteins were extracted in 8 mL Protein Extraction Buffer with 1% protease inhibitor. Cell debris was removed from the lysate by centrifugation at 12,000g for 10 min. The supernatant was incubated with 150 μL anti-MYC agarose beads (Sigma, Cat#A7470) for 2.5 h. Then the immunocomplexes were collected and washed with 1 mL low salt wash buffer (10 mM HEPES [pH 7.5], 100 mM NaCl, EDTA 1 mM, Glycerol 10%, 0.1% NP-40, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail) and high salt wash buffer (200 mM NaCl), each for four times. Immunocomplexes were eluted with Tris–glycine elution buffer. The total volume was concentrated to 30 μL with 3 kDa microspin columns. The 25 μL eluted proteins were loaded onto a single lane on a 10% SDS-PAGE gel for LC–MS (Beijing Protein Inovation, Beijing, China) assay, another 5 μL eluted proteins were used for immunoblotting.
Coimmunoprecipitation
The protoplasts were transfected with the purified plasmids and incubated overnight (16 h). Total proteins were extracted for CoIP with the extraction buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Triton-X 100, 1 mM DTT, proteinase inhibitor cocktail). For anti-GFP IP, total proteins were incubated with 10 μL agarose conjugated anti-GFP antibodies for 3 h and washed three times with the washing buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Triton-X 100, 1 mM DTT). The samples were boiled for 5 min with 6SDS loading buffer. The proteins were separated by SDS-PAGE and detected by anti-GFP, anti-MYC antibodies.
Firefly LCI assay
The full-length CDS sequence was fused to the C-terminus of GZnLUC and C-terminus of ccLUC (pCAMBIA). The constructed plasmids were transformed into Agrobacterium strain GV3101. Positive clones were incubated in 5 mL YEB liquid medium at 28°C for 16 h. The clones with different vectors were mixed equally (based on OD600). Agrobacteria were collected and resuspended in 2 mL of activity buffer (10 mM MES [pH 5.7], 10 mM MgCl2, 150 μM acetosyringone). After 2–5 h incubation at room temperature in darkness, the active Agrobacteria were infiltrated into expanded leaves of N. benthamiana and grown in the growth room for 48–72 h before the LUC activity measurement. For the CCD imaging and LUC activity measurement, infiltrated leaves were sprayed with 1 mM luciferin and then kept in the dark for 5 min. A precold CCD camera (1300B, Roper, Sarasota, FL, USA) was used to capture the LUC image and the exposure time for signal collection was 15 min.
Y2H assay
To validate the interaction between VDAL and PUB25/PUB26, and the interaction between MYB6 and PUB25/PUB26, full-length PUB25, PUB26, MYB6, or VDAL CDS was fused into pGBKT7 (Fradin et al., 2011) and pGADT7 (AD) vectors, respectively. These plasmids were cotransformed into yeast strain Gold using the lithium acetate method (Kong et al., 2015). Transformed yeast cells were separately plated on 2D synthetic dropout medium (Trp-/Leu-) selective medium and incubated at 28°C for 3–4 days. Then transformed yeast cells were transferred to fresh 2D and 4D media (Trp-/Leu-/His-/Ade-) with 10−1, 10−2, 10−3 and 10−4 dilution for interaction detecting, then the interactions were recorded after growing 3–4 days at 28°C. The interaction of BIK1 and PUB26 was used as the positive control.
In vitro ubiquitination assay
GST-PUB25, GST-PUB26, GST-VDAL, MBP-MYB6, HIS-PUB26, HIS-UBA1 (E1), or HIS-UBC10 (E2) was expressed in E. coli strain BL21 or Rosetta, respectively. GST-PUB25, GST-PUB26, or GST-VDAL protein was purified on Glutathione Sepharose (GE), respectively. MBP-MYB6 protein was purified on amylose resin (NEB) and HIS-PUB26, HIS-UBA1 (E1), or HIS-UBC10 (E2) protein was purified on Ni Sepharose (GE), respectively. For testing E3 ligase activity, 250 ng HIS-E1, 500 ng purified HIS-E2, 1.25 µg Flag-tagged Ub, 2 µg purified GST-PUB25 and GST-PUB26 or HIS-PUB26 were added to 30 µL of ubiquitination reaction buffer (50 mM Tris–HCl pH 7.5, 2 mM ATP, 5 mM MgCl2, 2 mM DTT). After incubation for 3 h at 30°C, all reactions were stopped by adding 6SDS loading buffer, and boiled for 5 min. For GST-VDAL and MBP-MYB6 ubiquitination assay, 1 µg purified GST-VDAL or MBP-MYB6 was added to above 30 µL of ubiquitination reaction buffer. After incubation for 3 h at 30°C, all reactions were stopped by adding 6SDS loading buffer, and boiled for 5 min. For competitive ubiquitination assays, 1 µg purified MBP-MYB6 and different concentration (1, 3, and 7 µg) of GST-VDAL, or 1 µg purified GST-VDAL and different concentration (1, 3, and 7 µg) of MBP-MYB6 were added to >30 µL ubiquitination reaction buffer. After incubation for 3 h at 30°C, all reactions were stopped by adding 6SDS loading buffer, and boiled for 5 min. The proteins were separated by 10% SDS-PAGE gel, and detected with anti-Ub, anti-GST, or anti-MBP antibodies.
In vivo ubiquitination assay
To determine the specificity for protein degradation of VDAL, in vivo ubiquitination assay was performed. Total proteins of 12 day VDAL-MYC in Col and pub25 pub26 dm were extracted after treatment with V. dahliae or V. dahliae combined MG132 for 12 h with the extraction buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Triton-X 100, 1 mM DTT, proteinase inhibitor cocktail), and incubated with 20 μL MYC beads coated with MYC antibodies for 3 h, and washed three times with washing buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Triton-X 100, 1 mM DTT). The samples were boiled for 5 min with 6SDS loading buffer. The proteins were separated by SDS-PAGE and probed with anti-Ub and anti-MYC antibodies. The ubiquitinated proteins were purified as described previously with slight modification (Kong et al., 2015). Total proteins of 12-day transgenic VDAL-MYC Col and pub25 pub26 dm were extracted after treatment with V. dahliae or V. dahliae combined MG132 for 12 h with the BI buffer (50 mM Tris–Cl (pH 7.5), 20 mM NaCl, 0.1% NP-40 and 5 mM ATP) in a prechilled mortar. The other experimental methods are as described previously (Kong et al., 2015).
Protein pull-down assay
To confirm the interaction between PUBs and VDAL, as well as PUBs and MYB6, the pull-down assay was performed as described previously (Wang et al., 2018b) with slight modifications. Briefly, GST, or GST-PUB26 protein (10 µg) was incubated with GE for 2 h with 1 mL pull-down binding buffer (150 mM NaCl, 10 mM Na2HPO4, 2 mM KH2PO4, 2.7 mM KCl, and 0.1% Nonidet P-40, pH 7.4) at 4°C. After a centrifugation at 1,000 g for 3 min at 4°C, the buffer was removed and 1 mL fresh binding buffer was added to the GE, then 2 µg VDAL-HIS, or MBP-MYB6 protein was added to the above fresh binding buffer (for competitive pull-down assay, GST-VDAL and MBP-MYB6 in Figure 6C, VDAL-HIS, and MBP-MYB6 in Figure 6D were added as indicated), along with Sepharose. The tube was rotated at 4°C for 2 h for protein binding. After a centrifugation at 1,000 g for 3 min at 4°C, the buffer was removed, and the Sepharose was washed five times with 1× PBS (150 mM NaCl, 10 mM Na2HPO4, 2 mM KH2PO4, and 2.7 mM KCl, pH 7.4) to remove nonspecifically bound proteins. All reactions were stopped by adding 50 μL 1× PBS and 10 µL 5× SDS loading buffer boiled for 5 min. The proteins were separated by 10% SDS-PAGE gel and detected with anti-Ub, anti-GST, or anti-MBP antibodies. All reactions were stopped by adding 50 μL 1× PBS and 10 µL 5× SDS loading buffer, and boiled for 5 min. The proteins were separated by 10% SDS-PAGE gel, and detected with anti-HIS, anti-GST, or anti-MBP antibodies.
ROS burst assay
ROS burst assay was performed as described previously with slight modifications (Wang et al., 2018a). Briefly, 5 mm-diameter leaf discs were collected from 4 to 5-week-old plants into 96-well plates, and floated overnight on sterile water in a 96-well plate. In the following day, the water was replaced with a solution of 20 mM luminol (Sigma) and 10 mg mL−1 horseradish peroxidase (Sigma) containing different concentrations of VDAL, then luminescence was recorded by the Tecan-i-control outfitted with the LUM module.
Cell-free protein degradation assay
To identify the protein stability of VDAL and MYB6, the cell-free protein degradation assay was performed as described previously (Kong et al., 2015) with slight modifications. For protein degradation in transgenic plants, the seeds of Pro35S:VDAL-GFP grown at 22°C for 10 days under long-day condition (16-h light/8-h dark cycles), then total proteins were extracted with native protein extraction buffer [50 mM Tris–MES (pH 8.0), 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA (pH 8.0), 5 mM DTT], the extracted supernatants were divided into two or four equal parts with addition of 50 µM MG132, 1 mM ATP or not, respectively, and the samples were cultured at 22°C with shaking for different times. For purified protein degradation, the seeds of Col and pub25 and pub26 grown at 22°C for 10 days under long-day conditions (16-h light/8-h dark cycles), then total proteins were extracted with native protein extraction buffer, 200 ng purified protein of GST-VDAL or GST-MYB6 from E. coli strain Rosetta was incubated in 600 μg total proteins for each reaction with addition of 50 µM MG132 and 1 mM ATP, or 1 mM ATP only, then the reactions were cultured at 22°C for different times. All reactions were stopped by adding 5× SDS loading buffer with boiled for 5 min. The proteins were separated by 10% SDS-PAGE gel and detected with anti-GFP or anti-GST antibodies.
Chitin-induced MAPK kinase activities assay
The MAPK kinase activity assay was performed as described previously with slight modification (Yamada et al., 2016; Wang et al., 2017a; Zhou et al., 2020a). Briefly, Arabidopsis seedlings were grown on MS medium for 7 days under 22-h light/2-h dark at 22°C and then treated with or without 500 μg mL−1 chitin 10 min. Total proteins were extracted with the extraction buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Triton-X 100, 1 mM DTT, proteinase inhibitor cocktail, 1× EDTA-free protease inhibitor cocktail, and 1× Phostop phosphatase inhibitor cocktail). The proteins were separated by SDS-PAGE and detected by anti-phospho-p44/42 MAPK antibodies (Cell Signaling #4370) and anti-actin antibodies, respectively.
Subcellular localization and GUS staining
To identify the subcellular localization, the full-length CDS of VDAL, PUB25, and PUB26 were cloned into pCAMBIA vector under control of 35S promoter and fused to GFP tag, respectively. To determine the subcellular localization of VDAL in cells, the protoplasts were transfected with each purified plasmid and incubated overnight (16 h). The GFP images were acquired at excitation of 488 nm and emission of 525 nm. To determine the subcellular localization of VDAL in the transgenic plants, the Pro35S:VDAL-GFP homozygous transgenic seedlings were obtained via the floral dip method (Clough and Bent, 1998). The GFP images were acquired at excitation of 488 nm and emission of 525 nm.
To identify the expression of PUB25, PUB26, and MYB6 in plants, the promoter of PUB25, PUB26, and MYB6 was cloned into pCAMBIA1391 vector, respectively. The plasmid was subsequently transformed into Agrobacterium GV3101 and transferred into plants via the floral dip method. T3 homozygous transgenic seedlings were used for the GUS staining assay.
VDAL zinc-binding assay
To determine the zinc-binding activity of VDAL, zinc-binding assay was performed as described previously (Citiulo et al., 2012) with slight modifications. About 4 ng GST or GST-VDAL purified protein was loaded onto 10 kDa microspin columns (Ambion, Austin, TX, USA). After two washes with 1 mL HS buffer (50 mM HEPES-KOH pH 7.5, 200 mM NaCl), supernatants were transferred to reaction tubes and incubated with 0.1 mM Zn2+ for 1 h at 37°C. The zinc-loaded proteins were then transferred to 10 kDa microspin columns and sequentially washed with 4 mL of HS buffer. Each flow-through was assayed for zinc content by 4-(2-pyridylazo) resorcinol (PAR) assay, until there was no Zn2+ in the buffer. Briefly, PAR was added to each sample to 0.1 mM and optical density was measured at 490 nm against a Zn2+ standard curve. Following 15 washes, undigested and digested (40 mg proteinase K, 50°C for 30 min) samples were again assayed for zinc content by PAR assay.
Biacore assay
To determine the affinity of HIS-VDAL to GST-PUB26, or MBP-MYB6 to GST-PUB26, the Biacore assay was employed (Yan et al., 2018; Supplemental Table S1). Biacore T200 evaluation 3.1 was used for data analysis. The generated signals were subtracted from the reference channel and then analyzed with kinetics fitting.
RT-quantitative PCR
Twelve-day-old seedlings were treated with or without 1 × 106 conidia mL−1 spore suspensions of V. dahliae 12 h. Total RNAs were extracted by HiPure Plant RNA Mini Kit RNeasy Plant Mini Kit (Magen; cat. R4151) with the kit instructions. The RNAs were reverse transcribed to cDNAs with MMLV reverse transcriptase (Thermo, Waltham, MA, USA). Quantitative PCR was performed with a 7300 Real-Time PCR system (Applied Biosystems Waltham, MA, USA) using SYBR Premix Ex Taq (TaKaRa, Shiga, Japan). ACTIN was employed as an internal control.
RNA-seq analysis
Arabidopsis seedlings were grown on MS medium for 10 days under 22-h light/2-h dark at 22°C and then treated with 1 × 106 conidia mL−1 spore suspensions of V. dahliae for 12 h. Total RNAs were extracted by HiPure Plant RNA Mini Kit RNeasy Plant Mini Kit (Magen; cat. R4151) following the manufacturer’s instructions. About 1 μg RNA for each sample was used for library construction with NEB Next Ultra RNA Library Prep Kit for Illumina according to the instructions and RNA-Seq was carried out on Illumina NovaSeq 6000 platform.
About 5.0 GB clean reads were generated for each sample. All reads were trimmed to 150-bp paired-end reads with high quality according to the base quality. The trimmed reads were mapped to the A. thaliana reference genome (TAIR10) using HISAT2 (Kim et al., 2019) with default settings. Differential gene expression analysis was performed using DESeq2 (Love et al., 2014).
Fold changes of the genes induced significantly by V. dahliae treatment were compared with the control (Col) sample in each group (using P < 0.05). Then Venn diagrams showing the differentially regulated genes (downregulated genes and upregulated genes) in common between the different samples were generated using an online tool jvenn (http://jvenn.toulouse.inra.fr/app/example.html). The heat map was drawn according to the expression levels in the myb6-Cas9, VDAL-OE and pub25 pub26 dm compared to Col using an online tool Heatmapper (http://www.heatmapper.ca/expression/). Gene enrichment analysis was performed with online tool AGRI GO (http://bioinfo.cau.edu.cn/agriGO/index.php).
Our raw data and the processed RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (PRJNA739851).
Virus-induced gene silencing
Agrobacterium cultures containing pTRV-CLA1, pTRV-GhMYB330, and pTRV-GhPUB26, pTRV-GFP, and pTRV-RNA1 were cultured in 2–3 mL LB at 37°C for 12–16 h, then transferred to 100 mL LB and cultured 12–16 h more, after which cultures were collected and resuspended to OD600 = 0.8 with infiltration buffer (10 mM MES, 10 mM MgCl2, 200 μM acetosyringone, and pH 5.4), and set in the dark for 3 h. GV3101 cells containing pTRV-RNA1 were mixed with cells harboring pTRV-GhMYB330, pTRV-GhPUB26, and pTRV-GFP and infiltrated into the true leaves of cotton seedlings. After 2 weeks, 2 mL spore suspension solution of V. dahliae prepared at 1 × 106 conidia mL−1 was infiltrated into the stem near the cotyledon. Disease indices were recorded and photographs were taken 15–25 days later.
Accession numbers
Sequence data from this article can be found in the GenBank under the following accession numbers: PUB25, AT3G19380; PUB26, AT1G49780; MYB6, AT4G09460; VDAL, AY524791, GhPUB26, GH_D13G0403; and GhMYB330, GH_A13G1430. All transgenic A. thaliana lines and T-DNA insertion mutants used in this study are in the Col-0 ecotype background. The T-DNA insertion mutants pub25 (SALK-147032C), pub26 (S351943 and GK308D07), myb6-1 (SALK_074789C), and myb6-2 (CS403018) were obtained from the ABRC. myb6 CRISPR Cas9 mutant was obtained through CRISPR Cas9 system. RNA-seq data (PRJNA739851).
Reagent and resources
Reagents and resources used in this study are listed in Supplemental Tables S2 and S3.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Characteristics of VDAL. Supports Figure 1.
Supplemental Figure S2 . Phylogenetic tree of the VDAL from different fungi. Supports Figure 1.
Supplemental Figure S3 . PUB25 and PUB26 have auto-ubiquitination activity and PUB26 can ubiquitinate VDAL in vitro. Supports Figures 2 and 3.
Supplemental Figure S4 . pub25 pub26 dm shows resistant to Verticillium wilt caused by V. dahliae. Supports Figures 3 and 4.
Supplemental Figure S5 . MYB6 is stable in pub25 pub26 dm. Supports Figure 5.
Supplemental Figure S6 . VDAL and MYB6 are competitors in PUB25 ubiquitination. Supports Figure 6.
Supplemental Figure S7 . The gene expression analyses and MAPK kinase activities in VDAL transgenic plants and different mutants. Supports Figure 7.
Supplemental Table 1 . Affinity and kinetics parameters of the target proteins from Biacore assay. Supports Figure 6.
Supplemental Table 2 . Reagents and resources used in this study.
Supplemental Table 3 . Primers used in this study.
Supplemental Data Set 1 . VDAL interaction protein candidates by LC–MS. Supports Figure 2.
Supplemental Data Set 2 . PUB26-ARM Y2H screen candidates. Supports Figure 4.
Supplemental Data Set 3 . Genes significantly induced by V. dahliae in myb6 Cas9 compared with Col (down, P < 0.05). Supports Figure 7.
Supplemental Data Set 4 . Overlapping genes among myb6 downregulated and pub25 pub26 and VDAL-OE upregulated genes. Supports Figure 7.
Supplemental File S1 . The sequence alignment of VDAL homologs from different fungi.
Supplementary Material
Acknowledgments
We thank Prof. Dingzhong Tang at Fujian Agriculture and Forestry University, Prof. Xianzhong Feng, Dr Jiantian Leng, and Yu Hui, at Northeast Geography and Agroecology for providing valuable advice on writing of the manuscript; Prof. Ruyu Li and Yongqing Li at South China Botanical Garden, Chinese Academy of Sciences for donating cotton cultivar; Prof. Zhaosheng Kong and Dr Zhidi Feng at Institute of Microbiology, Chinese Academy of Sciences for donating V. dahliae strain. Libo Shan at Texas A&M University for donating pSUC2 vector. We thank Prof. Jianmin Zhou at State Key Laboratory of Plant Genomics, China Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for providing NP-PUB25-Flag seeds and valuable advice. We thank Prof. Ziding Zhang for guidance in analyzing RNA-Sequence gene expression.
Funding
This research was financially supported by the National Key Research and Development Program of China (grant no. 2016YFD0101904); Major transgenic project Program of China (2016ZX08005-004 and 2014ZX08005-02B).
Conflict of interest statement. There are no conflicts of interest.
A.M., Z.G., and J.Q. designed the study. A.M. and D.Z. performed the experiments. A.M., H.L., Y.G., Y.H., Z.G., and J.Q. analyzed the data. A.M., Z.G., and J.Q. wrote the paper. All other authors participated in the discussion of the results and commented on the manuscript.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Zhizhong Gong (gongzz@cau.edu.cn) or Junsheng Qi (junshqi@cau.edu.cn)
Reference
- Afzal AJ, da Cunha L, Mackey D (2011) Separable fragments and membrane tethering of Arabidopsis RIN4 regulate its suppression of PAMP-triggered immunity. Plant Cell 23:3798–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983 [DOI] [PubMed] [Google Scholar]
- Betsuyaku S, Katou S, Takebayashi Y, Sakakibara H, Nomura N, Fukuda H (2018) Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol 59:8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8:521–539 [DOI] [PubMed] [Google Scholar]
- Block A, Toruno TY, Elowsky CG, Zhang C, Steinbrenner J, Beynon J, Alfano JR (2014) The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol 201:1358–1370 [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436 [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324:742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu B, Qiu D, Zeng H, Guo L, Yuan J, Yang X (2014) A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep 33:461–470 [DOI] [PubMed] [Google Scholar]
- Cao Y, Yang M, Ma W, Sun Y, Chen G (2018) Overexpression of SSBXoc, a single-stranded DNA-binding protein from Xanthomonas oryzae pv. oryzicola, enhances plant growth and disease and salt stress tolerance in transgenic Nicotiana benthamiana. Front Plant Sci 9:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castroverde CD, Nazar RN, Robb J (2016) Verticillium Ave1 effector induces tomato defense gene expression independent of Ve1 protein. Plant Signal Behav 11:e1245254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castroverde CD, Xu X, Nazar RN, Robb J (2017) Biotic factors that induce the tomato Ve1 R-gene. Plant Sci 265:61–69 [DOI] [PubMed] [Google Scholar]
- Chen H, Chen J, Li M, Chang M, Xu KM, Shang ZH, Zhao Y, Palmer I, Zhang YQ, McGill J. et al. (2017) A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22:777–788.e7 [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim W, Lee C, Oh CS (2013) Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Mol Plant Microbe Interact 26:1115–1122 [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186:757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citiulo F, Jacobsen ID, Miramon P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D (2012) Candida albicans scavenges host zinc via pra1 during endothelial invasion. PLoS Pathog 8:e1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66:487–511 [DOI] [PubMed] [Google Scholar]
- Deng CP, Wang Y, Huang FF, Lu SJ, Zhao LM, Ma XY, Kai GY (2020) SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J Integr Plant Biol 62:1688–1702 [DOI] [PubMed] [Google Scholar]
- Deng S, Wang CY, Zhang X, Wang Q, Lin L (2015) VdNUC-2, the Key Regulator of Phosphate Responsive Signaling Pathway, Is Required for Verticillium dahliae Infection. PLoS One 10:e0145190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11:539–548 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16:755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G, van der Linde K, Assmann D, Schwammbach D, Hof A, Mohanty A, Jackson D, Kahmann R (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog 5:e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM. et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581 [DOI] [PubMed] [Google Scholar]
- Fang A, Han Y, Zhang N, Zhang M, Liu L, Li S, Lu F, Sun W (2016) Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens. Mol Plant Microbe Interact 29:405–416 [DOI] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GCM, Joosten MHAJ, Thomma BPHJ (2011) Interfamily transfer of tomato Ve1 mediates Verticillium Resistance in Arabidopsis. Plant Physiol 156:2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CD, Nazar RN, Robb J, Liu CM, Thomma BP (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol 150:320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furniss JJ, Grey H, Wang Z, Nomoto M, Jackson L, Tada Y, Spoel SH (2018) Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity. PLoS Pathog 14:e1007447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao CY, Sun PW, Wang W, Tang DZ (2021) ArabidopsisE3 ligase KEG associates with and ubiquitinates MKK4 and MKK5 to regulate plant immunity. J Integr Plant Biol 63:327–339 [DOI] [PubMed] [Google Scholar]
- Gao F, Zhang BS, Zhao JH, Huang JF, Jia PS, Wang S, Zhang J, Zhou JM, Guo HS (2019) Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat Plants 5:1167–1176 [DOI] [PubMed] [Google Scholar]
- Gao XQ, Li FJ, Li MY, Kianinejad AS, Dever JK, Wheeler TA, Li ZH, He P, Shan LB (2013) Cotton GhBAK1 mediates verticillium wilt resistance and cell death. J Integr Plant Biol 55:586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakls V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19:423–429 [DOI] [PubMed] [Google Scholar]
- Goehre V, Spallek T, Haeweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18:1824–1832 [DOI] [PubMed] [Google Scholar]
- Han LB, Li YB, Wang FX, Wang WY, Liu J, Wu JH, Zhong NQ, Wu SJ, Jiao GL, Wang HY. et al. (2019) The cotton apoplastic protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen Verticillium dahliae. Plant Cell 31:520–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Yamaguchi K, Sakamoto K, Yoshimura S, Inoue K, Tsuge S, Kojima C, Kawasaki T (2014) Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat Commun 5:5430. [DOI] [PubMed] [Google Scholar]
- Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE (2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311:222–226 [DOI] [PubMed] [Google Scholar]
- Jiao L, Zhang Y, Lu J (2017) Overexpression of a stress-responsive U-box protein gene VaPUB affects the accumulation of resistance related proteins in Vitis vinifera ‘Thompson Seedless’. Plant Physiol Biochem 112:53–63 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2014) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- Jung C, Zhao P, Seo JS, Mitsuda N, Deng S, Chua NH (2015) PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 27:2016–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV (2009) Diversity, pathogenicity, and management of verticillium species. Annu Rev Phytopathol 47:39–62 [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, Thomma BP, Chen Z, Henrissat B, Lee YH, Park J. et al. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog 7:e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, Gong Z (2015) Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6:8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosarev P, Mayer KF, Hardtke C (2002) Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol 3:RESEARCH0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16:3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhou H, Zhang Y, Li Z, Yang Y, Guo Y (2020) The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev Cell 55:367–380e366 [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Y, Gao Z, Xu X, Wang Y, Lin Y, Ye P, Huang T (2021a) Plant U-box E3 ligases PUB25 and PUB26 control organ growth in Arabidopsis. New Phytol 229:403–413 [DOI] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y. et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15:329–338 [DOI] [PubMed] [Google Scholar]
- Li Q, Wang B, Yu JP, Dou DL (2021b) Pathogen-informed breeding for crop disease resistance. J Integr Plant Biol 63:305–311 [DOI] [PubMed] [Google Scholar]
- Liao D, Cao Y, Sun X, Espinoza C, Nguyen CT, Liang Y, Stacey G (2017) Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol 214:1646–1656 [DOI] [PubMed] [Google Scholar]
- Lin B, Zhuo K, Chen S, Hu L, Sun L, Wang X, Zhang LH, Liao J (2016) A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol 209:1159–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8:689–708 [DOI] [PubMed] [Google Scholar]
- Liu T, Song T, Zhang X, Yuan H, Su L, Li W, Xu J, Liu S, Chen L, Chen T. et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun 5:4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu J, Ning Y, Ding B, Wang X, Wang Z, Wang GL (2013) Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol Plant 6:605–620 [DOI] [PubMed] [Google Scholar]
- Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R (2015) Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66:513–545 [DOI] [PubMed] [Google Scholar]
- Love M, Anders S, Huber W (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332:1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhaes AMED, Ortiz-Morea FA, He P, Shan LB (2021) Plant plasma membrane-resident receptors: surveillance for infections and coordination for growth and development. J Integr Plant Biol 63:79–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, Pouzet C, Jauneau A, Roby D, Rivas S (2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat Commun 4:1476. [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50:1950–1964 [DOI] [PubMed] [Google Scholar]
- Miao S, Liu J, Guo J, Li JF (2020) Engineering plants to secrete affinity-tagged pathogen elicitors for deciphering immune receptor complex or inducing enhanced immunity. J Integr Plant Biol 62:761–776 [DOI] [PubMed] [Google Scholar]
- Modrof J, Becker S, Muhlberger E (2003) Ebola virus transcription activator VP30 is a zinc-binding protein. J Virol 77:3334–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Jones JDG (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592:110–115 [DOI] [PubMed] [Google Scholar]
- Njoroge SMC, Kabir Z, Martin FN, Koike ST, Subbarao KV (2009) Comparison of crop rotation for Verticillium wilt management and effect on Pythium species in conventional and organic strawberry production. Plant Dis 93:519–527 [DOI] [PubMed] [Google Scholar]
- Nomura K, DebRoy S, Lee YH, Pumplin N, Jones J, He SY (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313:220–223 [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee YN, Imboden LA, Chang JH, He SY (2011) Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc Natl Acad Sci U S A 108:10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V, Mucyn TS, Gimenez-lbanez S, Chapman HC, Gutierrez JR, Balmuth AL, Jones AME, Rathjen JP (2009) Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324:784–787 [DOI] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639–648 [DOI] [PubMed] [Google Scholar]
- Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JI, Liu HY, van Damme M. et al. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21:2928–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL (2003) Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol 10:250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT (1992) Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem 61:1053–1095 [DOI] [PubMed] [Google Scholar]
- Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M. et al. (2012) The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24:4748–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JL, Bao ZL, Ren HY, Wang JS, Dong HS (2004) Expression of harpin(xoo) in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phytopathology 94:1048–1055 [DOI] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40:979–995 [DOI] [PubMed] [Google Scholar]
- Pringa E, Martinez-Noel G, Muller U, Harbers K (2001) Interaction of the ring finger-related U-box motif of a nuclear dot protein with ubiquitin-conjugating enzymes. J Biol Chem 276:19617–19623 [DOI] [PubMed] [Google Scholar]
- Qi J, Song CP, Wang B, Zhou J, Kangasjarvi J, Zhu JK, Gong Z (2018) Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol 60:805–826 [DOI] [PubMed] [Google Scholar]
- Qin J, Wang K, Sun L, Xing H, Wang S, Li L, Chen S, Guo HS, Zhang J (2018) The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. Elife 7:e34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Liu SM, Zhang ZN, Sun LQ, He X, Lindsey K, Zhu LF, Zhang XL (2019) GhCyP3 improves the resistance of cotton to Verticillium dahliae by inhibiting the E3 ubiquitin ligase activity of GhPUB17. Plant Mol Biol 99:379–393 [DOI] [PubMed] [Google Scholar]
- Rajamuthiah R, Mylonakis E (2014) Effector triggered immunity. Virulence 5:697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput NA, Zhang M, Shen D, Liu T, Zhang Q, Ru Y, Sun P, Dou D (2015) Overexpression of a phytophthora cytoplasmic CRN effector confers resistance to disease, salinity and drought in Nicotiana benthamiana. Plant Cell Physiol 56:2423–2435 [DOI] [PubMed] [Google Scholar]
- Riechmann J, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe O, Samaha R (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110 [DOI] [PubMed] [Google Scholar]
- Roger WP, Li SF (1995) Isolation of two novel myb-like genes from Arabidopsis and studies on the DNA-binding properties of their products. Plant J 8:933–972 [DOI] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng LR, Brady JJ, Abramovitch RB, Xiao FM, Martin GB (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448:370–U313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarde SJ, Bouwmeester K, Venegas-Molina J, David A, Boland W, Dicke M (2019) Involvement of sweet pepper CaLOX2 in jasmonate-dependent induced defence against Western flower thrips. J Integr Plant Biol 61:1085–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenke D, Bottcher C, Scheel D (2011) Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ 34:1849–1864 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park CM (2010) MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol 186:471–483 [DOI] [PubMed] [Google Scholar]
- Sheikh AH, Eschen-Lippold L, Pecher P, Hoehenwarter W, Sinha AK, Scheel D, Lee J (2016) Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in Arabidopsis thaliana. Front Plant Sci 7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Yekondi S, Chen PW, Tsai CH, Yu CW, Wu K, Zimmerli L (2014) Environmental history modulates Arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-dependent manner. Plant Cell 26:2676–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Catanzariti AM, DeBoer K, Petre B, Gardiner DM, Singh KB, Dodds PN, Taylor JM (2017) LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep 7:44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Zarsky V, McDowell JM, Shirasu K, Trujillo M (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24:4703–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL (2014) The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front Plant Sci 5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Mitrousia GK, de Wit PJ, Fitt BD (2014) Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci 19:491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342:624–628 [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res :W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M, Kotur T, Liang W, Vogelmann K, Kleine T, Leister D, Brieske C, Yang S, Ludke D, Wiermer M. et al. (2017) E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol 215:1516–1532 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4:645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustun S, Sheikh A, Gimenez-Ibanez S, Jones A, Ntoukakis V, Bornke F (2016) The proteasome acts as a hub for plant immunity and is targeted by pseudomonas type III effectors. Plant Physiol 172:1941–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallad GE, Subbarao KV (2008) Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae. Phytopathology 98:871–885 [DOI] [PubMed] [Google Scholar]
- Veronese P, Narasimhan ML, Stevenson RA, Zhu JK, Weller SC, Subbarao KV, Bressan RA (2003) Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J 35:574–587 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang S, Stacey G (2004) Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol 5:125–135 [DOI] [PubMed] [Google Scholar]
- Wang C, Wang G, Zhang C, Zhu PK, Dai HL, Yu N, He ZH, Xu L, Wang ET (2017a) OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol Plant 10:619–633 [DOI] [PubMed] [Google Scholar]
- Wang H, Sun S, Ge W, Zhao L, Hou B, Wang K, Lyu Z, Chen L, Xu S, Kong L (2020a) Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 668:eaba5435. [DOI] [PubMed] [Google Scholar]
- Wang J, Dhroso A, Liu X, Baum TJ, Hussey RS, Davis EL, Wang X, Korkin D, Mitchum MG (2021) Phytonematode peptide effectors exploit a host post-translational trafficking mechanism to the ER using a novel translocation signal. New Phytol 229:563–574 [DOI] [PubMed] [Google Scholar]
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L. et al. (2018a) A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell 69:493–504.e496 [DOI] [PubMed] [Google Scholar]
- Wang K, He J, Zhao Y, Wu T, Zhou X, Ding Y, Kong L, Wang X, Wang Y, Li J. et al. (2018b) EAR1 negatively regulates ABA signaling by enhancing 2C protein phosphatase activity. Plant Cell 30:815–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Feng B, Zhou JM, Tang D (2020b) Plant immune signaling: advancing on two frontiers. J Integr Plant Biol 62:2–24 [DOI] [PubMed] [Google Scholar]
- Wang X, Ding Y, Li Z, Shi Y, Wang J, Hua J, Gong Z, Zhou JM, Yang S (2019) PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev Cell 51:222–235.e225 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu Y, Yu B, Yin Z, Xia Y (2017b) EXTRA-LARGE G PROTEINs interact with E3 ligases PUB4 and PUB2 and function in cytokinin and developmental processes. Plant Physiol 173:1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Johnson DA (2016) Verticillium dahliae infects, alters plant biomass, and produces inoculum on rotation crops. Phytopathology 106:602–613 [DOI] [PubMed] [Google Scholar]
- Xiao ZD, Yang C, Liu CL, Yang LM, Yang SH, Zhou J, Li FQ, Jiang LW, Xiao S, Gao CJ. et al. (2020) SINAT E3 ligases regulate the stability of the ESCRT component FREE1 in response to iron deficiency in plants. J Integr Plant Biol 62:1399–1417 [DOI] [PubMed] [Google Scholar]
- Xue J, Gong BQ, Yao XR, Huang XJ, Li JF (2020) BAK1-mediated phosphorylation of canonical G protein alpha during flagellin signaling in Arabidopsis. J Integr Plant Biol 62:690–701 [DOI] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T, Nakagami H, Mine A, Ishikawa K, Fujiwara M, Narusaka M, Narusaka Y, Ichimura K. et al. (2016) The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J 35:2468–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JB, Yao RF, Chen L, Li SH, Gu M, Nan FJ, Xie DX (2018) Dynamic perception of jasmonates by the F-box protein COI1. Mol Plant 11:1237–1247 [DOI] [PubMed] [Google Scholar]
- Yang LY, Zhang Y, Guan RX, Li S, Xu XW, Zhang SQ, Xu J (2020) Co-regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. J Integr Plant Biol 62:1780–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li C, Cai Z, Hu Y, Nolan T, Yu F, Yin Y, Xie Q, Tang G, Wang X (2017) SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev Cell 41:47–58.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q. et al. (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60:107–124 [DOI] [PubMed] [Google Scholar]
- Ye Q, Wang H, Su T, Wu WH, Chen YF (2018) The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low-Pi stress in Arabidopsis. Plant Cell 30:1062–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin ZY, Wang N, Pi L, Li L, Duan WW, Wang XD, Dou DL (2021) Nicotiana benthamiana LRR-RLP NbEIX2 mediates the perception of an EIX-like protein from Verticillium dahliae. J Integr Plant Biol 63:949–960 [DOI] [PubMed] [Google Scholar]
- You Q, Zhai K, Yang D, Yang W, Wu J, Liu J, Pan W, Wang J, Zhu X, Jian Y. et al. (2016) An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20:758–769 [DOI] [PubMed] [Google Scholar]
- Yu CJ, Dou K, Wang SQ, Wu Q, Ni M, Zhang TL, Lu ZX, Tang J, Chen J (2020) Elicitor hydrophobin Hyd1 interacts with Ubiquilin1-like to induce maize systemic resistance. J Integr Plant Biol 62:509–526 [DOI] [PubMed] [Google Scholar]
- Yu F, Wu Y, Xie Q (2016) Ubiquitin-proteasome system in ABA signaling: from perception to action. Mol Plant 9:21–33 [DOI] [PubMed] [Google Scholar]
- Yuan M, Ngou BPM, Ding P, Xin XF (2021a) PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62:102030. [DOI] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, XF Xin (2021b) Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16:2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Zhao YL, Jin Y, Zhang T, Guo HS (2014) Colonization process of Arabidopsis thaliana roots by a green fluorescent protein-tagged isolate of Verticillium dahliae. Protein Cell 5:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JG, Liu DR, Wang P, Ma XY, Lin WW, Cheng SX, Mishev K, Lu DP, Kumar R, Vanhoutte I. et al. (2018) Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc Natl Acad Sci U S A 115:E1906–E1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Zhang Y (2020) Plant immunity: danger perception and signaling. Cell 181:978–989 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Liu J, Wang JY, Chen SF, Chen LJ, Wang JF, Wang HB, Liu B (2020a) The juxtamembrane domains of Arabidopsis CERK1, BAK1, and FLS2 play a conserved role in chitin-induced signaling. J Integr Plant Biol 62:556–562 [DOI] [PubMed] [Google Scholar]
- Zhou SX, Zhu Y, Wang LF, Zheng YP, Chen JF, Li TT, Yang XM, Wang H, Li XP, Ma XC. et al. (2020b) Osa-miR1873 fine-tunes rice immunity against Magnaporthe oryzae and yield traits. J Integr Plant Biol 62:1213–1226 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.