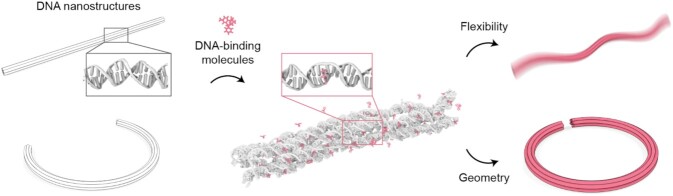
Abstract
Recent advances in DNA nanotechnology led the fabrication and utilization of various DNA assemblies, but the development of a method to control their global shapes and mechanical flexibilities with high efficiency and repeatability is one of the remaining challenges for the realization of the molecular machines with on-demand functionalities. DNA-binding molecules with intercalation and groove binding modes are known to induce the perturbation on the geometrical and mechanical characteristics of DNA at the strand level, which might be effective in structured DNA assemblies as well. Here, we demonstrate that the chemo-mechanical response of DNA strands with binding ligands can change the global shape and stiffness of DNA origami nanostructures, thereby enabling the systematic modulation of them by selecting a proper ligand and its concentration. Multiple DNA-binding drugs and fluorophores were applied to straight and curved DNA origami bundles, which demonstrated a fast, recoverable, and controllable alteration of the bending persistence length and the radius of curvature of DNA nanostructures. This chemo-mechanical modulation of DNA nanostructures would provide a powerful tool for reconfigurable and dynamic actuation of DNA machineries.
INTRODUCTION
The understanding and application of geometrical and mechanical responses of DNA with binding ligands have been one of the major challenges for decades (1,2). Small molecules non-covalently interacting with DNA have multiple binding mechanisms including intercalation and groove binding (3), and have been used for pharmaceutical (4), biological (5) and fluorescence applications (6). The underlying binding mechanisms of such ligands and corresponding nanomechanical characteristics of DNA have been widely studied by tweezer-based force spectroscopy (7–14) and atomic force microscope (AFM) imaging (15–17). However, most studies so far mainly focused on the response of an unstructured long DNA strand (7–13,15), while the remarkable recent advance of structural DNA nanotechnology enabled the utilization of structured DNA assemblies (18). A number of DNA nanostructures with planar (19) to 3D (20) shapes have been constructed and used as intracellular delivery carriers by containing DNA-binding drugs (21–23). Chemically-activated dynamic and reconfigurable mechanisms have also been utilized as nanomechanical components (24–26), molecular sensors (27) and connectors for higher-order assemblies (28–30). While chemo-mechanical actuation can provide a fast (up to seconds at strand level (2)) and repeatable geometric response to DNA assemblies, it is difficult to select a proper DNA-binding ligand and predict its working range for a targeted function of DNA nanostructures.
In this study, we used three intercalators, ethidium bromide (EtBr), doxorubicin (DOX) and dimeric cyanine dye oxazole yellow (YOYO-1), and two groove binders, 4′,6-diamidino-2-phenylindole (DAPI) and bisbenzimide H 33258 (H33258), to investigate how these DNA-binding molecules affect the mechanical and geometrical characteristics of structured DNA assemblies. Increased mechanical flexibility of the bundles in bending was commonly observed for all binders, but the softening effect was different depending on their intrinsic binding modes. EtBr and DAPI showed a substantial recovery of modulated flexibility by a simple buffer exchange process, thereby enabling repeatable chemo-mechanical actuation. Also, their ability to control the structural deformation was demonstrated for bent structures. Furthermore, we found that the bending stiffness of DNA origami bundles could be systematically varied by using ethidium intercalation together with engineered defects (31) (via short single-stranded gaps). Computational studies revealed a potential mechanism of how ethidium intercalation make the structured DNA bundles flexible.
MATERIALS AND METHODS
DNA materials and reagents
M13mp18 ssDNA (7249-nt-long) was purchased from Guild BioSciences (FOUNDATION M13mp18). Staple DNA oligonucleotides were provided from Bioneer Corporation (www.bioneer.co.kr). The yield and molecular weight of all staple strands were verified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) from the provider. EtBr (0.5 mg/ml), Doxorubicin hydrochloride (solid), DAPI (1 mg/ml) and H33258 (1 mg/ml) were purchased from Sigma-Aldrich, and YOYO-1 iodide (1 mM) was purchased from ThermoFisher Scientific. Other reagents including DI water, Tris-Aacetate-EDTA (TAE) and magnesium chloride (MgCl2) solution were purchased from Sigma-Aldrich.
Self-assembly and purification
100 ul of a folding mixture containing 20 nM of scaffold DNA, 100 nM of each staple strand, 1× TAE buffer (40 mM Tris-acetate and 1 mM EDTA) and 20 mM of MgCl2 was prepared. The mixture was subjected to the following temperature gradient by using a thermocycler (T100, Bio-Rad): heated up to 80°C at 1°C/s; 80°C to 65°C in one hour (2 min per −0.5°C); 65°C to 25°C in 40 h (30 min per −0.5°C); cooled down and hold at 4°C. Excessive staple strands were removed by centrifugal filtration (32), using Amicon Ultra Centrifugal Filter Units with 50 kDa cut-off filters (UFC505096, Merck KGaA). The concentration of the purified structures was set to 10 nM using a UV-VIS spectrophotometer (NanoDrop One, ThermoFisher Scientific).
Incubation of the samples and AFM measurement
DNA origami structures with 10 nM concentration containing folding buffer of 1× TAE and 20 mM MgCl2 (FOB20) were mixed with 1:19 ratio with DNA-binding molecules dissolved in FOB20 buffer to make 0.5 nM structure concentration with target binder concentration.
After 10 minu of incubation at room temperature, the prepared sample was deposited on a freshly cleaved mica substrate (highest grade V1 AFM Mica, Ted-Pella Inc.), and subsequently incubated for 5 min. The substrate was washed with DI water by three times and gently dried by N2 gun (<0.1 kgf/cm2) immediately. AFM images were taken by NX10 (Park Systems) by using a PPP-NCHR probe having a spring constant of 42 N/m (Nanosensors). The non-contact mode was used to measure typically 5 μm × 5 μm of the sample area in 1024 × 1024 pixel resolution by using SmartScan software. All measured images were flattened with linear and quadratic order using XEI 4.1.0 program (Park Systems) prior to further analysis.
Analysis of AFM images
Calculation of contour length and persistence length of DNA origami monomers from AFM images were done by custom scripts using MATLAB R2017b software (MathWorks Inc.). Refer to the previous report for a more detailed procedure (31). Briefly, collected individual monomer structures were converted into binary images to be thinned and skeletonized to obtain their contours. A parametric spline was used to fit the contour of each individual structure. The persistence length was measured with characteristic points of the fitting spline from every well-folded structure using a modified version of the open-source software tool Easyworm (33). Using the WLC model, the mean-square end-to-end distance (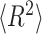 ) in two dimensions as a function of the distance along the contour (
) in two dimensions as a function of the distance along the contour (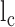 ) can be expressed as
) can be expressed as
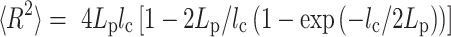 |
where 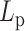 is the persistence length. Typically, the correlation coefficient of the data fitting is above 0.99 for each case. The standard deviation of the persistence length was calculated by a bootstrap method with subsets of randomly chosen contours with the same number of collected monomers with replacement and 1000 times of the repeating process.
is the persistence length. Typically, the correlation coefficient of the data fitting is above 0.99 for each case. The standard deviation of the persistence length was calculated by a bootstrap method with subsets of randomly chosen contours with the same number of collected monomers with replacement and 1000 times of the repeating process.
Molecular dynamics (MD) simulation
The starting atomic structure of the 6HB design was generated using caDNAno (34) and SNUPI (35). Each atomic structure was explicitly solvated using the TIP3P water model (36) with a distance >15 Å from the structure and boundary. It results in approximately 100 Å × 100 Å × 320 Å of cubic water box and neutralized to an ion concentration of 20 mM MgCl2. MD simulation was then performed using NAMD (37) with the CHARMM36 force field (38), periodic boundary conditions, the integration time step of 2 fs, and short-range electrostatic potentials with 12 Å cut-off. The long-range electrostatic interactions were computed using the Particle-Mesh-Ewald (PME) scheme (39) with the grid size of 1 Å. The potential energy of each system was minimized using the conjugate gradient method. Each structure was simulated >300 ns under the isobaric-isothermal (NPT) ensemble, and trajectories of the final 100 ns were used for further analysis. For the process of cross-sectional analysis and principal component analysis, refer to our previous report (31).
Finite element (FE) model and analysis
To predict the equilibrium shape and bending stiffness of DNA origami bundles, FE analysis was performed based on the modified CanDo (40) model. In brief, a dsDNA helix was modeled as beam elements having the regular B-form DNA geometry (diameter of 2.25 nm, axial rise of 0.34 nm and helicity of 10.5 bp/turn) and mechanical properties (stretch modulus of 1100 pN, bending rigidity of 230 pN nm2, and torsional rigidity of 460 pN nm2). The effect of different sequences, nicks, and gaps was not considered. Inter-helix crossovers were modeled by a beam element that connects two nodes that belong to two adjacent helices, respectively. To consider the effect of flexibility of crossover, their mechanical stiffnesses were defined by multiplying a scale factor to those of DNA duplex instead of using rigid beams (41). Normal mode analysis was performed at the equilibrated configuration to compute the bending stiffness of DNA origami bundles. Refer to the previous report for a detailed procedure (31).
RESULTS AND DISCUSSION
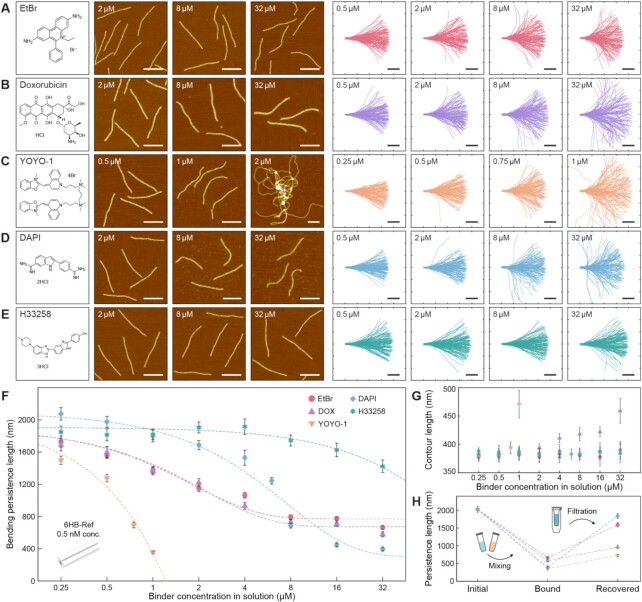
Molecules interacting with double-stranded DNA (dsDNA) can change the geometric and mechanical characteristics of structured DNA assemblies, which usually have a complex strand pathway and structural motifs (Figure 1). For example, scaffolded DNA origami, one of the most widely used techniques for folding DNA into a nanostructure, typically uses an M13mp18 circular single-stranded DNA (ssDNA) consisting of 7249 nucleotides (nt) as a scaffold and up to 200 short oligonucleotides (staples) which have complementary sequences to the scaffold (19,40). There exist multiple Holliday junctions (crossovers) and backbone breaks (nicks) in order to arrange DNA strands in a given pathway with inter-helical crosslinks to form a structure. Various curved and twisted structures can be constructed through designing lattice-packing rules, crossover intervals, and cross-sectional shapes (20,42,43). In this study, a simple six-helix-bundle (6HB) designed on a honeycomb lattice was selected to investigate the chemo-mechanical response to various DNA-binding molecules. Its geometric and mechanical characteristics have been well studied (Supplementary Figure S1) (31,44,45), and its slender profile is appropriate to measure the equilibrated shape and calculate the bending flexibility (31). First of all, we tested five representative DNA-binding molecules having different binding mechanisms to study the mechanical response of the 6HB reference (6HB-Ref) design (Figure 2). Assembled 6HB structures were diluted to 0.5 nM with folding buffer containing 20 mM MgCl2 and targeted binder concentration (Materials and Methods). Images of individual 6HB monomers incubated with different binders and concentrations were collected using AFM, and their mechanical flexibilities were characterized by measuring the bending persistence length from the extracted contours (Figure 2A–FSupplementary Figures S2–S44, and Supplementary Tables S1 and S2). Kurtosis values for representative 15 cases were converged to theoretical value of 3 implying that the deposited structures were equilibrated in 2D (Supplementary Figure S45) (33,46).
Figure 1.
Schematic illustration of the chemo-mechanical response of a DNA origami nanostructure with DNA-binding ligands. DNA-binding molecules such as ethidium can induce the deformation and the softening of DNA duplexes, leading to the changes in the flexibility and the geometry of an assembled structure. Gray and pink boxes indicate the equilibrated dsDNA without and with ethidium intercalation, respectively.
Figure 2.
Mechanical response of DNA origami bundles with different DNA binders. Chemical structure of binders, representative AFM images, and 120 aligned monomer contours of 6HB-Ref bundles incubated at the different concentrations with (A) EtBr, (B) DOX, (C) YOYO-1, (D) DAPI and (E) H33258. Refer to Supplementary Figures S2–S44 and Supplementary Table S1 for the results of the entire concentration range. Scale bars in the AFM images are 200 nm. Scale bars and ticks in the monomer contour graphs are 100 nm. (F) Calculated bending persistence lengths of the 6HB structures with different binders and concentrations. The molecular concentration of 6HB monomers was maintained at 0.5 nM for all cases. The dotted lines indicate the exponential fitting curves of experimental data for each case. Error bars indicate the standard deviation. (G) Measured monomer contour lengths of the 6HB-Ref bundles with different binders and concentrations. Error bars indicate the standard deviation. (H) Recovery test using buffer exchange method. Refer to Materials and Methods for details.
In order to confirm that the samples in our measurement condition are fully equilibrated in 2D substrate, we calculated the kurtosis, which is the ratio between the fourth moment and the square of the second moment for the angle distribution (46). Kurtosis around the value of 3 indicates that the angle distribution is Gaussian, therefore the structure is fully equilibrated in 2D. We performed this analysis to 15 cases with different DNA-binders and concentrations, and concluded that the kurtosis values were generally around 3 within a contour length range.
EtBr is a common DNA-binding fluorophore and known to increase the contour length, unwind the twist angle and decrease the persistence length of dsDNA by intercalation (7,10,47,48). We observed a gradual reduction in the bending stiffness of the 6HB-Ref design throughout the experimental ranges, and the maximum stiffness reduction was calculated as 67.3% in 32 μM concentration (Figure 2A and F, and Supplementary Figures S2–S11). The persistence length of DOX-intercalated 6HB-Ref showed a very similar trend with EtBr-intercalated cases (Figure 2B and F, and Supplementary Figures S12–S20). DOX is a type of anthracycline antibiotic and anticancer drugs, and known as an intercalating-dominant DNA binder. Geometrical perturbation by its intercalation is similar to that with EtBr (47), even though a complex force-dependent behavior regarding the persistence length was reported (49).
Besides the mono-intercalators, green-fluorescent DNA staining dye YOYO-1 was chosen as an example of bis-intercalating molecules (6). Due to the characteristics of bis-intercalation, its effect on elongation and unwinding of DNA are known to be stronger than that of mono-intercalators although contradictory results were reported regarding its effect on the persistence length (13,50). We observed a more drastic reduction in the bending stiffness up to 82.4% in only 1 μM YOYO-1 concentration. This is possibly due to the higher binding affinity of YOYO-1 than EtBr, and more significant extension and unwinding of dsDNA strands induced by bis-intercalation (Figure 2C and F, and Supplementary Figures S21–S25) (2,13). Highly stacked configurations by forming long chains were appeared in 2 μM concentration. Intermolecular crosslinking induced by YOYO-1 might be the reason for polymerization since the ratio of dye to DNA bp (∼1:1.8 in 2 μM concentration) largely exceeds the known maximum value (1:4) (6,51).
Lastly, the effect of minor groove binders was investigated. DAPI and H33258 are reported to preferentially interact with AT-rich regions, but the intercalative binding mode is also known to exist with GC-rich or mixed sequences (52–56). In general, minor groove binding mode of ligands is known to induce no significant geometric perturbation, but additional intercalating mode at different sites can influence the changes in geometrical and mechanical characteristics of dsDNA (3). To investigate the response by such effects at the DNA structure level, DAPI and H33258 were treated to the 6HB-Ref design. The results showed that the persistence length of DAPI-bound 6HB decreased slowly until 4 μM concentration, and then rapidly dropped at higher concentration levels (Figure 2D and F, and Supplementary Figures S26–S35). Unlike the intercalator-incubated samples, some of the structures incubated with DAPI at a higher concentration (>8 μM) have locally curved regions, which contribute largely to a drastic decrease of the bending persistence length (Figure 2D and Supplementary Figure S26). Meanwhile, H33258-bound 6HB maintained a similar level of persistence length until 8 μM, and then was getting slowly softer at higher concentrations (Figures 2E, F, and Supplementary Figures S36–S44).
Interestingly, EtBr, DAPI and H33258 ligands have almost no effect on the contour length of the bundle across the tested concentration range, whereas DOX and YOYO-1 showed its increase up to 19% and 22% in 32 and 1 μM, respectively (Figure 2G and Supplementary Table S1). Though it has been known that mono-intercalator EtBr tends to increase the contour length of dsDNA (10,47), that of our 6HB structure was not significantly changed by it. More confined configuration of dsDNA in the structured assembly might cause this difference.
The reversibility of chemo-mechanical modulation was tested for four DNA-binding molecules (Figure 2H and Supplementary Figure S46). Binder-incubated sample solutions were exchanged to pure folding buffer by three times during the centrifugal filtration (Materials and methods) (32). The recovery efficiency defined as the ratio between the average persistence lengths of initial and recovered 6HB monomers was measured. EtBr and DAPI showed a relatively high recovery rate (78.6% and 91.1%, respectively), whereas DOX and YOYO-1 showed an incomplete, partial recovery (36.1% and 47.5%, respectively). The recovery rate might be closely related to the binding affinity with DNA. It was reported that DOX-intercalated DNA origami structures could contain DOX up to few hours (21), and YOYO-1-DNA complex could maintain stable binding (13,51). One thing to note here is that the binding affinity and corresponding mechanical characteristics of DNA with binding ligands are reported to be influenced by surrounding buffer conditions as well (13). In contrast to many single-molecule experiments that use buffers with monovalent cations such as sodium chloride and sodium phosphate, divalent cations such as magnesium are typically used in DNA origami within 10 to 20 mM concentration range (40). Therefore, a careful selection of the type and concentration of binding ligands might be necessary depending on the purpose and working condition of DNA nanostructures.
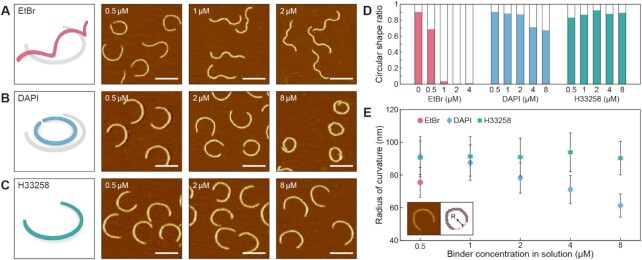
In addition to the mechanical flexibility, the effect of DNA-binders on the geometry of structured DNA assemblies was investigated by using a curved 6HB (6HB-Arc) design (Figure 3). The semi-circular geometry of the 6HB-Arc structure was designed by insertions and deletions of bps to the straight 6HB-Ref design (Supplementary Table S3) (43). When incubated with EtBr, structures with a smaller radius of curvature were observed in 0.5 μM concentration (Figure 3A and E). As the EtBr concentration increases (>1 μM), however, most structures were reconfigured into non-circular helical coils. It is because the gradual unwinding of constituting dsDNA strands due to the ethidium intercalation induces a cumulative out-of-plane rotation of the entire structure (Figure 3A and D, and Supplementary Figure S47). On the other hand, 6HB-Arc structures incubated with groove binders showed quite distinctive behaviors. The radius of curvature of DAPI-incubated 6HB-Arc gradually reduced up to 32% in 8 μM concentration without noticeable out-of-plane deformation, resulting in the formation of closed circles as shown in AFM images (Figure 3B and D–E, and Supplementary Figure S48). Similar to the decrement in the bending persistence length of DAPI-incubated straight 6HB structures, we attribute this behavior to the additional intercalating mode of DAPI that appears in higher molecular concentrations as well (57). Increased portion of the non-circular structures observed in 32 μM concentration supports this hypothesis that an increased amount of intercalation might have a negative impact on the structure to maintain its planar geometry (Supplementary Figure S48). The decreased radius of curvature of DAPI-incubated 6HB-Arc structures was successfully reverted by buffer exchange, though some of the highly distorted monomers seemed to fail to recover their original shapes (Supplementary Figure S49). On the contrary, H33258 did not induce the change in the radius of curvature until 8 μM concentration, as similar to its effect on the bending persistence length of the straight 6HB-Ref structure (Figures 3C–E and Supplementary Figure S50). But a large amount of non-circular structures was also observed in 32 μM concentration (Supplementary Figure S50).
Figure 3.
Geometrical response of curved DNA origami structures with different DNA binders. Schematic illustration of the initial (gray) and deformed (colored) shapes and representative AFM images of 6HB-Arc structures incubated at the different concentrations with (A) EtBr, (B) DAPI and (C) H33258. Scale bars in the AFM images are 200 nm. (D) The ratio of circular shape structures with different binder types and concentrations. (E) Measured radii of curvature of the 6HB-Arc structures with DAPI and H33258 binders. The molecular concentration of 6HB-Arc monomers was maintained at 0.5 nM for all cases. Error bars indicate the standard deviation.
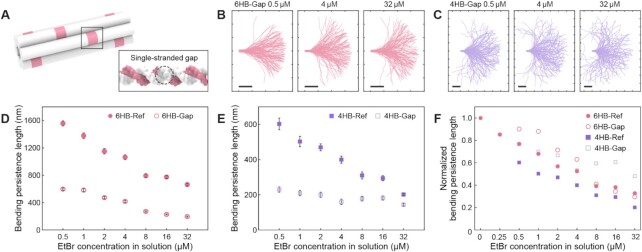
Next, we explored the possibility of modulating the mechanical stiffness of these bundles by both chemical (via binding molecules) and physical (via engineered defects (31)) modifications (Figure 4, Supplementary Figures S1 and S51–S74, and Supplementary Tables S2 and S4). In addition to the 6HB-Ref structure, four-helix-bundle designed on a square lattice (4HB-Ref) was tested (Supplementary Figure S1). Flexible versions of these structures achieved using multiple 5-nt-long single-stranded gaps (or engineered defects (31)) were included in the test (indicated as 6HB-Gap and 4HB-Gap) (Figure 4A). As we increased the concentration of EtBr, a similar trend of stiffness reduction was universally observed for both regular and defect-engineered structures, indicating that chemical and physical modulations can be used simultaneously (Figures 4B–E). Normalized bending stiffness, defined as the ratio of the persistence length to that of the unmodified structure, showed a similar rate of stiffness reduction for all cases (Figure 4F). Combined with engineered defects and EtBr intercalation, the maximum amounts of reduction in the bending stiffness were 90.8%, and 85.6% for 6HB and 4HB, respectively (Supplementary Table S1). These values are above the range of maximum softening effect that could be achieved using only engineering defects (67.5% and 70% in 6HB and 4HB, respectively) (31).
Figure 4.
Systematic modulation of bending persistence length using EtBr with different cross-sections and defect-engineered bundles. (A) Schematic illustration of defect-engineering. For 6HB-Gap and 4HB-Gap design, 5-nt-long ssDNA gaps were inserted in every nick position. Refer to Supplementary Figure S1 for details. (B) Representative 120 aligned monomer contours of 6HB-Gap bundles with different EtBr concentration. Ticks and scale bars: 100 nm. (C) Representative 120 aligned monomer contours of 4HB-Gap bundles with different EtBr concentration. Ticks and scale bars: 100 nm. (D) Calculated bending persistence lengths of 6HB-Ref and 6HB-Gap structures with different EtBr concentration. Error bars indicate the standard deviation. Refer to Supplementary Figures S51–S58 for detailed results. (E) Calculated bending persistence lengths of 4HB-Ref and 4HB-Gap structures with different EtBr concentration. Error bars indicate the standard deviation. Refer to Supplementary Figures S59–S74 for detailed results. (F) Normalized bending persistence lengths of all cases showing the decreasing behavior of bending persistence length as the increase of EtBr concentration.
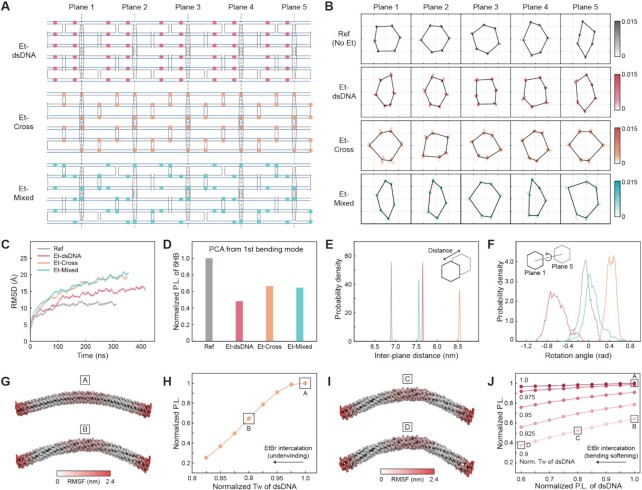
To understand the structural effect of DNA-binding molecules on structured DNA assemblies, we performed the MD simulation for 105-bp-long 6HB structure (Figures 5A–F and Supplementary Figures S75–S77). Considering the uncertainty of ethidium-binding positions on the DNA origami bundle, 60 ethidium molecules were initially distributed into the bundle structure in three different ways; all ethidiums to dsDNA region (Et-dsDNA), all ethidiums to crossover region (Et-Cross), and 30 ethidiums to dsDNA and 30 ethidiums to crossovers (Et-Mixed) (Figure 5A). According to the previous report (24), this corresponds to approximately 3 μM EtBr concentration when 0.5 nM DNA bundles exist in solution. During the equilibrium process, nine, four, and five ethidium molecules fell out from their initial positions in Et-dsDNA, Et-Cross, and Et-Mixed cases, respectively. The structure without intercalated ethidium was also simulated (indicated as Ref in Figure 5). For all cases, molecular trajectories of final 100 ns were used in analysis.
Figure 5.
MD simulation and computational analysis of ethidium-intercalated DNA origami bundles. (A) Cadnano (34) diagrams of 105-bp-long 6HB designs with different ethidium positions. For each case, ethidium molecules are initially located in dsDNA regions, crossovers, or evenly distributed in both regions, respectively. Refer to Supplementary Figures S75–S77 for MD simulation snapshots. (B) The time-average cross-sectional shape of five planes of 6HB structures with and without ethidium intercalators. The colored regions indicate the bp coordinates at each vertex. Refer to Supplementary Methods for the detailed process (ticks: 20 Å). (C) Root-mean-square deviation (RMSD) of MD trajectories for the whole simulation time. (D) Normalized bending persistence lengths of the ethidium-intercalated 6HBs calculated from PCA of MD trajectories. Refer to Supplementary Methods and Supplementary Figure S78 for more information. (E) Distribution of time-averaged inter-plane distance values. (F) Distributions of the time-averaged rotation angle between plane 1 and plane 5. A negative value indicates the counterclockwise rotation. (G) Schematic illustration of the first bending mode shape of the untwisted and twisted 6HB model. 210-bp-long 6HB was used for FE analysis. (H) The normalized persistence length of 6HB with respective to the normalized twist angle of dsDNA. (I) Schematic illustration of the first bending mode shape of the twisted 6HB models with different dsDNA bending stiffnesses. (J) The normalized persistence length of 6HB with respect to the normalized persistence length of dsDNA. P.L.: persistence length. Tw: twist angle.
The structural integrity of the bundles was analyzed from the coordinates of bps located at five cross-sectional planes with 21-bp-interval (Figures 5A, B). With ethidium intercalation, an increased amount of fluctuations was observed possibly due to the weakened basepairing interaction. Also, the root-mean-square deviation (RMSD) of ethidium-intercalated cases was higher than that of the reference (Figure 5C). The normalized bending persistence length was calculated using the eigenfrequency of the first bending mode from principal component analysis (Figure 5D and Supplementary Figure S78). Et-dsDNA, Et-Cross and Et-Mixed structures were 51.7%, 33.6% and 35.4% less stiff in bending, respectively, than the reference one without EtBr. From experiments, the corresponding reduction values ranged from 43% to 48% in 2–4 μM EtBr concentration, suggesting that ethidium might be bound to both dsDNA and crossover regions. According to the equilibrated shapes at the final time step, the inter-plane distance was increased for all three ethidium-intercalaed cases (Figure 5E). However, Et-dsDNA structure was left-twisted whereas Et-Cross design showed a right-handed twist (Figure 5F). Et-Mixed case showed almost no cross-sectional rotation, as the same number of ethidiums were intercalated to dsDNA and crossover regions in simulation. Since the portion of normal dsDNA regions is usually larger than that of crossovers in DNA origami designs, left-handed twist and coiling of ethidium-intercalated DNA origami bundles were reported in other experiments (24,58).
Coarse-grained simulation based on FE analysis was performed to see the effect of global twist of the bundle on its bending persistence length (Figure 5G–J) (41). First, we calculated the bending persistence length by changing the intrinsic twist angle of dsDNA to induce various levels of global twist while all the other geometric and mechanical properties were maintained. Overall, the bending stiffness decreased with the intrinsic twist angle of dsDNA (Figure 5G and H), implying that the global twist reduces the bending stiffness of the bundle. In addition, we changed the bending rigidity of dsDNA. Its effect on the softening of the bundle was bigger for more twisted bundles (Figure 5I and J). This result indicates that two effects of ethidium intercalation on dsDNA, geometric unwinding and mechanical softening, might synergistically increase the flexibility of structured DNA assemblies.
In summary, we investigated the chemo-mechanical response of structured DNA assemblies to DNA binding molecules, providing a simple and effective method to chemo-mechanically control the geometric and mechanical characteristics of DNA nanostructures. All intercalative ligands generally induced the softening of DNA origami structures in bending, while bis-intercalator YOYO-1 exhibited a more drastic effect. Minor groove binders showed a relatively irregular decreasing pattern than intercalators. In terms of controlling the curved shape, DAPI could be used to decrease the radius of curvature with suppressed out-of-plane deformation. Since EtBr and DAPI showed a good recovery rate of bending stiffness after buffer exchange, systematic and repeatable modulation of the shape and flexibility of structured DNA assemblies could be achieved by using them. Also, by exploiting the compatibility of chemo-mechanical modulation with the physical defect design, the controllable range of mechanical flexibility could be further expanded.
We expect that many other molecules including drugs, metal ions, and peptides that can interact with DNA and change its geometrical and mechanical characteristics, could be utilized as chemo-mechanical modulators of DNA nanostructures as well. Understanding and utilization of chemo-mechanical response of structured DNA assemblies may advance the method for the design and actuation of molecular machines and chemo-responsive nanomechanical components. Structural deformation and stiffness modulation induced by DNA-binding molecules can realize the unique working mechanism distinguished from DNA strand-based actuation system. For example, we recently demonstrated the threshold-based chemo-mechanical reconfiguration of the DNA origami ring structure (58). Cumulative internal stress induced by DNA-binding molecules were stored within a structure, and the structure was transformed to the other programmed state when the stress is over the threshold level. The results shown in this work can be utilized to develop more advanced reconfiguration mechanisms and control the working range of them. Also, our findings can provide an important information for the design of DNA-based molecular containers or DNA hydrogels that can be utilized to various biological and medical applications.
DATA AVAILABILITY
All data are available in the main text or the supplementary information.
Supplementary Material
Notes
Present address: Young-Joo Kim, Center for Nanomedicine, Institute for Basic Science (IBS), Seoul 03722, Korea.
Contributor Information
Chanseok Lee, Institute of Advanced Machines and Design, Seoul National University, Seoul 08826, Korea.
Young-Joo Kim, Institute of Advanced Machines and Design, Seoul National University, Seoul 08826, Korea.
Kyung Soo Kim, Department of Mechanical Engineering, Seoul National University, Seoul 08826, Korea.
Jae Young Lee, Institute of Advanced Machines and Design, Seoul National University, Seoul 08826, Korea.
Do-Nyun Kim, Institute of Advanced Machines and Design, Seoul National University, Seoul 08826, Korea; Department of Mechanical Engineering, Seoul National University, Seoul 08826, Korea; Institute of Engineering Research, Seoul National University, Seoul 08826, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Convergence Research of Scientific Challenges [NRF-2020M3F7A1094299]; Basic Research in Science & Engineering [NRF-2019R1A2C4069541] through the National Research Foundation of Korea (NRF) funded by Ministry of Science and ICT. Funding for open access charge: National Research Foundation of Korea (NRF).
Conflict of interest statement. None declared.
REFERENCES
- 1. Zimmer C., Wähnert U.. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog. Biophys. Mol. Biol. 1986; 47:31–112. [DOI] [PubMed] [Google Scholar]
- 2. Almaqwashi A.A., Paramanathan T., Rouzina I., Williams M.C.. Mechanisms of small molecule–DNA interactions probed by single-molecule force spectroscopy. Nucleic Acids Res. 2016; 44:3971–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Strekowski L., Wilson B.. Noncovalent interactions with DNA: an overview. Mutat. Res.-Fund. Mol. M. 2007; 623:3–13. [DOI] [PubMed] [Google Scholar]
- 4. Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002; 2:188–200. [DOI] [PubMed] [Google Scholar]
- 5. Werner M.H., Gronenborn A.M., Clore G.M.. Intercalation, DNA kinking, and the control of transcription. Science. 1996; 271:778–784. [DOI] [PubMed] [Google Scholar]
- 6. Glazer A.N., Rye H.S.. Stable dye–DNA intercalation complexes as reagents for high-sensitivity fluorescence detection. Nature. 1992; 359:859–861. [DOI] [PubMed] [Google Scholar]
- 7. Sischka A., Toensing K., Eckel R., Wilking S.D., Sewald N., Ros R., Anselmetti D. Molecular mechanisms and kinetics between DNA and DNA binding ligands. Biophys. J. 2005; 88:404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bazoni R.F., Lima C.H.M., Ramos E.B., Rocha M.S.. Force-dependent persistence length of DNA–intercalator complexes measured in single molecule stretching experiments. Soft Matter. 2015; 11:4306–4314. [DOI] [PubMed] [Google Scholar]
- 9. Murade C.U., Subramaniam V., Otto C., Bennink M.L.. Force spectroscopy and fluorescence microscopy of dsDNA–YOYO-1 complexes: implications for the structure of dsDNA in the overstretching region. Nucleic Acids Res. 2010; 38:3423–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lipfert J., Klijnhout S., Dekker N.H.. Torsional sensing of small-molecule binding using magnetic tweezers. Nucleic Acids Res. 2010; 38:7122–7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salerno D., Brogioli D., Cassina V., Turchi D., Beretta G.L., Seruggia D., Ziano R., Zunino F., Mantegazza F.. Magnetic tweezers measurements of the nanomechanical properties of DNA in the presence of drugs. Nucleic Acids Res. 2010; 38:7089–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y., Sischka A., Walhorn V., Tönsing K., Anselmetti D. Nanomechanics of fluorescent DNA dyes on DNA investigated by magnetic tweezers. Biophys. J. 2016; 111:1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Günther K., Mertig M., Seidel R.. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010; 38:6526–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bustamante C., Smith S.B., Liphardt J., Smith D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000; 10:279–285. [DOI] [PubMed] [Google Scholar]
- 15. Japaridze A., Benke A., Renevey S., Benadiba C., Dietler G.. Influence of DNA binding dyes on bare DNA structure studied with atomic force microscopy. Macromolecules. 2015; 48:1860–1865. [Google Scholar]
- 16. Berge T., Jenkins N.S., Hopkirk R.B., Waring M.J., Edwardson J.M., Henderson R.M.. Structural perturbations in DNA caused by bis-intercalation of ditercalinium visualised by atomic force microscopy. Nucleic Acids Res. 2002; 30:2980–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maaloum M., Muller P., Harlepp S.. DNA-intercalator interactions: structural and physical analysis using atomic force microscopy in solution. Soft Matter. 2013; 9:11233–11240. [Google Scholar]
- 18. Zhang F., Nangreave J., Liu Y., Yan H.. Structural DNA nanotechnology: state of the art and future perspective. J. Am. Chem. Soc. 2014; 136:11198–11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rothemund P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. [DOI] [PubMed] [Google Scholar]
- 20. Douglas S.M., Dietz H., Liedl T., Hogberg B., Graf F., Shih W.M.. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009; 459:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao Y.-X., Shaw A., Zeng X., Benson E., Nyström A.M., Högberg B.. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012; 6:8684–8691. [DOI] [PubMed] [Google Scholar]
- 22. Jiang Q., Song C., Nangreave J., Liu X., Lin L., Qiu D., Wang Z.-G., Zou G., Liang X., Yan H.et al.. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012; 134:13396–13403. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Q., Jiang Q., Li N., Dai L., Liu Q., Song L., Wang J., Li Y., Tian J., Ding B.et al.. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014; 8:6633–6643. [DOI] [PubMed] [Google Scholar]
- 24. Chen H., Zhang H., Pan J., Cha T.-G., Li S., Andréasson J., Choi J.H.. Dynamic and progressive control of DNA origami conformation by modulating DNA helicity with chemical adducts. ACS Nano. 2016; 10:4989–4996. [DOI] [PubMed] [Google Scholar]
- 25. Marras A.E., Shi Z., Lindell M.G., Patton R.A., Huang C.-M., Zhou L., Su H.-J., Arya G., Castro C.E.. Cation-activated avidity for rapid reconfiguration of DNA nanodevices. ACS Nano. 2018; 12:9484–9494. [DOI] [PubMed] [Google Scholar]
- 26. Gerling T., Wagenbauer K.F., Neuner A.M., Dietz H.. Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components. Science. 2015; 347:1446–1452. [DOI] [PubMed] [Google Scholar]
- 27. Ke Y., Meyer T., Shih W.M., Bellot G.. Regulation at a distance of biomolecular interactions using a DNA origami nanoactuator. Nat. Commun. 2016; 7:10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pal S., Zhang Y., Kumar S.K., Gang O.. Dynamic tuning of DNA-nanoparticle superlattices by molecular intercalation of double helix. J. Am. Chem. Soc. 2015; 137:4030–4033. [DOI] [PubMed] [Google Scholar]
- 29. Seo S.E., Wang M.X., Shade C.M., Rouge J.L., Brown K.A., Mirkin C.A.. Modulating the bond strength of DNA–nanoparticle superlattices. ACS Nano. 2016; 10:1771–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zadegan R.M., Lindau E.G., Klein W.P., Green C., Graugnard E., Yurke B., Kuang W., Hughes W.L.. Twisting of DNA origami from intercalators. Scientific Reports. 2017; 7:7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee C., Kim K.S., Kim Y.-J., Lee J.Y., Kim D.-N.. Tailoring the mechanical stiffness of DNA nanostructures using engineered defects. ACS Nano. 2019; 13:8329–8336. [DOI] [PubMed] [Google Scholar]
- 32. Wagenbauer K.F., Engelhardt F.A.S., Stahl E., Hechtl V.K., Stömmer P., Seebacher F., Meregalli L., Ketterer P., Gerling T., Dietz H.. How we make DNA origami. ChemBioChem. 2017; 18:1873–1885. [DOI] [PubMed] [Google Scholar]
- 33. Lamour G., Kirkegaard J.B., Li H., Knowles T.P., Gsponer J.. Easyworm: an open-source software tool to determine the mechanical properties of worm-like chains. Source Code Biol. Med. 2014; 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Douglas S.M., Marblestone A.H., Teerapittayanon S., Vazquez A., Church G.M., Shih W.M.. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009; 37:5001–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee J.Y., Lee J.G., Yun G., Lee C., Kim Y.-J., Kim K.S., Kim T.H., Kim D.-N.. Rapid computational analysis of DNA origami assemblies at near-atomic resolution. ACS Nano. 2021; 15:1002–1015. [DOI] [PubMed] [Google Scholar]
- 36. Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L.. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983; 79:926–935. [Google Scholar]
- 37. Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K.. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005; 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hart K., Foloppe N., Baker C.M., Denning E.J., Nilsson L., MacKerell A.D.. Optimization of the CHARMM Additive Force Field for DNA: Improved Treatment of the BI/BII Conformational Equilibrium. J. Chem. Theory Comput. 2012; 8:348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G.. A smooth particle mesh Ewald method. J. Chem. Phys. 1995; 103:8577–8593. [Google Scholar]
- 40. Castro C.E., Kilchherr F., Kim D.-N., Shiao E.L., Wauer T., Wortmann P., Bathe M., Dietz H.. A primer to scaffolded DNA origami. Nat. Methods. 2011; 8:221–229. [DOI] [PubMed] [Google Scholar]
- 41. Kim Y.-J., Lee C., Lee J.G., Kim D.-N.. Configurational design of mechanical perturbation for fine control of twisted DNA origami structures. ACS Nano. 2019; 13:6348–6355. [DOI] [PubMed] [Google Scholar]
- 42. Ke Y., Douglas S.M., Liu M., Sharma J., Cheng A., Leung A., Liu Y., Shih W.M., Yan H.. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 2009; 131:15903–15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dietz H., Douglas S.M., Shih W.M.. Folding DNA into twisted and curved nanoscale shapes. Science. 2009; 325:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kauert D.J., Kurth T., Liedl T., Seidel R.. Direct mechanical measurements reveal the material properties of three-dimensional DNA origami. Nano Lett. 2011; 11:5558–5563. [DOI] [PubMed] [Google Scholar]
- 45. Castro C.E., Su H.-J., Marras A.E., Zhou L., Johnson J.. Mechanical design of DNA nanostructures. Nanoscale. 2015; 7:5913–5921. [DOI] [PubMed] [Google Scholar]
- 46. Rivetti C., Guthold M., Bustamante C.. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996; 264:919–932. [DOI] [PubMed] [Google Scholar]
- 47. Cassina V., Seruggia D., Beretta G.L., Salerno D., Brogioli D., Manzini S., Zunino F., Mantegazza F.. Atomic force microscopy study of DNA conformation in the presence of drugs. Eur. Biophys. J. Biophys. Lett. 2011; 40:59–68. [DOI] [PubMed] [Google Scholar]
- 48. Ke Y., Bellot G., Voigt N.V., Fradkov E., Shih W.M.. Two design strategies for enhancement of multilayer–DNA-origami folding: underwinding for specific intercalator rescue and staple-break positioning. Chem. Sci. 2012; 3:2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silva E.F., Bazoni R.F., Ramos E.B., Rocha M.S.. DNA-doxorubicin interaction: new insights and peculiarities. Biopolymers. 2017; 107:e22998. [DOI] [PubMed] [Google Scholar]
- 50. Kundukad B., Yan J., Doyle P.S.. Effect of YOYO-1 on the mechanical properties of DNA. Soft Matter. 2014; 10:9721–9728. [DOI] [PubMed] [Google Scholar]
- 51. Rye H.S., Yue S., Wemmer D.E., Quesada M.A., Haugland R.P., Mathies R.A., Glazer A.N.. Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: properties and applications. Nucleic Acids Res. 1992; 20:2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson W.D., Tanious F.A., Barton H.J., Jones R.L., Strekowski L., Boykin D.W.. Binding of 4′,6-diamidino-2-phenylindole (DAPI) to GC and mixed sequences in DNA: intercalation of a classical groove-binding molecule. J. Am. Chem. Soc. 1989; 111:5008–5010. [Google Scholar]
- 53. Wilson W.D., Tanious F.A., Barton H.J., Jones R.L., Fox K., Wydra R.L., Strekowski L.. DNA sequence dependent binding modes of 4′,6-diamidino-2-phenylindole (DAPI). Biochemistry. 1990; 29:8452–8461. [DOI] [PubMed] [Google Scholar]
- 54. Bailly C., Colson P., Hénichart J.-P., Houssier C.. The different binding modes of Hoechst 33258 to DNA studied by electirc linear dichroism. Nucleic Acids Res. 1993; 21:3705–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Colson P., Bailly C., Houssier C.. Electric linear dichroism as a new tool to study sequence preference in drug binding to DNA. Biophys. Chem. 1996; 58:125–140. [DOI] [PubMed] [Google Scholar]
- 56. Silva E.F., Ramos E.B., Rocha M.S.. DNA interaction with Hoechst 33258: stretching experiments decouple the different binding modes. J. Phys. Chem. B. 2013; 117:7292–7296. [DOI] [PubMed] [Google Scholar]
- 57. Reis L.A., Rocha M.S.. DNA interaction with DAPI fluorescent dye: force spectroscopy decouples two different binding modes. Biopolymers. 2017; 107:e23015. [DOI] [PubMed] [Google Scholar]
- 58. Kim Y.-J., Park J., Lee J.Y., Kim D.-N.. Programming ultrasensitive threshold response through chemomechanical instability. Nat. Commun. 2021; 12:5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary information.