Abstract
Interaction between transcription and pre-mRNA processing via binding of polymerase II (Pol II) to factors involved in capping, splicing, and polyadenylation has recently been demonstrated. The C-terminal domain (CTD), a highly phosphorylated repeat sequence of the largest subunit of Pol II, has been implicated in this interaction because deletion of this domain affects downstream RNA processing events and because it is the binding site for numerous processing factors. Here we show that recombinant CTD, free of other components of Pol II, activated in vitro splicing and assembly of the spliceosome in nuclear extracts if, and only if, the assayed precursor RNA was recognized via exon definition, i.e., if the substrates contained complete exons with both 3′ and 5′ splice sites. Furthermore, depletion of intact Pol II inactivated splicing of this set of precursor RNAs and addition of recombinant CTD restored activity. The added recombinant CTD was quickly hyper- and hypophosphorylated in extract, became associated with the precursor RNA, and stimulated the association of U1 snRNPs but not ASF/SF2 with substrate RNA. These observations suggest that the mode of interaction between the CTD and splicing factors is integrally tied to exon definition and the mechanism whereby distal exons can be recognized and brought into juxtaposition during assembly of the spliceosome.
Recent reports have suggested the involvement of polymerase II (Pol II) in the splicing and polyadenylation steps of mRNA production (reviewed in references 3, 11, 18, 34, 42, and 43). Critical findings bolstering this hypothesis were observations that in vivo expression of a form of Pol II lacking the full-length C-terminal domain (CTD) of the largest subunit partially depressed capping, splicing, and polyadenylation of pre-mRNA (30, 31). The mammalian CTD, consisting of 52 imperfect repeats of the consensus sequence YSPTSPS (reviewed in reference 10), has the capacity to bind to known polyadenylation factors and has been shown to activate in vitro polyadenylation reactions (22, 30). Similarly, the CTD binds capping enzymes, snRNPs, and serine-arginine (SR)-like proteins (8, 9, 23, 26, 31, 33, 38, 47, 48). Even more striking, the ability of an SR protein to affect exon inclusion has been shown to be promoter dependent, suggesting a tight link between transcription and alternative splicing (12).
Purified Pol II activates splicing in an in vitro system uncoupled from transcription by using a cytoplasmic S100 extract supplemented with recombinant SR proteins (22), suggesting a direct role for Pol II in splicing in this system. A recombinant CTD fragment of the Pol II large subunit would not function in these assays, suggesting involvement of other portions of Pol II during interactions between transcription and splicing. The CTD itself, however, in the form of short peptides has been shown to inhibit both in vitro and in vivo splicing. Anti-CTD antibodies also inhibit splicing (15, 48), supporting a role for the CTD in the communication between Pol II and components of the splicing machinery.
A role for Pol II in pre-mRNA splicing must accommodate the need for multiple rounds of splicing per precursor RNA and the need to link exons located distally on the precursor. In mammalian pre-mRNAs, introns are usually quite large and exons are much smaller. Experiments with model precursor RNAs have suggested that the exon is used as the unit of initial recognition in such precursor RNAs via interactions between the factors that recognize the 3′ and 5′ splice sites of internal exons, the cap and 5′ splice site of 5′-terminal exons, and the 3′ splice site and poly(A) site of 3′-terminal exons in a process termed exon definition (reviewed in references 4, 6, and 40). The CTD becomes an attractive target as a mediator of exon definition by virtue of its uncommon repetitive structure and its ability to bind snRNPs and SR-related proteins (26, 48). To test this idea within a nuclear context, we asked if addition of recombinant mammalian CTD could affect in vitro splicing of substrate RNAs. The utilized precursor RNAs comprised a set containing the same 3′ and 5′ splice site sequences differing only in their intron-exon architecture and their ability to access exon definition because of the presence of intact exons. We observed a consistent three- to fivefold activation of spliceosome assembly and splicing activity by using substrates that contained complete internal exons and which therefore could be recognized by exon definition. In contrast, no activation was observed using substrates that lacked complete internal exons and in which pairs of splice sites could necessarily be found only in an intronic polarity. In support for a direct role for the CTD in exon recognition, partial depletion of Pol II from a nuclear extract depressed the splicing of the same exon-definition-dependent substrate RNAs but had little impact on substrates not accessing exon definition. Addition of recombinant CTD restored activity to the depleted extracts. Furthermore, the supplemental recombinant CTD became stably associated with the assembled spliceosome, suggesting a direct role in assembly. These results suggest that the CTD may be a stage for exon recognition and hold exons until subsequent exons are recognized and can be ligated.
MATERIALS AND METHODS
Plasmids and antibodies.
The utilized CTD–gluathathione S-transferase (CTD-GST) expression construct was kindly supplied by Bill Dynan (Medical College of Georgia). Recombinant CTD and GST were purified from Escherichia coli lysates by chromatography using glutathione columns (Pharmacia). Protein eluates of the recombinant CTD were concentrated to ∼3 mg/ml by centrifugation in a Biomax-Ultrafree filter device (Millipore). Monoclonal antibodies (MAbs) 8WG16, H5, and H14 were previously described by Thompson et al. (45) and Bregman et al. (7).
The precursor RNA constructs in this study contain sequences derived from the human adenovirus type 2 late transcription unit and are based on the MINX construct made in our laboratory (41). The two-intron construct Ad 100 and MT16 Short (MT16-S) substrates were described previously (44). The MT16 Long (MT16-L) construct is an extension of MT16-S at its incomplete third exon with the addition of an EcoRI-HindIII fragment from MINX that contains a 5′ splice site. Ad 600 is an intronic expansion at the HindIII site of Ad 100 with an insertion of 500 nucleotides derived from human hypoxanthine phosphoribosyltransferase intron 1.
In vitro RNA processing.
32P-labeled precursor RNAs were in vitro transcribed by SP6 RNA polymerase (Gibco-BRL). Precursors were capped during synthesis in the presence of diguanosine triphosphate. Standard in vitro splicing assays using HeLa nuclear extracts were described previously (41). To observe the maximum effect of the Pol II CTD, HeLa nuclear extract was reduced to a final concentration of 14% in all assays. Unless otherwise specified, reactions contained 100 pmol of purified recombinant full-length CTD-GST or GST. Assembled spliceosome complexes under splicing conditions were analyzed by the addition of heparin to a final concentration of 2 mg/ml and electrophoresis on native RNP gels.
Immunodepletion.
Three Pol II CTD-specific antibodies were combined for immunodepletion of endogenous Pol II from HeLa nuclear extracts. MAb 8WG16 (immunoglobulin G [IgG]) was directly bound to protein G-Sepharose beads. After a wash with phosphate-buffered saline, antibody and beads were cross-linked in the presence of dimethyl pimelidate (Pierce) for 30 min as previously described (19). To cross-link IgM antibodies H5 and H14, a rabbit anti-mouse IgM antibody was first bound to protein A-Sepharose beads and then H5 and H14 were added to react with anti-IgM. This sandwich binding was then conjugated to beads by addition of dimethyl pimelidate. Immobilized CTD-specific IgG and IgM antibodies were then mixed in a 1:1 ratio for immunodepletion. The same binding and cross-linking protocols were used in the mock depletion experiments to control for extract proteins capable of binding to both of the utilized IgG and IgM antibodies. Two sets of nonspecific antibodies containing both IgG and IgM in each group were cross-linked to protein G and protein A beads. In set 1, mouse anti-tubulin Tu9B (IgG) and 5H1 (IgM) were employed, and in set 2, purified mouse IgG (Jackson Immuno-Research) and IgM (Sigma) were used. For immunoabsorption, HeLa nuclear extract was diluted 1:1 with water and was then added to premixed antibody-beads (3:1, vol/vol) with gentle shaking at 4°C for 20 min. At the end of the depletion, absorption mixtures were allowed to sit in ice for sedimentation for at least 15 min and the supernatants were utilized as depleted nuclear extracts. The bead pellets were washed five times with NET buffer (0.15 M NaCl, 0.05 M Tris [pH 7.9], 0.05% NP-40, and 2 mM EDTA) and were used as antibody-bound fractions.
Immunoprecipitation.
For immunoprecipitation using MAbs against ASF/SF2 or U1 70-kDa protein, antibodies were prebound to protein G-Sepharose. Splicing reaction mixtures diluted with NET buffer were transferred to antibody-beads and incubated at 4°C with gentle rocking for 1.5 to 4 h. For immunoprecipitation using the antibody against the GST tag, purified anti-GST antibody (1 μg/μl; Zymed) was directly added to splicing reaction mixtures containing 32P-labeled precursors at required time points. After incubation in ice for 20 to 60 min, 15 μl of standardized Pansorbin cells (Calbiochem) was added to each reaction mixture for another 20 min of incubation in ice. The reaction mixture was then diluted with NET buffer to 400 μl and was gently rocked at 4°C for 1.5 to 4 h. Final immunocomplexes were extensively washed with NET buffer, and RNA was resolved on a 5% denaturing acrylamide gel. The precipitated RNA bands were quantified using PhosphorImager SI (Molecular Dynamics).
RESULTS
Full-length recombinant mammalian CTD activates in vitro splicing.
A few years ago it was demonstrated that short peptides containing eight CTD consensus repeat units were inhibitory to in vitro splicing (48). While trying to reproduce these studies using a full-length recombinant murine Pol II fragment containing the entire CTD sequence with its 52 consensus repeats linked to GST, we noticed an enhancement of in vitro splicing rather than inhibition (Fig. 1). Using standard in vitro splicing extracts from HeLa cells and a two-exon precursor RNA derived from adenovirus, a consistent three- to fivefold activation of splicing was observed in comparison with the control. Both products and intermediates of the splicing reaction were affected by addition of recombinant CTD. The addition of recombinant CTD did not shorten the time required for the initial appearance of lariat exon 2. It did, however, increase the amount of observable lariat exon 2, suggesting that the presence of the CTD facilitates recruitment of more precursor RNA into productive complexes. Enhancement by a fragment of Pol II suggested that the CTD can activate RNA processing, at least in part independently of the rest of the enzyme. Interestingly, instead of being an antagonist of native Pol II as was observed with short peptides, the recombinant full-length CTD fragment appears to provide a simplified substitute for Pol II during in vitro RNA splicing.
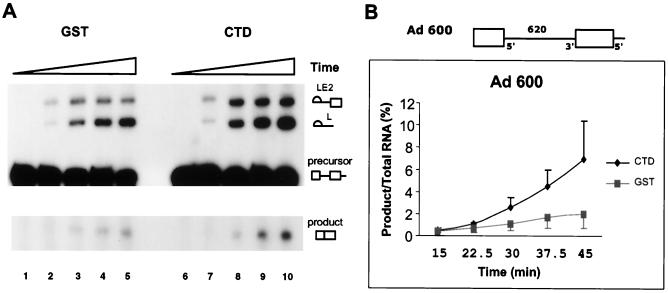
FIG. 1.
Supplemental recombinant CTD-GST stimulates in vitro splicing. (A) Gel of a time course of a standard splicing reaction using the Ad 600 substrate (diagrammed in panel B) and extract preincubated for 15 min with 100 pmol of recombinant CTD-GST or GST under splicing conditions at room temperature. At 15, 22.5, 30, 37.5, and 45 min, RNAs were removed and displayed on 5% denaturing gels. The intermediate and product RNAs of the splicing reaction mixture are indicated. Different vertical portions of the same gel are juxtaposed to permit display of a larger image. (B) Quantification of the appearance of spliced product RNA (y axis) at each time point (x axis) from three parallel experiments. Radioactivity in individual RNA bands was measured using the PhosphorImager, and the percentage of spliced product RNA was plotted. Bars, 1 standard deviation.
The experiment in Fig. 1 used recombinant GST as a control for the GST-tagged recombinant CTD. Similar results were observed using other proteins, such as bovine serum albumin, as a control. Ideally, one would like to use mutant CTD controls. However, given the repeated nature of the CTD, particularly the extremely high number of amino acid sites being potentially phosphorylated, construction of an appropriate mutant full-length CTD was problematic. As demonstrated below, the ability of recombinant GST-CTD to affect splicing activity was substrate specific. Thus, compared to the same control proteins, recombinant GST-CTD was able to affect the splicing activity of only some of the tested substrates. The utilized substrates were a related set with identical splice sites and intron and exon sequences that differed only in their architecture. Any potential artificial activation of splicing by recombinant CTD would not be predicted to differ among this set of substrates. Therefore, we considered the observed activation of splicing by the GST-CTD to be interesting enough to warrant further investigation.
Pol II is required for maximal in vitro splicing in nuclear extracts.
Observation of enhancement of splicing by addition of recombinant CTD suggested that native Pol II may actively participate in in vitro splicing via its CTD. To examine this possibility, we asked if addition of antibodies specific for endogenous Pol II could deplete splicing activity in a fashion that could be rescued by addition of recombinant CTD. We performed immunodepletion experiments targeting all forms of nuclear Pol II using a mixture of immobilized Pol II antibodies 8WG16, H5, and H14 (Fig. 2). As detected by immunoblotting, significant amounts of endogenous Pol II were removed from nuclear extract by this immunoabsorption protocol (Fig. 2B). Because antibody 8WG16 is IgG and both H5 and H14 are IgM, multiple control immunodepletions were performed using both IgG and IgM nonspecific antibodies. To ensure that the depletions were not removing constitutive splicing factors, we examined the levels of remaining proteins that were reactive with Sm- and SR-specific antibodies. Although slight depressions in the amounts of both Sm and SR proteins were detected, the bulk of these factors remained following immunoabsorption.
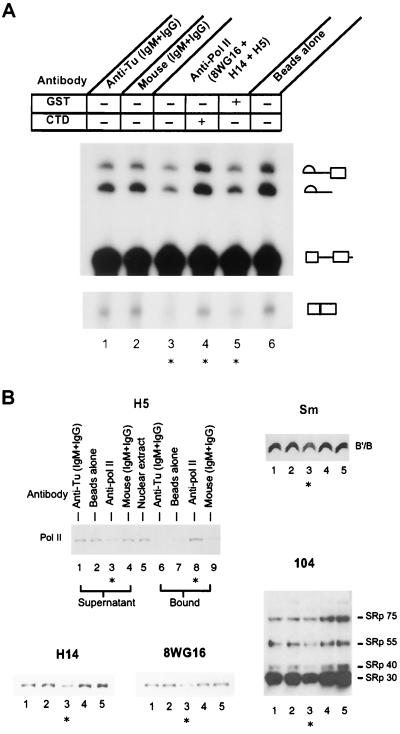
FIG. 2.
Partial depletion of Pol II depresses in vitro splicing, and activity can be rescued by addition of recombinant CTD-GST. (A) Splicing of Ad 600 after immunodepletion of Pol II from HeLa extracts and addition of complementing proteins. Extract was absorbed on Sepharose beads to which either Pol II antibodies or two different mixtures of control IgG and IgM antibodies were bound. To ensure removal of multiple forms of Pol II that differ in phosphorylation and therefore recognition by existing Pol II CTD-specific antibodies, multiple Pol II antibodies were used simultaneously (see Materials and Methods). The partially depleted nuclear extracts were assayed for splicing activity in the absence or presence of complementing recombinant CTD-GST or GST. ∗, experimental samples using extracts with a partial depletion of Pol II; lanes 1, 2, and 6, control depletions. Antibodies used in immunoabsorption are indicated above the panel: lane 1, mouse anti-tubulin antibodies Tu9B (IgG) and 5H1 (IgM); lane 2, commercial purified mouse IgG and IgM; lanes 3 to 5, mouse anti-Pol II antibodies 8WG16 (IgG), H5 (IgM), and H14 (IgM); lane 6, beads alone. Complementing CTD-GST or GST (100 pmol) was added to reaction mixtures in lane 4 or 5, respectively. Following a 15-min preincubation, Ad 600 splicing substrate RNA was added, and the reaction was continued for 40 min. Different vertical portions of the same gel are juxtaposed to permit display of a larger image. Reaction products and intermediates are indicated. (B) Detection of nuclear factors in immunodepleted extracts by immunoblotting. Extract was depleted using one of four antibody mixtures bound to beads as indicated above the H5 blot (the anti-Pol II antibodies contained the same mixture of antibodies as that in panel A). The supernatants from the depletions were tested for the presence of different proteins by Western blotting. The different forms of Pol II in the depleted extract were assayed using the H5, H14, or 8WG16 antibodies in separate blottings as indicated. Sm-containing snRNP proteins (the snRNP B and B′ protein region of the gel is shown) or SR proteins reactive with the 104 MAb were detected in the indicated blots. Lane numbers on each blot used the same depleted extracts as indicated on the H5 blot; thus, lane 3 (indicated by stars) on each blot corresponds to the supernatant from the immunodepletion using antibodies specific for Pol II. For the H5 blot, both supernatant and bound fractions were assayed by blotting. Lanes 1 to 4, nuclear extract supernatants after absorption on the indicated antibodies; lane 5, nuclear extract without immunoabsorption; lanes 6 to 9, proteins bound to the antibody beads. Equal amounts of nuclear extract were loaded in all lanes.
The partially depleted extracts were tested for their ability to support splicing of an adenovirus-based precursor RNA (Ad 600, the substrate utilized in Fig. 1) both in the presence and in the absence of supplemental recombinant CTD-GST or GST. As shown in Fig. 2A, splicing of Ad 600 significantly decreased using the partially depleted nuclear extract (Fig. 2, compare lane 3 to lanes 1 or 2). Supplementation of the depleted extract with recombinant CTD resulted in recovery of splicing activity to the level seen in the control depletions (compare lanes 3 and 4). Addition of GST alone had no ability to rescue splicing (compare lanes 3 and 5). These data provide supporting evidence that nuclear Pol II participates in splicing and that recombinant full-length CTD fragment can supply this function.
Enhancement of RNA processing by recombinant CTD is exon definition dependent.
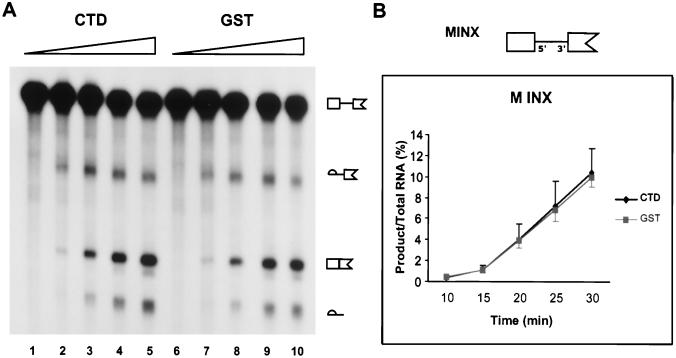
During this study, we tested the effect of supplementation of nuclear splicing extract with recombinant CTD by using a variety of precursor RNAs. Stimulation of in vitro splicing by the CTD was limited to only a subset of the utilized pre-mRNAs. For certain substrates, the CTD had no effect on splicing. One representative of the nonreactive substrates is MINX, shown in Fig. 3. This precursor contains the same splice sites as those in Ad 600 but has only one set of splice sites in an intronic configuration (i.e., exon 2 is incomplete, having only a 3′ splice site). No splicing activation was observed upon supplementation of nuclear extract with recombinant CTD (Fig. 3). Furthermore, partial immunodepletion of endogenous Pol II in an experiment similar to that in Fig. 2 resulted in little inhibition of splicing activity compared to the control depletion using nonspecific antibodies, and addition of recombinant CTD had minimal capacity to increase activity (data not shown).
FIG. 3.
Enhancement of RNA processing by recombinant CTD requires the presence of intact internal exons within the substrate RNA. (A) Splicing of the MINX precursor RNA containing an incomplete second exon (diagrammed in panel B) using regular nuclear extracts supplemented with recombinant CTD. Supplementation conditions were as described for Fig. 1. RNA was removed at 10 (lanes 1 and 6), 15 (lanes 2 and 7), 20 (lanes 3 and 8), 25 (lanes 4 and 9), and 30 (lanes 5 and 10) min and displayed on 5% denaturing gels. Reaction substrates, intermediates, and products are indicated. (B) Quantification of the splicing of the MINX substrate from three experiments similar to that shown in panel A. Percentage of product RNA was calculated for the duplicate experiments and displayed with error bars as in Fig. 1.
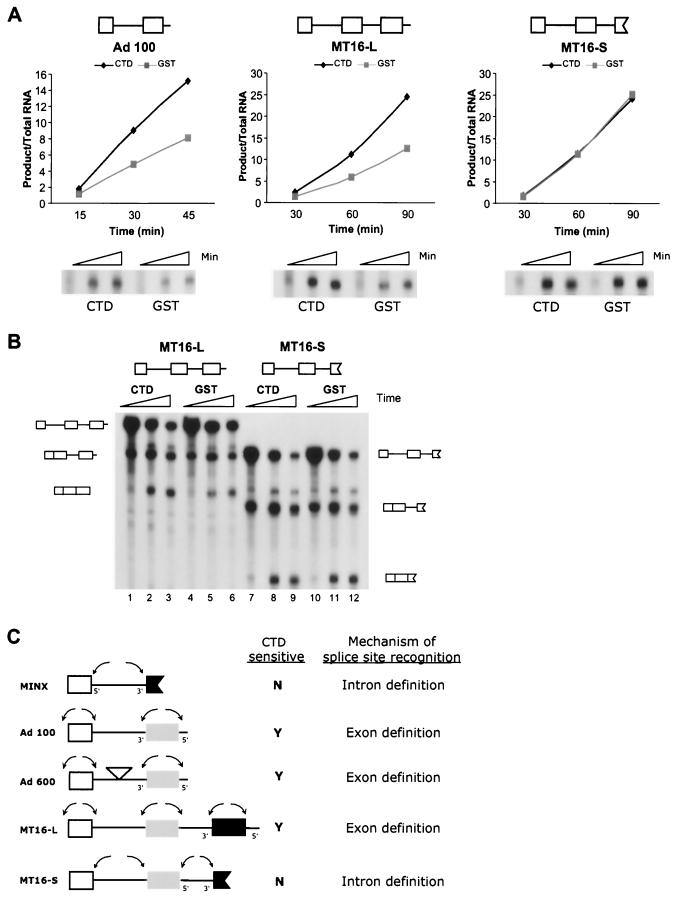
The above results strongly suggested that the precursor structural frame, i.e., the presence or absence of complete pairs of splice sites in either an intronic or an exonic polarity, played a critical role in the response of the substrates to recombinant CTD. As we collected examples of different substrates within a series of adenovirus-based constructs that contained identical splice sites but differed in the geometry of these splice sites, we saw a consistent pattern of sensitivity to the CTD. Figure 4 shows the CTD sensitivity of three additional substrates. Two of these three precursors were responsive (Ad 100 and MT16-L) and one was not responsive (MT16-S) to supplemental CTD. All 3′ splice sites in these precursor RNAs are identical, as are all 5′ splice sites. Intron lengths are also similar (diagrammed in Fig. 4C). Thus, the only difference among the set is the number and arrangement of the splice sites. For example, the Ad 100 and MINX precursor RNAs have identical introns and the 5′ splice site (with flanking exon and intron sequences) added at the end of Ad 100 is the same site present at the end of exon 1. A similar relationship exists between the two substrates, MT16-L and MT16-S, having two introns. The characteristic differentiating between the sensitive and insensitive precursor RNAs is the presence of matching pairs of splice sites in an exonic polarity in the former and their absence in the latter (replaced instead with matching pairs of splice sites in an intronic polarity). This difference suggests both that the CTD effect is a direct effect on the splicing of the responsive substrates and that something about the presence of an intact exon renders a substrate responsive to exogenous CTD.
FIG. 4.
Response to recombinant CTD is related to exon definition. (A) Splicing reactions using the precursor RNAs diagrammed above each gel in the presence of recombinant CTD. These precursor RNAs contain splice sites derived from adenovirus and are similar to the substrates used in Fig. 1 and 3 except for the geometry of the exons and introns. Supplementation with recombinant CTD and quantification of reactions were performed as described for Fig. 1. Below each graph is that portion of the gels that contained fully spliced product RNA created in these reactions. (B) The complete gel for the reactions of the three-exon substrates MT16-L and MT16-S shown in panel A. Substrates and fully spliced three-exon product RNAs are indicated. (C) Exon and intron structures of the adenovirus-based precursor RNAs compared in this study. The internal exon in all constructs is from exon 2 of the adenovirus major late transcription unit. Containing complete internal exons, Ad 100, Ad 600, and MT16-L can utilize exon definition recognition mechanisms (depicted by arrows). In contrast, MINX and MT16-S have incomplete exons and pairing of splice sites is only possible in an intronic polarity, leading to recognition via intron definition.
The exon definition hypothesis suggests that splice sites are paired either exonically to define exons or intronically to define introns. In vitro substrates with exonic pairs of splice sites can access exon definition via the cap-mediated recognition of the first exon and standard exon definition interactions across the subsequent exons (diagrammed in Fig. 4C). Substrates with an incomplete last exon can only pair splice sites across introns because of the absence of a final 5′ splice site after the last exon. Thus, the responsiveness of the substrates with complete exons and the lack of response of substrates with incomplete exons suggests that the CTD is involved during exon definition. In support of this hypothesis, we tested the ability of the CTD to stimulate the splicing of a precursor RNA that does not utilize exon definition pathways. For this purpose, we used substrates containing exons from the human alpha globin gene. In vivo, the splicing of this gene appears to use intron definition rather than exon definition because mutation of the 5′ splice site in intron 2 results in intron inclusion rather than exon skipping (32). The splicing of an in vitro substrate RNA containing the second alpha globin intron was refractory to the effect of the CTD (data not shown), again suggesting that utilization of exon definition is required for visualization of the CTD effect.
It should be noted that the unresponsive MT16-S substrate does contain an intact internal exon which might be expected to exon define. If it were to be recognized by this route, the 3′ splice site of the last exon would not have a partner 5′ splice site. Thus, this substrate could splice by two pathways: (i) exon definition to remove intron 1 and intron definition to remove intron 2 or (ii) intron definition to remove both introns. Analysis of the phenotype resulting from mutation of the 5′ splice site within intron 2 indicates that the substrate accesses both of these pathways (data not shown). Thus, one would predict an effect of the CTD on the splicing of intron 1 for those molecules undergoing splicing via pathway 1 but no effect on the removal of intron 2. None of the substrate molecules recognized via pathway 2 would be expected to be affected by the CTD for the removal of either intron. Because these substrates splice via an almost completely stepwise pathway in which intron 1 is removed before intron 2, it is possible to separate the effects on the splicing of the two introns. As shown in Fig. 4, the major effect observed was the removal of intron 2 which would occur by intron definition using either pathway. There is a small effect on the removal of intron 1 (observed at the earliest time point before appreciable intron 2 removal has occurred), suggesting that a subpopulation of the substrate molecules are being recognized by exon definition of exon 2 and are thus sensitive to the CTD.
CTD promotes assembly of spliceosomes.
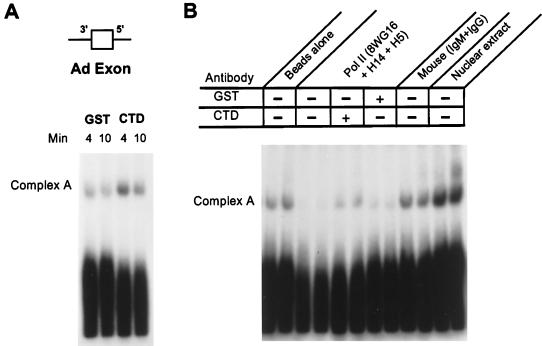
If the CTD is involved during exon definition, one predicts an effect during assembly of the ATP-dependent spliceosome. To test the effect of the CTD on assembly, we asked if the CTD would stimulate the assembly of a precursor RNA containing an intact internal exon with flanking splice sites but no complete intron (diagrammed in Fig. 5). Such substrates assemble only the first ATP-dependent spliceosome complex, complex A, and are dependent upon the entire exon and exon definition for maximal assembly (41). As demonstrated in Fig. 5A, supplemental CTD promotes in vitro spliceosome formation. Enhancement of assembly by recombinant CTD was also seen when other substrates containing complete exons were used, including Ad 600. In this case, the assembly of both complexes A and B was significantly increased by the addition of supplemental recombinant CTD (data not shown). As seen with splicing, addition of the CTD had minimal impact on the assembly of the MINX one-intron precursor that lacked a complete second exon (data not shown).
FIG. 5.
Supplemental recombinant CTD enhances initial ATP-dependent spliceosome assembly. (A) Assembly of a precursor RNA containing the intact adenovirus second exon and flanking splice sites (Ad exon) in the presence of recombinant GST or CTD-GST. Standard splicing reaction mixtures were supplemented with recombinant CTD proteins as described for Fig. 1. At the indicated times, spliceosome complexes were visualized by electrophoresis of reaction mixtures on native complex gels in the presence of 2 mg of heparin/ml. ATP-dependent spliceosomal complex A is denoted. (B) Depletion of Pol II and rescue of assembly activity in the depleted extract by recombinant CTD-GST. Immunodepletion conditions are identical to those in Fig. 2. The antibodies used for the depletion are indicated at the top. The complementing proteins added to the depleted extract are indicated for each lane. Reaction mixtures with depleted extracts were incubated for 4 or 10 min as shown in panel A to produce two sample lanes for each condition.
To confirm the involvement of endogenous Pol II during formation of splicing complexes, nuclear extract with a partial immunodepletion of Pol II as described for Fig. 2 was also used in assembly assays (Fig. 5B). As with splicing activity, reduction of the level of endogenous Pol II depressed spliceosome assembly. More importantly, addition of recombinant CTD-GST, but not GST, rescued assembly. These observations suggest a direct role of the Pol II CTD in promoting exon assembly during exon definition.
Recombinant CTD becomes associated with the spliceosome.
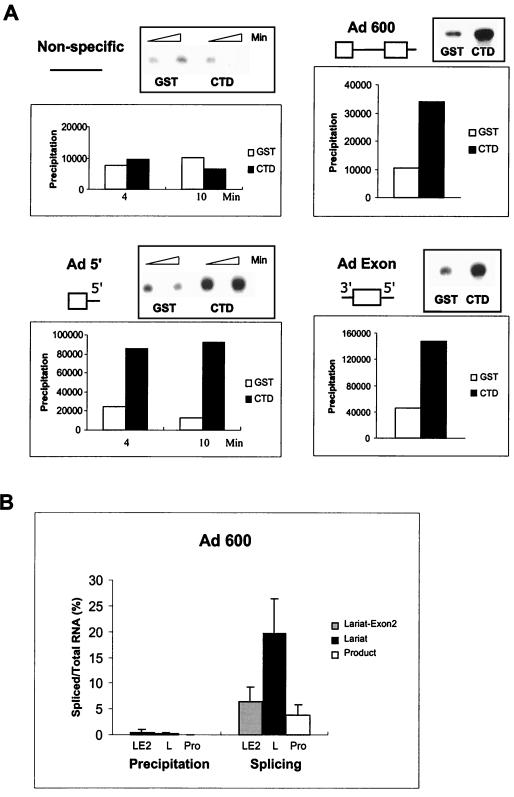
If supplemental CTD directly participates in in vitro splicing, it might be predicted to become incorporated into the spliceosome. To search for such an interaction, we asked if antibodies specific for the recombinant CTD could coimmunoprecipitate RNA substrates after their assembly into the spliceosome. Use of the CTD-specific antibodies 8WG16, H14, and H5 for this purpose was problematic because of their potential ability to prevent association of the CTD with interacting factors by virtue of their affinity for the multiple repetitive domains of the CTD. Indeed, we and others have observed that direct addition of the antibodies to in vitro reaction mixtures inhibits activities of splicing and polyadenylation (48; data not shown). Therefore, to do this experiment, we utilized an antibody against the GST tag to immunoprecipitate complexes containing recombinant CTD-GST (Fig. 6A). As diagrammed in Fig. 6A, three adenovirus-based substrate RNAs were employed—a precursor containing two exons (Fig. 1, Ad 600), an RNA containing only a complete internal exon (Fig. 5, Ad exon), and an RNA containing only a complete first exon (Ad 5′). An RNA containing no splicing signals was also used as a control. Three- to sevenfold more RNA was immunoprecipitated when recombinant CTD-GST was used to supplement the reaction mixtures than when GST alone had been added. The ability of an antibody directed towards the supplemental recombinant CTD to precipitate substrate RNA containing splicing signals but not a control RNA suggests incorporation of the recombinant CTD into assembled complexes.
FIG. 6.
Supplemental recombinant CTD associates with substrate RNA in the spliceosome. (A) Radiolabeled RNA substrates were incubated in an in vitro splicing assay supplemented with either GST-CTD or GST, and reaction mixtures were immunoprecipitated with an anti-GST antibody to detect the association of the CTD and substrate RNA. Immunoprecipitated RNAs were displayed on 5% denaturing acrylamide gels, and the radioactivity in each band was quantified in the phosphorimager. Substrates were as diagrammed. All sequences were derived from adenovirus exon 1 or 2. Antibodies were added to reaction mixtures at 4 and 10 min (nonspecific and Ad 5 RNAs), 15 min (Ad exon), or 19 min (Ad 600). (B) Relative immunoprecipitation of substrates, intermediates, and products using anti-GST antibodies from reaction mixtures supplemented with GST-CTD. Reaction RNAs were quantified in both the total reaction mixture and the immunoprecipitate by using the phosphorimager. The percentage of each RNA in the total reaction mixture is compared to the percentage of each species in the anti-GST antibody immunoprecipitate.
We also performed immunoprecipitations at later times during the splicing reaction to see if reaction intermediates and products were associated with the exogenously added CTD. Figure 6B shows a comparison of the relative amounts of splicing intermediates and products in the total reaction mixture and in the immunoprecipitates. From this figure, it is obvious that only small amounts of lariat exon 2 RNA were immunoprecipitable and that lariat and final spliced product RNAs were not. Thus, the interaction of the CTD appears to occur only during the earliest steps of precursor RNA recognition.
CTD facilitates binding of certain splicing factors to splice sites.
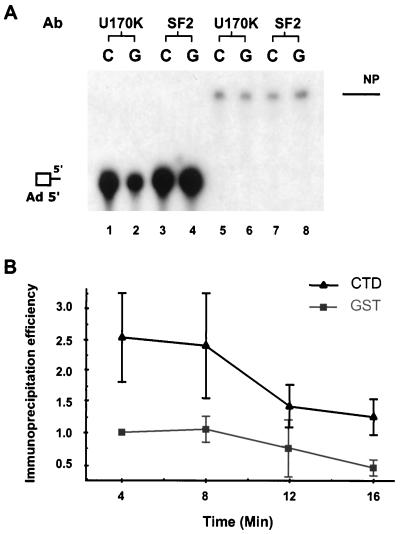
One hypothesis for the function of the CTD is that it recruits of splicing factors to the site of spliceosome assembly. It is already known that the CTD binds snRNPs (26, 33). We asked if the supplementation with recombinant CTD stimulated association of the U1 snRNP with substrate RNA (Fig. 7). A single first exon-Ad 5′ was incubated in a standard splicing reaction mixture supplemented with either recombinant CTD or GST and immunoprecipitated with an antibody specific for the U1 70-kDa (70k) protein. Compared to reactions with a nonspecific RNA without splice sites, a significant amount of Ad 5′ RNA was precipitated by the U1 70K antibody with or without supplemental CTD. However, two- to fourfold more RNA was precipitated in the reaction mixtures containing the CTD than in reaction mixtures containing the control GST (Fig. 7, compare lanes 1 and 2; quantified in Fig. 7B). Addition of the CTD did not prevent the natural dissociation of U1 snRNP from the complex as the reaction progressed, as observed by the decrease in immunoprecipitability after 10 min of reaction, suggesting that the CTD is only involved in stimulating early assembly but does not provide extra stabilization for complexes associated with CTD. Similar results were also seen when the two-exon-containing substrate Ad 600 was used (data not shown). These observations suggest that the CTD facilitated addition of the U1 snRNP to the spliceosome.
FIG. 7.
CTD facilitates early recruitment of U1 snRNPs but not the SR protein ASF/SF2 to cap-proximal exons. (A) Standard splicing reactions were performed in the presence of either CTD-GST (C) or GST (G) by using a capped substrate RNA containing a cap-proximal exon and its flanking splice site (Ad 5′) or a nonspecific RNA. At 8 min, reaction mixtures were immunoprecipitated using antibodies specific to U1 70K and ASF/SF2. (B) Quantification of the relative amount of immunoprecipitation using the U1 70K antibody. Relative radioactivity was determined in the phosphorimager for the precipitated RNA from three experiments and plotted versus reaction time. Error bars represent 1 standard deviation.
Recently, certain SR-rich proteins were demonstrated to associate with the Pol II CTD (48). SR proteins are known to facilitate association of U1 snRNPs with 5′ splice sites; therefore, it was possible that the ability of the CTD to facilitate U1 snRNP association with substrate was mediated by an interaction with SR proteins. One of the major SR proteins with a pronounced ability to stimulate U1 snRNP association with 5′ splice sites is the small SR protein ASF/SF2 (17, 24, 29, 46). To test if addition of exogenous CTD stimulated ASF/SF2 association with precursor RNA, we precipitated reaction mixtures with the Ad 5′ substrate RNA with an MAb specific for only ASF/SF2. As shown in Fig. 7A, anti-ASF/SF2 precipitated Ad 5′ RNA intensively, indicating a strong binding of this SR protein to the substrate. However, binding of ASF/SF2 to the substrate was not stimulated by supplemental CTD. Similar results were also seen using the substrate Ad 600 (data not shown), suggesting that the Pol II CTD only facilitates recruitment of certain splicing factors to splice sites and traditional SR proteins are probably not targets of CTD activation during spliceosome assembly.
Recombinant Pol II CTD is actively phosphorylated by the nuclear extract.
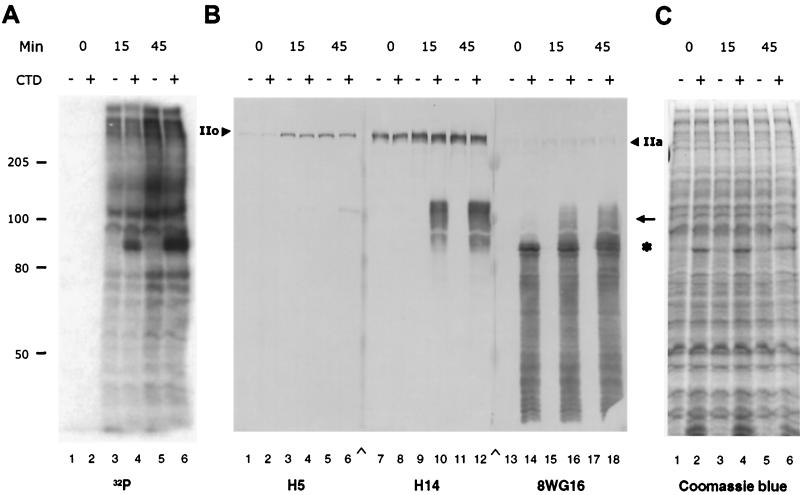
The phosphorylation state of the Pol II CTD has been implicated as important in interaction of the CTD with processing factors (21, 22, 30, 31, 33). The experiments in this study included a 15-min preincubation of the CTD with nuclear extract prior to the addition of precursor RNA to permit nuclear kinases to phosphorylate the CTD. As shown in Fig. 8, recombinant CTD was actively phosphorylated within these 15 min as detected by [32P]ATP incorporation or immunoblotting of phosphorylated forms by antibodies specific for the Pol II CTD. Three antibodies were used in immunoblotting—MAb 8WG16, which recognizes nonphosphorylated CTD epitopes (45), and MAbs H5 and H14, which recognize different phospho-epitopes (7, 26). All three antibodies recognized the large subunit of endogenous Pol II in nuclear extract (Fig. 8B, arrowhead). H5 and 8WG16 normally recognize the forms with the highest and lowest apparent molecular masses, respectively. H14 typically recognizes Pol II forms having intermediate states of phosphorylation and gel migration. For the recombinant CTD, both hyper- and hypophosphorylated fragments were formed during incubation in extract, as indicated by the appearance of higher-molecular-weight forms of the recombinant protein detectable with the CTD antibodies, indicating that hyperphosphorylation of CTD can occur without concomitant involvement in transcription. In addition, detection of a wide range of CTD forms with increased molecular weight after phosphorylation suggests that multiple phosphorylated forms of recombinant CTD were generated by kinases in the nuclear extract. However, the majority of the recombinant protein was hypophosphorylated, as revealed by [32P]phosphate labeling of the intact CTD-GST protein (Fig. 8A and C). Although our CTD preparation usually contained partial CTD peptide fragments, as detected in immunoblotting, these degraded peptides are only a small portion of the recombinant CTD as shown by Coomassie blue staining (Fig. 8) and they appeared not to be substrates for phosphorylation, suggesting both that the observed phosphorylation is not random and that the fragments are not probable participants in subsequent events.
FIG. 8.
The recombinant Pol II CTD fragment is in vitro phosphorylated to form multiple phosphorylated forms. (A) Incorporation of [32P]phosphate into recombinant CTD-GST in HeLa nuclear extract under splicing conditions. Splicing reaction mixtures contained 14% HeLa nuclear extract (lanes 1 to 6) and 20 pmol of GST (lanes 1, 3, 5) or 20 pmol of CTD-GST (lanes 2, 4, 6). Total incubation was for 0, 15, or 45 min. For the 15-min time point and the first 15 min of the 45-min time point, the reactions were performed at room temperature; the remainder of the incubation was at 30°C. Radioactive proteins were displayed on sodium dodecyl sulfate–6% polyacrylamide gels. The full-length CTD-GST fusion protein has an apparent molecular mass of 90 kDa. (B) Immunodetection of the CTD using Pol II antibodies H5 (lanes 1 to 6), H14 (lanes 7 to 12), and 8WG16 (lanes 13 to 18). Star, position of full-length recombinant CTD-GST; arrow, hyperphosphorylated CTD-GST; arrowheads, endogenous Pol II. IIo is the hyperphosphorylated form of Pol II; IIa is the non- or hypophosphorylated form of Pol II. Concentrations of CTD-GST or GST and incubation times are as in panel A. (C) Coomassie blue staining of the samples in panel A.
DISCUSSION
Participation of Pol II CTD in splicing.
The involvement of Pol II in splicing and the inherent communication between splicing and transcription indicated by this involvement has received recent interest (reviewed in references 3, 11, 18, 34, 42, and 43). We show here that just the CTD portion of the large subunit of Pol II can function to stimulate splicing in vitro in an assay uncoupled from transcription. Perhaps most interestingly, we observed that only substrate RNAs with intact exons (and their flanking splice sites) were subject to enhancement by the CTD. Combined with the observation that partial depletion of endogenous Pol II inhibits in vitro splicing such that readdition of a recombinant CTD fragment of Pol II restores activity, our results suggest a direct involvement of Pol II during splicing. Our observation is consistent with a recent study of Hirose et al. who reported stimulation of in vitro splicing and spliceosome assembly by purified RNA Pol II in a CTD-dependent manner (22). Compared to the 10-fold stimulation of splicing afforded by intact Pol II observed in their study, it is perhaps not surprising that we observed a weaker effect (three- to fivefold) using a recombinant CTD fragment.
In vitro approaches to the role of the CTD in splicing using reactions uncoupled from transcription present problems for the interpretation of results that indicate stimulation by the addition of intact Pol II or portions thereof to a reconstituted reaction. The potential exists for the CTD to bind splicing factors and thereby concentrate reagents in dilute extracts and indirectly enhance the splicing reaction via concentration effects. Although such effects may be an important part of the in vivo role of the CTD in splicing, they could also occur artificially in diluted in vitro extracts. Designing experiments to distinguish such effects from a more direct role of the CTD in splicing is nontrivial. We suggest that the substrate dependence observed in this study is difficult to reconcile with a concentration effect and implicates a more direct role for Pol II in exon recognition.
Exon definition and Pol II.
An important aspect of our results is that recombinant CTD free of other components of Pol II activated in vitro splicing and assembly of the spliceosome only if the assayed precursor RNA contained complete exons. This feature of the CTD effect may at least partially suggest why CTD length differs among species. Introns in Saccharomyces cerevisiae pre-mRNAs are small, and there are only two pre-mRNAs with more than one intron. Recognition of exons does not appear to operate in yeast; instead, introns are the recognition unit, with interactions spanning the intron (via intron definition; reviewed in references 4, 5, and 40). Yeast Pol II has the shortest number of CTD consensus sequence repeats (26 compared to 52 in mammals). In contrast, higher vertebrates have pre-mRNAs with multiple short exons separated by large introns and have been demonstrated to use the exon as the basic recognition unit (i.e., exon definition). Correspondingly, they have the longest CTD and the most variable CTD sequence.
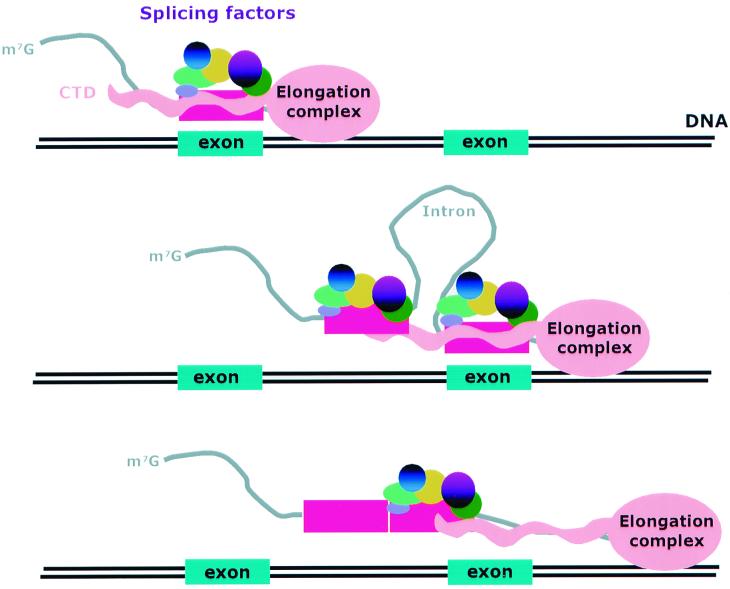
We suggest a model in which the CTD functions as a stage for exon assembly as exons are revealed by the transcriptional machinery (Fig. 9). Such a model easily fits with interpretations of microscopic studies that have suggested cotranscriptional splicing in vivo (2, 5). If correct, it would suggest a preferred removal of introns in a general 5′ to 3′ polarity within a large pre-mRNA. Although such a polarity has not always been observed in reverse transcription-PCR experiments on steady-state RNA populations (25), it remains possible that steady-state nuclear RNA populations may not represent a good model for the active transcription splicing machinery in which adjacent introns may only fleetingly exist in a single RNA. The models in Fig. 9 suggest that the CTD remains associated with a recently defined exon until the next exon in the precursor RNA is revealed during transcription and assembled. Although completely speculative at this point, such association might facilitate exon juxtaposition as well as exon recognition.
FIG. 9.
Proposed model of the involvement of the Pol II CTD in exon definition. The participation of the CTD is shown in two steps. In step one, the CTD mediates association of splicing factors with the newly transcribed exon. In step two, the polymerase translocates to the next exon, which also binds to the CTD close to the first exon, thereby facilitating splicing.
Use of exon definition in pre-mRNA recognition implies a two-step process during assembly of the active spliceosome—exon definition and exon juxtaposition. If the Pol II CTD acts as a platform for recruitment of splicing factors to exons, a logical inference is that CTD-bound exons facilitate exon juxtaposition. Exon definition is most commonly observed in vertebrate genes with small exons and large introns, occasionally extremely large introns. One unresolved problem in spliceosome assembly is how two distal exons can be juxtaposed over extremely long introns. The unique repetitive structure of CTD and its ability to bind exons make it an ideal candidate as the bridge for exon juxtaposition (Fig. 9).
Tying the role of the CTD to exon definition leads to several predictions. The first is that the role of the CTD in splicing may be limited or at least different in organisms that do not utilize exon-based recognition mechanisms, such as yeast. In yeast, therefore, one might predict a relationship between Pol II and capping but not between Pol II and splicing. Secondly, if the CTD recognizes exons, one might predict that the CTD also binds the pseudo-exons within introns that have recently been reported to function as splicing regulators or as intermediates in the splicing of long introns (20). Thus, CTD-spliceosome interactions could be part of the mechanism whereby higher eukaryotes handle extremely long introns. Also, using the CTD to recognize exons raises the possibility of an interplay between transcription and splicing factors during alternative processing, as has recently been suggested by the observation that SR-dependent alternative splicing is promoter-dependent (12). Thirdly, the CTD could play a role in recognition of three types of exons—cap-proximal first exons, internal exons, and 3′-terminal exons with poly(A) sites. Experimental evidence for an interaction between 3′-terminal exon definition and transcription has recently been reported (16, 21). Although not yet directly linked to exon definition, the binding of capping enzymes to the CTD (9, 23, 30, 47) and the need for caps in 5′-terminal exon definition (28) suggest a role here also.
CTD and splicing factors.
How might the CTD function during exon definition? We and others have viewed exon definition as an ATP-dependent process in which interactions between U2AF, U1 snRNPs, U2 snRNPs, and SR proteins bound to either constitutive splicing signals or enhancer elements serve to identify exons and create stable initial spliceosomal complexes. The relationship between this process and initial interactions that occur across introns is still not known but the former is thought to utilize the small subunit of U2AF and SR proteins whereas the latter usually invokes interactions of the large subunit of U2AF and SF1.
This study deliberately used precursor RNAs with strong vertebrate splice sites capable of both types of interactions. The 3′ splice site used in these substrates is capable of associating with U2 snRNPs and forming complex A in the absence of a 5′ splice site in either polarity although complex formation is assisted by the presence of a 5′ splice site in either an intronic or an exonic polarity. Addition of the CTD had no effect when a 5′ splice site was added upstream of the 3′ splice site but had an effect when the same 5′ splice site was placed downstream of the 3′ splice site. Thus, strong splicing substrates that bind splicing factors well and assemble with relatively equal efficiency into stable complexes responded differently to the CTD. We also observed that the addition of the CTD stimulated U1 snRNP binding to the substrate RNA but not the binding of either ASF/SF2 or U2AF. These observations suggest that the CTD affects the efficiency with which snRNPs bind to exons during exon definition. This model fits well with the pivotal role that 5′ splice sites and U1 snRNPs have been demonstrated to play in exon definition and suggests a fundamental difference between this process and the mechanism whereby U1 and U2 snRNPs interact across introns.
Phosphorylation of Pol II CTD during splicing.
CTD phosphorylation is closely related to the engagement of Pol II during transcription and a shift of the transcription machinery from initiation to elongation (reviewed in references 13 and 14). In sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the largest subunit of Pol II appears as two differently migrating species. The isoform with the slowest gel mobility, Pol IIo, is considered hyperphosphorylated and the isoform with the faster gel mobility, IIa, is considered to be hypo- or nonphosphorylated. In vitro phosphorylation experiments demonstrate that incorporation of 15 to 25 phosphates into the CTD is able to cause a mobility shift in electrophoresis (27, 49). A recent study has revealed that MAb H5 (binding Pol IIo specifically) and MAb H14 (binding Pol IIa) recognize phosphorylated serine 2 or serine 5, respectively, within the heptad CTD repeat, implying that in addition to the intensity of Pol II phosphorylation, IIo and IIa may differ in phosphorylation of preferred sites (39). This observation may also suggest that phosphorylation of serine 2 is important for CTD effects on splicing. However, both we and others have observed that in vitro RNA processing is inhibited by the CTD-specific antibody 8WG16, whose epitope is nonphosphorylated (47; data not shown).
Given the presence of multiple phosphorylation sites on the CTD, it is unclear which form of the phosphorylated CTD is actively involved in splicing. The presence of such a high number of phosphorylation sites is further complicated by the nonconsensus repeats present in the C-terminal portion of the vertebrate CTD. Recent reports have suggested that hyperphosphorylated Pol II has affinity for RNA processing factors involved in both capping and polyadenylation (2, 22, 30, 33); similarly purified Pol IIo but not IIa stimulates in vitro splicing in an S100 extract supplemented with SR proteins (22). In this study, using nuclear extract, we observed the rapid production of multiple bands of recombinant CTD with slower migration in electrophoresis, indicating the occurrence of both hyper- and hypophosphorylation and considerable diversity in the phosphorylation state of the final CTD protein. However, as indicated by [32P]phosphate incorporation (Fig. 8), the majority of the supplemental recombinant CTD was hypophosphorylated. We do not know which form of the CTD afforded stimulation in our experiments given the rapid changes in phosphorylation that occurred in extract. Our data do suggest that phosphorylation of the CTD is a dynamic process in nuclear extracts and that forms of the CTD can be generated that participate in the splicing process.
ACKNOWLEDGMENTS
We specially thank Bill Dynan for generously providing the CTD expression construct, Stephen L. Warren for Pol II antibodies H5 and H14, and Adrian Krainer for ASF/SF2 antibody.
This work was supported by NIH grant RO1 GM 38526 to S.M.B.
REFERENCES
- 1.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 2.Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 3.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 4.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 5.Beyer A L, Yvonne N, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 6.Black D L. Finding splice sites in a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 7.Bregman D B, Du L, van der Zee S, Warren S L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chabot B, Bisotto S, Vincent M. The nuclear matrix phosphoprotein p255 associates with splicing complexes as part of the [U4/U6.U5] tri-snRNP particle. Nucleic Acids Res. 1995;23:3206–3213. doi: 10.1093/nar/23.16.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corden J L. Tails of RNA polymerase II. Trends Biochem Sci. 1990;10:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 11.Corden J L, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 12.Cramer P, Caceres J F, Cazalla D, Kadener S, Muro A F, Baralle F E, Kornblihtt A R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 13.Dahmus M E. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- 14.Dahmus M E. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- 15.Du L, Warren S L. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dye M J, Proudfoot N J. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 17.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenleaf A L. A positive addition to a negative tail's tale. Proc Natl Acad Sci USA. 1993;90:10896–10897. doi: 10.1073/pnas.90.23.10896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlow E, Lane D, editors. Antibodies: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 521–523. [Google Scholar]
- 20.Hatton A R, Subramaniam V, Lopez A J. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by replacing at exon-exon junctions. Mol Cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- 21.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 22.Hirose Y, Tacke R, Manley J L. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 24.Jamison S F, Pasman Z, Wang J, Wil C, Luhrmann R, Manley J L, Garcia-Blanco M A. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler O, Jiang Y, Chasin L A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol. 1993;13:6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E, Du L, Bregman D B, Warren S L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J M, Greenleaf A L. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci USA. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 29.Lou H, Gagel R F, Berget S M. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- 30.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 31.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L Amgen EST Program. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCullough A J, Berget S M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neugebauer K M, Roth M B. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 35.Niwa M, Rose S D, Berget S M. Polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 36.Niwa M, Berget S M. Polyadenylation precedes splicing in vitro. Gene Exp. 1991;1:5–14. [PMC free article] [PubMed] [Google Scholar]
- 37.O'Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 38.Patturajan M, Wei X, Berezney R, Corden J L. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol Cell Biol. 1998;18:2406–2415. doi: 10.1128/mcb.18.4.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 40.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 41.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuman S. Origins of mRNA identity: capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 44.Talerico M, Berget S M. Effect of 5′ splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990;10:6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson N E, Steinberg T H, Aronson D B, Burgess R R. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- 46.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 47.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Corden J L. Phosphorylation causes a conformational change in the carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2297–2302. [PubMed] [Google Scholar]