Abstract
New pneumococcal conjugate vaccines covering a limited number of serotypes are likely to come into widespread use over the next few years. It is unknown what effect this will have on the relative importance of different serotypes as causes of pneumococcal infection. Hence, it will be important to monitor serotype prevalence before, during, and after the introduction of new vaccines. We have investigated the ability of a PCR method based on polymorphisms in two genes common to the different capsule loci to predict the serotype of 93 clinical isolates of Streptococcus pneumoniae submitted to the Central Public Health Laboratory in 1997. Of 70 isolates with vaccine serotypes, 65 were predicted to belong to the correct serotype; this number was improved to 69 with the inclusion of two additional patterns to the database. Of 23 isolates with other serotypes, 19 were correctly predicted as non-vaccine serotypes, the discrepancy lying with four isolates of 6A (non-vaccine serotype) that were indistinguishable from isolates of 6B (vaccine serotype). In situations in which culture of the organism is not feasible, this method could potentially be applicable directly to clinical specimens and could be a valuable aid to the surveillance of pneumococcal serotypes.
Respiratory disease has overtaken diarrheal disease as the major cause of death in children under the age of 5 years worldwide. Streptococcus pneumoniae is one of the most important pathogens involved. At the same time resistance to penicillin and other antimicrobials has dramatically increased, leading to serious problems in the treatment of pneumococcal disease. The case for primary prevention of pneumococcal infection by vaccination is now compelling.
New pneumococcal conjugate vaccines are currently undergoing extensive trials and are likely to be introduced in widespread vaccination programs in the near future (20). Since such vaccines can only cover up to 11 of the 90 pneumococcal serotypes, extensive surveillance of serotype prevalence in populations to be vaccinated will be needed. Furthermore, it is important that prevalence be monitored during and after implementation of vaccination programs, in case widespread vaccination leads to the selective increase of serotypes not covered by the vaccine.
Serotype prevalence is currently monitored by culture of the organism, followed by serology. A nonculture diagnostic method with the ability to determine serotype could have immense advantages in convenience and cost. One potential approach to a nonculture method is to use the PCR to detect pneumococcal DNA directly from a clinical specimen, focusing on a segment either responsible for or closely linked to production of the polysaccharide capsule (which determines serotype). At the present time sequences of the genes responsible for capsule production in several serotypes have been published, and serotype specific genes have been identified (1, 7, 9, 11–13, 15, 18, 19, 21, 22). However, an inconveniently large number of different PCR primers would be required to determine the serotype by this strategy. Furthermore, the large size of the serotype-specific region, at well over 10 kb in most cases, precludes the possibility of using a universal PCR with primers in conserved flanking regions as a method applicable directly to clinical specimens with current technology.
As an alternative approach which would be amenable to application on clinical material, we have investigated polymorphism within the first two genes of the serotype 19F capsule locus, cps19fA and cps19fB, which have homologous sequences in most if not all other serotypes (9, 19). These are physically the closest conserved genes to those actually determining serotype, and thus polymorphism within them would be expected to be tightly linked to capsule type. We have examined the potential to use this region in a PCR test as a marker for serotype among pneumococcal cultures submitted to the major reference center in the United Kingdom. Specifically, in the context of the introduction of a conjugate vaccine, can such a test predict (i) whether a pneumococcus is of a serotype contained in the vaccine and (ii), if so, which of the vaccine serotypes it is?
MATERIALS AND METHODS
Bacterial isolates.
Ten pneumococci from each of the eleven proposed conjugate vaccine serotypes (1, 3, 4, 5, 6B, 7F, 9V, 14, 18C, 19F, and 23F) were selected from among recent clinical isolates submitted to the reference laboratory from September to December 1997, along with a reference strain for each of serotypes 5 (NCTC 11887), 6B (NCTC 11888), 9V (NCTC 11897), and 23F (NCTC 11910). Uncharacterized pneumococci used for the prediction of serotype were submitted to the reference laboratory over a 2-week period in March 1998.
Pneumococci from the United Kingdom used in preliminary work were from the reference laboratory collections of 1990 to 1994. International isolates were from Japan (16 isolates), West Indies (8 isolates), Russia (37 isolates), and Norway (20 isolates). Other streptococci used to test the specificity of the primers included S. mutans (NCTC 10832), S. bovis type I (NCTC 8133), S. agalactiae (NCTC 12906), S. oralis (NCTC 11427), S. mitis (NCTC 12261), S. sanguis (NCTC 11085), S. salivarius (NCTC 8618), S. parasanguis (NCTC 12854), S. pyogenes (NCTC 12067), S. vestibularis (NCTC 12166), S. dysgalactiae (NCTC 4669), group G streptococcus (NCTC 9603), group K streptococcus (NCTC 10232), S. gordonii (97ID004000), and S. gallolyticus (97ID000570).
Serotyping of isolates.
Preliminary serotyping was performed by slide agglutination (5) with capsular typing sera and factor sera (Statens Serum Institute, Copenhagen, Denmark). Any discrepancies or equivocal results were rechecked by the Quellung reaction (16).
DNA extraction and PCR.
Streptococci of other species and pneumococci in preliminary experiments were cultured overnight in 20 ml of Todd-Hewitt broth (Oxoid). After sedimentation the cells were resuspended in 0.1 ml of TE (10 mM Tris HCl, 1 mM EDTA; pH 8.0). Lysozyme (10 μl at 50 mg/ml) was added, and cells were incubated for 30 min at 37°C, followed by the addition of 0.5 ml of GES (5 M guanidine thiocyanate, 0.1 M EDTA [pH 8.0], 0.5% Sarkosyl) and incubation at room temperature for 10 min. Ammonium acetate (0.25 ml at 7.5 M) was added, and the cells were mixed and placed on ice for 10 min. Chloroform-isoamyl alcohol (24:1) was added, mixed, and separated by centrifugation, and 0.7 ml of the aqueous phase was recovered. DNA was precipitated with 0.54 volumes of isopropanol and then redissolved and reprecipitated in 70% ethanol. The DNA was finally resuspended in 50 to 100 μl of TE.
In subsequent experiments, as specified in Results, pneumococcal DNA was prepared as a crude extract. A loopful of growth from a fresh plate was suspended in 100 μl of TE and heated to 100°C for 5 min.
Primers for PCR were CPSA3 (ATCCTTGTCAGCTCTGTGTC; GenBank accession no. U09239; positions 415 to 434 [9]) and CPSB2 (TCACTTGCAACTACATGAAC; positions 2191 to 2210 [9]). PCR was performed in 100-μl mixture containing 50 mM KCl, 10 mM Tris HCl (pH 8.3), 2.5 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphate, 100 ng of each primer, 1 μl of template DNA (purified or crude extract), and 1.2 U of AmpliTaq polymerase (Perkin-Elmer). Conditions for PCR were 1 cycle of 95°C for 3 min, followed by 30 cycles of 95°C for 1 min, 50°C for 2 min, and 72°C for 2 min in a Hybaid Thermal Reactor. PCR products were analyzed on 10-cm 0.9% agarose gels in Tris-borate-EDTA (TBE), stained with ethidium bromide, and viewed with UV illumination.
Restriction digests with AluI, HinfI, and RsaI (Promega) were performed on 10 to 18 μl of PCR product. Restriction fragments were separated on 4% polyacrylamide gels in TBE, stained with ethidium bromide, and viewed with UV illumination. Images were recorded with an IS 1000 Digital Imaging System (Flowgen). Fragment patterns were analysed with GelCompar, version 4.1 (Applied Maths, Kortrijk, Belgium), selecting the region of the gel between the 1,636- and 134-bp bands of the reference 1-kb ladder. Patterns were normalized by using three reference tracks per gel, a 400-point resolution, and a smoothing factor of 5. The bands were identified by the autosearch feature and artifacts on the images incorrectly identified as bands were manually deleted. No distinction was made for single bands that, on the basis of intensity, were likely to correspond to two comigrating fragments. Comparisons were made with a positional tolerance of 1%, a tolerance increase of 1.75%, and an optimization value of 0.5%. Patterns were clustered by the UPGMA method by using the Dice similarity coefficient. Restriction patterns of isolates of unknown serotype were compared with the database lists by using the identification feature (employing the above coefficient values and UPGMA clustering).
RESULTS
Specificity of primers.
In a test designed for direct application to clinical samples it is important to know whether an amplification product would be obtained with other species, particularly the related oral streptococci. DNA was extracted from isolates of different streptococcal species (see Materials and Methods) and used as a template for PCR. Only one isolate yielded a specific product that had a size close to that obtained with pneumococci. This was group K streptococcus NCTC 10232, which was determined to be a putative S. oralis strain on the basis of Rapid Strep ID32 API and the production of β-d-N-acetylglucosaminidase and β-d-N-acetylglucosaminidase (23).
Conservation of restriction profiles within serotypes.
A preliminary investigation of the correlation between restriction patterns in cpsA-cpsB and serotype was undertaken on a set of isolates in our laboratory collection from Norway, Japan, Russia, the United Kingdom, and the West Indies. PCR products amplified with primers CPSA3 and CPSB2 were digested with AluI and with HinfI, and the resulting patterns were compared visually. Upon combining the results with both restriction enzymes, 33 patterns were observed among 153 isolates of 25 serotypes. Each combination of AluI and HinfI restriction patterns was unique to a single serotype with the following exceptions: all 14 isolates of serotype 6A shared the same patterns as 8 of 16 6B isolates; 1 of 3 isolates of serotype 1 and 1 of 2 of 23A shared patterns with 9 of 11 23F isolates; 2 of 6 19A isolates shared patterns with all 3 7F isolates; 1 of 2 15B isolates and the sole 15C isolate shared patterns with all 3 serotype 22F isolates; the other 15B isolate shared patterns with 16 of 18 serotype 14 isolates. Representative isolates with patterns shared between serotypes were further analyzed by using additional restriction enzymes. It was observed that RsaI digestion could distinguish 19A from 7F isolates and some isolates of 6A from 6B. Twelve serotypes were represented by more than one type of pattern with one or both enzymes. Overall, the preliminary investigation suggested a close association between restriction profile and serotype.
The strategy of serotype prediction from the restriction profile needed to be tested further under conditions more relevant to the context in which it might be used, such as a national pneumococcal reference center. This required the establishment of a database of patterns from clinical isolates typical of the time and place and a computerized system for pattern storage and comparison. Thus, 10 recent clinical isolates (or 9 plus 1 reference strain) confirmed to belong to each of the 11 serotypes likely to be included in conjugate vaccines (serotypes 1, 3, 4, 5, 6B, 7F, 9V, 14, 18C, 19F, and 23F) were selected from among those recently submitted to the Central Public Health Laboratory. PCR with primers CPSA3 and CPSB2 was performed on a crude extract of each isolate. All isolates yielded a product corresponding to the predicted size of 1.8 kb.
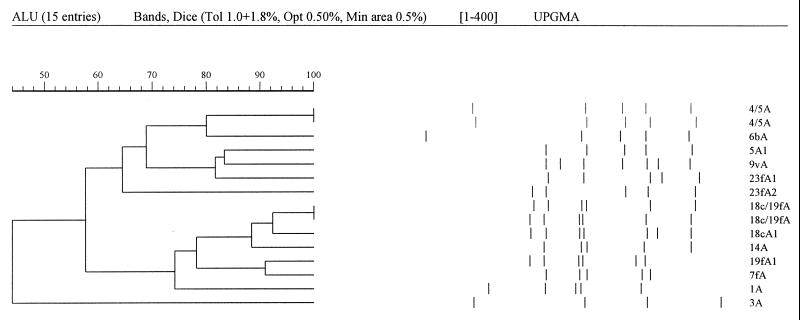
PCR products were digested with AluI, HinfI, and RsaI (three separate reactions), and the resultant fragments from each reaction were separated on polyacrylamide gels. The fragment patterns were recorded with a digital imager and analyzed by using GelCompar version 4.1 software as described in Materials and Methods. First, for each serotype the 10 isolates were compared to each other. For serotypes 1, 3, 4, 7F, and 9V the 10 isolates of the same serotype were indistinguishable with all three restriction enzymes. The restriction patterns obtained were given a code according to the serotype and enzyme used, e.g., the pattern obtained with AluI on all serotype 1 isolates was designated 1A, except for shared patterns as described below. For serotypes 5, 6B, 14, 18C, 19F, and 23F, two different sets of restriction patterns were obtained for the 10 isolates with, in most cases, a single isolate producing a distinct pattern from the other 9. These patterns were coded with an extra digit, e.g., the more common pattern obtained with AluI on serotype 23F isolates was designated 23fA1 and the less common pattern was designated 23fA2 (Table 1). Second, for each restriction enzyme in turn, isolates representing all the different restriction patterns for all the serotypes were compared. Figure 1 illustrates the comparison of all AluI patterns. The 15 patterns were derived from 11 different gels, normalized with reference ladders. Although normalization has not generated a visually perfect alignment, the program recognizes two pairs of matching patterns. Thus, with AluI all patterns were distinct except that one serotype 5 pattern was the same as the serotype 4 pattern and one 18C pattern was the same as one 19F pattern. (Apparent differences in the sum of fragment sizes, e.g., between 4A and 6bA, arise from comigrating fragments being scored as single bands and the discounting of bands below 134bp [see Materials and Methods].) Patterns were also shared between two pairs of serotypes for HinfI and three pairs of serotypes for RsaI (Table 1). Only one isolate, the reference serotype 5 isolate, shared patterns for two restriction enzymes with another serotype, and a combination of the three enzyme patterns (the restriction profile) was able to distinguish all 11 serotypes.
TABLE 1.
Restriction patterns obtained for 10 isolates of each of the 11 vaccine serotypes
| Serotype | No. of isolates | AluI pattern | HinfI pattern | RsaI pattern |
|---|---|---|---|---|
| 1 | 10 | 1A | 1H | 1/19fRa |
| 3 | 10 | 3A | 3H | 3R |
| 4 | 10 | 4/5Aa | 4H | 4/5Ra |
| 5 | 9 | 5A | 5H1 | 5R |
| 5 | 1 | 4/5A | 5H2 | 4/5R |
| 6B | 6 | 6bA | 6bH | 6bR |
| 6B | 4 | 6bA | 6bH | 6b/14Ra |
| 7F | 10 | 7fA | 7fH | 7fR |
| 9V | 10 | 9vA | 9vH | 9vR |
| 14 | 8 | 14A | 14/19fHa | 6b/14R |
| 14 | 2 | 14A | 14/19fH | 14R |
| 18C | 9 | 18cA | 18cH | 18cR |
| 18C | 1 | 18c/19fAa | 18cH | 18cR |
| 19F | 9 | 19fA | 14/19fH | 1/19fR |
| 19F | 1 | 18c/19fA | 19f/23fHa | 19fR |
| 23F | 9 | 23fA1 | 19f/23fH | 23fR1 |
| 23F | 1 | 23fA2 | 23fH | 23fR2 |
Patterns that are the same in isolates of different serotypes.
FIG. 1.
Comparison by GelCompar of AluI restriction digests representing all pattern types defined for the 11 vaccine serotypes. Normalized band patterns are represented on the right, and a dendrogram of relatedness of patterns by use of the Dice coefficient is on the left. Pattern matches of 100% (tolerance, 1.0 + 1.75%) are demonstrated between the 4/5A patterns produced by isolates of serotypes 4 and 5 and the 18c/19fA patterns from the 18C and 19F isolates.
Correlation of restriction profile to serotype in uncharacterized isolates.
To evaluate the ability of restriction profile to predict serotype, 93 consecutive isolates submitted to the Respiratory and Systemic Infection Laboratory (RSIL) over a 2-week period were examined. Restriction profile and serotype for these 93 isolates were determined independently by different experimenters. The three restriction patterns for each isolate were compared to the lists of patterns in the database described above. The criterion used for assignment of a predicted serotype from a restriction profile was that there must be ≥90% similarity to patterns of the same serotype from at least two of the three lists. This was established by comparing serotype assignment of the patterns from the first 20 isolates by visual inspection and GelCompar. An exception is applied in the case of the serotype 4/5 patterns: since the rare serotype 5 pattern for two enzymes is shared by serotype 4, all serotype 4 isolates would be expected to match serotype 5 in two lists; in this case the prediction of serotype 4 rather than 5 was made if there were matches to serotype 4 patterns in all three lists and to serotype 5 in two lists. On this basis isolates were assigned to 1 of the 11 vaccine serotypes or, if the above criterion was not met, they were considered to be “nonvaccine serotypes.” After analysis, the serotype predicted by PCR was compared to the serotype obtained by slide agglutination and the Quellung reaction.
The results of the prediction of serotype by restriction profile are given in Table 2. Sixty-five isolates with serotypes among the eleven vaccine types were correctly assigned, while one serotype 1 isolate was predicted to be either serotype 1 or serotype 19F. A further four isolates, two 6B and two 9V, were incorrectly predicted to have nonvaccine serotypes and had patterns that were clearly distinct from those in the database. Nineteen isolates with serotypes not included in the vaccine were correctly predicted to have nonvaccine serotypes, while four isolates, all serotype 6A (not in the vaccine), were incorrectly predicted to belong to the vaccine serotype 6B. Taking the inconclusive result of the serotype 1 isolate as an incorrect prediction, these results demonstrate that the assignment of serotype by DNA profile has a sensitivity of 0.93 and a specificity of 0.83. A further measure of the agreement between serotype predicted by DNA profile and actual serotype, which takes into account the actual frequency of different serotypes among the isolates investigated, is Cohen's kappa statistic. This can be computed as kappa = (po − pe)/(1 − pe), where po is the observed proportion of isolates where the two methods agree and pe is the expected proportion of agreement if the isolates were allocated to serotype at random while keeping the total number of isolates of each serotype fixed. With the results obtained, the kappa value was calculated to be 0.88. A hypothesis test of kappa equal to zero (i.e., if the serotypes were allocated at random) gives a P value of <0.0001.
TABLE 2.
Actual serotype and predicted serotype determined on the basis of restriction pattern matches for uncharacterized isolates
| Serotype | No. of isolates | AluI patterna | HinfI patterna | RsaI patterna | Predicted serotypeb |
|---|---|---|---|---|---|
| 1 | 1 | 1A | 14/19fH | 1/19fR | 1 or 19F |
| 3 | 2 | 3A | 3H | 3R | 3 |
| 4 | 1 | 4/5A | 4H | 4/5R | 4 |
| 5 | 1 | 5A | 5H1 | 5R | 5 |
| 6B | 7 | 6bA | 6bH | 6bR | 6B |
| 6B | 4 | 6bA | 6bH | 6b/14R | 6B |
| 6B | 2 | No IDc | No ID | No ID | NVd |
| 9V | 16 | 9vA | 9vH | 9vR | 9V |
| 9V | 2 | 9vA | 1H | No ID | NV |
| 9V | 1 | 9vA | No ID | 9vR | 9V |
| 9V | 1 | 9vA | 9vH | No ID | 9V |
| 14 | 15 | 14A | 14/19fH | 6b/14R | 14 |
| 14 | 2 | 18cA | 5H1, 14/19fH | 14R, 23fR1 | 14 |
| 18C | 2 | 18cA | 18cH | No ID | 18C |
| 18C | 1 | 18cA | 18cH | 18cR | 18C |
| 19F | 4 | 19fA | 14/19fH | 1/19fR | 19F |
| 19F | 4 | 18c/19fA | 14/19fH | 1/19fR | 19F |
| 19F | 1 | 19fA | 19f/23fH | No ID | 19F |
| 23F | 3 | 23fA1 | 19f/23fH | No ID | 23F |
| 6A (NV)d | 3 | 6bA | 6bH | 6b/14R | 6B |
| 6A (NV) | 1 | 6bA | 6bH | No ID | 6B |
| 6A (NV) | 1 | 6bA | 23fH | No ID | NV |
| 6A (NV) | 1 | No ID | No ID | No ID | NV |
| 8 (NV) | 3 | No ID | No ID | No ID | NV |
| 8 (NV) | 1 | No ID | 1H | No ID | NV |
| 9N (NV) | 1 | No ID | 5H2 | 3R | NV |
| 12 (NV) | 2 | 5A | No ID | No ID | NV |
| 12 (NV) | 1 | 5A | No ID | 14R | NV |
| 15 (NV) | 1 | 9vA | No ID | No ID | NV |
| 16F (NV) | 2 | 14A | 5H2 | No ID | NV |
| 20 (NV) | 3 | No ID | No ID | No ID | NV |
| 29 (NV) | 1 | NV | |||
| 35F (NV) | 2 | No ID | 9vH | No ID | NV |
Restriction patterns from the database created from isolates in Table 1 that match the new pattern by >90%.
Serotype predicted from restriction pattern matches with at least two enzymes.
No ID, no pattern matches by >90%.
NV, nonvaccine serotype.
DISCUSSION
Preliminary work on an international collection of isolates demonstrated a strong correlation between the cpsA-cpsB restriction profile and the serotype. The study was then focused to evaluate the potential of a method based on cpsA-cpsB polymorphism to predict the serotype among isolates submitted to the major pneumococcal reference center in the United Kingdom, targetting the 11 serotypes being considered for inclusion in conjugate vaccines.
Among 93 consecutive isolates submitted to RSIL over a 2-week period, correlation between the predicted serotype from the restriction profile and the actual serotype calculated on the basis of the proportions of each serotype in the collection (Cohen's kappa statistic) was 0.88 (P < 0.0001). Isolates that were incorrectly assigned were examined further to see whether the accuracy of prediction could be improved for future analyses. Two serotype 6B isolates were missed because they had patterns that differed from those in the database; work with other pneumococci in this laboratory has demonstrated that this additional 6B profile is quite common and specific to 6B; its inclusion in the database would improve the detection of 6B isolates. The additional 9V profile had not been observed previously, but its addition to the database would not compromise recognition of other serotypes in this study. Inclusion of these two additional patterns would improve the correlation, kappa, to 0.94.
The four isolates incorrectly assigned to vaccine serotypes were all serotype 6A but were predicted to be serotype 6B. It appears from this and other work that the cpsA-cpsB restriction profile often cannot distinguish between these subtypes, and we are currently investigating other strategies to address this. It is a matter of current debate whether these two subtypes are cross-protective. Published data on conjugate 6B vaccine suggests that cross-protection will be provided (6, 8), in which case it would not be necessary to distinguish them. However, this remains to be confirmed in more extensive trials.
Potential pitfalls in applying an indirect test to predict serotype need to be considered. First, such a test can never guarantee 100% accuracy because the genes examined do not themselves determine serotype and so would not be appropriate if the serotype of one particular isolate was critical. Since serotype does not influence treatment this is rarely if ever likely to be the case in clinical situations. Second, it is likely that serotype will sometimes become dislocated from the cpsA-cpsB restriction profile, either through mutation in restriction sites or through recombination. Recombination of capsule genes is now well documented (2, 3, 10, 14, 17), although in the cases characterized to date the point of cross-over occurs outside the locus so that serotype remains linked to cpsA-cpsB restriction profile (4). For these reasons it is essential to verify and if necessary modify the database in any potential application of the method and to continue to apply conventional serology in a proportion of cases.
Such pitfalls should be weighed against the possible advantages, chief of which is the potential to screen serotype directly from clinical samples, avoiding the necessity for isolation of the organism. Where the balance of advantages and difficulties lies depends on the situation in which the test is to be applied. We consider that a typical context could be the introduction of a conjugate pneumococcal vaccine targeted at all infants in a population. Serotype surveillance is needed both before and after introduction to monitor changes that might be induced in serotype prevalence by protection against a limited number of serotypes. A major rise in disease caused by previously unusual serotypes could necessitate a change in vaccine composition. In addition, it would be critical to determine whether infections occurring in vaccinated individuals were due to a vaccine serotype and, if so, to which one, in order to monitor the efficacy of the vaccine. In monitoring trends in serotype prevalence over time, the accuracy of an indirect test would probably be acceptable given the advantage of a nonculture system, and it is our present intention to apply this methodology to the detection of vaccine and nonvaccine serotype pneumococci in clinical specimens. However, in examining infections in vaccinated individuals a further test to distinguish serotypes 6A and 6B might be required and is currently under investigation.
ACKNOWLEDGMENTS
We are grateful for the support of the Special Trustees of the Royal London Hospital and the Medical Research Council (studentship to E.R.L.) and Colfuturo (scholarship to C.A.A.).
We thank Andre Charlett and the PHLS Statistics Unit for advice on statistical analysis and NHS and PHLS microbiologists for referring cultures. International isolates were kindly provided by N. Rikitomi, Nagasaki, Japan; P. Prabhakar, Caribbean Epidemiology Centre, Trinidad, West Indies; L. Strachunsky, Smolensk, Russia; and T. Bergen, Oslo, Norway.
REFERENCES
- 1.Arrecubieta C, Lopez R, Garcia E. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J Bacteriol. 1994;176:6375–6383. doi: 10.1128/jb.176.20.6375-6383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 3.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 4.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 5.Colman G, Cooke E M, Cookson B D, Cooper P G, Efstratiou A, George R C. Pneumococci causing invasive disease in Britain 1982 to 1990. J Med Microbiol. 1998;47:17–27. doi: 10.1099/00222615-47-1-17. [DOI] [PubMed] [Google Scholar]
- 6.Dagan R, Muallem M, Melamed R, Leroy O, Yagupsky P. Reduction of pneumococcal nasopharyngeal carriage in early infancy after immunization with tetravalent pneumococcal vaccines conjugated to either tetanus toxoid or diphtheria toxoid. Pediatr Infect Dis J. 1997;16:1060–1064. doi: 10.1097/00006454-199711000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Dillard J P, Yother J. Genetic and molecular characterisation of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol Microbiol. 1994;12:959–972. doi: 10.1111/j.1365-2958.1994.tb01084.x. [DOI] [PubMed] [Google Scholar]
- 8.Giebink G S, Meier J D, Quartey M K, Liebeler C L, Le C T. Immunogenicity and efficacy of Streptococcus pneumoniae polysaccharide-protein conjugate vaccines against homologous and heterologous serotypes in the chinchilla otitis media model. J Infect Dis. 1996;173:119–127. doi: 10.1093/infdis/173.1.119. [DOI] [PubMed] [Google Scholar]
- 9.Guidolin A, Morona J K, Morona R, Hansman D, Paton J C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect Immun. 1994;62:5384–5396. doi: 10.1128/iai.62.12.5384-5396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermans P W M, Sluijter M, Elzenaar K, van Veen A, Schonkeren J J M, Nooren F M, van Leeuwen W J, de Neeling A J, van Klingeren B, Verburgh H A, de Groot R. Penicillin-resistant Streptococcus pneumoniae in The Netherlands: results of a 1-year molecular epidemiologic survey. J Infect Dis. 1997;175:1413–1422. doi: 10.1086/516474. [DOI] [PubMed] [Google Scholar]
- 11.Iannelli F, Pearce B J, Pozzi G. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol. 1999;181:2652–2654. doi: 10.1128/jb.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolkman M A B, Morrison D A, van der Zeijst A M, Nuijten P J M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 14.Lefevre J C, Bertrand M A, Faucon G. Molecular analysis by pulsed-field gel electrophoresis of penicillin-resistant Streptococcus pneumoniae from Toulouse, France. Eur J Clin Microbiol Infect Dis. 1995;14:491–497. doi: 10.1007/BF02113426. [DOI] [PubMed] [Google Scholar]
- 15.Llull D, Lopez R, Garcia E, Munoz R. Molecular structure of the gene cluster responsible for the synthesis of the polysaccharide capsule of Streptococcus pneumoniae type 33F. Biochim Biophys Acta. 1998;1443:217–224. doi: 10.1016/s0167-4781(98)00213-9. [DOI] [PubMed] [Google Scholar]
- 16.Lund E, Henrichsen J. Laboratory diagnosis, serology and epidemiology of Streptococcus pneumoniae. Methods Microbiol. 1978;12:241–262. [Google Scholar]
- 17.McDougal L K, Facklam R, Reeves M, Hunter S, Swenson J M, Hill B C, Tenover F C. Analysis of multiply antimicrobial-resistant isolates of Streptococcus pneumoniae from the United States. Antimicrob Agents Chemother. 1992;36:2176–2184. doi: 10.1128/aac.36.10.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 20.Mulholland K. Strategies for the control of pneumococcal diseases. Vaccine. 1999;17:S79–S84. doi: 10.1016/s0264-410x(99)00112-7. [DOI] [PubMed] [Google Scholar]
- 21.Munoz R, Mollerach M, Lopez R, Garcia E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez M, Tomasz A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol. 1998;180:5273–5278. doi: 10.1128/jb.180.19.5273-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whiley R A, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol. 1998;13:195–216. doi: 10.1111/j.1399-302x.1998.tb00698.x. [DOI] [PubMed] [Google Scholar]